- 1Department of Pathobiology and Diagnostic Investigation, College of Veterinary Medicine, Michigan State University, East Lansing, MI, United States
- 2Department of Veterinary Public Health, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand
- 3National Animal Disease Center, USDA-Agricultural Research Service, Ames, IA, United States
- 4School of Veterinary Medicine and Biomedical Sciences, University of Nebraska, Lincoln, NE, United States
Intracellular iron concentration is tightly regulated to maintain cell viability. Iron plays important roles in electron transport, nucleic acid synthesis, and oxidative stress. A Mycobacterium avium subsp. paratuberculosis (MAP)-specific genomic island carries a putative metal transport operon that includes MAP3773c, which encodes a Fur-like protein. Although well characterized as a global regulator of iron homeostasis in multiple bacteria, the function of Fur (ferric uptake regulator) in MAP is unknown as this organism also carries IdeR (iron dependent regulator), a native iron regulatory protein specific to mycobacteria. Computational analysis using PRODORIC identified 23 different pathways involved in respiration, metabolism, and virulence that were likely regulated by MAP3773c. Thus, chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) was performed to confirm the putative regulon of MAP3773c (Fur-like protein) in MAP. ChIP-Seq revealed enriched binding to 58 regions by Fur under iron-replete and -deplete conditions, located mostly within open reading frames (ORFs). Three ChIP peaks were identified in genes that are directly related to iron regulation: MAP3638c (hemophore-like protein), MAP3736c (Fur box), and MAP3776c (ABC transporter). Fur box consensus sequence was identified, and binding specificity and dependence on Mn2+ availability was confirmed by a chemiluminescent electrophoresis mobility shift assay (EMSA). The results confirmed that MAP3773c is a Fur ortholog that recognizes a 19 bp DNA sequence motif (Fur box) and it is involved in metal homeostasis. This work provides a regulatory network of MAP Fur binding sites during iron-replete and -deplete conditions, highlighting unique properties of Fur regulon in MAP.
Introduction
Mycobacterium avium subsp. paratuberculosis (MAP) is the causative agent of Johne’s disease (JD) in ruminants, a chronic and incurable chronic enteritis characterized by persistent diarrhea that leads to malnutrition and muscular wasting (Rathnaiah et al., 2017). JD is present worldwide and imposes significant economic losses to the dairy industry (Garcia and Shalloo, 2015). Unfortunately, to date, reliable JD diagnostics are still lacking. Culture of MAP from feces has been the most reliable method for diagnosis of JD; however, MAP requires 8 to 16 weeks to produce colonies in culture, presenting a major hurdle to diagnosis (Bannantine et al., 2002).
Unlike other mycobacteria, MAP has special iron requirements. For optimal growth in vitro, it requires supplementation of the siderophore mycobactin J. Whole-genome sequencing of MAP K-10 provided a potential explanation for this dependency, revealing a truncation of the mbtA gene, with MAP making a protein that is 151–156 amino acids shorter than M. tuberculosis or M. avium (Li et al., 2005). It has been suggested that this truncation impairs the production of mycobactin from the mbtA–J operon (Li et al., 2005; Wang et al., 2014). Despite this truncation, Zhu et al. (2008) showed that MAP is still able to transcribe mycobactin synthesis genes inside macrophages. To corroborate these findings, Janagama et al. described the upregulation of several genes responsible for iron acquisition in infected tissues, including genes responsible for mycobactin biosynthesis (Janagama et al., 2010).
Iron is vital to fundamental biological processes, however, high intracellular concentrations of free iron are toxic to bacteria. As such, cells have developed tightly regulated processes for intracellular metal homeostasis (Eckelt et al., 2014). Bacteria control metal homeostasis by activating a set of genes regulated by metal-sensing transcription factors known as metalloregulatory proteins (Chandrangsu et al., 2017). In prokaryotes, there are two major families of metalloregulators: diphtheria toxin (DtxR) and ferric uptake regulator (Fur) (Hantke, 2001). In 2009, Janagama and others identified and characterized MAP2827, an iron-dependent regulator (IdeR) in MAP. A member of the DtxR protein family, IdeR is involved in regulatory mechanisms to acquire, store, or prevent excess accumulation of iron. The authors were able to confirm that MAP2827 was in fact IdeR and regulates genes involved in iron acquisition (mbtB) and iron storage (bfrA) (Janagama et al., 2009). However, in vitro iron stress showed that IdeR regulation is strain dependent, while IdeR from MAP cattle strain K-10 regulates mycobactin synthesis and storage genes similar to IdeR from M. tuberculosis. IdeR from MAP sheep strain S397 shows deficiency in iron storage function, resulting in a strain more sensitive to iron fluctuations (Janagama et al., 2010).
In addition to IdeR, MAP genome contains a putative metal transport MAP-specific operon and large genomic polymorphisms (LSPs), 15, that include a Fur-like transcriptional regulator, MAP3773c (Alexander et al., 2009). First identified in Escherichia coli, Fur has been shown to respond to iron-replete conditions to repress gene expression and allow sufficient concentration of intracellular iron for essential iron-related activities (Hantke, 1981; Bagg and Neilands, 1987; Lee and Helmann, 2007). Similar to several representatives of Fur family member, Fur protein requires binding of a divalent metal ion, either Fe2+ or Mn2+, for DNA-binding activation (Mills and Marletta, 2005; Lee and Helmann, 2007; Chandrangsu et al., 2017). Fur protein generally binds to a 19-bp inverted repeat sequence known as a “Fur box” (GATAATGATwATCATTATC; w = A or T), within the promoter of the regulated genes (Escolar et al., 1999). In MAP, functional genomics suggested three Fur boxes located in a 38-kb MAP-specific genomic island (LSP14) (Stratmann et al., 2004; Alexander et al., 2009). MAP genome includes a total of six specific genomic insertions: LSP4, LSP11, LSP12, LSP14, LSP15, and LSP16 (Alexander et al., 2009). As these islands are not presented in any other mycobacteria, it has been proposed and confirmed that they were acquired via horizontal gene transfer (Alexander et al., 2009; Wang et al., 2016). Furthermore, LSP14 and LSP15 encode several predicted genes involved in metal uptake systems.
To date, there have been characterization of the other Fur family members in MAP, FurA, and FurB, also known as Per (peroxidase stress response) and Zur (zinc uptake repressor), respectively, however, no information about the potential roles of Fur-like element has been described (Eckelt et al., 2014, Eckelt et al., 2015).
As a key virulence determinant, iron regulation in MAP and its role in pathogen survival and infection are important areas of research that may lead to advances in ability to improve culturing methods. To further elucidate the mechanisms of iron homeostasis in MAP, we investigated the putative function of the Fur-like gene (MAP3773c) in iron homeostasis in vitro. We applied in vivo ChIP-seq to confirm binding of MAP Fur as a transcription factor and to identify the regulon of genes under its control.
Materials and Methods
Bacterial Strains
MAP K-10 strain was grown at 37°C without shaking in Middlebrook 7H9 supplemented with 10% OADC (oleic acid, dextrose, catalase) enrichment medium (Thermo Fisher Scientific, Waltham, MA, United States), 0.05% Tween 80, and 2 mg of ferric mycobactin J (Allied Monitor Inc., Fayette, MO, United States) per liter. Antibiotics (μg/ml: kanamycin, 20; hygromycin, 100; streptomycin, 20) were added when necessary. Competent E. coli BL21(DE3) (EMD Biosciences, Madison WI, United States) and E. coli TOP10F cells (Invitrogen, Carlsbad, CA, United States) were grown in LB medium 37°C with shaking at 200 RPM.
Protein Expression
To express MAP Fur protein, competent E. coli BL21(DE3) (EMD Biosciences, Madison WI) carrying MAP3773c on pET-24b(+) were growing in LB medium with 30 μg/ml kanamycin. Cultures were kept at 37°C with shaking at 200 RPM for 4 h aerobic growth, until OD600 of 0.4 was obtained. Then, protein expression was induced with addition of 0.1 M IPTG and shaking at 37°C for an additional 2 h. The expressed MAP3773c was extracted using B-PER (Bacterial Protein Extraction Reagent; Pierce Biotechnology, Rockford, IL, United States), followed by purification using HisPur Ni-NTA resin columns per the manufacturer’s protocol (Pierce Biotechnology, Rockford, IL, United States). Purified protein was analyzed by SDS-PAGE and Western Blot using standard methods described previously (Bannantine and Paustian, 2006). The target band identified from the SDS-PAGE was excised for LC-MS/MS at Michigan State University Proteomics Facilities. Raw data were analyzed using Scaffold (Proteome Software, Portland, OR, United States).
Western Blotting
MAP K-10 were cultured as previously described until reaching an OD600 of ∼0.5. For iron starvation, cultures were treated with 2,2′-bipyridyl (DIP, 200 μM final) for 2 h shacking at 200 rpm at 37°C. Cells from iron-replete and -deplete conditions were washed with 1 × PBS and resuspended in freshly made buffer lysis buffer (20 mM HEPES; 50 mM KCl; 0.5 mM DTT; 10% glycerol; mini protease inhibitor), followed by cell lysing with MagNA Lyser (Roche Diagnostics, Sandhofer, Germany). For enrichment of Fur protein, samples were subjected to immunoprecipitation. Samples were incubated overnight with antibody for Fur detection at 4°C on a rotating platform followed by 2 h incubation [0.5 h at 4°C and 1.5 h at room temperature (RT)] on a rotating platform. Samples were washed two times with IPP150 buffer (10 mM Tris–HCl; 150 mM NaCl, 0.1% NP40) and two times with 1 × TE (0.05 M Tris–HCl; 10 mM EDTA) buffer. Beads were resuspended in elution buffer and incubated at 65°C for 15 min. The samples were subjected to SDS-PAGE and transferred to Nitrocellulose Membrane, 0.2 μm (Bio-Rad Laboratories, Hercules, CA, United States). Custom-made antibody that binds the MAP Fur protein (Genscript, Piscataway, NJ, United States) was used as primary antibody. Anti-rabbit IgG (whole molecule)–peroxidase antibody produced in goat (Sigma-Aldrich, St. Louis, MO, United States) was used as secondary antibody. The membrane was visualized with ChemiDoc MP Imaging System (Bio-Rad Laboratories, Hercules, CA, United States).
Computational Prediction of Fur-Regulated Genes
Virtual Footprint, part of The Prokaryotic Database of Gene Regulation (PRODORIC) (Münch et al., 2005), was used for prediction of Fur binding site. MAP K-10 genome was used as input DNA sequence, Fur box motif from E. coli was used as Position Weight Matrix, and searches were limited to -300 to +100 bases of each predicted ORF.
Chromatin Immunoprecipitation Followed by Sequencing (ChIP-Seq)
ChIP-enriched DNA samples were harvested following the protocol developed by Jaini et al. (2014) using a custom-made antibody that binds the MAP Fur protein (Genscript, Piscataway, NJ, United States). MAP K-10 culture with an OD600 of ∼0.5 was used to generate ChIP-DNA. In order to avoid false positive, input DNA was used as control, and this sample did not have ChIP enrichment. For iron starvation, cultures were treated with DIP (200 μM final) for 2 h shacking at 200 rpm at 37°C. Cells from iron-replete and -deplete conditions were washed with 1 × PBS. Formaldehyde was added at a final concentration of 1% and incubated at RT for 20 min in a platform rocker. Cross-linking was quenched by adding 250 mM of glycine and incubating for 15 min. Cells were washed two times with ice-cold 1 × PBS and resuspended in freshly made buffer lysis buffer (20 mM HEPES; 50 mM KCl; 0.5 mM DTT; 10% glycerol; mini protease inhibitor), followed by cell lysing with MagNA Lyser (Roche Diagnostics, Sandhofer, Germany). Cell suspensions were sonicated using Covaris M220 Focused-ultrasonicator (Covaris, Inc., Woburn, MA) for 18 min; 75.0 peak power; 20.0 duty factor, and 200 cycles/burst. Samples were incubated overnight with antibody for Fur detection at 4°C on a rotating platform followed by 2 h incubation (0.5 h at 4°C and 1.5 h at RT) on a rotating platform. Samples were washed two times with IPP150 buffer (10 mM Tris–HCl; 150 mM NaCl, 0.1% NP40) and two times with 1 × TE (0.05 M Tris–HCl; 10 mM EDTA) buffer. Beads were resuspended in elution buffer and incubated at 65°C for 15 min. 1 mg/ml of Proteinase K was added to each sample and incubated at 37°C for 2 h and transferred for 65°C for overnight incubation. DNA purification was performed using AmPurexp beads per the manufacturer’s protocol (Beckman Coulter, Indianapolis, IN, United States). Sample quality was analyzed by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States).
ChIP-Seq Library Construction and Sequencing
DNA fragments ∼300 bp were selected for library preparation and sequencing libraries were prepared using NEXTflexTM ChIP-seq kit (PerkinElmer, Austin, TX, United States) as per the manufacturer’s protocol. Pre- and post-library construction, chromatin immunoprecipitation products were quantified using a Qubit fluorometer (Invitrogen, Carlsbad, CA, United States) and an Agilent 2100 Bioanalyzer (Agilent technologies, Santa Clara, CA, United States). ChIP DNA replicates were pooled and sequenced. Approximately 20M reads per sample were generated, providing 150–1,000 depth of coverage. Sequencing was performed by ACGT, Inc. (Chicago, IL, United States).
ChIP-Seq Data Analysis
All analysis was done using CLC Genomics Workbench software 12.0 (QIAGEN, Aarhus, Denmark). Raw data generated from ChIP-seq were trimmed and mapped to the reference MAP K-10 genome (NCBI accession number NC_002944). Using CLC shape-based peak caller, ChIP-enriched DNA were aligned onto Input DNA (no ChIP enrichment); when the sequence coverage of a genomic region in the enriched DNA exceeded the Input DNA, a ChIP peak score was called. A list of all ChIP peaks with their respective P value was generated. The threshold for signal-to-noise ratio (ChIP-enriched DNA vs. no enriched) was set based on false discovery rate (FDR) value equal to or smaller than 10–50. FDR was calculated using Bonferroni correction on R software based on the P value generated by CLC.
Motif Detection
A Fur binding motif was generated using Find Individual Motif Occurrences (FIMO), part of the MEME suit (Grant et al., 2011), for all in vivo binding sites identified in ChIP-seq analyses. A P value of ≤ 0.001 was defined as statistical threshold for Fur binding motifs.
Electrophoretic Mobility Shift Assay (EMSA)
Physical binding of MAP3773c to the promoter sequences of MAP Fur box 1 (MAP3736c) was carried out by EMSA. Promoter sequences containing Fur box motifs were amplified using 5′ biotin-labeled primer via PCR. Purification of PCR products was done using the QIAquick PCR Purification kit (Qiagen, Germantown, MD, United States). Recombinant MAP Fur protein was expressed as stated above. Binding reaction: 1 × Binding Buffer (50 mM Tris–HCl; 25% glycerol; 10 μg/ml Salmon tests DNA; 250 nM NaCl; 5 mM DTT; 250 μg/ml BSA; nuclease free water), 10 mM MnCl2, 0–10 nM MAP Fur protein, 0–4 pmol Unlabeled DNA, and 20 fmol labeled DNA. The reactions were incubated for 30 min at RT followed by electrophoresis in a 5% native polyacrylamide gel [40% 19:1 Acrylamide; 50% Tris-Acetate (TA) buffer; 50% glycerol; 10% Ammonium persulfate (APS); 6% TEMED] using 1 × TA Buffer (1 M Tris acetate, 0.5 M Glacial acetic acid) as running buffer. After electrophoresis, gels were transferred onto a Biodyne B Nylon membrane (Pierce, Biotechnology, Rockford, IL, United States) and reactions were detected using chemiluminescence-based nucleic acid detection kit (Pierce, Biotechnology, Rockford, IL, United States).
Results
Genome-Wide Analysis of Fur Regulon
Using computational prediction, PRODORIC (Münch et al., 2005), 26 different pathways involved in respiration, metabolism, and virulence were identified as likely regulated by MAP3773c (Figure 1). To confirm the findings from the in silico analysis and determine which genes are regulated by Fur in MAP, chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) was performed. A custom-synthesized anti-Fur antibody capable of detecting the MAP Fur protein in its native form in MAP K-10 (Figure 2C) was used to generate ChIP binding profiles for MAP K-10 cultured under iron-replete and -deplete conditions (Figure 3).
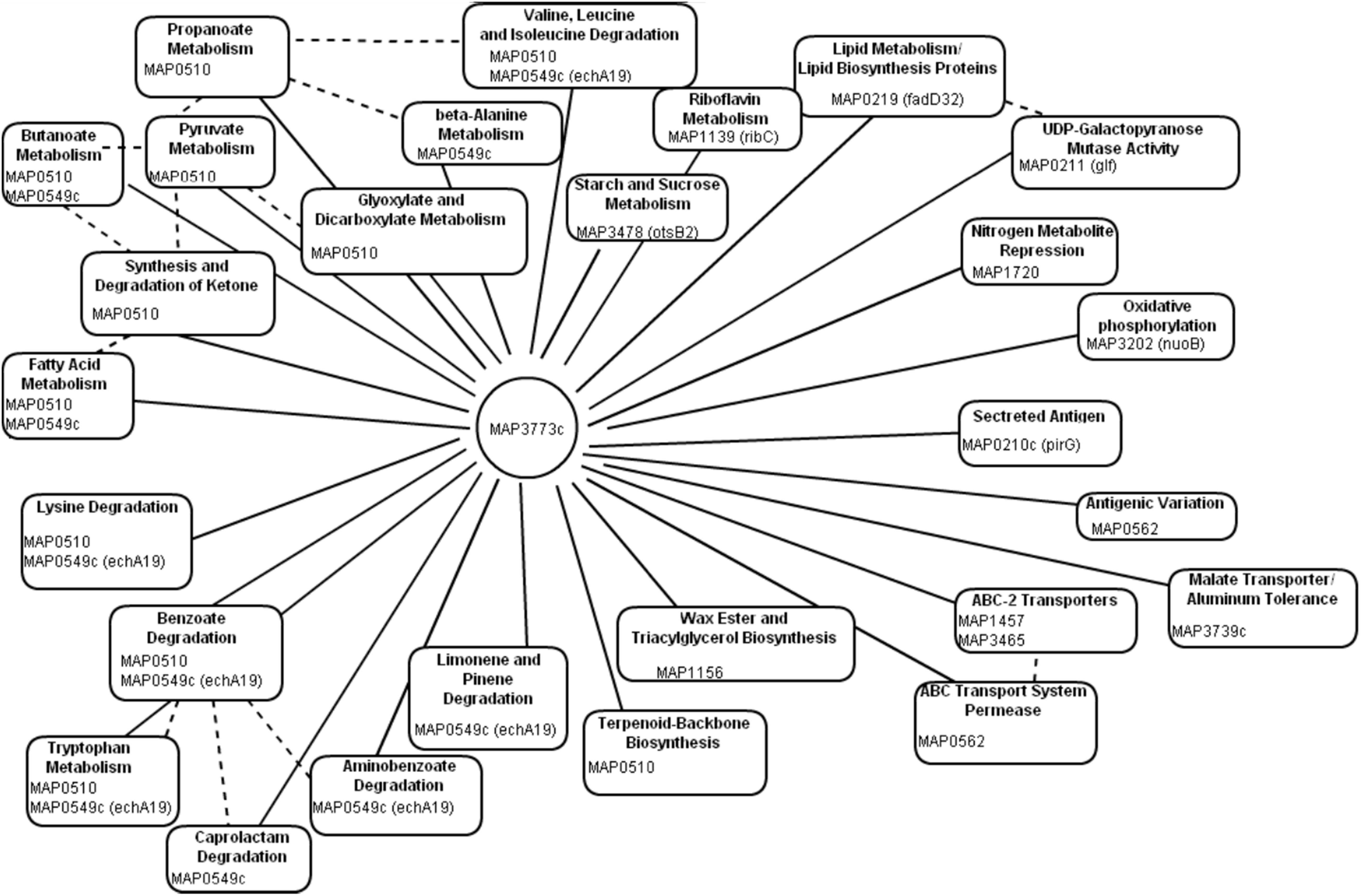
Figure 1. In silico analysis of Fur regulon. Using PRODORIC for MAP K-10 genome analysis to detect putative Fur binding and predict pathways regulated by MAP3773c. Solid lines represent pathways directly regulated by MAP3773c. Dashed lines indicated interrelated pathways.
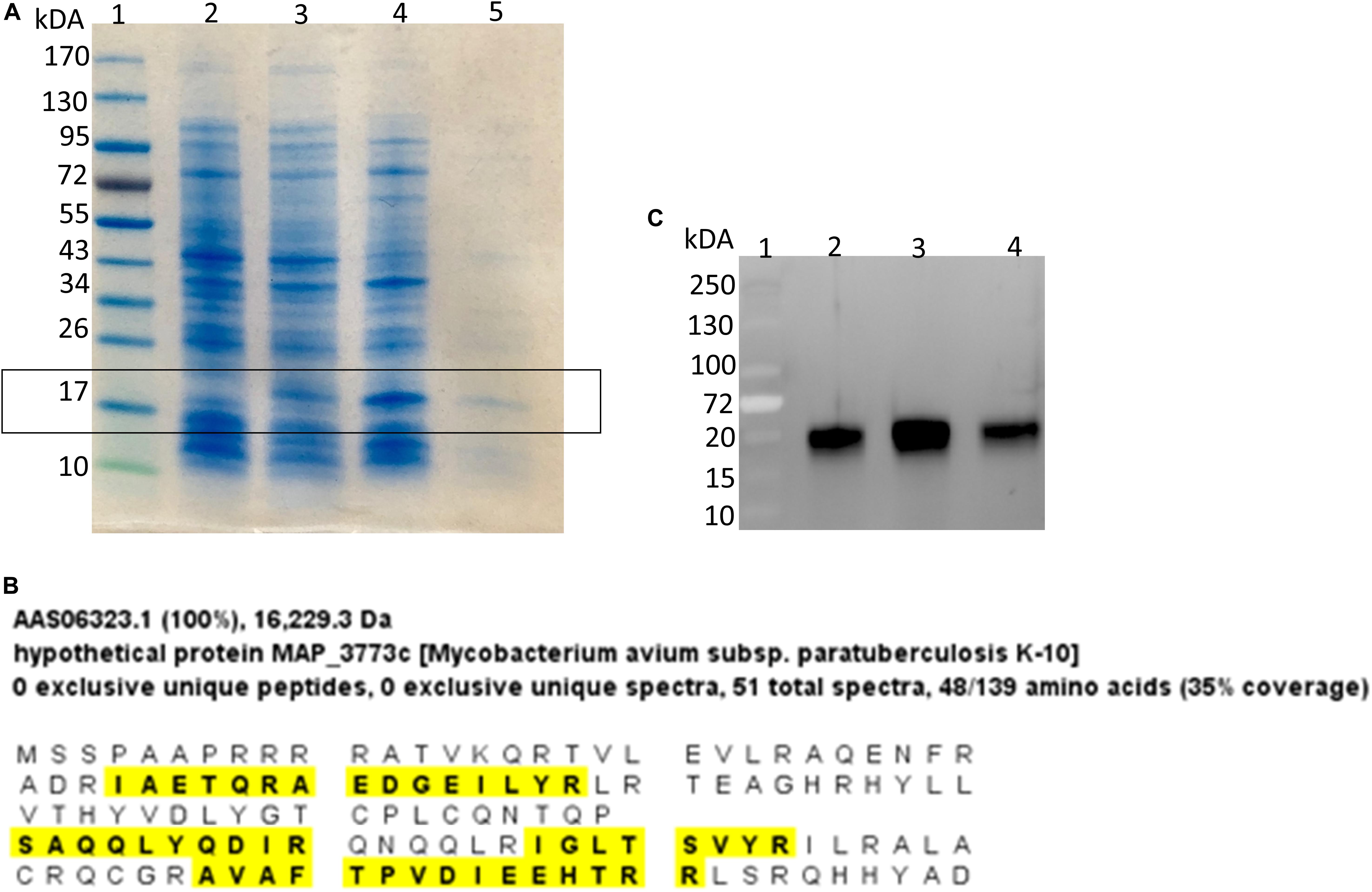
Figure 2. Identification of MAP Fur protein. (A) Coomassie stain of SDS-PAGE analysis of Fur expression in E. coli system. Lane 1, Protein ladder; Lane 2, BL21(DE3) carrying empty vector pET24b(+); Lane 3, BL21(DE3) carrying MAP3773c on pET24b(+); Lane 4, BL21(DE3) carrying MAP3773c on pET24b(+) with addition of IPTG; Lane 5, Purified recombinant MAP3773c protein. (B) Scaffold analysis of LC-MS/MS data from excised band from lane 5 showing peptide hits (yellow highlights) to 35% of complete MAP Fur sequence. (C) Western blot showing immunoprecipitation of Fur protein by anti-Fur antibody from MAP K-10 cultured under iron-replete and deplete condition. Lane 1, Protein ladder; Lane 2, Pull down of MAP K-10 cultured under iron-replete condition using 1 μg of anti-Fur antibody; Lane 3, Pull down of MAP K-10 cultured under iron-replete condition using 2 μg of anti-Fur antibody; Lane 4, Pull down of MAP K-10 cultured under iron-deplete condition using 1 μg of anti-Fur antibody.
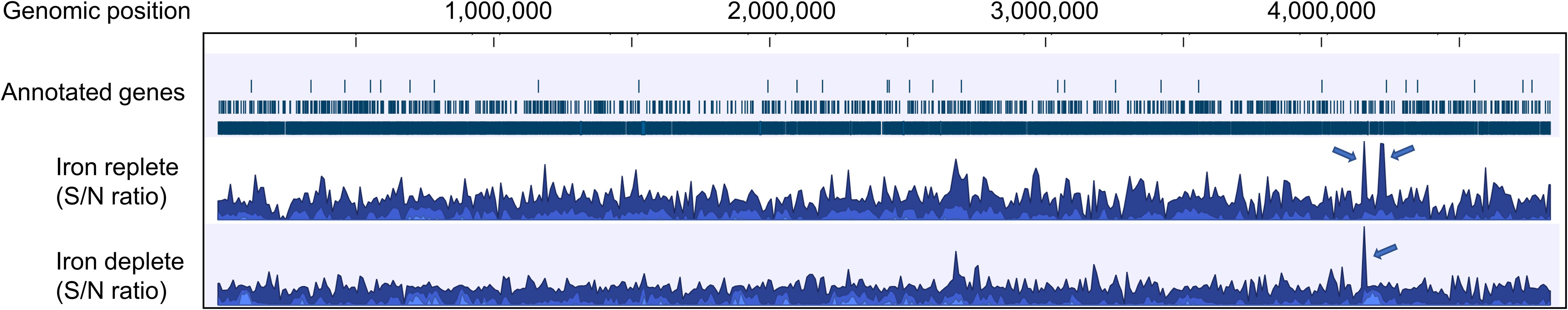
Figure 3. Overview of the mapped sequences within the reference genome MAP K-10 under iron-replete and -deplete conditions generated by CLC genomic workbench. After mapping onto the reference genome, iron-replete and -deplete samples were compared to control (input DNA), and signal-to-noise (S/N) ratio for peak calling was generated. Fur specifically binds various genomic loci under both conditions, but most of the ChIP peaks showed higher binding sites under iron-replete condition. Arrows indicate regions where ChIP peaks are associated with iron regulation.
ChIP peaks were called when the sequence coverage of genomic regions in the different treatments is enriched when compared to ChIP-seq control sample where the immunoprecipitation step was omitted (Strino and Lappe, 2016). Input DNA (no ChIP enrichment) had 34,907,295 (79.02% coverage against the MAP K-10 genome) uniquely mapped reads while ChIP-enriched DNA from iron-replete and iron-deplete conditions had 22,566,602 (55.66%) and 4,299,792 (14.73%) mapped reads, respectively.
Applying a P-value at ≤ 0.001, the ChIP-seq assay identified nine Fur binding sites out of 14 previously predicted by PRODORIC. ChIP-seq analysis revealed a total of 5,381 and 4,960 binding sites of Fur protein in the MAP K-10 genome (signal-to-noise ratio) under iron-replete and iron-deplete conditions, respectively (Figure 3).
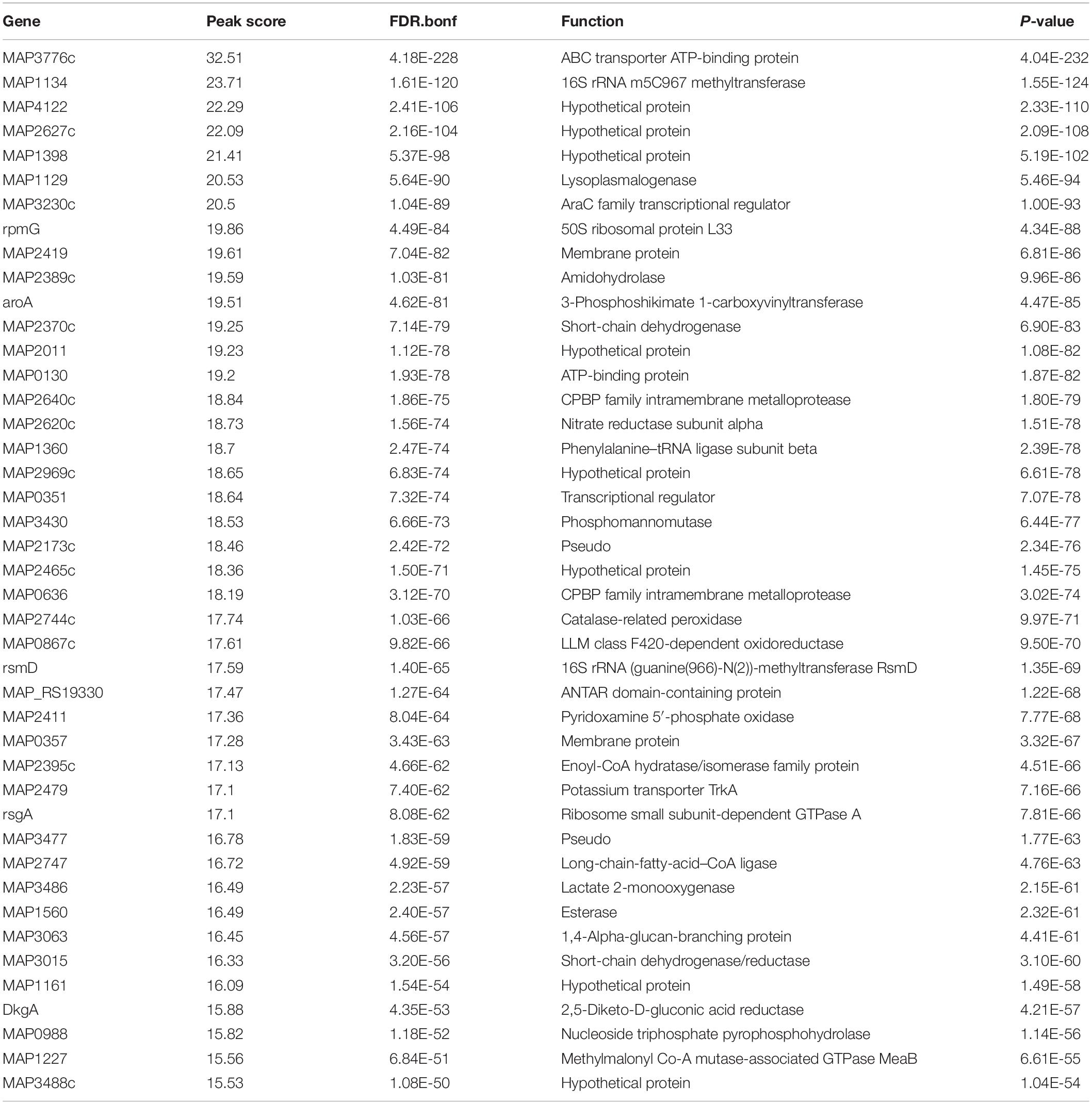
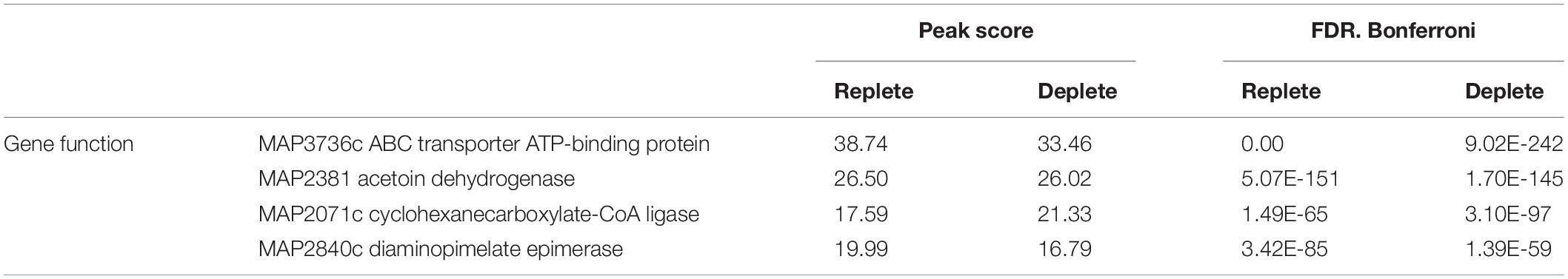
Applying a FDR at ≤ 10–50, under iron-replete conditions, a total of 43 significantly enriched regions were identified on the K-10 genome (Table 1). Peaks were either localized between open reading frames (ORFs) (27%; intergenic regions) and within annotated genes (73%). In contrast, under chelation treatment (iron depletion), 11 enriched regions were identified (Table 2), all showing binding sites within ORFs. Four ChIP peaks were present under both iron-replete and -deplete conditions simultaneously (Table 3). Diverse functions are encoded by genes where Fur bound on the MAP K-10 genome: cell wall synthesis, energy metabolism, respiration, and transcriptional/translation regulation. Out of 58 genes (FDR ≤ 10–50) from both conditions (Tables 1–3), 11 are annotated as hypothetical proteins, 2 are described as pseudogenes (Table 4), and three ChIP peaks are associated with iron regulation: MAP3638c, MAP3736c, and MAP3776c. Interestingly, Fur bound upstream of MAP3776c, an ABC transporter, only under iron-replete condition and binding to MAP3638c (hemophore-like protein) was identified only under iron-deplete conditions (Figures 4A,B).
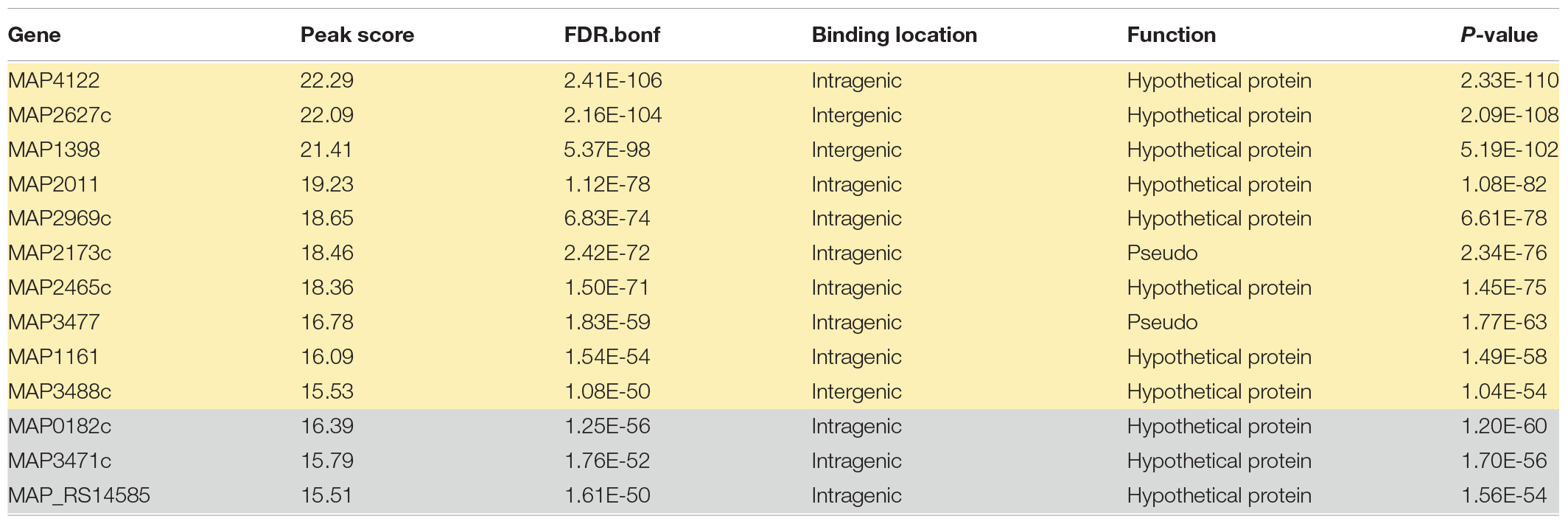
Table 4. List of genes regulated by Fur under iron-replete (yellow) and iron-deplete (gray) conditions with no function assigned, FDR ≤ 10–50.
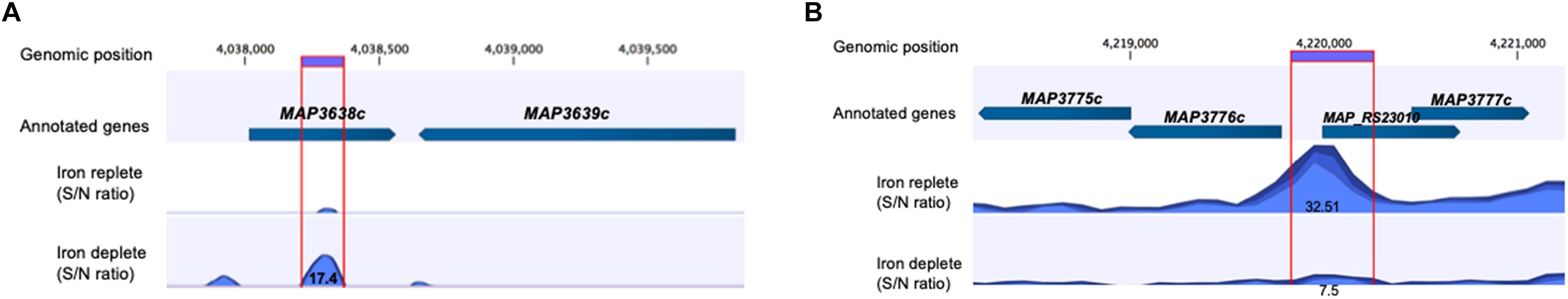
Figure 4. Applying FDR ≤ 10– 50, there are three ChIP peaks associated with iron regulation. (A) MAP Fur protein binds to the region of MAP3638c; however, only under iron-deplete condition is binding statically significant with a peak score of 17.4 (MAP3638c). (B) Under iron-replete conditions, there is a strong binding of MAP Fur to the region of MAP3776c represented by a peak score of 32.51.
Fur Binds to Fur Box Motif Under Iron-Replete or -Deplete Condition
Fur box consensus sequence was identified in ChIP-seq data using MEME-ChIP (Figure 5A). FIMO (Find Individual Motif Occurrences) analysis identified 15 occurrences of Fur box motif (P ≤ 0.001), 12 of them presented under iron-replete conditions and 3 under iron-deplete condition (Table 5).
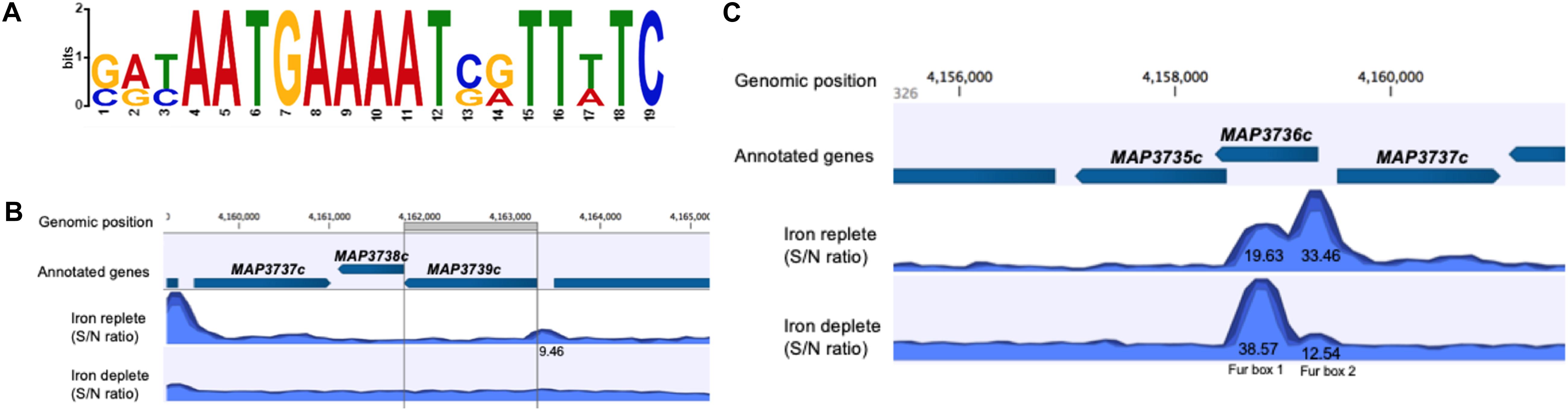
Figure 5. MAP Fur box analysis. (A) The most significant motif derived from ChIP-seq binding sequence using MEME. Height of each letter represents the relative frequency of each base at a different position in the consensus sequence. (B,C) A zoom-in of the MAP Fur boxes generated by CLC genomics. (B) Under both iron conditions, there is no binding of MAP Fur to the region of Fur box 3 (MAP3739c). ChIP peak (9.46) outside the ORF has FDR higher than the threshold of FDR ≤ 10– 50 (C) The enriched region of MAP Fur binding onto Fur Box 1 and 2 identified by ChIP-seq. ChIP peak showed higher occupancy under iron-deplete condition in the Fur Box 1 region. S/N denotes the signal-to-noise ratio for peak calling generated by CLC software.

Table 5. FIMO output. Most significant Fur box motif (SRYAATGAAAAT SRTTWTC) derived from ChIP-seq binding in iron-replete (yellow) and -deplete (gray) conditions.
From previous studies, it is known that the MAP K-10 genome contains three Fur box motifs (Stratmann et al., 2004). However, data from ChIP-seq showed that the Fur protein does not show significant binding (FDR ≥ 10–50) to the region of Fur box 3 (MAP3739c) (Figure 5B). The highest peak score from all ChIP-seq data was observed within and just upstream of MAP3736c, located on LSP14, MAP-specific genomic island (Alexander et al., 2009). Within MAP3736c (located between nucleotides 4158368 and 4159327), there are two putative Fur Boxes: Fur box 1 (located between nt 4158681 and 4158966 of the genome) and Fur box 2 (located between nt 4159132 and 4159456) (Stratmann et al., 2004). ChIP-seq analysis showed high binding in both regions, confirming the exact location (Figure 5C). When intracellular Fe2+ was depleted by the addition of 2,2-dipyridyl, MAP Fur bound with higher affinity to Fur box 1 region (peak score = 38.57) in contrast to a lower binding score for Fur box 2 (peak score = 12.54), while under replete conditions, where MAP was grown in complete media, the opposite was observed, a lower MAP Fur binding in the Fur box 1 region (peak score = 19.63) and a higher peak in Fur box 2 region (peak score = 33.46).
Validation of MAP Fur Binding
To confirm binding to the Fur promoter region, biotinylated or unlabeled PCR fragment including Fur box 1q identified by ChIP-seq was amplified and subjected to an electrophoretic mobility shift assay (EMSA) using purified MAP Fur protein (Figures 2A,B).
Titration of Fur protein in the presence of Mn2+ and 20 fmol of DNA showed that binding is dose-dependent, as the Fur concentration was increased, there was an increase of binding activity (Figure 6A). However, in the absence of Mn2+, Fur binding to DNA was not as efficient as in the presence of Mn2+ (Figure 6B).

Figure 6. EMSA analysis of MAP Fur binding to Fur box consensus DNA. Binding activity is represented by band intensity. Twenty femtomoles of MAP DNA including the Fur Box 1 consensus biotin labeled was run in a 5% native polyacrylamide gel with different concentrations of MAP Fur protein and Mn2+. (A) Protein–DNA binding is dose-dependent: titration of purified MAP Fur protein shows an increase of binding activity as more protein is added to the system. (B) Binding activity is more efficient in the presence of Mn2+: No addition of Mn2+ (Lane 5) binding occurs with a lower band intensity when compared to the sample with Mn2+ (Lanes 1–4). (C) Competitive EMSA. Fur protein was incubated with either biotin-labeled DNA probe or unlabeled DNA probe or with both. Biotin-labeled probe was detected using chemiluminescence-based nucleic acid detection kit. Addition of unlabeled DNA affects binding activity, showing binding specificity.
Furthermore, DNA–protein complex was specific to Fur binding site, as shown in the competition assay (Figure 6C), and adding a different concentration of excess unlabeled Fur box 1 probe competed with and abrogated labeled Fur box 1 probe binding to Fur protein.
Discussion
In this study, a full characterization of the Fur in MAP was performed. Fur and its involvement in iron homeostasis are well known in bacteria such as E. coli, Bacillus subtilis, and Salmonella Typhimurium. This protein has been shown to work as a repressor, by blocking RNA polymerase binding to the promoter region of genes involved in iron homeostasis by repressing transcription (Escolar et al., 1997), but can also work as an activator by positively regulating gene expression in response to iron through indirect mechanism involving repression of small regulatory RNA (Delany et al., 2001; Masse et al., 2005). The current study confirmed, by Western blot (Figure 2A) and mass spectrometry (Figure 2B), that MAP3773c encodes a Fur-like protein in MAP. A regulatory network of MAP Fur binding sites was identified using three independent approaches: (1) in silico (PRODORIC), (2) in vivo (ChIP-seq), and (3) in vitro (EMSA). In vivo and in vitro analyses established that Fur binding was responsive to iron availability.
ChIP-seq analysis expanded the number of MAP Fur binding sites, from 14 genes predicted by PRODORIC to 58 enriched binding regions (FDR ≤ 10–50). Binding locations were distributed almost evenly between intragenic and intergenic regions. While binding of Fur in intragenic regions refute the definition of a transcriptional factor (Browning and Busby, 2004), recent ChIP-seq studies with M. tuberculosis, E. coli, Salmonella, and Corynebacterium reported intragenic TF binding that play critical roles in transcription and significantly affect regulation of gene expression (Dillon et al., 2012; Fitzgerald et al., 2014; Knapp et al., 2015). Additionally, during characterization of the Fur regulon in Pseudomonas syringae, Butcher et al. (2011) did not observe general differences between Fur binding to intergenic and intragenic sites. Both showed comparable binding affinity in P. syringae, suggesting that, although 100% of MAP Fur binding under iron-deplete conditions are located in intragenic regions, MAP Fur can be biologically active and able to bind specific DNA sequences to control gene expression.
Iron regulation by Fur in MAP appears to be more complex than the classic model, where Fur acts as a repressor when sensing high intracellular Fe2+. It then forms the Fur–Fe2+ complex and binds to the Fur box sequence, which enables Fur transition from its inactive (apo-) to its activated (holo-) form (Hantke, 2001; Helmann, 2014). Additionally, data from the present study showed that MAP uses Fur in the absence of intracellular Fe2+, a process known as apo-regulation. In low-iron conditions, apo-Fur protein binds to the promoters of its target genes and regulates transcription (Miles et al., 2010).
The complexity of Fur regulation can be exemplified in the ChIP peak of MAP3736c, where apo-Fur binds to Fur box 1 under iron-deplete conditions and holo-Fur binds to Fur box 1 and 2 under iron-replete condition. The physiological significance of apo-Fur binding in MAP is unclear, however, previous studies with Helicobacter pylori showed that when iron levels are low, genes responsible for iron storage are repressed by apo-Fur (Bereswill et al., 2000). Furthermore, additional studies in Campylobacter jejuni showed that expression genes controlled by Fur was decreased in the wild-type strain under iron-deplete condition and, in a Fur knockout strain, expression was increased (Holmes et al., 2005), indicating that apo-Fur plays an important role in iron metabolism. Corroborating this result, ChIP-seq analysis identified apo-Fur binding to MAP3638c, only under iron starvation. MAP3638c is a hemophore-like protein, suggesting that MAP likely uses heme as an additional iron source as previously described in M. tuberculosis (Tullius et al., 2011).
Finally, to confirm and validate Fur-Fur box1 binding, an EMSA using PCR amplification of ChIP-seq-identified Fur box 1 and purified Fur-like protein (MAP3773c) was performed. The binding was dependent on the availability of Mn2+, a common surrogate metal that, unlike Fe2+, is stable in the presence of oxygen but promotes DNA binding and adopts the same coordination geometry as Fe2+ (Butcher et al., 2011). Additionally, a competitive gel shift assay confirmed specificity of MAP Fur binding to the Fur box 1 region. Taken together, the identification of consensus Fur box by ChIP-seq peaks combined with data from EMSA confirms that iron regulation in MAP is also mediated by a Fur homolog that recognizes the 19-bp DNA sequence, known as Fur box.
In this current study, we were not able to confirm Fur box 3 (MAP3739c) region as binding site for Fur protein as described by Stratmann et al. (2004). Computational methods as used by the group predicted binding sites relying on data available 15 years ago, which was likely incomplete. Further, most computational predictions of TF binding are prone to false discovery and need to be validated (Karimzadeh and Hoffman, 2018). By using directly and quantitatively sequencing in combination with specific antibody, as used in this currently study, ChIP-seq method provides a powerful strategy for identifying in vivo binding sites across entire genome (Collas, 2019).
Concluding Remark and Future Directions
In this work, we characterized MAP3773c, the Fur in MAP, using ChIP-seq. A genomic view of the MAP Fur regulatory network was identified, and several putative binding sites involved during iron-replete and -deplete conditions were discovered. Although this study is not a full description of the Fur regulon, our findings indicate that MAP Fur is a global regulator that recognizes many target sites in the genome, either by apo- or holo-Fur. Based on the proposed model by Lamont et al. (2013) where, in response to nitric oxide stress, MAP3737 (PPE family protein) acts as the iron sensor protein and promotes expression of MAP3734c-3736c, leading to activation of the iron uptake system, we hypothesize a stimulon regulatory pathway with two regulatory proteins (Fur and IdeR). In M. tuberculosis, genes from the PPE family are upregulated during iron limitation and are repressed by IdeR, suggesting possible involvement of MAP3737 in iron metabolism (Rodriguez et al., 2002). Thus, we proposed (Figure 7) that, during low-iron conditions, the iron sensor protein (MAP3737) activates a hypothetical master regulator (MR). The activation signal, which may or may not involve a phosphorylation cascade, leads to the transcription of apo-Fur that subsequently activates transcription of the iron uptake system. This leads to transport of carboxymycobactin (cMyco) into MAP. The cMyco + Fe3+ complex possesses FAD-binding activity, allowing it to interact with and activate the flavin iron reductase reducing Fe3+ to Fe2+. This is followed by disassociation of iron from the cMyco + Fe3+ complex and subsequent binding of liberated Fe2+ by Fur and IdeR. Fur–Fe2+ and Ide–Fe2+ can exert positive or negative regulation on the transcription of genes in their corresponding regulons. Further analysis of the complete MAP Fur regulon is underway; combining ChIP-seq data analysis from this work with another genome-scale experiment will provide a full understanding of direct or indirect roles of Fur in response to iron availability. To have a complete understanding of the MAP iron stimulon model, future studies will involve a basic understanding of Fur–IdeR interactions and how one or the other may be functional in MAP under a variety of in vivo and environmental conditions.
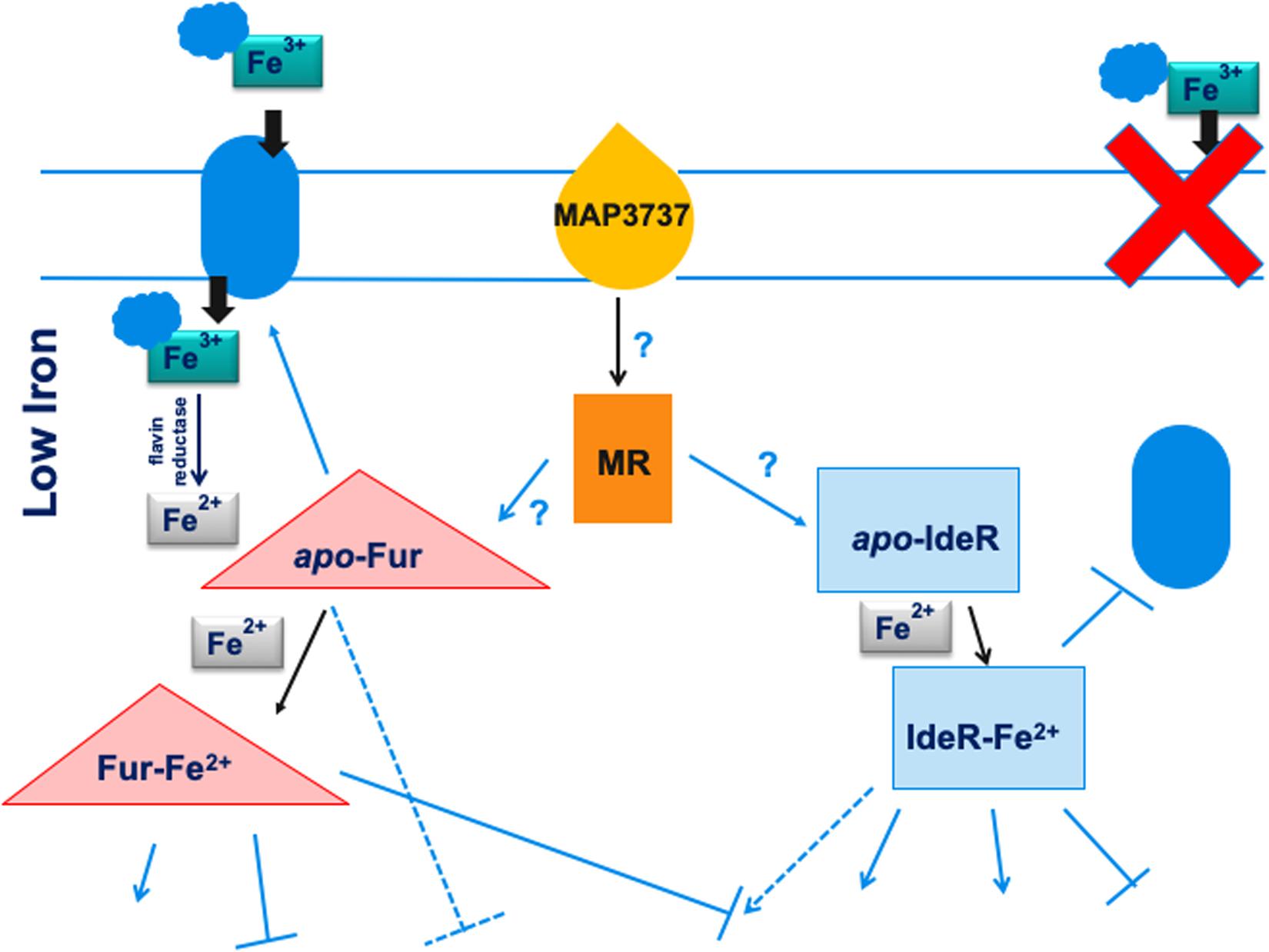
Figure 7. Model for the regulation of the iron stimulon in M. paratuberculosis. Under low-iron conditions, the iron sensor MAP3737 (yellow teardrop) initiates a signal transduction cascade activating a hypothetical master regulator (MR) of the stimulon (orange rectangle) leading to the transcription of apo-Fur and apo-IdeR. Under low iron, apo-Fur activates transcription of the iron uptake protein or system (blue oval) that is transported to the cell membrane and carries ferric iron bound to carboxymycobactin (cMyco) (blue cloud) into the bacterium. The cMyco + Fe3+ complex possesses FAD-binding activity, allowing interaction with an iron flavin reductase that converts Fe3+ to Fe2+ and disassociates the complex, liberating Fe2+. Apo-IdeR is inactive but, bound to iron (IdeR Fe2), represses transcription of iron import/export protein or system and iron transport is shut down (red X). In addition, bound to ferrous iron, either regulator can exert a positive (pointed blue arrows) or negative regulation (flat-headed arrows) on the transcription of genes in their corresponding regulons. Apo-Fur also exerts a regulatory effect on the Fur regulon. Some genes may be controlled by both Fur and IdeR in opposite ways (broken blue arrows). More speculative effects are depicted by arrows with question marks. Thus, in this model, both Fur and IdeR act in a coordinate fashion to regulate the iron stimulon composed of the Fur and IdeR regulons. Black pointed arrows are used for processes unrelated to transcription such as binding or signal transduction effects.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
SS conceived the idea, obtained funding, helped to develop a study design, and edited the manuscript. JB helped to develop the study design, served as a co-investigator on the USDA grant, developed the recombinant protein, and edited the manuscript. RB helped in the study design, served as a co-investigator on the grant, developed Figure 7 (a model for iron stimulon in MAP), and edited the manuscript. TJ analyzed the ChIP-seq data. FS developed the study design, performed all experiments, analyzed the data, and wrote the manuscript.
Funding
This work was funded by a NIFA grant (GRANT00169362) funded to SS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank John D. Helmann (Cornell University) for kindly providing us the EMSA protocol for Fur. We thank Evan P. Brenner (Michigan State University) for helpful discussions.
References
Alexander, D. C., Turenne, C. Y., and Behr, M. A. (2009). Insertion and deletion events that define the pathogen Mycobacterium avium subsp. paratuberculosis. J. Bacteriol. 191, 1018–1025. doi: 10.1128/JB.01340-08
Bagg, A., and Neilands, J. B. (1987). Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26, 5471–5477. doi: 10.1021/bi00391a039
Bannantine, J. P., Baechler, E., Zhang, Q., Li, L., and Kapur, V. (2002). Genome scale comparison of Mycobacterium avium subsp. paratuberculosis Reveals potential diagnostic sequences. J. Clin. Microbiol. 40, 1303–1310. doi: 10.1128/JCM.40.4.1303-1310.2002
Bannantine, J. P., and Paustian, M. L. (2006). Identification of Diagnostic Proteins in Mycobacterium avium subsp. paratuberculosis by a Whole Genome Analysis Approach. Diagnostic Bacteriology- Protocol – Second Edition. New Jersey: Humana Press.
Bereswill, S., Greiner, S., van Vliet, A. H., Waidner, B., Fassbinder, F., Schiltz, E., et al. (2000). Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182, 5948–5953. doi: 10.1128/jb.182.21.5948-5953.2000
Browning, D. F., and Busby, S. J. (2004). The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2, 57–65. doi: 10.1038/nrmicro787
Butcher, B. G., Bronstein, P. A., Myers, C. R., Stodghill, P. V., Bolton, J. J., Markel, E. J., et al. (2011). Characterization of the Fur Regulon in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 193, 4598–4611. doi: 10.1128/JB.00340-11
Chandrangsu, P., Rensing, C., and Helmann, J. (2017). Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350. doi: 10.1038/nrmicro.2017.15
Collas, P. (2019). Chromatin immunoprecipitation assays. Methods Mol. Biol. 567, 1–26. doi: 10.1007/978-1-60327-414-2
Delany, I., Spohn, G., Rappuoli, R., and Scarlato, V. (2001). The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42, 1297–1309. doi: 10.1046/j.1365-2958.2001.02696.x
Dillon, S. C., Espinosa, E., Hokamp, K., Ussery, D. W., Casadesus, J., and Dorman, C. J. (2012). LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 85, 1072–1089. doi: 10.1111/1462-2920.13838
Eckelt, E., Jarek, M., Fromke, C., Meens, J., and Goethe, R. (2014). Identification of a lineage specific zinc responsive genomic island in Mycobacterium avium subspecies paratuberculosis. BMC Genomics 15:1076. doi: 10.1186/1471-2164-15-1076
Eckelt, E., Meiβner, T., Meens, J., Laarmann, K., Nerlich, A., Jarek, M., et al. (2015). FurA contributes to the oxidative stress response regulation of Mycobacterium avium ssp. paratuberculosis. Front. Microbiol. 6:16. doi: 10.3389/fmicb.2015.00016
Escolar, L., de Lorenzo, V., and Pérez-Martíín, J. (1997). Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol. Microbiol. 26, 799–808. doi: 10.1046/j.1365-2958.1997.6211987.x
Escolar, L., Perez-Martin, J., and de Lorenzo, V. (1999). Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181, 6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999
Fitzgerald, D. M., Bonocora, R. P., and Wade, J. T. (2014). Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet. 10:e1004649. doi: 10.1371/journal.pgen.1004649
Garcia, A. B., and Shalloo, L. (2015). The economic impact and control of paratuberculosis in cattle. J. Dairy Sci. 98, 5019–5039. doi: 10.3168/jds.2014-9241
Grant, C. E., Bailey, T. L., and Noble, W.S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Hantke, K. (1981). Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182, 288–292. doi: 10.1007/bf00269672
Hantke, K. (2001). Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4, 172–177. doi: 10.1016/s1369-5274(00)00184-3
Helmann, J. D. (2014). Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J. Biol. Chem. 289, 28112–28120. doi: 10.1074/jbc.R114.587071
Holmes, K., Mulholland, F., Pearson, B. M., Pin, C., McNicholl-Kennedy, J., Ketley, J. M., et al. (2005). Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151, 243–257. doi: 10.1099/mic.0.27412-0
Jaini, S., Lyubetskaya, A., Gomes, A., Peterson, M., Park, S. T., Raman, S., et al. (2014). Transcription factor binding site mapping using ChIP-Seq. Microbiol. Spectr. 2. doi: 10.1128/microbiolspec.MGM2-0035-2013
Janagama, H. K., Lamont, E. A., George, S., Bannantine, J. P., Xu, W. W., Tu, Z. J., et al. (2010). Primary transcriptomes of Mycobacterium avium subsp. paratuberculosis reveal proprietary pathways in tissue and macrophages. BMC Genomics 11:561. doi: 10.1186/1471-2164-11-561
Janagama, H. K., Senthilkumar, T. M., Bannantine, J. P., Rodriguez, G. M., Smith, I., Paustian, M. L., et al. (2009). Identification and functional characterization of the iron- dependent regulator (IdeR) of Mycobacterium avium subsp. paratuberculosis. Microbiology 155, 3683–3690. doi: 10.1099/mic.0.031948-0
Karimzadeh, M., and Hoffman, M. M. (2018). Virtual ChIP-seq: predicting transcription factor binding by learning from the transcriptome. bioRvix
Knapp, G. S., Lyubetskaya, A., Peterson, M. W., Gomes, A. L. C., Ma, Z., Galagan, J. E., et al. (2015). Role of intragenic binding of cAMP responsive protein (CRP) in regulation of the succinate dehydrogenase genes Rv0249c-Rv0247c in TB complex mycobacteria. Nucleic Acids Res. 43, 5377–5393. doi: 10.1093/nar/gkv420
Lamont, E. A., Xu, W. W., and Sreevatsan, S. (2013). Host-Mycobacterium avium subsp. paratuberculosis interactome reveals a novel iron assimilation mechanism linked to nitric oxide stress during early infection. BMC Genomics. 14:694. doi: 10.1186/1471-2164-14-694
Lee, J. W., and Helmann, J. D. (2007). Functional specialization within the fur family metalloregulators. Biometals 20, 485–499. doi: 10.1007/s10534-006-9070-7
Li, L., Bannantine, J. P., Zhang, Q., Amonsin, A., May, B. J., Alt, D., et al. (2005). The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. U.S.A. 102, 12344–12349. doi: 10.1073/pnas.0505662102
Masse, E., Vanderpool, C. K., and Gottesman, S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005
Miles, S., Carpenter, B. M., Gancz, H., and Merrell, D. S. (2010). Helicobacter pylori apo-fur regulation appears unconserved across species. J. Microbiol. 48, 378–386. doi: 10.1007/s12275-010-0022-0
Mills, S. A., and Marletta, M. A. (2005). Metal binding characteristics and role of iron oxidation in the ferric uptake regulator from Escherichia coli. Biochemistry 44, 13553–13559. doi: 10.1021/bi0507579
Münch, R., Hiller, K., Grote, A., Scheer, M., Klein, J., Schobert, M., et al. (2005). Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21, 4187–4189. doi: 10.1093/bioinformatics/bti635
Rathnaiah, G., Zinniel, D. K., Bannantine, J. P., Stabel, J. R., Gröhn, Y. T., Collins, M. T., et al. (2017). Pathogenesis, molecular genetics, and genomics of Mycobacterium avium subsp. paratuberculosis, the etiologic Agent of Johne’s Disease. Front. Vet. Sci. 4:187. doi: 10.3389/fvets.2017.00187
Rodriguez, G. M., Voskull, M. I., Gold, B., Schoolnik, G. K., and Smith, I. (2002). IdeR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70, 3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002
Stratmann, J., Strommenger, B., Goethe, R., Dohmann, K., Gerlach, G. F., Stevenson, K., et al. (2004). A 38-kilobase pathogenicity island specific for Mycobacterium avium subsp. paratuberculosis encodes cell surface proteins expressed in the host. Infect. Immun. 72, 1265–1274. doi: 10.1128/iai.72.3.1265-1274.2004
Strino, F., and Lappe, M. (2016). Identifying in *-seq data using shape information. BMC Bioinformatics 17:S206. doi: 10.1186/s12859-016-1042-5
Tullius, M. V., Harmston, C. A., Owens, C. P., Chim, N., Morse, R. P., McMath, L. M., et al. (2011). Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U.S.A. 108, 5051–5056. doi: 10.1073/pnas.1009516108
Wang, J., Moolji, J., Dufort, A., Staffa, A., Domenech, P., Reed, M. B., et al. (2016). Iron acquisition in Mycobacterium avium subsp. paratuberculosis. J. Bacteriol. 198, 857–866. doi: 10.1128/JB.00922-15
Wang, J., Pritchard, J. R., Kreitmann, L., Montpetit, A., and Behr, M. A. (2014). Disruption of Mycobacterium avium subsp. paratuberculosis-specific gene impairs in vivo fitness. BMC Genomics. 15:415. doi: 10.1186/1471-2164-15-415
Keywords: Mycobacterium avium subsp. paratuberculosis, Fur, iron, regulon, ChIP-seq
Citation: Shoyama FM, Janetanakit T, Bannantine JP, Barletta RG and Sreevatsan S (2020) Elucidating the Regulon of a Fur-like Protein in Mycobacterium avium subsp. paratuberculosis (MAP). Front. Microbiol. 11:598. doi: 10.3389/fmicb.2020.00598
Received: 18 December 2019; Accepted: 18 March 2020;
Published: 23 April 2020.
Edited by:
Biswarup Mukhopadhyay, Virginia Tech, United StatesReviewed by:
Marcel Behr, McGill University, CanadaChristopher L. Hemme, The University of Rhode Island, United States
Hualiang Pi, Vanderbilt University Medical Center, United States
Copyright © 2020 Shoyama, Janetanakit, Bannantine, Barletta and Sreevatsan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Srinand Sreevatsan, c3JlZXZhdHNAbXN1LmVkdQ==
 Fernanda Miyagaki Shoyama
Fernanda Miyagaki Shoyama Taveesak Janetanakit2
Taveesak Janetanakit2 John P. Bannantine
John P. Bannantine Raul G. Barletta
Raul G. Barletta Srinand Sreevatsan
Srinand Sreevatsan