- Department of Life Sciences and Systems Biology, Mycotheca Universitatis Taurinensis, University of Torino, Turin, Italy
Fungi are widely distributed in the Oceans, interact with other organisms and play roles that range from pathogenic to mutualistic. The present work focuses on the characterization of the cultivable mycobiota associated with the seagrass Posidonia oceanica (L.) Delile collected off the Elba Island (Italy). We identified 102 taxa (mainly Ascomycota) by the mean of a polyphasic approach. Leaves, rhizomes, roots and matte were characterized by unique mycobiota revealing a “plant-part-specificity.” The comparison with the mycobiota associated with the green alga Flabellia petiolata and the brown alga Padina pavonica underlined a “substrate specificity.” Indeed, despite being part of the same phytocoenosis, these photosynthetic organisms recruit different fungal communities. The mycobiota seems to be necessary for the host’s defense and protection, playing, in this way, remarkable ecological roles. Among the 61 species detected in association with P. oceanica (including two species belonging to the newly introduced genus Paralulworthia), 37 were reported for the first time from the Mediterranean Sea.
Introduction
Posidonia oceanica (L.) Delile, otherwise known as Neptune grass, is the most important seagrass in the Mediterranean Sea (Personnic et al., 2014). This endemic long-lived plant (Magnoliophyta, kingdom Archaeplastida) relies mainly on vegetative reproduction, and its largest clones can spread over 15 km (Arnaud-Haond et al., 2012). Neptune grass meadows, or rather forests, are a spawning ground, a nursery and a permanent habitat for many species: the presence of over 400 different seaweeds species and several thousands of animal species makes this underwater habitat a unique biodiversity hotspot (Boudouresque, 2004).
Rhizomes, roots and senescent leaf sheaths of P. oceanica form a characteristic peat-like seabed layer (matte) that can be several meters thick and thousands of years old. Matte are extremely resistant to microbiological decay, however, herbivores can feed on them with the aid of symbiotic microorganisms such as fungi or bacteria capable of degrading organic matters (Vohnik et al., 2017). To this respect, it is important to underline that likewise other seagrasses, P. oceanica supports a complex ecosystem that supplies large amounts of nutrients to the organisms living within and in adjacent waters. Fungi play an important role in the functioning of this ecosystem: while parasites and patho ens can negatively affect the wellness of the meadows, mutualists may be essential for their health (Vohnik et al., 2019). Finally, saprobes transform dead plants into nutritive detritus through their degrading activity (Raghukumar, 2017). Overall, Posidonia forests are home to 20–25% of Mediterranean species and represent the climax community of the Mediterranean Sea (Minelli et al., 2008). However, in the aftermath of the anthropic impact, of the presence of exotic species and of the climate change, this climax community is seriously threatened and since 1992 is a priority in Annex I of the Council Directive 92/43/EEC on the conservation of natural habitats and of wild fauna and flora.
Marine fungi are a huge part of the microbial diversity hosted in the sea. Indeed, they have been retrieved worldwide from biotic and abiotic substrates such as algae, sediments, invertebrates, drift wood, etc. (Jones and Pang, 2012; Garzoli et al., 2014, 2015, 2018; Gnavi et al., 2017; Raghukumar, 2017; Bovio et al., 2018). Nevertheless, despite the total fungal diversity has been estimated to range between 10,000 and 12,500 taxa (Jones et al., 2019), only 1281 species have been described to date1. These organisms are important primary decomposers and living as mutualists (ecto- and endosymbionts), parasites, pathogens and saprobes, contribute greatly to nutrient cycling and food web (Amend et al., 2019; Grossart et al., 2019).
As part of a thorough investigation aimed to uncover the fungal diversity of the Mediterranean Sea, we focused on the isolation and identification of the cultivable mycobiota associated with P. oceanica, collected off the Elba Island (Tuscany). Once defined, the mycobiota was compared to those associated with the green and brown algae Flabellia petiolata (Gnavi et al., 2017) and Padina pavonica (Garzoli et al., 2018), two important components of the same phytocoenosis, collected during the same sampling and in close proximity. Finally, this paper introduces the novel genus Paralulworthia represented by two new species P. gigaspora and P. padinae.
Materials and Methods
Sampling
Plants of P. oceanica were collected from two sampling sites in the coastal waters of the Elba Island (Livorno, Italy): (1) Ghiaie (UTM WGS84 42°49′04″N, 10°19′20″E) and (2) Margidore (UTM WGS84 42°45′29″N, 10°18′24″E). Twelve plants (6 per sampling site) with the surrounding fluctuating senescent leaves (matte) were harvested at depth ranging from 5 to 15 m below sea level (bsl). Plants were maintained in sterile dark containers at 4°C and processed within 36 h from sampling. Specific permissions to operate in the protected area of “Le Ghiaie” (Ghiaie site) and in the freely accessible Margidore site, were obtained by the port authority of Portoferraio (Livorno, Italy).
Fungal Isolation
Each plant was divided into four parts: leaves, rhizomes, roots and matte (Figure 1). In order to remove unrefined sediments, each sample was individually sonicated (30” each time) and serially washed (three times) in sterilized artificial SeaWater (SW, 3.4% w/v Sea Salt mix – Sigma-Aldrich, St. Louis, MO, United States – in distilled water). Individual samples were homogenized in 20 mL of sterile filtered seawater by means of Ultra-Turrax (IKA, Staufen, Germany). One mL of homogenate was plated onto Petri dishes (15 cm Ø) containing the following culture media: Corn Meal Agar SeaWater (CMASW; 17g CMA – Sigma-Aldrich, St. Louis, MO, United States – in 1 L of filtered SW) or Agar Posidonia (AP; 20 g of P. oceanica heated in 100 mL seawater – 60°C, 30 min – were filtered prior to the addition of 18 g agar and SW up to 1L). Each medium was supplemented with antibiotics (Gentamicin 80 mg L–1, Piperacillin and Tazobactam 100 mg L–1 – Sigma-Aldrich, St. Louis, United States).

Figure 1. Posidonia oceanica medows in its natural habitat (A); a plant of P. oceanica with the four plant parts indicated (B).
Three replicates per medium and per sample were performed. A total of 288 plates were incubated at 15°C (spring average temperature of submerged meadows between 5 and 15 m bsl) for 15 days. Afterward, the temperatures was raised to 25°C for 45 days. This procedure allowed the isolation of both psychrophilic and mesophilic fungi. Colony forming units per gram of dry weight of each plant part (CFU g–1 dw) were recorded. Isolated fungi were maintained in pure culture.
Fungal Identification
Fungi were identified by combining morpho-physiological, molecular and phylogenetic studies. Following preliminary determination of genera according to macroscopic and microscopic features (Domsch et al., 1980; von Arx, 1981; Kiffer and Morelet, 1997), fungal isolates were transferred to the media recommended by the authors of selected genus monographs for species identification. Molecular identification was performed by amplifying and sequencing the appropriate genetic marker as previously described (Poli et al., 2016; Garzoli et al., 2018). PCR products were purified and sequenced at Macrogen Europe Laboratory (Madrid, Spain). The resulting ABI chromatograms were visually inspected, trimmed and assembled to obtain consensus sequences using Sequencer 5.0 (GeneCodes Corporation, Ann Arbor, MI, United States)2. The newly generated sequences were compared by BLASTn analyses (default settings) to those available in public nucleotide databases provided by the NCBI (Bethesda MD, United States) and by the Westerdijk Fungal Biodiversity Institute (Utrecht, The Netherlands). Similarity values equal or higher than 98% (e-value > e-100) were considered credible.
Representative strains of each species isolated in pure culture during this work are preserved at Mycotheca Universitatis Taurinensis (MUT)3. The accession numbers of the sequences deposited in GenBank are: MK578237 – MK578251; MK581058 – MK581072; MK626711 – MK626722; MN165499 – MN165504; MN173027 – MN173033; MN177616 – MN177627; MN173595 – MN173610.
Paralulworthia gigaspora and Paralulworthia posidoniae: Growth Condition, Morphological, and Molecular Study
Following pre-growth, MUT 435, MUT 672, MUT 5413, and MUT 5261 were inoculated in triplicate onto Petri dishes (9 cm Ø) containing MEASW and incubated at 21°C. The fungal growth, as well as macro- and microscopic features, was monitored at 4, 7, 14, 21 and 28 days from the inoculum. In an attempt to induce sporulation, sterile specimens of Quercus ruber cork and Pinus pinaster wood (species autochthonous to the Mediterranean area) were placed on 3 weeks old fungal colonies (Panebianco et al., 2002) which were further incubated for 4 weeks at 21°C. Colonized wood specimens were transferred into 50 mL tubes containing 20 mL sterile sea water and incubated at 21°C for 1 month. Mature reproductive fungal structures were observed and images captured with an optical microscope (Leica DM4500B, Leica Microsystems GmbH, Germany) equipped with a camera (Leica DFC320, Leica Microsystems GmbH, Germany). For a better inspection of ascomata growing inside wood tissues, scraps of colonized wood embedded in melted paraffin were dissected with a cryostat (Leica CM1850, Leica Microsystems GmbH, Germany).
Genomic DNA was extracted as mentioned above and the partial sequences of the internal transcribed spacers including the 5.8S rDNA gene (nrITS), 28S large ribosomal subunit (nrLSU) and 18S small ribosomal subunit (nrSSU) were amplified by PCR using the primer pairs ITS1/ITS4 (White et al., 1990), LR0R/LR7 (Vilgalys and Hester, 1990), NS1/NS4 (White et al., 1990). A dataset consisting of nrSSU, nrITS, and nrLSU, was assembled on the basis of BLASTn results and of the most recent phylogenetic study focused on Lulworthiales (Azevedo et al., 2017). Reference sequences were retrieved from GenBank (Table 2). Alignments were generated using MUSCLE (default conditions for gap openings and gap extension penalties), implemented in MEGA v. 7.0 (Molecular Evolutionary Genetics Analysis), visually inspected and trimmed by TrimAl v. 1.24 to delimit and discard ambiguously aligned regions. No incongruence was observed among single-loci phylogenetic trees and alignments were concatenated into a single data matrix with SequenceMatrix (Vaidya et al., 2011). The best evolutionary model under the Akaike Information Criterion (AIC) was determined with jModelTest 2 (Darriba et al., 2012). Phylogenetic inference was estimated using Maximum Likehood (ML) and Bayesian Inference (BI) criteria. The ML analysis was produced using RAxML v. 8.1.2 (Stamatakis, 2014) under GTR + I + G evolutionary model and 1,000 bootstrap replicates. BI was performed with MrBayes 3.2.2 (Ronquist et al., 2012) with the same substitution model (GTR + I + G). The alignment was run for 5 million generations with two independent runs each containing four Markov Chains Monte Carlo (MCMC) and sampling every 100 iterations. The first 25% of generated trees were discarded as “burn-in.” A consensus tree was generated using the “sumt” function of MrBayes and Bayesian posterior probabilities (BYPP) were calculated. Consensus trees were visualized in FigTree v. 1.4.25. Due to topological similarity of the two resulting trees, only BI analysis with BYPP and MLB values was reported.
Statistical Analysis
Differences among the total fungal loads (CFU g–1 dwt) of each plant portion were inferred by applying the analysis of variance (ANOVA), Bonferroni post hoc test (p < 0.05), using GraphPad Prism 5 for Windows, GraphPad Software (San Diego, CA, United States)6.
Significant differences among mycobiota of individual plant parts and organisms were evaluated by applying the PERmutational Multivariate ANalysis Of Variance (PERMANOVA; pseudo-F index; p < 0.05) and visualized by the Canonical Analysis of Principal Coordinates (CAP) or Principal Coordinate Analysis (PCO). The contribution of single species (in percentage) to the diversity observed within and between groups was assessed by SIMilarity PERcentage (SIMPER) analysis. These analyses were performed with the statistical package PRIMER 7 (Plymouth Routines in Multivariate Ecological Research, Albany Auckland, New Zealand).
Results
All plants of P. oceanica were colonized by fungi. The average fungal abundance (CFU g–1dw) of the four parts ranged from 5.0 × 102 CFU g–1dw to 1.4 × 104 CFU g–1dw (Figure 2). In Ghiaie, rhizomes displayed the highest fungal load, followed by matte, leaves and roots, whereas in Margidore leaves were the richest part, followed by rhizomes and matte evenly, and finally by roots. Considering the individual plant parts, only roots did not differ significantly between the two sampling sites.
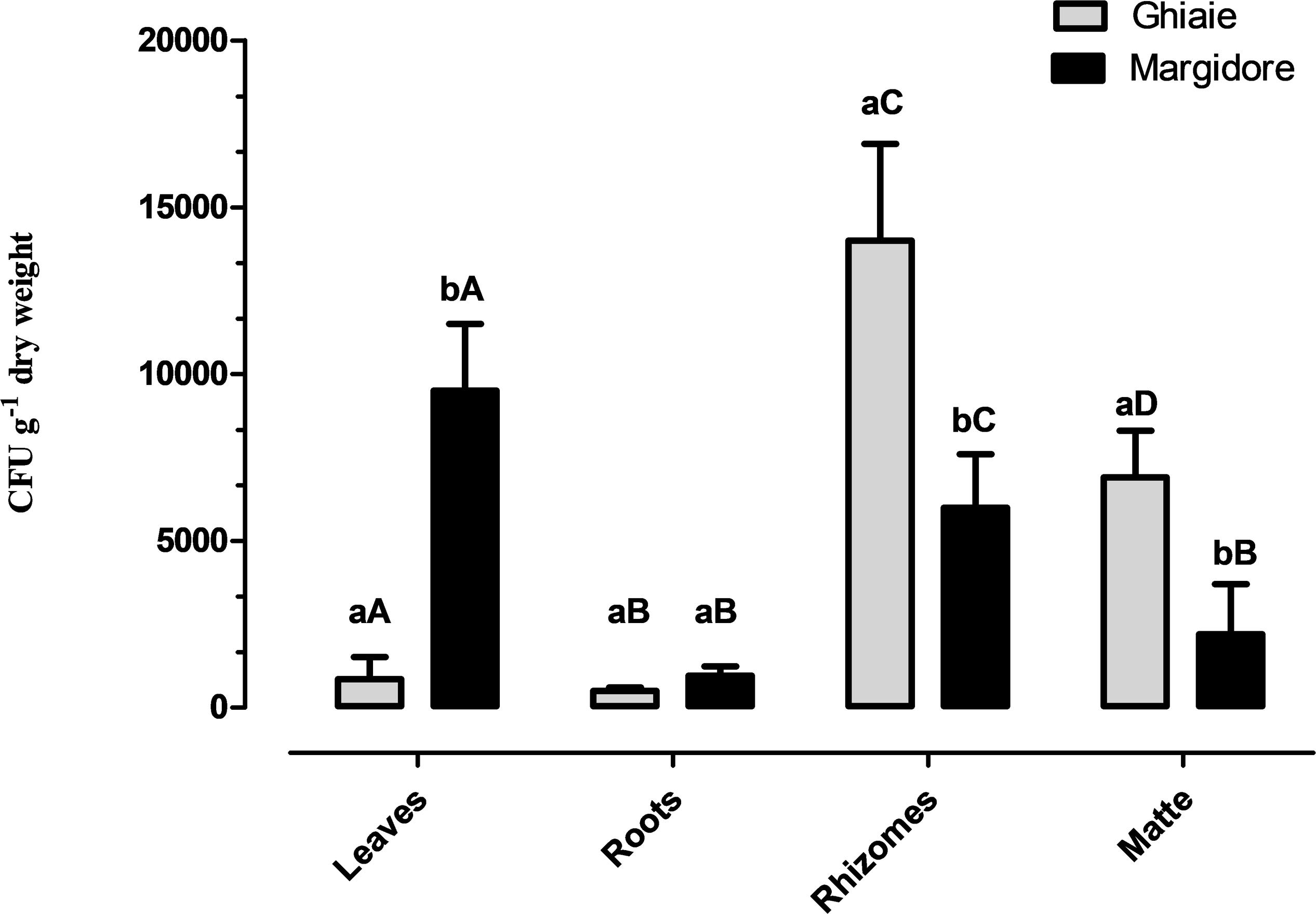
Figure 2. Fungal total load (CFU g– 1 dwt) detected in the four parts of P. oceanica (leaves, roots, rhizomes, and matte) in Ghiaie and Margidore. Results are expressed as mean ± SD and analyzed through two-way analysis of variance (ANOVA), Bonferroni post hoc (p < 0.05). Capital letters indicate significant differences within the same sampling site; lowercase letters indicate significant differences between sampling sites.
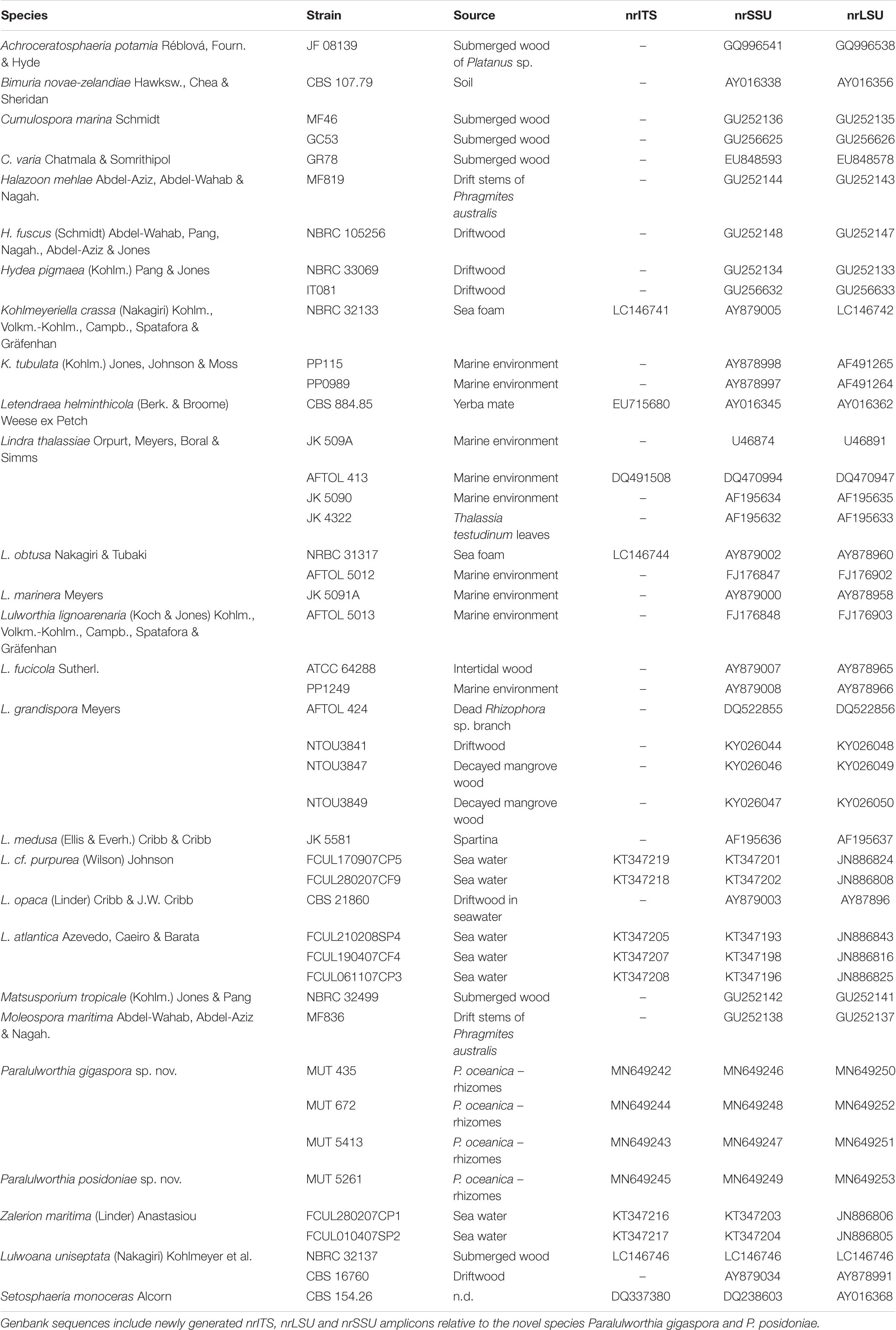
Overall, 287 isolates, representative of 102 taxa were retrieved: 43 from leaves, 70 from matte, 118 from rhizomes, and 56 from roots. The majority of taxa needed a specific medium and incubation temperature: 50 taxa were exclusively isolated from CMASW, 31 from AP and only 21 from both media; 76 taxa grew exclusively at 25°C and 11 at 15°C. In comparison to the plants sampled at Ghiaie, the number of taxa retrieved from the plants harvested at Margidore was almost double (Table 1).
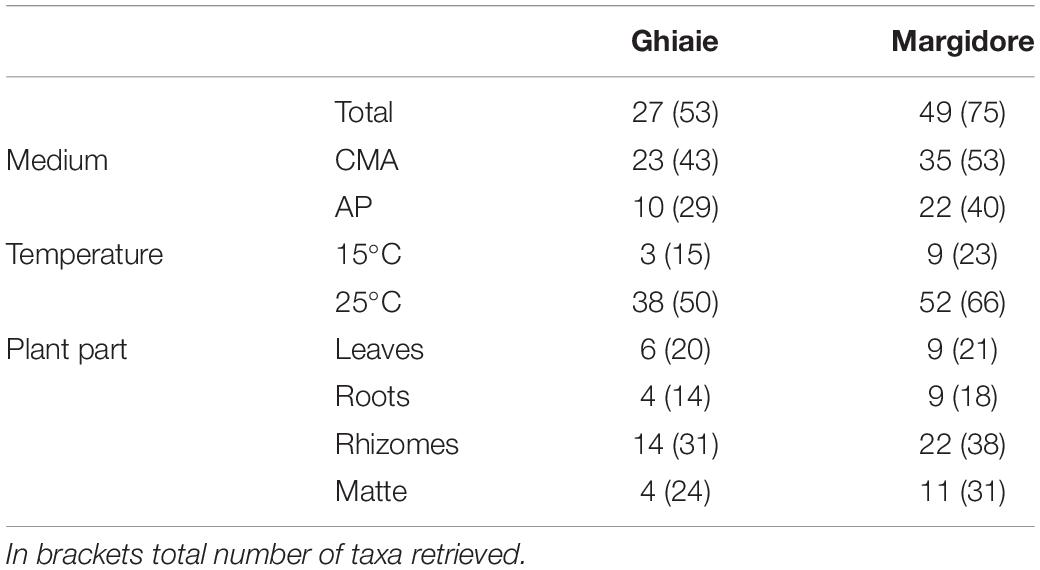
Table 1. Number of exclusive fungal taxa per site (Ghiaie and Margidore), medium (CMA and AP), temperature (15°C and 25°C) and plant part (leaves, roots, rhizomes, and matte).
Sixty-eight percent of the strains were identified at species level and 12% at genus level. Those fungi that remained identified at higher rank were sterile in axenic culture and did not accomplished a univocal molecular result. Most of the taxa belonged to Ascomycota (91) while Basidiomycota (8) and Mucoromycota (3) were scarcely represented (Table 3).
In general, 10 classes, 12 orders and 32 families were represented. With the exception of Leotiomycetes (3%), Ascomycota were homogenously distributed in three classes: Dothideomycetes (30%, mainly Pleosporales and Capnodiales – 18% and 12%, respectively), Sordariomycetes (29%, mainly Hypocreales and Microascales – 13% and 6%, respectively) and Eurotiomycetes (26%, mainly Eurotiales – 24%). As for Basidiomycota, the most represented classes were Agaricomycetes (4%), Tremellomycetes (1%), Ustilaginomycetes (1%) and Wallemiomycetes (1%). The single representative of Mucoromycota was Cunninghamella bertholletiae (Mucorales).
Penicillium and Cladosporium were the most abundant genera with 17 and 9 species, and were isolated from all sites and plant parts, regardless of the culture conditions used.
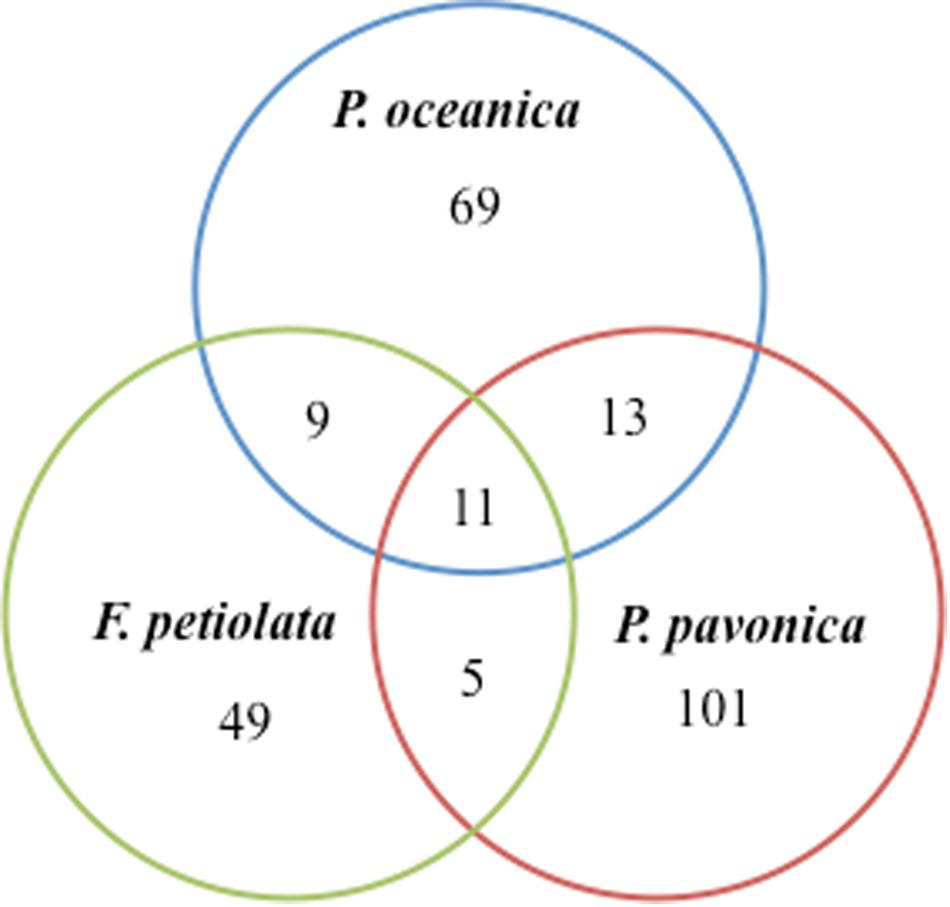
Out of the 102 taxa isolated, only Cladosporium cladosporioides and Penicillium commune were detected in all four plant parts; 7 and 18 taxa were isolated from three and two parts, respectively. The rest was exclusive to rhizomes (32), matte (15), leaves (14), and roots (13) (Figure 3).
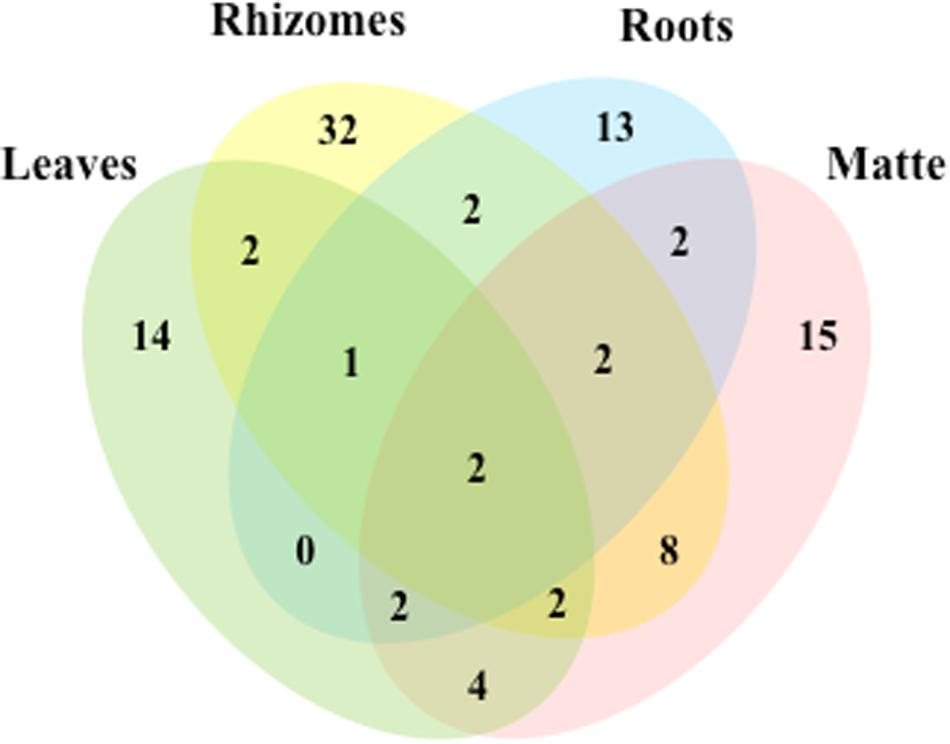
Figure 3. Venn diagram showing the total number of taxa and shared taxa among leaves, rhizomes, roots and matteof P. oceanica.
In terms of fungal species diversity, plant parts were significantly different (PERMANOVA; p ≤ 0.05; Figure 4), with the exception of leaves vs matte (PERMANOVA; p = 0.06). The most frequent species were Penicillium chrysogenum (72.5%) in leaves, Corollospora sp. (39.8%), Corollospora maritima (21.0%) and Penicillium chrysogenum (9.25%) in matte, Penicillium antarcticum (28.1%), Lulworthiales sp. (24.1%), Aspergillus sydowii (14.5%) and Microascales sp. (14.1%) in rhizomes, and finally Lulworthiales sp. (59.0%) and Penicillium antarcticum (23.8%) in roots (SIMPER analysis). No significant difference (PERMANOVA; p = 0.471) was observed between sampling sites.
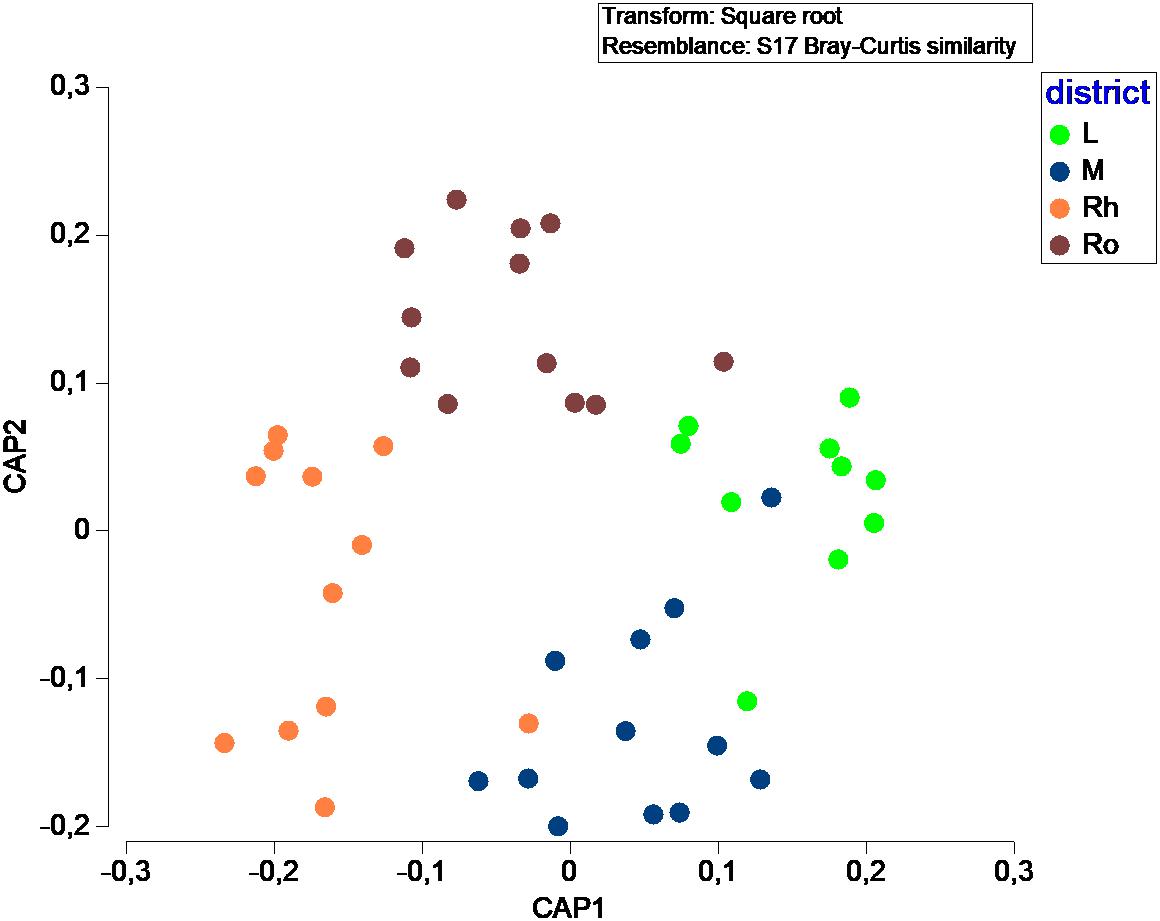
Figure 4. Canonical Analysis of Principal coordinates (CAP) illustrating the diversity of fungal communities among plant parts.
The mycobiota of P. oceanica included four strains of Lulworthiaceae (MUT 435, MUT 672, MUT 5413, and MUT 5261) that are representatives of a novel genus consisting of two species. The phylogenetic tree based on nrITS, nrSSU, and nrLSU (Figure 5), together with the morphological inspection confirmed the novelty of Paralulworthia gigaspora sp. nov. and of Paralulworthia posidoniae sp. nov.
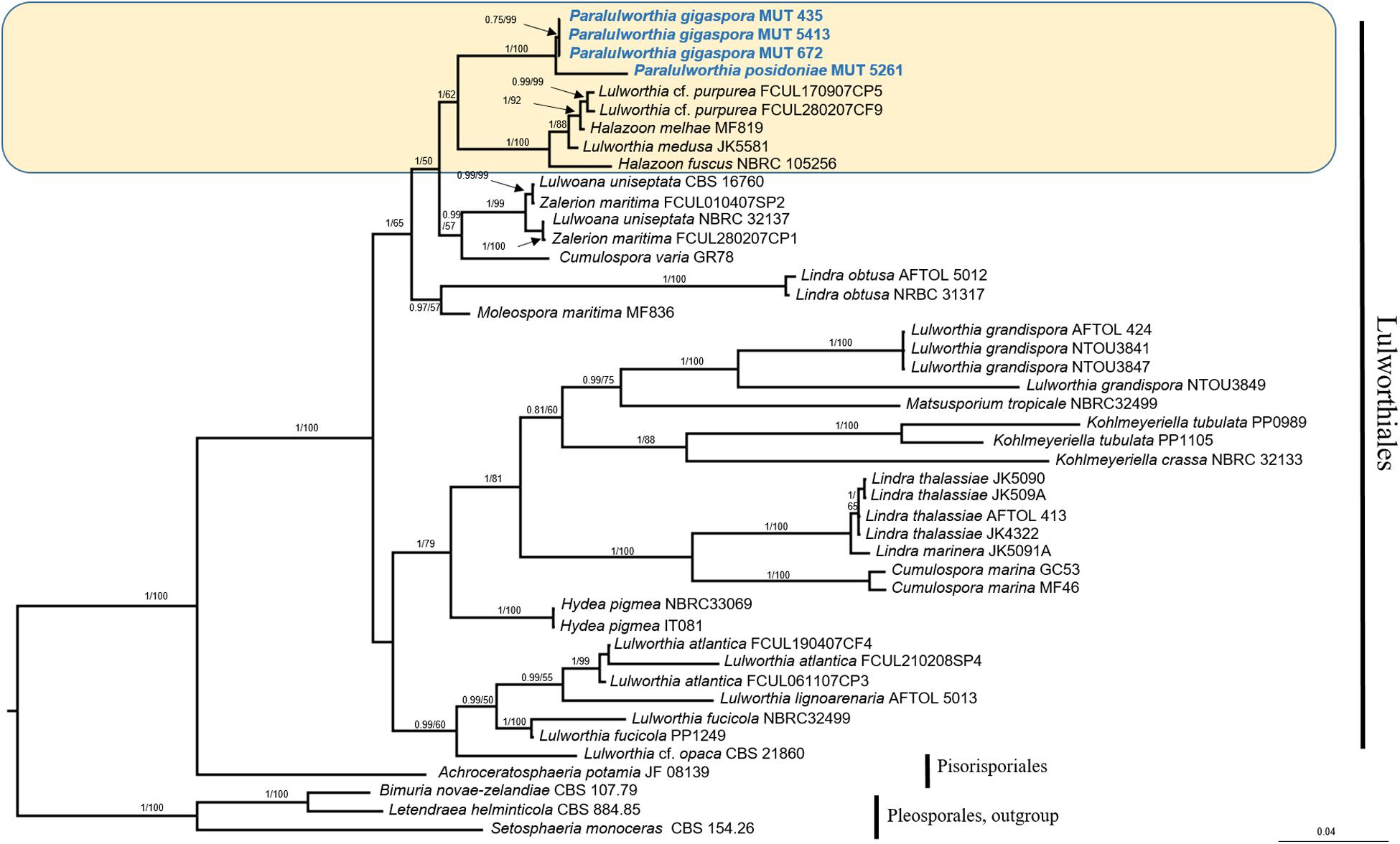
Figure 5. Bayesian phylogram of Lulworthiales based on a combined nrSSU, nrITS, and nrLSU dataset. The tree is rooted to Pleosporales. Branch numbers indicate BYPP/MLBvalues; Bar = expected changes per site (0.04).
Phylogenetic Inference and Taxonomy of Paralulworthia gigaspora and Paralulworthia posidoniae
Preliminary analyses carried out individually with nrITS, nrSSU, and nrLSU denoted no incongruence in the topology of the trees. The combined dataset, built on the basis of BLASTn results and of recent phylogenetic studies (Azevedo et al., 2017), consisted of 45 taxa (including MUT isolates) that represented 15 genera and 27 species. Twelve sequences (4 nrITS, 4 nrSSU, and 4 nrLSU) were newly generated, while 94 were retrieved from GenBank.
The combined dataset had an aligned length of 2135 characters, of which 1096 were constant, 352 were parsimony-uninformative and 687 parsimony informative.
The strains under investigation formed a strongly supported and separated clade (BYPP = 1.00; MLB = 100%) within Lulworthiaceae, unique family of the order Lulworthiales (Figure 5). Since this clade was close to the genus Lulworthia sensu latu, we introduced the novel genus Paralulworthia that includeed the two species Paralulworthia gigaspora and Paralulworthia posidoniae. The development of the sexual morphotype of MUT 435 and MUT 5261 on maritime pine wood confirmed the divergence between the two species.
Paralulworthia gen. nov.
MB 833285.
Type species: Paralulworthia gigaspora sp. nov.
Etymology: in reference to the phylogenetic closeness to the genus Lulworthia sensu latu.
Phylogenetic placement: Lulworthiaceae, Lulworthiales, Sordariomycetes, Ascomycota. The genus Paralulworthia gen. nov. clustered together with the genera Lulworthia sensu latu and Halazoon (Figure 5).
Description: growing efficiently on wood. Ascomata subglobose to ellipsoidal, single, scattered, immersed in the cortex or erumpent, brown to black at maturity; peridium dark brown composed of several layers of thick–walled cells of textura angularis. Asci containing 8 spores, dehiscent. Sterile elements not observed. Ascospores uni- to 3-septate at maturity, constricted at septa (particularly the central one), ellipsoid to fusiform, pointed at both ends, thin-walled, from light to dark brown at maturity, straight.
Paralulworthia gigaspora sp. nov.
MB 833288 (Figure 6).
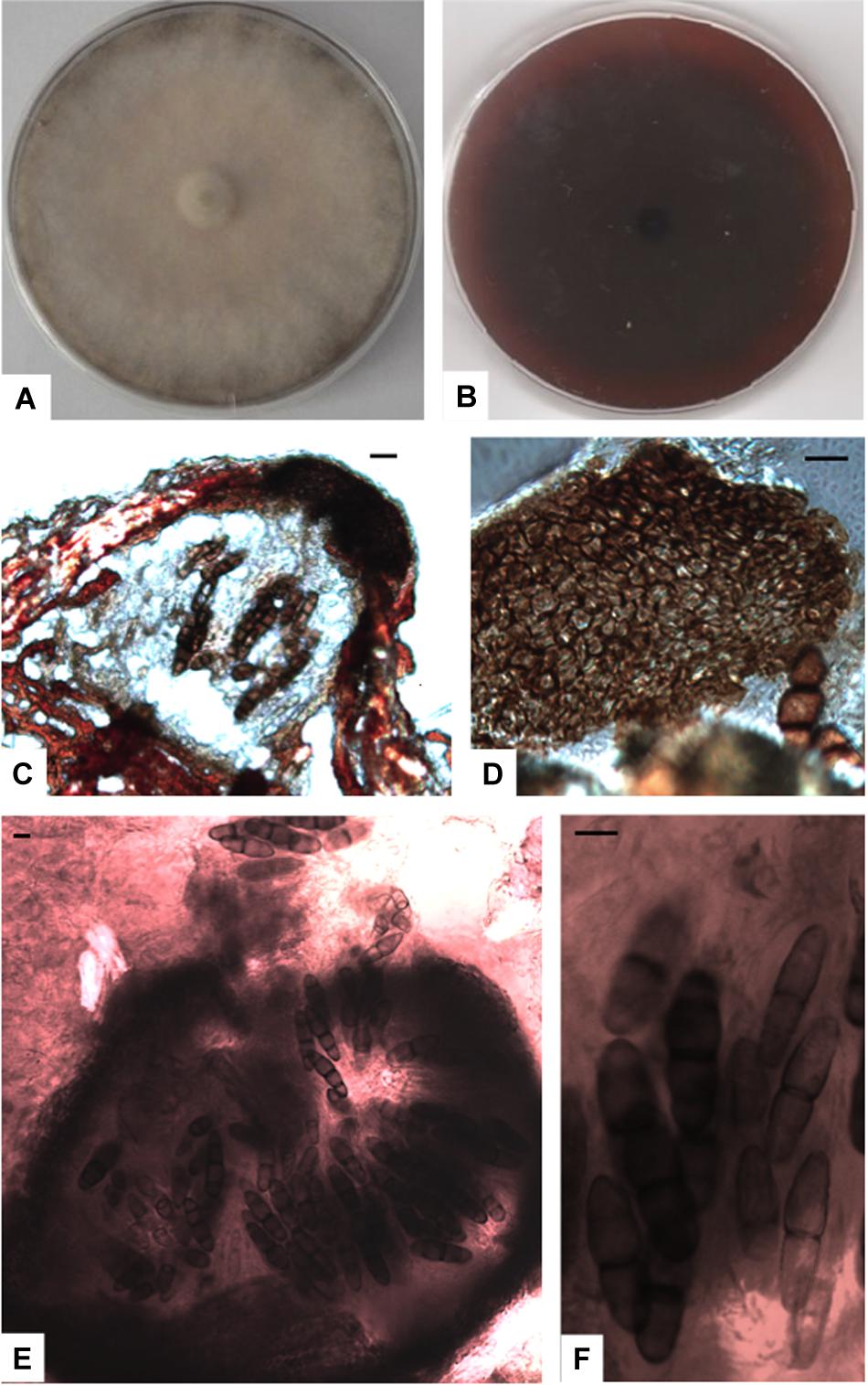
Figure 6. Paralulworthia gigaspora sp.nov. 28-days-old colony incubated at 21°C on MEASW (A) and reverse (B); ascomata (C,E), peridium with textura angularis (D); asci with eight ascospores at different degrees of maturity (F). Scale bars 20 μm.
Type: Europe, Italy, Tuscany, Mediterranean Sea, Elba Island (LI), Ghiaie ISL, 3–5 m depth, 42°49′04”N, 10°19′20″E March 2010, R. Mussat-Sartor and N. Nurra, on the sea grass P. oceanica rhizomes, MUT 435 holotype, living culture permanently preserved in metabolically inactively state by deep-freezing at MUT. MUT 672 and MUT 5413 = paratypes.
Etymology: in reference to the large size of the ascospores.
Description: growing actively on P. pinaster. Hyphae 3.2 μm wide, septate, from brownish to hyaline. Chlamydospores numerous, solitary and multicellular, from one to four celled, 2 × 3 μm diam., from light to dark brown, globose and subglobose.
Sexual morph: Ascomata 480-540 × 160–270 μm ( = 506.7 × 205 μm, n = 10), subglobose to ellipsoidal, single, scattered, immersed in the cortex or erumpent, brown to black at maturity; peridium dark brown 73-88 μm thick composed of several layers of thick–walled cells of textura angularis. Asci 115–130 × 30–40 μm ( = 118 × 31.8 μm, n = 10), fusiform to cylindrical, containing 8 spores, dehiscent, hyaline, vertically oriented to the host surface. Sterile elements not observed. Ascospores 76-83 × 19-25 μm ( = 79.3 × 21.7 μm, n = 10), uni- to 3-septate at maturity, constricted at septa (particularly the central one), ellipsoid to fusiform, pointed at both ends, thin-walled, from light to dark brown at maturity, straight.
Asexual morph not observed.
Colony description: growing on MEASW from 60 to 90 mm Ø after 28 days at 21°C, mycelium from feltrose to floccose, from whitish to ochre, reverse from light brown to bordeaux. A caramel colored diffusible pigment and exudates were often present.
Paralulworthia posidoniae sp. nov.
MB 833291 (Figure 7).
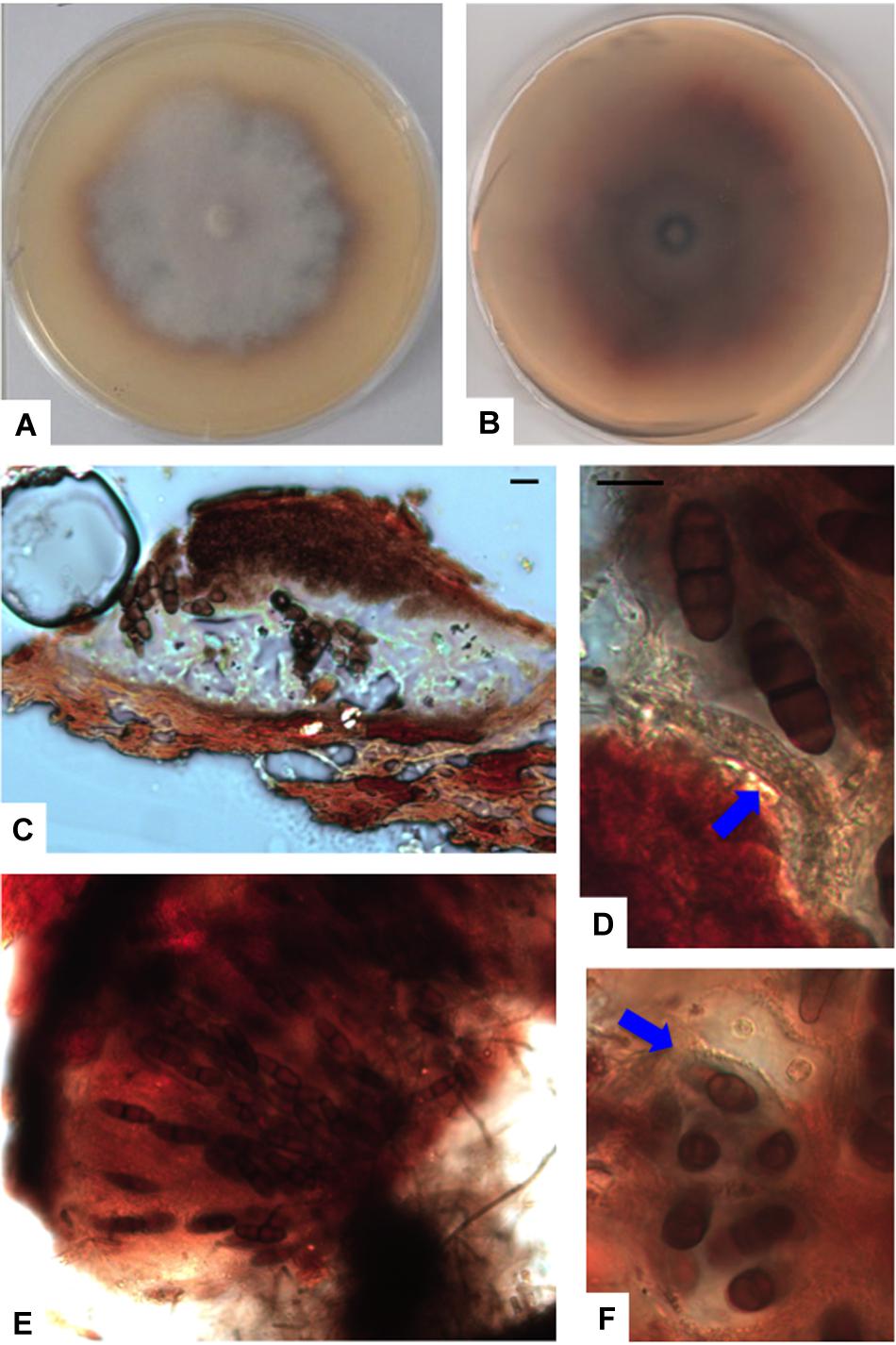
Figure 7. Paralulworthia posidoniae sp.nov. 28-days-old colony incubated at 21°C on MEASW (A) and reverse (B); ascoma (C), clavate young ascus (D, red arrow); mature asci in the ascoma (E), transversal section of asci with ascospores (F, red arrow). Scale bars 20 μm (C,E,F), 10 μm (D).
Type: Europe, Italy, Tuscany, Mediterranean Sea, Elba Island (LI), Ghiaie ISL, 3–5 m depth, 42°49′04″N, 10°19′20″E March 2010, R. Mussat-Sartor and N. Nurra, on the sea grass P.oceanica rhizomes, MUT 5261 holotype, living culture permanently preserved in metabolically inactively state by deep-freezing at MUT.
Etymology: in reference to the original substrate, the seagrass P. oceanica.
Description: growing actively on P. pinaster. Hyphae 2.5–3.5 μm ( = 2.9 μm, n = 10) wide, septate, from hyaline to dematiaceous.
Sexual morph: Ascomata 365–450 × 155–230 μm ( = 405 × 191.3 μm, n = 10), ellipsoidal, single, scattered, immersed in the cortex or erumpent, brown to black at maturity; peridium dark brown 23-50 μm thick composed of several layers of thick-walled cells of textura angularis. Asci 150–190 × 40–60 μm ( = 171.3 × 52 μm, n = 10), containing 8 spores, clavate and early dehiscent. Sterile elements not observed. Ascospores oblong, 32–43 × 12–16 μm ( = 38.4 × 13.2 μm, n = 10), uni- to 3-septate at maturity, constricted at septa (particularly the central one), ellipsoid to fusiform, pointed at both ends, thin-walled, from light to dark brown at maturity, straight.
Asexual morph not observed.
Colony description: growing on MEASW, reaching 42 mm Ø after 28 days at 21°C, mycelium feltrose with irregular edges, whitish with slight shades of purple in the middle, gray-green at the edges; reverse caramel. A caramel colored diffusible pigment and exudates were often present.
Discussion
Broadening the knowledge of marine fungi associated with a vast range of biotic and abiotic substrates is becoming more and more important. The reason behind this lies in the ecological relevance that these organisms may play in the oceans and in their possible exploitation as a source of potentially novel enzymes and bioactive compounds (Bovio et al., 2019; Grossart et al., 2019; Overy et al., 2019; Youssef et al., 2019).
Nowadays, P. oceanica is rapidly disappearing from the Mediterranean Sea, being seriously threatened by a number of factors7. Its mycobiota was partially studied in the past (Cuomo et al., 1985; Panno et al., 2013). Recently Vohnik and collaborators described the endophytes colonizing the roots of this endemic seagrass (Vohnik et al., 2017, 2019). Here, we investigated the cultivable mycobiota of P. oceanica with the intent of comparing it with the fungal communities of two photobionts of the same phytocoenosis.
Mycobiota of P. oceanica
The observed total fungal load ranged from 5 × 102 to 1.4 × 104 CFU g–1 dw, approximately one order of magnitude higher than what reported by Panno et al. (2013) (up to 1.6 × 103 CFU g–1 dw). Since the two sampling occurred in the same period of the year (March), we hypothesize that the climate factor did not affect the total load. The fungal abundance through the plant parts was also slightly different: in the present study, the fungal load followed the distribution rhizomes > leaves > matte > roots, whereas in the previous investigation matte and leaves were swapped (Panno et al., 2013). The use of specific culture media and of two incubation temperatures maximized the number of taxa isolated. Indeed, a lower temperature (15°C) allowed the growth of psychrotolerant/psychrophilic fungi such as four species of Penicillium (P. canescens, P. glabrum, P. paneum, P. vinaceum) (Table 2). Most probably, this is one of the reasons why we retrieved a number of taxa higher than what reported in 2013 by Panno (102 vs. 88).
As expected, Ascomycota prevailed, confirming once again the dominance of this phylum in the oceans8. In detail, Penicillium and Cladosporium, were widely represented in all plant parts and sampling sites, and were characterized by the highest number of species (17 and 9, respectively). Members of Penicillium and Cladosporium are commonly reported in marine environments worldwide, probably due to the high adaptation to the peculiar chemical-physical conditions (Raghukumar, 2017). On the other side, it is not unreasonable to relate the high abundance of these genera to their extraordinary sporulation rate that may end in an overestimation of the substrate colonization (Imhoff et al., 2011).
Interestingly, 37 species were retrieved for the first time from the Mediterranean Sea and 61 from P. oceanica (Table 2), while only 19 species were previously detected in P. oceanica (Cuomo et al., 1985; Panno et al., 2013; Gnavi et al., 2014). Several species, besides being isolated for the first time from the Mediterranean Sea and from P. oceanica, were never found in any marine habitat before. For instance, this is the case of Alfaria dandenongensis, Aspergillus pseudoglaucus, Cerrena unicolor, Didymella fabae, and Neoroussoella solani. A. pseudoglaucus is an opportunistic human pathogen (Siqueira et al., 2018) as well as N. solani, a causal agent of keratomycosis (Mochizuki et al., 2017), while their underwater ecological role is still obscure. On the contrary, we can hypothesize the role of the plant pathogen D. fabae and of the leaf litter saprobe A. dandenongensis9. Similarly, it is possible to assume an active role for white rot fungi (i.e., Bjerkandera adusta, C. unicolor and Schizophyllum commune) in transforming recalcitrant matrixes (Poli et al., 2018). Finally, entomopathogenic fungi like Cordyceps farinosa, due to their ability to degrade chitin, may be capable of attacking the shellfish carapace.
Distribution Among Plant Parts
The mycobiota of P. oceanica changed significantly among the four parts of the plant, with the exception of leaves vs. matte. This last observation could be ascribable to the similar structure of the substrates: the main components of matte are the decaying and fluctuating leaves (Vohnik et al., 2017). The presence of antagonistic organisms, both micro- and macro-, together with the specific localization of toxic metabolites, such as tannic acid in the leaves (Pergent et al., 2008), may affect the distribution among plant parts. It is not unexpected to retrieve unique mycobiota on differently organized and composed parts of a complex plant, as occurs in terrestrial phanerogams.
The only two species common to the four regions were C. cladosporioides and P. commune: the former is a usual inhabitant of the sea (Garzoli et al., 2015), the latter, although observed for the first time on P. oceanica, was already found associated with F. petiolata (Gnavi et al., 2017). P. chrysogenum dominated the mycobiota of the leaves, whereas the marine genus Corollospora prevailed in matte. With respect to rhizomes and roots, two taxa (Lulworthiales sp. and P. antarcticum) were the main components of the fungal communities, even though their proportions were different in the two plant parts. Vohník et al. (2016) found members of the order Lulworthiales in the rhizomes and roots of P. oceanica. Lulworthiales degrade wood and marsh plants in marine and estuarine environments and seem to be the most common seagrasses decomposers followed by Corollospora maritima. Indeed, C. maritima and Lulworthiales spp., are well-known cellulases producers and can break down complex ligno-cellulose compounds, making them available for other organisms nourishment (Raghukumar, 2017). Interestingly, two new species of Lulworthiales were found in rhizomes and described here for the first time.
The retrieval of P. chrysogenum in leaves and, to a lower extent in matte, may be related to defense mechanisms against predators and/or pathogens, as demonstrated in terrestrial plants, where the presence of this fungus stimulates the synthesis of secondary metabolites such as salycilic and jasmonic acids (Chen et al., 2018).
The Mycobiota of P. oceanica Compared With the Fungal Communities of F. petiolata and P. pavonica
F. petiolata and P. pavonica are two algae that characterize the Neptune forests. Since their mycobiota were recently described (Gnavi et al., 2017; Garzoli et al., 2018), we considered worthwhile to compare the fungal communities associated with these three organisms that grow only few centimeters apart. To this end, one must bear in mind that samplings occurred at the same time and in the same sites (Ghiaie and Margidore): it would be interesting to clarify whether a strict specificity between fungi and photobionts exists.
In terms of number of taxa, P. pavonica displayed the richest mycoflora, followed by P. oceanica and finally by F. petiolata, where the number of taxa was almost halved. As shown in Figure 8, P. oceanica and the two algae had 11 species in common: Acrostalagmus luteoalbus, Arthrinium arundinis, Cladosporium allicinum, C. cladosporioides, C. sphaerospermum, Gibellulopsis nigrescens, Massarina rubi, P. antarcticum, P. brevicompactum, P. roseopurpureum, and S. commune. Besides their broad diffusion in the oceans, these fungi, due to the secretion of secondary metabolites (e.g., antialgal, antifungal, and/or antibacterial) may help in deterring colonization of their hosts by other microbes, and in warding off predators (Raghukumar, 2017). On the other side, the three substrates show specific mycobiota, significantly different from each other (Figure 9), with high and equivalent percentages of dissimilarity (94.68–95.74%). The uniqueness of each community derives from the prevalence of exclusive taxa (Figure 8).
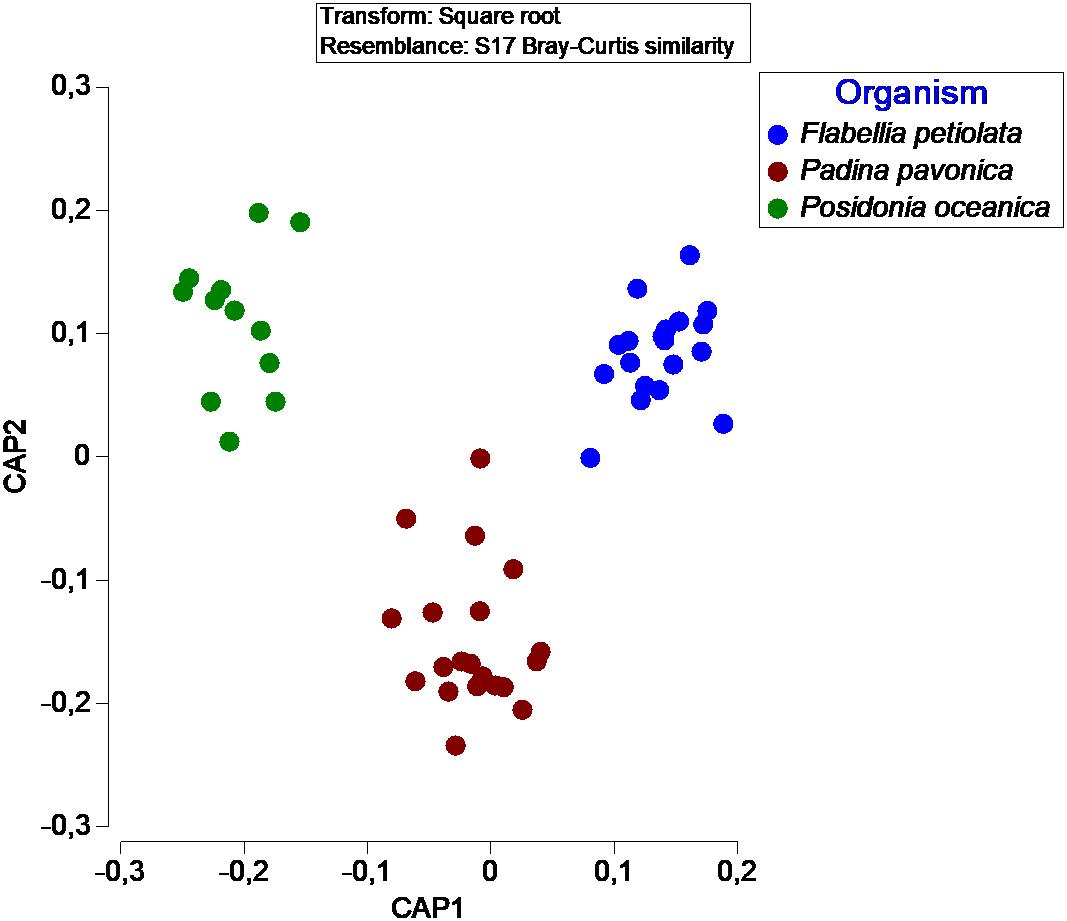
Figure 9. Canonical Analysis of Principal coordinates (CAP) illustrating the diversity of fungal communities associated with different organisms.
Confirming the “substrate specificity” is also the fact that there is a significant difference between the mycobiota of the three organisms even if the “non-photosynthesizing” parts of P. oceanica were excluded from the analysis. The leaves of P. oceanica are in fact characterized by a mycobiota significantly different from that of the other two photobionts (data not shown).
This divergence in “substrate specificity” may be due to the different composition and morphology of the hosts (e.g., P. pavonica with its hairy surface could entrap a high number of fungal propagules, contrary to the slippery texture of F. petiolata) and/or to the different metabolites produced by seagrasses and algae. Extracts of P. oceanica were proved active against bacteria, dermatophytes, and yeasts (Bernard and Pesando, 1989) as well as against Trypanosoma brucei rhodesiense and Leishmania donovani (Orhan et al., 2006). Likewise, the activity of P. pavonica extracts against bacteria and fungi was comparable to the efficiency showed by antibiotics and antimycotics used as control (Al-Enazi et al., 2018). Antibacterial, antiviral, antimitotic and antifungal properties were also detected in raw extracts of F. petiolata (Ballesteros et al., 1992). However, whether the macro-organisms, their microbiota, or the holobiont derived from their strict interaction is responsible for the production of the active compounds is still unclear. With the exclusion of cosmopolitan and/or widespread fungi, we can hypothesize the shaping of a relation between the microorganisms and their host, as recently demonstrated for three Atlantic sponges (Bovio et al., 2018). The establishment of a holobiont depends on both active (selection) and passive (drift) events. Due to the high connectivity of aquatic environments, differences in microbiota may be dependent on a combination of selection and drift. In most cases, fungi spread by passive dispersion of hyphal fragments and/or propagules that happen to adhere to the host. Their further development is consequent to the compatibility and interdependence between the individual organisms. In other words, the holobiont is an organism with its own features, modeled upon a mutual selection. The unimportance of the sampling site supports this idea. Ghiaie and Margidore are two extremely different sites both geographically (Ligurian Sea and Tyrrhenian Sea, respectively) and for the anthropic influence: while the former is located in a marine protected area, the latter is subjected to an intense anthropic disturbance. Nevertheless, no significant difference was detected in terms of fungal biodiversity.
An often-underappreciated ecological function of holobionts is their ability to restrain and nourish rare species: the host provides a niche favorable for the growth of specific communities different from those of the surrounding environment (Dittami et al., 2019). To this regard, we may define “rare” the two novel species here introduced.
Paralulworthia gigaspora and P. posidoniae
This investigation introduces two new species that belong to a novel genus of Lulworthiaceae, the sole family of the order Lulworthiales (Sordariomycetes). Lulworthiaceae spp. are strictly marine species found on a range of substrates (e.g., submerged wood, seaweed, seafoam, etc.). This group includes the following genera: Cumulospora, Halazoon, Hydea, Kohlmeyerella, Lulwoana, Lulworthia, Lindra, Matsusporium, and Moleospora. The new genus Paralulworthia accommodates the four strains MUT 435, MUT 672, MUT 5413, and MUT 5261. P. gigaspora and P. posidoniae formed a strongly supported cluster segregated from the genus Lulworthia sensu latu. The previously suggested polyphyletic nature of Lulworthia spp. (Azevedo et al., 2017), that discriminate Lulworthia sensu latu from Lulworthia sensu strictu is here confirmed and justifies the introduction of the new genus Paralulworthia. Besides, members of the genus Lulworthia are morphologically characterized by filamentous hyaline ascospores (Kohlmeyer et al., 2000), well distinct from the ellipsoidal brown ascospores of the new genus. Furthermore, beyond the phylogenetic distance, the dimension of the ascomata and ascospores allowed the recognition of two species. P. gigaspora and P. posidoniae were isolated exclusively from rhizomes that are charachterized by a high content of lignin (Kaal et al., 2016). Reproductive structures were detected only following colonization of P. pinaster wood specimens submerged in sea-water. These two observations indicate a possible lignicolous nature of these species.
Conclusion
The seagrass P. oceanica is home to a huge variety of fungi. The comparison of our data with those previously obtained on two algae collected in the same phytocoenosis underlines a substrate-specificity. Besides, we demonstrated a plant-part-specificity of the fungal communities associated with the host. Marine phanerogames, likewise their terrestrial counterparts, enroll distinct microbiomes in different regions (i.e., phylloplane, rhizoplane etc.). This selective distribution seems to increment the resistance of plant to pathogens and predators by producing chemo-attractants and anti-microbial compounds. This is even more important for Posidonia meadows that are seriously threatened by climate changes, allochthonous species and anthropic activities. Nonetheless, further studies will need to clarify the mechanisms (e.g., molecular, physiological, etc.) at the basis of the specific interaction between microorganisms and hosts responsible for the shaping of a holobiont.
Data Availability Statement
The sequences generated in this study can be found in GenBank. The accession numbers of the sequences deposited in GenBank are: MK578237–MK578251; MK581058–MK581072; MK626711–MK626722; MN165499–MN165504; MN173027–MN173033; MN177616–MN177627; MN173595–MN173610.
Author Contributions
AP conducted molecular studies, phylogenesis, data analysis, and manuscript writing. EB and LR carried morphological analysis. VP was responsible for morphological analysis and manuscript writing. GV supervised the work.
Funding
This study was funded by University of Torino (ex 60%) and Fondazione CRT (Turin, Italy).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the non-profit cooperative society Pelagosphera s.c.r.l. for collecting the seagrass samples.
Footnotes
- ^ www.marinefungi.org
- ^ http://www.genecodes.com
- ^ www.mut.unito.it
- ^ http://trimal.cgenomics.org
- ^ http://tree.bio.ed.ac.uk/software/figtree
- ^ www.graphpad.com
- ^ www.eea.europa.eu/data-and-maps/figures/distribution-of-the-marine-angiosperm-posidonia-oceanica-and-zostera-sp-in-the-mediterranean
- ^ www.marinefungi.org
- ^ www.FUNGuild.org
References
Abdel-Wahab, M. A., Nagahama, T., and Abdel-Aziz, F. A. (2009). Two new Corollospora species and one new anamorph based on morphological and molecular data. Mycoscience 50, 147–155. doi: 10.1007/s10267-008-0466-9
Al-Enazi, N. M., Awaad, A. S., Zain, M. E., and Alqasoumi, S. I. (2018). Antimicrobial, antioxidant and anticancer activities of Laurencia catarinensis, Laurencia majuscula and Padina pavonica extracts. Saudi Pharm. J. 26, 44–52. doi: 10.1016/j.jsps.2017.11.001
Amend, A., Burgaud, G., Cunliffe, M., Edgcomb, V. P., Ettinger, C. L., Gutierrez, M. H., et al. (2019). Fungi in the marine environment: open questions and unsolved problems. mBio 10:e01189-18. doi: 10.1128/mBio.01189-18
Arnaud-Haond, S., Duarte, C. M., Diaz-Almela, E., Marba, N., Sintes, T., and Serrao, E. A. (2012). Implications of extreme life span in clonal organisms: millenary clones in meadows of the threatened seagrass Posidonia oceanica. PLoS One 7:e30454. doi: 10.1371/journal.pone.0030454
Azevedo, E., Barata, M., Marques, M. I., and Caeiro, M. F. (2017). Lulworthia atlantica: a new species supported by molecular phylogeny and morphological analysis. Mycologia 109, 287–295. doi: 10.1080/00275514.2017.1302255
Ballesteros, E., Martin, D., and Uriz, M. J. (1992). Biological-activity of extracts from some mediterranean macrophytes. Bot. Mar. 35, 481–485. doi: 10.1515/botm.1992.35.6.481
Bernard, P., and Pesando, D. (1989). Antibacterial and antifungal activity of extracts from the rhizomes of the mediterranean seagrass Posidonia oceanica (L) Delile. Bot. Mar. 32, 85–88. doi: 10.1515/botm.1989.32.2.85
Boudouresque, C. F. (2004). Marine biodiversity in the Mediterranean: status of species, populations and communities. Trav. Sci. Parc Nat. Port Cros 20, 97–146.
Bovio, E., Garzoli, L., Poli, A., Luganini, A., Villa, P., Musumeci, R., et al. (2019). Marine fungi from the sponge Grantia compressa: biodiversity, chemodiversity, and biotechnological potential. Mar. Drugs 17:220. doi: 10.3390/md17040220
Bovio, E., Garzoli, L., Poli, A., Prigione, V., Firsova, D., McCormack, G., et al. (2018). The culturable mycobiota associated with three Atlantic sponges, including two new species: Thelebolus balaustiformis and T. spongiae. Fungal Syst. Evol. 1, 141–167. doi: 10.3114/fuse.2018.01.07
Bovio, E., Gnavi, G., Prigione, V., Spina, F., Denaro, R., Yakimov, M., et al. (2017). The culturable mycobiota of a Mediterranean marine site after an oil spill: isolation, identification and potential application in bioremediation. Sci. Total Environ. 576, 310–318. doi: 10.1016/j.scitotenv.2016.10.064
Chen, Z. Z., Wang, J. G., Li, Y., Zhong, Y., Liao, J. G., Lu, S. G., et al. (2018). Dry mycelium of Penicillium chrysogenum activates defense via gene regulation of salicylic acid and jasmonic acid signaling in Arabidopsis. Physiol. Mol. Plant Pathol. 103, 54–61. doi: 10.1016/j.pmpp.2018.04.006
Cuomo, V., Jones, E. B. G., and Grasso, S. (1988). Occurrence and distribution of marine fungi along the coast of the Mediterranean sea. Prog. Oceanogr. 21, 189–200. doi: 10.1016/0079-6611(88)90038-9
Cuomo, V., Vanzanella, F., Fresi, E., Cinelli, F., and Mazzella, L. (1985). Fungal flora of Posidonia oceanica and its ecological significace. Trans. Br. Mycol. Soc. 84, 35–40. doi: 10.1016/s0007-1536(85)80217-5
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Dittami, S. M., Arboleda, E., Auguet, J.-C., Bigalke, A., Briand, E., Cardenas, P., et al. (2019). A community perspective on the concept of marine holobionts: current status, challenges, and future directions. PeerJ 7:e27519v3.
Domsch, K. H., Gams, W., and Anderson, T. H. (1980). Compendium of Soil Fungi. London: Academic Press.
Furtado, S., and Jones, E. (1980). The colonization of selected naturally durable timbers by marine fungi and borers. Bull. Liaison Com. Int. Perm. Rech. Pres. Mat. Mil. Mar. 8, 63–93.
Garzoli, L., Gnavi, G., Tamma, F., Tosi, S., Varese, G. C., and Picco, A. M. (2015). Sink or swim: updated knowledge on marine fungi associated with wood substrates in the Mediterranean sea and hints about their potential to remediate hydrocarbons. Prog. Oceanogr. 137, 140–148. doi: 10.1016/j.pocean.2015.05.028
Garzoli, L., Gnavi, G., Varese, G. C., and Picco, A. M. (2014). Mycobiota associated with the rhodophyte alien species Asparagopsis taxiformis (Delile) trevisan de saint-leon in the Mediterranean sea. Mar. Ecol. Evol. Perspect. 36, 959–968. doi: 10.1111/maec.12189
Garzoli, L., Poli, A., Prigione, V., Gnavi, G., and Varese, G. C. (2018). Peacock’s tail with a fungal cocktail: first assessment of the mycobiota associated with the brown alga Padina pavonica. Fungal Ecol. 35, 87–97. doi: 10.1016/j.funeco.2018.05.005
Gnavi, G., Ercole, E., Panno, L., Vizzini, A., and Varese, G. C. (2014). Dothideomycetes and leotiomycetes sterile mycelia isolated from the Italian seagrass Posidonia oceanica based on rDNA data. Springerplus 3:508. doi: 10.1186/2193-1801-3-508
Gnavi, G., Garzoli, L., Poli, A., Prigione, V., Burgaud, G., and Varese, G. C. (2017). The culturable mycobiota of Flabellia petiolata: first survey of marine fungi associated to a Mediterranean green alga. PLoS One 12:e0175941. doi: 10.1371/journal.pone.0175941
Grasso, S., Laferla, R., and Jones, E. B. G. (1985). Lignicolous marine fungi in a harbor environment (Milazzo). Bot. Mar. 28, 259–264. doi: 10.1515/botm.1985.28.6.259
Grasso, S., Panebianco, C., and Laferla, R. (1990). Lignicolous marine fungi in the straits of Messina, Italy. Hydrobiologia 206, 149–154. doi: 10.1007/bf00018641
Greco, G., Capello, M., Cecchi, G., Cutroneo, L., Di Piazza, S., and Zotti, M. (2017). Another possible risk for the Mediterranean sea? Aspergillus sydowii discovered in the Port of Genoa (Ligurian sea, Italy). Mar. Pollut. Bull. 122, 470–474. doi: 10.1016/j.marpolbul.2017.06.058
Grossart, H. P., Van den Wyngaert, S., Kagami, M., Wurzbacher, C., Cunliffe, M., and Rojas-Jimenz, K. (2019). Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 17, 339–354. doi: 10.1038/s41579-019-0175-8
Imhoff, J. F., Labes, A., and Wiese, J. (2011). Bio-mining the microbial treasures of the ocean: new natural products. Biotechnol. Adv. 29, 468–482. doi: 10.1016/j.biotechadv.2011.03.001
Jones, E., Kuhne, H., Trussell, P., and Turner, R. (1972). Results of an international cooperative research programme on the biodeterioration of timber submerged in the sea. Mater. Organ. 7, 93–118.
Jones, E. B. G., and Pang, K. L. (2012). Tropical aquatic fungi. Biodivers. Conserv. 21, 2403–2423. doi: 10.1007/s10531-011-0198-6
Jones, E. B. G., Pang, K. L., Abdel-Wahab, M. A., Scholz, B., Hyde, K. D., Boekhout, T., et al. (2019). An online resource for marine fungi. Fungal Divers. 96, 347–433. doi: 10.1007/s13225-019-00426-5
Kaal, J., Serrano, O., Nierop, K. G. J., Schellekens, J., Cortizas, A. M., and Mateo, M. A. (2016). Molecular composition of plant parts and sediment organic matter in a Mediterranean seagrass (Posidonia oceanica) mat. Aqua. Bot. 133, 50–61. doi: 10.1016/j.aquabot.2016.05.009
Kiffer, E., and Morelet, M. (1997). Les Deuteromycetes: Classification et cles d’ Identification Generique. Paris: INRA Editions.
Kohlmeyer, J., Spatafora, J. W., and Volkmann-Kohlmeyer, B. (2000). Lulworthiales, a new order of marine Ascomycota. Mycologia 92, 453–458. doi: 10.2307/3761504
Minelli, A., Ruffo, S., and Stoch, F. (2008). Seagrass Meadows, Flowering Plants in the Mediterranean Sea. Udine: Mueseo Friulano di Storia Naturale.
Mochizuki, K., Nishida, T., Murata, K., Ishida, K., Sunada, A., Asari, S., et al. (2017). Roussoella solani causing keratomycosis, with an observed both sexual and asexual morphs. J. Infect. Chemother. 23, 651–654. doi: 10.1016/j.jiac.2017.03.005
Montemartini, A. C. (1979). La micoflora marina della baia di Portofino. Plant Biosyst. 113, 297–325. doi: 10.1080/11263507909426650
Orhan, I., Sener, B., Atici, T., Brun, R., Perozzo, R., and Tasdemir, D. (2006). Turkish freshwater and marine macrophyte extracts show in vitro antiprotozoal activity and inhibit FabI, a key enzyme of Plasmodium falciparum fatty acid biosynthesis. Phytomedicine 13, 388–393. doi: 10.1016/j.phymed.2005.10.010
Overy, D. P., Rama, T., Oosterhuis, R., Walker, A. K., and Pang, K. L. (2019). The neglected marine fungi, Sensu stricto, and their isolation for natural products’ discovery. Mar. Drugs 17:42. doi: 10.3390/md17010042
Panebianco, C., Tam, W. Y., and Jones, E. B. G. (2002). The effect of pre-inoculation of balsa wood by selected marine fungi and their effect on subsequent colonisation in the sea. Fungal Divers. 10, 77–88.
Panno, L., Bruno, M., Voyron, S., Anastasi, A., Gnavi, G., Miserere, L., et al. (2013). Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. New Biotechnol. 30, 685–694. doi: 10.1016/j.nbt.2013.01.010
Paz, Z., Komon-Zelazowska, M., Druzhinina, I. S., Aveskamp, M. M., Shnaiderman, A., Aluma, Y., et al. (2010). Diversity and potential antifungal properties of fungi associated with a Mediterranean sponge. Fungal Divers. 42, 17–26. doi: 10.1007/s13225-010-0020-x
Pergent, G., Boudouresque, C.-F., Dumay, O., Pergent-Martini, C., and Wyllie-Echeverria, S. (2008). Competition between the invasive macrophyte Caulerpa taxifolia and the seagrass Posidonia oceanica: contrasting strategies. BMC Ecol. 8:20. doi: 10.1186/1472-6785-8-20
Personnic, S., Boudouresque, C. F., Astruch, P., Ballesteros, E., Blouet, S., Bellan-Santini, D., et al. (2014). An ecosystem-based approach to assess the status of a Mediterranean ecosystem, the Posidonia oceanica seagrass meadow. PLoS One 9:e98994. doi: 10.1371/journal.pone.0098994
Poli, A., Lazzari, A., Prigione, V., Voyron, S., Spadaro, D., and Varese, G. C. (2016). Influence of plant genotype on the cultivable fungi associated to tomato rhizosphere and roots in different soils. Fungal Biol. 120, 862–872. doi: 10.1016/j.funbio.2016.03.008
Poli, A., Vizzini, A., Prigione, V., and Varese, G. C. (2018). Basidiomycota isolated from the Mediterranean sea – phylogeny and putative ecological roles. Fungal Ecol. 36, 51–62. doi: 10.1016/j.funeco.2018.09.002
Proksch, P., Putz, A., Ortlepp, S., Kjer, J., and Bayer, M. (2010). Bioactive natural products from marine sponges and fungal endophytes. Phytochem. Rev. 9, 475–489. doi: 10.1007/s11101-010-9178-9
Raghukumar, S. (2017). Fungi in Coastal and Oceanic Marine Ecosystems: Marine Fungi. Basel: Springer.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Siqueira, J. P. Z., Sutton, D. A., Gene, J., Garcia, D., Wiederhold, N., and Guarro, J. (2018). Species of Aspergillus section Aspergillus from clinical samples in the United States. Med. Mycol. 56, 541–550. doi: 10.1093/mmy/myx085
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Vohnik, M., Borovce, O., Kolarikova, Z., Sudova, R., and Reblova, M. (2019). Extensive sampling and high-throughput sequencing reveal Posidoniomyces atricolor gen. et sp. nov. (Aigialaceae, Pleosporales) as the dominant root mycobiont of the dominant Mediterranean seagrass Posidonia oceanica. Mycokeys 55, 59–86. doi: 10.3897/mycokeys.55.35682
Vohník, M., Borovec, O., and Kolaøík, M. (2016). Communities of cultivable root mycobionts of the seagrass Posidonia oceanica in the northwest Mediterranean sea are dominated by a hitherto undescribed pleosporalean dark septate endophyte. Microb. Ecol. 71, 442–451. doi: 10.1007/s00248-015-0640-5
Vohnik, M., Borovec, O., Zupan, I., Kolarik, M., and Sudova, R. (2017). Fungal root symbionts of the seagrass Posidonia oceanica in the central Adriatic sea revealed by microscopy, culturing and 454-pyrosequencing. Mar. Ecol. Prog. Ser. 583, 107–120. doi: 10.3354/meps12337
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: a Guide to Methods and Applications, Vol. 18, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, NY: Academic Press, Inc), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Keywords: marine fungi, Mediterranean Sea, seagrass, algae, phylogeny, Lulworthiales
Citation: Poli A, Bovio E, Ranieri L, Varese GC and Prigione V (2020) Fungal Diversity in the Neptune Forest: Comparison of the Mycobiota of Posidonia oceanica, Flabellia petiolata, and Padina pavonica. Front. Microbiol. 11:933. doi: 10.3389/fmicb.2020.00933
Received: 13 February 2020; Accepted: 20 April 2020;
Published: 26 May 2020.
Edited by:
Mohamed Hijri, Université de Montréal, CanadaReviewed by:
Dhanushka Nadeeshan Wanasinghe, Kunming Institute of Botany, ChinaZheng Wang, Yale University, United States
Copyright © 2020 Poli, Bovio, Ranieri, Varese and Prigione. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giovanna Cristina Varese, Y3Jpc3RpbmEudmFyZXNlQHVuaXRvLml0
 Anna Poli
Anna Poli Elena Bovio
Elena Bovio Lucrezia Ranieri
Lucrezia Ranieri Giovanna Cristina Varese
Giovanna Cristina Varese Valeria Prigione
Valeria Prigione