- 1Department of Dermatology and Venerology, Peking University First Hospital, Beijing, China
- 2Research Center for Medical Mycology, Peking University, Beijing, China
- 3National Clinical Research Center for Skin and Immune Diseases, Beijing, China
- 4Microbiology, Parasitology and Pathology Post-Graduation Program, Department of Pathology, Federal University of Paraná, Curitiba, Brazil
- 5Center of Expertise in Mycology of Radboud University Medical Center/Canisius Wilhelmina Hospital, Nijmegen, Netherlands
- 6Graduate Program in Bioprocess Engineering and Biotechnology, Federal University of Paraná, Curitiba, Brazil
- 7Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands
- 8Amsterdam Medical Center, Amsterdam, Netherlands
The two black yeasts Exophiala dermatitidis and Exophiala spinifera that are clinically considered as the most virulent species potentially causing disseminated infections are both producing extracellular capsule-like material, are compared. In this study, 10 genomes of E. spinifera and E. dermatitidis strains, including both clinical and environmental isolates, were selected based on phylogenetic analysis, physiology tests and virulence tests, sequenced on the Illumina MiSeq sequencer and annotated. Comparison of genome data were performed between intraspecific and interspecific strains. We found capsule-associated genes were however not consistently present in both species by the comparative genomics. The prevalent clinical species, E. dermatitidis, has small genomes containing significantly less virulence-associated genes than E. spinifera, and also than saprobic relatives. Gene OG0012246 and Myb-like DNA-binding domain and SANT/Myb domain, restricted to two strains from human brain, was shared with the neurotropic species Rhinocladiella mackenziei. This study indicated that different virulence profiles existed in the two capsule-producing black yeasts, and the absence of consistent virulence-associated profiles supports the hypothesis that black yeasts are opportunists rather than primary pathogens. The results also provide the key virulence genes and drive the continuing research forward pathogen–host interactions to explore the pathogenesis.
Introduction
Black yeasts in the ascomycete order Chaetothyriales are relatively frequent opportunistic agents of human disease. Species of Chaetothyriales producing budding cells in any stage of their life cycle are classified currently in the genus Exophiala. The Atlas of Clinical Fungi (de Hoog et al., 2019) lists 19 species that were proven to have been involved in infections of humans or cold-blooded vertebrates. Of these, Exophiala dermatitidis and Exophiala spinifera are the most common species in clinical settings, and are the only recurrent agents of severe, deep and disseminated infections in humans, then often with fatal outcome. In the past, patients were reportedly without significant underlying disease, but recent research has shown that such infections are mostly associated with an inherited defect in the dectin signaling pathway due to mutations in the CARD9 gene (Lanternier et al., 2015).
Virulence of black yeasts has been attributed to the presence of melanin in cell walls (Taborda et al., 2008), but since all members of the order Chaetothyriales are consistently melanized, this does not explain the difference in infective ability between species. Thermotolerance, for which marked differences are noted between species, is another important factor: 11 out of 19 opportunistic Exophiala species are at least weakly tolerant of 37°C (de Hoog et al., 2019), with E. dermatitidis and E. spinifera having the most pronounced thermotolerance. However, de Hoog et al. (2011) described a ‘waterborne clade’ of mesophilic species in the genus, comprising species commonly infecting fish, frogs, toads or crabs, often at epidemic proportions (Vicente et al., 2012; Pearson et al., 2019; Saraiva et al., 2019). Consequently, black yeasts lacking thermotolerance are also able to cause vertebrate infection. Gostincar et al. (2018) suggested polyextremotolerance as a prerequisite for opportunism, highlighting the interplay of several, independent factors enabling growth under non-optimal conditions of animal tissue. The black yeasts and relatives mitigate external stress of, e.g., dryness and irradiation by melanin, and detrimental effects of toxin can be compensated by pathways of the cytochrome P450 (Moreno et al., 2018) aiding degradation and co-assimilation of monoaromatic hydrocarbons. This combination of vitality factors provides a wide array of survival strategies, and it may be hypothesized that species with the most pronounced development of such pathways may have a higher ability of tissue invasion. In addition, Yurlova and de Hoog (2002) noted that E. dermatitidis and E. spinifera are unique in producing extracellular capsule-like structures during early exponential growth of budding cells. This may further enhance opportunism. Both species are able to disseminate in susceptible patients with formation of cutaneous acanthosis, but a difference in clinical predilection has been noted: E. dermatitidis has a tendency of neurotropism, whereas E. spinifera seems to be somewhat osteotropic (Song et al., 2017). This suggests that opportunism in these fungi might be fine-tuned.
The aim of the present paper is to compare the genomes of the above opportunists in Exophiala with each other and with species with other types or with no opportunistic potentials, in search of genes that might play a role in the above described differences. Several genomes were sequenced of each species, enabling to compare the intra-species variability in the genome in general and in potential virulence genes.
Materials and Methods
Strains, DNA Extraction, and Genome Sequencing
To extract genomic DNA, fungal mycelia of E. dermatitidis strains CBS 109144, CBS 115663, CBS 120473, CBS 132758, CBS 132754, and CBS 578.76, and E. spinifera strains CBS 101539, CBS 116557, CBS 123469, CBS 126013, and CBS 131564 were harvested from fresh cultures on Sabouraud’s Glucose Agar (GSA), washed using sterile Tris-EDTA buffer (TE), pH 8.0 in 2 mL vol screw-capped tubes, and then resuspended in 500 ml TE buffer. Fungal cell walls were disrupted using 0.5 mm glass beads in a BioSpec Mini-Beadbeater-16 (BioSpec) for 5 min and cooled on ice for an additional 5 min. DNA solutions were separated using two phenol/chloroform (1:24, pH 8.0) extractions. DNA was then precipitated by isopropanol, washed with 70% ethanol, dried at room temperature, and resuspended in 35 mL TE buffer, pH 8.0. DNA quantity and quality were determined using Qubit (Invitrogen, Applied BioSystems), and an Agilent Bio Analyzer 2100 using a 1000 DNA Chip (Agilent). Cardinal growth temperatures were determined on 2% malt extract agar (MEA; Difco). Plates were incubated at 37, 40, 42, 45°C in the dark for 2 weeks; plates contained double quantities of medium and were sealed to prevent drying out. Colony diameters were measured for studied strains.
Genome Sequencing
The extracted genome DNA of E. spinifera and E. dermatitidis strains were sequenced on the MiSeq (IlluminaTM, San Diego, CA, United States) sequencer using paired-end and mate-paired libraries. The library construction was done with Ion Plus Fragment Library Kit (Thermo Fisher ScientificTM) and Nextera XT (IlluminaTM) following the manufacturer instructions.
Genome Assembly
Quality control of the reads was performed using FastQC v0.11.81, and low-quality sequences were removed by BBMap2. High-quality reads were assembled by SPAdes v3.10.0 (Bankevich et al., 2012) using the respective reference genomes, E. dermatitidis CBS 525.76 = NIH/UT8656 and E. spinifera CBS 899.68, except to the Exophiala phaeomuriformis, that was made a de novo assembly. To finish the assemblies, the gaps were closed using FGAP (Piro et al., 2014). To identify de novo repeats across the genome assembly, the library produced by RepeatModeler was used as input for RepeatMasker v4.0.7. Genes were predicted by producing a training set using Genemark-ES v4.30 (Lomsadze et al., 2005). The predicted functional annotations and proteins it was performed by InterProScan v5.27-66.0 (Quevillon et al., 2005).
Read Mapping and SNP Calling
For SNP calling, high-quality sequencing reads of each of the genomes were mapped against the reference genome of the reference strains E. dermatitidis CBS 525.76 = NIH/UT8656 and E. spinifera CBS 899.68 deposited at GenBank, respectively, by using Burrows Wheeler Aligner (BWA) v0.7.17-r1188 mem (Li and Durbin, 2009) and sorted to the bam format using SAMtools v1.7. They were then marked duplicated using Picard v1.83 and indexed using SAMtools. Variants were identified using GATK HaplotypeCaller v3.4.9.0. SNP annotation was performed by VCFannotator4 to assess whether the SNP was found within an untranslated region, intron, or coding exon, and mutations were classified into synonymous (SYN), non-synonymous (NSY), read-through (RTH), and nonsense (STP).
Ortholog Detection
The protein sequences of the 10 strains of the two species E. dermatitidis and E. spinifera were clustered using OrthoFinder v2.1.2 (Emms and Kelly, 2015) to determine which proteins were shared or specific by the two species. In addition, the protein sequences of the five E. dermatitidis strains (CBS 109144, CBS 115663, CBS 120473, CBS 132758, and CBS 578.76) were also clustered to see which proteins were shared between isolates from brain (CBS 578.76, CBS 120473) and environmental isolates (CBS 109144, CBS 115663, CBS 132758).
Possible relationships of analyzed gene families with invasion of cerebral versus bone tissue which are critical virulence genes for the understanding of black yeast virulence, were searched in the literature, with functional confirmation in the online InterPro database5. Relevant genes and domains were blasted against the E. spinifera and E. dermatitidis genomes.
Results
Data Quality and Identification
Genomes of five strains of each species were sequenced (Table 1). Different techniques were applied, i.e., Pacbio and Illumina. Comparison of genome data of the same strain of E. spinifera, BMU00047 (JAAABH000000000) with the reference genome (CBS 899.68, JYBY00000000.1) provided perfect match with Illumina, but some translocations were noted compared to PacBio data (Figure 1). The genomes assembled with PacBio had less gaps and had more copies of the same gene. However, when single-genes were counted without duplication, the results of genome assembly are very similar to those assembled with Illumina reads (Figure 1). The reads of the newly sequenced strains of the two black yeasts E. dermatitidis and E. spinifera were quality-controlled. Low-quality reads were removed. The size of the forward/reverse sequencing reads of these strains ranged from 400 to 700M. The number of scaffolds obtained for each genome is presented in Table 2. The high-quality forward/reverse sequencing reads were assembled using de novo assembly. The genomes had sizes ranging between 25.88 and 34.3M. The genomes of E. dermatitidis on average were smaller than those of E. spinifera. Intraspecific variability was evaluated using Orthofinder, for strains of E. dermatitidis and of E. spinifera separately. In total, 55,750 clustered genes were found. Percentages of genomic variation ranged from 0.2 to 8.6% (1,626 genes) in E. dermatitidis genomes, while for E. spinifera, with a total of 108,201 genes, variation ranged from 0.5 to 3.2% (2,052 genes). In the genomic dendrogram (Figure 2), E. spinifera strains demonstrated a higher degree of branching, which is explained by a lower percentage (71.6%) of single-copy orthogroups (n = 8,979), while for E. dermatitidis this was 85.7% (n = 7,640). Estimation of gene gains and losses shows that E. spinifera has higher divergence than E. dermatitidis, with averages of 268.13 gains and 74.75 losses, versus 21 gains and 31 losses per speciation event, respectively. Compared to sibling species E. phaeomuriformis of E. dermatitidis and E. oligosperma of E. spinifera, considerable gene gains and expansions were noted (Figure 2), which deviated significantly (3109.8 genes on average) from intra-specific changes. Gene dynamics also differed between the species of study, i.e., on average 2111 genes (σ 314.3 in shared genes and 608.1 in specific genes) in E. dermatitidis, and on average 4107.8 (σ 121.18 in shared genes and 55.9 in specific genes) in E. spinifera.
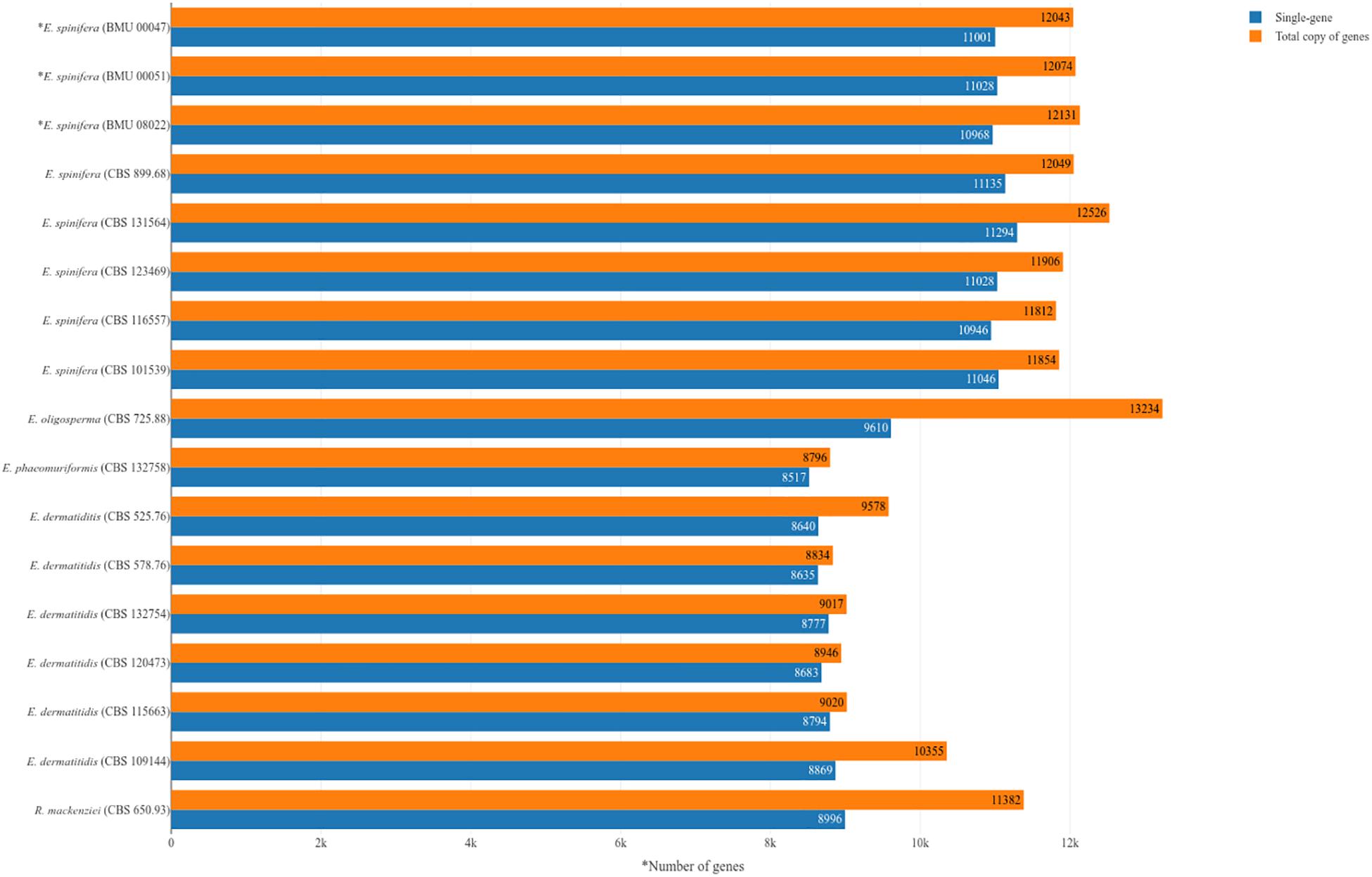
Figure 1. Numbers of genes in analyzed strains of Exophiala spinifera and E. dermatitidis, compared with reference genomes. ∗Strains analyzed with combined Illumina and PacBio approach; remaining strains analyzed with Illumina.
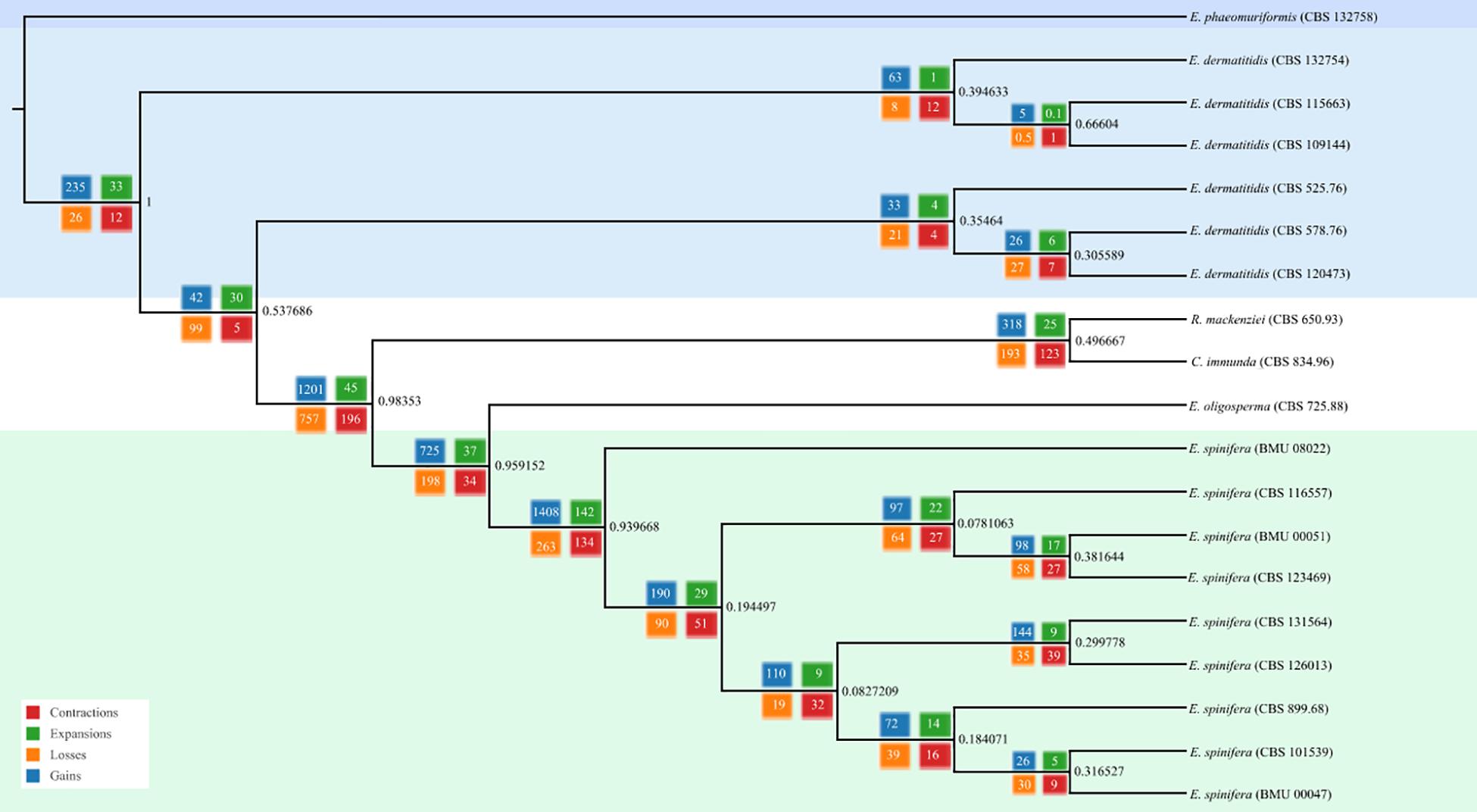
Figure 2. Phylogenomic tree based on whole genomic analysis using OrthoFinder; 5,374 single-copy orthogroups were detected, representing 41.86% of orthologous groups.
The core genomes of 6,812 genes were determined by comparing the shared genes in all Exophiala species sequenced to date (Figure 3). The accessory genomes of E. spinifera were considerably larger than those of E. dermatitidis, but CBS 115663 and particularly CBS 109144 in the latter species deviated considerably from remaining strains by having 580 and 1,586 unique genes, respectively. The use of long reads provides better resolution, which is particularly significant for repetitive regions of the genome. Whole-genome comparison of all genomes of the two species with reference genomes of E. phaeomuriformis (nearest neighbor of E. dermatitidis), E. oligosperma (nearest neighbor of E. spinifera), Cladophialophora immunda (saprobic hydrocarbon-assimilating species) and Rhinocladiella mackenziei (opportunistic neurotropic species) was based on detection of 5,374 single-copy orthogroups representing 41.86% of orthologous groups. The genomic dendrogram (Figure 2) showed correct clustering of all genomes, but with significant intra-specific variability, which in E. spinifera was twice that of E. dermatitidis.

Figure 3. Core and accessory genomes of E. spinifera and E. dermatitidis, showing the number of unique genes in each strain.
Protein Prediction and SNP Calling
Protein sequences of the new sequenced strains were predicted using the homology-based predictor InterProScan v5.27-66.0 (Quevillon et al., 2005). The numbers of the obtained genes for each of the strain are given in Table 2. Protein sequences of the 10 genomes of the two species were clustered to detect orthologs using OrthoFinder v2.1.2 (Emms and Kelly, 2015). A total of 12,690 clusters was detected. Among these, 7,291 clusters had protein sequences shared by E. dermatitidis and E. spinifera; 1,456 clusters were represented in the E. dermatitidis proteome only, and 3,253 clusters were exclusively present in E. spinifera.
In order to detect genomic variants, the high-quality sequencing reads of all genomes of the two species were aligned with the respective reference genomes, E. dermatitidis CBS 525.76 = NIH/UT8656 and E. spinifera CBS 899.68. The total numbers of genomic variants and non-synonymous SNPs of the E. dermatitidis and E. spinifera genomes are given in Table 3. The numbers of non-synonymous SNPs in E. dermatitidis varied from 8,887 to 30,126, and in E. spinifera from 17,978 to 43,282. The two brain-associated isolates of E. dermatitidis, CBS 528.76 and CBS 120473, shared two clusters containing genes with unique SNPs. The first cluster contained the genes HMPREF1120_03262 of E. dermatitidis and Z518_05210 of R. mackenziei that was linked to benzaldenhyde dehydrogenase. The second cluster contained genes HMPREF1120_08762 of E. dermatitidis and Z518_06196 of R. mackenziei.
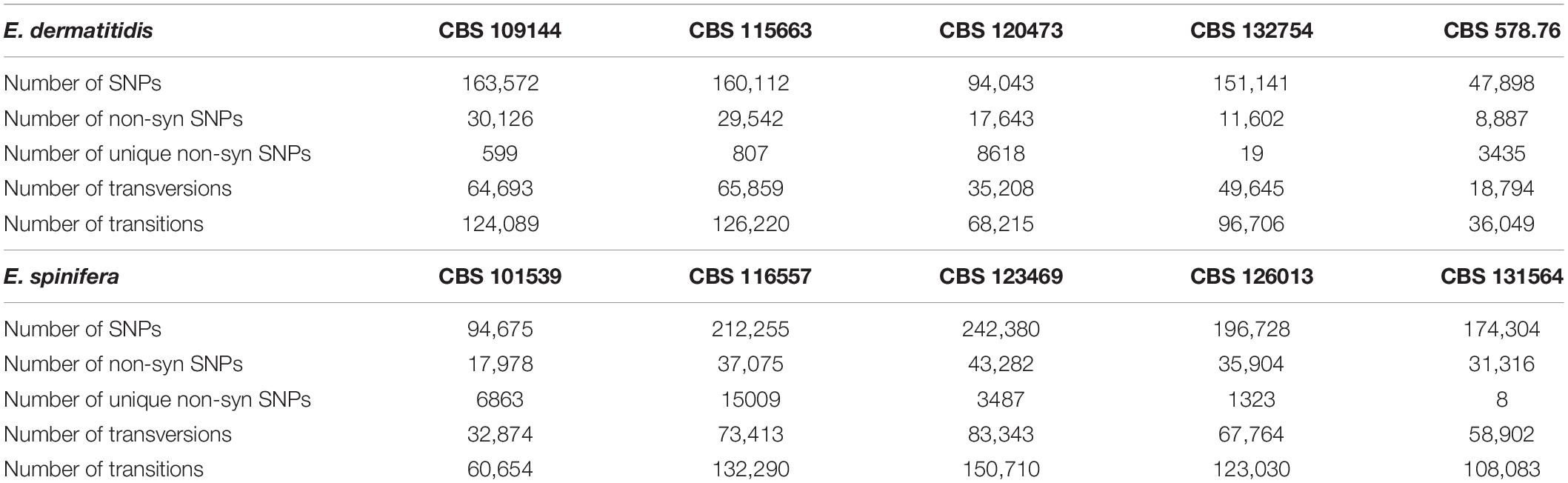
Table 3. Synonymous and non-synonymous SNP numbers of E. dermatitidis (CBS 525.76) and E. spinifera (CBS 899.68), compared with reference genomes.
Metabolism of Aromatic Compounds
To identify the presence of genes involved in pathways for the degradation of monoaromatic hydrocarbons (e.g., benzene, toluene, ethylbenzene, xylene, styrene, and other volatile pollutants), the genomes of reference strains of E. dermatitidis (CBS 525.76 = NIH/UT8656, AFPA00000000.1), E. spinifera (CBS 899.68, JYBY00000000.1), R. mackenziei (CBS 650.93, JYBU00000000.1), and C. immunda (CBS 834.96, JYBZ00000000.1) were assessed for the presence of genes required for this assimilation. The protein sequences of these strains clustered to 16,822 groups. No genes of the styrene degradation pathway were detected, but 16 clusters contained protein sequences required in the toluene pathway (Oelschlägel et al., 2018; Qu et al., 2019). Exophiala spinifera and E. dermatitidis differed significantly in the number of CYP450 genes, which were 123–133 and 61–72, respectively (Figure 4). In E. spinifera, dioxygenase (P340), which catalyzes the opening of the benzene ring, was missing in most of the strains. Several strains of both species lack β-carboxy-muconolactone hydrolase which is involved in degradation of the product of dioxygenase activity. Some variation was noted in the number of genes between strains of the same species, particularly in E. spinifera.
Virulence Genes
A total of 813 genes associated with virulence and 18 virulence domains were detected in E. dermatitidis and E. spinifera (Table 4). The highest number (n = 458) was found in C. immunda, CBS 834.96, the lowest (n = 267) in E. dermatitidis, CBS 578.76. The number of virulence genes in E. spinifera ranged between 389 and 432, and in E. dermatitidis between 267 and 303. Of the genes that are possibly linked with brain invasion (n = 192), OG0012246 was consistently detected in E. dermatitidis and R. mackenziei. Hypothetical proteins OG0012600 and OG0012602 were lacking in some E. dermatitidis. None of these proteins was present in E. spinifera. Thirty-four genes have been suggested to play a role in bone invasion (Supplementary Table S-1). Of these, 3 were detected in E. dermatitidis and E. spinifera, of which linker histone H1/H4 and proline racemase were present in both species. In E. spinifera strains, proline racemase was replaced by proline oxidase (McKenna et al., 2014).
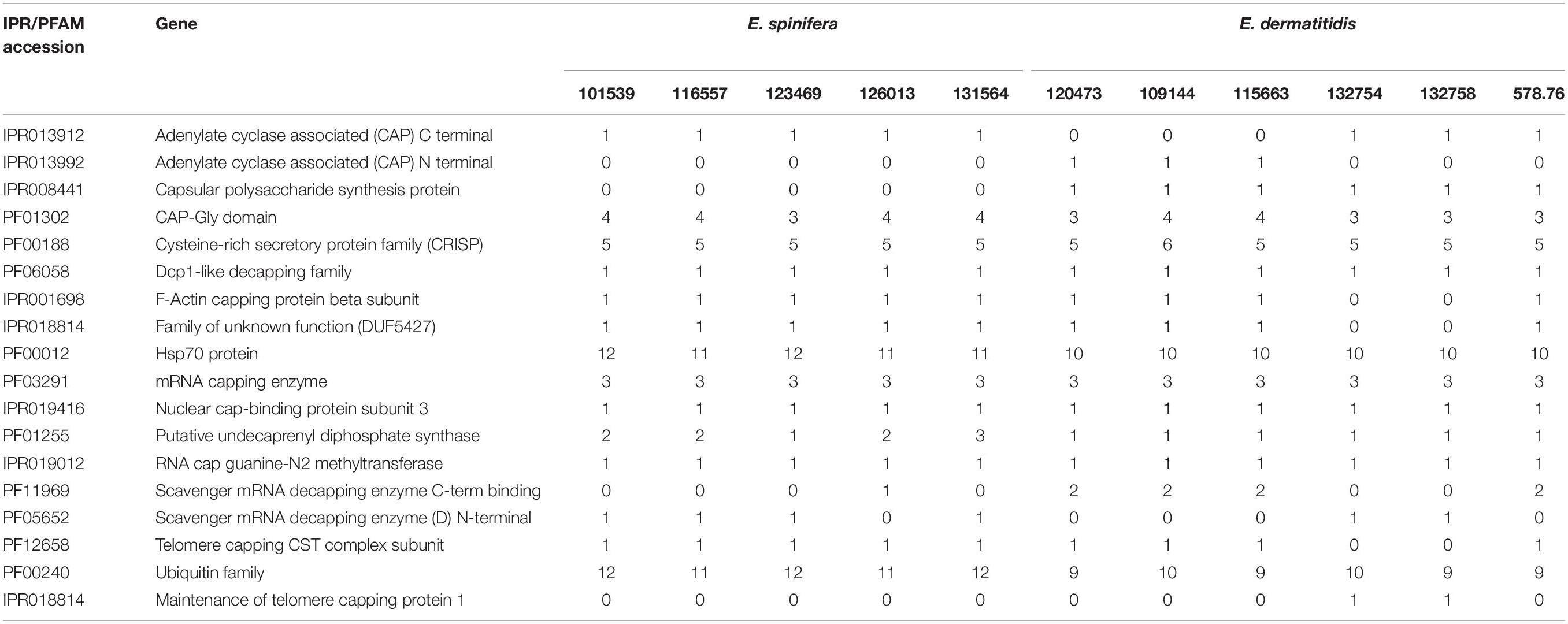
Table 4. Virulence domains detected in strains of Exophiala spinifera and E. dermatitidis (CBS accession numbers).
Gene families PFam and IPR in the literature have been associated with virulence; strains were found to differ rather considerably in the presence of these genes (Table 4). CAP Gly domain, cysteine-rich secretory protein family, Hsp70 protein, mRNA capping enzyme and ubiquitin family were consistently present with higher copy numbers (Choudhary and Schneiter, 2012; Chen et al., 2019). From the virulence gene prediction, the adenylate cyclase associated (CAP) Gly-domain was present in both species, while capsular polysaccharide synthesis protein and CAP N-terminal were present only in some of the E. dermatitidis strains. CAP C-terminal was consistently present in E. spinifera but lacking in three E. dermatitidis strains.
Two E. dermatitidis strains, CBS 578.76 and CBS 120473, were isolated from human brain. The species is known to be neurotropic upon dissemination (Sudhadham et al., 2008). The strains were found to be close to each other, clustering with strain CBS 132758 collected from a dishwasher (Figure 1). There were 18,283 unique SNPs present in the strains from brain, associated with 1,144 genes. To reveal possible differences between E. dermatitidis isolates from human brain and from the environment, the protein sequences of the five E. dermatitidis genomes were clustered; 9,151 clusters were obtained. Among these, 876 clusters contained protein sequences that were only found in the two brain isolates CBS 578.76 and CBS 120473. The proteins in CBS 578.76 and CBS 120473 shared two domains, i.e., Myb-like DNA-binding domain and SANT/Myb domain (OG0012246) (Supplementary Table S-2). The former protein has morphogenetic roles (Wieser and Adams, 1995), while the latter enhances development and pathogenicity (Kim et al., 2014). Myb-like DNA-binding domains of RNA polymerase III transcription factor IIIB (TFIIIB) have a function in the assembly of DNA complexes and recruitment of RNA polymerase to the promoter (Kassavetis et al., 1997).
Exophiala phaeomuriformis differs phenotypically from E. dermatitidis by its lower thermotolerance (de Hoog et al., 2019). This was confirmed in the analyzed set of strains: despite significant variation in growth velocity, E. dermatitidis strains all were able to grow at 45°C, while the maximum of E. phaeomuriformis was at 40°C (Figure 5).
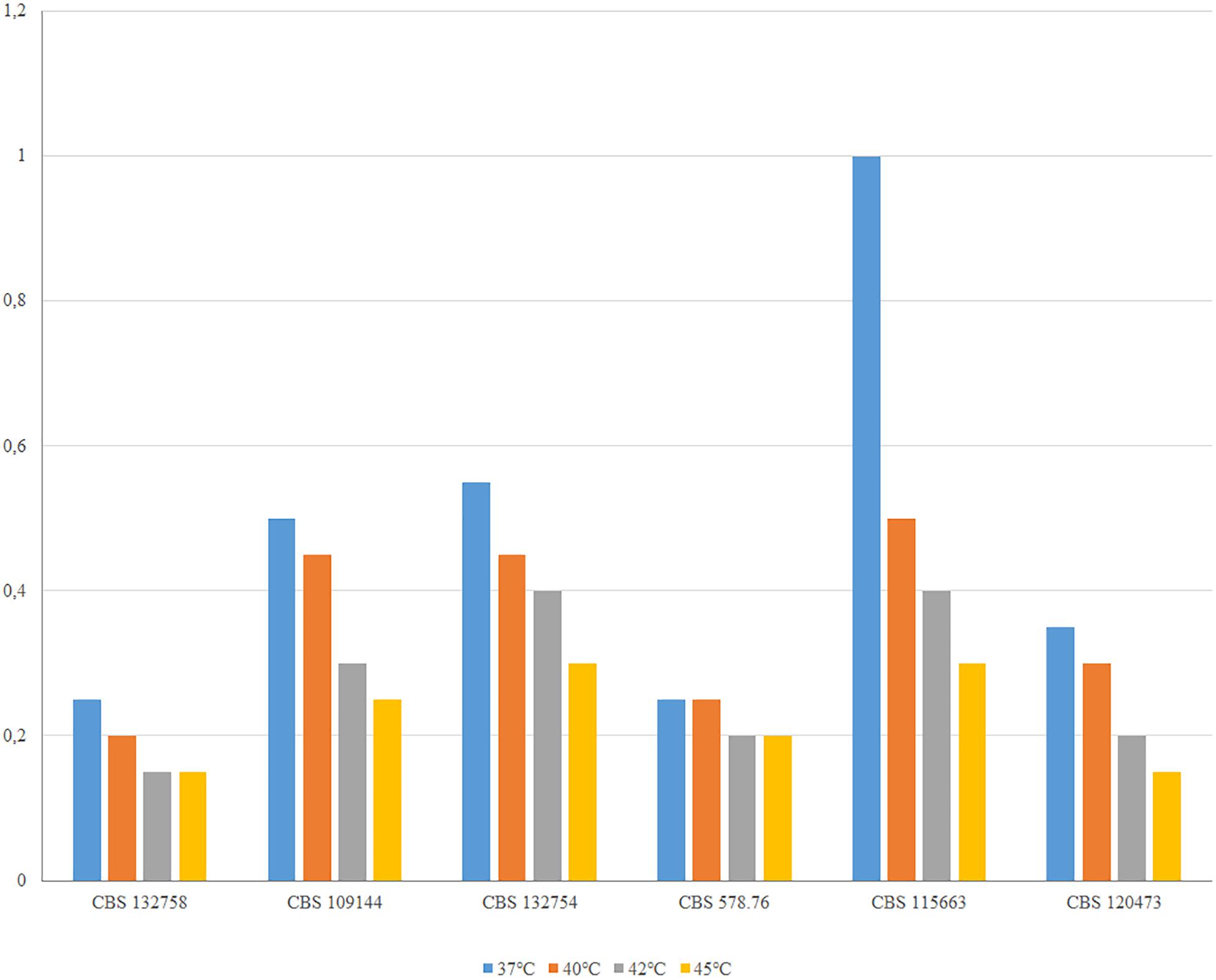
Figure 5. Thermotolerance of E. phaeomuriformis CBS 132758 and five E. dermatitidis strains measured after 2 weeks incubation on MEA medium.
Discussion
From clinical data, i.e., the number of published cases and their severity (Sudhadham et al., 2008; de Hoog et al., 2019) it has been concluded that E. dermatitidis and E. spinifera are the most virulent species in the genus Exophiala. In contrast, C. immunda has never been observed in clinical settings, which may either be explained by low virulence or its rarity. Remarkably, this fungus was found to have the highest number (n = 458) of genes that have been associated in the literature with virulence (Fisher et al., 1986; Chamond et al., 2003; Xu et al., 2012; Isola et al., 2013; Gümral et al., 2014; Zhang et al., 2014; Gelli, 2018; Bernardes et al., 2019). Exophiala dermatitidis is a common environmental species occurring in the domesticated environment (Isola et al., 2013; Gümral et al., 2014) and thus has a higher chance to of infection; it has a significantly lower number (n = 267–403) of virulence-associated genes than all species compared, including the equally common environmental species E. oligosperma (n = 424).
All virulence genes in this study can be summarized into the following four major classifications, the capsular polysaccharide synthesis, the CAP associated proteins (including decapping enzyme and capping protein), the Hsp70 protein, and ubiquitin family. Notably, the capsular polysaccharide synthesis protein and CAP N-terminal is present in E. dermatitidis strains only. Both species share the same cysteine-rich secretory proteins (CRISP), which can induce an increase of leukocytes in vivo, stimulating the production of cytokines and eicosanoids (Xu et al., 2012). Expression of a human CAP superfamily member, the cysteine-rich secretory protein 2 (CRISP2), rescues the phenotype of yeast mutants lacking Pry function and purified CRISP2 binds cholesterol in vitro, indicating that lipid binding is a conserved function of the CAP superfamily proteins playing an important role in lipid and sterol metabolism. These genes are also significant in Exophiala.
Exophiala dermatitidis and E. spinifera are the only chaetothyrialean black yeasts that possess capsule-like EPS around juvenile cells, while these reportedly are absent from sibling species E. phaeomuriformis and E. oligosperma, respectively. These genes are known in Cryptococcus, where the microtubule-associated CAP-glycine protein (Cgp1) governs growth, differentiation, and virulence of Cryptococcus neoformans (Gelli, 2018). Phenotypically, differences are known in the shape of black yeast capsules, which are regular in E. spinifera and irregular EPS in E. dermatitidis (Song et al., 2017). In addition, E. spinifera loses capsule formation at 37°C, while most E. dermatitidis strains maintain this capacity (Song et al., 2017). Different from Cryptococcus, Exophiala species may lose this purported virulence when it is needed at elevated temperature during host invasion.
One of the predicted genes found in both E. dermatitidis and E. spinifera, Cysteine-rich secretory protein (CRISP), has an effect on potassium channels and inflammatory processes (Bernardes et al., 2019). Pathogen-Related Yeast (PRY) genes belong to a large CAP protein superfamily (i.e., CRISP, antigen 5, and pathogenesis related 1 proteins). The conserved CAP domain of Pry1 is necessary and sufficient for lipid export and sterol binding (Choudhary and Schneiter, 2012).
There are two clusters belonging to the Myb-like DNA-binding domain and SANT/Myb domain only present in the neurotropic species E. dermatitidis and R. mackenziei, and absent from E. spinifera which was suggested to be somewhat osteotropic (Sudhadham et al., 2008). SANT/Myb-type genes are involved in conidiation of Cochliobolus carbonum (Zhang et al., 2014), while Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development (Wieser and Adams, 1995). The virulence gene MYT3 is required for pathogenesis and sexual development in Fusarium graminearum. The Multiprotein Transcription Factor TFIIIB is linked to RNA polymerase III-transcribed genes indirectly through interaction with DNA-bound TFIIIC or directly through DNA recognition by the TATA-binding protein, in turn recruits RNA polymerase III to the promoter. It is a key transcription factor in Saccharomyces cerevisiae (Kassavetis et al., 1997).
The genes possibly associated to osteotropy were shared by E. spinifera and E. dermatitidis, suggesting that the different predilection of disseminated strains might rather be linked to absence of neurotropism in E. spinifera rather than presence of bone-related genes. Linker histone H1 is an essential component of chromatin structure, linking nucleosomes into higher order structures and is eventually replaced by H5. Histone proteins have central roles in both chromatin organization as structural units of the nucleosome and gene regulation. Proline racemase catalyzes the interconversion of L- and D-proline (Fisher et al., 1986; Chamond et al., 2003). Three strains deviated by having proline oxidase instead of racemase. The family also contains several proteins that remain hypothetical. FAD-linked oxidoreductase was detected in a clinical E. spinifera strain. This concerns a family of bacterial oxidoreductases with covalently linked FAD (McNeil and Fineran, 2013; Bogachev et al., 2018).
Hsp70 proteins is a class of molecular chaperones that are shared by both species. Studies in Fusarium showed that knockout of an ER lumenal Hsp70 homolog FpLhs1 gene reduced growth, conidiation, and pathogenicity. FpLhs1 is likely to act on the development and virulence by regulating protein secretion (Chen et al., 2019). The ubiquitin family is also essential in both species. The ubiquitin-proteasome system plays an essential role in the regulation of intercellular protein degradation, and the biosynthetic gene cluster for himeic acid A has been proven to be a ubiquitin-activating enzyme (E1) inhibitor in Aspergillus japonicus (Hashimoto et al., 2018).
Black yeasts in general display remarkably diverse lifestyles, with a predilection for extreme and toxic environments such as those rich in aromatic compounds or heavy metals, or with high temperatures, increased salinity, and scarcity of nutrients (Paolo et al., 2006; Prenafeta-Boldu et al., 2006; Zhao et al., 2010). Moreno et al. (2018) noted that chaetothyrialean black fungi are exceptionally rich in cytochrome P450 genes enhancing toxin management. No genes involved in the styrene pathway, but comparative analysis of R. mackenziei against the aromatic hydrocarbon-degrading fungus C. immunda (Moreno et al., 2018) revealed the presence of orthologs that resemble the published fungal toluene degradation pathway (Figure 4) via protocatechuate (Parales et al., 2008; Blasi et al., 2017). Toluene was proven to be initially oxidized to benzyl alcohol by a membrane-bound CYP in toluene-growing cells of the closely related black fungus Cladophialophora saturnica CBS 114326 (previously confused with Cladosporium sphaerospermum) (Luykx et al., 2003). Exophiala spinifera has twice as many CYP as E. dermatitidis, a species having the smallest genomes of all members of the family Herpotrichiellaceae sequenced thus far (Teixeira et al., 2017). On the other hand, in most E. spinifera strains, dioxygenases needed for the opening of benzene rings via protocatechuate are missing, while they are consistently present in E. dermatitidis. Notably, p-hydroxybenzoate hydroxylase is present in both species, while it was absent from the toluene-degrading black fungus C. immunda (Blasi et al., 2017). It may be surmised that in E. spinifera the CYP P450 genes have other functions than toluene degradation. Nascimento et al. (2017) found rich populations of E. spinifera in degrading coconut shells rich in esters and hydrocarbons, while E. dermatitidis is unambiguously associated with monoaromatic pollutants, alkanes and creosotes (Isola et al., 2013; Gümral et al., 2014). Although this ability may enhance neurotropism in E. dermatitidis, the human brain is unlikely as a natural habitat for the species, which should be considered as an opportunist rather than a pathogen, as is exemplified by a recently described hydrocarbon-degrading in the Antarctic (Zhang et al., 2019). Other indications were found in the adverse response of E. spinifera to elevated temperature, and variability of essential cytochromes. A possible additional gene promoting neurotropism is OG0012246 which was consistently detected in E. dermatitidis and R. mackenziei but was absent from E. spinifera.
Data Availability Statement
The datasets generated for this study can be found in the GenBank Accessions:: JAAAJH000000000, JAAAJG000000000, JAAAJF000000000, JAAAJE000000000, JAAAJD000000000, JAAABF000000000, JAAABG000000000, JAAABH000000000, JYBY00000000, JYCA01000000, JAAAJI000000000, WXYG00000000, JAAAJK000000000, JAAAJJ000000000, JAAAJM000000000, JAAAJL000000000, AFPA00000000.1, and JYBU00000000.1.
Author Contributions
RL and GH designed the experiments and supervised the data analysis. YS performed the experiments and wrote the manuscript. NS, VW, and DV analyzed the data. LM and VV provided technical support. All authors discussed the results and commented on the manuscript.
Funding
The authors are grateful for the financial support of the International Cooperation and Exchanges Project (NSFC No. 81520108026), National Natural Science Foundation of China (NSFC No. 31670145) from National Natural Science Foundation of China. The authors also thank the Brazilian Federal Agency for Support and Evaluation of Graduate: Education Coordination for the Improvement of Higher Education Personnel-CAPES (Grant number: 88887.468196/2019-00) and National Counsel of Technological and Scientific Development (Grant number: 312811/2018-7).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful for the help of Zhe Wan in strains identification.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00586/full#supplementary-material
Footnotes
- ^ https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ https://sourceforge.net/projects/bbmap/
- ^ https://broadinstitute.github.io/picard/
- ^ http://vcfannotator.sourceforge.net/
- ^ http://www.ebi.ac.uk/interpro/
References
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bernardes, C. P., Menaldo, D. L., Zoccal, K. F., Boldrini-Franca, J., Peigneur, S., Arantes, E. C., et al. (2019). First report on BaltCRP, a cysteine-rich secretory protein (CRISP) from Bothrops alternatus venom: effects on potassium channels and inflammatory processes. Int. J. Biol. Macromol. 140, 556–567. doi: 10.1016/j.ijbiomac.2019.08.108
Blasi, B., Tafer, H., Kustor, C., Poyntner, C., Lopandic, K., and Sterflinger, K. (2017). Genomic and transcriptomic analysis of the toluene degrading black yeast Cladophialophora immunda. Sci. Rep. 7:11436. doi: 10.1038/s41598-017-11807-11808
Bogachev, A. V., Baykov, A. A., and Bertsova, Y. V. (2018). Flavin transferase: the maturation factor of flavin-containing oxidoreductases. Biochem. Soc. Trans. 46, 1161–1169. doi: 10.1042/bst20180524
Chamond, N., Gregoire, C., Coatnoan, N., Rougeot, C., Freitas-Junior, L. H., da Silveira, J. F., et al. (2003). Biochemical characterization of proline racemases from the human protozoan parasite Trypanosoma cruzi and definition of putative protein signatures. J. Biol. Chem. 278, 15484–15494. doi: 10.1074/jbc.m210830200
Chen, L., Geng, X., Ma, Y., Zhao, J., Chen, W., Xing, X., et al. (2019). The ER lumenal Hsp70 protein FpLhs1 is important for conidiation and plant infection in Fusarium pseudograminearum. Front. Microbiol. 10:1401. doi: 10.3389/fmicb.2019.01401
Choudhary, V., and Schneiter, R. (2012). Pathogen-related yeast (PRY) proteins and members of the CAP superfamily are secreted sterol-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 16882–16887. doi: 10.1073/pnas.1209086109
de Hoog, G. S., Vicente, V. A., Najafzadeh, M. J., Harrak, M. J., Badali, H., and Seyedmousavi, S. (2011). Waterborne Exophiala species causing disease in cold-blooded animals. Persoonia 27, 46–72. doi: 10.3767/003158511x614258
de Hoog, G. S., Guarro, J., Gené, J., Ahmed, S., Al-Hatmi, A. M. S., Figueras, M. J., et al. (2019). Atlas of Clinical Fungi,4th Edn Utrecht: Westerdijk Fungal Biodiversity Institute.
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome. Biol. 16, 157. doi: 10.1186/s13059-015-0721-2
Fisher, L. M., Albery, W. J., and Knowles, J. R. (1986). Energetics of proline racemase: racemization of unlabeled proline in the unsaturated, saturated, and oversaturated regimes. Biochemistry 25, 2529–2537. doi: 10.1021/bi00357a037
Gelli, A. (2018). A unique cytoskeleton-associated protein in Cryptococcus neoformans. Virulence 9, 752–753. doi: 10.1080/21505594.2018.1451185
Gostincar, C., Stajich, J. E., Zupancic, J., Zalar, P., and Gunde-Cimerman, N. (2018). Genomic evidence for intraspecific hybridization in a clonal and extremely halotolerant yeast. BMC Genomics 19:364. doi: 10.1186/s12864-018-4751-4755
Gümral, R., Tümgör, A., Saraçlı, M. A., Yıldıran, ŞT., Ilkit, M., and de Hoog, G. S. (2014). Black yeast diversity on creosoted railway sleepers changes with ambient climatic conditions. Microb. Ecol. 68, 699–707. doi: 10.1007/s00248-014-0459-455
Hashimoto, M., Kato, H., Katsuki, A., Tsukamoto, S., and Fujii, I. (2018). Identification of the biosynthetic gene cluster for himeic acid A: a ubiquitin-activating enzyme (E1) Inhibitor in Aspergillus japonicus MF275. Chembiochem 19, 535–539. doi: 10.1002/cbic.201700584
Isola, D., Selbmann, L., de Hoog, G. S., Fenice, M., Onofri, S., Prenafeta-Boldú, F. X., et al. (2013). Isolation and screening of black fungi as degraders of volatile aromatic hydrocarbons. Mycopathologia 175, 369–379. doi: 10.1007/s11046-013-9635-9632
Kassavetis, G. A., Bardeleben, C., Kumar, A., Ramirez, E., and Geiduschek, E. P. (1997). Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol. Cell. Biol. 17, 5299–5306. doi: 10.1128/mcb.17.9.5299
Kim, Y., Kim, H., Son, H., Choi, G. J., Kim, J. C., and Lee, Y. W. (2014). MYT3, a Myb-like transcription factor, affects fungal development and pathogenicity of Fusarium graminearum. PloS. One 9:e94359. doi: 10.1371/journal.pone.0094359
Lanternier, F., Barbati, E., Meinzer, U., Liu, L., Pedergnana, V., Migaud, M., et al. (2015). Inherited CARD9 deficiency in 2 unrelated patients with invasive Exophiala infection. J. Infect. Dis. 211, 1241–1250. doi: 10.1093/infdis/jiu412
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Lomsadze, A., Ter-Hovhannisyan, V., Chernoff, Y. O., and Borodovsky, M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic. Acids. Res. 33, 6494–6506. doi: 10.1093/nar/gki937
Luykx, D. M., Prenafeta-Boldu, F. X., and de Bont, J. A. (2003). Toluene monooxygenase from the fungus Cladosporium sphaerospermum. Biochem. Biophys. Res. Commun. 312, 373–379. doi: 10.1016/j.bbrc.2003.10.128
McKenna, R., Thompson, B., Pugh, S., and Nielsen, D. R. (2014). Rational and combinatorial approaches to engineering styrene production by Saccharomyces cerevisiae. Microb. Cell. Fact 13:123. doi: 10.1186/s12934-014-0123-122
McNeil, M. B., and Fineran, P. C. (2013). Prokaryotic assembly factors for the attachment of flavin to complex II. Biochim. Biophys. Acta 1827, 637–647. doi: 10.1016/j.bbabio.2012.09.003
Moreno, L. F., Ahmed, A. A. O., Brankovics, B., Cuomo, C. A., Menken, S. B. J., Taj-Aldeen, S. J., et al. (2018). Genomic understanding of an infectious brain disease from the desert. G3. 8, 909–922. doi: 10.1534/g3.117.300421
Nascimento, M. M. F., Vicente, V. A., Bittencourt, J. V. M., Gelinski, J. M. L., Prenafeta-Boldú, F. X., Romero-Güiza, M., et al. (2017). Diversity of opportunistic black fungi on babassu coconut shells, a rich source of esters and hydrocarbons. Fung. Biol. 121, 488–500. doi: 10.1016/j.funbio.2017.01.006
Oelschlägel, M., Zimmerling, J., and Tischler, D. (2018). A review: the styrene metabolizing cascade of side-chain oxygenation as biotechnological basis to gain various valuable compounds. Front. Microbiol. 9:490. doi: 10.3389/fmicb.2018.00490
Paolo, W. F. Jr., Dadachova, E., Mandal, P., Casadevall, A., Szaniszlo, P. J., and Nosanchuk, J. D. (2006). Effects of disrupting the polyketide synthase gene WdPKS1 in Wangiella [Exophiala] dermatitidis on melanin production and resistance to killing by antifungal compounds, enzymatic degradation, and extremes in temperature. BMC. Microbiol. 6:55. doi: 10.1186/1471-2180-6-55
Parales, R. E., Parales, J. V., Pelletier, D. A., and Ditty, J. L. (2008). Diversity of microbial toluene degradation pathways. Adv. Appl. Microbiol. 64, 1–73. doi: 10.1016/s0065-2164(08)00401-402
Pearson, M., Carcajona, D., Treasurer, J. W., et al. (2019). Exophiala angulospora infection in hatchery-reared lumpfish (Cyclopterus lumpus) broodstock. J. Fish. Dis. 42, 335–343.
Piro, V. C., Faoro, H., Weiss, V. A., Steffens, M. B. R., Pedrosa, F. O., Souza, E. M., et al. (2014). FGAP: an automated gap closing tool. BMC. Res. Notes 7:371. doi: 10.1186/1756-0500-7-371
Prenafeta-Boldu, F. X., Summerbell, R., and de Hoog, G. S. (2006). Fungi growing on aromatic hydrocarbons: biotechnology’s unexpected encounter with biohazard? FEMS. Microbiol. Rev. 30, 109–130. doi: 10.1111/j.1574-6976.2005.00007.x
Qu, H., Yi, J., Gao, X., Zhao, H., and Wang, Z. (2019). Anti-disuse osteoporosis activity of a complex of calcium-binding peptide from Auricularia auricula Protein Hydrolysates. J. Food. Sci. 84, 1909–1919. doi: 10.1111/1750-3841.14697
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucleic. Acids. Res. 33, W116–W120. doi: 10.1093/nar/gki442
Saraiva, M., Beckmann, M. J., Pflaum, S., Pearson, M., Carcajona, D., Treasurer, J. W., et al. (2019). Exophiala angulospora infection in hatchery-reared lumpfish (Cyclopterus lumpus) broodstock. J. Fish. Dis. 42, 335–343. doi: 10.1111/jfd.12940
Song, Y. G., Laureijssen-van de Sande, W. W. J., Moreno, L. F., Gerrits, van den Ende, B., Li, R. Y., et al. (2017). Comparative ecology of capsular exophiala species causing disseminated infection in humans. Front. Microbiol. 8:2514. doi: 10.3389/fmicb.2017.02514
Sudhadham, M., Prakitsin, S., Sivichai, S., Chaiyarat, R., Dorrestein, G. M., Menken, S. B., et al. (2008). The neurotropic black yeast Exophiala dermatitidis has a possible origin in the tropical rain forest. Stud. Mycol. 61, 145–155. doi: 10.3114/sim.2008.61.15
Taborda, C. P., da Silva, M. B., Nosanchuk, J. D., and Travassos, L. R. (2008). Melanin as a virulence factor of Paracoccidioides brasiliensis and other dimorphic pathogenic fungi: a minireview. Mycopathologia 165, 331–339. doi: 10.1007/s11046-007-9061-9064
Teixeira, M. M., Moreno, L. F., Stielow, B. J., Muszewska, A., Hainaut, M., Gonzaga, L., et al. (2017). Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales. Ascomycota). Stud. Mycol. 86, 1–28. doi: 10.1016/j.simyco.2017.01.001
Vicente, V. A., Orelis-Ribeiro, R., Najafzadeh, M. J., Sun, J., Guerra, R. S., Miesch, S., et al. (2012). Black yeast-like fungi associated with lethargic crab disease (LCD) in the mangrove-land crab, ucides cordatus (Ocypodidae). Vet. Microbiol. 158, 109–122. doi: 10.1016/j.vetmic.2012.01.031
Wieser, J., and Adams, T. H. (1995). flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes. Dev. 9, 491–502. doi: 10.1101/gad.9.4.491
Xu, X., Francischetti, I. M., Lai, R., Ribeiro, J. M., and Andersen, J. F. (2012). Structure of protein having inhibitory disintegrin and leukotriene scavenging functions contained in single domain. J. Biol. Chem. 287, 10967–10976. doi: 10.1074/jbc.M112.340471
Yurlova, N. A., and de Hoog, G. S. (2002). Exopolysaccharides and capsules in human pathogenic Exophiala species. Mycoses 45, 443–448. doi: 10.1046/j.1439-0507.2002.00807.x
Zhang, C., Sirijovski, N., Adler, L., and Ferrari, B. C. (2019). Exophiala macquariensis sp. nov., a cold adapted black yeast species recovered from a hydrocarbon contaminated sub-Antarctic soil. Fungal Biol. 123, 151–158. doi: 10.1016/j.funbio.2018.11.011
Zhang, J. X., Wu, Y. X., Ho, H., Zhang, H., He, P. F., and He, Y. Q. (2014). BZcon1, a SANT/Myb-type gene involved in the conidiation of Cochliobolus carbonum. G3. 4, 1445–1453. doi: 10.1534/g3.114.012286
Keywords: black yeasts, comparative genomics, intraspecific variability, interspecific differences, virulence profiles, opportunists
Citation: Song Y, da Silva NM, Weiss VA, Vu D, Moreno LF, Vicente VA, Li R and de Hoog GS (2020) Comparative Genomic Analysis of Capsule-Producing Black Yeasts Exophiala dermatitidis and Exophiala spinifera, Potential Agents of Disseminated Mycoses. Front. Microbiol. 11:586. doi: 10.3389/fmicb.2020.00586
Received: 31 January 2020; Accepted: 18 March 2020;
Published: 08 April 2020.
Edited by:
Martin Hoenigl, Medical University of Graz, AustriaReviewed by:
Macit Ilkit, Çukurova University, TurkeyYi Sun, Jingzhou Central Hospital, China
Aylin Döğen, Mersin University, Turkey
Copyright © 2020 Song, da Silva, Weiss, Vu, Moreno, Vicente, Li and de Hoog. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruoyu Li, bXljb2xhYkAxMjYuY29t; G. Sybren de Hoog, U3licmVuLmRlSG9vZ0ByYWRib3VkdW1jLm5s
 Yinggai Song
Yinggai Song Nickolas Menezes da Silva
Nickolas Menezes da Silva Vinicius Almir Weiss
Vinicius Almir Weiss Duong Vu
Duong Vu Leandro F. Moreno
Leandro F. Moreno Vania Aparecida Vicente
Vania Aparecida Vicente Ruoyu Li
Ruoyu Li G. Sybren de Hoog
G. Sybren de Hoog