- 1Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 2Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, Chiang Mai, Thailand
- 3World Agroforestry Centre, East and Central Asia, Kunming, China
- 4Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 5Centre of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 6AG Geobotany, Ruhr-University Bochum, Bochum, Germany
- 7Research Center of Microbial Diversity and Sustainable Utilization, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 8A.M.B, Circolo Micologico “Giovanni Carini”, Brescia, Italy
- 9A.M.B. Gruppo, Micologico Forlivese “Antonio Cicognani”, Forlì, Italy
- 10Academy of Science, The Royal Society of Thailand, Bangkok, Thailand
Helotiales is a polyphyletic order of Ascomycetes. The paucity of relevant molecular data and unclear connections of sexual and asexual morphs present challenges in resolving taxa within this order. In the present study, Patellariopsidaceae fam. nov., the asexual morph of Patellariopsis atrovinosa, and a new record of Cheirospora botryospora (Vibrisseaceae) on Fagus sylvatica (Fagaceae) from Italy are discussed based on morphology and molecular phylogeny. Phylogenetic analyses based on a combined sequence dataset of LSU and ITS were used to infer the phylogenetic relationships within the Helotiales. The results of this research provide a solid base to the taxonomy and phylogeny of Helotiales.
Introduction
The Leotiomycetes (Pezizomycotina) is a very diverse class and was erected when the super-class Leotiomyceta was split in to seven classes by Eriksson & Winka (Eriksson and Winka, 1997). Leotiomycetes currently comprises 13 orders, out of which eight are monotypic, while over 200 genera are represented by one species only (Baral, 2016; Wijayawardene et al., 2018; Ekanayaka et al., 2019; Johnston et al., 2019). Among the orders in Leotiomycetes, Helotiales consists of the highest number of genera, incertae sedis within the familial rank (ca. 90–151) (Baral, 2016; Quijada et al., 2018; Wijayawardene et al., 2018). Hawksworth (2001) estimated that Helotiales consists of 70,000 species. Only 2,334 species belonging to 423 genera in 25 families have been recorded in Helotiales. This constitutes half of all known species in Leotiomycetes (Ekanayaka et al., 2019).
Recent phylogenetic studies based on ribosomal DNA analyses have reported the polyphyletic nature of Helotiales (Ekanayaka et al., 2019; Johnston et al., 2019). The lack of knowledge between asexual and sexual morph connections complicates the systematics of this order (Wang et al., 2006b). Many helotialean fungi are known based on a sexual morph, with their asexual morphs being either undiscovered or assumed to have been lost in evolution (Wang et al., 2006b). On the other hand, it is suggested that asexual morphs from various environmental samples are members of Helotiales, with no mention of their sexual morphs (Sutton and Hennebert, 1994; Marvanova et al., 1997).
Helotiales is the largest group of non-lichen forming ascomycetes and occur in a wide range of niches (Ekanayaka et al., 2017; Wijayawardene et al., 2017). The members of Helotiales are recorded as plant pathogens, endophytes, nematode-trapping fungi, mycorrhizae, fungal parasites, terrestrial and aquatic saprobes, root symbionts and wood rot fungi (Wang et al., 2006a).
The objectives of this study are to introduce a new family with their sexual-asexual inter-connection and to provide a new host record for Cheirospora in Vibrisseaceae.
Materials and Methods
Plant Sample Collection, Morphological Studies and Isolation of Pure Culture
Dead aerial branches of Fagus sylvatica L. (Fagaceae) and Corylus avellana L. (Betulaceae) were collected from Passo la Calla, Stia (province of Arezzo [AR]) Italy and Fiumicello di Premilcuore (province of Forlì-Cesena [FC]) Italy, respectively. Specimens were preserved and observed following the method of Karunarathna et al. (2017). Hand-cut sections of the fruiting structures were mounted in water for microscopic studies and photomicrography. Specimens were examined with a Nikon ECLIPSE 80i compound microscope and photographed with a Canon EOS 600D digital camera fitted to the microscope. Measurements of morphological characteristics were made with the Tarosoft (R) Image Frame Work program and images used for figures were processed with Adobe Photoshop CS3 Extended version 10.0 (Adobe Systems, United States).
Single spore isolation was carried out following the method described in Chomnunti et al. (2014). Germinated spores were individually transferred to potato dextrose agar (PDA) plates and grown at 10–16°C. Colony color and other characteristics were observed and measured after 1 week and 3 weeks. The specimens were deposited in the Mae Fah Luang University Herbarium (MFLU), Chiang Rai, Thailand. Living cultures were deposited in Mae Fah Luang Culture Collection (MFLUCC). Facesoffungi (FoF) and Index Fungorum numbers (IF) were acquired as in Jayasiri et al. (2015) and Index Fungorum (2019).
DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was extracted from fresh fungal mycelium grown on PDA media at 16°C for 4 weeks using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux®, Hangzhou, China) following the instructions of the manufacturer.
The DNA amplification was performed by polymerase chain reaction (PCR). A partial sequence of the LSU rRNA gene region was amplified using the primer pair LR0R and LR5 (Vilgalys and Hester, 1990). The internal transcribed spacer regions (ITS1, 5.8S, ITS2) were amplified using the primer pair ITS5 and ITS4 (White et al., 1990). PCR was carried out following the protocol of Phookamsak et al. (2014). The quality of PCR products was checked by gel electrophoresis on 1% agarose gels stained with ethidium bromide. The amplified PCR fragments were sent to a commercial sequencing provider (Shanghai Sangon Biological Engineering Technology & Services Co., Shanghai, China). The sequence data acquired were deposited in GenBank (Table 1).
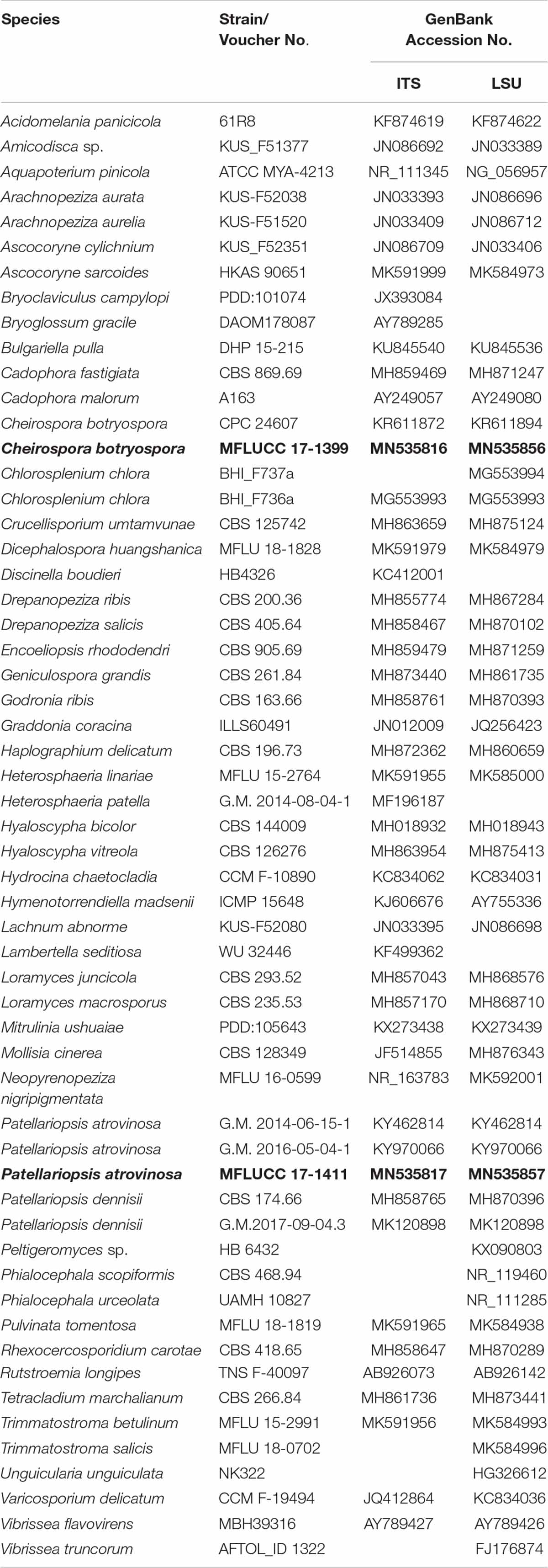
Table 1. Taxa used in the phylogenetic analyses and their corresponding GenBank numbers (Newly generated sequences are indicated in black bold).
Sequencing of the ITS region of strain MFLUCC 17-1411 was failed due to an intron, of about 1.4 kb in length, positioned between the binding site of primer ITS5 and the start of the ITS region. To obtain a double-stranded ITS sequence, a piece of sporodochium < 0.5 mm3 was removed from the specimen and added to a reaction tube with 5 μl of sterile distilled water (dH2O). The soaked specimen was frozen (−20°C) and thawn (+20°C) for five times and 0.5 μl of the solution was used for amplification. Based on initially obtained sequence information, a forward primer (Karu_F01: 5′-CAATGATCAAAGCAGTTGCG-3′) was designed, which has similar properties as the ITS4 primer and binds to the intron sequences close its 3′-end. The PCR reaction included 0.5 μl of the DNA-containing solution, 0.25 μl of each primer (Karu_F01 and ITS4; 10 μM, each), 5.25 μl of sterile dH2O and 6.25 μl of the GoTaq® G2 Hot Start Colorless Master Mix (PROMEGA; GoTaq® Hot Start Polymerase in 2 × Colorless GoTaq® Reaction Buffer (pH 8.5), 400 μM dNTPs, 4 mM MgCl2). The PCR commenced with 3 min denaturation at 95°C, followed by 33 amplification cycles (27 s at 94°C, 60 s at 56°C, and 90 s at 72°C) and a final elongation at 72°C for 7 min. The PCR products were cleaned by successive incubation at 37°C for 30 min and 80°C for 15 min after adding 0.2 μl exonuclease I (20.000 U/ml), 0.2 μl Shrimp-Alkaline-Phosphatase (1.000 U/ml; both New England Biolabs) and 1.6 μl sterile dH2O to 5 μl of PCR product. Purified PCR products were sequenced by the sequencing service of the Ruhr-Universität Bochum using a Genetic Analyzer 3130xl (Applied Biosystems).
Phylogenetic Analyses
Phylogenetic analyses were conducted separately based on LSU and ITS gene sequence data. Reference sequences (Table 1) of representative families in Leotiomycetes were retrieved from GenBank. The related sequences were obtained from a BLAST search and from recently published data (Ekanayaka et al., 2019). Individual datasets for LSU and ITS genes were aligned using the default settings of MAFFT V.7.0361 (Katoh et al., 2018) and improved manually where necessary using Bioedit. Aligned gene regions were concatenated using Bioedit v.7.2 (Hall, 1999) and analyzed.
Initial alignment of LSU region included 7163 base pairs and ITS region included 6619 base pairs. In the phylogenetic analysis, LSU and ITS regions consisted of ambiguously aligned regions. Hence, manual alignment was performed where necessary and some unambiguous regions were removed from the analysis. The removed regions of LSU data set are 0–2658, 2738–2888, 2897–2955, 3037–3220, 3285–3338, 3421–3464, 3723–3730, 4029–4103, 4129–4562, 4581–7163. The excluded regions of the ITS data set are 0–146, 149–173, 181–2164, 2199–2234, 2253–2307, 2714–2923, 2936–2971, 2986–3052, 3065–3113, 3132–3270, 3331–6619. In the final alignment, LSU and ITS regions consist of 898 and 587 bp, respectively.
Phylogenetic constructions of combined gene trees were performed using maximum likelihood (ML), maximum parsimony (MP) and bayesian inference (BI) criteria. Maximum likelihood trees were generated using the RAxML-HPC2 on XSEDE (8.2.8) (Stamatakis et al., 2008; Stamatakis, 2014) in the CIPRES Science Gateway platform (Miller et al., 2010) using the GTR+I+G model of evolution. The robustness of the most parsimonious tree was estimated based on 1000 bootstrap replications.
Maximum parsimony analysis was carried out in PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford, 2002) using the heuristic search option, random stepwise addition, and 1000 replicates, with maxtrees set at 1000. Descriptive tree statistics for parsimony such as Tree Length [TL], Consistency Index [CI], Retention Index [RI], Relative Consistency Index [RC] and Homoplasy Index [HI] were calculated for trees generated under different optimality criteria. The Kishino Hasegawa tests (Kishino and Hasegawa, 1989) were performed to determine whether the trees inferred under different optimality criteria were different.
Evolutionary models for phylogenetic analyses were selected independently for each locus using MrModeltest v. 3.7 (Nylander, 2004) under the Akaike Information Criterion (AIC) implemented in both PAUP v. 4.0b10 and MrBayes v. 3.
Bayesian inference analysis was conducted with MrBayes v. 3.1.2 (Huelsenbeck and Ronquist, 2001) to evaluate posterior probabilities (BYPP) (Rannala and Yang, 1996; Zhaxybayeva and Gogarten, 2002) by Markov Chain Monte Carlo sampling (BMCMC). Kimura 2-parameter model coupled with discrete gamma distribution with a proportion of invariant site (TrN+I+G) was applied for LSU gene region and symmetrical model with discrete gamma distribution coupled with a proportion of invariant sites (TIM2ef+I+G) was applied for ITS gene region. Two parallel runs were conducted, using the default settings, but with the following adjustments: Four simultaneous Markov chains were run for 2,000,000 generations and trees were sampled every 100th generation. The distribution of log-likelihood scores indicated the stationary phase for each search and were used to decide if extra runs were required to achieve convergence, using Tracer v. 1.6 (Rambaut et al., 2014). The first 20% of the generated trees represented the burn-in phase and were discarded. The remaining trees were used to calculate posterior probabilities of the majority rule consensus tree.
Phylograms were visualized with FigTree v1.4.0 (Rambaut, 2012) and edited in Microsoft Power Point (2016) and Adobe Illustrator CS5 (Version 15.0.0, Adobe, San Jose, CA, United States). The finalized alignment and the tree were deposited in TreeBASE, submission ID: 248922.
Results
Phylogenetic Analyses
Phylogenetic trees obtained from LSU and ITS single gene analyses as well as the combined gene analyses share similar overall topologies at the generic level and are in agreement with previous studies (Johnston et al., 2014; Crous et al., 2015; Ekanayaka et al., 2019). The concatenated LSU and ITS dataset consisted of 58 taxa.
The RAxML analysis of the LSU dataset yielded a best scoring tree (Figure 1) with a final ML optimization likelihood value of −16138.064322. The matrix had 758 distinct alignment patterns, with 19.46% of undetermined characters or gaps. Parameters for the GAMMA+P-Invar model of the LSU and ITS were as follows: Estimated base frequencies; A = 0.247456, C = 0.223115, G = 0.280101, T = 0.249328; substitution rates AC = 1.847670, AG = 2.484717, AT = 1.566081, CG = 1.283952, CT = 5.746161, GT = 1.000000; proportion of invariable sites I = 0.458978; gamma distribution shape parameter α = 0.633036. The maximum parsimony dataset consisted of 1490 characters, of which 860 were constant, 475 parsimony-informative and 155 parsimony-uninformative. The parsimony analysis of the data matrix resulted in four equally most parsimonious trees with a length of 948 steps (CI = 0.347, RI = 0.556, RC = 0.193, HI = 0.653) in the best tree.

Figure 1. RAxML tree based on a combined dataset of LSU and ITS partial sequence data. Bootstrap support values for maximum likelihood equal to or higher than 70%, maximum parsimony equal to or higher than 70%, and Bayesian posterior probabilities equal to or greater than 0.90 are displayed on the nodes, respectively. Newly generated sequences are indicated in white. The tree is rooted to Lambertella seditiosa and Rutstroemia longipes.
Taxonomy
In this section, Patellariopsidaceae Karun., Camporesi & K.D. Hyde, fam. nov. and the new record of Cheirospora botryospora are described and illustrated. Helotiales includes several families with sporodochial asexual morphs viz. Gelatinodiscaceae, Helotiaceae and Mollisiaceae. A morphological comparison among members of the families in Helotiales is given in Tables 2, 3.
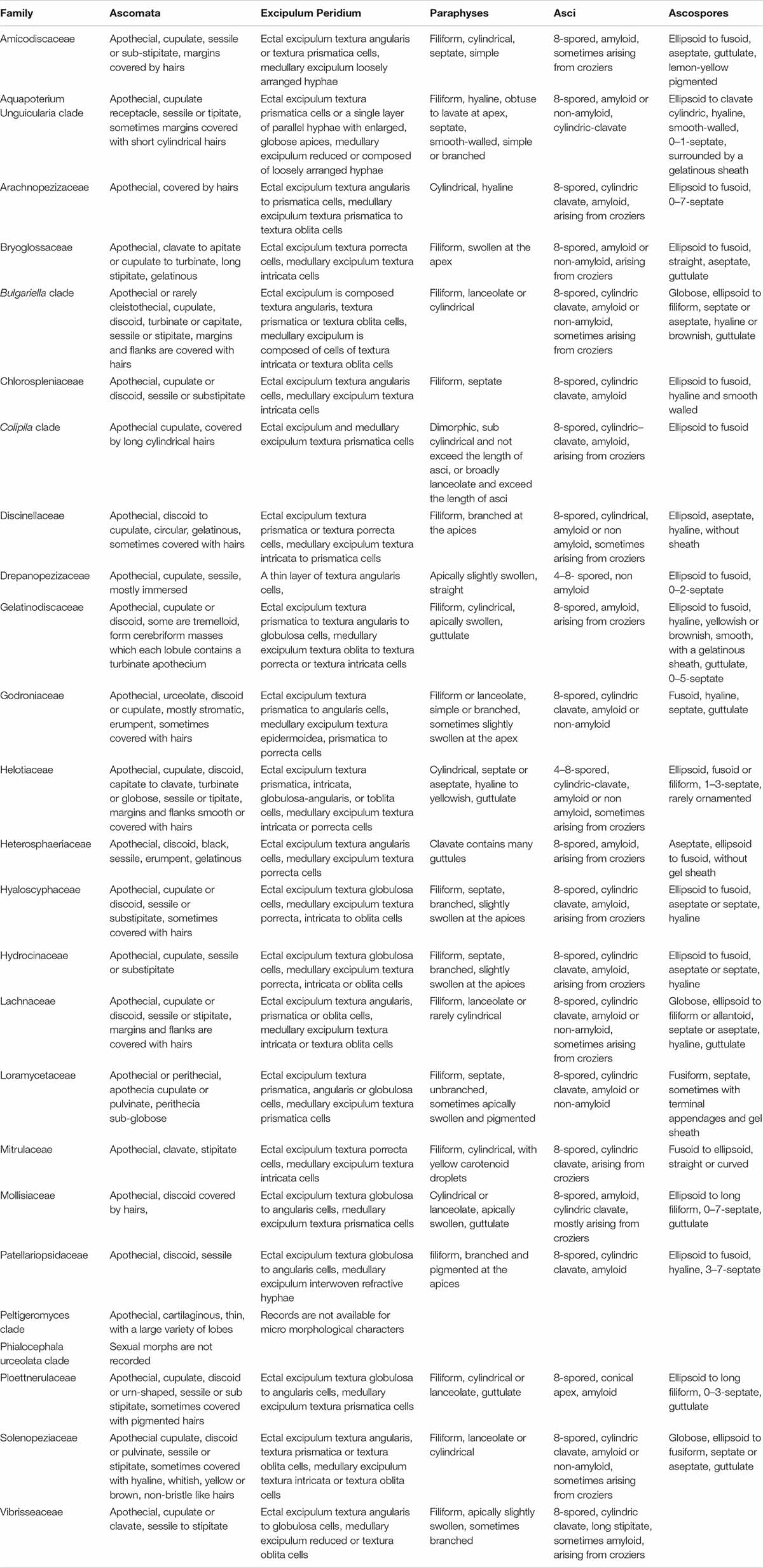
Table 3. Comparison of major sexual morph characteristics of families in order Helotiales based on Ekanayaka et al. (2019).
Patellariopsidaceae Karun., Camporesi and K.D. Hyde, Fam. Nov.
Index Fungorum number: IF556719, Facesoffungi number: FoF06573
Sexual Morph: Ascomata apothecial, discoid, sessile or stipitate. Ectal excipulum composed of cells of textura globulosa to angularis cells. Medullary excipulum composed of interwoven refractive hyphae. Paraphyses filiform branched and pigmented at the apices. Asci 8-spored, cylindric-clavate, amyloid. Ascospores ellipsoid to fusoid, hyaline, 3–7-septate. Asexual morphs: Saprobic on dead branch of Corylus avellana (Betulaceae). Asexual morph: Sporodochium, sub-epidermal or sub-peridermal, solitary. Conidiophores cylindrical, straight or slightly curved, branched over the conidiophore, septate, hyaline, expanding toward the apices, smooth. Conidiogenous cells holoblastic, polyblastic, cylindrical, integrated, hyaline, and smooth. Conidia, sphaerical, proliferating with several, short, lateral, acropetal, branched chains. Primary branches in turn develop secondary branches, which eventually form a globose to cylindrical mass of small, thick-walled, dark brown, septate, eguttulate, smooth, cheiroid, conidium-complex.
Notes
Patellariopsidaceae forms a well-supported (ML 74/BYPP 0.98) clade sister to Chlorospleniaceae, Loramycetaceae, Mollisiaceae and Vibrisseaceae. In Index Fungorum, Patellariopsis is included in Dermateaceae, but Wijayawardene et al. (2017) placed Patellariopsis in Helotiales genera incertae sedis based on morphology. Furthermore, in Ekanayaka et al. (2019), this clade was denoted as separate taxa based on phylogenetic analyses. Hence, we introduce this clade as a new family based on morphology and phylogeny.
Type Genus
Patellariopsis Dennis, Kew Bull. 19(1): 114 (1964)
Patellariopsis Dennis, Kew Bull. 19(1): 114 (1964)
Index Fungorum number: IF556217, Faces of Fungi number: FoF06575
The genus classified under Helotiales genera incertae sedis, Leotiomycetes (Wijayawardene et al., 2018). The type species is Patellariopsis clavispora (Berk. & Broome) Dennis. Five species are recorded in Index Fungorum (2019), P. atrovinosa (A. Bloxam ex Curr.) Dennis, P. carnea G.W. Beaton, P. clavispora (Berk. & Broome) Dennis, P. dennisii (E. Müll. & Hütter) Schläpf.-Bernh., and P. indica A. Pande. We were unable to find any reported described asexual morphs of Patellariopsis in the literature.
Patellariopsis atrovinosa (A. Bloxam ex Curr.) Dennis, Kew Bull. 29(1): 167 (1974)
Index Fungorum number: IF 319233, Facesoffungi number: FoF06574 Figure 2

Figure 2. Asexual morph of Patellariopsis atrovinosa (MFLU 16-2950). (A,B) Appearance of sporodochium on host substrate. (C) Longitudinal section of sporodochium. (D) Conidiophore attached to the host. (E–G) Various stages of conidiogenesis. (H–J) Conidia. (K) Germinated conidium. (L,M) Culture characteristics on PDA (L = from above, M = from below). Scale bars: C = 50 μm; D,E = 20 μm; F–K = 10 μm.
Saprobic on dead branch of Corylus avellana (Betulaceae). Sexual morph: Refer to Dennis (1974). Asexual morph: Sporodochium 33–37 μm high, 278–355 μm diam. ( = 35 × 324 μm, n = 5), sub-epidermal or sub-peridermal, solitary. Conidiophores 41–78 × 1–1.5 μm ( = 58 × 1.3 μm,
n = 20) cylindrical, straight or slightly curved, branched over the conidiophore, septate, hyaline, expanding toward the apices, smooth. Conidiogenous cells 1.5–2 × 1–1.6 μm ( = 1.8 × 1.4 μm, n = 20) holoblastic, polyblastic, cylindrical, integrated, hyaline, smooth. Conidia 1.5–2.7 × 1.5–2.5 μm ( = 2 × 2 μm, n = 40), sphaerical, proliferating with several, short, lateral, acropetal, branched chains. Primary branches in turn develop secondary branches, which eventually form a globose to cylindrical mass of small, thick-walled, dark brown, septate, eguttulate, smooth, cheiroid, conidium-complex.
Colonies growing on PDA becoming 2 cm within 10 days at 16°C, circular, flat, cottony, irregular margin, with less aerial mycelium, olivaceous green to gray from above and dark brown from below.
Material Examined
ITALY, Forlì-Cesena [FC], Fiumicello di Premilcuore, dead aerial branch of Corylus avellana L. (Betulaceae), 5 September 2015, E. Camporesi, IT 3178 (MFLU 16-2950), living cultures, MFLUCC 17-1411.
Cheirospora Moug. & Fr., in Fries, Syst. Orb. Veg. (Lundae) 1: 365 (1825)
Index Fungorum number: IF 7614, Faces of Fungi number: FoF06593
The genus is in Helotiales genera incertae sedis, Leotiomycetes (Wijayawardene et al., 2018). Ekanayaka et al. (2019) placed this genus under Vibrisseaceae. The type species is C. botryospora (Mont.) Berk. & Broome. There are four species in Index Fungorum (2019), C. alni Shabunin., C. betulina (P. Karst.) Kuntze., C. botryospora (Mont.) Berk. & Broome and C. oblonga (Fuckel) Kuntze.
Cheirospora botryospora (Mont.) Berk. & Broome, Ann. Mag. nat. Hist., Ser. 2 5: 455 (1850)
Index Fungorum number: IF 294800, Facesoffungi number: FoF06594 (Figure 3)
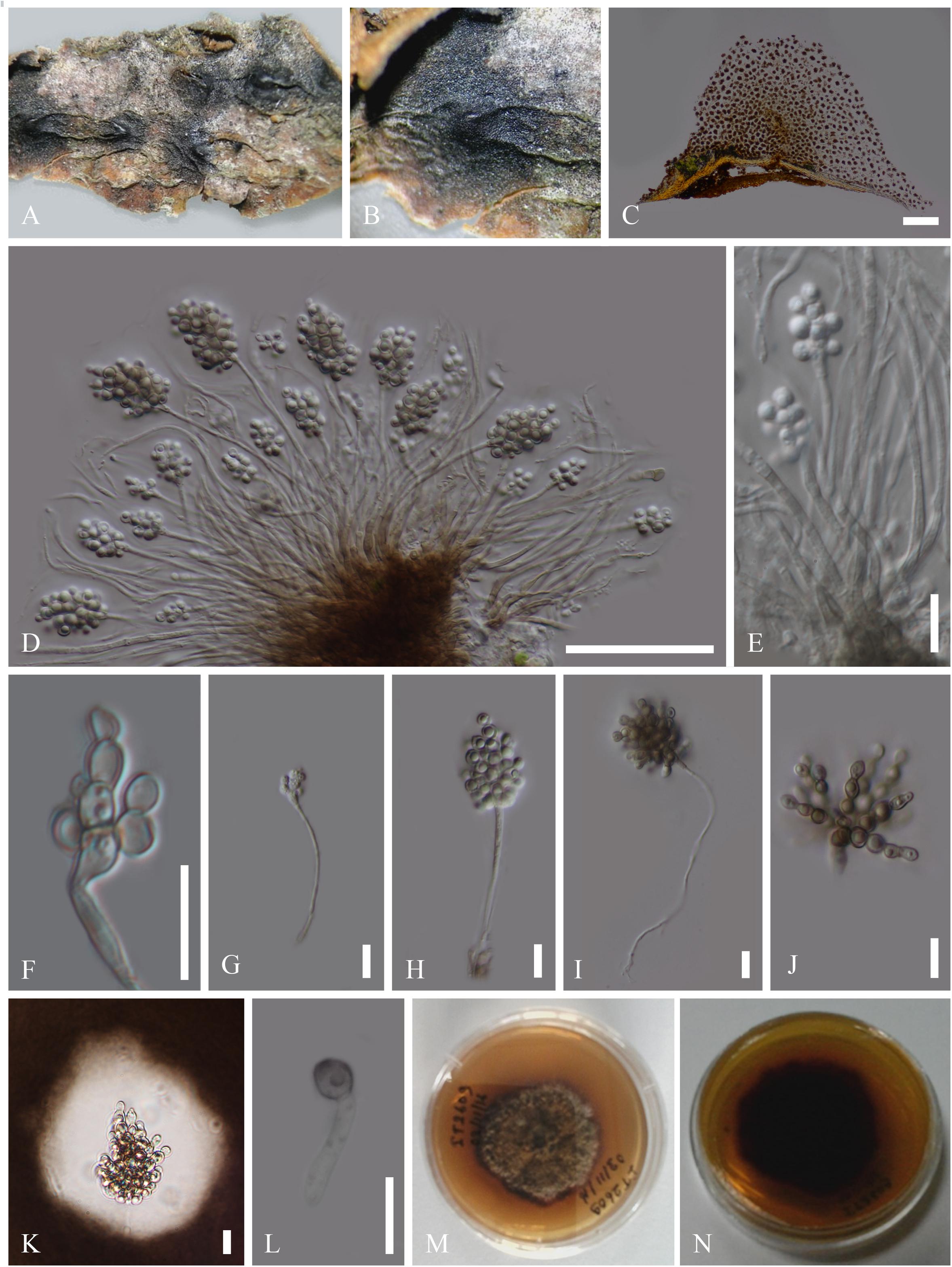
Figure 3. Asexual morph of C. botryospora (MFLU 15-2612). (A,B) Appearance of sporodochium on host substrate. (C) Longitudinal section of sporodochium. (D,E) Conidia attached to the conidiophores. (F–J) Conidia. (K) Conidia surrounded by mucilaginous sheath, stained with Indian ink. (L) Germinated conidium. (M,N) Culture characteristics on PDA (M = From above, N = From below). Scale bars: C = 20 μm; D,E = 50 μm; F–J = 10 μm.
Saprobic on dead branches of Fagus sylvatica L. Sexual morph: unidentified. Asexual morph: Sporodochium 1850–1854 μm high, 3728–3732 μm diam. ( = 1852 × 1730 μm, n = 5), sub-epidermal or sub-peridermal, solitary. Conidiophores 171–225 × 3–4 μm ( = 198 × 3.5 μm, n = 20) cylindrical, straight or slightly curved, branched only at the base, septate, hyaline, expanding toward the apices, smooth. Conidiogenous cells 8–7 × 10–11 μm ( = 7.5 × 10.5 μm, n = 20) holoblastic, polyblastic, cylindrical, integrated, hyaline, smooth. Conidia 10–11 × 9–11 μm ( = 10.5 × 10 μm, n = 40), sphaerical, proliferating with several, short, lateral, acropetal, branched chains. Primary branches in turn develop secondary branches which eventually form a globose to cylindrical mass of small, thick-walled, dark brown, septate, eguttulate, smooth, cheiroid, conidium-complex, enclosed in a gelatinous sheath.
Colonies growing on PDA to 2 cm diam. within 10 days at 16°C, circular, flat, cottony, irregular margin, with less aerial mycelium, olivaceous green to gray from above and dark brown from below.
Material Examined
ITALY, Province of Arezzo [AR], Passo la Calla - Stia, dead aerial branch of Fagus sylvatica (Fagaceae), 5 September 2015, E. Camporesi, IT 2609 (MFLU 15-2612), ex-type living culture, MFLUCC 17-1399.
Discussion
The highly divergent morphological, ecological and biological characteristics of Helotiales makes it a focus for taxonomic studies in the Leotiomycetes, as it is one of the most problematic groups for traditional classification and molecular phylogeny (Wang et al., 2006a). It is a poorly studied order, within which about 19–27% of the genera have an uncertain position at the family level (Baral, 2016). Hence, the taxa in Helotiales have already been subjected to several nomenclatural reinterpretations (Wang et al., 2006a). Lantz et al. (2011) revealed that some genera related to members in Helotiales were traditionally placed in Rhytismatales.
Patellariopsidaceae is established herein based on morphological and phylogenetic support. Comparisons of major sexual and asexual morph characteristics of families in Helotiales are provided in Tables 2 and 3. The asexual morph characteristics of this family are unique in having sporodichium with cheiroid conidium complex. The cheiroid conidium complexes are also present in C. botryospora in Vibrisseaceae. Patellariopsidaceae differs from Vibrisseaceae, in having highly branched conidiophores and thicker conidia complexes. Further, the sexual morph of the Patellariopsidaceae shows unique characteristics by having a sessile discoid apothecium, paraphyses with filiform, branched and pigmented apices, cylindric-clavate, amyloid asci and ellipsoid to hyaline septate ascospores. Patellariopsidaceae was further supported by phylogeny. Hence, herein we establish the Patellariopsidaceae under Helotiales.
Most of the Patellariopsis species were recorded from the United Kingdom with few exceptions (Beaton and Weste, 1978; Farr and Rossman, 2020). Patellariopsis atrovinosa on Prunus laurocerasus was also reported from the United Kingdom. In our study, we report the asexual morph of P. atrovinosa on Corylus avellana from Italy. Patellariopsis carnea on dead grass twigs was reported from Australia (Beaton and Weste, 1978). P. clavispora shows a wide host range, which includes Acer sp., Corylus sp., Crataegus sp., Fagus sp., Fraxinus sp., Ligustrum sp., Prunus sp., Quercus sp. and Symphoricarpos sp. from the United Kingdom (Dennis, 1978, 1986) and Mangifera indica from Pakistan (Ahmad, 1978).
Apart from ribosomal RNA sequence data, the use of protein-coding gene phylogenies involving helotialean fungi are slowly emerging (Wang et al., 2006b; Johnston et al., 2014). Most contemporary results suggest that the Helotiales and currently delimited families are not monophyletic and that the highly conserved small subunit (SSU) rRNA gene is not informative enough to resolve these lineages with confidence (Gernandt et al., 2001).
In our phylogenetic analyses, all the Patellariopsis strains available in the GenBank were included. Among them, the phylogenetic placement of P. dennisii (G.M. 2017 09 04.3) is ambiguous. No morphological descriptions are available in the literature for comparison (Ekanayaka et al., 2019; Vu et al., 2019) and the topology obtained in this study is similar to the topology obtained by Ekanayaka et al. (2019). Hence, we suggest the need for having more data to clarify the position of P. dennisii (G.M. 2017-09-04.3). The blast results for the Patellariopsis dennisii (CBS 174.66) strain include several other Ascomycetous fungi. Therefore, Patellariopsis dennisii (CBS 174.66) was excluded in our dataset after the preliminary phylogenetic analyses.
Genealogical Concordance Phylogenetic Species Recognition (GCPSR) analysis using multi-gene concatenated sequences is used to determine the recombination level within phylogenetically closely related species. Under this study three P. atrovinosa strains MFLUCC 17-1411, G.M. 2016-05-04.1, G.M. 2014-06-15.1, and P. dennisii G.M. 2017 09 04.3 were subjected to the GCPSR analysis. The analysis failed due to the lack of the informative characters in the highly similar sequences of P. atrovinosa strains MFLUCC 17-1411, G.M. 2016-05-04.1 and G.M. 2014-06-15.1.
Based on phylogenetic analyses, our strain MFLUCC 17-1411 forms a well-supported (ML 100/ MP 100/BYPP 1.00) clade with specimens G.M. 2016-05-04.1 and G.M. 2014-06-15.1 (both P. atrovinosa). The phylogenetic relatedness is supported by the 100% similarity between sequences. The asexual stage of Patellariopsis is not recorded in literature. Hence, no morphological comparison can be done between the strains MFLUCC 17-1411 and P. atrovinosa. However, Meyer & Carrières (Meyer and Carrières, 2007) have described an asexual hyphomycetous Periconia-like association for the sexual morph P. atrovinosa. Nevertheless, no scientific evidence was provided to confirm this association. Hence, we justify MFLUCC 17-1411 belongs to the P. atrovinosa.
Cheirospora botryospora MFLUCC 17-1399 forms a well-supported clade with (ML 100/MP 100/BYPP 1.00) C. botryospora CPC 24607 and this was further supported by morphology. C. botryospora CPC 24607 was isolated from Fagus sylvatica in Germany. Danti et al. (2002) identified an endophytic Cheirospora sp. on Fagus sylvatica from Italy. However, they were unable to identify it to the species level. Therefore, in this study, based on morphology and phylogeny the first report of C. botryospora on F. sylvatica from Italy is provided (Farr and Rossman, 2020).
In this study, the Patellariopsidaceae fam. nov. is introduced with an asexual morph. Furthermore, a new host record for the C. botryospora (Vibrisseaceae) and updated phylogenetic tree for Helotiales are provided.
Data Availability Statement
The datasets analyzed in this manuscript are not publicly available. Requests to access the datasets should be directed to YW51bWFuZHJhY2tAeWFob28uY29t.
Author Contributions
AK and KH designed the study. AK performed the morphological study and phylogenetic the data analyses with the help of DP, AE, KC, and RJ. DP did the primer design. SL and SK provided the grant. AK wrote the manuscript. RJ, DP, IG, KC, AE, SK, SL, RC, and KH reviewed and edited the manuscript. All authors reviewed and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer RJ declared a past co-authorship with the authors AK, DP, AE, RJ, KC, IG, RC, EC, KH, SL, SK to the handling editor.
Acknowledgments
We appreciate the kind support given by the laboratory staff of Center of Excellence in Microbial Diversity and sustainable Utilization, Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand; Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand and Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, China. The Plant Germplasm and Genomics Center in Germplasm Bank of Wild Species, Kunming Institute of Botany are thanked for the support in doing molecular work. KH would like to thank “the future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracena species” (Grant No. DBG6080013) and “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion” (RDG6130001). SK thanks Chiang Mai University for funding his postdoctoral research.
Footnotes
References
Baral, H.-O. (2016). “Inoperculate discomycetes,” in Syllabus of Plant Families: A. Engler’s Syllabus der Pflanzenfamilien Part 1/2, eds W. Jaklitsch, H. O. Baral, R. Lücking, H. T. Lumbsch, and W. Frey (Stuttgart: Schweizerbart Science Publishers) 157–205.
Beaton, G., and Weste, G. (1978). Four inoperculate discomycetes from Victoria, Australia. Trans. Br. mycol. Soc. 71, 215–221. doi: 10.1016/S0007-1536(78)80101-6
Butin, H., Pehl, L., de Hoog, G. S., and Wollenzien, U. (1996). Trimmatostroma abietis sp. nov. (hyphomycetes) and related species. Antonie Van Leeuwenhoek 69, 203–209. doi: 10.1007/BF00399607
Chomnunti, P., Hongsanan, S., Aguirre-Hudson, B., Tian, Q., Peršoh, D., Dhami, M. K., et al. (2014). The sooty moulds. Fungal Divers. 66, 1–36. doi: 10.1007/s13225-014-0278-5
Crous, P. W., Schumacher, R. K., Wingfield, M. J., Lombard, L., Giraldo, A., Christensen, M., et al. (2015). Fungal systematics and evolution: FUSE 1. Sydowia 67, 81–118. doi: 10.12905/0380.sydowia67-2015-0081
Danti, R., Sieber, T. N., and Sanguineti, G. (2002). Endophytic mycobiota in bark of European beech (Fagus sylvatica) in the Apennines. Mycol. Res. 106, 1343–1348. doi: 10.1017/S0953756202006779
Dennis, R. W. G. (1974). New or Interesting British Microfungi. II. Kew Bull. 29, 157–179. doi: 10.2307/4108382
Digby, S., and Goos, R. D. (1987). Morphology, development and taxonomy of Loramyces. Mycologia 79, 821–831. doi: 10.1080/00275514.1987.12025470
Duarte, S., Bärlocher, F., Pascoal, C., and Cássio, F. (2016). Biogeography of aquatic hyphomycetes: current knowledge and future perspectives. Fungal Ecol. 19, 169–181. doi: 10.1016/j.funeco.2015.06.002
Ekanayaka, A. H., Ariyawansa, H. A., Hyde, K. D., Jones, E. B. G., Daranagama, D. A., Phillips, A. J. L., et al. (2017). DISCOMYCETES: the apothecial representatives of the phylum Ascomycota. Fungal Divers. 87, 237–298. doi: 10.1007/s13225-017-0389-x
Ekanayaka, A. H., Hyde, K. D., Gentekaki, E., McKenzie, E. H. C., Zhao, Q., Bulgakov, T. S., et al. (2019). Preliminary classification of Leotiomycetes. Mycosphere 10, 310–489. doi: 10.5943/mycosphere/10/1/7
Farr, D. F., and Rossman, A. (2020). Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Available online at: https://nt.ars-grin.gov/fungaldatabases/ (accessed February 10, 2020).
Gernandt, D. S., Platt, J. L., Stone, J. K., Spatafor, J. W., Holst-Jensen, A., Hamelin, R. C., et al. (2001). Phylogenetics of Helotiales and Rhytismatales based on partial small subunit nuclear ribosomal DNA sequences. Mycologia 93, 915–933. doi: 10.1080/00275514.2001.12063226
Goh, T. K., and Hyde, K. D. (1998). A synopsis of and a key to Diplococcium species, based on the literature, with a description of a new species. Fungal Divers. 1, 65–83.
Goh, T. K., Hyde, K. D., and Umali, T. E. (1998). Two new species of Diplococcium from the tropics. Mycologia 90, 514–517. doi: 10.1080/00275514.1998.12026936
Gonçalves, V. N., Vaz, A. B. M., Rosa, C. A., and Rosa, L. H. (2012). Diversity and distribution of fungal communities in lakes of Antarctica. FEMS Microbiol. Ecol. 82, 459–471. doi: 10.1111/j.1574-6941.2012.01424.x
Gönczöl, J., and Révay, Á (2003). Treehole fungal communities: aquatic, aero-aquatic and ematiaceous hyphomycetes. Fungal Divers. 12, 19–34.
Goodwin, S. B. (2002). The barley scald pathogen Rhynchosporium secalis is closely related to the discomycetes Tapesia and Pyrenopeziza. Mycol. Res. 106, 645–654. doi: 10.1017/S0953756202006007
Gramaje, D., Mostert, L., and Armengol Fortí, J. (2011). Characterization of Cadophora luteo-olivacea and C. melinii isolates obtained from grapevines and environmental samples from grapevine nurseries in Spain. Phytopathol. Mediterr. 50, 112–126.
Grünig, C. R., Sieber, T. N., Rogers, S. O., and Holdenrieder, O. (2002). Genetic variability among strains of Phialocephala fortinii and phylogenetic analysis of the genus Phialocephala based on rDNA ITS sequence comparisons. Can. J. Bot. 80, 1239–1249. doi: 10.1139/b02-115
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 41, 95–98.
Hawksworth, D. L. (2001). The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105, 1422–1432. doi: 10.1017/S0953756201004725
Hernández-Restrepo, M., Gené, J., Castañeda-Ruiz, R. F., Mena-Portales, J., Crous, P. W., and Guarro, J. (2017). Phylogeny of saprobic microfungi from Southern Europe. Stud. Mycol. 86, 53–97. doi: 10.1016/J.SIMYCO.2017.05.002
Hernández-Restrepo, M., Silvera-Simón, C., Mena-Portales, J., Mercado-Sierra, Á, Guarro, J., and Gené, J. (2012). Three new species and a new record of Diplococcium from plant debris in Spain. Mycol. Prog. 11, 191–199. doi: 10.1007/s11557-011-0741-6
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Index Fungorum (2019). Available online at: http://www.indexfungorum.org/ (accessed July 25, 2019).
Iturriaga, T., and Israel, H. W. (1985). Studies in the genus Strossmayeria (Helotiales). 5. Conidia and conidiogenesis in Pseudospiropes: a light and electron microscope investigation. Can. J. Bot. 63, 195–200. doi: 10.1139/b85-022
Jaklitsch, W., Baral, H. O., Lucking, R., Lumbsch, H. T., and Frey, W. (2016). Syllabus of Plant Families - A. Engler’s Syllabus der Pflanzenfamilien Part 1/2. Stuttgart: Schweizerbart Science Publishers.
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, J., Buyck, B., Cai, L., et al. (2015). The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Johnston, P., Park, D., Baral, H.-O., Galán, R., Platas, G., and Tena, R. (2014). The phylogenetic relationships of Torrendiella and Hymenotorrendiella gen. nov. within the Leotiomycetes. Phytotaxa 177, 1–25. doi: 10.11646/phytotaxa.177.1.1
Johnston, P. R., Park, D., and Manning, M. A. (2010). Neobulgaria alba sp. nov. and its Phialophora-like anamorph in native forests and kiwifruit orchards in New Zealand. Mycotaxon 113, 385–396.
Johnston, P. R., Quijada, L., Smith, C. A., Baral, H.-O., Hosoya, T., Baschien, C., et al. (2019). A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10:1. doi: 10.1186/s43008-019-0002-x
Karunarathna, A., Senanayake, I., Jeewon, R., Phookamsak, R., Goonasekara, I., Wanasinghe, D., et al. (2017). Novel fungal species of Phaeosphaeriaceae with an asexual/sexual morph connection. Mycosphere 8, 1818–1834. doi: 10.5943/mycosphere/8/10/8
Katoh, K., Rozewicki, J., and Yamada, K. D. (2018). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
King, K. M., West, J. S., Brunner, P. C., Dyer, P. S., and Fitt, B. D. L. (2013). Evolutionary Relationships between Rhynchosporium lolii sp. nov. and Other Rhynchosporium Species on Grasses. PLoS One 8:e72536. doi: 10.1371/journal.pone.0072536
Kirschner, R., and Oberwinkler, F. (2001). Mycoparasitism by Three Species of Diplococcium (Hyphomycetes). Plant Biol. 3, 449–454. doi: 10.1055/s-2001-16457
Kishino, H., and Hasegawa, M. (1989). Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 29, 170–179. doi: 10.1007/BF02100115
König, H., Unden, G., and Fröhlich, J. (2017). “Biology of microorganisms on grapes,” in Must and in Wine, eds H. König, G. Unden, and J. Fröhlich (Cham: Springer International Publishing) doi: 10.1007/978-3-319-60021-5
Lantz, H., Johnston, P. R., Park, D., and Minter, D. W. (2011). Molecular phylogeny reveals a core clade of Rhytismatales. Mycologia 103, 57–74. doi: 10.3852/10-060
Legon, N. (2012). Cheirospora botryospora - a very common rarity? Field Mycol. 13, 128–130. doi: 10.1016/j.fldmyc.2012.10.006
Leuchtmann, A. (1987). Species of heterosphaeria discomycetes and their anamorphs. Mycotaxon 28, 261–284.
Marvanová, L., and Bärlocher, F. (2001). Hyphomycetes from Canadian streams. VI. Rare species in pure cultures. Czech Mycol. 53, 1–28.
Marvanova, L., Janardhanan, K. K., Rajendran, C., Natarajan, K., and Hawksworth, D. L. (1997). “Freshwater hyphomycetes: a survey with remarks on tropical taxa,” in Tropical Mycology, eds K. K. Janardhanan, C. Rajendran, K. Natarajan, and D. L. Hawksworth (Enfield: Science Publishers, Inc), 169–226.
Meyer, M., and Carrières, E. (2007). Inventaire de la Biodiversité dans la Forêt de Schnellert (Commune de Berdorf). Paris: Musée national d’histoire naturelle.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE) (IEEE), New Orleans, LA, 1–8. doi: 10.1109/GCE.2010.5676129
Peláez, F., Collado, J., Platas, G., Overy, D. P., Martín, J., Vicente, F., et al. (2011). Phylogeny and intercontinental distribution of the pneumocandin-producing anamorphic fungus Glarea lozoyensis. Mycology 2, 1–17. doi: 10.1080/21501203.2010.544334
Phookamsak, R., Liu, J. K., McKenzie, E. H. C., Manamgoda, D. S., Ariyawansa, H., Thambugala, K. M., et al. (2014). Revision of phaeosphaeriaceae. Fungal Divers. 68, 159–238. doi: 10.1007/s13225-014-0308-3
Quijada, L., Johnston, P. R., Cooper, J. A., and Pfister, D. H. (2018). Overview of Phacidiales, including Aotearoamyces gen. nov. on Nothofagus. IMA Fungus 9, 371–382. doi: 10.5598/imafungus.2018.09.02.08
Rambaut, A. (2012). FigTree version 1.4.0. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed February 8, 2020).
Rambaut, A., Drummond, A. J., and Suchard, A. (2014). Tracer v1.6. Available online at: http://beast.bio.ed.ac.uk/Tracer (accessed February 8, 2020).
Rannala, B., and Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Seaver, F. J. (1938). Photographs and descriptions of cup-fungi. XXIX Chloroscypha. Mycologia 30, 594–596. doi: 10.1080/00275514.1938.12017301
Shenoy, B. D., Jeewon, R., Wang, H., Kaur, A., Ho, W., Bhat, D. J., et al. (2010). Sequence data reveals phylogenetic affinities of fungal anamorphs Bahusutrabeeja, Diplococcium, Natarajania, Paliphora, Polyschema, Rattania and Spadicoides. Fungal Divers. 44, 161–169. doi: 10.1007/s13225-010-0059-8
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Sutton, B. C., and Ganapathi, A. (1978). Trimmatostroma excentricum sp.nov., on Eucalyptus from New Zealand and Fiji. N. Z. J. Bot. 16, 529–533. doi: 10.1080/0028825X.1978.10426880
Sutton, B. C., and Hennebert, G. L. (1994). “Interconnections amongst anamorphs and their possible contribution to ascomycete systematics,” in Ascomycete Systematics, ed. D. L. Hawksworth (Boston, MA: Springer), 77–100. doi: 10.1007/978-1-4757-9290-4_8
Swofford, D. L. (2002). Phylogenetic Analysis Using Parsimony Version 4.0 beta version. Sunderland: Sinauer Associates, doi: 10.1007/BF02198856
Travadon, R., Lawrence, D. P., Rooney-Latham, S., Gubler, W. D., Wilcox, W. F., Rolshausen, P. E., et al. (2015). Cadophora species associated with wood-decay of grapevine in North America. Fungal Biol. 119, 53–66. doi: 10.1016/j.funbio.2014.11.002
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/JB.172.8.4238-4246.1990
Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
Walsh, E., Duan, W., Mehdi, M., Naphri, K., Khiste, S., Scalera, A., et al. (2018). Cadophora meredithiae and C. interclivum, new species from roots of sedge and spruce in a western Canada subalpine forest. Mycologia 110, 201–214. doi: 10.1080/00275514.2017.1406748
Walsh, E., Luo, J., and Zhang, N. (2014). Acidomelania panicicola gen. et sp. nov. from switchgrass roots in acidic New Jersey pine barrens. Mycologia 106, 856–864. doi: 10.3852/13-377
Wang, Y.-Z. (2009). A new species of Arachnopeziza from Taiwan. Mycotaxon 108, 485–489. doi: 10.5248/108.485
Wang, Z., Binder, M., Schoch, C. L., Johnston, P. R., Spatafora, J. W., and Hibbett, D. S. (2006a). Evolution of helotialean fungi (Leotiomycetes, Pezizomycotina): a nuclear rDNA phylogeny. Mol. Phylogenet. Evol. 41, 295–312. doi: 10.1016/j.ympev.2006.05.031
Wang, Z., Johnston, P. R., Takamatsu, S., Spatafora, J. W., and Hibbett, D. S. (2006b). Toward a phylogenetic classification of the Leotiomycetes based on rDNA data. Mycologia 98, 1065–1075. doi: 10.1080/15572536.2006.11832634
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wijayawardene, N. N., Hyde, K. D., Lumbsch, H. T., Liu, J. K., Maharachchikumbura, S. S. N., Ekanayaka, A. H., et al. (2018). Outline of Ascomycota: 2017. Fungal Divers. 88, 167–263. doi: 10.1007/s13225-018-0394-8
Wijayawardene, N. N., Hyde, K. D., Rajeshkumar, K. C., Hawksworth, D. L., Madrid, H., Kirk, P. M., et al. (2017). Notes for genera: Ascomycota. Fungal Divers. 86, 1–594. doi: 10.1007/s13225-017-0386-0
Yoshikawa, M., and Yokoyama, T. (1992). Thedgonia ligustrina on Ligustrum japonicum and Cercospora kyotensis sp. nov. on Hydrangea serrata var. thunbergii. Nippon Kingakkai Kaiho 33, 177–184.
Keywords: Ascomycetes, Cheirospora botryospora, Leotiomycetes, Pezizomycotina, sporodochium
Citation: Karunarathna A, Peršoh D, Ekanayaka AH, Jayawardena RS, Chethana KWT, Goonasekara ID, Cheewangkoon R, Camporesi E, Hyde KD, Lumyong S and Karunarathna SC (2020) Patellariopsidaceae Fam. Nov. With Sexual-Asexual Connection and a New Host Record for Cheirospora botryospora (Vibrisseaceae, Ascomycota). Front. Microbiol. 11:906. doi: 10.3389/fmicb.2020.00906
Received: 17 February 2020; Accepted: 16 April 2020;
Published: 26 May 2020.
Edited by:
Gustavo Henrique Goldman, University of São Paulo, BrazilReviewed by:
Rajesh Jeewon, University of Mauritius, MauritiusZheng Wang, Yale University, United States
Copyright © 2020 Karunarathna, Peršoh, Ekanayaka, Jayawardena, Chethana, Goonasekara, Cheewangkoon, Camporesi, Hyde, Lumyong and Karunarathna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saisamorn Lumyong, c2Nib2kwMDlAZ21haWwuY29t; Samantha C. Karunarathna, c2FtYW50aGFrYXJ1bmFyYXRobmFAZ21haWwuY29t
 Anuruddha Karunarathna
Anuruddha Karunarathna Derek Peršoh
Derek Peršoh Anusha H. Ekanayaka
Anusha H. Ekanayaka Ruvishika S. Jayawardena
Ruvishika S. Jayawardena K. W. Thilini Chethana
K. W. Thilini Chethana Ishani D. Goonasekara
Ishani D. Goonasekara Ratchadawan Cheewangkoon
Ratchadawan Cheewangkoon Erio Camporesi8,9
Erio Camporesi8,9 Kevin D. Hyde
Kevin D. Hyde Saisamorn Lumyong
Saisamorn Lumyong Samantha C. Karunarathna
Samantha C. Karunarathna