
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 08 July 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01423
 Ding-Ding Guo1†
Ding-Ding Guo1† Li-Ming Luo1,2†
Li-Ming Luo1,2† Hai-Long Ma1
Hai-Long Ma1 Si-Ping Zhang2
Si-Ping Zhang2 Hang Xu2
Hang Xu2 Honghua Zhang1
Honghua Zhang1 Yong Wang2
Yong Wang2 Yongna Yuan3
Yongna Yuan3 Zhen Wang1
Zhen Wang1 Yong-Xing He2*
Yong-Xing He2*Pseudomonas aeruginosa is an opportunistic pathogen commonly infecting immunocompromised patients with diseases like cystic fibrosis (CF) and cancers and has high rates of recurrence and mortality. The treatment efficacy can be significantly worsened by the multidrug resistance (MDR) of P. aeruginosa, and there is increasing evidence showing that it is easy for this pathogen to develop MDR. Here, we identified a gene cluster, pltZ-pltIJKNOP, which was originally assumed to be involved in the biosynthesis of an antimicrobial pyoluteorin, significantly contributing to the antibiotic resistance of P. aeruginosa ATCC 27853. Moreover, the TetR family regulator PltZ binds to a semi-palindromic sequence in the promoter region of the pltIJKNOP operon and recognizes the antimicrobial 2,4-diacetylphloroglucinol (2,4-DAPG), which in turn induces the expression of the pltIJKNOP operon. Using quantitative proteomics method, it was indicated that the regulator PltZ also plays an important role in maintaining metabolic hemostasis by regulating the transporting systems of amino acids, glucose, metal ions, and bacteriocins.
Pseudomonas aeruginosa is an opportunistic pathogen that infects a wide range of hosts, including plants and animals (Rahme et al., 1995). As a plant pathogen, it was reported to infect lettuce (Paine and Branfoot, 1924; Elrod and Braun, 1942), sugarcane (Desai, 1935), and tobacco (Yu et al., 2008). Most prominently, infection of P. aeruginosa is common in immunocompromised patients with diseases like cystic fibrosis (CF) and cancers and has high rates of recurrence and mortality (Emerson et al., 2002). The treatment efficacy can be significantly worsened by the multidrug resistance (MDR) of P. aeruginosa, and there is increasing evidence showing that it is easy for this pathogen to develop MDR by acquiring transferable resistance genes via horizontal gene transfer (Hirsch and Tam, 2010; Botelho et al., 2019). Moreover, the genomes of P. aeruginosa encode several resistance–nodulation–cell division (RND)-type efflux systems, four of which, i.e., MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM, are characterized to be essential for MDR (Poole, 2011). Many clinical MDR isolates of P. aeruginosa have been shown to have upregulated expressions of these efflux systems, but it is unknown whether there are still yet-unidentified efflux pumps responsible for the MDR phenotypes of P. aeruginosa strains. Understanding the MDR mechanism is of primary priority for the development of new therapies for treating P. aeruginosa infection.
Interactions of P. aeruginosa with other co-colonizing microbes have been proven to be critical in driving its adaptation and evolutionary changes (McGuigan and Callaghan, 2015; Tognon et al., 2017; Flynn et al., 2019). In CF lungs, where P. aeruginosa and other pathogenic microbes such as Staphylococcus aureus and Haemophilus influenzae coexist, the interspecies interactions have been widely investigated, and it was revealed that the quorum sensing signals played an essential role in mediating the interspecies communication and formation of multispecies biofilm, which promoted survival and enhanced virulence (Mengeloglu et al., 2013; Cohen et al., 2016). Although the plant rhizosphere has been suggested to serve as the natural habitat and dissemination agent of P. aeruginosa strains, it is still largely unknown how P. aeruginosa interacts with other root-colonizing microorganisms and how these interactions shaped the important phenotypic traits, such as antibiotic resistance, in P. aeruginosa.
Certain strains of P. aeruginosa (M18, PACS88, and PACS171b) produce a broad-spectrum antibiotic named pyoluteorin (Huang et al., 2004; Kidarsa et al., 2011), which shows toxicity against bacteria and fungi, potentially providing an advantage in competitive colonization. The gene cluster related to pyoluteorin biosynthesis comprises two pairs of oppositely transcribed operons, pltRM-pltLABCDEFG and pltZ-pltIJKNOP. The pltLABCDEFG operon is responsible for pyoluteorin biosynthesis and is activated by the LysR-type regulator PltR (Li et al., 2012). The pltIJKNOP operon, which encodes an ATP-binding cassette (ABC) transporter, was assumed to be involved in pyoluteorin efflux (Brodhagen et al., 2005). Interestingly, pyoluteorin can also be produced by some strains of Pseudomonas protegens (Pf-5, CHA0, and H78) (Paulsen et al., 2005; Jousset et al., 2014; Huang et al., 2017), which are root-colonizing bacteria well known for the capability of producing a wide array of antimicrobials. In these bacterial strains, an antimicrobial, 2,4-diacetylphloroglucinol (2,4-DAPG), functions as a signaling molecule to repress the production of pyoluteorin through an unknown mechanism (Baehler et al., 2005). The gene cluster responsible for 2,4-DAPG biosynthesis also consists of two pairs of oppositely transcribed operons, phlF-phlACBDE and phlG-phlH. The 2,4-DAPG biosynthetic operon phlACBDE is transcriptionally repressed by the TetR-type regulator PhlF, which is released from DNA upon binding 2,4-DAPG (Delany et al., 2000). The phlG gene encodes a 2,4-DAPG hydrolase, whose expression is regulated by the other TetR-type regulator, PhlH, in response to 2,4-DAPG (Yan et al., 2017). Notably, PhlH shares moderate sequence identity (∼39%) with the PltZ regulator encoded in the pyoluteorin-related gene cluster pltZ-pltIJKNOP; however, the biological function and the mechanism of action are still elusive. In this work, we found that the gene cluster pltZ-pltIJKNOP is involved in the resistance of P. aeruginosa ATCC 27853 to chloramphenicol and ampicillin. Moreover, the TetR family regulator PltZ was shown to bind a semi-palindromic sequence in the promoter region of the pltIJKNOP operon and recognize the antimicrobial 2,4-DAPG, which in turn induces the expression of the pltIJKNOP operon.
Although P. aeruginosa M18, PACS171b, and PACS88A were shown to produce pyoluteorin, genomic analysis of P. aeruginosa LESB58, a highly virulent clinical strain isolated from CF patients, revealed that the pltB gene within the plt biosynthetic gene cluster pltRM-pltLABCDEFG had a 5-bp deletion resulting in a truncated PltB protein and a defect in pyoluteorin biosynthesis (Winstanley et al., 2009). The genomes of P. aeruginosa PAO1 and PA14 were also analyzed, but no plt gene clusters were found in these two strains. We next conducted a sequence analysis of the plt gene cluster in P. aeruginosa ATCC 27853, which was isolated from a blood specimen, and found that both pltR and pltB have premature stop codons (Figure 1A), suggesting that P. aeruginosa ATCC 27853 also lost the ability of producing pyoluteorin throughout evolution. By using high-performance liquid chromatography (HPLC), we confirmed that P. aeruginosa ATCC 27853 did not produce pyoluteorin under any of the conditions tested (Figure 1B). This suggested that the pltZ-pltIJKNOP operon was unlikely to be involved in transporting the endogenous pyoluteorin in P. aeruginosa ATCC 27853, and the biological function of this operon remains to be clarified.
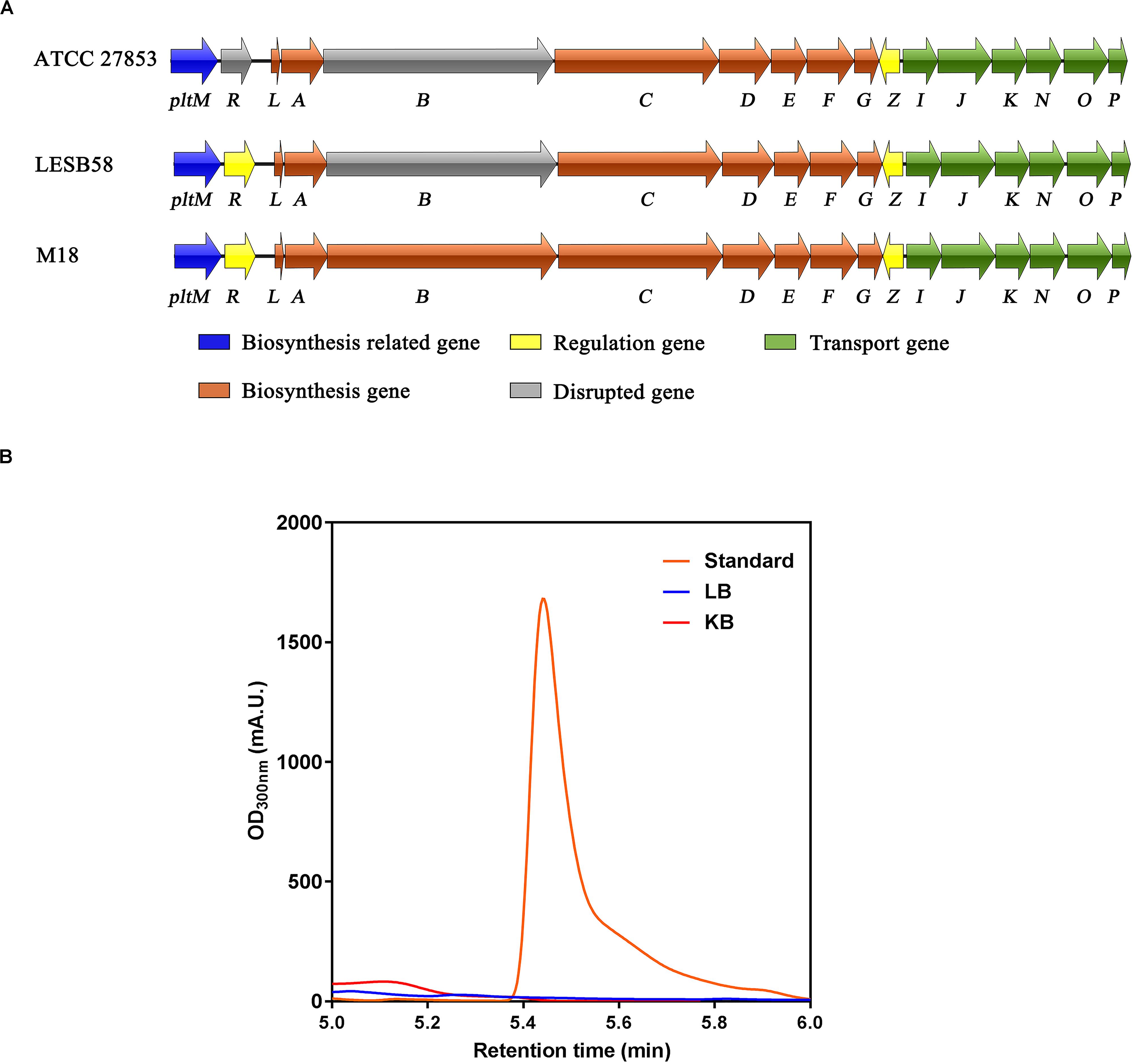
Figure 1. Biosynthetic gene cluster and production of pyoluteorin in Pseudomonas aeruginosa ATCC 27853. (A) Pyoluteorin biosynthetic gene cluster in P. aeruginosa ATCC 27853, P. aeruginosa LESB58, and P. aeruginosa M18. Arrows denote gene location and orientation and are colored according to molecular functions, except for the disrupted genes (in gray) caused by premature stop codons. (B) HPLC chromatograms of the supernatants of Luria–Bertani (blue) and King’s B (red) cultures inoculated with P. aeruginosa ATCC 27853 and the purchased pyoluteorin standard (orange).
To probe the function of the TetR family regulator PltZ, we constructed the ΔpltZ mutant and performed label-free quantitative proteomics to investigate the differentially expressed proteins between the ΔpltZ strain and the wild-type strain. In total, 2,478 proteins were identified. Student’s t-tests were performed with a threshold p < 0.05, resulting in 248 proteins quantified with high confidence. Using the criteria of a ΔpltZ/wild-type fold change <−2 or >2, we identified seven downregulated and 165 upregulated proteins in the ΔpltZ strain (Figure 2A). A Gene Ontology (GO) enrichment analysis revealed that these differentially expressed proteins were mainly involved in ATP binding, nucleotide binding, drug binding, and carbohydrate derivative binding (Figure 2B). Prominently, the expressions of PltI, PltJ, PltK, PltN, and PltO are significantly upregulated in the ΔpltZ strain, suggesting that PltZ acts as a repressor of the pltIJKNOP operon, whose function was previously suggested to be related with the efflux of pyoluteorin. The protein products of the biosynthetic pltLABCDEFG were not identified in the proteomic data, consistent with our finding that P. aeruginosa ATCC 27853 did not produce pyoluteorin (Huang et al., 2004). Sequence analysis indicated that the PltI, PltJ, and PltP proteins were similar to the three components of the type I secretion system, which comprised an ABC transporter, a membrane fusion protein (MFP), and an outer membrane protein (OMP). Structural prediction using the Phyre2 server (Kelley et al., 2015) further revealed that PltK is structurally similar to the transmembrane domain of the ABC transporter and that PltN together with PltO are structurally similar to the electron transporter CcdA belonging to the LysE superfamily (Zhou and Bushweller, 2018). However, it is currently unknown how the six translation products of the pltIJKNOP operon are assembled across the double membranes of P. aeruginosa ATCC 27853. Additionally, proteins involved in putrescine metabolism and transport were also upregulated in the ΔpltZ strain (Supplementary Table S1). Both Ga0133450_116399 and Ga0133450_113821 encode a glutamate–putrescine ligase, which is the first enzyme in the primary putrescine utilization pathway, while spuH (Ga0133450_11320) forms the transmembrane channel involved in putrescine uptake. Moreover, the upregulated proteins include a large number of components of transporting systems involved in the uptake of potassium (Ga0133450_114483), amino acids (Ga0133450_115924 and Ga0133450_113735), and glucose (Ga0133450_111819 and Ga0133450_111821) and the efflux of microcin (Ga0133450_113522) and cadmium (Ga0133450_113780), implying an essential role of PltZ in maintaining metabolic homeostasis. These findings indicated that the TetR family regulator PltZ exerts global influences over the physiological processes of P. aeruginosa ATCC 27853.
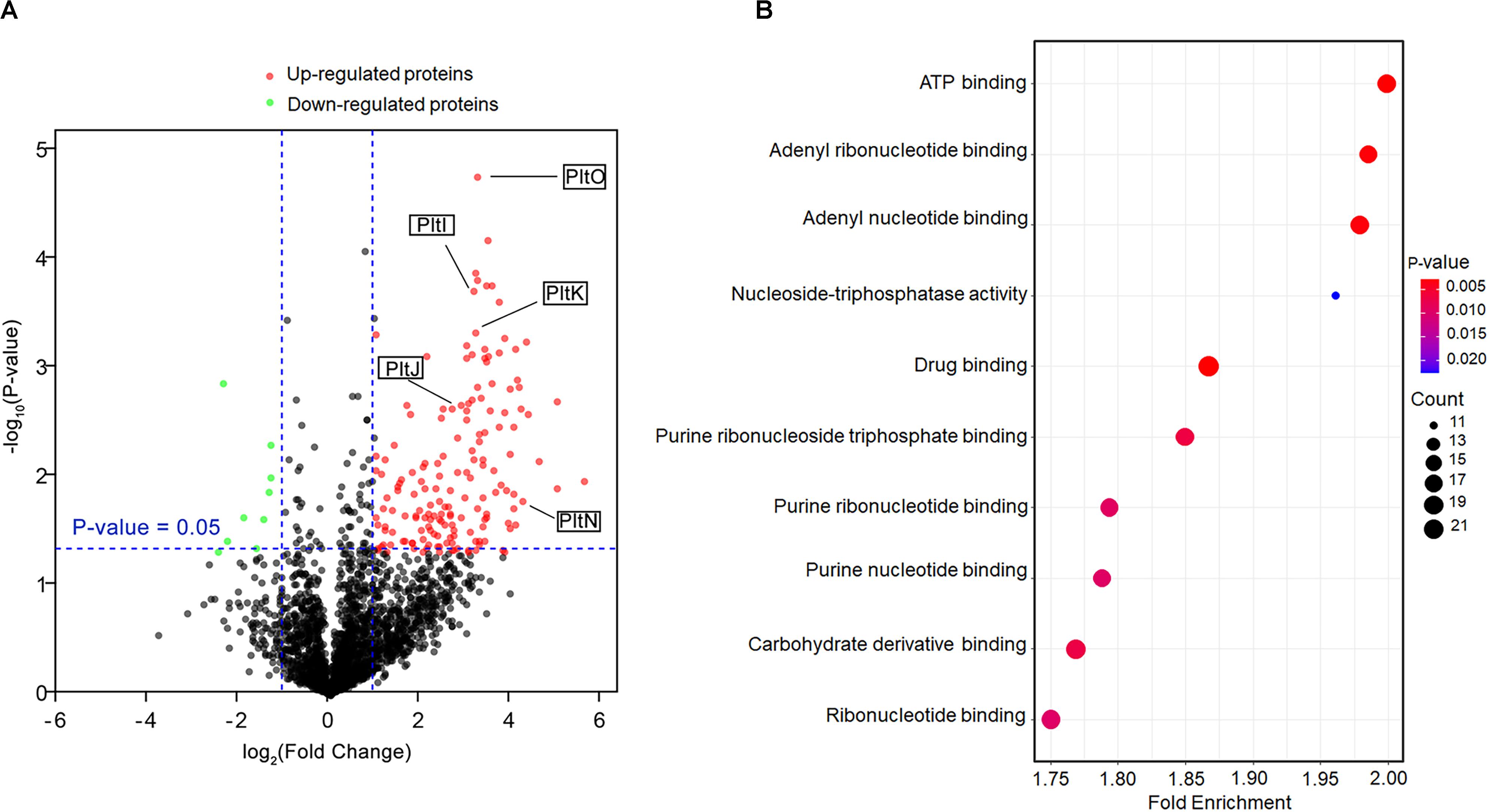
Figure 2. Comparative proteomic analysis of the wild-type and ΔpltZ strains. (A) Volcano plot representing the protein expression ratios of the wild-type vs. the ΔpltZ strain. For each protein, the –log10 (p-value) is plotted against its log2 (fold change). Proteins that were upregulated (p < 0.05, fold change > 2) in the ΔpltZ strain are in red while those downregulated (p < 0.05, fold change ≤ 2) are in green. (B) Top 10 hallmark pathways highlighting the differentially expressed protein pathways between the ΔpltZ and wild-type strains. The x-axis is the enrichment factor, the spot size denotes the protein number, and the spot color denotes the p-value.
Since the proteomic data indicated that PltZ negatively regulates the pltIJKNOP operon, we were wondering whether this regulation effect is through the direct physical interaction between PltZ and the promoter of the pltI gene. DNA fragments comprising the pltZ–pltI (designated as PpltI) intergenic region and the upstream sequence of pltR (designated as PpltR) were amplified by PCR and electrophoretic mobility shift assay (EMSA) was carried out to probe the PltZ–DNA interaction. It was shown that the PltZ bound to PpltI in a concentration-dependent manner, with a saturation concentration of PltZ as low as 250 nM, while no interaction was observed between PltZ and PpltR (Figure 3A). As PltZ shares high sequence identity with PhlH, we reasoned that these two proteins may recognize similar DNA motifs. Using the motif finding program MEME (Bailey and Elkan, 1994), we found that a 16-bp semi-palindromic DNA motif TNNAATTNNAATTNNA is shared among the upstream sequences of PltI and PhlG from P. aeruginosa ATCC 27853, P. protegens Pf-5, and Pseudomonas fluorescens F113 (Figure 3B). This PltZ binding motif was quite similar to the PhlH binding motif previously identified by Yan et al. (2017) and further confirmed by EMSA (Figure 3C). Mutating 10 out of the 16 sites in the predicted PltZ binding motif abolished the PltZ–DNA interaction, confirming the crucial role of the 16-bp semi-palindromic DNA motif in binding PltZ (Figure 3C). By using the BPROM program (Solovyey and Salamov, 2010), a -35/-10 motif (TTGATA/TTATTTACT) recognized by σ70 factors was predicted to be present in the upstream of pltI (Figure 3B). It is interesting to note that the −10 motif overlaps with the PltZ binding site, suggesting that PltZ represses the pltIJKNOP expression by competing with the σ70 factors. We further validated the pltZ-mediated regulation of pltIJKNOP expression by using quantitative real-time reverse transcription PCR (qRT-PCR), and it was revealed that deletion of pltZ led to a significant upregulation of the pltIJK transcription levels in P. aeruginosa ATCC 27853 (Figure 3D). Collectively, these results indicated that PltZ directly represses the expression of the pltIJKNOP operon through binding to the conserved semi-palindromic sequence upstream of pltI.
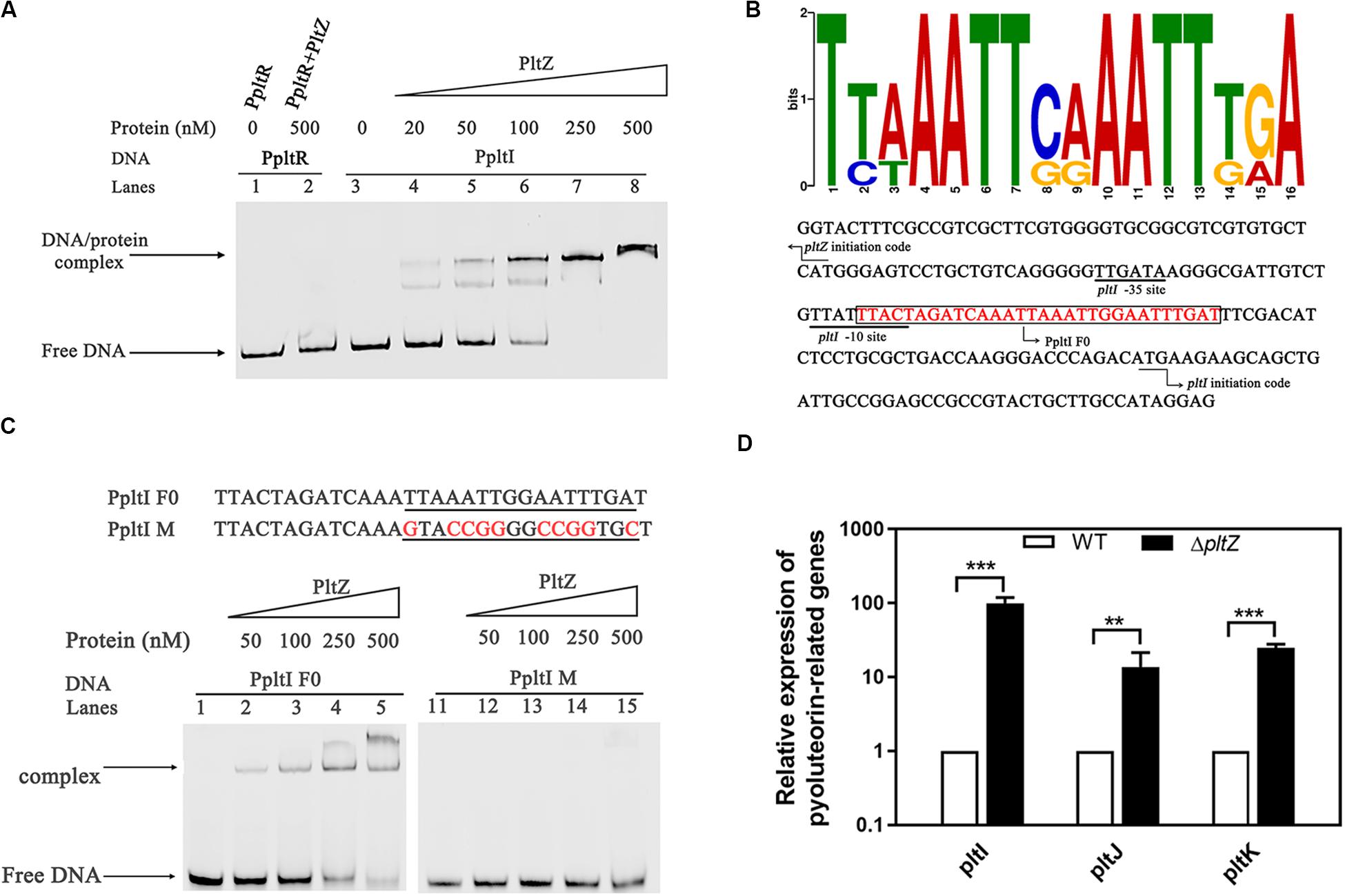
Figure 3. The transcription regulator PltZ binds to the upstream semi-palindromic sequence of the pltIJKNOP operon. (A) Electrophoretic mobility shift assay (EMSA) of PltZ with PpltI. The probe PpltI (20 ng) was incubated with different amounts of PltZ protein (20–500 nM) in total reaction mixtures of 20 μl each and the probe PpltR (20 ng) was used as the negative control. (B) The logo of the PltZ binding motif, which was generated from the conserved 16-nt sequences with the promoter regions of pltI and phlG from Pseudomonas aeruginosa ATCC 27853, Pseudomonas protegens Pf-5, and Pseudomonas fluorescens F113 using the program MEME (top). The pltI promoter region of P. aeruginosa ATCC 27853 was predicted by the BPROM program (bottom). Black lines denote the –35 and –10 promoter regions of pltI. The PltZ binding motif is shown in red. The DNA probe containing the PltZ binding motif (PpltI F0) used for the subsequent EMSA is boxed. Black arrows indicate the translation start sites of pltI and pltZ, respectively. (C) EMSA of PltZ with the probes PpltI F0 and PpltI M. A total of 20 ng DNA probes was incubated with increasing amounts of the PltZ protein (50–500 nM). PltZ was found to bind to PpltI F0, but not to PpltI M. (D) Quantitative real-time reverse transcription PCR (qRT-PCR) assays on pltI, pltJ, and pltK genes showing differences in expression between the wild-type and ΔpltZ strains. Error bars denote standard deviations (n = 3). **p < 0.05, ***p < 0.01.
The TetR family regulators typically recognize small-molecule ligands that modulate their DNA binding capacity. Since PltZ shares ∼39% sequence identity with another TetR family regulator, PhlH, which was shown to bind the antimicrobial 2,4-DAPG (Yan et al., 2017), we speculated that PltZ was likely to also interact with 2,4-DAPG. Using the isothermal titration calorimetry (ITC) assay, we confirmed that the purified PltZ protein could bind 2,4-DAPG with a binding constant of ∼0.19 μM (Figure 4A), which was in the range of the physiological concentration (Yan et al., 2017). Using the EMSA, we further showed that the PltZ/PpltI complex could be partially dissociated by 100 μM 2,4-DAPG (Figure 4B). Moreover, the transcription levels of pltI and pltJ were markedly increased in the presence of 100 μM 2,4-DAPG, confirming the induction effect of 2,4-DAPG in the pltIJKNOP expression (Figure 4C). However, in the ΔpltZ strain, there were no significant differences in the mRNA levels of pltI and pltJ after adding 2,4-DAPG (100 μM) to the medium (Figure 4C). These results confirmed that the induction effect of 2,4-DAPG on the pltIJKNOP expression is dependent on the interaction between PltZ and the antimicrobial 2,4-DAPG.
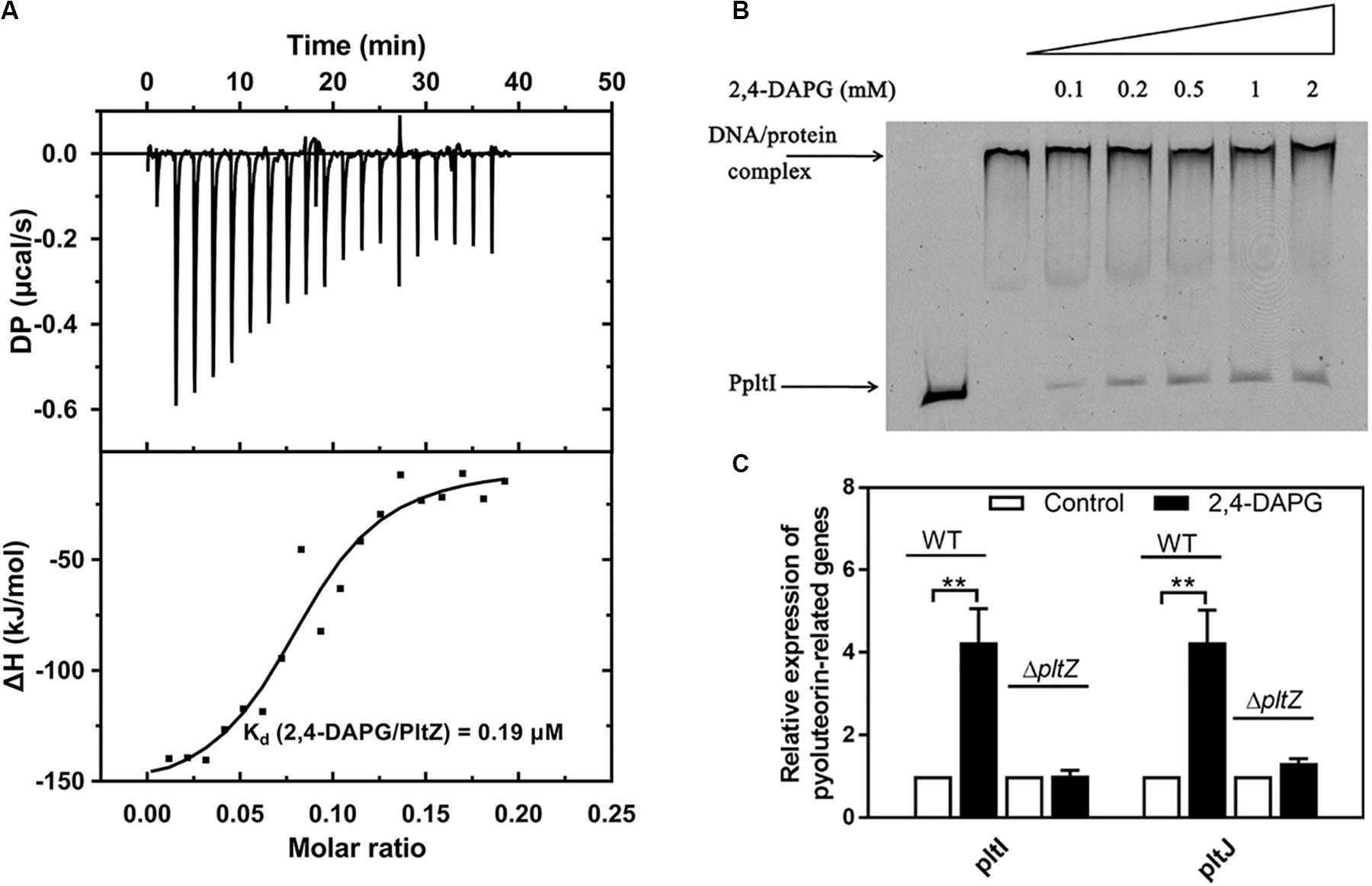
Figure 4. The antimicrobial 2,4-diacetylphloroglucinol (2,4-DAPG) releases the pltZ-mediated repression of pltIJKNOP expression. (A) Interaction between 2,4-DAPG and PltZ was evaluated by isothermal titration calorimetry (ITC). Data were analyzed using the MicroCal PEAQ-ITC software. (B) Different concentrations of 2,4-DAPG were added to explore the stability of the PltZ–PpltI complex using electrophoretic mobility shift assay (EMSA). The concentrations of PpltI and PltZ were 20 ng and 0.5 μM, respectively. From lanes 3–7, the ligand 2,4-DAPG was added at 0.1, 0.2, 0.5, 1, and 2 mM, respectively. (C) Quantitative real-time reverse transcription PCR (qRT-PCR) assays on pltI and pltJ genes of the wild-type and ΔpltZ strains after adding 100 μM 2,4-DAPG to the medium. P. aeruginosa cells were grown to logarithmic phase (8 h) for qRT-PCR assays. Error bars denote standard deviation (n = 3). **p < 0.05.
As the proteins PltI, PltJ, and PltP are similar to the three components of the type I secretion system, which is a two-membrane-spanning secretion apparatus similar to the RND efflux pumps commonly related to MDR (Dinesh and Ayush, 2013), we hypothesized that the pltIJKNOP operon may be involved in the antibiotic resistance of P. aeruginosa ATCC 27853. The wild-type strain showed resistance to a number of clinically relevant antibiotics, including chloramphenicol, ampicillin, tetracycline, kanamycin, and gentamicin. Deletion of pltI led to an increased susceptibility to chloramphenicol and ampicillin (Table 1), indicating that the pltIJKNOP operon contributes to the intrinsic antibiotic resistance of P. aeruginosa ATCC 27853. In contrast, the ΔpltZ strain showed a marked increase in the resistance to chloramphenicol and ampicillin (Table 1), consistent with its role in repressing pltIJKNOP expression. Interestingly, deletion of pltI had no effect on the susceptibility of P. aeruginosa ATCC 27853 to 2,4-DAPG (Table 1), suggesting that 2,4-DAPG was not the substrates of the PltIJKNOP efflux pump. Taken together, our data indicated that PltIJKNOP is a drug efflux pump contributing to the antibiotic resistance of P. aeruginosa, but the selectivity and action mechanism of this efflux pump await further investigation.
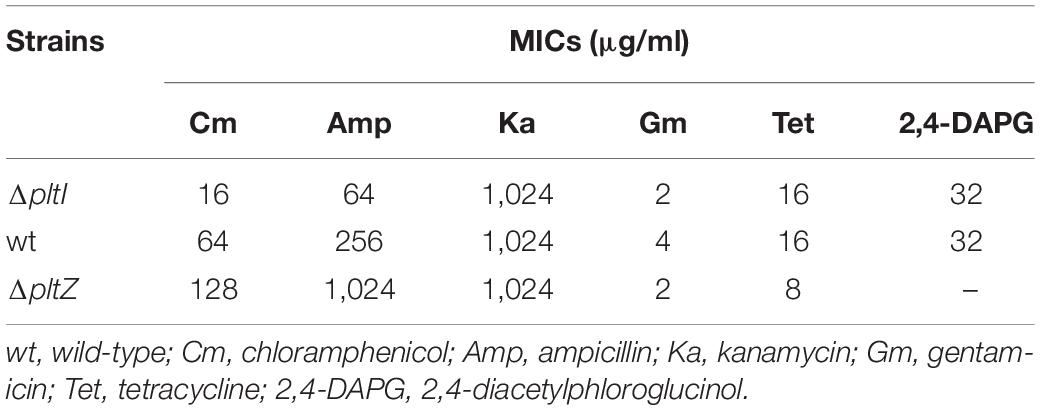
Table 1. Minimum inhibitory concentrations (MICs) of Pseudomonas aeruginosa ATCC 27853 and its mutant strains.
Antibiotics have been proven to be the most successful therapy to treat infection of microbial pathogens. Thus, the ecological function of antibiotics in nature has long been thought to be helping antibiotic producers outcompete the other microbes through bactericidal or bacteriostatic effects, which is a good example of the Darwinian struggle-for-life theory (Linares et al., 2006). However, in recent years, evidence began to accumulate that certain antibiotics with sublethal or sub-inhibitory concentrations can act as important signaling molecules facilitating intraspecies or interspecies interactions within microbial communities (Romero et al., 2011). One elegantly demonstrated example is that of the sub-inhibitory concentrations of the aminoglycoside tobramycin enhancing the biofilm formation of P. aeruginosa via the aminoglycoside response regulator (arr) (Hoffman et al., 2005). Here, we found that the antimicrobial 2,4-DAPG, which is produced by the rhizobacterium P. protegens, could be recognized by the TetR family regulator PltZ and induces the expression of the pltIJKNOP operon encoding a putative efflux pump. It was further shown that the pltIJKNOP operon is responsible for the resistance to chloramphenicol and ampicillin. As our proteomic data also revealed that PltZ influences multiple physiological processes of P. aeruginosa ATCC 27853, especially the transporting systems of amino acids, glucose, metal ions, and bacteriocins, it is probable that sub-inhibitory concentrations of 2,4-DAPG could serve as a signaling molecule modulating bacterial physiology through allosteric regulation of PltZ.
Pyoluteorin is an antibiotic produced by certain Pseudomonas species and suppresses plant diseases caused by the plant pathogen Pythium ultimum by inhibiting its growth (Howell, 1980). In P. protegens Pf-5, the biosynthesis of pyoluteorin was previously identified to be related to a 24-kb gene cluster named plt, within which 10 genes (pltMRLABCDEFG) are strictly required for pyoluteorin biosynthesis. Downstream of the pltG gene is the pltZ gene encoding a TetR family regulator, which was shown to negatively regulate the biosynthetic pltLABCDEFG operon (Huang et al., 2004). However, in P. aeruginosa ATCC 27853, there are premature stop codons within both pltR and pltB, making this strain incapable of producing pyoluteorin, which was further confirmed by HPLC. The inactivation of the pltR and pltB genes implies that the selective constraints imposed upon these two genes are relaxed in P. aeruginosa ATCC 27853, in which production of pyoluteorin may provide a few fitness advantages. This seems to be consistent with the clinical background of P. aeruginosa ATCC 27853, which was initially isolated from a blood specimen in Peter Bent Brigham Hospital in 1971 (Boston, United States) (Medeiros et al., 1971). Since pyoluteorin typically promotes the fitness of the producers in the rhizosphere where microbial competition is fierce, it is likely that, in clinical settings, pyoluteorin is not beneficial for the survival of P. aeruginosa ATCC27853 in human hosts but rather poses a metabolic burden on the producer cells, just as it did in P. protegens Pf-5 in laboratory cultivation (Yan et al., 2018).
Despite the biosynthetic pathway of pyoluteorin being disrupted in the ATCC 27853 strain, the pltIJKNOP operon, which is located downstream of the pltLABCDEFG operon, is fully functional. Sequence analysis indicated that PltI, PltJ, and PltP are highly similar to the three components of the type I secretion system, which spans the inner and outer membranes of Gram-negative microbes and translocates unfolded substrate proteins to the extracellular space in one step. Although it was unknown whether pltIJKNOP is involved in secreting certain substrate proteins, we have shown that the pltI gene contributes to resistance to several clinically relevant antibiotics. Deletion of the regulatory gene pltZ led to a significant increase in the antibiotic resistance of the ATCC 27853 strain, further supporting the role of pltZIJKNOP in antibiotic resistance. Since the highly virulent P. aeruginosa LESB58 strain also possesses pltZIJKNOP, which was found to be incorporated into its genome through horizontal gene transfer (Winstanley et al., 2009), it is possible that pltZIJKNOP could be a driving force for the acquired antibiotic resistance in certain clinical strains of P. aeruginosa. However, further work needs to be done to establish the substrate spectrum and action mechanism of the PltIJKNOP efflux pump in more detail.
Escherichia coli BL21 (DE3) cells were used to express recombinant proteins and E. coli DH5α cells were used for molecular cloning. The E. coli S17-1 strain was used for conjugation. E. coli was grown at 37°C at 220 rpm in Luria–Bertani (LB) broth or agar plates, and P. aeruginosa and its derivatives were grown at 37°C at 220 rpm in LB or King’s B (KB) medium (Wang et al., 2019). When required, the growth medium was supplemented with ampicillin (50 mg/L), kanamycin sulfate (50 mg/L), gentamicin (30 mg/L), sucrose (10%, m/v), and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal, 40 mg/L). Strains and plasmids are listed in Supplementary Table S2. The primers and DNA sequences for PCR amplification are listed in Supplementary Table S3.
The protein expression and purification of PltZ were performed as previously described (Wang et al., 2019). Briefly, the pltZ gene from P. aeruginosa ATCC 27853 was cloned into the pET-28b-derived vector and transformed into the E. coli BL21 (DE3) strain. When the optical density at 600 nm of the culture reached ∼0.8, 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added and then the bacteria were cultured for another 22 h at 16°C before harvesting. The cells were resuspended in buffer containing 10 mM imidazole, 20 mM Tris–HCl (pH 8.0), and 100 mM NaCl and then sonicated. The cell lysate was centrifuged at 12,000 × g for 30 min and then the supernatant was loaded onto a Ni-NTA affinity column. The target protein was eluted with buffer containing 20 mM Tris–HCl (pH 7.5), 100 mM NaCl, and 250 mM imidazole. The imidazole in the protein sample was then removed through multiple rounds of ultrafiltration. The protein was finally concentrated to ∼20 mg/ml and stored at −80°C for further usage.
The interaction between 2,4-DAPG and PltZ was measured using ITC at 25°C with a MicroCal PEAQ-ITC instrument (Malvern, United States). One millimolar 2,4-DAPG solution used for titration was prepared with the ITC buffer (20 mM Tris, pH 8.0, 100 mM NaCl). Protein samples were prepared by dialysis in the ITC buffer. The concentrations of PltZ and 2,4-DAPG were both 25 μM. The protein and ligand solutions were centrifuged at 12,000 × g for 30 min before titration. The titration was performed with a total of 19 injections of 40 μl 2,4-DAPG into the PltZ solution (200 μl). 2,4-DAPG was also titrated into the ITC buffer alone, and the resulting heat of dilution was subtracted from the experimental curve. Data analyses were performed with the MicroCal PEAQ-ITC Analyze software, and an independent binding model was used to calculate the thermodynamic parameters of interaction between PltZ and 2,4-DAPG.
Quantification of pyoluteorin was performed according to the method described previously (Yan et al., 2017). Briefly, HPLC was used to detect the pyoluteorin production of P. aeruginosa ATCC 27853. The culture samples were centrifuged at 12,000 × g for 10 min and the supernatant was subjected to HPLC analysis. The mobile phases were acetonitrile and distilled water containing 0.1% formic acid. Five percent acetonitrile was used to calibrate the Agilent ZORBAX Eclipse XDB-C18 column, 4.6 × 150 mm. The gradient was performed from 0 to 40% acetonitrile in 3 min and then from 40 to 100% acetonitrile in 3 min, at a flow rate of 1.5 ml/min. The retention time for pyoluteorin was 5.5 min.
The in-frame deletion mutants were constructed using a two-step homologous recombination method described previously (Wang et al., 2019). Briefly, DNA fragments approximately 1 kb of the upstream and 800 bp of the downstream regions of the target genes were amplified by PCR from P. aeruginosa ATCC 27853 genomic DNA using the primers described in Supplementary Table S3. The two fragments were digested with the corresponding restriction enzymes and cloned into the suicide vector pK18mobsacB-Gm. The resulting plasmids were conjugated from E. coli S17-1 into P. aeruginosa ATCC 27853 by biparental mating. The deletion stains were obtained by selection on LB plates containing Amp, Gm, and 10% sucrose and further confirmed by PCR amplification.
Pseudomonas aeruginosa ATCC 27853 wild-type and ΔpltZ strains were collected, lysed, reduced, and alkylated at OD600 = 1 as previously described (Wang et al., 2019). Briefly, the collected cells were resuspended in 100 μl of cell lysate (8 M urea, 10 mM TCEP, 40 mM CAA, and 10 mM Tris, pH 8.5), followed by boiling in a 95°C water bath for 5 min. The samples were diluted to 1 ml with the dilution buffer (10% ACN and 25 mM Tris, pH 8.5) and the proteins were digested with 10 μg/ml trypsin for 18 h at 37°C and further purified by C18 Stage Tips. Proteomics MS was analyzed by an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, Untied States) coupled online to an EASY-nLC 1200 system. Of the peptide sample, 4 μl was injected into a 15-cm-long, 75-μm inner diameter capillary analytic column packed with C18 particles of 2 μm diameter. The mobile phases for the LC include buffer A (0.1% FA) and buffer B (80% acetonitrile and 0.1% FA). The peptides were separated using a 90-min non-linear gradient consisting of 5–35% buffer B for 60 min, 35–80% buffer B for 20 min, and 100% buffer B for 10 min at a flow rate of 300 nl/min. The source voltage and current were set at 2.5 kV and 100 A, respectively. All mass spectrometry (MS) measurements were performed in the positive ion mode and acquired across the mass range of 300–1,800 m/z. The raw MS files were analyzed by Proteomics Discovery 2.2 software (Thermo Fisher Scientific, United States) and the MS/MS spectra were searched against the protein sequences of P. aeruginosa ATCC 27853. The methionine oxidation and cysteine carbamidomethylation were included as the variable modification and fixed modification, respectively. Other parameters were set up using the default values, and the false discovery rate was set to 0.01 for both peptide and protein identifications. Statistical analysis was done using the Perseus program (Tyanova et al., 2016). Student’s t-test was used to determine the significance of the differential expressions of proteins between the ΔpltZ and wild-type samples. Proteins within a statistical region of p < 0.05 and fold change >2 were considered as differentially expressed proteins. Biological pathway enrichment analysis was performed with the R package clusterProfiler (Yu et al., 2012) using the default settings, showing the 10 most descriptive categories.
The DNA fragments used for the EMSA were either amplified using PCR or synthesized (Supplementary Table S3). DNA probes were added at 40 ng and incubated at 4°C for 30 min with various amounts of proteins in a total volume of 20 μl mixtures. The mixture contained 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 10% (v/v) glycerol, 0.5 mM EDTA, and 50 mM KCl. The samples were subjected to 5% native PAGE with 1 × Tris borate–EDTA (TBE) buffer. Electrophoresis was performed at 120 V, 4°C in an ice-cold bath. The images were acquired by a PharosFX Gel imaging system (Bio-Rad, United States).
The qRT-PCR assays were performed as described previously (Yan et al., 2017). Briefly, total RNA of the wild-type and ΔpltZ mutants was isolated with Trizol and chloroform. First-strand cDNAs were synthesized using a PrimeScriptTM RT reagent kit with gDNA Eraser (TaKaRa, Dalian, China). For real-time PCR analysis, gene-specific primers were used, according to the manufacturer’s instructions (SYBR Premix Ex Taq II kit, TaKaRa, Dalian, China), using 100 ng of total RNA as the template (1.0 μl of cDNA samples). The qRT-PCR assays were performed on a CFX real-time PCR system (Bio-Rad, Hercules, CA, United States). The fold changes of the corresponding gene transcripts in the wild-type strain relative to the ΔpltZ strain were computed using the 2–ΔΔCT formula (Livak and Schmittgen, 2001). The 16S rRNA gene was used to normalize the data and at last three biological replicates were performed. Student’s t-test was used to compare the gene expression levels, and a p-value of 0.05 was considered statistically significant.
Minimum inhibitory concentration (MIC) analysis was carried out in 96-well microtiter plates with the standard broth microdilution method (Schwalbe et al., 2007). The bacteria used in the study were those harvested in the mid-exponential growth phase (absorbance at 600 nm was 0.6) and diluted 1,000-fold with the LB broth. Of the diluted bacterial culture, 100 μl was added to each well; the MIC values were determined as the lowest concentration that completely inhibited bacterial growth after 20 h of incubation at 37°C.
The datasets generated for this study can be found in the ProteomeXchange Consortium (Vizcaíno et al., 2016) with the data set identifier PXD016202 (http://www.proteomexchange.org).
Y-XH conceived and designed the experiments. L-ML performed the experiments. H-LM, S-PZ, HX, HZ, YW, YY, and ZW analyzed the data. Y-XH and D-DG wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31770535 and 31971422), the Natural Science Foundation of Gansu Province (Grant No. 17JR5RA208), and the Fundamental Research Funds for the Central Universities from Lanzhou University (Grant Nos. lzujbky-2019-cd05 and lzujbky-2017-151).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the mass spectrometry facility at the School of Life Sciences of Lanzhou University for support in proteomics data acquisition.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01423/full#supplementary-material
Baehler, E., Bottiglieri, M., Péchy-Tarr, M., Maurhofer, M., and Keel, C. (2005). Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 99, 24–38. doi: 10.1111/j.1365-2672.2005.02597.x
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Botelho, J., Grosso, F., and Peixe, L. (2019). Antibiotic resistance in Pseudomonas aeruginosa - Mechanisms, epidemiology and evolution. Drug Resist. Updat. 44:100640. doi: 10.1016/j.drup.2019.07.002
Brodhagen, M., Paulsen, I., and Loper, J. E. (2005). Reciprocal regulation of pyoluteorin production with membrane transporter gene expression in Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 71, 6900–6909. doi: 10.1128/aem.71.11.6900-6909.2005
Cohen, T. S., Hilliard, J. J., Jones-Nelson, O., Keller, A. E., O’Day, T., Tkaczyk, C., et al. (2016). Staphylococcus aureus α toxin potentiates opportunistic bacterial lung infections. Sci. Transl. Med. 8:329ra331. doi: 10.1126/scitranslmed.aad9922
Delany, I., Sheehan, M. M., Fenton, A., Bardin, S., Aarons, S., and O’Gara, F. (2000). Regulation of production of the antifungal metabolite 2,4-diacetylphloroglucinol in Pseudomonas fluorescens F113: genetic analysis of phlF as a transcriptional repressor. Microbiology 146(Pt 2), 537–546. doi: 10.1099/00221287-146-2-537
Dinesh, F., and Ayush, K. (2013). Resistance-nodulation-division multidrug efflux pumps in gram-negative bacteria: role in virulence. Antibiotics 2, 163–181. doi: 10.3390/antibiotics2010163
Elrod, R. P., and Braun, A. C. (1942). Pseudomonas aeruginosa: its role as a plant pathogen. J Bacteriol. 44, 633–645. doi: 10.1128/jb.44.6.633-645.1942
Emerson, J., Rosenfeld, M., McNamara, S., Ramsey, B., and Gibson, R. L. (2002). Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr. Pulmonol. 34, 91–100. doi: 10.1002/ppul.10127
Flynn, S., Reen, F. J., and O’Gara, F. (2019). Exposure to bile leads to the emergence of adaptive signaling variants in the opportunistic pathogen. Front. Microbiol. 10:2013. doi: 10.3389/fmicb.2019.02013
Hirsch, E. B., and Tam, V. H. (2010). Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 10, 441–451.
Hoffman, L. R., D’Argenio, D. A., MacCoss, M. J., Zhang, Z., Jones, R. A., and Miller, S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175. doi: 10.1038/nature03912
Howell, C. R. (1980). Suppression of Pythium ultimum-induced damping-off of cotton seedling by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 70, 712–715.
Huang, X., Wang, Z., Liu, Y., and Zhang, X. (2017). Complete Genome Sequence of H78, a Plant Growth-Promoting Rhizobacterium. Genome Announc. 5:e00233-17.
Huang, X., Zhu, D., Ge, Y., Hu, H., Zhang, X., and Xu, Y. (2004). Identification and characterization of pltZ, a gene involved in the repression of pyoluteorin biosynthesis in Pseudomonas sp. M18. FEMS Microbiol. Lett. 232, 197–202. doi: 10.1016/s0378-1097(04)00074-6
Jousset, A., Schuldes, J., Keel, C., Maurhofer, M., Daniel, R., Scheu, S., et al. (2014). Full-genome sequence of the plant growth-promoting bacterium Pseudomonas protegens CHA0. Genome Announc. 2:e00322-14. doi: 10.1128/genomeA.00322-314
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kidarsa, T. A., Goebel, N. C., Zabriskie, T. M., and Loper, J. E. (2011). Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol. Microbiol. 81, 395–414. doi: 10.1111/j.1365-2958.2011.07697.x
Li, S., Huang, X., Wang, G., and Xu, Y. (2012). Transcriptional activation of pyoluteorin operon mediated by the LysR-type regulator PltR bound at a 22 bp lys box in Pseudomonas aeruginosa M18. PLoS One 7:e39538. doi: 10.1371/journal.pone.0039538
Linares, J. F., Gustafsson, I., Baquero, F., and Martinez, J. L. (2006). Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. U.S.A. 103, 19484–19489. doi: 10.1073/pnas.0608949103
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
McGuigan, L., and Callaghan, M. (2015). The evolving dynamics of the microbial community in the cystic fibrosis lung. Environ. Microbiol. 17, 16–28. doi: 10.1111/1462-2920.12504
Medeiros, A. A., O’Brien, T. F., Wacker, W. E., and Yulug, N. F. (1971). Effect of salt concentration on the apparent in-vitro susceptibility of Pseudomonas and other gram-negative bacilli to gentamicin. J. Infect. Dis. 124(Suppl.), S59–S64. doi: 10.1093/infdis/124.supplement_1.s59
Mengeloglu, F. Z., Tas, T., Kocoglu, E., Bucak, Ö, and Tekin, D. (2013). Anti-Haemophilus influenzae activity of Pseudomonas aeruginosa. J. Microbiol. Res. Rev. 1, 24–29.
Paine, S. G., and Branfoot, J. M. (1924). Studies in bacteriosis xi. A bacterial disease of lettuce. Ann. Appl. Biol. 11, 312–317. doi: 10.1111/j.1744-7348.1924.tb05713.x
Paulsen, I. T., Press, C. M., Ravel, J., Kobayashi, D. Y., Myers, G. S. A., Mavrodi, D. V., et al. (2005). Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23, 873–878.
Poole, K. (2011). Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2:65. doi: 10.3389/fmicb.2011.00065
Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G., and Ausubel, F. M. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902. doi: 10.1126/science.7604262
Romero, D., Traxler, M. F., López, D., and Kolter, R. (2011). Antibiotics as signal molecules. Chem. Rev. 111, 5492–5505. doi: 10.1021/cr2000509
Schwalbe, R. S., Steele-Moore, L., and Goodwin, A. C. (2007). Antimicrobial Susceptibility Testing Protocols. Boca Raton, FL: CRC Press.
Solovyey, V., and Salamov, A. (2010). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, ed. R. W. Li (Hauppauge, NY: Nova Science Publishers), 61–78.
Tognon, M., Köhler, T., Gdaniec, B. G., Hao, Y., Lam, J. S., Beaume, M., et al. (2017). Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J. 11, 2233–2243. doi: 10.1038/ismej.2017.83
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., et al. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. doi: 10.1038/nmeth.3901
Vizcaíno, J. A., Csordas, A., del-Toro, N., Dianes, J. A., Griss, J., Lavidas, I., et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456. doi: 10.1093/nar/gkv1145
Wang, Y., Zhang, S.-P., Zhang, M.-Y., Kempher, M. L., Guo, D.-D., Han, J.-T., et al. (2019). The antitoxin MqsA homologue in Pseudomonas fluorescens 2P24 has a rewired regulatory circuit through evolution. Environ. Microbiol. 21, 1740–1756. doi: 10.1111/1462-2920.14538
Winstanley, C., Langille, M. G., Fothergill, J. L., Kukavica-Ibrulj, I., Paradis-Bleau, C., Sanschagrin, F., et al. (2009). Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 19, 12–23. doi: 10.1101/gr.086082.108
Yan, Q., Lopes, L. D., Shaffer, B. T., Kidarsa, T. A., Vining, O., Philmus, B., et al. (2018). Secondary Metabolism and interspecific competition affect accumulation of spontaneous mutants in the Gacs-gaca regulatory system. mBio 9:e01845-17.
Yan, X., Yang, R., Zhao, R.-X., Han, J.-T., Jia, W.-J., Li, D.-Y., et al. (2017). Transcriptional regulator PhlH modulates 2,4-diacetylphloroglucinol biosynthesis in response to the biosynthetic intermediate and end product. Appl. Environ. Microbiol. 83:e01419-17.
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287. doi: 10.1089/omi.2011.0118
Yu, L., Qin, X. Y., Du, J., Wang, A. Y., and Huang, Q. (2008). Bacterial leaf spot of tobacco caused by Pseudomonas aeruginosa in China. Plant Pathol. 57:774.
Keywords: antibiotic resistance, P. aeruginosa, pathogens, multidrug resistance, pyoluteorin, 2, 4-diacetylphloroglucinol, P. fluorescens, TetR regulator
Citation: Guo D-D, Luo L-M, Ma H-L, Zhang S-P, Xu H, Zhang H, Wang Y, Yuan Y, Wang Z and He Y-X (2020) The Regulator PltZ Regulates a Putative ABC Transporter System PltIJKNOP of Pseudomonas aeruginosa ATCC 27853 in Response to the Antimicrobial 2,4-Diacetylphloroglucinol. Front. Microbiol. 11:1423. doi: 10.3389/fmicb.2020.01423
Received: 15 January 2020; Accepted: 02 June 2020;
Published: 08 July 2020.
Edited by:
Aixin Yan, The University of Hong Kong, Hong KongReviewed by:
Yuji Morita, Meiji Pharmaceutical University, JapanCopyright © 2020 Guo, Luo, Ma, Zhang, Xu, Zhang, Wang, Yuan, Wang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Xing He, aGV5eEBsenUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.