- 1College of Agriculture and Biological Sciences, Dali University, Dali, China
- 2Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 3Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, Chiang Mai, Thailand
- 4Manaaki Whenua Landcare Research, Auckland, New Zealand
- 5Retired, Goa Velha, India
- 6Institute of Plant Health, Zhongkai University of Agriculture and Engineering, Guangzhou, China
During a study of diversity and taxonomy of lignicolous freshwater fungi in China, nine species of Acrogenospora were collected. Seven of these were new species and they are described and illustrated. With morphology, additional evidence to support establishment of new species is provided by phylogeny derived from DNA sequence analyses of a combined LSU, SSU, TEF1α, and RPB2 sequence dataset. Acrogenospora subprolata and A. verrucispora were re-collected and sequenced for the first time. The genus Acrogenospora is far more species rich than originally thought, with nine species found in a small area of Yunnan Province, China.
Introduction
Freshwater fungi are an ecological group that are defined by their presence in freshwater for the whole or part of their life cycle (Thomas, 1996; Wong et al., 1998), and include any species that grow on predominantly aquatic or semi-aquatic substrates (Goh and Hyde, 1996). Freshwater fungi play an important role in nutrient and carbon cycling, biological diversity and ecosystem functioning (Zhang et al., 2008; Swe et al., 2009). There have been many studies of freshwater fungi, especially on diversity, taxonomy and phylogeny (Tsui et al., 2000; Cai et al., 2002; Vijaykrishna et al., 2005; Vijaykrishna and Hyde, 2006; Hirayama et al., 2010; Ferrer et al., 2011; Barbosa et al., 2013; Raja et al., 2013, 2015) and recently from China (Hyde et al., 2016; Yang et al., 2017; Huang et al., 2018a, b; Su et al., 2018; Guo et al., 2019; Luo et al., 2019). In this study, we report nine Acrogenosporaceae species that were collected from freshwater habitats in China. Acrogenosporaceae was established by Jayasiri et al. (2018) to accommodate Acrogenospora within Minutisphaerales, with the latter being a freshwater ascomycetes order, comprising two families, Acrogenosporaceae and Minutisphaeraceae (Wijayawardene et al., 2020). Members of these two families are mostly reported from freshwater habitats (Goh et al., 1998; Raja et al., 2015; Bao et al., 2019; Hyde et al., 2019).
Acrogenospora was established by Ellis (1971) with two species, Acrogenospora sphaerocephala, and Farlowiella carmichaeliana (asexual morph). Ellis (1972) included two other species A. setiformis and F. australis in this genus. Hughes (1978) accepted the genus and added two additional species, A. gigantospora and A. novae-zelandiae. A taxonomic revision was provided by Goh et al. (1998) who accepted eight species, including two new combinations and two new species, and provided descriptions, illustrations and a key to species. Currently, 13 species are included in Acrogenospora (Hu et al., 2010; Ma et al., 2012; Hyde et al., 2019). Acrogenospora species are characterized by macronematous, mononematous, simple, brown, sometimes percurrently proliferating conidiophores; monoblastic, terminal or intercalary conidiogenous cells; and globose, ellipsoid or obovoid, olivaceous to brown conidia (Hughes, 1978; Goh et al., 1998).
The sexual morph of Acrogenospora has been linked with Farlowiella. Mason (1941) showed the connection between A. megalospora and Farlowiella armichaeliana based on cultural studies. Ellis (1971) reported the asexual morph of F. carmichaeliana as A. carmichaeliana. Ellis (1972) introduced A. australis as the asexual morph of F. australis based on morphological characters. Goh et al. (1998) accepted these two asexual morphs of Farlowiella and synonymized A. megalospora under F. carmichaeliana and A. altissima under F. australis. Jayasiri et al. (2018) carried out phylogenetic analyses with seven isolates of Acrogenospora and showed that A. sphaerocephala clustered with the sexual morph Farlowiella carmichaeliana. This confirmed the connection between Acrogenospora and Farlowiella. Hyde et al. (2019) also supported the asexual-sexual connection between these two genera based on a phylogenetic study. Based on recent nomenclatural changes with regards to one fungus one name, Acrogenospora was given priority (Wijayawardene et al., 2014; Rossman et al., 2015).
During our investigation of freshwater fungi on submerged wood along a north/south gradient in the Asian/Australasian region (Hyde et al., 2016), nine isolates of Acrogenospora were collected from freshwater habitats in China. Among them, two are identified as existing species, A. subprolata and A. verrucispora, and another seven are introduced as new species by comparing their morphology with known species of the genus, as well as performing phylogenetic analyses of on LSU, SSU, TEF1α, and RPB2 DNA sequence data. The objectives of this study are as follows: (i) describe and illustrate the newly collected Acrogenospora spp. from freshwater habitats in China; (ii) provide molecular data for Acrogenospora species and understand their phylogenetic relationships.
Materials and Methods
Isolation and Morphology
Samples of submerged wood were collected from Yunnan and Tibet provinces, China and taken to the laboratory in plastic bags. The samples were incubated in plastic boxes lined with moistened tissue paper at room temperature for one week. Specimen observations and morphological studies were conducted following the protocols provided by Luo et al. (2018).
Single spore isolations were carried out following the method described in Chomnunti et al. (2014). Germinating conidia were transferred aseptically to PDA and MEA plates supplemented with 100 mg of streptomycin and grown at room temperature in daylight. Colony color and other characters were observed and measured after 1 week and again after 3 weeks. The specimens were deposited in the Mae Fah Luang University (MFLU) Herbarium, Chiang Rai, Thailand. Living cultures are deposited in the Culture Collection at Mae Fah Luang University (MFLUCC). Facesoffungi numbers (FoF) were acquired as in Jayasiri et al. (2015) and Index Fungorum (2020). New species are established following recommendations outlined by Jeewon and Hyde (2016).
DNA Extraction, PCR Amplification, and Sequencing
Fungal mycelium was scraped from the surface of colonies grown on potato dextrose agar (PDA) or malt extract agar (MEA) at 25°C for 4 weeks, transferred into a 1.5 mL centrifuge tube and ground using liquid nitrogen. The EZ geneTM fungal gDNA kit (GD2416) was used to extract DNA from the ground mycelium according to the manufacturer’s instructions. Primers for PCR amplification used were LSUrDNA = LR0R/LR7 (Vilgalys and Hester, 1990), SSUrDNA = NS1/NS4 (White et al., 1990), (TEF1-α) = 983F/2218R and (RPB2) = fRPB2-5F/fRPB2-7cR (Liu et al., 1999). The PCR mixture was prepared as follows: 12.5 μl of 2 × Power Taq PCR MasterMix, 20 mM Tris-HCl pH 8.3, 100 Mm KCl, 3 mM MgCl2, stabilizer, and enhancer), 1 μl of each primer, 1 μl genomic DNA extract and 9.5 μl deionized water. The PCR of LSU, SSU and TEF1α gene was processed as follows: 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 50 s, elongation at 72°C for 1 min and a final extension at 72°C for 10 min, and finally kept at 4°C. The RPB2 gene region was amplified with an initial denaturation of 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 1 min, annealing at 54°C for 40 s, elongation at 72°C for 90 s, and the final extension at 72°C for 10 min. PCR amplification was confirmed on 1% agarose electrophoresis gels stained with ethidium bromide. Purification and sequencing of PCR products were carried out using the above-mentioned PCR primers at Beijing Tsingke Biological Engineering Technology and Services Co., Ltd. (Beijing, P.R. China).
Molecular Phylogenetic Analyses
Sequencing and Sequence Alignment
Sequences were assembled with BioEdit and those with high similarity indices were determined from a BLAST search to find the closest matches with taxa in Acrogenospora and from recently published data (Jayasiri et al., 2018; Hyde et al., 2019). All consensus sequences and the reference sequences were automatically aligned with MAFFT v. 7 and the strategy was using Auto (Katoh and Standley, 2013)1. Aligned sequences of each gene region (LSU, SSU, TEF1α and RPB2) were combined and manually improved using BioEdit v. 7.0.5.2 (Hall, 1999). Ambiguous regions were excluded from the analyses and gaps were treated as missing data. Phylogenetic analyses were performed using Maximum Likelihood (ML) and Bayesian tree building criteria.
Phylogenetic Analyses
Maximum likelihood analysis was performed at the CIPRES Science Gateway v.3.3 (Miller et al., 2010)2 using RAxML v. 8.2.8 as part of the “RAxML-HPC2 on XSEDE” tool (Stamatakis, 2006; Stamatakis et al., 2008). All model parameters were estimated by RAxML. The final ML search was conducted using the GTRGAMMA + I model which was estimated by using MrModeltest 2.2 (Nylander, 2004), Maximum likelihood bootstrap support was calculated from 1000 bootstrap replicates.
Bayesian analysis was performed using MrBayes v 3.1.2. (Ronquist and Huelsenbeck, 2003). The model of each genes was estimated using MrModeltest 2.2 (Nylander, 2004), GTR + I + G model was the best-fit model of LSU, SSU, TEF1α and RPB2 for Bayesian analysis. Posterior probabilities (PP) (Rannala and Yang, 1996) were performed by Markov chain Monte Carlo sampling (BMCMC) in MrBayes v.3.1.2 (Liu et al., 2012). Six simultaneous Markov chains were run for 50 million generations, and trees were sampled every 5000th generation (resulting in 10,000 trees). The first 2000 trees representing the burn-in phase of the analyses were discarded and the remaining 8000 (post burning) trees were used for calculating posterior probabilities (PP) in the majority rule consensus tree (Cai et al., 2006; Liu et al., 2012).
Maximum-parsimony analyses were performed using PAUP v.4.0b10 (Swofford, 2003). Gaps were treated as missing data with the heuristic search option with 1000 random sequence additions and tree bisection reconnection (TBR) branch-swapping. Maxtrees were unlimited, branches of zero length were collapsed and all parsimonious trees were saved. The consistency indices (CI), tree length (TL), homoplasy index (HI), rescaled consistency indices (RC), retention indices (RI) were calculated for each tree. Clade stability was assessed using a bootstrap (BT) analysis with 1000 replicates, each with 10 replicates of random stepwise addition of taxa. Other details are as provided by Jeewon et al. (2002, 2003)
Phylogenetic trees were represented by FigTree v. 1.4.4 (Rambaut, 2014) and edited in Microsoft Office PowerPoint 2016 (Microsoft Inc., United States). Newly generated sequences in this study were deposited in GenBank (Table 1) and the alignment used for the phylogenetic analyses were submitted to TreeBASE3 under the accession number: 26373.
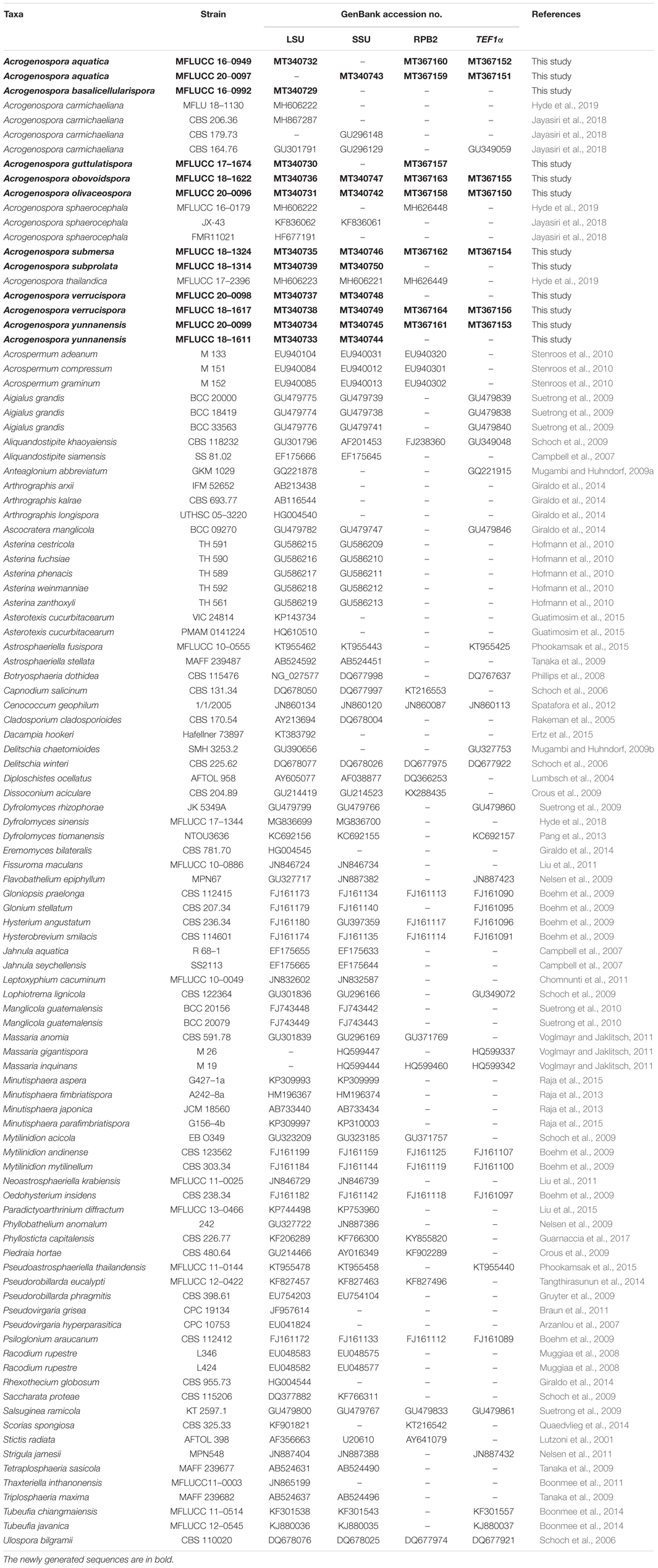
Table 1. GenBank numbers and culture collection accession numbers of species included in the phylogenetic study.
Results
Phylogenetic Analyses
The combined LSU, SSU, TEF, and RPB2 sequence dataset included 101 taxa (ingroup) and two outgroup taxa (Diploschistes ocellatus and Stictis radiata) with a total of 3853 characters (LSU: 867 bp; SSU:1020 bp; TEF1α: 915 bp; RPB2: 1051 bp) after alignment including the gaps. The RAxML and Bayesian analyses of the combined dataset resulted in phylogenetic reconstructions with largely similar topologies and the result of ML analysis with a final likelihood value of –49015.757408 is shown in Figure 1. The matrix had 1149 distinct alignment patterns, with 29.85% undetermined characters or gaps. Estimated base frequencies were: A = 0.253075, C = 0.235518, G = 0.278280, T = 0.194616; substitution rates AC = 1.387195, AG = 3.679854, AT = 1.133462, CG = 0.233127, CT = 7.472473, GT = 1.000000; gamma distribution shape parameter α = 0.303701. Bootstrap support values for RAxML and MP greater than 60% and Bayesian posterior probabilities greater than 0.95 are given at each node (Figure 1).
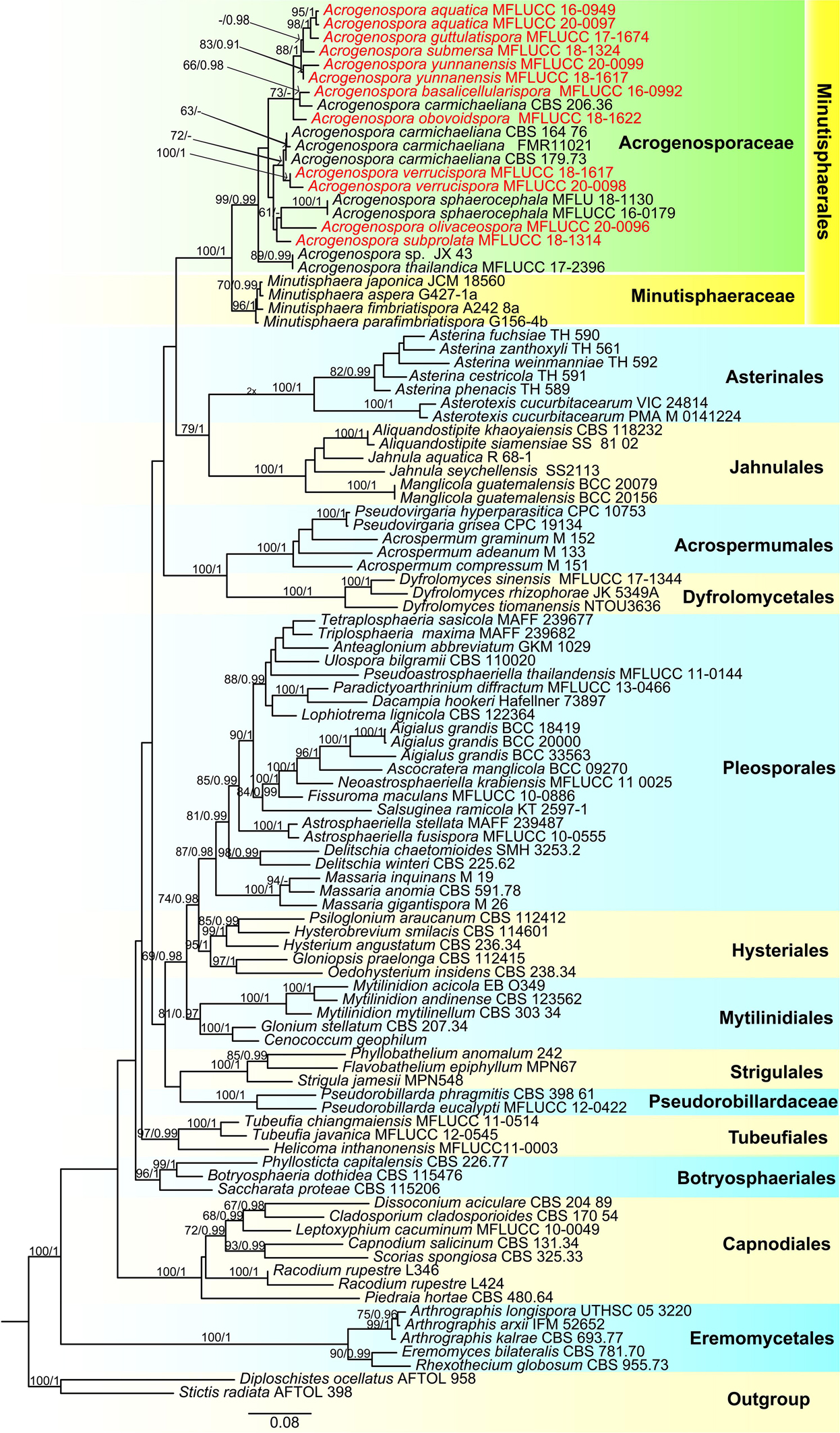
Figure 1. Phylogenetic tree based on RAxML analyses of a combined LSU, SSU, TEF1α and RPB2 dataset. Bootstrap support values for maximum likelihood ≥70% and Bayesian posterior probabilities ≥ 0.90 are indicated above the nodes as MLBS/PP. The tree is rooted with Diploschistes ocellatus (AFTOL 958) and Stictis radiata (AFTOL 398). The new isolates are in red.
In the phylogenetic analyses all the new strains grouped with members of Acrogenospora within Acrogenosporaceae with high support (99% ML and 0.99 BYPP). Acrogenospora aquatica, A. guttulatispora, A. submersa, A. yunnanensis grouped together, but separated in different clades. Two isolates of A. aquatica (MFLUCC 16–0949 and MFLUCC 20–0097) formed a distinct clade with high statistical support (95% ML and 1 BYPP). Acrogenospora guttulatispora was placed as a sister taxon to A. aquatica and A. submersa. Acrogenospora yunnanensis clustered with A. submersa. Acrogenospora basalicellularispora clustered with A. carmichaeliana (CBS 206.36) and sister to A. obovoidispora. Two strains of A. verrucispora (MFLUCC 20–0098 and MFLUCC 18–1617) clustered together with high statistical support (95% ML and 1 BYPP), and sister to A. carmichaeliana. Acrogenospora olivaceospora and A. subprolata grouped with A. sphaerocephala.
Acrogenospora aquatica D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov.
Index Fungorum number: IF 557599; Facesoffungi number: FoF 07984, Figure 2

Figure 2. Acrogenospora aquatica (MFLU 20–0291, holotype) (a,b) Colonies on wood. (c–f) Conidiophores, conidiogenous cells and conidia. (g–i) Conidiogenous cells and conidia. (j–m) Conidia. (n) Germinating conidium. Scale bars: (a,b) 200 μm, (c–f) 100 μm, (g–i) 30 μm, (j–n) 20 μm.
Holotype—MFLU 20–0291
Etymology—“Aquatica” in connection with the aquatic habitat from which it was recovered.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies effuse on natural substrate, hairy, dark brown. Mycelium mostly immersed, composed of septate, grayish brown, branched, smooth hyphae. Conidiophores 200–250 × 7.5–9.5 μm ( = 226 × 8.6 μm, n = 15), mononematous, macronematous, solitary, erect, straight or slightly flexuous, cylindrical, indeterminate, unbranched, brown to dark brown, paler toward apex, septate, smooth. Conidiogenous cells holoblastic, monoblastic, integrated, initially terminal, later becoming intercalary, cylindrical, smooth, pale brown, proliferating percurrently. Conidia 29–34.5 × 24–31 μm ( = 31.8 × 27.7 μm, n = 30), acrogenous, solitary, subprolate to broadly ellipsoidal, base truncate, dark brown to black, aseptate, lacking guttules, with a hyaline, globose to subglobose basal cell, smooth.
Material examined: CHINA, Yunnan Province, Dali, Cangshan Mountain, on decaying wood submerged in a stream, January 2016, Q.S. Zhou, S-763 (MFLU 20–0291, holotype), ex-type culture MFLUCC 20–0097. CHINA, Yunnan Province, Dali, Cangshan Mountain, on decaying wood submerged in Qingbixi Stream, March 2016, Z. L. Luo, S-282 (DLU 282, isotype), living culture MFLUCC 16-0949.
Notes: In our study, we found two species, A. basalicellularispora and A. aquatica with a hyaline, globose to subglobose basal cell. Acrogenospora aquatica can be distinguished from A. basalicellularispora by the size of conidiophores (259–395 × 8–12 vs. 202–250 × 7.8–9.3 μm). In addition, conidia of A. basalicellularispora are pale orange-brown to olivaceous-brown, with several small to large guttules, while conidia of A. aquatica are dark brown to black and lack guttules.
Acrogenospora aquatica is phylogenetically close to A. guttulatispora. Acrogenospora aquatica similar to A. guttulatispora in having mononematous, macronematous, unbranched conidiophores, holoblastic, monoblastic conidiogenous cells and acrogenous, dark brown to black conidia. However, A. aquatica differs from A. guttulatispora in having subprolate to broadly ellipsoidal conidia with a hyaline, globose to subglobose basal cell, lacking guttules, while conidia of A. guttulatispora are spherical or subspherical, with a large guttule, lacking basal cell.
Acrogenospora basalicellularispora D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov.
Index Fungorum number: IF 557596; Facesoffungi number: FoF 07981, Figure 3

Figure 3. Acrogenospora basalicellularispora (MFLU 20–0288, holotype) (a) Colony on wood. (b) Conidiophores, conidiogenous cells and conidia. (c) Conidiogenous cells and conidia. (d–h) conidia. (i) Germinating conidium. (j,k) Culture on MEA (upper and lower view). Scale bars: (a) 200 μm, (b) 50 μm, (c–i) 20 μm.
Holotype—MFLU 20–0288
Etymology—Referring to the conidia which have a basal cell.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies effuse on natural substrate, hairy, dark brown. Mycelium mostly immersed, composed of grayish brown, septate, branched, smooth hyphae. Conidiophores 260–395 × 8–12 μm ( = 327 × 10 μm, n = 20) wide, mononematous, macronematous, solitary, cylindrical, erect, straight or slightly flexuous, mostly unbranched, septate, brown to dark brown, slightly paler toward apex, smooth. Conidiogenous cells holoblastic, monoblastic, integrated, initially terminal, later becoming intercalary, cylindrical, smooth, pale brown, proliferating percurrently. Conidia 27.5–33.7 × 21.7–25.8 μm ( = 30.6 × 23.7 μm, n = 30) wide, acropleurogenous, solitary, dry, broadly obovoid to spherical, smooth, pale orange-brown to olivaceous brown, aseptate, with several small or large guttules, with a small, hyaline, subcylindrical to subglobose basal cell, germinating from basal cell.
Material examined: CHINA, Yunnan Province, Gaoligongshan Mountain, on decaying wood submerged in a stream, August 2015, A.L. Shi, S-431 (MFLU 20–0288, holotype); ex-type culture, MFLUCC 16–0992.
Notes: In the phylogenetic analysis, Acrogenospora basalicellularispora clustered with A. sphaerocephala (CBS 206.36) with low support (66% ML and 0.98 BYPP). Unfortunately, CBS 206.36 lacks a morphological description and only LSU sequence data is available in GenBank. Morphologically, our new isolate can be distinguished from other Acrogenospora species by its pale orange-brown to olivaceous brown, broadly obovoid to spherical conidia with several small to large guttules and a small, hyaline, subcylindrical to subglobose basal cell. In our study, A. aquatica also has conidia with a basal cell. However, we can distinguish them by the shape (broadly obovoid to spherical vs. subprolate to broadly ellipsoidal) and color (pale orange-brown to olivaceous brown vs dark brown to black) of conidia and size (260–395 × 8–12 vs. 200–250 × 7.5–9.5 μm) of conidiophores.
Acrogenospora guttulatispora D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov.
Index Fungorum number: IF 557597; Facesoffungi number: FoF 07982, Figure 4
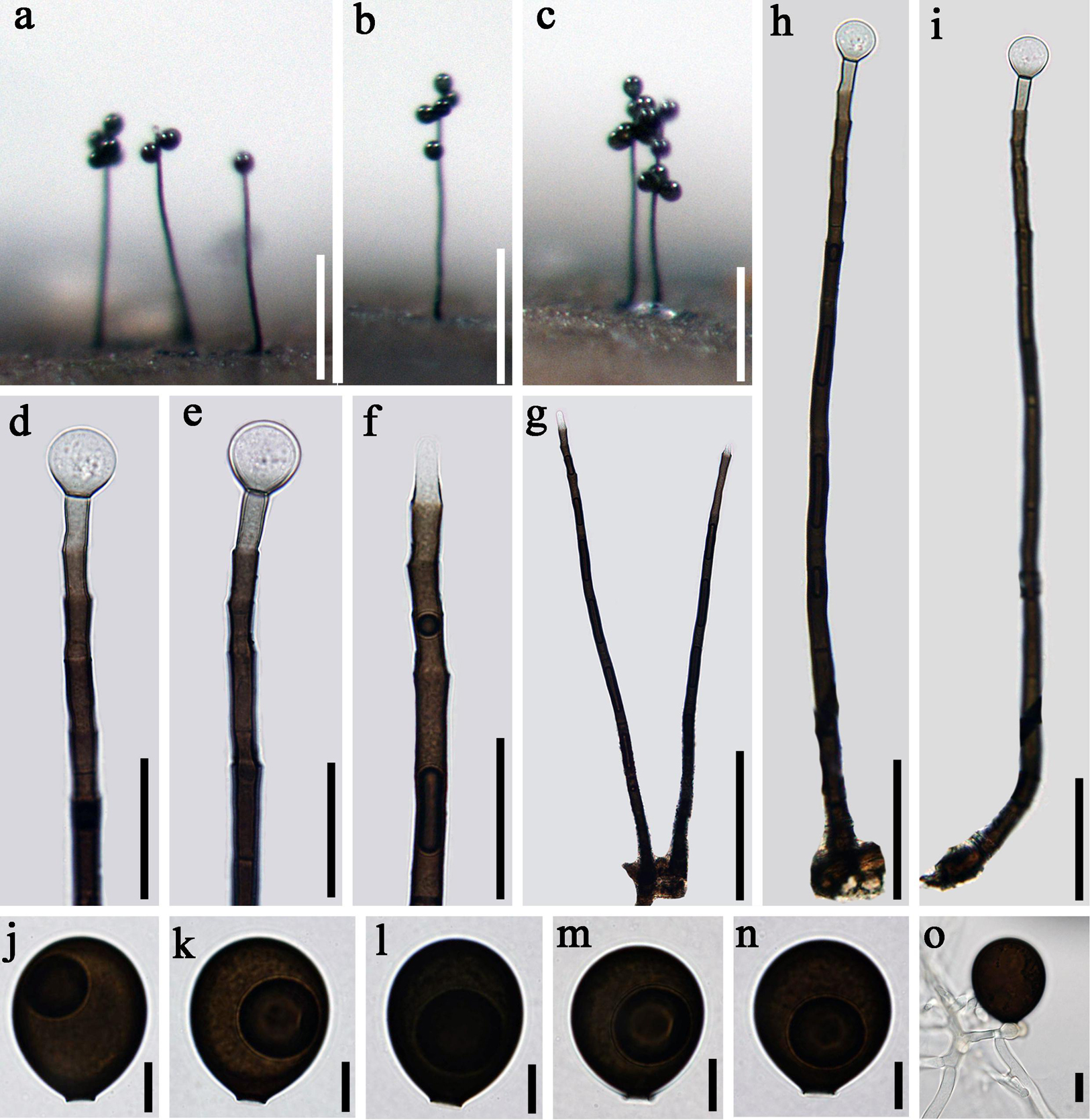
Figure 4. Acrogenospora guttulatispora (MFLU 20–0289, holotype) (a–c) Colonies on wood. (d–f) Conidiogenous cells and conidia. (g–i) Conidiophores. (j–n) conidia. (o) Germinating conidium. Scale bars: (a–c) 200 μm, (d–f) 30 μm, (g) 100 μm, (h,i) 50 μm, (j–o) 10 μm.
Holotype—MFLU 20–0289
Etymology—Referring to the large guttule in the conidia.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies effuse on natural substrate, hairy, dark brown. Mycelium mostly immersed, composed of septate, grayish brown, branched, smooth hyphae. Conidiophores 295–330 × 7.5–8.5 μm ( = 312.7 × 8 μm, n = 15), mononematous, macronematous, solitary, erect, straight or slightly flexuous, cylindrical, indeterminate, unbranched, dark brown, paler toward apex, pale brown to hyaline at apex, septate, guttulate, smooth. Conidiogenous cells holoblastic, monoblastic, integrated, initially terminal, later becoming intercalary, cylindrical, smooth, pale brown, proliferating percurrently. Conidia 30–33.5 × 26.5–28 μm ( = 34 × 27 μm, n = 30), acropleurogenous, solitary, spherical or subspherical, truncate at base, hyaline when young, dark brown when mature, aseptate, with a large guttule, smooth.
Material examined: CHINA, Yunnan Province, Dali, Cangshan Mountain, on decaying wood submerged in Heilongxi stream, June 2013, Z.L. Luo. S-189 (MFLU 20–0289, holotype), ex-type culture, MFLUCC 17–1674 = ICMP 21772.
Notes: Acrogenospora guttulatispora can be distinguished from other species by the large guttule in the conidia. In the phylogenetic analyses, A. guttulatispora is close to A. aquatica. However, the conidia of A. guttulatispora are spherical or subspherical with a large guttule, without a basal cell. While, those A. aquatica are subprolate to broadly ellipsoidal with a hyaline, globose to subglobose basal cell. In addition, there are 22 base pair differences in the RPB2 region between these two species.
Acrogenospora obovoidspora D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov.
Index Fungorum number: IF 557602; Facesoffungi number: FoF 04691, Figure 5

Figure 5. Acrogenospora obovoidispora (MFLU 20–0295, holotype) (a) Colony on wood. (b–e) Conidiophores with conidia. (f–i) Conidiogenous cells with conidia. (j–n) Conidia. Scale bars: (a) 200 μm, (b–e) 50 μm, (f–h) 30 μm, (i–n) 20 μm.
Holotype—MFLU 20–0295
Etymology—Referring to the broadly obovoid conidia of this fungus.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies effuse on natural substrate, hairy, dark brown. Mycelium partly immersed, partly superficial, composed of septate, brown to dark brown, branched, smooth hyphae. Conidiophores 209–277 × 7.5–10 μm ( = 243 × 8.8 μm, n = 15), mononematous, macronematous, solitary, erect, straight or slightly flexuous, cylindrical, unbranched, brown to dark brown, paler toward apex, septate, smooth. Conidiogenous cells holoblastic, monoblastic, integrated, initially terminal, later becoming intercalary, cylindrical, smooth, pale brown, proliferating percurrently. Conidia 32.5–37.5 × 27–32 μm ( = 35 × 29.6 μm, n = 30) wide, acrogenous, solitary, oval to broadly ellipsoidal, base truncate, aseptate, olivaceous brown to black, thick-walled, smooth.
Material examined: CHINA, Yunnan Province, Dali, Huadianba Mountain, saprobic on decaying wood submerged in a stream, 9 December 2017, Z.L. Luo, S-1614 (MFLU 20–0295, holotype); ex-type culture, MFLUCC 18–1622.
Notes: Acrogenospora obovoidispora is similar to A. gigantospora in having mononematous, macronematous, conidiophores and solitary, aseptate conidia. However, Acrogenospora obovoidspora differs from A. gigantospora in having solitary conidiophores and oval to broadly ellipsoidal, olivaceous brown to black conidia, while conidiophores of A. gigantospora are single or in groups of 2–4, and conidia are broadly obovoid to subspherical, dark brown to black (Ma et al., 2012).
Acrogenospora olivaceospora D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov.
Index Fungorum number: IF 557598; Facesoffungi number: FoF 07983, Figure 6
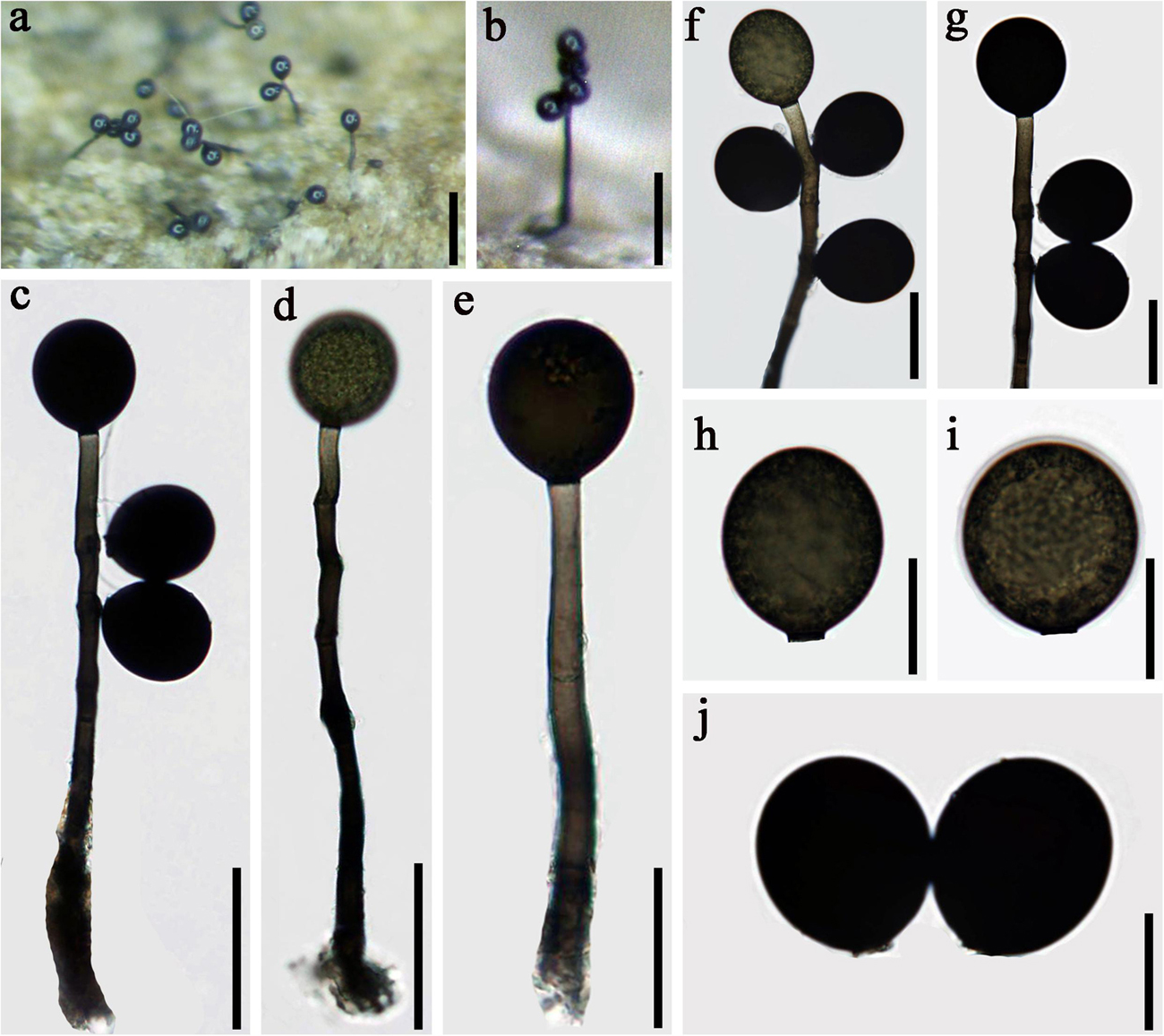
Figure 6. Acrogenospora olivaceospora (MFLU 20–0290, holotype) (a,b) Colonies on wood. (d,e) Conidiophores, conidiogenous cells, and conidia. (f,g) Conidiogenous cells with conidia. (a) 200 μm, (b) 100 μm, (h–j) Conidia. Scale bars: (c,d) 50 μm, (e–g) 30 μm, (h–j) 20 μm.
Holotype—MFLU 20–0290
Etymology—Referring to the conidia which are olive-green.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies effuse on natural substrate, hairy, dark brown. Mycelium mostly immersed, composed of grayish brown, septate, branched, smooth hyphae. Conidiophores 100–175 × 6–9 μm ( = 137 × 7.4 μm, n = 20), mononematous, macronematous, solitary, erect, straight or slightly flexuous, cylindrical, indeterminate, unbranched, dark brown to olive, paler toward apex, septate, smooth. Conidiogenous cells holoblastic, monoblastic, integrated, initially terminal, later becoming intercalary, cylindrical, smooth, pale brown, proliferating percurrently. Conidia 32–37 × 28–33 μm ( = 34.5 × 30.5 μm n = 30), acropleurogenous, solitary, subprolate to broadly ellipsoidal, base truncate, olive to black, aseptate, thick-walled, lacking guttules, smooth.
Material examined: CHINA, Yunnan Province, Dali, Cangshan Mountain, on decaying wood submerged in a stream, March 2016, H.W. Shen, S-715 (MFLU 20–0290, holotype), ex-type culture, MFLUCC 20–0096.
Notes: In the phylogenetic analyses, Acrogenospora olivaceospora clustered with A. sphaerocephala (MFLU 18-1130 and MFLUCC 16-0179). However, A. olivaceospora differs from A. sphaerocephala by the shape, color and size of conidia (Table 2). Acrogenospora olivaceospora has olive to black, subprolate to broadly ellipsoidal conidia, lacking guttules, while conidia of A. sphaerocephala are olive-green to brown, spherical or subspherical and guttulate.
Acrogenospora olivaceospora is most similar to A. subprolata in having subprolate to broadly ellipsoidal, olive to black, aseptate, thick-walled conidia. However, A. olivaceospora has solitary conidiophores whereas those of A. subprolata are sometimes in small groups. In addition, the conidiophores of A. olivaceospora are shorter (102–172 × 5.8–9 vs. 150–300 × 9–12 μm) and the conidia are smaller (32–36.9 × 28–32.8 vs. 39–46 × 30–39 μm).
Acrogenospora submersa D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov.
Index Fungorum number: IF 557601; Facesoffungi number: FoF 07986, Figure 7

Figure 7. Acrogenospora submersa (MFLU 20–0294, holotype) (a) Colonies on wood. (b–g) Conidiophores with conidia. (h,i) Conidiogenous cells with conidia. (j,k) conidia. (i) Germinating conidium. (m,n) Culture on MEA (upper and lower view). Scale bars: (a) 200 μm, (b–g) 50 μm, (h–l) 20 μm.
Holotype—MFLU 20–0294,
Etymology—Referring to the submerged habitat of this fungus.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies effuse on natural substrate, hairy, dark brown. Mycelium partly immersed, partly superficial, composed of septate, brown to dark brown, branched, smooth hyphae. Conidiophores 163–223 × 6.5–10 μm ( = 193 × 8.4 μm, n = 15), mononematous, macronematous, solitary, erect, straight or slightly flexuous, cylindrical, unbranched, brown to dark brown, paler toward apex, septate, smooth. Conidiogenous cells holoblastic, monoblastic, integrated, initially terminal, later becoming intercalary, cylindrical, smooth, pale brown, proliferating percurrently. Conidia 28–32.5 × 25–28 μm ( = 30.3 × 26.5 μm, n = 30), acropleurogenous, solitary, spherical or subspherical, base truncate, aseptate, hyaline when young, pale orange-brown to olivaceous brown when mature, smooth.
Material examined: CHINA, Yunnan Province, saprobic on decaying wood submerged in Lancang River, 9 December 2017, Z.L. Luo, S-1601 (MFLU 20–0294, holotype), ex-type culture, MFLUCC 18–1324.
Notes: Acrogenospora submersa is similar to A. hainanensis in having mononematous, macronematous, solitary, proliferating percurrently conidiophores, monoblastic, integrated, terminal conidiogenous cells and spherical or subspherical, aseptate conidia. However, A. submersa differs from A. hainanensis by having longer conidiophores (163–223 × 6.7–10 μm vs. 60–80 × 2–3.5 μm), and much larger conidia (28–32.5 × 25–28 μm vs. 7.5–9.5 × 7–8.5 μm), which are hyaline to pale orange-brown or olivaceous brown rather than brown. Phylogenetically, A. submersa is related to A. guttulatispora but in a distinct lineage. Therefore, we introduce it as a new species.
Acrogenospora subprolata Goh, K.D. Hyde & C.K.M. Tsui, Mycol. Res. 102(11): 1314 (1998) Figure 8

Figure 8. Acrogenospora subprolata (MFLU 20–0293) (a,b) Colonies on wood. (c–e) Conidiophores and conidia. (f–h) Conidiogenous cells with conidia. (i–l) conidia. Scale bars: (a,b) 200 μm, (c–e) 100 μm, (f,g) 30 μm, (h–l) 20 μm.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies effuse on natural substrate, hairy, dark brown. Mycelium partly immersed, partly superficial, composed of septate, brown to dark brown, branched, smooth hyphae. Conidiophores 212.5–348 × 8–10.5 μm ( = 280.5 × 9.4 μm, n = 15) wide, mononematous, macronematous, solitary, or in a small group, erect, straight or slightly flexuous, cylindrical, unbranched, brown to dark brown, paler toward apex, septate, smooth. Conidiogenous cells holoblastic, monoblastic, integrated, initially terminal, later becoming intercalary, cylindrical, smooth, pale brown, proliferating percurrently. Conidia 40–51 × 32–40 μm ( = 45 × 31.5μm, n = 30) wide, acrogenous, solitary, subprolate to broadly ellipsoidal, base truncate, aseptate, hyaline when young, olivaceous brown to black when mature, thick-walled, smooth.
Material examined: CHINA, Tibet Province, saprobic on decaying wood submerged in a stream, 2 May 2017, Z.L. Luo. S-1455 (MFLU 20–0293), living culture, MFLUCC 18–1314.
Notes: Acrogenospora subprolata is characterized by conidiophores that are macronematous, mononematous, solitary or in groups of 2–4 with multiple percurrent proliferations and by acrogenous, subprolate to broadly ellipsoidal, pale orange-brown to olivaceous brown, aseptate, thick-walled conidia. Our isolate fits well with the characters of A. subprolata as described by Goh et al. (1998). Therefore, we identify this collection as A. subprolata.
Acrogenospora verrucispora Hong Zhu, L. Cai & K.Q. Zhang [as ‘verrucospora’], Mycotaxon 92: 384 (2005) Figure 9

Figure 9. Acrogenospora verrucispora (MFLU 20–0287) (a,b) Colonies on wood. (c–h) Conidiophores with conidia. (n–p) Conidiogenous cells with conidia. (i–m) conidia. Scale bars: (a–f) 50 μm, (g,h,n–p) 20 μm, (i–m) 10 μm.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies effuse on natural substrate, hairy, dark brown. Mycelium partly immersed, partly superficial, composed of septate, brown to dark brown, branched, smooth hyphae. Conidiophores 103–149 × 5.6–7.4 μm ( = 125.8 × 6.5 μm, n = 15) wide, mononematous, macronematous, solitary or sometimes in a small group, erect, straight or slightly flexuous, cylindrical, unbranched, brown to dark brown, paler toward apex, septate, smooth. Conidiogenous cells holoblastic, monoblastic, integrated, initially terminal, later becoming intercalary, cylindrical, smooth, pale brown, proliferating percurrently. Conidia 21.3–26.5 × 20.6–25.5 μm ( = 24 × 23 μm, n = 30) wide, acrogenous, solitary, spherical or subspherical, base truncate, aseptate, hyaline when young, orange-brown to olivaceous brown when mature, distinctly verrucose.
Material examined: CHINA, Yunnan Province, Gaoligongshan Mountain, saprobic on decaying wood submerged in a stream, May 2017, H.W. Shen. S-1402 (MFLU 20–0287), living culture, MFLUCC 20–0098. S-1328 (DLU 1328), living culture, MFLUCC 18–1617.
Notes: Acrogenospora verrucispora was introduced by Zhu et al. (2005) with distinct verrucose conidia, it was collected from bamboo in a stream in Yunnan province, China. A. verrucispora is characterized by mononematous, macronematous, proliferating percurrently conidiophores, monoblastic, integrated conidiogenous cells and acrogenous, solitary, spherical or subspherical, verrucose conidia. Our isolate fits well with the original description of A. verrucispora. As the sequence data of A. verrucispora is not available in GenBank, we identify our isolate as A. verrucispora based on the morphological characters and provide sequence data for this species.
Acrogenospora yunnanensis D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov.
Index Fungorum number: IF 557600; Facesoffungi number: FoF 07985, Figure 10

Figure 10. Acrogenospora yunnanensis (MFLU 20–0292, holotype) (a) Colony on wood. (b–d) Conidiophores, conidiogenous cells and conidia. (e–g) Conidiogenous cells and conidia. (h–k) Conidia. Scale bars: (a) 200 μm, (b–d) 50 μm, (e–g) 30 μm, (h–j) 15 μm.
Holotype—MFLU 20–0292
Etymology—Referring to Yunnan province, China, where the fungus was collected.
Saprobic on submerged decaying wood. Sexual morph: Undetermined. Asexual morph: Colonies effuse on natural substrate, hairy, dark brown. Mycelium partly immersed, partly superficial, composed of septate, brown to dark brown, branched, smooth hyphae. Conidiophores 260–390 × 8.5–12 μm ( = 326 × 10.3 μm, n = 15), mononematous, macronematous, solitary, erect, straight or slightly flexuous, cylindrical, unbranched, brown to dark brown, paler toward apex, septate, smooth. Conidiogenous cells holoblastic, monoblastic, integrated, initially terminal, later becoming intercalary, cylindrical, smooth, pale brown, proliferating percurrently. Conidia 23–32.5 × 22–30 μm ( = 28 × 26 μm, n = 30), acropleurogenous, solitary, spherical or subspherical, truncate at base, aseptate, hyaline when young, dark brown to black when mature, smooth.
Material examined: CHINA, Yunnan Province, Laojunshan Mountain, on decaying wood submerged in a stream, August 2015, A.L. Shi, S-1114 (MFLU 20–0292, holotype); ex-type culture, MFLUCC 18–1611. CHINA, Gaoligingshan mountain, saprobic on decaying wood submerged in a stream, August 2015, A.L. Shi. S-774 (DLU 774, isotype), living culture, MFLUCC 20–0099.
Notes: In the phylogenetic analysis, Acrogenospora yunnanensis shares a sister relationship to A. submersa. Morphologically, A. yunnanensis can be distinguished from A. submersa by the longer conidiophores (Table 2) and color of conidia. Acrogenospora yunnanensis has dark brown to black conidia with a large guttule, while conidia of A. submersa are pale orange-brown to olivaceous brown at maturity and lack a guttule.
Morphologically, A. yunnanensis is most similar to A. gigantospora and A. subprolata in having similar conidial shape. However, they differ in size of conidiophores and conidia (Table 2).
Discussion
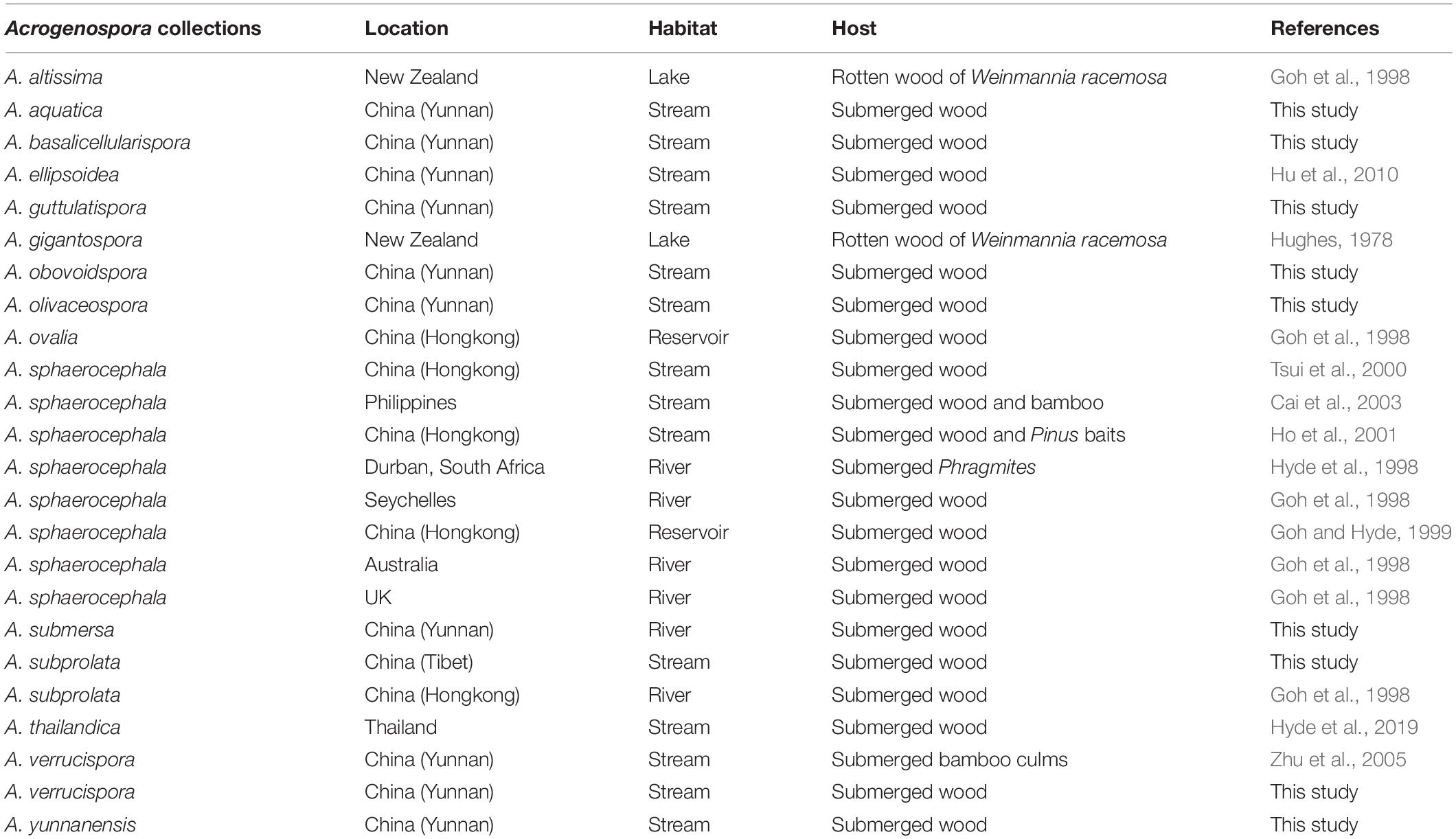
In this study, we provide new descriptions and illustrations for seven new species and two known species of Acrogenospora. This study contributes to a better taxonomic understanding and proposes that there could be a number of additional new species within the genus and its diversity could be much higher than anticipated. Acrogenospora species are cosmopolitan with worldwide distribution, they are mainly found on dead and submerged wood especially in freshwater habitats (Hughes, 1978; Goh et al., 1998; Ma et al., 2012; Hyde et al., 2019). Among the 20 Acrogenospora spp., 15 species were reported from freshwater habitats and only 5 of them were recovered from terrestrial habitats. Our study has shown that in a small area of Yunnan Province there are 14 species of Acrogenospora in streams alone and indicates that the genus is highly diverse, and has been found to occur with other genera in the region (Hyde et al., 2019). Previous studies have also reported that there could be an amazing fungal diversity hidden in the South East Asian region (Hyde et al., 2018).
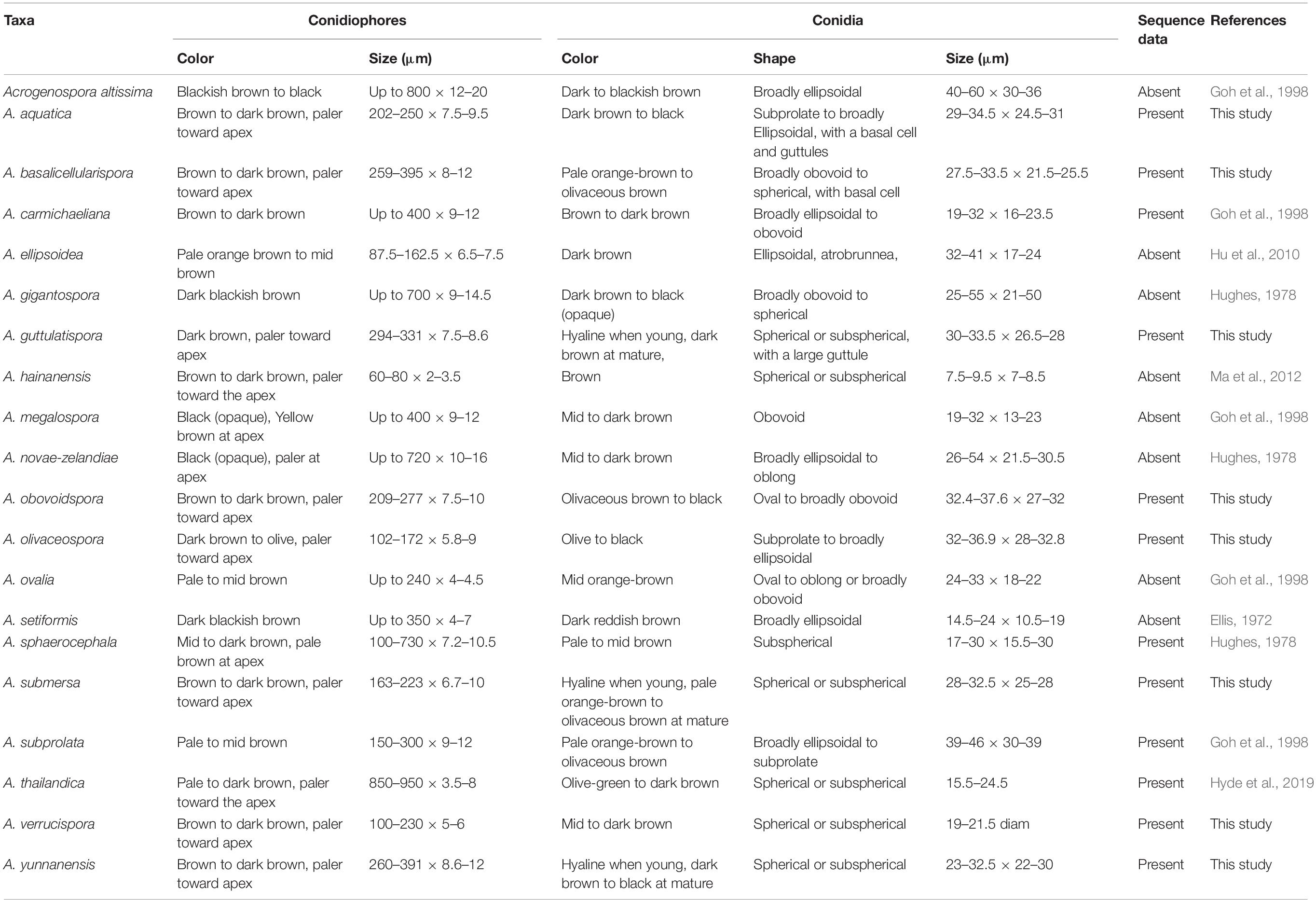
Acrogenospora species are quite similar to each other, and previous studies suggested to distinguish them based on conidial shape, size, and color and the degree of pigmentation of the conidiophores (Hughes, 1978, Table 2). We found that guttules and basal cells of conidia are also important characters to distinguish species and a morphological comparison of all Acrogenospora species is provided (Table 2).
Previous publications on submerged wood in freshwater have lumped several Acrogenospora collections and identified them based on morphology as A. sphaerocephala (Table 3) perhaps because of the difficulty of using morphs alone to delineate species and due to a lack of DNA sequence data. It is likely that the collections of A. sphaerocephala in older publications (Table 3) are wrongly named and further taxonomic work is necessary.
Before this study, there were 13 species of Acrogenospora but only three of them had sequence data available in GenBank, and there was no data for the ex-type strains. Our phylogenetic sampling included 12 strains of Acrogenospora (Acrogenosporaceae), and all strains grouped with four species of Minutisphaera (Minutisphaeraceae) within Minutisphaerales (99% ML and 0.99 PP, Figure 1). The results were similar to the analyses by Jayasiri et al. (2018). In our analyses, Acrogenospora carmichaeliana (CBS 206.36) did not cluster with other strains of A. carmichaeliana, instead clustered with our new isolate A. basalicellularispora with low statistical support. Unfortunately, there are no morphological descriptions for CBS 206.36, so we are unable to compare its morphology with our new isolate. Further collections and phylogenetic studies of Acrogenospora are needed to better understand the phylogenetic placement of those species which lack sequence data and, undoubtedly, many more novel species can be found.
The phylogenetic analysis provide clear resolution to the taxonomic complexities within this group. Protein-coding genes have been shown to be essential to identify a taxon up to species level (Tang et al., 2007, 2009; Jeewon et al., 2017). In our study, we sequenced the RPB2 and TEF1α sequence data to distinguish Acrogenospora species and the phylogenetic trees are provided in Supplementary Files 1, 2. In our phylogenetic tree (LSU + SSU + TEF1α + RPB2, Figure 1), Acrogenospora aquatica, A. guttulatispora, A. submersa and A. yunnanensis grouped together, but they constitute different clades based on phylogenies derived from the TEF and RPB2 data which clearly support that they are phylogenetically distinct species. Acrogenospora verrucispora clustered with A. carmichaeliana (Figure 1), but there are 9 bp differences in TEF1α gene region. In addition, they can be easily distinguished from each other by the shape, color and wall of conidia, (conidia of A. verrucispora are spherical or subspherical, orange-brown to olivaceous brown, distinctly verrucose-walled, while A. carmichaeliana has broadly ellipsoidal to obovoid, brown to dark brown, smooth-walled conidia). Acrogenospora olivaceospora is close to A. sphaerocephala, however, there are 12.3% nucleotide differences in RPB2 gene region between them. These results support our establishment of the new taxon as recommended by Jeewon and Hyde (2016). As for A. basalicellularispora and A. subprolata there are no DNA sequences available from protein-coding gene but they can be easily distinguished from other species based on morphological characters (Table 2).
Hyde et al. (2019) discussed whether Acrogenospora sphaerocephala was the asexual morph of Farlowiella carmichaeliana and whether A. megalospora was wrongly introduced as the asexual morph of F. carmichaeliana. In our phylogenetic analyses, Acrogenospora sphaerocephala did not cluster with A. carmichaeliana, forming different clades within Acrogenosporaceae. DNA sequences of Ex-type strains of both A. megalospora and Farlowiella carmichaeliana are unavailable in GenBank. Therefore, the connection of sexual and asexual morph of Farlowiella carmichaeliana is not clear and this needs further morpho-molecular evidence.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
D-FB conducted the experiments, analyzed the data, and wrote the manuscript. EM, DB, and KH revised the manuscript. Z-LL planned the experiments and analyzed the data. H-YS planned the experiments and funded the experiments. H-WS conducted the experiments. All authors revised the manuscript.
Funding
This work was mainly supported by the National Natural Science Foundation of China (Project IDs: 31860006 and 31660008) and Fungal Diversity Conservation and Utilization Innovation Team (ZKLX2019213). KH thanks the Thailand Research Fund for the grant entitled “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Sub-region (RDG6130001).”
Conflict of Interest
EM was employed by Manaaki Whenua Landcare Research.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
D-FB would like to thank Shaun Pennycook from Landcare Research, Auckland, New Zealand, for advising on the taxon names. Wen-Li Li and Yan-Mei Zhang are acknowledged for their help on DNA extraction and PCR amplification.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01606/full#supplementary-material
FILE S1 | Phylogenetic tree based on RAxML analysis of TEF1α sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (MP, red) higher than 75% are indicated above the nodes as MLBS/MPBS. The tree is rooted with Hysterographium fraxini (MFLU 15-3035 and MFLU 15-3681).
FILE S2 | Phylogenetic tree based on RAxML analysis of RPB2 sequence data. Bootstrap support values for maximum likelihood and maximum parsimony (MP, red) higher than 75% are indicated above the nodes as MLBS/MPBS. The tree is rooted with Aliquandostipite khaoyaiensis (AFTOL-ID 1364).
Footnotes
- ^ http://mafft.cbrc.jp/alignment/server/index.html
- ^ http://www.phylo.org/portal2/
- ^ https://www.treebase.org/
References
Arzanlou, M., Groenewald, J. Z., Gams, W., Braun, U., Shin, H. D., and Crous, P. W. (2007). Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud. Mycol. 58, 57–93. doi: 10.3114/sim.2007.58.03
Bao, D. F., Hyde, K. D., Luo, Z. L., Su, H. Y., and Nalumpang, S. (2019). Minutisphaera aquaticum sp. nov. increases the known diversity of Minutisphaeraceae. Asian J. Mycol. 2, 306–314. doi: 10.5943/ajom/2/1/21
Barbosa, F. R., Gusmao, L. F. P., Raja, H. A., and Shearer, C. A. (2013). New species and new records of freshwater ascomycetes from Brazil and Costa Rica. Mycologia 105, 335–343. doi: 10.3852/12-054
Boehm, E. W. A., Schoch, C. L., and Spatafora, J. W. (2009). On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycol. Res. 113, 461–479. doi: 10.1016/j.mycres.2008.12.001
Boonmee, S., Rossman, A. Y., Liu, J. K., Li, W. J., Dai, D. Q., Bhat, D. J., et al. (2014). Tubeufiales, ord. nov., integrating sexual and asexual generic names. Fungal Divers. 68, 239–298. doi: 10.1007/s13225-014-0304-7
Boonmee, S., Zhang, Y., Chomnunti, P., Chukeatirote, E., Tsui, C. K. M., Bahkali, A. H., et al. (2011). Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Divers. 51, 63–102. doi: 10.1007/s13225-011-0147-4
Braun, U., Crous, P. W., Groenewald, J. Z., Verkley, G. M., Groenewald, J. Z., and Crous, P. W. (2011). Pseudovirgaria, a fungicolous hyphomycete genus. IMA Fungus 2, 65–69. doi: 10.5598/imafungus.2011.02.01.09
Cai, L., Jeewon, R., and Hyde, K. D. (2006). Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Mycol. Res. 110, 137–150. doi: 10.1016/j.mycres.2005.09.014
Cai, L., Tsui, C. K. M., Zhang, K. Q., and Hyde, K. D. (2002). Aquatic fungi from Lake Fuxian. Yunnan, China. Fungal Divers. 9, 57–70.
Cai, L., Zhang, K. Q., McKenzie, E. H. C., and Hyde, K. D. (2003). Biodiversity of fungi on submerged wood in a stream and its estuary in the Tai Ho Bay, Hong Kong. Fungal Divers. 13, 205–220.
Campbell, J., Ferrer, A., Raja, H. A., Sivichai, S., and Shearer, C. A. (2007). Phylogenetic relationships among taxa in the Jahnulales inferred from 18S and 28S nuclear ribosomal DNA sequences. Can. J. Bot. 85, 873–882. doi: 10.1139/B07-080
Chomnunti, P., Hongsanan, S., Aguirre-Hudson, B., Tian, Q., Peršoh, D., Dhami, M. K., et al. (2014). The sooty moulds. Fungal Divers. 66, 1–36. doi: 10.1007/s13225-014-0278-5
Chomnunti, P., Schoch, C. L., Aguirre-Hudson, B., Ko-Ko, T. W., Hongsanan, S., Jones, E. B. G., et al. (2011). Capnodiaceae. Fungal Divers. 51, 103–134. doi: 10.1007/s13225-011-0145-6
Crous, P. W., Schoch, C. L., Hyde, K. D., Wood, R., Gueidan, C., de Hoog, G. S., et al. (2009). Phylogenetic lineages in the Capnodiales. Stud. Mycol. 64, 17–47. doi: 10.3114/sim.2009.64.02
Ertz, D., Diederich, P., Lawrey, J. D., Berger, F., Freebury, C. E., Coppins, B., et al. (2015). Phylogenetic insights resolve Dacampiaceae (Pleosporales) as polyphyletic: Didymocyrtis (Pleosporales, Phaeosphaeriaceae) with Phoma-like anamorphs resurrected and segregated from Polycoccum (Trypetheliales, Polycoccaceae fam. nov.). Fungal Divers. 74, 53–89. doi: 10.1007/s13225-015-0345-6
Ferrer, A., Miller, A. N., and Shearer, C. A. (2011). Minutisphaera and Natipusilla: two new genera of freshwater Dothideomycetes. Mycologia 103, 411–423. doi: 10.3852/10-177
Giraldo, A., Gené, J., Sutton, D. A., Madrid, H., Cano, J., Crous, P. W., et al. (2014). Phylogenetic circumscription of Arthrographis (Eremomycetaceae, Dothideomycetes). Persoonia 32, 102–114. doi: 10.3767/003158514X680207
Goh, T. K., and Hyde, K. D. (1996). Biodiversity of freshwater fungi. J. Ind. Microbiol. 17, 328–334. doi: 10.1007/BF01574764
Goh, T. K., and Hyde, K. D. (1999). Fungi on submerged wood and bamboo in the Plover Cove Reservoir, Hong Kong. Fungal Divers. 3, 57–85.
Goh, T. K., Hyde, K. D., and Tsui, K. M. (1998). The hyphomycete genus Acrogenospora, with two new species and two new combinations. Mycol. Res. 102, 1309–1315. doi: 10.1017/S0953756298006790
Gruyter, J. D., Aveskamp, M. M., Woudenberg, J. H. C., Verkley, G. J. M., Groenewald, J. Z., and Crous, P. W. (2009). Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycol. Res. 113, 508–519. doi: 10.1016/j.mycres.2009.01.002
Guarnaccia, V., Groenewald, J. Z., Li, H., Glienke, C., Carstens, E., Hattingh, V., et al. (2017). First report of Phyllosticta citricarpa and description of two new species, P. paracapitalensis and P. paracitricarpa, from citrus in Europe. Stud. Mycol. 87, 161–185. doi: 10.1016/j.simyco.2017.05.003
Guatimosim, E., Firmino, A. L., Bezerra, J. L., Pereira, O. L., Barreto, R. W., and Crous, P. W. (2015). Towards a phylogenetic reappraisal of Parmulariaceae and Asterinaceae (Dothideomycetes). Persoonia 35, 230–241. doi: 10.3767/003158515X688046
Guo, J. S., Zhang, Z., Qiao, M., and Yu, Z. F. (2019). Phalangispora sinensis sp. nov. from Yunnan, China and two new members of Wiesneriomycetaceae. Int. J. Syst. Evol. Microbiol. 69, 3217–3223. doi: 10.1099/ijsem.0.003612
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hirayama, K., Tanaka, K., Raja, H. A., Miller, A. N., and Shearer, C. A. (2010). A molecular phylogenetic assessment of Massarina ingoldiana sensu lato. Mycologia 102, 729–746. doi: 10.3852/09-230
Ho, W. H., Hyde, K. D., Hodgkiss, I. J., and Yanna. (2001). Fungal communities on submerged wood from streams in Brunei, Hong Kong, and Malaysia. Mycol. Res. 105, 1492–1501. doi: 10.1017/S095375620100507X
Hofmann, T. A., Kirschner, R., and Piepenbring, M. (2010). Phylogenetic relationships and new records of Asterinaceae (Dothideomycetes) from Panama. Fungal Divers. 43, 39–53. doi: 10.1007/s13225-010-0042-4
Hu, D. M., Cai, L., Chen, H., Bahkali, A. H., and Hyde, K. D. (2010). Four new freshwater fungi associated with submerged wood from Southwest Asia. Sydowia 62, 191–203.
Huang, S. K., Jeewon, R., Hyde, K. D., Bhat, J. D., Chomnunti, P., and Wen, T. C. (2018a). Beta-tubulin and Actin gene phylogeny supports Phaeoacremonium ovale as a new species from freshwater habitats in China. MycoKeys 41, 1–15. doi: 10.3897/mycokeys.41.27536
Huang, S. K., Jeewon, R., Hyde, K. D., Bhat, J. D., and Wen, T. C. (2018b). Novel taxa within Nectriaceae: Cosmosporella gen. nov. and Aquanectria sp. nov. from freshwater habitats in China. Crytogamie Mycol. 39, 169–192. doi: 10.7872/crym/v39.iss2.2018.169
Hughes, S. J. (1978). New Zealand Fungi 25, Miscellaneous species. N. Z. J. Bot. 16, 311–370. doi: 10.1080/0028825X.1978.10425143
Hyde, K. D., Fryar, S., Tian, Q., Bahkali, A. H., and Xu, J. C. (2016). Lignicolous freshwater fungi along a north–south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 19, 190–200. doi: 10.1016/j.funeco.2015.07.002
Hyde, K. D., Goh, T. K., and Steinke, T. O. (1998). Fungi on submerged wood in the Palmiet River, Durban, South Africa. S. Afr. J. Bot. 64, 151–162. doi: 10.1016/S0254-6299(15)30860-7
Hyde, K. D., Norphanphoun, C., Chen, J., Dissanayake, A. J., Doilom, M., Hongsanan, S., et al. (2018). Thailand’s amazing diversity: up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 93, 215–239. doi: 10.1007/s13225-018-0415-7
Hyde, K. D., Tennakoon, D. S., Jeewon, R., Bhat, D. J., Maharachchikumbura, S. S. N., and Rossi, W. (2019). Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 96, 1–242. doi: 10.1007/s13225-019-00429-2
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, D. J., Buyck, B., Cai, L., et al. (2015). The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Jayasiri, S. C., Hyde, K. D., Jones, E. B. G., and Persoh, D. (2018). Taxonomic novelties of hysteriform Dothideomycetes. Mycosphere 9, 803–837. doi: 10.5943/mycosphere/9/4/8
Jeewon, R., and Hyde, K. D. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Jeewon, R., Liew, E. C. Y., and Hyde, K. D. (2002). Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Mol. Phylogenet. Evol. 25, 378–392. doi: 10.1016/S1055-7903(02)00422-0
Jeewon, R., Liew, E. C. Y., Simpson, J. A., Hodgkiss, I. J., and Hyde, K. D. (2003). Phylogenetic significance of morphological characters in the taxonomy of Pestalotiopsis species. Mol. Phylogenet. Evol. 27, 372–383. doi: 10.1016/s1055-7903(03)00010-1
Jeewon, R., Wanasinghe, D. N., Rampadaruth, S., Puchooa, D., Zhou, L. G., Liu, A. R., et al. (2017). Nomenclatural and identification pitfalls of endophytic mycota based on DNA sequence analyses of ribosomal and protein genes phylogenetic markers: a taxonomic dead end? Mycosphere 8, 1802–1817. doi: 10.5943/mycosphere/8/10/7
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Liu, J. K., Hyde, K. D., Jones, E. B. G., Ariyawansa, H. A., Bhat, D. J., Boonmee, S., et al. (2015). Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 72, 1–197. doi: 10.1007/s13225-015-0324-y
Liu, J. K., Phookamsak, R., Doilom, M., Wikee, S., Li, Y. M., Ariyawansha, H., et al. (2012). Towards a natural classification of Botryosphaeriales. Fungal Divers. 57, 149–210. doi: 10.1007/s13225-012-0207-4
Liu, J. K., Phookamsak, R., Jones, E. B. G., Zhang, Y., Ko-Ko, T. W., Hu, H. L., et al. (2011). Astrosphaeriella is polyphyletic, with species in Fissuroma gen. nov., and Neoastrosphaeriella gen. nov. Fungal Divers. 51, 135–154. doi: 10.1007/s13225-011-0142-9
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lumbsch, H. T., Mangold, A., Lücking, R., García, M. A., and Martín, M. P. (2004). Phylogenetic position of the genera Nadvornikia and Pyrgillus (Ascomycota) based on molecular data. Acta Univ. Ups. Symb. Bot. 34, 10–17.
Luo, Z. L., Hyde, K. D., Liu, J. K., Bhat, D. J., Bao, D. F., Li, W. L., et al. (2018). Lignicolous freshwater fungi from China II: novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 9, 444–461. doi: 10.5943/mycosphere/9/3/2
Luo, Z. L., Hyde, K. D., Liu, J. K., Maharachchikumbur, S. S. N., Jeewon, R., Bao, D. F., et al. (2019). Freshwater Sordariomycetes. Fungal Divers. 99, 451–660. doi: 10.1007/s13225-019-00438-1
Lutzoni, F., Pagel, M., and Reeb, V. (2001). Major fungal lineages are derived from lichen symbiotic ancestors. Nature 411, 937–940. doi: 10.1038/35082053
Ma, J., Ma, L. G., Zhang, Y. D., Xia, J. W., and Zhang, X. G. (2012). Acrogenospora hainanensis sp. nov. and new records of microfungi from southern China. Mycotaxon 120, 59–66. doi: 10.5248/120.59
Mason, E. W. (1941). Annotated account of fungi received at the Imperial Mycological Institute. Mycol. Pap. 5, 101–144.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8.
Mugambi, G. K., and Huhndorf, S. M. (2009a). Molecular phylogenetics of Pleosporales: Melanommataceae and Lophiostomataceae re-circumscribed (Pleosporomycetidae, Dothideomycetes, Ascomycota). Stud. Mycol. 64, 103–121. doi: 10.3114/sim.2009.64.05
Mugambi, G. K., and Huhndorf, S. M. (2009b). Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi). Syst. Biodivers. 7, 453–464. doi: 10.1017/S147720000999020X
Muggiaa, L., Hafellnera, J., Wirtzb, N., Hawksworthc, D. L., and Grubea, M. (2008). The sterile microfilamentous lichenized fungi Cystocoleus ebeneus and Racodium rupestre are relatives of plant pathogens and clinically important dothidealean fungi. Mycol. Res. 112, 50–56. doi: 10.1016/j.mycres.2007.08.025
Nelsen, M. P., Lücking, R., Grube, M., Mbatchou, J. S., Muggia, L., RivasPlata, E., et al. (2009). Unravelling the phylogenetic relationships of lichenised fungi in Dothideomyceta. Stud. Mycol. 64, 135–144. doi: 10.3114/sim.2009.64.07
Nelsen, M. P., Lücking, R., Mbatchou, J. S., Andrew, C. J., Spielmann, A. A., and Lumbsch, H. T. (2011). New insights into relationships of lichen-forming Dothideomycetes. Fungal Divers. 51, 155–162. doi: 10.1007/s13225-011-0144-7
Nylander, J. A. A. (2004). MrModeltest v2 Program Distributed by the Author. Uppsala: Uppsala University.
Pang, K. L., Hyde, K. D., Alias, S. A., Suetrong, S., Guo, S. Y., Idid, R., et al. (2013). Dyfrolomycetaceae, a new family in the Dothideomycetes, Ascomycota. Cryptogam. Mycol. 34, 223–232. doi: 10.7872/crym.v34.iss3.2013.223
Phillips, A. J. L., Alves, A., Pennycook, S. R., Johnston, P. R., Ramaley, A., Akulov, A., et al. (2008). Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia 21, 29–55. doi: 10.3767/003158508X340742
Phookamsak, R., Norphanphoun, C., Tanaka, K., Dai, D. Q., Luo, Z. L., Liu, J. K., et al. (2015). Towards a natural classification of Astrosphaeriella-like species; introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellopsis, gen. nov. Fungal Divers. 74, 143–197. doi: 10.1007/s13225-015-0352-7
Quaedvlieg, W., Binder, M., Groenewald, J. Z., Summerell, B. A., Carnegie, A. J., Burgess, T. I., et al. (2014). Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 33, 1–40. doi: 10.3767/003158514X681981
Raja, H. A., El-Elimat, T., Oberlies, N. H., Shearer, C. A., Miller, A. N., Tanaka, K., et al. (2015). Minutisphaerales (Dothideomycetes, Ascomycota): a new order of freshwater ascomycetes including a new family, Minutisphaeraceae, and two new species from North Carolina, USA. Mycologia 107, 845–862. doi: 10.3852/15-013
Raja, H. A., Oberlies, N. H., Figueroa, M., Tanaka, K., Hirayama, K., Hashimoto, A., et al. (2013). Freshwater ascomycetes: Minutisphaera (Dothideomycetes) revisited, including one new species from Japan. Mycologia 105, 959–976. doi: 10.3852/12-313
Rakeman, J. L., Lafe, U. B. K., Chen, Y. C., Honeycutt, R. J., and Cookson, B. T. (2005). Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J. Clin. Microbiol. 43, 3324–3333. doi: 10.1128/JCM.43.7.3324-3333.2005
Rambaut, A. (2014). FigTree 1.4.2. Avaialble at: http://tree.bio.ed.ac.uk/software/figtree (accessed July 9, 2014).
Rannala, B., and Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rossman, A. Y., Crous, P. W., Hyde, K. D., Hawksworth, D. L., Aptroot, A., Bezerra, J. L., et al. (2015). Recommended names for pleomorphic genera in Dothideomycetes. IMA Fungus 6, 507–523. doi: 10.5598/imafungus.2015.06.02.14
Schoch, C. L., Crous, P. W., Groenewald, J. Z., Boehm, E. W. A., Burgess, T. I., de Gruyter, J., et al. (2009). A class wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 64, 1–15. doi: 10.3114/sim.2009.64.01
Schoch, C. L., Shoemaker, R. A., Seifert, K. A., Hambleton, S., Spatafora, J. W., and Crous, P. W. (2006). A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98, 1041–1052. doi: 10.1080/15572536.2006.11832632
Spatafora, J. W., Owensby, C. A., Douhan, G. W., Boehm, E. W. A., and Schoch, C. L. (2012). Phylogenetic placement of the ectomycorrhizal genus Cenococcum in Gloniaceae (Dothideomycetes). Mycologia 104, 758–765. doi: 10.3852/11-233
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 75, 758–771. doi: 10.1080/10635150802429642
Stenroos, S., Laukka, T., Huhtinen, S., Döbbeler, P., Myllys, L., Syrjänen, K., et al. (2010). Multiple origins of symbioses between ascomycetes and bryophytes suggested by a five-gene phylogeny. Cladistics 26, 281–300. doi: 10.1111/j.1096-0031.2009.00284.x
Su, X. J., Luo, Z. L., Jeewon, R., Bhat, D. J., Bao, D. F., Li, W. L., et al. (2018). Morphology and multigene phylogeny reveal new genus and species of Torulaceae from freshwater habitats in northwestern Yunnan, China. Mycol. Prog. 17, 531–545. doi: 10.1007/s11557-018-1388-3
Suetrong, S., Sakayaroj, J., Phongpaichit, S., and Jones, E. B. G. (2010). Morphological and molecular characteristics of a poorly known marine ascomycete, Manglicola guatemalensis (Jahnulales: Pezizomycotina; Dothideomycetes, Incertae sedis): new lineage of marine ascomycetes. Mycologia 102, 83–92. doi: 10.3852/07-147
Suetrong, S., Schoch, C. L., Spatafora, J. W., Kohlmeyer, J., Volkmann-Kohlmeyer, B., Sakayaroj, J., et al. (2009). Molecular systematics of the marine Dothideomycetes. Stud. Mycol. 64, 155–173. doi: 10.3114/sim.2009.64.09
Swe, A., Jeewon, R., Pointing, S. B., and Hyde, K. D. (2009). Diversity and abundance of nematode-trapping fungi from decaying litter in terrestrial, freshwater and mangrove habitats. Biodivers. Conserv. 18, 1695–1714. doi: 10.1007/s10531-008-9553-7
Swofford, D. L. (2003). PAUP∗. Phylogenetic Analysis Using Parsimony (∗and other Methods). Version 4. Sunderland, MA: Sinauer Associates.
Tanaka, K., Hirayama, K., Yonezawa, H., Hatakeyama, S., Harada, Y., Sano, T., et al. (2009). Molecular taxonomy of bambusicolous fungi: Tetraplosphaeriaceae, a new pleosporalean family with Tetraploa-like anamorphs. Stud. Mycol. 64, 175–209. doi: 10.3114/sim.2009.64.10
Tang, A. M. C., Jeewon, R., and Hyde, K. D. (2007). Phylogenetic utility of protein (RPB2, B-tubulin) and ribosomal (18S, 28S) gene sequences in the systematics of Sordariomycetes (Ascomycota, Fungi). Antonie Van Leeuwenhoek 91, 327–349. doi: 10.1007/s10482-006-9120-8
Tang, A. M. C., Jeewon, R., and Hyde, K. D. (2009). A re-evaluation of the evolutionary relationships within the Xylariaceae based on ribosomal and protein-coding gene sequences. Fungal Divers. 34, 127–155.
Tangthirasunun, N., Silar, P., Bhat, D. J., Chukeatirote, E., Chuketriote, E., Wijayawarden, E. D. N. N., et al. (2014). Morphology and phylogeny of Pseudorobillarda eucalypti sp. nov., from Thailand. Phytotaxa 176, 251–259. doi: 10.11646/phytotaxa.176.1.24
Thomas, K. (1996). “Australian freshwater fungi,” in Introductory Volume to the Fungi (Part2). Fungi of Australian, ed. C. A. Grgurinovic (Canberra: Australian Biological Resources Study), 1–37.
Tsui, C. K. M., Hyde, K. D., and Hodgkiss, I. J. (2000). Biodiversity of fungi on submerged wood in Hong Kong streams. Aquat. Microb. Ecol. 21, 289–298. doi: 10.3354/ame021289
Vijaykrishna, D., and Hyde, K. D. (2006). Inter and intra stream variation of lignicolous freshwater fungi in tropical Australia. Fungal Divers. 21, 203–224.
Vijaykrishna, D., Jeewon, R., and Hyde, K. D. (2005). Fusoidispora aquatica: new freshwater ascomycetes from Hong Kong based on morphology and molecules. Sydowia 57, 267–280.
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. Res. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Voglmayr, H., and Jaklitsch, W. M. (2011). Molecular data reveal high host specificity in the phylogenetically isolated genus Massaria (Ascomycota, Massariaceae). Fungal Divers. 46, 133–170. doi: 10.1007/s13225-010-0078-5
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide To Methods And Applications, eds G. M. Innis, D. Shinsky, and T. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Wijayawardene, N. N., Crous, P. W., Kirk, P. M., Hawksworth, D. L., Boonmee, S., Braun, U., et al. (2014). Naming and outline of Dothideomycetes–2014 including proposals for the protection or suppression of generic names. Fungal Divers. 69, 1–55. doi: 10.1007/s13225-014-0309-2
Wijayawardene, N. N., Hyde, K. D., Al-Ani, L. K. T., Tedersoo, L., Haelewaters, D., Rajeshkumar, K. C., et al. (2020). Outline of fungi and fungal-like taxa. Mycosphere 11, 1060–1456. doi: 10.5943/mycosphere/11/1/8
Wong, K. M. K., Goh, T. K., Hodgkiss, I. J., Hyde, K. D., Ranghoo, V. M., Tsui, C. K. M., et al. (1998). Role of fungi in freshwater ecosystems. Biodivers Conserv. 7, 1187–1206. doi: 10.1023/A:1008883716975
Yang, J., Liu, J. K., Hyde, K. D., Jones, E. B. G., and Liu, Z. Y. (2017). Two new species in Fuscosporellaceae from freshwater habitat in Thailand. Mycosphere 8, 1893–1903.
Zhang, Y., Jeewon, R., Fournier, J., and Hyde, K. D. (2008). Multi-gene phylogeny and morpho-taxonomy of Amniculicola lignicola: novel freshwater fungus from France and its relationships to the Pleosporales. Fungal Biol. 112, 1186–1194. doi: 10.1016/j.mycres.2008.04.004
Keywords: 7 new species, Acrogenosporaceae, molecular analyses, phylogeny, taxonomy
Citation: Bao D-F, McKenzie EHC, Bhat DJ, Hyde KD, Luo Z-L, Shen H-W and Su H-Y (2020) Acrogenospora (Acrogenosporaceae, Minutisphaerales) Appears to Be a Very Diverse Genus. Front. Microbiol. 11:1606. doi: 10.3389/fmicb.2020.01606
Received: 28 April 2020; Accepted: 19 June 2020;
Published: 24 July 2020.
Edited by:
Rajesh Jeewon, University of Mauritius, MauritiusReviewed by:
Huzefa A. Raja, University of North Carolina at Greensboro, United StatesAndrey M. Yurkov, German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), Germany
Copyright © 2020 Bao, McKenzie, Bhat, Hyde, Luo, Shen and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zong-Long Luo, bHVvem9uZ2xvbmdmdW5naUBxcS5jb20=; Hong-Yan Su, c3Vob25neWFuMTZAMTYzLmNvbQ==
 Dan-Feng Bao
Dan-Feng Bao Eric H. C. McKenzie4
Eric H. C. McKenzie4 Kevin D. Hyde
Kevin D. Hyde Zong-Long Luo
Zong-Long Luo Hong-Yan Su
Hong-Yan Su