
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 May 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01017
Azole resistance of the fungal pathogen Aspergillus fumigatus is an emerging problem. To identify novel mechanisms that could mediate azole resistance in A. fumigatus, we analyzed the transcriptome of a mitochondrial fission/fusion mutant that exhibits increased azole tolerance. Approximately 12% of the annotated genes are differentially regulated in this strain. This comprises upregulation of Cyp51A, the azole target structure, upregulation of ATP-binding cassette (ABC) superfamily and major facilitator superfamily (MFS) transporters and differential regulation of transcription factors. To study their impact on azole tolerance, conditional mutants were constructed of seven ABC transporters and 17 transcription factors. Under repressed conditions, growth rates and azole susceptibility of the mutants were similar to wild type. Under induced conditions, several transcription factor mutants showed growth phenotypes. In addition, four ABC transporter mutants and seven transcription factor mutants exhibited altered azole susceptibility. However, deletion of individual identified ABC transporters and transcription factors did not affect the increased azole tolerance of the fission/fusion mutant. Our results revealed the ability of multiple ABC transporters and transcription factors to modulate the azole susceptibility of A. fumigatus and support a model where mitochondrial dysfunctions trigger a drug resistance network that mediates azole tolerance of this mold.
Aspergillus fumigatus is an opportunistic fungal pathogen that causes life-threatening airborne infections in immunocompromised patients. The disease is called invasive aspergillosis and associated with high mortality rates of up to 95% (Kousha et al., 2011; Brown et al., 2012; Kosmidis and Denning, 2015). Treatment of invasive aspergillosis relies on the administration of antifungals. The azole class of antifungals is currently recommended as first line treatment for infections caused by A. fumigatus (Patterson et al., 2016). Azoles target the ergosterol biosynthesis pathway by inhibiting the lanosterol 14α-demethylase (CYP51) which is fungicidal for the mold A. fumigatus. However, in the recent years azole-resistant Aspergillus strains are emerging, thereby challenging the efficacy of current azole-based therapies (van der Linden et al., 2015; Perlin et al., 2017). As such, azole resistance was associated with therapy failure and increased mortality in patients suffering from invasive aspergillosis (van der Linden et al., 2015; Lestrade et al., 2019).
The majority of the azole-resistant clinical A. fumigatus isolates harbor resistance-mediating mutations in the target enzyme, the lanosterol 14α-demethylase (CYP51, also known as Erg11), or in its promoter region (Dudakova et al., 2017; Perlin et al., 2017). Unfortunately, the mechanistic nature facilitating azole resistance of the remaining clinical isolates – which may represent up to 50% of the azole resistant clinical isolates (Fraczek et al., 2013) – is largely unknown. Azole resistance of Candida species is often mediated by increased expression of efflux pumps (Morschhäuser, 2016; Prasad et al., 2016). It was therefore proposed that upregulation of drug efflux could represent the second most abundant azole resistance mechanism in A. fumigatus (Cowen et al., 2015; Dudakova et al., 2017; Perlin et al., 2017). Indeed, increased expression of several efflux pumps was observed in several azole resistant clinical A. fumigatus isolates without CYP51 mutations (Fraczek et al., 2013). However, experimental evidence that clearly demonstrates overexpression of these or other efflux pumps causes azole resistance of A. fumigatus is still lacking (Fraczek et al., 2013; Moye-Rowley, 2015; Dudakova et al., 2017). So far, only two ATP-binding cassette (ABC) transporters were shown to affect the azole tolerance of the mold under in vitro conditions (nicely summarized in Moye-Rowley, 2015).
Recently, we characterized the role of the mitochondrial fusion and fission machinery for viability, virulence and antifungal drug susceptibility of A. fumigatus (Neubauer et al., 2015). Surprisingly, we found that the disruption of mitochondrial fission and of mitochondrial fission and fusion results in Aspergillus mutants that exhibit increased azole tolerance. A role of mitochondrial function in azole tolerance was previously reported for pathogenic and non-pathogenic yeasts and linked to altered expression of efflux pumps (Traven et al., 2001; Tsai et al., 2006; Ferrari et al., 2011a, b; Shingu-Vazquez and Traven, 2011; Peng et al., 2012). But the exact mechanism responsible for the increased azole tolerance remained unresolved. Here we show that the expression of multiple ABC superfamily and major facilitator superfamily (MFS) transporters is upregulated in the mitochondrial fission and fusion mutant. In addition, expression of cyp51A, the gene that encodes one of two CYP51 orthologs, was upregulated and the expression of several transcription factors were up- or downregulated. Construction and phenotypic characterization of conditional mutants of the differentially regulated genes revealed the ability of several of the identified transcription factors and ABC transporters to affect the azole susceptibility of A. fumigatus. Our results suggest that mitochondrial dysfunction triggers increased expression of CYP51 and induction of efflux pumps in A. fumigatus which could mediate the increased azole tolerance observed in such mutants.
The non-homologous end joining-deficient A. fumigatus strain AfS35 was used as wild type in this study (Krappmann et al., 2006; Wagener et al., 2008). The conditional mutants mdu1tetOn (AFUA 8G07000), mdu2tetOn (AFUA_1G03800), mdu3tetOn (AFUA_5G01650), mdu4tetOn (AFUA_8G07280), mdu5tetOn (AFUA_2G15340), mdu6tetOn (AFUA_4G01470), mdu7tetOn (AFUA_4G00710), atfDtetOn (mdu8tetOn; AFUA_6G12150), mdu9tetOn (AFUA_2G09330), mdu10tetOn (AFUA_1G14860), mdu11tetOn (AFUA_2G14350), mdd1tetOn (AFUA_5G14290), mdd2tetOn (AFUA_1G15910), mdd3tetOn (AFUA_6G06535), gliZtetOn (mdd4tetOn; AFUA_6G09630), mdd5tetOn (AFUA_6G12160), mdd6tetOn (AFUA_4G06880), abc1tetOn (AFUA_3G01400), mdr1tetOn (abc2tetOn; AFUA_5G06070), abc3tetOn (AFUA_6G08020), abc4tetOn (AFUA_4G14130), abc5tetOn (AFUA_6G03080), abc6tetOn (AFUA_5G10510), and abc7tetOn (AFUA_3G07300) were constructed by inserting a doxycycline-inducible promoter cassette (oliC-tetOn; pJW128) before the coding regions of the indicated genes, essentially as described before (Helmschrott et al., 2013). The deletion mutants Δmdu2, Δmdu3, Δmdu11, Δmdd3, ΔgliZ (Δmdd4), Δmdd5, Δabc1, Δmdr1 (Δabc2), Δabc3, and Δabc4 in the parental strain Δdnm1 mgm1tetOn (Neubauer et al., 2015) were constructed using a phleomycin resistance cassette. The correct integration of the cassettes was verified using diagnostic PCRs, essentially as described before (Geißel et al., 2018). Conidia were harvested from cultures on Aspergillus minimal medium (AMM; Hill and Kafer, 2001) at 37°C. The conditional mutant mdd6tetOn was always raised under induced conditions on medium supplemented with 0.5 μg ml–1 doxycycline. Experiments were performed on or in Sabouraud medium [4% (w/v) d-glucose, 1%(w/v) peptone (#LP0034; Thermo Fisher Scientific; Rockford, IL, United States, pH 7.0]. When indicated, medium was supplemented with doxycycline (#631311; Clontech; Mountain View, CA, United States) or voriconazole (#A4320; Apexbt Technology LLC; Houston, TX, United States). Solid medium was supplemented with 2% (w/v) agar (214030; BD, Franklin Lakes, NJ, United States). Etest strips were obtained from bioMérieux (Marcyl’Etoile, France). To fully induce the conditional Tet-On promoter, medium was supplemented with 15 μg ml–1 if not stated differently (Helmschrott et al., 2013; Dichtl et al., 2015; Loiko and Wagener, 2017). Broth microdilution experiments were performed in 96 well plates. To this end, conidia were inoculated in 100 μl medium per well at a concentration of 5 × 104 conidia ml–1. Medium was supplemented with serial voriconazole dilutions (×0.75) starting with a concentration of 3 μg ml–1 voriconazole. Plates were incubated at 37°C and analyzed after 48 h.
For the gene expression analyses, three RNA samples were isolated and analyzed per strain (wild type and Δdnm1 mgm1tetOn). To this end, 2.5 × 107 conidia were inoculated in 50 ml Sabouraud liquid medium and cultured for 15 h at 37°C. Mycelium was separated from culture medium and washed with PBS buffer solution. RNA was extracted by pestling 100 mg mycelium in liquid nitrogen and immediately suspending the frozen ground powder in 450 μl RLT buffer (RNeasy Mini Kit (50); #142349288, QIAGEN GmbH, D-40724 Hilden). The RTL buffer suspension was subsequently processed according to the manufacturer’s instructions. RNA quality and quantity were analyzed with the Experion RNA StdSens Analysis Kit (#700-7103, Bio-Rad Laboratories, Hercules, CA, United States) and Experion System (100-240V, for RNA and DNA analyses, includes electrophoresis station, priming station, vortex station, software, USB2 cable, instructions) (#700-7001, Bio-Rad Laboratories, Hercules, California, United States). RNA sequencing was subsequently performed by GATC Biotech AG (InView Transcriptome Explorer; Konstanz, Germany) and the transcriptome mapped on the Af293 genome.
Three RNA samples were isolated and analyzed per strain (wild type, mdd3tetOn and mdu2tetOn) and condition (with and without 15 μg ml–1 doxycycline). To this end, 2.5 × 107 conidia were inoculated in 50 ml Sabouraud liquid medium with and without doxycycline and cultured for 12 h at 37°C. Mycelium was separated from culture medium and disrupted in Lysing-Matrix-C tubes (M.P. Biomedical; Irvine, CA, United States) supplemented with TRIzol Reagent (#15596-026, ambion/RNA, life technologies; Carlsbad, CA, United States) in a FastPrep-24 homogenizer (M.P. Biomedical; at 6.5 m s–1 for 60 s followed by cooling in an ice-cold water bath). RNA was subsequently purified following the instructions of the manufacturer of the TRIzol Reagent. The RNA pellet was resuspended with 60 μl DEPC-treated water (#AM9922, Ambion, Thermo Fisher Scientific; Rockford, IL, United States). Gene expression was then quantified with 100 ng RNA per PCR (100 ng μl–1) and specific primers and the Luna Universal One-Step-RT-qPCR Kit (#E3005; New England Biolabs, Ipswich, MA, United States) in a qTower3 (Analytik Jena, Jena, Germany). Expression data was normalized to act1 (actin; AFUA_6G04740) and analyzed with the ΔΔCt method described by Pfaffl (Pfaffl, 2001). Statistical analyses were performed with GraphPad Prism 5 (GraphPad Software, La Jolla, CA, United States). Statistical significance was calculated using ANOVA with post hoc Tukey.
RNA sequencing data were analyzed with Cuffdiff (Trapnell et al., 2010) using the web-based Galaxy platform (Afgan et al., 2018)1 to identify differentially expressed genes in the repressed Δdnm1 mgm1tetOn mutant in comparison to wild type (p = 0.007). Further analyses were performed with Gene Ontology Enrichment (Ashburner et al., 2000) of FungiDB (Basenko et al., 2018) by searching for over- and underrepresented GO terms (Gene ontology enrichment2; goslim_generic subset; molecular function/biological process; p = 0.05; 06.03.2018). The following GO term annotations were examined separately: mitochondrion (GO:0005739), mitochondrion organization (GO:0007005), transmembrane transport (GO:0055085), transmembrane transporter activity (GO:0022857), response to stress (GO:0006950), ergosterol biosynthetic process (GO:0006696), and regulation of ergosterol biosynthetic process (GO:0032443) (FungiDB; my strategies/genes/function prediction/GO term; 20.03.2019). For identification of the ABC and MFS transporters as well as the transcription factors which are up- and downregulated in Δdnm1 mgm1tetOn the differentially expressed genes were compared with the genes coding the indicated proteins using the free software Knime3.
To obtain DNA and protein sequences as well as ontology and protein function prediction the following databases were used for the respective species A. fumigatus: Aspergillus Genome Database4, Af293 (01.04.2016) (Cerqueira et al., 2014) and Fungi DB2, Af293 (28.03.2018) (Basenko et al., 2018); S. cerevisiae: Yeast Genome Database5, S288C (01.04.2016; 28.03.2018) (Engel et al., 2014); Candida albicans: Candida Genome Database6, SC5314/assembly 22 (28.03.2018) (Skrzypek et al., 2017). Pfam domains (PF000057, IPR003439 ABC_tran ABC transporter-like; PF01061, IPR013525 ABC2_membrane ABC-2 type; PF14510, ABC transporter extracellular N- terminal; PF06422, IPR010929 PDR_CDR CDR ABC transporter; PF00664, IPR001140 ABC_membrane ABC transporter, transmembrane domain; PF06472, IPR010509 ABC_membrane_2 ABC transporter, N-terminal; PF12848, ABC_tran_2 ABC transporter; PF00385, Chromo (Chromatin Organization Modifier) domain; PF04068, IPR007209 RLI RNase L inhibitor RLI, possible metal-binding domain; PF00037, IPR001450 Fer4 4Fe-4S binding domain; PF12661, IPR013032 hEGF EGF-like, conserved site; PF12698, ABC2_membrane_3; PF02492, IPR003495 cobW CobW/HypB/UreG domain) (Finn et al., 2016) of all known ABC transporters of S. cerevisiae (Klein et al., 2011) were used to analyze the genome of A. fumigatus to identify and classify the 52 ABC transporters. A similar approach was used to identify and classify the 211 MFS transporter in the genome of A. fumigatus (Pfam domain PF07690 (IPR011701 MFS_1 Major facilitator superfamily; Costa et al., 2014; Dos Santos et al., 2014). Putative transcription factors were identified using Gene Ontology (GO:0006355 – regulation of transcription, DNA-templated) and further classified with correlating Pfam domains (PF07716, Basic region leucine zipper; PF00172, Fungal Zn(2)-Cys(6) binuclear cluster domain; PF04082, Fungal specific transcription factor domain; PF05920, Homeobox KN domain; PF00170, bZIP transcription factor; PF11951, Fungal specific transcription factor domain; PF13086, AAA domain; PF13087, AAA domain).
Mitochondria continuously undergo fission and fusion processes. Two distinct and partially conserved machineries accomplish mitochondrial fission and fusion (Pernas and Scorrano, 2016). We previously studied the fission and fusion machineries of A. fumigatus by constructing and characterized conditional and deletion mutants of several core components of the respective machineries (Neubauer et al., 2015; Wagener, 2016). Of those the conditional Δdnm1 mgm1tetOn mutant was deleted for the mitochondrial fission dynamin Dnm1 (important for fission) whereas the mitochondrial fusion dynamin Mgm1 (important for fusion) is under control of a doxycycline-inducible Tet-On promoter. The Δdnm1 mgm1tetOn mutant exhibits a remarkable azole tolerance under induced conditions, very similar to the fission single deletion mutant Δdnm1 or mutants lacking other key components of the mitochondrial fission machinery such as Δmdv1 and Δfis1. Under repressed conditions, which means inactivation of fission and fusion, the azole tolerance is even further pronounced (Neubauer et al., 2015). To identify and explore potential mechanisms that contribute to the azole resistance of A. fumigatus, we compared the transcriptome of the Δdnm1 mgm1tetOn mutant with the wild type. To this end, freshly harvested conidia were inoculated in Sabouraud medium and cultured for 15 h at 37°C. RNA was extracted and subjected to RNA sequencing and differential expression analysis. As shown in Figure 1A and Supplementary Tables S1–S3, 1249 genes – approximately 12% of the annotated A. fumigatus genome – were differentially expressed in the Δdnm1 mgm1tetOn mutant under repressed conditions compared to wild type (p = 0.007). Of those, 621 genes were upregulated and 628 genes were downregulated.
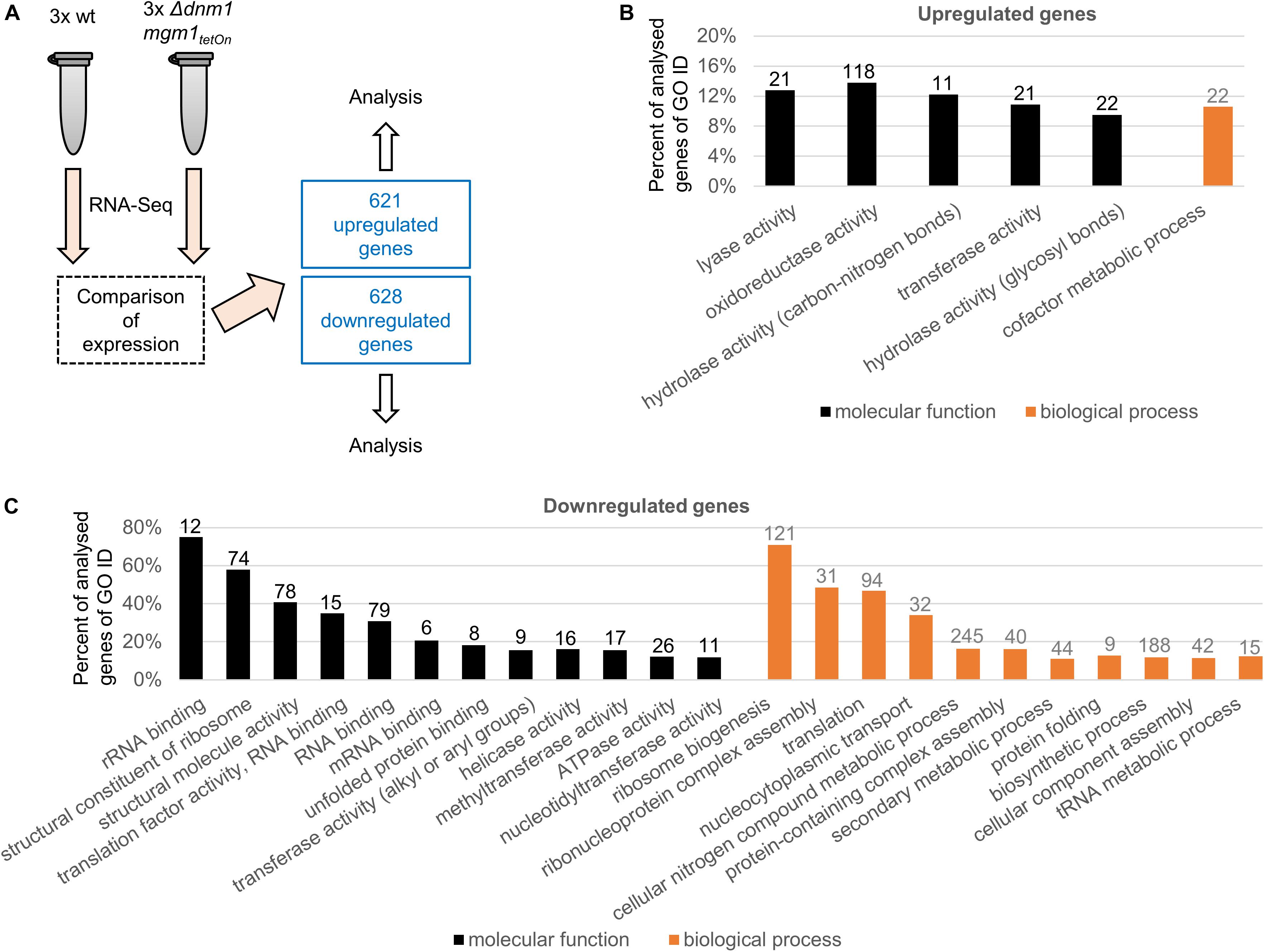
Figure 1. Analysis of differentially expressed genes in the Δdnm1 mgm1tetOn mutant. (A) The transcriptome of wild type (wt) and Δdnm1 mgm1tetOn were analyzed with RNA sequencing in triplicates. Differentially expressed genes were subsequently identified with Cuffdiff 32. 621 genes were upregulated and 628 genes were downregulated (p ≤ 0.007). (B,C) Significantly overrepresented (B) and underrepresented (C) GO terms (goslim_generic subset; p-value ≤ 0.05) for molecular function (black bars) and biological process (gray bars). The bars indicate the percentage of identified genes of all analyzed A. fumigatus genes associated with each GO slim term. The absolute number of identified genes is shown on top of each bar.
As expected, the gene with the highest negative fold-change was dnm1, the gene deleted in the double mutant. Surprisingly, no repression of mgm1 was detected even though the strain was cultured under repressive conditions (w/o doxycycline). To exclude mistaken identity of the analyzed strains we examined the RNA sequencing coverage (Supplementary Figure S1A). In contrast to the wild type where the mgm1 coverage already starts in the 5′ untranslated region, coverage in the Δdnm1 mgm1tetOn mutant begins with the start codon of mgm1 which is in line with the integration of the Tet-On promoter at this site. Since the repressibility of the analyzed strain is evident based on distinct growth phenotypes under repressed and induced conditions (Neubauer et al., 2015 and Supplementary Figure S1B), we concluded that the non-detected differential expression results from a combination of the leakiness of the conditional promoter system and potential heterogeneous expression of mgm1 in different vegetative/stationary hyphal compartments of the mycelium (Hewitt et al., 2016).
A genome ontology (GO) enrichment analysis was performed to identify which GO terms are over- and underrepresented (FungiDB database/gene ontology enrichment; goslim_generic subset; p = 0.05; Figures 1B,C; Ashburner et al., 2000; Basenko et al., 2018). Only, five molecular function and one biological process GO terms were overrepresented in the upregulated genes, those were primarily related to metabolism (Figure 1B and Supplementary Tables S4, S5). In contrast, multiple molecular function (12) and biological process (11) GO terms were enriched in the down regulated genes (Figure 1C and Supplementary Tables S6, S7). Interestingly, many of those (6 of 12 molecular function GO terms and 6 of 11 biological process GO terms) were related to ribosome and protein biogenesis.
We subsequently analyzed the number of genes that were either up- or downregulated with respects to specific GO terms related mitochondrial functions and known azole resistance mechanisms (Figures 2A–D). Of 731 genes associated with the mitochondrion (GO:0005739) or with mitochondrion organization (GO:0007005) 32 were upregulated and 42 downregulated (Figure 2A and Supplementary Table S8). Of 693 genes that were associated with transmembrane transport (GO:0055085) and transmembrane transporter activity (GO:0022857) 52 were upregulated and 42 downregulated (Figure 2B and Supplementary Table S9). Interestingly, only 16 of 693 genes associated with response to stress (GO:0006950) were upregulated and 45 downregulated (Figure 2C and Supplementary Table S10). Of the 30 genes that were annotated for being involved in ergosterol biosynthetic process (GO:0006696) or regulation of ergosterol biosynthetic process (GO:0032443) only two were up- and two downregulated. The two upregulated genes were AFUA_1G04540 (1.5-fold) and AFUA_1G17190 (1.6-fold) which encode the homologs of the Saccharomyces cerevisiae proteins ScMcr1, a mitochondrial NADH-cytochrome b5 reductase involved in ergosterol biosynthesis (YKL150W), and Pcs60, an oxalyl-CoA synthetase (YBR222C), respectively. The two downregulated genes were AFUA_3G10660 (0.6-fold) and AFUA_7G01220 (0.7-fold) which encode the homologs of S. cerevisiae Erg13, the HMG-CoA synthase (YML126C), and Erg9, the squalene synthase (YHR190W) (Figure 2D and Supplementary Table S11). Notably, cyp51A, the drug target of azole antifungals was not annotated for being involved in the ergosterol biosynthetic process (GO:0006696), even though it was 1.6-fold upregulated in the Δdnm1 mgm1tetOn mutant.
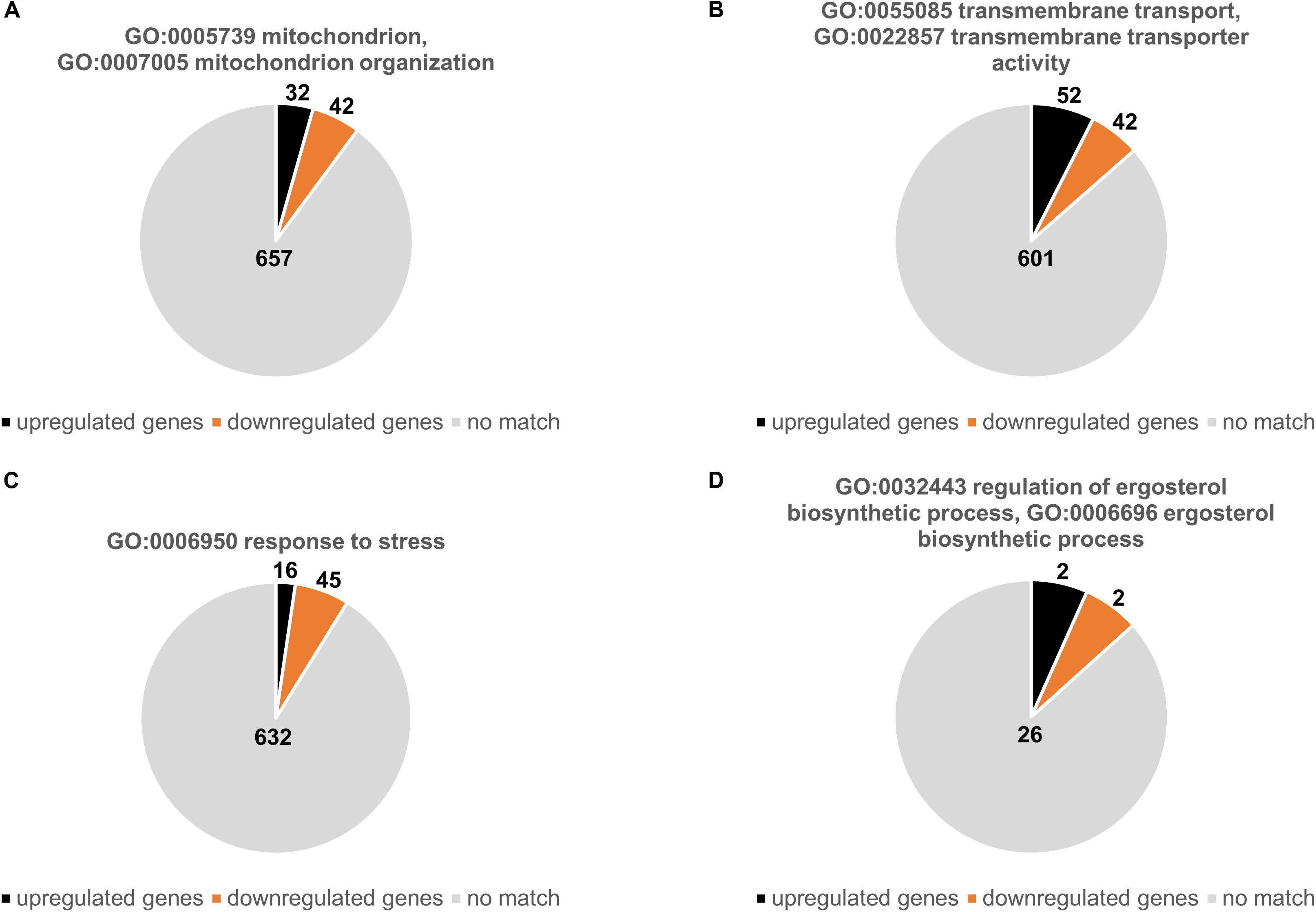
Figure 2. Up- and downregulated genes Δdnm1 mgm1tetOn mutant with a presumed role in mitochondrial biology, transmembrane transport, stress response and ergosterol metabolism. The charts show the number of up- and downregulated genes that belong to the indicated GO terms: (A) mitochondrion (GO:0005739) and mitochondrion organization (GO:0007005); (B) transmembrane transport (GO:0055085) and transmembrane transporter activity (GO:0022857); (C) response to stress (GO:0006950); (D) ergosterol metabolic process (GO:0008204). The absolute number of genes associated with the indicated GO terms is shown in charts.
Mitochondrial dysfunctions in baker’s yeast and in the opportunistic pathogen Candida glabrata cause the induction of a partially conserved pleiotropic drug resistance (PDR) network. This involves the increased expression of multiple efflux pumps (Traven et al., 2001; Tsai et al., 2006; Gulshan et al., 2008; Ferrari et al., 2009, 2011a,b; Shingu-Vazquez and Traven, 2011). We speculated that a similar mechanism exists in A. fumigatus which could explain the increased azole tolerance of the mitochondrial fission/fusion mutant. The genome of A. fumigatus encodes 52 ABC proteins and 211 MFS transporters, based on a Pfam database search for proteins annotated as ABC transporter (PF00005) or MFS (PF07690) (Supplementary Tables S12, S13). Of those, eight ABC proteins (15%) and 21 MFS transporters (10%) were upregulated in the fission/fusion mutant compared to wild type (Figures 3A,B and Supplementary Tables S14, S15). To our knowledge, none of these proteins has been characterized in A. fumigatus to date. A detailed analysis of the upregulated transporters revealed that several may have a role in drug efflux. One of the eight upregulated ABC proteins (encoded by Afu1g16440) belongs to the YEF3/RLi ABC protein family which is not implicated in transport and was therefore not considered as potential transporter. Four of the seven remaining upregulated putative ABC transporters have homologs in C. albicans or S. cerevisiae that were previously implicated in azole resistance (Supplementary Table S14). Similar, of the 21 upregulated MFS transporters nine have homologs in C. albicans or S. cerevisiae that were previously reported to mediate azole resistance (Supplementary Table S15).
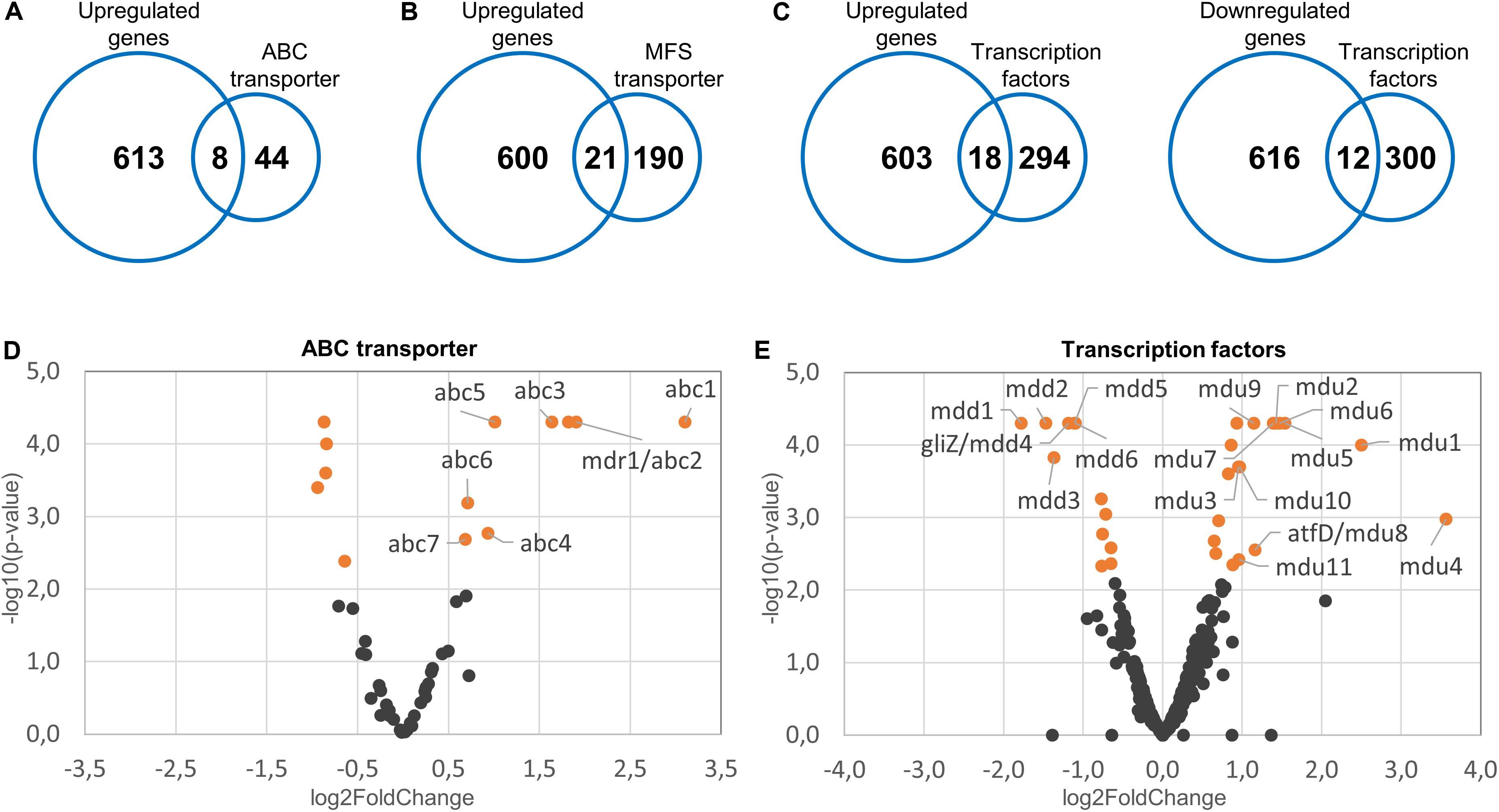
Figure 3. Upregulated efflux pumps and differentially regulated transcription factors in the Δdnm1 mgm1tetOn mutant. Fifty-two ABC transporters, 211 MFS transporters and 312 transcription factors were identified in the reference genome of A. fumigatus. The identified genes were subsequently compared to the list of genes upregulated (n = 612) or downregulated (n = 628) in the Δdnm1 mgm1tetOn mutant. The number of upregulated ABC transporters (A), upregulated MFS transporters (B), and up- or downregulated transcription factors (C) are indicated in the intersections of the Venn diagrams. (D,E) Volcano plots of the expressed ABC transporters (D) and transcription factors (E). The log2 fold changes of the individual genes (x-axis) were plotted against the corresponding adjusted p-values (-log10; y-axis). Differentially expressed genes (p = 0.007) are indicated in orange. Genes that were further analyzed by constructing conditional mutants thereof were labeled accordingly.
Induction of the pleiotropic drug response network in baker’s yeast and C. glabrata depends on a conserved zinc finger transcription factor that was named ScPdr3 or CgPdr1, respectively (Shingu-Vazquez and Traven, 2011). Unfortunately, due to the high number of highly similar transcription factors present in other species, functional orthologs cannot be easily identified with conventional homology searches. The genome of A. fumigatus encodes more than 300 potential transcription factors (GO:0006355; regulation of transcription, DNA-templated; Supplementary Table S16). Of those, 120 exhibit the Pfam domains PF00172 [fungal Zn(2)-Cys(6) binuclear cluster domain] and PF04082 (fungal specific transcription factor domain) which are the only two common domains that can be identified in ScPdr3 and CgPdr1 (Supplementary Table S16).
Interestingly, ScPdr3 as well as CgPdr1 were reported to regulate their own expression (Delahodde et al., 1995; Paul et al., 2011). We speculated that similar transcription factors could be involved in the increased azole tolerance of the fission/fusion mutant and that such functional orthologs would then also autoregulate their own expression. As shown in Figure 3C of the 312 potential transcription factors 18 were upregulated and 12 were downregulated in the fission/fusion mutant. Of these 30 differentially regulated genes, seven harbored the Pfam domain PF00172 in combination with PF04082 which are characteristic for ScPdr3 and CgPdr1 (Supplementary Table S17). Five genes harbored the Pfam domain PF00170 which is characteristic for bZip transcription factors (Supplementary Table S17). Notably, the bZip transcription factor Cap1 of C. albicans was previously shown to increase fluconazole tolerance by upregulating expression of the major multidrug transporter Cdr1 (Prasad et al., 2015).
Our data suggested that the overexpression of efflux pumps could be involved in the azole resistance of the fission/fusion mutant. To further investigate the potential role of efflux pumps we constructed conditional mutants of the differentially regulated ABC transporters (Figure 3D and Supplementary Table S14). AFUA_5G06070 (abc2) was already denominated mdr1 in current genome databases, and the respective name was used for consistency. To this end, a well-established doxycycline-inducible Tet-On promoter system was applied (Helmschrott et al., 2013). Under repressive conditions (no doxycycline), this system facilitated significant repression of genes in A. fumigatus which resulted in phenotypes similar to those of deletion mutants (Helmschrott et al., 2013; Dichtl et al., 2015; Loiko and Wagener, 2017). Upon induction (e.g., 15 μg ml–1 doxycycline) the same promoter enabled overexpression of genes (Helmschrott et al., 2013; Dichtl et al., 2015; Loiko and Wagener, 2017; Fischer et al., 2018). Of each gene the promoter was replaced, essentially as described before (Helmschrott et al., 2013). A technical limitation of this approach is that it cannot be guaranteed that repression and induction of the promoter is sufficient to cause a complete null or overexpression phenotype which may additionally depend on the individual genes. Thus, negative results in this screening do not rule out a role of a tested gene in azole resistance. At least two independent mutant clones were obtained and characterized for each gene. Repression of the Tet-On promoter (no doxycycline) had no significant impact on growth of any of the seven ABC transporter mutants (Figure 4 and Supplementary Figure S2). However, induction of the Tet-On promoter (+ doxycycline) resulted in slightly delayed conidiation compared to wild type, while the overall growth rate of the mycelium was not significantly affected (Figure 4 and Supplementary Figure S2).
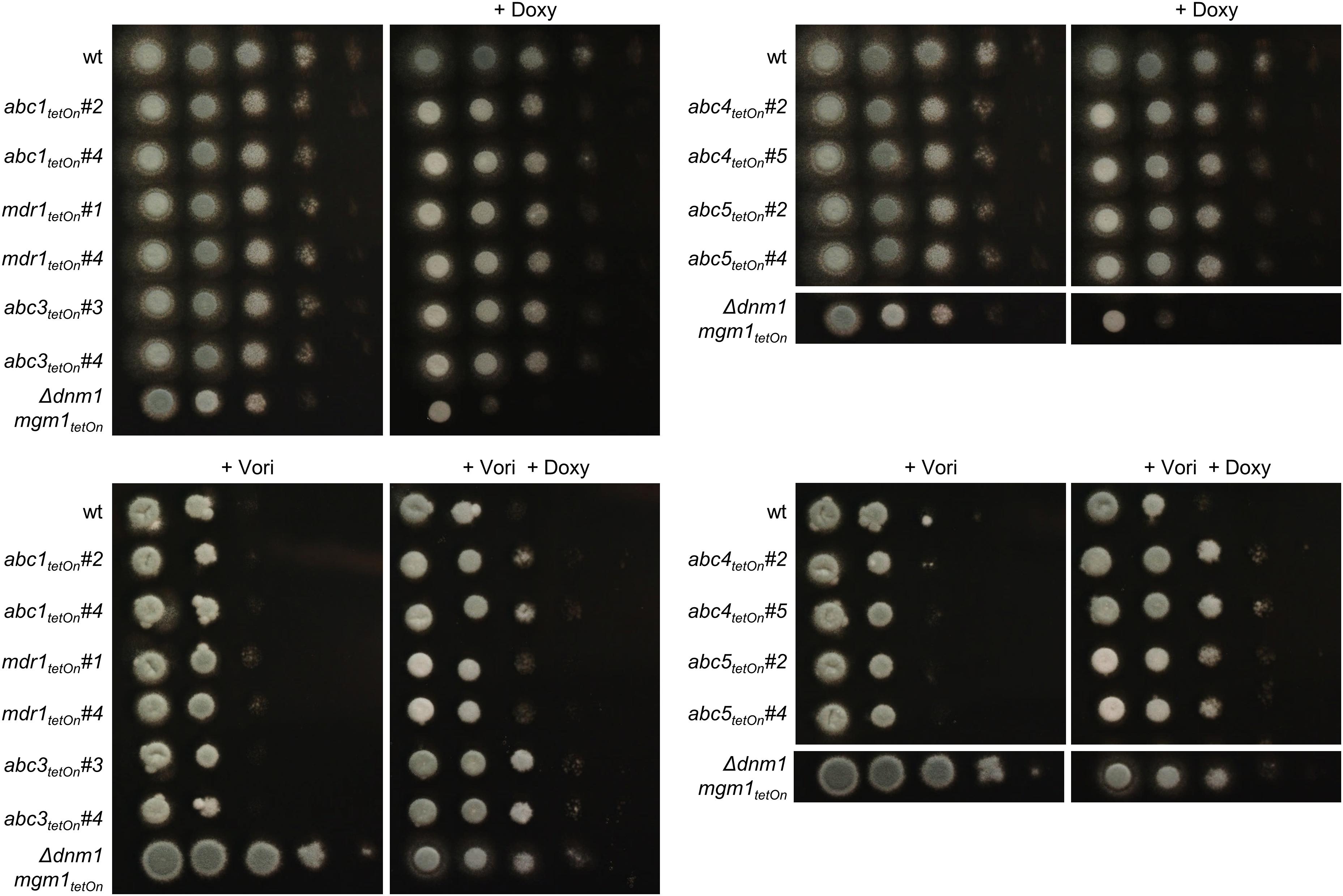
Figure 4. Induction of the conditional abc1tetOn, abc3tetOn, abc4tetOn, and abc5tetOn mutants increase azole tolerance of A. fumigatus. In a series of 10-fold dilutions derived from a starting suspension of 5 × 107 conidia ml– 1 of the indicated strains, aliquots of 3 μl were spotted on Sabouraud agar plates. Two independent conditional mutant clones per ABC transporter are shown. When indicated, medium was supplemented with doxycycline (15 μg ml– 1; + Doxy) to induce the Tet-On promoter, or with voriconazole (0.31 μg ml– 1; + Vori). Representative images were taken after 24 h (without Vori) or 36 h (+ Vori) incubation at 37°C.
To evaluate the impact of the ABC transporters on the azole susceptibility, we performed an initial screening of the obtained mutants with the broth microdilution method (two mutant clones per conditionally regulated gene; Supplementary Table S18). Mutants that appeared more or less susceptible to voriconazole were subsequently confirmed on solid agar. The wild type and conditional fission/fusion mutant Δdnm1 mgm1tetOn served as controls. Under repressed conditions all seven ABC transporter mutants exhibited a voriconazole susceptibility similar to wild type (data not shown). Under induced conditions, five of the seven ABC transporter mutants showed slightly altered minimal inhibitory concentrations (MICs) for voriconazole in the broth microdilution assay. Of those, four demonstrated also an increased voriconazole tolerance on solid agar (Figure 4 and Table 1): abc1tetOn, abc3tetOn, abc4tetOn and abc5tetOn.
Next, we used the same approach to study the role of the differentially regulated transcription factors for the azole susceptibility of A. fumigatus. Similar to the previously reported autoregulation of the PDR networks in S. cerevisiae and C. glabrata (Delahodde et al., 1995; Paul et al., 2011), the transcription factors that mediate azole resistance in A. fumigatus might show positive or negative regulation of their own expression. Conditional mutants of the 17 mostly affected transcription factors (cut-off log2 = 0.95/log2 = −0.95) were constructed by replacing the promoters of the respective genes with the doxycycline-inducible Tet-On promoter (Figure 3E and Supplementary Table S17). The genes were named mitochondrial dynamics upregulated (mdu) or mitochondrial dynamics downregulated (mdd) for transcription factors that are upregulated or downregulated in the mitochondrial fission/fusion mutant, respectively. AFUA_6G09630 (mdd4) was already denominated gliZ and AFUA_6G12150 (mdu8) atfD in current genome databases, and the respective names were used for consistency. Two independent mutant clones were obtained and analyzed for each gene. Of the eleven mdu mutants and six mdd mutants, only the mdd6tetOn mutant presented a remarkable phenotype under repressed conditions that was characterized by abolished conidiation (Figures 5, 6A and Supplementary Figure S3). Under induced conditions, the growth rates of the mdu3tetOn, mdu9tetOn, mdd2tetOn, and gliZtetOn (mdd4tetOn) mutants were moderately and of mdd6tetOn strikingly reduced. The mycelium of the mdd2tetOn mutant exhibited a brownish-yellow color. Besides this, conidiation of mdu2tetOn, mdu3tetOn, mdu5tetOn, mdu6tetOn, mdu7tetOn, mdu9tetOn, and mdd3tetOn were slightly and of mdd2tetOn, gliZtetOn (mdd4tetOn), and mdd6tetOn strongly reduced (Figures 5, 6A and Supplementary Figure S3).
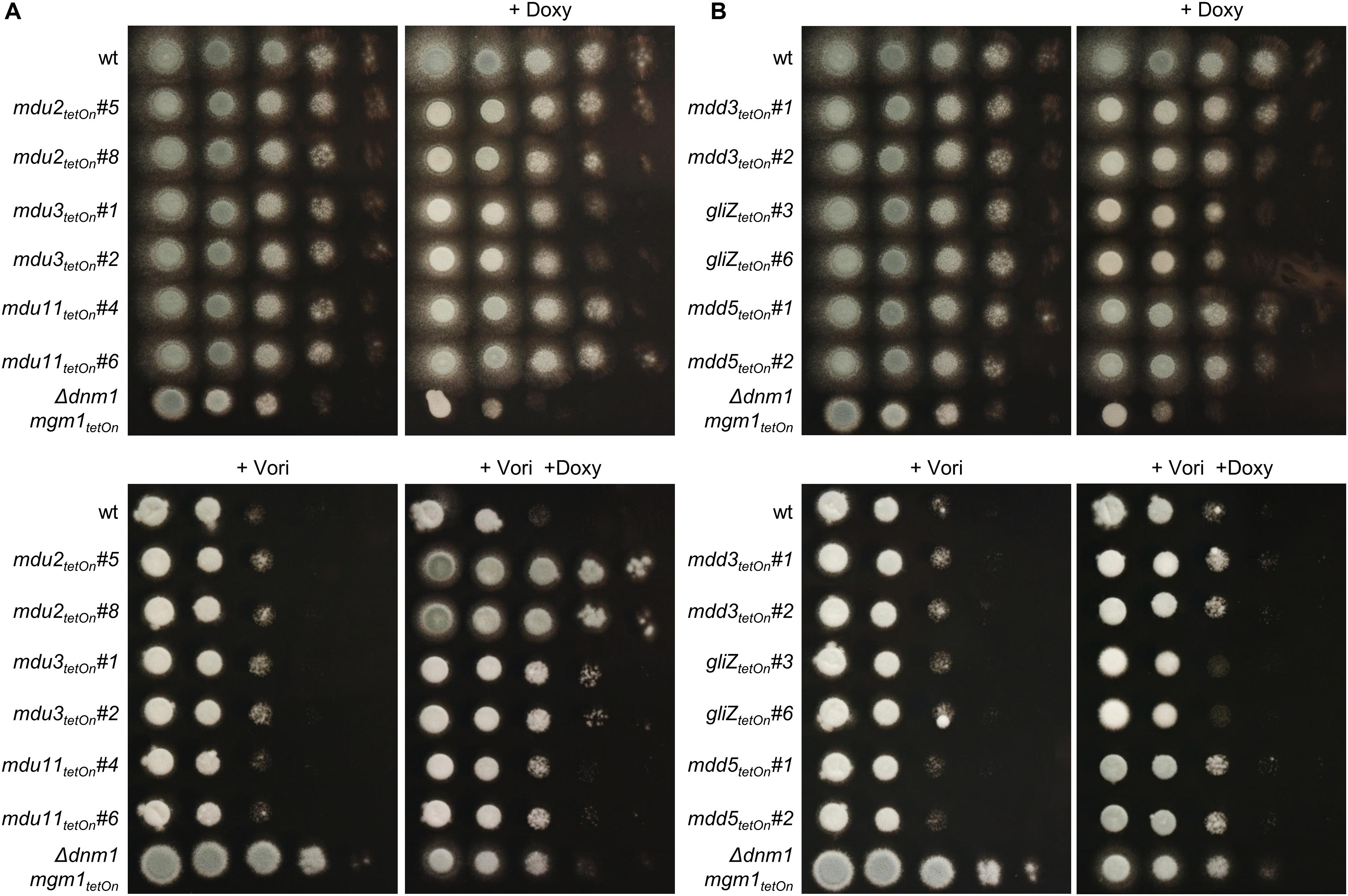
Figure 5. Induction of the conditional mdu2tetOn, mdu3tetOn, mdu11tetOn and of mdd3tetOn and mdd5tetOn mutants increase azole tolerance of A. fumigatus. In a series of 10-fold dilutions derived from a starting suspension of 5 × 107 conidia ml– 1 of the indicated strains, aliquots of 3 μl were spotted on Sabouraud agar plates. Two independent conditional mutant clones of transcription factors that are upregulated (A) or downregulated (B) in the mitochondrial fission/fusion mutant are shown. When indicated, medium was supplemented with doxycycline (15 μg ml– 1; +Doxy) to induce the Tet-On promoter, or with voriconazole (0.41 μg ml– 1; +Vori). Representative images were taken after 24 h (without Vori) or 36 h (+Vori) incubation at 37°C.
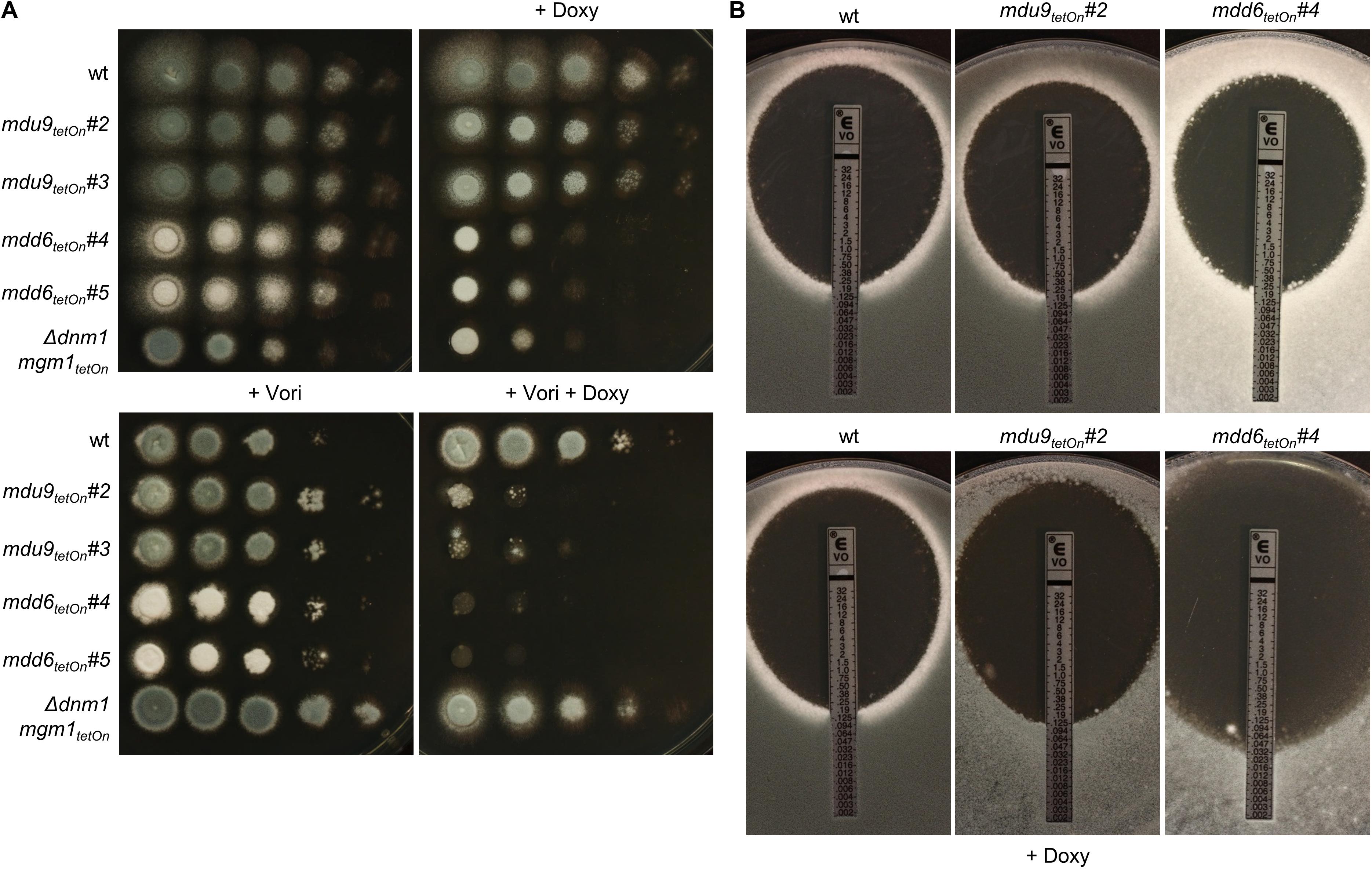
Figure 6. Induction of the conditional mdu9tetOn and mdd6tetOn mutants result in increased azole susceptibility. (A) In a series of 10-fold dilutions derived from a starting suspension of 5 × 107 conidia ml– 1 of the indicated strains, aliquots of 3 μl were spotted on Sabouraud agar plates. Two independent conditional mutant clones per transcription factor are shown. When indicated, medium was supplemented with doxycycline (15 μg ml– 1; +Doxy) to induce the Tet-On promoter, or with voriconazole (0.23 μg ml– 1; + Vori). Representative images were taken after 24 h (without Vori) or 36 h (+Vori) incubation at 37°C. (B) 1 × 106 conidia of the indicated strains were spread on Sabouraud agar plates. When indicated (+ Doxy), medium was supplemented with 15 μg ml– 1 doxycycline. Voriconazole Etest strips were applied and representative images were taken after 48 h incubation at 37°C.
The azole susceptibility of the mutants was then evaluated using the broth microdilution method (two mutant clones per conditionally regulated gene were tested; Supplementary Table S18). Under repressed conditions, the voriconazole MICs of all mdu and mdd mutants were similar to that of wild type. Under induced conditions, four mdu and four mdd mutants demonstrated slightly altered voriconazole MICs. Interestingly, while the MICs of six mutants were increased, two mutants showed slightly reduced MICs. The decreased azole susceptibilities of the three mdu mutants (mdu2tetOn, mdu3tetOn, and mdu11tetOn) and two of the three mdd mutants (mdd3tetOn and mdd5tetOn) under induced conditions were subsequently confirmed on solid agar (Figure 5 and Table 2). Similar, we could confirm the increased azole susceptibilities of mdu9tetOn and mdd6tetOn under induced conditions (Figure 6 and Table 2).
We identified four ABC transporters and six transcription factors that demonstrated the ability to increase the azole resistance of A. fumigatus. To check whether the increased azole tolerance correlates with an increased expression of the respective genes under induced conditions, the inducibility of two representative conditional mutants was analyzed using quantitative reverse transcription PCR (Supplementary Figure S4). We speculated that one of the identified transporters or transcription factors mediates the increased azole tolerance of the Δdnm1 mgm1tetOn mutant, similar to mitochondrial dysfunction-triggered azole resistance via ScPdr3 and CgPdr1 in S. cerevisiae and C. glabrata. We therefore deleted the genes encoding abc1, abc3, and abc4 as well as mdu2, mdu3, mdu11, mdd3, gliZ (mdd4), and mdd5 in the Δdnm1 mgm1tetOn mutant. Despite several attempts to delete abc5 in the Δdnm1 mgm1tetOn, we were not able to obtain any mutant clones. Since the mdr1tetOn (abc2tetOn) mutant showed minimally increased azole tolerance under induced conditions in the initial screen with the broth microdilution method, the respective gene (mdr1/abc2) was also deleted in the fission/fusion mutant. Deletion of these genes did not significantly affect the growth phenotype and azole susceptibility of the Δdnm1 mgm1tetOn mutant (Figure 7). However, in contrast to our expectation neither the deletion of the individual ABC transporters nor deletion of individual transcription factors did affect the azole susceptibility of the mitochondrial fission/fusion mutant (Figure 7).
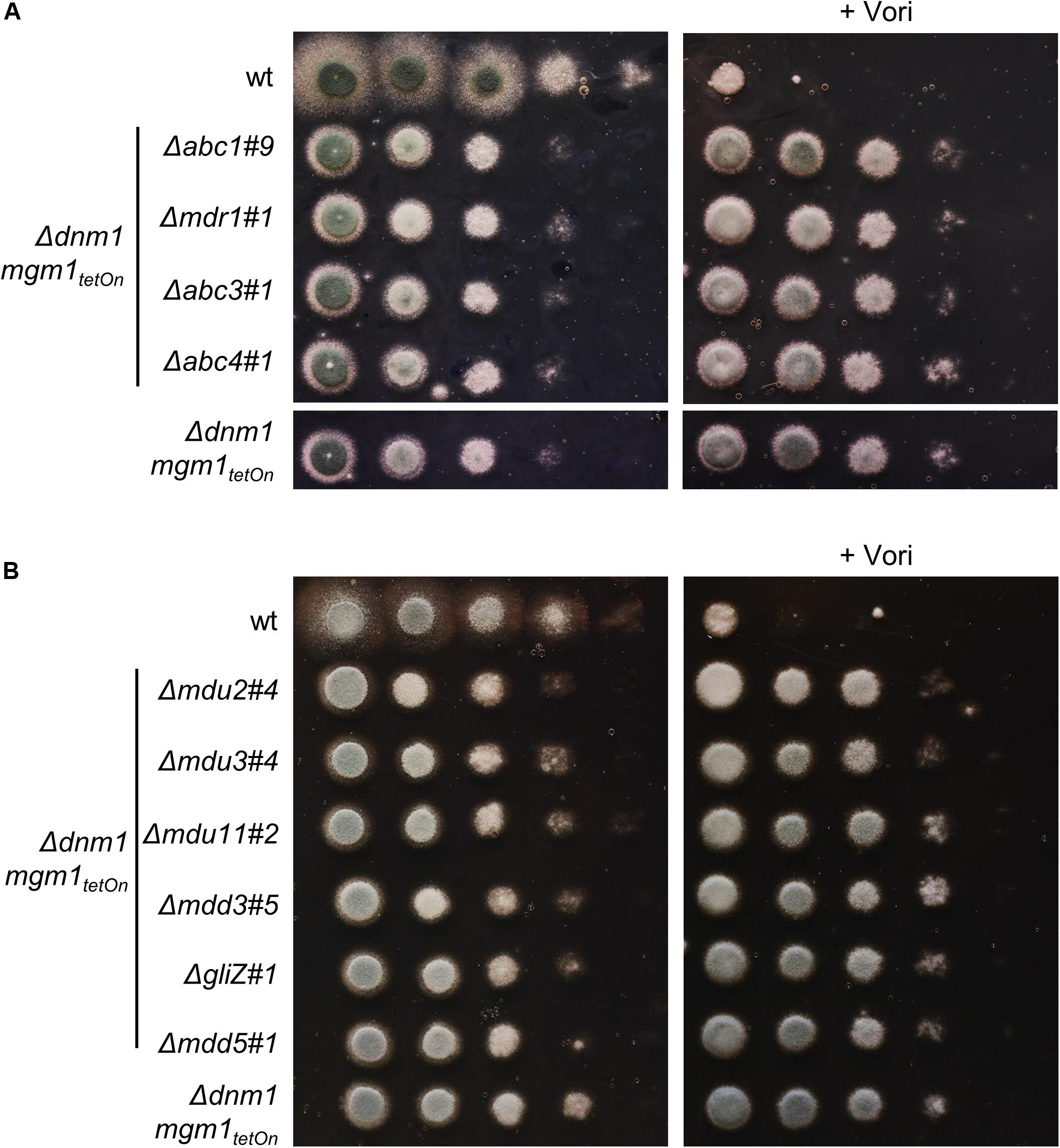
Figure 7. Deletion of genes encoding individual ABC transporters (A; abc1, mdr1 (abc2), abc3 and abc4) or transcription factors (B; mdu2, mdu3, mdu11, mdd3, gliZ (mdd4) and mdd5) in the Δdnm1 mgm1tetOn mutant does not affect azole susceptibility. In a series of 10-fold dilutions derived from a starting suspension of 5 × 107 conidia ml– 1 of the indicated strains, aliquots of 3 μl were spotted onto Sabouraud (SAB) agar plates. When indicated (+ 0.41 μg ml– 1 Vori), medium was supplemented with voriconazole. Plates without voriconazole were incubated for 24 h at 37°C and plates containing voriconazole were incubated for 36 h at 37°C.
Within our study we aimed on identifying mechanisms responsible for the increased azole tolerance of the mitochondrial fission/fusion mutant Δdnm1 mgm1tetOn. To this end, we performed RNA sequencing and analyzed the expression profile of this mutant. This revealed multiple potential explanations for the observed azole tolerance phenotype.
First, the RNA sequencing data indicate an increase in the expression of Cyp51A, the target enzyme of the azole antifungals. Although A. fumigatus expresses a second functionally redundant ortholog, Cyp51B (Mellado et al., 2005), mutations in cyp51A and in its promoter appear to represent the primary CYP51-related resistance mechanisms in this mold species (Perlin et al., 2017). Indeed, increased expression of cyp51A due to a tandem repeat in the promoter region is one of the few known mechanism that mediate azole resistance in A. fumigatus (Perlin et al., 2017). In a previous study, resistance of clinical A. fumigatus isolates was associated with a four to eightfold increase of cyp51A expression (Mellado et al., 2007). Compared to this, the 1,6-fold increase of cyp51A expression in the Δdnm1 mgm1tetOn mutant appears moderate. However, the azole tolerance of the Δdnm1 mgm1tetOn mutant is also moderate (MIC ˜1.27–1.69 μg ml–1, Neubauer et al., 2015) when compared to clinical resistant strains that are defined by a MIC > 2 μg ml–1 for voriconazole. Taken together, this supports a model where the upregulated cyp51A expression could be involved in the increased azole tolerance of the mitochondrial fission/fusion mutant.
A second potential explanation for the increased azole tolerance of the Δdnm1 mgm1tetOn mutant is the upregulation of efflux pumps. In fact, we found several ABC transporters (7) and MFS transporters (21) to be upregulated in the mutant. Overexpression of such transporters are known to cause clinically relevant azole resistance of Candida species (Morschhäuser, 2016; Prasad et al., 2016). Due to the large number of potentially involved MFS transporters we focused on the seven ABC transporters in this work. With the exception of Mdr1 (Abc2; Fraczek et al., 2013), none of them was previously characterized or linked to azole susceptibility in A. fumigatus. Our analysis of the seven conditional ABC transporters mutants under repressed conditions suggests that none of them significantly contributes to the azole tolerance of wild type. In case of Mdr1 (Abc2) this could be interpreted as being contradictory to the results of a previous study. Fraczek and colleagues reported that deletion of mdr1 (abc2; AFUA_5G06070) causes a twofold lower MIC for azoles (Fraczek et al., 2013). Considering this weak impact of mdr1 (abc2) deletion on the MIC (twofold decrease), we speculate that such a phenotype could potentially be masked in our mutant due to a low basal expression of the Tet-On promoter under repressed conditions (oliC-tetOn; Helmschrott et al., 2013). The induction of the conditional ABC transporter mutants revealed the general capability of four of the seven ABC transporters to increase the azole tolerance of the mold. Consequently, upregulation of these genes could contribute or even cause the increased azole tolerance of the mitochondrial fission/fusion mutant (Table 1).
Third, we analyzed the capability of 17 most up- or down-regulated transcription factors in the Δdnm1 mgm1tetOn mutant to modulate the azole tolerance of A. fumigatus. To our knowledge, only gliZ (mdd4; AFUA_6G09630) and atfD (mdu8; AFUA_6G12150) have been studied in A. fumigatus (Bok et al., 2006; Pereira Silva et al., 2017). gliZ was identified to be a major regulator of the gliotoxin biosynthesis gene cluster (Bok et al., 2006). An atfD deletion mutant was constructed and characterized by Silva and colleagues very recently (Pereira Silva et al., 2017). However, no specific function could be assigned to atfD, except for a moderate role in caspofungin susceptibility (Pereira Silva et al., 2017). Similar to the conditional ABC transporter mutants, none of the conditional mdd and mdu mutants demonstrated an altered azole susceptibility under repressed conditions. However, induction of the mutants revealed the capability of seven of the 17 transcription factors to affect the azole susceptibility of A. fumigatus (Table 2). Interestingly, five of the seven transcription factors increased and two decreased the azole tolerance upon induction of the Tet-On promoter.
Our transcriptional and experimental data argue for a model where mitochondrial dysfunction triggers the activations of a potentially conserved drug resistance network in A. fumigatus (Figure 8). Importantly, mitochondrial dysfunctions other than defects in mitochondrial dynamics were previously shown to also increase the azole tolerance in A. fumigatus (Bromley et al., 2016; Kroll et al., 2016; Geißel et al., 2018). A similar response, mediated by a conserved transcription factor which triggers a pleiotropic drug response network, was previously described for baker’s yeast and C. glabrata (Traven et al., 2001; Tsai et al., 2006; Gulshan et al., 2008; Ferrari et al., 2009, 2011a,b; Shingu-Vazquez and Traven, 2011; Peng et al., 2012). We therefore tested whether one of the multiple identified ABC transporters or transcription factors with the ability to modulate the azole susceptibility of A. fumigatus is specifically responsible for the increased azole tolerance of the mitochondrial fission/fusion mutant. To this end, triple mutants were constructed by deleting each candidate gene, with the exception of abc5, in the Δdnm1 mgm1tetOn mutant. However, deletion of the individual genes did not significantly affect the increased azole tolerance of the mitochondrial fission/fusion mutant. This indicates that the characterized genes alone are not essential for the observed phenotype. Functional redundancy of the identified factors could explain the absence of an apparent effect on the azole tolerance of the Δdnm1 mgm1tetOn mutant. However, to test this hypothesis multiple candidate genes would have to be deleted in the fission/fusion mutant at the same time.
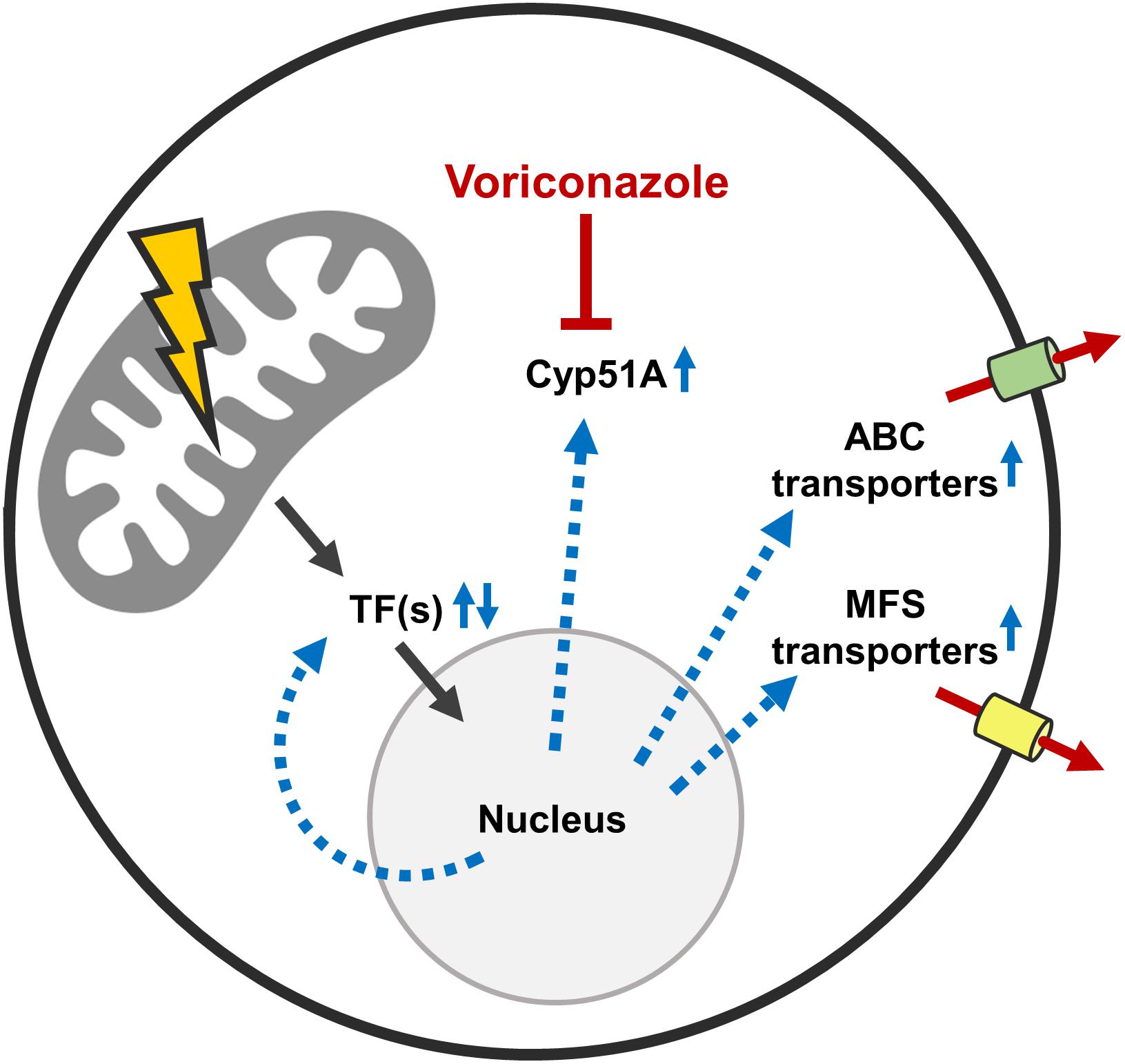
Figure 8. Possible mechanisms that mediate the increased azole tolerance of mitochondrial fission/fusion mutants. Mitochondrial dysfunction triggers altered expression of multiple azole tolerance-affecting genes (black arrows). Among the upregulated genes is the lanosterol 14α-demethylase Cyp51A which is the target enzyme that is inhibited by azole antifungals. Increased Cyp51A expression is a well-characterized azole resistance mechanism. Besides this, multiple transcription factors are up- and downregulated in the Δdnm1 mgm1tetOn mutant. At least five of these transcription factors are able to increase and two are able to decrease the azole susceptibility of A. fumigatus. In addition, multiple ABC and MFS transporters are upregulated in the Δdnm1 mgm1tetOn mutant. At least four of these ABC transporters are able to increase the azole tolerance of the mold. Deletion of individual identified transcription factors and ABC transporters does not diminish the increased azole tolerance of the Δdnm1 mgm1tetOn mutant. This suggests that the identified factors are functionally redundant or that other mechanisms are responsible for the azole tolerance of the mitochondrial fission/fusion mutant.
The datasets generated for this study can be found in the https://www.ncbi.nlm.nih.gov/bioproject/PRJNA627614.
JW conceived the study. LS, BG, and JW designed the experiments. LS, BG, and RM performed the experiments. JW and LS wrote the manuscript. All authors analyzed and discussed the data.
This work was in part supported by the Förderprogramm für Forschung und Lehre (FöFoLe) of the Medical Faculty of the Ludwig-Maximilians-Universität München and the German Research Foundation (DFG - WA 3016/2-1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01017/full#supplementary-material
Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Cech, M., et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Basenko, E. Y., Pulman, J. A., Shanmugasundram, A., Harb, O. S., Crouch, K., Starns, D., et al. (2018). FungiDB: an integrated bioinformatic resource for fungi and oomycetes. J. Fungi 4:39. doi: 10.3390/jof4010039
Bok, J. W., Chung, D., Balajee, S. A., Marr, K. A., Andes, D., Nielsen, K. F., et al. (2006). GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect. Immun. 74, 6761–6768. doi: 10.1128/IAI.00780-786
Bromley, M., Johns, A., Davies, E., Fraczek, M., Mabey Gilsenan, J., Kurbatova, N., et al. (2016). Mitochondrial complex I is a global regulator of secondary metabolism, virulence and azole sensitivity in fungi. PLoS One 11:e0158724. doi: 10.1371/journal.pone.0158724
Brown, G. D., Denning, D. W., Gow, N. A. R., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv13. doi: 10.1126/scitranslmed.3004404
Cerqueira, G. C., Arnaud, M. B., Inglis, D. O., Skrzypek, M. S., Binkley, G., Simison, M., et al. (2014). The Aspergillus genome database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 42, D705–D710. doi: 10.1093/nar/gkt1029
Costa, C., Dias, P. J., Sá-Correia, I., and Teixeira, M. C. (2014). MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Physiol. 5:197. doi: 10.3389/fphys.2014.00197
Cowen, L. E., Sanglard, D., Howard, S. J., Rogers, P. D., and Perlin, D. S. (2015). Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 5:a019752. doi: 10.1101/cshperspect.a019752
Delahodde, A., Delaveau, T., and Jacq, C. (1995). Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol. Cell. Biol. 15, 4043–4051. doi: 10.1128/mcb.15.8.4043
Dichtl, K., Samantaray, S., Aimanianda, V., Zhu, Z., Prévost, M.-C., Latgé, J.-P., et al. (2015). Aspergillus fumigatus devoid of cell wall β-1,3-glucan is viable, massively sheds galactomannan and is killed by septum formation inhibitors. Mol. Microbiol. 95, 458–471. doi: 10.1111/mmi.12877
Dos Santos, S. C., Teixeira, M. C., Dias, P. J., and Sá-Correia, I. (2014). MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: understanding their physiological function through post-genomic approaches. Front. Physiol. 5:180. doi: 10.3389/fphys.2014.00180
Dudakova, A., Spiess, B., Tangwattanachuleeporn, M., Sasse, C., Buchheidt, D., Weig, M., et al. (2017). Molecular tools for the detection and deduction of azole antifungal drug resistance phenotypes in Aspergillus species. Clin. Microbiol. Rev. 30, 1065–1091. doi: 10.1128/CMR.00095-16
Engel, S. R., Dietrich, F. S., Fisk, D. G., Binkley, G., Balakrishnan, R., Costanzo, M. C., et al. (2014). The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 4, 389–398. doi: 10.1534/g3.113.008995
Ferrari, S., Ischer, F., Calabrese, D., Posteraro, B., Sanguinetti, M., Fadda, G., et al. (2009). Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 5:e1000268. doi: 10.1371/journal.ppat.1000268
Ferrari, S., Sanguinetti, M., De Bernardis, F., Torelli, R., Posteraro, B., Vandeputte, P., et al. (2011a). Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice. Antimicrob. Agents Chemother. 55, 1852–1860. doi: 10.1128/AAC.01271-1210
Ferrari, S., Sanguinetti, M., Torelli, R., Posteraro, B., and Sanglard, D. (2011b). Contribution of CgPDR1-regulated genes in enhanced virulence of azole-resistant Candida glabrata. PLoS One 6:e17589. doi: 10.1371/journal.pone.0017589
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Fischer, J., Müller, S. Y., Netzker, T., Jäger, N., Gacek-Matthews, A., Scherlach, K., et al. (2018). Chromatin mapping identifies BasR, a key regulator of bacteria-triggered production of fungal secondary metabolites. eLife 7:40969. doi: 10.7554/eLife.40969
Fraczek, M. G., Bromley, M., Buied, A., Moore, C. B., Rajendran, R., Rautemaa, R., et al. (2013). The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 68, 1486–1496. doi: 10.1093/jac/dkt075
Geißel, B., Loiko, V., Klugherz, I., Zhu, Z., Wagener, N., Kurzai, O., et al. (2018). Azole-induced cell wall carbohydrate patches kill Aspergillus fumigatus. Nat. Commun. 9:3098. doi: 10.1038/s41467-018-05497-5497
Gulshan, K., Schmidt, J. A., Shahi, P., and Moye-Rowley, W. S. (2008). Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression. Mol. Cell. Biol. 28, 5851–5864. doi: 10.1128/MCB.00405-408
Helmschrott, C., Sasse, A., Samantaray, S., Krappmann, S., and Wagener, J. (2013). Upgrading fungal gene expression on demand: improved systems for doxycycline-dependent silencing in Aspergillus fumigatus. Appl. Environ. Microbiol. 79, 1751–1754. doi: 10.1128/AEM.03626-3612
Hewitt, S. K., Foster, D. S., Dyer, P. S., and Avery, S. V. (2016). Phenotypic heterogeneity in fungi: importance and methodology. Fungal Biol. Rev. 30, 176–184.
Hill, T. W., and Kafer, E. (2001). Improved protocols for Aspergillus salt stock solutions. Fungal Genet. Newsl. 48, 20–21.
Klein, C., Kuchler, K., and Valachovic, M. (2011). ABC proteins in yeast and fungal pathogens. Essays Biochem. 50, 101–119. doi: 10.1042/bse0500101
Kosmidis, C., and Denning, D. W. (2015). The clinical spectrum of pulmonary aspergillosis. Thorax 70, 270–277. doi: 10.1136/thoraxjnl-2014-206291
Kousha, M., Tadi, R., and Soubani, A. O. (2011). Pulmonary aspergillosis: a clinical review. Eur. Respir. Rev. 20, 156–174. doi: 10.1183/09059180.00001011
Krappmann, S., Sasse, C., and Braus, G. H. (2006). Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot. Cell 5, 212–215. doi: 10.1128/EC.5.1.212-215.2006
Kroll, K., Shekhova, E., Mattern, D. J., Thywissen, A., Jacobsen, I. D., Strassburger, M., et al. (2016). The hypoxia-induced dehydrogenase HorA is required for coenzyme Q10 biosynthesis, azole sensitivity and virulence of Aspergillus fumigatus. Mol. Microbiol. 101, 92–108. doi: 10.1111/mmi.13377
Lestrade, P. P., Bentvelsen, R. G., Schauwvlieghe, A. F. A. D., Schalekamp, S., van der Velden, W. J. F. M., Kuiper, E. J., et al. (2019). Voriconazole resistance and mortality in invasive Aspergillosis: a multicenter retrospective cohort study. Clin. Infect. Dis. 68, 1463–1471. doi: 10.1093/cid/ciy859
Loiko, V., and Wagener, J. (2017). The paradoxical effect of echinocandins in Aspergillus fumigatus relies on recovery of the β-1,3-Glucan synthase Fks1. Antimicrob. Agents Chemother. 61:1616. doi: 10.1128/AAC.01690-1616
Mellado, E., Garcia-Effron, G., Alcázar-Fuoli, L., Melchers, W. J. G., Verweij, P. E., Cuenca-Estrella, M., et al. (2007). A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51, 1897–1904. doi: 10.1128/AAC.01092-1096
Mellado, E., Garcia-Effron, G., Buitrago, M. J., Alcazar-Fuoli, L., Cuenca-Estrella, M., and Rodriguez-Tudela, J. L. (2005). Targeted gene disruption of the 14-alpha sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob. Agents Chemother. 49, 2536–2538. doi: 10.1128/AAC.49.6.2536-2538.2005
Morschhäuser, J. (2016). The development of fluconazole resistance in Candida albicans - an example of microevolution of a fungal pathogen. J. Microbiol. 54, 192–201. doi: 10.1007/s12275-016-5628-5624
Moye-Rowley, W. S. (2015). Multiple mechanisms contribute to the development of clinically significant azole resistance in Aspergillus fumigatus. Front. Microbiol. 6:70. doi: 10.3389/fmicb.2015.00070
Neubauer, M., Zhu, Z., Penka, M., Helmschrott, C., Wagener, N., and Wagener, J. (2015). Mitochondrial dynamics in the pathogenic mold Aspergillus fumigatus: therapeutic and evolutionary implications. Mol. Microbiol. 98, 930–945. doi: 10.1111/mmi.13167
Patterson, T. F., Thompson, G. R., Denning, D. W., Fishman, J. A., Hadley, S., Herbrecht, R., et al. (2016). Executive summary: practice guidelines for the diagnosis and management of Aspergillosis: 2016 update by the infectious diseases society of america. Clin. Infect. Dis. 63, 433–442. doi: 10.1093/cid/ciw444
Paul, S., Schmidt, J. A., and Moye-Rowley, W. S. (2011). Regulation of the CgPdr1 transcription factor from the pathogen Candida glabrata. Eukary. Cell 10, 187–197. doi: 10.1128/EC.00277-210
Peng, Y., Dong, D., Jiang, C., Yu, B., Wang, X., and Ji, Y. (2012). Relationship between respiration deficiency and azole resistance in clinical Candida glabrata. FEMS Yeast Res. 12, 719–727. doi: 10.1111/j.1567-1364.2012.00821.x
Pereira Silva, L., Alves de Castro, P., Dos Reis, T. F., Paziani, M. H., Von Zeska, Kress, M. R., et al. (2017). Genome-wide transcriptome analysis of Aspergillus fumigatus exposed to osmotic stress reveals regulators of osmotic and cell wall stresses that are SakAHOG1 and MpkC dependent. Cell Microbiol. 19:12681. doi: 10.1111/cmi.12681
Perlin, D. S., Rautemaa-Richardson, R., and Alastruey-Izquierdo, A. (2017). The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect. Dis. 17, e383–e392. doi: 10.1016/S1473-3099(17)30316-X
Pernas, L., and Scorrano, L. (2016). Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 78, 505–531. doi: 10.1146/annurev-physiol-021115-105011
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Prasad, R., Banerjee, A., Khandelwal, N. K., and Dhamgaye, S. (2015). The ABCs of Candida albicans multidrug transporter Cdr1. Eukary. Cell 14, 1154–1164. doi: 10.1128/EC.00137-115
Prasad, R., Rawal, M. K., and Shah, A. H. (2016). Candida efflux atpases and antiporters in clinical drug resistance. Adv. Exp. Med. Biol. 892, 351–376. doi: 10.1007/978-3-319-25304-6_15
Shingu-Vazquez, M., and Traven, A. (2011). Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukary. Cell 10, 1376–1383. doi: 10.1128/EC.05184-5111
Skrzypek, M. S., Binkley, J., Binkley, G., Miyasato, S. R., Simison, M., and Sherlock, G. (2017). The candida genome database (CGD): incorporation of assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 45, D592–D596. doi: 10.1093/nar/gkw924
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Traven, A., Wong, J. M., Xu, D., Sopta, M., and Ingles, C. J. (2001). Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. J. Biol. Chem. 276, 4020–4027. doi: 10.1074/jbc.M006807200
Tsai, H.-F., Krol, A. A., Sarti, K. E., and Bennett, J. E. (2006). Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50, 1384–1392. doi: 10.1128/AAC.50.4.1384-1392.2006
van der Linden, J. W. M., Arendrup, M. C., Warris, A., Lagrou, K., Pelloux, H., Hauser, P. M., et al. (2015). Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus. Emerg. Infect. Dis. 21, 1041–1044. doi: 10.3201/eid2106.140717
Wagener, J. (2016). Regulation of mitochondrial inner membrane fusion: divergent evolution with similar solutions? Curr. Genet. 62, 291–294. doi: 10.1007/s00294-015-0542-546
Wagener, J., Echtenacher, B., Rohde, M., Kotz, A., Krappmann, S., Heesemann, J., et al. (2008). The putative alpha-1,2-mannosyltransferase AfMnt1 of the opportunistic fungal pathogen Aspergillus fumigatus is required for cell wall stability and full virulence. Eukary. Cell 7, 1661–1673. doi: 10.1128/EC.00221-228
Keywords: Aspergillus fumigatus, mitochondrial dynamics, azole resistance, efflux pumps, transcription factors
Citation: Sturm L, Geißel B, Martin R and Wagener J (2020) Differentially Regulated Transcription Factors and ABC Transporters in a Mitochondrial Dynamics Mutant Can Alter Azole Susceptibility of Aspergillus fumigatus. Front. Microbiol. 11:1017. doi: 10.3389/fmicb.2020.01017
Received: 08 December 2019; Accepted: 24 April 2020;
Published: 26 May 2020.
Edited by:
Miguel Cacho Teixeira, University of Lisbon, PortugalReviewed by:
Scott Moye-Rowley, The University of Iowa, United StatesCopyright © 2020 Sturm, Geißel, Martin and Wagener. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johannes Wagener, ai53YWdlbmVyQGh5Z2llbmUudW5pLXd1ZXJ6YnVyZy5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.