- 1Faculty of Agriculture and Food, Kunming University of Science and Technology, Kunming, China
- 2Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, Chiang Mai, Thailand
- 3Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 4Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 5World Agroforestry Centre, East and Central Asia, Kunming, China
- 6Retired, Curca, India
- 7Department of Health Sciences, Faculty of Science, University of Mauritius, Reduit, Mauritius
- 8Faculty of Environmental Science and Engineering, Kunming University of Science and Technology, Kunming, China
- 9Department of Botany, University of British Columbia, Vancouver, BC, Canada
- 10Yunnan Key Lab of Soil Carbon Sequestration and Pollution Control, Kunming University of Science and Technology, Kunming, China
During our ongoing surveys of fungi on submerged wood in the Greater Mekong Subregion, we collected two new species similar to Bactrodesmium longisporum. Pseudobactrodesmium gen. nov. is introduced to accommodate the new species, P. aquaticum, P. chiangmaiensis and B. longisporum is transferred to this genus. Fasciculate conidiophores, enteroblastic conidiogenous cells and subulate to fusiform, phragmoseptate conidia with a tapering apical cell and sheath characterize the genus. Pseudobactrodesmium aquaticum has longer conidia than P. chiangmaiensis. The placement of Pseudobactrodesmium in Dactylosporaceae (Eurotiomycetes) is a novel finding based on analyses of combined LSU, SSU, ITS and RPB2 sequence data. Our study reveals that Pseudobactrodesmium is likely to be a speciose genus with different species in streams around the world.
Introduction
Dactylosporaceae accommodates ecologically and morphologically diverse genera, and was reinstated by Diederich et al. (2018) to replace Sclerococcaceae (Réblová et al., 2016). For example, the freshwater genus Cylindroconidiis has holoblastic conidiogenous cells (Yu et al., 2018), while the terrestrial genera Pseudosclerococcum and Rhopalophora are apothecial ascomycetes and dematiaceous phialidic hyphomycetes, respectively (Réblová et al., 2016; Olariaga et al., 2019). The terrestrial and marine genus Sclerococcum (= Dactylospora) has loose sporodochia with catenate conidia or apothecia-like ascomata often growing on lichens or decaying wood (Hawksworth, 1975; Jones et al., 1999; Pang et al., 2014; Pino-Bodas et al., 2017). Additionally, Fusichalara minuta, which is a dematiaceous phialidic hyphomycete, and some beetle-associated strains also cluster in this family (Vargasasensio et al., 2014; Tedersoo et al., 2017).
Aquatic hyphomycetes are a morphologically diverse and polyphyletic group (Shenoy et al., 2006; Baschien et al., 2013; Su et al., 2016). Species with similar morphological characters are difficult to identify without molecular data. Previously, identification was mostly carried out based on morphology and only a few asexual taxa have been subjected to phylogenetic studies (Goh and Hyde, 1996; Cai et al., 2002; Cai and Hyde, 2007). With more molecular data becoming available for phylogenetic analyses, numerous new combinations have been proposed to accommodate poorly documented hyphomycetous species (Lu et al., 2018; Yang et al., 2018a, b). Molecular data also demonstrated that some previously known congeneric species are now distributed in different families, e.g., Monodictys arctica in Leptosphaeriaceae (Day et al., 2006), M. capensis in Pleomonodictydaceae (Hernández-Restrepo et al., 2017), and some other Monodictys species in Parabambusicolaceae (Tanaka et al., 2015). Although the polyphyletic nature of some hyphomycetous genera were partially resolved, e.g., Dendryphion, Sporidesmium and torula-like species (Su et al., 2016), fresh collections with molecular data are still needed to obtain a natural classification of hyphomycetes.
Invalidly established by Berkeley and Broome (1865) with Sporidesmium abruptum as the type, the hyphomycetous genus Bactrodesmium was segregated from Sporidesmium, with B. abruptum as the lectotype (Hughes, 1958). Bactrodesmium is distributed worldwide with more than 48 species (Wijayawardene et al., 2017a; Index Fungorum database1). It was regarded as a member of Dothideomycetes based on the sexual-asexual morph connection between Bactrodesmium obliquum and Stuartella suttonii (Funk and Shoemaker, 1983; Wijayawardene et al., 2017b). However, with molecular evidence, Bactrodesmium was shown to be polyphyletic, as B. gabretae clustered within Helotiales, Leotiomycetes (Koukol and Kolárová, 2010), B. cubense had affinities to Morosphaeriaceae, Dothideomycetes (Hernández-Restrepo et al., 2017) and B. pallidum clustered in Savoryellaceae, Sordariomycetes (Hernández-Restrepo et al., 2017). Recently, Bactrodesmium fasciculare was transferred to a newly established genus Pleotrichocladium in Melanommataceae (Dothideomycetes) based on molecular data and morphology (Hernández-Restrepo et al., 2017). Moreover, the generic type B. abruptum was tentatively placed in Dothideomycetes based on morphological evidence but molecular data is still lacking (Pem et al., 2019). The phylogenetic position of other species still needs to be investigated.
We are studying the freshwater fungi on submerged wood along a north–south latitudinal gradient in the Asian/Australian region (Hyde et al., 2016) and have published several papers on the Greater Mekong Subregion ([citeskum]BR90,BR88,BR87,BR89,BR85,BR86[citeekum]Zhang et al., 2011, 2012, 2013, 2014, 2016, 2017; Dong et al., 2018; Wei et al., 2018; Yu et al., 2018; Wang et al., 2019). In this study, two taxa morphologically similar to Bactrodesmium longisporum were collected from submerged wood. To clarify the classification of the two new collections, we analyzed a combined LSU, SSU, ITS and RPB2 sequence dataset and compared their morphological characters. Pseudobactrodesmium, a new genus with two new species, and one new combination are introduced. Morphologically similar genera are compared with Pseudobactrodesmium and the taxonomic placements of Bactrodesmium species are discussed.
Materials and Methods
Isolation and Morphology
The decaying wood samples were collected from freshwater streams in Chiang Mai Province, Thailand and Yunnan Province, China. Specimens were placed in zip-lock plastic bags with moist cotton or tissue paper and taken to the laboratory. Morphological observations were carried out after incubation at room temperature for 1–2 weeks. Colonies were examined using a Nikon SMZ-171 dissecting microscope. Photomicrographs were made with a Nikon ECLIPSE Ni compound microscope fitted with a Canon EOS 600D digital camera. Measurements were made with the Tarosoft (R) Image Frame Work program. Images used for figures were processed with Adobe Photoshop CS5 software (Adobe Systems, United States). Single spore isolations were made from conidia onto potato dextrose agar (PDA) at room temperature, as detailed in Chomnunti et al. (2014) and cultured as outlined by Vijaykrishna et al. (2004) and Liu et al. (2010). Herbarium specimens (dry wood with fungal material) were deposited in the herbarium of Mae Fah Luang University (MFLU), Chiang Rai, Thailand and herbarium of Cryptogams, Kunming Institute of Botany Academia Sinica (HKAS), Kunming, China. Living cultures were deposited in Mae Fah Luang University Culture Collection (MFLUCC) and Kunming Institute of Botany Culture Collection (KUMCC). Facesoffungi and Index Fungorum numbers were registered as in Jayasiri et al. (2015) and Index Fungorum (2020), respectively.
DNA Extraction, PCR Amplification and Sequencing
Fungi were grown on PDA for 20–30 days at 25°C. A Biospin Fungus Genomic DNA Extraction Kit (Bioer Technology Co., Hangzhou, China) was used to extract total genomic DNA from fresh mycelia according to the manufacturer’s instructions. DNA amplification was performed by polymerase chain reaction (PCR). LSU, SSU, ITS and RPB2 gene regions were amplified using the primer pairs LR0R/LR5, NS1/NS4, ITS5/ITS4 and RPB2-5F/RPB2-7cR, respectively (Vilgalys and Hester, 1990; White et al., 1990; Rehner and Samuels, 1994; Liu et al., 1999). The amplifications were carried out in a 25 μL reaction volume containing 9.5 μL ddH2O, 12.5 μL 2 × PCR Master Mix, 1 μL DNA template, 1 μL each primer (10 μM). The PCR thermal cycles for the amplification of the gene regions followed the methods in Jeewon et al. (2004); Réblová et al. (2011), and Su et al. (2015). PCR products were checked on 1% agarose electrophoresis gels stained with Gel Red. The sequencing reactions were carried out by Shanghai Sangon Biological Engineering Technology and Services Co., Shanghai, China.
Phylogenetic Analyses
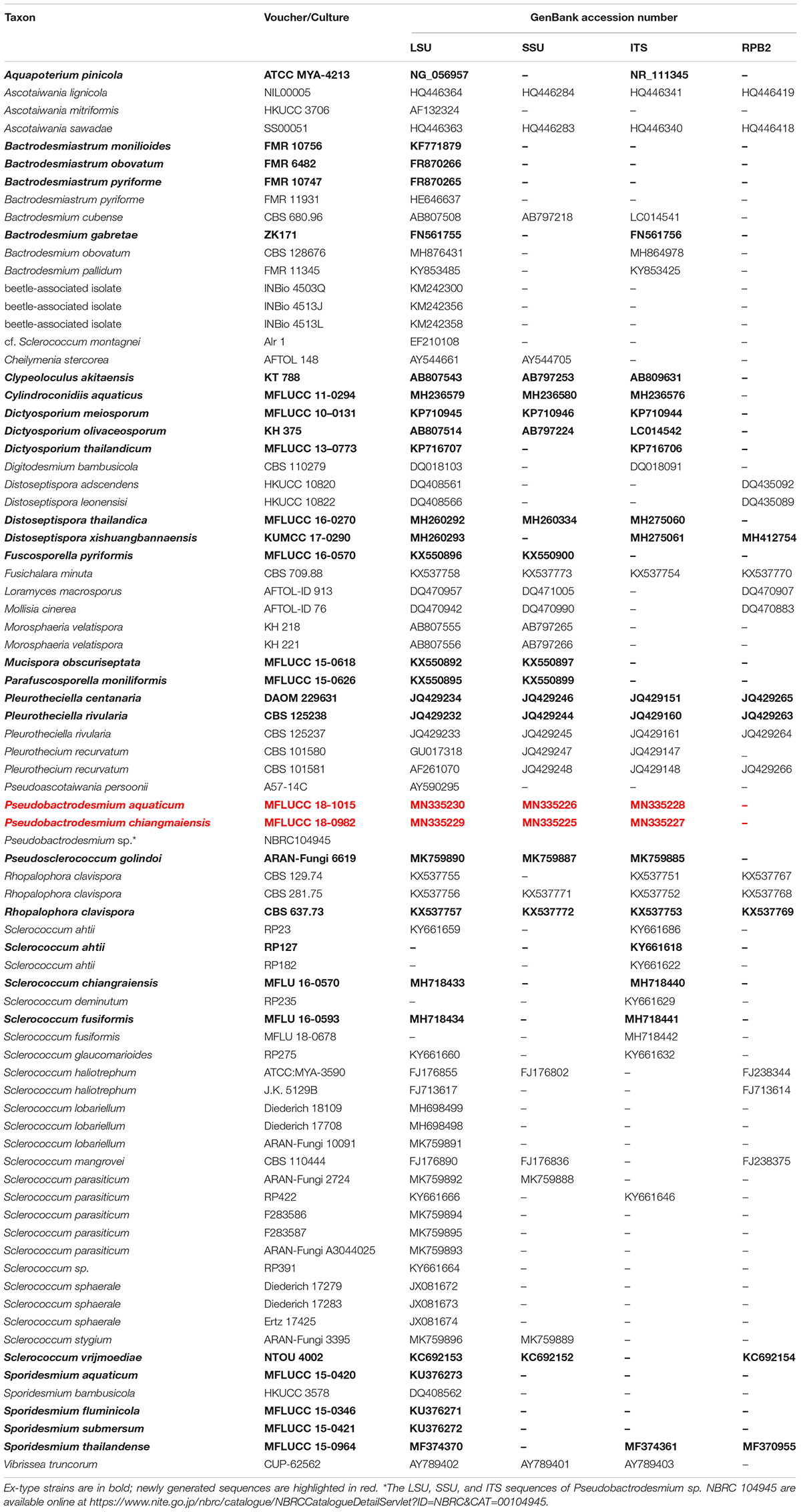
The qualities of raw sequences generated in this study were checked with Finch TV version 1.4.0. Based on nucleotide BLAST2 and previous publications (Raja et al., 2008; Koukol and Kolárová, 2010; Réblová et al., 2012, 2016; Diederich et al., 2013; Pang et al., 2014; Boonmee et al., 2016; Su et al., 2016; Yang et al., 2016; Hernández-Restrepo et al., 2017; Pino-Bodas et al., 2017; Yu et al., 2018; Dayarathne et al., 2019; Ekanayaka et al., 2019; Olariaga et al., 2019), related sequences together with newly generated ones were selected for constructing a phylogenetic tree. All sequences used in this study are listed in Table 1. The individual datasets of LSU, SSU, ITS and RPB2 were aligned using MAFFT v. 7.409 online version (Kazutaka and Standley, 2016) and manually verified with BioEdit v.7.2.5 Biological Sequence Alignment Editor (Ibis BioSciences, CA). Phylogenetic analyses of the combined dataset (LSU, SSU, ITS and RPB2) were inferred with maximum likelihood (ML) and Bayesian inference (BI) analyses.
A ML analysis was performed with RAxML-HPC v.8 on XSEDE in CIPRES Science Gateway (Miller et al., 2010, 2015) with 1000 rapid bootstrap replicates. The model selected for ML was GTRGAMMA. Maximum likelihood bootstrap values equal to or greater than 60% are given above or below the nodes (first value, Figure 1). Bayesian inference was conducted with MrBayes v. 3.1.2 (Huelsenbeck and Ronquist, 2001) to evaluate posterior probabilities (BPP) (Rannala and Yang, 1996) by Markov chain Monte Carlo (MCMC) sampling. The best-fit model was GTR + I + G for LSU, SSU and RPB2, and SYM + I + G for ITS. Six simultaneous Markov chains were run for one million generations and trees were sampled every 100 generation (resulting in 10000 trees). The first 2500 trees, representing the burn-in phase of the analyses, were discarded and the remaining 7500 trees were used for calculating posterior probabilities (PP) in the majority rule consensus tree (Larget and Simon, 1999). Bayesian posterior probabilities (BPP) equal to or greater than 0.95 are given above or below the nodes (second value, Figure 1).
Figure 1. RAxML tree generated from combined LSU, SSU, ITS, and RPB2 sequence data. Bootstrap support values for maximum likelihood (the first value) equal to or greater than 60% and Bayesian posterior probabilities (the second value) equal to or greater than 0.95 are given above or below the nodes. The tree is rooted to Cheilymenia stercorea (AFTOL 148) (Pezizomycetes). The ex-type strains are indicated in bold and newly generated sequences are indicated in blue. Four bactrodesmium-like species are highlighted in gray background. Symbols after generic names in Eurotiomycetes indicate the habitats of taxa as explained in the phylogram.
Phylogenetic trees were viewed with FigTree v1.4.03,4 and edited using Microsoft Office PowerPoint 2007 (Microsoft Corporation, WA, United States). The new sequences were deposited in GenBank (Table 1).
Results
Phylogenetic Analyses
Combined LSU, SSU, ITS, and RPB2 gene regions were employed to explore the taxonomy of new collections. The alignment comprised 79 strains (including two new strains) with an alignment length of 4381 total characters. The RAxML analysis resulted in a best scoring likelihood tree selected with a final value for the combined dataset ln L = −39819.188166. The matrix has 2564 distinct alignment patterns, with 62% of undetermined characters or gaps. Estimated base frequencies are as follows: A = 0.256432, C = 0.229269, G = 0.279558, T = 0.234740; substitution rates AC = 1.300864, AG = 2.616909, AT = 1.328213, CG = 1.028043, CT = 6.168383, GT = 1.000000; gamma distribution shape parameter α = 0.360140.
In the phylogenetic tree (Figure 1), the two new isolates are shown in Eurotiomycetes and distantly related to Bactrodesmium cubense (Dothideomycetes), B. gabretae (Leotiomycetes), and B. obovatum and B. pallidum (Sordariomycetes). The morphologically similar genera, e.g., Bactrodesmiastrum, Dictyosporium, Digitodesmium, Distoseptispora, and Sporidesmium, have phylogenetically unrelated relationships with our new strains (Figure 1). Pseudobactrodesmium aquaticum and P. chiangmaiensis constitute a distinct clade in the family Dactylosporaceae (Figure 1).
Taxonomy
Pseudobactrodesmium H. Zhang, W. Dong & K. D. Hyde, gen. nov.
Index Fungorum number: IF557247; Facesoffungi number: FoF07525
Etymology: in reference to bactrodesmium-like morphology
Saprobic on submerged wood in freshwater or decaying wood in terrestrial habitats. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies sporodochial, superficial, effuse, gregarious or scattered, brown, punctiform. Mycelium mostly immersed, composed of septate, branched, hyaline hyphae. Conidiophores macronematous, mononematous, fasciculate, compact, erect, subcylindrical, septate, usually unbranched, brown, smooth. Conidiogenous cells enteroblastic, with inconspicuous proliferations, integrated, terminal, subcylindrical, pale brown. Conidia acrogenous, solitary, dry, thin-walled, smooth-walled, clavate, subcylindrical, narrowly fusiform or subulate, euseptate, phragmoseptate, brown, often enveloped by a hyaline, spherical sheath at the apex. Apical cells elongated, tapering gradually toward the apex, with globose tuberculate apex.
Type species: Pseudobactrodesmium aquaticum W. Dong, H. Zhang & K.D. Hyde
Notes: Pseudobactrodesmium is characterized by enteroblastic conidiogenous cells and subulate or fusiform, evenly pigmented conidia with a tapering apical cell. In contrast, Bactrodesmium as typified by B. abruptum is quite distinct in producing holoblastic conidiogenous cells and clavate to fusiform, unevenly colored conidia which are mid or dark brown at the upper part and becoming paler toward the basal cell, 3- to multi-transversely septate, with very dark bands at the septa, the upper one thick and black, unequal cells, the penultimate cell much longer than any of the others (Ellis, 1971). Pseudobactrodesmium nests well within the family Dactylosporaceae in our phylogenetic tree of the combined sequence dataset (Figure 1). The unique combination of morphological characters of Pseudobactrodesmium stands apart from other existing genera in this family (Hawksworth, 1975; Ellis, 1976; Jones et al., 1999; Réblová et al., 2016; Yu et al., 2018; Olariaga et al., 2019). Pseudobactrodesmium is morphologically similar to a few dematiaceous hyphomycetous genera with long or short, septate conidia, e.g., Bactrodesmiastrum (Holubová-Jechová, 1984), Bactrodesmium (Ellis, 1971, 1976), Digitodesmium (Boonmee et al., 2016), Distoseptispora (Su et al., 2016), Scolecostigmina (Braun et al., 1999), and Sporidesmium (Su et al., 2016), but they are separated by molecular evidence (Figure 1).
Pseudobactrodesmium aquaticum W. Dong, H. Zhang & K.D. Hyde, sp. nov., Figure 2

Figure 2. Pseudobactrodesmium aquaticum (MFLU 18-1171, holotype). (a,b) Colonies on submerged wood. (c,d) Conidiophores bearing conidia. (e) Enteroblastic conidiogenous cells (arrow). (f) Conidiophore. (g–i) Conidia. (j) Conidial tips with sheaths. (k) Germinated conidium. (l) Colony on PDA (front view). (m) Colony on PDA (bottom view). Scale bars: (a) 200 μm, (b) 100 μm, (c,d,h,k) 20 μm, (e,g,i,j) 10 μm, (f) 5 μm.
Index Fungorum number: IF557248; Facesoffungi number: FoF07526
Etymology: aquaticum in reference to the aquatic habitat
Holotype: MFLU 18-1171
Saprobic on submerged wood in freshwater. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies sporodochial, superficial, effuse, gregarious or scattered, brown, punctiform. Mycelium mostly immersed, composed of septate, branched, hyaline hyphae. Conidiophores 26–38 × 3–4.5 μm ( = 34 × 3.8 μm, n = 10), macronematous, mononematous, fasciculate, compact, erect, subcylindrical, the apex slightly wider than the base, septate, slightly constricted at septa, usually unbranched, brown, smooth. Conidiogenous cells enteroblastic, with inconspicuous proliferations, integrated, terminal, subcylindrical, pale brown. Conidia (80–)90–105 × 6–8.5 μm ( = 95 × 7.5 μm, n = 20), acrogenous, solitary, dry, thin-walled, smooth-walled, clavate when young, subcylindrical to narrowly fusiform, or subulate when mature, straight or slightly curved, euseptate, (15–)16–19-phragmoseptate, slightly constricted and darker at septa, pale brown to brown, obscurely guttulate, wedge-shaped at basal cell, with tapering apical cells, often enveloped by a hyaline, spherical, thin, gelatinous sheath at the apex, 13–20 μm diam. Apical cells elongated, up to 6 μm long, tapering gradually toward the apex, easily becoming senescent, subhyaline, with obscured, subglobose tuberculate ends.
Culture characteristics: On PDA, colony circular, slow growing, reaching 10 mm in 50 days at 25°C, gray to brown from above, dark gray from below, surface rough, dry, raised, entire at edge.
Material examined: CHINA, Yunnan Province, Pingbian City, on submerged wood in a stream, 20 September 2017, W. Dong, WF-24A-1 (MFLU 18-1171, holotype), ex-type living culture MFLUCC 18-1015; ibid. WF-24A-2 (HKAS 101707, isotype), ex-isotype living culture KUMCC 18-0056.
Notes: Pseudobactrodesmium aquaticum is introduced as the type species of Pseudobactrodesmium having the typical euseptate, phragmoseptate conidia with apical sheath. The enteroblastic conidiogenous cells are particularly obvious in Figure 2e.
Pseudobactrodesmium chiangmaiensis X. D. Yu, W. Dong & K. D. Hyde, sp. nov., Figure 3

Figure 3. Pseudobactrodesmium chiangmaiensis (MFLU 18-0994, holotype). (a) Colonies on submerged wood. (b,c) Conidiophores bearing conidia. (d) Conidial tips with sheaths (arrow). (e) Apex of conidiophores. (f–h) Conidia. (i) Colony on PDA (front view). (j) Colony on PDA (bottom view). Scale bars: (a) 500 μm, (b,g,h) 20 μm, (c,d) 30 μm, (e) 10 μm, (f) 50 μm.
Index Fungorum number: IF557249; Facesoffungi number: FoF07527
Etymology: name reflects Chiang Mai, from where the species was collected
Holotype: MFLU 18-0994
Saprobic on submerged wood in freshwater. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies sporodochial, superficial, effuse, gregarious or scattered, dark brown to black, punctiform. Mycelium mostly immersed, composed of septate, branched, hyaline hyphae. Conidiophores 15–23 × 2.5–4 μm ( = 21.5 × 3.5 μm, n = 10), macronematous, mononematous, fasciculate, compact, erect, subcylindrical, septate, slightly constricted at the septa, usually unbranched, brown, smooth. Conidiogenous cells enteroblastic, with inconspicuous proliferations, integrated, terminal, subcylindrical, pale brown. Conidia 40–90 × 5.5–8.5 μm ( = 70 × 7 μm, n = 50), acrogenous, solitary, dry, thin-walled, smooth-walled, clavate when young, subcylindrical to narrowly fusiform, or subulate when mature, straight or slightly curved, euseptate, 6–19-phragmoseptate, slightly constricted and darker at septa, pale brown to brown, obscurely guttulate, wedge-shaped at basal cell, with tapering apical cells, often enveloped by a hyaline, spherical, thin, gelatinous sheath at the apex, 17–21 μm diam. Apical cells elongated, up to but rarely 16 μm long, tapering gradually toward apex, subhyaline, with subglobose tuberculate ends.
Culture characteristics: On PDA, colony circular, reaching 15 mm in 20 days at 25°C, dark gray to dark brown from above, dark gray to black from below, surface rough, dry, raised, margin entire.
Material examined: Thailand, Chiang Mai Province, on submerged wood in a stream, 9 February 2018, X.D. Yu, Y11 (MFLU 18-0994, holotype), ex-type living culture MFLUCC 18-0982.
Notes: Pseudobactrodesmium chiangmaiensis differs from P. aquaticum in having shorter conidia (40–90 × 5.5–8.5 μm vs. (80–)90–105 × 6–8.5 μm), longer apical cells (up to 16 μm vs. up to 6 μm), as well as darker colonies on the host (dark brown to black vs. brown). The conidial sheaths are obscure in P. chiangmaiensis when mounted in water, while they are easily observed in P. aquaticum. This is probably because the specimens were senescent which led the sheaths to deliquesce. In our phylogenetic tree, P. chiangmaiensis groups with P. aquaticum with strong bootstrap support (100% MLBS, 1.00 PP, Figure 1). However, a comparison of sequence data between P. chiangmaiensis and P. aquaticum shows a difference of 6, 7, 20, and 32 nucleotides in LSU, SSU, ITS, and TEF gene regions, respectively. This indicates that they are distinct species according to guidelines of Jeewon and Hyde (2016).
Pseudobactrodesmium longisporum (M.B. Ellis) W. Dong & K.D. Hyde, comb. nov.
Index Fungorum number: IF557250; Facesoffungi number: FoF07466
≡ Bactrodesmium longisporum M.B. Ellis, More Dematiaceous Hyphomycetes (Kew): 68 (1976)
≡ Stigmina longispora (M.B. Ellis) S. Hughes, N. Z. Jl Bot. 16(3): 353 (1978)
= Bactrodesmium stilboideum R. F. Castañeda & G. R. W. Arnold, Revta Jardín bot. Nac., Univ. Habana 6(1): 48 (1985)
≡ Stigmina longispora var. stilboidea (R. F. Castañeda & G. R. W. Arnold) J. Mena & Mercado, Reporte de Investigacion del Instituto de Ecología y Sistemática, Academia de Ciencias de Cuba, Ser. Bot. 17: 10 (1987)
Holotype: On dead wood of Alnus sp. in Great Britain (IMI 63746 B)
Known distribution: New Zealand (Hughes, 1978), Australia (Vijaykrishna and Hyde, 2006), Brazil (Barbosa and Gusmão, 2011; Barbosa et al., 2013; Santa Izabel and Gusmão, 2016, 2018), Cuba (Castañeda Ruiz and Arnold, 1985), Great Britain (Ellis, 1976), Hong Kong, China (Wong and Hyde, 2001), India (Prabhugaonkar, 2011), Venezuela (Castañeda Ruiz et al., 2009), México (Heredia et al., 2018), Peru (Shearer et al., 2015), Philippines (Cai et al., 2003), South Africa (Hyde et al., 1998), Thailand (Hu et al., 2010; this study), United States (Raja et al., 2007).
Notes: Bactrodesmium longisporum was described by Ellis (1976) with a line-drawing. It was subsequently synonymized with Stigmina longispora by Hughes (1978) who observed percurrently proliferating conidiophores in old specimens from New Zealand. Bactrodesmium stilboideum is another synonym listed in Index Fungorum database. However, they can be distinguished by the aggregation of conidiophores (synnematous in B. stilboideum vs. mononematous, fasciculate conidiophores in B. longisporum) (Ellis, 1976; Castañeda Ruiz and Arnold, 1985).
A Thai strain of B. longisporum (NBRC 104945) clustered with Pseudobactrodesmium chiangmaiensis (MFLUCC 18-0982) in our phylogenetic tree (Figure 1). A comparison of sequence data between NBRC 104945 and MFLUCC 18-0982 shows a difference of 2, 281, 5 nucleotides in LSU, SSU and ITS gene regions, respectively (NBRC 104945 has 3 major insertions spanning over 281 nucleotides in SSU gene). In this study, we name NBRC 104945 as Pseudobactrodesmium sp. until its morphological characters are established to formally name this isolate. Five additional strains with only ITS2 sequence data are named as Bactrodesmium longisporum in GenBank. However, their status should be treated with caution as they represent OTUS from a metagenomic study of a heap leaching system (Hu et al., 2015) and further evidence of conspecificity is needed.
Unfortunately, the holotype specimen of B. longisporum (IMI 63746 B), does not exist in herbarium IMI.5 According to protologue description of the holotype (Ellis, 1976), B. longisporum (IMI 63746 B) has similar conidial size to P. chiangmaiensis (MFLU 18-0994) (50–80 × 7–8 μm in former vs. 40–90 × 5.5–8.5 μm in latter). However, P. chiangmaiensis has elongated apical cells (up to 16 μm long) with subglobose tuberculate ends, which were not described and drawn in protologue of B. longisporum (Ellis, 1976). The conidiophores of B. longisporum are up to 50 μm long, but only 15–23 μm long in P. chiangmaiensis. The size of apical sheath of B. longisporum is also unclear. Thus, we treat them as different species and synonymize B. longisporum under Pseudobactrodesmium as the third species in the genus. Epitypification of Pseudobactrodesmium longisporum is needed using a collection from its type locality.
Discussion
Bactrodesmium longisporum has been recorded as having a worldwide distribution, however these records have not been verified with molecular data. The type of B. longisporum also appears to be lost and therefore its identity cannot be verified. We therefore designate our new species of Pseudobactrodesmium from China as the generic type, describe a second species from Thailand and transfer Bactrodesmium longisporum to the new genus. However, it is likely that many collections of this species have been misidentified and as more collections are made from different countries, we would expect Pseudobactrodesmium to become speciose.
Bactrodesmium is a complex genus in need of extensive taxonomic reassessment. Pem et al. (2019) reviewed the holotype material of Bactrodesmium abruptum (≡ Sporidesmium abruptum) and tentatively placed the generic type in Dothideomycetes incertae sedis based on morphology. Both B. cubense and B. obovatum produce clavate or obovate conidia with darker septa and unequal cells, similar to the type species B. abruptum (Ellis, 1971; Zucconi and Lunghini, 1997). However, our phylogenetic study shows that they belong to different classes, Dothideomycetes and Sordariomycetes, respectively (Figure 1). Bactrodesmium gabretae differs from B. abruptum by its transversely or occasionally oblique, distoseptate conidia, and phylogenetically clustered in Leotiomycetes (Figure 1). Bactrodesmium pallidum is different from B. abruptum but similar to our new genus Pseudobactrodesmium in conidial shape (Ellis, 1959), and phylogeny places this species in Sordariomycetes (Figure 1). Our phylogenetic study is in agreement with the studies of Koukol and Kolárová (2010) and Hernández-Restrepo et al. (2017).
Although molecular data of B. abruptum is still missing, the working hypothesis of Bactrodesmium sensu stricto in Dothideomycetes provides further evidence for the introduction of Pseudobactrodesmium. Pseudobactrodesmium shares some morphological characters with Digitodesmium in having acrogenous, long, transversely septate conidia with a hyaline sheath at the apex (Kirk, 1981; Boonmee et al., 2016). However, the semi-macronematous, moniliform conidiophores and digitate conidia of Digitodesmium are clearly distinguishable from the macronematous, subcylindrical conidiophores and subcylindrical to narrowly fusiform, or subulate conidia of Pseudobactrodesmium. Phylogeny also segregates them into different classes, viz. Pseudobactrodesmium in Eurotiomycetes, and Digitodesmium in Dothideomycetes (Tsui et al., 2006; Boonmee et al., 2016; this study). The conidia of Scolecostigmina are superficially similar to those of Pseudobactrodesmium, but the former is characterized by conspicuously annellate conidiogenous cells, thick-walled, smooth to verrucose conidia occasionally with a few longitudinal or oblique septa or a few intermixed distosepta, contrasting with inconspicuously proliferating conidiogenous cells and thin-walled, smooth, transversely phragmoseptate conidia with a hyaline, spherical sheath at the apex in Pseudobactrodesmium (Braun et al., 1999; Crous et al., 2013). Scolecostigmina, typified by S. mangiferae, clustered in Capnodiales (Dothideomycetes) (Crous et al., 2013), while Pseudobactrodesmium clustered in Dactylosporaceae (Eurotiomycetes). Pseudobactrodesmium longisporum is superficially similar to Gangliostilbe malabarica in the conidial shape and apical sheath, but the synnemata and apically rounded conidia of the latter can easily be separated from the former (Xia et al., 2015). These characters of G. malabarica are also distinguished from those in the collection of Castañeda Ruiz and Arnold (1985) bearing the name Bactrodesmium stilboideum.
It is challenging to reconstruct the phylogeny of Bactrodesmium considering lack of living cultures of B. abruptum. The species having clavate or obovate, long or short, transversely septate conidia with or without apical sheath are common and scattered in different groups (Ellis, 1976; Hughes, 1978; Holubová-Jechová, 1984; Braun et al., 1999; Koukol and Kolárová, 2010; Crous et al., 2013; Xia et al., 2015; Boonmee et al., 2016; Su et al., 2016; Hernández-Restrepo et al., 2017; Videira et al., 2017). These groups of fungi are morphologically similar and therefore molecular characters are of crucial importance to clarify their taxonomy. The sequence data of B. abruptum is needed in the future to clarify the natural classification of Bactrodesmium.
Data Availability Statement
The datasets generated for this study can be found in the NCBI GenBank: MN335230, MN335226, MN335228, MN335229, MN335225, and MN335227.
Author Contributions
WD conducted the experiments, analyzed the data, and wrote the manuscript. KH planned the experiments. MD analyzed the data. X-DY conducted the experiments. DB and RJ revised the manuscript. SB funded the experiments. G-NW conducted the experiments. SN planned the experiments. HZ planned the experiments, analyzed the data, and wrote the manuscript. All authors revised the manuscript.
Funding
This work was mainly supported by National Natural Science Foundation of China (Project ID: NSF 31500017 to HZ), Yunnan young and middle aged academic and technical leaders reserve talents (Project ID: 2018HB008). KH thanks the Foreign Experts Bureau of Yunnan Province, Foreign Talents Program (2018; grant no. YNZ2018002), Thailand Research grants entitled Biodiversity, phylogeny and role of fungal endophytes on above parts of Rhizophora apiculata and Nypa fruticans (grant no: RSA5980068), the future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species (grant no: DBG6080013), Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (grant no: RDG6130001). KH also thanks Chiang Mai University for the award of visiting Professor. MD would like to thank the 5th batch of Postdoctoral Orientation Training Personnel in Yunnan Province (grant no: Y934283261) and the 64th batch of China Postdoctoral Science Foundation (grant no: Y913082271). SB would like to thank the National Research Council of Thailand (No. 61215320023).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
WD would like to thank Prof. Eric H. C. McKenzie for correcting English on manuscript. RJ thanks Mae Fah Luang University and University of Mauritius for support.
Footnotes
- ^ http://www.indexfungorum.org/names/Names.asp
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://tree.bio.ed.ac.uk/
- ^ http://jebl.sourceforge.net/
- ^ http://www.herbimi.info/herbimi/results.htm?name=Bactrodesmium%20longisporum
References
Barbosa, F., and Gusmão, L. (2011). Conidial fungi from semi-arid Caatinga biome of Brazil. Rare freshwater hyphomycetes and other new records. Mycosphere 2, 475–485.
Barbosa, F. R., Raja, H. A., Shearer, C. A., and Gusmão, L. F. P. (2013). Some freshwater fungi from the Brazilian semi-arid region, including two new species of hyphomycetes. Cryptogam. Mycol. 34, 243–258. doi: 10.7872/crym.v34.iss2.2013.243
Baschien, C., Tsui, C. K. M., Gulis, V., Szewzyk, U., and Marvanová, L. (2013). The molecular phylogeny of aquatic hyphomycetes with affinity to the leotiomycetes. Fungal Biol. 117, 660–672. doi: 10.1016/j.funbio.2013.07.004
Berkeley, M. J., and Broome, C. E. (1865). Notices of British fungi (1038–1062). Ann. Mag. Nat. Hist. 15, 400–404.
Boonmee, S., D’souza, M. J., Luo, Z. L., Pinruan, U., Tanaka, K., Su, H. Y., et al. (2016). Dictyosporiaceae fam. nov. Fungal Divers. 80, 457–482. doi: 10.1007/s13225-016-0363-z
Braun, U., Mouchacca, J., and McKenzie, E. (1999). Cercosporoid hyphomycetes from New Caledonia and some other South Pacific islands. N.Z. J. Bot. 37, 297–327. doi: 10.1080/0028825X.1999.9512636
Cai, L., and Hyde, K. D. (2007). Anamorphic fungi from freshwater habitats in China: Dictyosporium tetrasporum and Exserticlava yunnanensis spp. nov., and two new records for Pseudofuscophialis lignicola and Pseudobotrytis terrestris. Mycoscience 48, 290–296. doi: 10.1007/S10267-007-0369-1
Cai, L., Zhang, K. Q., McKenzie, E. H. C., Ho, W. H., and Hyde, K. D. (2002). Acrodictys liputii sp. nov. and Digitodesmium bambusicola sp. nov. from bamboo submerged in the Liput River in the Philippines. Nova Hedwig. 75, 525–532. doi: 10.1127/0029-5035/2002/0075-0525
Cai, L., Zhang, K. Q., McKenzie, E. H. C., and Hyde, K. D. (2003). Freshwater fungi from bamboo and wood submerged in the Liput River in the Philippines. Fungal Divers. 13, 1–12.
Castañeda Ruiz, R. F., and Arnold, G. R. (1985). Deuteromycotina de Cuba. I. Hyphomycetes. Revista Jard. Bot. Nac. 6, 47–67.
Castañeda Ruiz, R. F., Guerrero, B., Adamo, G. M., Morillo, O., Minter, D. W., Stadler, M., et al. (2009). A new species of Selenosporella and two microfungi recorded from a cloud forest in Mérida, Venezuela. Mycotaxon 109, 63–74. doi: 10.5248/109.63
Chomnunti, P., Hongsanan, S., Aguirre-Hudson, B., Tian, Q., Peršoh, D., Dhami, M. K., et al. (2014). The sooty moulds. Fungal Divers. 66, 1–36. doi: 10.1007/s13225-014-0278-5
Crous, P. W., Braun, U., Hunter, G. C., Wingfield, M. J., Verkley, G., Shin, H.-D., et al. (2013). Phylogenetic lineages in Pseudocercospora. Stud. Mycol. 75, 37–114. doi: 10.3114/sim0005
Day, M. J., Gibas, C. F. C., Fujimura, K. E., Egger, K. N., and Currah, R. S. (2006). Monodictys arctica, a new hyphomycete from the roots of Saxifraga oppositifolia collected in the Canadian High Arctic. Mycotaxon 98, 261–272. doi: 10.7939/R3CF9J63X
Dayarathne, M. C., Maharachchikumbura, S. S. N., Jones, E. B. G., Dong, W., Devadatha, B., Yang, J., et al. (2019). Phylogenetic revision of Savoryellaceae and evidence for its ranking as a subclass. Front. Microbiol. 10:840. doi: 10.3389/fmicb.2019.00840
Diederich, P., Ertz, D., Lawrey, J. D., Sikaroodi, M., and Untereiner, W. A. (2013). Molecular data place the hyphomycetous lichenicolous genus Sclerococcum close to Dactylospora (Eurotiomycetes) and S. parmeliae in Cladophialophora (Chaetothyriales). Fungal Divers. 58, 61–72. doi: 10.1007/s13225-012-0179-4
Diederich, P., Lawrey, J. D., and Ertz, D. (2018). The 2018 classification and checklist of lichenicolous fungi, with 2000 non-lichenized, obligately lichenicolous taxa. Bryologist 121, 340–425. doi: 10.1639/0007-2745-121.3.340
Dong, W., Hyde, K. D., Bhat, D. J., and Zhang, H. (2018). Introducing Aculeata aquatica gen. et sp. nov., Minimelanolocus thailandensis sp. nov. and Thysanorea aquatica sp. nov. (Herpotrichiellaceae, Chaetothyriales) from freshwater in northern Thailand. Mycol. Prog. 17, 617–629. doi: 10.1007/s11557-018-1389-2
Ekanayaka, A. H., Jones, E. B. G., Hyde, K. D., and Zhao, Q. (2019). A stable phylogeny for Dactylosporaceae. Cryptogam. Mycol. 40, 23–44. doi: 10.5252/cryptogamie-mycologie2019v40a3
Ellis, M. B. (1959). Clasterosporium and some allied Dematiaceae-phragmosporae. II. Mycol. Pap. 72, 1–75.
Funk, A., and Shoemaker, R. (1983). Stuartella suttonii n. sp., the teleomorph of Bactrodesmium obliquum var. suttonii. Can. J. Bot. 61, 2277–2279. doi: 10.1139/b83-249
Goh, T. K., and Hyde, K. D. (1996). Cryptophiale multiseptata, sp. nov. from submerged wood in Australia, and keys to the genus. Mycol. Res. 100, 999–1004. doi: 10.1016/S0953-7562(96)80054-2
Hawksworth, D. L. (1975). A revision of lichenicolous fungi accepted by Keissler in Coniothecium. Trans. Br. Mycol. Soc. 65, 219–238. doi: 10.1016/S0007-1536(75)80005-2
Heredia, G., Arias-Mota, R. M., Mena-Portales, J., and Castañeda-Ruiz, R. F. (2018). Saprophytic synnematous microfungi. New records and known species for Mexico. Rev. Mex. Biodivers. 89, 604–618. doi: 10.22201/ib.20078706e.2018.3.2352
Hernández-Restrepo, M., Gené, J., Castañeda-Ruiz, R. F., Mena-Portales, J., Crous, P. W., and Guarro, J. (2017). Phylogeny of saprobic microfungi from Southern Europe. Stud. Mycol. 86, 53–97. doi: 10.1016/j.simyco.2017.05.002
Holubová-Jechová, V. (1984). Bactrodesmiastrum, a new genus of lignicolous Hyphomycetes. Folia Geobot. Phytotx. 19, 103–106. doi: 10.1007/BF02853338
Hu, D. M., Cai, L., Chen, H., Bahkali, A. H., and Hyde, K. D. (2010). Fungal diversity on submerged wood in a tropical stream and an artificial lake. Biodivers. Conserv. 19, 3799–3808. doi: 10.1007/s10531-010-9927-5
Hu, Q., Guo, X., Liang, Y. L., Hao, X. D., Ma, L. Y., Yin, H. Q., et al. (2015). Comparative metagenomics reveals microbial community differentiation in a biological heap leaching system. Res. Microbiol. 166, 525–534. doi: 10.1016/j.resmic.2015.06.005
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Hughes, S. J. (1958). Revisiones hyphomycetum aliquot cum appendice de nominibus rejiciendis. Can. J. Bot. 36, 727–836. doi: 10.1139/b58-067
Hughes, S. J. (1978). New Zealand fungi 25. Miscellaneous species. N.Z. J. Bot. 16, 311–370. doi: 10.1080/0028825X.1978.10425143
Hyde, K. D., Fryar, S., Tian, Q., Bahkali, A. H., and Xu, J. C. (2016). Lignicolous freshwater fungi along a north–south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 19, 190–200. doi: 10.1016/j.funeco.2015.07.002
Hyde, K. D., Goh, T. K., and Steinke, T. D. (1998). Fungi on submerged wood in the Palmiet River, Durban, South Africa. S. Afr. J. Bot. 64, 151–162. PMID:NOPMID
Index Fungorum (2020). Available online at: http://www.indexfungorum.org/names/names.asp (accessed January, 2020).
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, J., Buyck, B., Cai, L., et al. (2015). The Faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Jeewon, R., and Hyde, K. D. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Jeewon, R., Liew, E. C. Y., and Hyde, K. D. (2004). Phylogenetic evaluation of species nomenclature of Pestalotiopsis in relation to host association. Fungal Divers. 17, 39–55.
Jones, E. B. G., Abdel-Wahab, M. A., Alias, S. A., and Hsieh, S. Y. (1999). Dactylospora mangrovei sp. nov. (Discomycetes, ascomycota) from mangrove wood. Mycoscience 40, 317–320. doi: 10.1007/BF02463875
Kazutaka, K., and Standley, D. M. (2016). A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32, 1933–1942. doi: 10.1093/bioinformatics/btw108
Kirk, P. (1981). New or interesting microfungi II. Dematiaceous hyphomycetes from Esher Common, Surrey. Trans. Br. Mycol. Soc. 77, 279–297. doi: 10.1016/S0007-1536(81)80031-9
Koukol, O., and Kolárová, Z. (2010). Bactrodesmium gabretae (anamorphic Helotiales), a new sporodochial species described from spruce needles. Nova Hedwig. 91, 243–248. doi: 10.1127/0029-5035/2010/0091-0243
Larget, B., and Simon, D. L. (1999). Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol. Biol. Evol. 16, 750–759. doi: 10.1093/oxfordjournals.molbev.a026160
Liu, A.-R., Chen, S.-C., Wu, S.-Y., Xu, T., Guo, L.-D., Jeewon, R., et al. (2010). Cultural studies coupled with DNA based sequence analyses and its implication on pigmentation as a phylogenetic marker in Pestalotiopsis taxonomy. Mol. Phylogenet. Evol. 57, 528–535. doi: 10.1016/j.ympev.2010.07.017
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lu, Y.-Z., Liu, J.-K., Hyde, K. D., Jeewon, R., Kang, J.-C., Fan, C., et al. (2018). A taxonomic reassessment of tubeufiales based on multi-locus phylogeny and morphology. Fungal Divers. 92, 131–344. doi: 10.1007/s13225-018-0411-y
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE) (New Orleans, LA: IEEE), 1–8.
Miller, M. A., Schwartz, T., Pickett, B. E., He, S., Klem, E. B., Scheuermann, R. H., et al. (2015). A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinform. 11, 43–48. doi: 10.4137/EBO.S21501
Olariaga, I., Teres, J., Martín, J., Prieto, M., and Baral, H.-O. (2019). Pseudosclerococcum golindoi gen. et sp. nov., a new taxon with apothecial ascomata and a Chalara-like anamorph within the Sclerococcales (Eurotiomycetes). Mycol. Prog. 18, 895–905. doi: 10.1007/s11557-019-01500-7
Pang, K. L., Guo, S. Y., Alias, S. A., Hafellner, J., and Jones, E. B. G. (2014). A new species of marine Dactylospora and its phylogenetic affinities within the Eurotiomycetes, Ascomycota. Bot. Mar. 57, 315–321. doi: 10.1515/bot-2014-0025
Pem, D., Jeewon, R., Bhat, D. J., Doilom, M., Boonmee, S., Hongsanan, S., et al. (2019). Mycosphere notes 275-324: a morpho-taxonomic revision and typification of obscure Dothideomycetes genera (incertae sedis). Mycosphere 10, 1115–1246. doi: 10.5943/mycosphere/10/1/22
Pino-Bodas, R., Zhurbenko, M. P., and Stenroos, S. (2017). Phylogenetic placement within Lecanoromycetes of lichenicolous fungi associated with Cladonia and some other genera. Persoonia 39, 91–117. doi: 10.3767/persoonia.2017.39.05
Prabhugaonkar, A. (2011). Studies on Diversity and Activity of Microfungi Associated with Indigenous Palms of Western Ghats, India. Ph.D. thesis, Goa University, Goa].
Raja, H. A., Miller, A. N., and Shearer, C. A. (2008). Freshwater ascomycetes: Aquapoterium pinicola, a new genus and species of Helotiales (Leotiomycetes) from Florida. Mycologia 100, 141–148. doi: 10.1080/15572536.2008.11832506
Raja, H. A., Stchigel, A. M., Miller, A. N., Crane, J. L., and Shearer, C. A. (2007). Hyphomycetes from the Great Smoky Mountains National Park, including three new species. Fungal Divers. 26, 271–286.
Rannala, B., and Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Réblová, M., Gams, W., and Seifert, K. A. (2011). Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Stud. Mycol. 68, 163–191. doi: 10.3114/sim.2011.68.07
Réblová, M., Seifert, K. A., Fournier, J., and Stepánek, V. (2012). Phylogenetic classification of Pleurothecium and Pleurotheciella gen. nov. and its dactylaria-like anamorph (Sordariomycetes) based on nuclear ribosomal and protein-coding genes. Mycologia 104, 1299–1314. doi: 10.3852/12-035
Réblová, M., Untereiner, W. A., Štìpánek, V., and Gams, W. (2016). Disentangling Phialophora section Catenulatae: disposition of taxa with pigmented conidiophores and recognition of a new subclass, Sclerococcomycetidae (Eurotiomycetes). Mycol. Prog. 16, 27–46. doi: 10.1007/s11557-016-1248-y
Rehner, S. A., and Samuels, G. J. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 98, 625–634. doi: 10.1016/S0953-7562(09)80409-7
Santa Izabel, T. d. S., and Gusmão, L. F. P. (2016). Fungal succession on plant debris in three humid forests enclaves in the Caatinga biome of Brazil. Braz. J. Bot. 39, 1065–1076. doi: 10.1007/s40415-016-0305-8
Santa Izabel, T. d. S., and Gusmão, L. F. P. (2018). Richness and diversity of conidial fungi associated with plant debris in three enclaves of Atlantic Forest in the Caatinga biome of Brazil. Plant Ecol. Evol. 151, 35–47. doi: 10.5091/plecevo.2018.1332
Shearer, C. A., Zelski, S. E., Raja, H. A., Schmit, J. P., Miller, A. N., and Janovec, J. P. (2015). Distributional patterns of freshwater ascomycetes communities along an Andes to Amazon elevational gradient in Peru. Biodivers. Conserv. 24, 1877–1897. doi: 10.1007/s10531-015-0911-y
Shenoy, B. D., Jeewon, R., Wu, W. P., Bhat, D. J., and Hyde, K. D. (2006). Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycol. Res. 110, 916–928. doi: 10.1016/j.mycres.2006.06.004
Su, H. Y., Hyde, K. D., Maharachchikumbura, S. S. N., Ariyawansa, H. A., Luo, Z. L., Promputtha, I., et al. (2016). The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 80, 375–409. doi: 10.1007/s13225-016-0362-0
Su, H. Y., Udayanga, D., Luo, Z. L., Manamgoda, D. S., Zhao, Y. C., Yang, J., et al. (2015). Hyphomycetes from aquatic habitats in Southern China: species of Curvularia (Pleosporaceae) and Phragmocephala (Melannomataceae). Phytotaxa 226, 201–216. doi: 10.11646/phytotaxa.226.3.1
Tanaka, K., Hirayama, K., Yonezawa, H., Sato, G., Toriyabe, A., Kudo, H., et al. (2015). Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 82, 75–136. doi: 10.1016/j.simyco.2015.10.002
Tedersoo, L., Bahram, M., Puusepp, R., Nilsson, R. H., and James, T. Y. (2017). Novel soil-inhabiting clades fill gaps in the fungal tree of life. Microbiome 5:42. doi: 10.1186/s40168-017-0259-5
Tsui, C. K. M., Berbee, M. L., Jeewon, R., and Hyde, K. D. (2006). Molecular phylogeny of Dictyosporium and allied genera inferred from ribosomal DNA. Fungal Divers. 21, 157–166.
Vargasasensio, G., Pintotomas, A., Rivera, B., Hernandez, M., Hernandez, C., Sotomontero, S., et al. (2014). Uncovering the cultivable microbial diversity of Costa Rican beetles and its ability to break down plant cell wall components. PLoS One 9:e113303. doi: 10.1371/journal.pone.0113303
Videira, S., Groenewald, J., Nakashima, C., Braun, U., Barreto, R. W., de Wit, P. J., et al. (2017). Mycosphaerellaceae–chaos or clarity? Stud. Mycol. 87, 257–421. doi: 10.1016/j.simyco.2017.09.003
Vijaykrishna, D., and Hyde, K. D. (2006). Inter- and intra stream variation of lignicolous freshwater fungi in tropical Australia. Fungal Divers. 21, 203–224.
Vijaykrishna, D., Mostert, L., Jeewon, R., Gams, W., Hyde, K. D., and Crous, P. W. (2004). Pleurostomophora, an anamorph of Pleurostoma (Calosphaeriales), a new anamorph genus morphologically similar to Phialophora. Stud. Mycol. 50, 387–395.
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wang, G.-N., Yu, X.-D., Dong, W., Bhat, D. J., Boonmee, S., Zhang, D., et al. (2019). Freshwater hyphomycetes in Eurotiomycetes: a new species of Minimelanolocus and a new collection of Thysanorea papuana (Herpotrichiellaceae). Mycol. Prog. 18, 511–522. doi: 10.1007/s11557-019-01473-7
Wei, M. J., Zhang, H., Dong, W., Boonmee, S., and Zhang, D. (2018). Introducing Dictyochaeta aquatica sp. nov. and two new species of Chloridium (Chaetosphaeriaceae, Sordariomycetes) from aquatic habitats. Phytotaxa 362, 187–199. doi: 10.11646/phytotaxa.362.2.5
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Wijayawardene, N. N., Hyde, K. D., Rajeshkumar, K. C., Hawksworth, D. L., Madrid, H., Kirk, P. M., et al. (2017a). Notes for genera: Ascomycota. Fungal Divers. 86, 1–594. doi: 10.1007/s13225-017-0386-0
Wijayawardene, N. N., Hyde, K. D., Tibpromma, S., Wanasinghe, D. N., Thambugala, K. M., Tian, Q., et al. (2017b). Towards incorporating asexual fungi in a natural classification: checklist and notes 2012–2016. Mycosphere 8, 1457–1555. doi: 10.5943/mycosphere/8/9/10
Wong, M. K. M., and Hyde, K. D. (2001). Diversity of fungi on six species of Gramineae and one species of Cyperaceae in Hong Kong. Mycol. Res. 105, 1485–1491. doi: 10.1017/s0953756201004695
Xia, J.-W., Ma, Y.-R., Gao, J.-M., Li, Z., and Zhang, X.-G. (2015). Sporidesmiopsis malloti sp. nov. and new records from southern China. Mycotaxon 130, 827–833. doi: 10.5248/130.827
Yang, J., Liu, J. K., Hyde, K. D., Jones, E. B. G., and Liu, Z. Y. (2018a). New species in Dictyosporium, new combinations in Dictyocheirospora and an updated backbone tree for Dictyosporiaceae. Mycokeys 36, 83–105. doi: 10.3897/mycokeys.36.27051
Yang, J., Liu, N. G., Liu, J. K., Hyde, K. D., Jones, E. B. G., and Liu, Z. Y. (2018b). Phylogenetic placement of Cryptophiale, Cryptophialoidea, Nawawia, Neonawawia gen. nov. and Phialosporostilbe. Mycosphere 9, 1132–1150. doi: 10.5943/mycosphere/9/6/5
Yang, J., Maharachchikumbura, S. S. N., Bhat, D. J., Hyde, K. D., Mckenzie, E. H. C., Jones, E. B. G., et al. (2016). Fuscosporellales, a new order of aquatic and terrestrial hypocreomycetidae (Sordariomycetes). Cryptogam. Mycol. 4, 449–475. doi: 10.7872/crym/v37.iss4.2016.1
Yu, X. D., Dong, W., Bhat, D. J., Boonmee, S., Zhang, D., and Zhang, H. (2018). Cylindroconidiis aquaticus gen. et sp. nov., a new, lineage of aquatic hyphomycetes in Sclerococcaceae (Eurotiornycetes). Phytotaxa 372, 79–87. doi: 10.11646/phytotaxa.372.1.6
Zhang, H., Dong, W., Hyde, K. D., Bahkali, A. H., Liu, J. K., Zhou, D. Q., et al. (2016). Molecular data shows Didymella aptrootii is a new genus in Bambusicolaceae. Phytotaxa 247, 99–108. doi: 10.11646/phytotaxa.247.2.1
Zhang, H., Dong, W., Hyde, K. D., Maharachchikumbura, S. S. N., Hongsanan, S., Jayarama Bhat, D., et al. (2017). Towards a natural classification of Annulatascaceae-like taxa: introducing Atractosporales ord. nov. and six new families. Fungal Divers. 85, 75–110. doi: 10.1007/s13225-017-0387-z
Zhang, H., Hyde, K. D., Abdel-Wahab, M. A., Abdel-Aziz, F. A., Ariyawansa, H. A., Ko, T. W. K., et al. (2013). A modern concept for Helicascus with a Pleurophomopsis-like asexual state. Sydowia 65, 147–166.
Zhang, H., Hyde, K. D., Mckenzie, E. H. C., Bahkali, A. H., and Zhou, D. Q. (2012). Sequence data reveals phylogenetic affinities of Acrocalymma aquatica sp. nov., Aquasubmersa mircensis gen. et sp. nov. and Clohesyomyces aquaticus (freshwater coelomycetes). Cryptogam. Mycol. 33, 333–346. doi: 10.7872/crym.v33.iss3.2012.333
Zhang, H., Hyde, K. D., Zhao, Y. C., McKENZIE, E. H. C., and Zhou, D. Q. (2014). Freshwater ascomycetes: Lophiostoma vaginatispora comb. nov. (Dothideomycetes, Pleosporales, Lophiostomaceae) based on morphological and molecular data. Phytotaxa 176, 184–191. doi: 10.11646/phytotaxa.176.1.18
Zhang, H., Jones, E. B. G., Zhou, D. Q., Bahkali, A. H., and Hyde, K. D. (2011). Checklist of freshwater fungi in Thailand. Cryptogam. Mycol. 32, 199–217. doi: 10.7872/crym.v32.iss2.2011.199
Keywords: Bactrodesmium, multi-gene, sheath, submerged wood, taxonomy
Citation: Dong W, Hyde KD, Doilom M, Yu X-D, Bhat DJ, Jeewon R, Boonmee S, Wang G-N, Nalumpang S and Zhang H (2020) Pseudobactrodesmium (Dactylosporaceae, Eurotiomycetes, Fungi) a Novel Lignicolous Genus. Front. Microbiol. 11:456. doi: 10.3389/fmicb.2020.00456
Received: 30 September 2019; Accepted: 03 March 2020;
Published: 02 April 2020.
Edited by:
Martin G. Klotz, Washington State University, United StatesReviewed by:
Paul Diederich, National Museum of Natural History, LuxembourgGregorio Delgado, Eurofins EMLab P&K, United States
Copyright © 2020 Dong, Hyde, Doilom, Yu, Bhat, Jeewon, Boonmee, Wang, Nalumpang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huang Zhang, emhhbmdodWFuZzIwMDIxMTNAMTYzLmNvbQ==
 Wei Dong
Wei Dong Kevin D. Hyde
Kevin D. Hyde Mingkwan Doilom
Mingkwan Doilom Xian-Dong Yu1
Xian-Dong Yu1 Rajesh Jeewon
Rajesh Jeewon Saranyaphat Boonmee
Saranyaphat Boonmee Huang Zhang
Huang Zhang