- 1Division of Life Sciences, University of Northampton, Northampton, United Kingdom
- 2Healthcare Infection Society, London, United Kingdom
Lipooligosaccharide (LOS) is an integral component of the Campylobacter cell membrane with a structure of core oligosaccharides forming inner and outer core regions and a lipid A moiety. The gene content of the LOS core biosynthesis cluster exhibits extensive sequence variation, which leads to the production of variable cell surface LOS structures in Campylobacter. Some LOS outer core molecules in Campylobacter jejuni are molecular mimics of host structures (such as neuronal gangliosides) and are thought to trigger neuronal disorders (particularly Guillain–Barré syndrome and Miller Fisher syndrome) in humans. The extensive genetic variation in the LOS biosynthesis gene cluster, a majority of which occurs in the LOS outer core biosynthesis gene content present between lgtF and waaV, has led to the development of a classification system with 23 classes (A–W) and four groups (1–4) for the C. jejuni LOS region. This review presents an updated and simplified classification system for LOS typing alongside an overview of the frequency of C. jejuni LOS biosynthesis genotypes and structures in various C. jejuni populations.
Introduction
Campylobacter is a foodborne enteropathogen, which causes an acute, self-limiting gastroenteritis in humans with various non-specific symptoms including watery or bloody diarrhea, abdominal pain, headache, fever, chills, and dysentery (van Spreeuwel et al., 1985; Black et al., 1988; Perkins and Newstead, 1994). The annual estimated number for Campylobacter infection is 96 million worldwide (Havelaar et al., 2015). Campylobacter infection occurs in adults and children in developing countries and can also lead to death in young children (Janssen et al., 2008). In some cases, there can also be long-term, postinfection consequences of Campylobacter infection such as the neuronal disorders Guillain–Barré syndrome (GBS) and Miller Fisher syndrome (MFS), Reiter’s arthritis, and irritable bowel syndrome (IBS; Endtz et al., 2000; McCarthy and Giesecke, 2001). Cohort studies on confirmed Campylobacter cases estimate that GBS develops following infection in anywhere from 21 to 172 per 100,000 Campylobacter cases and that this rate is approximately 100-fold higher than in the general population (McCarthy and Giesecke, 2001; Tam et al., 2006; Scallan Walter et al., 2019). Case–control studies on patients diagnosed with GBS repeatedly show a significant association with Campylobacter, with an average rate of infection of 35.4% compared with 4.4% in controls (Poropatich et al., 2010). A recent systematic review of all factors contributing to the development of GBS concluded that Campylobacter infection was the most common trigger of the disease with a substantial evidence base (Wachira et al., 2019).
Campylobacter species, similar to Neisseria and Haemophilus, lack lipopolysaccharide (LPS) in the outer-cell membrane and instead possess lipooligosaccharide (LOS), which comprises of lipid A and core structures (Mandrell et al., 1992; Moran, 1997; Duncan et al., 2009). In comparison to LPS, LOS are low-molecular weight biological molecules lacking O chains (Moran, 1997). Other Campylobacter cell-surface structures include capsular polysaccharides (CPS), O-linked glycosylated flagellum, and N-linked glycoproteins. LOS, CPS, and O-linked glycans (mainly flagellar glycans) are variable among different strains, while N-linked glycoproteins remain conserved (Szymanski et al., 2003; Karlyshev et al., 2005; Day et al., 2012). The glycome that comprises these four types of carbohydrate containing conjugate molecules is synthesized by more than 8% of the genome in Campylobacter jejuni 11168 (Parkhill et al., 2000; Gundogdu et al., 2007).
No commercial vaccine has been developed for Campylobacter to date, and this is largely due to the versatile and diverse nature of Campylobacter physiology and genomics (Riddle and Guerry, 2016). Subunit vaccines formed with flagellum-secreted proteins (C. jejuni 81-176 FlaC, C. jejuni 81-176 FspA1, and C. jejuni CG8486 FspA2) and recombinant protein (ACE 393) have been experimentally tested in mice and healthy volunteers, respectively, but no promising candidates for human vaccines have been identified (Baqar et al., 2008; Poly et al., 2019). Glycoconjugate vaccines such as C. jejuni 81–176 conjugated CPS vaccine (CRM-197) has been tested in monkeys but remained unsuccessful, as it did not provide adequate immunity (Monteiro et al., 2009). A conjugated LOS vaccine has not been investigated yet for C. jejuni; however, LOS of two C. jejuni strains BH-01-0142 and CG8421 may be considered and utilized for vaccine development (Poly et al., 2019). LOS functions as a virulence determinant, immune modulator, and essential survival element, which make it a potential glycoconjugate vaccine candidate. However, prevalence of diverse LOS genotypes (due to variation within the gene content of LOS biosynthesis gene cluster) and presence of phase variation (a phenomenon where gene on/off switching varies the cell-surface LOS structures and functions) within the LOS biosynthesis genes are the two main features that make LOS less desirable as a vaccine candidate (Guerry et al., 2002; Gilbert et al., 2002; Parker et al., 2005, 2008). Furthermore, it is unclear how LOS genotypes are reflected in overall LOS biosynthetic structures. The prevalence of LOS genotypes vary from geographic region to region, while phase variation varies from strain to strain, and both types of LOS locus variation need to be investigated for vaccine design. The most prevalent LOS genotypes circulating regionally must be taken into account for maximal efficiency of LOS-conjugated vaccine, and therefore, the frequency of C. jejuni LOS genotypes in different countries including United States, United Kingdom, Netherlands, France, Belgium, Finland, Sweden, Japan, and Bangladesh has been investigated previously (Godschalk et al., 2004; Parker et al., 2005; Quiñones et al., 2007; Habib et al., 2009; Ellström et al., 2013, 2014, 2016; Islam et al., 2014, 2018; Ohishi et al., 2017; Elhadidy et al., 2018; Thépault et al., 2018). In this review, data from the literature relevant to the distribution of C. jejuni LOS locus genotypes in various geographical areas of the world will be analyzed to present an up-to-date picture of C. jejuni LOS genotype predominance, which may be important in vaccine design. This review also presents an updated and simplified classification system for the LOS biosynthesis locus to aid fellow researchers investigating increasingly complex levels of LOS variation and the role it plays in Campylobacter infection.
Campylobacter Los as a Virulence Determinant
LOS in Campylobacter does not only maintain the integrity of the cell membrane structure but also acts as a barrier for those molecules, which are transported through the cell membrane (Karlyshev et al., 2005). Deletion of LOS core in C. jejuni 11168 does not seem essential for viability (Marsden et al., 2009), but truncation of LOS can be lethal in C. jejuni strains other than 11168 (Phongsisay et al., 2007). For example, antibiotic permeability into the cell increases due to alteration in LOS structures, possibly because LOS structural changes decrease the cell membrane hydrophobicity. This is the reason that mutants of C. jejuni LOS genes are highly susceptible to some antibiotics, specifically to erythromycin (Kanipes et al., 2004; Jeon et al., 2009; Marsden et al., 2009). In addition to providing a barrier to antibiotics, LOS also confers resistance to Campylobacter cells against human serum proteins including α-defensins, cathelicidins, and bactericidal-/permeability-increasing proteins (Marsden et al., 2009; Keo et al., 2011). DNA uptake into a bacterial cell is an outer cell membrane-dependent process. Therefore, LOS modification in the outer cell membrane may also affect Campylobacter’s ability to uptake foreign DNA or its characteristic of natural transformation (Jeon et al., 2009; Marsden et al., 2009). Mutants of Campylobacter LOS genes, in comparison to their respective wild-type (WT) strains, have showed reduced adherence and invasion into host intestinal epithelial cells (Fry et al., 2000; Kanipes et al., 2004; Javed et al., 2012), which might be due to reduced interaction between host cell receptors and altered LOS structures. A caveat to these studies is that deletion of LOS biosynthesis genes or drastic changes in LOS structure may have a general destabilizing effect on the LOS. A mutant of C. jejuni 11168, lacking the core oligosaccharides in its LOS structures, was unable to invade Caco-2 cells, indicating the importance of LOS in Campylobacter invasion into host cells (Marsden et al., 2009). C. jejuni strains with sialylated LOS showed higher potential of adhesion, invasion, and translocation than those with non-sialylated LOS (Louwen et al., 2012). Two sialic acid biosynthesis genes (cgtB and wlaN) were found commonly present in highly invasive C. jejuni strains (Müller et al., 2007), and mutation of a sialic acid biosynthesis gene, cst-II, in a C. jejuni strain caused reduction in its invasion into epithelial cells (Louwen et al., 2008), supporting the role of LOS sialylation or sialic acid biosynthesis genes in C. jejuni invasion. However, C. jejuni mutants of other LOS biosynthesis genes (waaC and cj1136) also showed significant reduction in invasion into intestinal epithelial cells (Kanipes et al., 2008; Javed et al., 2012). Furthermore, only 23% of C. jejuni isolates from blood-borne infection or truly invasive strains contained sialic acid biosynthesis genes (Ellström et al., 2014). These studies indicate that not only sialic acid biosynthesis genes but also the overall presentation of LOS structure play an important role in adherence and invasion of C. jejuni into host cells. Complete cell-surface LOS structures in C. jejuni are also important for the optimum colonization of chick ceca, and this is linked to the increased hydrophobicity and susceptibility to bile of LOS mutants (Iwata et al., 2013). Thus, LOS is an important virulence determinant in C. jejuni. It may also be the case that since C. jejuni LOS are densely present on the cell surfaces and they are readily available to stimulate and interact with human immune cells such as macrophages, LOS could also contribute to the binding of other cell types. For example, the C. jejuni LOS terminal N-acetyl galactosamine residues bind to the human macrophage galactose-type lectin receptors (van Sorge et al., 2009). LOS containing sialic acid residues have particular significance for human disease due to their increased ability to bind to immune cells and their similarity to neuronal structures. C. jejuni LOS sialic acid residues bind to Toll-like receptor 4 (TLR-4) and sialoadhesin receptors present on the human macrophage cell surfaces (Klaas et al., 2012; Heikema et al., 2013; Stephenson et al., 2013). C. jejuni LOS sialic acid residues are also ligands of sialic acid binding immunoglobulin-like lectins present in human monocytes and natural killer cells (Avril et al., 2006). The LOS structures with variable epitopes presented on different C. jejuni cell surfaces can also mimic human neuronal gangliosides. For this reason, antibodies produced against the LOS structural epitopes do not only bind to LOS structures but also to human neuronal gangliosides. The cross-reactivity of anti-LOS antibodies with human gangliosides leads to the development of neuronal disorders (GBS and MFS) in humans (Yuki, 1997; Nachamkin et al., 1998; Endtz et al., 2000; McCarthy and Giesecke, 2001; Wakerley and Yuki, 2015). This is evident by the development of pathological changes in peripheral nerves and weakness in the limbs as well as production of anti-GM1 antibodies in rabbits upon sensitization with C. jejuni LOS (Yuki et al., 2004). Furthermore, knockout mutants of C. jejuni sialic acid biosynthesis genes (orf10 and cst-II) with truncated and non-sialylated LOS structures show reduced reactivity with GBS patient serum. In addition, administration of these mutated LOS structures into mice did not induce antiganglioside antibody responses (Godschalk et al., 2004). The allelic variation in the cst-II gene leads to the expression of either threonine (Thr) or asparagine (Asn) at position 51 of the sialyltransferase (Gilbert et al., 2002). This genetic polymorphism (and change in host-mimicking ganglioside epitopes) can further affect the development of autoimmune and clinical symptoms of GBS, supporting the role of LOS gene variations in GBS (Koga et al., 2005). In GBS, the cranial nerves extending from the brain to various areas of the head and neck are affected, which further develop difficulty in walking, muscle weakness, and muscle pain, while MFS, a variant of GBS, is characterized mainly by paralysis of eye muscles and problems with balance and coordination (Nachamkin et al., 1998). These postinfection complications are infrequent and typically appear in immune-compromised individuals, such as individuals with HIV infection (McCarthy and Giesecke, 2001; Janssen et al., 2008). They do not develop solely as a consequence of Campylobacter infection, and other bacterial or host-specific risk factors aid in stimulating the production of antiganglioside antibodies (Figure 1; Revez and Hänninen, 2012; Islam et al., 2014). This varies among C. jejuni strains and Campylobacter-infected individuals and contributes to a complex picture of GBS development and progression postinfection (Godschalk et al., 2007; Müller et al., 2007). Some C. jejuni strains do not produce ganglioside mimicking LOS structures at all despite the presence of sialic acid biosynthesis genes, and therefore, the presence of sialylated LOS biosynthesis genes do not always correspond with the C. jejuni potential for neural disease development (Houliston et al., 2011).
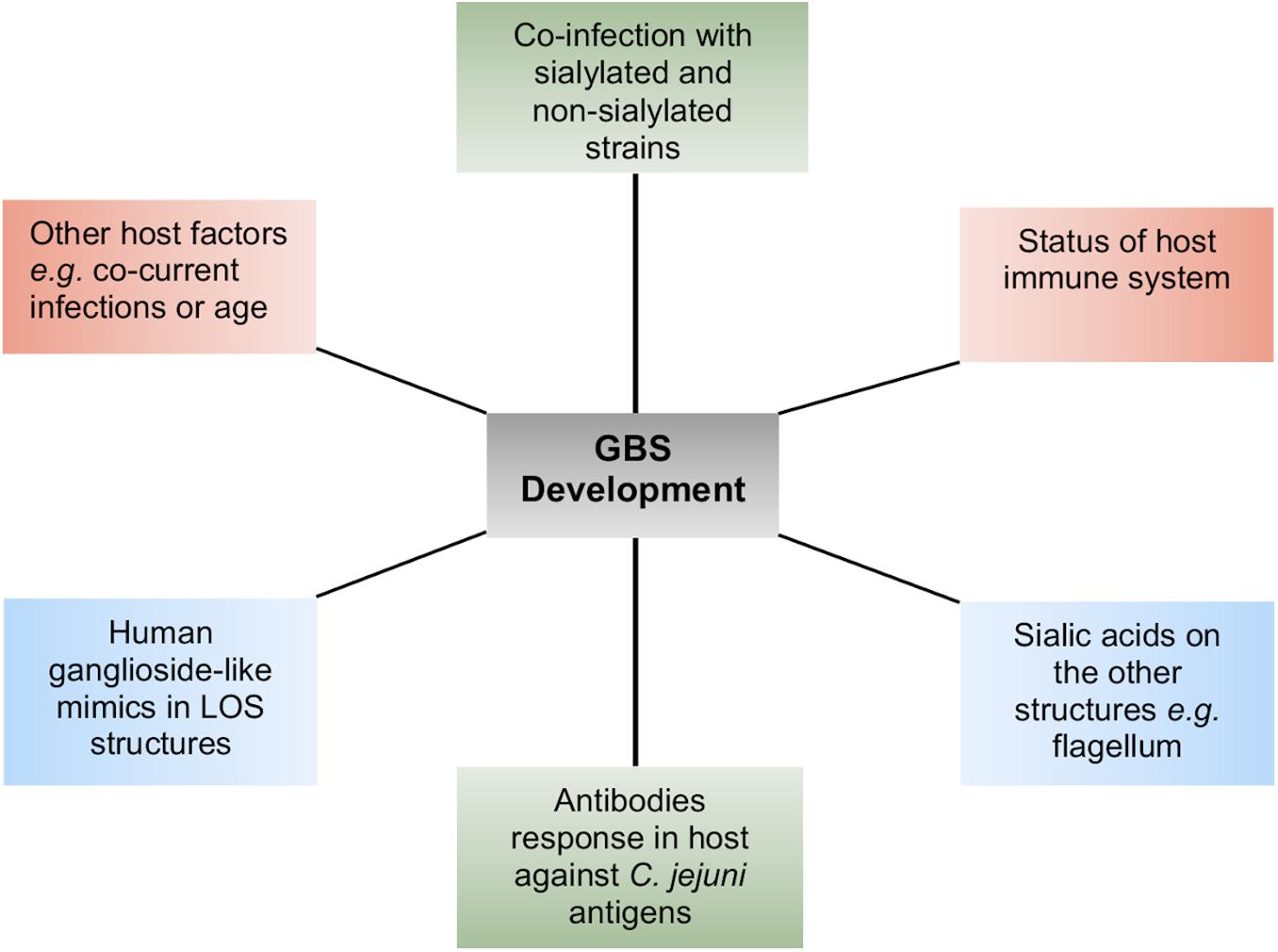
Figure 1. A summary of the different host–pathogen factors that contribute to the complexity of Guillain–Barré syndrome (GBS) development.
The Los Biosynthesis Locus in C. jejuni
In proposing a new classification for the LOS, it is important to appreciate the complexity of the LOS biosynthesis loci that has informed rationale for current and historic classification systems. The LOS lipid A backbone in C. jejuni contains a 3-diamino-2,3-dideoxy-D-glucopyranose linked to 2-amino-2-deoxy-D-glucose (GlcN), whereas the Campylobacter coli lipid A backbone consists of two GlcN (Culebro et al., 2016). The lipid A backbone in most of the Campylobacter strains is linked to six acyl chains (two hydroxyl-linked and four amide-linked; Moran, 1997). The LOS core biosynthesis in C. jejuni is achieved at the genetic level by a cluster of LOS biosynthesis genes (Figure 2). Each LOS biosynthesis gene produces an individual enzyme involved in either monosaccharide biosynthesis or addition of a particular monosaccharide to the LOS structure (Karlyshev et al., 2005; Parker et al., 2005, 2008; Iwata et al., 2013). The inner core of C. jejuni LOS has two heptose and two glucose units (Klena et al., 1998; Gilbert et al., 2002; Kanipes et al., 2004, 2006). The heptosyltransferase-I (waaC) adds the first heptose (Hep-I) to 3-deoxy-D-manno-octulosonic acid (KDO). Heptosyltransferase-II (waaF) catalyzes the addition of a second heptose (Hep-II) to Hep-I (Klena et al., 1998; Kanipes et al., 2004, 2006). In C. jejuni (strain 11168), Hep-1 and Hep-II are synthesized and added to the inner core of LOS by the phosphoheptose isomerase (gmhA), a D-glycero-beta-D-manno-heptose-7-phosphate kinase (waaE), an ADP-L-glycero-D-manno-heptose-6-epimerase (waaD), and a dephosphatase (gmhB; Karlyshev et al., 2005; Iwata et al., 2013). Unlike the inner core, the outer core of LOS varies extensively among C. jejuni strains (Linton et al., 2000; Godschalk et al., 2004; Houliston et al., 2011). The outer core of C. jejuni 11168 is synthesized by glycosyltransferases [cj1136 (orf4), cj1137 (orf14), and cj1138 (orf15)], N-acetyl galactosaminyl transferase [cgtA/neuA1 (orf5/10)], sialyltransferase [cst-III (orf7)], and galactosyltransferase (wlaN) and is illustrated as an example in Figure 2 (Gilbert et al., 2000, 2002; Linton et al., 2000; Guerry et al., 2002; Karlyshev et al., 2005; Javed et al., 2012). In addition to core synthesis, a LOS biosynthesis gene (waaM) is located in the cluster that encodes an enzyme (lipid A biosynthesis lauroyl acyltransferase) to catalyze the addition of a KDO molecule to the backbone of lipid A (Karlyshev et al., 2005). Campylobacter LOS structures are synthesized in the cytoplasmic side of the inner cell membrane from where they are flipped to the periplasmic side of the inner cell membrane and, finally, are integrated into the outer cell membrane (Whitfield and Trent, 2014; Simpson et al., 2015). Based on similarities to LPS assembly machinery in Escherichia coli, it is predicted that N-linked glycosylation glycosyltransferase (wlaM/pglG) and flippase (wlaB/pglK) can respectively facilitate cytoplasm-to-periplasm LOS flipping and periplasm-to-outer cell membrane LOS translocation in Campylobacter (Fry et al., 1998).
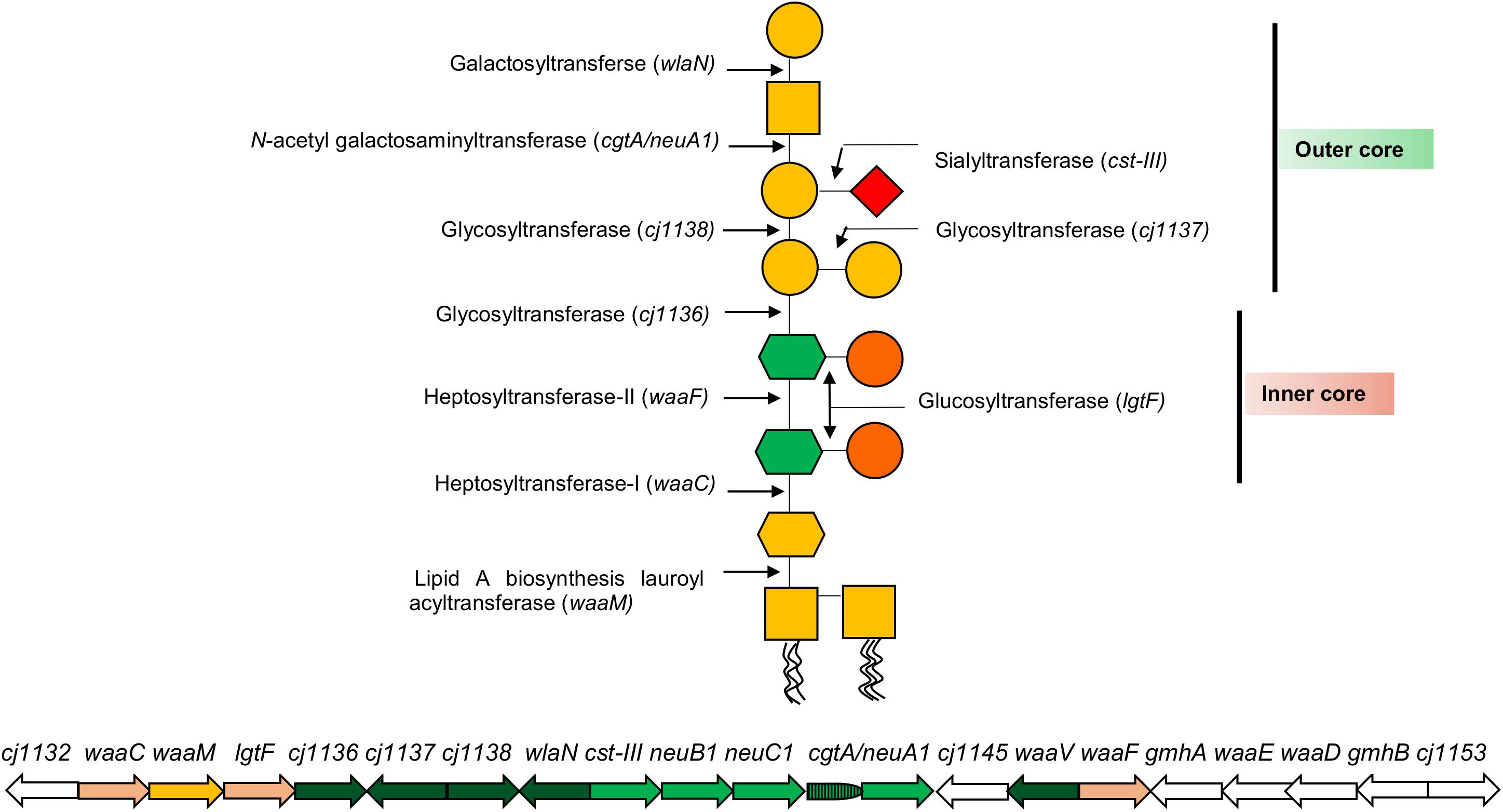
Figure 2. A representation of C. jejuni 11168 lipooligosaccharide (LOS) core biosynthesis gene cluster and its LOS structure. Each arrow represents an individual LOS core biosynthesis gene, and its direction indicates the direction of gene transcription. A LOS biosynthesis gene in yellow encodes an enzyme to catalyze the addition of a KDO molecule to lipid A. LOS biosynthesis genes in light pink encode enzymes for the synthesis of LOS inner core structure. LOS biosynthesis genes in green encode enzymes for the synthesis of LOS outer core structure, where LOS genes in light green (neuB1, neuC1, and cgtA/neuA1) synthesize sialic acid to incorporate into the outer core. LOS genes in white (gmhA, waaE, waaD, and gmhB) synthesize heptoses for inner core. Glycan structures were drawn according to the Symbol Nomenclature For Glycans (SNFG).
Variation in C. jejuni Los Biosynthesis Locus
The variation in the C. jejuni LOS biosynthesis gene region occurs either due to (i) mutations within the nucleotides of LOS biosynthesis gene sequences or (ii) the recombination between LOS biosynthesis gene/gene regions.
Variation at the Nucleotide Level
Nucleotide level variations within the LOS biosynthesis genes can occur due to phase variation, where slip strand mispairing during the replication of homopolymeric tracts can lead to insertions or deletions of single bases (Gilbert et al., 2002). The LOS gene wlaN in C. jejuni 11168, C. jejuni 331, and C. jejuni 2500 containing a homopolymeric tract of 8G produces the fully transcribed and functional gene product β-1,3-galactosyltransferase (Linton et al., 2000; Müller et al., 2007; Semchenko et al., 2012). A variant in these strains containing a 9G homopolymeric tract in wlaN results in a frameshift mutation and premature translational termination with a non-functional gene product, which cannot add the terminal galactose in the LOS structure and consequently converts a GM1-like LOS epitope into a GM2 mimic (Linton et al., 2000; Semchenko et al., 2012). Site-directed mutagenesis of the homopolymeric tract in C. jejuni 11168 wlaN from 8G to 11G increases the rate of phase variation ∼10-fold in this gene, and in general, the rate of phase variation increases with longer tract lengths in multiple genes (Bayliss et al., 2012). Phase variation of C. jejuni 11168 wlaN was not observed in vivo during colonization of chickens aged 2–4 weeks, but an increase in tract length from 8G to 9G to switch off expression of wlaN was detected after the passage of C. jejuni 224 and 331 in 5-day-old chicks, with C. jejuni 11168-O remaining unchanged (Bayliss et al., 2012; Semchenko et al., 2012). In the same study, C. jejuni 331 switched off wlaN after coculture with the intestinal cell line CaCo-2 and C. jejuni 224 switched off expression of the LOS genes Cj1144-45 after colonizing chicks, giving further evidence that strain- and host-specific factors can both influence phase variation of LOS genes (Semchenko et al., 2012). Phase variation in a number of other LOS biosynthesis genes has been observed at both the genotype and phenotype level in multiple strains including cst-II, cgtA, cgtD, orf23, and orf25 (Guerry et al., 2002; Parker et al., 2005; Godschalk et al., 2006, Godschalk et al., 2007; Houliston et al., 2011; Wanford et al., 2018). Multiple combinations of phase variable genes can also lead to novel LOS structures. Different combinations of on and off phenotypes in C. jejuni (strain GC149) are encoded by the cgtA and cgtD outer core glycosyltransferases and result in structural molecular mimics of either GD3 (cgtA off), GT1a (cgtA on/cgtD off), or ganglio/Pk (cgtA on/cgtD on) gangliosides (Houliston et al., 2011). Phase variation of LOS genes can therefore lead to mixed populations of LOS gene variants and increase the diversity of LOS structural epitopes within a single strain of Campylobacter (Guerry et al., 2002).
Sequence variation may also occur due to single nucleotide mutations, which can inactivate the LOS biosynthesis genes without involving the phenomenon of phase variation. For example, deletion of an A-base at position 1234 in lgtF (a LOS biosynthesis gene) alters the catalytic activity of its encoded enzyme, glycosyltransferase, in four C. jejuni strains (ATCC 43432, ATCC 43446, OH4382, and OH4384). As a result, the produced glycosyltransferase does not have the potential to catalyze the addition of ß-1,2-glucose to heptose-II during the LOS synthesis. Similarly, the base substitution of the final base in orf5/10 (cgtA/neuA1) in C. jejuni ATCC 43430 changes the amino acid (cysteine → tyrosine), which further leads to the production of a non-functional enzyme (Gilbert et al., 2002). The LOS gene, cgtA, with missing A-base at position 71 substitutes one amino acid in the cgtA-encoding enzyme, N-acetyl galactosaminyl transferase, which further leads to the inactivation of N-acetyl galactosaminyl transferase in C. jejuni OH4382 and OH4384 and truncates the LOS structure (Gilbert et al., 2002). Similarly, a five-base deletion from the cst-III gene of C. jejuni GB1 alters the number of amino acids (294→219) in sialyltransferase and eventually produces a non-sialylated LOS (Godschalk et al., 2007).
Variation at Allele or Gene Level and LOS Locus Classes in C. jejuni
In recent years, an alphabetical system of class organization for LOS genes within the C. jejuni LOS biosynthesis locus has been developed based on 23 C. jejuni LOS classes (A through W), which have been previously described (Gilbert et al., 2002; Parker et al., 2008; Richards et al., 2013). An insertion or deletion of a LOS biosynthesis gene or gene regions into the LOS locus can give rise to a different class type (Parker et al., 2005, 2008). Alterations of portions of the LOS biosynthesis genes or different alleles can also establish a new class or subclass; for example, allele variation in cgtA and wlaN genes generates A and B subclasses including A1, A2, B1, and B2 (Parker et al., 2005). In addition, disruption in resident LOS biosynthesis genes can also form a new class; for instance, disruption in class E orf26 establishes the LOS locus class P (Parker et al., 2005). The developed new locus type can be variable both in gene content and gene organization (Parker et al., 2005; Revez and Hänninen, 2012). C. jejuni acquires new genes in its LOS biosynthesis region by horizontal gene transfer. The horizontal transfer of LOS biosynthesis genes from C. jejuni O4 (GM1 strain) to C. jejuni 81116 (non-GM1 strain) changed it into a GM1-like LOS-producing strain (Phongsisay et al., 2006). Similarly, a C. jejuni GB11 strain possessing class C locus acquired a class A locus, identical to the LOS locus of C. jejuni ATCC 43446 while retaining the same sequence in the remainder of the genome (Gilbert et al., 2004).
Variation in LOS biosynthesis gene alleles causes alterations in the LOS structure. For instance, two cst-II gene alleles lead to the expression of either threonine (Thr) or asparagine (Asn) at position 51 of the translated enzyme. As a result, the enzyme retains either a monofunctional (Thr → 2,3-sialyltransferase activity) or a bifunctional (Asn → 2,3- and 2,8-sialyltransferase) activity and produces LOS with one and two sialic acids, respectively (Gilbert et al., 2002). Variation in LOS locus gene content can vary the carbohydrate content, linkages between the carbohydrate units, and core length in cell-surface LOS structures (Gilbert et al., 2002; Guerry et al., 2002). Variation in LOS locus gene content as well as in its gene organization varies the cell-surface LOS structurally and functionally. It is not always the case that LOS structures belonging to the same LOS locus type encode similar epitopes. C. jejuni 11168 and 520, both belong to class C, but C. jejuni 520 can produce a wider variety of human ganglioside mimics than C. jejuni 11168 (Semchenko et al., 2012). C. jejuni strains that contain a type A LOS locus frequently encode and express human ganglioside mimics on bacterial cell surfaces, which include GM1a, GM1b, GD1a, and GD1b (Nachamkin et al., 2002; Godschalk et al., 2004; Mortensen et al., 2009). For example, there is a GM1-like mimic in C. jejuni 11168 (class C), a GQ1b-like mimic in C. jejuni 81-176 (Class B), a Lewis type I-like mimic in C. jejuni RM1503 (class M), and a paragloboside/Pk-like antigens in C. jejuni RM1221 (class F; Godschalk et al., 2004; Mortensen et al., 2009; Houliston et al., 2011). C. jejuni GC149 (class R) contains sialic acid biosynthesis genes and may present ganglioside-like mimics (GT1a, GD3) as well as a hybrid form of ganglio and P-type antigens (Parker et al., 2008; Houliston et al., 2011). Other LOS classes such as D and E also possess human ganglioside-like LOS structures, but these are different to GM1, GD1, and GQ1b (Godschalk et al., 2004). Class P LOS have a lack of sialic acid and possess N-acetyl quinovosamine instead (Poly et al., 2008). The variable LOS structural epitopes presented by different C. jejuni LOS locus types are demonstrated in Table 1.
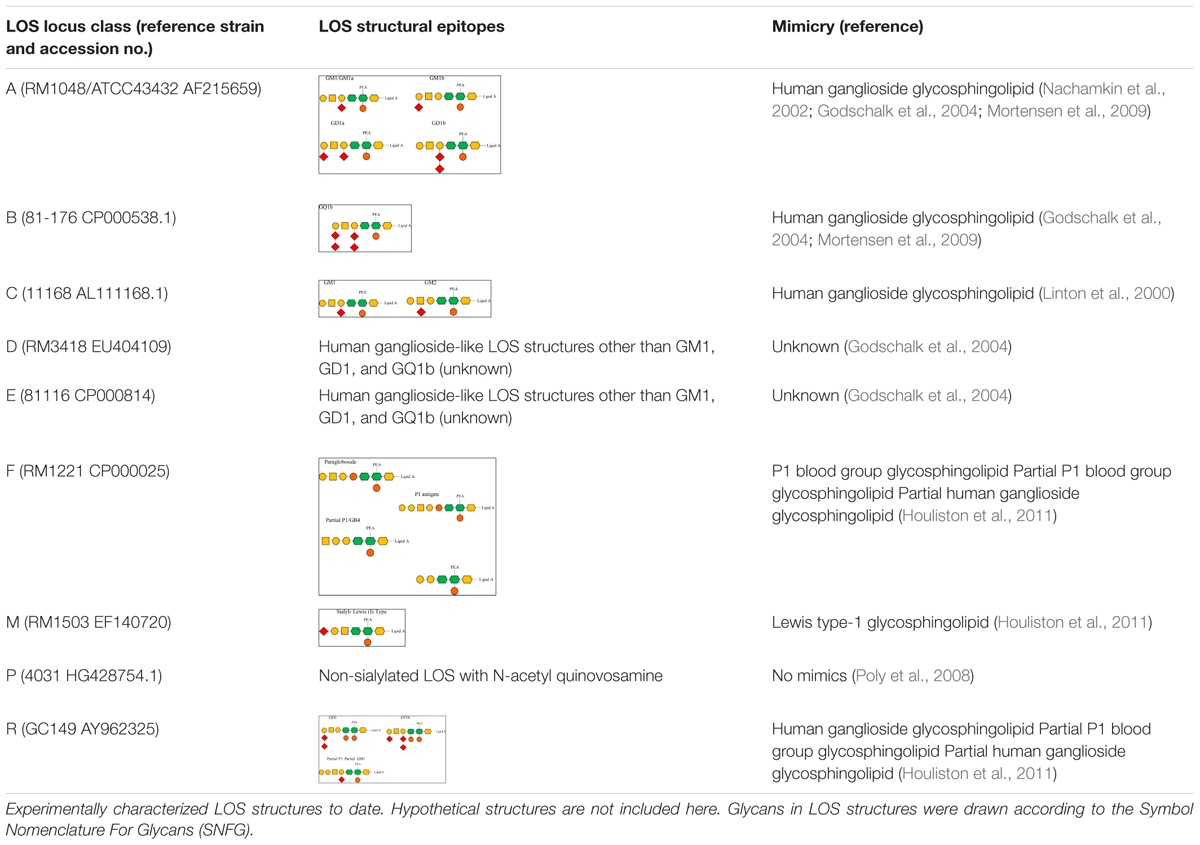
Table 1. Variable lipooligosaccharide (LOS) structures synthesized by different C. jejuni LOS locus types.
The expression of variable cell surface LOS structures and mimicry with human blood antigen glycosphingolipids or neuronal ganglioside glycosphingolipids as a consequence of gene variation in the LOS locus in C. jejuni is an important virulence factor and may have a direct link to the progression of specific neuronal disorders postinfection (Ang et al., 2002; Müller et al., 2007; Houliston et al., 2011; Semchenko et al., 2012). For example, C. jejuni strains with LOS locus class A and variable human ganglioside mimics (GM1a, GM1b, GD1a, and GD1b) trigger GBS in Campylobacter-infected patients (Nachamkin et al., 2002; Godschalk et al., 2004, 2007; Mortensen et al., 2009), whereas C. jejuni strains with LOS class B and corresponding GQ1b-like LOS structures are likely to develop MFS in Campylobacter-infected patients (Godschalk et al., 2007; Islam et al., 2014). Genetic diversity within the LOS locus plays an important role in the development of postinfection effects; however, this does not have any association with acute-phase symptoms such as diarrhea or abdominal pain (Poly et al., 2008; Mortensen et al., 2009; Ellström et al., 2013). This indicates that LOS sialylation is not required for human diarrheal disease and that both the sialylated and non-sialylated LOS can be used for vaccine design (Poly et al., 2008, 2019).
Simplification of C. jejuni Los Locus Classification
LOS classes A–H were initially described (Gilbert et al., 2002; Parker et al., 2005), and these known C. jejuni LOS classes were then primarily categorized into four groups and included LOS classes A, B, and C belonging to a group 1, LOS class E in group 2, LOS classes D and F in group 3, and LOS class G in group 4 (Karlyshev et al., 2005). Later, Parker et al. (2008) identified 11 more C. jejuni LOS classes including I–S. Subsequently, Richards et al. (2013) identified C. jejuni strains with novel LOS loci and established four more LOS classes including T, U, V, and W. The novel LOS loci identified in the latter two studies have never been assigned to the LOS groups. To better understand the prevalence of C. jejuni LOS groups and groups related to LOS classes, we propose a simplified LOS classification system (Figure 3) where various already known LOS classes have been assigned into the pre-established LOS groups (Karlyshev et al., 2005) on the basis of sharing similar LOS biosynthesis gene content. Group 1 includes all the LOS locus types (A, B, C, R, M, and V), which contain genes for sialic acid synthesis and translocation (orf7/cst-II/cst-III, orf8/neuB1, orf9/neuC1, and orf10/neuA1), whereas the other three groups have LOS loci with no sialic acid biosynthesis genes. Based on sequence similarity of LOS loci H, O, P, and W to locus E (orf21–orf34), these four classes are now assigned to group 2. Furthermore, K, Q, N, I, J, and S sharing orf17, orf18/cgtH, orf19/cgtG, and orf20/cgtE are assigned to LOS group 3, and L, G, T, and U sharing orf36, orf37, and orf38 are assigned to LOS group 4 classes.
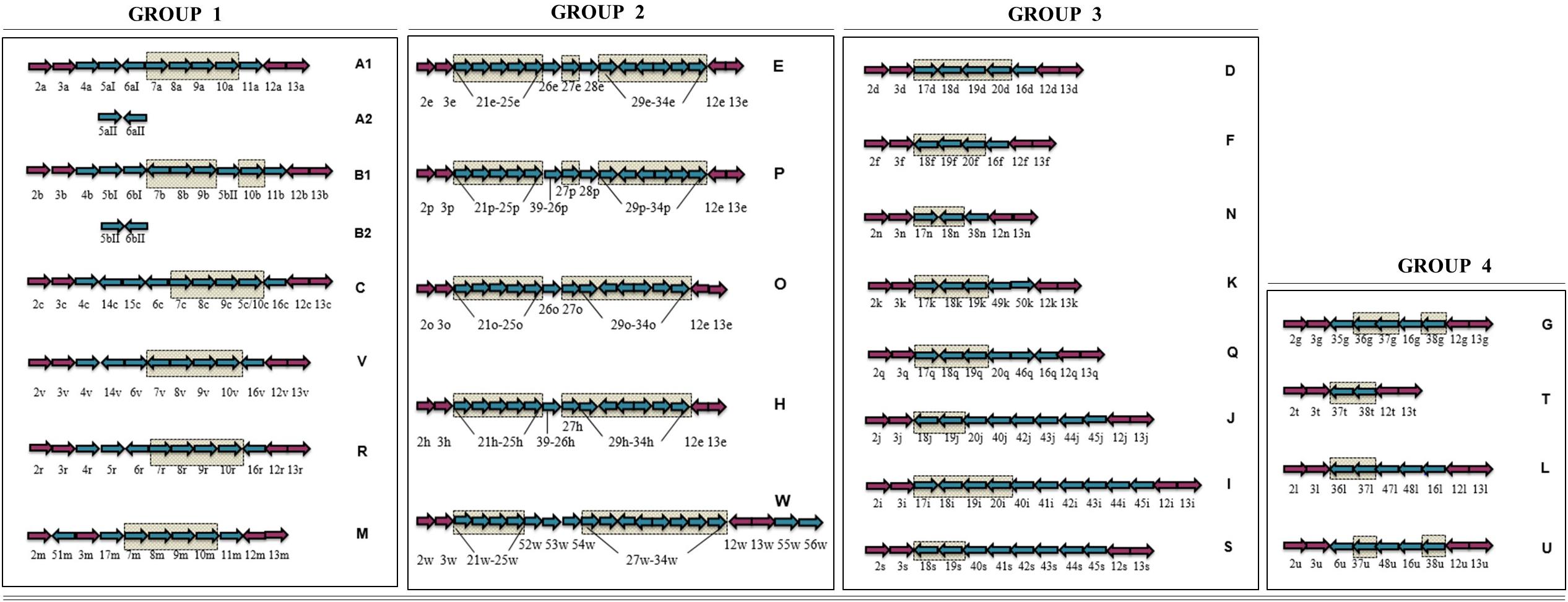
Figure 3. Simplified C. jejuni lipooligosaccharide (LOS) locus classification system. LOS classes are classified into the previously established four groups on the basis of sharing similar LOS biosynthesis gene content. Arrows in checked boxes: LOS biosynthesis gene content, shared between the classes within a LOS group; Blue arrows: Variable LOS biosynthesis genes, located between lgtF (orf3) and waaV (orf12); pink arrows: LOS biosynthesis genes, commonly present in all LOS classes. Genes are numbered according to the Parker et al. (2005) numbering system. Arrow direction represents the direction of gene transcription.
Prevalence of C. jejuni Los Locus Classes and Groups
A large number of available C. jejuni genomes with metadata have been deposited in recent years and have the potential to provide a full and comprehensive overview of the frequency of C. jejuni LOS genotypes in C. jejuni populations (from different isolation sources and various clonal complexes). However, much of these data remain unpublished and should be a focus of ongoing efforts. However, analysis of previously published studies examining the frequencies of C. jejuni LOS locus classes and groups present in enteritis, GBS, blood borne infection, and poultry-associated C. jejuni populations indicates that the hierarchy of LOS group (group 1 > group 2 > group 3 > group 4) is largely conserved among human- and poultry-derived C. jejuni isolates (Figure 4). This also indicates that the B and C LOS classes in clinical enteric disease and LOS class A in GBS-associated C. jejuni populations are found to be highly predominant. In comparison to the high prevalence of LOS class C (42%) in clinical isolates in Sweden (Ellström et al., 2016), a very small number of clinical strains (2%) in Bangladesh had association with LOS locus C (Islam et al., 2018), suggesting that C. jejuni LOS class distribution may vary geographically. Furthermore, when comparing the combined frequency of LOS ABC types in different populations of C. jejuni isolates (clinical, enteritis, and poultry), approximately 50–75% of strains in all C. jejuni populations belong to LOS classes A, B, or C. The only exception to these results was the data from Ellström et al. (2014) where C. jejuni were isolated from human blood-borne infections.
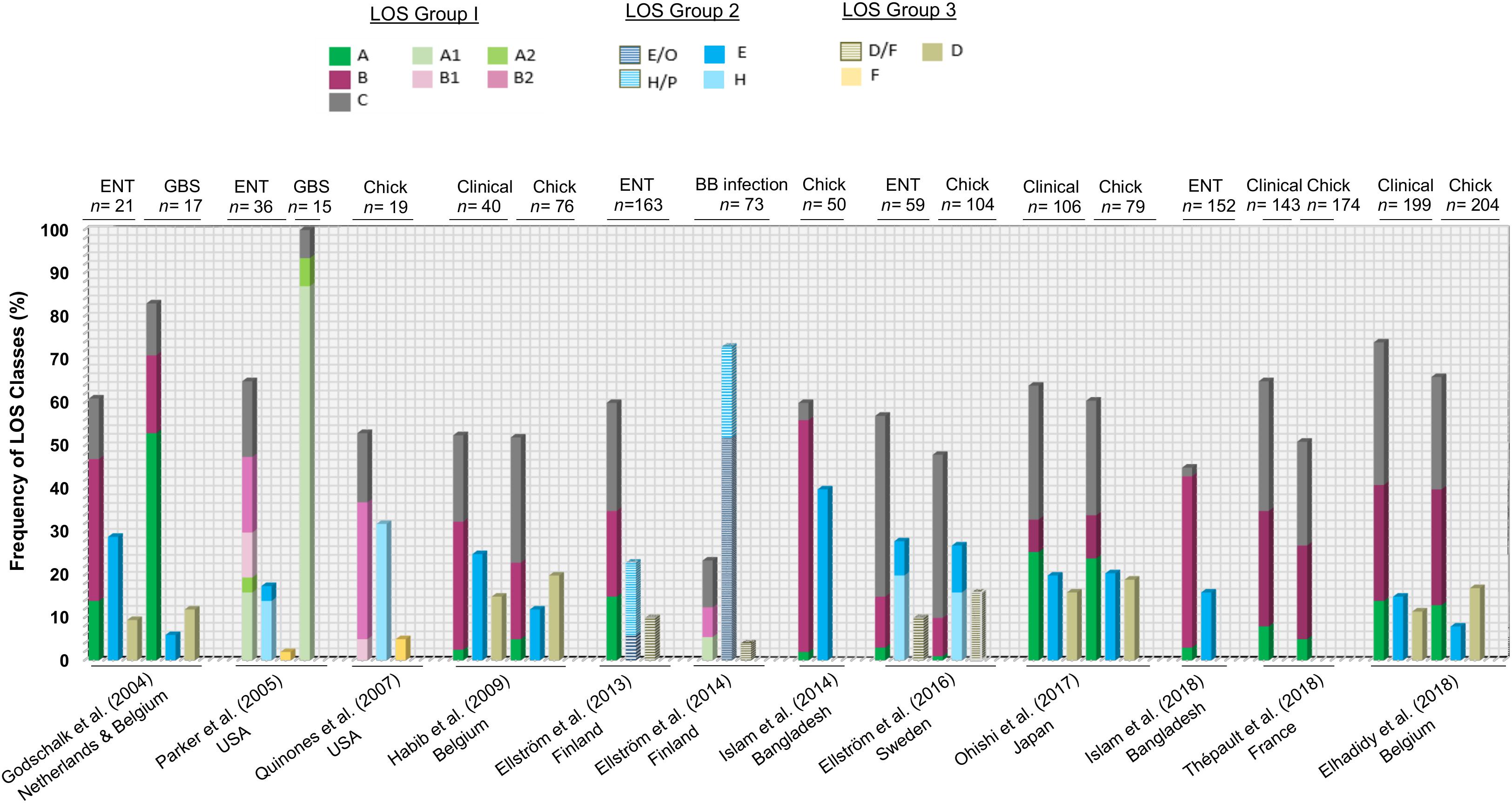
Figure 4. A comparison of previous findings of distribution of C. jejuni lipooligosaccharide (LOS) locus genotypes in various C. jejuni populations. ENT, C. jejuni isolates from enteritis patients; GBS, C. jejuni isolates from GBS patients; chick, C. jejuni isolates from chicken; BB infection, C. jejuni isolates from patients with blood-borne infections.
A definite correlation between C. jejuni LOS locus class prevalence and sequence type (ST) distribution has not been established yet due to the diverse population structure of C. jejuni (Habib et al., 2009; Islam et al., 2014). A few studies have shown concordance between specific STs and LOS classes. For example, LOS class B possessing GBS (50%) and enteritis (25%) C. jejuni isolates had ST-403 CC (Islam et al., 2009). Another study assigned class B diarrheal C. jejuni strains (3%) with ST-206 CC (Habib et al., 2009). LOS class C was associated with ST-21 CC in 14% of C. jejuni enteritis isolates (Revez and Hänninen, 2012), 11.2% of C. jejuni bacteremia isolates (Ellström et al., 2013), and 3.2% of diarrheal C. jejuni isolates (Habib et al., 2009). High frequency of LOS locus class C may be a contributor to the high predominance of clinical C. jejuni strains with ST-21 CC (Habib et al., 2009; Thépault et al., 2018). This ST-21 CC has also been found in LOS class A positive bacteremia (2.8%) and diarrheal C. jejuni (0.4%) isolates (Habib et al., 2009; Ellström et al., 2013; Ohishi et al., 2017), which might be due to the close phylogenetic relationship between C. jejuni isolates with LOS classes A and C (Gilbert et al., 2008). LOS group 2 classes (E, H, O, and P) are associated with ST-677 CC and ST-45 CC in C. jejuni bacteremia isolates (Ellström et al., 2013). However, another study found two other STs (ST-353 CC and ST-443 CC) for LOS group 2-related diarrheal C. jejuni strains in addition to ST-45 CC (Habib et al., 2009). Group 3 LOS class D have diarrheal C. jejuni strains (5%) that were assigned with ST-354 CC (Habib et al., 2009).
Concluding Remarks
This review extends the C. jejuni LOS locus classification system. Currently, genomic-based classification of the LOS region is incomplete and vague. By providing a more refined classification system, investigators will be more readily able to link genomic class to LOS biosynthetic structures as they become available. Full LOS structural characterization is currently limited, so it is hard to determine whether locus classes will readily align with LOS structures and is clearly a focus for future research and may aid vaccine design. We also provide an overview of the frequency of C. jejuni LOS genotypes in C. jejuni populations originated from different sources and reviews the association between C. jejuni LOS locus genotypes and different human ganglioside-mimicking sialylated LOS structures. This review summarizes the various contributing factors in GBS development post-Campylobacter infection and shows that LOS group 1 containing LOS locus classes A, B, and C are commonly present in almost every type of C. jejuni population studied to date, regardless of its originating source.
Author Contributions
AH wrote the first draft of the manuscript and prepared all figures. LM and AW made substantial and intellectual contributions to the work. All authors reviewed and/or edited the manuscript prior to submission and have approved the final version of manuscript as submitted.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ang, C. W., Noordzij, P. G., De Klerk, M. A., Endtz, H. P., Van Doorn, P. A., and Laman, J. D. (2002). Ganglioside Mimicry of Campylobacter jejuni lipopolysaccharides determines antiganglioside specificity in rabbits. Infect. Immun. 70, 5081–5085. doi: 10.1128/IAI.70.9.5081
Avril, T., Wagner, E. R., Willison, H. J., and Crocker, P. R. (2006). Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect. Immun. 74, 4133–4141. doi: 10.1128/IAI.02094-05
Baqar, S., Applebee, L. A., Gilliland, T. C., Lee, L. H., Porter, C. K., and Guerry, P. (2008). Immunogenicity and protective efficacy of recombinant Campylobacter jejuni flagellum-secreted proteins in mice. Infect. Immun. 76, 3170–3175. doi: 10.1128/IAI.00076-08
Bayliss, C. D., Bidmos, F. A., Anjum, A., Manchev, V. T., Richards, R. L., Grossier, J. P., et al. (2012). Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Res. 40, 5876–5889. doi: 10.1093/nar/gks246
Black, R. E., Levine, M. M., Clements, M. L., Hughes, T. P., and Blaser, M. J. (1988). Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157, 472–479. doi: 10.1093/infdis/157.3.472
Culebro, A., Revez, J., Pascoe, B., Friedmann, Y., Hitchings, M. D., Stupak, J., et al. (2016). Large sequence diversity within the biosynthesis locus and common biochemical features of Campylobacter coli lipooligosaccharides. J. Bacteriol. 198, 2829–2840. doi: 10.1128/JB.00347-16
Day, C. J., Semchenko, E. A., and Korolik, V. (2012). Glycoconjugates play a key role in Campylobacter jejuni infection?: interactions between host and pathogen. Front. Cell. Infect. Microbiol. 2:9. doi: 10.3389/fcimb.2012.00009
Duncan, J. A., Gao, X., Huang, M. T.-H., O’Connor, B. P., Thomas, C. E., Willingham, S. B., et al. (2009). Neisseria gonorrhoeae activates the proteinase cathepsin b to mediate the signaling activities of the NLRP3 and ASC-containing inflammasome. J. Immunol. 182, 6460–6469. doi: 10.4049/jimmunol.0802696
Elhadidy, M., Arguello, H., Álvarez-Ordóñez, A., Miller, W. G., Duarte, A., Martiny, D., et al. (2018). Orthogonal typing methods identify genetic diversity among Belgian Campylobacter jejuni strains isolated over a decade from poultry and cases of sporadic human illness. Int. J. Food Microbiol. 275, 66–75. doi: 10.1016/j.ijfoodmicro.2018.04.004
Ellström, P., Feodoroff, B., Hänninen, M. L., and Rautelin, H. (2013). Characterization of clinical Campylobacter jejuni isolates with special emphasis on lipooligosaccharide locus class, putative virulence factors and host response. Int. J. Med. Microbiol. 303, 134–139. doi: 10.1016/j.ijmm.2013.01.005
Ellström, P., Feodoroff, B., Hänninen, M. L., and Rautelin, H. (2014). Lipooligosaccharide locus class of Campylobacter jejuni: sialylation is not needed for invasive infection. Clin. Microbiol. Infect. 20, 524–529. doi: 10.1111/1469-0691.12382
Ellström, P., Hansson, I., Nilsson, A., Rautelin, H., and Olsson Engvall, E. (2016). Lipooligosaccharide locus classes and putative virulence genes among chicken and human Campylobacter jejuni isolates. BMC Microbiol. 16:116. doi: 10.1186/s12866-016-0740-5
Endtz, H. P., Ang, C. W., Van Den Braak, N., Duim, B., Rigter, A., Price, L. J., et al. (2000). Molecular characterization of Campylobacter jejuni from patients with Guillain-Barre and Miller Fisher syndromes. J. Clin. Microbiol. 38, 2297–2301.
Fry, B. N., Feng, S., Chen, Y. Y., Newell, D. G., Coloe, P. J., and Korolik, V. (2000). The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 68, 2594–2601. doi: 10.1128/IAI.68.5.2594-2601.2000
Fry, B. N., Korolik, V., ten Brinke, J. A., Pennings, M. T., Zalm, R., Teunis, B. J., et al. (1998). The lipopolysaccharide biosynthesis locus of Campylobacter jejuni 81116. Microbiology 144, 2049–2061. doi: 10.1099/00221287-144-8-2049
Gilbert, M., Brisson, J.-R., Karwaski, M.-F., Michniewicz, J., Cunningham, A.-M., Wu, Y., et al. (2000). Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. J. Biol. Chem. 275, 3896–3906. doi: 10.1074/jbc.275.6.3896
Gilbert, M., Godschalk, P. C., Karwaski, M. F., Ang, C. W., van Belkum, A., Li, J., et al. (2004). Evidence for acquisition of the lipooligosaccharide biosynthesis locus in Campylobacter jejuni GB11, a strain isolated from a patient with Guillain-Barr syndrome, by horizontal exchange. Infect. Immun. 72, 1162–1165. doi: 10.1128/iai.72.2.1162-1165.2004
Gilbert, M., Karwaski, M. F., Bernatchez, S., Young, N. M., Taboada, E., Michniewicz, J., et al. (2002). The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277, 327–337. doi: 10.1074/jbc.M108452200
Gilbert, M., Parker, C., Moran, A. (2008). “Campylobacter jejuni Lipooligosaccharides: Structures and Biosynthesis,” in Campylobacter, Third Edition eds Nachamkin, I., Szymanski, C., and Blaser, M. (Washington, DC: ASM Press), 483–504. doi: 10.1128/9781555815554.ch27
Godschalk, P. C. R., Heikema, A. P., Gilbert, M., Komagamine, T., Wim Ang, C., Glerum, J., et al. (2004). The crucial role of Campylobacter jejuni genes in anti-ganglioside antibody induction in Guillain-Barré syndrome. J. Clin. Invest. 114, 1659–1665. doi: 10.1172/JCI200415707
Godschalk, P. C. R., Kuijf, M. L., Li, J., St. Michael, F., Ang, C. W., Jacobs, B. C., et al. (2007). Structural characterization of Campylobacter jejuni lipooligosaccharide outer cores associated with Guillain-Barré and Miller Fisher syndromes. Infect. Immun. 75, 1245–1254. doi: 10.1128/IAI.00872-06
Godschalk, P. G., Gilbert, M., Jacobs, B. C., Kramers, T., Tio-Gillen, A. P., Wim Ang, C., et al. (2006). Co-infection with two different Campylobacter jejuni strains in a patient with Guillian-Barré syndrome. Microbes Infect. 8, 248–253. doi: 10.1016/j.micinf.2005.06.022
Guerry, P., Szymanski, C. M., Prendergast, M. M., Hickey, T. E., Ewing, C. P., Pattarini, D. L., et al. (2002). Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70, 787–793. doi: 10.1128/IAI.70.2.787-793.2002
Gundogdu, O., Bentley, S. D., Holden, M. T., Parkhill, J., Dorrell, N., and Wren, B. W. (2007). Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics 8:162. doi: 10.1186/1471-2164-8-162
Habib, I., Louwen, R., Uyttendaele, M., Houf, K., Vandenberg, O., Nieuwenhuis, E. E., et al. (2009). Correlation between genotypic diversity, lipooligosaccharide gene locus class variation, and caco-2 cell invasion potential of Campylobacter jejuni isolates from chicken meat and humans: contribution to virulotyping. Appl. Environ. Microbiol. 75, 4277–4288. doi: 10.1128/AEM.02269-08
Havelaar, A. H., Kirk, M. D., Torgerson, P. R., Gibb, H. J., Hald, T., Lake, R. J., et al. (2015). World health organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 12:e1001923. doi: 10.1371/journal.pmed.1001923
Heikema, A. P., Koning, R. I., Rico, S. D., dos, S., Rempel, H., Jacobs, B. C., et al. (2013). Enhanced, sialoadhesin-dependent uptake of guillain-barré syndrome-associated Campylobacter jejuni strains by human macrophages. Infect. Immun. 81, 2095–2103. doi: 10.1128/IAI.01437-12
Houliston, R. S., Vinogradov, E., Dzieciatkowska, M., Li, J., St Michael, F., Karwaski, M. F., et al. (2011). Lipooligosaccharide of Campylobacter jejuni: similarity with multiple types of mammalian glycans beyond gangliosides. J. Biol. Chem. 286, 12361–12370. doi: 10.1074/jbc.M110.181750
Islam, Z., Sarker, S. K., Jahan, I., Farzana, K. S., Ahmed, D., Faruque, A. S. G., et al. (2018). Capsular genotype and lipooligosaccharide locus class distribution in Campylobacter jejuni from young children with diarrhea and asymptomatic carriers in Bangladesh. Eur. J. Clin. Microbiol. Infect. Dis. 37, 723–728. doi: 10.1007/s10096-017-3165-7
Islam, Z., van Belkum, A., Wagenaar, J. A., Cody, A. J., de Boer, A. G., Sarker, S. K., et al. (2014). Comparative population structure analysis of Campylobacter jejuni from human and poultry origin in Bangladesh. Eur. J. Clin. Microbiol. Infect. Dis. 33, 2173–2181. doi: 10.1007/s10096-014-2184-x
Islam, Z., van Belkum, A., Wagenaar, J. A., Cody, A. J., de Boer, A. G., Tabor, H., et al. (2009). Comparative genotyping of Campylobacter jejuni strains from patients with Guillain-Barré syndrome in Bangladesh. PLoS One. 4:e7257. doi: 10.1371/journal.pone.0007257
Iwata, T., Chiku, K., Amano, K.-I., Kusumoto, M., Ohnishi-Kameyama, M., Ono, H., et al. (2013). Effects of lipooligosaccharide inner core truncation on bile resistance and chick colonization by Campylobacter jejuni. PLoS One 8:e56900. doi: 10.1371/journal.pone.0056900
Janssen, R., Krogfelt, K. A., Cawthraw, S. A., Van Pelt, W., Wagenaar, J. A., and Owen, R. J. (2008). Host-pathogen interactions in Campylobacter infections: the host perspective. Clin. Microbiol. Rev. 21, 505–518. doi: 10.1128/CMR.00055-07
Javed, M. A., Cawthraw, S. A., Baig, A., Li, J., McNally, A., Oldfield, N. J., et al. (2012). Cj1136 is required for lipooligosaccharide biosynthesis, hyperinvasion, and chick colonization by Campylobacter jejuni. Infect. Immun. 80, 2361–2370. doi: 10.1128/iai.00151-12
Jeon, B., Muraoka, W., Scupham, A., and Zhang, Q. (2009). Roles of lipooligosaccharide and capsular polysaccharide in antimicrobial resistance and natural transformation of Campylobacter jejuni. J. Antimicrob. Chemother. 63, 462–468. doi: 10.1093/jac/dkn529
Kanipes, M. I., Holder, L. C., Corcoran, A. T., Moran, A. P., and Guerry, P. (2004). A deep-rough mutant of Campylobacter jejuni 81-176 is noninvasive for intestinal epithelial cells. Infect. Immun. 72, 2452–2455. doi: 10.1128/IAI.72.4.2452
Kanipes, M. I., Papp-Szabo, E., Guerry, P., and Monteiro, M. A. (2006). Mutation of waaC, encoding heptosyltransferase I in Campylobacter jejuni 81-176, affects the structure of both lipooligosaccharide and capsular carbohydrate. J. Bacteriol. 188, 3273–3279. doi: 10.1128/jb.188.9.3273-3279.2006
Kanipes, M. I., Tan, X., Akelaitis, A., Li, J., Rockabrand, D., Guerry, P., et al. (2008). Genetic analysis of lipooligosaccharide core biosynthesis in Campylobacter jejuni 81-176. J. Bacteriol. 190, 1568–1574. doi: 10.1128/JB.01696-07
Karlyshev, A., Ketley, J., and Wren, B. (2005). The glycome. FEMS Microbiol. Rev. 29, 377–390. doi: 10.1016/j.femsre.2005.01.003
Keo, T., Collins, J., Kunwar, P., Blaser, M. J., and Iovine, N. M. (2011). Campylobacter capsule and lipooligosaccharide confer resistance to serum and cationic antimicrobials. Virulence 2, 30–40. doi: 10.4161/viru.2.1.14752
Klaas, M., Oetke, C., Lewis, L. E., Erwig, L. P., Heikema, A. P., Easton, A., et al. (2012). Sialoadhesin promotes rapid proinflammatory and type I IFN responses to a sialylated pathogen, Campylobacter jejuni. J. Immunol. 189, 2414–2422. doi: 10.4049/jimmunol.1200776
Klena, J. D., Gray, S. A., and Konkel, M. E. (1998). Cloning, sequencing, and characterization of the lipopolysaccharide biosynthetic enzyme heptosyltransferase I gene (waaC) from Campylobacter jejuni and Campylobacter coli. Gene 222, 177–185. doi: 10.1016/S0378-1119(98)00501-0
Koga, M., Takahashi, M., Masuda, M., Hirata, K., and Yuki, N. (2005). Campylobacter gene polymorphism as a determinant of clinical features of Guillain-Barré syndrome. Neurology 65, 1376–1381. doi: 10.1212/01.wnl.0000176914.70893.14
Linton, D., Gilbert, M., Hitchen, P. G., Dell, A., Morris, H. R., Wakarchuk, W. W., et al. (2000). Phase variation of a β-1, 3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 37, 501–514. doi: 10.1046/j.1365-2958.2000.02020.x
Louwen, R., Heikema, A., van Belkum, A., Ott, A., Gilbert, M., Ang, W., et al. (2008). The sialylated lipooligosaccharide outer core in Campylobacter jejuni is an important determinant for epithelial cell invasion. Infect. Immun. 76, 4431–4438. doi: 10.1128/IAI.00321-08
Louwen, R., Nieuwenhuis, E. E. S., van Marrewijk, L., Horst-Kreft, D., de Ruiter, L., Heikema, A. P., et al. (2012). Campylobacter jejuni translocation across intestinal epithelial cells is facilitated by ganglioside-like lipooligosaccharide structures. Infect. Immun. 80, 3307–3318. doi: 10.1128/IAI.06270-11
Mandrell, R. E., McLaughlin, R., Kwaik, Y. A., Lesse, A., Yamasaki, R., Gibson, B., et al. (1992). Lipooligosaccharides (LOS) of some Haemophilus species mimic human glycosphingolipids, and some LOS are sialylated. Infect. Immun. 60, 1322–1328. doi: 10.1128/iai.60.4.1322-1328.1992
Marsden, G. L., Li, J., Everest, P. H., Lawson, A. J., and Ketley, J. M. (2009). Creation of a large deletion mutant of Campylobacter jejuni reveals that the lipooligosaccharide gene cluster is not required for viability. J. Bacteriol. 191, 2392–2399. doi: 10.1128/JB.01397-08
McCarthy, N., and Giesecke, J. (2001). Incidence of Guillain-Barré syndrome following infection with Campylobacter jejuni. Am. J. Epidemiol. 153, 610–614. doi: 10.1093/aje/153.6.610
Monteiro, M. A., Baqar, S., Hall, E. R., Chen, Y. H., Porter, C. K., Bentzel, D. E., et al. (2009). Capsule polysaccharide conjugate vaccine against diarrheal disease caused by Campylobacter jejuni. Infect. Immun. 77, 1128–1136. doi: 10.1128/IAI.01056-08
Moran, A. P. (1997). Structure and conserved characteristics of Campylobacter jejuni lipopolysaccharides. J. Infect. Dis. 176(Suppl.), S115–S121.
Mortensen, N. P., Kuijf, M. L., Ang, C. W., Schiellerup, P., Krogfelt, K. A., Jacobs, B. C., et al. (2009). Sialylation of Campylobacter jejuni lipo-oligosaccharides is associated with severe gastro-enteritis and reactive arthritis. Microbes Infect. 11, 988–994. doi: 10.1016/j.micinf.2009.07.004
Müller, J., Meyer, B., Hänel, I., and Hotzel, H. (2007). Comparison of lipooligosaccharide biosynthesis genes of Campylobacter jejuni strains with varying abilities to colonize the chicken gut and to invade Caco-2 cells. J. Med. Microbiol. 56, 1589–1594. doi: 10.1099/jmm.0.47305-0
Nachamkin, I., Allos, B. M., and Ho, T. (1998). Campylobacter species and Guillain-Barre syndrome. Clin. Microbiol. Rev. 11, 555–567. doi: 10.1524/zkri.217.7.292.23647
Nachamkin, I., Liu, J., Li, M., Ung, H., Moran, A. P., Prendergast, M. M., et al. (2002). Campylobacter jejuni from patients with Guillain-Barré syndrome preferentially expresses a GD1a-like epitope. Infect. Immun. 70, 5299–5303. doi: 10.1128/IAI.70.9.5299-5303.2002
Ohishi, T., Aoki, K., Ishii, Y., Usui, M., Tamura, Y., Kawanishi, M., et al. (2017). Molecular epidemiological analysis of human- and chicken-derived isolates of Campylobacter jejuni in Japan using next-generation sequencing. J. Infect. Chemother. 23, 165–172. doi: 10.1016/j.jiac.2016.11.011
Parker, C. T., Gilbert, M., Yuki, N., Endtz, H. P., and Mandrell, R. E. (2008). Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: evidence of mosaic organizations. J. Bacteriol. 190, 5681–5689. doi: 10.1128/JB.00254-08
Parker, C. T., Horn, S. T., Gilbert, M., Miller, W. G., Woodward, D. L., and Mandrell, R. E. (2005). Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 43, 2771–2781. doi: 10.1128/JCM.43.6.2771-2781.2005
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Perkins, D. J., and Newstead, G. L. (1994). Campylobacter jejuni enterocolitis causing peritonitis, ileitis and intestinal obstruction. Aust. N. Z. J. Surg. 64, 55–58. doi: 10.1111/j.1445-2197.1994.tb02137.x
Phongsisay, V., Perera, V. N., and Fry, B. N. (2006). Exchange of lipooligosaccharide synthesis genes creates potential Guillain-Barre of Campylobacter jejuni. Infect. Immun. 74, 1368–1372. doi: 10.1128/IAI.74.2.1368
Phongsisay, V., Perera, V. N., and Fry, B. N. (2007). Expression of the htrB gene is essential for responsiveness of Salmonella typhimurium and Campylobacter jejuni to harsh environments. Microbiology 153, 254–262. doi: 10.1099/mic.0.29230-0
Poly, F., Noll, A. J., Riddle, M. S., Porter, C. K., Poly, F., Noll, A. J., et al. (2019). Update on Campylobacter vaccine development. Hum. Vaccin. Immunother. 15, 1389–1400. doi: 10.1080/21645515.2018.1528410
Poly, F., Read, T. D., Chen, Y. H., Monteiro, M. A., Serichantalergs, O., Pootong, P., et al. (2008). Characterization of two Campylobacter jejuni strains for use in volunteer experimental-infection studies. Infect. Immun. 76, 5655–5667. doi: 10.1128/IAI.00780-08
Poropatich, K. O., Fischer Walker, C. L., and Black, R. B. (2010). Quantifying the association between Campylobacter infection and Guillain-Barré syndrome: a systematic review. J. Health Popul. Nutr. 28, 545–552. doi: 10.3329/jhpn.v28i6.6602
Quiñones, B., Parker, C. T., Janda, J. M., Miller, W. G., and Mandrell, R. E. (2007). Detection and genotyping of Arcobacter and Campylobacter isolates from retail chicken samples by use of DNA oligonucleotide arrays. Appl. Environ. Microbiol. 73, 3645–3655. doi: 10.1128/AEM.02984-06
Revez, J., and Hänninen, M. L. (2012). Lipooligosaccharide locus classes are associated with certain Campylobacter jejuni multilocus sequence types. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2203–2209. doi: 10.1007/s10096-012-1556-3
Richards, V. P., Lefébure, T., Pavinski Bitar, P. D., and Stanhope, M. J. (2013). Comparative characterization of the virulence gene clusters (lipooligosaccharide [LOS] and capsular polysaccharide [CPS]) for Campylobacter coli, Campylobacter jejuni subsp. jejuni and related Campylobacter species. Infect. Genet. Evol. 14, 200–213. doi: 10.1016/j.meegid.2012.12.010
Riddle, M. S., and Guerry, P. (2016). Status of vaccine research and development for Campylobacter jejuni. Vaccine 34, 2903–2906. doi: 10.1016/j.vaccine.2016.02.080
Scallan Walter, E. J., Crim, S. M., Bruce, B. B., and Griffin, P. M. (2019). Incidence of Campylobacter-associated Guillian Barré syndrome estimated from health insurance data. Foodborne Pathog. Dis. 17, 23–28. doi: 10.1089/fpd.2019.2652
Semchenko, E. A., Day, C. J., Moutin, M., Wilson, J. C., Tiralongo, J., and Korolik, V. (2012). Structural heterogeneity of terminal glycans in Campylobacter jejuni lipooligosaccharides. PLoS One 7:e40920. doi: 10.1371/journal.pone.0040920
Simpson, D. J., Sacher, J. C., and Szymanski, C. M. (2015). Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr. Opin. Struct. Biol. 34, 69–77. doi: 10.1016/j.sbi.2015.07.006
Stephenson, H. N., John, C. M., Naz, N., Gundogdu, O., Dorrell, N., Wren, B. W., et al. (2013). Campylobacter jejuni lipooligosaccharide sialylation, phosphorylation, and amide/ester linkage modifications fine-tune human toll-like receptor 4 activation. J. Biol. Chem. 288, 19661–19672. doi: 10.1074/jbc.M113.468298
Szymanski, C. M., St. Michael, F., Jarrell, H. C., Li, J., Gilbert, M., Larocque, S., et al. (2003). Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from Campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278, 24509–24520. doi: 10.1074/jbc.M301273200
Tam, C. C., Rodrigues, L. C., Petersen, I., Islam, A., Hayward, A., and O’Brien, S. J. (2006). Incidence of Guillain-Barré Syndrome among patients with Campylobacter infection: a general practice research database study. J. Infect. Dis. 194, 95–97. doi: 10.1086/504294
Thépault, A., Guyard-Nicodème, M., Rose, V., Quesne, S., Queguiner, M., Houard, E., et al. (2018). A representative overview of the genetic diversity and lipooligosaccharide sialylation in Campylobacter jejuni along the broiler production chain in France and its comparison with human isolates. Int. J. Food Microbiol. 274, 20–30. doi: 10.1016/j.ijfoodmicro.2018.03.010
van Sorge, N. M., Bleumink, N. M. C., van Vliet, S. J., Saeland, E., van der Pol, W. L., van Kooyk, Y., et al. (2009). N-glycosylated proteins and distinct lipooligosaccharide glycoforms of Campylobacter jejuni target the human C-type lectin receptor MGL. Cell. Microbiol. 11, 1768–1781. doi: 10.1111/j.1462-5822.2009.01370.x
van Spreeuwel, J. P., Duursma, G. C., Meijer, C. J., Bax, R., Rosekrans, P. C., and Lindeman, J. (1985). Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut 26, 945–951. doi: 10.1136/gut.26.9.945
Wachira, V. K., Peixoto, H. M., and de Oliveira, M. R. F. (2019). Systematic review of factors associated with the development of Guillian-Barré syndrome 2007-2017: what has changed? Trop. Med. Int. Health 24, 132–142. doi: 10.1111/tmi.13181
Wakerley, B. R., and Yuki, N. (2015). Mimics and chameleons in Guillain–Barré and Miller Fisher syndromes. Pract. Neurol. 15, 90–99. doi: 10.1136/practneurol-2014-000937
Wanford, J. J., Lango-Scholey, L., Nothaft, H., Hu, Y., Szymanski, C. M., and Bayliss, C. D. (2018). Random sorting of Campylobacter jejuni phase variants due to a narrow bottleneck during colonization of broiler chickens. Microbiology 164, 896–907. doi: 10.1099/mic.0.000669
Whitfield, C., and Trent, M. S. (2014). Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128. doi: 10.1146/annurev-biochem-060713-035600
Yuki, N. (1997). Molecular Mimicry between Gangliosides and Lipopolysaccharides of Campylobacter jejuni Isolated from Patients with Guillain-Barré Syndrome and Miller Fisher Syndrome. J. Infect. Dis. 176, 150–153. doi: 10.1086/513800
Keywords: lipooligosaccharide, Campylobacter jejuni, ganglioside mimics, Guillain–Barré syndrome, Miller Fisher syndrome
Citation: Hameed A, Woodacre A, Machado LR and Marsden GL (2020) An Updated Classification System and Review of the Lipooligosaccharide Biosynthesis Gene Locus in Campylobacter jejuni. Front. Microbiol. 11:677. doi: 10.3389/fmicb.2020.00677
Received: 25 October 2019; Accepted: 24 March 2020;
Published: 19 May 2020.
Edited by:
Yuji Morita, Meiji Pharmaceutical University, JapanReviewed by:
Alejandra Kolehmainen, Thermo Fisher Scientific, FinlandKatell Rivoal, French Agency for Food Environmental and Occupational Health and Safety, France
Patrik Ellström, Uppsala University, Sweden
Copyright © 2020 Hameed, Woodacre, Machado and Marsden. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lee R. Machado, bGVlLm1hY2hhZG9Abm9ydGhhbXB0b24uYWMudWs=
 Amber Hameed
Amber Hameed Alexandra Woodacre
Alexandra Woodacre Lee R. Machado
Lee R. Machado Gemma L. Marsden
Gemma L. Marsden