- 1Department of Translational Research and New Technologies in Medicine and Surgery, University of Pisa, Pisa, Italy
- 2Center for Synthetic Microbiology (SYNMIKRO) and Department of Chemistry, Philipps University, Marburg, Germany
The multidomain (B-NG) protein FlhF, a flagellar biogenesis regulator in several bacteria, is the third paralog of the signal recognition particle (SRP)-GTPases Ffh and FtsY, which are known to drive protein-delivery to the plasma membrane. Previously, we showed that FlhF is required for Bacillus cereus pathogenicity in an insect model of infection, being essential for physiological peritrichous flagellation, for motility, and for the secretion of virulence proteins. Among these proteins, we found that the L2 component of hemolysin BL, one of the most powerful toxins B. cereus produces, was drastically reduced by the FlhF depletion. Herein, we demonstrate that B. cereus FlhF forms GTP-dependent homodimers in vivo since the replacement of residues critical for their GTP-dependent homodimerization alters this ability. The protein directly or indirectly controls flagellation by affecting flagellin-gene transcription and its overproduction leads to a hyperflagellated phenotype. On the other hand, FlhF does not affect the expression of the L2-encoding gene (hblC), but physically binds L2 when in its homodimeric form, recruiting the protein to the plasma membrane for secretion. We additionally show that FlhF overproduction increases L2 secretion and that the FlhF/L2 interaction requires the NG domain of FlhF. Our findings demonstrate the peculiar behavior of B. cereus FlhF, which is required for the correct flagellar pattern and acts as SRP-GTPase in the secretion of a bacterial toxin subunit.
Introduction
Bacillus cereus is a Gram-positive, motile, and spore-forming rod that is ubiquitously found in a wide range of environments including soil, water and foods. Long known as cause of two types of food-borne intoxications, the diarrheal and emetic syndromes, B. cereus is also responsible for local and systemic infections in humans (Ehling-Schulz et al., 2019). Its pathogenicity is associated with the bacterial ability to adhere to and colonize host surfaces through peritrichous flagella and flagellum-driven swimming and swarming (Senesi and Ghelardi, 2010). Beside motility, the secretion of tissue-destructive exoenzymes and toxins, e.g. hemolysins, phospholipases, trimeric toxins, cytotoxins and proteases (Ramarao and Sanchis, 2013; Ehling-Schulz et al., 2019), is of crucial importance in B. cereus pathogenicity. Among these factors, the tripartite hemolysin BL (HBL), consisting of a binding component (B) and two lytic components (L1 and L2), plays a primary role in diarrheal food poisoning and necrotizing infections such as endophthalmitis, due to its vascular permeability, hemolytic, and enterotoxic activity (Beecher et al., 1995a, b). Flagella, swarming differentiation, and HBL secretion are intrinsically linked in B. cereus. In fact, HBL export is increased in hyperflagellated swarm cells (Ghelardi et al., 2007; Salvetti et al., 2011) and mutations that reduce the number of flagella impact on toxin secretion (Senesi et al., 2002; Callegan et al., 2006; Salvetti et al., 2007; Mazzantini et al., 2016). In addition, failure to secrete HBL was demonstrated in aflagellated B. cereus natural isolates (Ghelardi et al., 2007) and in an acrystalliferous Bacillus thuringiensis (i.e. B. cereus) mutant for the Type-III flagellar export apparatus (Ghelardi et al., 2002).
The third signal recognition particle (SRP) GTPase, FlhF, is involved in controlling B. cereus flagellation and motility (Salvetti et al., 2007; Mazzantini et al., 2016). In many polarly flagellated species, this protein is required for establishing the correct place and/or quantity of flagella on the cell surface (Kazmierczak and Hendrixson, 2013; Altegoer et al., 2014; Gao et al., 2015; Schuhmacher et al., 2015; Navarrete et al., 2019), together with the MinD-like ATPase FlhG (Leipe et al., 2002). In the peritrichously flagellate Bacillus subtilis, FlhF and FlhG behave as antagonistic pair in regulating the symmetric grid-like pattern of flagellar basal body distribution, but do not control the number of flagella on the cell surface (Bange et al., 2011; Guttenplan et al., 2013). In B. cereus, no FlhG homolog was found and depletion of FlhF causes flagellar mislocalization and substantial reduction in the number of flagella (Salvetti et al., 2007).
Together with Ffh and FtsY, FlhF belongs to the SRP family of GTPases (Leipe et al., 2002). In the canonical SRP system, the Ffh/4.5S RNA complex attaches to the N-terminal signal sequence of nascent membrane protein at the ribosome, and then docks with its receptor (FtsY) on the membrane, thus resulting in the insertion of the protein into the membrane (Akopian et al., 2013; Mercier et al., 2017). SRP-GTPases are characterized by conserved N and G domains that form a structurally and functionally coupled unit (NG domain) (Grudnik et al., 2009). In all described SRP-GTPases, five nucleotide-binding signature elements (G1–G5) essential for GTP binding and hydrolysis and for SRPs dimerization have been identified in the G domain (Bange et al., 2007; Mazzantini et al., 2016). All three SRP-GTPases contain additional domains, which reflect the specific functional roles of these proteins. While Ffh presents a C-terminal methionine-rich domain (M) that is essential for signal peptide recognition and SRP-RNA binding (Janda et al., 2010; Hainzl et al., 2011), FtsY contains an unfolded N-terminal acidic domain (A) which ensures membrane attachment (de Leeuw et al., 2000; Stjepanovic et al., 2011; Altegoer et al., 2014). Similar to B. subtilis FlhF (Bange et al., 2007), the B. cereus protein carries a natively unfolded N-terminal lysine-rich basic (B) domain with unknown function (Mazzantini et al., 2016). GTP-bound FlhF forms homodimers in Xantomonas oryzae pv oryzae, B. subtilis, Pseudomonas aeruginosa, Shewanella oneidensis, Campylobacter jejuni, and Vibrio alginolyticus (Shen et al., 2001; Bange et al., 2007; Schniederberend et al., 2013; Gao et al., 2015; Gulbronson et al., 2016; Kondo et al., 2018).
In addition to the flagellar defects, FlhF depletion in B. cereus also causes an alteration in the amount of several extracellular proteins (Mazzantini et al., 2016). Among these proteins, the L2 component of HBL was found to be consistently reduced in the extracellular proteome of a B. cereus ΔflhF mutant (Mazzantini et al., 2016), suggesting a potential involvement of FlhF in L2 secretion mechanisms.
In the present study, the ability of B. cereus FlhF to form homodimers and the effect of point mutations in FlhF putative GTP-binding elements on homodimer formation were analyzed. Functional characterization of B. cereus FlhF as SRP protein for L2 is reported and the requirement of the NG domain for protein contact is defined. In addition, experiments were performed to investigate the effect of different FlhF levels on B. cereus flagellation and L2 secretion.
Materials and Methods
Bacterial Strains and Culture Conditions
All strains used in this study are described in Table 1. B. cereus ATCC 14579 wild-type (WT), its flhF mutant (ΔflhF, MP06), and flhF-overexpressed (MP08) derivatives (Salvetti et al., 2007) were grown at 37°C in Brain Heart Infusion (BHI) supplemented with 1% glucose (BHIG). When required, 5 μg/ml erythromycin for strain ΔflhF or 30 μg/ml kanamycin for strain MP08 selection were added. MP08 cultures were also supplemented with 4 mM isopropyl-β-D-1-tiogalattopiranoside (IPTG; Merck KGaA, Darmstadt, Germany) to induce pspac-dependent flhF expression. E. coli XL1-Blue was used for general cloning strategies, E. coli BTH101 was used as reporter strain in the bacterial adenylate cyclase two-hybrid system (BACTH) assays, and E. coli BL21 (DE3) for hexa-histidine pull-down experiments. E. coli strains were grown at 30°C or 37°C in Luria Bertani (LB) supplemented with 100 μg/ml of ampicillin, 50 μg/ml of kanamycin, or either, when required. E. coli BTH101 and BL21 (DE3) cultures were also supplemented with 0.5 and 1 mM IPTG, respectively, to induce plasmid gene expression.
Plasmids Construction
Plasmids and primers used in this study are described in Table 1 and Supplementary Table S1, respectively. For bacterial two-hybrid experiments, the complete ORFs of B. cereus flhF (GenBank ID: bc1670), hblC (GenBank ID: bc3104), bc1657, and the NG encoding fragment of flhF (flhFNG) were directly amplified from genomic DNA of B. cereus ATCC 14579 using primer pairs flhFF1/flhFR1, bc1657F1/bc1657R1, L2F2/L2R1, NGF1/flhFR1, respectively. To produce flhFT253Q and flhFD391A mutant variants, site-directed mutagenesis by combined overlap extension PCR (COE-PCR) was performed (Hussain and Chong, 2016). For each variant, two primers pairs (BcflhF-T253Q-F/flhFR1 and flhFF1/BcflhF-T253Q-R, and BcflhF-D391A-F/flhFR1 and flhFF1/BcflhF-D391A-R, respectively), were used to generate two flhF mutated fragments with overlapping ends. The full-length products were obtained directly joining the two fragments by PCR using flhFF1/flhFR1 as primer pair. All amplicons were digested with BamHI/KpnI (New England Biolabs, NEB, Ipswich, MA, United States), and ligated in pUT18, pUT18C, pKT25, and pKNT25 vectors, in frame with the isolated T18 and T25 domains of adenylate cyclase and under the control of the inducible plac promoter. For hexa-histidine pull-down assays, flhF and hblC were amplified using PDflhFF1/PDflhFR2 and PDL2F1/PDL2R1, digested with XbaI/XhoI and NsiI/XhoI (NEB), respectively, and ligated in pET303/CT-His, under the control of the T7 promoter. Recombinant vectors were propagated in E. coli XL1-Blue and analyzed by PCR, plasmids extraction, and sequencing.
Quantitative Real Time PCR
WT and ΔflhF cells were inoculated in 100 ml of BHIG broth and grown at 37°C until mid-exponential phase (OD600 of ∼1.0). Bacterial cultures were centrifuged at 4,000 × g at 4°C for 15 min. Cells were washed twice with cold diethylpyrocarbonate-treated water and suspended in 350 μl of RA1 lysis buffer (Macherey-Nagel, Düren, Germany) supplemented with 3.5 μl of β-mercaptoethanol and 0.35 g of zirconiabeads (diameter 0.1 mm; Biospec Products, Barltesville, Okla). Bacterial lysis was performed by shaking for 4 min with a mini-bead beater (Biospec Products), alternating 0.5 min of shake with 5 min in ice bath. Residual cells and debris were removed by centrifugation for 2 min at 12,000 × g and the aqueous phases were filtered through NucleoSpin® Filters. 350 μl of 70% absolute ethanol were added, and the mixture was applied to a NucleoSpin® RNA column. After being digested with 40 units of RNase-free DNase (Macherey-Nagel), total RNA was eluted from the column following the manufacturer’s instructions. 500 ng of the purified RNA were used as a template in one-step RT-PCR with the TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgenbiotech, Beijing, China), according to the manufacturers. qRT-PCR were performed on cDNA samples using the LightCyclerTM FastStart DNA Master SYBR Green I and analyzed in the LightCycler instrument (Roche, Basel, Switzerland). The rpoA (GenBank ID: bc0158) and the bc4306 genes (Reiter et al., 2011; Salvetti et al., 2011) were used as endogenous controls. qRT-PCRs were performed using the primer pairs bc1657 Fup/bc1657 Rdw, hblL2 up/hblL2 dw, rpoAup1/rpoAdw1, and gatB_Yqey up/gatB_Yqey dw (Supplementary Table S1) for bc1657, hblC, rpoA, and bc4306, respectively. The amplification conditions were optimized and the amplified fragments sequenced (Supplementary Table S1) before performing qRT-PCR experiments. Melting curve analysis was carried out in parallel to qRT-PCR to confirm the specificities of the amplification reactions. Data were analyzed using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Three biological replicates were performed and for each experiment three technical replicates were carried out.
Bacterial Adenylate Cyclase Two-Hybrid Assay
Bacterial adenylate cyclase two-hybrid assay experiments were performed according to Battesti and Bouveret (2012). The constructed BACTH vectors (Table 1) were co-transformed in chemical competent E. coli BTH101 cells and plates were incubated at 30°C for 48 h. For each transformation, colonies were inoculated in 3 ml of LB supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 0.5 mM IPTG, and grown overnight at 30°C. 10 μl of each culture were dropped on M63 synthetic medium (2 g/l (NH4)2SO4, 13.6 g/l KH2PO4, 0.5 mg/l FeSO4 × 7H2O, 1 mM MgSO4, 0.0001% thiamin, 0.2% maltose; pH 7.0) agar plates supplemented with 50 μg/ml ampicillin, 25 μg/ml kanamycin, 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Merck KGaA), and 0.5 mM IPTG. Plates were incubated at 30°C for 24–72 h. At least three biological replicates were performed and a representative result is shown.
Quantification of β-Galactosidase Activity
β-galactosidase assays were performed according to the Miller’s method (Miller, 1992). Overnight bacterial cultures were diluted in 10 ml of LB broth containing 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 0.5 mM IPTG and grown at 30°C to OD600 of ∼0.5. 1 ml of each culture was centrifuged at 8000 × g at 4 °C for 5 min and the pellet was suspended in an equal amount of chilled Z buffer (60 mM Na2HPO4 × 7H2O, 40 mM NaH2PO4 × H2O, 10 mM KCl, 1 mM MgSO4 × 7H2O, and 50 mM β-mercaptoethanol; pH 7.0). For cells permeabilization, 20 μl of 0.1% sodium dodecyl sulfate (SDS; Merck KGaA) and 40 μl of chloroform were added and the tubes were mixed by vortexing for 10 s. 100 μl of samples were diluted in 1 ml of chilled Z buffer and incubated with 200 μl of 4 mg/ml orto-nitrofenil-β-galactopyranoside (ONPG, Merck KGaA) at 28°C. The reaction was stopped by adding 250 μl of 1 M Na2CO3. β-galactosidase activity was calculated by the Miller formula (Miller units = 1000 × (OD420-(1.75 × OD550)/T × V × OD600); T, reaction time; V, volume of culture assayed in milliliter). Experiments were repeated three times in separate days and for each assay two technical replicates were carried out.
Hexa-Histidine Pull-Down Experiments
pET303/CT, pET303/flhFHis and pET303/hblC (Table 1) were independently transformed in chemical competent E. coli BL21 (DE3). Bacteria were grown at 37°C in 100 ml of LB broth to OD600 of ∼0.5 and genes expression was induced by adding 1 mM IPTG. Cultures were incubated for additional 2 h (OD600 of ∼2) and centrifuged at 5,000 × g for 5 min at 4°C. Pellets were suspended in 1:1 Tris Buffered Saline (TBS; 50 mM Tris, 150 mM NaCl, pH 7.2) and Pierce Lysis Buffer (Thermo Fisher Scientific, Waltham, MA, United States), and supplemented with Halt Protease Inhibitor Cocktail, EDTA-Free 1 × (Thermo Fisher Scientific). Soluble fractions were isolated by centrifugation at 12,000 × g for 5 min and stored at – 20°C until use. The correct production of FlhFHis and L2 in the respective E. coli lysates was assessed by immunoblot using mouse monoclonal Anti-His(C-term)-AP Antibody (a-His; Thermo Fisher Scientific) and rabbit polyclonal sera specific to L2 (Beecher et al., 1995b), respectively. The E. coli lysate expressing FlhFHis was incubated for 1 h at 4°C with 25 ml of settled HisPurTM Cobalt resin (Thermo Fisher Scientific), previously equilibrated according to the manufacturer’s instructions. After six washing steps with 1:1 TBS and Pierce Lysis Buffer containing 20 mM imidazole (Thermo Fisher Scientific), the E. coli lysate producing L2 was applied to the column for 2 h at 4°C. Eight wash steps with TBS containing 10 mM Imidazole were performed to remove E. coli proteins which non-specifically interact. Captured proteins were then eluted with 250 μl of 1:1 TBS and Pierce Lysis Buffer containing 290 mM Imidazole. FlhFHis and L2 identification was performed by 10% SDS-PAGE followed by immunodetection using α-His and α-L2 (Beecher et al., 1995b) antibodies, respectively. To rule out the possibility that L2 had an intrinsic affinity for the cobalt resin or non-specifically interacted with E. coli proteins able to bind the resin, the soluble fraction of cells expressing L2 was directly applied to the equilibrated resin and to the resin previously incubated with the lysate of E. coli cells containing the empty vector pET303/CT. Both control samples were treated as described above.
Flagella Staining and Flagellin Purification
WT and MP08 strains were grown to the late exponential growth phase in BHIG for 6 h at 37°C (OD600 of ∼2). For flagella observation, 10 μl of bacterial cells were directly stained with tannic acid and silver nitrate (Harshey and Matsuyama, 1994). Several samples were analyzed at 1,000 × magnification using an optical microscope (BH-2; Olympus, Tokyo, Japan). The extent of cell flagellation was analyzed as previously described (Calvio et al., 2005). Briefly, bacterial cultures were vigorously vortexed for 30 s, and harvested by centrifugation at 5,000 × g for 10 min at 4°C. Flagellar filaments were collected from supernatants by ultracentrifugation at 100,000 × g for 1 h at 4°C. Protein concentration was determined by PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific), samples were standardized accordingly, and were subjected to 10% SDS-PAGE followed by Comassie Blue staining. Experiments were performed in triplicate in separate days and a representative result is shown.
Preparation of B. cereus Supernatants
Protein samples were prepared by growing WT and MP08 cells to the late exponential growth phase in BHIG for 6 h at 37° C (OD600 of ∼2). Cells were normalized to the same OD600 and culture supernatants were collected by centrifugation at 10,000 × g, and added with Halt Protease Inhibitor Cocktail, EDTA-Free 1 × and 0.5 mM EDTA. Protein concentration was determined with PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific). To exclude cell lysis, the activity of the cytosolic marker fructose-1,6-bisphosphate aldolase in culture supernatants was spectrophotometrically determined (Warth, 1980). Culture supernatants were concentrated using Microcon® -10 centrifugal filter units (Merck KGaA). After being standardized for total proteins concentration, protein samples were subjected to 10% SDS-PAGE and electrotransferred on PVDF membranes for L2 immunodetection (Beecher et al., 1995b). All experiments were performed three times in separate days and a representative result is shown.
Image Analysis
Densitometric analysis of Comassie blue stained gels and immunodetected filters were performed using a ChemiDocTM XRS+ System with a software version 6.0.1.34 (Bio-Rad, Berkeley, California). Relative quantification was performed using the WT bands as reference.
In silico Analysis
Nucleotide and amino acid sequences in the FASTA format were retrieved from the European Nucleotide Archive1 and the UniProt databases (The UniProt Consortium, 2015), respectively. BLASTn2 was used for comparative analysis of nucleotide sequences obtained from sequencing. Compute pI/Mw tool (ExPASy Bioinformatics Resource Portal) was used for molecular weight prediction of amino acid sequences. The representation of B. cereus FlhF domains was realized using the Illustrator for Biological Sequences (IBS) tool (Liu et al., 2015).
Statistical Analysis
Data were expressed as the mean ± standard deviation (S.D). Statistical analysis was done on GraphPad Prism version 8.0.2. For β-galactosidase quantification, the one-way analysis of variance (ANOVA) followed by the Tukey HSD test for multiple comparisons was applied. For qRT-PCR experiments, ANOVA followed by Dunnett’s multiple comparisons test was used by setting the WT values as control. For densitometric data, the two-tailored Student’s t-test for unpaired data was used. A two-sided p-value (p) < 0.05 was considered significant.
Results
Self-Interaction of B. cereus FlhF
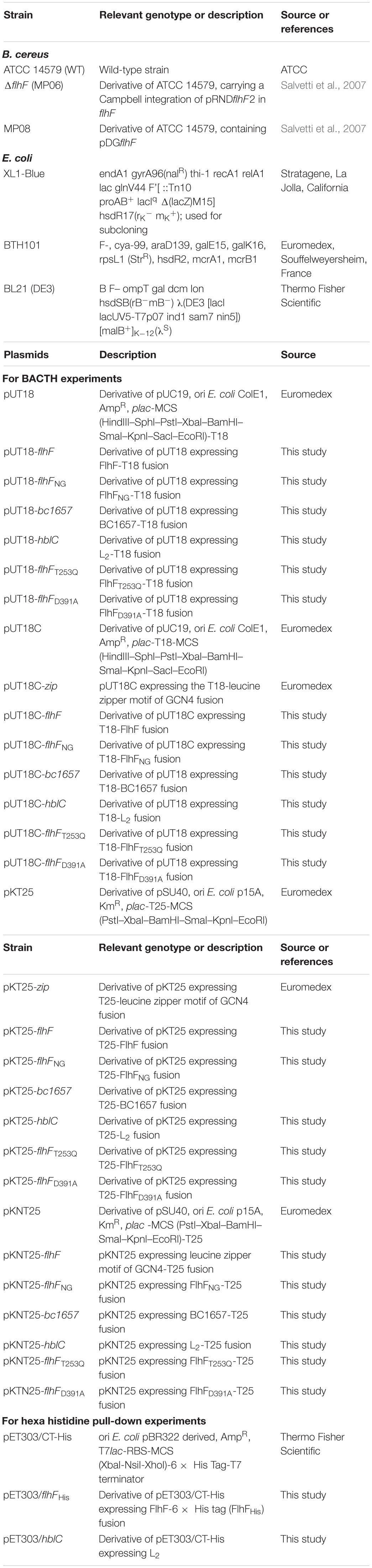
The bacterial adenylate cyclase two-hybrid (BACTH) approach was applied to investigate whether FlhF forms dimers in B. cereus. The complete ORF of B. cereus flhF was fused to the T18 and T25 encoding domains of Bordetella pertussis adenylate cyclase (Cya) contained in pUT18, pUT18C, pKT25, and pKNT25 (Table 1), under the control of the plac promoter. Expression of recombinant pUT18 and pKNT25 vectors lead to the production of hybrid proteins in which the adenylate cyclase domain is localized in a C-terminal position. Fusion proteins expressed by recombinant pUT18C and pKT25 possess the enzymatic domain in an N-terminal location. Four combinations of recombinant plasmids (pUT18-flhF/pKT25-flhF, pUT18-flhF/pKNT25-flhF, pUT18C-flhF/pKT25-flhF, and pUT18C-flhF/pKNT25-flhF) were co-transformed into the reporter strain E. coli BTH101, which is defective for Cya activity. Fusion proteins were tested for their ability to interact by growing co-transformed cells on M63 minimal medium plates containing maltose as unique carbon source and IPTG as inducer of protein expression. Only in the case of FlhF/FlhF interaction, Cya could be restored, cyclic adenosine monophosphate (cAMP) synthesized, and the maltose operon genes as well as the cAMP-reporter gene β-galactosidase expressed in the E. coli BTH101 strain. Colonies containing the combinations of vectors pUT18-flhF/pKT25-flhF and pUT18C-flhF/pKT25-flhF were able to grow, indicating that a physical interaction between FlhF monomers exists in vivo when fusion proteins carried the T25 domain in a N-terminal position. These qualitative observations were supported by quantification of the β-galactosidase activity that was significantly increased (p < 0.001) compared to the negative controls (Figure 1A).
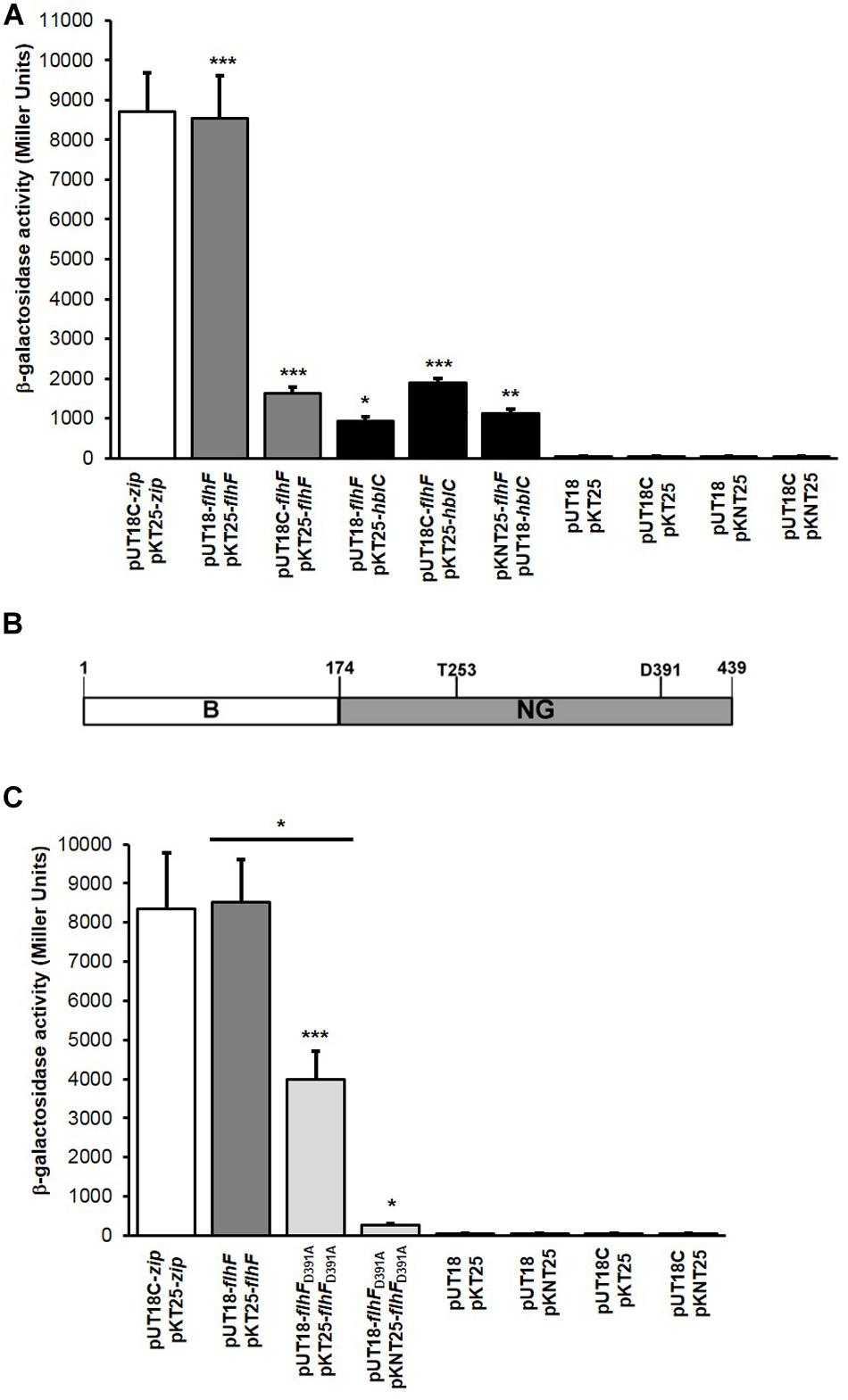
Figure 1. FlhF homodimerization, structure, and FlhF/L2 interaction. (A) Quantification of the β-galactosidase activity (reported in Miller Units) produced by positive clones. FlhF/FlhF interactions: gray bars; FlhF/L2 interactions: black bars; positive control: white bar; negative controls: dark gray bars. Values are expressed as the mean + S.D. from three independent experiments. ***p < 0.001, **p < 0.01, *p < 0.05, compared to the negative controls. (B) N-terminal B (white box, residues 1–173) and NG (light gray box, residues 174–439) domains of B. cereus FlhF. Threonine (T253) and aspartic acid (D391) residues potentially involved in protein homodimerization that were mutated by COE-PCR (T253Q and D391A) are shown. (C) Quantification of the β-galactosidase activity (reported in Miller Units) produced by E. coli clones grown on M63 medium. FlhFD391A/FlhFD391A interactions: light gray bars; FlhF/FlhF interaction: gray bar; positive control: white bar; negative controls: dark gray bars. Values are expressed as the mean + S.D. from three independent experiments. ***p < 0.001; **p < 0.01, *p < 0.05.
GTP-dependent homodimerization of B. subtilis FlhF has principally been attributed to a threonine (T184) and an aspartic acid residue (D320) found in the G1 and G4 elements of the G domain respectively, (Bange et al., 2007). To determine whether mutations in the corresponding residues of B. cereus FlhF (i.e. T253; D391; Figure 1B) altered self-dimerization, point mutations were generated by site-directed mutagenesis through combined overlap extension PCR (COE-PCR) to produce flhFT253Q and flhFD391A variants. BACTH experiments were separately performed with the two variants. No colonies were obtained on M63 plates with clones containing flhFT253Q, indicating that this residue is essential for FlhF dimerization (data not shown). In contrast, positive interactions were obtained with E. coli clones containing the combinations of vectors pUT18-flhFD391A/pKT25-flhFD391A and pUT18-flhFD391A/pKNT25-flhFD391A (data not shown). The β-galactosidase activity of these clones (Figure 1C) was higher than the negative controls (p < 0.001 for the combination pUT18-flhFD391A/pKT25-flhFD391A, and p < 0.05 for the combination pUT18-flhFD391A/pKNT25-flhFD391A), indicating that FlhF carrying the D391A substitution is still able to form dimers. However, β-galactosidase activity of E. coli clones containing pUT18-flhFD391A/pKT25-flhFD391A was reduced compared to cells carrying the unmutated flhF in the same combination of vectors (p < 0.05; Figure 1C). This result suggests that, although dimers can be formed when FlhF carries the mutation D391A, interaction among mutated monomers is less stable than among monomers of wild-type FlhF.
FlhF Is Required for Synthesis but Not for the Export of Flagellin
Since the FlhF depletion in B. cereus causes a drastical reduction in the number of flagella (Salvetti et al., 2007), we performed a comparative analysis of the expression of the flagellin gene bc1657 in the ΔflhF mutant and in the wild-type (WT) strain. As references, we used the genes rpoA and bc4306, encoding the DNA-directed RNA polymerase subunit alpha (Salvetti et al., 2011) and the Gatb_Yqey domain-containing protein (Reiter et al., 2011), respectively. For each strain, the threshold cycle (CT) of bc1657 was separately normalized to the CTs of the reference genes. The transcription level of bc1657 was lower in the ΔflhF mutant compared to the WT (p < 0.001) using both reference genes (Figure 2A). These findings indicate that FlhF is required for full bc1657 expression in B. cereus.
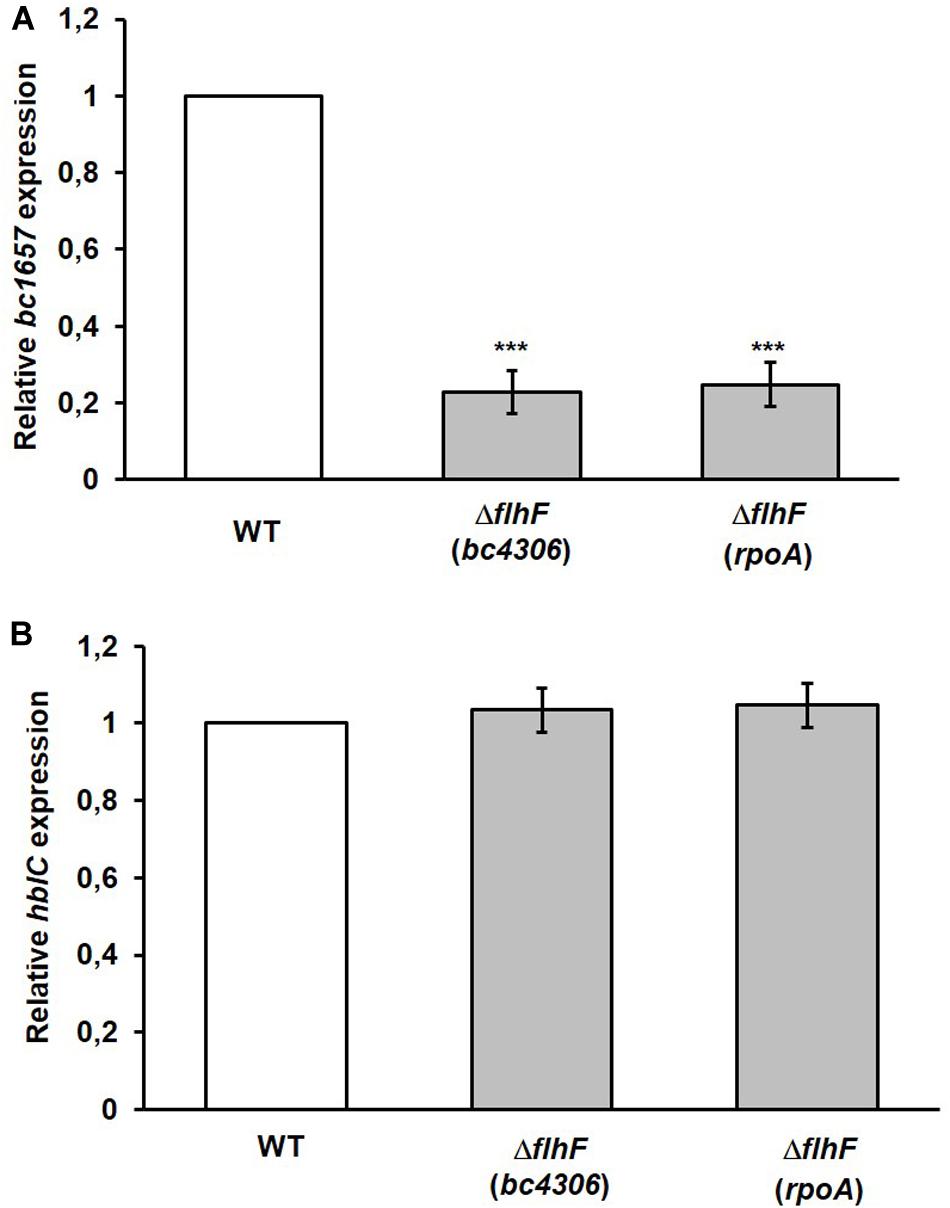
Figure 2. qRT-PCR analysis of the expression of flagellin (bc1657) and L2 (hblC) encoding genes in the ΔflhF mutant. (A) The level of bc1657 expression in the ΔflhF mutant (light gray bars) is presented as fold change relative to the corresponding gene in the WT strain (B. cereus ATCC 14579; white bar). (B) The level of hblC expression in the ΔflhF mutant (light gray bars) is presented as fold change relative to the corresponding gene in the WT strain (B. cereus ATCC 14579; white bar). For both figures, bc4306 and rpoA were independently used as reference genes for normalization. The relative bc1657 and hblC expression in the WT (white bars) is conventionally set to 1. Bars represent means ± S.D. from three independent experiments. ***p < 0.001 compared to the WT.
To evaluate whether FlhF could directly be involved in flagellin targeting/recruitment to the plasma membrane, a physical interaction between FlhF and flagellin was checked in vivo. BACTH experiments were applied using eight combinations of recombinant vectors carrying the complete ORFs of flhF and bc1657 (Table 1). No colonies were obtained on the M63 minimal medium indicating that FlhF is unable to interact with flagellin BC1657 in vivo (data not shown).
FlhF Is Dispensable for the Expression of the L2 Encoding Gene
The low amount of the L2 component of HBL found in the extracellular proteome of the B. cereus ΔflhF strain (Mazzantini et al., 2016) prompted us to assess whether the transcription level of the L2 encoding gene (hblC) was altered in such a strain. A comparative expression analysis of hblC was performed in the WT and the ΔflhF mutant. Also in this case, rpoA and bc4306 were used as reference genes. In contrast to flagellin, no significant difference (p > 0.05) in hblC expression was found using both reference genes, indicating that FlhF depletion does not alter hblC expression (Figure 2B). Therefore, the reduced amount of L2 secreted by the ΔflhF mutant (ratio ΔflhF/WT = 0.38) (Mazzantini et al., 2016) is not the result of an expression defect, but it likely seems the consequence of altered export.
FlhF Physically Interacts With the L2 Component of HBL
To investigate on whether a physical interaction between FlhF and L2 occurs, BACTH experiments were performed using the eight combinations of recombinant vectors carrying the complete ORFs of flhF and hblC of B. cereus (Table 1). Positive interactions on the M63 minimal medium were obtained with clones containing the combinations of vectors pUT18-flhF/pKT25-hblC, pUT18C-flhF/pKT25-hblC, and pKNT25-flhF/pUT18C-hblC. The β-galactosidase activity of positive clones (Figure 1A) was significantly higher than the negative controls (p < 0.05 for the combination pUT18-flhF/pKT25-hblC; p < 0.001 for the combination pUT18C-flhF/pKT25-hblC, and p < 0.01 for the combination pKNT25-flhF/pUT18C-hblC). These findings indicate that FlhF directly interacts with L2 in vivo.
Hexa-histidine (His) pull-down was used as independent in vitro method to confirm FlhF/L2 interaction. A C-terminal His-tagged version of B. cereus FlhF (FlhFHis) and an untagged version of L2 were expressed in E. coli BL21 (DE3) to form the bait and prey proteins, respectively. The soluble fraction of cells expressing FlhFHis was applied to the HisPurTM Cobalt Resin and used as bait. The resin was washed and directly treated with elution buffer to obtain the FlhFHis fraction. In parallel, FlhFHis-bound resin was incubated with the extract of cells expressing L2 and treated with the elution buffer to obtain the FlhFHis/L2 fraction. Both fractions were subjected to SDS-PAGE. A band having a molecular weight compatible with FlhFHis was found in the FlhFHis and FlhFHis/L2 fractions (Figure 3A). A second band with a molecular weight compatible with that predicted for L2 was observed only in the FlhFHis/L2 eluted fraction. The identification of L2 and FlhFHis in the FlhFHis/L2 fraction was performed by immunoblot analysis using anti-L2 (Beecher et al., 1995b) and anti-His antibodies, respectively. As shown in Figure 3B, immunoreactive bands corresponding to L2 and FlhFHis were found in this fraction. No FlhFHis and L2 immunoreactive bands were detected in controls samples (data not shown), indicating that L2 was pulled down only in the presence of FlhFHis. Taken together, these findings indicate a direct interaction between FlhF and the L2 protein.
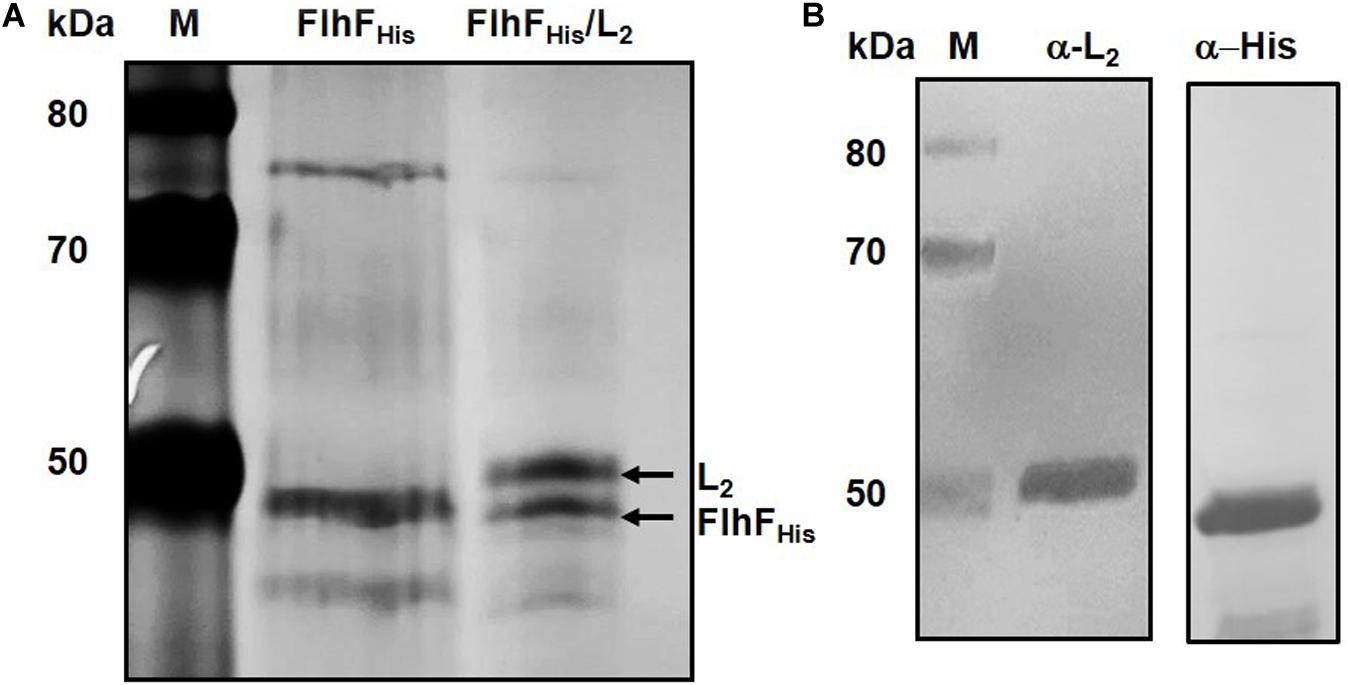
Figure 3. FlhF/L2 interaction tested by hexa-histidine pull-down. (A) SDS-PAGE of FlhFHis (lane 2) and FlhFHis/L2 (lane 3) eluted fractions. (B) Immunoblot analysis of the FlhFHis/L2 eluted fraction using anti-L2 (α-L2) and anti-His (α-His) antibodies. M = Thermo Scientific Spectra Multicolor Broad Range Protein Ladder (Thermo Fisher Scientific). Numbers on the left margins of the panels indicate the position of the molecular weight standards (kDa).
The NG Domain of FlhF Interacts With L2
To investigate if a physical contact between the NG domain and L2 can occur, the nucleotide sequence (flhFNG) encoding the NG domain of FlhF (Figure 1B) was amplified and cloned in the different vectors of the BACTH system (Table 1). The NG domain was tested for its ability to interact with L2 by co-transforming E. coli BTH101 with each of the above mentioned plasmids and each of the plasmids containing hblC (Table 1). The combinations of vectors pUT18-flhFNG/pKT25-hblC, pUT18-flhFNG/pKNT25-hblC, and pUT18C-flhFNG/pKNT25-hblC gave positive results on the M63 minimal medium. The β-galactosidase activity of these clones was significantly increased compared to the negative controls (p < 0.001; Figure 4). Based on these results, the NG domain of FlhF is able to interact with L2.

Figure 4. FlhFNG/L2 interactions tested by BACTH experiments. Quantification of the β-galactosidase activity (reported in Miller Units) produced by positive clones. FlhFNG/L2 interactions: black bars; positive control: white bar; negative controls: dark gray bars. Values are expressed as the mean + S.D. from three independent experiments. ***p < 0.001 compared to the negative controls.
Point Mutations in FlhF Impair L2 Binding
To identify a possible correlation between FlhF dimerization and L2 binding, BACTH experiments were performed. All the possible combinations of vectors containing flhFT253Q and hblC, and flhFD391A and hblC (Table 1) were used to transform E. coli BTH101.
None of the co-transformed E. coli cells grew on the M63 minimal medium (data not shown), indicating that no interaction of FlhFT253Q and FlhFD391A with L2 occurs and that both residues (T253 and D391) are essential for FlhF/L2 interaction. Taken together, our experiments suggest that the GTP-dependent homodimerization of FlhF is a prerequisite for its ability to interact with the L2 subunit of HBL.
Effect of FlhF Overproduction on Flagella and L2 Secretion
To evaluate the effect of FlhF overproduction on B. cereus flagellation and L2 secretion, a strain carrying a plasmid containing flhF under the control of the pspac promoter (MP08) was analyzed in comparison to the WT (Salvetti et al., 2007). Light microscopy of bacteria subjected to flagellar staining suggested an increase in the amount of flagella per cell in the MP08 strain compared to the WT (Figure 5A). Therefore, the extent of cell flagellation was analyzed by quantifying the amount of filament assembled flagellin in both strains. Density of the flagellin band was 1.83 ± 0.26-fold higher in the MP08 strain compared to the WT (p < 0.01; Supplementary Figure S1).
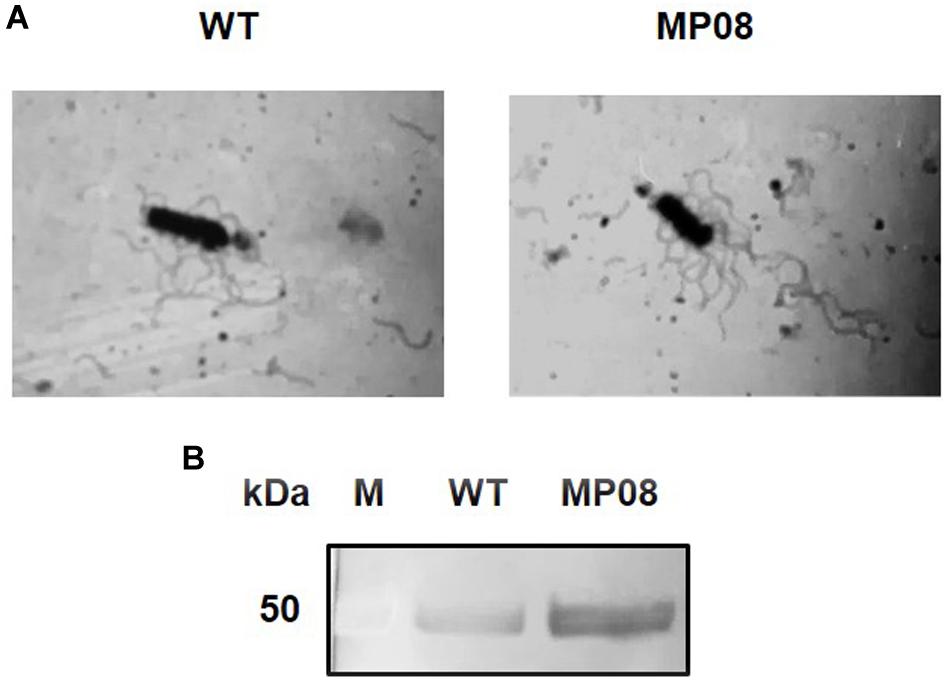
Figure 5. Effect of FlhF overexpression on B. cereus flagella and L2 secretion. (A) Light microscopy (1000×) of B. cereus ATCC 14579 (WT) and MP08 strain subjected to flagellar staining. (B) Immunodetection of the L2 component of HBL with rabbit polyclonal sera specific to L2. M = Thermo Scientific Spectra Multicolor Broad Range Protein Ladder (Thermo Fisher Scientific). Numbers on the left margins of the panels indicate the position of the molecular weight standards (kDa).
Normalized culture supernatants from the WT and MP08 were subjected to SDS-PAGE and immunoblot analysis for the detection of L2. The protein was more abundant (2.51 ± 0.171-fold higher) in the supernatant of the MP08 strain compared to the WT (p < 0.001; Figure 5B). Overall, these results indicate that FlhF overproduction increases the number of flagella and L2 secretion in B. cereus.
Discussion
In the present study, we found that B. cereus FlhF is able to self-dimerize in vivo. Two of the four combinations of recombinant plasmids used to test FlhF dimerization indicated a positive interaction. The absence of interaction observed with the other combinations could be the consequence of a misfolding or instability of the hybrid proteins that prevent the contact, as previously suggested (Bange et al., 2011; Battesti and Bouveret, 2012).
FlhF homodimers are stabilized by hydrogen bonds in trans that involve both FlhF monomers (Bange et al., 2007). A threonine residue, conserved in all FlhF proteins with the exception of P. aeruginosa FlhF (Mazzantini et al., 2016), is believed to be essential for dimer stabilization through the interaction with the side chains of a lysine and a glutamic acid residue of the G4 element (Bange et al., 2007). However, the impact of the replacement of this residue on FlhF dimerization has never been analyzed before. Herein we demonstrate that a T253Q substitution in B. cereus FlhF completely abrogates homodimerization in vivo, indicating that this residue is essential for the interaction of monomers and/or for dimer stabilization.
In B. subtilis FlhF, specificity toward the guanosine nucleotides GDP and GTP is established by the interaction between an aspartic acid residue (D320 in B. subtilis FlhF) of the G4 element and the guanine base of the nucleotide with two hydrogen bonds. This residue is conserved in all SRP-GTPases (Bange et al., 2007). When a mutation was introduced in a residue corresponding to D320 in FtsY, the ability of the protein to bind and hydrolyze GTP was impaired, and FtsY could not form a complex with Ffh (Shan et al., 2004). A point mutation of the same aspartic acid residue strongly reduced the FlhF GTPase activity in V. cholerae and S. oneidensis (Green et al., 2009; Gao et al., 2015), and abrogated GTP binding and attenuated self-dimerization in P. aeruginosa (Schniederberend et al., 2013). The same mutation also impaired GTP load and dimer formation of FlhF in V. alginolyticus (Kusumoto et al., 2009; Kondo et al., 2018). In the present study, we found that the replacement of the corresponding D391 residue in B. cereus FlhF attenuates homodimerization, suggesting that this residue is required but not essential in this process. This result is in line with previous findings showing that a threonine residue of the G5 element (T343 in B. subtilis), conserved only in FlhF homologs, forms a third hydrogen bond with GTP in the active site, enhancing FlhF nucleotide binding compared to other SRP-GTPases (Bange et al., 2007). Since this residue is also conserved in B. cereus FlhF (T414) (Mazzantini et al., 2016), it could exert the same function maintaining a basal degree of GTP binding and FlhF dimerization.
Functionally, the main role of FlhF is supposedly related to flagellar assembly. The protein has been described to, directly or indirectly, drive the synthesis of flagellar components, stimulating wild-type levels of flagellar genes expression (Niehus et al., 2004; Kim et al., 2012; Kazmierczak and Hendrixson, 2013; Schuhmacher et al., 2015; Ren et al., 2018), and/or to dictate the point of flagellar assembly by recruiting early flagellar components to the plasma membrane (Murray and Kazmierczak, 2006; Green et al., 2009; Guttenplan et al., 2013; Altegoer et al., 2014). In the ΔflhF mutant of B. cereus, the amount of flagellin BC1657 was drastically reduced (ratio ΔflhF/WT = 0.22) (Mazzantini et al., 2016). In this study, qRT-PCR experiments were performed to analyze the transcriptional level of bc1657 in the WT and the ΔflhF mutant using two housekeeping genes, rpoA and bc4306. rpoA has often been used as endogenous reference for qRT-PCR in B. cereus ATCC 14579 (Grande Burgos et al., 2009; Cadot et al., 2010; Salvetti et al., 2011). Since this gene has a high coefficient of variation (of about 43.0%), we also included the more stably expressed gene bc4306, which has a coefficient of variation of 8.41% (Bergman et al., 2006; Reiter et al., 2011). The finding that the transcription level of bc1657 was significantly reduced in the ΔflhF mutant, suggests that FlhF is required for the regulation of flagellar gene expression in B. cereus. In line with this hypothesis, no physical interaction between FlhF and BC1657 was found in vivo. However, we cannot exclude that B. cereus FlhF could be involved in the recruitment of early flagellar basal body components to the plasma membrane, as previously indicated for other microorganisms (Green et al., 2009; Guttenplan et al., 2013) thus indirectly influencing the expression of late flagellar genes.
The striking similarities between FlhF and Ffh/FtsY might reflect similar functions in the targeting of proteins to the plasma membrane before secretion. Herein we demonstrate that the reduction in the export of the L2 component of HBL by the ΔflhF mutant (Mazzantini et al., 2016) is not due to a reduced expression of hblC and that physical interaction between FlhF and L2 occurs in B. cereus. FlhF interacts with the flagellar export chaperone FliS in Helicobacter pylori (Rain et al., 2001), with FlhG in V. alginolyticus, B. subtilis and S. putrefaciens (Kusumoto et al., 2008; Bange et al., 2011; Rossmann et al., 2015), with the pilus-twitching motility regulator XooPilL in X. oryzae pv oryzae (Shen et al., 2001), with the C ring protein FliG and the polar scaffolding protein FimV in P. aeruginosa (Schniederberend et al., 2019). In addition, the protein is required for the recruitment of the MS ring early flagellar component FliF in V. cholerae (Green et al., 2009). This is the first report showing evidence that FlhF interacts with a protein that is not involved in flagellar biogenesis or bacterial motility.
Little is known on the role of FlhF domains and on how these domains are involved in the binding of FlhF to its interactors. The G domain of FlhF was found essential in the recruitment of FliF to the cell pole in V. cholerae (Green et al., 2009), while the entire NG domain mediates the interaction between FlhF and FlhG in B. subtilis (Bange et al., 2011). In this study, we describe the L2 component of HBL as novel binding partner of FlhF in B. cereus. FlhF can cycle between two mutually exclusive forms: a cytoplasmic, monomeric, and nucleotide-free or GDP-bound, and a GTP-dependent, membrane-bound homodimer which represents the “ON” state of FlhF (Bange et al., 2007; Schuhmacher et al., 2015; Kondo et al., 2018). The dimeric form of FlhF has been described to be required for FlhF/FlhG contact and for the recruitment of flagellar components to the plasma membrane (Kusumoto et al., 2009; Bange et al., 2011; Schuhmacher et al., 2015; Kondo et al., 2018). Our finding that the mutant variants FlhFT253Q and FlhFD391A completely lose the ability to interact with L2 suggests that stable homodimerization of FlhF, which requires both T253 and D391, is crucial for L2 binding in B. cereus. Thus, we favor the idea that the GTP-bound homodimer of FlhF serves during secretion of the L2 subunit of HBL.
The effect of flhF overexpression on bacterial flagellation has been investigated in several microorganisms. Similar to polar flagellates (Pandza et al., 2000; Correa et al., 2005; Kusumoto et al., 2008; Green et al., 2009) and different to B. subtilis (Guttenplan et al., 2013), FlhF overproduction in B. cereus results in a hyperflagellated phenotype. FlhF levels are also critical in defining the amount of one extracellularly secreted HBL component, being therefore the protein important in modulating the pathogenic potential of B. cereus.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
EG and GB designed the study. DMa and RF performed the experiments and prepared the figures. All authors analyzed the data and wrote the manuscript.
Funding
This research was partially supported by the Deutsche Forschungsgemeinschaft (DFG) project P11 with the collaborative research center 174, “Spatial-temporal dynamics of bacterial cells.”
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Michele Lai for densitometric analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00879/full#supplementary-material
FIGURE S1 | Effect of FlhF overexpression on B. cereus flagellar filaments. SDS-PAGE analysis of extracellular flagellin of the WT and MP08 strains.
TABLE S1 | Primers used for PCR amplification and/or sequencing.
Footnotes
References
Akopian, D., Shen, K., Zhang, X., and Shan, S. O. (2013). Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721. doi: 10.1146/annurev-biochem-072711-164732
Altegoer, F., Schuhmacher, J., Pausch, P., and Bange, G. (2014). From molecular evolution to biobricks and synthetic modules: a lesson by the bacterial flagellum. Biotechnol. Genet. Eng. Rev. 30, 49–64. doi: 10.1080/02648725.2014.921500
Bange, G., Kümmerer, N., Grudnik, P., Lindner, R., Petzold, G., Kressler, D., et al. (2011). Structural basis for the molecular evolution of SRP-GTPase activation by protein. Nat. Struct. Mol. Biol. 18, 1376–1380. doi: 10.1038/nsmb.2141
Bange, G., Petzold, G., Wild, K., Parltz, R., and Sinning, I. (2007). The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP. Proc. Natl. Acad. Sci. U.S.A. 104, 13621–13625. doi: 10.1073/pnas.0702570104
Battesti, A., and Bouveret, E. (2012). The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58, 325–334. doi: 10.1016/j.ymeth.2012.07.018
Beecher, D. J., Pulido, J. S., Barney, N. P., and Wong, A. C. (1995a). Extracellular virulence factors in Bacillus cereus endophthalmitis: methods and implication of involvement of hemolysin BL. Infect. Immun. 63, 632–639.
Beecher, D. J., Schoeni, J. L., and Wong, A. C. (1995b). Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect. Immun. 63, 4423–4428.
Bergman, N. H., Anderson, E. C., Swenson, E. E., Niemeyer, M. M., Miyoshi, A. D., and Hanna, P. C. (2006). Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J. Bacteriol. 188, 6092–6100. doi: 10.1128/JB.00723-726
Cadot, C., Tran, S. L., Vignaud, M. L., Buyser, M. L., Kolstø, A. B., Brisabois, A., et al. (2010). InhA1, NprA, and HlyII as candidates for markers to differentiate pathogenic from nonpathogenic Bacillus cereus strains. J. Clin. Microbiol. 48, 1358–1365. doi: 10.1128/JCM.02123-2129
Callegan, M. C., Novosad, B. D., Ramirez, R., Ghelardi, E., and Senesi, S. (2006). Role of swarming migration in the pathogenesis of Bacillus endophthalmitis. Invest. Ophthalmol. Vis. Sci. 47, 4461–4467. doi: 10.1167/iovs.06-0301
Calvio, C., Celandroni, F., Ghelardi, E., Amati, G., Salvetti, S., Ceciliani, F., et al. (2005). Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J. Bacteriol. 187, 5356–5366. doi: 10.1128/JB.187.15.5356-5366.2005
Correa, N. E., Peng, F., and Klose, K. E. (2005). Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol. 187, 6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005
de Leeuw, E., te Kaat, K., Moser, C., Menestrina, G., Demel, R., de Kruijff, B., et al. (2000). Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J. 19, 531–541. doi: 10.1093/emboj/19.4.531
Ehling-Schulz, M., Lereclus, D., and Koehler, T. M. (2019). The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol. Spectr. 7:10.1128/microbiolspec.GPP3-0032-2018.. doi: 10.1128/microbiolspec.GPP3-0032-2018
Gao, T., Shi, M., Ju, L., and Gao, H. (2015). Investigation into FlhFG reveals distinct features of FlhF in regulating flagellum polarity in Shewanella oneidensis. Mol. Microbiol. 98, 571–585. doi: 10.1111/mmi.13141
Ghelardi, E., Celandroni, F., Salvetti, S., Beecher, D. J., Gominet, M., Lereclus, D., et al. (2002). Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184, 6424–6433. doi: 10.1128/jb.184.23.6424-6433.2002
Ghelardi, E., Celandroni, F., Salvetti, S., Ceragioli, M., Beecher, D. J., Senesi, S., et al. (2007). Swarming behavior of and hemolysin BL secretion by Bacillus cereus. Appl. Environ. Microbiol. 73, 4089–4093. doi: 10.1128/AEM.02345-2346
Grande Burgos, M. J., Kovacs, A. T., Mironczuk, A. M., Abriouel, H., Galvez, A., and Kuipers, O. P. (2009). Response of Bacillus cereus ATCC 14579 to challenges with sublethal concentrations of enterocin AS-48. BMC Microbiol. 9:227. doi: 10.1186/1471-2180-9-227
Green, J. C. D., Kahramanoglou, C., Rahman, A., Pender, A. M. C., Charbonnel, N., and Fraser, G. (2009). Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J. Mol. Biol. 391, 679–690. doi: 10.1016/j.jmb.2009.05.075
Grudnik, P., Bange, G., and Sinning, I. (2009). Protein targeting by the signal recognition particle. Biol. Chem. 390, 775–782. doi: 10.1515/BC.2009.102
Gulbronson, C. J., Ribardo, D. A., Balaban, M., Knauer, C., Bange, G., and Hendrixson, D. R. (2016). FlhG employs diverse intrinsic domains and influences FlhF GTPase activity to numerically regulate polar flagellar biogenesis in Campylobacter jejuni. Mol. Microbiol. 99, 291–306. doi: 10.1111/mmi.13231
Guttenplan, S. B., Shaw, S., and Kearns, D. B. (2013). The cell biology of peritrichous flagella in Bacillus subtilis. Mol. Microbiol. 87, 211–229. doi: 10.1111/mmi.12103
Hainzl, T., Huang, S., Meriläinen, G., Brännström, K., and Sauer-Eriksson, A. E. (2011). Structural basis of signal-sequence recognition by the signal recognition particle. Nat. Struct. Mol. Biol. 18, 389–391. doi: 10.1038/nsmb.1994
Harshey, R. M., and Matsuyama, T. (1994). Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. U.S.A. 91, 8631–8635. doi: 10.1073/pnas.91.18.8631
Hussain, H., and Chong, N. F. (2016). Combined overlap extension PCR method for improved site directed mutagenesis. Biomed. Res. Int. 2016:8041532. doi: 10.1155/2016/8041532
Janda, C. Y., Li, J., Oubridge, C., Hernández, H., Robinson, C. V., and Nagai, K. (2010). Recognition of a signal peptide by the signal recognition particle. Nature 465, 507–510. doi: 10.1038/nature08870
Kazmierczak, B. I., and Hendrixson, D. R. (2013). Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol. Microbiol. 88, 655–663. doi: 10.1111/mmi.12221
Kim, S. M., Lee, D. H., and Choi, S. H. (2012). Evidence that the Vibrio vulnificus flagellar regulator FlhF is regulated by a quorum sensing master regulator SmcR. Microbiology 158, 2017–2025. doi: 10.1099/mic.0.059071-59070
Kondo, S., Imura, Y., Mizuno, A., Homma, M., and Kojima, S. (2018). Biochemical analysis of GTPase FlhF which controls the number and position of flagellar formation in marine Vibrio. Sci. Rep. 8:12115. doi: 10.1038/s41598-018-30531-30535
Kusumoto, A., Nishioka, N., Kojima, S., and Homma, M. (2009). Mutational analysis of the GTP-binding motif of FlhF which regulates the number and placement of the polar flagellum in Vibrio alginolyticus. J. Biochem. 146, 643–650. doi: 10.1093/jb/mvp109
Kusumoto, A., Shinohara, A., Terashima, H., Kojima, S., Yakushi, T., and Homma, M. (2008). Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154, 1390–1399. doi: 10.1099/mic.0.2007/012641-12640
Leipe, D. D., Wolf, Y. I., Koonin, E. V., and Aravind, L. (2002). Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72. doi: 10.1006/jmbi.2001.5378
Liu, W., Xie, Y., Ma, J., Luo, X., Nie, P., Zuo, Z., et al. (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. doi: 10.1093/bioinformatics/btv362
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Mazzantini, D., Celandroni, F., Salvetti, S., Gueye, S. A., Lupetti, A., Senesi, S., et al. (2016). FlhF is required for swarming motility and full pathogenicity of Bacillus cereus. Front. Microbiol. 7:1644. doi: 10.3389/fmicb.2016.01644
Mercier, E., Holtkamp, W., Rodnina, M. V., and Wintermeyer, W. (2017). Signal recognition particle binds to translating ribosomes before emergence of a signal anchor sequence. Nucleic Acids Res. 45, 11858–11866. doi: 10.1093/nar/gkx888
Miller, J. H. (1992). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Murray, T. S., and Kazmierczak, B. I. (2006). FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188, 6995–7004. doi: 10.1128/JB.00790-06
Navarrete, B., Leal-Morales, A., Serrano-Ron, L., Sarrió, M., Jiménez-Fernández, A., Jiménez-Díaz, L., et al. (2019). Transcriptional organization, regulation and functional analysis of flhF and fleN in Pseudomonas putida. PLoS One 14:e0214166. doi: 10.1371/journal.pone.0214166
Niehus, E., Gressmann, H., Ye, F., Schlapbach, R., Dehio, M., Dehio, C., et al. (2004). Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52, 947–961. doi: 10.1111/j.1365-2958.2004.04006.x
Pandza, S., Baetens, M., Park, C. H., Au, T., Keyan, M., and Matin, A. (2000). The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol. 36, 414–423. doi: 10.1046/j.1365-2958.2000.01859.x
Rain, J. C., Selig, L., De Reuse, H., Battaglia, V., Reverdy, C., Simon, S., et al. (2001). The protein–protein interaction map of Helicobacter pylori. Nature 409, 211–215. doi: 10.1038/35051615
Ramarao, N., and Sanchis, V. (2013). The pore-forming haemolysins of Bacillus cereus: a review. Toxins 5, 1119–1139. doi: 10.3390/toxins5061119
Reiter, L., Kolstø, A. B., and Piehler, A. P. (2011). Reference genes for quantitative, reverse-transcription PCR in Bacillus cereus group strains throughout the bacterial life cycle. J. Microbiol. Methods 86, 210–217. doi: 10.1016/j.mimet.2011.05.006
Ren, F., Lei, T., Song, Z., Yu, T., Li, Q., Huang, J., et al. (2018). Could FlhF be a key element that controls Campylobacter jejuni flagella biosynthesis in the initial assembly stage? Microbiol. Res. 207, 240–248. doi: 10.1016/j.micres.2017.12.006
Rossmann, F., Brenzinger, S., Knauer, C., Dörrich, A. K., Bubendorfer, S., Ruppert, U., et al. (2015). The role of FlhF and HubP as polar landmark proteins in Shewanella putrefaciens CN-32. Mol. Microbiol. 98, 727–742. doi: 10.1111/mmi.13152
Salvetti, S., Faegri, K., Ghelardi, E., Kolstø, A. B., and Senesi, S. (2011). Global gene expression profile for swarming Bacillus cereus bacteria. Appl. Environ. Microbiol. 77, 5149–5156. doi: 10.1128/AEM.00245-211
Salvetti, S., Ghelardi, E., Celandroni, F., Ceragioli, M., Giannessi, F., and Senesi, S. (2007). FlhF, a signal recognition particle-like GTPase, is involved in the regulation of flagellar arrangement, motility behavior and protein secretion in Bacillus cereus. Microbiology 153, 2541–2552. doi: 10.1099/mic.0.2006/005553-5550
Schniederberend, M., Abdurachim, K., Murray, T., and Kazmierczak, B. I. (2013). The GTPase activity of FlhF is dispensable for flagellar localization, but not motility, in Pseudomonas aeruginosa. J. Bacteriol. 195, 1051–1060. doi: 10.1128/JB.02013-2012
Schniederberend, M., Williams, J. F., Shine, E., Shen, C., Jain, R., Emonet, T., et al. (2019). Modulation of flagellar rotation in surface-attached bacteria: a pathway for rapid surface-sensing after flagellar attachment. PLoS Pathog. 15:e1008149. doi: 10.1371/journal.ppat.1008149
Schuhmacher, J. S., Thormann, K. M., and Bange, G. (2015). How bacteria maintain location and number of flagella? FEMS Microbiol. Rev. 39, 812–822. doi: 10.1093/femsre/fuv034
Senesi, S., Celandroni, F., Salvetti, S., Beecher, D. J., Wong, A. C. L., and Ghelardi, E. (2002). Swarming motility in Bacillus cereus and characterization of a fliY mutant impaired in swarm cell differentiation. Microbiology 148, 1785–1794. doi: 10.1099/00221287-148-6-1785
Senesi, S., and Ghelardi, E. (2010). Production, secretion and biological activity of Bacillus cereus enterotoxins. Toxins 2, 1690–1703. doi: 10.3390/toxins2071690
Shan, S. O., Stroud, R. M., and Walter, P. (2004). Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2:e320. doi: 10.1371/journal.pbio.0020320
Shen, Y., Chern, M. S., Silva, F. G., and Ronald, P. (2001). Isolation of a Xanthomonas oryzae pv. oryzae flagellar operon region and molecular characterization of flhf. Mol. Plant Microbe Interact. 14, 204–213. doi: 10.1094/MPMI.2001.14.2.204
Stjepanovic, G., Kapp, K., Bange, G., Graf, C., Parlitz, R., Wild, K., et al. (2011). Lipids trigger a conformational switch that regulates signal recognition particle (SRP)-mediated protein targeting. J. Biol. Chem. 286, 23489–23497. doi: 10.1074/jbc.M110.212340
The UniProt Consortium, (2015). UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212. doi: 10.1093/nar/gku989
Keywords: Bacillus cereus, flhF, L2, hemolysin BL, virulence, flagellin
Citation: Mazzantini D, Fonnesu R, Celandroni F, Calvigioni M, Vecchione A, Mrusek D, Bange G and Ghelardi E (2020) GTP-Dependent FlhF Homodimer Supports Secretion of a Hemolysin in Bacillus cereus. Front. Microbiol. 11:879. doi: 10.3389/fmicb.2020.00879
Received: 28 February 2020; Accepted: 15 April 2020;
Published: 06 May 2020.
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Monika Ehling-Schulz, University of Veterinary Medicine, Vienna, AustriaCatherine Duport, University of Avignon, France
Copyright © 2020 Mazzantini, Fonnesu, Celandroni, Calvigioni, Vecchione, Mrusek, Bange and Ghelardi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gert Bange, Z2VydC5iYW5nZUBzeW5taWtyby51bmktbWFyYnVyZy5kZQ==; Emilia Ghelardi, ZW1pbGlhLmdoZWxhcmRpQG1lZC51bmlwaS5pdA==
 Diletta Mazzantini
Diletta Mazzantini Rossella Fonnesu1
Rossella Fonnesu1 Francesco Celandroni
Francesco Celandroni Alessandra Vecchione
Alessandra Vecchione Devid Mrusek
Devid Mrusek Gert Bange
Gert Bange Emilia Ghelardi
Emilia Ghelardi