
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 July 2020
Sec. Microbe and Virus Interactions with Plants
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01601
 Xuyan Mo1†
Xuyan Mo1† Liyuan Zhang2,3†
Liyuan Zhang2,3† Yan Liu1
Yan Liu1 Xuan Wang1
Xuan Wang1 Jiaqi Bai1
Jiaqi Bai1 Kai Lu2
Kai Lu2 Shenshen Zou2,3
Shenshen Zou2,3 Hansong Dong1,2,3*
Hansong Dong1,2,3* Lei Chen2,3*
Lei Chen2,3*Type III (T3) proteic effectors occupy most of the virulence determinants in eukaryote-pathogenic Gram-negative bacteria. During infection, bacteria may deploy a nanomachinery called translocon to deliver T3 effectors into host cells, wherein the effectors fulfill their pathological functions. T3 translocon is hypothetically assembled by bacterial translocators, which have been identified as one hydrophilic and two hydrophobic proteins in animal-pathogenic bacteria but remain unclear in plant pathogens. Now we characterize Hpa2, HrpF, and XopN proteins as concomitant T3 translocators in rice bacterial blight pathogen by analyzing pathological consequences of single, double, and triple gene knockout or genetic complementation. Based on these genetic analyses, Hpa2, HrpF, and XopN accordingly contribute to 46.9, 60.3, and 69.8% proportions of bacterial virulence on a susceptible rice variety. Virulence performances of Hpa2, HrpF, and XopN were attributed to their functions in essentially mediating from-bacteria-into-rice-cell translocation of PthXo1, the bacterial T3 effector characteristic of transcription factors targeting plant genes. On average, 61, 62, and 71% of PthXo1 translocation are provided correspondingly by Hpa2, HrpF, and XopN, while they cooperate to support PthXo1 translocation at a greater-than-95% extent. As a result, rice disease-susceptibility gene SWEET11, which is the regulatory target of PthXo1, is activated to confer bacterial virulence and induce the leaf blight disease in rice. Furthermore, the three translocators also undergo translocation, but only XopN is highly translocated to suppress rice defense responses, suggesting that different components of a T3 translocon deploy distinct virulence mechanisms in addition to the common function in mediating bacterial effector translocation.
Many pathogenicity-determinative proteic effectors of Gram-negative bacteria, which are plant (Alfano and Collmer, 2004; Nissinen et al., 2007; White et al., 2009), or animal (Chatterjee et al., 2013; Domingues et al., 2016; Piscatelli et al., 2016) pathogens, are secreted by the bacterial type III (T3) secretion system. Subsequently, effectors must be translocated from bacterial cells into eukaryotic host cells in order to fulfill a virulent role in susceptible hosts and an avirulent role in resistant hosts (Bogdanove and Voytas, 2011; Zhang et al., 2015; Chen et al., 2017). T3 effector translocation follows the translocon-dependent or -independent model depending on variations in the genetic repertoire of bacterial T3 translocators (Julie and Guy, 2016; Shanmugam and Dalbey, 2019; Zhang et al., 2019a). The translocon-independent model emphasizes endocytosis of effectors through direct interaction with recognizing compounds situated in plasma membranes (PMs) of eukaryotic cells (Rüter et al., 2010; Santi-Rocca and Blanchard, 2017). The translocon-dependent model is conceived as a nanomachinery called “T3 translocon” (Büttner and Bonas, 2002), which is hypothetically formed by interactions of bacterial T3 translocators with each other and with specific recognizing constituents (Ji and Dong, 2015b; Büttner, 2016), both lipids (Büttner et al., 2002; Haapalainen et al., 2011), and proteins (Oh and Beer, 2007; Li et al., 2015, 2019), associated with eukaryotic PMs. In the translocon-dependent model, recognition of the hydrophilic translocator by a PM constituent is the first step toward the translocon assembly (Goure et al., 2004; Mueller et al., 2008). Lipid binding of hydrophobic translocators finalize the translocon with an inner channel to accommodate effector translocation (Mueller et al., 2008; Büttner, 2012; Ji and Dong, 2015b; Zhang et al., 2019a).
In relevance, a big problem is the paucity in knowledge about the T3-translocon composition or the number of T3 translocators that plant-pathogenic bacteria must possess (Ji and Dong, 2015a; Scheibner et al., 2017; Zhang et al., 2019b). It has been determined that T3 translocators of animal-pathogenic bacteria include one hydrophilic protein and two hydrophobic proteins (Ji and Dong, 2015b; Büttner, 2016). Whether a similar repertoire exists in plant-pathogenic bacteria remains unknown. Plant-pathogenic bacteria occupy more than 100 species in the current classification, but only 5 species have been studied in terms of T3 translocators. The first identified T3 translocator was the atypical hydrophobic protein HrpF in Xanthomonas campestris pv. vesicatoria (Xcv), the bacterial spot pathogen of pepper (Büttner et al., 2002; Noël et al., 2002). Then, HrpF homologs designated as PopF1 and PopF2 were shown to be concomitant translocators of the transcription activator-like effectors (TALE) AvrA in Ralstonia solanacearum, the pathogen that causes bacterial wilt in a variety of plant species (Meyer et al., 2006). The same pathogen has a different T3-translocator called PopA (Racape et al., 2005), one of harpin-group hydrophilic proteins that function as T3 accessory components (Choi et al., 2013; Ji and Dong, 2015b). Later, T3 translocators were characterized as four harpins (HrpK1, HrpZ1, HrpW1, and HopAK1) in Pseudomonas syringae pv. tomato (Pst), which causes bacterial speck in a variety of plant species (Kvitko et al., 2007), and the harpin HrpNEa in Erwinia amylovora, the bacterial pathogen that causes fire blight in rosaceous plants (Bocsanczy et al., 2017, 2008). Subsequently, HrpF and the harpin protein Hpa2 were found to be concomitantly required for T3 effector translocation from the rice bacterial stripe pathogen X. oryzae pv. oryzicola into cells of the host plant and the non-host tobacco (Li et al., 2011). To date, however, none of plant-pathogenic bacteria has been characterized with respect to the T3 translocon composition. In other words, it is unclear at present how many T3 translocators must be produced to ensure the translocation of T3 effectors by an infecting community (species, subspecies, or pathovar) of plant-pathogenic bacteria.
We have extensively studied pathological and physiological functions of the harpin protein Hpa1 from the rice bacterial pathogen Xanthomonas oryzae (Peng et al., 2004; Liu et al., 2006; Chen et al., 2008a, b; Sang et al., 2012; Li et al., 2013, 2014, 2015, 2019; Ji and Dong, 2015a, b; Dong et al., 2016; Wang et al., 2018; Zhang et al., 2019a). We previously characterized Hpa1 as a hydrophilic protein (Chen et al., 2008a) and recently demonstrated its role in T3 effector translocation from X. oryzae pv. oryzae (Xoo), the pathogen that causes bacterial blight in rice, into the host plant cells (Wang et al., 2018). Like all bacteria in the Xanthomonas genus, Xoo causes disease by the virulence role of T3 effectors, either TALEs, including Avr and Pth proteins, or non-TALEs, mainly Xanthomonas outer proteins (Xops) as nominated with the bacterial genus landmark (White et al., 2009; Bogdanove and Voytas, 2011). In general, TALEs and non-TALEs are secreted along with harpins by the T3 system in a chronological pattern (Roden et al., 2004; Wang et al., 2018) and then translocated into plant cells to play a virulent or avirulent role depending on plant varieties (Büttner, 2016). Moreover, Xoo is the most devastating bacterial pathogen of rice in China and other parts of East Asia, Southeast-Asian countries like Philippine, United States and other parts of North America, Oceanian countries including Australia, and West Africa as well (Mew, 1987). Also, Xoo is a model of plant bacterial pathogen used by the plant pathology community (Niño-Liu et al., 2006; Mansfield et al., 2012).
We use this bacterial model to determine the T3 translocator composition in plant bacterial pathogen. The major attempt is to know whether analogs of both hydrophobic and hydrophilic translocators already identified in animal-pathogenic bacteria (Büttner, 2012; Ji and Dong, 2015b) are required for plant-pathogenic bacteria to translocate their T3 effectors. By such studies, we will be able to judge how many translocators are required for the translocation of a T3-effector from a species of plant bacterial pathogen or a pathovar of the bacterial species. We have demonstrated that the function of Hpa1 as a T3 translocator essentially contributes to virulence of the Xoo strain PXO99A in the susceptible japonica rice variety Nipponbare (Wang et al., 2018). Another pivotal determinant of PXO99A virulence is the TALE PthXo1 (Yang et al., 2006). In Nipponbare, PthXo1 supports the bacterial virulence by activating the host susceptibility gene, OsSWEET11, synonym Os8N3 (Yang et al., 2006), which encodes a sugar transporter protein (Chen et al., 2010). Thus, the production and translocation of PthXo1 has the function of supporting sugar secretion from rice cells to provide potential nutrition for bacterial multiplication in the apoplastic space (Chen et al., 2010). In PXO99A-infected Nipponbare plants, Hpa1 serves as a translocator for PthXo1 and this function is indispensable for PthXo1 to activate OsSWEET11 expression (Wang et al., 2018).
The present study was devised to assess the possible role of Hpa2 and HrpF in PthXo1 translocation from PXO99A bacteria into rice cells because both proteins were previously identified as T3 translocators in other plant-pathogenic bacteria (Büttner et al., 2002; Li et al., 2011; Scheibner et al., 2017). Meanwhile, XopN was also considered due to its structural and functional characteristics. XopN is a pluripotent effector present in different Xanthomonas species, has the structural features of molecular adaptors, and therefore confers virulence by interacting with immunity-relevant proteins in plants (Roden et al., 2004; Taylor et al., 2012). After secreted by Xcv in infecting tomato, XopN localizes to the PM-cytoplasm interface, whereon XopN interacts with the plant receptor-like protein kinase TARK1 (Kim et al., 2009), and 14-3-3 proteins (Taylor et al., 2012) that also function as molecular adaptors (Camoni et al., 2018). As the PM localization is a common characteristic of T3 translocators (Mueller et al., 2008; Ji and Dong, 2015b; Zhang et al., 2019a), XopN is involved in the present study. We present evidence that the three proteins (Hpa2, HrpF, and XopN) secreted by PXO99A in infecting Nipponbare plants contribute to different proportions of PthXo1 translocation. We further show that XopN executes an additional function, inducing marked suppression of rice defense responses following a substantial translocation into rice cells.
In order to assess the individual roles of Hpa2, HrpF, and XopN in PXO99A-bacterial virulence on Nipponbare plants, virulence levels were compared when the hpa2, hrpF, and xopN genes were present canonically in the PXO99A genome, individually deleted from and then backfilled to the genome. Involved experimental operations were performed by conventional procedures (Yang et al., 2006; Wang et al., 2018). Gene knockout by double-crossover homologous recombination (Song and Yang, 2010) created the bacterial mutants ΔHpa2, ΔHrpF, and ΔXopN, respectively, while bringing the wild-type (WT) single genes back to the corresponding mutants resulted in the generation of genetically complementing strains Δhpa2/hpa2, ΔhrpF/hrpF, and ΔxopN/xopN (Supplementary Table 1). To assess the effects of Hpa2, HrpF, and XopN on bacterial multiplication, the different bacterial strains were subjected to 20-h culture in nutrient broth (NA) liquid medium. The gene-knockout mutants Δhpa2, ΔhrpF, and ΔxopN showed to be significantly (P < 0.001) inferior to the WT stain in bacterial populations, whereas, genetic complementation restored these mutants to the WT in extents of multiplication in the medium (Figure 1A). Evidently, Hpa2, HrpF, and XopN are necessary for full multiplication of bacteria under the culturing condition. Moreover, Hpa2, HrpF, and XopN provided substantial contributions to bacterial virulence, which was evaluated by using two recognized criteria: populations of antibiotics-labeled bacteria (Supplementary Table 1) propagated within inoculated leaves of the plants and severities of blight symptoms subsequently formed on the leaves.
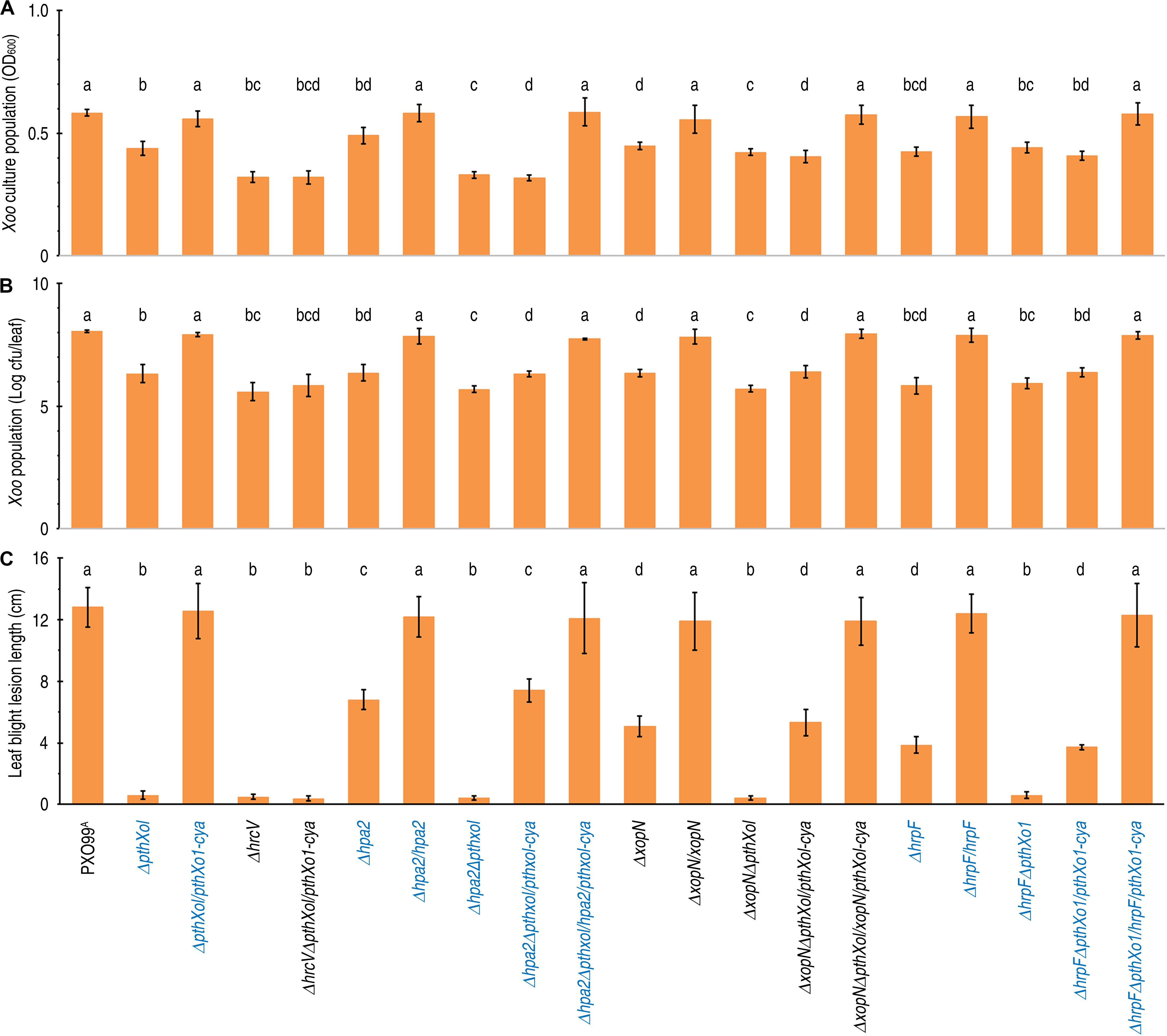
Figure 1. The effects of hpa1, hpa2, hrpF, and hrcV on Xoo bacterial multiplication in culture and virulence on the susceptible rice variety Nipponbare. (A–C) The Xoo WT strain PXO99A and its mutants generated by deleting the corresponding genes were tested in parallel experiments. Data shown are mean values ± standard deviation (SD) estimates. Number of biological repeats (n) = 6 independent experiments in (A); n = 30 inoculated leaves from 15 plants tested in 3 independent experiments in (B) and (C). Different letters on bar graphs indicate significant differences of data obtained from the experiments performed on the different bacterial strains (ANOVA and Duncan’s test, P < 0.001 in (A) and (C), P < 0.005 in B). (A) Bacterial populations of the different Xoo strains in a liquid medium after 20-h incubation. (B) Populations of bacteria multiplied in leaves. Leaves of 30-day-old Nipponbare rice seedlings were inoculated by leaf-top clipping with clinical scissors immediately dipped in the corresponding bacterial suspensions of the Xoo strains. Three days later, bacterial population in leaf tissues were determined. (C) Lesion length of bacterial blight on leaves 12 dpi.
To setup bacterial virulence assays, two expanded leaves of 30-day-old Nipponbare seedlings were inoculated by leaf-top clipping with clinical scissors immediately dipped in a bacterial suspension of the bacterial WT and recombinant strains. We first compared the WT strain PXO99A, single-gene-knockout mutants (Δhpa2, ΔhrpF, and ΔxopN), and subsequently complemented strains (Δhpa2/hpa2, ΔhrpF/hrpF, and ΔxopN/xopN) in terms of the two criteria for virulence assessment. When determined at the 3rd day post-inoculation (dpi), the bacterial WT and complementing strains displayed regular multiplication in leaf tissues, gaining bacterial populations around 108 colony formation unit (cfu) per leaf, but the Δhpa2, ΔhrpF, and ΔxopN mutants incurred significant reductions in cfu/leaf values (Figure 1B). In the subsequent 10 days, the blight symptoms developed in inoculated leaves with great variations in the visible severities as caused by the different bacterial genotypes, in which the Δhpa2, ΔhrpF, and ΔxopN mutants were much less aggressive than the WT and complementing strains (Figure 2). This difference was confirmed by digitizing the disease severity as leaf blight lesion length, which was significantly (P < 0.005) shorter in leaves inoculated with the bacterial mutant Δhpa2, ΔhrpF, or ΔxopN in contrast to the WT and complementing strains (Figure 1B). On average, 46.9, 60.3, and 69.8% reductions in leaf blight lesion length were caused by the corresponding gene deletion of Δhpa2, ΔhrpF, and ΔxopN from the PXP99A genome. These data suggest that Hpa2, HrpF, and XopN are integral virulence components of PXO99A, contributing to different proportions of PXO99A-bacterial virulence on the susceptible rice variety.
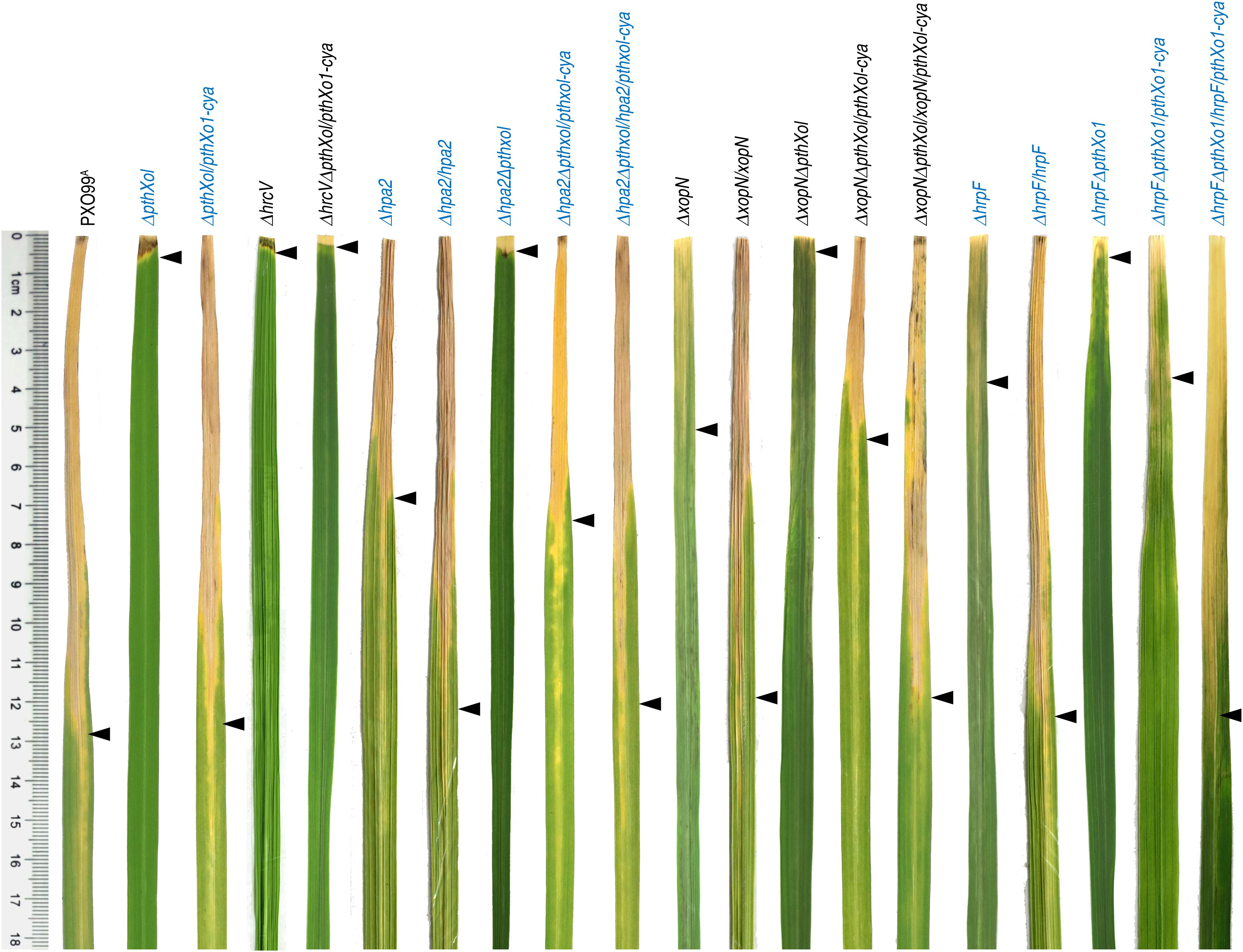
Figure 2. Bacterial blight symptoms on leaves of rice plants inoculated separately with PXO99A and mutants related to hpa1, hpa2, hrpF, and hrcV. Inoculation was performed on 30-day-old Nipponbare rice seedlings by using the leaf-clipping method. 12 h later, inoculated leaves were excised, spread on papers by the aid of sellotapes, and photographed. Leaf images were aligned, brightness/contrast was adjusted by a uniform extent, and background shadings were eliminated by using the magic wand and cut-off tools in Photoshop. Each leaf photo represents 30 leaves of 15 plants assessed in 3 independent experiment.
We looked for functional connections of Hpa2, HrpF, and XopN, which differentially contribute to PXO99A virulence on the rice variety Nipponbare (Figures 1, 2), with the bacterial TALE PthXo1, which determines the bacterial virulence (Yang et al., 2006) following secretion by the bacterial T3 pathway and translocation into Nipponbare cells (Li et al., 2019). Thus, we analyzed the effects of Hpa2, HrpF, and XopN on PthXo1 secretion and translocation. Both types of the molecular trafficking were analyzed by using calmodulin-dependent adenylate cyclase (Cya), a widely employed bacterial secretion reporter under culture conditions and a eukaryotic cytoplasmic import marker in plant and animal tests (Chakravarthy et al., 2017). We verified that the fused Cya did not affect the virulent role of PthXo1 in Nipponbare plants following inoculation by the leaf-top clipping with a bacterial suspension of the recombinant bacterial strain ΔpthXo1/pthXo1-cya in contrast to the WT strain PXO99A. Instead, ΔpthXo1/pthXo1-cya resembled PXO99A in multiplication within inoculated leaf tissues (Figure 1B) to cause leaf blight (Figure 2) with an equivalent severity shown as lesion length (Figure 1C). Immunoblotting analysis indicated that PthXo1-Cya was secreted by the bacterial T3 secretion pathway in the presence of HrcV (Figure 3), an inner membrane protein essential for substrate docking into the T3 system in Xoo (Sugio et al., 2005). In contrast to PthXo1, β-lactamase used as a lysis control was not secreted, by contrast, it remained inside bacterial cells no matter whether hrcV was present or absent (Figure 3), confirming the specificity in PthXo1 secretion by the T3 secretion pathway. Meanwhile, PthXo1-Cya was secreted equally well whether hpa2, hrpF, and xopN were deleted or remained in the PXO99A genome, suggesting that none of Hpa2, HrpF, and XopN is required for the effector secretion.
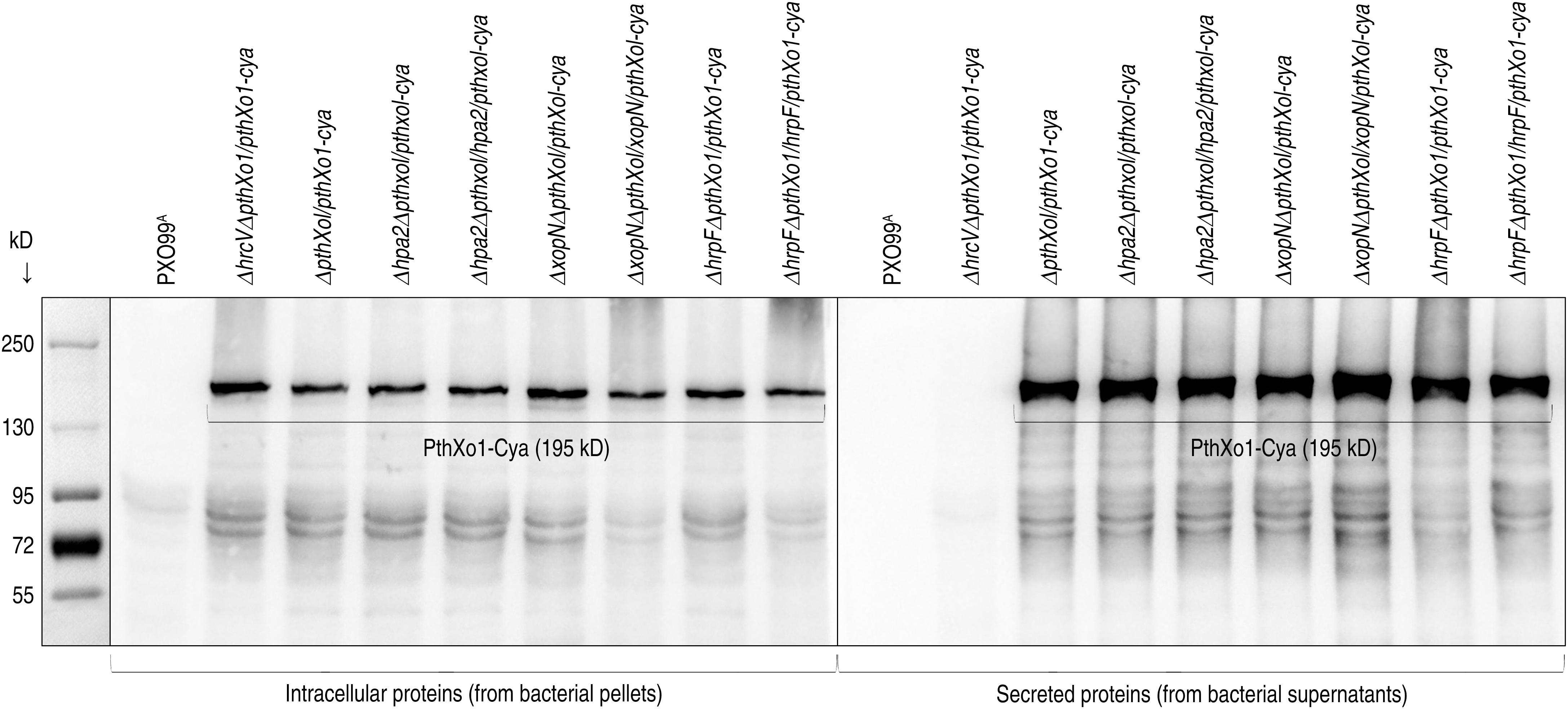
Figure 3. PthXo1 secretion by PXO99A and mutants defected in hpa1, hpa2, hrpF, or hrcV. The PthXo1 protein was fused to a Cya sequence and introduced in the fusion form into bacteria of the PXO99A mutants. Bacterial cultures of PXO99A and the mutants were used to isolate soluble proteins. Protein samples were analyzed by immunoblotting with the specific antibody against Cya.
In the Cya reporter assay using rice leaves inoculated by the leaf-center-infiltrating method as did usually (Kvitko et al., 2007; Chakravarthy et al., 2017; Li et al., 2019), Hpa2, HrpF, and XopN were found to partake in PthXo1 translocation from PXO99A-bacterial cells into the cytosol of Nipponbare cells. To initiate the assay, two elder leaves of 14-day-old Nipponbare seedlings were infiltrated with every bacterial suspension of pertinent Xoo strains. They were PXO99A, ΔpthXo1/pthXo1-cya, ΔhrcVΔpthXo1/pthXo1-cya, Δhpa2ΔpthXo1/pthXo1-cya, Δhpa2ΔpthXo1/hpa2/pthXo1-cya, ΔxopNΔpthXo1/pthXo1-cya, ΔxopNΔpthXo1/xopN/pthXo1-cya, ΔhrpFΔpthXo1/pthXo1-cya, and ΔhrpFΔpthXol/hrpF/pthXol-cya. After 12 h post-inoculation (hpi), leaves were sampled and used in determinations of TAL effector translocation of the in planta Xoo populations (Figure 4A) and PthXo1 translocation (Figure 4B). At 12 hpi, bacterial populations of the different strains in leaves were similar regardless of hpa1, hrpF, or xopN deletion (Figure 4A). In this case, the possible effect of bacterial population on quantitative changes in PthXo1 translocation from the different bacterial strains could be excluded. PthXo1 translocation was verified by immunoblotting of soluble proteins isolated from inoculated rice leaves and hybridized with αCya, the specific anti-Cya antibody (Figure 4B, inset). The Cya reporter system allows for accurate quantification of an effector moved into eukaryotic cells based on the cytosolic concentration of cAMP as an exclusive product of the effector-Cya activity (Chakravarthy et al., 2017). Thus, levels of PthXo1-Cya translocation were quantified as cAMP concentrations during the effector-Cya activity in leaf cells (Figure 4B, bar graph). In both analyses, little translocation was found with ΔhrcV/pthXo1-cya due to the absence of HrcV-mediated secretion (Figure 4).
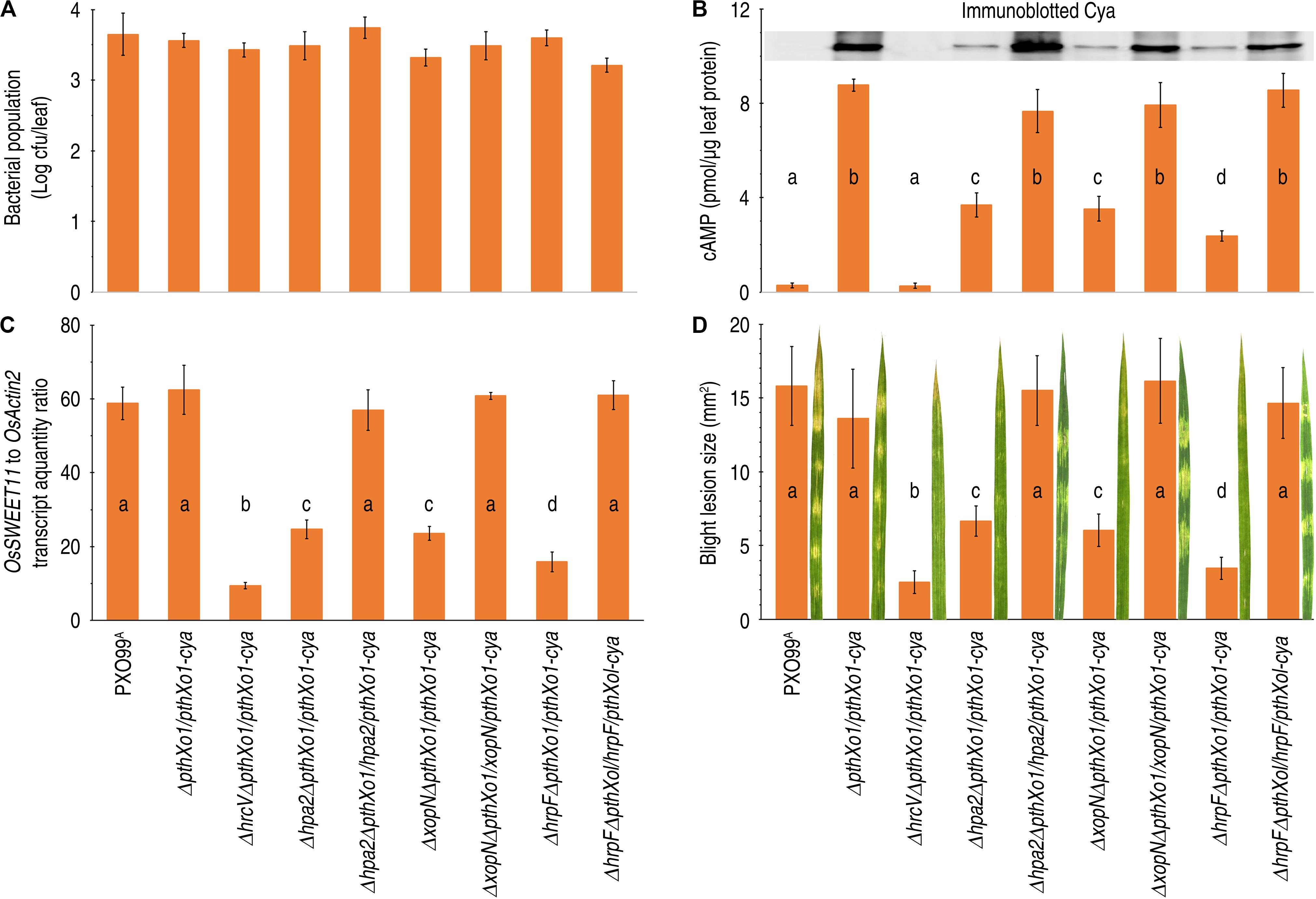
Figure 4. The effects of hpa1, hpa2, and hrpF on PthXo1 translocation and bacterial virulence. (A–D) Leaves of 14-day-old Nipponbare rice seedlings were inoculated by infiltration with bacterial suspensions of PXO99A and mutants defected in hpa1, hpa2, and hrpF, respectively. Inoculated leaves were excised and used in the different assays. Quantitative data are given as the means ± SDs. Different letters on bar graphs indicate significant differences of data obtained from the experiments performed on the different bacterial strains (ANOVA and Duncan’s test, P < 0.001, n = 3 independent experiment each involving 30 leaves of 15 plants). (A) Bacterial population in leaves 12 hpi. (B) PthXo1-Cya translocation levels shown as cytoplasmic cAMP concentrations in leaves 12 hpi. Inset shows immunoblotting of leaf proteins hybridized in the blot with specific anti-Cya antibody. (C) Rice OsSWEET11 gene expression in leaves 48 hpi. Gene expression was analyzed by RT-qPCR using the constitutively expressed OsActin2 gene as a reference. (D) Leaf blight severities and leaf images assessed at 7 dpi.
Immunoblotting clearly showed the αCya-hybridized protein bands indicating translocation of the PthXo1-Cya fusion protein from PXO99A cells into the cytosol of Nipponbare rice cells (Figure 4B, inset). However, the αCya-hybridized protein bands were detected from the Nipponbare plants inoculated with the bacterial strains ΔpthXo1/pthXo1-cya, Δhpa2ΔpthXo1/hpa2/pthXo1-cya, ΔxopNΔpthXo1/xopN/pthXo1-cya, and ΔhrpFΔpthXol/hrpF/pthXol-cya, which contain hpa2, hrpF, and xopN, respectively. In the case of genetic modification that had deleted any of the three genes, as did in the bacterial strains Δhpa2ΔpthXo1/pthXo1-cya, ΔxopNΔpthXo1/pthXo1-cya, or ΔhrpFΔpthXo1/pthXo1-cya, αCya-hybridized protein bands were not present in the protein blot (Figure 4B, inset). In the quantitative assay, high concentrations of cAMP were detected in leaves inoculated with the bacterial strains ΔpthXo1/pthXo1-cya, Δhpa2ΔpthXo1/hpa2/pthXo1-cya, ΔxopNΔpthXo1/xopN/pthXo1-cya, and ΔhrpFΔpthXol/hrpF/pthXol-cya, respectively. Cya activity was substantially decreased due to deletion of hpa1, hrpF, or xopN, as evidenced by a significant (P < 0.001) reduction in the cytoplasmic cAMP quantity in leaves inoculated with any of the Δhpa2ΔpthXo1/pthXo1-cya, ΔxopNΔpthXo1/pthXo1-cya, and ΔhrpFΔpthXo1/pthXo1-cya strains (Figure 4B, bar graph). Both qualitive and quantitative analysis results demonstrate that Hpa2, HrpF, and XopN share the concomitant function in PthXo1 translocation from bacteria into rice cells. Comparing cAMP concentrations in rice leaves inoculated with the bacterial strain ΔpthXo1/pthXo1-cya and the specific gene-knockout strains, Hpa2, HrpF, and XopN occupy average 60.5, 62.2, and 71.2% proportions of PthXo1 translocation under the condition in this study.
We assumed that the role of Hpa2, HrpF, and XopN in Pthxo1 translocation might cause OsSWEET11, the transcriptional target of PthXo1 (Yang et al., 2006), to be expressed in a manner of dependence on Hpa2, HrpF, and XopN present in the bacteria inoculated to rice plants. We examined this hypothesis by determining OsSWEET11 transcript quantities in Nipponbare leaves following inoculation by leaf infiltration with every bacterial suspension of the proper Xoo strains (Figure 4). Analyses by RT-qPCR, namely real-time reverse transcriptase (RT) polymerase chain reaction (PCR), indicated that OsSWEET11 was highly expressed in plants inoculated with the WT Xoo strain (PXO99A) or recombinant strains that concurrently contain pthxo1, hpa2, hrpF, and xopN (Figure 4C). RT-qPCR data also demonstrated that OsSWEET11 expression was significantly (P < 0.001) decreased in Nipponbare leaves inoculated with bacteria in which any of pthXo1, hpa2, hrpF, and xopN had been delete. Consistently, scoring leaf blight lesion size confirmed that recombinant Xoo strains that concurrently contain pthxo1, hpa2, hrpF, and xopN showed to be as aggressive as the WT strain in causing leaf blight symptoms, but the bacterial aggression incurred significant (P < 0.001) impairments by deleting any of these genes from the bacterial genome (Figure 4D). These results highlight the mechanistic sequence of pathological responses from the concomitant function of three translocators (Hpa2, HrpF, and XopN) in PthXo1 translocation, PthXo1-activated expression of OsSWEET11, and bacterial virulence performance with a result of leaf blight formation in the susceptible rice variety.
To elucidate whether Hpa2, HrpF, and XopN synergize in mediating PthXo1 translocation, we performed parallel experiments to compare the single, double, and triple gene-knockout PXO99A mutants with PXO99A and ΔpthXo1/pthXo1-cya in terms of PthXo1 translocation levels. Leaves of 14-day-old Nipponbare seedlings were inoculated by leaf infiltration with every bacterial suspension of the specific Xoo strains, in which PXO99A was used as a reference of background reading and ΔpthXo1/pthXo1-cya was assumed to provide the detectable maximal level of PthXo1 translocation. Based on cytoplasmic cAMP concentrations in leaves 12 hpi, PthXo1-Cya translocation incurred significant (P < 0.01) quantitative reductions in leaves of the seedlings inoculated with the single mutant Δhpa2ΔpthXo1/pthXo1-cya, ΔhrpFΔpthXo1/pthXo1-cya, or ΔXopNΔpthXo1/pthXo1-cya compared with ΔpthXo1/pthXo1-cya (Figure 5). Further decreases in quantities of translocated PthXo1-Cya were found in the plants inoculated with the double and triple gene-knockout mutants (Figure 5). On average, 61, 63, and 71% reductions in PthXo1-Cya translocation levels were caused by deleting the single genes hpa1, hrpF, and xopN, respectively, under background of ΔpthXo1/pthXo1-cya. Further reductions up to 27% and 30% were induced by double and triple gene knockout, respectively. Moreover, the triple mutant Δhpa2ΔhrpFΔxopNΔpthXo1/pthXo1-cya had the lowest amount of PthXo1-Cya translocation determined as cAMP content of 0.83 pmol/μg leaf protein, being 0.7% of the amount (cAMP 10.50 pmol/μg leaf protein) provided by ΔpthXo1/pthXo1-cya. Clearly, the concomitant presence of Hpa2, HrpF, and XopN is essential for the maximal translocation of PthoXo1, suggesting that the three translocators cooperate to mediate PthXol translocation from Xoo bacteria into rice cells.
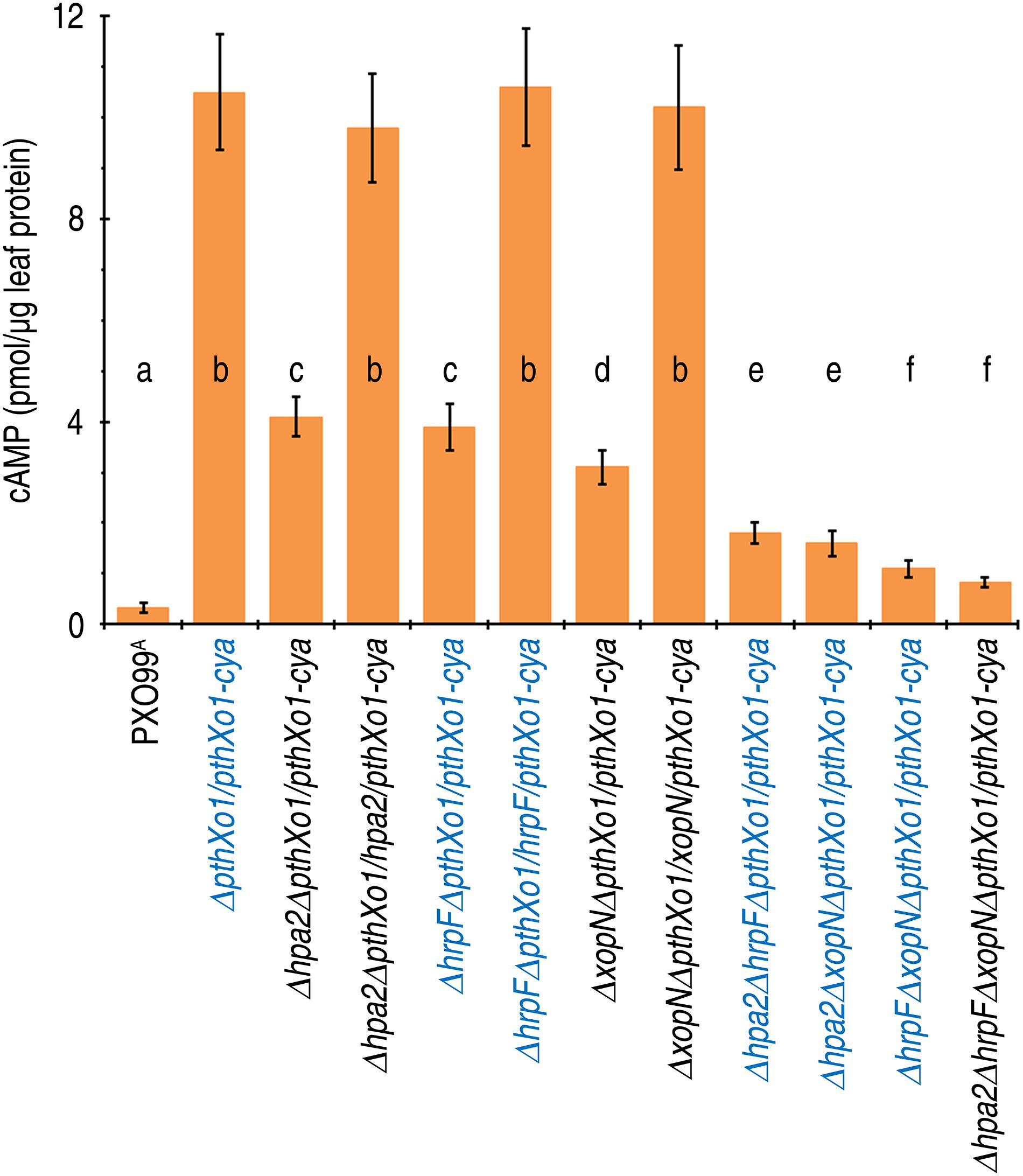
Figure 5. The cooperative effects of hpa1, hpa2, and hrpF on PthXo1 translocation levels quantified as cytoplasmic cAMP concentrations in rice leaves. Leaves of 14-day-old Nipponbare rice seedlings were inoculated by infiltration with bacterial suspensions of PXO99A and hpa1, hpa2, and hrpF single, double, and triple mutants, respectively. 12 h later, inoculated leaves were excised and used in measurements of cytoplasmic cAMP concentrations. Data shown are the mean values ± SDs. Different letters on bar graphs indicate significant differences of data obtained from the plants inoculated with the different bacterial strains (ANOVA and Duncan’s test, P < 0.001, n = 3 independent experiment each involving 30 leaves of 15 plants).
It is intriguing to know whether T3 translocators also undergo translocation as do T3 effectors. It is especially intriguing to know whether T3 translocators have additional pathological functions except for effector translocation. We addressed both questions by analyzing the correlation of bacterial virulence with secretion and translocation of the translocators from pertinent Xoo strains, including ΔhrcV/hpa2-cya, Δhpa2/hpa2-cya, ΔhrcV/xopN-cya, ΔxopN/xopN-cya, ΔhrcV/hrpF-cya, and ΔhrpF/hrpF-cya tested in comparison with PXO99A. By protein secretion analysis, we confirmed that Hap2-Cya, HrpF-Cya, and XopN-Cya were secreted by the bacterial T3 pathway with the requirement for HrcV (Figure 6). We further determined that Hap2-Cya, HrpF-Cya, and XopN-Cya underwent translocation from bacteria into rice cells (Figure 7A). In the assay, 30-day-old Nipponbare rice seedlings were inoculated by leaf-top clipping with clinical scissors dipped in every bacterial suspension of the pertinent Xoo strains. Based on cytoplasmic cAMP concentrations in inoculated rice leaves measured at 12 hpi, the three translocators were translocated by the bacteria whose hrcV gene was reserved and were translocated at different extents (Figure 7A). While Hap2-Cya and HrpF-Cya had small amounts of translocation (2.2 and 1.7 pmol cAMP per μg leaf protein, respectively), XopN-Cya was translocated at a significantly (P < 0.001) increased concentration (9.6 pmol cAMP per μg leaf protein), scored as 3.5-fold higher than Hpa1 and 4.6-fold higher than HrpF. By contrast, the three translocators were little translocated from the ΔhrcV/hpa2-cya, Δhpa2/hpa2-cya, and ΔhrcV/xopN-cya mutants generated by gene deletion and recombination under PXO99A background (Figure 7A). Meanwhile, the hrcV gene knockout highly arrested bacterial virulence, significantly (P < 0.001) decreased bacterial populations in leaves as determined at 3 dpi (Figure 7B), and significantly (P < 0.001) reduced lesion length of leaf blight as observed at 12 dpi (Figure 7C). When hrcV is present, however, ΔxopN/xopN-cya, Δhpa2/hpa2-cya, and ΔhrpF/hrpF-cya displayed similar levels of virulence, multiplied equally well in leaves (Figure 7B), and caused leaf blight with similar severities (Figure 7C). Furthermore, OsSWEET11 was expressed equally well in leaves pf plants inoculated with ΔxopN/xopN-cya, ΔhrcV/hrpF-cya, and ΔhrpF/hrpF-cya, respectively, (Figure 7D). In other words, levels of OsSWEET11 expression were similar (Figure 7D) although translocation levels of Hpa2-Cya and HrpF-Cya were small but the translocation amount of XopN-Cya was significantly (P < 0.001) higher (Figure 7A). Based on these results, we assumed that XopN might have more pathological roles than Hpa2 and HrpF in addition to the common function in mediating the bacterial effector translocation.
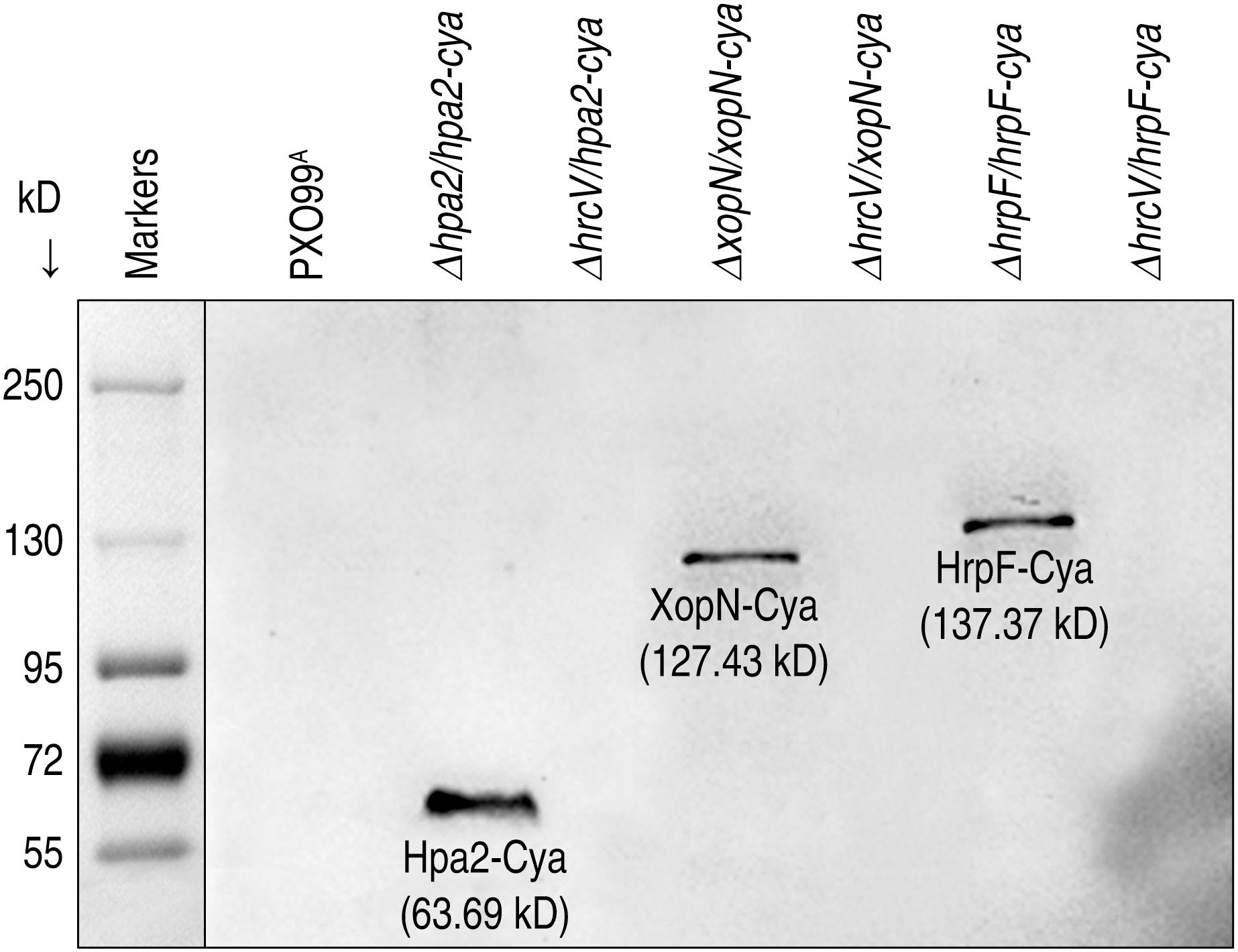
Figure 6. Hpa2, HrpF, and XopN translocation. Each of the three proteins was fused to a Cya sequence and introduced in the fusion form into bacteria of the PXO99A mutants. Bacterial cultures of PXO99A and the mutants were used to isolate soluble proteins. Protein samples were analyzed by immunoblotting with the specific antibody against Cya.
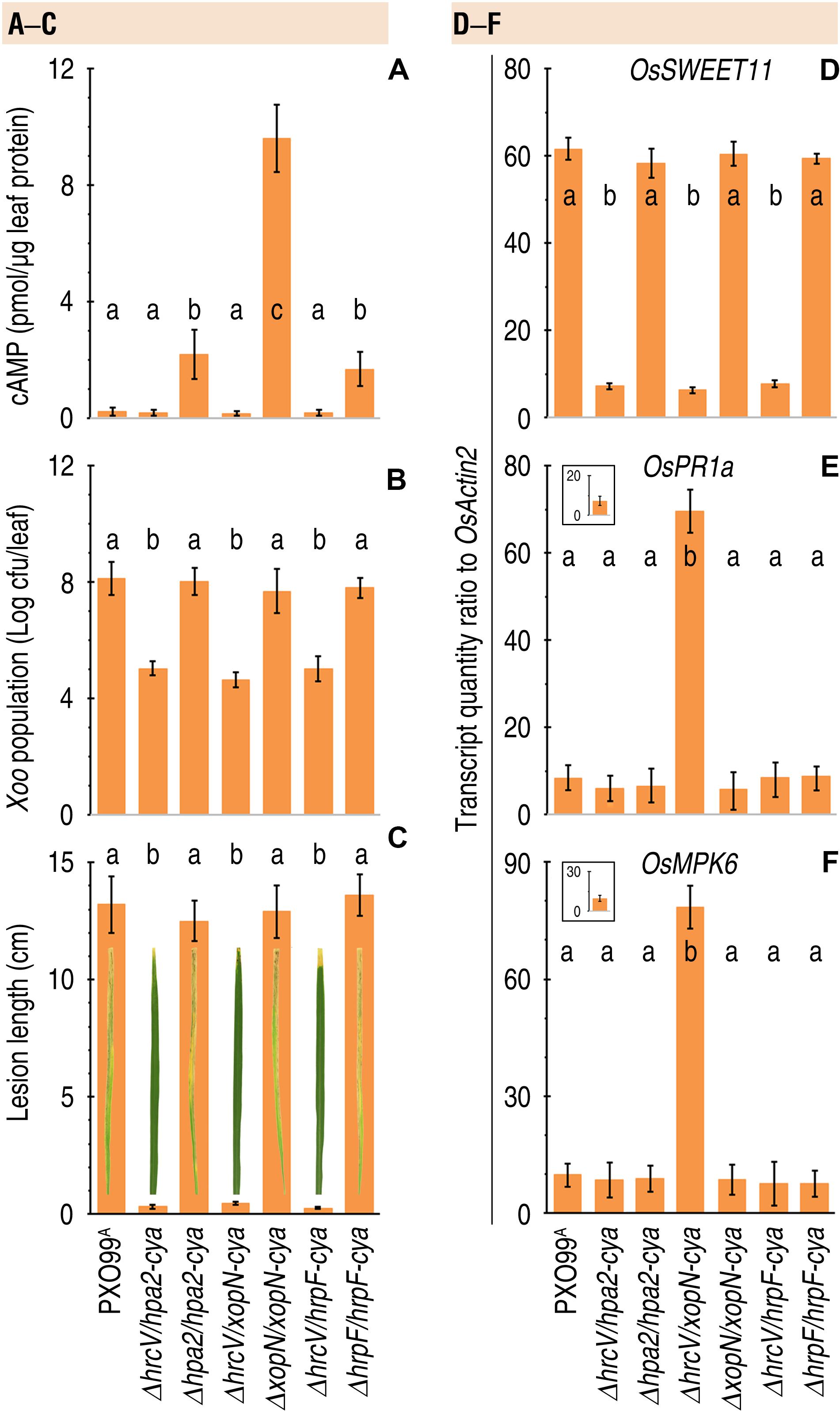
Figure 7. The coincident effects of Hpa2, HrpF, and XopN translocation on bacterial virulence and rice defense responses. (A–F) Leaves of 14-day-old Nipponbare rice seedlings were inoculated by infiltration with bacterial suspensions of PXO99A and the complementing strains with and without the hrcV gene, respectively. Inoculated leaves were excised and used in the different assays. Quantitative data are given as the means ± SDs. Different letters on bar graphs indicate significant differences of data obtained from the experiments performed on the different bacterial strains (ANOVA and Duncan’s test, P < 0.001, n = 3 independent experiment each involving 30 leaves of 15 plants). (A) Translocation levels of the Cya-fused proteins shown as cytoplasmic cAMP concentrations in leaves 12 hpi. (B) Bacterial population in leaves 3 dpi. (C) Leaf blight symptoms and severity qualification at 7 dpi. (D–F) Rice gene expression in leaves 48 hpi. Gene expression was analyzed by RT-qPCR using the constitutively expressed OsActin2 gene as a reference.
To validate this hypothesis, we elucidated whether defense responses occur in rice plants following translocation of the bacterial translocators. Considerable parts of plant defense responses are essentially subject to the ETI (effector-triggered immunity) and PTI (microbial pattern-triggered immunity) pathways (Kanyuka and Rudd, 2019). Therefore, we chose to analyze the foliar expression of OsPR1a and OsMPK6 genes, which are molecular markers of the ETI and PTI pathways, respectively, (Zhang et al., 2019b). In the assay, 30-day-old Nipponbare rice plants were divided into two groups. In the first group used as defense response gene expression control, plants remained free from inoculation. In the second group, plants were inoculated by leaf clipping with clinical scissors dipped separately in bacterial suspensions of Xoo strains PXO99A, ΔhrcV/hpa2-cya, Δhpa2/hpa2-cya, ΔhrcV/xopN-cya, ΔxopN/xopN-cya, ΔhrcV/hrpF-cya, and ΔhrpF/hrpF-cya. Subsequently, relative levels of OsPR1a and OsMPK6 expression in inoculated leaves were analyzed at 48 hpi. Low basal levels of OsPR1a (Figure 7E inset) and OsMPK6 (Figure 7F inset) expression were detected in leaves of plants that were not inoculated. In comparison, expression levels of both OsPR1a (Figure 7E) and OsMPK6 (Figure 7F) were highly increased in, and only in, the plants inoculated with the recombinant bacterial strain ΔxopN/xopN-cya. By contrast, the expression of OsPR1a (Figure 7E), and OsMPK6 (Figure 7F) were not evidently induced in the plants inoculated with PXO99A, ΔhrcV/hpa2-cya, Δhpa2/hpa2-cya, ΔhrcV/xopN-cya, ΔxopN/xopN-cya, ΔhrcV/hrpF-cya, and ΔhrpF/hrpF-cya, respectively. Instead, transcripts of both defense response genes remained around the basal levels in the plants following inoculation with any of the bacterial strains in which a functional XopN was present (Figures 7E,F main graphs vs. insets). Based on these results, we propose that XopN present in PXO99A acts to suppress defense responses in plants of the disease-susceptible rice variety under infection, but Hpa1 and HrpF do not have such a repressive effect on rice defense responses against Xoo infection.
Although the T3 translocon of eukaryote-pathogenic Gram-negative bacteria has been extensively studied in the past 20 years (Blocker et al., 2001; Büttner et al., 2002; Zhang et al., 2019a), characterization of the molecular mechanism that governs bacterial effector translocation is merely in the initial stage (Li et al., 2019; Picking and Barta, 2019). By browsing literatures (Table 1), we find that there was no study to demonstrate the composition of T3 translocon in different species of plant-pathogenic bacteria while infecting their host plants (Büttner, 2012; Ji and Dong, 2015b; Scheibner et al., 2017; Zhang et al., 2019a). Furthermore, in seven of eight previous reports that we have scrutinized with appreciation, bacterial effector translocation was measured by experiments performed on tobacco, the common non-host plant of almost all plant-pathogenic microbes, rather than host plants of the bacteria (Table 1). Evidently, looking for the composition of T3 translocon and demonstrating the functional connection between effector translocation and bacterial virulence are quite necessary especially for the study of plant-pathogenic bacteria.
We have identified Hpa2, HrpF, and XopN as concomitant translocators of the Xoo TALE PthXo1, which was well studied with respect to its function in determining virulence of the standard Xoo strain PXO99A on the susceptible variety Nipponbare (Yang et al., 2006; Wang et al., 2018; Bian et al., 2019; Li et al., 2019; Zhang et al., 2019b). Evidence that demonstrates the translocator role has been shown as quantitative changes of PthXo1 translocation in Nipponbare plants inoculated with the WT and genetically modified POX99A strains in the presence and absence of Hpa2, HrpF, and XopN, respectively. T3 translocators share the common functional characteristic of requirement for the translocation, rather than secretion, of bacterial effectors (Yang, 2017). In agreement with this criterion, Hpa2, HrpF, and XopN regulate the virulent role of PthXo1 by mediating its translocation from bacteria into rice cells, but the three translocators are not required for PthXo1 secretion through the bacterial T3 pathway. Shortly before this study, we have already characterized Hpa1 as a translocator of Xoo TALEs PthXo1 and AvrXa10 trafficking from the bacteria into rice cells, wherein PthXo1 plays a virulent role, but AvrXa10 executes its avirulent function (Wang et al., 2018; Bian et al., 2019). Therefore, Xoo possesses at least four T3-translocators that function concomitantly to facilitate effector translocation.
Our results detailed in this article highlight the classical question regarding the functional coordination of different components in the bacterial T3-translocator repertoire (Mueller et al., 2008; Büttner, 2012; Choi et al., 2013; Ji and Dong, 2015b). Multiple (known 3 or 4) T3-translocators present in an infecting community (a single species, subspecies, pathovar, or serovar) of eukaryote-pathogenic bacteria show to have overlapping and redundant functions (Kvitko et al., 2007; Mueller et al., 2008; Ji and Dong, 2015b). In Pst, for example, four harpins (HrpK1, HrpZ1, HrpW1, and HopAK1) serve as semiredudant concomitant translocators of the TALE AvrPto (Kvitko et al., 2007). Deleting any of the hrpK1, hrpZ1, hrpW1, and hopAK1 genes from the Pst genome causes a substantial reduction in AvrPto-Cya translocation, which is further diminished, but not eliminated, by the quadruple harpin gene polymutation (Kvitko et al., 2007). We show that Hpa2, HrpF, and XopN display a similar mode of functional redundancy. Deleting any of the hpa1, hrpF, and xopN genes from the PXO99A genome causes a substantial reduction in PthXo1 translocation, which is further diminished, but not eliminated, by the triple gene polymutation. The involved mechanism remains mysterious.
We propose that the molecular mechanism that governs the functional redundancy of multiple translocators in an infecting bacterial community is possibly related to the hypothetic regulatory modes of bacterial T3 effector translocation in the translocon-dependent or -independent manner. The translocon-dependent model emphasizes molecular interactions between bacterial T3 translocators and molecular interactions of T3 translocators with their receptors (Büttner and Bonas, 2002; Büttner et al., 2008; Büttner, 2012; Ji and Dong, 2015b), either lipids (Haapalainen et al., 2011; Li et al., 2011), or proteins (Oh and Beer, 2007; Li et al., 2015; Adam et al., 2017) situated in the eukaryotic PMs. The translocon-independent model hypothesizes that bacterial T3 effectors move into eukaryotic host cells by two routes (Zhang et al., 2019a). In the first, bacterial effectors characteristic of cell-penetrating peptide (RPP) make pore in the target PM (Scharnert et al., 2013; Rüter and Schmidt, 2017). The second route is the effector endocytosis through direct interaction with receptors located in eukaryotic PMs (Domingues et al., 2016). Such a molecular interaction may trigger the membrane trafficking mechanism (Allgood and Neunuebel, 2018) either by endoplasmic reticulum or vesicles (Wudick et al., 2015), providing a potential scheme for bacterial effector endocytosis (Cybulsky, 2017; Obacz et al., 2017).
Because PthXo1 does not belong to RPP, its translocation is not likely to use the pore formation mechanism. Instead, PthXo1 translocation needs typical T3 translocators Hpa1 (Wang et al., 2018), Hpa2 (Li et al., 2011), and HrpF (Büttner et al., 2002). Both Hpa1 (Chen et al., 2008a) and Hpa2 (Li et al., 2011) belong to hydrophilic proteins previously defined as T3 accessory proteins (Zhu et al., 2000; Choi et al., 2013). HrpF is an atypical hydrophobic protein, contains a hydrophobic group in the C-terminal sequence, but lacks hydrophilic groups in the N-terminal sequence (Büttner et al., 2002). HrpF is the T3 translocator first discovered in plant-pathogenic bacteria and is considered an essential component of the T3 translocon (Büttner et al., 2002; Scheibner et al., 2017). Therefore, it is very likely that PthXo1 is translocated in the translocon-dependent manner when the infecting bacteria can secrete all translocators required. Based on the functional redundancy of the concomitant translocators, we hypothesize that the translocon may deploy an adjustable mechanism to alter its functional levels according to variations in the translocator compositions. Such an adjustable mechanism is deployed to ensure effector translocation in the absence of any translocators, preventing the effector translocation from termination by loss-of-function mutation of any translocators. This hypothesis needs to be validated.
It is intriguing that T3 translocators also move into plant cells as do effectors as found so far in studies which have determined whether translocators undergo translocation (Kvitko et al., 2007; Mo et al., this study). In the current model, nevertheless, T3 translocators do not necessarily need to be translocated. Instead, they can fulfill their functions in mediating effector translocation if they interact each other according to the hierarchical sequence of secretion and meanwhile interact with the corresponding receptors situated in eukaryotic PMs (Büttner, 2012; Ji and Dong, 2015b). However, T3 translocators authentically undergo the transkingdom transport from bacteria into the cytosol of plant cells (Kvitko et al., 2007; Mo et al., this study). In this case, the translocators may execute more functions than mediating bacterial effector translocation. This notion has been partially validated by our finding about the dual functions of XopN in mediating PthXo1 translocation and in suppressing plant defense responses following the translocation by itself. At present, however, we do not have experimental evidence to show what responses, in addition to PthXo1 translocation, could be further induced by Hpa1, Hpa2, and HrpF translocated into rice cells. By contrast, it is convincing to propose that XopN is not only a T3 effector as previously defined, but also a T3 translocator as now appreciated. In conjecture, XopN executes the dual functions possibly relate to its trafficking route toward plant PMs (Kim et al., 2009). On the one hand, this subcellular localization satisfies the requirement for a T3 translocator to function by association with a specific receptor at eukaryotic PMs (Büttner, 2012; Ji and Dong, 2015b). On the other hand, following localization at the PM-cytoplasm interface, XopN interacts with TARK1 (Kim et al., 2009), and 14-3-3 proteins (Taylor et al., 2012) to induce suppression of the PTI signaling pathway (Taylor et al., 2012). Whether a similar signaling mechanism is employed by Xoo XopN translocated into rice cells remains to be studied.
Taken together, our findings in this study and previous works (Wang et al., 2018; Bian et al., 2019; Li et al., 2019; Zhang et al., 2019b) demonstrate that Hpa1, Hpa2, HrpF, and XopN are concomitant translocators of the TALE PthXo1 in the standard Xoo strain PXO99A while infecting the susceptible rice variety Nipponbare. We further show that in addition to mediate PthXo1 translocation, the three translocators themselves also move into rice cells, wherein XopN has a high quantity and induces suppression of rice defense responses. In the initial motivation, we did not intend to analyze whether the translocators function as specialists for PthXo1 only or generalists for more effectors. Instead, we have paid constant attentions to integral components of the T3 translocon and the assumed intricate process of the translocon assembly (Picking and Barta, 2019) particularly in plant-pathogenic bacteria (Ji and Dong, 2015b; Zhang et al., 2019a). Characterization of the translocon assembly has been a highly challengeable work for the scientific community. Identifying T3 translocators in Xoo is an indispensable step toward dissecting the bacterial T3 translocon assembly in the future.
Nipponbare seeds were initially provided by our colleague Professor Hongsheng Zhang (College of Agronomy, Nanjing Agricultural University) and were then reproduced and maintained in this lab. Seeds were germinated on filter papers immersed in sterile water in Petri dishes. After 3–5 days, the germinal seedlings were moved into 12-L pots (2–5 plants per pot) filled with a substrate containing peat, sand and vermiculite (1:1:1 v/v). Seeds were incubated and the plants were grown in environment-controlled chambers under 26 ± 1°C, 12-h light at 250 ± 50 μmol quanta/m2/s and a relative humidity of 85%.
Bacterial strains and plasmid vectors used and created in this study and information on antibiotic resistance and other elements are provided in Supplementary Table 1. Escherichia coli was grown at 37°C in Luria-Bertani broth (LB) or on LB agar (LA) plates with the appropriate antibiotics. Xoo strains were cultured at 28°C on nutrient broth (NB) or NA agar (NA) medium (Li et al., 2011). Bacteria were cultured on medium supplemented with 50 μg/mL ampicillin, 100 μg/mL spectinomycin, or 50 μg/mL kanamycin.
Gene deletion from and backfill to the PXO99A genome were performed by using the unmarked deletion method (Song and Yang, 2010). The hrcV and pthXo1 gene-knockout mutants and complementing strains were generated previously in our lab (Wang et al., 2018; Li et al., 2019). To create hpa2-, hrpF-, and xopN-knockout mutants, upstream and downstream flanking partial sequence fragments of each gene were amplified from the PXO99A genomic DNA and were connected by overlapped fusion-PCR using specific primers (Supplementary Table 2). Every PCR product was confirmed by sequencing and then cloned into the vector pK18sacB by digestion with the corresponding restriction enzymes (Supplementary Table 2) and ligation with T4 ligase (Thermo Scientific, Walsham, Massachusetts, United States). Every recombinant vector was introduced into PXO99A cells by electroporation, followed by single-colony selection on kanamycin-containing and sugar-absent NA plates. Colonies from single crossovers were transferred to NB broth, grown at 28°C for 12 h and then transferred onto plates containing NA and 10% sucrose. Sucrose-resistant colonies were replica streaked onto NA plates with and without kanamycin supplementation. Colonies resulting from double crossover events were selected based on kanamycin-negative and sucrose-positive traits, and unmarked mutants (Δhpa2, ΔhrpF, and ΔxopN) were confirmed by PCR amplification of hpa2, hrpF, and xopN, respectively. To create double and triple mutants, the recombinant vectors pK18sacB:Δhpa2, pK18sacB:ΔhrpF, and pK18sacB:ΔxopN were transferred correspondingly into one of the Δhpa2, ΔhrpF, and ΔxopN mutants or double mutants generated in the transformation process.
Different tags were attached to the 3’-terminus of the hpa2, hrpF, pthXo1, or xopN sequence in the pZWpthXo1 plasmid vectors (Wang et al., 2018). To create a cya-fused gene, a 1218-bp cya fragment encoding amino acids 2 to 406 of the Cya protein was amplified from plasmid pMS107 and prefixed with the last 51-bp region of hpa2, hrpF, xopN, or pthXo1 that contained a SacI recognition site (Supplementary Table 2). The recombinant sequence was inserted into pZWpthXo1 at the SacI site. Every recombinant vector was linearized with HindIII and cloned into the pHM1 vector for genetic complementation. The double complementary vectors, for example that for hpa1pthXo1, were constructed using two steps. First, the hpa1 sequence that was linked its own promoter was cloned into pHM1 between the PstI and KpnI sites. Second, pZWpthXo1 was linearized using HindIII and inserted into the HindIII site of pHM1hpa1. Complementation or transformation was performed by electroporation. Similar procedures were used to generate double complementation of other gene combinations.
Overnight Xoo NA cultures were washed twice and resuspended in sterile water to generate inoculum suspensions with an optical density of OD600 = 0.3. A bacterial suspension was inoculated on leaves of 14-day-old rice seedlings by infiltration with needleless syringes at 3 sites per leaf. Alternatively, a bacterial suspension was inoculated on leaves of 30-day-old rice plants by the leaf-clipping method (Li et al., 2011). Symptoms were scored by photographing or measuring lesion lengths or areas. Bacterial growth in rice leaves was measured by harvesting 10 leaves for each group of plants inoculated with a single bacterial strain.
Cya-containing PXO99A strains were grown in NB broth at 28°C with the appropriate antibiotics to logarithmic phase. Bacterial cells were harvested by centrifugation. The precipitated bacterial pellet was washed twice with sterile water and resuspended in 100 mL of type III-inducing XOM2 liquid media (Tsuge et al., 2002) to an of OD600 = 0.6. This XOM2 suspension was supplied with the appropriate antibiotics and incubated in a 28°C shaker at 220 rpm for 16 h. XOM2 cultures were then separated into cell pellet and supernatant fractions by centrifugation. The proteins in the pellet and supernatant were extracted by sonication and by precipitation with 12.5% trichloroacetic acid, respectively, (Li et al., 2011). Proteins were separated by 6% SDS-PAGE and transferred to Immobilon-P membranes (Millipore) for immunoblotting analyses using a Cya antibody (Santa Cruz) or β-lactamase antibody (Abcam). The ampicillin resistance protein β-lactamase protein is encoded by the pZWtal-cya vector (Supplementary Table 2), remains cell-bound unless non-specific cell leakage occurred, and was used as a control for non-specific cell lysis. Protein blots were incubated with the specific antibody and hybridized to horseradish peroxidase-conjugated goat antimouse immunoglobulin G from the BeyoECL Plus kit (Beyotime).
The Cya reporter is an accurate and complete roster of effector proteins translocated by pathogenic bacteria into host cells, offering a highly sensitive and robust assay for monitoring the translocation of the effectors (Chakravarthy et al., 2017). An effector is fused to the calmodulin-dependent adenylate-cyclase domain of CyaA. The increase in cAMP concentration in plant cells is then measured with an enzyme-linked immunosorbent assay kit. This assay was performed on 14- or 30-day-old rice seedlings inoculated with Cya-related Xoo strains. Bacterial suspensions were prepared from NA cultures and adjusted to an OD600 = 0.3. Each suspension was infiltrated into intercellular spaces of expanded leaves at three sites per leaf. At the designed time points, inoculated leaves were excised from inoculated leaves, weighted, and then frozen in liquid nitrogen in a mortar and ground with a ceramic pestle to a fine powder. The leaf powder was suspended in 350 μl of buffer A provided in the cAMP ELISA detection kit (GenScript Biotech Corp, Nanjing, China), followed by brief centrifugation at 10,000 g for 10 min. The supernatant was analyzed as per the manufacturer’s instruction book to determine intracellular cAMP concentrations. Total proteins in each sample for normalization were quantified by using a BCA protein assay kit (TransGen Biotech, Beijing, China).
Total RNA was isolated from leaves or protoplasts by using TRIzol (Invitrogen) and treated with DNase I (Invitrogen) to remove DNA. Northern blotting and RT-qPCR were performed as previously described (Li et al., 2019). The constitutively expressed OsActin2 gene was used as a reference. RT-qPCR analyses always employed specific primers (Supplementary Table 2) and included temple-absent controls. Relative expression level of a gene tested was quantified as the transcript quantity ratio to OsActin2.
All experiments were repeated at least three times with similar results as provided. Quantitative data were analyzed by using the commercial IBM SPSS19.0 software package (Shi, 2012) installed in the registered office computers. Homogeneity-of-variance in data was determined by Levene test. Formal distribution pattern of the data was confirmed by Kolmogorov-Smirnov test and P-P Plots. Analysis of variance (ANOVA) was performed along with Duncan’s test for multiple comparisons (Li et al., 2014) of data from at least three independent experiments each involving three repetitions at minimum unless specified elsewhere in the case that a leaf was treated as a statistical unit.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
XM and LZ designed and performed the experiments and wrote the manuscript. YL, XW, JB, and KL performed the experiments. SZ analyzed the data and commented on the manuscript. LC supervised the research and commented on the manuscript. HD conceived the research, designed the experiments, and wrote and finalized the manuscript.
Natural Science Foundation of China (Grant Number 31772247), Major Science and Technology Innovation Project of Shandong Province (2019JZZY020608), and Natural Science Fund for Outstanding Young Scholars of Shandong Province (JQ201807).
HD, LZ, and LC are the inventors on a provisional patent application 201911394862.6 that covers an Xoo T3 translocator.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Kristina Hedfalk for the technical support. This work was supported by Natural Science Foundation of Jiangsu Province, China National Key Research and Development Plan and Natural Science Foundation of China.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01601/full#supplementary-material
Adam, P. R., Barta, M. L., and Dickenson, N. E. (2017). “Characterization of type three secretion system translocator interactions with phospholipid membranes,” in Type 3 Secretion Systems: Methods and Protocols, Methods in Molecular Biology, Vol. 1531, eds M. L. Nilles and D. L. J. Condry (New York, NY: Springer Science + Business Media), 81–91.
Alfano, J. R., and Collmer, A. (2004). Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. doi: 10.1146/annurev.phyto.42.040103.110731
Allgood, S. C., and Neunuebel, M. R. (2018). The recycling endosome and bacterial pathogens. Cell Microbiol. 20:e12857. doi: 10.3389/fimmu.2016.00084
Bian, H., Zhang, L., Chen, L., Wang, W., Ji, H., and Dong, H. (2019). Real-time monitoring of translocation of selected type-III effectors from Xanthomonas oryzae pv. oryzae into rice cells. J. Biosci. 44:82. doi: 10.1007/s12038-019-9916-0
Blocker, A., Jouihri, N., Larquet, E., Gounon, P., Ebel, F., Parsot, C., et al. (2001). Structure and composition of the Shigella flexneri ‘needle complex’, a part of its type III secreton. Mol. Microbiol. 39, 652–663. doi: 10.1046/j.1365-2958.2001.02200.x
Bocsanczy, A. M., Huguet-Tapia, J. C., and Norman, D. J. (2017). Comparative Genomics of Ralstonia solanacearum identifies candidate genes associated with cool vrulence. Front. Plant. Sci. 8:1565. doi: 10.3389/fpls.2017.01565
Bocsanczy, A. M., Nissinen, R. M., Oh, C. S., and Beer, S. V. (2008). HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Mol. Plant Pathol. 9, 425–434. doi: 10.1111/j.1364-3703.2008.00471.x
Bogdanove, A. J., and Voytas, D. F. (2011). TAL effectors: customizable proteins for DNA targeting. Science 333, 1843–1846. doi: 10.1126/science.1204094
Büttner, C. R., Sorg, I., Cornelis, G. R., Heinz, D. W., and Niemann, H. H. (2008). Structure of the Yersinia enterocolitica type III secretion translocator chaperone SycD. J. Mol. Biol. 375, 997–1012. doi: 10.1016/j.jmb.2007.11.009
Büttner, D. (2012). Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. doi: 10.1128/MMBR.05017-11
Büttner, D. (2016). Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40, 894–937. doi: 10.1093/femsre/fuw026
Büttner, D., and Bonas, U. (2002). Port of entry – the type III secretion translocon. Trends Microbiol. 10, 186–192. doi: 10.1016/s0966-842x(02)02331-4
Büttner, D., Nennstiel, D., Klüsener, B., and Bonas, U. (2002). Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 184, 2389–2398. doi: 10.1128/jb.184.9.2389-2398.2002
Camoni, L., Visconti, S., Aducci, P., and Marra, M. (2018). 14-3-3 proteins in plant hormone signaling: doing several things at once. Front. Plant Sci. 9:297. doi: 10.3389/fpls.2018.00297
Chakravarthy, S., Huot, B., and Kvitko, B. H. (2017). Effector translocation: Cya reporter assay. Methods Mol. Biol. 1615, 473–487. doi: 10.1007/978-1-4939-7033-9_33
Chatterjee, S., Chaudhury, S., McShan, A. C., Kaur, K., and De Guzman, R. N. (2013). Structure and biophysics of type III secretion in bacteria. Biochemistry 52, 2508–2517. doi: 10.1021/bi400160a
Chen, H., Chen, J., Li, M., Chang, M., Xu, K., Shang, Z., et al. (2017). A bacterial Type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 22, 777–788. doi: 10.1016/j.chom.2017.10.019
Chen, L., Qian, J., Long, J., Yin, Q., Zhang, C., Wu, X., et al. (2008a). Identification of specific fragments of HpaGXooc, a harpin protein from Xanthomonas oryzae pv. oryzicola, that induces disease resistance and enhanced growth in rice. Phtopathology 98, 781–791. doi: 10.1094/PHYTO-98-7-0781
Chen, L., Zhang, S. J., Zhang, S. S., Qu, S., Long, J., Ren, H., et al. (2008b). A fragment of the Xanthomonas oryzae pv. oryzicola harpin HpaGXooc reduces disease and increases yield of rice in extensive grower plantings. Phytopathology 98, 792–802. doi: 10.1094/PHYTO-98-7-0792
Chen, L.-Q., Hou, B.-H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X.-Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Choi, M. S., Kim, W., Lee, C., and Oh, C. S. (2013). Harpins, multifunctional proteins secreted by Gram-negative plant-pathogenic bacteria. Mol. Plant Microbe Interact. 26, 1115–1122. doi: 10.1094/MPMI-02-13-0050-CR
Cybulsky, A. V. (2017). Endoplasmic reticulum stress, the unfolded protein response and autophagy in kidney diseases. Nat. Rev. Nephrol. 13, 681–696. doi: 10.1038/nrneph.2017.129
Domingues, L., Ismail, A., Charro, N., Rodríguez-Escudero, I., Holden, D. W., Molina, M., et al. (2016). The Salmonella effector SteA binds phosphatidylinositol 4-phosphate for subcellular targeting within host cells. Cell Microbiol. 18, 949–969. doi: 10.1111/cmi.12558
Dong, Y. L., Li, P., and Zhang, C. (2016). Harpin Hpa1 promotes flower development in Impatiens and Parochetus plants. Bot. Stud. 57:22. doi: 10.1186/s40529-016-0132-z
Goure, J., Pastor, A., Faudry, E., Chabert, J., Dessen, A., and Attree, I. (2004). The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72, 4741–4750. doi: 10.1128/IAI.72.8.4741-4750.2004
Haapalainen, M., Engelhardt, S., Küfner, I., Li, C. M., Nürnberger, T., Lee, J., et al. (2011). Functional mapping of harpin HrpZ of Pseudomonas syringae reveals the sites responsible for protein oligomerization, lipid interactions and plant defence induction. Mol. Plant Pathol. 12, 151–166. doi: 10.1111/j.1364-3703.2010.00655.x
Ji, H., and Dong, H. (2015b). Key steps in type III secretion system (T3SS) towards translocon assembly with potential sensor at plant plasma membrane. Mol. Plant Pathol. 16, 762–773. doi: 10.1111/mpp.12223
Ji, H., and Dong, H. (2015a). Biological significance and topological basis of aquaporin-partnering protein-protein interactions. Plant Signal. Behav. 10:e1011947. doi: 10.1080/15592324.2015.1011947
Julie, G., and Guy, T. V. N. (2016). Bacterial control of pores induced by the type III secretion system: mind the gap. Front. Immunol. 7:84.
Kanyuka, K., and Rudd, J. J. (2019). Cell surface immune receptors: the guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 50, 1–8. doi: 10.1016/j.pbi.2019.02.005
Kim, J. G., Li, X., Roden, J. A., Taylor, K. W., Aakre, C. D., Su, B., et al. (2009). Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21, 1305–1323. doi: 10.1105/tpc.108.063123
Kvitko, B. H., Ramos, A. R., Morello, J. E., Oh, H. S., and Collmer, A. (2007). Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J. Bacteriol. 189, 8059–8072. doi: 10.1128/JB.01146-07
Li, L., Wang, H., Gago, J., Cui, H., Qian, Z., Kodama, N., et al. (2015). Harpin Hpa1 interacts with aquaporin PIP1;4 to promote the substrate transport and photosynthesis in Arabidopsis. Sci. Rep. 10:1038. doi: 10.1038/srep17207
Li, P., Zhang, L., Mo, X., Ji, H., Bian, H., Hu, Y., et al. (2019). Aquaporin PIP1;3 of rice and harpin Hpa1 of bacterial blight pathogen cooperate in a type III effector translocation. J. Exp. Bot. 70, 3057–3073. doi: 10.1093/jxb/erz130
Li, X., Han, B., Xu, M., Han, L., Zhao, Y., Dong, H., et al. (2014). Plant growth enhancement and associated physiological responses are coregulated by ethylene and gibberellin in response to harpin protein Hpa1. Planta 239, 831–846. doi: 10.1007/s00425-013-2013-y
Li, X., Han, L., Zhao, Y., You, Z., Dong, H., and Zhang, C. (2013). Harpin Hpa1 needs nitroxyl terminus to promote vegetative growth and leaf photosynthesis in Arabidopsis. J. Biosci. 39, 127–137. doi: 10.1007/s12038-013-9408-6
Li, Y. R., Che, Y. Z., Zou, H. S., Cui, Y. P., Guo, W., Zou, L. F., et al. (2011). Hpa2 required by HrpF to translocate Xanthomonas oryzae transcriptional activator-like effectors into rice for pathogenicity. Appl. Environ. Microbiol. 77, 3809–3818. doi: 10.1128/AEM.02849-10
Liu, F., Liu, H., Jia, Q., Wu, X., Guo, X., Zhang, S., et al. (2006). The internal glycine-rich motif and cysteine suppress several effects of the HpaGXooc protein in plants. Phytopathology 96, 1052–1059. doi: 10.1094/PHYTO-96-1052
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Mew, T. W. (1987). Current status and future prospects of research on bacterial blight of rice. Annu. Rev. Phytopathol. 25, 359–382. doi: 10.1146/annurev.py.25.090187.002043
Meyer, D., Cunnac, S., Gueneron, M., Declercq, C., Van Gijsegem, F., Lauber, E., et al. (2006). PopF1 and PopF2, two proteins secreted by the type III protein secretion system of Ralstonia solanacearum, are translocators belonging to the HrpF/NopX family. J. Bacteriol. 188, 4903–4917. doi: 10.1128/JB.00180-06
Mueller, C. A., Broz, P., and Cornelis, G. R. (2008). The type III secretion system tip complex and translocon. Mol. Microbiol. 68, 1085–1095. doi: 10.1111/j.1365-2958.2008.06237.x
Niño-Liu, D. O., Ronald, P. C., and Bogdanove, A. J. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. doi: 10.1111/j.1364-3703.2006.00344.x
Nissinen, R. M., Ytterberg, A. J., Bogdanove, A. J., Van Wijk, K. J., and Beer, S. V. (2007). Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol. Plant Pathol. 8, 55–67. doi: 10.1111/j.1364-3703.2006.00370.x
Noël, L., Thieme, F., Nennstiel, D., and Bonas, U. (2002). Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184, 1340–1348. doi: 10.1128/jb.184.5.1340-1348.2002
Obacz, J., Avril, T., Le Reste, P. J., Urra, H., Quillien, V., Hetz, C., et al. (2017). Endoplasmic reticulum proteostasis in glioblastoma-From molecular mechanisms to therapeutic perspectives. Sci. Signal. 10:eaal2323. doi: 10.1126/scisignal.aal2323
Oh, C. S., and Beer, S. V. (2007). AtHIPM, an ortholog of the apple HrpN-interacting protein, is a negative regulator of plant growth and mediates the growth-enhancing effect of HrpN in Arabidopsis. Plant Physiol. 145, 426–436. doi: 10.1104/pp.107.103432
Peng, J., Bao, Z., Ren, H., Wang, J., and Dong, H. (2004). Expression of harpinXoo in transgenic tobacco induces pathogen defense in the absence of hypersensitive cell death. Phtopathology 94, 1048–1055. doi: 10.1094/PHYTO.2004.94.10.1048
Picking, W. D., and Barta, M. L. (2019). The tip complex: from host cell sensing to translocon formation. Curr. Top. Microbiol. Immunol. doi: 10.1007/82_2019_171 [Epub ahead of print].
Piscatelli, H. L., Li, M., and Zhou, D. (2016). Dual 4- and 5-phosphatase activities regulate SopB-dependent phosphoinositide dynamics to promote bacterial entry. Cell. Microbiol. 18, 705–719. doi: 10.1111/cmi.12542
Racape, J., Belbahri, L., Engelhardt, S., Lacombe, B., Lee, J., Lochman, J., et al. (2005). Ca-dependent lipid binding and membrane integration of PopA, a harpin-like elicitor of the hypersensitive response in tobacco. Mol. Microbiol. 58, 1406–1420. doi: 10.1111/j.1365-2958.2004.04910.x
Roden, J. A., Belt, B., Ross, J. B., Tachibana, T., Vargas, J., and Mudgett, M. B. (2004). A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. U.S.A. 101, 16624–16629. doi: 10.1073/pnas.0407383101
Rüter, C., Buss, C., Scharnert, J., Heusipp, G., and Schmidt, M. A. (2010). A newly identified bacterial cell-penetrating peptide that reduces the transcription of pro-inflammatory cytokines. J. Cell. Sci. 123, 2190–2198. doi: 10.1242/jcs.063016
Rüter, C., and Schmidt, M. A. (2017). Cell-penetrating bacterial effector proteins: better tools than targets. Trends Biotechnol. 35, 109–120. doi: 10.1016/j.tibtech.2016.08.002
Sang, S., Li, X., Gao, R., You, Z., Lü, B., Liu, P., et al. (2012). Apoplastic and cytoplasmic location of harpin protein Hpa1Xoo plays different roles in H2O2 generation and pathogen resistance in Arabidopsis. Plant Mol. Biol. 79, 375–391. doi: 10.1007/s11103-012-9918-x
Santi-Rocca, J., and Blanchard, N. (2017). Membrane trafficking and remodeling at the host-parasite interface. Curr. Opin. Microbiol. 40, 145–151. doi: 10.1016/j.mib.2017.11.013
Scharnert, J., Greune, L., Zeuschner, D., Lubos, M. L., Alexander Schmidt, M., and Rüter, C. (2013). Autonomous translocation and intracellular trafficking of the cell-penetrating and immune-suppressive effector protein YopM. Cell Mol. Life Sci. 70, 4809–4823. doi: 10.1007/s00018-013-1413-2
Scheibner, F., Marillonnet, S., and Büttner, D. (2017). The TAL effector AvrBs3 from Xanthomonas campestris pv. vesicatoria contains multiple export signals and can enter plant cells in the absence of the type III secretion translocon. Front. Microbiol. 8:2180. doi: 10.3389/fmicb.2017.02180
Shanmugam, S. K., and Dalbey, R. E. (2019). The conserved role of YidC in membrane protein biogenesis. Microbiol. Spectr. 7:1. doi: 10.1128/microbiolspec.PSIB-0014-2018
Shi, L. W. (2012). SPSS19.0 Statistical Analysis from Accidence to Conversance (in Chinese). Beijing: Tsinghua University Press, 109–143.
Song, C., and Yang, B. (2010). Mutagenesis of 18 type III effectors reveals virulence function of XopZPXO99 in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 23, 893–902. doi: 10.1094/MPMI-23-7-0893
Sugio, A., Yang, B., and White, F. F. (2005). Characterization of the hrpF pathogenicity peninsula of Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 18, 546–554. doi: 10.1094/MPMI-18-0546
Taylor, K. W., Kim, J. G., Su, X. B., Aakre, C. D., Roden, J. A., Adams, C. M., et al. (2012). Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector XopN from binding to TFT1 attenuate Xanthomonas virulence. PLoS Pathog. 8:e1002768. doi: 10.1371/journal.ppat.1002768
Tsuge, S., Furutani, A., Fukunaka, R., Oku, T., Tsuno, K., Ochiai, H., et al. (2002). Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J. Gen. Plant Pathol. 68, 363–371. doi: 10.1128/AEM.00513-16
Wang, X., Zhang, L., Ji, H., Mo, X., Li, P., Wang, J., et al. (2018). Hpa1 is a type III translocator in Xanthomonas oryzae pv. oryzae. BMC Microbiol. 18:105. doi: 10.1186/s12866-018-1251-3
White, F. F., Potnis, N., Jones, J. B., and Koebnik, R. (2009). The type III effectors of Xanthomonas. Mol. Plant Pathol. 10, 749–766. doi: 10.1111/j.1364-3703.2009.00590.x
Wudick, M. M., Li, X., Valentini, V., Geldner, N., Chory, J., Lin, J., et al. (2015). Subcellular redistribution of root aquaporins induced by hydrogen peroxide. Mol. Plant 8, 1103–1114. doi: 10.1016/j.molp.2015.02.017
Yang, B. (2017). Advances in Experimental Medicine and Biology 969 Aquaporins. Springer Science + Business Media B.V. doi: 10.1007/978-94-024-1057-0_17
Yang, B., Sugio, A., and White, F. F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 103, 10503–10508. doi: 10.1073/pnas.0604088103
Zhang, J., Yin, Z., and White, F. (2015). TAL effectors and the executor R genes. Front. Plant Sci. 6:641. doi: 10.3389/fpls.2015.00641
Zhang, L., Chen, L., and Dong, H. (2019a). Plant aquaporins in infection by and immunity against pathogens - a critical review. Front. Plant Sci. 10:632. doi: 10.3389/fpls.2019.00632
Zhang, L., Hu, Y., Li, P., Wang, X., and Dong, H. (2019b). Silencing of an aquaporin gene diminishes bacterial blight disease in rice. Aust. Plant Pathol. 48, 143–158. doi: 10.1007/s13313-018-0609-1
Keywords: Xanthomonas oryzae pv. oryzae, PthXo1, type III translocator, Hpa2, HrpF, XopN
Citation: Mo X, Zhang L, Liu Y, Wang X, Bai J, Lu K, Zou S, Dong H and Chen L (2020) Three Proteins (Hpa2, HrpF and XopN) Are Concomitant Type III Translocators in Bacterial Blight Pathogen of Rice. Front. Microbiol. 11:1601. doi: 10.3389/fmicb.2020.01601
Received: 27 March 2020; Accepted: 18 June 2020;
Published: 22 July 2020.
Edited by:
Li Yang, University of Georgia, United StatesReviewed by:
Meng Yuan, Huazhong Agricultural University, ChinaCopyright © 2020 Mo, Zhang, Liu, Wang, Bai, Lu, Zou, Dong and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hansong Dong, aHNkb25nQG5qYXUuZWR1LmNu; Lei Chen, Y2hlbmxlaUBzZGF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.