
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 February 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00080
 Maria Borowiak
Maria Borowiak Beatrice Baumann
Beatrice Baumann Jennie Fischer
Jennie Fischer Katharina Thomas
Katharina Thomas Carlus Deneke
Carlus Deneke Jens Andre Hammerl
Jens Andre Hammerl Istvan Szabo
Istvan Szabo Burkhard Malorny*
Burkhard Malorny*The polymyxin antibiotic colistin has been used in decades for treatment and prevention of infectious diseases in livestock. Nowadays, it is even considered as last-line treatment option for severe human infections caused by multidrug- and carbapenem-resistant Gram-negative bacteria. Therefore, the discovery of plasmid-mediated mobile colistin resistance (mcr) genes raised major public health concern. The aim of our study was to analyze colistin-resistant Salmonella enterica strains from animals, food, feed and the environment collected at the National Reference Laboratory for Salmonella in Germany on the presence of mcr-1 to mcr-9 genes. Altogether 407 colistin-resistant (MIC >2 mg/L) Salmonella isolates received between 2011 and 2018 were selected and screened by PCR using a published mcr-1 to mcr-5 as well as a newly developed mcr-6 to mcr-9 multiplex PCR protocol. 254 of 407 (62.4%) isolates harbored either mcr-1 (n = 175), mcr-4 (n = 53), mcr-5 (n = 18) or mcr-1 and mcr-9 (n = 8). The number of mcr-positive isolates ranged from 19 (2017) to 64 (2012) per year. WGS revealed that none of our isolates harbored the mcr-9.1 gene. Instead, two novel mcr-9 variants were observed, which both were affected by frameshift mutations and are probably non-functional. The mcr-harboring isolates were mainly derived from animals (77.2%) or food (20.1%) and could be assigned to ten different Salmonella serovars. Many of the isolates were multidrug-resistant. Co-occurrence of mcr-1 and AmpC or ESBL genes was observed in eight isolates. Our findings suggest that mcr genes are widely spread among colistin-resistant Salmonella isolates from livestock and food in Germany. Potential transfer of mcr-harboring isolates along the food chain has to be considered critically.
Non-typhoidal Salmonella (S.) enterica subsp. enterica are zoonotic pathogens which can cause severe human infections (salmonellosis) through the consumption of contaminated food. Spread of antibiotic resistance in S. enterica might impair successful antibiotic treatment of salmonellosis in patients where it is indispensable (Michael and Schwarz, 2016).
The polypeptide antibiotic colistin belongs to the group of polymyxins and has been used for decades in the animal production for treatment and prevention of infectious diseases (EMA, 2016). Comparing 30 European countries, Germany had the sixth highest polymyxin sale for food-producing species in 2016 (EMA and ESVAC, 2017). Especially colistin is frequently used in pig, cattle, sheep and poultry production, mainly for the treatment of gut infections caused by S. enterica and Escherichia (E.) coli (Emmerich and Drees, 2016). In human medicine, colistin played a minor role for long time, due to high side effects and the availability of better alternatives for antimicrobial treatment (Yu et al., 2015). However, in the last years, colistin was rediscovered for last-line treatment of severe human infections caused by multidrug- and carbapenem-resistant Gram-negative pathogens and is now considered as a critically important antimicrobial for human medicine (Falagas et al., 2005; Grégoire et al., 2017; World Health Organization [WHO], 2017). Therefore, the first discovery of a plasmid-encoded mobile colistin resistance gene (mcr-1) in 2015 in E. coli isolated from livestock animals, meat and patients as well as in Klebsiella pneumoniae from clinical settings in China raised major public health concern (Liu et al., 2016; Sun et al., 2018). Extensive follow-up screenings identified Moraxella spp. as potential origin of mcr-1 (Kieffer et al., 2017) and revealed that this gene can be found in various pathogenic and commensal Enterobacteriaceae from the environment, wildlife, livestock, food and patients worldwide (Sun et al., 2018; Lima et al., 2019). Due to the high prevalence, it is thought that food-producing animals are most likely the origin for mcr-1 mediated colistin resistance (Nordmann and Poirel, 2016; Poirel and Nordmann, 2016). These findings and the reclassification of colistin in human medicine led to a reassessment of colistin by the European Medicines Agency (EMA) suggesting to reduce the use of colistin in livestock production as much as possible (EMA, 2016).
After the detection of mcr-1, seven additional mcr genes (mcr-2 to mcr-8) were described (Xavier et al., 2016; Abuoun et al., 2017; Borowiak et al., 2017a; Carattoli et al., 2017; Yin et al., 2017; Wang et al., 2018; Yang et al., 2018). Recently, the inducible mcr-9 gene was added to the increasing number of mcr genes (Carroll et al., 2019). These results further enhanced public health concerns related to the emergence, persistence and spread of mobile colistin resistance and reinforced the call for ongoing extensive screenings in Enterobacteriaceae.
In this study we developed a gel-based multiplex PCR protocol which detects mcr-6, mcr-7, mcr-8, and mcr-9 for rapid screening purposes of bacterial isolates. We applied the novel mcr-6 to mcr-9 assay in combination with a previously published multiplex PCR for detection of mcr-1 to mcr-5 (Rebelo et al., 2018) on 407 colistin-resistant S. enterica isolates from the strain collection of the German National Reference Laboratory for Salmonella. These isolates were collected between 2011 and 2018 in the frame of routine diagnostics and national surveillance and monitoring programs and originated from livestock, animals, food, feed and the environment in Germany.
PCR screening was performed on 407 initially phenotypically colistin-resistant (MIC >2 mg/L) S. enterica isolates (with exception of serogroup D) obtained from livestock, animals, food, feed and the environment, collected in the frame of routine diagnostics as well as national monitoring and surveillance programs in the years 2011 to 2018 in Germany at the National Reference Laboratory for the Analysis and Testing of Zoonoses (NRL Salmonella). All strains selected have no epidemiological link (neither isolated at the same place, time, animal or food). All isolates were routinely subjected to serotyping by slide agglutination with antisera raised against O-, H1- and H2- antigens and MIC testing as described before (Borowiak et al., 2017a). For PCR screening, isolates were cultivated from stock cultures by inoculating liquid LB medium and subsequent incubation under shaking conditions (250 × rpm) for 16 h at 37°C. Thermal cell lysis preparations were produced as previously described (Borowiak et al., 2017a). DNA suitability for PCR amplification was confirmed by a standard PCR protocol for amplification of the housekeeping gene hemD using the primers recommended on Enterobase1. The hemD PCR was prepared in 25 μL including 12.5 μL 2x DreamTaq Green PCR Master Mix (Thermo Scientific, Vilnius, Lithuania), 0.5 μL of each 10 μM primer dilution, 9.5 μL PCR grade water and 2 μL of thermal cell lysis preparation as template DNA and carried out as follows: initial denaturation for 3 min at 95°C, 35 cycles denaturation for 30 s at 95°C, primer annealing for 30 s at 55°C and elongation for 30 s at 72°C followed by a final elongation step for 10 min at 72°C.
Positive controls for mcr-1, mcr-4, mcr-5, and mcr-9 were obtained using in-house strains S. Paratyphi B dT+ 08-00436 (mcr-1.1; BioSample: SAMN06645218), monophasic S. Typhimurium 15-SA02945 (mcr-4.2; BioSample: SAMEA104398406), S. Paratyphi B dT+ 13-SA01718 (mcr-5.1; BioSample: SAMN07420414) and S. Infantis 15-SA01028 (mcr-9.1; BioSample: SAMN08475067). Control strains for mcr-2 and mcr-3 were kindly provided by the National Food Institute, Technical University of Denmark (Copenhagen).
For mcr-6 to mcr-8 positive controls, DNA was synthesized (Integrated DNA Technologies, Leuven, Belgium) and subjected to PCR and cloning. Following gene regions were selected for DNA synthesis: mcr-6.1 (GenBank: NG_055781.1, bases 557-896), mcr-7.1 (GenBank: NG_056413.1:101-1720 bases 190-780), and mcr-8.1 (GenBank: NG_061399.1:101-1798, bases 630-1566).
All primer used in this study were synthesized by Eurogentec (Seraing, Belgium). PCR reactions for cloning were prepared in 50 μL volumes comprised of 2 μL synthetic DNA (final amount: 0.1 ng), 5 μL of 10× PCR Rxn Buffer, 1.5 μL of 50 mM MgCl2, 5 μL of 200 μM dNTP Mix (prepared from 100 mM dNTP solutions), 0.4 μL of Platinum® Taq DNA Polymerase (Invitrogen, Carlsbad, CA, United States), 5 μL of the respective 10 μM primer dilutions, and 26.1 μL PCR grade water. The primer used in the respective cloning PCR reactions were the same as used in the multiplex PCR protocol (Table 1). PCR was carried out in following steps: initial denaturation for 5 min at 95°C (all), 25 cycles denaturation for 30 s at 95°C (all), primer annealing for 30 s at 52°C (mcr-6), 50°C (mcr-7) or 53°C (mcr-8), elongation at 72°C for 20 s (mcr-6), 35 s (mcr-7) or 55 s (mcr-8) followed by a final elongation step for 5 min at 72°C (all). Freshly prepared PCR products were cloned into pCRTM 2.1-TOPO® vectors and transferred in One Shot® chemically competent E. coli TOP10F’ cells from the TOPO® TA Cloning® Kit (Invitrogen) following manufactures instructions. Positive transformants were selected on LB agar supplemented with 100 mg/L ampicillin (Sigma, Darmstadt, Germany) and confirmed by PCR. Plasmid DNA of transformants TOP10F’ + pCR2.1-mcr-6, TOP10F’ + pCR2.1-mcr-7, and TOP10F’ + pCR2.1-mcr-8 was extracted using the Invisorb® Spin Plasmid Mini Two Kit (Stratec, Berlin, Germany) and the insert sequence was confirmed by Sanger sequencing.
PCR screening for mcr-1 to mcr-5 was carried out using the multiplex PCR protocol published by Rebelo et al. (2018) with the deviation that the CLR5-F and CLR5-R primers (Liu et al., 2016) were used for amplification of mcr-1 resulting in a PCR product size of 309 bp.
Four primer pairs, one for each mcr-6 to mcr-9 gene, were designed in CLC Genomics Workbench 9.5.2 (Qiagen, Hilden, Germany) and subsequently adapted manually. The primer pairs were developed to amplify the four target genes, with a size ranging from 252 to 1011 bp. This allows the separation and visualization of PCR products on agarose gels. The mcr-6 to mcr-9 multiplex PCR was carried out in 25 μL PCR reactions comprised of 12.5 μL 2x DreamTaq Green PCR Master Mix, 0.5 μL of each 10 μM primer dilution (Table 1), 6.5 μL PCR grade water and 2 μL of thermal cell lysis preparation as template DNA. The PCR conditions were optimized as follows: initial denaturation for 3 min at 95°C, 30 cycles denaturation for 30 s at 95°C, primer annealing for 30 s at 55°C and elongation for 60 s at 72°C followed by a final elongation step for 10 min at 72°C. PCR products were electrophoretically separated on 1.5% agarose gels (Bioline, London, United Kingdom). The novel mcr-6 to mcr-9 multiplex protocol, which is in silico compatible with all known mcr-6 to mcr-9 variants (mcr-6.1, mcr-7.1, mcr-8.1, mcr-8.2, and mcr-9.1), was initially tested on different mcr-positive as well as mcr-negative control strains previously identified by whole-genome sequencing (Figure 1).
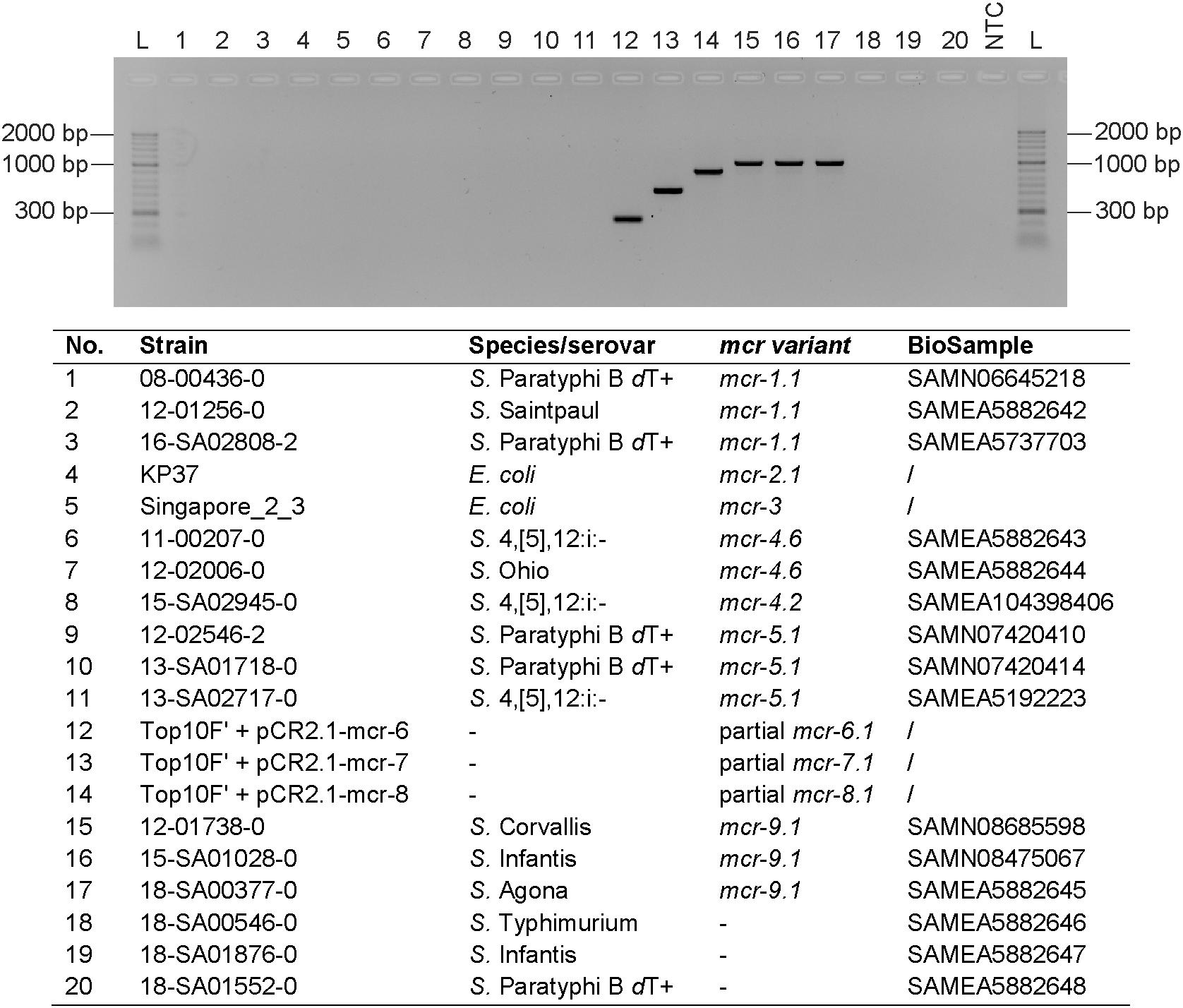
Figure 1. mcr-6 to mcr-9 multiplex PCR. The PCR was tested on DNA extractions of selected strains. PCR grade water was used as negative control (NTC) and HyperLadderII (Bioline) was used as molecular weight marker (L). The PCR products were electrophoretically separated on a 1.5% agarose gel (Bioline) supplemented with Midori Green (NIPPON Genetics Europe, Düren, Germany).
Non-serotypeable mcr-positive isolates, isolates showing phenotypic resistance to 3rd generation cephalosporins or isolates harboring the recently detected mcr-9 gene were subjected to WGS. DNA was extracted as previously described (Borowiak et al., 2017a), sequencing libraries were either prepared using the Nextera XT or the Nextera DNA Flex Kit (Illumina, San Diego, CA, United States) and sequencing was performed on a MiSeq or NextSeq benchtop sequencer (Illumina). Raw reads were trimmed using fastp v0.19.5 (Chen et al., 2018) and de novo assembled using unicycler v0.4.4 (Wick et al., 2017). Subsequently, the serovar was predicted from assembled sequencing data using SISTR (Yoshida et al., 2016). MLST2.0, ResFinder3.2 and PlasmidFinder2.1 provided by the Center of Genomic Epidemiology at DTU2 were further used to characterize the isolates. Whole-genome SNP analysis and clustering of isolates were performed as previously described (Borowiak et al., 2017a). As references, NCBI reference sequences NZ_CP026976.1 (S. Heidelberg) and NZ_CP019649 (S. Typhimurium) were used.
Since mcr-6, mcr-7, and mcr-8 isolates were, to our knowledge, not yet detected in Europe and the access of isolates was not possible, we cloned fragments of the particular genes into pCRTM 2.1-TOPO® vectors and transformed them into E. coli TOP10F’ cells. Thermal cell lysis preparations were subsequently used as positive controls. For mcr-9, three mcr-9.1 positive isolates previously identified by routine WGS analysis were selected as control strains. For each mcr-6 to mcr-9 control the multiplex protocol amplified a PCR product forming a distinct band with the expected size after agarose gel electrophoresis. Primer pairs of the particular mcr genes did not interfere with any other mcr gene present in selected mcr-1- to mcr-5-positive strains (Figure 1).
For PCR screening of colistin-resistant Salmonella isolates the mcr-1 to mcr-5 multiplex protocol published by Rebelo et al. (2018) as well as a newly developed multiplex PCR protocol for mcr-6 to mcr-9 was used. Results revealed that 254 out of 407 tested Salmonella isolates carried either mcr-1 (175 isolates), mcr-4 (53 isolates), mcr-5 (18 isolates) or mcr-1 and mcr-9 (8 isolates) (Table 2). Other mcr genes (mcr-2, mcr-3, mcr-6, mcr-7, and mcr-8) were not detected. The number of mcr-positive isolates ranged from 19 to 33 isolates per year with the exception of 2012 (detection of 64 mcr-positive isolates). The mcr-1- and mcr-4-positive isolates were observed in every year, while mcr-5-harboring isolates were only detected in 2011 to 2013 and 2016, and mcr-1- and mcr-9-positive isolates only in 2013 to 2015 (Supplementary Table S1). The mcr-positive Salmonella isolates belonged to various serovars and were derived from different isolation sources. Most mcr-harboring Salmonella isolates were associated with livestock (77.2%, mainly pig and poultry). However, 20.1% of the isolates were derived from food products (including beef, pork, poultry meat as well as minced meat and meat of unknown sources) and 1.2% were isolated from pet animals. The most abundant mcr-harboring serovars were S. Typhimurium (biphasic and monophasic variants) isolated from pig and pork (n = 159) followed by d-tartrate fermenting (dT+) S. Paratyphi B (formerly S. Java) isolated from poultry and poultry meat (n = 51) (Table 2). The mcr-1 gene was found in eight different Salmonella serovars and was mainly associated with pig/pork and poultry/poultry meat. However, the gene was also identified in cattle/beef. The mcr-4 gene was found in monophasic and biphasic S. Typhimurium, S. Rissen, and S. Ohio and was primarily associated with pigs. One mcr-4-harboring isolate was recovered from a quail. The mcr-5 gene was identified in S. Paratyphi B dT+ and monophasic and biphasic S. Typhimurium either associated with poultry/poultry meat or pig/pork. Similarly, mcr-9 was identified in monophasic S. Typhimurium from pig/pork or in S. Paratyphi B dT+ from poultry/poultry meat (Table 2 and Supplementary Table S1).
Altogether 98.8% of the mcr-harboring isolates detected in the study were resistant to three or more classes of antibiotics. Phenotypical resistance to ampicillin (96.1%), sulfamethoxazole (90.9%), streptomycin (89.7%, considering only isolates tested between 2011 and 2013 as streptomycin was removed from testing plates in 2014), tetracycline (81.5%), trimethoprim (63.4%), ciprofloxacin (34.3%), nalidixic acid (29.9%), chloramphenicol (26.3%), kanamycin (13.8%), gentamicin (14.2%), and cefotaxime/ceftazidime (3.1%) was observed (Supplementary Table S1).
By slide agglutination non-serotypeable mcr-positive isolates (n = 10), isolates showing phenotypic resistance to 3rd generation cephalosporins (n = 8) or isolates harboring the newly detected mcr-9 gene (n = 8) were subjected to WGS. Based on sequencing data, the serovar of all ten previously untypeable isolates could be inferred (Supplementary Tables S1, S2). Moreover, WGS data for the eight 3rd generation cephalosporin-resistant isolates revealed that two mcr-1-positive S. Heidelberg strains, isolated in 2017 and 2018 from poultry meat, carried the AmpC gene blaCMY–2 and six mcr-1-positive monophasic S. Typhimurium strains, isolated between 2013 and 2015 from pig and pork, harbored the ESBL gene blaSHV–12. The genetic relatedness of ESBL- or AmpC-producing isolates was further investigated using a SNP-based approach. Results revealed that the six monophasic S. Typhimurium were closely (0-11 SNPs) and the two S. Heidelberg isolates were distantly (46 SNP) related to each other.
The blaSHV–12 and mcr-1-harboring monophasic S. Typhimurium were also positive for the mcr-9 gene in PCR. WGS data analysis revealed that these isolates harbored a mcr-9 frameshift variant (cytosine deletion at position 1092, e.g., BioSample: SAMEA104398378). In two S. Paratyphi B dT+ isolates also tested positive for mcr-1 and mcr-9, another mcr-9 frameshift mutation (guanidine deletion at position 19, e.g., BioSample: SAMEA104396051) was observed (Supplementary Table S1).
We identified 254 mcr-harboring isolates in altogether 407 colistin-resistant S. enterica isolates obtained from livestock, food and pet animals collected in the frame of routine diagnostics as well as national monitoring and surveillance programs in the years 2011 to 2018 in Germany using a newly developed multiplex PCR assay detecting mcr-6 to mcr-9 genes in combination with a previously published mcr-1 to mcr-5 multiplex PCR assay (Rebelo et al., 2018). From this study, we conclude that in Germany, mcr-1 seems to be the most common mcr gene found in colistin-resistant S. enterica, followed by mcr-4 and mcr-5. These mcr genes have successfully emerged in at least ten Salmonella serovars. However, monophasic and biphasic S. Typhimurium from the pig production were the most prevalent mcr-harboring isolates, followed by S. Paratyphi B dT+ derived from poultry. This is in concordance with global observations reporting mono- and biphasic S. Typhimurium as the most prevalent mcr-positive serovars and poultry and swine as the most abundant sources of mcr-positive Salmonella isolates (Lima et al., 2019). Also other mcr-harboring Salmonella serovars including Heidelberg, Infantis, Newport, Paratyphi B dT+ (Java), Rissen, Saintpaul and Schwarzengrund have been previously reported. Most of them were described to harbor mcr-1 (Lima et al., 2019). However, distribution of other mcr genes in Salmonella might be underestimated to this timepoint, as extensive screenings of strain collections for novel mcr genes are still ongoing. In our study, mcr-4- and mcr-5-harboring isolates were frequently identified, whereas mcr-2, mcr-3, mcr-6, mcr-7, and mcr-8 could not be detected. The reason might be that those genes are restricted in their distribution in terms of bacterial host species or region.
In our collection, the mcr-9 gene was found in colistin-resistant isolates also positive for mcr-1. WGS analysis of the respective isolates revealed that two different frameshift mutations of mcr-9 occurred, leading probably to non-functional MCR-9 proteins. The ability of MCR-9 to cause colistin resistance was only investigated in few studies (Carroll et al., 2019; Kieffer et al., 2019). The mcr-9 gene was initially identified in a colistin-susceptible S. Typhimurium. When mcr-9 was expressed under an inducible promotor in E. coli NEB5α, a reduced colistin-susceptibility (MIC > 2.5 mg/L) was observed (Carroll et al., 2019). Results of a second study suggested that mcr-9 is inducible by sub-inhibitory concentrations of colistin. Inducibility was related to a two-component regulatory system located downstream of the mcr-9 gene (Kieffer et al., 2019). Altogether, the gene seems to be widely distributed. According to the NCBI Pathogen Detection Browser3, mcr-9 is present in various Enterobacteriaceae species including Klebsiella spp., E. coli, Enterobacter spp. and more than 40 different Salmonella serovars from human and animal origin collected worldwide. Screening in our NRL Salmonella WGS database for additional mcr-9 harboring isolates revealed the presence of mcr-9.1 in eight isolates belonging to four Salmonella serovars (Supplementary Table S3). All these isolates were initially tested to be colistin-susceptible (MIC ≤2 mg/L) and therefore not considered for the PCR screening. Among the mcr-9.1-positive isolates are the carbapenem-resistant VIM-1-producing S. Infantis strains 15-SA01028 (BioSample: SAMN08475067) isolated from a minced meat sample and the NDM-1-producing S. Corvallis 12-01738 (BioSample: SAMN08685598) derived from a wild bird previously sequenced using long-read sequencing (Fischer et al., 2013; Borowiak et al., 2017b, 2018; Hadziabdic et al., 2018). Both isolates harbored the mcr-9.1 gene on IncHI2/IncHI2A plasmids named pSE15-SA01028 and pSE12-01738-1, respectively. The two-component regulatory system associated with mcr-9.1 inducibility is present in pSE15-SA01028 but missing in pSE12-01738-1. In our PCR study, only S. enterica isolates with Minimum Inhibitory Concentration (MIC) values for colistin >2 mg/L were subjected to PCR screening. However, recent studies showed that colistin MIC values determined by the recommended broth microdilution method might vary and MIC values of ≤2 mg/L can be observed, not only for mcr-9, but also for other mcr-harboring isolates (Borowiak et al., 2019; Zając et al., 2019). Thus, additional strains from the collection of the NRL Salmonella exposing MIC values of ≤2 mg/L might encode mcr genes.
On the other hand, 153 of the 407 (37.6%) initially colistin-resistant Salmonella isolates were not positive for known mcr genes. These strains could harbor chromosomal point mutations or unknown plasmid-mediated colistin resistance determinants. However, in some of the isolates, naturally decreased colistin susceptibility and fluctuations in MIC measurement might be responsible for increased MIC values (>2 mg/L).
The occurrence of mcr genes in monophasic and biphasic S. Typhimurium, S. Infantis and S. Newport have to be considered critically, as these serovars belong to top five serovars leading to human salmonellosis in Europe (EFSA, 2018). mcr-harboring Salmonella isolates can spread to humans through the consumption of undercooked food products derived from livestock. Especially worrisome is the high number of isolates resistant to three or more classes of antibiotics, which are considered as multidrug-resistant (Magiorakos et al., 2012) and the detection of ESBL- and AmpC-producing isolates among mcr-positive Salmonella strains. ESBL-producing and mcr-harboring Salmonella isolates from the animal production were already described before (Wang et al., 2017; Carfora et al., 2018). Further spread of these isolates or resistance traits limits antimicrobial treatment options for infections in man and animal and represents a major public health concern.
For monitoring the spread of all known mcr genes in the food chain, the available mcr-1 to mcr-5 multiplex PCR was complemented with a novel mcr-6 to mcr-9 multiplex PCR assay. The protocol presented here can easily be extended to further mcr genes which might be detected in the future. Extensive PCR screening using a previously described and the newly developed mcr multiplex PCR protocol revealed that certain mcr genes are widely distributed among colistin-resistant Salmonella isolates from livestock and food in Germany. We could detect mcr-1, mcr-4 and mcr-5 genes as well as mcr-9 frameshift variants, which have emerged in various Salmonella serovars derived from different livestock animals, production lines and food products. The high rate of multidrug resistance as well as the co-occurrence of ESBL and AmpC genes with mcr-1 might threaten public health and have to be considered critically.
Raw sequence data can be found in the NCBI database under following BioSample accessions: SAMEA104396046, SAMEA104396051, SAMEA104396062, SAMEA104398375, SAMEA104398377, SAMEA104398378, SAMEA5192222, SAMEA5192223, SAMEA5737695, SAMEA5737696, SAMEA5737697, SAMEA5737699, SAMEA5737700, SAMEA5737701, SAMEA5737702, SAMEA5737703, SAMEA5737704, SAMEA5737705, SAMEA5737706, and SAMEA5757422.
MB and BM designed the study. JF and IS provided the samples and performed pre-analysis. MB, BB, KT, and JH performed the experiments. MB interpreted the results and wrote the draft manuscript. CD supported the bioinformatics analysis. JF, CD, JH, and BM were involved in manuscript revision prior submission of the manuscript.
This work was supported by the German Federal Institute for Risk Assessment (BfR 1322-715). Some of the strains were sequenced in the frame of the project “Establishing Next Generation sequencing Ability for Genomic analysis in Europe” (ENGAGE) funded by the European Food Safety Authority (EFSA, GP/EFSA/AFSCO/2015/01/CT1).
The conclusion, findings, and opinions expressed in this scientific paper reflect only the view of the authors and not the official position of the European Food Safety Authority.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Rene Hendriksen from the National Food Institute, Technical University of Denmark (Copenhagen), for providing mcr-2 and mcr-3 control strains.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00080/full#supplementary-material
Abuoun, M., Stubberfield, E. J., Duggett, N. A., Kirchner, M., Dormer, L., Nunez-Garcia, J., et al. (2017). mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015.. J. Antimicrob. Chemother. 72, 2745–2749. doi: 10.1093/jac/dkx286
Borowiak, M., Fischer, J., Baumann, B., Hammerl, J. A., Szabo, I., and Malorny, B. (2018). Complete genome sequence of a VIM-1-producing Salmonella enterica subsp. enterica serovar Infantis isolate derived from minced pork meat. Genome Announc. 6:e0327-18. doi: 10.1128/genomeA.00327-18
Borowiak, M., Fischer, J., Hammerl, J. A., Hendriksen, R. S., Szabo, I., and Malorny, B. (2017a). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 72, 3317–3324. doi: 10.1093/jac/dkx327
Borowiak, M., Hammerl, J. A., Deneke, C., Fischer, J., Szabo, I., and Malorny, B. (2019). Characterization of mcr-5-harboring Salmonella enterica subsp. enterica serovar Typhimurium isolates from animal and food origin in Germany. Antimicrob. Agents Chemother. 63:e063-19. doi: 10.1128/AAC.00063-19
Borowiak, M., Szabo, I., Baumann, B., Junker, E., Hammerl, J. A., Kaesbohrer, A., et al. (2017b). VIM-1-producing Salmonella Infantis isolated from swine and minced pork meat in Germany. J. Antimicrob. Chemother. 72, 2131–2133. doi: 10.1093/jac/dkx101
Carattoli, A., Villa, L., Feudi, C., Curcio, L., Orsini, S., Luppi, A., et al. (2017). Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro. Surveill. 22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589
Carfora, V., Alba, P., Leekitcharoenphon, P., Ballarò, D., Cordaro, G., Di Matteo, P., et al. (2018). Colistin resistance mediated by mcr-1 in ESBL-producing, multidrug resistant Salmonella Infantis in broiler chicken industry, Italy (2016–2017). Front. Microbiol. 9:1880. doi: 10.3389/fmicb.2018.01880
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:539361. doi: 10.1128/mBio.00853-19
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
EFSA (2018). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 16:5500.
EMA (2016). Updated Advice on the Use of Colistin Products in Animals Within the European Union: Development of Resistance and Possible Impact on Human and Animal Health. EMA/CVMP/CHMP/231573/2016. Amsterdam: EMA.
EMA and ESVAC (2017). Sales of Veterinary Antimicrobial Agents in 30 European Countries in 2015. Trends from 2010 to 2015. Seventh ESVAC report. EMA/184855/2017. London: EMA.
Emmerich, I. U., and Drees, M. (2016). Neubewertung von Colistin durch übertragbares Resistenzgen - ein Antibiotikum im Fokus. Deutsch. Tierärzteblatt 11, 1644–1648.
Falagas, M. E., Kasiakou, S. K., and Saravolatz, L. D. (2005). Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40, 1333–1341. doi: 10.1086/429323
Fischer, J., Schmoger, S., Jahn, S., Helmuth, R., and Guerra, B. (2013). NDM-1 carbapenemase-producing Salmonella enterica subsp. enterica serovar Corvallis isolated from a wild bird in Germany. J. Antimicrob. Chemother. 68, 2954–2956. doi: 10.1093/jac/dkt260
Grégoire, N., Aranzana-Climent, V., Magréault, S., Marchand, S., and Couet, W. (2017). Clinical pharmacokinetics and pharmacodynamics of colistin. Clin. Pharmacokinet. 56, 1441–1460. doi: 10.1007/s40262-017-0561-1
Hadziabdic, S., Borowiak, M., Bloch, A., Malorny, B., Szabo, I., Guerra, B., et al. (2018). Complete genome sequence of an avian native NDM-1-producing Salmonella enterica subsp. enterica serovar Corvallis strain. Genome Announc. 6:e00593-18. doi: 10.1128/genomeA.00593-18
Kieffer, N., Nordmann, P., and Poirel, L. (2017). Moraxella species as potential sources of MCR-like polymyxin resistance determinants. Antimicrob. Agents Chemother. 61:e00129-17. doi: 10.1128/AAC.00129-17
Kieffer, N., Royer, G., Decousser, J. W., Bourrel, A. S., Palmieri, M., Ortiz De La Rosa, J. M., et al. (2019). mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob. Agents Chemother. 63:e0965-19. doi: 10.1128/AAC.00965-19
Lima, T., Domingues, S., and Da Silva, G. J. (2019). Plasmid-mediated colistin resistance in Salmonella enterica: a review. Microorganisms 7:E55. doi: 10.3390/microorganisms7020055
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Michael, G. B., and Schwarz, S. (2016). Antimicrobial resistance in zoonotic nontyphoidal Salmonella: an alarming trend? Clin. Microbiol. Infect. 22, 968–974. doi: 10.1016/j.cmi.2016.07.033
Nordmann, P., and Poirel, L. (2016). Plasmid-mediated colistin resistance: an additional antibiotic resistance menace. Clin. Microbiol. Infect. 22, 398–400. doi: 10.1016/j.cmi.2016.03.009
Poirel, L., and Nordmann, P. (2016). Emerging plasmid-encoded colistin resistance: the animal world as the culprit? J. Antimicrob. Chemother. 71, 2326–2327. doi: 10.1093/jac/dkw074
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro. Surveill. 23:672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
Sun, J., Zhang, H., Liu, Y. H., and Feng, Y. (2018). Towards understanding MCR-like colistin resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006
Wang, J., Li, X., Li, J., Hurley, D., Bai, X., Yu, Z., et al. (2017). Complete genetic analysis of a Salmonella enterica serovar Indiana isolate accompanying four plasmids carrying mcr-1, ESBL and other resistance genes in China. Vet. Microbiol. 210, 142–146. doi: 10.1016/j.vetmic.2017.08.024
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7:122. doi: 10.1038/s41426-018-0124-z
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
World Health Organization [WHO] (2017). Critically Important Antimicrobials For Human Medicine – 5th rev. Licence: CC BY-NC-SA 3.0 IGO. Geneva: World Health Organization.
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro. Surveill. 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Yang, Y.-Q., Li, Y.-X., Lei, C.-W., Zhang, A.-Y., and Wang, H.-N. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1791–1795. doi: 10.1093/jac/dky111
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17
Yoshida, C. E., Kruczkiewicz, P., Laing, C. R., Lingohr, E. J., Gannon, V. P., Nash, J. H., et al. (2016). The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101
Yu, Z., Qin, W., Lin, J., Fang, S., and Qiu, J. (2015). Antibacterial mechanisms of polymyxin and bacterial resistance. Biomed. Res. Int. 2015:11.
Keywords: Salmonella, colistin resistance, mcr genes, multiplex PCR, multidrug resistance
Citation: Borowiak M, Baumann B, Fischer J, Thomas K, Deneke C, Hammerl JA, Szabo I and Malorny B (2020) Development of a Novel mcr-6 to mcr-9 Multiplex PCR and Assessment of mcr-1 to mcr-9 Occurrence in Colistin-Resistant Salmonella enterica Isolates From Environment, Feed, Animals and Food (2011–2018) in Germany. Front. Microbiol. 11:80. doi: 10.3389/fmicb.2020.00080
Received: 10 September 2019; Accepted: 15 January 2020;
Published: 04 February 2020.
Edited by:
Daniela Ceccarelli, Research Executive Agency, European Commission, BelgiumReviewed by:
Benoit Doublet, Institut National de la Recherche Agronomique (INRA), FranceCopyright © 2020 Borowiak, Baumann, Fischer, Thomas, Deneke, Hammerl, Szabo and Malorny. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Burkhard Malorny, QnVya2hhcmQuTWFsb3JueUBiZnIuYnVuZC5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.