- 1Biomedical Science Programme, Faculty of Health Sciences, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
- 2Faculty of Dentistry, Sir John Walsh Research Institute, University of Otago, Dunedin, New Zealand
- 3Bacteriology Unit, Institute for Medical Research, National Institute of Health, Setia Alam, Malaysia
- 4School of Biological Sciences, Universiti Sains Malaysia, Penang, Malaysia
In the fungal pathogen Aspergillus fumigatus, resistance to azole antifungals is often linked to mutations in CYP51A, a gene that encodes the azole antifungal drug target lanosterol 14α-demethylase. The aim of this study was to investigate whether similar changes could be associated with azole resistance in a Malaysian Fusarium solani species complex (FSSC) isolate collection. Most (11 of 15) clinical FSSC isolates were Neocosmospora keratoplastica and the majority (6 of 10) of environmental isolates were Neocosmospora suttoniana strains. All 25 FSSC isolates had high minimum inhibitory concentrations (MICs) for itraconazole and posaconazole, low MICs for amphotericin B, and various (1 to >32 mg/l) voriconazole susceptibilities. There was a tight association between a 23 bp CYP51A promoter deletion and high (>32 mg/l) voriconazole MICs; of 19 FSSC strains sequenced, nine isolates had voriconazole MICs > 32 mg/l, and they all contained the 23 bp CYP51A promoter deletion, although it was absent in the ten remaining isolates with low (≤12 mg/l) voriconazole MICs. Surprisingly, this association between voriconazole resistance and the 23 bp CYP51A promoter deletion held true across species boundaries. It was randomly distributed within and across species boundaries and both types of FSSC isolates were found among environmental and clinical isolates. Three randomly selected N. keratoplastica isolates with low (≤8 mg/l) voriconazole MICs had significantly lower (1.3–7.5 times) CYP51A mRNA expression levels than three randomly selected N. keratoplastica isolates with high (>32 mg/l) voriconazole MICs. CYP51A expression levels, however, were equally strongly induced (~6,500-fold) by voriconazole in two representative strains reaching levels, after 80 min of induction, that were comparable to those of CYP51B. Our results suggest that FSSC isolates with high voriconazole MICs have a 23 bp CYP51A promoter deletion that provides a potentially useful marker for voriconazole resistance in FSSC isolates. Early detection of possible voriconazole resistance is critical for choosing the correct treatment option for patients with invasive fusariosis.
Introduction
There is heightened concern about invasive mould infections (IMIs) as a consequence of the increasing number of immunocompromised patients (Lass-Flörl and Cuenca-Estrella, 2017; Rotjanapan et al., 2018). Fusaria are among the most frequent cause of IMIs after aspergilli (Nucci and Anaissie, 2007). They are also significant plant pathogens, causing severe vascular wilt and root rot disease in agriculturally important crops (Coleman, 2016; Kazan and Gardiner, 2018; Urbaniak et al., 2018). The Fusarium solani species complex (FSSC) comprises more than 60 species and accounts for ~60% of fusariosis cases worldwide (O'Donnell et al., 2010; Schroers et al., 2016). Fusariosis ranges from localised skin, nail, and eye lesions to disseminated infections (Al-Hatmi et al., 2018). In Singapore, an outbreak of contact lens associated keratitis caused by FSSC species occurred in 2005 involving 66 patients (Jureen et al., 2008). An epidemiological study in Japan, from 1998 to 2015, found that FSSC species accounted for 72.6% of all fusariosis cases, of which 77.8% were invasive fusariosis (IF) (Muraosa et al., 2017). An analysis of a collection of environmental Fusarium isolates from the Malaysian highlands found that 66.1% of 1,449 isolates belonged to the FSSC (Manshor et al., 2012), highlighting their predominance in Malaysia. Reports of clinical FSSC isolates in Malaysia are, however, rare. The first confirmed report was of a patient diagnosed with Fusarium keratitis in 1981 (Singh et al., 1981). Most other studies since then were reports of antifungal drug susceptibility testing of a few Fusarium isolates (Santhanam et al., 2008; Tzar et al., 2013, 2014, 2016). Historically, species of the FSSC were simply referred to as Fusarium solani. However, a Fusarium keratitis outbreak in the United States (Chang et al., 2006) changed that view through the application of multilocus sequence typing (MLST). “F. solani” species were divided into different “clades” based on translation elongation factor 1-alpha (TEF1-α) and RNA polymerase II (RPB2) sequences. The internal transcribed spacer (ITS) regions alone were insufficient to differentiate between species of the FSSC (O'Donnell and Cigelnik, 1997). More recently, Sandoval-Denis and Crous proposed renaming species of the FSSC because they actually belong to the related Neocosmospora genus (Sandoval-Denis and Crous, 2018); Fusarium petroliphilium (FSSC 1 clade) was renamed N. petroliphila, F. keratoplasticum (FSSC 2 clade) was renamed N. keratoplastica, F. falciforme (FSSC 3 clade) was renamed N. falciformis, and strains of the FSSC 7, 20 and 43 clades were named N. gamsii, N. suttoniana, and N. catenate, respectively (Sandoval-Denis and Crous, 2018).
Most Fusarium species have low susceptibilities to the majority of azole antifungals (Tupaki-Sreepurna et al., 2017; Rotjanapan et al., 2018; Herkert et al., 2019) and patients with IFs have high mortality rates (Esnakula et al., 2013; Silva et al., 2013; Okada et al., 2018). Voriconazole (VRC) and amphotericin B (AMB) are the recommended treatment options for localised infections and IFs (Efe İris et al., 2016; Okada et al., 2018). This is despite the fact that most FSSC isolates show relatively low VRC susceptibilities with the majority (369 of 555; 66%) exhibiting a minimum growth inhibitory concentration for VRC (MICVRC) of ≥8 mg/l (Espinel-Ingroff et al., 2016). Unlike most other Ascomycetes that have only one CYP51 gene, moulds of the Pezizomycotina clade have two CYP51 paralogues (CYP51A and CYP51B) (Brillowska-Dabrowska et al., 2015; Ruan et al., 2017; Abastabar et al., 2019), and Fusarium and Neocosmospora species also have a third CYP51 paralogue, CYP51C (Liu et al., 2011; Fan et al., 2013). In A. fumigatus, modifications of CYP51A are major contributors to azole resistance (Chowdhary et al., 2017). However, azole resistance mechanisms of species of the FSSC remain largely unknown.
The roles of the three CYP51 paralogues in growth, ascospore formation, azole resistance, and pathogenicity have been explored to some extent in the related plant fungal pathogen Fusarium graminearum (Becher et al., 2010; Liu et al., 2011; Fan et al., 2013). Although CYP51B accounts for most of the lanosterol 14α-demethylase activity under normal growth conditions, deletion of CYP51B had no effect on growth or viability because upregulation of CYP51A compensated for the loss of its activity (Fan et al., 2013). However, CYP51B deletion reduced ascospore formation and caused an increase in eburicol and 14-methylated sterol content in membranes (Fan et al., 2013). CYP51A was also upregulated in response to azole inhibition, and its deletion caused a 30-fold increase in azole sensitivities (Fan et al., 2013). It appears that, as in A. fumigatus, CYP51A is mainly responsible for the observed azole susceptibilities in F. graminearum (Liu et al., 2011; Fan et al., 2013). Cyp51C had no lanosterol 14α-demethylase activity but it was required for invasion of plant tissue (Fan et al., 2013).
The increasing incidence of IFs (Lortholary et al., 2010; Nucci et al., 2013; Tortorano et al., 2014) and reports of azole resistance caused by the widespread use of azole fungicides in agriculture (Chowdhary et al., 2013; Faria-Ramos et al., 2014; Vaezi et al., 2018) prompted us to study possible azole resistance mechanisms in Neocosmospora species of the FSSC.
Materials and Methods
Fungal Isolates
Fifteen Neocosmospora clinical FSSC isolates were obtained from the Hospital Canselor Tuanku Muhriz UKM & Institute for Medical Research, Malaysia. They had originally been collected from nail, skin, corneal scraping, and blood as part of routine diagnostic procedures. No identifying data from any humans were obtained or utilised in this study. A further 10 were environmental isolates from soil and plant debris (Table 1). All isolates were presumptively identified as Neocosmospora species based on conidia morphology (Leslie and Summerell, 2008). The isolates were grown on potato dextrose agar, PDA (Merck & Co., Kenilworth, USA) with incubation at 28°C for 4 to 7 days.
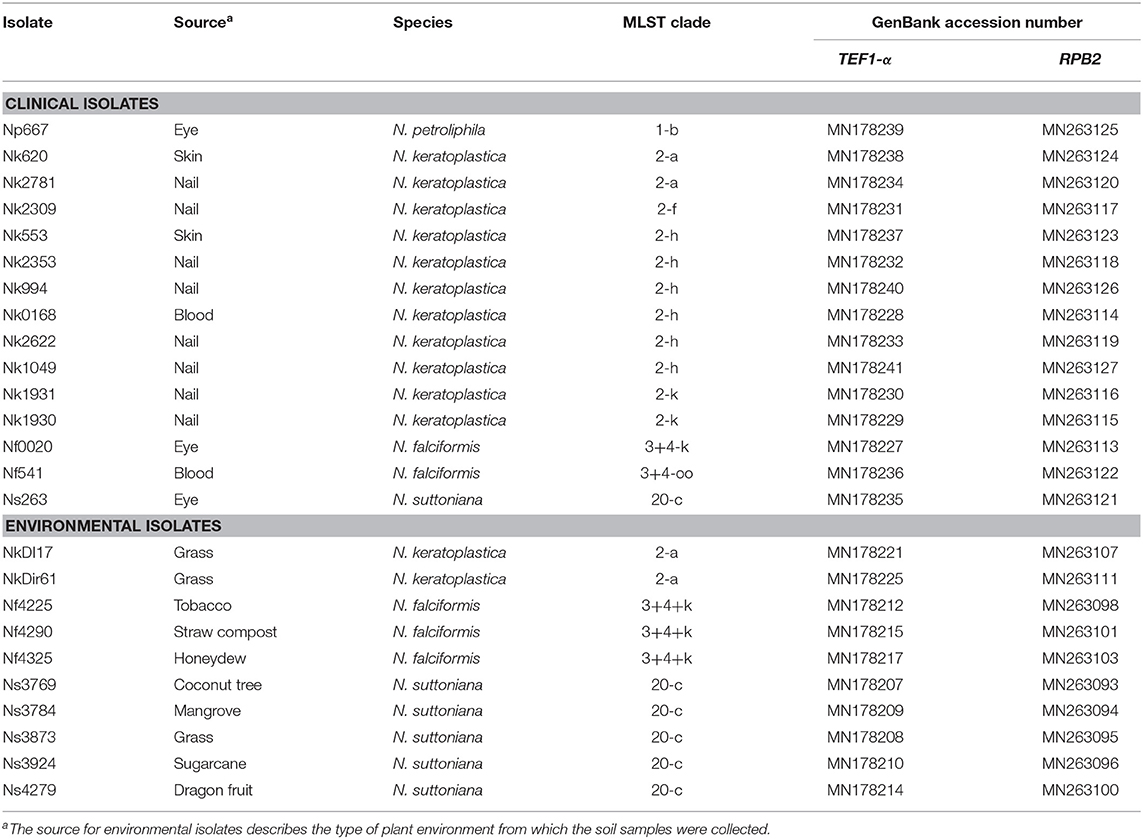
Table 1. List of 15 clinical and 10 environmental FSSC isolates from Malaysia, their MLST clade and GenBank accession numbers for TEF1-α and RPB2.
Molecular Identification and Antifungal Susceptibility Testing
Neocosmospora isolates were identified from their TEF1-α and RPB2 gene sequences (GenBank accession numbers are listed in Table 1) using the Fusarium MLST database (O'Donnell et al., 2008, 2012). Etest susceptibility testing was performed on 1.5% (w/v) agar (Merck & Co., Kenilworth, USA) plates containing 10.4 g/l RPMI 1640 medium R6504 (Sigma-Aldrich, St. Louis, USA), 2% (w/v) glucose (Merck & Co., Kenilworth, USA), and 165 mM MOPS (pH 7.0) (Sigma-Aldrich, St. Louis, USA). Etest strips for itraconazole (ITC), posaconazole (POS), VRC, and AMB were purchased from Biomerieux, Marcy I′Etoile (France). Conidial suspensions (~1–5 × 106 cfu/ml) were prepared according to the CLSI M38-A2 protocol (CLSI, 2008) with a slight modification. The mycelia of a 2-week old culture grown on PDA at 28°C was flooded with ~2 ml sterile 0.85% NaCl and scraped off using a pipette tip followed by filtration through a sterile double layer of gauze to remove large hyphae fragments. The densities of conidial cell suspensions were adjusted to an optical density (OD) of 0.16 measured at 530 nm. Susceptibilities were measured after inoculation of RPMI agar plates with a sterile cotton swab soaked in the conidial suspension, placing Etest strips on top of the cell layer, and incubating the plates at 35°C for 46–50 h. A. fumigatus ATCC 204305 and Candida parapsilosis ATCC 22019 were used as susceptible controls. The MICs were determined according to the Etest reading guide. Because clinical antifungal breakpoints have not yet been established for FSSC species, categorising individual isolates as susceptible, susceptible-dose-dependent, or resistant was not possible. Instead, we used the epidemiological cut-off values (ECVs) determined for FSSC species (Espinel-Ingroff et al., 2016) as a guide to interpret antifungal drug susceptibilities.
Genomic DNA Extraction
Cells of Neocosmospora isolates grown for a week on PDA plates at 28°C were collected in 1.5 ml 0.85% NaCl solution and homogenised in a 2 ml microcentrifuge tube with a mini pestle BioMasher-II (OPTIMA Inc., Itabashi-ku, Japan). The homogenised cell suspension was harvested by centrifugation at 15,339 g for 5 min, and genomic DNA (gDNA) was extracted from the cell pellet with a DNeasy Plant Extraction Kit (QIAGEN Inc., Valencia, USA) according to the manufacturer's instructions.
Amplification of CYP51 Genes
DNA oligomer primers used in this study are listed in Supplementary Table S1. Primers were initially designed using the only Neocosmospora genome sequence available at the time, the teleomorph Nectria haematococca mpVI 77-13-4 (Coleman et al., 2009). N. haematococca CYP51A, CYP51B, and CYP51C were identified with BLAST searches using the Cyp51A, Cyp51B, and Cyp51C (GenBank accession nos. XP_011321548, XP_011316750, and XP_011325340, respectively) (Cuomo et al., 2007) sequences of F. graminearum PH-1 as queries. CYP51A, CYP51B, and CYP51C, including parts of their 5′-upstream and 3′-downstream ORF sequences and parts of the housekeeping genes β-actin (ACT1) and GAPDH (GPD1), were amplified by PCR using gDNA extracts of N. keratoplastica Nk553, Nk2309, and Nk2781 as DNA templates and sequencing the DNA fragments. PCR amplifications were performed with HotStarTaq Master Mix (QIAGEN Inc., Valencia, USA) using 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 1 min on a T100™ thermal cycler (Bio-Rad, Hercules, USA). DNA sequences were submitted to GenBank and accession numbers for CYP51A, CYP51B, CYP51C, ACT1, and GPD1 are listed in Supplementary Table S2. N. keratoplastica CYP51A, CYP51B, CYP51C, ACT1, and GPD1 sequences were used to design species specific primers for qPCR amplification (Supplementary Table S1).
Phylogenetic Tree of N. keratoplastica CYP51 Orthologs
Introns of the CYP51 paralogues of N. keratoplastica Nk2781 were verified by PCR amplification and DNA sequencing of the cDNA. Protein alignments of related sequences were created with CLUSTALW (Thompson et al., 1994) and manually corrected if necessary. Phylogenetic inferences using two independent algorithms, Maximum Parsimony (MP) and Maximum Likelihood (ML), with 1,000 bootstrap (BS) replicates were performed using the Molecular Evolutionary Genetics Analysis (MEGA) software version 10.0.5 (Kumar et al., 2018). GenBank accession numbers for the Cyp51/Erg11 sequences that were used for the phylogenetic analysis are listed in Supplementary Table S3.
Total RNA Extraction
Conidial suspensions (10 μl of ~1 ×108 cfu/ml) of three N. keratoplastica isolates with MICVRCs > 32 mg/l (Nk2309, Nk2781, NkDI17) and three with MICVRCs ≤ 8 mg/l (NkDir61, Nk553, Nk994) were used to inoculate 50 ml potato dextrose broth (PDB) (Merck & Co., Kenilworth, USA) and incubated at 30°C for 21 h with shaking at 200 rpm. Logarithmic cells were harvested by filtration through a glass fibre round filter No. 6 (Schleicher & Schuell BioScience GmbH, Dassel, Germany) using a vacuum manifold and washed once with ~5 ml ice-cold distilled water. The “cell cake” was scraped off the filter with a scalpel, quickly transferred into a 1.5 ml microcentrifuge tube, snap frozen in liquid nitrogen, and stored at −80°C. Total RNA was extracted from frozen cell pellets (~100 mg) with a hot-phenol extraction protocol. In short, frozen cell pellets were dropped into a 15 ml Corex tube containing a mixture of 1 ml acid phenol (saturated with SAB buffer; 50 mM sodium acetate, 10 mM EDTA, pH 5.0), 2 ml SAB buffer, 100 μl 10% SDS, and ~1 g 0.5 mm acid-washed zirconia beads (BioSpec Products, Bartlesville, USA), at 65°C. The cells were broken and RNA released into the water phase by five cycles of 30 s thorough vortexing with 1.5 min incubation at 65°C in between each cycle. Liquid phases were separated by centrifugation at 10,000 g for 10 min and traces of phenol in the ~2 ml upper phase removed by chloroform extraction. Total RNA was ethanol precipitated, harvested by centrifugation at 30,000 g for 30 min, air dried for 10 min, and resuspended in 200 μl RNAse-free water. Traces of gDNA were removed by DNase treatment of samples with a PureLink DNase kit (Invitrogen Inc., Carlsbad, USA). Spectrophotometrically determined RNA concentrations and RNA integrity of total RNA extracts were confirmed by RNA gel electrophoresis (Supplementary Figure S1).
qPCR Quantification of mRNA Expression Levels
First strand cDNA was synthesised from 1 μg total RNA using the SuperScript IV VILO Master Mix (Invitrogen Inc., Carlsbad, USA) following the manufacturer's instructions. Quantification of CYP51A, CYP51B, and CYP51C cDNA by real-time PCR using Fast SYBR Green Master Mix (Applied Biosystems Inc., Foster City, USA) was performed with the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems Inc., Foster City, USA). The qPCR assay contained 1X Fast SYBR Green Master Mix, 400 nM each of forward and reverse primer, and 5 ng of cDNA template. Thermal cycling steps included a 95°C denaturation step for 20 s followed by 40 cycles of denaturation at 95°C for 3 s and DNA synthesis at 60°C for 30 s. Four 10-fold serial dilutions (5–0.005 ng) of first strand cDNA templates were used to determine the linear amplification range and the amplification efficiencies for each qPCR primer pair. A H2O negative control without cDNA template and a -RT control were included. An average quantification cycle (Cq) value for each sample was calculated from two technical replicates. mRNA transcript levels (2−ΔCq) were normalised to the referenced housekeeping genes ACT1 and GPD1. The fold-change of CYP51A mRNA levels relative to those in the strain with the lowest MICVRC (1 mg/l), Nk994, was calculated using the ΔΔCq method (2−ΔΔCq) (Livak and Schmittgen, 2001). Fold-change values between zero and one were expressed as fold-regulation [−1/(2−ΔΔCq)].
CYP51A, CYP51B, and CYP51C mRNA Expression Levels in Cells Grown in the Presence of VRC for 4 h or Exposed to Various Stress Conditions for 20 min
Total RNA was extracted from 50 ml logarithmic phase cell cultures of Nk2781 or Nk994 (i.e., cells were grown in PDB for 21 h at 30°C, as described above) that had been exposed for 0, 20, 40, 80, or 240 min to 16 mg/l VRC, or for 20 min to various stress conditions: 2 mM H2O2, 500 mM NaCl, pH 7.0 (165 mM MOPS), 37°C. A no-stress control was included by adding an equivalent volume of sterile H2O to the cells.
Results
Antifungal Susceptibilities of 15 Clinical and 10 Environmental Neocosmospora Isolates
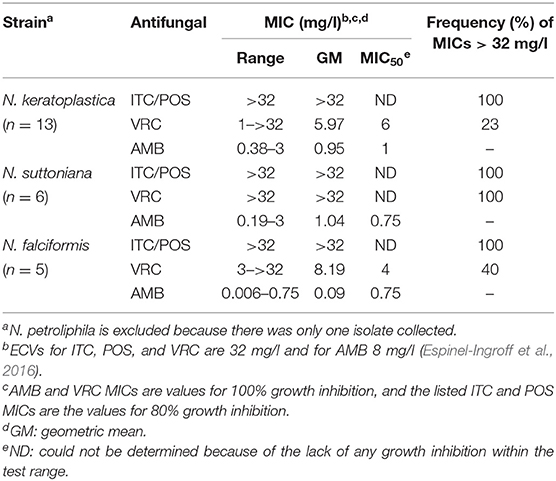
Thirteen of the 25 FSSC isolates were classified as N. keratoplastica (FSSC 2), five as N. falciformis (FSSC 3+4), six as N. suttoniana (FSSC 20), and one as N. petroliphila (FSSC 1) (Table 1). As expected (Espinel-Ingroff et al., 2016; Herkert et al., 2019), all 25 FSSC isolates had very high (>32 mg/l) MICs for the long-tailed azoles, ITC, and POS. Initially, the susceptibility of 15 clinical FSSC isolates to ITC was measured and all isolates showed very high MICITCs (>32 mg/l), and so we predicted the same or higher MICITCs in FSSC environmental isolates. Therefore, we measured the susceptibility of 10 environmental isolates to another long-tailed azole, POS, instead. All 25 FSSC isolates showed moderately low MICAMBs (≤3 mg/l) (Tables 2, 3, Supplementary Figure S2). However, the in vitro VRC susceptibilities divided into two groups: 14 group I isolates with MICVRCs ≤ 12 mg/l and 11 group II isolates that did not respond to VRC at all (MICs > 32 mg/l; Table 2). All clinical (1) and environmental (5) N. suttoniana (FSSC MLST clade 20-c) isolates had high MICVRCs (>32 mg/l). The five N. falciformis isolates divided into two groups with the three environmental isolates belonging to group I (MICs of 3 or 4 mg/l) and the two clinical isolates to group II (MICs > 32 mg/l), respectively. The majority (10 of 13; 77%) of N. keratoplastica isolates and the only N. petroliphila clinical isolate were group I isolates with MICVRCs ranging from 1 to 12 mg/l. However, two clinical N. keratoplastica isolates (Nk2781 and Nk2309) and one environmental isolate (NkDI17) were group II isolates with high MICVRCs > 32 mg/l (Tables 2, 3).
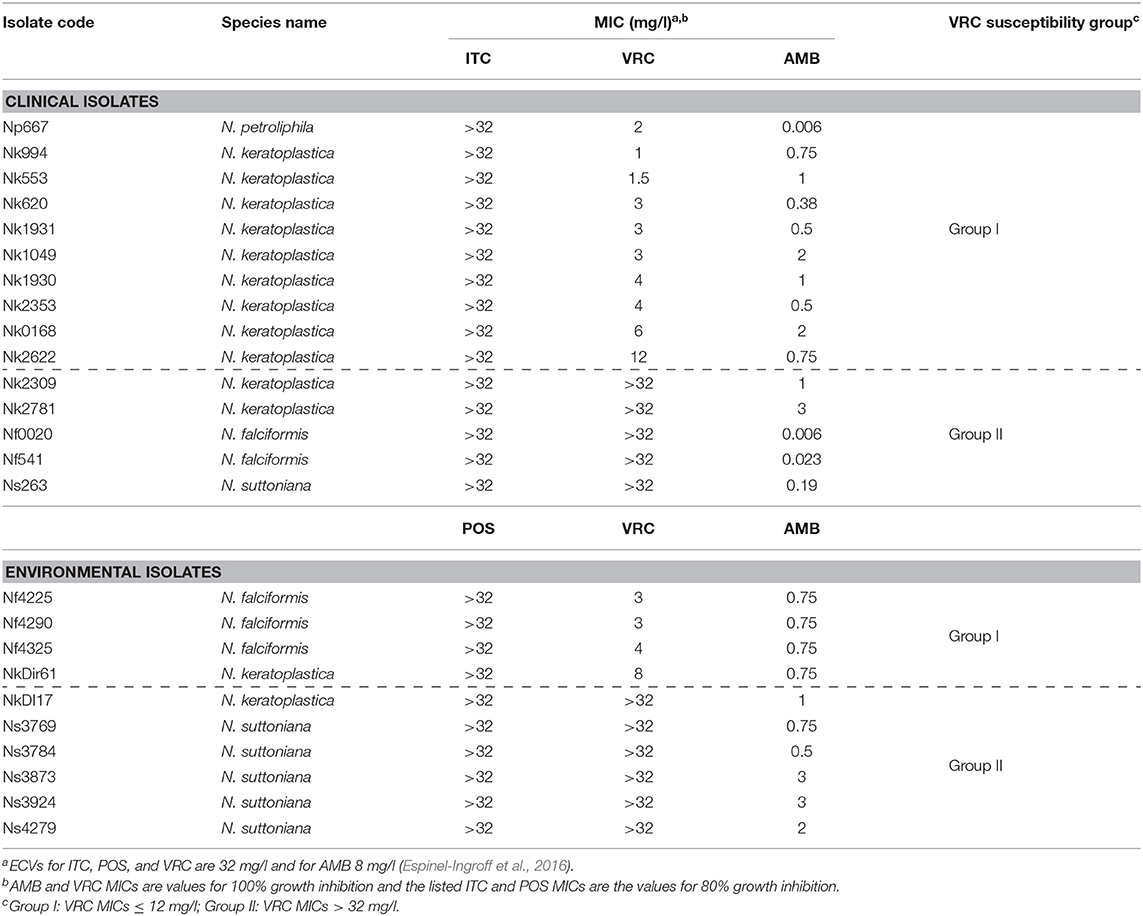
Table 2. Etest antifungal susceptibilities (MICs) of 15 clinical and 10 environmental FSSC isolates.
N. keratoplastica CYP51A, CYP51B, and CYP51C
All further investigations were performed with N. keratoplastica because these isolates were the most frequently isolated FSSC species in the clinic (11 of 15 strains; 73%, Table 2). Using N. haematococca mpVI 77-13-4 CYP51A, CYP51B, and CYP51C sequences as a guide, we isolated and sequenced the three orthologous ORFs of N. keratoplastica Nk2781, including parts of their 5′ upstream and 3′ downstream sequences. CYP51A, CYP51B, and CYP51C of N. keratoplastica Nk2781 consisted of 1,583, 1,753, and 1,651 nucleotides, respectively (Figure 1). CYP51A had one intron (62 bp) and CYP51B (114 bp and 55 bp) and CYP51C (47 bp and 53 bp) each had two introns (Figure 1). All introns were verified by sequencing the ORFs cDNA. Although the intron positions were conserved relative to N. haematococca, their sizes varied slightly: intron 1 and intron 2 of CYP51B were 4 bp and 2 bp larger and intron 1 of CYP51C was 2 bp larger than their N. haematococca counterparts. As in N. haematococca, CYP51A, CYP51B, and CYP51C encoded proteins of 506, 527, and 516 amino acids, respectively. N. keratoplastica Cyp51A was 56% identical to Cyp51B and 47% identical to Cyp51C. The relationship of the three Nk2781 Cyp51 paralogues with those of other Fusaria species and with those from plants and mammals is illustrated in Figure 2. The three paralogues formed three distinct phylogenetic branches with their respective N. haematococca, F. graminearum, F. oxysporum, and F. verticillioides orthologs (Figure 2).
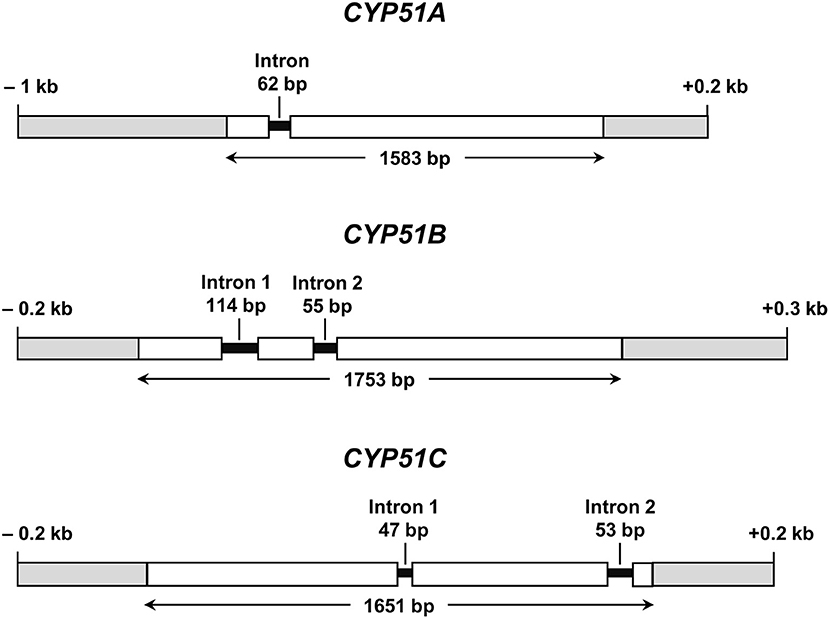
Figure 1. Graphical representations of N. keratoplastica CYP51A, CYP51B, and CYP51C (GenBank accession numbers MN296719, MN296724, and MN296725, respectively). Open boxes indicate ORF sequences, and grey boxes are upstream and downstream regions. Introns and their ORF positions are indicated with black lines. Bidirectional arrows underneath ORFs indicate the size of the entire ORF including introns.
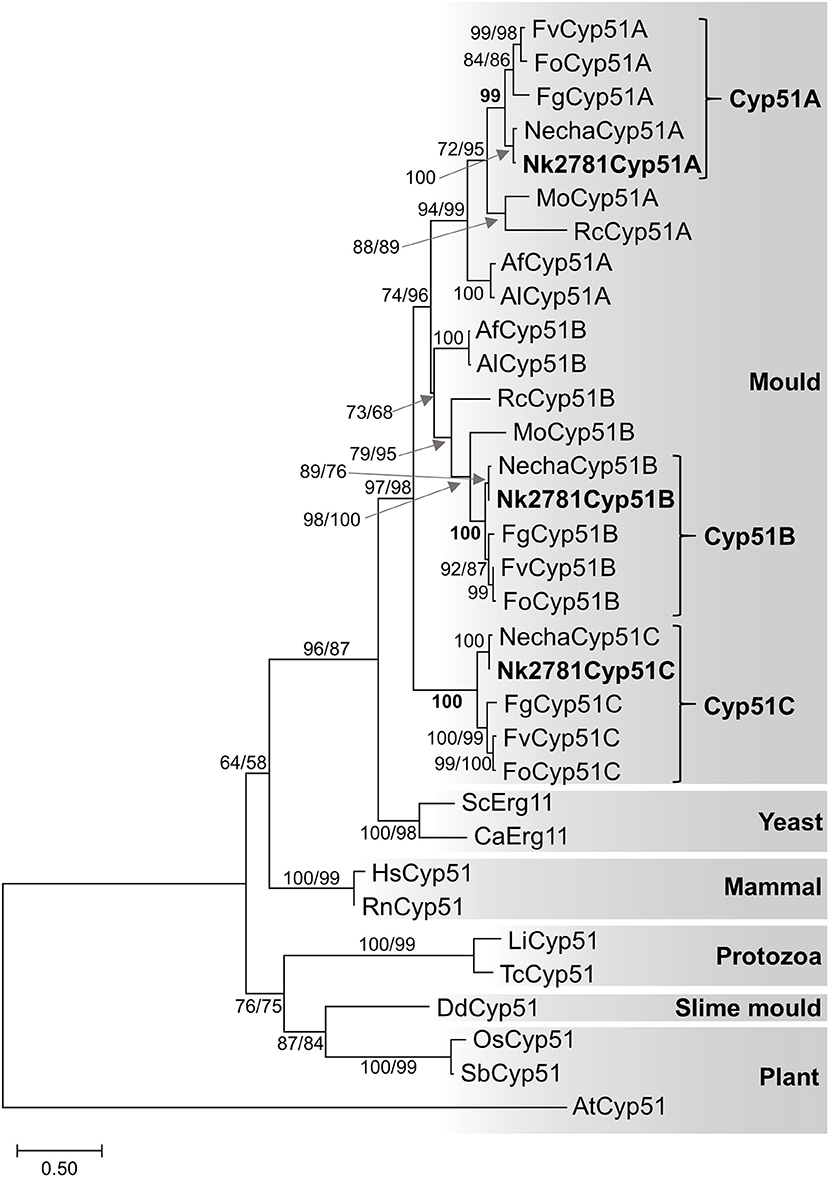
Figure 2. Maximum likelihood phylogram of Cyp51/Erg11 sequences of selected eukaryotes. Species names are abbreviated as: Af, A. fumigatus; Al, A. lentulus; Ca, Candida albicans; Fg, F. graminearum; Fo, F. oxysporum; Fv, F. verticillioides; Mo, Magnaporthe oryzae; Necha, Nectria haematococca; Rc, Rhynchosporium commune; Sc, Saccharomyces cerevisiae; protozoans Li, Leishmania infantum and Tc, Trypanosoma cruzi; slime mould Dd, Dictyostelium discoideum; mammals Hs, Homo sapiens and Rn, Rattus norvegicus; plants, AT Arabidopsis thaliana, Os, Oryza sativa and Sb, Sorghum bicolor. The A. thaliana Cyp51 sequence was used as the outgroup. Nk2781 Cyp51A, -B and -C are in bold letters. Numbers at internodes represent the percentage of maximum parsimony (MP) and maximum likelihood (ML) bootstrap support (MP-BS/ML-BS) of 1,000 replicates; a single number means both values were identical. The scale bar indicates the number of amino acid substitutions per position (the alignment contains 398 residues).
A Conserved Leucine at the Entry Gate of Cyp51A in Neocosmospora and Fusarium spp Possibly Contributes to Long-Tailed Azole Resistance
To identify possible changes in CYP51A that may explain the VRC resistance phenotype of group II N. keratoplastica isolates, we sequenced the entire ORF of eight clinical isolates: six group I strains, Nk994, Nk553, Nk1931, Nk620, Nk1049, and Nk2622 and two group II strains, Nk2309 and Nk2781. There were 12 non-synonymous SNPs, all in positions that have not been reported to be associated with azole resistance in ascomycetous fungi like A. fumigatus (Howard et al., 2009; Wiederhold et al., 2016; Moore et al., 2017) (see Supplementary Figure S3, Table S4). One notable exception—although obviously not the reason for VRC resistance because it was conserved across all eight N. keratoplastica isolates—was L218 in the equivalent position to M220 of A. fumigatus Cyp51A. Mutations of Cyp51A-M220 to I, K, T, or V residues were associated with azole resistance in A. fumigatus especially with resistance to the long-tailed azoles ITC and POS (Supplementary Table S4) (Howard et al., 2009; Wiederhold et al., 2016) and were predicted to affect the substrate entry gate (Snelders et al., 2010). The N. keratoplastica Cyp51A-L218 equivalent residues in N. keratoplastica Cyp51B and Cyp51C were M236 and V228, respectively. Interestingly, these three AfuCyp51A-M220 equivalent residues (L218, M236, and V228) were conserved across all three Fusarium and Neocosmospora Cyp51A, Cyp51B, and Cyp51C paralogs (Figure 3).
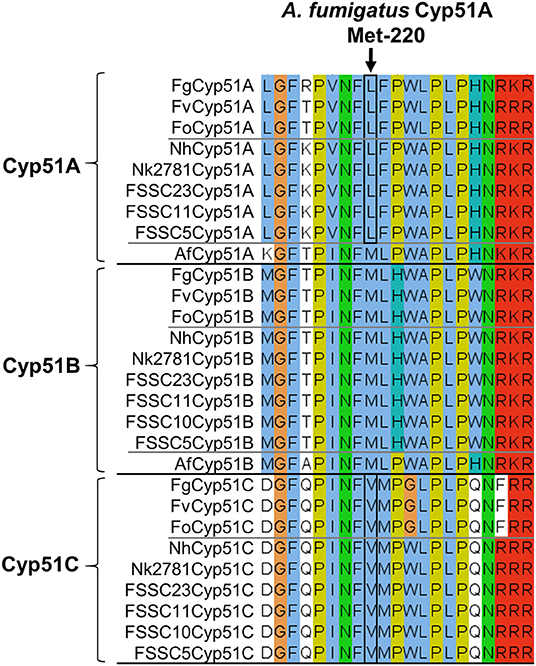
Figure 3. Alignment of Cyp51A, Cyp51B, and Cyp51C orthologs of three Fusarium species (top), six FSSC species including N. keratoplastica Nk2781 (centre) and A. fumigatus (it has only two Cyp51 orthologs, Cyp51A, and Cyp51B; bottom). FSSC10 also had only two Cyp51 orthologs, Cyp51B, and Cyp51C. The A. fumigatus Cyp51A-M220 equivalent residue is indicated with an arrow, and the Fusarium and FSSC equivalent Cyp51A-L and Cyp51C-V residues are encircled in black.
A 23 bp CYP51A Promoter Deletion Was Tightly Associated With VRC Resistance in 9 Group II FSSC Isolates
Sequencing the CYP51A promoter from ten N. keratoplastica isolates revealed a 23 bp promoter deletion (from −551 to −528) that was only present in the three group II (MICVRC > 32 mg/l) isolates (Figure 4, Table 3). There were no other group II specific sequences within the first 750 nucleotides upstream of the CYP51A ATG start codon. To our surprise, we found the same 23 bp promoter deletion in all six N. suttoniana FSSC isolates as well, while the three N. falciformis FSSC isolates with MICVRCs ≤ 4 mg/l had an almost identical 23 bp sequence as the seven group I N. keratoplastica isolates with MICVRCs ≤ 12 mg/l (one nucleotide difference was observed at position −539 “T” in N. keratoplastica and a “C” in N. falciformis; Figure 4). Thus, the tight association of VRC resistance (MIC > 32 mg/l) with a 23 bp CYP51A promoter deletion was conserved across species boundaries and their origin of isolation; group I (MICVRCs ≤ 12 mg/l) and group II (MICVRCs > 32 mg/l) isolates contained both clinical as well as environmental FSSC isolates. The CYP51A promoter sequences of FSSC5 and FSSC23, retrieved from the Joint Genome Institute (JGI) database, aligned reasonably well with the sequences of the group I FSSC isolates. It is also important to note that there were other consistent mutations within the group II isolates: a “T” to “C” nucleotide change and two nucleotide “AC” insertion at positions −521 and −516 to −517, respectively. However, those mutations are less likely to be involved in the altered VRC susceptibility as they were also present in FSSC5 and FSSC23 CYP51A promoter sequences which did not carry the 23 bp deletion.
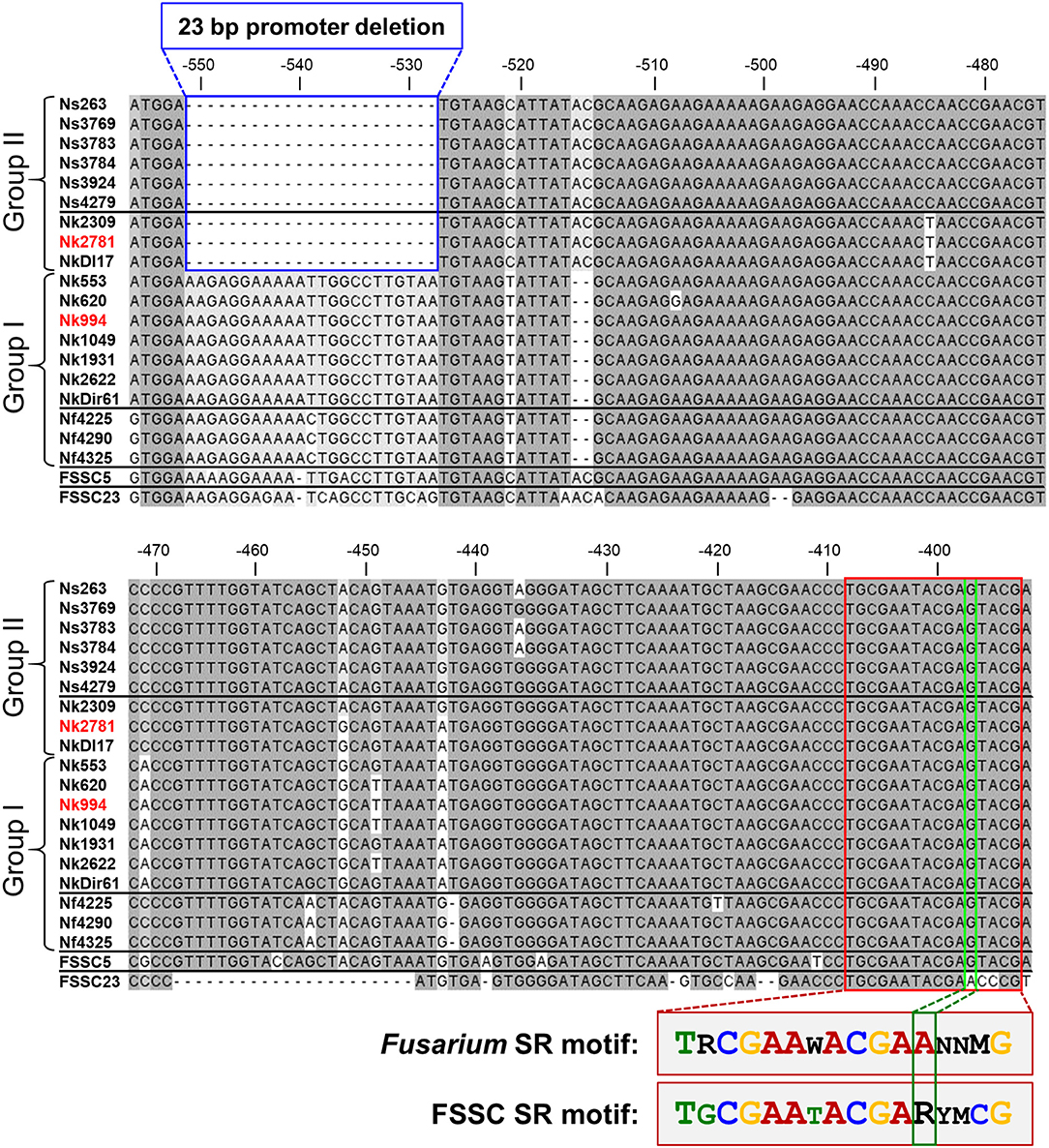
Figure 4. CYP51A promoter alignment (from −556 to −392 upstream of the Nk2781 CYP51A ATG start codon) of 19 FSSC isolates (6 N. suttoniana, 10 N. keratoplastica, 3 N. falciformis) and of FSSC5 and FSSC23 that were used as controls. The 23 bp CYP51A promoter deletion of FSSC isolates (Group II; 3 N. keratoplastica, 6 N. suttoniana) with high MICVRCs (>32 mg/l) are bordered in blue. FSSC isolates (Group I; 7 N. keratoplastica, 3 N. falciformis) with lower MICVRCs (≤12 mg/l) are underneath the group II sequences. The putative 16-bp sterol regulatory cis-element that was conserved in all FSSC isolates is bordered in red. There was one major change (bordered in green) to the core CGAA-NN-CGAA sterol regulatory (SR) binding motif of FgSR (Liu et al., 2019). FSSC23 had the conserved CGAA-NN-CGAA motif in the same position. A comparison of the conserved FSSC SR motif with the previously reported FgSR motif is provided underneath the alignment. The sequences of the red highlighted N. keratoplastica strains, Nk2781, and Nk994, were used for the VRC induction experiments.
To explore the possibility that the 23 bp CYP51A promoter deletion of group II FSSC isolates affected a potentially important transcription factor (TF) binding site(s), we searched the entire CYP51A promoter for the recently identified TF binding site of the major sterol regulatory protein of F. graminearum, FgSR (Liu et al., 2019). FgSR is a zinc-cluster family TF that binds as a homodimer to a 16-bp cis-element containing two conserved CGAA repeats separated by 2 nucleotides (Figure 4). All 19 FSSC isolates sequenced had one conserved, although slightly modified (i.e., the distal CGAA repeat element was changed to CGAG in most FSSC species; Figure 4), 16 bp sterol regulatory binding site ~120 bp downstream (−409–393) of the 23 bp CYP51A promoter deletion (Figure 4).
CYP51A and CYP51B Transcript Levels
The amount of total RNA obtained from 50 ml N. keratoplastica cell cultures ranged from 120 to 400 μg (260/280 ratio: 1.83–2.01). The qPCR assay amplification efficiencies using cDNA as template and qPCR primers designed according to N. keratoplastica sequences were between 100 and 108% for all amplicons. We selected GPD1 for transcript level normalisation (2−ΔCq) and to calculate fold changes (2−ΔΔCq). Although generally much lower than CYP51B, the normalised CYP51A mRNA expression levels of the three group II strains were 1.3, 2.4, and 2.6 times higher than in the strain with the lowest MICVRC (1 mg/l), Nk994 (Figure 5). The highest CYP51A mRNA expression level was observed in strain NkDI17 (group II) which was 7.5 times higher than the lowest, in strain NkDir61 (group I). The normalised CYP51B transcript levels, however, hardly varied at all between the six strains (Figure 5). As expected, N. keratoplastica CYP51B mRNA levels were much higher than CYP51A. They were, on average, 859 and 295 (Table 4) times higher than CYP51A mRNA levels in the three group I and the three group II strains, respectively (Figure 5). However, the difference between the mean mRNA level fold differences (CYP51B/CYP51A) for each group was not statistically significant (p-value = 0.20).
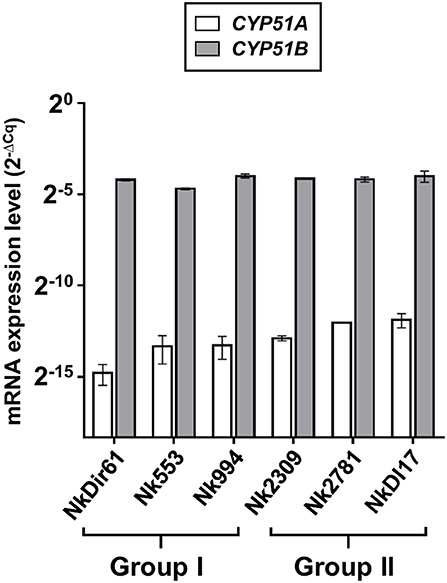
Figure 5. Quantification of CYP51A and CYP51B mRNA expression levels (2−ΔCq) in logarithmic phase cells of six N. keratoplastica isolates (group I: NkDir61, Nk553, Nk994; group II: Nk2309, Nk2781, NkDI17). The fold differences between the GPD1 normalised CYP51B and CYP51A mRNA levels and their group averages are listed in Table 4.
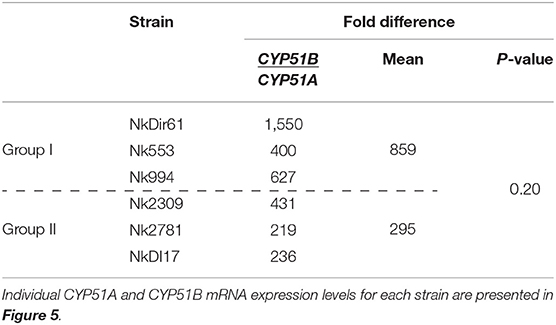
Table 4. Fold differences between GPD1 normalised CYP51A and CYP51B mRNA expression levels (2−ΔCq) of logarithmic phase cells of six N. keratoplastica isolates.
VRC Induced CYP51A mRNA Expression ~6,500-Fold Reaching Levels Similar to CYP51B
The amount of total RNA obtained from 50 ml N. keratoplastica cell cultures (harvested at t = 0, 20, 40, 80 or 240 min) ranged from 9 to 241 μg (260/280 ratio: 1.95–2.01). At time zero of the VRC induction experiment, the CYP51A mRNA levels of logarithmic phase Nk2781 cells were 230 and 5 times lower than those of CYP51B and CYP51C, respectively, while CYP51A mRNA levels of Nk994 cells were 802 and 11 times lower, respectively (Figure 6A). However, after 80 min VRC exposure, CYP51A mRNA levels reached levels that were comparable to CYP51B, but they remained higher (1.6- to 2.2-fold) in Nk2781 than in Nk994 throughout the 240 min induction period, although they were slightly lower (1.3-fold) at the 80 min time point (Figure 6A). The transcript levels of CYP51C remained lower than CYP51B throughout the same 240 min time period (i.e., 40 times lower at time zero and 200 times lower at 240 min in Nk2781; 20 and 90 times lower in Nk994). Although all three CYP51 paralogues of Nk2781 and Nk994 were induced by VRC, VRC induction was most pronounced for CYP51A (5,884 and 7,072-fold upregulation, respectively), less pronounced for CYP51B (22- and 9-fold upregulation, respectively), and even less so for CYP51C (6- and 3-fold upregulation, respectively; Figure 6B). Exposing logarithmic cells of the same two strains for 20 min to four different stress conditions, high salt (500 mM NaCl), high pH (pH 7.0), heat (37°C), and oxidative stress (2 mM H2O2), resulted in changes in CYP51 mRNA expression. There was moderate downregulation of all three CYP51 paralogues in both strains in response to salt (1.9- to 3.6-fold) and heat stress (1- to 15-fold). CYP51A transcript levels were increased (1.8- and 3-fold higher), however, when exposed to high pH. CYP51C transcript levels were particularly strongly affected (12- and 15-fold downregulation) by heat stress (Table 5). Exposing either N. keratoplastica strain to oxidative stress had little effect on all three CYP51 paralogue expression levels (Table 5).
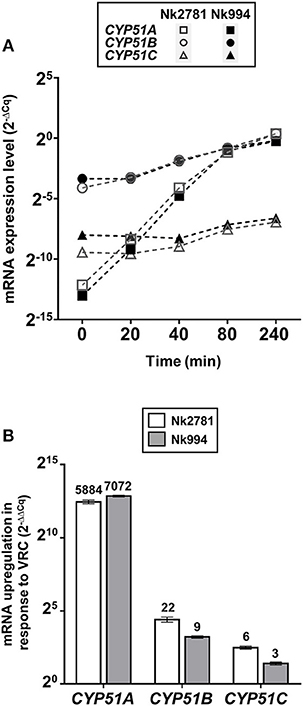
Figure 6. VRC induction of CYP51A, CYP51B and CYP51C. (A) GPD1 normalised CYP51A, CYP51B, and CYP51C mRNA expression levels (2−ΔCq) of Nk2781 (MICVRC > 32 mg/l) and Nk994 (MICVRC = 1 mg/l) N. keratoplastica strains harvested at the indicated times (0–240 min) after VRC induction (16 mg/l) of logarithmic phase cells grown for 21 h at 30°C in 50 ml PDB medium with shaking at 150 rpm. (B) Graph of the fold changes (2−ΔΔCq) of the GPD1 normalised CYP51A, CYP51B, and CYP51C mRNA expression levels for both strains after 240 min of VRC induction relative to the expression levels of uninduced logarithmic cells at time zero. Numbers on top of the bars indicate fold changes.
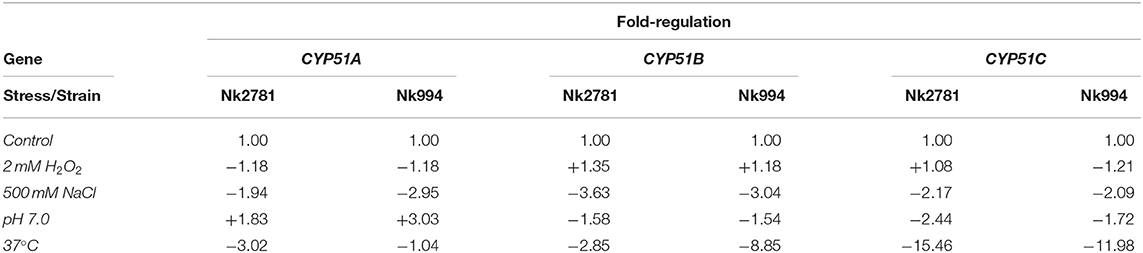
Table 5. Fold increased (+) or decreased (–) GPD1 normalised CYP51A, CYP51B, and CYP51C mRNA levels of Nk2781 (MICVRC > 32 mg/l) and Nk994 (MICVRC = 1 mg/l) cells exposed for 20 min to the indicated stress conditions.
Discussion
The 25 FSSC isolates from across Malaysia comprised 13 N. keratoplastica, six N. suttoniana, five N. falciformis, and one N. petroliphila isolate. N. falciformis strains were equally distributed among clinical (2) and environmental (3) isolates. However, most (11; 85%) N. keratoplastica and N. petroliphila strains were clinical isolates while most (6; 86%) N. suttoniana strains were of environmental origin. The species distribution of the 15 clinical FSSC isolates agreed with previous reports that identified N. petroliphila and N. keratoplastica (FSSC types 1 and 2) as the most frequent FSSC species isolated in the clinic (Chang et al., 2006; O'Donnell et al., 2012). N. keratoplastica was reported to cause invasive fusariosis in hematologic patients (Chiewchanvit et al., 2017), keratitis (Tupaki-Sreepurna et al., 2017), onychomycoses (Guevara-Suarez et al., 2016), and eumycetoma (Al-Hatmi et al., 2017a). It has also been reported to cause disease in equine and marine vertebrates and invertebrates (O'Donnell et al., 2016).
A high degree of agreement between the Etest, the EUCAST (96–100%), and the CLSI (90–100%) protocols for ITC, VRC, and AMB susceptibilities in Fusarium/Neocosmospora species has been reported (Tortorano et al., 2014; Al-Hatmi et al., 2017b). Although there are no clinical breakpoints for this fungal group, ECVs were established in 2016 (Espinel-Ingroff et al., 2016). The antifungal susceptibilities of our 25 FSSC isolates agreed with previous studies (Tortorano et al., 2014; Espinel-Ingroff et al., 2016) that reported high ITC (≥16 mg/l) and POS (≥8 mg/l) and variable VRC (0.5 to ≥16 mg/l) MICs for most FSSC isolates. The high (≥32 mg/l) MICVRCs of 44% of the isolates [5 clinical (33%) and 6 environmental (60%)] is of serious concern given that VRC is a recommended drug for the treatment of IF (Pascual et al., 2008; Efe İris et al., 2016). AMB appears to be the only effective treatment option for IF (Espinel-Ingroff et al., 2016). Although two recently introduced imidazoles, luliconazole, and lanoconazole, have shown promising in vitro activities against various Fusarium and Neocosmospora isolates (Abastabar et al., 2018; Todokoro et al., 2019) their application appears limited to topical treatment of superficial fungal infections (Scher et al., 2014; Gupta and Daigle, 2016). Combination antifungal therapy is a potentially useful alternative for difficult-to-treat invasive mould infections. In vitro synergies between VRC and micafungin (Heyn et al., 2005), VRC and terbinafine (Córdoba et al., 2008), and VRC and AMB antifungal drug combinations (Ho et al., 2007) have been reported for various Fusarium species. In addition, the successful treatment of a patient with IF using a VRC and liposomal AMB combination was recently reported (Efe İris et al., 2016).
We found no N. keratoplastica Cyp51A mutations that were associated with high MICVRCs (>32 mg/l) of group II isolates Nk2781 and Nk2309. However, the Cyp51A of all N. keratoplastica isolates (Supplementary Figure S3), including all other sequenced Fusarium and Neocosmospora FSSC species, had a conserved leucine in a position that was equivalent to M220 of A. fumigatus Cyp51A (Figure 3). Mutation of A. fumigatus Cyp51A-M220I caused ITC resistance and 4-fold increased MICVRCs in two separate ITC resistant clinical A. fumigatus isolates (Chen et al., 2005; Snelders et al., 2010). Previous studies proposed that mutating M220 in a loop region near the substrate entry channel into the central binding cavity of Cyp51A may block the access of larger azoles like ITC with its long hydrophobic tail (Howard et al., 2009; Snelders et al., 2010). The A. fumigatus Cyp51A-G54 and -M220 equivalent residues lining the substrate/inhibitor entry channel in the crystal structure of S. cerevisiae Cyp51 (Monk et al., 2014) bound to ITC is depicted in Supplementary Figure S4. Perhaps the conserved L218 residue at the substrate entry channel of N. keratoplastica Cyp51A, and all other Fusarium and Neocosmospora FSSC species (Figure 3), is one of the reasons why these moulds do not respond well to azole antifungals with long hydrophobic tails (ITC and POS).
Most interesting, however, was the discovery of a 23 bp CYP51A promoter deletion in all nine group II (MICVRCs > 32 mg/l) FSSC isolates (three N. keratoplastica and six N. suttoniana; Figure 4) that was absent in all ten group I (MICVRCs ≤ 12 mg/l) FSSC isolates (seven N. keratoplastica and three N. falciformis). Remarkably, this tight association of the 23 bp CYP51A promoter deletion with the high VRC resistance phenotype of group II isolates reached across species boundaries and was independent of their clinical (e.g., Nk2781, NkDI17) or environmental (e.g., Nk994, NkDir61) origin (Figure 4). Similar promoter mutations resulted in pan-azole, including VRC, resistance (Wiederhold et al., 2016) in clinical A. fumigatus isolates that were possibly selected for by excessive agricultural use of azole antifungals (Chowdhary et al., 2013; Faria-Ramos et al., 2014; Vaezi et al., 2018). Unfortunately, we were not able to confirm an association between VRC resistance and the 23 bp CYP51A promoter deletion in all N. falciformis isolates, for technical reasons. Despite a number of efforts, we simply could not amplify the CYP51A promoter from the two clinical isolates (Nf0020 and Nf541) which showed high MICVRCs > 32 mg/l. We suspect that their CYP51A 5′ upstream regions had sequence variations that prevented the primers designed against N. haematococca CYP51A to recognize their target sequence. Nevertheless, the tight association between VRC resistance and a 23 bp CYP51A promoter deletion preserved across species boundaries and independent of origin of isolation in the remaining 19 FSSC isolates was rather convincing. The collected evidence suggests two equally possible scenarios. Either the 23 bp CYP51A promoter deletion of group II FSSC isolates is a sequence variant of ancient origin (millions of years) that evolved before separation of individual FSSC species, or, alternatively, rare sexual recombination events between closely related FSSC species caused the exchange of the 23 bp CYP51A promoter deletion in the somewhat distant past (decades ago). Otherwise we would not expect to find the following species-specific SNPs: i) N. falciformis group I isolates had a C instead of a T inside the 23 bp promoter region that was deleted in group II isolates; ii) the three N. keratoplastica group II isolates had a unique T 45 bp downstream of the 23 bp promoter deletion, even though iii) they had identical sequences immediately surrounding the 23 bp promoter deletion with all other N. suttoniana group II isolates (Figure 4). Clearly, further investigations are necessary to distinguish between these two possibilities of: (i) an ancient CYP51A sequence variant shared between some closely related FSSC species, or (ii) a rather recent exchange of genetic material across species boundaries through rare sexual recombination events between closely related FSSC species that was perhaps selected for by the agricultural use of azoles over the past few decades.
Although VRC induced CYP51A mRNA expression in both Nk994 (group I) and Nk2781 (group II) to a similar extent, the CYP51A expression levels were consistently higher throughout the 4 h induction period (1.6- to 2.2-fold) in Nk2781 (Figure 6A). The ~6,500-fold increased CYP51A mRNA expression levels suggest that, like in A. fumigatus (Abastabar et al., 2019) and the plant fungal pathogen F. graminearum (Liu et al., 2011; Fan et al., 2013), CYP51A of N. keratoplastica is also a key player in the observed azole resistance phenotype of FSSC species. Sterol biosynthesis of Saccharomycotina species like S. cerevisiae or C. albicans is regulated by the sterol regulatory zinc-cluster TF Upc2 (Yang et al., 2015; Popp et al., 2017). Sterol biosynthesis of many other eukaryotes, including fungal species like A. fumigatus, C. neoformans and Schizosaccharomyces pombe, and also mammals including Homo sapiens, is regulated by a different type of sterol regulator: a TF called SREBP. A third type of sterol biosynthesis regulator, the zinc-cluster TF FgSR, has recently been discovered in the plant fungal pathogen Fusarium graminearum (Liu et al., 2019). This type of sterol regulatory network exists only in Sordariomycetes and Leotiomycetes fungi including Neocosmospora species of the FSSC. FgSR homodimers bind to a 16 bp cis-element of target gene promoters containing two conserved CGAA repeats. Sterol depletion triggers the activation of FgSR via MAP-kinase FgHog1 phosphorylation which, in turn, induces FgSR interaction with the chromatin remodelling complex SWI/SNF and the upregulation of target gene expression. As expected, we found an almost identical 16 bp cis-element ~400 bp upstream of the ATG start codon in the CYP51A promoter of all Necosmospora FSSC isolates and in all related Fusarium species. The only major difference was an A to G transition in the distal CGAA repeat (CGAG) that was conserved throughout the FSSC (Figure 4). However, the 16 bp cis-element was 120 bp downstream of the 23 bp deletion and present in all Neocosmospora isolates sequenced. We speculate that the 23 bp CYP51A promoter deletion affects CYP51A-chromatin-packing that causes a slightly (1.3–7.5-fold) increased basal CYP51A mRNA expression level in group II FSSC isolates (Figure 5). The presence of an unchanged sterol regulatory element in group II FSSC isolates explains why there was no significant difference in the regulation of CYP51A mRNA expression in group I and group II isolates in response to VRC.
Expression and/or upregulation of ATP-binding cassette (ABC) transporters is also frequently associated with azole resistance in fungi (Lamping et al., 2009; Panapruksachat et al., 2016; Watanasrisin et al., 2016; Paul et al., 2019). In A. fumigatus, the AtrR TF binds to, and regulates, both cyp51A and the ABC transporter abcG1. Binding of AtrR to a 34-bp tandem repeat element discovered in cyp51A promoters of azole resistant A. fumigatus clinical isolates increased cyp51A expression and azole resistance (Howard et al., 2009; Wiederhold et al., 2016; Paul et al., 2019). Whether the CYP51A promoter mutations alone caused ~2–32 times higher VRC MICs of group II strains or whether other factors (e.g., ABC transporters) also contribute to the observed azole resistance phenotype of FSSC isolates remains to be investigated. Nevertheless, our data suggest that the 23 bp CYP51A promoter deletion is a potentially valuable new VRC resistance marker to enable clinicians to choose the correct treatment option for the often lethal IF.
In summary, N. keratoplastica CYP51A mRNA levels are ~6,500-fold upregulated in response to azole antifungals to compensate for the loss of CYP51B function due to azole inhibition. There was a strong association of VRC resistance with a 23 bp CYP51A promoter deletion in all Neocosmospora FSSC isolates tested. The conserved “entry-gate” residue N. keratoplastica Cyp51A-L218 may possibly be related to the long-tailed azole resistance phenotype of FSSC species. A novel 16 bp sterol regulatory cis-element was present ~400 bp upstream of the ATG start codon of all sequenced CYP51A promoters. Taken together, our findings provide important first clues about possible azole resistance mechanisms in the medically, and agriculturally, important fungal pathogens of the FSSC.
Data Availability Statement
DNA sequences generated in this study have been made publicly available at GenBank. The datasets for phylogenetic analysis will be made available by the authors upon request, without undue reservation, to any qualified researcher.
Author Contributions
JJ performed the experiments and most of the data analysis. EL contributed to the experimental design and data analysis. JS, EL, and RC contributed to the conception and design of the study. LZ and MA provided the isolates used in this study and performed the morphological identification. TM supervised the qPCR experiments and mRNA expression analysis. JJ wrote the manuscript. EL, RC, TM, and JS contributed to manuscript revision. All authors read and approved the final manuscript.
Funding
This work was supported by Ministry of Education Malaysia under the Fundamental Research Grant Scheme (FRGS/1/2018/SKK11/UKM/02/1). JJ was the recipient of a Sir Thomas Kay Sidey Postgraduate Visiting Fellowship from the University of Otago Faculty of Dentistry.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Universiti Kebangsaan Malaysia Medical Centre and the Institute for Medical Research Malaysia for providing the clinical isolates. RC and EL acknowledge support from the Marsden Fund of the Royal Society of New Zealand (UOO1305).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00272/full#supplementary-material
References
Abastabar, M., Al-Hatmi, A. M., Moghaddam, M. V., de Hoog, G. S., Haghani, I., Aghili, S. R., et al. (2018). Potent activities of luliconazole, lanoconazole, and eight comparators against molecularly characterized Fusarium species. Antimicrob. Agents Chemother. 62:e00009-18. doi: 10.1128/AAC.00009-18
Abastabar, M., Hosseini, T., Valadan, R., Lagzian, M., Haghani, I., Aslani, N., et al. (2019). Novel point mutations in cyp51A and cyp51B genes associated with itraconazole and posaconazole resistance in Aspergillus clavatus isolates. Microb. Drug Resist. 25, 652–662. doi: 10.1089/mdr.2018.0300
Al-Hatmi, A. M., Bonifaz, A., Tirado-Sanchez, A., Meis, J. F., de Hoog, G. S., and Ahmed, S. A. (2017a). Fusarium species causing eumycetoma: report of two cases and comprehensive review of the literature. Mycoses 60, 204–212. doi: 10.1111/myc.12590
Al-Hatmi, A. M., Normand, A. C., Ranque, S., Piarroux, R., de Hoog, G. S., Meletiadis, J., et al. (2017b). Comparative evaluation of Etest, EUCAST, and CLSI methods for amphotericin B, voriconazole, and posaconazole against clinically relevant Fusarium species. Antimicrob. Agents Chemother. 61:e01671-16. doi: 10.1128/AAC.01671-16
Al-Hatmi, A. M. S., Bonifaz, A., Ranque, S., Sybren de Hoog, G., Verweij, P. E., and Meis, J. F. (2018). Current antifungal treatment of fusariosis. Int. J. Antimicrob. Agents 51, 326–332. doi: 10.1016/j.ijantimicag.2017.06.017
Becher, R., Hettwer, U., Karlovsky, P., Deising, H. B., and Wirsel, S. G. R. (2010). Adaptation of Fusarium graminearum to tebuconazole yielded descendants diverging for levels of fitness, fungicide resistance, virulence, and mycotoxin production. Phytopathology 100, 444–453. doi: 10.1094/PHYTO-100-5-0444
Brillowska-Dabrowska, A., Mroczynska, M., Nawrot, U., Wlodarczyk, K., and Kurzyk, E. (2015). Examination of cyp51A and cyp51B expression level of the first Polish azole resistant clinical Aspergillus fumigatus isolate. Acta Biochim. Pol. 62, 837–839. doi: 10.18388/abp.2015_1143
Chang, D. C., Grant, G. B., O'Donnell, K., Wannemuehler, K. A., Noble-Wang, J., Rao, C. Y., et al. (2006). Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA 296, 953–963. doi: 10.1001/jama.296.8.953
Chen, J., Li, H., Li, R., Bu, D., and Wan, Z. (2005). Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. J. Antimicrob. Chemother. 55, 31–37. doi: 10.1093/jac/dkh507
Chiewchanvit, S., Chongkae, S., Mahanupab, P., Nosanchuk, J. D., Pornsuwan, S., Vanittanakom, N., et al. (2017). Melanization of Fusarium keratoplasticum (F. solani species complex) during disseminated fusariosis in a patient with acute leukemia. Mycopathologia 182, 879–885. doi: 10.1007/s11046-017-0156-2
Chowdhary, A., Kathuria, S., Xu, J., and Meis, J. F. (2013). Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 9:e1003633. doi: 10.1371/annotation/4ffcf1da-b180-4149-834c-9c723c5dbf9b
Chowdhary, A., Sharma, C., and Meis, J. F. (2017). Azole-resistant aspergillosis: epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 216(Suppl. 3), S436–S444. doi: 10.1093/infdis/jix210
CLSI (2008). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard, 2nd Edn, CLSI Document M38-A2. Wayne, PA: Clinical and Laboratory Standards Institute.
Coleman, J. J. (2016). The Fusarium solani species complex: ubiquitous pathogens of agricultural importance. Mol. Plant Pathol. 17, 146–158. doi: 10.1111/mpp.12289
Coleman, J. J., Rounsley, S. D., Rodriguez-Carres, M., Kuo, A., Wasmann, C. C., Grimwood, J., et al. (2009). The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5:e1000618. doi: 10.1371/journal.pgen.1000618
Córdoba, S., Rodero, L., Vivot, W., Abrantes, R., Davel, G., and Vitale, R. (2008). In vitro interactions of antifungal agents against clinical isolates of Fusarium spp. Int. J. Antimicrob. Agents 31, 171–174. doi: 10.1016/j.ijantimicag.2007.09.005
Cuomo, C. A., Guldener, U., Xu, J. R., Trail, F., Turgeon, B. G., di Pietro, A., et al. (2007). The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317, 1400–1402. doi: 10.1126/science.1143708
Efe İris, N., Güvenç, S., Özçelik, T., Demirel, A., Koçulu, S., Çevik, E., et al. (2016). Successful treatment of disseminated fusariosis with the combination of voriconazole and liposomal amphotericin B. Turk. J. Haematol. 33, 363–364. doi: 10.4274/tjh.2016.0128
Esnakula, A. K., Summers, I., and Naab, T. J. (2013). Fatal disseminated Fusarium infection in a human immunodeficiency virus positive patient. Case Rep. Infect. Dis. 2013:379320. doi: 10.1155/2013/379320
Espinel-Ingroff, A., Colombo, A. L., Cordoba, S., Dufresne, P. J., Fuller, J., Ghannoum, M., et al. (2016). International evaluation of MIC distributions and epidemiological cutoff value (ECV) definitions for Fusarium species identified by molecular methods for the CLSI Broth microdilution method. Antimicrob. Agents Chemother. 60, 1079–1084. doi: 10.1128/AAC.02456-15
Fan, J., Urban, M., Parker, J. E., Brewer, H. C., Kelly, S. L., Hammond-Kosack, K. E., et al. (2013). Characterization of the sterol 14 alpha-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function. New Phytol. 198, 821–835. doi: 10.1111/nph.12193
Faria-Ramos, I., Farinha, S., Neves-Maia, J., Tavares, P. R., Miranda, I. M., Estevinho, L. M., et al. (2014). Development of cross-resistance by Aspergillus fumigatus to clinical azoles following exposure to prochloraz, an agricultural azole. BMC Microbiol. 14:155. doi: 10.1186/1471-2180-14-155
Guevara-Suarez, M., Cano-Lira, J. F., de Garcia, M. C., Sopo, L., de Bedout, C., Cano, L. E., et al. (2016). Genotyping of Fusarium isolates from onychomycoses in Colombia: detection of two new species within the Fusarium solani species complex and in vitro antifungal susceptibility testing. Mycopathologia 181, 165–174. doi: 10.1007/s11046-016-9983-9
Gupta, A. K., and Daigle, D. (2016). A critical appraisal of once-daily topical luliconazole for the treatment of superficial fungal infections. Infect. Drug Resist. 9, 1–6. doi: 10.2147/IDR.S61998
Herkert, P. F., Al-Hatmi, A. M. S., de Oliveira Salvador, G. L., Muro, M. D., Pinheiro, R. L., Nucci, M., et al. (2019). Molecular characterization and antifungal susceptibility of clinical Fusarium species from Brazil. Front. Microbiol. 10:737. doi: 10.3389/fmicb.2019.00737
Heyn, K., Tredup, A., Salvenmoser, S., and Müller, F.-M. (2005). Effect of voriconazole combined with micafungin against Candida, Aspergillus, and Scedosporium spp. and Fusarium solani. J. Antimicrob. Chemother. 49, 5157–5159. doi: 10.1128/AAC.49.12.5157-5159.2005
Ho, D. Y., Lee, J. D., Rosso, F., and Montoya, J. (2007). Treating disseminated fusariosis: amphotericin B, voriconazole or both? Mycoses 50, 227–231. doi: 10.1111/j.1439-0507.2006.01346.x
Howard, S. J., Cerar, D., Anderson, M. J., Albarrag, A., Fisher, M. C., Pasqualotto, A. C., et al. (2009). Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15, 1068–1076. doi: 10.3201/eid1507.090043
Jureen, R., Koh, T. H., Wang, G., Chai, L. Y., Tan, A. L., Chai, T., et al. (2008). Use of multiple methods for genotyping Fusarium during an outbreak of contact lens associated fungal keratitis in Singapore. BMC Infect. Dis. 8:92. doi: 10.1186/1471-2334-8-92
Kazan, K., and Gardiner, D. M. (2018). Transcriptomics of cereal–Fusarium graminearum interactions: what we have learned so far. Mol. Plant Pathol. 19, 764–778. doi: 10.1111/mpp.12561
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lamping, E., Ranchod, A., Nakamura, K., Tyndall, J. D. A., Niimi, K., Holmes, A. R., et al. (2009). Abc1p Is a multidrug efflux transporter that tips the balance in favor of innate azole resistance in Candida krusei. Antimicrob. Agents Chemother. 53, 354–369. doi: 10.1128/AAC.01095-08
Lass-Flörl, C., and Cuenca-Estrella, M. (2017). Changes in the epidemiological landscape of invasive mould infections and disease. J. Antimicrob. Chemother. 72(Suppl. 1), i5–i11. doi: 10.1093/jac/dkx028
Leslie, J. F., and Summerell, B. A. (2008). The Fusarium Laboratory Manual. Ames, IA: Blackwell Pub.
Liu, X., Yu, F., Schnabel, G., Wu, J., Wang, Z., and Ma, Z. (2011). Paralogous cyp51 genes in Fusarium graminearum mediate differential sensitivity to sterol demethylation inhibitors. Fungal Genet. Biol. 48, 113–123. doi: 10.1016/j.fgb.2010.10.004
Liu, Z., Jian, Y., Chen, Y., Kistler, H. C., He, P., Ma, Z., et al. (2019). A phosphorylated transcription factor regulates sterol biosynthesis in Fusarium graminearum. Nat. Commun. 10:1228. doi: 10.1038/s41467-019-09145-6
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ)CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lortholary, O., Obenga, G., Biswas, P., Caillot, D., Chachaty, E., Bienvenu, A. L., et al. (2010). International retrospective analysis of 73 cases of invasive fusariosis treated with voriconazole. Antimicrob. Agents Chemother. 54, 4446–4450. doi: 10.1128/AAC.00286-10
Manshor, N., Rosli, H., Ismail, N. A., Salleh, B., and Zakaria, L. (2012). Diversity of Fusarium species from highland areas in Malaysia. Trop. Life Sci. Res. 23, 1–15.
Monk, B. C., Tomasiak, T. M., Keniya, M. V., Huschmann, F. U., Tyndall, J. D. A., O'Connell, J. D., et al. (2014). Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc. Natl. Acad. Sci. U.S.A. 111, 3865–3870. doi: 10.1073/pnas.1324245111
Moore, C. B., Novak-Frazer, L., Muldoon, E., Dunn, K. W., Masania, R., Richardson, M. D., et al. (2017). First isolation of the pan-azole-resistant Aspergillus fumigatus Cyp51A TR46/Y121F/T289A mutant in a UK patient. Int. J. Antimicrob. Agents 49, 512–514. doi: 10.1016/j.ijantimicag.2017.01.004
Muraosa, Y., Oguchi, M., Yahiro, M., Watanabe, A., Yaguchi, T., and Kamei, K. (2017). Epidemiological study of Fusarium species causing invasive and superficial fusariosis in Japan. Med. Mycol. J. 58, E5–E13. doi: 10.3314/mmj.16-00024
Nucci, M., and Anaissie, E. (2007). Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20, 695–704. doi: 10.1128/CMR.00014-07
Nucci, M., Varon, A. G., Garnica, M., Akiti, T., Barreiros, G., Trope, B. M., et al. (2013). Increased incidence of invasive fusariosis with cutaneous portal of entry, Brazil. Emerg. Infect. Dis. 19, 1567–1572. doi: 10.3201/eid1910.120847
O'Donnell, K., and Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/mpev.1996.0376
O'Donnell, K., Humber, R. A., Geiser, D. M., Kang, S., Park, B., Robert, V. A., et al. (2012). Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia 104, 427–445. doi: 10.3852/11-179
O'Donnell, K., Sutton, D. A., Fothergill, A., McCarthy, D., Rinaldi, M. G., Brandt, M. E., et al. (2008). Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 46, 2477–2490. doi: 10.1128/JCM.02371-07
O'Donnell, K., Sutton, D. A., Rinaldi, M. G., Sarver, B. A., Balajee, S. A., Schroers, H. J., et al. (2010). Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 48, 3708–3718. doi: 10.1128/JCM.00989-10
O'Donnell, K., Sutton, D. A., Wiederhold, N., Robert, V. A., Crous, P. W., and Geiser, D. M. (2016). Veterinary fusarioses within the United States. J. Clin. Microbiol. 54, 2813–2819. doi: 10.1128/JCM.01607-16
Okada, K., Endo, T., Hashimoto, D., Saga, T., Ara, T., Ogasawara, R., et al. (2018). Disseminated fusariosis emerged from prolonged local genital infection after cord blood transplantation. J. Infect. Chemother. 24, 660–663. doi: 10.1016/j.jiac.2017.12.018
Panapruksachat, S., Iwatani, S., Oura, T., Vanittanakom, N., Chindamporn, A., Niimi, K., et al. (2016). Identification and functional characterization of Penicillium marneffei pleiotropic drug resistance transporters ABC1 and ABC2. Med. Mycol. 54, 478–491. doi: 10.1093/mmy/myv117
Pascual, A., Calandra, T., Bolay, S., Buclin, T., Bille, J., and Marchetti, O. (2008). Voriconazole therapeutic drug monitoring in patients with invasive mycoses improves efficacy and safety outcomes. Clin. Infect. Dis. 46, 201–211. doi: 10.1086/524669
Paul, S., Stamnes, M., Thomas, G. H., Liu, H., Hagiwara, D., Gomi, K., et al. (2019). AtrR is an essential determinant of azole resistance in Aspergillus fumigatus. MBio 10:e02563–e02518. doi: 10.1128/mBio.02563-18
Popp, C., Hampe, I. A. I., Hertlein, T., Ohlsen, K., Rogers, P. D., and Morschhäuser, J. (2017). Competitive fitness of fluconazole-resistant clinical Candida albicans strains. Antimicrob. Agents Chemother. 61:e00584-17. doi: 10.1128/AAC.00584-17
Rotjanapan, P., Chen, Y. C., Chakrabarti, A., Li, R. Y., Rudramurthy, S. M., Yu, J., et al. (2018). Epidemiology and clinical characteristics of invasive mould infections: a multicenter, retrospective analysis in five Asian countries. Med. Mycol. 56, 186–196. doi: 10.1093/mmy/myx029
Ruan, R., Wang, M., Liu, X., Sun, X., Chung, K.-R., and Li, H. (2017). Functional analysis of two sterol regulatory element binding proteins in Penicillium digitatum. PLoS ONE 12:e0176485. doi: 10.1371/journal.pone.0176485
Sandoval-Denis, M., and Crous, P. (2018). Removing chaos from confusion: assigning names to common human and animal pathogens in Neocosmospora. Persoonia 41, 109–129. doi: 10.3767/persoonia.2018.41.06
Santhanam, J., Teong, A. K., and Amran, F. (2008). Susceptibility testing of clinical mould isolates in Malaysia with the Etest and M38-A methods. J. Med. Sci. 8, 94–97. doi: 10.3923/jms.2008.94.97
Scher, R. K., Nakamura, N., and Tavakkol, A. (2014). Luliconazole: a review of a new antifungal agent for the topical treatment of onychomycosis. Mycoses 57, 389–393. doi: 10.1111/myc.12168
Schroers, H. J., Samuels, G. J., Zhang, N., Short, D. P., Juba, J., and Geiser, D. M. (2016). Epitypification of Fusisporium (Fusarium) solani and its assignment to a common phylogenetic species in the Fusarium solani species complex. Mycologia 108, 806–819. doi: 10.3852/15-255
Silva, G. M., Silveira, A. R., Betania, C. A., Macedo, D. P., and Neves, R. P. (2013). Disseminated fusariosis secondary to neuroblastoma with fatal outcome. Mycopathologia 176, 233–236. doi: 10.1007/s11046-013-9674-8
Singh, H., Jamal, F., Marahakim, M., and Song, C. (1981). Fusarium solani keratitis. first report from Malaysia. Med. J. Malaysia 36, 89–91.
Snelders, E., Karawajczyk, A., Schaftenaar, G., Verweij, P. E., and Melchers, W. J. (2010). Azole resistance profile of amino acid changes in Aspergillus fumigatus Cyp51A based on protein homology modeling. Antimicrob. Agents Chemother. 54, 2425–2430. doi: 10.1128/AAC.01599-09
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Todokoro, D., Suzuki, T., Tamura, T., Makimura, K., Yamaguchi, H., Inagaki, K., et al. (2019). Efficacy of luliconazole against broad-range filamentous fungi including Fusarium solani species complex causing fungal keratitis. Cornea 38, 238–242. doi: 10.1097/ICO.0000000000001812
Tortorano, A. M., Prigitano, A., Esposto, M. C., Arsic Arsenijevic, V., Kolarovic, J., Ivanovic, D., et al. (2014). European Confederation of Medical Mycology (ECMM) epidemiological survey on invasive infections due to Fusarium species in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1623–1630. doi: 10.1007/s10096-014-2111-1
Tupaki-Sreepurna, A., Al-Hatmi, A. M., Kindo, A. J., Sundaram, M., and de Hoog, G. S. (2017). Multidrug-resistant Fusarium in keratitis: a clinico-mycological study of keratitis infections in Chennai, India. Mycoses 60, 230–233. doi: 10.1111/myc.12578
Tzar, M., Suhaila, B., Shamsul, A., and Azizah, M. (2013). Epidemiology of fungal infections at an infectious disease reference centre in Malaysia. Int. Med. J. Malays. 12, 39–42.
Tzar, M., Binting, R. A. A., Saari, S. M., Kumar, T. V., Muhammad, M., Satim, H., et al. (2016). In vitro antifungal activities against moulds isolated from dermatological specimens. Malays. J. Med. Sci. 23, 32–39.
Tzar, M., Zetti, Z., Ramliza, R., Sharifah, A., and Leelavathi, M. (2014). Dermatomycoses in Kuala Lumpur, Malaysia. Sains Malays. 43, 1737–1742.
Urbaniak, C., Massa, G., Hummerick, M., Khodadad, C., Schuerger, A., and Venkateswaran, K. (2018). Draft genome sequences of two Fusarium oxysporum isolates cultured from infected Zinnia hybrida plants grown on the international space station. Genome Announc. 6:e00326-18. doi: 10.1128/genomeA.00326-18
Vaezi, A., Fakhim, H., Javidnia, J., Khodavaisy, S., Abtahian, Z., Vojoodi, M., et al. (2018). Pesticide behavior in paddy fields and development of azole-resistant Aspergillus fumigatus: should we be concerned? J. Mycol. Med. 28, 59–64. doi: 10.1016/j.mycmed.2017.12.007
Watanasrisin, W., Iwatani, S., Oura, T., Tomita, Y., Ikushima, S., Chindamporn, A., et al. (2016). Identification and characterization of Candida utilis multidrug efflux transporter CuCdr1p. FEMS Yeast Res. 16:fow042. doi: 10.1093/femsyr/fow042
Wiederhold, N. P., Gil, V. G., Gutierrez, F., Lindner, J. R., Albataineh, M. T., McCarthy, D. I., et al. (2016). First detection of TR34 L98H and TR46 Y121F T289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. J. Clin. Microbiol. 54, 168–171. doi: 10.1128/JCM.02478-15
Keywords: azole, Cyp51A, Neocosmospora, Fusarium, FSSC, sterol regulatory element
Citation: James JE, Lamping E, Santhanam J, Milne TJ, Abd Razak MF, Zakaria L and Cannon RD (2020) A 23 bp cyp51A Promoter Deletion Associated With Voriconazole Resistance in Clinical and Environmental Isolates of Neocosmospora keratoplastica. Front. Microbiol. 11:272. doi: 10.3389/fmicb.2020.00272
Received: 26 November 2019; Accepted: 06 February 2020;
Published: 31 March 2020.
Edited by:
Miguel Cacho Teixeira, University of Lisbon, PortugalReviewed by:
Akira Watanabe, Chiba University, JapanHamid Badali, The University of Texas Health Science Center at San Antonio, United States
Dominique Sanglard, Université de Lausanne, Switzerland
Copyright © 2020 James, Lamping, Santhanam, Milne, Abd Razak, Zakaria and Cannon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erwin Lamping, ZXJ3aW4ubGFtcGluZ0BvdGFnby5hYy5ueg==; Jacinta Santhanam, amFjaW50YUB1a20uZWR1Lm15
 Jasper Elvin James
Jasper Elvin James Erwin Lamping
Erwin Lamping Jacinta Santhanam
Jacinta Santhanam Trudy Jane Milne
Trudy Jane Milne Mohd Fuat Abd Razak3
Mohd Fuat Abd Razak3 Latiffah Zakaria
Latiffah Zakaria Richard David Cannon
Richard David Cannon