
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 February 2020
Sec. Food Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00184
Streptococcus thermophilus plays important roles in the dairy industry and is widely used as a dairy starter in the production of fermented dairy products. The genomes of S. thermophilus strains CS5, CS9, CS18, and CS20 from fermented milk in China were sequenced and used for biodiversity analysis. In the present study, the phylogenetic analysis of all 34 S. thermophilus genomes publicly available including these four strains reveals that the phylogenetic reconstruction does not match geographic distribution as strains isolated from the same continent are not even clustered on the nearby branches. The core and variable genes were also identified, which vary among strains from 0 to 202. CS9 strain contained 127 unique genes from a variety of distantly related species. It was speculated that CS9 had undergone horizontal gene transfer (HGT) during the long evolutionary process. The safety evaluation of these four strains indicated that none of them contains antibiotic resistance genes and that they are all sensitive to multiple antibiotics. In addition, the strains do not contain any pathogenic virulence factors or plasmids and thus can be considered safe. Furthermore, these strains were investigated in terms of their technological properties including milk acidification, exopolysaccharide (EPS) and γ-aminobutyric acid (GABA) production, and in vitro survival capacity in the gastrointestinal tract. CS9 possesses a special eps gene cluster containing significant traces of HGT, while the eps gene clusters of CS5, CS18, and CS20 are almost the same. The monosaccharide compositional analysis indicated that crude EPS-CS5, EPS-CS9, EPS-CS18, and EPS-CS20 contain similar monosaccharide compositions with different ratios. Furthermore, CS9 was one of a few GABA-producing strains that could ferment glutamate to produce GABA, which is beneficial for improving the acid tolerance of the strain. CS18 has the most potential for the production of fermented food among these four strains because of its fast growth rate, rapid acidifying capacity, and stronger acid and bile salt resistance capacity. This study focused on the genome analysis of the four new S. thermophilus strains to investigate the diversity of strains and provides a reference for selecting excellent strains by use of the genome data.
Streptococcus thermophilus is a necessary starter culture for yogurt manufacturing and widely used in yogurt and cheese production worldwide (Cui et al., 2016; Uriot et al., 2017). It has rapid acidifying capacity during milk fermentation and accelerates the coagulation of milk proteins. Meanwhile, this bacterium could synthesize some beneficial molecules, such as exopolysaccharide (EPS), aromatic compounds, and formic and folic acids, which contribute to many desirable production characteristics of dairy products including viscosity, texture, flavor, and water holding capacity, etc. (Hols et al., 2005; Cui et al., 2016, 2017). Furthermore, it has been confirmed that S. thermophilus has various health benefits such as anti-inflammatory activity, antimicrobial activity, antioxidant activity, as well as immunomodulation (Junjua et al., 2016; Cui et al., 2017; Uriot et al., 2017). These probiotic effects have a close relationship with the biologically active molecules produced by S. thermophilus such as EPS and bacteriocins (Hols et al., 2005; Junjua et al., 2016; Cui et al., 2017; Uriot et al., 2017).
This bacterium has been widely studied in the past few decades due to its widespread industrial application and attractive probiotic effects. In order to meet the demands of consumers for high-quality food products as well as the needs of the fermentation industry, finding new strains with outstanding processing and functional properties is a long-term and important focus for researchers. Additionally, new strains can contribute to natural diversity (Erkus et al., 2014).
In the last two decades, a number of S. thermophilus genomes have been published, and the whole genome information has improved the understanding of metabolic activities of this bacterium at the molecular level, including the biosynthesis of EPS and folate (Iyer et al., 2010; Cui et al., 2017; Li et al., 2018; Xiong et al., 2019), resistance to bacteriophage (Hao et al., 2018), proteolytic system (Tian et al., 2018), and carbohydrate metabolism (Prajapati et al., 2013), etc. Furthermore, the comparative genomic analysis of different S. thermophilus strains with various technological properties has advanced the development of knowledge about the relationship between genetic characters and phenotypic traits (Rasmussen et al., 2008; Vendramin et al., 2017).
Antibiotics started to be utilized to treat bacterial infections 60 years ago when natural strains did not contain drug resistance genes (Hughes and Datta, 1983). However, as the application of antibiotics becomes increasingly popular, resistance to antibiotics occurs during treatment. The excessive or wrong use of antibiotics coupled with the phenomenon of gene-level transfer between bacteria makes the strain gradually develop resistance in order to cope with pressure from the external environment (Polly, 1993; Levy, 1997). S. thermophilus is usually used as a natural food starter, thus these strains will inevitably enter the human body or animals. Once the strain carries a transferable resistance gene, it is likely to transfer the resistance gene to pathogens in the intestine, making it extremely difficult to prevent and kill them. Therefore, the analysis of strain antibiotic resistance and antibiotic resistance genes is crucially important when evaluating whether a strain can be used as a new food starter.
In order to obtain novel strains with outstanding functional/technological characteristics, 22 S. thermophilus strains were isolated from traditional yogurt samples, Inner Mongolia, China (Hu et al., 2018). Four strains named CS5, CS9, CS18, and CS20 with distinctive properties were selected by means of a series of evaluations in terms of their technological properties.
In this study, the whole genomes of these four strains were sequenced and used for biodiversity analysis of the S. thermophilus strains. The comparative analysis of genome sequences was carried out among all the S. thermophilus strains available in the public database. This kind of comparative phylogenetic analysis that is used for inspection of probiotic potentials has already been done for other bacterial species, such as Lactobacillus acidipiscis, Lactobacillus crispatus, Lactobacillus casei, and Lactobacillus helveticus (Ojala et al., 2014; Fontana et al., 2018, 2019; Kazou et al., 2018). Furthermore, important technological properties of the four strains were investigated including milk acidification, antibiotic resistance, EPS and γ-aminobutyric acid (GABA) production ability, and in vitro survival capacity in the gastrointestinal tract (GIT).
The results indicated that phylogenetic reconstruction does not match geographic distribution as strains isolated from the same continent are even not clustered on the nearby branches. It was found that the CS9 strain possessed a large number of unique genes, including a distinctive eps gene cluster and genes associated with replication, recombination, self-repair, etc. Therefore, CS9 could be used as a material for the research of evolution.
By screening and comparing the antibiotic resistance genes of these four strains and testing the sensitivity of the strains to 12 common antibiotics, it was found that these four strains did not contain any antibiotic resistance genes and that all of these 12 antibiotics showed a high degree of sensitivity. The results from the prediction and analysis of virulence factors and transposon indicate that the virulence factors in these four strains are mostly related to the synthesis and transport of active substances such as EPS as well as cell metabolism and thus are not pathogenic. Moreover, most of the transposase genes have been degenerated into pseudogenes due to frameshift mutations and therefore are unable to synthesize transposase which may transfer harmful genes. The remaining genes that can function are homologous to the genomes of known food-grade S. thermophilus. Based on the results of the safety evaluation, it can be concluded that all of these four strains of S. thermophilus can be considered safe; they do not cause antibiotic contamination in nature and do not contain or transport pathogenic genes. Thus, these four strains can be safely used as a natural food starter.
Furthermore, CS9 was one of the few GABA-producing strains that could ferment glutamate to produce GABA which can improve the acid tolerance of the strain. At the same time, among these four strains, CS18 has the most potential for the production of fermented food due to its fast growth rate, rapid acidifying ability, and stronger acid and bile salt resistance capacity.
Streptococcus thermophilus strains CS5, CS9, CS18, and CS20 were isolated from yogurt in our previous study (Hu et al., 2018). They were maintained in M17 medium along with 30% (v/v) glycerol at −80°C and propagated at 37°C for three generations before use. The cultures were transferred to sterilized skimmed milk (Fine Life, MCC Trading International GmbH, Germany) for fermentation experiments.
The whole genome sequencing of CS5, CS9, CS18, and CS20 was performed using a combined sequencing platform of Illumina platform and Pacbio RSII. Illumina PE library and PacBio library were constructed, respectively (Table 1). CS5 was sequenced by Illumina Miseq and Pacbio RSII. CS9, CS18, and CS20 were sequenced by Illumina Hiseq and Pacbio RSII. The whole genome map of the four strains was then completed by bioinformatics analysis after the quality control. The results were then assembled and reintegrated by Summer software. Genome annotation was performed using NCBI Prokaryotic Genome Annotation Pipeline1.
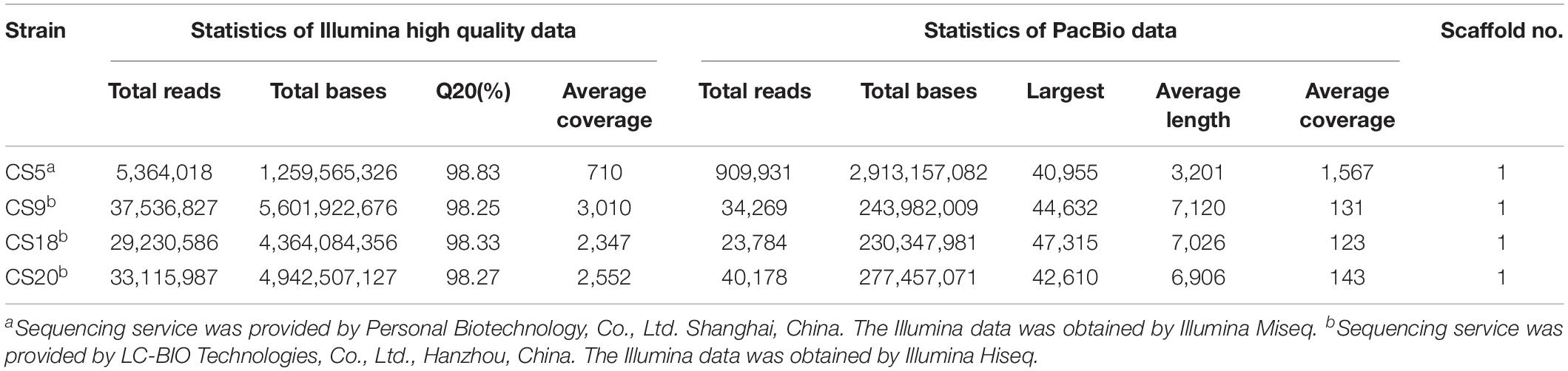
Table 1. The whole genome sequencing and assembly related information of Streptococcus thermophilus strains CS5, CS9, CS18, and CS20.
The ab initio gene prediction method was used to get gene models which were identified using Glimmer3 (Delcher et al., 1999). Then all gene models were blasted against the non-redundant (NR in NCBI) database, SwissProt2, KEGG3 (Kanehisa et al., 2012), and COG4 (Tatusov et al., 2000). Genomic islands (GIs) located on the genome were detected by software IslandViewer (Langille and Brinkman, 2009). The family distribution, conserved domain, and model structure of glycosyltransferase (GTF) were analyzed using the carbohydrate-active enzymes database5, the Conserved Domain Database6, and the SWISS-MODEL server7. The virulence genes in the S. thermophilus strains were predicted based on the VFDB (Virulence Factors of Pathogenic Bacteria) database. The similarities of antibiotic resistance genes and transposases were blast on the NCBI database, and the pseudogenes were determined by the NCBI annotation.
The sequencing service of CS5 was provided by Personal Biotechnology, Co., Ltd., Shanghai, China. The other three strains were sequenced by LC-BIO Technologies, Co., Ltd., Hanzhou, China. The nucleotide sequences of CS5, CS9, CS18, and CS20 genomes were submitted to GenBank and assigned accession numbers CP028896, CP030927, CP030928, and CP030250.
Thirty S. thermophilus available genomes were downloaded from the NCBI database in July 2018 (Supplementary Table S1). The phylogenetic tree was built based on the whole genome sequences by means of Neighbor-Joining within the MEGA 6.0 software (Tamura et al., 2013).
Antibiotic sensitivity determination was carried out according to Sorokulova et al. (2008) with some modifications: 1 ml of a suspension of 1 × 108 colony-forming units (CFUs)/ml was pipetted into 20 ml of melted solid M17 medium (55°C), then mixed and poured into a sterile dish. After the medium cooled and became solidified, wafers containing antibiotics (Bin He Microorganism Reagent, Co., Ltd.) were placed on the agar plate. Each plate was attached with three paper disks containing the same antibiotic. Then, those plates were incubated anaerobically for 48 h at an incubation temperature of 37°C before the diameters of the inhibition zones were measured and recorded. The diameter of the transparent circle around the antibiotic paper was compared to “Performance Standards for Antimicrobial Susceptibility Testing” (2013 edition) developed by the US Clinical and Laboratory Standards Institute to evaluate the sensitivity of bacteria to antibiotics. The antibiotics include streptomycin, ampicillin, erythromycin, clindamycin, oxacillin, rifampicin, amoxicillin, cefuroxime, tetracycline, chloramphenicol, penicillin, and vancomycin.
To determine the growth ability of strains, the cells were inoculated in M17 medium (1 L M17 liquid medium contains 5.0 g soya peptone, 2.5 g yeast extract, 5.0 g peptone, 0.5 g ascorbic acid, 5.0 g beef powder, 0.25 g magnesium sulfate, 5.0 g β-glycerol phosphate disodium salt, and 5.0 g lactose) and incubated anaerobically at 37°C overnight. The 200-μl volume of the suspension was then transferred to 10 ml M17 medium. The OD600 value and pH value were measured every hour for 8 h.
The acidifying activity of bacteria in milk was analyzed. The bacteria were incubated in M17 broth overnight and then were inoculated at 2% (v/v) to 250 ml sterile skimmed milk (Fine Life, MCC Trading International GmbH, Germany). The samples were then cultivated anaerobically at 42°C, and the pH was recorded every hour for 5 h. Each experiment was repeated at least three times.
Resistance to acidic conditions of strains was measured according to the method of Clark et al. (1997) with some modifications. Exponential phase cells were collected from M17 cultures with an OD600 value of 0.4–0.6. Pelleted cells were washed twice with 10 mM sterile phosphate buffer (pH 7.0). The pH of the M17 medium in the experimental group was adjusted to 3.5 with 1M HCl, and for the M17 medium in the control group, pH 7.0 was used. Cells were resuspended in an equal volume of acidic M17 broth and incubated for 1 h at 42°C. Acid-treated cells were harvested by centrifugation at 12,000 g for 2 min and were then inoculated in fresh M17 broth. The ability of acid resistance was evaluated by the duration of the growth retardation period.
Then the tolerance for bile salts was tested according to previous studies with some optimization (Gilliland et al., 1984). M17 medium with 0.3% (w/v) ox bile in the experimental group and without the addition of ox bile in the control group was inoculated with exponential phase cells. The next steps were the same as the method used in the evaluation of acid resistance. Three replicate experiments were carried out to evaluate the reproducibility of each test.
In order to detect the special genes in the eps gene cluster of CS9, primers were designed based on the sequences of CS9 (DR994_01980-DR994_01840, Supplementary Table S2). Amplification conditions were as follows: initial denaturation at 94°C for 2 min followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30–60 s. The final extension was carried out at 72°C for 2 min. PCR products were purified and sequenced.
Streptococcus thermophilus strains CS5, CS9, CS18, and CS20 were inoculated in 1 L M17 broth and cultivated at 37°C for 24 h. EPS was isolated according to the previous method with some modifications (Hu et al., 2018). In brief, the fermentation cultures were heated at 100°C for 15 min, and then the bacterial cells were removed by centrifugation (9,000 rpm, 20 min, 4°C). The supernatant was filtrated by a 0.22-μm membrane. The proteins in the supernatant were precipitated by adding 80% trichloroacetic acid (TCA) solution (w/v) in a final concentration of 4% (w/v) and were removed by centrifugation. The EPSs were precipitated by adding a three-time volume of precooled absolute ethanol. Next, the mixtures were stored at 4°C overnight. The precipitated EPS was then collected by centrifugation and dissolved in hot water (95°C). The sample was then dialyzed for 72 h at 4°C. Finally, the sample was freeze-dried before the concentrations of EPS, protein, and nucleic acid were determined. The sugar content was determined by the phenol-sulfuric acid method (Li and Shah, 2014).
The 10 mg of crude EPS was mixed with 2 ml of 2M trifluoroacetic acid (TFA) and hydrolyzed at 110°C for 6 h. The sample was added with methanol and dried three times, and the dried residue was dissolved in 1 ml 0.3M sodium hydroxide for derivatization. Standard glucose, galactose, mannose, rhamnose, arabinose, ribose, xylose, fucose, galacturonic acid, glucuronic acid, glucosamine, and galactosamine were prepared as comparison.
The high-performance liquid chromatography (HPLC) analysis of 1-phenyl-3-methyl-5-pyrazolone (PMP)-derived monosaccharides was carried out on an Agilent 1100 HPLC system with a C18 HPLC column (4.6 mm i.d. × 250 mm, 5 μm, Agilent, United States). The PMP derivatives elution was performed with a mixture of 0.1M phosphate buffer (pH 6.4, A) and acetonitrile (B) in gradient elution mode with a flow rate of 1 ml/min at 30°C. Gradient elution condition was 85% A and 15% B, 0–10 min; 83% A and 17% B, 10–25 min. The UV absorbance of the effluent was set at 245 nm. The PMP derivatives were quantified by comparing their values of peak area to calibrated standard curves.
DNA was extracted from cell culture using EZNA® Bacterial DNA Kit (Omega Bio-Tek, Inc., Norcross GA, United States) according to the manual of the kit. The gadB gene was detected with the primers gadF (5′-ATGAATGAGA AGCTATTCAGAGAGATT-3′) and gadR (5′-TTAATGATGGA AGCCACTGC-3′). Amplification conditions were as follows: initial denaturation at 94°C for 2 min followed by 30 cycles of 94°C for 30 s, 51°C for 30 s, and 72°C for 45 s. The final extension was carried out at 72°C for 2 min. PCR products were purified and sequenced.
GABA was quantified using HPLC (UltiMate3000, Thermo Fisher, United States) equipped with a HypersilTM BDS C18 column (125 mm × 4.6 mm; Thermo Fisher, United States) according to the method of Li et al. (2008) with some modifications.
S. thermophilus was cultivated in M17 medium with the supplement of 1% monosodium glutamate (MSG) for 24 h at 37°C. Cell-free supernatant is the determination for the quantification of GABA. The supernatant was passed through a 0.22-μm membrane filter before HPLC analysis.
The derivatization reagent was prepared as follows: 15 mg ortho-phthalaldehyde (OPA) was first added to 5 ml methyl alcohol with the addition of 200 μl 2-mercaptoethanol, stored at 4°C, avoiding light. The derivative system was as follows: 200 μl fermentation supernatant, 200 μl derivatization reagent, and 600 μl 0.5M boric acid buffer solution (pH 10.4). The mixture was reacted for 2 min and then injected. The injection volume is 10 μl. The mobile phase A was 0.0129M sodium acetate buffer (pH 7.20). And the mobile phase B was proportioned according to 0.0129M sodium acetate buffer (pH 7.20): acetonitrile: methanol at the ratio of 1:2:1. During the elution, phase B increased linearly from 25 to 50% before 25 min and then increased linearly from 50 to 85% during 25–30 min. The temperature of the column was maintained at 25°C, and the absorbance of the detector was set to 338 nm for detection. The standard solution of GABA was prepared with sterilized water in the concentration of 10 g/l and diluted before use. The GABA derivatives were quantified by comparing their values of peak area to calibrated standard curves.
The assays were conducted according to Seo et al. (2015) with some modifications. The experimental strain CS9 with gadB gene grown overnight was diluted into fresh M17 media (pH 7.0) in 1% inoculation and then cultivated until the OD600 values reached 0.3–0.5 in order to ensure that the cells were in the logarithmic growth phase. These adapted cell cultures were inoculated into M17 media at pH 2.0 and pH 2.5 with or without 1% MSG. The initial cell density inoculated for acid treatment was nearly 5 × 106 CFU per milliliter. The acid-treated cultures were then incubated at 37°C without shaking, and the samples were collected after being cultivated for 2 h. Aliquots were serially diluted, and triplicates were plated onto M17 agar plates. Colonies were counted after 24 h.
Survival rate was calculated as follows: [(CFU per ml at time 2 h)/(CFU per ml at time 0)] × 100%. The results presented are the averages of triplicate experiments with the standard deviations included. Strain CS20 without the gadB gene was used as the control.
Extracellular pH (pHex) of cultures was measured first by the pH meter (Sartorius pB10, Germany). And the fluorescent probe, 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyflurescein, acetoxymethyl ester (BCECF AM, Beyotime, China) was used to label the prepared cells for the measurement of intracellular pH (pHin) (Huang et al., 2016). Cultures were incubated in the dark at 37°C for 30 min with the addition of 3 μM BCECF AM. Bacterial cells were washed three times with phosphate-buffered saline (PBS) buffer and were then resuspended in normal saline with 3 × 106 cells. Fluorescence intensities of the stained cells were measured in the Multi-Mode Microplate Readers (SpectraMax iD3, United States) at the excitation wavelengths and emission of 488 and 535 nm, respectively (Hansen et al., 2016).
For the construction of the calibration curve, the stained cells were suspended in physiological saline having different pH values (2, 3, 4, 5, 6, 7, and 8) adjusted by hydrochloric acid (HCl). The ionophore nigericin was added to the final concentrations of 4 μM followed by incubation at 37°C for 10 min resulting in the equilibration of both potassium and proton ions across the cell membrane. The fluorescence intensities were measured the same as described above.
Statistical treatment of data was performed by means of the SPSS software (SPSS, Inc., Chicago, IL, United States). The homogeneous subsets were defined by means of Duncan. Subsets were established for α = 0.05.
In this study, S. thermophilus strains CS5, CS9, CS18, and CS20 isolated from Chinese fermented milk were sequenced to investigate the biodiversity of the S. thermophilus strains (Figure 1). The genome sequencing and assembly-related information was shown in Table 1. The genome map of the S. thermophilus strain was completed by Illumina platform combined with Pacbio RSII. The whole genome map of the four strains was then completed by bioinformatics analysis after the quality control. The genomes of CS5, CS9, CS18, and CS20 were identified to contain 1,857,164, 1,860,988, 1,858,890, and 1,936,216 bases, respectively.
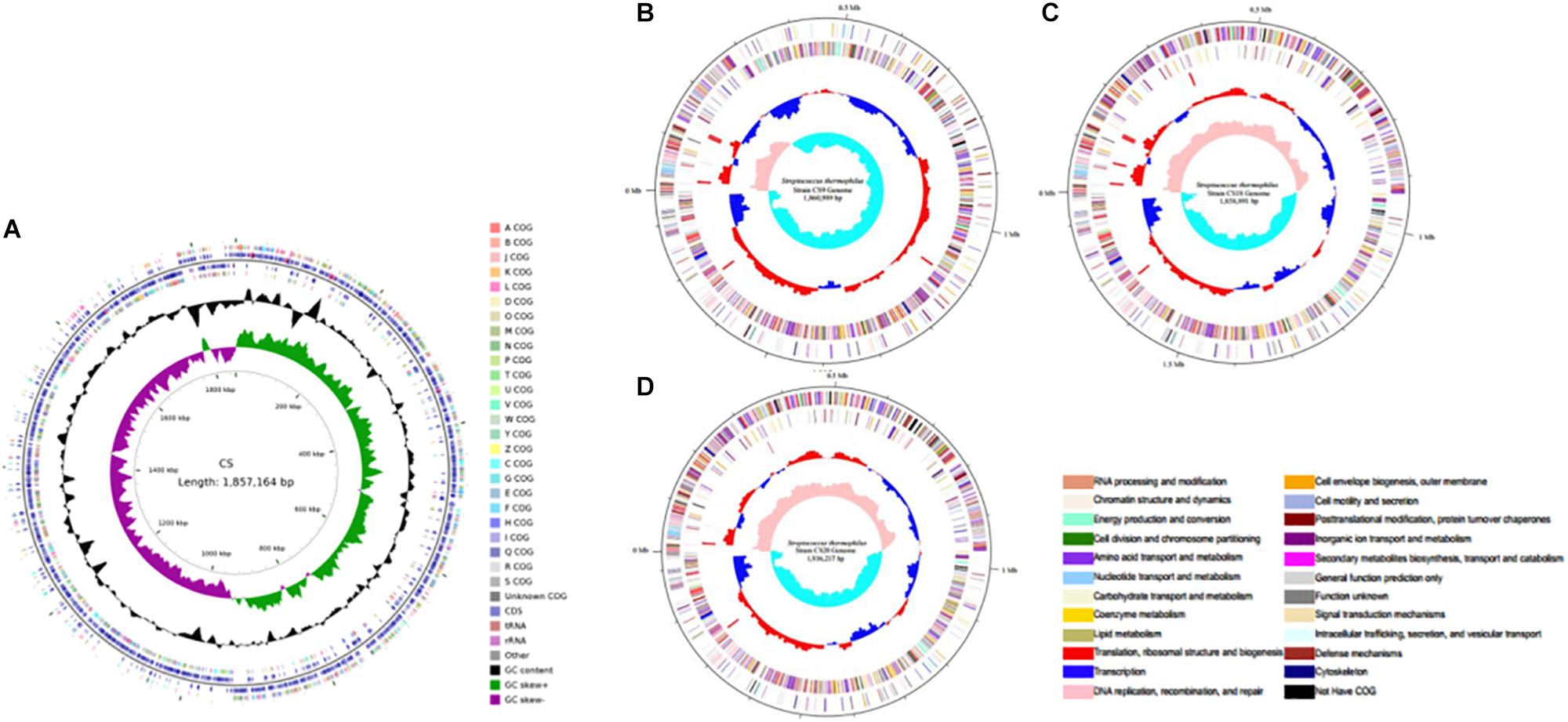
Figure 1. Circular genome map of the four new sequenced Streptococcus thermophilus chromosomes. (A) CS5; (B) CS9; (C) CS18; (D) CS20.
A comparative analysis of 34 S. thermophilus genomes available in public databases including these four new sequenced genomes was performed (Supplementary Table S1). It was found that the length of S. thermophilus genomes ranged from 1.73 to 2.07 Mb, while the G + C% contents were all around 39%. It is worth mentioning that CS20 is the second-largest genome of these 34 S. thermophilus strains just after strain M17PTZA496.
Most strains were isolated from fermented milk or other related milk products except the strains APC 151 and ST3, which were acquired from the digestive tract of a marine fish and commercial dietary supplements, respectively. The phylogenetic analysis was performed based on the conserved proteins detected in all tested genome sequences (Figure 2). It is clear that S. thermophilus strains from China distributed in various branches. They are from different provinces or areas of China; their growth environment and conditions are quite different resulting in their distinctive characters during the process of evolution. Both B59671 and LMD-9 are from the United States, but they are located so far away from each other. The similar situation also occurs in CNRZ1066 and JIM8232, which are from France. Thus, it could be concluded that the phylogenetic reconstruction based on the sequenced genomes of strains was not consistent with their geographic distribution: strains from different areas may be clustered on the same branch, while those from the same country are not necessarily located on the nearby branch. The result is consistent with that of a previous study (Vendramin et al., 2017).
In addition, CS5, CS18, and CS20 show a high proximity, while CS9 is far away from them. Specifically, CS9 is phylogenetically close to European and United States strains especially to the strain 1F8CT isolated from Italian Grana Padano Dop cheese. It reveals that the strains might not be clustered together even though they are isolated from the same region and source. It could be suggested that the long-term evolutionary process probably contributed to the difference of CS9 from the other strains.
Venn diagram analysis indicated that the 34 strains of S. thermophilus contain 963 core genes and specific genes from 0 to 202 in different strains (Figure 3A and Supplementary Table S3). Thirteen, 127, 12, and 61 unique genes were identified on CS5, CS9, CS18, and CS20 genomes, respectively. Among all tested strains, CS9 possessed the second-largest amount of unique genes, following strain M17PTZA496 harboring 202 special genes.
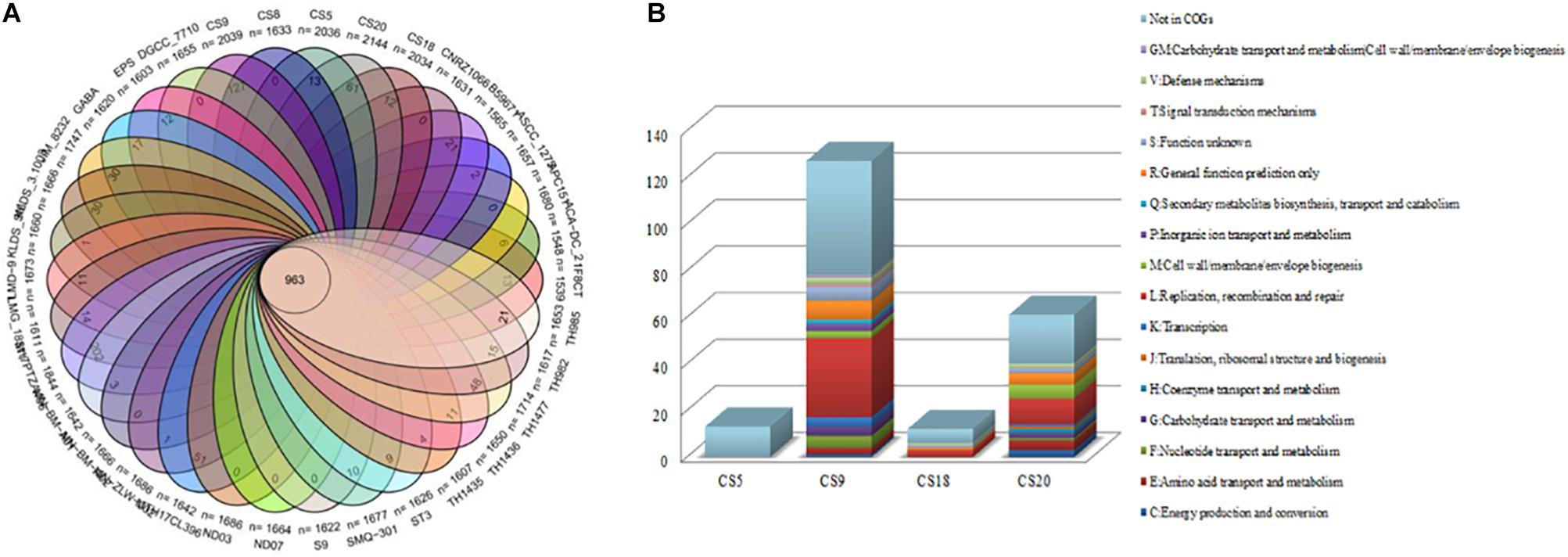
Figure 3. Core genes and unique genes of Streptococcus thermophilus strains. (A) Venn diagram analysis of 34 S. thermophilus strains. (B) Unique genes of four new sequenced S. thermophilus strains into COG categories.
The unique genes of CS5, CS9, CS18, and CS20 were found to be involved in diverse functions according to the COGs annotations (Figure 3B). Most of the specific genes of CS9 belonged to the family L which is related to replication, recombination, and repair of the cells. The abundant genes associated with cell replication and modification could increase the survival ability of strains when facing the change in environmental stress during the long-term evolutionary process. The results indicated that CS9 represents the robust and most evolved among all the four strains.
Horizontal gene transfer is defined as the transmission of genetic materials between distantly related organisms in phylogenetic evolution (Goldenfeld and Woese, 2007). These unique genes were probably acquired by HGT. For instance, a heavy metal transporter CzcA (DR994_03030) predicted in CS9 genome is responsible for the transport of heavy metal ions, and it shows 100% similarity to a gene of Streptococcus pneumoniae. It was speculated that CzcA may be obtained from S. pneumoniae via HGT. HGT could play a key role in improving technological properties of S. thermophilus strains by acquiring new genes during the process of evolutionary and environmental adaptation (Hols et al., 2005). A lot of HGT events have been found in the S. thermophilus strains, which give strains new characteristics and environmental adaptability (Hols et al., 2005; Liu et al., 2009; Eng et al., 2011).
Genomic islands might predict the regions of HGT in the bacterial chromosome (Juhas et al., 2009). GIs located on these four genomes were identified by the IslandViewer software (Figure 4). It was found that CS5, CS9, CS18, and CS20 contained 20, 19, 17, and 19 integrated GIs, respectively. The integrated GIs of these four strains harbored 255, 288, 286, and 306 genes, respectively.
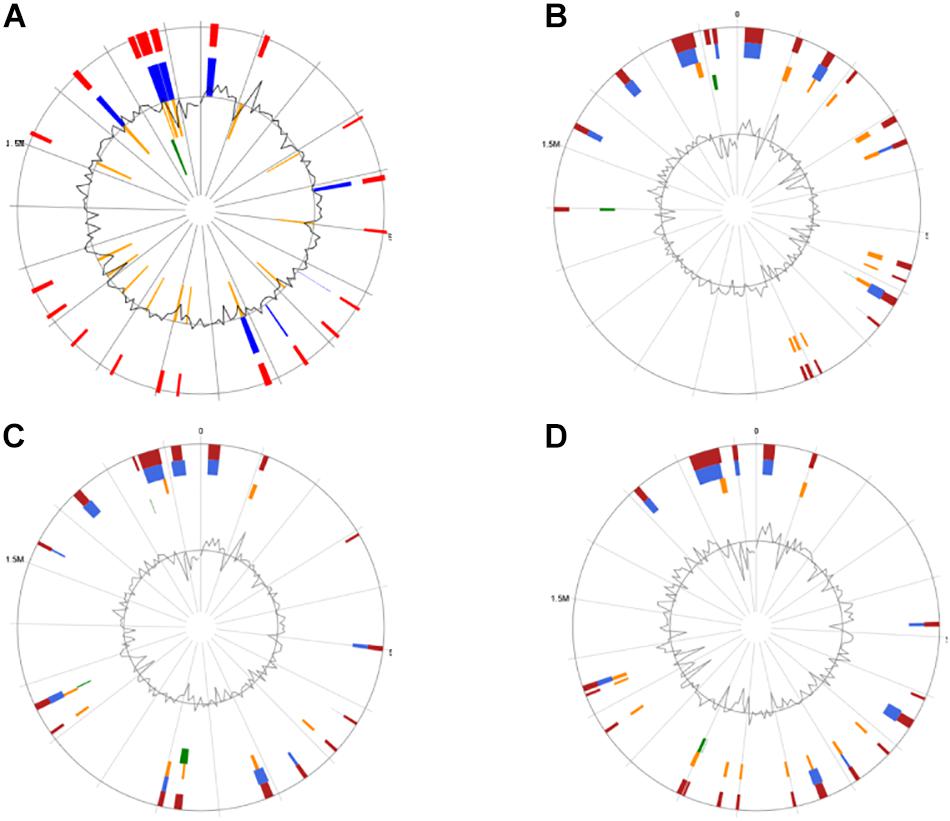
Figure 4. Circular map of the four Streptococcus thermophilus genome as generated by IslandViewer. Highlighted regions correspond to genomic islands (GIs). (A) CS5; (B) CS9; (C) CS18; (D) CS20.
The safety of strains is very important for their application. In the present study, presence or absence of antibiotic resistance genes or virulence factors, mobilome (number of transposons and plasmids) has been discussed.
The sequences of the S. thermophilus CS5, CS9, CS18, and CS20 genomes have shown that none of the four strains contains any antibiotic-resistant genes. Although there is a tetracycline transporter (multidrug-efflux transporter) on the antibiotic gene islands of CS5 (C1A39_09480), CS18 (DTA40_09485), and CS20 (DTA54_09390), tetracycline resistance genes have not been detected. Therefore, it could be suggested that CS5, CS9, CS18, and CS20 do not contain any antibiotic resistance and that they do not cause antibiotic contamination when used as a food starter (Toomey et al., 2010).
In order to further verify the results of the analysis of the gene level, 12 common antibiotics were selected to study the resistance of these four S. thermophilus strains, and the results were shown in Table 2. As can be seen from the results, S. thermophilus CS5, CS9, CS18, and CS20 are all sensitive to these 12 common antibiotics, which is consistent with the results of the genomic analysis. This has confirmed that these four strains will not cause antibiotic contamination if used in food production.
The protein sequences of the four strains were compared against the VFDB database to obtain data on potential virulence factors on the genome (Table 3 and Supplementary Table S4). Comparison rate higher than 60% can be considered as a credible prediction. It can be seen that the number of potential virulence genes in S. thermophilus CS20 was the most, up to 68. The full genome length and number of genes of CS20 were the most among the four strains of S. thermophilus, which indicated that the genetic diversity of the CS20 genome was the most. The numbers of virulence genes in CS5, CS9, and CS18 were similar, with 48, 49, and 49 virulence genes, respectively. In general, most of these predicted potential virulence factors have a credible similarity to the S. thermophilus genome, with a comparison rate up to 95%. Most of these genes are associated with the synthesis and transport of EPS. S. thermophilus is a widely recognized food-grade microorganism; therefore, it can be inferred that the virulence genes homologous to S. thermophilus are not pathogenic. Studies have shown that S. thermophilus is different from other pathogenic streptococci, and there are generally three reasons why it is not pathogenic. Firstly, compared to other pathogenic streptococci, some of the genes in S. thermophilus which are involved in the sugar transport system have been degraded in the process of evolution, resulting in their inability to utilize a large number of carbohydrates (Tettelin, 2001). The ability to utilize a wide range of carbohydrates has been reported to be essential for the virulence of pathogenic streptococci, and the observed impairment of this function in S. thermophilus is likely to reduce its virulence potential. Also, it could be found that many streptococcal virulence-related genes (VRGs) are absent from these four S. thermophilus genomes or are present as only to encode basic cellular functional proteins (Bolotin et al., 2004).
Transposases are small mobile elements that are usually found in bacterial genome and only contain their own transposition genes. Transposases are normally 0.8–2.5 kb in length and possess inverted repeats at their termini. In addition, transposases are the important factors for horizontal gene transfer (HGT) between strain genomes (Guédon et al., 1995). Thus, the vitality and similarity of transposases as well as the virulence or resistance genes around them have become important indexes for the safety evaluation.
The total numbers of transposases in S. thermophilus CS5, CS9, CS18, and CS20 were 132, 95, 139, and 116, respectively. But it was predicted that more than 80% of them were pseudogenes in each of them. The real numbers of transposases with a real potential function in S. thermophilus CS5, CS9, CS18, and CS20 were only 23, 15, 25, and 19, respectively.
Moreover, the gene blast results showed that these available transposases represent the high similarity to other established S. thermophilus genomes (Supplementary Table S5). As S. thermophilus is a recognized food-grade microorganism, these transposases are not potentially dangerous.
Plasmid is a circular DNA molecule apart from the cell chromosome as well as nuclear area DNA and is able to replicate itself in the host cell. Some plasmids could be transferred between strains, which might lead to the HGT. Therefore, the presence of plasmids in S. thermophilus is also an uncertain safety hazard. The detection of plasmids in the four S. thermophilus strains is displayed in Supplementary Figure S1. It could be seen that no plasmid was detected in these four strains. It also confirmed that all of the four strains could be safely used as natural food starters.
In summary, S. thermophilus CS5, CS9, CS18, and CS20 do not contain antibiotic resistance genes, virulence factors with significant pathogenicity, or plasmids that can replicate independently. In addition, these four strains contain few transposon genes. This indicates that the four strains CS5, CS9, CS18, and CS20 are not potentially toxic, and therefore they can be safely used as a novel natural food starter.
Exopolysaccharide biosynthetic pathways contain the transportation and phosphorylation of sugars, the formation of sugar nucleotides, the synthesis of repeating units, the polymerization of repeating units, and the exportation of EPS (Laws et al., 2001).
The carbohydrate utilization and sugar transport systems of CS5, CS9, CS18, and CS20 were analyzed (Figure 5). S. thermophilus takes up carbohydrates through a phosphoenolpyruvate (PEP)-phosphotransferase system (PTS) or a secondary transport system (Cui et al., 2016). The PEP-PTS consists of a PEP-dependent phosphotransferase (enzyme I, EI), a histidine-containing phosphocarrier protein (HPr, heat-stable protein), and a sugar-specific permease (transporter), which is a membrane-bound complex known as enzyme II (EII). These four genomes all harbor a PEP-dependent phosphotransferase and the HPr (Figure 5). Furthermore, genes responsible for sucrose, mannose, and fructose PTS transporter sugar-specific permease enzymes were detected in these genomes (Figure 5). Therefore, sucrose, mannose, and fructose could be transported by specific PEP-PTSs.
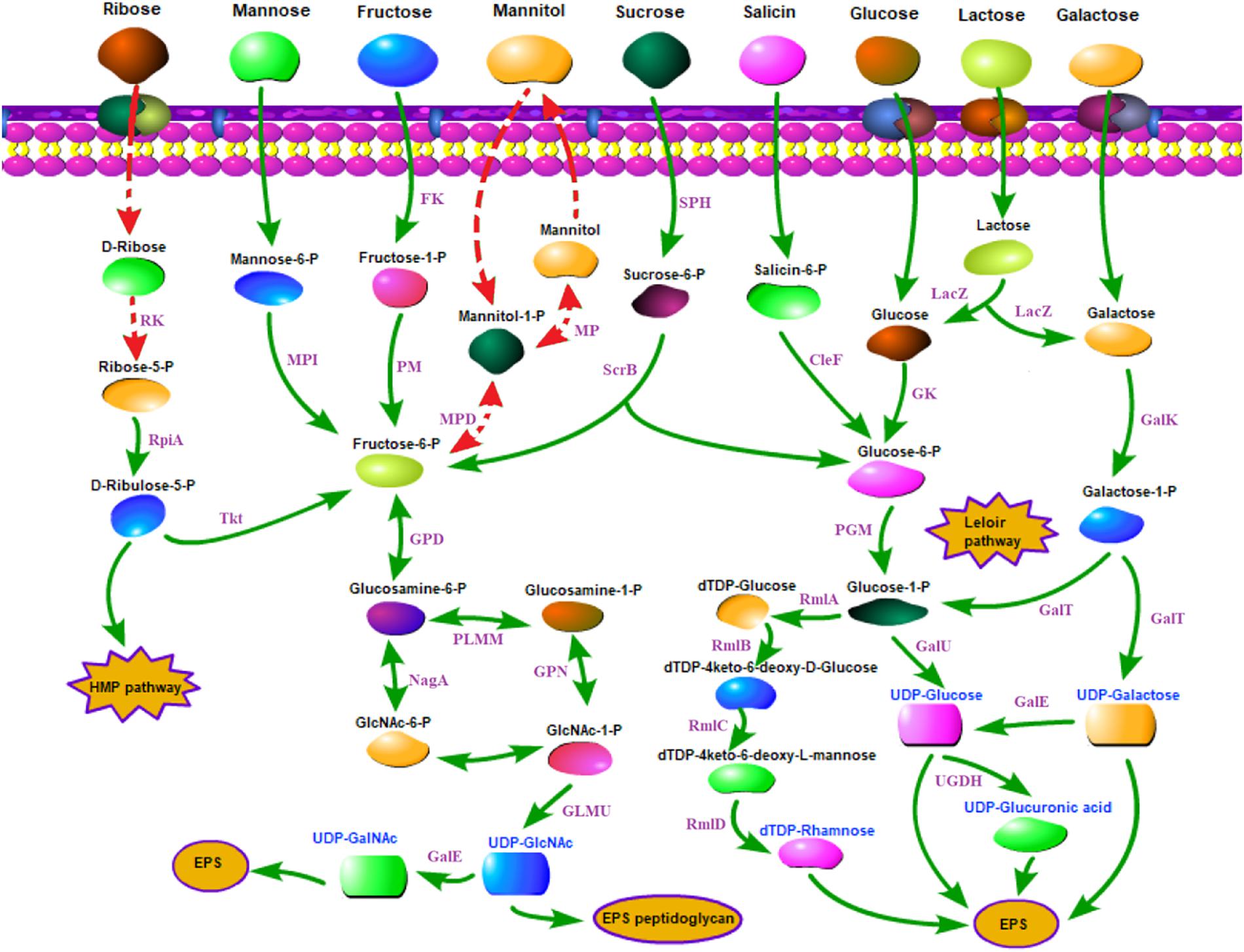
Figure 5. Carbohydrate utilization and sugar transport system in Streptococcus thermophilus strains CS5, CS9, CS18, and CS20. CelF, 6-phospho-beta-glucosidase; Fk, fructokinase; GalE, UDP-galactose-4-epimerase; GalK, galactokinase; GalT, UTP-galactose-1-phosphate uridylyltransferase; GalU, UDP-glucose pyrophosphorylase; Gk, glucokinase; GLMU, N-acetylglucosamine-1-phosphate uridyltransferase; GPD, glucosamine-6-phosphate deaminase; HMP pathway, pentose phosphate pathway; LacZ, beta-galactosidase; MP, mannitol-1-phosphatase; MPD, mannitol-1-phosphate 5-dehydrogenase; MPI, mannose-6 phosphate isomerase; NagA, N-acetyl-alpha-D-galactosaminidase; PGM, α-phosphoglucomutase; PlMM, phosphoglucosamine mutase; RK, ribokinase; RmlA, glucose-1-phosphate thymidylyltransferase; RmlB, dTDP-glucose 4, 6-dehydratase; RmlC, dTDP-4-dehydrorhamnose 3,5-epimerase; RmlD, dTDP-4-keto-L-rhamnose reductase; RpiA, ribose-5-phosphate isomerase; ScrB, sucrose6-phosphate hydrolase; SPH, sucrose-6-phosphate hydrolase; Tkt, transketolase; UDP-GalNAc, UDP-N-acetylgalactosamine; UDP-GlcNAc, UDP-N-acetylglucosamine; UGDH, UDP-glucose 6-dehydrogenase. Double-headed arrow indicates the bidirectional reaction. Arrow indicates a reaction. Red dashed arrows indicate incomplete or inactive pathways in S. thermophilus. Purple fonts indicate enzyme. Blue fonts indicate nucleotide sugars in S. thermophilus.
It was identified previously that CS5, CS18, and CS20 showed the ability to ferment a broad range of carbohydrates; they could metabolize glucose, fructose, lactose, sucrose, maltose, D (+) mannose, mannitol, D (−) salicin, melezitose, ribose, and sorbitol (Hu et al., 2018). Additionally, CS18 and CS20 could utilize D (+) galactose. CS9 can only use five kinds of sugars, including glucose, fructose, lactose, D (+) mannose, and melezitose (Hu et al., 2018).
For sugars without a specialized PEP-PTS transport system, bacteria transport them through non-PEP-PTS transport systems, i.e., primary and secondary transport systems (Laws et al., 2001). In S. thermophilus, sugars are mainly transported by means of permease. Genes encoding lactose/galactose permease and glucose permease were identified in these four genomes (Figure 5).
Lactose is the only carbohydrate in milk, and its content is up to 4.2% in milk, and is the preferred carbon source of S. thermophilus because of the long-term adaptation of strains in milk environment. Lactose is converted to glucose and galactose by β-galactosidase (LacZ) in S. thermophilus. The LacZ and the related transport of lactose have been identified in all the four strains in the present studies (Figure 5). S. thermophilus Gal-positive strains are used to reduce the browning defects even to inhibit the growth of undesirable strains (Mukherjee, 1994; de Vin et al., 2005). However, S. thermophilus Gal-positive strains are not common (de Vin et al., 2005; Hou et al., 2017). de Vin et al. (2005) had reported that only eight strains among the 49 tested S. thermophilus strains were Gal-positive. Hou et al. (2017) also found only 8% of 51 strains of S. thermophilus isolated from the Moorepark culture collection could utilize galactose. Therefore, the screening and identification of new Gal-positive S. thermophilus are desirable for food fermentation. As CS18 and CS20 could utilize galactose, they could be used in the production of cheese in order to eliminate or reduce the adverse influences of galactose accumulation.
The Leloir pathway is the most common route for galactose utilization in S. thermophilus (Hutkins and Morris, 1987). Galactose was converted into glucose 1-phosphate by the galRKTEM gene cluster and then entered the glycolysis process or other anabolic pathways (Hutkins and Morris, 1987). It is usually recognized that gal promoter plays an important role in the galactose catabolism (Vaughan et al., 2001).
The DNA fragments about 437 bp containing both of the galR and galK promoter regions of CS5, CS9, CS18, and CS20 were picked out from their genome sequences. Most of these sequences in the galR-galK intergenic region are completely the same except for some mutations of few bases (Figure 6). The four fragments could be classified into type I and type II. CS5, CS9, and CS18 all contain the type I sequence, while CS20 belongs to type II. This result is consistent with that of our previous research (Hu et al., 2018).
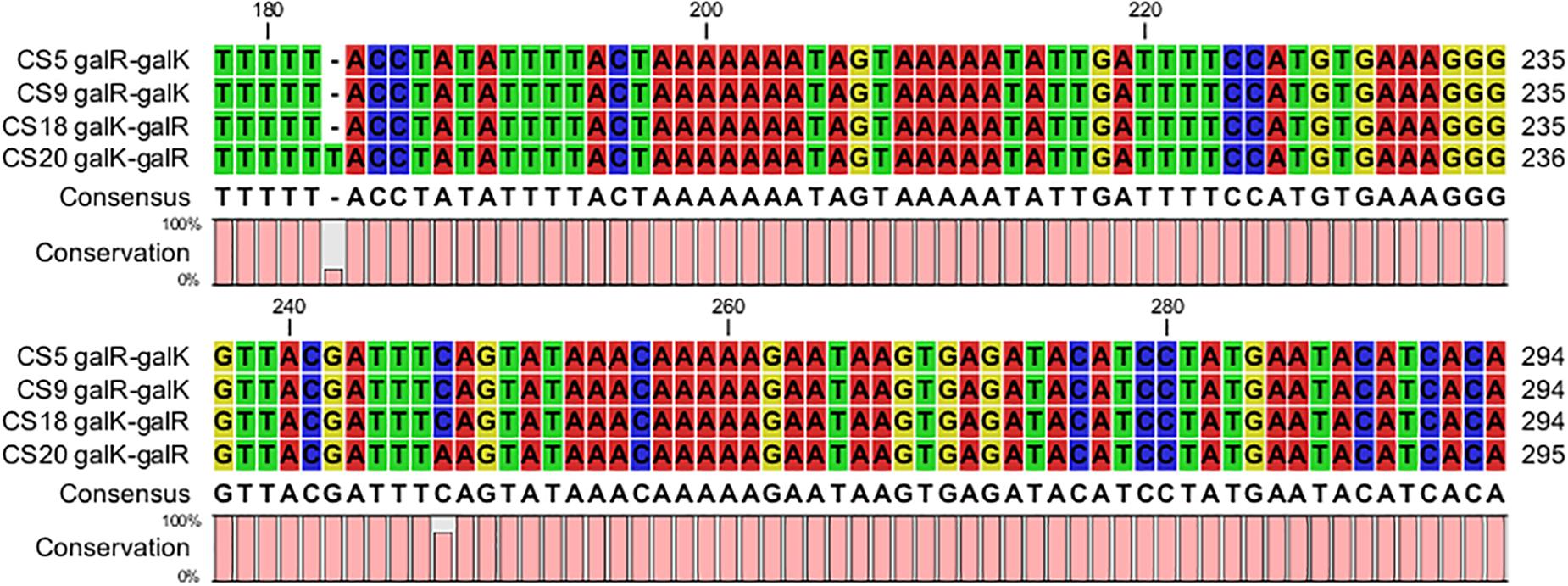
Figure 6. Alignment of the nucleotide sequences in the galR–galK intergenic region of Streptococcus thermophilus strains CS5, CS9, CS18, and CS20.
Eight different types of DNA fragments types (NS1–NS8) were detected in the galR-galK intergenic region of 49 S. thermophilus (de Vin et al., 2005). In the present study, the type I nucleotide sequences are exactly the same as the NS8 type which can be found in Gal-negative strains. Type II located on the Gal-positive strain CS20, however, is beyond the types NS1–NS8. Gal-negative strains CS5 and CS9 contain type I. The result is consistent with that of a previous study (de Vin et al., 2005). However, the Gal-positive strain CS18 also belongs to type I. This indicated that although the gal promoter played a key role in the galactose utilization, it did not exactly determine the Gal-positive phenotype of S. thermophilus.
Furthermore, CS5, CS18, and CS20 can utilize salicin except CS9. Salicin is converted to salicin-6-P by the sugar-specific IIA component of the PTS system, which was detected in all the strains except CS9 (Figure 5). The ribose-positive S. thermophilus strains were rarely found (Erkus et al., 2014). Interestingly, CS5, CS18, and CS20 were all able to utilize ribose. Generally, D-ribose has to be phosphorylated by ribokinase before being utilized. However, the ribokinase gene was not detected in all the three genomes. It suggested that S. thermophilus might utilize ribose by other non-homologous pathways, and it requires further genetic characterization.
The eps gene clusters determining the EPS biosynthesis were found in CS5, CS9, CS18, and CS20 (Figure 7A). They could be divided into four parts: (1) the regulatory genes of EPS production (epsA, epsB); (2) genes for controlling the EPS chain length (epsC, epsD); (3) the EPS repeat units forming genes; (4) the genes related to polymerization and exportation of repeating units (Cui et al., 2017).
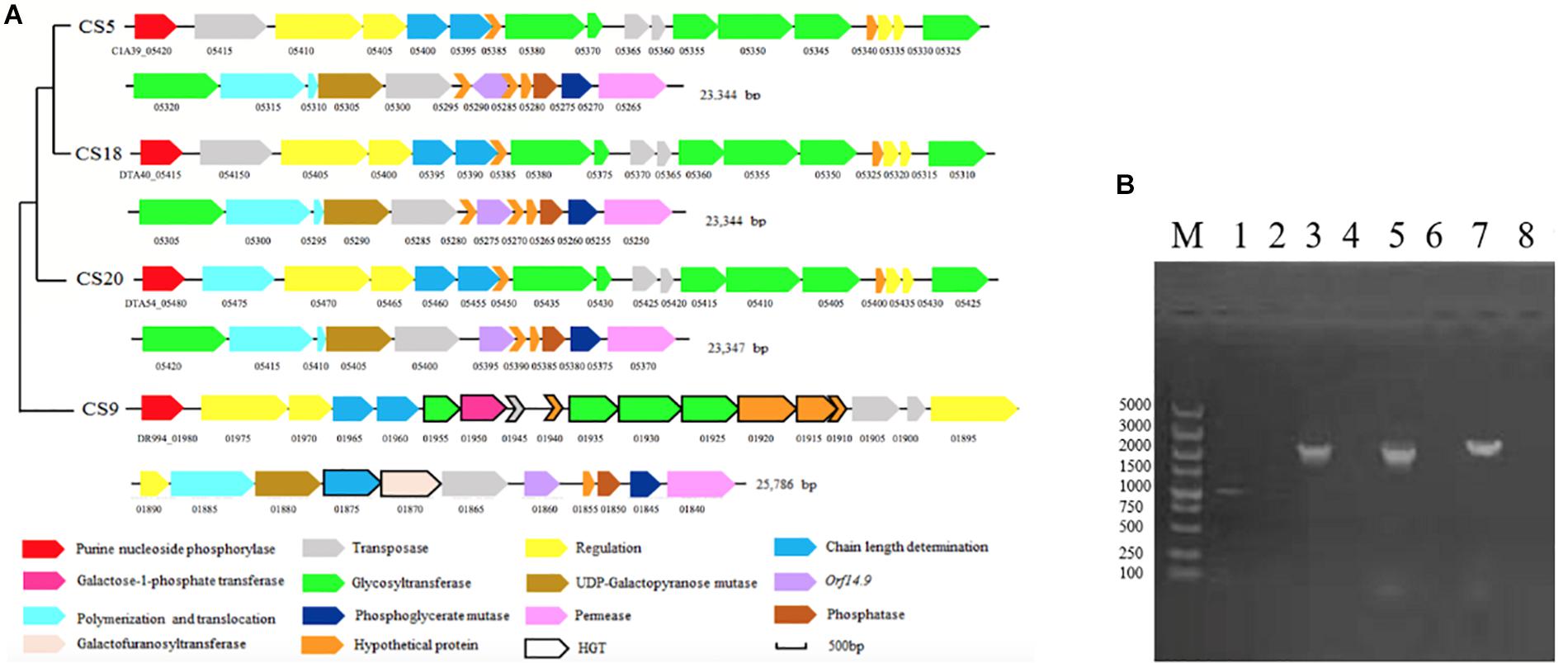
Figure 7. The eps gene cluster analysis of Streptococcus thermophilus strains CS5, CS9, CS18, and CS20. (A) Organization of eps gene cluster among S. thermophilus strains CS5, CS9, CS18, and CS20. Genes encoding the related proteins are marked with a different color. (B) Detection of special genes in the eps cluster of S. thermophilus CS9. M: DL5000; 1, 2: 1950-F/1950-R; 3, 4: 1920-1925-F/1920-1925-R; 5, 6:1880-1885-F/1880-1885-R; 7, 8:1930-1935-F/1930-1935-R; 1, 3, 5, 7: CS9; 2, 4, 6, 8: CS20 was used as negative control.
There was no significant difference among the eps gene clusters of CS5, CS18, and CS20 except a few bases distinctions. This result further confirmed that three S. thermophilus strains have close relationships. In addition, these eps clusters belonged to a common type and were found in some S. thermophilus strains, including ASCC1275, Sfi39, KLDS MS, MN-BM-A02, DGCC7710, C106, KLDS 3.1003, TH982, FI9186, and MTC310 (Cui et al., 2017). Furthermore, there was also an additional gene unit eps2C-eps2D in the eps clusters of CS5, CS18, and CS20 which could affect the length of the EPS unit.
Glycosyltransferases can take part in the transportation of various glyconucleotides. Six genes encoding GTFs were detected in the eps gene clusters of CS5, CS18, and CS20. They could transport UDP-glucose, UDP-galactose, dTDP-rhamnose, UDP-N-acetyl-glucosamine (UDP-GlcNAc), and UDP-galacofuranose to form various EPS units by glycosidic bonds.
Interestingly, the eps gene cluster of CS9 is unique and different from those of any known S. thermophilus (Figure 7A). Two regulatory EPS gene units (eps1A-eps1B and eps2A-eps2B) were found in CS9. Therefore, it was speculated that the additional eps2A-eps2B could improve the EPS production. To our knowledge, it has never been found in the other S. thermophilus eps gene clusters (Cui et al., 2017).
Especially, four GTFs of the eps gene cluster of CS9 have no homology to those of S. thermophilus. These GTFs (DR994_01925, DR994_01930, DR994_01935, and DR994_01955) originated from Clostridium butyricum, Eubacteriaceae bacterium, Streptococcus equinus, and Lactococcus lactis, respectively. DR994_01925 was predicted to belong to GTF family 2 which could transfer sugar from UDP-glucose, UDP-N-acetyl-galactosamine (UDP-GalNAc), or GDP-mannose to a range of substrates. DR994_01930 was a member of the GTB-type superfamily. DR994_01935 contains a conserved L-rhamnosyltransferase domain. It was speculated that EPS-CS9 might contain rhamnose. DR994_01955, however, was found similar to galactosyl transferase CpsE which could catalyze the addition of galactose to an oligosaccharide precursor or to a lipid intermediate.
At the same time, a relatively rare galactofuranose transferase (DR994_01870) was found in CS9, which was detected only in S. thermophilus strains 488, DSM 20617, JIM8232, and TH1436 (Delorme et al., 2011; Treu et al., 2014). It showed a high similarity to a galactofuranosyltransferase (WP_039693594.1) of Streptococcus gallolyticus (100% coverage, 92% identity). The results suggested that there was HGT between CS9 and S. gallolyticus, resulting in the exchange and integration of functional genes.
In addition, there were some other vestiges of HGT presenting in the eps gene cluster of CS9. For example, a gene encoding capsular biosynthesis protein (DR994_01885) was highly similar to the partial genome sequence of Streptococcus infantarius subsp. infantarius CJ18 (100% coverage, 98.11% identity); a gene encoding galactose-1-phosphate transferase (DR994_01950) showed high homology with the EPS synthesis-related gene of L. lactis subsp. cremoris NIZO B35 (100% coverage, 99.07% identity).
One IS256 transposase and four hypothetical proteins showed similarity with the dairy starter strains (Lactobacillus delbrueckii and L. lactis subsp. lactis), Streptococcus macedonicus, and Bacillus sp. In a word, the eps gene cluster of CS9 contained 12 specific genes with high homology to a variety of distantly related species, and all of them had never been detected in S. thermophilus strains (Supplementary Table S6). Most of these genes are located in the GIs. The GC contents of these genes are quite different from those of the CS9 genome (GC content is 38.92%). On the contrary, the GC contents of these non-S. thermophilus genes are similar to those of the original strains (Supplementary Table S6). It was speculated that CS9 had undergone HGT in a complex environment containing many other species during the long evolutionary process. These special genes were detected by PCR to confirm their real existence (Figure 7B). The results indicated that these specific genes predicted in CS9 genome exactly exist.
The EPS production ability of all the four strains in M17 medium was assayed. The crude EPS-CS5, EPS-CS9, EPS-CS18, and EPS-CS20 were 173.8, 168.3, 178.2, and 161.2 mg/ml, respectively (Figure 8A). All the four strains have similar EPS production ability in M17 medium. However, our previous studies indicated that CS9 has high EPS yield in milk (333.77 mg/ml) (Hu et al., 2018). The EPS yield of the other three strains in milk is 224.97, 213.40, and 171.95 mg/ml, respectively (Hu et al., 2018). EPS production of S. thermophilus is varied from 50 to 1,029 mg/L (Cui et al., 2017). Generally, more than 200 mg/L is considered high EPS production.
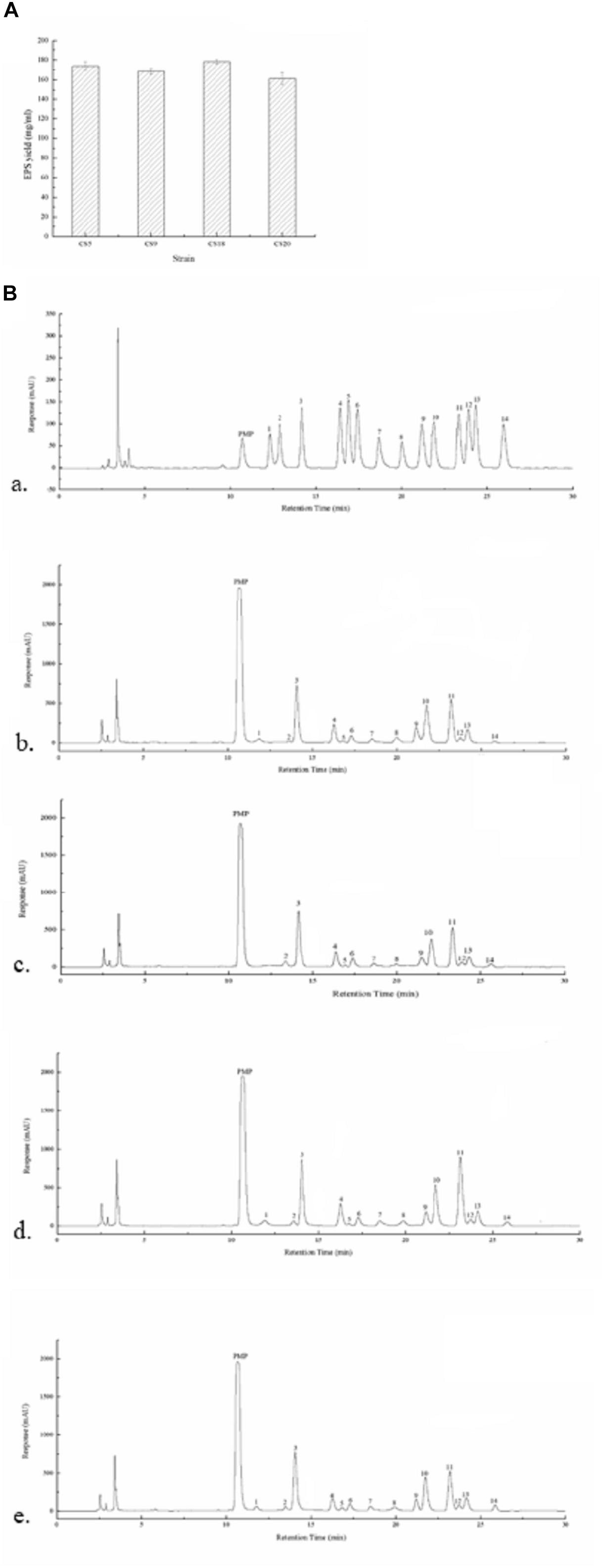
Figure 8. Exopolysaccharides (EPSs) produced by Streptococcus thermophilus strains CS5, CS9, CS18, and CS20. (A) The EPS yield of all the four strains. (B) The high-performance liquid chromatography (HPLC) chromatograms of 1-phenyl-3-methyl-5-pyrazolone (PMP) derivatives of 12 standard monosaccharides (a) and component monosaccharides released from EPS-CS5 (b); from EPS-CS9 (c); from EPS-CS18 (d); from EPS-CS20 (e). (Peaks: (1) gulouronic acid; (2) mannosuronic acid; (3) mannose; (4) ribose; (5) rhamnose; (6) glucosamine; (7) glucuronic acid; (8) galacturonic acid; (9) galactosamine; (10) glucose; (11) galactose; (12) xylose; (13) arabinose; (14) fucose.)
The monosaccharide compositions of crude EPS-CS5, EPS-CS9, EPS-CS18, and EPS-CS20 were analyzed using HPLC. Galactose, glucose, and mannose were the main monosaccharides in these crude EPSs with different ratios, and a small amount of rhamnose, ribose, xylose, fucose, galacturonic acid, glucuronic acid, glucosamine, and galactosamine were also found (Figure 8B). The results are consistent with the genetic analysis of nucleotide sugars and eps cluster. The eps gene clusters in CS5, CS18, and CS20 are almost the same, thus the monosaccharide compositions of crude EPS-CS5, EPS-CS18, and EPS-CS20 are similar. While there was no gulouronic acid detected in EPS-CS9. In detail, the composition of mannose, glucose, and galactose in EPS-CS5, EPS-CS9, EPS-CS18, and EPS-CS20 were 1.2:1:1, 0.9:0.7:1, 1.3:0.9:1, and 1.3:1:1, respectively.
The CS9 eps gene cluster is relatively special among these four S. thermophilus strains as there are some genes belonging to the non-S. thermophilus genomes. Usually, HGT transfers are linked to the acquisition of beneficial traits. Compared with the other three strains, the EPS production of CS9 has obvious advantage in milk. The connection between specific genes of eps gene cluster and EPS production as well as the high number of HGT events in CS9 will be further studied in the future.
Growth dynamics of strains could reflect their technological behavior, such as the capability of milk fermentation. The growth abilities of CS5, CS9, CS18, and CS20 were measured at M17 medium (Figure 9A and Supplementary Tables S7, S8). The growth rates of CS5 and CS18 were similar. They are higher than those of CS9 and CS20 during the growth from 1 to 3 h. Among these four strains, the growth rate of CS9 was lowest. And the final biomasses of CS5 and CS18 were higher than those of CS20 and CS9 after the growth of 8 h.
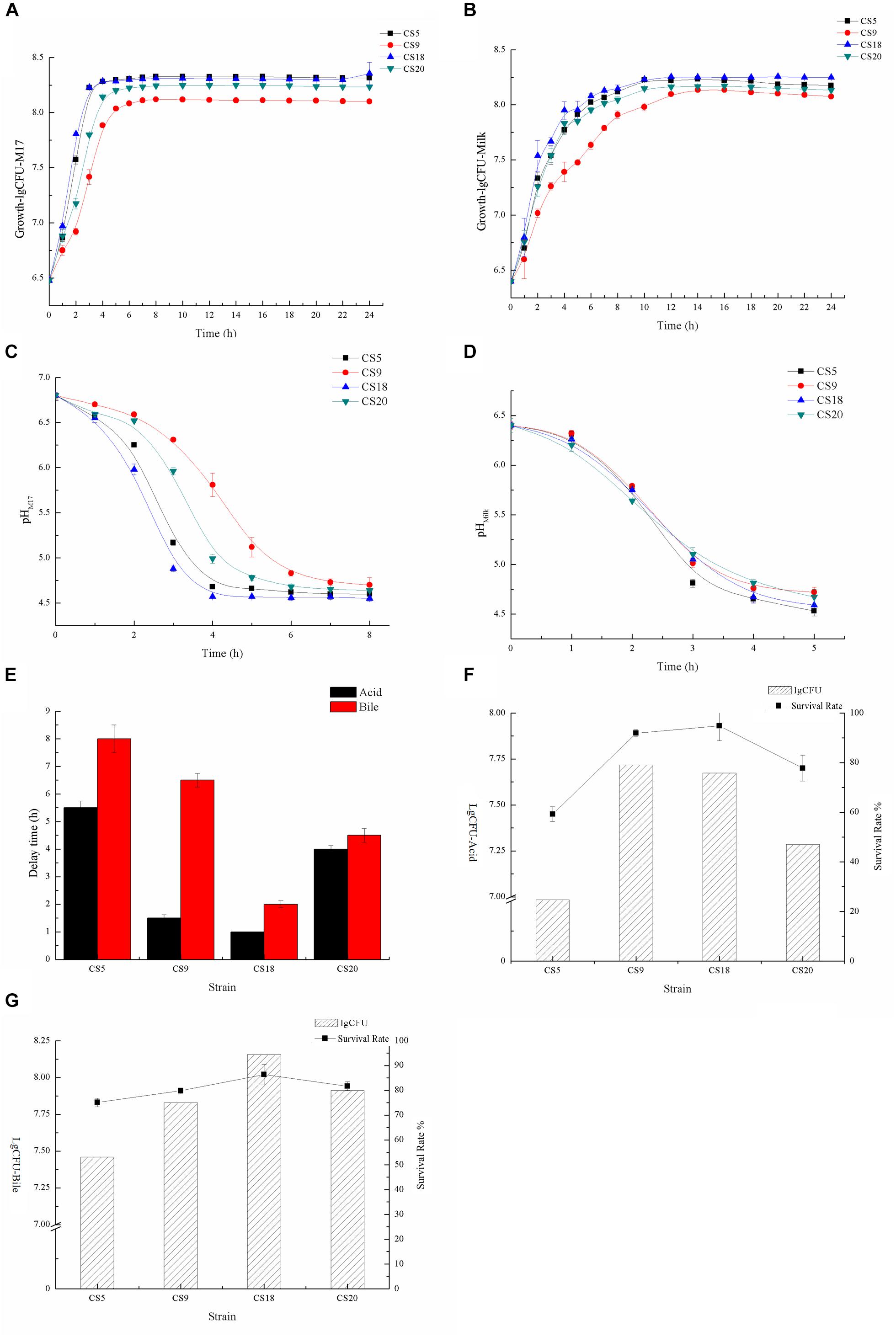
Figure 9. Identification of fermented characters (A). Growth curves of CS5, CS9, CS18, and CS20 in M17 liquid broth (B). Growth curves of CS5, CS9, CS18, and CS20 in milk (C). Acidification kinetics of CS5, CS9, CS18, and CS20 in M17 liquid broth (D). Acidification kinetics of CS5, CS9, CS18, and CS20 in milk (E). Growth retardation period of CS5, CS9, CS18, and CS20 after stress treatment (F). The viable count (lgCFU) and survival rate of CS5, CS9, CS18, and CS20 under the acid treatment (G). The viable count (lgCFU) and survival rate of CS5, CS9, CS18, and CS20 under the bile treatment.
Compared with the growth condition in M17, the four strains grew poorly in skimmed milk, but the growth trend of the strains was consistent with that of M17 (Figure 9B and Supplementary Tables S9, S10). The growths of S. thermophilus CS5, CS18, and CS20 are similar, but the growth of CS9 is obviously at a disadvantage. This might be because the key protease PrtS that could utilize casein in milk is not detected in the CS9 genome, so the growth condition of CS9 in milk was obviously weaker than the three strains CS5, CS18, and CS20 containing the prtS gene.
CS5 has the highest acid-producing capacity among these four strains. The pH value of the CS5 culture can reach 4.6 in less than 4 h, while the CS20 culture takes the longest time to reach pH 4.6. This result was consistent with the growth ability of strains (Figure 9C). However, the acidification rates of these four strains were similar in skimmed milk medium, and all strains reached pH 4.6 which is the attributes required for milk fermentation end point after 5 h (Figure 9D). It was speculated that milk is better for the growth of strains than M17.
The rapid acidification capability of starter culture strains was essential to guarantee the excellent characteristic of dairy products and to improve food safety by preventing the growth of pathogenic bacteria (Gaden et al., 1992). It has been known that pH 4.6 is the value required for casein coagulation and the inhibition of pathogen growth (Vendramin et al., 2017).
The growth retardation period of strain after stress treatment could reflect the acidity and bile salt resistance of the strains. Contrary to the growth situation, the tolerance of CS5 to acid and bile salt is the weakest among these four strains, and the growth retardation period were 5.5 and 8 h, respectively (Figure 9E). CS18 had the strongest adaptability to acid and bile salt stress because CS18 resumed normal growth condition only after 1 or 2 h.
It could be seen from the Supplementary Table S11 that the survival rates of these four strains were different under the same stress treatment. The acid and bile salt tolerance of the strain was measured by the value of lgCFU (Figures 9F,G and Supplementary Table S11). After the acid treatment, the number of viable bacteria of S. thermophilus CS18 was the highest and so was the survival rate, indicating that CS18 was the most tolerant to acid among the four strains of S. thermophilus. The acid tolerance of strains CS5 and CS20 was relatively low, with the survival rates of 24.67 and 47.03%, respectively. In terms of bile salt tolerance, CS18 was also the most resistant to bile salt, with a survival rate of 94.40%, while CS5 had the weakest bile salt resistance, with a survival rate as low as 53.17% after incubation with 0.3% bile salt. The results of viable counts were consistent with the results of growth lag time: the strain with a higher survival rate after stress treatment had a shorter growth delay time. The results can reflect the stress tolerance of the strains and help screen out the strains with a strong potential for colonization.
In general, CS18 had a fast growth rate, short fermentation time, and strong acid and bile salt resistance, indicating that CS18 has the potential to be used in food fermentation and can survive better in the GIT of mammals.
GABA represents several beneficial physiological activities in humans, and some GABA-rich products have already been used as food additives or functional food supplements (Foster and Kemp, 2006). GABA-producing S. thermophilus strains are uncommon in the food industry. Brasca et al. (2016) investigated 191 S. thermophilus strains isolated from different origins, and only a total of 20 strains (10%) harbored the gadB gene. Among all the strains investigated in this study apart from CS5, CS9, CS18, and CS20 strains, only seven strains (Supplementary Table S1) were found to contain gadB gene, including B59671, APC151, GABA, KLDS_3.1003, TH1435, ND03, and ACA-DC 2 (Sun et al., 2011; Treu et al., 2014; Alexandraki et al., 2017; Evivie et al., 2017; Linares et al., 2017; Renye et al., 2017). In these seven strains, the GABA synthesis ability of only a few strains was evaluated.
GABA was synthesized by bacteria through the glutamic acid decarboxylase (GAD) system, which includes a Glu/GABA antiporter GadC, and a GAD GadA or GadB. In the present study, the genome analysis indicated that only CS9 contains a GAD system among these four strains (Figure 10A). It showed 100% similarity to those in other S. thermophilus genomes containing gadB genes (Figure 10A). In order to display the gadB gene in CS9 more intuitively, it was amplified (Figure 10B).
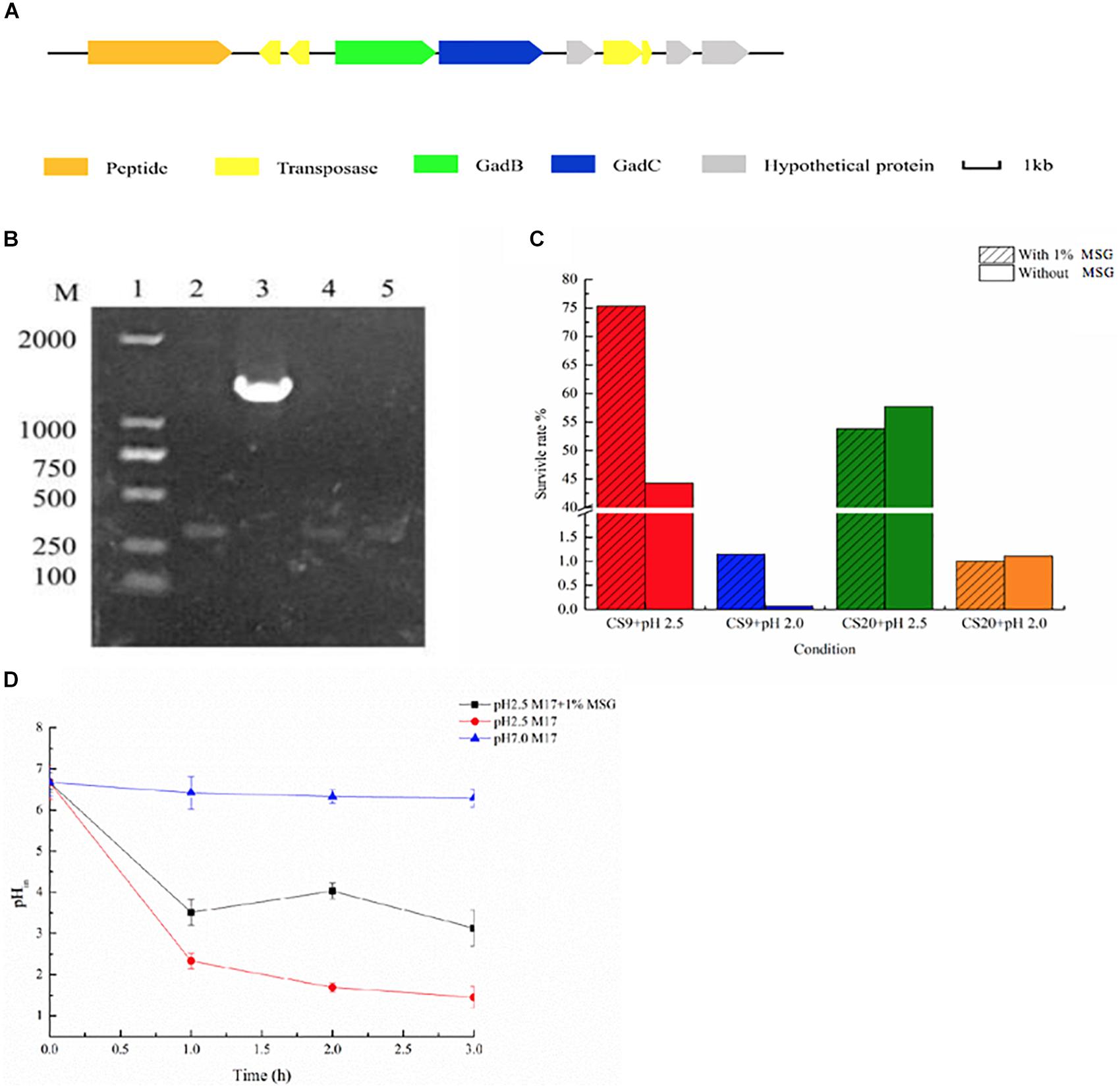
Figure 10. γ-Aminobutyric acid (GABA) produced by CS9. (A) Glutamic acid decarboxylase (GAD) system detected in CS9. (B) The detection of gadB gene of CS5, CS9, CS18, and CS20. (C) Effect of GABA production on the acid resistance ability of the strain (12-h cultures; acid-adapted cells) during the acid resistance assay (37°C and 2-h incubation) carried out in MRS medium (pH 2.5). Survival rate of CS9 cells under different growth conditions. (D) Intracellular pH (pHin) value of CS9 cells under different growth conditions.
Furthermore, the GABA production ability of CS9 was analyzed. The amount of GABA produced by CS9 reached 950.36 mg/L after 24 h fermentation. It showed that CS9 could produce a competitive GABA yield comparing with other S. thermophilus GABA-producing strains, such as Y2 (7984.75 ± 293.33 mg/l), APC151 (2 mg/ml), and ST110 (655 μM) (Yang et al., 2008; Somkuti et al., 2012; Linares et al., 2016). Therefore, the GABA-producing S. thermophilus CS9 could be a new potential starter used in the production of functional foods rich in GABA by means of optimization of fermentation conditions.
Furthermore, GABA synthesis by glutamate decarboxylation in bacteria has already been found associated with acid resistance (Feehily and Karatzas, 2013; Teixeira et al., 2014). Therefore, the effect of GABA production on the acid resistance ability of strain CS9 was evaluated. The result indicated that the high-yield GABA-producing strain CS9 has a high survival rate under the serious acid treatment in the medium supplemented with MSG (Figure 10C). When cultivated in pH 2.5, the survival rate of CS9 reached 75.34% because of the production of GABA, while the rate was only 44.30% without GABA production. As for pH 2.0, CS9 could barely grow, but the survival rate of CS9 could reach 1.15% with the addition of MSG. However, there were no obvious differences in the survival rate of CS20 without gadB gene under the condition of pH 2.0 and pH 2.5 with the supplement of MSG.
In addition, the results showed that the pHin value of CS9 cells with the supplement of 1% MSG was significantly higher than in M17 liquid medium (pH 2.5) (Figure 10D). After adding 1% MSG to the acid-treated medium, the cells consumed protons by fermenting MSG and increasing the pHin value of the cells, so that the strain CS9 could adapt to the extremely acidic conditions. Furthermore, it was found that different treatment times also had an effect on the pHin value of cells. The pHin value of cells was the highest after 2 h incubation with 1% MSG addition. While in the first hour the strain used a few MSG to consume protons, therefore, the pHin value did not change significantly. Also, the pHin value decreased again after 3 h, which is probably because the overall activity of the strain was getting lower and lower when CS9 was incubated in the acidic environment for too long. The pHin value of the cultures without MSG addition presented a declining trend with the accumulation of protons in cells. Obviously, CS9 could eliminate intracellular protons during the decarboxylation of glutamate by the GAD system to maintain the pHin homeostasis under acidic conditions. It was speculated that CS9 could better survive in the GIT of mammals, allowing it to serve certain functions.
In the present study, a comparative analysis of 34 S. thermophilus genomes was performed, and the influence of geographical origin on genetic variability was assessed. Results indicated that strains isolated in the same continent infrequently cluster together. Four S. thermophilus strains isolated from Chinese traditional fermented milk with excellent physiological and biochemical characteristics were first sequenced and analyzed. In addition, the results from the safety evaluation show that the four newly isolated S. thermophilus strains cannot cause antibiotic contamination in nature since they do not contain antibiotic resistance genes and are highly sensitive to many common antibiotics. Moreover, the number of virulence genes of these four strains is much fewer than that of pathogenic Streptococcus. There are no obvious pathogenic factors, and most of the transposon genes are gradually degenerated into pseudogenes during the evolution. The remaining transposon genes that can express normally are homologous to food-grade S. thermophilus, which also proves that these four strains do not have the potential to transfer harmful factors to other strains. That is to say, the safety evaluation shows that these four strains can be safely applied as a natural food starter without any adverse effects on the environment and the human body. CS9 contains a number of unique genes from the distantly related species, has a larger genome size, and represents a good candidate for the evolution research. Furthermore, CS9 can produce a high yield of GABA by the fermentation of glutamate; therefore, CS9 had a higher potential in improving human health. At the same time, it was found that CS9 could eliminate intracellular protons during the decarboxylation of glutamate by the GAD system and then maintain the pHin homeostasis under acidic conditions and result in its higher survival rate. CS18 showed the fastest growth rate, the shortest fermentation time, and the strongest acid and bile salt resistance. It was suggested that CS18 has potential for food fermentation and could better survive in the GIT of mammals. At the same time, this study provides preliminary data for the study of the connection between a genotype and the phenotype (Table 4). Further research is needed in the future.
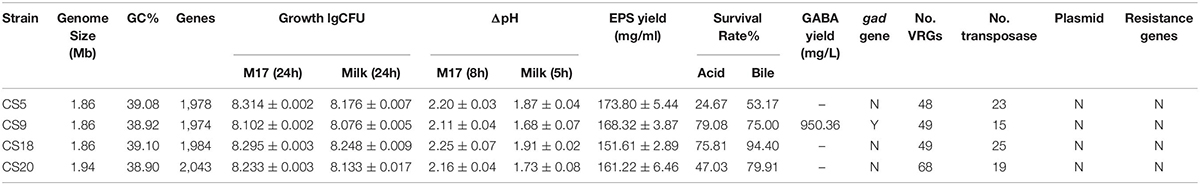
Table 4. Some genotype and the phenotype traits of the four S. thermophilus strains CS5, CS9, CS8, and CS20.
The datasets generated for this study can be found in the nucleotide sequences of CS5, CS9, CS18 and CS20 genomes were submitted to GenBank and assigned accession numbers CP028896, CP030927, CP030928, and CP030250.
YC conceived the idea of the study and designed the experiments. TH carried out most experiments, analyzed the data, and wrote the manuscript. YZ and CZ carried out some experiments. YC, YZ and XQ revised the manuscript.
This work was supported by the National Natural Science Foundation of China (Grant Numbers 31471712 and 31371827).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00184/full#supplementary-material
FIGURE S1 | The detection of plasmids in the CS5, CS9, CS18, and CS20 strains. M: DL15000; 1: CS5; 2: CS9; 3: CS18; 4: CS20.
TABLE S1 | The information of Streptococcus thermophilus strains used in this study.
TABLE S2 | The primers used in the detection of the special genes in the eps gene cluster of CS9.
TABLE S3 | Core genes and unique genes of S. thermophilus strains used in this study.
TABLE S4 | Prediction of virulence-related genes (VRGs) in CS5, CS9, CS18, and CS20.
TABLE S5 | Number of transposases in the four S. thermophilus strains.
TABLE S6 | The analysis of specific genes in the eps cluster of CS9.
TABLE S7 | The growth curve of the four S. thermophilus strains in M17∗.
TABLE S8 | The specific growth rates and final biomasses of strains of CS5, CS9, CS18, and CS20 in M17∗.
TABLE S9 | The growth curve of the four S. thermophilus strains in Milk.
TABLE S10 | The specific growth rates and final biomasses of strains of CS5, CS9, CS18, and CS20 in Milk∗.
TABLE S11 | Resistance ability of the four S. thermophilus strains.
Alexandraki, V., Kazou, M., Blom, J., Pot, B., Tsakalidou, E., and Papadimitriou, K. (2017). The complete genome sequence of the yogurt isolate Streptococcus thermophilus ACA-DC 2. Stand. Genomic Sci. 12:18. doi: 10.1186/s40793-017-0227-5
Bolotin, A., Quinquis, B., Renault, P., Sorokin, A., Ehrlich, S. D., Kulakauskas, S., et al. (2004). Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22, 1554–1558. doi: 10.1038/nbt1034
Brasca, M., Hogenboom, J. A., Morandi, S., Rosi, V., D’Incecco, P., Silvetti, T., et al. (2016). Proteolytic activity and production of γ-aminobutyric acid by Streptococcus thermophilus cultivated in microfiltered pasteurized milk. J. Agric. Food Chem. 64, 8604–8614. doi: 10.1021/acs.jafc.6b03403
Clark, P. A., Cotton, L. N., and Martin, J. H. (1997). Selection of Bifidobacteria for use as delivery adjuncts in cultured dairy foods. II. Tolerance to stimulated pH of human stomachs. Cult. Dairy Prod. J. 28, 11–14.
Cui, Y., Jiang, X., Hao, M., Qu, X., and Hu, T. (2017). New advances in exopolysaccharides production of Streptococcus thermophilus. Arch. Microbiol. 199, 799–809. doi: 10.1007/s00203-017-1366-1
Cui, Y. H., Xu, T., Qu, X., Hu, T., Jiang, X., and Zhao, C. (2016). New insights into various production characteristics of Streptococcus thermophilus strains. Int. J. Mol. Sci. 17:1701. doi: 10.3390/ijms17101701
de Vin, F., Rådström, P., Herman, L., and De Vuyst, L. (2005). Molecular and biochemical analysis of the galactose phenotype of dairy Streptococcus thermophilus strains reveals four different fermentation profiles. Appl. Environ. Microbiol. 71, 3659–3667. doi: 10.1128/aem.71.7.3659-3667.2005
Delcher, A. L., Harmon, D., Kasif, S., White, O., and Salzberg, S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641. doi: 10.1093/nar/27.23.4636
Delorme, C., Bartholini, C., Luraschi, M., Pons, N., Loux, V., Almeida, M., et al. (2011). Complete genome sequence of the pigmented Streptococcus thermophilus strain JIM8232. J. Bacteriol. 193, 5581–5582. doi: 10.1128/JB.05404-11
Eng, C., Thibessard, A., Danielsen, M., Rasmussen, T. B., Mari, J. F., and Leblond, P. (2011). In silico prediction of horizontal gene transfer in Streptococcus thermophilus. Arch. Microbiol. 193, 287–297. doi: 10.1007/s00203-010-0671-8
Erkus, O., Okuklu, B., Yenidunya, A. F., and Harsa, S. (2014). High genetic and phenotypic variability of Streptococcus thermophilus strains isolated from artisanal Yuruk yoghurts. LWT – Food Sci. Technol. 58, 348–354. doi: 10.1016/j.lwt.2013.03.007
Evivie, S. E., Li, B., Ding, X., Meng, Y., Yu, S., Du, J., et al. (2017). Complete genome sequence of Streptococcus thermophilus KLDS 3.1003, a strain with high antimicrobial potential against foodborne and vaginal pathogens. Front. Microbiol. 8:1238. doi: 10.3389/fmicb.2017.01238
Feehily, C., and Karatzas, K. A. G. (2013). Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 114, 11–24. doi: 10.1111/j.1365-2672.2012.05434.x
Fontana, A., Falasconi, I., Molinari, P., Treu, L., Basile, A., Vezzi, A., et al. (2019). Genomic comparison of Lactobacillus helveticus strains highlights probiotic potential. Front. Microbiol. 10:1380. doi: 10.3389/fmicb.2019.01380
Fontana, A., Zacconi, C., and Morelli, L. (2018). Genetic signatures of dairy Lactobacillus casei group. Front. Microbiol. 9:2611. doi: 10.3389/fmicb.2018.02611
Foster, A. C., and Kemp, J. A. (2006). Glutamate- and GABA-based CNS therapeutics. Curr. Opin. Pharmacol. 6, 7–17. doi: 10.1016/j.coph.2005.11.005
Gaden, E. J., Bokanga, M., Harlander, S., Hesseltine, C., and Steinkraus, K. (1992). Applications of Biotechnology in Traditional Fermented Foods. Washington, DC: National Academies Press.
Gilliland, S. E., Staley, T. E., and Bush, L. J. (1984). Importance of bile tolerance of Lactobacillus acidophilus used as dietary adjunct. J. Dairy Sci. 67, 3045–3051. doi: 10.3168/jds.s0022-0302(84)81670-7
Goldenfeld, N., and Woese, C. (2007). Biology’s next revolution. Nature 445:369. doi: 10.1038/445369a
Guédon, G., Bourgoin, F., Pébay, M., Roussel, Y., Colmin, C., Simonet, J. M., et al. (1995). Characterization and distribution of two insertion sequences, IS1191 and iso-IS981, in Streptococcus thermophilus: does intergeneric transfer of insertion sequences occur in lactic acid bacteria co-cultures? Mol. Microbiol. 16, 69–78. doi: 10.1111/j.1365-2958.1995.tb02392.x
Hansen, G., Johansen, C. L., Marten, G., Wilmes, J., Jespersen, L., and Arneborg, N. (2016). Influence of extracellular pH on growth, viability, cell size, acidification activity, and intracellular pH of Lactococcus lactis in batch fermentations. Appl. Microbiol. Biotechnol. 100, 5965–5976. doi: 10.1007/s00253-016-7454-3
Hao, M., Cui, Y., and Qu, X. (2018). Analysis of CRISPR-Cas system in Streptococcus thermophilus and its application. Front. Microbiol. 9:257. doi: 10.3389/fmicb.2018.00257
Hols, P., Hancy, F., Fontaine, L., Grossiord, B., Prozzi, D., Leblond-Bourget, N., et al. (2005). New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29, 435–463. doi: 10.1016/j.femsre.2005.04.008
Hou, J., Hannon, J. A., McSweeney, P. L. H., Beresford, T. P., and Guinee, T. P. (2017). Effect of galactose metabolising and non-metabolising strains of Streptococcus thermophilus as a starter culture adjunct on the properties of Cheddar cheese made with low or high pH at whey drainage. Int. J. Dairy 65, 44–55. doi: 10.1016/j.idairyj.2016.10.003
Hu, T., Zhang, Y., Cui, Y., Zhao, C., Jiang, X., Zhu, X., et al. (2018). Technological properties assessment and two component systems distribution of Streptococcus thermophilus strains isolated from fermented milk. Arch. Microbiol. 200, 567–580. doi: 10.1007/s00203-017-1468-9
Huang, S., Ai, Z. W., Sun, X. M., Liu, G. F., Zhai, S., Zhang, M., et al. (2016). Influence of arginine on the growth, arginine metabolism and amino acid consumption profiles of Streptococcus thermophilus T1C2 in controlled pH batch fermentations. J. Appl. Microbiol. 121, 746–756. doi: 10.1111/jam.13221
Hughes, V. M., and Datta, N. (1983). Conjugative plasmids in bacteria of the ‘pre-antibiotic’ era. Nature 302, 725–726. doi: 10.1038/302725a0
Hutkins, R. W., and Morris, H. A. (1987). Carbohydrate metabolism by Streptococcus thermophilus: a review. J. Food Prot. 50, 876–884. doi: 10.4315/0362-028X-50.10.876
Iyer, R., Tomar, S. K., Maheswari, T. U., and Singh, R. (2010). Streptococcus thermophilus strains: multifunctional lactic acid bacteria. Int. Dairy J. 20, 133–141. doi: 10.1016/j.idairyj.2009.10.005
Juhas, M., van der Meer, J. R., Gaillard, M., Harding, R. M., Hood, D. W., and Crook, D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. doi: 10.1111/j.1574-6976.2008.00136.x
Junjua, M., Kechaou, N., Chain, F., Awussi, A. A., and Dary-Mourot, A. (2016). A large scale in vitro screening of Streptococcus thermophilus strains revealed strains with a high anti-inflammatory potential. LWT – Food Sci. Technol. 70, 78–87. doi: 10.1016/j.lwt.2016.02.006
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., and Tanabe, M. (2012). KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 40, 109–114. doi: 10.1007/978-1-62703-107-3_17
Kazou, M., Alexandraki, V., Blom, J., Pot, B., Tsakalidou, E., and Papadimitriou, K. (2018). Comparative genomics of Lactobacillus acidipiscis ACA-DC 1533 isolated from traditional Greek kopanisti cheese against species within the Lactobacillus salivarius clade. Front. Microbiol. 9:1244. doi: 10.3389/fmicb.2018.01244
Langille, M. G., and Brinkman, F. S. (2009). IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25, 664–665. doi: 10.1093/bioinformatics/btp030
Laws, A., Gu, Y. C., and Marshall, V. (2001). Biosynthesis characterisation and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol. Adv. 19, 597–625. doi: 10.1016/s0734-9750(01)00084-2
Levy, S. B. (1997). Antibiotic resistance: an ecological imbalance. Ciba Found. Symp. 207, 1–9; discussion 9–14.
Li, B., Ding, X., Evivie, S. E., Jin, D., Meng, Y., Huo, G., et al. (2018). Genomic and phenotypic analyses of exopolysaccharides produced by Streptococcus thermophilus KLDS SM. J. Dairy Sci. 101, 106–112. doi: 10.3168/jds.2017-13534
Li, H., Gao, D., Cao, Y., and Xu, H. (2008). A high γ-aminobutyric acid-producing Lactobacillus brevis isolated from Chinese traditional paocai. Ann. Microbiol. 58, 649–653. doi: 10.1007/bf03175570
Li, S., and Shah, N. P. (2014). Antioxidant and antibacterial activities of sulphated polysaccharides from Pleurotus eryngii and Streptococcus thermophilus ASCC 1275. Food Chem. 165, 262–270. doi: 10.1016/j.foodchem.2014.05.110
Linares, D. M., Arboleya, S., Ross, R. P., and Stanton, C. (2017). Complete genome sequence of the gamma-aminobutyric acid-producing strain Streptococcus thermophilus APC151. Genome Announc. 5:e00205-17. doi: 10.1128/genomeA.00205-17
Linares, D. M., O’Callaghan, T. F., O’Connor, P. M., Ross, R. P., and Stanton, C. (2016). Streptococcus thermophilus APC151 strain is suitable for the manufacture of naturally GABA-enriched bioactive yogurt. Front. Microbiol. 7:1876.
Liu, M., Siezen, R. J., and Nauta, A. (2009). In silico prediction of horizontal gene transfer events in Lactobacillus bulgaricus and Streptococcus thermophilus reveals protocooperation in yogurt manufacturing. Appl. Environ. Microbiol. 75, 4120–4129. doi: 10.1128/AEM.02898-08
Mukherjee, K. K. (1994). Isolation of galactose-fermenting thermophilic cultures and their use in the manufacture of low browning Mozzarella cheese. J. Dairy Sci. 77, 2839–2849. doi: 10.3168/jds.s0022-0302(94)77223-4
Ojala, T., Kankainen, M., Castro, J., Cerca, N., Edelman, S., Westerlund-Wikström, B., et al. (2014). Comparative genomics of Lactobacillus crispatus suggests novel mechanisms for the competitive exclusion of Gardnerella vaginalis. BMC Genomics 15:1070. doi: 10.1186/1471-2164-15-1070
Polly, S. M. (1993). The antibiotic paradox: how miracle drugs are destroying the miracle. JAMA. 270, 384–385. doi: 10.1001/jama.1993.03510030108048
Prajapati, J. B., Nathani, N. M., Patel, A. K., Senan, S., and Joshi, C. G. (2013). Genomic analysis of dairy starter culture Streptococcus thermophilus MTCC 5461. J. Microbiol. Biotechnol. 23, 459–466. doi: 10.4014/jmb.1210.10030
Rasmussen, T. B., Danielsen, M., Ondrej, V., Garrigues, C., Johansen, E., and Pedersen, M. B. (2008). Streptococcus thermophilus core genome: comparative genome hybridization study of 47 strains. Appl. Environ. Microbiol. 74, 4703–4710. doi: 10.1128/AEM.00132-08
Renye, J. A., Needleman, D. S., Somkuti, G. A., and Steinberg, D. H. (2017). Complete genome sequence of Streptococcus thermophilus strain B59671, which naturally produces the broad-spectrum bacteriocin thermophilin 110. Genome Announc. 5:e01213-17. doi: 10.1128/genomeA.01213-17
Seo, S. W., Kim, D., O’Brien, E. J., Szubin, R., and Palsson, B. O. (2015). Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat. Commun. 6:7970. doi: 10.1038/ncomms8970
Somkuti, G. A., Renye, J. A., and Steinberg, D. H. (2012). Molecular analysis of the glutamate decarboxylase locus in Streptococcus thermophilus ST110. J. Ind. Microbiol. Biot. 39, 957–963. doi: 10.1007/s10295-012-1114-0
Sorokulova, I. B., Pinchuk, I. V., Denayrolles, M., Osipova, I. G., Huang, J. M., Cutting, S. M., et al. (2008). The safety of two Bacillus probiotic strains for human use. Dig. Dis. Sci. 53, 954–963. doi: 10.1007/s10620-007-9959-1
Sun, Z., Chen, X., Wang, J., Zhao, W., Shao, Y., Wu, L., et al. (2011). Complete genome sequence of Streptococcus thermophilus strain ND03. J. Bacteriol. 193, 793–794. doi: 10.1128/JB.01374-10
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Teixeira, J. S., Seeras, A., Sanchez-Maldonado, A. F., Zhang, C., Su, M. S., and Gänzle, M. G. (2014). Glutamine, glutamate, and arginine-based acid resistance in Lactobacillus reuteri. Food Microbiol. 42, 172–180. doi: 10.1016/j.fm.2014.03.015
Tettelin, H. (2001). Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506. doi: 10.1126/science.1061217
Tian, H., Li, B., Evivie, S. E., Sarker, S. K., Chowdhury, S., Lu, J., et al. (2018). Technological and genomic analysis of roles of the cell-envelope protease PrtS in yoghurt starter development. Int. J. Mol. Sci. 19:E1068. doi: 10.3390/ijms19041068
Toomey, N., Bolton, D., and Fanning, S. (2010). Characterization and transferability of antibiotic resistance genes from lactic acid bacteria isolated from Irish pork and beef abattoirs. Res. Microbiol. 161, 127–135. doi: 10.1016/j.resmic.2009.12.010
Treu, L., Vendramin, V., Bovo, B., Campanaro, S., Corich, V., and Giacomini, A. (2014). Whole-genome sequences of Streptococcus thermophilus strains TH1435 and TH1436, isolated from raw goat milk. Genome Announc. 2:e01129-13. doi: 10.1128/genomeA.01129-13
Uriot, O., Denis, S., Junjua, M., Roussel, Y., Dary-Mourot, A., and Blanquet-Diot, S. (2017). Streptococcus thermophilus: from yogurt starter to a new promising probiotic candidate? J. Funct. Foods 37, 74–89. doi: 10.1016/j.jff.2017.07.038
Vaughan, E. E., Van Den Bogaard, P. T., Catzeddu, P., Kuipers, O. P., and de Vos, W. M. (2001). Actiation of silent gal genes in the lac-gal regulon of Streptococcus thermophilus. J. Bacteriol. 183, 1184–1194.
Vendramin, V., Treu, L., Campanaro, S., Lombardi, A., Corich, V., and Giacomini, A. (2017). Genome comparison and physiological characterization of eight Streptococcus thermophilus strains isolated from Italian dairy products. Food Microbiol. 63, 47–57. doi: 10.1016/j.fm.2016.11.002
Xiong, Z. Q., Kong, L. H., Lai, P. F., Xia, Y. J., Liu, J. C., Li, Q. Y., et al. (2019). Genomic and phenotypic analyses of exopolysaccharide biosynthesis in Streptococcus thermophilus S-3. J. Dairy Sci. 102, 4925–4934. doi: 10.3168/jds.2018-15572
Keywords: Streptococcus thermophilus, genome comparison, technological property, antibiotic resistance, acid resistance capacity, exopolysaccharide (EPS) production, γ-aminobutyric acid (GABA) biosynthesis
Citation: Hu T, Cui Y, Zhang Y, Qu X and Zhao C (2020) Genome Analysis and Physiological Characterization of Four Streptococcus thermophilus Strains Isolated From Chinese Traditional Fermented Milk. Front. Microbiol. 11:184. doi: 10.3389/fmicb.2020.00184
Received: 26 June 2019; Accepted: 24 January 2020;
Published: 28 February 2020.
Edited by:
Lisa Solieri, University of Modena and Reggio Emilia, ItalyReviewed by:
Nuno Pereira Mira, University of Lisbon, PortugalCopyright © 2020 Hu, Cui, Zhang, Qu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhua Cui, eWhjdWlAaGl0LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.