
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 May 2020
Sec. Biology of Archaea
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01066
 Nienke van der Kolk1
Nienke van der Kolk1 Alexander Wagner1,2
Alexander Wagner1,2 Michaela Wagner1,3
Michaela Wagner1,3 Bianca Waßmer1
Bianca Waßmer1 Bettina Siebers2
Bettina Siebers2 Sonja-Verena Albers1*
Sonja-Verena Albers1*The thermophilic archaeon Sulfolobus acidocaldarius can use different carbon sources for growth, including the pentoses D-xylose and L-arabinose. In this study, we identified the activator XylR (saci_2116) responsible for the transcriptional regulation of the pentose transporter and pentose metabolizing genes in S. acidocaldarius. A xylR deletion mutant showed growth retardation on D-xylose/L-arabinose containing media and the lack of transcription of the respective ABC transporter. In contrast to so far used promoters for expression in S. acidocaldarius, the xylR responsive promoters have a very low background activity. Finally, two XylR dependent promoters next to the long-established maltose inducible promotor were used to construct a high-throughput expression vector system for S. acidocaldarius to efficiently clone and express proteins in S. acidocaldarius.
Although metabolically less versatile than Saccharolobus solfataricus, the thermophilic archaeon S. acidocaldarius grows on different sugar substrates, including dextrin, sucrose, D-glucose, D-xylose, and L-arabinose (Grogan, 1989; Hatzinikolaou et al., 2001). Recently, the transporter responsible for pentose uptake in S. acidocaldarius was identified: XylF (saci_2122), XylG (saci_2121), XylH (saci_2120) (Wagner et al., 2018). Also, the pentose metabolizing genes involved in the aldolase independent Weimberg pathway were characterized. The promiscuous glucose dehydrogenase 1 (GDH1, Saci_1079), D-xylonate/L-arabinoate dehydratase (D-KDXD/L-KDAD, Saci_1039), the 2-keto-deoxy-D-xylonate/L-arabinoate dehydratase (D-KDXD/L-KDAD, Saci_1939), and the α-ketoglutarate semialdehyde dehydrogenase (α-KGSADH, Saci_1938) degrade the metabolites to α-ketoglutarate, a central intermediate of the citric acid cycle. Our studies demonstrated that S. acidocaldarius strain MW001 utilizes only the Weimberg pathway and not the aldolase-dependent Dahms pathway (Wagner et al., 2018). Previous 13C labeling studies in S. acidocaldarius (DSM 639) grown on D-xylose revealed that both routes, the Weimberg and Dahms pathway, are operative in vivo at similar ratios (Nunn et al., 2010).
Like most characterized sugar transporters in Archaea, the pentose transporter in S. acidocaldarius belongs to the ATP-binding cassette (ABC) transporter systems. Whereas the other characterized archaeal sugar ABC transporters fall into the carbohydrate uptake transporters 1 (CUT1) family, this pentose transporter belongs to the carbohydrate uptake transporter 2 (CUT2) family and was obtained via horizontal gene transfer from bacteria (Wagner et al., 2018).
The pentose transporter operon encodes for a 10-transmembrane-spanning subunit and only a single nucleotide-binding domain, probably forming a homodimer (Wagner et al., 2018).
The promotor region of the genes encoding the S. acidocaldarius pentose transporter as well as pentose degrading enzymes contain a cis-regulatory element the so-called “Ara-box,” which is conserved among different Sulfolobales and has been studied intensively and been used in Sulfolobus expression systems (Brouns et al., 2006; Lubelska et al., 2006; Peng et al., 2009). The palindromic motif can be found in the promotor regions of genes upregulated in the presence of L-/D-arabinose or D-xylose (Brouns et al., 2006; Peng et al., 2009; Wagner et al., 2018). Notably, although S. acidocaldarius is in contrast to Saccharolobus solfataricus not able to utilize D-arabinose as carbon source, induction of the sugar-binding protein (SBP) promoter (saci_2122) was observed in the presence of D-arabinose (Brouns et al., 2006). Although the Ara-box has been known for a while, the respective Ara-box binding transcription factor (TF) has not been identified yet. In Archaea, multiple components of the RNA polymerase (RNAP) show homology to the eukaryotic RNAPII in both structure and organization (Zillig et al., 1979; Langer et al., 1995; Werner, 2007), and they are often considered as a simplified version of the eukaryotic machinery (Bell and Jackson, 2001). However, the eukaryal-like transcription machinery is regulated by transcription factors (TF) that resemble those in bacteria (Aravind et al., 2005; Ouhammouch et al., 2005). Like in bacteria, archaeal TFs are often homodimers recognizing specific semi-palindromic DNA motifs in the promotor region (Peeters et al., 2004, 2013; Keese et al., 2010). The largest fraction of TFs contain an HTH (helix-turn-helix) domain, although RHH (ribbon-helix-helix) motif and the zinc ribbon are not uncommon (Aravind and Koonin, 1999; Aravind et al., 2005).
Here, we present the identification of the transcriptional activator XylR (saci_2116), which is essential for the transcription of genes in the xylose/arabinose uptake and degradation pathway in S. acidocaldarius. Furthermore, we established a high-throughput expression system driven by carbon source induction. The new expression system provides functional expression plasmids based on the FX-cloning principle (Geertsma and Dutzler, 2011), which were optimized to facilitate high-throughput expression in the thermophile S. acidocaldarius. Like other expression systems in Sulfolobales (Albers et al., 2006; Peng et al., 2012), expression can be induced by addition of different carbon sources; two pentose (i.e., D-xylose, L-/D-arabinose)-inducible promotors (Psaci_1938 and Psaci_2122) and one with either maltose or dextrin induction (Psaci_1165). In this new set of vectors genes can be cloned from an entry vector with a ligation independent method in one step in various different expression vectors. In addition, traditional restriction site dependent cloning can be performed. This will facilitate efficient screening for optimal expression conditions of homologous and recombinant proteins in S. acidocaldarius.
Sulfolobus acidocaldarius was grown aerobically in a 75°C incubator in basal Brock medium at pH 3.5 (Brock et al., 1972). The medium was supplemented with 0.1% w/v NZ-Amine and with different carbon sources e.g., 0.2% dextrin w/v, 0.2% D-arabinose w/v, 0.2% L-arabinose w/v, 0.2% D-xylose w/v, 0.2% sucrose w/v or 0.2% D-glucose w/v, unless specified otherwise. MW001 and other deletion mutants (Supplementary Table S1) were supplemented with 10 μg/ml uracil for growth. The growth was monitored by determination of the optical density at 600 nm. Escherichia coli Top10 and ER1821 (New England Biolabs, Frankfurt am Main, Germany), used for cloning and methylation of the plasmids, were grown in Lysogenic broth medium (10 g/L tryptone, 5 g/L yeast, 10 g/L NaCl) at 37°C supplemented with the respective antibiotics.
To obtain a markerless deletion of saci_2116 plasmid pSVA595 was constructed. With a PCR the last ∼500 bp of saci_2116 were amplified and cloned into the MCSII of pSVA431 using restriction enzymes KpnI and NcoI. Approximately 500 bp region upstream and downstream of the target gene were PCR amplified. The two fragments were annealed by overlap extension PCR and cloned with PstI and BamHI into the MCSI site of the previously obtained plasmid. The two most outward primers (upstream and downstream) were used to create a linear fragment via PCR, which was used for transformation to competent MW001 cells as described in Wagner et al. (2012).
MW001 cells were made competent according to (Wagner et al., 2012). Methylated plasmids or linear DNA fragments were transformed by using a Gene Pulser Xcell (BioRad, München, Germany) at 2000 V, 600 Ω, 25 μF in 1 mm cuvettes. After recovery in Brock medium at 75°C, as described in Wagner et al. (2014), the cells were plated on gelrite plates lacking uracil but containing 0.2% Dextrin and 0.1% N-Z-Amine and grown for 5 days at 75°C in a plastic box to prevent the plates from drying-out. For in genome deletions the transformed cells were grown in liquid medium without uracil and plated on second selection gelrite plates containing 200 μg/ml 5-FOA (5-Fluoroorotic acid) and 10 μg/ml uracil.
The transformed cells were grown in Brock with 0.2% NZ-Amine, to prevent pre-adaptation till an OD600 nm between 0.3 and 0.5. The next day 5 ml Brock with 0.1% NZ-Amine or 0.1% NZ-Amine supplemented with 0.2% L-arabinose, 0.2% D-xylose or 0.2% glucose were inoculated with cells to an OD600 nm of 0.01. The optical density was measured over a course of 72 h. All cultures were grown in biological triplicates and technical triplicates.
For the complementation of the ΔxylR strain (MW413), pSVA-xylR was constructed by replacing the Para (Psaci_2122) and the LacZ gene in pSVAaraFX-stop by xylR, including its own promoter. The fragment was amplified by PCR and cloned into the pSVAaraFX-stop with SacII and SalI, creating pSVA-xylR.
One of the proteins of S. islandicus (m1627_2327) showed homology with XylR (54% identity, 81% positives) and in trans complementation was attempted with this gene. m1627_2327 was amplified by PCR. To make sure the start site was recognized by S. acidocaldarius, the xylR promoter (Psaci_2116) was cloned upstream of m1627_2327 by overlap PCR. The fragment was cloned into pSVAaraFX-stop with SacII and XhoI, resulting in pSVA-S.isl-xylR. The ΔxylR strain (MW413) was transformed with pSVA-xylR, pSVA-S.isl-xylR, and pSVAaraFX-empty, the latter plasmid serving as negative control. pSVAaraFX-empty was created by replacing the lacZ gene by Stop codons in all three frames. Complementation was assessed by growth comparison in 5 ml Brock medium supplemented with 0.1% N-Z-Amine or 0.1% N-Z-Amine and 0.2% L-arabinose in glass tubes covered with lid.
MW001 and MW413 were grown in triplicate in medium supplemented with 0.1% N-Z-Amine or 0.1% N-Z-Amine and 0.2% sugar (L-arabinose, D-arabinose, D-xylose, dextrin, sucrose or glucose). Cell cultures (14 ml) were harvested at mid-exponential phase (OD600 ∼ 0.4). RNA was isolated as described previously (Lassak et al., 2012). cDNA synthesis was carried out with QuantiNovaTM Reverse Transcription Kit (Qiagen, Venlo, Netherlands) according to the manufactures protocol. The quantitative RT-PCR was performed in the Magnetic Inducible Cycler (MIC) (Bio molecular systems, Upper Coomera, Australia) with 2xqPCRBIO SyGreen mix Lo-ROX (PCRBiosystems, London, United Kingdom) by following the manufacturer’s instruction. Probes were activated by heating to 95°C for 2 min, followed by 40 cycles of 95°C for 5 s and 60°C for 20 s. The transcription levels were analyzed with gene-specific primers (Supplementary Table S3) for the sugar binding protein (saci_2122). The cq-values were normalized to the cq-values of the housekeeping gene secY (saci_0574).
pSVAaraFX-lacS-xylR is based on the expression vector pSVAaraFX-Stop. This vector contains lacS, cloned into the expression cassette as reporter gene via FX cloning and additionally the xylR regulator gene under its own promoter, inserted in the backbone via restriction with PstI, XmaI. In this vector two different point mutations in the Ara-box of the ara-promoter were generated via round PCR, resulting in pSVAaraBox1 and pSVAaraBox2. Prior to transformation into S. acidocaldarius plasmids were methylated in E. coli ER1821 to prevent restriction by the SuaI restriction enzyme.
The transformed cells were grown in Brock with 0.2% NZ-Amine, to prevent pre-adaptation. The next day 5 ml Brock with 0.1% NZ-Amine and 0.2% L-arabinose or 0.2% dextrin were inoculated from a pre-culture and grown till OD600 nm 0.3–0.4. Cell pellets were collected by spinning down 2 ml at 16,000 × g for 5 min. The pellets were stored at −20°C. The experiment was performed in triplicate.
Cell pellets were lysed in Z-buffer (pH 7) (10 mM KCl, 1 mM MgSO4, 60 mM Na2HPO4 and 40 mM NaH2PO4) supplemented with 1% Triton-X to a theoretical OD of 3.2. The assay was performed at 56°C in a 96-well plate with 20 μl cell lysate, 170 μl Z-buffer and 10 μl ONPG (σ-nitrophenyl-β-d-galactopyranoside) solution (40 μM). The conversion rate of ortho-nitrophenol was measured at 410 m in a microplate reader (CLARIOstar, BMG Labtech, Ortenberg, Germany). Additionally, the protein concentration of the samples was measured with the BCA kit according to manufactures protocol (Serva Electrophoresis GmbH, Heidelberg, Germany).
The galactosidase activity was calculated in Miller Units with the equation:
For a detailed description (see Supplementary Table S2). In brief, the pSVAmZ-SH10 plasmid was used as backbone for all expression plasmids (Wagner et al., 2014). Two silent single base pair mutations were introduced to remove the SapI restriction sites from the backbone, which would spoil FX-cloning. In return the cloning cassette was modified and two SapI restriction sites compatible to the FX-cloning system were integrated. PCR products of ∼100 bp base pairs upstream of saci_2122 (pSVAaraFX-) or saci_1938 (pSVAxylFX-) were generated to create the pentose inducible expression plasmids, and a PCR product of ∼150 bp base pairs upstream of saci_1165 (pSVAmalFX-) for the maltose/dextrin inducible promoter. With an overlap PCR the N-terminal tags were fused to the different promoters and cloned into the backbone with SacII and NcoI. The overlapping primers containing the C-terminal tags were annealed and cloned into the backbone with ApaI and XhoI.
To measure the promotor activity in presence of different sugars, a PCR fragment of lacS (sso_3019) was cloned into pSVAmalFX-Stop, pSVAxylFX-Stop, and pSVAaraFX-Stop according to the FX-method with SapI, creating pSVAmalFX-lacS, pSVAxylFX-lacS, and pSVAaraFX-lacS respectively.
Transformed MW001 cells were grown on Brock with 0.1% NZ-Amine and 0.3% sucrose. The next day 5 ml Brock was inoculated with 0.1% NZ-Amine and 0.3% of the respective sugar: L-arabinose, dextrin, D-glucose, maltose, sucrose or D-xylose. After 18 h cell pellets were collected by spinning down 2 ml at full speed for 5 min in a table top centrifuge. The pellets were stored at −20°C until used for the ONPG assay.
Recently, we identified the pentose transporter in S. acidocaldarius (Wagner et al., 2018). This ABC-transporter from the CUT-2 family is most homologous to bacterial transporters. Interestingly, the promotor region comprises a palindromic motif known as the Ara-box (Wagner et al., 2018). The Ara-box is conserved among different Sulfolobales species and shown to be essential for the D-arabinose inducible promoter activity (Supplementary Figure S1) (Brouns et al., 2006; Peng et al., 2009). However, the transcription factor regulating this promoter was still unidentified. Downstream of the S. acidocaldarius pentose transporter (saci_2120–2122), the gene saci_2116 is located, which is a predicted transcriptional regulator containing a helix-turn-helix (HTH) domain (Figure 1A). This gene will now be referred to as xylR.
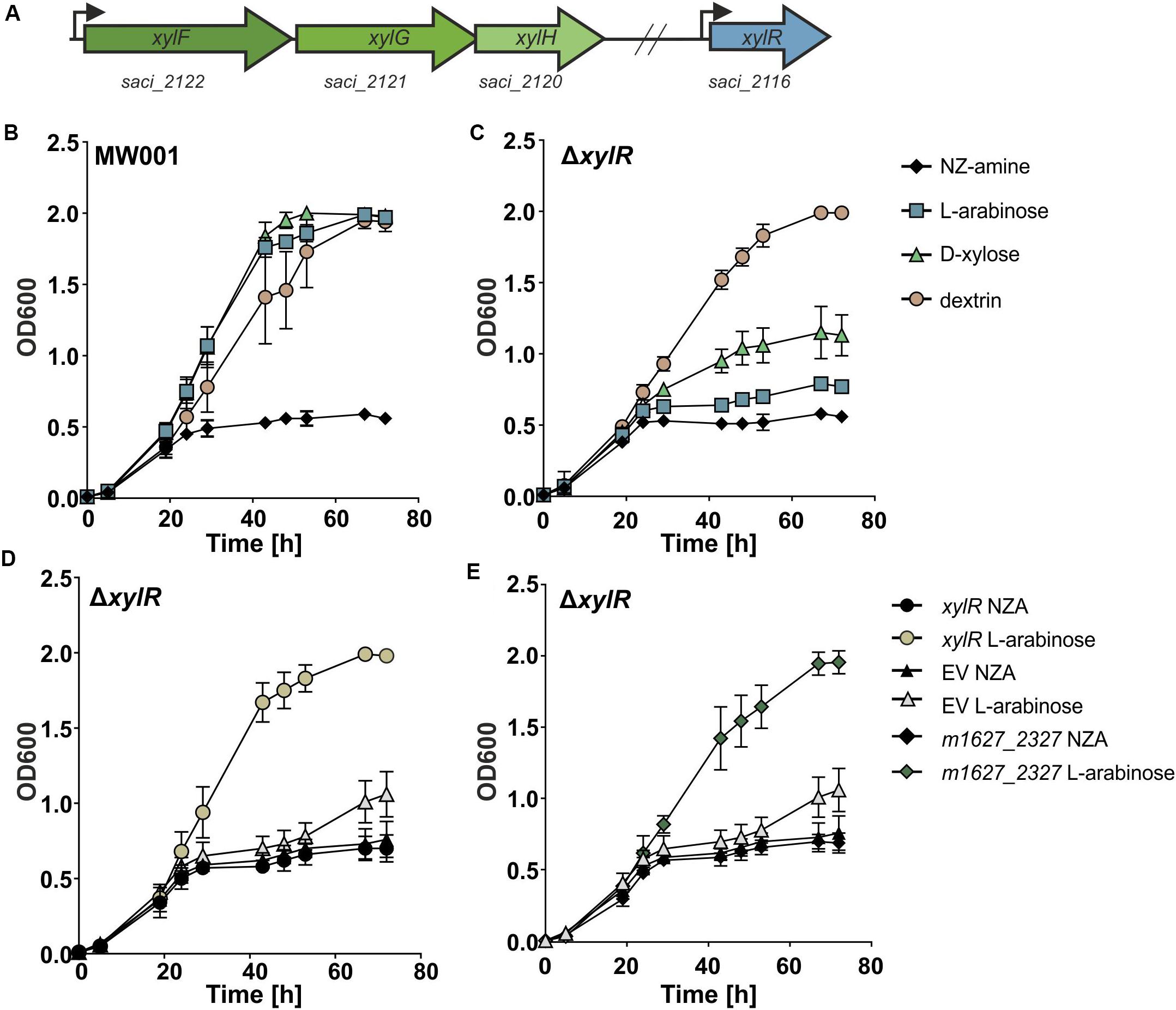
Figure 1. (A) A schematic representation of the genes encoding the pentose transporter (in green) and xylR (in blue) S. acidocaldarius. The transcription start sites are depicted with an arrow. (B) Growth comparison of parental (MW001) and (C) xylR (Δsaci_2116) deletion strain on 0.1% N-Z-Amine and 0.1% N-Z-Amine supplemented with 0.1% L-arabinose, 0.1% D-xylose or 0.1% glucose. Complementation of the ΔxylR strain with S. acidocaldarius xylR (D) and with the S. islandicus homolog of xylR (m1627_2327) (E).
To elucidate the importance of XylR for the growth of S. acidocaldarius on pentoses, a markerless deletion of xylR was made in the uracil auxotroph parent strain MW001 (Wagner et al., 2012). The effect of the knock-out of xylR on the growth of S. acidocaldarius was tested by growth experiments on different carbon sources (Figures 1B,C). When grown on L-arabinose or D-xylose the xylR deletion mutant exhibited a strong growth reduction. However, the ΔxylR strain showed no phenotype when grown on NZ-Amine alone or NZ-Amine and dextrin, excluding a general growth defect of the mutant. Also, growth of the deletion mutant could be fully restored when complemented with a plasmid expressing xylR from its own promoter (pSVA-xylR) (Figure 1D). This showed that xylR was essential for growth of S. acidocaldarius on pentoses like L-arabinose and D-xylose.
As mentioned before, the Ara-box is conserved in the promoter region of a variety of genes among different Sulfolobales species and shown to be essential for their promoter activity (Peng et al., 2009). Therefore, it is very likely that the putative regulator(s) binding to this DNA motif should be conserved among the different Sulfolobus species. The protein sequence of XylR showed homology with predicted regulators in the genomes of S. islandicus and S. solfataricus P1 with around 50% identity. Therefore, xylR of S. islandicus (m1627_2327) was expressed in the S. acidocaldarius deletion strain ΔxylR to test whether it can complement and thereby fully restore growth of the ΔxylR deletion strain on pentoses. The promoter of saci_2116 was cloned upstream of m1627_2327 in a plasmid (pSVA-S.isl-xylR) and transformed into competent ΔxylR S. acidocaldarius cells. The transformed cells were grown in Brock medium supplemented with 0.1% N-Z-Amine or 0.1% N-Z-Amine and 0.2% L-arabinose. From the growth curve it was clear that while the cells complemented with the empty plasmid (pSVAaraFX-empty) showed no growth stimulation in media supplemented with L-arabinose, the cells expressing M1627_2327 grew to higher ODs (Figure 1E). These results suggest that M1627_2327 is the XylR homolog in S. islandicus.
To confirm the role of XylR as an activator of the xyl/ara promoter, transcription levels of D-xylose/D-/L-arabinose inducible genes were tested in MW001 and the xylR deletion strain. Expression levels of the sugar binding protein of the pentose importer – XylF – (saci_2122) were tested in cells grown on different carbon sources for both MW001 and ΔxylR (Figure 2). In the parental strain, xylF transcription is highly upregulated in the presence of pentoses (i.e., L-arabinose, D-xylose, and also D-arabinose, where the latter does not serve as carbon source for S. acidocaldarius) but not in the presence of hexoses. However, in the absence of xylR no significant induction can be detected in any condition. The same results were obtained when the expression levels of the KDXD/KDAD encoding gene (saci_1939), essential for the degradation of pentoses, were tested (Supplementary Figure S2).
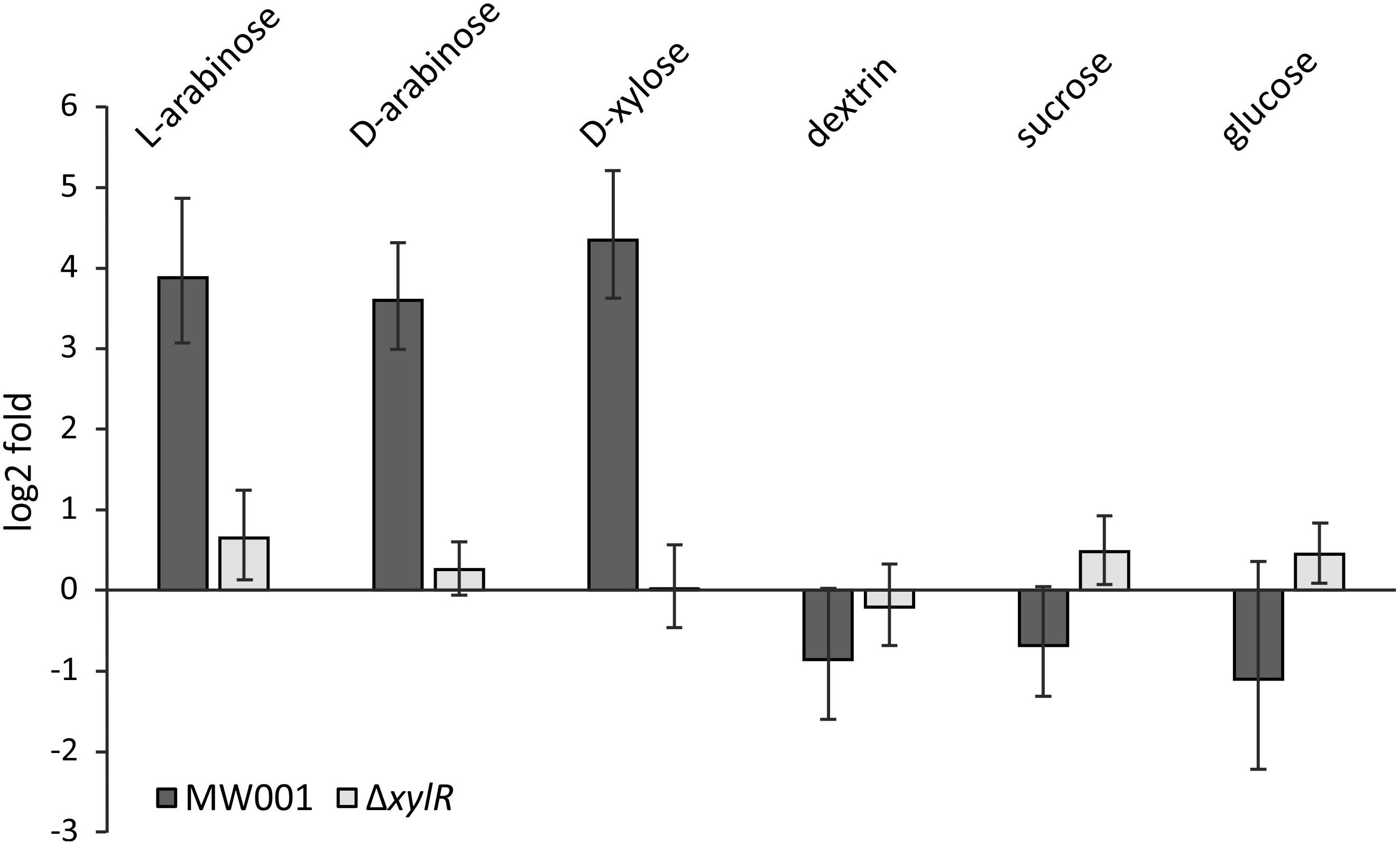
Figure 2. Transcription levels of xylF (saci_2122) in the parental strain MW001 and the ΔxylR deletion mutant when grown on different sugars. The strains were cultured on 0.1% N-Z-Amine supplemented with the different sugars (0.2% L-arabinose, 0.2% D-arabinose, 0.2% D-xylose, 0.2% dextrin, 0.2% sucrose or 0.2% D-glucose). Bars indicate the sugar-specific transcription compared to cells only grown on N-Z-Amine on log2-fold.
To verify that XylR indeed binds to the pentose inducible promoters, DNA binding assays with purified protein would be the method of choice. However, any attempt to express XylR in E. coli or in S. acidocaldarius failed as the addition of an affinity tag seemed to destabilize the protein.
Therefore, we used an ONPG assay, to verify that XylR is responsible for the induction of the pentose promoter. LacS was expressed in MW001 and MW413 (ΔxylR) from pSVAaraLacS (without xylR) or pSVAaraFX-LacS-xylR (with xylR). The cell extracts of the transformants grown on L-arabinose or dextrin were tested for hydrolysis of ONPG (Figure 3). When xylR was present, either in the genome or on the plasmid, the promotor shows high activity in the presence of L-arabinose and limited activity when the cells were grown on dextrin. However, activity is completely abolished in the absence of xylR. To confirm the basic necessity of the Ara-box the last two bases of the Ara-box were exchanged in pSVAaraFX-LacS-xylR, creating pSVAaraBox1 (Figure 3D) and pSVAaraBox2 (Figure 3E), respectively. These single point mutations in the Ara-box nearly abolished all promoter activity (Figure 3C) and therefore show the importance of the conserved motif.
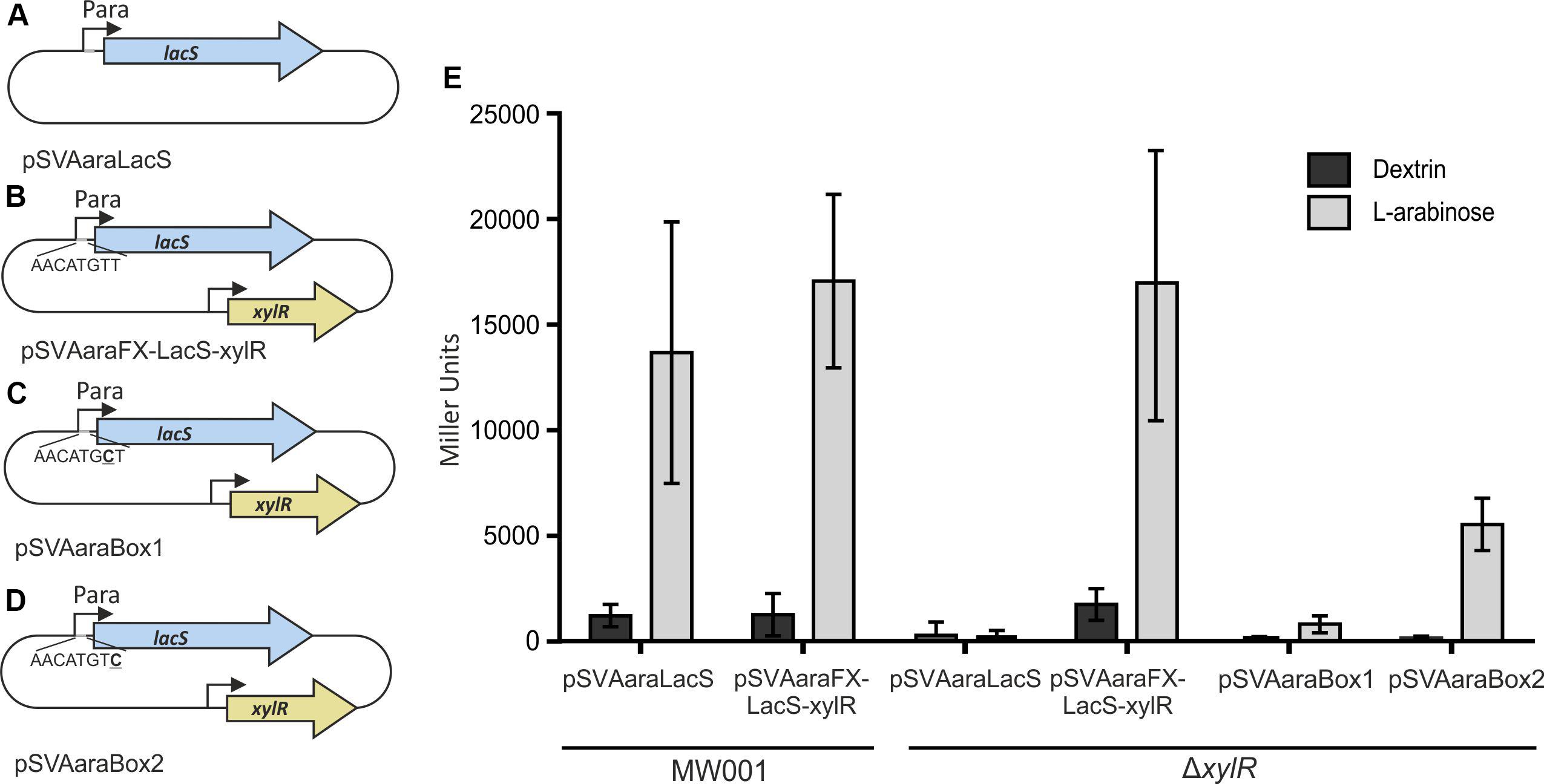
Figure 3. Schematic illustration of the different vector constructs (A–D). pSVAaraLacS (A) and pSVAaraFX-LacS-xylR (B) expressing lacS under control of the ara-promotor (Psaci_2122) and the Ara-box mutants pSVAaraBox1 (C) and pSVAaraBox2 (D) are depicted. pSVAaraFX-LacS-xylR (B) contains in addition xylR under control of its own promotor. The respective mutation in the Ara-box (C,D) is underlined. (E) β-galactosidase activity of MW001 and ΔxylR transformed with the different vector constructs. The strains were cultured in 0.1% N-Z-Amine and supplemented with either 0.2% L-arabinose or 0.2% dextrin. The β-galactosidase activity (E) is shown in Miller Units.
Although a lot of progress is made in establishing a genetic toolbox for S. acidocaldarius a high-throughput expression system is lacking. To optimize high-throughput protein expression different plasmids were constructed that provide easy expression of proteins with different promotors and tags (Table 1, detailed vector map shown in Supplementary Figure S4). This toolbox is based on the FX-cloning (Geertsma and Dutzler, 2011). The new expression system allows to insert any sequence of interest into different expression vectors with a variety of N- and C-terminal tags, which have proven to work well in S. acidocaldarius.
In difference to the established and general available FX-cloning system at Addgene repository, the suicide selection cassette was replaced by a blue/white selection cassette based on the E. coli lacZ gene. The gene of interest can be amplified using the primers designed on fxcloning.org and cloned into the target plasmid by simultaneous SapI restriction, followed by ligation and transformation (see Supplementary Figure S3).
Like other expression systems for Sulfolobales (Albers et al., 2006; Peng et al., 2012), these expression plasmids can be induced by addition of different carbohydrates. The system provides two different pentose-inducible promotors (Pxyl/Psaci_1938) and (Para/Psaci_2122) both inducible by D-xylose, L-arabinose, which serve as carbon sources for S. acidocaldarius and D-arabinose as artificial inducer and a third promoter, which is inducible with either maltose and dextrin (Pmal/Psaci_1165). In addition, the system allows for expression with different C-terminal and N-termial tags (Table 1) and is suitable for both cloning via the class IIS restriction enzyme SapI, or standard cloning with NcoI in combination with another restriction enzyme of the MCS. To compare the promoter activities on different carbon sources LacS was cloned behind all three different promotors and measured in an beta-galactosidase assay (Figure 4). The plasmids with Pmal show highest β-galactosidase activity upon maltose and dextrin induction, whereas the Pxyl and Para plasmids show induced activity upon induction with the pentoses D-xylose and L-arabinose. Both show relatively low basal activities for other sugars.
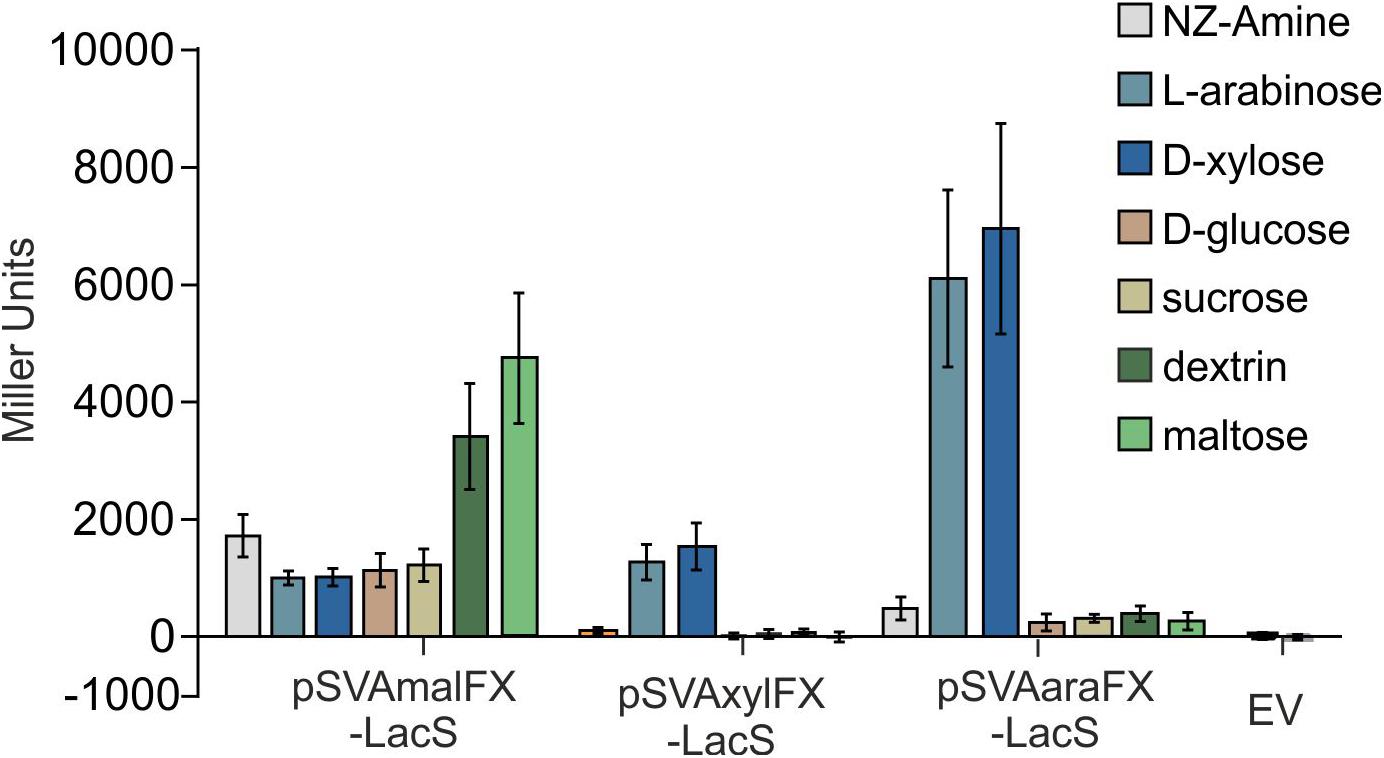
Figure 4. β-galactosidase activity of MW001 transformed with pSVAmalFX-LacS, pSVAxylFX-LacS and pSVAaraFX-LacS grown on different sugars. The strains were cultured on 0.1% N-Z-Amine (gray) and supplemented with 0.2% L-arabinose (light blue), 0.2% D-xylose (dark blue), 0.2% D-glucose (orange), 0.2% sucrose (yellow), 0.2% dextrin (dark green) or 0.2% maltose (light green). The empty vector (EV) was grown on NZ-Amine and NZ-Amine with 0.2% L-arabinose. Activity is shown in Miller Units.
Notably, by using the SapI cloning strategy for simultaneous restriction of the insert and the backbone vector in S. acidocaldarius, two additional amino acids alanine and serine are inserted behind the start methionine in the expressed proteins. To analyze for the influence of this modification at the translation start, LacS was cloned again into pSVAaraFX-Stop via classic double restriction using NcoI and XhoI, avoiding the addition of the two extra amino acids. In the subsequent ONPG assay the different vector constructs were compared and the vector pSVAaraFX-LacS(NcoI) showed a significant higher expression than pSVAaraFX-LacS (SapI cloning strategy) (Figure 5), indicating that the additional two amino acids interfere with transcription/translation and thus reduce expression of the lacS gene. However, for other genes this effect has not been observed (data not shown). Therefore, it has to be tested for each target gene whether this causes a problem or not. As shown in Figure 5 the more than 3.5kb smaller pSVAaraFX-LacS(NcoI) reaches the same expression level for lacS like the maltose inducible pSVA1450, which carries an extra copy of the regulator malR.
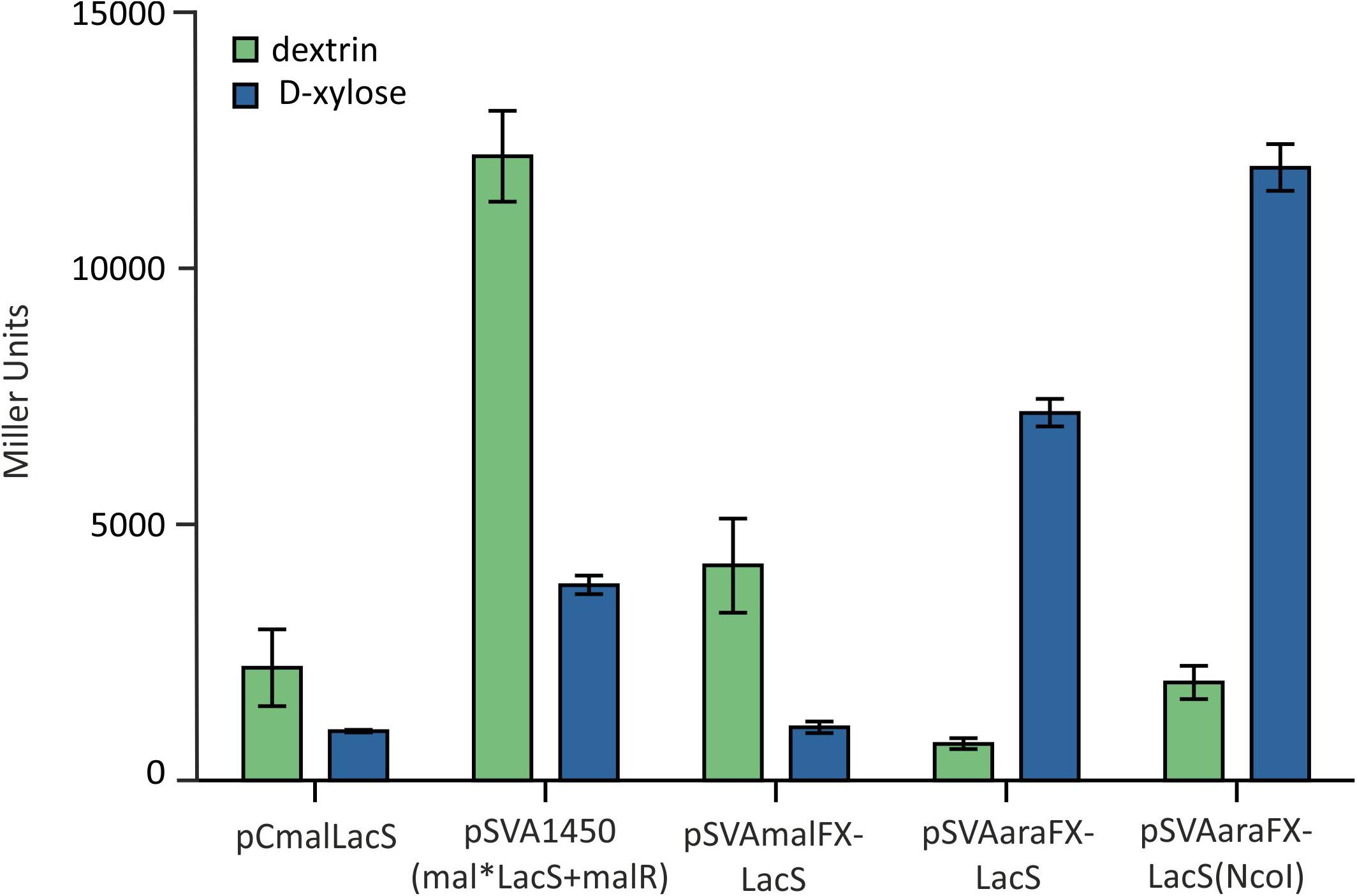
Figure 5. β-galactosidase activity of MW001 transformed with the indicated different expression vectors. The strains were cultured on 0.1% N-Z-Amine and supplemented with 0.3% dextrin (green) or 0.2% D-xylose (blue).
In order to confirm the functionality of the system we cloned the previously described esterase saci_1116 (Zweerink et al., 2017) via the NcoI and XhoI restriction site in pSVAaraFX-SH10 and pSVAaraFX-NtH10S with C-terminal Strep-tagII-10xHis-tag and N-terminal 10xHis-Strep-tagII, respectively. Cells were grown in Brock medium supplemented with 0.1% N-Z-Amine and dextrin or D-xylose, respectively (Supplementary Figure S5). For saci_1116 expression the C-terminal tag showed higher expression. In addition, only in D-xylose induced cells the expression was significantly enhanced whereas in presence of dextrin almost no expression was observed. This demonstrates again that the pentose-inducible vectors show only very low basal expression.
In this study, we identified a positive regulator responsible for controlling the expression of the genes involved in pentose import as well as pentose degradation in S. acidocaldarius: XylR. In S. acidocaldarius, xylR (saci_2116) can be found in the genetic neighborhood of the ABC-transporter genes (saci_2120–2122) responsible for pentose (i.e., D-xylose and L-arabinose) uptake. Based on the data presented in this paper, we propose that XylR is an activator that binds the Ara-box in the promotor region of genes involved in pentose uptake and degradation. In the absence of XylR, S. acidocaldarius shows significant growth retardation when grown on pentoses, and qRT-PCR studies showed significantly less transcription of the genes involved in uptake of pentose sugars [XylF, SBP encoding gene (saci_2122)]. However, transcription of xylR itself does not seem to be regulated at transcriptional levels, suggesting the existence of a different mechanism of xylR activation, e.g., via posttranslational modification or further alternative TFs acting at a higher hierarchical level.
The Ara-box motif AACATGTT was first described in S. solfataricus in the upstream region of D-arabinose-induced genes (Brouns et al., 2006). Later, the motif was found to be conserved among different Sulfolobales species and shown to be essential for the promotor activity (Peng et al., 2009, 2011, 2012; Wagner et al., 2018) and successfully used in early Sulfolobus expression systems (Albers et al., 2006). Mutagenesis of the Ara-box in combination with the LacS reporter gene assay confirmed that the conserved Ara sequence motif as well as XylR is required for L-arabinose induced expression in S. acidocaldarius. Using growth studies as well as the lacS reporter gene assay we demonstrate that the S. islandicus XylR homolog could indeed complement the S. acidocaldarius deletion mutant. Therefore, we postulate that we have identified XylR (saci_2116) as transcriptional activator of the pentose regulon in Sulfolobales. It is also interesting to note, that the regulatory elements have been conserved among the Sulfolobales, although S. acidocaldarius was shown to have acquired the L-arabinose/D-xylose ABC transporter from bacteria (Wagner et al., 2018).
To further improve the S. acidocaldarius expression system, we combined the previously established vector system with the newly identified pentose inducible promoters as well as FX cloning and constructed a whole cassette of different vectors from which genes can be expressed in S. acidocaldarius. Next to the standard His- and Strep-tags for immunodetection and simple purification in any scale, a HA-tag was chosen because of its high sensitivity and selectivity with very low background in S. acidocaldarius cell extracts. Importantly, we could show that in contrast to the so far used maltose inducible promoter, the pentose (D-xylose/L-arabinose) inducible promoters are less leaky in conditions where the inducing sugars are not added, therefore allowing for the expression of toxic gene products. Further on, since S. acidocaldarius is not able to utilize D-arabinose as carbon source, D-arabinose can be used as steady inducer similar to isopropyl-β-D-thiogalactopyranosid (IPTG). This is especially interesting for mutational analysis/complementation studies of genes involved in sugar metabolism as well as expression studies e.g., overexpression of regulatory proteins, where the induction of protein expression should not induce the utilization of alternative carbon sources and thus changes in the central carbohydrate metabolism of S. acidocaldarius. This new tool kit will further help to establish S. acidocaldarius as a model organism for research in crenarchaea and its use biotechnology (Quehenberger et al., 2017).
All datasets presented in this study are included in the article/Supplementary Material.
NK, MW, and BW executed all experiments related to XylR. AW planned and cloned the FX cloning cassette vectors and performed all test involving these. S-VA and BS conceived, supervised, and analyzed the experiments. All authors wrote the manuscript.
NK and AW were supported by the Federal Ministry of Education and Research (BMBF) [e:Bio initiative, HotSysApp, 031L0078B (S-VA), 031L0078A (BS)]. S-VA and BS acknowledge support from the HotAcidFACTORY grant [031B0848C (S-VA), 031B0848A (BS)].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01066/full#supplementary-material
Albers, S.-V., Jonuscheit, M., Dinkelaker, S., Urich, T., Kletzin, A., Tampé, R., et al. (2006). Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microbiol. 72, 102–111. doi: 10.1128/AEM.72.1.102-111.2006
Aravind, L., Anantharaman, V., Balaji, S., Babu, M., and Iyer, L. (2005). The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262. doi: 10.1016/j.femsre.2004.12.008
Aravind, L., and Koonin, E. V. (1999). DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 27, 4570–4658. doi: 10.1093/nar/27.23.4658
Bell, S. D., and Jackson, S. P. (2001). Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4, 208–213. doi: 10.1016/S1369-5274(00)00190-199
Brock, T. D., Brock, K. M., Belly, R. T., and Weiss, R. L. (1972). Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84, 54–68. doi: 10.1007/BF00408082
Brouns, S. J. J., Walther, J., Snijders, A. P. L., van de Werken, H. J. G., Willemen, H. L. D. M., Worm, P., et al. (2006). Identification of the missing links in prokaryotic pentose oxidation pathways. J. Biol. Chem. 281, 27378–27388. doi: 10.1074/jbc.M605549200
Geertsma, E. R., and Dutzler, R. (2011). A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry 50, 3272–3278. doi: 10.1021/bi200178z
Grogan, D. W. (1989). Phenotypic characterization of the Archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171, 6710–6719. doi: 10.1128/jb.171.12.6710-6719.1989
Hatzinikolaou, D. G., Kalogeris, E., Christakopoulos, P., Kekos, D., and Macris, B. J. (2001). Comparative growth studies of the extreme thermophile Sulfolobus acidocaldarius in submerged and solidified substrate cultures. World J. Microbiol. Biotechnol. 17, 229–234. doi: 10.1023/A:1016675924997
Keese, A. M., Schut, G. J., Ouhammouch, M., Adams, M. W. W., and Thomm, M. (2010). Genome-wide identification of targets for the archaeal heat shock regulator Phr by Cell-Free Transcription of Genomic DNA. J. Bacteriol. 192, 1292–1298. doi: 10.1128/JB.00924-929
Langer, D., Thuriaux, P., Zillig, W., and Hain, J. (1995). Transcription in Archaea Similarity to that in Eucarya. Proc. Natl. Acad. Sci. U.S.A. 92, 5768–5772. doi: 10.1073/pnas.92.13.5768
Lassak, K., Neiner, T., Ghosh, A., Klingl, A., Wirth, R., and Albers, S.-V. (2012). Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83, 110–124. doi: 10.1111/j.1365-2958.2011.07916.x
Lubelska, J. M., Jonuscheit, M., Schleper, C., Albers, S.-V., and Driessen, A. J. M. (2006). Regulation of expression of the arabinose and glucose transporter genes in the thermophilic archaeon Sulfolobus solfataricus. Extremophiles 10, 383–391. doi: 10.1007/s00792-006-0510-517
Nunn, C. E. M., Johnsen, U., Schönheit, P., Fuhrer, T., Sauer, U., Hough, D. W., et al. (2010). Metabolism of pentose sugars in the hyperthermophilic archaea Sulfolobus solfataricus and Sulfolobus acidocaldarius. J. Biol. Chem. 285, 33701–33709. doi: 10.1074/jbc.M110.146332
Ouhammouch, M., Langham, G. E., Hausner, W., Simpson, A. J., El-Sayed, N. M. A., and Geiduschek, E. P. (2005). Promoter architecture and response to a positive regulator of archaeal transcription. Mol. Microbiol. 56, 625–637. doi: 10.1111/j.1365-2958.2005.04563.x
Peeters, E., Peixeiro, N., and Sezonov, G. (2013). Cis -regulatory logic in archaeal transcription. Biochem. Soc. Trans. 41, 326–331. doi: 10.1042/BST20120312
Peeters, E., Thia-Toong, T.-L., Gigot, D., Maes, D., and Charlier, D. (2004). Ss-LrpB, a novel Lrp-like regulator of Sulfolobus solfataricus P2, binds cooperatively to three conserved targets in its own control region. Mol. Microbiol. 54, 321–336. doi: 10.1111/j.1365-2958.2004.04274.x
Peng, N., Ao, X., Liang, Y. X., and She, Q. (2011). Archaeal promoter architecture and mechanism of gene activation. Biochem. Soc. Trans. 39, 99–103. doi: 10.1042/BST0390099
Peng, N., Deng, L., Mei, Y., Jiang, D., Hu, Y., Awayez, M., et al. (2012). A Synthetic Arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic Archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 78, 5630–5637. doi: 10.1128/AEM.00855-812
Peng, N., Xia, Q., Chen, Z., Liang, Y. X., and She, Q. (2009). An upstream activation element exerting differential transcriptional activation on an archaeal promoter. Mol. Microbiol. 74, 928–939. doi: 10.1111/j.1365-2958.2009.06908.x
Quehenberger, J., Shen, L., Albers, S. V., Siebers, B., and Spadiut, O. (2017). Sulfolobus - A potential key organism in future biotechnology. Front. Microbiol. 8:2474. doi: 10.3389/fmicb.2017.02474
Wagner, M., Shen, L., Albersmeier, A., van der Kolk, N., Kim, S., Cha, J., et al. (2018). Sulfolobus acidocaldarius transports pentoses via a carbohydrate uptake transporter 2 (CUT2)-type ABC transporter and metabolizes them through the aldolase-independent Weimberg pathway. Appl. Environ. Microbiol. 84:AEM.01273-17. doi: 10.1128/AEM.01273-1217
Wagner, M., van Wolferen, M., Wagner, A., Lassak, K., Meyer, B. H., Reimann, J., et al. (2012). Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front. Microbiol. 3:214. doi: 10.3389/fmicb.2012.00214
Wagner, M., Wagner, A., Ma, X., Kort, J. C., Ghosh, A., Rauch, B., et al. (2014). Investigation of the malE promoter and MalR, a positive regulator of the maltose regulon, for an improved expression system in Sulfolobus acidocaldarius. Appl. Environ. Microbiol. 80, 1072–1081. doi: 10.1128/AEM.03050-3013
Werner, F. (2007). Structure and function of archaeal RNA polymerases. Mol. Microbiol. 65, 1395–1404. doi: 10.1111/j.1365-2958.2007.05876.x
Zillig, W., Stetter, K. O., and Janekovic, D. (1979). DNA-Dependent RNA Polymerase from the Archaebacterium Sulfolobus acidocaldarius. Eur. J. Biochem. 96, 597–604. doi: 10.1111/j.1432-1033.1979.tb13074.x
Keywords: genetics, genetic tools, transcriptional regulator, xylose metabolism, archaea, Sulfolobus
Citation: van der Kolk N, Wagner A, Wagner M, Waßmer B, Siebers B and Albers S-V (2020) Identification of XylR, the Activator of Arabinose/Xylose Inducible Regulon in Sulfolobus acidocaldarius and Its Application for Homologous Protein Expression. Front. Microbiol. 11:1066. doi: 10.3389/fmicb.2020.01066
Received: 28 March 2020; Accepted: 29 April 2020;
Published: 26 May 2020.
Edited by:
Qunxin She, Shandong University, ChinaReviewed by:
Changyi Zhang, University of Illinois at Urbana–Champaign, United StatesCopyright © 2020 van der Kolk, Wagner, Wagner, Waßmer, Siebers and Albers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sonja-Verena Albers, c29uamEuYWxiZXJzQGJpb2xvZ2llLnVuaS1mcmVpYnVyZy5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.