- State Key Laboratory of Microbial Technology, Institute of Microbial Technology, Shandong University, Qingdao, China
Myxococcus xanthus DK1622 has two RecA genes, recA1 (MXAN_1441) and recA2 (MXAN_1388), with unknown functional differentiation. Herein, we showed that both recA genes were induced by ultraviolet (UV) irradiation but that the induction of recA1 was more delayed than that of recA2. Deletion of recA1 did not affect the growth but significantly decreased the UV-radiation survival, homologous recombination (HR) ability, and induction of LexA-dependent SOS genes. In contrast, the deletion of recA2 markedly prolonged the lag phase of bacterial growth and increased the sensitivity to DNA damage caused by hydrogen peroxide but did not change the UV-radiation resistance or SOS gene inducibility. Protein activity analysis demonstrated that RecA1, but not RecA2, catalyzed DNA strand exchange (DSE) and LexA autocleavage in vitro. Transcriptomic analysis indicated that RecA2 has evolved mainly to regulate gene expression for cellular transportation and antioxidation. This is the first report of functional divergence of duplicated bacterial recA genes. The results highlight the evolutionary strategy of M. xanthus cells for DNA HR and genome sophistication.
Introduction
RecA, an ATP-dependent recombinase, is the core enzyme for DNA homologous recombination (HR), as well as being a promotion agent for LexA autolysis in bacteria (Lusetti and Cox, 2002). RecA also contributes to the repair of stalled and collapsed DNA replication forks by postreplication repair pathways (translesion DNA synthesis or template switching), playing an important role in DNA lesion tolerance pathways (Bichara et al., 2011; Quinet et al., 2017; Prado, 2018; Jaszczur et al., 2019). In addition, RecA participates in horizontal gene transfer between different strains (Lawrence and Retchless, 2009; Herrero-Fresno et al., 2015; García-Solache et al., 2016; He et al., 2016), which also causes genetic diversity. Thus, HR delicately balances genomic stability and diversity (Carr and Lambert, 2013; Greene, 2016). After binding to ssDNA, the RecA/ssDNA filament complex may serve as a signal of DNA damage, resulting in the self-cleavage of LexA, which activates the SOS response, increasing the expression of LexA-repressed genes. In the best characterized Escherichia coli SOS response, LexA autolysis derepresses the expression of more than 40 genes involved in DNA repair, mutagenesis, and many other cellular processes (Cox, 2003, 2007; Maslowska et al., 2019). Because of its pros and cons in genomic stability and variability, the functions of RecA are strictly regulated; for example, the function of RecA in E. coli is regulated at the gene transcription and protein activity levels. In the gene transcription induced by the SOS response, particularly, there is a 10–20 times difference in gene expression before and after induction (Cox, 1999, 2007).
Most bacteria, including E. coli, have a single recA gene, while some bacteria possess duplicate recA genes; however, duplicate recA genes have been investigated only in Bacillus megaterium and Myxococcus xanthus. In B. megaterium, duplicate recA genes were found to both be damage-inducible and similarly showed some DNA repair ability in E. coli (Nahrstedt et al., 2005). In the model strain of myxobacteria, M. xanthus DK1622, both RecA1 (MXAN_1441) and RecA2 (MXAN_1388) can partly restore the UV resistance of the E. coli recA mutant, and recA2, but not recA1, was found to be inducible by mitomycin or nalidixic acid (Norioka et al., 1995; Campoy et al., 2003). It is unclear how the duplicate RecA proteins play divergent functions in the DNA recombination and SOS induction in this organism.
In this study, we genetically and biochemically investigated the functions of RecA1 and RecA2 in M. xanthus. We found that both recA genes were inducible by UV irradiation but in different periods. The recA1 deletion had no significant effects on cellular growth but reduced the UV-radiation resistance and induction ability of the SOS gene. In contrast, the absence of recA2 did not affect irradiation resistance but significantly reduced bacterial growth and resistance to oxidative damage. In vitro protein activity analysis indicated that RecA1, but not RecA2, had the homologous strand exchange activity and was able to promote LexA autolysis. Transcriptomic analysis indicated that the recA2 gene was crucial for intracellular substance transport and antioxidant capacity. We discuss the molecular mechanisms for the functional divergence of the RecA1 and RecA2 proteins.
Materials and Methods
Strains, Media, and DNA Substrates
The bacterial strains and plasmids used in this study are described in Supplementary Table S1. The E. coli strains were routinely grown on Luria-Bertani (LB) agar or in LB liquid broth at 37°C. The M. xanthus strains were grown in CYE liquid medium with shaking at 200 rpm or grown on agar plates with 1.5% agar at 30°C (Bretscher and Kaiser, 1978). When required, a final concentration of 40 μg/ml kanamycin (Kan) or 100 μg/ml ampicillin (Amp) was added to the solid or liquid medium.
Single-stranded viral DNA was isolated from M13mp18, and its 3 kb linear dsDNA was amplified by PCR and purified by a DNA purification kit (Tiangen, Beijing, China). A 60-nt oligomer from the M13 genome, 5′-CTG TCA ATG CTG GCG GCG GCT CTG GTG GTG GTT CTG GTG GCG GCT CTG AGG GTG GTG GCT-3′, was obtained from Tsingke Biotech (Qingdao). The 60-nt oligomer was 32P-labelled using a polynucleotide kinase (Ausubel et al., 1995) and stored in TE buffer (10 mM Tris–HCl, pH 7.0, and 0.5 mM EDTA).
Growth and Resistance Analysis
Myxococcus xanthus strains were grown in CYE medium with shaking at 200 rpm at 30°C to an optical density of 0.5 at 600 nm (OD600). Cells were then collected by centrifugation at 8000 rpm for 10 min, washed with 10 mM phosphate buffer (pH 7.0), and diluted to 1 OD600 in the same buffer.
For the radiation damage assay, cells in 10 mM phosphate buffer (pH 7.0) were irradiated at room temperature with a gradient dose from 0 to 200 J/m2 using a UV Crosslinker (Fisher Scientific). Then, the cells were resuspended in fresh CYE medium and incubated at 30°C for 4 h. After incubation, cells were harvested by centrifugation and either used for a further assay or stored at −80°C.
For the oxidative damage assay, cells were suspended in phosphate buffer (pH 7.0) with a concentration of 1 OD, and hydrogen peroxide (H2O2) was added to a final concentration from 1 to 5 mM. The bacterial suspension was incubated for 20 min at room temperature with gentle shaking. After treatment, the suspension was immediately 10-fold diluted in the same phosphate buffer to end the oxidative damage reaction. Then, cells in the suspension were collected for further assay.
The growth assay was determined by growing cells in liquid medium at 30°C. Strains were inoculated at 0.02 OD600 and grown in CYE media for 84 h with shaking at 200 rpm. The OD600 was read every 12 h.
The survival rate was determined by a soft agar colony formation assay. Briefly, to determine the cell survival rate, M. xanthus cells were grown to the early exponential growth stage (OD ≈ 0.5). The cells were treated with UV or H2O2 as described above and were then diluted with fresh medium and mixed at a 1:2 ratio with melted 0.6% soft agar (50°C). The mixture was then spread on CYE plates. After a few minutes for medium solidification, the cultures were incubated at 30°C until clone formation. The survival percentage was calculated as the number of colony-forming units (CFUs) (damaged) divided by the total number of CFUs (undamaged).
Homologous Recombination Assay
According to a previously reported method (Sheng et al., 2005), the recombination rate in M. xanthus was determined by measuring the probability of a resistance gene inserted into the genome through HR. The selected insertion site was located in the noncoding sequence between the MXAN_4466 and MXAN_4467 genes. Then, 500-bp fragments upstream and downstream of the insertion site were amplified with primers (UpF 5′-cacgggctacacgcaggtgcgggg-3′/UpR 5′-ttaagctttcgtttcagcggggactgcctgg-3′ and DownF 5′-caaagcttccaggcagtccccgctgaaacga-3′/DownR 5′-ggcatcgtccctggcggcggcgtgg-3′). The Kan resistance gene (kanR) with its promoter was simultaneously amplified from plasmid pZJY41 with primers 5′-gctgaagcttgtgctgaccccgggtgaat-3′/5′-agaagcttccagagtcccgctcagaagaac-3′. Then, the three DNA segments were linked by the HindIII site according to the arrangement of the upstream segment, resistance gene, and downstream segment. The linked DNA fragment was amplified with primers (UpF 5′-cacgggctacacgcaggtgcgggg-3′ and DownR 5′-ggcatcgtccctggcggcggcgtgg-3′) and quantitatively introduced into M. xanthus via electroporation (1.25 kV, 300 W, 50 mF, and 1 mm cuvette gap). A serial dilution was spread on CYE plates with or without Kan and incubated at 32°C for 72 h to count CFUs. The recombination ability was calculated by the following formula: recombination efficiency (%) = (CFUs with Cam/CFUs without Cam) × 100.
Genetic Manipulations
Escherichia coli plasmids were isolated by the alkaline lysis method, and the chromosomal DNA of E. coli or M. xanthus was extracted using a bacterial genome DNA extraction kit (Tiangen, Beijing, China). Cloning of the genes recA1, recA2, and lexA from M. xanthus was performed according to the general steps (Ausubel et al., 1995). The genes were amplified by PCR and ligated into the pET15b expression plasmid. The primers used here are listed in Supplementary Table S2.
Mutant construction was performed using the markerless mutation in M. xanthus DK1622, with the pBJ113 plasmid using the Kan-resistant cassette for the first round of screening and the galK gene for the negative screening (Ueki et al., 1996). Briefly, the up- and downstream homologous arms were amplified with primers (listed in Supplementary Table S2) and ligated at the BamHI site. The ligated fragment was inserted into the EcoRI/HindIII site of pBJ113. The resulting plasmid was introduced into M. xanthus via electroporation (1.25 kV, 300 W, 50 mF, and 1 mm cuvette gap). The second round of screening was performed on CYE plates containing 1% galactose (Sigma). The recA1 (named RA1) and recA2 (named RA2) mutants were identified and verified by PCR amplification and sequencing.
We attempted to construct the recA1/recA2 double mutant from the single deletion mutant (RA1 or RA2) using the same procedure as described above, but all failed.
RNA Extraction, RT-PCR, and RNA-Seq Assay
Total RNA of M. xanthus cells was extracted using RNAiso Plus reagent following the manufacturer’s protocol (Takara, Beijing, China). cDNA synthesis was performed using the PrimeScript RT Reagent Kit with random primers. The synthesized cDNA samples were diluted five times prior to RT-PCR. The primers were designed for lexA, recA1, and recA2 (Supplementary Table S2). RT-PCR was accomplished using the SYBR Premix Ex Taq Kit (Takara, China) on an ABI StepOnePlus Real-Time PCR System (ThermoFisher Scientific, United States). Gene expression was normalized to the gapA expression and calculated using the equation: change (x-fold) = 2–ΔΔCt (Schefe et al., 2006).
RNA sequencing was conducted by Vazyme (Beijing, China). Three independent repeats are set for each sample. All the up- and downregulated genes were obtained by comparing the expression of the genes with that of the control, and their gene functions were annotated using the NR, GO, and KEGG databases.
Protein Expression, Purification, and Characterization
The constructed expression plasmids with recA1, recA2, or lexA were introduced into E. coli BL21(DE3) competent cells. Protein expression was induced with 1 mM IPTG and purified with Ni-NTA agarose according to the manual of the Ni-NTA purification system (Invitrogen). After overnight dialysis with storage buffer [20 mM Tris–HCl (pH 7.2), 150 mM NaCl, 0.1 mM DTT, 0.1 mM EDTA, and 50% glycerol], the purified proteins were quantified and stored at −80°C.
The ATPase activity of RecA protein was determined in the presence or absence of DNA according to the methods described previously (Sheng et al., 2005). The final reaction mixture in a 2-ml volume contained: 20 mM Tris–HCl (pH 7.4), 10 mM NaCl, 5 mM MgCl2, 2 mM KCl, 3 mM ATP (Sigma), 1 mM CaCl2, 1 mM DTT, and 2% glycerol. The mixture was preheated to 32°C before the addition of RecA and DNA. ATPase activity was determined by measuring the free phosphate ion (Pi) released from ATP using an ultramicro ATPase activity detection kit (Nanjing Jiancheng Bioengineering, Nanjing, China).
In vitro LexA cleavage analysis was performed as described previously (Sheng et al., 2005).
D-loop assays for strand assimilation were performed according to the previously described methods (Cloud et al., 2012; Huang et al., 2017) with some modifications. Briefly, 0.2 μM RecA and 10 nM 32P-labelled ssDNA was combined in 9 μl of reaction buffer containing 25 mM Tris–HCl (pH 7.5), 75 mM NaCl, 5 mM MgCl2, 3 mM ATP, 1 mM DTT, and 1 mM CaCl2 and incubated at 37°C for 5 min. Then, 1 μl of RF M13 plasmid was added to a final concentration of 1 μM, and the incubation at 37°C was continued for 20 min. The reaction was stopped by adding sodium dodecyl sulfate to 0.5% and proteinase K to 1 mg/ml. The deproteinated reaction products were run on a 0.9% agarose 1× TAE gel and visualized using autoradiography with phosphor screen.
In vitro DNA strand-exchange reactions were performed as described previously (Sheng et al., 2005).
Results
Duplicate recA Genes in M. xanthus Are Both Induced by UV Irradiation
The two RecA proteins of M. xanthus DK1622 are highly conserved and are both homologous to the RecA protein of E. coli K12 (EcRecA). The two RecA coding genes have high G+C contents (66 and 65%, respectively); the amino acid identity of RecA1 and RecA2 is 64.6%, and they are 59.36 and 62.04% to EcRecA, respectively. Similar to EcRecA (Story et al., 1992; Lee and Wang, 2009), RecA1 and RecA2 consist of three structural domains, a small N-terminal domain (NTD), a core ATPase domain (CAD), and a large C-terminal domain (CTD). CAD contains the conserved ATPase Walker A and Walker B domains and L1 and L2 DNA-binding domains (Figure 1A). The CAD of RecA1 and RecA2 are highly conserved, while the NTD and CTD are varied. Compared with EcRecA, the two RecA proteins of M. xanthus have more basic amino acids, and the theoretical isoelectric points [pI, calculated by online software (ExPASy – Compute pI/Mw tool)] of RecA1 and RecA2 are 7.04 and 6.5, respectively; EcRecA is more acidic, with a theoretical pI of 5.09 (Figure 1B). Differences in the amino acid composition suggested that the RecA1 and RecA2 proteins might vary in their functions.
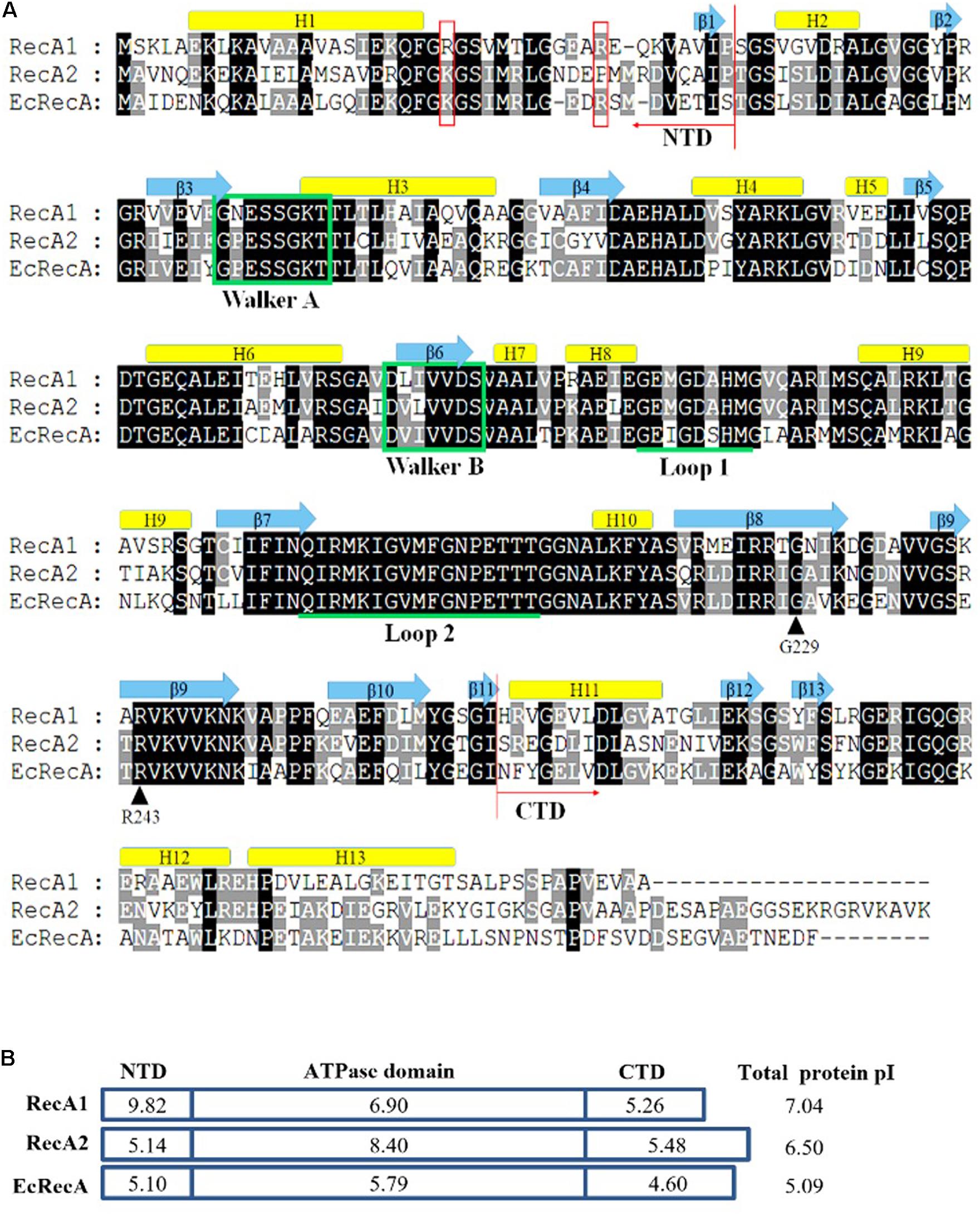
Figure 1. Amino acid sequence comparison of RecA proteins. (A) Alignment of M. xanthus RecA1 and RecA2 and E. coli RecA (EcRecA, b2699). Positions of the N-terminus (NTD) and the C-terminus (CTD) domains are indicated with red arrows, respectively. Their secondary structures all contain 13 alpha-helixes and 13 beta-sheets, which are indicated above their corresponding amino acid sequences. The ATP binding Walker A and B motifs are marked in green frames, and the putative DNA binding sites Loop L1 and L2 are indicated by underlines of the corresponding amino acid sequences. Two reported LexA binding sites (G229 and R243) are indicated by black arrows. K23 and R33 in the N-terminal region of EcRecA are labeled with red boxes. (B) The pI features of the domains of the three RecA proteins. The theoretical pI values were computed using ExPASy online tools (Compute pI/Mw).
The SOS response of M. xanthus cells to DNA damage can be divided into LexA-dependent and -independent types (Campoy et al., 2003). The LexA-dependent SOS genes, e.g., lexA, typically possess a LexA-box sequence in their promoters. Each of the two recA genes of M. xanthus has its own promoter and is not a part of an operon. A typical LexA-box sequence was found in the promoter of recA2 but not in the recA1 promoter (Figure 2A). Previous studies reported that recA2 was obviously induced by nalidixic acid and mitomycin C but that recA1 was not induced by mitomycin C (Norioka et al., 1995; Campoy et al., 2003). We treated M. xanthus cells with 15 J/m2 UV irradiation, which is also a normal induction agent for investigating the bacterial SOS response (Courcelle et al., 2001; Rastogi et al., 2010; Richa et al., 2015). RT-PCR revealed that lexA and recA2 were upregulated by 8.3 times and 10.7 times, respectively, 4 h after UV irradiation at 15 J/m2 (Figure 2B). Interestingly, the recA1 gene was also UV-induced by 6.4 times. The basal expression level of recA1 was very low and was less than one-tenth that of recA2. The low expression level of recA1 might be the reason why the expression of recA1 was not detected by Northern blotting (Norioka et al., 1995). The generation time of M. xanthus cells is about 3–4 h in the exponential growth stage. We found that the induction of recA2 peaked at approximately 3 h after UV treatment, whereas the induction time of recA1 was delayed and peaked 5 h after the treatment (Figure 2C). The different expression levels and induction time points implied that the two RecA proteins participate in the repair of different types of DNA damage caused by UV irradiation.
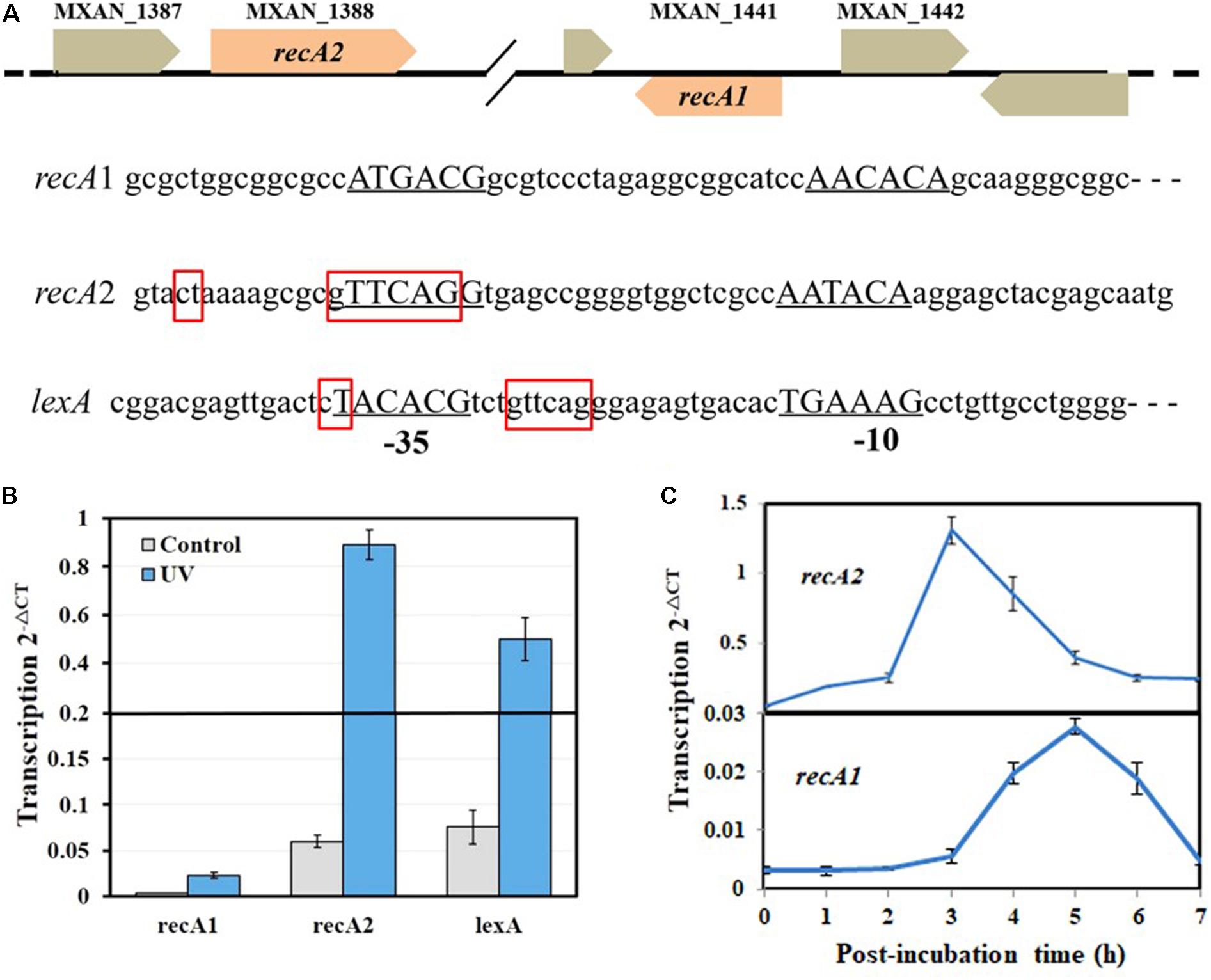
Figure 2. Organization and UV inducibility of the recA1 and recA2 genes of M. xanthus DK1622. (A) Schematic gene location and promoter alignment of M. xanthus recA1 and recA2. RNA polymerase binding sites (–10 and –35 regions) are underlined, and the corresponding nucleotide sequences are in capitals. The SOS box regions are framed in red squares, and the sequence in the promoter of the lexA gene (MXAN_4446) was used as a control. (B) UV inducibility of recA1 and recA2. The strains were incubated for 4 h after UV irradiation treatment with a dose of 15 J/m2 in a UV crosslinking machine and used to detect transcription of recA1 and recA2 by RT-PCR. lexA was used as a control. (C) The induction time points of recA1 and recA2. After being exposed to UV irradiation at a dose of 15 J/m2, the cell cultures were post-incubated at 30°C, sampled at certain intervals to extract the total RNA for RT-PCR. The error bars in panels B and C represent means ± SEM (n = 3, p < 0.05 versus inner reference).
Inactivation of recA2 Compromises the Growth of M. xanthus Cells
In previous studies, recA2 deletion mutants were not obtained in either M. xanthus or B. megaterium (Norioka et al., 1995; Campoy et al., 2003; Nahrstedt et al., 2005). However, in this study, we successfully obtained the deletion mutant of both recA1 and recA2 in M. xanthus, named RA1 and RA2, respectively (Figure 3A). According to the two-step screening method employed, the acquisition probability from reverse screening was ∼10–6 for the deletion of recA1 and ∼3.3 × 10–10 for the deletion of recA2, and this may be the reason why it is difficult to make a recA2 mutant. At present, there is no evidence of a suppressor mutation, which may be required to achieve the deletion of recA2. However, although more evidence is needed, we speculate that the difficulty in the screening of RA2 is probably related to the function of RecA2 in growth. We also tried to construct the double knockout mutant of recA1 and recA2 but failed, and this might be because the double mutation had a serious impact on cell survival and was synthetically lethal. recA1 deletion had no significant effects on cellular growth, but deletion of recA2 caused the mutant to have a long lag phase. After the lag phase, growth of the RA2 mutant did not slow down significantly in the logarithmic phase, and the mutant culture reached a similar density as wild-type DK1622 (Figures 3B,C).
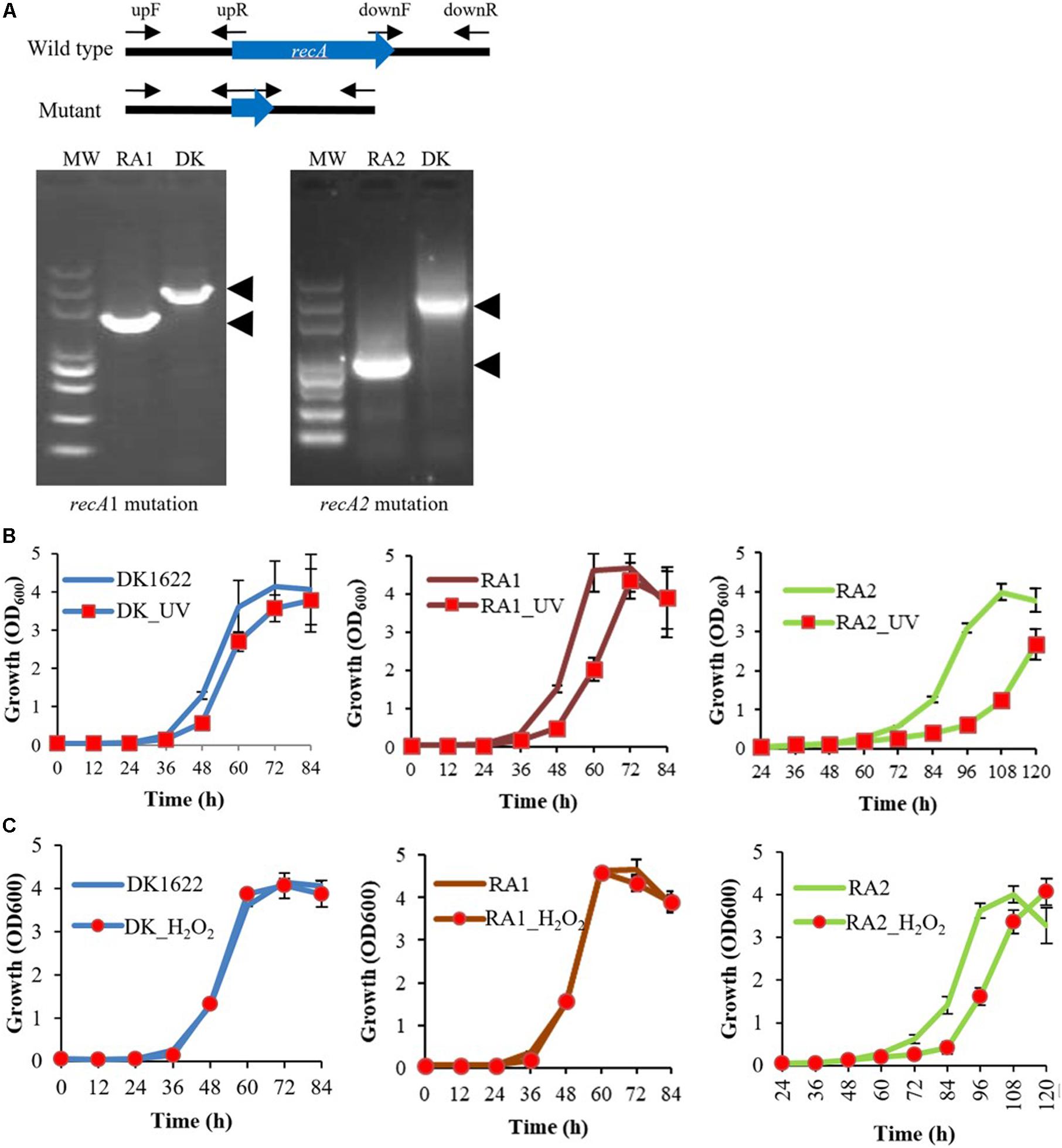
Figure 3. Mutations of recA1 and recA2, and their effects on the growth of M. xanthus. (A) Deletion of recA1 or recA2 in M. xanthus DK1622, using the markerless knockout plasmid pBJ113, producing the RA1 or RA2 mutants. The deletion was verified by PCR using their primer pairs (upF/downR) and sequencing. (B) Separate growth comparisons of DK1622, RA1, and RA2 with and without UV treatment at a dose of 15 J/m2. (C) Separate growth comparisons of DK1622, RA1, and RA2 with and without the H2O2 treatment at a final concentration of 3 mM for 15 min. The error bars indicate the SEM for six replicates.
When treated with 15 J/m2 UV irradiation, compared with those without UV treatment, the growth abilities were delayed in DK1622, RA1, and RA2 cells, and the growth delay was more notable in RA2 (Figure 3B). When treated with 3 mM H2O2 for 15 min, DK1622 and RA1 cells showed almost the same growth curve, while the growth of RA2 cells was delayed significantly compared with that of the strains without the treatment (Figure 3C). The results demonstrated that recA2, but not recA1, is an important factor for cell growth after UV irradiation and oxidation damage.
recA1 and recA2 Are Separately Crucial for UV Resistance and H2O2 Resistance
We measured the survival rates of the wild-type strain and the recA deletion mutants treated with different dosages of UV irradiation (0–25 J/m2) and H2O2 (1–5 mM). All three strains had decreased survival rates with increasing UV irradiation or H2O2 concentration. Interestingly, the survival rate of RA1 cells decreased more significantly than that of RA2 at each UV-radiation dosage, which had a highly similar survival curve to the wild-type strain (Figure 4A). In addition, the survival rate of RA2 cells decreased more significantly at each H2O2 concentration than that of RA1 and DK1622 cells, which showed similar survival curves when treated with hydrogen peroxide (Figure 4B). Thus, RecA1 is needed for the survival of M. xanthus cells under UV irradiation, which is similar to that of EcRecA (Alexseyev et al., 1996), whereas RecA2 is involved in tolerance to H2O2 damage in cells.
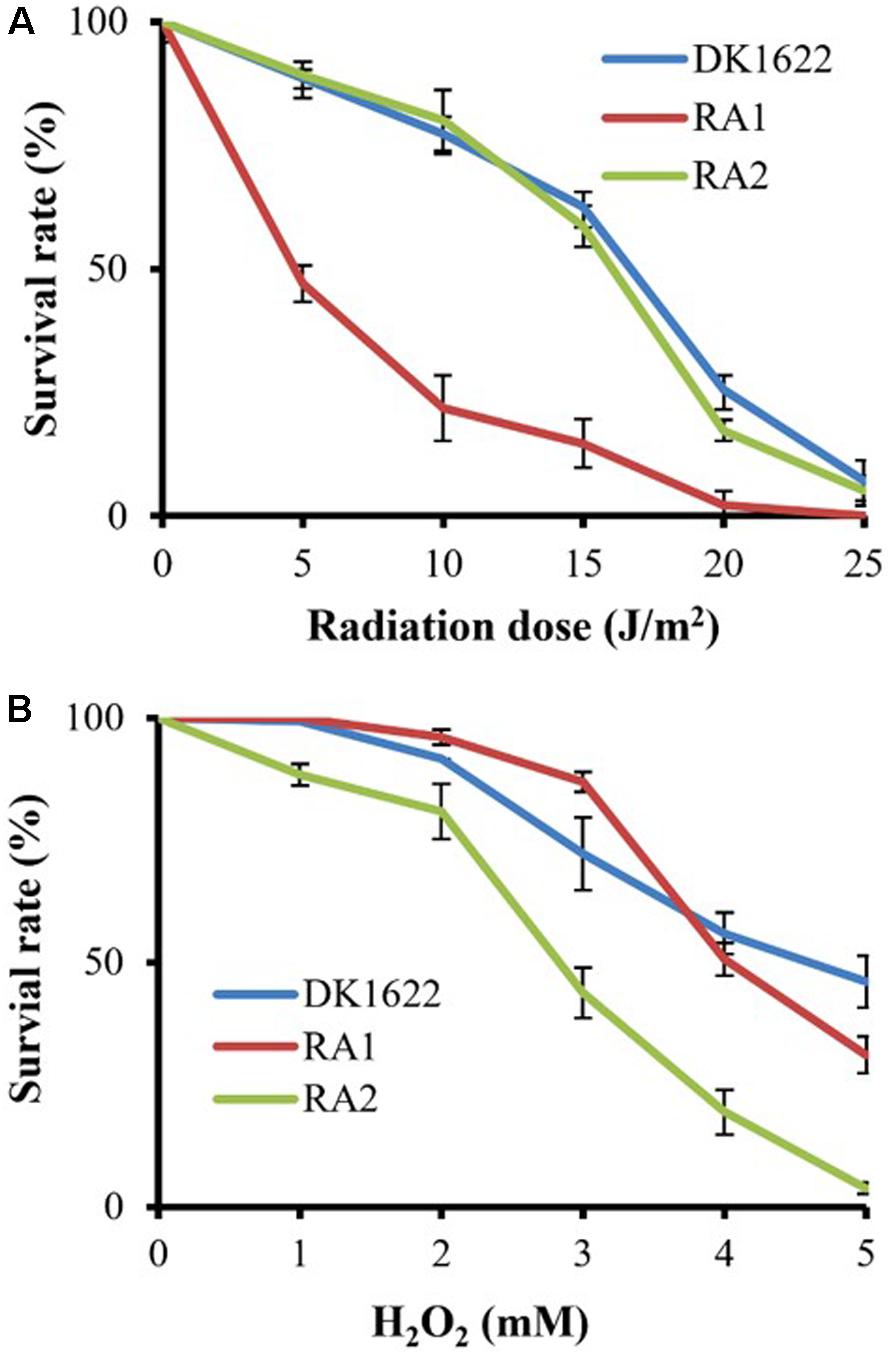
Figure 4. Survival of M. xanthus wild-type strain DK1622 and the mutants RA1 and RA2. (A) Survival curves after exposure to UV irradiation at different dosages (0–25 J/m2). (B) Survival curves after hydrogen peroxide treatment at different concentrations (0–5 mM). The percentage of surviving cells was calculated by comparing with the corresponding non-treated cells. The error bars indicate the SEM for six replicates.
RecA1, Not RecA2, Is Responsible for HR and LexA-Dependent SOS Induction
DNA HR and SOS induction are the two main cellular functions of the RecA proteins (Cox, 2003). We analyzed the in vivo integration abilities of an antibiotic resistance gene into the genomes of the DK1622, RA1, and RA2 strains. Calculated from the appearance of resistant colonies, the recombination rate of RA1 cells was significantly lower than that in either DK1622 (p = 0.0088) or RA2 (p = 0.0157) cells, while the differences between the recombination rates of RA2 and DK1622 cells were not significant (p = 0.1049) (Figure 5A). The results showed that recA1 is important for the recombination process in M. xanthus.
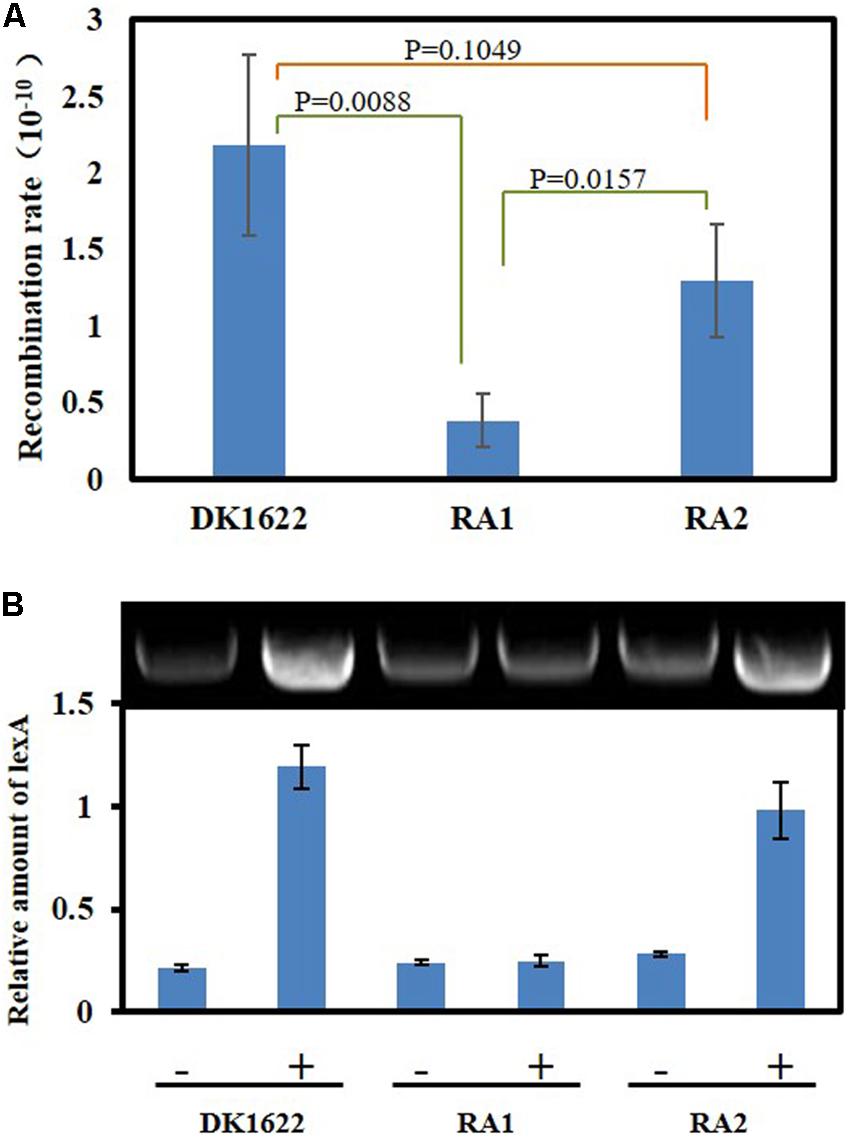
Figure 5. Intracellular DNA recombination rate and induction analysis of the lexA gene. (A) The cellular recombination rate of DK1622, RA1, and RA2. The two ends of the resistance gene (kan) have homologous DNA sequences with the two ends of the insertion site, respectively, and the kan gene is transferred into the M. xanthus through electrical transformation. The recombination rate was calculated by measuring the proportion of kanamycin-resistant colonies. (B) The inducibility of the lexA gene. Myxococcus lexA is a known SOS gene induced through LexA-dependent SOS response, and herein, its UV inducibility represents the activation of LexA-dependent SOS response. The strains were irradiated with 15 J/m2 UV irradiation (+) or mock treatment (–), and the transcription of lexA was determined by RT-PCR.
Previous studies indicated that the expression of lexA is induced by the LexA-dependent SOS response in M. xanthus (Norioka et al., 1995). We compared the transcription of lexA in the M. xanthus DK1622, RA1, and RA2 strains in response to the 15 J/m2 UV irradiation treatment. The RT-PCR results showed that lexA could be induced by UV in both DK1622 and RA2 but not in the RA1 mutant (Figure 5B). Thus, the deletion of recA1, rather than recA2, affected the UV-induction of lexA, i.e., RecA1 is responsible for LexA-dependent SOS induction.
RecA1 and RecA2 Both Have ss- and ds-DNA Promoted ATPase Activities
We further expressed and purified the RecA1 and RecA2 proteins (Figure 6A) and measured their in vitro ATPase activities by the quantitative analysis of inorganic phosphorus released from ATP hydrolysis (Figure 6B). In the reaction mixture without the addition of DNA, RecA1 and RecA2 both exhibited low ATPase activities, and the ATPase activity of RecA2 was somewhat higher than that of RecA1. For example, a microgram of purified RecA2 released 0.1428 nanomole Pi in an hour, which is approximately 2.44 times the hydrolysis capacity of RecA1 on ATP (0.0586 nmol Pi/μg/h). The addition of DNA, especially ssDNA, markedly promoted the ATPase activity of both RecA1 and RecA2, and this is consistent with the functionality of classic RecA proteins (Cox, 2003; Greene, 2016). Thus, RecA1 and RecA2 are both DNA-dependent (more dependent on ssDNA) ATPase enzymes. In the presence of DNA (dsDNA or ssDNA), the increase in the ATPase activity of RecA1 was higher than that of RecA2. For example, the ATPase activity of 1 ng RecA1 increased by 10.69 times (from 0.0586 to 0.6265 nmol Pi/μg/h) with the addition of ssDNA, while the increase of that in RecA2 was only double (from 0.1428 to 0.2857 nmol Pi/μg/h). Similarly, the addition of dsDNA increased the ATPase activities of RecA1 and RecA2 by 6.89 times (from 0.0586 to 0.4038 nmol Pi/μg/h) and 1.86 times (from 0.1428 to 0.2658 nmol Pi/μg/h), respectively.
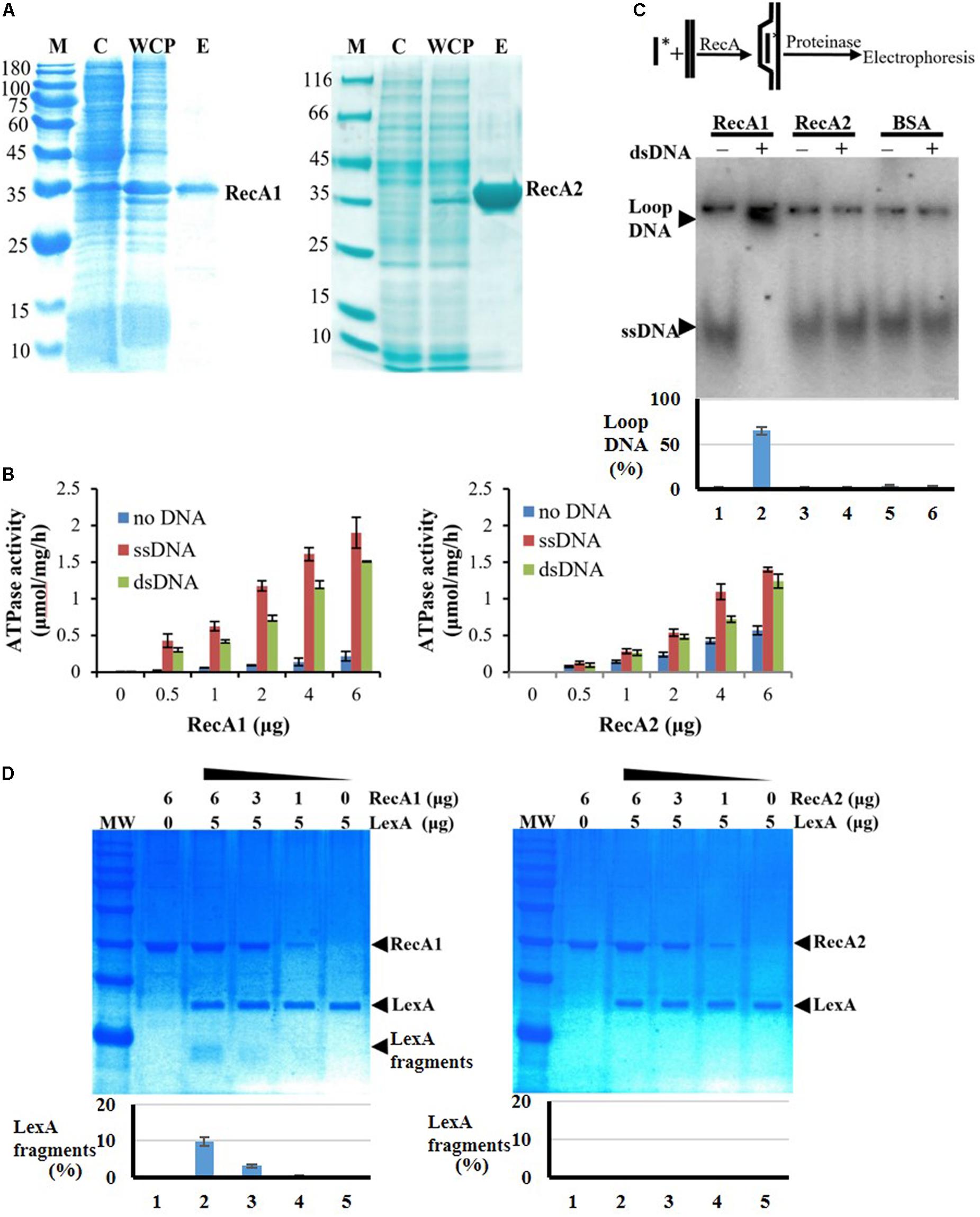
Figure 6. Expression and activity analysis of RecA proteins. (A) Expression and purification of RecA1 and RecA2. M, marker; C, control; WCP, whole-cell protein; E, eluent of purified protein. (B) Assays of ATPase activities. The ATPase activity was determined by measuring free phosphate ion (Pi) released from enzymolysis of ATP. The error bar is calculated from three independent repeats. (C) D-loop assay. A 60-nt 32P-labeled ssDNA fragment and a superhelical dsDNA (RF M13) sequence were mixed and incubated with and without the addition of purified RecA1 or RecA2 proteins. If the protein has HR activity, the homologous pairing reaction will be initiated, thus forming the ssDNA–dsDNA complex. Bovine serum albumin (BSA) was used as a control. Relative DNA-labeled intensities of the bands were quantified by a Gel-Doc image analysis system (Bio-Rad). The percentage of loop-DNA-labeled intensity in the total labeled intensity (including the labeled strength in ssDNA, loop-DNA, and residual DNA in the origin) was used to quantitate the RecA activity. (D) The promotion ability of RecA1 (left) or RecA2 (right) on the cleavage of LexA proteins. The MxLexA protein was incubated with gradient concentrations of RecA1 or RecA2 proteins in the presence of ssDNA and ATP. Reactions were stopped and visualized on a 1.2% SDS-PAGE gel stained with Coomassie brilliant blue. The bands were quantified by computerized image analysis (Bio-Rad), and the percentage of LexA fragments in the total LexA signal in every lane was used to quantify the ability of RecA to stimulate LexA autocleavage.
RecA1, but Not RecA2, Has in vitro HR Capacity and Activates LexA Autolysis
Strand invasion or D-loop formation is a central step in HR and is one of the most common biochemical assays for characterizing the activity of RecA-type recombinase (Cox, 2003; Greene, 2016; Huang et al., 2017). We analyzed the in vitro recombination activities of RecA1 and RecA2 in a DNA strand recombination reaction system containing 32P-ssDNA and homologous plasmid dsDNA. The reaction products were separated by agarose gel electrophoresis, and a lagged radiolabeling band appeared in the lane containing purified RecA1 but that with not RecA2 (Figure 6C). Furthermore, DNA strand exchange (DSE), another characteristic reaction for RecA-catalyzed HR with single-stranded circular DNA and its homologous double-stranded linear DNA, was used to identify the recombination activity of RecA1 and RecA2 (Supplementary Figure S5). The joined molecule DNA (jmDNA) appeared and increased as a recombinant product with the gradient addition of the RecA1 protein but not with that of the RecA2 protein. The above results indicated that RecA1, but not RecA2, has HR activity in M. xanthus, and these results are consistent with the in vivo recombination results (Figure 5A).
RecA promotes LexA autolysis at a specific site, thereby enabling the expression of SOS genes inhibited by LexA (Janion, 2008; Kovačič et al., 2013). We monitored the LexA cleavage activity promoted by RecA1 and RecA2, using the M. xanthus LexA protein as a substrate. The results showed that the LexA autolysis fragments were detected in the reaction with RecA1 but not that with RecA2 (Figure 6D). Thus, RecA1 participated in the LexA-dependent SOS induction reaction, and this is also consistent with the RA1 mutant losing the induction ability of the SOS gene lexA (Figure 5B).
Discussion
RecA is an ATP-dependent recombinase central to DNA HR and activation of the LexA-dependent SOS response. Although the recA gene is duplicated in some bacterial cells, its functions have not been investigated. In M. xanthus DK1622, the expression of recA1 is very low and is less than one-tenth of that of recA2. The two recA genes are both inducible by UV irradiation, but the induction of recA2 was significantly earlier than that of recA1 and recN (Supplementary Figure S6). Generally, the DNA repair genes expressed in the early and late stages of SOS are responsible for the error−free repair and maintenance processes and error-prone DNA synthesis against serious DNA damage, respectively (Kuzminov, 1999; Janion, 2008; Maslowska et al., 2019). Thus, the two RecA proteins are both involved in UV resistance, probably for different lesions caused by UV irradiation (Sinha and Häder, 2002); RecA2 is involved in the early repair processes, and RecA1 is involved in serious DNA-damage repair, i.e., post-replication repair. The deletion of recA2 caused the mutant to have a long lag phase, but the recA1 deletion had no significant effect on cellular growth. It is known that the growth lag phase is an adaptation period of bacterial cells to changes in temperature and nutrients in new environments (Monod, 1949; Vermeersch et al., 2019), macromolecule damage repair, and protein misfolding accumulated during cell arrest (Dukan and Nyström, 1998; Saint-Ruf et al., 2007; Rolfe et al., 2012; Bertrand, 2019), and enzyme preparation for rapid growth in the logarithmic phase (Monod, 1949; Rolfe et al., 2012). Thus, RecA2, instead of RecA1, plays a crucial role in the repair process required for cellular growth. Similar to the classic bacterial RecA, RecA1 possesses DNA recombination activity and SOS gene induction ability, which are required for survival under UV irradiation. However, RecA2 has lost its HR and SOS gene induction abilities but has evolved to play roles in the regulation of gene expression for cellular growth as well as cellular survival under oxidation pressure by hydrogen peroxide. This is the first study to clearly determine the divergent functions of duplicated recA genes in bacterial cells.
To obtain more clues about the potential mechanisms of RecA2 in M. xanthus, we compared the transcriptomes of the recA2 mutant strain (RA2) and wild-type strain DK1622. Overall, 79 genes were found to be differentially expressed (padj. < 0.05) by the deletion of recA2, and this included 60 upregulated genes and 19 downregulated genes (Figure 7A; for details, refer to Supplementary Table S3). Gene ontology (GO) enrichment analysis based on the KEGG database showed that the differentially expressed genes (DEGs) were assigned to 30 GO terms in the categories of biological process, cellular component, and molecular function (Figure 7B). Obviously, the biological process DEGs formed the largest group and included 17 GO terms, followed by the molecular function (10 GO terms) and cellular component (three GO terms) groups (Figure 7B). The DEGs were mainly enriched in two functional regions. One is related to transport and location, including the categories of transport (14 genes), localization and establishment of localization (28 genes), transmembrane transport (five genes), and protein transmembrane transport (three genes). The other category is related to antioxidation and includes the categories of oxidoreductase activity (three genes), peroxiredoxin activity (two genes), ferric iron binding (two genes), antioxidant capacity (three genes), and catalase (one gene). These DEGs were significantly enriched in ABC transporters and several metabolism-related pathways, such as methane metabolism, biosynthesis of secondary metabolites, and metabolic pathways (Figure 7C). Combining this with the experimental results presented in this study, we propose that the function of recA2 is mainly related to cellular transportation and antioxidation, which are required for the normal growth of cells.
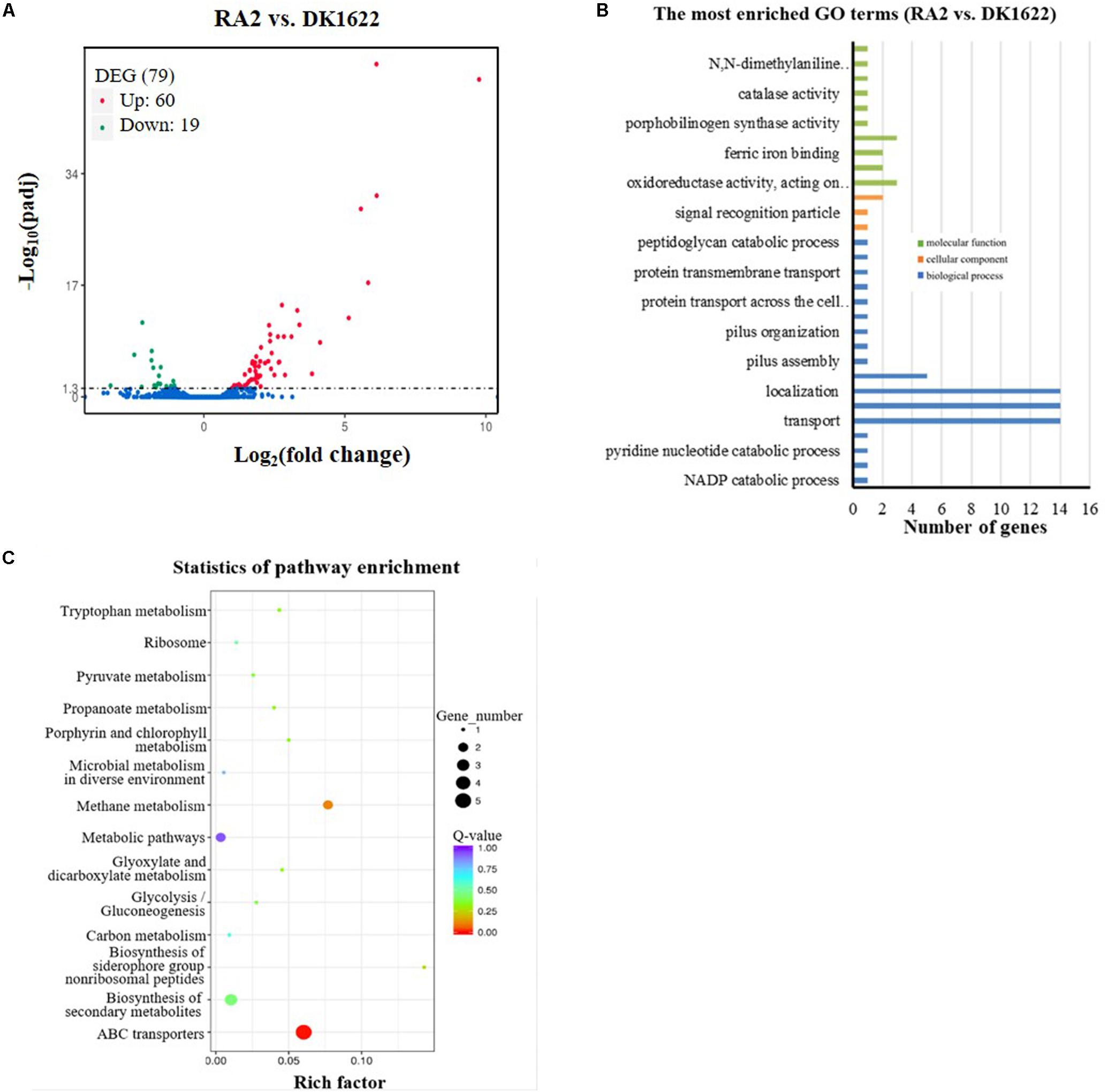
Figure 7. Comparison of transcriptomes between the recA2 mutant (RA2) and the wild-type strain (DK1622). (A) Volcano plot of differentially expressed gene (DEG) distributions. Red dots and green dots represent the up- and down-regulated genes with significant differences, respectively (padj. < 0.05). The blue dots represent the genes that have not changed significantly. (B) The distribution of GO category and (C) pathways of DGEs between RA2 and DK1622. Enriched GO is shown in three categories: biological process (blue), molecular function (green), and cellular component (orange).
RecA1 and RecA2 are both homologous proteins with E. coli RecA. They both retain the DNA-dependent ATPase activity and self-aggregation ability of E. coli RecA (Figure 6B and Supplementary Figure S7), and the interaction between RecA1 and RecA2 may even be due to the conserved self-aggregation sites. On the other hand, these two proteins also showed significantly different activities in DNA recombination and LexA autocleavage. Amino acid sequence alignment showed that the RecA1 and RecA2 amino acid sequences are highly similar in the CAD, and are mainly varied in the NTD and CTD (Figure 1A). Lys23 and Arg33 in the N-terminal region are both necessary for the nucleoprotein filament of RecA–ssDNA to capture homologous dsDNA (Lee and Wang, 2009). The corresponding amino acids at the two sites are both alkaline arginine residues in RecA1, which is consistent with that in EcRecA. In RecA2, however, the amino acids at the two sites are arginine and proline, respectively (Figure 1A). We aligned the N-terminal sequences of 11 reported bacterial RecA proteins. The amino acids at the corresponding 23rd site are all alkaline amino acids but are less conserved at the 33rd site (Supplementary Figure S1). Nine RecAs, including RecA1 of M. xanthus, have positively charged residues (Arg or Lys) at the 33rd site, while three RecAs, including RecA2 from M. xanthus, RecA from Prevotella ruminicola (Aminov et al., 1996), and RecA from Borrelia burgdorferi (Huang et al., 2017), did not have the positively charged residues at this site. RecAs with an alkaline amino acid at the 33rd site all have DNA recombination activity (Sano and Kageyama, 1987; Nussbaumer and Wohlleben, 1994; Umelo et al., 1996; Cox, 1999; Kim et al., 2002; Orillard et al., 2011; Grove et al., 2012; Carrasco et al., 2019). However, similar to RecA2, RecAs from P. ruminicola and B. burgdorferi were reported to have no anti-ultraviolet radiation ability (Aminov et al., 1996; Huang et al., 2017). The results presented in this study demonstrate that RecA2 of M. xanthus evolved to affect the genes for cellular transportation and antioxidation, which are obviously related to damage repair for cellular growth.
As in E. coli RecA (Lusetti et al., 2003; Kovačič et al., 2013), RecA 1 and RecA2 both have conserved LexA binding sites, including Gly229 and Arg243, in their C-terminal regions and 10 neighboring amino acids (Figure 1A), which, however, does not explain the differences between the two proteins in promoting LexA autolysis. Notably, while the three domains of EcRecA are all acidic, the NTD of RecA1 and the CAD of RecA2 are alkaline, with pI values of 9.82 and 8.40, respectively. Accordingly, RecA1 forms more negative charges on the outer side of the polymer, while RecA2 forms more negative charges on the inner side of the helical structure (Supplementary Figure S2). We noted that, unlike the E. coli LexA (EcLexA) protein, which is an acidic protein [theoretical pI is 6.23, calculated by online software (ExPASy Compute pI/Mw tool)], M. xanthus LexA (MxLexA) is a basic protein, and its theoretical pI is 8.77. EcLexA and MxLexA are conserved in their catalytic and DNA binding domains, and the differences between the two proteins lie mainly in the linker region (Supplementary Figure S3). The EcLexA linker contains more acidic amino acids (theoretical pI = 3.58), while the linker of MxLexA contains more basic amino acids (theoretical pI = 8.75). In addition, MxLexA has two more fragments flanking the linker sequence. The additional fragment at the N-terminal side destroys the β2 folding structure and further lengthens the irregular linker of MxLexA, leading to a long irregular chain containing more basic amino acids (theoretical pI = 12.01). According to the binding mode between EcLexA and EcRecA (Kovačič et al., 2013), the linker region of LexA is close to the inner groove of the RecA protein filament (Supplementary Figure S4). The inner helix part of RecA2 (in the CAD) is alkaline (Supplementary Figure S2B), which hinders MxLexA binding to the RecA filament and thus hinders its ability to promote MxLexA self-cleavage.
Myxobacteria has a relatively large genome size (9–14 Mbp) and contains many DNA repeats (Goldman et al., 2006; Andersson and Hughes, 2009; Han et al., 2013). These repetitive DNA fragments are potential substrates for RecA-catalyzed HR. Functional divergence of duplicate RecA proteins and low expression of the recombination enzyme RecA1 reduce the DNA recombinant activity without affecting other cellular repair functions in M. xanthus (such as the functions carried out by RecA2). In the sequenced myxobacterial genomes (Supplementary Table S4), all the strains, except Anaeromyxobacter, harbor two recA genes, and their amino acid sequences are highly conserved. For example, the amino acid identities of RecA1 and RecA2 of all Myxococcus are >89.4 and 93.6%, respectively. The Anaeromyxobacter strains have a single recA gene in their genomes; however, their genomes are small (5.0–5.2 Mbp) and possess few repetitive sequences. We propose that the functional divergence and expression regulation of duplicate RecA proteins might be a strategy for bacteria with a large number of repetitive sequences in their large genomes to avoid incorrect recombination.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
D-HS and Y-ZL designed the experiments. D-HS, Y-XW, MQ, and J-YZ performed the experiments. D-HS, X-JY, and Y-ZL analyzed the data. D-HS and Y-ZL wrote the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (NSFC) (Grant Nos. 31670076 and 31471183), the National Key Research and Development Program (2018YFA0901700), and the Key Program of Shandong Natural Science Foundation (Grant No. ZR2016QZ002) to Y-ZL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This manuscript has been released as a Pre-Print at bioRxiv.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00140/full#supplementary-material
References
Alexseyev, A. A., Bakhlanova, I. V., Zaitsev, E. N., and Lanzov, V. A. (1996). Genetic characteristics of new recA mutants of Escherichia coli K-12. J. Bacteriol. 178, 2018–2024. doi: 10.1128/jb.178.7.2018-2024.1996
Aminov, R. I., Nagamine, T., Ogata, K., Sugiura, M., Tajima, K., and Benno, Y. (1996). Cloning, sequencing and complementation analysis of the recA gene from Prevotella ruminicola. FEMS Microbiol. Lett. 144, 53–59.
Andersson, D. I., and Hughes, D. (2009). Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43, 167–195. doi: 10.1146/annurev-genet-102108-134805
Ausubel, F. M., Brent, R., and Kingston, R. E. (1995). Short Protocals in Molecular Biology, 3rd Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Bertrand, R. L. (2019). Lag phase is a dynamic, organized, adaptive, and evolvable period that prepares bacteria for cell division. J. Bacteriol. 201:e00697-618. doi: 10.1128/JB.00697-18
Bichara, M., Meier, M., Wagner, J., Cordonnier, A., and Lambert, I. B. (2011). Postreplication repair mechanisms in the presence of DNA adducts in Escherichia coli. Mutat. Res 727, 104–122. doi: 10.1016/j.mrrev.2011.04.003
Bretscher, A. P., and Kaiser, D. (1978). Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 133, 763–768. doi: 10.1128/jb.133.2.763-768.1978
Campoy, S., Fontes, M., Padmanabhan, S., Cortés, P., Llagostera, M., and Barbé, J. (2003). LexA-independent DNA damage-mediated induction of gene expression in Myxococcus xanthus. Mol. Microbiol. 49, 769–781. doi: 10.1046/j.1365-2958.2003.03592.x
Carr, A. M., and Lambert, S. (2013). Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J. Mol. Biol. 425, 4733–4744. doi: 10.1016/j.jmb.2013.04.023
Carrasco, B., Serrano, E., Martín-González, A., Moreno-Herrero, F., and Alonso, J. C. (2019). Bacillus subtilis MutS modulates RecA-mediated DNA strand exchange between divergent dna sequences. Front Microbiol. 10:237. doi: 10.3389/fmicb.2019.00237
Cloud, V., Chan, Y. L., Grubb, J., Budke, B., and Bishop, D. K. (2012). Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225. doi: 10.1126/science.1219379
Courcelle, J., Khodursky, A., Peter, B., Brown, P. O., and Hanawalt, P. C. (2001). Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 58, 41–64.
Cox, M. M. (1999). Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic. Acid. Res. Mol. Biol. 63, 311–366. doi: 10.1016/s0079-6603(08)60726-6
Cox, M. M. (2003). The bacterial RecA protein as a motor protein. Annu Rev Microbiol. 57, 551–577. doi: 10.1146/annurev.micro.57.030502.090953
Cox, M. M. (2007). Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 42, 41–63. doi: 10.1080/10409230701260258
Dukan, S., and Nyström, T. (1998). Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 12, 3431–3441. doi: 10.1101/gad.12.21.3431
García-Solache, M., Lebreton, F., McLaughlin, R. E., Whiteaker, J. D., Gilmore, M. S., and Rice, L. B. (2016). Homologous recombination within large chromosomal regions facilitates acquisition of β-lactam and vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 60, 5777–5786. doi: 10.1128/aac.00488-16
Goldman, B. S., Nierman, W. C., Kaiser, D., Slater, S. C., Durkin, A. S., Eisen, J. A., et al. (2006). Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U.S.A. 103, 15200–15205. doi: 10.1073/pnas.0607335103
Greene, E. C. (2016). DNA sequence alignment during homologous recombination. J. Biol. Chem. 291, 11572–11580. doi: 10.1074/jbc.R116.724807
Grove, D. E., Anne, G., Hedayati, M. A., and Bryant, F. R. (2012). Stimulation of the Streptococcus pneumoniae RecA protein-promoted three-strand exchange reaction by the competence-specific SsbB protein. Biochem. Biophys. Res. Commun. 424, 40–44. doi: 10.1016/j.bbrc.2012.06.059
Han, K., Li, Z. F., Peng, R., Zhu, L. P., Zhou, T., Wang, L. G., et al. (2013). Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci. Rep. 3:2101. doi: 10.1038/srep02101
He, S., Chandler, M., Varani, A. M., Hickman, A. B., Dekker, J. P., and Dyda, F. (2016). Mechanisms of evolution in high-consequence drug resistance plasmids. mBio 7:e01987-16.. doi: 10.1128/mBio.01987-16
Herrero-Fresno, A., Rodicio, R., Montero, I., Garcaí, P., and Rodicio, M. R. (2015). Transposition and homologous recombination drive evolution of pUO-StVR2, a multidrug resistance derivative of pSLT, the virulence plasmid specific of Salmonella enterica serovar Typhimurium. Infect Genet Evol. 29, 99–102. doi: 10.1016/j.meegid.2014.11.010
Huang, S. H., Hart, M. A., Wade, M., Cozart, M. R., McGrath, S. L., and Kobryn, K. (2017). Biochemical characterization of Borrelia burgdorferi’s RecA protein. PLoS One 12:e0187382. doi: 10.1371/journal.pone.0187382
Janion, C. (2008). Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int. J. Biol. Sci. 4, 338–344. doi: 10.7150/ijbs.4.338
Jaszczur, M. M., Vo, D. D., Stanciauskas, R., Bertram, J. G., Sikand, A., Cox, M. M., et al. (2019). Conformational regulation of Escherichia coli DNA polymerase V by RecA and ATP. PLoS Genet. 15:e1007956. doi: 10.1371/journal.pgen.1007956
Kim, J. I., Sharma, A. K., Abbott, S. N., Wood, E. A., Dwyer, D. W., Jambura, A., et al. (2002). RecA Protein from the extremely radioresistant bacterium Deinococcus radiodurans: expression, purification, and characterization. J Bacteriol. 184, 1649–1660. doi: 10.1128/jb.184.6.1649-1660.2002
Kovačič, L., Paulič, N., Leonardi, A., Hodnik, V., Anderluh, G., Podlesek, Z., et al. (2013). Structural insight into LexA-RecA∗ interaction. Nucleic Acids Res. 41, 9901–9910. doi: 10.1093/nar/gkt744
Kuzminov, A. (1999). Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63, 751–813. doi: 10.1128/mmbr.63.4.751-813.1999
Lawrence, J. G., and Retchless, A. C. (2009). The interplay of homologous recombination and horizontal gene transfer in bacterial speciation. Methods Mol Biol. 532, 29–53. doi: 10.1007/978-1-60327-853-9_3
Lee, C. D., and Wang, T. F. (2009). The N-terminal domain of Escherichia coli RecA have multiple functions in promoting homologous recombination. J. Biomed. Sci. 16:37. doi: 10.1186/1423-0127-16-37
Lusetti, S. L., and Cox, M. M. (2002). The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71, 71–100. doi: 10.1146/annurev.biochem.71.083101.133940
Lusetti, S. L., Wood, E. A., Fleming, C. D., Modica, M. J., Korth, J., Abbott, L., et al. (2003). C-terminal deletions of the Escherichia coli RecA protein. Characterization of in vivo and in vitro effects. J. Biol. Chem. 278:1637216380.
Maslowska, K. H., Makiela-Dzbenska, K., and Fijalkowska, I. J. (2019). The SOS system: a complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen. 60, 368–384. doi: 10.1002/em.22267
Nahrstedt, H., Schröder, C., and Meinhardt, F. (2005). Evidence for two recA genes mediating DNA repair in Bacillus megaterium. Microbiology 151(Pt 3), 775–787. doi: 10.1099/mic.0.27626-0
Norioka, N., Hsu, M. Y., Inouye, S., and Inouye, M. (1995). Two recA genes in Myxococcus xanthus. J. Bacteriol. 177, 4179–4182. doi: 10.1128/jb.177.14.4179-4182.1995
Nussbaumer, B., and Wohlleben, W. (1994). Identification, isolation and sequencing of the recA gene of Streptomyces lividans TK24. FEMS Microbiol. Lett. 118, 57–63. doi: 10.1016/0378-1097(94)90596-7
Orillard, E., Radicella, J. P., and Marsin, S. (2011). Biochemical and cellular characterization of Helicobacter pylori RecA, a protein with high-level constitutive expression. J. Bacteriol. 193, 6490–6497. doi: 10.1128/JB.05646-11
Quinet, A., Lemaçon, D., and Vindigni, A. (2017). Replication fork reversal: players and guardians. Mol. Cell 68, 830–833. doi: 10.1016/j.molcel.2017.11.022
Rastogi, R. P., Richa Kumar, A., Tyagi, M. B., and Sinha, R. P. (2010). Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids 2010:592980. doi: 10.4061/2010/592980
Richa, Sinha, R. P., and Häder, D. P. (2015). Physiological aspects of UV-excitation of DNA. Top Curr. Chem. 356, 203–248. doi: 10.1007/128_2014_531
Rolfe, M. D., Rice, C. J., Lucchini, S., Pin, C., Thompson, A., Cameron, A. D., et al. (2012). Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J. Bacteriol. 194, 686–701. doi: 10.1128/JB.06112-11
Saint-Ruf, C., Pesut, J., Sopta, M., and Matic, I. (2007). Causes and consequences of DNA repair activity modulation during stationary phase in Escherichia coli. Crit. Rev. Biochem. Mol. Biol. 42, 259–270. doi: 10.1080/10409230701495599
Sano, Y., and Kageyama, M. (1987). The sequence and function of the recA gene and its protein in Pseudomonas aeruginosa PAO. Mol. Gen. Genet. 208, 412–419. doi: 10.1007/bf00328132
Schefe, J. H., Lehmann, K. E., Buschmann, I. R., Unger, T., and Funke-Kaiser, H. (2006). Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. J. Mol. Med 84, 901–910. doi: 10.1007/s00109-006-0097-6
Sheng, D., Liu, R., Xu, Z., Singh, P., Shen, B., and Hua, Y. (2005). Dual negative regulatory mechanisms of RecX on RecA functions in radiation resistance, DNA recombination and consequent genome instability in Deinococcus radiodurans. DNA Repair. 4, 671–678. doi: 10.1016/j.dnarep.2005.02.006
Sinha, R. P., and Häder, D. P. (2002). UV-induced DNA damage and repair: a review. Photochem. Photobiol. Sci. 1, 225–236.
Story, R. M., Weber, I. T., and Steitz, T. A. (1992). The structure of the E. coli RecA protein monomer and polymer. Nature 355, 318–325. doi: 10.1038/355318a0
Ueki, T., Inouye, S., and Inouye, M. (1996). Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183, 153–157. doi: 10.1016/s0378-1119(96)00546-x
Umelo, E., Noonan, B., and Trust, T. J. (1996). Cloning, characterization and expression of the recA gene of Aeromonas salmonicida. Gene 175, 133–136. doi: 10.1016/0378-1119(96)00138-2
Keywords: RecA, duplicate genes, Myxococcus xanthus, DNA recombination, antioxidation, functional divergence, SOS response
Citation: Sheng D-H, Wang Y-X, Qiu M, Zhao J-Y, Yue X-J and Li Y-Z (2020) Functional Division Between the RecA1 and RecA2 Proteins in Myxococcus xanthus. Front. Microbiol. 11:140. doi: 10.3389/fmicb.2020.00140
Received: 16 October 2019; Accepted: 21 January 2020;
Published: 12 February 2020.
Edited by:
Torsten Waldminghaus, University of Marburg, GermanyReviewed by:
Juan Carlos Alonso, Centro Nacional de Biotecnología (CNB), SpainAndrei Kuzminov, University of Illinois at Urbana–Champaign, United States
Copyright © 2020 Sheng, Wang, Qiu, Zhao, Yue and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue-Zhong Li, bGlsYWJAc2R1LmVkdS5jbg==
†ORCID: Yue-Zhong Li, orcid.org/0000-0001-8336-6638
 Duo-Hong Sheng
Duo-Hong Sheng Yi-Xue Wang
Yi-Xue Wang Miao Qiu
Miao Qiu Jin-Yi Zhao
Jin-Yi Zhao Yue-Zhong Li
Yue-Zhong Li