
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 June 2020
Sec. Microbiotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01258
Increasing evidence has shown that DNA methylation is involved in gene regulation in prokaryotes. However, there have been very limited reports about the role of DNA methylation in regulation of gene expression and physiological functions in cyanobacteria. In Synechocystis sp. PCC 6803, four genes on the plasmid pSYSX are predicted to encode the type I restriction-methylation system, slr6095 and slr6096 for the M subunit, slr6097 for the S subunit and slr6102 for the R subunit. Compared to the wild type, slr6095, slr6096, and slr6097 mutants lacked the GGm6AN7TTGG/CCAm6AN7TCC methylation in genomic DNA. Transcriptomic analysis indicated that 171 genes were reproducibly up- or down-regulated in all three mutants relative to the wild type. The changed expression of some genes, including sll1356 for glycogen phosphorylase (GlgP), was associated with the loss of GGm6AN7TTGG/CCAm6AN7TCC methylation in the coding regions or the upstream non-coding sequences. Inactivation of slr6095, slr6096, or slr6097 increased the expression of sll1356 and the GlgP activity but lowered the glycogen content. These results indicated that the DNA methylation by a type I RM system could alter the expression of certain genes and physiological functions in Synechocystis sp. PCC 6803.
DNA methylation as an epigenetic signal is found in both eukaryotes and prokaryotes. In bacteria, a methyl group from S-adenosyl-L-methionine (AdoMet) is transferred to cytosine or adenine, resulting in N4-methyl-cytosine, C5-methyl-cytosine, or N6-methyl-adenine on DNA (Sánchez-Romero and Casadesús, 2020). Because methyl groups located in the major groove of double helix alter protein-DNA interactions and DNA curvature, DNA methylation may protect DNA from cleavage by restriction enzymes and regulate gene expression, cell cycle, chromosome replication, phase variation, virulence, and mismatch repair (Sánchez-Romero and Casadesús, 2020). DNA methylation is catalyzed by DNA methyltransferases (MTases), which include those of restriction-modification (RM) systems and orphan methylases (such as Dam and Dcm in Escherichia coli; Blow et al., 2016).
Cyanobacteria are a group of prokaryotes that perform oxygenic photosynthesis (Castenholz, 2001). Restriction-methylation systems have been predicted in genomes of most cyanobacteria. In addition, some orphan methylases were identified. In Anabaena sp. PCC 7120, a heterocyst-forming filamentous cyanobacterium, there are four predicted orphan MTases, denoted as Dmt A to D, in addition to the MTases of RM systems (Matveyev et al., 2001). These orphan MTases modifiy DNA at GATC (DmtA), GGCC (DmtB), CGATCG (DmtC), or RCCGGY (DmtD). In Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803), a unicellular cyanobacterium, there are at least five different methylation activities toward specific DNA sequences: M.Ssp6803I for m5CGATCG (Scharnagl et al., 1998), M.Ssp6803II for GGm4CC, M.Ssp6803III for Gm6ATC, M.Ssp6803IV for GAm6AGGC, and M.Ssp6803V for the bipartite motif GGm6AN7TTGG/CCAm6AN7TCC (Hagemann et al., 2018). DNA methylation may play roles in cellular and physiological functions. For an example, the GGm4CC methylation is involved in regulation of gene expression, fine-tuning of DNA replication and DNA repair in Synechocystis sp. PCC 6803 (Gärtner et al., 2019).
In cyanobacteria, glycogen is the most important storage polysaccharide. It is an α-1,4 linked α-1,6 branched glucose polymer. The synthesis of glycogen depends on ADP glucose pyrophosphorylase, glycogen synthase, and a branching enzyme (Preiss, 1984), whereas the degradation of glycogen depends on glycogen phosphorylase (GlgP), the debranching enzyme, α-1,4-glucanotransferase, and maltodextrin phosphorylase (Ball and Morell, 2003). GlgP is the key enzyme involved in glycogen digestion. In Synechocystis 6803, there are two GlgP genes, glgP1 (sll1356), and glgP2 (slr1367), which are functionally diverged (Fu and Xu, 2006) and differently regulated (Osanai et al., 2014). Under autotrophic growth conditions, glgP1 plays a more important role in glycogen digestion (Fu and Xu, 2006).
When we analyzed the previously reported DNA methylase M.Ssp6803V (slr6095), we realized that it is probably not an orphan methylase as proposed, because slr6095 (N-terminal portion of M subunit, for methylation), slr6096 (C-terminal portion of M subunit), slr6097 (S subunit, for recognition specificity), and slr6102 (R subunit, for restriction) are predicted to encode a type I RM system. In this type of RM system, M and S subunits are required for the restriction activity as well (Murray, 2000), therefore their encoding genes can be inactivated in the presence of a functional R subunit gene. We generated mutants for slr6095, slr6096, and slr6097, and analyzed differences in genome-wide gene expression and DNA methylation between mutants and the wild type strain. Inactivation of these genes consistently abolished DNA methylation at GGm6AN7TTGG/CCAm6AN7TCC motifs in the genome and indirectly altered the expression of many genes and the glycogen catabolism in Synechocystis 6803.
Synechocystis sp. PCC 6803 and derivatives were cultured in BG11 in flasks on a rotary shaker at 30°C under continuous illumination of 30 μE m–2 s–1. Kanamycin (25 μg mL–1) was added as needed.
Genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980) with modifications. The cyanobacterial cells were broken by grinding in liquid nitrogen and suspended in CTAB solution. The broken cell suspension was treated with proteinase K (100 μg mL–1) and β-mercaptoethanol (2%, v/v) at 65°C for 1 h and centrifuged at 17,226 g for 30 min. The supernatant was extracted with phenol:chloroform:isoamyl alcohol (25:24:1), followed by chloroform:isoamyl alcohol (24:1). Nucleic acids were precipitated with isopropanol, washed with 70% ethanol, dissolved in TE buffer. After removal of RNA with RNase A (100 μg mL–1), DNA was extracted with phenol and chloroform, precipitated with ethanol and dissolved in TE buffer. The quality of DNA was assessed by agarose-gel electrophoresis and OD260/OD280 (1.8∼2.0).
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. After removal of DNA with DNase RQ1 (Promega, Madison, WI, United States), the sample was extracted with Trizol reagent again. RNA was precipitated with isopropanol, washed with 70% ethanol and dissolved in nuclease-free ddH2O. RNA integrity was verified on Bioanalyzer (Agilent, Santa Clara, CA, United States) using RNA 6000 nano chip, and the RNA integrity numbers (RIN) were shown to be >6.5.
Molecular manipulations were performed according to manufacturers’ instructions. Clones of PCR products were confirmed by sequencing. Construction of plasmids and primers used are described in Supplementary Table S1 in the Supplementary Material.
Using overlap PCR, an XhoI site was introduced into slr6095, slr6096, and slr6097, and the DNA fragment containing one of these genes, interrupted by the XhoI site, was cloned into pMD18-T, then a kanamycin-resistance (Kmr) cassette was inserted into the XhoI site, producing plasmids pHB6515, pHB6520, and pHB6517. These plasmids were used to inactivate slr6095, slr6096, and slr6097 in Synechocystis 6803.
Transformation of Synechocystis 6803 was performed as described by Williams (1988). Briefly, 100 μl of cell suspension was mixed with DNA (10 μg mL–1) and incubated for 4 to 6 h at 30°C in the light of 30 μE m–2 s–1, spread onto membrane filters resting on BG11 agar plates and allowed to grow for 20 h. The filters were then transferred to plates supplemented with glucose (5 mM) and kanamycin (25 μg mL–1). Transformants usually appeared within 4 days. After streaking on plates, the transformants were cultured in liquid BG11 with kanamycin. The complete segregation of mutants was confirmed by PCR. Synechocystis strains and primers used are described in Supplementary Table S1 in the Supplementary Material.
DNA methylation was analyzed by whole-genome SMRT sequencing. Briefly, libraries with 10 kb inserts were prepared according to the PacBio Sequel Microbiological sequencing protocol, and the genome DNA was sequenced with a PacBio Sequel platform. DNA methylation was then analyzed with the SMRT raw data using the SMRT LINK 6.0.0 software1. Base modifications and motifs were analyzed using P_ModificationDetection and P_MotifFinder module from the SMRT Analysis software. Because SMRT sequencing is not suitable for m5C methylation (Murray et al., 2012), we collected the data for m6A and m4C only (consistent between two biological repeats). Raw sequence read data has been deposited in the NCBI Sequence Read Archive (SRA) with the study identifier SRP256040.
Synechocystis 6803 and mutants were cultured in BG11 to an OD730 of 1.0 at 30°C in the light of 30 μE m–2 s–1. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, United States). DNA was removed with RNase-free DNase I (Promega, Madison, WI, United States). RNA was precipitated with isopropanol and stored in 70% ethanol. Strand-specific RNA-seq libraries were prepared using the Illumina small RNA sample preparation kit. RNA sequencing was carried out using the HiSeq 2500 sequencing instrument (Illumina, San Diego, CA, United States) to generate paired-end reads with a length of 150 bp. Reads were aligned against the Cyanobase2 genome sequence for Synechocystis 6803 using TopHat (Trapnell et al., 2009). Differential gene expression analysis was performed by using the DESeq package (Anders and Huber, 2010). Fold changes of >2 or <0.5 with P-value of <0.01 were defined as differentially expressed. Data are means ± SD produced from 3 biological replicates. Raw sequence read data has been deposited in the NCBI SRA with the study identifier SRP258660.
Reverse transcription and the RT-qPCR analysis were performed according to Gao and Xu (2012). rnpB (RNase P subunit B; Vioque, 1992) was used as the internal control. PCR primers (indicated with “RT” in name) are listed in Supplementary Table S1. Data are means ± SD produced from 3 technical repeats.
Synechocystis 6803 and mutants were cultured in BG11 to an OD730 of 1.0. Glycogen contents and GlgP activities were determined according to previously described (Fu and Xu, 2006). Data are means ± SD produced from 3 biological replicates.
The type I RM system was predicted according to the REBASE3 and the Cyanobase4. Structural domains were predicted using SMART analysis5. Similarity searches were performed using Blastp6. Multiple sequence alignment was performed using the ClustalW7.
slr6095 is located in a gene cluster on the plasmid pSYSX (Figure 1). According to the REBASE and the Cyanobase, slr6095, and slr6096 are predicted to encode two portions of the M subunit of a type I RM system, slr6097 encodes the S subunit, slr6102 the R subunit. Alignment with N6-Mtases from Rhizobiales bacterium and Thiorhodovibrio sp. 970 shows that Slr6095 and Slr6096 are similar to the N-terminal and C-terminal portions of the M subunit (for methylation), respectively (Supplementary Figure S1). Slr6097 is predicted to contain a methylase-S domain and shows high similarity to the S subunit HsdS (for sequence recognition, GenBank accession no. AFWS02000001.1) of a predicted type I RM system in Thiorhodovibrio sp. 970 (69% similarity; e-value, 4e-108). In the same gene cluster, Slr6012 contains an HSDR_N domain and is similar to the R subunit HsdR (for restriction) in Thiorhodovibrio sp. 970 (59% similarity; e-value, 0.0). We tentatively call these proteins M. Ssp6803V-n, M. Ssp6803V-c (n and c, N-terminal, or C-terminal portion), S. Ssp6803V, and R.Ssp6803V.
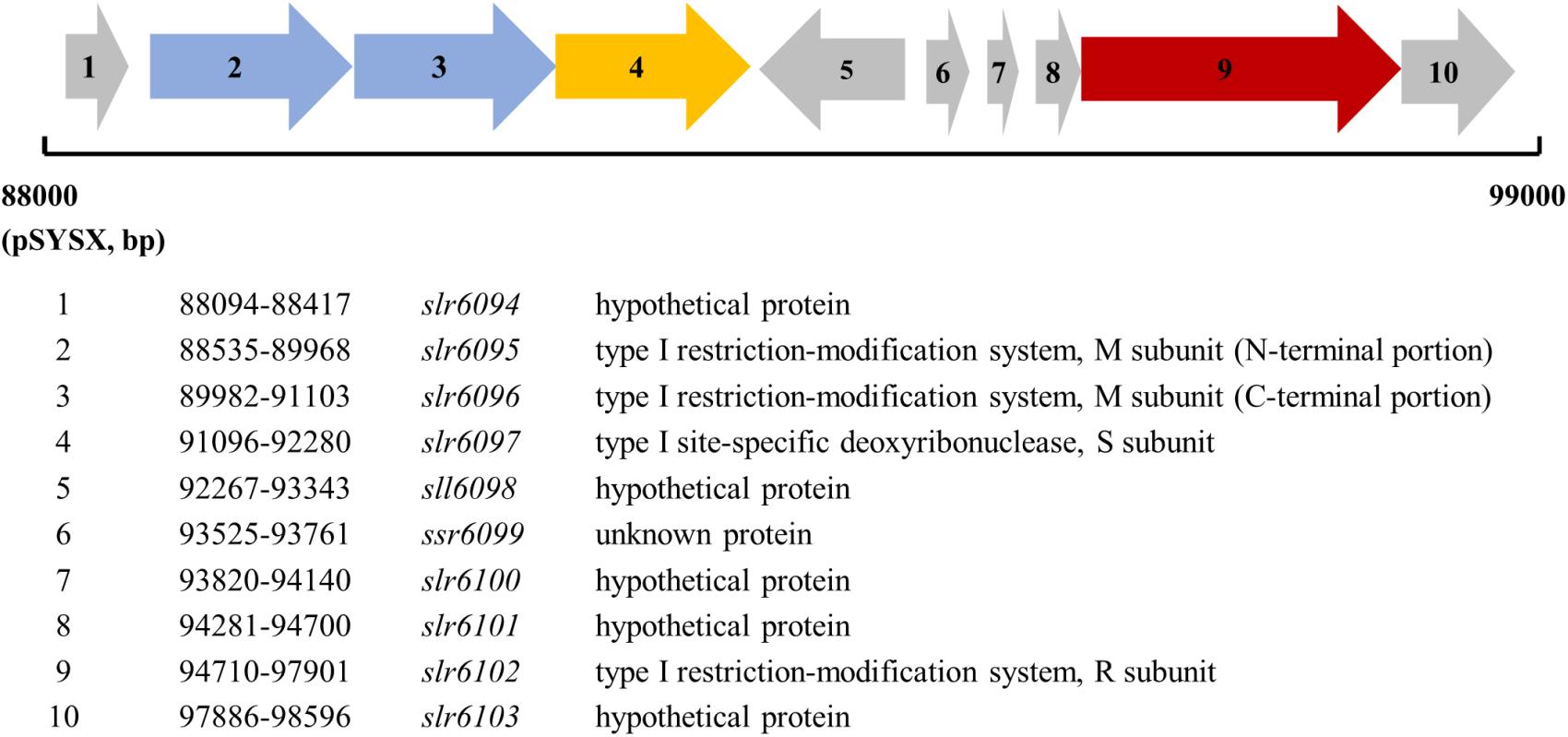
Figure 1. Genes encoding the type I restriction-modification system on the plasmid pSYSX in Synechcystis 6803.
To investigate the functions of slr6095, slr6096, and slr6097, we generated mutant strains for each of them (Figure 2A). A Kmr cassette was inserted into the target genes via homologous double crossover, and the complete segregation of mutants was confirmed with PCR (Figure 2B). Because M and S subunits of a type I RM system are involved in the restriction activity, the complete segregation of the three mutants does not imply that the predicted R subunit (Slr6102) is not functional.
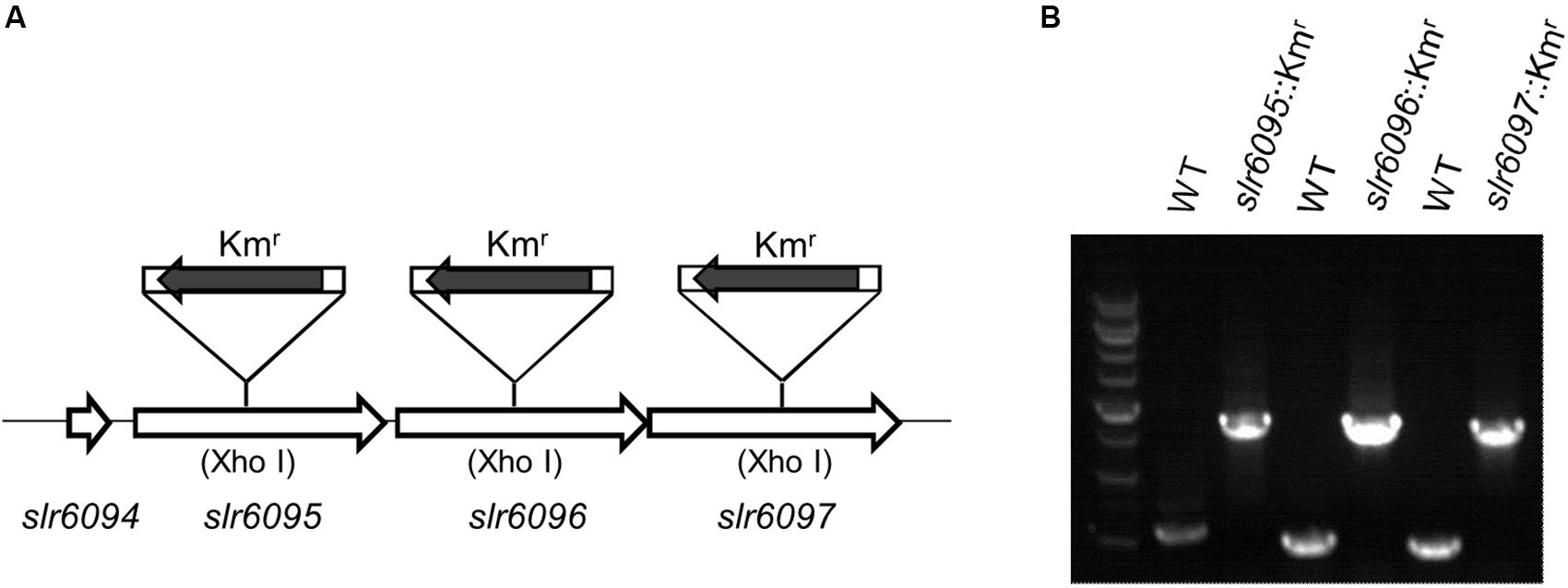
Figure 2. Inactivation of genes in Synechocystis 6803. (A) Schematic diagram showing insertion of target genes with the Kmr cassette. (B) Agarose electrophoresis of PCR products for examination of mutants.
We examined the whole-genome methylation landscape using SMRT sequencing technology. This approach is modestly sensitive to m5C methylation but detects m6A and m4C with high accuracy and sensitivity. As shown in Table 1, in the wild-type genome there are methylations at motifs Gm6ATC, GGm4CC, GAm6AGGC, and GGm6AN7TTGG/CCAm6AN7TCC. Unlike the wild type, mutant strains slr6095::Kmr, slr6096::Kmr, and slr6097::Kmr showed no bipartite methylation at GGm6AN7TTGG/CCAm6AN7TCC (hereafter Ssp6803V methylation). This is consistent with the predicted functions of the three genes. We also noticed that the GGm4CC methylation decreased in slr6096::Kmr and slr6097::Kmr in comparison to the wild type level. This could be due to an indirect effect of the two genes on the efficiency of GGm4CC methylation.
DNA methylation may alter gene expression. We performed RNA-seq analyses to compare gene expression in the wild type and mutants (Supplementary Tables S3–S5). Relative to gene expression in the wild type, 143 genes were up-regulated and 160 genes down-regulated in slr6095::Kmr, 137 genes up-regulated, and 172 genes down-regulated in slr6096::Kmr, 109 genes up-regulated, and 150 genes down-regulated in slr6097::Kmr. There are 73 genes up-regulated and 98 genes down-regulated in all three mutants (Table 2). These 171 genes (73 + 98) are involved in photosynthesis, ATP synthesis, pigment syntheses, carbon metabolism, nutrient transport, stress tolerance, signal transduction, and gene regulation, etc.
To understand the effects of DNA methylation on gene expression in Synechocystis 6803, we attempted to find out the relationship between DNA methylation and gene expression. The Ssp6803V methylation sites are distributed in the encoding sequences of 869 genes (23.74% of all in the genome) and the sequences (1 kb) upstream of 270 genes (7.38% of all). Of the 171 genes with changed expression, 75 have no recognition sequence for the Ssp6803V methylation, 40 have the motifs only in their encoding regions (Table 2 and Supplementary Table S2). It is not possible for these genes to be directly regulated by the DNA methylation. However, there are some genes, such as sll1286 (transcriptional regulator), sll1883 (argJ), sll1926 (hypothetical protein), and slr0093 (dnaJ), whose Ssp6803V methylation sites may overlap regulatory sequences (see transcription start sites and methylation positions in Table 2). The expression of these genes could be modulated by methylation due to altered interactions between regulatory proteins and recognition sites.
Among genes that showed changed expression in the mutants relative to the wild type, slr0653 encodes the principal RNA polymerase sigma factor (SigA), sll1356 (glgP1) encodes GlgP. Both were upregulated in the mutants. RT-qPCR analysis confirmed the increased expression of sigA and glgP1 in three mutants; glgP2, however, showed no or slight change in mRNA level (Figure 3).
Because GlgP1 plays an important role in glycogen turnover (Fu and Xu, 2006), we further measured the GlgP activities and total glycogen contents in these strains under photoautotrophic conditions. As shown in Figure 4A, all three mutants showed higher GlgP activities than the wild type. Higher GlgP activity implies more rapid digestion of glycogen. Consistently, the mutants showed lower glycogen contents than the wild type (Figure 4B).
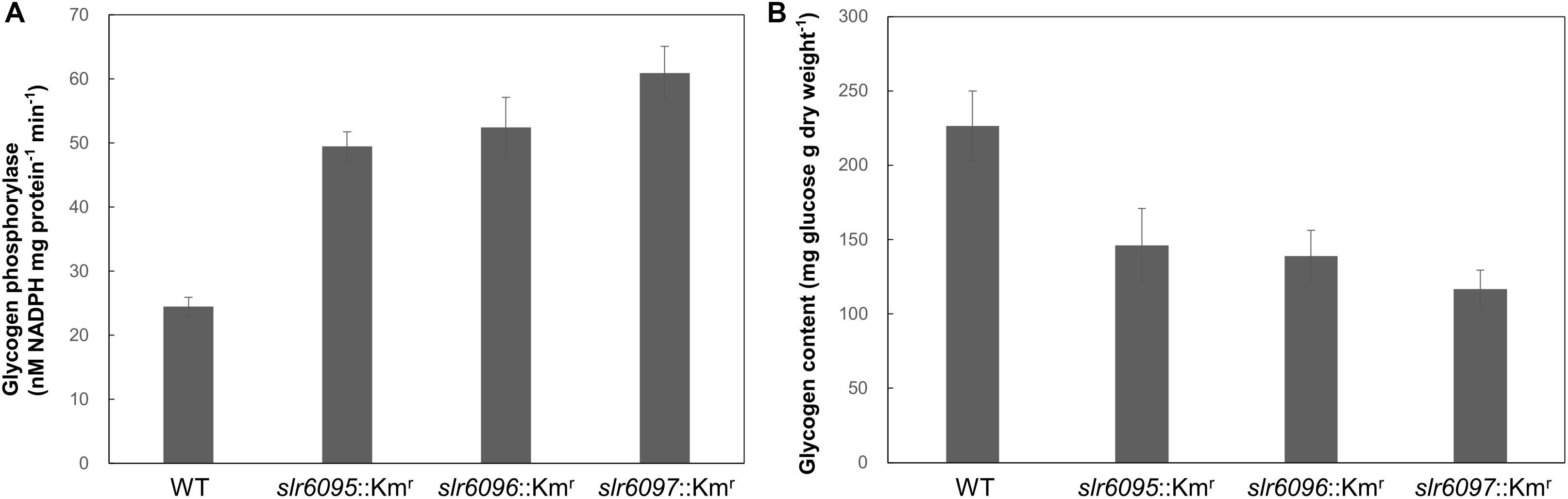
Figure 4. Glycogen phosphorylase activities (A) and glycogen contents (B) in Synechocystis 6803 and mutants.
Some genes involved in photosynthesis (phycobilisome, photosystem I, photosystem II, and CO2 concentrating mechanism), electron transfer (petC2, petF), or ATP synthesis were downregulated in the mutants (Table 2), but petE and rbcX (Rubisco chaperonin) were significantly upregulated. Some heat shock protein/molecular chaperone genes were also downregulated in the mutants (Table 2). The mutants showed no or very slight change in growth at 30°C but reduced growth at 40°C compared to the wild type (Supplementary Figure S2 and data no shown).
In Synechocystis 6803, the role of GGm4CC methylation in regulation of gene expression has been reported (Gärtner et al., 2019). Now we show that the GGm6AN7TTGG/CCAm6AN7TCC methylation, catalyzed by the type I RM system Ssp6803V, is also involved in regulation/modulation of gene expression.
Genes slr6095-slr6096-slr6097 and slr6102 on the plasmid pSYSX are predicted to encode a type I RM system, but the ancestor M subunit gene split into slr6095 and slr6096 in the evolution. Unlike a type II RM system, in which the restriction activity is only dependent on the restriction enzyme, cleavage of the target DNA by a type I RM system is dependent on S, R, and M subunits (Murray, 2000). Therefore, successful inactivation of genes for M and S subunits does not imply that the R subunit gene is not functional. In the RNA-seq data, the mRNA abundance of slr6102 is similar to slr6095-slr6096-slr6097. It is possible that this type I RM system is fully functional.
In Synechocystis 6803 mutants slr6095::Kmr, slr6096::Kmr, and slr6097::Kmr, the Ssp6803V methylation was completely eliminated. Downstream of slr6097 is sll6098, which is oriented 3’ to 3’ relative to slr6097 (Figure 1). Therefore, the phenotype of slr6097::Kmr could not be attributed to a polar effect. Insertion of slr6095 and slr6096 may have polar effects on the downstream gene(s), but these two and slr6097 are very closely related in function, their phenotypes are supposed to be similar to each other. The very similar phenotypes of the three mutants excluded the possibility that a second mutation might have caused the elimination of DNA methylation and physiological effects, if any.
To analyze the effects of Ssp6803V methylation on gene expression in Synechocystis 6803, we compared the mRNA expression profiles of the wild type and mutants and identified 171 genes that showed up- or down-regulated expression in all three mutants. Most of these genes are apparently indirectly regulated by the Ssp6803V methylation (Table 2). glgP1 is one of those upregulated genes (Table 2 and Figure 3), suggesting that the type I RM system may tune glycogen catabolism via gene regulation. Differences in GlgP activities and glycogen contents between the wild type and mutants (Figure 4) confirmed such a physiological effect of the Ssp6803 methylation. Apparently, the DNA methylation reduces the expression of GlgP1 for glycogen turnover and enhances the storage of glycogen in Synechocystis 6803. The mutants also showed reduced growth at 40°C. This phenotype may reflect an integrated effect of changed gene expression (such as expression of genes for photosynthesis, stress responses, etc.) on the high temperature tolerance. In addition, the changed expression of heat shock protein genes (Table 2) could affect salt and oxidative stress tolerance (Asadulghani et al., 2004; Sakthivel et al., 2009; Kim et al., 2014) as well.
The impact of type I RM systems on gene expression has been reported in other bacteria (De Ste Croix et al., 2017). For an example, in Streptococcus pneumoniae, a human pathogen, the virulence was shown to be regulated by a type I RM system (SpnD39III) with phase variable recognition specificities (Manso et al., 2014). SpnD39III variants showed distinct genome methylation patterns and gene expression profiles. However, there are no SpnD39III sites in promoters or regulatory regions of those differentially expressed genes. Similarly, no Ssp6803V sites can be found upstream of most genes differentially expressed in the wild type and methylation mutants. Possibly, addition of methyl groups to target sequences can alter protein-DNA interactions that are involved in maintaining regional DNA conformation, therefore indirectly modulates the expression of glgP1 and many other genes. The underlying mechanism for type I RM systems to impact gene expression in bacteria should be a further question to be tackled in the future.
Raw sequence read data for DNA methylation analysis has been deposited in the NCBI SRA with the study identifier SRP256040 (runs: SRR11528407-SRR11528414). Raw sequence read data for RNA-seq has been deposited in the NCBI SRA with the study identifier SRP258660 (runs: SRR11614230-SRR11614241).
XX designed and supervised the project. DW performed the experiments and bioinformatic analyses. YW performed bioinformatic analyses. XX, DW, and YW analyzed the data. DW and XX wrote the manuscript. XX edited the manuscript.
This research was supported by the National Natural Science Foundation of China (31470203), the State Key Laboratory of Freshwater Ecology and Biotechnology at IHB, CAS (2019FBZ09), and the Knowledge Innovation Project of Hubei Province (2017CFA021).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01258/full#supplementary-material
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Asadulghani, N. K., Kaneko, Y., Kojima, K., Fukuzawa, H., Kosaka, H., and Nakamoto, H. (2004). Comparative analysis of the hspA mutant and wild-type Synechocystis sp. strain PCC 6803 under salt stress: evaluation of the role of hspA in salt-stress management. Arch. Microbiol. 182, 487–497. doi: 10.1007/s00203-004-0733-x
Ball, S. G., and Morell, M. K. (2003). From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54, 207–233. doi: 10.1146/annurev.arplant.54.031902.134927
Blow, M. J., Clark, T. A., Daum, C. G., Deutschbauer, A. M., Fomenkov, A., Fries, R., et al. (2016). The epigenomic landscape of prokaryotes. PLoS Genet. 12:e1005854. doi: 10.1371/journal.pgen.1005854
Castenholz, R. W. (2001). “Oxygenic photosynthetic bacteria,” in Bergey’s Manual of Systematic Bacteriology, 2nd Edn, Vol. 1, eds D. R. Boone and R. W. Castenholz (New York, NY: Springer-Verlag), 474–599.
De Ste Croix, M., Vacca, I., Kwun, M. J., Ralph, J. D., Bentley, S. D., Haigh, R., et al. (2017). Phase-variable methylation and epigenetic regulation by type I restriction-modification systems. FEMS Microbiol. Rev. 41(SuppL. 1), S3–S15. doi: 10.1093/femsre/fux025
Fu, J., and Xu, X. (2006). The functional divergence of two glgP homologues in Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 260, 201–209. doi: 10.1111/j.1574-6968.2006.00312.x
Gao, H., and Xu, X. (2012). The cyanobacterial NAD kinase gene sll1415 is required for photoheterotrophic growth and cellular redox homeostasis in Synechocystis sp. strain PCC 6803. J. Bacteriol. 194, 218–224. doi: 10.1128/JB.05873-11
Gärtner, K., Klähn, S., Watanabe, S., Mikkat, S., Scholz, I., Hess, W. R., et al. (2019). Cytosine N4-Methylation via M.Ssp6803II is involved in the regulation of transcription, fine-tuning of DNA replication and DNA repair in the cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 10:1233. doi: 10.3389/fmicb.2019.01233
Hagemann, M., Gärtner, K., Scharnagl, M., Bolay, P., Lott, S. C., Fuss, J., et al. (2018). Identification of the DNA methyltransferases establishing the methylome of the cyanobacterium Synechocystis sp. PCC 6803. DNA Res. 25, 343–352. doi: 10.1093/dnares/dsy006
Kim, J., Ahn, M. S., Park, Y. M., Kim, S. W., Min, S. R., Jeong, W. J., et al. (2014). Synechocystis PCC6803 and PCC6906 dnaK2 expression confers salt and oxidative stress tolerance in Arabidopsis via reduction of hydrogen peroxide accumulation. Mol. Biol. Rep. 41, 1091–1101. doi: 10.1007/s11033-013-2955-y
Manso, A. S., Chai, M. H., Atack, J. M., Furi, L., De Ste Croix, M., Haigh, R., et al. (2014). A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun. 5:5055. doi: 10.1038/ncomms6055
Matveyev, A. V., Young, K. T., Meng, A., and Elhai, J. (2001). DNA methyltransferases of the cyanobacterium Anabaena PCC 7120. Nucleic Acids Res. 29, 1491–1506. doi: 10.1093/nar/29.7.1491
Murray, I. A., Clark, T. A., Morgan, R. D., Matthew, B., Anton, B. P., Khai, L., et al. (2012). The methylomes of six bacteria. Nucleic Acids Res. 40, 11450–114362. doi: 10.1093/nar/gks891
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325.
Murray, N. E. (2000). Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 64, 412–434. doi: 10.1128/mmbr.64.2.412-434.2000
Osanai, T., Oikawa, A., Numata, K., Kuwahara, A., Iijima, H., Doi, Y., et al. (2014). Pathway-level acceleration of glycogen catabolism by a response regulator in the cyanobacterium Synechocystis species PCC 6803. Plant Physiol. 164, 1831–1841. doi: 10.1104/pp.113.232025
Preiss, J. (1984). Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 38, 419–458. doi: 10.1146/annurev.mi.38.100184.002223
Sakthivel, K., Watanabe, T., and Nakamoto, H. (2009). A small heat-shock protein confers stress tolerance and stabilizes thylakoid membrane proteins in cyanobacteria under oxidative stress. Arch. Microbiol. 191, 319–328. doi: 10.1007/s00203-009-0457-z
Sánchez-Romero, M. A., and Casadesús, J. (2020). The bacterial epigenome. Nat. Rev. Microbiol. 18, 7–20. doi: 10.1038/s41579-019-0286-2
Scharnagl, M., Richter, S., and Hagemann, M. (1998). The cyanobacterium Synechocystis sp. strain PCC 6803 expresses a DNA methyltransferase specific for the recognition sequence of the restriction endonuclease PvuI. J. Bacteriol. 180, 4116–4122. doi: 10.1074/jbc.M112.379339
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). Tophat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Vioque, A. (1992). Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 20, 6331–6337. doi: 10.1093/nar/20.23.6331
Keywords: Synechocystis, type I RM system, gene regulation, glgP, glycogen catabolism
Citation: Wu D, Wang Y and Xu X (2020) Effects of a Type I RM System on Gene Expression and Glycogen Catabolism in Synechocystis sp. PCC 6803. Front. Microbiol. 11:1258. doi: 10.3389/fmicb.2020.01258
Received: 01 April 2020; Accepted: 18 May 2020;
Published: 09 June 2020.
Edited by:
Lei Chen, Tianjin University, ChinaReviewed by:
Guodong Luan, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), ChinaCopyright © 2020 Wu, Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xudong Xu, eHV4QGloYi5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.