- 1Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, United States
- 2Center for Genome Research and Biocomputing, Oregon State University, Corvallis, OR, United States
Rhodococcus is a genus of Gram-positive bacteria with species that can cause growth deformations to a large number of plant species. This ability to cause disease is hypothesized to be dependent on a cluster of three gene loci on an almost 200 kb-sized linear plasmid. To reevaluate the roles of some of the genes in pathogenicity, we constructed and characterized deletion mutants of fasR and four fas genes. Findings confirmed that fasR, which encodes a putative transcriptional regulator, is necessary for pathogenesis. However, three of the fas genes, implicated in the metabolism of plant growth promoting cytokinins, are dispensable for the ability of the pathogen to cause disease. We also used long-read sequencing technology to generate high quality genome sequences for two phytopathogenic strains in which virulence genes are diverged in sequence and/or hypothesized to have recombined into the chromosome. Surprisingly, findings showed that the two strains carry extremely diverse virulence plasmids. Ortholog clustering identified only 12 genes present on all three virulence plasmids. Rhodococcus requires a small number of horizontally acquired traits to be pathogenic and the transmission of the corresponding genes, via recombination and conjugation, has the potential to rapidly diversify plasmids and bacterial populations.
Introduction
Plasmids are major drivers in the evolution of bacterial pathogens of plants and animals. Acquired plasmids can innovate recipients by conferring novel functions such as the capacity to remodel host cells and suppress immunity (e.g., Reuter et al., 2014; The et al., 2016; Barton et al., 2018). Acquired plasmids can also coopt chromosomal genes and reprogram them to cause a change in lifestyle (Letek et al., 2010). Understanding plasmid ecology and evolution is critical but their study is challenging (Orlek et al., 2017). Plasmids are typically mobile, dispensable, and enriched in repeated sequences. As a consequence, plasmids are often variable in gene composition and their sequences can be difficult to identify in draft genome assemblies.
Rhodococcus is a genus of non-motile, non-sporulating, high G+C, mycolic acid containing Gram-positive bacteria (Larkin et al., 1998). Its members are cosmopolitan and can be found in terrestrial and aquatic environments, as pathogens of mammals, and as epiphytic and endophytic bacteria of plants. The genus is diverse, and, in a phylogeny, plant associated Rhodococcus strains are found in a clade separated from others by a long branch (Savory et al., 2017). Evidence suggests that plants may benefit from their symbioses with Rhodococcus. Strains have been associated to biocontrol and plant growth promoting activities (Francis et al., 2016; Hong et al., 2016; Savory et al., 2017). Moreover, Rhodococcus has been repeatedly identified in the microbiota of diverse species of plants, including Arabidopsis, rice, clover, and soybean (Delmotte et al., 2009; Bulgarelli et al., 2012; Knief et al., 2012; Lundberg et al., 2012; Vorholt, 2012; Bai et al., 2015). However, plant associated Rhodococcus bacteria can also be pathogenic. Strains carrying a virulence plasmid cause proliferations of buds or shoots, called leafy galls or witch’s brooms, to monocots, dicots, herbaceous, and some woody plants (Murai et al., 1980; Crespi et al., 1992; Putnam and Miller, 2007).
A virulence plasmid characterized from D188, a Rhodococcus fascians strain, is thought to be representative of those across the different species-level groups of plant pathogenic Rhodococcus (Savory et al., 2017). Plasmid pFiD188 is a linear replicon of approximately 200 kb (Francis et al., 2012). This plasmid has a cluster of genes that are either necessary for, or implicated in, virulence. The fasR and fas loci are necessary for virulence (Crespi et al., 1992; Temmerman et al., 2000). FasR is predicted to be a member of the AraC-type transcriptional regulator family. Despite its name, FasR does not appear to have a role in inducing the transcription of the fas operon (Temmerman et al., 2000). Of the fas operon, fasA, fasD, fasE, and fasF were concluded to be necessary for pathogenicity (Pertry et al., 2010). In vitro activity and functional annotations suggest three of these proteins are involved in the metabolism of cytokinins, which are best known as plant growth promoting hormones (Kieber and Schaller, 2014). FasD shows isopentenyltransferase (IPT) activity, one of the steps in cytokinin biosynthesis (Crespi et al., 1992). FasF exhibits phosphoribohydrolase activity and is a member of the Lonely Guy (LOG) family, a group of proteins that in plants catalyze the formation of free base forms of cytokinins (Pertry et al., 2010). FasE shows cytokinin dehydrogenase activity and can cleave and irreversibly inactivate cytokinins (Pertry et al., 2010). FasA is annotated as a putative cytochrome P450 monooxygenase. Also on pFiD188 and adjacent to fasR is the 10 gene att locus (Crespi et al., 1992; Maes et al., 2001). Mutants of att are reported to be attenuated in virulence and work suggested products of att are involved in signaling (Maes et al., 2001). Last, between fasR and fas are mtr1 and mtr2, which encode methyltransferases (Francis et al., 2012; Radhika et al., 2015). Their predicted roles in virulence are described in a following section.
A population genomics study of phytopathogenic Rhodococcus suggested that there is low diversity among virulence plasmids (Savory et al., 2017). A total of 64 of 66 sequenced pathogenic strains have a plasmid that is similar in sequence and structure as pFiD188. Two exceptions are strains A21d2 and A25f, which because of the absence of homologs of most genes on pFiD188, were predicted to have virulence genes in their chromosomes (Creason et al., 2014). Additionally, the virulence genes of A21d2 are more diverged in sequence. The fasRA21d2 allele has a high number of single nucleotide polymorphisms and small insertion/deletion polymorphisms, relative to fasRD188. A21d2 also lacks homologs of mtr1, mtr2, as well as fas genes, and encodes a putative IPT-LOG hybrid protein (Creason et al., 2014). A25f, in contrast, has homologs of the cluster spanning from att, fasR, mtr1, mtr2, and fas on a single large contig. None of the other genes on the contig are homologous to those on pFiD188.
Even prior to the discovery of the fas genes, cytokinins were implicated in the virulence of Rhodococcus (Klämbt et al., 1966; Thimann and Sachs, 1966). One of the extant models predicts that a mixture of three cytokinin types, isopentenyladenine (iP), trans-zeatin (tZ), and cis-zeatin (cZ), as well as modified variants, are synthesized in a fas-dependent manner and secreted into plant cells to cause disease (Pertry et al., 2010; Stes et al., 2011). However, tZ is not reproducibly detected, cZ does not accumulate in a fas-dependent manner, and many of the cytokinin types accumulate in infected plants in patterns inconsistent with being causative (Eason et al., 1996; Galis et al., 2005; Creason et al., 2014; Jameson et al., 2019). A second model predicts that methyltransferases encoded by mtr1 and mtr2 function upstream of FasD to yield methylated cytokinins (Radhika et al., 2015). These novel cytokinins are predicted to be responsible for the ability of Rhodococcus to cause disease (Jameson et al., 2019). However, methylated cytokinins accumulate to low levels in bacteria and their levels do not vary in a plasmid-dependent manner (Pertry et al., 2009, 2010). A third model suggests that iP is the only type that is synthesized in a fas dependent manner but whether it directly causes disease has also been questioned (Creason et al., 2014; Jameson et al., 2019; Thapa et al., 2019).
Here, we used two approaches to generate resources to understand how Rhodococcus plasmids confer pathogenicity. We constructed and tested deletion mutants of fasR and four fas genes in strain D188. We also examined the role of fasR in A21d2. Results suggested that models of Rhodococcus virulence may have been informed by polar mutants of fas. We also generated higher quality genome sequences for pathogenic strains A21d2 and A25f. Resequencing uncovered two novel virulence plasmids in A21d2 and A25f that are distinct in structure and composition relative to pFiD188. Findings show that virulence of plant pathogenic Rhodococcus requires surprisingly few plasmid-encoded functions.
Materials and Methods
Bacterial Strains and Growth Conditions
Strains of Rhodococcus spp. were maintained on solid LB medium at 28°C or grown overnight in liquid LB medium with appropriate antibiotics at 28°C with shaking. Escherichia coli DH5α, 10-beta, and ccdb- were maintained on solid LB medium at 37°C or grown overnight in liquid LB medium at 37°C with shaking. Antibiotics were used at final concentrations of 25 μg/ml chloramphenicol, 25 μg/ml gentamicin, and 30 μg/ml kanamycin.
Extraction of Nucleic Acids
To extract genomic DNA from Rhodococcus, the Wizard Genomic DNA Purification Kit was used, following the instructions of the manufacturer for Gram-positive bacteria (Promega Corporation, Madison, WI, United States). To extract plasmids from E. coli, the Qiagen QIAprep Miniprep Kit was used, following the instructions of the manufacturer (Qiagen Company, Hilden, Germany). To extract RNA from Rhodococcus, cells were pelleted and resuspended in 500 μl Tri-Reagent (Sigma-Aldrich) in 2 ml tubes containing Lysing Matrix B (MP Biomedicals, Irvine, CA, United States). A FastPrep24 (MP Biomedicals, Irvine, CA, United States) was used to lyse cells for 1 min at 6.0 M/s. After, 100 μl of chloroform was added to each tube. Tubes were vortexed briefly and centrifuged for 10 min at 4°C at maximum speed. The upper phase was transferred to a new tube and 350 μl of isopropanol was added. Samples were incubated at −20°C overnight and then centrifuged for 10 min at 4°C at maximum speed. The supernatant was decanted, samples were washed two times with 70% ethanol, and the RNA was resuspended in DEPC-treated H2O.
DNA Cloning
Primers used for cloning are described in Supplementary Table S1. Over the course of the study, various methods, including Gateway (Invitrogen, Carlsbad, CA, United States), Gibson (NEB, Ipswich, MA, United States), and restriction enzyme digestion and ligation were used for cloning. Unless stated, DNA modifying enzymes were from NEB (Ipswich, MA, United States).
Gateway cloning was employed to make constructs used for generating deletion mutants. The pSelAct plasmid was linearized with SmaI and treated with CIP (van der Geize et al., 2008). The Gateway Cassette C.1 was released from pBluescript via an EcoRV digest. The two fragments were ligated by T4 DNA ligase to yield the plasmid, pSelAct-GW. Fragments corresponding to approximately 1.5 kb of the regions flanking the genes of interest were amplified in a two-step PCR. First, fragments were amplified using sequence-specific primers with partial attB sequences. Equimolar amounts of the two fragments were assembled in a second reaction and PCR with primers B1 and B2 were used to link the two fragments and complete the attB sequences. Assembled fragments were cloned via BP reaction into pDONR207 and cloned into pSelAct-GW using LR Clonase (Invitrogen, Carlsbad, CA, United States).
The fasR genes were cloned downstream of the L5 promoter by replacing either the tdTomato or GFP-encoding genes in vectors pJDC60 or pJDC165, respectively. For fasRA21d2 and fasRD188 (+ATG1), the coding sequences were amplified, products were digested with EcoRI and BamHI and ligated to pJDC60 digested with the same restriction enzymes. Gibson cloning was used to generate the shorter variants of fasRD188 and the fasRD188 frameshift construct. The NEBuilder Assembly Tool v.1 (NEB, Ipswich, MA, United States) was used to design primers that amplified both pSelAct, the fasRD188 fragments, or the full length that also included a thymine to cause a frameshift. Fragments were amplified using KAPA HIFI, following the manufacturer’s instructions (Kapa Biosystems, St. Louis, MO, United States). Equimolar amounts of the two fragments were assembled using Gibson Assembly Mastermix and incubated for 30 min at 50°C.
PCR, restriction enzyme digestion, and Sanger sequencing were used to verify all constructs.
Bacterial Transformation and Generation of Deletion Mutants
Rhodococcus fascians competent cells were prepared from overnight-grown 3 ml cultures. Cells were pelleted and washed twice with sterile, cold dH2O, followed by one wash with sterile, cold 10% glycerol. Cells were resuspended in 50 μl of 10% glycerol. Plasmid DNA (0.5–1 μg) was added to the cells, incubated on ice for 30 min and cells were electroporated in 1.0 mm gap cuvettes at 2.2 kV (Bio-Rad Micropulser, Hercules, CA, United States). Cells were resuspended in LB medium and incubated overnight at 28°C prior to plating on LB medium with appropriate antibiotics. For generating deletion mutants, merodiploids were selected for on LB plates with apramycin (50 μg/ml) and verified by PCR. Merodiploids were grown in liquid LB without antibiotics for 16–24 h at 28°C with shaking. Recombinants were selected for by growing cells for 5–10 days on mineral acetate medium with 5-fluorocytosine (100 μg/ml; van der Geize et al., 2008). PCR and Sanger sequencing of products were used to screen for and verify mutants.
Quantitative Real-Time PCR
RNA was resuspended in a solution of 2 μl 10 × DNAse I buffer, 1 μl DNase I (Invitrogen, Carlsbad, CA, United States), 1 μl RNaseOUT (Invitrogen, Carlsbad, CA, United States) and 16 μl DEPC-treated H2O. Samples were incubated at room temperature for 15 min. DNase I was inactivated by adding 1 μl of 25 mM EDTA and heating samples for 10 min at 65°C. A total of 1 μl of DNAse-treated RNA from each sample was used directly in PCR to verify that there was no residual DNA sufficient for amplification. RNA was quantified using the Qubit RNA BR Assay Kit and a Qubit 4 Fluorimeter (Invitrogen, Carlsbad, CA, United States).
First-strand cDNA was synthesized from 1.0 μg total RNA, using qScript cDNA Supermix and following the manufacturer’s instructions (Quantabio, Beverly, MA, United States). cDNA equivalent to 25 ng of starting RNA template was used in 20 μl reaction volumes consisting of 10 μl Supermix, 0.5 μl each forward and reverse primers, 1 μl cDNA and 8 μl sterile RNase-free H2O. Reaction conditions were 95°C for 3 min followed by 40 cycles of 95°C, 10 s, 55°C 10 s, 72°C for 1 min on a Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States). Bio-Rad CFX Meistro software was used to analyze the data.
Plant Inoculation Assays
Nicotiana benthamiana seeds were sterilized in a 20% bleach solution with a drop of polyoxyethylene sorbitan monolaurate (Tween 20) and shaken for 20–30 min. Seeds were washed twice with distilled water and resuspended in 1 ml sterile dH2O. Sterilized seeds were plated on MS agar medium (half-strength Murashige and Skoog medium, 0.5 M MES) and maintained vertically, at room temperature and constant light, for 3 days prior to inoculation. Overnight cultures of Rhodococcus were prepared in LB medium with appropriate antibiotics. Cells were pelleted, washed with sterile dH2O, and their concentration measured using a spectrophotometer. Suspensions were adjusted to OD600 = 0.5 in 100 mM MgCl2 prior to inoculation. Each seedling was inoculated with ∼3–5 μl of bacterial suspension. For each experiment, a negative control of an equivalent volume of 100 mM MgCl2 was included. Plates were allowed to dry briefly before being returned to the growing conditions described previously. Images of plants were taken 7 days post inoculation (dpi). Roots were measured via ImageJ (Schneider et al., 2012). All experiments were repeated at least three times with 40–50 seedlings inoculated per treatment. Data were analyzed, using one-way ANOVA followed by Tukey’s multiple comparisons test, as described in Savory et al. (2017). GraphPad Prism v. 7 was used for analyses (GraphPad Software, La Jolla, CA, United States).
For leafy gall assays, N. benthamiana seeds were sterilized as described above, plated on MS agar medium, and grown at room temperature under a 10:14 h light:dark cycle for 4 weeks. Bacteria were prepared as described above and 10 μl of OD600 = 0.5 of bacteria were dropped onto meristems of plants that had been pinched with forceps. Plants were returned to the growth chamber and inspected up to 28 dpi when images were taken. Infections included a mock treatment in which 10 μl of 100 mM MgCl2 was dropped onto meristems that had been pinched with forceps. At least three replicates were performed with 12 individuals per experiment.
DNA Sequencing
For Sanger sequencing, PCR products were treated with 2.5 units of Exonuclease I and 0.25 U of Shrimp Alkaline Phosphatase and incubated at 37°C for 40 min and 80°C for 20 min. Products were sequenced on an ABI3730 capillary sequencing machine at the Oregon State University Center for Genome Research and Biocomputing (CGRB).
Strains A21d2 and A25f were multiplexed and sequenced with 1D chemistry (LSK109) on an Oxford Nanpore MinION Mk1b, run using a MinIT coprocessor. Native Ligation kit (SQK-LSK109) libraries were prepared according to the recommended native barcoding protocol and sequenced in multiplex. Fast basecalling and demultiplexing was performed live using Guppy v. 3.2.6 (Oxford Nanopore, Oxford, United Kingdom). Demultiplexing was either performed using Guppy during sequencing for A21d2 or using qcat v. 1.1.0 with the options “–trim –kit ‘NBD104/NBD114”’ for A25f (Oxford Nanopore, Oxford, United Kingdom). Illumina short reads were trimmed for adapters and quality, using the BBTools program bbduk v. 35.82 with Illumina barcode sequences and the options “ktrim = r k = 23 mink = 9 hdist = 1 minlength = 100 tpe tbo” (Bushnell, 2020). Long reads were de novo assembled using Canu v. 1.8 with the options “overlapper = mhap utgReAlign = true corMinCoverage = 0 useGrid = false corThreads = 4 maxThreads = 4 genomeSize = (Illumina-only assembly size) “–nanopore-raw” for A21d2 and with Flye v. 2.6 with the options “” for A25f (Koren et al., 2017; Kolmogorov et al., 2019). Unicycler v. 0.4.7 with options “–mode bold” and “–existing_long_read_assembly” was used to combine long-read assemblies with quality-trimmed Illumina reads to generate hybrid assemblies (Creason et al., 2014; Wick et al., 2017). Bandage v. 0.8.1 was used to assess the assembly graphs for completeness (Wick et al., 2015). Final assemblies were annotated using Prokka v. 1.14.0 “–gram pos –addgenes –rfam” (Seemann, 2014).
Genome Analyses
Get_homologues v. 3.2.3 with MCL clustering was used to cluster orthologous genes between strains D188, A21d2, and A25f (Contreras-Moreira and Vinuesa, 2013). Each virulence plasmid gene was linked to a particular ortholog group (orthogroup) and used to identify homologs of genes on virulence plasmids.
The translated sequences of attB, fasR, and fasD were used in BLAST searches (default search parameters) against the NCBI nr database on February 10, 2020 to identify homologous sequences. The top 500 hits to full-length sequences for each were downloaded and aligned using MAFFT v. 7.402 e-ins-i (Katoh and Standley, 2013). Maximum likelihood phylogenies were constructed using IQ-TREE v. 1.6.12 with the options “–bb 1000 –alrt 1000” (Nguyen et al., 2015).
Genome sequences of Rhodococcus sp. Leaf225 and Williamsia sp. Leaf354 (accessions GCF_001426145.1 and GCF_001424365.1) were downloaded from NCBI. Sequences from pFiD188 and plasmids of A21d2 and A25f were used as queries in searches against these genome assemblies.
Results
Reevaluating the Role of fas Genes in Pathogenicity of Rhodococcus
The mechanistic basis by which Rhodococcus cause disease is unresolved, as findings from different research groups are incongruent (Stes et al., 2011; Creason et al., 2014; Jameson et al., 2019). In earlier attempts to repeat findings on the accumulation of cytokinins in culture grown Rhodococcus, we raised the possibility that the use of polar mutants may have contributed to inconsistencies between models (Creason et al., 2014). To address this, we constructed deletion mutants of fasA, fasD, fasE, and fasF in strain D188 to reevaluate the necessity of these genes in the pathogenicity of Rhodococcus.
Phytopathogenic Rhodococcus causes symptoms to root systems and meristems of N. benthamiana. Seedlings infected with strain D188 were significantly stunted in root growth and terminally arrested at the cotyledon stage, relative to mock inoculated plants (Figures 1A,B). Meristems of plants directly inoculated with strain D188 had a proliferation of buds, typical of leafy gall disease (Figure 1A). Unexpectedly, N. benthamiana inoculated with ΔfasA, ΔfasE, or ΔfasF consistently exhibited symptoms similar to those on plants infected with pathogenic strain D188 (Figure 1). The roots of seedlings were stunted, and their lengths were not significantly different relative to the root lengths of plants infected with D188 (p-value ≤ 0.05). The three deletion mutants also reproducibly caused leafy galls on plants. Data suggest that fasA, fasE, and fasF are not individually necessary for Rhodococcus strain D188 to cause disease.
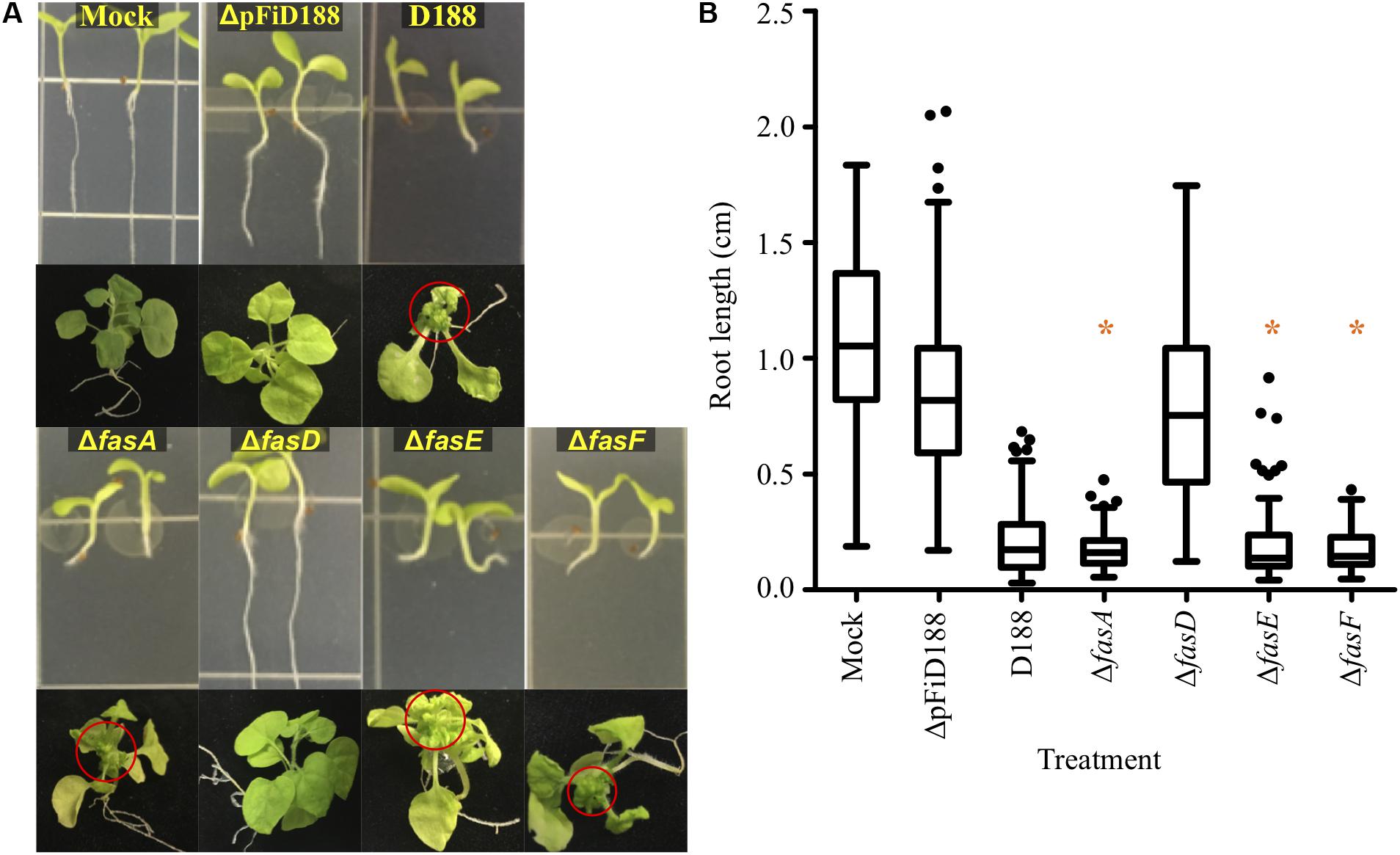
Figure 1. Most fas deletion mutants are pathogenic. (A) Wild type and deletion mutants of Rhodococcus strain D188 were inoculated onto seedlings or meristems of N. benthamiana. Red circles circumscribe leafy galls. This is a composite figure in which photographs were taken of plants treated in different experiments. Images of representative plants are shown. In all experiments, positive and negative control treatments were included. (B) At 7 dpi, roots were imaged and measured. Differences were compared relative to the corresponding D188 control group (* is not significant with a p-value threshold ≤0.05). Similar results were reproduced in at least two additional repeats of the experiments.
In contrast, plants inoculated with ΔfasD developed similarly to mock- and ΔpFiD188-inoculated plants (Figure 1). The latter treatment is with a non-pathogenic strain of D188 that lacks the virulence plasmid (Savory et al., 2017). Roots of ΔfasD-infected seedlings grew normally and were significantly longer than roots of D188-infected seedlings. Plants infected with fasD failed to show leafy gall disease symptoms when meristems were inoculated. However, quantitative reverse-transcriptase PCR (qRT-PCR) analyses showed that expression of the entire fas locus was compromised in the ΔfasD mutant (Supplementary Figure S1). The fasA, fasE, and fasF genes were expressed at significantly lower levels than in strain D188. It is also the case that the fasD coding sequence overlaps with fasE and in generating ΔfasD, we also compromised fasE. But as shown, the ΔfasE mutant is still pathogenic and dispensable for virulence (Figure 1). We also quantified expression of the fas genes in the original fasD insertion mutant relative to their expression in strain D188. Expression of fasA, fasE, and fasF is severely affected (Supplementary Figure S1). These data confirm that the fas operon is necessary for virulence, but we cannot unequivocally conclude that the fasD gene is necessary, as both an insertion within and deletion of, compromises expression of all examined fas genes at the locus.
Refining the Annotation of fasR
Comparisons of the sequences of the 64 pFiD188-like plasmids identified a sequencing mistake in the original annotation of fasR that was carried forward in analyses (Temmerman et al., 2000; Francis et al., 2012). A guanine base was inserted upstream of the annotated ATG of fasRD188. Correcting the error revealed two in frame ATG codons 213 (ATG1) and 168 (ATG2) nucleotides 5′ to the originally annotated start codon (ATG3; Figure 2A; Creason et al., 2014). Automated annotation programs identified ATG2 as the start because it has a strong ribosome binding sequence. Of the 12 nucleotides immediately upstream of ATG2, 11 are guanine or adenine bases. We cloned the three potential coding sequences, starting from each of the first three ATG codons, downstream of a mycobacteriophage L5 promoter that demonstrably constitutively expresses in Rhodococcus. We also generated a new mutant allele of fasRD188 (fs fasRD188) by introducing a nucleotide into the coding sequence to cause a frameshift mutation because previous studies used a deletion mutant that removes a portion of the neighboring attH gene (Figure 2A; Temmerman et al., 2000). On plants, fs fasRD188 failed to inhibit root development and failed to cause leafy gall disease, confirming the necessity of fasR for pathogenicity (Figures 2B,C).
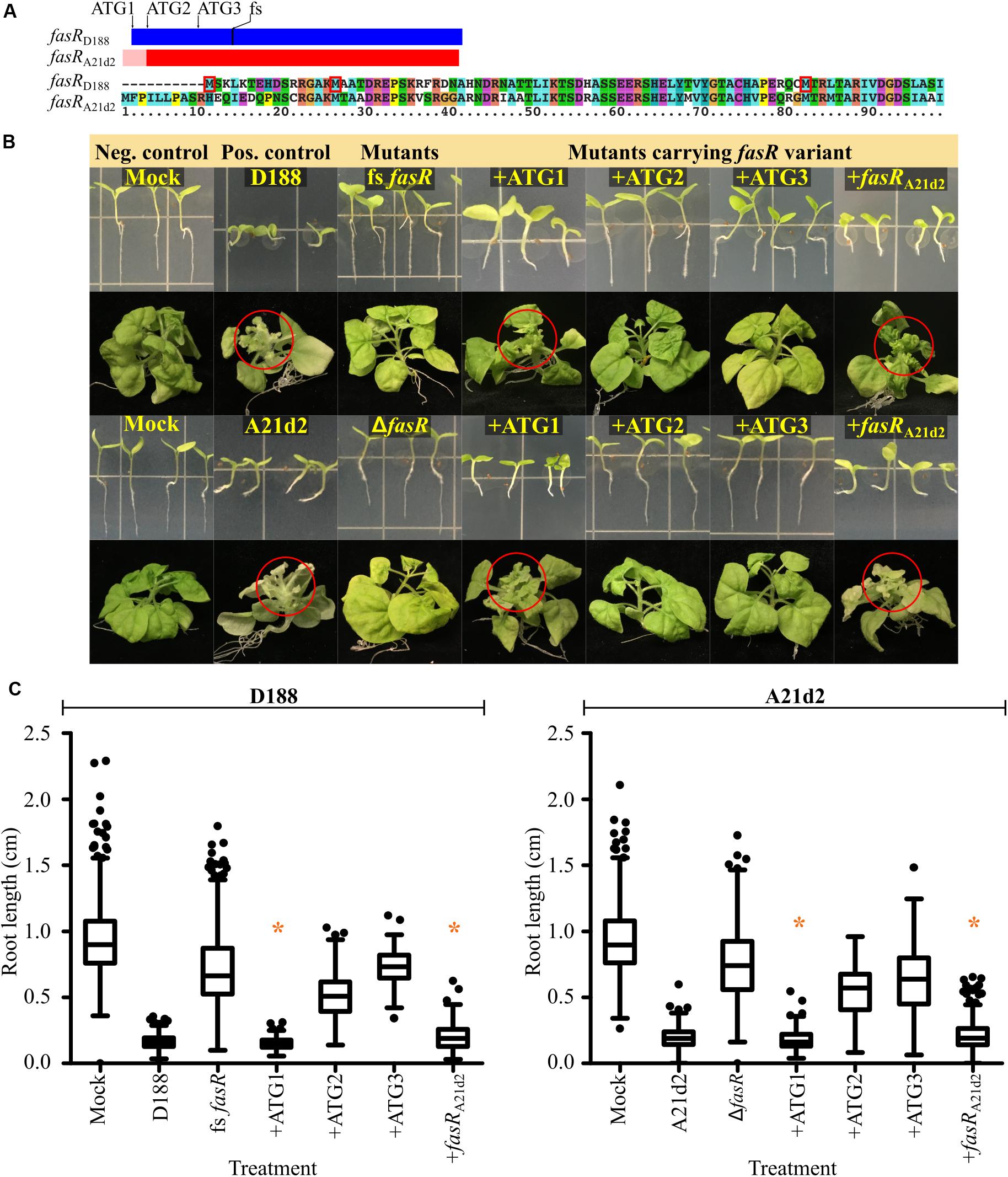
Figure 2. The fasR gene is necessary for virulence of D188 and A21d2. (A) Scaled schematic of the fasR coding sequences of D188 (blue) and A21d2 (red). The first three in-frame ATG codons of fasRD188 are shown. The location of the single base insertion introduced to cause a frameshift (fs) in fasRD188 is also indicated. A possible 5′ extension to fasRA21d2 is depicted by a pink-colored box. An alignment of the first ∼100 amino acids of the FasR variants of D188 and A21d2 is shown below the schematic. The first three methionine residues of FasR of D188 are boxed in red. (B) Wild type and mutant strains of D188 and A21d2 were inoculated onto seedlings or meristems of N. benthamiana. Red circles circumscribe leafy galls. Images of representative plants are shown. This is a composite figure in which photographs were taken of plants treated in different experiments. Images of representative plants are shown. In all experiments, positive and negative control treatments were included. (C) At 7 dpi, roots were imaged and measured. Differences were compared relative to the corresponding D188 or A21d2 control groups (* is not significant with a p-value threshold ≤0.05). Similar results were reproduced in at least two additional repeats of the experiments.
We next tested each of the three possible fasRD188 coding sequences to determine which is sufficient to complement the mutant. The mutant strain carrying the longest coding sequence (+ATG1) inhibited root growth to the same degree as strain D188 (Figures 2B,C). In contrast, inoculations with strains carrying the fasR coding sequence starting from the second (+ATG2) or third ATG (+ATG3) codon resulted in root lengths that were intermediate to those of mock- and D188-inoculated plants. Importantly, on meristems of plants, the mutant strain carrying +ATG1 caused leafy gall symptoms whereas fs fasRD188 carrying either of the shorter variants caused no visible disease symptoms. Either ATG1 is the start of the coding sequence or sequences upstream of ATG2 or ATG3 are necessary for stable expression of fasR.
Of the 66 alleles of fasR that have been sequenced, the A21d2 allele is the most diverged, and automated programs predicted it to be 18 codons shorter relative to the D188 allele (Figure 2A; Creason et al., 2014). There is another in frame ATG 75 nucleotides upstream from the ATG originally annotated as the start codon (pink region in Figure 2A). This aligned to another ATG further 5′ but out of frame to ATG1-3 of fasRD188. We relied on the originally annotated start codon of fasRA21d2. We generated and tested a fasR deletion mutant in A21d2 and confirmed its necessity in pathogenicity (Figures 2B,C). The ΔfasR mutant of A21d2 could be complemented with fasRA21d2 expressed from the L5 promoter. We next tested whether fasRA21d2 and fasRD188 alleles could complement fasR mutants in either strain. Despite the diversity, fasRA21d2 could complement the fasR frameshift mutant of D188. Similarly, +ATG1 could complement ΔfasR of A21d2 but neither +ATG2 or +ATG3 could do so.
Discovering Two Novel Virulence Plasmids in Phytopathogenic Rhodococcus
To gain more insights into the genomic context of virulence loci in strains A21d2 and A25f, we used Oxford Nanopore sequencing to generate long reads and combined them with the original Illumina-derived short reads. Nanopore sequencing produced 418,684 and 45,552 reads, respectively, with a mean read length of 7.8 and 4.0 kb. The longest reads for A21d2 and A25f were 112 and 106 kb, respectively. The A21d2 chromosome is 5,667,798 bp, and the A25f chromosome is 5,706,031 bp. Both strains have plasmids. Based on the assembly graph, two of the three plasmids in A21d2 are circular molecules (Figure 3A). The single plasmid in A25f is fragmented into three contigs (Figure 3A). Analysis suggested contig 3, which is 0.6 kb in length, has a repeated sequence that misassembled into one. The sequences are predicted to flank the other small contig and join to the largest plasmid contig to form a single circular molecule.
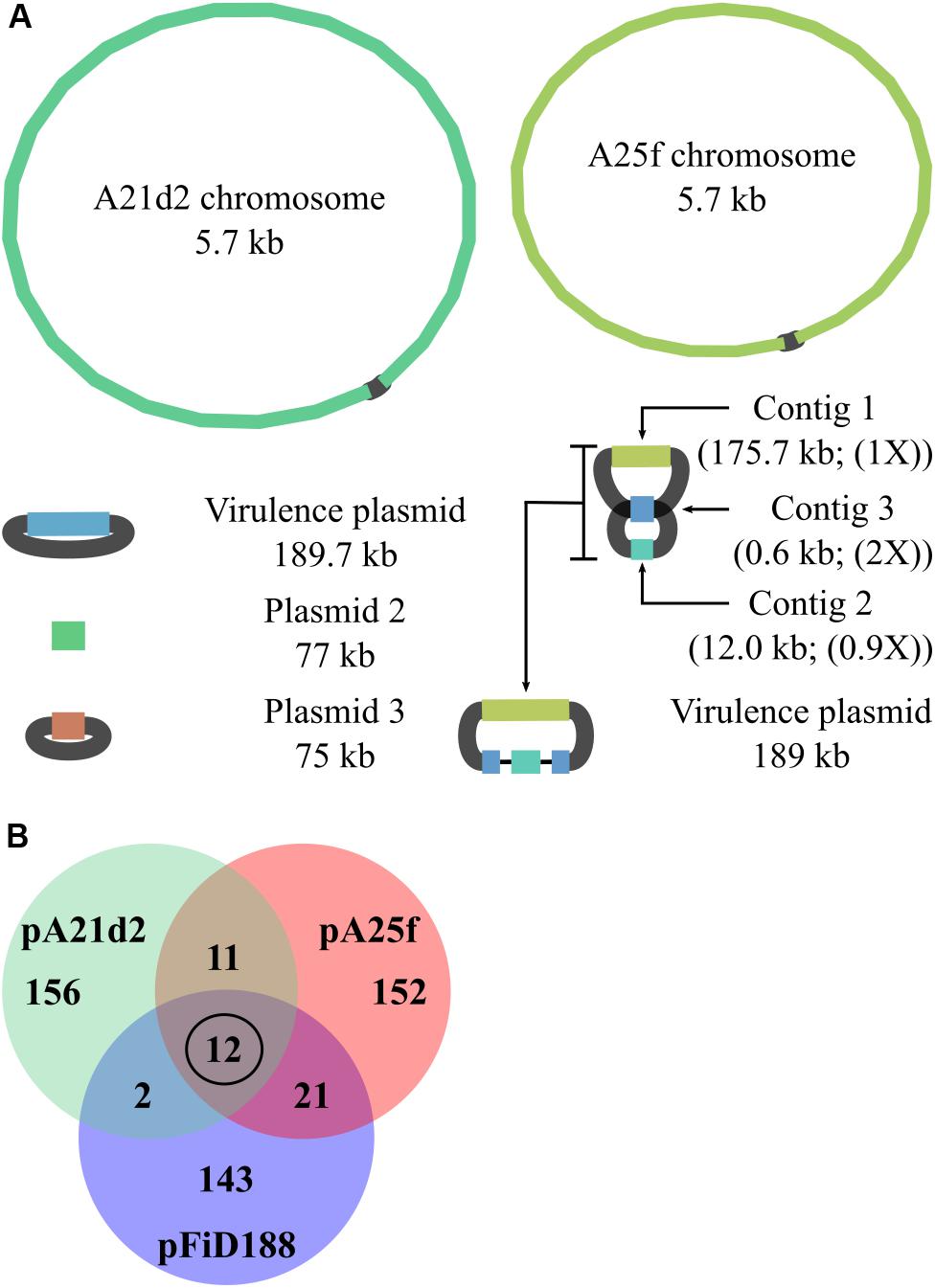
Figure 3. Pathogenic strains A21d2 and A25f have novel virulence plasmids. (A) Graphs of hybrid assembled genome sequences of A21d2 and A25f. Sizes of the replicons are shown. For the pA25f, a predicted structure is presented. The relative depth of coverage of plasmid contigs is shown in parentheses. (B) Venn diagram of gene homolog similarity. The sequences of all genes on the plasmids of A21d2, A25f, and D188 were translated and clustered on the basis of homology.
To our surprise, the virulence loci in each of these strains are located on circular plasmids. The virulence plasmid of A21d2, referred to as pA21d2, is 189,682 bp and has a cluster, from A21d2_05402 to A21d2_0514, of genes that are homologs of the 10 att genes, fasR, and IPT-LOG-encoding gene. The latter is annotated as miaA. MiaA is a tRNA-isopentenyltransferase (tRNA-IPT) that modifies base 37 on a subset of tRNAs (Schweizer et al., 2017). The annotations for tRNA-IPTs and IPTs are frequently misassigned by automated annotation programs (Takei et al., 2001; Lindner et al., 2014). Consistent with previous reports, there are no homologs of fasA, fasB, fasC, fasE, mtr1, or mtr2 downstream of fasR or anywhere else on any of the replicons of A21d2. Instead, adjacent and downstream of fasR on pA21d2 is a gene that encodes a putative radical SAM enzyme (Creason et al., 2014). In pA25f, from A25f_05486 to A25f_05504, there are homologs of att, fasR, and fasA-F that are conserved in sequence and order to the corresponding region of pFiD188.
We constructed phylogenetic trees for the translated sequences of attB, fasR, and fasD (Figure 4). AttB of pA21d2 and A25f grouped with those of plant pathogenic Rhodococcus, suggesting attB and likely all att genes are derived from a recent common ancestor (Figure 4A). In the FasR and FasD trees, the sequences from A21d2 are more distant and their corresponding genes likely emerged from a more ancestral source or diverged via recombination (Figures 4B,C; Creason et al., 2014). In contrast, the pA25f variants of these two virulence protein sequences grouped with others, supporting a scenario in which a homologous fragment spanning from A25f_05486 to A25f_05504 (att through fas) is a recombinogenic unit that can be shared among diverse Rhodococcus plasmids.
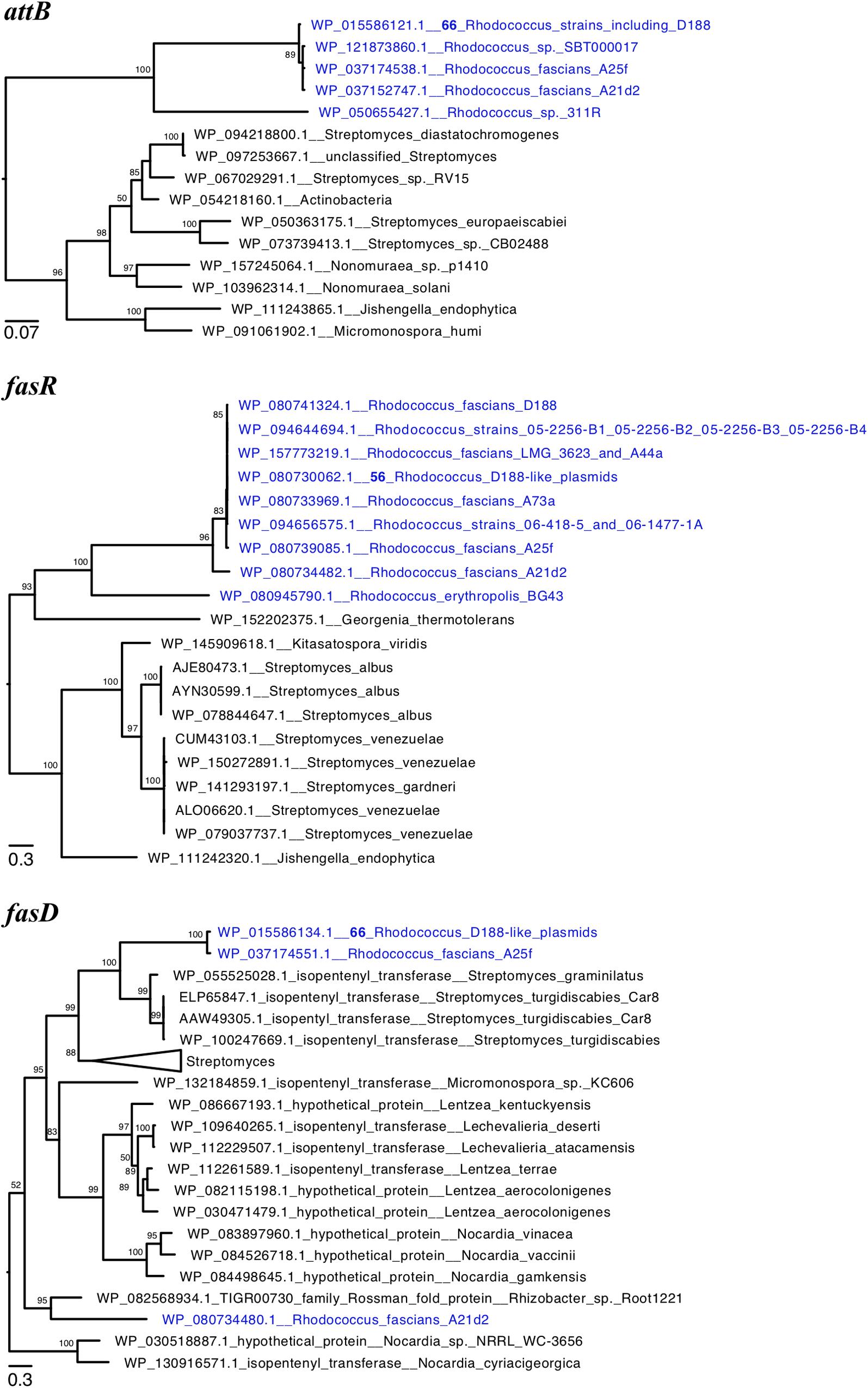
Figure 4. Phylogeny of virulence genes of plant pathogenic Rhodococcus. Maximum likelihood phylogenetic trees of translated sequences of (A) attB, (B) fasR, and (C) fasD. The trees were midpoint rooted and a subset of clades of Rhodococcus virulence proteins are shown. Tips with identical sequences are indicated. Large clades were collapsed and labeled. Bootstrap values greater than 50 are shown.
Two publicly available genome sequences of non-plant associated Rhodococcus also have homologs of attB (Figure 4A). Strain 311R was isolated from hydrocarbon-contaminated soils and strain SBT000017 was isolated from marine organisms (Macintyre et al., 2014; Ehsani et al., 2015). In a species phylogeny, strain 311R clustered with Rhodococcus erythropolis, distant from the major clade with plant pathogenic strains (Savory et al., 2017). In contrast, strain SBT000017 clusters with plant pathogenic strains and is in the same clade as strain D188. Both 311R and SBT000017 have a complete att locus but no homologs of fasR, mtr1, mtr2, fas, or homologs that encode the putative radical SAM enzyme or IPT-LOG and are inferred to be non-pathogenic. In strain 311R, the translated attB sequence is divergent (71% amino acid identity) relative to the D188 sequence. Its att locus is adjacent to integrase-encoding genes and followed by genes of unknown function and located in the chromosome between aspC_1 and pur. In strain D188, aspC_1 and pur are immediately adjacent to one another, suggesting that strain 311R acquired a small island that circumscribes the att locus. In strain SBT000017, the translated att sequences are very similar to those of pFiD188 and range from 97.7 to 99.5% amino acid identity. The att locus is located on yet another novel plasmid.
The percentage of genes with homologs among pFiD188, pA21d2, and pA25f is low and suggest that they are unrelated plasmids that acquired the capacity to confer virulence to Rhodococcus. We translated the gene sequences of the three plasmids and clustered the sequences on the basis of homology. We identified only 12 gene sequences that are common to all three plasmids. Ten corresponded to the genes of the att locus and one is fasR (Figure 3B and Supplementary Table S2). The last encodes a predicted hypothetical protein (AMY56290.1). Other than these 12 genes, the number of other genes with homologs common to pairs of virulence plasmids is low, ranging from 2 to 20. Given that pFiD188 and A25f are identical in gene composition from att through fas, it was not surprising that there is slightly higher similarity between these two plasmids. No gene on pFiD188, implicated in plasmid maintenance or conjugation, had a homolog on the other two plasmids, indicating that pFiD188-like, pA21d2, and pA25f plasmids have separate origins (Francis et al., 2012).
Mining Genome Sequences for Virulence Plasmids
Last, we reexamined the genome sequences of Rhodococcus sp. Leaf225 and Williamsia sp. Leaf354 for evidence of virulence plasmids (Bai et al., 2015). These two strains, despite one belonging to a different bacterial genus, were used to support the model that pathogenic Rhodococcus synthesizes novel methylated cytokinins and should therefore minimally have homologs of mtr1, mtr2, and fasD (Jameson et al., 2019). Previous work using att, fasR, or fas sequences from pFiD188 in BLAST queries failed to identify homologs in their genome sequences (Savory et al., 2017). To test the possibility that strains Leaf225 and Leaf354 have homologs of the novel plasmids uncovered in this study, we used all gene sequences from all three plasmids as queries in BLAST searches against the genome sequences of Leaf225 and Leaf354 (Supplementary Table S3). In addition to little evidence for virulence plasmids, we found no evidence that Leaf225 and Leaf354 have homologs of the genes core to all three plasmids, genes encoding Mtr1, Mtr2, the putative radical SAM enzyme or the IPT-LOG hybrid. When we lowered the thresholds in searches, we identified hits (<34–40% amino acid similarity) to Mtr1 and Mtr2 in Leaf225 (WP_027503960.1 and WP_056447883.1) and Leaf354 (WP_055787211.1, WP_055786245.1, WP_055790396.1, WP_055793823, WP_082501997.1, and WP_156378120.1 of Leaf354). However, use of these eight sequences as queries in reciprocal searches against the D188 genome sequence yielded better hits to genes in the chromosome, not pFiD188. Data suggested that Leaf225 and Leaf354 lack homologs of genes necessary for, or associated to, virulence.
Discussion
Virulence plasmids are essential for Rhodococcus to cause leafy gall disease to plants (Crespi et al., 1992; Pertry et al., 2009, 2010; Stes et al., 2011; Creason et al., 2014; Savory et al., 2017). Using long-read sequencing technology, we discovered two new virulence plasmids in plant pathogenic Rhodococcus (Figure 3A). Comparisons showed that there are very few conserved genes among the three known plasmids and our genetic analysis of mutants suggested these plasmids require very few plasmid-encoded functions to confer virulence. However, it has been suggested that additional pFiD188-encoded genes are required for pathogenicity of strain D188. A cosmid that covers att and fas regions from pFiD188 is insufficient for pathogenicity and it has also been suggested that fas expression requires another plasmid-encoded transcription factor for full expression (Crespi et al., 1992; Temmerman et al., 2000). Our analyses suggest if other plasmid-encoded functions are required for pathogenicity, that these functions are conferred by genes highly diverged in sequence.
Clustering of a small number of genes necessary for pathogenicity is a low barrier for recombination to convert non-virulence plasmids to virulence plasmids. The discovery of a novel plasmid bearing an att locus in SBT000017 raises the potential that the att locus can mediate recombination among distinct plasmids. This is one plausible mechanism to explain how pA25f and pFiD188, plasmids that differ dramatically in composition, exchange virulence loci (Figure 4). Spread of virulence loci among plasmids and spread of virulence plasmids among strains, combine to accelerate the diversification of pathogenic Rhodococcus populations (Savory et al., 2017). This has significant implications on disease ecology because diversification can drive the emergence of disease on novel hosts, increased aggressiveness, and further increased spread (McDonald and Stukenbrock, 2016).
Of the functions inferred to be necessary for virulence, fas-derived cytokinins have long been central to models (Stes et al., 2011; Creason et al., 2014; Jameson et al., 2019). It has not been definitively demonstrated that fasD is necessary for pathogenicity. Nonetheless, the data are consistent with IPT being encoded on all three plasmids and collectively point to IPT activity as the fas-encoded function that is necessary for virulence (Figure 1). It remains unresolved as to whether the products of IPT are synthesized to be secreted to manipulate plants. One source of disagreement among virulence models is the use of polar mutants, which unknowingly impacted conclusions regarding the role of each fas gene in virulence and the synthesis of specific cytokinin types (Desomer et al., 1991; Crespi et al., 1992; Pertry et al., 2010). The original fasD insertion mutant has polar effects and there is also potential that other previously published fas mutants have polar effects (Supplementary Figure S1). The fasA mutant was identified from the same insertion library as the fasD mutant and was carried forward in subsequent studies. The fasE and fasF mutants were made via insertional mutagenesis (Pertry et al., 2010).
Another source of disagreement stems from the challenges with directly testing virulence models. During infection, it is difficult to show cytokinins originate from the microbe. Both partners in the symbiosis produce the hormone. Unless the pathogen is able to synthesize overwhelming quantities of cytokinins, the majority of detected cytokinins are likely derived from the host because the biomass of the plant vastly exceeds that of the pathogen. In culture, Rhodococcus cells produce picomolar to nanomolar amounts of cytokinins (Pertry et al., 2009, 2010; Creason et al., 2014). It is also the case that cytokinins are involved in plant immunity and levels may change because of responses of the plant, not directly because of the pathogen (Choi et al., 2010; Großkinsky et al., 2011; Jiang et al., 2013; Albrecht and Argueso, 2017). Further confounding analyses is that plants and bacteria have alternative sources of cytokinins. A subset of tRNAs in plants and bacteria are modified with cytokinins, hypothesized to promote more efficient translation (Schweizer et al., 2017). The degradation of tRNAs is a source of cytokinins that have yet to be accounted for to understand how fas-derived cytokinins contribute to virulence (Matsubara et al., 1968; Cherayil and Lipsett, 1977; Murai et al., 1980; Kasahara et al., 2004).
Findings here also cloud the role of methylated cytokinins in the virulence of Rhodococcus. In contrast to the conclusion that all pathogenic strains have mtr1 and mtr2, we confirmed that A21d2 lacks mtr genes (Jameson et al., 2019). However, A21d2 encodes a member of the radical SAM enzyme family, and it is possible that this enzyme could contribute to the synthesis of methylated cytokinins (Zhang et al., 2012). Regardless, while levels of methylated cytokinins of plants change in response to infection, increases do not strictly correlate to infection by strains carrying homologs of genes demonstrably necessary for causing disease, or homologs of mtr genes implicated in the synthesis of methylated cytokinins (Supplementary Table S3). Strains Leaf225 and Leaf354 were originally isolated from asymptomatic Arabidopsis plants (Bai et al., 2015). Furthermore, Rhodococcus Leaf225 clusters in a clade with plant associated Rhodococcus strains in which no pathogenic isolates have been uncovered. Williamsia sp. Leaf354 is not a member of the Rhodococcus genus.
There are several examples that suggest microbe-synthesized cytokinins may have roles other than in virulence. Claviceps purpurea encodes an IPT-LOG hybrid protein and its corresponding mutant is unaffected in virulence and is altered in development (Hinsch et al., 2015). Some oncogenic plasmids of Agrobacterium carry tzs, which encodes an IPT that localizes to the bacterial membrane and has been implicated in virulence gene signaling (Aly et al., 2008; Hwang et al., 2013). Recent results showed that genes other than the IPT-encoding etz gene of Pantoea, are sufficient to cause non-pathogenic strains to elicit gall formation on plants (Lichter et al., 1995; Nissan et al., 2019). Finally, it is particularly noteworthy that cytokinins regulate development of the slime mold, Dictyostelium discoideum (Anjard and Loomis, 2008).
The att locus represents 10 of the core genes on the virulence plasmids and has been implicated in virulence (Maes et al., 2001). Inconsistent with the reported attenuated phenotype, pFiD188 carrying a non-functional att locus is sufficient to transform multiple strains of Rhodococcus into being pathogenic (Savory et al., 2017). In addition, Streptomyces turgidiscabies strain Car8 can also cause leafy galls on plants (Joshi and Loria, 2007). This strain lacks an att locus but has a cluster that includes fasR, mtr1, mtr2, and the fas genes. The strict conservation of att on the three plasmids among plant pathogenic Rhodococcus suggests either att is maintained because of physical linkage to virulence genes or there is a selective advantage to maintaining the locus. Data are consistent with the possibility that the advantage is unrelated to virulence.
The low number of functions conserved on virulence plasmids of plant pathogenic Rhodococcus is consistent with a mechanism of virulence gene cooption (Letek et al., 2010). This model predicts that horizontally acquired genes encode regulators of gene expression. They function to coopt chromosomal genes core to the genus and misregulate their expression to reprogram the recipient genome for virulence. The strongest candidate for cooptive virulence is fasR because of its necessity for virulence (Figure 2; Temmerman et al., 2000). Furthermore, the D188 and A21d2 alleles of fasR could complement either mutant, and there is potential that FasR recognizes and binds sequences that are conserved among plant associated Rhodococcus. Identifying the fasR-regulon is important and findings have potential to provide insights into the mechanisms by which virulence plasmids transition Rhodococcus to pathogens.
Data Availability Statement
Biosample IDs for the genome sequences are SAMN02569990 for A21d2 and SAMN0256991 for A25f.
Author Contributions
ES, AW, AC, and JC conceived and developed the experiments. ES, AW, DS, AC, SF, and EP performed the experiments and analyzed the data. ES, AW, DS, and JC wrote and edited the manuscript.
Funding
ES was supported by a grant from USDA-NIFA, 2013-67012-21139. AW was supported by a grant from USDA-NIFA, 2017-67012-26126. We thank the Department of Botany and Plant pathology for their support of the computing cluster and SF. DS was supported by a SURE science award and the College of Agricultural Sciences Undergraduate Research Award.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Jeff Cirillo for providing pJDC160 and pJDC165. We also thank Melodie Putnam and Dr. Taifo Mahmud for their comments and insights.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01022/full#supplementary-material
FIGURE S1 | Characterization of the fasD mutants. qRT-PCR of the insertion and deletion mutants of fasD. The relative-relative expression of fasA, fasD, fasE, and fasF was determined against expression of the conserved and abundantly expressed Elongation factor Tu (Ef-Tu)-encoding gene and the corresponding gene of D188. The experiment was repeated with similar results.
TABLE S1 | Oligonucleotides used in this study.
TABLE S2 | Orthogroups containing at least one gene from two or more virulence plasmids.
TABLE S3 | TBLASTN results of Leaf str. homologs.
References
Albrecht, T., and Argueso, C. T. (2017). Should I fight or should I grow now? The role of cytokinins in plant growth and immunity and in the growth-defence trade-off. Ann. Bot. 119, 725–735. doi: 10.1093/aob/mcw211
Aly, K. A., Krall, L., Lottspeich, F., and Baron, C. (2008). The type IV secretion system component VirB5 binds to the trans-zeatin biosynthetic enzyme Tzs and enables its translocation to the cell surface of Agrobacterium tumefaciens. J. Bacteriol. 190, 1595–1604. doi: 10.1128/JB.01718-07
Anjard, C., and Loomis, W. F. (2008). Cytokinins induce sporulation in Dictyostelium. Development 135, 819–827. doi: 10.1242/dev.018051
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. doi: 10.1038/nature16192
Barton, I. S., Fuqua, C., and Platt, T. G. (2018). Ecological and evolutionary dynamics of a model facultative pathogen: Agrobacterium and crown gall disease of plants. Environ. Microbiol. 20, 16–29. doi: 10.1111/1462-2920.13976
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren, van Themaat, E., Ahmadinejad, N., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Bushnell, B. (2020). BBMap. SourceForge. Available online at: https://sourceforge.net/projects/bbmap/ (accessed February 19, 2020).
Cherayil, J. D., and Lipsett, M. N. (1977). Zeatin ribonucleosides in the transfer ribonucleic acid of Rhizobium leguminosarum, Agrobacterium tumefaciens, Corynebacterium fascians, and Erwinia amylovora. J. Bacteriol. 131, 741–744.
Choi, J., Huh, S. U., Kojima, M., Sakakibara, H., Paek, K.-H., and Hwang, I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 19, 284–295. doi: 10.1016/j.devcel.2010.07.011
Contreras-Moreira, B., and Vinuesa, P. (2013). GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696–7701. doi: 10.1128/AEM.02411-13
Creason, A. L., Vandeputte, O. M., Savory, E. A., Davis, E. W., Putnam, M. L., Hu, E., et al. (2014). Analysis of genome sequences from plant pathogenic Rhodococcus reveals genetic novelties in virulence loci. PLoS ONE 9:e101996. doi: 10.1371/journal.pone.0101996
Crespi, M., Messens, E., Caplan, A. B., Van Montagu, M., and Desomer, J. (1992). Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 11, 795–804.
Delmotte, N., Knief, C., Chaffron, S., Innerebner, G., Roschitzki, B., Schlapbach, R., et al. (2009). Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 16428–16433. doi: 10.1073/pnas.0905240106
Desomer, J., Crespi, M., and Van Montagu, M. (1991). Illegitimate integration of non-replicative vectors in the genome of Rhodococcus fascians upon electrotransformation as an insertional mutagenesis system. Mol. Microbiol. 5, 2115–2124. doi: 10.1111/j.1365-2958.1991.tb02141.x
Eason, J., Morris, R., and Jameson, P. (1996). The relationship between virulence and cytokinin production by Rhodococcus fascians (Tilford 1936) Goodfellow 1984. Plant Pathol. 45, 323–331.
Ehsani, E., Jauregui, R., Geffers, R., Jareck, M., Boon, N., Pieper, D. H., et al. (2015). Draft genome sequence of Rhodococcus sp. Strain 311R. Genome Announc. 3:e378-15. doi: 10.1128/genomeA.00378-15
Francis, I., De Keyser, A., De Backer, P., Simón-Mateo, C., Kalkus, J., Pertry, I., et al. (2012). pFiD188, the linear virulence plasmid of Rhodococcus fascians D188. Mol. Plant-Microbe Interact. 25, 637–647. doi: 10.1094/MPMI-08-11-0215
Francis, I. M., Stes, E., Zhang, Y., Rangel, D., Audenaert, K., and Vereecke, D. (2016). Mining the genome of Rhodococcus fascians, a plant growth-promoting bacterium gone astray. N. Biotechnol. 33, 706–717. doi: 10.1016/j.nbt.2016.01.009
Galis, I., Bilyeu, K., Wood, G., and Jameson, P. E. (2005). Rhodococcus fascians: shoot proliferation without elevated cytokinins? Plant Growth Regul. 46, 109–115.
Großkinsky, D. K., Naseem, M., Abdelmohsen, U. R., Plickert, N., Engelke, T., Griebel, T., et al. (2011). Cytokinins mediate resistance against Pseudomonas syringae in tobacco through increased antimicrobial phytoalexin synthesis independent of salicylic acid signaling. Plant Physiol. 157, 815–830. doi: 10.1104/pp.111.182931
Hinsch, J., Vrabka, J., Oeser, B., Novák, O., Galuszka, P., and Tudzynski, P. (2015). De novo biosynthesis of cytokinins in the biotrophic fungus Claviceps purpurea. Environ. Microbiol. 17, 2935–2951. doi: 10.1111/1462-2920.12838
Hong, C. E., Jeong, H., Jo, S. H., Jeong, J. C., Kwon, S.-Y., An, D., et al. (2016). A leaf-inhabiting endophytic bacterium, Rhodococcus sp. KB6, enhances sweet potato resistance to black rot disease caused by Ceratocystis fimbriata. J. Microbiol. Biotechnol. 26, 488–492. doi: 10.4014/jmb.1511.11039
Hwang, H.-H., Yang, F.-J., Cheng, T.-F., Chen, Y.-C., Lee, Y.-L., Tsai, Y.-L., et al. (2013). The Tzs protein and exogenous cytokinin affect virulence gene expression and bacterial growth of Agrobacterium tumefaciens. Phytopathology 103, 888–899. doi: 10.1094/PHYTO-01-13-0020-R
Jameson, P. E., Dhandapani, P., Song, J., Zatloukal, M., Strnad, M., Remus-Emsermann, M. N. P., et al. (2019). The cytokinin complex associated with Rhodococcus fascians: which compounds are critical for virulence? Front. Plant Sci. 10:674. doi: 10.3389/fpls.2019.00674
Jiang, C.-J., Shimono, M., Sugano, S., Kojima, M., Liu, X., Inoue, H., et al. (2013). Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Mol. Plant-Microbe Interact. 26, 287–296. doi: 10.1094/MPMI-06-12-0152-R
Joshi, M. V., and Loria, R. (2007). Streptomyces turgidiscabies possesses a functional cytokinin biosynthetic pathway and produces leafy galls. Mol. Plant-Microbe Interact. 20, 751–758. doi: 10.1094/MPMI-20-7-0751
Kasahara, H., Takei, K., Ueda, N., Hishiyama, S., Yamaya, T., Kamiya, Y., et al. (2004). Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J. Biol. Chem. 279, 14049–14054. doi: 10.1074/jbc.M314195200
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kieber, J. J., and Schaller, G. E. (2014). Cytokinins. Arabidopsis Book 12:e0168. doi: 10.1199/tab.0168
Klämbt, D., Thies, G., and Skoog, F. (1966). Isolation of cytokinins from Corynebacterium fascians. Proc. Natl. Acad. Sci. U.S.A. 56, 52–59.
Knief, C., Delmotte, N., Chaffron, S., Stark, M., Innerebner, G., Wassmann, R., et al. (2012). Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6, 1378–1390. doi: 10.1038/ismej.2011.192
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Larkin, M. J., De Mot, R., Kulakov, L. A., and Nagy, I. (1998). Applied aspects of Rhodococcus genetics. Antonie van Leeuwenhoek 74, 133–153.
Letek, M., González, P., Macarthur, I., Rodríguez, H., Freeman, T. C., Valero-Rello, A., et al. (2010). The genome of a pathogenic Rhodococcus: cooptive virulence underpinned by key gene acquisitions. PLoS Genet. 6:e1001145. doi: 10.1371/journal.pgen.1001145
Lichter, A., Barash, I., Valinsky, L., and Manulis, S. (1995). The genes involved in cytokinin biosynthesis in Erwinia herbicola pv. gypsophilae: characterization and role in gall formation. J. Bacteriol. 177, 4457–4465. doi: 10.1128/jb.177.15.4457-4465.1995
Lindner, A.-C., Lang, D., Seifert, M., Podlešáková, K., Novák, O., Strnad, M., et al. (2014). Isopentenyltransferase-1 (IPT1) knockout in Physcomitrella together with phylogenetic analyses of IPTs provide insights into evolution of plant cytokinin biosynthesis. J. Exp. Bot. 65, 2533–2543. doi: 10.1093/jxb/eru142
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Macintyre, L., Zhang, T., Viegelmann, C., Martinez, I. J., Cheng, C., Dowdells, C., et al. (2014). Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar. Drugs 12, 3416–3448. doi: 10.3390/md12063416
Maes, T., Vereecke, D., Ritsema, T., Cornelis, K., Thu, H. N., Van Montagu, M., et al. (2001). The att locus of Rhodococcus fascians strain D188 is essential for full virulence on tobacco through the production of an autoregulatory compound. Mol. Microbiol. 42, 13–28. doi: 10.1046/j.1365-2958.2001.02615.x
Matsubara, S., Armstrong, D. J., and Skoog, F. (1968). Cytokinins in tRNA of Corynebacterium fascians. Plant Physiol. 43, 451–453.
McDonald, B. A., and Stukenbrock, E. H. (2016). Rapid emergence of pathogens in agro-ecosystems: global threats to agricultural sustainability and food security. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20160026. doi: 10.1098/rstb.2016.0026
Murai, N., Skoog, F., Doyle, M. E., and Hanson, R. S. (1980). Relationships between cytokinin production, presence of plasmids, and fasciation caused by strains of Corynebacterium fascians. Proc. Natl. Acad. Sci. U.S.A. 77, 619–623. doi: 10.1073/pnas.77.1.619
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nissan, G., Chalupowicz, L., Sessa, G., Manulis-Sasson, S., and Barash, I. (2019). Two Pantoea agglomerans type III effectors can transform nonpathogenic and phytopathogenic bacteria into host-specific gall-forming pathogens. Mol. Plant Pathol. 20, 1582–1587. doi: 10.1111/mpp.12860
Orlek, A., Stoesser, N., Anjum, M. F., Doumith, M., Ellington, M. J., Peto, T., et al. (2017). Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front. Microbiol. 8:182. doi: 10.3389/fmicb.2017.00182
Pertry, I., Václavíková, K., Depuydt, S., Galuszka, P., Spíchal, L., Temmerman, W., et al. (2009). Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc. Natl. Acad. Sci. U.S.A. 106, 929–934. doi: 10.1073/pnas.0811683106
Pertry, I., Václavíková, K., Gemrotová, M., Spíchal, L., Galuszka, P., Depuydt, S., et al. (2010). Rhodococcus fascians impacts plant development through the dynamic fas -mediated production of a cytokinin mix. Mol. Plant-Microbe Interact. 23, 1164–1174. doi: 10.1094/MPMI-23-9-1164
Putnam, M. L., and Miller, M. L. (2007). Rhodococcus fascians in herbaceous perennials. Plant Dis. 91, 1064–1076. doi: 10.1094/PDIS-91-9-1064
Radhika, V., Ueda, N., Tsuboi, Y., Kojima, M., Kikuchi, J., Kudo, T., et al. (2015). Methylated cytokinins from the phytopathogen Rhodococcus fascians mimic plant hormone activity. Plant Physiol. 169, 1118–1126. doi: 10.1104/pp.15.00787
Reuter, S., Connor, T. R., Barquist, L., Walker, D., Feltwell, T., Harris, S. R., et al. (2014). Parallel independent evolution of pathogenicity within the genus Yersinia. Proc. Natl. Acad. Sci. U.S.A. 111, 6768–6773. doi: 10.1073/pnas.1317161111
Savory, E. A., Fuller, S. L., Weisberg, A. J., Thomas, W. J., Gordon, M. I., Stevens, D. M., et al. (2017). Evolutionary transitions between beneficial and phytopathogenic Rhodococcus challenge disease management. Elife 6:e30925. doi: 10.7554/eLife.30925
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schweizer, U., Bohleber, S., and Fradejas-Villar, N. (2017). The Modified Base Isopentenyladenosine and its derivatives in tRNA. RNA Biol. 14, 1197–1208. doi: 10.1080/15476286.2017.1294309
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Stes, E., Vandeputte, O. M., El Jaziri, M., Holsters, M., and Vereecke, D. (2011). A successful bacterial coup d’état: how Rhodococcus fascians redirects plant development. Annu. Rev. Phytopathol. 49, 69–86. doi: 10.1146/annurev-phyto-072910-095217
Takei, K., Sakakibara, H., and Sugiyama, T. (2001). Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 276, 26405–26410. doi: 10.1074/jbc.M102130200
Temmerman, W., Vereecke, D., Dreesen, R., Van Montagu, M., Holsters, M., and Goethals, K. (2000). Leafy gall formation is controlled by fasR an AraC-type regulatory gene in Rhodococcus fascians. J. Bacteriol. 182, 5832–5840. doi: 10.1128/jb.182.20.5832-5840.2000
Thapa, S. P., Davis, E. W., Lyu, Q., Weisberg, A. J., Stevens, D. M., Clarke, C. R., et al. (2019). The evolution, ecology, and mechanisms of infection by gram-positive, plant-associated bacteria. Annu. Rev. Phytopathol. 57, 341–365. doi: 10.1146/annurev-phyto-082718-100124
The, H. C., Thanh, D. P., Holt, K. E., Thomson, N. R., and Baker, S. (2016). The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat. Rev. Microbiol. 14, 235–250. doi: 10.1038/nrmicro.2016.10
Thimann, K. V., and Sachs, T. (1966). The role of cytokinins in the” fasciation” disease caused by Corynebacterium fascians. Am. J. Bot. 53, 731–739.
van der Geize, R., de Jong, W., Hessels, G. I., Grommen, A. W. F., Jacobs, A. A. C., and Dijkhuizen, L. (2008). A novel method to generate unmarked gene deletions in the intracellular pathogen Rhodococcus equi using 5-fluorocytosine conditional lethality. Nucleic Acids Res. 36, e151. doi: 10.1093/nar/gkn811
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Keywords: virulence plasmids, evolution, Gram-positive, Rhodococcus, cytokinins
Citation: Savory EA, Weisberg AJ, Stevens DM, Creason AL, Fuller SL, Pearce EM and Chang JH (2020) Phytopathogenic Rhodococcus Have Diverse Plasmids With Few Conserved Virulence Functions. Front. Microbiol. 11:1022. doi: 10.3389/fmicb.2020.01022
Received: 19 March 2020; Accepted: 27 April 2020;
Published: 25 May 2020.
Edited by:
Clay Fuqua, Indiana University Bloomington, United StatesReviewed by:
Ana Valero-Rello, Independent Researcher, Valencia, SpainAlexia Hapeshi, University of Warwick, United Kingdom
Copyright © 2020 Savory, Weisberg, Stevens, Creason, Fuller, Pearce and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeff H. Chang, Y2hhbmdqQHNjaWVuY2Uub3JlZ29uc3RhdGUuZWR1
†These authors have contributed equally to this work
‡Present address: Elizabeth A. Savory, Oregon Department of Agriculture, Salem, OR, United States Danielle M. Stevens, Integrative Genetics and Genomics Graduate Group, University of California, Davis, Davis, CA, United States Allison L. Creason, Computational Biology Program, Oregon Health & Science University, Portland, OR, United States
 Elizabeth A. Savory
Elizabeth A. Savory Alexandra J. Weisberg
Alexandra J. Weisberg Danielle M. Stevens
Danielle M. Stevens Allison L. Creason1‡
Allison L. Creason1‡ Emma M. Pearce
Emma M. Pearce Jeff H. Chang
Jeff H. Chang