- 1Department of Biotechnology, Alagappa University, Karaikudi, India
- 2Department of Periodontology, College of Dentistry, University of Florida, Gainesville, FL, United States
Methicillin-resistant Staphylococcus aureus (MRSA) is a serious human pathogen which has been listed as a high-priority multi-drug resistance pathogen by the World Health Organization (WHO). Persistent MRSA infections are often associated with biofilm formation and resistance to conventional antimicrobial therapy. Inhibiting the surface adherence and the virulence of the bacterium is the current alternative approach without affecting growth to reduce the possibility of resistance development. Although numerous antibiofilm agents have been identified, their mode of action remains unclear. Combining two drugs with different modes of action will improve the efficiency of the treatment strategy against MRSA. The present study was aimed to decipher the molecular mechanism underlying the antibiofilm activity of thymol against MRSA and assess the ability of thymol to improve the antibacterial activity of rifampicin. Thymol significantly inhibited 88% of MRSA biofilm formation at 100 μg/ml and reduced the surface adherence of MRSA on glass, stainless steel, and titanium surface coated with human plasma as evidenced by microscopic analyses. qPCR analysis of global virulence regulatory genes and biofilm assay with S. aureus wild type, ΔsarA, and Δagr strains revealed the sarA-mediated antibiofilm activity of thymol and inhibition of sarA-controlled virulence factors. Congo red assay and erythrocyte lysis assay further confirmed the reduction in polysaccharide intracellular adhesin and hemolysin. Importantly, thymol enhanced the antibacterial and the biofilm eradication efficiency of rifampicin against MRSA and also reduced the formation of persisters. Thus, the present study reveals the sarA-dependent antibiofilm efficacy of MRSA and suggests thymol as the promising combinatorial candidate in potentiating the antibacterial activity of rifampicin against persistent MRSA infections.
Introduction
Staphylococcus aureus, a Gram-positive human commensal bacterium of the nasal epithelium turns virulent when the individual is immune-compromised. S. aureus not alone causes simple skin infections but also causes lethal infections such as pulmonary infections, invasive endocarditis, septic arthritis, osteomyelitis, and peri-implantitis (Taylor and Unakal, 2020). The treatment of these lethal conditions is a challenging task due to evolving multi-drug-resistant strains such as methicillin-resistant S. aureus (MRSA). MRSA is majorly involved in community-associated, hospital-associated as well as livestock-associated infections (Palavecino, 2014). Numerous virulence traits of MRSA make it stubborn against the conventional antibiotic therapy. Biofilm formation is one such trait which enables the bacterium to survive against various physiological stresses. Bacterial cells adhered to a surface with the help of slimy polymeric substances called biofilm, and this biofilm provides multicellular behavior to the unicellular bacterial cells. The rate of gene transfer inside the biofilm is higher than that between planktonic cells (Kumar et al., 2019). Bacterial cells especially living in biofilm mode are having a peculiar behavior of synchronized virulence gene expression, and many enzymes present in biofilm matrix are reported to cleave the antibiotics. In addition, the biofilm blocks the penetration of antibiotics as it is slimy and mucus in nature. Altogether the bacterial cells residing in biofilm are much more resistant to antibiotics than their planktonic counterparts (Olsen, 2015; Hall and Mah, 2017).
Bacterium has an intricate regulatory network to coordinate the synthesis of virulence factors. Although various global regulatory systems have been identified in S. aureus, the accessory gene regulatory (agr) system and the staphylococcal accessory regulatory system (sarA) are prototypes in nature. More specifically, both systems are well known to regulate biofilm formation and virulence factor production in a reciprocal way (Jenul and Horswill, 2018). That is, the active agr system negatively regulates the adhesion genes responsible for biofilm formation and leads to biofilm dispersal, whereas the active sarA system enhances the biofilm formation. Hence, agr and sarA systems act as molecular switches in regulating biofilm formation in S. aureus (Vasudevan, 2019).
As biofilm plays a critical role in the development of antibiotic resistance, targeting biofilm formation has become an alternative strategy to antibiotics (Buommino et al., 2014). Notably, antibiofilm agents are reported to potentiate the efficacy of antibiotics against bacterial biofilm when combined with antibiotics. Plenty of compounds with antibiofilm activity have been identified from various natural resources so far (Abraham et al., 2012; Sethupathy et al., 2016, 2017; Selvaraj et al., 2019; Valliammai et al., 2019). Thymol is a major constituent in the essential oil of thyme plant (Thymus vulgaris), and it is known to have various biological properties such as antibacterial, antifungal, antioxidant, and cognitive-enhancing activities (Braga, 2005; Tohidpour et al., 2010; Azizi et al., 2012). Few reports are available on the antibiofilm activity of thymol against S. aureus, but the molecular mechanism underlying the biofilm inhibitory potential of thymol remains unclear (Nostro et al., 2007; García-Salinas et al., 2018). Henceforth, the goal of the present study is to unravel the molecular mechanism of the antibiofilm efficacy of thymol and to find out the ability of thymol to improve the efficacy of rifampicin.
Materials and Methods
Bacterial Strains and Growth Conditions
The S. aureus strains used in the present study are listed in Table 1. The reference MRSA strain used throughout the study was obtained from the American Type Culture Collection (ATCC). The clinical isolates were collected from pharingitis patients at the Rajaji Government Hospital, Madurai, Tamil Nadu for our earlier work (Gowrishankar et al., 2012). Newman wild-type and mutant strains of S. aureus were gifted by Dr. Christiane Wolz, a professor at the Institute for Medical Microbiology and Hygiene, University of Tubingen, Germany. For the biofilm assays, all the bacterial strains were cultured in tryptone soya broth (TSB) supplemented with 1% sucrose (TSBS) and kept at 37°C in the shaking incubator for 24 h. For maintenance, the bacterial cultures were grown in TSB and stored as glycerol stocks at −80°C.
Stock Solution
Thymol was purchased from Sigma-Aldrich, India. Ten milligrams of thymol dissolved in 1 ml of methanol was used as stock solution (10 mg/ml). Methanol alone was used as vehicle control in all the assays.
Determination of Biofilm Inhibitory Concentration
A 24-well polystyrene microtiter plate (MTP) containing 1 ml of TSBS with various concentrations of thymol (20–200 μg/ml) was inoculated with 1% of 6-h cultures of S. aureus strains (1 × 108 cells) and incubated at 37°C for 24 h. Methanol was used as vehicle control. For biofilm quantification, planktonic cells were carefully removed and biofilm cells were washed with sterile saline solution to remove unbound cells, and the plate was air-dried. Then, the biofilm cells were stained with 0.4% crystal violet solution for 20 min, washed to remove the excess stain, and air-dried. For quantification, the biofilm cells were destained with 30% glacial acetic acid solution, and absorbance was read at 570 nm using a multi-label reader (Spectramax M3, United States). The percentage of biofilm inhibition was calculated using the formula:
The lowest concentration of thymol which exhibited maximum biofilm inhibition was set as the biofilm inhibitory concentration (BIC) (Valliammai et al., 2019).
Colony-Forming Unit and Alamar Blue Assay
Two milliliters of MRSA cultures grown in the absence and the presence of thymol (25, 50, and 100 μg/ml) for 24 h was measured for absorbance at 600 nm. The control and the treated cultures were serially diluted and spread on a plate for colony-forming unit (CFU) counting. Then, the control and the treated cells were pelletized by centrifugation and dispersed in 2 ml of phosphate-buffered saline (PBS). A total of 900 μl of this suspension was aliquoted into wells of a 24-well MTP and 100 μl of Alamar blue solution (6.5 mg/ml) was added to each well. This plate was kept in the dark for 4 h, and fluorescence was measured at 530 and 590 nm for excitation and emission, respectively (Sarker et al., 2007).
Light and Confocal Laser Scanning Microscopic Analyses
For microscopic analysis, biofilm was allowed to form on glass/stainless steel slides (1 cm × 1 cm) immersed in TSBS in the absence and the presence of thymol (25, 50, and 100 μg/ml) for 24 h at 37°C. After 24 h, the glass slides were taken out and washed with sterile saline solution. For the light microscopic analysis, the glass slides were stained with 0.4% crystal violet for 20 min and washed. After drying, the glass slides were observed under a light microscope (Nikon Eclipse 80i, United States) at ×400 magnification. For confocal laser scanning microscopic analysis, stainless steel slides were stained with 0.1% acridine orange in the dark for 20 min and washed. After drying, the stainless steel slides were observed under CLSM (LSM 710, Carl Zeiss, Germany) at ×200 magnification (Valliammai et al., 2019).
Plasma Coating on Titanium Surface and Scanning Electron Microscopy Analysis
Plasma extracted from healthy human blood was diluted to 20% concentration using 50 mM sodium bicarbonate solution. Titanium slides (1 cm × 1 cm) placed in a 24-well MTP were covered with 1 ml of the prepared plasma solution and incubated at 4°C overnight. After the incubation period, the plasma solution was removed and the titanium slides were washed with sterile distilled water. Then, biofilm was allowed to form on the titanium surface in the absence and the presence of thymol (25, 50, and 100 μg/ml) as mentioned earlier. After 24 h of biofilm formation, the titanium slides were taken out and washed. For SEM analysis, the titanium slides were fixed with 2% glutaraldehyde at 4°C overnight and dehydrated with increasing concentrations of ethanol (20, 40, 60, 80, and 100%). After drying, the titanium slides were subjected to gold sputtering and observed under SEM (VEGA 3 TESCAN, Czech Republic) (Walker and Horswill, 2012).
qPCR Analysis
Total RNA from 24-h control and thymol (100 μg/ml)-treated MRSA cultures was extracted using the TRIzol method of RNA extraction and converted to cDNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, United States). The quantification of gene expression in the control and the thymol-treated samples was performed in triplicate on a thermal cycler (7500 Sequence Detection System, Applied Biosystems Inc., Foster, CA, United States) using a PCR mix (SYBR Green Kit, Applied Biosystems, United States) at a predefined ratio. The fold change in gene expression was calculated by the 2–ΔΔCt method with gyrB as the housekeeping gene (Livak and Schmittgen, 2001). The details of the primer sequences of the genes (agrA, agrC, sarA, icaA, icaD, fnbA, fnbB, and hla) used in this study are given in Table 2.
Biofilm Assay With Mutant Strains
As stated earlier, a 24-well MTP assay was performed with the S. aureus wild-type strain and isogenic ΔsarA and Δagr strains. All these strains were treated with increasing concentrations of thymol (20–100 μg/ml) for 24 h at 37°C and observed for biofilm inhibition using crystal violet as mentioned.
Congo Red Agar Assay
Tryptone soya broth supplemented with 1% sucrose containing 2% bacteriological agar and 0.08% Congo red was prepared and sterilized. After sterilization, thymol (25, 50, and 100 μg/ml) or methanol was mixed with the media and poured into Petri plates. The MRSA culture was streaked on the agar plates with and without thymol and incubated at 37°C for 24 h. After 24 h of incubation, the plates were photographed (Knobloch et al., 2002).
Extracellular Polysaccharide Quantification
Biofilm formation assay was done in the absence and the presence of thymol (25, 50, and 100 μg/ml) as stated earlier. After 24 h of biofilm formation, planktonic cells were discarded and the biofilm matrix was scraped and collected using sterile PBS. To the collected biofilm, an equal volume of 5% phenol and five volumes of concentrated sulfuric acid containing 0.2% hydrazine sulfate were added and mixed well. This mixture was incubated in the dark at room temperature for 1 h. Then, the samples were centrifuged at 10,000 rpm for 10 min to collect the supernatants, and absorbance was measured at 490 nm (Dubois et al., 1951).
Erythrocyte Lysis Assay
The 48-h MRSA cultures grown in the absence and the presence of thymol (25, 50, and 100 μg/ml) were centrifuged to collect cell-free culture supernatant (CFCS). Then, 100 μl of CFCS was mixed with 900 μl of 2% human red blood cell suspension prepared in PBS. This mixture was incubated at 37°C for 1 h and centrifuged at 10,000 rpm for 10 min. The collected supernatant was read for absorbance at 540 nm (Bernheimer, 1988).
Determination of Minimum Inhibitory Concentration and Combinatorial Growth Inhibition Assay
Test tubes containing 2 ml of TSB with 1% MRSA cells (1 × 108 cells) were treated with either thymol (0–200 μg/ml) or rifampicin (0–1 μg/ml) for 24 h at 37°C. After incubation, growth optical density (OD) at 600 nm was measured to determine the BIC. For the combinatorial assay, MRSA cells were treated with rifampicin (0.015, 0.03, and 0.06 μg/ml) in the presence and the absence of thymol (100 μg/ml) and incubated at 37°C for 24 h. After incubation along with growth OD at 600 nm, metabolic viability was also measured using Alamar blue assay as mentioned previously (Sarker et al., 2007).
Disc Diffusion Assay
Methicillin-resistant Staphylococcus aureus cells (5 × 108 cells) were spread on the surface of tryptic soya agar (TSA) plates without and with thymol (100 μg/ml) using sterile cotton swabs. The sterile discs were placed in the center of the agar surface and loaded with increasing concentrations of rifampicin (0.015, 0.03, and 0.06 μg/ml). The agar plates were incubated at 37°C for 24 h and photographed.
Time Killing Assay
Methicillin-resistant Staphylococcus aureus cultures (1 × 108 cells) were treated with rifampicin (0.06 μg/ml) and/or thymol (100 μg/ml), and 5 μl of the control and the treated cultures was spotted on the TSA plates every 1 h. In addition, 100 μl of the control and the treated cultures was spread on TSA plates every 2 h after serial dilution for CFU counting. The agar plates were incubated for 24 h at 37°C and photographed (Poonacha et al., 2017).
Persister Formation Assay
An overnight culture of MRSA was diluted to 1:1,000 ratio using TSB and incubated at 37°C in the shaking incubator at 250 rpm to reach an exponential phase (0.8 OD at 600 nm). Then, the exponential-phase MRSA cultures were challenged with ×100 concentration of rifampicin (6 μg/ml) in the absence and the presence of thymol (25, 50, and 100 μg/ml) and incubated at 37°C in the shaking incubator at 250 rpm for 3 h. After incubation, 100 μl of the culture was spread on TSA, and persister cell survival was determined by counting the number of colonies formed on TSA after incubation for 24 h at 37°C (Lee et al., 2016).
Biofilm Eradication Assay
Methicillin-resistant Staphylococcus aureus cells were allowed to form a biofilm in a 24-well MTP for 24 h without any treatment. The preformed biofilm was then treated with rifampicin (0.06 μg/ml) and/or thymol (100 μg/ml) for 12 h. After treatment, the planktonic cells were discarded and the biofilm cells were scraped out using sterile PBS and assessed for viability using Alamar blue as mentioned earlier. Additionally, the biofilm cells were serially diluted and plated for CFU counting.
Statistics
All the experiments were carried out in three biological replicates with at least two experimental replicates, and the results were expressed as mean ± standard deviation. Statistical analysis was performed using SPSS 17.0 software package (SPSS Inc., Chicago, IL, United States), and one-way ANOVA followed by Duncan’s post hoc test was used to assess the significance. A p-value ≤ 0.05 was set as statistically significant.
Results
Thymol Inhibits MRSA Biofilm Formation Without Affecting Growth
The crystal violet quantification of biofilm formed in the absence and the presence of various concentrations of thymol (20–200 μg/ml) exhibited a dose-dependent antibiofilm activity with maximum biofilm inhibition of 88% at 100 μg/ml without affecting growth. Beyond this concentration, thymol started to affect the growth of MRSA and biofilm inhibition was not much increased. Hence, 100 μg/ml of thymol was considered as the BIC and taken for further assays (Figure 1). The same concentration of thymol was also found to inhibit the biofilm formation of clinical isolates, namely, MSSA-51, MSSA-46, and MRSA-44 (Supplementary Figure S1). CFU analysis and growth OD demonstrated the non-antibacterial activity of thymol at 25, 50, and 100 μg/ml (Supplementary Figure S2A). In the Alamar blue assay, the metabolic viability of thymol-treated MRSA cells was comparable to that of the control cells, confirming that thymol did not affect the metabolic viability of MRSA (Supplementary Figure S2B).
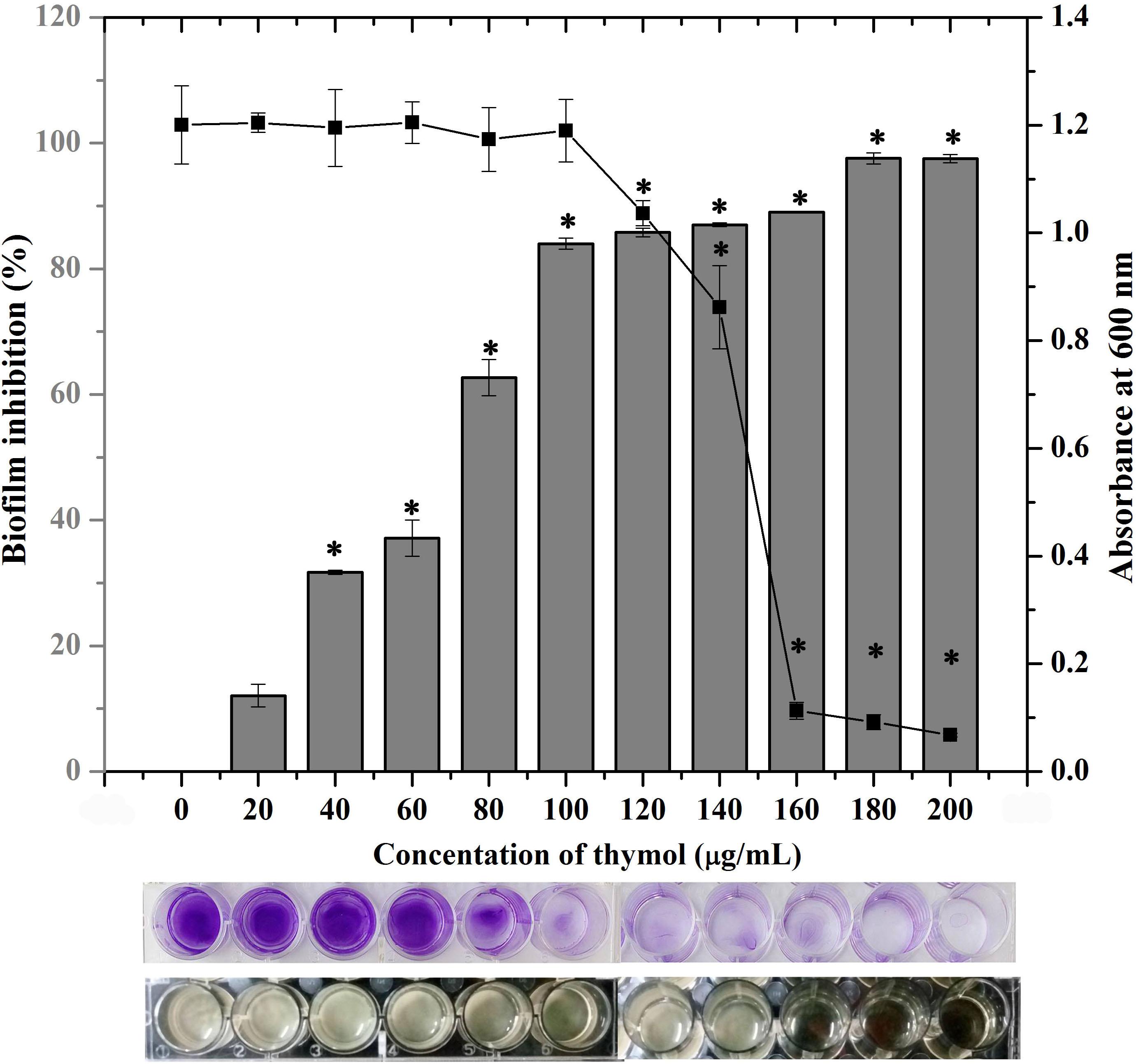
Figure 1. Effect of thymol on the biofilm formation of methicillin-resistant Staphylococcus aureus (MRSA) as assessed by crystal violet staining. The bar graph indicates the percentage of biofilm inhibition and the line graph indicates the growth at OD600 nm. The bottom images show the corresponding crystal violet-stained biofilm and the growth of MRSA in microtitre plate. The error bars indicate standard deviations. The asterisks represent statistical significance (p < 0.05).
Thymol Impedes the Adherence of MRSA on Glass, Stainless Steel Slides, and Titanium Slide Coated With Human Plasma
After confirming the non-antibacterial nature of thymol, the antibiofilm efficacy of thymol was further assessed by microscopic analysis. Light micrographs showed the gradual reduction in surface coverage with increasing concentrations of thymol. Furthermore, CLSM micrographs confirmed the ability of thymol to inhibit the surface adherence of MRSA, and notably, the thickness of the biofilm was also reduced upon increasing concentrations of thymol treatment (Figure 2). The human-plasma-coated titanium slides were subjected to MRSA biofilm formation in the absence and the presence of thymol and observed under SEM. The control titanium surface was observed to be completely covered by three-dimensional MRSA biofilm, whereas a monolayer of dispersed MRSA cells was observed in the thymol-treated titanium surface (Figure 3).
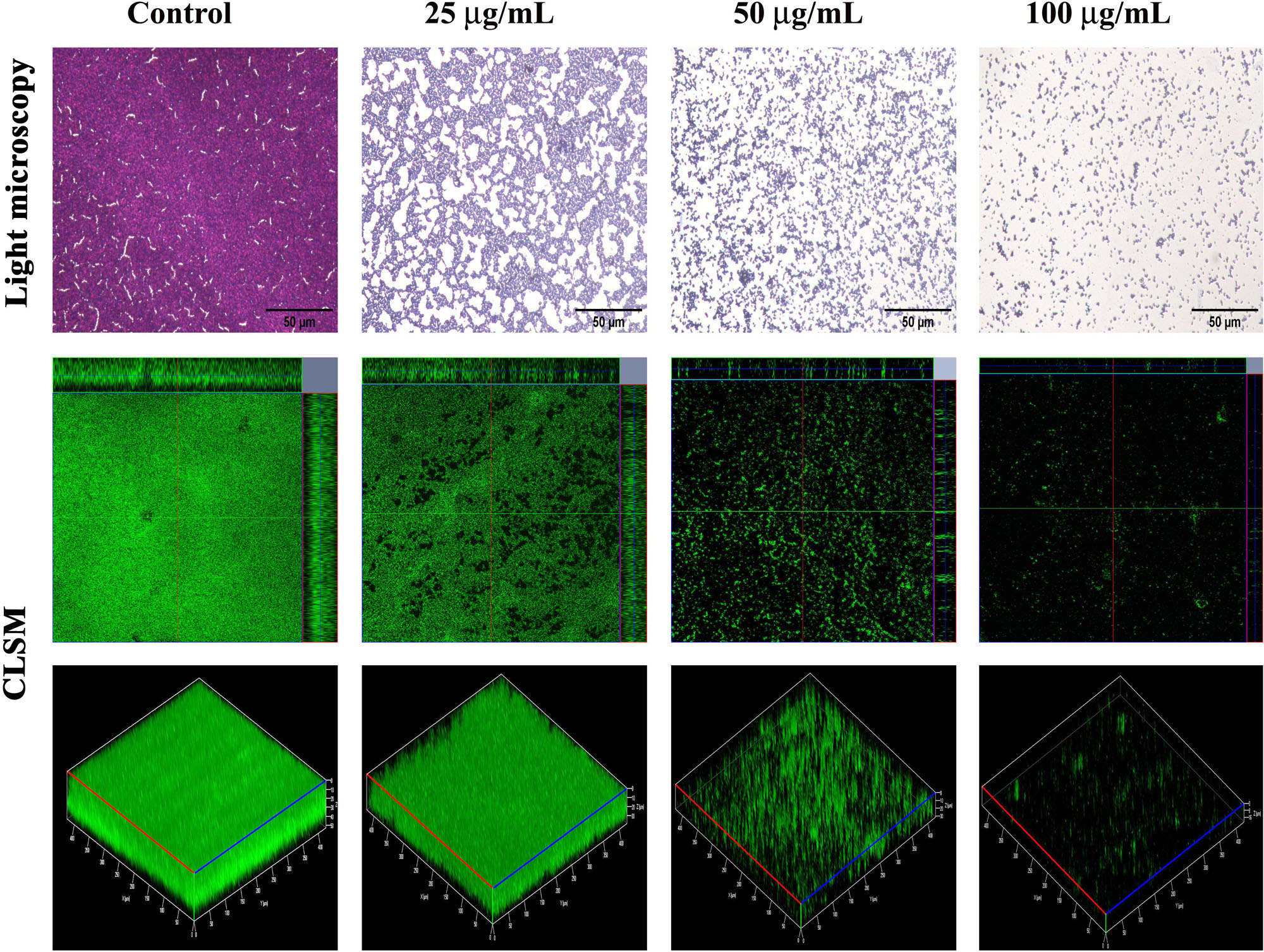
Figure 2. Light microscopic images (400×) and ortho- and three-dimensional confocal laser scanning microscopy images (200×) depicting the dose-dependent antibiofilm potential of thymol against methicillin-resistant Staphylococcus aureus. Scale bar = 50 μm in the light micrographs.
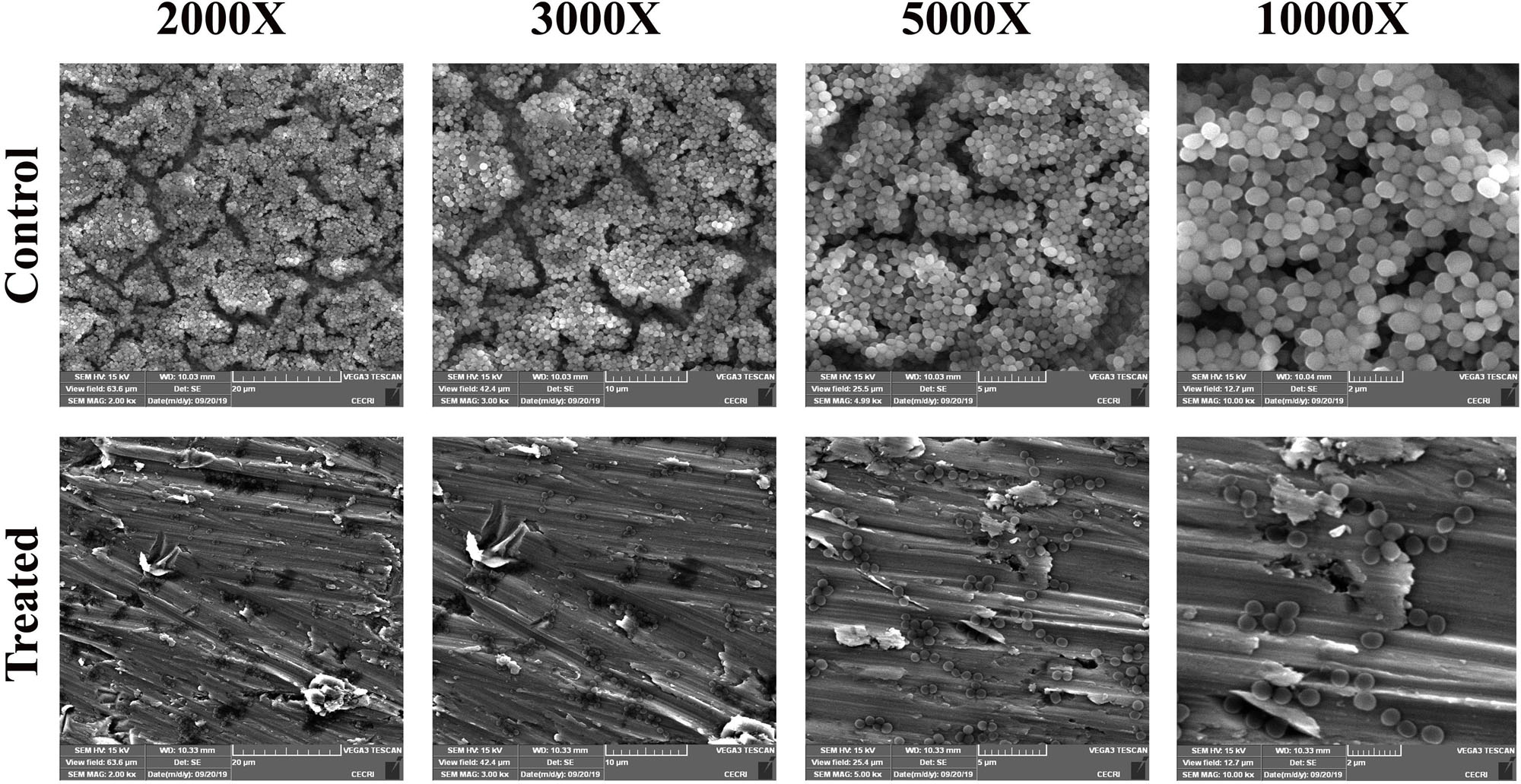
Figure 3. SEM images showing the reduction in adherence of methicillin-resistant Staphylococcus aureus on plasma-coated titanium surface upon thymol treatment (100 μg/ml) at different magnifications.
sarA-Dependent Antibiofilm Activity of Thymol
The expression of important regulatory genes involved in biofilm formation in the presence of thymol was examined by qPCR analysis. The expression of sarA was found to be decreased by twofold, whereas the expression of agrA and agrC was found to be unaltered by thymol treatment (Figure 4). Furthermore, the decreased expression of sarA-regulated virulence genes such as fnbA, fnbB, icaA, icaD, and hla was observed. The expression of fnbA and fnbB was slightly reduced by 0.02- and 0.11-fold, respectively, and nearly onefold down-regulation was observed in the expression of other virulence genes [icaA (1.08), icaD (0.89), and hla (1.23), respectively]. In order to identify the molecular-level target, the effect of thymol on biofilm formation of S. aureus wild type and ΔsarA and Δagr strains was examined by crystal violet quantification assay as mentioned earlier. Biofilm quantification using OD570 nm revealed that thymol was able to inhibit the biofilm formation of S. aureus wild type and Δagr strains at 100 μg/ml. However, the biofilm formation by ΔsarA was found to be unaffected by thymol treatment (Figures 5A,B).
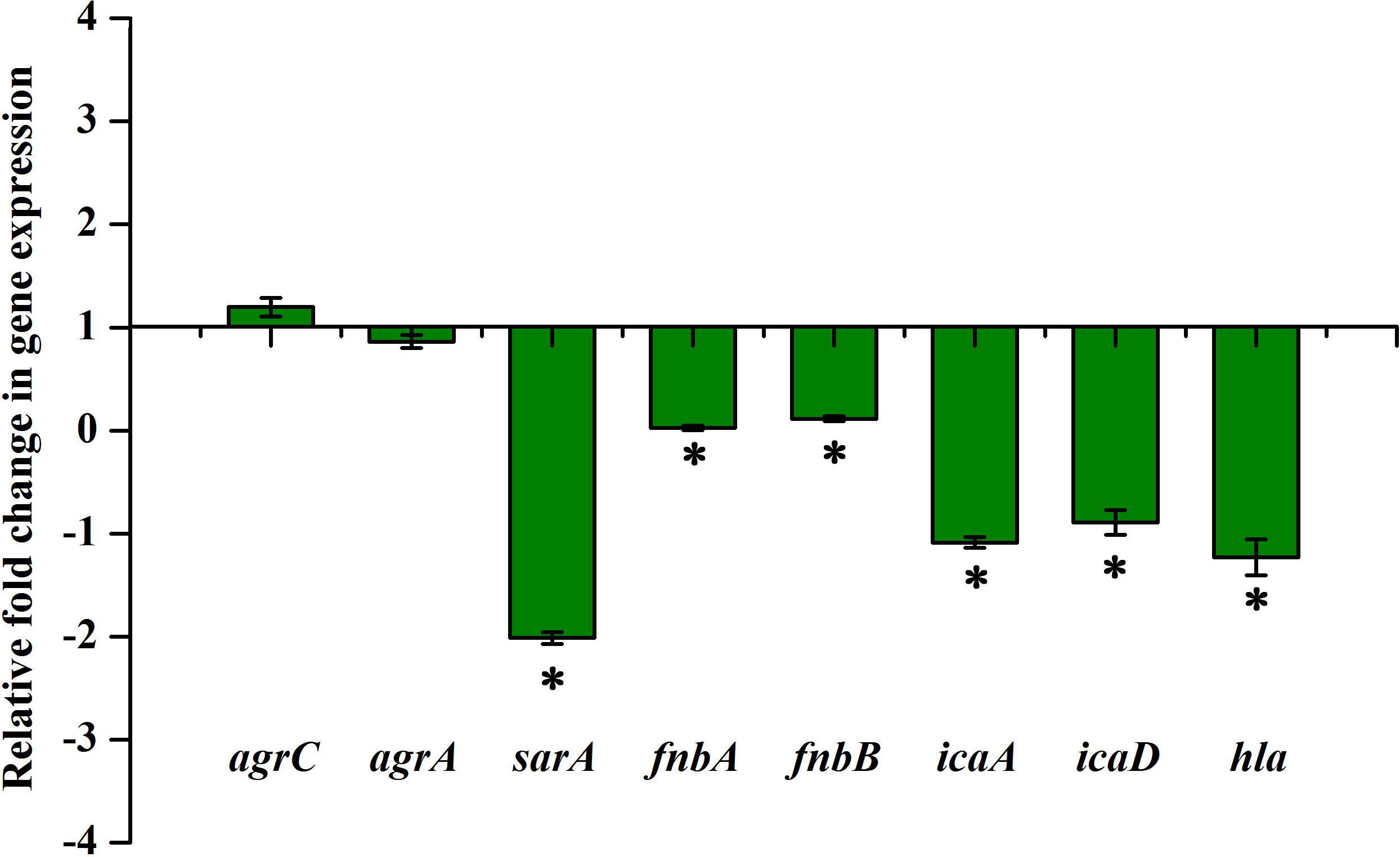
Figure 4. qPCR analysis of the expression of biofilm and virulence-associated genes after 24 h of thymol treatment. The error bars indicate standard deviations. The asterisks represent statistical significance (p < 0.05).
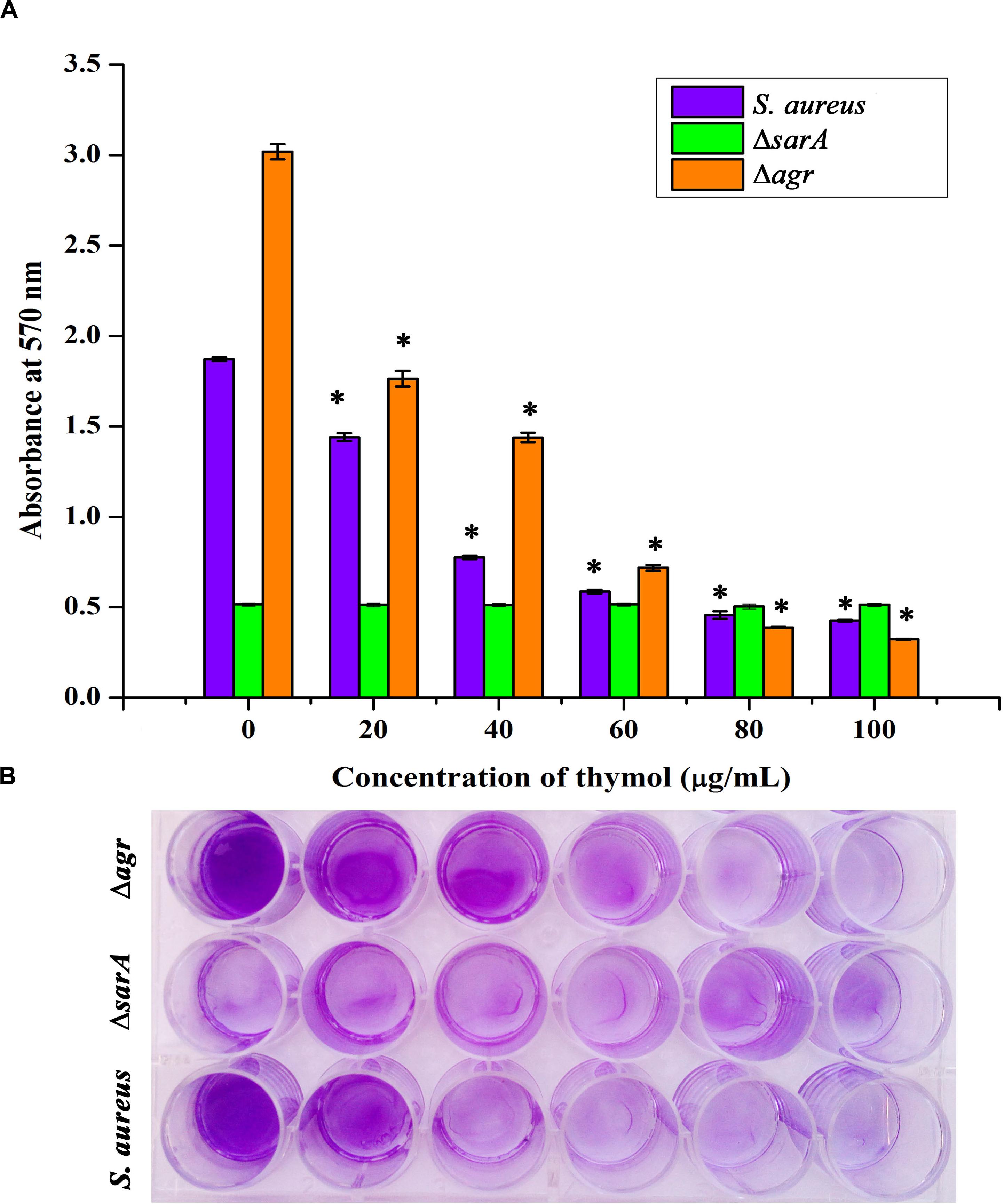
Figure 5. (A) Antibiofilm efficacy of thymol on wild-type Staphylococcus aureus and Δagr (affected) and on ΔsarA (not affected) as depicted by crystal violet quantification of biofilm. (B) The 24-well microtiter plate assay depicting the effect of thymol on the biofilm formation of wild-type S. aureus, Δagr, and ΔsarA. The error bars indicate standard deviations. The asterisks represent statistical significance (p < 0.05).
Thymol Hampers sarA-Controlled Virulence in MRSA
In the congo red agar (CRA) assay, the MRSA colonies appeared black in the absence of thymol; thymol treatment gradually inhibited the black coloration, and at 100 μg/ml, the MRSA cells appeared white, with complete inhibition of polysaccharide intracellular adhesion (PIA) (Figure 6A). Extracellular polysaccharide (EPS) present in MRSA biofilm was quantified by phenol-sulfuric acid method of carbohydrate quantification, and a significant reduction in EPS was observed at OD490 nm in the thymol-treated samples when compared to the control sample (Figure 6B). In the erythrocyte lysis assay, the production of hemolysin was assessed by mixing CFCS of the control and the thymol-treated MRSA with human erythrocytes. The thymol treatment resulted in the complete inhibition of erythrocyte lysis when compared to the red-colored erythrocyte lysis in the control sample (Figure 6C). On the whole, thymol significantly reduced the virulence factors under the control of sarA which is clearly depicted in the schematic representation (Figure 7).
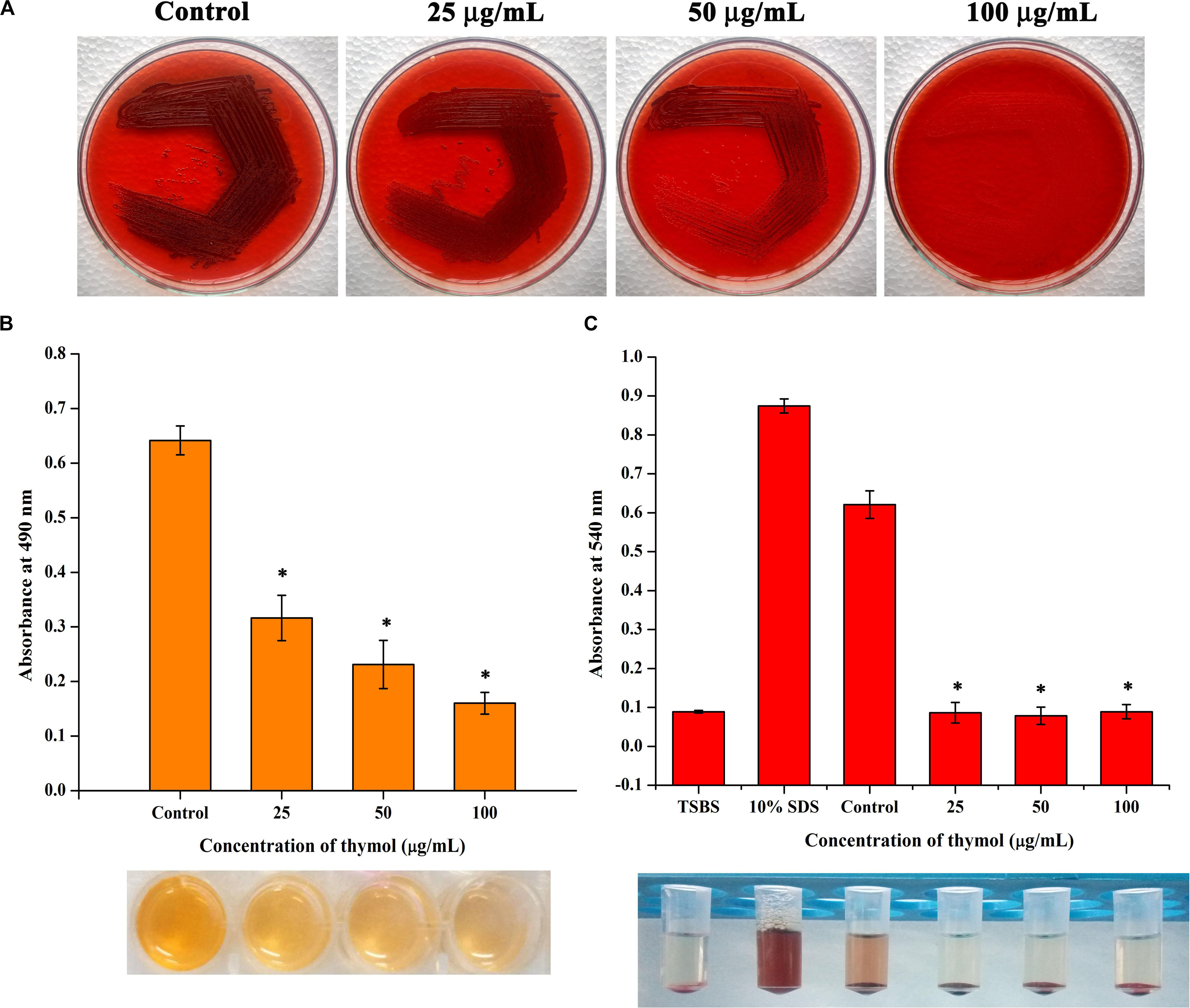
Figure 6. (A) Congo red assay representing the reduction in polysaccharide intracellular adhesion synthesis upon thymol treatment. (B) Thymol inhibits extracellular polysaccharide synthesis as revealed by phenol sulfuric acid quantification. (C) Thymol greatly inhibits the production of hemolysin as exhibited by erythrocyte lysis assay. The bottom image shows the corresponding assay tubes. The error bars indicate standard deviations. The asterisks represent statistical significance (p < 0.05).
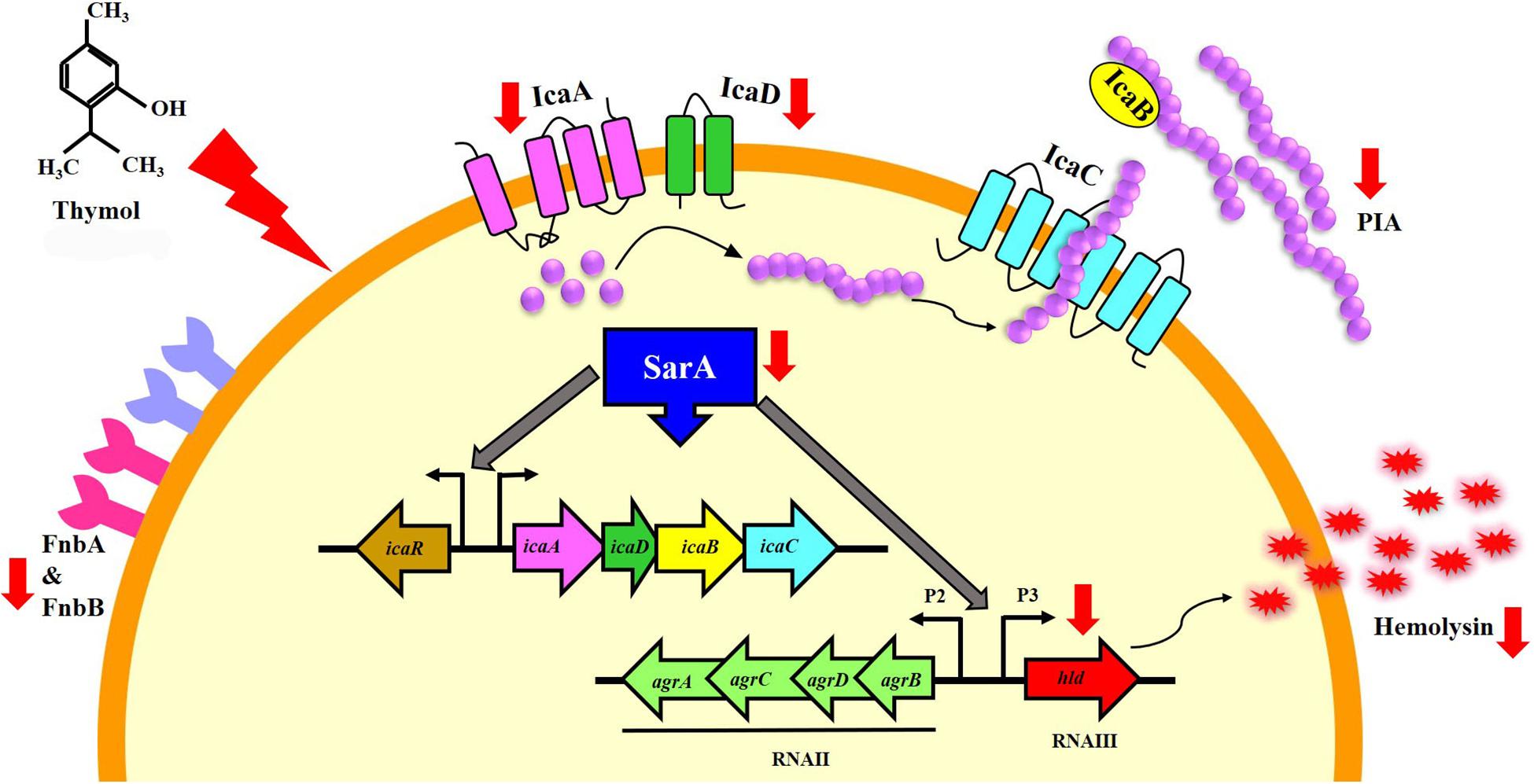
Figure 7. Schematic representation of the molecular mechanism of thymol mediated by sarA. The red arrows indicate the down-regulation of sarA-regulated virulence genes by thymol.
Thymol Improves the Antibacterial Activity of Rifampicin Against Planktonic Cells, Mature Biofilm, and Persister Formation of MRSA
Antibacterial Activity of Rifampicin on Planktonic Cells
The minimum inhibitory concentration (MIC) of thymol and rifampicin was determined as 160 and 0.06 μg/ml, respectively (Supplementary Figures S3, S4), as maximum growth was inhibited at the minimum concentrations. Hence, for the combinatorial assay, the BIC of thymol (100 μg/ml) was selected as it does not affect the growth of MRSA. The antibacterial activity of rifampicin at MIC (0.06 μg/ml) and at sub-MIC (0.015 and 0.03 μg/ml) concentrations in the presence of thymol was examined by growth OD measurement (Figure 8A) and Alamar blue-based viability assay (Figure 8B). The results demonstrated the enhanced reduction in growth as well as viability upon combinatorial treatment with rifampicin and thymol than treatment with rifampicin alone. In addition, images of disc diffusion assay (Figure 8C) clearly displayed the increased zone of clearance at all the tested concentrations of rifampicin in the presence of thymol. The zone of clearance (mm) of each plate was measured and presented as bar graph (Figure 8D). Interestingly, the MRSA cells appeared white in color in the TSA plates containing thymol because of staphyloxanthin inhibition, in contrast to the yellow-colored cells on the control TSA plates.
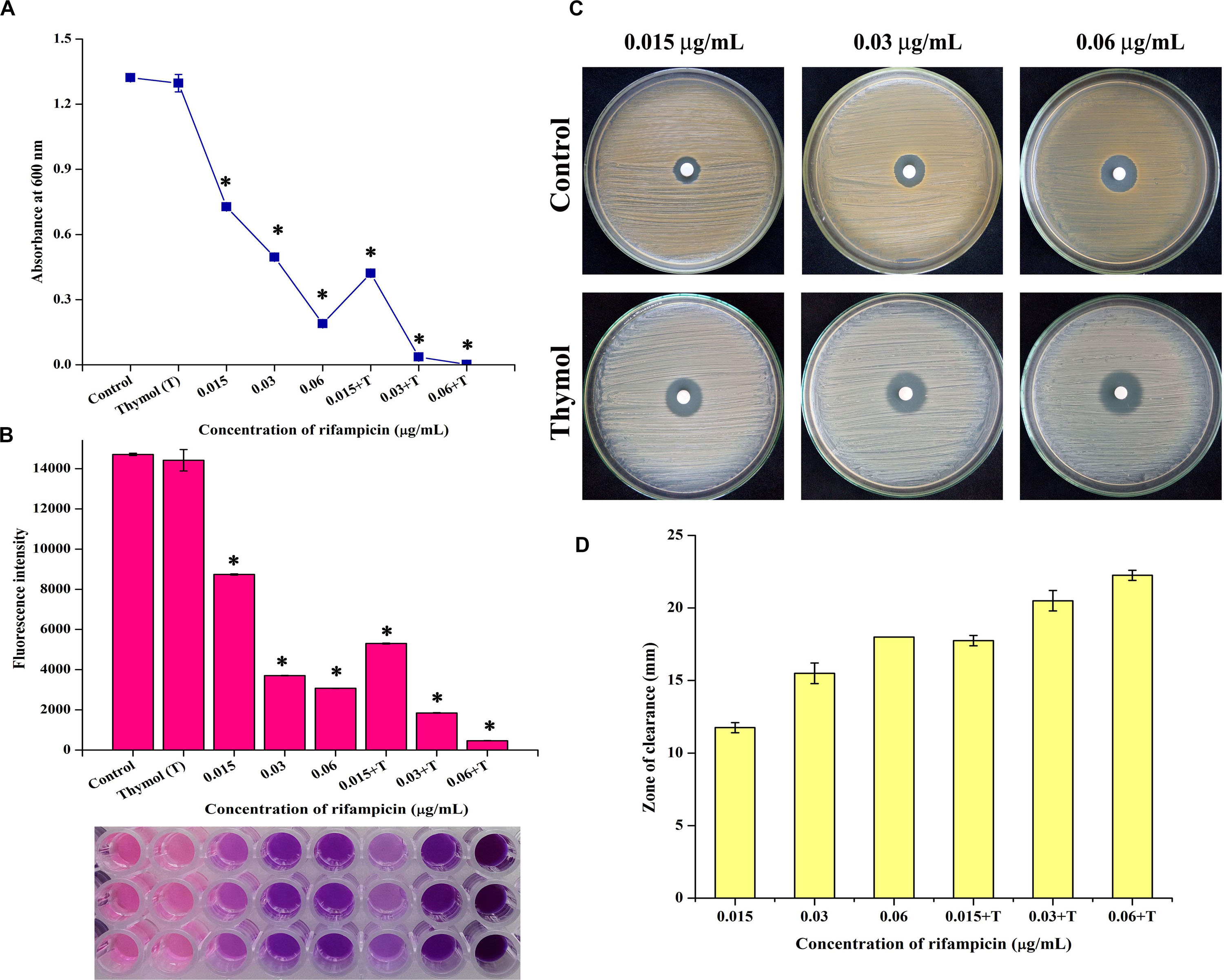
Figure 8. Thymol enhances the antibacterial activity of rifampicin as shown by growth optical density measurement (A), Alamar blue assay (B), disc diffusion assay (C), and its zone of clearance (D). The error bars indicate standard deviations. The asterisks represent statistical significance (p < 0.05).
Time Killing Kinetics and Persister Formation of Rifampicin
The killing efficacy of rifampicin in the absence and the presence of thymol on overnight-grown MRSA culture was measured for 12 h by spot assay and CFU counting (Figures 9A,B). From the results, it is clear that although rifampicin reduced the number of surviving cells for first 6 h, a certain subpopulation of cells remained viable for 12 h. In the case of the combinatorial treatment with rifampicin and thymol, the entire population was killed within 6 h. Furthermore, a concentration-dependent reduction of persister formation was observed upon combinatorial treatment when compared to the persisters formed upon treatment with ×100 concentration of rifampicin alone (Figure 9C). The 100-μg/ml concentration of thymol especially completely inhibited the persister formation by rifampicin.
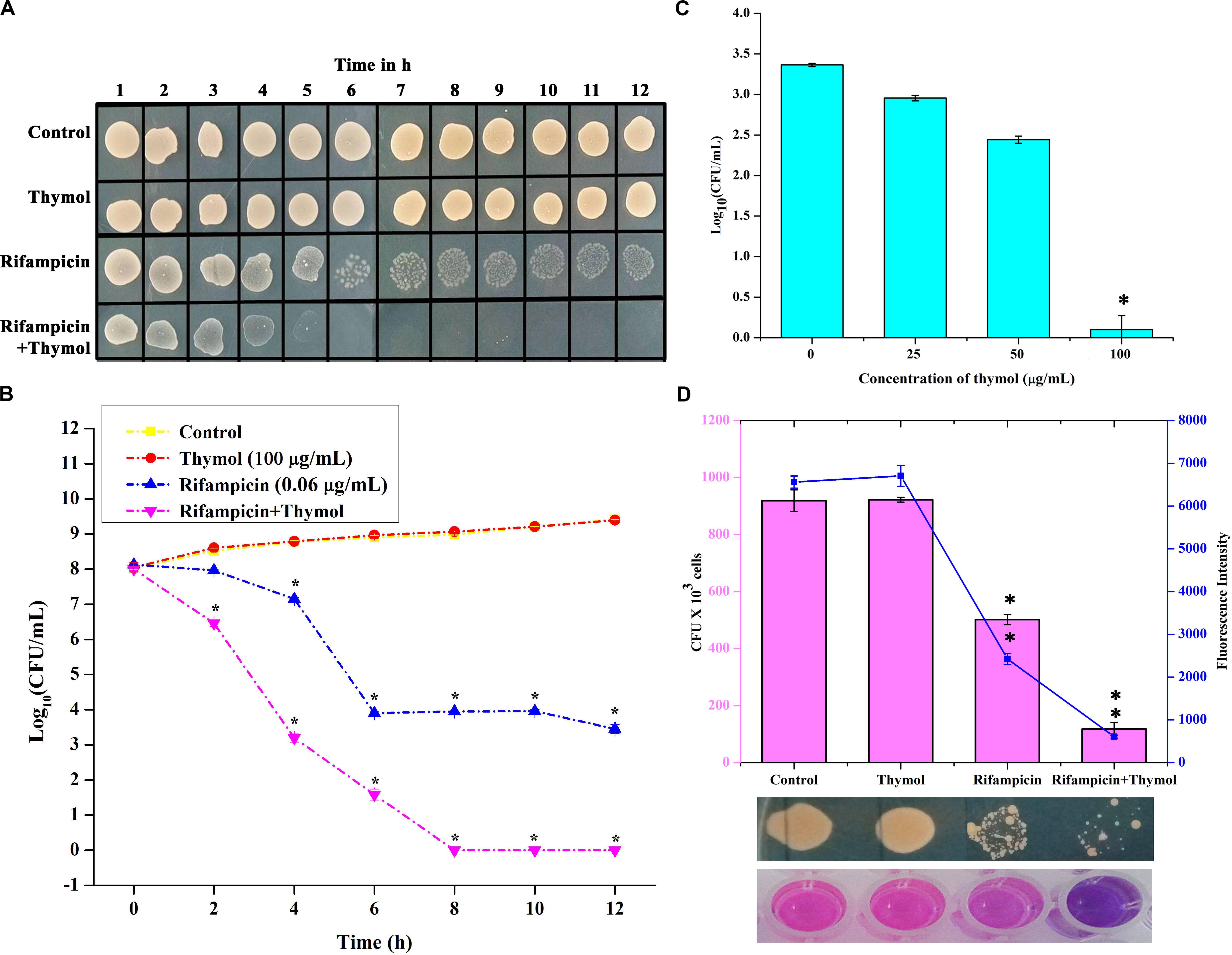
Figure 9. Thymol potentiates the killing efficacy of rifampicin as shown by time killing kinetics studied by spot assay (A) and colony-forming unit (CFU) counting (B) and reduces persister formation (C). Thymol improves the biofilm eradication ability of rifampicin as shown by CFU analysis and Alamar blue assay (D). The error bars indicate standard deviations. The asterisks represent statistical significance (p < 0.05).
Antibacterial Activity of Rifampicin on Mature Biofilm
The effect of thymol on the antibacterial activity of rifampicin against the mature biofilm of MRSA was assessed. The viability of 24-h mature biofilm cells after a 12-h treatment with rifampicin and thymol alone and in combination was examined by CFU and Alamar blue assay (Figure 9D). The results of both assays unveiled that thymol treatment enhanced the antibacterial activity of rifampicin on preformed MRSA biofilm.
Discussion
Thymol drastically inhibited the in vitro biofilm formation of S. aureus strains (MRSA ATCC strain and clinical isolates) at 100 μg/ml without affecting the growth and the metabolic viability. This is more advantageous in excluding the possibility of drug resistance as thymol did not apply any selection pressure on MRSA. Surface adherence in the form of biofilm provides a protective stay to MRSA from adverse conditions. Light and CLSM microscopic analyses further confirmed the ability of thymol to interfere with surface adherence of MRSA on glass and stainless steel slides. MRSA was reported to be the predominant human pathogen involved in infections associated with various implantable medical devices (Pinto et al., 2019). The adhesion of host matrix proteins on implant surfaces serves as an initiative factor for biofilm formation. MRSA produces specialized adhesive proteins that are named as microbial surface components recognizing adhesive matrix molecules, which could interact with host proteins (Foster and Höök, 1998). Thus, titanium surface coated with human plasma was taken for biofilm assays to mimic this natural phenomenon. As evidenced by SEM micrographs, a multilayered biofilm, with three-dimensional microcolonies, was observed in the control surface coated with plasma, whereas thymol treatment efficiently inhibited the adherence of MRSA even on plasma-coated titanium surface. The antibiofilm efficacy observed on various surfaces such as polystyrene, glass, stainless steel, and titanium irrespective of the nature of the surface makes thymol a better antibiofilm therapeutic candidate.
In order to find out the molecular mechanism of thymol at the transcriptional level, the expression of important biofilm regulatory genes such as sarA, agrA, and agrC was examined in the presence of thymol. SarA is basically a DNA binding protein which regulates the expression of genes involved in pathogenesis and stands as the global regulator of virulence in S. aureus (Bayer et al., 1996). On the other hand, agr is a two-component regulatory system which regulates toxin production and adhesion according to the quorum of bacteria present in a particular environment. An active agr system turns on the toxin gene expression and suppresses the adhesion gene expression (Boles and Horswill, 2008). Previous research on DNA binding sites of SarA revealed that SarA binds to the intergenic region between P2 and P3 promoters of the agr system and activates the expression of hemolysin via the RNAIII transcript. Apart from this, SarA binds to the upstream promoter regions of several target genes encoding fibronectin binding proteins (FnbA & FnbB), protein A (Spa), enterotoxin C, and PIA synthesis proteins (Chan and Foster, 1998; Chien et al., 1999). It is well known that sarA regulates the virulence of MRSA in both agr-dependent and agr-independent manner and serves as the therapeutic target to attenuate the virulence. Notably, previous studies have shown the sarA based repression of biofilm formation by various drugs in MRSA (Arya and Princy, 2013; Arya et al., 2015; Balamurugan et al., 2015, 2017). In the present study, thymol reduced the expression of sarA but did not alter the expression of agrA and agrC. This result leads to the hypothesis that the antibiofilm activity of thymol could be sarA-mediated. To verify this hypothesis, the antibiofilm activity of thymol was assessed on ΔsarA and Δagr strains. It was already reported that sarA mutation reduces the ability of S. aureus to form a biofilm, whereas agr mutation induces biofilm formation. In the present study also, ΔsarA formed less biofilm than that of the wild-type strain. As predicted, thymol was found to be ineffective on ΔsarA, and interestingly, thymol effectively inhibited the biofilm formation by Δagr. The inefficacy of thymol on ΔsarA validated the sarA-dependent antibiofilm activity of thymol.
To further prove this mechanism, the effect of thymol on the transcription of sarA-controlled virulence genes was investigated. The result revealed the down-regulation of sarA-controlled virulence genes such as icaA, icaD, fnbA, fnbB, and hla. PIA produced by ica operon plays a vital role in MRSA biofilm formation. It is well reported that the expression of the ica locus is positively regulated by sarA in S. aureus (Cerca et al., 2008). The PIA polymer is synthesized in the cytoplasm and transported outside the cell, which then mediates the intracellular adhesion and thereby provides the structural stability to biofilm (O’Gara, 2007). The reduction in the expression of icaA and icaD was further confirmed by CRA assay as it is an important method to identify the PIA production and ica-positive S. aureus by means of black coloration. The results of the CRA assay revealed that, as a downstream impact of sarA inhibition by thymol, the treated cells appeared white in color due to the complete inhibition of PIA synthesis. Phenol-sulfuric acid quantification of EPS also evidenced that thymol inhibited the EPS present in the biofilm matrix. Hemolysin is an important pore-forming virulence factor produced by S. aureus and plays a crucial role in invasive staphylococcal infections. The synthesis of hemolysin was affected by sarA inhibitors in previous studies (Arya et al., 2015; Balamurugan et al., 2017). In the present study also, the inhibition of sarA expression by thymol ultimately reduced the production of hemolysin, as exhibited by the erythrocyte lysis assay. The in vitro assays confirmed that the reduction in sarA expression could be the central mechanism involved in the antibiofilm activity of thymol, and a schematic representation of the molecular mechanism of thymol is depicted in Figure 7. Rifampicin, also known as rifampin, is the broad-spectrum antibiotic used to treat several microbial infections and commonly used to treat skin infections, prosthetic joint infections, and biofilm-associated infections caused by S. aureus (Lebeaux et al., 2014). Mostly, rifampicin is included in the combinatorial treatment strategy because of its ability to easily penetrate cells, and the development of resistance against rifampicin can also be minimized (Chang et al., 2020). Thus, the present study evaluated the efficacy of thymol to enhance the antibacterial efficacy of rifampicin against MRSA and found a great reduction in the growth and the viability of MRSA upon combinatorial treatment. The increased zone of clearance in the disc diffusion assay and the enhanced killing efficacy with less time span (6 h) confirmed the potential of the combinatorial treatment. Persisters are antibiotic-tolerant bacterial cells but different from antibiotic-resistant mutants since the antibiotic tolerance of persisters is not inheritable and reversible. Persisters play a critical role in the recurrence of a bacterial infection after the course of an antibiotic treatment, and this condition is clinically challenging (Lechner et al., 2012). Interestingly, thymol was found to be effective in reducing persister formation upon rifampicin treatment, and it would be of great advantage in the clinical settings. The eradication of preformed biofilm on medical devices is another hard task which could not be done with the available antibiotics, and possibly the combinatorial treatment of antibiotics and antibiofilm agents could solve this critical issue (Roy et al., 2013). Thus, the effect of combinatorial treatment on mature biofilm was evaluated in the present study. The number of viable bacteria in the mature biofilm after treatment with the rifampicin and thymol combination was greatly reduced than the rifampicin-alone-treated sample as revealed by CFU analysis and Alamar blue viability assay. Thus, all these results strongly suggest that thymol has the ability to boost the antibacterial activity of rifampicin in the form of combinatorial treatment against planktonic, biofilm, and persister cells of MRSA, and this property of thymol is highly appreciable in terms of clinical applications.
Conclusion
The present study unveiled the sarA-dependent antibiofilm activity and inhibition of other virulence factors such as PIA and hemolysin synthesis by thymol. The inhibition of MRSA adherence on various surfaces such as polystyrene, glass, stainless steel, and plasma-coated titanium advocates the potential of thymol as a surface-independent antibiofilm candidate in clinical context. The possibility of resistance development is also meager as thymol exerts non-antibacterial antibiofilm activity at 100 μg/ml. Thymol potentiated the antibacterial activity of rifampicin on planktonic as well as biofilm cells and reduced the persister formation. The ability of thymol to enhance the antibacterial and the biofilm eradication efficiency of rifampicin makes it a promising therapeutic candidate for combinatorial treatment strategy.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Ethical Committee, Alagappa University, Karaikudi (IEC Ref No: IEC/AU/2016/1/4). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AV and SK designed the study. AV, AS, and UY performed the experiments. AV analyzed the data, prepared the figures and tables, and wrote the manuscript. AS performed the statistical analysis. SK and CA revised the manuscript. All authors have read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors sincerely acknowledge the computational and bioinformatics facility provided by the Alagappa University Bioinformatics Infrastructure Facility (funded by DBT, GOI; File No. BT/BI/25/012/2012, BIF). The authors also acknowledge DST-FIST [Grant No. SR/FST/LSI-639/2015(C)], UGC-SAP [Grant No. F.5-1/2018/DRS-II(SAP-II)], and DST-PURSE [Grant No. SR/PURSE Phase 2/38(G)] for providing instrumentation facilities. The authors thank RUSA 2.0 [F.24-51/2014-U, Policy (TN Multi-Gen), Dept. of Edn, GoI]. SK is thankful to UGC for Mid-Career Award [F.19-225/2018(BSR)]. AV is thankful to RUSA 2.0 for financial assistance rendered in the form of RUSA Ph.D. Fellowship. The authors are grateful to Dr. Prof. Christiane Wolz, Institute for Medical Microbiology and Hygiene, University of Tubingen, Germany, for providing the S. aureus wild type, Δagr and ΔsarA strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01744/full#supplementary-material
Abbreviations
agr, accessory gene regulator; BIC, biofilm inhibitory concentration; CFCS, cell-free culture supernatant; CFU, colony forming unit; CLSM, confocal laser scanning microscope; CRA, congo red agar; EPS, extracellular polysaccharide; MRSA, methicillin-resistant Staphylococcus aureus; MTP microtiter plate; PIA, polysaccharide intracellular adhesion; sarA, staphylococcal accessory regulator A; SEM, scanning electron microscopy; TSBS, tryptone soya broth supplemented with 1% sucrose; WHO, World Health Organization.
References
Abraham, K. P., Srieenivas, J., Venkateswarulu, T. C., Indira, M., Babu, D. J., Diwakar, T., et al. (2012). Investigation of the potential anti biofilm activities of plant extracts. Int. J. Pharm. Pharm. Sci. 4, 282–285.
Arya, R., and Princy, S. A. (2013). Computational approach to design small molecule inhibitors and identify SarAas a potential therapeutic candidate. Med. Chem. Res. 22, 1856–1865. doi: 10.1007/s00044-012-0185-9
Arya, R., Ravikumar, R., Santhosh, R. S., and Princy, S. A. (2015). SarAbased novel therapeutic candidate against Staphylococcus aureusassociated with vascular graft infections. Front. Microbiol. 6:416. doi: 10.3389/fmicb.2015.00416
Azizi, Z., Ebrahimi, S., Saadatfar, E., Kamalinejad, M., and Majlessi, N. (2012). Cognitive-enhancing activity of thymol and carvacrol in two rat models of dementia. Behav. Pharmacol. 23, 241–249. doi: 10.1097/FBP.0b013e3283534301
Balamurugan, P., Hema, M., Kaur, G., Sridharan, V., Prabu, P. C., Sumana, M. N., et al. (2015). Development of a biofilm inhibitor molecule against multidrug resistant Staphylococcus aureusassociated with gestational urinary tract infections. Front. Microbiol. 6:832. doi: 10.3389/fmicb.2015.00832
Balamurugan, P., Praveen Krishna, V., Bharath, D., Lavanya, R., Vairaprakash, P., and Adline Princy, S. (2017). Staphylococcus aureusQuorum regulator sarAtargeted compound, 2-[(Methylamino) methyl] phenol inhibits biofilm and down-regulates virulence genes. Front. Microbiol. 8:1290. doi: 10.3389/fmicb.2017.01290
Bayer, M. G., Heinrichs, J. H., and Cheung, A. L. (1996). The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178, 4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996
Bernheimer, A. W. (1988). Assay of hemolytic toxins. Methods Enzymol. 165, 213–217. doi: 10.1016/s0076-6879(88)65033-6
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated dispersal of Staphylococcus aureusbiofilms. PLoS Pathog. 4:e1000052. doi: 10.1371/journal.ppat.1000052
Braga, P. C. (2005). Thymol: antibacterial, antifungal and antioxidant activities. G. Ital. Ostet. Ginecol. 27, 267–272.
Buommino, E., Scognamiglio, M., Donnarumma, G., Fiorentino, A., and D’Abrosca, B. (2014). Recent advances in natural product-based anti-biofilm approaches to control infections. Mini Rev. Med. Chem. 14, 1169–1182. doi: 10.2174/1389557515666150101095853
Cerca, N., Brooks, J. L., and Jefferson, K. K. (2008). Regulation of the intercellular adhesin locus regulator (icaR) by SarA, σB, and IcaR in Staphylococcus aureus. J. Bacteriol. 190, 6530–6533. doi: 10.1128/JB.00482-08
Chan, P. F., and Foster, S. J. (1998). Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180, 6232–6241. doi: 10.1128/.180.23.6232-6241.1998
Chang, A. T., Cosimi, R. A., and Bochan, M. R. (2020). Treatment of Staphylococcal Device infections: synergistic daptomycin with ceftaroline versus rifampin-adjunct therapy. Open Forum Infect. Dis. 7:ofaa072. doi: 10.1093/ofid/ofaa072
Chien, Y., Manna, A. C., Projan, S. J., and Cheung, A. L. (1999). SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274, 37169–37176. doi: 10.1074/jbc.274.52.37169
Dubois, M., Gilles, K., Hamilton, J. K., Rebers, P. A., and Smith, F. (1951). A colorimetric method for the determination of sugars. Nature 168:167. doi: 10.1038/168167a0
Foster, T. J., and Höök, M. (1998). Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6, 484–488. doi: 10.1016/s0966-842x(98)01400-0
García-Salinas, S., Elizondo-Castillo, H., Arruebo, M., Mendoza, G., and Irusta, S. (2018). Evaluation of the antimicrobial activity and cytotoxicity of different components of natural origin present in essential oils. Molecules 23:1399. doi: 10.3390/molecules23061399
Gowrishankar, S., Duncun Mosioma, N., and Karutha Pandian, S. (2012). Coral-associated bacteria as a promising antibiofilm agent against methicillin-resistant and-susceptible Staphylococcus aureusbiofilms. Evid. Based Complementary Altern. 2012:862374. doi: 10.1155/2012/862374
Hall, C. W., and Mah, T.-F. (2017). Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 41, 276–301. doi: 10.1093/femsre/fux010
Jenul, C., and Horswill, A. R. (2018). Regulation of Staphylococcus aureusvirulence. Microbiol. Spectr. 6:1. doi: 10.1128/microbiolspec.GPP3-0031-2018
Knobloch, J. K.-M., Horstkotte, M. A., Rohde, H., and Mack, D. (2002). Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med. Microbiol. Immunol. 191, 101–106. doi: 10.1007/s00430-002-0124-3
Kumar, S., Chandra, N., Singh, L., Hashmi, M. Z., and Varma, A. (2019). Biofilms in Human Diseases: Treatment and Control. Cham: Springer.
Lebeaux, D., Ghigo, J.-M., and Beloin, C. (2014). Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78, 510–543. doi: 10.1128/MMBR.00013-14
Lechner, S., Lewis, K., and Bertram, R. (2012). Staphylococcus aureuspersisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol. 22, 235–244. doi: 10.1159/000342449
Lee, J.-H., Kim, Y.-G., Gwon, G., Wood, T. K., and Lee, J. (2016). Halogenated indoles eradicate bacterial persister cells and biofilms. AMB Express 6:123. doi: 10.1186/s13568-016-0297-6
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Nostro, A., Sudano Roccaro, A., Bisignano, G., Marino, A., Cannatelli, M. A., Pizzimenti, F. C., et al. (2007). Effects of oregano, carvacrol and thymol on Staphylococcus aureus and Staphylococcus epidermidisbiofilms. J. Med. Microbiol. 56, 519–523. doi: 10.1099/jmm.0.46804-0
O’Gara, J. P. (2007). ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188. doi: 10.1111/j.1574-6968.2007.00688.x
Olsen, I. (2015). Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 34, 877–886. doi: 10.1007/s10096-015-2323-z
Palavecino, E. L. (2014). Clinical, epidemiologic, and laboratory aspects of methicillin-resistant Staphylococcus aureusinfections. Methods Mol. Biol. 1085, 1–24. doi: 10.1007/978-1-62703-664-1_1
Pinto, R. M., Lopes-de-Campos, D., Martins, M. C. L., Van Dijck, P., Nunes, C., and Reis, S. (2019). Impact of nanosystems in Staphylococcus aureusbiofilms treatment. FEMS Microbiol. Rev. 43, 622–641. doi: 10.1093/femsre/fuz021
Poonacha, N., Nair, S., Desai, S., Tuppad, D., Hiremath, D., Mohan, T., et al. (2017). Efficient killing of planktonic and biofilm-embedded coagulase-negative Staphylococci by bactericidal protein P128. Antimicrob. Agents Chemother. 61:e00457-17. doi: 10.1128/AAC.00457-17
Roy, V., Meyer, M. T., Smith, J. A. I., Gamby, S., Sintim, H. O., Ghodssi, R., et al. (2013). AI-2 analogs and antibiotics: a synergistic approach to reduce bacterial biofilms. Appl. Microbiol. Biotechnol. 97, 2627–2638. doi: 10.1007/s00253-012-4404-6
Sarker, S. D., Nahar, L., and Kumarasamy, Y. (2007). Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitroantibacterial screening of phytochemicals. Methods 42, 321–324. doi: 10.1016/j.ymeth.2007.01.006
Selvaraj, A., Jayasree, T., Valliammai, A., and Pandian, S. K. (2019). Myrtenol attenuates MRSA biofilm and virulence by suppressing sarA expression dynamism. Front. Microbiol. 10:2027. doi: 10.3389/fmicb.2019.02027
Sethupathy, S., Ananthi, S., Selvaraj, A., Shanmuganathan, B., Vigneshwari, L., Balamurugan, K., et al. (2017). Vanillic acid from Actinidia deliciosaimpedes virulence in Serratia marcescensby affecting S-layer, flagellin and fatty acid biosynthesis proteins. Sci. Rep. 7:16328. doi: 10.1038/s41598-017-16507-x
Sethupathy, S., Prasath, K. G., Ananthi, S., Mahalingam, S., Balan, S. Y., and Pandian, S. K. (2016). Proteomic analysis reveals modulation of iron homeostasis and oxidative stress response in Pseudomonas aeruginosaPAO1 by curcumin inhibiting quorum sensing regulated virulence factors and biofilm production. J. Proteomics 145, 112–126. doi: 10.1016/j.jprot.2016.04.019
Taylor, T. A., and Unakal, C. G. (2020). Staphylococcus Aureus. Treasure Island, FL: StatPearls Publishing.
Tohidpour, A., Sattari, M., Omidbaigi, R., Yadegar, A., and Nazemi, J. (2010). Antibacterial effect of essential oils from two medicinal plants against Methicillin-resistant Staphylococcus aureus(MRSA). Phytomedicine 17, 142–145. doi: 10.1016/j.phymed.2009.05.007
Valliammai, A., Sethupathy, S., Priya, A., Selvaraj, A., Bhaskar, J. P., Krishnan, V., et al. (2019). 5-Dodecanolide interferes with biofilm formation and reduces the virulence of Methicillin-resistant Staphylococcus aureus(MRSA) through up regulation of agr system. Sci. Rep. 9:13744. doi: 10.1038/s41598-019-50207-y
Vasudevan, R. (2019). Agr/sarA: molecular switches of biofilm regulation in Staphylococcus aureus.J. Microbiol. Exp. 7, 17–18. doi: 10.15406/jmen.2019.07.00233
Keywords: MRSA, thymol, rifampicin, sarA, biofilm inhibition, biofilm eradication
Citation: Valliammai A, Selvaraj A, Yuvashree U, Aravindraja C and Karutha Pandian S (2020) sarA-Dependent Antibiofilm Activity of Thymol Enhances the Antibacterial Efficacy of Rifampicin Against Staphylococcus aureus. Front. Microbiol. 11:1744. doi: 10.3389/fmicb.2020.01744
Received: 08 April 2020; Accepted: 03 July 2020;
Published: 31 July 2020.
Edited by:
Israel Castillo-Juárez, Colegio de Postgraduados (COLPOS), MexicoReviewed by:
Adline Princy Solomon, SASTRA University, IndiaAyaz Ahmed, University of Karachi, Pakistan
Yan Q. Xiong, David Geffen School of Medicine at UCLA, United States
Copyright © 2020 Valliammai, Selvaraj, Yuvashree, Aravindraja and Karutha Pandian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shunmugiah Karutha Pandian, c2tfcGFuZGlhbkByZWRpZmZtYWlsLmNvbQ==
 Alaguvel Valliammai
Alaguvel Valliammai Anthonymuthu Selvaraj
Anthonymuthu Selvaraj Udayakumar Yuvashree
Udayakumar Yuvashree Chairmandurai Aravindraja
Chairmandurai Aravindraja Shunmugiah Karutha Pandian
Shunmugiah Karutha Pandian