- 1School of Medical Instrument and Food Engineering, University of Shanghai for Science and Technology, Shanghai, China
- 2The Biotechnology Research Institute, Shanghai Academy of Agricultural Sciences, Shanghai, China
- 3Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Shandong, China
Listeria monocytogenes (LM) is a gram-positive facultative intracellular pathogen that could stimulate host to produce inflammatory response, cell-mediated immunity, and humoral immunity. In this study, an attenuated live vector vaccine for Aeromonas hydrophila (AH) named EGDeABdd-dat-ompW was successfully constructed using an attenuated vector named EGDeABdd, in which dal, dat, actA, and inlB genes were deleted from wild-type LM-EGDe. To construct EGDeABdd-dat-ompW, a recombinant plasmid pERL3-dat-ompW obtained by inserting the dat gene from EGDe and outer membrane protein gene ompW from AH into pERL3 plasmid was transformed into EGDeABdd cell. The safety and immunogenicity of EGDeABdd-dat-ompW as an attenuated vector vaccine for delivery of OMPW were assessed through analyzing invasion to Caco-2 cells and mice, cytokine production of macrophagocyte and mouse splenocytes, and T-cell proliferation of mouse splenocytes. Serum titers against AH and the immunoprotective effect of the vaccine to mice were also measured after intravenous injection with vaccine for four times. The results showed that the live vector vaccine EGDeABdd-dat-ompW for AH exhibited high attenuation in invading Caco-2 cells and mice than did EGDe. Real-time PCR (RT-PCR) showed that cytokines (e.g., TNF-α, IL-6, and IL-1β from macrophages; and IL-6 and IFN-γ from mouse splenocytes) had significantly increased after immunization by EGDeABdd-dat-ompW. Meanwhile, the vaccine could induce the production of CD3+CD4+ and CD3+CD8+ T-cell proliferation of mice and generate effective immunoprotection against lethal challenge of 20 × LD50 AH. All these results indicated that the attenuated EGDeABdd-dat could be used as a live vector for the delivery of the exogenous gene, not only possessing safety but also providing high immunogenicity. The successful application in the AH vaccine further showed that it could be used in other fields such as vaccines in cancer or infectious diseases.
Introduction
Listeria monocytogenes (LM), a gram-positive facultative intracellular pathogen, usually causes listeriosis. Upon invading a host cell, LM could escape from the phagocytic cells such as macrophages and dendritic cells (Alberti-Segui et al., 2007) by producing a pore-forming protein, listeriolysin O (LLO), which lyses the vesicular membrane, allowing LM to enter the cytoplasm, where the secreted proteins by LM are degraded to peptides accessed to major histocompatibility complex (MHC) class I molecules of antigen processing and presentation to CD8+ T cells. The majority of engulfed bacteria are killed by phagolysosomes, and the degraded proteins access directly to MHC class II molecules of antigen presentation to CD4+ T cells (Chen et al., 2014). Meanwhile, LM enhances the antigen presentation effect through the binding of the bacterial wall surface protein and toll-like receptors to antigen-presenting cells. Based on the unique properties mentioned above, it is a promising candidate as a live vector for virus, bacterial disease, or cancer vaccine (Wood and Paterson, 2014).
Aeromonas hydrophila (AH) is widely distributed in various water bodies in nature, which is a typical human–animal–fish comorbid pathogen (Janda and Abbott, 2010). AH could produce highly toxic exotoxins, such as hemolysin, tissue toxins, necrotic toxins, enterotoxins, and proteases, which are responsible for diseases such as hemorrhagic septicemia, dropsy, ulceration, asymptomatic septicemia, and exophthalmos, resulting in high mortality in infected animals and huge economic losses in warm water aquaculture. The preferred way is the use of antibiotic to control the disease in aquaculture, which is presently not recommended owing to the risk involved in the development of resistance in pathogens and the transfer of resistance genes to other animals and human pathogens (Karunasagar et al., 1994). Hence, there is an urgent need to use preventative measures. Vaccination is considered as an alternative strategy to protect fish or animals against diseases. However, one of the major problems in the development of vaccine is how to select an effective antigen.
The bacterial outer membrane proteins (OMPs) play a key role in the virulence of the bacteria and are considered potential vaccine candidates (Maiti et al., 2012). The gene (ompW) of outer membrane protein W is widely present in gram-negative bacteria, such as Vibrio spp. (Jalajakumari and Manning, 1990), Escherichia coli (Li et al., 2016), Salmonella typhimurium (Morales et al., 2012), Aeromonas spp. (Liu et al., 2011), and Cronobacter sakazakii (Ye et al., 2018). It is relatively conservative and has good immunogenicity (Maiti et al., 2010), which is inferred to have the cross-immunoprotective effect.
Because of its the strong ability to survive in wide pH and temperature (Chen et al., 2016) and its potential damage to intestinal epithelial cells and the brain (Salazar et al., 2018), LM used for vector should be attenuated in virulence for safety consideration. In this study, attenuated LM named EGDeABdd, in which the actA, inlB, dal, and dat genes were deleted from wild-type EGDe, was used as the vaccine vector. A recombinant plasmid pERL3-dat-ompW was constructed by cloning the dat and ompW genes into SmaI/SalI and SalI/SacI sites of the pERL3 plasmid, and a LLO promoter and secreted signal peptide sequences were cloned in the upstream of the ompW gene. The nucleic acid vaccine for AH named EGDeABdd-dat-ompW was successfully constructed by electroporation of the recombinant plasmid pERL3-dat-ompW into EGDeABdd. The safety and immunogenicity of the EGDeABdd-dat-ompW as a vaccine was analyzed.
Materials and Methods
Materials
Attenuated strain EGDeABdd based on wild-type LM genetic background EGDe (BAA-679, serotype 1/2a) was constructed in our laboratory. Wild-type Aeromonas hydrophila (CPBC-Ah-0003) was a gift from Dr. Xincang Li (East China Sea Fisheries Research Institute). Plasmid pERL3 was a gift from Professor Qin Luo (Huazhong University of Science and Technology). Primers for gene amplification were synthesized by Sangon (Shanghai, China) listed in Table 1. Six- to 8-week-old Balb/c (in safety evaluation assay) and C57BL/6 mice (in immunity evaluation assay) were purchased from JieSijie (Shanghai, China). All animal studies have been approved by China Ethics Committee and performed in accordance with the ethical standards. Macrophage cell lines Raw264.7 were purchased from Shanghai Cell Bank (Shanghai, China).
Fluorescence-conjugated anti-mouse antibodies CD3-PerCP-Cy5.5, CD4-FITC, and CD8-PE were from Becton Dickinson (Franklin Lakes, NJ, United States). Brain heart infusion (BHI) medium was purchased from Land Bridge (Beijing, China). Real-time PCR (RT-PCR)-related reagents and rapid cloning kit were from TaKaRa (Dalian, China). PCR and RT-PCR thermal cycler were from Applied Biosystems (United States). Electroporation instrument was from Bio-Rad (United States). Other chemicals were of analytical reagent grade.
Strain Construction
Construction of EGDeABdd-t Strain
The gene dat of LM was amplified by PCR using the primers T1 and T2 and cloned into pERL3 at the SmaI and SalI sites as shown in Figure 1. The plasmid named pERL3-dat was electroporated into EGDeABdd to construct the strain named EGDeABdd-t. Positive clones were picked from BHI agar plates and identified by PCR using the primers T3 and T4.
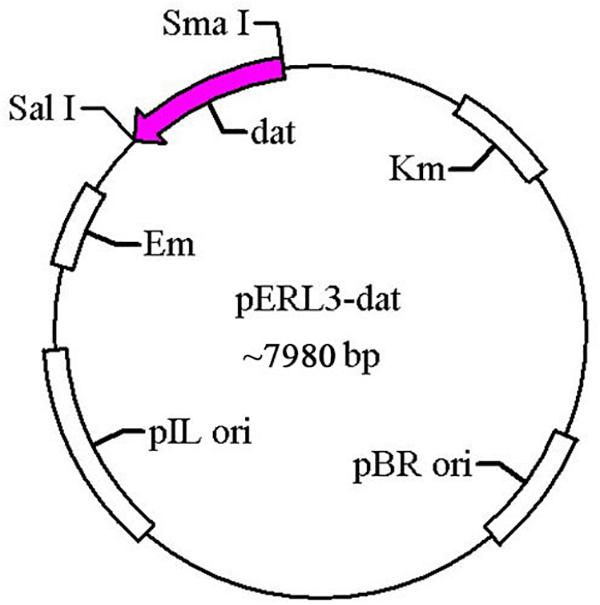
Figure 1. Schematic diagram of recombinant plasmid pERL3-dat. The gene dat from EGDe was amplified using primers T1 and T2 and inserted at SmaI/SalI sites.
Growth Kinetics of EGDeABdd-t
To study the growth kinetics of the attenuated strain EGDeABdd-t in the absence of D-alanine, the mutant cultured in the presence of 100 μg/ml of D-alanine was washed to remove extracellular D-alanine and suspended in BHI medium without D-alanine. Optical density values (OD values) of bacteria at 600 nm were measured per hour using microplate reader (SpectraMax M2; Molecular Devices, Sunnyvale, CA, United States).
Construction of EGDeABdd-dat-ompW Strain
The gene ompW of AH and LLO promoter (truncated LLO, containing 45 amino acids) sequences amplified by PCR using the W1/W2 and L1/L2 primers were spliced by overlap extension PCR, following by insertion at of the SalI/SacI sites of recombinant plasmid pERL3-dat as shown in Figure 2. The new recombinant plasmid named pERL3-dat-ompW was electroporated into EGDeABdd to construct a vaccine strain named EGDeABdd-dat-ompW. Clones identified as positive by PCR using primers W1 and W2 were also determined by sequence analysis.
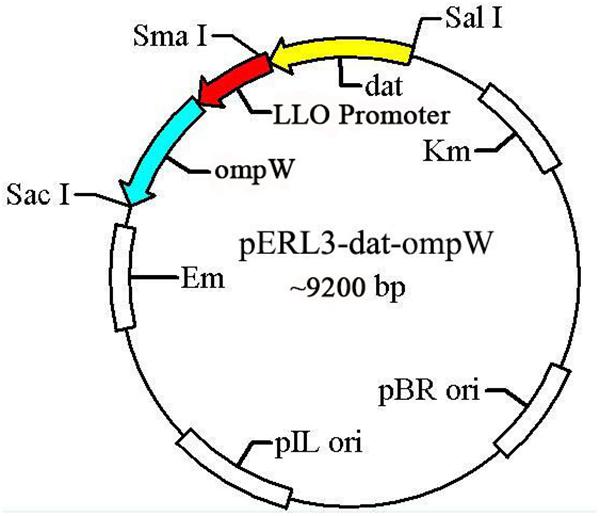
Figure 2. Schematic diagram of recombinant plasmid pERL3-dat-ompW. Listeriolysin O (LLO) promoter sequences of hly and ompW of AH were amplified using primers L1/L2 and W1/W2, respectively. Two fragments were linked using overlap extension PCR, following by insertion at SalI and SacI sites.
Analysis of Protein Secreted by EGDeABdd-dat-ompW
The expression and secretion of the truncated LLO and OMPW (tLLO-OMPW, containing 204 amino acids of OMPW, 45 amino acids of tLLO, and 6 histidine tag) fusion protein was confirmed in the culture supernatants by western blot using anti-His label antibody according to the related literatures. EGDeABdd-dat was the control, which was constructed from EGDeABdd containing recombinant plasmid pERL3-dat-tLLO.
Gene Expression of Strain Detection in Transcription Level
To analyze the difference of the constructs in gene expression, 12 virulence genes and 2 genes related to D-alanine synthesis were analyzed by RT-PCR. Five milliliters of saturated bacterial solution was used to extract RNA. According to kit instructions, the RNA was reverse transcribed into cDNA. RT-PCR was performed using cDNA as a template. The data were processed by 2–ΔΔ Ct method for relative quantitative analysis, with the gene expression level of EGDe as a control and 16S rRNA as an internal reference. The calculation formula is as follows. ΔCt = Ct mean value of strain target gene – Ct mean value of corresponding 16S rRNA, and ΔΔCt = ΔCt of mutant target gene −ΔCt of the EGDe corresponding target gene. 2–ΔΔ Ct is the relative expression level of mutant virulence gene to EGDe.
Safety Assessment
In vitro Infection to Caco-2 Cells
The human Caco-2 cell lines were used to evaluate the invasiveness of vaccine strains. Cells cultured in a 12-well plates at 105 cells/well were infected by EGDe, EGDeAB-dat, and EGDeABdd-dat-ompW at multiplicity of infection (MOI) = 1:100. After incubation at 37°C for 2 h, the free or adherent bacteria were removed and killed by phosphate-buffered saline (PBS) washing for two times following by 200 μg/ml of penicillin incubation at 37°C for 30 min. The infected cells were lysed in 1% Triton X-100 (Sigma, St. Louis, MO, United States) for 15 min and then gradient diluted to culture in BHI plates containing 200 μg/ml of D-alanine and 5 μg/ml of erythromycin. Colony-forming units (CFUs) were counted 24 h later.
The Ability-Induced Caco-2 Cell Apoptosis
After co-culture with EGDe, EGDeAB-dat, and EGDeABdd-dat-ompW for 3 h, Caco-2 cells were harvested and stained with propidium iodide (PI) staining buffer to identify late apoptotic and necrotic cells. The cells were also stained with fluorescein isothiocyanate (FITC)-conjugated annexin V for 15 min to evaluate early apoptosis. Cells of different periods were then analyzed using flow cytometry (Becton Dickinson, Franklin Lakes, NJ, United States).
In vivo Infection to Mice
A dose of 2 × 107 CFUs EGDe, EGDeAB-dat, and EGDeABdd-dat-ompW were intravenously injected into 6- to 8-week-old Balb/c mice (n = 3). On the first day after injection, the spleen and liver were collected from the mice. The single-cell suspension was serially diluted and cultured on chromogenic agar plates. The CFUs per gram for organs were calculated to determine the reduced virulence of a vaccine to mice.
Mouse Liver Slices
Three days after intravenous injection of EGDeABdd-dat-ompW with a dose of 5 × 107 CFUs (0.1 × 50% lethal dose, LD50 about 5 × 108 CFUs), the livers were collected and fixed in formalin for 24 h followed by paraffin embedding, slicing, and hematoxylin and eosin (H&E) staining. Histological morphology of the livers was observed under the microscope. PBS with equal volume was the control.
Immunological Evaluation
Cytokine Measurement
Infections by pathogenic microorganisms elicit host immune responses, especially including inflammation, as innate immunity is responsible typically for initial infection-directed responses. After co-culture at MOI = 100:1 in the presence of EGDeABdd-dat or EGDeABdd-dat-ompW for 3 h, macrophages Raw264.7 were harvested. RNA was extracted, and RT-PCR was performed to analyze the relative expression of cytokines. The C57BL/6 mice were intravenously immunized with EGDeABdd-dat or EGDeABdd-dat-ompW of 5 × 107 CFUs (0.1 × LD50, LD50 about 5 × 108 CFUs), and PBS was the control. On the first day after immunization, mouse spleens were collected, and the RNA was extracted. After reverse transcription of RNA to cDNA, RT-PCR was carried out. The cytokine expression in PBS group was considered to be 1, which was used as control.
Cytokine detection kit based on sandwich ELISA was used for quantitative analysis of IL-6 from macrophages and INF-γ from mice according to instructions. The C57BL/6 mice were intravenously injected with EGDeABdd-dat or EGDeABdd-dat-ompW of 5 × 107 CFUs, and PBS was the control. On the third day after immunization, mouse serum was collected as antigen for sandwich ELISA.
CD4+ and CD8+ T-Cell Detection
Before T-cell detection, the mice were immunized for three times; 5 × 107 CFUs of EGDeABdd-dat and EGDeABdd-dat-ompW were intravenously injected into mice (Yang et al., 2014) at 0, 7, and 14 days. PBS with equal volume was the control. One week after the third immunization, the splenocytes were harvested from the mice and stained by the fluorescence-conjugated anti-mouse antibodies CD3-PerCP-Cy5.5, CD4-FITC, and CD8-PE for 15 min. Before detection, unstained and single-stained CD3, CD4, and CD8 cells were used to set machine parameters and compensation. Then, the lymphocyte population is gated according to the cell size on the scatter plot. Finally, cells simultaneously stained with three antibodies flowed through the detector and are displayed on the CD4/CD8 scatter plot.
Mouse Immunization and Aeromonas hydrophila Challenge
The mice were immunized by intravenous injection with EGDeABdd-dat-ompW or EGDeABdd-dat strain of 5 × 107 CFUs (0.1 × LD50), and PBS was the blank control. One week after the fourth immunization, the blood of three mice in each group was collected to detect the antibody produced against AH using the indirect ELISA, and other mice were challenged with AH of 2 × 106 CFUs (20 × LD50, LD50 about 105 CFUs). The immune interval was 1 week, and the immunization plan is shown in Figure 3.
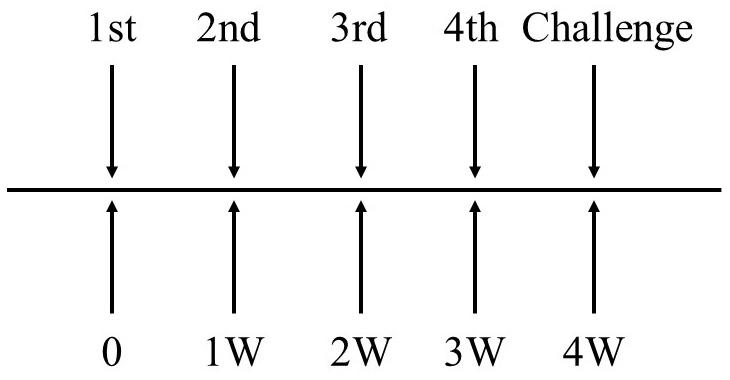
Figure 3. The schematic diagram of immunization plan to mice. Immune dose of EGDeABdd-dat-ompW or EGDeABdd-dat strain was 5 × 107 CFUs (about 0.1 × LD50), and immune interval was 1 week. The mice were immunized four times. One week after the fourth immunization, mice were challenged with AH or EGDe of 20 × LD50.
The same experiment was performed, and the mice were challenged with LM EGDe of 2 × 107 CFUs (20 × LD50).
Statistical Analysis
Results of each experiment are analyzed by GraphPad Prism 5 software (San Diego, CA, United States) to compare the differences between groups by one-way analysis of variance followed by an unpaired t-test. Values of P < 0.05,P < 0.01, or P < 0.001 were considered significant (∗), highly significant (∗∗), or extremely significant (∗∗∗). Data represent means ± standard deviation (SD).
Results
Strain Construction and Analysis
Strain Construction of EGDeABdd-t
The growth curve of plasmid-supplemented strain was determined by microplate reader at 600 nm every 1 h, and the results are shown in Figure 4. The nutrient mutant EGDeABdd could not survive in normal medium, whereas the plasmid-supplemented strain EGDeABdd-t restored the abilities of growth and reproduction without exogenous addition of D-alanine. The results indicated the gene dat in plasmid successfully expressed the D-alanine transaminase protein to supplement the D-alanine and regulated bacterial growth.
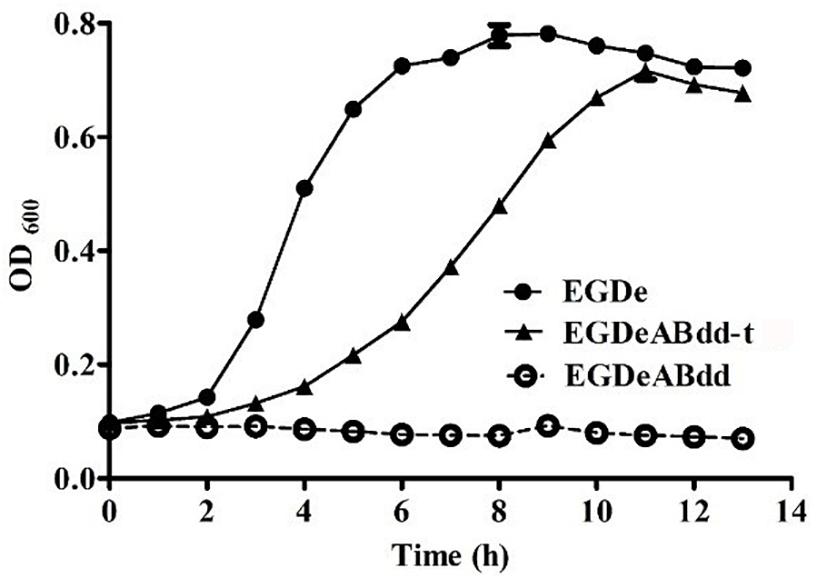
Figure 4. Growth curve of bacteria in normal brain heart infusion (BHI) medium. EDGe was wild-type Listeria monocytogenes (LM). EDGeABdd was a mutant in which the actA, inlB, dal, and dat genes were deleted. EDGeABdd-t was constructed from EDGeABdd containing a pERL3-dat plasmid. EDGeABdd-t could survive in normal BHI without D-alanine.
Protein Expression of the Vaccine Strain EGDeABdd-dat-ompW
The BHI supernatant of the vaccine strain was collected after overnight culture, and the protein in the supernatant was precipitated using pre-cooled ethanol. The western blot method was used to detect the level of protein expression. As shown in Figure 5, the fusion protein (tLLO-OMPW) was observed in about 28 kDa (lane 2), which was consistent with estimated size. The control EGDeABdd-dat has no observed band (lane 1). The results indicated the exogenous gene ompW was successfully expressed and secreted to the supernatant of the medium.
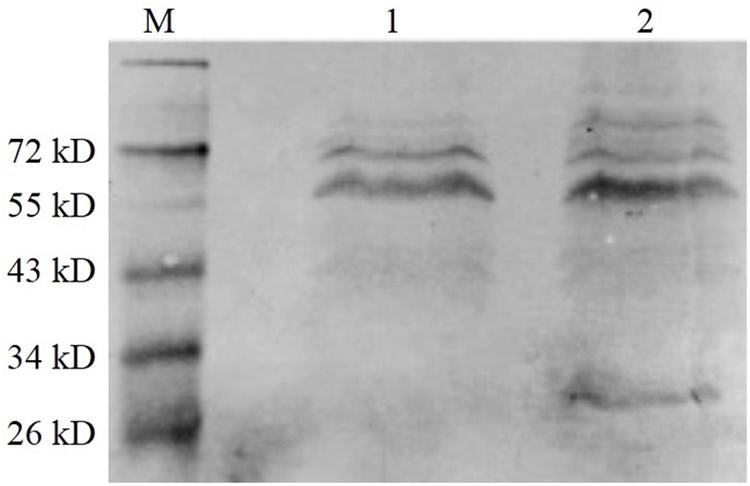
Figure 5. Western blot analysis of exogenous OMPW expression. M was the marker. (1) The control strain EGDeABdd-dat containing recombinant plasmid pERL3-dat-tLLO. (2) The vaccine strain EGDeABdd-dat-ompW containing recombinant plasmid pERL3-dat-ompW. The fusion protein containing 204 AA of OMPW, 45 AA of tLLO, and 6 His-tag has about 28 kDa. Proteins in culture supernatant were collected using cold ethanol precipitation.
Gene Expression Detection in Transcription Levels
The expression levels of virulence gene were detected by qRT-PCR (Ma’ayeh et al., 2018). As shown in Figure 6, the genes including inlA, prfA, and sigB had no significant difference between four strains, whereas the plcA and plcB genes were significantly increased (value > 1) in strains of EGDeABdd, EGDeABdd-dat, and EGDeABdd-dat-ompW compared with EGDe. Meanwhile, the gene hly in EGDeABdd was significantly increased compared with the other three strains. The gene dat in EGDeABdd-dat and EGDeABdd-dat-ompW and the gene ompW in EGDeABdd-dat-ompW had a high expression. No detection signals were observed in the actA, inlB, dal, and dat genes of EGDeABdd, which was further indicated that these genes were successfully deleted.
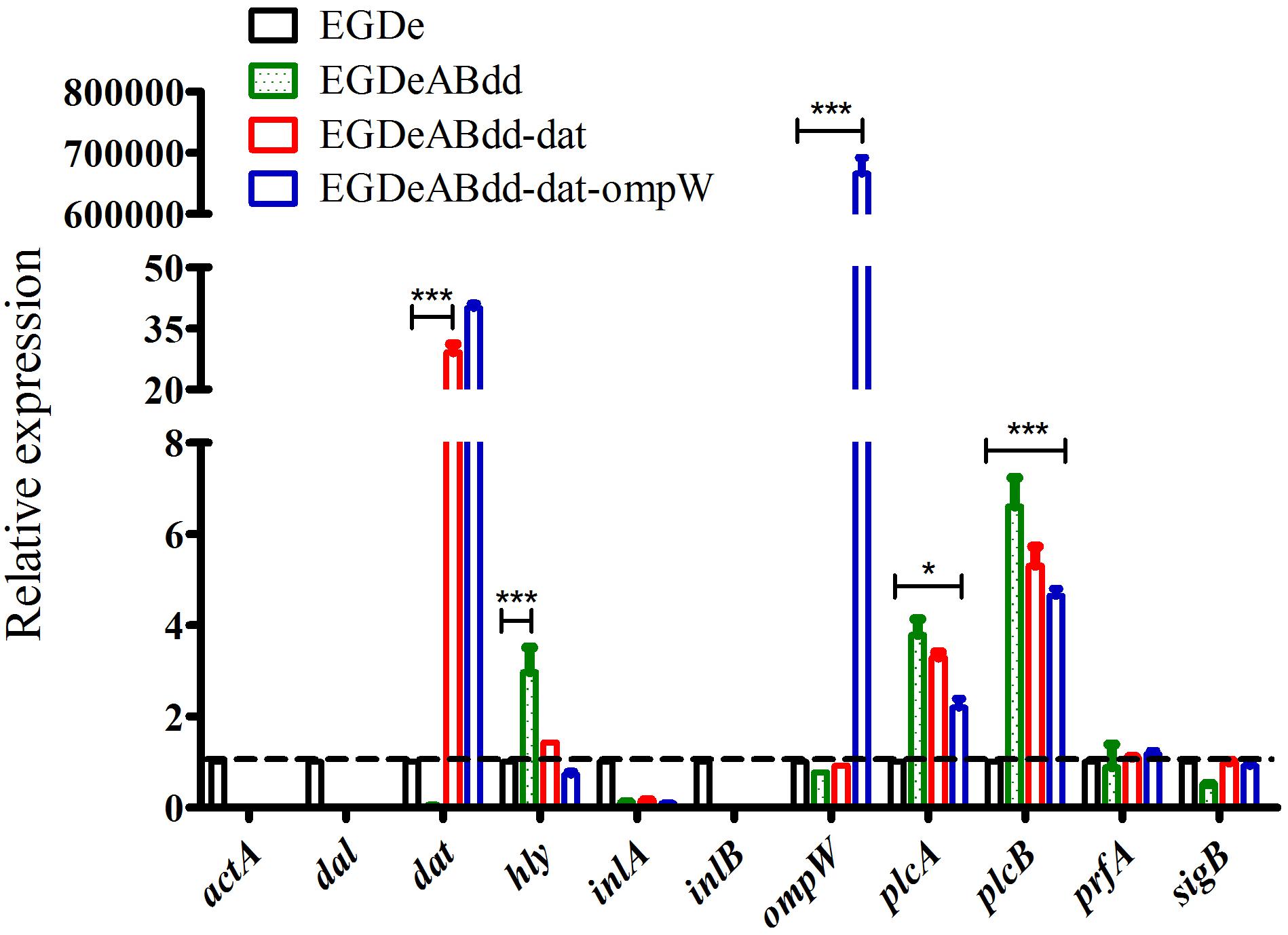
Figure 6. RT-PCR detection of virulence gene relative expression levels. Gene transcription of EGDe was considered to be 1, and others were compared with it. Value > 1 indicated gene high expression; otherwise, it represented low expression. *P < 0.05; ***P < 0.001.
Safety Assessment
Cell Infection and Induced Caco-2 Apoptosis
Caco-2 is colorectal cancer cell line and usually used to simulate the bacterial infection ability of penetrating the intestinal barrier. After incubation with EGDe, EGDeABdd-dat, and EGDeABdd-dat-ompW at MOI = 1:100 for 2 h, extracellular bacteria were killed by penicillin, and then the intracellular bacteria were obtained by plate counting. As shown in Figure 7A, the invasion ability to cells of the vaccine strain EGDeABdd-dat-ompW decreased significantly compared with EGDe. Meanwhile, results indicated that introducing the ompW gene could not cause changes in toxicity, especially in an increase in virulence to cells than could EGDeABdd-dat.
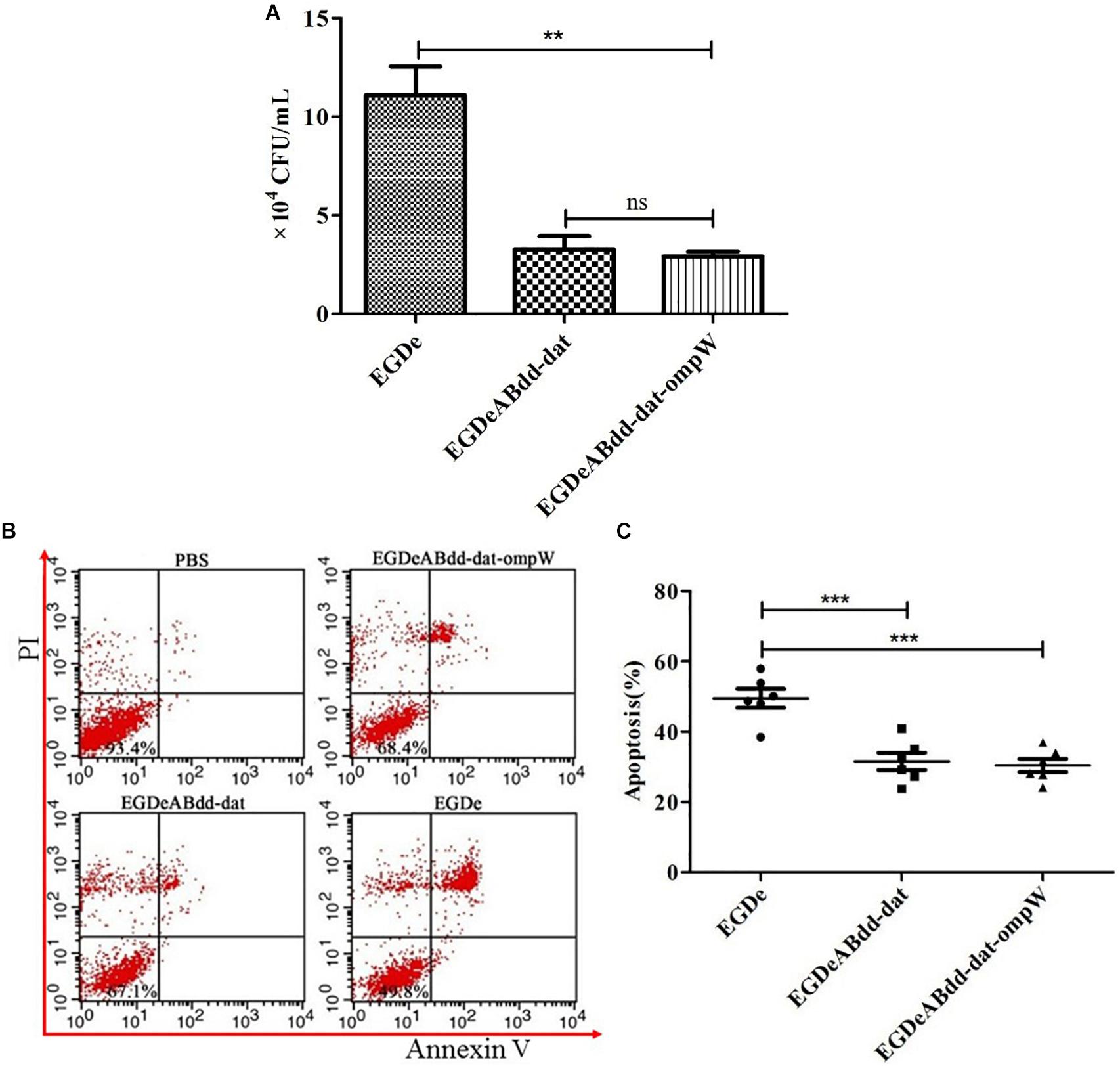
Figure 7. Cytotoxicity of EGDe, EGDeABdd-dat, and EGDeABdd-dat-ompW to Caco-2 cells. (A) Number of bacteria invading into cells. The Caco-2 cells were cultured in 12-well plates at 1.0 × 105 cells/well and infected with EGDe, EGDeABdd-dat, and EGDeABdd-dat-ompW at multiplicity of infection (MOI) = 1:100. The infected cells were lysed in 1% Triton X-100 and diluted. The dilutions were counted in brain heart infusion (BHI) plates. (B) Impact of EGDe, EGDeABdd-dat, and EGDeABdd-dat-ompW on apoptosis of Caco-2 cells assessed by flow cytometry. Caco-2 cells were invaded by strains for 3 h. The cells were stained with propidium iodide (PI) and fluorescein isothiocyanate (FITC)-conjugated annexin V for 15 min. (C) The rates of apoptosis of EGDe, EGDeABdd-dat, and EGDeABdd-dat-ompW to Caco-2 cells. EGDeABdd-dat and EGDeABdd-dat-ompW had a lower rate of apoptosis than EGDe. **P < 0.01; ***P < 0.001.
Annexin V-FITC/PI apoptosis detection kit and flow cytometry were used for cell staining and signal detection, respectively. From Figure 7B, living cells, early apoptotic cells, late apoptotic cells, and necrotic cells were distributed in annexin V−/PI−, annexin V+/PI−, annexin V+/PI+, and annexin V-/PI + regions, respectively. Results of apoptosis are shown in Figure 7C. From Figures 7B,C, the trend of bacteria inducing Caco-2 cell apoptosis was consistent with infection ability to cells. All the results indicated that the vaccine strain with low cytotoxicity was much safer than EGDe.
In vivo Infection
After invasion by bacteria for 24 h, the spleen and liver of mice were harvested, and the CFUs per gram were obtained as shown in Figure 8A. The EGDeABdd-dat and EGDeABdd-dat-ompW strains had significantly lower distribution levels in the spleen and liver, which further indicated that the virulence of the vaccine strain was significantly weakened. Liver section and H&E staining results are shown in Figure 8B, there was only a very mild inflammatory response, and no tissue damage appeared.

Figure 8. Safety evaluation of vaccine strain to mice. (A) Organ distribution of EGDeABdd-dat and EGDeABdd-dat-ompW. Strains [2 × 107 colony-forming units (CFUs)] were intravenously administered to mice, and CFUs per gram in the spleen and liver were counted on the first day after injection. (B) Histological morphology of liver from mice immunized by EGDeABdd-dat-ompW. Immunized mice were sacrificed 3 days after intravenous injection of 5.0 × 107 CFUs EGDeABdd-dat-ompW. The liver was collected for H&E staining. Slight inflammatory responses were observed in some hepatocytes, and no tissue necrosis or structural damage was seen. **P < 0.01; ***P < 0.001.
Immunization Evaluation
Cytokine Analysis
Results of cytokines of macrophages Raw264.7 and mice detected by RT-PCR are shown in Figures 9A,B. TNF-α, IL-6, and IL-1β from macrophages had significantly increased after co-culture with strains for 3 h. One day after immunization of mice, IL-6 and IFN-γ of splenocytes had significantly increased. The quantitative analysis of IL-6 of macrophages and IFN-γ of mouse splenocytes using sandwich ELISA are shown in Figure 9C. The concentration of IL-6 and IFN-γ could reach to 90 and 130 pg/ml.

Figure 9. Cytokine analysis of macrophages Raw264.7 and mouse splenocytes by RT-PCR and ELISA. (A) Cytokines of macrophages detected by RT-PCR. The multiplicity of infection (MOI) of bacteria and cells was 100:1, and incubation time was 3 h. (B) Cytokines of mouse splenocytes detected by RT-PCR. A dose of 5 × 107 colony-forming units (CFUs) of bacteria was injected into C57BL/6 mice, and phosphate-buffered saline (PBS) was the control. On the first day after immunization, the splenocytes were collected for RNA extraction. (C) Quantitative analysis of IL-6 from macrophages and IFN-γ from mouse splenocytes using sandwich ELISA. The mice were immunized by 5 × 107 CFUs of bacteria. On the third day after immunization, mouse serum was collected as antigen for sandwich ELISA. **P < 0.01; ***P < 0.001.
T-Cell Immune Responses Analyzed by Flow Cytometry
Mice’s CD3+CD4+ and CD3+CD8+ T-cell immune responses to bacteria were analyzed by flow cytometry. As shown in Figure 10, the percentage of CD4+ AND CD8+ T cells in EGDeABdd-dat-ompW group was significantly increased than those in PBS group, which indicated that the vaccine with low cytotoxicity could induce T-cell immune response to promote T-cell proliferation and activate the MHC class I and MHC class II pathways to present antigens.
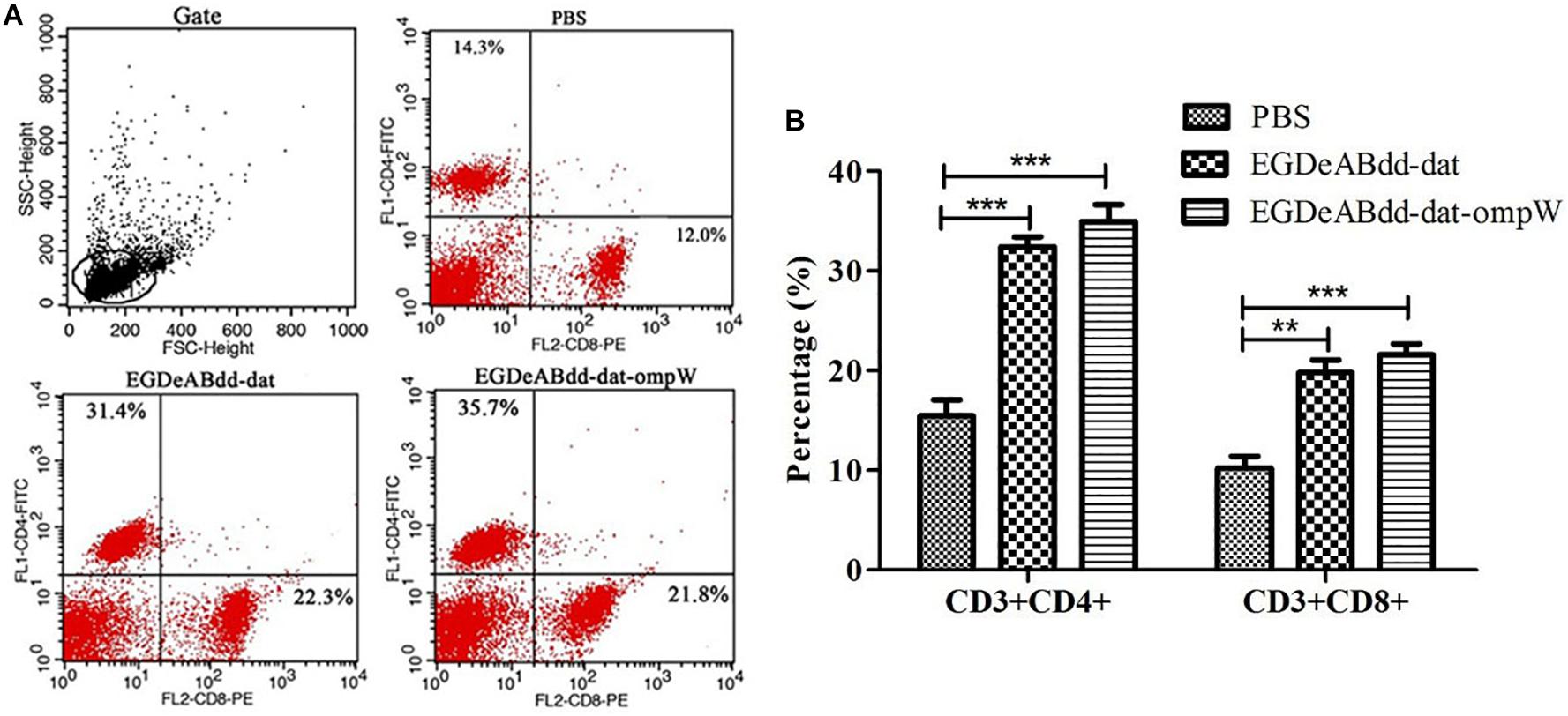
Figure 10. Mouse T-cell immune responses to different bacteria analyzed by flow cytometry. (A) Flow cytometry analysis of splenocyte proliferation. One week after immunization with EGDeABdd-dat and EGDeABdd-dat-ompW for three times using a dose of 5.0 × 107 CFUs (0.1 LD50), the splenocytes were collected and stained with CD3, CD4, and CD8 antibodies. (B) The percentage of CD4+ and CD8+ T cells. An increase of T cells appeared in EGDeABdd-dat and EGDeABdd-dat-ompW groups. **P < 0.01; ***P < 0.001.
Mouse Serum Titers and Immune Protection
One week after the fourth immunization, the serum titers from different groups were measured by indirect ELISA. Indirect ELISA was performed as follows (Zeng et al., 2016). Gradient serum was added to a 96-well plate coated with antigen (AH) for incubation at 37°C for 1 h. After being washed for three times, horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody was added and reacted for 1 h. After being washed for three times, the TMB substrate was added for 15 min. The reaction was terminated by 2 M of sulfuric acid. Absorbance values were measured at 450 nm. When the values were higher than 2.1 × negative values, the results were judged to be positive, and the maximum dilution of serum was the titer of the mouse. As shown in Figure 11A, serum titers of EGDeABdd-dat-ompW group could reach to 1:100, whereas the titers of EGDeABdd-dat group were 1:10.

Figure 11. Analysis of humoral immune response and immunoprotective effect. (A) Serum titers of mice against Aeromonas hydrophila (AH) immunized with EGDeABdd-dat and EGDeABdd-dat-ompW. T1–T3, mice immunized with EGDeABdd-dat for four times using a dose of 5 × 107 colony-forming units (CFUs), and the immune interval was 1 week. W1–W3, mice immunized with EGDeABdd-dat-ompW for four times using a dose of 5 × 107 CFUs, and the interval was 1 week. (B) Immunoprotective effect of phosphate-buffered saline (PBS), EGDeABdd-dat, and EGDeABdd-dat-ompW groups to AH. One week after the fourth immunization, serum collection and challenge were performed. The mice in PBS, EGDeABdd-dat, and EGDeABdd-dat-ompW groups were challenged with AH of 2 × 106 CFUs. (C) Mice were challenged with Listeria monocytogenes (LM) EGDe using a dose of 2 × 107 CFUs. The EGDeABdd-dat and EGDeABdd-dat-ompW groups could provide a 100% protective effect.
On the same day, the immunized mice in PBS, EGDeABdd-dat, and EGDeABdd-dat-ompW groups were challenged with AH with a dose of 2 × 106 CFUs (20 × LD50), and the results are shown in Figure 11B. All the mice in PBS group died within 2 days, whereas the EGDeABdd-dat and EGDeABdd-dat-ompW groups could provide an immunoprotective effect of 18 and 58%, respectively, after immunization for four times.
The immunized mice in PBS, EGDeABdd-dat, and EGDeABdd-dat-ompW groups were also challenged with LM EGDe with a dose of 2 × 107 CFUs (20 × LD50), and the results are shown in Figure 11C. All the mice in PBS group died within a week, whereas the EGDeABdd-dat and EGDeABdd-dat-ompW groups could provide a 100% immunoprotective effect.
Discussion
LM has been shown to have an exceptional capacity to induce anti-tumor immunity to tumor-associated antigens, such as HPV16 E7 (Jia et al., 2017), influenza virus NP3 (Pan et al., 1999), HIV-1 gag (Okuda et al., 1995; Jiang et al., 2007), tuberculosis (Yin et al., 2017), metastatic pancreatic cancer (Le et al., 2016), and canine osteosarcoma (Mason et al., 2016). In this study, an AH vaccine using attenuated LM as delivery vector was described, and the safety and the immunogenicity of the vaccine were assessed. The attenuated LM vector of the dal, dat, actA, and inlB gene deletions harbors a plasmid to express antigens, and in this study, the dat gene from LM and AH antigen OMPW was successfully implemented. By plasmid supplementation of the dat gene in auxotrophic mutant EGDeABdd, recombinant plasmid could stably exist in a strain without antibiotic screening. Owing to the extensive distribution in a variety of bacteria, such as LM, Escherichia coli, and Bacillus subtilis, the dal gene was considered more important than the dat gene in LM. The construction concept of the carrier was to replenish the dal gene of B. subtilis in previous studies (Yang et al., 2014), whereas the gene dat of LM selected for plasmid carrying was successfully implemented in this study.
The results of in vivo and in vitro studies provided evidence demonstrating that the vaccine strain had less virulence than had EGDe. The results of immunization experiments suggested that the vaccine had the ability to present antigen to the immune system and could provide 58% immunoprotective effect higher than 18% of the vector group. Meanwhile, the vaccine could induce the expansion of CD3+CD4+ T cells and CD3+CD8+ T cells; thus, it could induce immune responses to kill cells infected by bacteria. All the results indicated that the EGDeABdd-dat-ompW strain used as a vaccine for AH was safe and highly immunogenic.
An ideal LM vector vaccine designed for animal or human use has been achieved, which the bacterium is less virulent yet still retains its ability to present antigen to the immune system. Owing to the reduced virulence and increased safety, the dal, dat, actA, and inlB deletion strains were superior to the dal/dat deletion or actA/inlB deletion strain as a vaccine vector. In conclusion, the EGDeABdd-dat-ompW vaccine could stably secrete the OMPW, an OMP of AH, as a protective antigen to elicit a specific immune response. EGDeABdd-dat-ompW strain is a safe and effective vaccine that could stimulate host to produce antibody in serum against AH and improve survival when challenged with AH. The successful application of strain EGDeABdd-dat in an AH vaccine, indicating EGDeABdd-dat strain as a vaccine vector in future clinical use, will be realized soon. The broader goal of the study is to extend the application of the live attenuated vector to designing potential constructs expressing other single or multiple heterologous antigens.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
All animal studies have been approved by the China Ethics Committee and performed in accordance with the ethical standards.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. HZ was responsible for experimental operation and article writing. MX, CD, and JM gave experimental help. DX, XW, and JQ gave article modification help. QL designed the experiment.
Funding
The research was supported by the Agricultural Science Promotion Plan of Shanghai (2017, no. 4-4), the Key Supporting Program of Science and Technology in Shanghai (13430502400), and the National Natural Science Foundation of China (no. 31871897).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alberti-Segui, C., Goeden, K. R., and Higgins, D. E. (2007). Differential function of Listeria monocytogenes listeriolysin O and phospholipases C in vacuolar dissolution following cell-to-cell spread. Cell. Microbiol. 9, 179–195.
Chen, Y., Allard, E., Wooten, A., Hur, M., Sheth, I., Laasri, A., et al. (2016). Recovery and growth potential of Listeria monocytogenes in temperature abused milkshakes prepared from naturally contaminated ice cream linked to a listeriosis outbreak. Front. Microbiol. 7:764. doi: 10.3389/fmicb.2016.00764
Chen, Z., Ozbun, L., Chong, N., Wallecha, A., Berzofsky, J. A., and Khleif, S. N. (2014). Episomal expression of truncated listeriolysin O in LmddA-LLO-E7 vaccine enhances antitumor efficacy by preferentially inducing expansions of CD4+FoxP3- and CD8+ T cells. Cancer Immunol. Res. 2:911. doi: 10.1158/2326-6066.CIR-13-0197
Jalajakumari, M. B., and Manning, P. A. (1990). Nucleotide sequence of the gene, ompW, encoding a 22kDa immunogenic outer membrane protein of Vibrio cholerae. Nucleic Acids Res. 18:2180.
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Jia, Y. Y., Tan, W. J., Duan, F. F., Pan, Z. M., Chen, X., Yin, Y. L., et al. (2017). A genetically modified attenuated listeria vaccine expressing HPV16 E7 kill tumor cells in direct and antigen-specific manner. Front. Cell. Infect. Microbiol. 7:279. doi: 10.3389/fcimb.2017.00279
Jiang, S., Rasmussen, R. A., Nolan, K. M., Frankel, F. R., Lieberman, J., Mcclure, H. M., et al. (2007). Live attenuated Listeria monocytogenes expressing HIV Gag: immunogenicity in rhesus monkeys. Vaccine 25, 7470–7479.
Karunasagar, I., Pai, R., Malathi, G. R., and Karunasagar, I. (1994). Mass mortality of Penaeus monodon larvae due to antibiotic-resistant Vibrio harveyi infection. Aquaculture 128, 203–209.
Le, D. T., Wanggillam, A., Picozzi, V., Greten, T. F., Crocenzi, T., Springett, G., et al. (2016). Safety and survival with GVAX pancreas prime and Listeria Monocytogenes–expressing mesothelin (CRS-207) boost vaccines for metastatic pancreatic cancer. J. Clin. Oncol. 33, 1325–1333. doi: 10.1200/JCO.2014.57.4244
Li, W., Wen, L., Li, C., Chen, R., Ye, Z., Zhao, J., et al. (2016). Contribution of the outer membrane protein OmpW in Escherichia coli to complement resistance from binding to factor H. Microb. Pathog. 98, 57–62. doi: 10.1016/j.micpath.2016.06.024
Liu, M. Z., Ye, X., and Deng, G. C. (2011). Expression and immunogenicity analysis of the outer membrane protein W gene of Aeromonas hydrophila. Microbiol. China 38, 437–445.
Ma’ayeh, S. Y., Knörr, L., Sköld, K., Granham, A., Ansell, B. R. E., Jex, A. R., et al. (2018). Responses of the differentiated intestinal epithelial cell line caco-2 to infection with the Giardia intestinalis GS Isolate. Front. Cell. Infect. Microbiol. 8:244. doi: 10.3389/fcimb.2018.00244
Maiti, B., Raghunath, P., Karunasagar, I., and Karunasagar, I. (2010). Cloning and expression of an outer membrane protein OmpW of Aeromonas hydrophila and study of its distribution in Aeromonas spp. J. Appl. Microbiol. 107, 1157–1167. doi: 10.1111/j.1365-2672.2009.04296.x
Maiti, B., Shetty, M., Shekar, M., Karunasagar, I., and Karunasagar, I. (2012). Evaluation of two outer membrane proteins, Aha1 and OmpW of Aeromonas hydrophila as vaccine candidate for common carp. Vet. Immunol. Immunopathol. 149, 298–301. doi: 10.1016/j.vetimm.2012.07.013
Mason, N., Gnanandarajah, J., Engiles, J., Gray, F., Laughlin, D., Gaurnier-Hausser, A., et al. (2016). Immunotherapy with a HER2 targeted listeria induces HER2-specific immunity and demonstrates potential therapeutic effects in a phase I trial in canine osteosarcoma. Clin. Cancer Res. 22, 4380–4390.
Morales, E. H., Calderón, I. L., Collao, B., Gil, F., Porwollik, S., Mcclelland, M., et al. (2012). Hypochlorous acid and hydrogen peroxide-induced negative regulation of Salmonella enterica serovar Typhimurium ompW by the response regulator ArcA. BMC Microbiol. 12:63. doi: 10.1186/1471-2180-12-63
Okuda, K., Bukawa, H., Hamajima, K., Kawamoto, S., Sekigawa, K., Yamada, Y., et al. (1995). Induction of potent humoral and cell-mediated immune responses following direct injection of DNA encoding the HIV type 1 env and rev gene products. Aids Res. Hum. Retrovirus. 11, 933–943.
Pan, Z. K., Weiskirch, L. M., and Paterson, Y. (1999). Regression of established B16F10 melanoma with a recombinant Listeria monocytogenes vaccine. Cancer Res. 59, 5264–5269.
Salazar, J. K., Bathija, V. M., Carstens, C. K., Narula, S. S., Shazer, A., Stewart, D., et al. (2018). Listeria monocytogenes growth kinetics in milkshakes made from naturally and artificially contaminated ice cream. Front. Microbiol. 9:62. doi: 10.3389/fmicb.2018.00062
Wood, L. M., and Paterson, Y. (2014). Attenuated Listeria monocytogenes: a powerful and versatile vector for the future of tumor immunotherapy. Front. Cell. Infect. Microbiol. 4:51. doi: 10.3389/fcimb.2014.00051
Yang, Y., Hou, J., Lin, Z., Zhuo, H., Chen, D., Zhang, X., et al. (2014). Attenuated Listeria monocytogenes as a cancer vaccine vector for the delivery of CD24, a biomarker for hepatic cancer stem cells. Cell. Mol. Immunol. 11:184. doi: 10.1038/cmi.2013.64
Ye, Y., Ling, N., Gao, J., Zhang, M., Zhang, X., Tong, L., et al. (2018). Short communication: roles of outer membrane protein W (OmpW) on survival and biofilm formation of Cronobacter sakazakii under neomycin sulfate stress. J. Dairy Sci. 101, 2927–2931. doi: 10.3168/jds.2017-13517
Yin, Y., Lian, K., Zhao, D., Tao, C., Chen, X., Tan, W., et al. (2017). A promising listeria-vectored vaccine induces Th1-type immune responses and confers protection against tuberculosis. Front. Cell. Infect. Microbiol. 7:407. doi: 10.3389/fcimb.2017.00407
Keywords: Listeria monocytogenes, attenuation, live vector vaccine, Aeromonas hydrophila, safety
Citation: Zeng H, Xie M, Ding C, Ma J, Xu D, Wang X, Qiu J and Liu Q (2020) Attenuated Listeria monocytogenes as a Vaccine Vector for the Delivery of OMPW, the Outer Membrane Protein of Aeromonas hydrophila. Front. Microbiol. 11:70. doi: 10.3389/fmicb.2020.00070
Received: 29 January 2019; Accepted: 14 January 2020;
Published: 21 February 2020.
Edited by:
Ian Marriott, University of North Carolina at Charlotte, United StatesReviewed by:
Hridayesh Prakash, Amity University, IndiaCarmen Alvarez-Dominguez, Marqués de Valdecilla Health Research Institute (IDIVAL), Spain
Copyright © 2020 Zeng, Xie, Ding, Ma, Xu, Wang, Qiu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Liu, bGl1cUB1c3N0LmVkdS5jbg==
 Haijuan Zeng1,2
Haijuan Zeng1,2 Qing Liu
Qing Liu