- 1State Key Laboratory of Materials-Oriented Chemical Engineering, Nanjing Tech University, Nanjing, China
- 2College of Food Science and Light Industry, Nanjing Tech University, Nanjing, China
- 3College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing, China
Due to the adverse effect on the environment caused by excessive use of chemical fertilizers, the development of sustainable agriculture attracts a growing demand of biological based fertilizers composed of living microorganisms. In this study, an Actinobacteria Streptomyces lydicus M01 was isolated from the rhizosphere soil of Pyrus calleryana. This strain effectively promoted the plant growth and suppressed a foliar disease caused by Alternaria alternata on cucumbers. S. lydicus M01 exhibited growth promoting characteristics such as phosphate solubilization, IAA secretion, siderophore and ACC deaminase production. Through Illumina sequencing of the 16S rRNA gene and ITS gene of the soil microbes, we found that the application of S. lydicus M01 altered the composition of the microbial community by promoting beneficial groups, including bacteria genera Pseudarthrobacter, Sphingomonas, Rhodanobacter, and Pseudomonas, fungi genera Fusicolla, Humicola, Solicoccozyma, and Paraphaeosphaeria. Most of these bacteria and eukaryotes exhibit positive effects on growth promotion, such as nutrient accumulation, auxin secretion, abiotic stress alleviation, biological control, or bioremediation. Furthermore, studies on the reactive oxygen species (ROS) level and antioxidants of cucumber leaves revealed that S. lydicus M01 treatment reduced the ROS accumulation and increased the activities of antioxidases related with ROS scavenging, which indicated an enhanced disease resistance of cucumbers under biotic stress. Thus, our results suggest that the application of S. lydicus M01 can systemically affect plant microbiome interactions and represent a promising sustainable solution to improve agricultural production instead of chemical fertilizers.
Introduction
Chemically synthesized fertilizers composed of nitrogen, phosphorus, and potassium are being used in large amounts to increase the output in a variety of crop plants. At the same time, their excess utilizations adversely affect the environment, causing pollution to soil, water and air (Galloway et al., 2008; Youssef and Eissa, 2014). Thus, sustainable agriculture attracts a growing demand of bio-based fertilizers composed of living microorganisms alternative to the agro-chemicals. Plants and rhizosphere harbor a variety of microbial cells, and the plant-associated microbes can be classified into beneficial, deleterious and neutral groups on the basis of their effects on plant growth (Beneduzi et al., 2012). Beneficial bacteria that promote plant growth are usually referred to as plant growth-promoting rhizobacteria (PGPR) (Ahmad et al., 2008; Bhattacharyya and Jha, 2012).
Plant growth-promoting rhizobacteria affect plant growth in direct and indirect ways. The direct promotion of plant growth by PGPR entails either providing the plant with a compound that is synthesized by the bacterium, for example, siderophore and phytohormones such as auxin, cytokinin, and gibberellins, or facilitating the uptake of certain nutrients from the environment such as phosphate solubilization, nitrogen fixation, and iron chelation (Glick, 1995; Olanrewaju et al., 2017). The indirect promotion of plant growth occurs when PGPR decrease the detrimental effects of phytopathogenic organisms or induced systemic resistance (ISR) (Raupach et al., 1996; Pieterse et al., 2014).
Streptomyces constitute a major clade of the phylum Actinobacteria, these filamentous prokaryotes are ubiquitous in soils and are commonly found in the rhizosphere or inside plant roots. Streptomyces are known to rich in secondary metabolites, they produce over two-thirds of the clinically useful antibiotics of natural origin and are highlighted in medical industry (Watve et al., 2001; Bibb, 2013). The applications of Streptomyces in agriculture are mainly as biocontrol agents, several biocontrol products from this genus are already being marketed and the discovery of natural products is still ongoing (Yuan and Crawford, 1995; Law et al., 2017). However, as a PGPR, the plant growth promotion effects of Streptomyces have not been adequately studied. Some study reported that Streptomyces promote plant growth by the producing indole-3-acetic acid (IAA), siderophores and 1-aminocyclopropane-1-carboxylate deaminase (ACCd) to reduce stress in plants (El-Tarabily, 2008; Rungin et al., 2012; Sadeghi et al., 2012). Streptomyces reside in rhizosphere and interact with other microorganisms in the microbial community, but little is known on their effects in soil microbial community (Wu et al., 2019). Cucumber (Cucumis sativus L.) is one of the most common vegetables worldwide, many studies reported that strains such as Bacillus subtilis, Bacillus amyloliquefaciens, and Pseudomonas spp. affect soil microbial community in the cucumber rhizosphere(Yamamoto-Tamura et al., 2011; Li et al., 2016; Qin et al., 2017), however, as a promising sources of biocontrol agents, how Streptomyces affect soil microbial community in cucumber rhizosphere remains unknown.
Leaf blight caused by the fungus Alternaria alternata is a common foliar disease in a variety of fruits and vegetables including pears, cucumber, and watermelon. The pathogen also develops during the cold storage of fruits, becoming visible during the marketing period thereby causing large losses (Timmer et al., 2003; Wang et al., 2008). In this study, an Actinobacteria isolated from the rhizosphere soil of Callery pear (Pyrus calleryana) in Nanjing Laoshan Forest exhibited strong inhibition effects against A. alternata. Based on 16S rRNA gene and phylogenetic analysis, the strain was identified as S. lydicus M01. We found that this strain significantly promote the growth of cucumbers without obvious plant pathogen present. The growth promoting mechanisms were evaluated from the perspective of the phytohormone production of S. lydicus M01 and its influence on the microbial community in cucumber rhizosphere. In addition, the alleviation of foliar disease caused by A. alternata on cucumbers was also evaluated, the reduction of reactive oxygen species (ROS) accumulation and increased antioxidase activities in cucumber leaves indicated indirect effects of S. lydicus M01 on plant growth promotion and disease suppression. To our knowledge, this is the first report of using Streptomyces to treat a foliar disease on cucumbers and its effects in cucumber rhizosphere microbial community.
Materials and Methods
Isolation and Identification of Streptomyces Strain
Soil samples were collected from the rhizosphere of Callery pear (P. calleryana) in a hillside located in Laoshan Forest, Nanjing, China (32°11′N 118°62′E). For preparation of the soil suspension sample, 5 g of soil was mixed with 45 ml sterile distilled water in a sterile bottle and shaken for 30 min. Then, 200 μl of the suspension was spread on PDA medium supplemented with 20 mg/l nalidixic acid and 50 mg/l cycloheximide. For preliminary classification of bacterial population, morphological features such as colony pigmentation were used. After incubation at 28°C for 21 days, colonies with suspected actinomycetes morphology were selected and further purified on International Streptomyces Project (ISP) 2 medium and stored as glycerol stock (20%, v/v) at −80°C.
The genomic DNA of M01 was extracted from cells grown in potato dextrose broth (PDB) medium at 28°C for 24 h using a bacterial genomic DNA extraction kit (Takara) according to standard protocol. The universal bacterial primers 27F and 1492R (27F: 5′-AGAGTTTGATCCTGGCTCAG-3′; 1492R: 5′-GGTTACCTTGTTACGACTT-3′) were used to amplify the 16S rRNA gene. The PCR reaction volume was 50 μl, which contains 25 μl Taq DNA Polymerase and reaction buffer mix, 1 μl primer forward (10 μM), 1 μl primer reverse (10 μM), 1 μl genomic DNA template and 22 μl nuclease free water. The reaction conditions were 95°C for 5 min, then 30 cycles of 95°C for 30 s and 55°C for 30 s followed by 72°C for 90 s. A final step was performed at 72°C for 5 min. The PCR products were separated and sequenced by Genscript (Nanjing). The assembled 16S rRNA sequences were subjected to a NCBI Nucleotide BLAST. The neighbor-joining tree of the 16S rRNA was generated using MEGA ver. 7 (Saitou and Nei, 1987). Parameters for developing neighbor-joining tree included the use of the Poisson substitution model and a pairwise deletion method to treat gaps in the program.
Antagonistic Effects of Streptomyces lydicus M01 on Pathogens
The antifungal activity of M01 against Leaf blight fungus A. alternata was evaluated using Petri dish assays. Briefly, S. lydicus M01 was cultured on ISP 2 plate at 28°C for 14 days. The plant pathogen A. alternata was cultured on PDA plates at 28°C for 4 days. A square inoculum of A. alternata was using sterile scoop and placed on a new PDA plate. At the same time, S. lydicus M01 was restreaked on the four sides around the square inoculum with a distance of 2 cm. Plates inoculated with A. alternata in the absence of M01 served as control experiments. All samples were incubated at 28°C for 3 days.
Determination of Siderophore Production, Phosphate Solubilization and Indole-3-Acetic Acid
Chrome-Azurol S (CAS) agar was used to determine siderophore production qualitatively (Alexander and Zuberer, 1991). Paper disc soaked with the strain culture was placed on the CAS agar and incubated for 3 days. The change of the mixture color around the colonies after incubation indicated the presence of siderophores.
Streptomyces lydicus M01 was inoculated on agars containing precipitated tricalcium phosphate to determine phosphate solubilization according to Rao (1999). After 24 h incubation, presence of clear zones around the M01 colonies was used as evidence for positive phosphate solubilization.
Streptomyces lydicus M01 grown on ISP 2 agar was inoculated into PDB and incubated at 28°C for 5 days with shaking at 200 rpm, mycelia were removed by filtration. The IAA content was determined based on the method described previously (Patten and Glick, 2002). One milliliter supernatant of the S. lydicus M01 culture was mixed with 4 ml of Salkowski’s reagent vigorously and incubated at room temperature for 25 min absorbance at 535 nm. The concentration of IAA was determined by absorbance at 535 nm and comparison with standard curve.
Determination of ACC Deaminase Activity
Streptomyces lydicus M01 grown on ISP 2 agar was streaked on minimal medium agar free of nitrogen (MM) supplemented with either 3 mM ACC or 2 g/L–1 (NH4)2SO4. The agar was incubated at 28°C in the dark. Growth on MM agar supplemented with ACC indicates ACC deaminase activity (Jaemsaeng et al., 2018).
Pot Experimental Design and Rhizosphere Soil Sampling
The S. lydicus M01 culture was grown in PDB on a rotary shaker (200 rpm) for 4 days at 28°C. The cell suspension (2 × 108 CFU/ml) was used as inoculum for the pot experiment.
Cucumber Seed Shaoshi No. 1 was purchased from the Qingxian Aisen Vegetable Seed Development Center. The seeds were sown in plastic pots contained a mixed substrate of vermiculite, peat soil, and perlite at a ratio of 1:1:0.5 (v:v:v). Then 50, 100, and 150 ml of the S. lydicus M01 cell suspensions were applied as a root drench. Equivalent volume of sterile PDB was applied as control treatments (CKs) and half-strength Hoagland’s nutrient solution was supplied every week. Three plants were planted per pot. The growth of cucumber seedlings were assessed 21 days after inoculation. The rhizosphere soil samples of cucumber seedlings were collected by vigorously shaking the roots from the 100 ml S. lydicus M01 cell suspension treatment and control for high-throughput sequencing. The shoot length, root length, and fresh weights of cucumber seedlings were measured. Each experiment was repeated three times.
For the investigation of S. lydicus M01’s effects on foliar disease caused by A. alternata, 8-week-old plants were sprayed with a fresh spore suspension of A. alternata at 1 × 108 CFU/ml using a hand atomizer. The plants were maintained in a light growth chamber and were supplied with half-strength Hoagland’s nutrient solution every 7 days. Disease incidence was assessed 30 days after inoculation with A. alternata. The disease severity index was rated 0–4 (0 = healthy; 1 = less than 25% of leaves wilted; 2 = 25–50% of leaves wilted; 3 = 50–75% of leaves wilted; 4 = 75–100% of leaves wilted). The disease index was calculated using the formula: Disease index = [Σ(rating × number of plants rated)/(total number of plants × highest rating)] × 100 (Shi et al., 2017). Cucumber leaves were frozen using liquid nitrogen and stored at −20°C for later determination of antioxidases activities.
Soil DNA Extraction, PCR Amplification and Illumina Sequencing
Microbial community genomic DNA was extracted from soil samples using the E.Z.N.A® soil DNA Kit (Omega Bio-tek, Norcross, GA, United States). The hypervariable region V3–V4 of the bacterial 16S rRNA gene were amplified with primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) whilst the fungal ITS sequence gene were amplified with primer pairs ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′), and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′). The PCR amplification was performed as follows: initial denaturation at 95°C for 3 min, followed by 27 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 45 s, and final extension at 72°C for 10 min, end at 4°C. PCR reactions were performed in triplicate. PCR products was purified and quantified using QuantusTM Fluorometer (Promega, United States).
High-throughput sequencing was carried out on the Illumina MiSeq platform by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered by Trimmomatic and merged by FLASH. The quality trimming process are as follows: (1) The 300 bp reads were truncated at any site receiving an average quality score <20 over a 50 bp sliding window, discarding the truncated reads that were shorter than 50 bp. Reads containing N-bases are also removed. (2) The pair-end reads are merged into a sequence according to their overlap relationship, and the minimum overlap length is 10 bp. (3) The maximum mismatch ratio allowed in the overlap area of the merged sequence is 0.2. (4) Distinguish samples and correcting the direction of the sequence based on the barcode and primer sequences at both ends of the sequence, the number of mismatches allowed in the barcode is 0, and the maximum number of mismatched primers is 2. And the sequence data was subsampled to equal sequencing depth for each sample. Operational taxonomic units (OTUs) with 97% similarity cutoff (Li et al., 2016) were clustered and chimeric sequences were identified and removed using UPARSE (version 7.11). The taxonomy of each OTU representative sequence was analyzed by RDP Classifier2 against the 16S rRNA database (Silva 132/16S_bacteria) and ITS database (unite7.2/its_fungi) using confidence threshold of 0.7, relative abundance below 1% were combined as others.
3,3′-Diaminobenzidine and Nitro Blue Tetrazolium Staining
The content of H2O2 was estimated qualitatively in cucumber leaves using diaminobenzidine (DAB) staining and O2– in the leaves was detected by nitro blue tetrazolium (NBT) staining (Kaur et al., 2016). Three leaves were detached and assayed for each treatment. To detect H2O2, leaves were dipped into DAB solution (1 mg/ml) prepared in double distilled water (pH = 3.8) in a Petri dish using tweezers. To detect O2–, the leaves were immersed in 6 mM NBT solution prepared in sodium citrate (pH = 6.0). Then dipped samples were vacuum infiltrated for 10 min at 60 KPa pressure and incubate at room temperature for 10 min under room light. After incubation, samples were boiled in absolute ethanol till chlorophyll is removed from samples completely. Finally, samples were cooled and stored in 20% glycerol. Images were captured using a light microscope.
Assay of Antioxidases and MDA Concentration
Malondialdehyde (MDA) concentration of cucumber leaves was measured following methods described previously with some modifications (Hartman et al., 2004). Half a gram fresh leaves were homogenized in 5% (w/v) trichloroacetic acid (TCA) and the homogenate was centrifuged at 10,000 × g for 10 min. The supernatant was mixed with 5 ml of 0.5% thiobarbituric acid and heated at 100°C for 25 min, then was cooled to room temperature and centrifuged at 8,500 × g for 5 min. The absorbance was measured at 450, 532, and 600 nm. The MDA concentration was calculated using the formula: MDA = [6.45 (A532−A600) −0.56 A450]/fresh weight.
Superoxide dismutase (SOD) enzyme activity was determined following the method described previously (García-Limones et al., 2002). Briefly, 1 g fresh leaves were homogenized and centrifuged to prepare the supernatant. The crude enzyme extract was added into a reaction mixture to start the reaction and maintained under 4000 l× of light for 20 min. In the control reaction, the crude enzyme extract was replaced with phosphate buffer. The reaction was quenched by switching off the light, the absorbance at 560 nm was measured. The amount of enzyme that inhibited NBT reduction by 50% was defined as a unit of SOD activity. Results are expressed as units mg–1 of protein.
Peroxidase (POD) activity was assayed in a reaction mixture contained 2.9 ml of 0.05 M phosphate buffer, 0.5 ml of 2% H2O2, 0.1 ml of 2% guaiacol, and 0.1 ml of crude enzyme extract. The change of absorbance was measured at 470 nm. POD activity was defined as the amount of guaiacol oxidized per minute, and is expressed as nanomoles per minute per mg of protein.
Polyphenol oxidase (PPO) activity was measured following Tao et al. (2007) with some modifications. One gram fresh leaves were homogenized in phosphate buffer (pH 6.5) containing 0.20 g polyethylene glycol and 10 mM citric acid. The mixture was centrifuged at 10,000 × g for 15 min at 4°C. The activity assay was performed in a mixture containing 0.01 M substrate catechol, 0.1 M phosphate buffer and crude enzyme. Changes in absorbance at 420 nm were measured for 5 min. One unit of enzyme activity was defined as the change in absorbance at 420 nm for 1 g fresh weight per minute.
GenBank Accession Number
The 16S ribosomal RNA sequence of S. lydicus M01 was deposited in NCBI under the GenBank accession numbers MN744679. The raw reads of Illumina MiSeq sequencing were deposited at NCBI’s Sequence Read Archive (SRA) database (accession number PRJNA598072).
Statistical Analysis
Statistical analysis of the data was conducted by one-way ANOVA followed by Duncan’s test (p < 0.05) using SPSS 25.0 software.
Results
Isolation and Identification of the Pathogen Resistant Train Streptomyces lydicus M01
With the supplement of cycloheximide and nalidixic acid, six bacterial isolates were isolated from the rhizosphere soil of Callery pear (P. calleryana). Among the isolated strains, M01 showed highest antifungal activity against the leaf blight plant pathogen A. alternata (Figures 1a,b). 16S rRNA gene analysis showed that M01 shared 99.93, 99.86, and 99.57% sequence identities with S. lydicus strain GS93 isolate 23, S. lydicus strain ATCC 25470 and S. lydicus strain NBRC 13058, respectively. Combined with morphological features of the colonies and mycelia (Figures 1c,d), M01 was classified as S. lydicus and the 16S rRNA gene sequence of M01 was submitted to GeneBank, the accession number is MN744679. And S. lydicus M01 was deposited in China general microbiological culture collection center (CGMCC NO: 16840). Neighbor-joining tree based on 16S ribosomal RNA showed that S. lydicus M01 formed a subclade with the nearest neighbor S. lydicus strain GS93, S. lydicus WYEC 108 and S. lydicus ATCC 25470 were separated in different clades (Supplementary Figure S1).
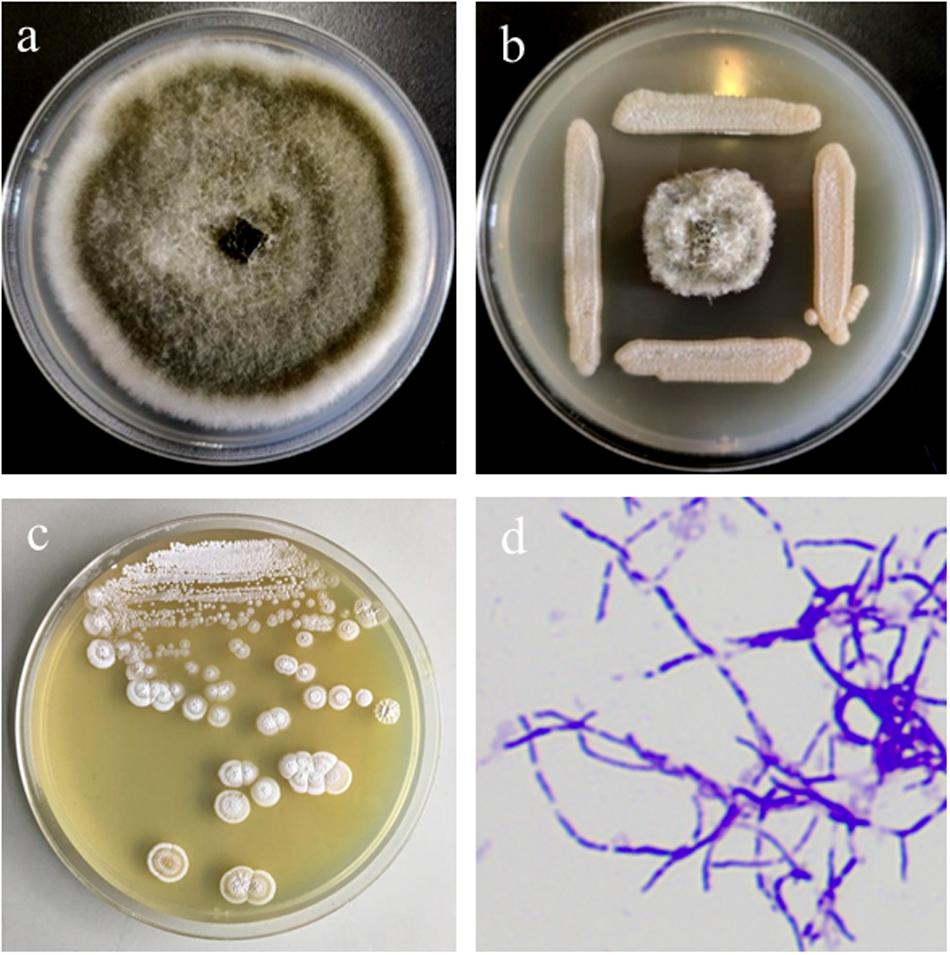
Figure 1. Antifungal activity of Streptomyces lydicus M01 against plant pathogen Alternaria alternata and morphological features of M01 colonies and mycelia. (a) Plant pathogen A. alternata grown on PDA plate as control. (b) Antagonistic effects of S. lydicus M01 against A. alternata. (c) Morphological features of the S. lydicus M01colonies. (d) Mycelia characteristics of S. lydicus M01 stained with crystal violet by light microscopy at 100× magnification.
Effect of S. lydicus M01 on the Growth of Cucumber Seedlings
The effects of S. lydicus M01 inoculation on plant growth promotion were investigated in cucumber plants. After 21 days of inoculation, growth promotion effects on cucumber plants treated with 50 ml M01 culture were not significant. However, when the inoculation amount was increased to 100 ml, the plant height, root status and leaf number and expansion of cucumber seedlings were all significantly enhanced compared to the CK (Figures 2A,B). Further increase in the inoculation amount reduced the growth promotion effects. With the treatment of 100 ml M01 culture, the shoot length, root length, and fresh weight were increased by 18.71, 26.53, and 49.74%, respectively, compared to CK (Figures 2C–E).
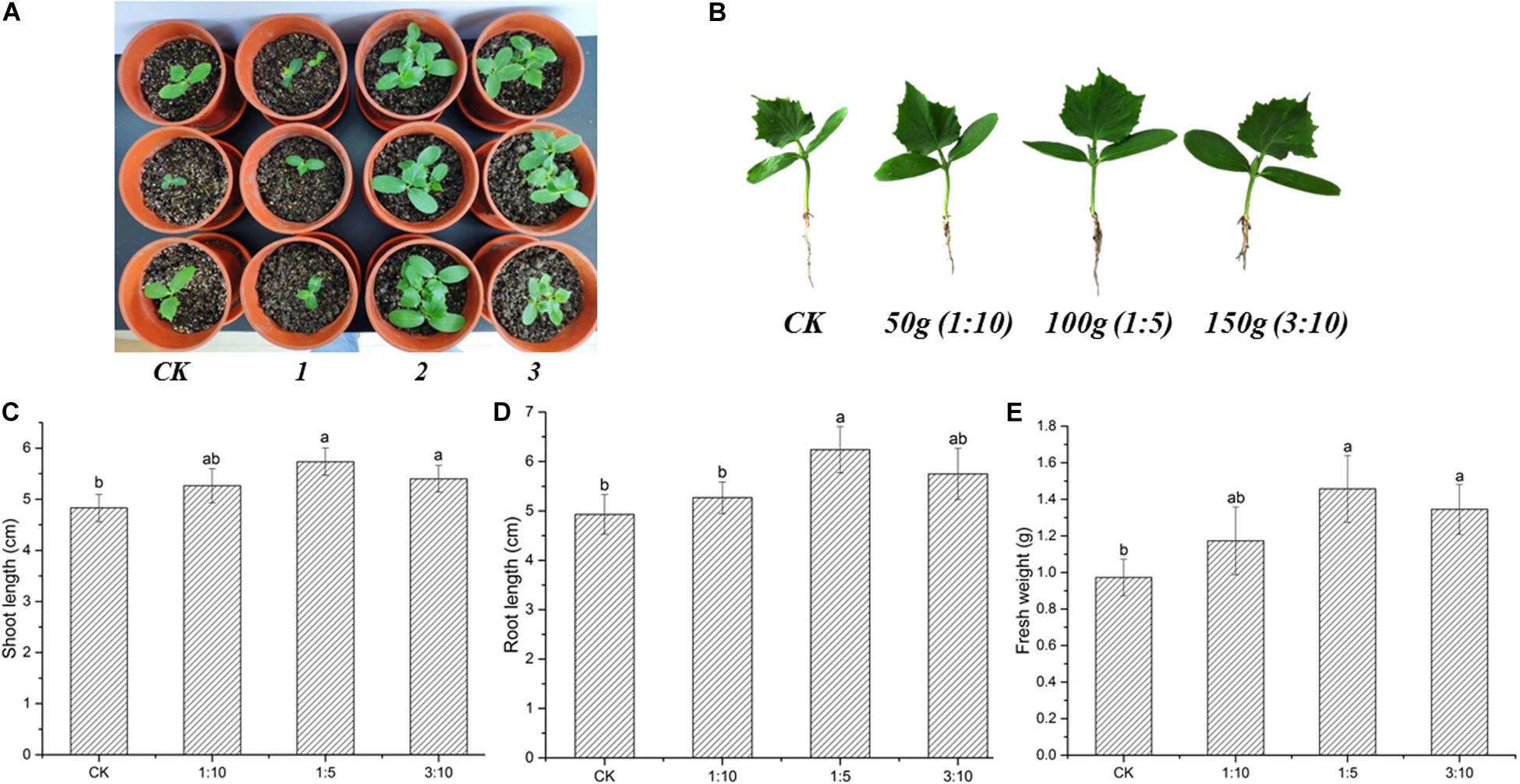
Figure 2. Effects of S. lydicus M01 on the growth of cucumber seedlings. (A) Effects of S. lydicus M01 on the cucumber seedlings growth in pot experiments. CK, control; 1, 2, and 3, S. lydicus M01 culture were mixed with the soil at a ratio of 1:10, 1:5, and 3:10 (w:w), respectively. (B) Effects of S. lydicus M01 on phenotype of cucumber seedlings under different inoculum amount. (C) Shoot length, (D) root length and (E) fresh weight of cucumber plants grown in soil treated or not (CK) with S. lydicus M01. Statistical analysis was conducted by one-way ANOVA followed by Duncan’s test (p < 0.05) using SPSS 25.0 software. Different letters on columns indicate significant difference (p < 0.05).
To characterize of plant growth promoting traits of S. lydicus M01, the IAA production, phosphate solubilization potential, siderophore production and ACC deaminase activity were determined. The IAA produced was determined to be 19.81 ± 1.7 μg/ml. The presence of a clear zone around the colonies indicated that S. lydicus M01 has the ability to solubilize phosphate and produces siderophore (Supplementary Figure S2). ACC deaminase activity was also qualitatively evaluated, successful growing on the MM agar supplemented with ACC indicated ACC deaminase activity.
Effects of S. lydicus M01 on Soil Microbial Community Composition
In this study, analysis based on Illumina sequencing of the six rhizosphere soil samples generated 289,648 16S rRNA gene sequences and 407,987 fungal ITS sequences. The average amplicon length of the six bacterial samples was 414 bp, the average amplicon length of the six eukaryotic samples was 229 bp. After quality trimming, an average of 39,747 reads per sample was generated for the bacteria analyses and 67,544 reads per sample for the eukaryotic sample. The subsampling depth was equalized to the depth of the smallest sample (32,213 reads for the bacteria analyses and 56,672 reads for the eukaryotic sample). A total of 1864 OTUs for bacteria and 623 OTUs for fungi were identified. OTUs with abundance greater than 1% were defined as dominant, while OTUs with abundance less than 1% were combined as “others.”
Analysis against 16S rRNA database and ITS database revealed that 43 dominant bacterial and 17 dominant fungal genera were identified at the genus level in samples from different treatments (Figure 3). The 43 dominant bacterial genera included Pseudarthrobacter, Limnobacter, Sphingomonas, Pseudolabrys, Pseudomonas, Bradyrhizobium, Bryobacter, Rhodanobacter, Streptomyces, Nocardioides, Granulicella, Devosia, Gemmatimonas, Occallatibacter, Ramlibacter, Marmoricola, Massilia, Mesorhizobium, Lactobacillus and 24 unclassified bacteria (Figure 3A). Analysis based on the relative abundance of classified bacterial genera revealed that treatment of S. lydicus M01 influenced the bacterial and fungal composition of the soil compared with non-treated soil. The abundance of species increased included Pseudarthrobacter, Sphingomonas, Rhodanobacter, Pseudomonas, Bryobacter, and Streptomyces, the abundance of species decreased included Limnobacter, Bradyrhizobium, and Pseudolabrys (Figure 4A and Supplementary Table S1). The 17 dominant fungal genera included Mortierella, Penicillium, Naganishia, Acephala, Ascobolus, Fusicolla, Humicola, Pseudogymnoascus, Solicoccozyma, Paraphaeosphaeria, Thelonectria, Fusarium and five unclassified fungi (Figure 3B). The species Mortierella occupied a large portion in the total population in all samples, and the abundance was significantly increased in the treated samples. Except for Mortierella, the abundance of several other species such as Solicoccozyma, Paraphaeosphaeria, Humicola, and Fusicolla were also enriched. In the contrast, the abundance of Fusarium, Ascobolus, and Thelonectria which contain potential fungal pathogen species were significantly reduced (Figure 4B and Supplementary Table S1).
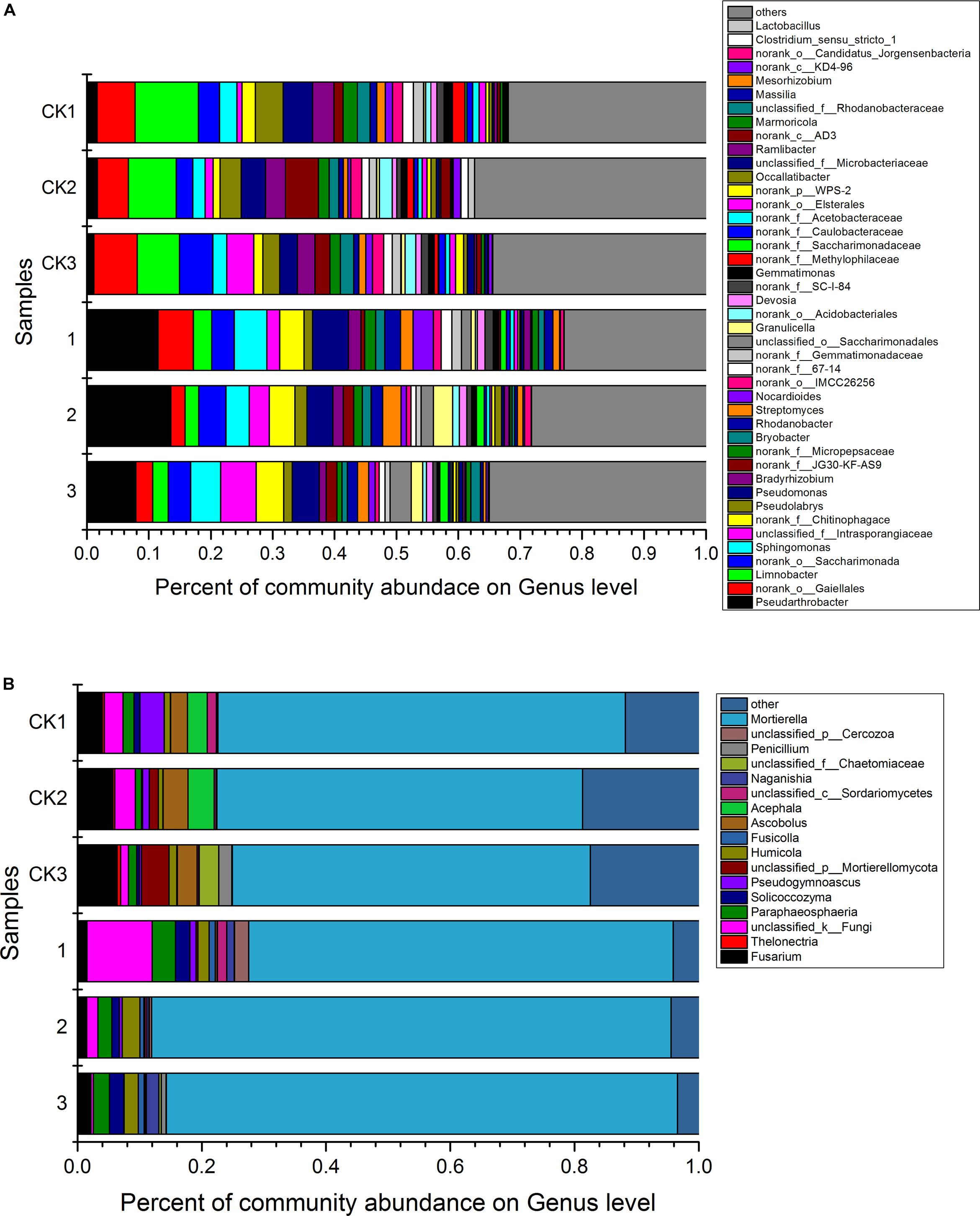
Figure 3. The relative abundance of dominant bacterial genera (A) and fungal genera (B) for soil samples collected from soil treated or not with Streptomyces lydicus M01. CK1, CK2, and CK3, untreated plants; 1,2, and 3, treated plants. The relative abundance was based on the proportional frequencies of DNA sequences that could be classified at the genus level. The different numbers (1, 2, and 3) indicate the three replications.
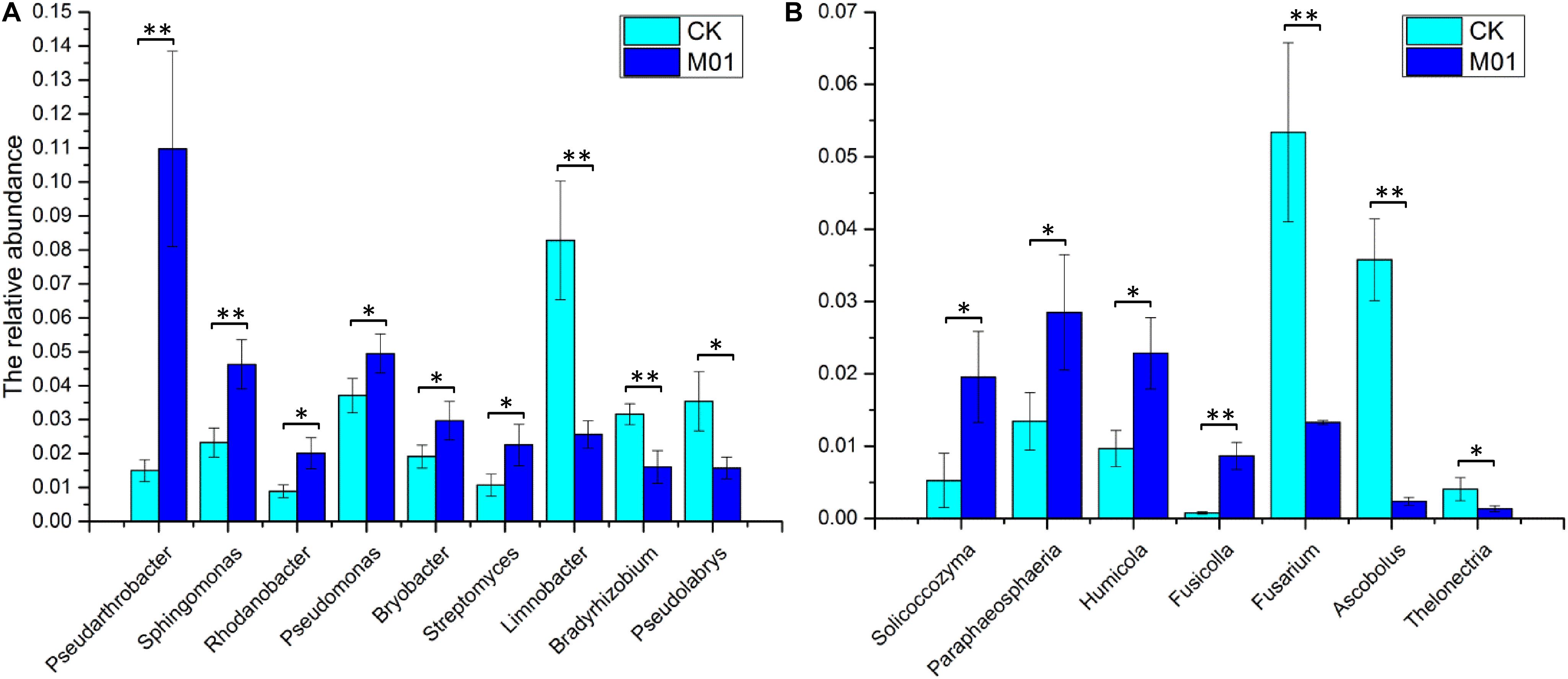
Figure 4. The relative abundance of the main bacterial (A) and fungal genera (B) with significant differences for the soil samples collected from soil treated or not with S. lydicus M01. CK, untreated plants; M01, treated plants. Each histogram represents the mean ± SE of three independent experiments. An independent sample t-test was implemented for statistical analysis of the significance between the M01-treated and the CK group (* for 0.01 < p < 0.05; ** for p < 0.01).
Streptomyces lydicus M01 Alleviates Foliar Disease Caused by Alternaria alternata
In contrast to soil microorganisms, microbial resources from the plant phyllosphere that have the potential to suppress diseases and support plant growth are limited (Rastogi et al., 2013). Therefore, it is tempting to take advantage of soil-borne microbes to develop foliar treatments of crops to fight economically relevant diseases. Thus, S. lydicus M01’s effects on foliar disease caused by A. alternata were investigated.
After inoculating with A. alternata, disease symptoms appeared on cucumber plants including yellow and wilted leaves. The disease incidence reached 80.25%. However, pretreatment with S. lydicus M01 significantly reduced disease incidence, with less yellowing and wilting of the cucumber leaves (Figure 5A). Compared to the control, the application S. lydicus M01 reduced the disease incidence by 52.4% (Figure 5B and Supplementary Table S2). ROS were postulated to be an integral part of the defense response of the plant, which are involved in the interaction of plants with pathogenic fungi. In this study, the production of hydrogen peroxide in leaves was detected by DAB staining, and the superoxide accumulation was analyzed by NBT staining. Leaves of plants treated with A. alternata showed the deepest brown color than control and other treatments, however, leaves of plants pretreated with S. lydicus M01 showed lighter brown color after the perception of pathogens. Similarly, blue staining was observed in leaves of plants treated with A. alternata, but pretreatment of S. lydicus M01 significantly reduced the staining intensity (Figure 6 and Supplementary Figure S4).
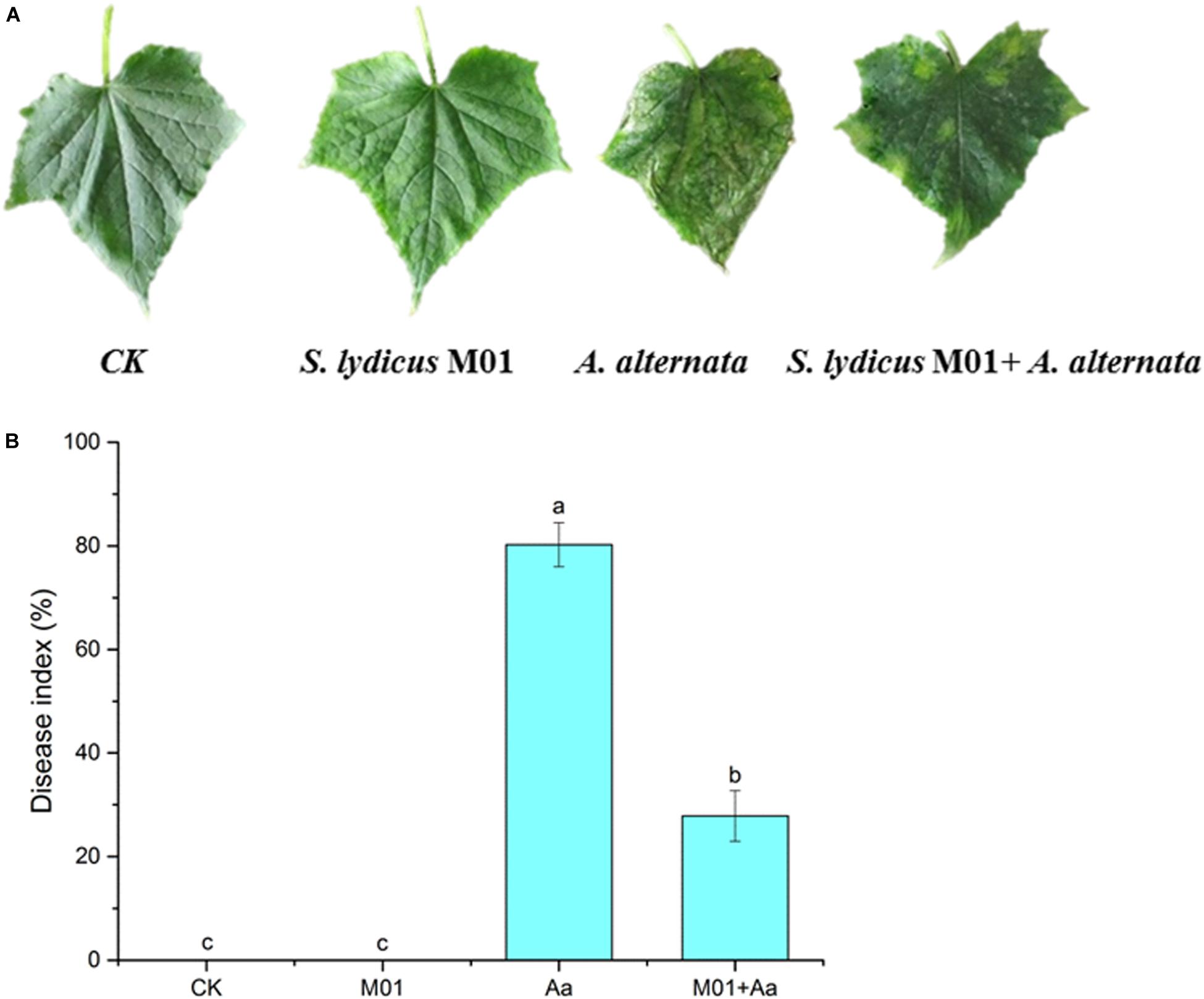
Figure 5. Symptom (A) and disease index (B) on cucumber leaves 30 days after inoculation with A. alternata following different treatments. CK, untreated plants; S. lydicus M01, plants treated with S. lydicus M01; A. alternata, plants treated with A. alternata; S. lydicus M01+ A. alternata, plants pretreated with S. lydicus M01 then with A. alternata. The histogram represents the mean ± SE of three independent experiments. Statistical analysis was conducted by one-way ANOVA followed by Duncan’s test (p < 0.05) using SPSS 25.0 software. Different letters on columns indicate significant difference (p < 0.05).
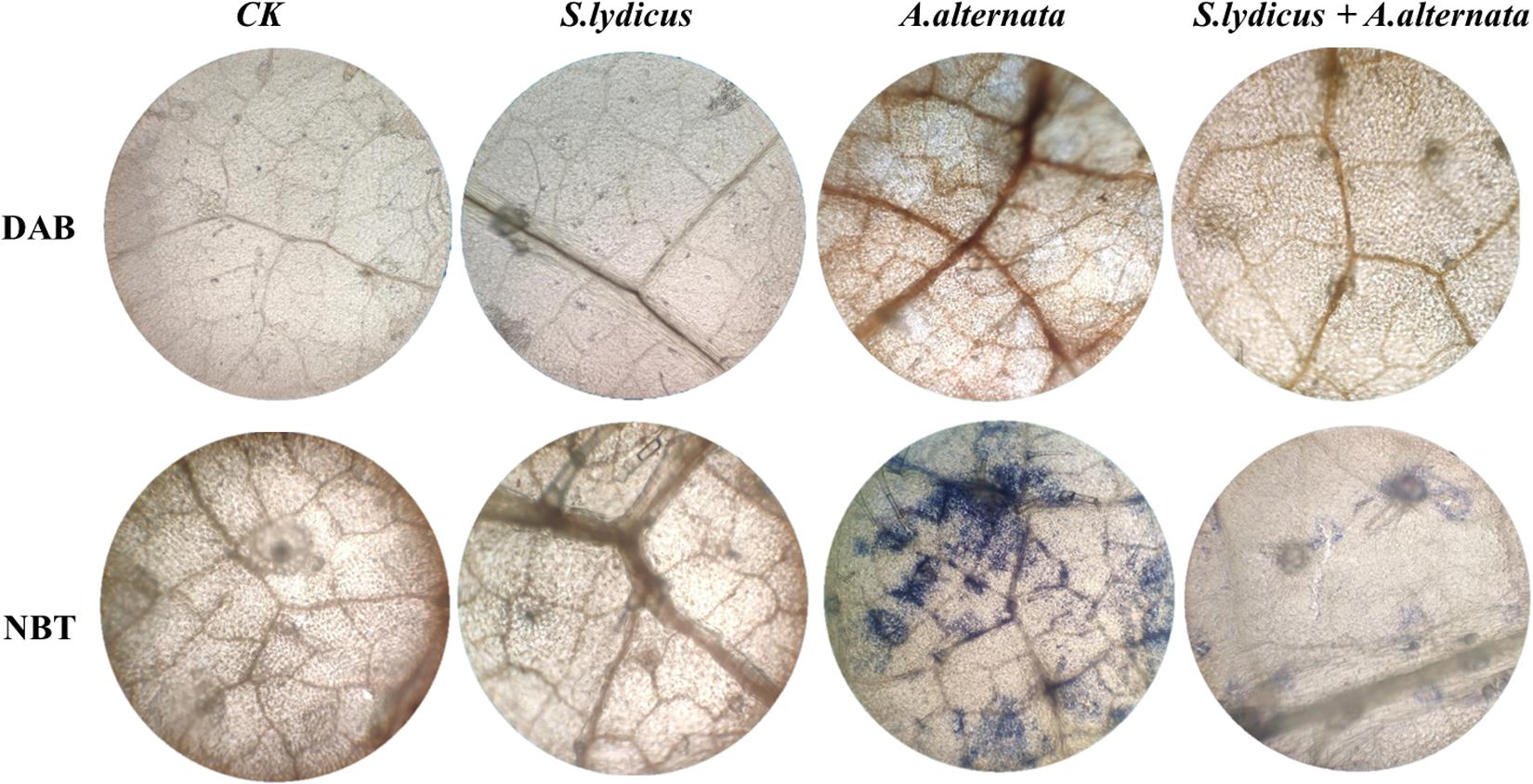
Figure 6. Analysis of ROS accumulation in cucumber leaves following different treatments. DAB, H2O2 accumulation was detected by DAB staining. NBT, O2– accumulation was detected by NBT staining.
Malondialdehyde content is considered as an index of membranous lipid peroxidation (Morsy et al., 2007) and stress-related enzymes PPO, POD, and SOD are commonly used to assess the physiological and biochemical responses of plants to biotic stresses (Gechev et al., 2003). The MDA content of plant leaves treated with A. alternata was significantly increased compared to the control, but leaves of plant pretreated with S. lydicus M01 was lower (Figure 7A). A significant increase in the PPO activity was also found in plant treated with the plant pathogen, but a higher activity was produced with the pretreatment of S. lydicus M01 (Figure 7B). Similar trends were observed in the POD and SOD activity determination, the highest activity was found in the plant treated with S. lydicus M01 then the plant pathogen (Figures 7C,D). Notably, an increase of PPO activity was observed in plant treated with S. lydicus M01 without invasion of plant pathogen.
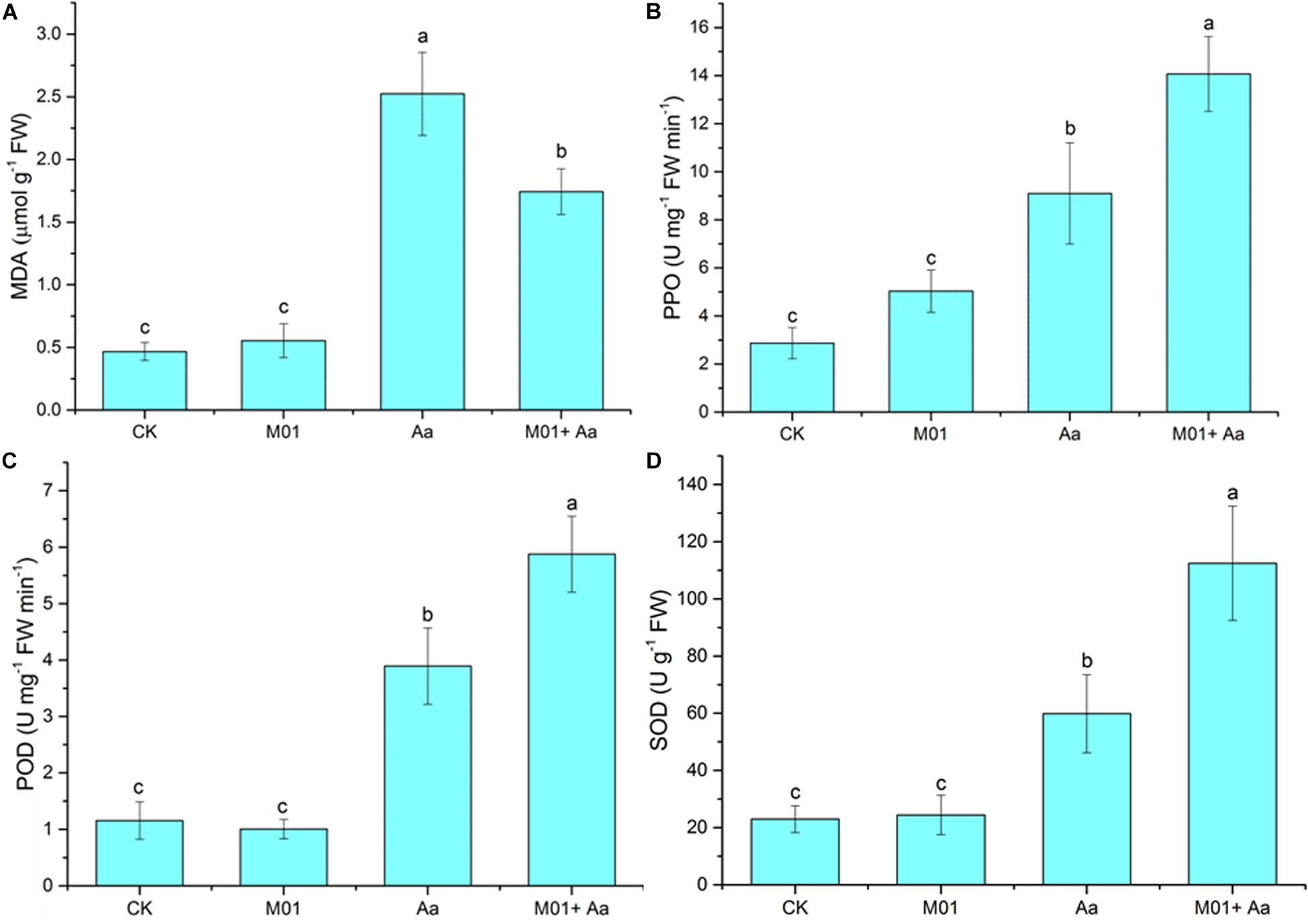
Figure 7. Activities of defense-related enzyme and concentrations of malondialdehyde measured in leaves. (A) Malondialdehyde (MDA). (B) Polyphenol oxidase (PPO). (C) Peroxidase (POD). (D) Superoxide dismutase (SOD). Statistical analysis was conducted by one-way ANOVA followed by Duncan’s test (p < 0.05) using SPSS 25.0 software. Different letters on columns indicate significant difference (p < 0.05).
Discussion
The green revolution of the past century has significantly improved crop yield by extensive use of mineral fertilizers and chemically synthesized pesticides. Despite immediate benefits, this strategy is reaching its limits in maintaining food security for a growing population (Bosch et al., 2014). Alternative strategies for plant growth promotion and protection against pests are associated with soil microbes for nutrition gains and disease suppression (Rey and Schornack, 2013). Among the various soil-borne bacteria and fungi associated with agriculture, species in the Streptomyces genus, which has been highlighted in medical drug discoveries, are major actors in soil suppression of diseases (Cordovez et al., 2015; Cha et al., 2016) owing to the numerous metabolites they produce which inhibit the development of microbial pathogens. Despite their popularity as biocontrol agents, understanding of Streptomyces’s function as biological fertilizers is limited, especially their influence in soil microbial community.
In the present study, S. lydicus M01 distributed in the rhizosphere soil of Callery pear (P. calleryana) was isolated and exhibited strong activity against leaf blight plant pathogen A. alternata. Phylogenetic analysis revealed that S. lydicus M01 was clustered in a different clade compared with the commercialized S. lydicus WYEC 108 (Yuan and Crawford, 1995) and other Streptomyces strains, which indicated potential differences. In addition, comparative genomic analyses showed that closely related Streptomyces strains have high diversity in the secondary metabolite gene clusters they contain (Hwang et al., 2014; Jia et al., 2017). In the pot experiments, the application of S. lydicus M01 had a positive effect on the growth of cucumber seedlings, compared to the CK treatment, the plant height, fresh weight, and root status were all increased to different degrees. This result is in agreement with our previous study when S. lydicus M01 promoted cucumber seedlings growth in hydroponic experiments (Supplementary Figure S3). These results, in combination with the production of IAA, ACC desaminase, and siderophores demonstrate that S. lydicus M01 has the capacity to stimulate plant growth. Then, the underlying mechanism was investigated in the aspect of changes in soil microbial community following the application of S. lydicus M01. To minimize error caused by microbe culture, all cell suspension used in the treatment were from the same batch and sterile nutrient broth of the same composition were used as control. Thus, their influence the microbial community should be minor.
Analysis of microbial community composition and structure revealed that a higher relative abundance of Pseudarthrobacter, Sphingomonas, Rhodanobacter, Pseudomonas, Bryobacter, and Streptomyces genera were identified in the S. lydicus M01 treated soil. Species from Pseudarthrobacter are capable of degrading high concentrations of environmental contaminants such as 4-chlorophenol and phenanthrene (Westerberg et al., 2000; Kim et al., 2008). As such, they were used in bioremediation. Sphingomonas are widely distributed in nature, having been isolated from plant root systems and other sources. Reports showed that Sphingomonas stimulates the growth plants through phytohormone production, nitrogen fixation and driving developmental plasticity in the roots (Yang et al., 2014; Luo et al., 2019). Additionally, Sphingomonas was also shown to alleviate salinity stress in plants (Khan et al., 2017). Rhodanobacter are known denitrifying bacteria that are used for bioremediation (Prakash et al., 2012), interestingly, several strains showed biological control activity toward root-rot plant pathogen (De Clercq et al., 2006). Pseudomonas is one of the most common and widely used biocontrol and PGPR strains due to their pathogen inhibition and systemic resistance inducing properties (Chin-A-Woeng et al., 2000; Haas and Défago, 2005). Bryobacter inhabit in peat, but the function is unknown (Kulichevskaya et al., 2010). The relative abundance of Streptomyces was also significantly increased 21 days after inoculation which may indicate an effective colonization of S. lydicus M01. Most of the bacteria from the genera listed above have characteristics associated with pathogen inhibition, plant auxin biosynthesis or bioremediation. However, the abundance of Bradyrhizobium and Pseudolabrys that both function in nitrogen fixation (Hennecke, 1990; Rangel et al., 2017; Wongdee et al., 2018) were significantly reduced. Furthermore, the abundance of Limnobacter species that have the ability to oxidize thiosulfate (Lu et al., 2011; Vedler et al., 2013) was also reduced. Overall, most of the bacterial communities significantly altered by S. lydicus M01 treatment had a positive effect on growth promotion, biological control, alleviation of abiotic stress, or bioremediation.
The use of S. lydicus M01 in the plant growth promotion of cucumber seedlings occurs not only through shifting the rhizospheric bacterial community but also through altering the composition of the fungal community. Previous studies showed that Fusicolla sp. is a putative plant-growth-promoting rhizobacteria that correlated positively with canola yield (Lay et al., 2018) and Humicola is a fungus with potential for biological control of plant diseases (Ko et al., 2011). In this study, abundance of the two species were significantly increased in the presence of S. lydicus M01. Furthermore, the fungal genera Solicoccozyma and Paraphaeosphaeria were also enriched in comparison with the control soil. Solicoccozyma are yeasts isolated from soil that function in biodegradation (Stosiek et al., 2019). Species in Paraphaeosphaeria were found to enhance plant temperature tolerance in Arabidopsis (McLellan et al., 2007), thus the enrichment of Paraphaeosphaeria may contribute to the alleviation of abiotic stress of cucumbers. Notably, Fusarium was identified in the all soil samples and its abundance was significantly reduced in the presence of S. lydicus M01. Since this fungus is widely distributed in soil and most species are harmless, plant pathogenic species were not considered in the present study. Some species of Ascobolus were found on rotting Brassica stems (Buczacki and Wilkinson, 1992) and Thelonectria was reported to produce glycosidase inhibitors (Miyauchi et al., 2016), both genera were decreased in the presence of S. lydicus M01. Plants are tightly associated with complex microbial communities, modification of the plant microbiome play an important role to reduce the occurrence of plant disease, increase agricultural production, and reduce chemical inputs. Thus, overall shift in soil microbial community by M01 or other microbes is important in agriculture, resulting in more sustainable agricultural practices.
Plant growth-promoting rhizobacteria affect plant growth in direct and indirect ways. The indirect promotion of plant growth occurs when PGPR decrease the deleterious effects of phytopathogenic organisms or ISR. Foliar disease caused by A. alternata develops on leaves and often spread through air. However, colonization of PGPR on the aerial parts of plants remains a technical bottleneck (Samac et al., 2003). Thus, in this study, the alleviation of foliar disease by S. lydicus M01 around the cucumber rhizosphere reflected indirect effects of PGPR on plant growth promotion and disease suppression. The ROS production is one of the earliest cellular responses following successful pathogen recognition and accumulation of ROS were consistently observed in the plant after the perception of pathogens (Torres et al., 2006). However, excessive production of ROS causes progressive oxidative damage and ultimately cell death (Das and Roychoudhury, 2014). The reduced staining intensity of ROS in cucumber leaves indicated that the S. lydicus M01 treatment reduced ROS accumulation. Further investigation revealed that antioxidants related with ROS scavenging were influenced by S. lydicus M01 treatment under biotic stress. MDA content is usually increased under stressed conditions such as pathogenic infection (Heber et al., 1996). In this study, pretreatment with S. lydicus M01 significantly reduced the MDA content, which reflects a lower level of membranous lipid peroxidation. This result agrees with previous report that the content of MDA in leaves treated with biological fertilizer was much lower than the control plants (Wu et al., 2008). Antioxidases such as SOD and POD are associated with eliminating ROS (Asada, 1992), whereas PPO may be induced to produce lignin and other phenolics to strengthen the cell walls (Avdiushko et al., 1993). In this study, the POD, SOD, and PPO enzyme activities were increased significantly when plants were pretreated with S. lydicus M01 before the invasion of plant pathogen, but the activities of these enzymes increased less in cucumber only treated with A. alternata. Similar results were also observed in previous report when antagonistic bacterium B. subtilis strain AR12 was used to induce defense enzymes toward Ralstonia solanacearum in tomato (Li et al., 2008). These results suggest that S. lydicus M01 enhanced disease resistance of cucumber, at least in part, by increasing activities of defense related enzymes. Further studies are required for investigation of ISR and related signaling pathways.
Conclusion
In conclusion, the application of S. lydicus M01 effectively promoted the plant growth and suppressed foliar disease caused by A. alternata in cucumbers. S. lydicus M01 may systemically affect plant-soil interactions by producing auxins, siderophores and ACC deaminase and increasing the abundance of certain plant beneficial microbes, such as bacteria genera Pseudarthrobacter, Sphingomonas, Rhodanobacter, and Pseudomonas and fungi genera Fusicolla and Humicola. Furthermore, S. lydicus M01 enhanced plant disease resistance through induced antioxidases activity, possibly together with other beneficial microbes influenced around rhizosphere. The beneficial plant microbiome interactions represent a promising sustainable solution to improve agricultural production instead of chemical fertilizers.
Data Availability Statement
The 16S ribosomal RNA sequence of Streptomyces lydicus M01 was deposited in NCBI under the GenBank accession numbers MN744679. The raw reads of Illumina MiSeq sequencing were deposited at NCBI’s Sequence Read Archive (SRA) database (accession number PRJNA598072).
Author Contributions
MW, JX, and JM performed the experiments. MW analyzed the sequencing data. XF provided the technical assistance. HX and MW designed the experiments. MW wrote the manuscript. HX and HY revised the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31900030), the University Natural Science Research Project in Jiangsu Province (19KJB530009), the Key R&D Projects in Jiangsu Province (BE2019390), and Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (XTB1804).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00942/full#supplementary-material
Footnotes
References
Ahmad, F., Ahmad, I., and Khan, M. S. (2008). Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 163, 173–181.
Alexander, D. B., and Zuberer, D. A. (1991). Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12, 39–45. doi: 10.1007/BF00369386
Asada, K. (1992). Ascorbate peroxidase – a hydrogen peroxide-scavenging enzyme in plants. Physiol. Plant. 85, 235–241. doi: 10.1111/j.1399-3054.1992.tb04728.x
Avdiushko, S. A., Ye, X. S., and Kuc, J. (1993). Detection of several enzymatic activities in leaf prints of cucumber plants. Physiol. Mol. Plant Pathol. 42, 441–454.
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. P. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35(Suppl. 4), 1044–1051. doi: 10.1590/s1415-47572012000600020
Bhattacharyya, P. N., and Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Bibb, M. J. (2013). Understanding and manipulating antibiotic production in actinomycetes. Biochem. Soc. Trans. 41, 1355–1364. doi: 10.1042/bst20130214
Bosch, F. V. D., Oliver, R., Berg, F. V. D., and Paveley, N. (2014). Governing principles can guide fungicide-resistance management tactics. Annu. Rev. Phytopathol. 52, 175–195. doi: 10.1146/annurev-phyto-102313-050158
Buczacki, S. T., and Wilkinson, J. (1992). Collins Guide: Mushrooms and Toadstools of Britain and Europe. New York, NY: HarperCollins.
Cha, J.-Y., Han, S., Hong, H.-J., Cho, H., Kim, D., Kwon, Y., et al. (2016). Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 10, 119–129. doi: 10.1038/ismej.2015.95
Chin-A-Woeng, T. F. C., Bloemberg, G. V., Mulders, I. H. M., Dekkers, L. C., and Lugtenberg, B. J. J. (2000). Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant Microbe Interact. 13, 1340–1345. doi: 10.1094/mpmi.2000.13.12.1340
Cordovez, V., Carrion, V. J., Etalo, D. W., Mumm, R., Zhu, H., van Wezel, G. P., et al. (2015). Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front. Microbiol. 6:1081. doi: 10.3389/fmicb.2015.01081
Das, K., and Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2:53. doi: 10.3389/fenvs.2014.00053
De Clercq, D., Van Trappen, S., Cleenwerck, I., Ceustermans, A., Swings, J., Coosemans, J., et al. (2006). Rhodanobacter spathiphylli sp. nov., a gammaproteobacterium isolated from the roots of Spathiphyllum plants grown in a compost-amended potting mix. Int. J. Syst. Evol. Microbiol. 56, 1755–1759. doi: 10.1099/ijs.0.64131-0
El-Tarabily, K. A. (2008). Promotion of tomato (Lycopersicon esculentum Mill.) plant growth by rhizosphere competent 1-aminocyclopropane-1-carboxylic acid deaminase-producing streptomycete actinomycetes. Plant Soil 308, 161–174.
Galloway, J. N., Townsend, A. R., Erisman, J. W., Bekunda, M., Cai, Z., Freney, J. R., et al. (2008). Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892. doi: 10.1126/science.1136674
García-Limones, C., Hervás, A., Navas-Cortés, J. A., Jiménez-Díaz, R. M., and Tena, M. (2002). Induction of an antioxidant enzyme system and other oxidative stress markers associated with compatible and incompatible interactions between chickpea (Cicer arietinum L.) and Fusarium oxysporum f. sp.ciceris. Physiol. Mol. Plant Pathol. 61, 325–337.
Gechev, T., Willekens, H., Van Montagu, M., Inzé, D., Van Camp, W. I. M., Toneva, V., et al. (2003). Different responses of tobacco antioxidant enzymes to light and chilling stress. J. Plant Physiol. 160, 509–515. doi: 10.1078/0176-1617-00753
Glick, B. R. (1995). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117. doi: 10.1139/m95-015
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Hartman, G. L., Huang, Y. H., and Li, S. (2004). Phytotoxicity of Fusarium solani culture filtrates from soybeans and other hosts assayed by stem cuttings. Australas. Plant Pathol. 33, 9–15. doi: 10.1071/AP03072
Heber, U., Miyake, C., Mano, J., Ohno, C., and Asada, K. (1996). Monodehydroascorbate radical detected by electron paramagnetic resonance spectrometry is a sensitive probe of oxidative stress in intact leaves. Plant Cell Physiol. 37, 1066–1072. doi: 10.1093/oxfordjournals.pcp.a029055
Hennecke, H. (1990). Nitrogen fixation genes involved in the Bradyrhizobium japonicum-soybean symbiosis. FEBS Lett. 268, 422–426.
Hwang, K.-S., Kim, H. U., Charusanti, P., Palsson, B. Ø., and Lee, S. Y. (2014). Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 32, 255–268. doi: 10.1016/j.biotechadv.2013.10.008
Jaemsaeng, R., Jantasuriyarat, C., and Thamchaipenet, A. (2018). Molecular interaction of 1-aminocyclopropane-1-carboxylate deaminase (ACCD)-producing endophytic Streptomyces sp. GMKU 336 towards salt-stress resistance of Oryza sativa L. cv. KDML105. Sci. Rep. 8:1950. doi: 10.1038/s41598-018-19799-9
Jia, N., Ding, M.-Z., Luo, H., Gao, F., and Yuan, Y.-J. (2017). Complete genome sequencing and antibiotics biosynthesis pathways analysis of Streptomyces lydicus 103. Sci. Rep. 7:44786. doi: 10.1038/srep44786
Kaur, N., Sharma, I., Kirat, K., and Pati, P. K. (2016). Detection of reactive oxygen species in Oryza sativa L. (Rice). Bio Protoc. 6:e2061. doi: 10.21769/BioProtoc.2061
Khan, A. L., Waqas, M., Asaf, S., Kamran, M., Shahzad, R., Bilal, S., et al. (2017). Plant growth-promoting endophyte Sphingomonas sp. LK11 alleviates salinity stress in Solanum pimpinellifolium. Environ. Exp. Bot. 133, 58–69.
Kim, K. K., Lee, K. C., Oh, H.-M., Kim, M. J., Eom, M. K., and Lee, J.-S. (2008). Arthrobacter defluvii sp. nov., 4-chlorophenol-degrading bacteria isolated from sewage. Int. J. Syst. Evol. Microbiol. 58, 1916–1921. doi: 10.1099/ijs.0.65550-0
Ko, W. H., Yang, C. H., Lin, M. J., Chen, C. Y., and Tsou, Y. J. (2011). Humicola phialophoroides sp. nov. from soil with potential for biological control of plant diseases. Bot. Stud. 52, 197–202.
Kulichevskaya, I. S., Suzina, N. E., Liesack, W., and Dedysh, S. N. (2010). Bryobacter aggregatus gen. nov., sp. nov., a peat-inhabiting, aerobic chemo-organotroph from subdivision 3 of the Acidobacteria. Int. J. Syst. Evol. Microbiol. 60, 301–306. doi: 10.1099/ijs.0.013250-0
Law, J. W.-F., Ser, H.-L., Khan, T. M., Chuah, L.-H., Pusparajah, P., Chan, K.-G., et al. (2017). The potential of Streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Front. Microbiol. 8:3. doi: 10.3389/fmicb.2017.00003
Lay, C.-Y., Bell, T. H., Hamel, C., Harker, K. N., Mohr, R., Greer, C. W., et al. (2018). Canola root–associated microbiomes in the Canadian prairies. Front. Microbiol. 9:1188. doi: 10.3389/fmicb.2018.01188
Li, L., Ma, J., Ibekwe, A. M., Wang, Q., and Yang, C.-H. (2016). Cucumber rhizosphere microbial community response to biocontrol agent Bacillus subtilis B068150. Agriculture 6, 2–15.
Li, S.-M., Hua, G.-G., Liu, H.-X., and Guo, J.-H. (2008). Analysis of defence enzymes induced by antagonistic bacterium Bacillus subtilis strain AR12 towards Ralstonia solanacearum in tomato. Ann. Microbiol. 58:573. doi: 10.1007/bf03175560
Lu, H., Sato, Y., Fujimura, R., Nishizawa, T., Kamijo, T., and Ohta, H. (2011). Limnobacter litoralis sp. nov., a thiosulfate-oxidizing, heterotrophic bacterium isolated from a volcanic deposit, and emended description of the genus Limnobacter. Int. J. Syst. Evol. Microbiol. 61, 404–407. doi: 10.1099/ijs.0.020206-0
Luo, Y., Wang, F., Huang, Y., Zhou, M., Gao, J., Yan, T., et al. (2019). Sphingomonas sp. Cra20 increases plant growth rate and alters rhizosphere microbial community structure of Arabidopsis thaliana under drought stress. Front. Microbiol. 10:1221. doi: 10.3389/fmicb.2019.01221
McLellan, C. A., Turbyville, T. J., Wijeratne, E. M. K., Kerschen, A., Vierling, E., Queitsch, C., et al. (2007). A rhizosphere fungus enhances Arabidopsis thermotolerance through production of an HSP90 inhibitor. Plant Physiol. 145, 174–182. doi: 10.1104/pp.107.101808
Miyauchi, R., Ono, C., Ohnuki, T., and Shiba, Y. (2016). Nectrisine biosynthesis genes in Thelonectria discophora SANK 18292: identification and functional analysis. Appl. Environ. Microbiol. 82, 6414–6422. doi: 10.1128/AEM.01709-16
Morsy, M. R., Jouve, L., Hausman, J.-F., Hoffmann, L., and Stewart, J. M. (2007). Alteration of oxidative and carbohydrate metabolism under abiotic stress in two rice (Oryza sativa L.) genotypes contrasting in chilling tolerance. J. Plant Physiol. 164, 157–167. doi: 10.1016/j.jplph.2005.12.004
Olanrewaju, O. S., Glick, B. R., and Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33:197. doi: 10.1007/s11274-017-2364-9
Patten, C. L., and Glick, B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. doi: 10.1128/aem.68.8.3795-3801.2002
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Wees, S. C. M. V., and Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Prakash, O., Green, S. J., Jasrotia, P., Overholt, W. A., Canion, A., Watson, D. B., et al. (2012). Rhodanobacter denitrificans sp. nov., isolated from nitrate-rich zones of a contaminated aquifer. Int. J. Syst. Evol. Microbiol. 62, 2457–2462. doi: 10.1099/ijs.0.035840-0
Qin, Y., Shang, Q., Zhang, Y., Li, P., and Chai, Y. (2017). Bacillus amyloliquefaciens L-S60 reforms the rhizosphere bacterial community and improves growth conditions in cucumber plug seedling. Front. Microbiol. 8:2620. doi: 10.3389/fmicb.2017.02620
Rangel, W. D. M., de Oliveira Longatti, S. M., Ferreira, P. A. A., Bonaldi, D. S., Guimarães, A. A., Thijs, S., et al. (2017). Leguminosae native nodulating bacteria from a gold mine As-contaminated soil: multi-resistance to trace elements, and possible role in plant growth and mineral nutrition. Int. J. Phytoremediation 19, 925–936. doi: 10.1080/15226514.2017.1303812
Rastogi, G., Coaker, G. L., and Leveau, J. H. J. (2013). New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 348, 1–10. doi: 10.1111/1574-6968.12225
Raupach, G. S., Liu, L., Murphy, J. F., Tuzun, S., and Kloepper, J. W. (1996). Induced systemic resistance in cucumber and tomato against cucumber mosaic cucumovirus using plant growth-promoting rhizobacteria (PGPR). Plant Dis. 80, 891–894.
Rey, T., and Schornack, S. (2013). Interactions of beneficial and detrimental root-colonizing filamentous microbes with plant hosts. Genome Biol. 14:121. doi: 10.1186/gb-2013-14-6-121
Rungin, S., Indananda, C., Suttiviriya, P., Kruasuwan, W., Jaemsaeng, R., and Thamchaipenet, A. (2012). Plant growth enhancing effects by a siderophore-producing endophytic streptomycete isolated from a Thai jasmine rice plant (Oryza sativa L. cv. KDML105). Antonie Van Leeuwenhoek 102, 463–472. doi: 10.1007/s10482-012-9778-z
Sadeghi, A., Karimi, E., Dahaji, P. A., Javid, M. G., Dalvand, Y., and Askari, H. (2012). Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World J. Microbiol. Biotechnol. 28, 1503–1509.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Samac, D. A., Willert, A. M., McBride, M. J., and Kinkel, L. L. (2003). Effects of antibiotic-producing Streptomyces on nodulation and leaf spot in alfalfa. Appl. Soil Ecol. 22, 55–66.
Shi, L., Du, N., Shu, S., Sun, J., Li, S., and Guo, S. (2017). Paenibacillus polymyxa NSY50 suppresses Fusarium wilt in cucumbers by regulating the rhizospheric microbial community. Sci. Rep. 7:41234. doi: 10.1038/srep41234
Stosiek, N., Terebieniec, A., Za̧bek, A., Młynarz, P., Cieśliński, H., and Klimek-Ochab, M. (2019). N-phosphonomethylglycine utilization by the psychrotolerant yeast Solicoccozyma terricola M 3.1.4. Bioorg. Chem. 93:102866. doi: 10.1016/j.bioorg.2019.03.040
Tao, F., Zhang, M., and Yu, H.-Q. (2007). Effect of vacuum cooling on physiological changes in the antioxidant system of mushroom under different storage conditions. J. Food Eng. 79, 1302–1309.
Timmer, L. W., Peever, T. L., Solel, Z., and Akimitsu, K. (2003). Alternaria diseases of citrus – Novel pathosystems. Phytopathol. Mediterr. 42, 99–112.
Torres, M. A., Jones, J. D. G., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Vedler, E., Heinaru, E., Jutkina, J., Viggor, S., Koressaar, T., Remm, M., et al. (2013). Limnobacter spp. as newly detected phenol-degraders among Baltic Sea surface water bacteria characterised by comparative analysis of catabolic genes. Syst. Appl. Microbiol. 36, 525–532. doi: 10.1016/j.syapm.2013.07.004
Wang, Y., Bao, Y., Shen, D., Feng, W., Yu, T., Zhang, J., et al. (2008). Biocontrol of Alternaria alternata on cherry tomato fruit by use of marine yeast Rhodosporidium paludigenum Fell & Tallman. Int. J. Food Microbiol. 123, 234–239. doi: 10.1016/j.ijfoodmicro.2008.02.002
Watve, M. G., Tickoo, R., Jog, M. M., and Bhole, B. D. (2001). How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176, 386–390. doi: 10.1007/s002030100345
Westerberg, K., Elväng, A. M., Stackebrandt, E., and Jansson, J. K. (2000). Arthrobacter chlorophenolicus sp. nov., a new species capable of degrading high concentrations of 4-chlorophenol. Int. J. Syst. Evol. Microbiol. 50, 2083–2092. doi: 10.1099/00207713-50-6-2083
Wongdee, J., Boonkerd, N., Teaumroong, N., Tittabutr, P., and Giraud, E. (2018). Regulation of nitrogen fixation in Bradyrhizobium sp. strain DOA9 involves two distinct NifA regulatory proteins that are functionally redundant during symbiosis but not during free-living growth. Front. Microbiol. 9:1644. doi: 10.3389/fmicb.2018.01644
Wu, H.-S., Yang, X.-N., Fan, J.-Q., Miao, W.-G., Ling, N., Xu, Y.-C., et al. (2008). Suppression of Fusarium wilt of watermelon by a bio-organic fertilizer containing combinations of antagonistic microorganisms. BioControl 54:287.
Wu, Q., Lu, C., Ni, M., Wang, H., Liu, W., and Chen, J. (2019). Streptomyces lydicus A01 affects soil microbial diversity, improving growth and resilience in tomato. J. Integr. Plant Biol. 61, 182–196. doi: 10.1111/jipb.12724
Yamamoto-Tamura, K., Ohno, M., Fujii, T., Kataoka, S., Numata, S., Nakajima, M., et al. (2011). Assessment of the effects of genetically modified Pseudomonas spp. expressing chitinase on the soil microbial community in the cucumber rhizosphere. Jpn. Agric. Res. Q. 45, 377–383. doi: 10.6090/jarq.45.377
Yang, S., Zhang, X., Cao, Z., Zhao, K., Wang, S., Chen, M., et al. (2014). Growth-promoting Sphingomonas paucimobilis ZJSH1 associated with Dendrobium officinale through phytohormone production and nitrogen fixation. Microb. Biotechnol. 7, 611–620. doi: 10.1111/1751-7915.12148
Youssef, M., and Eissa, M. (2014). Biofertilizers and their role in management of plant parasitic nematodes. A review. J. Biotechnol. Pharm. Res. 5, 1–6.
Keywords: plant growth promotion, Streptomyces lydicus M01, rhizospheric microbial community, Alternaria alternata, reactive oxygen species, disease resistance
Citation: Wang M, Xue J, Ma J, Feng X, Ying H and Xu H (2020) Streptomyces lydicus M01 Regulates Soil Microbial Community and Alleviates Foliar Disease Caused by Alternaria alternata on Cucumbers. Front. Microbiol. 11:942. doi: 10.3389/fmicb.2020.00942
Received: 30 December 2019; Accepted: 20 April 2020;
Published: 15 May 2020.
Edited by:
Paulo José Pereira Lima Teixeira, University of São Paulo, BrazilReviewed by:
Amit K. Jaiswal, Purdue University, United StatesDerek Lundberg, Max Planck Institute for Developmental Biology, Germany
Copyright © 2020 Wang, Xue, Ma, Feng, Ying and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Xu, eHVoQG5qdGVjaC5lZHUuY24=
 Mingxuan Wang1,2
Mingxuan Wang1,2 Hanjie Ying
Hanjie Ying Hong Xu
Hong Xu