
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 May 2020
Sec. Virology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00851
This article is part of the Research Topic Novel Insights into Plant-Geminivirus Interactions View all 13 articles
 Zhenggang Li1,2
Zhenggang Li1,2 Zhenguo Du1
Zhenguo Du1 Yafei Tang1
Yafei Tang1 Xiaoman She1
Xiaoman She1 Xiaomei Wang1
Xiaomei Wang1 Yanhua Zhu1
Yanhua Zhu1 Lin Yu1
Lin Yu1 Guobing Lan1
Guobing Lan1 Zifu He1,2*
Zifu He1,2*Tomato leaf curl Guangdong virus (ToLCGdV) is a begomovirus associated with a Tomato yellow leaf curl disease (TYLCD) epidemic in Guangdong province, China. Being the least conserved protein among geminivirus proteins, the function of C4 during ToLCGdV infection has not been elucidated. In this study, the infectious clones of ToLCGdV and a ToLCGdV mutant (ToLCGdVmC4) with disrupted C4 ORF were constructed. Although ToLCGdV and ToLCGdVmC4 could infect Nicotiana benthamiana and tomato plants, ToLCGdVmC4 elicited much milder symptoms compared with ToLCGdV. To further verify the role of C4 in viral pathogenesis, C4 was expressed in N. benthamiana from Potato virus X (PVX) vector. The results showed that ToLCGdV C4 enhanced the pathogenicity of PVX and induced more severe developmental abnormalities in plants compared with PVX alone or PVX-mC4. In addition, ToLCGdV C4 suppresses systemic gene silencing in the transgenic N. benthamiana line 16c, but not local gene silencing induced by sense GFP in wild-type N. benthamiana plants. Moreover, C4 suppresses transcriptional gene silencing (TGS) by reducing the DNA methylation level of 35S promoter in 16c-TGS N. benthamiana plants. Furthermore, C4 could also interact with the receptor-like kinase (RLK) BARELY ANY MERISTEM 1 (BAM1), suggesting that C4 may suppress gene silencing by interfering with the function of BAM1 in the cell-to-cell spread of RNAi. All these results suggest that C4 is a pathogenic determinant of ToLCGdV, and C4 may suppress post-transcriptional gene silencing (PTGS) by interacting with BAM1.
Geminiviridae constitutes a large family of plant viruses with small, single-stranded and circular DNA (sscDNA) genomes encompassed within characteristic twinned icosahedral particles (Hanley-Bowdoin et al., 1999; Fauquet et al., 2008). Geminiviruses, which are transmitted by insects and could infect both dicots and monocots (John et al., 2001), have emerged as one of the most important groups of plant pathogens worldwide, especially in tropical and subtropical regions (Boulton, 2003; Varma and Malathi, 2003; Rojas et al., 2005; Mansoor et al., 2006; Fauquet et al., 2008; Navas-Castillo et al., 2011; Sattar et al., 2013). Geminiviridae includes nine genera Becurtovirus, Begomovirus, Capulavirus, Curtovirus, Eragrovirus, Grablovirus, Mastrevirus, Topocuvirus, and Turncurtovirus with different genome organization, host range, and insect vectors (Zerbini et al., 2017). Except the genus Begomovirus, geminivirus consists of monopartite component with 2.5∼3.0 kb in length (Zerbini et al., 2017).
Begomoviruses, transmitted by the whitefly vector Bemisia tabaci, are either monopartite or bipartite and infect only dicots. The DNA A virion-sense strand of bipartite begomovirus encodes the coat protein CP (AV1) and movement protein (AV2), which may both be involved in virus movement. The complementary-sense strand of DNA A component encodes the replication-associated protein (Rep, ORF AC1), transcriptional activator protein (TrAP, ORF AC2), replication enhancer protein (REn, ORF AC3) and C4 protein (ORF AC4). The DNA B component of bipartite begomovirus encodes two proteins, the nuclear shuttle protein (NSP, ORF BV1) on the virion-sense strand and the movement protein (MP, ORF BC1) on the complementary-sense strand (Hanley-Bowdoin et al., 1999). For both DNA A and DNA B, the bidirectionally arranged ORFs are separated by a long intergenic region (IR) which carries key elements for initiating viral replication and transcription (Hanley-Bowdoin et al., 1999). The genome of monopartite begomovirus encodes all information required for the viral infection and consists of a single ssDNA molecule which resembles the DNA A of bipartite begomovirus. However, an increasing number of monopartite begomoviruses have been found to require betasatellites for successful infection and symptom induction in some host plants (Zhou, 2013; Yang et al., 2019).
As one of the least conserved proteins among geminivirus proteins, the function of AC4/C4 protein varies among different geminiviruses. C4 is a major pathogenic determinant in curtoviruses and some monopartite begomoviruses, disruption of C4 reduces viral infections and symptom development (Stanley and Latham, 1992; Teng et al., 2010; Li et al., 2018). Transient expression of Beet curly top virus (BCTV) C4 or Tomato leaf curl Yunnan virus (TLCYnV) C4 contributes to PVX infection, and induces severe developmental abnormalities similar with phenotypes induced by virus infection in both N. benthamiana and Arabidopsis (Latham et al., 1997; Piroux et al., 2007; Mills-Lujan and Deom, 2010; Mills-Lujan et al., 2015; Mei et al., 2018b). However, AC4 of the bipartite begomovirus Tomato golden mosaic virus (TGMV) has no obvious effect on symptom development (Piroux et al., 2007). More and more studies have found that AC4/C4 induces plant abnormal development by regulating brassinosteroid (BR) signaling pathway through interaction with members of Arabidopsis SHAGGY-like protein kinase (AtSK) family (Piroux et al., 2007; Mills-Lujan and Deom, 2010; Deom and Mills-Lujan, 2015; Mills-Lujan et al., 2015; Bi et al., 2017; Mei et al., 2018a, b).
AC4/C4 also functions as a viral suppressor of RNA silencing (VSR) to suppress both post-transcriptional gene silencing (PTGS) and transcriptional gene silencing (TGS) (Vanitharani et al., 2004; Gopal et al., 2007; Yang et al., 2011; Fondong, 2013; Xie et al., 2013). AC4 encoded by African cassava mosaic virus (ACMV) and Sri Lankan cassava mosaic virus (SLCMV), but not East African cassava mosaic Cameroon virus (EACMCV) nor Indian cassava mosaic virus (ICMV), have been shown to suppress RNA silencing by specifically binding to the single-stranded miRNAs and siRNAs to block the miRNA-mediated cleavage of target mRNAs (Chellappan et al., 2004, 2005; Vanitharani et al., 2004; Fondong et al., 2007). A recent study also shows that Tomato yellow leaf curl virus (TYLCV) C4 interacts with the receptor-like kinases (RLKs) BARELY ANY MERISTEM 1 (BAM1) and its homolog BAM2, interfering with the function of BAM1 and BAM2 in the intercellular spread of RNAi (Rosas-Diaz et al., 2018). Moreover, Mungbean yellow mosaic virus (MYMV) AC4 also interacts with BAM1 at plasma membrane (PM), and the PM location of MYMV AC4 depends on S-palmitoylation (Carluccio et al., 2018). AC4/C4 can also reverse methylation-mediated TGS. Cotton leaf curl Multan virus (CLCuMuV) C4 have been shown to repress both PTGS and TGS by interacting with S-adenosyl methionine synthetase (SAMS) and inhibiting SAMS activity (Ismayil et al., 2018).
A group of begomoviruses, most of which are monopartite geminiviruses, infect tomato plants and are considered to be responsible for a devastating disease called tomato yellow leaf curl disease (TYLCD) which is a major limiting factor for tomato cultivation worldwide (Navas-Castillo et al., 2011). A new begomovirus which might be responsible for the epidemic of TYLCD in Guangdong province of China during the year 2004–2006 was isolated and named Tomato leaf curl Guangdong virus (ToLCGdV) (He et al., 2005). ToLCGdV is a monopartite begomovirus with 2744nt in length and encodes 6 proteins as the other monopartite geminiviruses. As expected, ToLCGdV CP encoded by virion-sense strand shares the highest amino acid identity compared with other begomoviruses, while C4 protein shares the least amino acid identity (He et al., 2005). As the huge diversity of AC4/C4, the function of ToLCGdV C4 is still unknown.
In this study, the infectious clones of ToLCGdV and ToLCGdVmC4 were constructed. ToLCGdV C4 is a pathogenic determinant by contributing to ToLCGdV and PVX infection. ToLCGdV C4 is also a VSR by suppressing systemic gene silencing but not local gene silencing. Moreover, ToLCGdV C4 could reverse methylation-mediated TGS. In addition, ToLCGdV C4 suppresses PTGS maybe by interacting with BAM1. These results demonstrate that ToLCGdV C4 plays an important role in virus infection by functioning as both pathogenic determinant and VSR.
To construct ToLCGdV infectious clones pGreenII-1.3A-ToLCGdV which contains a 1.3-mer tandem repeat of ToLCGdV, the full-length sequence and a 0.8-kb fragment of ToLCGdV were amplified with primer pairs ToLCGDV-F1/R1 and ToLCGDV-F2/R2 (Supplementary Table 1), respectively, and introduced into SmaI-digested pGreenII (Hellens et al., 2000) by Seamless Cloning and Assembly (Takara). pGreenII-1.3A-ToLCGdVmC4 was generated by QuickChange® site-directed mutagenesis (Agilent Technologies) with pGreenII-1.3A-ToLCGdV as templates.
C4 and the mutated C4 nucleotide sequences were amplified from pGreenII-1.3A-ToLCGdV or pGreenII-1.3A-ToLCGdVmC4 and introduced into pGR107 (Lu et al., 2003) to obtain PVX-C4 and PVX-mC4. pGD-C4-Myc was generated by inserting C4 sequence into pGD-Myc (Goodin et al., 2002). Subsequently, the sequence of C4-Myc was amplified and introduced into pGR107 to get PVX-C4-Myc.
For bimolecular fluorescence complementation (BiFC) assays, C4 was amplified and cloned into pSPYNE-35S and pSPYCE-35S split YFP destination vectors (Walter et al., 2004). For subcellular localization experiments, C4 was amplified, followed by cloning into pGDGm (Goodin et al., 2002; Fan et al., 2014) with standard protocols.
Primers used for plasmid construction in this study are listed in Supplementary Table 1.
Nicotiana benthamiana and tomato plants used in this study were grown in a climate chamber with a 13h/11h light/dark photoperiod at 24°C. N. benthamiana were agroinfiltrated at 5–6 leaf stage, and tomato plants were inoculated at 6–7 leaf stage.
For western blot detection, samples were taken and weighed, followed by ground in liquid nitrogen. Two volumes of 1 × SDS loading buffer was added and boiled for 10 min. After centrifugation for 10 min at 12 000 rpm, 20 μl supernatant was loaded per lane on the SDS-PAGE gel. Then proteins were transferred to the nitrocellulose membranes. The secondary antibody was purchased from Sigma-Aldrich, Inc., Alkaline phosphatase was visualized with NBT/BCIP (Sangon Biotech, Shanghai, China).
AC4/C4 protein sequences were obtained by searching National Center for Biotechnology Information (NCBI) GenBank using the protein sequence of ToLCGdV C4. Phylogenetic trees were constructed with MEGA7 (Kumar et al., 2016) software using neighbor-joining method based on AC4/C4 protein sequences. 1000 bootstrap replicates were performed to obtain support for the identified phylogenetic relationships.
Total RNA was extracted with Trizol reagent (Takara) and treated with RNase-free rDNase I (Takara) before reverse transcription reaction. Equal amount of treated RNA was subjected to reverse transcription using PrimeScript RT Reagent Kit (Takara). Real-time RT-PCR was performed with SYBR Premix Ex Taq II (Takara) using CFX96 Real-Time System (Bio-Rad, United States).
To identify the suppressor activity of C4, 16c transgenic N. benthamiana plants (provided by Dawei Li lab) and 16c-TGS plants were used. 16c-TGS plants were generated as described (Buchmann et al., 2009). Agrobacterium infiltration was performed as described previously (Hamilton et al., 2002). Tomato bushy stunt virus (TBSV) p19 and an empty vector (pGD) were used as positive and negative controls, respectively. GFP fluorescence was observed under a long-wave UV lamp (Black Ray model B 100A; UV Products) and photographed using a Nikon D70 digital camera with a Y48 yellow filter.
Agrobacterium strains harboring the corresponding plasmids were co-infiltrated into 5–6 leaf stage N. benthamiana plants. Samples were taken at 3 dpi and visualized under Zeiss LSM710 confocal microscope (Carl Zeiss 710, Germany). Excitation wavelengths were as follows: EYFP, 514 nm; RFP, 561 nm.
Genomic DNA was extracted with a cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). Cytosine methylation level of the 35S promoter region was assayed by next generation sequencing-based BSP method as described previously (Gao et al., 2014, 2015; Pan et al., 2018). Primer pair used to amplify 35S promoter was described previously (Ismayil et al., 2018). PCR products of 35S promoter from different samples were quantified by Qubit 3.0 (Thermo Fisher), followed by bisulfite treatment with EZ DNA Methylation Gold Kit (Zymo Research). Then the products were subjected to adapter ligation by PCR amplification to generate barcoded libraries. Barcoded libraries from different samples were pooled together equally for standard pair-end sequencing with Illumina HiSeq PE150. The acquired data were processed with standard protocols (Li et al., 2010).
Co-immunoprecipitation assays were performed according to the previously published protocols (Rubio et al., 2005; Hu et al., 2015). Inoculated N. benthamiana leaves were collected at 3 dpi to perform Co-IP assays with anti-c-Myc agarose affinity gel (Sigma Aldrich, United States).
Tomato leaf curl Guangdong virus was firstly reported and named by He et al. (2005), and no satellite molecules were found to be associated with ToLCGdV. ToLCGdV contains 2744 nucleotides and encodes 6 proteins, V1, V2, C1, C2, C3, and C4 (Figure 1A). As the least conserved protein among geminiviruses, the variety of C4 is one of the main reasons that responsible for the diversity of geminiviruses. To explore the relationship of ToLCGdV C4 with other begomoviruses AC4/C4, multiple AC4/C4 protein sequences of different begomoviruses (Table 1) were obtained by searching NCBI GenBank using the protein sequence of ToLCGdV C4. Phylogenetic analysis shows that ToLCGdV C4 shares the highest amino acids similarity (92.94%) with AC4 of Euphorbia leaf curl virus (EuLCuV) (Figure 1B), an Euphorbia pulcherrima-infecting monopartite begomovirus firstly identified in China (Ma et al., 2004). ToLCGdV C4 is also close to AC4 proteins encoded by other weed-infecting begomoviruses like Asystasia begomovirus 2 (ABgV2), Allamanda leaf curl virus (AllLCV), and Pouzolzia mosaic Guangdong virus (PouMGDV) (Figure 1B). In contrast, ToLCGdV C4 has relatively farther genetic distance with other tomato-infecting begomoviruses (Figure 1B). Thus, it is speculated that ToLCGdV may be a recombinant begomovirus originated from the weed-infecting begomoviruses.
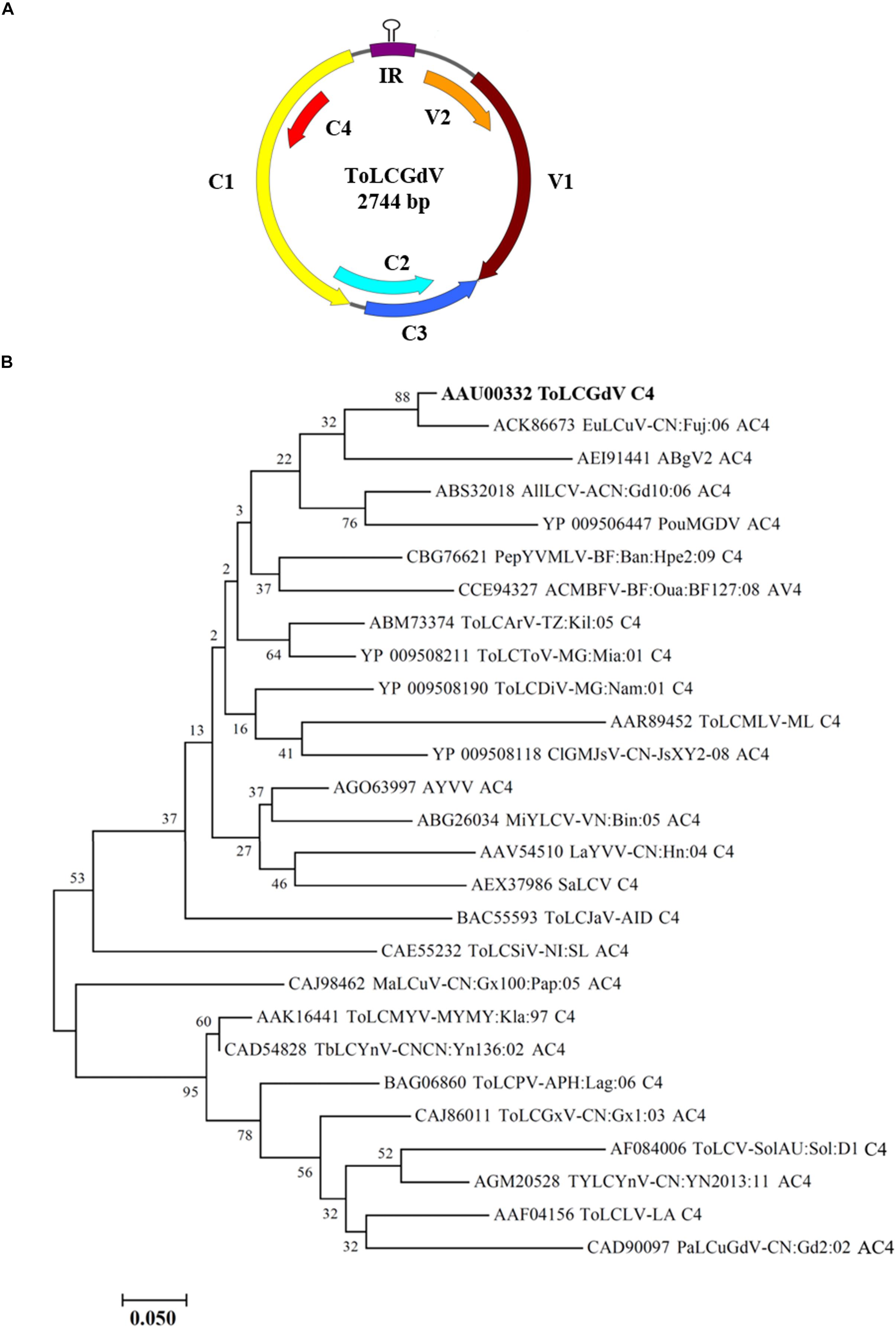
Figure 1. Schematic diagram of ToLCGdV genome and phylogenetic tree constructed from AC4/C4 protein sequences by neighbor-joining method. (A) Schematic diagram of ToLCGdV genome. Viral genes are indicated by filled arrows. IR, intergenic region. (B) Phylogenetic tree constructed from AC4/C4 protein sequences by neighbor-joining method. Twenty-seven AC4/C4 protein sequences were acquired from the GenBank. Full names and the origins of the begomoviruses used in the phylogenetic analyses are listed in Table 1. Numbers on the branches are bootstrap obtained from 1000 replicates. ToLCGdV C4 are marked in bold.
To study the molecular characteristic of ToLCGdV, the infectious clones of ToLCGdV and ToLCGdVmC4 which contains an unexpressed C4 were constructed. pGreenII-1.3A-ToLCGdV (Figure 2A) and pGreenII-1.3A-ToLCGdVmC4 (Figure 2B) were transformed to Agrobacterium tumefaciens strain GV3101 and then infiltrated into 5–6 leaf stage N. benthamiana plants. Comparing with control plants, plants agroinfiltrated with pGreenII-1.3A-ToLCGdV showed symptoms with stunting, newly leaves curly, and yellowing at 10 days post inoculation (dpi) (Figure 2C), while plants agroinfiltrated with pGreenII-1.3A-ToLCGdVmC4 showed visible symptoms with newly leaves curly until 15 dpi (Figure 2C). Western blot detection also confirmed that though the mutant virus can still move systemically to the upper leaves, the virus accumulation reduced significantly due to the loss of C4 (Figures 2D,E). In summary, inoculation of N. benthamiana verified the validity of the infectious clones and demonstrate that loss of C4 remarkably delay and weaken disease symptoms of ToLCGdV in N. benthamiana.
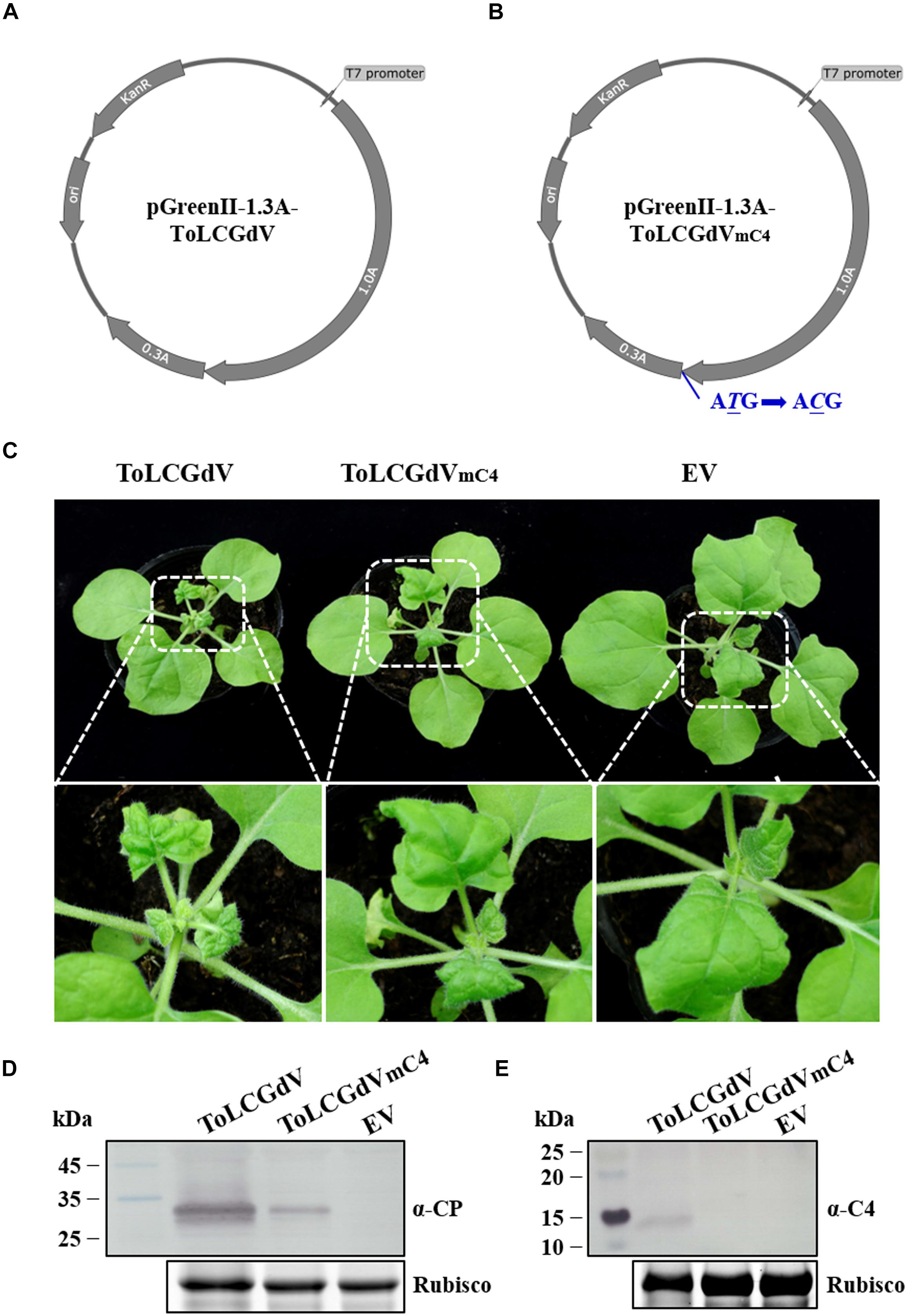
Figure 2. Construction of ToLCGdV and ToLCGdVmC4 infectious clones. (A) Schematic depiction of pGreenII-1.3A-ToLCGdV. Full-length and 0.3-time sequence of ToLCGdV were amplified and cloned into pGreenII vector (Hellens et al., 2000). KanR, kanamycin resistance. ori, replication initial origin. (B) Schematic depiction of pGreenII-1.3A-ToLCGdVmC4. Loss expression of C4 was made by replacing the start codon ATG with ACG, which has no effect on the expression of C1. KanR, kanamycin resistance. ori, replication initial origin. (C) Symptoms of N. benthamiana plants agroinfiltrated with pGreenII-1.3A-ToLCGdV or pGreenII-1.3A-ToLCGdVmC4. Agrobacterium strains harboring pGreenII-1.3A-ToLCGdV or pGreenII-1.3A-ToLCGdVmC4 were infiltrated into the leaves of 5–6 leaf-stage N. benthamiana. Photos were taken at 13 dpi. The bottom panel shows the magnification of the white dotted frame. EV, empty vector. (D) Western blot detection of virus accumulation in the infected plants. The upper leaves were taken at 13 dpi and subjected to western blot detection with anti-ToLCGdV AV1 (CP) antibody. Rubisco shows equal sample loading. Numbers on the left indicate molecular weight. (E) Western blot detection of C4 in the upper infected leaves. Leaves were taken at 13 dpi to perform western blot with anti-C4 antibody. Rubisco is used as equal loading. Numbers on the left indicate molecular weight.
To explore the function of C4 during ToLCGdV infection in its natural host tomato, Agrobacterium strain containing pGreenII-1.3A-ToLCGdV or pGreenII-1.3A-ToLCGdVmC4 was infiltrated into tomato plants. Tomato plants agroinfiltrated with pGreenII-1.3A-ToLCGdV developed obvious symptoms including leaf curling and yellowing at 30 dpi (Figure 3A), indicating the monopartite nature of ToLCGdV. However, tomato plants infected by ToLCGdVmC4 showed much milder symptoms compared with plants infected by ToLCGdV or mock-treated (Figure 3A). In addition, ToLCGdVmC4-infected tomato plants were not as shortened as ToLCGdV-infected tomato plants (Figure 3B). Quantitative PCR also confirmed that loss of C4 significantly reduced viral accumulation (Figure 3C). These results implied that though ToLCGdV can still move systemically without C4 expression, C4 plays an important role in ToLCGdV symptom development.
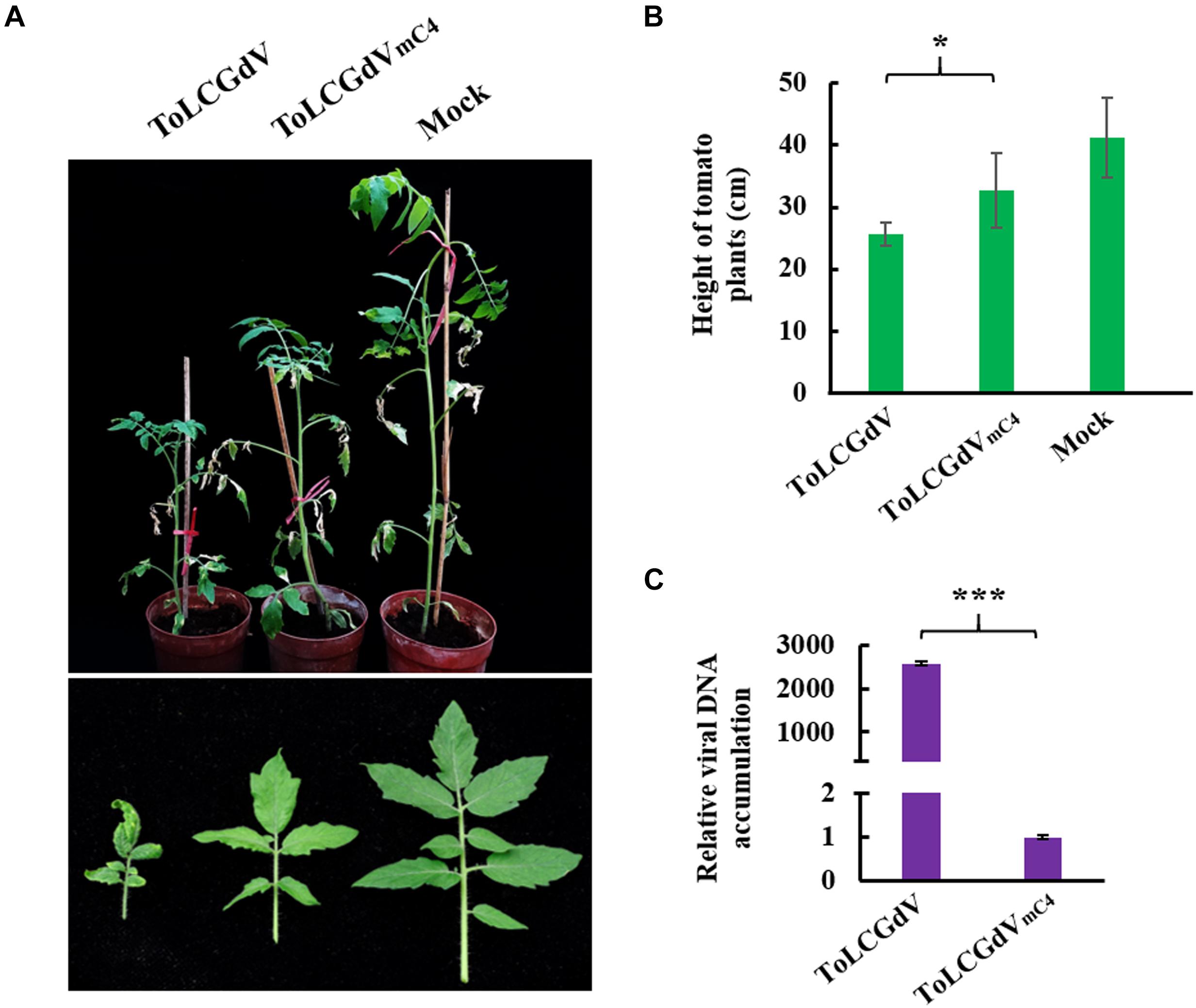
Figure 3. Effects of C4 on ToLCGdV pathogenicity in tomato. (A) Symptoms of tomato plants infected by ToLCGdV or ToLCGdVmC4. Agrobacterium strains harboring pGreenII-1.3A-ToLCGdV or pGreenII-1.3A-ToLCGdVmC4 were infiltrated into 4–6-week-old tomato plants. Photos were taken at 30 dpi. (B) Statistical analysis of the height of tomato plants infected by ToLCGdV and ToLCGdVmC4. Ten plants of each treatment were performed to measure the height. *p < 0.05. (C) Quantitative PCR to detect the viral DNA accumulation. Samples were taken at 30 dpi and subjected to DNA extraction. 0.2 μg DNA was used to perform real-time PCR. ***p < 0.001 (extremely significant).
To further substantiate the role of C4 in pathogenesis independent of ToLCGdV, C4 fused with Myc tag was cloned into the Potato virus X (PVX) vector pGR107 (Lu et al., 2003) to yield PVX-C4-Myc. Moreover, mC4-Myc with start codon mutation was also cloned into pGR107 to make PVX-mC4-Myc. Agrobacterium containing PVX-C4-Myc, PVX-mC4-Myc, or PVX was infiltrated into 5–6 leaf-stage N. benthamiana. Mild mosaic symptoms on the top leaves of N. benthamiana infected by PVX-C4-Myc were first observed at 5 dpi, while no obvious symptoms were observed on plants inoculated with PVX-mC4-Myc or PVX at this time (Supplementary Figure 1). At 10 dpi, severe mosaic and curling of the leaves were observed on plants infected by PVX-C4-Myc, while only mild mosaic was observed in plants infected by PVX-mC4-Myc or PVX (Figure 4A). At later time, symptoms on the newly emerged leaves of plants inoculated with PVX or PVX-mC4-Myc became very weak and hardly visible (Supplementary Figure 1). However, symptoms including mosaic, curling, and distortion caused by PVX-C4-Myc sustained throughout the life of the plants. Western blot experiments performed at 10 dpi with anti-PVX CP antibody and anti-Myc antibody confirmed that C4 could remarkably increase PVX accumulation (Figures 4B,C), further implying that C4 may be a pathogenic determinant independent of ToLCGdV infection.
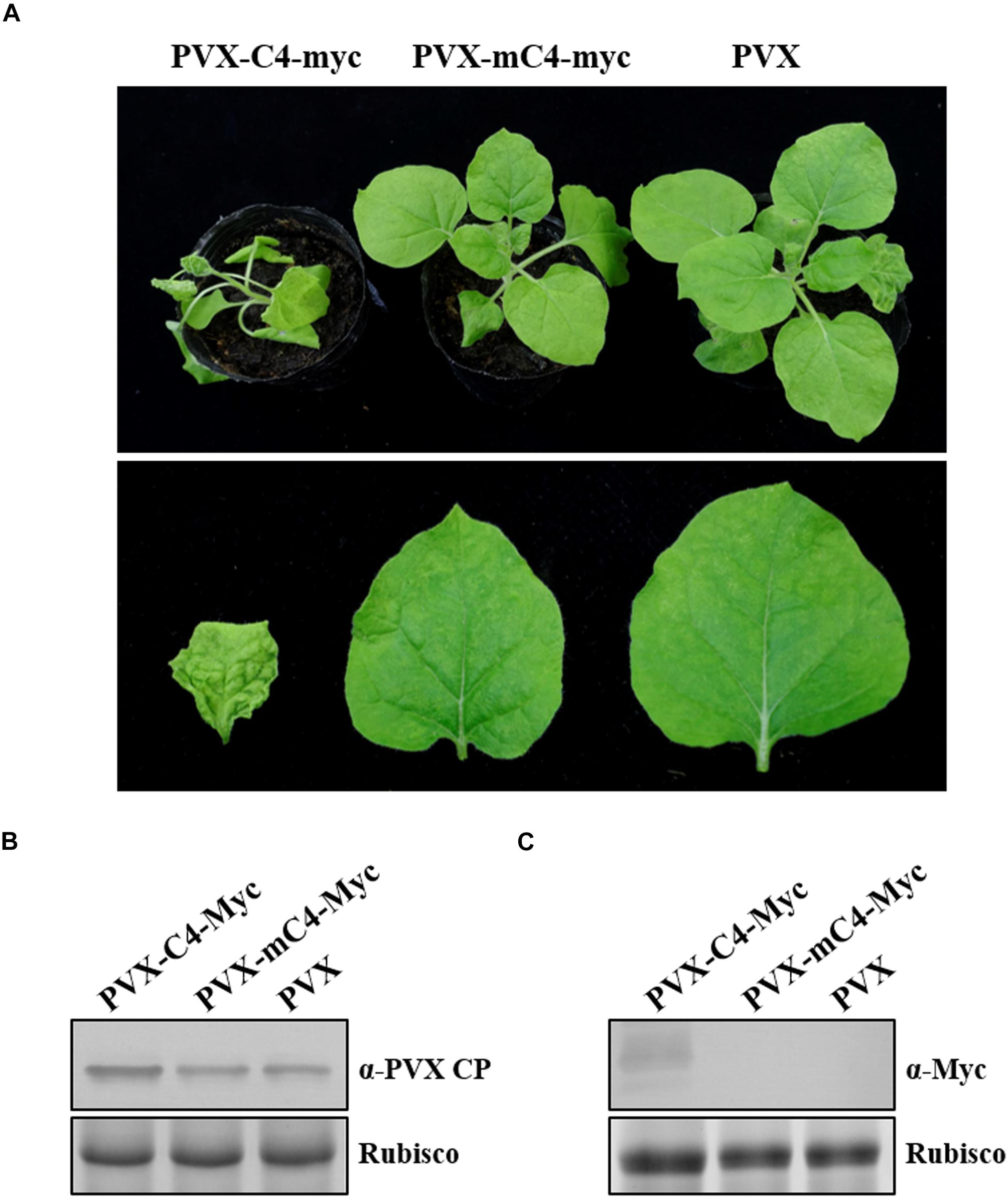
Figure 4. Enhancement of C4 on PVX symptom development. (A) Symptoms of N. benthamiana plants infected by PVX-C4-Myc, PVX-mC4-Myc, and PVX, respectively. Agrobacterium strain harboring PVX-C4-Myc, PVX-mC4-Myc, or PVX was infiltrated into 5–6 leaf-stage N. benthamiana plants. Photos were taken at 10 dpi. (B) Western blot detection of the upper infected leaves. Samples were taken at 10 dpi and subjected to western blot with anti-PVX CP antibody. Rubisco indicates equal sample loading. (C) Western blot to detect C4 expression. Samples were taken at 10 dpi and western blot was performed with anti-Myc antibody. Rubisco indicates equal sample loading.
AC4/C4 encoded by some begomoviruses have been reported to function as a VSR (Vanitharani et al., 2004; Bisaro, 2006; Gopal et al., 2007). To explore the function of ToLCGdV C4 in RNA silencing suppression, a widely adopted method based on Agrobacterium infiltration was utilized (Johansen, 2001). Agrobacterium strains harboring C4-Myc and the positive sense-GFP (sGFP) (Bragg and Jackson, 2004; Dong et al., 2016; Zhang et al., 2017, 2018) were mixed equally and co-infiltrated into N. benthamiana leaves. Co-infiltration of p19 and sGFP was used as positive control, while co-infiltration of empty vector (EV) and sGFP was used as negative control. Three days after agroinfiltration, strong GFP fluorescence was observed in regions co-infiltrated with sGFP and p19 (Figure 5A). However, no obvious or little fluorescence was observed in leaf patches co-infiltrated with sGFP and C4-Myc or sGFP and EV (Figure 5A). The results were the same at 6 dpi. Western blot detection confirmed that C4 had no effect on GFP protein accumulation compared with p19 and EV (Figure 5B).
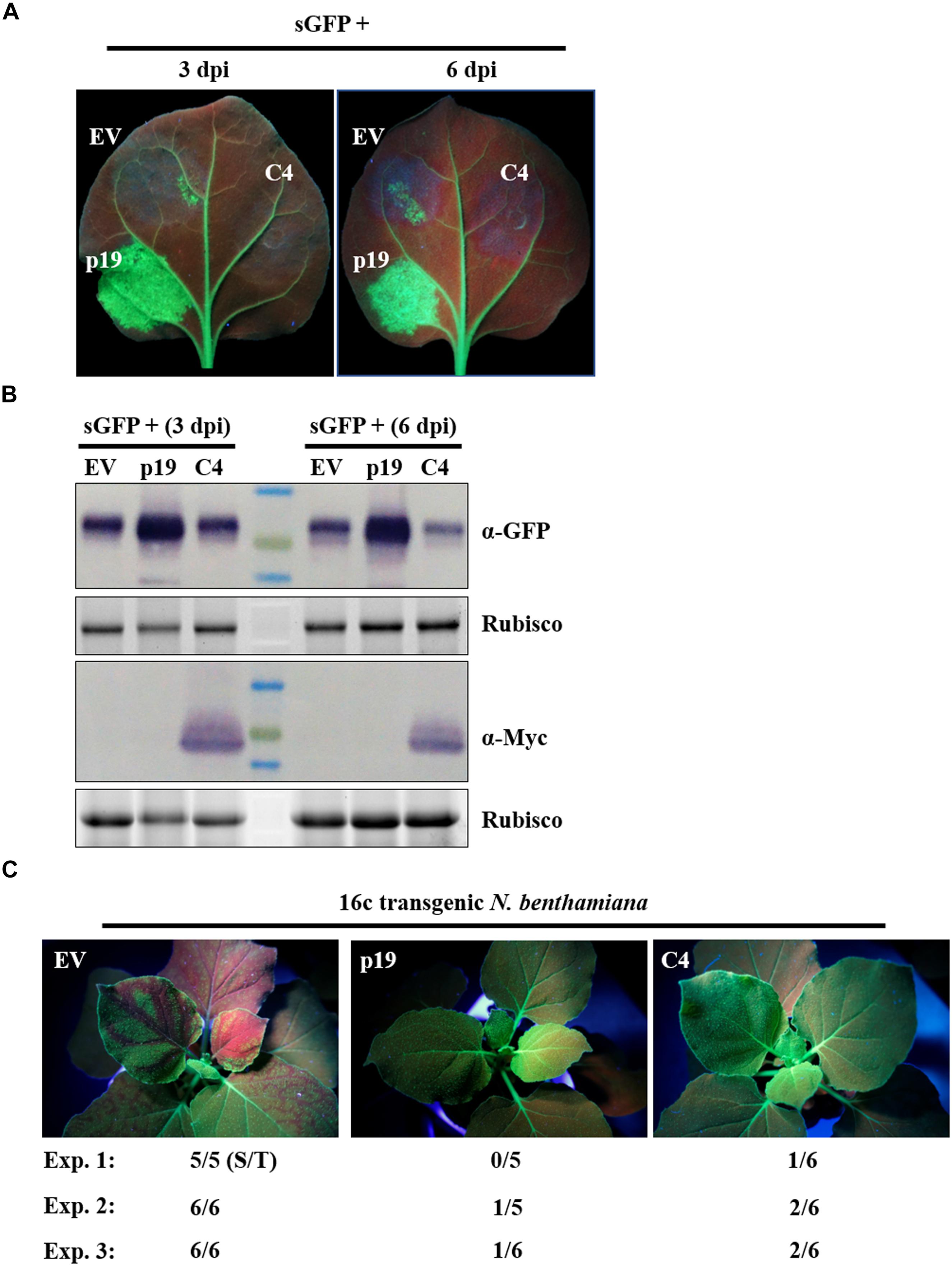
Figure 5. Suppression of ToLCGdV C4 in PTGS. (A) Effect of C4 in local gene silencing. Agrobacterium strains containing sGFP and p19, sGFP and EV, or sGFP and C4-Myc were co-infiltrated into the same leaf of N. benthamiana. EV and p19 were used as negative and positive controls, respectively. Leaves were photographed under UV light at 3 and 6 dpi. (B) Western blot assays of GFP and C4 expression in co-infiltrated leaves. Samples taken at 3 and 6 dpi were subjected to western blot with anti-GFP and anti-Myc antibody. Rubisco indicates equal sample loading. (C) Suppression of C4 in systemic gene silencing. Agrobacterium strains containing sGFP and p19, sGFP and EV, or sGFP and C4-Myc were co-infiltrated into 16c transgenic N. benthamiana plants. Inoculated plants were observed under UV light at 12 dpi. Three independent experiments were repeated. The number ratio (S/T) indicates the systemic silencing (S) among the total number of infiltrated plants (T).
Next, we tested whether C4 suppresses systemic movement of RNA silencing signals. C4-Myc, p19, or EV were mixed with sGFP and co-infiltrated into leaves of 16c transgenic N. benthamiana (Voinnet and Baulcombe, 1997). Three independent experiments were repeated to detect the systemic movement of GFP silencing signals. As expected, transient expression of p19, but not the EV, suppressed the movement of GFP silencing signals from infiltrated leaves to upper leaves at 12 dpi (Figure 5C). C4 could also suppressed systemic movement of GFP silencing signals, though not as effective as p19 (Figure 5C).
In summary, these results demonstrate that C4 functions as a VSR by repressing systemic gene silencing but not local gene silencing.
To identify whether ToLCGdV C4 could suppress methylation-mediated TGS, 16c-TGS transgenic N. benthamiana plants were used. In 16c-TGS plants, GFP was silenced by TGS, and TGS was induced by Tobacco rattle virus (TRV) vector which contains a portion of the 35S promoter sequence (Buchmann et al., 2009; Raja et al., 2010). Agrobacterium harboring PVX-C4-Myc or PVX was infiltrated into 16c-TGS plants, and the plants were photographed under white light and UV lamp at 12 dpi. 16c-TGS plants infiltrated with PVX showed no visible GFP fluorescence, while plants infiltrated with PVX-C4-Myc showed obvious GFP fluorescence (Figure 6A). 16c and 16c-TGS N. benthamiana plants were used as positive and negative controls, respectively. Western blot and real-time RT-PCR confirmed that GFP protein and mRNA expressions were significantly increased in 16c-TGS plants infiltrated with PVX-C4-Myc compared with plants infiltrated with PVX (Figures 6B,C).
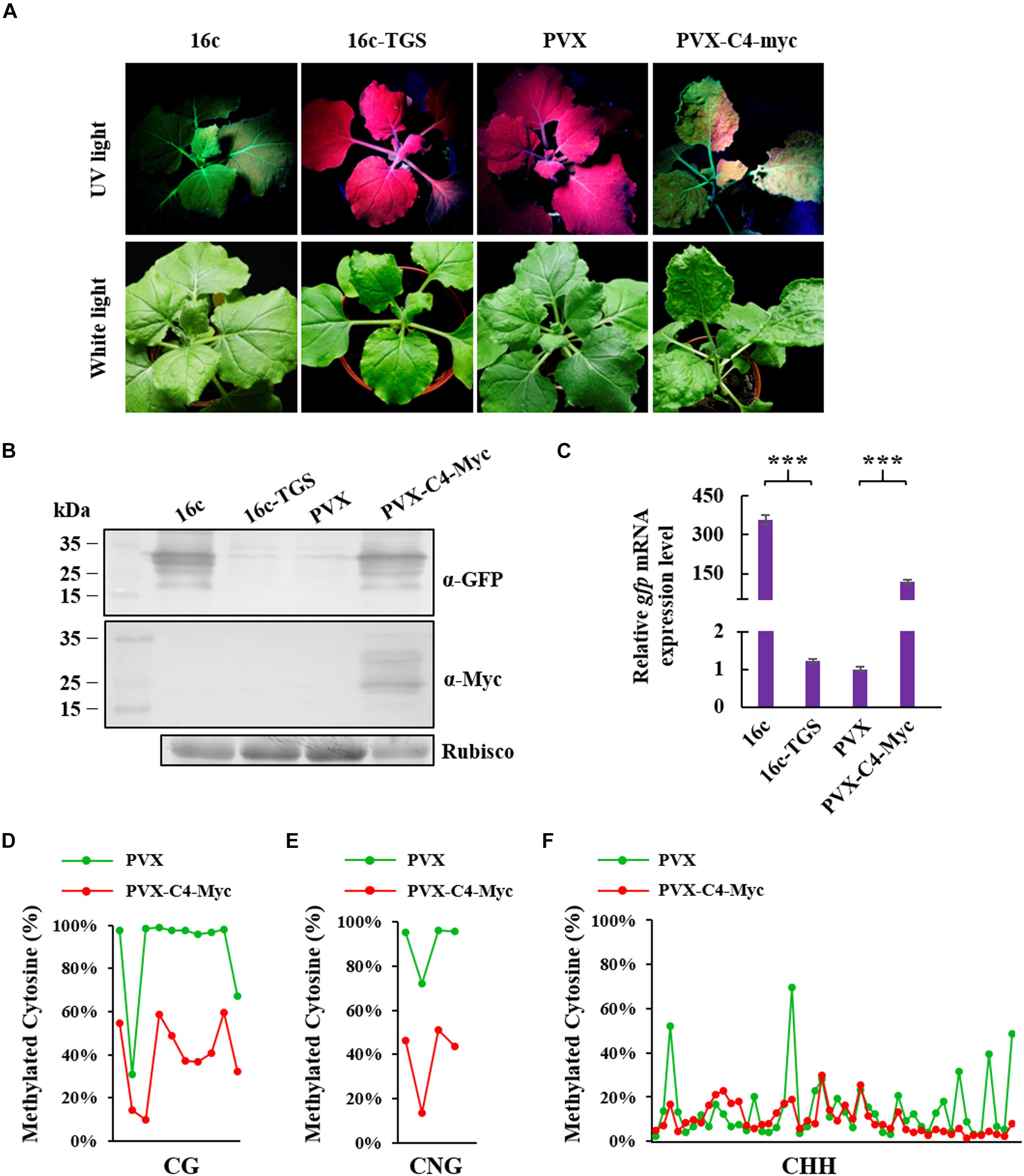
Figure 6. Suppression of ToLCGdV C4 in TGS. (A) PVX-based expression of C4 suppressed TGS. Agrobacterium strains containing sGFP and p19, sGFP and EV, or sGFP and C4-Myc were co-infiltrated into 16c-TGS N. benthamiana plants. At 12 dpi, the infiltrated plants were observed and photographed under UV light and white light. (B) Western blot assays to detect the expressions of GFP and C4 in infiltrated plants. Samples were taken at 12 dpi and subjected to western blot with anti-GFP and anti-Myc antibody, respectively. Rubisco was used as a loading control. Numbers on the left indicate molecular weight. (C) Real-time RT-PCR detection of GFP mRNA accumulation in infiltrated 16c-TGS plants. Samples were taken at 12 dpi and 2 μg RNA was used to perform detection. ∗∗∗p < 0.001 (extremely significant). Cytosine methylation level of CG sites (D), CNG sites (E), and CHH sites (F) in 35S promoter region. Cytosine methylation level was measured by next generation sequencing-based bisulfite sequencing PCR (BSP). Green dots represent the cytosine residues of 35S promoter in 16c-TGS plants infiltrated with PVX, and red dots represents the cytosine residues of 35S promoter in plants infiltrated with PVX-C4-Myc. The detailed methylation level of each CG, CNG, and CHH sites are listed in Supplementary Table 2.
To further confirm the ability of C4 to reverse methylation-mediated TGS, next generation sequencing-based bisulfite sequencing PCR (BSP) was performed to assess the cytosine methylation level of 35S promoter. Cytosine methylation level of ten CG, four CNG, and forty-eight CHH sites in the 35S promoter region were analyzed. C4 expressed from the PVX vector reduced cytosine methylation remarkably at CG (48%), CNG (51%), and CHH (5%) sites (Figures 6D–F and Supplementary Table 2). These results indicated that ToLCGdV C4 is able to suppress methylation-mediated TGS.
Tomato yellow leaf curl virus C4 and MYMV AC4 have been reported to interact with BAM1 in PM and plasmodesmata (PD) to inhibit the intercellular spread of RNAi (Carluccio et al., 2018; Rosas-Diaz et al., 2018), the interaction between ToLCGdV C4 and BAM1 was also investigated. Agrobacterium strains containing C4-EYFP and BAM1-RFP were co-infiltrated into N. benthamiana leaves. C4 co-localizes with BAM1 both in PM and PD (Figure 7A). BiFC assays found that C4 interacts with BAM1 at PM (Figure 7B). Co-IP assays further confirmed the interaction between C4 and BAM1 (Figure 7C). These results imply that ToLCGdV C4 may adopt the same strategy as TYLCV C4 to suppress PTGS (Rosas-Diaz et al., 2018).
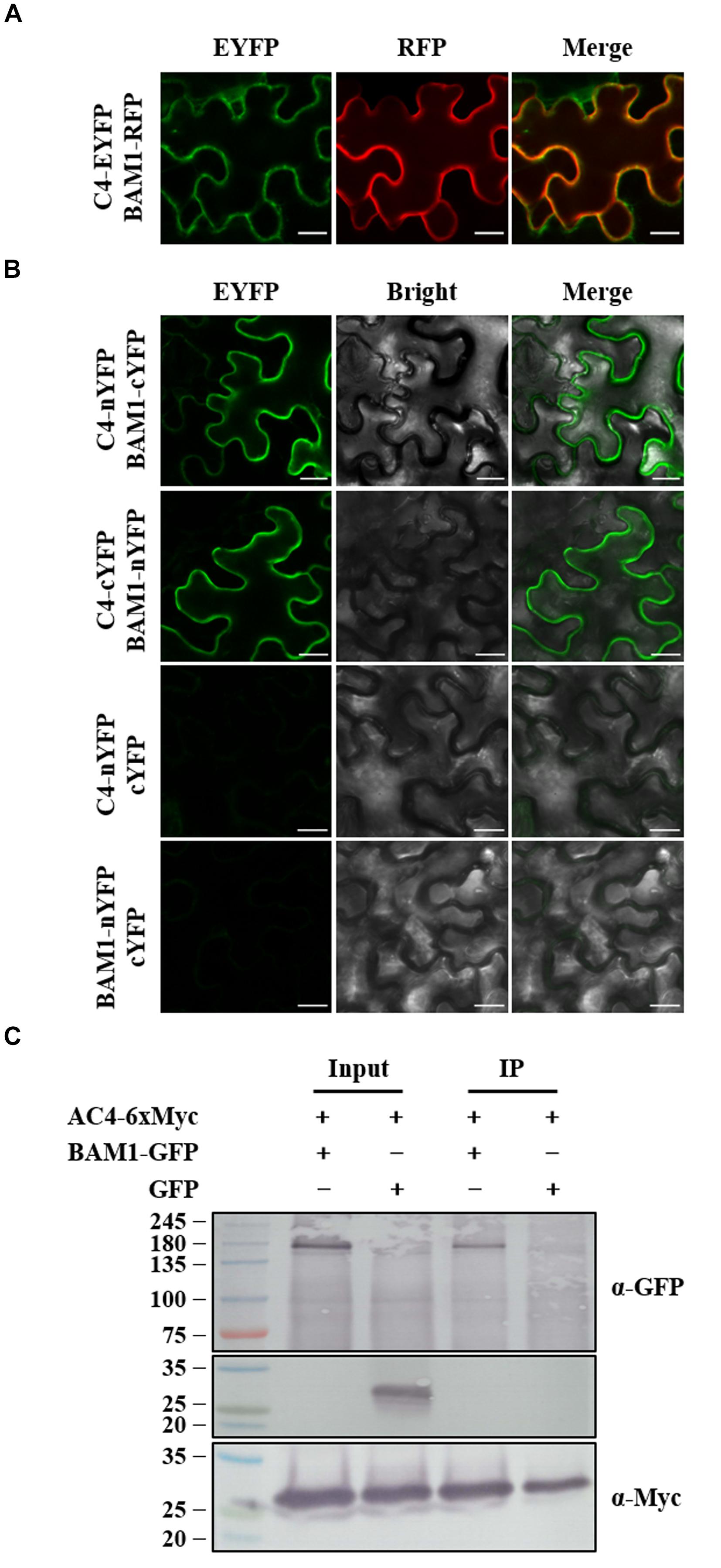
Figure 7. Co-localization and interaction between C4 and BAM1. (A) Co-localization of C4 and BAM1. Agrobacterium strains harboring C4-EYFP and BAM1-RFP were co-infiltrated into N. benthamiana leaves. At 3 dpi, the infiltrated leaves were taken and observed under confocal microscope. Bars = 20 μm. (B) Interaction between C4 and BAM1 by BiFC. Agrobacterium strains harboring C4-nYFP and BAM1-cYFP, or C4-cYFP and BAM1-nYFP were co-infiltrated into N. benthamiana leaves and subjected to confocal microscopy observation at 3 dpi. C4-nYFP and cYFP, and BAM1-nYFP and cYFP were used as negative controls. Bars = 20 μm. (C) Co-IP assays to detect the interaction between C4 and BAM1. Agrobacterium strains containing AC4-6xMyc and BAM1-GFP, or AC4-6xMyc and GFP were co-infiltrated into N. benthamiana leaves. Samples were taken at 3 dpi to perform Co-IP assays. Western blot detections were conducted with anti-GFP and anti-Myc antibodies, respectively. Numbers on the left indicate molecular weight.
In this study, we report that ToLCGdV may be a recombinant begomovirus originated from some weed-infecting begomoviruses based on phylogenetic analyses of multiple AC4/C4 protein sequences. ToLCGdV C4 is closely related to the AC4 of Euphorbia leaf curl virus (EuLCuV), Asystasia begomovirus 2 (ABgV2), Allamanda leaf curl virus (AllLCV), and Pouzolzia mosaic Guangdong virus (PouMGDV). All these begomoviruses are firstly isolated from wild plant hosts (He et al., 2009; Tang et al., 2014; Wyant et al., 2015), demonstrating the frequent recombination between wild hosts and crops (Lefeuvre and Moriones, 2015; Garcia-Arenal and Zerbini, 2019). Recombination is a key process in the evolution of many viruses, especially for geminiviruses which contain a highly compacted genome (∼2.7 kb) (Garcia-Andres et al., 2007; Varsani et al., 2008; Martin et al., 2011). It has been highly speculated that recombination between different geminiviruses in even different hosts is one of the main reasons for the diversification of geminiviruses (Padidam et al., 1999; Briddon et al., 2010). One of the best studied examples is the recombination between TYLCD-associated begomoviruses. Begomovirus contains over 320 species and is one of the largest plant virus genera (Zerbini et al., 2017). Begomoviruses are transmitted by whitefly between wild hosts and crops (Perefarres et al., 2012; Rey et al., 2012), which increases the opportunities of mix infection and recombination (Lima et al., 2012). Tomato leaf curl Yunnan virus (TLCYnV), a begomovirus firstly isolated from Malvastrum coromandelianum, evolved from Tomato yellow leaf curl China virus (TYLCCNV) by recombination and is highly infectious to a range of host plants by acquiring a more virulent C4 (Xie et al., 2013). Moreover, a recombinant virus named tomato yellow leaf curl Malaga virus (TYLCMalV) is the recombination of Tomato yellow leaf curl Sardinia virus (TYLCSV) and TYLCV (Monci et al., 2002). TYLCMalV exhibits a novel pathogenic phenotype and has an enlarge host range, which contribute to the prevalence in the region where it was detected (Monci et al., 2002). Thus, we speculate that ToLCGdV may be a recombinant virus evolved from two or more wild plant hosts, acquiring more virulence and becoming prevalent in Guangdong province where ToLCGdV was isolated.
We also found that disruption of C4 delayed ToLCGdV symptom development in N. benthamiana and significantly reduced viral DNA accumulation in tomato plants. However, the detailed function of C4 during ToLCGdV infection remains unknown. C4 has long been demonstrated to be required for monopartite begomovirus infection (Stanley and Latham, 1992; Teng et al., 2010; Li et al., 2018). Recently, AC4 of EACMCV has been shown to be involved in virus infection, knockout mutation in AC4 ORF delayed virus symptom development and plants recovered from the mutant virus infection (Chen et al., 2019). In addition, AC4 of ACMV (Hipp et al., 2016) and of MYMV (Carluccio et al., 2018) were also required for virus infection.
Tomato leaf curl Guangdong virus C4 expressed from the PVX vector significantly increased PVX concentration, and induced leaf curling and severe mosaic in plants inoculated with PVX-C4-Myc compared with plants inoculated with PVX or PVX-mC4-Myc. This result implies that C4 may be a pathogenic determinant essential for abnormalities in infected plants. Induction of hyperplasia and tumorigenic growth in infected plant tissues by AC4/C4 has been extensively studied (Mills-Lujan et al., 2015). The nature of hyperplasia and tumorigenic growth is uncontrolled DNA replication and loss function of host cell cycle regulators. AC4/C4 is considered to be the “oncogene” that manipulates the host cell cycle to stimulate DNA replication (Nikitin and Luftig, 2012). However, the mechanism about how AC4/C4 induces abnormalities in infected plants may be variable according to the diversity of AC4/C4. A recent study found that TLCYnV C4 interacts and relocates the glycogen synthase kinase 3 (GSK3)/SHAGGY like kinase, named NbSKη, from the nucleus to membrane. Relocalization of NbSKη affects the degradation of Cyclin D1.1, thereby inducing the cell division (Mei et al., 2018b). Another study found that Beet severe curly top virus (BSCTV) C4 induces the expression of a RING finger E3 ligase, RKP, which antagonizes with an inhibitor of cyclin-dependent kinase (CDK) to induce cell cycle (Cheng et al., 2013).
Tomato leaf curl Guangdong virus C4 is a VSR which inhibits both TGS and PTGS. TGS and PTGS are the main pathways exploited by plants to counter virus infection by inducing viral genome methylation or sequence-specific mRNA degradation. TGS acts as defenses against DNA viruses, however, geminiviruses have evolved to encode proteins to interfere with these processes. VSR encoded by begomovirus is often able to suppress DNA methylation-mediated TGS, like βC1 of Tomato yellow leaf curl China virus betasatellite (TYLCCNB) (Yang et al., 2011), C4 of CLCuMuV (Ismayil et al., 2018), and C4 of Tomato leaf curl Yunnan virus (Xie et al., 2013). TYLCCNB βC1 interacts and inhibits the activity of a key enzyme required for maintenance of the methyl cycle, S-adenosyl homocysteine hydrolase (SAHH) (Yang et al., 2011).
Tomato leaf curl Guangdong virus C4 inhibits systemic gene silencing but not local gene silencing, this is also the case for TYLCV C4 (Luna et al., 2012). AC4/C4 of TYLCV or MYMV has been found to suppress intercellular spread of RNAi by interacting with BAM1 (Rosas-Diaz et al., 2018). We found that ToLCGdV C4 also colocalizes and interacts with BAM1. However, AC4/C4 encoded by TYLCV, MYMV, and ToLCGdV shares low amino acids identity (Supplementary Figure 2), implying that different begomoviruses may adopt the same strategy to suppress PTGS.
All datasets generated for this study are included in the article/Supplementary Material.
ZL, ZD, and ZH conceived and designed the experiments. ZL, ZD, YT, and LY conducted the experiments. XS, XW, GL, and YZ analyzed the data. ZL and ZD wrote the manuscript. All authors read and approved the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (31871937 and 31801712), the Key Research and Development Program of Guangdong Province (2018B020202006), the National Key Research and Development Program of China (2018YFD0201209), and Special Fund for Scientific Innovation Strategy-Construction of High-Level Academy of Agriculture Science (R2017YJ-YB3004, R2019PY-QF003, and R2018QD-058).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to RL-D (Chinese Academy of Sciences, Shanghai, China) who generously provided BAM1 related agrobacterium strains. We would also thank Dr. Yule Liu (Tsinghua University, Beijing, China) for his kind help in the generation of 16c-TGS N. benthamiana plants.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00851/full#supplementary-material
FIGURE S1 | Symptoms of N. benthamiana infected by PVX-C4-myc, PVX-mC4-myc, and PVX at different times.
FIGURE S2 | Amino acids alignment of AC4/C4 of ToLCGdV, TYLCV, and MYMV.
TABLE S1 | Primers used in this study.
TABLE S2 | Cytosine methylation level of each CG, CNG, and CHH site.
Bi, H., Fan, W., and Zhang, P. (2017). C4 protein of Sweet potato leaf curl virus regulates brassinosteroid signaling pathway through interaction with ATBIN2 and affects male fertility in Arabidopsis. Front. Plant Sci. 8:1689. doi: 10.3389/fpls.2017.01689
Bisaro, D. M. (2006). Silencing suppression by geminivirus proteins. Virology 344, 158–168. doi: 10.1016/j.virol.2005.09.041
Boulton, M. I. (2003). Geminiviruses: major threats to world agriculture. Ann. Appl. Biol. 142, 143–143. doi: 10.1111/j.1744-7348.2003.tb00239.x
Bragg, J. N., and Jackson, A. O. (2004). The C-terminal region of the Barley stripe mosaic virus γb protein participates in homologous interactions and is required for suppression of RNA silencing. Mol. Plant Pathol. 5, 465–481. doi: 10.1111/j.1364-3703.2004.00246.x
Briddon, R. W., Patil, B. L., Bagewadi, B., Nawaz-ul-Rehman, M. S., and Fauquet, C. M. (2010). Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol. Biol. 10:97. doi: 10.1186/1471-2148-10-97
Buchmann, R. C., Asad, S., Wolf, J. N., Mohannath, G., and Bisaro, D. M. (2009). Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83, 5005–5013. doi: 10.1128/JVI.01771-1778
Carluccio, A. V., Prigigallo, M. I., Rosas-Diaz, T., Lozano-Duran, R., and Stavolone, L. (2018). S-acylation mediates Mungbean yellow mosaic virus AC4 localization to the plasma membrane and in turns gene silencing suppression. PLoS Pathog. 14:e1007207. doi: 10.1371/journal.ppat.1007207
Chellappan, P., Vanitharani, R., and Fauquet, C. M. (2005). MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. U.S.A. 102, 10381–10386. doi: 10.1073/pnas.0504439102
Chellappan, P., Vanitharani, R., Pita, J., and Fauquet, C. M. (2004). Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J. Virol. 78, 7465–7477. doi: 10.1128/JVI.78.14.7465-7477.2004
Chen, K., Khatabi, B., and Fondong, V. N. (2019). The AC4 protein of a cassava geminivirus is required for virus infection. Mol. Plant Microbe Interact. 32, 865–875. doi: 10.1094/MPMI-12-18-0354-R
Cheng, Y., Cao, L., Wang, S., Li, Y., Shi, X., Liu, H., et al. (2013). Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 75, 642–655. doi: 10.1111/tpj.12228
Deom, C. M., and Mills-Lujan, K. (2015). Toward understanding the molecular mechanism of a geminivirus C4 protein. Plant Signal. Behav. 10, e1109758. doi: 10.1080/15592324.2015.1109758
Dong, K., Wang, Y., Zhang, Z., Chai, L.-X., Tong, X., Xu, J., et al. (2016). Two amino acids near the N-terminus of Cucumber mosaic virus 2b play critical roles in the suppression of RNA silencing and viral infectivity. Mol. Plant Pathol. 17, 173–183. doi: 10.1111/mpp.12270
Doyle, J., and Doyle, J. (1987). Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull. 19, 11–15.
Fan, H., Sun, H., Wang, Y., Zhang, Y., Wang, X., Li, D., et al. (2014). Deep sequencing–based transcriptome profiling reveals comprehensive insights into the responses of Nicotiana benthamiana to Beet necrotic yellow vein virus infections containing or lacking RNA4. PLoS One 9:e85284. doi: 10.1371/journal.pone.0085284
Fauquet, C. M., Briddon, R. W., Brown, J. K., Moriones, E., Stanley, J., Zerbini, M., et al. (2008). Geminivirus strain demarcation and nomenclature. Arch. Virol. 153, 783–821. doi: 10.1007/s00705-008-0037-36
Fondong, V. N. (2013). Geminivirus protein structure and function. Mol. Plant Pathol. 14, 635–649. doi: 10.1111/mpp.12032
Fondong, V. N., Reddy, R. V., Lu, C., Hankoua, B., Felton, C., Czymmek, K., et al. (2007). The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol. Plant Microbe Interact. 20, 380–391. doi: 10.1094/MPMI-20-4-0380
Gao, F., Liang, H., Lu, H., Wang, J., Xia, M., Yuan, Z., et al. (2015). Global analysis of DNA methylation in hepatocellular carcinoma by a liquid hybridization capture-based bisulfite sequencing approach. Clin. Epigenet. 7:86. doi: 10.1186/s13148-015-0121-121
Gao, F., Zhang, J., Jiang, P., Gong, D., Wang, J. W., Xia, Y., et al. (2014). Marked methylation changes in intestinal genes during the perinatal period of preterm neonates. BMC Genomics 15:716. doi: 10.1186/1471-2164-15-716
Garcia-Andres, S., Accotto, G. P., Navas-Castillo, J., and Moriones, E. (2007). Founder effect, plant host, and recombination shape the emergent population of begomoviruses that cause the tomato yellow leaf curl disease in the Mediterranean basin. Virology 359, 302–312. doi: 10.1016/j.virol.2006.09.030
Garcia-Arenal, F., and Zerbini, F. M. (2019). Life on the edge: geminiviruses at the interface between crops and wild plant hosts. Annu Rev Virol. 6, 411–433. doi: 10.1146/annurev-virology-092818-15536
Goodin, M. M., Dietzgen, R. G., Schichnes, D., Ruzin, S., and Jackson, A. O. (2002). pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31, 375–383. doi: 10.1046/j.1365-313X.2002.01360.x
Gopal, P., Pravin Kumar, P., Sinilal, B., Jose, J., Kasin Yadunandam, A., and Usha, R. (2007). Differential roles of C4 and βC1 in mediating suppression of post-transcriptional gene silencing: evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 123, 9–18. doi: 10.1016/j.virusres.2006.07.014
Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679.
Hanley-Bowdoin, L., Settlage, S. B., Orozco, B. M., Nagar, S., and Robertson, D. (1999). Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18, 71–106. doi: 10.1080/07352689991309162
He, Z. F., Mao, M. J., Yu, H., Li, H. P., and Chen, X. (2009). Molecular characterization of a distinct begomovirus infecting Allamanda cathartica in Guangdong. China. Arch. Virol. 154, 1199–1202. doi: 10.1007/s00705-009-0445-442
He, Z. F., Yu, H., and Luo, F. F. (2005). Molecular characteristics of DNA A of Tomato leaf curl Guangdong virus isolate G2. Wei Sheng Wu Xue Bao. 45, 48–52.
Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S., and Mullineaux, P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. doi: 10.1023/A:1006496308160
Hipp, K., Rau, P., Schafer, B., Pfannstiel, J., and Jeske, H. (2016). Translation, modification and cellular distribution of two AC4 variants of African cassava mosaic virus in yeast and their pathogenic potential in plants. Virology 498, 136–148. doi: 10.1016/j.virol.2016.07.011
Hu, Y., Li, Z., Yuan, C., Jin, X., Yan, L., Zhao, X., et al. (2015). Phosphorylation of TGB1 by protein kinase CK2 promotes Barley stripe mosaic virus movement in monocots and dicots. J. Exp. Bot. 66, 4733–4747. doi: 10.1093/jxb/erv237
Ismayil, A., Haxim, Y., Wang, Y., Li, H., Qian, L., Han, T., et al. (2018). Cotton Leaf Curl Multan virus C4 protein suppresses both transcriptional and post-transcriptional gene silencing by interacting with SAM synthetase. PLoS Pathog. 14:e1007282. doi: 10.1371/journal.ppat.1007282
Johansen, L. K. (2001). Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126, 930–938. doi: 10.1104/pp.126.3.930
John, S., Margaret, I. B., and Davies, J. W. (2001). “Geminiviridae,” in eLS, eds Wiley Online Library (Hoboken, NJ: John Wiley & Sons, Ltd).
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Latham, J. R., Saunders, K., Pinner, M. S., and Stanley, J. (1997). Induction of plant cell division by Beet curly top virus gene C4. Plant J. 11, 1273–1283. doi: 10.1046/j.1365-313X.1997.11061273.x
Lefeuvre, P., and Moriones, E. (2015). Recombination as a motor of host switches and virus emergence: geminiviruses as case studies. Curr. Opin. Virol. 10, 14–19. doi: 10.1016/j.coviro.2014.12.005
Li, H., Zeng, R., Chen, Z., Liu, X., Cao, Z., Xie, Q., et al. (2018). S-acylation of a geminivirus C4 protein is essential for regulating the CLAVATA pathway in symptom determination. J. Exp. Bot. 69, 4459–4468. doi: 10.1093/jxb/ery228
Li, Y., Zhu, J., Tian, G., Li, N., Li, Q., Ye, M., et al. (2010). The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 8:e1000533. doi: 10.1371/journal.pbio.1000533
Lima, A. T. M., Sobrinho, R. R., Gonzalez-Aguilera, J., Rocha, C. S., Silva, S. J. C., Xavier, C. A. D., et al. (2012). Synonymous site variation due to recombination explains higher genetic variability in begomovirus populations infecting non-cultivated hosts. J. Gen. Virol. 94(Pt 2), 418–431. doi: 10.1099/vir.0.047241-47240
Lu, R., Malcuit, I., Moffett, P., Ruiz, M. T., Peart, J., Wu, A. J., et al. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. doi: 10.1093/emboj/cdg546
Luna, A. P., Morilla, G., Voinnet, O., and Bejarano, E. R. (2012). Functional analysis of gene-silencing suppressors from tomato yellow leaf curl disease viruses. Mol. Plant Microbe Interact. 25, 1294–1306. doi: 10.1094/MPMI-04-12-0094-R
Ma, X. Y., Cai, J. H., Li, G. X., Qin, B. X., and Zhou, X. P. (2004). Molecular characterization of a distinct begomovirus infecting Euphorbia pulcherrima in China. J. Phytopathol. 152, 215–218. doi: 10.1111/j.1439-0434.2004.00832.x
Mansoor, S., Zafar, Y., and Briddon, R. W. (2006). Geminivirus disease complexes: the threat is spreading. Trends Plant Sci. 11, 209–212. doi: 10.1016/j.tplants.2006.03.003
Martin, D. P., Biagini, P., Lefeuvre, P., Golden, M., Roumagnac, P., and Varsani, A. (2011). Recombination in eukaryotic single stranded DNA viruses. Viruses 3, 1699–1738. doi: 10.3390/v3091699
Mei, Y., Wang, Y., Hu, T., Yang, X., Lozano-Duran, R., Sunter, G., et al. (2018a). Nucleocytoplasmic shuttling of geminivirus C4 protein mediated by phosphorylation and myristoylation is critical for viral pathogenicity. Mol Plant 11, 1466–1481. doi: 10.1016/j.molp.2018.10.004
Mei, Y., Yang, X., Huang, C., Zhang, X., and Zhou, X. (2018b). Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKη-mediated phosphorylation in Nicotiana benthamiana. PLoS Pathog. 14:e1006789. doi: 10.1371/journal.ppat.1006789
Mills-Lujan, K., Andrews, D. L., Chou, C. W., and Deom, C. M. (2015). The roles of phosphorylation and SHAGGY-like protein kinases in geminivirus C4 protein induced hyperplasia. PLoS One 10:e0122356. doi: 10.1371/journal.pone.0122356
Mills-Lujan, K., and Deom, C. M. (2010). Geminivirus C4 protein alters Arabidopsis development. Protoplasma 239, 95–110. doi: 10.1007/s00709-009-0086-z
Monci, F., Sánchez-Campos, S., Navas-Castillo, J., and Moriones, E. (2002). A natural recombinant between the geminiviruses Tomato yellow leaf curl Sardinia virus and Tomato yellow leaf curl virus exhibits a novel pathogenic phenotype and is becoming prevalent in Spanish populations. Virology 303, 317–326. doi: 10.1006/viro.2002.1633
Navas-Castillo, J., Fiallo-Olive, E., and Sanchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Nikitin, P. A., and Luftig, M. A. (2012). The DNA damage response in viral-induced cellular transformation. Br. J. Cancer 106, 429–435. doi: 10.1038/bjc.2011.612
Padidam, M., Sawyer, S., and Fauquet, C. M. (1999). Possible emergence of new geminiviruses by frequent recombination. Virology 265, 218–225. doi: 10.1006/viro.1999.0056
Pan, X., Gong, D., Nguyen, D. N., Zhang, X., Hu, Q., Lu, H., et al. (2018). Early microbial colonization affects DNA methylation of genes related to intestinal immunity and metabolism in preterm pigs. DNA Res. 25, 287–296. doi: 10.1093/dnares/dsy001
Perefarres, F., Thierry, M., Becker, N., Lefeuvre, P., Reynaud, B., Delatte, H., et al. (2012). Biological invasions of geminiviruses: case study of TYLCV and Bemisia tabaci in Reunion Island. Viruses 4, 3665–3688. doi: 10.3390/v4123665
Piroux, N., Saunders, K., Page, A., and Stanley, J. (2007). Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKη, a component of the brassinosteroid signalling pathway. Virology 362, 428–440. doi: 10.1016/j.virol.2006.12.034
Raja, P., Wolf, J. N., and Bisaro, D. M. (2010). RNA silencing directed against geminiviruses: post-transcriptional and epigenetic components. Biochim. Biophys. Acta 1799, 337–351. doi: 10.1016/j.bbagrm.2010.01.004
Rey, M. E. C., Ndunguru, J., Berrie, L. C., Paximadis, M., Berry, S., Cossa, N., et al. (2012). Diversity of dicotyledenous-infecting geminiviruses and their associated DNA molecules in Southern Africa, including the south-west Indian ocean islands. Viruses 4, 1753–1791. doi: 10.3390/v4091753
Rojas, M. R., Hagen, C., Lucas, W. J., and Gilbertson, R. L. (2005). Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43, 361–394. doi: 10.1146/annurev.phyto.43.040204.135939
Rosas-Diaz, T., Zhang, D., Fan, P., Wang, L., Ding, X., Jiang, Y., et al. (2018). A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl. Acad. Sci. U.S.A. 115, 1388–1393. doi: 10.1073/pnas.1715556115
Rubio, V., Shen, Y., Saijo, Y., Liu, Y., Gusmaroli, G., Dinesh-Kumar, S. P., et al. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41, 767–778. doi: 10.1111/j.1365-313X.2004.02328.x
Sattar, M. N., Kvarnheden, A., Saeed, M., and Briddon, R. W. (2013). Cotton leaf curl disease - an emerging threat to cotton production worldwide. J. Gen. Virol. 94(Pt 4), 695–710. doi: 10.1099/vir.0.049627-49620
Stanley, J., and Latham, J. R. (1992). A symptom variant of beet curly top geminivirus produced by mutation of open reading frame C4. Virology 190, 506–509. doi: 10.1016/0042-6822(92)91243-n
Tang, Y. F., Du, Z. G., He, Z. F., Brown, J. K., and She, X. M. (2014). Identification and molecular characterization of two begomoviruses from Pouzolzia zeylanica (L.) Benn. exhibiting yellow mosaic symptoms in adjacent regions of China and Vietnam. Arch. Virol. 159, 2799–2803. doi: 10.1007/s00705-014-2049-2048
Teng, K., Chen, H., Lai, J., Zhang, Z., Fang, Y., Xia, R., et al. (2010). Involvement of C4 protein of Beet severe curly top virus (family Geminiviridae) in virus movement. PLoS One 5:e11280. doi: 10.1371/journal.pone.0011280
Vanitharani, R., Chellappan, P., Pita, J. S., and Fauquet, C. M. (2004). Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 78, 9487–9498. doi: 10.1128/JVI.78.17.9487-9498.2004
Varma, A., and Malathi, V. G. (2003). Emerging geminivirus problems: a serious threat to crop production. Ann. Appl. Biol. 142, 145–164. doi: 10.1111/j.1744-7348.2003.tb00240.x
Varsani, A., Shepherd, D. N., Monjane, A. L., Owor, B. E., Erdmann, J. B., Rybicki, E. P., et al. (2008). Recombination, decreased host specificity and increased mobility may have driven the emergence of maize streak virus as an agricultural pathogen. J. Gen. Virol. 89(Pt 9), 2063–2074. doi: 10.1099/vir.0.2008/003590-3590
Voinnet, O., and Baulcombe, D. C. (1997). Systemic signalling in gene silencing. Nature 389, 553–555. doi: 10.1038/39215
Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/j.1365-313X.2004.02219.x
Wyant, P., Strohmeier, S., Fischer, A., Schafer, B., Briddon, R. W., Krenz, B., et al. (2015). Light-dependent segregation of begomoviruses in Asystasia gangetica leaves. Virus Res. 195, 225–235. doi: 10.1016/j.virusres.2014.10.024
Xie, Y., Zhao, L., Jiao, X., Jiang, T., Gong, H., Wang, B., et al. (2013). A recombinant begomovirus resulting from exchange of the C4 gene. J. Gen. Virol. 94(Pt 8), 1896–1907. doi: 10.1099/vir.0.053181-53180
Yang, X., Guo, W., Li, F., Sunter, G., and Zhou, X. (2019). Geminivirus-associated betasatellites: exploiting chinks in the antiviral arsenal of plants. Trends Plant Sci. 24, 519–529. doi: 10.1016/j.tplants.2019.03.010
Yang, X., Xie, Y., Raja, P., Li, S., Wolf, J. N., Shen, Q., et al. (2011). Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 7:e1002329. doi: 10.1371/journal.ppat.1002329
Zerbini, F. M., Briddon, R. W., Idris, A., Martin, D. P., Moriones, E., Navas-Castillo, J., et al. (2017). ICTV virus taxonomy profile: geminiviridae. J. Gen. Virol. 98, 131–133. doi: 10.1099/jgv.0.000738
Zhang, K., Zhang, Y., Yang, M., Liu, S., Li, Z., Wang, X., et al. (2017). The Barley stripe mosaic virus γb protein promotes chloroplast-targeted replication by enhancing unwinding of RNA duplexes. PLoS Pathog. 13:e1006319. doi: 10.1371/journal.ppat.1006319
Zhang, X., Dong, K., Xu, K., Zhang, K., Jin, X., Yang, M., et al. (2018). Barley stripe mosaic virus infection requires PKA-mediated phosphorylation of γb for suppression of both RNA silencing and the host cell death response. New Phytol. 218, 1570–1585. doi: 10.1111/nph.15065
Keywords: Tomato yellow leaf curl Guangdong virus, C4, pathogenic determinant, PTGS, TGS, BAM1
Citation: Li Z, Du Z, Tang Y, She X, Wang X, Zhu Y, Yu L, Lan G and He Z (2020) C4, the Pathogenic Determinant of Tomato Leaf Curl Guangdong Virus, May Suppress Post-transcriptional Gene Silencing by Interacting With BAM1 Protein. Front. Microbiol. 11:851. doi: 10.3389/fmicb.2020.00851
Received: 12 December 2019; Accepted: 09 April 2020;
Published: 05 May 2020.
Edited by:
Rosa Lozano-Durán, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Pablo A. Manavella, CONICET Santa Fe, ArgentinaCopyright © 2020 Li, Du, Tang, She, Wang, Zhu, Yu, Lan and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zifu He, aGV6ZkBnZHBwcmkuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.