
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 07 July 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01565
This article is part of the Research Topic Stress Response and Effectors as Therapeutic Targets View all 4 articles
Given that a subpopulation of most bacterial cells becomes dormant due to stress, and that the resting cells of pathogens can revive and reconstitute infections, it is imperative to find methods to treat dormant cells to eradicate infections. The dormant bacteria that are not spores or cysts are known as persister cells. Remarkably, in contrast to the original report that incorrectly indicated indole increases persistence, a large number of indole-related compounds have been found in the last few years that kill persister cells. Hence, in this review, along with a summary of recent results related to persister cell formation and resuscitation, we focus on the ability of indole and substituted indoles to combat the persister cells of both pathogens and non-pathogens.
Persisters are stress tolerant cells that arise due to metabolic inactivity (Hobby et al., 1942; Bigger, 1944; Kwan et al., 2013; Pontes and Groisman, 2019) and without genetic change (Lewis, 2010). This dormancy was established by the original work with persisters showing non-growing Staphylococcus aureus cells are tolerant to penicillin (Hobby et al., 1942; Bigger, 1944). In contrast to persistence, which occurs in a small sub-population of cells, resistance occurs when mutations arise that allow growth in the presence of the antibiotic, and tolerance occurs when slow growth (e.g., stationary-phase cells) makes the entire population less susceptible to the antibiotic (Kaldalu et al., 2016; Kudrin et al., 2017). We have tried to clarify these terms to reduce the confusion in the persister-related literature (Wood et al., 2013; Kim and Wood, 2016, 2017; Kim et al., 2018a; Wood and Song, 2020) and tried to indicate how mistakes are being made in the persister literature by not waiting for a true plateau in the classic graph of the remaining viable cells during stress conditions that indicates the presence of persister cells (i.e., “biphasic” cell graph) (Song and Wood, 2020a). In addition, there is another term for the dormant state, “viable but non-culturable,” but we have demonstrated that the viable fraction of these cells is the same as persisters cells, at least for Escherichia coli and enterohemorrhagic E. coli (EHEC; Kim et al., 2018a).
Persisters have been shown to form from nutrient, antibiotic, acid, and oxidative stress (Hong et al., 2012; Kim et al., 2018a). Since nearly all cells starve (Song and Wood, 2018), persistence is likely a universal resting state of Bacteria and Archaea (Song and Wood, 2020b). Although persistence occurs to a small extent spontaneously (Balaban et al., 2004), it primarily arises as a highly regulated response to the environment (Dörr et al., 2010; Möker et al., 2010; Vega et al., 2012; Kwan et al., 2013, 2015; Hu et al., 2015; Song and Wood, 2020a; Wood and Song, 2020). This environmental response results in a small sub-population of stress-tolerant cells (∼1% or less) in biofilms and in stationary-phase cultures (Lewis, 2007, 2008).
As expected from a universal trait, persistence has been seen in all bacterial species tested (Van den Bergh et al., 2017). Strikingly, chronic infections are probably caused by resuscitated persister cells (Lewis, 2010; Van den Bergh et al., 2017); hence, they are important for cystic fibrosis (Lewis, 2007) and tuberculosis (Jayaraman, 2008). Therefore, understanding persistence is vital for developing more effective treatments for bacterial infections.
ppGpp has been linked to persistence (Korch et al., 2003; Nguyen et al., 2011; Chowdhury et al., 2016a; Svenningsen et al., 2019); hence, there is near consensus (Korch et al., 2003; Nguyen et al., 2011; Chowdhury et al., 2016a) for a role of the alarmone ppGpp for forming persisters (Svenningsen et al., 2019). However, until recently, the mechanism by which ppGpp leads to the formation of persister cells has been enigmatic.
To understand the link between ppGpp and persistence, it is informative to understand how ppGpp slows metabolism. To weather stressful conditions, cells reduce replication, transcription, and translation by synthesizing guanosine tetraphosphate and guanosine pentaphosphate (henceforth, ppGpp) (Gaca et al., 2015). ppGpp slows DNA replication by inhibiting DNA primase (Gaca et al., 2015), and ppGpp slows transcription by stimulating RpoS (sigmaS, the stress response sigma factor for the stationary phase) and RpoE (sigmaE, the stress response sigma factor for misfolded proteins in the periplasm) (Dalebroux and Swanson, 2012). ppGpp also inhibits the synthesis of purine nucleotides (Wang et al., 2019) and regulates purine homeostasis through its activation of nucleosidase PpnN (Zhang Y. E. et al., 2019). ppGpp slows translation by reducing the production of ribosomes (Shimada et al., 2013b).
The activity of specific proteins is also reduced directly by ppGpp; for example, ppGpp binds and inhibits GTPases (Gaca et al., 2015). ppGpp also binds to GTPase HflX, the protein that activates dormant 100S ribosomes (Zhang et al., 2018), to prevent reactivation of inactivated ribosomes (Corrigan et al., 2016; Zhang et al., 2018) (Figure 1A). In addition, ppGpp inhibits the ribosome-associated GTPase Era that is involved in the biogenesis of 30S ribosome subunits (Wood et al., 2019).
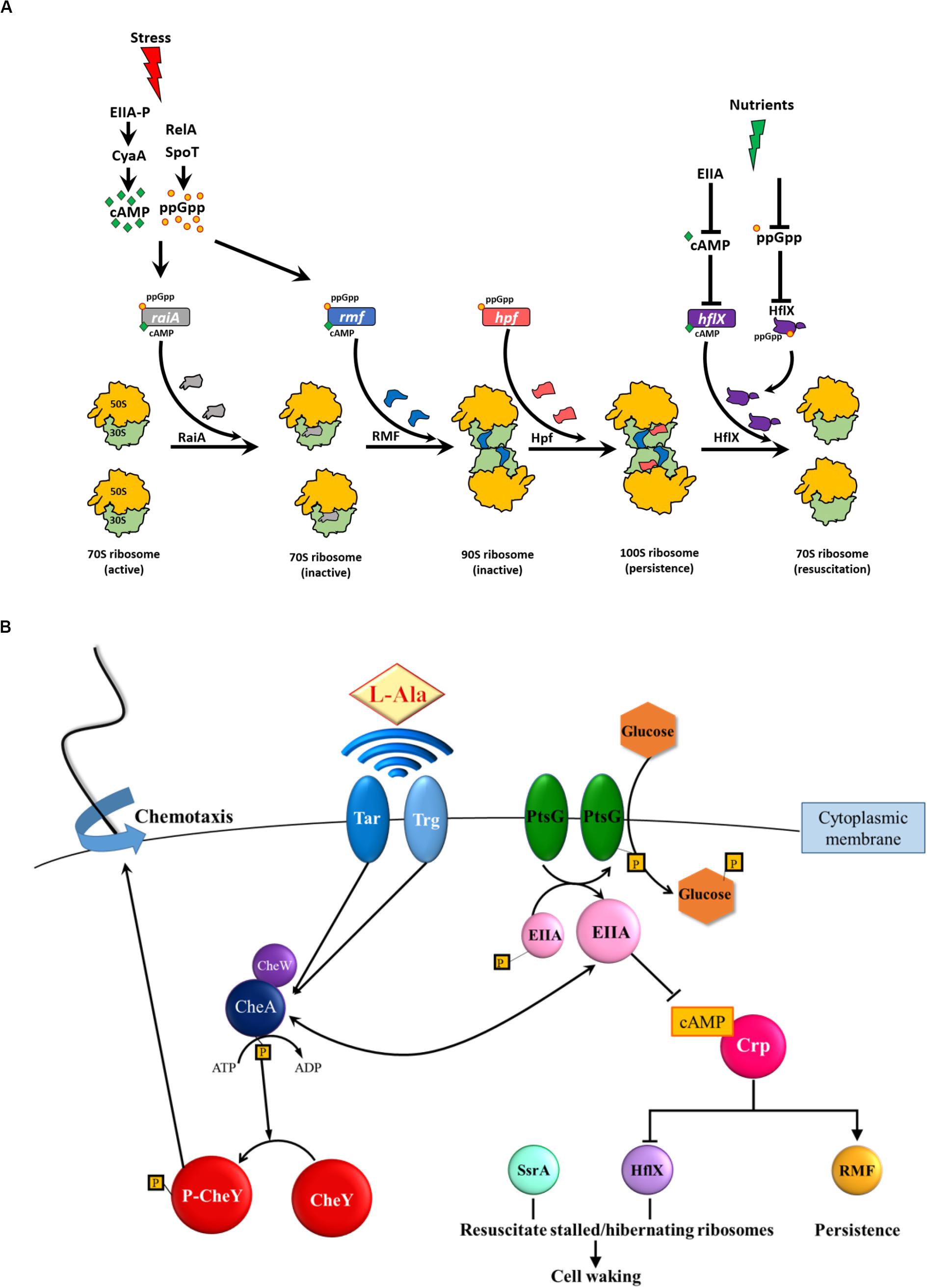
Figure 1. (A) ppGpp ribosome dimerization persister (PRDP) model for generating and resuscitating persister cells (Wood and Song, 2020). Myriad stresses (e.g., antibiotics, nutrient limitation, osmotic stress, and acid stress) induce the stringent response which results in (p)ppGpp (henceforth ppGpp) formation by RelA/SpoT in E. coli and generation of cAMP (e.g., upon glucose depletion via the phosphorylated glucose phosphotransfer enzyme, EIIA-P). ppGpp induces the genes encoding ribosome inactivation proteins, raiA, hpf, and hpf and cAMP induces raiA and rmf. RaiA inactivates 70S ribosomes, RMF converts 70S ribosomes into (inactive 90S ribosomes, and Hpf converts inactive 90S ribosomes into 100S ribosomes. At the protein level, ppGpp binds GTPase HflX to likely inactivate it (by blocking GTP binding), and cAMP represses hflX. With removal of the stress and the addition of nutrients, cAMP levels decrease (due to unphosphorylated EIIA) which stimulates HflX production; HflX dissociates 100S ribosomes into active 70S ribosomes and growth resumes. Used with permission. (B) Schematic of persister cell waking via alanine and glucose (Yamasaki et al., 2020). For alanine resuscitation, methyl-accepting chemotaxis proteins Tar and Trg sense the amino acid and relay this to chemotaxis response regulators CheA and CheY, which stimulate chemotaxis. For glucose resuscitation, phosphotransferase protein PtsG imports the sugar, which results in dephosphorylation of EIIA, reduction in cAMP, activation of chemotaxis, and ribosome rescue via HflX and SsrA. Spheres indicate proteins, diamonds indicate amino acids, hexagons indicate glucose, boxed P indicates phosphate, → indicates induction, and -∣ indicates repression. Used with permission from Elsevier (license #4807600114542).)
Critically, for persister cell formation, ppGpp inactivates ribosomes (Figure 1A) by (i) inducing rmf (Izutsu et al., 2001), which encodes the ribosome modulation factor (RMF) that inactivates 70S ribosomes, (ii) inducing hpf (Prossliner et al., 2018), which encodes the hibernation promoting factor (Hpf), and (iii) inducing raiA (Prossliner et al., 2018), which encodes the ribosome-associated inhibitor (RaiA).
Others have focused on determining how ppGpp activates toxins of toxin/antitoxin (TA) systems and leads to persistence, but these works have been retracted (Maisonneuve et al., 2018a, b; Maisonneuve et al., 2019). Instead, we have proposed the simpler ribosome dimerization persister (PRDP) model (Figure 1A) in which ppGpp generates persister cells directly; i.e., without TA systems, by inactivating ribosomes by converting 70S ribosomes into inactive 100S ribosomes (Song and Wood, 2020b; Wood and Song, 2020). In support of this model, we found (Song and Wood, 2020b) that (i) most ribosomes in persister cells are inactive as 100S ribosomes, (ii) inactivation of RMF, Hpf, and RaiA leads to the formation of fewer persister cells and increases single-cell persister resuscitation substantially, and (iii) single-cell persister resuscitation is not affected by ppGpp levels. This model does not rely on TA systems for persister cell formation as their link to persistence is unconvincing (Conlon, 2016; Goormaghtigh et al., 2018; Pontes and Groisman, 2019).
Since persistence occurs without ppGpp, although at much lower levels (Chowdhury et al., 2016a), the PRDP model also includes a role for cAMP in activating RMF and Hpf without ppGpp, which leads to the formation of inactive and 100S ribosomes (Figure 1A). Specifically, starvation (e.g., glucose depletion) leads to elevated cAMP, induces rmf (Shimada et al., 2013b) and induces raiA (Prossliner et al., 2018). In addition, cAMP represses hflX (Lin et al., 2011). Therefore, cAMP plays a similar role to ppGpp for persister cell formation, since increased concentrations of both cell signals lead to ribosome inactivation and persistence in a sub-population of cells.
For persister cell resuscitation (Figure 1B), using single cells, we were the first to demonstrate persister cells resuscitate in an heterogeneous manner as they recognize external nutrients; the rate of resuscitation depends on the number of active ribosomes (Kim et al., 2018b). This heterogeneous nature of persister cell resuscitation was subsequently verified by others (Goormaghtigh and Van Melderen, 2019; Pu et al., 2019). Using single cells and searches over all E. coli proteins, we determined that persister cell resuscitation is initiated by recognizing external nutrients through receptors for chemotaxis (for amino acids) and phosphotransferase membrane proteins (for glucose) and does not require proteins specialized for persistence (Figure 1B) (Yamasaki et al., 2020). Resuscitation is also not primarily spontaneous but instead is based on the recognition of nutrients (Yamasaki et al., 2020). The presence of external nutrients (i.e., signals) is propagated to the cytosol by reducing concentrations of the secondary messenger cAMP; reduction in cAMP allows ribosomes stalled on mRNA to be rescued and inactive 100S ribosomes to be activated by HflX (Figure 1B) (Yamasaki et al., 2020). The resuscitating cells also initiate chemotaxis toward fresh nutrients, which is logical since nutrient depletion triggered persistence in the first place (Yamasaki et al., 2020). Therefore, we discovered specific signals for resuscitation, how those signals are detected by the exterior of the cell, how that external signal is propagated inside the cell via a second messenger, and that the cell initiates chemotaxis to nutrients upon waking (Yamasaki et al., 2020).
The PRDP model (Figure 1A) suggests that persister cell formation is an elegantly regulated response to stress. Experimental support for this idea is that spontaneous persisters are rare (Balaban et al., 2004) but various environmental forms of stress (e.g., antibiotics, hydrogen peroxide, acid) can convert almost the whole exponentially growing population into persister cells (Hong et al., 2012; Kwan et al., 2013). Similarly, the PRDP model suggests persister cell resuscitation is also an elegant environmental response rather than a spontaneous event, and our data with resuscitation with the amino acid alanine supports this (Yamasaki et al., 2020). Since nearly all cells face nutrient limitations and need dormant states to weather this stress, it is reasonable that cells require elegant regulation for both persister cell formation and resuscitation. Critically, the PRDP model suggests the “phenotypic switch” for persistence is predicated on the number of ribosomes inactivated; hence, only a small sub-population of stressed cells become persistent since they are the cells with a threshold level of ribosomes inactivated (Song and Wood, 2020a; Wood and Song, 2020); i.e., not all stationary cells are persisters since not all of these cells have a large enough percentage of ribosomes inactivated.
The PRDP model is general in that it is applicable to how persister cells form from various stresses since RMF has been shown to increase persistence dramatically in E. coli for myriad stresses including (i) ampicillin (Song and Wood, 2020b), ciprofloxacin (Song and Wood, 2020b), netilmicin (Tkachenko et al., 2017), gentamicin (McKay and Portnoy, 2015), acid (El-Sharoud and Niven, 2007), osmotic stress (Shcherbakova et al., 2015), and nutrient limitation (Yamagishi et al., 1993; Bubunenko et al., 2007). Furthermore, since RMF (Prossliner et al., 2018) and HflX (Basu and Yap, 2017) are conserved in bacteria, and Hpf is distributed in several kingdoms (i.e., prokaryotes and plants) (Akiyama et al., 2018), the PRDP model is probably applicable for the formation of the persister cells of many species. For example, persister cell formation of the opportunistic pathogen Pseudomonas aeruginosa also requires ppGpp (Nguyen et al., 2011) and both Hpf and ppGpp (but not RMF) are necessary for protecting ribosomes and ensuring the long term survival of P. aeruginosa during nutrient limitation (Akiyama et al., 2017). Furthermore, ppGpp plays a role in hpf expression in P. aeruginosa (Akiyama et al., 2018). Critically, for cysts of Rhodospirillum centenum, the first genes activated for waking encode for ribosomes and translation machinery (initiation, elongation, and release factors) (Ashok and Bauer, 2020); hence, it appears the PRDP model holds for many species and resting states.
Indole, a product of tryptophan metabolism, is a multi-tiered signal in that it is an intra-species, inter-species, and interkingdom signal. As an intra-species signal, indole controls the quorum-sensing of E. coli (Lee et al., 2007a) primarily at low temperatures (Lee et al., 2008). As an interspecies signal, indole reduces the virulence of P. aeruginosa, which does not synthesize it, by reducing the virulence factors pyocyanin, rhamnolipid, 2-heptyl-3-hydroxy-4(1H)-quinolone, and pyoverdine (Lee et al., 2009a); this leads to increased competitiveness of commensal E. coli with P. aeruginosa (Chu et al., 2012). Also as an interspecies signal, indole reduces the virulence of EHEC by repelling it (negative chemotaxis), and by reducing its biofilm formation, motility, and attachment to HeLa cells (Bansal et al., 2007). Hence, we have suggested indole may be used as an anti-virulence compound (Lee et al., 2009a, 2015), and, indeed, indole was used successfully to reduce the virulence of P. aeruginosa in guinea pigs by reducing pulmonary colonization and increasing clearance in the lungs (Lee et al., 2009a). Twelve years later, the Sperandio group confirmed that indole reduces EHEC virulence in the gastrointestinal (GI) tract (Kumar and Sperandio, 2019). Furthermore, indole reduces the pathogenicity of S. aureus (Lee et al., 2013).
Strikingly, indole is an interkingdom signal, too. In the GI tract, indole produced by commensal bacteria tightens human epithelial cell junctions which reduces invasion by pathogens (Bansal et al., 2010; Shimada et al., 2013a). Also in the GI tract, we hypothesized that indole is probably hydroxylated by oxygenases to become an even more potent signal; for example, 7-hydroxyindole diminishes the virulence of P. aeruginosa more effectively than indole (Lee et al., 2007a). Furthermore, since many human and plant hormones are indole derivatives (e.g., indole-3-acetic acid, serotonin, melatonin, epinephrine), indole may be the archetype for cell hormones (Lee et al., 2007b). Further evidence showing indole in an interkingdom signal includes that for some plants (e.g., maize), indole is emitted to warn other plants of herbivores like the beet armyworm (Frey et al., 2000; Erb et al., 2015).
Moreover, indole reduces E. coli biofilm formation (Domka et al., 2006, 2007; Lee et al., 2007a, b, 2009b) and its production is reduced in biofilms (Domka et al., 2007). Also, by investigating the TA system YafQ/DinJ (Hu et al., 2015) and the phosphodiesterase DosP (Kwan et al., 2015), it was discovered that indole reduces E. coli persister cell formation.
Although there is one report claiming indole increases persistence with E. coli (Vega et al., 2012), consistent and overwhelming evidence has shown indole and substituted indoles reduce persistence in both Bacteria and Archaea (Hu et al., 2015; Kwan et al., 2015; Lee et al., 2016; Megaw and Gilmore, 2017; Li et al., 2019; Song et al., 2019; Manoharan et al., 2020; Masuda et al., 2020; Sun et al., 2020; Yam et al., 2020). For years, this was perplexing but it seems the most-probable reason for this different result lies in the solvent utilized to solubilize indole. Indole is relatively insoluble so to reach physiological concentrations (about 1 mM), a diluent must be used; dimethyl sulfoxide is the preferred solvent (Song et al., 2019) given it has little effect on cells if kept at less than 0.2 volume percent. In contrast, ethanol is not preferred due to its toxicity. Therefore, inconsistent results are most likely due to solvent effects. Hence, experiments with indole should include (i) suitable negative controls (i.e., solvent addition without indole) and (ii) multiple indole stock solutions to keep solvent addition uniform as indole concentrations are varied. In this way, indole is studied rather than the diluent.
We previously organized chemicals used to combat persister cells into three categories: (i) preventing persister cell formation, (ii) killing dormant cells, and (iii) resuscitating dormant cells followed by killing by traditional antibiotics (Figure 2) (Wood, 2016). As we show in this section, indigoids primarily inhibit persistence by killing dormant cells as a result of membrane damage.
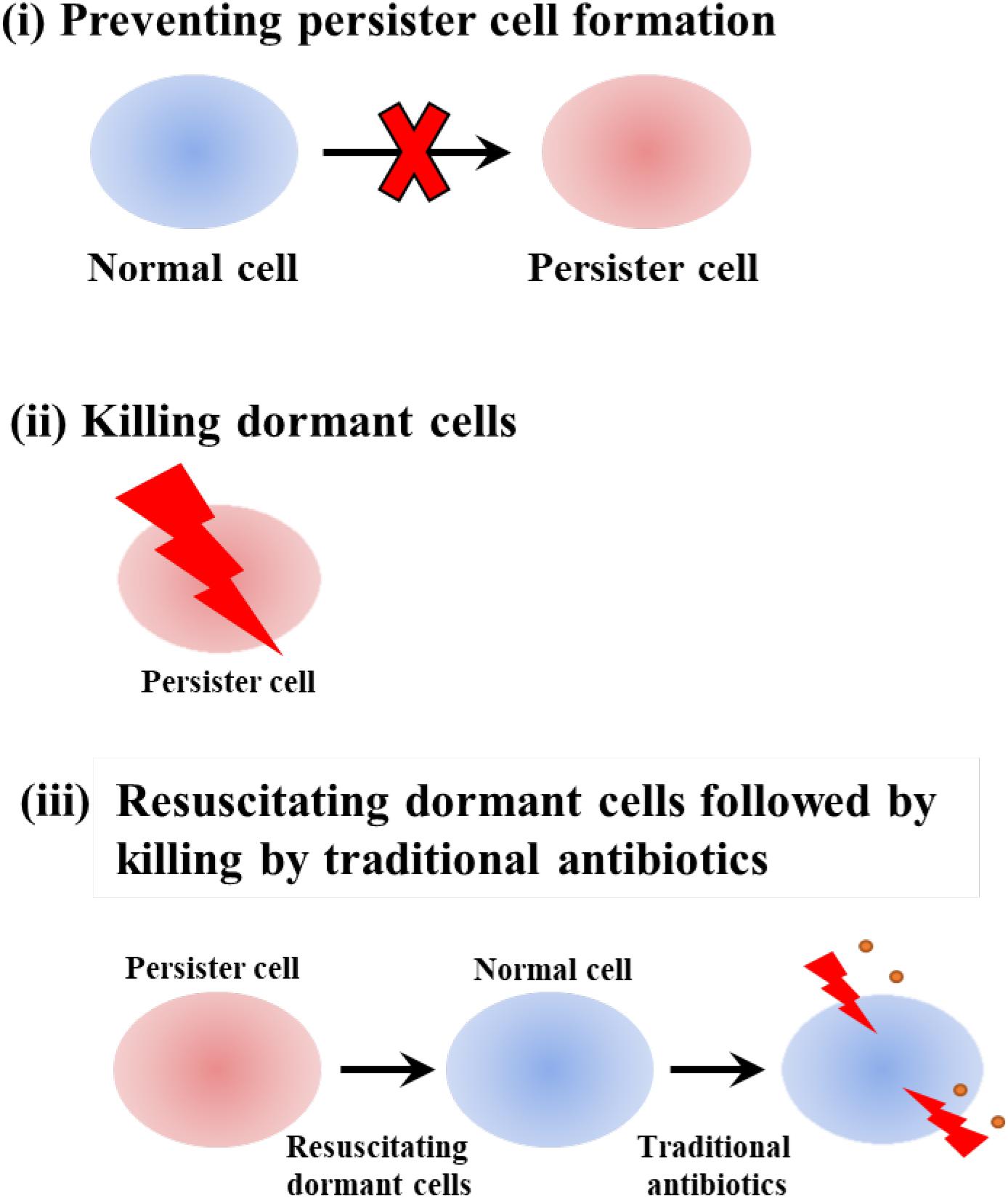
Figure 2. Schematic of combatting persister cells by (i) preventing persister cell formation, (ii) killing dormant cells, and (iii) resuscitating dormant cells followed by killing by traditional antibiotics.
For E. coli, we discovered 2 mM indole reduces persistence (Hu et al., 2015; Kwan et al., 2015) and found the effect with ampicillin to be about 52-fold. A corroboration of the reduction of persistence by indole with the same strain was published recently along with the interesting result that indole also reduces heat tolerance in E. coli (Masuda et al., 2020). The ability of indole to kill a wide range of persister cells is illustrated by its ability to also kill the persister cells of the archaeal strain Haloferax volcanii (up to 188-fold increase in killing) (Megaw and Gilmore, 2017).
However, substituted indoles are even more active in killing persister cells. For example, by using our method to convert nearly the whole E. coli bacterial population into persister cells (Kwan et al., 2013; Kim et al., 2018b), so compounds may be more readily screened for persister killing, 36 indole derivatives were assayed for persister killing including halogenated-, methoxy-, methyl-, and nitro-indoles. From this screen, it was found that halogenated indoles such as 4-fluoroindole, 7-chloroindole, 7-bromoindole, and 5-iodoindole (Figure 3) eradicate E. coli persisters. Moreover, 5-iodoindole was the most effective indigoid with 1500-fold greater activity than unsubstituted indole with E. coli (Lee et al., 2016). 5-Iodoindole also eradicated S. aureus persister cells but was not effective with P. aeruginosa (Lee et al., 2016). Hence, a new class of powerful anti-persister compounds was discovered based on indole that eradicates both Gram negative and Gram positive cells.
Recently, a substituted indole was found that is effective in killing P. aeruginosa persister cells: 5-nitro-3-phenyl-1H-indol-2-yl-methylamine hydrochloride (NPIMA, Figure 3) (Song et al., 2019). NPIMA was discovered by converting the E. coli exponential cell population into persister cells by pre-treating with rifampicin to stop transcription (Kwan et al., 2013; Kim et al., 2018b), then performing the first, direct, high-throughput screening of persister cells (Song et al., 2019); a 10,000-member library of druglike compounds was utilized. It was found that NPIMA was more effective than 5-iodoindole (Lee et al., 2016) and cisplatin (Chowdhury et al., 2016b) in killing E. coli persisters. Importantly, NPIMA also eradicated both P. aeruginosa and S. aureus persisters. Critically, the mechanism of NPIMA persister killing was determined and found to be due to membrane damage (Song et al., 2019). Furthermore, E. coli resistance to NPIMA did not occur in a week, and NPIMA was found effective in a wound model with P. aeruginosa and S. aureus (Song et al., 2019).
Indole derivatives have also been combined both with antibiotics and metals to increase their effectiveness in persister cell killing. For example, 5-methylindole (Figure 3) combined with tobramycin kills methicillin-resistant S. aureus and Staphylococcus epidermidis persisters (Sun et al., 2020). In addition, 5-nitroindole (Figure 3) kills E. coli, P. aeruginosa, and Enterobacter tabaci persister cells, and its effectiveness was increased by combining it with copper and zinc nanoparticles (Manoharan et al., 2020).
Since tuberculosis kills 1.5 million people every year (Yang et al., 2017), it is imperative that compounds that eradicate persister cells related to mycobacteria be identified. Critically, a substituted indole, N-[(6-trifluoromethyl)-1H-indol-2-yl)methyl]cycloocctanamine (IMA6, Figure 3), has been identified that kills Mycobacterium abscessus persister cells (Yam et al., 2020). In addition, 4-fluoro and 6-methoxyindoles combined with a cationic amphiphilic motif (e.g., lipophilic n-octyl side chain at position 1 and a positively charged azepanyl or 1,4-dioxa-8-azaspiro[4.5]decane moiety at position 3, Figure 3 for compound 74a) have been identified that kill Mycobacterium tuberculosis and kill Mycobacterium bovis persister cells by damaging the membrane (Yang et al., 2017). No resistance was found to compound 74a in 8 weeks, and the compound was active on S. aureus but had no activity on E. coli (Yang et al., 2017). Hence, substituted indoles are effective against some of the most dangerous pathogens that are often in non-replicating states and require treatments for 1 year with current antibiotics.
In addition to killing persister cells, indole also has another remarkable trait: it selectively allows E. coli cells to resuscitate from dormancy while preventing other cells from resuscitating (Zhang W. et al., 2019). Specifically, indole has no effect on E. coli resuscitation, but indole prevents P. aeruginosa persisters from waking (Zhang W. et al., 2019). Furthermore, indole allows E. coli to outcompete P. aeruginosa (Zhang W. et al., 2019). Critically, indole has no toxicity with non-dormant and dormant P. aeruginosa cells at physiological levels (Zhang W. et al., 2019) so the inhibition of resuscitation is not due to toxicity and not due to a difference in the number of P. aeruginosa persister cells that are formed. Unfortunately, the mechanism of indole inhibition has not been determined.
This indole phenotype likely gives E. coli a fitness advantage over its competitors and may be one of the main reasons indole is secreted from E. coli at such high levels, around 0.7 mM (Domka et al., 2006). These results are also physiologically relevant since both E. coli and P. aeruginosa are found together in the GI tract as P. aeruginosa is present in up to 12% of healthy individuals (Bodey et al., 1983) and is found sometimes in the GI tract of critically ill surgical patients (Marshall et al., 1993). Since indole from E. coli also reduces many of the quorum-sensing-related virulence factors of P. aeruginosa as an inter-species signal (Lee et al., 2009a), these new results (Zhang W. et al., 2019) indicate indole from E. coli both reduces P. aeruginosa virulence as well as prevents its resuscitation from the persister state.
It seems the most important aspect of indole secretion by E. coli is not related to the control of its own gene expression as a quorum-sensing signal but instead lies in the influence of indole on its neighbors as an interkingdom and interspecies signal. For example, indole controls few genes (Wang et al., 2001; Lee et al., 2008); in contrast, indole is clearly beneficial to the host of E. coli (e.g., by tightening epithelial cell junctions to prevent sepsis) (Bansal et al., 2010) and indole is beneficial for controlling the competitors of commensal E. coli since indole both reduces the virulence (Lee et al., 2009a) and the resuscitation of the pathogen P. aeruginosa (Zhang W. et al., 2019) as well as reduces the virulence of EHEC (Bansal et al., 2007; Lee et al., 2007a).
Making use of our discovery that indole reduces persistence (Hu et al., 2015; Kwan et al., 2015), many labs now have independently identified indigoids that are potent for killing persister cells. Future work on the ability of these substituted indoles to enter host cells and kill intracellular persisters would be interesting; note that indole itself is actively transported in E. coli by Mtr but has some less-efficient diffusion into the bacterial cell (Vega et al., 2012). Hence, one can be sanguine about the future and bringing some of these compounds to market to treat recalcitrant infections.
Both authors contributed to the article and approved the submitted version. TW conceived the review. SS and TW authored the manuscript.
This work was supported by funds derived from the Biotechnology Endowed Professorship for TW at the Pennsylvania State University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figure 1A is reprinted from Biofilm 2: 100018, SS and TK, “Forming and waking dormant cells: The ppGpp ribosome dimerization persister model,” Figure 1, Copyright (2020), with permission from Elsevier. Figure 1B is reprinted from iScience 23: 100792, R. Yamasaki, SS, M. J. Benedik, and TK, “Persister Cells Resuscitate Using Membrane Sensors that Activate Chemotaxis, Lower cAMP Levels, and Revive Ribosomes,” Figure 7, Copyright (2020), with permission from Elsevier.
Akiyama, T., Williamson, K. S., and Franklin, M. J. (2018). Expression and regulation of the Pseudomonas aeruginosa hibernation promoting factor. Mol. Microbiol. 110, 161–175. doi: 10.1111/mmi.14001
Akiyama, T., Williamson, K. S., Schaefer, R., Pratt, S., Chang, C. B., and Franklin, M. J. (2017). Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc. Natl. Acad. Sci. U.S.A. 114, 3204–3209. doi: 10.1073/pnas.1700695114
Ashok, N., and Bauer, C. E. (2020). Evidence of defined temporal expression patterns that lead a gram-negative cell out of dormancy. PLoS Genet. 16:e1008660. doi: 10.1371/journal.pgen.1008660
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L., and Leibler, S. (2004). Bacterial persistence as a phenotypic switch. Science 305, 1622–1625. doi: 10.1126/science.1099390
Bansal, T., Alaniz, R. C., Wood, T. K., and Jayaraman, A. (2010). The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 228–233. doi: 10.1073/pnas.0906112107
Bansal, T., Englert, D., Lee, J., Hegde, M., Wood, T. K., and Jayaraman, A. (2007). Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect. Immun. 75, 4597–4607. doi: 10.1128/iai.00630-07
Basu, A., and Yap, M.-N. F. (2017). Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc. Natl. Acad. Sci. U.S.A. 114, E8165–E8173. doi: 10.1073/pnas.1709588114
Bigger, J. W. (1944). Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244, 497–500.
Bodey, G. P., Bolivar, R., Fainstein, V., and Jadeja, L. (1983). Infections caused by Pseudomonas aeruginosa. Rev. Infect Dis. 5, 279–313.
Bubunenko, M., Baker, T., and Court, D. L. (2007). Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J. Bacteriol. 189, 2844–2853. doi: 10.1128/jb.01713-06
Chowdhury, N., Kwan, B. W., and Wood, T. K. (2016a). Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci. Rep. 6:20519. doi: 10.1038/srep20519
Chowdhury, N., Wood, T. L., Martínez-Vázquez, M., García-Contreras, R., and Wood, T. K. (2016b). DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol. Bioeng. 113, 1984–1992. doi: 10.1002/bit.25963
Chu, W., Zere, T. R., Weber, M. M., Wood, T. K., Whiteley, M., Hidalgo-Romano, B., et al. (2012). Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl. Environ. Microbiol. 78, 411–419. doi: 10.1128/aem.06396-11
Conlon, B. P., Rowe, S. E., Gandt, A. B., Nuxoll, A. S., Donegan, N. P., Zalis, E. A., et al. (2016). Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 1:16051. doi: 10.1038/nmicrobiol.2016.51
Corrigan, R. M., Bellows, L. E., Wood, A., and Gründling, A. (2016). ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 113, E1710–E1719. doi: 10.1073/pnas.1522179113
Dalebroux, Z. D., and Swanson, M. S. (2012). ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212.
Domka, J., Lee, J., Bansal, T., and Wood, T. K. (2007). Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9, 332–346.
Domka, J., Lee, J., and Wood, T. K. (2006). YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72, 2449–2459.
Dörr, T., Vulić, M., and Lewis, K. (2010). Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317. doi: 10.1371/journal.pbio.1000317
El-Sharoud, W. M., and Niven, G. W. (2007). The influence of ribosome modulation factor on the survival of stationary-phase Escherichia coli during acid stress. Microbiology 153, 247–253. doi: 10.1099/mic.0.2006/001552-0
Erb, M., Veyrat, N., Robert, C. A. M., Xu, H., Frey, M., Ton, J., et al. (2015). Indole is an essential herbivore-induced volatile priming signal in maize. Nat. Commun. 6:6273. doi: 10.1038/ncomms7273
Frey, M., Stettner, C., Paré, P. W., Schmelz, E. A., Tumlinson, J. H., and Gierl, A. (2000). An herbivore elicitor activates the gene for indole emission in maize. Proc. Natl. Acad. Sci. U.S.A. 97, 14801–14806. doi: 10.1073/pnas.260499897
Gaca, A. O., Colomer-Winter, C., and Lemos, J. A. (2015). Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 197, 1146–1156. doi: 10.1128/jb.02577-14
Goormaghtigh, F., and Van Melderen, L. (2019). Single-cell imaging and characterization of Escherichia coli persister cells to ofloxacin in exponential cultures. Sci. Adv. 5:eaav9462. doi: 10.1126/sciadv.aav9462
Goormaghtigh, F., Fraikin, N., Putrinš, M., Hallaert, T., Hauryliuk, V., Garcia-Pino, A., et al. (2018). Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli Type II persister cells. mBio 9:e00640-18. doi: 10.1128/mBio.00640-18
Hobby, G. L., Meyer, K., and Chaffee, E. (1942). Observations on the mechanism of action of penicillin. Exp. Biol. Med. 50, 281–285. doi: 10.3181/00379727-50-13773
Hong, S. H., Wang, X., O’Connor, H. F., Benedik, M. J., and Wood, T. K. (2012). Bacterial persistence increases as environmental fitness decreases. Microbial. Biotechnol. 5, 509–522. doi: 10.1111/j.1751-7915.2011.00327.x
Hu, Y., Kwan, B. W., Osbourne, D. O., Benedik, M. J., and Wood, T. K. (2015). Toxin YafQ increases persister cell formation by reducing indole signalling. Environ. Microbiol. 17, 1275–1285. doi: 10.1111/1462-2920.12567
Izutsu, K., Wada, A., and Wada, C. (2001). Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp. Genes Cells 6, 665–676. doi: 10.1046/j.1365-2443.2001.00457.x
Jayaraman, R. (2008). Bacterial persistence: some new insights into an old phenomenon. J. Biosci. 33, 795–805.
Kaldalu, N., Hauryliuk, V., and Tenson, T. (2016). Persisters—as elusive as ever. Appl. Microbiol. Biotechnol. 100, 6545–6553. doi: 10.1007/s00253-016-7648-8
Kim, J.-S., Chowdhury, N., Yamasaki, R., and Wood, T. K. (2018a). Viable but non-culturable and persistence describe the same bacterial stress state. Environ. Microbiol. 20, 2038–2048. doi: 10.1111/1462-2920.14075
Kim, J.-S., Yamasaki, R., Song, S., Zhang, W., and Wood, T. K. (2018b). Single cell observations show persister cells wake based on ribosome content. Environ. Microbiol. 20, 2085–2098. doi: 10.1111/1462-2920.14093
Kim, J.-S., and Wood, T. K. (2016). Persistent persister misperceptions. Front. Microbiol. 7:2134. doi: 10.3389/fmicb.2016.02134
Kim, J.-S., and Wood, T. K. (2017). Tolerant, growing cells from nutrient shifts are not persister cells. mBio 8:e00354-17. doi: 10.1128/mBio.00354-17
Korch, S. B., Henderson, T. A., and Hill, T. M. (2003). Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50, 1199–1213.
Kudrin, P., Varik, V., Oliveira, S. R. A., Beljantseva, J., Del Peso Santos, T., Dzhygyr, I., et al. (2017). Sub-inhibitory concentrations of bacteriostatic antibiotics induce relA-dependent and relA-independent tolerance to β-lactams. Antimicrob. Agents Chemother. 61:e02 173-16. doi: 10.1128/aac.02173-16
Kumar, A., and Sperandio, V. (2019). Indole signaling at the host-microbiota-pathogen interface. mBio 10:e01031-19. doi: 10.1128/mBio.01031-19
Kwan, B. W., Osbourne, D. O., Hu, Y., Benedik, M. J., and Wood, T. K. (2015). Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol. Bioeng. 112, 588–600. doi: 10.1002/bit.25456
Kwan, B. W., Valenta, J. A., Benedik, M. J., and Wood, T. K. (2013). Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 57, 1468–1473.
Lee, J., Attila, C., Cirillo, S. L. G., Cirillo, J. D., and Wood, T. K. (2009a). Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microbial. Biotech. 2, 75–90. doi: 10.1111/j.1751-7915.2008.00061.x
Lee, J., Bansal, T., Jayaraman, A., Bentley, W. E., and Wood, T. K. (2007a). Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl. Environ. Microbiol. 73, 4100–4109.
Lee, J., Jayaraman, A., and Wood, T. K. (2007b). Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42.
Lee, J., Maeda, T., Hong, S. H., and Wood, T. K. (2009b). Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl. Environ. Microbiol. 75, 1703–1716.
Lee, J., Zhang, X.-S., Hegde, M., Bentley, W. E., Jayaraman, A., and Wood, T. K. (2008). Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2, 1007–1023.
Lee, J.-H., Cho, H. S., Kim, Y.-G., Kim, J.-A., Banskota, S., Cho, M. H., et al. (2013). Indole and 7-benzyloxyindole attenuate the virulence of Staphylococcus aureus. Appl. Microbiol. Biotechnol. 97, 4543–4552. doi: 10.1007/s00253-012-4674-z
Lee, J.-H., Kim, Y.-G., Gwon, G., Wood, T. K., and Lee, J. (2016). Halogenated indoles eradicate bacterial persister cells and biofilms. AMB Express 6:123. doi: 10.1186/s13568-016-0297-6
Lee, J.-H., Wood, T. K., and Lee, J. (2015). Roles of Indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 23, 707–718. doi: 10.1016/j.tim.2015.08.001
Lewis, K. (2008). Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322, 107–131.
Lewis, K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. doi: 10.1146/annurev.micro.112408.134306
Li, Y., Liu, B., Guo, J., Cong, H., He, S., Zhou, H., et al. (2019). L-Tryptophan represses persister formation via inhibiting bacterial motility and promoting antibiotics absorption. Future Microbiol. 14, 757–771. doi: 10.2217/fmb-2019-0051
Lin, H. H., Hsu, C. C., Yang, C. D., Ju, Y. W., Chen, Y. P., and Tseng, C. P. (2011). Negative effect of glucose on ompA mRNA stability: a potential role of cyclic AMP in the repression of hfq in Escherichia coli. J. Bacteriol. 193, 5833–5840. doi: 10.1128/jb.05359-11
Maisonneuve, E., Castro-Camargo, M., and Gerdes, K. (2018a). Retraction notice to: (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 172:1135. doi: 10.1016/j.cell.2018.02.023
Maisonneuve, E., Shakespeare, L. J., Jørgensen, M. G., and Gerdes, K. (2018b). Retraction for Maisonneuve et al Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 115, E2901–E2901. doi: 10.1073/pnas.1803278115
Maisonneuve, E., Roghanian, M., Gerdes, K., and Germain, E. (2019). Retraction for Germain et al. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Natl. Acad. Sci. U.S.A. 116:11077. doi: 10.1073/pnas.1906160116
Manoharan, R. K., Mahalingam, S., Gangadaran, P., and Ahn, Y.-H. (2020). Antibacterial and photocatalytic activities of 5-nitroindole capped bimetal nanoparticles against multidrug resistant bacteria. Colloids Surf. B Biointerf. 188:110825. doi: 10.1016/j.colsurfb.2020.110825
Marshall, J. C., Christou, N. V., and Meakins, J. L. (1993). The gastrointestinal tract. The “undrained abscess” of multiple organ failure. Ann. Surg. 218, 111–119.
Masuda, Y., Sakamoto, E., Honjoh, K.-I., and Miyamoto, T. (2020). Role of toxin-antitoxin-regulated persister population and indole in bacterial heat tolerance. Appl. Environ. Microbiol. doi: 10.1128/aem.00935-20
CrossRef Full Text [Epub ahead of print].
McKay, S. L., and Portnoy, D. A. (2015). Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob. Agents Chemother. 59, 6992–6999. doi: 10.1128/aac.01532-15
Megaw, J., and Gilmore, B. F. (2017). Archaeal persisters: persister cell formation as a stress response in haloferax volcanii. Front. Microbiol. 8:1589. doi: 10.3389/fmicb.2017.01589
Möker, N., Dean, C. R., and Tao, J. (2010). Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 192, 1946–1955. doi: 10.1128/jb.01231-09
Nguyen, D., Joshi-Datar, A., Lepine, F., Bauerle, E., Olakanmi, O., Beer, K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986.
Pontes, M. H., and Groisman, E. A. (2019). Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci. Signal. 12:eaax3938. doi: 10.1126/scisignal.aax3938
Prossliner, T., Winther, K. S., Sørensen, M. A., and Gerdes, K. (2018). Ribosome Hibernation. Annu. Rev. Genet. 52, 321–348. doi: 10.1146/annurev-genet-120215-035130
Pu, Y., Li, Y., Jin, X., Tian, T., Ma, Q., Zhao, Z., et al. (2019). ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell. 73, 143–156. doi: 10.1016/j.molcel.2018.10.022
Shcherbakova, K., Nakayama, H., and Shimamoto, N. (2015). Role of 100S ribosomes in bacterial decay period. Genes Cells 20, 789–801. doi: 10.1111/gtc.12273
Shimada, Y., Kinoshita, M., Harada, K., Mizutani, M., Masahata, K., Kayama, H., et al. (2013a). Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 8:e80604. doi: 10.1371/journal.pone.0080604
Shimada, T., Yoshida, H., and Ishihama, A. (2013b). Involvement of cyclic AMP receptor protein in regulation of the rmf gene encoding the ribosome modulation factor in Escherichia coli. J. Bacteriol. 195, 2212–2219. doi: 10.1128/jb.02279-12
Song, S., Gong, T., Yamasaki, R., Kim, J.-S., and Wood, T. K. (2019). Identification of a potent indigoid persister antimicrobial by screening dormant cells. Biotechnol. Bioeng. 116, 2263–2274. doi: 10.1002/bit.27078
Song, S., and Wood, T. K. (2018). Post-segregational killing and phage inhibition are not mediated by cell death through toxin/antitoxin systems. Front. Microbiol. 9:814. doi: 10.3389/fmicb.2018.00814
Song, S., and Wood, T. K. (2020a). Are we really studying persister cells? Environ. Microbiol. Rep. doi: 10.1111/1758-2229.12849
CrossRef Full Text. [Epub ahead of print].
Song, S., and Wood, T. K. (2020b). ppGpp ribosome dimerization model for bacterial persister formation and resuscitation. Biochem. Biophys. Res. Com. 523, 281–286. doi: 10.1101/663658
Sun, F., Bian, M., Li, Z., Lv, B., Gao, Y., Wang, Y., et al. (2020). 5-methylindole potentiates aminoglycoside against gram-positive bacteria including Staphylococcus aureus persisters under hypoionic conditions. Front. Cell. Infect. Microbiol. 10:84. doi: 10.3389/fcimb.2020.00084
Svenningsen, M. S., Veress, A., Harms, A., Mitarai, N., and Semsey, S. (2019). Birth and resuscitation of (p)ppGpp induced antibiotic tolerant persister cells. Sci. Rep. 9:6056. doi: 10.1038/s41598-019-42403-7
Tkachenko, A. G., Kashevarova, N. M., Tyuleneva, E. A., and Shumkov, M. S. (2017). Stationary-phase genes upregulated by polyamines are responsible for the formation of Escherichia coli persister cells tolerant to netilmicin. FEMS Microbiol. Lett. 364:fnx084. doi: 10.1093/femsle/fnx084
Van den Bergh, B., Fauvart, M., and Michiels, J. (2017). Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 41, 219–251. doi: 10.1093/femsre/fux001
Vega, N. M., Allison, K. R., Khalil, A. S., and Collins, J. J. (2012). Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 8, 431–433.
Wang, B., Dai, P., Ding, D., Del Rosario, A., Grant, R. A., Pentelute, B. L., et al. (2019). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150. doi: 10.1038/s41589-018-0183-4
Wang, D., Ding, X., and Rather, P. N. (2001). Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183, 4210–4216.
Wood, A., Irving, S. E., Bennison, D. J., and Corrigan, R. M. (2019). The (p)ppGpp-binding GTPase Era promotes rRNA processing and cold adaptation in Staphylococcus aureus. PLoS Genet. 15:e1008346. doi: 10.1371/journal.pgen.1008346
Wood, T. K. (2016). Combatting bacterial persister cells. Biotechnol. Bioeng. 113, 476–483. doi: 10.1002/bit.25721
Wood, T. K., Knabel, S. J., and Kwan, B. W. (2013). Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 79, 7116–7121. doi: 10.1128/AEM.02636-13
Wood, T. K., and Song, S. (2020). Forming and waking dormant cells: the ppGpp ribosome dimerization persister model. Biofilm 2:100018. doi: 10.1016/j.bioflm.2019.100018
Yam, Y.-K., Alvarez, N., Go, M.-L., and Dick, T. (2020). Extreme drug tolerance of mycobacterium abscessus “Persisters”. Front. Microbiol. 11:359. doi: 10.3389/fmicb.2020.00359
Yamagishi, M., Matsushima, H., Wada, A., Sakagami, M., Fujita, N., and Ishihama, A. (1993). Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase-and growth ratedependent control. EMBO J. 12, 625–630.
Yamasaki, R., Song, S., Benedik, M. J., and Wood, T. K. (2020). Persister cells resuscitate using membrane sensors that activate chemotaxis. Lower cAMP levels, and revive ribosomes. iScience 23:100792. doi: 10.1016/j.isci.2019.100792
Yang, T., Moreira, W., Nyantakyi, S. A., Chen, H., Aziz, D. B., Go, M.-L., et al. (2017). Amphiphilic indole derivatives as antimycobacterial agents: structure–activity relationships and membrane targeting properties. J. Med. Chem. 60, 2745–2763. doi: 10.1021/acs.jmedchem.6b01530
Zhang, W., Yamasaki, R., Song, S., and Wood, T. K. (2019). Interkingdom signal indole inhibits Pseudomonas aeruginosa persister cell waking. J. Appl. Microbiol. 127, 1768–1775. doi: 10.1111/jam.14434
Zhang, Y. E., Bærentsen, R. L., Fuhrer, T., Sauer, U., Gerdes, K., and Brodersen, D. E. (2019). (p)ppGpp Regulates a Bacterial Nucleosidase by an allosteric two-domain switch. Mol. Cell. 74, 1239.e4–1249.e4. doi: 10.1016/j.molcel.2019.03.035
Keywords: persisters, indole, substituted indole, resuscitation, formation
Citation: Song S and Wood TK (2020) Combatting Persister Cells With Substituted Indoles. Front. Microbiol. 11:1565. doi: 10.3389/fmicb.2020.01565
Received: 09 April 2020; Accepted: 16 June 2020;
Published: 07 July 2020.
Edited by:
Weihui Wu, Nankai University, ChinaReviewed by:
Shiwei Wang, Northwest University, ChinaCopyright © 2020 Song and Wood. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas K. Wood, dHdvb2RAZW5nci5wc3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.