- 1State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 2College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3School of Minerals Processing and Bioengineering, Central South University, Changsha, China
- 4Key Laboratory of Biometallurgy of Ministry of Education, Central South University, Changsha, China
- 5Department of Pathogen Biology, School of Basic Medical Science, Xi’an Medical University, Xi’an, China
Members of the genus Metallosphaera are widely found in sulfur-rich and metal-laden environments, but their physiological and ecological roles remain poorly understood. Here, we sequenced Metallosphaera tengchongensis Ric-A, a strain isolated from the Tengchong hot spring in Yunnan Province, China, and performed a comparative genome analysis with other Metallosphaera genomes. The genome of M. tengchongensis had an average nucleotide identity (ANI) of approximately 70% to that of Metallosphaera cuprina. Genes sqr, tth, sir, tqo, hdr, tst, soe, and sdo associated with sulfur oxidation, and gene clusters fox and cbs involved in iron oxidation existed in all Metallosphaera genomes. However, the adenosine-5′-phosphosulfate (APS) pathway was only detected in Metallosphaera sedula and Metallosphaera yellowstonensis, and several subunits of fox cluster were lost in M. cuprina. The complete 3-hydroxypropionate/4-hydroxybutyrate cycle and dicarboxylate/4-hydroxybutyrate cycle involved in carbon fixation were found in all Metallosphaera genomes. A large number of gene family gain events occurred in M. yellowstonensis and M. sedula, whereas gene family loss events occurred frequently in M. cuprina. Pervasive strong purifying selection was found acting on the gene families of Metallosphaera, of which transcription-related genes underwent the strongest purifying selection. In contrast, genes related to prophages, transposons, and defense mechanisms were under weaker purifying pressure. Taken together, this study expands knowledge of the genomic traits of Metallosphaera species and sheds light on their evolution.
Introduction
Extremely acidophilic archaea of the genus Metallosphaera belong to the order Sulfolobales within the Crenarchaeota. Together with the genera Acidianus and Sulfolobus, physiologically versatile Metallosphaera spp. contribute significantly to biogeochemical element cycling and biomining processes. Metallosphaera spp. stand out due to their ability for facultative autotrophic growth and tolerance to high concentrations of metal ions (Ai et al., 2016; Wheaton et al., 2016). Members of the genus Metallosphaera grow aerobically at low pH values (<4) and high temperatures (>60°C). They are found in sulfur-rich and metal-laden environments including solfataric areas (Metallosphaera sedula DSM 5348 and strains from laboratory evolution) (Huber et al., 1989; Ai et al., 2016, 2017), hot springs (such as Metallosphaera hakonensis HO1-1 = JCM8857, Metallosphaera cuprina Ar-4, Metallosphaera tengchongensis Ric-A, Metallosphaera yellowstonensis MK-1, Metallosphaera sp. UBA165, and Metallosphaera sp. Obs4) (Takayanagi et al., 1996; Kozubal et al., 2008, 2011; Liu et al., 2011a; Peng et al., 2015; Parks et al., 2017), and bioleaching-related heaps (Metallosphaera prunae Ron 12/II) (Fuchs et al., 1995). Metallosphaera species are of great potential in the extraction of base and precious metals from ores exploiting their ability to oxidize reduced inorganic sulfur compounds (RISCs) and ferrous ion, a process mediated by a set of terminal oxidases that are attached to their cell membranes (Auernik and Kelly, 2008, 2010; Orell et al., 2010). Previous studies showed that application of thermophilic archaea in chalcopyrite bioleaching could achieve faster dissolution rates and higher copper leaching yields in comparison to widely used mesophilic bioleaching bacteria because the formation of the surface passivation and diffusion layer of chalcopyrite was significantly reduced at high temperature (Rawlings, 2005; Urbieta et al., 2015; Castro and Donati, 2016).
Metallosphaera species can grow on peptides in a heterotrophic mode, fix carbon dioxide using reduced sulfur compounds as reductant in an autotrophic mode, or grow on casamino acids and FeSO4 or metal sulfides in a mixotrophic mode (Peeples and Kelly, 1995; Ulrike et al., 2005; Alber et al., 2006; Auernik and Kelly, 2008, 2010; Michael et al., 2010; Han and Kelly, 2015). Autotrophic carbon fixation through the 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle in Metallosphaera has been confirmed by genomics, transcriptomics, proteomics, and biochemical assays (Sebastian et al., 2011; Jiang et al., 2014). Reduced Fe and S (Fe2+, HS–, S0, S4O62+, and S2O32+) are important electron donors for iron or sulfur-oxidizing microorganism. Unlike Fe2+ oxidation by the bacteria Acidithiobacillus ferrooxidans and Leptospirillum ferrooxidans, which require the blue copper protein rusticyanin and various c-type cytochromes (Rohwerder et al., 2003; Singer et al., 2008), the Fe oxidation mechanism of iron-oxidizing archaea is yet unconfirmed, although potential proteins were noticed in the genomes of Ferroplasma, Sulfolobus, and Metallosphaera species (Dopson et al., 2005; Bathe and Norris, 2007; Auernik et al., 2008). The genes involved in iron oxidation (Kozubal et al., 2011) such as fox, cbs, rusticyanin, and sulfocyanin have been detected in the genomes of M. sedula and M. yellowstonensis, but their pervasiveness in Metallosphaera is yet unknown. RISC oxidation mechanisms are complex and diverse in extremely thermoacidophilic archaea. Sulfur oxygenase reductase genes are present in the genus Acidianus, in the species Sulfolobus tokadaii, and in bacteria, but are absent from the genomes of M. sedula and M. cuprina (Auernik et al., 2008; Liu et al., 2011b), which raised the question how sulfur oxidation is initiated in Metallosphaera. Of the species within the genus Metallosphaera, genome sequences and genomic analyses have been reported for four isolates (M. cuprina Ar-4, M. hakonensis HO1-1 = JCM_8857, M. sedula DSM_5348, and M. yellowstonensis MK1) (Auernik et al., 2008; Liu et al., 2011b). M. tengchongensis Ric-A, the newest member of Metallosphaera, which was isolated from a sulfuric hot spring in Tengchong, Yunnan Province, China, showed an excellent performance in copper extraction from chalcopyrite (Peng et al., 2015). To better understand the unique metabolism, adaptation for extreme thermal and acidic conditions, roles in biogeochemical cycling, and the evolutionary history of the genus Metallosphaera, we performed genome sequencing of M. tengchongensis Ric-A and compared its genome with 18 available Metallosphaera genomes, of which 12 are from Genbank database of National Center for Biotechnology Information (NCBI) (M. sedula DSM5348, ARS50-1, ARS50-2, ARS120-1, ARS120-2, SARC-M1, and CuR1; M. hakonensis HO1-1 and JCM8857; M. cuprina Ar-4; M. yellowstonensis MK-1; Metallosphaera sp. UBA165) and six scaffold genomes are from the Integrated Microbial Genomes and Microbiomes (IMG/M) system of DOE Joint Genome Institute (JGI) (Metallosphaera spp. My-r02, My-r05, My-r06, YNP_08, YNP_14, and Obs4) (Table 1). In this work, we performed comprehensive analyses of genome-based phylogenetic relationships, metabolic pathway and gene function, heavy metal resistance, adhesion and motility, as well as mobile genetic elements (MGEs) and selective pressure. These findings will improve our understanding of the adaptive strategies of the organisms to their harsh environment and provide clues to design biomining or bioremediation processes in the future.
Materials and Methods
Sample Collection and Sequencing
Metallosphaera tengchongensis strain Ric-A was isolated from the muddy water samples of sulfuric hot springs (24.57 N and 98.26 E, with the temperature range of 55–96°C and a pH range of 2.5–7.5, dissolved oxygen range of 0.01–1.00 mg/L) in Tengchong county of Yunnan Province, China. The hot springs are rich in S (SO42–, 701.20–22.46 mg/L), Fe (13.89–0.05 mg/L), Ca (71.55–1.12 mg/L), K (22.64–63.01 mg/L), Al (42.43–0.13 mg/L), and other elements (Liu et al., 2011a; Peng et al., 2015; Qin et al., 2019). The sample was concentrated by tangential flow ultrafiltration through a hollow fiber membrane (Tianjin MOTIMO Membrane Technology, China). An aerobic enrichment culture in the flask with filtration membrane was established by inoculating the concentrate in basal salts medium (BSM) with elemental sulfur as energy source. The compositions of BSM were L–1: (NH4)2SO4, 3 g; K2HPO4 ⋅ 3H2O, 0.5 g; MgSO4 ⋅ 7H2O, 0.5g; KCl, 0.1 g; Ca(NO3)2, 0.01 g, added with 1 ml of trace element solution (FeCl3 ⋅ 6H2O, 1.1 g; CuSO4 ⋅ 5H2O, 0.05 g; H3BO3, 0.2 g; MnSO4 ⋅ H2O, 0.2 g; Na2MoO4 ⋅ 2H2O, 0.08 g; CoCl2 ⋅ 6H2O, 0.06 g; ZnSO4 ⋅ 7H2O, 0.09 g in 1 L of distilled water). After static culture for 5–7 days at 65°C, samples of the grown culture were spread on BSM solid plates with potassium tetrathionate (K2S4O6, 10 mmol/L) or yeast extract (1 g/L) as energy source. The plates were incubated for 7 days at 65°C. Colonies were picked and purified by re-plating. The purified strain Ric-A was grown at 70°C in BSM (pH 2.0) supplemented with 1 g/L yeast extract. The stationary-phase cells were harvested by centrifugation. The genomic DNA was extracted from the concentrated cells according to the instruction of “JGI Bacterial DNA isolation CTAB-2012.”1 After checking its quality, DNA was fragmented and the fragments were end-repaired and polyadenylated, and then ligated to sequencing adapter. SMRTbell DNA library was constructed by using Blue Pippin Size-Selection System; library quality was evaluated by Qubit 3.0 Fluorometer (Life Technologies, Grand Island, NY, United States) and sequenced by PacBio Biosciences (PacBio) RSII and Genome Analyzer IIx sequence platforms at Chinese National Human Genome Center at Shanghai (CHGC). After sequencing, the low-quality reads were filtered by the SMRT 2.3.0 (Chin et al., 2013), and the filtered reads were assembled to generate one contig without gaps. The hierarchical genome-assembly process (HGAP.3) pipeline implemented in SMRT Analysis 2.3.02 was used to correct for random errors in the long seed reads (seed length threshold 6 Kb) by aligning shorter reads from the same library against them. The resulting corrected, preassembled reads were used for de novo assembly. Genome data of 18 previously sequenced strains belonging to Metallosphaera were collected from NCBI and IMG-M database. The detailed genomic statistics information and source of 19 Metallosphaera strains used in this study were summarized in Table 1.
Average Nucleotide Identity and Whole Genome Alignments
Comparisons of average nucleotide identity (ANI) based on Blast algorithm were conducted using the pyani module3 with default parameters. We applied the “progressive Mauve program” within Mauve v 2.3.0 (Darling et al., 2004) for constructing and visualizing multiple genome alignments of M. tengchongensis Ric-A with four other available complete genome sequences of M. sedula DSM 5348, M. hakonensis HO1-1, M. cuprina Ar-4, and M. yellowstonensis MK1. BlastN-based whole-genome comparison of strains M. cuprina Ar-4, M. hakonensis HO1-1, Metallosphaera sp. My-r06, M. sedula DSM 5348, M. tengchongensis Ric-A, and M. yellowstonensis MK1 were performed and represented with BRIG-0.95 (Alikhan et al., 2011), and these strains were used as references, respectively. GC content and GC skew of each genome were also indicated.
Phylogenomic Analyses
We constructed a phylogenetic tree of the 19 Metallosphaera spp. genomes based on whole-genome sequences with CVTree3 (Xu and Hao, 2009). The phylogenetic tree of the 19 genomes based on concatenation of the 85 core genes in a genome was constructed with the neighbor-joining (NJ), UPGMA, and maximum-likelihood (ML) method using MEGA-X (Kumar et al., 2018) with 1000 bootstrap replicates.
Genome Annotation
We applied Prokka (Seemann, 2014) and IMG Annotation Pipeline v.4.16.0 (Chen et al., 2017) for genome annotation and putative horizontally transferred gene detection. We performed Diamond BlastP v0.9.24 (Buchfink et al., 2015) with a cutoff e-value of 1e–5 together with the dbCAN database v2.0 (Le et al., 2018) for identification of genes related to carbohydrate activity enzyme (CAZymes). The BacMet v2.0 database (Pal et al., 2014) that contained genes with experimentally confirmed metal resistance function was used to identify the genes associated with metal resistance in Metallosphaera genomes. Gene annotations based on COG (Tatusov et al., 2001), Pfam (Finn et al., 2014), and TIGRFAM (Selengut et al., 2007) databases were performed via WebMGA (Wu et al., 2011) using Blast with a cutoff e-value of 1e–5.
Prediction of Mobile Genetic Elements
We applied the ISFinder (Siguier et al., 2006)4 to predict and classify insertion sequences (IS) and transposases within Metallosphaera genomes with Blastp v0.9.24 (cutoff e-value of 1e–5). We applied the IslandViewer 4 (Bertelli et al., 2017),5 which integrated prediction methods including IslandPath-DIMOB and SIGI-HMM that analyzed sequence composition, and another comparative genomic islands (GIs) prediction method IslandPick, to detect putative GIs distributed over Metallosphaera genomes. We applied PHASTER (Phage Search Tool Enhanced Release) (Arndt et al., 2016)6 for detection and annotation of prophage and prophage remnant sequences within Metallosphaera genomes. We also applied CrisprCasFinder (Couvin et al., 2018)7 for detection of CRISPRs and Cas genes within Metallosphaera genomes.
Comparative Genomic Analyses of Metallosphaera
The Bacterial Pan Genome Analyses tool (BPGA) pipeline (Chaudhari et al., 2016) was used to perform pan/core-genome analyses and calculation applying default parameters. The size of the Metallosphaera pan-genome was fitted into a power law regression function Ps = κnγ with a built-in program of BPGA (Chaudhari et al., 2016), in which Ps was the total number of gene families, n stood for the number of tested genomes, and γ was free parameters. If exponent γ < 0, then the pan-genome of Metallosphaera was suggested to be “closed.” In this case, the size of the pan-genome is relatively constant, even if new genomes were added into the analysis. On the contrary, the pan-genome was suggested to be “open” in the case of 0 < γ < 1. In addition, the size of the core-genome of Metallosphaera was fitted into an exponential decay function Fc = κcexp(−n/τc) with a built-in program of BPGA pipeline (Chaudhari et al., 2016), in which Fc stood for the number of core gene families, while κc and τc were free parameters. Gene family clustering followed by genome-wide comparisons of five Metallosphaera-type strains including M. tengchongensis Ric-A, M. sedula DSM 5348, M. hakonensis HO1-1, M. cuprina Ar-4, and M. yellowstonensis MK1 together with UniProt search, Gene Ontology (GO) Slim annotation, and GO enrichment analyses (default cutoff p-value is 0.05) were performed via OrthoVenn (Wang et al., 2015)8 with default parameters.
Gene Family Evolution Analyses
Count is a software designed to analyze numerical profiles of homologous gene families on a phylogeny, which can execute ancestral reconstructions and predict family- and lineage-specific characteristics along the evolutionary tree (Miklós, 2010). We used Count software, combined BPGA v2.0 pipeline, and Wagner parsimony algorithm (Farris, 1970) for gene family clustering, ancestor genome size estimation, and detecting gene family gain and loss events, together with family expansion and contraction events with penalty ratio set to 1. We conducted the analyses only on five Metallosphaera-type strains, including M. tengchongensis Ric-A, M. sedula DSM 5348, M. hakonensis HO1-1, M. cuprina Ar-4, and M. yellowstonensis MK1 taken into consideration, due to the estimation requiring complete sets of testing gene families available only in species with high-quality genome databases.
Selective Pressure Analyses
We detected the numbers of sites under negative (purifying) or positive (diversifying) selection and estimated global dN/dS values of each gene family that contained more than three non-identical sequences (due to limitation of HyPhy) based on multiple alignments of orthologous codon sequences and a tree topology by means of HyPhy package (Pond and Muse, 2005) using the Fixed Effects Likelihood (FEL) method (Kosakovsky Pond and Frost, 2005) (applied a likelihood ratio test with default cutoff p-value: 0.1) via datamonkey server (Weaver et al., 2018).9 The coding sequences of five Metallosphaera-type strains, including M. tengchongensis Ric-A, M. sedula DSM 5348, M. hakonensis HO1-1, M. cuprina Ar-4, and M. yellowstonensis MK1, were aligned with muscle codon alignment module implemented in MEGACC (Kumar et al., 2012) to obtain final codon alignments.
Results and Discussion
General Genome Features of M. tengchongensis and Other Strains
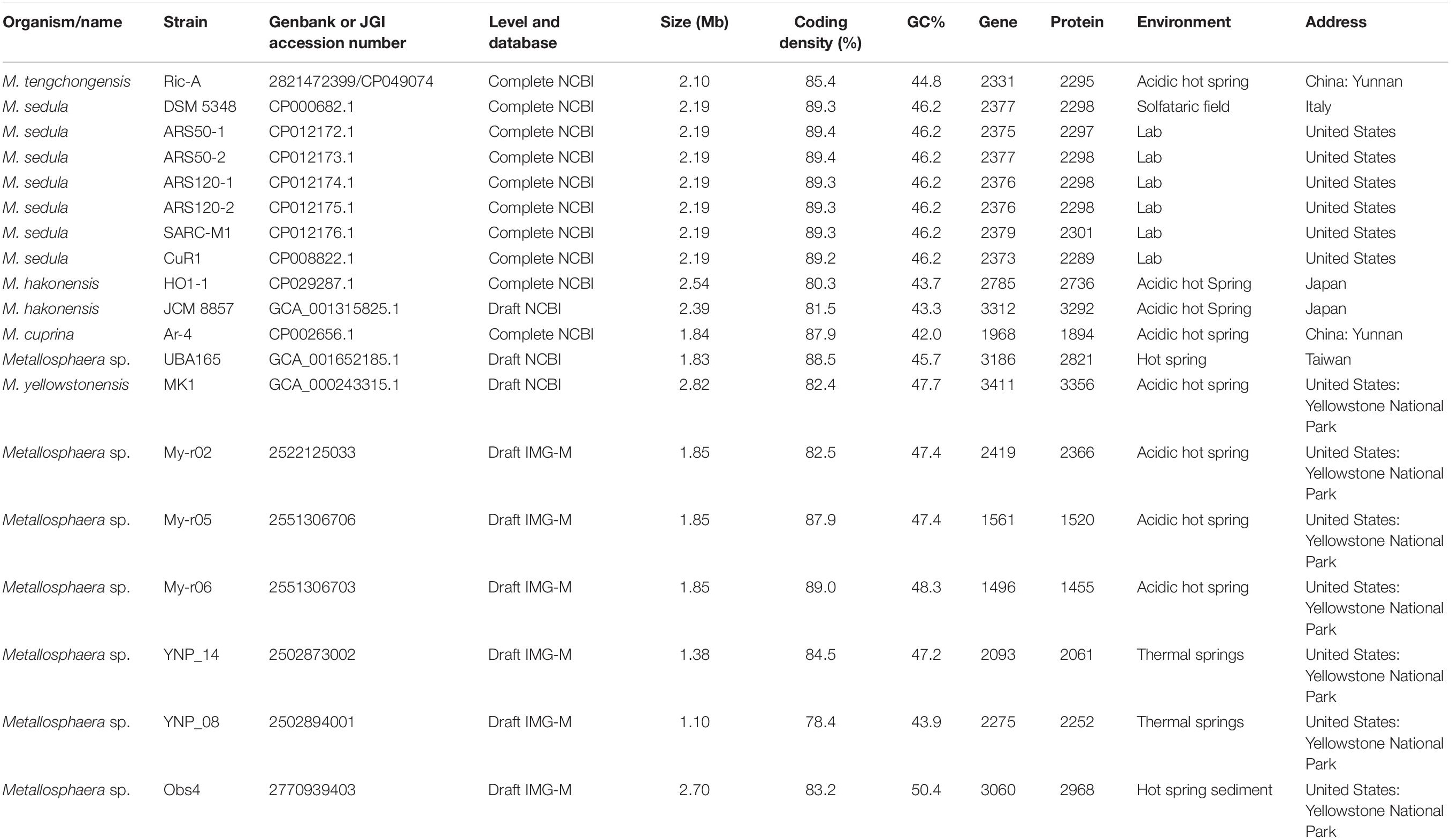
A total of 2331 CDS, including 32 tRNA and 4 rRNA, were predicted in the genome of M. tengchongensis strain Ric-A using Prokka (Seemann, 2014). Whole-genome BLASTN-based ANI analyses showed only an ANI of 72.3% compared with the closest Metallosphaera genome (Supplementary Table S1). A summary of features for the 19 Metallosphaera genomes is listed in Table 1. M. yellowstonensis MK1 possessed the largest genome (2.82 Mb). The G + C contents of the 19 genomes ranged from 42.0 to 50.4%. These genomes varied in coding density from 78.4 to 89.4%, indicating substantial intra-genus differences. We determined that the previously unclassified strain UBA165 was a member of M. sedula, and strains My-r02, My-r05, My-r06, YNP_08, and YNP_14 were members of M. yellowstonensis based on an ANI cutoff of 96% (Richter and Rossello-Mora, 2009), as supported by further phylogenetic analyses (Figure 1, Supplementary Figure S1, and Supplementary Table S1). The genome alignment of M. tengchongensis Ric-A with four other complete genomes of Metallosphaera strains using Mauve (Darling et al., 2004) indicated that the chromosomal alignments of Metallosphaera genomes were non-conserved, as shown by the presence of hundreds of poorly organized collinear blocks and numerous inversed and rearranged regions (Supplementary Figure S2). Each Metallosphaera species harbored genomic regions that were not commonly shared; most of these harbored poorly characterized proteins as revealed by whole genome comparison of Metallosphaera spp. using BRIG (Alikhan et al., 2011) (Supplementary Figures S2, S3).
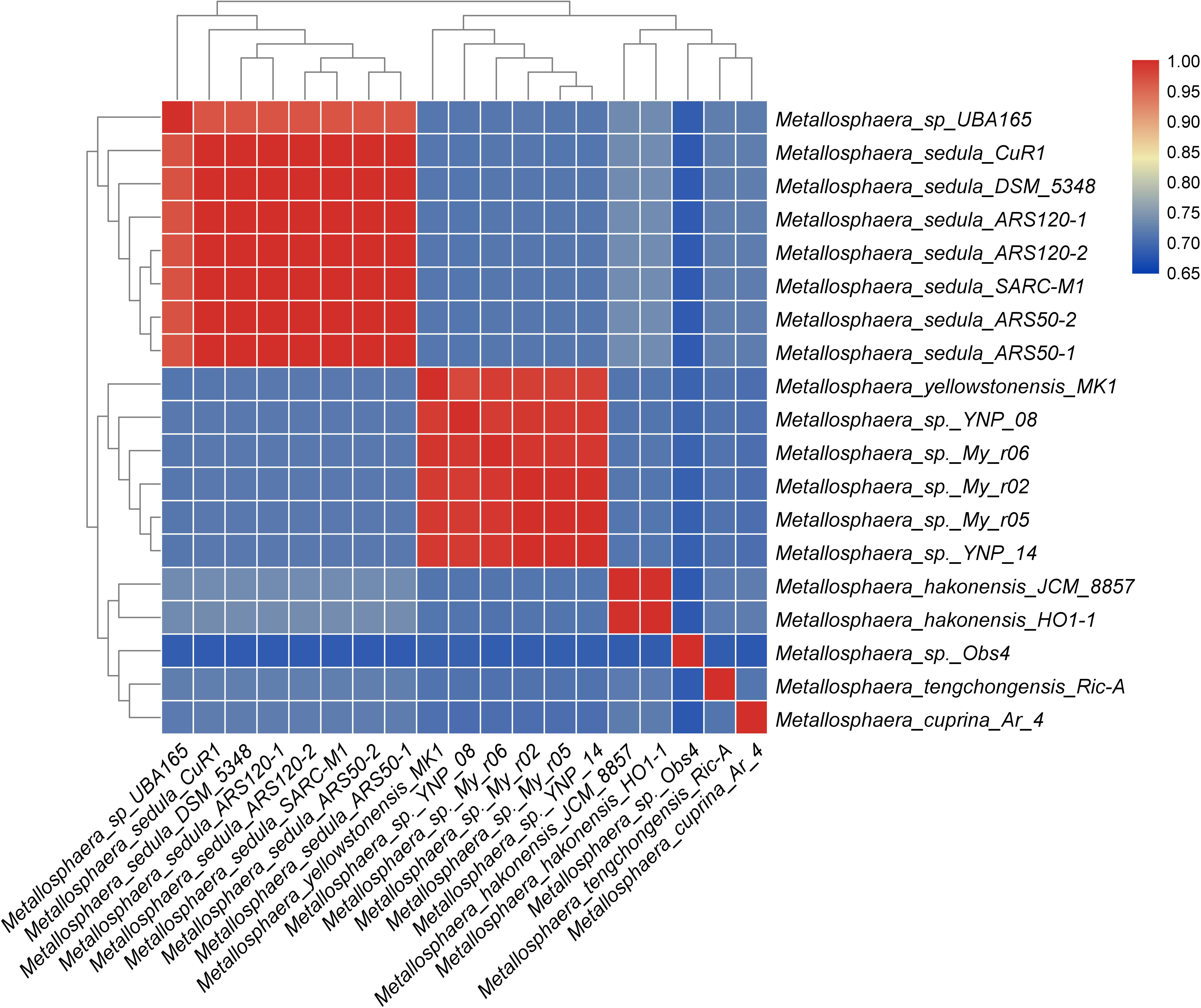
Figure 1. Heat map showing species boundary of genus Metallosphaera based on ANI value (cutoff value = 96%), which is a universally accepted genomic measure for prokaryotic species delineation (Richter and Rossello-Mora, 2009).
Core- and Pan-Genome and Phylogenomic Analysis of Metallosphaera
The phylogenetic trees based on the concatenated alignment of 85 core genes inferred with NJ, ML, MP, and UPGMA methods were congruent with each other, and the phylogenetic tree based on whole-genome analyses was congruent with the core-gene phylogenetic tree (Supplementary Figures S4, S5). Strain Obs4 is located on a clade apart from other Metallosphaera strains (Supplementary Figures S1, S4, S5). However, strain Obs4 should still be considered a member of genus Metallosphaera based on percentage of conserved proteins (POCP). Strain Obs4 had a POCP of 50.2% against M. yellowstonensis MK1, within the genus cutoff value of 50% (Qin et al., 2014). The pan-genome of 19 Metallosphaera strains possessed 6499 gene families, while the core-genome possessed only 85 gene families. Core- and pan-genome analyses of the 19 Metallosphaera genomes revealed an “open” pan-genome fitted into a power law regression function [Ps (n) = 1955.63 n0.372819] with a parameter (γ) of 0.372819 falling into the range 0 < γ < 1. The core-genome was fitted into an exponential regression [Fc (n) = 1886.87 e–0.16335 n], which had a steep slope, reaching a minimum of 85 gene families after the 19th genome was added (Figure 2B). The result of COG annotation revealed that the core-genome had a higher proportion of genes involved in COG categories that are associated with central biological functions translation, ribosomal structure, and biogenesis (J); posttranslational modification, protein turnover, chaperones (O); and coenzyme transport and metabolism (H) than the accessory genome and strain-unique genome. In contrast, the accessory genome had a higher proportion of genes related to COG energy production and conversion (C) and lipid transport and metabolism (I). We found that strain-specific gene families had a higher proportion of genes categorized in COG replication, recombination and repair (L), cell wall/membrane/envelope biogenesis (M), inorganic ion transport and metabolism (P), and carbohydrate transport and metabolism (G) (Figure 2). We propose that these genes are associated with adaptive evolution within the genus Metallosphaera. The oligotrophic, metal-laden, and extremely acidic environments select for a highly efficient DNA injury repair system and a flexible trophic mode in Metallosphaera. The genomic diversity and specificity of different Metallosphaera strains are reflecting their distinct survival strategies in different environments.
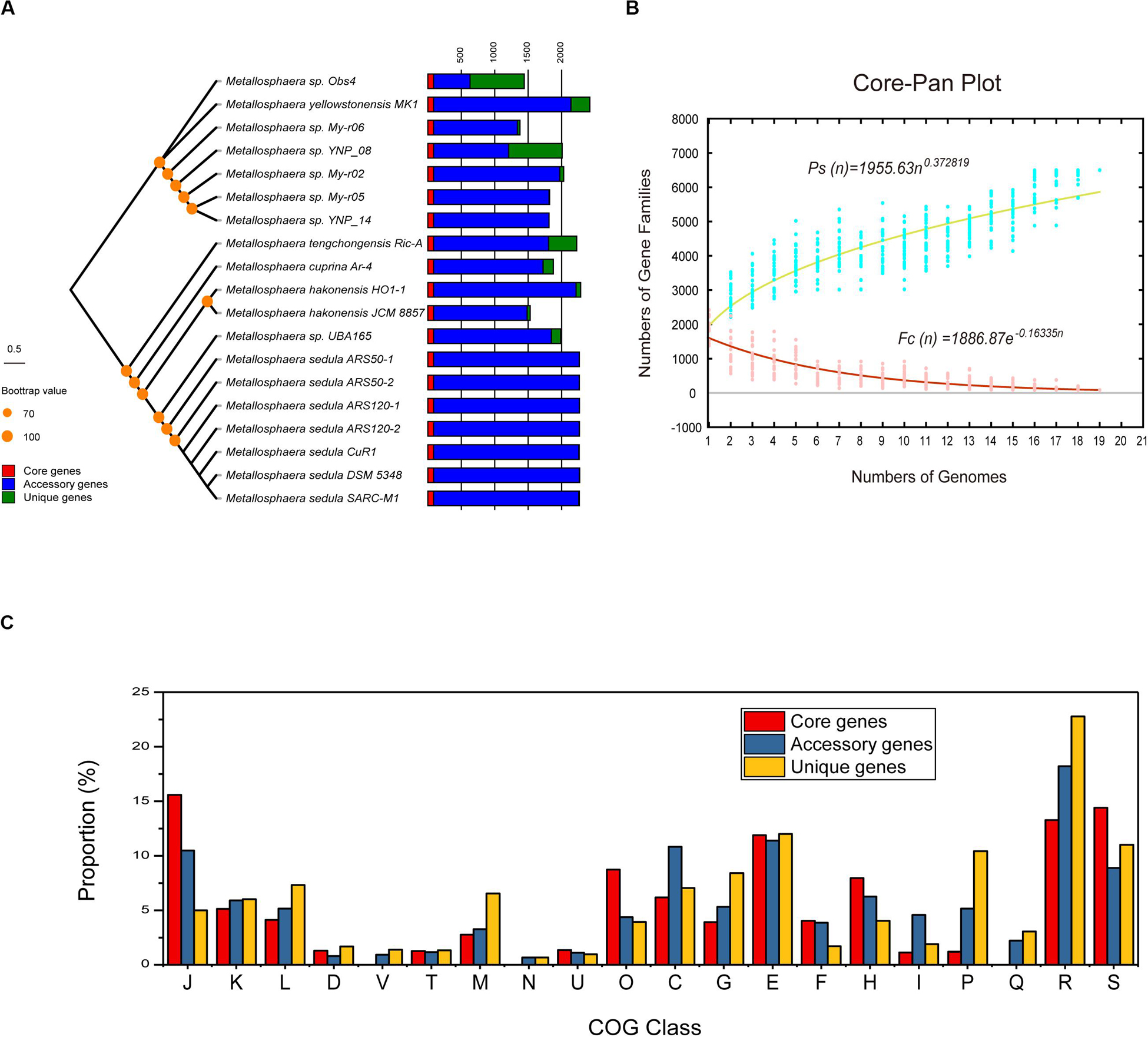
Figure 2. Pan-genome analyses of strains in the genus Metallosphaera. (A) NJ phylogenetic tree based on concatenated 85 core genes (1000 bootstrap replicates) of 19 Metallosphaera strains (left) and stack bar diagram showing sizes of orthologous genes shared by all strains (i.e., the core-genome), number of orthologous genes shared by partial strains (i.e., the accessory genome), and number of strain-specific gene families (i.e., the unique gene) in each strain. (B) The curve of the fitting function and the estimation of pan-genome and core-genome sizes of Metallosphaera. Ps and Fc stand for the total number of gene families and the number of core gene families, respectively. More details for modeling approaches are presented in Section 2.6 “Comparative Genomic Analyses of Metallosphaera”. (C) Bar chart showing proportions of COG categories of the different parts of Metallosphaera pan-genome (i.e., core, accessory, unique). COG categories description (J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; D, cell cycle control, cell division, chromosome partitioning; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, Cell motility; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown).
Metabolic and Functional Potential
Sulfur Metabolism
Sulfur and hydrogen are important in energy flow in thermal environments, such as marine hydrothermal systems, continental solfataras, and hot springs, where many bacteria and archaea can grow by oxidizing hydrogen, sulfide, elemental sulfur, and thiosulfate (Amend and Shock, 2001; Zeldes et al., 2019). It was reported that Metallosphaera could utilize different sulfur compounds as energy sources for growth (Peng et al., 2015). The genes sqr encoding for sulfide:quinone oxidoreductase (SQR), which catalyzes the oxidation of hydrogen sulfide forming polysulfide, were detected in all 19 genomes of Metallosphaera. Unlike the genus Acidianus and certain members of Sulfolobus, the genes encoding the homologs of sulfur oxygenase/reductase (SOR), a key enzyme for archaeal sulfur oxidation (Kletzin, 1992; Chen et al., 2005; Urich et al., 2006; Dai et al., 2016), were not found in the genomes of all strains of Metallosphaera. However, sulfur dioxygenases (SDO, sdo) were encoded in all strains except for M. cuprina and Metallosphaera sp. Obs4 (Figure 3). In addition, sulfite-acceptor oxidoreductases (SAOR, saor) genes were also detected in all strains (Figure 3). Metallosphaera spp. also harbored genes encoding for tetrathionate hydrolase (TTH, tth), sulfite reductase (SIR, sir), and genomic clusters encoding for the thiosulfate:quinone oxidoreductase (TQO) subunits (doxA/doxD) and heterodisulfide reductase (HDR, hdrC1-hdrB1A-hyp-hdrC2-hdrB2) complex. The genes encoding for thiosulfate sulfurtransferase (TST, tst) were also detected in all species (Figure 3). According to the genome composition, we speculate that the pathway of sulfur metabolism for Metallosphaera might be SDO oxidized sulfur to sulfite, SAOR oxidized sulfite to sulfate, and TQO was responsible for the transformation of thiosulfate to tetrathionate, whereas TTH catalyzed the tetrathionate hydrolysis into thiosulfate, sulfur, and sulfate. However, the function of some proteins, especially for SDO and SAOR, had not been verified in Metallosphaera members. TST and HDR catalyzed the mutual conversion of thiol proteins (RSH) and sulfane sulfates (RSSH) in the cytoplasm (Chen et al., 2012), and HDR was also implicated in transferring electrons to reduce Fdox (Hua et al., 2018). Besides, tusA-dsrE2-dsrE3A gene clusters that functioned in cytoplasmic sulfur trafficking and dissimilatory tetrathionate oxidation were found located next to the hdr clusters in all Metallosphaera genomes, similar to other Sulfolobales members (Urbieta et al., 2017). The subunits SoeAB of the heterotrimeric membrane-bound sulfite-oxidizing enzyme complex SoeABC were detected in all Metallosphaera genomes, but the subunit SoeC was not identified in Metallosphaera sp. UBA165, Metallosphaera sp. Obs4, and other strains of My-r02, YNP-08, My-r06, My-r05, and YNP-14, probably resulting from the incompleteness of these genomes. Genes coding ATP sulfurylase (SAT, sat), adenosine-5′-phosphosulfate reductase (APR, apr) subunit AprA, and phosphoadenosine phosphosulfate reductase (PAPSr, papsr) that are involved in adenosine-5′-phosphosulfate (APS) pathway were only detected in M. sedula and M. yellowstonensis (Figure 3). Genes encoding for soxABC and doxBCE complex involved in sulfur oxidation and electron transfer were also annotated in Metallosphaera genomes. It was reported that sulfite was readily oxidized to sulfate through the direct SoeABC pathway and/or the indirect APS pathway presented in the cytoplasm in the purple sulfur bacterium Allochromatium vinosum (Christiane et al., 2013). Whether these pathways work in other phototrophic sulfur bacteria and sulfur-oxidized archaea remains unknown. Genes (e.g., hynS, hynL, hoxM, hypC, hypD, and isp 1) encoded for the structural and auxiliary proteins of Ni/Fe hydrogenase that are potentially associated with electron transfer between hydrogen and sulfur-containing compounds were only detected in M. sedula and M. yellowstonensis; however, genes hypE, hynY, and isp 2 were presented in all analysis strains. No genes of hypZ, hynZ, and hypY were found on the genomes of all strains (Laska et al., 2003) (Supplementary Figure S6).
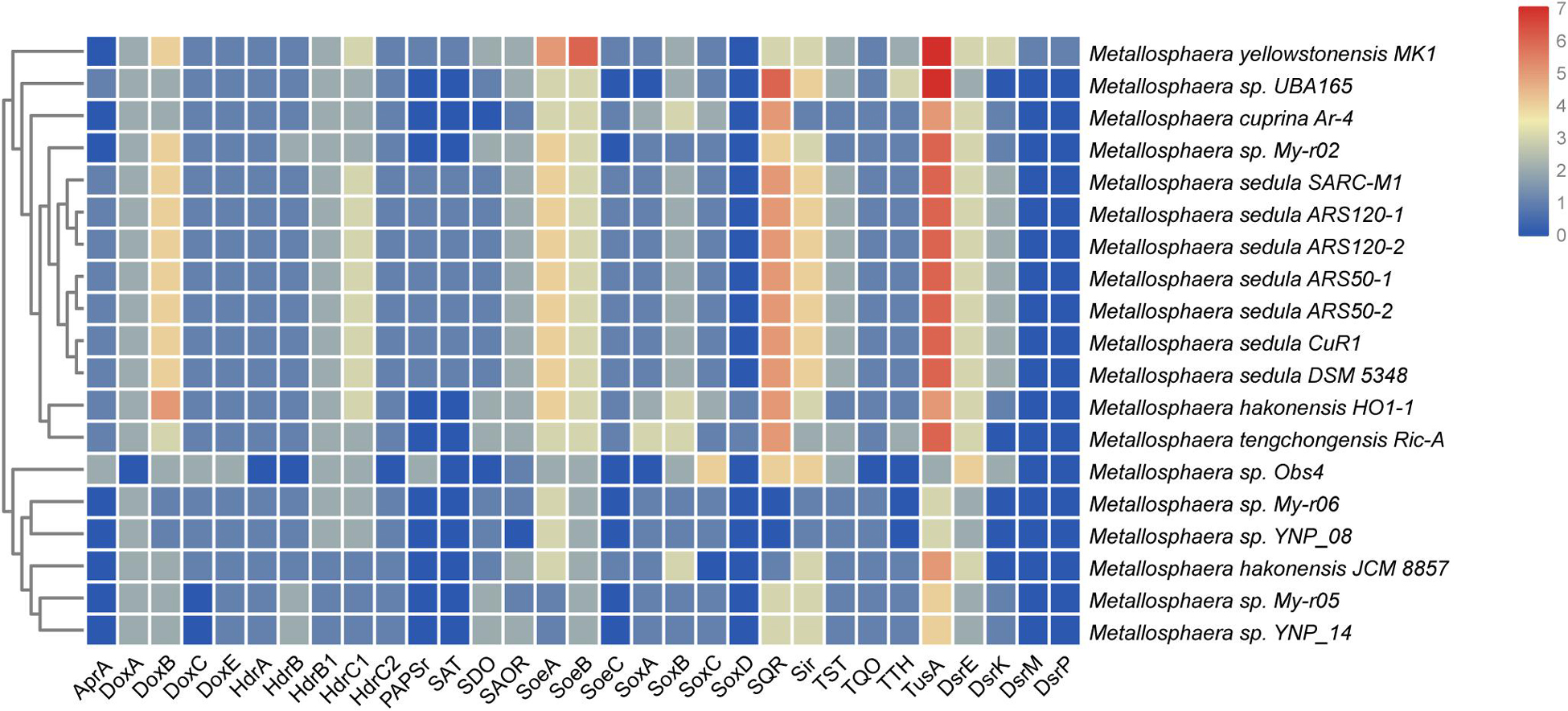
Figure 3. Heat map showing the distribution and numbers of genes encoding putative proteins in sulfur metabolism in various Metallosphaera strains. AprA, APS reductase; SAT, ATP sulfurylase; TST, thiosulfate sulfurtransferase; SDO, sulfur dioxygenase; DoxA/DoxD, thiosulfate:quinone oxidoreductase subunits; Sir, sulfite reductase; TTH, tetrathionate hydrolase; HdrA/HdrB/HdrC, heterodisulfide reductase complex subunits; PAPSr, phosphoadenosine phosphosulfate reductase; SQR, sulfide:quinone oxidoreductase; SoeABC, sulfite-oxidizing enzyme; CbsAB/SoxNL, terminal oxidase complex; DoxE, sulfocyanin; MCO, multicopper blue protein; doxBCE, terminal oxidase complex; TusA/DsrE, cytoplasmic sulfur trafficking proteins.
Iron Metabolism
The fox genes involved in iron oxidation (Kozubal et al., 2011) were detected in all Metallosphaera-type species, but M. cuprina may have lost the subunits foxD, foxE, foxF, and foxI (Supplementary Figure S7). The arrangement of fox genes was similar in Metallosphaera spp. following the pattern foxA-A′-I-B-C-D-E-F-J-G-H, but it is different from other members of iron-oxidizing Sulfolobales (Figure 4), and in M. hakonensis HO1-1, a third copy of foxA was found within this cluster, probably resulting from gene duplication (Figure 4). The transcription initiation directions of these fox genes were not consistent, and their open reading frames (ORFs) were separated by spacers (Figure 4). Genes encoding for terminal oxidase complex cbsAB/soxNL were found downstream of fox cluster in M. sedula and M. yellowstonensis, while in strain M. tengchongensis Ric-A, these two clusters were separated by a genome region of about 180 kb (Figure 4). A gene for sulfocyanin (SoxE), a blue copper-containing protein that may function as a temporary electron storage or electron carrier in the iron-oxidizing electron transport chain (Kozubal et al., 2011), occurred in all Metallosphaera genomes in this study. Genes encoding for multicopper blue protein (mco) that contained two plastocyanin type I copper domains were only detected in M. yellowstonensis and M. sedula (Kozubal et al., 2011). These oxidases may couple the reduction of oxygen to proton translocation in cooperation with Fox complex mentioned-above (Auernik and Kelly, 2008; Kozubal et al., 2011).
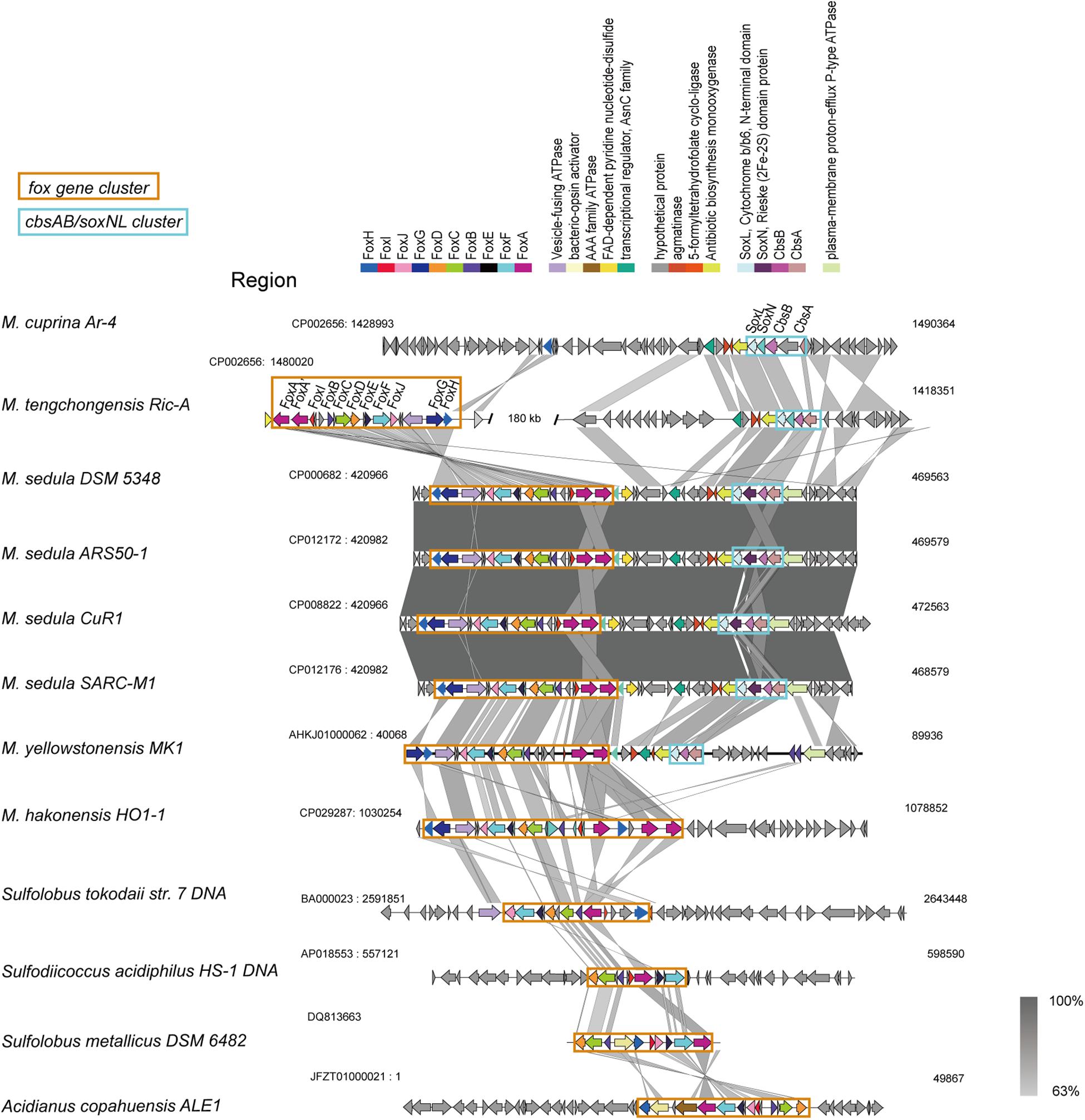
Figure 4. Synteny analysis of fox gene clusters and soxNL/cbsAB gene clusters derived from representative Metallosphaera strains and other representative species in Sulfolobales.
Carbon Metabolism
Metallosphaera spp. contained an abundant repertoire of carbohydrate-active enzymes (CAZymes) including carbohydrate esterases (CEs), carbohydrate binding molecules (CBMs), glycosyltransferases (GTs), glycoside hydrolases (GHs), auxiliary activity proteins (AAs), and a small number of polysaccharide lyases (PLs), of which GTs were most abundant, and Metallosphaera sp. Obs4 possessed the most carbohydrate active enzymes (Supplementary Figure S7). The diversity of encoding genes for CAZymes possibly reflected different carbohydrate metabolism strategies in Metallosphaera. A complete set of genes encoding for glycolysis, gluconeogenesis, the archaeal pentose phosphate pathway, as well as an atypical TCA cycle (replacing genes encoding the alpha-ketoglutarate dehydrogenase with those encoding 2-oxoacid:ferredoxin oxidoreductase) were detected in all Metallosphaera species. Metallosphaera spp. also contained complete non-phosphorylative and semi-phosphorylative Entner–Doudoroff (ED) pathways. Genes encoding homologous enzymes for the complete 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle and the dicarboxylate/4-hydroxybutyrate (DC/HB) cycle involved in autotrophic carbon fixation (Berg et al., 2007, 2010; Jiang et al., 2014) were also found in all Metallosphaera genomes (Supplementary Figure S9). It is reported that carbon monoxide was ubiquitous in hydrothermal habitats (King and Weber, 2007). All members of Metallosphaera in this study possessed putative type I carbon monoxide dehydrogenase (CODH) encoded by gene cluster coxLSM; however, the typical active site motif (VAYRCSFR) of CODH (Dobbek et al., 2002) was not observed in these proteins, which indicated that these CODH probably do not use CO; alternatively, they could possess a novel active site motif (Supplementary Figure S9).
Nitrogen Metabolism
All Metallosphaera strains possessed genes encoding for nitrate reductase and nitrite reductase involved in assimilatory nitrate reduction. Except for narG genes in seven strains of M. sedula, the narGHJI operon encoding for dissimilatory nitrate reductase was only found in M. yellowstonensis MK1 (Supplementary Figure S10). This operon was located on a GI in M. yellowstonensis MK1, suggesting that it might be acquired through horizontal gene transfer (HGT) events (Supplementary Table S2; Bertelli et al., 2017). Urease gene clusters consisting of the functional subunits (ureAB and ureC) and accessory proteins (ureE, ureF, and ureG) were found in all Metallosphaera species except for M. yellowstonensis (Supplementary Figure S10), indicating that urea assimilation was employed by most Metallosphaera strains to provide sufficient ammonia by converting one urea molecule into one carbon dioxide molecule plus two ammonia molecules. Genes encoding for nitrilase and formamidase, metabolizing organic nitrogen to ammonia, and genes encoding for ammonium transport (amt) and ammonia-dependent biosyntheses, such as carbamoyl-phosphate synthases (carAB), glutamate dehydrogenases (gdhA), and glutamine synthetases (glnA), were found in the genomes of all Metallosphaera species (Supplementary Figure S10). There is also a complete set of genes involved in arginine synthesis in all Metallosphaera species (Supplementary Figure S10). Polyamine derived from arginine can stabilize DNA by protecting DNA from free radical attacks and thermal denaturation (Abby et al., 2018). Polyamine biosynthesis-related genes that encoded for agmatinase (speB), S-adenosylmethionine decarboxylase (speD), and spermidine synthase (speE) were found in the genomes of all Metallosphaera species, while the arginine decarboxylase gene (speA) for the first two steps of putrescine biosynthesis was not detected; its function may have been substituted by S-adenosylmethionine decarboxylase (speD) (Giles and Graham, 2008).
Heavy Metal Resistance
Metallosphaera can survive natural and anthropogenic metal-rich environments, and BacMet database annotations revealed an abundant repertoire of heavy metal resistance genes in Metallosphaera spp., exhibiting diverse strategies to avert the deleterious effect of toxic metals on biological function (Supplementary Figure S11). Most of the genes related to arsenic resistance (aioAB: arsenite oxidase; arsABR: arsenical pump-driving ATPase; arsM: methyltransferase), divalent-cation resistance (copARZ, cueA, cutA, czcD, corRC, nccN, nikABCDER, and mntRH), mercury resistance (merA: mercuric reductase), and iron regulation (fecDE, furA, fbpC) were found in all tested Metallosphaera genomes. Alkylmercury lyase (encoded by gene merB) that cleaved mercury-alkyl bonds for mercury detoxification (Melnick and Gerard, 2007) was only detected in M. yellowstonensis. Genes encoding for “DNA-binding protein from starved cells” (Dps) (Blake et al., 2005), which can physically shield DNA against oxidative damage, were only detected in strains of M. sedula and Metallosphaera sp. UBA1654. Dps proteins were also able to control Fenton reaction through storing ferric oxide as a mineral core on their interior cage surface (Blake et al., 2005).
Adhesion and Motility
Etracellular polysaccharides (EPS) play a significant role in cell adhesion and biofilm formation, which is closely related to colonization, mineral solubilizing ability, and protection against adverse environmental conditions (Basak et al., 2014; Marino et al., 2018; Yu et al., 2019). Gene clusters encoding for d-TDP-glucose pyrophosphorylase (rfbA), d-TDP-glucose 4,6-dehydratase (rfbB), d-TDP-4-dehydrorhamnose 3,5-epimerase (rfbC), and d-TDP-4-dehydrorhamnose reductase (rfbD), which can convert glucose-1-phosphate to the EPS precursor d-TDP-rhamnose via a series of reactions, were found in all Metallosphaera genomes (Supplementary Figure S9). Motility conferred by flagella can provide a competitive advantage for microorganisms to move toward beneficial conditions. From the genomic and functional analysis, it is found that the sequenced archaea maintain a unique flagellum composition and mode of assembly, distinct from the bacteria. Archaeal flagellin commonly anchors on a fla locus, which was encoded by 7–13 flagella-related genes. Studies of the flagella system of the crenarchaeal model strain Sulfolobus acidocaldarius identified seven fla genes called flaB, flaX, flaG, flaF, flaH, flaI, and flaJ, and confirmed that all seven genes were essential for assembly and motility (Thomas et al., 2001; Lassak et al., 2012; Dominik et al., 2016). However, in the genomes of 19 strains of Metallosphaera, genes encoding for crenarchaeal flagellin (flaB) and flagella accessory proteins (flaH, flaI, flaG, flaF, and flaJ) were only detected in strains of M. sedula, M. yellowstonensis, and Metallosphaera sp. UBA165. Further, a unique crenarchaeal gene flaX, coding for a structural part of the archaeal flagellum assembly apparatus only present in Metallosphaera sp. Obs4 and Metallosphaera sp. My-r06 (Supplementary Figure S12). The roles of FlaX and other accessory proteins in Metallosphaera members need further experimental verification. Genome comparative results indicated that the flagellum assembly apparatus did not present in every genome of these strains, which is consistent with the motility difference of Metallosphaera strains known by their physiological research (Peng et al., 2015). The genomes of M. tengchongensis Ric-A and M. hakonensis DSM 7519 only contain Fla L. Both species do not possess flagella and do not have motility (Peng et al., 2015).
Mobile Genetic Elements and CRISPR-Cas Systems
Mobile genetic elements, including GIs, IS, transposons, and phages, are genome segments that display intra- and/or extracellular translocation abilities associated with HGT (Springael and Top, 2004). The prokaryotic CRISPR (clustered, regularly, interspaced, short, palindromic repeats)-Cas (CRISPR-associated genes) systems are attested to confer resistance to viral attack and mediate interactions between the host and phage. The results showed that in Metallosphaera, the number of transposon sequences per genome ranged from 65 (M. cuprina Ar-4) to 283 (M. yellowstonensis MK1) and the number of GI-related sequences per genome ranged from 21 (M. cuprina Ar-4) to 487 (M. yellowstonensis MK1) (Supplementary Tables S2, S3). GI regions of Metallosphaera spp. harbored genes related to the formation of extracellular polysaccharides (EPS) that account for ∼2.4% of all sequences, defense and DNA repair systems (∼2.7%), and stress resistance (∼2.0%). These additional functionalities endowed by HGT events may facilitate adaptive survival and protect Metallosphaera against DNA damage and protein denaturation in its acidic hot spring habitat (Yu et al., 2019). The number of prophages and prophage remnants ranged from 0 to 27 (M. sedula DSM5348) (Supplementary Table S3). Type I-A and/or type III-A/D CRISPR-Cas systems were found in all the genomes in this study except for Metallosphaera sp. My-r06. M. tengchongensis Ric-A contained the most (410) CRISPR-Cas-related genes or spacers. CRISPR-Cas-related sequences were detected in predicted GI regions of all Metallosphaera spp. except for M. cuprina Ar-4, reflecting the mobility of CRISPR-Cas systems (Koonin and Makarova, 2017; Krupovic et al., 2017; Peters et al., 2017).
Gene Ontology Enrichment and Evolutionary Analyses of Five Metallosphaera-Type Strains
We further applied the software OrthoVenn (Yi et al., 2015) and Count (Miklós, 2010) for gene clustering, GO enrichment analyses, and gene family evolutionary analyses of five Metallosphaera-type strains including M. tengchongensis Ric-A, M. sedula DSM 5348, M. hakonensis HO1-1, M. cuprina Ar-4, and M. yellowstonensis MK1. Results showed that 1614 (68.6%) out of 2353 identified gene families were shared by all species. M. sedula DSM 5348 had the most gene families (2133) in its genome, whereas M. cuprina Ar-4 had the fewest (1827), and M. yellowstonensis MK1 possessed the most strain-specific gene families (42) followed by M. tengchongensis Ric-A (18) (Figure 5). GO enrichment analyses showed that in significantly enriched (p-value < 0.05) commonly shared gene families, GO terms were associated with basic biological functions including translation (GO:0006412), rRNA binding (GO:0019843), and ribosome-related function (GO:0005840; GO:0003735). Functions significantly enriched (p-value < 0.05) in accessory genes families were related to electron transport chain (GO:0022900; GO:0008137; GO:0042773; GO:0048038; GO:0006741; GO:0008121; GO:0003951), lipid metabolism-related long-chain fatty acid-CoA ligase activity (GO:0004467), and nutrient transport and catabolic process of nitrogen sources such as urea (GO:0043419; GO:0009039) and carbon sources such as lactate (GO:0035873), mandelate (GO:0019596), glycolate (GO:0097339), benzoylformate (GO:0050695), and maleylacetate (GO:0018506) related to heterotrophic lifestyle. Proton symporter activity (GO:0015295; GO:0015538) involved in pH homeostasis, amino acid biosynthesis-related denitrification pathway (GO:0019333), and sulfur metabolism-related functions including sulfate assimilation (GO:0000103), hydrogen sulfide biosynthetic process (GO:0070814), SIR (GO:0050311) together with hydrogenase-related functions (GO:0033748; GO:0008901), arginine metabolism (GO:0016990; GO:0019547), and cysteine biosynthesis process (GO:0019344) were also enriched (p-value < 0.05) in accessory gene families (Figure 5). These functions and pathways probably reflected the adaptation of Metallosphaera spp. to acidic, sulfur-rich, heat, and metal-laden environments. Metallosphaera may take advantage of hydrogen sulfide for cysteine biosynthesis. Disulfide bonds in thermophilic proteins are omnipresent in thermophiles and help stabilize proteins against the harsh conditions (Beeby et al., 2005). In addition, cysteine-rich proteins such as disulfide oxidoreductase can be used by microorganisms to chelate heavy metal ions in the cytoplasm so as to reduce metal-induced reactive oxygen species (ROS), which is supported by previous results that Cu2+ exposure induced assimilatory sulfur metabolism for cysteine biosynthesis in Metallosphaera (Wheaton et al., 2016; Meslé et al., 2017). Interestingly, functions enriched (p-value < 0.05) in strain-specific gene families of M. yellowstonensis MK1 were related to aromatic compound metabolism such as catabolic process of benzoate (GO:0018623; GO:0043640), phthalate (GO:0046239; GO:0018796), naphthoate (GO:0018582), toluene (GO:0042203), and phenanthrene (GO:0042216) (Figure 5), which suggested that M. yellowstonensis MK1 was apt at utilizing a broader spectrum of organic carbon sources possessing great potential in bioremediation. Strain-specific gene families of M. tengchongensis Ric-A were enriched (p-value < 0.05) in methyltransferase-related functions (Figure 5); these genes are probably involved in arsenic detoxification by catalyzing the formation of volatile trimethylarsine from arsenite (Qin et al., 2006; Ai et al., 2017).
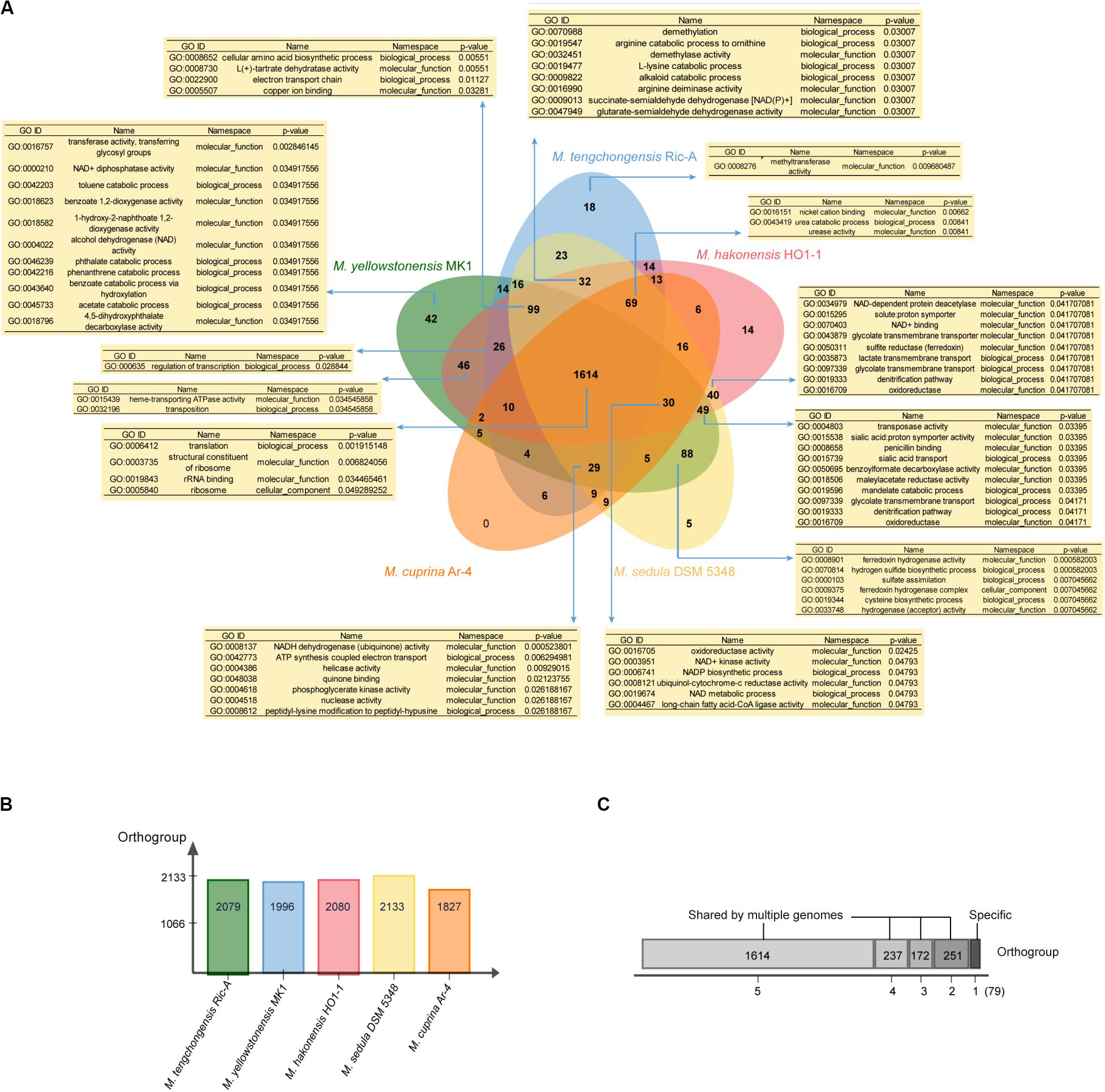
Figure 5. (A) Venn diagram showing the numbers of orthogroup including the core-genome (shared by all five Metallosphaera type strains), the accessory genome (shared by some but not at all of these strains), and strain-specific gene families in individual genomes; enriched Gene ontology (GO) terms (p < 0.05) in respective orthogroups are shown in the tables. (B) Bar chart showing the total numbers of orthogroup identified in each genome of the five Metallosphaera-type strains. (C) The total numbers of orthogroups of strain-specific gene families (=1) or the accessory genome (shared by 2, 3, and 4 strains) and core-genome of five Metallosphaera-type strains.
To decipher the evolutionary histories of the Metallosphaera species, gene family gain, loss, expansion, and contraction events were predicted by mapping the identified gene families onto the core-gene tree (Figure 6). A large number of gene family gain events occurred at node 1, accounting for ∼6% of gene families, and at the branches of M. yellowstonensis and M. sedula, accounting for ∼10 and 6% of gene families, respectively. By means of gene acquisition, members of Metallosphaera have largely expanded their genetic diversity, resulting in functional divergence, which was similar to other Archaea (Brügger et al., 2002). Of gene families undergoing gain events, about half of them were poorly characterized, and a considerable proportion of them were related to COG (X) Mobilome: prophages, transposons (∼6%), COG (K) Transcription (∼3%), and COG (V) Defense mechanisms (∼3%, which mostly were associated with CRISPR-Cas system) (Figure 6). Several sulfate assimilation and archaeal flagella biosynthesis-related genes were gained at the branches of M. yellowstonensis and M. sedula, indicating that sulfate assimilation and flagella biosynthesis were derived features of these strains (Supplementary Table S4). Several DNA damage repair genes (e.g., uve and spl) that helped maintain DNA fidelity were gained at the branches of M. hakonensis, M. tengchongensis, M. sedula, M. yellowstonensis, and node 1, and genes associated with EPS synthesis (e.g., rfa and gal) that enhanced colonization were gained in the branches of M. sedula and M. yellowstonensis (Supplementary Table S4). Several oxidoreductase encoding genes that associated with carbohydrate metabolism (e.g., porAB and acoAB) were gained at the branches of M. sedula and M. tengchongensis. A few of oxidoreductase genes related to aromatic compound degradation (e.g., hca and nfn) were gained at the branches of M. yellowstonensis, M. sedula, and node 2. Genes encoding for putative type I CODH were expanded in M. sedula but contracted in M. yellowstonensis (Supplementary Table S4). Sulfocyanin encoding genes were expanded in M. hakonensis, M. tengchongensis, and Heme/copper-type cytochrome/quinol oxidase encoding genes associated with sulfur and iron oxidation (Supplementary Table S4) were expanded in the branches of node 1 and M. yellowstonensis. Indicating their importance in niche adaption, several genes involved in nitrogen transport and metabolism were expanded in several species: ammonia permease encoding gene amtB in M. cuprina and M. tengchongensis, nitrite reductase encoding gene nirD in M. hakonensis and M. sedula, and adenosylmethionine decarboxylase encoding gene speD involved in polyamine biosynthesis in M. yellowstonensis. However, the urease encoding gene operon ureABCDEF was lost at the branch of M. yellowstonensis (Supplementary Table S4). In contrast to conspicuous gene acquisitions in other Metallosphaera strains, gene family loss events occurred frequently in M. cuprina taking up ∼6% of gene families, which mostly compromised genes belonging to COGs Energy production and conversion (C) and Carbohydrate transport and metabolism (G) such as permease of fucose, sugar, arabinose, dehydrogenase of succinate, tartrate, malate/L-lactate, aldehyde and altronate, and oxidase of sulfite (Supplementary Table S4). Genomic streamlining in adaptation to the acidic, thermal, and oligotrophic environment may be the main reason for gene family losses in M. cuprina; previous studies showed that growth temperature was negatively correlated with genome size in bacteria (Sabath et al., 2013) and that deletion of dispensable sequences from bacterial genomes led to dose-dependent growth (Kurokawa et al., 2016; Zhang et al., 2017; Ren et al., 2019).
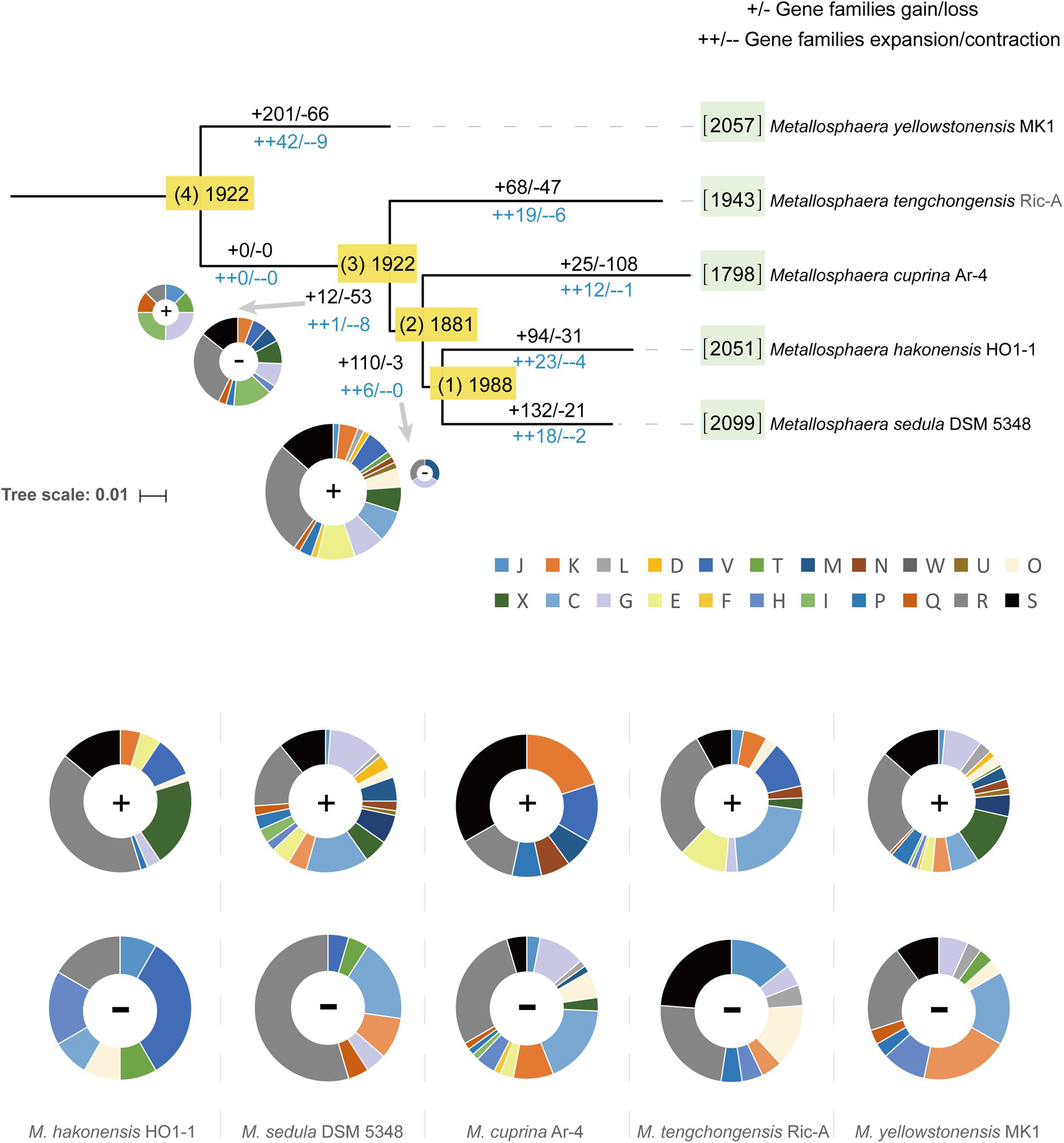
Figure 6. Ancestral genome content reconstruction of Metallosphaera species with COUNT software. The numbers of gene families of each strain were shown in brackets before the names of strains. The numbers of gain and loss events were marked at each lineage of the tree. “+”s represent gain events and “–”s represent loss events. The pie chart shows the numbers of gained genes by COG categories. A list of gained and lost genes for the Metallosphaera-type strains, and nodes 1 and 2 are shown in Supplementary Table S4. COG categories description (J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown).
Selective Pressure Analyses
Functions undergoing rapid evolution can be distinguished, taking into account the functional categories and selective pressure (Carretero-Paulet et al., 2015; Zhong et al., 2018). To gain insight into the conservation and evolution of various gene families in Metallosphaera, evolutionary pressure on each gene family that contained at least three non-identical sequences in five species of Metallosphaera was measured by calculating global substitution rates (dN/dS) of non-synonymous (dN) to synonymous (dS), as well as numbers of mutation sites under significant negative or positive selection in each gene family. Results indicated that there was pervasive strong purifying (negative) selection (dN/dS < 1) acting on the gene families of Metallosphaera with no gene families showing positive selection (dN/dS > 1), among them 86.6% gene families showing a dN/dS ratio lower than 0.1 (Figure 7 and Supplementary Table S5), which emphasized purifying selection contributing largely to the long-term stability of Metallosphaera genomes by removing deleterious mutations. Gene families of Metallosphaera exhibit different degrees of purifying selection pressure as shown by COG annotation. Genes undergoing the strongest purifying selection were those related to Transcription (K), indicating that these functions were highly conserved, while genes related to mobilome: prophages, transposons (X) and Defense mechanisms (V) were under weaker purifying pressure since they showed relatively higher dN/dS ratios and more sites under diversifying (positive) selection, which indicated that these genes could gain specific adaptive mutations associated with the acquisition of new or adaptive functions (Figure 7 and Supplementary Table S5).
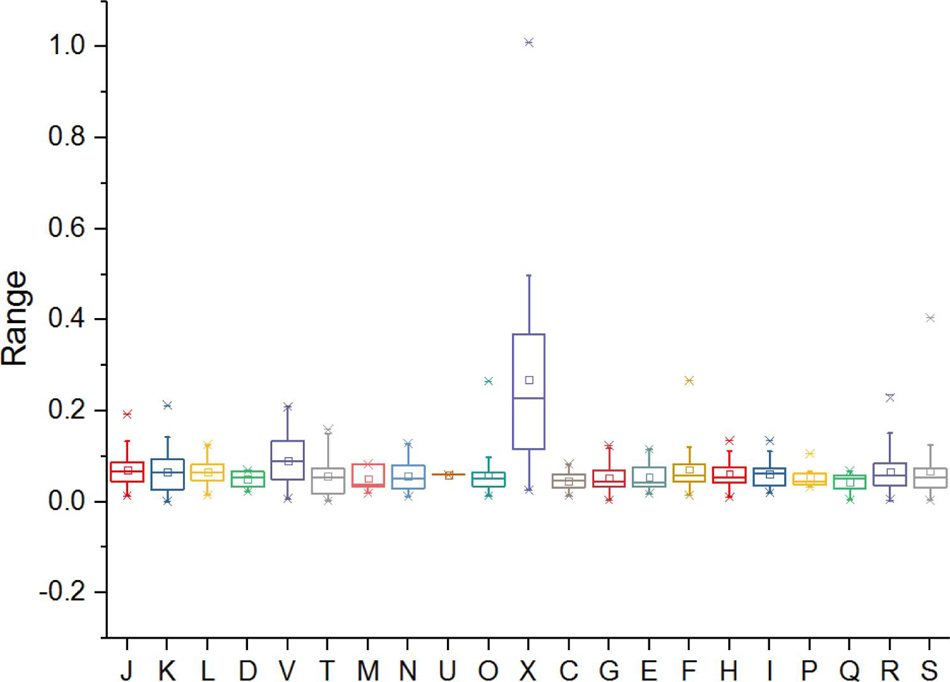
Figure 7. Distribution of selection pressures (indicated by values of dN/dS) on various gene families of Metallosphaera spp. based on COG categories. dN, non-synonymous (amino acid-changing) substitutions; dS, synonymous (amino acid-unchanging) substitutions. dN:dS < 1 indicates the action of purifying selection (negative selection), which purges deleterious mutations to conserve the protein structure and function (Carretero-Paulet et al., 2015).
Genome Expansion Through HGT
Horizontal gene transfer is one of the important engines that drive genomic diversity and adaptive evolution of microbes, especially in the case of those that inhabit extreme environments (Olga et al., 2009; Zhang et al., 2017; Li et al., 2019), which in this case also contributed considerably to genome contents of Metallosphaera. Based on COG annotation, the transferred genes in Metallosphaera spp. comprised mostly defensive and metabolic functions, with approximately 6.7% defense mechanisms (V), 6.4% energy production and conversion (C), 6.4% carbohydrate transport and metabolism (G), 4.9% amino acid transport and metabolism (E), 3.9% coenzyme transport and metabolism (H), 3.2% inorganic ion transport and metabolism (P), 2.1% lipid transport and metabolism (I), and 1.1% secondary metabolites biosynthesis, transport, and catabolism (Q) (Supplementary Figure S13). Informational proteins such as ribosomal proteins and RNA processing proteins experienced fewer HGT events in comparison with other gene families (Supplementary Figure S13). Most HGT events appeared to be acquired from the same domain (Archaea), which is typical among the identified HGTs (Figure 8 and Supplementary Table S6). Bacteria also contributed largely to the emerging genetic diversity of Metallosphaera through cross-domain HGT, and acquired genes such as hydrogenase and benzoate/toluate dioxygenase also appeared to facilitate adaption to different niches, resulting in the functional divergence within the genus of Metallosphaera (Figure 8 and Supplementary Table S6). However, it should be noted that though metagenome-assembled genomes (MAGs) included in this study have supplemented available genomic data of Metallosphaera and expanded sample size, these MAGs may contain contamination generated during the binning process. Thus, the results of MAGs should be interpreted carefully.
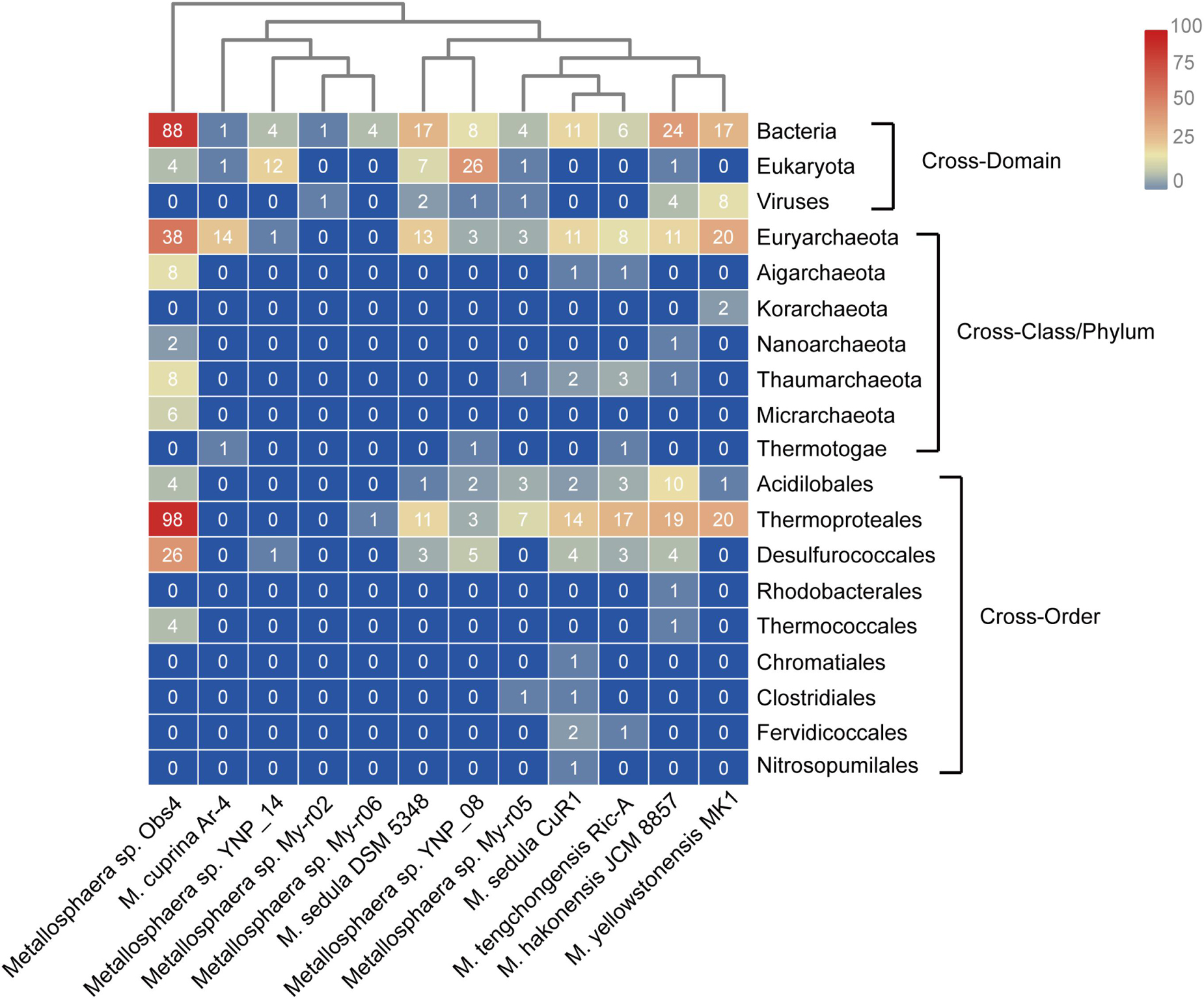
Figure 8. Heat map showing the number and distribution of predicted horizontally transferred genes in different Metallosphaera strains and their predicted donors (right).
Conclusion
In this study, the genome of a Metallosphaera species (M. tengchongensis) was comparatively analyzed with 18 other genomes from Metallosphaera strains to enrich our understanding of the genetic traits, metabolism, and environmental adaption mechanism of the Metallosphaera and shed light on their evolutionary history. Metallosphaera microbes were widely found in sulfur-rich and metal-burdened environments, and all Metallosphaera strains analyzed in this study contained sulfur and iron oxidation genes. However, the APS pathway was only detected in M. sedula and M. yellowstonensis, and certain subunits of the fox cluster were lost in M. cuprina. Gene deficiency of SOR hinted that new unusual enzymes might undertake the S0 oxidation role. Complete TCA cycles and ED pathways coexisted with HP/HB and DC/HB cycles in all genomes of these strains and supported their heterotrophic, autotrophic, and mixotrophic growth modes. The genes for assimilatory nitrate reduction were present in all genomes of 19 strains; however, a complete dissimilatory nitrate reductase gene cluster was only found in M. yellowstonensis, which demonstrated that Metallosphaera species and strains have different abilities to use inorganic nitrogen. Genes encoding for flagellin and flagella accessory proteins were only detected in strains of M. sedula, M. yellowstonensis, Metallosphaera sp. Obs4, Metallosphaera sp. UBA165, and Metallosphaera sp. My_r06. We also found that functions related to assimilatory sulfur metabolism and cysteine biosynthesis associated with ROS reduction were significantly enriched in accessory gene families. Evolutionary analyses showed that massive gene family gain events occurred at the branches of M. yellowstonensis and M. sedula, whereas considerable gene family loss events occurred in M. cuprina and pervasive strong purifying selection was found acting on the gene families of Metallosphaera. We also found that HGT played an important role in shaping the genetic and functional diversity of Metallosphaera. These findings provide a data basis for subsequent studies of metabolism and environmental adaption mechanisms in thermophilic Archaea, and for strategies to design cellular biocatalysts for the biomining process.
Data Availability Statement
The datasets generated for this study can be found in the genome sequence of Metallosphaera tengchongensis strain Ric-A has been deposited at JGI IMG-ER database under the IMG Taxon OID 2821472399, and Genbank database under accession number CP049074.
Author Contributions
C-YJ, S-JL, and HY designed and coordinated the study. PW, LZL, and LJL performed the bioinformatics analysis. YQ, ZL, and XL carried out the experiments and interpreted data for the work. PW and LZL wrote the manuscript. C-YJ and S-JL edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (grants 31670124 and 31600040), the Major Research Plan of National Natural Science Foundation of China (grant 91851206), Key Research Program of Chinese Academy of Sciences (ZDRW-ZS-2018-1), Basic Work of Science and Technology in the Ministry of Science and Technology (grant 2015FY110100), and the National Basic Research Program of China (2014CB846000).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01192/full#supplementary-material
FIGURE S1 | Phylogenetic tree of Metallosphaera spp. with (A) Maximum Likelihood (ML) method; (B) Minimum Evolution (ME) method; (C) UPGMA method; (D) Maximum Parsimony (MP) method based on 16S rRNA sequences. Bootstrap values are indicated at each node based on a total of 1000 bootstrap replicates.
FIGURE S2 | Whole genome alignment with Mauve showing syntenic blocks between type strains M. tengchongensis Ric-A, M. cuprina Ar-4, M. sedula DSM 5348, and M. hakonensis HO1-1 (from top to bottom).
FIGURE S3 | BlastN-based whole genome comparison of strains M. cuprina Ar-4, M. hakonensis HO1-1, Metallosphaera sp. My-r06,M. sedula DSM 5348, M. tengchongensis Ric-A, and M. yellowstonensis MK1 (from inside to outside) with BRIG, and these strains were used as reference, respectively. GC content and GC skew (inner rings 1 and 2) of each genome were also indicated.
FIGURE S4 | Phylogenetic tree of Metallosphaera spp. rooted by Encephalitozoon cuniculi GB M1 and Prochlorococcus marinus MIT 9301 based on whole genome sequences using CVTree 3.
FIGURE S5 | Phylogenetic tree of Metallosphaera spp. and other members of Sulfolobales based on whole genome sequences using CVTree 3.
FIGURE S6 | Heat map showing the distribution and number of genes encoding putative hydrogenase related proteins in different Metallosphaera strains.
FIGURE S7 | Heat map showing the distribution and numbers of genes encoding putative proteins involved in iron oxidation in different Metallosphaera strains.
FIGURE S8 | Heat map showing the distribution and numbers of genes encoding carbohydrate active enzymes (CAZymes) in different Metallosphaera strains. Abbreviations in the chart: AA, auxiliary activities proteins; CBM, carbohydrate-binding molecules; CE, carbohydrate esterases; GH, glycoside hydrolases; GT, glycosyltransferases; PL, polysaccharide lyases.
FIGURE S9 | Heat map showing the distribution and numbers of genes encoding putative enzymes involved in carbon metabolism in different Metallosphaera strains. Abbreviation and KO term descriptions in the chart: CoxL/M/S, Carbon monoxide dehydrogenase large/medium/small chain; GalE, UDP-glucose 4-epimerase; RfbA, d-TDP-glucose pyrophosphorylase; RfbB, d-TDP-glucose 4;6-dehydratase; RfbC, d-TDP-4-dehydrorhamnose 3;5-epimerase; RfbD, d-TDP-4-dehydrorhamnose reductase; K01895, acetyl-CoA synthetase; K00172, pyruvate ferredoxin oxidoreductase gamma subunit; K00626, acetyl-CoA C-acetyltransferase; K01681 aconitate hydratase; K01007, pyruvate; water dikinase (EC:2.7.9.2); K01964 acetyl-CoA/propionyl-CoA carboxylase (EC:6.4.1.2); K15016, enoyl-CoA hydratase; K15017, malonyl-CoA/succinyl-CoA reductase; K01595, phosphoenolpyruvate carboxylase; K15039 3-hydroxypropionate dehydrogenase (NADP+) (EC:1.1.1.298); K15018, 3-hydroxypropionyl-coenzyme A synthetase; K15019 3-hydroxypropionyl-coenzyme A dehydratase; K14534, -hydroxybutyryl-CoA dehydratase; K00031, isocitrate dehydrogenase; K00024, malate dehydrogenase; K15020, acryloyl-coenzyme A reductase; K14466, 4-hydroxybutyrate-CoA ligase; K05606, methylmalonyl-CoA; K00175, 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase; K01678, fumarate hydratase; K01848 methylmalonyl-CoA mutase; K18859, succinate dehydrogenase/fumarate reductase; K01902, succinyl-CoA synthetase.
FIGURE S10 | Heat map showing the distribution and numbers of genes encoding putative proteins involved in nitrogen metabolism in different Metallosphaera strains. Abbreviation descriptions in the chart: GdhA, glutamate dehydrogenase [NAD(P)+] (EC 1.4.1.3); GlnA, glutamine synthetase (EC 6.3.1.2); CarA, carbamoyl-phosphate synthase small subunit (EC 6.3.5.5); CarB, carbamoyl-phosphate synthase large subunit (EC 6.3.5.5); NarG, nitrate reductase/nitrite oxidoreductase, alpha subunit (EC 1.7.5.1; 1.7.99.4); NarH, nitrate reductase/nitrite oxidoreductase, beta subunit (EC 1.7.5.1; 1.7.99.4); NarJ, nitrate reductase delta subunit; NarI, nitrate reductase gamma subunit (EC 1.7.5.1; 1.7.99.4); Nrt, nitrate/nitrite transporter; Amt, ammonium transporter; UreAB, UreCD, urease functional subunits; UreE, UreF, UreG, urease accessory proteins; ArgH, argininosuccinate lyase; ArgI, ornithine carbamoyltransferase; ArcA, arginine decarboxylase; ArgG, argininosuccinate synthase; SpeB, agmatinase; SpeD, S-adenosylmethionine decarboxylase; SpeE, spermidine synthase.
FIGURE S11 | Heat map showing patterns of the distribution and numbers of genes encoding putative proteins involved in heavy metal resistance in different Metallosphaera strains.
FIGURE S12 | Heat map showing patterns of the distribution and numbers of genes encoding putative proteins involved in flagella biosynthesis in different Metallosphaera strains.
FIGURE S13 | The COG proportions of predicted horizontal transferred genes in Metallosphaera spp.
TABLE S1 | Average nucleotide identity (ANI) based on whole genome alignments of Metallosphaera spp.
TABLE S2 | Statistics of genomic islands in the genomes of Metallosphaera spp.
TABLE S3 | Statistics of Mobile genetic elements (MGEs) (A), prophages and prophage remnants (B), Transposons (C), CRISPR-Cas related sequences (D) present in the genomes of Metallosphaera spp.
TABLE S4 | Gene families undergoing gain (A), loss (B), expansion (C), and contraction (D) events in Metallosphaera spp. detected with COUNT.
TABLE S5 | Selection pressure on gene families of Metallosphaera spp.
TABLE S6 | Putative horizontally transferred genes of Metallosphaera spp.
Footnotes
- ^ https://jgi.doe.gov/user-programs/pmo-overview/protocols-sample-preparation-information/
- ^ https://www.pacb.com/support/software-downloads/
- ^ https://pypi.org/project/pyani/
- ^ http://www-is.biotoul.fr
- ^ https://www.pathogenomics.sfu.ca/islandviewer/
- ^ http://phaster.ca/
- ^ https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index
- ^ https://orthovenn2.bioinfotoolkits.net/home
- ^ http://www.datamonkey.org/fel/
References
Abby, S. S., Melcher, M., Kerou, M., Krupovic, M., Stieglmeier, M., Rossel, C., et al. (2018). Candidatus Nitrosocaldus cavascurensis, an ammonia oxidizing, extremely thermophilic archaeon with a highly mobile genome. Front. Microbiol. 9:28. doi: 10.3389/fmicb.2018.00028
Ai, C., McCarthy, S., Eckrich, V., Rudrappa, D., Qiu, G., and Blum, P. (2016). Increased acid resistance of the archaeon, Metallosphaera sedula by adaptive laboratory evolution. J. Ind. Microbiol. Biotechnol. 43, 1–11.
Ai, C., McCarthy, S., Liang, Y., Rudrappa, D., Qiu, G., and Blum, P. (2017). Evolution of copper arsenate resistance for enhanced enargite bioleaching using the extreme thermoacidophile Metallosphaera sedula. J. Ind. Microbiol. Biotechnol. 44, 1613–1625. doi: 10.1007/s10295-017-1973-5
Alber, B., Olinger, M., Rieder, A., Kockelkorn, D., Jobst, B., Hügler, M., et al. (2006). Malonyl-coenzyme a reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp. J. Bacteriol. 188, 8551–8559. doi: 10.1128/jb.00987-06
Alikhan, N. F., Petty, N. K., Zakour, N. L. B., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Amend, J. P., and Shock, E. L. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and bacteria. FEMS Microbiol. Rev. 25, 175–243. doi: 10.1111/j.1574-6976.2001.tb00576.x
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21.
Auernik, K. S., and Kelly, R. M. (2008). Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 74, 7723–7732. doi: 10.1128/aem.01545-08
Auernik, K. S., and Kelly, R. M. (2010). Impact of molecular hydrogen on chalcopyrite bioleaching by the extremely thermoacidophilic archaeon Metallosphaera sedula. Appl. Environ. Microbiol. 76, 2668–2672. doi: 10.1128/aem.02016-09
Auernik, K. S., Maezato, Y., Blum, P. H., and Kelly, R. M. (2008). The genome sequence of the metal-mobilizing, extremely thermoacidophilic Archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 74, 682–692. doi: 10.1128/aem.02019-07
Basak, G., Lakshmi, V., Chandran, P., and Das, N. (2014). Removal of Zn(ii) from electroplating effluent using yeast biofilm formed on gravels: batch and column studies. J. Environ. Health Sci. Eng. 12:8.
Bathe, S., and Norris, P. R. (2007). Ferrous iron- and sulfur-induced genes in Sulfolobus metallicus. Appl. Environ. Microbiol. 73, 2491–2497. doi: 10.1128/aem.02589-06
Beeby, M., O’Connor, B. D., Ryttersgaard, C., Boutz, D. R., and Yeates, T. O. (2005). The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 3:e309. doi: 10.1371/journal.pbio.0030309
Berg, I. A., Daniel, K., Wolfgang, B., and Georg, F. (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318, 1782–1786. doi: 10.1126/science.1149976
Berg, I. A., Ramos-Vera, W. H., Petri, A., Huber, H., and Fuchs, G. (2010). Study of the distribution of autotrophic CO2 fixation cycles in crenarchaeota. Microbiology 156, 256–269. doi: 10.1099/mic.0.034298-0
Bertelli, C., Laird, M. R., Williams, K. P., Lau, B. Y., Hoad, G., Winsor, G. L., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, 30–35.
Blake, W., Jesse, M., Deborah, W., Mark, Y., Dryden, K. A., Young, M., et al. (2005). An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc. Natl. Acad. Sci. U.S.A. 102, 10551–10556. doi: 10.1073/pnas.0501497102
Brügger, K., Redder, P., She, Q. X., Confalonieri, F., Zivanovic, Y. V., and Garrett, R. A. (2002). Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 206, 131–141. doi: 10.1016/s0378-1097(01)00504-3
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Carretero-Paulet, L., Chang, T. H., Librado, P., Ibarra-Laclette, E., Herrera-Estrella, L., Rozas, J., et al. (2015). Genome-wide analysis of adaptive molecular evolution in the carnivorous plant Utricularia gibba. Genome Biol. Evol. 7, 444–456. doi: 10.1093/gbe/evu288
Castro, C., and Donati, E. (2016). Effects of different energy sources on cell adhesion and bioleaching of a chalcopyrite concentrate by extremophilic archaeon Acidianus copahuensis. Hydrometallurgy 162, 49–56. doi: 10.1016/j.hydromet.2016.02.014
Chaudhari, N. M., Kumar, G. V., and Chitra, D. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373.
Chen, I. A., Markowitz, V. M., Chu, K., Palaniappan, K., Szeto, E., Huang, J., et al. (2017). IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D573.
Chen, L. X., Ren, Y. L., Lin, J. Q., Liu, X. M., Pang, X., Lin, J. Q., et al. (2012). Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS One 7:e39470. doi: 10.1371/journal.pone.0039470
Chen, Z. W., Jiang, C. Y., She, Q., Liu, S. J., and Zhou, P. J. (2005). Key role of cysteine residues in catalysis and subcellular localization of sulfur oxygenase reductase of Acidianus tengchongensis. Appl. Environ. Microbiol. 71, 621–628. doi: 10.1128/aem.71.2.621-628.2005
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read smrt sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Christiane, D., Bettina, F., Daniela, H., Anne, K., and Renate, Z. (2013). Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159, 2626–2638. doi: 10.1099/mic.0.071019-0
Couvin, D., Bernheim, A., Toffanonioche, C., Touchon, M., Michalik, J., Néron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251.
Dai, X., Wang, H., Zhang, Z., Li, K., Zhang, X., Mora-López, M., et al. (2016). Genome sequencing of Sulfolobus sp. A20 from costa rica and comparative analyses of the putative pathways of carbon, nitrogen, and sulfur metabolism in various Sulfolobus strains. Front. Microbiol 7:1902. doi: 10.3389/fmicb.2016.01902
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Dobbek, H., Gremer, L., Kiefersauer, R., Huber, R., and Meyer, O. (2002). Catalysis at a dinuclear [cusmo(=o)oh] cluster in a co dehydrogenase resolved at 1.1-a resolution. Proc. Natl. Acad. Sci. U.S.A. 99, 15971–15976. doi: 10.1073/pnas.212640899
Dominik, E., Lena, H., Khoa, P. T., Bräsen, C., Wen, Q., Wright, P. C., et al. (2016). Protein phosphorylation and its role in archaeal signal transduction. FEMS Microbiol. Rev. 40, 625–647. doi: 10.1093/femsre/fuw020
Dopson, M., Baker-Austin, C., and Bond, P. L. (2005). Analysis of differential protein expression during growth states of Ferroplasma strains and insights into electron transport for iron oxidation. Microbiology 151, 4127–4137. doi: 10.1099/mic.0.28362-0
Farris, J. S. (1970). Methods for computing Wagner trees. Syst. Zool. 19, 83–92. doi: 10.1093/sysbio/19.1.83
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, 222–230.
Fuchs, T., Huber, H., Teiner, K., Burggraf, S., and Stetter, K. O. (1995). Metallosphaera prunae, sp. nov. a novel metal-mobilizing, thermoacidophilic archaeum, isolated from a uranium mine in Germany. Syst. Appl. Microbiol. 8, 560–566. doi: 10.1016/s0723-2020(11)80416-9
Giles, T. N., and Graham, D. E. (2008). Crenarchaeal arginine decarboxylase evolved from an s-adenosylmethionine decarboxylase enzyme. J. Biol. Chem. 283, 25829–25838. doi: 10.1074/jbc.m802674200
Han, C. J., and Kelly, R. M. (2015). Biooxidation capacity of the extremely thermoacidophilic archaeon Metallosphaera sedula under bioenergetic challenge. Biotechnol. Bioeng. 58, 617–624. doi: 10.1002/(sici)1097-0290(19980620)58:6<617::aid-bit7>3.0.co;2-l
Hua, Z. S., Qu, Y. N., Zhu, Q., Zhou, E. M., Qi, Y. L., Yin, Y. R., et al. (2018). Genomic inference of the metabolism and evolution of the archaeal phylum Aigarchaeota. Nat. Commun. 9:2832.
Huber, G., Spinnler, C., Gambacorta, A., and Stetter, K. O. (1989). Metallosphaera sedula gen, and sp. nov. represents a new genus of aerobic, metal-mobilizing, Thermoacidophilic Archaebacteria. Syst. Appl. Microbiol. 12, 38–47. doi: 10.1016/s0723-2020(89)80038-4
Jiang, C. Y., Liu, L. J., Guo, X., You, X. Y., Liu, S. J., and Poetsch, A. (2014). Resolution of carbon metabolism and sulfur-oxidation pathways of Metallosphaera cuprina Ar-4 via comparative proteomics. J. Proteomics 109, 276–289. doi: 10.1016/j.jprot.2014.07.004
King, G. M., and Weber, C. F. (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5, 277–310.
Kletzin, A. (1992). Molecular characterization of the sor gene, which encodes the sulfur oxygenase/reductase of the thermoacidophilic Archaeum Desulfurolobus ambivalens. J. Bacteriol. 174, 5854–5859. doi: 10.1128/jb.174.18.5854-5859.1992
Koonin, E. V., and Makarova, K. S. (2017). Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. Genome Biol. Evol. 9, 2812–2825. doi: 10.1093/gbe/evx192
Kosakovsky Pond, S. L., and Frost, S. D. (2005). Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22, 1208–1222. doi: 10.1093/molbev/msi105
Kozubal, M., Macur, R. E., Korf, S., Taylor, W. P., Ackerman, G. G., Nagy, A., et al. (2008). Isolation and distribution of a novel iron-oxidizing crenarchaeon from acidic geothermal springs in Yellowstone national park. Appl. Environ. Microbiol. 74, 942–949. doi: 10.1128/aem.01200-07
Kozubal, M. A., Dlakic, M., Macur, R. E., and Inskeep, W. P. (2011). Terminal oxidase diversity and function in “Metallosphaera yellowstonensis”: gene expression and protein modeling suggest mechanisms of Fe(II) oxidation in the Sulfolobales. Appl. Environ. Microbiol. 77, 1844–1853. doi: 10.1128/aem.01646-10
Krupovic, M., Béguin, P., and Koonin, E. V. (2017). Casposons: mobile genetic elements that gave rise to the CRISPR-Cas adaptation machinery. Curr. Opin. Microbiol. 38, 36–43. doi: 10.1016/j.mib.2017.04.004
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, S., Stecher, G., Peterson, D., and Tamura, K. (2012). MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28, 2685–2686. doi: 10.1093/bioinformatics/bts507
Kurokawa, M., Seno, S., Matsuda, H., and Ying, B. W. (2016). Correlation between genome reduction and bacterial growth. DNA Res. 23, 517–525. doi: 10.1093/dnares/dsw035
Laska, S., Lottspeich, F., and Kletzin, A. (2003). Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology 149, 2357–2371. doi: 10.1099/mic.0.26455-0
Lassak, K., Neiner, T., Ghosh, A., Klingl, A., Wirth, R., and Albers, S. V. (2012). Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83, 110–124. doi: 10.1111/j.1365-2958.2011.07916.x
Le, H., Han, Z., Wu, P., Entwistle, S., Li, X., Tanner, Y., et al. (2018). DbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 46, 516–521.
Li, L., Liu, Z., Meng, D., Liu, X., Li, X., Ming, Z., et al. (2019). Comparative genomic analysis reveals the distribution, organization, and evolution of metal resistance genes in the genus Acidithiobacillus. Appl. Environ. Microbiol. 85:e02153-18. doi: 10.1128/AEM.02153-18
Liu, L. J., You, X. Y., Guo, X., Liu, S. J., and Jiang, C. Y. (2011a). Metallosphaera cuprina sp. nov., an acidothermophilic, metal-mobilizing archaeon. Int. J. Syst. Evol. Microbiol. 61, 2395–2400. doi: 10.1099/ijs.0.026591-0
Liu, L. J., You, X. Y., Zheng, H., Wang, S., Jiang, C. Y., and Liu, S. J. (2011b). Complete genome sequence of Metallosphaera cuprina, a metal sulfide-oxidizing archaeon from a hot spring. J. Bacteriol. 193, 3387–3388. doi: 10.1128/jb.05038-11
Marino, M., Maifreni, M., and Baggio, A. (2018). Innocente N1 inactivation of foodborne bacteria biofilms by aqueous and gaseous ozone. Front. Microbiol. 28:2024. doi: 10.3389/fmicb.2018.02024
Melnick, J. G., and Gerard, P. (2007). Cleaving mercury-alkyl bonds: a functional model for mercury detoxification by MerB. Science 317, 225–227. doi: 10.1126/science.1144314
Meslé, M., Beam, J. P., Jay, Z. J., Bodle, B., Bogenschutz, E. L., and Inskeep, W. P. (2017). Hydrogen peroxide cycling in high-temperature acidic geothermal springs and potential implications for oxidative stress response. Front. Mar. Sci. 4:130. doi: 10.3389/fmars.2017.00130
Michael, H., Krieger, R. S., Martina, J., and Georg, F. (2010). Characterization of acetyl-CoA/propionyl-CoA carboxylase in Metallosphaera sedula. Carboxylating enzyme in the 3-hydroxypropionate cycle for autotrophic carbon fixation. Eur. J. Biochem. 270, 736–744. doi: 10.1046/j.1432-1033.2003.03434.x
Miklós, C. (2010). Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26, 1910–1912. doi: 10.1093/bioinformatics/btq315
Olga, Z., Swithers, K. S., Pascal, L., Fournier, G. P., Bickhart, D. M., DeBoy, R. T., et al. (2009). On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc. Natl. Acad. Sci. U.S.A. 106, 5865–5870. doi: 10.1073/pnas.0901260106
Orell, A., Navarro, C. A., Arancibia, R., Mobarec, J. C., and Jerez, C. A. (2010). Life in blue: copper resistance mechanisms of bacteria and Archaea used in industrial biomining of minerals. Biotechnol. Adv. 28, 839–848. doi: 10.1016/j.biotechadv.2010.07.003
Pal, C., Bengtsson-Palme, J., Rensing, C., Kristiansson, E., and Larsson, D. G. (2014). BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 42, 737–743.
Parks, D. H., Rinke, C., Chuvochina, M., Chaumeil, P. A., Woodcroft, B. J., Evans, P. N., et al. (2017). Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542. doi: 10.1038/s41564-017-0012-7
Peeples, T. L., and Kelly, R. M. (1995). Bioenergetic response of the extreme thermoacidophile Metallosphaera sedula to thermal and nutritional stresses. Appl. Environ. Microbiol. 61, 2314–2321. doi: 10.1128/aem.61.6.2314-2321.1995
Peng, T. J., Liu, L. J., Liu, C., Yang, Z. F., Liu, S. J., and Jiang, C. Y. (2015). Metallosphaera tengchongensis sp. nov., an acidothermophilic archaeon isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 65, 537–542. doi: 10.1099/ijs.0.070870-0
Peters, J. E., Makarova, K. S., Shmakov, S., and Koonin, E. V. (2017). Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc. Natl. Acad. Sci. U.S.A. 114, 7358–7366.
Pond, S. L. K., and Muse, S. V. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. doi: 10.1093/bioinformatics/bti079
Qin, J., Rosen, B. P., Zhang, Y., Wang, G., Franke, S., and Rensing, C. (2006). Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103, 2075–2080. doi: 10.1073/pnas.0506836103
Qin, Q. L., Xie, B. B., Zhang, X. Y., Chen, X. L., Zhou, B. C., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/jb.01688-14
Qin, Y. L., Liang, Z. L., Song, Y., Wang, B. J., Liu, S. J., and Jiang, C. Y. (2019). Amplicon-based high-throughput sequencing reveals the microbial diversity in Rehai hot springs, Tengchong, Yunnan Province, (in chinese). Microbiol. China 46, 2482–2493.
Rawlings, D. E. (2005). Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microbial. Cell Fact. 4, 1–15.
Ren, M., Feng, X., Huang, Y., Wang, H., Hu, Z., Clingenpeel, S., et al. (2019). Phylogenomics suggests oxygen availability as a driving force in Thaumarchaeota evolution. ISME J. 13, 2150–2161. doi: 10.1038/s41396-019-0418-8
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rohwerder, T., Gehrke, T., Kinzler, K., and Sand, W. (2003). Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 63, 239–248.
Sabath, N., Ferrada, E., Barve, A., and Wagner, A. (2013). Growth temperature and genome size in bacteria are negatively correlated, suggesting genomic streamlining during thermal adaptation. Genome Biol. Evol. 5, 966–977. doi: 10.1093/gbe/evt050
Sebastian, E., Michael, H., Wolfgang, E., Katharina, W., Berg, I. A., Ramos-Vera, W. H., et al. (2011). Labeling and enzyme studies of the central carbon metabolism in Metallosphaera sedula. J. Bacteriol. 193, 1191–1200. doi: 10.1128/jb.01155-10
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Selengut, J. D., Haft, D. H., Tanja, D., Anurhada, G., Michelle, G. G., Nelson, W. C., et al. (2007). TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 35, D260–D264.
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36.
Singer, S. W., Chan, C. S., Zemla, A., VerBerkmoes, N. C., Hwang, M., Hettich, R. L., et al. (2008). Characterization of cytochrome 579, an unusual cytochrome isolated from an iron-oxidizing microbial community. Appl. Environ. Microbiol. 74, 4454–4462. doi: 10.1128/aem.02799-07
Springael, D., and Top, E. M. (2004). Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol. 12, 53–58. doi: 10.1016/j.tim.2003.12.010
Takayanagi, S., Kawasaki, H., Sugimori, K., Yamada, T., Sugai, A., Ito, T., et al. (1996). Sulfolobus hakonensis sp. nov., a novel species of acidothermophilic archaeon. Int. J. Syst. Bacteriol. 46, 377–382. doi: 10.1099/00207713-46-2-377
Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., et al. (2001). The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29, 22–28. doi: 10.1093/nar/29.1.22
Thomas, N. A., Bardy, S. L., and Jarrell, K. F. (2001). The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25, 147–174. doi: 10.1111/j.1574-6976.2001.tb00575.x
Ulrike, K., Sly, L. I., and Mcewan, A. G. (2005). Respiratory gene clusters of Metallosphaera sedula - differential expression and transcriptional organization. Microbiology 151, 35–43. doi: 10.1099/mic.0.27515-0
Urbieta, M. S., Donati, E. R., Chan, K. G., Shahar, S., Sin, L. L., and Goh, K. M. (2015). Thermophiles in the genomic era: biodiversity, science, and applications. Biotechnol. Adv. 33, 633–647. doi: 10.1016/j.biotechadv.2015.04.007
Urbieta, M. S., Rascovan, N., Vázquez, M. P., and Donati, E. (2017). Genome analysis of the thermoacidophilic archaeon Acidianus copahuensis focusing on the metabolisms associated to biomining activities. BMC Genomics 18:445. doi: 10.1186/s12864-017-3828-x
Urich, T., Gomes, C. M., Kletzin, A., and Frazao, C. (2006). X-ray structure of a self-compartmentalizing sulfur cycle metalloenzyme. Science 311, 996–999.
Wang, Y., Coleman-Derr, D., Chen, G., and Gu, Y. Q. (2015). OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 43, 78–84.
Weaver, S., Shank, S. D., Spielman, S. J., Li, M., Muse, S. V., and Kosakovsky-Pond, S. L. (2018). Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 35, 773–777. doi: 10.1093/molbev/msx335
Wheaton, G. H., Mukherjee, A., and Kelly, R. M. (2016). Transcriptomes of the extremely thermoacidophilic archaeon Metallosphaera sedula exposed to metal “shock” reveal generic and specific metal responses. Appl. Environ. Microbiol. 82, 4613–4627. doi: 10.1128/aem.01176-16
Wu, S., Zhu, Z., Fu, L., Niu, B., and Li, W. (2011). WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics 12:444. doi: 10.1186/1471-2164-12-444
Xu, Z., and Hao, B. (2009). CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 37, W174–W178.
Yi, W., Colemanderr, D., Chen, G., and Gu, Y. Q. (2015). OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 43, 78–84.
Yu, R. L., Liu, Z. H., Yu, Z. J., Wu, X. L., and Zeng, W. M. (2019). Relationship among the secretion of extracellular polymeric substances, heat resistance, and bioleaching ability of Metallosphaera sedula. Int. J. Miner. Metall. Mater. 26, 1504–1511. doi: 10.1007/s12613-019-1851-4
Zeldes, B. M., Loder, A. J., Counts, J. A., Haque, M., and Kelly, R. M. (2019). Determinants of sulfur chemolithoautotrophy in the extremely thermoacidophilic Sulfolobales. Environ. Microbiol. 21, 3696–3710. doi: 10.1111/1462-2920.14712
Zhang, X., Liu, X., Liang, Y., Guo, X., Xiao, Y., Ma, L., et al. (2017). Adaptive evolution of extreme acidophile Sulfobacillus thermosulfidooxidans potentially driven by horizontal gene transfer and gene loss. Appl. Environ. Microbiol. 83, e3016–e3098.
Keywords: comparative genomics, Metallosphaera, metabolic potential, evolution, horizontal gene transfer
Citation: Wang P, Li LZ, Qin YL, Liang ZL, Li XT, Yin HQ, Liu LJ, Liu S-J and Jiang C-Y (2020) Comparative Genomic Analysis Reveals the Metabolism and Evolution of the Thermophilic Archaeal Genus Metallosphaera. Front. Microbiol. 11:1192. doi: 10.3389/fmicb.2020.01192
Received: 16 January 2020; Accepted: 11 May 2020;
Published: 19 June 2020.
Edited by:
Andreas Teske, The University of North Carolina at Chapel Hill, United StatesReviewed by:
Hongchen Jiang, China University of Geosciences, ChinaZackary J. Jay, Montana State University, United States
Copyright © 2020 Wang, Li, Qin, Liang, Li, Yin, Liu, Liu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuang-Jiang Liu, bGl1c2pAaW0uYWMuY24=; Cheng-Ying Jiang, amlhbmdjeUBpbS5hYy5jbg==
†These authors have contributed equally to this work
 Pei Wang1,2†
Pei Wang1,2† Liang Zhi Li
Liang Zhi Li Li Jun Liu
Li Jun Liu Shuang-Jiang Liu
Shuang-Jiang Liu Cheng-Ying Jiang
Cheng-Ying Jiang