- 1Shenzhen Key Laboratory of Microbial Genetic Engineering, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 2Key Laboratory of Functional Inorganic Material Chemistry (MOE), School of Chemistry and Materials Science, Heilongjiang University, Harbin, China
The sucrose non-fermenting 1/AMP-activated protein kinase (SNF1/AMPK) is a central regulator of carbon metabolism and energy production in the eukaryotes. In this study, the functions of the Podospora anserina SNF1 (PaSNF1) ortholog were investigated. The ΔPaSNF1 mutant displays a delayed development of mycelium and fruiting bodies and fails to form ascospores. The expression of the PaSNF1 gene in the strain providing female organs in a cross is sufficient to ensure fertility, indicating a maternal effect. Results of environmental stress showed that ΔPaSNF1 was hypersensitive to stress, such as osmotic pressure and heat shock, and resistant to fluconazole. Interestingly, the knockout of PaSNF1 significantly promoted sterigmatocystin (ST) synthesis but suppressed cellulase [filter paperase (FPA), endoglucanase (EG), and β-glucosidase (BG)] activity. Further, transcriptome analysis indicated that PaSNF1 made positive regulatory effects on the expression of genes encoding cellulolytic enzymes. These results suggested that PaSNF1 may function in balancing the operation of primary and secondary metabolism. This study suggested that SNF1 was a key regulator concerting vegetative growth, sexual development, and stress tolerance. Our study provided the first genetic evidence that SNF1 was involved in the ST biosynthesis and that it may also be a major actor of lignocellulose degradation in P. anserina.
Introduction
Lignocellulose is one of the most abundant renewable resources on earth, mainly composed of cellulose, hemicellulose, and lignin (Jonsson et al., 2013). It can be converted into different carbohydrates, including glucose, xylose, and aromatic monomer compounds, which could be applied for the production of ethanol, carbohydrates, and aromatic products (Ragauskas et al., 2014; Kawaguchi et al., 2016; Wu et al., 2018). It has promising applications in food, paper, and textile industries as well as the production of biofuels and other chemicals (Paulova et al., 2015; Kubicek and Kubicek, 2016; Yenkie et al., 2016). However, the commercial applications of lignocellulosic degradation in the production of biofuels and chemicals were limited by its complex structure, biological obstinacy, low efficiency, and high cost, thus hindering the effective utilization of the raw material in green technology (Weng et al., 2008; Paulova et al., 2015). Biodegradation of lignocellulose into fermentable sugars with cellulase [including Carbohydrate-Active Enzymes (CAZy)] has been an environmental-friendly approach (Gupta et al., 2016). Filamentous fungi were considered as promising candidates for producing lignocellulolytic enzymes (Couturier et al., 2016).
Filamentous fungi and their enzymes show great potential in the degradation of tough lignocellulose (Bomble et al., 2017). Some ascomycetes such as Trichoderma reesei and Aspergillus nidulans have been commonly applied in the industrial production of cellulase and hemicellulase (de Assis et al., 2015; Bischof et al., 2016). Many cellulose and lignin degradation-associated enzymes were found in the filamentous fungus Podospora anserina, making this fungus a model to elucidate the enzymatic decomposition of plant biomass (Couturier et al., 2016). Recent studies have shown that catalase, laccase, and bilirubin oxidase participated in the degradation of cellulose in P. anserina (Bourdais et al., 2012; Xie et al., 2014, 2015). The regulation mechanism of cellulase and hemicellulase gene expression has been extensively investigated (Benocci et al., 2017). In cellulolytic fungi, several key transcriptional activators as well as repressors have being involved in this regulation. For example, the transcription factor XlnR/XYR1 regulated the expression of cellulase and hemicellulase genes in Aspergillus niger, Neurospora crassa, and T. reesei, respectively (Stricker et al., 2008; Sun et al., 2012; Dos Santos, Castro et al., 2016). In addition, the transcription factors CLR-1 and CLR-2 were essential for growth on cellulose in N. crassa (Coradetti et al., 2012). Recent studies have shown that the CLR-4 acted as a positive activator of cellulase expression by directly binding to the promoters of CLR-1 and CLR-2 (Liu et al., 2019). CLR-3 repressed the cellulolytic response through CLR-1 (Huberman et al., 2017). The conserved zinc-finger transcription factor Cre1/CreA was the key factor in mediating carbon catabolite repression (CCR), an important mechanism for suppressing cellulase production during growth on favorably metabolized carbon source, such as glucose (Sun and Glass, 2011; Lichius et al., 2014). Deletion of Cre1/CreA alleviated some aspects of CCR for cellulolytic enzyme expression in Penicillium decumbens and N. crassa (Sun and Glass, 2011; Liu et al., 2013). Therefore, transcriptional activation or de-repression via nutrient sensing pathways has also been essential for inducing the lignocellulolytic response.
Sucrose non-fermenting 1/AMP-activated protein kinase (SNF1/AMPK kinases) was regarded as an important regulator that participated in the development of fungi and activation of catabolite-repressed genes (Lee et al., 2009; Ghillebert et al., 2011). In yeast, as a heterotrimer kinase complex, SNF1 contained α-subunit encoded by SNF1; β-subunits encoded by GAL83, SIP1, and SIP2; and γ-subunit encoded by SNF4. The SNF1 complex can be activated by three upstream kinases (Sak1, Elm1, and Tos3) via phosphorylation (Hedbacker and Carlson, 2008), whereas it could also be inactivated by Reg1-Glc7 protein phosphatase (Sanz et al., 2000). The regulation of SNF1 activity played a central role in the alleviation of CCR, for both Saccharomyces cerevisiae and the majority of filamentous fungi (Brown et al., 2013). In yeast, under the glucose starvation, Mig1 was phosphorylated by SNF1, inducing the de-repression of glucose repressed genes. Studies have shown that the CCR mechanism was different in filamentous fungi and yeast. CreA, a Mig1 homolog, was a critical regulator of carbon catabolism inhibition in the lignocellulolytic fungus A. nidulans and the orthologous SNF1 kinase was essential for CreA/Cre1 de-repression (Brown et al., 2013). However, homologous proteins of Cre1 could not be phosphorylated by homologous proteins of SNF1 in T. reesei (Cziferszky et al., 2003). Overall, it was unclear whether there was a conserved mechanism for SNF1 to regulate carbon catabolite in filamentous fungi. Little was known about the functions of SNF1 in the fungal development and lignocellulosic degradation.
Besides, SNF1 complex participated in a wide variety of physiological processes, such as fungal meiosis, spore formation, filamentous growth, and biofilm formation (Lee et al., 2009; Ghillebert et al., 2011). It was also involved in the response to environmental stresses, such as salt stress, heat shock, alkaline pH, and endoplasmic reticulum stress (Hahn and Thiele, 2004; Hong and Carlson, 2007; Casamayor et al., 2012; Ferrer-Dalmau et al., 2015). For many pathogenic filamentous fungi, SNF1 genes showed critical roles in fungal development and virulence, such as Magnaporthe oryzae (Yi et al., 2008), Gibberella zeae (Lee et al., 2009), Verticillium dahlia (Tzima et al., 2011), Penicillium digitatum (Zhang et al., 2013), Beauveria bassiana (Wang et al., 2014), Leptosphaeria maculans (Feng et al., 2014), and Fusarium virguliforme (Islam et al., 2017). The lack of SNF1 led to inhibitory expression of cell-wall-degrading enzymes, damage of sporulation, and spore germination; thus, its virulence on the hosts would be impaired.
Biosynthesis of secondary metabolites (SMs) may be tightly regulated. Their expression was often triggered by chemical or environmental stimuli and coordinated with the development and morphogenesis of the producing organism. Sterigmatocystin (ST) was one of the ultimate precursors in the biosynthesis of the aflatoxins. The gene cluster responsible for the ST biosynthesis was identified in A. nidulans by Brown et al. (1996). A similar cluster of genes was found in P. anserina (Slot and Rokas, 2011). ST was detected in cultures of a freshly isolated P. anserina strain (Bills and Gloer, 2016). The ST biosynthetic gene cluster was regulated by the cluster-specific AflR transcription factor, which functioned downstream of glucose sensing (Fernandes et al., 1998). Overexpression of PaAflR led to a ST overproduction and a female sterility phenotype in P. anserina (Shen et al., 2019). Additionally, LaeA also regulated the ST biosynthetic gene cluster in an AflR-dependent manner. The ΔLaeA mutation abolishes AflR expression and subsequently the production of ST and other SMs (Bok and Keller, 2004). Interestingly, Cre1, which regulated cellulase production, was also involved in the synthesis of SMs (Monroy et al., 2017). However, the importance of SNF1 and their functional connections in regulating SMs have been unknown.
Elucidating the physiological functions of SNF1 contributed to understanding the mechanisms of filamentous fungi sense nutrition and cellular energetic status, which was paramount for the industrial production of cellulase. In this study, we identified a homolog of SNF1 in the genome of P. anserina. With the use of a homologous recombination strategy, the disruption mutant of P. anserina SNF1 (PaSNF1) was constructed, and the effects of PaSNF1 in fungal development and lignocellulosic degradation were characterized. ΔPaSNF1 was characterized for defects in sexual reproduction (e.g., fruiting body formation), colony formation, ascospore germination, and stress responses. Our study provided the first genetic evidences that SNF1 was involved in the ST biosynthesis. Enzymology and transcriptome analysis confirmed that PaSNF1 played important roles in cellulose degradation.
Materials and Methods
Strains and Culture Conditions
All strains of Podospora anserina applied in this study were derived from the “S” wild-type (WT) strain, which was sequenced for obtaining the genome of the P. anserina (Espagne et al., 2008; Grognet et al., 2014; Silar et al., 2019). The standard culture protocol of P. anserina could be found at http://podospora.igmors.u-psud.fr/methods.php. The Δmus51:Hygro mutant strain differed from the S WT reference strain by a single deletion of the mus-51 gene, in which frequency of targeted gene replacement was increased (El-Khoury et al., 2008).
Phylogenetic Analysis
The putative SNF1 genes of P. anserina were identified by BLAST at the P. anserina website by retrieving various fungal SNF11. In this study, the putative SNF1 encoded by Pa_2_770 was named as PaSNF1. The alignment was manually refined, and sites with ambiguous alignment were removed from the dataset. The phylogenetic tree was reconstructed using a neighbor-joining method, and an alignment was produced with the CLUSTALW algorithm in the MEGA5 software (Wang et al., 2014), in which the numbers at nodes indicated bootstrap values with 1,000 replications.
Generation of Deletion Mutants and Complementation
A deletion cassette for the SNF1 (Pa_2_770) gene was constructed with a geneticin resistance cassette as previously reported (Xie et al., 2014, 2015, 2018). The Δmus51:Hygro protoplasts were transformed by the PCR products with geneticin cassette. Transformants were screened on selective medium containing 100 μg/ml of geneticin and 75 μg/ml of hygromycin. Putative deleted mutants were firstly selected by PCR. The PCR amplified specific junctions to the replaced locus, with two primers that annealed at one end of the selectable resistance gene and upstream of the proximal flank in the deletion cassette (Supplementary Table S1). Homologous recombination of the deletion cassette allowed amplification of a predictable fragment on each side of the selectable resistance gene. Three candidate transformants would be genetically purified by crossing them with WT. It aimed to eliminate the potential untransformed nuclei and then segregate out the Δmus-51 mutation. Mat- and mat + geneticin-R and hygromycin-S strains were identified from each progeny, and one transformant was subjected to Southern blot analysis for final validation (Supplementary Figure S2). Only purified transformants verified by Southern blot would be selected as stock deletion mutants for subsequent studies.
To ensure that the relevant PaSNF1 gene was inactivated in the derived phenotypes, the complementation procedure was done by ectopic insertion of corresponding gene under the control of its native promoter. The WT with complete CDS and the 5′ (500 bp) and 3′ (200 bp) flanking regions were amplified by PCR using a high-fidelity polymerase and primers of PaSNF1-cF and PaSNF1-cR (Supplementary Table S1). The DNA fragment was cloned into the pBC-Nourseo (Lalucque et al., 2012) at the NotI and BamHI sites to yield pBC-Nours-PaSNF1 plasmid. This plasmid was transformed into the protoplasts of the ΔPaSNF1 mutant strain. Transformants (CPPaSNF1) were screened on selective medium with 40 μg/ml of nourseothricin (Xie et al., 2018). The presence of the WT allele was also verified by PCR.
Growth and Development in Standard Conditions
To determine the roles of PaSNF1 in fungal physiology, both the deleted mutant and WT were incubated separately on M2 medium at 27°C. Colony morphology, pigmentation, perithecium formation, ascospore production, ascospore dispersal, and germination were observed during the vegetative growth and sexual cycle. Three plates were set for each treatment, and the experiment was repeated once.
Stress Sensitivity Assays
In this study, mat + mating type strains were used unless specified. For evaluating the stress sensitivity, WT, ΔPaSNF1, and CPPaSNF1 were incubated separately on M2 medium, supplemented with osmotic stress reagents (0.5 M of NaCl, 0.5 M of KCl, 0.5 M of sorbitol, and 0.75 M of glycerol), oxidative reagents (0.01% H2O2 and 50 μM of menadione), cell wall inhibitors [0.02% Congo red and 0.05% sodium dodecyl sulfate (SDS)], and cell membrane inhibitor (2.5 μg/ml of fluconazole). The menadione was obtained from Macklin, and fluconazole was purchased from Aladdin. Fresh mycelium was incubated at 45°C in water bath for 2 h. Equal volume of mycelium was inoculated on M2. Equal volume of mycelium without heat shock served as control. All cultures were incubated at 27°C for 3 days, colony size was measured, three plates were set for each treatment, and the experiment was repeated once.
Detection of Secondary Metabolites by High-Performance Liquid Chromatography
To analyze the SMs, both WT and ΔPaSNF1 (mat + mating type) strains were incubated separately in 100 ml of liquid medium (in 250-ml triangle bottle) at 27°C, shaking at 150 rpm for 7 days. Mycelium and the liquid phase were separated by centrifugation. The supernatant was extracted for three times with an equal volume of ethyl acetate. The organic phase was transferred for evaporation to yield a concentrated residue. The residue was re-suspended in 1 ml of methanol and then centrifuged at 12,000 rpm for 10 min. The obtained supernatant was filtered with 0.22-μm Millipore filter and then analyzed with high-performance liquid chromatography (HPLC) (Waters e2695 separations Module) with 2998 PDA detector with ultimate XB-C18 column (4.6 × 250 mm, Welch Materials, Inc.). The conditions for HPLC were as follows: flow rate was 1 ml/min, column temperature was 30°C, and the solvent gradients were solvent A (H2O) and solvent B (methanol). The sample was detected for 20 min using a linear gradient of 10–90% (0–20 min), 100% MeOH (20.01–30 min), and 10% MeOH (30.01–30 min). The wavelength of UV detector was set at 254 nm.
Characterization of Sterigmatocystin Synthesis in ΔPaSNF1 Mutant
For the purification of these compounds, ΔPaSNF1 mutant (mat + mating type) were cultured in 5 l of liquid medium at 27°C, shaken at 150 rpm for 7 days. Culture media were Solution 1 0.5% (v/v), Solution 2 0.5% (v/v), Solution 3 0.5% (v/v), Solution 4 0.5% (v/v), biotin 0.05% (v/v), thiamine 0.05% (v/v), microelement 0.1% (v/v), yeast extract 0.5% (m/v), and dextrin blonde 0.55% (m/v). The standard culture protocol of P. anserina could be found at http://podospora.igmors.u-psud.fr/methods.php. The culture was extracted with ethyl acetate for three times. The organic phase was evaporated to obtain the crude residue (1.3 g). The residue was separated on silica gel column with a stepped gradient elution of CH2Cl2–MeOH (100:0–0:100) to yield 6 subfractions (Fr. 1–Fr. 6). Fr. 1 (a total of 105.4 mg) was further purified by semi-preparative HPLC, and ST was gained (at 18.5 min). Nuclear magnetic resonance (NMR) spectra were performed on a Bruker Avance III 500 spectrometer (600 MHz). DMSO-d6 was applied as the solvent for analyzing ST (Pachler et al., 1976; Oleinikova et al., 2012).
Cellulose Degradation and Enzyme Assays
For evaluating the activity of cellulase, both WT and ΔPaSNF1 (mat + mating type) were cultured. The mycelium agar pieces (2 × 2 mm) from actively growing WT and mutant strains were transferred to Petri dishes with cellophane placed on M2 medium, and then the mycelia were incubated at 27°C for 2 days (Xie et al., 2015). The harvested mycelium was smashed in 200 μl of extraction buffer (sodium citrate buffer 50 mM, pH7.0) by FastPrep (4.0) for 20 s. All the samples were centrifuged at 10,000 rpm for 10 min to obtain the supernatants. The activity of FPA and EG were measured by the 3,5-dinitrosalicylic acid (DNS) method with filter paper and carboxymethyl cellulose as substrates (Liao et al., 2018). The activity of BG and exo-1,4-β-glucanase (CBH) was determined using p-nitrophenyl-β-D-glucopyranoside (pNPG) and p-nitrophenol-D-cellobioside (pNPC) as the substrates, respectively (Li et al., 2016). Protein concentration was quantified with Bradford method (Bradford, 1976). All the determinations were performed in triplicate. To evaluate the cellulose degradation, the M2 standard minimal synthetic medium contains dextrin (0.5%) as carbon source. Dextrins were replaced with microcrystalline cellulose (MCC; 0.5%) from Sigma and 0.5 g/plate straw materials. Fragmented mycelia from mat + and mat- strains of WT or ΔPaSNF1 were mixed and then inoculated in the center of the plate under constant light illumination. Photographs were taken after 7 days of incubation.
Transcriptome Analysis
The mycelia of WT and ΔPaSNF1 grown in cellulose (3 days) were harvested by filtration and immediately frozen in liquid nitrogen. The culture medium was a fungal liquid culture medium. An equivalent amount of carbon source dextrin was replaced with microcrystalline cellulose. The composition of the medium was as follows: Solution 1 0.5% (v/v), Solution 2 0.5% (v/v), Solution 3 0.5% (v/v), Solution 4 0.5% (v/v), biotin 0.05% (v/v), thiamine 0.05% (v/v), microelement 0.1% (v/v), yeast extract 0.5% (m/v), and Avicel (Sigma) 0.55% (m/v). Total RNA was extracted using TRIzol® RNA reagent (Invitrogen, Carlsbad, CA, United States) and further purified using DNase I (RNeasy Mini kit, Qiagen, Hilden, Germany). NanoDrop and agarose gel electrophoresis were used to check RNA integrity and concentration.
Qualified RNA with an OD260/OD280 > 1.8 and RNA integrity number (RIN) > 7.0 was prepared according to Majorbio (Shanghai, China) standard protocols and sequenced on an Illumina HiSeq 2000/2500 platform (San Diego, CA, United States). All data in this study were generated by sequencing two independent duplicate samples. Total clean reads were mapped against predicted transcripts from the WT P. anserina strain genome2,3 to assess sequence homology and functional annotation. Read mapping and counting using TopHat (version 2.1.1) and hisat2 (version 2.1.0) were performed. RSEM (version 1.3.1) software was used to analyze gene expression levels with the fragments per kilobase of exon per million mapped reads (FPKM) method. Differentially expressed genes (DEGs) were screened using the DESeq2 (version 1.24.0) with fold change of more than 1.5 and p-value ≤ 0.05 as thresholds. The dataset was also manually screened for the genes encoding carbohydrate active enzymes. Hierarchical clustering analysis was performed using the pheatmap package in R (version 3.6.1)2. RNA-Seq data were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number PRJNA597840.
Quantitative RT-PCR
The mycelia of WT and ΔPaSNF1 grown in cellulose (3 days) were harvested by filtration and immediately frozen in liquid nitrogen. Total RNA was extracted using TRIzol® RNA reagent (Invitrogen, Carlsbad, CA, United States) for total RNA extraction. All total RNA samples were reversely transcribed into cDNAs and then assessed for the transcript levels of 20 genes via RT-qPCR with paired primers (Supplementary Table S1). All RT-qPCR experiments with three cDNA samples of each strain were performed under the action of SYBR® Premix Ex TaqTM (TaKaRa, Dalian, China). Initially, three reference genes were used for normalization: GPD (glyceraldehyde 3-phosphate dehydrogenase, formally Pa_3_5110), PDF2 (protein phosphatase PP2A regulatory subunit A, Pa_7_6690), and UBC (ubiquitin, Pa_4_7790). All three were stably expressed; comparatively speaking, PDF2 has the best stability, and in RNA-Seq data, the difference between the FPKM of PaPDF2 in WT and ΔPaSNF1 was not significant. Thus, only the PaPDF2 gene was used as a reference gene to normalize gene expression results. The ratio of each gene transcript in ΔPaSNF1 over that in WT was converted to log2 for comparison with the corresponding log2 ratio from the RNA-Seq data.
Microscopic Observations
The perithecia of mycelia grown on agar were observed and captured with a homemade microscope, combining a camera [monochromatic 1/3′′ complementary metal–oxide–semiconductor (CMOS) sensor with a resolution of 1,280 × 960 pixels] and a telecentric objective (4 × magnification and 65-mm focal length). Pictures of squashed perithecia and ascospore were captured with a Nikon eclipse Ni-V microscope and analyzed with ImageJ.
Results
Identification of SNF1 Homolog in Podospora anserina and Phylogenetic Analysis
The genome of Podospora anserina has been sequenced4. SNF1 protein sequences of yeast and two filamentous fungi Neurospora crassa and Hypocrea jecorina were used as a query to identify the homolog in P. anserina genome. A homologous protein was found in P. anserina, presenting the highest identity with N. crassa, H. jecorina, and yeast, which was 75.64, 83.63, and 41%, respectively, and designated as SNF1 (Protein ID: CAP72646.1; Pa_2_770). The open reading frame (ORF) of SNF1 in P. anserina was predicted to be 2,127 bp in length, encoding a peptide of 708 amino acids with a predicted molecular weight of ∼79.91 kDa (Supplementary Figure S1A). SMART analysis5 of SNF1 suggested that this protein contained a serine/threonine (Ser/Thr) kinase catalytic domain (KD) (residues 67–318) and a two C-terminal domain (CTD) (residues 532–628 and residues 633–704). KD with the identity of 78.17% exhibited a high similarity to the SNF1 Ser/Thr catalytic domain in Saccharomyces cerevisiae. The degree of similarity was even lower at the CTD (25% identity), and only a few blocks showed identical residues (Supplementary Figures S1,C). Maximum likelihood method was applied to analyze the similarities among above 11 proteins, and a phylogenetic tree was constructed (Figure 1). The predicted amino acid sequence of PaSNF1 shared an identity of 75.64% with NcSNF1 of N. crassa (CAD70761), 64.67% with HjSNF1 of H. jecorina (AF291845) (Cziferszky et al., 2003), 67.73% with FoSNF1 of Fusarium oxysporum (AAN32715) (Ospina-Giraldo et al., 2003), 66.06% with BbSNF1 of Beauveria bassiana (EJP66968) (Wang et al., 2014), 63.41% with PmSNF1 of Pestalotiopsis microspora (AUW40141) (Wang et al., 2018), 57.53% with MgSNF1 of Magnaporthe grisea (ABF48563) (Yi et al., 2008), 57.94% with SsSNF1 of Sclerotinia sclerotiorum (CAB40826) (Vacher et al., 2003), 44.35% with PdSNF1 of Penicillium digitatum (AFS18464) (Zhang et al., 2013), and 43.43% with CcSNF1 of Cochliobolus carbonum (AAD43341) (Tonukari et al., 2000). PaSNF1 protein showed lower homology with SNF1 protein from S. cerevisiae, which was 42%. SNF1 orthologs showed that PaSNF1 from different fungi clustered together, whereas yeast homologs formed a separate cluster (Figure 1).
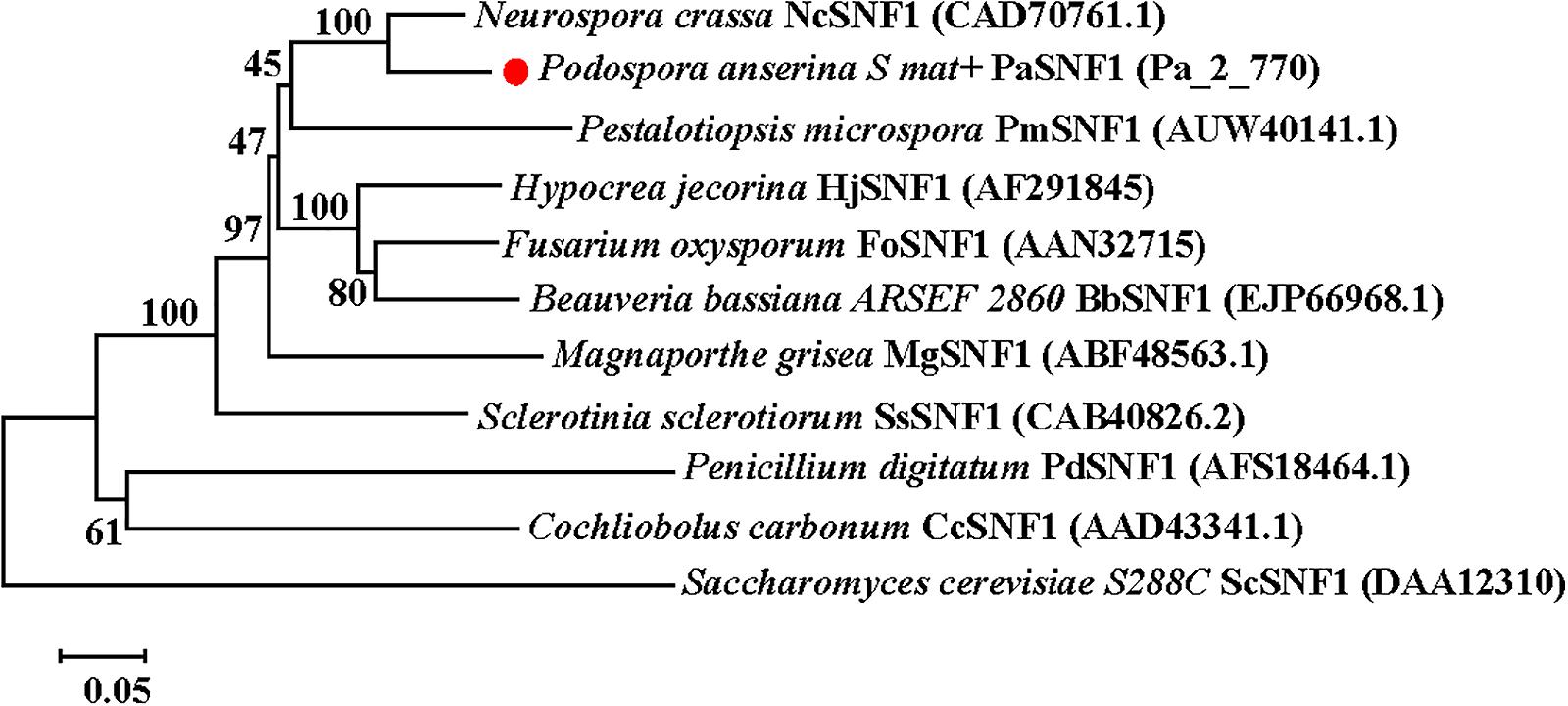
Figure 1. Phylogenetic analysis of PaSNF1 and SNF1 orthologs identified in other organisms. The phylogenetic tree was reconstructed using a neighbor-joining method. Numbers at nodes indicate bootstrap values with 1,000 replications. GenBank accession number of a given SNF1-coding gene was parenthesized following the name of each fungal species. Distance scale: scale that represents the number of differences between sequences (e.g., 0.05 means 5% differences between two sequences). SNF1, sucrose non-fermenting 1; PaSNF1, Podospora anserina SNF1.
Creation of PaSNF1 Knockout and Complementation Mutants
PaSNF1 disruption mutant was generated by replacing a coding sequence of PaSNF1 with the genetic resistance gene. The PaSNF1 disruption mutant without additional ectopic integrations of the replacement cassette sequence mutants was confirmed by PCR and Southern blot (Supplementary Figure S2). For the complementation CPPaSNF1, a functional PaSNF1 ORF was introduced into the ΔPaSNF1 mutant, and the obtained complementation mutants were screened on a selective medium containing nourseothricin and geneticin and then confirmed by PCR.
Effects of PaSNF1 on Growth and Development of Podospora anserina in Standard Conditions
To investigate the effects of SNF1 in the development of P. anserina, the PaSNF1 mutant (ΔPaSNF1), complementation mutants (CPPaSNF1), and the WT strains were cultured under the optimal conditions for growth and sexual reproduction (on M2 minimal medium at 27°C, in the presence of light). Then the detailed phenotypic analysis was performed. The knockout of PaSNF1 gene led to the growth and developmental defects of P. anserina. Compared with that of WT, the growth of the mutant ΔPaSNF1 was significantly decreased. Colony size of ΔPaSNF1 was 77% of WT after 3 days of incubation (Figures 2A,F). The fruiting bodies of WT strain began to develop after 5 days. There were numerous viable ascospores expelled after 2 days (Figure 2B). ΔPaSNF1 mat + x ΔPaSNF1 mat- led to a defect in perithecium production and ascospore formation. Compared with WT, there was a 62% reduction in the production of perithecia (Figure 2G). Surprisingly, no ascospore was expelled from perithecia in ΔPaSNF1 mutant (Figures 2B,E). Microscopic observation of perithecia showed that the ascus shells produced by WT and mutants were similar, without significant difference in diameter. The ascus shell of the mutant also differentiated into a normal neck (Figure 2C). Microscopic observations of squashed perithecia were made in order to observe produced ascospores and asci. Typical rosettes of asci with four ascospores were observed in WT strain crosses. Usual rosettes were composed of mature ascospores, which were dark green owing to accumulation of melanin, and not fully ripened ascospores, which were light green. However, transparent asci were observed and failed to form intact ascospores in the ΔPaSNF1 (Figure 2D). These observations indicate that the lack of ascospore release in the ΔPaSNF1 was due to a defect in ascospore formation rather than a defect in the mechanism of forcible ascospore discharge.
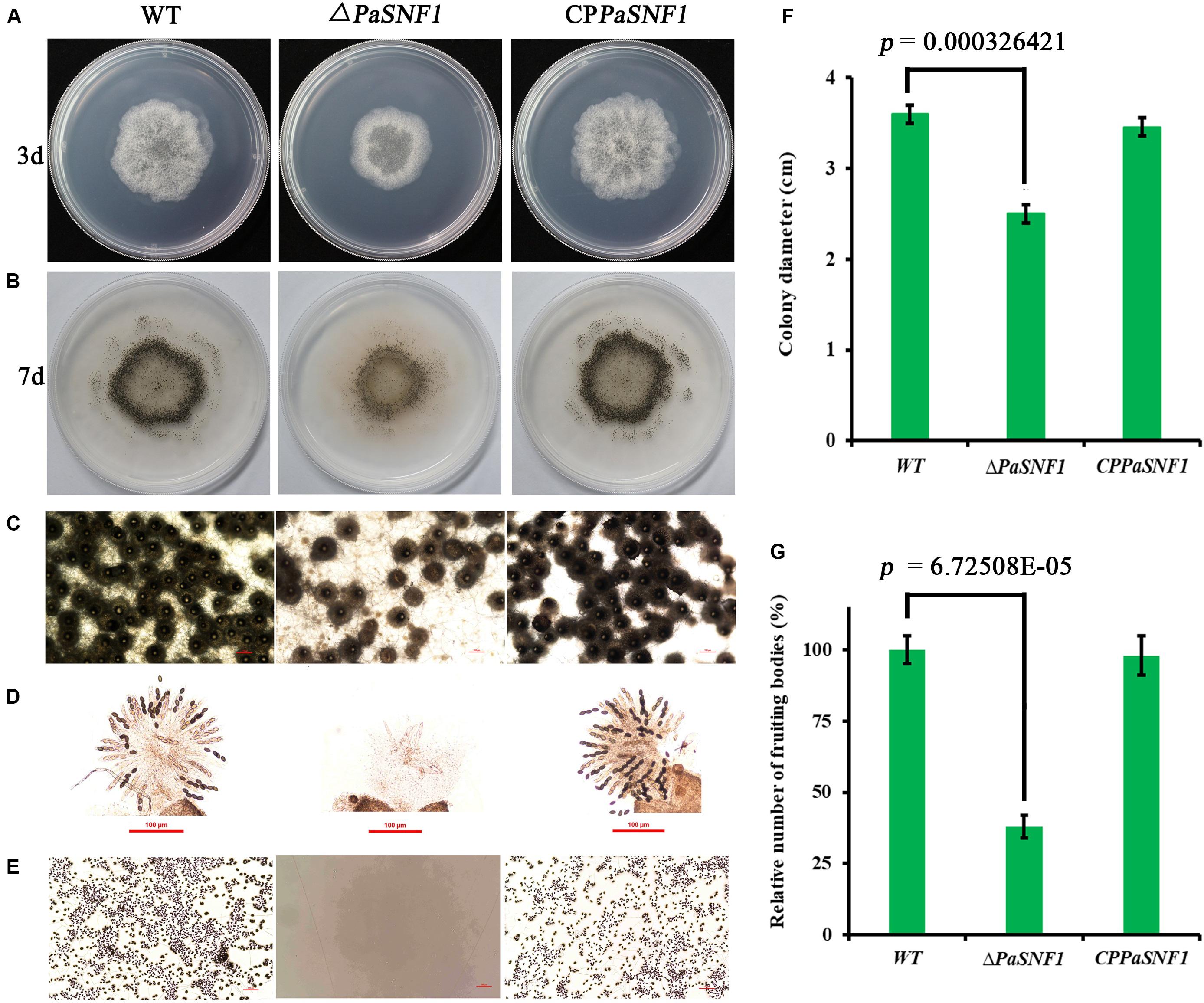
Figure 2. Growth and development of WT and mutants on M2 medium. (A) Colony size of WT and ΔPaSNF1 after 3 days of incubation. (B) Fertility on M2 medium developed after 5 days. Perithecia are visible as small black dots. All the strains produced the usual ring of mature perithecia in the center of the Petri dish. (C) Microscopic observation of perithecia of WT and ΔPaSNF1. (D) Microscopic observation of ascospores inside perithecia. (E) Microscopic observation of ascospores erupting from fruiting bodies. (F,G) Colony diameter and fruiting bodies number, respectively; fruiting body number was expressed with respect to that of WT considered as 100%. Error bars are standard deviations of triplicate samples. Differences in the data were assessed by the T-test. WT, wild type; PaSNF1, Podospora anserina SNF1.
To further analyze the role of the PaSNF1 gene in sexual development, we investigated the behavior of ΔPaSNF1 strain as male and female partners in crosses with a WT strain. Plates with ΔPaSNF1 mat + and WT mat- mycelia were covered with 1.5 ml of sterile water, which spread male gametes from one mycelium on the other and allowed these spermatia to fertilize female organs (Figure 3). WT mat- female organs fertilized by ΔPaSNF1 mat + spermatia produced ascospores, indicating that the deletion of PaSNF1 is not dominant and can be complemented by the functional PaNSF1 gene present in the WT female strain. In contrast, ΔPaSNF1 mat + female organs fertilized by WT mat- spermatia did not produce ascospores, indicating that ΔPaSNF1 has a maternal effect; that is, the presence of a functional PaSNF1 gene in the male nuclei cannot complement the absence of this gene in the female organ. Crosses on plates with ΔPaSNF1 mat- and WT mat + indicated that the maternal effect of ΔPaSNF1 is not mating-type dependent. In addition, SNF1 knockout resulted in a red–orange pigment deposition of the mutant ΔPaSNF1, compared with WT on solid M2 plates (Figure 2B). To further confirm that the phenotypic changes observed in the mutant resulted from the deletion of PaSNF1 instead of other unknown genetic mutations, we conducted complementation analyses of ΔPaSNF1. The defects of ΔPaSNF1 in mycelial growth, sexual development, and pigmentation were complemented by genetic complementation. Our results indicated that PaSNF1 not only affected mycelial growth but also regulated fruit body formation and developmental maturation.
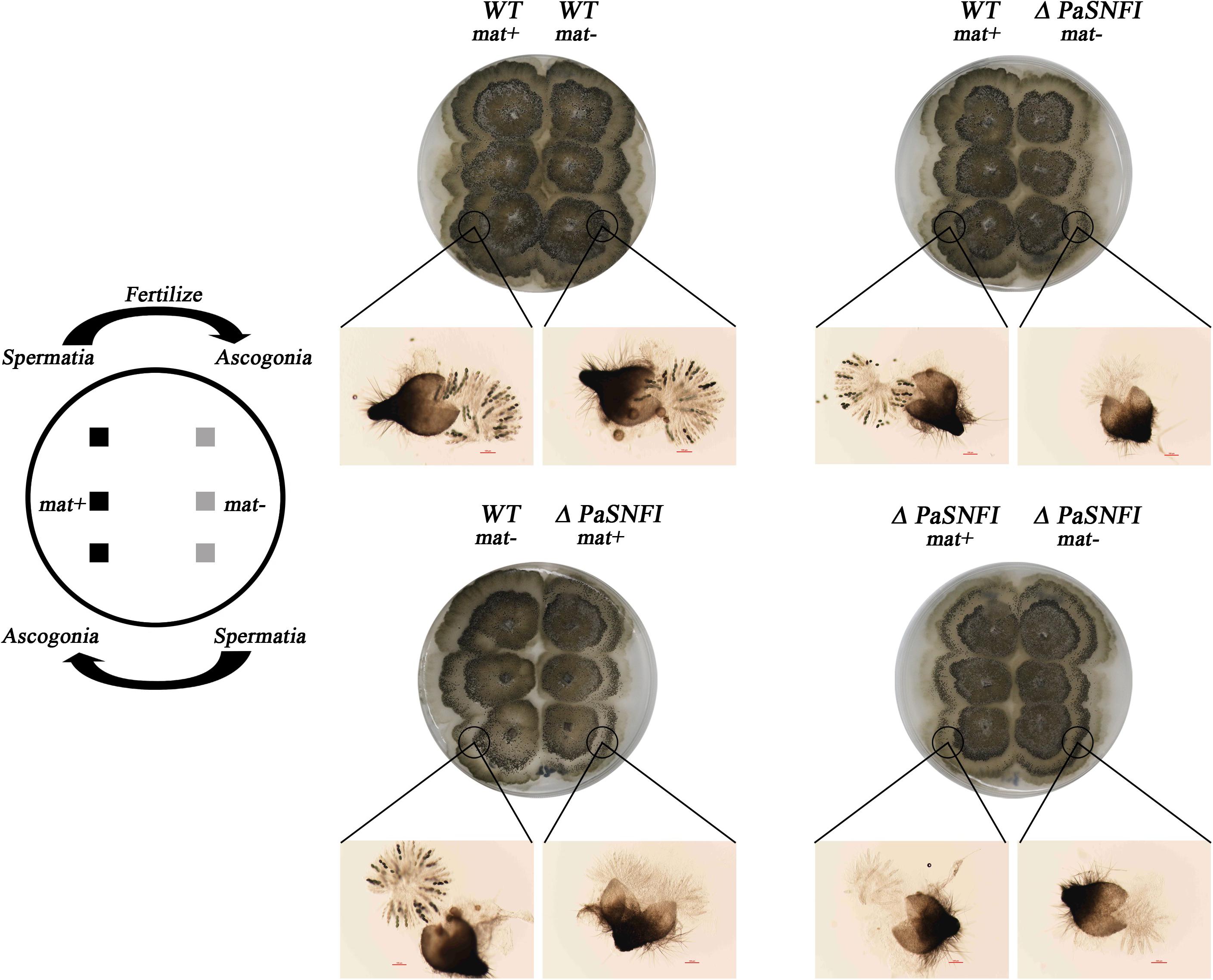
Figure 3. Spreading experiments. Podospora anserina is a heterothallic species with two mating types called mat+ and mat-. Each mating type differentiates male and female organs. Male organs (spermatia) are single cells that can be spread by water all over the plate. They are recognized by female organs of opposite mating type, which fuse with the male cells and subsequently differentiate in mature fruiting bodies. Crosses were made by inoculating the strains 1.5 cm apart. After 3 days of growth, 1.5 ml of water was added and spread all over the plate. The pictures were taken 4 days after fertilization.
The Roles of SNF1 in the Environmental Stress Tolerance
To determine the roles of PaSNF1 in response to stress conditions, the growth of the strains under different stress conditions was analyzed (Figure 4). The growth of the ΔPaSNF1 mutant was significantly inhibited by 0.5 M of NaCl, 0.5 M of KCl, and 0.75 M of glycerol, compared with that of WT and CPPaSNF1, whereas the result was different for sorbitol. This result suggested that PaSNF1 may be necessary for the cellular responses of P. anserina to osmotic pressure. In contrast, the ΔPaSNF1 mutant was insensitive to 0.01% H2O2. However, on plates supplemented with 50 μM of oxidant menadione, the growth rate of ΔPaSNF1 mutant was obviously slower, implying the roles of SNF1 in its tolerance to this stress. The ΔPaSNF1 mutant showed similar sensitivity to heat shock, presented with slower growth, and significantly increased inhibition rate. Particularly interestingly, the resistance of the ΔPaSNF1 mutant to the fungicide fluconazole was significantly increased after knocking out of the SNF1 gene; compared with WT and CPPaSNF1 strains, the growth of ΔPaSNF1 mutant was significantly increased while the inhibition rate was reduced. These results indicated that the PaSNF1 gene in P. anserina played an important role in its response to osmotic stress, oxidative stress, and heat shock.
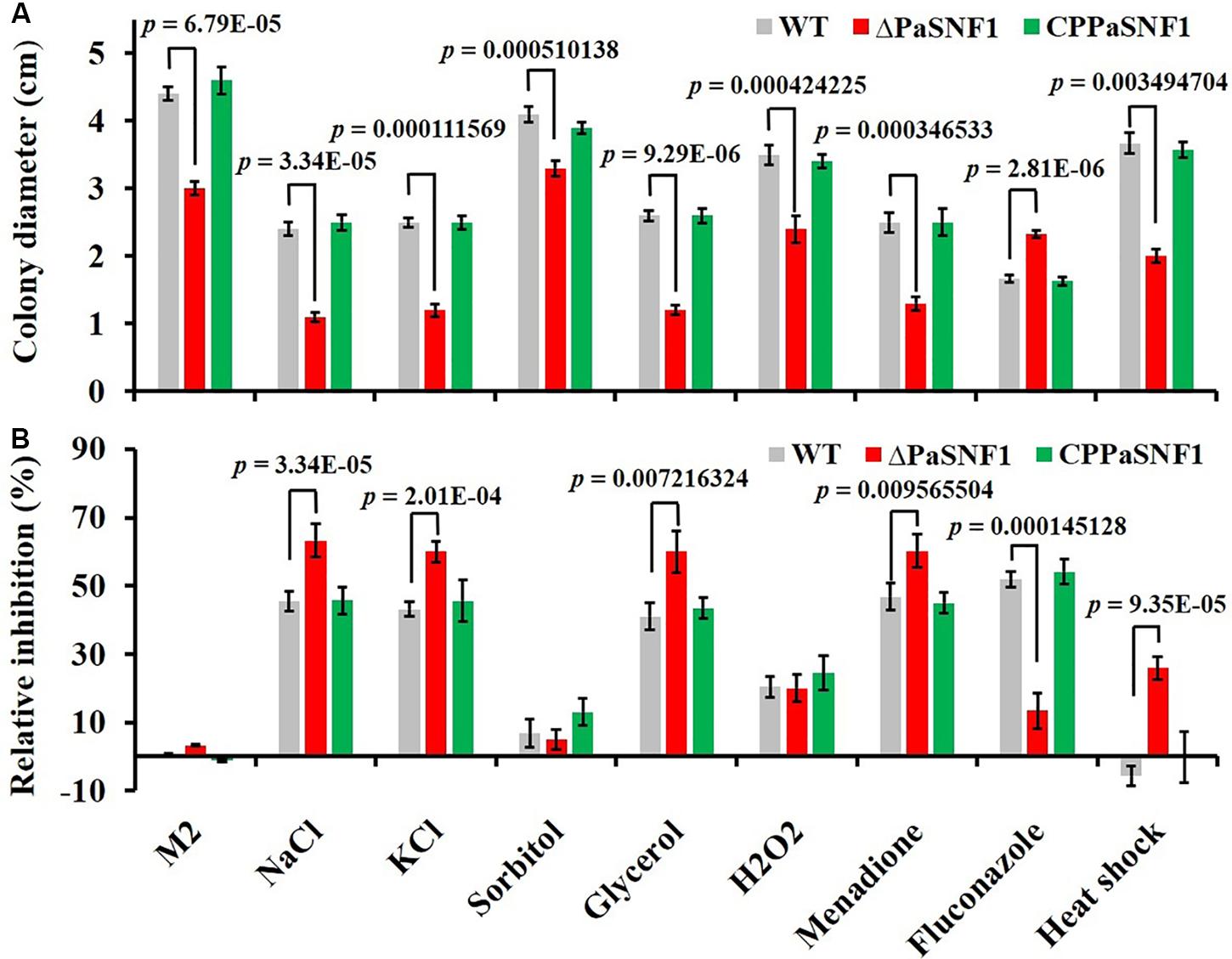
Figure 4. Tolerance analysis to environmental stresses. (A) Colony sizes of WT, ΔPaSNF1, and CPPaSNF1 were incubated separately on M2 medium, supplemented with one of the osmotic stress reagents (0.5 M of NaCl, 0.5 M of KCl, 0.5 M of sorbitol, and 0.75 M of glycerol), oxidative reagents (0.01% H2O2 and 50 μM of menadione), and 2.5 μg/ml of fluconazole; and fresh mycelium was incubated at 45°C in water bath for 2 h. M2 medium without the supplements was served as the control. (B) Relative inhibition rate. All cultures were incubated at 27°C for 3 days, and colony size was measured. Three plates were set for each treatment, and the experiment was repeated once. Error bars are standard deviations of triplicate samples. Differences in the data were assessed by the T-test. WT, wild type; PaSNF1, Podospora anserina SNF1.
The Regulation of SNF1 for Sterigmatocystin Biosynthesis
In fungi, SMs were generally associated with the developmental processes in response to various abiotic or biotic external stimuli (Yin et al., 2012). To examine the regulation effects of SNF1 in biosynthesis of SMs in P. anserina, the profile of secondary compounds in ΔPaSNF1 cultures was analyzed by HPLC. We found that the profile of SMs was significantly altered after the deletion of PaSNF1 (Figure 5A). Based on HPLC profiling, peak of SM 1 (at 18.5 min) in ΔPaSNF1 was 50% higher than that of WT (Figures 5A,C). To further identify the compound at 18.5 min, large-scale fermentation was performed; the metabolites were further isolated and purified. The compound was identified as ST by NMR (Figure 5B and Supplementary Figures S3, S4). The result suggested that the biosynthesis of ST was negatively regulated by PaSNF1. The above results demonstrated that PaSNF1 was a regulator of SMs and may be involved in either activation or silencing of certain gene clusters in P. anserina.
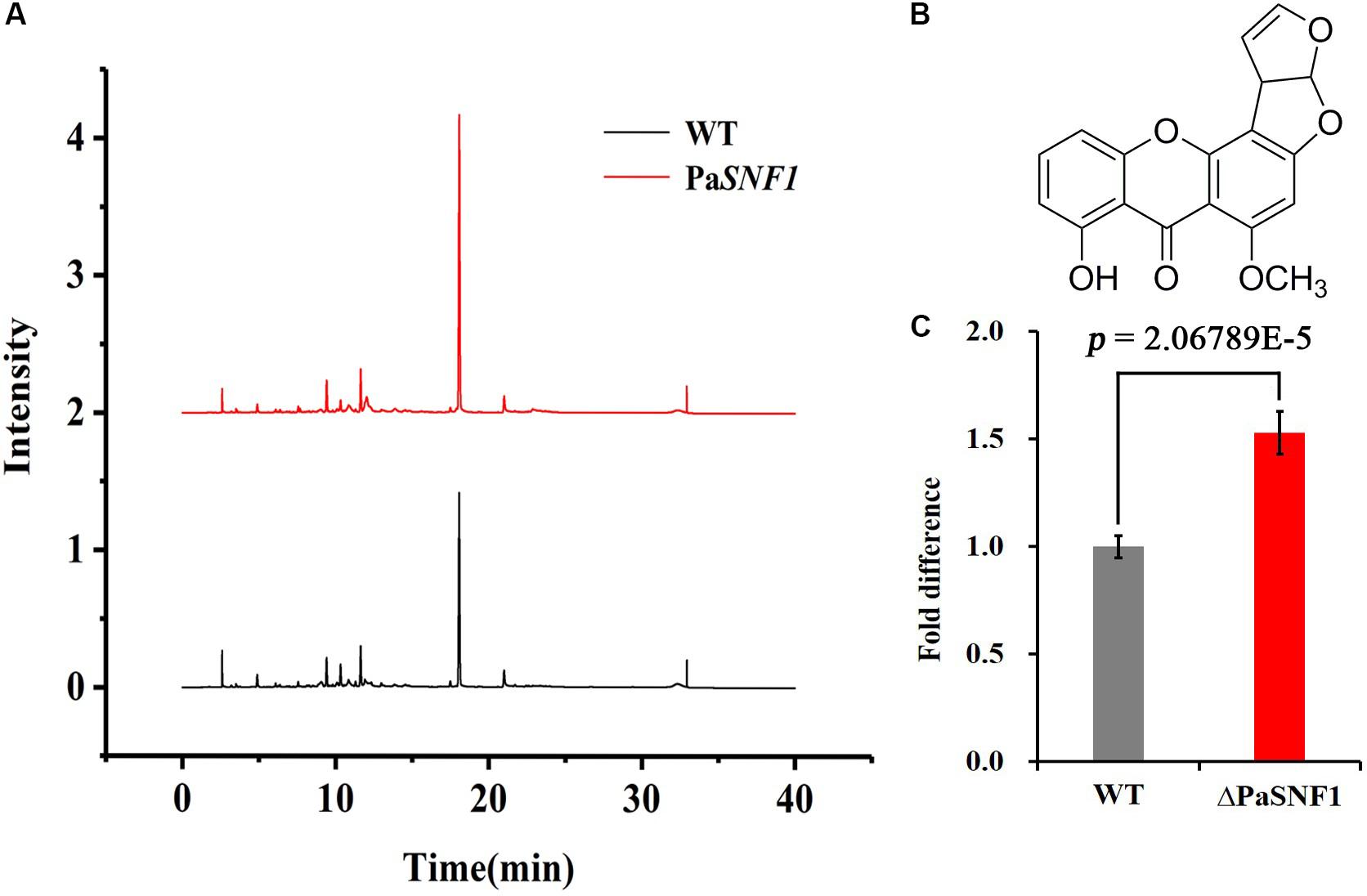
Figure 5. Identification and analysis of ST. (A) HPLC analysis of SM production with WT and ΔPaSNF1 mutant. The strains were grown on M2 media for 7 days. (B) The chemical structure of ST by NMR. (C) Relative amounts of ST in ΔPaSNF1 strain compared with WT. Error bars are standard deviations of triplicate samples. Differences in the data were assessed by the T-test. ST, sterigmatocystin; HPLC, high-performance liquid chromatography; SM, secondary metabolite; WT, wild type; PaSNF1, Podospora anserina SNF1; NMR, nuclear magnetic resonance.
Knockout of SNF1 Led to Impaired Cellulose-Utilizing Ability
To investigate whether the PaSNF1 kinase participated in the production of cellulase, the cellulase activity and cellulose-utilizing ability of WT and ΔPaSNF1 were detected. The cellulase activity of WT and ΔPaSNF1 was further compared, including FPA, EG, CBH, and BG (Figure 6A). After 3 days of culture on M2 medium, the activities of FPA, EG, and BG were significantly lower than those of WT, which were reduced to 17.2, 27.4, and 50.3%, respectively, of WT (Figure 6A). After 7 days of culture on MCC agar medium (with microcrystalline cellulose as carbon source instead of dextrin), the fruit body formation of ΔPaSNF1 mutant was significantly reduced, and similar phenotypes were observed on straw medium (Figure 6B). Therefore, PaSNF1 was considered as critical for the cellulose-utilizing ability of P. anserina.
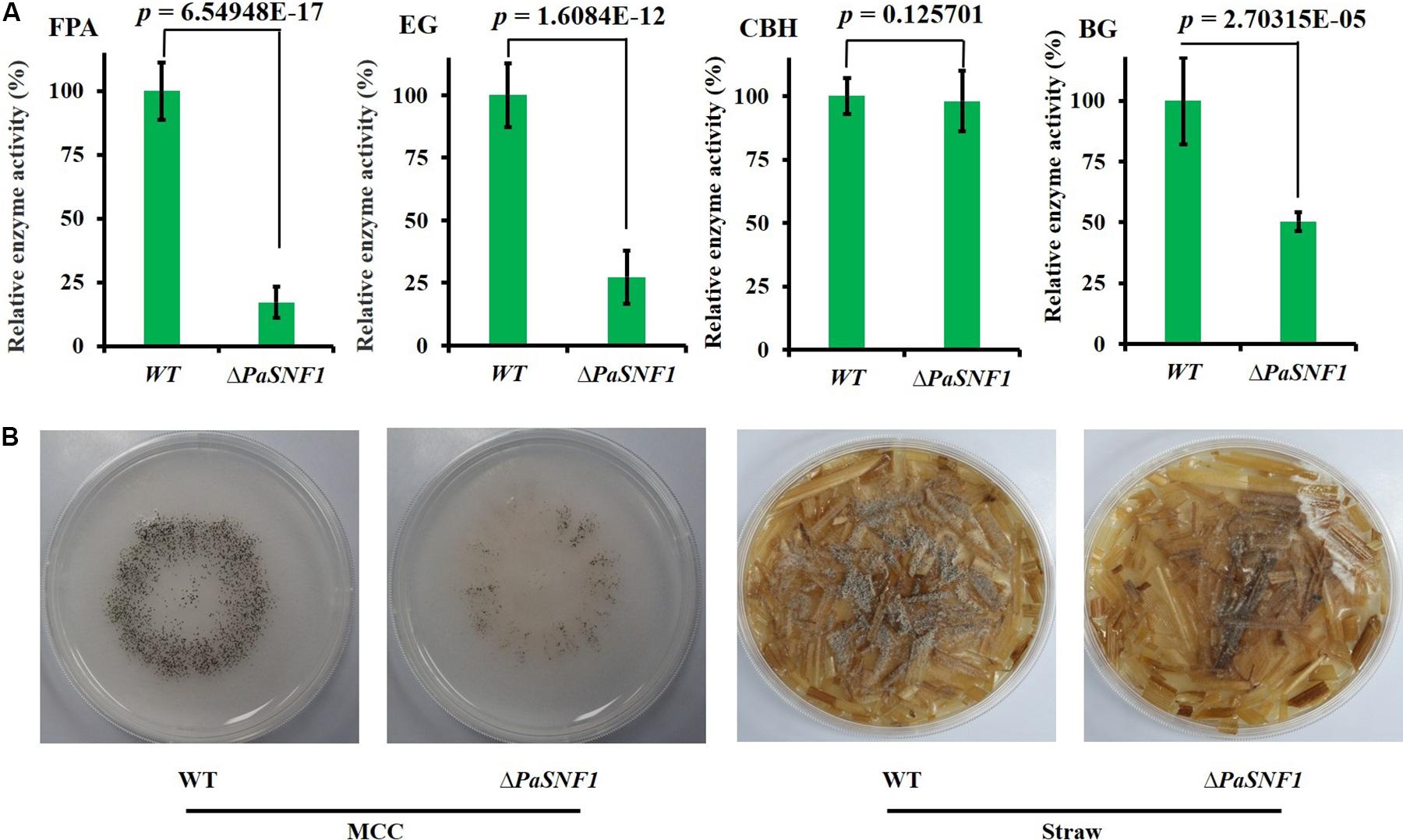
Figure 6. Knockdown of PaSNF1 led to defects in cellulose-utilizing ability. (A) Enzyme assays in WT and ΔPaSNF1 mutant. The mycelia of 2-day-old cultures grown on M2 medium of the indicated strains were harvested and smashed in proper extraction buffer. Activity was expressed with respect to that of WT considered as 100%. Error bars are standard deviations of triplicate samples. Differences in the data were assessed by the T-test. (B) Fertility of WT and ΔPaSNF1 on medium with MCC and straw medium. Fertility was measured by counting the number of mature perithecia on plates containing MCC and straw medium as sole carbon source. Pictures of mycelia of WT and ΔPaSNF1 mutant were taken after 7 days of growth. Fruiting bodies (perithecia) are visible as small black dots. PaSNF1, Podospora anserina SNF1; WT, wild type; MCC, microcrystalline cellulose.
SNF1 Regulated the Expression of the Carbohydrate-Active Enzymes Genes and Transporters in Podospora anserina
The results described above confirmed that SNF1 made effects on cellulase and hemicellulase expression of P. anserina; however, these effects could not be validated solely from growth data. To determine whether the defect in cellulase secretion and activity in the ΔPaSNF1 mutant was due to the failure to induce cellulase gene expression versus a defect in cellulase secretion, we assessed genome-wide expression differences via RNA-Seq between the WT and ΔPaSNF1 strains in medium containing Avicel. Transcriptome results showed that significant expression differences in 468 genes were observed in ΔPaSNF1 compared with WT; DEGs accounted for 4.3% of all genetic data, among which 129 genes were up-regulated and 339 genes were down-regulated (Figure 7A and Supplementary Table S2). Among these genes, 41 of 471 CAZyme encoding genes were detected (Carbohydrate Active Enzymes database6), 24 genes were significantly down-regulated, 17 genes were significantly up-regulated (Figure 7B and Supplementary Table S3), and the DEGs accounted for 8.7% of all CAZyme-encoding genes. To verify whether the CAZyme-encoding genes were enriched and overexpressed, we used Log2FC(ΔPaSNF1/WT) to perform statistical analysis (T-test) on up-regulated and down-regulated genes between CAZyme-encoding genes and non-CAZyme-encoding genes in DEGs. The results indicate that the differences of up-regulated (p value = 0.219811) and down-regulated genes (p-value = 0.740564) between CAZyme-encoding genes and non-CAZyme-encoding genes were not significant. However, the proportion (41/468 × 100% = 8.76%) of CAZyme-encoding genes in DEGs was doubled as compared with the proportion (471/10,888 × 100% = 4.33%) of all CAZyme-encoding genes in the genome, and these results suggest to some extent that more CAZyme-encoding genes were induced to be activated, although CAZyme-encoding genes are not overrepresented in the DEGs in ΔPaSNF1. These down-regulated genes encoded CAZymes in the glycoside hydrolase family (GH2, GH3, GH5, GH13, GH17, GH37, GH55, GH76, GH81, and GH94), laccase (AA1 and Lac), cellobiose dehydrogenases (AA3 and CDH), and lytic polysaccharide monooxygenases (LPMO, AA9, and AA11); however, in the ΔPaSNF1 mutant, FPKM of LPMOs was very small, and the FPKM of AA9 and AA11 is 0.665 and 3.83, respectively (Figure 7C and Supplementary Table S3). The majority of which comprised glycosyl hydrolases (GHs) participated in cellulose and hemicellulose degradation. These findings implied that expression of most of the cellulolytic and hemicellulolytic genes was affected by the deletion of PaSNF1. Down-regulated genes also contained some glycosidase activity against various side chains in hemicellulose, such as endo-1,6-alpha-mannosidase (PODANS_1_19460), β-galactosidase (PODANS_1_18330), chitin deacetylase (PODANS_1_4780), and α,α-trehalase (PODANS_1_2050). The reduced expression of various glycosidase genes was also reflected in the decreased ability to grow on their oligosaccharide substrates (Supplementary Figure S5). The majority of up-regulated genes encoded CAZymes are the glycoside hydrolase family (GH6, GH7, GH11, GH18, and GH53) and LPMOs (AA9: PODANS_1_21900, PODANS_2_4860, PODANS_3_2580, PODANS_5_10760, PODANS_6_11370, and PODANS_6_11470). FPKM of LPMOs was significantly increased, which reached 69% in up-regulated genes encoded CAZymes in ΔPaSNF1, 2.5 times that of WT (Figure 7D), which may be related to the increase of H2O2 in the mutant. In addition, 23 transporters were found to be down-regulated in the ΔPaSNF1 strain compared with WT, and nine of these genes encoded proteins that belong to the major facilitator superfamily (MFS) permeases (Figure 7E and Supplementary Table S4). Four sugar transporters were included: PODANS_5_4520 (SPT1) encoded sugar and polyol transporter 1 (SPT1) (Schilling and Oesterhelt, 2007), PODANS_4_9820 (Hgt-1) was identified as a high-affinity glucose transporter (Baruffini et al., 2006), and PODANS_5_480 and PODANS_1_22840 (MstA) showed the capability of transporting monosaccharides including xylose (Jorgensen et al., 2007). However, none of the transcription factors known to directly regulate cellulase gene expression exhibited altered expression. Interestingly, two genes (PODANS_3_3440 and PODANS_5_12360) encoding secondary metabolic regulators LAE1 were significantly down-regulated (Supplementary Table S2), which was a homolog of methyltransferase LAE1 of Trichoderma reesei. It was involved in cellulase gene expression regulations (Seiboth et al., 2012), suggesting an indirect relationship between SNF1 and cellulase expression. To validate the DEGs identified from RNA-Seq data, transcript levels of 10 up-regulated (PODANS_1_9380, PODANS_1_5230, PODANS_1_12700, PODANS_4_5380, PODANS_5_9670, PODANS_6_2530, PODANS_7_8775, PODANS_4_5370, PODANS_2_7185, and PODANS_1_990) and 10 down-regulated genes (PODANS_3_1840, PODANS_6_9965, PODANS_7_150, PODANS_5_3595, PODANS_3_2688, PODANS_3_780, PODANS_6_10085, PODANS_5_11880, PODANS_1_19460, and PODANS_7_8100) in the mycelium cells of ΔPaSNF1 and WT were assessed via RT-qPCR with paired primers (Supplementary Table S1) and compared with those in the RNA-Seq data. As a result, all the gene transcripts assessed in RT-qPCR showed the same up-regulated or down-regulated trends as determined by the RNA-Seq analysis for DEGs identification (Figure 7F).
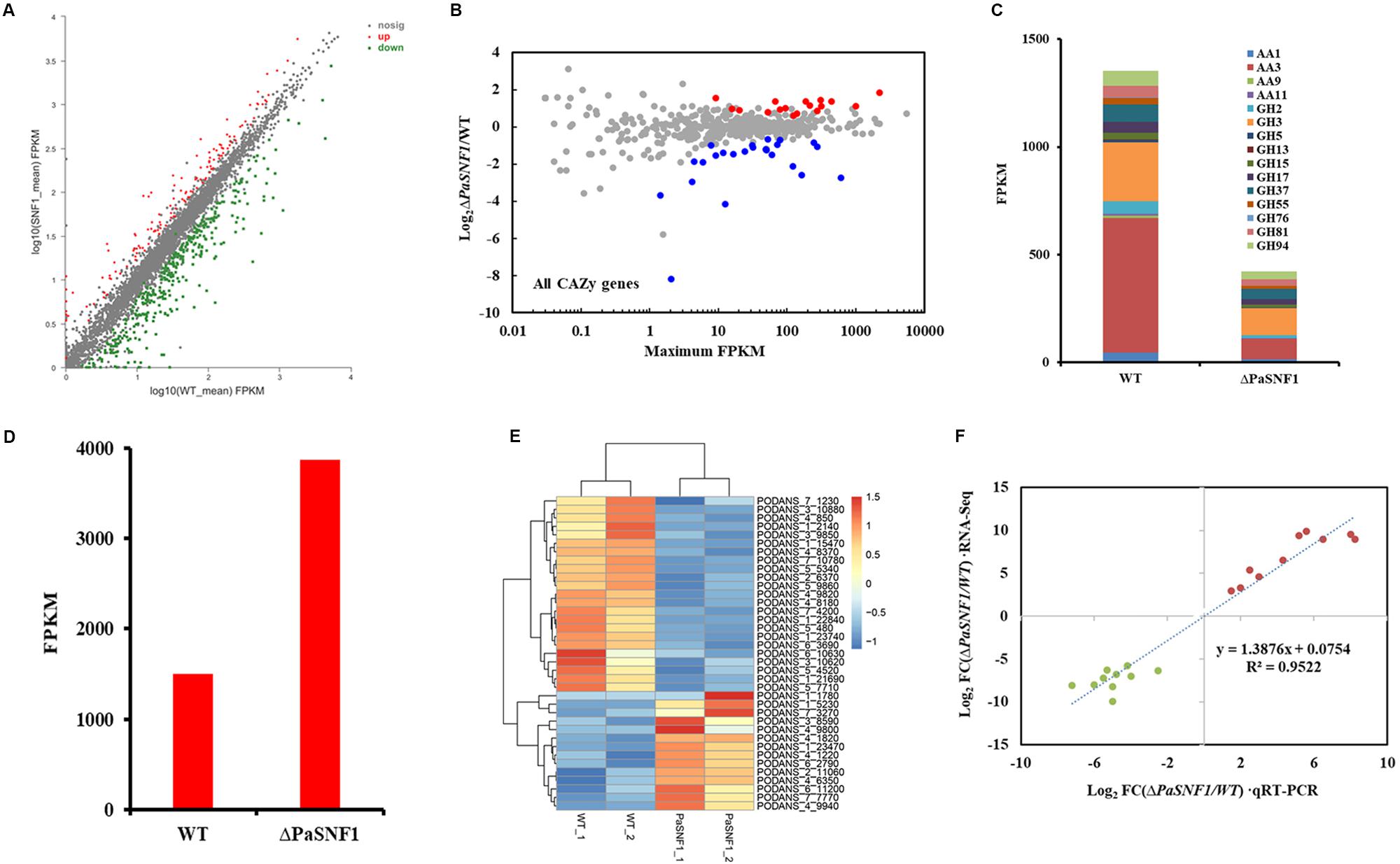
Figure 7. Comparative transcriptomics analysis between ΔPaSNF1 and WT grown in Avicel medium. (A) Differentially expressed genes in the mutant ΔPaSNF1 relative to WT. The expression profile was calculated for conditions including Avicel. Genes that showed differential expression identified by DESeq2 package are shown. Up-regulated and down-regulated genes are shown in red and green, respectively. (B) Relative mRNA abundance of all CAZy genes in ΔPaSNF1 mutant versus WT. The significantly up-regulated genes are shown in red, whereas down-regulated genes are shown in blue (based on adjusted p-value). (C) Total expression of genes encoding major cellulases from RNA-Seq data in WT and ΔPaSNF1 strains. (D) Total expression of genes encoding LPMO from RNA-Seq data in WT and ΔPaSNF1 strains. (E) Regulation of putative transporter genes by PaSNF1. Values in heatmaps were calculated by log2 (Gene_FPKM in ΔPaSNF1/Gene_FPKM in WT). FPKM mapped. (F) Expression level validation of DEGs using RT-qPCR. Comparisons of log2 ratios of 10 up-regulated and 10 down-regulated genes from the RNA-Seq data and RT-qPCR experiments with paired primers (Supplementary Table S1). PaSNF1, Podospora anserina SNF1; WT, wild type; CAZy, Carbohydrate-Active Enzymes; DEG, differentially expressed genes.
Discussion
Organisms would suffer a variety of nutritional conditions during their life cycle. Complex signaling networks have been evolved in microbes to identify and prioritize the utilization of available energy sources, such as the CCR system. The CCR system would prevent the utilization of difficultly metabolizable carbon sources for ensuring the preferential utilization of easily metabolizable carbon sources such as D-glucose. For many fungi, several genes involved in the carbon catabolism were regulated by CCR, including the genes encoding carbohydrate-degrading enzymes such as cellulase, amylase, and xylanase. The SNF1/AMPK was highly conserved among eukaryotic organisms, which was a key regulator in both carbon metabolism and energy homeostasis. The regulation of SNF1 activity played a central role in the alleviation of CCR (Brown et al., 2013). During the absence of glucose, SNF1 regulated the expression of metabolic genes by controlling transcriptional activators and repressors (Carlson, 1999). In Saccharomyces cerevisiae, SNF1 acted as an activator for more than 400 glucose-repressing genes, some of which were involved in alternative carbon source utilization pathways (Young et al., 2003). In this study, the function of PaSNF1 was explored. Our results demonstrated that PaSNF1 played important roles in regulating vegetative growth, sexual development, stress responses, and biosynthesis of SMs in Podospora anserina and then confirmed that it was a major actor of lignocellulose degradation in P. anserina.
The PaSNF1 protein consisted of a serine/threonine (Ser/Thr) kinase catalytic domain (KD) (residues 67–318) and a two CTD (residues 532–628 and 633–704), showing the same structure with the SNF1s identified in many other organisms (Zhang et al., 2013; Islam et al., 2017). In addition, the PaSNF1 protein generally shared a high sequence identity with the SNF1s of other organisms, especially the catalytic domain (Cziferszky et al., 2003; Ospina-Giraldo et al., 2003; Wang et al., 2014, 2018). Phylogenetic analysis of SNF1 homologous proteins from filamentous fungi and yeasts formed a separate cluster. Results of phylogenetic analysis indicated that PaSNF1 was much more closely related to the SNF1 orthologs from the filamentous ascomycetous fungi than those from ascomycetous yeasts, plants, or mammals (Figure 1). Among the SNF1s in the filamentous fungi, the PaSNF1 was closest to the SNF1 ortholog identified in Neurospora crassa, Hypocrea jecorina, Fusarium oxysporum, and Beauveria bassiana (Figure 1).
The essential role of SNF1 in filamentous fungi was the regulation of vegetative growth and sexual development. The ability to form the perithecia and ascospores was affected in GzSNF1 deletion mutants (Lee et al., 2009), conidiation was also affected by the loss of PdSNF (Zhang et al., 2013), and the ablation of LmSNF1 would result in impaired sporulation and spore germination (Feng et al., 2014). In ΔMoSNF1 mutant, the sporulation ability was abolished (Yi et al., 2008), and the PmSNF1 also played a critical role in conidiation and vegetative growth in Pestalotiopsis microspora NK17 (Wang et al., 2018). Deletion of SNF1 in Fusarium graminearum led to defects in vegetative growth and sexual reproduction (Yu et al., 2014). Our observations were consistent with the above results; the knockout of PaSNF1 gene led to the growth and developmental defects of P. anserina. The PaSNF1 did not only affect mycelial growth but also regulate fruit body formation and developmental maturation (Figure 2). This finding suggested that PaSNF1 could trigger the expression of genes required for the formation of fruit body and ascospores. Mechanisms underlying these functions may be associated with the inadequate or unbalanced nutrients (Lee et al., 2009).
In yeast, the SNF1 protein kinase has been necessary for its response to various environmental stresses such as sodium and lithium salts, alkaline pH, heat shock, and hyperosmolarity (Hong and Carlson, 2007). Treatment with high concentrations of salts (0.8 M of KCl and 1 M of NaCl), 1 M of sorbitol, and 2 mM of oxidant H2O2 could dramatically inhibit vegetative growth of the ΔPmSNF1 mutant strain (Wang et al., 2018). However, the response of the SNF1 mutant of Cryptococcus neoformans to osmotic, salt, and oxidative stresses was not significantly different from that of the WT (Hu et al., 2008). Our results showed that the growth of the ΔPaSNF1 mutant would be significantly inhibited by 0.5 M of NaCl, 0.5 M of KCl, and 0.75 M of glycerol; however, ΔPaSNF1 mutant was not sensitive to 0.5 M of sorbitol. A comparable function for SNF1 in the C. neoformans JEC21 has been reported (Yang et al., 2010). Our data clearly demonstrated the crucial roles of PaSNF1 in the tolerance of fungus to heat shock; the mycelium of the ΔPaSNF1 mutant was sensitive to heat shock of 45°C. This function of SNF1 has been previously reported in other fungi, such as C. neoformans and Pestalotiopsis microspora (Yang et al., 2010; Wang et al., 2018). Moreover, after the knockout of SNF1 gene, the resistance of the ΔPaSNF1 mutant to the fungicide fluconazole would be significantly increased, whereas the sensitivity to fluconazole was not varied in ΔCnSNF1 mutant (Hu et al., 2008). It indicated the remarkable functional deviation between PaSNF1 and CnSNF1, although the underlying mechanism remained to be identified.
Ascomycete genomes code for on average 40 biosynthetic gene clusters with crucial importance in SM synthesis (Pusztahelyi et al., 2015). Genome analyses in aspergilli suggest between 39 and 80 biosynthetic gene clusters per species (Inglis et al., 2013). Sequence analysis of the P. anserina genome revealed its ability to produce 40 putative SMs including 18 polyketide synthases (PKSs) (Espagne et al., 2008). However, until now, only limited chemical investigations have been conducted, and plenty of SMs were yet to be discovered. Recent studies have shown that ABR1 was involved in biosynthesis of DHN melanin, which colored ascospores; and knockout of ABR1 led to decreased ascospore pigmentation accompanied by lowered ascospore germination rate (Xie et al., 2018). Shen et al. (2019) reported that PaStcA and PaAflR were key genes necessary for the biosynthesis of ST in P. anserina, deletion of PaStcA that blocked the ST pathway, and overexpression of PaAflR that resulted in the excessive accumulation of ST, thus leading to a female sterility phenotype. In this study, to identify the metabolites regulated by PaSNF1, the large-scale fermentation of the ΔPaSNF1 mutant was performed, and the structures of metabolites with unique peaks were characterized. This result suggested that PaSNF1 may have negative effects on the biosynthesis of ST. The general pattern of the peaks for ST was significantly increased in the HPLC profile of ΔPaSNF1 mutant compared with WT (Figure 5). Correspondingly, the deletion of PaSNF1 led to a defect in perithecium production and ascospore formation. Moreover, OE-PaAflR displayed overproduction of ST, and overexpression of PaAflR also led to sterility (Shen et al., 2019). These results indicated that ST was closely related to the development of fungi and spore formation. To our knowledge, this has been the first report of SNF’s regulation effects on ST biosynthesis.
In addition to regulating ST synthesis, SNF1 also played a critical role in regulating cellulose degradation. We showed that a ΔPaSNF1 mutant displayed severe growth defects on cellulose, and similar phenotypes were observed on straw medium (Figure 6B), which was correlated with a lack of cellulolytic enzyme activity. Indeed, the enzyme activities of FPA, EG, and BG in ΔPaSNF1 mutant were significantly reduced (Figure 6A). The defects in PaSNF1 would lead to the reduced (or impaired) production of such enzymes, thus inhibiting the degradation of lignocellulose and utilization of carbon sources. Consistent with this phenotype, the expression of the many CAZymes genes of P. anserina was significantly decreased in the ΔPaSNF1 mutant compared with WT (Figure 7). Following the extracellular degradation of the lignocellulose, the uptake of the soluble breakdown products has been a key process to regulate the transcription of cellulases and related genes. The transporters played an important role, some of which exhibited the ability to sense the breakdown products during their passage through the cell membrane (Novy et al., 2019). Transcriptome analysis showed that the genes coding for MFS permeases were also highly repressed in the ΔPaSNF1 mutant, including four putative sugar transporters. These findings suggested that PaSNF1 played an important role in positively regulating the expression of genes encoding cellulolytic enzymes. Interestingly, our data revealed that the main transcription factors that regulated the expression of cellulases in P. anserina showed no change of expression in the mutant strain relative to WT strain. For example, the expression of XYR1, CLR1, and CLR2 was not significantly varied in mutant strains, especially CRE1, which was the key protein of CCR and the direct target protein of SNF1. The expression of cellulosic biomass degrading enzyme genes was regulated by various global regulators. The protein methyltransferase LAE1 was one global regulator of SM gene clusters such as ST formation, which regulated sexual and asexual developmental processes in Ascomycota (Tani et al., 2014). LAE1 also played an important role in regulating the expression of cellulase, polysaccharide hydrolases, and xyr1 in Trichoderma reesei (Seiboth et al., 2012). Our results showed that two putative LAE1 (PODANS_3_3440; PODANS_5_12360) expressions were significantly down-regulated in the ΔPaSNF1. This result suggested a refined dual mechanism that controlled the expression of cellulase genes through the modulation of transcription factors.
SNF1 promoted feeding on lignocellulose and reduced the synthesis of ST. There may be a balance between feeding and fighting competitors on the basis of anticorrelated effects on each pathway. In our study, negative correlation was observed based on the data of enzyme activity and metabolite map. In a previous study, analysis of the genome revealed that CAZymes of T. reesei were often observed in clusters along with genes involved in SM (Martinez et al., 2008). It was considered as a means to fend off competitors for nutrients. Fungi constantly faced the challenge to outcompete other organisms in complex ecosystems. Therefore, they developed powerful enzyme systems for degradation of substrates, which provided for fast growth and efficient colonization of their environment (van den Brink and de Vries, 2011). However, fungi also evolved the ability to kill, or at least inhibit the growth of their competitors, by producing a versatile array of SMs (Demain and Fang, 2000). The application of these different survival utilities should be tightly controlled in order to balance the assignment of resources for feeding to succeed by superior growth or fighting to decrease the chances of survival for competitors. This study supported this hypothesis of coordination of substrate degradation and competition. This may be a mechanism via which fungi evolved to balance the operation of primary and secondary metabolism during its life cycle. Thus, an economic distribution of resources for feeding (enzyme production) and fighting (SM production) would be reasonable.
Conclusion
In conclusion, in this work, the roles of PaSNF1 in vegetative growth, sexual development, and lignocellulose degradation in Podospora anserina were investigated. Our study clearly suggested that SNF1 played a critical regulation role in growth, development, and stress response. Importantly, SNF1 can function as a critical global regulator for the SM biosynthesis and cellulase production. In the future, proteomics analysis and ChIP-seq on PaSNF1 would be performed to identify the interaction partners of PaSNF1. This will be crucial to develop deeper understanding on the cellulase-regulating signal transduction processes and the SNF1-mediated regulation mechanism of lignocellulose degradation in P. anserina, as well as its effects on growth and development.
Data Availability Statement
The datasets generated for this study can be found in the SRA database: https://www.ncbi.nlm.nih.gov/sra/PRJNA597840, accession number PRJNA597840.
Author Contributions
YL, PY, and NX designed the study. YL, XL, YQ, and SL performed the experiments. YL, GL, SL, and LM analyzed the data. YL and NX wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Funding
This study was supported jointly by the National Natural Science Foundation of China (No. 31601014), the Shenzhen Science and Technology Key Project (No. JSGG20171013091238230), the Science and Technology Project of Shenzhen City, the Shenzhen Bureau of Science, Technology and Information (No. JCYJ20180305123659726), and Science and Technology Application Demonstration Project (Shenzhen Government, No. KJYY20180201180253571).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01038/full#supplementary-material
Footnotes
- ^ http://podospora.i2bc.paris-saclay.fr/
- ^ https://www.ncbi.nlm.nih.gov/genome/?term=txid5145[orgn],
- ^ http://www.r-project.org
- ^ http://podospora.i2bc.paris-saclay.fr/index.php
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.cazy.org/
References
Baruffini, E., Goffrini, P., Donnini, C., and Lodi, T. (2006). Galactose transport in Kluyveromyces lactis: major role of the glucose permease Hgt1. FEMS Yeast Res. 6, 1235–1242. doi: 10.1111/j.1567-1364.2006.00107.x
Benocci, T., Aguilar-Pontes, M. V., Zhou, M., Seiboth, B., and de Vries, R. P. (2017). Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol. Biofuels 10:152. doi: 10.1186/s13068-017-0841-x
Bills, G. F., and Gloer, J. B. (2016). Biologically active secondary metabolites from the fungi. Microbiol. Spectr. 4:FUNK-0009-2016.
Bischof, R. H., Ramoni, J., and Seiboth, B. (2016). Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact. 15:106. doi: 10.1186/s12934-016-0507-6
Bok, J. W., and Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3, 527–535. doi: 10.1128/ec.3.2.527-535.2004
Bomble, Y. J., Lin, C. Y., Amore, A., Wei, H., Holwerda, E. K., Ciesielski, P. N., et al. (2017). Lignocellulose deconstruction in the biosphere. Curr. Opin. Chem. Biol. 41, 61–70. doi: 10.1016/j.cbpa.2017.10.013
Bourdais, A., Bidard, F., Zickler, D., Berteaux-Lecellier, V., Silar, P., and Espagne, E. (2012). Wood utilization is dependent on catalase activities in the filamentous fungus Podospora anserina. PLoS One 7:e29820. doi: 10.1371/journal.pone.0029820
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brown, D. W., Yu, J. H., Kelkar, H. S., Fernandes, M., Nesbitt, T. C., Keller, N. P., et al. (1996). Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. U.S.A. 93, 1418–1422. doi: 10.1073/pnas.93.4.1418
Brown, N. A., de Gouvea, P. F., Krohn, N. G., Savoldi, M., and Goldman, G. H. (2013). Functional characterisation of the non-essential protein kinases and phosphatases regulating Aspergillus nidulans hydrolytic enzyme production. Biotechnol. Biofuels 6:91. doi: 10.1186/1754-6834-6-91
Carlson, M. (1999). Glucose repression in yeast. Curr. Opin. Microbiol. 2, 202–207. doi: 10.1016/s1369-5274(99)80035-6
Casamayor, A., Serrano, R., Platara, M., Casado, C., Ruiz, A., and Arino, J. (2012). The role of the Snf1 kinase in the adaptive response of Saccharomyces cerevisiae to alkaline pH stress. Biochem. J. 444, 39–49. doi: 10.1042/BJ20112099
Coradetti, S. T., Craig, J. P., Xiong, Y., Shock, T., Tian, C., and Glass, N. L. (2012). Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc. Natl. Acad. Sci. U.S.A. 109, 7397–7402. doi: 10.1073/pnas.1200785109
Couturier, M., Tangthirasunun, N., Ning, X., Brun, S., Gautier, V., Bennati-Granier, C., et al. (2016). Plant biomass degrading ability of the coprophilic ascomycete fungus Podospora anserina. Biotechnol. Adv. 34, 976–983. doi: 10.1016/j.biotechadv.2016.05.010
Cziferszky, A., Seiboth, B., and Kubicek, C. P. (2003). The Snf1 kinase of the filamentous fungus Hypocrea jecorina phosphorylates regulation-relevant serine residues in the yeast carbon catabolite repressor Mig1 but not in the filamentous fungal counterpart Cre1. Fungal Genet. Biol. 40, 166–175. doi: 10.1016/s1087-1845(03)00082-3
de Assis, L. J., Ries, L. N., Savoldi, M., Dos Reis, T. F., Brown, N. A., and Goldman, G. H. (2015). Aspergillus nidulans protein kinase A plays an important role in cellulase production. Biotechnol. Biofuels 8:213. doi: 10.1186/s13068-015-0401-1
Demain, A. L., and Fang, A. (2000). The natural functions of secondary metabolites. Adv. Biochem. Eng. Biotechnol. 69, 1–39.
Dos Santos, Castro, L., de Paula, R. G., Antonieto, A. C., Persinoti, G. F., Silva-Rocha, R., et al. (2016). Understanding the role of the master regulator XYR1 in Trichoderma reesei by global transcriptional analysis. Front. Microbiol. 7:175. doi: 10.3389/fmicb.2016.00175
El-Khoury, R., Sellem, C. H., Coppin, E., Boivin, A., Maas, M. F., Debuchy, R., et al. (2008). Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet. 53, 249–258. doi: 10.1007/s00294-008-0180-3
Espagne, E., Lespinet, O., Malagnac, F., Da Silva, C., Jaillon, O., Porcel, B. M., et al. (2008). The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9:R77. doi: 10.1186/gb-2008-9-5-r77
Feng, J., Zhang, H., Strelkov, S. E., and Hwang, S. F. (2014). The LmSNF1 gene is required for pathogenicity in the canola blackleg pathogen Leptosphaeria maculans. PLoS One 9:e92503. doi: 10.1371/journal.pone.0092503
Fernandes, M., Keller, N. P., and Adams, T. H. (1998). Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 28, 1355–1365. doi: 10.1046/j.1365-2958.1998.00907.x
Ferrer-Dalmau, J., Randez-Gil, F., Marquina, M., Prieto, José, A., and Casamayor, A. (2015). Protein kinase Snf1 is involved in the proper regulation of the unfolded protein response in Saccharomyces cerevisiae. Biochem. J. 468, 33–47. doi: 10.1042/bj20140734
Ghillebert, R., Swinnen, E., Wen, J., Vandesteene, L., Ramon, M., Norga, K., et al. (2011). The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS J. 278, 3978–3990. doi: 10.1111/j.1742-4658.2011.08315.x
Grognet, P., Bidard, F., Kuchly, C., Tong, L. C., Coppin, E., Benkhali, J. A., et al. (2014). Maintaining two mating types: structure of the mating type locus and its role in heterokaryosis in Podospora anserina. Genetics 197, 421–432. doi: 10.1534/genetics.113.159988
Gupta, V. K., Kubicek, C. P., Berrin, J. G., Wilson, D. W., Couturier, M., Berlin, A., et al. (2016). Fungal enzymes for bio-products from sustainable and waste biomass. Trends Biochem. Sci. 41, 633–645. doi: 10.1016/j.tibs.2016.04.006
Hahn, J.-S., and Thiele, D. J. (2004). Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J. Biol. Chem. 279, 5169–5176. doi: 10.1074/jbc.M311005200
Hedbacker, K., and Carlson, M. (2008). SNF1/AMPK pathways in yeast. Front. Biosci. 13, 2408–2420. doi: 10.2741/2854
Hong, S. P., and Carlson, M. (2007). Regulation of snf1 protein kinase in response to environmental stress. J. Biol. Chem. 282, 16838–16845. doi: 10.1074/jbc.M700146200
Hu, G., Cheng, P. Y., Sham, A., Perfect, J. R., and Kronstad, J. W. (2008). Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol. Microbiol. 69, 1456–1475. doi: 10.1111/j.1365-2958.2008.06374.x
Huberman, L. B., Coradetti, S. T., and Glass, N. L. (2017). Network of nutrient-sensing pathways and a conserved kinase cascade integrate osmolarity and carbon sensing in Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 114, E8665–E8674. doi: 10.1073/pnas.1707713114
Inglis, D. O., Binkley, J., Skrzypek, M. S., Arnaud, M. B., Cerqueira, G. C., Shah, P., et al. (2013). Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 13:91. doi: 10.1186/1471-2180-13-91
Islam, K. T., Bond, J. P., and Fakhoury, A. M. (2017). FvSNF1, the sucrose non-fermenting protein kinase gene of Fusarium virguliforme, is required for cell-wall-degrading enzymes expression and sudden death syndrome development in soybean. Curr. Genet. 63, 723–738. doi: 10.1007/s00294-017-0676-9
Jonsson, L. J., Alriksson, B., and Nilvebrant, N. O. (2013). Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol. Biofuels 6:16. doi: 10.1186/1754-6834-6-16
Jorgensen, T. R., vanKuyk, P. A., Poulsen, B. R., Ruijter, G. J., Visser, J., and Iversen, J. J. (2007). Glucose uptake and growth of glucose-limited chemostat cultures of Aspergillus niger and a disruptant lacking MstA, a high-affinity glucose transporter. Microbiology 153(Pt 6), 1963–1973. doi: 10.1099/mic.0.2006/005090-0
Kawaguchi, H., Hasunuma, T., Ogino, C., and Kondo, A. (2016). Bioprocessing of bio-based chemicals produced from lignocellulosic feedstocks. Curr. Opin. Biotechnol. 42, 30–39. doi: 10.1016/j.copbio.2016.02.031
Kubicek, C. P., and Kubicek, E. M. (2016). Enzymatic deconstruction of plant biomass by fungal enzymes. Curr. Opin. Chem. Biol. 35, 51–57. doi: 10.1016/j.cbpa.2016.08.028
Lalucque, H., Malagnac, F., Brun, S., Kicka, S., and Silar, P. (2012). A non-Mendelian MAPK-generated hereditary unit controlled by a second MAPK pathway in Podospora anserina. Genetics 191, 419–433. doi: 10.1534/genetics.112.139469
Lee, S. H., Lee, J., Lee, S., Park, E. H., Kim, K. W., Kim, M. D., et al. (2009). GzSNF1 is required for normal sexual and asexual development in the ascomycete Gibberella zeae. Eukaryot. Cell 8, 116–127. doi: 10.1128/EC.00176-08
Li, C., Lin, F., Li, Y., Wei, W., Wang, H., Qin, L., et al. (2016). A beta-glucosidase hyper-production Trichoderma reesei mutant reveals a potential role of cel3D in cellulase production. Microb. Cell Fact. 15:151. doi: 10.1186/s12934-016-0550-3
Liao, G. Y., Zhao, S., Zhang, T., Li, C. X., Liao, L. S., Zhang, F. F., et al. (2018). The transcription factor TpRfx1 is an essential regulator of amylase and cellulase gene expression in Talaromyces pinophilus. Biotechnol. Biofuels 11:276. doi: 10.1186/s13068-018-1276-8
Lichius, A., Seidl-Seiboth, V., Seiboth, B., and Kubicek, C. P. (2014). Nucleo-cytoplasmic shuttling dynamics of the transcriptional regulators XYR1 and CRE1 under conditions of cellulase and xylanase gene expression in Trichoderma reesei. Mol. Microbiol. 94, 1162–1178. doi: 10.1111/mmi.12824
Liu, G., Zhang, L., Qin, Y., Zou, G., Li, Z., Yan, X., et al. (2013). Long-term strain improvements accumulate mutations in regulatory elements responsible for hyper-production of cellulolytic enzymes. Sci. Rep. 3:1569. doi: 10.1038/srep01569
Liu, Q., Li, J., Gao, R., Li, J., Ma, G., and Tian, C. (2019). CLR-4, a novel conserved transcription factor for cellulase gene expression in ascomycete fungi. Mol. Microbiol. 111, 373–394. doi: 10.1111/mmi.14160
Martinez, D., Berka, R. M., Henrissat, B., Saloheimo, M., Arvas, M., Baker, S. E., et al. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26, 553–560. doi: 10.1038/nbt1403
Monroy, A. A., Stappler, E., Schuster, A., Sulyok, M., and Schmoll, M. (2017). A CRE1- regulated cluster is responsible for light dependent production of dihydrotrichotetronin in Trichoderma reesei. PLoS One 12:e0182530. doi: 10.1371/journal.pone.0182530
Novy, V., Nielsen, F., Seiboth, B., and Nidetzky, B. (2019). The influence of feedstock characteristics on enzyme production in Trichoderma reesei: a review on productivity, gene regulation and secretion profiles. Biotechnol. Biofuels 12:238. doi: 10.1186/s13068-019-1571-z
Oleinikova, G., Denisenko, V., Slinkina, N., and Afiyatullov, S. H. (2012). Secondary metabolites of the marine fungus Aspergillus ustus KMM 4640. Chem. Nat. Compd. 48, 467–469. doi: 10.1007/s10600-012-0276-3
Ospina-Giraldo, M. D., Mullins, E., and Kang, S. (2003). Loss of function of the Fusarium oxysporum SNF1 gene reduces virulence on cabbage and Arabidopsis. Curr. Genet. 44, 49–57. doi: 10.1007/s00294-003-0419-y
Pachler, K. G., Steyn, P. S., Vleggaar, R., and Wessels, P. L. (1976). Carbon-13 nuclear magnetic resonance assignments and biosynthesis of aflatoxin B 1 and sterigmatocystin. J. Chem. Soc. Perkin 1 11, 1182–1189.
Paulova, L., Patakova, P., Branska, B., Rychtera, M., and Melzoch, K. (2015). Lignocellulosic ethanol: technology design and its impact on process efficiency. Biotechnol. Adv. 33, 1091–1107. doi: 10.1016/j.biotechadv.2014.12.002
Pusztahelyi, T., Holb, I. J., and Pócsi, I. (2015). Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 6:573. doi: 10.3389/fpls.2015.00573
Ragauskas, A. J., Beckham, G. T., Biddy, M. J., Chandra, R., Chen, F., Davis, M. F., et al. (2014). Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. doi: 10.1126/science.1246843
Sanz, P., Alms, G. R., Haystead, T. A. J., and Carlson, M. (2000). Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20, 1321–1328. doi: 10.1128/Mcb.20.4.1321-1328.2000
Schilling, S., and Oesterhelt, C. (2007). Structurally reduced monosaccharide transporters in an evolutionary conserved red alga. Biochem. J. 406, 325–331. doi: 10.1042/bj20070448
Seiboth, B., Karimi, R. A., Phatale, P. A., Linke, R., Hartl, L., Sauer, D. G., et al. (2012). The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol. Microbiol. 84, 1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x
Shen, L., Poree, F. H., Gaslonde, T., Lalucque, H., Chapeland-Leclerc, F., and Ruprich-Robert, G. (2019). Functional characterization of the sterigmatocystin secondary metabolite gene cluster in the filamentous fungus Podospora anserina: involvement in oxidative stress response, sexual development, pigmentation and interspecific competitions. Environ. Microbiol. 21, 3011–3026. doi: 10.1111/1462-2920.14698
Silar, P., Dauget, J. M., Gautier, V., Grognet, P., Chablat, M., Hermann-Le Denmat, S., et al. (2019). A gene graveyard in the genome of the fungus Podospora comata. Mol. Genet. Genomics 294, 177–190. doi: 10.1007/s00438-018-1497-3
Slot, J. C., and Rokas, A. (2011). Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr. Biol. 21, 134–139. doi: 10.1016/j.cub.2010.12.020
Stricker, A. R., Mach, R. L., and de Graaff, L. H. (2008). Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl. Microbiol. Biotechnol. 78, 211–220. doi: 10.1007/s00253-007-1322-0
Sun, J., and Glass, N. L. (2011). Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS One 6:e25654. doi: 10.1371/journal.pone.0025654
Sun, J., Tian, C., Diamond, S., and Glass, N. L. (2012). Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot. Cell 11, 482–493. doi: 10.1128/EC.05327-11
Tani, S., Kawaguchi, T., and Kobayashi, T. (2014). Complex regulation of hydrolytic enzyme genes for cellulosic biomass degradation in filamentous fungi. Appl. Microbiol. Biotechnol. 98, 4829–4837. doi: 10.1007/s00253-014-5707-6
Tonukari, N. J., Scott-Craig, J. S., and Walton, J. D. (2000). The Cochliobolus carbonum SNF1 gene is required for cell wall-degrading enzyme expression and virulence on maize. Plant Cell 12, 237–247. doi: 10.1105/tpc.12.2.237
Tzima, A. K., Paplomatas, E. J., Rauyaree, P., Ospina-Giraldo, M. D., and Kang, S. (2011). VdSNF1, the sucrose nonfermenting protein kinase gene of Verticillium dahliae, is required for virulence and expression of genes involved in cell-wall degradation. Mol. Plant Microbe Interact. 24, 129–142. doi: 10.1094/MPMI-09-09-0217
Vacher, S., Cotton, P., and Fèvre, M. (2003). Characterization of a SNF1 homologue from the phytopathogenic fungus Sclerotinia sclerotiorum. Gene 310, 113–121. doi: 10.1016/s0378-1119(03)00525-0
van den Brink, J., and de Vries, R. P. (2011). Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbiol. Biotechnol. 91, 1477–1492. doi: 10.1007/s00253-011-3473-2
Wang, D., Li, Y., Wang, H., Wei, D., Akhberdi, O., Liu, Y., et al. (2018). The AMP-activated protein kinase homolog Snf1 concerts carbon utilization, conidia production and the biosynthesis of secondary metabolites in the taxol-producer Pestalotiopsis microspora. Genes 9:E59. doi: 10.3390/genes9020059
Wang, X. X., He, P. H., Feng, M. G., and Ying, S. H. (2014). BbSNF1 contributes to cell differentiation, extracellular acidification, and virulence in Beauveria bassiana, a filamentous entomopathogenic fungus. Appl. Microbiol. Biotechnol. 98, 8657–8673. doi: 10.1007/s00253-014-5907-0
Weng, J. K., Li, X., Bonawitz, N. D., and Chapple, C. (2008). Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19, 166–172. doi: 10.1016/j.copbio.2008.02.014
Wu, X., Fan, X., Xie, S., Lin, J., Cheng, J., Zhang, Q., et al. (2018). Solar energy-driven lignin-first approach to full utilization of lignocellulosic biomass under mild conditions. Nat. Catal. 1, 772–780. doi: 10.1038/s41929-018-0148-8
Xie, N., Chapeland-Leclerc, F., Silar, P., and Ruprich-Robert, G. (2014). Systematic gene deletions evidences that laccases are involved in several stages of wood degradation in the filamentous fungus Podospora anserina. Environ. Microbiol. 16, 141–161. doi: 10.1111/1462-2920.12253
Xie, N., Ruprich-Robert, G., Silar, P., and Chapeland-Leclerc, F. (2015). Bilirubin oxidase-like proteins from Podospora anserina: promising thermostable enzymes for application in transformation of plant biomass. Environ. Microbiol. 17, 866–875. doi: 10.1111/1462-2920.12549
Xie, N., Ruprich-Robert, G., Silar, P., Herbert, E., Ferrari, R., and Chapeland-Leclerc, F. (2018). Characterization of three multicopper oxidases in the filamentous fungus Podospora anserina: a new role of an ABR1-like protein in fungal development? Fungal Genet. Biol. 116, 1–13. doi: 10.1016/j.fgb.2018.04.007
Yang, J., Li, D., Liu, X., Pan, J., Yan, B., and Zhu, X. (2010). Regulation of virulence factors, carbon utilization and virulence by SNF1 in Cryptococcus neoformans JEC21 and divergent actions of SNF1 between cryptococcal strains. Fungal Genet. Biol. 47, 994–1000. doi: 10.1016/j.fgb.2010.08.002
Yenkie, K. M., Wu, W., Clark, R. L., Pfleger, B. F., Root, T. W., and Maravelias, C. T. (2016). A roadmap for the synthesis of separation networks for the recovery of bio-based chemicals: matching biological and process feasibility. Biotechnol. Adv. 34, 1362–1383. doi: 10.1016/j.biotechadv.2016.10.003
Yi, M., Park, J. H., Ahn, J. H., and Lee, Y. H. (2008). MoSNF1 regulates sporulation and pathogenicity in the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 45, 1172–1181. doi: 10.1016/j.fgb.2008.05.003
Yin, W. B., Amaike, S., Wohlbach, D. J., Gasch, A. P., Chiang, Y. M., Wang, C. C., et al. (2012). An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol. Microbiol. 83, 1024–1034. doi: 10.1111/j.1365-2958.2012.07986.x
Young, E. T., Dombek, K. M., Tachibana, C., and Ideker, T. (2003). Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278, 26146–26158. doi: 10.1074/jbc.M301981200
Yu, J., Son, H., Park, A. R., Lee, S. H., Choi, G. J., Kim, J. C., et al. (2014). Functional characterization of sucrose non-fermenting 1 protein kinase complex genes in the Ascomycete Fusarium graminearum. Curr. Genet. 60, 35–47. doi: 10.1007/s00294-013-0409-7
Keywords: Podospora anserina, sucrose non-fermenting 1, sexual development, stress tolerance, secondary metabolism, lignocellulose degradation
Citation: Li Y, Yan P, Lu X, Qiu Y, Liang S, Liu G, Li S, Mou L and Xie N (2020) Involvement of PaSNF1 in Fungal Development, Sterigmatocystin Biosynthesis, and Lignocellulosic Degradation in the Filamentous Fungus Podospora anserina. Front. Microbiol. 11:1038. doi: 10.3389/fmicb.2020.01038
Received: 29 December 2019; Accepted: 27 April 2020;
Published: 10 June 2020.
Edited by:
Guodong Liu, Shandong University, ChinaReviewed by:
Shuai Zhao, Guangxi University, ChinaRobert Debuchy, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2020 Li, Yan, Lu, Qiu, Liang, Liu, Li, Mou and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning Xie, bmluZy54aWVAc3p1LmVkdS5jbg==
 Yuanjing Li
Yuanjing Li Pengfei Yan2
Pengfei Yan2