- 1Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 2State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Villosiclava virens is the prevalent causative pathogen of rice false smut, a destructive rice disease. Mating-type genes play a vital role in the evolution of mating systems in fungi. Some fungi have lost MAT1-1-3, one of the mating-type genes, during evolution, whereas others still retain MAT1-1-3. However, how MAT1-1-3 regulates the sexual development of heterothallic V. virens remains unknown. Here, we generated the MAT1-1-3 mutants, which exhibited defects in vegetative growth, stress response, pathogenicity, sclerotia formation and fruiting body maturation. An artificial outcrossing inoculation assay showed that the Δmat1-1-3 mutant was unable to produce sclerotia. Unexpectedly, the Δmat1-1-3 mutant could form immature fruiting bodies without mating on potato sucrose agar medium (PSA) compared with the wild-type strain, most likely by activating the truncated MAT1-2-1 transcription to regulate the sexual development. Moreover, RNA-seq data showed that knockout of MAT1-1-3 results in misregulation of a subset of genes involved in sexual development, MAPK signaling, cell wall integrity, autophagy, epigenetic modification, and transcriptional regulation. Collectively, this study reveals that MAT1-1-3 is required for asexual and sexual development, and pathogenicity of V. virens, thereby provides new insights into the function of mating-type genes in the fungi life cycle and infection process.
Introduction
Villosiclava virens (Anamorph, Ustilaginoidea virens) is a plant pathogen that causes rice false smut (RFS), which results in substantial rice yield losses worldwide (Brooks et al., 2010; Ladhalakshmi et al., 2012; Jecmen and Tebeest, 2015; Fan et al., 2016). V. virens infects rice florets to produce the RFS balls, which can generate sclerotia on their surface (Ashizawa et al., 2012; Tang et al., 2013; Hu et al., 2014; Song et al., 2016; Yong et al., 2018). Additionally, V. virens directly affects the rice food safety by producing mycotoxins, which are harmful to humans and animals (Li et al., 2008; Fu et al., 2017; Wang et al., 2017). In addition to the completion of genome sequencing and the available transcriptome data (Zhang et al., 2014), the recently established clustered regularly interspaced short palindromic repeats (CRISPR) mediated gene knockout system provides an effective tool to study the gene function in V. virens (Liang et al., 2018; Fang et al., 2019; Guo et al., 2019; Yu et al., 2019). During the pathogen’s sexual life cycle, V. virens can produce ascospores by sexual reproduction (Zhang et al., 2014; Yong et al., 2018), which, through invasion of rice spikelets, are considered to be one of the primary infection sources of rice false smut (RFS) (Ikegami, 1960; Wang, 1995). Genetic recombination occurs during sexual reproduction when two compatible strains mate, thereby playing a vital role in expanding V. virens genetic diversity (Sun et al., 2013; Wang et al., 2014). Thus, the sexual reproduction plays an important role in the prevalence of RFS and genetic diversity in V. virens.
Sexual reproduction is a key step for fungi to complete their life cycle (Zhang et al., 2014). In ascomycetous fungi, sexual reproduction, controlled by mating type alleles/idiomorphs (MAT) (Yun et al., 1999; Zheng and Wang, 2013), is divided into three major modes: heterothallic, homothallic and pseudohomothallic (or secondary homothallic) (Whittle et al., 2011; Zheng and Wang, 2013). The two opposite idiomorphs are named MAT1-1 (or MATA) and MAT1-2 (or MATa) (Alby et al., 2009; Whittle et al., 2011). Homothallic (self-compatible) species carry both MAT1-1 and MAT1-2 idiomorph genes in a single nucleus, usually closely linked or fused (Nelson, 1996; Wilken et al., 2012). In contrast, heterothallic (self-incompatible) species have alternate MAT idiomorph in different nuclei (Coppin et al., 1997; Yun et al., 1999). Extensive evidences showed that these mating modes could exist in different fungi belong to the same genus and play an important role in evolutionary biology. For example, the Neurospora species employ these three mating modes to complete the process of sexual reproduction, and multiple switches in the mating systems have occurred in the genus’ evolutionary history (Nygren et al., 2011; Whittle et al., 2011). For ascomycete Cordyceps sensu lato (sl), there are also reproductive switches between homothallic and heterothallic modes (Hu et al., 2013; Zheng et al., 2013). In addition, some studies supported the hypothesis that the sexual reproduction of Aspergillus genus may evolve from homothallic to heterothallic mode (Galagan et al., 2005; Paoletti et al., 2005), but still need more evidences to prove in the future. However, some studies supported the hypothesis that the ancestral mating system in Neurospora was heterothallic (Gioti et al., 2012). Compared with heterothallism, homothallism is more likely to accumulate deleterious genomic mutations (Whittle et al., 2011). To avoid self-crossing, genetic barriers have evolved sexual dimorphism to prevent selfing (Klix et al., 2010). Therefore, as one of the factors of species evolution, these studies indicate that heterothallism plays a more important role than homothallism during species evolution (Lee et al., 2010; Zheng and Wang, 2013).
Villosiclava virens has a heterothallic sexual reproduction system controlled by MAT1-1 and MAT1-2 idiomorphs (Yu J. J. et al., 2015). The MAT1-1 idiomorph includes MAT1-1-1, MAT1-1-2, MAT1-1-3, and a truncated MAT1-2-1, and the MAT1-2 idiomorph includes MAT1-2-1 and MAT1-2-8 (Yu J. J. et al., 2015). Extensive studies show that MAT1-1 and MAT1-2 in fungi have different functions to regulate sexual reproduction. For example, in heterothallic N. crassa, matA-1 (MAT1-1-1) mutant is sterile but matA-2 (MAT1-1-2) and matA-3 (MAT1-1-3) mutants have slightly reduced fertility (Ferreira et al., 1998). In heterothallic Podospora anserina, SMR2 (MAT1-1-3) mutant can produce asci, but the number is reduced (Zickler et al., 1995). However, in homothallic Fusarium graminearum, mat1-1-2 and mat1-1-3 mutants are fertile, mat1-1-1 and mat1-2-1 mutants display male- and female-specific defects, respectively (Zheng et al., 2015). In addition, in homothallic Sordaria macrospora, SmtA-3 (MAT1-1-3) is not required for fruiting body development and also has no effect on vegetative morphology (Klix et al., 2010). In filamentous ascomycetes, MAT1-1-1 and MAT1-2-1 encode α-box domain proteins and high mobility group (HMG) domain transcription factors, respectively (Turgeon et al., 1993b). MAT1-1-3 also encodes an HMG domain transcription factor (Coppin et al., 1997; Debuchy and Turgeon, 2006). In contrast to SmtA-3 (MAT1-1-3) and SMR2 (MAT1-1-3) proteins, the HMG motif is lacking in the putative SmtA-3 protein (Klix et al., 2010). Different functional requirements for MAT1-1-3 in sexual development between homothallic F. graminearum and S. macrospora imply that some of the regulatory networks controlled by MAT proteins may not be conserved across filamentous ascomycetes (Kim et al., 2012). Thus, the function of MAT1-1-3 homologs in sexual reproduction is variable (Yokoyama et al., 2005), and little is known about MAT1-1-3 function in V. virens.
In this study, MAT1-1-3 was characterized using Δmat1-1-3 mutant generated with the CRISPR/Cas9 system. We performed phylogenetic comparisons of V. virens with other fungi and the MAT1-1-3 proteins from different species. We have demonstrated that the transcription factor MAT1-1-3 is required for vegetative growth, stress response, pathogenicity and sexual development in V. virens. The Δmat1-1-3 mutant could form an immature fruiting body without mating in PSA medium plate. Transcriptome analysis of Δmat1-1-3 mutant and wild-type strains revealed differential expression of a subset of genes. In addition, the expression level of truncated MAT1-2-1 was up-regulated in Δmat1-1-3 mutant. Collectively, our study demonstrates that MAT1-1-3 plays a vital role in sexual and asexual development in V. virens.
Materials and Methods
Strains and Growth Conditions
Villosiclava virens WT strains Uv1-56 (MAT1-1 mating-type) and Uv2-51 (MAT1-2 mating-type) and all transformants generated in this study were cultured and maintained on potato sucrose agar (PSA) plates (200 g/L potato, 20 g/L sucrose, 15 g/L agar) at 28°C in the dark. Agrobacterium tumefaciens strain AGL1 was incubated on LB medium (0.5% yeast extract, 1% tryptone, 1% NaCl) at 28°C. Yeast strain AH109 was incubated on YPDA medium (1% yeast extract, 2% peptone, 2% glucose, 0.003% adenine hemisulfate, 2% agar) at 30°C. All strains used are listed in Supplementary Table S1. For DNA extraction, hyphae were harvested after growth in PSA at 28°C for 7 days in the dark. Conidiation in potato sucrose broth (PSB) medium (200 g/L potato and 20 g/L sucrose) and fungal growth on PSA plates were measured using protocols from a previous study (Xie et al., 2019). Colony diameters of Uv1-56, indicated mutant and complemented strains were measured on PSA medium after 15 days at 28°C.
For conidiation, Uv1-56, mutant or complemented strains were cultured in PSB medium with shaking at 150 rpm at 28°C for 7 days. Then, the cultured mixtures were filtered, and the concentrations of conidia were measured using a hemocytometer. For the germination test, conidial suspension droplets were added to water agar plates, incubated at 28°C for 12 h and photographed with an Olympus BX-53 microscope. For stress tests, Uv1-56, mutant and complemented strains were incubated on PSA at 28°C for 15 days with different concentrations of stress agents, including 0.5 M NaCl, 0.7 M Sorbitol, 0.07% H2O2, 600 μg/ml congo red (CR), 600 μg/ml calcofluor white (CFW) and 0.03% sodium dodecyl sulfate (SDS). The inhibition rates were calculated as described previously (Xie et al., 2019). Three biological replicates were performed to assess conidiation, germination and stress tests.
Orthology and Phylogenetic Analysis
A multisequence phylogenetic tree was constructed using the concatenated sequences of core genes identified by the default parameters of the CEGMA (v2.5) pipeline. Briefly, the CEGMA software contains the core eukaryotic genes dataset, which has 458 core genes. We searched the homologous genes in different fungal genome database using CEGMA software based on these 458 core genes. The phylogenetic tree was constructed using the homologous proteins encoded by the core homologous genes. Multiple sequence alignments of proteins were made by muscle, and then a neighbor-joining tree was built using MEGA7 with 1000 bootstrap repeats for distance estimation. The similarity of MAT1-1-3 homologs from different fungi was analyzed with MEGA7 software. The sequences of MAT1-1-3 homologous proteins in different fungi proteome database was obtained from National Center for Biotechnology Information (NCBI) with specific accession number as below: Villosiclava virens (Ustilaginoidea virens, AKE48501.1), Metarhizium anisopliae (BAE93596.1), Metarhizium robertsii (XP_007819908.2), Ephelis japonica (BAD72606.2), Epichloe typhina (BAD72610.2), Claviceps purpurea (BAD72602.2), Ophiocordyceps sinensis (AGW27558.1), Tolypocladium inflatum (BAE93600.1), Pyrenopeziza brassicae (CAA06846.1), Fusarium graminearum (AAG42812.1), Fusarium fujikuroi (AAC71053.1), Fusarium oxysporum (AEO15073.1), Sordaria macrospora (KAA8630682.1), Neurospora crassa (AAC37476.1), Podospora anserina (CAA52051.1).
Plasmid Construction and Transformant Generation
For constructing gene replacement plasmids, the MAT1-1-3 gene replacement vector (pMD19-MAT1-1-3) was obtained using previously reported methods (Li et al., 2019). Briefly, upstream and downstream flanking sequences (∼1 kb) of MAT1-1-3 were cloned using primers MAT1-1-3-upF/upR and MAT1-1-3-doF/doR, respectively. The hygromycin B-resistance (HYG) gene fragment was amplified from the SK1044-hyg plasmid using primers F3/R3. The upstream, downstream and HYG fragments were inserted into vector T-Vector pMD19. To generate the Cas9-gRNA vector, gene gRNA spacers were designed with the gRNA designer website for best on-target scores1. Synthetic sgRNA oligos were annealed and inserted into BsmBI-digested pmCas9:tRp-gRNA (Liang et al., 2018) to generate pmCas9:tRp-gRNA-MAT1-1-3 constructs. The pMD19-MAT1-1-1, pMD19-MAT1-1-2, pmCas9:tRp-gRNA-MAT1-1-1 and pmCas9:tRp-gRNA-MAT1-1-2 constructs were generated in a similar manner. All constructs were confirmed by sequencing.
To generate knockout mutants, replacement pMD19-MAT1-1-3 vector and pmCas9:tRp-gRNA-MAT1-1-3 were co-transformed into strain Uv1-56 protoplasts using the PEG-mediated method as described previously (Li et al., 2019). HYG gene was used to screen for knockout transformants. MAT1-1-3 was verified by PCR with primer pair MAT1-1-3-F2/R2, and the HYG gene was verified by PCR with primers F1/R3 and F3/R1. Primers F1/R1 were used to amplify the inserted fragment by PCR, and sequencing traces of junction regions confirmed that MAT1-1-3 was replaced by the HYG gene in mutants. The MAT1-1-1 and MAT1-1-2 knockout mutants were also generated using the same method.
For complementation assays, the MAT-1-1-3 gene, including its 1.5 kb promoter, was amplified with primers C-MAT-1-1-3-F/R. The amplified fragment was cloned into BamHI and HindIII digested pKO1-NEO-Uv vector and was verified by sequencing analysis. The complementary vector was transformed into A. tumefaciens strain AGL1 and complementation transformants generated by the Agrobacterium-mediated (ATMT) were screened with geneticin selection using methods described previously (Yu M. N. et al., 2015). All primers used in this study are listed in Supplementary Table S2.
Pathogenicity and Sclerotia Formation Assays
To detect pathogenicity, the rice susceptible cultivar Liangyoupeijiu was artificially inoculated with V. virens as described previously (Tang et al., 2013; Yu J. J. et al., 2015) with minor modifications. Briefly, the rice plants grown in a greenhouse were used to inoculate with V. virens. Uv1-56, Uv2-51, complemented and mutant strains were cultured in PSB at 28°C for 7 days with rotary shaking at 150 rpm/min. Mixtures of hyphae and conidia were created with a blender, and the conidia concentration was adjusted to 1 × 106/mL with PSB. Seven days before the heading stage, about 1–2 mL of inoculum suspension was injected into 15 swollen flag leaf sheaths of rice using sterilized syringes. Inoculated rice was cultured in the greenhouse for 6 days at 25°C at 95%–100% humidity (RH) and then were placed at 28°C and about 80% RH. The rice false smut balls were counted after 30 dpi. The average diseased grain rate was calculated as previously described (Hu et al., 2014) and modified as follows: average diseased grain rate = (average of the number of false smut balls per panicle/average of the total number of rice grains per panicle) × 100%.
For sclerotia formation assays, the hyphae and conidia suspension of Uv1-56 and Uv2-51 prepared as above were mixed in a 1:1 ratio to generate the mixed inoculum suspension. The mixed inoculum suspension of Δmat1-1-3 and Uv2-51 (or complemented strain and Uv2-51) was also prepared using the same method. The mixed inoculum suspension was inoculated the rice swollen flag leaf sheath of rice on the seventh day before heading stage using a needle syringe. The inoculated rice plants were grown in a greenhouse at 25°C and 95%–100% RH for 6 days. Then these plants were transferred to another growth room under normal conditions (25°C-30°C, 80% RH, and 12-hr light/12-hr dark photoperiod) for 24 days of growth. Finally, these plants were further transferred to a growth incubator at a day-and-night temperature of 25°C/15°C and 70% RH for another 15 days of growth before counting the sclerotia. The sclerotium formation rate was calculated as follows: sclerotium formation rate = (average of the number of sclerotia/average of the number of false smut balls) × 100%. The sclerotia germination assay was performed according to previous methods (Yong et al., 2018). The statistical analyses were performed by Student’s t-test. All experiments were repeated three times.
Confrontation Assay
The WT Uv1-56, Uv2-51 and mutant strains were cultured on PSA plates for 15 days. The mycelium plugs from 15-day-old PSA cultures were placed onto the right and left sides of the PSA plates, respectively. Thirty plates replicates were performed in each experimental treatment. Hyphae of Uv1-56 and Δmat1-1-3 were harvested at 10 and 15 days for RNA extraction. Meanwhile, the cell fusion of opposing mating type strains was observed and photographed with an Olympus BX-53 microscope. In order to observe the formation of fruiting bodies, the strain culture time was extended to 60 days and photographed with a digital camera. Three biological replicates were performed.
qRT-PCR and RNA-seq Analysis
Gene expression was evaluated by quantitative real-time PCR with specific primers. For the confrontation assay, Uv1-56 or Δmat1-1-3 was crossed with Uv2-51. Samples of confrontation culture strains were harvested at 10 and 15 days. RNA was extracted with the BioTeKe RNA reagent Kit and cDNA reverse transcription was performed using the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time, Takara). An ABI PRISM 7000 Sequence Detection System (Applied Biosystems, United States) with SYBR® Premix Ex Taq (Tli RNaseH Plus) (Takara, Japan) was used. The reactions were conducted as described previously (Fang et al., 2019). β-tubulin was used as the internal reference for measuring gene expression. Relative expression was determined using the 2–ΔΔCt method (Livak and Schmittgen, 2001). For detecting the transcript level of MAT1-1-3 and truncated MAT1-2-1 in wild-type Uv1-56, mycelia were harvested after incubation in PSB at 28°C for 7 days for RNA extraction. Transcript amounts were calculated using the 2–ΔCT method in ABI 7000 System Sequence Detection Software. Three biological replicates were performed and the results showed similar trends.
RNA from wild-type Uv1-56 and Δmat1-1-3 mutant, which were harvested after incubation on PSB at 28°C for 7 days, was extracted using Trizol® Reagent. RNA-seq libraries were prepared with the ScriptSeq v2 kit (Epicentre SSV21124) following a published method (Jiao et al., 2018). Transcriptome sequencing was performed by Illumina HiSeq 2500 with the paired-end 2 × 150 bp model at the Majorbio biomedical technology Co., Ltd. (Shanghai, China). RNA-seq data from each sample were aligned to version 1 of the Villosiclava virens reference genome (Zhang et al., 2014) using Hisat2. Transcript assembly was performed using Cufflinks (Trapnell et al., 2012). Analysis of the difference in gene expression was performed using DEGseq. The cutoff for differential expression was transcripts per million reads (TPM) adjusted p < 0.05 and |log2FC| ≥ 1.
Generation of MAT1-1-3-GFP Fusion Transformants and Subcellular Localization
To obtain the MAT1-1-3-GFP construct, a cDNA fragment of the MAT1-1-3 gene was amplified from strain Uv1-56 cDNA template with primers MAT1-1-3-GFP-F/R and cloned into BamHI and SmaI digested pKD1GFP with the ClonExpress II One Step Cloning Kit (Vazyme, China). The resulting plasmid was confirmed by sequencing analysis. The pKD1GFP plasmid was transformed into A. tumefaciens strain AGL1 using the freeze-thaw method (Weigel and Glazebrook, 2006) and was then transformed into V virens via ATMT transformation as described previously (Yu M. N. et al., 2015). After 7 days of A. tumefaciens strain and V. virens co-culturing, transformants were picked and observed under a fluorescence microscope. MAT1-1-3-GFP fusion transformants were incubated on PSB at 28°C for 7 days. The hyphae and conidia were harvested and stained with 4′, 6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, United States) to visualize nuclei as described previously (Li et al., 2015). A rotary laser confocal microscope (Ultra View VoX, PerkinElmer, United States) was used to observe MAT1-1-3 colonization.
4′,6-Diamidino-2-Phenylindole (DAPI) and Calcofluor White Staining
The conidia and hyphae of indicated strains were harvested after 7 days in PSB. Staining was conducted as described previously (Li et al., 2015). To observe the septum of hyphae, the indicated strains were stained with 20 μg/ml calcofluor white (CFW), which was used to stain the cellulose and chitin in the cell wall (Roncero and Durán, 1985), for 5–15 min, and the pictures were taken by a fluorescence microscope (Nikon Eclipse 80i, Japan). The hyphae and conidia were and stained with 20 μg/mL 4′, 6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, United States) for 1 h to visualize nucleus under the PerkinElmer UltraView VoX confocal (PerkinElmer, United States).
Yeast Two-Hybrid Assay
Protein-protein interactions were performed by the yeast two-hybrid system (Clontech, Laboratories, Inc.). MAT1-1-3 ORF was amplified using primers BD113-F/R, from the cDNA of Uv1-56, and then was cloned into the SmaI-digested pGBKT7 vector using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The same method was used to clone MAT1-1-1 and MAT1-1-2 ORFs into the SmaI-digested pGADT7 vector to make prey constructs. The resulting bait and prey constructs were confirmed by sequencing analysis and co-transformed into yeast strain AH109. The transformants were grown on the SD-Trp-Leu and SD-Trp-Leu-His-Ade medium plates at 30°C for 3–5 days. This experiment was repeated three times.
Results
Phylogenetic Analyses of V. virens and MAT1-1-3 Proteins With Other Fungi
To explore the evolutionary relationships of V. virens with other fungi, we selected 31 representative members of fungi, including heterothallic, homothallic and combined species. Phylogenetic analysis established that V. virens is more closely related to Clavicipitaceae although received weak bootstrap support (i.e., Metarhizium spp. Epichloe typhina, Claviceps purpurea), followed by Ophiocordycipitaceae (i.e., Ophiocordyceps and Tolypocladium), then Cordycipitaceae (i.e., Cordyceps spp. Beauveria bassiana) (Figure 1A). The Clavicipitaceae, Ophiocordycipitaceae and Cordycipitaceae all belong to Hypocreales (Bushley et al., 2013). In Figure 1A, fungi indicated with red undergo homothallic sexual reproduction, fungi indicated with green undergo homothallic and heterothallic sexual reproduction, and the remaining fungi are heterothallic (Figure 1A). However, the sexual reproductive mode of Ophiocordycipitaceae switches between homothallism and heterothallism. For example, Tolypocladium inflatum is heterothallic, but Ophiocordyceps sinensis is homothallic (Hu et al., 2013). As shown by the blue circle in Figure 1A, some fungi have lost the MAT1-1-3 gene during Hypocreales evolution. For example, E. typhina and O. sinensis have the MAT1-1-3 gene, while C. tenuipes, C. militaris and B. bassiana have lost the MAT1-1-3 gene and their sexual reproduction is heterothallic (Figure 1A). V. virens also has MAT1-1-3, suggesting that the gene plays an important role in the evolution of sexual reproduction. Phylogenetic analysis further showed that the MAT1-1-3 protein from V. virens is clustered with those from Metarhizium spp. (Figure 1B). In addition, all of the sequences from 15 diverse fungi were found to contain the high mobility group (HMG) domain, except SmtA-3 (Figure 1B). Thus, we further compared the HMG domain sequences of 14 fungi species using WebLogo. Multiple sequence alignment of these proteins showed that the HMG domain is conserved among different fungi (Figure 1C). Collectively, the results suggest that MAT1-1-3 is a conserved protein among different fungi.
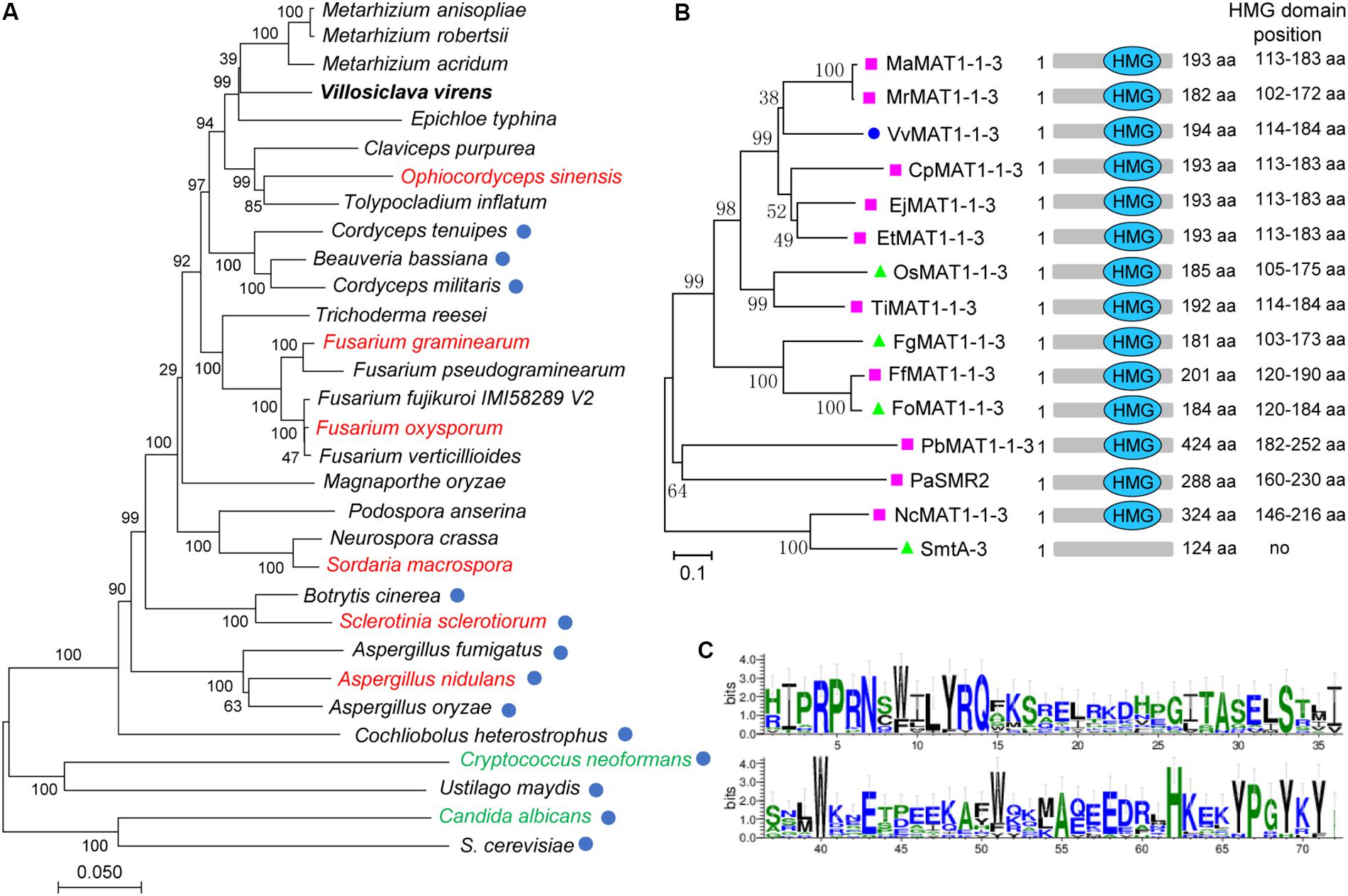
Figure 1. Phylogenetic analysis of Villosiclava virens with other fungi and VvMAT1-1-3 protein with MAT1-1-3 from various fungi. (A) The multisequence phylogenetic tree was constructed by MEGA7 with core eukaryotic genes identified using the CEGMA (v2.5) pipeline to show the evolutionary relationship of V. virens with different fungal species. Blue circles indicate species that do not have a MAT1-1-3 gene. The red names indicate fungi that undergo homothallic sexual reproduction, the green names indicate fungi that undergo sexual reproduction that is homothallic and heterothallic. The black names indicated fungi that are heterothallic and possess a MAT1-1-3 gene. (B) Phylogenetic tree of fungal MAT1-1-3 genes and the position of the high mobility group (HMG) domain in different fungal MAT1-1-3 genes. Pink squares indicate heterothallic fungi; green triangles indicate homothallic fungi; the blue circle indicates V. virens. (C) The weblogo depicts the multiple sequence alignment of HMG domains from the 14 protein sequences in (B). The logo shows the conservation of various residues. The y-axis represents the bit score. A score of 4 on the y-axis means 100% conservation. The x-axis displays the amino acid position.
MAT1-1-3 Affects V. virens Mycelial Growth, Conidial Morphology and Production
To investigate the function of V. virens MAT1-1-3, we generated Δmat1-1-3 mutants in strain Uv1-56 (wild type) by using the CRISPR/Cas9 system (Liang et al., 2018; Li et al., 2019) to replace MAT1-1-3 with an HYG gene (hygromycin B-resistance) (Supplementary Figure S1A). Genome DNA PCR and sequencing analysis of three Δmat1-1-3 mutants (#8, #16 and #20) confirmed that MAT1-1-3 was successfully replaced (Supplementary Figures S1B,C). These mutants had similar phenotypes, so mutant #16 was selected for additional study. A complementation assay was carried out with Δmat1-1-3#16 to generate the CΔmat1-1-3#16 complemented strain (Supplementary Figures S1B,C). The MAT1-1-3 is located at the MAT1-1 idiomorph of V. virens, which also contains two other mating type genes MAT1-1-1 and MAT1-1-2. Thus, to assess whether MAT1-1-1 and MAT1-1-2 have the same phenotype with MAT1-1-3, we also generated Δmat1-1-1 and Δmat1-1-2 mutants using the same method.
The Δmat1-1-3 and Δmat1-1-1 mutants, but not Δmat1-1-2 mutant, exhibited a reduction in mycelial growth rate compared with those of the WT Uv1-56 and complemented strain (CΔmat1-1-3#16) (Figures 2A,B). Moreover, the colony of Δmat1-1-3 mutant exhibited deeper pigmentation on PSA medium plate compared with those of the WT Uv1-56, Δmat1-1-1, Δmat1-1-2 mutants and complemented strain (Figures 2A,B). Based on the difference in colony morphology between Δmat1-1-3 mutant and the other two mutants (Δmat1-1-1 and Δmat1-1-2), we only focused on studying the function of MAT1-1-3 in this study. The Δmat1-1-3 mutant produced more globose conidia compared with WT and complemented strain, which produced more oval conidia (Figures 2C–E). The conidia produced by Δmat1-1-3 mutant geminated slower than that of produced by WT and complemented strain (Figure 2C). CFW staining showed that the hyphal septa of the Δmat1-1-3 mutant increased and chitin accumulated on the tip of mycelium in the Δmat1-1-3 mutant compared with WT (Figure 2F), indicating that MAT1-1-3 involved in regulating cell wall chitin and cellulose synthesis. Mycelial growth and hyphal septa defects of the mutant were rescued in the complemented strain CΔmat1-1-3#16 (Figure 2). Together, these results indicated that MAT1-1-3 was required for conidial morphogenesis, conidia production, germination and cell wall integrity.
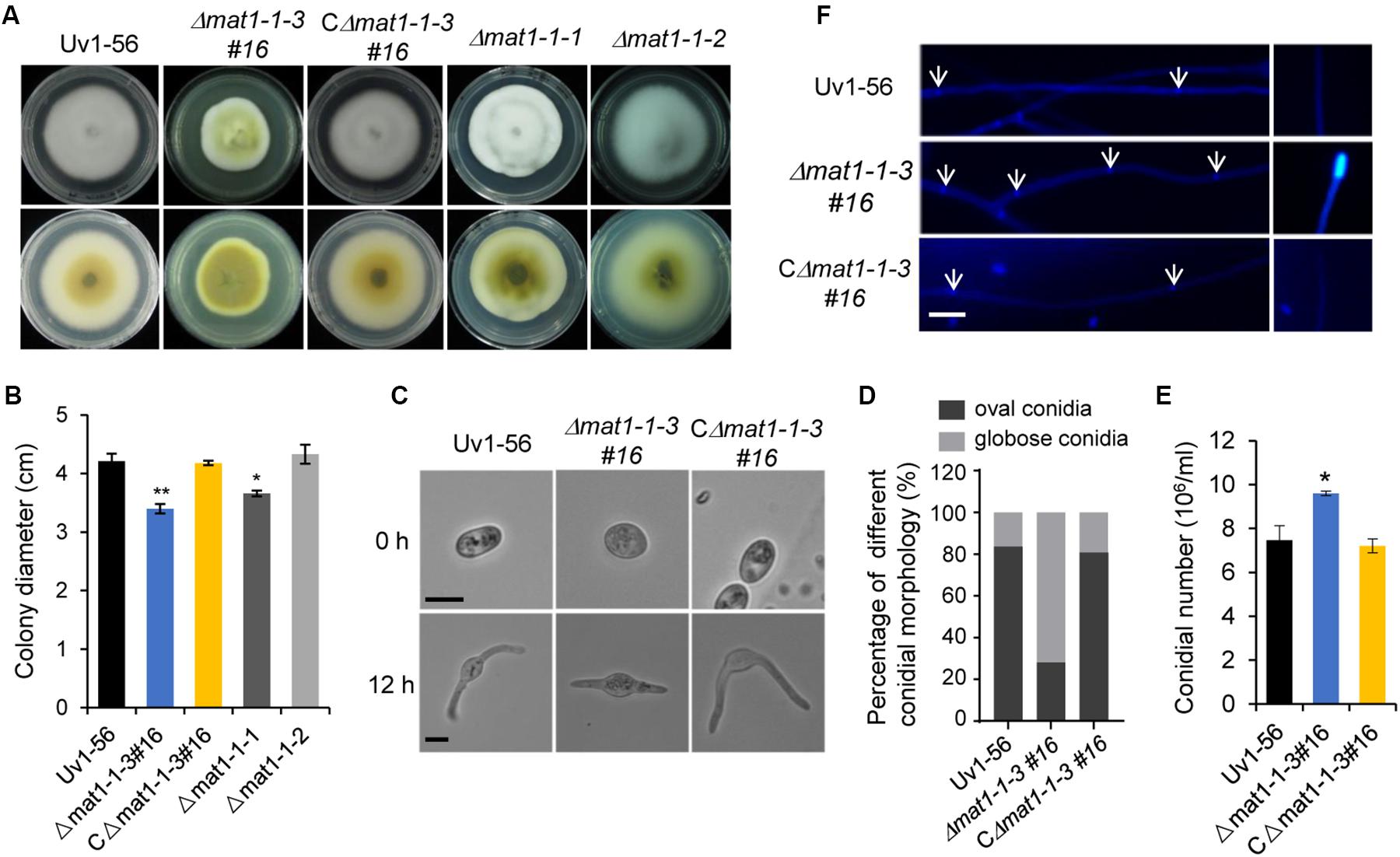
Figure 2. MAT1-1-3 affects mycelial growth, conidial morphology and production. (A) Colony morphology of wild-type (Uv1-56), Δmat1-1-3#16 deletion mutant, complemented strain CΔmat1-1-3#16, Δmat1-1-1 and Δmat1-1-2 deletion mutants on PSA medium after 15 days of incubation at 28°C. (B) Quantified growth rate of wild type, mutant and complemented strain in (A). Colony diameter was statistically analyzed, and error bars represent SD. The asterisks indicate significant differences (one-way ANOVA, *p < 0.05, **p < 0.01). (C) The conidial morphology and germination of wild-type, mutant and complemented strains were photographed after culturing on water agar plates for 0 h and 12 h. Scale bars, 5 μm. (D) Statistical analysis of the percentage of different conidial morphology in wild-type, mutant and complemented strains. (E) Statistical analysis of conidia production on PSB medium after 7 days of culturing with 150 rpm shaking at 28°C. Data are shown as mean ± SD from three independent replicates. Asterisks indicate significant differences (one-way ANOVA, *p < 0.05). (F) Hyphae of wild-type, mutant and complemented strains were stained with calcofluor white, which detects chitin and cellulose in the cell wall, and fluorescence is shown in blue color. The hyphal septum of the Δmat1-1-3 mutant was shorter than that of wild type and complemented strains. Deletion of MAT1-1-3 altered the distribution of chitin in the cell wall. Arrows indicate the hyphal septa. Scale bars, 10 μm.
MAT1-1-3 Regulates V. virens in Response to Different Abiotic Stresses
To test whether knockout of MAT1-1-3 affects V. virens responses to the osmotic, oxidative stress and cell wall integrity. The WT, mutant and complemented strains were cultured on PSA medium containing different stress agents. The growth inhibition rate of mycelial growth was calculated after 15 days of culture at 28°C. Our results demonstrated that the growth inhibition rate of mycelial growth of mutant displays significant difference, the Δmat1-1-3 mutant exhibited increases in the tolerance to NaCl, Sorbitol, H2O2, CR and CFW, but more sensitive to SDS compared with WT and complemented strain CΔmat1-1-3#16 (Figure 3). Taken together, these results indicated that MAT1-1-3 is required for regulating the V. virens responses to osmotic stress and oxidation stress as well as cell wall integrity.
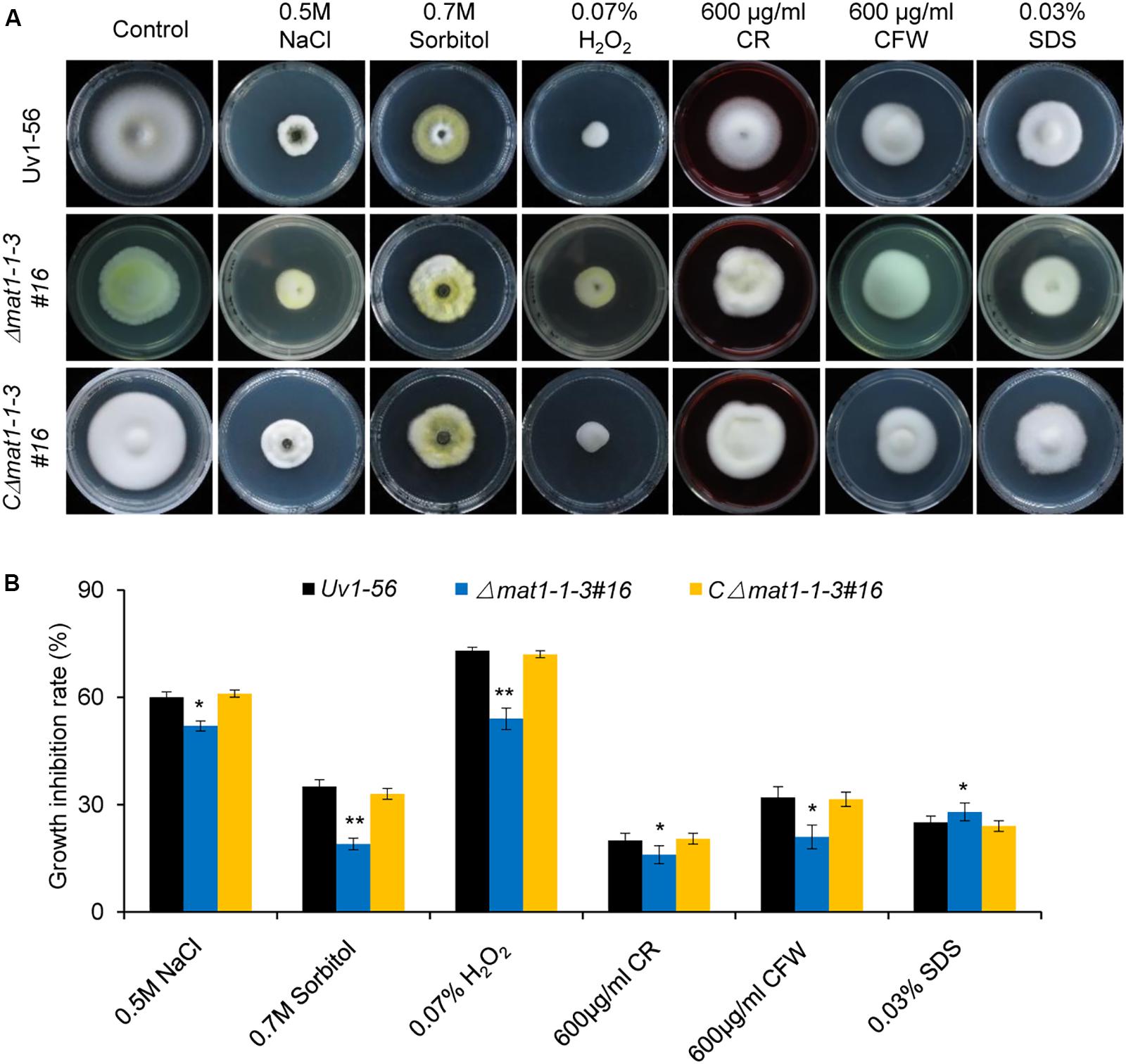
Figure 3. MAT1-1-3 is involved in regulating pathogen stress responses. (A) Mycelial radial growth of the indicated wild-type, Δmat1-1-3#16 and complemented CΔmat1-1-3#16 strains on PSA medium supplemented with salt stress agent (0.5 M NaCl), osmotic stress agent (0.7 M Sorbitol), oxidative-stress agent (0.07% H2O2), and cell wall disturbing agents Congo Red (CR, 600 μg/mL), calcofluor white (CFW, 600 μg/mL) and sodium dodecyl sulfate (0.03% SDS). Photographs were taken after 15 days of incubation at 28°C. (B) Statistical analysis of the indicated strains growth inhibition rate under different stress conditions. Colony diameters of the indicated strains were measured. Data are shown as mean ± SD of three independent replicates. Asterisks indicate significant differences (one-way ANOVA, *p < 0.05, **p < 0.01).
MAT1-1-3 Is Required for the Pathogenicity of V. virens
To further evaluate the role of MAT1-1-3 in fungal pathogenicity, suspensions of shattered hyphae and conidia from various strains were individually inoculated into booting stage rice panicles. At 30 days post-inoculation (dpi), the false smut balls produced on rice spikelets were counted to evaluate V. virens pathogenicity. Rice panicles infected by the Δmat1-1-3 mutant produced significantly fewer false smut balls than those infected by WT and complemented CΔmat1-1-3#16 strains (Figure 4). This result suggested that MAT1-1-3 was required for the pathogenicity of V. virens.
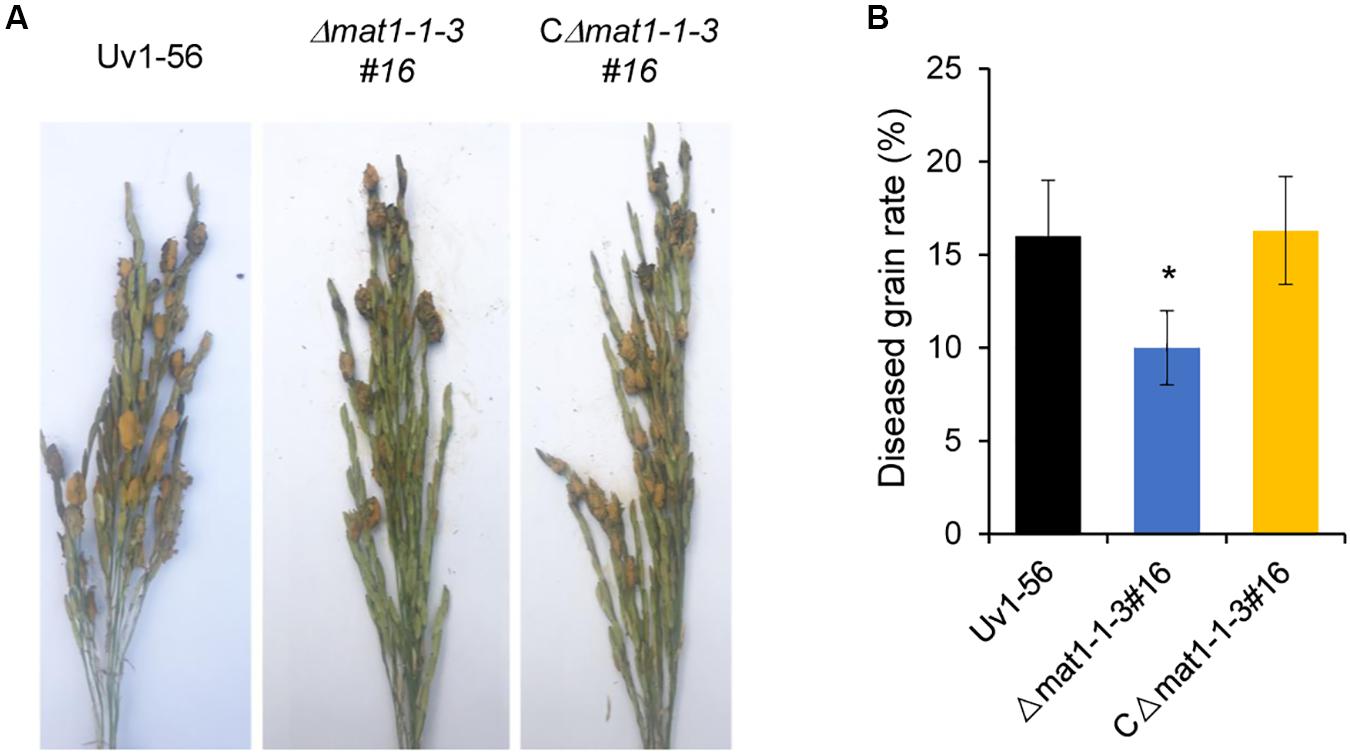
Figure 4. MAT1-1-3 is required for Villosiclava virens full pathogenicity in rice. (A) Rice spikelets were infected with an inoculum of wild-type Uv1-56, Δmat1-1-3#16 mutant and complemented strains. Pictures were taken 30 days post-inoculation. (B) Statistical analysis of the average diseased grains rate in the infected spikelets. False smut balls were counted from each single rice panicle as described in materials and methods. Data represent means ± SD from three independent experiments. Asterisks indicate significant differences (one-way ANOVA, *p < 0.05).
MAT1-1-3 Is Required for Sclerotia Formation
We next investigated whether MAT1-1-3 regulated sclerotia formation of V. virens. On the seventh day before heading stage, the swollen flag leaf sheath of rice was inoculated with the mixed suspension of conidia and hyphae of Uv1-56 & Uv2-51, Δmat1-1-3 & Uv2-51, and CΔmat1-1-3#16 & Uv2-51. As shown in Figure 5, Δmat1-1-3 mutant could not produce the sclerotia, whereas the wild-type Uv1-56 could produce the sclerotia on the surface of false smut balls, which were marked by the red arrows. Apparently, the complemented strain CΔmat1-1-3#16 restored the sclerotia production to the WT level (Figure 5). Together, the results suggested that MAT1-1-3 was required for sclerotia formation and sexual development of V. virens.
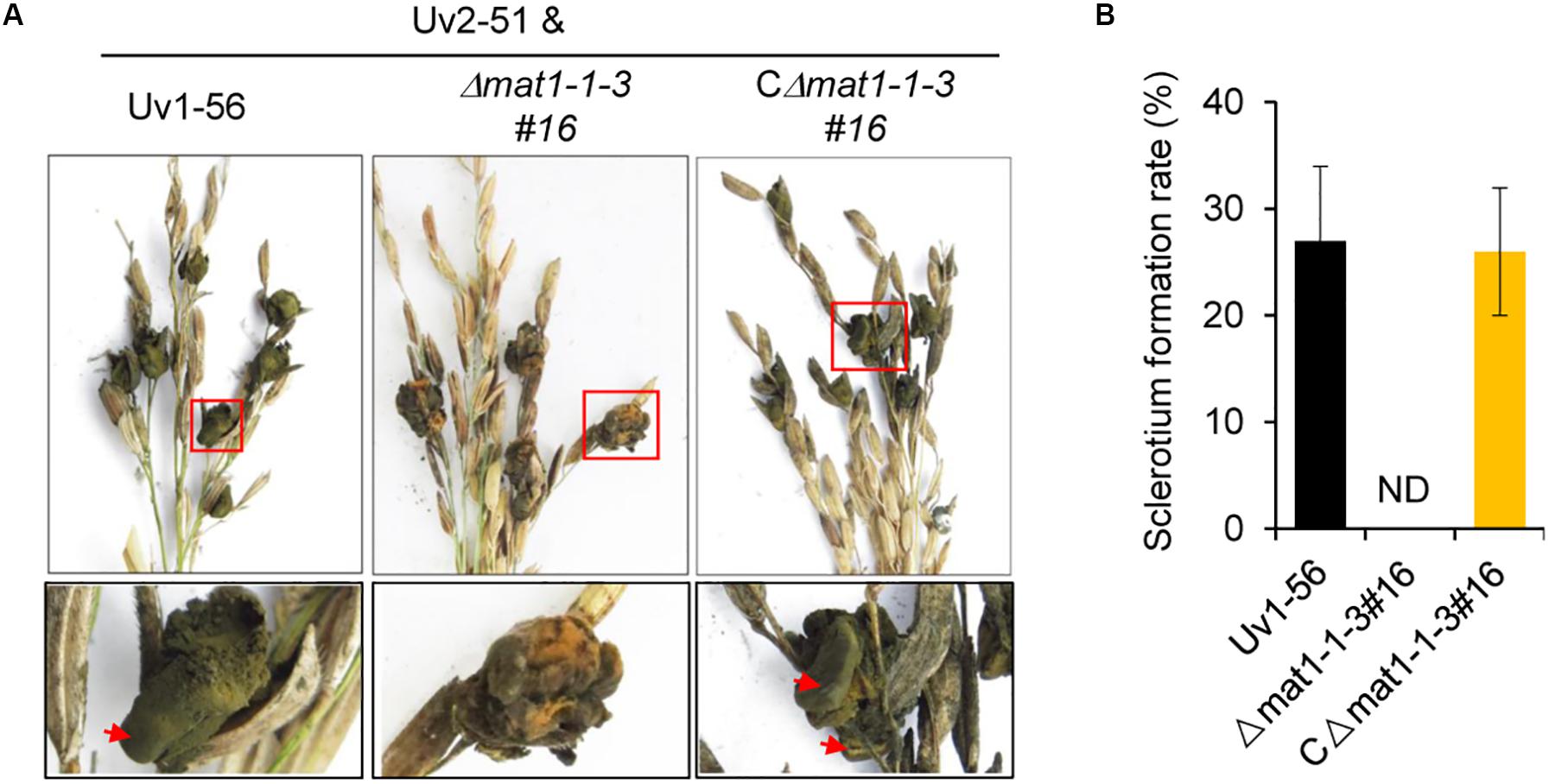
Figure 5. MAT1-1-3 is indispensable for Villosiclava virens sclerotia formation and sexual development. (A) In contrast with Uv1-56 and complemented strains, the Δmat1-1-3#16 mutant was unable to produce sclerotia. Pictures were photographed 45 days post-inoculation (dpi). The enlarged regions marked with squares were shown at the bottom. (B) Statistical analysis of sclerotium formation rates of wild-type, mutant or complemented strains crossed with Uv2-51 at 45 dpi. Data are shown as means ± SD of three independent replicates. ND, not detection.
MAT1-1-3 Is a Negative Regulator of the Immature Fruiting Body in V. virens
The fertile sclerotia play an important role in the sexual cycle and development of V. virens, but current research shows that it is only produced in rice spikelets infected by two opposite mating-type strains in the field. The sclerotia can germinate and form different sexual structures, such as primordia, fruiting body, asci and ascospores. The germination of sclerotia collected from the field was observed in the laboratory. Germination assays showed that the sclerotia began to germinate at 50 days, primordia emerged at 55 to 65 days, and mature fruiting bodies, filled with asci containing ascospores, appeared at 75 days (Figure 6A). Although Δmat1-1-3 mutant could not produce sclerotia after outcrossing with Uv2-51 in the field (Figure 5), the Δmat1-1-3 mutant alone formed the fruiting body primordium and germinated on the PSA medium (Figure 6B). The mycelium of Δmat1-1-3 mutant began to grow densely at 20 days and a large number of hyphae differentiated into filaments at 30 days (Figure 6B). Then the primordia appeared at 35 days (Figure 6B). At 40 days, the primordia of fruiting body appeared (Figure 6B). However, the fruiting body could not reach maturity and the immature stromata had no ascospores (marked with red squares), even though extended the growth time in PSA medium plate (Figure 6B), indicating that the Δmat1-1-3 mutant alone can form similar sexual structures. Conversely, wild-type Uv1-56 and CΔmat1-1-3#16 could not form fruiting body primordia and the colonies began to dry and shrink after 40 days on PSA medium (Figure 6C). After crossing with Uv2-51, only the Δmat1-1-3 mutant could form the primordia at 35 days compared with WT and CΔmat1-1-3#16 (Figure 6D). Taken together, these results indicate that MAT1-1-3 negatively regulates the formation and development of immature fruiting bodies during the sexual development of V. virens.
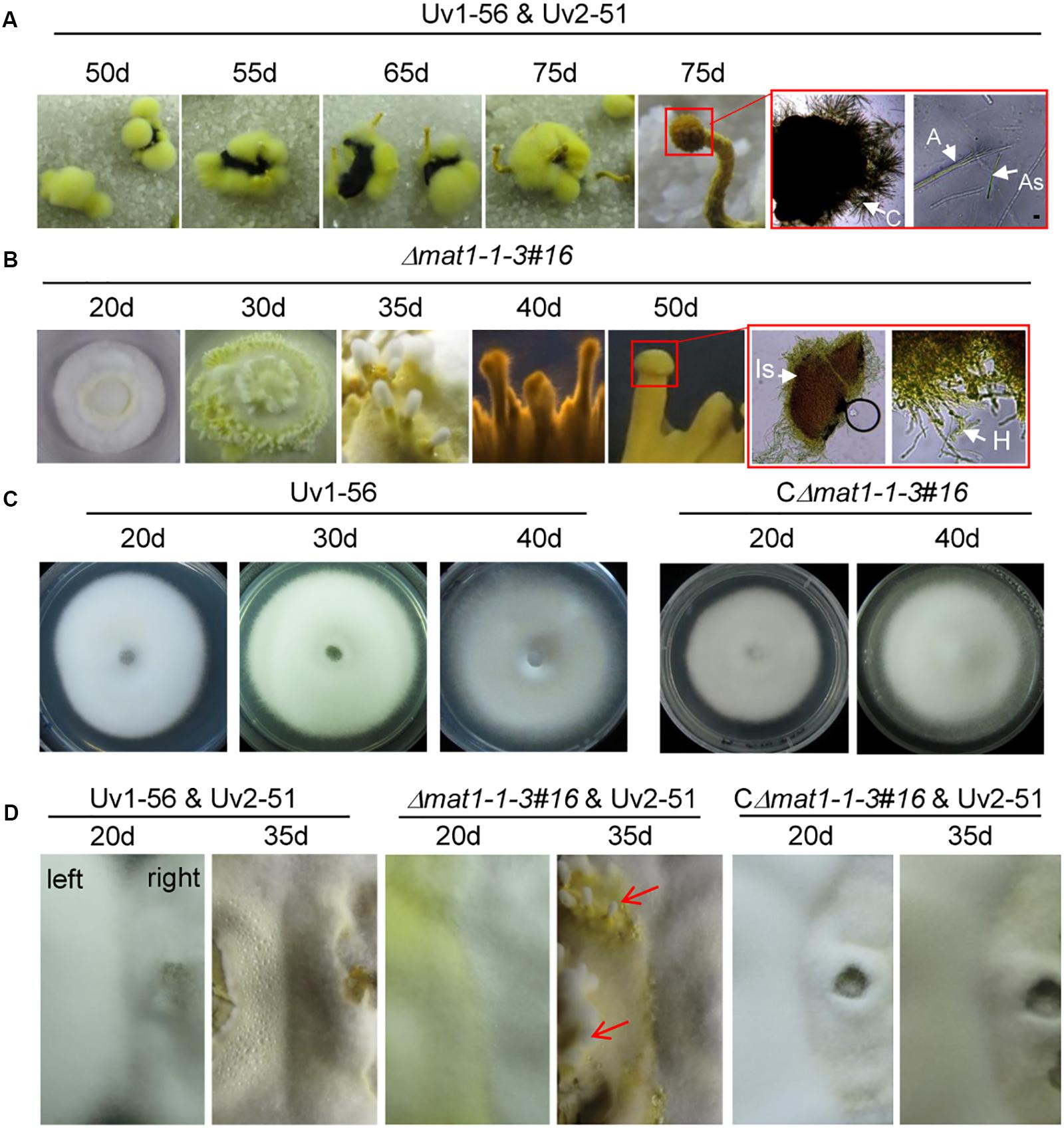
Figure 6. MAT1-1-3 negatively regulates the formation of fruiting bodies in Villosiclava virens. (A) Inoculum of wild-type Uv1-56 mixed with Uv2-51 infected the rice panicle to form fertile sclerotia. Wild-type sclerotia germination formed different sexual structures at different periods. The magnified red square region indicates that mature fruiting bodies could form a cavity and ascospores. (B) Sexual structures for the Δmat1-1-3 mutant formed after different periods on the PSA medium. The magnified red square region shows that the fruiting body was immature, no ascospores were produced and only hypha formed. (C) Wild-type Uv1-56 and complemented CΔmat1-1-3#16 strains could not form the sexual structure on PSA plates. (D) For the confrontation assay, wild-type Uv1-56, complemented strain or mutant Δmat1-1-3 were confronted with wild-type Uv2-51 and cultured at 28°C for 20 days and 35 days on PSA medium. The colony growing on the right in the plate represents the Uv2-51 strain. The Δmat1-1-3 mutant, but not WT Uv1-56 and complemented strains, formed sexual structures at 35 days after confronting culture with Uv2-51. Is, Immature stroma; H, hypha; C, cavity; A, asci; As, ascospores.
Deletion of MAT1-1-3 Affects Expression of Pheromone Precursor and Pheromone Receptor Genes and Mating Hyphae Growth
The expression level of pheromone precursor and pheromone receptor genes are strictly regulated during sexual reproduction. Yu et al. (2016) identified several genes homologous to pheromone precursor (PPG1, a homolog of ccg-4 in N. crassa) and pheromone receptors (PRE1 and PRE2, homolog of pre-1 and pre-2 in N. crassa, respectively) in V. virens (Yu et al., 2016). Given that MAT1-1-3 regulated the sexual development of V. virens, we investigated the expression of pheromone precursor and pheromone receptors genes after confronting mutant or WT strains with the MAT1-2 strain Uv2-51 for 10 days or 15 days. Compared with the WT, qRT-PCR results showed decreased PPG1 expression and increased PRE1 and PRE2 expression in the Δmat1-1-3 mutant at 10 days and 15 days, and the expression level of PRE1 and PRE2 at 10 days was higher than at 15 days (Figure 7A), suggesting that MAT1-1-3 can regulate transcription of these genes. It has been known that pheromone-related genes also regulate the cell fusion of opposing mating type strains before mating (Jones and Bennett, 2011). To test if the cell fusion was affected in the Δmat1-1-3 mutant, we employed a confrontation assay (Snetselaar et al., 1996). In the control, mating hyphae formed normally when Uv1-56 was confronted with Uv2-51 at 10 days (Figure 7B). However, when the Δmat1-1-3 mutant was confronted with Uv2-51, both strains had longer mating hyphae than those in the control, and the hyphae of Δmat1-1-3 mutant were more frequently fused with Uv2-51 than with WT strain at 15 days (Figure 7B). At 20 days, the Δmat1-1-3 mutant colony fused better with Uv2-51 than with the wild-type strain (Figure 7B). Taken together, these results demonstrate that MAT1-1-3 is likely to regulate the expression of PPG1, PRE1 and PRE2 to affect mating hyphae growth and hyphae fusion.
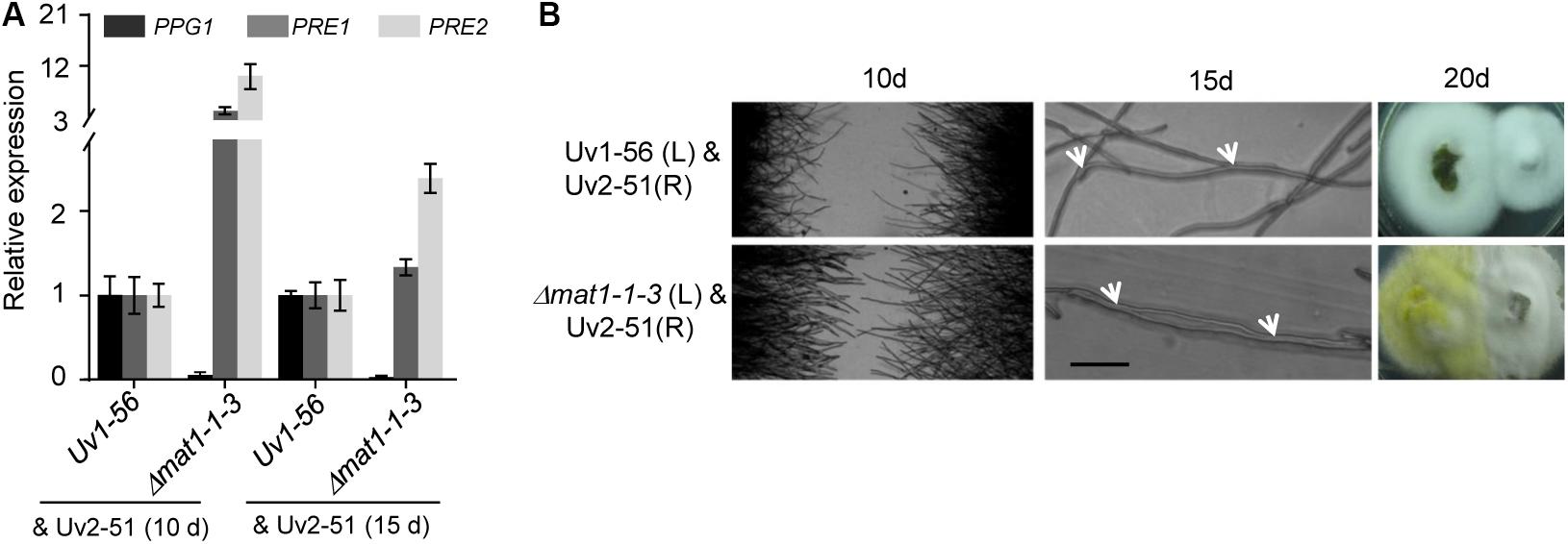
Figure 7. MAT1-1-3 affects the expression of pheromone and pheromone receptor genes. (A) qRT-PCR analysis of PPG1, PRE1 and PRE2 in wild-type Uv1-56 and Δmat1-1-3 mutant. The wild-type and mutant strains were confronted with the MAT1-2 type strain Uv2-51 after 10 days or 15 days on PSA medium. (B) Mating hyphae formation assay with wild-type and mutant strains confronted with Uv2-51. After 10 days, when Δmat1-1-3 crossed with Uv2-51, both strains formed mating hyphae that were longer than those of control Uv1-56 crossed with Uv2-51. After 15 days, hyphal fusions, indicated by arrows, appeared between Uv1-56 and Δmat1-1-3 mutant crossed with Uv2-51. The Δmat1-1-3 mutant fused earlier than wild-type with the Uv2-51 strain at 20 days. Scale bars, 10 μm.
Subcellular Localization and the Interacting Protein of MAT1-1-3
To better understand the biological function of MAT1-1-3, we examined the localization of MAT1-1-3 fused with green fluorescence protein (GFP) by a confocal microscope. The transformants expressing MAT1-1-3-GFP were generated in the background of WT Uv1-56. We observed that MAT1-1-3-GFP signal accumulated in the nucleus and cytoplasm of the conidia and hyphae (Figure 8A). The nuclear signal was confirmed by nuclear dye DAPI. The proteins encoded by mating-type genes can form a heterodimer, which is involved in morphological changes and spore development in fungi (Kämper et al., 1995; Lee et al., 2010; Kües et al., 2011). To investigate the relationship between the MAT1-1-3 and other mating-type proteins in V. virens, we performed a yeast two-hybrid (Y2H) assay to examine the interaction. Y2H assay showed that MAT1-1-3 interacted with MAT1-1-1, but could not interact with MAT1-1-2 (Figure 8B), indicating that MAT1-1-3 and MAT1-1-1 may form a complex to regulate sexual reproduction of V. virens.
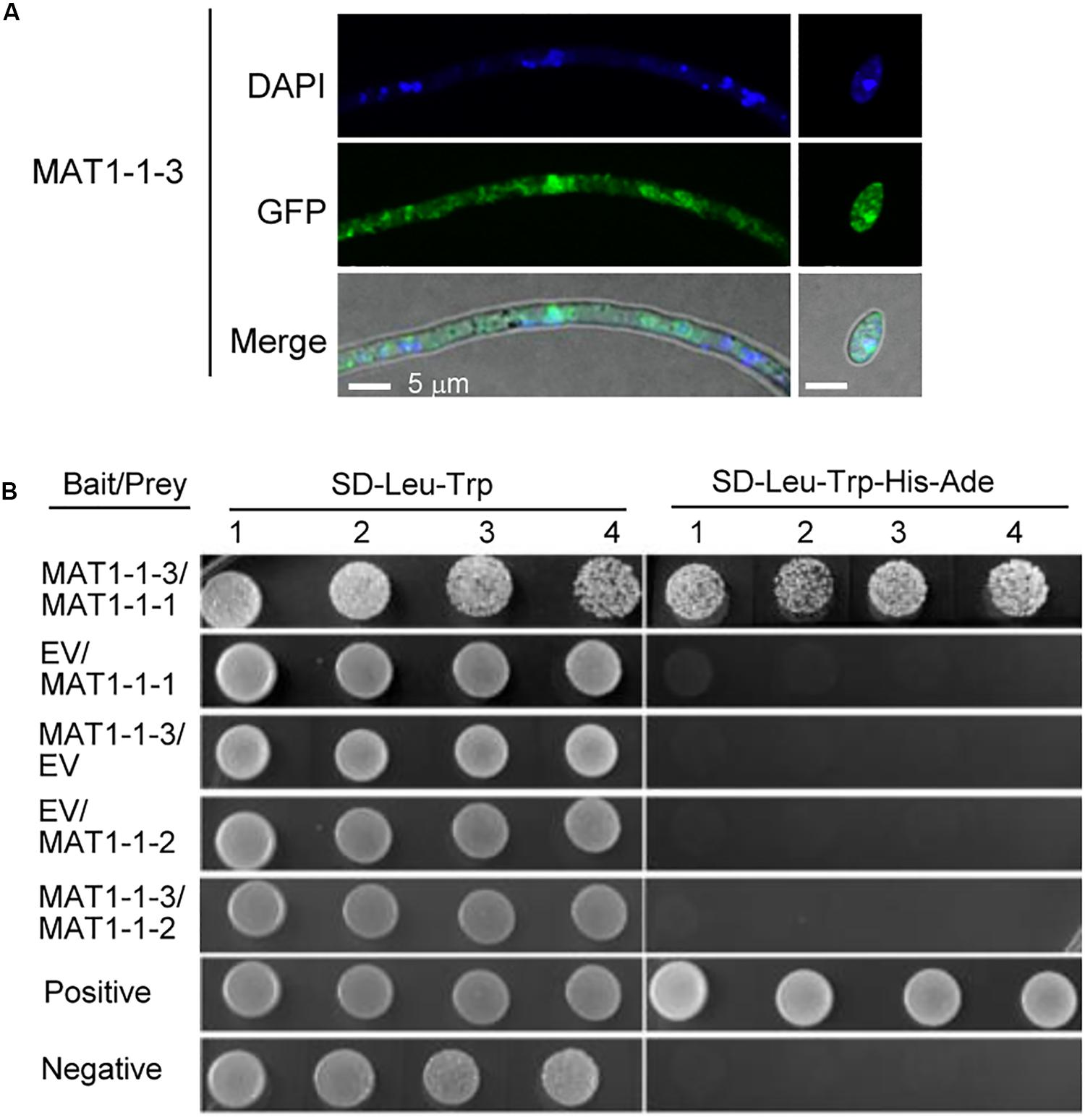
Figure 8. Subcellular localization and the interacting protein of MAT1-1-3 in Villosiclava virens. (A) The subcellular localization of MAT1-1-3-GFP in the hyphae and conidia of V. virens. The hyphae and conidia were stained with DAPI before the signal was observed with a confocal microscope. Scale bars, 5 μm. (B) The yeast two-hybrid (Y2H) assay was used to examine interactions between MAT1-1-1, MAT1-1-2 and MAT1-1-3. EV, empty vector; SD/-Leu-Trp-His-Ade, synthetic dropout (SD) media lacking leucine, tryptophan, histidine and Adenine; SD/-Leu-Trp, SD media lacking leucine and tryptophan. Numbers indicate repeats using different single colonies.
MAT1-1-3 Deletion Affects the Transcription of a Subset of Genes
To further study MAT1-1-3 regulation mechanisms in V. virens, we performed transcriptome sequencing (RNA-seq) analysis of the Δmat1-1-3 mutant. The transcripts of MAT1-1-3 could be detected when WT cultured in potato sucrose broth (PSB) medium (Supplementary Figure S2A). Thus, the samples for RNA-seq were extracted from mycelia of Δmat1-1-3 mutant and WT cultured in PSB for 7 days. We prepared three biological replicates samples for Δmat1-1-3 (Δmat1-1-3_1/2/3) mutant and WT (WT_1/2/3), respectively, though one sample from Δmat1-1-3 and another sample from WT were contaminated and discarded. The correlation coefficients (0.8954 and 0.8098) for the expression profiles of all transcripts between two wild-type (WT-1 and WT-3) or two mutant (Δmat1-1-3_1 and Δmat1-1-3_3) samples, respectively (Figure 9A), suggested that the RNA-seq data were credible. Analysis of the differentially expressed genes (DEG) revealed that the MAT1-1-3 knockout affected the transcription of a subset of genes (Supplementary Table S3). In comparison with WT, 1001 genes showed increased expression and 651 genes showed decreased expression in the Δmat1-1-3 mutant (Figures 9B,C and Supplementary Table S4). Enrichment analysis of Gene Ontology (GO) categories indicated that all significantly differentially expressed genes involved in regulation of molecular function, biosynthetic process, response to stress, regulation of cellular process, cellular metabolic and cell cycle process were significantly enriched (Figure 9D).
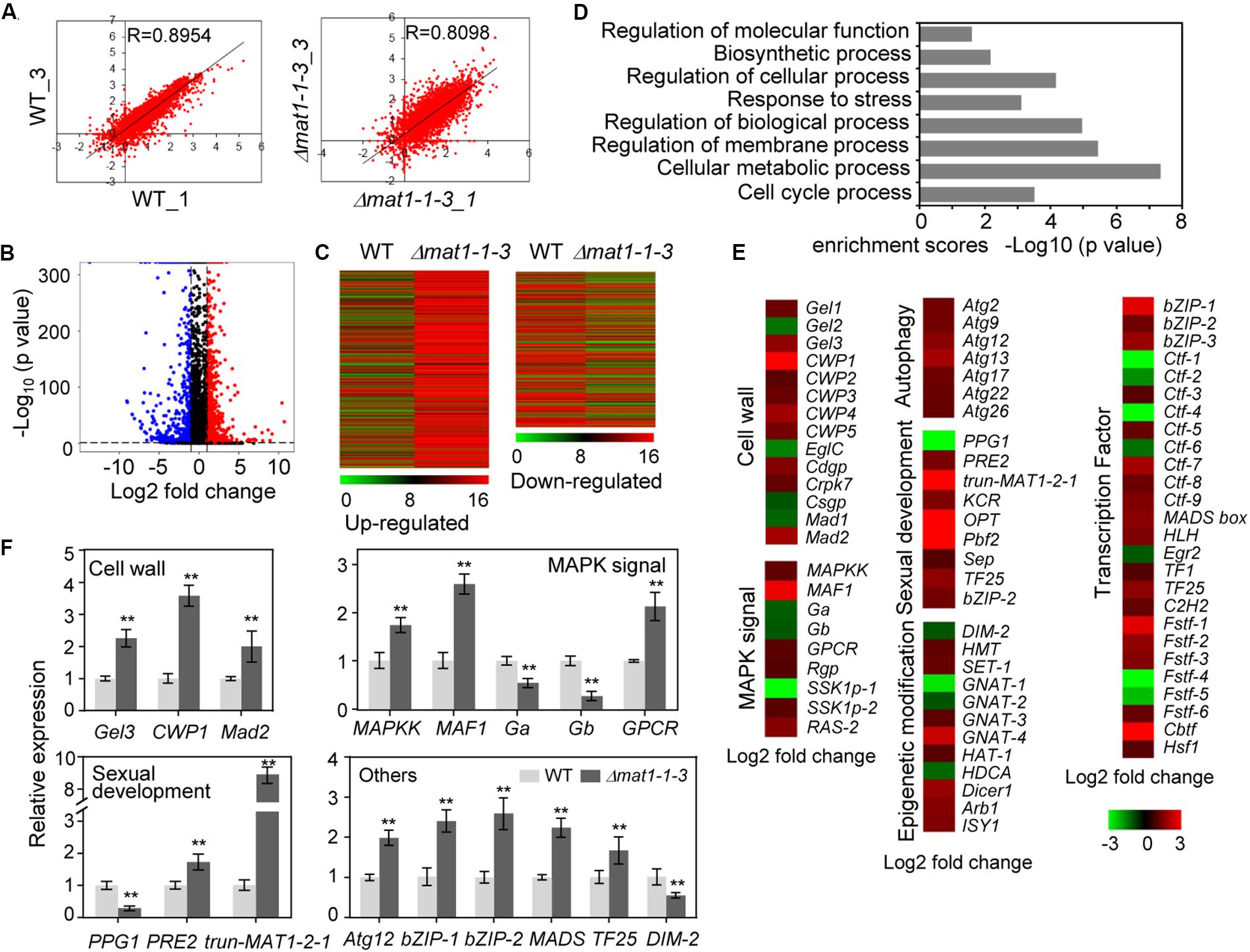
Figure 9. MAT1-1-3 regulates the expression of a subset of genes. (A) Scatter plots showing correlation of whole-genome expression measured as transcripts per million reads (TPM) between two WT samples (WT_1 versus WT_3, left) or two Δmat1-1-3 mutant samples (Δmat1-1-3_1 versus Δmat1-1-3_3, right). Gene expression levels were measured for mycelium grown in PSB for 7 days. The x-axis indicates gene expression in WT_1 or Δmat1-1-3_1, and the y-axis indicates gene expression in WT_3 or Δmat1-1-3_3 mutant. (B) Volcano map of differently expressed genes between WT and Δmat1-1-3 mutant. The x-axis indicates relative gene expression (log2 fold change between WT and Δmat1-1-3 mutant), and the y-axis indicates the mean of normalized counts [–log10 (p-value)]. Genes with significantly up-regulated expression (p < 0.05 and log2 fold change ≥ 1) are shown in red. Genes with significantly down-regulated expression (p < 0.05 and log2 fold change ≤ 1) are shown in blue. (C) Heatmaps of genes up-regulated and down-regulated in the Δmat1-1-3 mutant compared with WT. The original means of reading counts were adjusted by log2 transformation using MeV4.0. (D) Gene ontology (GO) enrichment analysis of significantly differentially expressed genes in Δmat1-1-3 compared with WT. The x-axis indicates enrichment scores [–log10 (p-value)] (the P-value indicates the possibility of significant enrichment) for each GO item on the y-axis. The fold enrichment was calculated based on the frequency of genes annotated to the term compared to their frequency in all transcripts detected. (E) Differentially expressed genes in the Δmat1-1-3 mutant might indicate involvement in pathogenesis and sexual development. Regulated genes include genes involved in cell wall integrity, MAPK signaling, sexual development, autophagy, and epigenetic modification. Some genes encoding transcription factors were also regulated by MAT1-1-3. The full names of abbreviated genes are shown in Supplementary Table S5. (F) qRT-PCR analysis of MAT1-1-3 regulated genes. Relative expression levels were normalized with β-tubulin as the internal standard and presented as means ± SD from three biological replicates. Asterisks indicate significant differences (Student’s t-test, *p < 0.05, **p < 0.01).
Extensive evidence has shown that genes involved in fungal cell wall integrity, MAPK signaling, sexual development, autophagy process, transcription factors and epigenetic modification are involved in fungal pathogenicity, growth and development. We further analyzed 1652 up- and down-regulated genes and found that a subset of genes involved in these processes was misregulated in the Δmat1-1-3 mutant (Figure 9E and Supplementary Table S5). The cell wall is essential for growth and participates in morphogenetic and differentiation processes in fungi (de Medina-Redondo et al., 2008). qRT-PCR results confirmed increased expression in Δmat1-1-3 mutant of genes encoding beta-1,3-glucanosyltransferase (Gel3), cell wall protein (CWP1) and adhesin protein Mad2 (Mad2) (Figure 9F). Several genes involved in the MAPK signaling pathway, which is essential for mating (Herskowitz, 1995; Banuett, 1998), including encoding the mitogen-activated protein kinase (MAF1), mitogen-activated protein kinase kinase (MAPKK) and G-protein coupled receptor (GPCR) increased expression in Δmat1-1-3 mutant; however, the expression was reduced for two genes encoding G protein alpha and beta subunit (Ga and Gb) (Figure 9F and Supplementary Table S5). Additionally, in the mutant, the expression levels of several genes involved in sexual development increased, like PRE2, or decreased, like PPG1 (Figure 9F), which is consistent with our previous results (Figure 7A). Studies have reported that genes encoding transcription factors (bZIP-1, bZIP-2, MADS-box and TF25) involved in fruiting body development (Zheng and Wang, 2013; Yu et al., 2016), with significantly increased expression in the Δmat1-1-3 mutant (Figures 9E,F). In addition, all seven autophagy-related genes were up-regulated in the mutant, and several epigenetic modification related genes, such as DNA methyltransferase (DIM-2), histone methyltransferase (HMT) and GNAT family acetyltransferase, were misregulated in the mutant (Figures 9E,F and Supplementary Table S5). Taken together, these results suggest that MAT1-1-3 regulates the expression of a subset of genes to control related functions in V. virens.
Our previous study showed that the truncated MAT1-2-1 down-stream of the MAT1-1-3 is part of the MAT1-2-1 located on MAT1-2 idiomorph (Supplementary Figure S2B) (Yu J. J. et al., 2015). The length of truncated MAT1-2-1 is 273 bp without ATG, which shares 87% identities with the 273 bp at 3′ end of MAT1-2-1 in the nucleotide sequence (Supplementary Figure S2C). The encoding protein with an HMG domain was predicted according to the 3′ end of MAT1-2-1 sequence, which shares 77% identities with the corresponding sequences in MAT1-2-1 (Supplementary Figure S2). RNA-seq data showed that the expression of truncated MAT1-2-1 gene increased about 9-fold in the Δmat1-1-3 mutant, which was confirmed by qRT-PCR (Figures 9E,F). However, fewer transcripts of truncated MAT1-2-1 are detected in WT compared with MAT1-1-3 (Supplementary Figure S2A). The results suggest that MAT1-1-3 negatively regulates the expression level of truncated MAT1-2-1.
Discussion
Sexual cycle plays a crucial role in V. virens overwintering and genetic diversity (Sun et al., 2013; Wang et al., 2014; Deng et al., 2015; Yong et al., 2018). The sclerotia produced during the sexual cycle can survive in winter and produce a large number of ascospores in the coming year, which are considered as the primary infection source (Yong et al., 2018). Genetic recombination occurs during sexual reproduction and expands V. virens genetic diversity (Sun et al., 2013; Wang et al., 2014). Mating type genes play crucial roles in sexual reproduction (Zheng and Wang, 2013). Here, to better understand the genetic basis of sexual reproduction in V. virens, we characterized a mating type gene MAT1-1-3 in V. virens. We found that MAT1-1-3 is a negative regulator of the immature fruiting body in V. virens, meanwhile, it is required for the asexual development and pathogenicity.
The Evolution of V. virens and MAT1-1-3 Proteins
The study of evolutionary relationships contributes to better understand the fungal origin and the relationships between different fungi. Previously, Zhang et al. (2014) systematically analyzed the evolutionary relationship of V. virens with other eleven fungi (ten Ascomycota and one Basidiomycota outgroup), including Metarhizium spp. via comparative genomic analyses and proteome comparisons. This study showed that V. virens is more closely to Metarhizium spp. than to other species including the plant pathogen Claviceps purpurea and the insect pathogenic fungus Cordyceps militaris. Our phylogenetic analysis data also showed that V. virens and Metarhizium spp. are in the same cluster, implying that the evolutionary relationship is close. The sexual reproduction mode of Metarhizium spp. is heterothallic (Zheng et al., 2011), which is consistent with V. virens. In addition, phylogenetic analysis data also showed that MAT1-1-3 proteins of V. virens and Metarhizium spp. are also in the same cluster. Among 31 fungi that we analyzed, some fungi (C. tenuipes, C. militaris and B. bassiana) have lost the MAT1-1-3 gene, however, most fungi still have it. Several researches showed that MAT1-1-3 may have been lost during the shift from plant to insect hosts, and could be a good indicator of Clavicipitaceae evolution (Yokoyama et al., 2006; Zheng et al., 2013). Our results showed that MAT1-1-3 of V. virens is required for sclerotia formation and sexual development.
The Relationship Between Truncated Mating Type Genes and Mating Type Genes
Increasing studies show that unequal recombination/crossover and transposable elements (TE)-mediated translocations play an important role in the evolution of mating loci (Gioti et al., 2012; Dyer et al., 2016). In Grosmannia clavigera, the MAT1-2 idiomorph contained a truncated MAT1-1-1 located up-stream of the MAT1-2-1 gene, which is homologous to the MAT1-1-1 gene in the MAT1-1 idiomorph (Tsui et al., 2013). Moreover, this truncated MAT1-1-1 is present in multiple G. clavigera isolates. And they found that the unequal recombination/crossover event likely accounted for the truncated MAT1-1-1. Because the unequal recombination/crossover events have occurred in many fungal MAT idiomorphs. For example, the truncated MAT1-1-1 gene was found in the MAT1-2 idiomorph of Hypocrea jecorina (Seidl et al., 2009) and also was identified in the MAT1-2 idiomorph of at least five Phialocephala species (Zaffarano et al., 2010). In contrast, a partial MAT1-2-1 sequence (360bp) located in the MAT1-1 idiomorph of Aspergillus fumigatus (Paoletti et al., 2005). Our study here also showed that a truncated MAT1-2-1 was found in MAT1-1 idiomorph of V. virens. The length of truncated MAT1-2-1 is 273 bp without start codon, but contains stop codon, which shares 87% identities with the 273 bp at 3′ end of MAT1-2-1 in the nucleotide sequence. The encoding protein with an HMG domain was predicted according to the 3′ end of MAT1-2-1 sequence. The encoding protein shares 77% identities with the corresponding amino acid sequences in MAT1-2-1. Thus, we also think that the truncated MAT1-2-1 organization is due to unequal recombination/crossover events during sexual reproduction.
Several studies showed that recombination/crossover event caused the truncated genes (MAT1-1-1 or MAT1-2-1) to be non-functional or inactive in mating, thereby being called pseudogenes. For example, the truncated MAT1-1-1 gene in Ophiostoma montium is highly eroded and possible is a pseudogene (Tsui et al., 2013). Similarly, the truncated MAT1-1-1 gene in Cordyceps takamontana (known as Isaria tenuipes) was a pseudogene due to accumulated mutations and stop codons (Yokoyama et al., 2003). Our previous study showed that the truncated MAT1-2-1 of V. virens was also named a pseudogene due to no start codon and very low detectable transcripts. However, the transcripts of truncated MAT1-2-1 increased about 9-fold after knockout MAT1-1-3, indicating that it may have function, thereby being named truncated MAT1-2-1 here. These data also suggest that MAT1-1-3 suppresses the expression of truncated MAT1-2-1 to regulate the mating of V. virens. Consistently, the transgenic strains, carrying both mating type genes, are unable to produce progeny in isolates of N. crassa (Glass et al., 1990), Podospora anserina (Coppin and Debuchy, 2000) and Cochliobolus heterostrophus (Turgeon et al., 1993a). These results, including our data, suggest that the truncated genes may interfere or compete with the signal from the “resident/original” HMG domain at the same locus. It is also possible that the truncated genes have evolved new functions through adaptive evolution (Tsui et al., 2013). Therefore, it will be important for future studies to investigate whether the truncated genes have functions and the mechanisms of how it regulates sexual reproduction.
Function of MAT1-1-3 in Asexual Development, Sexual Cycle and Pathogenicity
In heterothallic N. crassa, the matA-3 (MAT1-1-3) is dispensable for vegetative growth and sexual reproduction, such as asci production (Ferreira et al., 1998). However, the mutant AIIRIP, which contains mutations in both mat A-2 and mat A-3 (Glass and Lee, 1992; Ferreira et al., 1996), normally mates as a mat A strain, but produces very few asci with ascospores (Ferreira et al., 1998). The FMR1, SMR1 and SMR2 (MAT1-1-3) are required for the development of fertilized female organs in P. anserine, SMR2 (MAT1-1-3) was not essential for vegetative growth (Debuchy et al., 1993). However, the homothallic S. macrospora SmtA-3 (MAT1-1-3) is not essential for fertility and vegetative growth (Klix et al., 2010), and F. graminearum MAT1-1-3 is not essential for perithecium formation and vegetative growth, but is required for ascosporogenesis in self-crosses (Zheng et al., 2015). In our study, we demonstrated that knockout of MAT1-1-3 caused obvious growth and conidial morphology defects compared to wild type. Additionally, the Δmat1-1-3 mutant could not form sclerotia after infecting rice spikelets. Meanwhile, MAT1-1-3 is required for mating hyphae growth. A recent study showed that the MAT1-1-3 of F. graminearum is not required for virulence (Zheng et al., 2015). However, MAT1-1-3 was essential for the full pathogenicity of V. virens. Fungal sclerotia, which usually form in response to adverse environments like winter, germinate to form hyphae that serve as specific structures to start sexual reproduction under suitable conditions, thereby acting as the primary inoculum in the disease cycle (Yong et al., 2018; Liu et al., 2019). In outcrosses, the Δmat1-1-3 mutant was unable to produce sclerotia and could not initiate sexual reproduction. In addition, MAT1-1-3 interacted with MAT1-1-1, which is consistent with the MAT proteins in Saccharomyces cerevisiae and Phaffia rhodozyma to form complexes to regulate function (Haber, 2012; David-Palma et al., 2016). Thus, the MAT1-1-3 plays a key role in asexual development, sexual reproduction and pathogenicity of V. virens. Fungal fruiting bodies are important structures for fungi development. The fruiting bodies formed by Cordyceps. spp. have an important value as edible and medicinal mushrooms (Zheng et al., 2011, 2013). Several studies reported that the mechanisms of regulating fungal fruiting body formation are different. For example, the MAT1-2 single mating-type strain of Ophiocordyceps xuefengensis could form sterile fruiting bodies, with a MAT1-2 locus that is only MAT1-2-1 (Zou et al., 2017). The heterothallic C. militaris still can produce perithecia and ascospores although the loss of MAT1-1-3 in the genome in outcrosses (Zheng et al., 2011). The mature fruiting bodies were formed after being crossed with two opposite strains. However, this is the first ascomycete species reported that the C. militaris Cm01 stain with only a MAT1-1 single mating-type idiomorph also can produce stroma (fruiting body) without mature perithecia and ascospores on caterpillar pupae (Zheng et al., 2011). This is consistent with our result that the Δmat1-1-3 mutant can produce fruiting body primordium on PSA medium plate, but the fruiting body could not reach maturity and produce ascospores. Altogether, the result indicates that MAT1-1-3 is required for regulating fruiting bodies development in V. virens. Furthermore, a recent study showed that during outcrossing in C. militaris, MAT1-2-1 is required for fruiting body formation, and MAT1-1-2 mutant could produce fruiting body but with sterile perithecia (Lu et al., 2016). These results suggest that the fruiting body also can be regulated by other mating-type genes, which provides an important basis for us to study the function of other mating-type genes in different fungi.
MAT1-1-3 Regulates the Expression of a Subset Genes Involved in MAPK Pathway, Cell Wall Integrity, Epigenetic Modification and Autophagy
Mitogen-activated protein kinase (MAPK) cascades are a conserved signal pathway that has been shown to play a key role in transduction extracellular signals to cellular responses (Gustin et al., 1998; Goldsmith and Dhanasekaran, 2007). Meanwhile, the G proteins perceive the extracellular signal through cell surface receptors, such as G-protein-coupled receptor (GPCR), which are critically involved in the regulation of different MAPK networks (Goldsmith and Dhanasekaran, 2007). MAPK modules regulate many important signaling pathways including cell proliferation, differentiation and apoptosis (Chang and Karin, 2001). Extensive studies showed that MAPK encoding genes are required for sexual reproduction in fungi (Herskowitz, 1995; Banuett, 1998). For example, the MAPK genes are required for fruiting body formation in Aspergillus (AN1017) and Neurospora (NC02393) (Pöggeler et al., 2006). In yeast, MAPK Slt2p, also called Mpk1p, involved in the regulation of many cellular events including the sexual cycle (Gustin et al., 1998). Moreover, a recent study showed that the blue-light receptor gene white collar-1 (CmWC-1) from C. militaris mediated the fruiting body development most likely via regulating G protein-coupled receptors (Yang et al., 2016). Strikingly, our RNA-seq and qRT-PCR data showed that the expression levels of MAF1, encoding a mitogen-activated protein kinase, MAPKK and GPCR increased in Δmat1-1-3 mutant. Furthermore, Ga and Gb, encoding G protein alpha and beta subunit respectively, exhibited reduced expression in Δmat1-1-3 mutant. Thus, the results indicate that the MAPK signal pathway also participates in the V. virens sexual reproduction, although the mechanism of MAPK regulation remains unclear. In addition, MAT1-1-3 of V. virens is also required for regulating the expression of PRE2 and PPG1, encoding pheromone receptor and pheromone precursor, respectively, which are important for fungal sexual reproduction (Pöggeler et al., 2006). Thus, it will contribute to better understand the V. virens sexual cycle via studying the function of these genes regulated by MAT1-1-3.
The cell wall is essential for growth, signal transduction and participates in morphogenetic and differentiation processes in yeast and fungi (de Medina-Redondo et al., 2008). We found that knockout of MAT1-1-3 affected the V. virens cell wall integrity. Furthermore, MAT1-1-3 also regulated abiotic stress, such as salt, osmotic- and oxidatives tress. Notably, many genes involved in cell wall integrity and stress response were misregulated in Δmat1-1-3 mutant. The Gel1, encoding the 1, 3-b-glucanosyltransferase, is required for maintaining the cell wall integrity of F. graminearum (Bolouri et al., 2017). Similarly, Gel2 is required for cell wall morphogenesis and virulence in Aspergillus fumigatus (Mouyna et al., 2005). Apparently, these two genes are up-regulated and down-regulated in Δmat1-1-3 mutant, respectively. In addition, several genes involved in epigenetic modification can be regulated by MAT1-1-3, such as DIM-2, HMT, SET-1 (encoding SET domain-containing protein), HAT-1 (encoding histone acetyltransferase) and HDCA (encoding histone deacetylase A). SET-2 (a homolog of HMT in V. virens) is essential for normal growth and development in Neurospora (Adhvaryu et al., 2005). Zheng et al. (2011) reported that C. militaris fruiting without mating also induced expression of several epigenetic genes. Epigenetic modification plays important roles in silencing and activating different gene clusters, including virulence genes, secondary metabolism and development related genes (Cichewicz, 2010). Thus, the future research on how MAT1-1-3 regulates these genes expression will expand the sexual development networks. Our RNA-seq and qRT-PCR data also show that the expression of all 7 autophagy-related genes, including ATG2, ATG9, ATG12, ATG13, ATG17, ATG22 and ATG26, was increased in Δmat1-1-3 mutant. The autophagy is an evolutionarily conserved cellular transport pathway involved in many cellular processes (Mizushima et al., 2011). For example, Atg22 plays a vital role in the development and virulence of F. oxysporum (Khalid et al., 2019). However, the function of these autophagy-related genes in V. virens is still unknown. Taken together, the RNA-seq data indicated that MAT1-1-3 may regulate developmental processes and virulence by regulating the function of epigenetic modification and autophagy-related genes, which provides a clue for future research on how MAT1-1-3 regulates the function of these genes.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
MYo, JY, and YL designed the study. MYo, XP, MYu, HC, and ZQ performed the experiments. YD, RZ, TS, XY, and ZC helped to analyze the data. MYo, WL, and YL wrote the manuscript. YL contributed to reagents, materials, and analysis tools. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31571961) and the China Postdoctoral Science Foundation (2018M632257).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jin-Rong Xu (Purdue University, United States) for providing the CRISPR-Cas9 system plasmid. We also thank Dr. Liang Kong for helpful advice.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01337/full#supplementary-material
Footnotes
References
Adhvaryu, K. K., Morris, S. A., Strahl, B. D., and Selker, E. U. (2005). Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot. Cell 4, 1455–1464. doi: 10.1128/EC.4.8.1455-1464.2005
Alby, K., Schaefer, D., and Bennett, R. J. (2009). Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460, 890–893. doi: 10.1038/nature08252
Ashizawa, T., Takahashi, M., Arai, M., and Arie, T. (2012). Rice false smut pathogen, Ustilaginoidea virens, invades through small gap at the apex of a rice spikelet before heading. J. Gen. Plant Pathol. 78, 255–259. doi: 10.1007/s10327-012-0389-3
Banuett, F. (1998). Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 62, 249–274.
Bolouri, M. M. R., Vilcinskas, A., and Rahnamaeian, M. (2017). The insect-derived antimicrobial peptide metchnikowin targets Fusarium graminearum β (1,3)glucanosyltransferase Gel1, which is required for the maintenance of cell wall integrity. Biol. Chem. 398, 491–498. doi: 10.1515/hsz-2016-0295
Brooks, S. A., Anders, M. M., and Yeater, K. M. (2010). Effect of furrow irrigation on the severity of false smut in susceptible rice varieties. Plant Dis. 94, 570–574. doi: 10.1094/pdis-94-5-0570
Bushley, K. E., Li, Y., Wang, W. J., Wang, X. L., Jiao, L., Spatafora, J. W., et al. (2013). Isolation of the MAT1-1 mating type idiomorph and evidence for selfing in the Chinese medicinal fungus Ophiocordyceps sinensis. Fungal Biol. 117, 599–610. doi: 10.1016/j.funbio.2013.06.001
Chang, L., and Karin, M. (2001). Mammalian MAP kinase signalling cascades. Nature 410, 37–40. doi: 10.1038/35065000
Cichewicz, R. H. (2010). Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 27, 11–22. doi: 10.1039/b920860g
Coppin, E., and Debuchy, R. (2000). Co-expression of the mating-type genes involved in internuclear recognition is lethal in Podospora anserina. Genetics 155, 657–669.
Coppin, E., Debuchy, R., Arnaise, S., and Picard, M. (1997). Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61, 411–428.
David-Palma, M., Sampaio, J. P., and Gonçalves, P. (2016). Genetic dissection of sexual reproduction in a primary homothallic basidiomycete. PLoS Genet. 12:e1006110. doi: 10.1371/journal.pgen.1006110
Debuchy, R., Arnaise, S., and Lecellier, G. (1993). The mat- allele of Podospora anserina contains three regulatory genes required for the development of fertilized female organs. Mol. Gen. Genet. 241, 667–673. doi: 10.1007/BF00279909
Debuchy, R., and Turgeon, B. G. (2006). “Mating-type structure, evolution, and function in euascomycetes,” in Growth, Differentiation and Sexuality. The Mycota (A Comprehensive Treatise On Fungi As Experimental Systems For Basic And Applied Research), Vol. 1, eds U. Kües and R. Fischer (Berlin: Springer), 293–323.
de Medina-Redondo, M., Arnáiz-Pita, Y., Fontaine, T., Del Rey, F., Latgé, J. P., and Vázquez De Aldana, C. R. (2008). The β-1,3-glucanosyltransferase gas4p is essential for ascospore wall maturation and spore viability in Schizosaccharomyces pombe. Mol. Microbiol. 68, 1283–1299. doi: 10.1111/j.1365-2958.2008.06233.x
Deng, Q. D., Yong, M. L., Li, D. Y., Lai, C. H., Chen, H. M., Fan, J., et al. (2015). Survey and examination of the potential alternative hosts of Villosiclava virens, the pathogen of rice false smut, in China. J. Integr. Agr. 14, 1332–1337. doi: 10.1016/S2095-3119(15)61030-9
Dyer, P. S., Inderbitzin, P., and Debuchy, R. (2016). “14 Mating-type structure, function, regulation and evolution in the pezizomycotina,” in Growth, Differentiation and Sexuality. The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research), Vol. I ed. J. Wendland (Cham: Springer).
Fan, J., Yang, J., Wang, Y. Q., Li, G. B., Li, Y., Huang, F., et al. (2016). Current understanding on Villosiclava virens, a unique flower-infecting fungus causing rice false smut disease. Mol. Plant Pathol. 17, 1321–1330. doi: 10.1111/mpp.12362
Fang, A. F., Gao, H., Zhang, N., Zheng, X. H., Qiu, S. S., Li, Y. J., et al. (2019). A novel effector gene SCRE2 contributes to full virulence of Ustilaginoidea virens to rice. Front. Microbiol. 10:845. doi: 10.3389/fmicb.2019.00845
Ferreira, A. V., An, Z., Metzenberg, R. L., and Glass, N. L. (1998). Characterization of mat A-2, mat A-3 and deltamatA mating-type mutants of Neurospora crassa. Genetics 148, 1069–1079.
Ferreira, A. V. B., Saupe, S., and Glass, N. L. (1996). Transcriptional analysis of the mt A idiomorph of Neurospora crassa identifies two genes in addition to mt A-1. Mol. Gen. Genet. 250, 767–774. doi: 10.1007/bf02172989
Fu, X., Xie, R., Wang, J., Chen, X., Wang, X., Sun, W., et al. (2017). Development of colloidal gold-based lateral flow immunoassay for rapid qualitative and semiquantitative analysis of ustiloxins A and B in rice samples. Toxins 9:79. doi: 10.3390/toxins9030079
Galagan, J. E., Calvo, S. E., Cuomo, C., Ma, L. J., Wortman, J. R., Batzoglou, S., et al. (2005). Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438, 1105–1115. doi: 10.1038/nature04341
Gioti, A., Mushegian, A. A., Strandberg, R., Stajich, J. E., and Johannesson, H. (2012). Unidirectional evolutionary transitions in fungal mating systems and the role of transposable elements. Mol. Biol. Evol. 29, 3215–3226. doi: 10.1093/molbev/mss132
Glass, N. L., Grotelueschen, J., and Metzenberg, R. L. (1990). Neurospora crassa A mating-type region. Proc. Natl. Acad. Sci. U.S.A. 87, 4912–4916. doi: 10.1073/pnas.87.13.4912
Glass, N. L., and Lee, L. (1992). Isolation of Neurospora crassa A mating type mutants by repeat induced point (Rip) mutation. Genetics 132, 125–133.
Goldsmith, Z., and Dhanasekaran, D. N. (2007). G Protein regulation of MAPK networks. Oncogene 26, 3122–3142. doi: 10.1038/sj.onc.1210407
Guo, W., Gao, Y., Yu, Z., Xiao, Y., Zhang, Z., and Zhang, H. (2019). The adenylate cyclase UvAc1 and phosphodiesterase UvPdeH control the intracellular cAMP level, development, and pathogenicity of the rice false smut fungus Ustilaginoidea virens. Fungal Genet. Biol. 129, 65–73. doi: 10.1016/j.fgb.2019.04.017
Gustin, M. C., Albertyn, J., Alexander, M., and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264–1300.
Haber, J. E. (2012). Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191, 33–64. doi: 10.1534/genetics.111.134577
Herskowitz, I. (1995). MAP kinase pathways in yeast: for mating and more. Cell 80, 187–197. doi: 10.1016/0092-8674(95)90402-6
Hu, M. L., Luo, L. X., Wang, S., Liu, Y. F., and Li, J. Q. (2014). Infection processes of Ustilaginoidea virens during artificial inoculation of rice panicles. Eur. J. Plant Pathol. 139, 67–77. doi: 10.1007/s10658-013-0364-7
Hu, X., Zhang, Y. J., Xiao, G. H., Zheng, P., Xia, Y. L., Zhang, X. Y., et al. (2013). Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin. Sci. Bull. 58, 2846–2854. doi: 10.1007/s11434-013-5929-5
Ikegami, H. (1960). Studies on the false smut of rice IV. Infection of the false smut due to inoculation with chlamydospores and ascospores at the booting stage of rice plants. Res. Bull. Fac. Agric. Gifu Univ. 12, 45–51.
Jecmen, A. C., and Tebeest, D. O. (2015). First report of the occurrence of a white smut infecting rice in Arkansas. J. Phytopathol. 163, 138–143. doi: 10.1111/jph.12263
Jiao, Y. Q., Lee, Y. K., Gladman, N., Chopra, R., Christensen, S. A., Regulski, M., et al. (2018). MSD1 regulates pedicellate spikelet fertility in sorghum through the jasmonic acid pathway. Nat. Commun. 9:822. doi: 10.1038/s41467-018-03238-4
Jones, S. K. Jr., and Bennett, R. J. (2011). Fungal mating pheromones: choreographing the dating game. Fungal Genet. Biol. 48, 668–676. doi: 10.1016/j.fgb.2011.04.001
Kämper, J., Reichmann, M., Romeis, T., Bolker, M., and Kahmann, R. (1995). Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81, 73–83. doi: 10.1016/0092-8674(95)90372-0
Khalid, A. R., Zhang, S., Luo, X., Mehmood, K., Rahim, J., Shaheen, H., et al. (2019). Role of autophagy-related gene atg22 in developmental process and virulence of Fusarium oxysporum. Genes 10:365. doi: 10.3390/genes10050365
Kim, H. K., Cho, E. J., Lee, S., Lee, Y. S., and Yun, S. H. (2012). Functional analyses of individual mating-type transcripts at MAT loci in Fusarium graminearum and Fusarium asiaticum. FEMS Microbiol. Lett. 337, 89–96. doi: 10.1111/1574-6968.12012
Klix, V., Nowrousian, M., Ringelberg, C., Loros, J. J., Dunlap, J. C., and Pöggeler, S. (2010). Functional characterization of MAT1-1-specifc mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot Cell 9, 894–905. doi: 10.1128/EC.00019-10
Kües, U., James, T., and Heitman, J. (2011). “6 Mating type in basidiomycetes: unipolar, bipolar, and tetrapolar patterns of sexuality,” in Evolution of Fungi and Fungal-Like Organisms, eds S. Pöggeler and J. Wöstemeyer (Berlin: Springer), 97–160. doi: 10.1007/978-3-642-19974-5_6
Ladhalakshmi, D., Laha, G. S., Singh, R., Karthikeyan, A., Mangrauthia, S. K., Sundaram, R. M., et al. (2012). Isolation and characterization of Ustilaginoidea virens and survey of false smut disease of rice in India. Phytoparasitica 40, 171–176. doi: 10.1007/s12600-011-0214-0
Lee, S. C., Ni, M., Li, W. J., Cecelia, S., and Heitman, J. (2010). The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 74, 298–340. doi: 10.1128/MMBR.00005-10
Li, C. H., Melesse, M., Zhang, S. J., Hao, C. F., Wang, C. F., Zhang, H. C., et al. (2015). FgCDC14 regulates cytokinesis, morphogenesis, and pathogenesis in Fusarium graminearum. Mol. Microbiol. 98, 770–786. doi: 10.1111/mmi.13157
Li, P. X., Evans, C. D., Wu, Y. Z., Cao, B., Hamel, E., and Joullié, M. M. (2008). Evolution of the total syntheses of ustiloxin natural products and their analogues. J. Am. Chem. Soc. 130, 2351–2364. doi: 10.1021/ja710363p
Li, Y. J., Wang, M., Liu, Z. H., Zhang, K., Cui, F. H., and Sun, W. X. (2019). Towards understanding the biosynthetic pathway for ustilaginoidin mycotoxins in Ustilaginoidea virens. Environ. Microbiol. 21, 2629–2643. doi: 10.1111/1462-2920.14572
Liang, Y. F., Han, Y., Wang, C. F., Jiang, C., and Xu, J. R. (2018). Targeted deletion of the USTA and UvSLT2 genes efficiently in Ustilaginoidea virens with the CRISPR-Cas9 system. Front. Plant Sci. 9:699. doi: 10.3389/fpls.2018.00699
Liu, Y., Liu, J. K., Li, G. H., Zhang, M. Z., Zhang, Y. Y., Wang, Y. Y., et al. (2019). A novel Botrytis cinerea-specific gene BcHBF1 enhances virulence of the grey mould fungus via promoting host penetration and invasive hyphal development. Mol. Plant Pathol. 20, 731–747. doi: 10.1111/mpp.12788
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Y. Z., Xia, Y. L., Luo, F. F., Dong, C. H., and Wang, C. S. (2016). Functional convergence and divergence of mating-type genes fulfilling in Cordyceps militaris. Fungal Genet. Biol. 88, 35–43. doi: 10.1016/j.fgb.2016.01.013
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Mouyna, I., Morelle, W., Vai, M., Monod, M., and Latgé, J. P. (2005). Deletion of gel2 encoding for a β (1-3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus. Mol. Microbiol. 56, 1675–1688. doi: 10.1111/j.1365-2958.2005.04654.x
Nelson, M. A. (1996). Mating systems in ascomycetes: a romp in the sac. Trends Genet. 12, 69–74. doi: 10.1016/0168-9525(96)81403-x
Nygren, K., Strandberg, R., Wallberg, A., Nabholz, B., Gustafsson, T., García, D., et al. (2011). A comprehensive phylogeny of Neurospora reveals a link between reproductive mode and molecular evolution in fungi. Mol. Phylogenet. Evol. 59, 649–663. doi: 10.1016/j.ympev.2011.03.023
Paoletti, M., Rydholm, C., Schwier, E., Anderson, M. J., Szakacs, G., Lutzoni, F., et al. (2005). Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15, 1242–1248. doi: 10.1016/j.cub.2005.05.045
Pöggeler, S., Nowrousian, M., Ringelberg, C., Loros, J. J., Dunlap, J. C., and Kück, U. (2006). Microarray and real-time PCR analyses reveal mating type-dependent gene expression in a homothallic fungus. Mol. Genet. Genom. 275, 492–503. doi: 10.1007/s00438-006-0107-y
Roncero, C., and Durán, A. (1985). Effect of calcofluor white and congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163, 1180–1185.
Seidl, V., Seibel, C., Kubicek, C. P., and Schmoll, M. (2009). Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci. U.S.A. 106, 13909–13914. doi: 10.1073/pnas.0904936106
Snetselaar, K. M., Bölker, M., and Kahmann, R. (1996). Ustilago maydis mating hyphae orient their growth toward pheromone sources. Fungal Genet. Biol. 20, 299–312. doi: 10.1006/fgbi.1996.0044
Song, J. H., Wei, W., Lv, B., Lin, Y., Yin, W. X., Peng, Y. L., et al. (2016). Rice false smut fungus hijacks the rice nutrients supply by blocking and mimicking the fertilization of rice ovary. Environ. Microbiol. 18, 3840–3849. doi: 10.1111/1462-2920.13343
Sun, X. Y., Kang, S., Zhang, Y. J., Tan, X. Q., Yu, Y. F., He, H. Y., et al. (2013). Genetic diversity and population structure of rice pathogen Ustilaginoidea virens in China. PLoS One 8:e76879. doi: 10.1371/journal.pone.0076879
Tang, Y. X., Jin, J., Hu, D. W., Yong, M. L., Xu, Y., and He, L. P. (2013). Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets. Plant Pathol. 62, 1–8. doi: 10.1111/j.1365-3059.2012.02629.x
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Tsui, C. K., DiGuistini, S., Wang, Y., Feau, N., Dhillon, B., Bohlmann, J., et al. (2013). Unequal recombination and evolution of the mating-type (MAT) loci in the pathogenic fungus Grosmannia clavigera and relatives. G3 3, 465–480. doi: 10.1534/g3.112.004986
Turgeon, B. G., Bohlmann, H., Ciuffetti, L. M., Christiansen, S. K., Yang, G., Schäfer, W., et al. (1993a). Cloning and analysis of the mating type genes from Cochliobolus heterostrophus. Mol. Gen. Genet. 238, 270–284. doi: 10.1007/bf00279556
Turgeon, B. G., Christiansen, S. K., and Yoder, O. C. (1993b). “Mating type genes in ascomycetes and their imperfect relatives,” in The Fungal Holomorph: Mitotic, Meiotic And Pleomorphic Speciation In Fungal Systematics, eds D. R. Reynolds and J. W. Taylor (Wallingford: CABI Publishing), 199–215.
Wang, F., Zhang, S., Liu, M. G., Lin, X. S., Liu, H. J., Peng, Y. L., et al. (2014). Genetic diversity analysis reveals that geographical environment plays a more important role than rice cultivar in Villosiclava virens population selection. Appl. Environ. Microbiol. 80, 2811–2820. doi: 10.1128/AEM.03936-13
Wang, G. (1995). The sexual stage of Ustilaginoidea virens and the infection process of ascospores on rice. J. Zhejiang Wanli Univ. Z 1, 3–9.
Wang, X., Wang, J., Lai, D., Wang, W., Dai, J., Zhou, L., et al. (2017). Ustiloxin G, a new cyclopeptide mycotoxin from rice false smut balls. Toxins 9:54. doi: 10.3390/toxins9020054
Weigel, D., and Glazebrook, J. (2006). Transformation of Agrobacterium using the freeze-thaw method. CSH Protoc. 7, 1031–1036. doi: 10.1101/pdb.prot4666
Whittle, C. A., Nygren, K., and Johannesson, H. (2011). Consequences of reproductive mode on genome evolution in fungi. Fungal Genet. Boil. 48, 661–667. doi: 10.1016/j.fgb.2011.02.005
Wilken, P. M., Steenkamp, E. T., Hall, T. A., Beer, Z. W. D., Wingfield, M. J., and Wingfield, B. D. (2012). Both mating types in the heterothallic fungus Ophiostoma quercus contain MAT1-1 and MAT1-2 genes. Fungal Biol. 116, 427–437. doi: 10.1016/j.funbio.2012.01.002
Xie, S. L., Wang, Y. F., Wei, W., Li, C. Y., Liu, Y., Qu, J. S., et al. (2019). The Bax inhibitor UvBI-1, a negative regulator of mycelial growth and conidiation, mediates stress response and is critical for pathogenicity of the rice false smut fungus Ustilaginoidea virens. Curr. Genet. 65, 1185–1197. doi: 10.1007/s00294-019-00970-2
Yang, T., Guo, M., Yang, H., Guo, S., and Dong, C. (2016). The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 100, 743–755. doi: 10.1007/s00253-015-7047-6
Yokoyama, E., Arakawa, M., Yamagishi, K., and Hara, A. (2006). Phylogenetic and structural analyses of the mating-type loci in Clavicipitaceae. FEMS Microbiol. Lett. 264, 182–191. doi: 10.1111/j.1574-6968.2006.00447.x
Yokoyama, E., Yamagishi, K., and Hara, A. (2003). Structures of the mating-type loci of Cordyceps takaomontana. Appl. Environ. Microbiol. 69, 5019–5022. doi: 10.1128/AEM.69.8.5019-5022.2003
Yokoyama, E., Yamagishi, K., and Hara, A. (2005). Heterothallism in Cordyceps takaomontana. FEMS Microbiol. Lett. 250, 145–150. doi: 10.1016/j.femsle.2005.07.004
Yong, M. L., Deng, Q. D., Fan, L. L., Miao, J. K., Lai, C. H., Chen, H. M., et al. (2018). The role of Ustilaginoidea virens sclerotia in increasing incidence of rice false smut disease in the subtropical zone in China. Eur. J. Plant Pathol. 150, 669–677. doi: 10.1007/s10658-017-1312-8
Yu, J., Yu, M., Song, T., Cao, H., Pan, X., Yong, M., et al. (2019). A homeobox transcription factor UvHOX2 regulates chlamydospore formation, conidiogenesis, and pathogenicity in Ustilaginoidea virens. Front. Microbiol. 10:1071. doi: 10.3389/fmicb.2019.01071
Yu, J. J., Sun, W. X., Yu, M. N., Yin, X. L., Meng, X. K., Zhao, J., et al. (2015). Characterization of mating-type loci in rice false smut fungus Villosiclava virens. FEMS Microbiol. Lett. 362:fnv014. doi: 10.1093/femsle/fnv014
Yu, J. J., Yu, M. N., Nie, Y. F., Sun, W. X., Yin, X. L., Zhao, J., et al. (2016). Comparative transcriptome analysis of fruiting body and sporulating mycelia of Villosiclava virens reveals genes with putative functions in sexual reproduction. Curr. Genet. 62, 575–584. doi: 10.1007/s00294-015-0563-1
Yu, M. N., Yu, J. J., Hu, J. K., Huang, L., Wang, Y. H., Yin, X. L., et al. (2015). Identification of pathogenicity-related genes in the rice pathogen Ustilaginoidea virens through random insertional mutagenesis. Fungal Genet. Biol. 76, 10–19. doi: 10.1016/j.fgb.2015.01.004
Yun, S. H., Berbee, M. L., Yoder, O. C., and Turgeon, B. G. (1999). Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. U.S.A. 96, 5592–5597. doi: 10.1073/pnas.96.10.5592
Zaffarano, P. L., Duò, A., and Grünig, C. R. (2010). Characterization of the mating type (MAT) locus in the Phialocephala fortinii s. l. – Acephala applanata species complex. Fungal Genet. Biol. 47, 761–772. doi: 10.1016/j.fgb.2010.06.001
Zhang, Y., Zhang, K., Fang, A. F., Han, Y. Q., Yang, J., Xue, M. F., et al. (2014). Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat. Commun. 5:3849. doi: 10.1038/ncomms4849
Zheng, P., and Wang, C. S. (2013). Sexuality control and sex evolution in fungi. Sci. Sin. Vitae. 43, 1090–1097. doi: 10.1360/052013-317
Zheng, P., Xia, Y., Xiao, G., Xiong, C., Hu, X., Zhang, S., et al. (2011). Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional chinese medicine. Genome Biol. 12:R116. doi: 10.1186/gb-2011-12-11-r116
Zheng, P., Xia, Y. L., Zhang, S. W., and Wang, C. S. (2013). Genetics of Cordyceps and related fungi. Appl. Microbiol. Biotechnol. 97, 2797–2804. doi: 10.1007/s00253-013-4771-7
Zheng, Q., Hou, R., Zhang, J. Y., Ma, J. W., Wu, Z. S., Wang, G. H., et al. (2015). The MAT locus genes play different roles in sexual reproduction and pathogenesis in Fusarium graminearum. PLoS One 10:e0131623. doi: 10.1371/journal.pone.0131623
Zickler, D., Arnaise, S., Coppin, E., Debuchy, R., and Picard, M. (1995). Altered mating-type identity in the fungus Podospora anserina leads to selfish nuclei, uniparental progeny, and haploid meiosis. Genetics 140, 493–503.
Keywords: mating type gene, Villosiclava virens, MAT1-1-3, sexual development, pathogenicity
Citation: Yong M, Yu J, Pan X, Yu M, Cao H, Qi Z, Du Y, Zhang R, Song T, Yin X, Chen Z, Liu W and Liu Y (2020) MAT1-1-3, a Mating Type Gene in the Villosiclava virens, Is Required for Fruiting Bodies and Sclerotia Formation, Asexual Development and Pathogenicity. Front. Microbiol. 11:1337. doi: 10.3389/fmicb.2020.01337
Received: 27 February 2020; Accepted: 25 May 2020;
Published: 25 June 2020.
Edited by:
Laure Ries, University of São Paulo, BrazilReviewed by:
Kin-Ming (Clement) Tsui, Weill Cornell Medicine - Qatar, QatarJing Fan, Sichuan Agricultural University, China
Zide Jiang, South China Agricultural University, China
Copyright © 2020 Yong, Yu, Pan, Yu, Cao, Qi, Du, Zhang, Song, Yin, Chen, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongfeng Liu, bGl1eWZAamFhcy5hYy5jbg==
 Mingli Yong
Mingli Yong Junjie Yu
Junjie Yu Xiayan Pan
Xiayan Pan Mina Yu
Mina Yu Huijuan Cao1
Huijuan Cao1 Rongsheng Zhang
Rongsheng Zhang Xiaole Yin
Xiaole Yin Wende Liu
Wende Liu