- 1Jiangsu Key Laboratory of Zoonosis, Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, China
- 2Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality, Ministry of Agriculture of China, Yangzhou, China
- 3Department of Infection Biology, Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom
- 4Joint International Research Laboratory of Agriculture and Agri-product Safety, Ministry of Education of China, Yangzhou, China
FlhF is a key protein required for complete flagellar synthesis, and its deletion results in the complete absence of a flagella and thus motility in Campylobacter jejuni. However, the specific mechanism still remains unknown. In this study, RNA-Seq, EMSAs, ChIP-qPCR and β-Galactosidase assays were performed to elucidate the novel interactions between FlhF and genes involved in flagellar synthesis. Results showed that FlhF has an overall influence on the transcription of flagellar genes with an flhF mutant displaying down-regulation of most flagellar related genes. FlhF can directly bind to the flgI promoter to regulate its expression, which has significant expression change in an flhF mutant. The possible binding site of FlhF to the flgI promoter was explored by continuously narrowing the flgI promoter region and performing further point mutations. Meanwhile, FlhF can directly bind to the promoters of rpoD, flgS, and fliA encoding early flagellin regulators, thereby directly or indirectly regulating the synthesis of class I, II, and III flagellar genes, respectively. Collectively, this study demonstrates that FlhF may directly regulate the transcription of flagellar genes by binding to their promoters as a transcriptional regulator, which will be helpful in understanding the mechanism of FlhF in flagellar biosynthetic and bacterial flagellation in general.
Introduction
Campylobacter jejuni flagella are considered the main virulence factor playing a key role in many important biological activities, such as motility, chemotaxis, adhesion, secreting virulence and colonization factors (Beeby, 2015; Burnham and Hendrixson, 2018; Subramanian and Kearns, 2019). C. jejuni is a microaerophilic, Gram-negative bacterium, and is the leading cause of foodborne related gastroenteritis worldwide (Flint et al., 2016; Burnham and Hendrixson, 2018). It generates a single unsheathed flagellum at one or both poles of the cell (Hendrixson and Dirita, 2003; Matsunami et al., 2016; Liang and Connerton, 2018). Flagellar biosynthesis is complicated requiring expression of more than 50 genes highly regulated by a complex regulatory network that ensures the coordinate expression to construct an intact flagella organelle (Balaban et al., 2009; Grinnage-Pulley et al., 2016; Liang and Connerton, 2018). Given the importance of flagella, a thorough understanding of its assembly is necessary.
In many bacteria, flagella genes are grouped together into operons and are controlled by global regulatory factors (Chilcott and Hughes, 2000; Prouty et al., 2001; Dasgupta et al., 2003; Liu and Ochman, 2007). In C. jejuni however, scattered flagellar genes lack a global regulatory factor, such as FlhDC in E. coli, creating a challenge for exploring the regulation mechanism on the flagellar synthesis. The formation of flagella is divided into three cascades (Balaban et al., 2009). In the early stage, σ70 factor-dependent class I genes are synthesized, including flagellar export apparatus (FEA, consisting of FlhA, FlhB, FliF, FliO, FliP, FliQ, and FliR), σ28, σ54 factors and FlgSR TCS. Then class II genes and class III genes are synthesized in sequence (Wösten et al., 2004; Joslin and Hendrixson, 2009; Lertsethtakarn et al., 2011). Although flagella have long been extensively studied, there remains a gap in our knowledge as to the regulation mechanisms of flagellar proteins synthesis (Gao et al., 2014).
One identified protein that primarily affects flagellar biosynthesis is FlhF. In other species, the disruption of flhF can lead to a range of different phenotypes, including reduced flagellar gene expression, decreased or absent motility, decreased virulence, abnormal flagella assembly and number, and even no flagellation (Kazmierczak and Hendrixson, 2013; Burnham and Hendrixson, 2018). In C. jejuni, an flhF mutant leads to a complete loss of motility and a non-flagellar phenotype. Despite FlhF having a crucial influence on the flagellar synthesis, the specific genetic regulatory mechanisms are unclear (Kim et al., 2012; Kazmierczak and Hendrixson, 2013; Schuhmacher et al., 2015). Thus far, many studies of FlhF have focused on its role in determining the position and number of flagella substructure. FlhF is a member of the signal recognition particle (SRP) associated GTPase family, however the exact function is not well defined (Kim et al., 2012; Guttenplan et al., 2013; Schniederberend et al., 2013; Schuhmacher et al., 2015; Gulbronson et al., 2016). Other studies have identified the influence on flagellar gene expression, nevertheless, the results reported have not always been in alignment (Niehus et al., 2004; Correa et al., 2005; Murray and Kazmierczak, 2006; Balaban et al., 2009; Kim et al., 2012). In C. jejuni, the specific mechanisms of FlhF still need to be explored in depth.
Transcriptional regulators are important for biological response and their adaptability to different conditions in the organism (Galán-Vásquez et al., 2016). Some organisms have many transcriptional regulators, for example E. coli has seven σ factors, Bacillus subtilis has 19, Streptomyces coelicolor over 60 and more than 100 in Sorangium cellulosum (Bervoets and Charlier, 2019). However, the C. jejuni genome carries only three sigma factors, RpoD, RpoN, and FliA (Hwang et al., 2011). Meanwhile, there are approximately 34 other transcription factors in C. jejuni (Wösten et al., 2004; Nachamkin et al., 2008; Grinnage-Pulley et al., 2016). Genome-wide analysis indicates that C. jejuni strains contain between approximately 1,650 and 1,800 genes (Parkhill et al., 2000; Hofreuter et al., 2006; Parker et al., 2006). So this indicates that all C. jejuni biological functions, including bacterial replication, adaptation to environments and bacterial pathogenicity are largely controlled by a limited number (∼2% of the total) of C. jejuni proteins (Nachamkin et al., 2008). Therefore, the discovery of new transcriptional factors and transcriptional regulation mechanisms is essential to better analyze C. jejuni biology.
Considering the crucial influence of FlhF on the flagellar synthesis, we speculate that FlhF may directly regulate flagellar genes expression like a transcription factor. We have applied EMSA and ChIP-qPCR to explore the transcriptional function of FlhF here. The overall influence of FlhF on flagellar gene expression was analyzed by RNA-Seq. We further explored whether FlhF directly regulates early flagellar regulatory factors including RpoD, RpoN, FliA, FlgSR two-component system (TCS) (Petersen et al., 2003; Wösten et al., 2004). Collectively, our results firstly reveal that FlhF may directly regulate flagellar genes transcription by binding the promoters of specific genes (Huffman and Brennan, 2002). Moreover, we proposed a pattern for the feasible transcriptional regulatory pathways of FlhF in flagellar synthesis which will be helpful in understanding the C. jejuni flagella biosynthetic pathway and bacterial flagellation in general.
Materials and Methods
Bacterial Strains and Plasmids
All strains and plasmids used in this study are listed in Supplementary Table S1. Briefly, C. jejuni 81–176 strain and its derivatives were typically grown on Campylobacter blood-free selective agar containing charcoal cefoperazone deoxycholate (CCDA) (Oxoid, Basingstoke, United Kingdom) at 42°C under microaerobic conditions (85% N2, 10% CO2, and5% O2). Escherichia coli DH5α and BL21 strains were grown at 37°C in Luria-Bertani (LB) broth or on LB agar (Ren et al., 2018). As required, antibiotics were added to the medium for C. jejuni or E. coli at the following concentrations: 100 μg ml–1 ampicillin, 50 μg ml–1 kanamycin, or 20 μg ml–1 chloramphenicol. The plasmid pMD-19T (simple) (TaKaRa, Dalian, China) was used as a suicide vector in cloning and strain construction. Plasmid pRK2013 (Biomedal, Seville, Spain) is a helper plasmid for triparental mating conjugation, while pUOA18 is a C. jejuni shuttle vector courtesy of Qijing Zhang (Iowa State University, Ames, United States).
Construction of C. jejuni Mutant and Complemented Strains
To inactivate flhF, its flanking regions and the KanR cassette were amplified from C. jejuni genome and pRY107, then, ligated into pMD-19T (simple) using T4 ligase to obtain a suicide plasmid. The primers used for strain construction are listed in Supplementary Table S2. The suicide plasmid was electroporated into C. jejuni competent cells, and the resulting transformants were selected on CCDA agar containing 50 μg ml–1 kanamycin. The flhF complement strain was constructed by the shuttle vector pUOA18 as previously described (Ren et al., 2018). The target gene was amplified and ligated directly downstream of the promoter Pmetk in the shuttle vector. The recombinant plasmid was mobilized into the flhF mutant strain by triparental mating using E. coli DH5α transformant containing pUOA18-Pmetk-flhF plasmid as the donor strain and DH5α (pRK2013) as the helper strain, by the method described by Miller et al. (2000). The cultures of flhF mutant strain were removed and resuspended in PBS to an OD600 of 1.0. Overnight cultures of the donor and helper E. coli strains were subcultured into Luria-Bertani (LB) broth and grown to an OD600 of 1.2. Cells were mixed at a ratio of 1:1:10 (donor/helper/recipient), spotted onto the Mueller-Hinton (MH) agar plate (BD, United States), and incubated overnight at 42°C under microaerophilic conditions. The mating spot was then resuspended in Mueller-Hinton (MH) broth and plated onto CCDA plate amended with Polymyxin B (6.7 μg/ml), Rifampicin (10 μg/ml), Trimethoprim (5 μg/ml), and chloramphenicol (20 μg/ml). The plates were examined after 3–5 days for the appearance of C. jejuni colonies, and verified by polymerase chain reaction (PCR).
Expression and Purification of FlhF-His6, CmeR-His6
FlhF and CmeR proteins were expressed in E. coli DE3 system containing pET-30-FlhF and pCold I-CmeR, respectively. The flhF,cmeR genes were amplified from C. jejuni genome, then, ligated into pET-30a (between BamHI and XhoI sites) and pCold I (between XhoI and SalI sites), respectively, using the ClonExpress II one-step cloning kit (Vazyme, Nanjing, China) to generate the recombinant expressing plasmids used to transform E. coli DE3. Then they were cultured on a LB plate containing 50 μg/ml kanamycin and verified by PCR and nucleotide sequencing. The FlhF-His6, CmeR-His6 protein were expressed and purified from the soluble extract by affinity chromatography using a HiTrap Ni2+-chelating column. The purification procedure followed the instructions of the manufacturer of the His Bind Purification Kit (Novagen, EMO Millipore corp, Billerica, MA, United States). Purified Protein were analyzed by SDS-PAGE (Supplementary Figure S5) and stored at −70°C.
Construction of Promoter-lacZ Transcriptional Fusions
The promoter regions of interest were amplified from C. jejuni genome and ligated into pMW10 to obtain promoter-lacZ transcriptional fusion plasmids (Wosten et al., 1998). With the aid of plasmid pRK2013, the transcriptional fusion plasmids were introduced into WT and the flhF mutant strain by amphiphilic mating conjugation, which was cultured on a CCDA agar containing 50 μg/ml kanamycin, and verified by PCR. The primers used for strain construction are listed in Supplementary Table S2.
RNA Isolation and Quantitative Real-Time PCR
Campylobacter jejuni 81-176, FlhF-kan mutant strain was grown on CCDA plates and suspended in Mueller-Hinton (MH) broth (BD, United States) with an initial OD540 of 0.07, cultured for 8 h with 42°C, 120 rpm, and total RNA was extracted by using an RNeasy plus mini kit (Qiagen, Hilden, Germany) obeying the instructions of manufacturer. cDNA was synthesized by a total of 500 ng of RNA using RT reagent kit (TaKaRa, Dalian, China), which was subjected to quantitative real-time PCR (qRT-PCR) or stored at −70°C until use. qRT-PCR was carried out in an ABI PRISM 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) using a FastStart Universal SYBR Green Master (ROX) (Roche Diagnostics, Mannheim, Germany). Cycling conditions were as follows: 2 min at 50°C, then 40 cycles of 30 s at 95°C and 34 s at 60°C. As previously described, relative genes expression was calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001). All specific primers are listed in Supplementary Table S2, in which the glyA gene was used as an endogenous control. A series of 10-fold diluted cDNA were used as templates and the standard curves were generated for each candidate genes. The PCR efficiency (E) was calculated using the following formula (Pfaffl, 2001):
RNA-Seq
To analyze the transcriptome, RNA-Seq libraries were generated for six bacterial samples [2 bacterial strains (WT, ΔflhF) × 3] from cDNA using instructions according to the TruSeqTM RNA sample preparation Kit (Illumina, San Diego, CA, United States). As previously described, the quality control of the total RNA samples was performed using a 2100 Bioanalyzer (Agilent) and the ND-2000 (NanoDrop Technologies). Only high-quality RNA samples (OD260/280 = 1.8∼2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5) were used to construct sequencing library (Zhang et al., 2019). The cDNA was then synthesized according to the SuperScript double-strand cDNA synthesis kit (Invitrogen, Carlsbad, CA, United States). The RNA-Seq libraries were subjected to quality inspection using an Agilent 2100 Bioanalyzer (Agilent Technologies) and sequenced on an Illumina HiSeq4000 (Illumina Inc., San Diego, CA, United States), which was biologically replicated in a separate experiment by Majorbio (Shanghai, China). Sequence reads were processed and mapped as previously described (Garber et al., 2011). Gene expression (FPKM) and differential expression levels were analyzed using Rsem1 and edgeR software2. For functional annotation of mRNA, we used Blastx with the NCBI-NR database, String, Swissprot and the Kyoto encyclopedia of genes and genomes (KEGG) database. Statistical analysis according to the method described in the previous period, P-value < 0.05 was considered to be statistically significant. All RNA-Seq data was uploaded to the EBI ENA databased (Accession number PRJEB34440).
Electrophoretic Mobility Shift Assay (EMSAs)
The EMSAs were performed as follows: PCR fragments encompassing the promoters of genes with FAM-labeled were amplified using genomic DNA of C. jejuni 81-176 as a template. The DNA fragments were gel-purified using MiniBEST Agarose Gel DNA Extraction Kit (Takara, Japan). Each PCR product (≈5 ng) was mixed with increasing concentrations of purified FlhF-His6 in a final volume of 20 μl buffer containing 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 1 mM DTT, 0.5 mM EDTA and 5% glycerol. The reactions were incubated for 30 min at 25°C and then loaded with 10×EMSA loading buffer on 6% polyacrylamide non-denaturing gels in 0.5 × Tris-borate-EDTA buffer. Each reaction was verified to be specific by adding 10-fold non-specific competitor [Poly(dI:dC)]. For a negative control, synthesized His-tag was incubated with flgI promoter, denoted as negative control (NC). For positive controls, cmeA promoter was incubated with the purified CmeR protein, and cmeA promoter alone, denoted as positive control (PC) (Cagliero et al., 2007).
ChIP-qPCR
The 3 × FLAG-tagged strain (WT flhF-FLAG) was grown under microaerobic conditions and then pelleted by centrifugation. As described previously (Blasco et al., 2012), however, with some variation, ChIP was performed based on established methods as follows. Formaldehyde was added to bacterial cells (1% final concentration) for cross-linking and then incubated at room temperature for 25 min. Reactions were quenched with 0.5 M glycine, and samples were pelleted and washed three times with PBS. The samples were then used for ChIP following the Chromatin Immunoprecipitation kit (Millipore, United States) protocol. The antibody used was the anti-FLAG mouse monoclonal antibody (Sigma). For ChIP-qPCR experiments, untreated chromatin was de-cross-linked by boiling for 10 min and purified for use as the “input” control. The relative enrichment of candidate gene promoters was performed with qRT-PCR and represents the value of the immunoprecipitated DNA divided by the input unprecipitated DNA. These values were normalized to the values obtained for each promoter precipitated using untagged wild-type in order to account for non-specific enrichment. The results represent the mean enrichment measured via qPCR in at least three biological replicate experiments.
β-Galactosidase Assay
Campylobacter jejuni cells carrying the transcriptional fusion plasmids were grown on CCDA plates and suspended in MH broth with the same OD600. The cells with centrifugation were suspended thoroughly with 1 ml Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0) and shaken vigorously to lyse the cells with adding 30 μl of chloroform and 0.1% SDS. The assays were performed at 37°C with 200 μl ONPG (O-nitrophenol-β-D-galactopyranoside, 4 mM, Sigma) and monitored at 420 nm (Cagliero et al., 2007). β-Galactosidase activities were calculated in Miller Units using the formula given below: β-Galactosidase activity = A420 × 1000 × min–1 × ml–1 × A600–1. The results were reported as the mean of three biological replicates.
Results
FlhF Has an Overall Impact on the Transcription of Flagella Components
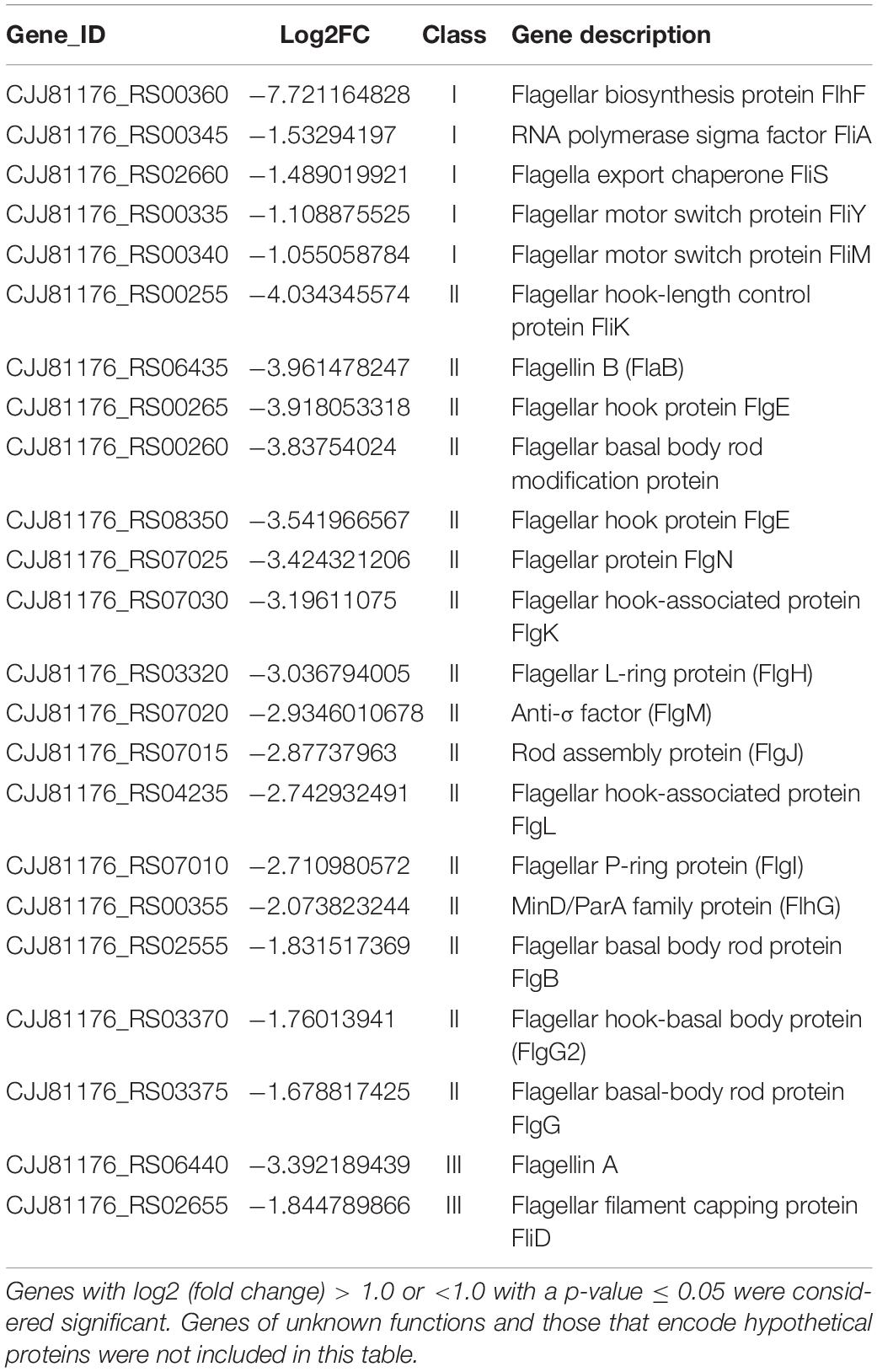
To investigate which genes are regulated by FlhF on the transcriptional level, we performed high-throughput RNA sequencing based on the genetic background (flhF mutant vs. wild-type). RNA-Seq data (Supplementary Table S3) showed all the modulated flagellar related genes are down-regulated in the mutant, suggesting FlhF has a positive role in flagellar gene transcription. Among these down-regulated genes, 26 genes are involved in the process of flagellar biosynthesis. Grouping these genes according to their function and substructure affiliation showed a general trend of down-regulation from flagella export apparatus, motor/switch components, an unknown function (hypothetical genes with unknown function), flagellin glycosylation, transcription regulators, flagella basal body, the filament, to the most down-regulated which was the flagella hook (Figure 1A). Flagellar genes are classified into three cascades, class I genes are σ70 dependent, while class II and class III genes require σ54 and σ28 factor, respectively (Ren et al., 2018). A one-way ANOVA analysis showed that class I, III, and II genes were significantly modified when comparing the flhF mutant to the respective wild-type strain (Figure 1B). qRT-PCR was performed to verify the results of the RNA-Seq data. Seven flagellar genes that belong to different cascades were randomly selected (Figure 2). The amplification efficiency of each pair of primers were close to 2 (Supplementary Figure S1) and the reference gene glyA was constantly expressed under this experimental condition due to the relatively stable CT values (Supplementary Table S4).
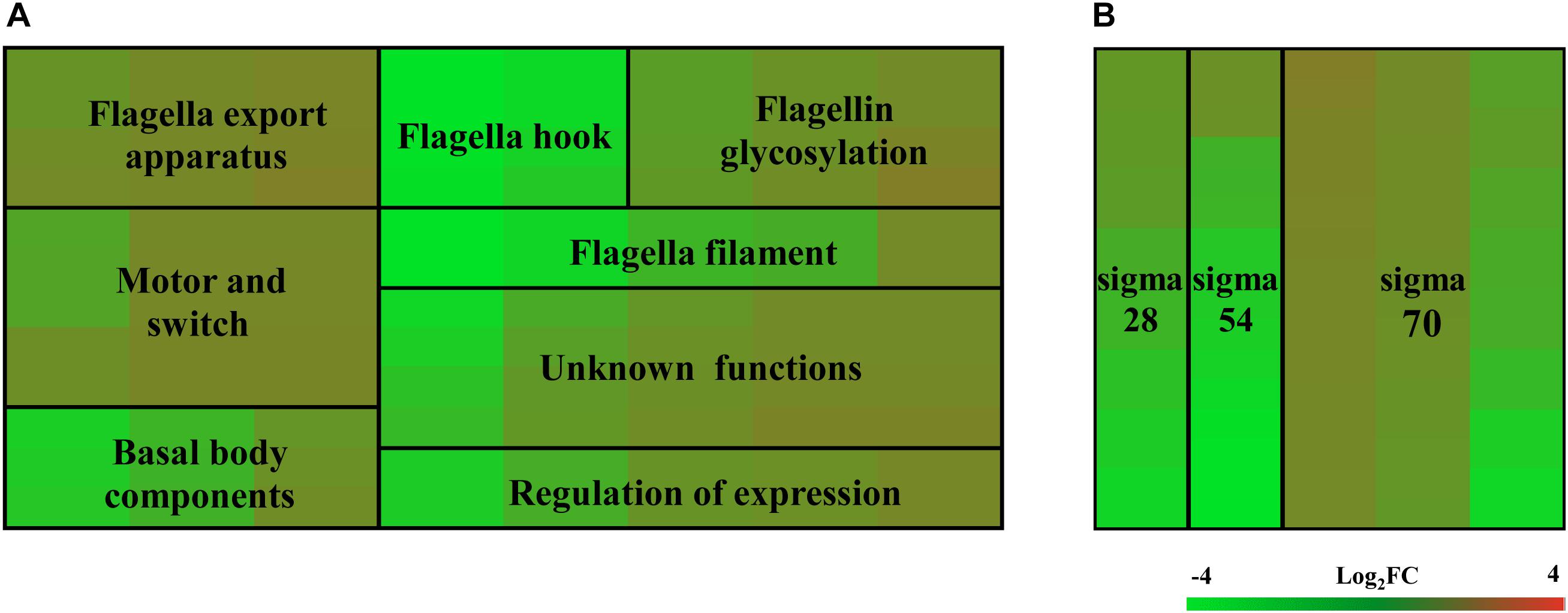
Figure 1. Heat map of relative mRNA expression of C. jejuni flagella genes. The transcription of flagella genes in the RNA-Seq were analyzed according to their function and substructure affiliation (A), as well as transcription cascade (B). Flagella hook genes and class II genes were significantly affected according to the ANOVA analysis at P < 0.05. Green and red in the heat map represent down-regulation and up-regulation of genes in the flhF mutant relative to the wild-type strain, with more saturated colors representing a larger differential effect as listed in the bar.
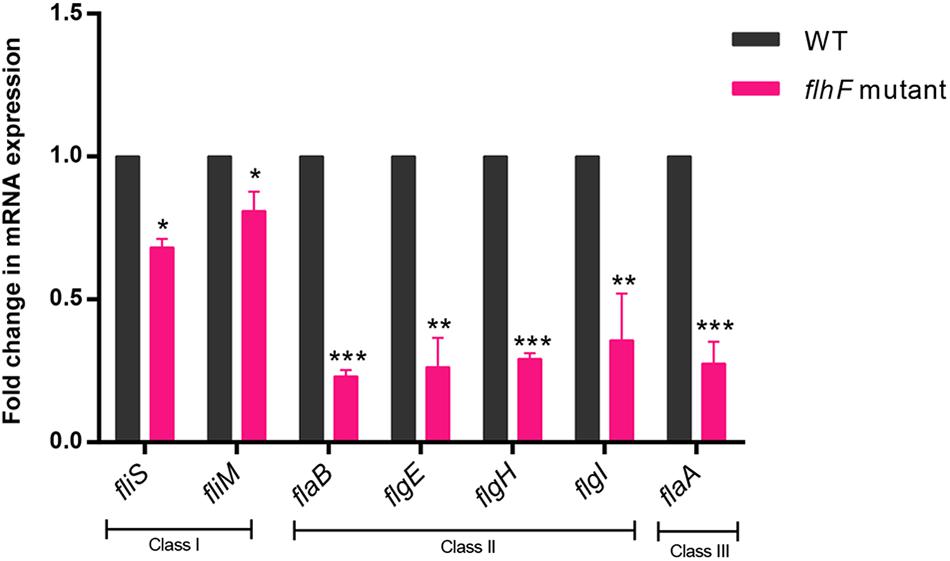
Figure 2. qRT-PCR for verification of RNA-Seq data. qRT-PCR data demonstrated correlation with the RNA-Seq data when investigating the transcript fold-change difference between WT strain and the flhF mutant strain. Each sample was examined in three biological replicates and was repeated with three technical replicates. Data are presented as mean ±SD. Data were analyzed by using a one-sample t-test (* P < 0.05, ** P < 0.01, *** P < 0.001).
FlhF May Directly Regulate flgI Transcription by Binding Its Promoter
To determine the transcriptional function of FlhF, six genes with significantly different expression were randomly selected from the RNA-Seq results, including fliK, flaB, flgE, flaA, flgL, flgI, to explore whether FlhF binds their promoters by Electrophoretic mobility shift assay (EMSA). Our results demonstrated that the purified FlhF-His6 bound to the promoter of flgI (Figure 3), the flagellar P-ring component, but did not bind to the promoters of other genes (Supplementary Figures S2A–E). The results were further verified by Chromatin Immunoprecipitation quantitative PCR (ChIP-qPCR) analysis (Figure 4). We selected flgI and flaB to perform ChIP-qPCR, which showed that the promoter of flgI was extremely enriched in FlhF-ChIP samples, and the relative quantity was significantly higher than in the IgG control samples (Figure 4), while the promoter of flaB was not enriched in the FlhF-ChIP samples (Supplementary Figure S3). In summary, all results demonstrated that FlhF may directly regulate flgI as a positive transcriptional regulator.
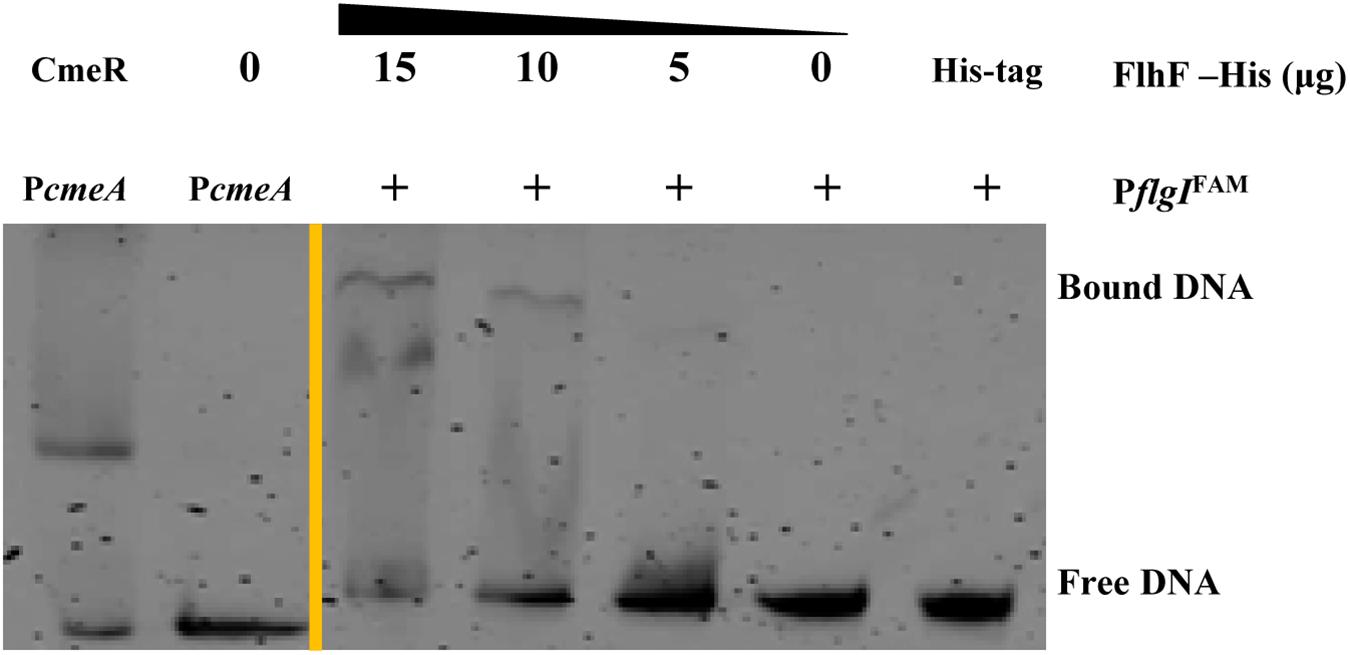
Figure 3. FlhF directly interact with the flgI promoter. EMSA demonstrated that the fluorescently labeled DNA probes of the flgI promoter were incubated with purified FlhF-6His at different concentrations. Each reaction was verified to be specific by adding 10-fold non-specific competitor [Poly(dI:dC)]. For a negative control, synthesized His-tag was incubated with flgI promoter, denoted as negative control (NC). For positive controls, cmeA promoter was incubated with the purified CmeR protein, and cmeA promoter alone, denoted as positive control (PC). The “+” symbol indicates the presence of FlhF-6His.
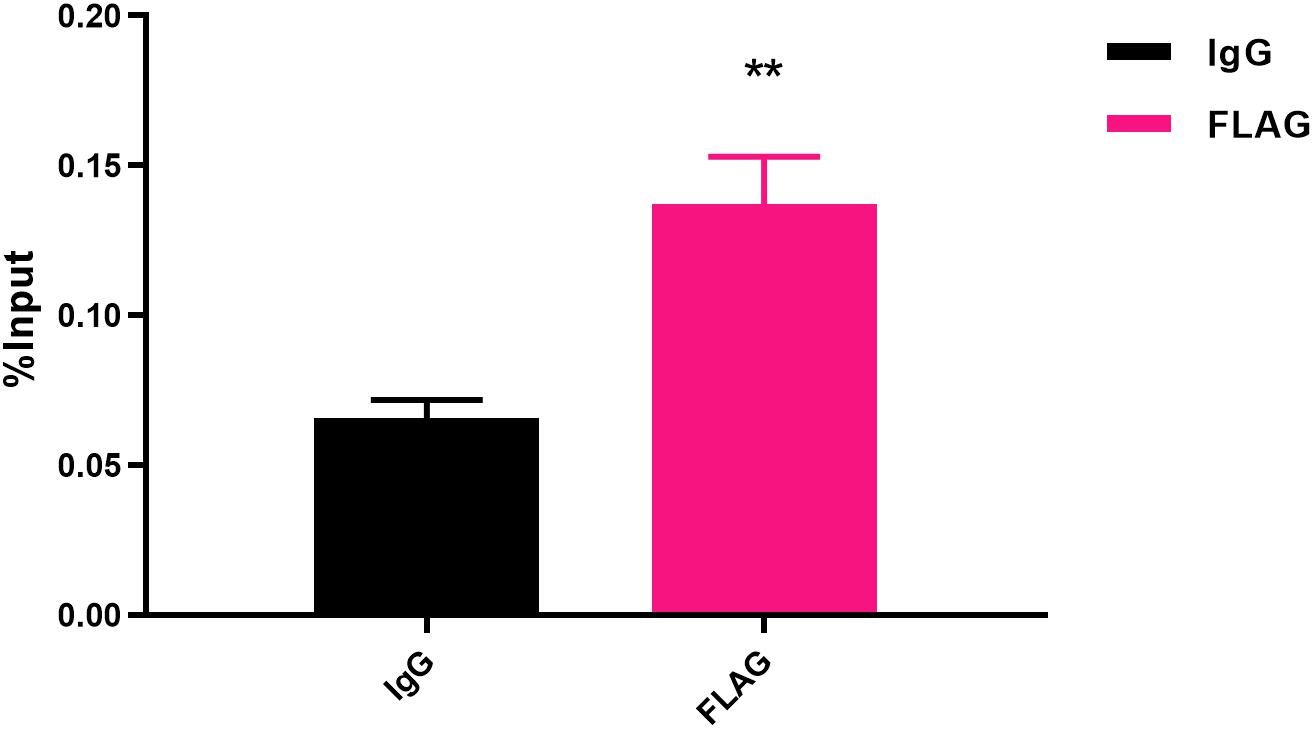
Figure 4. Fold enrichment of the flgI promoter in ChIP samples measured via ChIP-qPCR. The results show the promoter of flgI was extremely enriched in FlhF-ChIP samples, and the relative quantity was significantly higher than in the IgG control samples. Genome fragments isolated from wild-type flhF-FLAG strain were immunoprecipitated with corresponding antibodies, and analyzed by real-time PCR using primer sets corresponding to transcriptional start site regions of the flgI. For ChIP-qPCR experiments, the sample was cross-linked and sonicated to generate small DNA fragments, and then divided into three parts. The first was used as an input control. The second was incubated with normal rabbit IgG that will not bind to nuclear proteins to generate immuno-precipitated DNA (IgG). The third was as ChIP-FLAG immunoprecipitated sample (FLAG) (O’Geen et al., 2010). Results represent mean enrichment as measured by qPCR in at least three biological replicate experiments. Data are presented as mean ±SD. Data were analyzed by one-sample t-test (** P values are < 0.01, but > 0.001) to estimate the significance of fold change between FlhF-ChIP samples and IgG control samples.
FlhF-Binding Site in the flgI Promoter
To delineate the contribution of portions of the flgI promoter for binding of FlhF, the flgI promoter was divided into six fragments (Figure 5A), which were amplified with FAM-labeling and ligated into pMW10 to perform EMSA and β-Galactosidase assay here. EMSA results showed that the purified FlhF-His6 bound to the fragments 1–4 of the flgI promoter, but did not bind to the fragments 5–6 (Figure 5B), which demonstrated that the putative binding site of FlhF in the flgI promoter was between 101–125 bp. β-Galactosidase assays performed to verify the results from 5B and showed that the fragments 1–4 of the flgI promoter had notable differences between the WT and flhF mutant strain, but fragment 5 has no difference with negligible activity, which was consistent with the EMSAs (Figure 5C). We used strains containing pMW10 served as a negative control. Therefore, we speculated the FlhF-binding site (FBS) in flgI promoter is the region “−76 to −51” (AAGAAATTTGGATCAACTAGCTTAAG) (Figure 5D). To further investigate the necessity of this motif for the binding of FlhF to the flgI promoter (PflgI), we selected a point mutation every 5 bp on the possible FlhF-binding site to generate five different point mutation fragments which were amplified with FAM-labeling (Figure 5E). EMSA results showed that the purified FlhF-His6 bound to the 26 bp possible FlhF-binding site of the flgI promoter, but did not bind to the five different point mutation fragments (Figure 5F), which demonstrated that the five-point mutations abolish binding of FlhF to PflgI.
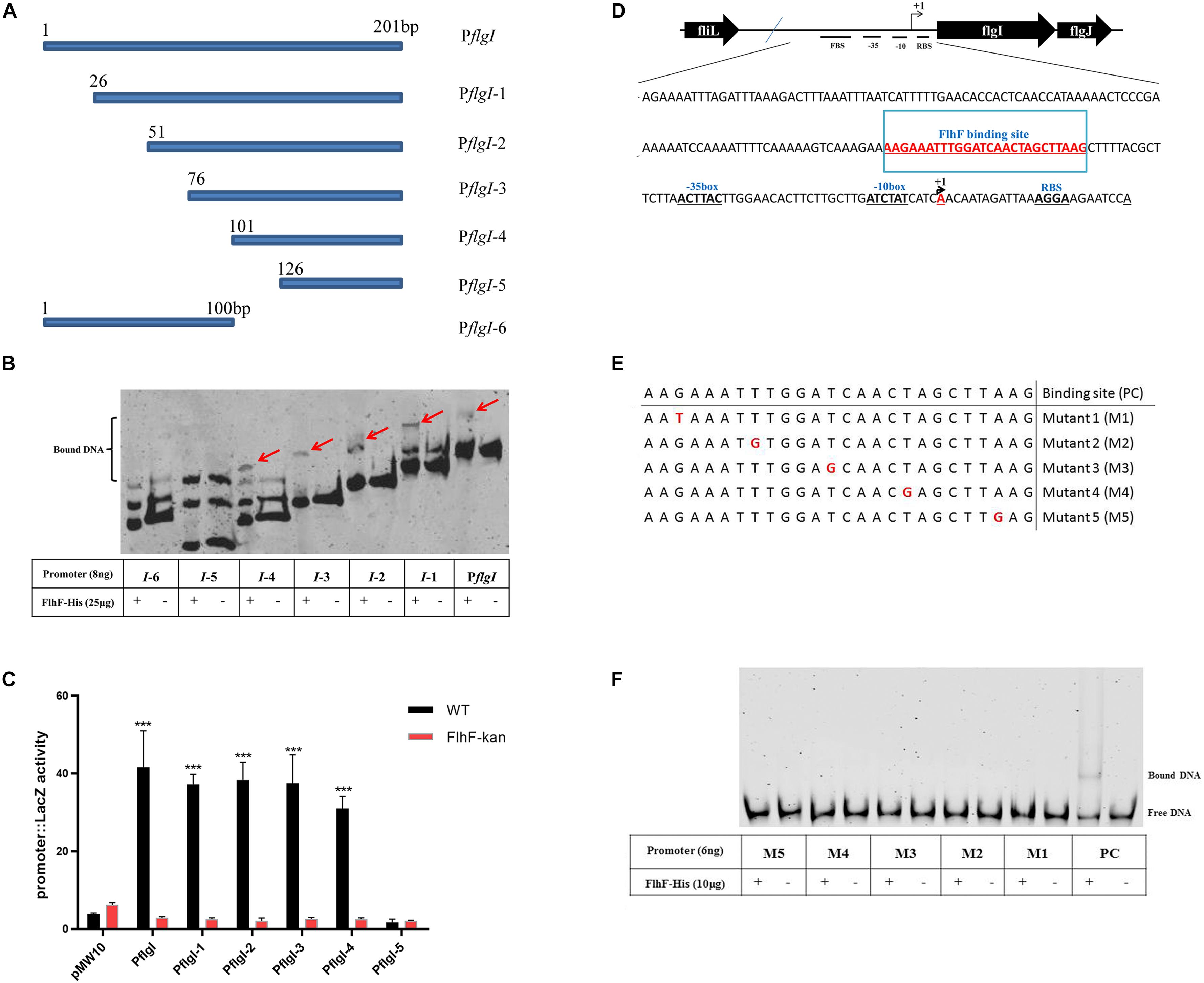
Figure 5. FlhF- binding site in the flgI promoter. (A) A diagram of the flgI promoter (a 201 bp fragment of the promoter adjacent to the start codon “ATG”) and the six regions carried on the DNA fragments used in this assay. Fragment names are displayed in figure. (B) EMSA analysis of FlhF specifically binding to the flgI promoter. All seven fragments (8 ng) with FAM labeled in (A) were incubated with purified FlhF-His6 (25 μg). The “+” and “–” symbols indicate cultures incubated with FlhF-His6 and without FlhF-His6, respectively. The red arrows represent the bound DNA. Each reaction was verified to be specific by adding 10-fold non-specific competitor [Poly(dI:dC)]. (C) β-Galactosidase assay for verification of EMSA results. Expression of the β-Galactosidase activities in WT and flhF mutant strains containing different fragments. The activity is expressed as the mean ± SE from three biological experiments. The strains containing pMW10 served as negative control. Data were analyzed by t-test to estimate the significance of fold change between WT and FlhF-kan samples. The symbol “***” means P < 0.001. (D) Diagram showing the promoter region of the flgI gene. The ribosome-binding site (RBS) are underlined and the transcription start sites are labeled as +1 and marked in red. The −35/−10 motif were located directly upstream of the transcriptional start site +1A. We speculated that the FlhF-binding site was underlined and marked in red (−76 to −51). (E) A diagram of five different point mutations of possible FlhF-binding site on PflgI (a 26 bp fragment) and the six DNA fragments used in this assay. Fragment names are displayed in figure. PC signifies positive control. (F) EMSA analysis of the binding of FlhF to the five different point mutations. All six fragments (6 ng) with FAM labeled in (E) were incubated with purified FlhF-His6 (10 μg). The “+” and “–” symbols indicate cultures incubated with FlhF-His6 and without FlhF-His6, respectively. Each reaction was verified to be specific by adding 10-fold non-specific competitor [Poly(dI:dC)].
FlhF Directly Regulates Flagellar Gene Regulators rpoD, fliA, flgS
RNA-Seq results demonstrated that FlhF has an overall impact on the transcription of flagellar components. We hypothesize that FlhF may regulate flagellar gene expression by directly regulating key regulatory factors rpoD, rpoN, fliA, flgSR TCS during flagellar synthesis. Hence, we explored whether FlhF regulates them directly by binding their promoters by EMSA. Results showed that the purified FlhF-His6 bound to the promoters of rpoD, fliA, flgS (Figures 6A–C), but did not bind to the promoters of rpoN and flgR (Supplementary Figures S4A,B).
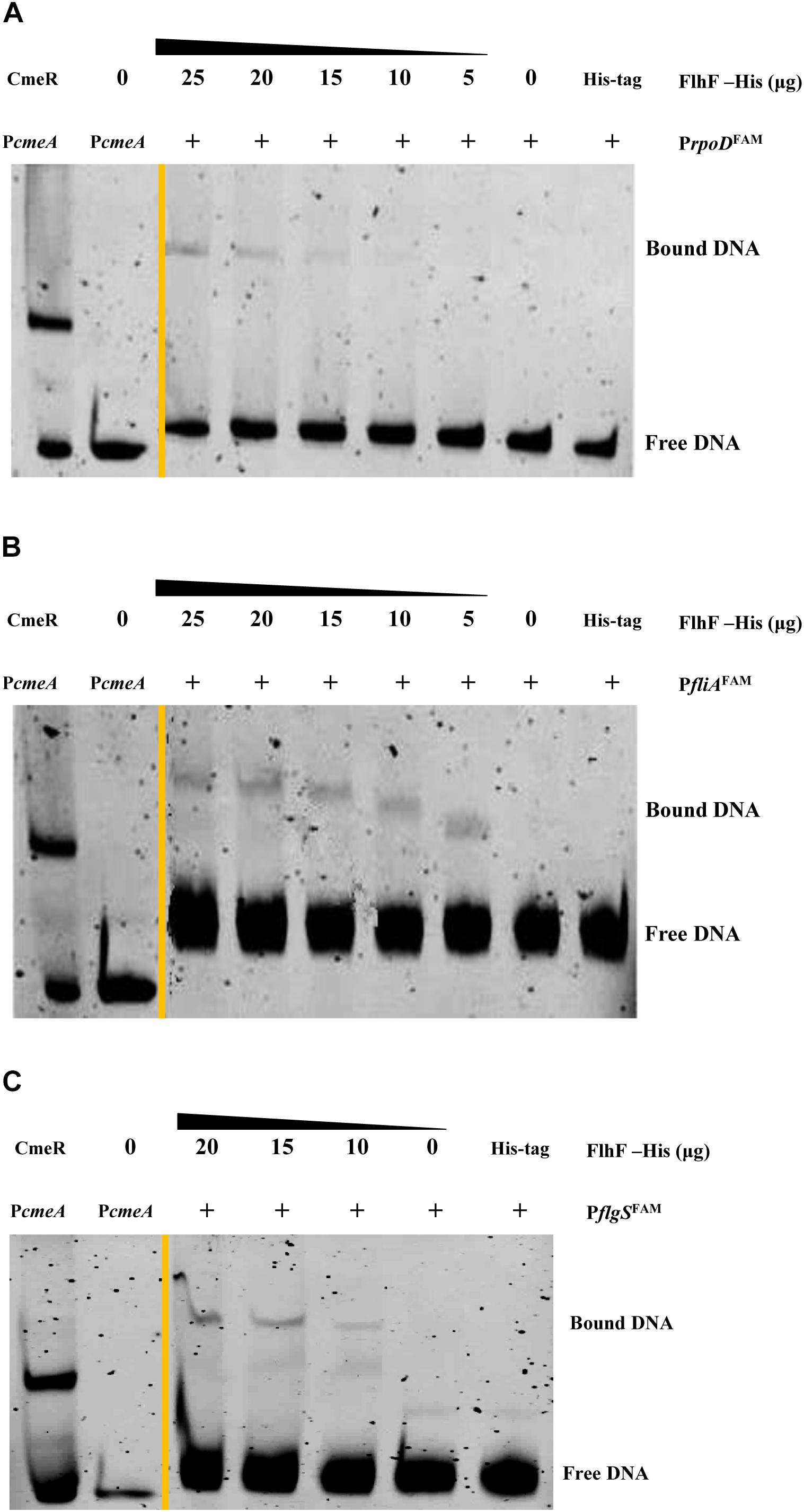
Figure 6. FlhF directly interact with the promoter of rpoD, fliA, flgS. The fluorescently labeled DNA probes of the promoter were incubated with purified FlhF-HIS6 at different concentrations and promoters (A) rpoD; (B) fliA and (C) flgS respectively. For a negative control, synthesized His-tag was incubated with flgI promoter, denoted as negative control (NC). Each reaction was verified to be specific by adding 10-fold non-specific competitor Poly(dI:dC)]. For positive controls, cmeA promoter was incubated with the purified CmeR protein, and cmeA promoter alone, denoted as positive control (PC). The “+” symbol indicates the presence of FlhF-6His.
Discussion
In C. jejuni, flagella is a major virulence factor with a complex synthesis process. FlhF is one of several key proteins that influence flagellar biosynthesis. Deletion of flhF results in a non-motile and non-flagellar phenotype. Despite FlhF having a crucial influence on flagellar synthesis, the specific mechanism of its role remains unclear (Kim et al., 2012; Kazmierczak and Hendrixson, 2013; Schuhmacher et al., 2015). In this study, RNA-Seq was performed to investigate the role of FlhF further. Previous studies involved in functionality of FlhF influencing the flagellar genes expression are varied depending on the bacteria of choice (Niehus et al., 2004; Correa et al., 2005; Murray and Kazmierczak, 2006; Lertsethtakarn et al., 2011; Kim et al., 2012). In Helicobacter pylori, FlhF was found to primarily affect class II and class III flagella genes expression. For Pseudomonas aeruginosa, a mutation of FlhF resulted in decreased transcription of the class IV gene fliC. In Vibrio cholerae and Vibrio vulnificus, FlhF positively affected the transcription of class III and class IV flagella genes. However, our results revealed that FlhF may act as an activator of flagellar genes and thus an overall influence on flagellar gene expression in C. jejuni (Figure 2).
In order to further explore how FlhF affects flagellar expression, in our study we investigated the putative function of FlhF directly influencing flagella synthesis by binding the promoter of flagellar genes. FlhF may positively control flgI expression by binding to promoter of flgI (Figures 4–5), which is the flagellar P-ring component (Boll and Hendrixson, 2013). The P-ring together with L-ring are thought to be required for smooth rotation, functioning as a sleeve in many motile bacteria (Hizukuri et al., 2006, 2008). We found flgI and flgH had 6.6-fold and 8-fold reduction in expression respectively after the deletion of flhF using RNA-Seq (Supplementary Figure S3), which indirectly supported our hypothesis that flhF has a potential regulatory role. Additionally, the protein-binding site and DNA binding site are important for transcriptional regulators. Our results have revealed that the possible binding site of FlhF in the flgI promoter is “AAGAAATTTGGATCAACTAGCTTAAG” (Figure 5). Five different point mutations were generated to further investigate that the complete promoter site may be necessary for binding of FlhF to PflgI. Meanwhile, since FlhF mainly affects class II genes, we speculated that there may be other genes besides flgI that can be directly regulated by FlhF. ChIP-seq will be performed to identify further hits in the future.
In addition, we also found FlhF can directly bind to the promoters of rpoD, flgS, and fliA genes respectively (Figure 6). These genes are key regulatory factors during flagellar synthesis. Balaban proposed that FlhF may directly or indirectly influence the FEA-FlgSR pathway to initiate σ54-dependent genes expression in C. jejuni (Balaban et al., 2009). We propose FlhF may bind PflgS to stimulate flgS phosphorylation of flgR, cooperating with σ54 factor indirectly, to initiate class II gene transcription synthesis. Meanwhile, FlhF can also directly regulate specific class II gene flgI. In addition, FlhF may directly influence the synthesis of class I and III flagellar genes by binding to the rpoD and fliA promoters respectively. However, the expression of fliA is inhibited by FlgM, and interestingly both fliA and flgM were significantly downregulated in the absence of FlhF (Table 1). Thus, one possible hypothesis is that FlhF directly regulates fliA in an independent pathway to promote class III genes synthesis (FlgM being an anti-sigma factor that possibly does not affect fliA expression, but its activity). Finally, we proposed a hypothetical model for the regulatory pathway of FlhF in the flagellar system (Figure 7). In addition, in order to further investigate whether there are similar sites between the binding promoters, we compared the putative 26 bp binding sequence in PflgI with the PrpoD, PfliA, and PflgS promoters through the MEME website (Bailey, 2002). The MEME analysis identified a similar sequence, an AT-rich region (data not shown). We will explore the conservation of FlhF binding sequences in the future.
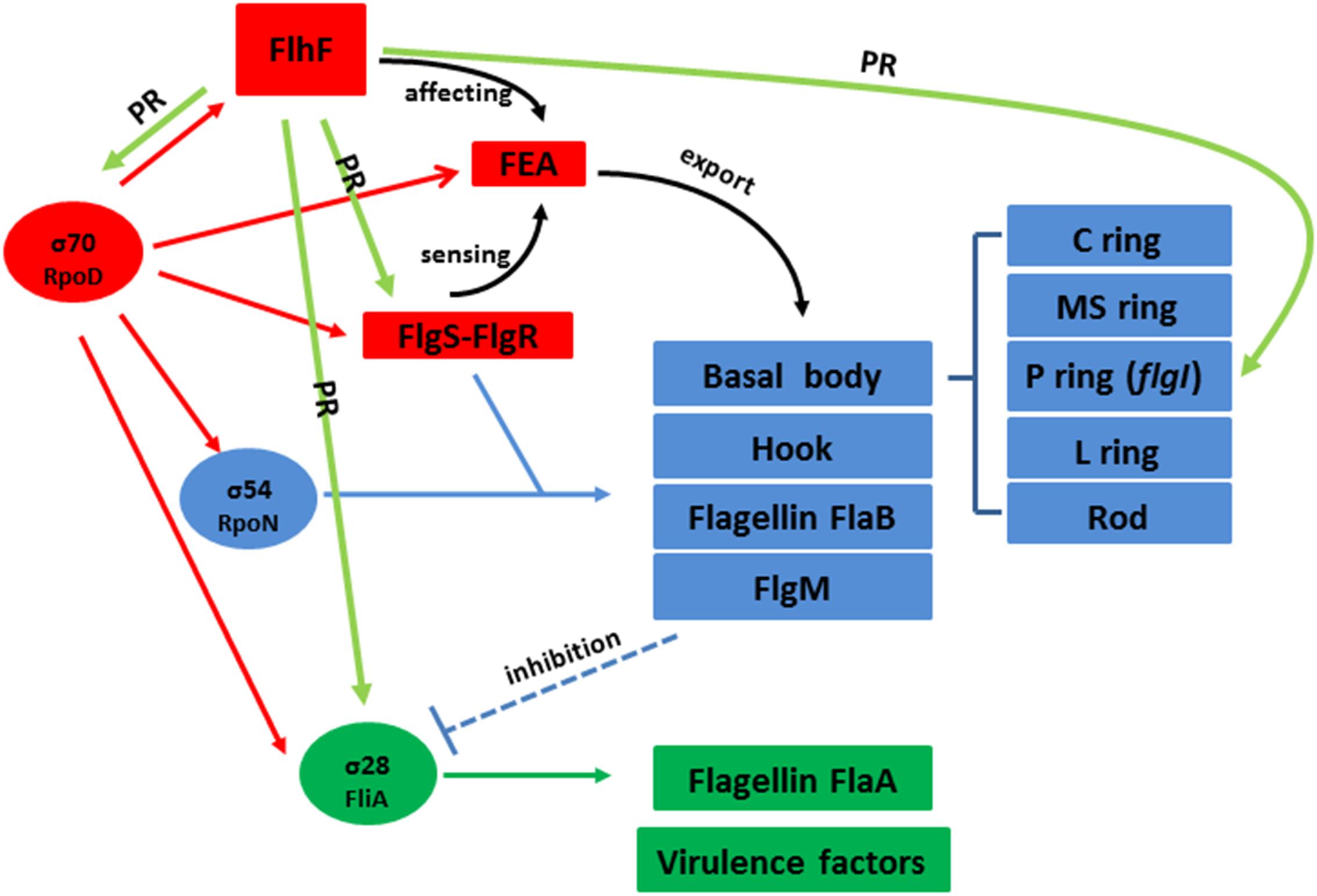
Figure 7. Hypothetical model for the regulatory pathway of FlhF in the C. jejuni flagellar system. C. jejuni flagellar genes are classified into three cascades, class I genes (red) are σ70 (rpoD) dependent, while class II (blue) and class III (green) genes require σ54 (rpoN) and σ28 (fliA) factor, respectively. The σ70 factor-dependent class I genes are constitutively expressed. FlhF acts before the flagella export apparatus (FEA) formation. The FEA formation will also be detected by the FlgSR two-component system (TCS) and activates the σ54 factor-dependent class II genes. After FlgM is secreted from the cell, the inhibition of the σ28 factor-dependent class III genes is relieved and they are expressed. In our study, we speculate that FlhF may directly regulate the synthesis of class I and III flagellar genes by binding to the rpoD and fliA promoters, activating σ70 and σ28 factor respectively. For the class II flagellar genes, FlhF may bind PflgS to stimulate flgS phosphorylation of flgR, cooperating with σ54 factor, indirectly to initiate class II gene transcription synthesis. In addition FlhF may also directly regulate the specific class II gene flgI by binding to its promoter. “PR” signifies “positive regulating” and is presented with a green arrow. The black curved arrow represents the physiological function and the dotted arrow indicates inhibition.
So far, a number of studies have reported that FlhF is a member of the signal recognition particle (SRP)-related GTPase family regulating the number and position of flagella (Green et al., 2009; Guttenplan et al., 2013). However, no FlhF homologs or functionally similar protein with DNA-binding activity has been reported. In our study, we hypothesis that in addition to being an SPR GTPase, FlhF can also directly influence flagella synthesis by binding to the promoters of flagellar genes in C. jejuni. In addition, the GTPase activity of FlhF is not required for flagellar gene transcription in C. jejuni (Gulbronson et al., 2016). Therefore, we speculate that the GTPase activity of FlhF may have little to do with the proposed regulation of FlhF here.
In summary, this study demonstrates that FlhF may directly regulate the transcription of flagellar genes by binding to their promoters as a transcriptional regulator. This will help in our attempts to understand the mechanistic role of FlhF in flagellar biosynthetic and bacterial flagellation. We hope this study will be used as foundation for future studies on FlhF function.
Data Availability Statement
The data is on EBI ENA website with accession number PRJEB34440 (https://www.ebi.ac.uk/ena/data/search?query=PRJEB34440).
Author Contributions
XL, FR, OG, JH, and XJ conceived and designed the experiments. XL, GC, PH, and QC performed the experiments. XL analyzed the data. XL, JH, and XJ contributed reagents, materials, and analysis tools. XL wrote the manuscript. OG and JH reviewed the manuscript.
Funding
This work was supported by the NSFC (31872493), National Key Research and Development Program of China (2018YFD0500500), Priority Academic Program Development of Jiangsu Higher Education Institutions, Yangzhou University High-end Talent Support Program and International Academic Exchange.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00460/full#supplementary-material
Footnotes
- ^ http://deweylab.biostat.wisc.edu/rsem/
- ^ http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html
References
Bailey, T. L. (2002). Discovering novel sequence motifs with MEME. Curr. Protoc. Bioinfor. 00, 2.4.1–2.4.35. doi: 10.1002/0471250953.bi0204s00
Balaban, M., Joslin, S. N., and Hendrixson, D. R. (2009). FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni. J. Bacteriol. 191, 6602–6611. doi: 10.1128/JB.00884-889
Beeby, M. (2015). Motility in the epsilon-proteobacteria. Curr. Opin. Microbiol. 28, 115–121. doi: 10.1016/j.mib.2015.09.005
Bervoets, I., and Charlier, D. (2019). Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol. Rev. 43, 304–339. doi: 10.1093/femsre/fuz001
Blasco, B., Chen, J. M., Hartkoorn, R., Sala, C., Uplekar, S., Rougemont, J., et al. (2012). Virulence regulator EspR of mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 8:e1002621. doi: 10.1371/journal.ppat
Boll, J. M., and Hendrixson, D. R. (2013). A regulatory checkpoint during flagellar biogenesis in Campylobacter jejuni initiates signal transduction to activate transcription of flagellar genes. mBio. 4, e432–e413. doi: 10.1128/mBio.00432-413
Burnham, P. M., and Hendrixson, D. R. (2018). Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol. 16, 551–565. doi: 10.1038/s41579-018-0037-39
Cagliero, C., Maurel, M.-C., Cloeckaert, A., and Payot, S. (2007). Regulation of the expression of the CmeABC efflux pump in Campylobacter jejuni: identification of a point mutation abolishing the binding of the CmeR repressor in an in vitro-selected multidrug-resistant mutant. FEMS Microbiol. Lett. 267, 89–94. doi: 10.1111/j.1574-6968.2006.00558.x
Chilcott, G. S., and Hughes, K. T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708. doi: 10.1128/mmbr.64.4.694-708.2000
Correa, N. E., Peng, F., and Klose, K. E. (2005). Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol. 187, 6324–6332. doi: 10.1128/JB.187.18.6324
Dasgupta, N., Wolfgang, M. C., Goodman, A. L., Arora, S. K., Jyot, J., Lory, S., et al. (2003). A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50, 809–824. doi: 10.1046/j.1365-2958.2003.03740.x
Flint, A., Stintzi, A., and Saraiva, L. M. (2016). Oxidative and nitrosative stress defences of Helicobacter and Campylobacter species that counteract mammalian immunity. FEMS Microbiol. Rev. 40, 938–960. doi: 10.1093/femsre/fuw025
Galán-Vásquez, E., Sánchez-Osorio, I., and Martínez-Antonio, A. (2016). Transcription factors exhibit differential conservation in bacteria with reduced genomes. PLoS One 11:e0146901. doi: 10.1371/journal.pone.0146901
Gao, B., Lara-Tejero, M., Lefebre, M., Goodman, A. L., and Galán, J. E. (2014). Novel components of the flagellar system in epsilonproteobacteria. mBio 5, 1–13. doi: 10.1128/mBio.01349-1314
Garber, M., Grabherr, M. G., Guttman, M., and Trapnell, C. (2011). Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 8, 469–477. doi: 10.1038/nmeth.1613
Green, J. C. D., Kahramanoglou, C., Rahman, A., Pender, A. M. C., Charbonnel, N., and Fraser, G. M. (2009). Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J. Mol. Biol. 391, 679–690. doi: 10.1016/j.jmb.2009.05.075
Grinnage-Pulley, T., Mu, Y., Dai, L., and Zhang, Q. (2016). Dual repression of the multidrug efflux pump CmeABC by CosR and CmeR in Campylobacter jejuni. Front. Microbiol. 7:1097. doi: 10.3389/fmicb.2016.01097
Gulbronson, C. J., Ribardo, D. A., Balaban, M., Knauer, C., Bange, G., and Hendrixson, D. R. (2016). FlhG employs diverse intrinsic domains and influences FlhF GTPase activity to numerically regulate polar flagellar biogenesis in Campylobacter jejuni. Mol. Microbiol. 99, 291–306. doi: 10.1111/mmi.13231
Guttenplan, S. B., Shaw, S., and Kearns, D. B. (2013). The cell biology of peritrichous flagella in Bacillus subtilis. Mol. Microbiol. 87, 211–229. doi: 10.1111/mmi.12103
Hendrixson, D. R., and Dirita, V. J. (2003). Transcription of σ54 -dependent but not σ28 -dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50, 687–702. doi: 10.1046/j.1365-2958.2003.3731.x
Hizukuri, Y., Kojima, S., Yakushi, T., Kawagishi, I., and Homma, M. (2008). Systematic Cys mutagenesis of FlgI, the flagellar P-ring component of Escherichia coli. Microbiology 154, 810–817. doi: 10.1099/mic.0.2007/013854-13850
Hizukuri, Y., Yakushi, T., Kawagishi, I., and Homma, M. (2006). Role of the intramolecular disulfide bond in FlgI, the flagellar P-ring component of Escherichia coli. J. Bacteriol. 188, 4190–4197. doi: 10.1128/JB.01896-1895
Hofreuter, D., Tsai, J., Watson, R. O., Novik, V., Altman, B., Benitez, M., et al. (2006). Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74, 4694–4707. doi: 10.1128/IAI.00210-216
Huffman, J. L., and Brennan, R. G. (2002). Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 12, 98–106. doi: 10.1016/S0959-440X(02)00295-296
Hwang, S., Jeon, B., Yun, J., and Ryu, S. (2011). Roles of RpoN in the resistance of Campylobacter jejuni under various stress conditions. BMC Microbiol. 11:207. doi: 10.1186/1471-2180-11-207
Joslin, S. N., and Hendrixson, D. R. (2009). Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J. Bacteriol. 191, 2656–2667. doi: 10.1128/JB.01689-1688
Kazmierczak, B. I., and Hendrixson, D. R. (2013). Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol. Microbiol. 88, 655–663. doi: 10.1111/mmi.12221
Kim, S. M., Lee, D. H., and Choi, S. H. (2012). Evidence that the Vibrio vulnificus flagellar regulator FlhF is regulated by a quorum sensing master regulator SmcR. Microbiology 158, 2017–2025. doi: 10.1099/mic.0.059071-59070
Lertsethtakarn, P., Ottemann, K. M., and Hendrixson, D. R. (2011). Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65, 389–410. doi: 10.1146/annurev-micro-090110-102908
Liang, L., and Connerton, I. F. (2018). FlhF(T368A) modulates motility in the bacteriophage carrier state of Campylobacter jejuni. Mol. Microbiol. 110, 616–633. doi: 10.1111/mmi.14120
Liu, R., and Ochman, H. (2007). Stepwise formation of the bacterial flagellar system. Proc. Natl. Acad. Sci. U.S.A. 104, 7116–7121. doi: 10.1073/pnas.0700266104
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Matsunami, H., Barker, C. S., Yoon, Y. H., Wolf, M., and Samatey, F. A. (2016). Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat. Commun. 7, 1–10. doi: 10.1038/ncomms13425
Miller, W. G., Bates, A. H., Horn, S. T., Brandl, M. T., Wachtel, M. R., and Mandrell, R. E. (2000). Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66, 5426–5436. doi: 10.1128/aem.66.12.5426-5436.2000
Murray, T. S., and Kazmierczak, B. I. (2006). FlhF Is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188, 6995–7004. doi: 10.1128/JB.00790-796
Nachamkin, I., Szymanski, C. M., and Blaser, M. J. (eds) (2008). Campylobacter, 3rd Edn. Washington DC: ASM Press.
Niehus, E., Gressmann, H., Ye, F., Schlapbach, R., Dehio, M., Dehio, C., et al. (2004). Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52, 947–961. doi: 10.1111/j.1365-2958.2004.04006.x
O’Geen, H., Frietze, S., and Farnham, P. J. (2010). Using ChIP-seq technology to identify targets of zinc finger transcription factors. Methods Mol. Biol. 649, 437–455. doi: 10.1007/978-1-60761-753-2_27
Parker, C. T., Quinones, B., Miller, W. G., Horn, S. T., and Mandrell, R. E. (2006). Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J. Clin. Microbiol. 44, 4125–4135. doi: 10.1128/JCM.01231-1236
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Petersen, L., Larsen, T. S., Ussery, D. W., On, S. L. W., and Krogh, A. (2003). Rpo D promoters in Campylobacter jejuni exhibit a strong periodic signal instead of a - 35 box. J. Mol. Biol. 326, 1361–1372. doi: 10.1016/S0022-2836(03)00034-32
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Prouty, M. G., Correa, N. E., and Klose, K. E. (2001). The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39, 1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x
Ren, F., Lei, T., Song, Z., Yu, T., Li, Q., Huang, J., et al. (2018). Could FlhF be a key element that controls Campylobacter jejuni flagella biosynthesis in the initial assembly stage? Microbiol. Res. 207, 240–248. doi: 10.1016/j.micres.2017.12.006
Schniederberend, M., Abdurachim, K., Murray, T. S., and Kazmierczaka, B. I. (2013). The GTPase activity of FlhF is dispensable for flagellar localization, but not motility, in Pseudomonas aeruginosa. J. Bacteriol. 195, 1051–1060. doi: 10.1128/JB.02013-2012
Schuhmacher, J. S., Thormann, K. M., and Bange, G. (2015). How bacteria maintain location and number of flagella? FEMS Microbiol. Rev. 39, 812–822. doi: 10.1093/femsre/fuv034
Subramanian, S., and Kearns, D. B. (2019). Functional regulators of Bacterial Flagella. Annu. Rev. Microbiol. 73, 1–22. doi: 10.1146/annurev-micro-020518-115725
Wosten, M. M., Boeve, M., Koot, M. G., van Nuenen, A. C., and van der Zeijst, B. A. (1998). Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180, 594–599.
Wösten, M. M. S. M., Wagenaar, J. A., and Van Putten, J. P. M. (2004). The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279, 16214–16222. doi: 10.1074/jbc.M400357200
Keywords: Campylobacter jejuni, FlhF, transcriptional regulator, flagellar biosynthesis, pathogenesis
Citation: Li X, Ren F, Cai G, Huang P, Chai Q, Gundogdu O, Jiao X and Huang J (2020) Investigating the Role of FlhF Identifies Novel Interactions With Genes Involved in Flagellar Synthesis in Campylobacter jejuni. Front. Microbiol. 11:460. doi: 10.3389/fmicb.2020.00460
Received: 14 September 2019; Accepted: 04 March 2020;
Published: 24 March 2020.
Edited by:
Paloma López, Center for Biological Research (CSIS), SpainReviewed by:
Ian F. Connerton, University of Nottingham, United KingdomManuel Zúñiga Cabrera, Institute of Agrochemistry and Food Technology (IATA), Spain
Copyright © 2020 Li, Ren, Cai, Huang, Chai, Gundogdu, Jiao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinlin Huang, amlubGluQHl6dS5lZHUuY24=
 Xiaofei Li
Xiaofei Li Fangzhe Ren
Fangzhe Ren Guoqiang Cai2
Guoqiang Cai2 Ozan Gundogdu
Ozan Gundogdu Xinan Jiao
Xinan Jiao