- 1Department of Infectious Diseases, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 2Anhui Center for Surveillance of Bacterial Resistance, Hefei, China
- 3Institute of Bacterial Resistance, Anhui Medical University, Hefei, China
- 4Department of Neurology, Xiangya Hospital Central South University, Changsha, China
- 5Department of Infectious Diseases, The Chaohu Affiliated Hospital of Anhui Medical University, Hefei, China
Objective: The aims of this study were to describe azithromycin (AZM) susceptibility patterns among Shigella isolates in Anhui, China and identify predictors of resistance with mobile element-mediated genes.
Methods: A total of 517 non-duplicate Shigella isolates (449 S. flexneri and 68 S. sonnei) were collected in the Anhui Province of China from September 2011–September 2015, and screened for the plasmid-mediated genes of decreased susceptibility to AZM (DSA), using polymerase chain reaction amplification and sequencing. Conjugation experiments and pulsed-field gel electrophoresis were conducted for all mphA-positive DSA isolates.
Results: The DSA rate for 449 S. flexneri isolates was 33.6%, compared with 39.7% for 68 S. sonnei isolates. Among 161 DSA S. flexneri isolates, 93 (57.8%) carried the mphA gene. Among 27 DSA S. sonnei isolates, 11 (40.7%) carried the mphA gene. However, other plasmid-mediated DSA genes were not found in these isolates. A total of 89 transconjugants (95.7%) were obtained from 93 mphA-positive S. flexneri isolates through conjugation, and 10 transconjugants (90.9%) were obtained from 11 mphA-positive S. sonnei isolates. Furthermore, the minimum inhibitory concentrations (MICs) of AZM among 89 S. flexneri transconjugants ranged from 4 to 128 μg/mL, with an MIC50 of 8 μg/mL and MIC90 of 32 μg/mL. The MICs of AZM among 10 S. sonnei transconjugants ranged from 4 to 256 μg/mL, with an MIC50 of 8 μg/mL and MIC90 of 64 μg/mL. Thirteen clusters were found for mphA-positive S. flexneri, and five clusters were found for mphA-positive S. sonnei. Furthermore, 10 homologous isolates among 13 mphA-positive S. flexneri isolates with high-level DSA were from Sixian county and were multidrug-resistant strains. Of the 10 homologous S. flexneri isolates, eight were from children (≤5 years old), and two from the elderly (>60 years old).
Conclusion: Our study demonstrates that the DSA for Shigella isolates was severe, and the plasmid-mediated mphA gene was the most common macrolide resistance gene detected in Shigella isolates collected in Anhui, China. The mphA-positive S. flexneri isolates with high-level DSA facilitated clonal spread in children and the elderly. This finding is noteworthy and warrants further study.
Introduction
Shigellosis is an acute invasive enteric infection caused by bacteria of the genus Shigella, including S. dysenteriae, S. flexneri, S. boydii, and S. sonnei. The symptoms of shigellosis usually include fever, headache, vomiting, and abdominal cramps, followed by watery diarrhea. All species of Shigella could be spread by direct contact with an infected person or via contaminated objects, food, or water, as well as close personal or sexual contact with fecal exposure (Kotloff et al., 2018).
Although shigellosis is typically self-limiting, treatment with appropriate antimicrobial therapy can shorten the duration of illness and prevent transmission. In guidelines published by the WHO in 2005, ciprofloxacin (CIP) was considered the first-line treatment for shigellosis, with ceftriaxone (CRO) and azithromycin (AZM) listed as alternative options. However, widespread use of CIP and third-generation cephalosporins have led to increasing resistance to these antimicrobials among Shigella isolates, especially in Asia (Liu et al., 2012, 2017; Zhang et al., 2014; Kotloff et al., 2018). The American Academy of Pediatrics and the Infectious Diseases Society of America have recommended the use of AZM to treat multidrug-resistant shigellosis (Erdman et al., 2008; Pickering et al., 2009; Shane et al., 2017). In China, AZM is infrequently used to treat shigellosis. Thus, there is a relative lack of published research on decreased susceptibility to AZM (DSA) for Shigella in China.
As a macrolide antibiotic, AZM has been used primarily to treat Gram-positive bacterial infections, and has shown favorable effects against various Gram-negative organisms (Gomes et al., 2013b). However, there are currently no interpretive criteria for AZM susceptibility testing of Shigella. According to the Clinical and Laboratory Standards Institute guidelines (CLSIs) published in 2019, epidemiological cut-off values (ECVs) of minimum inhibitory concentrations (MICs) ≥ 16 μg/mL (for S. flexneri) and MIC ≥ 32 μg/mL (for S. sonnei) were considered indicative of DSA. Previous studies on DSA have been focused mainly on punctual mutations in the rplD (encoding the L4 ribosomal protein), rplV (encoding the L22 ribosomal protein), and rrlH (23S rRNA) genes, and chromosomal efflux pumps (such as OmpA and OmpW) (Gomes et al., 2013a; Campbell et al., 2019).
More recently, studies have demonstrated that DSA-Shigella isolates in many areas are closely associated with the plasmid-borne mphA gene, which encodes macrolide-2′-phosphotransferase mediating macrolide resistance (Gaudreau et al., 2014; Darton et al., 2018; Yousfi et al., 2019). Besides the mphA gene, other mobile genetic elements also play an important role in the development of DSA, including mphB, ermA, ermB, ermC, ereA, ereB, mefA, msrA, ermF, ermT, and ermX) (Belkacem et al., 2016; Salah et al., 2019). Owing to the capacity for horizontal transmission among bacteria, the prevalence of mobile element-located resistance genes is a considerable challenge in controlling the spread of DSA.
To the best of our knowledge, the susceptibility of Shigella to AZM has not yet been recorded in Anhui, China. Therefore, the aims of this study were to describe AZM susceptibility patterns among Shigella isolates in Anhui, China and identify predictors of resistance with the mobile element-mediated genes.
Materials and Methods
Bacteria Isolates
A total of 517 non-duplicate Shigella isolates (449 S. flexneri and 68 S. sonnei) were collected from the fecal samples of various patients at 34 hospitals in the Anhui Province of China, between September 2011 and September 2015. Among 517 patients with shigellosis, 506 patients lived in Anhui, eight patients lived in another province, and the original location of three patients was unknown. None of these patients had taken AZM within the previous 3 months. Individual isolates were identified by using standard microbiological and biochemical methods. All Shigella isolates were confirmed by using an API-20E system (bioMérieux, Marcy l’ Étoile, France) and serotyped by using commercial antisera (Denka Seiken Co., Ltd., Tokyo, Japan). Strains of Escherichia coli ATCC 25922, Salmonella H9812, and sodium azide-resistant Escherichia coli J53 (E. coli J53AzR) were stored at the Anhui Center for Surveillance of Bacterial Resistance (Hefei, Anhui, China).
The study was conducted in accordance with the guidelines of the Declaration of Helsinki, the principles of Good Clinical Practice, and Chinese regulatory requirements, and was approved by the local Ethics Committees of the First Affiliated Hospital of Anhui Medical University (Hefei, China). All patients gave written informed consent.
Antimicrobial Agents
All antibiotics were obtained from Sigma-Aldrich China (Shanghai, China). Bacteria were cultured to the log-phase (approximately 1.5 × 108 CFU/mL). The MICs of AZM, CIP, CRO, and ampicillin (AMP) were determined by the agar dilution method using Mueller-Hinton agar (Oxoid Ltd., Basingstoke, United Kingdom) containing a series of twofold diluted antibiotics. The antibiotic plates with bacterial colonies were incubated at 37°C for 18–20 h. The results were interpreted according to CLSI breakpoints published in 2019. The susceptibility data of Shigella isolates were accepted only if the MIC for quality control strains, tested in parallel, was within the acceptable ranges given in the CLSI guidelines. The quality control strain was E. coli ATCC 25922.
Detection of Plasmid-Mediated Resistance Genes
Bacterial DNA was extracted by the boiling method and plasmid DNA was extracted using the Qiagen Plasmid Purification kit (QIAGEN, Hilden, Germany). The plasmid-mediated DSA genes (mphA/B, ermA/B/C/F/T/X, ereA/B, mefA, and msrA) for DSA-Shigella isolates were identified through the polymerase chain reaction (PCR). Primer sequences are shown in Supplementary Table S1. Cycle conditions for polymerase activation were as follows: initial denaturation at 95°C for 3 min; then 30 cycles of amplification, including denaturation at 95°C for 30 s; annealing for 30 s (annealing temperature is presented in Supplementary Table S1); extension at 72°C for 1 min; and a single final extension at 72°C for 10 min. All PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany) and directly sequenced. Sequence alignments were compared with the GenBank nucleotide database to determine the plasmid-mediated AZM resistance genotype.
Conjugation Experiments and Transfer of Drug Resistance
Conjugation experiments were conducted for all isolates positive for mphA, with E. coli J53AzR as the recipient. Transconjugants were selected on MacConkey agar plates supplemented with sodium azide (200 μg/mL) (Sigma Chemical Co., St. Louis, MO, United States), and AMP (32 μg/mL). Transconjugants were tested by the biochemical method and confirmed as E. coli using the API-20E system (bioMérieux). Plasmid DNA extraction from donors and transconjugants was performed using the Qiagen Plasmid Purification kit (QIAGEN, Hilden, Germany). The transconjugants were examined through PCR for the presence of the mphA gene using plasmid DNA as the template. The MICs of AZM, AMP, CIP, and CRO were determined for the recipient, donors, and transconjugants. The assays were conducted according to the methods described above.
Pulsed-Field Gel Electrophoresis
The DNA fingerprinting profiles were analyzed by pulsed-field gel electrophoresis (PFGE) after digestion with the restriction enzyme, XbaI (Takara, Inc., Dalian, China), according to the procedures developed by the US Centers for Disease Control and Prevention (CDC) Pulse Net program. Electrophoresis was performed on 1% agarose gels in 0.5 M Tris/borate/EDTA buffer on a CHEF Mapper® XA PFGE system (Bio-Rad, Hercules, CA, United States) for 22 h at 14°C, with run conditions of 6 V/cm, a pulse angle of 120°, and pulse times from 2.16 to 63.8 s. Salmonella H9812 was used as a molecular mass marker. The band profiles of PFGE were analyzed using the BioNumerics software (version 7.6, Applied Maths, sint-martens-latem, Belgium). Dendrograms of the band profiles were produced using the unweighted pair group method with mathematical averaging. The relatedness of isolates was calculated using the Dice coefficient with a band position tolerance setting of 1–1.5%. Isolates were defined as the same PFGE type (clonal) if the Dice coefficient was ≥ 85%.
Statistical Analysis
Data were analyzed using the SPSS version 16.0 software (SPSS Inc., Chicago, IL, United States). Univariate analysis was performed using the chi-squared test or Fisher’s exact test, as appropriate. P-values were based on two-tailed test results, and P < 0.05 were considered statistically significant.
Results
Antimicrobial Susceptibility
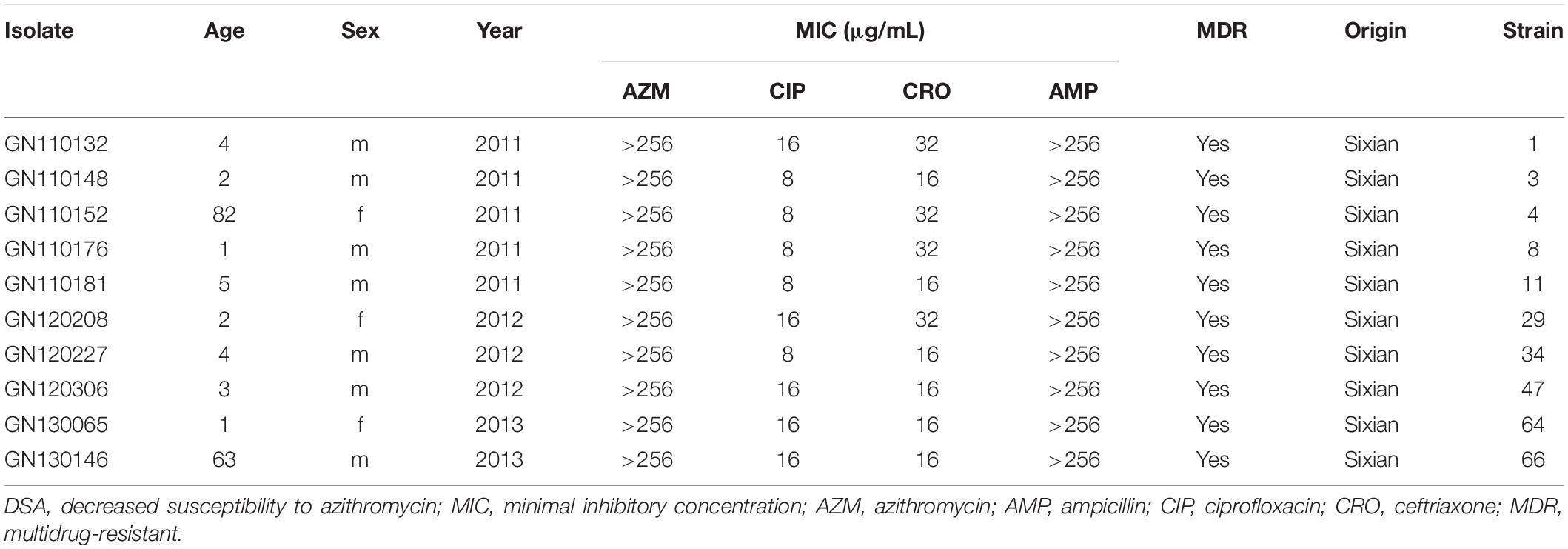
The characteristics of patients with shigellosis and the antimicrobial resistance of 517 Shigella isolates are presented in Table 1. The DSA rate for 449 S. flexneri isolates was 33.6%, compared with 39.7% for 68 S. sonnei isolates (χ2 = 0.966, P = 0.33). No significant differences were noted in gender or age between S. flexneri and S. sonnei isolates with DSA. The CIP resistance rate for 449 S. flexneri isolates was 49.7%, compared with 19.1% for 68 S. sonnei isolates (χ2 = 22.213, P < 0.0001). Regarding CIP resistance, no significant differences were noted between S. flexneri and S. sonnei among male participants and all age groups, and similarly, no significant differences were noted among female participants. The CRO resistance rate for 449 S. flexneri isolates was 45.7%, compared with 33.8% for 68 S. sonnei isolates (χ2 = 3.355, P = 0.07). No significant differences were noted between S. flexneri and S. sonnei, regarding gender, age, and CRO resistance. However, significant difference regarding CRO resistance was noted in children under 5 years of age.
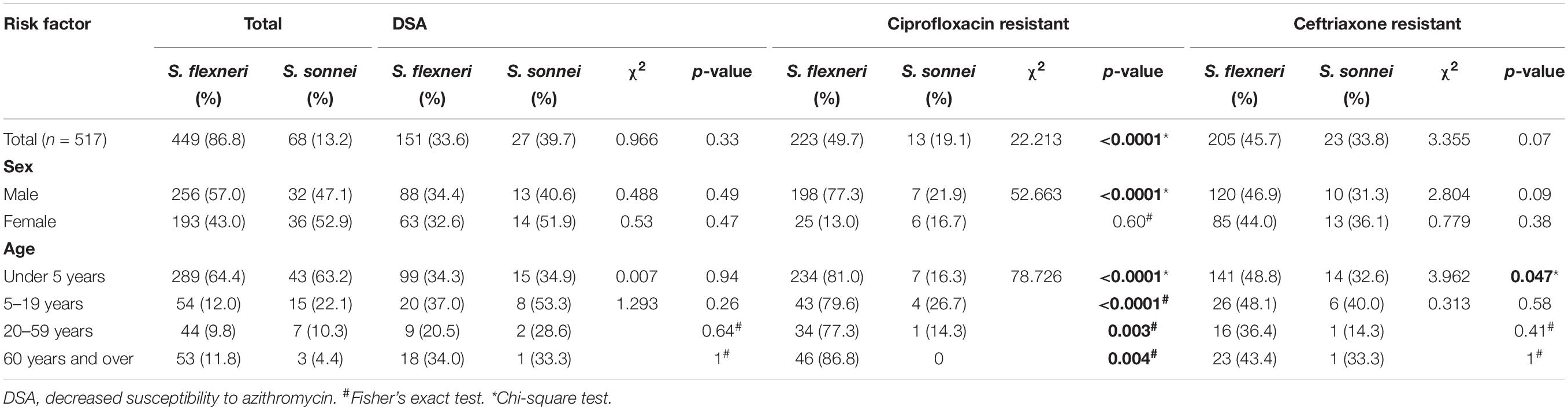
Table 1. Characteristics of patients with shigellosis and antimicrobial resistance of 517 Shigella isolates.
The MIC frequencies of AZM for S. flexneri and S. sonnei are presented in Figure 1. The MICs of AZM among 449 S. flexneri isolates ranged from < 1 to > 256 μg/mL, with an MIC50 of 4 μg/mL and MIC90 of 128 μg/mL. The MICs among 68 S. sonnei isolates ranged from 4 to > 256 μg/mL, with an MIC50 of 16 μg/mL and MIC90 of > 256 μg/mL.
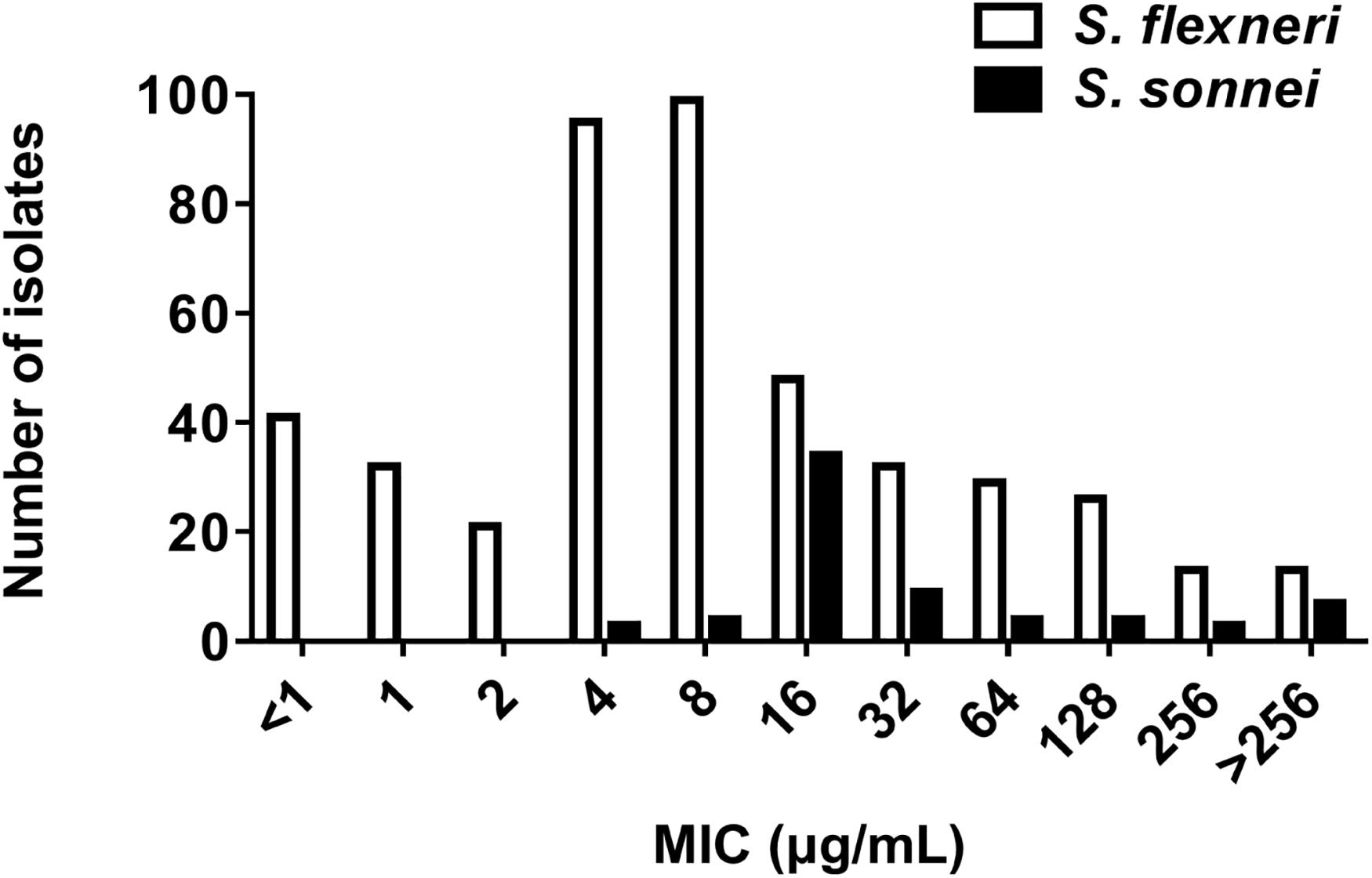
Figure 1. Azithromycin MIC by species. MIC frequency of azithromycin for S. flexneri (449 isolates, MIC50 = 4 μg/mL; MIC90 = 128 μg/mL) and S. sonnei (68 isolates, MIC50 = 16 μg/mL; MIC90 > 256 μg/mL).
Prevalence of Plasmid-Mediated DSA Genes
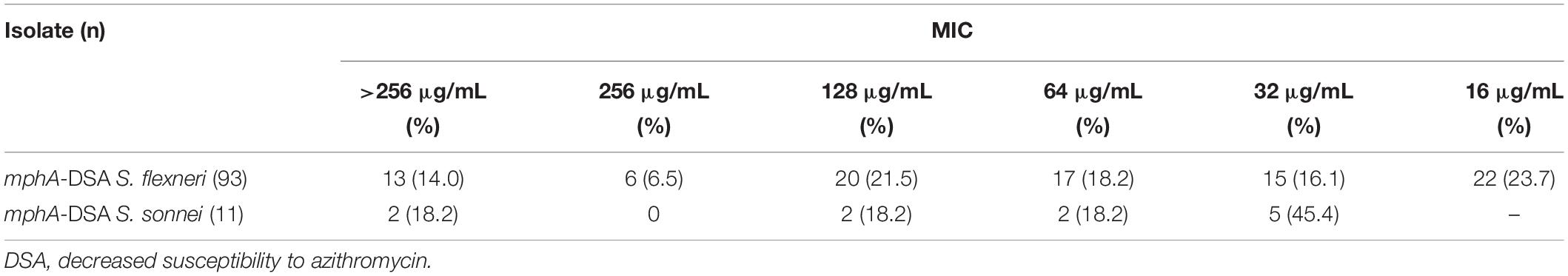
Among 161 DSA S. flexneri isolates, 93 (57.8%) carried the mphA gene. Among 27 DSA S. sonnei isolates, 11 (40.7%) carried the mphA gene. However, other plasmid-mediated DSA genes (such as mphB, ermA, ermB, ermC, ermF, ermT, ermX, ereA, ereB, mefA, and msrA) were not detected among these isolates. The MIC frequency of AZM for mphA-positive Shigella isolates are presented in Table 2. Of the 93 mphA-positive S. flexneri isolates, 13 (14%) exhibited high-level DSA (MIC > 256 μg/mL). Of the 11 mphA-positive S. sonnei isolates, two (18.2%) exhibited high-level DSA.
Transferability of Resistance Genes and Antimicrobial Susceptibility for Transconjugants
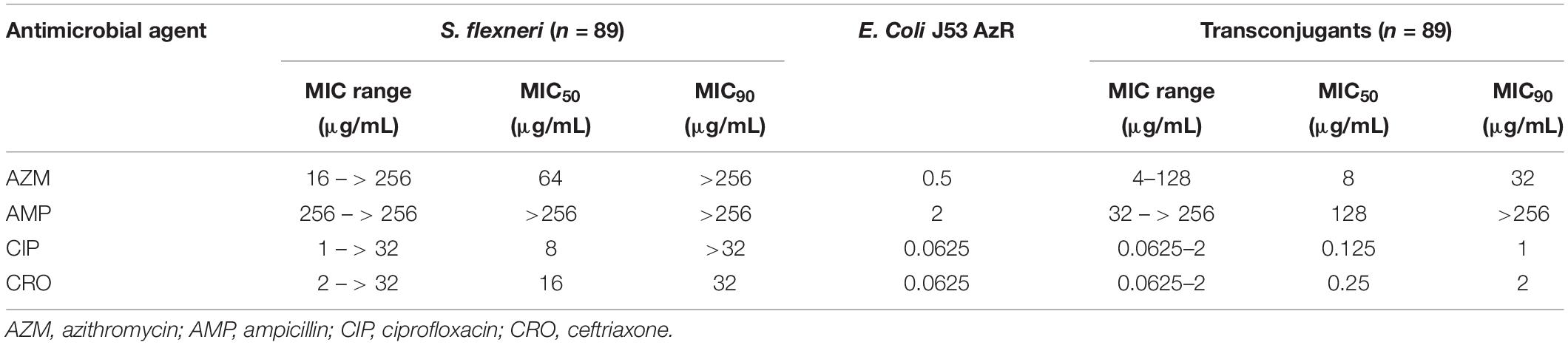
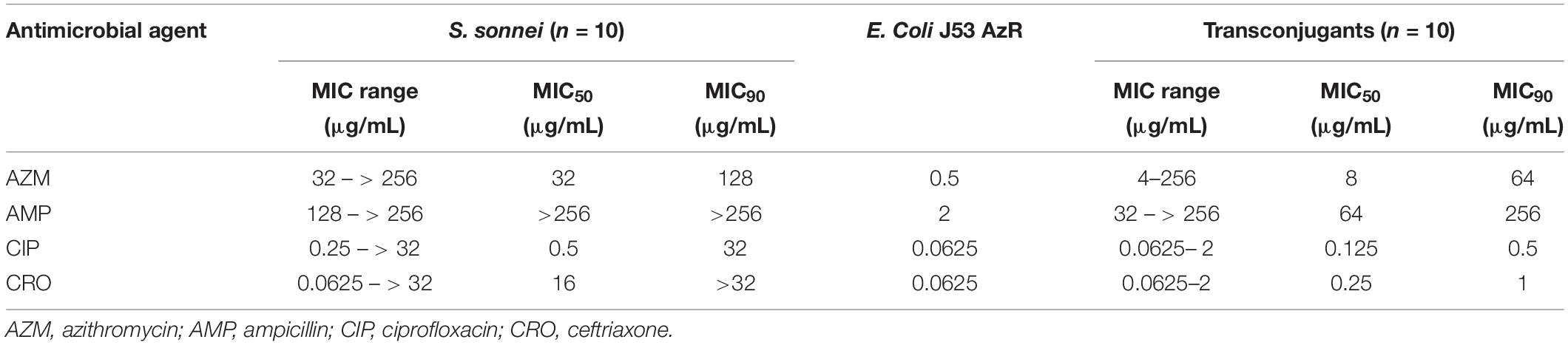
A total of 89 transconjugants (95.7%) were obtained from 93 mphA-positive S. flexneri isolates by conjugation, while 10 transconjugants (90.9%) were obtained from 11 mphA-positive S. sonnei isolates. The MICs of AZM among 89 transconjugants from S. flexneri isolates ranged from 4 to 128 μg/mL, with an MIC50 of 8 μg/mL and MIC90 of 32 μg/mL (Table 3). The MICs of AZM among 10 transconjugants from S. sonnei isolates ranged from 4 to 256 μg/mL, with an MIC50 of 8 μg/mL and MIC90 of 64 μg/mL (Table 4).
PFGE Analysis
The PFGE analysis was performed to determine whether clonal dissemination of mphA-positive Shigella isolates was responsible for the DSA in 104 isolates (93 S. flexneri and 11 S. sonnei). Figures 2, 3 show the dendrogram with PFGE images of mphA-positive S. flexneri and mphA-positive S. sonnei isolates, respectively. Thirteen clusters were detected for mphA-positive S. flexneri isolates and five clusters were detected for mphA-positive S. sonnei isolates. The predominant clones for mphA-positive S. flexneri isolates were P1 (44.1%, 41/93); P2 (30.1%, 28/93); and P5 (6.5%, 6/93). Clones P3, P4, and P7 of mphA-positive S. flexneri comprised three isolates (3.2%) each. Clones P11 and P13 of mphA-positive S. flexneri comprised two isolates (2.2%) each. Clones P6, P8, P9, and P12 of mphA-positive S. flexneri comprised one isolate (1.1%) each. The predominant clone for mphA-positive S. sonnei was P1 (63.6%; 7/11). Clones P2, P3, P4, and P5 of mphA-positive S. sonnei comprised one isolate (9.1%) each.
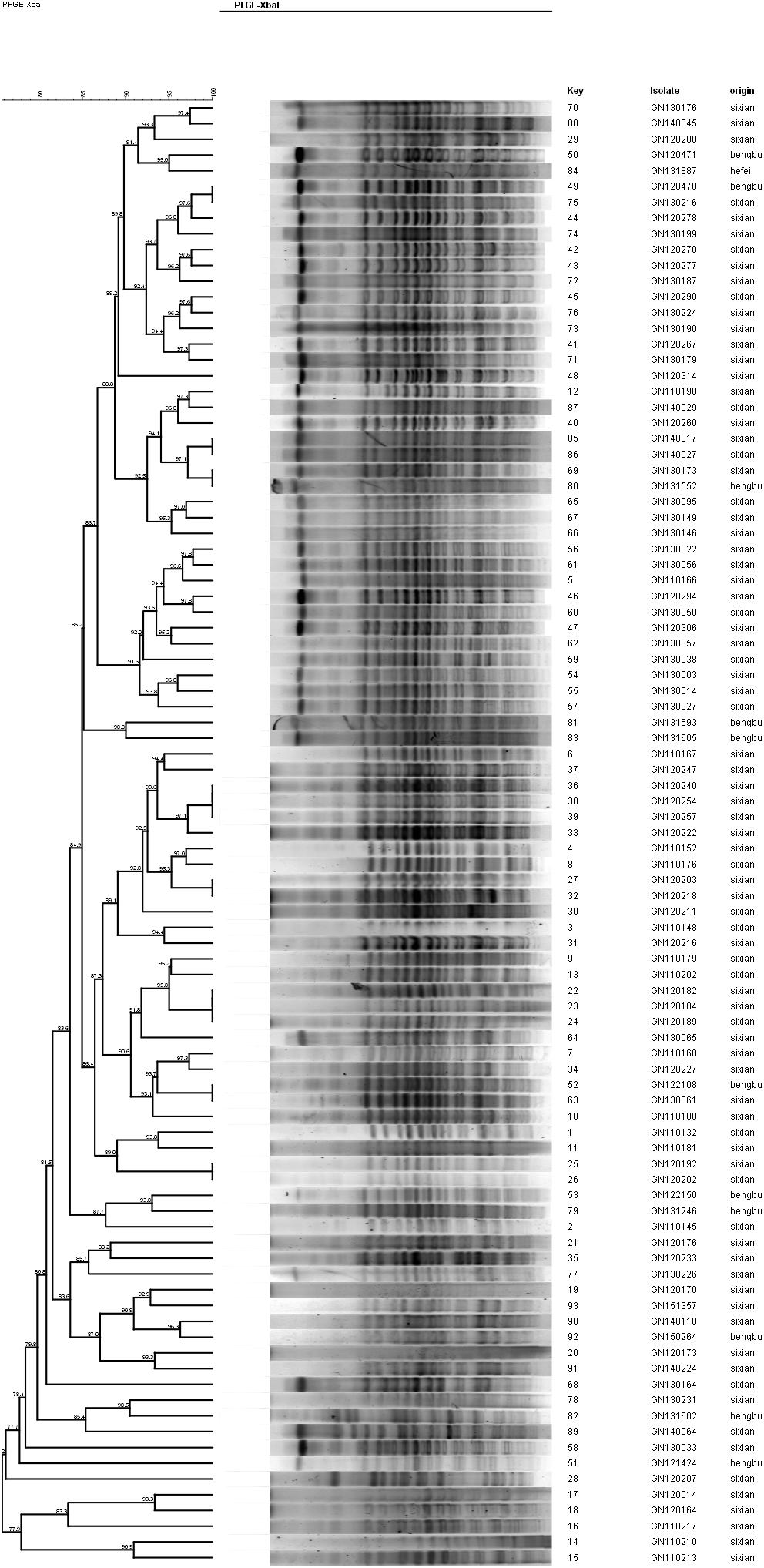
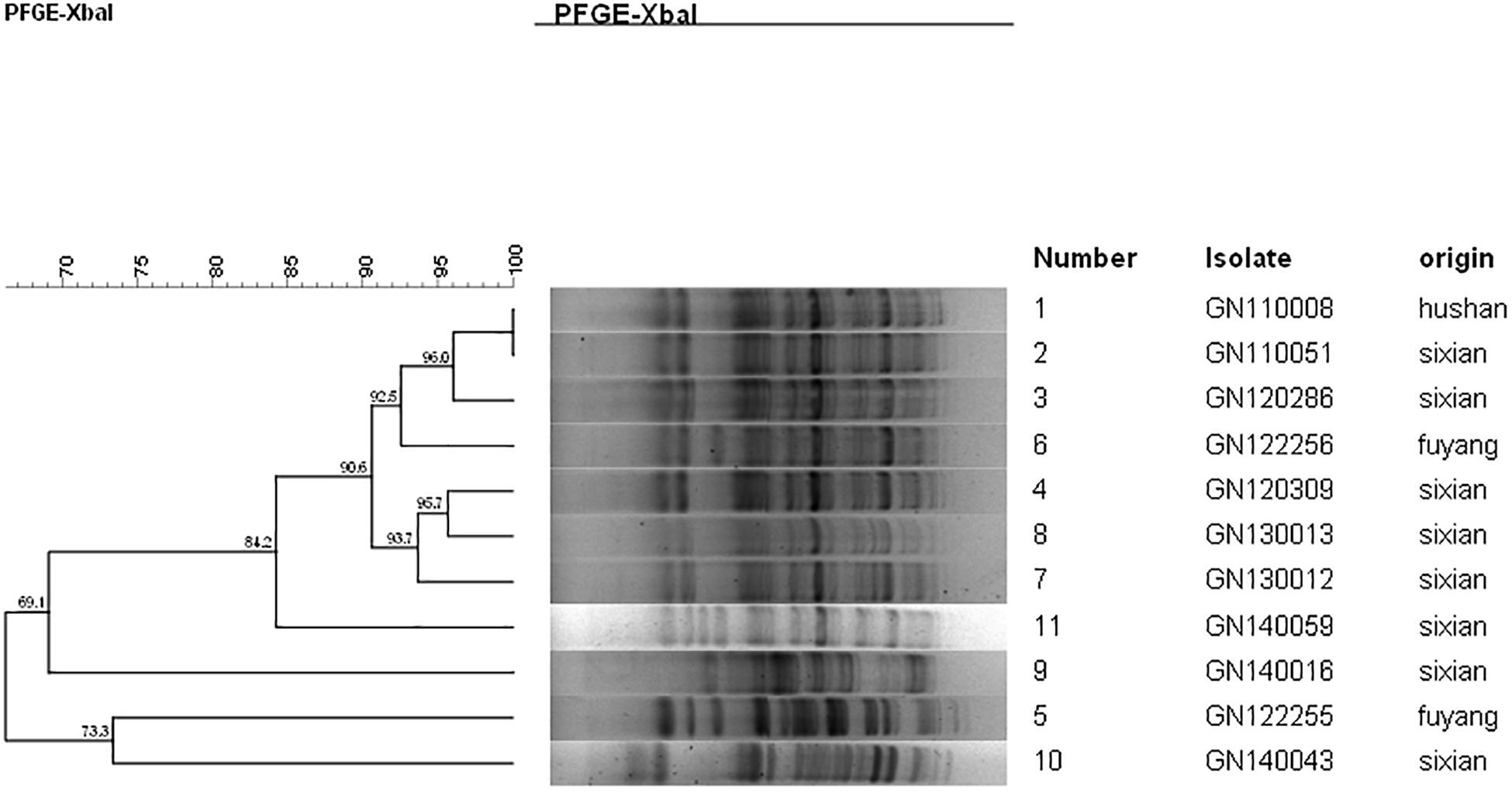
Ten isolates showed homology among 13 mphA-positive S. flexneri isolates with high-level DSA (Figure 4). The homology findings for the 10 S. flexneri isolates are presented in Table 5. All 10 homologous S. flexneri isolates were from Sixian county and were multidrug-resistant (MDR) strains. Of the 10 homologous S. flexneri isolates, eight were from children (≤5 years old), and two were from the elderly (>60 years old).
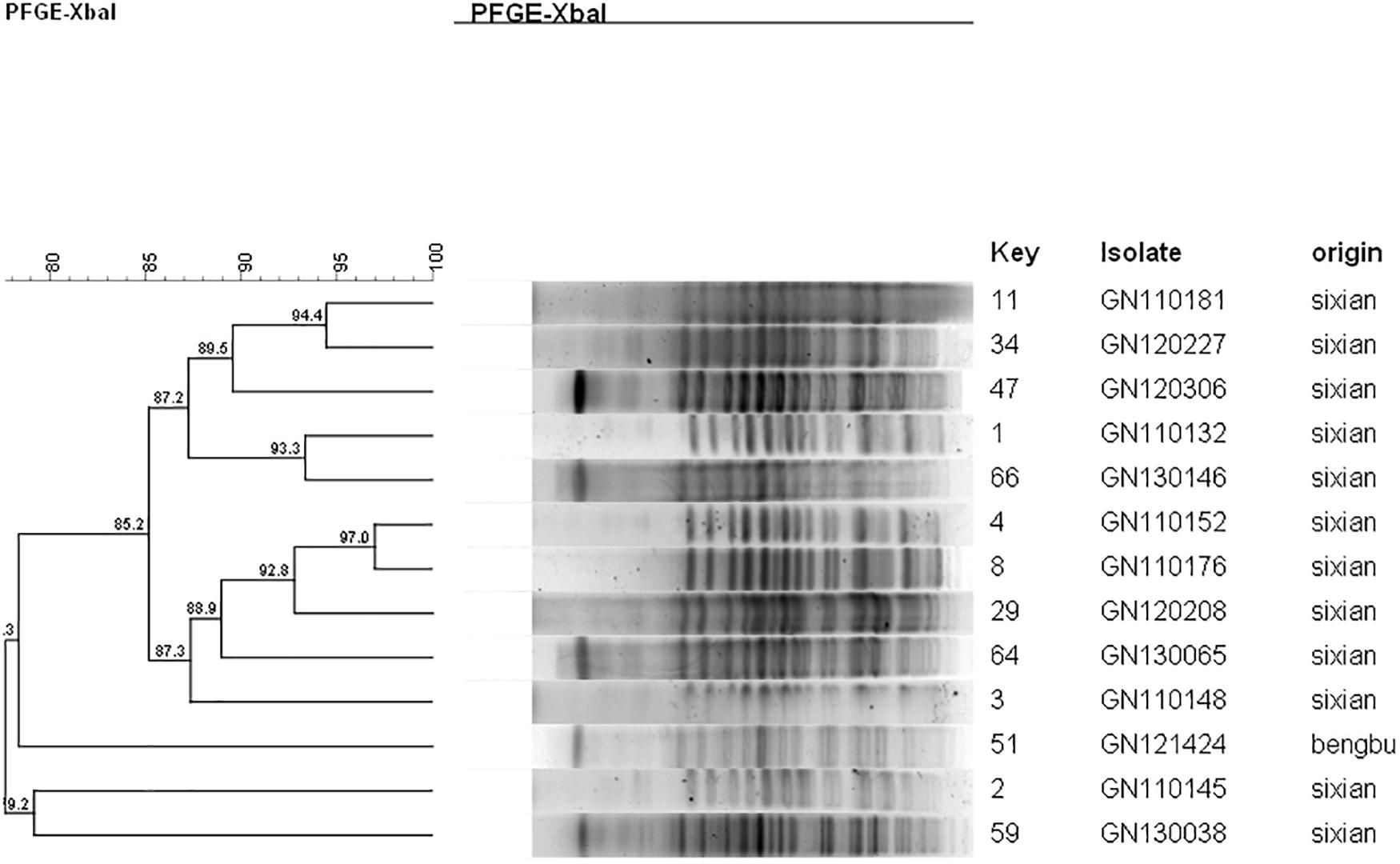
Figure 4. Dendogram of pulsed-field gel elecrophoresis for S. flexneri with high-level decreased susceptibility to azithromycin.
Discussion
In the current study, 449 S. flexneri isolates showed a high resistance pattern to CIP (49.7%) and CRO (45.7%), as well as DSA (33.6%); compared with 68 S. sonnei isolates showing resistance to CIP (19.1%) and CRO (33.8%), as well as DSA (39.7%). These results indicate that antimicrobial resistance in Shigella isolates is a serious challenge in Anhui, China. It is notable that DSA has increased globally and antimicrobial resistance among Shigella isolates can vary from region to region. For example, in India, the DSA rate was 48% in Shigella isolates from 2006 to 2011 (Bhattacharya et al., 2014). In Palestine, the DSA rate was 42% in Shigella isolates from 2004 to 2014 (Salah et al., 2019). However, in Australia, the DSA rate was 13.1% in Shigella isolates from 2013 to 2014, and the DSA has been reported more frequently in men who have sex with men (MSM) (Brown et al., 2017). In the United States, the DSA rate of Shigella isolates was 4.2% in 2012 (Sjölund et al., 2013).
In the current study, no significant differences were noted in gender and age between S. flexneri and S. sonnei isolates with DSA (Table 1). However, some studies have shown that males are more likely to be infected with a DSA strain than females, especially the MSM (Baker et al., 2015; Liao et al., 2016; Brown et al., 2017). The most likely explanation for this findings is the fact that Shigella isolates in the current study were isolated mainly from children < 5 years old. There are no clinical breakpoints for AZM in Shigella species in the CLSI guidelines. According to the CLSI (2019), the ECVs of MICs ≥ 16 μg/mL (for S. flexneri) and ≥ 32 μg/mL (for S. sonnei) are considered indicative of DSA, which is supported by the current results. In our data, S. flexneri (MIC50 4 μg/mL) had a lower MIC distribution than S. sonnei (MIC50 16 μg/mL).
Acquired macrolide resistance may arise from various mechanisms. The plasmid-mediated resistance determinants play an important role in the prevalence of DSA isolates because they are readily transferable. In the current study, 93 isolates (57.8%) carried the mphA gene among 161 DSA S. flexneri isolates, and 11 isolates (40.7%) carried the mphA gene among 27 DSA S. sonnei isolates. No other plasmid-mediated DSA genes were detected. Interestingly, all S. flexneri isolates with an MIC > 256 μg/mL carried the mphA gene. This indicates that the mphA gene is closely related to a high level of DSA (MIC > 256 μg/mL).
Furthermore, the mphA gene is normally associated with mobile elements (Gaudreau et al., 2014; Darton et al., 2018; Yousfi et al., 2019). Therefore, it was interesting to determine whether mphA-positive Shigella isolates would be able to transfer their macrolide resistance genes to recipients, and if the resulting transconjugants would be macrolide-resistant. In the current study, 89 transconjugants for mphA-positive S. flexneri and 10 transconjugants for mphA-positive S. sonnei were obtained by conjugation. This indicates that the mphA gene could be transferred between bacteria by horizontal exchange.
Furthermore, the transconjugants of mphA-positive S. flexneri isolates showed a 32-fold increase in the MIC50 and 128-fold increase in the MIC90 of AZM, compared with the recipient (Table 3). The transconjugants of mphA-positive S. sonnei isolates showed a 16-fold increase in the MIC50 and 128-fold increase in the MIC90 of AZM, compared with the recipient (Table 4). The above results are consistent with those of E. coli transconjugants with elevated MICs of AZM, and the confirmed location of the mphA gene on a plasmid (Sjölund et al., 2013).
Despite the high diversity, 80.7% of the mphA-positive DSA S. flexneri isolates were represented by three dominant PFGE clusters (P1, P2, and P5), and 63.6% of the mphA-positive DSA S. sonnei isolates were represented by one dominant PFGE cluster (P1). A genetic similarity value of 85% could indicate clonal spread of mphA-positive DSA-Shigella isolates in the community. Furthermore, genetic similarity was noted between the mphA-positive Shigella isolates with high-level DSA that was transmitted during the years 2011–2013.
Notably, all 10 homologous mphA-positive S. flexneri isolates with high-level DSA were MDR strains and were from children and the elderly. These findings might be attributed to a poor immune status in children and the elderly. These results also indicate that the mphA-positive S. flexneri isolates with high-level DSA were associated with clonal spread in children and the elderly.
Recently, researchers found that shigellosis can be transmitted among MSM patients, in whom oroanal sex and concurrent infection with HIV can significantly increase the infection rate (Baker et al., 2015; Liao et al., 2016; Yousfi et al., 2019). Baker et al. (2015) described the intercontinental dissemination of DSA shigellosis in MSM through sexual transmission. However, this concept was not analyzed in the current study, because data on MSM patients was not collected from an early stage. In future research, we will pay attention to the spread of DSA shigellosis in MSM in Anhui, China. Shigella isolates with DSA remains a topic of great concern. A provincial surveillance program should be established to determine the prevalence of Shigella with DSA. Furthermore, studies should be extended to elucidate the transmission mechanisms associated with DSA, in order to limit and prevent its emergence.
Conclusion
The current study demonstrates that the DSA for Shigella isolates is serious. We also confirmed that the plasmid-mediated mphA gene is the most common macrolide resistance gene in Shigella isolates collected in Anhui, China. The plasmid-mediated mphA gene can be transferred horizontally, to yield either the same or different strains with DSA. Our results also suggest the presence of a high diversity of mphA-positive DSA-Shigella isolates. It was noteworthy that the mphA-positive S. flexneri isolates with high-level DSA was associated with clonal spread in children and the elderly. This finding warrants further attention.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the local Ethics Committees of the First Affiliated Hospital of Anhui Medical University (Hefei, China). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
JL designed the experiments. YL, HL, and NL performed the experiments. XX, YZ, YY, and YG analyzed the results. All authors involved in the design of the experiments and reviewed the manuscript.
Funding
This research was supported by the research grants from the National Natural Science Foundation of China (Nos. 81673242 and 81973983).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01181/full#supplementary-material
Abbreviations
AMP, ampicillin; AZM, azithromycin; CIP, ciprofloxacin; CRO, ceftriaxone; DSA, decreased susceptibility to azithromycin; MDR, multidrug-resistant; MIC, minimal inhibitory concentration.
References
Baker, K. S., Dallman, T. J., Ashton, P. M., Day, M., Hughes, G., Crook, P. D., et al. (2015). Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect. Dis. 15, 913–921. doi: 10.1016/S1473-3099(15)00002-X
Belkacem, A., Jacquier, H., Goubard, A., Mougari, F., La Ruche, G., Patey, O., et al. (2016). Molecular epidemiology and mechanisms of resistance of azithromycin-resistant Neisseria gonorrhoeae isolated in France during 2013-14. J. Antimicrob. Chemother. 71, 2471–2478. doi: 10.1093/jac/dkw182
Bhattacharya, D., Bhattacharya, H., Thamizhmani, R., Sayi, D. S., Reesu, R., Anwesh, M., et al. (2014). Shigellosis in Bay of Bengal Islands, India: clinical and seasonal patterns, surveillance of antibiotic susceptibility patterns, and molecular characterization of multidrug-resistant Shigella strains isolated during a 6-year period from 2006 to 2011. Eur. J. Clin. Microbiol. Infect. Dis. 33, 157–170. doi: 10.1007/s10096-013-1937-2
Brown, J. D., Willcox, S. J., Franklin, N., Hazelton, B., Howard, P., Reinten, T., et al. (2017). Shigella species epidemiology and antimicrobial susceptibility: the implications of emerging azithromycin resistance for guiding treatment, guidelines and breakpoints. J. Antimicrob. Chemother. 72, 3181–3186. doi: 10.1093/jac/dkx268
Campbell, D., Bowen, A., Bhatnagar, A., McCullough, A., Grass, J., Chen, J., et al. (2019). Identification and characterization of Shigella with decreased susceptibility to azithromycin in the United States, 2005 to 2014. J. Glob. Antimicrob. Resist. 21, 417–419. doi: 10.1016/j.jgar.2019.12.005
Darton, T. C., Tuyen, H. T., Newton, P. N., Dance, D., and Phetsouvanh, R. (2018). Azithromycin Resistance in Shigella spp. in Southeast Asia. Antimicrob. Agents Chemother. 62:e001748-17. doi: 10.1128/AAC.01748-17
Erdman, S. M., Buckner, E. E., and Hindler, J. F. (2008). Options for treating resistant Shigella species infections in children. J. Pediatr. Pharmacol. Ther. 13, 29–43. doi: 10.5863/1551-6776-13.1.29
Gaudreau, C., Barkati, S., Leduc, J. M., Pilon, P. A., Favreau, J., Bekal, S., et al. (2014). Shigella spp. with reduced azithromycin susceptibility, Quebec, Canada, 2012-2013. Emerg. Infect. Dis. 20, 854–856. doi: 10.3201/eid2005.130966
Gomes, C., Martinez-Puchol, S., Durand, D., Lluque, A., and Mosquito, S. (2013a). Which mechanisms of azithromycin resistance are selected when efflux pumps are inhibited? Int. J. Antimicrob. Agents 42, 307–311. doi: 10.1016/j.ijantimicag.2013.05.012
Gomes, C., Pons, M. J., Magallon-Tejada, A., Durand, D., and Lluque, A. (2013b). In vitro development and analysis of Escherichia coli and Shigella boydii azithromycin-resistant mutants. Microb. Drug Resist. 19, 88–93. doi: 10.1089/mdr.2012.0036
Kotloff, K. L., Riddle, M. S., Platts-Mills, J. A., Pavlinac, P., and Zaidi, A. (2018). Shigellosis. Lancet 391, 801–812. doi: 10.1016/S0140-6736(17)33296-8
Liao, Y. S., Liu, Y. Y., Lo, Y. C., and Chiou, C. S. (2016). Azithromycin-Nonsusceptible Shigella flexneri 3a in Men who have Sex with Men, Taiwan, 2015-2016. Emerg. Infect. Dis. 23, 345–346. doi: 10.3201/eid2302.161260
Liu, Y., Cheng, Y., Yang, H., Hu, L., Cheng, J., Ye, Y., et al. (2017). Characterization of Extended-spectrum beta-Lactamase genes of Shigella flexneri isolates with fosfomycin resistance from patients in china. Ann. Lab. Med. 37, 415–419. doi: 10.3343/alm.2017.37.5.415
Liu, Y., Hu, L., Pan, Y., Cheng, J., Zhu, Y., Ye, Y., et al. (2012). Prevalence of plasmid-mediated quinolone resistance determinants in association with beta-lactamases, 16S rRNA methylase genes and integrons amongst clinical isolates of Shigella flexneri. J. Med. Microbiol. 61, 1174–1176. doi: 10.1099/jmm.0.042580-0
Pickering, L. K., Baker, C. J., Freed, G. L., Gall, S. A., Grogg, S. E., Poland, G. A., et al. (2009). Immunization programs for infants, children, adolescents, and adults: clinical practice guidelines by the Infectious diseases society of America. Clin. Infect. Dis. 49, 817–840. doi: 10.1086/605430
Salah, M., Shtayeh, I., Ghneim, R., Al-Qass, R., Sabateen, A., Marzouqa, H., et al. (2019). Evaluation of Shigella species azithromycin CLSI epidemiological cutoff values and macrolide resistance genes. J. Clin. Microbiol. 57:e01422-18. doi: 10.1128/JCM.01422-18
Shane, A. L., Mody, R. K., Crump, J. A., Tarr, P. I., Steiner, T. S., Kotloff, K., et al. (2017). 2017 infectious diseases society of america clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin. Infect. Dis. 65, 1963–1973. doi: 10.1093/cid/cix959
Sjölund, K. M., Bowen, A., Reporter, R., Folster, J. P., Grass, J. E., Howie, R. L., et al. (2013). Outbreak of infections caused by Shigella sonnei with reduced susceptibility to azithromycin in the United States. Antimicrob. Agents Chemother. 57, 1559–1560. doi: 10.1128/AAC.02360-12
Yousfi, K., Gaudreau, C., Pilon, P. A., Lefebvre, B., Walker, M., Fournier, E., et al. (2019). Genetic mechanisms behind the spread of reduced susceptibility to azithromycin in Shigella strains isolated from men who have sex with men in quebec, Canada. Antimicrob. Agents Chemother. 63:e01679-18. doi: 10.1128/AAC.01679-18
Keywords: Shigella, plasmid, azithromycin, multidrug-resistant, PFGE
Citation: Liu Y, Li H, Lv N, Zhang Y, Xu X, Ye Y, Gao Y and Li J (2020) Prevalence of Plasmid-Mediated Determinants With Decreased Susceptibility to Azithromycin Among Shigella Isolates in Anhui, China. Front. Microbiol. 11:1181. doi: 10.3389/fmicb.2020.01181
Received: 28 February 2020; Accepted: 08 May 2020;
Published: 30 June 2020.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaCopyright © 2020 Liu, Li, Lv, Zhang, Xu, Ye, Gao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiabin Li, bGlqaWFiaW5AYWhtdS5lZHUuY24=
†These authors have contributed equally to this work
 Yanyan Liu
Yanyan Liu Hongru Li
Hongru Li Na Lv
Na Lv Yalong Zhang
Yalong Zhang Xihai Xu
Xihai Xu Ying Ye
Ying Ye Yufeng Gao
Yufeng Gao Jiabin Li
Jiabin Li