- 1Key Laboratory of Biomaterials of Guangdong Higher Education Institutes, Department of Biomedical Engineering, Jinan University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Food Quality and Safety, College of Food Science, South China Agricultural University, Guangzhou, China
- 3Guangdong Laboratory of Lingnan Modern Agriculture, South China Agricultural University, Guangzhou, China
- 4Guangdong Shunde Innovative Design Institute, Foshan, China
Vibrio parahaemolyticus is a significant seafood-borne pathogen, leading to serious acute gastrointestinal diseases worldwide. In this study, a reliable 4-plex droplet digital PCR (ddPCR) was successfully established and evaluated for the simultaneous detection of V. parahaemolyticus based on tlh, tdh, ureR, and orf8 in food samples using single intact cells. The targets tlh and ureR were labeled with 6-Carboxyfluorescein (FAM), and the targets tdh and orf8 were labeled with 5’-Hexachlorofluorescein (HEX). Due to reasonable proration of primers and probes corresponding into the two fluorescence channels of the ddPCR detecting platforms, the clearly separated 16 (24) clusters based on fluorescence amplitude were obtained. For better results, the sample hot lysis time and the cycle number were optimized. The results showed that the minimum number of “rain” and maximum fluorescence amplification were presented for precise detection in the condition of 25 min of the sample hot lysis time and 55 cycles. The sensitivity of this 4-plex ddPCR assay was 39 CFU/mL, which was in accordance with that of the conventional plate counting and was 10-fold sensitive than that of qPCR. In conclusion, the 4-plex ddPCR assay presented in this paper was a rapid, specific, sensitive, and accurate tool for the detection of V. parahaemolyticus including pandemic group strains and could be applied in the differentiation of V. parahaemolyticus in a wide variety of samples.
Introduction
Vibrio parahaemolyticus, a gram-negative and halophilic foodborne bacterium, which was first discovered in 1953 and commonly found in marine or estuarine all over the world, has been a leading causal agent of acute bacterial gastroenteritis with consumption of raw, mishandled or undercooked seafood (Fujino et al., 1953; Baross and Liston, 1968; Krantz et al., 1969). Up to now, V. parahaemolyticus has been acknowledged as the foremost foodborne pathogen that has caused outbreaks of foodborne illnesses worldwide. Take some examples: during May to September 2013, an outbreak associating with V. parahaemolyticus resulted in 104 illnesses along the U.S. Atlantic Coast (Newton et al., 2014); In India, 56.8% samples from diarrhoeal cases were identified for V. parahaemolyticus (Guin et al., 2019); In China, the number of the foodborne poisoning outbreak due to V. parahaemolyticus was increased from 31.1% between 1991 and 2001 to over 70% between 1998 and 2013. According to the data from the China National Center for Food Safety Risk Assessment (CFSA), there were 4.95 million people infected by V. parahaemolyticus per year (Pang et al., 2017). Therefore, it is crucial to develop rapid, accurate, reliable, and convenient identification methods for pathogenic V. parahaemolyticus. On the other hand, most of V. parahaemolyticus strains isolated from the environment are non-pathogenic, only a small part of them can cause food poisoning outbreaks (Velazquez-Roman et al., 2014). Hence, classification and identification of the pathogenic and non-pathogenic strains are also necessary.
Most of the conventional quantitative and identifying techniques, such as colony counting, have presented certain drawbacks with their inherent slowness, lots of workload and materials (Velusamy et al., 2010). In contrast to the conventional methods, rapid detection method such as PCR is convenient, sensitive and enables overcoming the disadvantages. PCR can be categorized into conventional PCR, quantitative PCR (qPCR), and (ddPCR). The ddPCR is an emerging technology for the detection and quantification of single or multiple targets simultaneously (Ottesen et al., 2006; Tadmor et al., 2011). Compared with other PCR techniques, ddPCR is an endpoint and absolute quantification approach based on the limiting dilution and Poisson distribution, unlike analog qPCR that relies on rate-based measurements (Ct values) and calibration curves (Hindson et al., 2011). It is not only more sensitive but also more accurate than qPCR in the quantification of rare target molecules at low target copy numbers (Bahar et al., 2018). Besides, ddPCR is remarkably efficient and high-throughput for processing 96 samples at a time (Hindson et al., 2013).
Effective detection of pathogenic V. parahaemolyticus requires the identification of several specific targets. The target of the thermolabile hemolysin gene (tlh) is used for the identification of total V. parahaemolyticus (Taniguchi et al., 1986; Bej et al., 1999). In contrast, thermostable direct hemolysin (TDH) encoded by tdh and TDH-related hemolysin (TRH) encoded by trh are two major virulence factors leading to V. parahaemolyticus-mediated disease (Nishibuchi and Kaper, 1995). However, the trh gene has two variants (trh1, trh2), which only share 84% sequence identity (Kishishita et al., 1992; Raghunath et al., 2010; Niu et al., 2018). Classification errors are emerged due to the lack of sequence identity of the trh gene and the relatedness of the hemolysin toxins (Suthienkul et al., 1995; Niu et al., 2018). The trh gene cluster is immediately downstream of ureR, so these two genes are genetically-linked. Using a more highly conserved ureR gene as a target is a useful tool for proxy detection of trh and can overcome difficulties associated with trh variation (Nilssona and Turner, 2016). The V. parahaemolyticus pandemic group strains including O3:K6 and its derivatives (the O4:K68, O1:K25, and O1:KUT serotypes) have emerged since 1996 (Matsumoto et al., 2000; Han et al., 2015). The orf8 gene, a peculiar open reading frame of the filamentous phage f237, is their common genetic marker (Nasu et al., 2000; Myers et al., 2003). A putative adherence protein encoded by orf8 may increase the virulence of O3:K6 (Ceccarelli et al., 2013; Letchumanan et al., 2014). Therefore, the tlh, tdh, ureR, and orf8 genes are generally used as targets to determine whether a strain is pathogenic V. parahaemolyticus (Su and Liu, 2007; Letchumanan et al., 2014).
To our knowledge, most researchers have developed qPCR for detecting V. parahaemolyticus (Su and Liu, 2007; Letchumanan et al., 2014; Zhang et al., 2015a,b). However, there were few studies on the detection of V. parahaemolyticus based on ddPCR. In previous work, we have developed a 3-plex ddPCR method for detecting and genotyping V. parahaemolyticus (Lei et al., 2020). However, the orf8 gene is not included in the study. In this paper, we established a 4-plex ddPCR method for the first time which could detect V. parahaemolyticus with single intact cells, as illustrated in Figure 1. In brief, the suspension of bacterial cells in ddPCR mixture was dispersed into droplets with a microfluidic chip, followed by the release of gDNA by hot lysis, ddPCR reaction and analysis. As four probes of these genes were labeled by only two fluorescent dyes, the probe concentrations were proportionally distributed to form a matrix with 16 (24) clusters, which explicitly displayed and quantified droplets containing one/two/three/four certain genes, corresponding to gene types of single intact bacterial cells. This method was beneficial for identifying V. parahaemolyticus encoded by different virulence genes accurately, rapidly, selectively, sensitively and reproducibly, which could offer precise supports to clinical diagnosis and treatments in bromatoxism, as well as in the monitoring of seafood contamination.
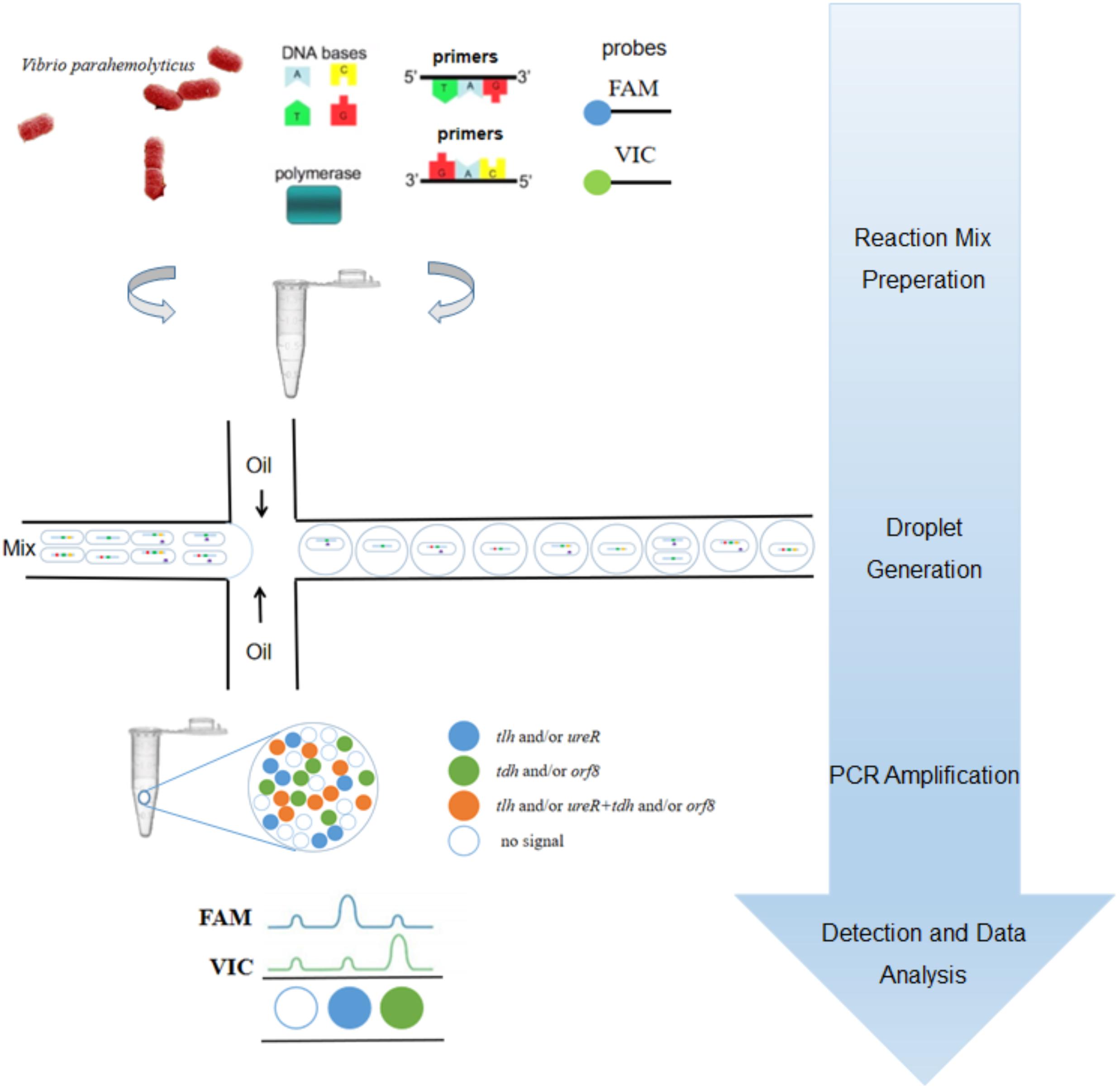
Figure 1. The procedural framework of the 4-plex ddPCR for specific detection of intact V. parahaemolyticus cells. The bacterial solution containing intact cells were added to the PCR reaction. The cells were dispersed into droplets prior to PCR-amplification. The targets of tlh (green), tdh (red), ureR (yellow), and orf8 (purple) were determined based on fluorescent probes.
Materials and Methods
Bacterial Strains and Culture Conditions
The strains used in this study are shown in Table 1. All Vibrio strains were cultured in tryptic soy broth (TSB; Beijing Land Bridge Technology Company Ltd., Beijing, China) with 3% (w/w) sodium chloride (NaCl) on a rotary shaker (YiHeng, ShangHai, China) at 150 rpm and 37°C for 16 h. All other strains were cultured in Luria-Bertani (LB) broth and kept in a shake incubator (150 rpm) at 37°C for 16 h. The bacterial cells were 10-fold serial diluted for ddPCR detection. The plate count method was used for comparing with the ddPCR method, where bacteria were grown on TCBS (hopebio, QingDao, China) or LB agar plates at 37°C for 24 h.
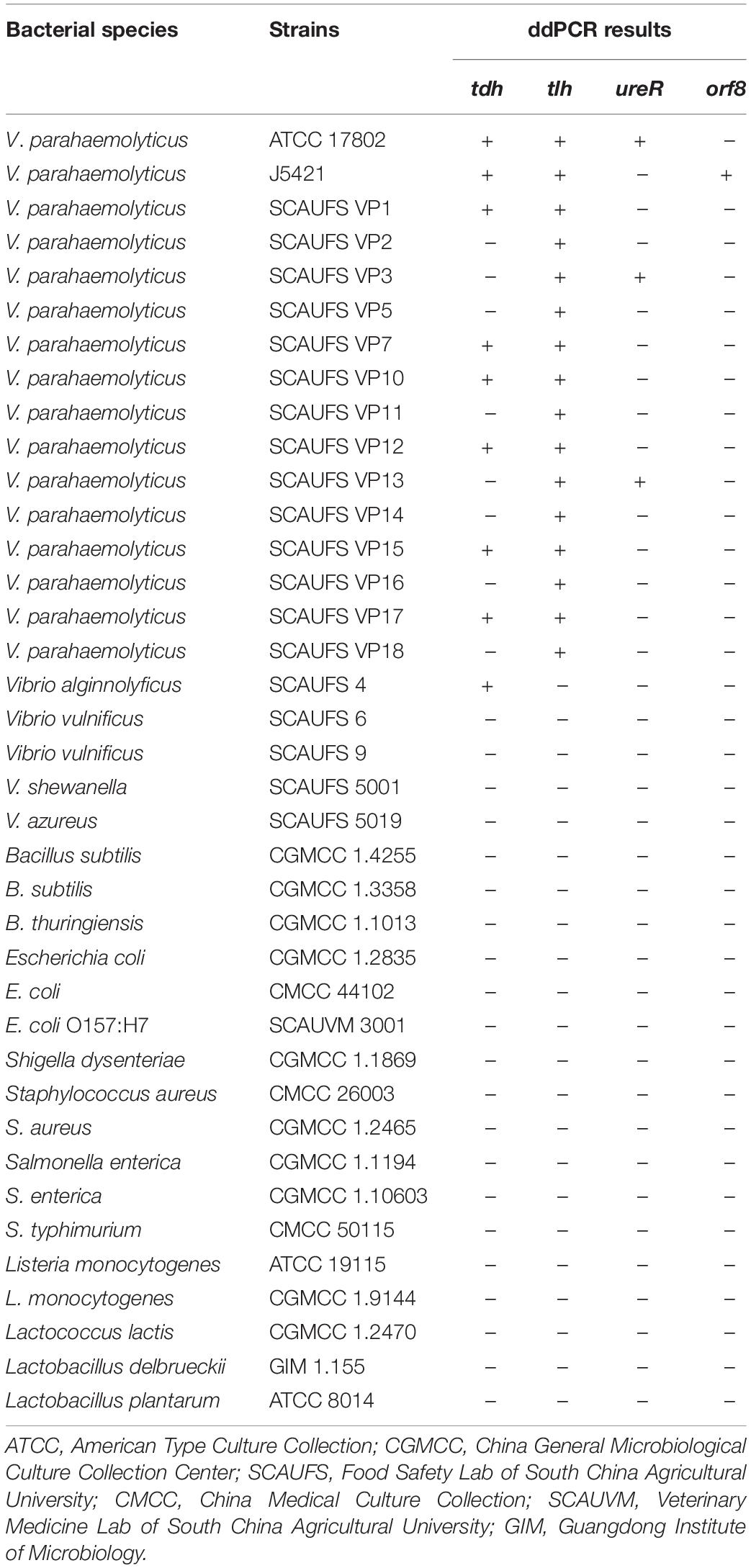
Table 1. Specificity of the ddPCR for tlh, tdh, ureR, and orf8 target genes for different bacterial strains.
Primers and Probes
According to V. parahaemolyticus strain ATCC17802 (CP014046.2, CP014047.2) and V. parahaemolyticus plasmid pO3K6 DNA (NC_002473.1; tdh, tlh, and trh/ureR are located in chromosome 2, orf8 is usually located in plasmid), multiplex primers and probe sets for tlh, tdh, ureR, and orf8 genes were designed by the Primer Premier 5.0 (Premier Biosoft, Canada). Specificity analysis of amplicons was carried out using nucleotide BLAST (National Center for Biotechnology Information, Bethesda, MD, United States). The sequences of the primers and probes in this study were listed in Table 2.
Optimization of the Sample Hot Lysis Time and the Cycle Number
In order to increase the detection accuracy and effectiveness, both the sample hot lysis time and the cycle number were optimized. Four sample hot lysis time lengths (15, 20, 25, and 30 min) and four different cycle numbers (45, 50, 55, and 60 cycles) were tested, respectively.
Quantitative PCR
The qPCR reaction was conducted in a total reaction volume of 20 μL containing 0.5 μM of each primer F/R and 0.125 μM of the probe, 10 μL of 2 × SGExcel GoldStar TaqMan Mixture (Sangon Biotech, Shanghai, China), 2 μL of cell template, and nuclease-free distilled water up to the final volume. The reaction process started by hot lysis treatment at 95°C for 30 min, followed by 60 cycles of 30 s at 95°C for denaturation, 1 min at 54°C for annealing and extension.
4-plex Droplet Digital PCR
Droplet digital PCR (ddPCR) was carried out on the MicroDropTM (MD) ddPCR system (Forever Gene, Shunde, Guangdong, China), which was consist of Microchip, Creator, and Detector for microdrop operations. The ddPCR mixture was performed in a 20 μL reaction volume composed of 2 μL of cell suspension, 10 μL of 2 × Supermix for Probes (Forever Gene), 900 nM of each primer, 0.125 μM of tlh, 0.625 μM of tdh, 0.25 μM of ureR, and 1.25 μM of orf8 probes, nuclease-free distilled water up to the final volume. The cell suspensions were serially diluted to ensure the majority of droplets were ideally either contained single cell or empty. Approximate 80 thousand water-in-oil droplets were produced using MD Microchip and MD Creator, then transferred to a conventional 96-well PCR plate followed by heat-sealing. In each droplet, an independent reaction was conducted to end point using the ABI Veriti FAST PCR (United States). The protocol was as follow: 25 min at 95°C for the cell hot lysis and Taq hot start, followed by 55 cycles of 30 s at 95°C (denaturation), 60 s at 54°C (annealing and extension), and a final hold of 10 min at 98°C (droplet stabilization), then cooling to 25°C. Following amplification, the samples were transferred to a MD Detector (Forever Gene), and the fluorescence amplitudes of four genes in every cell were read. The MD software then fit the fraction of positive droplets to the Poisson distribution algorithm to determine the number of targets and identify different strains of V. parahaemolyticus.
Comparing ddPCR With Plate Counting and qPCR
To evaluate the sensitivity of the 4-plex ddPCR assays, the same samples were simultaneously detected by plate counting, qPCR, and ddPCR. The 10-fold serially diluted V. parahaemolyticus solutions (3.9 × 101–3.9 × 107 CFU/mL) were grown on TCBS agar plates at 37°C for 24 h, as well as 2 μL solution of each sample was detected directly by qPCR and ddPCR. All assays were triplicated and repeated for three times. The standard curves were constructed using ten-fold serial dilutions of the mixed bacterial solution containing V. parahaemolyticus ATCC17802 and V. parahaemolyticus J5421 (2:1) covering the range from 3.9 × 107 to 3.9 × 101 CFU/mL.
Assessment of the 4-plex ddPCR Method With Complex Cell Templates
In this assay, eight types of cell templates (genes) were prepared for the 4-plex ddPCR evaluations for mimic complex detections: template 1 (tlh), template 2 (tlh and ureR), template 3 (tlh and tdh), template 4 (tlh and orf8), template 5 (tlh, ureR, and tdh), template 6 (tlh, tdh, and orf8), template 7 (tlh, ureR, and orf8), and template 8 (tlh, tdh, ureR, and orf8).
Detection of V. parahaemolyticus in the Background of High Concentrations of Non-target Bacteria
To study the impact of high levels of non-target bacteria (Escherichia coli and Listeria monocytogenes), mixtures of V. parahaemolyticus (3.9 × 103 CFU/mL) with different concentrations (101–107 CFU/mL) of non-target bacteria were prepared. In this assay, the V. parahaemolyticus at 3.9 × 103 CFU/mL was chosen as control. Meanwhile, the suspensions containing 106 CFU/mL of non-target bacteria and V. parahaemolyticus cells at different concentrations (3.9 × 101–3.9 × 107 CFU/mL) were prepared for detection of V. parahaemolyticus at serial concentrations in the background of high levels of certain non-target bacteria.
Verifying Assay With Artificial Contaminated White Calms
White clams were obtained from supermarket and washed by ddH2O. Twenty-five grams of clam tissues were inoculated with 1 mL bacterial solution at a concentration of 108 CFU/g and mixed with 225 mL 0.1% aseptic peptone water (PW). Then the mixture was homogenized for 5 min and centrifuged for 1 min (1000 rpm, 4°C). The supernatant was filtered to remove large particles and centrifuged at 12,000 rpm for 10 min to obtain the bacterial cells. Finally, the bacterial cells were suspended in 250 μL ddH2O and were serially diluted as templates for the 4-plex ddPCR.
Results
Specificity of Primers and Probes
Ten strains of V. parahaemolyticus and 22 strains of non-V. parahaemolyticus were tested to determine the specificity of the 4-plex ddPCR assays. As shown in Table 1, only the target bacterial strains are positively observed, while no positive signal occurred for non-target strains, confirming 100% inclusivity and exclusivity for V. parahaemolyticus by these assays. Here we know that strain ATCC17802 is tlh+tdh+ureR+, and strain J5421 is tlh+tdh+orf8+. These two strains were adopted as sample strains in subsequent works.
Optimization of the Heat Lysis Time of the Sample
The samples were treated at 95°C for different time lengths: 15, 20, 25, and 30 min. When the heat lysis time is 20 min or 25 min, positive and negative droplets are well separated. The fluorescence amplitude is highest at 25 min (Figure 2A). Compared with 30 min (Figure 2B), every cluster is concentrated and can be well distinguished at 25 min (Figure 2C). In summary, the sample treated at 95°C for 25 min displays superior 2D plots.
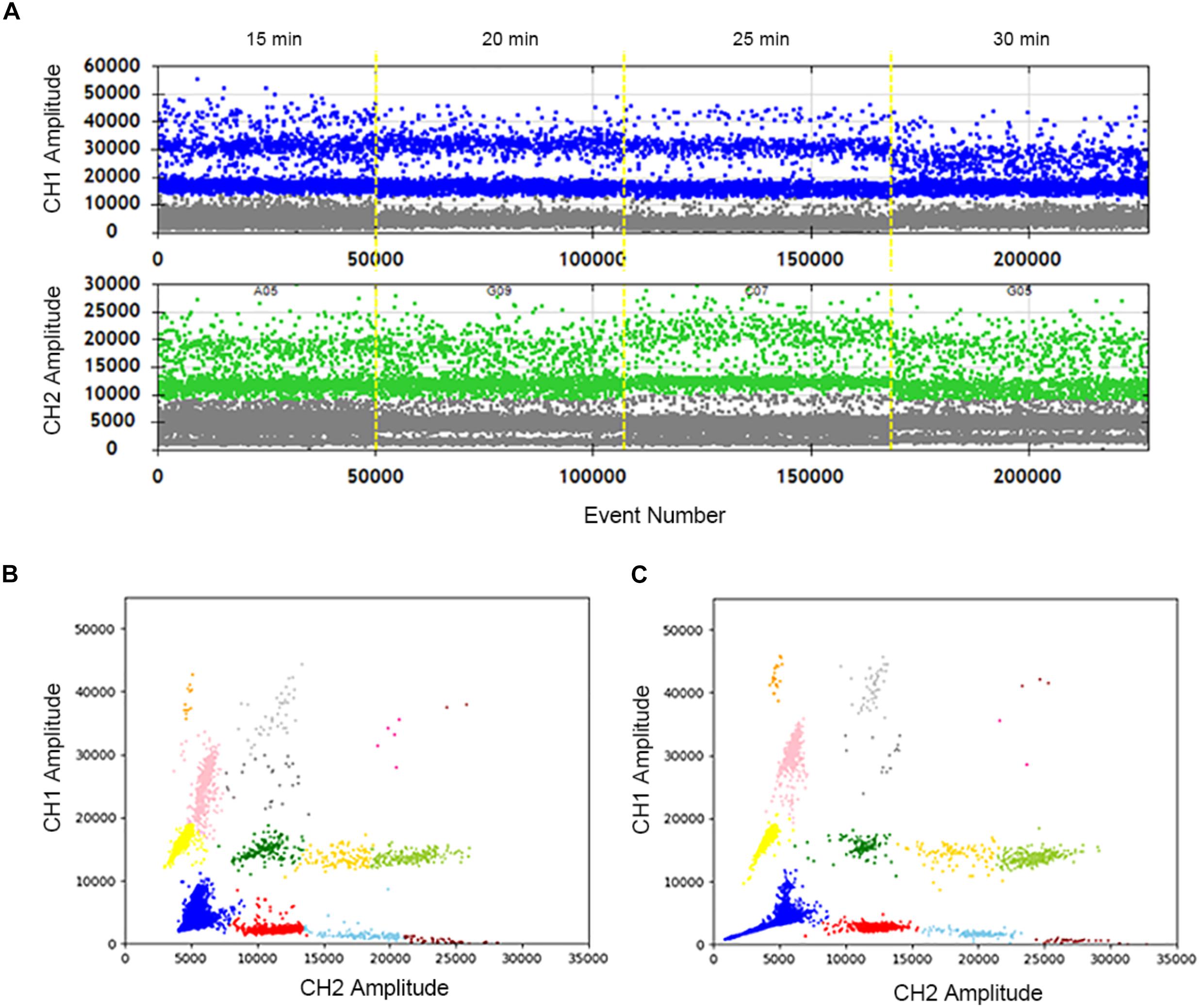
Figure 2. Optimization of the sample hot lysis time in two channels and 2D plot. (A) The positive droplets were represented as blue and the negative droplets were represented as gray in FAM. The positive droplets were represented as green and the negative droplets were represented as gray in VIC. (B) The 2D plot was obtained in 30 min of the hot lysis time. (C) The 2D plot was obtained in 25 min of the sample hot lysis time.
Optimization of Cycle Number
The effects of cycle numbers at 45, 50, 55, and 60 were investigated. In Figure 3A, the vertical axis shows the fluorescence amplitude values. In the FAM channel (CH1), significant differences among 4 wells are observed. The total droplet number of the four wells are similar, indicating that cycle numbers don’t impact the stabilization of droplets. However, when the cycle number is 55, the clusters are divided clearly with high fluorescence amplitudes, and droplets in the “rain” between clusters are least. The similar conclusions can easily be drawn, which shows the highest fluorescence amplitude and separated clusters in the HEX/VIC channel (CH2). Meanwhile, a comparison of the 2D plot of 60 cycles (Figure 3B) with that of 55 cycles (Figure 3C) illustrate that the clusters are separated more completely, droplets in the “rain” decrease and the fluorescence amplitude increases in the condition of 55 cycles. Therefore, 55 cycles would be better for this assay. Hence, not only detection time is reduced, but also the accuracy is improved after optimizations.
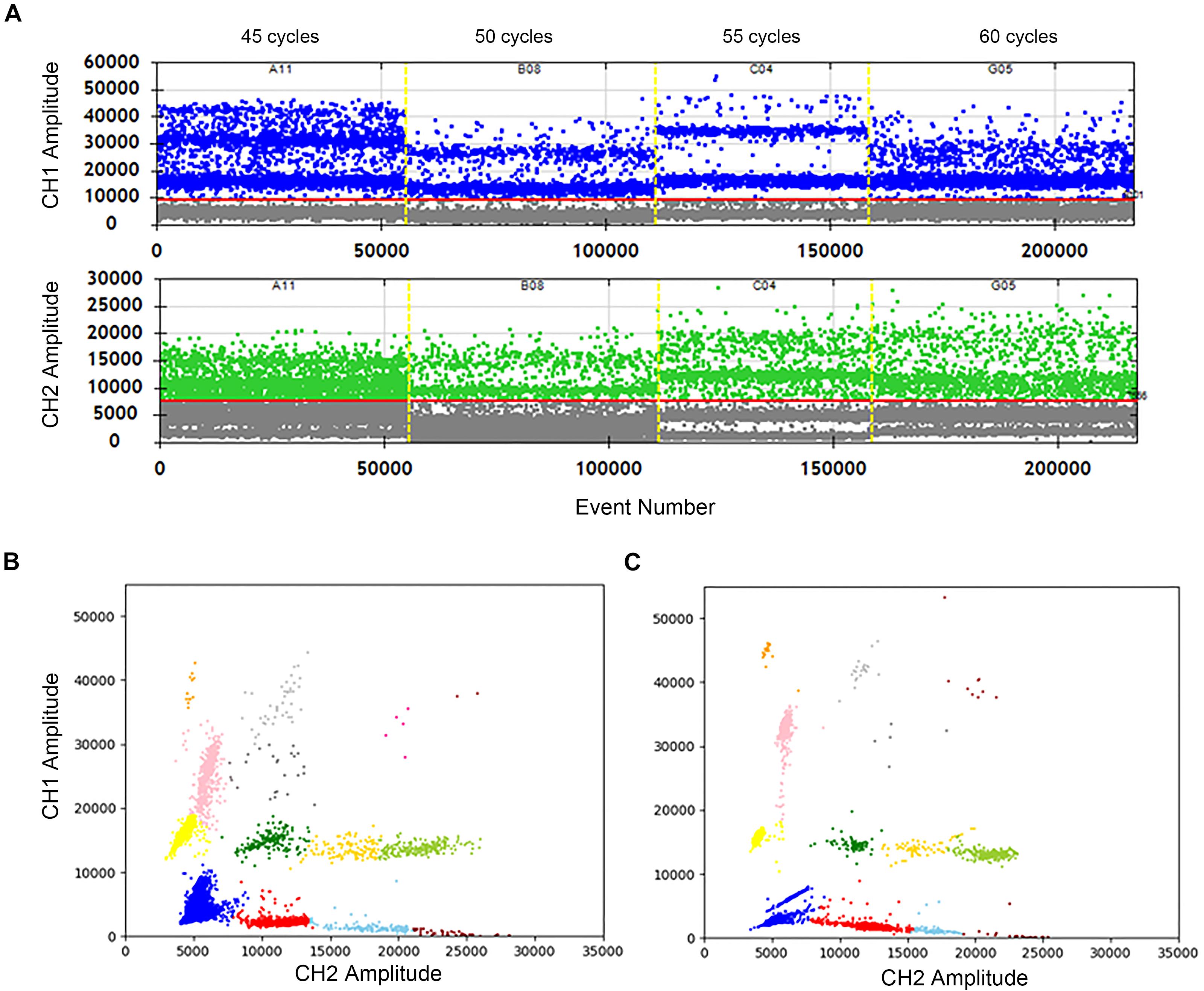
Figure 3. Optimization of the cycle number in two channels and 2D plot. (A) The positive droplets were represented as blue and the negative droplets were represented as gray in FAM. The positive droplets were represented as green and the negative droplets were represented as gray in VIC. (B) The 2D plot was obtained in 60 cycles of the amplification. (C) The 2D plot was obtained in 55 cycles of the amplification.
Performance of Plate Counting, qPCR and ddPCR
To evaluate the accuracy of the 4-plex ddPCR assays, we compared this method with the standard plate counting method. The 10-fold diluted V. parahaemolyticus solutions (3.9 × 101–3.9 × 107 CFU/mL) were detected for plate counting, qPCR and ddPCR. As shown in Figures 4A–D, the results of plate counting on TCBS agar are agreed with ddPCR results ranging from 3.9 × 107–3.9 × 101 CFU/mL, illustrating a good linear relationship of the detection with a correlation coefficient (R2) of 0.9982 (tlh), 0.9965 (tdh), 0.9882 (ureR), and 0.9784 (orf8), respectively. We noted that absolute quantifications by ddPCR corresponded to 87.0–137.1% of the theoretical cell numbers by plate counting, indicating that absolute detection by ddPCR is credible. Additionally, the measured concentration by ddPCR is always higher than that of the traditional plate counting method. It exhibited that our 4-plex ddPCR provided an accurate and efficient tool for the detection of V. parahaemolyticus. Figures 4E,F present the typical results of detecting different concentrations of V. parahaemolyticus based on the ddPCR and qPCR. It indicates that when the concentration of V. parahaemolyticus decreases to 3.9 × 101 CFU/mL, the fluorescence curve of qPCR disappears; meanwhile, the corresponded data of ddPCR can be presented, and the limit of detection (LOD) of ddPCR is defined as 39 CFU/mL. Compared with qPCR, the ddPCR method showed greater potential for sensitive detection of V. parahaemolyticus, especially at lower concentrations.
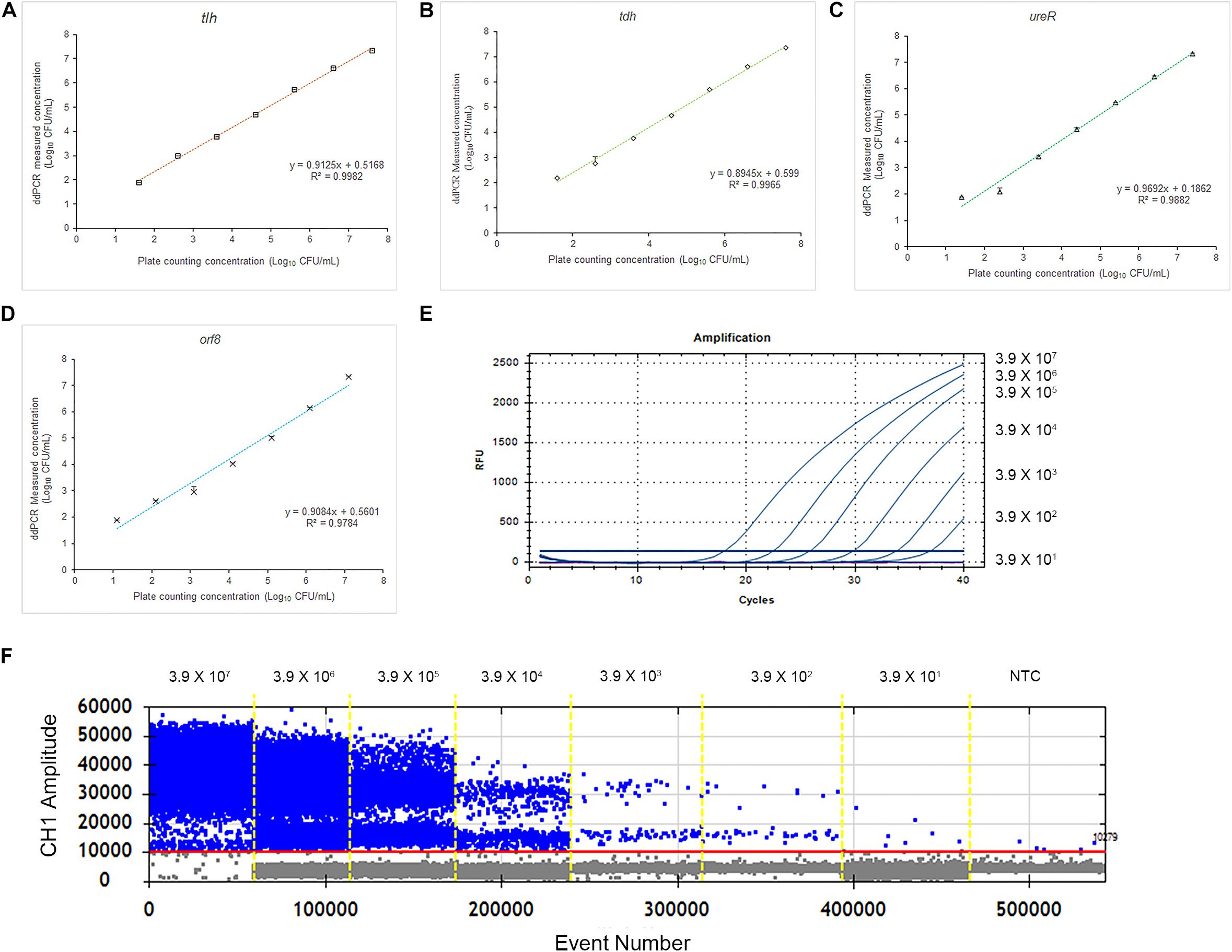
Figure 4. Quantification of V. parahaemolyticus by plate counting, qPCR, and ddPCR. (A) Standard curves of tlh in 4-plex ddPCR. Comparative analysis of diluted series of indicated bacterial solution. The horizontal axis was plate counting detection results for V. parahaemolyticus. (B) Standard curves of tdh in 4-plex ddPCR. (C) Standard curves of ureR in 4-plex ddPCR. (D) Standard curves of orf8 in 4-plex ddPCR. (E) Fluorescence curves were obtained based on qPCR by using the same templates. (F) The picture of FAM channel of the 4-plex ddPCR by using the same templates. Plotted values represented the mean value and standard deviations obtained from three triplicate tests. NTC = non-template control.
4-plex ddPCR Detecting Different Strains of V. parahaemolyticus
The results of eight detection systems using different templates were shown in Figure 5. It has to be underlined that a better quality in terms of clusters separation had been obtained after optimizations. Moreover, when the templates are positive for tlh, tdh, ureR, and/or orf8, the specific clusters are presented in the 2D-plot. According to the cluster position, we could directly distinguish the bacteria with different genes. Therefore, the 4-plex ddPCR is a feasible and accurate method for the classification of different strains of V. parahaemolyticus.
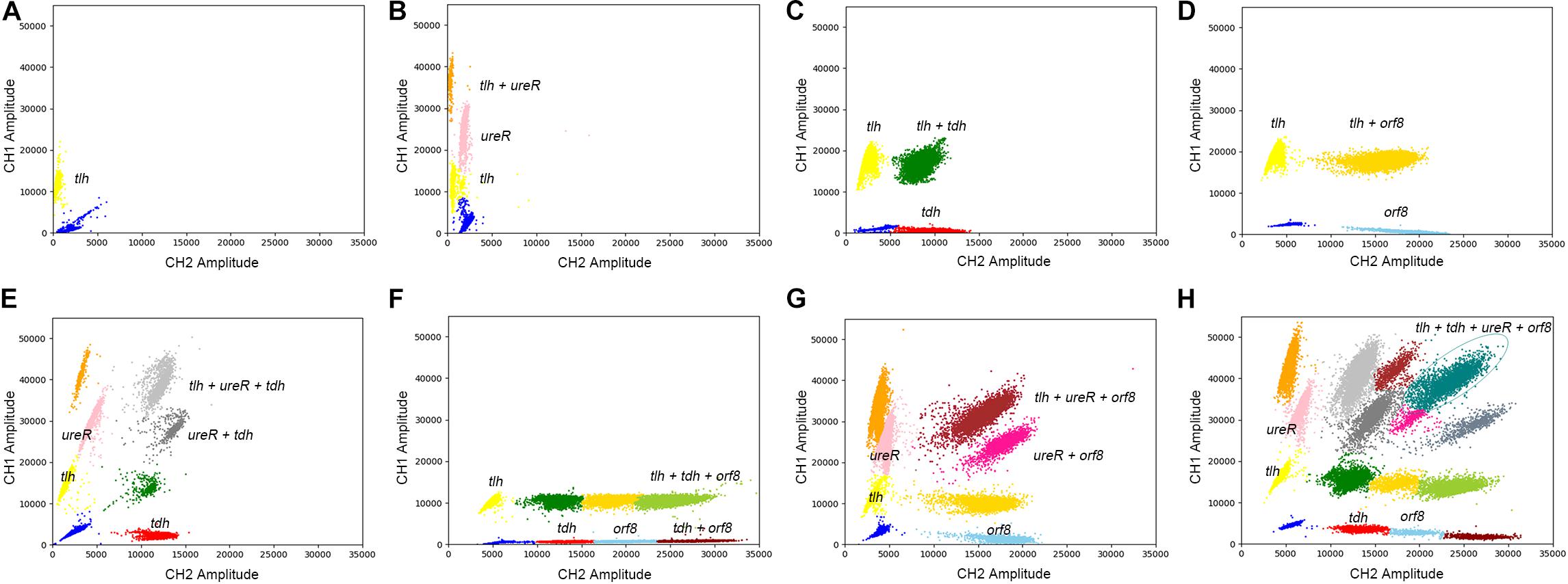
Figure 5. The 2D plot of different targets, the target contains (A) tlh; (B) tlh, ureR; (C) tlh, tdh; (D) tlh, orf8; (E) tlh, ureR, and tdh; (F) tlh, tdh, and orf8; (G) tlh, ureR, and orf8; (H) tlh, tdh, ureR, and orf8.
Distinguishing V. parahaemolyticus From High Levels of Escherichia coli and Listeria monocytogenes
In order to evaluate the selectivity of the 4-plex ddPCR assay for V. parahaemolyticus detection, other pathogenic bacteria including E. coli and L. monocytogenes at a concentration from 101–106 CFU/mL, were examined under the same detecting conditions. The V. parahaemolyticus cultures were diluted to approximately 3.9 × 103 CFU/mL. Figure 6A reveals that the measured concentrations by our method were close to that of the control, while background bacteria are at different concentrations (101–106 CFU/mL). The results indicate that the 4-plex ddPCR method displays good selectivity for V. parahaemolyticus, and the detections of V. parahaemolyticus are not disturbed by other bacteria no matter how high the non-target bacteria concentrations (101–106 CFU/mL) are. In Figures 6B–E, when the concentration of background bacteria is 106 CFU/mL, the detections of V. parahaemolyticus (3.9 × 101–3.9 × 107 CFU/mL) have a good linear and are in consistent with the results without background bacteria. The R2 are 0.9978 (tlh), 0.998 (tdh), 0.9898 (ureR), and 0.9739 (orf8). The results indicate that either in the absence or presence of high concentrations of non-target bacteria, the detections of V. parahaemolyticus at different concentrations (101–107 CFU/mL) are not impacted.
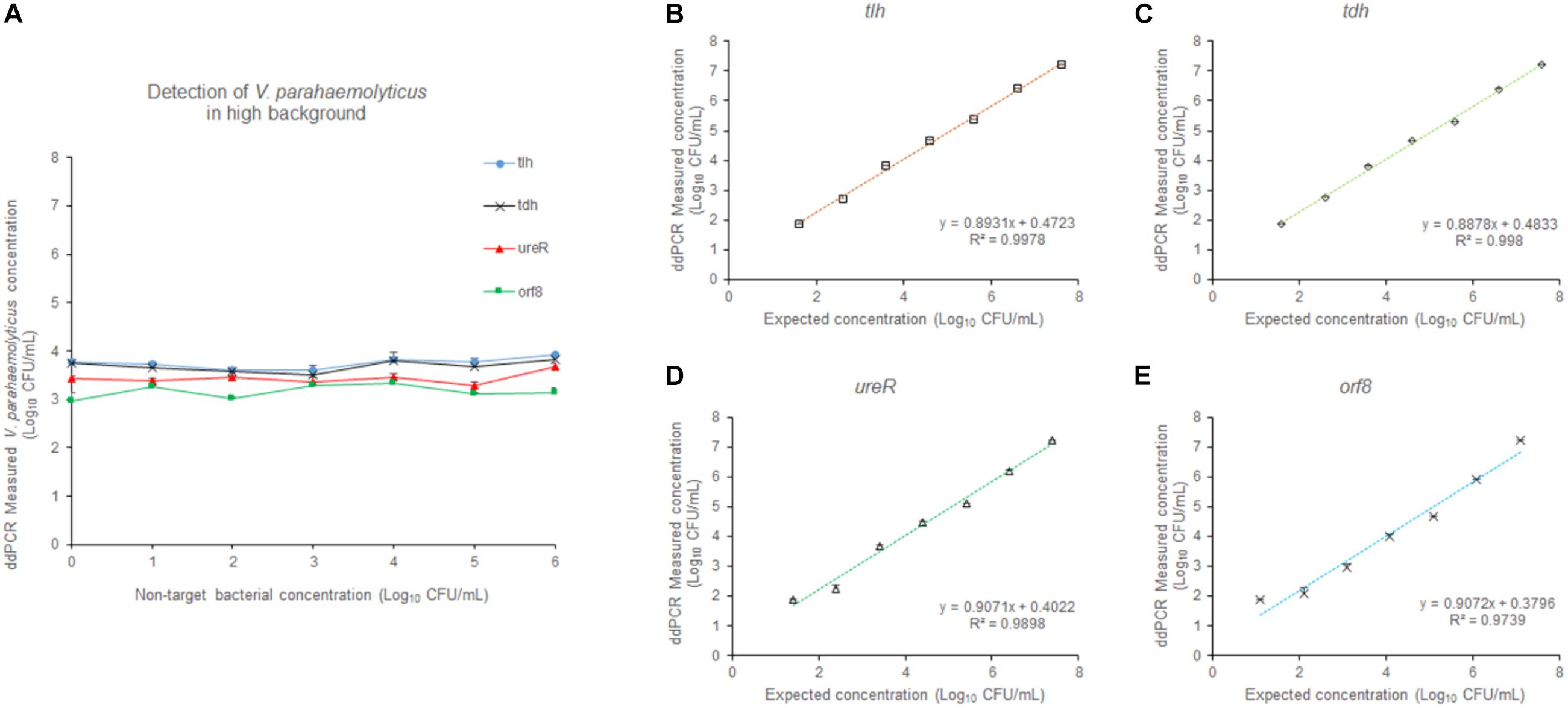
Figure 6. The effect of the high concentration of non-target bacteria on the detection. (A) At different concentrations (101–106 CFU/mL) of non-target bacteria, V. parahaemolyticus at the concentration of 103 CFU/mL was detected. At the concentration (106 CFU/mL) of non-target bacteria, V. parahaemolyticus samples containing tlh (B), tdh (C), ureR (D), and orf8 (E) at different concentrations (101–107 CFU/mL) were detected. Plotted values represented the means of 3 independent replicates.
Performance of the 4-plex ddPCR Assay in Detecting Artificial Contaminated Seafood Samples
To assess the efficiency of the multiplex ddPCR, both quantitative bacterial solution and artificial contaminated white clam tissue containing the equal concentration of bacterial cells were used as the templates for the detection of V. parahaemolyticus. As shown in Figure 7, there is no significant difference between the results of the two templates (p > 0.05). It confirmed that the established method was applicable for the determination of V. parahaemolyticus in a wide range of concentrations in complex food samples.
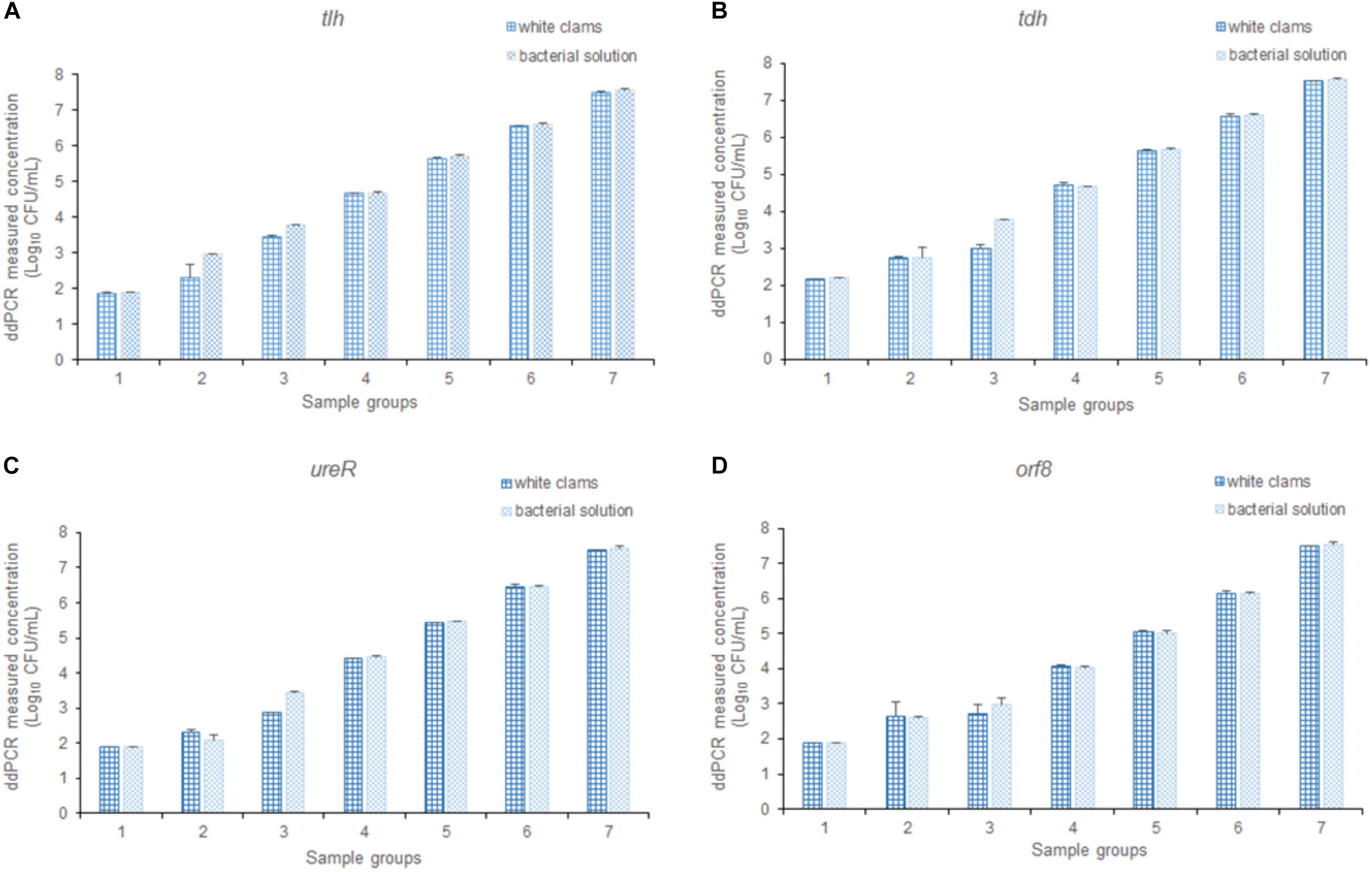
Figure 7. The detection results of V. parahaemolyticus in solution and spiked white clams, analyzing (A) tlh, (B) tdh, (C) ureR, and (D) orf8, respectively. Plotted values represented the mean value and standard deviations obtained from three triplicate tests.
Discussion
Vibrio parahaemolyticus is an enteric pathogen found in a variety of seafood, especially clams, shrimps, and oysters (Liu et al., 2018; Cao et al., 2019). Although the presence of V. parahaemolyticus is extensive in marine and estuarine environments, not all strains of V. parahaemolyticus result in illness (Colwell et al., 1977). Only a small subset of V. parahaemolyticus strains are pathogenic (Velazquez-Roman et al., 2014). In molecular epidemiological studies, the strains with tdh+, and/or trh+ were generally distinguished as virulent strains, which were up to 90% of the isolated clinical strains (Zhang and Austin, 2005; Chao et al., 2010). Either TDH, TRH, or both can be produced by pathogenic V. parahaemolyticus (Honda and Iida, 1993; Nishibuchi and Kaper, 1995). Hence, the target genes tdh and trh, encoding TDH hemolysin and TRH hemolysin, respectively, have been used for the classification of pathogenic isolates of V. parahaemolyticus (Shirai et al., 1990; Yoh et al., 1995). The transcriptional regulator for the urease gene cluster was encoded by the ureR gene, immediately upstream of trh. Replacing trh with ureR as a target gene could circumvent trh sequence variation (Nilssona and Turner, 2016). In February 1996, an atypical increase in diarrheal infections caused by V. parahaemolyticus occurred in Kolkata and India. Then, this clone rapidly spread throughout the majority of Southeast Asian, Atlantic, and Gulf coasts of the United States (Okuda et al., 1997; Chowdhury et al., 2000; Matsumoto et al., 2000), Europe (Martinez-Urtaza et al., 2005), and Africa (Ansaruzzaman et al., 2005). This epidemiology was related to V. parahaemolyticus O3:K6 serotype, which was pandemic strain generally contain the orf8 gene. All in all, it is of great importance to apply tlh, tdh, ureR, and orf8 to facilitate accurate quantification of V. parahaemolyticus in samples. Based on this method, we can take more genes such as trh, toxR, and toxS into account in the future.
What kind of genes the bacteria carried determined the bacterial characteristics. The 4-plex ddPCR method was very effective for detecting tlh, tdh, ureR, and orf8 genes simultaneously. It was beneficial to find linkage among the 4 targets for classifying V. parahaemolyticus. When gDNA extracts were used as the template, the fragments would be segregated into different droplets. However, when the intact cell was used as a template, a single cell was contained in a droplet and the linkage among four genes in one cell was presented. In the previous report, the multiplexed single intact cell droplet digital PCR (MuSIC ddPCR) method was developed for the detection of Enterohemorrhagic E. coli (EHEC; McMahon et al., 2017). However, it was only 2-plex and displayed low amplification efficiency. In this paper, the 4-plex ddCR method based on the single intact cell for the detection of V. parahaemolyticus was first established. The results indicated that our method not only could identify pathogenic V. parahaemolyticus conveniently, precisely and reliably, but also was more sensitive than qPCR.
For determining the influence of non-target bacteria, two experiments were conducted. On one hand, V. parahaemolyticus (3.9 × 103 CFU/mL) mixed with different concentrations of non-target bacteria (101–106 CFU/mL) were detected. On the other hand, different concentrations (3.9 × 101–3.9 × 107 CFU/mL) of V. parahaemolyticus in the high background of non-target bacteria (106 CFU/mL) were tested. The V. parahaemolyticus detection results of the samples with non-target bacteria were consistent with that of the sample without non-target. The presence of non-target bacteria showed less impact on detection. The result presented strong evidence that our method displayed high sensitivity and specificity. Different from other ddPCR detection which had narrow range (Wang et al., 2018; Zhong et al., 2018; Jahne et al., 2019; Nshimyimana et al., 2019), our method presented a wider detection range from 3.9 × 107 to 3.9 × 101 CFU/mL. Owing to approximate 80 thousand droplets produced, a narrowed detection range of ddPCR was circumvented. In addition, our detection method required a small amount of sample (2 μL), which provided new ideas for the detection of rare samples.
Conclusion
In conclusion, the 4-plex ddPCR method was developed for the rapid, sensitive, convenient, and reliable detection of V. parahaemolyticus. Four characteristic target genes of V. parahaemolyticus including tlh, tdh, ureR, and orf8 were successfully determined. It could not only absolutely quantify V. parahaemolyticus cells but also effectively help classify different strains of V. parahaemolyticus. This 4-plex ddPCR method would greatly facilitate the detection of different serotypes of V. parahaemolyticus and provide a new method for detecting other bacteria.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
SL performed the experiments and prepared the manuscript. XG designed the experiments and analyzed the data. WX contributed to manuscript revision. ZR contributed analysis tools. ZW provided reagents. SC performed parts of the experiments. QZ designed the experiments and contributed to manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by National Natural Science Foundation of China (Grant No. 31972046), National Key R&D Program of China (Grant No. 2017YFC1601203), and Key-Area Research and Development Program of Guangdong Province (Grant No. 2018B020206001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ansaruzzaman, M., Lucas, M., Deen, J. L., Bhuiyan, N. A., Wang, X., Safa, A., et al. (2005). Pandemic serovars (O3 : K6 and O4 : K68) of Vibrio parahaemolyticus associated with diarrhea in mozambique: spread of the pandemic into the African continent. J. Clin. Microbiol. 43, 2559–2562. doi: 10.1128/JCM.43.6.2559-2562.2005
Bahar, M. H., Wist, T. J., Bekkaoui, D. R., Hegedus, D. D., and Olivier, C. Y. (2018). Aster leafhopper survival and reproduction, and aster yellows transmission under static and fluctuating temperatures, using ddPCR for phytoplasma quantification. Sci. Rep. 8:227. doi: 10.1038/s41598-017-18437-0
Baross, J., and Liston, J. (1968). Isolation of Vibro parahaemolyticus from the Northwest Pacific. Nature 217, 1263–1264. doi: 10.1038/2171263a0
Bej, A. K., Patterson, D. P., Brasher, C. W., Vickery, M. C. L., Jones, D. D., and Kaysner, C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36, 215–225. doi: 10.1016/S0167-7012(99)00037-8
Cao, X., Zhao, L., Zhang, J., Chen, X., Shi, L., Fang, X., et al. (2019). Detection of viable but nonculturable Vibrio parahaemolyticus in shrimp samples using improved real-time PCR and real-time LAMP methods. Food Control 103, 145–152. doi: 10.1016/j.foodcont.2019.04.003
Ceccarelli, D., Hasan, N. A., Huq, A., and Colwell, R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 3:97. doi: 10.3389/fcimb.2013.00097
Chao, G., Jiao, X., Zhou, X., Wang, F., Yang, Z., Huang, J., et al. (2010). Distribution of genes encoding four pathogenicity islands (VPaIs), T6SS, biofilm, and type I pilus in food and clinical strains of Vibrio parahaemolyticus in China. Foodborne Pathog. Dis. 7, 649–658. doi: 10.1089/fpd.2009.0441
Chowdhury, N. R., Chakraborty, S., Ramamurthy, T., Nishibuchi, M., Yamasaki, S., Takeda, Y., et al. (2000). Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg. Infect. Dis. 6, 631–636. doi: 10.3201/eid0606.000612
Colwell, R. R., Kaper, J., and Joseph, S. W. (1977). Vibrio cholerae, Vibrio parahaemolyticus, and other vibrios: occurrence and distribution in Chesapeake Bay. Science 198, 394–396. doi: 10.1126/science.198.4315.394-a
Fujino, T., Okuno, Y., Nakada, D., Aoyama, A., Fukai, K., Mukai, T., et al. (1953). On the bacteriological examination of shirasu-food poisoning. Med. J. Osaka Univ. 4, 299–304.
Guin, S., Murugan, S., Anjay Goutam, C., Gururaja, P. P., Ramamurthy, T., and Suresh, C. D. (2019). Pathogenic Vibrio parahaemolyticus indiarrhoeal patients, fish and aquatic environments and their potential for inter-source transmission. Heliyon 5:e1743. doi: 10.1016/j.heliyon.2019.e01743
Han, H., Li, F., Yan, W., Guo, Y., Li, N., Liu, X., et al. (2015). Temporal and spatial variation in the abundance of total and pathogenic Vibrio parahaemolyticus in Shellfish in China. PLoS One 10:130302. doi: 10.1371/journal.pone.0130302
Hindson, B. J., Ness, K. D., Masquelier, D. A., Belgrader, P., Heredia, N. J., Makarewicz, A. J., et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analyt. Chem. 83, 8604–8610. doi: 10.1021/ac202028g
Hindson, C. M., Chevillet, J. R., Briggs, H. A., Gallichotte, E. N., Ruf, I. K., Hindson, B. J., et al. (2013). Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 10:1003. doi: 10.1038/nmeth.2633
Honda, T., and Iida, T. (1993). The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev. Med. Microbiol. 4, 106–113. doi: 10.1097/00013542-199304000-00006
Jahne, M. A., Brinkman, N. E., Keely, S. P., Zimmerman, B. D., Wheaton, E. A., and Garland, J. L. (2019). Droplet digital PCR quantification of norovirus and adenovirus in decentralized wastewater and graywater collections: implications for onsite reuse. Water Res. 169:213. doi: 10.1016/j.watres.2019.115213
Kishishita, M., Matsuoka, N., Kumagai, K., Yamasaki, S., Takeda, Y., and Nishibuchi, M. (1992). Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 58, 2449–2457. doi: 10.1128/AEM.58.8.2449-2457.1992
Krantz, G. E., Colwell, R. R., and Lovelace, E. (1969). Vibrio parahaemolyticus from the blue crab Callinectes sapidus in Chesapeake Bay. Science 164, 1286–1287. doi: 10.1126/science.164.3885.1286
Lei, S., Gu, X., Zhong, Q., Duan, L., and Zhou, A. (2020). Absolute quantification of Vibrio parahaemolyticus by multiplex droplet digital PCR for simultaneous detection of tlh, tdh and ureR based on single intact cell. Food Control 114, 1–9. doi: 10.1016/j.foodcont.2020.107207
Letchumanan, V., Chan, K., and Lee, L. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Liu, Y., Zhong, Q., Wang, J., and Lei, S. (2018). Enumeration of Vibrio parahaemolyticus in VBNC state by PMA-combined real-time quantitative PCR coupled with confirmation of respiratory activity. Food Control 91, 85–91. doi: 10.1016/j.foodcont.2018.03.037
Martinez-Urtaza, J., Simental, L., Velasco, D., DePaola, A., Ishibashi, M., Nakaguchi, Y., et al. (2005). Pandemic Vibrio parahaemolyticus O3:K6. Europe. Emerg. Infect. Dis. 11, 1319–1320. doi: 10.3201/eid1108.050322
Matsumoto, C., Okuda, J., Ishibashi, M., Iwanaga, M., Garg, P., Rammamurthy, T., et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38, 578–585. doi: 10.1128/JCM.38.2.578-585.2000
McMahon, T. C., Blais, B. W., Wong, A., and Carrillo, C. D. (2017). Multiplexed single intact cell droplet digital PCR (MuSIC ddPCR) method for specific detection of Enterohemorrhagic E. coli (EHEC) in food enrichment cultures. Front. Microbiol. 8:332. doi: 10.3389/fmicb.2017.00332
Myers, M. L., Panicker, G., and Bej, A. K. (2003). PCR detection of a newly emerged pandemic Vibrio parahaemolyticus O3:K6 pathogen in pure cultures and seeded waters from the Gulf of Mexico. Appl. Environ. Microbiol. 69, 2194–2200. doi: 10.1128/AEM.69.4.2194-2200.2003
Nasu, H., Iida, T., Sugahara, T., Yamaichi, Y., Park, K. S., Yokoyama, K., et al. (2000). A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38, 2156–2161. doi: 10.1128/.38.6.2156-2161.2000
Newton, A. E., Garrett, N., Stroika, S. G., Halpin, J. L., Turnsek, M., and Mody, R. K. (2014). Increase in Vibrio parahaemolyticus infections associated with consumption of atlantic coast shellfish-2013. MMWR Morb. Mort. Wkly Rep. 63, 335–336. doi: 10.1186/1745-6673-9-16
Nilssona, W. B., and Turner, J. W. (2016). The thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus: sequence variation and implications for detection and function. J. Microbiol. Methods 126, 1–7. doi: 10.1016/j.mimet.2016.04.007
Nishibuchi, M., and Kaper, J. B. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63, 2093–2099. doi: 10.1128/IAI.63.6.2093-2099.1995
Niu, B., Hong, B., Zhang, Z., Mu, L., Malakar, P. K., Liu, H., et al. (2018). A novel qPCR method for simultaneous detection and quantification of viable pathogenic and non-pathogenic Vibrio parahaemolyticus (tlh+, tdh+, and ureR+). Front. Microbiol. 9:1747. doi: 10.3389/fmicb.2018.01747
Nshimyimana, J. P., Cruz, M. C., Wuertz, S., and Thompson, J. R. (2019). Variably improved microbial source tracking with digital droplet PCR. Water Res. 159, 192–202. doi: 10.1016/j.watres.2019.04.056
Okuda, J., Ishibashi, M., Hayakawa, E., Nishino, T., Takeda, Y., Mukhopadhyay, A. K., et al. (1997). Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35, 3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997
Ottesen, E. A., Hong, J. W., Quake, S. R., and Leadbetter, J. R. (2006). Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 314, 1464–1467. doi: 10.1126/science.1131370
Pang, B., Ding, X., Wang, G., Zhao, C., Xu, Y., Fu, K., et al. (2017). Rapid and quantitative detection of Vibrio parahemolyticus by the mixed-dye-based loop-mediated isothermal amplification assay on a self-priming compartmentalization microfluidic chip. J. Agric. Food Chem. 65, 11312–11319. doi: 10.1021/acs.jafc.7b03655
Raghunath, P., Maiti, B., Shekar, M., Karunasagar, I., and Karunasagar, I. (2010). Clinical isolates of Aeromonas veronii biovar veronii harbor a nonfunctional gene similar to the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. FEMS Microb. Lett. 307, 151–157. doi: 10.1111/j.1574-6968.2010.01974.x
Shirai, H., Ito, H., Hirayama, T., Nakamoto, Y., Nakabayashi, N., Kumagai, K., et al. (1990). Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58, 3568–3573. doi: 10.1128/IAI.58.11.3568-3573.1990
Su, Y., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Suthienkul, O., Ishibashi, M., Lida, T., Nettip, N., Supavej, S., Eampokalap, B., et al. (1995). Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J. Infect. Dis. 172, 1405–1408. doi: 10.1093/infdis/172.5.1405
Tadmor, A. D., Ottesen, E. A., Leadbetter, J. R., and Phillips, R. (2011). Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science 333, 58–62. doi: 10.1126/science.1200758
Taniguchi, H., Hirano, H., Kubomura, S., Higashi, K., and Mizuguchi, Y. (1986). Comparison of the nucleotide sequences of the genes for the thermostable direct hemolysin and the thermolabile hemolysin from Vibrio parahaemolyticus. Microb. Pathog. 1, 425–432. doi: 10.1016/0882-4010(86)90004-5
Velazquez-Roman, J., Leon-Sicairos, N., Hernandez-Diaz, L., and Canizalez-Roman, A. (2014). Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell. Infect. Microbiol. 3:110. doi: 10.3389/fcimb.2013.00110
Velusamy, V., Arshak, K., Korostynska, O., Oliwa, K., and Adley, C. (2010). An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol. Adv. 28, 232–254. doi: 10.1016/j.biotechadv.2009.12.004
Wang, M., Yang, J., Gai, Z., Huo, S., Zhu, J., Li, J., et al. (2018). Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk. Intern. J. Food Microbiol. 266, 251–256. doi: 10.1016/j.ijfoodmicro.2017.12.011
Yoh, M., Kawakami, N., Funakoshi, Y., Okada, K., and Honda, T. (1995). Evaluation of two assay kits for thermostable direct hemolysin (TDH) as an indicator of TDH-Related hemolysin (TRH) produced by Vibrio parahaemolyticus. Microbiol. Immunol. 39, 157–159. doi: 10.1111/j.1348-0421.1995.tb02183.x
Zhang, X. H., and Austin, B. (2005). Haemolysins in Vibrio species. J. Appl. Microbiol. 98, 1011–1019. doi: 10.1111/j.1365-2672.2005.02583.x
Zhang, Z., Liu, H., Lou, Y., Xiao, L., Liao, C., Malakar, P. K., et al. (2015a). Quantifying viable Vibrio parahaemolyticus and Listeria monocytogenes simultaneously in raw shrimp. Appl. Microbiol. Biotechnol. 99, 6451–6462. doi: 10.1007/s00253-015-6715-x
Zhang, Z., Xiao, L., Lou, Y., Jin, M., Liao, C., Malakar, P. K., et al. (2015b). Development of a multiplex real-time PCR method for simultaneous detection of Vibrio parahaemolyticus, Listeria monocytogenes and Salmonella spp. in raw shrimp. Food Control 51, 31–36. doi: 10.1016/j.foodcont.2014.11.007
Keywords: Vibrio parahaemolyticus, 4-plex ddPCR, qPCR, single intact cell, precise detection
Citation: Lei S, Gu X, Xue W, Rong Z, Wang Z, Chen S and Zhong Q (2020) A 4-plex Droplet Digital PCR Method for Simultaneous Quantification and Differentiation of Pathogenic and Non-pathogenic Vibrio parahaemolyticus Based on Single Intact Cells. Front. Microbiol. 11:1727. doi: 10.3389/fmicb.2020.01727
Received: 17 April 2020; Accepted: 01 July 2020;
Published: 12 August 2020.
Edited by:
Julio Parra-Flores, University of the Bío Bío, ChileReviewed by:
Ariadnna Cruz-Córdova, Hospital Infantil de México Federico Gómez, MexicoYong Zhao, Shanghai Ocean University, China
Copyright © 2020 Lei, Gu, Xue, Rong, Wang, Chen and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Zhong, emhvbmdxcEBzY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Shuwen Lei2,3†
Shuwen Lei2,3† Xiaokui Gu
Xiaokui Gu Qingping Zhong
Qingping Zhong