
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 July 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01572
 Zhuhua Wu1†
Zhuhua Wu1† Wenjing Wei1†
Wenjing Wei1† Ying Zhou2
Ying Zhou2 Huixin Guo1
Huixin Guo1 Jiao Zhao3
Jiao Zhao3 Qinghua Liao1
Qinghua Liao1 Liang Chen1
Liang Chen1 Xiaoli Zhang2*
Xiaoli Zhang2* Lin Zhou1*
Lin Zhou1*In recent years, the treatment of tuberculosis is once again facing a severe situation because the existing antituberculosis drugs have become weaker and weaker with the emergence of drug-resistant Mycobacterium tuberculosis (Mtb). The studies of cell division and cell cycle-related factors in Mtb are particularly important for the development of new drugs with broad-spectrum effects. Mycobacterium smegmatis (Msm) has been used as a model organism to study the molecular, physiological, and drug-resistant mechanisms of Mtb. Bioinformatics analysis has predicted that MSMEG_6171 is a MinD-like protein of the septum site-determining protein family associated with cell division in Mycobacterium smegmatis. In our study, we use ultrastructural analysis, proteomics, metabolomics, and molecular biology techniques to comprehensively investigate the function of MSMEG_6171. Overexpression of MSMEG_6171 in Msm resulted in elongated cells, suggesting an important role of MSMEG_6171 in regulating cell wall morphology. The MSMEG_6171 overexpression could enhance the bacterial resistance to vancomycin, ethionamide, meropenem, and cefamandole. The MSMEG_6171 overexpression could alter the lipid metabolism of Msm to cause the changes on cellular biofilm property and function, which enhances bacterial resistance to antibiotics targeting cell wall synthesis. MSMEG_6171 could also induce the glyceride and phospholipid alteration in vivo to exhibit the pleiotropic phenotypes and various cellular responses. The results showed that amino acid R249 in MSMEG_6171 was a key site that can affect the level of bacterial drug resistance, suggesting that ATPase activity is required for function.
Mycobacterial diseases are a major public health concern caused by mycobacteria species including TB, leprosy, and atypical mycobacterial infections (Russell et al., 2010; Tiwari et al., 2018; Katale et al., 2019). The TB treatment faces a serious situation because of the multidrug-resistant tuberculosis (MDR-TB) (Pontali et al., 2017). The atypical mycobacterial infections are mainly caused by opportunistic pathogens from non-tuberculous mycobacteria. Non-tuberculous mycobacteria are inherently resistant to most commonly used antimicrobials and many anti-TB drugs (Falkinham, 2018). Thus, it is urgent to eradicate pathogenic mycobacteria from infection sources and hosts.
Mycobacterial species, especially Mtb, have a characteristic lipid-rich (up to 60%) cell wall that confers low permeability to many common antibiotics and chemotherapeutic agents (Bhat et al., 2017). The cell wall lipids also allow proliferation in host by inducing the formation of foamy macrophages (Santucci et al., 2016). Many antimicrobial drugs target this well-organized barrier of mycobacterial cell envelope (Rombouts et al., 2012; Lee et al., 2017; Rodriguez-Rivera et al., 2018). This well-organized barrier is composed of plasma membrane, cell wall core, and outermost layer (Hett and Rubin, 2008; Grzegorzewicz et al., 2016). The plasma membrane forms the bulk of the inner leaflet (Brennan, 2003). A variety of soluble lipids and proteins make up the outermost layer, including lipids with some long- and short-chain fatty acids complementing each other (Kaur and Kaur, 2017). The cell wall core is composed of peptidoglycan, which is covalently linked to heteropolysaccharide arabinogalactan via P-GlcNAc-Rha. A large variety of unusual lipids in the formation of cell walls are thought to enhance cell wall impermeability and to increase the intrinsic resistance of mycobacteria to antimicrobial drugs (Rens et al., 2016). Enzymes involved in lipid biosynthesis, intracellular transport, and utilization become the excellent therapeutic intervention targets (Bailo et al., 2015). Recent studies have shown that an increasing number of genes are involved in lipid synthesis, modification, and transport of cell wall components (Ripoll et al., 2007; Pang et al., 2013; Grzegorzewicz et al., 2015; Aguilar-Ayala et al., 2017).
Mycobacterium smegmatis (Msm) shares more than 2000 homologous genes with Mtb (Li et al., 2016). The unique Msm cell wall structure is similar to that of Mtb and many other mycobacterial species (King, 2003). Therefore, Msm is often used to study the molecular, physiological, and drug resistance mechanisms of Mtb. MSMEG_6171 is an uncharacterized protein in Msm; although it is similar to the MinD-like protein, it belongs to the MinD/Ssd family (England et al., 2011). Previous studies have shown that MinD and MinC jointly constitute the conservative center of Min system (Conti et al., 2015). Through recruiting MinC to the cell membrane, MinD can interact with MinC to inhibit the polymerization of FtsZ, thus restricting the assembly of Z-ring to the middle of cells (Lutkenhaus and Sundaramoorthy, 2003; Conti et al., 2015; Schuhmacher et al., 2015). The homolog of MSMEG_6171 in Mtb, Rv3660c, shares 61% amino acid sequence identity, and it characterizes as a putative septum site determining protein eliciting bacterial filamentation (England et al., 2011). It also elicits responses including the dormancy regulon and alternative sigma factors involved in adaptive metabolism (England et al., 2011). These conclusions are based primarily on transcriptome data and bioinformatic prediction without sufficient experimental data. Therefore, we undertook a study to experimentally determine the function of MSMEG-6171.
Here, we present a thorough investigation of MSMEG_6171 in Msm. First, we found that MSMEG_6171 overexpression can affect the bacterial biofilm properties and can resist the antibiotics targeting the bacterial cell wall. Secondly, our results showed that MSMEG_6171 can induce the glyceride and phospholipid alteration to display the pleiotropic phenotypes and various cellular responses in vivo. Thirdly, MSMEG_6171 overexpression can enhance the bacterial resistance to some antibiotics involved in cell wall synthesis. Finally, we also found that the amino acid R249 in MSMEG_6171 was a key site that can affect the level of bacterial resistance. Taken together, we proved that MSMEG_6171 overexpression can alter the lipid metabolism of Msm to change the structure and function of the mycobacterial cell envelope and to enhance bacterial resistance to antibiotics.
Msm and its recombinant strains were cultured as described by Parish et al. (2001). A Mycobacterium smegmatis mc2155 mutant without msmeg_6171 was constructed by performing allelic exchange using the p1NIL/pGOAL19-based flexible cassette (Parish and Stoker, 2000). Briefly, the 855-bp upstream and 849-bp downstream sequences of the msmeg_6171 gene were amplified using oligonucleotides 6171UF/6171UR and 6171DF/6171DR. Msm:6171, NTD, and CTD of msmeg_6171 were constructed by subcloning the EcoRI-HindIII fragment from the Msm genome into the corresponding sites in the pMV261 vector. msmeg_6171 was mutated by the site-directed mutagenesis method using three pairs of specific primers, respectively (Supplementary Table S1). The eGFP gene was cloned into pMV261-MSMEG_6171 to generate the plasmid pMV261-MSMEG_6171-eGFP (see Supplementary Materials and Methods). All bacterial strains and PCR oligonucleotides used in this study are listed in Supplementary Table S1.
The total RNA of Mycobacterium smegmatis mc2155 was extracted and converted to cDNA using commercial kits (FastRNATM Pro Blue Kit, MP Biomedicals, United States; SuperScript III First-Strand Synthesis Kit, Thermo Fisher Scientific, United States). The mRNA expression of 7 genes (msmeg_6171, mps2, msmeg_0400, msmeg_4757, msmeg_6761, pks, msmeg_3375) were analyzed using SYBR Green (Maeda et al., 2003). SigA served as the internal reference. Mean ± SEM was calculated from three independent experiments, and significant difference analysis was determined by Student’s unpaired t-test.
Msm and its mutant strains were used for growth curve analysis, sliding motility assay, Congo Red assay, and biofilm assay. Bacteria culture were adjusted to an optical density at 600 nm (OD600) of 0.2, and growth curves were obtained from OD600 detected at intervals of 3 h. 2 μL culture of each bacterial strain was dispensed on the medium (LB medium supplemented with 100 μg/mL Congo Red (Sigma) and 20 μg/ml kanamycin, Sauton liquid medium, and 7H9 medium supplemented with 0.3% agar) and then was used for corresponding Congo Red assay, biofilm assay, and sliding motility assay (Zanfardino et al., 2016; Lai et al., 2018). Cells were incubated at 37°C for 3 days.
Scanning electron microscopy was performed as described previously (Dahl, 2004). Msm strains were grown to stationary phase. Culture aliquots were concentrated by centrifugation, resuspended, and fixed with 2.5% glutaraldehyde. Then, sequential ethanol dehydrations were performed. After permutation with isopentylosacetate, critical point drying was determined using a Hitachi HCP-2 Critical Point Dryer (Hitachi High-Technologies Corp., Tokyo, Japan). Samples were gold sputter coated (Technics Hummer V; Anatech), and ultrastructure examination was performed using a Zeiss Ultra 55 electron microscope.
Cells were processed for TEM as described previously (Vijay et al., 2012). In brief, Msm strains were harvested by centrifugation and fixed in 1% (vol/vol) osmium tetroxide (Sigma-Aldrich) and 0.15 M sodium cacodylate buffer (pH 7.2) (Sigma-Aldrich) for 1 h at room temperature and postfixed for 2 h at room temperature in 0.15 M cacodylate buffer (pH 7.2) containing 2% (wt/vol) tannic acid and 2% (vol/vol) glutaraldehyde. Samples were dehydrated in a graded ethanol series and embedded in Spurr resin. Thin sections (70 nm) were cut and stained with 1% uranyl acetate and Reynold’s lead citrate and observed under a Tecnai Spirit (FEI) transmission electron microscope at 100 kV. Images were captured at a magnification of 98000×; analyses of cell wall thickness were performed using ImageJ.
The standard broth dilution method (Mengatto et al., 2006) was used to determine the MICs of Msm strains to different antibiotics (isoniazid, rifampicin, streptomycin, vancomycin, ethionamide, cycloserine, meropenem, cefoxitin, cefamandole). MICs were determined at 2 or 3 days post-incubation. Three independent experiments were performed. The detailed description is shown in Supplementary Materials and Methods.
Proteins interacting with MSMEG_6171 were analyzed by Flag-based pull-down assay according to previously published procedures with some modifications (Meniche et al., 2014). The protein complexes were separated by SDS-PAGE, visualized with silver staining, and then submitted to LC-MS analysis (see Supplementary Materials and Methods).
Proteins were extracted, digested, and labeled using the iTRAQ(R) Reagents Multiplex Kit (sigma) following the manufacturer’s instructions. Chromatography–mass spectrometry (LC-MS) and bioinformatics analysis were performed according to previously published procedures with some modifications (Sharma et al., 2015). Three biological replicates were provided for each group (see Supplementary Materials and Methods).
The MSMEG_6171-interacting prey proteins Were cloned in pGADT7AD, and the gene for the bait protein MSMEG_6171 Was cloned Into pGBKT7 (Matchmaker Gold Yeast Two-Hybrid System, Clontech, United States) to identify MSMEG_617–protein interactions according to the manufacturer’s instructions (Fields and Song, 1989).
Extraction and purification of fatty acids were performed as previously described (Liu and Nikaido, 1999). Cells were cultured and pelleted by centrifugation and resuspended in a saponification mixture (5% KOH in 88% methyl cellosolve), and the mixture was heated at 121°C for 1 h and cooled to room temperature. The mixture was acidified by adding 1.5 ml of 6 N HCl and was extracted with CHCl3. The CHCl3 layer was transferred into a glass tube, and the solvent was evaporated in a water bath at 80°C. The fatty acids were determined by analytical TLC on Silica Gel 60 (Macherey-Nagel) using dichloromethane/hexane (3:1; v/v) and revealed by spraying the plates with molybdophosphoric acid (10% in ethanol). Fatty acid samples were dissolved in diethyl ether and subjected to ESI/MS analysis in the negative ion mode. Mass spectra were determined on an Agilent 6500 Series Q-TOF B.08.00 (B8058.3SP1) mass spectrometer equipped with an ESI source. Data acquisition and analysis were performed using Qualitative Navigator B.08.00 software.
Bacterial strains were centrifuged, washed, and stored at −80°C for subsequent extraction of metabolites. For intracellular metabolites, samples were reconstituted and disrupted by FastPrep-24 (MP Biomedicals) (Carette et al., 2018). The intracellular metabolites were analyzed in the positive ion mode on an AB 5600 + TripleTOF mass spectrometer system. The relevant parameters were referred to previous studies (Khan et al., 2017). MS-DIAL version 2.24 and MONA were used for further data process and analysis (Horai et al., 2010). For lipid extraction, the chloroform/methanol solution (2:1, v:v) was added, mixed, and carried out by an ultrasonic cleaner. The supernatant was dried by N2 and then was dissolved in chloroform/methanol solution (2:1, v:v). For lipid samples, Dionex UltiMate 3000 (UHPLC)–Thermo Orbitrap Elite was used for LC-MS analysis. The chromatographic column used was Waters UPLC BEH C18 (1.7 μm 100∗2.1 mm). Mass spectrometry was operated in both positive and negative ion modes with the parameters optimized according to previously published procedures with some modifications (Layre et al., 2014). Raw data were converted to mz. data format by Agilent MassHunter Qualitative Analysis B.08.00 software (Agilent Technologies, United States). The XCMS program was used to further process and analyze the data. Visualization matrices containing the sample name, m/z-RT pair, and peak area were obtained after the internal standard normalization and weight normalization. The data matrices were imported into SIMCA-P 14.0 (Bjerrum et al., 2015) (Umetrics, Umea, Sweden), mean-centered, and scaled to Pareto variance. MetaboAnalyst software was used for multidimensional statistical analysis. The significantly different metabolites were determined with variable influence of P-values less than 0.05.
Bioinformatics analysis indicated that MSMEG_6171 is a MinD-like protein, and its adjacent proteins are involved in the secretion system of cell membranes, morphological differentiation, and metabolism (Figure 1A). To experimentally evaluate the physiological role of MSMEG_6171 in Msm, we constructed a recombinant msmeg_6171 overexpression strain (Msm:6171), a msmeg_6171 deleted strain (MsmΔ6171), and an MsmΔ6171 strain complemented with msmeg_6171 (MsmΔ6171:6171) (Supplementary Figure S1). Consistent with the previous study of Ssd protein in Mtb (England et al., 2011), Msm:6171 strains were also observed with significantly increased cell lengths (3.6 ± 0.42 μm) and MsmΔ6171 were significantly decreased (1.35 ± 0.51 μm), compared to wild-type strains (2.50 ± 0.11 μm). We also found that the cell lengths were partly recovered in the compensation strains (3.30 ± 0.42 μm) (Figure 1B). The viability of MsmΔ6171 strains showed that MSMEG_6171 was non-essential for the Msm culture in vitro. Using Vancomycin-BODIPY assay, we did not observe any inhibition or enhancement of the septum formation indicating that the filamentation may not relate to suppression or enhancement of septum formation in the merodiploid and null strains (Supplementary Figure S2).
Interestingly, overexpression of MSMEG_6171 resulted in a drastic change in the colony morphology from a rough to a smooth phenotype on solid medium [Figure 1C(a)] and exhibited an altered capacity to assemble into biofilms [Figure 1C(b)]. In addition, the Msm:6171 strains also showed a significantly increased sliding motility [Figure 1C(c)]. This phenomenon may be the result of the highly hydrophobic surface of the mutant cells, which plays an important role in the sliding motility (Maya-Hoyos et al., 2015). The MSMEG_6171-GFP fusion protein was constructed and introduced into MsmΔ6171 strains to detect the localization of MSMEG_6171 in Msm. Fluorescence microscopy analysis showed that the fluorescence signal was detected in the cytosol, indicating the probable localization of MSMEG_6171 (Supplementary Figure S3). The subcellular localization of MinD to division sites reported by Marston et al. (1998) showed similar results: Polar localization was not observed as seen in B. subtilis.
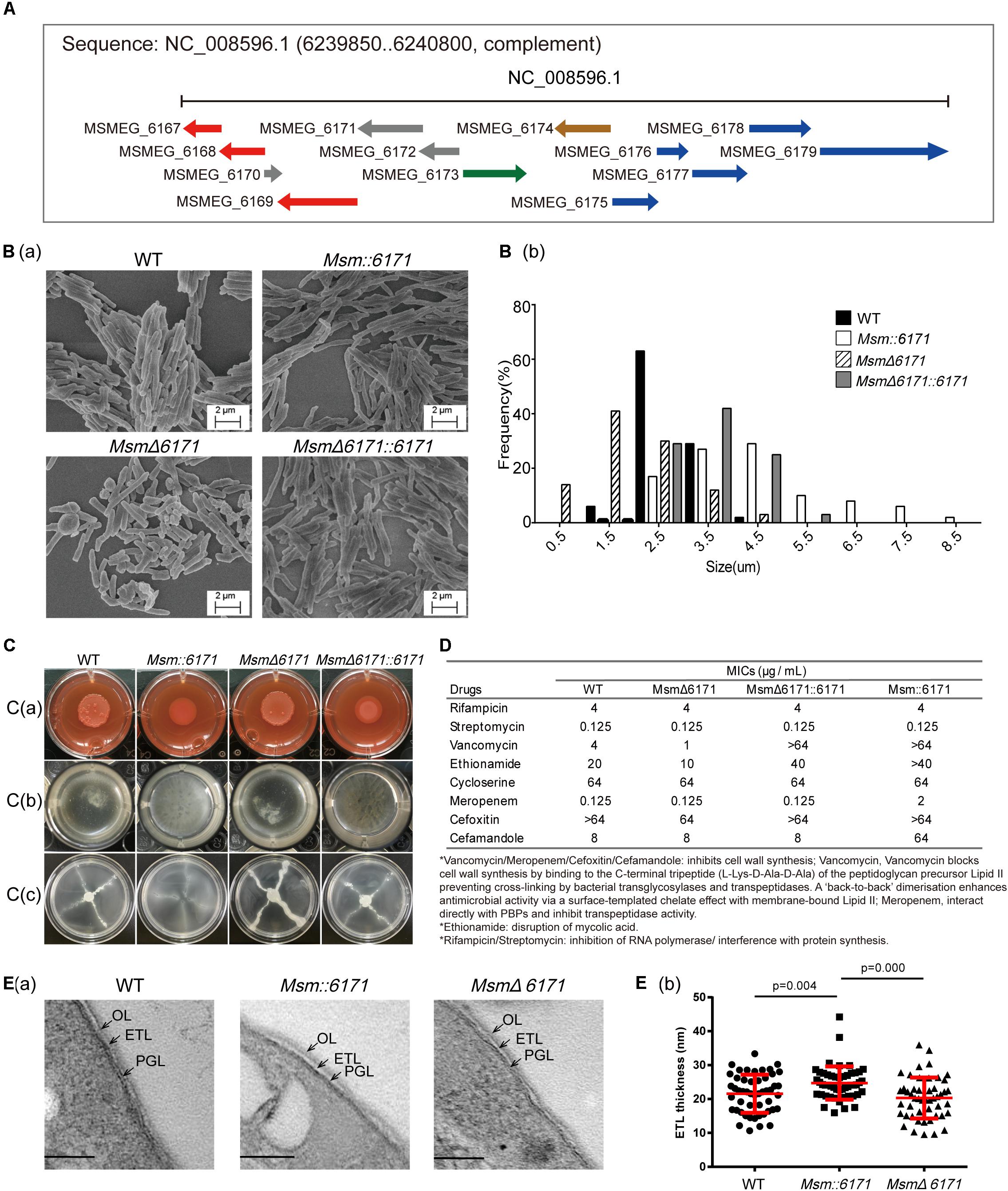
Figure 1. Phenotypic analysis of recombination Msm strains. (A) Schematic representation of the msmeg_6171 gene cluster and biosynthetic locus on the chromosome. The genes involved in the secretion system of cell membranes (red), morphological differentiation (green), and metabolism (blue) are indicated. (B) Msm strains were visualized by scanning electron microscopy (SEM). (B)(a) Images are representative of different fields of bacteria from exponentially growing cultures at 37°C. (B)(b) Lengths of the bacterial cells were calculated from the coordinates of both ends of the cell as measured from representative fields as visualized by scanning electron microscopy. Multiple fields were examined and values calculated in 0.5–1-mm increments from multiple fields of over 100 cells. (C) Compared with the WT strain, Msm:6171, MsmΔ6171, and complemented (MsmΔ6171:6171) strains were tested with the following phenotypes: (C)(a) Colony morphology on Congo Red, (C)(b) biofilm production at the air–liquid interface, and (C)(c) sliding motility. (D) Drug susceptibility of Msm:6171, MsmΔ6171, and MsmΔ6171:6171 compared with WT strains. ∗MIC in μg/mL. (E) Ultrastructures of cell envelopes of Msm. (E)(a) TEM images of Msm cells with detailed ultrastructure of cell envelope layers OL, ETL, and PGL. OL, outer layer; ETL, electron transparent layer; PGL, peptidoglycan layer. (B)(b) Cell envelope ETL thickness measurement between WT, Msm:6171, and MsmΔ6171. 50 cells of each strain were measured. Scale bar = 100 nm.
To investigate whether MSMEG_6171 could affect the drug susceptibility of Msm, we measured the minimum inhibitory concentration (MIC) of eight different antibiotics (rifampicin, streptomycin, vancomycin, ethionamide, cycloserine, meropenem, cefoxitin, and cefamandole) with the four strains. For all the strains, we found that the MICs of rifampicin, streptomycin, and cycloserine were at the same level. Notably, the Msm:6171 recombinant strains were 16-fold more resistant to vancomycin than the WT strains. Consistently, MSMΔ6171 strains were more susceptible to vancomycin. A similar phenomenon was obtained using ethionamide. In addition, the MSM:6171 strains had significantly increased resistance to meropenem and cefamandole (Figure 1D). Since vancomycin, meropenem, or cefamandole can inhibit cell wall synthesis, ethionamide is involved in disruption of mycolic acid; these data revealed that dramatic changes in the properties of the cell envelope in Msm:6171 recombinant strains could affect the integrity and permeability of the cell envelope, as well as the organization and motility of the bacterial populations. Additionally, the null strains had little change on the envelope integrity and permeability, while MsmΔ6171:6171 generally resulted in intermediate phenotypes, indicating that these phenomena are directly related to overexpression of MSMEG_6171.
To further investigate the effects of MSMEG_6171 on bacteria at the cell envelope, we investigated cell envelope ultrastructural features in WT and recombinant strains by transmission electron microscopy. The ultrastructure of these cells displayed a triple-layered cell envelope which are composed of the electron dense outer layer (OL), electron transparent layer (ETL), and peptidoglycan layer (PGL). The ETL of the cell envelope had an average thickness of 24.70 nm (±4.91 nm) in Msm:6171 and 21.57 nm (±5.61 nm) in WT) (Figure 1E).
To further gain new insights into how MSMEG_6171 affects bacterial cell envelope properties, we performed global protein expression profiling on the Msm:6171 strains, MsmΔ6171 strains, and the WT strains. Compared to the WT strains, 440 proteins were differentially expressed with fold changes of 1.5 or more (p values < 0.05), including 279 upregulated proteins and 161 downregulated proteins in the Msm:6171 strains. In the MsmΔ6171 strains, 70 downregulated proteins and 126 upregulated proteins exhibited fold changes greater than 1.5 (Supplementary Table S2). Gene Ontology (GO) enrichment analysis and KEGG pathway analysis were also performed to classify the function of those significantly altered proteins. We conducted a protein–protein interaction network analysis for proteins encoded by those potential MSMEG_6171 target genes using STRING 10.014. Among the upregulated proteins in the MSM:6171 strains, we found that a large number of candidates, MSMEG_6171-regulated proteins, are involved in glycerolipid metabolism, fatty acid metabolism, glycerophospholipid metabolism, and ABC transporter (Figures 2A,B). Interestingly, among the downregulated proteins in the MSM:6171 strains, 22 ribosomal proteins and 19 proteins associated with amino acid metabolism were enriched, which may be related to the slower growth of MSM:6171 strains (Supplementary Figure S4).
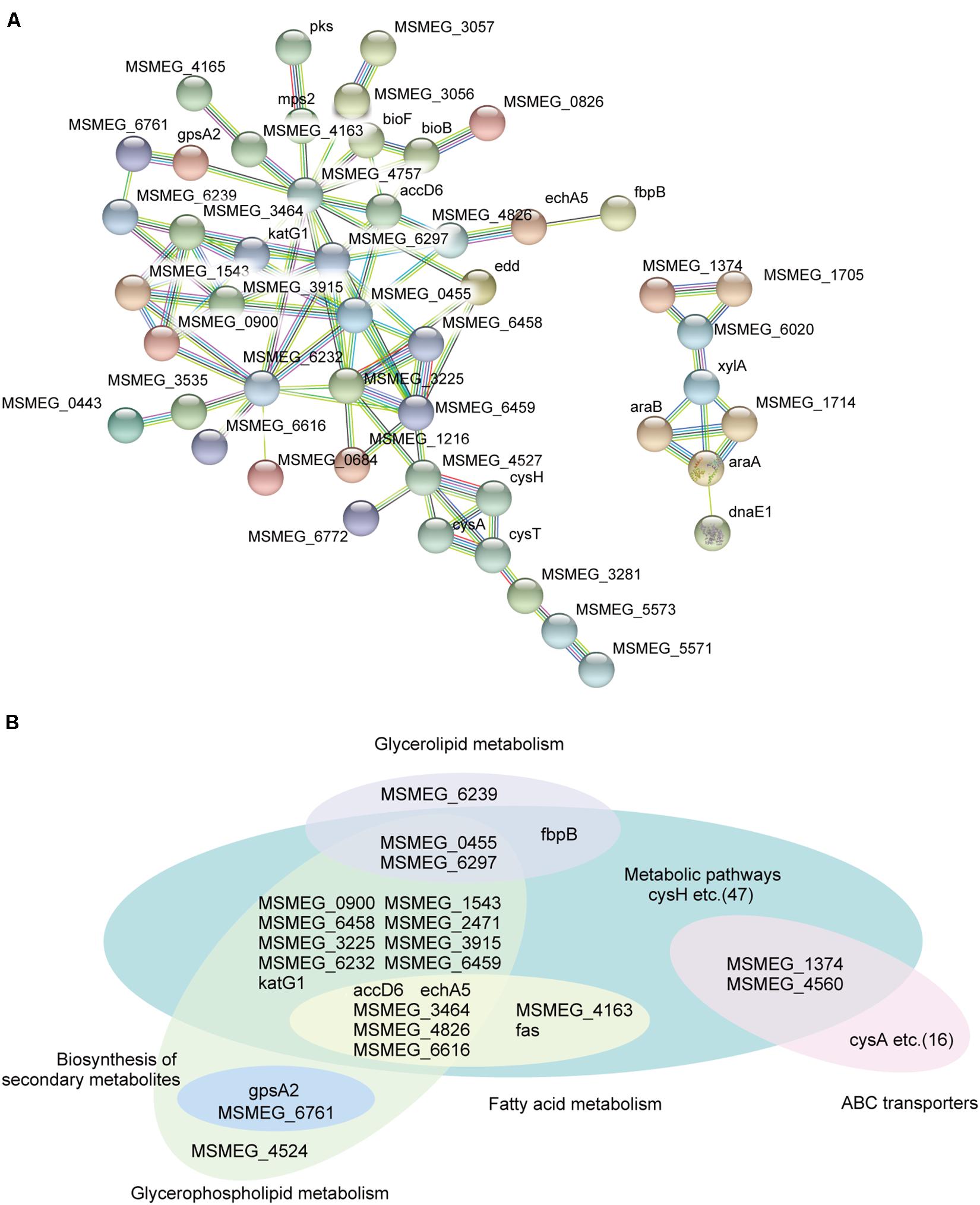
Figure 2. Network analysis of proteins potentially regulated by MSMEG_6171. (A) A subset of candidate MSMEG_6171-regulated proteins involved in the signaling pathways of the metabolic pathways and ABC transporters. (B) Classification of candidate MSMEG_6171 regulated proteins, including biosynthesis of secondary metabolites, fatty acid metabolism, glycerolipid metabolism, and glycerophospholipid metabolism.
We next focused on the proteins which can interact with MSMEG_6171 (Figure 3A). Seventeen proteins were found using pull-down assay (Figures 3B,C). Among these proteins, six proteins were significantly upregulated in the Msm:6171 recombinant strains compared with WT (Figure 4A). Quantitative RT-PCR analysis was also performed to verify the differentially expressed proteins at the level of their mRNA (Figure 4B). The results showed that MSMEG_4757, Mps2, Pks, MSMEG_0400, and MSMEG_6761 could increase with the overexpression of MSMEG_6171, suggesting that these proteins could interact directly or indirectly with MSMEG_6171. Notably, these proteins were involved in the activation of the lipid synthesis and metabolism (Figure 4D), indicating that MSMEG_6171 may play a role in the synthesis and metabolism of the lipid portion of cell envelope in Msm. We next tried to verify in vitro which proteins can directly interact with MSMEG_6171. We expressed MSMEG_6171 in Escherichia coli (E. coli), but we found that E. coli-overexpressed MSMEG_6171 could not survive. It indicated that MSMEG_6171 is toxic to E. coli. Thus, it was difficult to determine whether these proteins directly interacted with MSMEG_6171 or not in vitro. Using the yeast two-hybrid screen with MSMEG_6171 as bait, we tested these proteins obtained from pull-down assay except MSMEG_4757, Mps2, Pks, and MSMEG_0400 which failed to construct into yeast. The screen identified MSMEG_6367, a galactofuranosyltransferase involved in cell wall biosynthesis, to be an interactor of MSMEG_6171 (Figure 4C). Besides, MSMEG_4757, Mps2, Pks, MSMEG_0400, and MSMEG_6761 showed no significant change in protein level in MSMΔ6171 strains (Supplementary Table S2), although some corresponding mRNA decreased, such as pks and MSMEG_6761 (Figure 4B). As mentioned above, there were no significant differences in biofilm formation and Congo Red staining experiments between MsmΔ6171 strains and WT strains. However, the Msm:6171 strains became as smooth and moist as E. coli (Figure 1C). These phenotypic changes are most likely related to these upregulated proteins associated with the overexpression of MSMEG_6171. Based on those abovementioned data, we speculated that MSMEG_6171 may play a role in the lipid synthesis, metabolism, and biofilm formation in Msm.
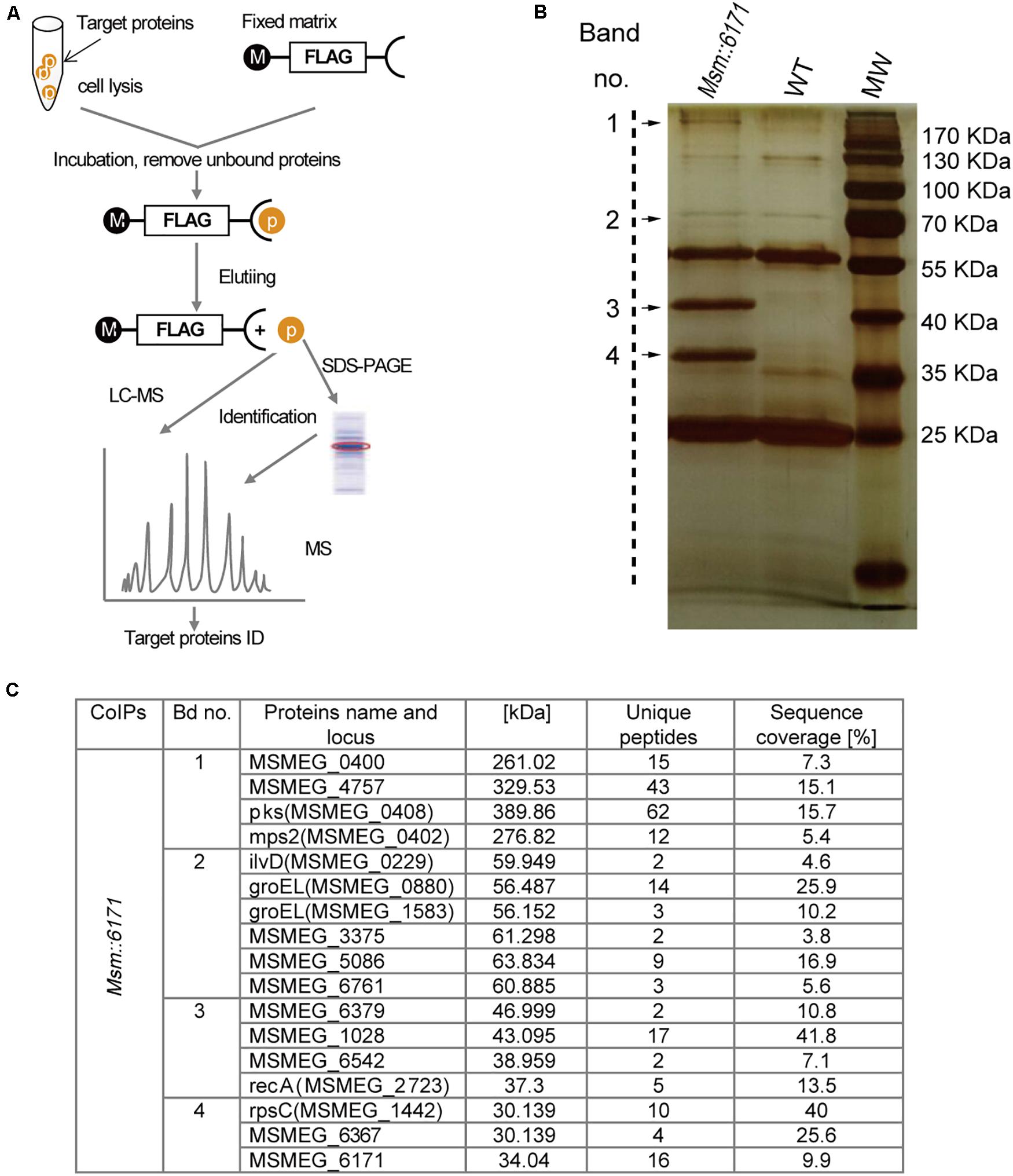
Figure 3. MSMEG_6171 interacts with proteins involved in lipid synthesis and metabolism. (A) FLAG-based pull-down scheme. Pull-down assay was performed with anti-FLAG M2 affinity gel, and the eluate was separated by SDS PAGE, and visualized with silver staining. (B) Then, the target bands were excised and subjected to in-gel tryptic digestion for LC-MS analysis. Tandem mass spectrometry of specific proteins in the target bands were shown in (C). WT, wild-type Msm mc2155 strains; MW, molecular weight standards.
To further investigate the effects of MSMEG_6171 on bacteria at the metabolomics level, the intracellular metabolites of Msm:6171, MsmΔ6171, and WT strains were extracted and determined by LC-MS under the same conditions to acquire the metabolomics profiles. To identify altered metabolites in Msm:6171, MsmΔ6171 strains compared to WT strains, PLS-DA, and PCA was carried out for analysis. The score plot of the PCA obtained from the LC-MS data clearly discriminated between samples Msm:6171 and WT strains, while not in MsmΔ6171 (Supplementary Figures S5A,B). A total of 597 putative metabolites that were differentially expressed (FDR < 0.05) were identified in Msm:6171 strains compared with the WT strains (Supplementary Table S3). Most of the lipids are glycerophospholipid and glycerolipid, viz. PA, PE, PC, PS, PI, PG and DG, MGDG, DGDG, and SQDG (Supplementary Figure S5C). Moreover, the changed metabolites mostly consisted of lipids and amino acids (Supplementary Figure S5D). KEGG pathway enrichment is shown in Supplementary Figure S5E, and glycerolipid metabolism and the glycerophospholipid metabolism pathway were the drastically altered metabolic activities through overexpression of MSMEG_6171 with considerable enrichment (P < 0.05). Besides, some amino acids, such as phenylalanine, tyrosine, and tryptophan biosynthesis; glycine, serine, and threonine metabolism; phenylalanine metabolism; cysteine and methionine metabolism; and aminoacyl-tRNA biosynthesis, were also significantly enriched, as well as nucleotide-related pathways—purine metabolism and riboflavin metabolism.
The results of the above proteome and total metabolome showed that the function of MSMEG_6171 was related to lipid metabolism. Next, we conducted the lipid metabolome to further study the lipid composition with specific changes (Supplementary Table S3). The results of PLS-DA and PCA showed that the lipid profile of Msm:6171 could be clearly discriminated with WT strains, but MsmΔ6171 did not show such difference (Figures 5A,B). We found that many lipids were significantly decreased in Msm:6171 strains, which are major constituents of the cell membrane (Figure 5C). It suggested that there was a difference of the lipid membrane structure between the Msm:6171 strain and WT strain. The significant changes of lipids were associated with the biosynthesis of lipoteichoic acid. For example, TGs, PGs, DGDGs, and DGs were drastically decreased, which could alter LTA biosynthesis in Msm:6171 strains. Besides, the lipids with significant differences between the two groups were not only the precursor of LTA but also the main components of the Msm bacterial membrane. The balance of lipid components was critical for proper membrane–protein structure, transport function, DNA replication, and cell division. Changes in the composition of neutral lipids may disturb the permeability of the cell membranes by altering the surface charge of the membrane, such as TGs, DGDGs, and DGs, the major zwitterionic phospholipid PEs, and the major anionic phospholipids, phosphatidylglycerols (PGs) and CLs. Differentially expressed lipids were identified in the MSM:6171 strains which were involved in the synthesis of membrane lipids (Figure 5D). Furthermore, we extracted and analyzed the fatty acids from the WT and MSM:6171 strains to detect the change of fatty acids by TLC and ESI/MS. TLC analysis of fatty acid composition after saponification showed that these bands from WT and MSM:6171 strains migrated at a similar rate, but the abundance of mycotic acid extracted from MSM:6171 strains is higher than that of WT strains (Figure 5E). Consistently, ESI/MS profiles of free fatty acids of MSM:6171 strains (Figure 5F) exhibited a relatively higher intensity of [M-H]– ions ranging from m/z 261 to 361 than WT. In a nutshell, we speculated that MSMEG_6171 may play a role in the lipid synthesis, metabolism, and biofilm formation in Msm according to the above data.
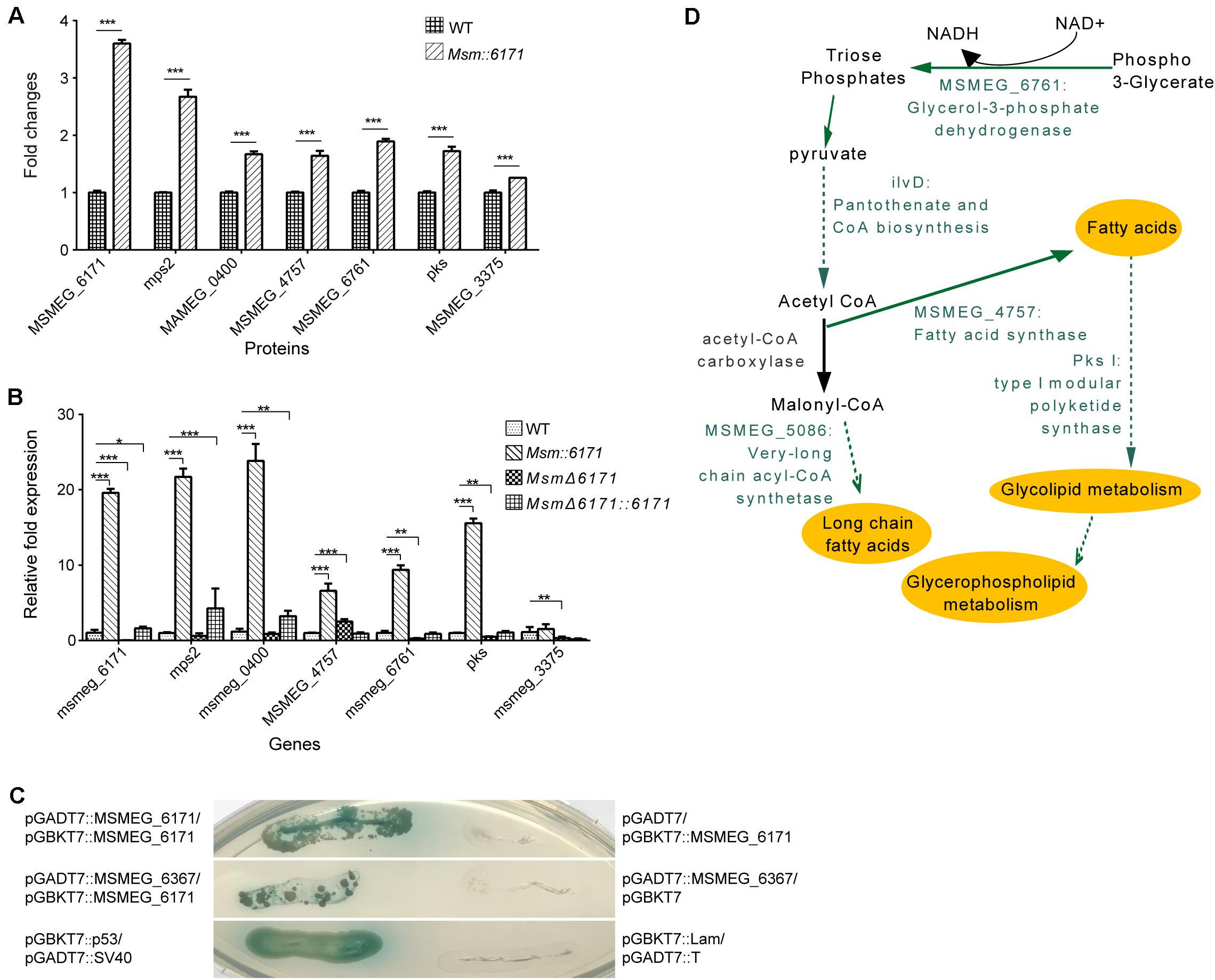
Figure 4. Identification of proteins interacting with MSMEG_6171. (A) Fold change of protein expression levels in Msm:6171 (including MSMEG_6171, mps2, MSMEG_0400, MSMEG_4757, MSMEG_6761, pks, MSMEG_3375) were calculated relative to WT control. ∗∗∗ represents P-value < 0.01. (B) Semi-quantitative RT-PCR analysis was performed to verify the expression of genes corresponding to the proteins in Figure 3C. Data were analyzed using the ΔΔCT method with the M. smegmatis reference gene sigA as the control. Data were mean values ± SD from independent biological samples. ∗∗∗ represents P-value < 0.01, and ∗ represents P-value < 0.05. (C) The GAL4-based Matchmaker Gold Yeast Two-Hybrid system was used to identify interacting partners of MSMEG_6171, which was cloned into the pGBKT7 vector as a fusion to the GAL4 DNA-binding domain (pGBKT7:MSMEG_6171). The prey proteins of MSMEG_6171 in the pull-down assay were expressed as fusions to the Gal4 activation domain using pGADT7AD. The screen revealed that MSMEG_6171 interacts with MSMEG_6367 and itself. P53 and SV40 were exploited to serve as the positive control. pGBKT7:MSMEG_6171/pGADT7AD, pGBKT7/pGADT7AD:MSMEG_6367, and pGBKT7:Lam/pGADT7:T represent negative controls. (D) Schematic summary of the specific proteins involved in lipid metabolism pathways. The green characters represent the significantly upregulated candidate MSMEG_6171-regulated proteins.
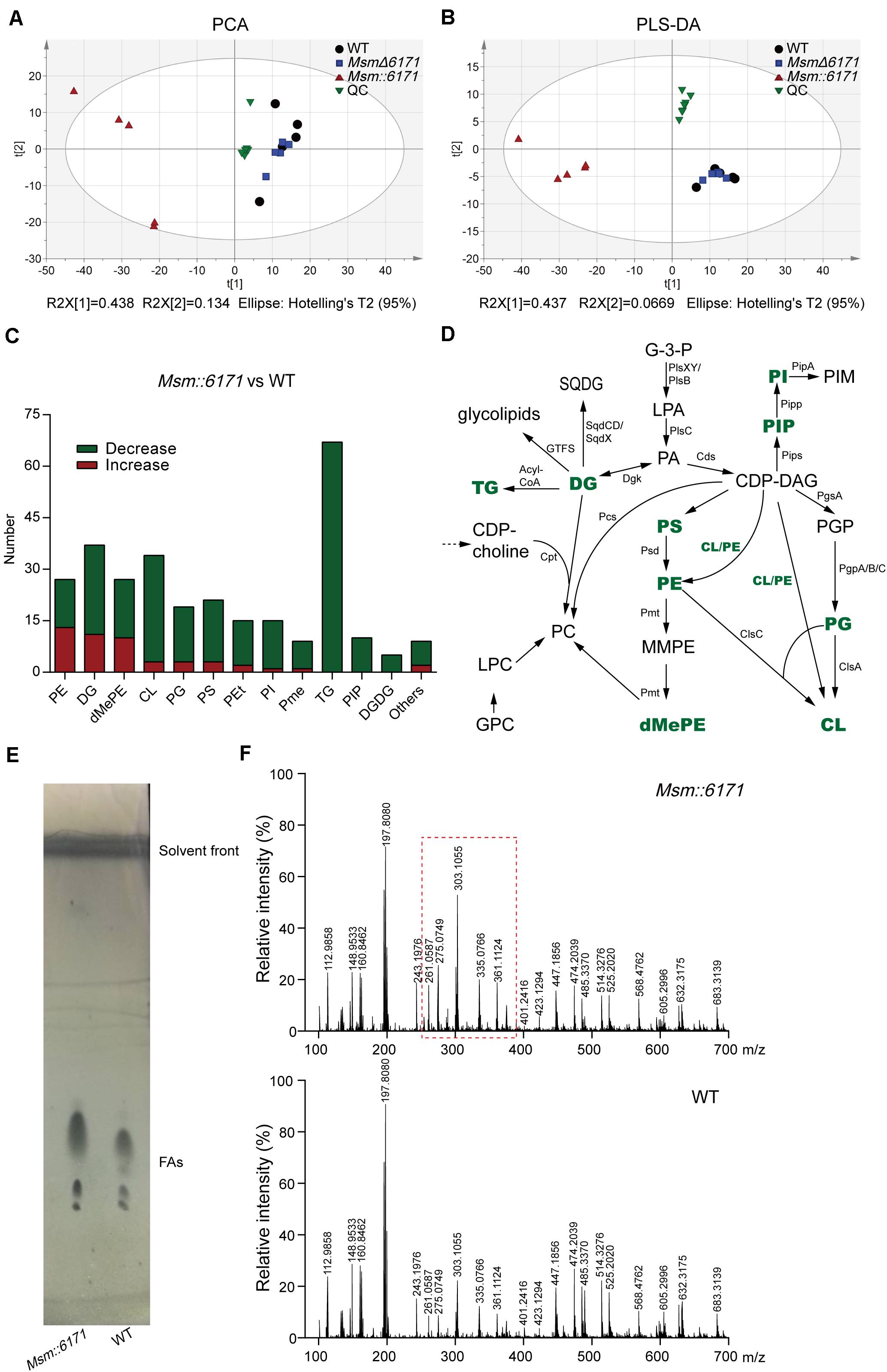
Figure 5. LC/MS-based metabolomics analysis of lipid metabolome. (A) PCA score and (B) PLS-DA score scatter plots acquired from LC-MS data in the ESI– and ESI+ modes. R2, the total explained variation of the model; Q2, the predictability of the model. WT, wild-type Msm mc2155 strain; QC, quality control. (C) The number of drastically altered lipids in the Msm:6171 strains compared with the WT strains. Red, increased lipids; green, decreased lipids. (D) Schematic representation of enriched pathways involved in the synthesis of membrane lipids. Green characters represent the differentially expressed lipids identified in the MSM:6171 strains. CL, cardiolipin; G3P, glycerol-3-phosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; GPC, glycerophosphocholine; PGP, phosphatidylglycerol phosphate; PG, phosphatidylglycerol; PIP, phosphatidylinositol phosphate; PI, phosphatidylinositol; PIM, phosphatidylinositol mannoside; SQDG, sulfoquinovosyl diacylglycerol; GTF, glycosyltransferase; DG, diacylglycerol; CDP, DG-cytidine diphosphate-diacylglycerol; PS, phosphatidylserine; MMPE, monomethyl PE; DMPE, dimethyl PE; PC, phosphatidylcholine; LPC, lysophosphatidylcholine; PE, phosphatidylethanolamine. (E) TLC analysis of fatty acids extracted from Msm:6171 and WT. Visualization was by phosphomolybdic acid spray. (F) ESI-MS-TOF mass spectra analysis of organic extracts from Msm:6171 and WT displaying the [M-H]– ions.
Using Phyre2 analysis, we acquired a three-dimensional model of MSMEG_6171 and showed the potential binding sites of ligands (Mg2+, ATP, and ADP) (Figure 6A). MSMEG_6171 presented two domains, an NTD (from Met1 to Gly80) and a CTD (G81 to A316). The CTD of MSMEG_6171 is predicted to be an ATPase that is highly similar to the bacterial cell division-related ATPase MinD (PDB code: 1g3r,% i.d. = 19) of Pyrococcus furiosus and the chromosome segregation protein Soj (PDB code: 2bek,% i.d. = 23). In contrast to the canonical motif (GGxxGxGK[ST]), MinD/Soj-like ATPases contain a deviant Walker-A (p-loop) motif (xKGGxGK[ST]), with two lysines as a subgroup of ATPases. One of the lysines is near the carboxyl end of the motif, which is common to all Walker-A motifs. The other one is at the beginning of the motif (Lutkenhaus and Sundaramoorthy, 2003). The Walker-A motif of a pilus assembly protein, ErTadZ of Eubacterium rectale, is further degraded (132PCGGVGTS139) with the loss of both lysines but shows weak ATPase activity. The p-loop of MSMEG_6171 (93KGGAGASV100) contains an alanine at the position corresponding to the second lysine but has the first conserved lysine. All of the above findings suggest that MSMEG_6171 may be an ATPase that plays an important role in cell length in accordance with the phenotype of overexpression or knockout of msmeg_6171. In the nucleic acid-binding pocket of the model of MSMEG_6171, the conserved P-loop (93KGGAGASV100) is similar to a clamp holding the ATP or ADP molecule. R249 participates in a polar interaction with the purine of ATP. D280 interacts with the 2′-OH of the ribose of ATP. The side chain of L279 acts as a scaffold to support the plane formed by the ribose of ATP.
Based on the predicted results mentioned above, we constructed two deletion mutants (MsmΔ6171:6171Δ 81–316 and MsmΔ6171:6171Δ 1–80) and three site-directed mutants of MSMEG_6171 (MsmΔ6171:6171S99A,V100A, MsmΔ6171:6171R249A, and MsmΔ6171:6171L279A,D280A) and then introduced these mutants into the MsmΔ6171 strains to test the effects of the mutations on the bacterial cell surface and envelope properties in vivo. As shown in Figures 6B–D, we found that all the mutants especially MsmΔ6171:6171R249A exhibited significant changes in colony morphology, biofilm formation, and sliding motility compared to the WT strains, except for the mutant MsmΔ6171:6171L279A,D280A. Furthermore, the MsmΔ6171:6171Δ 1–80 and MsmΔ6171:6171R249A mutant strains were more susceptible to vancomycin than the WT strains (Figure 6E). Surprisingly, the resistance of Msm to vancomycin was significantly enhanced when only the CTD was expressed. The NTD was not sufficient for vancomycin resistance. Taken together, our results indicated that R249 plays an important role in the resistance to vancomycin.
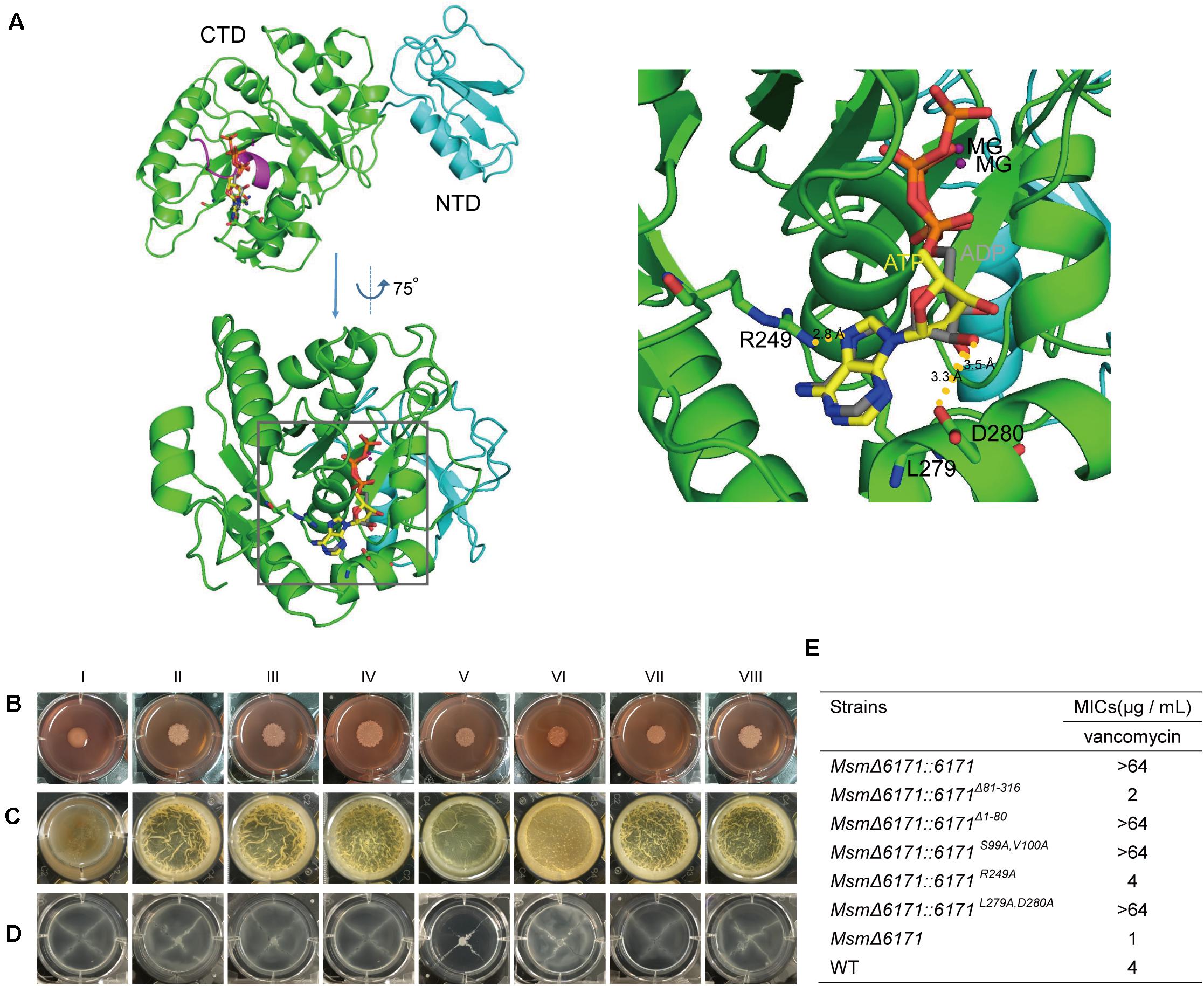
Figure 6. The msmeg_6171 mutants can affect bacterial colony morphology, surface hydrophobicity, biofilm formation, and spreading. (A) 3D-structural model of MSMEG_6171 predicted by Phyre 2. The monomer of MSMEG_6171 consists of two domains: an N-terminal domain (NTD) (cyan cartoons) and a C-terminal domain (CTD) (green cartoons). The p-loop (93KGGAGASV100) is shown as a magenta cartoon, and the ligands (ATP and ADP) are shown as sticks. Interfaces between MSMEG_6171 and the ligands (ATP and ADP) predicted by Phyre 2. The ATP and ADP are shown as sticks, and the nucleic acid-binding pocket is marked with the black frame. (G) With enlargement of the region of the binding pocket, R249 interacts with the purine of ATP, and D280 shows the interaction with 2′-OH of ribose of ATP, and the side chain of L279 likes the scaffold to support the plane formed by the ribose of ATP. (B) Colony morphology on Congo Red, (C) biofilm formation, and (D) sliding motility. (I) MsmΔ6171:6171, (II) MsmΔ6171:6171Δ 81– 316, (III) MsmΔ6171:6171Δ 1– 80, (IV) MsmΔ6171:6171S99A,V100A, (V) MsmΔ6171:6171R249A, (VI) MsmΔ6171:6171L279A, D280A, (VII) MsmΔ6171, and (VIII) WT. (E) Analysis of the susceptibility of the msmeg_6171 mutant strains to vancomycin.
Based on bioinformatics analysis, England et al. (2011) previously found that Rv3660c, the homologous protein of MSMEG_6171, plays an important role in cell morphology and adaptive metabolism. In this study, we experimentally characterized the role of MSMEG_6171 in Msm. Our results showed that the overexpression of MSMEG_6171 has an impact on bacterial physiology, from the increase in biofilm formation to the change in cell size and properties. These changes could alter the drug sensitivity of Msm to the antibiotics involved in cell wall synthesis, such as vancomycin. We found that filamentation possibly does not relate to Z-ring formation in the merodiploid and null strains of our experiments (Supplementary Figure S2). However, this result conflicted with England et al.’s (2011) previous report that the filamentation of Mtb rv3660c merodiploid strains had been caused by inhibition of FtsZ polymerization and Z-ring formation. Integrated proteome and metabolome results have shown that overexpression of MSMEG_6171 could regulate lipid biosynthesis and metabolism by inducing the alteration of glyceride and phospholipid to exhibit the pleiotropic phenotypes and various cellular responses. The results of this study enrich our understanding of the function of the MinD family proteins.
A combination of Flag-based pull-down, proteomics, and bioinformatics analyses led us to discover many proteins involved in lipid biosynthesis and metabolism enriched in MSMEG_6171 merodiploid strains, such as MSMEG_4757, Mps2, Pks, MSMEG_0400, and MSMEG_6761. MSMEG_4757 is a fatty acid synthase (fas) that catalyzes fatty acid synthesis. MSMEG_6761, a glycerol-3-phosphate dehydrogenase (GPDH), can catalyze the reversible redox conversion of glycerone phosphate to sn-glycerol 3-phosphate (Ou et al., 2006). Pks belongs to a family of multidomain enzymes that produce polyketides as a category of lipids (Jenke-Kodama et al., 2005). The mps2 gene encodes the mycobacterial peptide synthetase required for the formation of the tripeptide-amino-alcohol moiety, which is then linked to the 3-hydroxy/methoxy C26–C34 acyl chain, the lipid moiety of glycopeptidolipids (Jaspersen et al., 2006; Park et al., 2015). Our metabolomics results further showed that many glyceride and phospholipids were drastically decreased in merodiploid strains compared to wild strains and null strains, such as TGs, CLs, and other phospholipids. In Msm, the basic skeleton of the cell membrane showed the presence of glycerophospholipids including CL/PG, PE, PI, and PIMs. These glycerophospholipids isolated the cell from the external environment and ensured the independence of biochemical reactions as well as the formation of internal compartments in which different biological functions can be completed without interference (Pan et al., 2018).
It has been demonstrated that membrane lipid composition was considered to be a critical factor in determining cell size and shape, as well as a basic morphological parameter to ensure bacterial cell surface adhesion and biofilm formation. For example, the CL-lacking E. coli cells exhibit a filamentous phenotype in the M9 minimal medium. The elimination of CLs can result in a decrease in growth rate in the M9 medium, suggesting that CLs act as a crucial role in the maintenance of cell size and homeostasis and in proper cell division (Rowlett et al., 2017). A similar phenomenon emerged in this study that CL was significantly decreased when MSMEG_6171 was overexpressed, along with a lower growth rate and increased cell length. It is well known that DGs and TGs mainly exist in the outer membrane (mycomembrane) and inclusion bodies in the cytoplasm of Msm (Bansal-Mutalik and Nikaido, 2014). Baek et al. (2011) had demonstrated that TGs could play a causal role in governing growth, metabolic rate, and antibiotic susceptibility by reducing the carbon flux through the tricarboxylic acid cycle. Therefore, we hypothesized that the sharply reduced TG in MSMEG_6171-overexpressed strains was associated with bacterial phenotypic changes and drug sensitivity. The specific mechanism of its action required further study.
Notably, the significant changes in lipid profile and proteins associated with lipid biosynthesis and metabolism were only found in MSMEG_6171 merodiploid strains, not in MsmΔ6171 strains. It indicated that MSMEG_6171 may not be a key regulatory protein involved in lipid metabolism. It is also possible that other proteins with similar functions can compensate for its function when MSMEG_6171 was deleted.
Our TEM result revealed that MSMEG_6171 alters envelope ultrastructure varies between Msm:6171 and WT. The ETL is mainly composed of lipids like mycolic acids (Mdluli et al., 1998). Our proteomics and transcriptomics results showed that proteins involved in mycolic acid biosynthesis (fas) which interacts with MSMEG_6171 were significantly upregulated, and metabolomics results further showed that many glyceride and phospholipids were decreased in merodiploid strains compared to wild strains and null strains. All of these results indicate that active synthesis of mycolic acids may increase ETL thickness; consistently, TLC and ESI/MS analysis of mycolic acids is a reminder that MSMEG_6171 overexpression shows a positive correlation with mycolic acid content. Further investigations are needed to understand the role of MSMEG_6171 in regulating lipid synthesis and metabolism.
Vancomycin is an antibiotic, which targets lipid biosynthesis by specifically binding to lipid II to hinder subsequent building blocks from the penicillin-binding proteins, thereby inhibiting transglycosylation and transpeptidation. We found that overexpression of MSMEG_6171 could decrease the susceptibility of Msm to vancomycin. The isogenic deletion of MSMEG_6171 increased the susceptibility to vancomycin. Furthermore, the expression of vancomycin-responsive genes, such as MSMEG_1764, MSMEG_3945, and MSMEG_0586 (Provvedi et al., 2009; Zeng et al., 2016) were decreased in MSMEG_6171 multiploid strains, following increased vancomycin resistance.
Some reports have also shown that the lipid composition of bacterial cells could correlate with antibiotic resistance, because they were involved in the biosynthesis of major membrane lipids and lipoteichoic acid. Hewelt-Belka et al. (2016) found that MGDG, PG, and DGDG were upregulated in antibiotic-resistant S. aureus strains, whereas the levels of DG lipid groups were downregulated. They also found that PG was increased while CL and Lys-PG levels were decreased in methicillin-sensitive strains, compared with methicillin-resistant S. aureus. Another study showed that the expression of plasmid-mediated resistance to fusidic acid in a S. aureus strain was associated with changes in the molar ratio of PG to lysl-PG (Chopra, 1976). Provvedi et al. (2009) also reported that treatment with 10X-MIC vancomycin induced strong repression of genes required for lipid metabolism (32%) in Mtb. Therefore, differences in the lipid patterns between MSMEG_6171 multiploid strains and wild strains suggested that vancomycin susceptibility may be associated with the lipid composition of bacterial cells.
In this study, through integrating scanning electron microscopy analysis, transmission electron microscopy, proteomics, metabolomics, and molecular biology techniques, we reveal that MSMEG_6171 could function in cell envelope organization via regulating the lipid metabolism of Msm; this may further enhance the bacterial antibiotic resistance and result in other different important cellular phenotypes. MSMEG_6171 is predicted to be a MinD/Soj-like ATPase by bioinformatics analysis. We also analyzed its ATPase active site and found that R249 could play an important role in the bacterial phenotype and vancomycin resistance. The ATPase activity of MSMEG_6171 and its regulations of drug sensitivity and lipid metabolism still need to be confirmed in vitro by substantial experiments.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD014779 and 10.6019/PXD014779.
WW and ZW conceived, designed and supervised the overall study. XZ and LZ acquired funding and supervised and administered the project. WW and ZW provided the mutated mycobacterial strains. HG, QL, and LC coordinated the experiments to proceed smoothly. ZW, WW, YZ, and JZ processed the samples and performed the experiments. ZW, WW, and YZ analyzed the data. WW and ZW wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by A Major Infectious Disease Prevention and Control of the National Science and Technique Major Project (2018ZX10715004 and 2017ZX10201302), the Open Project of the State Key Laboratory for Infectious Diseases Prevention and Control (Grant No. 2019SKLID314), the National Natural Science Foundation of China (Grant No. 31800129), the Science and Technology Planning Project of Guangzhou (Grant No. 201904010149), and the Medical Scientific Research Foundation of Guangdong Province (Grant Nos. A2020611 and A2020072).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01572/full#supplementary-material
CL, cardiolipin; CTD, C-terminal domain; DG, diglyceride; DGDG, digalactosyl diglyceride; MGDG, monogalactosyl diglyceride; Msm, Mycobacterium smegmatis; Mtb, Mycobacterium tuberculosis; NTD, N-terminal domain; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidyl glycerin; PI, phosphatidylinositol; PIM, phosphatidyl-myo-inositol mannosides; pks, polyketide synthase; PS, phosphatidylserine; SQDG, sulfoquinovosyl diacylglycerol; TB, tuberculosis; TGs, triacylglycerols.
Aguilar-Ayala, D. A., Tilleman, L., Van Nieuwerburgh, F., Deforce, D., Palomino, J. C., Vandamme, P., et al. (2017). The transcriptome of Mycobacterium tuberculosis in a lipid-rich dormancy model through RNAseq analysis. Sci. Rep. 7:17665. doi: 10.1038/s41598-017-17751-x
Baek, S. H., Li, A. H., and Sassetti, C. M. (2011). Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 9:e1001065. doi: 10.1371/journal.pbio.1001065
Bailo, R., Bhatt, A., and Ainsa, J. A. (2015). Lipid transport in Mycobacterium tuberculosis and its implications in virulence and drug development. Biochem. Pharmacol. 96, 159–167. doi: 10.1016/j.bcp.2015.05.001
Bansal-Mutalik, R., and Nikaido, H. (2014). Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc. Natl. Acad. Sci. U.S.A. 111, 4958–4963. doi: 10.1073/pnas.1403078111
Bhat, Z. S., Rather, M. A., Maqbool, M., Lah, H. U., Yousuf, S. K., and Ahmad, Z. (2017). Cell wall: a versatile fountain of drug targets in Mycobacterium tuberculosis. Biomed. Pharmacother. 95, 1520–1534. doi: 10.1016/j.biopha.2017.09.036
Bjerrum, J. T., Wang, Y., Hao, F., Coskun, M., Ludwig, C., Gunther, U., et al. (2015). Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn’s disease and healthy individuals. Metabolomics 11, 122–133. doi: 10.1007/s11306-014-0677-3
Brennan, P. J. (2003). Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis 83, 91–97. doi: 10.1016/s1472-9792(02)00089-86
Carette, X., Platig, J., Young, D. C., Helmel, M., Young, A. T., Wang, Z., et al. (2018). Multisystem analysis of Mycobacterium tuberculosis reveals kinase-dependent remodeling of the pathogen-environment interface. mBio 9:e02333-17. doi: 10.1128/mBio.02333-17
Chopra, I. (1976). Mechanisms of resistance to fusidic acid in Staphylococcus aureus. J. Gen. Microbiol. 96, 229–238. doi: 10.1099/00221287-96-2-229
Conti, J., Viola, M. G., and Camberg, J. L. (2015). The bacterial cell division regulators MinD and MinC form polymers in the presence of nucleotide. FEBS Lett. 589, 201–206. doi: 10.1016/j.febslet.2014.11.047
Dahl, J. L. (2004). Electron microscopy analysis of Mycobacterium tuberculosis cell division. FEMS Microbiol. Lett. 240, 15–20. doi: 10.1016/j.femsle.2004.09.004
England, K., Crew, R., and Slayden, R. A. (2011). Mycobacterium tuberculosis septum site determining protein, Ssd encoded by rv3660c, promotes filamentation and elicits an alternative metabolic and dormancy stress response. BMC Microbiol. 11:79. doi: 10.1186/1471-2180-11-79
Falkinham, J. O. III (2018). Challenges of NTM drug development. Front. Microbiol. 9:1613. doi: 10.3389/fmicb.2018.01613
Fields, S., and Song, O. K. (1989). A novel genetic system to detect protein-protein interactions. Nature 340, 245–246. doi: 10.1371/journal.pcbi.1000659
Grzegorzewicz, A. E., de Sousa-d’Auria, C., McNeil, M. R., Huc-Claustre, E., Jones, V., Petit, C., et al. (2016). Assembling of the Mycobacterium tuberculosis cell wall core. J. Biol. Chem. 291, 18867–18879. doi: 10.1074/jbc.M116.739227
Grzegorzewicz, A. E., Eynard, N., Quemard, A., North, E. J., Margolis, A., Lindenberger, J. J., et al. (2015). Covalent modification of the Mycobacterium tuberculosis FAS-II dehydratase by isoxyl and thiacetazone. ACS Infect. Dis. 1, 91–97. doi: 10.1021/id500032q
Hett, E. C., and Rubin, E. J. (2008). Bacterial growth and cell division: a mycobacterial perspective. Microbiol. Mol. Biol. Rev. 72, 126–156. doi: 10.1128/mmbr.00028-07
Hewelt-Belka, W., Nakonieczna, J., Belka, M., Baczek, T., Namiesnik, J., and Kot-Wasik, A. (2016). Untargeted lipidomics reveals differences in the lipid pattern among clinical isolates of Staphylococcus aureus resistant and sensitive to antibiotics. J. Proteome Res. 15, 914–922. doi: 10.1021/acs.jproteome.5b00915
Horai, H., Arita, M., Kanaya, S., Nihei, Y., Ikeda, T., Suwa, K., et al. (2010). MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714. doi: 10.1002/jms.1777
Jaspersen, S. L., Martin, A. E., Glazko, G., Giddings, T. H. Jr., Morgan, G., Mushegian, A., et al. (2006). The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell. Biol. 174, 665–675. doi: 10.1083/jcb.200601062
Jenke-Kodama, H., Sandmann, A., Muller, R., and Dittmann, E. (2005). Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22, 2027–2039. doi: 10.1093/molbev/msi193
Katale, B. Z., Mbugi, E. V., Keyyu, J. D., Fyumagwa, R. D., Rweyemamu, M. M., van Helden, P. D., et al. (2019). One Health approach in the prevention and control of mycobacterial infections in Tanzania: lessons learnt and future perspectives. One Health Outlook 1:2. doi: 10.1186/s42522-019-0002-1
Kaur, G., and Kaur, J. (2017). Multifaceted role of lipids in Mycobacterium leprae. Future Microbiol. 12, 315–335. doi: 10.2217/fmb-2016-0173
Khan, M. Z., Bhaskar, A., Upadhyay, S., Kumari, P., Rajmani, R. S., Jain, P., et al. (2017). Protein kinase G confers survival advantage to Mycobacterium tuberculosis during latency-like conditions. J. Biol. Chem. 292, 16093–16108. doi: 10.1074/jbc.M117.797563
King, G. M. (2003). Uptake of carbon monoxide and hydrogen at environmentally relevant concentrations by mycobacteria. Appl. Environ. Microbiol. 69, 7266–7272. doi: 10.1128/aem.69.12.7266-7272.2003
Lai, L. Y., Lin, T. L., Chen, Y. Y., Hsieh, P. F., and Wang, J. T. (2018). Role of the Mycobacterium marinum ESX-1 secretion system in sliding motility and biofilm formation. Front. Microbiol. 9:1160. doi: 10.3389/fmicb.2018.01160
Layre, E., Al-Mubarak, R., Belisle, J. T., and Branch Moody, D. (2014). Mycobacterial lipidomics. Microbiol. Spectr. 2, 341–360. doi: 10.1128/microbiolspec.MGM2-0033-2013
Lee, S. Y., Kim, H. Y., Kim, B. J., Kim, H., Seok, S. H., Kim, B. J., et al. (2017). Effect of amikacin on cell wall glycopeptidolipid synthesis in Mycobacterium abscessus. J. Microbiol. 55, 640–647. doi: 10.1007/s12275-017-6503-7
Li, Q., Ge, F., Tan, Y., Zhang, G., and Li, W. (2016). Genome-Wide Transcriptome Profiling of Mycobacterium smegmatis MC2 155 cultivated in minimal media supplemented with cholesterol, androstenedione or glycerol. Int. J. Mol. Sci. 17:689. doi: 10.3390/ijms17050689
Liu, J., and Nikaido, H. (1999). A mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids accumulates meromycolates. Proc. Natl. Acad. Sci. U.S.A. 96, 4011–4016. doi: 10.1073/pnas.96.7.4011
Lutkenhaus, J., and Sundaramoorthy, M. (2003). MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol. Microbiol. 48, 295–303. doi: 10.1046/j.1365-2958.2003.03427.x
Maeda, H., Fujimoto, C., Haruki, Y., Maeda, T., Kokeguchi, S., and Petelin, et al. (2003). Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 39, 81–86. doi: 10.1016/s0928-8244(03)00224-4
Marston, A. L., Thomaides, H. B., Edwards, D. H., Sharpe, M. E., and Errington, J. (1998). Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12, 3419–3430. doi: 10.1101/gad.12.21.3419
Maya-Hoyos, M., Leguizamon, J., Marino-Ramirez, L., and Soto, C. Y. (2015). Sliding motility, biofilm formation, and glycopeptidolipid production in Mycobacterium colombiense strains. Biomed. Res. Int. 2015:419549. doi: 10.1155/2015/419549
Mdluli, K., Swanson, J., Fischer, E., Lee, R. E., and Barry, C. E. I. I. I. (1998). Mechanisms involved in the intrinsic isoniazid resistance of Mycobacterium avium. Mol. Microbiol. 27, 1223–1233. doi: 10.1046/j.1365-2958.1998.00774.x
Mengatto, L., Chiani, Y., and Imaz, M. S. (2006). Evaluation of rapid alternative methods for drug susceptibility testing in clinical isolates of Mycobacterium tuberculosis. Mem. Inst. Oswaldo Cruz 101, 535–542. doi: 10.1590/s0074-02762006000500009
Meniche, X., Otten, R., Siegrist, M. S., Baer, C. E., Murphy, K. C., Bertozzi, C. R., et al. (2014). Subpolar addition of new cell wall is directed by diviva in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 111, E3243–E3251. doi: 10.1073/pnas.1402158111
Ou, X., Ji, C., Han, X., Zhao, X., Li, X., Mao, Y., et al. (2006). Crystal structures of human glycerol 3-phosphate dehydrogenase 1 (GPD1). J. Mol. Biol. 357, 858–869. doi: 10.1016/j.jmb.2005.12.074
Pan, J., Hu, C., and Yu, J.-H. (2018). Lipid biosynthesis as an antifungal target. J. Fungi 4:50. doi: 10.3390/jof4020050
Pang, L., Tian, X., Pan, W., and Xie, J. (2013). Structure and function of mycobacterium glycopeptidolipids from comparative genomics perspective. J. Cell. Biochem. 114, 1705–1713. doi: 10.1002/jcb.24515
Parish, T., Lewis, J., and Stoker, N. G. (2001). Use of the mycobacteriophage L5 excisionase in Mycobacterium tuberculosis to demonstrate gene essentiality. Tuberculosis 81, 359–364. doi: 10.1054/tube.2001.0312
Parish, T., and Stoker, N. G. (2000). Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146(Pt 8), 1969–1975. doi: 10.1099/00221287-146-8-1969
Park, I. K., Hsu, A. P., Hervé, T., Shallom, S. J., and Zelazny, A. M. (2015). Clonal diversification, changes in lipid traits and colony morphology in Mycobacterium abscessus clinical isolates. J. Clin. Microbiol. 53, 3438–3447. doi: 10.1128/JCM.02015-15
Pontali, E., D’Ambrosio, L., Centis, R., Sotgiu, G., and Migliori, G. B. (2017). Multidrug-resistant tuberculosis and beyond: an updated analysis of the current evidence on bedaquiline. Eur. Resp. J. 49:1700146. doi: 10.1183/13993003.00146-2017
Provvedi, R., Boldrin, F., Falciani, F., Palu, G., and Manganelli, R. (2009). Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 155(Pt 4), 1093–1102. doi: 10.1099/mic.0.024802-0
Rens, C., Laval, F., Daffe, M., Denis, O., Frita, R., Baulard, A., et al. (2016). Effects of lipid-lowering drugs on vancomycin susceptibility of mycobacteria. Antimicrob. Agents Chemother. 60, 6193–6199. doi: 10.1128/aac.00872-16
Ripoll, F., Deshayes, C., Pasek, S., Laval, F., Beretti, J. L., Biet, F., et al. (2007). Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC Genomics 8:114. doi: 10.1186/1471-2164-8-114
Rodriguez-Rivera, F. P., Zhou, X., Theriot, J. A., and Bertozzi, C. R. (2018). Acute modulation of mycobacterial cell envelope biogenesis by front-line tuberculosis drugs. Angew. Chem. Int. Ed. Engl. 57, 5267–5272. doi: 10.1002/anie.201712020
Rombouts, Y., Brust, B., Ojha, A. K., Maes, E., Coddeville, B., Elass-Rochard, E., et al. (2012). Exposure of mycobacteria to cell wall-inhibitory drugs decreases production of arabinoglycerolipid related to Mycolyl-arabinogalactan-peptidoglycan metabolism. J. Biol. Chem. 287, 11060–11069. doi: 10.1074/jbc.M111.327387
Rowlett, V. W., Mallampalli, V., Karlstaedt, A., Dowhan, W., Taegtmeyer, H., Margolin, W., et al. (2017). Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 199:e00849-16. doi: 10.1128/jb.00849-16
Russell, D. G., Barry, C. E. III, and Flynn, J. L. (2010). Tuberculosis: what we don’t know can, and does, hurt us. Science 328, 852–856. doi: 10.1126/science.1184784
Santucci, P., Bouzid, F., Smichi, N., Poncin, I., Kremer, L., De Chastellier, C., et al. (2016). Experimental models of foamy macrophages and approaches for dissecting the mechanisms of lipid accumulation and consumption during dormancy and reactivation of Tuberculosis [J]. Front. Cell. Infect. Microbiol. 6:122. doi: 10.3389/fcimb.2016.00122
Schuhmacher, J. S., Rossmann, F., Dempwolff, F., Knauer, C., Altegoer, F., Steinchen, W., et al. (2015). MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc. Natl. Acad. Sci. U.S.A. 112, 3092–3097. doi: 10.1073/pnas.1419388112
Sharma, S., Rajmani, R. S., Kumar, A., Bhaskar, A., Singh, A., Manivel, V., et al. (2015). Differential proteomics approach to identify putative protective antigens of Mycobacterium tuberculosis presented during early stages of macrophage infection and their evaluation as DNA vaccines. Indian J. Exp. Biol. 53, 429–439.
Tiwari, A., Suryawanshi, P., Raikwar, A., Arif, M., and Richardus, J. H. (2018). Household expenditure on leprosy outpatient services in the Indian health system: a comparative study. PLoS Negl. Trop. Dis. 12:e0006181. doi: 10.1371/journal.pntd.0006181
Vijay, S., Anand, D., and Ajitkumar, P. (2012). Unveiling unusual features of formation of septal partition and constriction in mycobacteria–an ultrastructural study. J. Bacteriol. 194, 702–707. doi: 10.1128/JB.06184-11
Vizcaino, J. A., Csordas, A., Del-Toro, N., Dianes, J. A., Griss, J., Lavidas, I., et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44:11033. doi: 10.1093/nar/gkw880
Zanfardino, A., Migliardi, A., D’Alonzo, D., Lombardi, A., Varcamonti, M., and Cordone, A. (2016). Inactivation of MSMEG_0412 gene drastically affects surface related properties of Mycobacterium smegmatis. BMC Microbiol. 16:267. doi: 10.1186/s12866-016-0888-z
Keywords: MSMEG_6171, lipid metabolism, Mycobacterium smegmatis, drug resistance, multi-omics
Citation: Wu Z, Wei W, Zhou Y, Guo H, Zhao J, Liao Q, Chen L, Zhang X and Zhou L (2020) Integrated Quantitative Proteomics and Metabolome Profiling Reveal MSMEG_6171 Overexpression Perturbing Lipid Metabolism of Mycobacterium smegmatis Leading to Increased Vancomycin Resistance. Front. Microbiol. 11:1572. doi: 10.3389/fmicb.2020.01572
Received: 22 July 2019; Accepted: 17 June 2020;
Published: 24 July 2020.
Edited by:
Satoshi Tsuneda, Waseda University, JapanReviewed by:
Vijay Srinivasan, University of Oxford, United KingdomCopyright © 2020 Wu, Wei, Zhou, Guo, Zhao, Liao, Chen, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Zhang, emhhbmd4aWFvbGkwMTA1QDE2My5jb20=; Lin Zhou, emhvdWxpbmdqa0AxNjMuY29t
†Co-first authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.