
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 July 2020
Sec. Fungi and Their Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01633
This article is part of the Research Topic Fungal Genetics in Plant Biomass Conversion View all 11 articles
Lignocellulose is an abundant waste resource and has been considered as a promising material for production of biofuels or other valuable bio-products. Currently, one of the major bottlenecks in the economic utilization of lignocellulosic materials is the cost-efficiency of converting lignocellulose into soluble sugars for fermentation. One way to address this problem is to seek superior lignocellulose degradation enzymes or further improve current production yields of lignocellulases. In the present study, the lignocellulose degradation capacity of a thermophilic fungus Chaetomium thermophilum was firstly evaluated and compared to that of the biotechnological workhorse Trichoderma reesei. The data demonstrated that compared to T. reesei, C. thermophilum displayed substantially higher cellulose-utilizing efficiency with relatively lower production of cellulases, indicating that better cellulases might exist in C. thermophilum. Comparison of the protein secretome between C. thermophilum and T. reesei showed that the secreted protein categories were quite different in these two species. In addition, to prove that cellulases in C. thermophilum had better enzymatic properties, the major cellulase cellobiohydrolase I (CBH1) from C. thermophilum and T. reesei were firstly characterized, respectively. The data showed that the specific activity of C. thermophilum CBH1 was about 4.5-fold higher than T. reesei CBH1 in a wide range of temperatures and pH. To explore whether increasing CBH1 activity in T. reesei could contribute to improving the overall cellulose-utilizing efficiency of T. reesei, T. reesei cbh1 gene was replaced with C. thermophilum cbh1 gene by integration of C. thermophilum cbh1 gene into T. reesei cbh1 gene locus. The data surprisingly showed that this gene replacement not only increased the cellobiohydrolase activities by around 4.1-fold, but also resulted in stronger induction of other cellulases genes, which caused the filter paper activities, Azo-CMC activities and β-glucosidase activities increased by about 2.2, 1.9, and 2.3-fold, respectively. The study here not only provided new resources of superior cellulases genes and new strategy to improve the cellulase production in T. reesei, but also contribute to opening the path for fundamental research on C. thermophilum.
Plant biomass from agriculture and forestry is one of the most abundant resources on the earth and it has been considered as a promising material for producing renewable biofuels and other value-added bio-products (Himmel et al., 2007). One of the bottlenecks of this process is the enzymatic degradation of biomass-derived polysaccharides. Currently, the filamentous fungi Trichoderma, Aspergillus, and Penicillium species are the most commonly used producer for lignocellulases in industry (Kubicek et al., 2014). However, the optimum temperature of enzymes from these species is normally from 30 to 50°C at which the efficiency of biomass polysaccharides saccharification is very low. Many thermophilic fungi can secrete thermostable biomass-degrading enzymes including lignocellulases, proteases, amylases, laccases, chitinases, lipases, and esterases, which holds a great promise in industrial applications (DeCastro et al., 2016). However, due to the lack of genetic tools for strain engineering in thermophilic species, progress toward the improvement of their enzyme production has been hampered. One way to overcome these drawbacks is the introduction of lignocellulases encoding genes from thermophilic fungi into mesophilic species, in which a variety of genetic tools have been developed, to combine the advantages of mesophilic and thermophilic properties.
The soft-rot fungus Trichoderma reesei is a model organism for plant biomass degradation and widely used in industry for the production of cellulases and xylanases due to the large capacity of hydrolytic enzymes secretion (Bischof et al., 2016). After rounds of random mutagenesis, several industrial strains can secrete lignocellulases over 100 g/L culture (Cherry and Fidantsef, 2003). Besides its large protein secretion capacity, T. reesei naturally has the ability to form disulfide bonds and to glycosylate proteins (Singh et al., 2015). In addition, T. reesei can grow on inexpensive substrate such as lignocellulosic waste materials and many secreted enzymes from T. reesei are under the GRAS status (Generally Recognized as Safe) (Sewalt et al., 2016). These properties have promoted efforts to develop T. reesei as an excellent host for production of lignocellulases and recombinant proteins. Many studies have shown that the lignocellulose degradation ability could be improved by recombinant expression of biomass degrading enzymes from different organisms in T. reesei (Zhang et al., 2010; Dashtban and Qin, 2012; Treebupachatsakul et al., 2015).
Chaetomium thermophilum, an ascomycete thermophilic species, commonly exists in the heating phase of composts with an optimum growth temperature around 50°C (Kellner et al., 2016). The release of C. thermophilum genome database (Bock et al., 2014) and several biotechnological studies indicate that there are valuable biomass degradation associated enzymes existed in C. thermophilum (Voutilainen et al., 2008; Li et al., 2010). However, the secreted enzyme components and their properties in C. thermophilum have been rarely studied. In the present study, the capability of lignocellulose degradation of C. thermophilum was evaluated and compared to that of T. reesei. Besides, the enzyme properties of the major cellulase cellobiohydrolase I (CBH1) from T. reesei and C. thermophilum were compared. Moreover, to explore whether increasing CBH1 activities in T. reesei cellulases complex could contribute to improving the cellulose degradation efficiency, the native T. reesei cbh1 gene was replaced with C. thermophilum cbh1 gene in T. reesei. The CBH activities, filter paper activities, endoglucanases activities, and β-glucosidase activities, along with the transcriptional levels of several cellulases genes including eg1, eg2, cbh2, and bgl1, as well as the transcriptional factors (TFs) xyr1, cre1, and ace3 of the resultant strains have been investigated. This study may provide a new resource of superior cellulases genes and a new strategy to improve the efficiency of biomass saccharification by T. reesei.
Escherichia coli Trans1-T1 used for recombinant plasmid construction was bought from TransGen Biotech. T. reesei Tu6 strain (ATCC MYA-256) was used as control to evaluate lignocellulose degradation efficiency of C. thermophilum. T. reesei Tu6Δku70 strain used as host strain to construct indicated recombinant strains was kindly provided by Dr. Monika Schmoll (AIT Austrian Institute of Technology, Austria). T. reesei strains Tr-cTcbh1, Tr-cCcbh1, and Tr-Ctcbh1 were constructed in this study and described below. C. thermophilum (CGMCC 3.17990) was provided by China general microbiological culture collection center. T. reesei strains were grown for 5 to 6 days at 28°C on potato dextrose agar plates (PDA) or PDA supplemented with 5 mM uridine when necessary. Minimal medium (Penttilä et al., 1987) was applied as a selective medium for the screening of T. reesei transformants. C. thermophilum strain was grown for 5–7 days at 50°C on PDA slants.
For protein secretion and enzyme activities analysis, T. reesei conidia were firstly inoculated into 100 mL liquid minimal medium with 2% glucose as the carbon source (MM+2% glucose) at 106 conidia/mL and grown at 28°C on a rotary shaker for 48 h at 200 rpm, the mycelia were then filtered through 200 mesh sifter (30 μm pore diameter) and washed twice with carbon-free medium. About 2.2 g of this wet mycelia was added into 50 mL of fresh cellulase-inducing minimal medium in which 2% glucose (w/v) was substituted with 2% Avicel or 2% sugarcane bagasse and grown at 200 rpm at 28°C in consistent dark. For analysis of C. thermophilum, 7 days of C. thermophilum culture grown on a slant (18 mm × 180 mm) was scraped and first inoculated into Neurospora Vogel’s minimal medium (VMM) (Vogel, 1956) with 2% glucose as carbon source and grown at 50°C on a rotary shaker (200 rpm) for 36 h. The cultures were then filtered and about 2.2 g of the wet mycelia was respectively transferred into 50 mL VMM media with 2% Avicel or 2% sugarcane bagasse as carbon source and grown at 50°C on a rotary shaker (200 rpm) for indicated time in the natural light.
The electroporation transformation method was based on the protocol described in Schuster et al. (2012) with some optimization. In brief, fresh T. reesei conidia (within 7 days) grown on a 90 mm agar plate was harvested and washed three times using 1.1 M ice cold sorbitol to make conidia suspension at the concentration of 108 conidia/mL. A total of 1–2 μg of transformed DNA in a final volume of 10 and 90 μL of sorbitol washed conidia suspension was put into pre-chilled electroporation cassette. Gene Pulser Xcell electroporation system (Bio-Rad, United States) was used for electroporation with the set of 1.6 kV, 600 Ω, 25 μF. After electroporation, 900 μL of 1.1 M icy cold sorbitol was immediately added into the electroporation reaction mix before transferring into 9 mL of YPD (1% yeast extract, 2% peptone, 1% dextrose). The whole mixture was incubated for 12 h at 30°C and centrifuged at 2000 × g for 5 min. The electroporated conidia were re-suspended in 500 μL of YPD and added with 14 mL of top MM agar media. The whole mixture was spread over 2–3 bottom MM agar plates and incubated at 30°C for 5–7 days.
Cpyr4 series strains were constructed by transforming strain Tu6Δku70 with plasmid pSK-pyr4 (Qin et al., 2012).
Tr-cTcbh1 was constructed by transforming strain Tu6Δku70 with a DNA fragment including pyr4 gene expression cassette and cDNA1 promoter (936 bp upstream of the start codon of cDNA1 gene) flanked with 2 kb upstream of cbh1 promoter (1.5 kb upstream of the start codon of cbh1 gene) and 2 kb downstream of cbh1 promoter. The plasmid pTrcTcbh1 used for generating Tr-cTcbh1 transformants was constructed by ligation of the above fragments into a backbone from plasmid pBluescript SK (+). The primer pair pTrcTcbh1-F1F/R was used for amplifying 2 kb fragment of the upstream of cbh1 promoter. The primer pair pTrcTcbh1-F2F/R was used to amplify the pyr4 gene expression cassette. The primer pair pTrcTcbh1-F3F/R was used for amplifying cDNA1 promoter. The primer pair pTrcTcbh1-F4F/R was used for amplifying 2 kb downstream of cbh1 promoter. The primer pair pTrcTcbh1-F5F/R was used for amplifying the vector backbone from plasmid pBluescript SK (+).
Tr-cCcbh1 strain was generated by transforming strain Tu6Δku70 with a fragment containing the expression cassette of pyr4 gene, cDNA1 promoter and cDNA sequence of C. thermophilum cbh1 gene (Accession No: AM711862.1), flanked with 2 kb upstream of the cbh1 promoter and 2 kb downstream of T. reesei cbh1 gene (Accession No: XM_006969162). The plasmid pTrcCcbh1 used for generating Tr-cCcbh1 transformants was constructed by ligation of the fragments of cDNA sequence of C. thermophilum cbh1 gene and 2 kb downstream of T. reesei cbh1 gene into the vector backbone from the plasmid pTrcTcbh1. The primer pair pTrcCcbh1-F1F/R was used for amplifying cDNA sequence of C. thermophilum cbh1 gene. The primer pair pTrcCcbh1-F2F/R was used for amplifying the fragment of 2 kb downstream of T. reesei cbh1 gene. The primer pair pTrcCcbh1-F3F/R was used for amplifying the vector backbone which included 2 kb fragment of the upstream of cbh1 promoter, pyr4 gene expression cassette, and cDNA1 promoter.
Tr-Ctcbh1 was created by co-transforming strain Tu6Δku70 with linearized plasmid pSK-pyr4 (Qin et al., 2018) and a fragment including cDNA sequence of C. thermophilum cbh1 gene, flanked with 2 kb upstream and downstream of T. reesei cbh1 gene. The plasmid pTrCtcbh1 used for generating Tr-Ctcbh1 transformants was constructed by ligation of 2 kb upstream of T. reesei cbh1 gene and cDNA sequence of C. thermophilum cbh1 gene into the vector backbone from plasmid pTrcCcbh1. The primer pair pTrCtcbh1-F1F/R was used for amplifying 2 kb upstream of T. reesei cbh1 gene. The primer pair pTrCtcbh1-F2F/R was used for amplifying cDNA sequence of C. thermophilum cbh1 gene. The primer pair pTrCtcbh1-F3F/R was used for amplifying the vector backbone from plasmid pTrcCcbh1 which included 2 kb downstream of T. reesei cbh1 gene.
All types of transformants were selected on minimal media without adding uridine and tested for genotypes by diagnostic PCR. The sequence of all the primers used in this study was listed in the Supplementary Table S1.
As described before (Stappler et al., 2017), the abundance of biomass accumulation in the culture containing insoluble cellulose was determined by the measurement of the intracellular protein concentration of mycelia. The mycelia in culture were filtered using a vacuum filter system and added 1 mL 0.1 M NaOH and lysed using a mini-bead beater (Biospec Products, Bartlesville, OK, United States) with 0.5 mm diameter glass beads. The mixture was incubated for 1 h at room temperature and centrifuged for 5 min at 12,000 rpm and the supernatants were transferred to new tubes. The protein concentration in the supernatants was measured by using the Bradford Protein Assay.
Protein concentration in supernatants of all cultures was determined by Bradford method by following the instruction (Bio-Rad Protein Assay). Cellobiohydrolase (CBH) activities of Tr-cTcbh1, Tr-cCcbh1, and Cpyr4 series strains under glucose were measured using 1 mM soluble 4-methylumbelliferyl-b-D-cellobiose (MUC, Sigma) as a substrate following methods described in Wu et al. (2017). In brief, 10 μL of culture supernatant and 40 μL of 1 mM MUC in 50 mM sodium acetate buffer (pH 5) were incubated together at 50°C for 15 min by using thermocycler. After 15 min, the temperature was raised to 95°C for 5 min to deactivate the enzyme. After cooling down to room temperature, the fluorescence was measured at 445 nm with excitation with 365 nm. CBH activities of Tr-Ctcbh1 and Cpyr4 series strains under cellulose were measured using the same substrate, but the method was as described as in Bailey and Tähtiharju (2003). CBH1 activity was calculated by subtraction of β-glucosidase and endoglucanase activity. One unit of CBH activity was defined as the amount of enzyme necessary to release 1 μmol methyl umbelliferone in one minute. The total cellulase activities in the supernatants were measured by filter paper activity using Whatman No. 1 filter paper as the substrate (Ghose, 1987). Endoglucanase activity was determined by using azo-carboxymethyl cellulose (Azo-CM-Cellulose; Megazyme) as the substrate and following the manufacturer’s specifications. β-glucosidase activity was measured by using 4-nitrophenyl -D-glucopyranoside (pNPG) as the substrate and following the method described in Tian et al. (2009). In brief, 50 μL of 10 × diluted culture supernatant was added into 200 μL of 500 μM pNPG in 50 mM sodium acetate buffer, pH 5.0. The mixture was incubated at 50°C for 10 min and terminated by adding 750 μL of 1 M Na2CO3. After cooling down to room temperature, the absorbance at 400 nm was measured. One unit of β-glucosidase activity was defined as the amount of enzyme necessary to release 1 μmol p-nitrophenol in 1 min.
The optimum pH was determined by measuring the enzyme activities using Glycine-HCl (pH 2.0–3.0, 50 mM), Citric buffer (pH 3.0–6.0, 50 mM), phosphate buffer (pH 6.0–8.0, 200 mM) and Glycine-NaOH (pH 9.0–10.0, 50 mM) at 50°C. A total of 25 μL of filtered culture supernatant was added into 75 μL of the each of the above buffers with 1 mM MUC as substrate and incubated at 50°C for 1 h and then transferred to 95°C for 5 min to deactivate the enzyme. The enzyme activities were calculated by the absorbance under 445 nm emission and 365 nm excitation after incubation. The pH stability was assayed by incubating the enzyme at different pH range from pH 2.0–9.0 for 2 h at room temperature. Then the samples were diluted 10-fold in 50 mM citric buffer, pH 4.0 before measuring the residual activity. The optimum temperature of recombinant CBH1 was determined between 20 and 80°C in 50 mM citric buffer, pH 4.0. Thermal stability was investigated by incubating the enzyme at a different temperature ranging from 20 to 80°C for 30 min in 50 mM citric buffer, pH 4.0. The remaining activities were measured under standard conditions.
The supernatants of 120 h of C. thermophilum and T. reesei culture under Avicel were used to run SDS-PAGE firstly. The SDS-PAGE gels were subsequently cut and digested by trypsin and then analyzed using an EASY-nLC 1000 liquid chromatograph which was connected in line with an Orbitrap Fusion Tribrid mass spectrometer equipped with a nanoelectrospray ionization (nanoESI) source (Thermo Fisher Scientific, Waltham, MA, United States). The peptides were loaded onto a trap column (C18, 5 μm particles, 100 μm ID, 3 cm length, Dr. Maisch GmbH) and separated using an analytical column (C18, 3 μm particles, 75 μm ID, 15 cm length, Dr. Maisch GmbH) at a flow rate of 400 nL/min with a 60 min LC gradient composed of Solvent A [0.1% formic acid (v/v)] and Solvent B [acetonitrile, 0.1% formic acid (v/v)]. The gradient was 3–8% B for 3 min, 8–23% B for 45 min, 23–35% B for 9 min, 35–80% B for 2 min, and finally 80% B for 1 min. Full-scan mass spectra were acquired in the positive-ion mode over the m/z range from 350 to 1550 using the Orbitrap mass analyzer in profile format with a mass resolution setting of 120,000. In the data-dependent mode, the top-speed mode was selected with the most intense ions exceeding an intensity threshold of 5000 counts from each full-scan mass spectrum for tandem mass spectrometry (MS/MS) analysis using HCD activation type with 33% collision energy. MS/MS spectra were acquired in centroid format using an ion trap detector. Real-time charge state screening was enabled to exclude unassigned and 1+ charge states from MS/MS analysis. The exclusion duration in dynamic exclusion was set as 20 s. Raw data were searched against the amino acid sequences of T. reesei or C. thermophilum using Mascot software (version 2.5).
A total of 500 μL of appropriately diluted hydrolysates from filter paper was analyzed on a Thermo Scientific Dionex ICS-5000 high-performance liquid chromatography (Dionex Corporation, Sunnyvale, CA, United States) instrument equipped with a CarboPac PA100 guard column (4 × 50 mm) and an analytical column (4 × 250 mm) with a flow rate as 1 mL/min at 22°C. The samples were resolved in a mobile phase of 100 mM NaOH. Glucose and cellobiose were used as standards.
An aliquot of 2 × 106 mL of T. reesei conidia were first inoculated into 100 mL MM+2% glucose media and grown at 28°C on a rotary shaker for 48 h at 200 rpm, the mycelia were then filtered through 200 mesh sifter (30 μm pore diameter) and washed twice with carbon-free MM media. A total of 2.2 g of the filtered wet mycelia were transferred to MM+1% Avicel medium and cultured at 28°C in the constant dark at 200 rpm for 48 h. Biological triplicates were prepared for each strain. Total RNA was isolated from samples taken at 12, 24, and 48 h of shift using zirconia/silica beads and a Mini-Beadbeater with 1 mL TRIzol reagent (from Invitrogen). RNA purification and DNase I treatment were performed using Easy Pure RNA Purification Kit (from TransGen Biotech). qRT-PCR was performed using TransScript Green One-Step qRT-PCR SuperMixKit (from TransGen Biotech) on the LightCycler 96 instrument (from Roche Life Science). Primer sequences used for amplification are described in Supplementary Table S1. The relative transcriptional levels of the tested genes were normalized to the expression level of the actin gene and calculated by 2–ΔCt as a relative expression level.
For all the experiments, three biologically replicated strains and three technical replicates for each strain were set for statistical analysis. Statistical significance was determined by t-test analysis by the false discovery rate (FDR) approach by using the Prism GraphPad software.
To search for thermophilic lignocellulases with outstanding enzyme properties, the cellulose degradation capacity of the thermophilic fungus C. thermophilum was evaluated and compared to that of T. reesei. To alleviate potential effects caused by differential spore germination efficiency, C. thermophilum CGMCC 3.17990 and T. reesei strain Tu6 were firstly grown in medium containing glucose as the sole carbon source and the same amount of wet mycelia (about 2.2 g) were then transferred into medium containing Avicel or sugarcane bagasse as the carbon source. The biomass accumulation, the extracellular protein concentration, and the filter paper activities in the supernatant were measured at different time points. The abundance of biomass accumulation in the culture containing insoluble carbon source Avicel or sugarcane was determined by the measurement of the intracellular protein concentration of mycelia.
The data shown in Figure 1A demonstrated that the biomass accumulation in C. thermophilum was significantly faster than that in T. reesei under both Avicel and sugarcane bagasse. Under 48 h of the after-transferring culture, C. thermophilum showed obvious growth phenotype under both Avicel and sugarcane bagasse compared to T. reesei (Figure 1B), suggesting higher utilization efficiency of cellulose materials in C. thermophilum relative to that in T. reesei. Further analysis of the extracellular protein levels (Figure 1C) in the culture supernatants of C. thermophilum and T. reesei showed that under Avicel, the levels of secreted proteins in C. thermophilum were slightly higher than that in T. reesei in the first 3 days. However, after 3 days, the extracellular protein levels in T. reesei were significantly higher than those in C. thermophilum. Under sugarcane bagasse, the extracellular protein levels in T. reesei were higher than that in C. thermophilum in all the tested time points and similar to the observed phenotype under Avicel, the difference of secreted protein levels between C. thermophilum and T. reesei significantly enlarged after 3 days. These data suggested that the secreted protein levels in the supernatants of C. thermophilum were considerably less than those in T. reesei when response to both Avicel and sugarcane bagasse. However, analysis of the filter paper activities in the culture supernatants (Figure 1D) showed that consistent with the growth phenotype, C. thermophilum exhibited significantly higher filter paper activities compared to T. reesei in all the tested time points and under both celluloses carbon sources. This observed phenomenon indicated that C. thermophilum might contain cellulases with better specific activities or have better enzyme complex system for cellulose degradation.
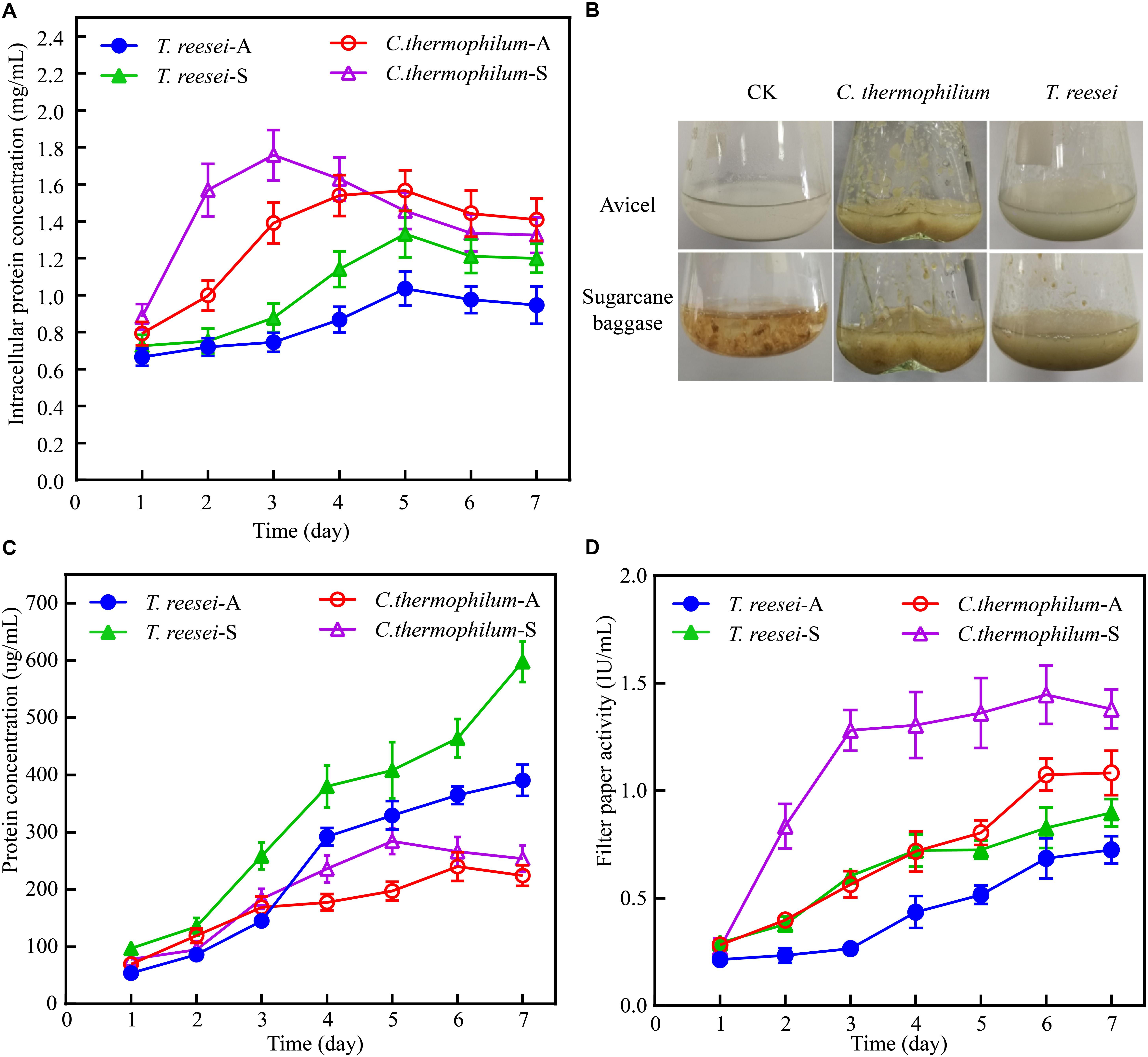
Figure 1. Phenotypic analysis of C. thermophilum and T. reesei when response to cellulose. (A) The intracellular protein concentration of C. thermophilum and T. reesei mycelia collected from the culture under Avicel or sugarcane bagasse after transferring from the glucose culture. (B) Growth phenotype of C. thermophilum and T. reesei under Avicel or sugarcane bagasse. About 2.2 g wet mycelia of C. thermophilum and T. reesei were collected and transferred into Avicel or sugarcane bagasse media from glucose contained media and cultivated for 48 h. (C) Total secreted protein levels in the supernatants of C. thermophilum and T. reesei in Avicel or sugarcane bagasse media at different time points. (D) Filter paper activities of the supernatants of C. thermophilum and T. reesei in Avicel or sugarcane bagasse media at different time points. Shown in (A,C,D) are the mean values of three biological replicates. Error bars show the standard deviations between these replicates. T. reesei-A, T. reesei-S, C. thermophilum-A, and C. thermophilum-S respectively indicates T. reesei grown on Avicel (A) or sugarcane bassage (S) and C. thermophilum grown on Avicel (A) or sugarcane bassage (S). Shown in (B) is one of the three represents.
To further investigate the mechanism behind the higher cellulose degradation efficiency in C. thermophilum, the supernatants of C. thermophilum and T. reesei cultures under Avicel were analyzed by SDS-PAGE. The data shown in Figure 2A indicated that the bands pattern of the secreted proteins in C. thermophilum was quite different from that in T. reesei. Although the total extracellular protein levels in T. reesei were more than that in C. thermophilum, it was worth noting that compared to T. reesei, there were more protein bands in the supernatants of C. thermophilum cellulose culture, indicating C. thermophilum might secrete more protein categories.
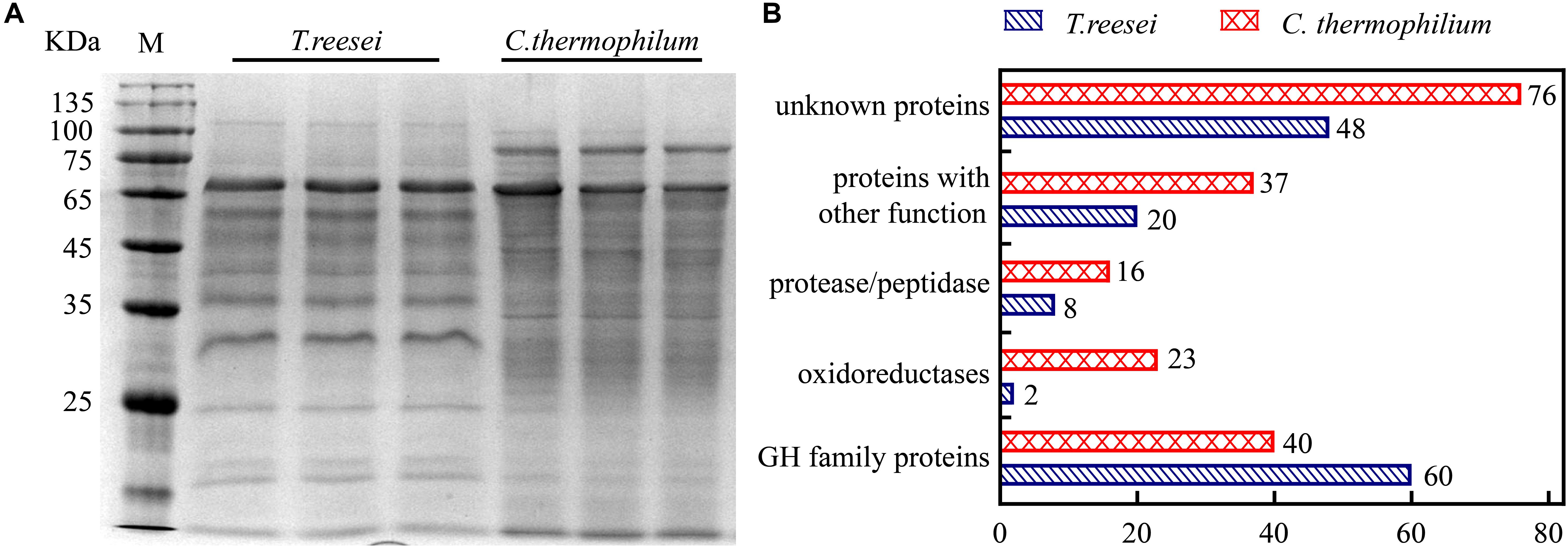
Figure 2. The extracellular protein categories in the supernatants of cellulose cultures were different between C. thermophilum and T. reesei. (A) SDS-PAGE analysis of the secreted proteins in the supernatant of C. thermophilum and T. reesei in 120-h Avicel cultures after transferring from glucose cultures. (B) Major protein categories of the secretome of C. thermophilum and T. reesei under Avicel. The data was collected by UPLC-MS/MS analysis and the detailed information was listed in Supplementary Tables S2–S7.
Based on the SDS-PAGE analysis, the components and the relative abundances of the extracellular proteins in the supernatants were subsequently evaluated by performing ultra-high performance liquid chromatography coupled tandem mass spectroscopy (UPLC-MS/MS). The data shown in Figure 2B and Supplementary Table S2–S7 further confirmed that the extracellular protein categories were quite different between C. thermophilum and T. reesei. In C. thermophilum, 192 proteins were identified from the UPLC-MS/MS analysis, including 40 glycoside hydrolase proteins, 23 oxidoreductases, 16 protein degradation associated proteins (protease or peptidase) as well as 113 proteins with unknown function or other functions. While in T. reesei, 138 proteins were identified including 60 glycoside hydrolase proteins, 8 protease and peptidases, 2 oxidoreductases, and 68 proteins with unknown function or other functions. Unlike T. reesei, in which most of the secreted proteins were glycoside hydrolase proteins, C. thermophilum contained fewer glycoside hydrolase proteins and much more oxidoreductases, proteases, and peptidases. Considering that most lignocellulases belongs to glycoside hydrolase family proteins, C. thermophilum might contain fewer lignocellulases compared to T. reesei. Beside containing cellulases with higher specific activities, the different secreted protein categories in C. thermophilum might also play an important role in its high-efficiency of degrading celluloses.
To investigate if C. thermophilum contain superior cellulases relative to T. reesei, one of the major cellulases, CBH1, from C. thermophilum and T. reesei were first characterized and compared. To prevent the effects caused by other cellulases on the measurement of CBH1 activities, a cellulase-free background system (Uzbas et al., 2012) was applied to respectively express C. thermophilum CBH1 (Ct-CBH1) and T. reesei CBH1 (Tr-CBH1) using cDNA1 promoter in T. reesei strain Tu6Δku70 under glucose. For the construction of Tr-CBH1 expression strain, the promoter of cbh1 gene in Tu6Δku70 was replaced with cDNA 1 promoter via homologous recombination (Figure 3A), while for expressing Ct-CBH1, the DNA region including both cbh1 promoter and the ORF of cbh1 gene was replaced with cDNA 1 promoter and the cDNA of Ct-CBH1 (Figure 3B). The generated strains were named as Tr-cTcbh1 and Tr-cCcbh1, respectively. Since in the transformants of Tr-cTcbh1 and Tr-cCcbh1, the uridine auxotrophy of Tu6Δku70 strains were complemented by the pyr4 gene, meanwhile, a uridine complementary strain Cpyr4 was thus constructed by transforming the plasmid pSK-pyr4 (Qin et al., 2018) into Tu6Δku70 and used as a control for analysis of CBH1 expression.
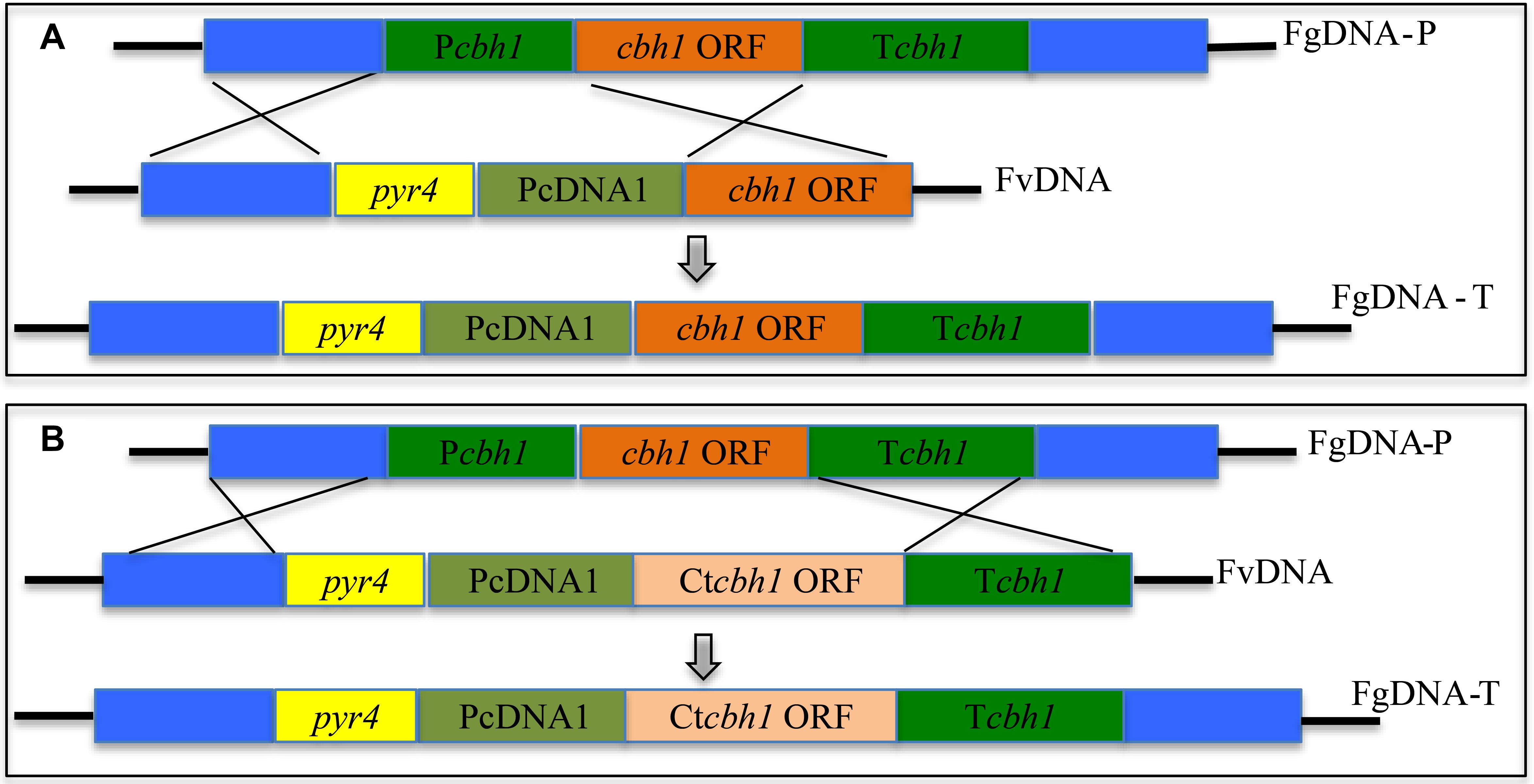
Figure 3. Expression of Tr-CBH1 and Ct-CBH1 in T. reesei using cellulase-free background system. (A) Schematic diagram of the construction of recombinant strain Tr-cTcbh1. (B) Schematic diagram of the construction of recombinant strain Tr-cCcbh1. FgDNA-P indicates fragments in the parental strain. FvDNA indicates Fragment from plasmid vector constructed for the generation of the indicated recombinant strain. FgDNA-T indicates DNA fragments in the generated transformant strain.
Three independent strains of each Tr-cTcbh1, Tr-cCcbh1, and Cpyr4 transformants were respectively cultivated in shake flasks with glucose as the only carbon source. The supernatants of 48 h post-inoculation culture were used for analyzing the secreted protein concentration, CBH activities, as well as SDS-PAGE analysis. As shown in Figure 4A, compared to Cpyr4 strains, Tr-cTcbh1 and Tr-cCcbh1 strains showed a significantly additional band about 65 kDa. The protein concentration in the supernatants of Tr-cTcbh1 and Tr-cCcbh1 cultures was also significantly higher (Figure 4B). Additional enzyme activity analysis (Figure 4B) further confirmed that the recombinant cbh1 gene from both C. thermophilum and T. reesei were successfully expressed in T. reesei under glucose, in which the expression of the native cellulases genes was largely repressed due to the regulation of carbon catabolite repression (CCR). It is worth noting that the expression levels of Ct-CBH1 in Tr-cCcbh1 strains were slightly higher than Tr-CBH1 in Tr-cTcbh1 strains (Figure 4A). However, the CBH activity in Tr-cCcbh1 strains was about 4.7-fold higher than that in Tr-cTcbh1 strains (Figure 4B). The data that CBH1 activities normalized by total proteins in the supernatant indicated that the relative specific activities of CBH1 from C. thermophilum were about 4.5-fold higher than CBH1 from T. reesei. Although whether other cellulases in C. thermophilum have higher specific activities still needed to be characterized, the existence of CBH1 with higher specific activity at least confirmed that C. thermophilum indeed contains superior cellulase.
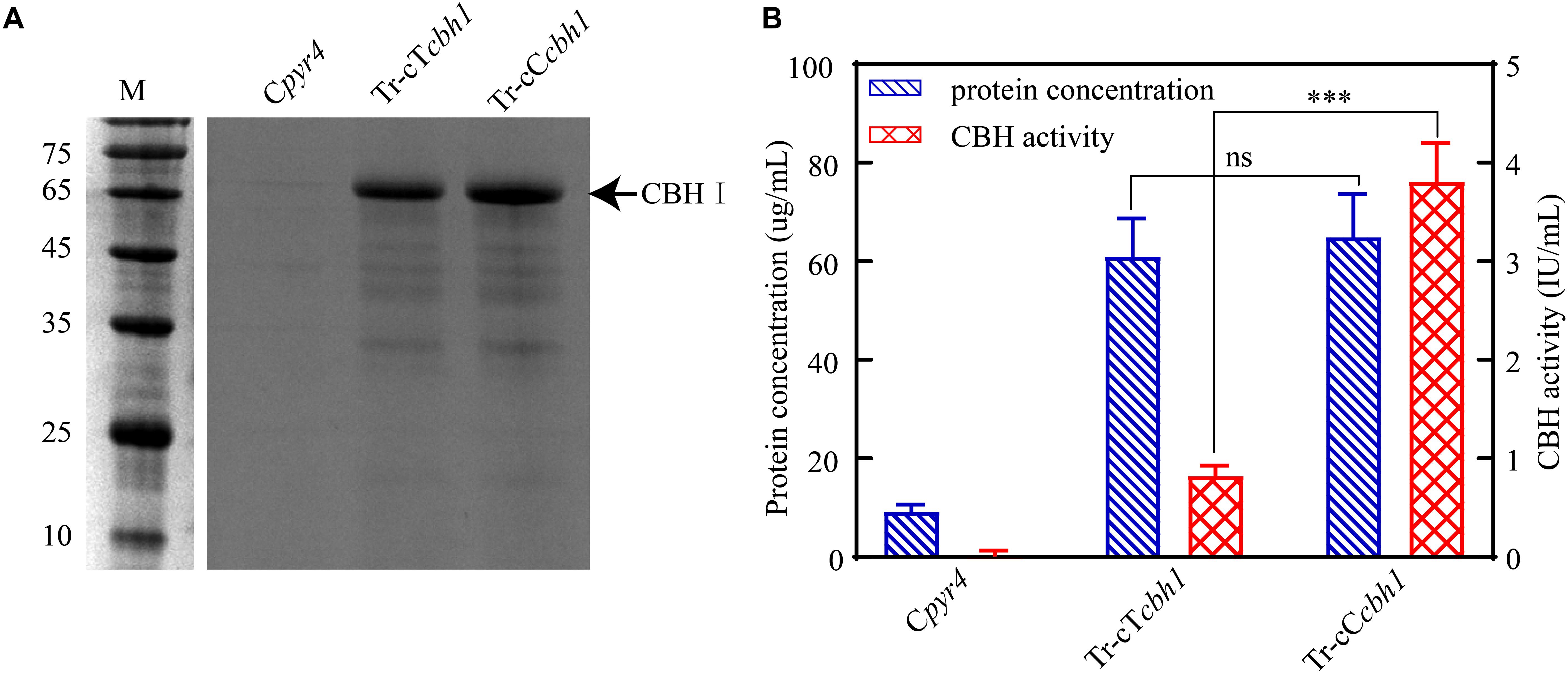
Figure 4. Ct-CBH1 showed higher specific activity compared to Tr-CBH1. (A) SDS-PAGE analysis of the secreted proteins in the supernatant of the recombinant T. reesei strains Cpyr4, Tr-cTcbh1, and Tr-cCcbh1. A suspension of 106 conidia/mL was inoculated into MM media with glucose as the only carbon source and cultivated for 48 h. A total of 20 μL of the supernatant was used for SDS-PAGE analysis. (B) Total secreted protein levels and CBH activities in the supernatants of the indicated T. reesei strains. The samples used in (B) were as same as in (A). Shown are the mean values of three biological replicates. Error bars show the standard deviations between these replicates. The significance of differences between Tr-cTcbh1 and Tr-cCcbh1 strains was based on t-test analysis by the FDR approach. Asterisks indicate significant differences (∗∗∗P < 0.001). ns, not significant.
Considering that C. thermophilum is a thermophilic fungus, whose optimum growth temperature is around 50°C (Kellner et al., 2016), the optimum temperature of Ct-CBH1 and its temperature stability were subsequently measured and compared with Tr-CBH1. To make sure that the assay for temperature activity curve was carried under the appropriate pH condition, the optimum pH and pH stability for both Ct-CBH1 and Tr-CBH1 were first determined at 50°C. As shown in Figures 5A,B, the optimal pH for both Ct-CBH1 and Tr-CBH1 was pH 4.0, and both Ct-CBH1 and Tr-CBH1 displayed the best residual activities at pH 4.0, suggesting pH 4.0 was the optimum condition for CBH1 activity measurement. The assays for optimum temperature were thus carried at pH 4.0, and at seven different temperatures (20–80°C), by using MUC as the substrate. The result shown in Figure 5C demonstrated that the optimum temperature of Ct-CBH1 was 70°C, while the optimum temperature of Tr-CBH1 was 60°C, and among all the temperatures, Ct-CBH1 displayed higher activities than Tr-CBH1. For the temperature stability analysis (Figure 5D), Tr-CBH1 was stable between 50 and 60°C, the remaining activity dropped dramatically below 50°C or above 60°C. However, Ct-CBH1 was more stable at most of the tested temperatures, which was consistent with reports at previous study (Voutilainen et al., 2008). Taken together, these data indicated that although the enzymatic characteristics of other cellulase genes in C. thermophilum still need to be identified, the above results are at least proved that better cellulases composition system in C. thermophilum was one of the reasons for its higher cellulose-utilizing efficiency relative to T. reesei.
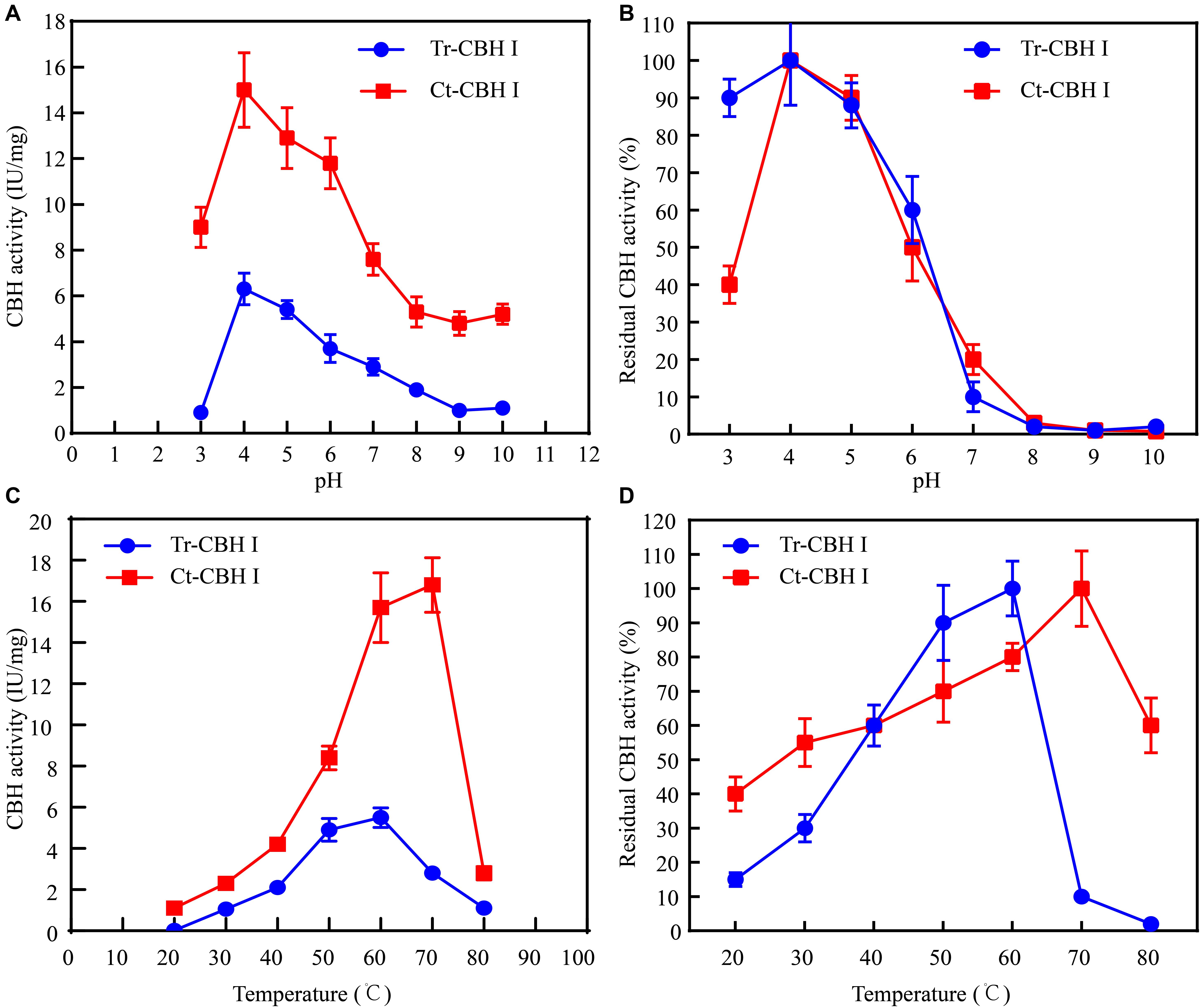
Figure 5. Ct-CBH1 showed higher optimum temperature and better temperature stability compared to Tr-CBH1. (A) Optimum pH determination of Tr-CBH1 and Ct-CBH1. (B) pH stability analysis of Tr-CBH1 and Ct-CBH1. (C) Optimum temperature determination of Tr-CBH1 and Ct-CBH1. (D) Temperature stability analysis of Tr-CBH1 and Ct-CBH1. Shown are the mean values of three biological replicates. Error bars show the standard deviations between these replicates.
Since the specific activity of Ct-CBH1 was significantly higher than Tr-CBH1, cellobiohydrolase activity in T. reesei would be increased if Ct-CBH1 could be expressed in T. reesei as a similar level as its native Tr-CBH1. To investigate whether increased cellobiohydrolase activities in T. reesei could improve its cellulose utilization, recombinant T. reesei strain in which the native cbh1 gene was replaced with cbh1 gene from C. thermophilum was created and named as Tr-Ctcbh1 (Figure 6A).
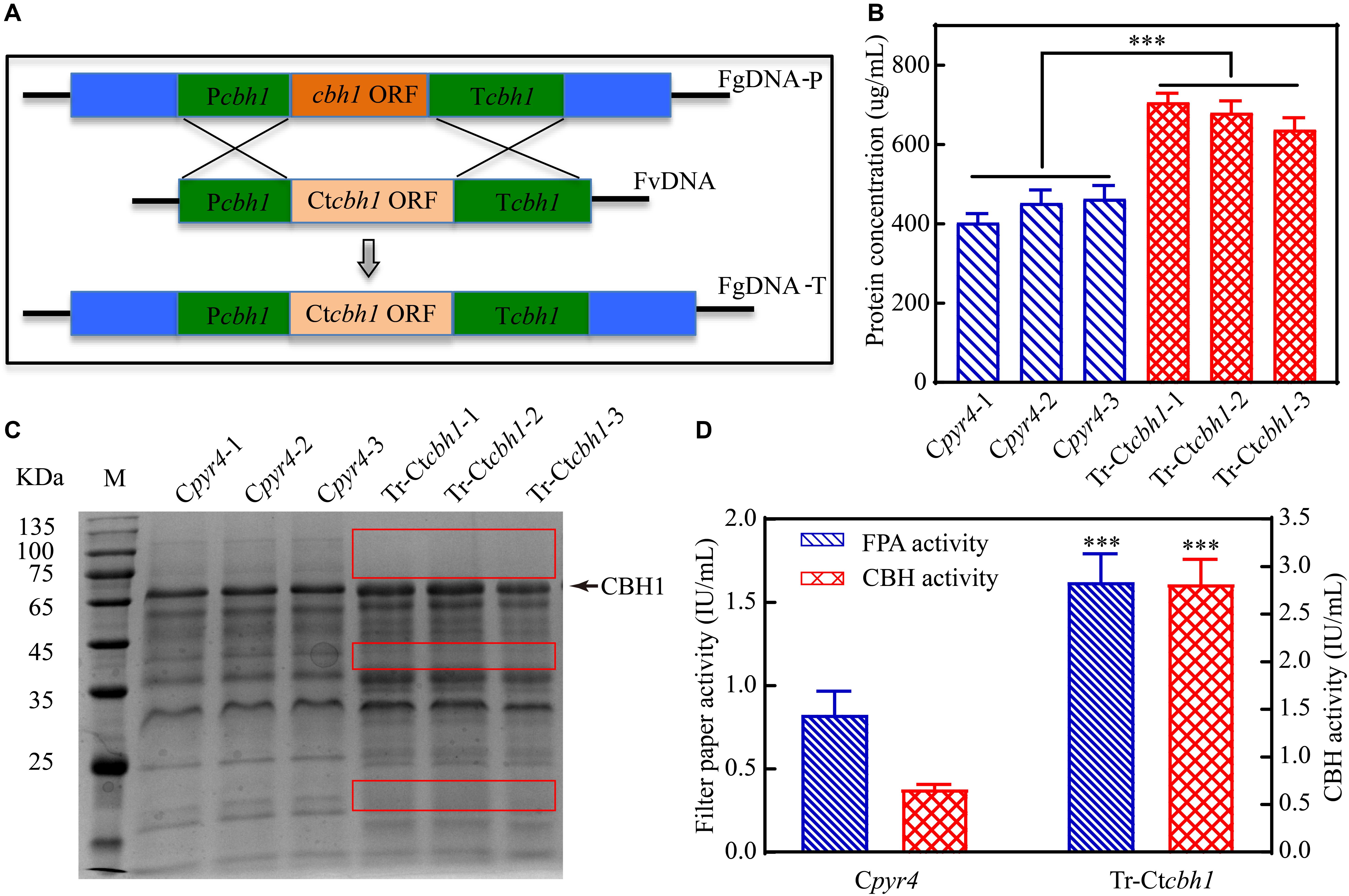
Figure 6. Replacement of Tr-cbh1 with Ct-cbh1 gene in T. reesei increased the production of cellulases. (A) Schematic diagram of the construction of recombinant T. reesei strain Tr-Ctcbh1. FgDNA-P indicates the fragment in the parental strain. FvDNA indicates the fragment from plasmid vector constructed for generation of the indicated recombinant strain. FgDNA-T indicates DNA fragment in the generated transformant strain. (B) Total secreted protein levels of three independent Cpyr4 and Tr-Ctcbh1 transformants in the supernatant of 120-h Avicel cultures after transferring from glucose cultures. Shown are the mean values of three technical replicates. Error bars show the standard deviations between these replicates. (C) SDS-PAGE analysis of the extracellular proteins of Cpyr4 and Tr-Ctcbh1 strains. The protein bands highlighted with the red frame line indicates proteins in Tr-Ctcbh1 strains were less than that in Cpyr4 strains. (D) Filter paper activities and CBH activities of Cpyr4 and Tr-Ctcbh1 strains. The samples used in (C,D) were as same as in (B). Error bars in (D) show the standard deviations between the biological replicates shown in (C). The significance of differences between Cpyr4 and Tr-Ctcbh1 strains was based on t-test analysis by the FDR approach. Asterisks indicate significant differences (∗∗∗P < 0.001). ns, not significant.
Three independent Tr-Ctcbh1 and Cpyr4 recombinant strains were selected for the measurement of the secreted protein levels, CBH activities, and the filter paper activities under Avicel. It was surprisingly demonstrated that the total secreted protein levels in Tr-Ctcbh1 strains were about 1.8-fold higher than those in Cpyr4 strains (Figure 6B) and the expression levels of CBH1 in Tr-Ctcbh1 strains were also significantly higher compared to that in Cpyr4 strains (Figure 6C). The cellobiohydrolase activities in Tr-Ctcbh1 strains were about 4.1-fold higher than those in Cpyr4 strains due to the higher specific activity of Ct-CBH1. In addition, Tr-Ctcbh1 strains also displayed around 2.2-fold of increased filter paper activities compared to Cpyr4 strains (Figure 6D).
Increased CBH1 activities in Tr-Ctcbh1 strains without any change of ß-glucosidase might cause the accumulation of cellobiose when degrading celluloses. Considering that filter paper activity assays were based on the measurement of the abundance of reducing sugars by DNS method (Xiao et al., 2004), thus the increased filter paper activities in Tr-Ctcbh1 strains might be caused by the accumulation of cellobiose. To prove this, the amount of glucose and cellobiose in the solution of filter paper activity assay (FPA) was determined by high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD). The data shown in Figure 7A unexpectedly revealed that there was no cellobiose accumulation in FPA solutions of both Tr-Ctcbh1 and Cpyr4 strains, indicating that ß-glucosidase in Tr-Ctcbh1 strains seemed to be enough to convert the extra cellobiose into glucose. The possible interpretation for this was that more expression of ß-glucosidases might be triggered by the increased CBH1 activity. To investigate whether increased CBH1 activity in T. reesei could trigger more expression of other cellulases, besides cellobiohydrolases activities, the activities of ß-glucosidases and endoglucanases of Tr-Ctcbh1 and Cpyr4 strains under Avicel were measured. As expected, consistent with the above FPA activities data (Figure 6D), the activities of endoglucanase and ß-glucosidase of Tr-Ctcbh1 strains in 120 h Avicel cultures were 1.9 and 2.3-fold greater than that of Cpyr4 strains (Figure 7B). Taken together, these data suggested that replacement of Tr-CBH1 with Ct-CBH1 in T. reesei not only resulted in increasing CBH1 activities, but also lead to more production of other cellulases. Thus we concluded that increased CBH1 activities in T. reesei could contribute to improving the cellulose degradation ability in T. reesei.
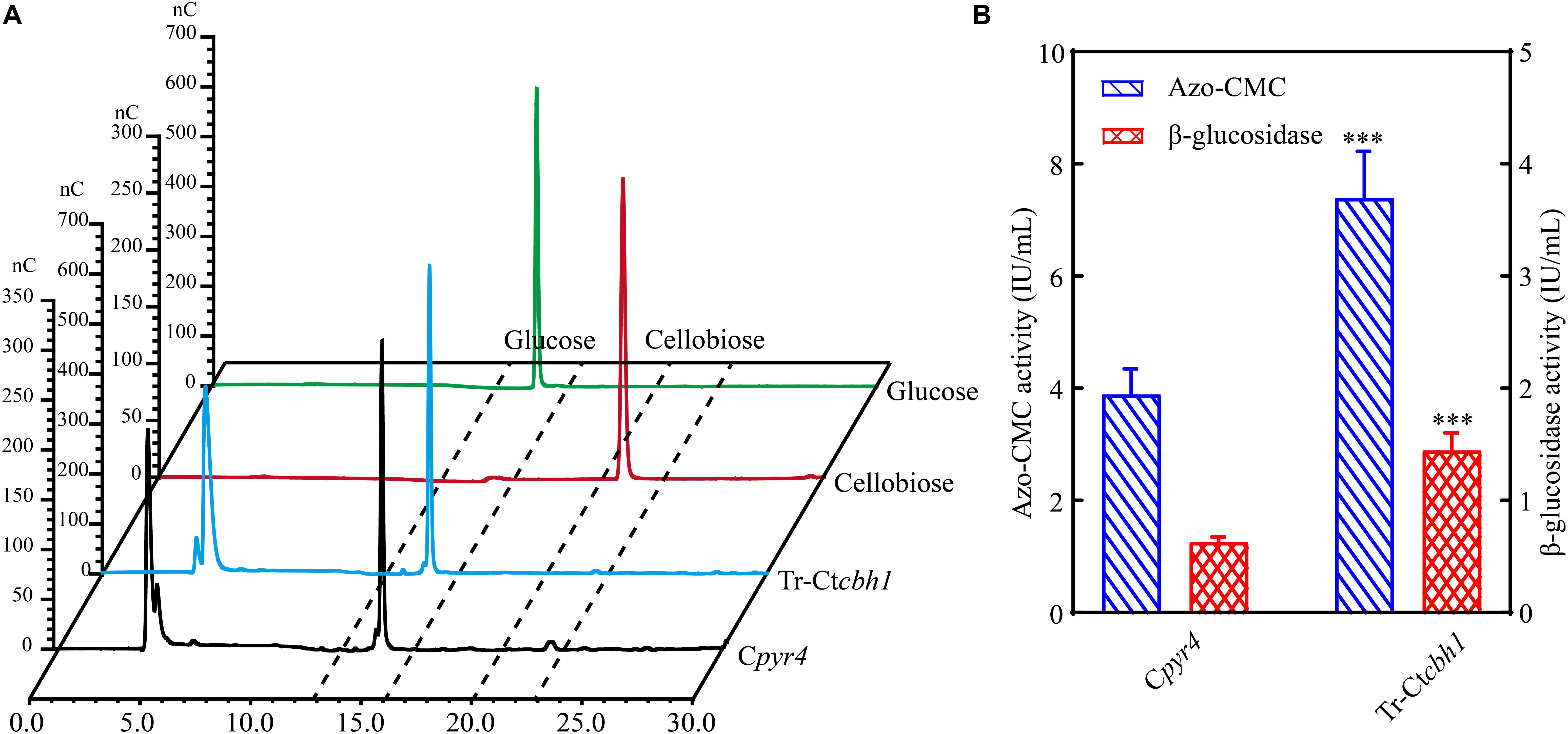
Figure 7. The activities of other cellulases were also increased in Tr-Ctcbh1 strains. (A) HPAEC-PAD analysis of glucose and cellobiose content in the solution of filter paper activity assay (FPA). nC, nano counts, indicating the intensity (abundance) of the signal for each peak. Glucose and cellobiose indicate 100 μM glucose and cellobiose standard sample, respectively. Tr-Ctcbh1 and Cpyr4 indicate sample taken from the solution of FPA assay with the supernatants of Cpyr4 and Tr-Ctcbh1 strains under Avicel cultures, respectively. (B) Measurements of the endoglucanase activities and β-glucosidase activities of Cpyr4 and Tr-Ctcbh1 strains in the supernatant of 120-h MM+2% Avicel cultures after transferring from 48 h of MM+2% glucose cultures. The significance of differences between Cpyr4 and Tr-Ctcbh1 strains was based on t-test analysis by the FDR approach. Asterisks indicate significant differences (∗∗∗P < 0.001). ns, not significant.
To further explore the mechanism behind the higher cellulase production in Tr-Ctcbh1 strains, the transcriptional levels of several major cellulases genes including eg1 (transcript ID: 122081), eg2 (transcript ID: 120312), cbh2 (transcript ID: 72567), and bgl1 (transcript ID: 76672) under Avicel at different time points were measured by qRT-PCR. As shown in Figure 8, at the time point of 12 h, the expression levels of eg1 and eg2 in Tr-Ctcbh1 strains were slightly lower than that in Cpyr4 strains, and the expression levels of the other two tested genes in Tr-Ctcbh1 strains were similar as in Cpyr4 strains. However, after induction in cellulose condition for 48 h, all these tested genes in Tr-Ctcbh1 strains were increased at different extent. The mRNA levels of eg1 and eg2 genes in Tr-Ctcbh1 strains were increased by 1.7 (Figure 8A) and 1.5-fold (Figure 8B) compared to that in Cpyr4 strains, while the mRNA levels of cbh2 and bgl1 genes in Tr-Ctcbh1 strains were 1.8 (Figure 8C) and 2.1-fold (Figure 8D) higher than that in Cpyr4 strains. These data suggested that the more expression of cellulases genes in Tr-Ctcbh1 strains occurred at transcriptional levels.
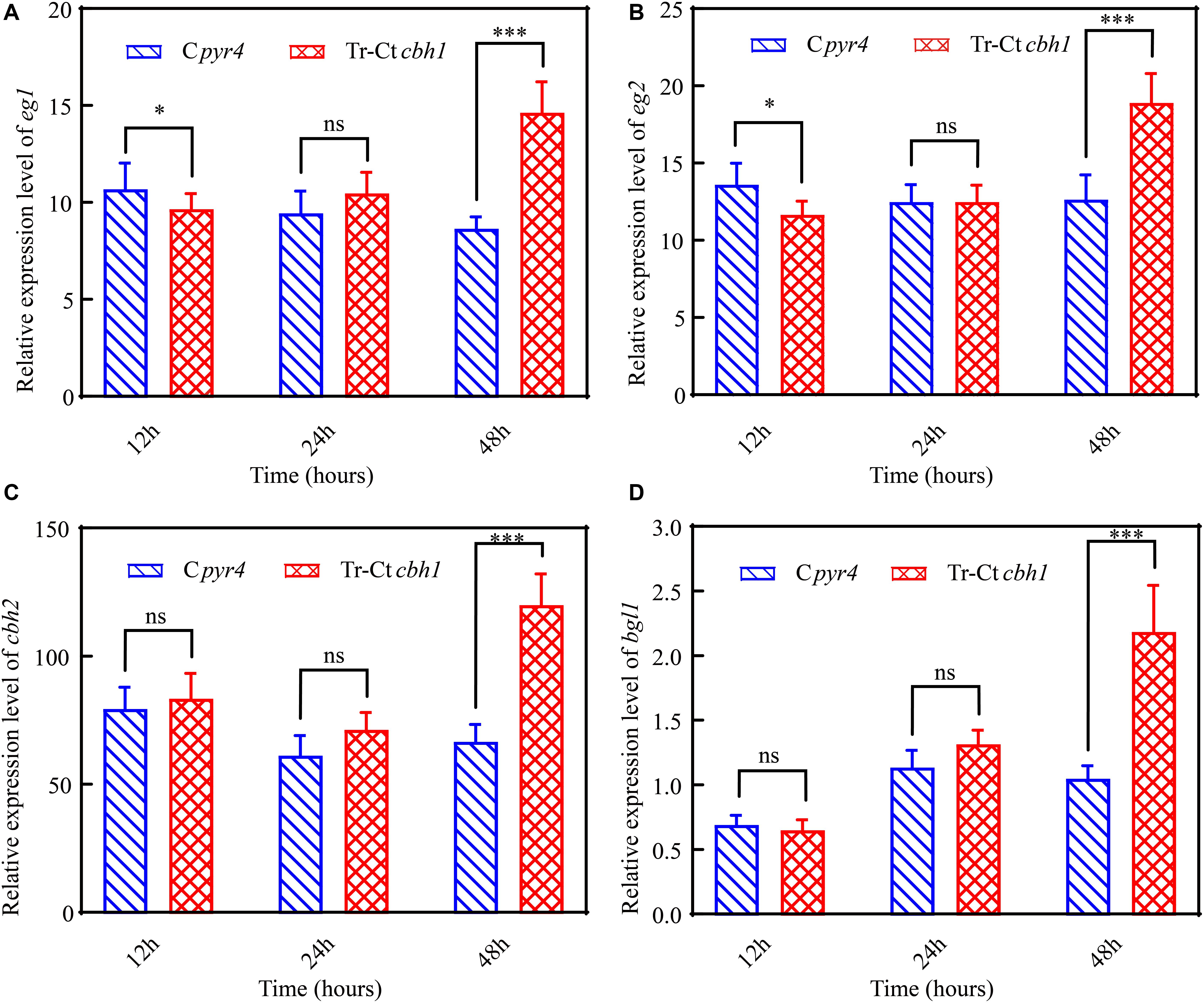
Figure 8. Increased CBH1 activities in T. reesei enhanced the induction and expression of the major cellulases genes. Relative quantification of mRNA levels of eg1 (A), eg2 (B), cbh2 (C), and bgl1 (D) genes of Cpyr4 and Tr-Ctcbh1 strains by qRT-PCR. RNA samples were obtained from the culture of MM+2% Avicel at the indicated time point after a shift of 48 h of MM+2% glucose cultures. The significance of differences between Cpyr4 and Tr-Ctcbh1 strains was based on t-test analysis by the FDR approach. Asterisks indicate significant differences (∗P < 0.05; ∗∗∗P < 0.001). ns, not significant.
In T. reesei, several major TFs including three activators xyr1, ace2 and ace3, and two repressors cre1 and ace1 were involved in the regulation of the expression of genes encoding lignocellulases genes (Benocci et al., 2017). To investigate if these TFs were involved in the regulation of the increased production of cellulases in Tr-Ctcbh1 strains, the expression levels of these TFs in Tr-Ctcbh1 and Cpyr4 strains under Avicel were also measured by qRT-PCR. The data shown in Figure 9 demonstrated that the relative mRNA levels of xyr1, cre1, and ace3 in Tr-Ctcbh1 strains were significantly increased over time compared to that in Cpyr4 strains. At the time point of 48 h, the expression levels of xyr1 and cre1 in Tr-Ctcbh1 strains were about 17.4 and 4.5-fold higher than that in Cpyr4 strains (Figures 9A,B), while the expression levels of ace3 were only slightly increased by 1.5-fold (Figure 9E). However, the expression levels of ace1 and ace2 in Tr-Ctcbh1 strains exhibited similar levels as that in Cpyr4 strains at all of the time points (Figures 9C,D). Considering that the induction of xyr1 requires cre1 (Portnoy et al., 2011), it was reasonable that the expression of cre1 increased along with xyr1 here. However, the reason for that the expression level of activator ace2 was not increased in Tr-Ctcbh1 strains still needs to be investigated. The data here at least indicated that the major regulator xyr1 was involved in higher cellulase production in Tr-Ctcbh1 strains.
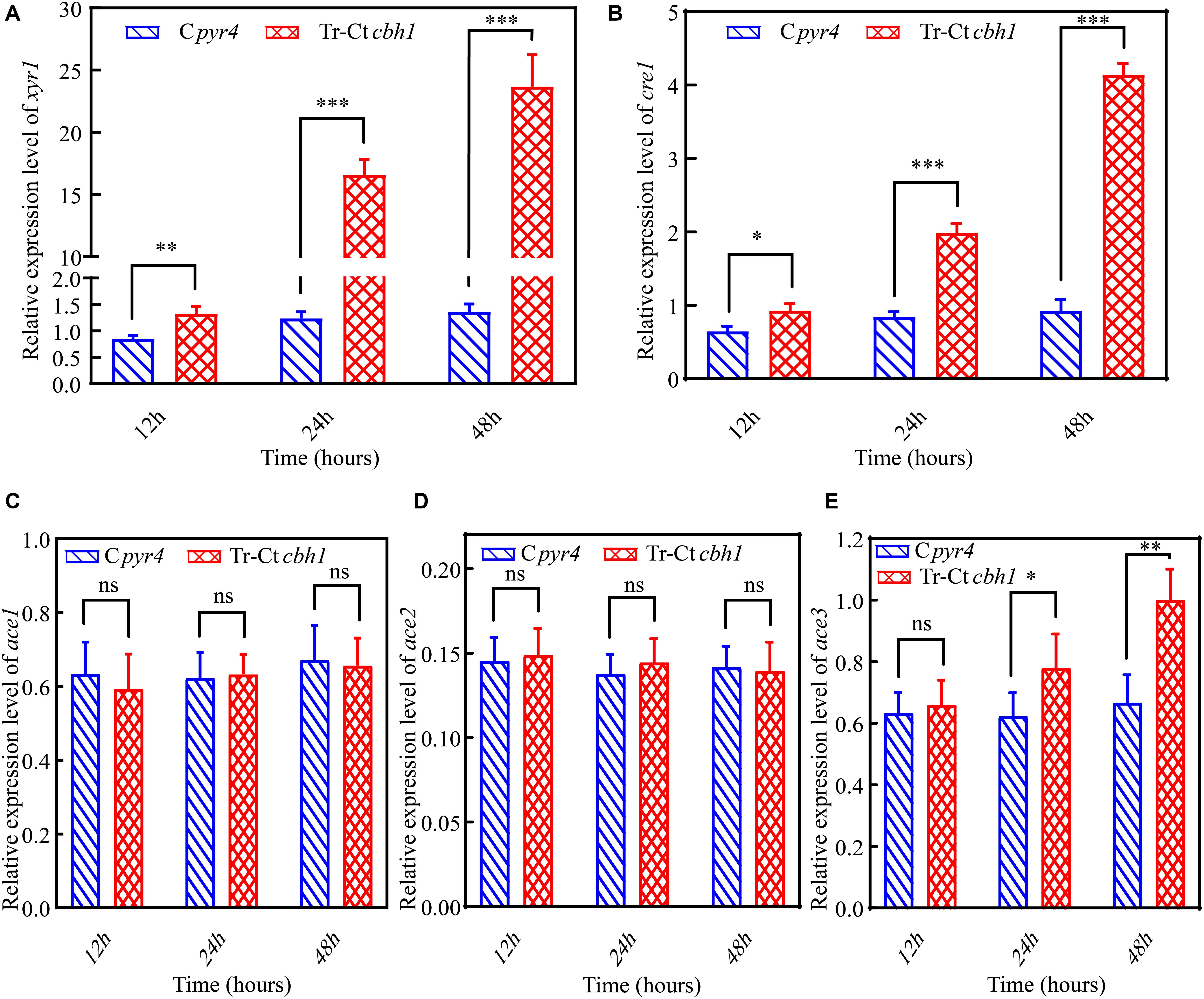
Figure 9. The higher production of cellulases in Tr-Ctcbh1 strains was caused by increased expression level of xyr1. Relative quantification of mRNA levels of xyr1 (A), cre1 (B), ace1 (C), ace2 (D), and ace3 (E) genes of Cpyr4 and Tr-Ctcbh1 strains by qRT-PCR. RNA samples were obtained from 12, 24, and 48 h of cultures of MM+2% Avicel after a shift of 48 h of MM+2% glucose cultures. The significance of differences between Cpyr4 and Tr-Ctcbh1 strains was based on t-test analysis by the FDR approach. Asterisks indicate significant differences (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). ns, not significant.
Biorefineries include the sustainable processing of plant biomass into a variety of bioproducts and specialty chemicals. Many of these processes require enzymes that are stable at high temperature to enhance mass transfer and reduce substrate viscosity, with a lower risk of contamination. As natural plant-biomass degraders, thermophilic fungi must contain thermostable biomass-degrading enzymes. Therefore, in this study, we evaluated the cellulose-utilizing efficiency of a thermophilic fungus C. thermophilum and surprisingly found that compared to T. reesei, although C. thermophilum secreted fewer cellulases, it showed higher efficiency to degrade celluloses. This finding indicated that C. thermophilum might contain cellulases with higher specific activities. The characterization and comparison of Ct-CBH1 and Tr-CBH1 further confirmed this hypothesis. However, UPLC-MS/MS analysis of C. thermophilum supernatants demonstrated there were many oxidoreductases included in C. thermophilum protein secretome. Considering that the certain oxidants could make lignocellulose more susceptible for enzymatic degradation (Zhang et al., 2016; Den et al., 2018), the existence of such amounts of oxidoreductases in the supernatants of C. thermophilum culture implied that in addition to contain higher efficient cellulases, C. thermophilum might display a different mode of action to degrade lignocelluloses.
In T. reesei, it has been generally believed that β-glucosidase is a barrier for its cellulose degradation capacity because the extracellular β-glucosidase comprises only about 1% of the total T. reesei cellulases complex (Karkehabadi et al., 2014), based on which, many efforts have been conducted to increase enzyme efficiency of hydrolyzing cellulosic substrates by increasing β-glucosidase amount in the cellulases complex from T. reesei (Banerjee et al., 2010; Treebupachatsakul et al., 2016). In the opposite, because of the large amount, studies about increasing enzyme efficiency of cellobiohydrolase in T. reesei have been rarely reported. In this study, we first reported that increased cellobiohydrolase activities in T. reesei could result in a significant increase of its production of other cellulases and cellulose-utilizing efficiency.
It was worth noting that SDS-PAGE analysis of the supernatants of the Tr-Ctcbh1 and Cpyr4 strains (Figure 6C) showed variations in specific protein levels rather than an overall increase or decrease in the levels of all secreted proteins in Tr-Ctcbh1 strains compared to that in Cpyr4 strains. Relative to Cpyr4 strains, the expression levels of only a few proteins (highlighted with red frame line in Figure 6C) were decreased, most of the secreted proteins were increased in Tr-Ctcbh1 strains. Since most of the extracellular proteins in the supernatant of T. reesei under cellulose were cellulases and hemi-cellulases (Herpoël-Gimbert et al., 2008), these data suggested that only improved the function of CBH1 in T. reesei might lead to increasing the expression levels of most lignocellulose degradation associated genes. The results that the FPA activities (Figure 6D), the Azo-CMC activities and β-glucosidase activities (Figure 7B) in Tr-Ctcbh1 strains were significantly higher than those in Cpyr4 strains, and besides, the transcriptional levels of cbh2, eg1, eg2, and bgl1 genes were also higher in Tr-Ctcbh1 strains (Figure 8), which further proved that the induction and expression of most cellulases genes in T. reesei could be improved by only enhancing CBH1 activities.
The increased expression levels of xyr1, cre1, and ace3 in Tr-Ctcbh1 strains (Figure 9) suggested that an extra inducer might be generated in T. reesei when response to cellulose with more cellobiohydrolases in its cellulases system. Cellobiohydrolase is an exocellulase, which cleaves cellulose to release cellobiose from the non-reducing ends. Theoretically, increased cellobiohydrolase in T. reesei should result in an accumulation of cellobiose when response to cellulose if the expression level of ß-glucosidase was not correspondingly increased. The higher expression levels of most cellulase genes in Tr-Ctcbh1 strains could explain why there was no cellobiose accumulation. A possible interpretation for the increased production levels of cellulases in Tr-Ctcbh1 strains was that the extra cellobiose generated by the increased cellobiohydrolase activities at the very beginning might function as a possible inducer to trigger more expression of cellulases genes (Figure 10). It has been previously reported that cellobiose could induce a low level of cellulase gene expression in T. reesei (Vaheri et al., 1979). In addition, sophorose, which is widely viewed as the potent inducer of cellulases in T. reesei, also could be generated by the transglycosylation of cellobiose by ß-glucosidase (Sternberg and Mandels, 1979). Furthermore, the growth of Neurospora crassa under cellobiose could not induce the expression of celluases genes, however, the deletion of three major β-glucosidase genes in N. crassa could result in the expression of cellulases under cellobiose (Znameroski et al., 2012). Moreover, deletion of two intracellular β-glucosidase genes (cel1a and cel1b) and one extracellular β-glucosidase (cel3a) gene in T. reesei also could increase the expression levels of cellulases genes when response to cellobiose (Zhou et al., 2012). All of these previous reports suggested that cellobiose played an important role in the induction of cellulases genes, especially when the relative ratio of the cellobiohydrolase and ß-glucosidase was enlarged, which was consistent with the phenomenon observed in this study. Therefore, we speculated that the potential temporary accumulation of cellobiose resulted from the disruption of the synergetic effect of cellobiohydrolase and ß-glucosidase might be able to function as an inducer to trigger the extra induction of cellulases genes in T. reesei (Figure10).
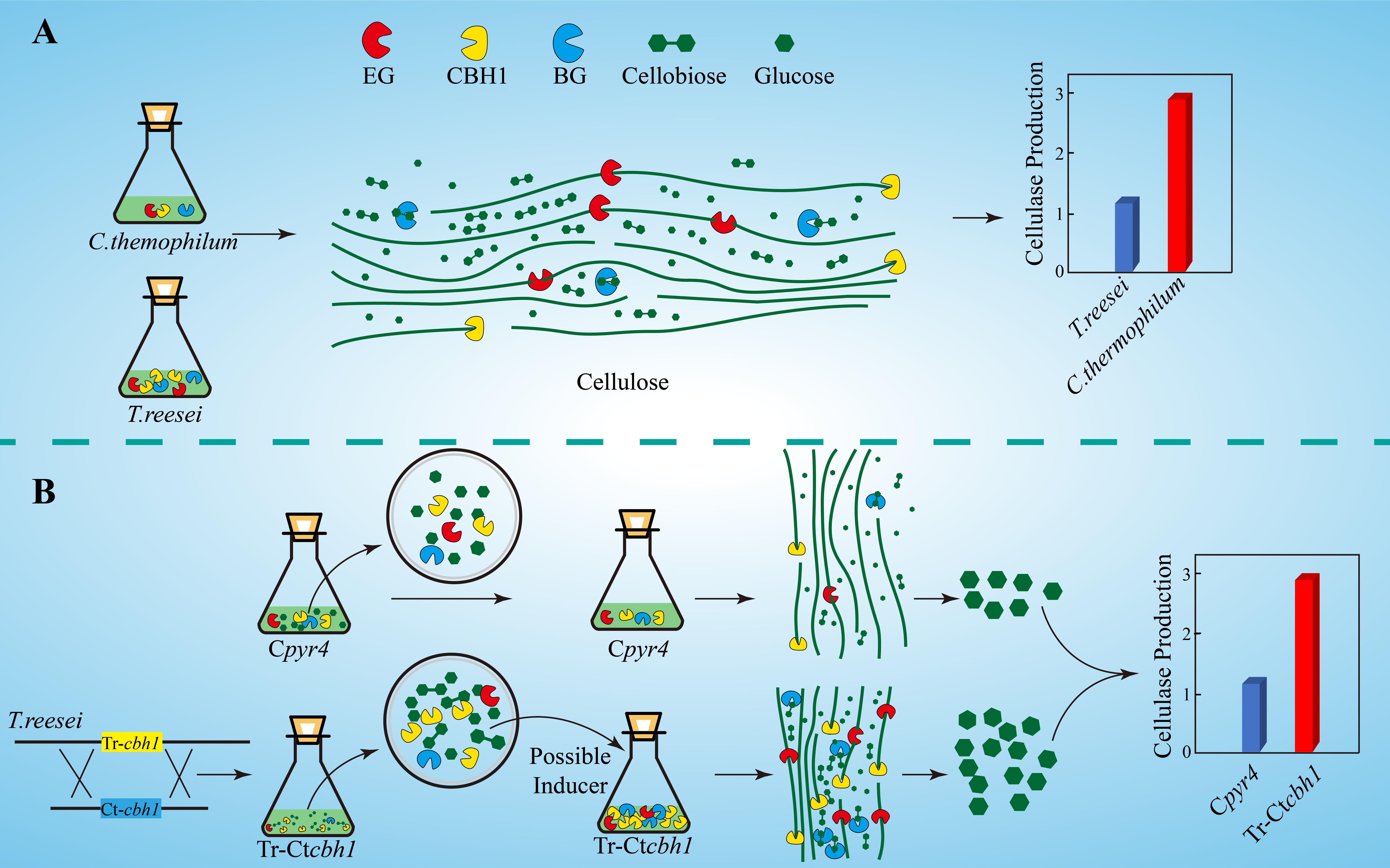
Figure 10. Graphic summary of this study. (A) Compared to T. reesei, C. thermophilum degrades celluloses more efficiently with lower production of cellulases. (B) The proposed model of the mechanism for higher production of cellulases in Tr-Ctcbh1 strains. Replacement of Tr-cbh1 with Ct-cbh1 gene in T. reesei first increased the CBH1 activity, which could cause instantaneous accumulation of cellobiose. This extra cellobiose might function as a possible inducer to trigger more induction and expression of cellulases genes.
To search for new resources of lignocellulases for plant biomass degradation, here we reported that a thermophilic fungus C. thermophilum could degrade celluloses more efficiently with less secreted cellulases compared to T. reesei, implying the existence of excellent cellulases genes in its genome. Comparison of the enzyme properties of CBH1 from C. thermophilum and T. reesei further confirmed this hypothesis. More interesting, our data showed that only raising the function of CBH1 in T. reesei could lead to a marked increase in the production levels of other cellulases. Based on the observed phenomenon, we speculated that a certain amount of temporary cellobiose accumulation might play an important role in the process of cellulases genes induction. This study not only provided a new strategy to improve the cellulases yields in T. reesei, but also opened the path for fundamental research on thermophilic fungi such as C. thermophilum, as well as stimulating investigations into their potential application as sources of commercially important enzymes.
All datasets generated for this study are included in the article/Supplementary Material.
XJ and JD participated in the conception of the study and carried out the majority of the experiments. RH and ZZ were involved in evaluating the cellulose-utilizing efficiency of C. thermophilum and T. reesei. FQ was involved in mass spectrometry analysis. JH was involved in the project leadership and participated in editing the manuscript. LQ was involved in the conception of the study and participated in the guidance with experimental strategies and technical direction. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31800060 and 31741002); Natural Science Foundation of Fujian Province in China (2019I0009); and by the Scientific Research Innovation Program “Xiyuanjiang River Scholarship” of College of Life Sciences, Fujian Normal University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Zhiyang Dong and Dr. Xiuzhen Chen (Institute of Microbiology, Chinese Academy of Sciences) for their assistance with the UPLC-MS/MS experiment. We also thank Xiaoyun Su (Feed Research Institute, Chinese Academy of Agricultural Sciences) for his assistance with HPAEC-PAD experiment.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01633/full#supplementary-material
TABLE S1 | Primers used in this study.
TABLE S2 | UPLC-MS/MS analysis of protein secretome of T. reesei under Avicel (Processed data).
TABLE S3 | UPLC-MS/MS analysis of protein secretome of C. thermophilum under Avicel (Processed data).
TABLE S4 | UPLC-MS/MS analysis of protein secretome of T. reesei under Avicel (Raw data of sample 1).
TABLE S5 | UPLC-MS/MS analysis of protein secretome of T. reesei under Avicel (Raw data of sample 2).
TABLE S6 | UPLC-MS/MS analysis of protein secretome of C. thermophilum under Avicel (Raw data of sample 1).
TABLE S7 | UPLC-MS/MS analysis of protein secretome of C. thermophilum under Avicel (Raw data of sample 2).
ATCC, American type culture collection; CBH1, cellobiohydrolase I gene; CGMCC, China general microbiological culture collection center; FDR, false discovery rate; FPA, filter paper assay; HPAEC-PAD, ultra-high performance liquid chromatography coupled tandem mass spectroscopy; MM, minimal medium; pNPG, 4-nitrophenyl -D-glucopyranoside; SD, standard deviation; TFs, transcriptional factors; UPLC-MS/MS, ultra-high performance liquid chromatography coupled tandem mass spectroscopy; VMM, Vogel’s minimal medium; YPD, media containing 1% yeast extract 2% peptone 1% dextrose.
Bailey, M. J., and Tähtiharju, J. (2003). Efficient cellulase production by Trichoderma reesei in continuous cultivation on lactose medium with a computer-controlled feeding strategy. Appl. Microbiol. Biotechnol. 62, 156–162. doi: 10.1007/s00253-003-1276-9
Banerjee, G., Car, S., Scott-Craig, J. S., Borrusch, M. S., Aslam, N., and Walton, J. D. (2010). Synthetic enzyme mixtures for biomass deconstruction: production and optimization of a core set. Biotechnol. Bioeng. 106, 707–720. doi: 10.1002/bit.22741
Benocci, T., Aguilar-Pontes, M. V., Zhou, M., Seiboth, B., and De Vries, R. P. (2017). Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol. Biofuels 10:152. doi: 10.1186/s13068-017-0841-x
Bischof, R. H., Ramoni, J., and Seiboth, B. (2016). Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact. 15:106. doi: 10.1186/s12934-016-0507-6
Bock, T., Chen, W. H., Ori, A., Malik, N., Silva-Martin, N., Huerta-Cepas, J., et al. (2014). An integrated approach for genome annotation of the eukaryotic thermophile Chaetomium thermophilum. Nucleic Acids Res. 42, 13525–13533. doi: 10.1093/nar/gku1147
Cherry, J. R., and Fidantsef, A. L. (2003). Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 14, 438–443. doi: 10.1016/s0958-1669(03)00099-5
Dashtban, M., and Qin, W. (2012). Overexpression of an exotic thermotolerant beta-glucosidase in Trichoderma reesei and its significant increase in cellulolytic activity and saccharification of barley straw. Microb. Cell Fact. 11:63. doi: 10.1186/1475-2859-11-63
DeCastro, M. E., Rodriguez-Belmonte, E., and Gonzalez-Siso, M. I. (2016). Metagenomics of thermophiles with a focus on discovery of novel thermozymes. Front. Microbiol. 7:1521. doi: 10.3389/fmicb.2016.01521
Den, W., Sharma, V. K., Lee, M., Nadadur, G., and Varma, R. S. (2018). Lignocellulosic biomass transformations via greener oxidative pretreatment processes: access to energy and value-added chemicals. Front. Chem. 6:141. doi: 10.3389/fchem.2018.00141
Ghose, T. K. (1987). Measurement of cellulase activities. Pure Appl. Chem. 59, 257–268. doi: 10.1351/pac198759020257
Herpoël-Gimbert, I., Margeot, A., Dolla, A., Jan, G., Mollé, D., Lignon, S., et al. (2008). Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol. Biofuels 1:18. doi: 10.1186/1754-6834-1-18
Himmel, M. E., Ding, S.-Y., Johnson, D. K., Adney, W. S., Nimlos, M. R., Brady, J. W., et al. (2007). Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807. doi: 10.1126/science.1137016
Karkehabadi, S., Helmich, K. E., Kaper, T., Hansson, H., Mikkelsen, N. E., Gudmundsson, M., et al. (2014). Biochemical characterization and crystal structures of a fungal family 3 beta-glucosidase, Cel3A from Hypocrea jecorina. J. Biol. Chem. 289, 31624–31637. doi: 10.1074/jbc.M114.587766
Kellner, N., Schwarz, J., Sturm, M., Fernandez-Martinez, J., Griesel, S., Zhang, W., et al. (2016). Developing genetic tools to exploit Chaetomium thermophilum for biochemical analyses of eukaryotic macromolecular assemblies. Sci. Rep. 6:20937. doi: 10.1038/srep20937
Kubicek, C. P., Starr, T. L., and Glass, N. L. (2014). Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 52, 427–451. doi: 10.1146/annurev-phyto-102313-045831
Li, A. N., Yu, K., Liu, H. Q., Zhang, J., Li, H., and Li, D. C. (2010). Two novel thermostable chitinase genes from thermophilic fungi: cloning, expression and characterization. Bioresour. Technol. 101, 5546–5551. doi: 10.1016/j.biortech.2010.02.058
Penttilä, M., Nevalainen, H., Rättö, M., Salminen, E., and Knowles, J. (1987). A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61, 155–164. doi: 10.1016/0378-1119(87)90110-7
Portnoy, T., Margeot, A., Seidl-Seiboth, V., Le Crom, S., Chaabane, F. B., Linke, R., et al. (2011). Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase. Eukaryot. Cell 10, 262–271. doi: 10.1128/EC.00208-10
Qin, L., Jiang, X., Dong, Z., Huang, J., and Chen, X. (2018). Identification of two integration sites in favor of transgene expression in Trichoderma reesei. Biotechnol. Biofuels 11:142. doi: 10.1186/s13068-018-1139-3
Qin, L. N., Cai, F. R., Dong, X. R., Huang, Z. B., Tao, Y., Huang, J. Z., et al. (2012). Improved production of heterologous lipase in Trichoderma reesei by RNAi mediated gene silencing of an endogenic highly expressed gene. Bioresour. Technol. 109, 116–122. doi: 10.1016/j.biortech.2012.01.013
Schuster, A., Bruno, K. S., Collett, J. R., Baker, S. E., Seiboth, B., Kubicek, C. P., et al. (2012). A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol. Biofuels 5:1. doi: 10.1186/1754-6834-5-1
Sewalt, V., Shanahan, D., Gregg, L., La Marta, J., and Carrillo, R. (2016). The generally recognized as safe (GRAS) process for industrial microbial enzymes. Ind. Biotechnol. 12, 295–302. doi: 10.1089/ind.2016.0011
Singh, A., Taylor, L. E. II, Vander Wall, T. A., Linger, J., Himmel, M. E., Podkaminer, K., et al. (2015). Heterologous protein expression in Hypocrea jecorina: a historical perspective and new developments. Biotechnol. Adv. 33, 142–154. doi: 10.1016/j.biotechadv.2014.11.009
Stappler, E., Walton, J. D., Beier, S., and Schmoll, M. (2017). Abundance of secreted proteins of Trichoderma reesei is regulated by light of different intensities. Front. Microbiol. 8:2586. doi: 10.3389/fmicb.2017.02586
Sternberg, D., and Mandels, G. R. (1979). Induction of cellulolytic enzymes in trichoderma reesei by sophorose. J. Bacteriol. 139, 761–769. doi: 10.1128/jb.139.3.761-769.1979
Tian, C., Beeson, W. T., Iavarone, A. T., Sun, J., Marletta, M. A., Cate, J. H., et al. (2009). Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 106, 22157–22162. doi: 10.1073/pnas.0906810106
Treebupachatsakul, T., Nakazawa, H., Shinbo, H., Fujikawa, H., Nagaiwa, A., Ochiai, N., et al. (2016). Heterologously expressed Aspergillus aculeatus beta-glucosidase in Saccharomyces cerevisiae is a cost-effective alternative to commercial supplementation of beta-glucosidase in industrial ethanol production using Trichoderma reesei cellulases. J. Biosci. Bioeng. 121, 27–35. doi: 10.1016/j.jbiosc.2015.05.002
Treebupachatsakul, T., Shioya, K., Nakazawa, H., Kawaguchi, T., Morikawa, Y., Shida, Y., et al. (2015). Utilization of recombinant Trichoderma reesei expressing Aspergillus aculeatus beta-glucosidase I (JN11) for a more economical production of ethanol from lignocellulosic biomass. J. Biosci. Bioeng. 120, 657–665. doi: 10.1016/j.jbiosc.2015.04.015
Uzbas, F., Sezerman, U., Hartl, L., Kubicek, C. P., and Seiboth, B. (2012). A homologous production system for Trichoderma reesei secreted proteins in a cellulase-free background. Appl. Microbiol. Biotechnol. 93, 1601–1608. doi: 10.1007/s00253-011-3674-8
Vaheri, M. P., Vaheri, M. E., and Kauppinen, V. S. (1979). Formation and release of cellulolytic enzymes during growth of Trichoderma reesei on cellobiose and glycerol. Eur. J. Appl. Microbiol. Biotechnol. 8, 73–80. doi: 10.1007/bf00510268
Vogel, H. J. (1956). A convenient growth medium for Neurospora (Medium N). Microb. Genet Bull. 13, 42–43.
Voutilainen, S. P., Puranen, T., Siika-Aho, M., Lappalainen, A., Alapuranen, M., Kallio, J., et al. (2008). Cloning, expression, and characterization of novel thermostable family 7 cellobiohydrolases. Biotechnol. Bioeng. 101, 515–528. doi: 10.1002/bit.21940
Wu, V. W., Dana, C. M., Iavarone, A. T., Clark, D. S., and Glass, N. L. (2017). Identification of glutaminyl cyclase genes involved in pyroglutamate modification of fungal lignocellulolytic enzymes. mBio 8:e2231-16. doi: 10.1128/mBio.02231-16
Xiao, Z., Storms, R., and Tsang, A. (2004). Microplate-based filter paper assay to measure total cellulase activity. Biotechnol. Bioeng. 88, 832–837. doi: 10.1002/bit.20286
Zhang, J., Presley, G. N., Hammel, K. E., Ryu, J.-S., Menke, J. R., Figueroa, M., et al. (2016). Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proc. Natl. Acad. Sci. U.S.A. 113, 10968–10973. doi: 10.1073/pnas.1608454113
Zhang, J., Zhong, Y., Zhao, X., and Wang, T. (2010). Development of the cellulolytic fungus Trichoderma reesei strain with enhanced beta-glucosidase and filter paper activity using strong artificial cellobiohydrolase 1 promoter. Bioresour. Technol. 101, 9815–9818. doi: 10.1016/j.biortech.2010.07.078
Zhou, Q., Xu, J., Kou, Y., Lv, X., Zhang, X., Zhao, G., et al. (2012). Differential involvement of β-glucosidases from Hypocrea jecorina in rapid induction of cellulase genes by cellulose and cellobiose. Eukaryot. Cell 11, 1371–1381. doi: 10.1128/ec.00170-12
Keywords: Trichoderma reesei, thermophilic fungi, Chaetomium thermophilum, cellobiohydrolase I, lignocellulase, biomass, cellulase
Citation: Jiang X, Du J, He R, Zhang Z, Qi F, Huang J and Qin L (2020) Improved Production of Majority Cellulases in Trichoderma reesei by Integration of cbh1 Gene From Chaetomium thermophilum. Front. Microbiol. 11:1633. doi: 10.3389/fmicb.2020.01633
Received: 14 March 2020; Accepted: 23 June 2020;
Published: 14 July 2020.
Edited by:
Jiwei Zhang, University of Minnesota, Twin Cities, United StatesReviewed by:
Xu Fang, Shandong University, ChinaCopyright © 2020 Jiang, Du, He, Zhang, Qi, Huang and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianzhong Huang, aGp6QGZqbnUuZWR1LmNu; Lina Qin, cWlubG5AZmpudS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.