
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 May 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00865
This article is part of the Research Topic Role Of Small Colony Variants (SCVs) And Persisters In Staphylococcus aureus Pathogenesis And Development Of Therapeutic Approaches View all 9 articles
 Yongpeng Shang1†
Yongpeng Shang1† Xiaofei Wang1†
Xiaofei Wang1† Zhong Chen1†
Zhong Chen1† Zhihui Lyu1
Zhihui Lyu1 Zhiwei Lin2
Zhiwei Lin2 Jinxin Zheng2
Jinxin Zheng2 Yang Wu1
Yang Wu1 Qiwen Deng2
Qiwen Deng2 Zhijian Yu2*
Zhijian Yu2* Ying Zhang3*
Ying Zhang3* Di Qu1*
Di Qu1*PhoU homologs are one of the determinant factors in the regulation of persister formation and phosphate metabolism in many bacterial species; however, the functions of PhoU homologs exhibit species-specific characteristics. The pathogenesis of Staphylococcus aureus is closely correlated with persister formation and virulence factors. The functions of two PhoU homologs, PhoU1 and PhoU2, in S. aureus are unclear yet. In this study, single- and double-deletion mutants of phoU1 and phoU2 were generated in strain USA500 2395. The ΔphoU1 or ΔphoU2 mutants displayed a change in persister formation and virulence compared to the parent strain; the persisters to vancomycin and levofloxacin were decreased at least 1,000-fold, and the number of intracellular bacteria surviving in the A549 cells for 24 h decreased to 82 or 85%. The α-hemolysin expression and activity were increased in the ΔphoU2 mutants. Transcriptome analysis revealed that 573 or 285 genes were differentially expressed by at least 2.0-fold in the ΔphoU1 or ΔphoU2 mutant vs. the wild type. Genes involved in carbon and pyruvate metabolism were up-regulated, and virulence genes and virulence regulatory genes were down-regulated, including type VII secretion system, serine protease, leukocidin, global regulator (sarA, rot), and the two-component signal transduction system (saeS). Correspondingly, the deletion of the phoU1 or phoU2 resulted in increased levels of intracellular pyruvate and ATP. Deletion of the phoU2, but not the phoU1, resulted in the up-regulation of inorganic phosphate transport genes and increased levels of intracellular inorganic polyphosphate. In conclusion, both PhoU1 and PhoU2 in S. aureus regulate virulence by the down-regulation of multiple virulence factors (type VII secretion system, serine protease, and leucocidin) and the persister generation by hyperactive carbon metabolism accompanied by increasing intracellular ATP. The results in S. aureus are different from what we have previously found in Staphylococcus epidermis, where only PhoU2 regulates biofilm and persister formation. The different functions of PhoU homologs between the two species of Staphylococcus warrant further investigation.
Staphylococcus aureus is a human pathogen that colonizes human skin and mucous membranes (Otto, 2010; Tong et al., 2015). It can invade phagocytic, epithelial, or endothelial cells and allows for the formation of persisters that can cause chronic and recurrent infections (Conlon, 2014). Moreover, the pathogenicity of S. aureus is closely correlated to its virulence factors (such as hemolysins, leukotoxins, enterotoxin, and coagulase) and biofilm formation (Dinges et al., 2000; Otto, 2014). Persisters are a subpopulation of bacterial cells that are tolerant to antibiotics without changes in minimum inhibitory concentration (MIC) values in the whole population and are one of the most important factors in the failure of antibiotic therapy (Wilmaerts et al., 2019). Persister formation is often explained by multiple mechanisms such as the reduction of cellular energy, cessation of DNA replication, blocked transcription and translation, decreased intracellular antibiotic concentrations, and antibiotic-induced damage (El-Halfawy and Valvano, 2015; Fisher et al., 2017; Wilmaerts et al., 2019). PhoU homologs are associated with persister formation in species including Escherichia coli, Pseudomonas aeruginosa, Mycobacterium tuberculosis, and Staphylococcus epidermidis (Li and Zhang, 2007; Shi and Zhang, 2010; Wang et al., 2017). The biological functions of the PhoU homologs in S. aureus are unclear and still require further investigation.
PhoU orthologs are found in many species of bacteria, but not in humans, and have been identified as phosphate-specific transport system accessory proteins (Morohoshi et al., 2002; Wang et al., 2013; de Almeida et al., 2015). In E. coli, PhoU protein is involved in the response to environmental Pi levels by interacting with PhoR and PstB (Gardner et al., 2014). A crystal structure analysis of PhoU protein in Thermotoga maritima revealed multinuclear iron clusters by a conserved E(D)XXXD motif pair (Liu et al., 2005). Little is known about the function of PhoU besides being a phosphate regulator. One phoU homolog is in the pst operon of E. coli and P. aeruginosa, whereas two phoU homologs are found in M. tuberculosis, Mycobacterium marinum, S. epidermidis, and S. aureus. In 2007, Li and Zhang (2007) showed that the inactivation of phoU in E. coli resulted in persister reduction and an up-regulated transcription level of some functional genes involved in energy production, nutrient transportation, flagellar synthesis, and chemotaxis. In P. aeruginosa, a phoU mutant displayed increased levels of intracellular guanosine tetraphosphate (ppGpp) and polyphosphate (polyP), impacted antibiotic susceptibility, and decreased growth rate of planktonic bacteria; however, there was no effect on biofilm formation (de Almeida et al., 2015). Two phoU homologs from M. tuberculosis, phoY1 and phoY2, showed distinct functions in different strains. In the M. tuberculosis H37Rv strain, the phoY2 mutant, but not the phoY1 mutant, increased the susceptibility to rifampicin and pyrazinamide and decreased persister formation, while in the M. tuberculosis Erdman strain, phoY1 and phoY2 double mutants, but not the single mutants (deletion of phoY1 or phoY2), increased the susceptibility to rifampicin and decreased persister formation (Shi and Zhang, 2010; Namugenyi et al., 2017). Thus, the biological functions of PhoU homologs show species-specific characteristics, which require individual investigations.
Our previous studies found that in S. epidermidis, the biological functions and regulation of PhoU homologs are different from those of some other bacterial species in specific ways (Wang et al., 2017). In S. epidermidis, the genome contains two PhoU homologs: phoU1, in the same operon as that pst and it has high homology with the phoU of E. coli, and phoU2, which is located in the pit operon (Wang et al., 2017). PhoU2, but not PhoU1, is an important regulator of biofilm formation and of tolerance to multiple stresses (Wang et al., 2017). The deletion of phoU2 resulted in growth retardation, decreased persister formation, and biofilm reduction in S. epidermidis, while phoU1 deletion had no effect on the bacterial phenotypes tested (Wang et al., 2017). PhoU2 deletion alters cellular metabolic processes such as inorganic phosphate metabolism, galactose metabolism, the pentose phosphate pathway, and the tricarboxylic acid cycle (Wang et al., 2017). In the genus Staphylococcus, both S. aureus and S. epidermidis are important pathogens, but their pathogenic mechanisms differ. The main pathogenic mechanisms of S. aureus are secretions of a variety of toxins to destroy host cells, invasion and survival in cells, and biofilm formation (Fraunholz and Sinha, 2012; Otto, 2014; Moormeier and Bayles, 2017). By comparison, the pathogenesis of S. epidermidis is mainly due to the formation of biofilms on materials used for medical interventions that resist clearance by the immune system and antibiotics. Persisters are generated during S. aureus or S. epidermidis infection (Grassi et al., 2017). A genome analysis of S. aureus USA500 2395 allowed for the identification of two phoU homologs, phoU1 and phoU2, located in the pst operon and pit operon, respectively. Overton et al. (2011) analyzed the transcriptomes and the proteomics of the S. aureus ATCC 8325 strain and suggested that phoU1 is involved in persister formation in the presence of the cationic bacitracin (ranalexin) (Overton et al., 2011). The pit operon of the S. aureus HG003 strain contains the pitA and the phoU2 (pitR) genes. A single-point mutation in pitA (downstream of phoU2) resulted in high tolerance to daptomycin, and phoU2 (pitR) was required for the expression of this phenotype (Mechler et al., 2016). We therefore speculated that the regulatory functions of the two phoU homologs (phoU1 and phoU2) of S. aureus may differ from those in S. epidermidis. In S. aureus, the regulation of persister formation and virulence, and their interconnections, may be associated with the phoU homologs and requires further investigation.
In the present study, we generated phoU single mutants of S. aureus strain USA500 2395, named ΔphoU1 and ΔphoU2. The effects of these deletions on bacterial growth, persister formation, and metabolism were investigated. Comparisons of the transcriptome profiles of ΔphoU1 vs. the parent strain and of ΔphoU2 vs. the parent strain allowed for differentially expressed genes (DEGs) that were involved in phosphate metabolism, carbon and pyruvate metabolism, and virulence gene expression to be identified. We analyzed intracellular inorganic phosphate (Pi), polyP, glucose, pyruvate, ATP, bacterial survival in cells, and hemolysis, respectively. The deletion of phoU1 or phoU2 of S. aureus increased carbon metabolism and intracellular ATP levels, which may be associated with the decreased antibiotic tolerance of bacteria and the reduced intracellular survival of bacteria in human lung epithelial A549 cells. The results suggest that both PhoU1 and PhoU2 of S. aureus are involved in the regulation of persister generation and virulence.
The bacterial strains and plasmids used for cloning are listed in Supplementary Table S1. The S. aureus strain USA500 2395 was used for the construction of gene knockout and complementation. S. aureus strain USA300 FPR3757 and SA113 were used for gene silencing. E. coli DC10B was used for staphylococcal cloning host. S. aureus strains were grown at 37°C in tryptic soya broth (TSB) (OXOID, Basingstoke, United Kingdom). E. coli was grown at 37°C in Luria broth (1% tryptone, 0.5% NaCl, and 0.5% yeast extract). B2 media (2.5% yeast extract, 1% tryptone, 0.5% glucose, 2.5% NaCl, and 0.1% K2HPO4) was used for preparing and recovering the electrocompetent cells of S. aureus after electroporation. The antibiotics were used at the following concentrations: ampicillin at 100 μg/ml, chloramphenicol at 10 μg/ml, anhydrotetracycline at 50 ng/ml, and erythromycin at 10 μg/ml (Sigma, United States).
The phoU1 and phoU2 deletion mutants of S. aureus USA500 2395 were constructed using the temperature-sensitive plasmid pKOR1 (Bae and Schneewind, 2006). The upstream and downstream fragments of phoU1 or phoU2 were amplified by PCR, ligated by T4 DNA ligase, and cloned into vector pKOR1, resulting in recombinant pKOR1-ΔphoU1 or pKOR1-ΔphoU2. The plasmids pKOR1-ΔphoU1andpKOR1-ΔphoU2 were transferred into E. coli DC10Band then into USA500 2395.ΔphoU1andΔphoU2were generated by the homologous recombination method of allelic exchange, as described (Bae and Schneewind, 2006). By transferring pKOR1-ΔphoU2 vector into ΔphoU1, the double-deletion mutant of phoU1 and phoU2 (ΔphoU1ΔphoU2) was constructed. The gene deletion mutants were verified by PCR, quantitative reverse transcription-PCR (qRT-PCR), and sequencing.
Complementation of the ΔphoU1 and the ΔphoU2was achieved by the E. coli–Staphylococcus shuttle vector pCN51 and pRB473. The phoU1 or the phoU2, with their promoter regions, was amplified by PCR and inserted into pCN51. The pCN51-phoU1 or pCN51-phoU2 plasmid was transferred into the corresponding deletion mutants by electroporation. The pRB473-phoU2 plasmid was constructed according to the abovementioned method. The complemented ΔphoU1ΔphoU2 was constructed by transferring the pCN51-phoU1 and pRB473-phoU2 by electroporation.
The silencing strains of phoU1 or phoU2 were constructed by the shuttle plasmid pMX6 (Helle et al., 2011; Xu et al., 2017). The hairpin structure formed by the plasmid pMX6 was used for constructing antisense RNA (asRNA) expression. The plasmid of asRNA phoU1 or asRNA phoU2 was constructed by firstly amplifying a sequence of about 200 nt containing the start codon of the corresponding gene. Then, the sequence was inserted in the reverse direction downstream of the anhydrotetracycline-inducible promoter in pMX6. The plasmid of asRNA phoU1 or asRNA phoU2 was transferred into USA500 2395, USA300 FPR3757, and SA113 by electroporation, resulting in the silencing strains. Primers are listed in the Supplementary Tables S3, S4.
S. aureus strains were grown to stationary phase (12 h) and then diluted (1:200) in TSB medium. For bacterial growth curves, bacteria were grown at 37°C with shaking at 220 rpm and monitored by measuring the OD600 at 1 h intervals for 24 h by Bioscreen C (Turku, Finland). For viable bacterial count, bacteria were grown at 37°C with shaking, and the cells were plated in serial dilutions on TSB agar at 4 and 12 h, then the colony-forming units (CFU) were counted.
According to the CLSI National Committee for Clinical Laboratory, the MICs were determined by using serial twofold dilutions of the antibiotics (vancomycin, levofloxacin, gentamicin, and daptomycin) in Mueller–Hinton broth (MH, OXOID). The initial cell density was 105 CFU/ml. Then, the initial bacteria were inoculated into MH broth for 16–20 h. The MIC is defined as the lowest concentration of antibiotics that inhibited the visible growth of bacteria. MH broth without antibiotic served as the control. The minimal bactericidal concentration (MBC) values were identified by plating 100-μl samples, from tubes with no visible bacterial growth in MIC tests, onto MHB agar plates. The concentration that reduced the viability of the initial bacterial inoculum by =99.9% was the MBC.
Persisters were determined as described (Li and Zhang, 2007). S. aureus strains were grown in TSB for 12 h to reach the stationary phase. Different 25 × MIC antibiotics were added to the cultures (final concentrations: levofloxacin at 12.5 mg/L and vancomycin at 25 mg/L). At each time point (0, 4, 8, 12, 24, 48, and 72 h), 1 ml of bacteria was collected by centrifugation (6,000 rpm), washed twice with cold saline, serially diluted 10-fold, and plated on TSB agar. Then, the CFU were counted.
Overnight cultures (12 h) of S. aureus strains were diluted 1:200 into 7 mM H2O2 or 0.005% sodium dodecyl sulfate (SDS) TSB and incubated at 37°C, and the OD600 was measured. Overnight cultures of S. aureus strains were serially diluted 10-fold. Five microliters of the diluted samples was spotted onto TSA plates containing 7 mM H2O2 or 0.005% SDS and incubated at 37°C overnight. The plates with bacterial colonies were photographed.
Total RNA for RNA-Seq and qRT-PCR was extracted by RNeasy Mini kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. In brief, S. aureus and the derivative strains were diluted 1:200 into 20 mM TSB and incubated at 37°C. At 12 h, 8 ml of bacteria was collected at 6,000 rpm and washed twice times with cold saline. With 0.5 ml of 0.1 mm zirconia-silica beads, the cells were homogenized for five rounds using a Mini-Bead beater (Biospec, Bartlesville, OK, United States) at 4,800 rpm for 1 min and were cooled on ice for 1 min. Then, the samples were centrifuged at 12,000 rpm, and RNA in the supernatant was extracted using the silica-based filter of Neasy Mini kit.
The samples were prepared according to the Illumina RNA Sequencing Sample Preparation Guide. In brief, three biological replicates for each of the S. aureus were treated with RNase-free DNase I (Takara) to remove the genomic DNA. The BioAnalyzer 2100 system was used to evaluate RNA quality. The samples were treated with the RiboZero rRNA removal kit (gram-positive organisms) to remove ribosomal RNA. Fragmented RNA was reverse-transcribed using random primers. The cDNA library included fragment sizes of 200–300 bp, which were prepared by the mRNA-Seq Sample Prep kit and verified on the BioAnalyzer 2100 system. Then, the fragment size is amplified by Illumina cBot and sequenced by Illumina HiSeq 2500.
Quality control involves discarding of rRNA reads, sequencing adapters, short fragments, and other low-quality reads. The remaining reads were multi-mapped to the genome of S. aureus USA500 2395 at the NCBI website with the Bowtie2 software. BED Tools software was used to count the transcript expression levels. Per kilobase of gene per million mapped reads (RPKM) reported the RNA-seq gene expression values. Integrated Genomics Viewer was used to visualize the date. DEGseq software was used to quantify the differential expression of different transcripts. Significant differences in expression ratios were defined as at least 2.0- or 0.5-fold change in transcript level. The P-values cutoff was calculated (0.05). IPA Software was used to analyze differentially expressed genes in the canonical pathway. The number of genes mapped to the pathway vs. the total genes present in the canonical pathway determined the significance of the pathway.
A pool of 5 μg of total RNA for S. aureus strains was DNA-digested and reverse-transcribed into cDNA using PrimeScript TM RT reagent kit. Then, 100 ng/μl cDNA was used for qRT-PCR with TB green PCR reagents. All reagents were from Takara Biotechnology. The reactions were performed in a Mastercycler realplex system (Eppendorf AG, Hamburg, Germany) and normalized using gyrB (DNA gyrase subunit B) as the housekeeping gene. Each gene qRT-PCR was performed in triplicate.
Cytoscape software was used to construct a protein–protein interaction network (PPI) according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database1.
Intracellular Pi was quantified with a commercially available kit (no. ab65622; Abcam), which was modified for S. aureus (Mechler et al., 2015). In brief, overnight cultures of S. aureus strains were diluted 1:200 into TSB and incubated at 37°C. After 12 h, the cells were harvested by centrifugation at 4,000 rpm for 10 min at 4°C. The cells were washed twice with ice-cold, double-distilled water and adjusted to OD600 ≈1; then, they were lysed with 0.1 mm glass-silica beads in a BeadBeater apparatus (BioSpec), followed by centrifugation to get the supernatant. The supernatant was used to measure OD650 (Thermo VARIOSKAN LUX). The Pi levels were determined according to the manufacturer’s instructions.
Intracellular polyP levels were determined using 4’-6-diamidino-2-phenylindole (DAPI; Sigma) as described previously (Aschar-Sobbi et al., 2008). Overnight cultures of S. aureus strains were diluted 1:200 into TSB and incubated at 37°C. After 12 h, the cells were washed twice with Tris-HCl buffer (100 mM Tris, pH 7.4) and adjusted to OD600 ≈ 1. DAPI was added to a final concentration of 20 μM. After 15 min of agitation at room temperature in the dark, the fluorescence signal was determined using a microplate reader with excitation at 415 nm and emission at 550 nm (Thermo VARIOSKAN LUX).
Glucose and pyruvate were measured using the Glucose Assay Kit (ab65333; Abcam) and Pyruvate Assay Kit (ab65342; Abcam) according to the manufacturer’s instructions (Liang et al., 2018; Schnack et al., 2019). S. aureus strains were diluted 1:200 into TSB and incubated at 37°C. After 12 h, culture supernatant and cells were harvested. The cells were washed twice with phosphate-buffered saline (PBS) and adjusted to OD600 ≈ 1 with assay buffer and then were lysed with 0.1-mm glass-silica beads in a BeadBeater apparatus (BioSpec), followed by centrifugation to obtain the supernatant. The fluorescence signal of the supernatant was determined using a microplate reader with excitation at 535 nm and emission at 587 nm (Thermo VARIOSKAN LUX).
The ATP levels of S. aureus strains were measured using a Promega BacTiter Glo kit according to the manufacturer’s instructions (Bonora et al., 2015). S. aureus strains were diluted 1:200 into TSB and incubated at 37°C. After 12 h, the cells were washed twice with PBS and adjusted to OD600 ≈1. A volume of BacTiter-GloTM Reagent was added equal to the volume of the cell culture medium present in each well. The complexes were mixed briefly and incubated for 5 min. Luminescence was determined using a microplate reader (Thermo VARIOSKAN LUX).
An assay of bacterial invasion was performed as previously described (Liang et al., 2006; Sharma et al., 2013; Mechler et al., 2015). A549 human lung epithelial cells were cultured in Dulbecco’s modified Eagle’s medium/F-12 medium supplemented with 10% fetal calf serum, streptomycin (100 μg/ml), and penicillin (100 μg/ml) in a 5% CO2 incubator at 37°C. The cells were passaged and expanded every 2 days. At 1 day prior to infection, 2 × 105 cells were seeded in 24-well plates with an antibiotic-free cell culture medium and incubated at 37°C in a 5% CO2 incubator. S. aureus strains were grown for 4 and 12 h in TSB. One milliliter of the bacterial culture was washed with ice-cold saline and resuspended in 1 ml of cell culture medium. Approximately 2 × 106 CFU/ml of S. aureus strains were seeded in a 24-well plate. The plates were centrifuged at 1,000 rpm for 5 min to synchronize infection and then incubated for 1 h at 37°C in a 5% CO2 incubator. The culture medium was removed. Extracellular bacteria were treated with 100 μg/ml gentamicin and 20 μg/ml lysostaphin (Sangon Biotech) for 30 min. The monolayer cells were washed three times with PBS (pH 7.4) and incubated for an additional 1 h (invasion capacity) or 24 h (intracellular survival). Intracellular bacteria were counted for the CFU by lysis of the host cells with 0.01% Triton X-100.
As rabbit erythrocytes were exquisitely sensitive to alpha-toxin (Bernheimer et al., 1968), assessment of the alpha-hemolysin activity was done by the analysis of rabbit erythrocytes lysis as Kernodle DS et al. had previously described (Sharma-Kuinkel et al., 2012). Briefly, the S. aureus strains were diluted 1:200 into TSB and incubated at 37°C. After 12 h, the culture supernatant was harvested. The supernatant was removed with a 0.22 μm filter (Millipore). Commercial 4% rabbit erythrocytes (SBJ-RBC-RAB003, Sbjbio, China) stored in Alsevers solution were four times diluted with PBS. The supernatant was added, equal to the volume of 1% of rabbit erythrocytes, and then incubated at 37°C for 30 min. The OD550 of each well was measured by a spectrophotometer. Then, 0.1% Triton X-100 served as the 100% hemolysis control (positive control), and PBS was the negative control. All experiments were performed in triplicate.
The collected supernatant was added 5 × loading buffer and the mixture was heated at 100°C for 5 min. Equal volumes of the mixture were separated using SDS-PAGE (10%) and transferred to polyvinylidene fluoride membrane (pore size, 0.45 μm; Millipore) by electrotransfer. The membranes were blocked with 5% skim milk for 2 h at room temperature and then incubated with alpha-hemolysin polyclonal antibody (Sigma S7531) overnight at 4°C. After washing three times with PBST, the membranes were incubated with HRP-conjugated goat anti-rabbit IgG (Santa Cruz, Santa Cruz, CA, United States). The immunoreactive bands were detected by visualization using an enhanced chemiluminescence Western blotting system (Thermo Fisher Scientific, Waltham, MA, United States).
All of the data were analyzed with SPSS (version 16.0) and compared using the independent-samples t-test. Differences with P-value < 0.05 were considered as statistically significant.
The RNA-Seq data were submitted to the Gene Expression Omnibus database. The accession number was GSE139071.
In the genome of S. aureus, strain USA500 2395 (GenBank accession number CP007499), two PhoU homologs are present: phoU1 is located in the pst operon and phoU2 is located in the pit operon. An alignment analysis of the PhoU homologs showed a high identity at the nucleotide level (>99%) and at the amino acid level (100%) in S. aureus strains (Supplementary Table S1). The identity of S. aureus PhoU1 or PhoU2, when compared with S. epidermidis, was 70 and 87%, respectively, at the nucleotide level and 95% for both proteins at the amino acid level (Supplementary Table S1).
Single or double mutants of phoU1 and phoU2 were constructed in the USA500 2395 strain using the temperature-sensitive plasmid pKOR1 and were named ΔphoU1, ΔphoU2, and ΔphoU1ΔphoU2. The mutants were verified by PCR, qRT-PCR, and sequencing (the data not shown). Complementation of the ΔphoU1 and the ΔphoU2 mutants was achieved using the vectors pCN51 and pRB473, which were named C-ΔphoU1 and C-ΔphoU2, respectively. We then determined the growth curve and the viable bacterial counts of the 3 phoU mutants and the wild-type strain USA500 2395. Both the growth curve and the viable bacterial count of ΔphoU1 or ΔphoU2 were similar to USA500 2395 (Supplementary Figures S1A,B).
Previous reports indicated that PhoU2, but not PhoU1, impacted antibiotic tolerance in S. epidermidis (Wang et al., 2017); therefore, we investigated the effects of deleting the phoU homolog on S. aureus antibiotic tolerance. The antibiotic tolerance of ΔphoU1, ΔphoU2, and USA500 2395 was determined using a modified procedure described by Li and Zhang. The ΔphoU1, ΔphoU2, and USA500 2395 strains were incubated in TSB for 12 h (to reach stationary growth phase), followed by 5 days of incubation with 25 × MIC vancomycin (25 μg/ml) or levofloxacin (12.5 μg/ml). The surviving CFU were counted at different time points. The antibiotic tolerance of both ΔphoU1 and ΔphoU2 showed 4-log reductions when compared with USA500 2395, and no viable bacteria were detected in either ΔphoU1 or ΔphoU2 after vancomycin exposure with 25 × MIC for 48 h, whereas in USA500 2395, 3.8 × 104 CFU were detected (Table 1). Moreover, no viable bacteria were detected in either ΔphoU1 or ΔphoU2 after exposure to 25 × MIC levofloxacin for 24 h, compared to USA500 2395 where 5 × 104 CFU were detected (Table 1). The complemented strains C-phoU1 and C-phoU2 restored the antibiotic tolerance, while the P-phoU1 and P-phoU2 strains with empty plasmids performed the same as the phoU mutants. The MIC and MBC of ΔphoU1 and ΔphoU2 were similar to that of USA500 2395 (Supplementary Table S2).
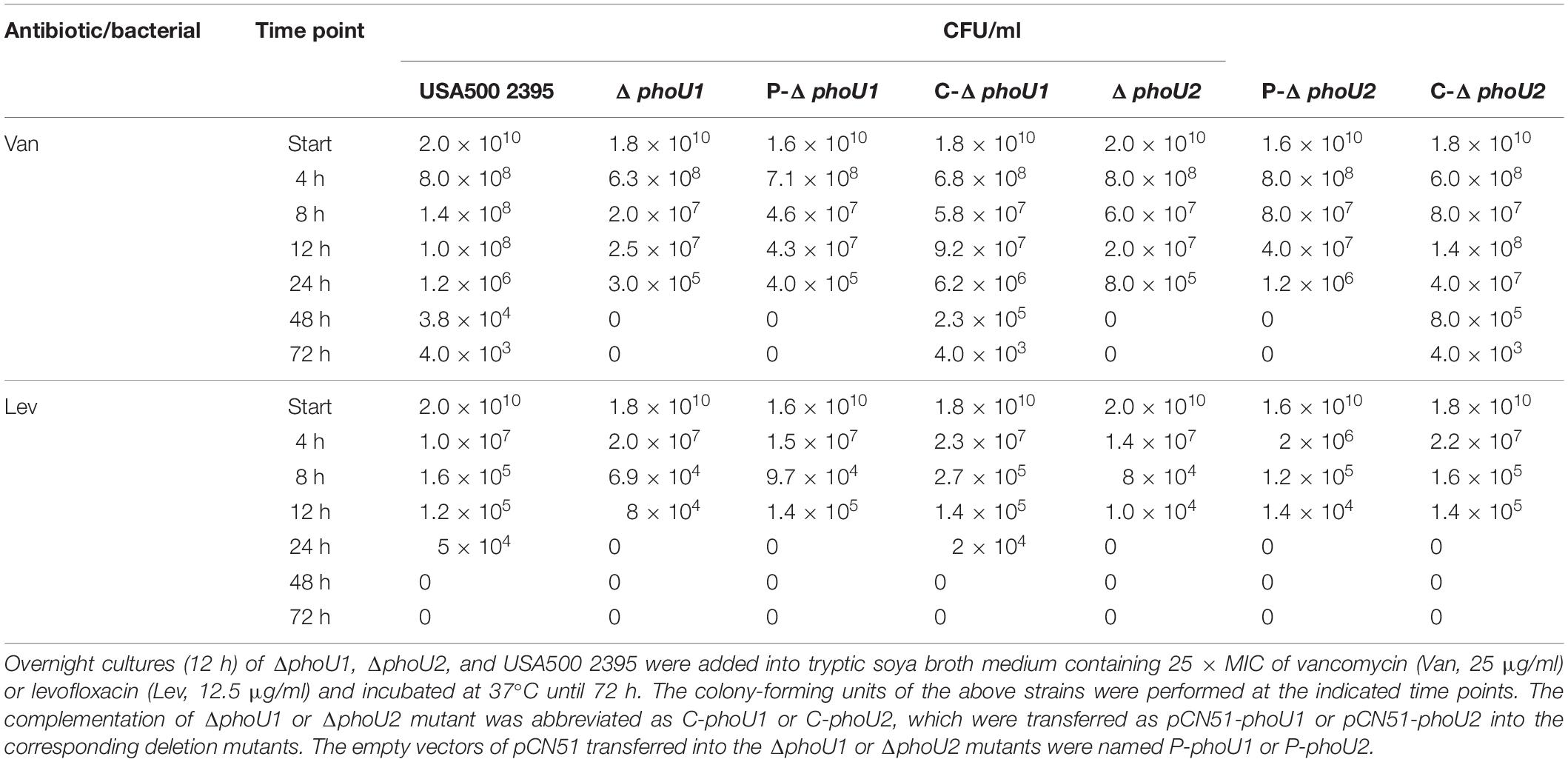
Table 1. Deletion of phoU1 or phoU2 decreased the persister formation in Staphylococcus aureus under the pressure of antibiotics.
We evaluated the tolerance of ΔphoU1 and ΔphoU2 to stresses (SDS and H2O2) and found that, when exposed in TSB containing 7 mmol H2O2, the lag phase of ΔphoU1 and ΔphoU2 was extended by 2 and 4 h, respectively, in comparison to USA500 2395 (Figure 1A). When cultured in 0.005% SDS TSB for 12 h, the OD600 values of ΔphoU1 and ΔphoU2 were 0.96 ± 0.013 and 0.84 ± 0.007, respectively, which were all lower than that of USA500 2395 (1.15 ± 0.023) (Figure 1C). When cultured in 7 mmol H2O2 (or 0.005% SDS) TSB for 6 h, bacteria were collected and plated on TSB agar plates in serial dilutions, and CFU were counted. The CFU of ΔphoU1 and ΔphoU2 were log10 7.92 ± 1.74 (or log10 7.6 ± 0.82) and log10 7.48 ± 0.53 (or log10 7. 20 ± 0.43), respectively, which were all lower than that of USA500 2395 log10 8.6 ± 0.72 (or log10 8.5 ± 0.53) (Figures 1B,D). The complemented strains C-phoU1 and C-phoU2 restored the tolerance to H2O2 and SDS.
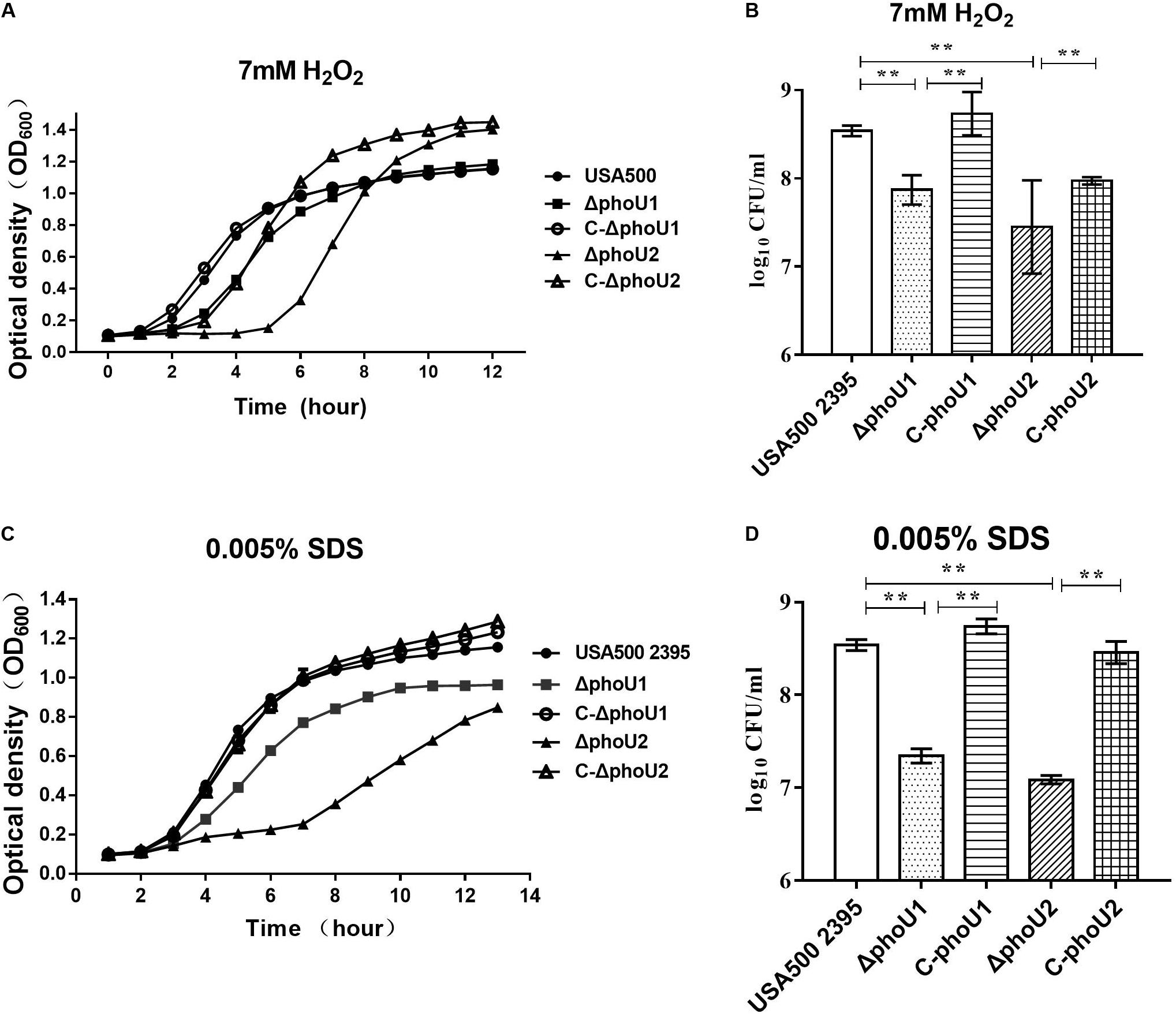
Figure 1. Deletion of phoU1 or phoU2 increased the sensitivity to H2O2 and sodium dodecyl sulfate (SDS) of Staphylococcus aureus. Overnight cultures (12 h) of ΔphoU1, ΔphoU2, C- phoU1, C- phoU2, and USA500 2395 strains were diluted at 1:200 into tryptic soya broth (TSB) containing either (A) hydrogen peroxide (H2O2; 7 mM) or (C) SDS (0.005%), then grown at 37°C with shaking at 220 rpm, and monitored by measuring the OD600 at the indicated time points until 12 h. (B) H2O2 (7 mM) or (D) SDS (0.005%), collecting (A,C) 6 h culture; bacteria in serial dilutions were plated on TSB agar plates and colony-forming units were counted. The experiments were repeated three times, and error bars indicate the standard deviation. Mutant strains exhibited significant differences (**P < 0.01) when compared with the wild-type and the complemented strains C-phoU1 or C-phoU2.
The ΔphoU1, ΔphoU2, and USA500 2395 strains were exposed onto TSB agar containing either H2O2 (7 mmol) or SDS (0.005%) and then incubated for 24 h at 37°C. The number of colonies on the plates displayed the final impact on the growth under the pressure of H2O2 and SDS. The ΔphoU1 and ΔphoU2 mutants displayed higher sensitivity to SDS than that of the parent strain, when 103 CFU was spotted onto TSB agar containing SDS (0.005%) (Supplementary Figure S4). However, the sensitivity of the ΔphoU1 and ΔphoU2 mutants to H2O2 was similar to that of the parent strain (Supplementary Figure S4).
For the analysis of ΔphoU1, ΔphoU2, and USA500 2395 transcriptomes, bacterial RNA was extracted at the stationary phase (after 12 h of growth) and analyzed by RNA-Seq. Transcriptome analysis indicated 573 differentially expressed genes between ΔphoU1 and USA500 2395, including 456 up-regulated and 117 down-regulated genes. Between ΔphoU2 and USA500 2395, 285 differentially expressed genes were identified, including 53 up-regulated and 232 down-regulated genes. We selected 65 DEGs for validation by RT-qPCR and established that 61 DEGs were consistent, following RNA-Seq. A PPI was constructed based on the KEGG database1. This suggested that the DEGs between ΔphoU1 and USA500 2395 were widely involved in various metabolic pathways (including iron transport and the metabolism of carbon, pyruvate, urease, and fatty acids) and that a majority of these genes were up-regulated (Table 2 and Figure 2). Furthermore, the transcription levels of the DEGs between ΔphoU2 and USA500 2395 that were linked to phosphate metabolism were up-regulated, whereas the majority of those linked to carbon and pyruvate metabolism were down-regulated (Table 3 and Figure 2). In both ΔphoU1 and ΔphoU2 mutants, the transcription levels of virulence-related factors (including the type VII secretion system operon esxA, esaA, essA, essB, and essC), serine-like proteinases (sspA, splF, splE, splD, splC, splB, and splA), leukocidin (lukG, lukH, and lukD), and hemolysin (hlgA, hlgC, hlgB, and hlα) were down-regulated compared with those of USA500 2395 (Supplementary Table S5). The transcription levels of fibrinogen-binding genes and sarA (a regulator of fibronectin-binding proteins, hemolysins, and serine proteases) between ΔphoU1 and USA500 2395 were down-regulated, whereas those of codY (a repressor of the Agr system) and graS/graR were up-regulated. The transcription levels of phoP/phoR between ΔphoU2 and USA500 2395 were up-regulated, while those of sarA, sarR, sarZ, codY, and rot were down-regulated.
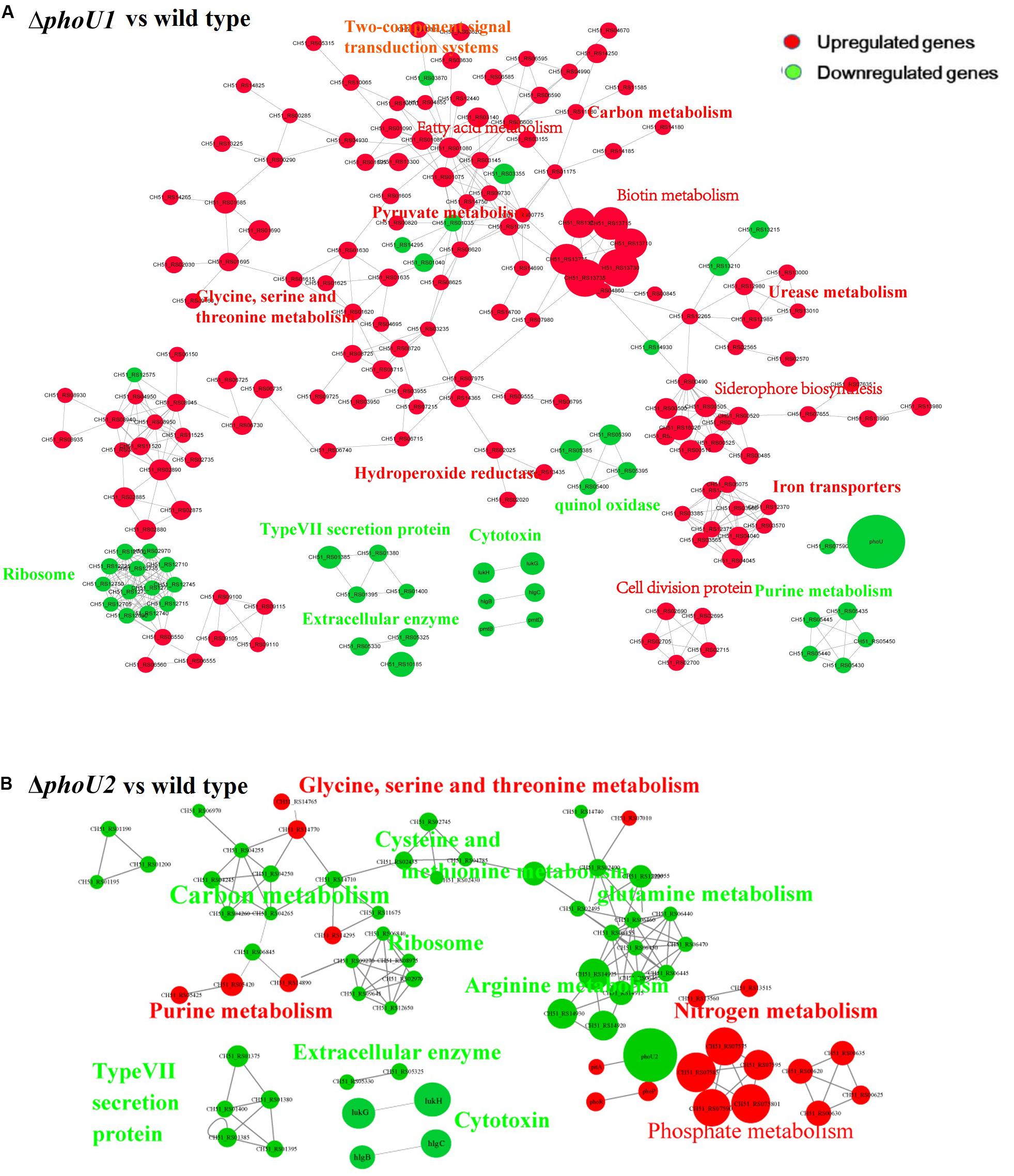
Figure 2. Protein–protein interaction network. Up-regulated or down-regulated differentially expressed genes are indicated in red or green, respectively. The size of the node indicates multiple gene expression. The lines represent protein–protein interactions, including binding/association, phosphorylation, activation, and inhibition. Interaction networks of proteins encoded by between DEGs identified by transcriptome comparison between ΔphoU1 and wild type (A) and ΔphoU2 and wild type (B).
Based on PPI analysis, we investigated the pathways of phosphate, carbon, and pyruvate metabolism using assays of Pi, polyP, glucose, pyruvate, and ATP, respectively.
PhoU homologs have been identified as phosphate-specific transport system accessory proteins. We quantified the Pi concentration of USA500 2395, ΔphoU1, and ΔphoU2 strains when grown in culture for 12 h. The intracellular Pi concentration of ΔphoU1 and ΔphoU2 was similar to that observed in USA500 2395 (Figure 3A). The intracellular polyP level was determined using a DAPI-based fluorescence approach. The ΔphoU2 mutant accumulated a significantly higher level of polyP (1.5-fold) than the parent strain after 12 h of growth (Figure 3B). The complemented strains of C-phoU2 exhibited restored intracellular polyP levels.
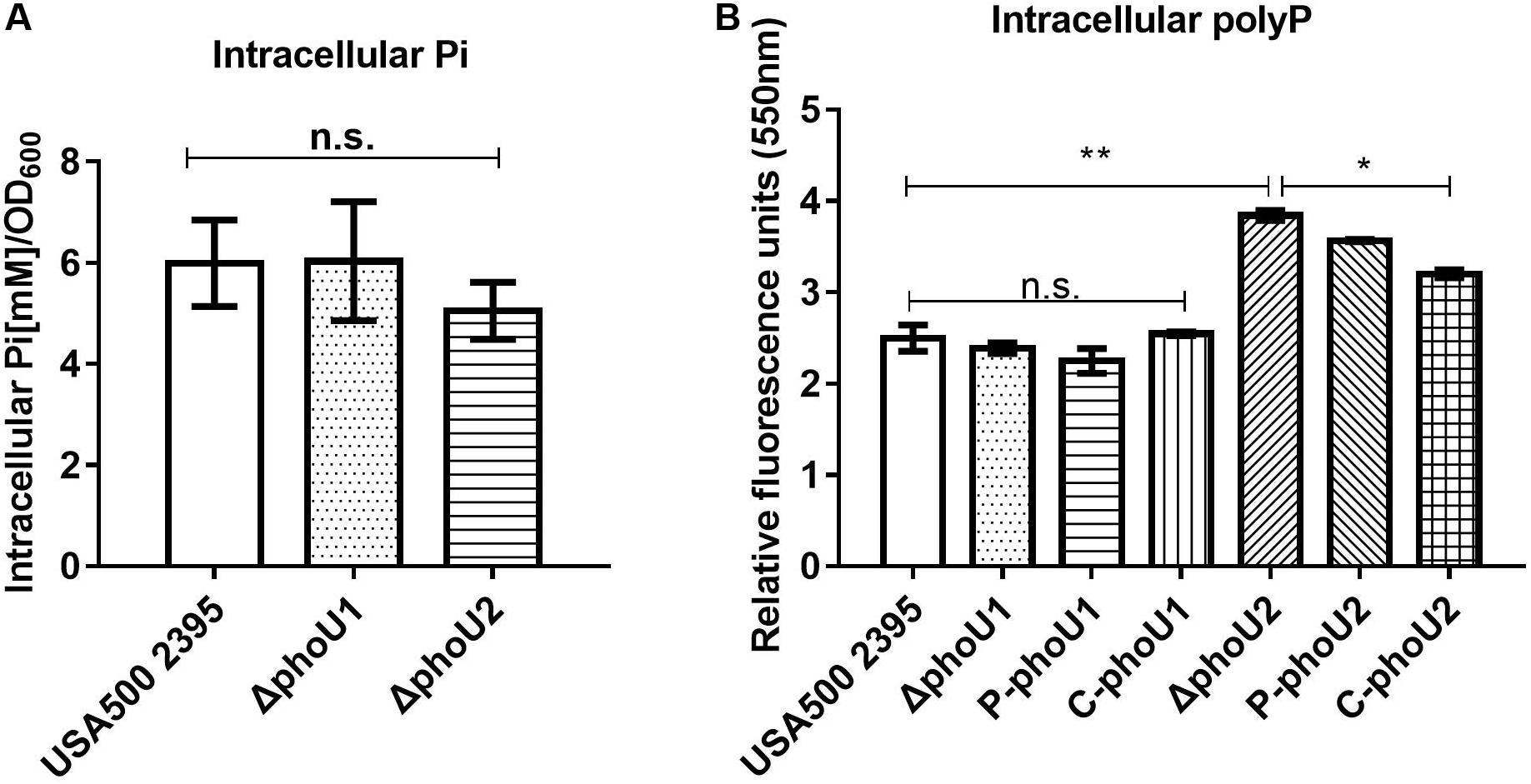
Figure 3. Intracellular Pi and polyP levels in ΔphoU1 and ΔphoU2 of Staphylococcus aureus. (A) Intracellular Pi. Bacteria were grown for 12 h; then, the cells were homogenized by OD600 and lysed with 0.1-mm glass-silica beads in a BeadBeater apparatus. Centrifugation to get the supernatant. The supernatant was used to measure OD650. Pi levels were determined according to the manufacturer’s instructions. (B) Intracellular polyP. Bacteria were grown for 12 h; then, the cells were homogenized by OD600 and incubated for 15 mins with 4’-6-diamidino-2-phenylindole. The fluorescence signal was determined at an excitation of 415 nm and an emission of 550 nm. The experiments were repeated three times, and error bars indicate the standard deviation. ΔphoU2 exhibited significant differences (**P < 0.01) when compared with the wild-type and the complemented strains (C-phoU2, *P < 0.05) in intracellular polyP. The complementation of ΔphoU1 or ΔphoU2 mutant was abbreviated as C-phoU1 or C-phoU2. The empty vectors of pCN51 transferred into the ΔphoU1 or ΔphoU2 mutant were named P-phoU1 or P-phoU2. n.s., no significance.
Glucose and pyruvate levels in the mutants ΔphoU1 and ΔphoU2 were determined using an Abcam assay kit after growth for 12 h. Our data indicated that the extracellular glucose levels of ΔphoU1 (68.05 ± 6.84) and ΔphoU2 (66.86 ± 8.34) were reduced when compared to those of USA500 2395 (182.93 ± 23.17; P < 0.01) (Figure 4A). The intracellular pyruvate level of ΔphoU1 (184.26 ± 15.70) and ΔphoU2 (109.3 ± 3.55) was significantly increased when compared to the parent strain (30.99 ± 6.11) (Figure 4B). The complemented strains of C-phoU1 exhibited restored glucose and pyruvate levels, but C-phoU2 had no difference with the wild-type strain.
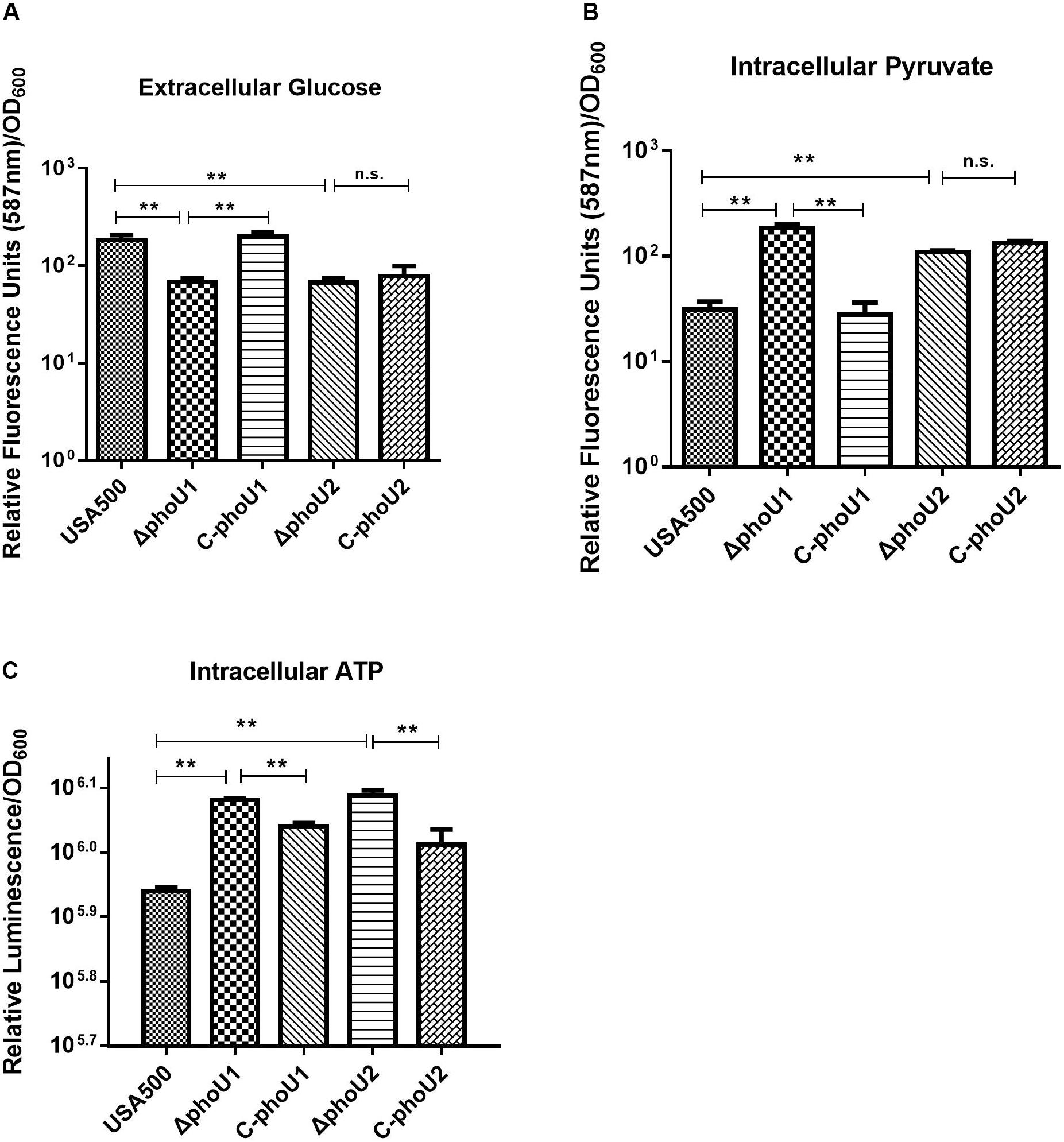
Figure 4. Extracellular glucose, intracellular pyruvate, and ATP in ΔphoU1 and ΔphoU2 of Staphylococcus aureus. (A) Extracellular glucose. Bacteria were grown for 12 h, and then the culture supernatant was harvested. Fluorescence signal was determined at an excitation of 535 nm and an emission of 587 nm. (B) Intracellular pyruvate. Bacteria were grown for 12 h; then, the cells were homogenized by OD600 and lysed with 0.1-mm glass-silica beads in a BeadBeater apparatus. Centrifugation to get the supernatant. The fluorescence signal was determined at an excitation of 535 nm and an emission of 587 nm. (C) Intracellular ATP. Bacteria were grown for 12 h; then, the cells were homogenized by OD600 and added equal to the volume of BacTiter-GloTM Reagent. Luminescence was determined. The experiments were repeated three times, and error bars indicate the standard deviation. ΔphoU1 and ΔphoU2 exhibited significant differences (**P < 0.01) when compared with the wild type in extracellular glucose, intracellular pyruvate, and ATP. The complemented strains of C-phoU1 exhibited restored glucose, pyruvate, and ATP levels, and C-phoU2 exhibited restored ATP levels.
The intracellular ATP level of ΔphoU1 and ΔphoU2 after growth for 12 h was determined using a Promega BacTiter Glo kit. The results showed that the ΔphoU1 and ΔphoU2 mutant cells accumulated a significantly higher level of ATP (1.4-fold) than the parent strain (Figure 4C). The complemented strains of C-phoU1 and C-phoU2 exhibited restored ATP levels.
The differentially expressed genes of the ΔphoU1 and ΔphoU2 mutants that were related to virulence, including the type VII secretion system operon (that promotes long-term bacterial persistence) and hemolysin, were investigated by assays of invasion and survival in A549 human lung epithelial cells and by measuring the activity and the expression of α-hemolysin in culture supernatants.
For S. aureus, which is an important opportunistic pathogen that causes community-acquired pneumonia, assays of invasion and survival were detected using A549 human lung epithelial cells as previously described (Liang et al., 2006; Sharma et al., 2013; Mechler et al., 2015). After culturing for 4 h (log phase) or 12 h (stationary phase), wild-type, ΔphoU1, and ΔphoU2 mutants were inoculated into A549 cells at a multiplicity of infection of 10:1. After incubation for 1 h, the viable bacterial count showed no significant difference between ΔphoU1, ΔphoU2, and USA500 2395, whereas after 24 h of incubation, the 4- or 12-h viable bacterial count of ΔphoU1 (4.079 ± 0.10 log10)/(3.65 ± 0.09 log10) and ΔphoU2 (3.90 ± 0.12 log10)/(3.53 ± 0.12 log10) showed 15–18% significant decrease compared with that observed in USA500 2395 (4.55 ± 0.18 log10)/(4.05 ± 0.03 log10) (Figure 5).
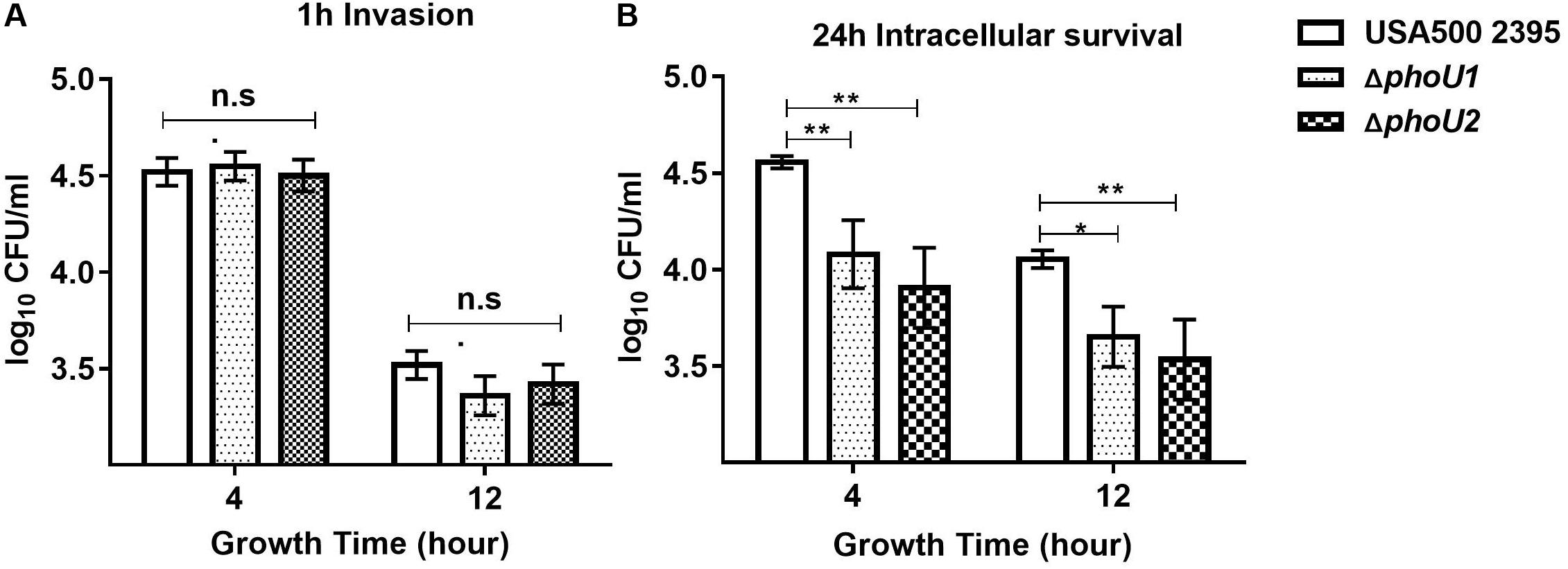
Figure 5. Invasion and intracellular survival of USA500 2395, ΔphoU1, and ΔphoU2 in the human lung epithelial cell line A549. Bacteria grown for 4 and 12 h were used to inoculate the A549 cells at a multiplicity of infection of 10:1. (A) Intracellular bacteria were counted for the colony-forming units after 1 h to determine the invasion capacity and (B) after 24 h to assay intracellular survival. The experiments were repeated three times, and error bars indicate the standard deviation. The 4- and 12-h-culture ΔphoU1 and ΔphoU2 exhibited significant differences (**P < 0.01; *P < 0.05) when compared with the wild type after 24 h of intracellular survival.
The hemolytic activity of the phoU homolog mutants at 12 h (stationary phase) was determined using rabbit erythrocytes. The α-hemolysin activity of ΔphoU2 (94.02%), was significantly enhanced compared with that of USA500 2395 (70.96%) (P < 0.01), whereas the α-hemolysin activity of ΔphoU1 (66.25%) was lower than that in USA500 2395 (70.96%) (Figure 6A). Western blot analysis indicated that the α-hemolysin levels in ΔphoU2 were significantly higher than in USA500 2395; however, no significant difference was found between the α-hemolysin level in ΔphoU1 and USA500 2395 (Figure 6B). Complementation of the ΔphoU1 and ΔphoU2 mutants restored the expression and the activity of α-hemolysin to the wild type’s level.
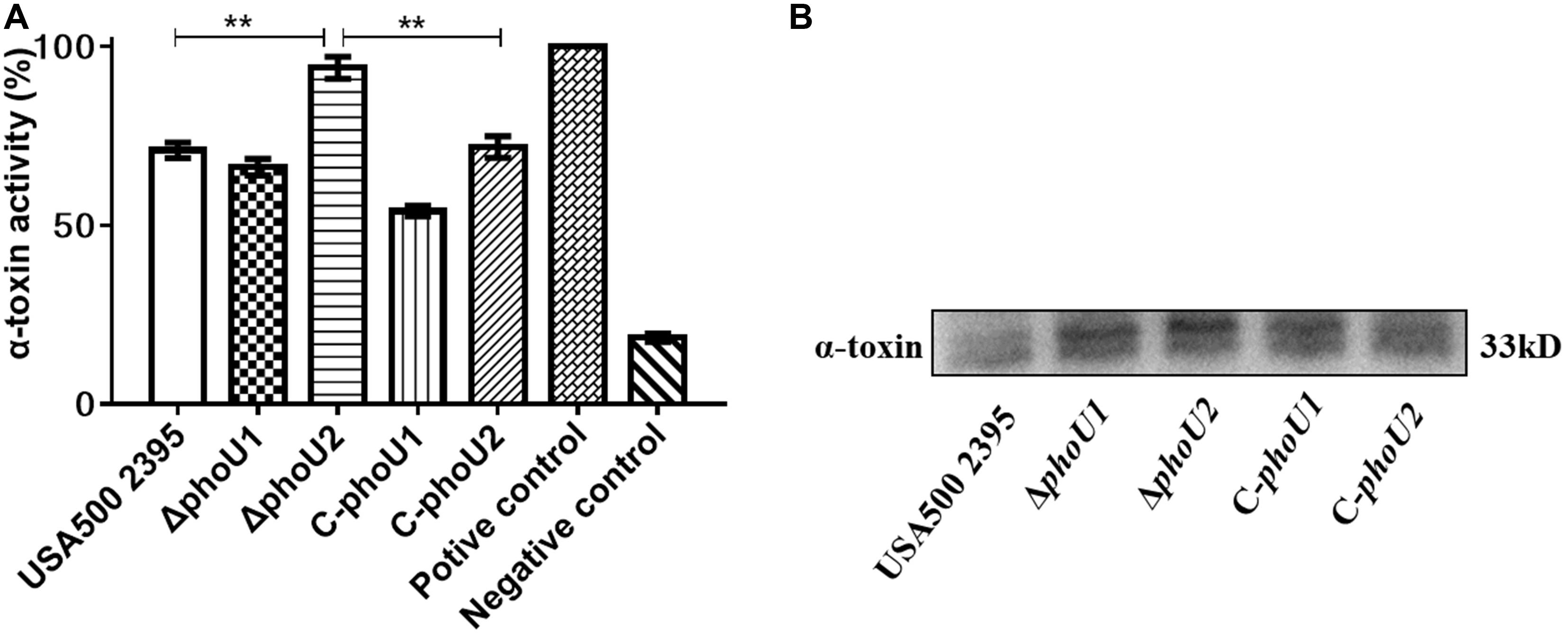
Figure 6. Extracellular alpha-hemolysin activity and secretion of USA500 2395, ΔphoU1, and ΔphoU2. Bacteria were grown for 12 h; then, the culture supernatant was harvested and filtered through a 0.22 μm filter. (A) Extracellular alpha-hemolysin activity. The supernatant was added equal to the volume of 1% of rabbit erythrocytes and then incubated at 37°C for 30 min, followed by centrifugation to obtain the supernatant. The supernatant was used to measure OD550; 0.1% triton X-100 served as the 100% hemolysis control (positive control), and 1 × phosphate-buffered saline was the negative control. (B) Western blot analysis of extracellular alpha-hemolysin. The experiments were repeated three times, and error bars indicate the standard deviation. ΔphoU2 exhibited significant differences (**P < 0.01) when compared with the wild-type and the complemented strain C-phoU2. The complementation of ΔphoU1 or ΔphoU2 mutant was abbreviated as C-phoU1 or C-phoU2. The empty vector of pCN51 transferred into the ΔphoU1 or ΔphoU2 mutant was named P-phoU1 or P-phoU2.
PhoU is a negative regulator of persister formation in most bacteria (Li and Zhang, 2007; Shi and Zhang, 2010). Mounting evidence has indicated the species-specific functions of PhoU homologs in bacteria; however, the impact of these proteins on bacterial growth, persister production, biofilm formation, and virulence of S. aureus is poorly understood. In this study, deletion of either phoU1or phoU2 of S. aureus resulted in persister reduction in the presence of antibiotics (vancomycin or levofloxacin). The reduced persister formation in the ΔphoU1 and ΔphoU2 mutants is not due to a decreased intrinsic resistance to antibiotics as the MIC/MBCs of mutants show no change in antibiotic susceptibility tests. Our results suggest that both phoU1 and phoU2 are required for antibiotic tolerance in S. aureus, whereas a previous study showed that phoU2, but not phoU1, deletion resulted in persister reduction in S. epidermidis (Wang et al., 2017). Inactivation of phoU in E. coli and P. aeruginosa has been shown to reduce antibiotic susceptibility and persister formation (Li and Zhang, 2007; de Almeida et al., 2015). Deletion of phoY2 in M. tuberculosis H37Rv increased the susceptibility to pyrazinamide and rifampicin and reduced persister formation (Shi and Zhang, 2010). However, single deletions of phoY1 and phoY2 mutants of the M. tuberculosis Erdman strain showed no impact on drug susceptibility and persister formation (Namugenyi et al., 2017). By comparison, the double-deletion phoY1 and phoY2 mutants in the M. tuberculosis Erdman strain exhibited a decrease in persister formation and specifically enhanced the susceptibility to rifampicin, but not to other antimycobacterial drugs. Growth curve and survival assays were both measured in the presence of the cell wall-disrupting detergent sodium dodecyl sulfate and the reactive oxygen hydrogen peroxide (H2O2). Our results suggest that both phoU1 and phoU2 are required for resistance to SDS and H2O2 in S. aureus. Inactivation of phoU in E. coli, P. aeruginosa, and S. epidermidis has been shown to reduce tolerance to H2O2. This suggests that the functions and the regulatory mechanisms of phoU homologs in relation to antimicrobial susceptibility and persister formation are distinct in different bacteria.
Persisters are often slow growing or non-growing with reduced metabolism (Maisonneuve and Gerdes, 2014; Prax and Bertram, 2014; Cabral et al., 2018; Kaldalu and Tenson, 2019). Deletion of phoU in E. coli and P. aeruginosa slowed the growth rates but increased the metabolism (Li and Zhang, 2007; de Almeida et al., 2015). Single ΔphoY1 or ΔphoY2 mutants of M. tuberculosis had no significant effects on bacterial growth but decreased the viable bacteria count of ΔphoY1ΔphoY2 double mutant at the stationary phase (Wang et al., 2013; Namugenyi et al., 2017). The S. epidermidis phoU2 deletion resulted in growth retardation, whereas the present study demonstrated that the growth curve and the viable bacterial count of S. aureus ΔphoU1 and ΔphoU2 were the same as that of the parent strain, which were confirmed by silenced phoU1 or phoU2 in USA500 2395, USA300, and SA113 strains (Supplementary Figure S2).
The mechanisms of persister formation are complex. Intracellular polyP level is associated with antibiotic tolerance and persister formation. PolyP accumulation has been associated with increased persister frequency in both E. coli and M. tuberculosis (Thayil et al., 2011; Singh et al., 2013; Chuang et al., 2015; Germain et al., 2015). Differently, the enhanced polyP accumulation, resulting from the inactivation or the deletion of phoU in M. tuberculosis, E. coli, P. aeruginosa, or S. epidermidis, was accompanied by a decreased sensitivity to antibiotics and persister generation (Morohoshi et al., 2002; Li and Zhang, 2007; de Almeida et al., 2015; Namugenyi et al., 2017; Wang et al., 2017). These data were consistent with our current study in terms of intracellular polyP accumulation and persister generation reduction in the ΔphoU1 and the ΔphoU2 strains of S. aureus.
The bacterial metabolic state is a major determinant of persister. This view can be used to explain the bacteria in the biofilm. Cells in the periphery of the biofilm, where metabolic activity is highest, indicate that antibiotic efficacy is highest. The cells in the center of the biofilm exhibit a marked dormancy and low antibiotic efficiency (Walters et al., 2003). Another example is starvation stress, which induces changes in the expression of metabolic pathways such as energy metabolism, amino acid metabolism, and lipid metabolism, while long-term starvation of M. tuberculosis reduces susceptibility to rifampicin, isoniazid, and metronidazole (Betts et al., 2002). Most antibiotics, such as quinolones, aminoglycosides, and β-lactams, kill bacteria by corrupting targets which are energy dependent. Quinolones inhibit the ligase activity of gyrase and topoisomerase and release DNA with single- and double-strand breaks that lead to cell death (Hooper, 2001). Aminoglycoside acts by producing toxic misfolded peptides (Davis, 1987). Beta-lactams exert their antibacterial effect by irreversibly binding to the Ser residue of the penicillin-binding protein active site, forcing a futile cycle of peptidoglycan synthesis (Cho et al., 2014). Those targets require ATP to function. Recent studies showed that lowering intracellular ATP with arsenate treatment could increase the level of persister formation in E. coli and S. aureus under fluoroquinolones (Conlon et al., 2016; Shan et al., 2017). Persister formation is associated with ATP depletion in E. coli and S. aureus (Conlon et al., 2016; Shan et al., 2017). ATP can be produced by multiple cellular pathways, including oxidative phosphorylation, ATP synthase, and polyP, which can be converted to ATP (Resnick and Zehnder, 2000; Bonora et al., 2012). The change of metabolism pathways may be an association with various factors, and more comprehensive considerations are required. Hence, the results of metabolic products would become more convincing. Our result showed that the S. aureus ΔphoU1 or ΔphoU2 mutant decreased the level of persister formation by hyperactive carbon metabolism (decreased extracellular glucose and intracellular pyruvate) accompanied by increasing intracellular ATP. In the S. aureus ΔphoU1 mutant, the transcriptome analysis revealed that 26 genes up-regulated by twofold and three genes down-regulated by two- to threefold, identified to be involved in carbon, pyruvate, and glycolysis metabolism, consistent with the hyperactive carbon metabolism and increasing intracellular ATP. In the S. aureus ΔphoU2 mutant, our analysis showed that 13 genes up-regulated by three- to 24-fold of transported phosphate. The increasing intracellular polyP was consistent with the high intracellular ATP content in the ΔphoU2 mutant. However, three genes (D-lactate dehydrogenase, pyruvate formate lyase, and alcohol dehydrogenase) were up-regulated by threefold and 10 genes were down-regulated by twofold in carbon, pyruvate, and glycolysis metabolism. Lactate dehydrogenase (catalyzes the interconversion of pyruvate and lactate), pyruvate formate lyase (regulates the formation of acetyl-CoA, which is important in the production of energy), and alcohol dehydrogenase (converts pyruvate to acetaldehyde and involved in the production of ATP) play an important part in carbon, pyruvate, and glycolysis metabolism (Garvie, 1980; Lamed and Zeikus, 1981; Hasona et al., 2004). It is possible that the result of the ΔphoU2 mutant presented decreased extracellular glucose and intracellular pyruvate (Wang et al., 2017). Those results suggest that, in S. aureus ΔphoU1 or ΔphoU2, decreased tolerance to antibiotics or stresses of SDS and H2O2 may be associated with higher levels of carbon metabolism and intracellular ATP. Complementary strains C-phoU1, C-phoU2, and C-phoU1phoU2 restored the majority of phenotypes, and only the C-phoU2 strains failed to restore the phenotypes of glucose and pyruvate, indicating that the results were not due to a secondary mutation elsewhere on the chromosome. The reason may be due to the PhoU, a global metabolic repressor. Regulation has a dual function which is reflected in the induced response to phosphate limitation but also inhibited when Pi is in excess (Gardner et al., 2014; Lubin et al., 2015; diCenzo et al., 2017). The expression of the phoU1 or the phoU2 in the complementary strains C-phoU1 and C-phoU2 were around three- or fivefold higher than that in the wild-type strain. Excessive phoU2 expression may cause diminished compensation function in glucose and pyruvate phenotypes. This may not be a unique reason as bacteria have a complex metabolism regulatory system and thus need further study.
The intracellular invasion and the survival rates of S. aureus are crucial for the pathogenicity of chronic infection (Rollin et al., 2017; Tan et al., 2019). Several virulence factors play a critical role in the pathogenic procession. We found that either phoU1 or phoU2 is required for the intracellular survival of S. aureus in human lung epithelial A549 cells. In the method of invasion and intracellular survival assays, gentamicin (10 μg/ml) was added in the culture medium to restrict the extracellular growth of bacteria when S. aureus and A549 cells were co-incubated. Gentamicin and erythromycin may have an antagonizing effect (Penn et al., 1982), so we could not perform the invasion and the intracellular survival assays on the complemented strains of C-phoU1 and C-phoU2. However, a previous report, as a piece of supporting evidence, showed that a single-point mutation in pitA (downstream of phoU2) resulted in a decrease in the intracellular survival of S. aureus in human epithelial cells (Mechler et al., 2015). The deletion of phoU1 or phoU2 in S. aureus resulted in the down-regulation of multiple virulence systems, including the type VII secretion system, serine protease, and leucocidin, which could explain the decreased survival of the phoU1 and the phoU2 mutants.
The α-hemolysin is a major virulence factor in S. aureus infections (Kebaier et al., 2012; Vandenesch et al., 2012). We observed that the α-hemolysin activity and its expression were increased in ΔphoU2, while there was no effect noted in ΔphoU1. This suggests that phoU2, but not phoU1, is a negative regulator of α-hemolysin. However, the transcription of α-hemolysin in ΔphoU2 was reduced after 12 h of growth when assessed by RNA-Seq and RT-PCR.
The virulence regulation mechanisms of S. aureus are very complex (Bronner et al., 2004). The virulence regulatory systems include two-component systems, including AgrAC, SaeRS, ArlRS, etc., and the global regulator, including cytoplasmic SarA-family, CodY, Rot, etc. (Haag and Bagnoli, 2017). Among them, AgrAC, SarA, and SaeRS are positive regulators, whereas CodY and Rot are negative regulators of virulence genes (Pragman and Schlievert, 2004). The deletion of phoU1 resulted in a decreasing expression of sarA and saeS and increasing expressions of codY. The deletion of phoU2 resulted in decreasing expressions of sarA family (sarA, sarR, and sarZ), codY, and rot and an increasing expression of phoRP, the function of which is unknown. Therefore, the regulatory mechanisms of phoU1 and phoU2 in relation to virulence require further study.
S. aureus biofilm formation also plays an important role in persister production during chronic infections (Archer et al., 2011; Conlon, 2014). S. aureus SA113 is a biofilm positive strain characterized by rsbU, tcaR, and agr mutants (Herbert et al., 2010). Our results showed that silencing phoU2, but not phoU1, can reduce biofilm formation in the S. aureus SA113 strain (Supplementary Figure S3). This is consistent with the previous finding in S. epidermidis that PhoU2, but not PhoU1, is an important regulator of biofilm formation.
In summary, both PhoU1 and PhoU2 of S. aureus regulate persister generation and bacterial virulence. The deletion of phoU1 or phoU2 of S. aureus resulted in a decrease in intracellular bacterial survival rate in human epithelial cells by down-regulation of multiple virulence factors, including the type VII secretion system, serine protease, and leucocidin. In a follow-up work, the double mutant of ΔphoU1ΔphoU2 was constructed, and the metabolic variation was similar to the single mutant in extracellular glucose, intracellular pyruvate, ATP, and intracellular polyP levels (Supplementary Figure S5). The results in S. aureus are different from what we have previously found in S. epidermidis, where only PhoU2 regulates biofilm and persister formation. In S. aureus, transcriptome analysis revealed that 573 or 285 genes were differentially expressed in the ΔphoU1 or the ΔphoU2 mutant vs the wild type. In S. epidermidis, deletion of phoU1 just led to 92 differentially expressed genes, while deletion of phoU2 could result in 945 differentially expressed genes (Wang et al., 2017). In S. aureus, both phoU1 and phoU2 regulate virulence by the global regulator (sarA, rot, and codY) and persister generation by the hyperactive carbon metabolism accompanied by increasing intracellular ATP. In the S. epidermidis ΔphoU2 mutant, the reduction of biofilm and persister can be explained by the down-regulated expression of several important genes involved in growth (including yycFG, rsuU, pflA, and nrdD) and increased ATP by up-regulated ATP synthase (Dubrac et al., 2008; Wang et al., 2017). In the previous studies, we found that the regulatory mechanisms of some global regulators (SaeRS, YycFG, SrrAB, and ArlRS) have different roles between S. aureus and S. epidermidis (Lou et al., 2011; Wu et al., 2012, 2015; Xu et al., 2017). These observations hint that different mechanisms occur in the regulation of S. aureus and S. epidermidis, even though they belong to the staphylococcus. Therefore, the different regulation mechanisms of PhoU homologs in S. aureus and S. epidermidis warrant further investigation in the future.
All datasets generated for this study are included in the article/Supplementary Material.
DQ, YZ, and ZY designed the research. YS, XW, and ZC participated in most of the experiments. ZLy, ZLi, JZ, YW, and QD analyzed the data. YS drafted the manuscript. DQ, YZ, and ZY revised the manuscript.
This work was supported by the National Natural Science Foundation of China (81871622, 81271791, 81571955, 81671982, and 81991532), the National Major Scientific and Technological Projects in “Infectious Disease” (2018ZX10734401), “Development of Key Technologies and Drugs for the Prevention of Drug-Resistant Bacteria” (2019ZX09721001), and “Research on Biosafety Technology of High-Level Biosafety Laboratory and Important Pathogen Laboratory” (2018ZX10734401-04), and the Sanming Project of Medicine in Shenzhen (SMGC201705029).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We gratefully acknowledge Dr. Tao Xu (Department of Infectious Diseases, Huashan Hospital, Fudan University, Shanghai, China) for his suggestions on the gene knockout.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00865/full#supplementary-material
Archer, N. K., Mazaitis, M. J., Costerton, J. W., Leid, J. G., Powers, M. E., and Shirtliff, M. E. (2011). Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2, 445–459. doi: 10.4161/viru.2.5.17724
Aschar-Sobbi, R., Abramov, A. Y., Diao, C., Kargacin, M. E., Kargacin, G. J., French, R. J., et al. (2008). High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J. Fluoresc. 18, 859–866. doi: 10.1007/s10895-008-0315-4
Bae, T., and Schneewind, O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63. doi: 10.1016/j.plasmid.2005.05.005
Bernheimer, A. W., Avigad, L. S., and Grushoff, P. (1968). Lytic effects of staphylococcal alpha-toxin and delta-hemolysin. J. Bacteriol. 96, 487–491. doi: 10.1128/jb.96.2.487-491.1968
Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi: 10.1046/j.1365-2958.2002.02779.x
Bonora, M., Patergnani, S., Rimessi, A., De Marchi, E., Suski, J. M., Bononi, A., et al. (2012). ATP synthesis and storage. Purinergic Signal. 8, 343–357. doi: 10.1007/s11302-012-9305-8
Bonora, M., Wieckowsk, M. R., Chinopoulos, C., Kepp, O., Kroemer, G., Galluzzi, L., et al. (2015). Molecular mechanisms of cell death: central implication of ATP synthase in mitochondrial permeability transition. Oncogene 34:1608. doi: 10.1038/onc.2014.462
Bronner, S., Monteil, H., and Prévost, G. (2004). Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28, 183–200. doi: 10.1016/j.femsre.2003.09.003
Cabral, D. J., Wurster, J. I., and Belenky, P. (2018). Antibiotic persistence as a metabolic adaptation: stress, metabolism, the host, and new directions. Pharmaceuticals 11;14. doi: 10.3390/ph11010014
Cho, H., Uehara, T., and Bernhardt, T. G. (2014). Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311. doi: 10.1016/j.cell.2014.11.017
Chuang, Y.-M., Bandyopadhyay, N., Rifat, D., Rubin, H., Bader, J. S., and Karakousis, P. C. (2015). Deficiency of the novel exopolyphosphatase Rv1026/PPX2 leads to metabolic downshift and altered cell wall permeability in Mycobacterium tuberculosis. mBio 6:e02428-14. doi: 10.1128/mBio.02428-14
Conlon, B. P. (2014). Staphylococcus aureus chronic and relapsing infections: evidence of a role for persister cells: an investigation of persister cells, their formation and their role in S. aureus disease. Bioessays 36, 991–996. doi: 10.1002/bies.201400080
Conlon, B. P., Rowe, S. E., Gandt, A. B., Nuxoll, A. S., Donegan, N. P., Zalis, E. A., et al. (2016). Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 1:16051. doi: 10.1038/nmicrobiol.2016.51
Davis, B. D. (1987). Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51, 341–350. doi: 10.1128/mmbr.51.3.341-350.1987
de Almeida, L. G., Ortiz, J. H., Schneider, R. P., and Spira, B. (2015). phoU inactivation in Pseudomonas aeruginosa enhances accumulation of ppGpp and polyphosphate. Appl. Environ. Microbiol. 81, 3006–3015. doi: 10.1128/AEM.04168-14
diCenzo, G. C., Sharthiya, H., Nanda, A., Zamani, M., and Finan, T. M. (2017). PhoU allows rapid adaptation to high phosphate concentrations by modulating PstSCAB transport rate in Sinorhizobium meliloti. J. Bacteriol. 199, e00143–00117. doi: 10.1128/jb.00143-17
Dinges, M. M., Orwin, P. M., and Schlievert, P. M. (2000). Exotoxins of Staphylococcus aureus. Clin.Microbiol. Rev. 13;16. doi: 10.1128/CMR.13.1.16
Dubrac, S., Bisicchia, P., Devine, K. M., and Msadek, T. (2008). A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70, 1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x
El-Halfawy, O. M., and Valvano, M. A. (2015). Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207. doi: 10.1128/CMR.00058-14
Fisher, R. A., Gollan, B., and Helaine, S. (2017). Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15:453. doi: 10.1038/nrmicro.2017.42
Fraunholz, M., and Sinha, B. (2012). Intracellular Staphylococcus aureus: live-in and let die. Front. Cell. Infect. Microbiol. 2:43. doi: 10.3389/fcimb.2012.00043
Gardner, S. G., Johns, K. D., Tanner, R., and McCleary, W. R. (2014). The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J. Bacteriol. 196, 1741–1752. doi: 10.1128/JB.00029-14
Garvie, E. I. (1980). Bacterial lactate dehydrogenases. Microbiol. Rev. 44, 106–139. doi: 10.1128/mmbr.44.1.106-139.1980
Germain, E., Roghanian, M., Gerdes, K., and Maisonneuve, E. (2015). Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 112, 5171–5176. doi: 10.1073/pnas.1423536112
Grassi, L., Di Luca, M., Maisetta, G., Rinaldi, A. C., Esin, S., Trampuz, A., et al. (2017). Generation of persister cells of Pseudomonas aeruginosa and Staphylococcus aureus by chemical treatment and evaluation of their susceptibility to membrane-targeting agents. Front. Microbiol. 8:1917. doi: 10.3389/fmicb.2017.01917
Haag, A. F., and Bagnoli, F. (2017). The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr. Top. Microbiol. Immunol. 409, 145–198. doi: 10.1007/82_2015_5019
Hasona, A., Kim, Y., Healy, F. G., Ingram, L. O., and Shanmugam, K. T. (2004). Pyruvate formate lyase and acetate kinase are essential for anaerobic growth of Escherichia coli on xylose. J. Bacteriol. 186, 7593–7600. doi: 10.1128/JB.186.22.7593-7600.2004
Helle, L., Kull, M., Mayer, S., Marincola, G., Zelder, M. E., Goerke, C., et al. (2011). Vectors for improved Tet repressor-dependent gradual gene induction or silencing in Staphylococcus aureus. Microbiology 157(Pt 12), 3314–3323. doi: 10.1099/mic.0.052548-0
Herbert, S., Ziebandt, A. K., Ohlsen, K., Schafer, T., Hecker, M., Albrecht, D., et al. (2010). Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 78, 2877–2889. doi: 10.1128/IAI.00088-10
Hooper, D. C. (2001). Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin. Infect. Dis. 32(Suppl. 1), S9–S15. doi: 10.1086/319370
Kaldalu, N., and Tenson, T. (2019). Slow growth causes bacterial persistence. Sci. Signal. 12:eaay1167. doi: 10.1126/scisignal.aay1167
Kebaier, C., Chamberland, R. R., Allen, I. C., Gao, X., Broglie, P. M., Hall, J. D., et al. (2012). Staphylococcus aureus α-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J. Infect. Dis. 205, 807–817. doi: 10.1093/infdis/jir846
Lamed, R. J., and Zeikus, J. G. (1981). Novel NADP-linked alcohol–aldehyde/ketone oxidoreductase in thermophilic ethanologenic bacteria. Biochem. J. 195, 183–190. doi: 10.1042/bj1950183
Li, Y., and Zhang, Y. (2007). PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51, 2092–2099. doi: 10.1128/AAC.00052-07
Liang, X., Yee, S. W., Chien, H. C., Chen, E. C., Luo, Q., Zou, L., et al. (2018). Organic cation transporter 1 (OCT1) modulates multiple cardiometabolic traits through effects on hepatic thiamine content. PLoS Biol. 16:e2002907. doi: 10.1371/journal.pbio.2002907
Liang, X., Yu, C., Sun, J., Liu, H., Landwehr, C., Holmes, D., et al. (2006). Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 74, 4655–4665. doi: 10.1128/IAI.00322-06
Liu, J., Lou, Y., Yokota, H., Adams, P. D., Kim, R., and Kim, S. H. (2005). Crystal structure of a PhoU protein homologue: a new class of metalloprotein containing multinuclear iron clusters. J. Biol. Chem. 280, 15960–15966. doi: 10.1074/jbc.M414117200
Lou, Q., Zhu, T., Hu, J., Ben, H., Yang, J., Yu, F., et al. (2011). Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiol. 11:146. doi: 10.1186/1471-2180-11-146
Lubin, E. A., Henry, J. T., Fiebig, A., Crosson, S., and Laub, M. T. (2015). Identification of the PhoB regulon and role of phou in the phosphate starvation response of Caulobacter crescentus. J. Bacteriol. 198, 187–200. doi: 10.1128/JB.00658-15
Maisonneuve, E., and Gerdes, K. (2014). Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548. doi: 10.1016/j.cell.2014.02.050
Mechler, L., Bonetti, E.-J., Reichert, S., Flötenmeyer, M., Schrenzel, J., Bertram, R., et al. (2016). Daptomycin tolerance in the Staphylococcus aureus pitA6 mutant is due to upregulation of the dlt operon. Antimicrob. Agents Chemother. 60, 2684–2691. doi: 10.1128/AAC.03022-15
Mechler, L., Herbig, A., Paprotka, K., Fraunholz, M., Nieselt, K., and Bertram, R. (2015). A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 59, 5366–5376. doi: 10.1128/aac.00643-15
Moormeier, D. E., and Bayles, K. W. (2017). Staphylococcus aureus biofilm: a complex developmental organism. Mol. Microbiol. 104, 365–376. doi: 10.1111/mmi.13634
Morohoshi, T., Maruo, T., Shirai, Y., Kato, J., Ikeda, T., Takiguchi, N., et al. (2002). Accumulation of inorganic polyphosphate in mutants of Escherichia coli and Synechocystis sp. Strain PCC6803. Appl. Environ. Microbiol. 68:4107. doi: 10.1128/AEM.68.8.4107-4110.2002
Namugenyi, S. B., Aagesen, A. M., Elliott, S. R., and Tischler, A. D. (2017). Mycobacterium tuberculosis PhoY proteins promote persister formation by mediating pst/SenX3-RegX3 phosphate sensing. mBio 8:e00494-17. doi: 10.1128/mBio.00494-17
Otto, M. (2010). Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev. Dermatol. 5, 183–195. doi: 10.1586/edm.10.6
Otto, M. (2014). Staphylococcus aureus toxins. Curr. Opin. Microbiol. 17, 32–37. doi: 10.1016/j.mib.2013.11.004
Overton, I. M., Graham, S., Gould, K. A., Hinds, J., Botting, C. H., Shirran, S., et al. (2011). Global network analysis of drug tolerance, mode of action and virulence in methicillin-resistant S. aureus. BMC Syst. Biol. 5:68. doi: 10.1186/1752-0509-5-68
Penn, R. L., Ward, T. T., and Steigbigel, R. T. (1982). Effects of erythromycin in combination with penicillin, ampicillin, or gentamicin on the growth of Listeria monocytogenes. Antimicrob. Agents Chemother. 22, 289–294. doi: 10.1128/aac.22.2.289
Pragman, A. A., and Schlievert, P. M. (2004). Virulence regulation in Staphylococcus aureus: the need for in vivo analysis of virulence factor regulation. Pathog. Dis. 42, 147–154. doi: 10.1016/j.femsim.2004.05.005
Prax, M., and Bertram, R. (2014). Metabolic aspects of bacterial persisters. Front. Cell. Infect. Microbiol. 4:148. doi: 10.3389/fcimb.2014.00148
Resnick, S. M., and Zehnder, A. J. (2000). In vitro ATP regeneration from polyphosphate and AMP by polyphosphate:AMP phosphotransferase and adenylate kinase from Acinetobacter johnsonii 210A. Appl. Environ. Microbiol. 66, 2045–2051. doi: 10.1128/aem.66.5.2045-2051.2000
Rollin, G., Tan, X., Tros, F., Dupuis, M., Nassif, X., Charbit, A., et al. (2017). Intracellular survival of Staphylococcus aureus in endothelial cells: a matter of growth or persistence. Front. Microbiol. 8:1354. doi: 10.3389/fmicb.2017.01354
Schnack, L., Sohrabi, Y., Lagache, S. M. M., Kahles, F., Bruemmer, D., Waltenberger, J., et al. (2019). Mechanisms of trained innate immunity in oxLDL primed human coronary smooth muscle cells. Front. Immunol. 10:13. doi: 10.3389/fimmu.2019.00013
Shan, Y., Brown Gandt, A., Rowe, S. E., Deisinger, J. P., Conlon, B. P., and Lewis, K. (2017). ATP-dependent persister formation in Escherichia coli. mBio 8:e02267-16. doi: 10.1128/mBio.02267-16
Sharma, P., Lata, H., Arya, D. K., Kashyap, A. K., Kumar, H., Dua, M., et al. (2013). Role of pilus proteins in adherence and invasion of Streptococcus agalactiae to the lung and cervical epithelial cells. J. Biol. Chem. 288, 4023–4034. doi: 10.1074/jbc.M112.425728
Sharma-Kuinkel, B. K., Ahn, S. H., Rude, T. H., Zhang, Y., Tong, S. Y., Ruffin, F., et al. (2012). Presence of genes encoding panton-valentine leukocidin is not the primary determinant of outcome in patients with hospital-acquired pneumonia due to Staphylococcus aureus. J. Clin. Microbiol. 50, 848–856. doi: 10.1128/jcm.06219-11
Shi, W., and Zhang, Y. (2010). PhoY2 but not PhoY1 is the PhoU homologue involved in persisters in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65, 1237–1242. doi: 10.1093/jac/dkq103
Singh, R., Singh, M., Arora, G., Kumar, S., Tiwari, P., and Kidwai, S. (2013). Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. J. Bacteriol. 195:2839. doi: 10.1128/JB.00038-13
Tan, X., Coureuil, M., Ramond, E., Euphrasie, D., Dupuis, M., Tros, F., et al. (2019). Chronic Staphylococcus aureus lung infection correlates with proteogenomic and metabolic adaptations leading to an increased intracellular persistence. Clin. Infect. Dis. 69, 1937–1945. doi: 10.1093/cid/ciz106
Thayil, S. M., Morrison, N., Schechter, N., Rubin, H., and Karakousis, P. C. (2011). The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence. PLoS One 6:e28076. doi: 10.1371/journal.pone.0028076
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. Jr. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Vandenesch, F., Lina, G., and Henry, T. (2012). Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: a redundant arsenal of membrane-damaging virulence factors? Front. Cell. Infect. Microbiol. 2:12. doi: 10.3389/fcimb.2012.00012
Walters, M. C. III, Roe, F., Bugnicourt, A., Franklin, M. J., and Stewart, P. S. (2003). Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47, 317–323. doi: 10.1128/aac.47.1.317-323.2003
Wang, C., Mao, Y., Yu, J., Zhu, L., Li, M., Wang, D., et al. (2013). PhoY2 of mycobacteria is required for metabolic homeostasis and stress response. J. Bacteriol. 195, 243–252. doi: 10.1128/jb.01556-12
Wang, X., Han, H., Lv, Z., Lin, Z., Shang, Y., Xu, T., et al. (2017). PhoU2 but Not PhoU1 as an important regulator of biofilm formation and tolerance to multiple stresses by participating in various fundamental metabolic processes in Staphylococcus epidermidis. J. Bacteriol. 199:e00219-17. doi: 10.1128/jb.00219-17
Wilmaerts, D., Windels, E. M., Verstraeten, N., and Michiels, J. (2019). General mechanisms leading to persister formation and awakening. Trends Genet. 35, 401–411. doi: 10.1016/j.tig.2019.03.007
Wu, Y., Wang, J., Xu, T., Liu, J., Yu, W., Lou, Q., et al. (2012). The two-component signal transduction system ArlRS regulates Staphylococcus epidermidis biofilm formation in an ica-dependent manner. PLoS One 7:e40041. doi: 10.1371/journal.pone.0040041
Wu, Y., Wu, Y., Zhu, T., Han, H., Liu, H., Xu, T., et al. (2015). Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions. J. Bacteriol. 197, 459–476. doi: 10.1128/jb.02231-14
Keywords: persisters, PhoU homolog, Staphylococcus aureus, virulence, phosphate metabolism, ATP
Citation: Shang Y, Wang X, Chen Z, Lyu Z, Lin Z, Zheng J, Wu Y, Deng Q, Yu Z, Zhang Y and Qu D (2020) Staphylococcus aureus PhoU Homologs Regulate Persister Formation and Virulence. Front. Microbiol. 11:865. doi: 10.3389/fmicb.2020.00865
Received: 30 October 2019; Accepted: 14 April 2020;
Published: 26 May 2020.
Edited by:
Hui Wu, University of Alabama at Birmingham, United StatesReviewed by:
Friedrich Götz, University of Tübingen, GermanyCopyright © 2020 Shang, Wang, Chen, Lyu, Lin, Zheng, Wu, Deng, Yu, Zhang and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijian Yu, eXV6aGlqaWFuc211QDE2My5jb20=; Ying Zhang, eXpoYW5nQGpoc3BoLmVkdQ==; Di Qu, ZHF1QHNobXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.