
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 25 June 2020
Sec. Food Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01412
This article is part of the Research TopicIntegrating Whole Genome Sequencing into Source Attribution and Risk Assessment of Foodborne Bacterial PathogensView all 7 articles
 Monika Kurpas1
Monika Kurpas1 Jacek Osek1
Jacek Osek1 Alexandra Moura2,3,4
Alexandra Moura2,3,4 Alexandre Leclercq2,3
Alexandre Leclercq2,3 Marc Lecuit2,3,4,5
Marc Lecuit2,3,4,5 Kinga Wieczorek1*
Kinga Wieczorek1*Listeria monocytogenes is one of the major foodborne pathogens. Isolates of PCR-serogroups IIb (n = 17) and IVb (n = 31) recovered from food (n = 33) and food processing environment (n = 15) in Poland were characterized using whole genome sequencing. Most isolates belonged to Multi-Locus Sequence Type (MLST) ST2 (31.3%) and ST5 (22.9%). Core genome MLST (cgMLST) analysis classified isolates into seven sublineages (SL) and 25 different cgMLST types (CT). Consistent with the MLST results, most sublineages were SL2 and SL5. Eleven isolates harbored aacA4 encoding resistance to aminoglycosides, three isolates harbored emrC (n = 3) and one brcABC (n = 1) encoding tolerance to benzalkonium chloride. Isolates belonging to SL5 CT2323 carried a so far unreported inlB allele with a deletion of 141 nucleotides encoding the β-repeat sheet and partially the GW1 domain of InlB. Comparison with publicly available genome sequences from L. monocytogenes isolated from human listeriosis cases in Poland from 2004 to 2013 revealed five common CTs, suggesting a possible epidemiological link with these strains. The present study contributes to characterize the diversity of L. monocytogenes in ready-to-eat (RTE) meat and meat processing environments in Poland and unravels previously unnoticed links with clinical cases in Europe.
Listeria monocytogenes is one of the most common foodborne zoonotic pathogens and the cause of listeriosis in human (Buchanan et al., 2017). The disease mostly occurs in elderly people, immunosuppressed patients, and pregnant women and their fetus/newborns (Swaminathan and Gerner-Smidt, 2007; Charlier et al., 2017). In immunosuppressed and older individuals, L. monocytogenes can cause septicaemia and meningitis, and in pregnant women induce miscarriage and neonatal listeriosis (Ramaswamy et al., 2007). L. monocytogenes may cause either sporadic cases or outbreaks and many of them have been linked to the consumption of contaminated food of animal origin (Buchanan et al., 2017; Glebícová et al., 2018). Different food products may be contaminated with L. monocytogenes, either at the production stage or during processing (Maury et al., 2019). According to a recent EFSA zoonotic report, depending on the ready-to-eat food category, from 0 to 3.18% samples were contaminated with L. monocytogenes in 2018 (European Food Safety Authority [EFSA], 2019). In the same year, 2,459 listeriosis cases were reported in the European Union, with a hospitalisation rate of 97.0% and a high fatality of 15.6% (European Food Safety Authority [EFSA], 2019). However, these values may not reflect the actual situation because European countries have different surveillance systems and the exhaustiveness of case notification and reporting also varies.
Molecular typing of L. monocytogenes is important for detecting clusters of human listeriosis cases as well as identifying the source of food contamination. Rapid, standardized and cheap methods are needed for screening many bacterial isolates as a first line for subsequent detailed characterization. PCR-serogrouping enables classification of L. monocytogenes into molecular serogroups which cover particular PCR-serogroups: IIa (including serovars 1/2a, 3a), IIb (1/2b, 3b, 7), IIc (1/2c, 3c), IVb (4b, 4d, 4e), and L (including serovars 4a, 4ab, 4c and other species of Listeria sensu stricto), respectively (Doumith et al., 2004). Most human infections are due to isolates of PCR-serogroups IVb, IIa, and IIb, which are responsible for over 95% of listeriosis cases, with L. monocytogenes of PCR-serogroup IVb causing more than half of these cases (Bergholz et al., 2018). Another molecular typing method is the multilocus sequence typing (MLST), which is based on the sequence variants of 7 housekeeping genes (Ragon et al., 2008). Recent advances in high-throughput sequence sequencing have enabled analysis of bacterial isolates at the whole genome level (Luth et al., 2018). The core genome MLST (cgMLST) typing method takes into account the sequence variation of 1,748 L. monocytogenes core genes, improving isolate discrimination and allowing a standardized comparison with isolate databases for outbreak investigations and surveillance (Moura et al., 2016, 2017).
Besides its increased discrimination power as compared to previous methods such as PFGE, whole genome sequencing enables simultaneous identification of antimicrobial resistance and virulence genes as well as other genetic determinants playing a role in L. monocytogenes infection (Moura et al., 2016).
Virulence of L. monocytogenes is associated with its ability to invade, multiply, and survive within host cells (Portnoy et al., 2002). Clones of L. monocytogenes differ in their pathogenic potential (Maury et al., 2016, 2019). Some L. monocytogenes are hypervirulent for humans are more often isolated from infection cases, whereas other isolates are hypovirulent, in part due to loss-of-function mutations in virulence genes (Glebícová et al., 2015; Maury et al., 2016, 2019). One of the most important virulence markers identified in L. monocytogenes is Listeria Pathogenicity Island-1 (LIPI-1), composed of six virulence genes regulated by PrfA, a transcriptional activator for more than 140 genes. These include inlA and inlB genes. InlA and InlB interaction with their respective receptor E-cadherin and c-Met, expressed by human epithelial cells, mediate L. monocytogenes internalisation into non-phagocytic cells (Lingnau et al., 1995). Premature stop codons (PMSC) in the inlA gene attenuate virulence in L. monocytogenes (Jacquet et al., 2004; Maury et al., 2016). Other proteins belonging to the internalin family are also engaged in L. monocytogenes virulence activity (Bierne et al., 2007).
It has been also shown that some L. monocytogenes clones survive better in the environment than others. This is the result of biofilm development and expression of tolerance to sanitizers, oxidative stress, alkaline or acid conditions and fridge temperature (Colegiogri et al., 2017). L. monocytogenes contains genomic islands Survival Stress Islet 1 (SSI-1) and Survival Stress Islet 2 (SSI-2) which are responsible for survival of the bacteria in suboptimal conditions commonly present in food processing environments (Hein et al., 2011). Tolerance to sanitizers and disinfectants such as benzalkonium chloride, is often encoded by emrC and bcrABC genes (Dutta et al., 2013), present on mobile genetic elements that may be easily transmitted between different L. monocytogenes (Dutta et al., 2013; Kremer et al., 2017; Maury et al., 2019).
In Poland, limited information is available regarding L. monocytogenes diversity in food and food processing environment. The objectives of the present study were: (i) the molecular characterisation of L. monocytogenes of PCR-serogroups IIb and IVb isolated from ready-to-eat meat and meat processing environment in Poland using WGS and (ii) the comparison of the obtained sequences with genomes of L. monocytogenes isolated from human listeriosis cases.
A total of 48 L. monocytogenes isolates classified by PCR-serogrouping (as detailed below) into PCR-serogroup IIb (n = 17) and PCR-serogroup IVb (n = 31) were selected for the whole genome sequencing and genomic analyses. They were isolated in official laboratories between 2014 and 2017 from different kinds of ready-to-eat (RTE) food of animal origin (e.g., ham, sausages or meat) (n = 33) and from food processing environment (n = 15), using the standard ISO 11290-1:1996+A1:2004 (ISO, 1996). The isolates were originated from 9 voivodeships (administrative regions) of Poland and sent to the laboratory of the Department of Hygiene of Food of Animal Origin, National Veterinary Research Institute in Pulawy. Subsequently, the isolates were cultured on TSYEA (tryptone soya yeast extract agar; Oxoid, United Kingdom) at 37 ± 1°C for 18–4 h and the isolates were identified biochemically at species level using API Listeria (Biomerieux, France) according to the manufacturer’s instructions. The confirmed L. monocytogenes were stored at −70°C for further analysis.
L. monocytogenes were cultured on TSYEA at 37 ± 1°C for 18–24 h and a loopful of bacteria was transferred into 100 μl of TRIS buffer (A&A Biotechnology, Poland). DNA was extracted using the Genomic Mini protocol (A&A Biotechnology) with the modification by adding 15 μl of lysozyme (10 mg/ml; Sigma-Aldrich, United States) for 30 min at 37°C. L. monocytogenes isolates were molecularly typed for PCR-serogroups using multiplex PCR as previously described (Doumith et al., 2004). All isolates used in the study are listed in Supplementary Table S1.
DNA extraction was performed according to the modified Genomic Mini protocol as described above with additional modification by suspension of DNA in 100 μl of DNAse, RNAse free water (MP Biomedicals, United States) at 75°C. DNA quality and concentration were measured by NanoDrop and Qubit three (Thermo Fisher Scientific, United States). DNA library was prepared by the Nextera XT DNA Library Preparation Kit (Illumina, United States) according to the manufacturer’s instruction. DNA was sequenced on either Illumina Mi-seq (n = 40) or NextSeq500 (n = 8) sequencing platforms, respectively. All sequences trimmed with fqCleaner v.3.0 (Alexis Criscuolo, Institut Pasteur) and assembled with SPAdes v.3.11 with the automatic kmer selection (Bankevich et al., 2012). Assembly quality was assessed using the number of contigs, N50 and L50 metrics.
MLST (7 loci; Ragon et al., 2008) and cgMLST profiles (1,748 loci; Moura et al., 2016) were extracted from the assemblies using the BLASTN algorithm (Altschul et al., 1990) as previously described (Moura et al., 2016). MLST profiles were classified into sequence types (ST) and grouped into clonal complexes (CCs) as previously described (Ragon et al., 2008). cgMLST profiles were grouped into cgMLST types (CTs) and sublineages (SLs), using the cut-offs of 7 and 150 allelic mismatches, respectively, as previously described (Moura et al., 2016). The cgMLST profiles obtained in this study were also compared with those from 55 PCR-serogroup IVb and 8 PCR-serogroup IIb publicly available genome sequences of L. monocytogenes isolated from patients in Poland (Kuch et al., 2018). All calculations were made with the method described above. Allele numbers, STs, CCs, CTs, and SLs were determined according to the Listeria sequence typing database and its tools available on BIGSdb-Lm platform1 (Jolley and Maiden, 2010; Moura et al., 2016). MLST and cgMLST profile comparisons were done using Bionumerics 7.6 software (Applied Maths, Belgium) with single linkage algorithm ignoring missing values in pairwise comparisons. Dendrograms were visualized using iTOL2 (Letunic and Bork, 2016).
Identification of antimicrobial resistance, virulence and benzalkonium chloride tolerance genes (listed in Supplementary Table S1) was done using BIGSdb-Lm platform. Single gene alignments were performed using BLAST3 and MEGA7 softwares (Kumar et al., 2015).
Eight different MLST sequence types were identified: ST2 (31.2%, n = 15 isolates), ST5 (23.0%, n = 11), ST145 (12.5%, n = 6), ST1 (10.4%, n = 5), ST6 (10.4%, n = 5), ST3 (8.3%, n = 4), ST187 (2.1%, n = 1), and ST191 (2.1%, n = 1). L. monocytogenes belonging to PCR-serogroup IIb contained isolates classified to four sequence types (ST3, ST5, ST87, and ST191) whereas isolates of PCR-serogroup IVb were classified to 4 other STs (ST1, ST2, ST6, and ST145) (Figure 1 and Supplementary Table S1). Moreover, L. monocytogenes were grouped into seven clonal complexes (CC1, CC2, CC3, CC5, CC6, CC87, and CC191). Isolates from food processing environment mostly belonged to ST5/CC5 (8 out of 15 isolates) whereas the isolates of food origin were mainly classified to ST2/CC2 (14 out of 33 isolates) (Supplementary Table S1).
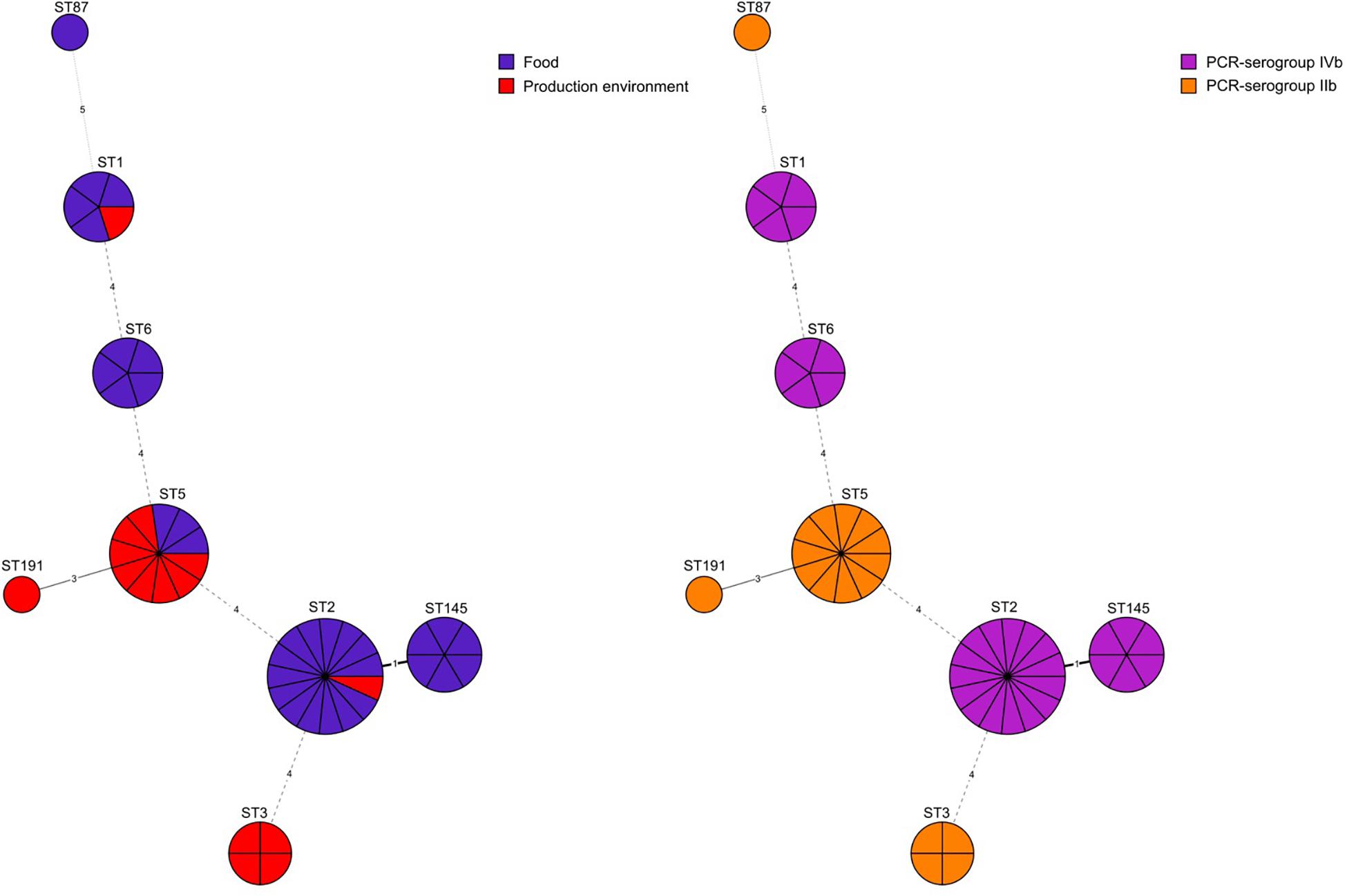
Figure 1. Minimum spanning tree analysis based on MLST allelic profiles of 48 L. monocytogenes isolates. Each circle represents a sequence type (ST). The numbers on the connecting lines illustrate the numbers of target genes with different alleles. The sources of isolates and their serogroups are distinguished by the colors.
Based on the cgMLST analysis, L. monocytogenes isolates were classified into 25 different CTs and grouped into seven SLs: SL2 (43.7%, n = 21 isolates), SL5 (22.9%, n = 11), SL1 (10.4%, n = 5), SL6 (10.4%, n = 5), SL3 (8.3%, n = 4), SL87 (2.1%, n = 1), SL191 (2.1%, n = 1) (Figure 2 and Supplementary Table S1).

Figure 2. Dendrogram representing clustering of L. monocytogenes PCR-serogroups IIb and IVb by cgMLST using single linkage clustering method. Degeneracy cut off value were 97% of similarity. Column: 1 – strain ID, 2 – source, 3 – PCR-serogroup, 4 – ST type, 5 – cgMLST type, 6 – year of isolation, 7 – place of isolation, 8 – detected genes. Color squares show presence of Listeria Pathogenicity Islands 1 and 2 (blue), internalins (orange), benzalkonium chloride resistance (green) and aminoglycosides resistance genes (yellow), respectively. Black squares indicate the presence of genes with truncation. Full metadata information are available in Supplementary Table S1. F, food, FPE, food production environment.
Among isolates originating only from food (n = 19), 6 different cgMLST types comprising more than one isolate were identified. The most prevalent one, cgMLST type L1-SL2-ST145-CT375, covered 6 isolates obtained from 4 different regions (kujawsko-pomorskie, lubuskie, śla̧skie, wielkopolskie voivodeships). The remaining cgMLST types were L1-SL2-ST2-CT4382 (4 isolates), L1-SL2-ST2-CT4380 (3 isolates), L1-SL5-ST5-CT3136 (2 isolates), L1-SL2-ST2-CT2416 (2 isolates), and L1-SL6-ST6-CT1955 (2 isolates) (Figure 2 and Supplementary Table S1).
Isolates exclusively from food processing environment (n = 7) were classified into seven different cgMLST types (Supplementary Table S1), mostly to SL5 and SL3.
Two CTs comprised isolates from both food and food processing environment. The first cgMLST type, L1-SL5-ST5-CT2323 (n = 8), consisted of one isolate from meat and 7 environmental isolates, including 4 isolates from floor drains in meat production areas. They originated from 3 voivodeships (Polish administrative regions) and were collected during two years (2015–2016). The second cgMLST type, L1-SL2-ST2-CT4325 (n = 4), consisted of 3 isolates from food and one isolate from food processing environment, isolated in 3 voivodeships during two-year period (2016, 2017) (Supplementary Table S1).
When comparing the present isolates with publicly available ones obtained from human listeriosis cases in Poland collected from 2004 to 2013, it was noticed that L. monocytogenes responsible for infections mostly belonged to sublineages SL3, SL1 and SL6, while the majority of the isolates of food and food production environment origins were classified into SL2 and SL5 sublineages (Figure 3). Moreover, five cgMLST types, belonging to sublineages SL1, SL2, and SL6, were found among L. monocytogenes isolated from both humans and food (Figure 4). In detail, isolate 741 recovered in 2017 from food (cgMLST type L1-SL1-ST1-CT1385) displayed 2–7 allelic difference to a cluster of seven isolates of human origin isolated during 2011–2013. Within cgMLST type L1-SL2-ST2-CT4382, 4 isolates (ID539, ID531, ID412, and ID253) collected from food between 2015 and 2016 also showed 2–7 allelic difference to the clinical isolate (ID34354) obtained in 2011. Moreover, among L. monocytogenes strains of L1-SL6-ST6-CT461 and L1-SL1-ST1-CT322 cgMLST types, isolated from food between 2015 and 2016 as well as from clinical cases in 2012, five and six allelic mismatches were also identified. Additionally, one food isolate (ID70, cgMLST L1-SL6-CT443 type from 2014) did not reveal any allelic differences with the isolate of clinical origin (ID41667) from 2013 (Figure 5).
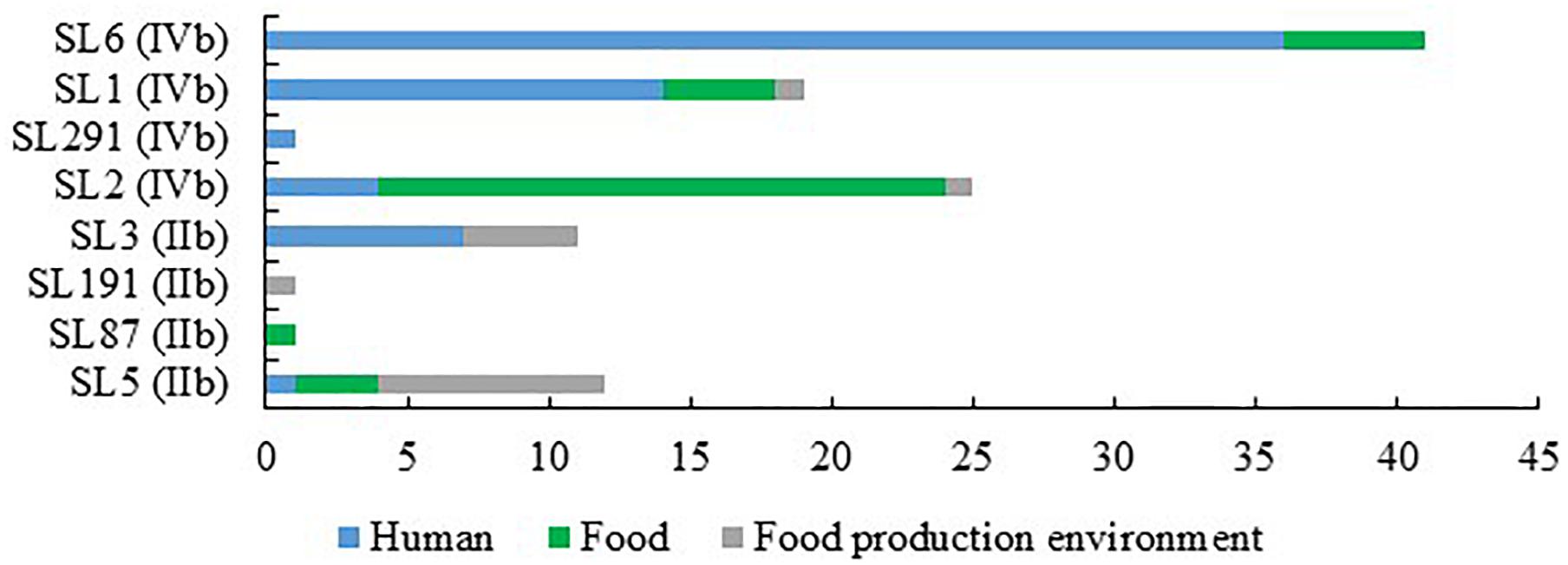
Figure 3. Number of L. monocytogenes sublineages among isolates of PCR-serogroups IIb and IVb tested in the study.
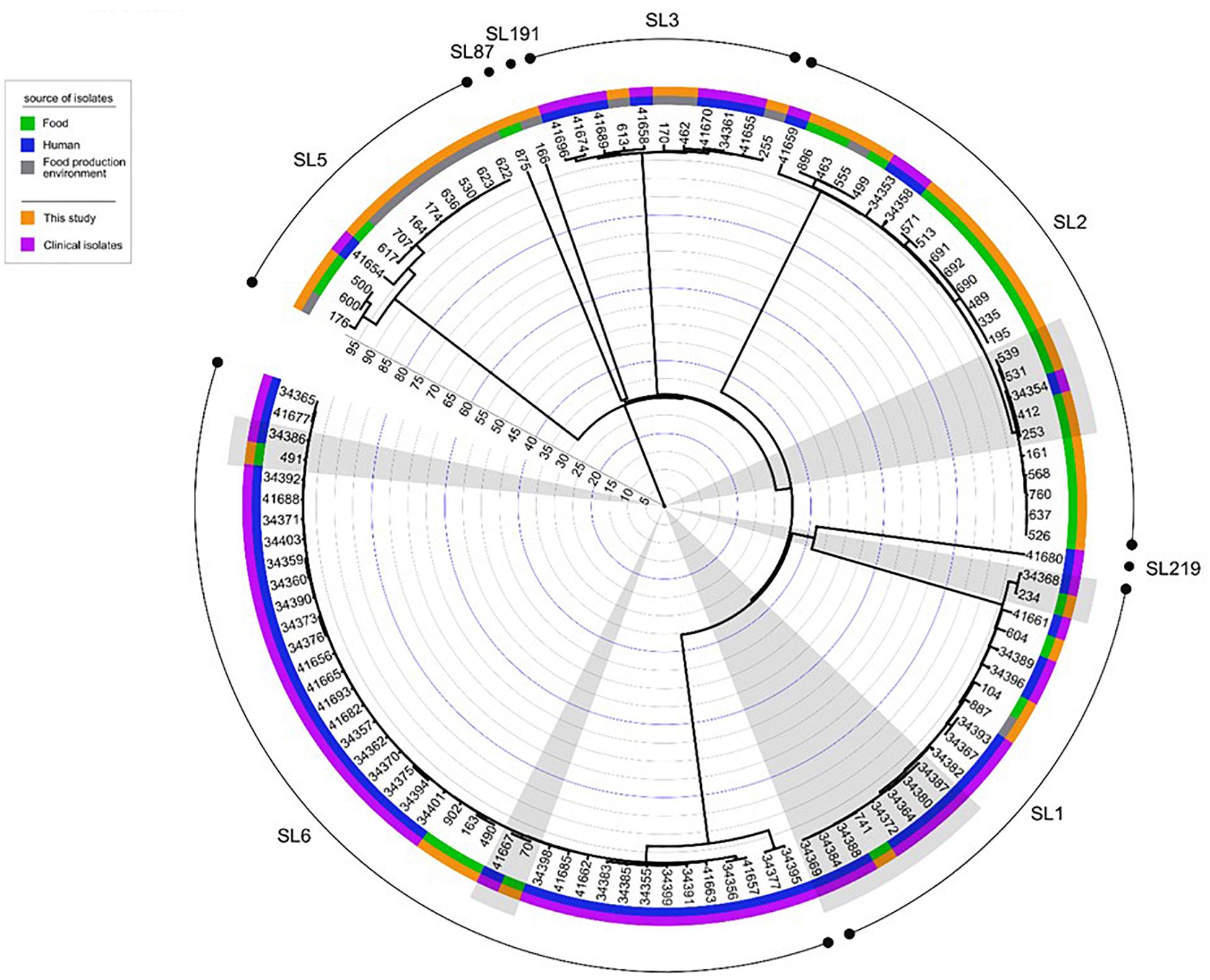
Figure 4. Dendrogram representing the comparison of L. monocytogenes isolates analyzed in this study with the published genomes of strains of PCR-serogroups IIb and IVb recovered in Poland from humans with listeriosis. Analysis was made on the base of cgMLST profiles using a single linkage clustering method. Gray shadows highlight the five groups of isolates with the 99.6% similarity, isolated from both humans and food. Clinical isolates were described by Kuch et al. (2018). Rings info, from the outer side: sublineages, source of isolates, number of isolates, the scale. Full metadata information is available in Supplementary Table S1.

Figure 5. Detailed view of the five cgMLST types identified in this study common to Polish clinical cases described by Kuch et al. (2018). Dendrograms were built based on cgMLST profiles using a single linkage clustering method. In the nodes, the number of allelic distances is indicated. F – food; H – humans.
Identification of selected virulence genes showed that the pathogenicity islands LIPI-1 and LIPI-3 were present in 48 (100%) and 15 (31.3%) isolates tested, respectively. LIPI-3 was identified in 10 isolates of PCR-serogroup IVb (5 SL1 and 5 SL6) and 5 isolates classified to PCR-serogroup IIb (4 SL3 and 1 SL191). None of the isolates was positive for LIPI-4 genes. The internalin gene family members inlA, inlB, inlC, inlE, inlF, and inlJ were found in all isolates tested, whereas the inlH and inlK genes were present in 47 (97.9%) and 46 (95.8%) isolates, respectively. inlG was found only in 5 isolates of PCR-serogroup IVb classified to SL6 (Supplementary Table S1).
Further analysis revealed that almost all L. monocytogenes SL5 isolates (10 out of 11; 90.9%, belonging to two different cgMLST types), harbored a premature stop codon (PMSC) mutation in inlA (Figure 3). In L1-SL5-ST5-CT2323 (8 isolates), the inlA PMSC type 1 (T1818A; Nightingale et al., 2005) was detected, whereas in L1-SL5-ST5-CT3136 (2 isolates) a new PMSC was identified (new allele inlA_302, termed PMSC type 31). This new mutation was caused by the deletion of adenosine in position 2209, resulting in a shorter peptide of 753 amino acids lacking InlA anchoring motif to peptidoglycan (Figure 6).
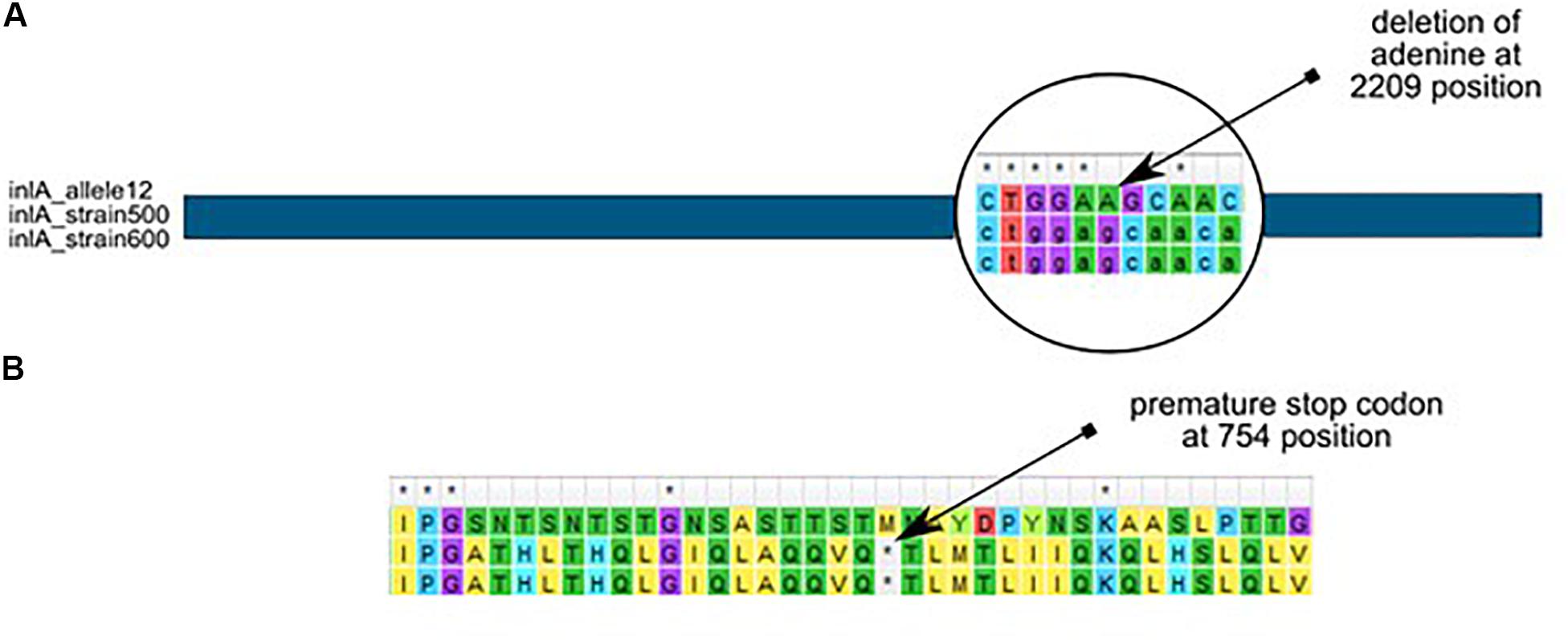
Figure 6. Adenine deletion in the newly identified inlA PMSC type 31 mutation in L. monocytogenes L1-SL5-ST5-CT3136 (PCR-serogroup IIb), as compared to inlA allele 12 (from SL5) retrieved from BIGSdb-Lm. (A) – nucleotide alignment, (B) – amino acid alignment.
In addition to an inlA PMSC, isolates belong to L1-SL5-ST5-CT2323 cgMLST type also carried a new inlB allele (allele 170), with a deletion of 141 nucleotides encoding the β-repeat sheet and partially the GW1 domain of the InlB protein.
Genes conferring benzalkonium chloride tolerance (bcrABC or emrC) were detected in 4 (8.3%) out of 48 analyzed L. monocytogenes (Supplementary Table S1). The bcrABC gene cassette was noticed in one isolate (L1-SL5-ST5-CT2050) originating from a meat processing plant. The emrC gene was harbored by 3 isolates (all L1-SL5-ST5-CT2323) obtained from the floor drains in slaughterhouses.
The SSI-1 gene cluster encoding the stress factor was detected among isolates belonging to L1-SL3-ST3 (4 isolates from CT1297, CT4803, CT2432, CT4327), L1-SL5-ST5 (3 isolates from CT2050, CT3136), and L1-SL191-ST191-CT2428 (1 isolate) cgMLST types, respectively. None of the examined isolates harbored the SSI-2 stress marker (Supplementary Table S1). Additionally, the isolates had the genes responsible for resistance to lysozyme (pdgA) and bile salts (bsh), which belong to core genes present in all L. monocytogenes (Supplementary Table S1).
WGS identification of antimicrobial resistance genes revealed that in all 48 L. monocytogenes isolates tested, four intrinsic resistance genes were present, as expected for core genes: fosX (resistance to fosfomycin), lmo0919 (lincosamides), norB (quinolones), and sul (sulfonamides). The acquired aacA4 gene (encoding resistance to aminoglycosides) was found in some serogroup IIb isolates, belonging to 4 different cgMLST types: L1-SL5-ST5-CT2323 (8 isolates), L1-SL3-ST3-CT2432, L1-SL3-ST3-CT4327, and L1-SL3-ST3-CT4803 (1 isolate each), mainly obtained from food production environment in 4 voivodeships (Figure 2).
A total of 48 L. monocytogenes isolates originating from ready-to-eat meat and meat processing environment were typed and characterized for the presence of virulence, antimicrobial resistance and stress response genetic determinants. MLST typing revealed that L. monocytogenes isolates were classified to 6 clonal complexes. Among them, there were CC1, CC2, and CC6 (total of 31, 64.6% isolates), previously described as human infection-associated hypervirulent clones in France (Maury et al., 2016). Several other studies have also shown that L. monocytogenes of clonal complexes CC1, CC2, CC4, and CC6 are often isolated from patients with listeriosis (Bergholz et al., 2018; Kuch et al., 2018). In the present investigation most of the isolates classified to these hypervirulent clonal complexes were recovered from RTE food. This confirms the previous findings that meat products may be a potential source of L. monocytogenes associated with human infections (Buchanan et al., 2017). Furthermore, the present analysis also showed that isolates belonging to five cgMLST types found in food (LI-SL1-ST1-CT1385, LI-SL1-ST1-CT322, LI-SL2-ST2-CT4382, LI-SL6-ST6-CT461, LI-SL6-ST6-CT443) were previously identified among human listeriosis isolates in Poland (Kuch et al., 2018) and other European countries (Denmark and France) (Moura et al., 2016, 2017). The close genetic distance between isolates within each cgMLST suggests a possible epidemiological link.
Most of L. monocytogenes isolates of PCR-serogroup IIb (11 of 17, 64.7%) belonged to clonal complex CC5 and were mainly of food processing environment origin (8 isolates). Moreover, 4 of these strains belonged to L1-SL5-ST5-CT2323 cgMLST type, were collected from floor drains at different environmental conditions present in cooling rooms and slaughterhouses. It was previously shown that floor drains may be “hotspots” on the spreading pathways of L. monocytogenes in food processing plants (Dzieciol et al., 2016). Several studies demonstrated that isolates belonging to CC5 and sublineage SL5 survive better in food processing environment than others, probably due to the presence of pLM80 plasmid with the bcrABC gene cassette responsible for tolerance to benzalkonium chloride, a very common compound of sanitizers using in food industry (Muhterem-Uyar et al., 2018). In the present study, the bcrABC marker was identified only in one isolate classified to LI-SL5-ST5-CT2050 cgMLST type of PCR-serogroup IIb. Another gene connected with benzalkonium chloride resistance (emrC) was found in 3 L. monocytogenes isolates, all belonging to LI-SL5-ST5-CT2323. These results suggest that some L. monocytogenes strains tested in the present study harbor resistance to adverse environmental conditions and may survive in food processing plants.
The results of the present study also showed that 11 L. monocytogenes strains from PCR-serogroup IIb, assigned to cgMLST types L1-SL5-ST5-CT2323, L1-SL3-ST3-CT4327, and L1-SL3-ST3-CT2432, harbored the aac4A gene which encodes resistance to aminoglycosides. It has been previously shown that this gene was present in different bacterial species and can be transferred between bacteria, including L. monocytogenes identified in food and food processing environment (Van et al., 2007; Sacha et al., 2012).
In the current study, the differences in the presence of virulence factors inlA and inlB among L. monocytogenes isolates belonging to PCR-serogroups IIb and IVb were found. The premature stop codon in the inlA gene was detected in 10 isolates classified to PCR-serogroup IIb and sequence type ST5 whereas this gene marker was not identified in any of the strains of PCR-serogroup IVb. The distribution of PMSC dependent on L. monocytogenes PCR-serogroups were also described by other authors (Jennison et al., 2017; Su et al., 2019). It has been previously shown that L. monocytogenes with this mutation are less virulent for humans, because truncation of this gene leads to the lack of the anchoring motif which is necessary for the cross-linking of InlA to peptidoglycan (Lebrun et al., 1996; Jonquieres et al., 1998). As described before, strains with cgMLST type L1-SL5-ST5-CT2323 have type 1 mutation in the inlA gene which was also confirmed in the present study (Glebícová et al., 2015). Moreover, a novel type of mutation identified in this gene was found in two isolates belonging to ST5 but classified to L1-SL5-ST5-CT3136.
The virulence gene inlB was identified in all 48 L. monocytogenes tested in the present study, including the isolates of cgMLST L1-SL5-ST5-CT2323 type with the deletion of 141 nucleotides. However, the impact of this mutation in InlB function has not been tested, but it may alter InlB association to L. monocytogenes cell surface. Strains with the same deletion, isolated from mushroom production environment, have been identified previously (Muruesan et al., 2015). It has been described that different isoforms of the inlB gene have an impact on signaling pathways and invasiveness of L. monocytogenes (Wałecka-Zacharska et al., 2015; Sobyanin et al., 2017; Chalenko et al., 2019). Of note, differences in the sequences of the inlB gene among strains of wild animals and humans were observed, which may suggest that mutations in this gene may have an impact on the invasiveness of L. monocytogenes (Zaytseva et al., 2007). Moreover, all our L. monocytogenes with deletion in the inlB gene also had PMSC mutation within the inlA marker which may suggest that these isolates are hypovirulent. Furthermore, among 8 isolates classified to ST3, ST5, and ST191 sequence types, the SSI-1 marker responsible for the adaptation of isolates to the extreme environmental conditions (resistance to bile salts and acids) was detected (Hein et al., 2011).
Whole genome sequencing of L. monocytogenes of PCR-serogroups IIb and IVb isolated from food and food processing environment allowed the characterization of the isolates and the ability to look at genetic traits associated with virulence and resistance to antimicrobials and environmental conditions. MLST and cgMLST analyses allowed determination of the CCs, SLs and CTs for molecular characterization and comparison of the present isolates with strains originated from different sources and countries. It was shown that some L. monocytogenes from food and from human listeriosis cases in Poland were classified to 5 common cgMLST types. Our study confirms food of animal origin as a source of pathogenic L. monocytogenes for humans.
The datasets generated for this study can be found in the NCBI database under BioProject PRJNA629756 and in BIGSdb-Lm.
MK, JO, and KW contributed to the conception and design of the study. KW and JO planned the study. MK and AM performed the experiments and performed the bioinformatic analyses. MK, JO, AM, AL, ML, and KW analyzed the data and drafted the manuscript. All authors critically read and approved the final version of the manuscript.
The research was funded by KNOW (Leading National Research Centre) Scientific Consortium Healthy Animal Safe Food allocated on the basis of the decision of the Ministry of Science and Higher Education No. 05-1/KNOW2/2015.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Department of Omics Analyses, National Veterinary Research Institute, Pulawy, Poland for samples’ sequencing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01412/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1006/jmbi.1990.9999
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its application to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bergholz, T., Shah, M., Burall, L., Rakic Martinez, M., and Datta, A. (2018). Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 102, 3475–3485. doi: 10.1007/s00253-018-8852-5
Bierne, H., Sabet, C., Personnic, N., and Cossart, P. (2007). Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9, 1156–1166. doi: 10.1016/j.micinf.2007.05.003
Buchanan, R., Gorris, L. G. M., Hayman, M. M., Jackson, T. C., and Whiting, R. C. (2017). A review of Liseria monocytogenes: an udate on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 75, 1–13. doi: 10.1016/j.foodcont.2016.12.016
Chalenko, Y., Kalinin, E., Marchenkov, V., Sysolyatina, E., Surin, A., Sobyanin, K., et al. (2019). Phylogenetically defined isoforms of Listeria monocytogenes invasion factor InlB differently activate intracellular signaling pathways and interact with the receptor gC1q-R. Intern. J. Mol. Sci. 20:4138. doi: 10.3390/ijms20174138
Charlier, C., Perrodeau, É, Leclercq, A., Cazenave, B., Pilmis, B., Henry, B., et al. (2017). Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect. Dis. 17, 510–519. doi: 10.1016/S1473-3099(16)30521-7
Colegiogri, A., Bruini, I., Di Ciccio, P. A., Zanardi, E., Ghidini, S., and Ianeri, A. (2017). Listeria monocytogenes biofilms in the wonderland of food industry. Pathogenes 6, 1–9.
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004
Dutta, V., Elhanafi, D., and Kathariou, S. (2013). Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 79, 6067–6074. doi: 10.1128/AEM.01751-13
Dzieciol, M., Schornsteiner, E., Muhterem-Uyar, M., Stessl, B., Wagner, M., and Schmitz-Esser, S. (2016). Bacterial diversity of floor drain biofilms and drain waters in a Listeria monocytogenes contaminated food processing environment. Intern. J. Food Microbiol. 223, 33–40. doi: 10.1016/j.ijfoodmicro.2016.02.004
European Food Safety Authority [EFSA] (2019). Scientific report on the European Union One Health 2018 zoonoses report. EFSA J. 17:5926. doi: 10.2903/j.efsa.2019.5926
Glebícová, T., Kolácková, I., Pantucek, R., and Karpíšková, R. (2015). A novel mutation leading to a premature stop codon in inlA of Listeria monocytogenes isolated from neonatal listeriosis. New Microbiol. 38, 293–296.
Glebícová, T., Zobanikova, M., Tomasticova, Z., Van Walle, I., Rupitsh, W., and Karpíšková, R. (2018). An outbreak of listerisosis linked to turkey meat products in Czech Republic, 2012–2016. Epidemiol. Infect. 146, 1407–1412. doi: 10.1017/S0950268818001565
Hein, I., Klinger, S., Dooms, M., Flekna, G., Stess, B., Leclercq, A., et al. (2011). Stress Survival Islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in Sequence Type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 77, 2169–2173.
ISO (1996). 11290-1:1996+A1:2004 Microbiology of Food and Animal Feeding Stuffs - Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes - Part 1: Detection Method. Geneva: International Organization for Standardization.
Jacquet, C., Doumith, M., Gordon, J. I., Martin, P. M. V., Cossart, P., and Lecuit, M. (2004). A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189, 2094–2100. doi: 10.1086/420853
Jennison, A. V., Masson, J. J., Fang, N.-X., Graham, R. M., Bradbury, M. I, Fegan, N., et al. (2017). Analysis of the Listeria monocytogenes population structure among isolates from 1931 to 2015 in Australia. Front. Microbiol. 8:603. doi: 10.3389/fmicb.2017.00603
Jolley, K., and Maiden, M. C. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595
Jonquieres, R., Bierne, H., Mengaud, J., and Cossart, P. (1998). The inlA gene of Listeria monocytogenes LO28 harbours a nonsense mutation resulting in release of internalin. Infect. Immun. 66, 3420–3422. doi: 10.1128/iai.66.7.3420-3422.1998
Kremer, P. H., Lees, J. A., Koopmans, M. M., Ferwerda, B., Arnds, A. W., Feller, M. M., et al. (2017). Benzalkonium tolerance genes and outcome in Listeria monocytogenes meningitis. Clin. Microbiol. Infect. 23, 265.e1–265.e7. doi: 10.1016/j.cmi.2016.12.008
Kuch, A., Goc, A., Belkiewicz, K., Filipello, V., Ronkiewicz, P., Gołêbiewska, A., et al. (2018). Molecular diversity and antimicrobial susceptibility of Listeria monocytogenes isolates from invasive infections in Poland (1997–2013). Sci. Rep. 8:14562. doi: 10.1038/s41598-018-32574-0
Kumar, S., Stecher, G., and Tamura, K. (2015). MEGA7: molecular evolutionary genetics analysis version 7.0. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lebrun, M., Mengaud, J., Ohayon, H., Nato, F., and Cossart, P. (1996). Internalin must be on the bacterial surface to mediate entry of Listeria monocytogenes into epithelial cells. Mol. Microbiol. 21, 579–592. doi: 10.1111/j.1365-2958.1996.tb02566.x
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, 242–245.
Lingnau, A., Domann, E., Hudel, M., Bock, M., Nichterlein, T., Wehland, J., et al. (1995). Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by prfA-dependent and -independent mechanisms. Infect. Immun. 63, 3896–3903. doi: 10.1128/iai.63.10.3896-3903.1995
Luth, S., Kleta, S., and Al Dahouk, S. (2018). Whole genome sequencing as a typing tool for foodborne pathogenes lika Listeria monocytogenes – The way toward global harmonisation and data exchange. Trends Food Sci. Technol. 73, 67–75. doi: 10.1016/j.tifs.2018.01.008
Maury, M. M., Bracq-Dieye, H., Huang, L., Vales, G., Lavina, M., Thouvenot, P., et al. (2019). Hypervirulent Listeria monocytogenes clones’ adapting to mammalian gut accounts for their association with dairy products. Nat. Commun. 10:2488. doi: 10.1038/s41467-019-10380-0
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Liseria monocytogenes hypervirulence by harnessine its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tar, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2:16185. doi: 10.1038/nmicrobiol.2016.185
Moura, A., Tourdjman, M., Leclercq, A., Hamelin, E., Laurent, E., Fredriksen, N., et al. (2017). Real-time Whole-Genome Sequencing for survelliance of Liseria monocytogenes, France. Emerg. Infect. Dis. 23, 1462–1470.
Muhterem-Uyar, M., Luminita, C., Wagner, K. H., Wagner, M., Schmitz-Esser, S., and Stessl, B. (2018). New aspects on Listeria monocytogenes ST5-ECVI predominance in a heavily contaminated cheese processing environment. Front. Microbiol. 9:64. doi: 10.3389/fmicb.2018.00064
Muruesan, L., Kucerova, Z., Knabel, S., and LaBorde, L. (2015). Predominance and distribution of a persistent Listeria monocytogenes clone in a commercial fresh mushroom processing environment. J. Food Prot. 78, 1988–1998. doi: 10.4315/0362-028X.JFP-15-195
Nightingale, K. K., Windham, K., Martin, K. E., Yeung, M., and Wiedmann, M. (2005). Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. App. Environ. Microbiol. 71, 8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005
Portnoy, D. A., Auerbuch, V., and Glomski, I. J. (2002). The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell mediated immunity. J. Cell Biol. 158, 409–414.
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Ramaswamy, V., Cresence, V., Rejitha, J., Lekshmi, M., Dharsana, K., Prasad, S., et al. (2007). Listeria - review of epidemiology and pathogenesis. J. Microbiol. Immunol. Infect. 40, 4–13.
Sacha, P., Jaworowska, J., Ojdana, D., Wieczorek, P., Czaban, S., and Tryniszewska, E. (2012). Occurrence of the aacA4 gene among multidrug resistant strains of Pseudomonas aeruginosa isolated form bronchial secretions obtained from the Intensive Therapy Unit at University Hospital in Bialystok, Poland. Folia Histichem. Cytobiol. 5, 322–324. doi: 10.5603/fhc.2012.0043
Sobyanin, K., Sysolyatina, E., Krivozubov, M., Chalenko, Y., Karyagina, A., and Ermolaeva, S. (2017). Naturally occurring InlB variants that support intragastric Listeria monocytogenes infection in mice. FEMS Microbiol. Lett. 364:fnx011. doi: 10.1093/femsle/fnx011
Su, X., Cao, G., Zhang, J., Pan, H., Zhang, D., Kuang, D., et al. (2019). Characterization of internalin genes in Listeria monocytogenes from food and humans, and their association with the invasion of Caco-2 cells. Gut Pathog. 11:30.
Swaminathan, B., and Gerner-Smidt, P. (2007). The epidemiology of human listeriosis. Microbes Infect. 9, 1236–1243. doi: 10.1016/j.micinf.2007.05.011
Van, T. T. H., Moutafis, G., Tran, L. T., and Coloe, P. J. (2007). Antibiotic resistance in food-borne bacterial contaminants in Vietnam. Appl. Environ. Microbiol. 24, 7906–7911. doi: 10.1128/AEM.00973-07
Wałecka-Zacharska, E., Kosek-Paszkowska, K., Bania, J., Staroniewicz, Z., Bednarski, M., and Wieliczko, A. (2015). Invasiveness of Listeria monocytogenes strains isolated from animals in Poland. Pol. J. Vet. Sci. 18, 697–702. doi: 10.1515/pjvs-2015-0090
Keywords: Listeria monocytogenes, food, meat, cgMLST, WGS
Citation: Kurpas M, Osek J, Moura A, Leclercq A, Lecuit M and Wieczorek K (2020) Genomic Characterization of Listeria monocytogenes Isolated From Ready-to-Eat Meat and Meat Processing Environments in Poland. Front. Microbiol. 11:1412. doi: 10.3389/fmicb.2020.01412
Received: 21 April 2020; Accepted: 01 June 2020;
Published: 25 June 2020.
Edited by:
Frederique Pasquali, University of Bologna, ItalyReviewed by:
Dara Leong, Teagasc Food Research Centre, IrelandCopyright © 2020 Kurpas, Osek, Moura, Leclercq, Lecuit and Wieczorek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kinga Wieczorek, a2luZ2Eud2llY3pvcmVrQHBpd2V0LnB1bGF3eS5wbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.