- 1School of Pharmacy, University College London, London, United Kingdom
- 2Liverpool School of Tropical Medicine, Liverpool, United Kingdom
- 3Wellcome Sanger Institute, Hinxton, Cambridge, United Kingdom
- 4Faculty of Science, Prince of Songkla University, Songkhla, Thailand
- 5European Bioinformatics Institute, European Molecular Biology Laboratory, Hinxton, Cambridge, United Kingdom
- 6London School of Hygiene and Tropical Medicine, London, United Kingdom
- 7Faculty of Allied Health Sciences, Thammasat University, Pathumtanee, Thailand
- 8Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
Antibiotic resistant strains of Acinetobacter baumannii are responsible for a large and increasing burden of nosocomial infections in Thailand and other countries of Southeast Asia. New approaches to their control and treatment are urgently needed and an attractive strategy is to remove the bacterial polysaccharide capsule, and thus the protection from the host’s immune system. To examine phylogenetic relationships, distribution of capsule chemotypes, acquired antibiotic resistance determinants, susceptibility to complement and other traits associated with systemic infection, we sequenced 191 isolates from three tertiary referral hospitals in Thailand and used phenotypic assays to characterize key aspects of infectivity. Several distinct lineages were circulating in three hospitals and the majority belonged to global clonal group 2 (GC2). Very high levels of resistance to carbapenems and other front-line antibiotics were found, as were a number of widespread plasmid replicons. A high diversity of capsule genotypes was encountered, with only three of these (KL6, KL10, and KL47) showing more than 10% frequency. Almost 90% of GC2 isolates belonged to the most common capsule genotypes and were fully resistant to the bactericidal action of human serum complement, most likely protected by their polysaccharide capsule, which represents a key determinant of virulence for systemic infection. Our study further highlights the importance to develop therapeutic strategies to remove the polysaccharide capsule from extensively drug-resistant A. baumanii during the course of systemic infection.
Introduction
A. baumannii is an opportunistic pathogen that can cause potentially lethal nosocomial infections (Howard et al., 2012). These are frequently a result of trauma, surgery, catheterization or endotracheal intubation (Chopra et al., 2014), and A. baumannii can escape the local immune reaction by evading neutrophils, macrophages and complement (C') (Russo et al., 2008; García-Patiño et al., 2017). This immune escape therefore necessitates the use of antimicrobials, and the key determinant of clinical outcome of A. baumannii infection is treatment failure due to the high number of antibiotic resistant strains (Wong et al., 2017).
Multidrug resistant (MDR) strains of A. baumannii have spread rapidly over recent decades (Zarrilli et al., 2013; Hamidian and Nigro, 2019). The high prevalence of strains resistant to nearly all antibiotics, especially well-tolerated cephalosporins and carbapenems, has led to the revival of drugs considered to be of last resort such as polymyxins (Falagas and Kasiakou, 2005; Sahbudak Bal et al., 2018) for systemic administration. However, resistance to colistin is now more prevalent and polymyxins are now used less widely due to serious side effects associated with these agents (Sahbudak Bal et al., 2018). As a consequence, the World Health Organization has identified carbapenem-resistant A. baumannii (CRAB) as the greatest bacterial threat to global human health and the top priority pathogen for development of new antibiotics (Tacconelli et al., 2018).
Recent surveillance data indicates that A. baumannii causes under 2% of healthcare associated infections in the United States (Sievert et al., 2013; Bulens et al., 2018) but prevalence is much higher in Southern and South Eastern Asia, where it is frequently the major nosocomial infectious agent (Suwantarat and Carroll, 2016). The burden of A. baumannii infection is particularly severe in Thailand, with isolates accounting for 15–16% of hospital-acquired bacteremia cases and displaying very high levels (70–88%) of carbapenem resistance, and mortality rates in excess of 60% due to MDR A. baumannii bacteremia (Chaisathaphol and Chayakulkeeree, 2014; Hongsuwan et al., 2014; Suwantarat and Carroll, 2016; Hsu et al., 2017; Sirijatuphat et al., 2018). Presence of the over-expressed carbapenemase blaOXA–23, or blaOXA–51 in combination with IS elements, account for most of the CRAB phenotypes (Figueiredo et al., 2009; Teo et al., 2015; Wong et al., 2017). Molecular typing identified three European clones; two have spread globally and are now identified as GC1 and GC2 (Higgins et al., 2010; Hamidian and Nigro, 2019) and the majority of isolates from Asia belong to global clone 2 (GC2) (Kim et al., 2013; Kamolvit et al., 2015).
The large majority of A. baumannii strains produce a substantial capsular polysaccharide that protects them from external threats (Kenyon and Hall, 2013), and an attractive treatment option is enzymatic removal of the protective capsules (Mushtaq et al., 2004; Lin et al., 2014; Negus et al., 2015); capsule-free mutants were highly susceptible to C'-mediated attack (Lees-Miller et al., 2013), in marked contrast to their encapsulated parent strains. A major advantage of this approach is that it circumvents the accumulation of antibiotic resistance determinants, but has the potential disadvantage that variation of the capsular polysaccharide may limit the utility of individual depolymerases as found in bacteriophages or other organisms, which typically hydrolyze only one or a limited number of capsular types (Oliveira et al., 2017; Hernandez-Morales et al., 2018; Lin et al., 2018; Singh et al., 2018).
We report a detailed characterization of 191 recent isolates from three major hospitals in Thailand using whole-genome sequencing and functional assays, with particular reference to their surface properties and antibiotic resistance profiles. We also sought to identify factors that contribute to the capacity of GC2 isolates to cause infection through increased virulence (Zarrilli et al., 2013), using genomic data and bioassays, in relation to the role of the capsule in the determination of resistance to C'-mediated attack.
Materials and Methods
Bacterial Isolates
A total of 191 A. baumannii isolates were cultured from wound pus, sputum, urine, blood, and excised tissue at the clinical microbiology laboratories of three tertiary referral hospitals in Thailand (Figure 1A). Bacteria were initially identified by routine biochemical tests implemented for identification of Gram-negative bacteria. Species were further confirmed by whole-genome sequencing and sequence typing as below. The hospitals were Thammasat University Hospital, Pathum Thani Province (47 isolates; April 2016), Siriraj Hospital, Bangkok (84 consecutive isolates; April 2016) and Songklanagarind Hospital, Hat Yai, Songkhla Province (60 isolates; August 2016). Siriraj is the largest hospital in Thailand with 2,300 beds, 1,000,000 outpatients per annum and 80,000 inpatients per annum; equivalent figures for Songklanagarind are 846, 1,019,375, and 40,936 and for Thammasat 601, 384,088, and 40,745 (data from 2017). Details of these isolates are given in Supplementary Table S1. Susceptibilities to clinically relevant antibiotics were determined using the Vitek 2 system (Bosshard et al., 2006).
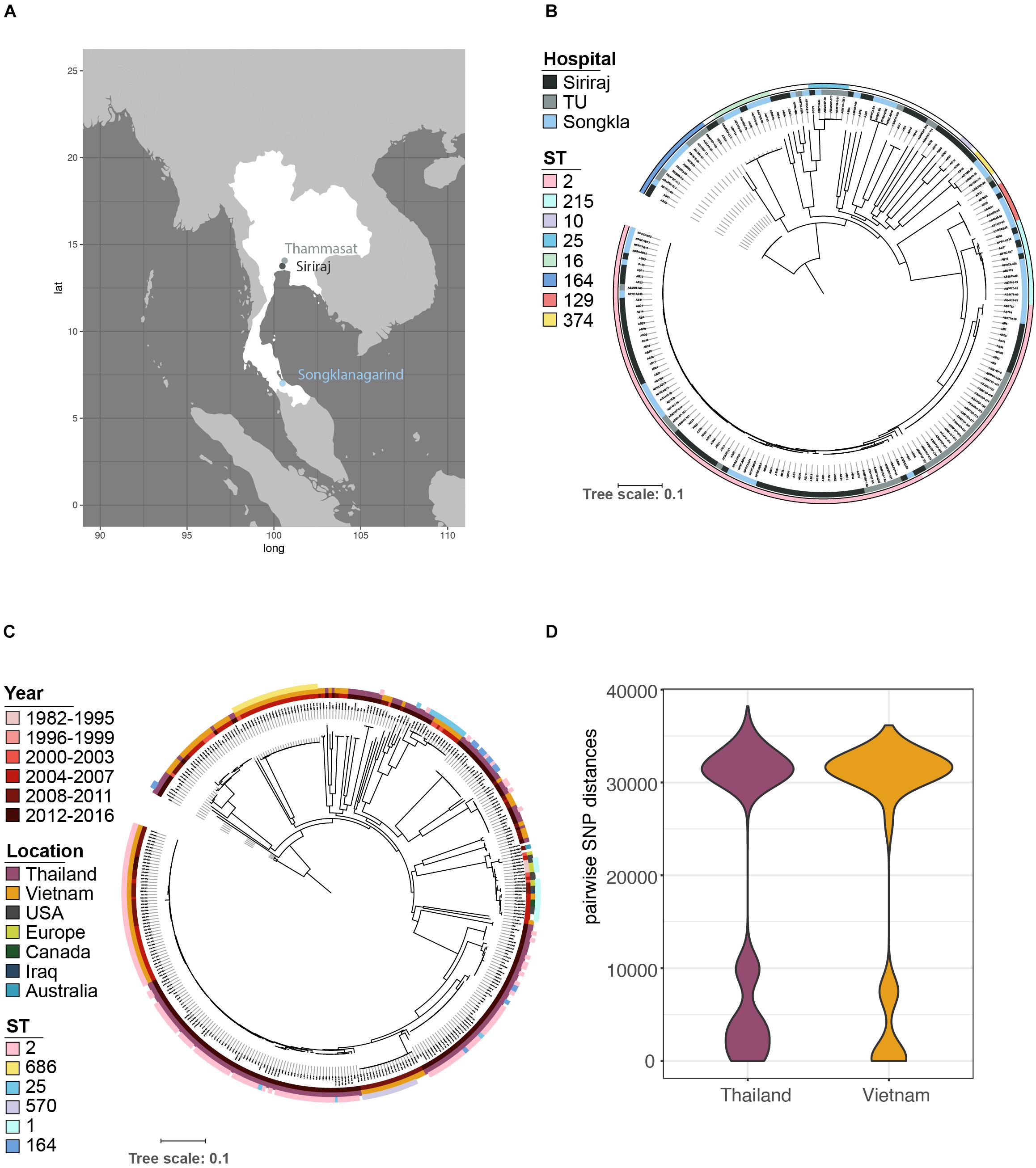
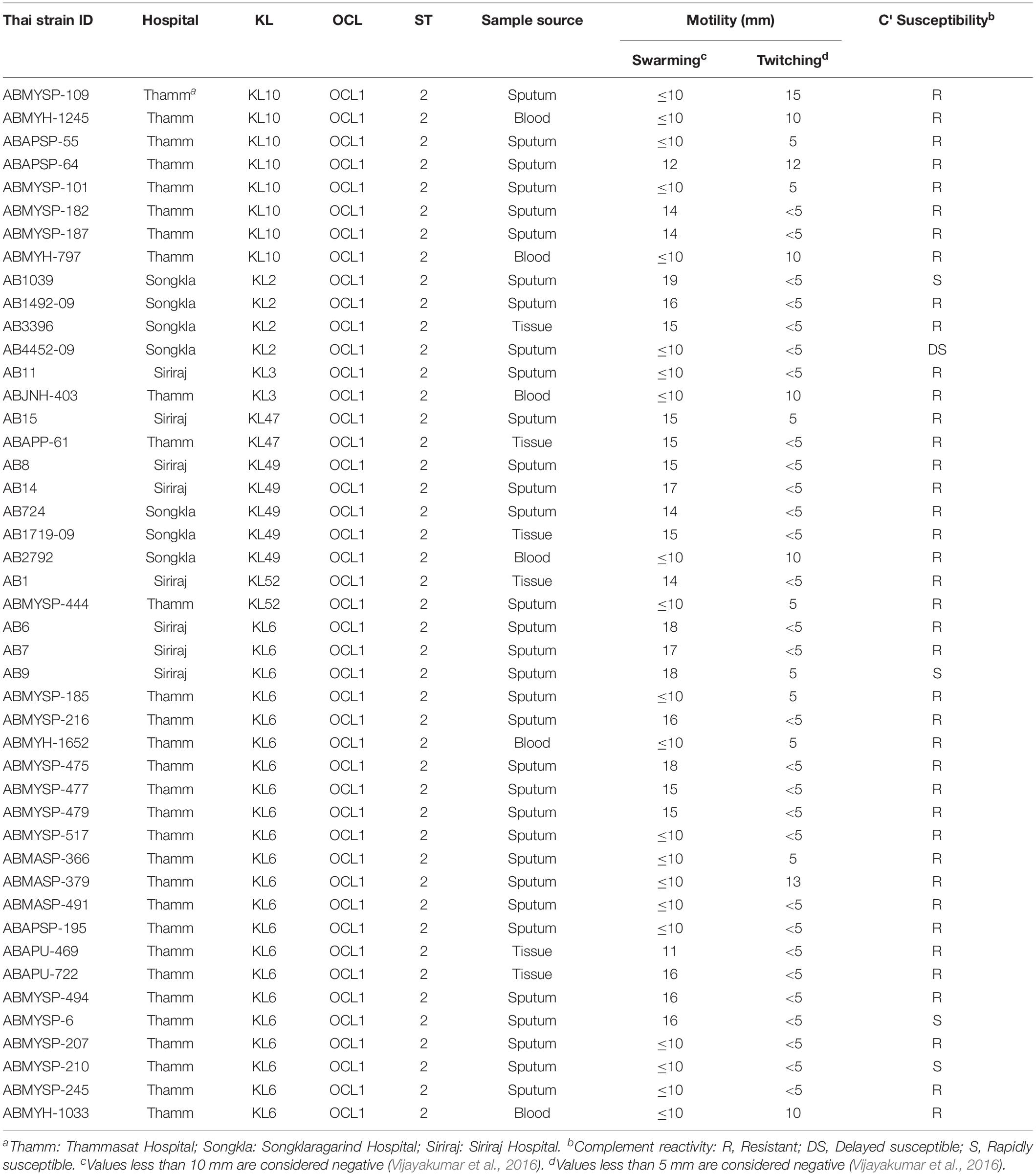
Figure 1. The population structure of A. baumannii isolated from a variety of infections in April 2016 at three major Thai hospitals. (A) Samples were obtained from geographically distinct regions of the country. (B) Core gene phylogeny showed that the bacterial populations were circulating amongst the three hospitals; no single lineage dominated at any one location. (C) Our data in context with the global population structure based on published data. (D) A more detailed comparison of the data structure of pairwise SNP distances shows a similar distribution between our samples and a recent study from one hospital in Vietnam (Schultz et al., 2016), with a similarly high prevalence of ST2 (C), but also a considerable number of more distantly related isolates from other regions.
Genome Sequencing, Assembly, and Annotation
Genomic DNA was extracted and sequenced using Illumina-B HiSeq X paired-end sequencing. Annotated assemblies were produced according to (Page et al., 2016a). Sequence reads were assembled de novo with Velvet v1.2 (Zerbino and Birney, 2008) and VelvetOptimiser v2.2.5 (Gladman and Seemann, 2008). Reads were annotated using PROKKA v1.11 (Seemann, 2014). The stand-alone scaffolder SSPACE (Boetzer et al., 2011) was used to refine contig assembly; sequence gaps were filled using GapFiller (Boetzer and Pirovano, 2012). Genomes with greater than 5% contamination levels as determined by Kraken (Wood and Salzberg, 2014), fully assembled genomes of less than 4.5 Mpb or comprising 500 or more contigs were removed. Putative genomes with less than 60% sequence similarity with the reference genome were assessed with CheckM (Parks et al., 2015) for genome completeness and contamination; isolates with greater than 3% contamination levels were excluded from the study. SNPs were called against the A. baumannii reference genome to identify heterozygous SNPs, and isolates with more than 2% were removed from further analysis (Page et al., 2016a), resulting in the 191 genomes analyzed in this study. As we could also observe several gdhB duplicate sequences, a known problem of the Oxford MLST scheme (Bartual et al., 2005; Gaiarsa et al., 2019), sequence types were assigned and are reported only based on the Pasteur scheme (Diancourt et al., 2010; Page et al., 2016). Novel sequence types were assigned for non-typeable isolates through the PubMLST (Supplementary Table S1), three isolates could not be assigned as the assemblies were missing one allele.
Phylogenetic Analyses
The pan genome for the global and Thai isolate analyses was determined with Roary (Page et al., 2015) using a Protein BLAST identity of 95% and a core definition of 99%. SNPs were extracted from the core gene alignment using SNP sites (Page et al., 2016b) and the output used to run RAxML v8.2.8 (Stamatakis, 2014) to calculate the phylogenetic tree with 100 bootstraps under the GTR time-reversible model. The resulting alignment for the global dataset was also used to determine pairwise SNP distances with the dist.gene function from the ape package in R (Paradis et al., 2004). To place our isolates in a broader context, we compared them with recently published sequence data of A. baumannii causing ventilator-associated pneumonia in the intensive care unit of a Vietnamese hospital, in addition to data from several other published studies (Supplementary Table S2).
Antibiotic Resistance and Traits Associated With Infection
Antibiotic resistance genes were detected with the curated version of the ARG-ANNOT database available at the SRST2 site (Gupta et al., 2014; Inouye et al., 2014), rpoB SNP mutations were assessed comparing the sequences against described resistance mutations (Giannouli et al., 2012; Pérez-Varela et al., 2017), and virulence factors with VFDB (Chen et al., 2016), using the read-based search program ARIBA (Hunt et al., 2017). Plasmid replicons were detected with a custom database composed of 30 genes involved in plasmid replication, stabilization and mobilization from Acinetobacter plasmids (Bertini et al., 2010; Salto et al., 2018); some additional plasmids (Gao et al., 2011; Hamidian et al., 2012, 2016; Zhang et al., 2013; Jones et al., 2014; Blackwell and Hall, 2017; Hamidian et al., 2017) were also included (full database Supplementary Dataset S1); and analyses were undertaken using ARIBA software v2.12.1 (Hunt et al., 2017). To account for potential variation in surface proteins or other virulence factors, a custom-made collection of A. baumannii virulence factors (Supplementary Table S6) was searched against our isolates using phmmer (Eddy, 2011; Eijkelkamp et al., 2011, 2014; Harding et al., 2013; Scott et al., 2014; Weber et al., 2015; Lee et al., 2017). Representations of trees and metadata were performed using iTOL (Letunic and Bork, 2016) and the ggplot2 and ggtree packages in R (Wickham, 2009; Yu et al., 2018). KL and OCL genotypes of our isolates were identified using the capsule identification program kaptive, based on a curated A. baumannii specific database (Wyres et al., 2019; Supplementary Table S1).
C' Susceptibility
Commercial (MP Biomedicals, United Kingdom) pooled human serum was stored and used to determine susceptibility to C', essentially as previously described (Loraine et al., 2018). Early mid-logarithmic-phase Luria-Bertani (LB) broth cultures of A. baumannii were washed three times with 200 μl of gelatin-veronal-buffered saline containing Mg2+ and Ca2+ (GVB++; pH 7.35) and suspended in 400 μl of GVB++. The suspensions (200?μl) were mixed with 390 μl of pre-warmed (37°C) normal human serum to give a final concentration of ∼1 × 106 CFU, the mixtures incubated at 37°C for 3 h and bacteria quantified by serial dilution and overnight incubation on LB agar (see Supplementary Table S3 for all raw data). The 45 GC2 isolates were exposed to 66% normal human serum and enumerated bacterial survivors over a 3 h incubation period (Malke, 1986). Isolates were assigned to one of three categories: resistant (R), showing no (or only transient) reduction in viable count during the incubation period; delayed susceptible (DS), displaying significant (∼90%) survival after 1 h and low survival (<10%) after 3 h incubation; the inocula of rapidly susceptible (S) isolates were reduced to below 10% after 1 h incubation. All experiments were performed in duplicate and results expressed as percent survival over this time period. Pre-warmed, heat-inactivated human serum (56°C, 30 min) served as control. All raw data is given in Supplementary Table S3.
Capsule Measurements
The size of the capsule for each isolate was determined by negative staining with India ink, microscopic imaging and calculation of the area occupied by the capsule using CellProfiler image analysis software (v3.1.9; Lamprecht et al., 2007). One bacterial colony was resuspended in PBS and mixed in a 1:1 ratio with India Ink stain (BD India Ink Reagent Dropper) and applied to a microscope slide with a coverslip. Microscopic imaging with a Zeiss Axiostar plus transmitted light microscope fitted with an Olympus SC30 digital camera and using a 100× oil immersion lens and embedded scale bar. All raw data is given in Supplementary Table S4.
Motility
Swarming and twitching motility were assayed by the subsurface agar method (Clemmer et al., 2011) using LB broth containing either 0.4 or 0.8% agar. Briefly, freshly grown cultures of A. baumannii were stabbed to enable spread of bacteria on the surface of 0.4% agar plates for swarming motility and the interphase between the bottom of the Petri dish and the 0.8% agar layer for twitching motility. The plates were incubated at 37°C for 48 h: positive swarming motility was defined as a zone greater than 10 mm around the site of inoculation. For twitching motility at the interstitial surface between the agar and the petri dish, the agar was discarded, and bacteria visualized by staining stained with 0.2% crystal violet. Positive twitchers were defined as those cultures that showed a zone diameter greater than 5 mm. Assays were performed a minimum of three times for each isolate. All raw data is provided in Supplementary Table S5.
Results
Major Lineages Are Circulating in the Region
Phylogenetic analysis identified several lineages circulating in all the three hospitals (Figures 1A,B). The majority of isolates belong to GC2 (n = 106/191), represented exclusively by sequence type 2 (ST2) of the Pasteur scheme. No isolates belonging to GC1 were identified, a key clonal group in the evolution of multi-drug resistance in A. baumannii (Holt et al., 2016). Non-GC2 isolates belonged to ST164 (n = 14; 7.3%), ST215 (n = 13; 6.8%), ST16 (n = 9; 4.7%), ST25 (n = 6; 3.1%), ST129 (n = 6; 3.1%), ST374 (n = 4; 2.1%), and ST10 (n = 2; 1.0%); three isolates could not be sequence-typed, most likely due to low-quality genomes, and thus missing one of the MLST alleles. The high prevalence of GC2 and lack of GC1 of our dataset from 2016 closely resembles the population structure from the Vietnamese hospital outbreak (Schultz et al., 2016) over the period 2009-12 (Figure 1C); both datasets include a considerable number of deep branching lineages. These similarities in population structure are also mirrored when comparing the distribution of pairwise single-nucleotide polymorphisms (SNPs) between the datasets from Vietnam and Thailand (Figure 1D).
Antimicrobial Resistance
Phenotypic resistance profiles for 115 of the strains confirmed the very high levels of antibiotic resistance encountered with clinical isolates of A. baumannii, especially against β-lactam agents (e.g., ceftriaxone: 115/115, 100%), including carbapenems (Figure 2A; 98/115, 85.2%), but also against other major antibiotic classes: fluoroquinolones [98/115, 85.2% resistant/intermediate (R/I)], aminoglycosides (79/115, 68.7% R/I) and trimethoprim (76/115, 66.1% R/I), and multidrug resistance was, as expected, associated with a high number of acquired resistance genes (Figures 2A,C and Supplementary Figures S1, S2) indicating either gain through larger elements carrying several genes as previously described as a key driver for A. baumannii resistance (Bonomo and Szabo, 2006; Post and Hall, 2009). blaOXA–23, the most prominent carbapenem resistance gene, is present in 85.2% of imipenem resistant strains (Figure 2C). Few isolates carried the blaNDM–1 gene and a low number of acquired ampC genes were detected (Figure 2C and Supplementary Figure S2). We also note the presence of the arr gene, as well as rpoB mutations, conferring rifampicin resistance, one of the last line antimicrobials used against CRAB (Thapa et al., 2009; Durante-Mangoni et al., 2014).
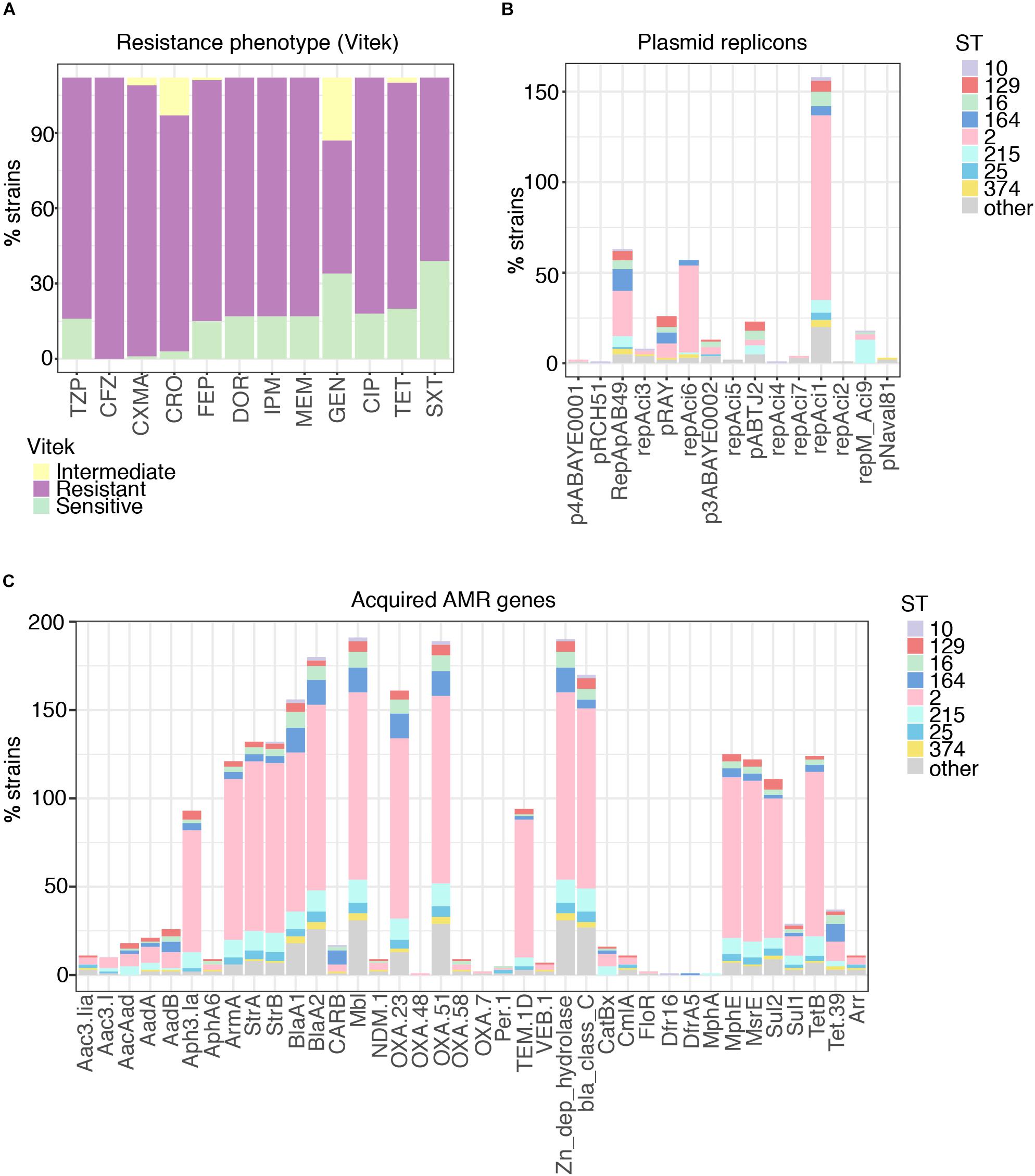
Figure 2. Phenotypic resistance of A. baumannii at high levels for all antimicrobial classes. (A) Resistance phenotypes measured on site at time of isolation clearly demonstrate the highly problematic levels of resistance in A. baumannii, with > 70% non-sensitive against all tested classes. TZP, piperacillin-tazobactam; CFZ, cefazolin; CXMA, cefuroxime axetil; CRO, ceftriaxone; FEP, cefepime; DOR, doripenem; IPM, imipenem; MEM, meropenem; GEN, gentamicin; CIP, ciprofloxacin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole. (B) Distribution of plasmids carried by A. baumannii in relation to sequence type (ST). (C) Distribution of acquired antimicrobial resistance genes carried by A. baumannii in relation to sequence type (ST).
Mobile Elements
All but ten isolates contained at least one of the plasmid replicons (Supplementary Dataset S1) and 121 contained two to maximal five (Figure 2B). The three plasmid replicons detected at highest frequency were RepAci1, RepAci6, and RepApAB49. Each of these plasmid types were found across a number of STs, although RepAci1 plasmids were present in almost all the ST2 isolates (102/106) and RepApAB49 was found in 12/14 ST164 isolates. Recently, a RepAci1 plasmid was shown to be mobilized by a co-residing conjugative RepAci6 plasmid (Blackwell and Hall, 2019), and these two replicons co-occur in the genomes of 56 isolates; RepAci6 only was detected in one isolate, and RepAci1 only in 46. RepAci6 plasmids were the most common self-transmissible plasmids detected. Plasmid replicons detected frequently included those matching pRAY∗, which is often associated with the aadB gene (Hamidian et al., 2012), RepAci3, p3ABAYE, pABTJ2, and RepAci9. RepMAci9 was detected in all thirteen ST215 isolates. Seven plasmid types were present in low frequency (Figure 2B) and an additional fifteen plasmid sequences were not detected in the Thai collection (Supplementary Dataset S1).
Genes Associated With Capsules and Outer Core
A. baumannii does not contain genes involved in lipopolysaccharide (LPS) O-antigen ligase activity (Kenyon and Hall, 2013; Weber et al., 2015), synthesizing instead a lipooligosaccharide (LOS) consisting of an outer core oligosaccharide (OCL) linked to Lipid A (Kenyon and Hall, 2013; Kenyon et al., 2014a); at least twelve distinct OCL structures have been inferred from genomic data (Kenyon et al., 2014b). We mapped all Thai isolates against an A. baumannii specific databases for capsular and LOS loci (KL and OCL, resp.; Supplementary Figure S3; Wyres et al., 2019). In similar fashion to the Vietnam study (Schultz et al., 2016), we noted a high diversity of KL within both GC2 and non-GC2 isolates. KL6 (15.2%), KL10 (15.7%), KL47 (11.0%), KL2 (8.4%), KL52 (7.9%), KL3 (7.3%), KL49 (6.3%), KL24 (5.8%), KL14 (3.1%), and KL28 (2.1%) were frequently encountered and KL32, KL63, KL57, KL8, KL108, KL19, KL113, KL116, KL60, KL43, KL37, KL9, KL125, and KL7 were represented in 2% or fewer isolates. KL could not be determined in 15 isolates (7.9%). KL2 and KL49 were found at least twice in the Vietnam isolates although we did not detect KL58, strongly represented in Schultz et al. (2016). Eight distinct capsule loci in our GC2 isolates were detected in isolates from all three hospitals during April 2016 and provide a challenge for novel therapies targeting bacterial cell surfaces. Furthermore, seven distinct LOS loci were detected amongst the Thai isolates (Supplementary Figure S3 and Supplementary Table S1). The majority of GC2 isolates carried genes for OCL1 biosynthesis (91 isolates, 85.8% of all GC2 isolates, 61.8% total), whilst the other types, OCL2 (6.8%), OCL3 (4.2%), OCL4 (1.6%), OCL5 (15.7%), OCL6 (4.7%), and OCL7 (5.2%) were also widely distributed amongst our isolates; there was, however, no clear association between K- and LOS-types (Figure 3).
Figure 3. Properties of 46 GC2 A. baumannii Thai isolates belonging to the common capsule genotypes encountered in this study. (A) Motility measurements, capsule size, and C' susceptibility in phylogenetic context. (B) C' resistance profiles stratified by capsule type.
Linking Virulence-Associated Phenotype, Site of Isolation, and Genotype in GC2
We examined 45 GC2 isolates belonging to the major capsule types identified in the Thai collection: KL10 (eight isolates), KL2 (4), KL3 (2), KL47 (2), KL49 (5), KL52 (2), and KL6 (22). Although A. baumannii strains lack flagella, the species displays type IV-mediated twitching motility that facilitates spreading on abiotic surfaces (Vijayakumar et al., 2016), and it has been linked to the capacity of strains to cause systemic infection (Harding et al., 2018). Only six of our 45 GC2 isolates were derived from blood samples but all displayed twitching motility (Table 1, Supplementary Table S5, and Figure 3A). In contrast, none of six tissue isolates and only a minority of sputum isolates (10/33) were motile in this fashion. The capacity to swarm on semi-solid agar (surface-associated motility; Harding et al., 2018) can also be linked to a more virulent phenotype (Eijkelkamp et al., 2011; Tipton and Rather, 2017). 24/45 of the Thai GC2 isolates displayed surface-associated (swarming) motility; 6/6 of these were from tissue samples and 18/33 from sputum (Table 1 and Figure 3A). Three isolates from sputum exhibited both forms of motility.
Many loci that have been linked to the capacity of A. baumannii to colonize, invade and disseminate within the host, such those encoding adhesins, capsules, quorum sensors, iron sequestering systems and other nutrient scavengers (Harding et al., 2018), are essential or advantageous for survival in its natural habitat, predominantly soil and water (Baumann et al., 1968). The distribution of genes based on a publicly available virulence factor database is shown in Supplementary Figure S4, but whilst there are clear differences, no trend (for example increased prevalence in GC2) could be observed. As expected, siderophores, adhesins involved in biofilm formation and maintenance, and a variety of genes determining capsule biosynthesis are widely distributed among the isolates.
GC2 Capsule Size Correlates With Survival in Human Serum
A large proportion (40/45, 88.9%) were refractory to C'-mediated killing; of the remainder, only four were categorized as S (Table 1). All KL10, KL3, KL47, KL49, and KL52 isolates belonged to the R group, with only KL2 (2/4) and KL6 (3/23) capsule types displaying any degree of C' susceptibility (Figures 3A,B). All 45 GC2 isolates examined were encapsulated. The C' susceptible isolates elaborated significantly smaller capsules than R A. baumannii (R, mean 1.62 mm2; DS, 0.31 mm2; S, 0.81 mm2); all capsule locus predictions however showed a perfect or almost perfect match, emphasizing that the capsule biosynthesis locus is likely intact (Figure 3A). Capsules containing sialic acids protect Gram-negative bacteria from C' attack (Rautemaa and Meri, 1999), and N-acetylneuramininc acid and related non-ulosonic and sialic acid structures have recently been found as repeat-unit constituents or as modifications of capsule structures in hypermucoviscous K. pneumoniae (Lin et al., 2014) and A. baumannii (Vinogradov et al., 2014; Kenyon et al., 2015; Singh et al., 2018), and associated with increased infectivity. Biosynthesis of sialic acids begins with the conversion of UDP-N-acetylglucosamine to UDP and N-acetylmannosamine by the hydrolyzing 2-epimerase NeuC; a homolog of this enzyme has been described for A. baumannii and its crystal structure determined (Ko et al., 2018). In our set of genomes, the neuC homolog (A0A154EJU5_ACIBA) was found only in the genomes of the five C' resistant isolates carrying genes for biosynthesis of the K49 capsular polysaccharide and is indeed a component of the KL49 locus and should thus correctly be annotated as lgaC; the repeat unit of the K49 capsular polysaccharide is composed of α-L-fucosamine, α-D-glucosamine and the non-ulosonic acid α-8-epi-legionaminic acid (Vinogradov et al., 2014).
OmpA, one of most abundant porins, is also known to bind factor H in human serum (Kim et al., 2009), and implicated to prevent C' mediated killing; it is however present in all our GC2 strains (Supplementary Figure S4). A more detailed analysis of putative factors explaining the phenotypes (type IV pili, surface proteins, secretion systems, biofilm formation (Weber et al., 2015; Lee et al., 2017; Supplementary Table S6) of the 47 GC2 isolates showed no differences that correlated with any of the phenotypes tested. We also included sequence analyses of PilA, which has been shown to influence twitching motility (Ronish et al., 2019), however, the sequences from all phenotyped isolates were identical.
Discussion
Multi-drug resistant A. baumannii infections are rapidly increasing and require the use of last-line treatments such as colistin. An additional challenge further narrowing the spectrum of available options for highly resistant A. baumannii infections is that last-line treatments available often overlap with other highly problematic infections. One example is the use of rifampicin in combination with colistin against CRAB, which is also one of the last options to treat the increasing number of multi-drug resistant tuberculosis (MDR TB) cases, and use of rifampicin is therefore restricted in use against organisms other than MDR TB (Thapa et al., 2009; Durante-Mangoni et al., 2014; Leite et al., 2016; Seijger et al., 2019). There is therefore a growing interest in the potential of non-antibiotic therapeutic approaches including bacteriophage-derived capsule depolymerases as treatment alternative to antimicrobial chemotherapy (Waldor et al., 2005; García-Quintanilla et al., 2013; Seijger et al., 2019).
We present the analysis of a set of 191 A. baumannii clinical isolates from three major hospitals in Thailand with very high levels of drug resistance. The population structure is biased toward the major clone GC2, as has been observed in other studies in geographic proximity (Schultz et al., 2016). However, the inter-mixed origins of closely related isolates from all three hospitals clearly indicates that both GC2 as well as less dominant sequence types are circulating in the region, and are frequently (re)introduced into hospitals, as opposed to a clonal outbreak within one hospital. In addition to the phylogenetic diversity (almost 50% non-GC2 isolates) and the even spread across the three hospitals, we show that there is a high degree of strain-to-strain capsule variability, and development of depolymerase therapeutics will need to account for the challenge of a wide range of capsule types. Nevertheless, a recent study has demonstrated the potential of capsule depolymerase against A. baumannii in a Galleria mellonella (wax moth) larvae infection model and protection of both normal and immunocompromised mice from lethal peritoneal sepsis (Liu et al., 2019a).
The enzyme also sensitized the C'-resistant isolate to serum (Liu et al., 2019b), which is highly relevant as the large majority of our GC2 isolates (40/45) were C' resistant, in similar proportion to other recent studies (Sanchez-Larrayoz et al., 2017; Skerniškytë et al., 2019). LPS O-side chains prevent assembly of the C5b-9 complex by steric hindrance; A. baumannii however does not decorate its LOS with O-side chains but is able to modify the lipid A moiety of LOS by acylation, resulting in increased survival in blood (Bartholomew et al., 2019), which could prevent C5b-9 intercalation into the bilayer. Alternatively, there is some evidence that A. baumannii may prevent C' activation: resistant clinical isolates bound fH, a key inhibitor of the alternative C' pathway (Kim et al., 2009), preventing C5b-9 generation. King et al. (2009) found that clinical isolates did not bind fH but circumvented C3b deposition, again preventing C5b-9-mediated bacterial killing. Cell surface-located sialic acids are potent recruiters of fH and we therefore examined Thai GC2 isolates for evidence of neuC-dependent sialyl biosynthesis. The neuC homolog is part of the KL49 locus, however, non-ulosonic acid sugars are also found in the K2 and K6 types (Kenyon and Hall, 2013), which have C' sensitive as well as resistant phenotypes.
Current evidence indicates that C' killing of susceptible A. baumannii proceeds predominantly through the activation of the alternative pathway (Kim et al., 2009; Jacobs et al., 2010; Sanchez-Larrayoz et al., 2017). The lack of classical pathway killing may be due to the absence of C'-activating IgG or IgM directed against A. baumannii surface structures in normal human serum, suggesting that the predominant means to avoid bactericidal effects is prevention or subversion of activation of the alternative pathway. It is likely that the polysaccharide capsule is the predominant macromolecule facilitating C' resistance (Harding et al., 2018) and the four fully C' susceptible isolates in the current study elaborated less capsule than the resistant group. Capsule depolymerases as an alternative means of resolving A. baumannii systemic infections would thus be worth exploring but may be limited by the wide diversity of capsule types likely to be encountered in current clinical isolates.
Whilst the current focus is placed on GC2, it is important to point out that GC1 and GC2 seem to follow different strategies for interacting with the immune system and hospital environment. Whilst we report low motility and high C' resistance for GC2 and the associated genetic background, GC1 seems to follow a very different route, with high motility profiles and different adherence profiles than GC2 (Skerniškytë et al., 2019). It is thus crucial to increase active surveillance of A. baumannii epidemiology, as different high-risk lineages may need different approaches to reduce their burden in the clinic.
Data Availability Statement
The datasets generated for this study can be found in the European Nucleotide Archive ERS1930151–ERS1930323.
Author Contributions
PT, NT, and RAS conceived the study. JL, EH, GB, and PT designed experimental procedures. JL, EH, RS, and GB performed the experiments, analyzed and curated the data. SV, PS, and PK assembled the bacterial collection. PT and EH wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This study was funded by the Newton Fund through Medical Research Council award MR/N012542/1. Assembly of the A. baumannii clinical isolate collection was enabled by National Science and Technology Development Agency award FDA-CO-2559-1448-TH. RS was supported by the Thailand Research Fund through the Royal Golden Jubilee PhD Programme (Grant No. PHD/0106/2559) funded by the Newton Fund delivered by the British Council Thailand. The funders played no role in the study or in the preparation of this article or in the decision to publish.
Acknowledgments
The National Institute for Health Research University College London Hospitals Biomedical Research Centre provided infrastructural support. We acknowledge expert informatics support from the Pathogen Informatics team at the Wellcome Sanger Institute and thank Elisa Gonzalez de Herrero Martinez for assistance with DNA preparation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00548/full#supplementary-material
FIGURE S1 | Resistance genes and phenotypic resistance. The strains were grouped according to the number of agents in the Vitek screen the respective strains were resistant to, along the x-axis. The y-axis shows the number of strains in the relevant class, the color of the bars shows resistance (dark purple), intermediate (yellow), or sensitive (green) against the respective antimicrobial of the subplot. This shows that almost all strains are resistant against 12 reagents, sensitivity of the highly-resistant ones on the far end of the x-scale is only occasionally in sulfonamides or tetracycline, but all are fully resistant against the β-lactam class.
FIGURE S2 | Presence of genes encoding antibiotic resistance in Thai A. baumannii isolates. The guidance tree is shown in Figure 3A. Bla, β-lactamases; AGly, aminoglycosides; MLS, macrolides; Phe, chloramphenicol; Rif, rifampin; Sul, sulfonamides; Tet, tetracycline; Tmt, trimethoprim. AMR genes were sourced from the curated version of the ARG-ANNOT database available at the SRST2 site. Isolates from Thammasat University Hospital, Siriraj Hospital, and Songklanagarind Hospital are designated TU, Siriraj, and Songkla, respectively. Sequence types are shown, as indicated in the legend. Chromosomal mutations for RpoB are also shown, we could detect potential resistance-conferring changes (Giannouli et al., 2012; Pérez-Varela et al., 2017) D525N, H535Q, and S540F.
FIGURE S3 | Cell surface polysaccharide diversity of A. baumannii Thai isolates. Capsular (KL) and outer core loci (OCL) in silico typing of Thai isolates. A. baumannii shows considerable variation in K-type and a more conserved distribution of OCL-types.
FIGURE S4 | Virulence genes associated with Thai A. baumannii isolates. The guidance tree is shown in Figure 3A. Antibiotic resistance genes were detected with the curated version of the ARG-ANNOT database available at the SRST2 site using ARIBA. Isolates from Thammasat University Hospital, Siriraj Hospital, and Songklanagarind Hospital are designated TU, Siriraj, and Songkla, respectively. Sequence types are shown, as indicated in the legend.
TABLES S1–S6 | Properties of A. baumannii isolates.
DATASET S1 | The custom-made plasmid replicon collection used to assign plasmid types to A. baumannii.
References
Bartholomew, T. L., Kidd, T. J., Sá Pessoa, J., Conde, Álvarez, R., and Bengoechea, J. A. (2019). 2-Hydroxylation of Acinetobacter baumannii Lipid A Contributes to Virulence. Infect. Immun. 87:e00066-19.. doi: 10.1128/IAI.00066-19
Bartual, S. G., Seifert, H., Hippler, C., Luzon, M. A. D., Wisplinghoff, H., and Rodríguez-Valera, F. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005
Baumann, P., Doudoroff, M., and Stanier, R. Y. (1968). A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter). J. Bacteriol. 95, 1520–1541. doi: 10.1128/jb.95.5.1520-1541.1968
Bertini, A., Poirel, L., Mugnier, P. D., Villa, L., Nordmann, P., and Carattoli, A. (2010). Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4168–4177. doi: 10.1128/AAC.00542-10
Blackwell, G. A., and Hall, R. M. (2017). The tet39 Determinant and the msrE-mphE Genes in Acinetobacter plasmids are each part of discrete modules flanked by inversely oriented pdif (XerC-XerD) sites. Antimicrob. Agents Chemother. 61:e00780-17. doi: 10.1128/AAC.00780-17
Blackwell, G. A., and Hall, R. M. (2019). Mobilisation of a small Acinetobacter plasmid carrying an oriT transfer origin by conjugative RepAci6 plasmids. Plasmid 103, 36–44. doi: 10.1016/j.plasmid.2019.04.002
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and Pirovano, W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579. doi: 10.1093/bioinformatics/btq683
Boetzer, M., and Pirovano, W. (2012). Toward almost closed genomes with GapFiller. Genome Biol. 13:R56. doi: 10.1186/gb-2012-13-6-r56
Bonomo, R. A., and Szabo, D. (2006). Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43(Suppl. 2), S49–S56. doi: 10.1086/504477
Bosshard, P. P., Zbinden, R., Abels, S., Böddinghaus, B., Altwegg, M., and Böttger, E. C. (2006). 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 44, 1359–1366. doi: 10.1128/JCM.44.4.1359-1366.2006
Bulens, S. N., Yi, S. H., Walters, M. S., Jacob, J. T., Bower, C., Reno, J., et al. (2018). Carbapenem-Nonsusceptible Acinetobacter baumannii, 8 US Metropolitan Areas, 2012-2015. Emerging Infect. Dis. 24, 727–734. doi: 10.3201/eid2404.171461
Chaisathaphol, T., and Chayakulkeeree, M. (2014). Epidemiology of infections caused by multidrug-resistant gram-negative bacteria in adult hospitalized patients at Siriraj Hospital. J. Med. Assoc. Thai. 97(Suppl. 3), S35–S45.
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Chopra, T., Marchaim, D., Johnson, P. C., Awali, R. A., Doshi, H., Chalana, I., et al. (2014). Risk factors and outcomes for patients with bloodstream infection due to Acinetobacter baumannii-calcoaceticus complex. Antimicrob. Agents Chemother. 58, 4630–4635. doi: 10.1128/AAC.02441-14
Clemmer, K. M., Bonomo, R. A., and Rather, P. N. (2011). Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157, 2534–2544. doi: 10.1099/mic.0.049791-0
Diancourt, L., Passet, V., Nemec, A., Dijkshoorn, L., and Brisse, S. (2010). The Population Structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034
Durante-Mangoni, E., Utili, R., and Zarrilli, R. (2014). Combination therapy in severe Acinetobacter baumannii infections: an update on the evidence to date. Future Microbiol. 9, 773–789. doi: 10.2217/fmb.14.34
Eddy, S. R. (2011). Accelerated Profile HMM Searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Eijkelkamp, B. A., Stroeher, U. H., Hassan, K. A., Papadimitrious, M. S., Paulsen, I. T., and Brown, M. H. (2011). Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol. Lett. 323, 44–51. doi: 10.1111/j.1574-6968.2011.02362.x
Eijkelkamp, B. A., Stroeher, U. H., Hassan, K. A., Paulsen, I. T., and Brown, M. H. (2014). Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020. doi: 10.1186/1471-2164-15-1020
Falagas, M. E., and Kasiakou, S. K. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40, 1333–1341. doi: 10.1086/429323
Figueiredo, S., Poirel, L., Papa, A., Koulourida, V., and Nordmann, P. (2009). Overexpression of the naturally occurring blaOXA-51 Gene in Acinetobacter baumannii mediated by novel insertiON SEQuence ISAba9. Antimicrobial. Agents Chemother. 53, 4045–4047. doi: 10.1128/AAC.00292-09
Gaiarsa, S., Batisti Biffignandi, G., Esposito, E. P., Castelli, M., Jolley, K. A., Brisse, S., et al. (2019). Comparative analysis of the two Acinetobacter baumannii multilocus sequence typing (MLST) schemes. Front. Microbiol. 10:930. doi: 10.3389/fmicb.2019.00930
Gao, F., Wang, Y., Liu, Y.-J., Wu, X.-M., Lv, X., Gan, Y.-R., et al. (2011). Genome sequence of Acinetobacter baumannii MDR-TJ. J. Bacteriol. 193, 2365–2366. doi: 10.1128/JB.00226-11
García-Patiño, M. G., García-Contreras, R., and Licona-Limón, P. (2017). The Immune Response against Acinetobacter baumannii, an Emerging Pathogen in Nosocomial Infections. Front. Immunol. 8:441. doi: 10.3389/fimmu.2017.00441
García-Quintanilla, M., Pulido, M. R., López-Rojas, R., Pachón, J., and McConnell, M. J. (2013). Emerging therapies for multidrug resistant Acinetobacter baumannii. Trends Microbiol. 21, 157–163. doi: 10.1016/j.tim.2012.12.002
Giannouli, M., Di Popolo, A., Durante-Mangoni, E., Bernardo, M., Cuccurullo, S., Amato, G., et al. (2012). Molecular epidemiology and mechanisms of rifampicin resistance in Acinetobacter baumannii isolates from Italy. Int. J. Antimicrob. Agents 39, 58–63. doi: 10.1016/j.ijantimicag.2011.09.016
Gladman, S., and Seemann, T. (2008). Velvet Optimiser. Available online at: https://github.com/tseemann/VelvetOptimiser (accessed January, 2019).
Gupta, S. K., Padmanabhan, B. R., Diene, S. M., Lopez-Rojas, R., Kempf, M., Landraud, L., et al. (2014). ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220. doi: 10.1128/AAC.01310-13
Hamidian, M., Ambrose, S. J., and Hall, R. M. (2016). A large conjugative Acinetobacter baumannii plasmid carrying the sul2 sulphonamide and strAB streptomycin resistance genes. Plasmid 8, 43–50. doi: 10.1016/j.plasmid.2016.09.001
Hamidian, M., Nigro, S. J., and Hall, R. M. (2012). Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J. Antimicrob. Chemother. 67, 2833–2836. doi: 10.1093/jac/dks318
Hamidian, M., Nigro, S. J., Hartstein, R. M., and Hall, R. M. (2017). RCH51, a multiply antibiotic-resistant Acinetobacter baumannii ST103IP isolate, carries resistance genes in three plasmids, including a novel potentially conjugative plasmid carrying oxa235 in transposon Tn6252. J. Antimicrob. Chemother. 72, 1907–1910. doi: 10.1093/jac/dkx069
Hamidian, M., and Nigro, S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 5:e000242. doi: 10.1099/mgen.0.000306
Harding, C. M., Hennon, S. W., and Feldman, M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. doi: 10.1038/nrmicro.2017.148
Harding, C. M., Tracy, E. N., Carruthers, M. D., Rather, P. N., Actis, L. A., and Munson, R. S. (2013). Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e0360-13. doi: 10.1128/mBio.00360-13
Hernandez-Morales, A. C., Lessor, L. L., Wood, T. L., Migl, D., Mijalis, E. M., Cahill, J., et al. (2018). Genomic and biochemical characterization of Acinetobacter podophage petty reveals a novel lysis mechanism and tail-associated depolymerase. Activity. J. Virol. 92, e1064-17. doi: 10.1128/JVI.01064-17
Higgins, P. G., Dammhayn, C., Hackel, M., and Seifert, H. (2010). Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65, 233–238. doi: 10.1093/jac/dkp428
Holt, K., Kenyon, J. J., Hamidian, M., Schultz, M. B., Pickard, D. J., Dougan, G., et al. (2016). Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb. Genom. 2:e000052. doi: 10.1099/mgen.0.000052
Hongsuwan, M., Srisamang, P., Kanoksil, M., Luangasanatip, N., Jatapai, A., Day, N. P., et al. (2014). Increasing incidence of hospital-acquired and healthcare-associated bacteremia in northeast Thailand: a multicenter surveillance study. PLoS One 9:e109324. doi: 10.1371/journal.pone.0109324
Howard, A., O’Donoghue, M., Feeney, A., and Sleator, R. D. (2012). Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 3, 243–250. doi: 10.4161/viru.19700
Hsu, L.-Y., Apisarnthanarak, A., Khan, E., Suwantarat, N., Ghafur, A., and Tambyah, P. A. (2017). Carbapenem-Resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin. Microbiol. Rev. 30, 1–22. doi: 10.1128/CMR.00042-16
Hunt, M., Mather, A. E., Sánchez-Busó, L., Page, A. J., Parkhill, J., Keane, J. A., et al. (2017). ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 3:e000131. doi: 10.1099/mgen.0.000131
Inouye, M., Dashnow, H., Raven, L.-A., Schultz, M. B., Pope, B. J., Tomita, T., et al. (2014). SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6:90. doi: 10.1186/s13073-014-0090-6
Jacobs, A. C., Hood, I., Boyd, K. L., Olson, P. D., Morrison, J. M., Carson, S., et al. (2010). Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun. 78, 1952–1962. doi: 10.1128/IAI.00889-09
Jones, L. S., Toleman, M. A., Weeks, J. L., Howe, R. A., Walsh, T. R., and Kumarasamy, K. K. (2014). Plasmid carriage of bla NDM-1 in clinical Acinetobacter baumannii isolates from India. Antimicrob. Agents Chemother. 58, 4211–4213. doi: 10.1128/AAC.02500-14
Kamolvit, W., Sidjabat, H. E., and Paterson, D. L. (2015). Molecular epidemiology and mechanisms of carbapenem resistance of Acinetobacter spp. in Asia and Oceania. Microb. Drug Resist. 21, 424–434. doi: 10.1089/mdr.2014.0234
Kenyon, J. J., and Hall, R. M. (2013). Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. doi: 10.1371/journal.pone.0062160
Kenyon, J. J., Holt, K. E., Pickard, D., Dougan, G., and Hall, R. M. (2014a). Insertions in the OCL1 locus of Acinetobacter baumannii lead to shortened lipooligosaccharides. Res. Microbiol. 165, 472–475. doi: 10.1016/j.resmic.2014.05.034
Kenyon, J. J., Nigro, S. J., and Hall, R. M. (2014b). Variation in the OC locus of Acinetobacter baumannii genomes predicts extensive structural diversity in the lipooligosaccharide. PLoS One 9:e107833. doi: 10.1371/journal.pone.0107833
Kenyon, J. J., Marzaioli, A. M., Hall, R. M., and De Castro, C. (2015). Structure of the K12 capsule containing 5,7-di-N-acetylacinetaminic acid from Acinetobacter baumannii isolate D36. Glycobiology 25, 881–887. doi: 10.1093/glycob/cwv028
Kim, D. H., Choi, J.-Y., Kim, H. W., Kim, S. H., Chung, D. R., Peck, K. R., et al. (2013). Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob. Agents Chemother. 57, 5239–5246. doi: 10.1128/AAC.00633-13
Kim, S. W., Choi, C. H., Moon, D. C., Jin, J. S., Lee, J. H., Shin, J.-H., et al. (2009). Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol. Lett. 301, 224–231. doi: 10.1111/j.1574-6968.2009.01820.x
King, L. B., Swiatlo, E., Swiatlo, A., and McDaniel, L. S. (2009). Serum resistance and biofilm formation in clinical isolates of Acinetobacter baumannii. FEMS Immunol. Med. Microbiol. 55, 414–421. doi: 10.1111/j.1574-695X.2009.00538.x
Ko, T.-P., Lai, S.-J., Hsieh, T.-J., Yang, C.-S., and Chen, Y. (2018). The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase from Acinetobacter baumannii: a comparative study with human GNE. J. Biol. Chem. 293, 10119–10127. doi: 10.1074/jbc.RA118.001971
Lamprecht, M. R., Sabatini, D. M., and Carpenter, A. E. (2007). CellProfiler: free, versatile software for automated biological image analysis. BioTechniques 42, 71–75. doi: 10.2144/000112257
Lee, C.-R., Lee, J. H., Park, M., Park, K. S., Bae, I. K., Kim, Y. B., et al. (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol 7:55. doi: 10.3389/fcimb.2017.00055
Lees-Miller, R. G., Iwashkiw, J. A., Scott, N. E., Seper, A., Vinogradov, E., Schild, S., et al. (2013). A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 89, 816–830. doi: 10.1111/mmi.12300
Leite, G. C., Oliveira, M. S., Perdigão-Neto, L. V., Rocha, C. K. D., Guimarães, T., Rizek, C., et al. (2016). Antimicrobial combinations against pan-resistant Acinetobacter baumannii isolates with different resistance mechanisms. PLoS One 11:e0151270. doi: 10.1371/journal.pone.0151270
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Lin, H., Paff, M. L., Molineux, I. J., and Bull, J. J. (2018). Antibiotic therapy using phage depolymerases: robustness across a range of conditions. Viruses 10:E622. doi: 10.3390/v10110622
Lin, T.-L., Hsieh, P.-F., Huang, Y.-T., Lee, W.-C., Tsai, Y.-T., Su, P.-A., et al. (2014). Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J. Infect. Dis. 210, 1734–1744. doi: 10.1093/infdis/jiu332
Liu, Y., Leung, S. S. Y., Guo, Y., Zhao, L., Jiang, N., Mi, L., et al. (2019a). The capsule depolymerase Dpo48 Rescues Galleria mellonella and mice from Acinetobacter baumannii Systemic Infections. Front. Microbiol. 10:545. doi: 10.3389/fmicb.2019.00545
Liu, Y., Mi, Z., Mi, L., Huang, Y., Li, P., Liu, H., et al. (2019b). Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ 7:e6173. doi: 10.7717/peerj.6173
Loraine, J., Heinz, E., De Sousa Almeida, J., Milevskyy, O., Voravuthikunchai, S. P., Srimanote, P., et al. (2018). Complement susceptibility in relation to genome sequence of recent Klebsiella pneumoniae isolates from thai hospitals. mSphere 3:e0537-18. doi: 10.1128/mSphere.00537-18
Malke, H. (1986). M. Sussman (Editor). The Virulence of Escherichia coli — Reviews and Methods. XV + 473 S., 45 Abb., 3S Tab. London-Orlando-San Diego-New. York-Toronto-Montreal Sydney-Tokyo 1985. Academic Press. $ 75.00. ISBN: 0-12-677520-6. J Basic Microbiol. 26, 127–128. doi: 10.1002/jobm.3620260219
Mushtaq, N., Redpath, M. B., Luzio, J. P., and Taylor, P. W. (2004). Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob. Agents Chemother. 48, 1503–1508. doi: 10.1128/aac.48.5.1503-1508.2004
Negus, D., Vipond, J., Hatch, G. J., Rayner, E. L., and Taylor, P. W. (2015). Parenteral administration of capsule depolymerase EnvD prevents lethal inhalation anthrax infection. Antimicrob. Agents Chemother. 59, 7687–7692. doi: 10.1128/AAC.01547-15
Oliveira, H., Costa, A. R., Konstantinides, N., Ferreira, A., Akturk, E., Sillankorva, S., et al. (2017). Ability of phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains: specific genomic pattern variation of phages. Environ. Microbiol. 19, 5060–5077. doi: 10.1111/1462-2920.13970
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Page, A. J., De Silva, N., Hunt, M., Quail, M. A., Parkhill, J., Harris, S. R., et al. (2016a). Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb. Genom. 2:e000083. doi: 10.1099/mgen.0.000083
Page, A. J., Taylor, B., Delaney, A. J., Soares, J., Seemann, T., Keane, J. A., et al. (2016b). SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2:e000056. doi: 10.1099/mgen.0.000056
Page, J. A., Taylor, B., and Keane, J. A. (2016). Multilocus sequence typing by blast from de novo assemblies against PubMLST. JOSS 1:118. doi: 10.21105/joss.00118
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Pérez-Varela, M., Corral, J., Vallejo, J. A., Rumbo-Feal, S., Bou, G., Aranda, J., et al. (2017). Mutations in the β-Subunit of the RNA polymerase impair the surface-associated motility and virulence of Acinetobacter baumannii. Infect. Immun. 85:e0327-17. doi: 10.1128/IAI.00327-17
Post, V., and Hall, R. M. (2009). AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 2667–2671. doi: 10.1128/AAC.01407-08
Rautemaa, R., and Meri, S. (1999). Complement-resistance mechanisms of bacteria. Microbes Infect. 1, 785–794. doi: 10.1016/s1286-4579(99)80081-1
Ronish, L. A., Lillehoj, E., Fields, J. K., Sundberg, E. J., and Piepenbrink, K. H. (2019). The structure of PilA from Acinetobacter baumannii AB5075 suggests a mechanism for functional specialization in Acinetobacter type IV pili. J. Biol. Chem. 294, 218–230. doi: 10.1074/jbc.RA118.005814
Russo, T. A., Beanan, J. M., Olson, R., MacDonald, U., Luke, N. R., Gill, S. R., et al. (2008). Rat pneumonia and soft-tissue infection models for the study of Acinetobacter baumannii biology. Infect. Immun. 76, 3577–3586. doi: 10.1128/IAI.00269-08
Sahbudak Bal, Z., Kamit Can, F., Yazici, P., Berna Anil, A., Duyu, M., Yilmaz Ciftdogan, D., et al. (2018). The evaluation of safety and efficacy of colistin use in pediatric intensive care unit: results from two reference hospitals and review of literature. J. Infect. Chemother. 24, 370–375. doi: 10.1016/j.jiac.2017.12.017
Salto, I. P., Torres Tejerizo, G., Wibberg, D., Pühler, A., Schlüter, A., and Pistorio, M. (2018). Comparative genomic analysis of Acinetobacter spp. plasmids originating from clinical settings and environmental habitats. Sci. Rep. 8:7783. doi: 10.1038/s41598-018-26180-3
Sanchez-Larrayoz, A. F., Elhosseiny, N. M., Chevrette, M. G., Fu, Y., Giunta, P., Spallanzani, R. G., et al. (2017). Complexity of Complement Resistance Factors Expressed by Acinetobacter baumannii needed for survival in human serum. J. Immunol. 199, 2803–2814. doi: 10.4049/jimmunol.1700877
Schultz, M. B., Pham Thanh, D., Tran, Do Hoan, N., Wick, R. R., Ingle, D. J., et al. (2016). Repeated local emergence of carbapenem-resistant Acinetobacter baumannii in a single hospital ward. Microb. Genom. 2:e000050. doi: 10.1099/mgen.0.000050
Scott, N. E., Kinsella, R. L., Edwards, A. V. G., Larsen, M. R., Dutta, S., Saba, J., et al. (2014). Diversity within the O -linked protein glycosylation systems of Acinetobacter Species. Mol Cell Proteomics 13, 2354–2370. doi: 10.1074/mcp.M114.038315
Seemann, T., and Gladman, S. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seijger, C., Hoefsloot, W., Bergsma-de Guchteneire, I., Te Brake, L., van Ingen, J., Kuipers, S., et al. (2019). High-dose rifampicin in tuberculosis: experiences from a Dutch tuberculosis centre. PLoS One 14:e0213718. doi: 10.1371/journal.pone.0213718
Sievert, D. M., Ricks, P., Edwards, J. R., Schneider, A., Patel, J., Srinivasan, A., et al. (2013). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2009-2010. Infect. Control Hosp. Epidemiol. 34, 1–14. doi: 10.1086/668770
Singh, J. K., Adams, F. G., and Brown, M. H. (2018). Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front. Microbiol. 9:3301. doi: 10.3389/fmicb.2018.03301
Sirijatuphat, R., Sripanidkulchai, K., Boonyasiri, A., Rattanaumpawan, P., Supapueng, O., Kiratisin, P., et al. (2018). Implementation of global antimicrobial resistance surveillance system (GLASS) in patients with bacteremia. PLoS One 13:e0190132. doi: 10.1371/journal.pone.0190132
Skerniškytë, J., Krasauskas, R., Péchoux, C., Kulakauskas, S., Armalytë, J., and Sužiedëlienë, E. (2019). Surface-related features and virulence among Acinetobacter baumannii Clinical isolates belonging to international clones I and II. Front. Microbiol. 9:3116. doi: 10.3389/fmicb.2018.03116
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Suwantarat, N., and Carroll, K. C. (2016). Epidemiology and molecular characterization of multidrug-resistant Gram-negative bacteria in Southeast Asia. Antimicrob. Resist Infect. Control 5, 15. doi: 10.1186/s13756-016-0115-6
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Teo, J., Lim, T.-P., Hsu, L.-Y., Tan, T.-Y., Sasikala, S., Hon, P.-Y., et al. (2015). Extensively drug-resistant Acinetobacter baumannii in a Thai hospital: a molecular epidemiologic analysis and identification of bactericidal Polymyxin B-based combinations. Antimicrob. Resist Infect. Control 4, 2. doi: 10.1186/s13756-015-0043-x
Thapa, B., Tribuddharat, C., Rugdeekha, S., Techachaiwiwat, W., Srifuengfung, S., and Dhiraputra, C. (2009). Rifampin resistance in carbapenem-resistant Acinetobacter baumannii in Siriraj Hospital, Thailand. Nepal. Med. Coll J. 11, 232–237.
Tipton, K. A., and Rather, P. N. (2017). An ompR-envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075. J. Bacteriol. 199:e0705-16. doi: 10.1128/JB.00705-16
Vijayakumar, S., Rajenderan, S., Laishram, S., Anandan, S., Balaji, V., and Biswas, I. (2016). Biofilm formation and motility depend on the nature of the Acinetobacter baumannii Clinical Isolates. Front. Public Health 4:105. doi: 10.3389/fpubh.2016.00105
Vinogradov, E., Maclean, L., Xu, H. H., and Chen, W. (2014). The structure of the polysaccharide isolated from Acinetobacter baumannii strain LAC-4. Carbohydr. Res. 390, 42–45. doi: 10.1016/j.carres.2014.03.001
Waldor, M. K., Friedman, D. I., and Adhya, S. L. (eds) (2005). Phages: Their role in Bacterial Pathogenesis and Biotechnology. Washington, D.C: ASM Press.
Weber, B. S., Harding, C. M., and Feldman, M. F. (2015). Pathogenic Acinetobacter: from the Cell Surface to Infinity and Beyond. J. Bacteriol. 198, 880–887. doi: 10.1128/JB.00906-15
Wong, D., Nielsen, T. B., Bonomo, R. A., Pantapalangkoor, P., Luna, B., and Spellberg, B. (2017). Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 30, 409–447. doi: 10.1128/CMR.00058-16
Wood, D. E., and Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46. doi: 10.1186/gb-2014-15-3-r46
Wyres, K. L., Cahill, S. M., Holt, K. E., Hall, R. M., and Kenyon, J. J. (2019). Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microbiology[Epub ahead of print],
Yu, G., Lam, T. T.-Y., Zhu, H., and Guan, Y. (2018). Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 35, 3041–3043. doi: 10.1093/molbev/msy194
Zarrilli, R., Pournaras, S., Giannouli, M., and Tsakris, A. (2013). Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41, 11–19. doi: 10.1016/j.ijantimicag.2012.09.008
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Keywords: Acinetobacter baumannii, antibiotic resistance, phylogenomics, surface structures, complement, global clone 2
Citation: Loraine J, Heinz E, Soontarach R, Blackwell GA, Stabler RA, Voravuthikunchai SP, Srimanote P, Kiratisin P, Thomson NR and Taylor PW (2020) Genomic and Phenotypic Analyses of Acinetobacter baumannii Isolates From Three Tertiary Care Hospitals in Thailand. Front. Microbiol. 11:548. doi: 10.3389/fmicb.2020.00548
Received: 26 November 2019; Accepted: 13 March 2020;
Published: 06 April 2020.
Edited by:
Benjamin Andrew Evans, University of East Anglia, United KingdomReviewed by:
Nabil Karah, Umeå University, SwedenJohanna J. Kenyon, Queensland University of Technology, Australia
Raffaele Zarrilli, University of Naples Federico II, Italy
Copyright © 2020 Loraine, Heinz, Soontarach, Blackwell, Stabler, Voravuthikunchai, Srimanote, Kiratisin, Thomson and Taylor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter W. Taylor, cGV0ZXIudGF5bG9yQHVjbC5hYy51aw==
†These authors have contributed equally to this work
 Jessica Loraine1†
Jessica Loraine1† Eva Heinz
Eva Heinz Rosesathorn Soontarach
Rosesathorn Soontarach Grace A. Blackwell
Grace A. Blackwell Richard A. Stabler
Richard A. Stabler Pattarachai Kiratisin
Pattarachai Kiratisin Peter W. Taylor
Peter W. Taylor