
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 August 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01839
This article is part of the Research Topic Insights into New Strategies to Combat Biofilms View all 29 articles
Staphylococcus epidermidis is frequently associated with biofilm-related infections. Biofilms drastically reduce the efficacy of conventional antibiotics and the host immune system. In S. epidermidis biofilm formation, a major role is played by the YycG/YycF two-component system, and previous findings have indicated that inhibitors targeting the cytoplasmic HATPase_c domain of YycG kinase in S. epidermidis exhibit bactericidal and biofilm-killing activities. Therefore, we hypothesized that monoclonal antibodies (mAbs) against YycG extracellular (YycGex) domain would block the signal transduction and influence the biofilm formation of S. epidermidis. In this study, we screened out two YycGex-specific mAbs showing the highest affinity for the target, mAbs 2F3 and 1H1. These mAbs inhibited S. epidermidis biofilm formation in a dose-dependent manner, and at a concentration of 160 μg/mL, mAbs 2F3 and 1H1 caused 78.3 and 93.1% biofilm reduction, respectively, relative to normal mouse IgG control. When co-cultivated with YycGex mAbs, S. epidermidis cells showed diminished initial-adherence capacity, and the antibody treatment further led to a marked decrease in the synthesis of polysaccharide intercellular adhesin and in the transcriptional level of genes encoding proteins involved in biofilm formation. Lastly, we determined that the epitopes recognized by the two YycGex mAbs are located within aa 59–70 of the YycGex domain. It indicates that the YycGex domain may be a potential candidate as a vaccine for the prevention of S. epidermidis biofilm infections.
Staphylococcus epidermidis is part of the normal bacterial flora colonizing the skin and mucous membranes of the human body (Yao et al., 2006; Fey and Olson, 2010). The main pathogenicity associated with S. epidermidis is the formation of thick, multilayered, highly structured biofilms on artificial surfaces (Vuong and Otto, 2002; McCann et al., 2008). Biofilm formation is a highly complex process that involves multiple regulatory factors, such as two-component systems (TCSs) (Sauer, 2003; Mikkelsen et al., 2011; Rasamiravaka et al., 2015; Wolska et al., 2016). The YycG/YycF TCS is conserved in low-G + C gram-positive bacteria (Fabret and Hoch, 1998; Fukuchi et al., 2000), and the TCS regulates bacterial growth, cell-wall metabolism, cell division, virulence-factor expression, and biofilm formation (Ng et al., 2005; Senadheera et al., 2005; Bisicchia et al., 2007; Fukushima et al., 2008). Because YycG/YycF TCS plays an essential role in bacterial biological functions and because no YycG/YycF homologs or structurally similar proteins are expressed in humans, the YycG/YycF TCS represents a potential antibacterial target (Winkler and Hoch, 2008; Gotoh et al., 2010).
The YycG histidine kinase of S. epidermidis prototypically contains two transmembrane sections, an extracytoplasmic loop (YycGex domain), cytoplasmic HAMP and Per-Arnt-Sim (PAS) domains, and a catalytic core of HisKA and HATPase_c domains (Ng and Winkler, 2004; Dubrac et al., 2008). Our previous work demonstrated that inhibitors targeting the HATPase_c domain exhibit not only bactericidal activity but also potent activity against staphylococcal biofilms (Qin et al., 2006; Huang et al., 2012; Liu et al., 2014; Lv et al., 2017). Thus, we hypothesized that monoclonal antibodies (mAbs) against the YycGex domain would block the signal transduction and thereby influence the phenotypic properties of S. epidermidis RP62A. Analysis of the YycG sequence revealed that the YycGex domain contains aa 142–154 flanked by two transmembrane domains that adopt a PAS or PhoQ-DcuS-CitA fold (Santelli et al., 2007; Chang et al., 2010; Shah et al., 2013). The folding motif is regarded as a sensor that receives environmental signals and modulates YycG kinase activity (Ng and Winkler, 2004; Szurmant et al., 2008). Accordingly, we generated mAbs against the YycGex domain and investigated their effects on bacterial functions.
Out of nine YycGex-specific mAbs (2A9, 2B3, 2B4, 2F1, 2F3, 2E3, 2G9, 1H1, and 1H3) generated using hybridoma technology, two YycGex mAbs exhibiting the highest affinity for YycGex protein, mAbs 2F3 and 1H1, were selected for further experiments. Here, we investigated the anti-biofilm activities of these YycGex mAbs and identified the epitope recognized by the antibodies. Our results showed that YycGex mAbs inhibit S. epidermidis biofilm formation and that their target epitope is located within aa 59–70 of YycGex domain. We report, for the first time, that mAbs against the extracellular domain of a TCS can affect biofilm formation. Our findings indicate that the use of YycGex mAbs represents a potential treatment strategy against biofilm infection and, further, that the epitope recognized by YycGex mAbs could serve as a vaccine candidate for preventing S. epidermidis biofilm infections.
Staphylococcus epidermidis RP62A was obtained from American Type Culture Collection (ATCC 35984), and S. epidermidis clinical isolates were collected from the Department of Laboratory Medicine, Affiliated Gulou Hospital, Medical College of Nanjing University. Staphylococcal strains were cultured in tryptic soy broth (TSB; Oxoid Ltd., Basingstoke, United Kingdom), and Escherichia coli strains DH5α and BL21(DE3) were cultured in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl). Kanamycin sulfate was obtained from Sigma Aldrich (St. Louis, MO, United States).
As noted in the introduction section, the S. epidermidis essential histidine kinase YycG contains these domains: two transmembrane sections, an extracytoplasmic domain, cytoplasmic HAMP and PAS domains, and the catalytic core of HisKA and HATPase_c domains. Our previous work showed that inhibitors targeting the HATPase_c domain, developed using a structure-based virtual-screening approach, exhibit both bactericidal activity and potent anti-biofilm activity in the case of staphylococci. Because the extracytoplasmic domain of the protein (YycGex domain) is considered to play roles in sensing external stimuli, we hypothesized that mAbs targeting the YycGex domain might block signal transduction by binding to this domain and thus influence bacterial biofilm formation. A 453-bp NcoI/XhoI DNA fragment encoding the YycGex domain (aa 34–184 of YycG) was amplified from the chromosomal DNA of S. epidermidis RP62A (GenBank accession number NC_002976) by using the primers yycGex-F and yycGex-R (Table 1). The yycGex DNA fragment was ligated into the pET28a (+) expression vector, and the resulting plasmid was transformed into E. coli DH5α competent cells. The YycGex protein was expressed in E. coli BL21(DE3) cells and induced (for 14 h at 25°C) by adding isopropyl β-D-1-thiogalactopyranoside (IPTG; Thermo Fisher Scientific, Waltham, MA, United States) to a final concentration of 0.8 mM. The bacterial cells were lysed by sonication and centrifuged, and the supernatant was used for protein purification by using Ni-NTA agarose (Qiagen, Los Angeles, CA, United States) according to the manufacturer’s instructions. Purified YycGex protein was dialyzed against Tris buffer-15% glycerol and then sterilized by passage through a 0.22-μm-pore-size filter (Millipore, Bedford, MA, United States). The recombinant YycGex protein was used for immunizing BALB/c mice, and YycGex-specific mAbs were generated using the hybridoma technology by Abmart Inc. (Shanghai, China)1.
Overnight cultures of S. epidermidis RP62A were subcultured (1:200) in fresh TSB supplemented with 160 μg/mL YycGex mAbs and incubated at 37°C for 12 h with shaking. The effect of mAbs on bacterial growth was automatically monitored every 0.5 h by measuring the optical density at 600 nm (OD600) by using a BioScreen Growth Curve machine (Growth Curves USA, Piscataway, NJ, United States); normal mouse IgG (mIgG) served as the negative control. The experiments were performed in triplicate and the means and standard error of the mean (SEM) were calculated.
The viable cell counts of bacteria incubated with YycGex mAbs were determined through plate counts on nutrient agar. Overnight cultures of S. epidermidis RP62A were diluted 1:200 into TSB containing 160 μg/mL YycGex mAbs and incubated at 37°C with shaking for 6 or 24 h; subsequently, 1-mL aliquots of bacterial cells were washed twice with ice-cold 0.9% NaCl solution and pelleted by centrifugation for 3 min at 4000 rpm. The pellets were resuspended in 1 mL of ice-cold 0.9% NaCl solution and serially diluted (10-fold), and then 5-μL aliquots from each dilution were spotted onto tryptic soy agar plates and the CFU/mL values were calculated. Three independent experiments were performed for this assay.
Inhibition of biofilm formation by YycGex mAbs was detected using a semiquantitative microtiter plate assay. Overnight planktonic cultures of S. epidermidis RP62A were diluted 1:200 in fresh TSB containing twofold serial dilutions of mAbs (20, 40, 80, and 160 μg/mL), and then microtiter plates containing the samples were initially incubated at 4°C for 2 h and later placed in a 37°C incubator for 24 h. After the incubation and the removal of non-adherent cells by washing thrice with phosphate-buffered saline (PBS), adherent biofilms were fixed with 95% methanol and stained with 2% (w/v) crystal violet, and then excess stain was removed by washing in distilled water. To quantify biofilm formation, OD570 was determined using an xMarkTM Microplate Absorbance Spectrophotometer (Bio-Rad Laboratories, Hercules, CA, United States). Bacteria treated with mIgG served as the control. The experiments were performed using three replicates and repeated thrice independently.
Overnight cultures of S. epidermidis RP62A (1:200 dilution) were inoculated into 1 mL of TSB containing 80 μg/mL YycGex mAbs in FluoroDishes (FD35-100; WPI, Sarasota, FL, United States) and incubated statically at 37°C for 24 h. Planktonic cells were removed by washing with 0.9% NaCl solution, and biofilms were stained using a Live/Dead BacLight Viability kit (Thermo Fisher Scientific, Waltham, MA, United States) for 20 min at room temperature, protected from light exposure. We obtained a series of Z-section images by using a Leica confocal microscope (TCS-SP5, Leica, Wetzler, Germany) with a 63 × oil-immersion objective and then generated three-dimensional images by using IMARIS 7.0.0 software package (Bitplane, Zurich, Switzerland). Three independent experiments were performed.
Overnight-grown S. epidermidis RP62A cells were diluted 1:200 in TSB containing 160 μg/mL YycGex mAbs and incubated at 37°C with shaking at 220 rpm for 6 h. The bacteria were harvested by centrifugation and resuspended in PBS to an OD600 of 0.1 or 1.0 and then loaded in triplicate into either 6-well or 96-well polystyrene microtiter plates, respectively (Nunc GmbH, Wiesbaden, Germany). After incubation for 2 h at 37°C, the plates were washed thrice with PBS to remove non-adherent bacteria, and the adherent bacteria were stained with 0.5% crystal violet as described previously (Lei et al., 2011) and visualized microscopically using an Axiovert 200 inverted microscope (Carl Zeiss, Thornwood, NY, United States) or stained with 2% crystal violet and used for OD570 measurement. The experiments were performed using three replicates and repeated thrice independently.
Polysaccharide intercellular adhesin was detected by performing a lectin dot-blot assay with horseradish peroxidase (HRP)-conjugated wheat germ agglutinin (WGA-HRP, Biotium, Inc. Hayward, CA, United States) as previously described (Wu et al., 2015). Briefly, overnight cultures of S. epidermidis RP62A were diluted 1:200 in TSB containing 160 μg/mL YycGex mAbs and incubated at 37°C with shaking at 220 rpm for 6 h. The bacteria were harvested and resuspended in 50 μL of 0.5 M EDTA (pH 8.0) and then incubated for 5 min at 100°C. After centrifugation (13,000 × g, 5 min), the supernatant was treated with proteinase K (20 mg/mL; Roche) at 37°C for 2 h, after which the enzyme was inactivated at 100°C for 10 min. Next, 1-μL aliquots of serially diluted PIA extracts were spotted onto nitrocellulose membranes (Millipore) by using a 96-well dot-blot vacuum manifold (Biometra GmbH, Niedersachsen, Germany), and after air-drying, the membranes were blocked for 2 h with 3% (w/v) skim milk in Tris-buffered saline containing 0.1% Tween-20 (TBST). Subsequently, the membranes were incubated with WGA-HRP (2 μg/mL) for 1 h at room temperature, and after washing thrice with TBST, the PIA signal was visualized using an enhanced chemiluminescence (ECL) Western blotting detection kit (Thermo Fisher Scientific, Waltham, MA, United States).
Overnight cultures of S. epidermidis RP62A were diluted 1:200 into fresh TSB containing 160 μg/mL YycGex mAbs and incubated at 37°C with shaking for 24 h. Bacterial cells were harvested by centrifugation at 13,000 rpm for 10 min, and the resulting supernatants were filtered through 0.22-μm-pore-size filters (Millipore, Bedford, MA, United States) to obtain cell-free supernatants. The eDNA from the supernatants was isolated using an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1, v/v/v) and then ethanol-precipitated. The precipitated eDNA was washed in 70% ethanol, air-dried, resuspended in TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 8.0), and visualized on gel-red-stained 1% agarose gels.
To measure autolysis, overnight-grown S. epidermidis RP62A was diluted 1:200 into TSB containing 1 M NaCl plus 160 μg/mL YycGex mAbs and incubated at 37°C for 6 h. Next, the bacterial cells were pelleted by centrifugation, and the pellets were washed thrice with ice-cold water and resuspended to an OD600 of 1.0 in 50 mM glycine buffer, pH 8.0, containing 0.05% Triton X-100. Lastly, the bacterial cells were incubated at 37°C with shaking and the OD600 was measured at 0.5-h intervals for 3 h. All experiments were repeated thrice.
Overnight cultures of S. epidermidis RP62A were diluted 1:200 into TSB containing 160 μg/mL YycGex mAbs and incubated at 37°C with shaking at 220 rpm for 6 h. The bacteria were collected by centrifugation at 4000 rpm for 10 min and then washed twice with ice-cold 0.9% NaCl solution. The cell pellets were homogenized by using 0.1-mm zirconia-silica beads in a Mini-BeadBeater (Biospec Products, Bartlesville, OK, United States), operated at a speed of 4800 rpm for 60 s; 5 cycles of homogenization were performed with 1-min intervals on ice. Total RNA was extracted using a RNeasy kit (Qiagen, Los Angeles, CA, United States) according to the manufacturer’s instruction. After RNA extraction, residual DNA was removed from the total RNA samples, and cDNA was synthesized by using a PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio Inc., Otsu, Japan) for manual description. qRT-PCR was performed on an Applied Biosystems 7500 Real Time PCR system (Applied Biosystems, Foster City, CA, United States) by using SYBR Premix Ex Taq (Takara Bio Inc., Otsu, Japan) and these cycling conditions: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. Melting curves were recorded after 40 cycles to confirm primer specificity by heating from 60°C to 95°C. All sample reactions were performed in triplicate with housekeeping gyrase B subunit gene (gyrB) used as an internal standard. Real-time PCR primers for each tested gene are listed in Table 1.
To locate the epitopes recognized by YycGex mAbs, YycGex protein was truncated using the following method. DNA fragments encoding truncated YycGex proteins were subcloned into the BamHI and XhoI sites of a pET SUMO vector, and nine plasmids containing inserts of the desired length were verified through sequencing. All primers used in this study are listed in Table 1. E. coli strain BL21(DE3) carrying the aforementioned recombinant plasmids were incubated in LB medium at 37°C with shaking (at 220 rpm) until an OD600 of 0.6, and then 0.8 mM IPTG was added to induce expression at 25°C for 14 h; subsequently, cells were harvested by centrifugation and boiled in 400 μL of 1 × SDS sample buffer and stored at −20°C.
A KOD -Plus- Mutagenesis Kit (Toyobo, Osaka, Japan) was used to introduce alanine substitutions into the YycGex domain according to the manufacturer’s instructions. The plasmid pET SUMO expressing the truncated YycGex fragment TF34–83 was used as a template. The amplification conditions included an initial denaturation step of 2 min at 94°C, followed by 10 cycles of 10 s at 98°C, 30 s at 55°C, and 6 min at 68°C. The mutant plasmids were verified using DNA sequencing before use in transformation of E. coli strain BL21(DE3). The mutagenesis primers used in the assay are listed in Table 1.
Cell pellets obtained from 5 mL of LB medium were boiled in 400 μL of 1 × SDS sample buffer for 10 min, and total proteins were separated using 10% (w/v) SDS-PAGE. Gels were either stained with Coomassie Brilliant Blue R-250 or the proteins were electrophoretically transferred onto 0.45-μm PVDF membranes (Millipore, Bedford, MA, United States). After blocking for 2 h with 5% skim milk in PBST (PBS containing 0.05% Tween-20), the membranes were incubated with YycGex mAbs (1:1000 in PBST) at room temperature for 3 h. Binding was probed using HRP-conjugated anti-mouse IgG, and after washing for 60 min with PBST, band intensity was measured by staining with an ECL detection reagent (Thermo Fisher Scientific, Waltham, MA, United States), according to the manufacturer’s instructions, and exposing the membranes to X-OMAT film (Kodak, Rochester, NY, United States).
To identify the function of a key amino acid residue recognized by YycGex mAbs in S. epidermidis RP62A, we next constructed a yycG (K65V) mutant in the bacterial chromosome by means of allelic replacement, performed using the temperature-sensitive plasmid pKOR1 as described previously (Bae and Schneewind, 2006). Briefly, a 1.0-kb DNA fragment (1∼1000 bp of yycG) was PCR-amplified from S. epidermidis RP62A genomic DNA (GenBank accession number NC_002976) by using the primers yycG-attB1/2 and then cloned into pKOR1 by using the Gateway BP cloning method, which yielded the plasmid pKOR1-yycG1000. We next used primers K65V-F (5′-GCTGATAAAGATAAAGGTTCAGTCAAC-3′) and K65V-R (5′-ATAAACCTTTTCAATATTGACGTCTAA-3′) harboring the K65V mutation together with the plasmid pKOR1-yycG1000 as the template and used a KOD -Plus- Mutagenesis Kit (Toyobo, Osaka, Japan) according to the manufacturer’s instructions. The mutated plasmid was first transformed into E. coli DC10B and then into S. epidermidis RP62A, and this was followed by allelic replacement as described previously (Zhu et al., 2010; Lou et al., 2011). The presence of the mutation (K65V) in the S. epidermidis genome was verified by sequencing the PCR product of YycGex amplified from the mutant genome.
Experiments were performed using three replicates and repeated thrice independently. Results are expressed as means ± standard deviation (SD) or SEM. For statistical comparisons, Student’s t-tests were performed using GraphPad Prism version 7 (GraphPad Software Inc., San Diego, CA, United States). P < 0.05 was considered statistically significant.
YycG/YycF TCS regulates S. epidermidis biofilm formation (Xu et al., 2017). Because inhibitors targeting the cytoplasmic HATPase_c domain of YycG exhibit anti-biofilm activity (Qin et al., 2006; Huang et al., 2012; Liu et al., 2014; Lv et al., 2017), we generated mAbs against the extracellular domain of YycG (YycGex, aa 34–184) of S. epidermidis RP62A, and from nine hybridoma lines, we selected for further investigation the two mAbs (2F3 and 1H1) that exhibited the highest affinity for the protein.
To test the effect of YycGex mAbs on the biofilm formation of S. epidermidis RP62A, 20–160 μg/mL YycGex mAbs were incubated with the bacteria for 24 h at 37°C, and then biofilm formation was evaluated using a microtiter plate assay; here, mIgG-treated and untreated groups were used as controls. YycGex mAbs inhibited S. epidermidis biofilm formation in a dose-dependent manner (Figure 1A), and mAbs 2F3 and 1H1 at 20 μg/mL reduced biofilm formation by 27.3 and 49.2%, respectively, relative to the control groups. At 160 μg/mL, mAbs 2F3 and 1H1 caused 78.3 and 93.1% biofilm reduction, respectively, whereas the mIgG-treated group only showed 5.9% inhibition. After incubation with YycGex mAbs (80 μg/mL) for 24 h at 37°C, S. epidermidis RP62A formed rough and loose biofilms, as observed by means of Live/Dead staining and CLSM, whereas the biofilms in the mIgG-treated and untreated groups were smooth and evenly distributed. However, the proportions of dead bacteria in the biofilm were similar in all groups (Figure 1B). YycGex mAbs 2F3 and 1H1 did not affect the growth and survival of S. epidermidis RP62A at the highest concentration tested (160 μg/mL) (Supplementary Figure 1).
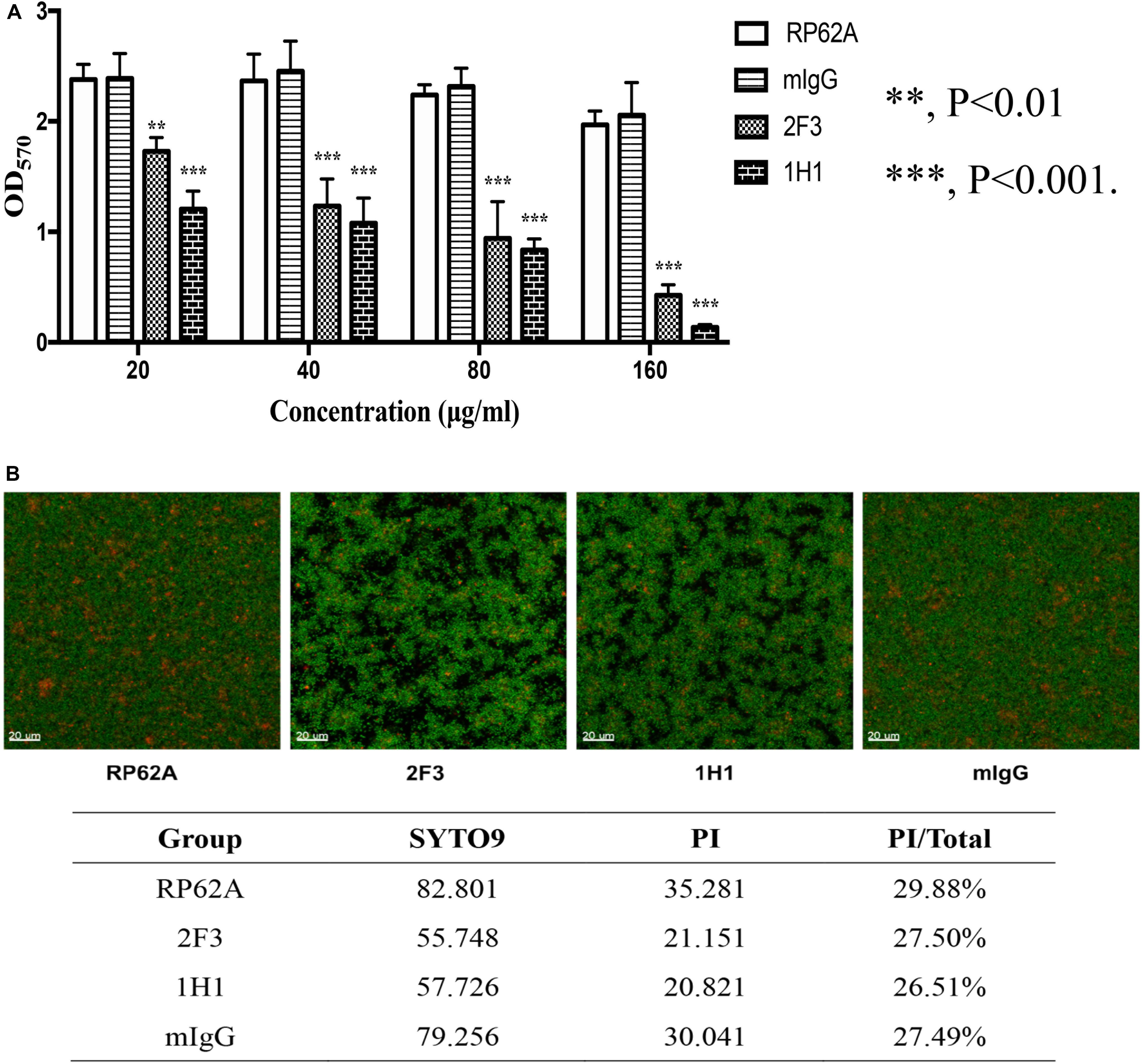
Figure 1. Effects of YycGex mAbs on the biofilm formation of S. epidermidis RP62A. (A) Inhibition of biofilm formation by YycGex mAbs was detected using a microtiter plate assay. S. epidermidis RP62A was incubated with twofold serial dilutions of YycGex mAbs (160–20 μg/mL) at 37°C for 24 h. Biofilms were stained with 2% (w/v) crystal violet and the OD570 was read. Statistical analyses were performed using Student’s t-test. (B) Effect of YycGex mAbs on biofilm morphology of S. epidermidis RP62A, examined using CLSM. S. epidermidis RP62A was incubated with 80 μg/mL YycGex mAbs at 37°C for 24 h. Viable and dead cells fluoresced green and red, respectively. PI/Total values indicated the proportion of dead cells within the biofilms. RP62A, untreated; mIgG, normal mouse IgG treated.
The first step in biofilm formation is bacterial adherence to a surface; thus, we determined how this initial attachment of S. epidermidis is affected by YycGex mAbs, and we used a semiquantitative microtiter plate assay to assess cell attachment. Under the microscope, no cell clusters were detected when S. epidermidis RP62A was cultured in the presence of YycGex mAbs for 6 h at 37°C, whereas clusters were observed in the mIgG-treated and untreated groups (Figure 2A). The adherent bacteria were quantitated by staining with crystal violet and measuring the OD570: Both mAb 2F3 and mAb 1H1 significantly reduced the number of attached cells in the wells (OD570 = 0.118 ± 0.012 and 0.115 ± 0.005, respectively) as compared to that in the untreated control group (OD570 = 0.302 ± 0.013) (P < 0.001) (Figure 2B).
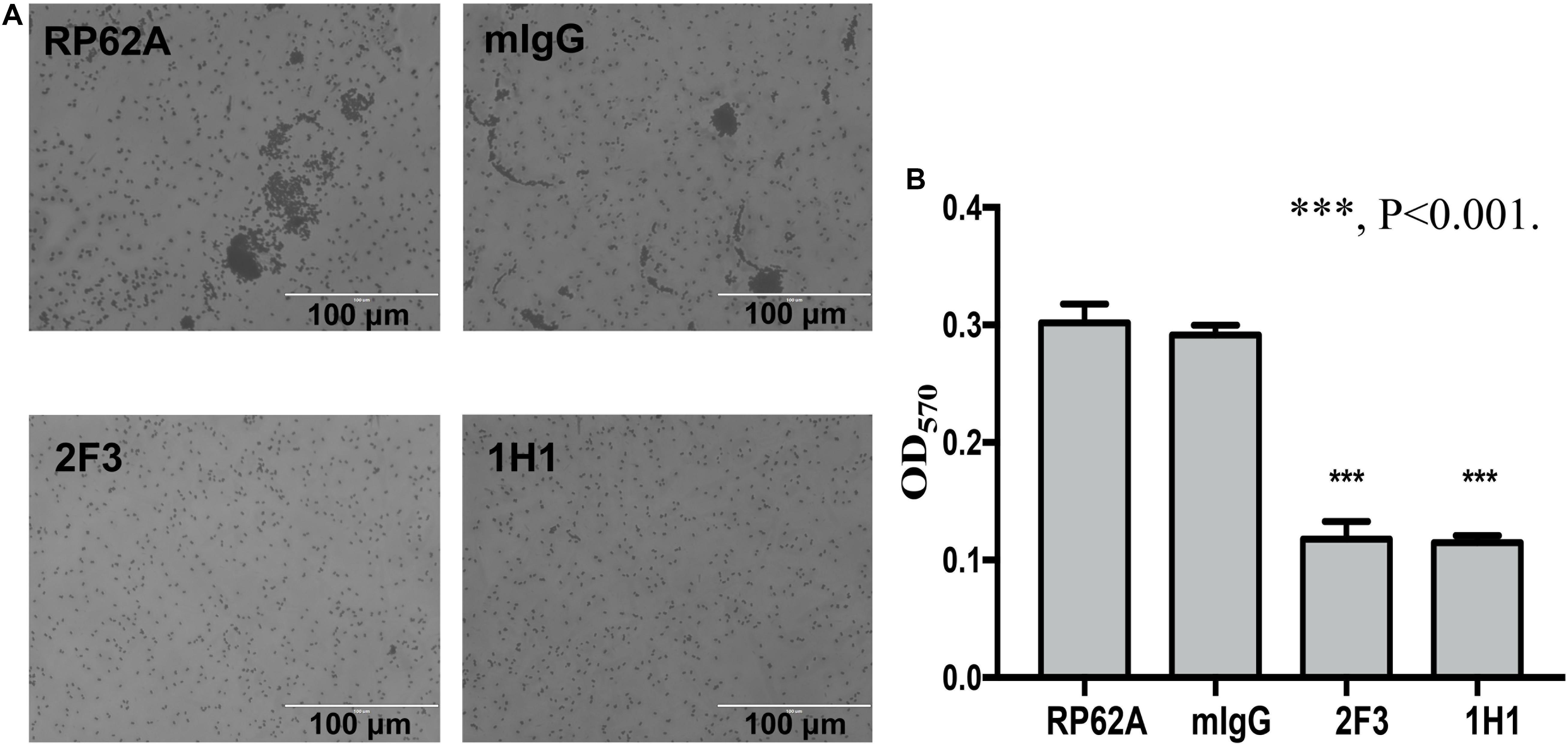
Figure 2. Effect of YycGex mAbs on initial attachment of S. epidermidis RP62A. Bacterial cells were diluted in TSB containing 160 μg/mL YycGex mAbs and incubated at 37°C with shaking at 220 rpm for 6 h. (A) Adherence ability visualized microscopically. (B) Cell attachment measured as OD570 after crystal violet staining. Experiments were performed using three replicates and repeated thrice independently. RP62A: untreated; mIgG: normal mouse IgG treated. Statistical analyses were performed using Student’s t-test. ∗∗∗P < 0.001.
To investigate the effect of YycGex mAbs on EPS production, PIA biosynthesis and eDNA release were determined in the case of S. epidermidis RP62A cultured with 160 μg/mL YycGex mAbs at 37°C for 6 h. Almost no PIA was detected in the presence of 160 μg/mL YycGex mAbs at dilutions up to 1:50 by using WGA-HRP in a lectin dot-blot assay, whereas a considerably stronger signal was detected in the mIgG-treated and untreated groups at dilutions up to 1:200 (Figure 3A). The release of eDNA into the supernatant of cells treated with YycGex mAbs was similar to that in the untreated S. epidermidis RP62A cells, as visualized on gel-red-stained 1% agarose gels (Figure 3B).
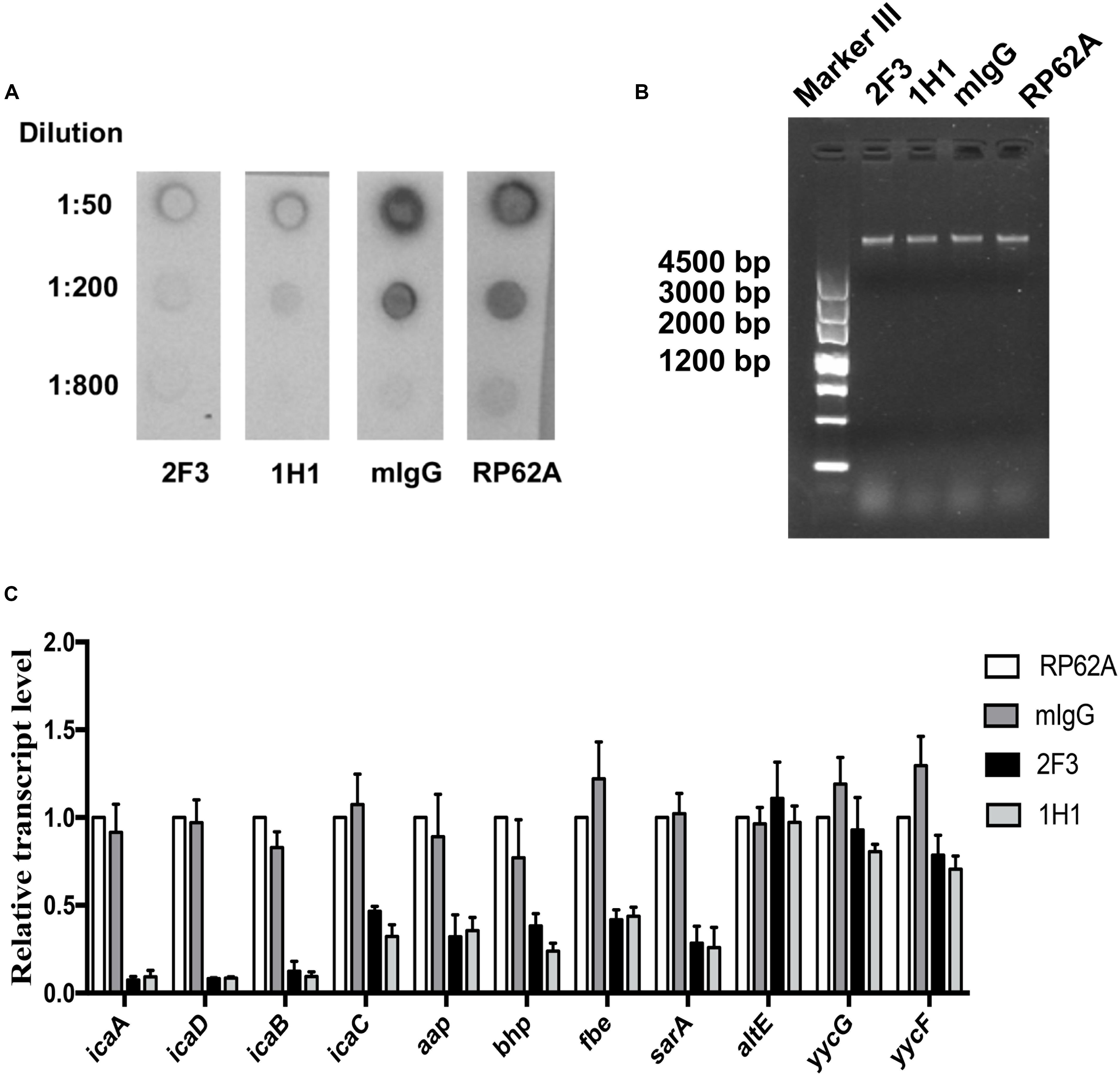
Figure 3. Effect of YycGex mAbs on EPS synthesis and transcription of biofilm-related genes of S. epidermidis RP62A. Overnight cultures of S. epidermidis RP62A were diluted in TSB containing 160 μg/mL YycGex mAbs and incubated at 37°C for 6 h. (A) Effect of YycGex mAbs on PIA synthesis. (B) Effect of YycGex mAbs on eDNA release. eDNA was isolated and analyzed on 1% agarose gels. (C) Effect of YycGex mAbs on transcriptional levels of biofilm-related genes. All reactions were performed in triplicate, with housekeeping gyrase B subunit gene (gyrB) used as an internal standard. RP62A, untreated; mIgG, normal mouse IgG treated.
We next assessed the transcriptional levels of the biofilm-related genes icaADBC, aap, bhp, fbe, and sarA by performing qRT-PCR with RNA samples extracted from S. epidermidis RP62A incubated with 160 μg/mL mAbs 2F3 and 1H1 at 37°C for 6 h. All of these genes were downregulated in the presence of YycGex mAbs, with a marked (8–13-fold) reduction in transcript levels being measured for icaA, icaD, and icaB, which encode PIA; by contrast, the expression of altE, yycG, and yycF did not differ between the mAb-treated and untreated groups (Figure 3C).
Localization of the B-cell epitopes recognized by YycGex-specific mAbs can help in identifying the main antigenic determinants of the YycGex domain and developing peptide vaccines for S. epidermidis biofilm infections. Thus, we mapped the epitopes within YycGex domain by using protein truncation combined with Western blotting analysis. Moreover, by performing alanine-scanning mutagenesis of YycGex proteins, we identified multiple residues that are essential for YycGex mAb binding.
To locate the epitopes recognized by YycGex mAbs, we generated YycGex truncation fragments (TFs) of distinct sizes that were N-terminally fused to a SUMO-tag, and we used Western blotting to examine the binding of YycGex mAbs to these fragments. First, YycGex domain was truncated into three fragments: TF34–83, TF84–133, and TF134–184. Both mAb 2F3 and mAb 1H1 recognized TF34–83 (Figure 4A). Next, TF34–83 was further truncated into TF34–58, TF47–71, and TF59–83; mAbs 2F3 and 1H1 both reacted with TF47–71 and TF59–83, which indicated that the YycGex mAbs recognize the region containing aa 47–83 (Figure 4B). Lastly, the fragment containing aa 47–83 of the YycGex domain was truncated into TF47–58, TF59–70, and TF71–83, and YycGex mAbs were found to bind TF59–70 (Figure 4C).
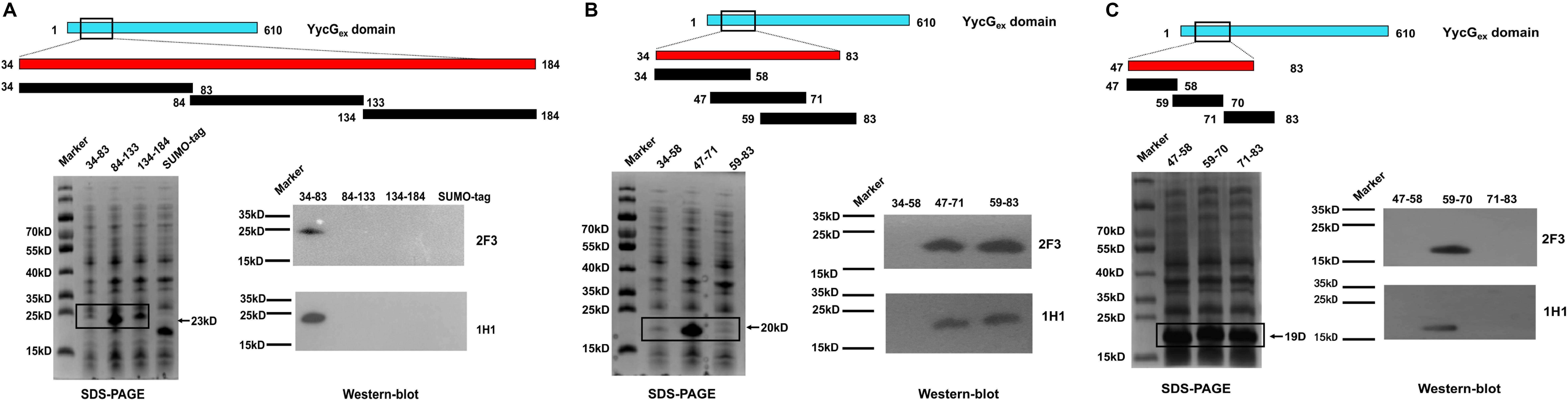
Figure 4. Epitope mapping of YycGex mAbs within YycGex domain. YycGex domain was truncated into fragments of distinct sizes harboring an N-terminally fused SUMO-tag, as shown in the schematic diagrams; the specific recognition of the truncation fragments (TFs) by YycGex mAbs was assessed through Western blotting. Binding of YycGex mAbs to (A) TF34–83, TF84–133, and TF134–184; (B) TF34–58, TF47–71, and TF59–83; and (C) TF47–58, TF59–70, and TF71–83.
To identify the amino acid residues in the epitope that are essential for binding by YycGex mAbs, we substituted every three amino acid residues with alanine individually within aa 47–70 by using site-directed mutagenesis; we constructed eight mutants and used Western blotting to examine their binding by YycGex mAbs. Binding of the epitope by both mAb 2F3 and mAb 1H1 was abolished by the mutations K62V/V63L/Y64V, K65V/D66V/K67V, and D68V/K69V/G70V, which indicated that the key amino acid residues in the epitope recognized by YycGex mAbs are located within aa 62–70: KVYKDKDKG (Figure 5A). Consequently, we mutated these nine amino acid residues individually to alanine and then examined antibody binding; the binding of mAbs 2F3 and 1H1 to the epitope was abolished by four single mutations, Y64V, K65V, K67V, and K69V, which suggested that the key amino acid residues in the YycGex mAb epitope were Tyr64, Lys65, Lys67, and Lys69 (Figure 5B).
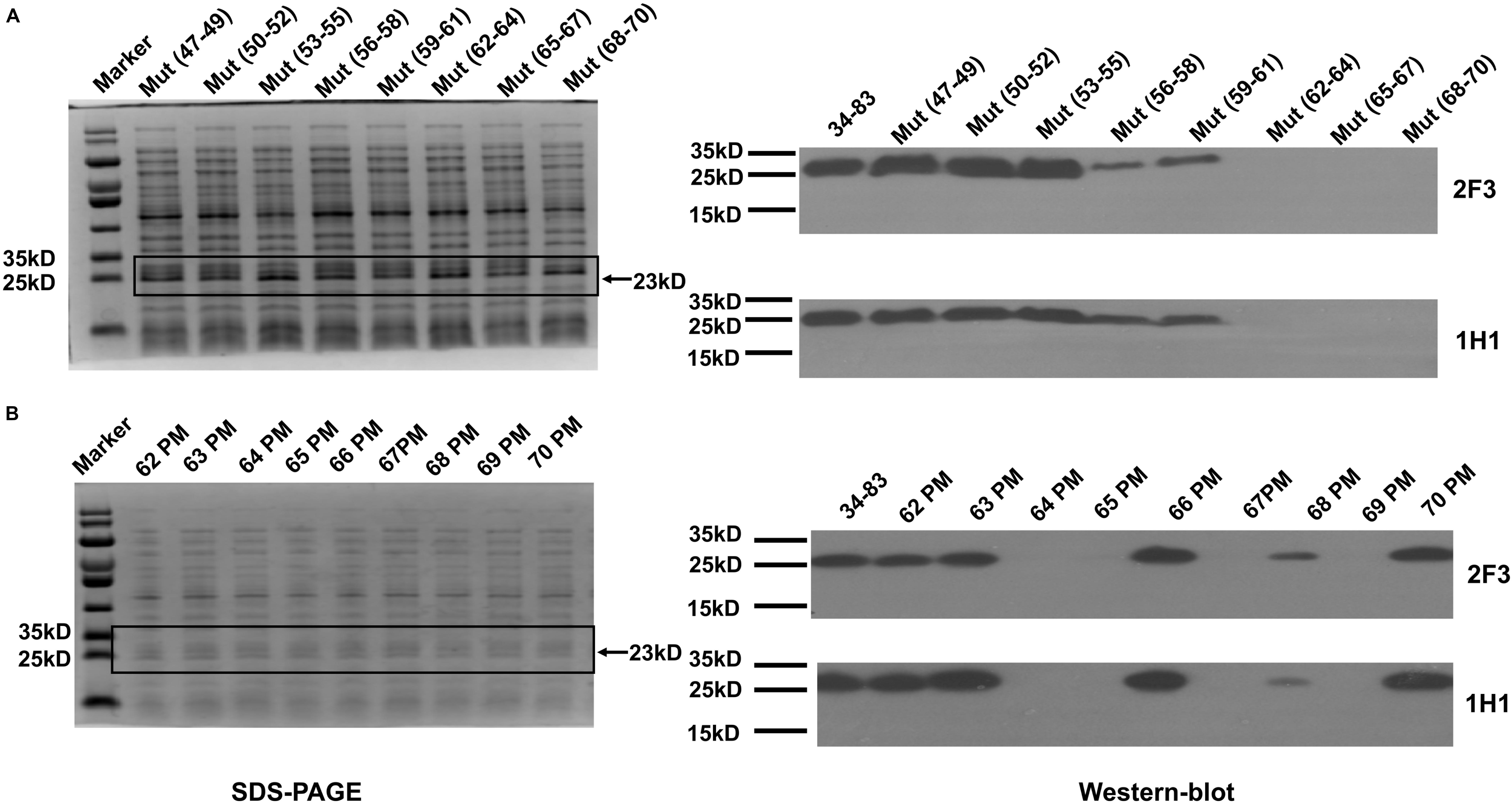
Figure 5. Identification of critical amino acid residues in YycGex domain epitope. Specific recognition by YycGex mAbs of TF34–83 harboring alanine substitutions was assessed using Western blotting. Binding of YycGex mAbs to TF34–83 carrying (A) triple-alanine substitutions and (B) single-alanine substitutions.
Lastly, to evaluate how the key amino acids in the mAb epitope (Tyr64, Lys65, Lys67, and Lys69) affect biofilm inhibition by YycGex mAbs, we generated yycG mutations in S. epidermidis RP62A (Y64V/K65V/K67V/K69V, Y64V, and K65V) through allelic replacement by using the temperature-sensitive plasmid pKOR1, but only the yycG (K65V)-mutant strain was screened out; viable mutant strains were not obtained in the case of the other mutations mentioned. The yycG (K65V) mutant was verified by sequencing the PCR product of YycGex amplified from the mutant genome, and pKOR1 plasmid loss from mutant strains was confirmed through PCR amplification and agarose-gel analysis (Supplementary Figure 2A).
Growth curves and biofilm formation were no different between the yycG (K65V) mutant strain and the parental strain (Supplementary Figures 2B,C). However, the inhibitory effect of YycGex mAbs on biofilm formation of the yycG (K65V) mutant was significantly decreased with increasing concentrations of the antibodies (20, 40, and 80 μg/mL) as compared with the inhibition in the parental RP62A strain (P < 0.05) (Figure 6). At 80 μg/mL, mAbs 2F3 and 1H1 reduced the biofilm formation of RP62A strain by 68.5 and 70.9%, respectively, but caused only 50.0 and 33.5% reduction in the yycG (K65V) mutant.
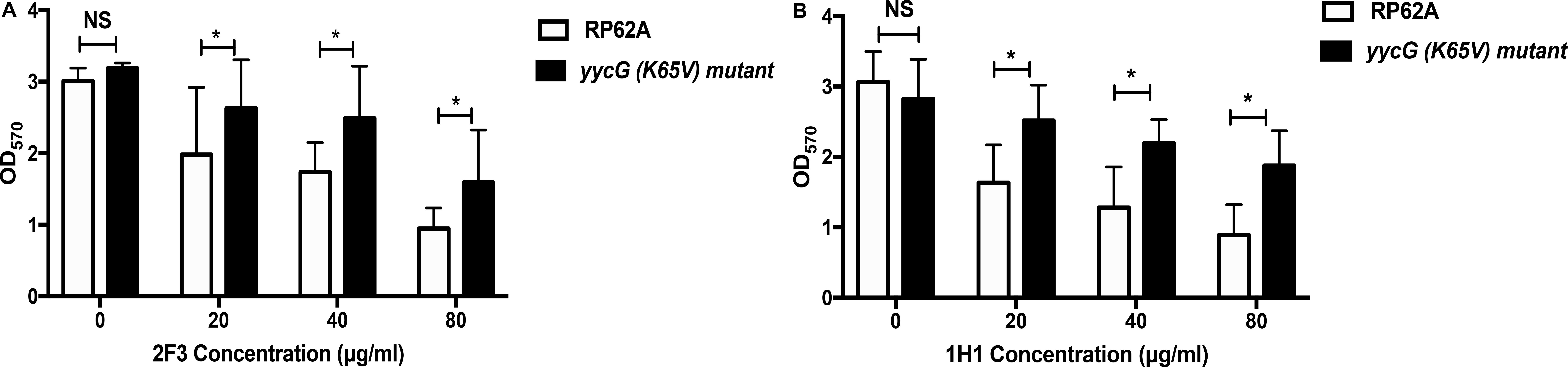
Figure 6. Effect of YycGex mAbs on biofilm formation of the yycG (K65V) mutant. Inhibition of biofilm formation by YycGex mAbs was detected using a semiquantitative plate assay. Overnight cultures were diluted in fresh TSB containing twofold serial dilutions of YycGex mAbs (80–20 μg/mL) and incubated at 37°C for 24 h. Biofilms were stained with 2% crystal violet, and OD570 was measured. Effect of mAb 2F3 (A) and mAb 1H1 (B) on the biofilm formation of the yycG (K65V) mutant and S. epidermidis RP62A. Experiments were performed using three replicates and repeated thrice independently. Statistical analysis was performed using Student’s t-test. *P < 0.05; NS, not significant.
Staphylococcus epidermidis has emerged as one of the most critical opportunistic pathogens of nosocomial infections in recent years because of the increase in frequency of invasive procedures (Vuong et al., 2003; Chessa et al., 2015). The main pathogenicity associated with S. epidermidis involves the formation of biofilms on implanted medical devices (Rupp, 2014; Hamad et al., 2015). The presence of the bacteria in biofilms drastically reduces the efficacy of conventional antibiotics and the host immune system, and this leads to persistent or recurrent infections (Wolcott et al., 2010; Romling and Balsalobre, 2012). The YycG/YycF TCS is essential for bacterial growth and plays a major role in biofilm formation (Fukuchi et al., 2000; Howell et al., 2003; Mohedano et al., 2005; Ahn and Burne, 2007; Bisicchia et al., 2007). YycG histidine kinase is a transmembrane protein, and we have shown that inhibitors that target the cytoplasmic HATPase_c domain of S. epidermidis exhibit bactericidal and biofilm-killing activities (Qin et al., 2006; Huang et al., 2012; Liu et al., 2014; Lv et al., 2017). Previous studies have found that the extracellular domain of the protein (YycGex domain) adopts a PAS fold predicted as a sensor of YycG by sensing environmental signals such as oxygen, light, redox potential, or the presence of specific ligands (Taylor and Zhulin, 1999; Santelli et al., 2007; Chang et al., 2010; Shah et al., 2013; Kim et al., 2016), but little is known of the nature of the signals and factors sensed by YycG. However, whether the blockade of YycGex domain would influence the signal transduction and biofilm formation of the bacterium has remained unclear. Therefore, in this study, we investigated the effect of mAbs against the YycGex domain on the biological functions of S. epidermidis RP62A.
We selected two mAbs showing the highest affinity for YycGex protein, mAbs 2F3 and 1H1, and we found that both mAbs reduced the biofilm formation of S. epidermidis RP62A in a dose-dependent manner under static conditions without affecting bacterial growth. Furthermore, both mAb 2F3 and mAb 1H1 reduced the biofilm formation of five S. epidermidis clinical isolates by 54.1–74.7% at the concentration of 160 μg/mL (Supplementary Table 1), which suggests that YycGex mAbs represent potential candidates for use in clinical treatments to prevent biofilm formation. However, the cell wall of S. epidermidis is formed by a thick layer (20–80 nm) composed mainly of glycine-rich peptidoglycan that might impede the binding of YycGex mAbs to the YycGex domain, and this could be responsible for the high concentration of YycGex mAbs (160 μg/mL) required for producing their effect. Thus, further investigation is necessary to develop methods to reduce the treatment concentration of YycGex mAbs while retaining their strong biofilm-inhibition activity.
We found that YycGex mAbs diminished the capacity of S. epidermidis for initial attachment, which is the first step in biofilm formation. Biofilm formation is commonly divided into three steps: initial adhesion, accumulation, and maturation (Sauer, 2003; Wolska et al., 2016). After attaching to a surface, bacteria produce the EPS, a complex mixture of biopolymers primarily consisting of polysaccharides (PIA), proteins, and nucleic acids (eDNA) (Costerton et al., 1987; Vu et al., 2009). We detected the expression of genes (aap, bhp, fbe, and sarA) encoding proteins related to the initial attachment and accumulation of the S. epidermidis biofilm (Mack et al., 1994; Cho et al., 2002; Bowden et al., 2005; Vuong et al., 2005; Schaeffer et al., 2015), and all of these genes were downregulated after treatment with YycGex mAbs. The downregulation of icaADB expression was in accord with the decrease in PIA synthesis: The ica operon encodes PIA-synthesis enzymes, and PIA is a poly-β (1–6)-N-acetylglucosamine that plays critical roles in intercellular adhesion and in the structural integrity of biofilms (Heilmann et al., 1996; Gerke et al., 1998; Lister and Horswill, 2014; Arciola et al., 2015). Another key EPS component of biofilms is the eDNA released after bacterial autolysis, and this eDNA is related to initial cell adhesion and subsequent intracellular accumulation (Mann et al., 2009; Tang et al., 2013; Regina et al., 2014; Ibanez de Aldecoa et al., 2017). The release of eDNA into the supernatant and the Triton X-100-induced autolysis of the cells that were treated with YycGex mAbs were similar to those of untreated S. epidermidis RP62A cells (Supplementary Figure 3). The signals in the environment induce auto-phosphorylation of the cytoplasmic domain HATPase_c of YycG, and the phosphoryl group is then transferred from histidine residue to an aspartate residue in the receiver domain of the YycF response regulator, which further results in the conformation change of YycF (Mohedano et al., 2005; Cronan and Thomas, 2009). Phosphorylated YycF is usually involved in the DNA-binding and transcription activation of the target genes. The previous study in our lab showed that in S. epidermidis, YycF not only regulates biofilm-associated regulators such as arlRS, sarA, and sarX but also binds to the promoters of icaADBC to directly modulate PIA production, which related with bacterial biofilm formation (Xu et al., 2017). Thus, YycGex mAbs might inhibit biofilm formation by affecting the signal transduction from YycG to YycF, which led to the downregulated expression level of icaADBC and diminished initial-adherence capacity and, thus, likely influenced the subsequent accumulation step.
Next, we determined that the epitope recognized by YycGex mAbs is located within aa 59–70, and by analyzing the antibody-binding region harboring alanine substitutions, we identified four critical amino acids in the epitope: Tyr64, Lys65, Lys67, and Lys69. To further evaluate how these key amino acids affect biofilm inhibition by YycGex mAbs, we generated yycG mutations in the S. epidermidis chromosome (Y64V/K65V/K67V/K69V, Y64V, and K65V). Ultimately, we screened out the yycG (K65V) mutant, and we found that YycGex mAbs more weakly affected the biofilm formation of the yycG (K65V) mutant as compared with that of the parental RP62A strain. This finding indicated that the residue K65 might be directly involved in the binding of YycGex mAbs to YycGex domain. However, we failed to obtain the mutants yycG (Y64V/K65V/K67V/K69V) and yycG (Y64V) in S. epidermidis RP62A, which indicates that some of these residues likely play a crucial role in maintaining bacterial growth and survival.
In summary, YycGex mAbs showed anti-biofilm activity in the case of S. epidermidis and the epitope recognized by the mAbs was located in aa 59–70 of the YycGex domain. The key amino acids within the epitope were identified as Tyr64, Lys65, Lys67, and Lys69, and the importance of Lys65 was confirmed in S. epidermidis RP62A through single-point chromosome mutagenesis. Our findings suggest that mAbs against the YycGex domain could be used in new treatment strategies targeting biofilm-associated infections caused by S. epidermidis. Moreover, the epitope recognized by YycGex mAbs could be used to further study the structure of the YycGex domain and might represent a vaccine candidate for the prevention of S. epidermidis biofilm infections. However, further investigation is necessary to elucidate the mechanism by which YycGex mAbs block S. epidermidis signal transduction. Moreover, the effect of YycGex mAbs against biofilm-associated infections of S. epidermidis needs to be further investigated in a stable animal model.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
All mice experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of School of Basic Medical Sciences, Fudan University (IACUC Animal Project Number 20150119-081) according to Regulations for the Administration of Affairs Concerning Experimental Animals, China.
ZL and DQ conceived and designed the experiments. ZL, YS, XW, YW, JZ, HL, TG, LY, and DQ performed the experiments. ZL, YS, and DQ created the figures, analyzed the data, and wrote the manuscript. All the authors contributed to the article and approved the submitted version.
This work was supported by the National Mega-project for Innovative Drugs (2019ZX09721001), the National Natural Science Foundation of China (81871622 and 81571955), and the National Mega-project for Infectious Diseases (2018ZX10734401).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Dr. Li Chen, Fudan University, for kindly providing the pET SUMO vector.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01839/full#supplementary-material
Ahn, S. J., and Burne, R. A. (2007). Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J. Bacteriol. 189, 6293–6302. doi: 10.1128/jb.00546-07
Arciola, C. R., Campoccia, D., Ravaioli, S., and Montanaro, L. (2015). Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front. Cell. Infect. Microbiol. 5:7. doi: 10.3389/fcimb.2015.00007
Bae, T., and Schneewind, O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63. doi: 10.1016/j.plasmid.2005.05.005
Bisicchia, P., Noone, D., Lioliou, E., Howell, A., Quigley, S., Jensen, T., et al. (2007). The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65, 180–200. doi: 10.1111/j.1365-2958.2007.05782.x
Bowden, M. G., Chen, W., Singvall, J., Xu, Y., Peacock, S. J., Valtulina, V., et al. (2005). Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 151(Pt 5), 1453–1464. doi: 10.1099/mic.0.27534-0
Chang, C., Tesar, C., Gu, M., Babnigg, G., Joachimiak, A., Pokkuluri, P. R., et al. (2010). Extracytoplasmic PAS-like domains are common in signal transduction proteins. J. Bacteriol. 192:1156. doi: 10.1128/JB.01508-09
Chessa, D., Ganau, G., and Mazzarello, V. (2015). An overview of Staphylococcus epidermidis and Staphylococcus aureus with a focus on developing countries. J. Infect. Dev. Ctries 9, 547–550. doi: 10.3855/jidc.6923
Cho, S. H., Naber, K., Hacker, J., and Ziebuhr, W. (2002). Detection of the icaADBC gene cluster and biofilm formation in Staphylococcus epidermidis isolates from catheter-related urinary tract infections. Int. J. Antimicrob. Agents 19, 570–575. doi: 10.1016/s0924-8579(02)00101-2
Costerton, J. W., Cheng, K. J., Geesey, G. G., Ladd, T. I., Nickel, J. C., Dasgupta, M., et al. (1987). Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41, 435–464. doi: 10.1146/annurev.mi.41.100187.002251
Cronan, J. E., and Thomas, J. (2009). Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 459, 395–433. doi: 10.1016/S0076-6879(09)04617-5
Dubrac, S., Bisicchia, P., Devine, K. M., and Msadek, T. (2008). A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70, 1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x
Fabret, C., and Hoch, J. A. (1998). A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180, 6375–6383. doi: 10.1128/.180.23.6375-6383.1998
Fey, P. D., and Olson, M. E. (2010). Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 5, 917–933. doi: 10.2217/fmb.10.56
Fukuchi, K., Kasahara, Y., Asai, K., Kobayashi, K., Moriya, S., and Ogasawara, N. (2000). The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146(Pt 7), 1573–1583. doi: 10.1099/00221287-146-7-1573
Fukushima, T., Szurmant, H., Kim, E. J., Perego, M., and Hoch, J. A. (2008). A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis. Mol. Microbiol. 69, 621–632. doi: 10.1111/j.1365-2958.2008.06308.x
Gerke, C., Kraft, A., Sussmuth, R., Schweitzer, O., and Gotz, F. (1998). Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273, 18586–18593. doi: 10.1074/jbc.273.29.18586
Gotoh, Y., Eguchi, Y., Watanabe, T., Okamoto, S., Doi, A., and Utsumi, R. (2010). Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 13, 232–239. doi: 10.1016/j.mib.2010.01.008
Hamad, T., Hellmark, B., Nilsdotter-Augustinsson, A., and Soderquist, B. (2015). Antibiotic susceptibility among Staphylococcus epidermidis isolated from prosthetic joint infections, with focus on doxycycline. Apmis 123, 1055–1060. doi: 10.1111/apm.12465
Heilmann, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D., and Gotz, F. (1996). Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x
Howell, A., Dubrac, S., Andersen, K. K., Noone, D., Fert, J., Msadek, T., et al. (2003). Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49, 1639–1655. doi: 10.1046/j.1365-2958.2003.03661.x
Huang, R. Z., Zheng, L. K., Liu, H. Y., Pan, B., Hu, J., Zhu, T., et al. (2012). Thiazolidione derivatives targeting the histidine kinase YycG are effective against both planktonic and biofilm-associated Staphylococcus epidermidis. Acta Pharmacol. Sin. 33, 418–425. doi: 10.1038/aps.2011.166
Ibanez de Aldecoa, A. L., Zafra, O., and Gonzalez-Pastor, J. E. (2017). Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front. Microbiol. 8:1390. doi: 10.3389/fmicb.2017.01390
Kim, T., Choi, J., Lee, S., Yeo, K. J., Cheong, H. K., and Kim, K. K. (2016). Structural studies on the extracellular domain of sensor histidine kinase YycG from Staphylococcus aureus and its functional implications. J. Mol. Biol. 428, 3074–3089. doi: 10.1016/j.jmb.2016.06.019
Lei, M. G., Cue, D., Roux, C. M., Dunman, P. M., and Lee, C. Y. (2011). Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J. Bacteriol. 193, 5231–5241. doi: 10.1128/jb.05454-11
Lister, J. L., and Horswill, A. R. (2014). Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 4:178. doi: 10.3389/fcimb.2014.00178
Liu, H., Zhao, D., Chang, J., Yan, L., Zhao, F., Wu, Y., et al. (2014). Efficacy of novel antibacterial compounds targeting histidine kinase YycG protein. Appl. Microbiol. Biotechnol. 98, 6003–6013. doi: 10.1007/s00253-014-5685-8
Lou, Q., Zhu, T., Hu, J., Ben, H., Yang, J., Yu, F., et al. (2011). Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiol. 11:146. doi: 10.1186/1471-2180-11-146
Lv, Z., Zhao, D., Chang, J., Liu, H., Wang, X., Zheng, J., et al. (2017). Anti-bacterial and anti-biofilm evaluation of thiazolopyrimidinone derivatives targeting the histidine kinase YycG protein of Staphylococcus epidermidis. Front. Microbiol. 8:549. doi: 10.3389/fmicb.2017.00549
Mack, D., Nedelmann, M., Krokotsch, A., Schwarzkopf, A., Heesemann, J., and Laufs, R. (1994). Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62, 3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994
Mann, E. E., Rice, K. C., Boles, B. R., Endres, J. L., Ranjit, D., Chandramohan, L., et al. (2009). Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. doi: 10.1371/journal.pone.0005822
McCann, M. T., Gilmore, B. F., and Gorman, S. P. (2008). Staphylococcus epidermidis device-related infections: pathogenesis and clinical management. J. Pharm. Pharmacol. 60, 1551–1571. doi: 10.1211/jpp/60.12.0001
Mikkelsen, H., Sivaneson, M., and Filloux, A. (2011). Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 13, 1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x
Mohedano, M. L., Overweg, K., de la Fuente, A., Reuter, M., Altabe, S., Mulholland, F., et al. (2005). Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187, 2357–2367. doi: 10.1128/jb.187.7.2357-2367.2005
Ng, W. L., Tsui, H. C., and Winkler, M. E. (2005). Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J. Bacteriol. 187, 7444–7459. doi: 10.1128/jb.187.21.7444-7459.2005
Ng, W. L., and Winkler, M. E. (2004). Singular structures and operon organizations of essential two-component systems in species of Streptococcus. Microbiology 150(Pt 10), 3096–3098. doi: 10.1099/mic.0.27550-0
Qin, Z., Zhang, J., Xu, B., Chen, L., Wu, Y., Yang, X., et al. (2006). Structure-based discovery of inhibitors of the YycG histidine kinase: new chemical leads to combat Staphylococcus epidermidis infections. BMC Microbiol. 6:96. doi: 10.1186/1471-2180-6-96
Rasamiravaka, T., Labtani, Q., Duez, P., and El Jaziri, M. (2015). The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed. Res. Int. 2015:759348. doi: 10.1155/2015/759348
Regina, V. R., Lokanathan, A. R., Modrzynski, J. J., Sutherland, D. S., and Meyer, R. L. (2014). Surface physicochemistry and ionic strength affects eDNA’s role in bacterial adhesion to abiotic surfaces. PLoS One 9:e105033. doi: 10.1371/journal.pone.0105033
Romling, U., and Balsalobre, C. (2012). Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272, 541–561. doi: 10.1111/joim.12004
Rupp, M. E. (2014). Clinical characteristics of infections in humans due to Staphylococcus epidermidis. Methods Mol. Biol. 1106, 1–16. doi: 10.1007/978-1-62703-736-5_1
Santelli, E., Liddington, R. C., Mohan, M. A., Hoch, J. A., and Szurmant, H. (2007). The crystal structure of Bacillus subtilis YycI reveals a common fold for two members of an unusual class of sensor histidine kinase regulatory proteins. J. Bacteriol. 189, 3290–3295. doi: 10.1128/jb.01937-06
Sauer, K. (2003). The genomics and proteomics of biofilm formation. Genome Biol. 4:219. doi: 10.1186/gb-2003-4-6-219
Schaeffer, C. R., Woods, K. M., Longo, G. M., Kiedrowski, M. R., Paharik, A. E., Buttner, H., et al. (2015). Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect. Immun. 83, 214–226. doi: 10.1128/iai.02177-14
Senadheera, M. D., Guggenheim, B., Spatafora, G. A., Huang, Y. C., Choi, J., Hung, D. C., et al. (2005). A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 187, 4064–4076. doi: 10.1128/jb.187.12.4064-4076.2005
Shah, N., Gaupp, R., Moriyama, H., Eskridge, K. M., Moriyama, E. N., and Somerville, G. A. (2013). Reductive evolution and the loss of PDC/PAS domains from the genus Staphylococcus. BMC Genomics 14:524. doi: 10.1186/1471-2164-14-524
Szurmant, H., Bu, L., Brooks, C. L., and Hoch, J. A. (2008). An essential sensor histidine kinase controlled by transmembrane helix interactions with its auxiliary proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 5891–5896. doi: 10.1073/pnas.0800247105
Tang, L., Schramm, A., Neu, T. R., Revsbech, N. P., and Meyer, R. L. (2013). Extracellular DNA in adhesion and biofilm formation of four environmental isolates: a quantitative study. FEMS Microbiol. Ecol. 86, 394–403. doi: 10.1111/1574-6941.12168
Taylor, B. L., and Zhulin, I. B. (1999). PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. MMBR 63, 479–506. doi: 10.1128/mmbr.63.2.479-506.1999
Vu, B., Chen, M., Crawford, R. J., and Ivanova, E. P. (2009). Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554. doi: 10.3390/molecules14072535
Vuong, C., Gerke, C., Somerville, G. A., Fischer, E. R., and Otto, M. (2003). Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188, 706–718. doi: 10.1086/377239
Vuong, C., Kidder, J. B., Jacobson, E. R., Otto, M., Proctor, R. A., and Somerville, G. A. (2005). Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187, 2967–2973. doi: 10.1128/jb.187.9.2967-2973.2005
Winkler, M. E., and Hoch, J. A. (2008). Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190, 2645–2648. doi: 10.1128/jb.01682-1687
Wolcott, R. D., Rhoads, D. D., Bennett, M. E., Wolcott, B. M., Gogokhia, L., Costerton, J. W., et al. (2010). Chronic wounds and the medical biofilm paradigm. J. Wound Care 19, 45–52. doi: 10.12968/jowc.2010.19.2.46966
Wolska, K. I., Grudniak, A. M., Rudnicka, Z., and Markowska, K. (2016). Genetic control of bacterial biofilms. J. Appl. Genet. 57, 225–238. doi: 10.1007/s13353-015-0309-2
Wu, Y., Wu, Y., Zhu, T., Han, H., Liu, H., Xu, T., et al. (2015). Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions. J. Bacteriol. 197, 459–476. doi: 10.1128/jb.02231-14
Xu, T., Wu, Y., Lin, Z., Bertram, R., Gotz, F., Zhang, Y., et al. (2017). Identification of genes controlled by the essential YycFG two-component system reveals a role for biofilm modulation in Staphylococcus epidermidis. Front. Microbiol. 8:724. doi: 10.3389/fmicb.2017.00724
Yao, Y., Vuong, C., Kocianova, S., Villaruz, A. E., Lai, Y., Sturdevant, D. E., et al. (2006). Characterization of the Staphylococcus epidermidis accessory-gene regulator response: quorum-sensing regulation of resistance to human innate host defense. J. Infect. Dis. 193, 841–848. doi: 10.1086/500246
Keywords: Staphylococcus epidermidis, biofilm, YycG histidine kinase, two-component system, epitope mapping
Citation: Lyu Z, Shang Y, Wang X, Wu Y, Zheng J, Liu H, Gong T, Ye L and Qu D (2020) Monoclonal Antibodies Specific to the Extracellular Domain of Histidine Kinase YycG of Staphylococcus epidermidis Inhibit Biofilm Formation. Front. Microbiol. 11:1839. doi: 10.3389/fmicb.2020.01839
Received: 30 April 2020; Accepted: 14 July 2020;
Published: 07 August 2020.
Edited by:
Luis Cláudio Nascimento da Silva, Universidade Ceuma, BrazilReviewed by:
Rodolfo García-Contreras, National Autonomous University of Mexico, MexicoCopyright © 2020 Lyu, Shang, Wang, Wu, Zheng, Liu, Gong, Ye and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Di Qu, ZHF1QHNobXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.