- 1Department of Immunology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
- 2Department of Biosciences, University of Exeter, Exeter, United Kingdom
- 3Siriraj Research Group in Immunobiology and Therapeutic Sciences, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
- 4Laboratory of Biotechnology, Chulabhorn Research Institute, Bangkok, Thailand
- 5The Roslin Institute, The Royal (Dick) School of Veterinary Studies, University of Edinburgh, Edinburgh, United Kingdom
- 6Defence Medical and Environmental Research Institute, DSO National Laboratories, Singapore, Singapore
Burkholderia pseudomallei, the causative agent of melioidosis, can survive and replicate in macrophages. Little is known about B. pseudomallei genes that are induced during macrophage infection. We constructed a B. pseudomallei K96243 promoter trap library with genomic DNA fragments fused to the 5′ end of a plasmid-borne gene encoding enhanced green fluorescent protein (eGFP). Microarray analysis showed that the library spanned 88% of the B. pseudomallei genome. The recombinant plasmids were introduced into Burkholderia thailandensis E264, and promoter fusions active during in vitro culture were removed. J774A.1 murine macrophages were infected with the promoter trap library, and J774A.1 cells containing fluorescent bacteria carrying plasmids with active promoters were isolated using flow cytometric-based cell sorting. Candidate macrophage-induced B. pseudomallei genes were identified from the location of the insertions containing an active promoter activity. A proportion of the 138 genes identified in this way have been previously reported to be involved in metabolism and transport, virulence, or adaptation. Novel macrophage-induced B. pseudomallei genes were also identified. Quantitative reverse-transcription PCR analysis of 13 selected genes confirmed gene induction during macrophage infection. Deletion mutants of two macrophage-induced genes from this study were attenuated in Galleria mellonella larvae, suggesting roles in virulence. B. pseudomallei genes activated during macrophage infection may contribute to intracellular life and pathogenesis and merit further investigation toward control strategies for melioidosis.
Introduction
Burkholderia pseudomallei is a saprophytic bacterial pathogen that causes melioidosis, primarily in Southeast Asia and Northern Australia (Wuthiekanun et al., 2005; Wiersinga et al., 2006). However, the global distribution of melioidosis is now believed to be much broader, and it is predicted that there are 45 countries where the disease is underreported and 34 countries where the disease is likely to be present (Limmathurotsakul et al., 2016). Humans often acquire melioidosis through wounds or by the inhalation of contaminated dust or water droplets (Wiersinga et al., 2006). Melioidosis in humans can appear as a rapidly fatal septicemia, acute pneumonia, or subacute disease. B. pseudomallei is resistant to many antibiotics, making it difficult to treat, and currently, there is no vaccine to protect against melioidosis (Dance, 2000). In the United States, B. pseudomallei is a Tier 1 select agent, owing to its potential to cause a mass casualty event after a deliberate release (Wagar, 2016). Additionally, B. pseudomallei is listed in Schedule five pathogens and toxins controlled under the Anti-Terrorism, Crime and Security Act (ATCSA) in the United Kingdom.
Burkholderia pseudomallei deploys a variety of virulence factors during its interactions with host cells, and these contribute to immune evasion and pathogenesis (Wiersinga et al., 2006; Galyov et al., 2010). After the bacteria are taken up by phagocytic cells, a Type III secretion system (T3SS-3) mediates the escape of the bacteria from the phagosome (Stevens et al., 2002; Gong et al., 2011). Once they are free in the cytoplasm, actin-based motility allows the bacteria to spread intracellularly and intercellularly (Stevens M.P. et al., 2005) and a Type VI secretion system (T6SS-5) promotes cell fusion, enabling B. pseudomallei to form multinucleated giant cells (MNGCs) and spread from cell to cell to evade immune surveillance (Burtnick et al., 2011; Schwarz et al., 2014).
To gain insight into how B. pseudomallei survives and to establish the infection in host cells, a range of techniques have been used. A comprehensive DNA microarray has been used to investigate the transcription profile of B. pseudomallei within human U937 macrophages (Chieng et al., 2012). RNA sequencing has been also applied to study the transcriptome of B. pseudomallei in the RAW264.7 murine macrophage cell line and during acute respiratory infection of inbred mice (Ooi et al., 2013). Transposon mutagenesis has also been used to identify genes required for virulence and the intracellular lifestyle of B. pseudomallei (Cuccui et al., 2007; Moule et al., 2015).
In vivo expression technology (IVET) has also been used to study bacterial gene expression (Valdivia and Falkow, 1997). IVET is a promoter-trapping technique that reveals bacterial promoters which are active during the interaction with the host by screening for expression of a reporter gene. Previously, an IVET screen using a chloramphenicol resistance gene as the reporter has been employed to identify B. pseudomallei promoters activated in RAW264.7 cells. This revealed that a promoter driving the T6SS-5 gene cluster was induced following uptake by macrophages (Shalom et al., 2007). However, when using antibiotic resistance genes as reporters, weakly expressed promoters may not be identified by this methodology because they fail to activate sufficient gene expression to confer phenotypic resistance. This drawback is circumvented by using differential fluorescence induction (DFI), in which fluorescence-activated cell sorting (FACS) is used to isolate host cells harboring fluorescent bacteria that carry active promoter fusions to a fluorescent reporter protein (Bumann and Valdivia, 2007). This technique allows high-throughput screening for positive clones that can be isolated directly from infected cells, or animal tissues, under various conditions.
In this study, we constructed a promoter library of B. pseudomallei using a plasmid containing a promoterless gene encoding enhanced green fluorescent protein (eGFP) and then introduced this into the closely related bacterium Burkholderia thailandensis to allow screening of infected macrophages by FACS under biosafety Level 2 conditions. The library was depleted of fusions that were active during growth on laboratory medium. After selection of clones activated during infection of macrophages, DNA sequencing identified B. pseudomallei promoters that were induced during infection. A summary of the method used to isolate clones showing differential eGFP expression in macrophages is shown in Figure 1. This identified promoters upstream of genes encoding proteins with a broad range of functions, including virulence-associated proteins and proteins which have not previously been shown to play roles in B. pseudomallei–host cell interactions. These gene products can now be evaluated as targets for the development of vaccines or novel therapeutics.
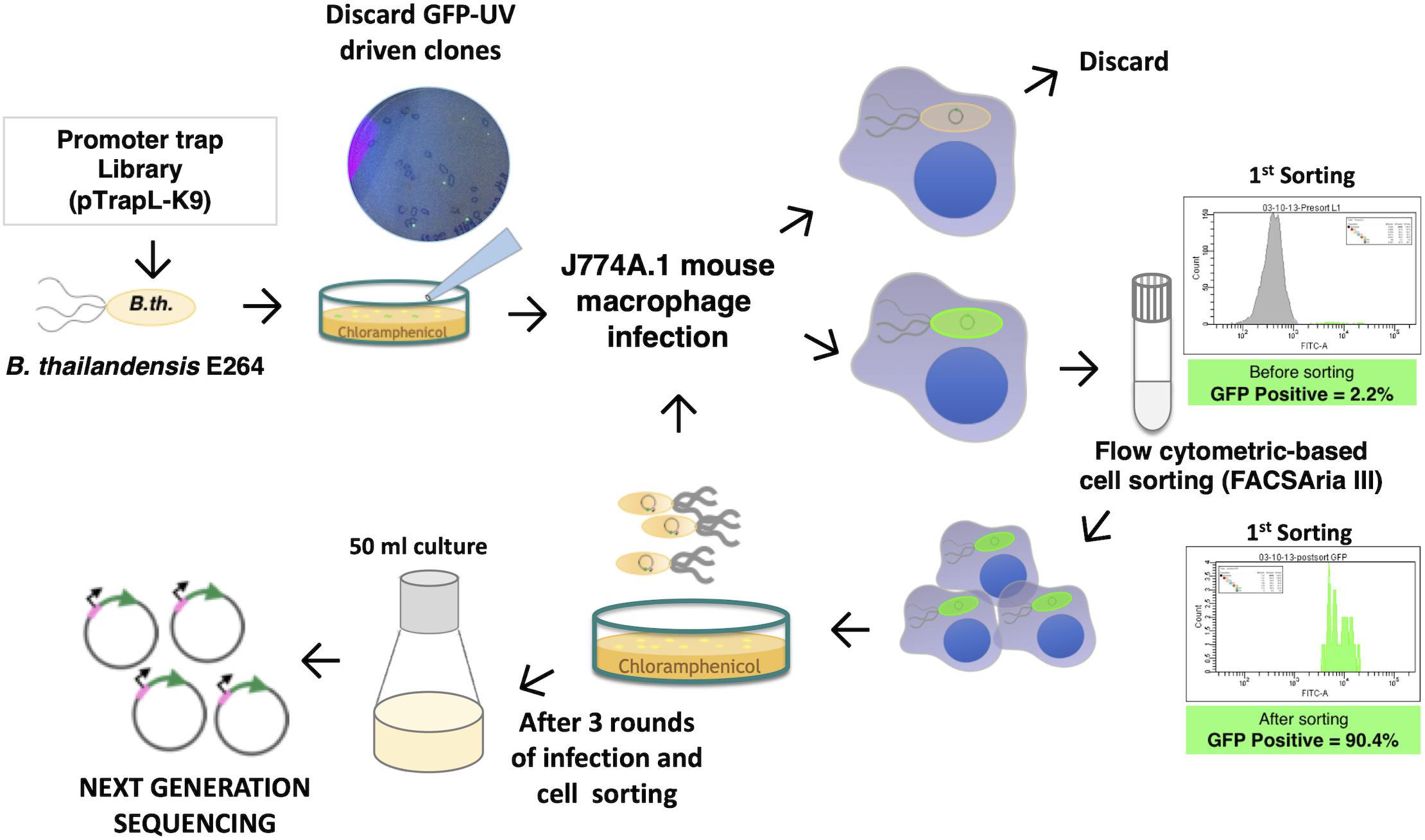
Figure 1. Scheme for identification of macrophage-induced genes of Burkholderia pseudomallei K96243 in Burkholderia thailandensis E264 using differential fluorescence induction (DFI). DNA fragments of B. pseudomallei were ligated into a promoterless-eGFP plasmid, resulting in a pTrapL-K9 library that was then transformed into B. thailandensis E264. Clones that fluoresced on the Luria-Bertani (LB) agar medium under UV light were discarded, and only non-fluorescent colonies on LB agar were pooled and subjected to infected J774A.1 cells. Enhanced green fluorescent protein (eGFP)-positive clones were isolated by cell sorting. Intracellular bacteria were released from sorted cells for reinfection. In a total of three rounds of infection, the eGFP-expressing bacteria were cultured for plasmid extraction and sequencing.
Materials and Methods
Bacterial Strains, Mouse Macrophages, and Culture Conditions
Burkholderia pseudomallei strain K96243 was isolated from a human melioidosis patient (Holden et al., 2004). B. thailandensis E264 has been genome sequenced (Yu et al., 2006) and is widely used as a surrogate for B. pseudomallei in cell-based assays and has been confirmed to deploy many of the same virulence factors during interaction with macrophages. Bacterial stocks were kept in 15% (v/v) glycerol at −70°C. The bacteria were cultured in Luria-Bertani (LB) broth or LB agar (Criterion) with or without 50 μg/ml of chloramphenicol (Sigma). Bacteria were grown at 37°C. All experiments working with B. pseudomallei were conducted at Mahidol University in a biosafety level 3 laboratory with approval by the Technical Biosafety Committee of National Center for Genetic Engineering and Biotechnology (BIOTEC). Genetic modification for the introduction of B. pseudomallei DNA fragments (700–1,500 bp) into B. thailandensis was approved by the Siriraj Biosafety Risk Management Taskforce (Approval No. SI2017-007).
The J774A.1 mouse macrophage cell line was obtained from the American Type Culture Collection (ATCC). The cells were routinely grown and maintained in Dulbecco’s modified Eagle medium (DMEM; Gibco), supplemented with 10% heat-inactivated (30 min, 56°C) fetal bovine serum (FBS; HyClone).
Construction of Promoterless-eGFP Plasmid Vector
The pBHR4-gfp plasmid was modified from a variant of broad-host-range plasmid pBHR4-groS-RFP (Wand et al., 2011) where the gene encoding red fluorescent protein (RFP) had been replaced with one encoding eGFP. The eGFP gene was amplified by PCR using a forward primer (Trap-F) that anneals to the start of the eGFP gene with added restriction sites for SacI, BglII, and SpeI and a reverse primer (Trap-R) that anneals to the end of the eGFP gene with an added BamHI restriction site. The constitutive groS promoter (PgroS) and rfp of the original pBHR4-groS-RFP plasmid was removed by restriction enzyme digestion using SacI and BamHI, which resulted in the pBHR4-backbone fragment and a smaller groS-RFP fragment. The linearized backbone was then ligated together with SacI and BamHI restricted eGFP gene fragment PCR products to create the promoterless-eGFP plasmid pBHR4-gfp. A constitutive eGFP expression plasmid (pBHR4-groS-GFP) was constructed for use as a positive control when macrophages were infected with eGFP-expressing bacteria. The groS promoter was removed from the original pBHR4-groS-RFP plasmid by restriction enzyme digestion using SacI and SpeI. The resulting groS promoter fragment was then cloned into SacI/SpeI restricted plasmid pBHR4-gfp to create plasmid pBHR4-groS-eGFP.
B. pseudomallei Genomic DNA Preparation
A single colony of B. pseudomallei K96243 was inoculated into 10 ml of LB broth and cultured with shaking for 16 h. Two milliliters of the culture was transferred to a microcentrifuge tube and centrifuged at 14,000 × g for 1 min. DNA was extracted from the pelleted cells using a DNA extraction step according to the manufacturer’s instructions (Geneaid Biotech). The DNA pellet was resuspended in an elution buffer and stored at −20°C until used. The yield and purity of the DNA were determined by spectrophotometry (NanoDrop Technologies).
Promoter Trap Library Construction
To construct the promoter trap library, B. pseudomallei K96243 genomic DNA was partially digested with the restriction enzyme Sau3AI. After agarose gel electrophoresis, DNA fragments in the range of 700–1,500 bp were eluted from the gel. The purified DNA fragments were ligated to dephosphorylated BglII-digested plasmid pBHR4-gfp5′ of the promoterless-eGFP gene. The resulting library of cloned B. pseudomallei K96243 genomic fragments (pTrapL-K9) was introduced into B. thailandensis E264 by electroporation. The recombinant clones (approximately 40,000 colonies) were selected on LB agar supplemented with 50 μg/ml of chloramphenicol and pooled. An aliquot of the B. thailandensis E264 pTrapL-K9 library was subcultured in LB broth supplemented with chloramphenicol for plasmid extraction. To assess library coverage, the extracted plasmids were labeled with Cy3 (green fluorescent dye) and hybridized to a high-density tiling microarray based on the B. pseudomallei K96243 genome, essentially as described (Jitprasutwit et al., 2014). The array comprises 384,926 50-mer probes representing both sense and antisense strands at an average resolution of 35 bp (with a mean overlap between probes of 15 bp) and represents 95.1% of the B. pseudomallei K96243 genome, including intergenic regions (Jitprasutwit et al., 2014). To identify B. pseudomallei genes that were preferentially induced in macrophages, B. thailandensis E264 clones from the pTrapL-K9 library with promoters active during culture on an LB agar medium (based on green fluorescence under UV light) were discarded. Only non-fluorescent colonies on LB agar were pooled and used to infect J774A.1 cells.
J774A.1 Mouse Macrophage Infection and Cell Sorting
J774A.1 cell monolayers in 75-cm2 tissue culture flasks (Costar) were infected with either B. pseudomallei or B. thailandensis. Various multiplicities of infection (MOIs) were tested to determine the optimal MOI for screening of the promoter trap library. J774A.1 macrophages were infected with an overnight culture of B. thailandensis E264 carrying the pTrapL-K9 library using MOIs of 15, 20, and 25 and incubated at 37°C under 5% CO2 atmosphere for 1 h to bring bacteria into contact with the cells and allow bacterial entry. Cell monolayers were washed thrice with pre-warmed phosphate-buffered saline (PBS) to remove the non-adherent bacteria and overlaid with medium containing 1 ml of 250 μg/ml of kanamycin (Gibco) to kill extracellular bacteria for 2 h. At 3 h post infection, the monolayers were washed with pre-warmed PBS to remove antibiotic and overlaid with DMEM. The monolayers were further incubated for 3 h. After 6 h post infection, the monolayers were washed with pre-warmed PBS. The infected cells were treated with 0.05% trypsin–EDTA (Gibco), washed with PBS, and subjected to cell sorting. Briefly, the infected cells were suspended in PBS containing 2% FBS and immediately transferred into sample tubes that were compatible for the BD FACSAria III cell sorter (BD Biosciences). A doublet discrimination gating strategy was applied to identify a population of singlet cells. Fluorescence due to eGFP was detected using the channel for fluorescein isothiocyanate (FITC). Fluorescence intensity from uninfected macrophages was used as a cutoff for fluorescence background. A sorted population inferred to comprise eGFP-positive cells was identified on a two-dimensional dot plot presenting forward-scatter signal versus FITC fluorescent signal. The eGFP-positive cells were sorted into a collection tube containing PBS with 2% FBS and used for DFI screening. After cell sorting, intracellular bacteria were released using 0.1% Triton X-100 (Sigma) in PBS to lyse the sorted cells. These bacteria were then used to repeat the infection and screening process, with a total of three rounds of macrophage infection prior to recovery of bacterial clones for sequence analysis.
Plasmid DNA Extraction and Sequencing
Plasmid DNA was extracted from B. thailandensis eGFP-positive clones after three passages in macrophages using the plasmid DNA isolation kit (Geneaid Biotech). In brief, a pool of B. thailandensis was inoculated into 50 ml of LB broth supplemented with 50 μg/ml chloramphenicol and cultured at 37°C with shaking for 16 h. The plasmid DNA was extracted according to the manufacturer’s instructions and the yield and purity was determined using a spectrophotometer (NanoDrop Technologies). DNA inserts in the sorted pTrapL-K9 clones were amplified by PCR using primers Trap-F and Trap-R (Supplementary Table 1), which anneal to the plasmid backbone flanking the inserts. The amplified PCR products were used directly as input for Illumina sequencing. Sequencing libraries were prepared using a NEB Next DNA library preparation kit according to the manufacturer’s instructions (New England Biolabs). Sequencing was performed on an Illumina MiSeq flow cell generating 250-bp paired end reads. Illumina adapters were removed and sequences quality trimmed using ea-utils (Aronesty, 2010). The resulting reads were remapped onto the concatenated chromosomes I and II of B. pseudomallei K96243 sequence using the BWA aligner (Li and Durbin, 2009). The position of the mapped reads were extracted from the bam files, and reads were visualized using the Artemis genome viewer (Rutherford et al., 2000).
Bacterial RNA Extraction and cDNA Synthesis
To validate candidate in vivo induced promoters from the library screen, gene expression of B. pseudomallei K96243 during macrophage infection was compared with bacteria grown in cell culture media. An overnight culture of B. pseudomallei was diluted in DMEM supplemented with 10% FBS to give an MOI of 20. This bacterial suspension was used to infect J774A.1 macrophages. Infected macrophages were incubated at 37°C under 5% CO2 atmosphere for 1 h. As above, the extracellular bacteria were removed by washing and killed by adding of 250 μg/ml of kanamycin. In parallel, the same bacterial suspension was grown in DMEM supplemented with 10% FBS and incubated at 37°C under 5% CO2 atmosphere. At each time point (2 and 4 h post infection), bacterial RNA was extracted from infected J774A.1 cells by a differential lysis method with modifications (Chieng et al., 2012). Macrophage monolayers were lysed in 0.1% Triton X-100 in PBS (Sigma) for 5 min at 37°C. Lysates were collected and subjected to differential centrifugation; first at 800 × g for 5 min to sediment macrophage cells and cellular debris, and secondly at 8,000 × g for 10 min to pellet bacterial cells. Similarly, the bacteria in DMEM supplemented with 10% FBS were harvested by centrifugation at the same time points. Pellets were immediately subjected to RNA extraction using the Total RNA Mini Kit according to the manufacturer’s instructions (Geneaid). To remove traces of genomic DNA, the RNA samples were treated with DNase I (Promega). SuperScript III First-Strand Synthesis System (Invitrogen) was used to convert total RNA into cDNA according to the manufacturer’s instructions. The yield and purity of the RNA was determined by spectrophotometry (NanoDrop Technologies). The absence of DNA contamination was confirmed by PCR using primers specific to 23S ribosomal RNA genes (23S rRNA) of B. pseudomallei before proceeding to cDNA synthesis.
Quantitative Real-Time PCR
Quantitative real-time PCR (qRT-PCR) was performed on 50-ng cDNA in a final volume of 20 μl of LightCycler® FastStart DNA Master PLUS SYBR Green I (Roche). All experiments were conducted with three biological replicates and relative expression ratios were calculated by comparing the Ct value of the in vivo condition (infected macrophages) to the in vitro control (growth in cell culture medium). The genes coding for 23S rRNA and cytochrome d ubiquinol oxidase subunit II (cydB) were used as the reference genes for normalization. These genes were chosen as references because their expression does not differ significantly between culture in vitro and in infected macrophages (Chieng et al., 2012). The primers for amplification of specific genes are described in Supplementary Table 2. The Livak method (Livak and Schmittgen, 2001) was used to determine the relative expression of selected genes in macrophages (in vivo) compared to during growth in cell culture medium. After normalization, the expression of each selected gene against the reference gene compensates for any difference in the amount of cDNA per sample, and a differential gene expression value was calculated from the fold change of gene expression in B. pseudomallei infecting macrophages relative to those grown in cell culture medium at the specified time points.
Construction of B. pseudomallei Mutants
Deletion mutants were generated using a positive-selection suicide replicon as described in a previous study, with some modifications (Logue et al., 2009). Briefly, primers were designed to amplify 400-bp upstream and downstream regions flanking the target genes, bpss1622 and bpss2104. The upstream and downstream amplicons were ligated using ApaI and XbaI restriction sites for bpss1622 and bpss2104, respectively. The amplicons fused in this way were ligated with pJCB12 suicide vectors at SpeI and XbaI restriction sites for Δbpss1622 or SpeI and SphI restriction sites for Δbpss2104 and then transformed into Escherichia coli MFD pir+. Next, the donor E. coli harboring pJCB12Δbpss1622 or pJCB12Δbpss2104 were conjugated into B. pseudomallei K96243 by filter mating (Smith and Guild, 1980). Resulting B. pseudomallei merodiploid strains were selected on LB agar supplemented with 50 μg/ml of chloramphenicol and 100 μg/ml of diaminopimelic acid (Sigma). The selected positive merodiploid strains were subcultured in the absence of chloramphenicol and plated on LB agar lacking NaCl and containing 20% (w/v) sucrose and incubated at room temperature to select double recombinants in which the pJCB12 replicons had excised from the chromosome. Candidate mutants were identified on the basis of sucrose resistance and sensitivity to chloramphenicol. Deletion of the target genes in strains that had undergone successful allelic exchange was validated by PCR.
Virulence in Galleria mellonella
Groups of 10 G. mellonella larvae were used for virulence studies. Overnight cultures of B. pseudomallei wild type, Δbpss1622, or Δbpss2104 mutants were diluted in PBS. Groups of larvae were challenged by injection of an approximately 1000 colony-forming unit (CFU) of each strain at the posterior proleg and incubated in the dark at 37°C (Harding et al., 2013). Another group of larvae were injected with 10 μl of PBS as a control. At intervals between 24 and 60 h after injection, the larvae were scored for death, evidenced as no response to gentle pushing with a pipette tip, and any color change to brown or black was recorded periodically. This experiment was performed in triplicate.
Statistical Analysis
Data from three independent experiments were collected and analyzed using the student’s unpaired t-test using the GraphPad Prism 7 software. For G. mellonella virulence studies, a log-rank (Mantel–Cox) test was used to compare survival curves using the GraphPad Prism 7 software. A P-value of <0.05 was considered significant.
Results
Construction of a B. pseudomallei K96243 Promoter Trap Library
We first constructed a library of size-selected (700–1,500 bp) DNA fragments of B. pseudomallei, which were ligated into a broad-host-range vector upstream of a promoterless gene encoding eGFP. The resulting pTrapL-K9 library was transformed into B. thailandensis E264 and approximately 40,000 colonies were obtained. To test the library was adequately diverse and comprehensively spanned the B. pseudomallei genome, plasmid DNA extracted from the complete library was labeled with Cy3 fluorescent dye and hybridized to a high-density B. pseudomallei K96243 tiling microarray. Analysis of the hybridization pattern indicated that 88% of B. pseudomallei K96243 genome was represented in the plasmid library with good coverage across both chromosomes (Figure 2).
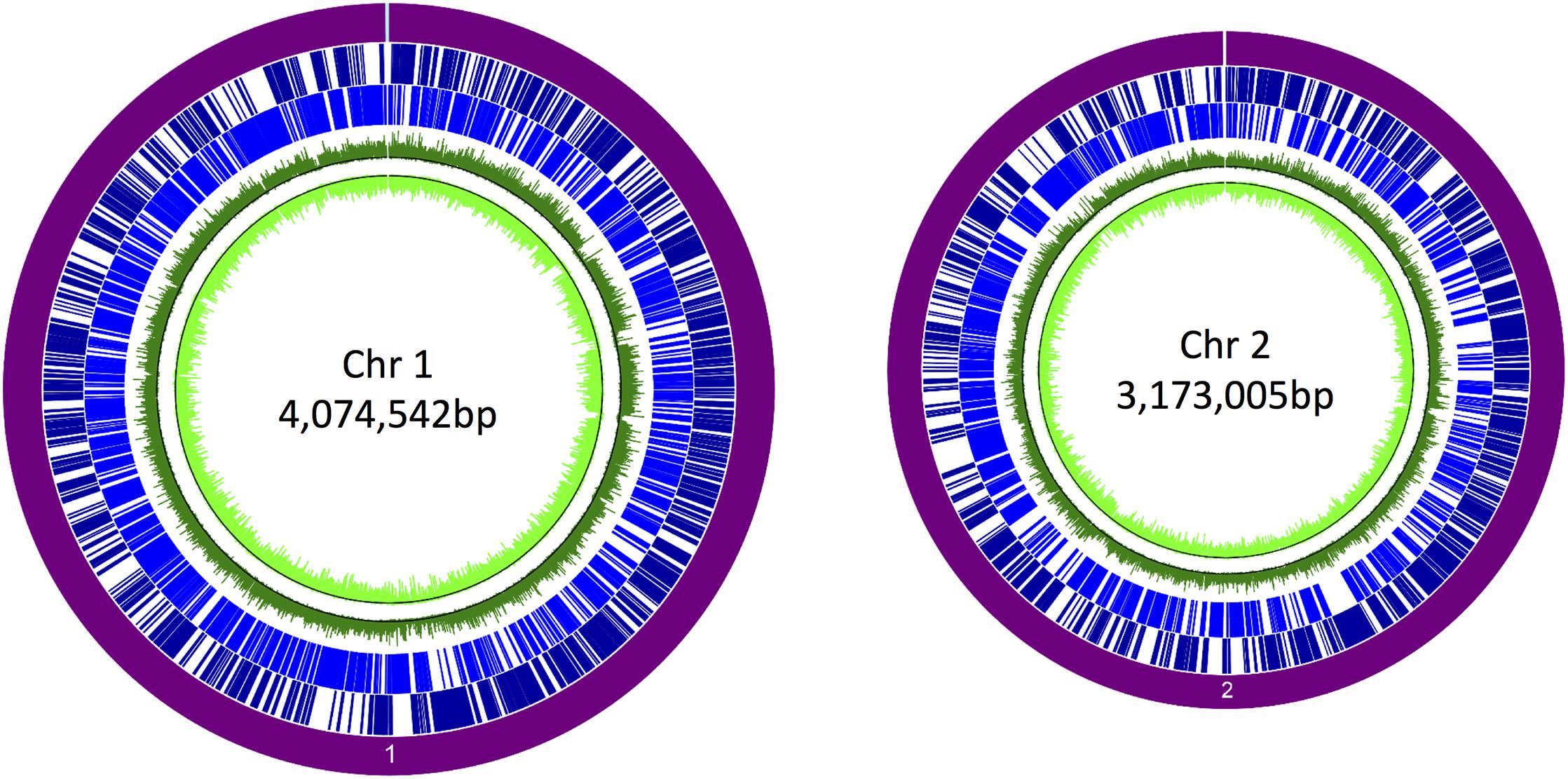
Figure 2. Coverage of the promoter trap library in Burkholderia pseudomallei K96243. The purple circle denotes each chromosome. Dark blue lines denote genes on the forward strand, and light blue lines denote genes on the reverse strand. The corresponding signals obtained by hybridization of the pTrapL-K9 library to the tiling array are indicated by the green circles. The black line on the green tracks is the cutoff for background. Percentage coverage of the genome by the pTrapL-K9 library (88%) has been corrected for the fact that the microarray probes cover 95.1% of the B. pseudomallei genome.
Isolation of Clones Showing Differential eGFP Expression in Macrophages
Following depletion of clones expressing eGFP during culture on LB agar, J774A.1 murine macrophages were infected with the remaining B. thailandensis E264 pTrapL-K9 library using an optimal MOI. Internalization efficiencies of B. thailandensis E264 were increased when using a higher MOI and the number of bacteria recovered from infected macrophages was significantly lower (P < 0.05) when using an MOI of 15, compared to using an MOI of 20 or 25. However, there was no significant difference between the numbers of bacteria recovered from macrophages after infection at an MOI of 20 or 25 (Supplementary Figure 1). Therefore, for subsequent library screening, J774A.1 cells were infected with the B. thailandensis E264 pTrapL-K9 library at an MOI of 20. With this inoculum, 5.97 × 106 CFU were recovered from approximately 8 × 106 infected cells, indicating that on average there were 0.7 bacteria per cell at the point of analysis.
J774A.1 cells infected with the B. thailandensis E264 pTrapL-K9 library were screened for a fluorescent signal at 6 h after macrophage infection. The rationale for screening the library at 6 h post infection was that we could maximize the number of cells that were infected but before cell-cell fusion occurred (Wand et al., 2011). The time is sufficient for internalized bacteria to escape phagosomes and display actin-based motility (Stevens J.M. et al., 2005; Wand et al., 2011). A sequential gating strategy was used to minimize false positives. Macrophages infected with B. thailandensis carrying the promoterless-eGFP plasmid were included as a negative control. Only 0.19% of the macrophage population exhibited a dim-auto-fluorescence signal from uninfected macrophages. This data was used to determine the background fluorescence intensity for further analysis (Figure 3A, Supplementary Figure 2). B. thailandensis harboring pBHR4-groS-eGFP was used as a positive control for eGFP-positive cells (Figure 3B, Supplementary Figure 2). Levels of fluorescence of these controls were measured in order to set the appropriate gates to distinguish uninfected cells and GFP-negative cells from the fluorescent macrophages inferred to be infected with B. thailandensis carrying in vivo induced promoters.
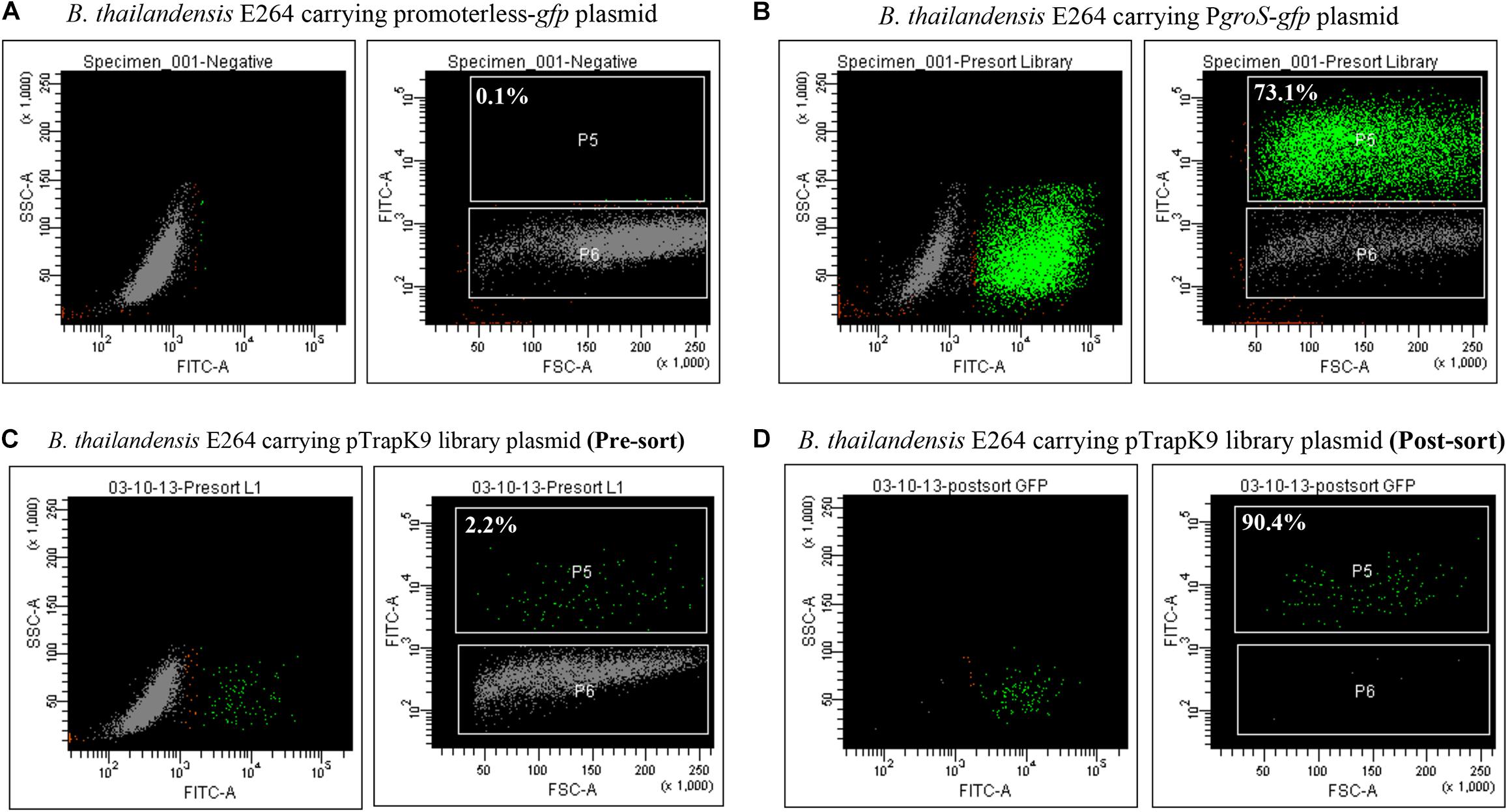
Figure 3. Enhanced green fluorescent protein (eGFP) expression in J774A.1 macrophages. (A) Gating for eGFP-positive cells was set according to the fluorescent background of macrophages infected with Burkholderia thailandensis carrying a promoterless-gfp control plasmid. (B) eGFP expression was observed in macrophages infected with B. thailandensis carrying PgroS-eGFP. (C) Macrophages infected with the B. thailandensis promoter trap library (pre-sorted). (D) The post-sorted macrophage infected with B. thailandensis promoter trap library. The percentages of eGFP-positive cells are indicated.
In the first round, macrophages infected with the B. thailandensis pTrapL-K9 library were sorted and 2.2% of the infected cells were eGFP positive (Figure 3C, Supplementary Figure 2). The sorted eGFP-positive cells were reanalyzed by flow cytometry and 90% of the collected cells were eGFP positive (Figure 3D, Supplementary Figure 2). To further enrich eGFP-positive clones, bacteria were released from the sorted cells by gentle lysis and used to re-infect J774A.1 macrophages. In total, we carried out three rounds of macrophage passage of the library. Finally, bacteria were recovered and plated onto LB agar. Each colony was visualized under a UV light illuminator to confirm that they did not fluoresce when cultured in vitro and the resulting colony exhibiting green fluorescence was not detected (data not shown). These clones were collected and stored.
Bacterial Virulence and Oxidative Stress Response Genes Were Induced in Infected Macrophages
Colonies of the bacteria that fluoresced in macrophages but not on agar were pooled and plasmids extracted. Amplicons for the inserts in the recovered pTrapL-K9 plasmids were sequenced by Illumina MiSeq analysis. In total 552,458 reads were obtained, of which 503,624 reads (91.16%) mapped onto the B. pseudomallei K96243 reference genome and this revealed 138 different regions distributed across the B. pseudomallei K96243 genome, with 60% mapped to chromosome 2 (Figure 4). The length of Illumina mapped regions ranged from 33 to 567 bp (134 bp on average). Within the 138 regions identified, 2 to 8,452 sequence reads mapped to the B. pseudomallei K96243 genome. The locations of mapped regions within the genome were visualized using Artemis and genes at the 3′ end of the cloned fragments were identified. The B. pseudomallei genes associated with macrophage-induced promoters were categorized into 2 groups, according to the distance between the 5′ end of the mapped regions and the predicted start codon of the associated gene. Mapped regions upstream of the associated gene or overlapping with the predicted start codon (n = 27) were assigned to Group A and mapped regions within a predicted coding sequence (n = 111) were assigned to Group B (Supplementary Table 2). The possibility that the genes associated with the mapped fragment were part of an operon was assessed from previously reported condition-dependent transcriptome data for B. pseudomallei (Ooi et al., 2013). We found that 33% of genes associated with mapped regions were not part of an operon, 22% were the first gene in a predicted operon, 17% were the last gene of the predicted operon and 28% of associated genes were located at another position within the operon. The proportion of promoters identified that are predicted to control transcription of single genes is broadly consistent with the predicted number of monocistronic transcripts from sequencing of the genome (Holden et al., 2004) and whole-genome transcriptome profiling (Ooi et al., 2013).
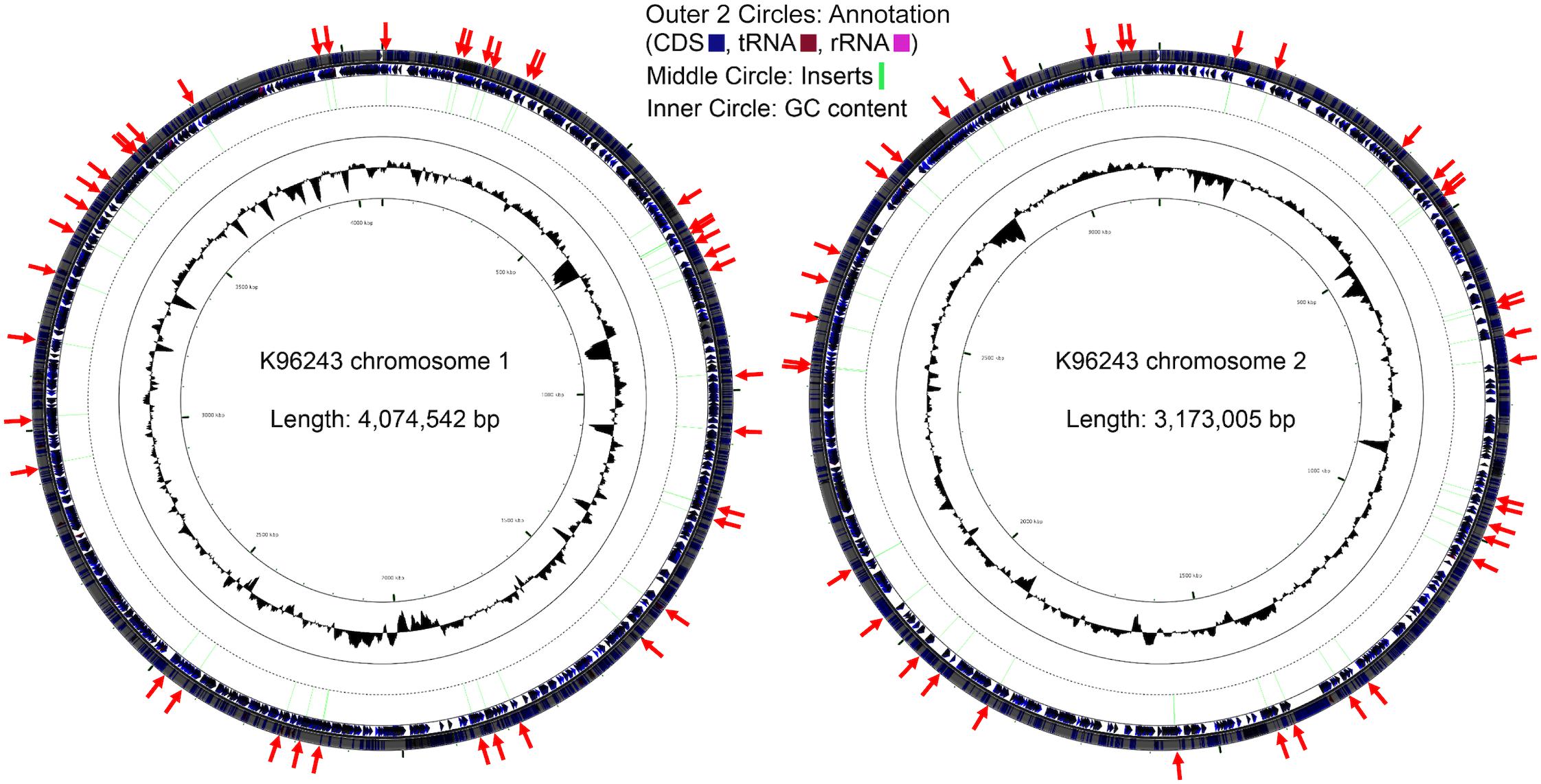
Figure 4. Distribution of the Burkholderia pseudomallei genes located downstream of the identified putative inducible promoters during macrophage infection. Locations of the genes immediately downstream of the identified macrophage putative promoters (red arrows) across the respective chromosomes.
The functions of genes 3′ end of cloned fragments inferred to contain macrophage-induced promoters were predicted from annotation in GenBank and EMBL or published literature. Known virulence factors were identified, including flagellin (BPSL3319), the T3SS effector protein BapC (BPSS1526), the adhesin BoaA (BPSS0796), and catalase-peroxidase KatG that is predicted to be involved in resistance to oxidative stress (BPSL2865). Moreover, we identified predicted regulatory systems as macrophage-induced, including the trans-membrane invasion-related two-component sensor IrlS protein (BPSS1039) and a metal-related two-component system IrlS2 (BPSS1995). Some of the genes we identified are predicted to be involved in responding to environmental signals inside macrophages (Supplementary Table 3). Additionally, the genes we identified that are involved in LPS biosynthesis and RNA modification have been reported to be upregulated in B. thailandensis grown in anoxic conditions (Peano et al., 2014).
We compared our gene list with previously reported datasets, for example from mutagenesis with in vivo negative selection (Moule et al., 2015), the transcriptome under oxidative stress (Jitprasutwit et al., 2014), in silico prediction of virulence genes (Schell et al., 2008), and screening for serodiagnostic antigens (Felgner et al., 2009). We found that nearly one third of the genes identified from our promoter trap study had not previously been reported to be associated with virulence or as in vivo-induced antigens (Supplementary Table 3).
Validation of the Putative Macrophage-Induced B. pseudomallei Genes by RT-PCR and Virulence Study
As the study used B. thailandensis as a surrogate host for screening of the K96243 promoter trap library owing to constraints on cell sorting at biosafety Level 3, we sought to verify that candidate macrophage-induced genes are differentially transcribed during B. pseudomallei infection of J774A.1 cells. Additionally, the virulence of selected mutants was tested in G. mellonella larvae. A total of 15 genes were selected for validation by qRT-PCR or virulence in G. mellonella (Table 1).
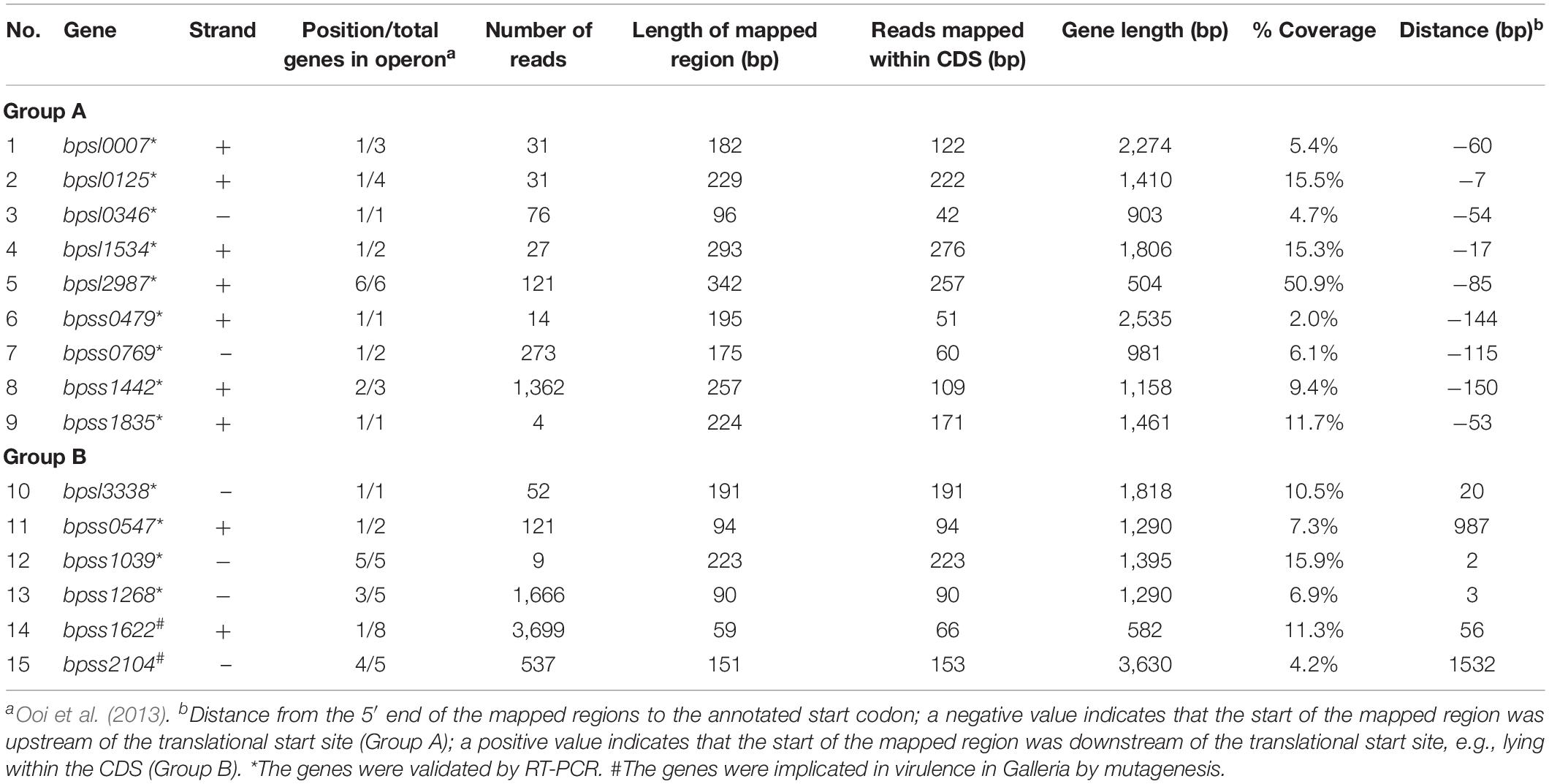
Table 1. Burkholderia pseudomallei genes located downstream of putative macrophage-induced promoters and selected for validation using qRT-PCR and mutagenesis.
Transcription of 13 genes downstream of putative macrophage-induced promoters (nine in Group A and four in Group B) was examined 2 or 4 h after infection of J774A.1 macrophages by B. pseudomallei K96243 at an MOI of 20 relative to expression in LB broth after 2 or 4 h of culture by qRT-PCR. B. pseudomallei RNA was collected at 2 or 4 h rather than 6 h post infection with B. thailandensis, because of the faster growth of B. pseudomallei K96243 in macrophages, compared with B. thailandensis E264 (Wand et al., 2011; Kovacs-Simon et al., 2019). Fold changes in gene expression were calculated by normalizing the level of cDNA with a reference gene (23S rRNA or cydB) prior to comparison of gene expression in macrophages against in vitro growth (Table 2). We selected genes from Group A and Group B by choosing from their gene organization. Genes that were not a part of an operon, or were the first, middle, or last gene of a predicted operon, were selected randomly. We included bpss1498, encoding a Type VI secretion system protein, as a positive control as the gene is known to be upregulated in macrophages (Chieng et al., 2012).
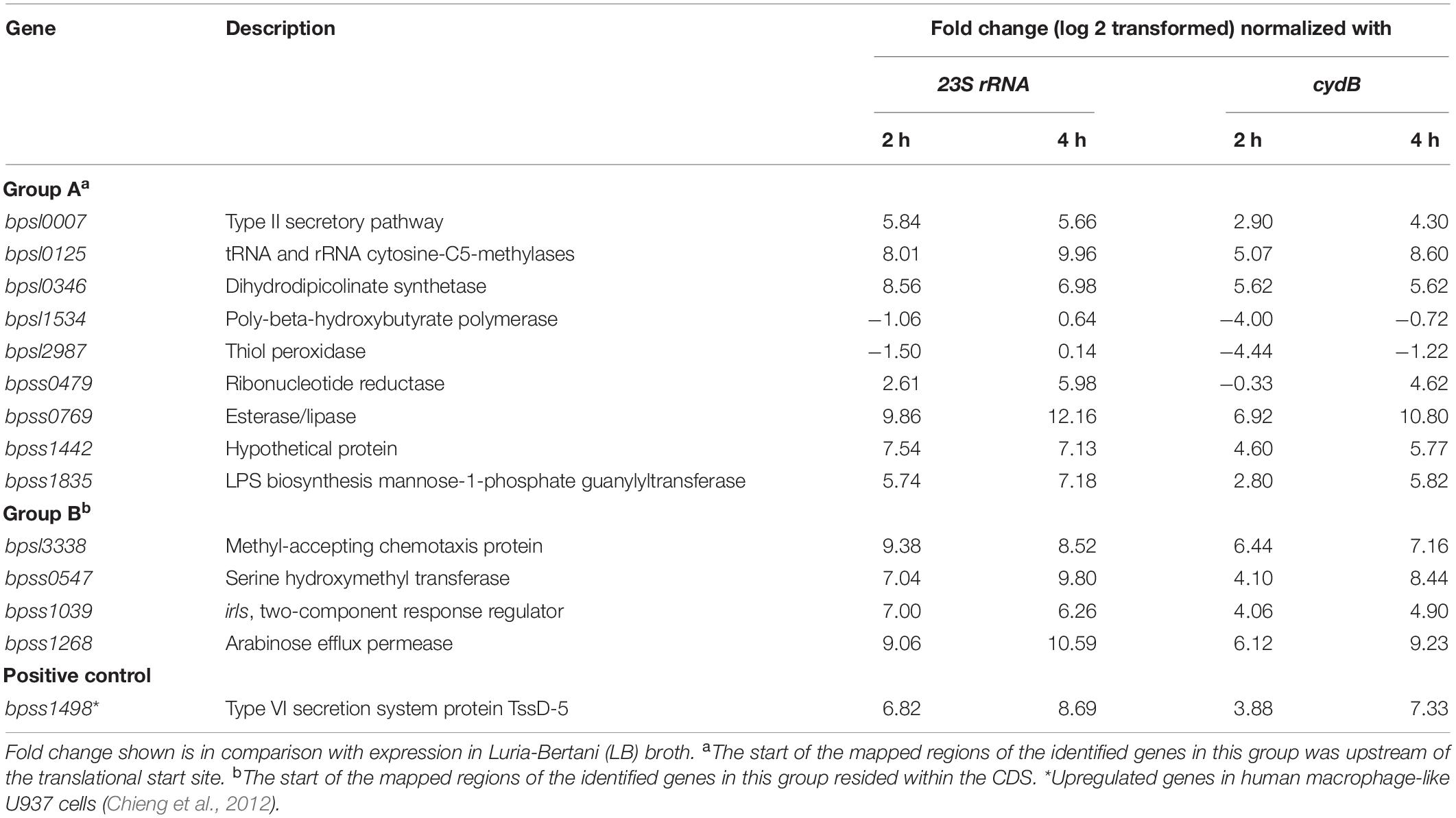
Table 2. Expression of 13 genes measured using qRT-PCR in Burkholderia pseudomallei during infection of J774A.1 macrophages.
As expected, we also found increased expression of bpss1498 after infection of J774A.1 cells with B. pseudomallei. Of the 13 genes identified from our promoter trap study, 11 showed greater than twofold increases in expression in bacteria isolated from macrophages compared to bacteria cultured in LB broth. Two genes in Group A, bpsl1534 and bpsl2987 encoding poly-beta-hydroxybutyrate polymerase and thiol peroxidase, respectively, showed reduced expression in macrophages compared to LB depending on the time and reference gene used, albeit the magnitude of the change was limited and the direction of the effect was inconsistent. The data obtained for bpsl1534 and bpsl2987 by qRT-PCR were not consistent with specific induction of transcription of the genes suggested by the fluorescence-based screen. This may reflect the different sampling times (qRT-PCR at 2 and 4 h post infection vs. 6 h post infection for the DFI screen). Also, it may be that these genes were expressed transiently or that the encoding mRNAs had atypical half-lives.
We selected two genes in Group B that had not previously been mutated, bpss1622 (an inner rod component of Type III secretion system cluster 2) and bpss2104 (a component of basal secretion apparatus of Type IV secretion system cluster 3) as targets for deletion in B. pseudomallei K96243 because these genes have not previously been investigated for a role in virulence. Double-recombinants lacking the targeted regions were validated by PCR. The wild-type strain and the mutants were then tested for virulence in G. mellonella larvae. No deaths were observed when larvae were injected with PBS (Figure 5). At 32 h post-challenge, wild-type B. pseudomallei K96243 caused 100% mortality but the B. pseudomalleiΔbpss1622 or Δbpss2104 mutants caused only 10% mortality (Figure 5). While this implicates the targeted macrophage-induced regions in virulence, both genes are part of operons and further research will be required to determine if observed phenotypes are attributable to the genes per se, or polar effects.
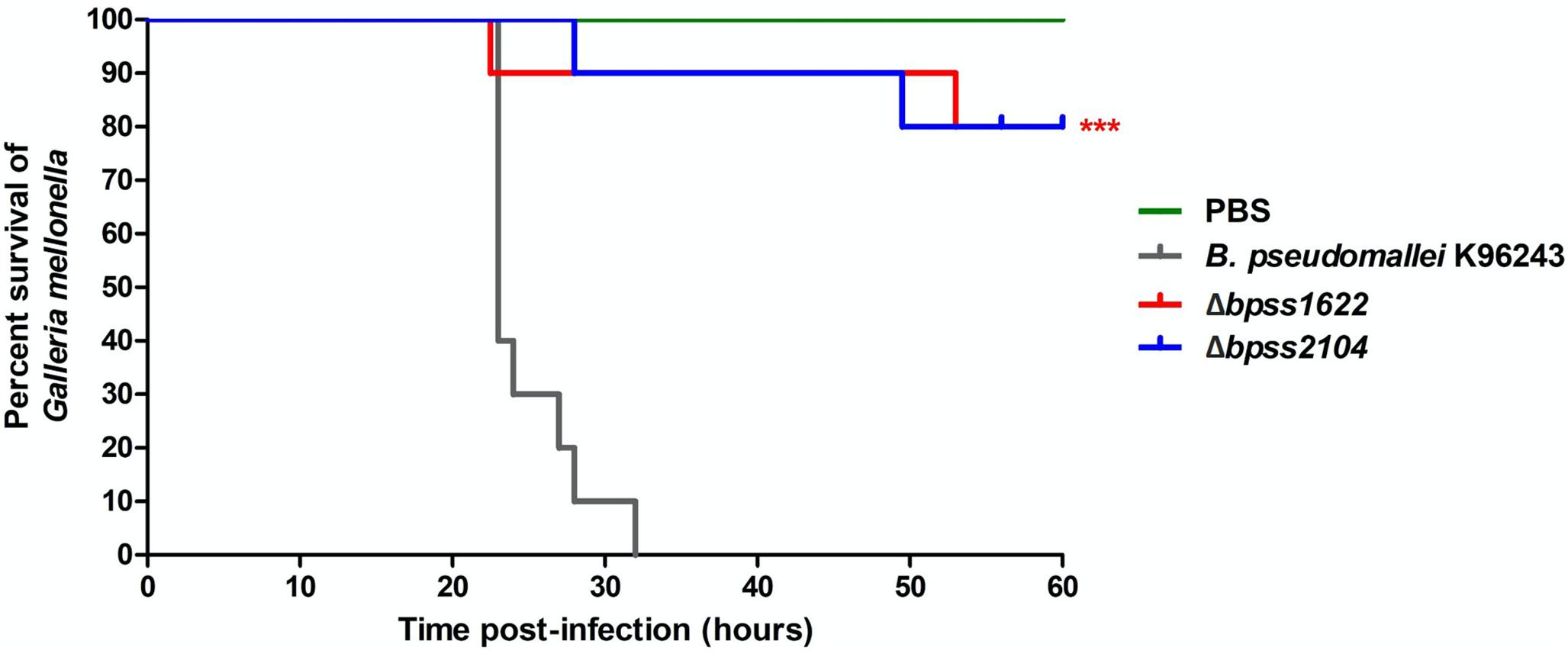
Figure 5. Virulence of Burkholderia pseudomallei strains in Galleria mellonella larvae. Groups of 10 larvae were challenged with the strains indicated. The experiment was performed in triplicate and showed similar results. A representative result is shown as the percentage of surviving larvae from 24 to 60 h after infection with B. pseudomallei K96243 wild type (gray), Δbpss1622 mutant, or Δbpss2104 mutant. P-values were determined by the log rank (Mantel–Cox) test, and triple asterisks denote a significant difference of P < 0.0001 between the wild type and mutants.
Discussion
We have established a promoter trap library and used DFI to identify B. pseudomallei genes induced during macrophage infection. Previously, B. pseudomallei genes which are differentially expressed in broth and in human macrophage-like cells as measured using a DNA microarrays, has been reported (Chieng et al., 2012). Both microarray and DFI technologies detect differential gene expression at the level of transcription. Microarrays measure the levels of mRNA directly whereas the approach we have used measures promoter activity using a fluorescent reporter. Each approach has advantages and limitations. A limitation of microarrays is the difficulty of detecting low level gene expression. Additionally, because mRNA is often unstable, genes which show temporal upregulation may not be identified using methods which directly measure mRNA levels, unless samples are taken frequently. In contrast, the use of a stable fluorescent reporter allows promoter activity to be measured retrospectively. Finally, reporter-based methods are of value where there is a need to study gene expression in individual infected cells (Valdivia and Falkow, 1997).
Our library showed 88% genome coverage of the B. pseudomallei K96243 genome, but, because the DNA fragments were generated after partial cleavage with restriction endonuclease, some promoters may not have been cloned or obtained in the correct orientation relative to the reporter gene. As a result, some macrophage-induced promoters were likely not detected in this study. Also, our strategy for screening for promoter activity in B. thailandensis assumes that gene regulation is similar in B. thailandensis and B. pseudomallei and this may not be the case for all genes. For example, the arabinose assimilation operon is differentially represented in these two species, with consequences for the expression of the Bsa Type III secretion system (Moore et al., 2004).
To identify promoters induced in host cells, we screened B. thailandensis-infected macrophages at a time point in the infection cycle at which phagosome escape and actin-based motility have been reported (Stevens J.M. et al., 2005; Wand et al., 2011), but prior to cell fusion resulting in the formation of multinucleate giant cells (MNGCs). To avoid the possibility that fluorescent macrophages were infected by multiple bacterial clones we used three cycles of infection and cell sorting. After that, the recovered B. thailandensis eGFP-positive clones were propagated by culturing in LB broth for plasmid preparation. This in vitro growth stage might affect the abundance of some clones leading to the enrichment of false negatives but not false positives. Our microarray data, which were derived from the library grown in vitro, showed comprehensive coverage of the genome. Any selection imposed by growth in LB broth was therefore not obvious from our analysis of library diversity.
After sequencing the clones recovered from fluorescent macrophages, the short reads obtained were aligned with the B. pseudomallei K96243 genome. This revealed whether the 5′ end of the fragment was upstream or within the predicted open reading frame (ORF) of a gene. Typically, a promoter would be located upstream of the ORF. In this study, most of the promoters were mapped within an annotated gene. However, it is possible that the gene annotation is incorrect or that an internal promoter is present. Internal promoters have been reported in a range of bacteria (Miyata et al., 2013; Namprachan-Frantz et al., 2014; Perault and Cotter, 2018). For example, the Streptococcus pyogenes salivaricin (sal) operon is regulated by a promoter upstream of the operon and also by a second promoter located within the operon (Namprachan-Frantz et al., 2014). Internal promoters are also found in genes encoding immunity proteins of Vibrio cholerae, which are independently expressed of other proteins encoded within the same operon (Miyata et al., 2013). Another example found in Burkholderia dolosa where a gene internal promoter is involved in expression of the Burkholderia-type contact-dependent growth inhibition (CDI) system-encoding locus (bcpAIOB). The region upstream of the B. dolosa bcp-3 operon does not show promoter activity and fails to drive the expression of the downstream genes. However, promoter activity was detected when the 500 bp distal region of the first gene in the bcp-3 operon was cloned into a lacZ expression vector, indicating that a promoter resides at the 3′ end of the first gene of the operon (Perault and Cotter, 2018).
In this study, we identified 138 genomic regions inferred to possess promoter activity during J774A.1 macrophage infection. A previous microarray study carried out at 1, 2, 4, or 6 h after infection of U937 macrophages revealed 25 B. pseudomallei genes that were upregulated (Chieng et al., 2012). Only bpss0143, encoding a transcriptional regulator, was identified in both studies. However, our qRT-PCR analysis also showed differences compared to the previously reported microarray based study (Chieng et al., 2012). For example, the increased expression of bpss1835, encoding an LPS biosynthesis mannose-1-phosphate guanylyl transferase, was confirmed by qRT-PCR in our study but this gene showed reduced expression at 6 h post infection in human U937 cells, compared to in vitro culture (Chieng et al., 2012). This could reflect differences in the intracellular environment in U937 human macrophages and in J774A.1 mouse macrophages. Or it may indicate differences in gene regulation in B. thailandensis and B. pseudomallei. Alternatively, the fundamental differences in microarray and qRT-PCR methodologies could be responsible for these differences.
A greater proportion of the macrophage-induced promoters identified in this study were located on chromosome 2 of B. pseudomallei K96243. This chromosome encodes many accessory functions, including functions associated with adaptation and survival in different niches and well-characterized virulence genes (Holden et al., 2004). Thirteen genes downstream of the promoters we identified are proximal to genes which have previously been shown to be involved in oxidative stress responses (Jitprasutwit et al., 2014). These include genes upregulated when B. pseudomallei K96243 is exposed to hydrogen peroxide (H2O2) and which might play a role in survival in macrophages (Jitprasutwit et al., 2014). It was also noticeable that a fifth of the genes we identified in this study (28 genes) have previously been reported in a transposon-directed insertion site sequencing (TraDIS) library as genes essential for the in vitro growth of B. pseudomallei K96243 (Moule et al., 2014). Although in vitro induced B. pseudomallei promoters should be excluded in our study, we found that bpss0547, which was from this in vitro TraDIS study, showed increased expression after macrophage infection in our study. Therefore, our findings suggest that some essential genes may also play a role in virulence. Approximately one fifth of the macrophage-induced promoters we identified (27 promoters) have been reported to co-locate with virulence-related genes shared by B. pseudomallei and Burkholderia mallei (Schell et al., 2008) although four of the genes (bpsl1057, bpss0796, bpss0822, and bpss1268) are absent in B. thailandensis (Schell et al., 2008). The roles of these genes have already been reported. For example, bpss0796 encoding a putative trimeric autotransporter protein (BoaA) plays a role in survival of B. pseudomallei in macrophages (Balder et al., 2010), and a boaA mutant was attenuated in BALB/c mice (Lazar Adler et al., 2015). In addition, BPSS0796 is also identified as an immunogenic protein in melioidosis patient serum (Tiyawisutsri et al., 2007). Collectively, these data support our proposal that the genes co-located with the promoters we have identified play a role in the virulence of B. pseudomallei. Our proposal is also supported by the finding that bpss1622 or bpss2104 deletion mutants were attenuated in wax moth larvae.
Our promoter trap library and DFI screening strategy enabled us to identify B. pseudomallei promoters induced during infection of mouse macrophages. A number of novel B. pseudomallei genes induced during macrophage infection that have not been reported in the previous literature were found. In comparison with antibiotic resistance-based IVET that requires an appropriate concentration to isolate promoters that are active at a specific level, promoters with transient or weak activity can be detected using DFI (Rediers et al., 2005). A promoter trap screen using fusions to a promoterless chloramphenicol resistance gene integrated into the B. pseudomallei genome identified 15 different genomic loci after four rounds of RAW264.7 macrophage infection using an MOI of 100 (Shalom et al., 2007). Among these were three genes (tssH-5, tssI-5, and tssM-5) located within the same Type VI protein secretion system cluster (tss-5), mntH, encoding a natural resistance-associated macrophage protein (NRAMP)-like manganese ion transporter, and bhuT, a heme acquisition gene (Shalom et al., 2007). No overlap was detected in the genes identified in the present study. Our dataset complements other datasets of potential virulence-associated genes in B. pseudomallei and opens new opportunities to investigate the roles of the genes we have identified in intracellular life and disease.
Data Availability Statement
The datasets GENERATED for this study can be found in the SNBI SRA accession PRJNA599542.
Author Contributions
SJ, RT, and SK contributed to the conception and design of the study. SJ, CH, NO, PW, VM, PV, JS, and CO performed the experiments. SJ and NJ analyzed the data and wrote the manuscript. PV, JS, and MS provided technical guidance and suggested and commented on the design of the experiments. RT, MS, and SK edited the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work has been funded through a Siriraj Grant for the Research and Development (R016033016), Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand. SJ (PHD/0270/2551) and VM (PHD/0190/2560) were supported by the Royal Golden Jubilee Ph.D. Program, Thailand Research Fund, Thailand.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Exeter Sequencing Service facility, which received generous support from the Wellcome Trust Institutional Strategic Support Fund (WT097835MF), Wellcome Trust Multi User Equipment Award (WT101650MA), Medical Research Council Clinical Infrastructure Funding (MR/M008924/1), and BBSRC LOLA award (BB/K003240/1). JS and MS gratefully acknowledge support from the Biotechnology and Biological Sciences Research Council (BBS/E/D/20002173).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00072/full#supplementary-material
References
Aronesty, E. (2010). Command-Line Tools for Processing Biological Sequencing Data. Durham, NC: EA-Utils.
Balder, R., Lipski, S., Lazarus, J. J., Grose, W., Wooten, R. M., Hogan, R. J., et al. (2010). Identification of Burkholderia mallei and Burkholderia pseudomallei adhesins for human respiratory epithelial cells. BMC Microbiol. 10:250. doi: 10.1186/1471-2180-10-250
Bumann, D., and Valdivia, R. H. (2007). Identification of host-induced pathogen genes by differential fluorescence induction reporter systems. Nat. Protoc. 2, 770–777. doi: 10.1038/nprot.2007.78
Burtnick, M. N., Brett, P. J., Harding, S. V., Ngugi, S. A., Ribot, W. J., Chantratita, N., et al. (2011). The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79, 1512–1525. doi: 10.1128/IAI.01218-10
Chieng, S., Carreto, L., and Nathan, S. (2012). Burkholderia pseudomallei transcriptional adaptation in macrophages. BMC Genomics 13:328. doi: 10.1186/1471-2164-13-328
Cuccui, J., Easton, A., Chu, K. K., Bancroft, G. J., Oyston, P. C., Titball, R. W., et al. (2007). Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect. Immun. 75, 1186–1195. doi: 10.1128/IAI.01240-06
Dance, D. A. (2000). Melioidosis as an emerging global problem. Acta Trop. 74, 115–119. doi: 10.1016/s0001-706x(99)00059-5
Felgner, P. L., Kayala, M. A., Vigil, A., Burk, C., Nakajima-Sasaki, R., Pablo, J., et al. (2009). A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc. Natl. Acad. Sci. U.S.A. 106, 13499–13504. doi: 10.1073/pnas.0812080106
Galyov, E. E., Brett, P. J., and DeShazer, D. (2010). Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu. Rev. Microbiol. 64, 495–517. doi: 10.1146/annurev.micro.112408.134030
Gong, L., Cullinane, M., Treerat, P., Ramm, G., Prescott, M., Adler, B., et al. (2011). The Burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS One 6:e17852. doi: 10.1371/journal.pone.0017852
Harding, C. R., Schroeder, G. N., Collins, J. W., and Frankel, G. (2013). Use of Galleria mellonella as a model organism to study Legionella pneumophila infection. J. Vis. Exp. 8:e50964. doi: 10.3791/50964
Holden, M. T., Titball, R. W., Peacock, S. J., Cerdeno-Tarraga, A. M., Atkins, T., Crossman, L. C., et al. (2004). Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U.S.A. 101, 14240–14245. doi: 10.1073/pnas.0403302101
Jitprasutwit, S., Ong, C., Juntawieng, N., Ooi, W. F., Hemsley, C. M., Vattanaviboon, P., et al. (2014). Transcriptional profiles of Burkholderia pseudomallei reveal the direct and indirect roles of Sigma E under oxidative stress conditions. BMC Genomics 15:787. doi: 10.1186/1471-2164-15-787
Kovacs-Simon, A., Hemsley, C. M., Scott, A. E., Prior, J. L., and Titball, R. W. (2019). Burkholderia thailandensis strain E555 is a surrogate for the investigation of Burkholderia pseudomallei replication and survival in macrophages. BMC Microbiol. 19:97. doi: 10.1186/s12866-019-1469-8
Lazar Adler, N. R., Stevens, M. P., Dean, R. E., Saint, R. J., Pankhania, D., Prior, J. L., et al. (2015). Systematic mutagenesis of genes encoding predicted autotransported proteins of Burkholderia pseudomallei identifies factors mediating virulence in mice, net intracellular replication and a novel protein conferring serum resistance. PLoS One 10:e0121271. doi: 10.1371/journal.pone.0121271
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Limmathurotsakul, D., Golding, N., Dance, D. A., Messina, J. P., Pigott, D. M., Moyes, C. L., et al. (2016). Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat. Microbiol. 1:15008. doi: 10.1038/nmicrobiol.2015.8
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-(CT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Logue, C. A., Peak, I. R., and Beacham, I. R. (2009). Facile construction of unmarked deletion mutants in Burkholderia pseudomallei using sacB counter-selection in sucrose-resistant and sucrose-sensitive isolates. J. Microbiol. Methods 76, 320–323. doi: 10.1016/j.mimet.2008.12.007
Miyata, S. T., Unterweger, D., Rudko, S. P., and Pukatzki, S. (2013). Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathog. 9:e1003752. doi: 10.1371/journal.ppat.1003752
Moore, R. A., Reckseidler-Zenteno, S., Kim, H., Nierman, W., Yu, Y., Tuanyok, A., et al. (2004). Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72, 4172–4187. doi: 10.1128/IAI.72.7.4172-4187.2004
Moule, M. G., Hemsley, C. M., Seet, Q., Guerra-Assuncao, J. A., Lim, J., Sarkar-Tyson, M., et al. (2014). Genome-wide saturation mutagenesis of Burkholderia pseudomallei K96243 predicts essential genes and novel targets for antimicrobial development. mBio 5:e00926-e13. doi: 10.1128/mBio.00926-13
Moule, M. G., Spink, N., Willcocks, S., Lim, J., Guerra-Assuncao, J. A., Cia, F., et al. (2015). Characterization of new virulence factors involved in the intracellular growth and survival of Burkholderia pseudomallei. Infect. Immun. 84, 701–710. doi: 10.1128/IAI.01102-15
Namprachan-Frantz, P., Rowe, H. M., Runft, D. L., and Neely, M. N. (2014). Transcriptional analysis of the Streptococcus pyogenes salivaricin locus. J Bacteriol 196, 604–613. doi: 10.1128/JB.01009-13
Ooi, W. F., Ong, C., Nandi, T., Kreisberg, J. F., Chua, H. H., Sun, G., et al. (2013). The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet. 9:e1003795. doi: 10.1371/journal.pgen.1003795
Peano, C., Chiaramonte, F., Motta, S., Pietrelli, A., Jaillon, S., Rossi, E., et al. (2014). Gene and protein expression in response to different growth temperatures and oxygen availability in Burkholderia thailandensis. PLoS One 9:e93009. doi: 10.1371/journal.pone.0093009
Perault, A. I, and Cotter, P. A. (2018). Three distinct contact-dependent growth inhibition systems mediate interbacterial competition by the cystic fibrosis pathogen Burkholderia dolosa. J. Bacteriol. 200:e00428-18. doi: 10.1128/JB.00428-18
Rediers, H., Rainey, P. B., Vanderleyden, J., and De Mot, R. (2005). Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69, 217–261. doi: 10.1128/MMBR.69.2.217-261.2005
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Schell, M. A., Lipscomb, L., and DeShazer, D. (2008). Comparative genomics and an insect model rapidly identify novel virulence genes of Burkholderia mallei. J. Bacteriol. 190, 2306–2313. doi: 10.1128/JB.01735-07
Schwarz, S., Singh, P., Robertson, J. D., LeRoux, M., Skerrett, S. J., Goodlett, D. R., et al. (2014). VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82, 1445–1452. doi: 10.1128/IAI.01368-13
Shalom, G., Shaw, J. G., and Thomas, M. S. (2007). In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153(Pt 8), 2689–2699. doi: 10.1099/mic.0.2007/006585-0
Smith, M. D., and Guild, W. R. (1980). Improved method for conjugative transfer by filter mating of Streptococcus pneumoniae. J. Bacteriol. 144, 457–459. doi: 10.1128/jb.144.1.457-459.1980
Stevens, J. M., Ulrich, R. L., Taylor, L. A., Wood, M. W., Deshazer, D., Stevens, M. P., et al. (2005). Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J. Bacteriol. 187, 7857–7862. doi: 10.1128/JB.187.22.7857-7862.2005
Stevens, M. P., Stevens, J. M., Jeng, R. L., Taylor, L. A., Wood, M. W., Hawes, P., et al. (2005). Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 56, 40–53. doi: 10.1111/j.1365-2958.2004.04528.x
Stevens, M. P., Wood, M. W., Taylor, L. A., Monaghan, P., Hawes, P., Jones, P. W., et al. (2002). An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46, 649–659. doi: 10.1046/j.1365-2958.2002.03190.x
Tiyawisutsri, R., Holden, M. T., Tumapa, S., Rengpipat, S., Clarke, S. R., Foster, S. J., et al. (2007). Burkholderia Hep_Hag autotransporter (BuHA) proteins elicit a strong antibody response during experimental glanders but not human melioidosis. BMC Microbiol. 7:19. doi: 10.1186/1471-2180-7-19
Valdivia, R. H., and Falkow, S. (1997). Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277, 2007–2011. doi: 10.1126/science.277.5334.2007
Wagar, E. (2016). Bioterrorism and the role of the clinical microbiology laboratory. Clin. Microbiol. Rev. 29, 175–189. doi: 10.1128/CMR.00033-15
Wand, M. E., Muller, C. M., Titball, R. W., and Michell, S. L. (2011). Macrophage and Galleria mellonella infection models reflect the virulence of naturally occurring isolates of B. pseudomallei, B. thailandensis and B. oklahomensis. BMC Microbiol. 11:11. doi: 10.1186/1471-2180-11-11
Wiersinga, W. J., van der Poll, T., White, N. J., Day, N. P., and Peacock, S. J. (2006). Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4, 272–282. doi: 10.1038/nrmicro1385
Wuthiekanun, V., Mayxay, M., Chierakul, W., Phetsouvanh, R., Cheng, A. C., White, N. J., et al. (2005). Detection of Burkholderia pseudomallei in soil within the Lao People’s Democratic Republic. J. Clin. Microbiol. 43, 923–924. doi: 10.1128/JCM.43.2.923-924.2005
Keywords: Burkholderia pseudomallei, intracellular, macrophage, gene expression, promoter trap library, screen, differential fluorescence induction
Citation: Jitprasutwit S, Jitprasutwit N, Hemsley CM, Onlamoon N, Withatanung P, Muangsombut V, Vattanaviboon P, Stevens JM, Ong C, Stevens MP, Titball RW and Korbsrisate S (2020) Identification of Burkholderia pseudomallei Genes Induced During Infection of Macrophages by Differential Fluorescence Induction. Front. Microbiol. 11:72. doi: 10.3389/fmicb.2020.00072
Received: 30 October 2019; Accepted: 14 January 2020;
Published: 21 February 2020.
Edited by:
Axel Cloeckaert, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Natalie Lazar Adler, University of Leicester, United KingdomPaolo Landini, University of Milan, Italy
Copyright © 2020 Jitprasutwit, Jitprasutwit, Hemsley, Onlamoon, Withatanung, Muangsombut, Vattanaviboon, Stevens, Ong, Stevens, Titball and Korbsrisate. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard W. Titball, Ui5XLlRpdGJhbGxAZXhldGVyLmFjLnVr; Sunee Korbsrisate, c3VuZWUua29yQG1haGlkb2wuZWR1
†These authors have contributed equally to this work
 Siroj Jitprasutwit1†
Siroj Jitprasutwit1† Niramol Jitprasutwit
Niramol Jitprasutwit Claudia M. Hemsley
Claudia M. Hemsley Nattawat Onlamoon
Nattawat Onlamoon Patoo Withatanung
Patoo Withatanung Veerachat Muangsombut
Veerachat Muangsombut Joanne M. Stevens
Joanne M. Stevens Mark P. Stevens
Mark P. Stevens Richard W. Titball
Richard W. Titball Sunee Korbsrisate
Sunee Korbsrisate