- 1Department of Epidemiology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 2Curriculum in Genetics and Molecular Biology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 3Department of Microbiology and Immunology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Emerging coronaviruses (CoV) are constant global public health threats to society. Multiple ongoing clinical trials for vaccines and antivirals against CoVs showcase the availability of medical interventions to both prevent and treat the future emergence of highly pathogenic CoVs in human. However, given the diverse nature of CoVs and our close interactions with wild, domestic and companion animals, the next epidemic zoonotic CoV could resist the existing vaccines and antivirals developed, which are primarily focused on Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS CoV). In late 2019, the novel CoV (SARS-CoV-2) emerged in Wuhan, China, causing global public health concern. In this review, we will summarize the key advancements of current vaccines and antivirals against SARS-CoV and MERS-CoV as well as discuss the challenge and opportunity in the current SARS-CoV-2 crisis. At the end, we advocate the development of a “plug-and-play” platform technologies that could allow quick manufacturing and administration of broad-spectrum countermeasures in an outbreak setting. We will discuss the potential of AAV-based gene therapy technology for in vivo therapeutic antibody delivery to combat SARS-CoV-2 outbreak and the future emergence of severe CoVs.
Introduction
The zoonotic transmission and subsequent adaptation to humans of emerging RNA viruses is a global public health concern. In the 21st century alone, coronaviruses (CoV) have been responsible for two separate endemics, the severe acute respiratory syndrome (SARS) and Middle East Respiratory Syndrome (MERS) CoVs (de Wit et al., 2016). In late Dec 2019, a novel SARS-like CoV designated 2019 nCoV emerged in Wuhan China, causing > 60,000 cases and over 1350 deaths in an ongoing epidemic (Hui et al., 2020). Other highly pathogenic threat viruses that have emerged in the 21st century include influenza viruses, Ebola viruses, flaviviruses and paramyxoviruses (Mackenzie and Jeggo, 2013). The high mutation and recombination rate of RNA viruses drives the emergence of new viral strains that can rapidly adapt to new and changing ecologies (Drake and Holland, 1999; Lauring and Andino, 2010). Furthermore, industrialization, globalization and traditional cultural habits potentiate the likelihood of zoonotic transmission and facilitate the spread of viruses in the human population. While new outbreaks from emergent viruses are inevitable, scientists, epidemiologists, and the health care industry are racing to develop new technologies to better predict and minimize the impact of an outbreak by employing global viral surveillance programs and developing vaccines and antivirals (Lipkin and Firth, 2013). A major challenge of vaccine and antiviral development is the elusive nature of the emerging viruses, which oftentimes emerge from highly heterogeneous populations of virus strains that circulate in animal reservoirs (Lauring and Andino, 2010). Therefore, to prepare for future outbreaks, vaccines and antivirals will need to be both potent and broadly effective against multiple potential emerging viruses within and across virus families. Additionally, in order to control and prevent viral spread, treatments must be readily available to affected populations and have a fast response time. In this review, we will focus our discussion on the challenges, as well as current development, of vaccines and antivirals for SARS-CoV and MERS-CoV. At the end, we will also discuss the potential use of AAV-based gene therapy as a quick response to prevent and treat emerging viral infections in the current SARS-CoV-2 and future outbreak situations.
Endemic and Emerging Coronaviruses
Coronaviruses are a diverse group of positive-stranded RNA viruses which infect a wide range of animals from birds to mammals, causing a variety of diseases (Perlman and Netland, 2009; Woo et al., 2009; Peck et al., 2015). Based on sequence identity of the spike protein or the non-structural proteins (nsp), CoVs are classified into four different sub-groups, alphacoronaviruses, betacoronaviruses, gammacoronaviruses, and deltacoronaviruses (Figure 1). Human coronaviruses (hCoVs), such as 229E, OC43, NL-63 and HKU-1 are highly transmissible respiratory viruses which are responsible for around 10-20% of common cold cases annually (McIntosh et al., 1970; Cabeça et al., 2013). HCoV-related illness is often self-limited in immune competent individuals but may cause more severe upper and lower respiratory tract infections in the young and elderly population (Woo et al., 2005; Lau et al., 2006). In addition, highly pathogenic CoVs may emerge through zoonotic reservoirs. In the past two decades, SARS-CoV and MERS-CoV emerged from bats and spread to humans through intermediate hosts including civet cats and camels, respectively (Raj et al., 2014). SARS-CoV and MERS-CoV belong to the sub-groups 2b and 2c of the Betacoronavirus genus (Peck et al., 2015). The latest CoV outbreak is the SARS-CoV-2, a Betacoronavirus 2b which emerged from bats and spread to humans (Lu et al., 2020). The mortality rate of these viruses range from 10 to 40% but can exceed 50% in the elderly (Min et al., 2004; Li et al., 2005; Bolles et al., 2011b; Raj et al., 2014; Sharif-Yakan and Kanj, 2014; World Health Organization [WHO], 2018). The unusually high mortality rate is linked to disease progression leading to acute respiratory distress syndrome (ARDS) which causes hypoxemia, pulmonary edema, and infiltration of inflammatory immune cells in the lung (Cabeça et al., 2013; Gralinski and Baric, 2015). If unresolved, the diseases progress to late phase ARDS, leading to end-stage lung disease and death (Ding et al., 2003). Currently, no vaccines or antiviral drugs are approved to prevent or treat severe CoV infection.
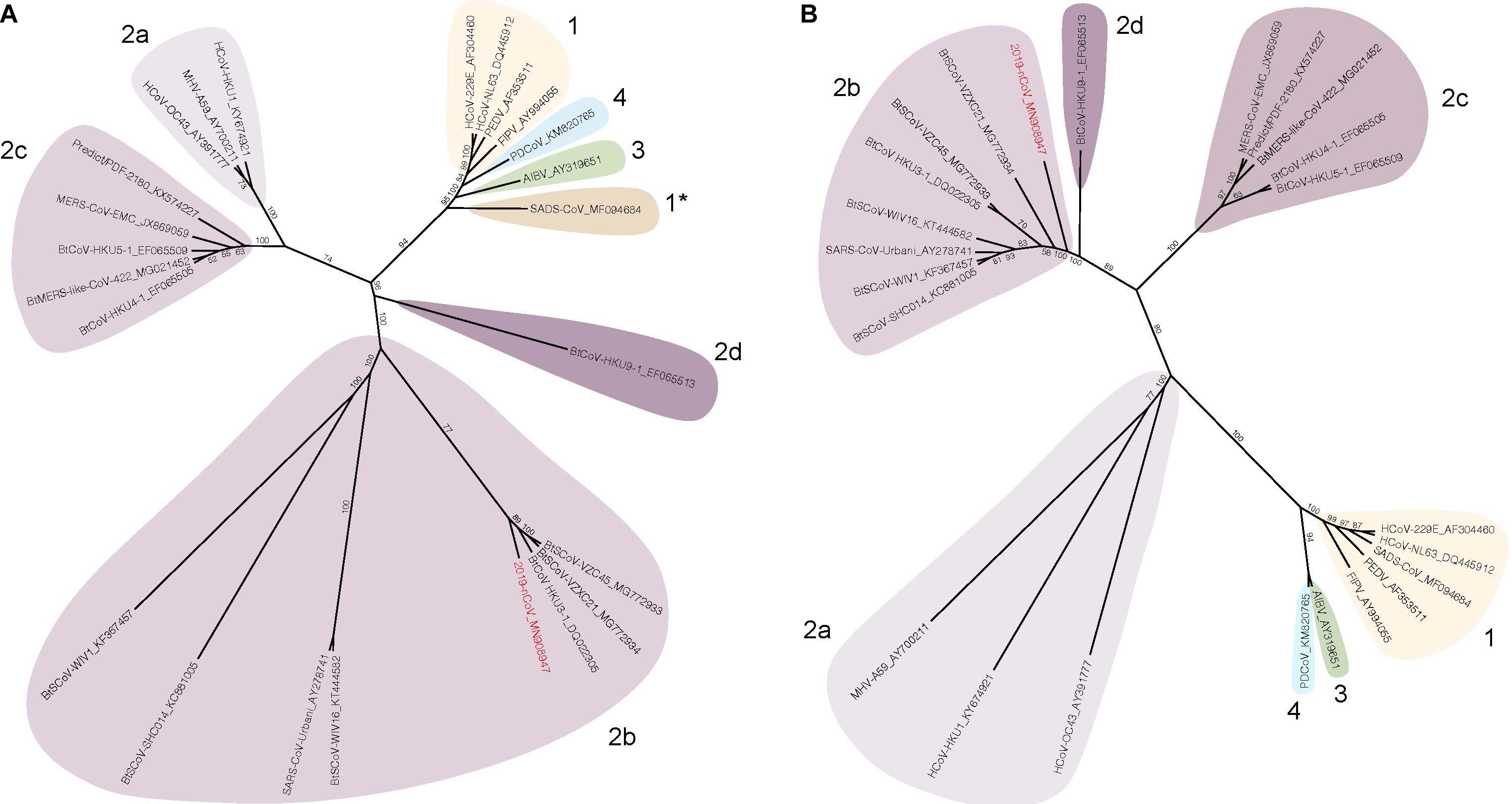
Figure 1. Spike and nsp12 phylogeny of representative coronaviruses. The Spike (A) and nsp12 (B) protein sequences of selected coronaviruses were aligned and phylogenetically compared. Coronavirus genera are grouped by classic subgroup designations (1, 2a-d, 3, and 4). In the Spike tree in (A), SADS-CoV is designated as 1* because of its distinctive grouping compared with more conserved proteins (e.g., nsp12, see (B)). Branches in each tree are labeled with consensus support values (in %). Sequences were aligned using free end gaps with the Blosum62 cost matrix, and the tree was constructed using the neighbor-joining method based on the multiple sequence alignment in Geneious Prime. Numbers following the underscores in each sequence correspond to the GenBank Accession number. The SARS-CoV-2 is highlighted in red. The radial phylogram was exported from Geneious and then rendered for publication using Adobe Illustrator CC 2020.
The Challenge for Vaccine Development
The CoV S-protein is the major envelope glycoprotein and the main determinant of protective immunity. The S-protein is composed of two principle subunits, S1 and S2; S1 governs receptor binding and S2 is responsible for membrane fusion (Li, 2016). Similar to other class I fusion proteins, S-protein undergoes a major conformational change between pre-fusion and post-fusion which also presents different antigenic epitopes. While able to bind to both conformations, Abs targeting the post-fusion form are not necessary neutralizing; in contrast, Abs targeting the pre-fusion form of the S-protein correlate better with neutralization. In particular, vaccines and neutralizing antibodies (nAbs) which target the receptor binding domain (RBD) of the S-protein can effectively neutralize the virus (Zhu et al., 2007; Tang et al., 2014). However, due to high selective pressure and tropism determinants, S-protein is the most diverse region of CoV. For instance, the S-proteins from SARS-CoV and MERS-CoV share only 44% sequence identity (Kandeel, 2018). The majority of differences between S-proteins is in the S1 region, which is further separated into the N-terminal Domain (NTD) and RBD. The diverse nature of the RBD between SARS-CoV and MERS-CoV is reflected in the use of different entry receptors, either angiotensin I converting enzyme 2 (ACE2) or dipeptidyl peptidase 4 (DPP4), respectively (Li et al., 2003; Raj et al., 2013). The diversity of S-protein also renders vaccines and nAbs unlikely to be cross-protective between existing and emerging CoVs. Surveillance and experimental data have identified multiple animal SARS- and MERS-like CoVs that have significant diversity in S-protein and are able to replicate in human cells without adaptation (Menachery et al., 2015, 2016; Luo et al., 2018). Once transmitted to human population, such variation between S-protein between the previous and the new emerging CoVs will pose a major challenge on the progress of vaccine development.
Criteria for Generating Effective Vaccine for CoVs
Vaccine Criteria for SARS and MERS-CoV Viruses
The surface glycoproteins are the main target for vaccine development. In CoV infection, Abs against S-protein were shown to be protective in multiple animal studies. Furthermore, in a passive immunization study in camel, the nAb level is directly correlated to lung pathology and survival (Zhao et al., 2015). As such, one of the main goals for CoV vaccines in humans is the ability to elicit a strong humoral immune response against the S-protein. Particularly, the pre-fusion form of the S-protein is an attractive target for Abs to confer protective immunity. In order to “lock” the S-protein in its antigenic optimal pre-fusion form, two mutations, V1060P and L1061P, were introduced into the MERS-CoV S-protein (Pallesen et al., 2017). The resulting MERS S-2P is able to elicit both RBD and non-RBD binding nAbs (Pallesen et al., 2017). The same strategy was also shown to work in the SARS-CoV-2 S-protein (Wrapp et al., 2020). Other structural proteins such as E and M and the nsp N also contribute to viral protection and clearance (Channappanavar et al., 2014; Zhao et al., 2016; Deng et al., 2018). Similarly to other respiratory viruses such as influenza, mucosal IgA plays a major role in disease protection and has a synergistic effect with IgG (Belshe et al., 2000; Plotkin, 2010). In order to elicit a strong mucosal IgA immunity, the route of vaccine administration is important. Studies have shown that intranasal inoculation of a recombinant RBD vaccine can elicit greater mucosal IgA production than intramuscular or subcutaneous injection (Ma et al., 2014a). However, the duration of mucosal antibodies is typically shorter lived than systemic IgG responses and the longest longitudinal study of MERS-CoV IgA responses ended after 6 months (Hapfelmeier et al., 2010; Ma et al., 2014a). In comparison, in a natural infection case study, SARS-targeting systemic memory B cells were present up to 6 years for SARS-CoV (Oh et al., 2012; Tang et al., 2019); and up to 34 months post infection for MERS-CoV (Payne et al., 2016). Another important consideration for SARS-CoV and MERS-CoV vaccination is the T cell response against the virus, specifically the N proteins, which is important for viral clearance (Zhao et al., 2010; Channappanavar et al., 2014). A study has shown that adoptive transfer of viral specific T-cells to SCID mice enhances survival and reduces lung titer after SARS-CoV infection (Zhao et al., 2010). Moreover, intranasal vaccination of N protein using the VEEV replicon system elicits CD4+ memory T-cells responses in the airway. Upon challenge, the airway CD4+ memory T cells secrete IFN-γ, which subsequently enhances the innate immune response as well as coordinates the CD8+ T cell priming and migration which protects mice from lethal disease, but not weight loss or virus titers under carefully controlled conditions (Zhao et al., 2016). Interestingly, Rag−/− mice are able to clear SARS-CoV infection, suggesting innate immunity is sufficient for viral clearance (Zhao et al., 2010). However, the mechanism of this viral clearance is still unknown. Another important aspect of emerging CoV vaccine is the breadth of protection. As mentioned previously, the antigenic variation in the S-protein between CoVs limits the breadth of cross protection against multiple emerging CoVs, and is especially true for S-protein only vaccines (Wang et al., 2018, 2019).
Learning from natural infection, MERS-CoV specific CD4+ and CD8+ T cells are detected in PBMCs in MERS-CoV infected survivors (Zhao et al., 2017). Therefore, a balance of B cell and T cell responses is generally considered the gold standard to prevent and resolve MERS-CoV and SARS-CoV infection. Multiple strategies have been developed to elicit long lasting B and T cell responses for SARS-CoV and MERS-CoV. These include the traditional live attenuated, inactivated, and subunit vaccines, and newer development of nanoparticles, vectorized vaccines, and RNA/DNA vaccines. In this review, we have selected only the vaccine studies that have an in vivo challenge model and have summarized the different parameters, including vaccine components, dosage, challenge conditions, animal models and the study outcome in Table 1. We will also discuss each type of vaccine strategy and focus on the finished clinical trial targeting SARS-CoV and the 3 ongoing clinical trials targeting MERS-CoV using DNA (Martin et al., 2008; Modjarrad et al., 2019) and vectorized vaccines. Other comprehensive reviews on CoV vaccine development can be found elsewhere (Zhang et al., 2014; Du and Jiang, 2015; Perlman and Vijay, 2016; Schindewolf and Menachery, 2019; Yong et al., 2019).
Current Vaccine Strategies
Inactivated vaccines are the quickest option for vaccine development in an outbreak situation. Multiple chemical and physical methods have been applied singly or in combination to inactivate CoVs, including β-propiolactone, formalin, formaldehyde and UV. While multiple studies have shown the efficacy of inactivated vaccines in hamster, ferret and multiple mouse challenge models (Stadler et al., 2005; See et al., 2006, 2008; Spruth et al., 2006; Roberts et al., 2010), one study suggested a potential vaccine enhanced pathologies (Bolles et al., 2011a). In this study, double inactivation (DIV) of SARS-CoV using formalin and UV+alum elicits a Th2 skewed response and is only partially protective to young mice (6–8 weeks-old) and not protective to experimentally aged mice (12–14 weeks-old) (Bolles et al., 2011a). Furthermore, upon challenge, DIV vaccinated mice show increased infiltration of eosinophils, neutrophils and other inflammatory cell populations in the lung, likely due to the N specific immune response (Bolles et al., 2011a). Further studies have suggested that by replacing alum with TLR agonists such as Poly I:C, Poly U or LPS as adjuvants, the skewed Th2 responses can be alleviated and may reduce the infiltration of eosinophils in the lung (Iwata-Yoshikawa et al., 2014).
Gamma-ray inactivated whole MERS-CoV (WIV) with alum or M59 also suffers from the Th2 skewed immune response and pulmonary eosinophilia upon challenge, suggesting a potential risk of using inactive virus for CoV vaccination (Agrawal et al., 2016). Interestingly, formaldehyde inactivated MERS-CoV co-administered with alum and CpG shows a more balanced Th1/Th2 response and is able to protect mice from a challenge model (Ad5 transduced hDPP4) with reduction in lung viral titer and improved lung pathology. There is no observable vaccine-induced lung pathology or infiltration of eosinophils upon challenge (Deng et al., 2018). The difference in vaccine outcomes indicates an incomplete understanding of the effect of adjuvants on the inactivated SARS and MERS-CoV vaccines. In respiratory syncytial virus (RSV) vaccines, formalin inactivated vaccines presents the post-fusion form predominately and fail to elicit protective immune responses. Whether the same phenomenon exists in inactivated CoV is still unknown, although it could explain the discrepancy between reports. The inconsistent results from different vaccine models also underscores the host genetic elements affecting vaccine outcome. Nevertheless, inactivated vaccine is still one of the most straightforward methods for vaccine development and has the quickest response time in an outbreak situation.
Live attenuated viral vaccines are the closest mimic of natural infection and generally elicit strong B and T cell responses (Zhao et al., 2017). Multiple strategies have been used to genetically attenuate SARS-CoV and MERS-CoV by either deleting or mutating structural, non-structural or accessory proteins. Intranasal immunization of a SARS-CoV lacking E protein (rSARS-CoV-ΔE) has shown complete protection from pulmonary replication in a Golden Syrian hamster model (Lamirande et al., 2008). A similar virus has been generated in the MERS backbone, creating a conditional mutant that requires trans expression of E for productive replication (Almazán et al., 2013). Mutations of the DEDD motif of the 3′ to 5′ exonuclease (ExoN-nsp14) “proof-reading protein” on a mouse-adapted SARS-CoV attenuated the virus both in vitro and in vivo. A single intranasal immunization is able to elicit strong nAbs (>6-fold protective titers) and completely protect against lethal challenges in an aged mouse model (12 months-old BALB/c) (Graham et al., 2012). Mutations in the nsp 16 (NSP16), a 2′O-methyltransferase, in both SARS-CoV and MERS-CoV have also been shown to attenuate the viruses and to protect BALB/c and CRISPR-Cas humanized DPP4-288-330 mice from lethal challenge (Menachery et al., 2014, 2017, 2018). Moreover, these attenuation strategies can be multiplexed, leading to highly stable live attenuated vaccines with limited capability to undergo recombination and reversion repair (Graham et al., 2018; Menachery et al., 2018). Although the live attenuated vaccine is effective in small animal models, there remain safety concerns about potential revertants and recombination with natural CoVs which hinders their usage in the clinical setting. Furthermore, live attenuated vaccines often require greater time for development and safety testing which lessens utility in an outbreak situation.
Protein-based subunit vaccines are considered the safest format of vaccine. However, the low immunogenicity of subunit vaccines dictates a heavily reliance on adjuvants. Different forms of the S-protein, including the S1 RBD, RBD-Fc (RBD with human IgG Fc fusion), and N-terminal domain (NTD), have demonstrated various degrees of nAb responses and protection in multiple animal models including non-human primates (NHP) (Lan et al., 2015; Zhang et al., 2016; Jiaming et al., 2017; Wang et al., 2017; Deng et al., 2018; Nyon et al., 2018; Adney et al., 2019). For instance, the SARS-CoV S1 subunit vaccine produced from sf9 cells and with the adjuvant saponin or protollin are able to reduce lung viral titer in young or aged mice after challenge, respectively (Bisht et al., 2005; Hu et al., 2007). An RBD subunit of SARS-CoV produced from Chinese hamster ovarian (CHO) cells with Freund’s adjuvant is able to protect young BALB/c mice from infection (Du et al., 2010). MERS-CoV-S1 with adjuvants MF59 or Advax HCXL is able to protect alpacas and dromedary camels against MERS-CoV challenge (Adney et al., 2019). Adjuvant selection can affect the vaccine outcome, and combinations of adjuvants can have synergistic effects on the strength of the response. For instance, rRBD with adjuvants alum and CpG ODN together elicits a stronger humoral and cellular T cell response (Lan et al., 2014). Additionally, rNTD with alum is able to reduce lung pathology in a non-lethal MERS-CoV challenge (Jiaming et al., 2017). Instead of adjuvants alone, immune enhancers such as an Fc fragment, which increases the protein half-life when fused with the RBD, can also elicit a stronger IgG nAb and cellular immune response in multiple experimental animals (Du et al., 2013; Ma et al., 2014b; Tang et al., 2015; Nyon et al., 2018). RBD-Fc fusion subunit vaccine is able to protect a lethal challenge of MERS-CoV in adenovirus transduced hCD26/DPP4 mice (Zhang et al., 2016; Wang et al., 2017). While a large amount of work concerning the subunit vaccines has been done in conjunction with different adjuvants, the effect of each adjuvant is not well understood and multi-adjuvant systems (combinatorial admixes) have not been rigorously tested. A more systematic method of studying the effect of different adjuvants on CoV vaccines will be valuable for vaccine development, perhaps using genetic reference populations that more accurately phenocopy human genetic variation (Leist and Baric, 2018).
Trimeric forms of the S-protein and RBD have been developed using the T4 trimerization domain to mimic the native conformation of the spike RBD (Kam et al., 2007; Tai et al., 2016). The trimeric RBD antigens are able to elicit a robust nAb response and protect 80% of hDPP4 transgenic mice from lethal MERS-CoV challenge, although most animals still experienced slight weight loss (Tai et al., 2016). Alternatively, the MERS-CoV S-protein has been structurally designed to remain in perfusion state by mutating V1060 and L1061 at the tip of the central helix to proline (S-2P) (Kirchdoerfer et al., 2016; Pallesen et al., 2017). The MERS S-2P protein retains the receptor binding properties of the wild-type S and elicits nAbs against at least 3 different S domains, including RBD, NTD, and S2. Intramuscular injection of the MERS S-2P elicits nAb responses in mice comparable to the monomeric S1 and trimeric S-protein antigens. (Pallesen et al., 2017).
Similar to subunit vaccines are the viral like particle (VLP) and nanoparticle vaccines. VLP and nanoparticles provide multivalent binding similar to actual viruses without the potential safety concerns. In SARS-CoV, multiple systems were used to generate S-protein VLPs, including the mouse hepatitis virus (MHV) and influenza matrix 1 (M1). In the chimeric MHV system, the SARS-CoV protein is co-expressed with the MHV E, M and N proteins to produce MHV-S VLP. Mice vaccinated with the MHV-S VLP and alum have inhibited viral replication in lung after a homologous strain challenge (Lokugamage et al., 2008). Instead of using MHV structural proteins, the influenza system express the SARS-CoV S-protein with influenza virus M1 proteins in Sf9 cells to create the M1-S VLP. Immunization of M1-S VLP with aluminum hydroxide can protect mice from a lethal challenge of SARS-CoV (Liu et al., 2011). In MERS-CoV, expression of S, E, and M proteins using the baculovirus system produces VLPs that are morphologically similar to the actual virus (Coleman et al., 2014; Wang et al., 2017b). Other methods such as CPV-based (Wang et al., 2017a), rabies virus (MV)-based (Wirblich et al., 2017), ferritin-based (Seong, 2018), and S-protein aggregates (Coleman et al., 2014) are all able to elicit immune responses and reduce viral replication in a mouse model when co-administered with Matrix M1 adjuvant (Coleman et al., 2017). Although all show different degree of immune response in animal, only one study showed a reduction of viral titer in vivo via an adenovirus transduced hCD26/DPP4 mouse model (Coleman et al., 2017).
Clinical Trials for SARS- and MERS-CoV Vaccines
A finished phase 1 clinical trial for a SARS-CoV vaccine is a DNA vaccine that encodes the ectodomain of the SARS-CoV S-protein (NCT00099463). DNA vaccines rely on a continuous expression of antigen from a DNA plasmid that is injected intramuscularly and electroporated (Muthumani et al., 2015; Wang et al., 2015; Chi et al., 2017). In pre-clinical studies, 3 doses of an intramuscular plasmid injection was able to reduce viral titer in both lung and nasal turbinate in a BALB/c challenge model (Yang et al., 2004). The phase 1 trial showed favorable results; after 3 doses of DNA vaccine, all subjects showed CD4+ T cell responses, while 80% of subjects had nAbs and 20% of subjects showed CD8+ T cell responses. However, there has been no follow-up in the vaccine development, likely due to the end of the SARS-CoV outbreak (Martin et al., 2008). One MERS-CoV vaccine that is currently undergoing clinical trials (NCT03721718) is a DNA-based vaccine (Modjarrad et al., 2019). GLS-5300 is a DNA vaccine based on a consensus full-length S-protein from MERS-CoV under the control of a CMV promoter. In preclinical studies, the vaccine was electroporated into mice, camel and rhesus monkeys three times within 1 month. The vaccine elicited B cell responses in all animals at 1 month post vaccination, and extracted IgG was able to neutralize multiple strains of MERS-CoV including England/2/2013, and Al-Hasa/1/2013 and, surprisingly, a group 1b CoV NL63 and a group 2a CoV HKU1 using the pseudotype neutralization assay (Muthumani et al., 2015). T cell responses were assessed in mice and monkeys, with both demonstrating T cell responses as indicated by the presence of IFN-γ, TNFα and IL2-secreting CD4+ and CD8+ T cells after peptide stimulation. Rhesus monkeys were also protected from challenge of the vaccine strain with lower viral titers and lung pathology as assessed by radiography and pathology studies (Muthumani et al., 2015). Currently, GLS-5300 has completed Phase I clinical trials (safety). Three doses (0.67, 2, and 6 mg) of GLS-5300 were electroporated intramuscularly at weeks 0, 4, and 12. Ninety-four percent of subjects were seroconverted and nAbs were detected in 50% of the individuals. Seventy-six percent of subjects developed T-cell responses against peptides derived from MERS-CoV S-proteins (Modjarrad et al., 2019). Other than a full S-protein DNA vaccine, different designs also show promising results in preclinical mouse models. Notably, a DNA vaccine composed of only the S1 domain showed efficacy when paired with different adjuvants (Chi et al., 2017). Hybrid strategies using a DNA vaccine paired with a protein booster also showed promising results in eliciting more balanced Th1 and Th2 responses (Wang et al., 2015; Al-Amri et al., 2017).
Vector-Based Vaccines
Two out of the three clinical trials for MERS vaccines are vectorized vaccines. Viral vector-based vaccines have multiple advantages over the generic protein or DNA-based subunit vaccines. Firstly, as viral vectors utilize a defined viral entry mechanism, they are more efficient at delivering DNA into cells. Second, the vector itself can serve as an adjuvant which in turn elicits both B- and T-cell responses (Ura et al., 2014). Finally, a wide variety of vector systems including measles viruses (Malczyk et al., 2015; Bodmer et al., 2018), Venezuelan equine encephalitis virus (VEEV) replicon system (Deming et al., 2006; Agnihothram et al., 2014; Zhao et al., 2016), adeno-associated virus (AAV) (Du et al., 2008), parainfluenza type 3 (BHPIV3) (Buchholz et al., 2004; Bukreyev et al., 2004), human and chimpanzee adenovirus (hAd5 and ChAdOx1) (See et al., 2006, 2008; Kobinger et al., 2007; Kim et al., 2014; Guo et al., 2015; Hashem et al., 2019) and modified vaccinia virus Ankara (MVA) (Bisht et al., 2004; Czub et al., 2005; Kapadia et al., 2005; Haagmans et al., 2016) have been previously established for use as vaccine platforms for multiple infectious diseases. Herein, we will focus on the three systems that have been or are currently under clinical trial; all others are summarized in Table 1.
Replication-defective adenovirus vectors are one of the most effective choices to deliver vaccine antigens. Human adenovirus 5 (hAd5) and enteric adenovirus type 41 (Ad41) have both been used to deliver MERS-CoV S or S1 proteins. Intramuscular inoculation of the vaccine elicits both B-cell (nAb titer) and T-cell (IFN-γ secreting splenocytes and pulmonary lymphocytes) responses (Guo et al., 2015). However, pre-existing nAbs against hAd5 and 41 in the human population have limited their usage to dromedary camels instead of humans (Chirmule et al., 1999). The pre-existing nAb problem against hAd5 can be circumvented by using a chimpanzee adenovirus. One such platform is ChAdOx1, wherein the adenovirus E1 gene is replaced by a MERS-CoV S-protein with an N-terminal secretion peptide from human plasminogen activator (tPA) driven by a CMV promoter. Intramuscular inoculation of the vector successfully elicits nAbs against S-protein as quickly as 14 days post infection. Splenic CD8+ T-cells secreting IFN-γ, TNF-α and IL-17 are also present at 28 days post infection (Alharbi et al., 2017). A single intranasal or intramuscular inoculation of ChAdOx1-MERS is able to protect a human DPP4 transgenic mouse from lethal challenge by MERS-CoV, and the vaccine (Munster et al., 2017) is currently under phase 1 clinical trial (NCT03399578). CHAdOx1 has also shown effective protection for Rift Valley Fever Virus in dromedary camels (Warimwe et al., 2016).
The modified vaccinia virus Ankara (MVA) vector is another effective platform for MERS-CoV vaccine development. S-protein is inserted into the MAV genome at deletion site III driven by the viral P11 promoter. After a single intramuscular injection, BALB/c mice produce nAbs against both the RBD and S2, as tested in vitro (Song et al., 2013). A follow-up study identified IFN-γ secreting splenocytes after peptide S291 stimulation at 56 days post infection, suggesting the vaccine is able to elicit memory CD8+ T cell responses. The vaccine is also able to protect a hDDP4-transduced BALB/c mouse model. RNA genomes in the lungs and lung pathology are drastically reduced compared to mock vaccination (Volz et al., 2015). The MAV based MERS-CoV vaccine has also been tested on dromedary camels and elicits induction of nAbs in sera and nasal swabs. Vaccinated camels also show reduced RNA genomes and gross pathology. The Phase I clinical trial has just been completed and the results are pending (NCT03615911).
Antivirals
The Challenge for Treatment Windows
The average incubation period for SARS-CoV and MERS-CoV is around 5 days (Zumla et al., 2015; de Wit et al., 2016) and the main site of viral replication is the lower respiratory tract (Corman et al., 2015; Petrovsky, 2016). At 7–10 days after symptomatic onset, viral RNA titer peaks in the upper respiratory tract (Drosten et al., 2004; Corman et al., 2015). For terminal cases, disease lasts for an average of 12 days post symptomatic onset for MERS-CoV and 24 days for SARS-CoV (Zumla et al., 2015). Interestingly, severe symptoms begin as the viral titer is decreasing, suggesting that severe CoV pathogenesis is due to immune complications and the inability to resolve inflammation (Peiris et al., 2003a; Wang et al., 2004). SARS-CoV upregulates pro-inflammatory cytokine production in the lung, including IL1, IL6, IL8, IL10, CXCL10 and TNF-α production (Binnie et al., 2014). Compared to SARS-CoV infected patients with mild diseases, patients with ARDS fail to induce interferon (IFN) expression and the subsequent IFN-stimulated genes (ISGs) that are indicative of adaptive immune responses (Cameron et al., 2007; Binnie et al., 2014). The inability to switch from an innate immune response to the adaptive immune response may lead to uncontrollable inflammation and severe end stage lung disease. Given the rapid progression of symptoms to terminal illness, there is only approximately a 1 week treatment window after the onset of symptoms for antiviral and medical intervention (Widagdo et al., 2017). This treatment window could further compromised by delays in virus diagnosis, causing a challenge for timely medical intervention administration when the virus titers and pathological symptom are relatively mild (Corman et al., 2015; Ahmed, 2019). Unlike humans, experimental animal models have a compressed disease course (<7 days), and it is difficult to differentiate between early and late phase of infection. It would be beneficial for clinical studies to separate the early- (<10 days) and late-phase (>10 days) patients to determine differences in patient response between the groups.
The Challenge for Therapeutic Development
Despite of the presence of a 3′ to 5′ exoribonuclease (exoN) proofreading enzyme, their large genome size (28–30 kb) means that CoVs remain in the category of highly mutating viruses (Eckerle et al., 2007, 2010; Denison et al., 2011). The high mutation rate poses a considerable challenge for antiviral development as drug resistant viruses could arise or already exist within the quasispecies in nature or from an infected individual (Briese et al., 2014). For instance, after a prolonged period of nAb or drug treatments, the CoV can acquire mutations which confer resistance to the therapeutics (Zumla et al., 2016a). The appearance of these “escape viruses” has been confirmed in multiple mouse model studies in well-contained laboratory settings as well as in nature (Neuman et al., 2006; Ter Meulen et al., 2006; Huang et al., 2008; Jiang et al., 2014; Tang et al., 2014; Tai et al., 2017; Kleine-Weber et al., 2019). Fortunately, some of the mutations that confer drug resistance also compromise viral fitness and attenuate the virus (Deng et al., 2014; Agostini et al., 2018). Another therapeutic intervention is the use of immune modulators, such as IFN-α2a/2b, IFN-β1b and corticosteroids in treating CoVs (Loutfy et al., 2003; Peiris et al., 2003b; Sung et al., 2004). Although multiple studies have shown efficacy of IFN-α2a/2b and IFN-β1b singly or in combination with off-labeled antivirals such as ribavirin, lopinavir (LPV) and ritonavir (RTV) in treating SARS- and MERS-CoV in mouse, rhesus monkey and marmoset models (Haagmans et al., 2004; Barnard et al., 2006b; Falzarano et al., 2013b; Chan et al., 2015), clinical studies have shown inconclusive results (Momattin et al., 2013; Mo and Fisher, 2016). Currently, a randomized controlled trial is underway to determine the efficacy of a combination of LPV/RTV and IFN-β1b in treating MERS-CoV infection (NCT02845843).
Convalescent Plasma and Monoclonal Antibodies
Although there is no approved drug to treat severe CoV infection, multiple strategies have shown promising results in an experimental setting (Figure 2). Convalescent plasma (CP) is derived from patients who have recovered from SARS-CoV and MERS-CoV infection and contains high titer nAbs (Hsueh et al., 2004; Al Kahlout et al., 2019; Shin et al., 2019). Some data suggest that CP use against SARS-CoV infection is safe and, when administered at an early time point, may reduce mortality (Mair-Jenkins et al., 2015). CP neutralizing titers > 1:80 may have a positive impact on infected MERS-CoV patients with respiratory failure (Ko et al., 2018). Unfortunately, CP from convalescent patients is difficult to obtain in large quantities and the Ab titer is too low to have beneficial effects, making it difficult to be used as a main stream therapeutic or for clinical testing (Arabi et al., 2016). As such, no systematic, well-designed clinical trial has formally demonstrated the efficacy of CP in emerging CoV infection. Rather, a clinical trial for testing CP against MERS-CoV infection was withdrawn prior to patient enrollment (NCT02190799).
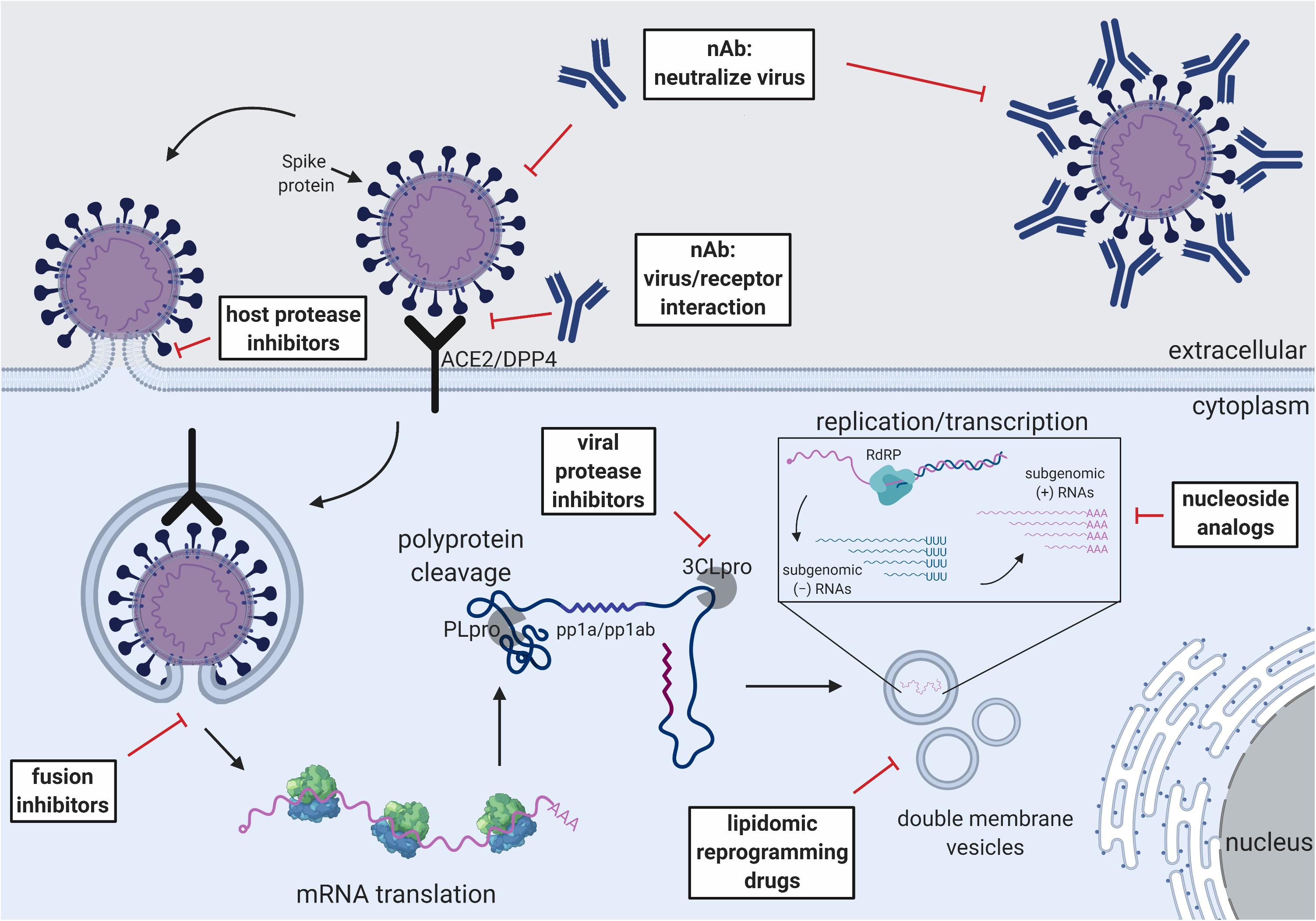
Figure 2. Schematic of the CoV replication cycle and key steps for antiviral targets. White text boxes indicate the subtype of antivirals that work either extracellularly or intracellularly. Different steps of the CoV replication cycle are illustrated in cartoon form, including receptor binding, membrane fusion, viral RNA replication, sub-genomic RNA transcription and translation.
Although CP from convalescent patients is difficult to obtain, the active component (neutralizing antibodies) can be isolated and subsequently produced in large quantity using recombinant technology (Corti et al., 2016). Passive infusion of neutralizing monoclonal Abs (mAbs) has been used for several diseases with success, including RSV and Ebola virus (Graham and Ambrosino, 2015; Zumla et al., 2016b). Numerous highly potent mAbs against SARS-CoV and MERS-CoV have been isolated by multiple groups all over the world using different methods such as phage display and direct B cell cloning from immunized animals or convalescent patients (Sui et al., 2004; Traggiai et al., 2004; Greenough et al., 2005; van den Brink et al., 2005; Zhu et al., 2007; Corti et al., 2016). While all of them shows protecting activity in vitro, several of them have also shown efficacy in mouse and NHP models (Ter Meulen et al., 2004; Johnson et al., 2016; Chen et al., 2017; van Doremalen et al., 2017; de Wit et al., 2018; Xu et al., 2019). The plethora of potent nAbs have also provided insight into the major antigenic sites on which vaccine development should focus. A list of mAbs that show efficacy in vivo against SARS-CoV and MERS-CoV are summarized in Table 2. Due to the differences in testing conditions, direct comparison of mAb efficacy should be avoided. Multiple comprehensive review articles on the subject can also be found (Prabakaran et al., 2009; Coughlin and Prabhakar, 2012; Xu et al., 2019).
The binding epitopes of some well-studied mAbs provide valuable information for the neutralization mechanism and clinical implications. The majority of the mAbs target the RBD of the spike protein and prevent viral attachment. For example, mAb 80R is able to protect both in vitro and a 16 weeks old mouse model against SARS-CoV Urbani. However, it is unable to protect other strains due to amino acid variations in the RBD (Zhu et al., 2007). On the other hand, S230.15 mimics receptor binding and triggers conformational changes in the SARS S-protein, completely protecting young and old mice from SARS-CoV challenge against multiple SARS-CoV, including Urbani, GD03 and SZ16 (Rockx et al., 2008; Walls et al., 2019a). Similar to SARS-CoV, the majority of mAbs targeting MERS-CoV, such as MERS-4, MERS-27 (Jiang et al., 2014), m336 (Ying et al., 2014, 2015) and humanized Ab 4C2 and 2E6 (Li et al., 2015) all target the RBD and prevent the virus from binding to DPP4 with high potency. Interestingly, the mAb LCA60, isolated from a MERS-CoV infected patient, binds to RBD region and confers a broader neutralizing breadth, and is able to neutralize EMC2012 and London1 strains of MERS-CoV (Corti et al., 2015). Non-RBD targeting Abs G2 and G4 recognize the non-RBD region of S1 and S2 of MERS-CoV, respectively, showing cross-reactivity with multiple MERS-CoV variants and can protect hDPP4 transduced mice from challenge (Wang et al., 2015, 2018). Two nanobodies isolated from camelids, NbMS10 and HCAb-83, show potency in hDPP4 transgenic mice by reducing weight loss and increasing survival after challenge. Interestingly, NbMS10 is able to protect mice as a therapeutic treatment 3 days post infection (Raj et al., 2018; Zhao et al., 2018).
While many mAbs show promising properties for clinical use, two mAbs, REGN3048 and REGN3051, have completed phase 1 clinical trials (NCT03301090). These two mAbs were isolated from VelocImmune mice (expressing the variable regions of human Ig heavy and kappa light chain) immunized with MERS S-protein. Both mAbs show picomolar binding and inhibition of MERS pseudo particles transduction on Huh7 cells. REGN3048 is able to neutralize seven natural isolates of S variants, and IP injection of the mAb at 1 day before or after challenge reduces MERS-CoV RNA levels and lung pathologies in an hDPP4 transgenic mouse model (Pascal et al., 2015). Given the acute severe phase that is associated with emerging CoV infections, a major hurdle for therapeutic antibodies and drugs is that early administration will likely prove most efficacious in clinical care, as has also been shown with influenza virus, Ebola virus and RSV immunotherapeutics (Olinger et al., 2012; Qiu et al., 2012; Fry et al., 2014; Rezaee et al., 2017).
Fusion and Viral Protease Inhibitors
Another critical step for CoV life cycle is membrane fusion and the subsequent release of the RNA genome for replication (Millet and Whittaker, 2018). Membrane fusion of CoVs is governed by the S2 domain of the S-protein (Pallesen et al., 2017; Tortorici and Veesler, 2019). The S2 stem undergoes a major conformational change at the two heptad repeat regions (HR1 and HR2) to bring the host and viral membrane in close proximity for fusion pore formation (Yuan et al., 2017; Walls et al., 2019b). Multiple peptidomimetic fusion inhibitors that mimic the HR1 and HR2 of either SARS-CoV or MERS-CoV block the formation of the helical core and efficiently inhibit membrane fusion in the micromolar range (Gao et al., 2013; Lu et al., 2014; Xia et al., 2019). A single report has shown treatment of HR2 peptide 5h prior to MERS-CoV challenge in an Ad5-hDPP4 transduction mouse model reduces viral lung titer (Channappanavar et al., 2015). The ability of these drugs to protect in lethal, high titer mouse models has yet to be proven.
CoV nsp3 and nsp5 genes encode the papain-like cysteine protease (PLpro) and 3C-like serine protease (3CLpro), respectively (Perlman and Netland, 2009). PLpro cleaves the polyprotein and separates it into nsp1 to 4 while 3CLpro separates nsp4 to 16 (Ziebuhr et al., 2000; Harcourt et al., 2004). Since polyprotein processing is a critical step for CoV replication and transcription, viral proteases are high priority drug targets. Originally developed as HIV protease inhibitors, LPV and RTV have low micromolar activity against 3CLpro of both SARS-CoV and MERS-CoV in vitro (Wu et al., 2004; De Wilde et al., 2014). Testing in SARS-CoV infected patients has shown beneficial outcomes, including lowering the viral load, reducing the onset of ARDS, and lowering mortality rates with LPV and RTV (see nucleoside analogs) treatments (Chu et al., 2004). However, most of the drug studies were performed using retrospective control, sometimes with unbalanced gender ratios, and no treatment has proven efficacious in a randomized control trial (Zumla et al., 2016a). In a marmoset model, LPV/RTV treatment suggested a modest improvement in clinical and pathological outcome as well as reduction of viral load (Chan et al., 2015). However, due to different pathological consequences of treated and untreated group, viral titers were measured at different time points post-infection and all the experiments were performed with a relatively small number (n = 3) of single gender (male) animals (Chan et al., 2015). SARS-CoV and MERS-CoV infection has been shown to be heavily biased by age and gender, where elders and males experience more severe complications than females in clinical cases and mouse models (Karlberg et al., 2004; Alghamdi et al., 2014; Channappanavar et al., 2017). A single subject in a case study, an elderly patient, survived a severe MERS-CoV infection using a combination therapy of LPV/RTV, IFN1α, and ribavirin (Kim et al., 2016). An ongoing clinical trial in Saudi Arabia has begun to use a combination of LPV/RTV and IFNβ-1b for laboratory-confirmed MERS-CoV infection (NCT02845843). Given the importance of 3CLpro I viral life cycle, it is an attractive target for novel drug development (Kumar et al., 2017).
Nucleoside Analogs and RdRp Inhibitors
Ribavirin is a guanosine analog that targets RNA dependent RNA polymerases (RdRp) and has a broad efficacy against RNA viruses (Loustaud-Ratti et al., 2016). The drug inhibits viral RNA synthesis and has shown efficacy against HCV and RSV (De Clercq et al., 2016). Ribavirin inhibits SARS-CoV and MERS-CoV replication at high concentration in vitro (Tan et al., 2004; Falzarano et al., 2013a), However, ribavirin enhances viral replication in the mouse lung and prolongs viral persistence in a SARS-CoV mouse model (Barnard et al., 2006a). Although not effective as a monotherapy, ribavirin shows a synergistic effect against SARS-CoV and MERS-CoV when combine with IFN (Momattin et al., 2013). In a rhesus macaque model, MERS-CoV infected monkeys show improvement after treating with ribavirin and IFN-α2b (Falzarano et al., 2013b). Furthermore, ribavirin has been used in clinical settings during SARS-CoV and MERS-CoV outbreaks (Omrani et al., 2014; Khalid et al., 2015; Shalhoub et al., 2015). While some studies report positive outcome after treatment with ribavirin + IFN, others suggest no significant improvement (Dicaro et al., 2004; Khalid et al., 2015). Due to the variation in dosage and time of administration and the lack of control, the efficacy of using ribavirin in patients is inconclusive. Mechanistically, studies show that recombinant CoV with a deleted proof-reading exonucleases N (ExoN) shows higher sensitivity toward ribavirin. These results indirectly suggest that the low activity of ribavirin in CoV could be explained by the presence of a proofreading exonucleases N (ExoN, nsp14) which can excise the drug from the viral mRNA in CoVs (Smith et al., 2013; Ferron et al., 2017).
Two nucleoside analogs, β-D-N4-Hydroxycytidine (NHC) and GS-5734 (remdesivir), have shown high efficacy against CoVs and are less sensitive to ExoN (Sheahan et al., 2017, 2020; Agostini et al., 2018, 2019). NHC is a cytidine analog and has recently been shown to inhibit multiple viruses, including influenza virus, RSV and Ebola virus (Reynard et al., 2015; Urakova et al., 2017; Yoon et al., 2018). It has micro-molar EC50 against both alpha and beta CoVs including SARS-CoV and MERS-CoV (Barnard et al., 2004; Pyrc et al., 2006; Agostini et al., 2019). Currently under clinical development for Ebola viruses, remdesivir is a nucleoside prodrug that is effective against multiple other RNA viruses including Nipah viruses, RSV, and CoVs (Lo et al., 2017; Sheahan et al., 2017). Remdesivir has sub-micromolar inhibition concentrations in a broad range of CoVs including SARS-CoV, MERS-CoV, and hCoV-NL63, as well as pre-pandemic bat-CoVs WIV1 and SHC014 in an in vitro human airway epithelial (HAE) model (Sheahan et al., 2017; Agostini et al., 2018). Prophylactic administration (one day pre-infection) of remdesivir can mitigate disease by reducing the viral titer and lung pathology in lethal mouse models challenged with a mouse adapted SARS-CoV MA15. Remdesivir also shows therapeutic activity when administered early at one day post-infection (corresponding to 7–10 days after the onset of symptoms in human infection). However, treatment initiated two days post infection does not improve disease outcomes, although the murine disease model is more compressed than in humans (Sheahan et al., 2017). Importantly, remdesivir shows excellent prophylactic protection. Rhesus macaques are completely protected from MERS-CoV infection as scored by lung pathology and clinical score as well as inhibited viral growth. Therapeutic treatment 12-h post infection shows moderate improvement of clinical outcomes on NHPs (de Wit et al., 2020). The parental nucleoside of remdesivir, GS-441524, has also shown to be effective for treating FIP, a disease caused by the α-CoV FIPV (Murphy et al., 2018; Pedersen et al., 2019). Currently, NHC and remdesivir are the only broadly effective antiviral drugs against all SARS-like, MERS-like, human contemporary, and animal CoVs (Sheahan et al., 2017; Agostini et al., 2018; Murphy et al., 2018; Pedersen et al., 2019). Some antiviral drugs, such as chloroquine and T-705, also show efficacy in vitro and are under consideration for the current COVID-19 outbreak (Wang et al., 2020).
Host Factor Inhibitors and Immuno-Modulators
Host Protease Inhibitors
Like all class I viral fusion proteins, CoV S glycoproteins require proteolytic cleavage by host proteases for membrane fusion and viral entry (Belouzard et al., 2012; White and Whittaker, 2016; Millet and Whittaker, 2018). Two cleavage events have been characterized in SARS-CoV and MERS-CoV. The first cleavage event separates the head (S1) and the fusion stem (S2) by cutting the S1/S2 junction (Millet and Whittaker, 2015). The second cleavage event occurs at the S2’ site which is usually located immediately upstream of the fusion peptide (Belouzard et al., 2009). Multiple proteases have been shown to be involved in the cleavage events, including cathepsin-L, trypsin-like serine proteases, transmembrane serine proteases (TTSP) and proprotein convertases such as furin (Millet and Whittaker, 2015). Cathepsin-L inhibitors MDL28170 and SSAA09E1 block SARS-CoV pseudotyped particle infection in pre-treated 293T cells (Simmons et al., 2005; Adedeji et al., 2013). While a peptidomimetic furin substrate, decanoyl-RVKR-chloromethylketone, has been shown to inhibit cleavage of MERS S-protein and block infection in multiple cell lines including normal human bronchial epithelial cells (NHBE), the in vivo potency of this approach is less certain (Gierer et al., 2014; Millet and Whittaker, 2014; Matsuyama et al., 2018).
Host Receptor Inhibitors
The blocking of receptor interactions is also a target of antiviral development. N-(2-aminoethyl)-1 aziridine-ethanamine (NAAE) blocks the interaction of SARS S-protein and ACE2 and inhibits S-mediated cell-to-cell fusion at millimolar concentrations (Huentelman et al., 2004). Similarly, an Ab blocking DPP4 can also inhibit MERS-CoV infection on primary bronchial epithelial cells (Raj et al., 2013). Importantly, the S-proteins of SARS-CoVs and MERS-CoV interact with their receptors outside of the active sites. Therefore, it would be of interest to develop inhibitors that do not affect the normal function of the host proteins but abolish the interaction of the CoV and receptor. Otherwise, long term inhibition of cellular proteins could have adverse effects on the host. For instance, inhibition of ACE2 may cause hypertension (Danilczyk and Penninger, 2006). Additionally, DPP4 is also responsible for multiple cellular functions including immune homeostasis, stem cell development, metabolism and T-cell regulation, and hence is not an ideal target for MERS-CoV infection (Matteucci and Giampietro, 2016; Ou et al., 2019).
Other Host Factor Inhibitors
Another attractive target to inhibit CoV infection is the host metabolic pathways essential for CoV life cycles. CoVs replicate and transcribe in membrane-bound vesicles derived from the host’s rough ER (Snijder et al., 2006; Stertz et al., 2007; Reggiori et al., 2010). MERS-CoV infection upregulates the biosynthetic pathways of multiple major lipogenic enzymes, including fatty acid synthase (FAS), acetyl-CoA carboxylase (ACC) and HMG-CoA synthase (HMGCS) (Yuan et al., 2019). AM580, which targets the major lipid biosynthesis transactivator n-SREBPs by interfering with and downregulating global lipid synthesis (Goldstein et al., 2006; Yuan et al., 2019), inhibits the replication of multiple viruses including influenza virus, Zika virus, Enterovirus-A71 and MERS-CoV (Yuan et al., 2019). Remarkably, MERS-CoV viral titer is reduced by 1,000- to 1,000,000-fold in the presence of AM580 in Huh7 cells or a human intestinal organoid model, respectively, and IP injection of AM580 for 3 days protects a human hDPP4 transgenic mouse model from MERS-CoV lethal challenge (Yuan et al., 2019).
Immune Modulators
Corticosteroids
SARS-CoV and MERS-CoV both cause lung inflammation and can progress into severe respiratory syndrome. Lacking direct antiviral effect, corticosteroids are an immune suppressor which is administrated to severe patients to alleviate lung inflammation. However, immune suppression could also facilitate viral replication. Therefore, corticosteroids are often administrated with antiviral or other immune modulators, such as IFN which can activate the immune system. Retrospective and prospective clinical studies have shown mixed observation in treating SARS-CoV and MERS-CoV patients with corticosteroids, IFN and ribavirin; while some showed a positive effect, others showed no difference.
IFNs
SARS-CoV and MERS-CoV are able to suppress the induction of IFN synthesis by multiple mechanisms including inhibition of the IFN signal transduction pathways and evading detection by pattern recognition receptors (PRRs) and toll-like receptors (TLRs) (Frieman et al., 2008; Lim et al., 2016; Mubarak et al., 2019). Therefore, external administration of IFN regiments could re-initiate the antiviral immune response in the host. During the SARS-CoV and MERS-CoV outbreaks, IFN-α, IFN-β, and IFN-γ were used in combination with various antiviral drugs, including LPV/RTV, ribavirin, corticosteroids and poly I:C (Zumla et al., 2016a). Although most of the clinical reports are positive, there is no consensus on the efficacy of IFN treatment in CoV infection. In animal model studies, IFN treatment is only effective when administrated at early time point for both SARS-CoV and MERS-CoV (Channappanavar et al., 2016, 2019). Furthermore, many of the clinical studies were confounded by multiple factors, including IFN dosages, combination of different antivirals, and the stage of infection (Mo and Fisher, 2016). For instance, some studies targeted patients in the late stage of infection and show a worse survival rate than average (Al-Tawfiq et al., 2014; Khalid et al., 2014). Based on multiple research reports, IFN-β shows the best efficacy in treating MERS-CoV infection (Chan et al., 2013; Hart et al., 2014; Kim et al., 2016). Currently, an open labeled, well controlled clinical study is aiming to test a set dose of LPV/RTV and IFN-β in treating MERS-CoV. The trial will probably provide insight into treating MERS-CoV infection in human population.
The Current Sars-CoV-2 Outbreak: a Challenge, an Opportunity
In December 2019, a new CoV outbreak started in Wuhan, China, a megacity with a population of 11 million. With an estimated basic reproduction number (R0) of 1.4 – 2.5, SARS-CoV-2 quickly spread to every province in China and began to spread globally by the end of January 2020 (Zhou et al., 2020), and was declared a worldwide pandemic in March 2020 by the World Health Organization. SARS-CoV-2 is capable of human-to-human transmission via either symptomatic or asymptomatic patients (Rothe et al., 2020). The high mutation rate of CoVs and the possibility of super-spreader could potentiate the continuous spreading globally. Originally from bat, the SARS-CoV-2 Is 96% identical to a bat CoV designated RaTG13, isolated from a cave in Yunnan Province, China and belongs to the betacoronavirus 2b family, as does SARS-CoV (Zhou et al., 2020). However, the S-proteins between SARS-CoV and 2019 nCoV share only 76–78% sequence similarity, rendering the current experimental vaccines and antivirals unlikely to be fully protective against SARS-CoV-2. As many group 2b SARS-like CoV have pre-epidemic potential, vaccines and countermeasures should be targeted against all of these strains to maximally protect against current and future threats to the global health and economy (Menachery et al., 2015, 2016).
After two CoV outbreaks in the last two decades, the public health officials and clinicians have experience in preventing spread and treating SARS-CoV-2 patients. Prompt communication between governments, quick quarantine procedures, and rapid viral detection assays have helped minimize SARS-CoV-2 cases in other countries thus far. Additionally, the scientific community has developed novel vaccine strategies and experimental antivirals to fight emerging CoV. New vaccines that specifically targeting SARS-CoV-2 are under development. Moderna had started a phase 1 clinical trial on an RNA vaccine (mRNA-1273) that encodes the prefusion stabilized form of SARS-CoV-2 S-protein (NCT04283461). Although there are no published studies of a RNA vaccine platform against CoV, the RNA platform has shown efficacy against multiple other viral infectious diseases including influenza, rabies, Flavivirus, and Ebola viruses in experimental animal models (Zhang et al., 2019). Furthermore, pre-clinical data of the Zika virus mRNA vaccine (mRNA-1893) from Moderna has shown protection against Zika virus and abrogated maternal transmission of Zika virus in pregnant mice (Richner et al., 2017; Jagger et al., 2019). Broad-spectrum antivirals which have shown efficacy against multiple CoVs have the potential to treat SARS-CoV-2 (Sheahan et al., 2017, 2020). Given the high number of infected patients and at risk individuals, the current situation provides an opportunity to initiate clinical trials for (a) vaccine formulation designed to protect uninfected people, (b) broad spectrum antivirals to treat infected individuals and (c) formulation of immune modulators to alleviate clinical pathologies. A well-designed clinical trial could not only ameliorate the current situation, but also lay the foundation for future CoV outbreaks. Supported by a recent study on SARS-CoV-2 (Wang et al., 2020) and multiple research studies on SARS and MERS-CoVs (Sheahan et al., 2017, 2020; de Wit et al., 2020) and a single clinical report (Holshue et al., 2020), remdesivir hold promises as treatment for SARS-CoV-2. The Chinese government has started a clinical trial using remdesivir for treating SARS-CoV-2 patients with mild to severe symptoms (NCT04252664, NCT04257656). On the clinical side, if time and resources permit, a centralized repository that records and digitizes infection cases would aid future medical and epidemiology studies through machine learning programs. Finally, the current situation also provides an opportunity to develop unconventional treatments, such as gene therapy, to target infectious diseases.
Unconventional Vaccines and Therapeutics – Gene Therapies
The field of gene therapy has undergone rapid growth in the last 10 years. Although mainly focused on rare genetic diseases, its potential in treating infectious diseases should not be discounted. One of the leading vectors, adeno-associated virus (AAV), has proven to be safe for human use and multiple AAV-based gene therapy drugs are approved by the FDA and the EMA (Kay et al., 2000; Kaplitt et al., 2007). So far, the only report of use of AAV in CoV is as a DNA vaccine to deliver SARS-CoV spike protein for immunization (Du et al., 2008). Given the recent developments in human antibody cloning technologies, AAV holds a promising potential to be a hybrid of vaccine and therapeutic which acts as a passive immunization vector to provide protection for the outbreak and as a therapeutic in early time scales.
AAV as a Vector for Passive Immunization Against Emerging CoV
The Vectors – Safety
AAV is a non-pathogenic, non-enveloped, 4.7 kb single-stranded DNA virus belonging to the Dependoparvovirus genus within the family Parvoviridae (Cotmore et al., 2014). AAV infects a wide variety of animals, from bearded dragons to humans. Natural AAV isolates have different tissue tropisms in humans and can be reverse engineered to better fulfill various medical needs. To target SARS-CoV, MERS-CoV and other respiratory virus infections, a human airway tropic AAV is needed. Multiple reports have demonstrated that natural isolates AAV5 (Zabner et al., 2000), AAV6 (Limberis et al., 2009), and AAV9 (Adam et al., 2014) as well as engineered vectors AAV2.5 (Li et al., 2009) and AAV2.5T (Excoffon et al., 2009) are able to transduce human lung epithelial cells, including primary human airway epithelial (HAE) cultures. Although there is a high prevalence of nAbs against AAVs in the human population, new technologies have been developed to engineer AAV to evade humoral immune responses (Tse et al., 2015, 2017). These developments potentially allow for the delivery of CoV vaccines or immunotherapeutic directly to the mucosal compartments of the lung.
The Package – Flexibility
Passive immunization of AAV can be developed as a platform technology in which the nAb can be quickly exchanged to target specific pathogens. Multiple studies have shown passive immunization using AAV is effective against viral infectious diseases such as HIV, (Balazs et al., 2012; Lin and Balazs, 2018) Ebola, (Limberis et al., 2016) influenza, (Balazs et al., 2013; Laursen et al., 2018), and others (Nieto and Salvetti, 2014). The package for delivery is extremely flexible, from authentic immunoglobulins (IgG) to immunoadhesins (IA) to single chain variable fragments (scFv) to bi-specific antibodies (Naso et al., 2017). Furthermore, a combination of Abs, small antiviral peptides and immuno-modulators can be co-delivered at the same time to achieve multidimensional therapy. Although AAV-based passive immunization has not yet been tested as therapeutic, it could serve as a fast-acting prophylactic alternative to traditional vaccines.
The Timing – Quick
The most important aspect to control an outbreak is to reduce the spread by protecting the population from infection. However, there is a lag time between the beginning of an outbreak and the development of an effective vaccine in which the population is completely vulnerable. Prophylactic treatments, such as infusion of Abs and antivirals, could protect individual for a short duration. However, these prophylactic treatments require constant intake to stay effective and are toxic as well as financially impractical for long-term use (Hansel et al., 2010). AAV-based passive immunization could perfectly fill the vacuum by protecting the population before the arrival of a vaccine. Since AAV is a platform technology, anti-viral packages can be swapped and tested quickly, within a month (Strobel et al., 2019). After administration in animals, protection can be achieved in less than a week, faster than any vaccine strategies. In an influenza study, AAV9 delivery of IA via intranasal inoculation protected animals from lethal influenza challenges including H5N1, H1N1 and H1N1 1918 within 3 days of AAV administration (Limberis et al., 2013). Unlike traditional gene therapy in which the transgene lasts for long periods of time, the natural turnover rate of airway epithelia means that the nAb introduction is not permanent, reducing the chance that the host will produce antibodies targeting the therapeutic antibody delivered by AAV (anti-drug antibody responses) (Nieto and Salvetti, 2014). Therefore, AAV-based passive immunization is a quick and excellent option to deploy in an outbreak situation for emerging infectious diseases.
The Challenges
AAV-based gene therapy has great potential for treating viral infectious diseases. However, there are multiple hurdles for AAV-based gene therapy to achieve its full potential. (1) Low transduction efficiency, (2) pre-existing nAb against AAVs, (3) transgene toxicity and loss of expression, and (4) extremely high price tag (Colella et al., 2018; Kaemmerer, 2018). Fortunately, multiple strategies have been developed to address these hurdles (Tse et al., 2015). For instance, through vector engineering, a new generation of AAV vectors can evade pre-existing nAbs while retaining a good transduction profile in the respiratory system (Li et al., 2009; Tse et al., 2017). For rare genetic diseases, life-long gene expression is important for therapeutic purposes but could cause toxicity. On the contrary, for emerging CoVs, the goal is a short-term protection from the virus. Therefore, AAV can target epithelial cells that have a regular turn-over rate, hence providing short-term protection and preventing transgene toxicity. AAV-based therapies are known for their extremely high prices, sometime up to a million US dollar per treatment. However, the prices are inflated due to the small market size of rare genetic diseases and the cost for drug development. Given the huge market size of infectious diseases, the price for AAV-based therapy for infectious diseases should be more reasonable. Unlike rare genetic diseases and other infectious diseases, an emerging CoV outbreak is uniquely suitable for AAV-based passive immunization as a short-term protection therapy before vaccine deployment.
Conclusion
The continuous development of vaccines, antivirals, and hopefully gene therapies will provide an arsenal for combating and controlling emerging CoV diseases. For vaccine development, understanding the antigenicity and neutralizing antibody footprints of different CoVs could aid the development of broad-spectrum vaccines and, ultimately, the possibility of a universal CoV vaccine. Vaccine formulation, including dosage and adjuvants, should be tested systemically and, if possible, on animal models that reflects the genetic variation of the human population. A deeper understanding of the basic biology of CoV and host-virus interaction can lead to the discovery of druggable targets. Importantly, we should not overlook the potential of the existing broad-spectrum antivirals and should start clinical trials on these drugs in a timely fashion. Prevention is always the best treatment, and constant viral surveillance of wild animals for potential emerging CoVs is extremely important. Testing the outbreak potential of heterologous SARS- and MERS-like viruses with different spike proteins could better prepare society from the next outbreak. A centralized digital database that collect public health and clinical information of the current outbreak would allow global retrospective studies in the future using machine learning and other big data analysis. On the research side, quick, reliable and easily employed viral testing kits should be developed. Innovative technologies such as gene therapy should be adequately explored for their potential to combat CoVs and act as another line of defense against elusive emerging viral diseases. The current SARS-CoV-2 poses a huge challenge for society; however, given experience with emerging CoVs and global effort, it is hoped that the impact of the outbreak will be minimal.
Author Contributions
LT wrote the review. RM, RG, and RB reviewed and revised the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
The manuscript presented here were supported by grants from the National Institute of Allergy and Infectious Disease of the United States National Institutes of Health (NIH) by awards AI108197, AI110700, AI132178, and AI149644 (RB). Figure 2 is generated using biorender (https://biorender.com/).
Acknowledgments
We thank Dr. Timothy Sheahan and Dr. Yixuan Hou for critical reading of this manuscript and all members in the RB laboratory for helpful discussions.
References
Adam, V. S., Crosariol, M., Kumar, S., Ge, M. Q., Czack, S. E., Roy, S., et al. (2014). Adeno-associated virus 9-mediated airway expression of antibody protects old and immunodeficient mice against influenza virus. Clin. Vaccine Immunol. 21, 1528–1533. doi: 10.1128/CVI.00572-514
Adedeji, A. O., Severson, W., Jonsson, C., Singh, K., Weiss, S. R., and Sarafianos, S. G. (2013). Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 87, 8017–8028. doi: 10.1128/jvi.00998-913
Adney, D. R., Wang, L., Van Doremalen, N., Shi, W., Zhang, Y., Kong, W., et al. (2019). Efficacy of an adjuvanted middle east respiratory syndrome coronavirus spike protein vaccine in dromedary camels and alpacas. Viruses 11:212. doi: 10.3390/v11030212
Agnihothram, S., Gopal, R., Yount, B. L. J., Donaldson, E. F., Menachery, V. D., Graham, R. L., et al. (2014). Evaluation of serologic and antigenic relationships between middle eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. J. Infect. Dis. 209, 995–1006. doi: 10.1093/infdis/jit609
Agostini, M. L., Andres, E. L., Sims, A. C., Graham, R. L., Sheahan, T. P., Lu, X., et al. (2018). Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 9, 1–15. doi: 10.1128/mBio.00221-218
Agostini, M. L., Pruijssers, A. J., Chappell, J. D., Gribble, J., Lu, X., Andres, E. L., et al. (2019). Small-molecule antiviral β-d-N4-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 93:e01348-19. doi: 10.1128/JVI.01348-1319
Agrawal, A. S., Tao, X., Algaissi, A., Garron, T., Narayanan, K., Peng, B. H., et al. (2016). Immunization with inactivated Middle East Respiratory Syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum. Vaccines Immunother. 12, 2351–2356. doi: 10.1080/21645515.2016.1177688
Ahmed, A. E. (2019). Diagnostic delays in Middle East respiratory syndrome coronavirus patients and health systems. J. Infect. Public Health 12, 767–771. doi: 10.1016/j.jiph.2019.04.002
Al Kahlout, R. A., Nasrallah, G. K., Farag, E. A., Wang, L., Lattwein, E., Müller, M. A., et al. (2019). Comparative serological study for the prevalence of Anti-MERS coronavirus antibodies in high- and low-risk groups in qatar. J. Immunol. Res. 2019:1386740. doi: 10.1155/2019/1386740
Al-Amri, S. S., Abbas, A. T., Siddiq, L. A., Alghamdi, A., Sanki, M. A., Al-Muhanna, M. K., et al. (2017). Immunogenicity of candidate MERS-CoV DNA vaccines based on the spike protein. Sci. Rep. 7:44875. doi: 10.1038/srep44875
Alghamdi, I. G., Hussain, I. I., Almalki, S. S., Alghamdi, M. S., Alghamdi, M. M., and El-Sheemy, M. A. (2014). The pattern of Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive epidemiological analysis of data from the Saudi Ministry of Health. Int. J. Gen. Med. 7, 417–423. doi: 10.2147/IJGM.S67061
Alharbi, N. K., Padron-Regalado, E., Thompson, C. P., Kupke, A., Wells, D., Sloan, M. A., et al. (2017). ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine 35, 3780–3788. doi: 10.1016/j.vaccine.2017.05.032
Almazán, F., Dediego, M. L., Sola, I., Zuñiga, S., Nieto-torres, J. L., Marquez-jurado, S., et al. (2013). A Vaccine candidate east respiratory syndrome coronavirus as a vaccine candidate. mBio 4, 1–11. doi: 10.1128/mBio.00650-13.Editor
Al-Tawfiq, J. A., Momattin, H., Dib, J., and Memish, Z. A. (2014). Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int. J. Infect. Dis. 20, 42–46. doi: 10.1016/j.ijid.2013.12.003
Arabi, Y. M., Hajeer, A. H., Luke, T., Raviprakash, K., Balkhy, H., Johani, S., et al. (2016). Feasibility of using convalescent plasma immunotherapy for MERS-CoV infection, Saudi Arabia. Emerg. Infect. Dis. 22, 1554–1561. doi: 10.3201/eid2209.151164
Balazs, A. B., Bloom, J. D., Hong, C. M., Rao, D. S., and Baltimore, D. (2013). Broad protection against influenza infection by vectored immunoprophylaxis in mice. Nat. Biotechnol. 31:647. doi: 10.1038/nbt.2618
Balazs, A. B., Chen, J., Hong, C. M., Rao, D. S., Yang, L., and Baltimore, D. (2012). Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481, 81–84. doi: 10.1038/nature10660
Barnard, D. L., Day, C. W., Bailey, K., Heiner, M., Montgomery, R., Lauridsen, L., et al. (2006a). Enhancement of the infectivity of SARS-CoV in BALB/c mice by IMP dehydrogenase inhibitors, including ribavirin. Antiviral Res. 71, 53–63. doi: 10.1016/j.antiviral.2006.03.001
Barnard, D. L., Day, C. W., Bailey, K., Heiner, M., Montgomery, R., Lauridsen, L., et al. (2006b). Evaluation of immunomodulators, interferons and known in vitro SARS-CoV inhibitors for inhibition of SARS-CoV replication in BALB/c mice. Antivir. Chem. Chemother. 17, 275–284. doi: 10.1177/095632020601700505
Barnard, D. L., Hubbard, V. D., Burton, J., Smee, D. F., Morrey, J. D., Otto, M. J., et al. (2004). Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and β-D-N4-hydroxycytidine. Antivir. Chem. Chemother. 15, 15–22. doi: 10.1177/095632020401500102
Belouzard, S., Chu, V. C., and Whittaker, G. R. (2009). Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 106, 5871–5876. doi: 10.1073/pnas.0809524106
Belouzard, S., Millet, J. K., Licitra, B. N., and Whittaker, G. R. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4, 1011–1033. doi: 10.3390/v4061011
Belshe, R. B., Gruber, W. C., Mendelman, P. M., Mehta, H. B., Mahmood, K., Reisinger, K., et al. (2000). Correlates of immune protection induced by live, attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine. J. Infect. Dis. 181, 1133–1137. doi: 10.1086/315323
Binnie, A., Tsang, J. L. Y., and Dos Santos, C. C. (2014). Biomarkers in acute respiratory distress syndrome. Curr. Opin. Crit. Care 20, 47–55. doi: 10.1097/MCC.0000000000000048
Bisht, H., Roberts, A., Vogel, L., Bukreyev, A., Collins, P. L., Murphy, B. R., et al. (2004). Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U.S.A. 101, 6641–6646. doi: 10.1073/pnas.0401939101
Bisht, H., Roberts, A., Vogel, L., Subbarao, K., and Moss, B. (2005). Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology 334, 160–165. doi: 10.1016/j.virol.2005.01.042
Bodmer, B. S., Fiedler, A. H., Hanauer, J. R. H., and Mühlebach, M. D. (2018). Live-attenuated bivalent measles virus-derived vaccines targeting Middle East respiratory syndrome coronavirus induce robust and multifunctional T cell responses against both viruses in an appropriate mouse model. Virology 521, 99–107. doi: 10.1016/j.virol.2018.05.028
Bolles, M., Deming, D., Long, K., Agnihothram, S., Whitmore, A., Ferris, M., et al. (2011a). A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 85, 12201–12215. doi: 10.1128/jvi.06048-6011
Bolles, M., Donaldson, E., and Baric, R. (2011b). SARS-CoV and emergent coronaviruses: viral determinants of interspecies transmission. Curr. Opin. Virol. 1, 624–634. doi: 10.1016/j.coviro.2011.10.012
Briese, T., Mishra, N., Jain, K., East, M., Syndrome, R., Quasispecies, C., et al. (2014). Dromedary camels in Saudi Arabia include homologues of human isolates revealed through whole-genome analysis etc. mBio 5, 1–5. doi: 10.1128/mBio.01146-14.Editor
Buchholz, U. J., Bukreyev, A., Yang, L., Lamirande, E. W., Murphy, B. R., Subbarao, K., et al. (2004). Contributions of the structural proteins of severe respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. U.S.A. 101, 9804–9809. doi: 10.1073/pnas.0403492101
Bukreyev, A., Lamirande, E. W., Buchholz, U. J., Vogel, L. N., Elkins, W. R., St Claire, M., et al. (2004). Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet 363, 2122–2127. doi: 10.1016/S0140-6736(04)16501-X
Cabeça, T. K., Granato, C., and Bellei, N. (2013). Epidemiological and clinical features of human coronavirus infections among different subsets of patients. Influenza Other Respir. Viruses 7, 1040–1047. doi: 10.1111/irv.12101
Cameron, M. J., Ran, L., Xu, L., Danesh, A., Bermejo-Martin, J. F., Cameron, C. M., et al. (2007). Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 81, 8692–8706. doi: 10.1128/jvi.00527-527
Chan, J. F. W., Chan, K. H., Kao, R. Y. T., To, K. K. W., Zheng, B. J., Li, C. P. Y., et al. (2013). Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 67, 606–616. doi: 10.1016/j.jinf.2013.09.029
Chan, J. F. W., Yao, Y., Yeung, M. L., Deng, W., Bao, L., Jia, L., et al. (2015). Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERSCoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 212, 1904–1913. doi: 10.1093/infdis/jiv392
Channappanavar, R., Fehr, A. R., Vijay, R., Mack, M., Zhao, J., Meyerholz, D. K., et al. (2016). Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19, 181–193. doi: 10.1016/j.chom.2016.01.007
Channappanavar, R., Fehr, A. R., Zheng, J., Wohlford-Lenane, C., Abrahante, J. E., Mack, M., et al. (2019). IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 129, 3625–3639. doi: 10.1172/JCI126363
Channappanavar, R., Fett, C., Mack, M., Ten Eyck, P. P., Meyerholz, D. K., and Perlman, S. (2017). Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J. Immunol. 198, 4046–4053. doi: 10.4049/jimmunol.1601896
Channappanavar, R., Lu, L., Xia, S., Du, L., Meyerholz, D. K., Perlman, S., et al. (2015). Protective effect of intranasal regimens containing peptidic middle east respiratory syndrome coronavirus fusion Inhibitor against MERS-CoV infection. J. Infect. Dis. 212, 1894–1903. doi: 10.1093/infdis/jiv325
Channappanavar, R., Zhao, J., and Perlman, S. (2014). T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 59, 118–128. doi: 10.1007/s12026-014-8534-z
Chen, Z., Bao, L., Chen, C., Zou, T., Xue, Y., Li, F., et al. (2017). Human neutralizing monoclonal antibody inhibition of Middle East respiratory syndrome coronavirus replication in the common marmoset. J. Infect. Dis. 215, 1807–1815. doi: 10.1093/infdis/jix209
Chi, H., Zheng, X., Wang, X., Wang, C., Wang, H., Gai, W., et al. (2017). DNA vaccine encoding Middle East respiratory syndrome coronavirus S1 protein induces protective immune responses in mice. Vaccine 35, 2069–2075. doi: 10.1016/j.vaccine.2017.02.063
Chirmule, N., Propert, K. J., Magosin, S. A., Qian, Y., Qian, R., and Wilson, J. M. (1999). Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 6, 1574–1583. doi: 10.1038/sj.gt.3300994
Chu, C. M., Cheng, V. C. C., Hung, I. F. N., Wong, M. M. L., Chan, K. H., Chan, K. S., et al. (2004). Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 59, 252–256. doi: 10.1136/thorax.2003.012658
Colella, P., Ronzitti, G., and Mingozzi, F. (2018). Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 8, 87–104. doi: 10.1016/j.omtm.2017.11.007
Coleman, C. M., Liu, Y. V., Mu, H., Taylor, J. K., Massare, M., Flyer, D. C., et al. (2014). Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine 32, 3169–3174. doi: 10.1016/j.vaccine.2014.04.016
Coleman, C. M., Venkataraman, T., Liu, Y. V., Glenn, G. M., Smith, G. E., Flyer, D. C., et al. (2017). MERS-CoV spike nanoparticles protect mice from MERS-CoV infection. Vaccine 35, 1586–1589. doi: 10.1016/j.vaccine.2017.02.012
Corman, V. M., Albarrak, A. M., Omrani, A. S., Albarrak, M. M., Farah, M. E., Almasri, M., et al. (2015). Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 62, 477–483. doi: 10.1093/cid/civ951
Corti, D., Passini, N., Lanzavecchia, A., and Zambon, M. (2016). Rapid generation of a human monoclonal antibody to combat Middle East respiratory syndrome. J. Infect. Public Health 9, 231–235. doi: 10.1016/j.jiph.2016.04.003
Corti, D., Zhao, J., Pedotti, M., Simonelli, L., Agnihothram, S., Fett, C., et al. (2015). Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. U.S.A. 112, 10473–10478. doi: 10.1073/pnas.1510199112
Cotmore, S. F., Agbandje-McKenna, M., Chiorini, J. A., Mukha, D. V., Pintel, D. J., Qiu, J., et al. (2014). The family Parvoviridae. Arch. Virol. 159, 1239–1247. doi: 10.1007/s00705-013-1914-1911
Coughlin, M. M., and Prabhakar, B. S. (2012). Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: target, mechanism of action, and therapeutic potential. Rev. Med. Virol. 22, 2–17. doi: 10.1002/rmv.706
Czub, M., Weingartl, H., Czub, S., He, R., and Cao, J. (2005). Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine 23, 2273–2279. doi: 10.1016/j.vaccine.2005.01.033
Danilczyk, U., and Penninger, J. M. (2006). Angiotensin-converting enzyme II in the heart and the kidney. Circ. Res. 98, 463–471. doi: 10.1161/01.RES.0000205761.22353.5f
De Clercq, E., Li, G., Clercq, E., and De (2016). Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 29, 695–747. doi: 10.1128/CMR.00102-15.Address
De Wilde, A. H., Jochmans, D., Posthuma, C. C., Zevenhoven-Dobbe, J. C., Van Nieuwkoop, S., Bestebroer, T. M., et al. (2014). Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 58, 4875–4884. doi: 10.1128/AAC.03011-3014
de Wit, E., Feldmann, F., Cronin, J., Jordan, R., Okumura, A., Thomas, T., et al. (2020). Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U.S.A. 117, 6771–6776. doi: 10.1073/pnas.1922083117
de Wit, E., Feldmann, F., Horne, E., Okumura, A., Cameroni, E., Haddock, E., et al. (2019). Prophylactic efficacy of a human monoclonal antibody against MERS-CoV in the common marmoset. Antiviral Res. 163, 70–74. doi: 10.1016/j.antiviral.2019.01.016
de Wit, E., Feldmann, F., Okumura, A., Horne, E., Haddock, E., Saturday, G., et al. (2018). Prophylactic and therapeutic efficacy of mAb treatment against MERS-CoV in common marmosets. Antiviral Res. 156, 64–71. doi: 10.1016/j.antiviral.2018.06.006
de Wit, E., Van Doremalen, N., Falzarano, D., and Munster, V. J. (2016). SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14, 523–534. doi: 10.1038/nrmicro.2016.81
Deming, D., Sheahan, T., Heise, M., Yount, B., Davis, N., Sims, A., et al. (2006). Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 3:e0030525. doi: 10.1371/journal.pmed.0030525
Deng, X., StJohn, S. E., Osswald, H. L., O’Brien, A., Banach, B. S., Sleeman, K., et al. (2014). Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J. Virol. 88, 11886–11898. doi: 10.1128/jvi.01528-1514
Deng, Y., Lan, J., Bao, L., Huang, B., Ye, F., Chen, Y., et al. (2018). Enhanced protection in mice induced by immunization with inactivated whole viruses compare to spike protein of middle east respiratory syndrome coronavirus. Emerg. Microbes Infect. 7:60. doi: 10.1038/s41426-018-0056-57
Denison, M. R., Graham, R. L., Donaldson, E. F., Eckerle, L. D., and Baric, R. S. (2011). Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 8, 270–279. doi: 10.4161/rna.8.2.15013
Dicaro, A., Li, Y., Strong, J. E., Aoki, F., Plummer, F., Jones, S. M., et al. (2004). Severe acute respiratory syndrome - related coronavirus is inhibited by interferon- a. J. Infect. Dis. 189, 1164–1167. doi: 10.1086/382597
Ding, Y., Wang, H., Shen, H., Li, Z., Geng, J., Han, H., et al. (2003). The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 200, 282–289. doi: 10.1002/path.1440
Drake, J. W., and Holland, J. J. (1999). Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U.S.A. 96, 13910–13913. doi: 10.1073/pnas.96.24.13910
Drosten, C., Chiu, L. L., Panning, M., Leong, H. N., Preiser, W., Tam, J. S., et al. (2004). Evaluation of advanced reverse transcription-PCR assays and an alternative PCR target region for detection of severe acute respiratory syndrome-associated Coronavirus. J. Clin. Microbiol. 42, 2043–2047. doi: 10.1128/JCM.42.5.2043-2047.2004
Du, L., and Jiang, S. (2015). Middle East respiratory syndrome: current status and future prospects for vaccine development. Expert Opin. Biol. Ther. 15, 1647–1651. doi: 10.1517/14712598.2015.1092518
Du, L., Kou, Z., Ma, C., Tao, X., Wang, L., Zhao, G., et al. (2013). A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses?: implication for developing therapeutics and vaccines. PLoS One 8:e0081587. doi: 10.1371/journal.pone.0081587
Du, L., Zhao, G., Chan, C. C. S., and Li, L. (2010). A 219-mer CHO-Expressing Receptor-Binding Domain. Viral Immunol. 23, 211–219. doi: 10.1089/vim.2009.0090
Du, L., Zhao, G., Lin, Y., Sui, H., Chan, C., Ma, S., et al. (2008). Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-. J. Immunol. 180, 948–956. doi: 10.4049/jimmunol.180.2.948
Du, L., Zhao, G., Yang, Y., Qiu, H., Wang, L., Kou, Z., et al. (2014). A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J. Virol. 88, 7045–7053. doi: 10.1128/jvi.00433-14
Eckerle, L. D., Becker, M. M., Halpin, R. A., Li, K., Venter, E., Lu, X., et al. (2010). Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6:e1000896. doi: 10.1371/journal.ppat.1000896
Eckerle, L. D., Lu, X., Sperry, S. M., Choi, L., and Denison, M. R. (2007). High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 81, 12135–12144. doi: 10.1128/jvi.01296-1297
Excoffon, K. J. D. A., Koerber, J. T., Dickey, D. D., Murtha, M., Keshavjee, S., Kaspar, B. K., et al. (2009). Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc. Natl. Acad. Sci. U.S.A. 106, 3865–3870. doi: 10.1073/pnas.0813365106
Falzarano, D., De Wit, E., Martellaro, C., Callison, J., Munster, V. J., and Feldmann, H. (2013a). Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci. Rep. 3, 1–6. doi: 10.1038/srep01686
Falzarano, D., de Wit, E., Rasmussen, A. L., Feldmann, F., Okumura, A., Scott, D. P., et al. (2013b). Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat. Med. 19, 1313–1317. doi: 10.1038/nm.3362
Ferron, F., Subissi, L., Theresa, A., Morais, S. De, Thi, N., Le, T., et al. (2017). Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U.S.A. 115, E162–E171. doi: 10.1073/pnas.1718806115
Frieman, M., Heise, M., and Baric, R. (2008). SARS coronavirus and innate immunity. Virus Res. 133, 101–112. doi: 10.1016/j.virusres.2007.03.015
Fry, A. M., Goswami, D., Nahar, K., Sharmin, A. T., Rahman, M., Gubareva, L., et al. (2014). Efficacy of oseltamivir treatment started within 5 days of symptom onset to reduce infl uenza illness duration and virus shedding in an urban setting in Bangladesh?: a randomised placebo-controlled trial. Lancet Infect. Dis. 14, 109–118. doi: 10.1016/S1473-3099(13)70267-70266
Gao, J., Lu, G., Qi, J., Li, Y., Wu, Y., Deng, Y., et al. (2013). Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of middle east respiratory syndrome coronavirus. J. Virol. 87, 13134–13140. doi: 10.1128/jvi.02433-2413
Gierer, S., Müller, M. A., Heurich, A., Ritz, D., Springstein, B. L., Karsten, C. B., et al. (2014). Inhibition of proprotein convertases abrogates processing of the middle eastern respiratory syndrome coronavirus spike protein in infected cells but does not reduce viral infectivity. J. Infect. Dis. 211, 889–897. doi: 10.1093/infdis/jiu407
Goldstein, J. L., DeBose-Boyd, R. A., and Brown, M. S. (2006). Protein sensors for membrane sterols. Cell 124, 35–46. doi: 10.1016/j.cell.2005.12.022
Graham, B. S., and Ambrosino, D. M. (2015). History of passive antibody administration for prevention and treatment of infectious diseases. Curr. Opin. HIV AIDS 10, 129–134. doi: 10.1097/COH.0000000000000154
Graham, R. L., Becker, M. M., Eckerle, L. D., Bolles, M., Denison, M. R., and Baric, R. S. (2012). A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 18, 1820–1826. doi: 10.1038/nm.2972
Graham, R. L., Deming, D. J., Deming, M. E., Yount, B. L., and Baric, R. S. (2018). Evaluation of a recombination-resistant coronavirus as a broadly applicable, rapidly implementable vaccine platform. Commun. Biol. 1:179. doi: 10.1038/s42003-018-0175-177
Gralinski, L. E., and Baric, R. S. (2015). Molecular pathology of emerging coronavirus infections. J. Pathol. 235, 185–195. doi: 10.1002/path.4454
Greenough, T. C., Babcock, G. J., Roberts, A., Hernandez, H. J., and Thomas, W. D. Jr. (2005). Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J. Infect. Dis. 191, 507–514. doi: 10.1086/427242
Guo, X., Deng, Y., Chen, H., Lan, J., Wang, W., Zou, X., et al. (2015). Systemic and mucosal immunity in mice elicited by a single immunization with human adenovirus type 5 or 41 vector-based vaccines carrying the spike protein of Middle East respiratory syndrome coronavirus. Immunology 145, 476–484. doi: 10.1111/imm.12462
Haagmans, B. L., Kuiken, T., Martina, B. E., Fouchier, R. A. M., Rimmelzwaan, G. F., van Amerongen, G., et al. (2004). Pegylated interferon-α protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 10, 290–293. doi: 10.1038/nm1001
Haagmans, B. L., van den Brand, J. M. A., Raj, V. S., Volz, A., Wohlsein, P., Smits, S. L., et al. (2016). An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science 351, 77–81. doi: 10.1126/science.aad1283
Hansel, T. T., Kropshofer, H., Singer, T., Mitchell, J. A., and George, A. J. T. (2010). The safety and side effects of monoclonal antibodies. Nat. Rev. Drug Discov. 9, 325–338. doi: 10.1038/nrd3003
Hapfelmeier, S., Lawson, M. A. E., Slack, E., Kirundi, J. K., Stoel, M., Heikenwalder, M., et al. (2010). Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709. doi: 10.1126/science.1188454
Harcourt, B. H., Jukneliene, D., Kanjanahaluethai, A., Bechill, J., Severson, K. M., Smith, C. M., et al. (2004). Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78, 13600–13612. doi: 10.1128/JVI.78.24.13600
Hart, B. J., Dyall, J., Postnikova, E., Zhou, H., Kindrachuk, J., Johnson, R. F., et al. (2014). Interferon-β and mycophenolic acid are potent inhibitors of middle east respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 95, 571–577. doi: 10.1099/vir.0.061911-61910
Hashem, A. M., Algaissi, A., Agrawal, A. S., Al-Amri, S. S., Alhabbab, R. Y., Sohrab, S. S., et al. (2019). A highly immunogenic, protective, and safe adenovirus-based vaccine expressing middle east respiratory syndrome coronavirus S1-CD40L fusion protein in a transgenic human dipeptidyl peptidase 4 mouse model. J. Infect. Dis. 220, 1558–1567. doi: 10.1093/infdis/jiz137
Holshue, M. L., DeBolt, C., Lindquist, S., Lofy, K. H., Wiesman, J., Bruce, H., et al. (2020). First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382, 929–936. doi: 10.1056/nejmoa2001191
Houser, K. V., Gretebeck, L., Ying, T., Wang, Y., Vogel, L., Lamirande, E. W., et al. (2016). Prophylaxis with a Middle East respiratory syndrome coronavirus (MERS-CoV)-specific human monoclonal antibody protects rabbits from MERS-CoV infection. J. Infect. Dis. 213, 1557–1561. doi: 10.1093/infdis/jiw080
Hsueh, P. R., Huang, L. M., Chen, P. J., Kao, C. L., and Yang, P. C. (2004). Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 10, 1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x
Hu, M. C., Jones, T., Kenney, R. T., Barnard, D. L., Burt, D. S., and Lowell, G. H. (2007). Intranasal Protollin-formulated recombinant SARS S-protein elicits respiratory and serum neutralizing antibodies and protection in mice. Vaccine 25, 6334–6340. doi: 10.1016/j.vaccine.2007.06.017
Huang, J. D., Zheng, B. J., and Sun, H. Z. (2008). Helicases as antiviral drug targets. Hong Kong Med. J. 14, 36–38.
Huentelman, M. J., Zubcevic, J., Hernández Prada, J. A., Xiao, X., Dimitrov, D. S., Raizada, M. K., et al. (2004). Sructure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension 44, 903–906. doi: 10.1161/01.HYP.0000146120.29648.36
Hui, D. S., Azhar, I. E., Madani, T. A., Ntoumi, F., Kock, R., Dar, O., et al. (2020). The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — The latest 2019 novel coronavirus outbreak in Wuhan. China Int. J. Infect. Dis. 91, 264–266. doi: 10.1016/j.ijid.2020.01.009
Iwata-Yoshikawa, N., Uda, A., Suzuki, T., Tsunetsugu-Yokota, Y., Sato, Y., Morikawa, S., et al. (2014). Effects of toll-like receptor stimulation on eosinophilic infiltration in lungs of BALB/c mice immunized with UV-inactivated severe acute respiratory syndrome-related coronavirus vaccine. J. Virol. 88, 8597–8614. doi: 10.1128/jvi.00983-914
Jagger, B. W., Dowd, K. A., Chen, R. E., Desai, P., Foreman, B., Burgomaster, K. E., et al. (2019). Protective efficacy of nucleic acid vaccines against transmission of zika virus during pregnancy in mice. J. Infect. Dis. 220, 1577–1588. doi: 10.1093/infdis/jiz338
Jiaming, L., Yanfeng, Y., Yao, D., Yawei, H., Linlin, B., Baoying, H., et al. (2017). The recombinant N-terminal domain of spike proteins is a potential vaccine against Middle East respiratory syndrome coronavirus (MERS-CoV) infection. Vaccine 35, 10–18. doi: 10.1016/j.vaccine.2016.11.064
Jiang, L., Wang, N., Zuo, T., Shi, X., Poon, K.-M. M. V., Wu, Y., et al. (2014). Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 6:234ra59. doi: 10.1126/scitranslmed.3008140
Johnson, R. F., Bagci, U., Keith, L., Tang, X., Mollura, D. J., Zeitlin, L., et al. (2016). 3B11-N, a monoclonal antibody against MERS-CoV, reduces lung pathology in rhesus monkeys following intratracheal inoculation of MERS-CoV Jordan-n3/2012. Virology 490, 49–58. doi: 10.1016/j.virol.2016.01.004
Kaemmerer, W. F. (2018). How will the field of gene therapy survive its success? Bioeng. Transl. Med. 3, 166–177. doi: 10.1002/btm2.10090
Kam, Y. W., Kien, F., Roberts, A., Cheung, Y. C., Lamirande, E. W., Vogel, L., et al. (2007). Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine 25, 729–740. doi: 10.1016/j.vaccine.2006.08.011
Kandeel, M. (2018). Bioinformatics analysis of the recent MERS-CoV with special reference to the virus-encoded Spike protein. Mol. Enzymol. Drug Targets 01, 1–10. doi: 10.21767/2572-5475.10001
Kapadia, S. U., Rose, J. K., Lamirande, E., Vogel, L., Subbarao, K., and Roberts, A. (2005). Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology 340, 174–182. doi: 10.1016/j.virol.2005.06.016
Kaplitt, M. G., Feigin, A., Tang, C., Fitzsimons, H. L., Mattis, P., Lawlor, P. A., et al. (2007). Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson’s disease: an open label, phase I trial. Lancet 369, 2097–2105. doi: 10.1016/S0140-6736(07)60982-60989
Karlberg, J., Chong, D. S. Y., and Lai, W. Y. Y. (2004). Do men have a higher case fatality rate of severe acute respiratory syndrome than women do? Am. J. Epidemiol. 159, 229–231. doi: 10.1093/aje/kwh056
Kay, M. A., Manno, C. S., Ragni, M. V., Larson, P. J., Couto, L. B., McClelland, A., et al. (2000). Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat. Genet. 24, 257–261. doi: 10.1038/73464
Khalid, M., Al Rabiah, F., Khan, B., Al Mobeireek, A., Butt, T. S., and Al Mutairy, E. (2015). Ribavirin and interferon-alpha2b as primary and preventive treatment for Middle East respiratory syndrome coronavirus: a preliminary report of two cases. Antivir. Ther. 20, 87–91. doi: 10.3851/IMP2792
Khalid, M., Khan, B., Al Rabiah, F., Alismaili, R., Saleemi, S., Rehan-Khaliq, A. M., et al. (2014). Middle eastern respiratory syndrome corona virus (MERS CoV): case reports from a tertiary care hospital in Saudi Arabia. Ann. Saudi Med. 34, 396–400. doi: 10.5144/0256-4947.2014.396
Kim, E., Okada, K., Kenniston, T., Raj, V. S., AlHajri, M. M., Farag, E. A. B. A. B. A., et al. (2014). Immunogenicity of an adenoviral-based Middle East Respiratory syndrome coronavirus vaccine in BALB/c mice. Vaccine 32, 5975–5982. doi: 10.1016/j.vaccine.2014.08.058
Kim, U. J., Won, E.-J., Kee, S.-J., Jung, S.-I., and Jang, H.-C. (2016). Combination therapy with lopinavir/ritonavir, ribavirin and interferon-alpha for Middle East respiratory syndrome. Antivir. Ther. 21, 455–459. doi: 10.3851/IMP3002
Kirchdoerfer, R. N., Cottrell, C. A., Wang, N., Pallesen, J., Yassine, H. M., Turner, H. L., et al. (2016). Pre-fusion structure of a human coronavirus spike protein. Nature 531, 118–121. doi: 10.1038/nature17200
Kleine-Weber, H., Elzayat, T., Wang, L., Elzayat, M. T., Wang, L., Graham, B. S., et al. (2019). Mutations in the spike protein of Middle East respiratory syndrome coronavirus transmitted in Korea increase. J. Virol. 93, 1–14. doi: 10.1128/JVI.01381-1318
Ko, J.-H., Seok, H., Cho, S. Y., Ha, Y. E., Baek, J. Y., Kim, S. H., et al. (2018). Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: a single centre experience. Antivir. Ther. 23, 617–622. doi: 10.3851/IMP3243
Kobinger, G. P., Figueredo, J. M., Rowe, T., Zhi, Y., Gao, G., Sanmiguel, J. C., et al. (2007). Adenovirus-based vaccine prevents pneumonia in ferrets challenged with the SARS coronavirus and stimulates robust immune responses in macaques. Vaccine 25, 5220–5231. doi: 10.1016/j.vaccine.2007.04.065
Kumar, V., Shin, J. S., Shie, J. J., Ku, K. B., Kim, C., Go, Y. Y., et al. (2017). Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLPro inhibitors. Antiviral Res. 141, 101–106. doi: 10.1016/j.antiviral.2017.02.007
Lamirande, E. W., DeDiego, M. L., Roberts, A., Jackson, J. P., Alvarez, E., Sheahan, T., et al. (2008). A live attenuated severe acute respiratory syndrome coronavirus is immunogenic and efficacious in golden syrian hamsters. J. Virol. 82, 7721–7724. doi: 10.1128/jvi.00304-308
Lan, J., Deng, Y., Chen, H., Lu, G., Wang, W., Guo, X., et al. (2014). Tailoring subunit vaccine immunity with adjuvant combinations and delivery routes using the middle east respiratory coronavirus (MERS-CoV) receptor-binding domain as an antigen. PLoS One 9:e0112602. doi: 10.1371/journal.pone.0112602
Lan, J., Yao, Y., Deng, Y., Chen, H., Lu, G., Wang, W., et al. (2015). Recombinant receptor binding domain protein induces partial protective immunity in rhesus macaques against Middle East respiratory syndrome coronavirus challenge. EBioMedicine 2, 1438–1446. doi: 10.1016/j.ebiom.2015.08.031
Lau, S. K. P., Woo, P. C. Y., Yip, C. C. Y., Tse, H., Tsoi, H. W., Cheng, V. C. C., et al. (2006). Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 44, 2063–2071. doi: 10.1128/JCM.02614-2615
Lauring, A. S., and Andino, R. (2010). Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 6:e1001005. doi: 10.1371/journal.ppat.1001005
Laursen, N. S., Friesen, R. H. E., Zhu, X., Jongeneelen, M., Blokland, S., Vermond, J., et al. (2018). Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science 362, 598–602. doi: 10.1126/science.aaq0620
Leist, S. R., and Baric, R. S. (2018). Giving the genes a shuffle: using natural variation to understand host genetic contributions to viral infections. Trends Genet. 34, 777–789. doi: 10.1016/j.tig.2018.07.005
Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3, 237–261. doi: 10.1146/annurev-virology-110615-142301
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. doi: 10.1038/nature02145
Li, W., Shi, Z., Yu, M., Ren, W., Smith, C., Epstein, J. H., et al. (2005). Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–680. doi: 10.1126/science.1118391
Li, W., Zhang, L., Johnson, J. S., Zhijian, W., Grieger, J. C., Ping-Jie, X., et al. (2009). Generation of novel aav variants by directed evolution for improved cftr delivery to human ciliated airway epithelium. Mol. Ther. 17, 2067–2077. doi: 10.1038/mt.2009.155
Li, Y., Wan, Y., Liu, P., Zhao, J., Lu, G., Qi, J., et al. (2015). A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 25, 1237–1249. doi: 10.1038/cr.2015.113
Lim, Y., Ng, Y., Tam, J., and Liu, D. (2016). Human coronaviruses: a review of virus-host interactions. Diseases 4, 26. doi: 10.3390/diseases4030026
Limberis, M. P., Adam, V. S., Wong, G., Gren, J., Kobasa, D., Ross, T. M., et al. (2013). Intranasal antibody gene transfer in mice and ferrets elicits broad protection against pandemic influenza. Sci. Transl. Med. 5:187ra72. doi: 10.1126/scitranslmed.3006729
Limberis, M. P., Tretiakova, A., Nambiar, K., Wong, G., Racine, T., Crosariol, M., et al. (2016). Adeno-associated virus serotype 9-expressed zmapp in mice confers protection against systemic and airway-acquired ebola virus infection. J. Infect. Dis. 214, 1975–1979. doi: 10.1093/infdis/jiw460
Limberis, M. P., Vandenberghe, L. H., Zhang, L., Pickles, R. J., and Wilson, J. M. (2009). Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol. Ther. 17, 294–301. doi: 10.1038/mt.2008.261
Lin, A., and Balazs, A. B. (2018). Adeno-associated virus gene delivery of broadly neutralizing antibodies as prevention and therapy against hiv-1 11 medical and health sciences 1103 clinical sciences 11 medical and health sciences 1108 medical microbiology marit van Gils, m.j.vangils@amc. Retrovirology 15, 1–17. doi: 10.1186/s12977-018-0449-447
Lipkin, W. I., and Firth, C. (2013). Viral surveillance and discovery. Curr. Opin. Virol. 3, 199–204. doi: 10.1016/j.coviro.2013.03.010
Liu, Y. V., Massare, M. J., Barnard, D. L., Kort, T., Nathan, M., Wang, L., et al. (2011). Chimeric severe acute respiratory syndrome coronavirus (SARS-CoV) S glycoprotein and influenza matrix 1 efficiently form virus-like particles (VLPs) that protect mice against challenge with SARS-CoV. Vaccine 29, 6606–6613. doi: 10.1016/j.vaccine.2011.06.111
Lo, M. K., Jordan, R., Arvey, A., Sudhamsu, J., Shrivastava-Ranjan, P., Hotard, A. L., et al. (2017). GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 7, 1–7. doi: 10.1038/srep43395
Lokugamage, K. G., Yoshikawa-Iwata, N., Ito, N., Watts, D. M., Wyde, P. R., Wang, N., et al. (2008). Chimeric coronavirus-like particles carrying severe acute respiratory syndrome coronavirus (SCoV) S protein protect mice against challenge with SCoV. Vaccine 26, 797–808. doi: 10.1016/j.vaccine.2007.11.092
Loustaud-Ratti, V., Debette-Gratien, M., Jacques, J., Alain, S., Marquet, P., Sautereau, D., et al. (2016). Ribavirin: Past, present and future. World J. Hepatol. 8, 123–130. doi: 10.4254/wjh.v8.i2.123
Loutfy, M. R., Blatt, L. M., Siminovitch, K. A., Ward, S., Wolff, B., Lho, H., et al. (2003). Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 290, 3222–3228. doi: 10.1001/jama.290.24.3222
Lu, L., Liu, Q., Zhu, Y., Chan, K. H., Qin, L., Li, Y., et al. (2014). Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 5:3067. doi: 10.1038/ncomms4067
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 6736, 1–10. doi: 10.1016/S0140-6736(20)30251-30258
Luo, C.-M., Wang, N., Yang, X.-L., Liu, H.-Z., Zhang, W., Li, B., et al. (2018). Discovery of novel bat coronaviruses in south china that use the same receptor as middle east respiratory syndrome coronavirus. J. Virol. 92:e00116-18. doi: 10.1128/JVI.00116-118
Ma, C., Li, Y., Wang, L., Zhao, G., Tao, X., Tseng, C.-T. K., et al. (2014a). Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: implication for designing novel mucosal MERS vaccines. Vaccine 32, 2100–2108. doi: 10.1016/j.vaccine.2014.02.004
Ma, C., Wang, L., Tao, X., Zhang, N., Yang, Y., Tseng, C.-T. K., et al. (2014b). Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments—The importance of immunofocusing in subunit vaccine design. Vaccine 32, 6170–6176. doi: 10.1016/j.vaccine.2014.08.086
Mackenzie, J. S., and Jeggo, M. (2013). Reservoirs and vectors of emerging viruses. Curr. Opin. Virol. 3, 170–179. doi: 10.1016/j.coviro.2013.02.002
Mair-Jenkins, J., Saavedra-Campos, M., Baillie, J. K., Cleary, P., Khaw, F. M., Lim, W. S., et al. (2015). The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 211, 80–90. doi: 10.1093/infdis/jiu396
Malczyk, A. H., Kupke, A., Prüfer, S., Scheuplein, V. A., Hutzler, S., Kreuz, D., et al. (2015). A highly immunogenic and protective Middle East respiratory syndrome coronavirus vaccine based on a recombinant measles virus vaccine platform. J. Virol. 89, 11654–11667. doi: 10.1128/jvi.01815-15
Martin, J. E., Louder, M. K., Holman, L. S. A., Gordon, I. J., Enama, M. E., Larkin, B. D., et al. (2008). A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine 26, 6338–6343. doi: 10.1016/j.vaccine.2008.09.026
Matsuyama, S., Shirato, K., Kawase, M., Terada, Y., Kawachi, K., Fukushi, S., et al. (2018). Middle East Respiratory syndrome coronavirus spike protein is not activated directly by cellular furin during viral entry into target cells. J. Virol. 92:e00683-18. doi: 10.1128/JVI.00683-618
Matteucci, E., and Giampietro, O. (2016). Dipeptidyl peptidase-4 (CD26): knowing the function before inhibiting the enzyme. Curr. Med. Chem. 4, 80–102. doi: 10.2174/9781681082493116090005
McIntosh, K., Kapikian, A. Z., Turner, H. C., Hartley, J. W., Parrott, R. H., and Chanock, R. M. (1970). Seroepidemiologic studies of coronavirus infection in adults and children. Am. J. Epidemiol. 91, 585–592. doi: 10.1093/oxfordjournals.aje.a121171
Menachery, V. D., Gralinski, L. E., Mitchell, H. D., Dinnon, K. H., Leist, S. R., Yount, B. L., et al. (2017). Middle East respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere 2, 1–12. doi: 10.1128/msphere.00346-317
Menachery, V. D., Gralinski, L. E., Mitchell, H. D., Dinnon, K. H., Leist, S. R., Yount, B. L., et al. (2018). Combination attenuation offers strategy for live attenuated coronavirus vaccines. J. Virol. 92, 1–15. doi: 10.1128/jvi.00710-718
Menachery, V. D., Yount, B. L., Debbink, K., Agnihothram, S., Gralinski, L. E., Plante, J. A., et al. (2015). A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 21, 1508–1513. doi: 10.1038/nm.3985
Menachery, V. D., Yount, B. L., Josset, L., Gralinski, L. E., Scobey, T., Agnihothram, S., et al. (2014). Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-o-methyltransferase activity. J. Virol. 88, 4251–4264. doi: 10.1128/jvi.03571-3513
Menachery, V. D., Yount, B. L., Sims, A. C., Debbink, K., Agnihothram, S. S., Gralinski, L. E., et al. (2016). SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U.S.A. 113, 3048–3053. doi: 10.1073/pnas.1517719113
Millet, J. K., and Whittaker, G. R. (2014). Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 111, 15214–15219. doi: 10.1073/pnas.1407087111
Millet, J. K., and Whittaker, G. R. (2015). Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 202, 120–134. doi: 10.1016/j.virusres.2014.11.021
Millet, J. K., and Whittaker, G. R. (2018). Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology 517, 3–8. doi: 10.1016/j.virol.2017.12.015
Min, J., Yu, D., Liang, W., Xu, R., Wang, Z., Fang, L., et al. (2004). Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303, 1666–1669. doi: 10.1126/science.1092002
Mo, Y., and Fisher, D. (2016). A review of treatment modalities for Middle East Respiratory Syndrome. J. Antimicrob. Chemother. 71, 3340–3350. doi: 10.1093/jac/dkw338
Modjarrad, K., Roberts, C. C., Mills, K. T., Castellano, A. R., Paolino, K., Muthumani, K., et al. (2019). Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 19, 1013–1022. doi: 10.1016/S1473-3099(19)30266-X
Momattin, H., Mohammed, K., Zumla, A., Memish, Z. A., and Al-Tawfiq, J. A. (2013). Therapeutic Options for Middle East respiratory syndrome coronavirus (MERS-CoV) - possible lessons from a systematic review of SARS-CoV therapy. Int. J. Infect. Dis. 17, e792–e798. doi: 10.1016/j.ijid.2013.07.002
Mubarak, A., Alturaiki, W., and Hemida, M. G. (2019). Middle east respiratory syndrome coronavirus (mers-cov): infection, immunological response, and vaccine development. J. Immunol. Res. 2019, 1–11. doi: 10.1155/2019/6491738
Munster, V. J., Wells, D., Lambe, T., Wright, D., Fischer, R. J., Bushmaker, T., et al. (2017). Protective efficacy of a novel simian adenovirus vaccine against lethal MERS-CoV challenge in a transgenic human DPP4 mouse model. NPJ Vaccines 2, 1–3. doi: 10.1038/s41541-017-0029-21
Murphy, B. G., Perron, M., Murakami, E., Bauer, K., Park, Y., Eckstrand, C., et al. (2018). The nucleoside analog GS-441524 strongly inhibits feline infectious peritonitis (FIP) virus in tissue culture and experimental cat infection studies. Vet. Microbiol. 219, 226–233. doi: 10.1016/j.vetmic.2018.04.026
Muthumani, K., Falzarano, D., Reuschel, E. L., Tingey, C., Flingai, S., Villarreal, D. O., et al. (2015). A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci. Transl. Med. 7:301ra132. doi: 10.1126/scitranslmed.aac7462
Naso, M. F., Tomkowicz, B., Perry, W. L., and Strohl, W. R. (2017). Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 31, 317–334. doi: 10.1007/s40259-017-0234-235
Neuman, B. W., Stein, D. A., Kroeker, A. D., Bestwick, R. K., Iversen, P. L., Moulton, H. M., et al. (2006). Inhibition and escape of SARS-CoV treated with antisense morpholino oligomers. Adv. Exp. Med. Biol. 581, 567–571. doi: 10.1007/978-0-387-33012-9_103
Nieto, K., and Salvetti, A. (2014). AAV vectors vaccines against infectious diseases. Front. Immunol. 5:5. doi: 10.3389/fimmu.2014.00005
Nyon, M. P., Du, L., Tseng, C.-T. K. T. K., Seid, C. A., Pollet, J., Naceanceno, K. S., et al. (2018). Engineering a stable CHO cell line for the expression of a MERS-coronavirus vaccine antigen. Vaccine 36, 1853–1862. doi: 10.1016/j.vaccine.2018.02.065
Oh, H. J., Gan, S. K., Bertoletti, A., and Tan, Y. (2012). Understanding the T cell immune response in SARS coronavirus infection. Emerg. Microbes Infect. 1:e23. doi: 10.1038/emi.2012.26
Olinger, G. G., Pettitt, J., Kim, D., Working, C., Bohorov, O., Bratcher, B., et al. (2012). Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc. Natl. Acad. Sci. U.S.A. 109, 18030–18035. doi: 10.1073/pnas.1213709109
Omrani, A. S., Saad, M. M., Baig, K., Bahloul, A., Abdul-Matin, M., Alaidaroos, A. Y., et al. (2014). Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 14, 1090–1095. doi: 10.1016/S1473-3099(14)70920-X
Ou, X., Leary, H. A. O., and Broxmeyer, H. E. (2019). Review Article Implications of DPP4 modi fi cation of proteins that regulate stem / progenitor and more mature cell types. Blood 122, 161–170. doi: 10.1182/blood-2013-02-487470.X.O
Pallesen, J., Wang, N., Corbett, K. S., Wrapp, D., Kirchdoerfer, R. N., Turner, H. L., et al. (2017). Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U.S.A. 114, E7348–E7357. doi: 10.1073/pnas.1707304114
Pascal, K. E., Coleman, C. M., Mujica, A. O., Kamat, V., Badithe, A., Fairhurst, J., et al. (2015). Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. U.S.A. 112, 8738–8743. doi: 10.1073/pnas.1510830112
Payne, D. C., Iblan, I., Rha, B., Alqasrawi, S., Haddadin, A., Nsour, M., et al. (2016). Persistence of antibodies against Middle East respiratory syndrome coronavirus. Emerg. Infect. Dis. 22, 1824–1826.
Peck, K. M., Burch, C. L., Heise, M. T., and Baric, R. S. (2015). Coronavirus host range expansion and middle east respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Annu. Rev. Virol. 2, 95–117. doi: 10.1146/annurev-virology-100114-155029
Pedersen, N. C., Perron, M., Bannasch, M., Montgomery, E., Murakami, E., Liepnieks, M., et al. (2019). Efficacy and safety of the nucleoside analog GS-441524 for treatment of cats with naturally occurring feline infectious peritonitis. J. Feline Med. Surg. 21, 271–281. doi: 10.1177/1098612X19825701
Peiris, J. S. M., Chu, C. M., Cheng, V. C. C., Chan, K. S. H., Hung, I. F. N., Poon, L. L. M., et al. (2003a). Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361, 1767–1772. doi: 10.1016/S0140-6736(03)13412-13415
Peiris, J. S. M., Lai, S. T., Poon, L. L. M., Guan, Y., Yam, L. Y. C., Lim, W., et al. (2003b). Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361, 1319–1325. doi: 10.1016/S0140-6736(03)13077-13072
Perlman, S., and Netland, J. (2009). Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7, 439–450. doi: 10.1038/nrmicro2147
Perlman, S., and Vijay, R. (2016). Middle East respiratory syndrome vaccines. Int. J. Infect. Dis. 47, 23–28. doi: 10.1016/j.ijid.2016.04.008
Petrovsky, N. (2016). SARS coronavirus infections of the lower respiratory tract and their prevention. Microbiol. Respir. Syst. Infect. 2016, 45–53. doi: 10.1016/B978-0-12-804543-5.00003-8
Plotkin, S. A. (2010). Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 17, 1055–1065. doi: 10.1128/CVI.00131-110
Prabakaran, P., Zhu, Z., Xiao, X., Biragyn, A., Dimitrov, A. S., Broder, C. C., et al. (2009). Potent human monoclonal antibodies against SARS CoV, Nipah and Hendra viruses. Expert Opin. Biol. Ther. 9, 355–368. doi: 10.1517/14712590902763755
Pyrc, K., Bosch, B. J., Berkhout, B., Jebbink, M. F., Dijkman, R., Rottier, P., et al. (2006). Inhibition of human coronavirus NL63 infection at early stages of the replication cycle. Antimicrob. Agents Chemother. 50, 2000–2008. doi: 10.1128/AAC.01598-1595
Qiu, H., Sun, S., Xiao, H., Feng, J., Guo, Y., Tai, W., et al. (2016). Single-dose treatment with a humanized neutralizing antibody affords full protection of a human transgenic mouse model from lethal Middle East respiratory syndrome (MERS)-coronavirus infection. Antiviral Res. 132, 141–148. doi: 10.1016/j.antiviral.2016.06.003
Qiu, X., Audet, J., Wong, G., Pillet, S., Bello, A., Cabral, T., et al. (2012). Successful treatment of ebola virus - infected cynomolgus macaques with monoclonal antibodies. Sci. Transl. Med. 4, 1–12.
Raj, V. S., Mou, H., Smits, S. L., Dekkers, D. H. W., Müller, M. A., Dijkman, R., et al. (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254. doi: 10.1038/nature12005
Raj, V. S., Okba, N. M. A., Gutierrez-Alvarez, J., Drabek, D., van Dieren, B., Widagdo, W., et al. (2018). Chimeric camel/human heavy-chain antibodies protect against MERS-CoV infection. Sci. Adv. 4, 1–10. doi: 10.1126/sciadv.aas9667
Raj, V. S., Osterhaus, A. D. M. E., Fouchier, R. A. M., and Haagmans, B. L. (2014). MERS: emergence of a novel human coronavirus. Curr. Opin. Virol. 5, 58–62. doi: 10.1016/j.coviro.2014.01.010
Reggiori, F., Monastyrska, I., Verheije, M. H., Calì, T., Ulasli, M., Bianchi, S., et al. (2010). Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7, 500–508. doi: 10.1016/j.chom.2010.05.013
Reynard, O., Nguyen, X. N., Alazard-Dany, N., Barateau, V., Cimarelli, A., and Volchkov, V. E. (2015). Identification of a new ribonucleoside inhibitor of ebola virus replication. Viruses 7, 6233–6240. doi: 10.3390/v7122934
Rezaee, F., Linfield, D. T., Harford, T. J., and Piedimonte, G. (2017). Ongoing developments in RSV prophylaxis?: a clinician’s analysis. Curr. Opin. Virol. 112781, 70–78. doi: 10.1016/j.coviro.2017.03.015
Richner, J. M., Himansu, S., Dowd, K. A., Butler, S. L., Salazar, V., Fox, J. M., et al. (2017). Modified mRNA vaccines protect against zika virus infection. Cell 168, 1114.e10–1125.e10. doi: 10.1016/j.cell.2017.02.017
Roberts, A., Lamirande, E. W., Vogel, L., Baras, B., Goossens, G., Knott, I., et al. (2010). Immunogenicity and protective efficacy in mice and hamsters of a β-propiolactone inactivated whole virus SARS-CoV vaccine. Viral Immunol. 23, 509–519. doi: 10.1089/vim.2010.0028
Rockx, B., Corti, D., Donaldson, E., Sheahan, T., Stadler, K., Lanzavecchia, A., et al. (2008). Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J. Virol. 82, 3220–3235. doi: 10.1128/jvi.02377-2377
Rothe, C., Schunk, M., Sothmann, P., Bretzel, G., Froeschl, G., Wallrauch, C., et al. (2020). Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 382, 970–971. doi: 10.1056/nejmc2001468
Schindewolf, C., and Menachery, V. D. (2019). Middle east respiratory syndrome vaccine candidates: cautious optimism. Viruses 11:74. doi: 10.3390/v11010074
See, R. H., Petric, M., Lawrence, D. J., Mok, C. P. Y., Rowe, T., Zitzow, L. A., et al. (2008). Severe acute respiratory syndrome vaccine efficacy in ferrets: whole killed virus and adenovirus-vectored vaccines. J. Gen. Virol. 89, 2136–2146. doi: 10.1099/vir.0.2008/001891-1890
See, R. H., Zakhartchouk, A. N., Petric, M., Lawrence, D. J., Mok, C. P. Y., Hogan, R. J., et al. (2006). Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. J. Gen. Virol. 87, 641–650. doi: 10.1099/vir.0.81579-81570
Seong, B. L. (2018). Chaperna-mediated assembly of ferritin-based middle east respiratory syndrome-coronavirus nanoparticles. Front. Immunol. 9:1093. doi: 10.3389/fimmu.2018.01093
Shalhoub, S., Farahat, F., Al-Jiffri, A., Simhairi, R., Shamma, O., Siddiqi, N., et al. (2015). IFN-α2a or IFN-β1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J. Antimicrob. Chemother. 70, 2129–2132. doi: 10.1093/jac/dkv085
Sharif-Yakan, A., and Kanj, S. S. (2014). Emergence of MERS-CoV in the middle east: origins, transmission, treatment, and perspectives. PLoS Pathog. 10:e1004457. doi: 10.1371/journal.ppat.1004457
Sheahan, T. P., Sims, A. C., Graham, R. L., Menachery, V. D., Gralinski, L. E., Case, J. B., et al. (2017). Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9, 1–11. doi: 10.1126/scitranslmed.aal3653
Sheahan, T. P., Sims, A. C., Leist, S. R., Schäfer, A., Won, J., Brown, A. J., et al. (2020). Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 11:222. doi: 10.1038/s41467-019-13940-13946
Shin, H. S., Kim, Y., Kim, G., Lee, J. Y., Jeong, I., Joh, J. S., et al. (2019). Immune responses to middle east respiratory syndrome coronavirus during the acute and convalescent phases of human infection. Clin. Infect. Dis. 68, 984–992. doi: 10.1093/cid/ciy595
Simmons, G., Gosalia, D. N., Rennekamp, A. J., Reeves, J. D., Diamond, S. L., and Bates, P. (2005). Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 102, 11876–11881. doi: 10.1073/pnas.0505577102
Smith, E. C., Blanc, H., Surdel, M. C., Vignuzzi, M., and Denison, M. R. (2013). Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 9:e1003565. doi: 10.1371/journal.ppat.1003565
Snijder, E. J., van der Meer, Y., Zevenhoven-Dobbe, J., Onderwater, J. J. M., van der Meulen, J., Koerten, H. K., et al. (2006). Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80, 5927–5940. doi: 10.1128/jvi.02501-2505
Song, F., Fux, R., Provacia, L. B., Volz, A., Eickmann, M., Becker, S., et al. (2013). Middle east respiratory syndrome coronavirus spike protein delivered by modified vaccinia virus ankara efficiently induces virus-neutralizing antibodies. J. Virol. 87, 11950–11954. doi: 10.1128/jvi.01672-1613
Spruth, M., Kistner, O., Savidis-Dacho, H., Hitter, E., Crowe, B., Gerencer, M., et al. (2006). A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine 24, 652–661. doi: 10.1016/j.vaccine.2005.08.055
Stadler, K., Roberts, A., Becker, S., Vogel, L., Eickmann, M., Kolesnikova, L., et al. (2005). SARS vaccine protective in mice [2]. Emerg. Infect. Dis. 11, 1312–1314. doi: 10.3201/eid1108.041003
Stertz, S., Reichelt, M., Spiegel, M., Kuri, T., Martínez-Sobrido, L., García-Sastre, A., et al. (2007). The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361, 304–315. doi: 10.1016/j.virol.2006.11.027
Strobel, B., Zuckschwerdt, K., Zimmermann, G., Mayer, C., Eytner, R., Rechtsteiner, P., et al. (2019). Standardized, Scalable, and Timely Flexible Adeno-Associated Virus Vector Production Using Frozen High-Density HEK-293 Cell Stocks and CELLdiscs. Hum. Gene Ther. Methods 30, 23–33. doi: 10.1089/hgtb.2018.228
Sui, J., Li, W., Murakami, A., Tamin, A., Matthews, L. J., Wong, S. K., et al. (2004). Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 101, 2536–2541. doi: 10.1073/pnas.0307140101
Sung, J. J. Y., Wu, A., Joynt, G. M., Yuen, K. Y., Lee, N., Chan, P. K. S., et al. (2004). Severe acute respiratory syndrome: report of treatment and outcome after a major outbreak. Thorax 59, 414–420. doi: 10.1136/thx.2003.014076
Tai, W., Wang, Y., Fett, C. A., Zhao, G., Li, F., Perlman, S., et al. (2017). Recombinant receptor-binding domains of multiple middle east respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J. Virol. 91:e01651-16. doi: 10.1128/JVI.01651-1616
Tai, W., Zhao, G., Sun, S., Guo, Y., Wang, Y., Tao, X., et al. (2016). A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology 499, 375–382. doi: 10.1016/j.virol.2016.10.005
Tan, E. L. C., Ooi, E. E., Lin, C. Y., Tan, H. C., Ling, A. E., Lim, B., et al. (2004). Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg. Infect. Dis. 10, 581–586. doi: 10.3201/eid1004.030458
Tang, F., Quan, Y., Xin, Z., Wrammert, J., Ma, M., Lv, H., et al. (2019). Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 186, 7264–7268. doi: 10.4049/jimmunol.0903490
Tang, J., Zhang, N., Tao, X., Zhao, G., Guo, Y., Tseng, C. K., et al. (2015). Optimization of antigen dose for a receptor- binding domain-based subunit vaccine against MERS coronavirus. Hum. Vaccines Immunother. 11, 1244–1250. doi: 10.1080/21645515.2015.1021527
Tang, X. C., Agnihothram, S. S., Jiao, Y., Stanhope, J., Graham, R. L., Peterson, E. C., et al. (2014). Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. U.S.A. 111, E2018–E2026. doi: 10.1073/pnas.1402074111
Ter Meulen, J., Bakker, A. B. H., Van Den Brink, E. N., Weverling, G. J., Martina, B. E. E., Haagmans, B. L., et al. (2004). Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet 363, 2139–2141. doi: 10.1016/S0140-6736(04)16506-16509
Ter Meulen, J., Van Den Brink, E. N., Poon, L. L. M., Marissen, W. E., Leung, C. S. W., Cox, F., et al. (2006). Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 3:e237. doi: 10.1371/journal.pmed.0030237
Tortorici, M. A., and Veesler, D. (2019). Structural Insights into Coronavirus Entry, 1st Edn. Amsterdam: Elsevier.
Traggiai, E., Becker, S., Subbarao, K., Kolesnikova, L., Uematsu, Y., Gismondo, M. R., et al. (2004). An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 10, 871–875. doi: 10.1038/nm1080
Tse, L. V., Klinc, K. A., Madigan, V. J., Castellanos Rivera, R. M., Wells, L. F., Havlik, L. P., et al. (2017). Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc. Natl. Acad. Sci. U.S.A. 114, E4812–E4821. doi: 10.1073/pnas.1704766114
Tse, L. V., Moller-Tank, S., and Asokan, A. (2015). Strategies to circumvent humoral immunity to adeno-associated viral vectors. Expert Opin. Biol. Ther. 15, 845–855. doi: 10.1517/14712598.2015.1035645
Ura, T., Okuda, K., and Shimada, M. (2014). Developments in viral vector-based vaccines. Vaccines 2, 624–641. doi: 10.3390/vaccines2030624
Urakova, N., Kuznetsova, V., Crossman, D. K., Sokratian, A., Guthrie, D. B., Kolykhalov, A. A., et al. (2017). β-D- N 4 -hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J. Virol. 92, 1–22. doi: 10.1128/jvi.01965-1917
van den Brink, E. N., ter Meulen, J., Cox, F., Jongeneelen, M. A. C., Thijsse, A., Throsby, M., et al. (2005). Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J. Virol. 79, 1635–1644. doi: 10.1128/jvi.79.3.1635-1644.2005
van Doremalen, N., Falzarano, D., Ying, T., de Wit, E., Bushmaker, T., Feldmann, F., et al. (2017). Efficacy of antibody-based therapies against Middle East respiratory syndrome coronavirus (MERS-CoV) in common marmosets. Antiviral Res. 143, 30–37. doi: 10.1016/j.antiviral.2017.03.025
Volz, A., Kupke, A., Song, F., Jany, S., Fux, R., Shams-Eldin, H., et al. (2015). Protective efficacy of recombinant modified vaccinia virus ankara delivering middle east respiratory syndrome coronavirus spike glycoprotein. J. Virol. 89, 8651–8656. doi: 10.1128/jvi.00614-615
Walls, A. C., Xiong, X., Park, Y. J., Tortorici, M. A., Snijder, J., Quispe, J., et al. (2019a). Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176, 1026.e15–1039.e15. doi: 10.1016/j.cell.2018.12.028
Walls, A. C., Xiong, X., Rey, A., Park, Y., Corti, D., Veesler, D., et al. (2019b). Activation of coronavirus fusion unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176, 1026–1039.
Wang, C., Zheng, X., Gai, W., Wong, G., Wang, H., Jin, H., et al. (2017a). Novel chimeric virus-like particles vaccine displaying MERS-CoV receptor-binding domain induce specific humoral and cellular immune response in mice. Antiviral Res. 140, 55–61. doi: 10.1016/j.antiviral.2016.12.019
Wang, C., Zheng, X., Gai, W., Zhao, Y., Gao, Y., Yang, S., et al. (2017b). MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget 8, 12686–12694.
Wang, Y., Tai, W., Yang, J., Zhao, G., Sun, S., Tseng, C.-T. K., et al. (2017). Receptor-binding domain of MERS-CoV with optimal immunogen dosage and immunization interval protects human transgenic mice from MERS-CoV infection. Hum. Vaccin. Immunother. 13, 1615–1624. doi: 10.1080/21645515.2017.1296994
Wang, L., Shi, W., Chappell, J. D., Joyce, M. G., Zhang, Y., Kanekiyo, M., et al. (2018). Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the middle east respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape. J. Virol. 92, 1–21. doi: 10.1128/jvi.02002-2017
Wang, L., Shi, W., Joyce, M. G., Modjarrad, K., Zhang, Y., Leung, K., et al. (2015). Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 6, 1–11. doi: 10.1038/ncomms8712
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271. doi: 10.1038/s41422-020-0282-280
Wang, N., Rosen, O., Wang, L., Turner, H. L., Stevens, L. J., Corbett, K. S., et al. (2019). Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD. Cell Rep. 28, 3395.e6–3405.e6. doi: 10.1016/j.celrep.2019.08.052
Wang, W., Chen, S., Liu, I., Kao, C., Chen, H., Chiang, B., et al. (2004). Temporal relationship of viral load, ribavirin, interleukin (IL)-6, IL-8, and clinical progression in patients with severe acute respiratory syndrome. Clin. Infect. Dis. 39, 1071–1075. doi: 10.1086/423808
Warimwe, G. M., Gesharisha, J., Carr, B. V., Otieno, S., Otingah, K., Wright, D., et al. (2016). Chimpanzee adenovirus vaccine provides multispecies protection against rift valley fever. Sci. Rep. 6, 1–7. doi: 10.1038/srep20617
White, J. M., and Whittaker, G. R. (2016). Fusion of enveloped viruses in endosomes. Traffic 17, 593–614. doi: 10.1111/tra.12389
Widagdo, W., Okba, N. M. A., Stalin Raj, V., and Haagmans, B. L. (2017). MERS-coronavirus: from discovery to intervention. One Heal. 3, 11–16. doi: 10.1016/j.onehlt.2016.12.001
Wirblich, C., Coleman, C. M., Kurup, D., Abraham, T. S., Bernbaum, J. G., Jahrling, P. B., et al. (2017). One-health: a safe, efficient, dual-use vaccine for humans and animals against middle east respiratory syndrome coronavirus and rabies virus. J. Virol. 91, 1–15.
Woo, P. C. Y., Lau, S. K. P., Huang, Y., and Yuen, K. Y. (2009). Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 234, 1117–1127. doi: 10.3181/0903-MR-94
Woo, P. C. Y., Lau, S. K. P., Tsoi, H., Huang, Y., Poon, R. W. S., Chu, C., et al. (2005). Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J. Infect. Dis. 192, 1898–1907. doi: 10.1086/497151
World Health Organization [WHO] (2018). WHO MERS Global Summary and Assessment of Risk. Geneva: WHO.
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 1263, 1260–1263. doi: 10.1126/science.abb2507
Wu, C. Y., Jan, J. T., Ma, S. H., Kuo, C. J., Juan, H. F., Cheng, Y. S. E., et al. (2004). Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U.S.A. 101, 10012–10017. doi: 10.1073/pnas.0403596101
Xia, S., Yan, L., Xu, W., Agrawal, A. S., Algaissi, A., Tseng, C. T. K., et al. (2019). A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 5:eaav4580. doi: 10.1126/sciadv.aav4580
Xu, J., Jia, W., Wang, P., Zhang, S., Shi, X., Wang, X., et al. (2019). Antibodies and vaccines against Middle East respiratory syndrome coronavirus. Emerg. Microbes Infect. 8, 841–856. doi: 10.1080/22221751.2019.1624482
Yang, Z. Y., Kong, W. P., Huang, Y., Roberts, A., Murphy, B. R., Subbarao, K., et al. (2004). A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428, 561–564. doi: 10.1038/nature02463
Ying, T., Du, L., Ju, T. W., Prabakaran, P., Lau, C. C. Y., Lu, L., et al. (2014). Exceptionally potent neutralization of middle east respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 88, 7796–7805. doi: 10.1128/jvi.00912-914
Ying, T., Prabakaran, P., Du, L., Shi, W., Feng, Y., Wang, Y., et al. (2015). Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 6:8223. doi: 10.1038/ncomms9223
Yong, C. Y., Ong, H. K., Yeap, S. K., Ho, K. L., and Tan, W. S. (2019). Recent advances in the vaccine development against middle east respiratory syndrome-coronavirus. Front. Microbiol. 10:1781. doi: 10.3389/fmicb.2019.01781
Yoon, J. J., Toots, M., Lee, S., Lee, M. E., Ludeke, B., Luczo, J. M., et al. (2018). Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob. Agents Chemother. 62, 1–18. doi: 10.1128/AAC.00766-718
Yuan, S., Chu, H., Chan, J. F. W., Ye, Z. W., Wen, L., Yan, B., et al. (2019). SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat. Commun. 10:120. doi: 10.1038/s41467-018-08015-x
Yuan, Y., Cao, D., Zhang, Y., Ma, J., Qi, J., Wang, Q., et al. (2017). Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 8:15092. doi: 10.1038/ncomms15092
Zabner, J., Seiler, M., Walters, R., Kotin, R. M., Fulgeras, W., Davidson, B. L., et al. (2000). Adeno-associated virus type 5 (AAV5) but not AAV2 binds to the apical surfaces of airway epithelia and facilitates gene transfer. J. Virol. 74, 3852–3858. doi: 10.1128/JVI.74.8.3852-3858.2000
Zhang, C., Maruggi, G., Shan, H., and Li, J. (2019). Advances in mRNA vaccines for infectious diseases. Front. Immunol. 10:594. doi: 10.3389/fimmu.2019.00594
Zhang, N., Channappanavar, R., Ma, C., Wang, L., Tang, J., Garron, T., et al. (2016). Identification of an ideal adjuvant for receptor-binding domain-based subunit vaccines against Middle East respiratory syndrome coronavirus. Cell. Mol. Immunol. 13, 180–190. doi: 10.1038/cmi.2015.03
Zhang, N., Jiang, S., and Du, L. (2014). Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev. Vaccines 13, 761–774. doi: 10.1586/14760584.2014.912134
Zhao, G., He, L., Sun, S., Qiu, H., Tai, W., Chen, J., et al. (2018). A novel nanobody targeting middle east respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J. Virol. 92:e00837-18. doi: 10.1128/JVI.00837-818
Zhao, J., Alshukairi, A. N., Baharoon, S. A., Ahmed, W. A., Bokhari, A. A., Nehdi, A. M., et al. (2017). Recovery from the Middle East respiratory syndrome is associated with antibody and T cell responses. Sci. Immunol. 5393, 1–11.
Zhao, J., Perera, R. A. P. M., Kayali, G., Meyerholz, D., Perlman, S., and Peiris, M. (2015). Passive immunotherapy with dromedary immune serum in an experimental animal model for middle east respiratory syndrome. J. Virol. 89, 6117–6120. doi: 10.1128/JVI.00446-415
Zhao, J., Zhao, J., and Perlman, S. (2010). T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice ? J. Virol. 84, 9318–9325. doi: 10.1128/JVI.01049-1010
Zhao, J. J. J., Zhao, J. J. J., Mangalam, A. K., Channappanavar, R., Fett, C., Meyerholz, D. K., et al. (2016). Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 44, 1379–1391. doi: 10.1016/j.immuni.2016.05.006
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. doi: 10.1038/s41586-020-2012-2017
Zhu, Z., Chakraborti, S., He, Y., Roberts, A., Sheahan, T., Xiao, X., et al. (2007). Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. U.S.A. 104, 12123–12128. doi: 10.1073/pnas.0701000104
Ziebuhr, J., Snijder, E. J., and Gorbalenya, A. E. (2000). Virus-encoded proteinases and proteolytic processing in the nidovirales. J. Gen. Virol. 81(Pt 4), 853–879.
Zumla, A., Chan, J. F. W. W., Azhar, E. I., Hui, D. S. C. C., and Yuen, K. Y. (2016a). Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 15, 327–347. doi: 10.1038/nrd.2015.37
Zumla, A., Hui, D. S., Perlman, S., To, C., and Stanley, P. (2015). Middle east respiratory syndrome HHS public access. Lancet 386, 995–1007. doi: 10.1016/S0140-6736(15)60454-60458
Keywords: coronavirus (CoV), vaccine, antivirals, adeno-associate virus, passive immunization strategy, MERS- and SARS-CoV, 2019 nCoV
Citation: Tse LV, Meganck RM, Graham RL and Baric RS (2020) The Current and Future State of Vaccines, Antivirals and Gene Therapies Against Emerging Coronaviruses. Front. Microbiol. 11:658. doi: 10.3389/fmicb.2020.00658
Received: 11 November 2019; Accepted: 23 March 2020;
Published: 24 April 2020.
Edited by:
Lu Lu, Fudan University, ChinaReviewed by:
Jasper Fuk Woo Chan, The University of Hong Kong, Hong KongSusan Baker, Loyola University Chicago, United States
Copyright © 2020 Tse, Meganck, Graham and Baric. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralph S. Baric, cmJhcmljQGVtYWlsLnVuYy5lZHU=
 Longping V. Tse
Longping V. Tse Rita M. Meganck2
Rita M. Meganck2 Rachel L. Graham
Rachel L. Graham