- 1Department of Infectious Diseases, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Microbial Technology and Bioinformatics of Zhejiang Province, Hangzhou, China
- 3Department of Clinical Laboratory, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 4Department of Laboratory Medicine, The Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, China
Acinetobacter pittii is increasingly recognized as a clinically important species. Here, we identified a carbapenem-non-resistant A. pittii clinical isolate, A1254, harboring blaOXA–499, blaOXA–826, and blaADC–221. The blaOXA–499 genetic environment in A1254 was identical to that of another OXA-499-producing, but carbapenem-resistant, A. pittii isolate, YMC2010/8/T346, indicating the existence of phenotypic variation among OXA-499-producing A. pittii strains. Under imipenem-selective pressure, the A1254 isolate developed resistance to carbapenems in 60 generations. Two carbapenem-resistant mutants (CAB009 and CAB010) with mutations in the blaOXA–499 promoter region were isolated from two independently evolved populations (CAB001 and CAB004). The CAB009 mutant, with a mutation at position −14 (A to G), exhibited a four-fold higher carbapenem minimum inhibitory concentration (MIC) and a 4.53 ± 0.19 log2 fold change higher expression level of blaOXA–499 than the ancestor strain, A1254. The other mutant, CAB010, with a mutation at position −42 (G to A), showed a two-fold higher carbapenem MIC and a 1.65 ± 0.25 log2 fold change higher blaOXA–499 expression level than the ancestor strain. The blaOXA–499 gene and its promoter region were amplified from the wild-type strain and two mutant isolates and then individually cloned into the pYMAb2-Hygr vector and expressed in Acinetobacter baumannii ATCC 17978, A. pittii LMG 1035, and A. pittii A1254. All the transformed strains were resistant to carbapenem, irrespective of whether they harbored the initial or an evolved promoter sequence, and transformed strains expressing the promoter from the most resistant mutant, CAB009, showed the highest carbapenem MICs, with values of 32–64 μg/ml for imipenem and 128 μg/ml for meropenem. RNA sequencing was performed to confirm the contribution of blaOXA–499 to the development of carbapenem resistance. Although the CAB009 and CAB010 transcriptional patterns were different, blaOXA–499 was the only differentially expressed gene shared by the two mutants. Our results indicate that carbapenem-non-resistant Acinetobacter spp. strains carrying blaOXA genes have the potential to develop carbapenem resistance and need to be further investigated and monitored to prevent treatment failure due to the development of resistance.
Introduction
Acinetobacter spp. are increasingly raising serious concern because of their ability to rapidly develop resistance to a wide range of antimicrobials. Among these species, Acinetobacter baumannii, Acinetobacter nosocomialis, and Acinetobacter pittii are the most frequently isolated in hospitals globally (Weber et al., 2015). In the last few decades, relatively few studies have investigated non-baumannii Acinetobacter spp., likely owing to their low prevalence and resistant rates (Chen et al., 2019). However, non-baumannii Acinetobacter spp. are increasingly being found in clinical specimens and deserve more attention. This is true of A. pittii, previously called Acinetobacter genomic species three, which is increasingly found in food, clinical patients, and healthy individuals (Yang et al., 2012; Al Atrouni et al., 2016; Silva et al., 2018). Among Acinetobacter spp., A. pittii is the most commonly identified causative agent of nosocomial infections in patients hospitalized both in general ward and in intensive care units (ICUs) in Germany and is the most prevalent species identified among hospital-acquired A. calcoaceticus–A. baumannii (ACB) complex bloodstream isolates in France (Schleicher et al., 2013; Pailhories et al., 2018).
The ability of bacteria to rapidly acquire resistance poses crucial challenges to clinical treatment. We have previously shown that A. baumannii strains developed greater resistance with within-host evolution, thereby limiting the treatment options (Hua et al., 2017). Similar effects have also been observed for other species. For example, Pseudomonas aeruginosa showed rapid and large increases in resistance to carbapenem during antibiotic therapy that were likely due to de novo evolution and/or the selection of resistant subpopulations, indicating a potential risk for the rapid spread of antimicrobial resistance (Tueffers et al., 2019). Development of carbapenem resistance in sequential clinical isolates of Raoultella ornithinolytica carrying blaOXA–232 in a hospitalized patient during the course of ertapenem therapy was recently reported, highlighting the diagnostic challenges posed by strains producing inefficient types of carbapenemase (Iovleva et al., 2019). A more comprehensive understanding of resistance development would provide a molecular basis for improving the treatment of infections. However, to the best of our knowledge, no study to date has investigated the potential for the development of carbapenem resistance in A. pittii.
The emergence of carbapenem-resistant A. pittii has been reported worldwide. The rate of carbapenem resistance in A. nosocomialis and A. pittii isolates increased from 7.5% in 2010 to 22% in 2014 (Chen et al., 2019). Carbapenem resistance in A. pittii is mainly associated with the production of the carbapenem-hydrolyzing class D β-lactamases (CHDLs), such as OXA-23, OXA-58, OXA-72, OXA-143, and their variants (Zander et al., 2014a; D’Souza et al., 2017; Singkham-In and Chatsuwan, 2018; Chen et al., 2019). Besides, NDM-1-producing A. pittii isolates have also been reported in several cases (Yang et al., 2012; Hammerum et al., 2015; Pailhories et al., 2017; Deglmann et al., 2019). The emergence of carbapenem resistance in A. pittii reflects its ability to acquire and spread resistance genes, posing a challenge to the management of carbapenem-resistant non-baumannii Acinetobacter spp.
In this study, we characterized the A. pittii A1254 clinical isolate, which carries blaOXA–499, intrinsic blaOXA–826, and blaADC–221, but is susceptible to imipenem and intermediate susceptible to meropenem. The carbapenem resistance profile of A1254 was different from that of another OXA-499 producing A. pittii strain, YMC2010/8/T346, even though the blaOXA–499 genetic environment was identical in the two strains, indicating the existence of phenotypic variation among OXA-499-producing A. pittii strains. Thus, we investigated the effect of imipenem exposure in the A1254 isolate to reveal its potential to develop carbapenem resistance.
Materials and Methods
Bacterial Isolates and Culture Conditions
Acinetobacter pittii A1254 is a clinical strain isolated from the sputum sample of a patient with chronic obstructive pulmonary disease from the People’s Hospital of Quzhou, Zhejiang Province, China, in 2010. The A1254 isolate was initially described in our previous study on the prevalence of carbapenem-hydrolyzing class D β-lactamase genes in Acinetobacter spp. isolates (Ji et al., 2014). A. baumannii reference strain ATCC 17978, A. pittii reference strain LMG1035, and Escherichia coli DH5α were employed for cloning experiments. The liquid medium used was Luria–Bertani (LB) broth (Oxford, United Kingdom). The medium was supplemented with imipenem (0.75 μg/ml) as required.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of imipenem and meropenem were evaluated using the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2018). Simultaneously, carbapenem MICs against A1254 were evaluated with Etest strips (bioMérieux, Marcy-l’Étoile, France). E. coli strain ATCC 25922 was used as quality control. The results were interpreted in accordance with the CLSI breakpoints.
Experimental Evolution Under Imipenem Selective Pressure
Four single colonies of the A. pittii A1254 ancestor strain were inoculated in LB broth supplemented with imipenem (0.75 μg/ml). This concentration was maintained throughout the experimental evolution. All the evolved lineages were passaged daily and independently with shaking (200 rpm) at 37°C. A 20-μl volume of overnight culture was collected and inoculated at a 1:100 dilution daily for 9 days, and the generations were calculated (∼6.64 generations a day) with reference to Nicoloff et al. (2019). The four evolved populations were designated as CAB001, CAB002, CAB003, and CAB004. A different single colony was passaged daily in antibiotic-free LB broth as a blank control.
Whole-Genome Sequencing and Sequence Analysis
Genomic DNA of the four evolved populations (CAB001–CAB004) was extracted on day 9 using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA, United States) following the manufacturer’s recommendations. The quality and quantity of genomic DNA were determined by agarose gel electrophoresis and a NanoDrop spectrophotometer. A 300-bp library for Illumina paired-end sequencing was constructed from 5 μg of DNA using a Paired-End DNA Sample Prep Kit (Illumina, San Diego, CA, United States). The ancestor strain, A. pittii A1254, was sequenced by both long-read nanopore sequencing (Oxford Nanopore Technologies, Oxford, United Kingdom) and Illumina paired-end sequencing. Reads obtained from Illumina paired-end sequencing were used to correct the result of the nanopore sequencing using the Unicycler assembly pipeline (Wick et al., 2017). Paired-end sequence reads were assembled by SPAdes (Bankevich et al., 2012) and the de novo assemblies were subsequently annotated using the Prokka pipeline (Seemann, 2014). The genome of A1254 was input to the CGE web server for detection of resistance genes (selected% ID threshold, 90%; selected minimum length, 60%)1 (Zankari et al., 2012). Breseq was used to find mutations in evolved populations compared with A1254 (Deatherage and Barrick, 2014). Detected mutations were confirmed by PCR and Sanger sequencing. The primers used are listed in Table 1.
Cloning and Transformation
Fragments of blaOXA–499 with or without the promoter region were amplified from the wild-type A. pittii A1254 strain and the CAB009 and CAB010 mutants. Additionally, blaOXA–826 was also amplified, with or without its promoter region, but only from A. pittii A1254. The products were cloned into the BamHI and SalI-digested shuttle vector PYMAb2-Hygr using the ClonExpress® II One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, China) following the manufacturer’s recommendations. Cloning was performed based on recombination. Briefly, PYMAb2-Hygr was digested with BamHI and SalI. Using the primer pairs shown in Table 1, PCR products comprising the target fragment flanked by the recombination sequences were obtained. Then, the linearized vector, purified PCR product, buffer, and enhanced recombinase (Exnase II) were mixed and incubated for 30 min, yielding the recombinant vectors. The recombinant vectors were transformed into A. baumannii ATCC 17978, A. pittii LMG1035, and A1254 by electroporation.
Quantitative Reverse Transcription PCR
Quantitative reverse transcription PCR (RT-qPCR) was performed to measure the expression level of blaOXA–499 and blaOXA–826 in A. pittii A1254 and the CAB009 and CAB010 mutants submitted or not to imipenem selection. Total RNA was extracted using the RNeasy Mini Kit (Qiagen). RNA was reverse transcribed using random hexamers from Invitrogen (Carlsbad, CA, United States) and a reverse transcriptase from Takara Bio (Ôtsu, Japan), according to the manufacturer’s instructions. Quantitative PCR was performed using the SYBR® Premix Ex TaqTM PCR Kit (Takara Bio) in a LightCycler 480 system (Roche Molecular Diagnostics, Rotkreuz, Switzerland). The Ct value of each sample was measured under the following conditions: 95°C for 5 min, followed by 40 amplification cycles at 95°C for 10 s, 52°C for 30 s, and 72°C for 30 s. The rpoB gene was used as an internal reference. The primers used are listed in Table 1. Triplicate samples were included in each run, and RT-qPCR was performed three times independently. Data were calculated based on the ΔΔCt method (Livak and Schmittgen, 2001). Log2 fold change was used to evaluate the expression levels. Genes were identified as differentially expressed when the |log2 fold change| was >1.5 (Wright et al., 2017). Differences in expression levels were assessed by two-tailed Student’s t-tests. P < 0.05 was considered significant.
RNA-Sequencing (RNA-Seq)
Three single colonies of each isolate (A1254, CAB009, and CAB010) were grown overnight in LB broth, diluted 1:100 in 100 ml of fresh LB medium, and harvested at the mid-log growth phase. The subsequent RNA extraction, library construction, and transcriptomic analysis were performed by staff at MAGIGENE (Guangzhou, China). RNA was extracted using TRIzol Reagent (Invitrogen) and treated with DNase. rRNA was removed using a Ribo-Zero rRNA Removal Kit (Illumina). Paired-end RNA-sequencing (RNA-Seq) libraries were constructed with the NEBNext® Ultra IITM Directional RNA Library Prep Kit (New England Biolabs, Inc., Ipswich, MA, United States) and sequenced on the Illumina HiSeq/MiSeq NextSeq platform (Illumina). Raw reads were filtered by fastp (version 0.19.7) (Chen et al., 2018). After quality control, the reads were compared with ribosomal RNA (rRNA) sequences in the Rfam database, and unmapped reads were used for subsequent analysis. Filtered reads were mapped to the A1254 genome (GenBank accession number: CP049806-CP049810) using Hisat2 (version 2.1.0) (Kim et al., 2015). The read counts were calculated by RSEM (version 1.3.1) (Li and Dewey, 2011). The output data were analyzed by edgeR (version 3.20.2) (Robinson et al., 2010) and differences in the expression profiles of CAB009 and CAB010 were assessed by the expression ratio of each gene between the mutant and A1254. Genes were considered to be differentially expressed if their false discovery rate (FDR) was <0.05 and the |log2 fold change| was >1.5.
Results
Resistance Genes and the Genetic Environment of blaOXA–499 in A. pittii A1254
Based on CLSI guidelines, A. pittii A1254 was susceptible to imipenem (MIC, 2 μg/ml) and intermediate susceptible to meropenem (MIC, 4 μg/ml) using the broth microdilution method. However, when determined by Etest strips, A1254 was susceptible to both imipenem (MIC, 1.5 μg/ml) and meropenem (MIC, 2 μg/ml), consistent with the result reported for when A1254 was first described (Ji et al., 2014). Although there was a one-fold difference, the MIC of meropenem evaluated by the two methods was the breakpoint for susceptibility and intermediate susceptibility, respectively. Because in this study we also employed the broth microdilution method to determine the MICs for the other strains, we adopted 4 μg/ml as the meropenem MIC against A1254 and determined the strain to be intermediate susceptible to meropenem.
A1254 carries four plasmids in addition to its 4,065,905-bp-long chromosome. We identified two oxacillinase (OXA) genes in A1254: blaOXA–499 and a variant of blaOXA–500. blaOXA–499 is a variant of blaOXA–143 identified in 2017 and is reported to confer resistance to carbapenem (D’Souza et al., 2017). The blaOXA–500 variant was submitted to National Center for Biotechnology information (NCBI) as a member of the OXA-213 family, and NCBI designated it as blaOXA–826. The phylogenetic tree of relative OXA variants is shown in Figure 1. However, according to a recent study, it might be more appropriate to classify OXA-826 into the OXA-272-like family, which may be the intrinsic A. pittii OXA (Kamolvit et al., 2014; D’Souza et al., 2017). We also found and submitted a variant of blaADC–25, which was designated as blaADC–221 by NCBI. blaOXA–499, blaOXA–826, and blaADC–221 are all located on the chromosome. No other resistance genes were detected in A1254.
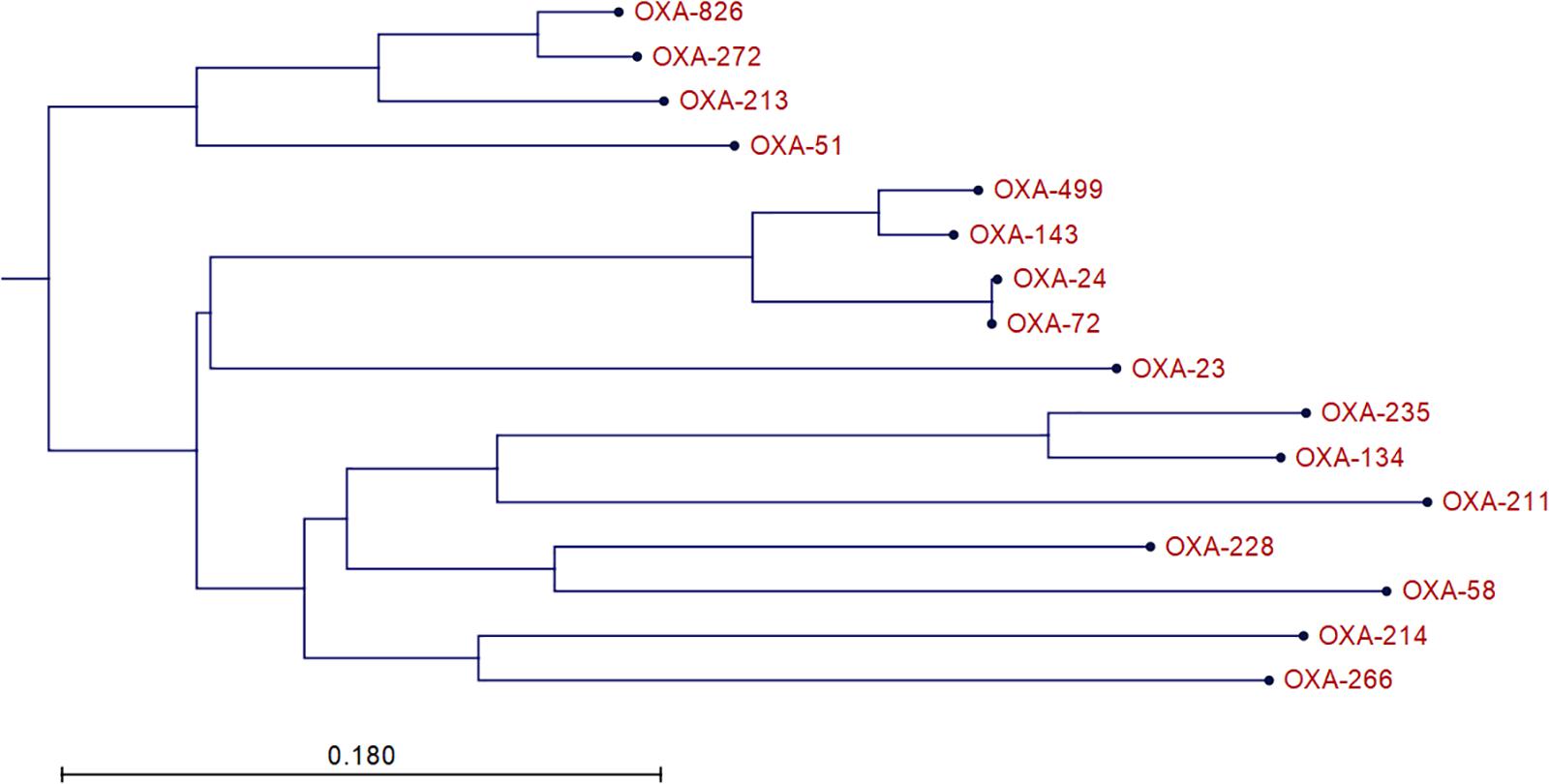
Figure 1. Phylogenetic tree of relative OXA variants. The neighbor-joining tree was generated using CLC Main Workbench 8.5.1 (QIAGEN Bioinformatics).
We compared the genetic environment of blaOXA–499 in A1254 with that in the A. pittii isolate YMC2010/8/T346 in which blaOXA–499 was first identified (GenBank accession number: CP017938). In the YMC2010/8/T346 isolate, blaOXA–499 is located within a 4,085-bp genomic fragment insertion consisting of a putative peptidase gene, blaOXA–499, and a TonB-dependent receptor plug domain, along with Xer C/D like recombination sites (D’Souza et al., 2017). The same fragment was identified in A1254 (100% identity).
Carbapenem-Resistant Evolved Populations and Mutations
We serially passaged four populations independently for 9 days (∼6.64 generations per day). On day 9 (∼60 generations), we found that two of the four populations (CAB001 and CAB004) had become resistant to both imipenem and meropenem. For population 1, CAB001, the imipenem MIC increased to 32 μg/ml (a four-fold increase) and that of meropenem increased to 64 μg/ml (also a four-fold increase). For population 4, CAB004, the imipenem MIC increased to 8 μg/ml (a two-fold increase) while that of meropenem increased to 32 μg/ml (a three-fold increase).
To identify the mutations responsible for the increased MICs (at least two-fold) against CAB001 and CAB004, we compared the genome of each evolved population to the genome sequence of the A1254 ancestor strain. Mutations with a frequency >70% are listed in Supplementary Table S1. Mutations occurring in population two (CAB002) and population three (CAB003) where the imipenem MIC showed only a one-fold increase (4 μg/ml) were excluded from further analysis. Therefore, the mutation in the promoter region of blaOXA–499, with a frequency of 100%, was the only mutation in CAB001 remaining for further analysis. A colony from the CAB001 population harboring this mutation was isolated and designated as CAB009.
Based on the evidence from CAB001, we suspected that the mutation in the promoter region of blaOXA–499 might be responsible for the increased carbapenem MICs in the population. To explore whether this mutation also existed in other populations, we isolated single colonies from CAB002–CAB004 and analyzed the sequence of the blaOXA–499 promoter region in each colony by PCR and Sanger sequencing. We identified another mutation in the promoter region of blaOXA–499 in a CAB004-derived colony, and we designated this mutant as CAB010. No mutations were found in the promoter region of blaOXA–499 in CAB002- and CAB003-derived colonies. Carbapenem MICs against the ancestor strain and the mutants are listed in Table 2.

Table 2. Minimum inhibitory concentrations (MICs) for strains and transformants harboring recombinant vectors.
Thus, we identified two mutations in the promoter region of blaOXA–499 in CAB009 and CAB010 that might mediate carbapenem resistance. The promoter region of blaOXA–499 was defined based on a previous study (Zander et al., 2014a). In the A. pittii CAB009 mutant from population one, we identified an A to G base substitution located one base upstream of the −10 region (position −14) (Figure 2), yielding a 5′-TG-3′ motif in the extended −10 element. In the A. pittii CAB010 mutant from population 4, we identified a G to A base substitution upstream of the −35 hexamer (position −42) within the upstream (UP) element (Figure 2).
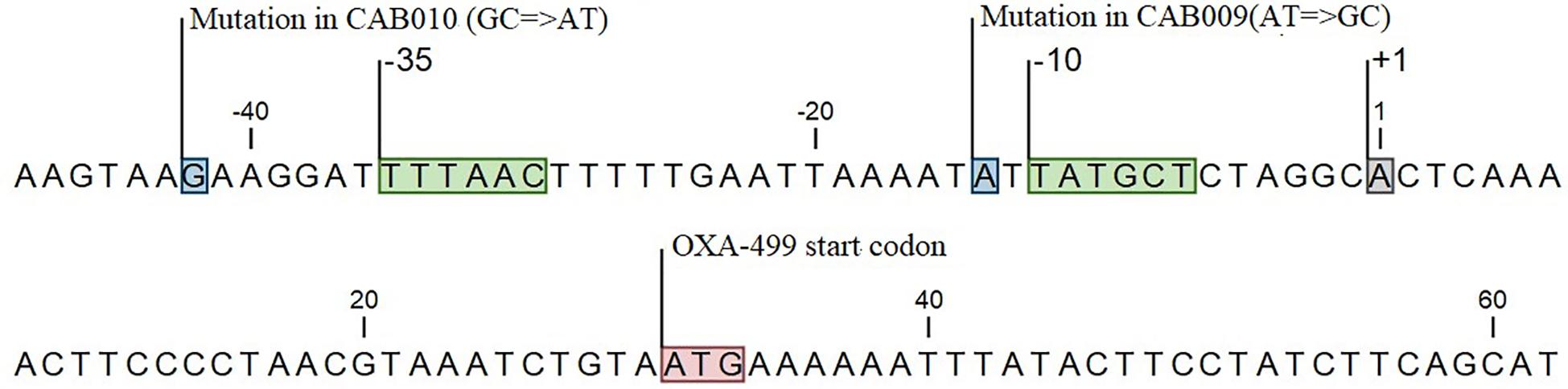
Figure 2. Locations of mutations in the carbapenem-resistant mutants CAB009 and CAB010 (boxed in blue). The start codon (boxed in red), the transcription initiation site (boxed in gray), and the −10 and −35 regions (boxed in green) are also indicated.
Relative Expression Levels of blaOXA–499 and blaOXA–826 in the Mutants
We next performed RT-qPCR to determine how the mutations affected the expression levels of blaOXA–499 and blaOXA–826. The results revealed that the expression level of blaOXA–499 increased significantly in CAB009, both in antibiotic-free LB broth (4.53 ± 0.19) and in LB broth supplemented with 0.75 μg/ml imipenem (4.68 ± 0.60). However, the difference between the two conditions was not significant (P = 0.6931) (Figure 3A). In CAB010, a more significant increase in blaOXA–499 expression was observed under 0.75 μg/ml imipenem pressure (2.54 ± 0.15) compared with that in antibiotic-free LB broth (1.65 ± 0.25) (P = 0.006) (Figure 3A). The higher expression level of blaOXA–499 in CAB009 might explain why it was more resistant to carbapenem than CAB010 (Table 2). Under imipenem selection pressure, there was a significant decrease in the expression level of blaOXA–826 in CAB010 but not CAB009 (Figure 3B). However, based on the |log2 fold-change| threshold (>1.5), only blaOXA–499 was identified as being differentially expressed in both CAB009 and CAB010. The RT-qPCR results indicated that blaOXA–499, but not blaOXA–826, played an important role in the development of carbapenem resistance. Supplementary Table S2 depicts the fold changes normalized by rpoB and blaOXA–826, respectively.
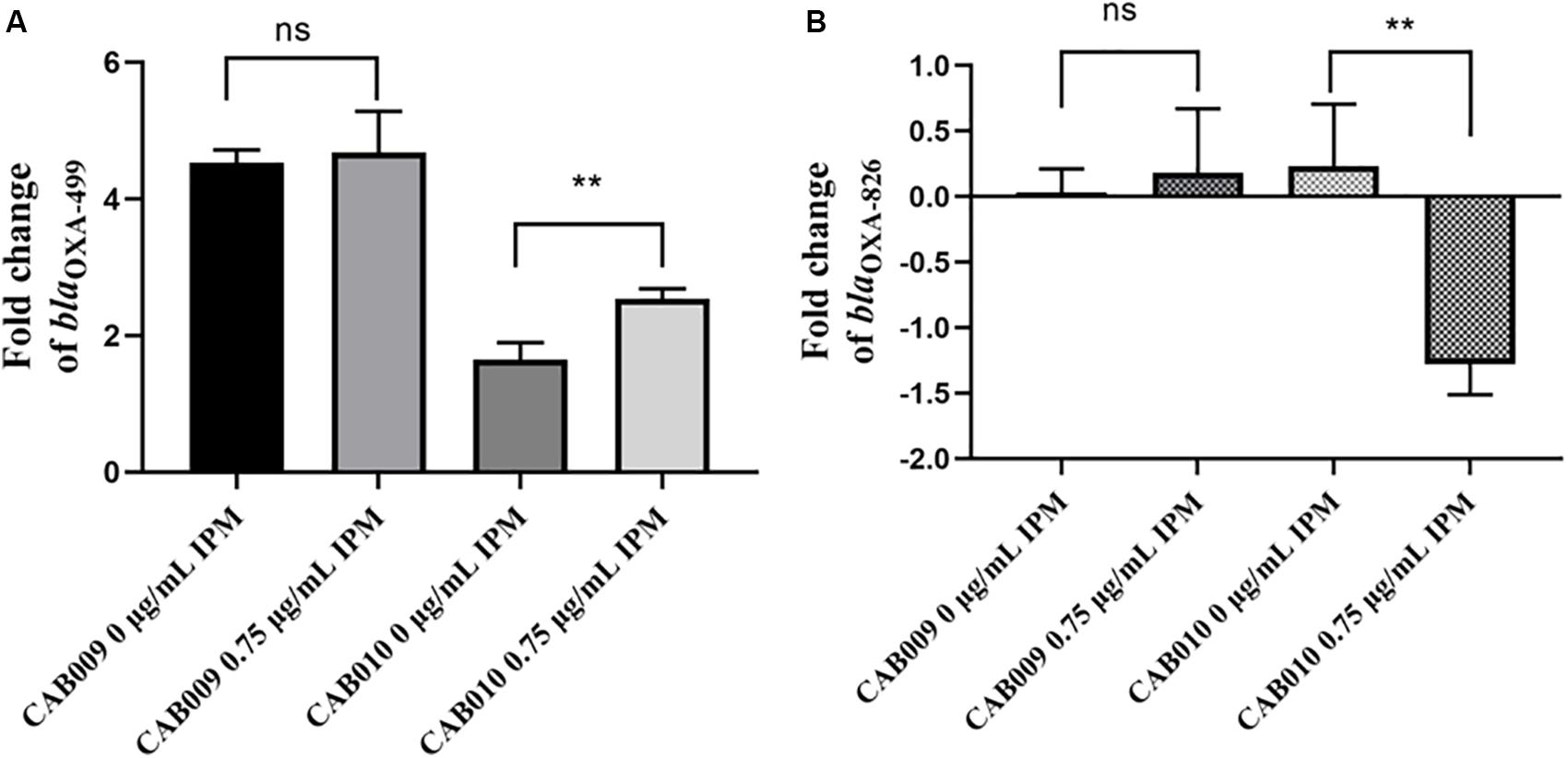
Figure 3. Log2 fold changes of (A) blaOXA–499 and (B) blaOXA–826 expression levels in mutants undergoing or not imipenem selection pressure. RT-qPCR analysis was performed with three biological and technical replicates per experiment. The 2–ΔΔCt method was applied to calculate the fold change using rpoB as the reference gene and Acinetobacter pittii A1254 as the reference strain. The expression level was calculated as the log2 fold change. The bars represent means ± SD from triplicate biological repeats. The P-values (each mutant undergoing or not imipenem selection pressure) were determined by two-tailed Student’s t-tests. **P < 0.008; ns, not significant. IPM, imipenem.
The Results of Cloning and Transformation
To examine how the mutations affect carbapenem resistance, we amplified a sequence containing either blaOXA–499 alone or blaOXA–499 together with an upstream 114-bp segment that included the promoter region, from A1254, CAB009, and CAB010. The sequence of blaOXA–826 with or without an upstream segment (446 bp in total, comprising a putative promoter region) was amplified from A1254. All the amplified products were cloned into the pYMAb2-Hygr vector. The recombinant vectors were electroporated into A. baumannii ATCC 17978, A. pittii LMG 1035, and A. pittii A1254 for expression analysis. The strains carrying the corresponding recombinant vectors and their MICs are listed in Table 2.
The results were similar among the transformed strains with different genetic backgrounds. blaOXA–499 could not confer resistance to carbapenem without the upstream sequence. However, all the recombinant strains harboring blaOXA–499 with its original promoter became resistant to imipenem (MIC, 16 μg/ml) and meropenem (MIC, 64–128 μg/ml). Similar MICs were observed for recombinant strains harboring blaOXA–499 containing the promoter from CAB010 (P010), except for OXA-499_P010-transformed A. baumannii ATCC 17978, showing a one-fold higher imipenem MIC (32 μg/ml) than OXA-499_P-transformed strains (16 μg/ml). Among the A. baumannii and A. pittii strains, blaOXA–499 containing the CAB009 promoter (P009) conferred the highest imipenem MICs (32–64 μg/ml, one- to two-fold change) when compared with the recombinant strains harboring the original promoter. Increased meropenem MICs were only observed for A. pittii A1254 and LMG 1035 (128 μg/ml, one-fold).
Transcriptomic Analysis
Differentially expressed genes between A1254 and CAB009 or CAB010 were identified based on the defined threshold. Overall, although the transcriptional patterns of CAB009 and CAB010 were different, blaOXA–499 was the only differentially expressed gene shared by the two mutants.
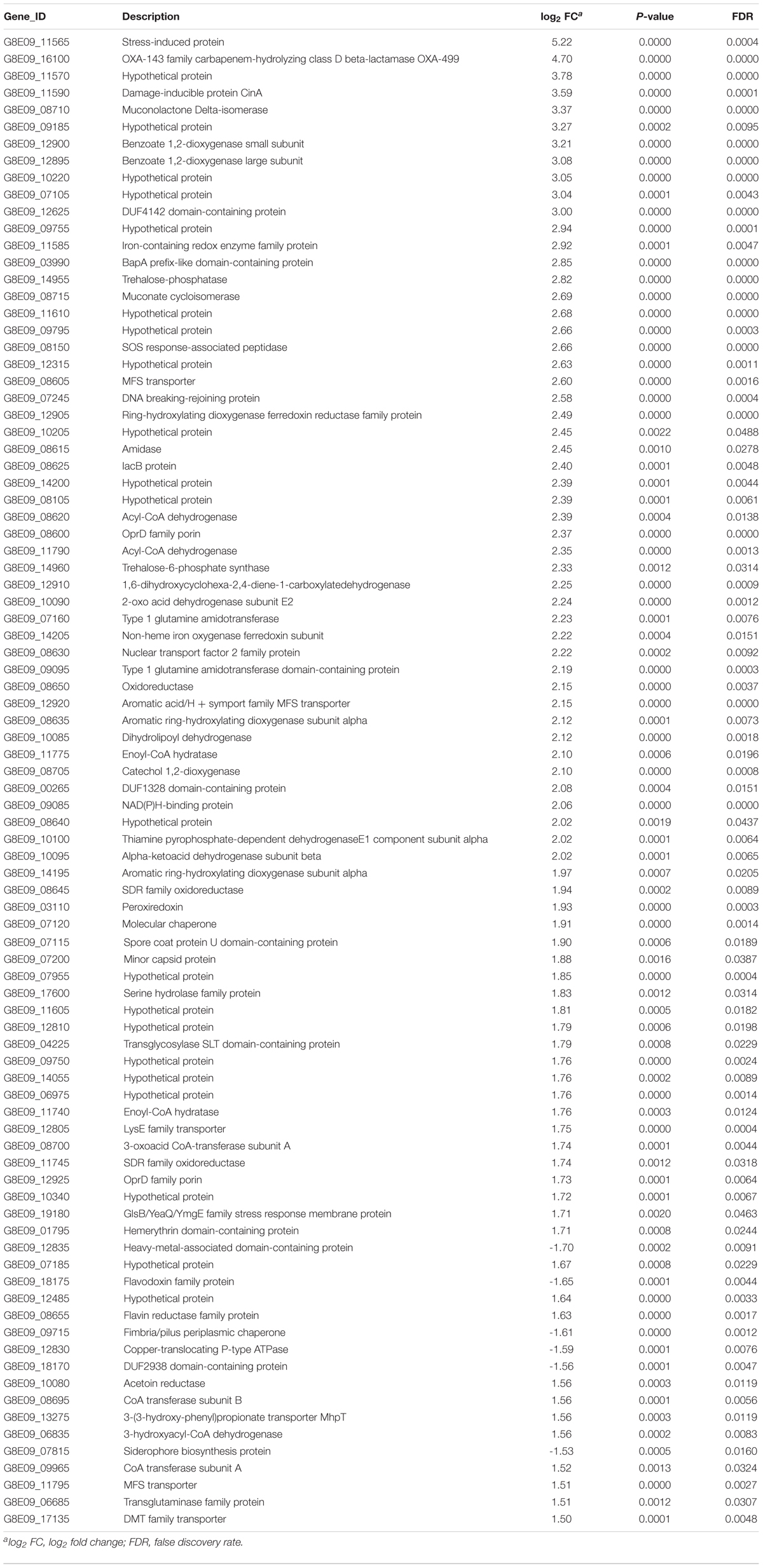
Eighty-eight differentially expressed genes were identified in CAB009 (Table 3). Among them, 82 were upregulated and six downregulated. A stress-induced protein-encoding gene G8E09_11565 showed a 5.22 log2 fold change higher expression level in CAB009 than in A1254, ranking first in the expression profile. This was followed by blaOXA–499, which showed a 4.70 log2 fold change higher expression level. We only identified three genes that were differentially expressed between CAB010 and A1254 (Table 4). The expression of blaOXA–499 was 2.04 log2 fold change higher in CAB010 than in A1254. G8E09_06800 (encoding a LysR family transcriptional regulator) and G8E09_06805 (encoding a type one glutamine amidotransferase domain-containing protein) showed 1.73 and 1.62 log2 fold change higher expression levels than A1254, respectively.
Discussion
Acinetobacter pittii, a member of the ACB complex, is increasingly recognized as a clinically important species following an improvement in identification methods that can better discriminate between A. pittii and A. baumannii (Yang et al., 2012). Here, we identified a carbapenem-non-resistant A. pittii clinical isolate, A1254, carrying blaOXA–499, intrinsic blaOXA–826, and blaADC–221. OXA-499 was first identified in a carbapenem-resistant A. pittii clinical isolate, YMC2010/8/T346, recovered from a patient in South Korea in 2010 and reported in 2017 (D’Souza et al., 2017). A1254 was also isolated in 2010. YMC2010/8/T346 is susceptible to imipenem (MIC, 2 μg/ml) but resistant to meropenem (MIC, 16 μg/ml). We compared the 4,085-bp blaOXA–499 genetic environment between A1254 and YMC2010/8/T346 and found them to be identical. However, A1254 is susceptible to imipenem and intermediate-susceptible to meropenem based on the microbroth dilution method and even susceptible to meropenem when the Etest is used. The different MICs among A. pittii strains sharing an identical blaOXA–499 genetic environment indicate that blaOXA–499 expression might vary according to host and result in phenotypic variation, which might be due to the presence of additional related mechanisms that influence the phenotype.
However, when cloned into a vector, blaOXA–499 containing either the initial or mutated promoter could confer carbapenem resistance in host strains. A similar result was reported by Zander et al. (2014a), who found that, although OXA-255 did not confer carbapenem resistance to the A. pittii clinical isolate AF726, OXA-255-transformed A. baumannii ATCC 17978 and A. pittii SH024 were resistant to carbapenem. Similar to OXA-499, OXA-255 is also a member of the OXA-143 family and the 2,239-bp genomic fragment containing blaOXA–255 (GenBank accession number, KC479325) in A. pittii AF726 is similar to that of blaOXA–499 in A1254 and YMC2010/8/T346 (99% identities). Considering that the genes were cloned into a shuttle vector and transformed into different Acinetobacter spp. strains by electroporation, we suspect that, apart from the genetic background of transformed strains, the introduction of the vector might also have influenced the expression level by providing multiple copies of the gene (dose effect), whereas in A1254, blaOXA–499 was located on the chromosome and as a single copy.
RT-qPCR and RNA-Seq results (log2 fold changes) for blaOXA–499 showed the same trend, although RNA-Seq data showed higher values. There was a significant difference in the number of differently expressed genes between CAB009 and CAB010 when compared with A1254. As CAB009 and CAB010 were isolated from two independently evolved populations, both mutants might harbor additional mutations besides the base substitutions in the promoter region of blaOXA–499. The different genotypes of CAB009 and CAB010 might explain the differences in the transcriptional profiles.
Subinhibitory concentrations of antibiotics can lead to gene expression changes and promote resistance (Bernier and Surette, 2013; Chen et al., 2017). In this study, we demonstrated that a carbapenem-non-resistant strain, A. pittii A1254, which tends to be neglected in the clinical microbiology laboratory, can rapidly develop resistance to carbapenems when cultured in broth containing a low concentration of imipenem. Here, we evaluated the possible mechanisms underlying the increased expression level of blaOXA–499 by analyzing the sequence of the promoter region. The mutant CAB009 isolate harbored an A to G transition at position −14, causing a base combination transformation from 5′-TA-3′ to 5′-TG-3′ one base upstream of the −10 region. The importance of the 5′-TG-3′ motif in the extended -10 element has been demonstrated in E. coli (Prost and Cozzone, 1999; Burr et al., 2000; Mitchell et al., 2003). It is well known that the −10 and −35 regions are where RNA polymerase (RNAP) contacts, resulting in promoter recognition and initiation of transcription. There is an additional promoter element, the “extended −10 element,” located one base upstream of the −10 region, with the major 5′-TG-3′ determinant positioned at −15/−14 with respect to the transcription start site (Mitchell et al., 2003). The 5′-TG-3′ motif is an important determinant of promoter activity (Voskuil and Chambliss, 2002; Prost and Cozzone, 1999; Burr et al., 2000). In our study, we suspect that it was the presence of the 5′-TG-3′ motif in the extended −10 element of the blaOXA–499 promoter of the CAB009 mutant that led to the increased expression of blaOXA–499, resulting in resistance to carbapenem. Another mutant, CAB010, exhibited a G to A transition at position −42, upstream of the −35 region. The UP element, located upstream of the −35 element (from approximately −40 to −60), can be recognized by RNAP and facilitates its initial binding as well as the subsequent steps in transcription initiation (Estrem et al., 1999; Presnell et al., 2019). A consensus UP element sequence consists almost exclusively of A and T residues and leads to increased promoter activity (Estrem et al., 1998). For the wild-type strain, A. pittii A1254, the proximal site of the UP element, contains a near-perfect A tract from position −39 to −44, interrupted only by a G at position −42. Following the transition from G to A at position −42, the proximal site in the CAB010 mutant became a perfect A tract.
In this study, we employed whole-genome sequencing (WGS) to identify putative resistance genes in the ancestor strain and mutations in the populations obtained from the experimental evolution. For clinical use, comprehensive databases of known resistant genes and related mutations are necessary for the successful prediction of antibiotic-resistance phenotypes. It is known that the accumulation of one or more single-nucleotide variants (SNVs) in genes encoding antibiotic targets or transposon insertions can lead to antibiotic resistance (Schurch and van Schaik, 2017). Published carbapenem resistance-related mechanisms in A. pittii include plasmid-borne blaOXA–23, blaOXA–72, or blaOXA–58; AbaR4-located blaOXA–23 on the chromosome; plasmid-borne class I integron containing blaIMP–1 (Montealegre et al., 2012; Silva et al., 2018; Chen et al., 2019); and a composite transposon containing blaNDM–1 (Yang et al., 2012). In addition, it has been proposed that A. pittii may be a resistance reservoir for the dissemination of NDM-1 (Bogaerts et al., 2013; Huang et al., 2015). Overexpression of blaOXA is typically mediated through promoters provided by insertion sequence (IS) elements, although OXA-40 and OXA-143 appear to be exceptions to this (Higgins et al., 2009). The association with IS elements and frequent presence in plasmids highlight the potential of blaOXA genes to spread within Acinetobacter spp. via transposition events and horizontal gene transfer (Zander et al., 2014b). For OXA-143-like and OXA-40-like, blaOXA–499 is flanked by XerC/XerD-like recombinase sites in both A. pittii A1254 and YMC2010/8/T346, suggesting that this gene was acquired through recombination. This recombination system is exploited by mobile DNA elements to integrate into the host genome (Midonet and Barre, 2014). A similar genetic context was also reported for other A. pittii isolates, in which the resistance gene was flanked by XerC/XerD-like recombinase sites (Cayo et al., 2014; Ruan et al., 2017; Brasiliense et al., 2019). This suggests that A. pittii may be an important source of resistance genes and contribute to their dissemination among species.
Here, we report for the first time that mutations in the promoter region of blaOXA–499 can contribute to the development of carbapenem resistance, complementing other known carbapenem resistance mechanisms in A. pittii. Enhanced resistance to β-lactam resulting from promoter mutations has also been reported in Staphylococcus aureus (Basuino et al., 2018). Additionally, loss of pncA expression due to promoter mutation conferred pyrazinamide resistance in multidrug-resistant tuberculosis isolates (Pang et al., 2017). This indicated that predicting resistance phenotypes based on the presence of resistance genes may be inaccurate in some circumstances; mutations on non-coding regions such as the promoter region should also be taken into consideration. One limitation of our study was that we failed to introduce a mutation in the wild-type A1254 strain due to technical restrictions. Moreover, we did not elucidate the specific mechanisms underlying the different carbapenem MICs between A. pittii strains harboring blaOXA–499.
In conclusion, to the best of our knowledge, our study represents the first investigation on the development of carbapenem resistance in an OXA-499-harboring, but carbapenem-non-resistant, A. pittii isolate. The genetic environment of blaOXA–499 was identical to that of a previously reported carbapenem-resistant A. pittii strain, indicating the existence of phenotypic variation in OXA-499-producing strains. We demonstrated that carbapenem-non-resistant A. pittii A1254 could become resistant to carbapenem under imipenem selective pressure and that a single-base substitution in the promoter region of blaOXA–499 contributed to the carbapenem-resistance phenotype. This highlights the need to monitor the potential development of carbapenem resistance when treating infections caused by non-resistant strains. The potential risk of resistance development requires that more attention be paid to the type, courses, and doses of antibiotics prescribed.
Data Availability Statement
The GenBank accession number of blaOXA–826 is MK810442 (https://www.ncbi.nlm.nih.gov/nuccore/MK810442). The GenBank accession number of blaADC–221 is MN654470 (https://www.ncbi.nlm.nih.gov/nuccore/MN654470.1). The GenBank accession number of A. pittii A1254 is CP049806-CP049810 (BioProject: PRJNA610163). The SRA accession numbers for populations CAB001–CAB004 are SRR11306746, SRR11306745, SRR11306744, and SRR11306743, respectively (BioProject: PRJNA610163). The SRA accession numbers for raw reads of RNA-Seq are SRR11648396–SRR11648404 (BioProject: PRJNA610163).
Author Contributions
YC, YY, and XHu designed the study. LZ, YF, XHa, and QX performed the experiments. XH, SW, BY, and LL analyzed the bioinformatics data. LZ and XHa wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of Zhejiang Province (grant number LQ17H190003) and the National Natural Science Foundation of China (grant numbers 81702041 and 31970128).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mrs. Haiping Wang (Zhejiang University) and Mrs. Ping Zhang (Zhejiang University) for kindly experiment assisting in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01134/full#supplementary-material
Footnotes
References
Al Atrouni, A., Joly-Guillou, M. L., Hamze, M., and Kempf, M. (2016). Reservoirs of non-baumannii Acinetobacter species. Front. Microbiol. 7:49. doi: 10.3389/fmicb.2016.00049
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Basuino, L., Jousselin, A., Alexander, J. A. N., Strynadka, N. C. J., Pinho, M. G., Chambers, H. F., et al. (2018). PBP4 activity and its overexpression are necessary for PBP4-mediated high-level beta-lactam resistance. J. Antimicrob. Chemother. 73, 1177–1180. doi: 10.1093/jac/dkx531
Bernier, S. P., and Surette, M. G. (2013). Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 4:20. doi: 10.3389/fmicb.2013.00020
Bogaerts, P., Huang, T. D., Rezende de Castro, R., Bouchahrouf, W., and Glupczynski, Y. (2013). Could Acinetobacter pittii act as an NDM-1 reservoir for Enterobacteriaceae? J. Antimicrob. Chemother. 68, 2414–2415. doi: 10.1093/jac/dkt201
Brasiliense, D. M., Lima, K. V. B., Perez-Chaparro, P. J., Mamizuka, E. M., de Oliveira Souza, C., Dutra, L. M. G., et al. (2019). Emergence of carbapenem-resistant Acinetobacter pittii carrying the blaOXA-72 gene in the Amazon region. Brazil. Diagn. Microbiol. Infect. Dis. 93, 82–84. doi: 10.1016/j.diagmicrobio.2018.07.017
Burr, T., Mitchell, J., Kolb, A., Minchin, S., and Busby, S. (2000). DNA sequence elements located immediately upstream of the -10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 28, 1864–1870. doi: 10.1093/nar/28.9.1864
Cayo, R., Merino, M., Ruiz Del Castillo, B., Cano, M. E., Calvo, J., Bou, G., et al. (2014). OXA-207, a novel OXA-24 variant with reduced catalytic efficiency against carbapenems in Acinetobacter pittii from Spain. Antimicrob. Agents Chemother. 58, 4944–4948. doi: 10.1128/AAC.02633-13
Chen, F. J., Huang, W. C., Liao, Y. C., Wang, H. Y., Lai, J. F., Kuo, S. C., et al. (2019). Molecular epidemiology of emerging carbapenem resistance in acinetobacter nosocomialis and acinetobacter pittii in Taiwan, 2010 to 2014. Antimicrob. Agents Chemother. 63:e02007-18. doi: 10.1128/AAC.02007-18
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, X., Meng, X., Gao, Q., Zhang, G., Gu, H., and Guo, X. (2017). Meropenem selection induced overproduction of the intrinsic carbapenemase as well as phenotype divergence in Acinetobacter baumannii. Int. J. Antimicrob. Agents 50, 419–426. doi: 10.1016/j.ijantimicag.2017.04.015
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing; 28th Informational Supplement. CLSI Document M100. Wayne, PA: CLSI.
Deatherage, D. E., and Barrick, J. E. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188. doi: 10.1007/978-1-4939-0554-6_12
Deglmann, R. C., Kobs, V. C., Oliveira, D., Burgardt, P., Franca, P. H. C., and Pillonetto, M. (2019). Earliest identification of New Delhi metallo-beta-lactamase 1 (NDM-1) in Acinetobacter pittii in Brazil. Rev. Soc. Bras. Med. Trop. 52:e20180348. doi: 10.1590/0037-8682-0348-2018
D’Souza, R., Pinto, N. A., Higgins, P. G., Hwang, I., Yong, D., Choi, J., et al. (2017). First report of the carbapenemase gene blaOXA-499 in Acinetobacter pittii. Antimicrob. Agents Chemother. 61:e02676-16. doi: 10.1128/AAC.02676-16
Estrem, S. T., Gaal, T., Ross, W., and Gourse, R. L. (1998). Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. U.S.A. 95, 9761–9766. doi: 10.1073/pnas.95.17.9761
Estrem, S. T., Ross, W., Gaal, T., Chen, Z. W., Niu, W., Ebright, R. H., et al. (1999). Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Genes Dev. 13, 2134–2147. doi: 10.1101/gad.13.16.2134
Hammerum, A. M., Hansen, F., and Littauer, P. (2015). Use of whole-genome sequencing for characterisation of a ST119 NDM-1-producing Acinetobacter pittii from a patient in Denmark with no history of recent travel. Int. J. Antimicrob. Agents 46, 351–352. doi: 10.1016/j.ijantimicag.2015.05.005
Higgins, P. G., Poirel, L., Lehmann, M., Nordmann, P., and Seifert, H. (2009). OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 5035–5038. doi: 10.1128/AAC.00856-09
Hua, X., Zhou, Z., Yang, Q., Shi, Q., Xu, Q., Wang, J., et al. (2017). Evolution of Acinetobacter baumannii in vivo: international clone II, more resistance to ceftazidime, mutation in ptk. Front. Microbiol. 8:1256. doi: 10.3389/fmicb.2017.01256
Huang, T. W., Lauderdale, T. L., Liao, T. L., Hsu, M. C., Chang, F. Y., Chang, S. C., et al. (2015). Effective transfer of a 47 kb NDM-1-positive plasmid among Acinetobacter species. J. Antimicrob. Chemother. 70, 2734–2738. doi: 10.1093/jac/dkv191
Iovleva, A., Mettus, R. T., McElheny, C. L., Griffith, M. P., Mustapha, M. M., Pasculle, A. W., et al. (2019). High-level carbapenem resistance in OXA-232-producing raoultella ornithinolytica triggered by ertapenem therapy. Antimicrob. Agents Chemother. 64:e01335-19. doi: 10.1128/AAC.01335-19
Ji, S., Chen, Y., Ruan, Z., Fu, Y., Ji, J., Fu, Y., et al. (2014). Prevalence of carbapenem-hydrolyzing class D beta-lactamase genes in Acinetobacter spp. isolates in China. Eur. J. Clin. Microbiol. Infect. Dis. 33, 989–997. doi: 10.1007/s10096-013-2037-z
Kamolvit, W., Higgins, P. G., Paterson, D. L., and Seifert, H. (2014). Multiplex PCR to detect the genes encoding naturally occurring oxacillinases in Acinetobacter spp. J. Antimicrob. Chemother. 69, 959–963. doi: 10.1093/jac/dkt480
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Midonet, C., and Barre, F. X. (2014). Xer site-specific recombination: promoting vertical and horizontal transmission of genetic information. Microbiol. Spectr. 2, 163–182. doi: 10.1128/microbiolspec.MDNA3-0056-2014
Mitchell, J. E., Zheng, D., Busby, S. J., and Minchin, S. D. (2003). Identification and analysis of ‘extended -10’ promoters in Escherichia coli. Nucleic Acids Res. 31, 4689–4695. doi: 10.1093/nar/gkg694
Montealegre, M. C., Maya, J. J., Correa, A., Espinal, P., Mojica, M. F., Ruiz, S. J., et al. (2012). First identification of OXA-72 carbapenemase from Acinetobacter pittii in Colombia. Antimicrob. Agents Chemother. 56, 3996–3998. doi: 10.1128/aac.05628-11
Nicoloff, H., Hjort, K., Levin, B. R., and Andersson, D. I. (2019). The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 4, 504–514. doi: 10.1038/s41564-018-0342-0
Pailhories, H., Hadjadj, L., Mahieu, R., Crochette, N., Rolain, J. M., and Kempf, M. (2017). Fortuitous diagnosis of NDM-1-producing Acinetobacter pittii carriage in a patient from France with no recent history of travel. J. Antimicrob. Chemother. 72, 942–944. doi: 10.1093/jac/dkw505
Pailhories, H., Tiry, C., Eveillard, M., and Kempf, M. (2018). Acinetobacter pittii isolated more frequently than Acinetobacter baumannii in blood cultures: the experience of a French hospital. J. Hosp. Infect. 99, 360–363. doi: 10.1016/j.jhin.2018.03.019
Pang, Y., Zhu, D., Zheng, H., Shen, J., Hu, Y., Liu, J., et al. (2017). Prevalence and molecular characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates from Southern China. BMC Infect. Dis. 17:711. doi: 10.1186/s12879-017-2761-6
Presnell, K. V., Flexer-Harrison, M., and Alper, H. S. (2019). Design and synthesis of synthetic UP elements for modulation of gene expression in Escherichia coli. Synth. Syst. Biotechnol. 4, 99–106. doi: 10.1016/j.synbio.2019.04.002
Prost, J. F., and Cozzone, A. J. (1999). Detection of an extended-10 element in the promoter region of the pckA gene encoding phosphoenolpyruvate carboxykinase in Escherichia coli. Biochimie 81, 197–200. doi: 10.1016/s0300-9084(99)80052-2
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Ruan, Z., Chen, Y., and Wang, J. (2017). Glimpse into the genome sequence of a multidrug-resistant Acinetobacter pittii ST950 clinical isolate carrying the blaOXA-72 and blaOXA-533 genes in China. Mem. Inst. Oswaldo Cruz. 112, 723–727. doi: 10.1590/0074-02760170019
Schleicher, X., Higgins, P. G., Wisplinghoff, H., Korber-Irrgang, B., Kresken, M., and Seifert, H. (2013). Molecular epidemiology of Acinetobacter baumannii and Acinetobacter nosocomialis in Germany over a 5-year period (2005–2009). Clin. Microbiol. Infect. 19, 737–742. doi: 10.1111/1469-0691.12026
Schurch, A. C., and van Schaik, W. (2017). Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann. N. Y. Acad. Sci. 1388, 108–120. doi: 10.1111/nyas.13310
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Silva, L., Mourao, J., Grosso, F., and Peixe, L. (2018). Uncommon carbapenemase-encoding plasmids in the clinically emergent Acinetobacter pittii. J. Antimicrob. Chemother. 73, 52–56. doi: 10.1093/jac/dkx364
Singkham-In, U., and Chatsuwan, T. (2018). Mechanisms of carbapenem resistance in Acinetobacter pittii and Acinetobacter nosocomialis isolates from Thailand. J. Med. Microbiol. 67, 1667–1672. doi: 10.1099/jmm.0.000845
Tueffers, L., Barbosa, C., Bobis, I., Schubert, S., Hoppner, M., Ruhlemann, M., et al. (2019). Pseudomonas aeruginosa populations in the cystic fibrosis lung lose susceptibility to newly applied beta-lactams within 3 days. J. Antimicrob. Chemother. 74, 2916–2925. doi: 10.1093/jac/dkz297
Voskuil, M. I., and Chambliss, G. H. (2002). The TRTGn motif stabilizes the transcription initiation open complex. J. Mol. Biol. 322, 521–532. doi: 10.1016/s0022-2836(02)00802-1
Weber, B. S., Harding, C. M., and Feldman, M. F. (2015). Pathogenic acinetobacter: from the cell surface to infinity and beyond. J. Bacteriol. 198, 880–887. doi: 10.1128/JB.00906-15
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wright, M. S., Jacobs, M. R., Bonomo, R. A., and Adams, M. D. (2017). Transcriptome remodeling of Acinetobacter baumannii during infection and treatment. mBio 8:e02193-16. doi: 10.1128/mBio.02193-16
Yang, J., Chen, Y., Jia, X., Luo, Y., Song, Q., Zhao, W., et al. (2012). Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin. Microbiol. Infect. 18, E506–E513. doi: 10.1111/1469-0691.12035
Zander, E., Bonnin, R. A., Seifert, H., and Higgins, P. G. (2014a). Characterization of blaOXA-143 variants in Acinetobacter baumannii and Acinetobacter pittii. Antimicrob. Agents Chemother. 58, 2704–2708. doi: 10.1128/aac.02618-13
Zander, E., Fernandez-Gonzalez, A., Schleicher, X., Dammhayn, C., Kamolvit, W., Seifert, H., et al. (2014b). Worldwide dissemination of acquired carbapenem-hydrolysing class D beta-lactamases in Acinetobacter spp. other than Acinetobacter baumannii. Int. J. Antimicrob. Agents 43, 375–377. doi: 10.1016/j.ijantimicag.2014.01.012
Keywords: Acinetobacter pittii, carbapenem resistance, OXA-499, oxacillinase, carbapenemase, phenotypic variation
Citation: Zhang L, Fu Y, Han X, Xu Q, Weng S, Yan B, Liu L, Hua X, Chen Y and Yu Y (2020) Phenotypic Variation and Carbapenem Resistance Potential in OXA-499-Producing Acinetobacter pittii. Front. Microbiol. 11:1134. doi: 10.3389/fmicb.2020.01134
Received: 28 January 2020; Accepted: 05 May 2020;
Published: 09 June 2020.
Edited by:
Benjamin Andrew Evans, University of East Anglia, United KingdomReviewed by:
Martina Barchitta, University of Catania, ItalyIgnasi Roca Subirà, Instituto Salud Global Barcelona (ISGlobal), Spain
Copyright © 2020 Zhang, Fu, Han, Xu, Weng, Yan, Liu, Hua, Chen and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Chen, chenyan@zju.edu.cn; Yunsong Yu, yvys119@zju.edu.cn
†These authors have contributed equally to this work
 Linyue Zhang
Linyue Zhang Ying Fu2,3†
Ying Fu2,3† Xinhong Han
Xinhong Han Shanshan Weng
Shanshan Weng Xiaoting Hua
Xiaoting Hua Yan Chen
Yan Chen Yunsong Yu
Yunsong Yu