
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 February 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00282
Genome-editing CRISPR/Cas9 technology has led to the development of artificial transcriptional repressors, also known as CRISPR interference (CRISPRi). The deactivated Cas9 (dCas9) protein guided by crRNA can specifically bind to target DNA sequences, including promoters and operators, without DNA cleavage. Protospacer adjacent motif (PAM) sequence dependence may be disadvantageous in the design of target-specific CRISPRi, as the PAM sequence is essential for DNA cleavage by the CRISPR/Cas9 system. We constructed a chromosomally integrated dCas9 system (ΔaraBAD:dcas9) in Escherichia coli under the control of the L-arabinose-inducible PBAD promoter. Plasmids carrying various crRNAs with target sequences specific for the gal promoter (−10 region), and the galETK structural genes in the gal operon, were transformed into dCas9-expressing E. coli. Cellular growth and/or galactose metabolic rates were monitored in the presence or absence of gratuitous L-arabinose. D-galactose consumption and cell growth rates were partially retarded by targeting transcriptional elongation but were fully inhibited by targeting transcriptional initiation. Moreover, RT-qPCR analysis showed that CRISPRi with several modified PAM sequences can repress the transcription of target DNAs. These results indicate that cellular metabolic rates and cell growth can be controlled by targeting structural genes or regulatory regions using CRISPRi; also, a loose PAM sequence dependence can expand the DNA targets of CRISPRi.
The biological role of CRISPR has been identified as a prokaryotic adaptive immune system (Mojica et al., 2005; Pourcel et al., 2005). Since then, the application of CRISPR has been expanding explosively in the field of biotechnology (Doudna and Charpentier, 2014), especially because the components of CRISPR nucleases (i.e. crRNAs and Cas proteins) are separated functionally in the recognition of target nucleotide sequences and breakage of phosphodiester bonds in nucleic acids (Dagdas et al., 2017). Moreover, a specific protospacer adjacent motif (PAM) (∼3–5 bp) sequence must be located near the target sequence for the proper operation of the CRISPR/Cas system (Rath et al., 2015). In addition, the 5′-NGG sequence should be located right after target DNA sequences in the case of CRISPR/Cas9 (O’connell et al., 2014).
The CRISPR/Cas9 nuclease has been widely used in gene knockout studies in eukaryotic cells by the highly specific digestion of target DNA sequences followed by non-homologous end joining, resulting in the insertion or deletion of target genes (Zhang et al., 2014b). In addition, site-specific genome editing can be achieved by the introduction of single-stranded oligonucleotides and/or double-stranded DNA fragments flanking homologous regions into prokaryotic or eukaryotic cells, respectively, which can be negatively selected by the CRISPR/Cas9 nuclease (Knott and Doudna, 2018).
In addition to genome-editing tools, deactivated or dead Cas9 (dCas9) was created to be applied to artificial gene regulators (Larson et al., 2013). dCas9 has inactivated HNH and RuvC domains responsible for nuclease activity. It has been known that the target DNA sequence cannot be digested but is occupied by dCas9-crRNA complexes, which can affect the expression of target genes, and this is called CRISPR interference (CRISPRi) (Marraffini and Sontheimer, 2010a). When the dCas9-crRNA complex binds to the promoter of target genes, the initiation of transcription can be inhibited. If the complex binds to the coding region of the target gene, the elongation of transcription can be blocked (Qi et al., 2013). In prokaryotic cells, gene regulation is hardly achieved by RNA interference. Therefore, CRISPRi is very helpful in the engineering of microbial cells (Qiu et al., 2018; Xu and Qi, 2019).
Since the bacterial adaptive immune system, CRISPR, has evolved to distinguish self and non-self target sequences (Marraffini and Sontheimer, 2010b), the presence of the PAM sequence near the target DNA is essential for the DNA double-strand breakage generated by the Cas9-crRNA complex (Anders et al., 2014). Such PAM dependence can narrow down the possible target sequences in the Cas9-crRNA complex. However, only binding to the target is necessary for the purpose of dCas9-crRNA, which suggests that PAM dependence is lower than Cas9-crRNA dependence.
In this study, we evaluated how strongly the dCas9-crRNA complex inhibits initiation and elongation in the transcription of the gal operon. Also, we tested whether dCas9-crRNA can recognize DNA targets with modified PAM sequences (NNG and NGN). Our results demonstrate that cellular metabolic rates can be controlled with CRISPRi and that a loose PAM sequence dependence can expand the DNA targets of dCas9-crRNA.
Escherichia coli K-12 DH5α and MG1655 strains were used as a cloning host and a dCas9 expression host, respectively (Table 1). E. coli cells were grown routinely in LB medium. When needed, antibiotics such as ampicillin (50 μg/ml), kanamycin (25 μg/ml), and spectinomycin (75 μg/ml) were used.
The primers used in this study are listed in Table 2. In order to make a dcas9-KmR cassette, the dcas9 gene was amplified using plasmid pdCas9 [a gift from Luciano Marraffini, Addgene plasmid #46569 (Bikard et al., 2013)] as a template and fused with a kanamycin resistance gene by overlap PCR. The PCR products of the dcas9-KmR cassette with a homologous region for recombineering at the araBAD promoter in the chromosome were electroporated into E. coli K-12 MG1655 cells harboring plasmid pKD46 (Datsenko and Wanner, 2000) to generate E. coli HK1060 (ΔaraBAD:PBAD-dcas9-KmR). Then, plasmid pKD46 was cured at 42°C. The disruption of the araBAD genes and the integration of the dcas9 gene in HK1060 cells were confirmed by PCR.
To generate the crRNA expression plasmids, pTargetF (a gift from Sheng Yang, Addgene plasmid #62226) was used as a template to amplify 0.9 and 1.2-kb fragments with primer pairs of Cas-galK-WF and Sm_ATG_R and Sm_TAA_F and Cas-galK-WR, respectively. These two fragments were purified for isothermal assembly using Gibson Assembly® Master Mix (NEB, MA, United States) to make pHK459 plasmid (Table 1). Other crRNA plasmids carrying the spectinomycin resistance gene as a selection marker were constructed by Gibson Assembly in the same manner. Target sequences recognized by crRNAs were designed in the DNA sequences (i.e. the gal promoter and the galE, galT, and galK genes) in the gal operon. The extended −10 region was selected as the DNA target in the gal promoter. For the experiments with modified PAM sequences, we constructed additional crRNA plasmids targeting the gal promoter and the galK gene. All the crRNA plasmids were confirmed by Sanger sequencing and transformed into HK1060 cells for the growth assay.
HK1060 cells carrying each crRNA plasmid were grown in LB broth containing spectinomycin at 37°C overnight as starter cultures. Next, 1.0% of cells were inoculated in 25 ml of M9 minimal medium containing sodium succinate (final 20 mM) as a carbon source and spectinomycin in 250-ml flasks. crRNAs were expressed constitutively, and gratuitous L-arabinose (final 1 mM) was added for the expression of dCas9 proteins to form the dCas9-crRNA complex in HK1060 cells carrying crRNA plasmids. To determine whether the transcription of the gal operon could be negatively regulated by the dCas9-crRNA complex, D-galactose (final 20 mM) was added as an additional carbon source 6 h after the beginning of flask cultures. Optical density at 600 nm was measured every 3 h to monitor cellular growth using an Ultrospec 8000 spectrophotometer (GE Healthcare, Uppsala, Sweden).
Expression levels of galE and galK genes were monitored using RT-qPCR. HK1060 cells carrying each crRNA plasmid were grown M9 minimal medium containing sodium succinate (final 20 mM) as a carbon source with or without L-arabinose (final 1 mM). D-galactose (final 20 mM) was added 6 h after the beginning of flask cultures. RNAs were isolated using the RNeasy® Mini kit (Cat. No. 74104; Qiagen, Hilden, Germany) 3 h after the addition of D-galactose. PCR primer sequences for the target genes were designed at the Universal Probe Library Assay Design Center1 and listed in Table 2. Quantitative real-time PCR (RT-qPCR) reactions were carried out on a LightCycler 96 (Roche Diagnostics, Mannheim, Germany) using the RealHelixTM qPCR kit (Nanohelix, South Korea). Five nanograms of each total RNA were used in RT-qPCR reactions under the following conditions: cDNA synthesis (50°C, 40 min); denaturation (95°C, 12 min); amplification for 50 cycles (95°C, 20 s; 60°C, 1 min). The raw fluorescence data were normalized against the expression level of 16S ribosomal RNA. In order to calculate the relative abundance of galE (374–453 base region from +1 start codon) and galK (895–963 base region) transcripts, mRNA levels of galE and galK genes in the presence of L-arabinose were divided by mRNA levels of the corresponding genes in the absence of L-arabinose.
The concentrations of D-galactose in the culture medium were determined by high-performance liquid chromatography (HPLC, RID-10A RI monitor, Shimadzu, Japan) with an Aminex HPX-87H column (300 × 7.8 mm, Hercules, BioRad), as described previously (Kim et al., 2013). After centrifugation of the cell culture broth, the supernatant was filtered through a 0.2-μm syringe filter. The column was isocratically eluted at 47°C with a flow rate of 0.5 ml min–1 using 0.01 N H2SO4.
Cellular growth and D-galactose utilization were monitored to assess how well CRISPRi interferes with transcription of the gal operon. Since CRISPRi can inhibit the initiation and elongation of transcription, the gal promoter and the gal structural genes (galE, galT, and galK) were used as the DNA targets of the dCas9-crRNA complex (Figure 1A). Cells carrying the L-arabinose-inducible dcas9 gene were transformed with various crRNA plasmids. The transformed cells were grown in succinate minimal media in the presence or absence of L-arabinose, which can serve as a gratuitous inducer. Succinate was used as a carbon source to prevent carbon catabolite repression. At the early exponential phase (∼OD600 of 0.3), D-galactose was added to the cell culture at 6 h after inoculation. If the gal transcription was not inhibited by dCas9-crRNAs, the increase of cellular growth and the consumption of D-galactose could be observed.
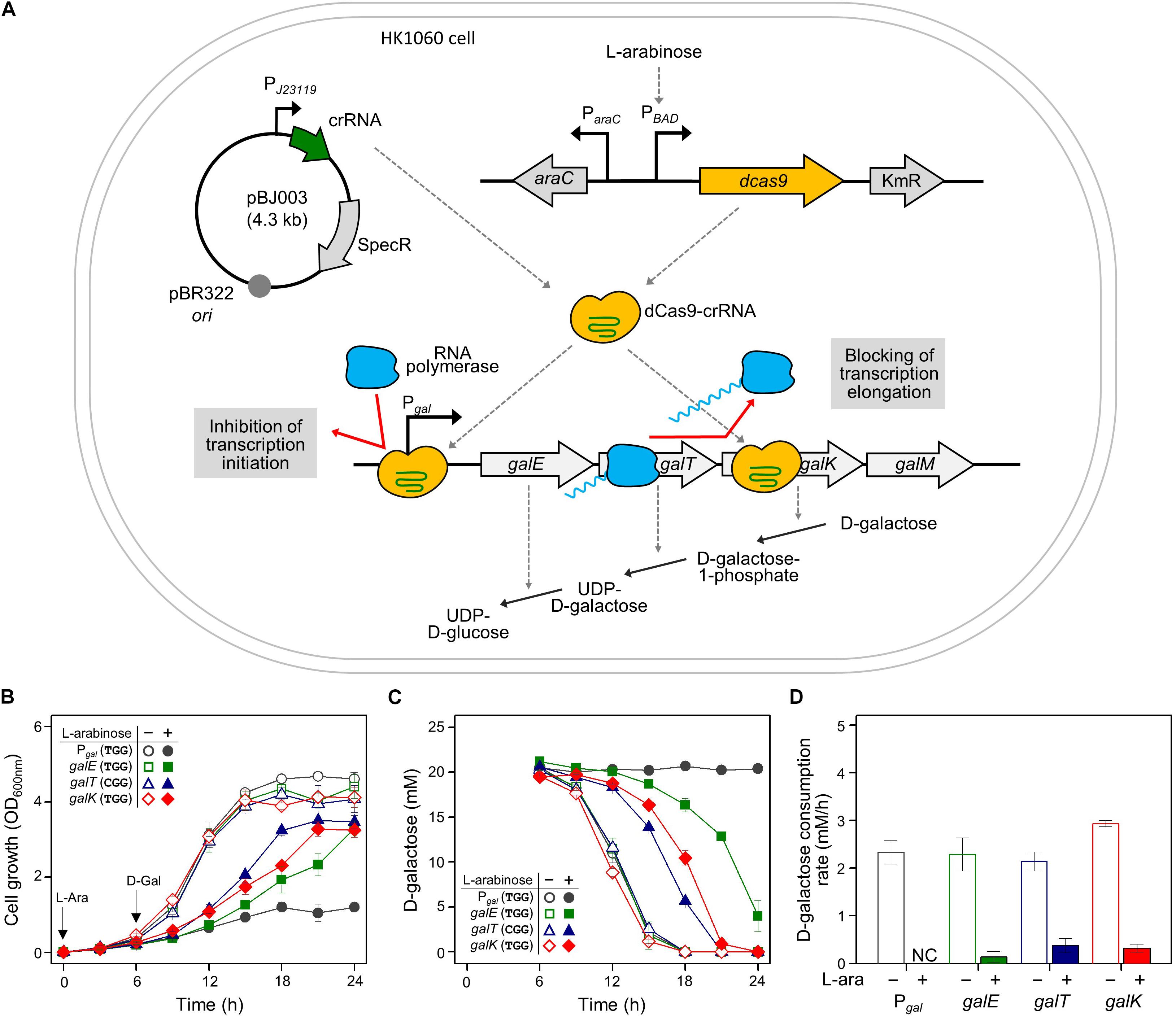
Figure 1. Regulation of gal operon and control of D-galactose metabolic rates by CRISPRi. (A) Expression of gal promoter and structural genes regulated by dCas9-crRNAs. The control of the gal promoter and structural galETK genes was used as a model system for comparing the transcriptional initiation and elongation by CRISPRi. The target DNAs in the gal operon can be attached by active dCas9-crRNA complexes, which are formed by dCas9 proteins expressed in the presence of non-metabolizable L-arabinose, and crRNAs transcribed constitutively in cells. D-galactose metabolism depends on the expression of D-galactose metabolic enzymes in the gal operon. (B) Growth profiles of HK1060 cells carrying specific crRNAs targeting gal promoter and structural galETK genes. Retarded cell growth was observed due to the repression of galactose enzymes by L-arabinose. (C) Change of residual D-galactose concentration in culture media. D-galactose was added at 6 h and monitored until the late stationary growth phase. (D) D-galactose consumption rates affected by CRISPRi. The concentration changes of residual D-galactose between 9 and 12 h were used for the calculation of D-galactose consumption rates. Arrows indicate when L-arabinose (if needed) and D-galactose were added. NC, not consumed.
HK1060 cells were transformed with crRNA plasmids expressing crRNAs recognizing the DNA targets with the original PAM sequence (5′-NGG). As a result, in the absence of L-arabinose, all the transformant cells reached an OD600 of 4, and consumed D-galactose (total 20 mM) within 18 h (Figures 1B,C). However, cell growth rates were decreased in the presence of non-metabolizable L-arabinose. This means L-arabinose-induced dCas9 can combine with crRNA to make dCas9-crRNA, which can interfere with the transcription of D-galactose metabolic genes.
In the case of the gal promoter, D-galactose was not consumed at all until the end of the experiment (Figure 1C). The gal promoter is composed of two overlapped extended −10 promoters (Lewis et al., 2015), which may be covered thoroughly by dCas9-crRNA complex. These results indicate that transcriptional initiation can be completely inhibited by CRISPRi.
In the case of structural genes as the inhibition targets, the cell growth and D-galactose consumption were differentially retarded. This means that the expression of gal genes was repressed partially by the blocking of transcriptional elongation. In the profiles of HPLC, the level of gratuitous L-arabinose was not changed in any of the samples, because the ara operon was disrupted by the integration of the dcas9 gene in HK1060 cells. It was confirmed that our model CRISPRi system operated fully and partially in the promoter and structural genes of D-galactose metabolism, respectively.
In order to determine whether CRISPRi follows the PAM sequence rule (5′-NGG) of CRISPR/Cas9, various crRNA plasmids were designed, as shown in Table 1. Modified PAM sequences (5′-NGN and 5′-NNG) were tested in the gal promoter as the DNA target of the dCas9-crRNA complex (Figures 2A,B).
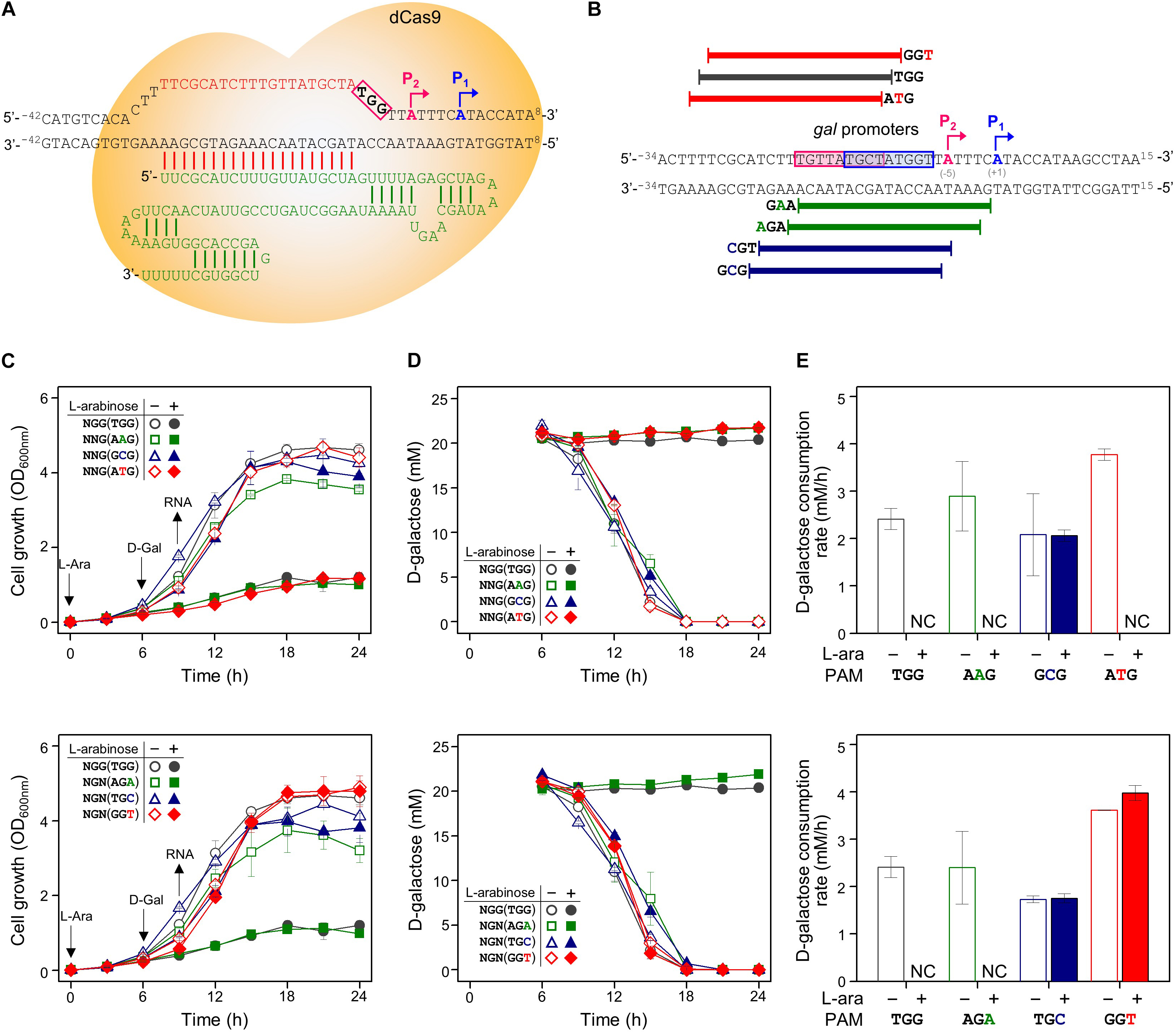
Figure 2. Tight repression of the gal promoter by CRISPRi with modified PAM sequences. (A) The inability of the RNA polymerase to recognize and attach to the gal promoter region, which is occupied by the dCas9-crRNA complex, inhibits the initiation of transcription. Blue and red boxes indicate overlapped –10 regions of P1 and P2 promoters, respectively. The gal promoter-specific crRNA was transcribed from plasmid pBJ005. (B) Design of target DNA sequences with modified PAM sequences in gal promoter. The gal P1 and P2 promoters overlapped each other. The modified PAM sequences of NNG (top) and NGN (bottom) were evaluated by growth profiles (C), utilization of D-galactose (D), and comparison of D-galactose consumption rates (E). D-galactose was added at 6 h, and the concentration changes of residual D-galactose between 9 and 12 h were used to calculate D-galactose consumption rates. NC, not consumed. Arrows indicate when L-arabinose (if needed) and D-galactose were added, and when samples were taken for RNA extraction.
In addition to in NGG (TGG), the growth retardation was observed in the case of NAG (AAG), NTG (ATG), and NGA (AGA) (Figure 2C). Moreover, D-galactose consumption was not observed with the expanded PAM sequences until the end of the experiments (Figure 2D). The D-galactose consumption rates (between 9 and 12 h in graphs) in the cases of NCG (GCG), NGC (TGC), and NGT (GGT) were not changed in the presence (dCas9 expressed) or absence (dCas9 not expressed) of L-arabinose (Figure 2E). Transcript analysis showed that the differential repression of the gal promoter by dCas9-crRNA complexes with the modified PAM sequences. The order of repression was as follows: NGG (TGG) > NAG (AAG) > NGA (AGA) > NTG (ATG) (Figure 3A). In the cases of NCG (GCG), NGC (TGC), and NGT (GGT), the repression of gal promoter could not be observed in the RT-qPCR experiment. These data showed that even weakly attached dCas9-crRNA complex with modified PAM sequences did not result in sufficient gal transcription for galactose metabolism.
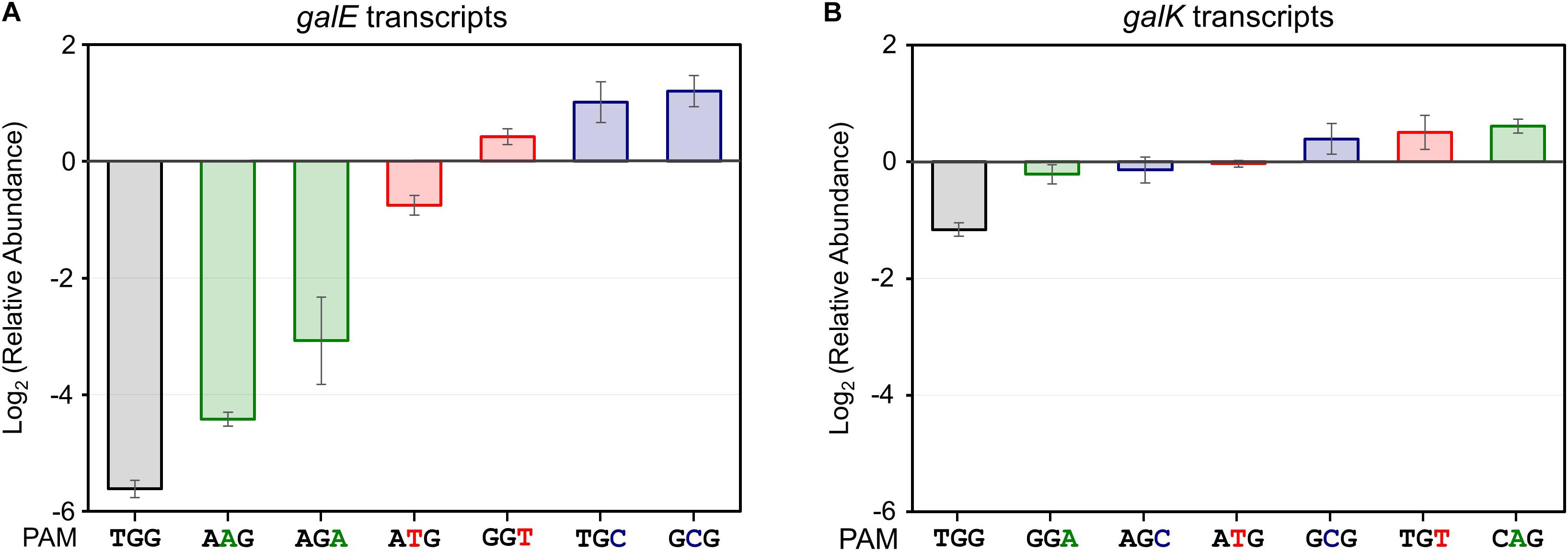
Figure 3. Relative transcript analysis of galE (A) and galK (B) genes affected by the presence of dCas9-crRNA complexes. galE and galK transcripts were analyzed for monitoring of transcriptional initiation and elongation, respectively. The relative abundance of galE and galK transcripts was obtained by calculating the normalized transcript data of galE and galK genes in the presence of L-arabinose (dcas9 induced) divided by those in the absence of L-arabinose (dcas9 uninduced).
The same set of modified PAM sequences (5′-NGN and 5′-NNG) was tested in the galK structural gene as the DNA target of the dCas9-crRNA complex (Figures 4A,B). As a result, cell growth was affected slightly in the cases of NGG (TGG), NTG (ATG), and NGA (GGA) as PAM sequences (Figure 4C). According to the D-galactose consumption rate, the order of interference was as follows: NGG (TGG) > NGA (GGA) > NTG (ATG) (Figures 4D,E). The D-galactose consumption rate (between 9 and 12 h in graphs) in the case of ATG was decreased from ∼3 mM/h (dCas9 not expressed due to the absence of L-arabinose) to ∼1 mM/h (dCas9 expressed by L-arabinose).
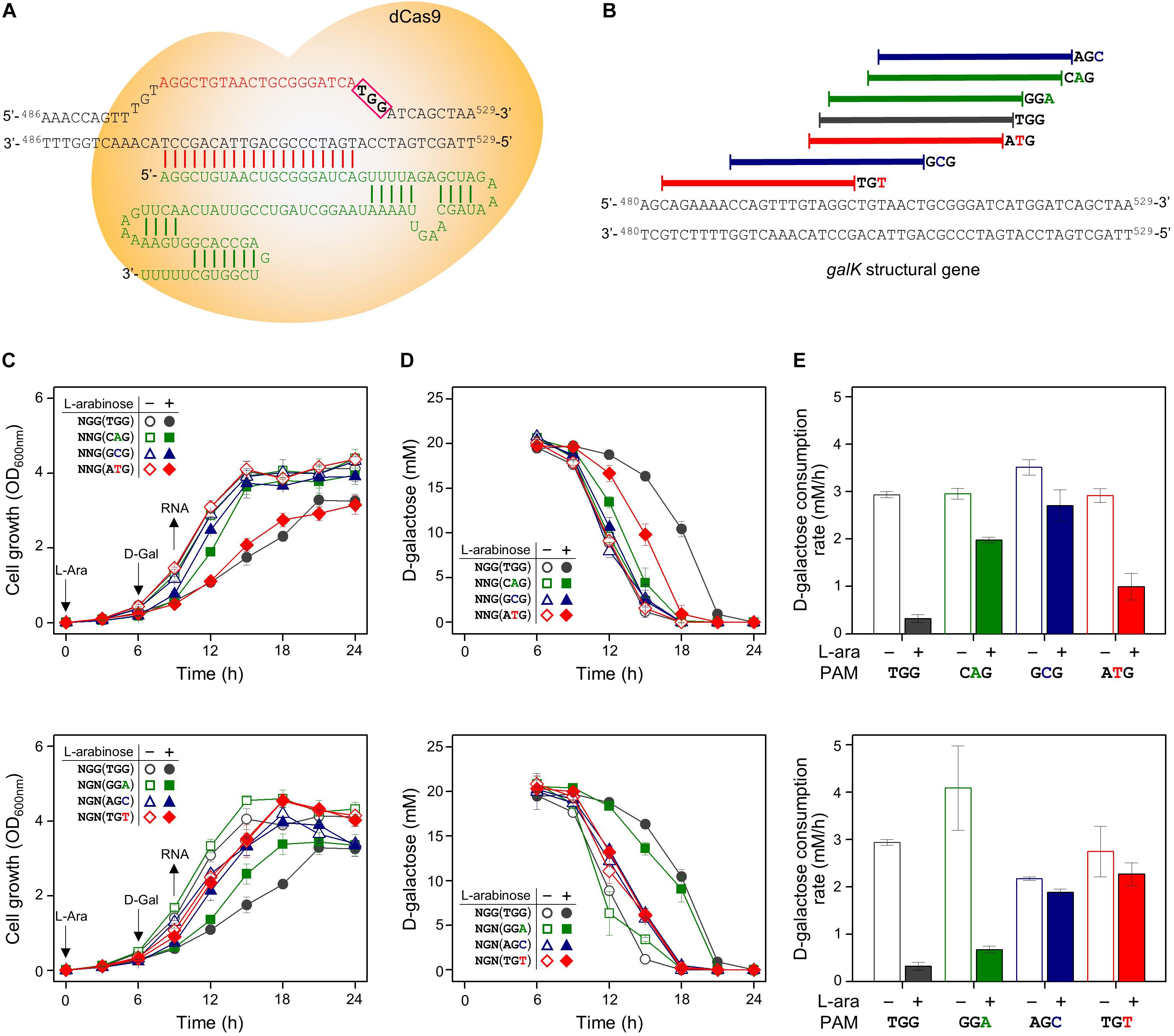
Figure 4. Differential repression of galK structural gene by dCas9-crRNAs recognizing modified PAM sequences. (A) Precise target recognition and proper PAM sequences are required for the blocking of transcriptional elongation in the galK gene by CRISPRi. The galK-specific crRNA was transcribed from plasmid pHK459. (B) Design of target DNA sequences with modified PAM sequences in galK gene. The modified PAM sequences of NNG (top) and NGN (bottom) were evaluated by growth profiles (C), utilization of D-galactose (D), and comparison of D-galactose consumption rates (E). D-galactose was added at 6 h, and the concentration changes of residual D-galactose between 9 and 12 h were used to calculate D-galactose consumption rates. Arrows indicate when L-arabinose (if needed) and D-galactose were added, and when samples were taken for RNA extraction.
We analyzed the abundance of mRNA of the galK gene in the presence or absence of dCas9-crRNA complexes (Figure 3B). The galK transcripts in downstream region of dCas9-crRNA complex-binding sites were monitored by RT-qPCR. In case of NGG (TGG) as a PAM sequence, the transcriptional elongation of the galK gene was affected by CRISPRi. However, any significant interference in the transcriptional elongation was not clearly observed in other PAM sequences using RT-qPCR analysis.
CRISPR interference can regulate the expression of DNA targets by inhibiting the transcription process. Designed target recognition sequences of modular crRNAs can be used to bind any DNA target to dCas9-crRNA complexes (Hawkins et al., 2015). While nonsense point mutation can affect cells permanently by deleting the function of the target gene, dCas9-mediated gene expression control allows the selection of desired time points (Quebatte and Dehio, 2017). Endogenous competing pathway genes can be repressed by CRISPRi in E. coli for metabolic engineering (Kim et al., 2017). Even if the genes are essential for growth, metabolic engineering with CRISPRi provides the necessary metabolites without disturbing cell growth (Cress et al., 2017).
Gene regulation using CRISPRi is carried out in two ways. First, binding of the dCas9-crRNA complex in the promoter region can fully repress transcriptional initiation. Also, elongation of transcription can be blocked by positioning the dCas9-crRNA complex in the middle of the open reading frame of the target gene (Cao et al., 2017). In our study, the gal operon and galactose metabolism were used as a model to demonstrate how dCas9-crRNA can precisely regulate the expression of target genes (gal promoter and structural galE, galT, and galK genes) and the corresponding metabolism (Figure 1A).
Non-metabolizable L-arabinose-induced dCas9 proteins can combine with target-specific crRNAs to make the dCas9-crRNA complex, which can bind to the DNA targets in the galactose promoter, preventing cellular consumption of D-galactose (Figure 1D). When the gal structural genes (galE, galT, and galK) were designed as the DNA targets of the dCas9-crRNA complex, we observed differential D-galactose consumption rates. RT-qPCR analysis showed that the initiation of galE transcription was significantly repressed, and the elongation of galK transcription was slightly blocked the dCas9-crRNAs with TGG as PAM sequence (Figure 3). These results might be explained by that RNA polymerase cannot bind to the gal promoter occupied by the dCas9-crRNA complex, but RNA polymerase can pass through some of the roadblocks of dCas9-crRNA during transcriptional elongation. Although all enzymes encoded by galE, galT, and galK genes are essential in galactose metabolism, the most retarded cell growth was observed in galE-targeted cells (Figure 1B), presumably because galE mutant cells cannot grow well due to the depletion of pyrimidine nucleotides (Lee et al., 2009).
The PAM sequence should be considered in CRISPR technologies when the target DNA sequence is designed. The biological role of the PAM sequence is to discriminate between non-self DNA from outside cells and self DNA stored in the chromosome in bacterial adaptive immune system. It has been known that the PAM sequence is indispensable for DNA cleavage by the Cas9-crRNA complex (Mojica et al., 2009). It has been reported that the hydrogen bonds between nucleotides in the PAM sequence (5′-NGG) and the two arginine residues (Arg1333 and Arg1335) in the Cas9 protein are important in DNA cleavage (Anders et al., 2014). The PAM dependence in the CRISPR/Cas9 system has been studied for expanding the target sequence and reducing off-target activity. Reportedly, the 5′-NGA sequence can function as PAM, although the modified PAM sequence exhibits less gfp gene cleavage activity (Zhang et al., 2014a). It is still unclear whether the PAM sequence is essential in the recognition of target sequences in CRISPR/dCas9 in bacterial cells.
The PAM dependence in CRISPRi was investigated using regulation of the gal promoter (Figure 2) and the galK gene (Figure 4) by dCas9-crRNA complex. The differential repressions of gal promoter by modified PAM sequences were observed by RT-qPCR analysis (Figure 3A). The order of gal promoter repression showed that target binding affinities of dCas9-crRNAs with modified PAM sequences might be weaker than those with the original PAM sequence (NGG). However, the CRISPRi-induced growth retardation and totally impaired D-galactose consumption suggest that NAG, NTG, and NGA can function as the original PAM sequence (NGG) in the gal transcription initiation (Figure 2). This means that the gal promoter even weakly occupied by dCas9-crRNA complex cannot be properly recognized by the RNA polymerase in the cells.
In the case of the galK structural gene as the DNA target of CRISPRi, transcriptional elongation can be clearly modulated by CRISPRi with the original PAM sequence (NGG) based on the results of cellular growth, metabolic rates, and transcript abundance. Since the original PAM sequence occurs frequently in the open reading frame of common genes, the expression of essential genes can be attenuated by CRISPRi with the original PAM sequence.
These results showed that the DNA target sequence for CRISPRi can be expanded, allowing the design of more specific target sequences such as narrow promoter regions. Our results also show that cellular growth and metabolic rates can be controlled by dCas9-crRNAs. The information about CRISPRi with expanded PAM sequences may be useful in the design of DNA targets and crRNAs for fine gene regulation in microbial biotechnology.
All datasets generated for this study are included in the article/supplementary material.
BK, HK, and SL designed the research, analyzed the data and wrote the manuscript. BK and HK performed the experiments.
This study was supported by the National Research Foundation of Korea (NRF-2019R1A4A1024764, 2018R1A6A3A11051083, 2017R1E1A1A01075124, and 2015R1A2A2A0105402).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Anders, C., Niewoehner, O., Duerst, A., and Jinek, M. (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573. doi: 10.1038/nature13579
Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., and Marraffini, L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437. doi: 10.1093/nar/gkt520
Cao, Y., Li, X., Li, F., and Song, H. (2017). CRISPRi-sRNA: transcriptional-translational regulation of extracellular electron transfer in Shewanella oneidensis. ACS Synth. Biol. 6, 1679–1690. doi: 10.1021/acssynbio.6b00374
Cress, B. F., Leitz, Q. D., Kim, D. C., Amore, T. D., Suzuki, J. Y., Linhardt, R. J., et al. (2017). CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production. Microb. Cell Fact. 16:10. doi: 10.1186/s12934-016-0623-3
Dagdas, Y. S., Chen, J. S., Sternberg, S. H., Doudna, J. A., and Yildiz, A. (2017). A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 3:eaao0027. doi: 10.1126/sciadv.aao0027
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346;1258096. doi: 10.1126/science.1258096
Hawkins, J. S., Wong, S., Peters, J. M., Almeida, R., and Qi, L. S. (2015). Targeted transcriptional repression in bacteria using CRISPR interference (CRISPRi). Methods Mol. Biol. 1311, 349–362. doi: 10.1007/978-1-4939-2687-9_23
Kim, H. J., Hou, B. K., Lee, S. G., Kim, J. S., Lee, D. W., and Lee, S. J. (2013). Genome-wide analysis of redox reactions reveals metabolic engineering targets for D-lactate overproduction in Escherichia coli. Metab. Eng. 18, 44–52. doi: 10.1016/j.ymben.2013.03.004
Kim, S. K., Seong, W., Han, G. H., Lee, D. H., and Lee, S. G. (2017). CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb. Cell Fact. 16:188. doi: 10.1186/s12934-017-0802-x
Knott, G. J., and Doudna, J. A. (2018). CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869. doi: 10.1126/science.aat5011
Larson, M. H., Gilbert, L. A., Wang, X., Lim, W. A., Weissman, J. S., and Qi, L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196. doi: 10.1038/nprot.2013.132
Lee, S. J., Trostel, A., Le, P., Harinarayanan, R., Fitzgerald, P. C., and Adhya, S. (2009). Cellular stress created by intermediary metabolite imbalances. Proc. Natl. Acad. Sci. U.S.A. 106, 19515–19520. doi: 10.1073/pnas.0910586106
Lewis, D. E. A., Le, P., and Adhya, S. (2015). Differential role of base pairs on gal promoters strength. J. Mol. Biol. 427, 792–806. doi: 10.1016/j.jmb.2014.12.010
Marraffini, L. A., and Sontheimer, E. J. (2010a). CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11, 181–190. doi: 10.1038/nrg2749
Marraffini, L. A., and Sontheimer, E. J. (2010b). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571. doi: 10.1038/nature08703
Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J., and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. doi: 10.1099/mic.0.023960-0
Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
O’connell, M. R., Oakes, B. L., Sternberg, S. H., East-Seletsky, A., Kaplan, M., and Doudna, J. A. (2014). Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266. doi: 10.1038/nature13769
Pourcel, C., Salvignol, G., and Vergnaud, G. (2005). CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663. doi: 10.1099/mic.0.27437-0
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., and Arkin, A. P. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Qiu, H., Gong, J., Butaye, P., Lu, G., Huang, K., Zhu, G., et al. (2018). CRISPR/Cas9/sgRNA-mediated targeted gene modification confirms the cause-effect relationship between gyrA mutation and quinolone resistance in Escherichia coli. FEMS Microbiol. Lett. 365:fny127. doi: 10.1093/femsle/fny127
Quebatte, M., and Dehio, C. (2017). Systems-level interference strategies to decipher host factors involved in bacterial pathogen interaction: from RNAi to CRISPRi. Curr. Opin. Microbiol. 39, 34–41. doi: 10.1016/j.mib.2017.08.002
Rath, D., Amlinger, L., Rath, A., and Lundgren, M. (2015). The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie 117, 119–128. doi: 10.1016/j.biochi.2015.03.025
Xu, X., and Qi, L. S. (2019). A CRISPR-dCas toolbox for genetic engineering and synthetic biology. J. Mol. Biol. 431, 34–47. doi: 10.1016/j.jmb.2018.06.037
Zhang, Y., Ge, X., Yang, F., Zhang, L., Zheng, J., Tan, X., et al. (2014a). Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 4:5405. doi: 10.1038/srep05405
Keywords: metabolic regulation, PAM dependence, interference, dCas9, CRISPR
Citation: Kim B, Kim HJ and Lee SJ (2020) Regulation of Microbial Metabolic Rates Using CRISPR Interference With Expanded PAM Sequences. Front. Microbiol. 11:282. doi: 10.3389/fmicb.2020.00282
Received: 07 October 2019; Accepted: 07 February 2020;
Published: 28 February 2020.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
Soo Rin Kim, Kyungpook National University, South KoreaCopyright © 2020 Kim, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang Jun Lee, c2FuZ2psZWVAY2F1LmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.