- 1Department of Genetics, Ribeirão Preto Medical School, University of São Paulo, São Paulo, Brazil
- 2Department of Microbiology, Institute of Biological Sciences, Federal University of Minas Gerais, Belo Horizonte, Brazil
- 3Department of Biotechnology, University of Ribeirão Preto, Ribeirão Preto, Brazil
The ability of fungi to sense environmental stressors and appropriately respond is linked to secretory system functions. The dermatophyte infection process depends on an orchestrated signaling regulation that triggers the transcription of genes responsible for adherence and penetration of the pathogen into host-tissue. A high secretion system is activated to support the host-pathogen interaction and assures maintenance of the dermatophyte infection. The gateway of secretion machinery is the endoplasmic reticulum (ER), which is the primary site for protein folding and transport. Current studies have shown that ER stress that affects adaptive responses is primarily regulated by UPR and supports fungal pathogenicity; this has been assessed for yeasts and Aspergillus fumigatus, in regard to how these fungi cope with host environmental stressors. Fungal UPR consists of a transmembrane kinase sensor (Ire1/IreA) and a downstream target Hac1/HacA. The active form of Hac is achieved via non-spliceosomal intron removal promoted by endonuclease activity of Ire1/IreA. Here, we assessed features of HacA and its involvement in virulence and susceptibility in Trichophyton rubrum. Our results showed that exposure to antifungals and ER-stressing agents initiated the activation of HacA from T. rubrum. Interestingly, the activation occurs when a 20 nt fragment is removed from part of the exon-2 and part of intron-2, which in turn promotes the arisen of the DNA binding site motif and a dimer interface domain. Further, we found changes in the cell wall and cellular membrane composition in the ΔhacA mutant as well as an increase in susceptibility toward azole and cell wall disturbing agents. Moreover, the ΔhacA mutant presented significant defects in important virulence traits like thermotolerance and growth on keratin substrates. For instance, the development of the ΔhacA mutant was impaired in co-culture with keratinocytes or human nail fragments. Changes in the pro-inflammatory cytokine release were verified for the ΔhacA mutant during the co-culture assay, which might be related to differences in pathogen-associated molecular patterns (PAMPs) in the cell wall. Together, these results suggested that HacA is an integral part of T. rubrum physiology and virulence, implying that it is an important molecular target for antidermatophytic therapy.
Introduction
The superficial infections of the skin and nails represent the most common human mycoses, and it is estimated to affect about 1.7 billion of the population (Brown et al., 2012). These infections are mainly caused by dermatophytes, in which Trichophyton rubrum followed by Trichophyton interdigitale have been described as the predominant species isolated in dermatophytosis cases worldwide (Almeida et al., 2019).
During the dermatophyte-host interaction, a complex signaling network enables infection establishment. A profound metabolic change is required to overcome the hostile host environment, in which fungi cope with acidic skin pH, shortages of nutrients, skin desquamation, the action of phagocytic cells, and antimicrobial peptides (Leal et al., 2009; Martinez-Rossi et al., 2016b). Besides, a highly efficient secretion system is triggered to support the host attachment and nutrient acquisition by the pathogen (Martinez-Rossi et al., 2016b; Mercer and Stewart, 2019).
The endoplasmic reticulum (ER) represents the gateway of the secretory pathway, where most of the plasma membrane and secreted proteins undergo proper folding and post-translation modifications (Schwarz and Blower, 2016). The secretory system is used by different pathogens to express virulence factors, and to cope with stress conditions, which ultimately might favor their adaptation to specific biological niches (Krishnan and Askew, 2014).
When the ER capacity is overwhelmed by high concentrations of proteins inside the milieu, which exceed the ER folding competence, there is an accumulation of misfolded proteins that compromise cellular physiology. In order to mitigate this ensuing status of ER stress, a series of adaptive responses collectively termed unfolded protein responses (UPR) is initiated (Moore and Hollien, 2012). The UPR pathway is activated to restore ER homeostasis by enhancing the folding ability and controlling misfolded proteins disposal. In fungi, UPR consists of an ER-transmembrane sensor Ire1/IreA (Ser/Thr kinase) with an endonuclease domain, and the transcription factor Hac1/HacA. Upon ER stress, the IreA is activated and cleaves in a non-canonical way to the Hac mRNA. The splicing sites are recognized through a conserved RNA secondary structure that flanks the cleavage sites. The splicing of cytosolic Hac1/HacA mRNA shifts the open reading frame, and the spliced form is translated to a potent bZIP transcription factor that is transported to the nucleus, where it regulates the UPR target genes (Saloheimo et al., 2003).
The UPR has been reported to be a therapeutic vulnerability target in pathogenic fungi. It is assumed to be an essential regulator of Aspergillus fumigatus pathogenicity (Feng et al., 2011; Richie et al., 2011). Moreover, in Cryptococcus neoformans and Candida albicans, UPR affected virulence traits as deletions in these genes were associated with impairment of the ability to switch from yeast to hyphal form, compromised cell wall adhesive proteins, and reduction of thermotolerance (Matsumoto et al., 2005; Wimalasena et al., 2008; Blankenship et al., 2010; Krishnan and Askew, 2014).
Notably, consistent divergences of domain structure from mammalian Hac1/HacA ortholog, termed Xbp1, denote HacA as an attractive target for antifungal therapy (Zhang et al., 2016). Further, the current paradigm is that ER-stress response pathways are involved in the expression of different virulence traits that may be necessary for pathogen-host interaction (Krishnan and Askew, 2014). Thus, we have addressed the scope of HacA in the dermatophyte Trichophyton rubrum. Here, we characterized HacA from T. rubrum and confirmed its involvement in T. rubrum pathogenicity and adaptive responses to different stressors, and the immune modulation of keratinocyte cells.
Materials and Methods
Strain and Culture Conditions
Trichophyton rubrum CBS118892 strain (Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Netherlands) was cultivated in malt extract at 28°C, as previously described (Peres et al., 2016). Conidia suspension was obtained from 20-days-old plates, and the concentration was estimated by using a Newbauer chamber. Approximately 1 × 106 conidia were added in 50 mL of liquid Sabouraud followed by incubation at 28°C for 96 h under continuous shaking. The resulting mycelia were then transferred to 100 mL of Sabouraud in the presence of sublethal doses of acriflavine (ACR), caspofungin (CASP), griseofulvin (GRS), terbinafine (TRB), or undecanoic acid (UDA), and in the absence of drugs (control), followed by incubation at 28°C with shaking (120 rpm) for 12 h. The concentrations used for each drug were 70% of their minimum inhibitory concentration values, and obtained in accordance with the CLSI (M-38A2) (Clinical Laboratory Standards Institute [CLSI], 2008) with the following modifications, in Sabouraud media corresponding to 5.46 μg/mL for ACR, 87.5 μg/mL for CASP, 2.76 μg/mL for GRS, 0.014 μg/mL for TRB, and 35 μg/mL for UDA. Dithiothreitol (DTT) and tunicamycin (TUN), in concentrations of 10 mM and 21.87 μg/mL, respectively, were used as a positive control for the UPR response. The cytotoxic compounds were purchased from Sigma-Aldrich (St. Louis, MO, United States), with the exception of CASP, which was purchased from Merck (Kenilworth, NJ, United States).
Total RNA Extraction and cDNA Synthesis
The mycelia were ground by mechanical pulverization with a pestle and mortar in liquid nitrogen, and total RNA isolation was carried out using TRIzol (Thermo Fisher Scientific, Carlsbad, CA, United States) following the manufacturer’s instructions. RNA samples were treated with RNase-free DNAse I (Sigma-Aldrich). Complementary DNA was synthesized from each condition containing 1000 ng of total RNA in a 20 μL reaction volume using a High Capacity cDNA Synthesis kit (Thermo Fisher Scientific #4368814).
RT-PCR and qRT-PCR
For qualitative expression analysis, primer pairs that yielded PCR products around the predicted hacA excision region (Table 1) were used to amplify the two isoform products of the gene coding for HacA in T. rubrum (TERG_05396). For the PCR reaction, approximately 140 ng of cDNA and 0.2 pmol/μL of each oligonucleotide were used. Thermocycler conditions were 95°C for 2 min, followed by 35 cycles at 95°C for 30 s, 53°C for 45 s, and 72°C for 1 min, and the last cycle at 72°C for 10 min.
The qPCR assays were performed using SYBR green PCR master mix (Applied Biosystems), 70 ng cDNA, forward and reverse primers used in concentrations previously determined (Table 1) in a 12.5 μL reaction mixture. The rpb2 gene was used as a reference control, and all the analyses were carried out as previously described (Jacob et al., 2012). A set of genes potentially regulated by HacA was analyzed.
Identification and Characterization of T. rubrum hacA
A Blastx search using the S. cerevisiae hac1 sequence as a query identified TERG_05396 as a putative ortholog for this gene. Thereafter, the prediction of the non-canonical intron excision site was determined by Infernal software (Nawrocki et al., 2009). From a multiple alignment file of hacA RNAs in the Stockholm format, a covariance model was established. From this model, screening of the T. rubrum genome was carried out and resulted in a region of 62 nt corresponding to fractions of exon-2 and intron-2. Next, oligonucleotides surrounding this predicted region were used for products amplification of cDNAs from T. rubrum following exposure to antifungal and ER stress compounds, and the sequencing of these products showed the intron removed from T. rubrum hacA mRNA.
hacA Gene Deletion
The inactivation gene cassette was obtained using a split-marker approach (Kuck and Hoff, 2010). The 5′ UTR and 3′ UTR were fused with parts of a hygromycin resistance gene (hph) from pCSN43. The Overlap PCR was used to join the PCR products. Sets of primers were used for the split-marker approach, as described in Supplementary Table S1.
The protoplasts transformation was carried out as previously described (Fachin et al., 2006). The transformants were selected in Cove’s medium with 1 M sucrose and 500 μg/mL hygromycin. PCR screened the prominent colonies through analysis of amplification of the hacA gene. Thereafter, the loss of hacA gene and integration of hph gene were confirmed by PCR and Southern blot analysis (Supplementary Figure S1).
Biochemical Assays
The quantification of ergosterol content was carried out as previously described (Arthington-Skaggs et al., 1999) with modifications. Mycelia of wild type and ΔhacA were inoculated in 50 mL liquid Sabouraud medium and incubated at 28°C for 24 h under shaking (200 rpm). The mycelia were then harvested and transferred to 20 mL fresh liquid Sabouraud medium and incubated for an extra 48 h under the same conditions described above. Further, the biomass was harvested through vacuum filtration and dried under sterile filter papers. The dried mycelium was weighed prior to saponification. Ergosterol was extracted as previously described (Arthington-Skaggs et al., 1999) and quantified spectrophotometrically based on a standard curve of different concentrations of ergosterol (Sigma-Aldrich #45480). Values were presented as μg ergosterol per g dry weight.
The keratinolytic activity was determined, as previously described (Gradisar et al., 2005). Keratin was used as a substrate (pH 8.0), and 1.0 mL of the culture supernatant was utilized as the enzyme. Conidia suspension (5 × 105) of each strain, wild type and ΔhacA was inoculated into 25 mL of water that contained keratin (2.5 g/L) at pH 5.0, as the sole carbon and nitrogen sources, and incubated at 28°C for 7 days under shaking conditions (120 rpm). The mycelia were then harvested, and the supernatant was collected. The mycelial dry weight was obtained, and the pH of the supernatant was determined. Thereafter, the keratinolytic activity was estimated and expressed as units per gram of dry weight.
Phenotypic Assays
The analysis of growth rates was performed in different culture media: (i) agar malt extract (MEA), pH 5.7, (ii) potato dextrose agar (PDA), pH 5.7, (iii) Sabouraud, pH 5.7, (iv) MM (Cove’s) containing nitrate (70 mM) and glucose (55 mM) pH 5.0 (Cove, 1966), and (v) MMK corresponding to MM supplemented with 5% of powder keratin, pH 5.0. A mycelium plug (0.8 cm) was inoculated into the plate center and incubated for 9 days at 28°C.
A microculture of strains was also carried out to assess differences in hyphal formation. The microculture was performed in Sabouraud agar and incubated for 6 days at 28°C.
Susceptibility and Thermotolerance Assays
A serial drop dilution assay was performed to analyze the susceptibility of strains toward antifungals and an ER stress compound DTT. Plates were inoculated with different concentrations of a conidia suspension in the range of 106–102 cell/mL. After 7 days of incubation at 28°C, images were taken using Image J software (Schneider et al., 2012). The compounds assessed were: DTT (5 and 10 mM), ketoconazole – KTC (3.90, 1.95 and 0.98 μg/mL), GRS (0.98 μg/mL) and TRB (0.005 μg/mL). The antifungal compounds were used as sublethal doses (1/4 of MIC value for GRS and TRB, and 1/2, 1/4 and 1/8 of MIC value for KTC), as preliminarily determined through microdilution assays in T. rubrum (data not shown).
In order to evaluate the susceptibility toward compounds that act on cell wall, a conidia suspension (approximately 1 × 105) was inoculated into the center of each well of a multi-well plate containing Sabouraud agar supplemented with concentrations of calcofluor white – CFW (in the range of 0–80 μg/mL) or CASP (in the range of 0–200 μg/mL) and incubated for 7 days at 28°C.
The tolerance to thermal stress conditions was also evaluated. In this sense, the number of colonies grown (CFU) after exposure to conidia (104 cells and 103 cells) at different temperatures (37 and 42°C) for 30 and 60 min were determined.
Coculture and Nail Infection
Nail interaction assay was performed as previously described (Peres et al., 2016) with minor modifications. Human nail fragments (approximately 25 cm2) were sterilized by autoclaving. Nail fragments were then soaked with conidia (1 × 104/mL) from T. rubrum strains for 1 h, followed by the addition of 200 μL of distilled water. The plates were incubated at 28°C for 72 h, and fungal growth was assessed by light microscopy (Leica DMI3000B). The human keratinocytes (HaCaT) cell line was cultured, as previously described (Komoto et al., 2015). The coculture assay was performed using 2 × 106 conidia/mL and 2.5 105 keratinocyte cells/mL. The coculture was incubated for 24 h at 37°C in 5% CO2. HaCaT and conidia cells were grown on RPMI medium used as controls for qPCR and cytokine evaluation assays. The cytokine levels in the supernatant cells were determined by Elisa (Peprotech, NJ, United States) according to the manufacturer’s recommendation. Committee of Ethics in Human Research of the Ribeirão Preto Medical School at the University of São Paulo approved all experiments involving the use of human nail fragments provided by healthy adults (protocol No.8330/2009).
Results
Identification and Characterization of T. rubrum hacA
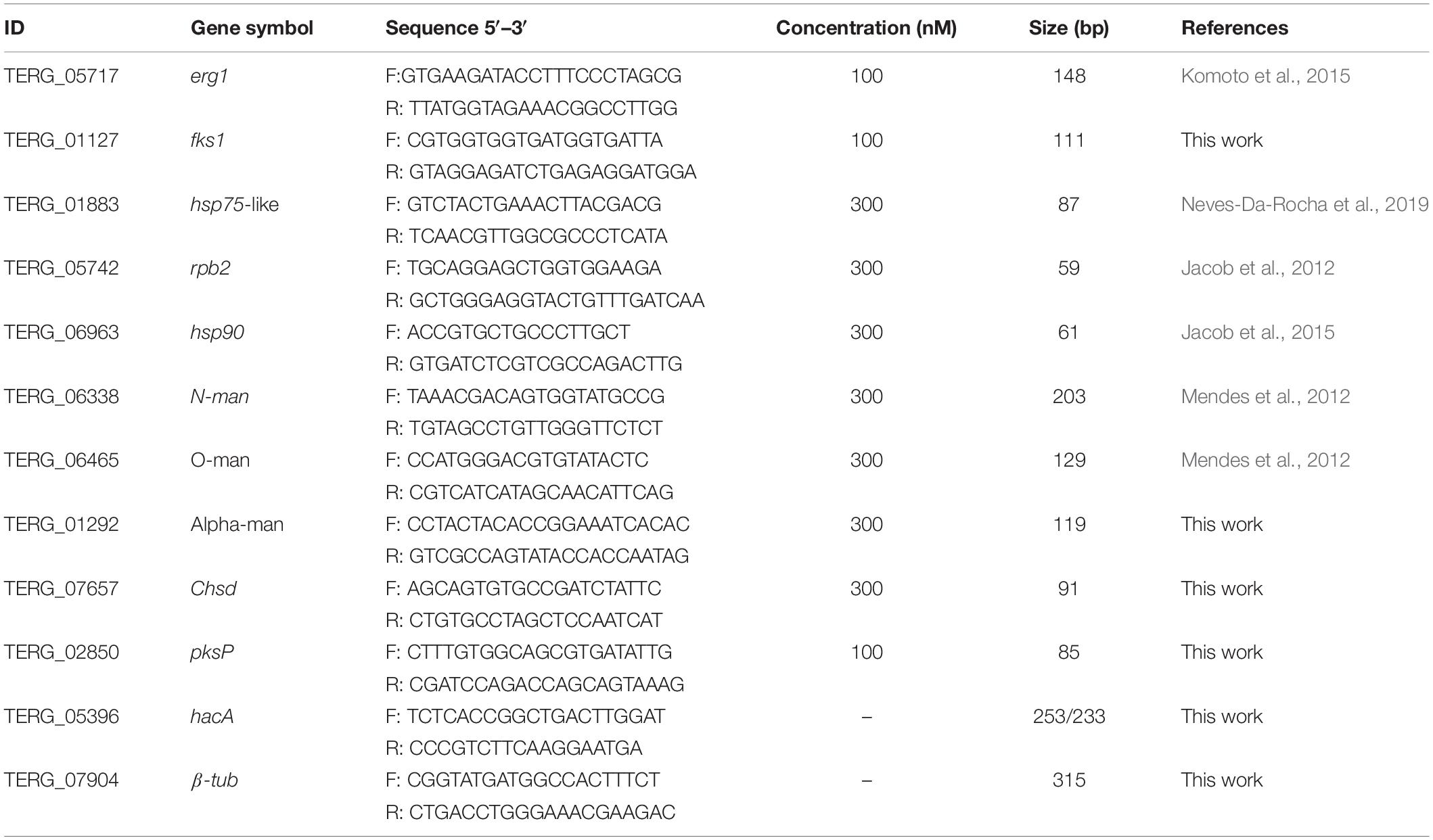
BlastX using hac-1 from Saccharomyces cerevisiae as a query revealed TERG_05396 as a putative ortholog for this gene in T. rubrum. The predicted hac-1/hacA gene from T. rubrum contains 1281 nucleotides. The comparison among mRNA secondary structures of hac-1/hacA sequences from different fungal species evidenced a region of intron excision for prompt hac-1/hacA activation. Following sequencing of the hacA amplified products from T. rubrum, we showed that under ER stress, a fragment of 20 nt was removed from hacA mRNA. We also demonstrated that some antifungal compounds (griseofulvin and terbinafine) were responsible for hacA activation through the 20 nt fragment removal as well as the positive controls DTT and tunicamycin (Figure 1A). Curiously this fragment corresponds to portions of both exon-2 and intron-2, and even upon excision, part of intron-2 (47 nt) was retained (Figure 1B). Further, the consensus of this excision region and the surrounding regions generated a typical secondary structure of hac-1/hacA, in which the boundaries surrounding the intron/exon of excision made two little hairpins (Figure 1C). The removal of the fragment of 20 nucleotides changes the open reading frame arising a DNA binding site and dimer interface residues, which are considered crucial for UPR function in the homologs of HacA as already assessed in other fungi (Oh et al., 2010). The resultant protein is 395 amino acids long and contains a conserved bZIP domain, plus a coiled domain. The uninduced form is 402 amino acids long, and shows the bZIP domain and coiled domain, but neither the DNA binding site nor the dimer interface residues are presented (Figure 1D). Moreover, we ascertained a different pattern in the induced form of HacA from T. rubrum in comparison to the filamentous fungi such as Aspergillus nidulans, A. fumigatus, and Neurospora crassa, where the main changes between the induced and uninduced form were located on the 3′ portion of the protein (Figure 1E).
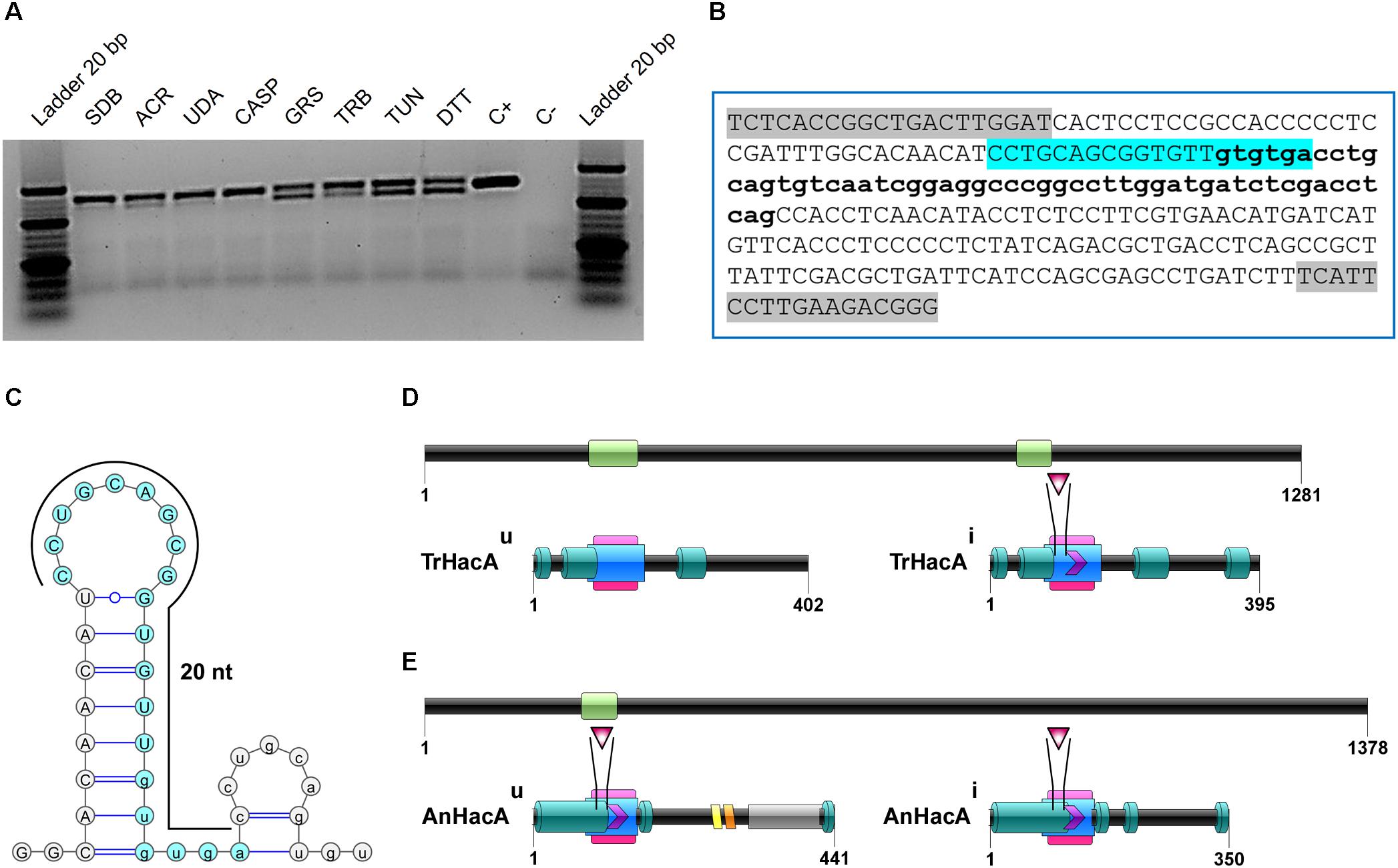
Figure 1. The hacA from T. rubrum is processed under antifungals exposure and ER stress-inducing agents. (A) RT-PCR analysis of hacA mRNA was used to detect the processing under antifungals and chemical exposure. (B) Schematic representation of an unconventional fragment removal of parts of the exon-2 and intron-2 from hacA mRNA, primers used for RT-PCR are indicated by gray boxes, the position of the excised fragment is shown by the color contoured box. (C) Prediction of the twin stem-loop structure of an unconventional splicing of T. rubrum and fungi from hacA mRNA carried out by mFOLD (Zuker, 2003) and the drawing of structures with VARNA (Darty et al., 2009). (D) Schematic representation of HacA induced isoform (hacAi) with 395 aa and uninduced isoform (hacAu) with 402 aa from T. rubrum. (E) Schematic representation of HacA induced (with 350 aa) and non-induced (with 441 aa) isoforms from A. nidulans. The introns in the hacA gene are represented by green boxes, and directly below both of the HacA isoforms are shown. The bZIP domain is shown in a pink box, the coil domain is shown in a blue rectangle, the mobiDB-lite domains are shown as dark green cylinders, a red triangle shows the DNA binding site, and the dimer interface residues are represented by a purple arrow. The transmembrane domain is shown in an orange lozenge and in a yellow lozenge is shown the non-cytoplasmatic domain, and the cytoplasmatic domain is shown as a gray box.
Deletion of hacA Gene
Deletion of the hacA gene from T. rubrum was carried out by replacing the entire coding region with a hygromycin resistance cassette (hph) using a split-marker approach (Kuck and Hoff, 2010). The two overlapping fragments corresponded to 5′ UTR joined to hph, and 3′ UTR joined to hph, and were confirmed by PCR generating the expected products of 2611 and 3733 bp, respectively. Enzyme restriction using EcoRI (Thermo Scientific) also made the expected fragments of 452 and 2159 bp (5′UTR fragment), and 684, 1283, and 1766 bp (3′ UTR fragment). The PCR confirmed the loss of the hacA gene, and homologous integration was identified by genomic Southern blot analysis (Supplementary Figure S1).
hacA Involvement in T. rubrum Susceptibility Toward Cytotoxic Compounds, and Thermotolerance
The growth of the ΔhacA strain was healthy under non-stressful conditions. However, we evidenced differences in growth rates under chemically-induced ER stress and antifungal exposure. The mutant strain showed an enhanced sensitivity to DTT (a reducing agent that leads to ER stress), compared to the wild-type strain (Figure 2) and a reduced growth was observed following KTC exposure, whereas under TRB exposure the mutant strain presented a slight increase in growth (Figure 2). In regard to compounds that act on the cell wall, slight differences in the growth rate were noticed for the ΔhacA strain during CFW exposure, primarily in the higher concentrations of 40 and 80 μg/mL, while CASP promoted a slight reduction in colony growth at all tested levels (Figure 3).
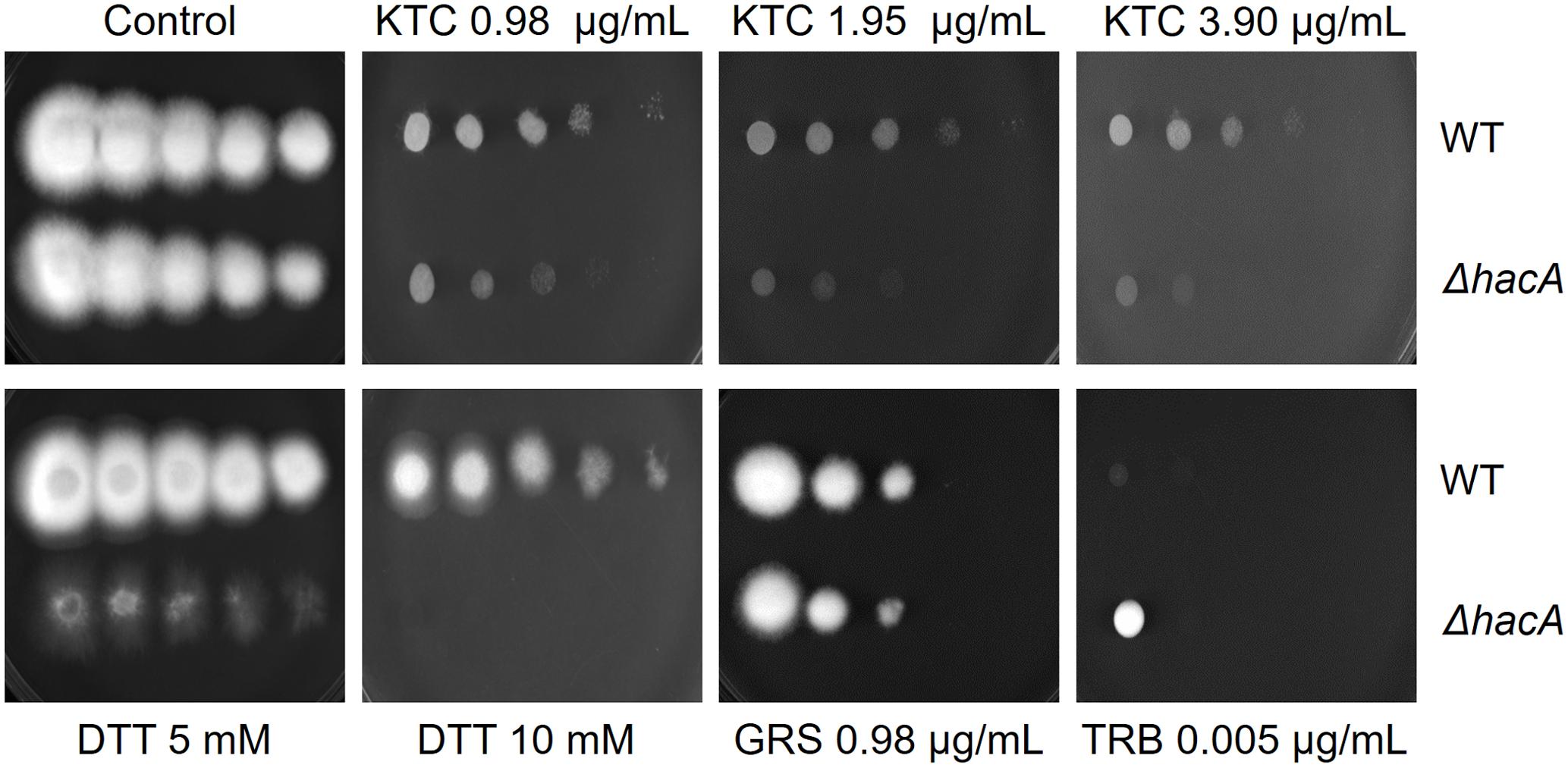
Figure 2. hacA plays a role in T. rubrum susceptibility toward antifungals and DTT. Susceptibility of T. rubrum strains to ketoconazole (KTC), in concentrations of 0.98, 1.95, and 3.90 μg/mL; DTT (5 and 10 mM); griseofulvin (GRS) in concentration of 0.98 μg/mL, and terbinafine (TRB), in a concentration of 0.005 μg/mL. Plates were inoculated, from left to right in each panel, with 106, 105, 104, 103, and 102 conidia/mL and incubated at 28°C for 7 days.
Figure 3. hacA affects T. rubrum susceptibility to cell wall inhibitor agents. Susceptibility of T. rubrum strains to CASP (0, 12.5, 25, 50, 100, and 200 μg/mL), and CFW (0, 5, 10, 20, 40, and 80 μg/mL). Plates were inoculated with approximately 1 × 105 cells, and the drugs were added in corresponding concentrations from left to right, respectively, and incubated for 7 days at 28°C.
The activation of the UPR enhances the protein folding and contributes to protein secretion, thus favoring the adaptation of pathogenic fungi for intracellular survival. Notably, fast-paced proliferation at mammalian body temperature is an important virulence attribute that favors adaptation. HacA seems to play a pivotal role in orchestrating adaptive responses to thermal stress (Richie et al., 2011; Boppidi et al., 2018). Since higher temperatures induce protein conformational changes and alter protein secretion (Mori et al., 2000), we assessed the ability of conidia of both strains to withstand different temperatures (37 and 42°C). Indeed, the mutant was found to be thermosensitive to a higher extent. Analysis of the number of colonies showed a decrease when the mutant was cultivated at 37°C (approximately 15 and 33% for 30 and 60 min of exposure, respectively) and a marked reduction was observed at 42°C (approximately 50 and 70% for 30 and 60 min, respectively). Meanwhile, the wild type strain showed less than a 5% reduction in the number of the colonies for both temperatures assessed (Figure 4). These data suggest that hacA was of paramount importance for T. rubrum growth at high temperatures.
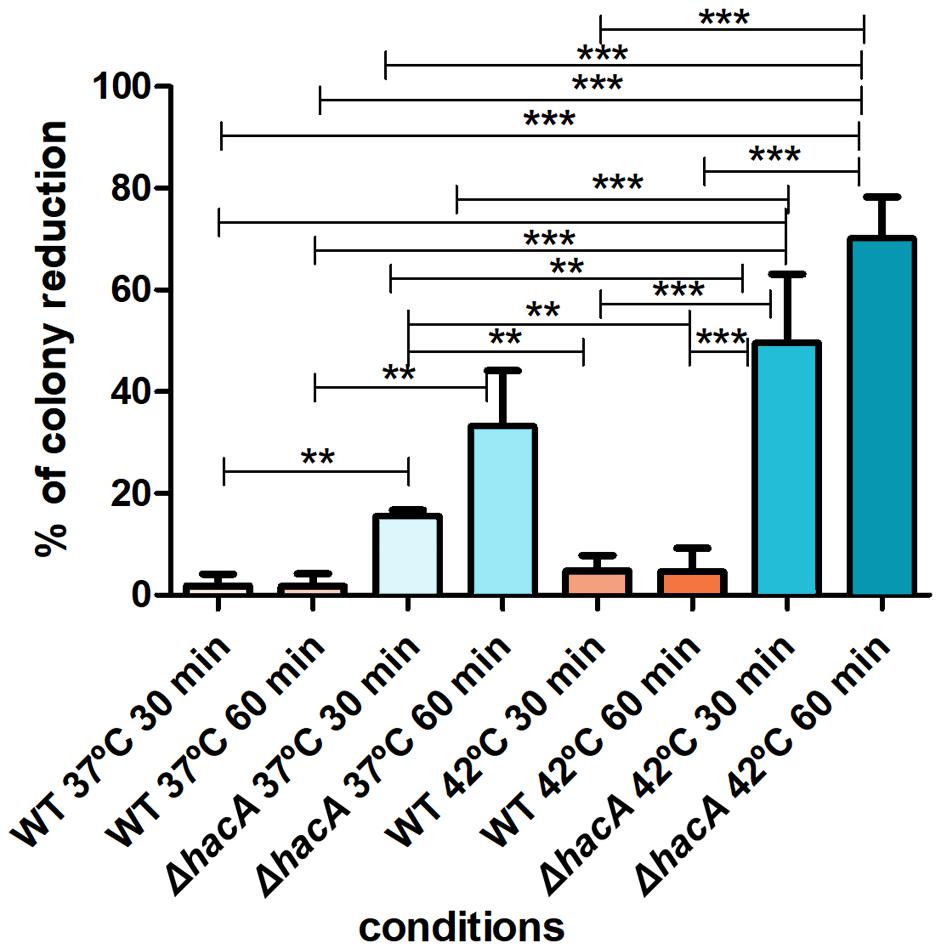
Figure 4. hacA affects thermotolerant growth. Equal concentrations of conidia from each strain were exposed to different temperatures (37 and 42°C) for 30 and 60 min. The control consisted of conidia from both strains without thermal exposition. Thereafter, conidia were inoculated in Sabouraud medium. The percentage reduction for the number of colonies is depicted in this graph. Significantly different values are shown by asterisks, and were determined using ANOVA followed by Tukey’s ad hoc test (**P < 0.01; ***P < 0.001).
hacA Association With Nutritional Versatility
During host-pathogen interaction, T. rubrum encounters a stressing environment, and to stablish the infection the fungus employs metabolic reprogramming to make productive use of the keratin from the host as a nutrient source (Martinez-Rossi et al., 2016b; Mercer and Stewart, 2019; Monod and Mehul, 2019). In order to determine the importance of hacA to nutritional versatility different media were tested like standard media for fungal growth such as Sabouraud, MEA, and PDA (representing rich substrates of pre-digested proteins), or Cove’s medium (Cove, 1966) (as a restrictive substrates source), or Cove’s medium supplemented with keratin (representing a complex protein source that mimics the dermatophyte infection). Although both strains showed equivalent growth rates on less complex substrates, consistent differences were observed for growth on the keratin substrate. The growth of mutant strain on keratin was impaired (Figure 5). Notably, differences in pigmentation were found between the wild type and the mutant colonies during growth on Sabouraud, MEA, and PDA (Supplementary Figure S2). These results hinted that a compromise in pathways related to keratin utilization exists, and also identified changes in secondary metabolite production.
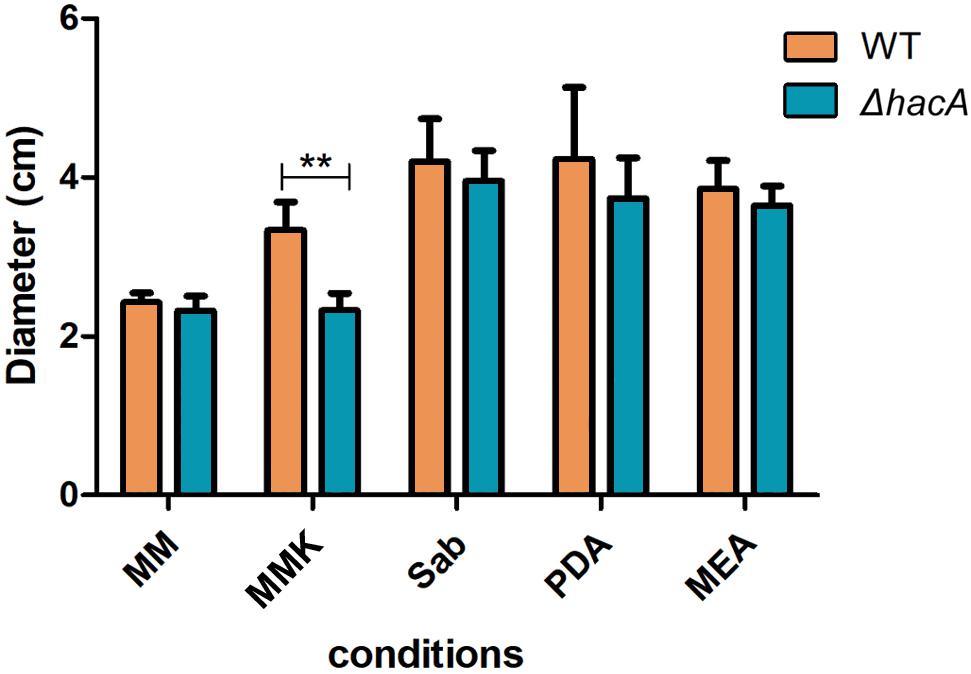
Figure 5. hacA supports T. rubrum growth on keratin sources. An equal plug from each indicated strain was spotted onto plates with a different culture medium. The growth rate (diameter in cm) was calculated for each culture, and each strain after 9 days at 28°C. MM, minimal medium; MMK, minimal medium contained keratin; Sab, Sabouraud; PDA, Potato dextrose agar; MEA, agar malt extract. Statistical significance was determined using Two-Way RM ANOVA followed by Bonferroni’s post-test (**P < 0.01).
hacA Regulates Membrane Homeostasis and Cell Wall Composition
The cell wall is the primary interface between fungus and host. As described previously in this work, ΔhacA showed differences in the growth rate in comparison to the wild type after exposure to the cell wall stressing agents. Further, under microscopic observation of the T. rubrum strains, a marked curling of the hyphae was observed for the mutant strain which suggested a defect in the hyphae directionality (Figure 6). Additionally, a marked impairment in protoplast regeneration was assessed for the ΔhacA mutant (data not shown). As ergosterol is the principal sterol within the cellular membrane with roles in fluidity, membrane protein assembly (Lv et al., 2016), and also in cell wall composition, we assessed the ergosterol content in both strains. The results showed a slight reduction in this content for the mutant strain (Figure 7).
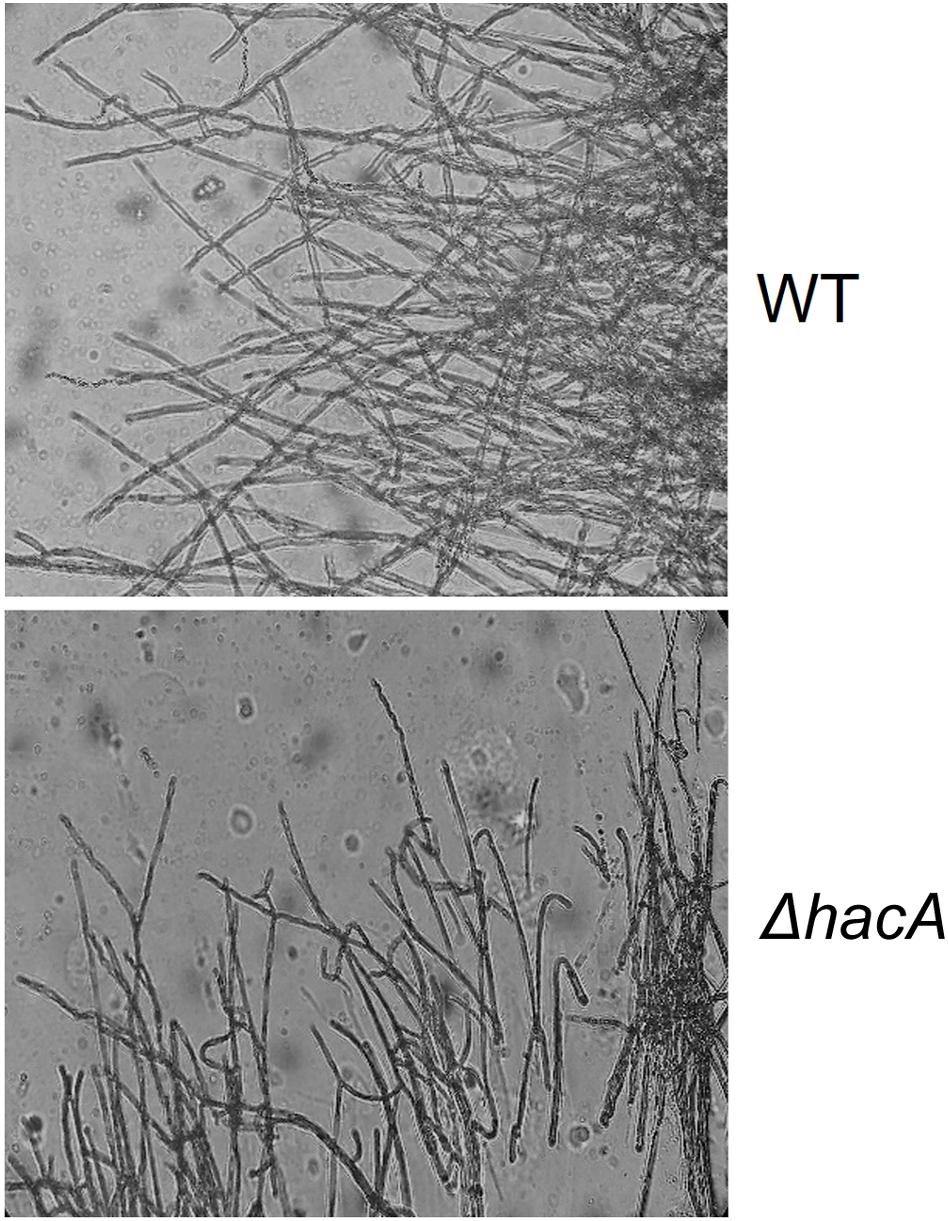
Figure 6. hacA supports hyphae directionality in T. rubrum. A microculture from both strains after 6 days at 28°C under microscopy light at magnification ×200.
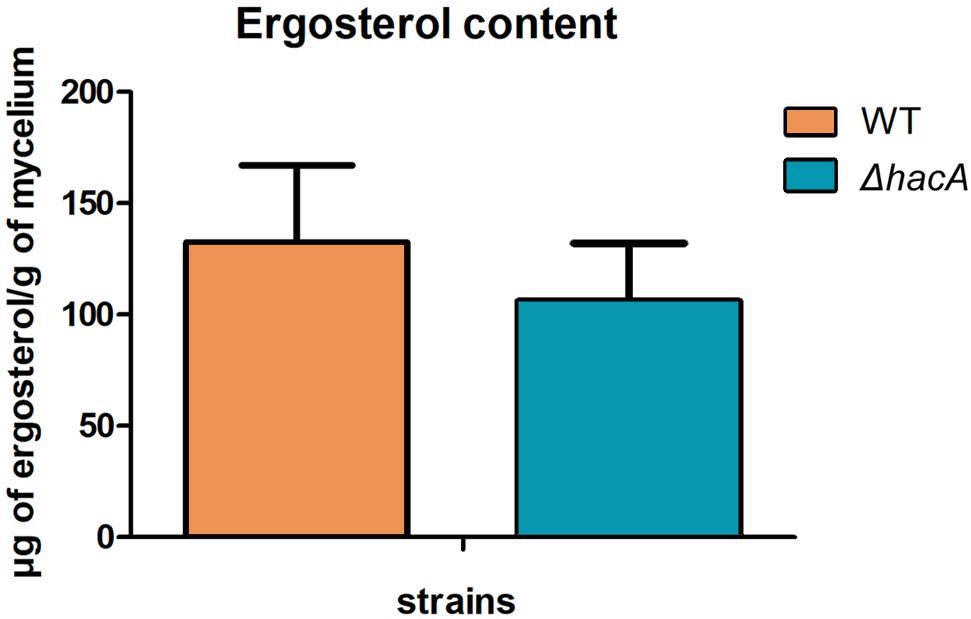
Figure 7. hacA contributes to ergosterol biosynthesis in T. rubrum. Ergosterol content in wild type and mutant strains assessed per gram of mycelium dry weight.
hacA Participates in Fungus-Host Interaction
We performed an interaction between conidia from both strains, wild type, and ΔhacA, with human nail fragments as well as in co-cultures with keratinocytes to assess virulence traits. These results showed decreased ability of the hacA mutant to grow on human nail fragments. Further, the directionality of hyphae seemed to be impaired, and the curling hyphae were evidenced (Figure 8A and Supplementary Figure S3). Moreover, the co-culture assay demonstrated a decrease in hyphal development for the ΔhacA mutant in comparison to the wild type (Figure 8B), which correlated with reduction of growth on the keratin media (Figure 9A). Otherwise, the activity of the secreted keratinolytic proteases was increased in the hacA mutant in comparison to the wild-type when both strains were cultivated on keratin powder (150 and 90 Units/g dry weight of mycelium, respectively (Figure 9B). This uncorrelated data might be due to a metabolic rearrangement as an attempt to use this complex protein substrate. Previous work reported that T. rubrum under undecanoic acid exposure presented a decrease in keratin growth and an increase in keratinolytic activity, which might be a consequence of changes in enzymes by post-translation modifications (Mendes et al., 2018).
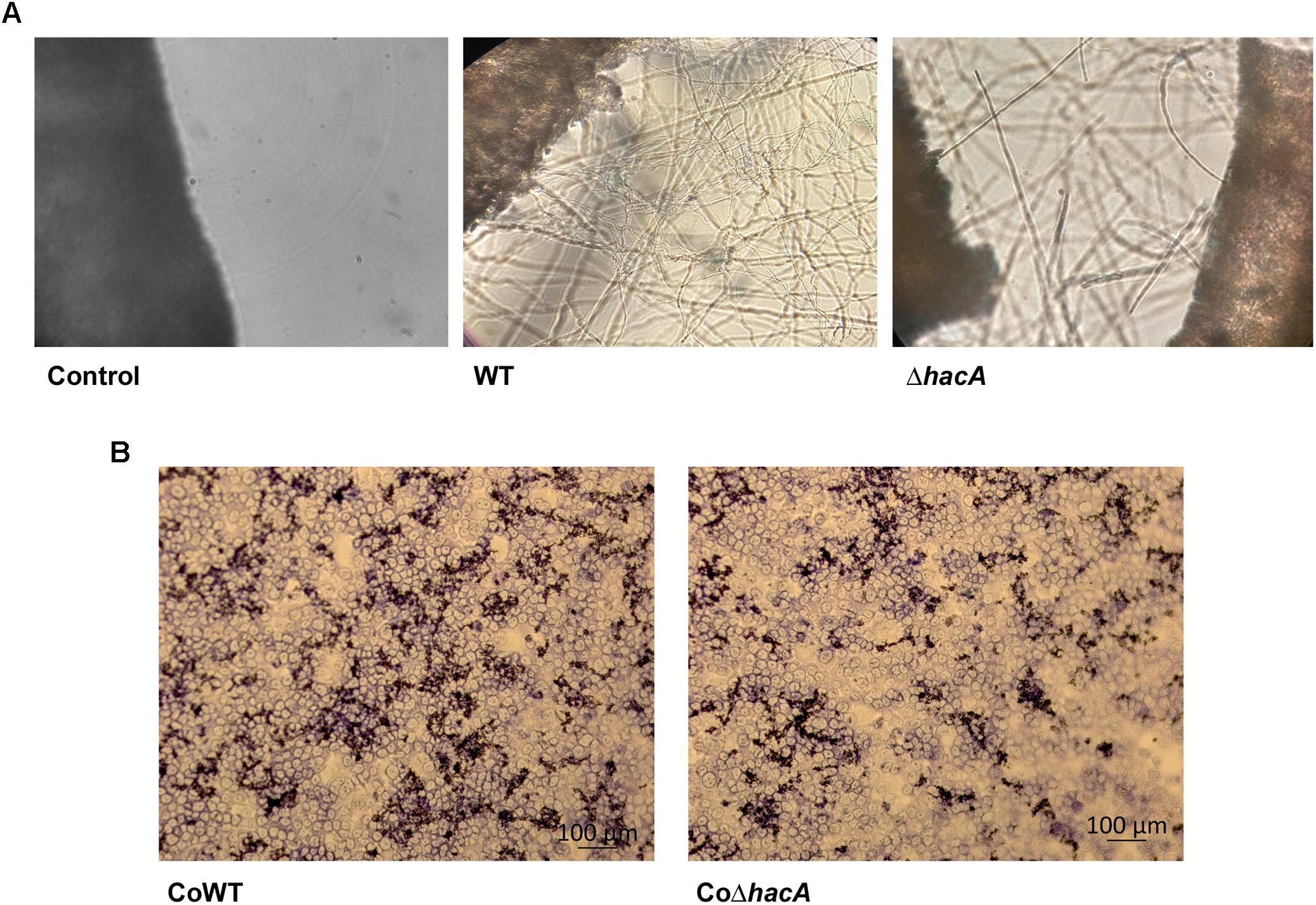
Figure 8. hacA contributes to host-fungi interaction. Effect of the hacA gene deletion on the growth of T. rubrum on human molecules. (A) Conidia from wild type and mutant strain were incubated on human nails for 72 h at 28°C. Fungal growth was observed by light microscopy. The black objects seen on the left side are related to nail fragments. (B) Coculture of T. rubrum conidia from both strains with keratinocyte cell type HaCaT for 24 h at 37°C.
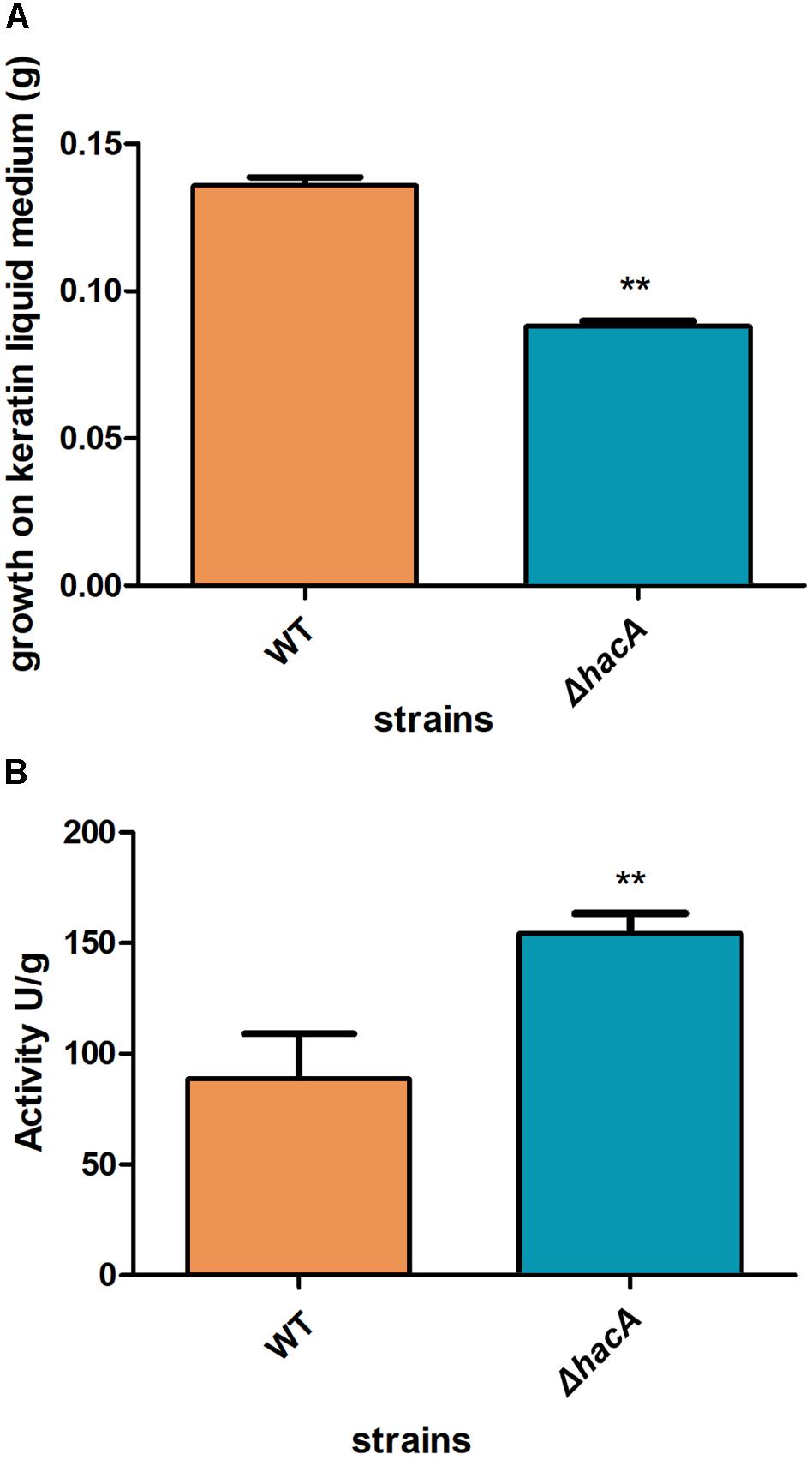
Figure 9. hacA supports keratinolytic feature of T. rubrum. (A) Dry weight mycelium from wild type and mutant strains expressed per gram of dry weight mycelium. (B) Keratinolytic activity from both strains was determined as mycelium-specific activities in units per gram of dry weight mycelium. Statistical significance determined using Unpaired t-test (**P < 0.01).
During fungus-host interaction, the fungal cell wall was the first line of contact with the host. Thus, the composition of the cell wall offers a wide array of molecules that act as patterns for recognition by host immune system and profoundly impact the relationship between host and pathogen. The receptors activated by these molecules are called pattern recognition receptors (PRRs), and their activation triggers intracellular signaling that leads to the production of pro-inflammatory mediators, like cytokines. In this work, we assessed the levels of IL-1β, IL-8, and TNFα after 24 h of co-culture of the human keratinocyte line (HaCaT) with conidia from both strains, the hacA mutant promoted differences in cytokine production, were related to an increase in TNFα and a decrease in IL-8; however, no significant differences were assessed for IL-1β (Figure 10). This result highlights the differences in cell wall patterns for the mutant strain.
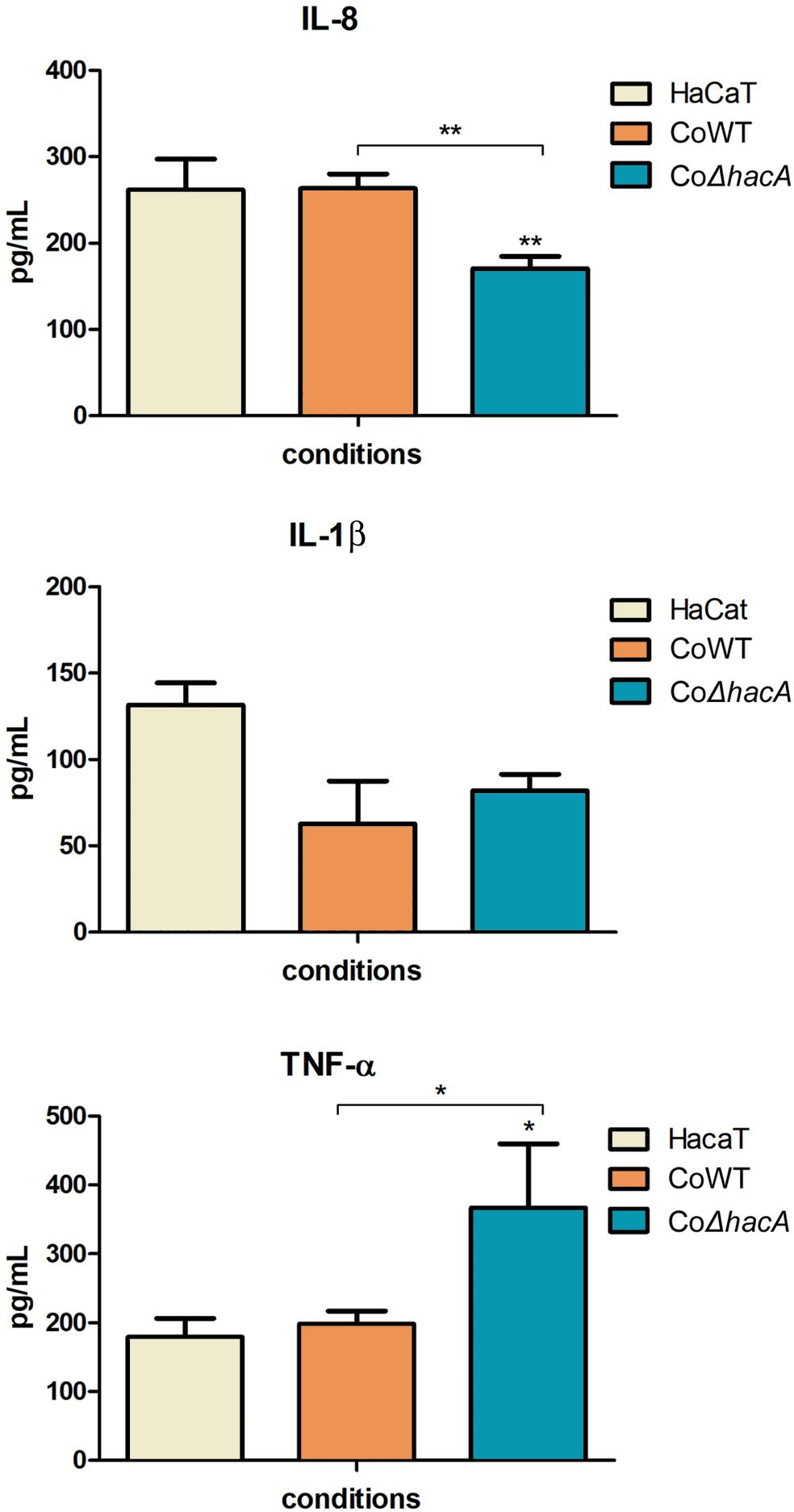
Figure 10. hacA participates in immune modulation in keratinocyte cells. Levels of pro-inflammatory cytokines output by HaCaT after coculture with conidia from the wild type and mutant strain for 24 h. Comparison of IL-8, IL-1β, and TNF-α secretion by HaCaT. Significantly different values are shown by asterisks, and were determined using ANOVA followed by Tukey’s ad hoc test (*P < 0.05; **P < 0.01).
hacA Regulates the Expression of Genes Belonging to Different Biological Processes
To assess genes that are directly or indirectly regulated by HacA, we evaluated the modulation of genes related to cell wall synthesis, ergosterol biosynthesis, pigmentation, heat shock proteins, and the genes coding for mannosyltransferase enzymes (Figures 11, 12).
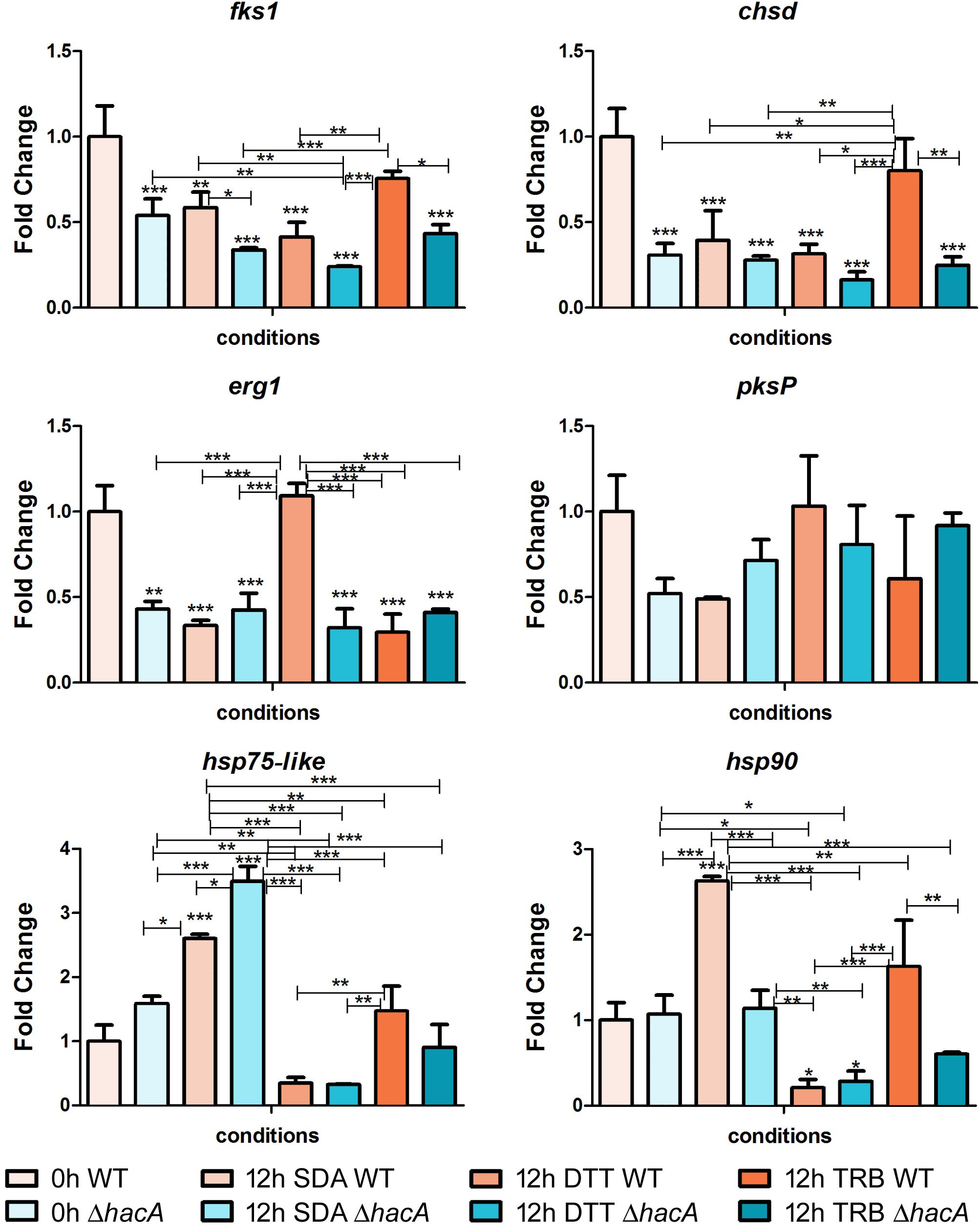
Figure 11. hacA regulates genes belonging to the different metabolic processes. Transcriptional levels of encoding genes of chsD, fks1, hsp75-like, hsp90, erg1, and pkP evaluated by qPCR for 12 h of both T. rubrum strains (wild type and ΔhacA) growth on Sabouraud (SDA), or SDA with DTT (10 mM), or Terbinafine (TRB in 0.014 μg/mL) compared to 0 h (control). Significantly different values are shown by asterisks, and were determined using ANOVA followed by Tukey’s ad hoc test (*P < 0.05; **P < 0.01; ***P < 0.001). Asterisks close to the bars are related to the comparison with the calibrator (0 h WT).
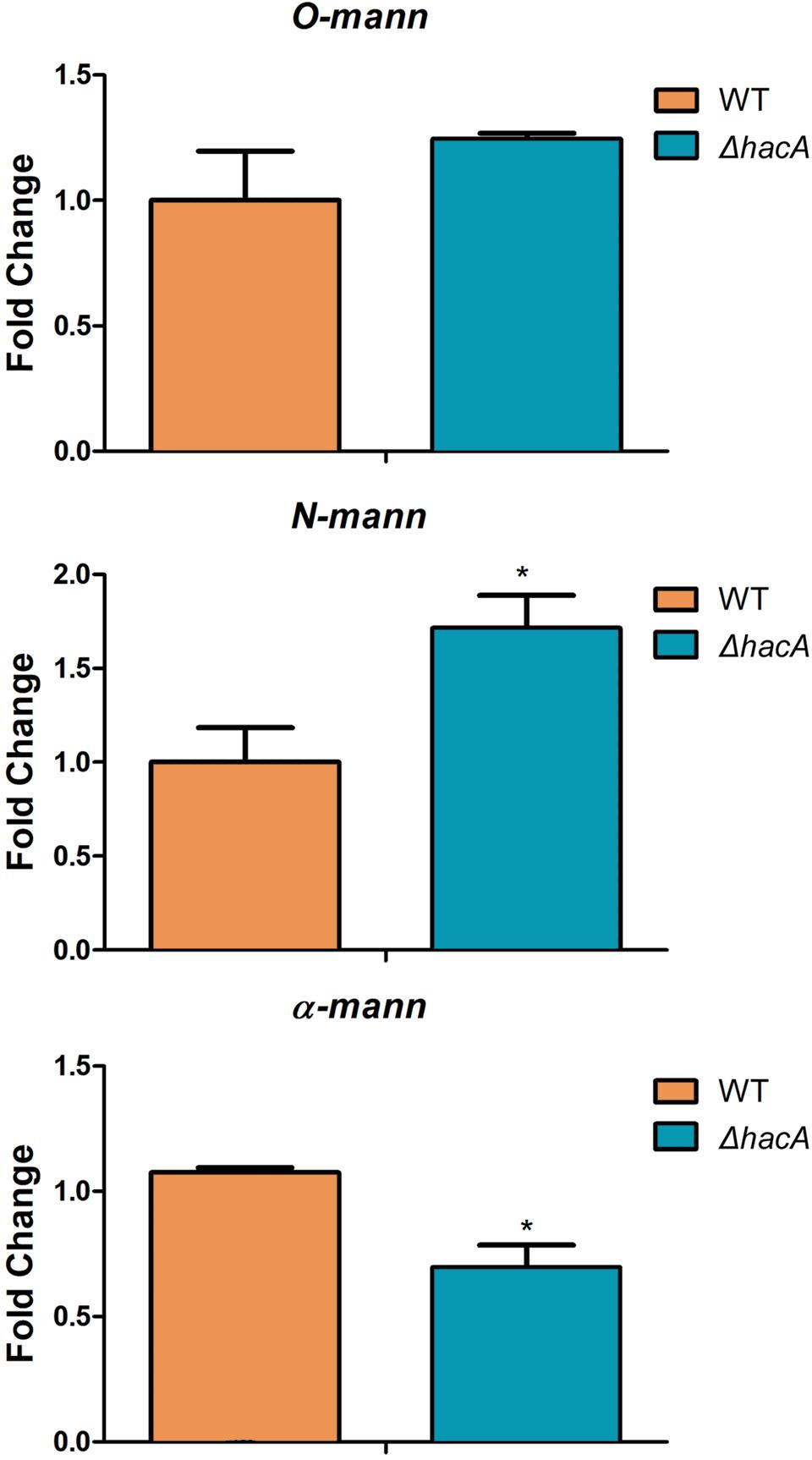
Figure 12. hacA regulates genes coding for mannosyltransferase in T. rubrum. Transcriptional levels of N-mann, O-mann, and α-mann from T. rubrum conidia cocultured with human keratinocyte cell line HaCaT for 24 h. Statistical significance was determined using Unpaired t-test (*P < 0.05).
Among cell wall encoding enzymes, we assessed the gene modulation of fks1 and chsD, and we observed marked differences in control (0 h) between the wild type and mutant, with a significant decrease in transcript levels for the mutant strain. Further, over time (additional 12 h of fungal growth), the mutant strain exhibited reduced transcription levels of fks1, compared to the wild type, whereas no differences in transcript levels were shown for chsD among the strains at the same time point. We also demonstrated that in response to terbinafine exposure, an increase in chsD transcriptional levels was assessed for the wild type, probably as an adaptive response leading to cell wall remodeling. Several lines of evidence point to the UPR impact on the fungal wall and cell membrane, suggesting potential connections between cell wall integrity (CWI) pathway and UPR (Malavazi et al., 2014). Moreover, previous work showed the relevance of chsB and chsD genes for the cell to cope with cell wall stress caused by pkcA deletion in A. fumigates (Rocha et al., 2015).
The erg1 transcript levels showed a marked decrease for the mutant in comparison to the wild type control (0 h). Chemical treatment with DTT promoted an up-regulation of erg1 from the wild type strain, while no significant differences were observed for the mutant. DTT induces an ER stress and as this organelle is linked to lipid synthesis; thus, could be possible that induction on the erg1 transcript levels was an attempt to cope with the ER perturbation. Conversely, no differences were found in the transcript levels of erg1 were verified for the TRB treatment at this time point.
The analyses of gene modulation for genes coding for Hsp90 and Hsp75-like exhibited different profiles among the studied strains. After 12 h of fungal growth, the wild type showed a prominent induction at the hsp90 levels, whereas at this same time point the mutant strain prompted a rise in the hsp75-like levels. These results might reflect a compensatory modulation in the ΔhacA strain. However, when exposed to terbinafine, only the wild type maintained elevated transcript levels for both Hsp encoding genes. It is already known that Hsp is involved in the sensing and adaptation of pathogens to thermal conditions as well as diverse conditions. In this context, previous reports have suggested Hsp90 as a potential therapeutic target as it is an abundant Hsp (corresponding to approximately 2% of the protein repertoire) which contributes to cell wall integrity, germination, pigmentation of conidia, drug resistance, and virulence (Jacob et al., 2015; Martinez-Rossi et al., 2016a, 2018). Remarkably, impairment of regulation in the mutant strain might be correlated with a compromise in cell wall composition, thermal tolerance, drug resistance, and changes in colony pigmentation previously described in this work.
In an attempt to analyze differences in pigmentation, the pksP gene modulation was assessed, and a slight difference in regulation was verified for the control (0 h) between strains. Notwithstanding, the DHN-melanin pathway consists of other genes that could be directly or indirectly regulated by HacA like pksP, arp1, and arp2. Another study has also reported that differences in the melanin of A. fumigatus within the conidial cell wall was found to be related to both stress tolerance and virulence (Hagiwara et al., 2017).
We also assessed the modulation of alpha-mannosyltranferase, N-mannosyltransferase, and O-mannosyltransferase genes because these results would be linked to differences in the immune responses triggered by each strain as was described previously in this article. We evaluated their modulation using RNA extracted from co-cultures. These results revealed significant changes in expression levels of the N-mannosyltranferase and the alpha-mannosyltranferase encoded genes within the ΔhacA mutant (Figure 12).
Together, these results are consistent with the analysis of the promoter region of the T. rubrum genes which matched the consensus motif of UPRE-1, UPRE-2, or UPRE-3, which are known as unfolded protein response elements. Our analysis showed that approximately 25% of T. rubrum genome might be potentially regulated by HacA (Supplementary Table S2). Among them are the mannosyltransferase enzymes, Hsps, fatty acid biosynthetic enzymes, cell wall enzymes, and proteases (Supplementary Table S2). The functional enrichment of these genes showed some main categories, such as fatty acid biosynthetic processes, methyltransferase activity, membrane composition and transport, oxidoreductase, and pyrimidine nucleotide biosynthetic processes (Figure 13).
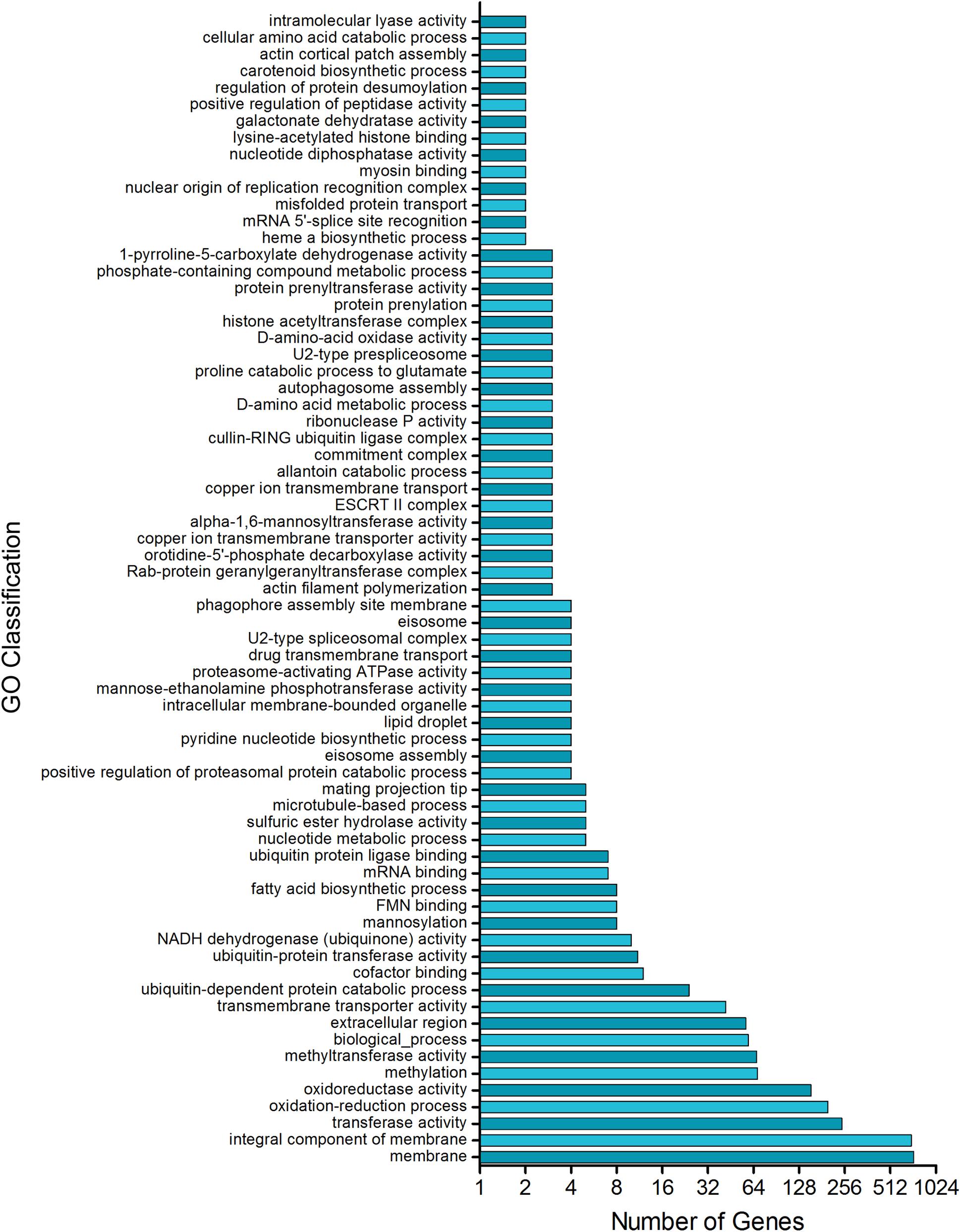
Figure 13. Putative HacA target genes in T. rubrum genome. Functional enrichment of genes in T. rubrum genome with a recognition site for UPRE-1 or UPRE-2 or UPR-3 motif consensus.
Discussion
Endoplasmic reticulum (ER) is the gateway for the secretory pathway and is the center for post-translational modification, accurate folding, and assembly of up to 30% of the cellular proteome (Schwarz and Blower, 2016). Fungi are organisms specialized for secretion and the utilization of this mechanism is entwined to the ability to sense environmental stress, thus leading to adequate responses (Krishnan and Askew, 2014). As homeostasis in the ER secretory capacity is followed by UPR activation, and current research has reported that the UPR genes are important vulnerability points to be exploited in fungal therapy, we decided to address the scope of hacA in the dermatophyte T. rubrum.
A non-canonical splicing of hacA mRNA mediates the activated form of HacA through IreA, The RNase domain of IreA is responsible for catalyzing the intron removal from hacA in a spliceosome independent manner, which in turn leads to a shift in the open reading frame, and the arisen of bZIP domain in activated HacA (Mori et al., 2000). Here, we demonstrated that the fragment which is removed from T. rubrum hacA mRNA is part of exon-2 and part of intron-2, and removal of this fragment prompt a shift in the open reading frame and the arisen in the DNA binding site, and then this activated form would be ready to go to the nucleus to bind with the target genes.
Noteworthy, peculiarities in this pathway were described herein, and they were related to differences in the hacA gene sequence arrangement since within this dermatophyte this gene is composed of 2 introns whereas in A. nidulans, A. fumigatus, and N. crassa present only one intron. In addition, features of the induced form of HacA from T. rubrum are also different in comparison to these other proteins from Aspergillus spp. and N. crassa. In these species, the changes in induced form occur in the 3′ portion of the protein, with no differences observed in the DNA binding site or in the dimer interface residues (Figures 1D,E). Remarkably, particularities in this well-characterized process were previously described (Montenegro-Montero et al., 2015), and they strengthen the importance of investigating different systems to address its involvement in fungal lifestyle and niche association preferences.
In filamentous fungi such as A. fumigatus, approximately 10% of the whole genome may be potentially regulated by IreA-HacA, whereas in Saccharomyces cerevisiae it is estimated that HacA might have regulated 5% through the UPRE-1 recognition sites (Fordyce et al., 2012; Krishnan and Askew, 2014). In this sense, another work showed that although all known bZIP transcription factors adopted, in general, one binding mode, HacA might present more binding sites, which correlated with the vast target repertoire (Fordyce et al., 2012). Indeed, it is assumed that normal growth conditions also require UPR activation. Bioinformatic analyses carried out here included searches for UPREs within the target gene promoters of T. rubrum identified a total of 2,178 target genes. These genes belong to diverse categories such as genes coding for the cell wall components, enzymes involved in fatty acid synthesis, Hsps, proteases, mannosyltransferase enzymes, and genes related to transcription and translation processes (Supplementary Table S2).
Thereafter, we hypothesized the requirement of UPR is dependent on HacA under different stressors, and we verified its traits of thermotolerance and in drug susceptibility. The results exhibited a marked sensitivity for the mutant strain during fungal growth at 37°C, and the most prominent effect occurred at 42°C. Although dermatophytes typically infect the skin, hair, and nails, there are some case reports of invasion into the dermis, subcutaneous tissue, or internal tissues (Marconi et al., 2010; Dai et al., 2019), which require adaptation to higher temperatures. Further, differences in the pigmentation of the colonies may be correlated to increased thermal sensitivity, since there is a link between melanin biosynthesis, thermal environment, and stress tolerance as was demonstrated for conidia from A. fumigatus (Hagiwara et al., 2017). In addition, this thermal sensitivity could be related to a reduction in ergosterol content, but we did not find compelling evidence for the T. rubrum mutant strain. Nonetheless, even a small reduction in ergosterol may destabilize the cellular membrane, and high temperatures can affect membrane fluidity and permeability (Simonin et al., 2007). Herein, we assumed that both correlated conditions contribute to inability of conidia survival under thermal stress. Also, changes in ergosterol levels and impairment in the cellular membrane might be entwined to sensibilization of the mutant strain toward ketoconazole. In A. fumigatus mutants (ΔireA and ΔhacA), an increase in azole susceptibility was also assessed (Feng et al., 2011).
In regard to cell wall impairment, there was evidence that this effect correlated with defects in the cellular membrane composition, and potentially a link between impairment of membrane composition and a decrease in the protoplast regeneration ability (Bitencourt et al., 2013). Indeed, the deletion of hacA in T. rubrum affected the hyphae directionality, compromised protoplast regeneration, and enhanced its susceptibility to cell wall inhibitors. The involvement of UPR in hyphae formation and cell wall integrity pathways were previously described (Krysan, 2009; Rocha et al., 2015). Beyond this, differences in cell wall patterns in the mutants for UPR were intimately linked to differences in virulence and host-pathogen interactions in other fungi (Cheon et al., 2011; Feng et al., 2011; Richie et al., 2011). These traits might be related to the difficulty of these mutants to grow at 37°C, as well as due to the involvement of UPR in host attachment. Indeed, our data showed a decrease in ΔhacA growth at the HaCaT cell line and human nail fragments. Further, we also demonstrated differences in the production of pro-inflammatory cytokines after infection with the wild type and ΔhacA strains. In summary, our data suggest a different pattern in cell wall organization, which might contribute to different pro-inflammatory cytokine release profiles. Previous works have shown that post-translation modification conducted in molecules from the cell wall had prompted recognition by different PRRs, and finally triggered differences in the immune response pattern via the keratinocyte surface (Figueiredo et al., 2011; Snarr et al., 2017). Furthermore, differences in cytokine release by keratinocytes are reportedly associated with the acute or chronic lesions caused by zoophilic or anthropophilic dermatophytes, respectively (Hau et al., 2015). While zoophilic species lead to a broad spectrum of cytokine release, including members from Th1, Th2, and Th17 response, anthropophilic species only induced the expression of IL-6, IL-8, IL-1β, and eotaxin-2 (Shiraki et al., 2006).
Our findings demonstrated that the mutant strain from T. rubrum showed decreased growth on keratin as the sole substrate. Previous studies have described the role of the UPR genes in supporting the growth of A. fumigatus in the lungs and that of A. niger in maltose as the growth substrate (Jorgensen et al., 2009; Feng et al., 2011). Involvement of the integrated pathways ERAD and UPR in the utilization of complex substrates was confirmed in the ΔhacA/derA mutant from A. fumigatus grown on lung and skim milk (Richie et al., 2011). Other studies have reported that the growth of filamentous fungi on complex substrates required high activity from the secretory pathway for this condition needed to elicit higher amounts of extracellular enzymes, which was accompanied by activation of UPR (Feng et al., 2011; Richie et al., 2011). Curiously, enhanced keratinolytic activity was exhibited for the ΔhacA mutant strain. We hypothesized that this strategy was due to a metabolic rearranging as an attempt to compensate for difficulties in the utilization of this complex protein substrate.
Conclusion
Our data unveil for the first time the involvement of HacA in dermatophytes physiology, response to stress, and host-pathogen interaction. Notwithstanding, a deeper understanding of the complex cross-talking of HacA with different metabolic pathways is paramount in importance to widen the knowledge about this potent transcriptional regulation. Regardless of how this will be addressed in the future, this work underscores the critical roles of HacA in T. rubrum virulence and adaptive responses. Together, these results provide valuable information about the efficacy of the use of HacA as a potential molecular target for novel antifungal therapy.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
ETHICS STATEMENT
The Committee of Ethics in Human Research of the Ribeirão Preto Medical School at the University of São Paulo approved all experiments involving the use of human nail fragments provided by healthy adults (Protocol No. 8330/2009). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
NM-R, TB, and AR conceived the study and wrote the manuscript. TB performed the experimental design and laboratory experiments. AF and EL contributed with deletion cassette construction. PS performed the bioinformatics analysis. NP assisted in the immunological assays. VO assisted in the microbiological assays.
Funding
This work was supported by grants from the Brazilian Agencies: São Paulo Research Foundation – FAPESP (Proc. No. 2014/03847-7, and Fellowship No. 2015/23435-8 to TB), National Council for Scientific and Technological Development – CNPq (Grant Nos. 305797/2017-4 and 304989/2017-7), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) – Finance Code 001, and Fundação de Apoio ao Ensino, Pesquisa e Assistência –FAEPA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Bruna A. M. Cantelli for assistance in coculture assays, M. Mazucato, and M. D. Martins for technical support, and P. P. Júnior for microscopic analysis support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00193/full#supplementary-material
References
Almeida, F., Rodrigues, M. L., and Coelho, C. (2019). The still underestimated problem of fungal diseases worldwide. Front. Microbiol. 10:214. doi: 10.3389/fmicb.2019.00214
Arthington-Skaggs, B. A., Jradi, H., Desai, T., and Morrison, C. J. (1999). Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J. Clin. Microbiol. 37, 3332–3337. doi: 10.1128/jcm.37.10.3332-3337.1999
Bitencourt, T. A., Komoto, T. T., Massaroto, B. G., Miranda, C. E., Beleboni, R. O., Marins, M., et al. (2013). Trans-chalcone and quercetin down-regulate fatty acid synthase gene expression and reduce ergosterol content in the human pathogenic dermatophyte Trichophyton rubrum. BMC Comple. Altern. Med. 13:229. doi: 10.1186/1472-6882-13-229
Blankenship, J. R., Fanning, S., Hamaker, J. J., and Mitchell, A. P. (2010). An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752. doi: 10.1371/journal.ppat.1000752
Boppidi, K. R., Ribeiro, L. F. C., Iambamrung, S., Nelson, S. M., Wang, Y., Momany, M., et al. (2018). Altered secretion patterns and cell wall organization caused by loss of PodB function in the filamentous fungus Aspergillus nidulans. Sci. Rep. 8:11433. doi: 10.1038/s41598-018-29615-z
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv113.
Cheon, S. A., Jung, K. W., Chen, Y. L., Heitman, J., Bahn, Y. S., and Kang, H. A. (2011). Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog 7:e1002177. doi: 10.1371/journal.ppat.1002177
Clinical Laboratory Standards Institute [CLSI] (2008). “Refeference method for broth dilution antifungal susceptibility testing of filamentous fungi,” in Clinical Laboratory Standards Institute (CLSI) Approved -, 2nd Edn, (Wayne, PA: CLSI).
Cove, D. J. (1966). The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113, 51–56. doi: 10.1016/s0926-6593(66)80120-0
Dai, Y., Xia, X., and Shen, H. (2019). Multiple abscesses in the lower extremities caused by Trichophyton rubrum. BMC Infect. Dis. 19:271. doi: 10.1186/s12879-019-3897-3
Darty, K., Denise, A., and Ponty, Y. (2009). VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975. doi: 10.1093/bioinformatics/btp250
Fachin, A. L., Ferreira-Nozawa, M. S., Maccheroni, W. Jr., and Martinez-Rossi, N. M. (2006). Role of the ABC transporter TruMDR2 in terbinafine, 4-nitroquinoline N-oxide and ethidium bromide susceptibility in Trichophyton rubrum. J. Med. Microbiol. 55, 1093–1099. doi: 10.1099/jmm.0.46522-0
Feng, X., Krishnan, K., Richie, D. L., Aimanianda, V., Hartl, L., Grahl, N., et al. (2011). HacA-independent functions of the ER stress sensor IreA synergize with the canonical UPR to influence virulence traits in Aspergillus fumigatus. PLoS Pathog 7:e1002330. doi: 10.1371/journal.ppat.1002330
Figueiredo, R. T., Carneiro, L. A., and Bozza, M. T. (2011). Fungal surface and innate immune recognition of filamentous fungi. Front. Microbiol. 2:248. doi: 10.3389/fmicb.2011.00248
Fordyce, P. M., Pincus, D., Kimmig, P., Nelson, C. S., El-Samad, H., Walter, P., et al. (2012). Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses. Proc. Natl. Acad. Sci. U.S.A. 109, E3084–E3093. doi: 10.1073/pnas.1212457109
Gradisar, H., Friedrich, J., Krizaj, I., and Jerala, R. (2005). Similarities and specificities of fungal keratinolytic proteases: comparison of keratinases of Paecilomyces marquandii and Doratomyces microsporus to some known proteases. Appl. Environ. Microbiol. 71, 3420–3426. doi: 10.1128/aem.71.7.3420-3426.2005
Hagiwara, D., Sakai, K., Suzuki, S., Umemura, M., Nogawa, T., Kato, N., et al. (2017). Temperature during conidiation affects stress tolerance, pigmentation, and trypacidin accumulation in the conidia of the airborne pathogen Aspergillus fumigatus. PLoS One 12:e0177050. doi: 10.1371/journal.pone.0177050
Hau, C. S., Tada, Y., Kanda, N., and Watanabe, S. (2015). Immunoresponses in dermatomycoses. J. Dermatol. 42, 236–244. doi: 10.1111/1346-8138.12718
Jacob, T. R., Peres, N. T. A., Martins, M. P., Lang, E. A., Sanches, P. R., Rossi, A., et al. (2015). Heat Shock Protein 90 (Hsp90) as a Molecular Target for the Development of Novel Drugs Against the Dermatophyte Trichophyton rubrum. Front. Microbiol. 6:10. doi: 10.3389/fmicb.2015.01241
Jacob, T. R., Peres, N. T. A., Persinoti, G. F., Silva, L. G., Mazucato, M., Rossi, A., et al. (2012). rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med. Mycol. 50, 368–377. doi: 10.3109/13693786.2011.616230
Jorgensen, T. R., Goosen, T., Hondel, C. A., Ram, A. F., and Iversen, J. J. (2009). Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway. BMC Genomics 10:44. doi: 10.1186/1471-2164-10-44
Komoto, T. T., Bitencourt, T. A., Silva, G., Beleboni, R. O., Marins, M., and Fachin, A. L. (2015). Gene expression response of Trichophyton rubrum during coculture on keratinocytes exposed to antifungal agents. Evid. Based. Complement. Alternat. Med. 2015:180535. doi: 10.1155/2015/180535
Krishnan, K., and Askew, D. S. (2014). Endoplasmic reticulum stress and fungal pathogenesis. Fungal. Biol. Rev. 28, 29–35. doi: 10.1016/j.fbr.2014.07.001
Krysan, D. J. (2009). The cell wall and endoplasmic reticulum stress responses are coordinately regulated in Saccharomyces cerevisiae. Commun. Integr. Biol. 2, 233–235. doi: 10.4161/cib.2.3.8097
Kuck, U., and Hoff, B. (2010). New tools for the genetic manipulation of filamentous fungi. Appl. Microbiol. Biotechnol. 86, 51–62. doi: 10.1007/s00253-009-2416-7
Leal, J., Squina, F. M., Freitas, J. S., Silva, E. M., Ono, C. J., Martinez-Rossi, N. M., et al. (2009). A splice variant of the Neurospora crassa hex-1 transcript, which encodes the major protein of the Woronin body, is modulated by extracellular phosphate and pH changes. FEBS Lett. 583, 180–184. doi: 10.1016/j.febslet.2008.11.050
Lv, Q. Z., Yan, L., and Jiang, Y. Y. (2016). The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn. Virulence 7, 649–659. doi: 10.1080/21505594.2016.1188236
Malavazi, I., Goldman, G. H., and Brown, N. A. (2014). The importance of connections between the cell wall integrity pathway and the unfolded protein response in filamentous fungi. Briefings Func. Genomics 13, 456–470. doi: 10.1093/bfgp/elu027
Marconi, V. C., Kradin, R., Marty, F. M., Hospenthal, D. R., and Kotton, C. N. (2010). Disseminated dermatophytosis in a patient with hereditary hemochromatosis and hepatic cirrhosis: case report and review of the literature. Med. Mycol. 48, 518–527. doi: 10.3109/13693780903213512
Martinez-Rossi, N. M., Bitencourt, T. A., Peres, N. T. A., Lang, E. A. S., Gomes, E. V., Quaresemin, N. R., et al. (2018). Dermatophyte resistance to antifungal drugs: mechanisms and prospectus. Front. Microbiol. 9:1108. doi: 10.3389/fmicb.2018.01108
Martinez-Rossi, N. M., Jacob, T. R., Sanches, P. R., Peres, N. T. A., Lang, E. A. S., Martins, M. P., et al. (2016a). Heat shock proteins in dermatophytes: current advances and perspectives. Curr. Genomics 17, 99–111. doi: 10.2174/1389202917666151116212437
Martinez-Rossi, N. M., Peres, N. T. A., and Rossi, A. (2016b). Pathogenesis of dermatophytosis: sensing the host tissue. Mycopathologia 182, 215–227. doi: 10.1007/s11046-016-0057-9
Matsumoto, R., Akama, K., Rakwal, R., and Iwahashi, H. (2005). The stress response against denatured proteins in the deletion of cytosolic chaperones SSA1/2 is different from heat-shock response in Saccharomyces cerevisiae. BMC Genomics 6:141. doi: 10.1186/1471-2164-6-141
Mendes, N. S., Bitencourt, T. A., Sanches, P. R., Silva-Rocha, R., Martinez-Rossi, N. M., and Rossi, A. (2018). Transcriptome-wide survey of gene expression changes and alternative splicing in Trichophyton rubrum in response to undecanoic acid. Sci. Rep. 8:14. doi: 10.1038/s41598-018-20738-x
Mendes, N. S., Trevisan, G. L., Cruz, A. H. S., Santos, R. S., Peres, N. T. A., Martinez-Rossi, N. M., et al. (2012). Transcription of N- and O-linked mannosyltransferase genes is modulated by the pacC gene in the human dermatophyte Trichophyton rubrum. FEBS Open Bio 2, 294–297. doi: 10.1016/j.fob.2012.09.005
Mercer, D. K., and Stewart, C. S. (2019). Keratin hydrolysis by dermatophytes. Med. Mycol. 57, 13–22. doi: 10.1093/mmy/myx160
Monod, M., and Mehul, B. (2019). Recent findings in onychomycosis and their application for appropriate treatment. J. Fungi. 5:20. doi: 10.3390/jof5010020
Montenegro-Montero, A., Goity, A., and Larrondo, L. F. (2015). The bzip transcription factor HAC-1 Is Involved in the Unfolded Protein Response And Is Necessary For Growth On Cellulose in Neurospora crassa. PLoS One 10:e0131415. doi: 10.1371/journal.pone.0131415
Moore, K. A., and Hollien, J. (2012). The unfolded protein response in secretory cell function. Annu. Rev. Genet. 46, 165–183. doi: 10.1146/annurev-genet-110711-155644
Mori, K., Ogawa, N., Kawahara, T., Yanagi, H., and Yura, T. (2000). mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci.U.S. A. 97, 4660–4665. doi: 10.1073/pnas.050010197
Nawrocki, E. P., Kolbe, D. L., and Eddy, S. R. (2009). Infernal 1.0: inference of RNA alignments. Bioinformatics 25, 1335–1337. doi: 10.1093/bioinformatics/btp157
Neves-Da-Rocha, J., Bitencourt, T. A., Oliveira, V. M., Sanches, P. R., Rossi, A., and Martinez-Rossi, N. M. (2019). Alternative splicing in heat shock protein transcripts as a mechanism of cell adaptation in Trichophyton rubrum. Cells 8:E1206. doi: 10.3390/cells8101206
Oh, M. H., Cheon, S. A., Kang, H. A., and Kim, J. Y. (2010). Functional characterization of the unconventional splicing of Yarrowia lipolytica HAC1 mRNA induced by unfolded protein response. Yeast 27, 443–452. doi: 10.1002/yea.1762
Peres, N. T. A., Silva, L. G., Santos Rda, S., Jacob, T. R., Persinoti, G. F., Rocha, L. B., et al. (2016). In vitro and ex vivo infection models help assess the molecular aspects of the interaction of Trichophyton rubrum with the host milieu. Med. Mycol. 54, 420–427. doi: 10.1093/mmy/myv113
Richie, D. L., Feng, X., Hartl, L., Aimanianda, V., Krishnan, K., Powers-Fletcher, M. V., et al. (2011). The virulence of the opportunistic fungal pathogen Aspergillus fumigatus requires cooperation between the endoplasmic reticulum-associated degradation pathway (ERAD) and the unfolded protein response (UPR). Virulence 2, 12–21. doi: 10.4161/viru.2.1.13345
Rocha, M. C., Godoy, K. F., De Castro, P. A., Hori, J. I., Bom, V. L., Brown, N. A., et al. (2015). The Aspergillus fumigatus pkcA G579R mutant is defective in the activation of the cell wall integrity pathway but is dispensable for virulence in a neutropenic mouse infection model. PLoS One 10:e0135195. doi: 10.1371/journal.pone.0135195
Saloheimo, M., Valkonen, M., and Penttila, M. (2003). Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 47, 1149–1161. doi: 10.1046/j.1365-2958.2003.03363.x
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schwarz, D. S., and Blower, M. D. (2016). The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol. Life Sci. 73, 79–94. doi: 10.1007/s00018-015-2052-6
Shiraki, Y., Ishibashi, Y., Hiruma, M., Nishikawa, A., and Ikeda, S. (2006). Cytokine secretion profiles of human keratinocytes during Trichophyton tonsurans and Arthroderma benhamiae infections. J. Med. Microbiol. 55, 1175–1185. doi: 10.1099/jmm.0.46632-0
Simonin, H., Beney, L., and Gervais, P. (2007). Cell death induced by mild physical perturbations could be related to transient plasma membrane modifications. J. Membr. Biol. 216, 37–47. doi: 10.1007/s00232-007-9027-y
Snarr, B. D., Qureshi, S. T., and Sheppard, D. C. (2017). Immune recognition of fungal polysaccharides. J. Fungi. 3:E47.
Wimalasena, T. T., Enjalbert, B., Guillemette, T., Plumridge, A., Budge, S., Yin, Z., et al. (2008). Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet. Biol. 45, 1235–1247. doi: 10.1016/j.fgb.2008.06.001
Zhang, L., Zhang, C., and Wang, A. (2016). Divergence and conservation of the major UPR branch IRE1-bZIP signaling pathway across eukaryotes. Sci. Rep. 6:27362. doi: 10.1038/srep27362
Keywords: mycoses, secretory system, unfolded protein response, dermatophytes, endoplasmic reticulum, host-pathogen interaction
Citation: Bitencourt TA, Lang EAS, Sanches PR, Peres NTA, Oliveira VM, Fachin AL, Rossi A and Martinez-Rossi NM (2020) HacA Governs Virulence Traits and Adaptive Stress Responses in Trichophyton rubrum. Front. Microbiol. 11:193. doi: 10.3389/fmicb.2020.00193
Received: 15 November 2019; Accepted: 27 January 2020;
Published: 20 February 2020.
Edited by:
Hector Mora Montes, University of Guanajuato, MexicoReviewed by:
Vishukumar Aimanianda, Institut Pasteur, FranceRoberta Gaziano, University of Rome Tor Vergata, Italy
Copyright © 2020 Bitencourt, Lang, Sanches, Peres, Oliveira, Fachin, Rossi and Martinez-Rossi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nilce M. Martinez-Rossi, bm1tcm9zc2lAdXNwLmJy
 Tamires A. Bitencourt
Tamires A. Bitencourt Elza A. S. Lang
Elza A. S. Lang Pablo R. Sanches
Pablo R. Sanches Nalu T. A. Peres
Nalu T. A. Peres Vanderci M. Oliveira
Vanderci M. Oliveira Ana Lúcia Fachin
Ana Lúcia Fachin Antonio Rossi
Antonio Rossi Nilce M. Martinez-Rossi
Nilce M. Martinez-Rossi