
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 07 February 2020
Sec. Extreme Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00047
 Sohaib H. Mazhar1,2†
Sohaib H. Mazhar1,2† Martin Herzberg3†
Martin Herzberg3† Ibtissem Ben Fekih2
Ibtissem Ben Fekih2 Chenkang Zhang2,4
Chenkang Zhang2,4 Suleiman Kehinde Bello2
Suleiman Kehinde Bello2 Yuan Ping Li2
Yuan Ping Li2 Junming Su2
Junming Su2 Junqiang Xu2
Junqiang Xu2 Renwei Feng2
Renwei Feng2 Shungui Zhou1*
Shungui Zhou1* Christopher Rensing1,2*
Christopher Rensing1,2*The highly heavy metal resistant strain Cupriavidus metallidurans BS1 was isolated from the Zijin gold–copper mine in China. This was of particular interest since the extensively studied, closely related strain, C. metallidurans CH34 was shown to not be only highly heavy metal resistant but also able to reduce metal complexes and biomineralizing them into metallic nanoparticles including gold nanoparticles. After isolation, C. metallidurans BS1 was characterized and complete genome sequenced using PacBio and compared to CH34. Many heavy metal resistance determinants were identified and shown to have wide-ranging similarities to those of CH34. However, both BS1 and CH34 displayed extensive genome plasticity, probably responsible for significant differences between those strains. BS1 was shown to contain three prophages, not present in CH34, that appear intact and might be responsible for shifting major heavy metal resistance determinants from plasmid to chromid (CHR2) in C. metallidurans BS1. Surprisingly, the single plasmid – pBS1 (364.4 kbp) of BS1 contains only a single heavy metal resistance determinant, the czc determinant representing RND-type efflux system conferring resistance to cobalt, zinc and cadmium, shown here to be highly similar to that determinant located on pMOL30 in C. metallidurans CH34. However, in BS1 another homologous czc determinant was identified on the chromid, most similar to the czc determinant from pMOL30 in CH34. Other heavy metal resistance determinants such as cnr and chr determinants, located on megaplasmid pMOL28 in CH34, were shown to be adjacent to the czc determinant on chromid (CHR2) in BS1. Additionally, other heavy metal resistance determinants such as pbr, cop, sil, and ars were located on the chromid (CHR2) and not on pBS1 in BS1. A diverse range of genomic rearrangements occurred in this strain, isolated from a habitat of constant exposure to high concentrations of copper, gold and other heavy metals. In contrast, the megaplasmid in BS1 contains mostly genes encoding unknown functions, thus might be more of an evolutionary playground where useful genes could be acquired by horizontal gene transfer and possibly reshuffled to help C. metallidurans BS1 withstand the intense pressure of extreme concentrations of heavy metals in its environment.
Cupriavidus metallidurans BS1 was isolated from a gold copper mine in China and was found to have 99.9–100% 16S rRNA gene identity to available genomes in the National Center for Biotechnology Information (NCBI) database of C. metallidurans strains (CH34, NBRC 102507, NDB3NO24, Ni-2, NA1, NA4, NE12 and H1130) using the Geneious alignment (global alignment; cost matrix, identity) (Kearse et al., 2012). The complete genome of C. metallidurans BS1 was sequenced because it might give additional insights into the interplay of gold (Au) and copper (Cu) handling in gold biomineralization. C. metallidurans CH34 was shown to accumulate toxic Au(I/III) complexes from solution which in turn induced Cu resistance gene clusters, to probably promote cellular defense against oxidative stress (Nies, 1999; Wiesemann et al., 2013). Au(III)-complexes are rapidly reduced to intermediate Au(I)-species by C. metallidurans cells and Au(I)-species are subsequently imported into the bacterial cytoplasm (Wiesemann et al., 2013; Zammit et al., 2016) where they exert toxic effects (Jian et al., 2009; Reith et al., 2009).
The strain C. metallidurans CH34 was shown to be a facultative chemolitho-autotrophic β-proteobacterium belonging to the family Burkholderiaceae/order Burkholderiales and has been found to be highly resistant to Zn2+, Cd2+, Ni2+, AsO43– CrO42–, Hg2+, Ag+, Cu1+/2+, Pb2+, and Co2+ (Vandamme and Coenye, 2004; Monchy et al., 2007; Janssen et al., 2010). C. metallidurans strains are Gram negative and aerobic bacteria that are able to grow on different nutrient sources such as Tris gluconate minimal media, blood agar, tryptic soy agar, and R2A nutrient mediums (Mergeay et al., 1985; Diels and Mergeay, 1990; Gent et al., 2001). Different strains of these bacteria have evolved novel strategies and mechanisms to survive metal-stressed conditions via the exchange of genetic materials through transposons and other mobile genetic elements. Cupriavidus metallidurans was shown to survive in various mesophilic metal-contaminated environments such as mining sites for gold, copper and zinc because it possesses a an abundance of transition metal efflux systems and other heavy metal resistance determinants (Diels and Mergeay, 1990; Mergeay et al., 2003). The environmental impacts of mining range from ecosystems destruction with accompanying loss of bio-diversity resources to the accumulation of heavy metals and other pollutants (Mergeay et al., 2003). Gold-rich environments, such as gold mining sites, weathering Au deposits, auriferous soils and sediments embody highly toxic mobile Au-complexes as well as a challenging assortment of transition metals (Mergeay et al., 2003; Rea et al., 2016; Wiesemann et al., 2017).
In C. metallidurans CH34, 12 metal-transporting RND (Resistance-Nodulation-Cell division) systems of the heavy metal efflux RND (HME) protein family are known (Tseng et al., 1999; Nies et al., 2006). The plasmid-encoded cobalt–zinc–cadmium resistance pumps CzcCBA and the cobalt–nickel pump CnrCBA have been extensively studied and shown to confer resistance to Co, Zn, Cd, and Ni (Mergeay et al., 1985; Nies et al., 1987, 1989; Collard et al., 1993; Nies, 1995). The cnr operon has been reported to have the ability to mutate to additional zinc resistance. There exist extensive homologies in the cnr and czc encoded structural proteins and the ranges of the exported cations suggest that the two operons have evolved from a common ancestor operon (Collard et al., 1993). This could be due to the ability of both operons to encode determinants for cobalt efflux. CzcD, an important member of the CDF (Cation Diffusion Facilitators) protein family, is part of the czc resistance system that transports Zn2+, Cd2+, and Co2+ (Nies, 1992; Anton et al., 2004). Despite the presence of CzcCBA efflux complex, the absence of CzcD decreased cobalt resistance and leads to an extreme sensitive cobalt phenotype when the second CDF protein, DmeF, is also absent (Munkelt et al., 2004; Scherer and Nies, 2009). Hence, this study is targeted at highlighting the genetic rearrangements involved in heavy metal resistance and metals detoxification systems found in a new strain C. metallidurans BS1, isolated from a gold–copper mine in China and compare it with the well-studied strain C. metallidurans CH34.
The heavy metal resistant bacterium, C. metallidurans BS1 was isolated from soil samples collected at the mid altitude (590 m) of the gold–copper mine located at the geography coordinates N25° 11.027′ and E116° 24.414′. The bacterium was isolated by inoculating the collected soil samples into Reasoner’s 2A (R2A) agar medium (Qingdao Hope Bio-Technology Co. Ltd., Qingdao, China). There were three replicates of each sample and prior to inoculation of the soil samples on the R2A medium, 1 g of each sample, was mixed with 1000 μL double distilled water (ddw) and incubated for 30 min at 30°C with 150 revolutions per minute (rpm). After incubation, 100 μL of the supernatant was used for the inoculation in R2A medium. The broth culture was incubated at 30°C with shaking at 250 rpm while the agar plates were incubated in the dark at 30°C overnight. The bacterial strain was purified by continuously growing the strain on R2A agar solid plates, without any heavy metal addition, until single colonies were obtained. Finally, the isolates were stored at −80°C in 30% sterile glycerol for long term storage and in −20°C in 30% sterile glycerol for continuous laboratory experiments.
The single colonies were later used to examine the minimal inhibitory concentration (MIC) on CuSO4, ZnSO4 7 × H2O, CdCl2 5 × 2H2O, NaAsO2, NiCl2 × 6H2O, HAuCl4 4 × H2O, CoCl2 × 6H2O and Pb(NO3)2 supplemented mineral salts media 284 (gluconate 2g/L as sole carbon source) cultures plates. Mineral salts medium 284 contained (1000 ml of DD water) Tris 6.06 g pH 7.0, NaCl 4.65 g, KCl 1.49 g, NH4Cl 1.07 g, Na2SO4 0.43 g, MgCl2 6∗H2O 0.20 g, CaCl2 2H2O 0.03 g, Na2HPO4 2H2O 0.23 g, Fe(III)NH4 citrate 0.005 g, sodium gluconate 2 g, 1 ml of the trace element solution SL 7 and Difco BactoAgar 15 g L–1 (Nies et al., 2006). Medium 284 is a Tris-buffered and low phosphate medium designed to reduce metal complexation and determine accurate MICs (Mergeay et al., 1985).
The bacterium strain was grown aerobically in 40 ml R2A broth culture incubated at 30°C with shaking at 250 rpm. After 24 h of growth, the genomic DNA of the bacterium strain was extracted using the TIANamp Bacteria DNA Isolation Kit following the standard protocol provided by the manufacturer (TianGen Biotech, Beijing Co., Ltd.). The quantity and purity of genomic DNA were assessed using an UV spectrophotometry (Nanodrop ND-1000, J & H Technology Co., Ltd.). The OD260/280 value of the genomic DNA higher than 1.80 was picked to check the intact ones via agarose gel electrophoresis (0.8%). Samples containing greater than 25 μg of intact genomic DNA were sent out to perform complete genome sequencing. Molecular identification of the strain was carried out by partial sequencing of the 16S rRNA gene. The 27F/1492R primer pair was used to amplify 16S sequence and the PCR product was subsequently sequenced. Amplification of the 16S rRNA gene was performed using the polymerase chain reaction (PCR). BIO-RAD S1000TM Thermal Cycler (Foster City, CA, United States) machine was used for the PCR and PCR cycle used for amplification was as follows: 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 54°C, 1 min at 72°C and a final extension of 7 min at 72°C. The machine was finally set at 16°C for 2 min for final cooling before the storage of the amplified products. The PCR products were sequenced by Biosune Company (Shanghai, China) using the Sanger method. API®ZYM system test kit (bioMérieux, Inc. SA- 69280 Marcy-l’Etoile, France) was used for biochemical characterization of C. metallidurans strain BS1 according to manufacturer’s protocol.
The C. metallidurans strain BS1 genome was sequenced using a PacBio RS II platform and Illumina HiSeq 4000 platform at the Beijing Genomics Institute (BGI, Shenzhen, China). Four SMRT cells Zero-Mode Waveguide arrays of sequencing, were used by the PacBio platform to generate the sub reads set. PacBio sub reads (length < 1 kb) were removed. The detailed methods of library construction and sequencing can be found at Illumina’s official website1. The program Pbdagcon was used for self-correction2. Draft genome contigs, which are uncontested groups of fragments, were assembled using the Celera Assembler against a high quality corrected circular consensus sequence subreads set. To improve the accuracy of the genome sequences, GATK3 and SOAP tool packages (SOAP2, SOAPsnp, SOAPindel) were used to make single-base corrections. To trace the presence of any plasmid, the filtered Illumina reads were mapped using SOAP to the bacterial plasmid database4 (last accessed July 8, 2016).
Close relative and phylogenetic affiliation of the obtained 16S rRNA sequences were determined by using the BLAST search program at the NCBI website5. The 16S rRNA gene sequences were submitted for comparison and identification to the GenBank databases using the NCBI Blastn algorithm and to the EMBL databases using the Fasta algorithm. The phylogenetic tree of 16S rRNA gene sequences and the amino acid sequences of compared resistance determinants were inferred by Geneious prime 2020 0.4. (Geneious Tree Builder) Geneious prime 2020 0.4. (Geneious Tree Builder)6, global alignment, gap open penalty 12, gap extension penalty 3, Blosum62 cost matrix, Jukes-Cantor, Neighbor-Joining, Resampling Method – Bootstrap; with 100 replicates). Morphological and physiological experiments were done to further characterize the bacteria.
The complete genome sequence of C. metallidurans BS1 was submitted to NCBI Prokaryotic Genome Annotation Pipeline (Annotation Software revision 4.6) and The RAST Server: Rapid Annotation using Subsystem Technology (version 2.0) for gene identification following the standard operating procedures. The genes were predicted using Glimmer 3.02 (Delcher et al., 2007) as part of the RAST annotation pipeline (Overbeek et al., 2014). EggNOG 4.5.1 server was used to get the information about genes associated with COG for each predicted protein (Huerta-Cepas et al., 2016), PFAM domains were identified using PFAM 31.0 (Finn et al., 2016). Signal peptides were identified using the SignalP server 4.1 (Petersen et al., 2011), transmembrane helices were inferred using the TMHMM server v. 2.0 (Krogh et al., 2001) and CRISPR repeats were analyzed using CRISPRFinder (Grissa et al., 2007). RAST was used to identify functional genes of interest involved in heavy metal efflux, diffusion and binding systems. Circos software was used to display the circular representation of three replicons of genome of C. metallidurans BS1.
The complete genome was submitted to NCBI and was released on 21st of March, 2019 with GenBank assembly accession number GCA_003260185.2, under BioProject: PRJNA224116 and BioSample: SAMN08974431.
Cupriavidus metallidurans BS1 is a Gram negative, motile bacterium in the form of short rods (Figure 1) very similar to the description by Mergeay et al. (1985) about C. metallidurans CH34. The colonies of C. metallidurans BS1 were creamy in color when grown on R2A agar plates and were also found to exhibit some enzyme activities such as alkaline phosphatase, esterase, esterase lipase, leucine aryl amidase, acid phosphatase and Naphthol-AS-BI-phosphohydrolase. C. metallidurans BS1 is free living and can resist high concentration of heavy metals in its natural environment. Species of C. metallidurans are reported to be group of short rods (0.8 × 1.2–2.2 μm) bacteria that are oxidase- and catalase-positive, produce indole from tryptophan, assimilate histidine, D-gluconate, adipate and L-malate but not D-glucose, L-arabinose, D-fructose, D-mannose, D-mannitol, N-Acetyl-D-glucosamine or maltose as C-source (Mergeay, 2015). The 16S rRNA sequence analysis of BS1 from complete genome assembly showed close relatedness (99.9 – 100% of 1,533 bp) to other strains of C. metallidurans, displayed in the phylogenetic tree (Supplementary Figure S1). Phylogram constructed from Genome clustering (Microscope) using Mash (Pairwise Genome Distance) and ANI (Average Nucleotide Identity) represented the high similarities between C. metallidurans strains CH34, BS1 and NA4 in Figure 2. Circular representation of the three replicons of genome of C. metallidurans BS1 using ncRNA, repetitive sequences, methylation, GC content, GC skew and other information on the genome. GC skew analysis was performed using (G - C)/(G + C) calculations based on Genomic sequences of C. metallidurans BS1, the results of gene distribution, ncRNA distribution and gene annotation are also shown on this Figure 3 at the same time. However, at genome level, strain NA4 shows a higher degree of kinship to type strain CH34 than strain BS1. This could be an indication of a genetic drift correlating with global distribution and habitat adaptation (Mergeay et al., 1978; Ali et al., 2019). C. metallidurans BS1 conferred resistance to Zn2+ displaying a MIC of 20 mM, Cd2+ (2.5 mM), Co2+ (20mM), Ni2+ (8 mM), As3+ (3.5 mM), Cu2+ (5 mM), Au3+ (1 μM) and Pb2+ (1.7 mM) on Mineral salts medium 284 plates. The MICs were compared to those obtained with C. metallidurans CH34: Zn2+ (25 mM), Cd2+ (3.5 mM), Co2+ (35mM), Ni2+ (7 mM), As3+ (2.5 mM), Cu2+ (3 mM), Au3+ (0.5 μM) and Pb2+ (1.6 mM) respectively (Table 1). The MICs for zinc, cobalt and cadmium was higher in C. metallidurans CH34 while the MICs for nickel, arsenite and copper was higher in C. metallidurans BS1 than in C. metallidurans CH34. In C. metallidurans BS1, nccCB”B’A nreB mmrQ displayed 100% amino acid (A.A) similarity to C. metallidurans CH34 but the cnr determinants were significantly different (Supplementary Table S6). For example, the nickel diffusion facilitator cnrT in C. metallidurans BS1 was found having 100% A. A similarity with Cupriavidus nantongensis X1 (Fang et al., 2016). The different cnr operon might be the reason of the higher MIC for nickel in C. metallidurans BS1 compared to C. metallidurans CH34.
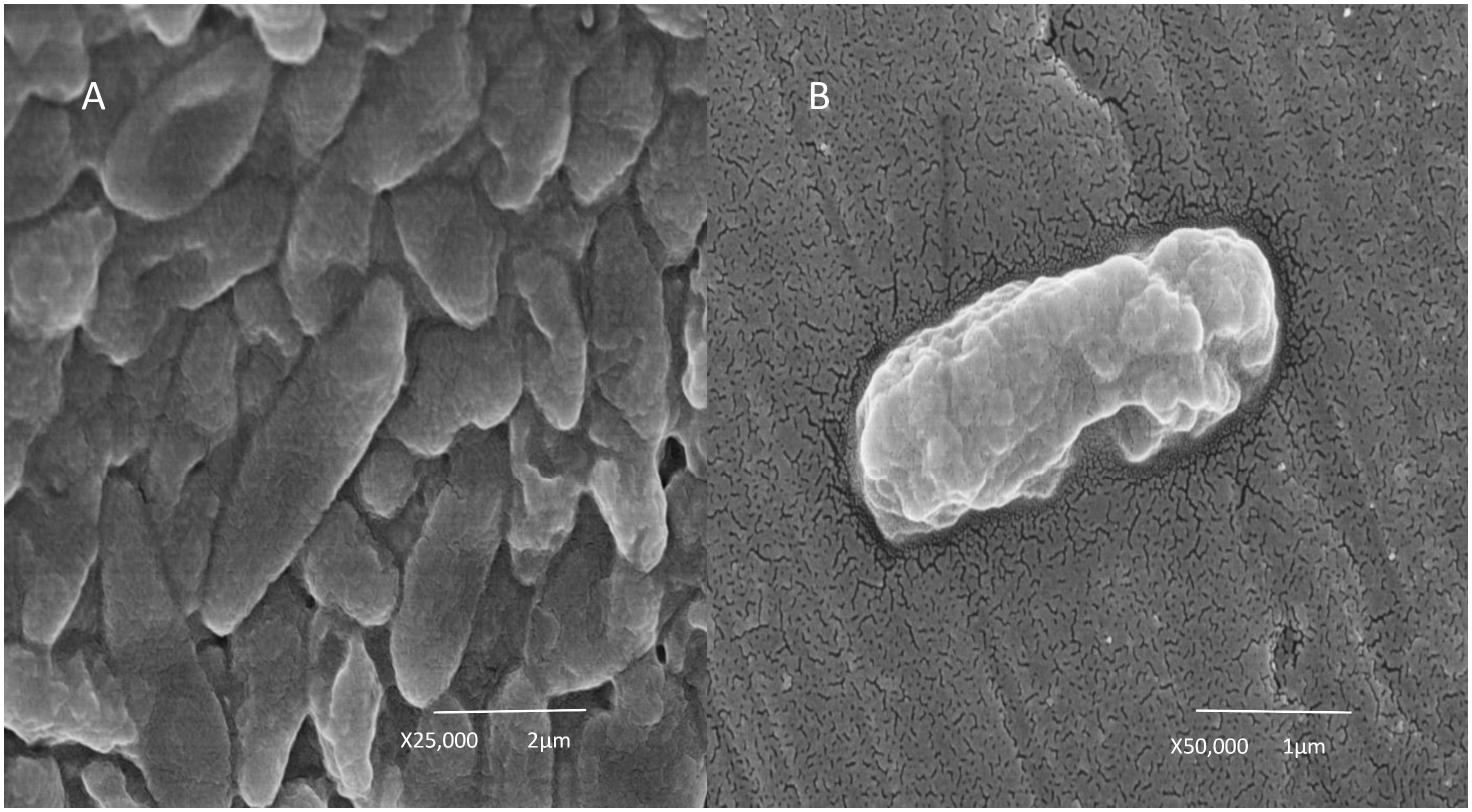
Figure 1. Scanning electron microscopy image of Cupriavidus metallidurans BS1. (A) Combined growths of bacterial cells and (B) Single bacterial cell growth.
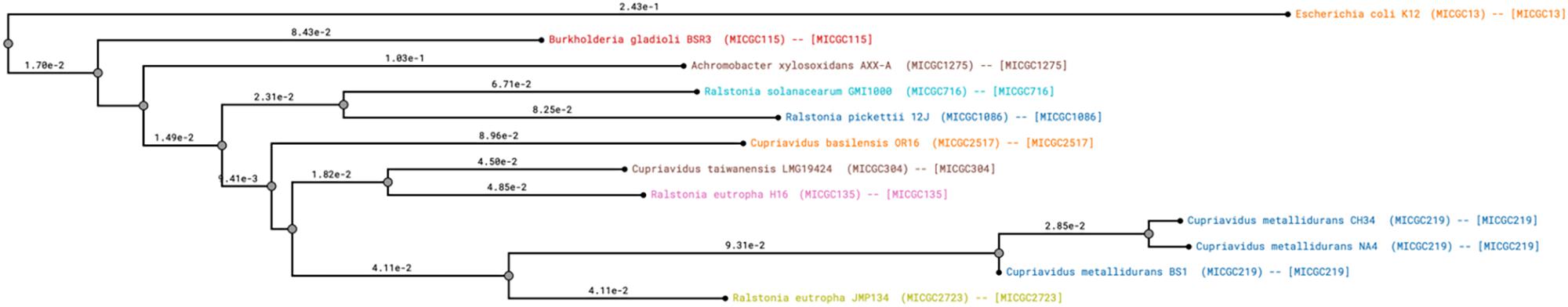
Figure 2. Phylogram - Genome clustering (Microscope) using Mash (Pairwise Genome Distance) and ANI (Average Nucleotide Identity) of Cupriavidus metallidurans strains CH34, BS1 and NA4. Names of the analyzed species and the MICGC clusters (MicroScope Genome Cluster) are displayed. A progenome species cluster is defined by computed Mash distances below 0.06 (i.e., ANI > 94%). Cupriavidus metallidurans CH34, NA4 and BS1 cluster together in MICGC219 (CH34 _ BS1 = 0.030; CH34 _ NA4 = 0.014) (Konstantinidis and Tiedje, 2005; Blondel et al., 2008; Ondov et al., 2016).
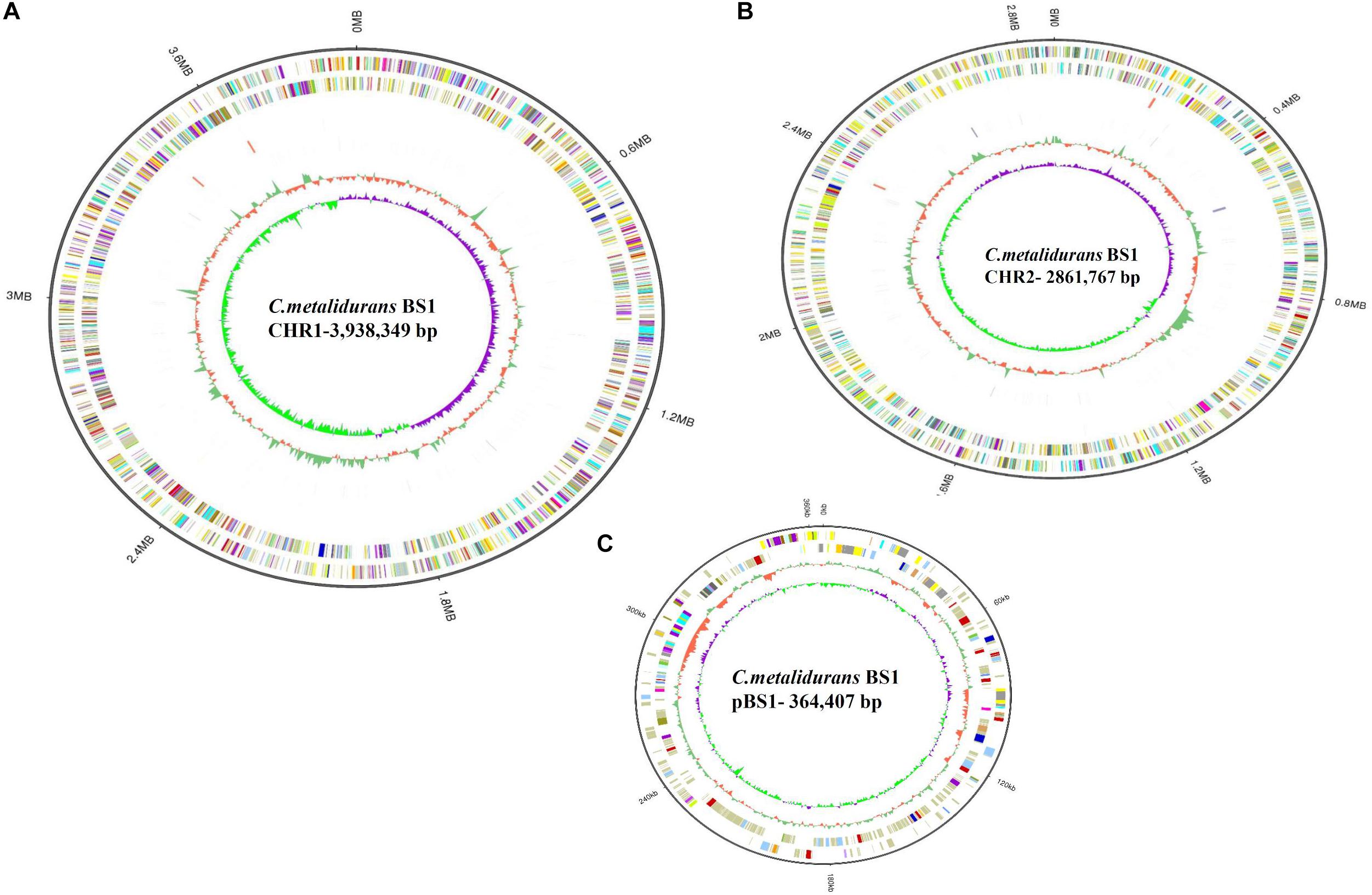
Figure 3. Circular representation of the three replicons of C. metallidurans BS1. (A – Chromosome, CHR1); (B – Chromid, CHR2); (C – Plasmid). Circles display from the outwards to inwards, (ring 1): Genome size in MB; (ring 2): Forward strand gene, COG classification; (ring 3): Reverse strand gene, COG classification; (ring 4): Forward strand ncRNA; (ring 5): Reverse strand ncRNA; (ring 6): repeat; (ring 7): GC-content; circle 8: GC-skew (G-C/G + C ratio).
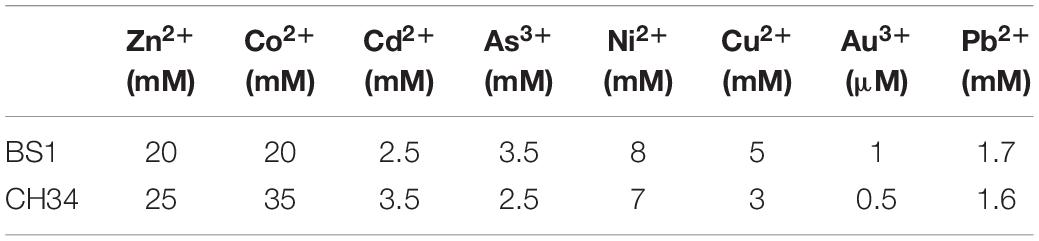
Table 1. Minimal inhibitory concentration (MIC) of selected heavy metals for Cupriavidus metallidurans BS1 and Cupriavidus metallidurans CH34 in mineral salts media.
The complete genome of C. metallidurans BS1 was shown to be 7,164,523 bp in total with 3 replicons and 63.5% G + C content, while the genome of the type strain C. metallidurans CH34 is comprised of 4 replicons (chromosome + chromid + pMOL28 + pMOL30) with 6,91335 bp of size in total and identical 63.5% of G + C content. C. metallidurans BS1 contained one chromosome (CHR1 – 3,938,349 bp), one chromid (CHR2 – 2,580,084 bp) and one megaplasmid – pBS1 (364,407 bp) close in size to the combined two megaplasmids, pMOL28 and pMOL30 (405,179 bp) but missing almost 40,000 bp, present in type strain C. metallidurans CH34 (Janssen et al., 2010). Comparative genomic features between BS1, CH34 and NA4 are shown in Table 2 and general genome statistics are given in Table 3. Several genome sequences of different C. metallidurans strains have been analyzed and studied further, including C. metallidurans H1130 (102 contigs), C. metallidurans NA1 (238 contigs), C. metallidurans NA4 (chromosome + chromid + pNA4_A-_D), C. metallidurans NE12 (87 contigs), C. metallidurans NBRC101272 (63 contigs), C. metallidurans NDB3NO24 (38 contigs) and C. metallidurans Ni-2 (chromosome + pNi-2_1-3) (Janssen et al., 2010; Van Houdt et al., 2012; Monsieurs et al., 2013, 2014; Mergeay, 2015; Ali et al., 2019). The analyzed strains possess the capability to maintain viability over a wide range of metal (Vandamme and Coenye, 2004; Scherer and Nies, 2009; Van Houdt et al., 2018; Lee et al., 2019). Rfam7 predicted that both C. metallidurans CH34 and C. metallidurans BS1 contained four sets of 5S, 16S, 23S r RNA genes. A difference was found as BS1 was shown to contain 22 sRNA and 63 tRNA genes with 1,900 and 4,927 bp respectively, specifying all 20 amino acids, while CH34 contains 62 tRNA and 12 sRNA (Janssen et al., 2010). RNA analysis for C. metallidurans BS1 was performed and predicted using RNAmmer software (V:1.2), compared with rRNA database and Rfam database (Lowe and Eddy, 1997; Lagesen et al., 2007; Gardner et al., 2009; Nawrocki et al., 2015; Chan and Lowe, 2019). C. metallidurans BS1 was shown to have 6855 coding sequences (CDS) (Table 2) while MaGe system predicted 6717 coding sequences in type strain C. metallidurans CH34 out of which 4,518 CDS are being assigned with functions while the rest of the CDS are hypothetical or conserved hypothetical (Janssen et al., 2010). The clusters of orthologous groups of proteins (COGs) were accessed and 5,008 (74.15%) CDS were assigned to one or more COG functional classes, which were then sorted into 25 groups as shown in Figure 4 (Galperin et al., 2015; Makarova et al., 2015). CDS assigned to COG of type strain CH34 are 5,133 predicted, COGnitor using MaGe system of annotation (Tatusov, 2001). COG analysis assigned 560 genes (maximum) to transcription functions and one gene (minimum) to cytoskeleton and also to RNA processing and modification function. Following transcription, 541 genes were assigned to general function prediction, 501 genes to energy production and conversion, and 465 genes to amino acid transport and metabolism. RAST subsystem analysis placed 28% of the protein coding genes into subsystem categories with the largest percentage assigned to amino acids (AA) and derivatives.
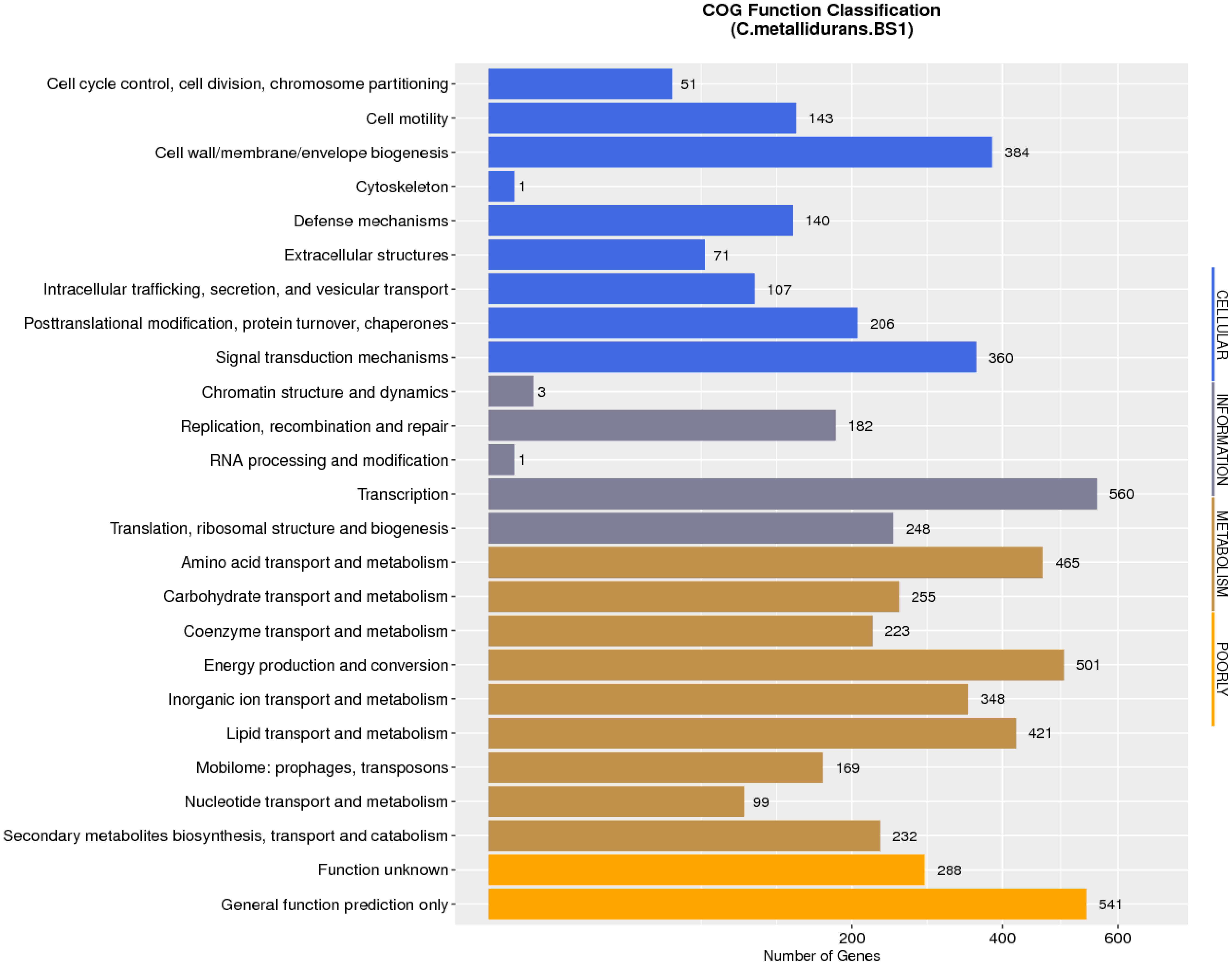
Figure 4. Number of genes associated with general COG functional categories of C. metallidurans strain BS1. COG: Cluster of Orthologous Groups of proteins. This protein database is created and maintained by NCBI. The database is based on the evolutionary relationships of protein systems between bacteria, algae and eukaryotes. Protein sequence are classified into one kind of COG part and each kind of COG part is composed of homologs sequences which are used to deduce the function of the protein. The clusters of orthologous groups of proteins (COGs) were accessed and 5,008 (74.15%) CDS of C. metallidurans BS1 were assigned to one or more COG functional classes COG database is divided into twenty parts by their functions. The statistics is listed above.
The chromid in both CH34 and BS1 harbors genes specialized in the activities of sulfonate/taurine transport and utilization, sulfite oxidation, polyhydroxy alkanoic acid (PHA) synthesis and conversion, biofilm formation, and exopolysaccharide synthesis squalene/hopene synthesis, tannin degradation (Janssen et al., 2010). The chromosome (CHR1) was shown to contain genes encoding functions related to DNA replication, DNA repair, translation and transcription and protein processing. The chromid (CHR2) of strain BS1 contained genes encoding determinants putatively responsible for an adaptive response comprising of genes encoding functions such as carotenoid biosynthesis - carX (Rmet_5644) is 97%, crtB (Rmet_4149) is 99%, tolerance and utilization of acetone - acxR (Rmet_4104) 97% (acxABC (Rmet_4105-7) absent) similar to the chromid of CH34. A translation initiation factor IF-1 infA2 (Rmet_5176) in BS1 was 100% similar to the one present in CH34.
Genome analysis by synteny plot of the C. metallidurans strains CH34, BS1 and NA4is showing a high genome plasticity (Figure 5 and Supplementary Figure S2). This genome plasticity is characterized by deletion, insertion and duplication as well as rearrangements on the genomic level caused by horizontal gene transfer, (HGT) transposons, IS elements and prophages. Insertion sequence elements distribution in C. metallidurans BS1 was accessed from ISFinder and described in Supplementary Table S1. The BS1 genome showed high similarities to C. metallidurans CH34 and other genomes of related Cupriavidus strains, particularly related to the chromosome (CHR1) (Millacura et al., 2018). All CMGIs (catabolic-metabolic genomic islands) are missing, probably acquired in C. metallidurans CH34 by horizontal gene transfer, on which the genes encoding the hydrogenase, enzymes responsible for the Calvin cycle and metal resistance determinants are also located (Van Houdt et al., 2009) (Table 4). This absence is shown by gaps in the chromosome of strain BS1 (Figure 5). These genomic gaps also included CMGI-2 and CMGI-3, which carry the genes for hydrogen-dependent facultative chemolithotrophy coupled with carbon dioxide fixation (Mergeay et al., 1985; Van Houdt et al., 2009; Herzberg et al., 2015). This leads to a main characteristic in the phenotypic distinction of these two strains. Another missing island is CMGI-1, harboring the gene for an anabolic copper PIB1-type ATPase (CtpA), the gene locus encoding a zinc and cadmium PIB2-type ATPase (CadA) as well as one mercury resistance cluster (Legatzki et al., 2003; Scherer and Nies, 2009; González-Guerrero et al., 2010; Raimunda et al., 2011). Furthermore, the synteny plot shows transfers of several larger gene regions from the pMOL30 and smaller regions of the pMOL28 to the chromid in the BS1 strain. The majority of these regions are loci whose gene products code for metal resistance determinants, especially metal efflux systems. Surprisingly, a large part of these loci were brought into spatial proximity by mobile elements, and two metal-resistance super clusters were formed as newly sorted genome islands (Figure 6 and Supplementary Table S6). The major difference between C. metallidurans BS1 and CH34 is that C. metallidurans BS1 contained three prophages while it does not have the structural and accessory genes for the soluble and membrane-bound hydrogenase present in C. metallidurans CH34 (Mergeay et al., 1985; Janssen et al., 2010; Herzberg et al., 2015). Presence of prophages were also confirmed in C. metallidurans NA1, C. metallidurans NA4, C. metallidurans H1130 and C. metallidurans Ni-2 via PHAST8 and/or PHASTER (Van Houdt et al., 2018). Strain BS1 contained predicted intact prophages [Bacill_SP_15_ (NC_031245; 7), Burkho_phiE255 (NC_009237; 32), and Ralsto_RSA1 (NC_009382; 29)] as well as NA4, H1130 and Ni-2, detailed information of prophages is mentioned Table 5. Previous studies also provided evidence for the presence of prophages in other strains of C. metallidurans than strain CH34 (Van Houdt et al., 2018; Ali et al., 2019).
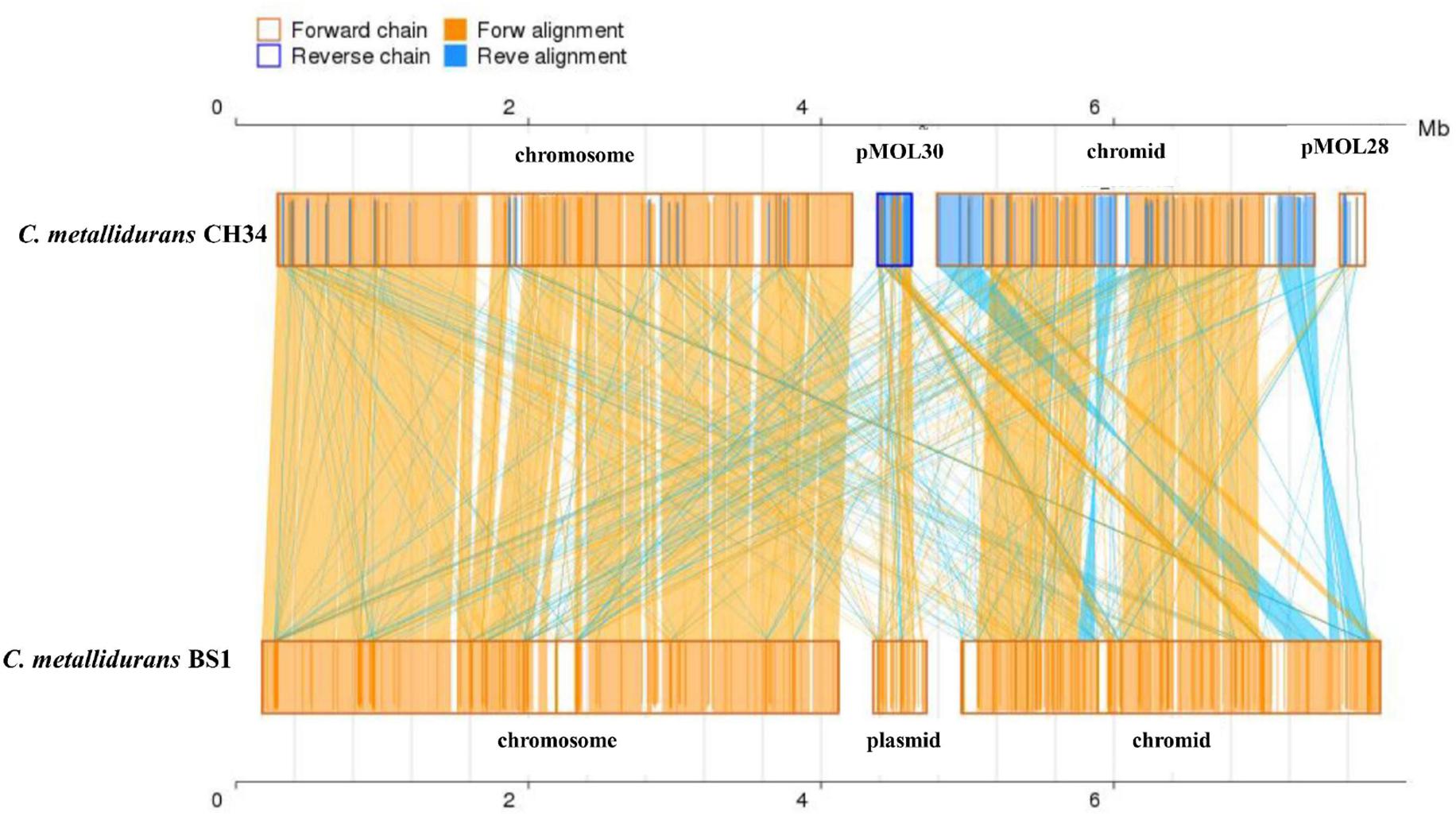
Figure 5. Synteny plot of C. metallidurans strain CH34 and BS1 on nucleic acid level. Synteny was constructed using MCScanX toolkit. The chromosome, chromid and plasmids of both strains were compared. Yellow box represents the forward chain and blue box represents the reverse chain within the upper and following sequence region. In the box of sequence, the yellow region represents the nucleic acid sequence in the forward chain of this genome sequence and the blue region represents the nucleic acid sequence in the reverse chain of this genome sequence. In the middle region of two sequences, the yellow line represents the forward alignment and the blue line represents the reverse complementary alignment.
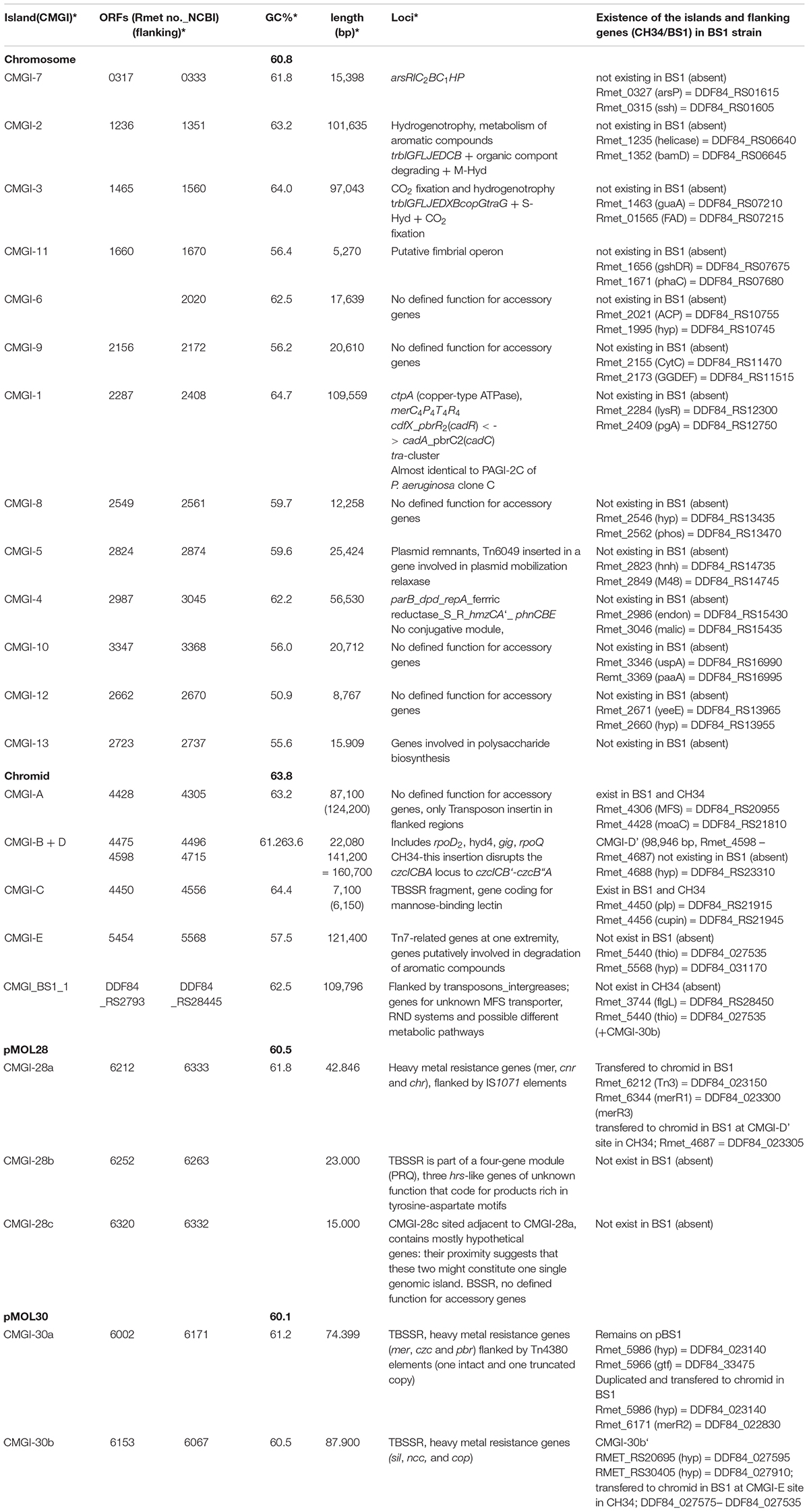
Table 4. Comparison and characteristics of the genomic islands of C. metallidurans strain CH34 and BS1; (*) (Van Houdt et al., 2012; Ali et al., 2019).
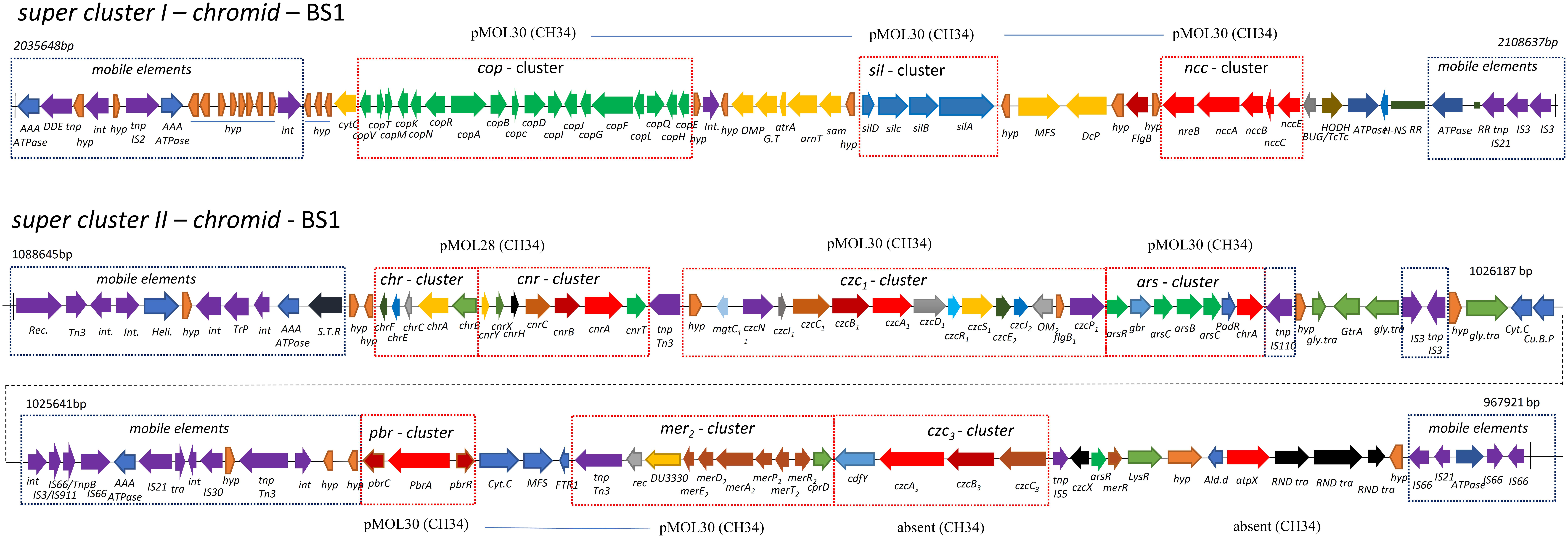
Figure 6. Heavy metals resistance determinant superclusters associated with chromid in Cupriavidus metallidurans BS1. Same scale was used for all three lines. Above, the dashed frames are the localization on the corresponding replicon of CH34 as indicated. Super cluster I is comprised of Cop, Sil and Ncc determinants adjacent to each other on chromid between 2,035,648 bp and 2,108,637 bp. Super cluster II includes determinants of Chr–, Cnr–, Czc1–, Pbr–, Mer– cluster, CdfY, Czc3-cluster and AtpX determinates on chromid between 1,088,645 bp and 967,921 bp. – int, integrase; STP, site specific tyrosine recombinase; hyp, hypothetical protein; C.H.P, Conserved Hypothetical Protein; R.R, Repeat region; flgB, Flagellar basal-body rod protein; tnp, transposase; P.I, Phage integrase; OMP, Outer Membrane Protein; Glycosyl Transferase; gtrA, trA family Protein; arnT, Polymyxin resistance protein ArnT; sam, SAM dependent methyltransferase; DCP, Diguanylate cyclase/phosphodiesterase domain 2; SDM, S-adenosylmethionine-dependent methyltransferase; MSF, MFS-type transporter; BUG/TcTc, BUG/TctC family periplasmic protein; HODH, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase; Cyt.C, Cytochrome C; FTR, Iron permease; ald.d, Aldehyde dehydrogenase; CuBP, copper binding protein.
In order to maintain metal homeostasis in the presence of multiple heavy metals, the transportome contains the metal uptake systems in addition to the metal efflux systems as the first pillar of defense (Nies, 2016). The strain BS1 shows a high degree of conservation on gene and amino acid sequence level as well as the gene loci synteny of the metal uptake systems encoding genome regions in comparison to strain CH34. In contrast to the transportome, the knowledge about the direct interaction with the metal handling proteins of the cytoplasm, cytoplasmic membrane and the periplasm, which form the metal repositories as the second pillar of bacterial metal homeostasis (Herzberg et al., 2014b), is underrepresented but further investigation is in progress (Herzberg et al., 2014a, 2016; Bütof et al., 2017; Chandrangsu et al., 2019). In order to adapt the cellular metal homeostasis to the specific environmental conditions, its regulation is of central importance as a third pillar. Besides the one- and two-component regulatory systems, sigma factors of the extracytoplasmic function (ECF) family are of high importance to adapt the metal transport and metal handling to the specific cellular requirements and to avoid toxic effects of the various heavy metals (Missiakas and Raina, 1998; Heimann, 2002; Mascher, 2013; Große et al., 2019). The ECFs and ICFs (intracytoplasmic function), as well as the metal uptake systems, show a high degree of similarity on the gene and amino acid sequence level as well as the gene loci synteny. One exception is the ECF CnrH, which was transferred from the plasmid to the chromid with the entire cnr cluster by mobile elements and is part of the metal super cluster I structure (Grass et al., 2000). The alternative RNA polymerase sigma-70, ICF – RpoD2 (Rmet_4661, on chromid in CH34), and ECF RpoQ (Rmet_4686, on chromid in CH34) are not present on the genome of strain BS1 due to the lack of CMGI-D. Instead of the missing sigma factor, strain BS1 harbors one additional (SigX) with 74.5% similarity on amino acid level to RpoO (Supplementary Table S2). This metallophilic strain BS1, harbors numerous gene clusters encoding metal-resistance determinants enabling detoxification of transition metal ions and complexes (Nies, 2003, 2016). With regard to synteny and genome plasticity, mediated by mobile elements, C. metallidurans strain BS1 displayed a transfer of the metal resistance super cluster I from plasmid (pMOL30 in CH34) to chromid (Supplementary Table S4 and Figures 6, 7). The generation of an extended cluster II on chromid could also be observed. In this second cluster, the chr - cnr, czc1, pbr, czc3 as well as cdfY (encoding an not exist CDF family protein in CH34) and atpX (encoding an not existing P-type ATPase family protein in CH34) were brought into spatial proximity on the chromid (Supplementary Tables S5, S6 and Figures 6, 7).
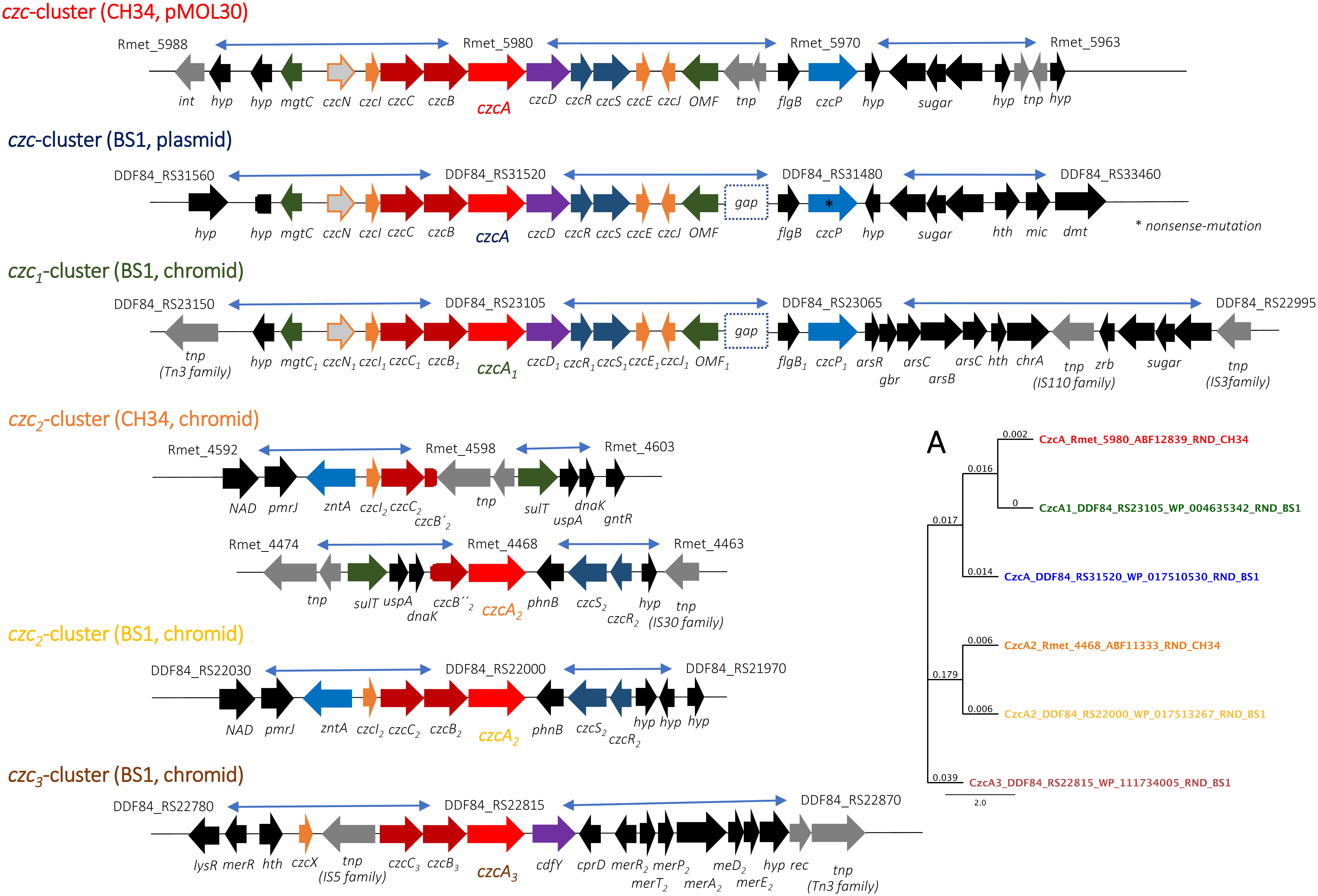
Figure 7. Comparison of the czc cluster - synteny of the C. metallidurans strains BS1 and CH34. Shown are the loci of the czc cluster of C. metallidurans CH34 on pMOL30 and the chromid as well as of BS1 on the plasmid and chromid, as well as the synteny of these gene regions. In bright red and highlighted are czcA genes as well as in dark red the structural genes of the tripartite RND efflux system, in orange genes encoding for associated periplasmic resistance proteins, in violet genes coding for CDF proteins, in blue genes encoding P-type ATPases, in dark green genes encoding transporters of the outer membrane, in gray genes encoding transposases, dark blue genes encoding the associated 2-component systems and in black genes encoding putative functions of the flanking region. Int, integrase; hyp, hypothetical gene; tnp, transposase; OMF, outer membrane factor; hth, helix-turn-helix transcriptional regulator; mic, mechanosensitive ion channel family protein; dmt, DMT family transporter; gbr, glyoxalase/bleomycin resistance/dioxygenase family protein; zrb, zinc ribbon domain-containing protein; sugar, glycosyltransferase family 2 protein, GtrA family protein, glycosyl transferase; NAD, NAD-dependent dehydratase; rec, recombinase family; CprD, cupredoxin like protein. (A) Molecular Phylogenetic tree based on the full amino acid sequences of CzcA encoded on pMOL30 of C. metallidurans CH34 and the homologues as well as paralogous CzcA (HME-RND) proteins of both strains. In addition to the gene names, the respective locus tags and NCBI GenBank protein ID numbers of both strains are indicated. In addition, in color matching the synteny representation was marked. The evolutionary history was inferred by using the Geneious prime 2019.2.1 (Geneious Tree Builder) (https://www.geneious.com), global alignment, gap open penalty 12, gap extension penalty 3, Blosum62 cost matrix, Jukes-Cantor, Neighbor-Joining. The tree was drawn to scale, with branch lengths calculated using the average pathway method; the scale bar corresponds to the number of substitutions per site (Kearse et al., 2012).
The encoded determinants of heavy metal resistance that were found on the genome of BS1 included chemiosmotic efflux of cations with proton antiporters of the HME-RND family (Heavy Metal Efflux – Resistances, Nodulation and Cell Division, TC_2.A.6.3) having three-component cation efflux systems such as a well-studied CzcCBA system (cobalt zinc cadmium), CnrCBA (cobalt nickel), CusCBA (copper, silver), NccCBA (nickel, cobalt and cadmium) and plenty of additional genes encoding cation diffusion facilitators (CDF family, TC_2.A.4) such as czcD, dmeF and fieF, a member of the DMT (permeases of the drug/metabolite transporter, TC_2.A.7) family such as cnrT, major facilitator superfamily (MFS super family) such as nccT/nreB and also cation P-type ATPases for cytoplasmic detoxification (zntA, pbrA, cupA/copF, and czcP) (Aziz et al., 2008) detoxifying Cu(I)/Ag(I), Pb(II)/Cd(II)/Zn(II) and Hg(II) ATPases (Silver et al., 1993). Members of the P-type ATPases and CDF protein family transport excess heavy metals such as Zn2+, Cd2+ or Co2+ from the cytoplasm to the periplasm (Silver et al., 1993; Fagan and Saier, 1994; Ashburner et al., 2000; Montanini et al., 2007; Nies, 2016) and the proton-driven efflux systems such as CzcCBA export these metals from the periplasm to the outside of the cell (Paulsen and Saier, 1997; Nies, 2003) as a two stage mode of action to prevent toxic effects by metal excess. The genes encoding chr (Cr), czc (Co, Zn, Cd) and ncc (Ni, Co, Cd) in C. metallidurans BS1 could be identified with up to 100% AA sequence similarities to those found in C. metallidurans CH34. This high number of different resistance determinants enable multiple resistance mediation and ability to handle the following metals; Zn2+, Cd2+, Ni2+, Ag+, Cu1+/2+, Pb2+, and Co2+, from the micromolar to the millimolar concentration range.
At the first level of detoxification, the cytoplasmic efflux, the two strains BS1 and CH34 showed a high degree of similarity between the metal-transporting ATPases (Figure 8). In both genomes existed gene loci coding for the PIB2/4-type ATPases ZntA, PbrA and CzcP, which are primarily involved in Zinc, cadmium, cobalt and lead efflux, with a 97–100% conservation at the amino acid level (Wang et al., 2014). This is also reflected in the copper-transporting PIB1-type ATPases CupA, CopF, and RdxI, homologous of the E. coli CopA (Rensing et al., 2000; Fan and Rosen, 2002; Raimunda et al., 2011; Wiesemann et al., 2013, 2017). Exceptions are found in the non-existent P-type ATPases CadA and CtpA, due to the lack of CMGI-1 in the genome of strain BS1. In contrast, the locus tag DDF84_RS22765 encodes a P-type ATPase (AtpX) not present in strain CH34, which displayed a higher similarity on amino acid-level to ZntA of E. coli than to ZntA of BS1 or CH34 (Rensing et al., 1997; Wang et al., 2014). The ATPases involved in the uptake of potassium (K+-type) or calcium (Ca+-type) are present in both strains with a similarity of 98-99% (Maguire, 2006; Cromie and Groisman, 2010) (Figure 8). The family of MerR regulators includes members involved in the gene regulation of zinc-, cadmium-, cobalt-, lead- and copper-exporting P-type ATPases, of mercury resistance systems, and are involved in the oxidative stress response (Brown et al., 2003; Hobman et al., 2005; Tottey et al., 2005; Molina-Henares et al., 2009; Ibáñez et al., 2015).
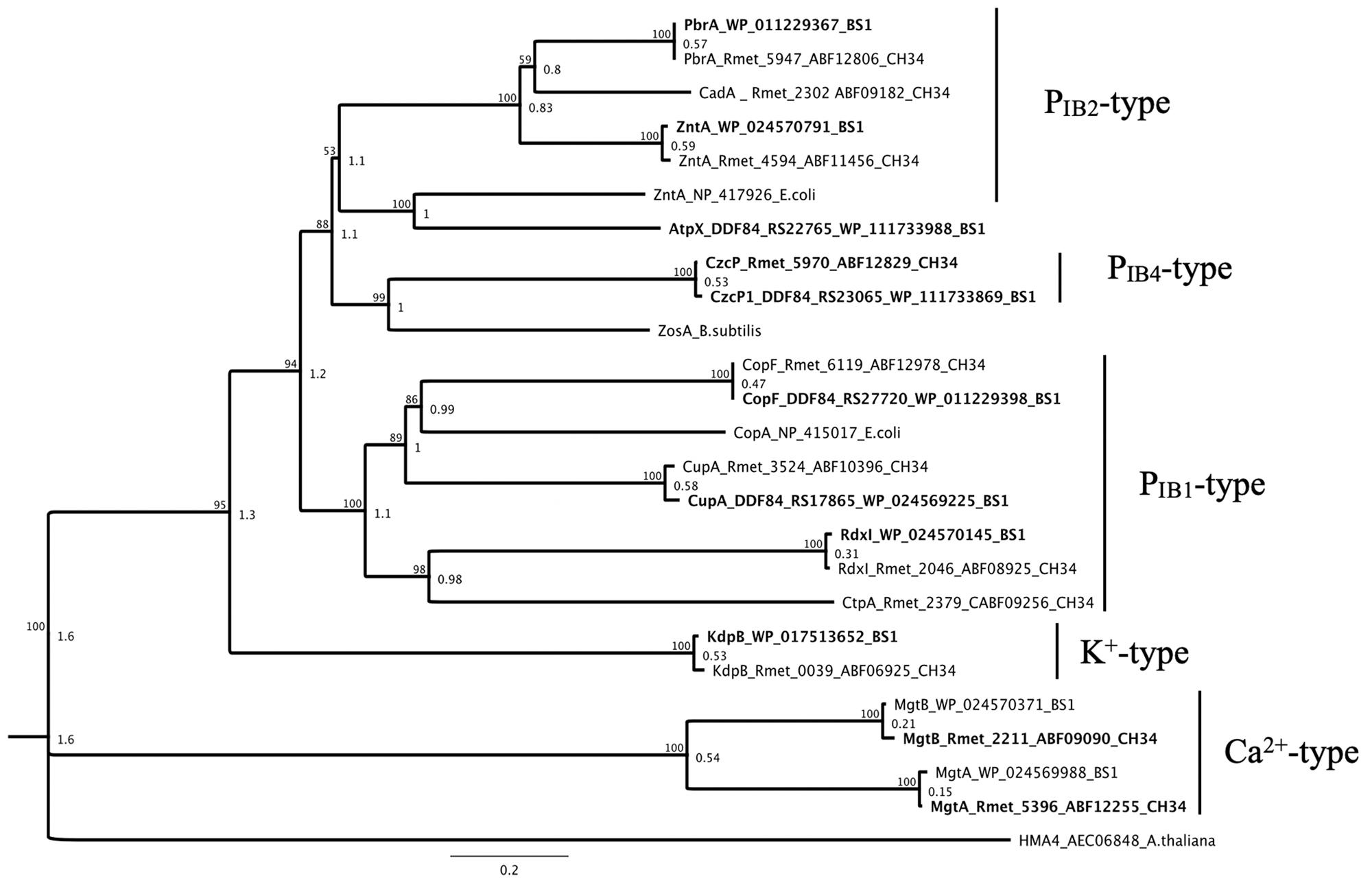
Figure 8. Molecular Phylogenetic tree based on the full amino acid sequences of P-type ATPases encoded by the genome of C. metallidurans CH34 and the homologs as well as paralogous of both strains. In addition to the gene names, the respective locus tags and NCBI GenBank protein ID numbers of both strains are indicated, BS1 are bold. The evolutionary history was inferred by using the Geneious prime 2020 0.4. (Geneious Tree Builder) (https://www.geneious.com), global alignment with free gaps, Identity (1.0/0.0), gap open penalty 12, gap extension penalty 3, 2 refinement iterations, Genetic Distance Model _ Jukes-Cantor, Neighbor-Joining, Resampling Method – Bootstrap; 100. The ATPase subfamilies are indicated (Nies, 2016). The ZntAE. coli, ZosAB. subtilis, HME4A. thaliana, and CopAE. coli (P-type ATPase family) amino acid sequences from Escherichia coli strain K-12 substr. MG1655, Bacillus subtilis and Arabidopsis thaliana were used as reference (Rensing et al., 1997, 2000; Fan and Rosen, 2002; Okkeri and Haltia, 2006; Wang et al., 2014). The tree is drawn to scale, with branch lengths calculated using the average pathway method; the scale bar corresponds to the number of substitutions per site, the branch labels are consensus Support (%) and Node Heights (Kearse et al., 2012).
Genes for the three more closely characterized members of the CDF protein family from C. metallidurans CH34, CzcD, DmeF and FieF, could also be found with a high degree of conservation on the genome of the C. metallidurans BS1 studied here (Scherer and Nies, 2009). The czcD gene occured two times (czcD/czcD1) by duplication of the czc cluster on plasmid and chromid in BS1 and the plasmid encoded CDF protein showed a lower similarity with 91.5% in comparison to the chromid encoded version with 100% similarity on the amino acid level to CzcD of CH34 (Figure 7 and Supplementary Table S4). CzcD clustered with ZitB of E. coli in one separated branch in the phylogenetic tree and could be sorted to the Zn-CDFs (Anton et al., 2004; Montanini et al., 2007). DmeF is mainly involved in the cobalt homeostasis and occured in both strain with 96.6% similarity (Munkelt et al., 2004; Scherer and Nies, 2009). A CdfX (locus tag: Rmet_2299) homolog was absent in strain BS1 due to the lack of CMGI-1 (Table 4). The other homolog to the CdfX is known from Paraburkholderia xenovorans LB400 and indicated a common origin and acquisition through horizontal gene transfer (Van Houdt et al., 2009). The Fe/Zn-CDF member FieF (Rmet_3406) encoded on chromosome of C. metallidurans CH34 shared an amino acid similarity of 99.7% with his homologous in C. metallidurans BS1 and cluster with FieF (YiiP) of E. coli as well as with an unknown and uncharacterized CDF (CdfY) member only encoded on chromid in BS1 (Munkelt et al., 2004; Grass et al., 2005; Lu and Fu, 2007) (Figure 9).
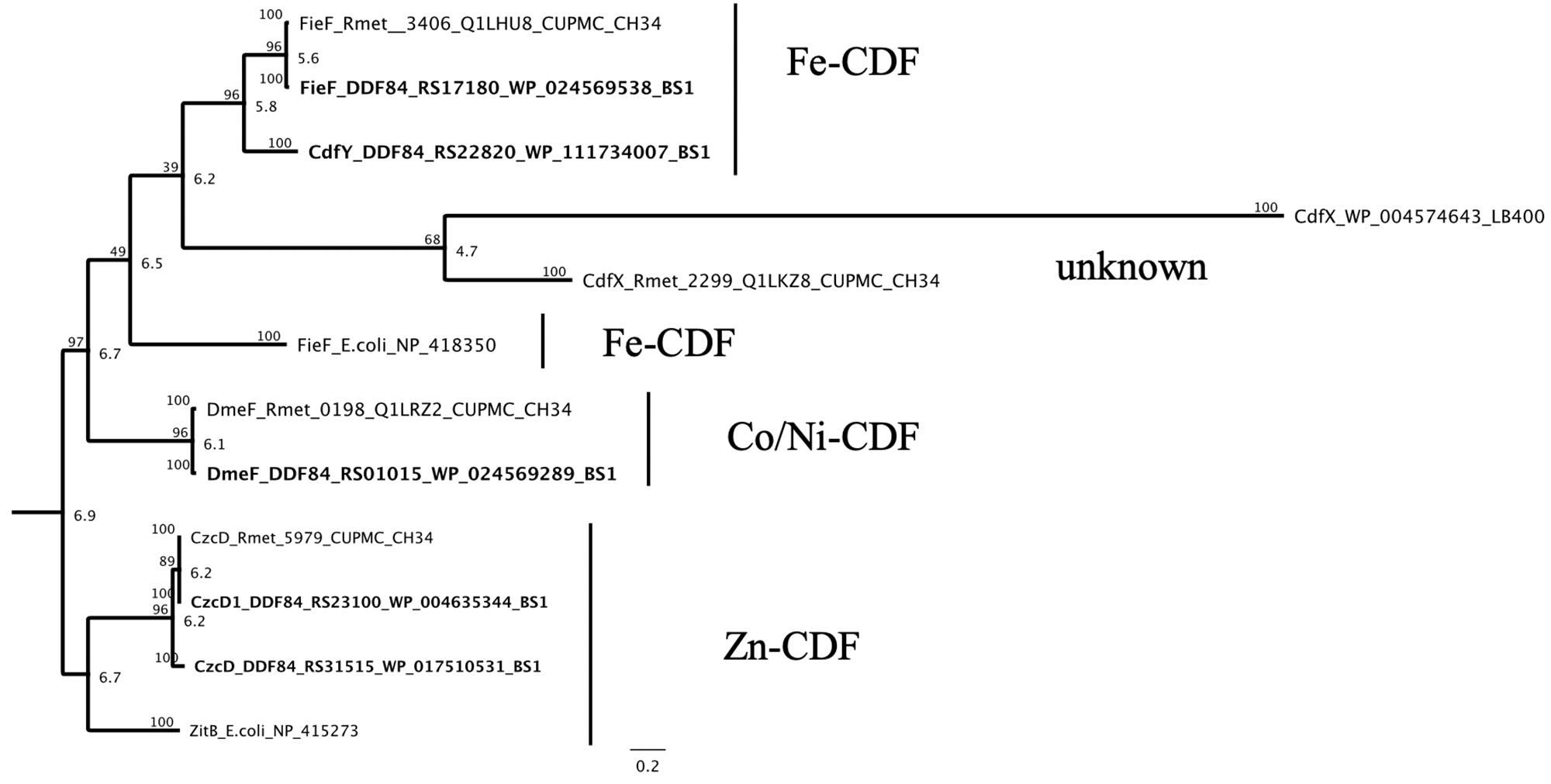
Figure 9. Molecular Phylogenetic tree based on the full amino acid sequences of the CDF (cation diffusion facilitator family) protein encoded on the genome of C. metallidurans CH34 and the homologs as well as paralogous of both strains. In addition to the gene names, the respective locus tags and NCBI GenBank protein ID numbers of both strains are indicated, proteins encoded on the genome of BS1 are displayed in bold. The evolutionary history was inferred by using the Geneious prime 2020 0.4. (Geneious Tree Builder) (https://www.geneious.com), global alignment with free gaps, Identity (1.0/0.0), gap open penalty 12, gap extension penalty 3, 2 refinement iterations, Genetic Distance Model _ Jukes-Cantor, Neighbor-Joining, Resampling Method – Bootstrap; 100. The CDF subfamilies are indicated (Montanini et al., 2007). The ZitBE. coli, FieFE. coli and CdfXP. xenovorans (CDF family) amino acid sequences from Escherichia coli strain K-12 substr. MG1655 and Paraburkholderia xenovorans LB400 were used as reference (Lee, 2002; Van Houdt et al., 2009). The tree is drawn to scale, with branch lengths calculated using the average pathway method; the scale bar corresponds to the number of substitutions per site, the branch labels are consensus Support (%) and Node Heights (Kearse et al., 2012).
The bacterial tripartite RND systems are very powerful metal efflux pumps, known from the best studied members czc, cnr and cus (Kim et al., 2011; Long et al., 2012; Nikaido, 2018). The presence of genes encoding 14 members associated with MFP (membrane fusion protein) and OMF (outer membrane factor) in synteny was detected and these could be sorted to the HME-RND efflux systems in BS1 which have been shown to be involved in heavy metal resistance/tolerance (Figure 10). In this study, we found homologs of the HME1 [CzcA and HmuA (CzcA2)], HME2 (CnrA and NccA), HME3a (ZniA, ZneA, and HmvA), HME3b (NimA and HmzA), HME3c (HmyA) and HME4 (CusA and SilA) group (Nies et al., 2006; Edward et al., 2013; Nies, 2016). The gene loci of the three component efflux systems (Sil, Cnr, Ncc) were encoded on chromid in C. metallidurans BS1 while on type strain C. metallidurans CH34, they were shown to be present on different plasmids (pMOL28 and pMOL30) (Janssen et al., 2010) (Supplementary Table S6). In addition, genes encoding two-component-regulatory system in C. metallidurans BS1 were annotated as “DNA binding heavy metal response regulator,” for CzcR and as “heavy metal sensor histidine kinase” for CzcS. These genes were found adjacent to a gene encoding CzcP, characterized as cation efflux P1B4-type-ATPase involved in Zn homeostasis and part of czcNICBADRSJE_czcP cluster (Nies, 1992; Grosse et al., 2007; Fairbrother et al., 2013).
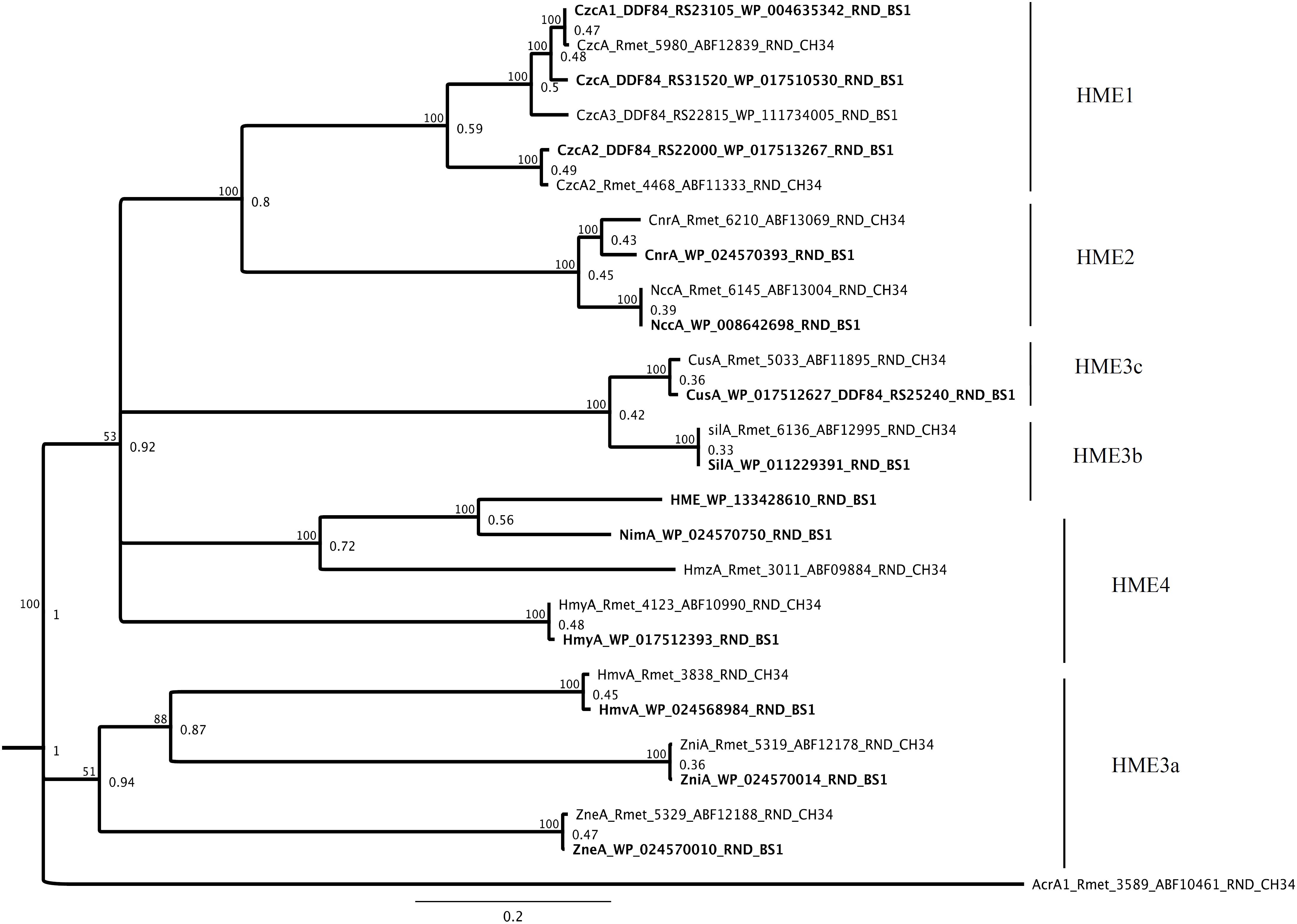
Figure 10. Molecular Phylogenetic tree based on the full amino acid sequences of the HME-RND members encoded on the genome of C. metallidurans CH34 and the homologs as well as paralogous of both strains. In addition to the gene names, the respective locus tags and NCBI GenBank protein ID numbers of both strains are indicated. The HME subfamilies are indicated (Nies, 2016). The evolutionary history was inferred by using the Geneious prime 2020 0.4. (Geneious Tree Builder) (https://www.geneious.com), global alignment, gap open penalty 12, gap extension penalty 3, Blosum62 cost matrix, Jukes-Cantor, Neighbor-Joining, Resampling Method – Bootstrap; 100. AcrA1 (Rmet_3589_ABF10461) from CH34 is used as an outgroup as a member of the HAE-RND family. The tree is drawn to scale, with branch lengths calculated using the average pathway method; the scale bar corresponds to the number of substitutions per site, the branch labels are consensus Support (%) and Node Heights (Kearse et al., 2012).
The annotation of C. metallidurans BS1 genome revealed a high degree of similarity of metal resistant factors to the well-studied C. metallidurans strain CH34. Overall, the genome of this bacterium contained 42 genes coding for proteins indirectly or directly involved in copper homeostasis including Cu resistance proteins encoded by the copS1R1A1C1B1D1 gene cluster as well as another homolog but extended gene cluster copHMKNSRACBDIJGFLQHE (Supplementary Table S3), four multicopper oxidases (CopA, CopA1, CopAX, and CopAY) (Figure 11), the RND system CusFCBA and SilDCBA (Supplementary Table S6) and three copper-translocating PIB1-type ATPases (CupA, CopF, and RdxI) (Figure 8). The fourth, anabolic, PIB1-type ATPases (CtpA) from the genome of strain CH34 encoded on CMGI-D is not present due to the absence of this genomic island in strain BS1 (Nies, 2016) (Table 4). Data exist for several of these systems that show their direct involvement in copper homeostasis and their induction of gene expression under increasing copper concentrations in strain CH34, including the two cop clusters, the divergon cupCAR and the cus operon (Nies et al., 2006; Wiesemann et al., 2013). Furthermore, parallels are known regarding the induction of these clusters in the comparison between copper and gold. With increasing gold concentrations and, as a result, increased uptake of Au(III) complexes, the induction of the cop clusters and cup divergon occurs in addition to the induction of mer genes (part of the mercury detoxification by reduction and export of Hg(II) (Reith et al., 2009; Rojas et al., 2014). The e MerR-type transcriptional regulator CupR/CueR (Rmet_3523) from C. metallidurans was shown to be activated by Au-complexes (Stoyanov and Brown, 2003; Checa et al., 2007; Jian et al., 2009; Wiesemann et al., 2013; Ta et al., 2014) (Supplementary Figure S3) leading to formation of the CupA (PIB1-type ATPase) efflux pump. Many predicted protein homologies were found in both C. metallidurans BS1 and the well characterized strain C. metallidurans CH34. The cupR and cupA gene products from C. metallidurans CH34 have 99 and 98% amino acid (AA) sequences identity, respectively, to those found on the genome of C. metallidurans BS1 (Figure 8) (Altschul, 1997). These genes were located downstream of cupC, encoding a putative copper chaperone, which has 100% AA sequence similarity with cupC encoded on chromosome of CH34. However, the strongest upregulation under this inducing conditions observed (Reith et al., 2009) was that of a genomic region (Rmet_4685 - Rmet_4682) renamed “gig” for “gold-induced genes” (Wiesemann et al., 2013). Upstream in the opposite direction are genes for the extracytoplasmic function (ECF) sigma factor RpoQ (Grosse et al., 2007), followed by the gene for a putative anti-sigma factor (Rmet_4686/7). Surprisingly, neither the gig cluster nor the sigma factor RpoQ are present in strain BS1 due to the lack of CMGI-D, although this strain is an isolate from a copper-gold mine and strain CH34 was isolated from a Belgian zinc decantation tank in 1976 (Mergeay et al., 1978, 1985). C. metallidurans was able to biomineralizes Au nanoparticles (Au at valence level of zero) by reductive precipitation from Au(I/III) -complexes and provided evidence for the bacterial contribution to the authigenic formation of secondary bacterioform gold grains and nuggets (Reith et al., 2007, 2009). The genes are coding for the periplasmic copper oxidases CopA/CopA1, which functions as an oxygen consuming Au(I)-oxidase that detoxifies gold complexes by a reduction pathway of Au(III)-complexes via Au(I)-intermediates to Au(0) nanoparticles in the periplasm are highly conserved in both strains (Bütof et al., 2018) (Supplementary Table S3 and Figure 11). This genome endowment in terms of copper and gold resistance factors should allow the C. metallidurans strain BS1, just like strain CH34, to survive in high copper and gold contaminated environments.
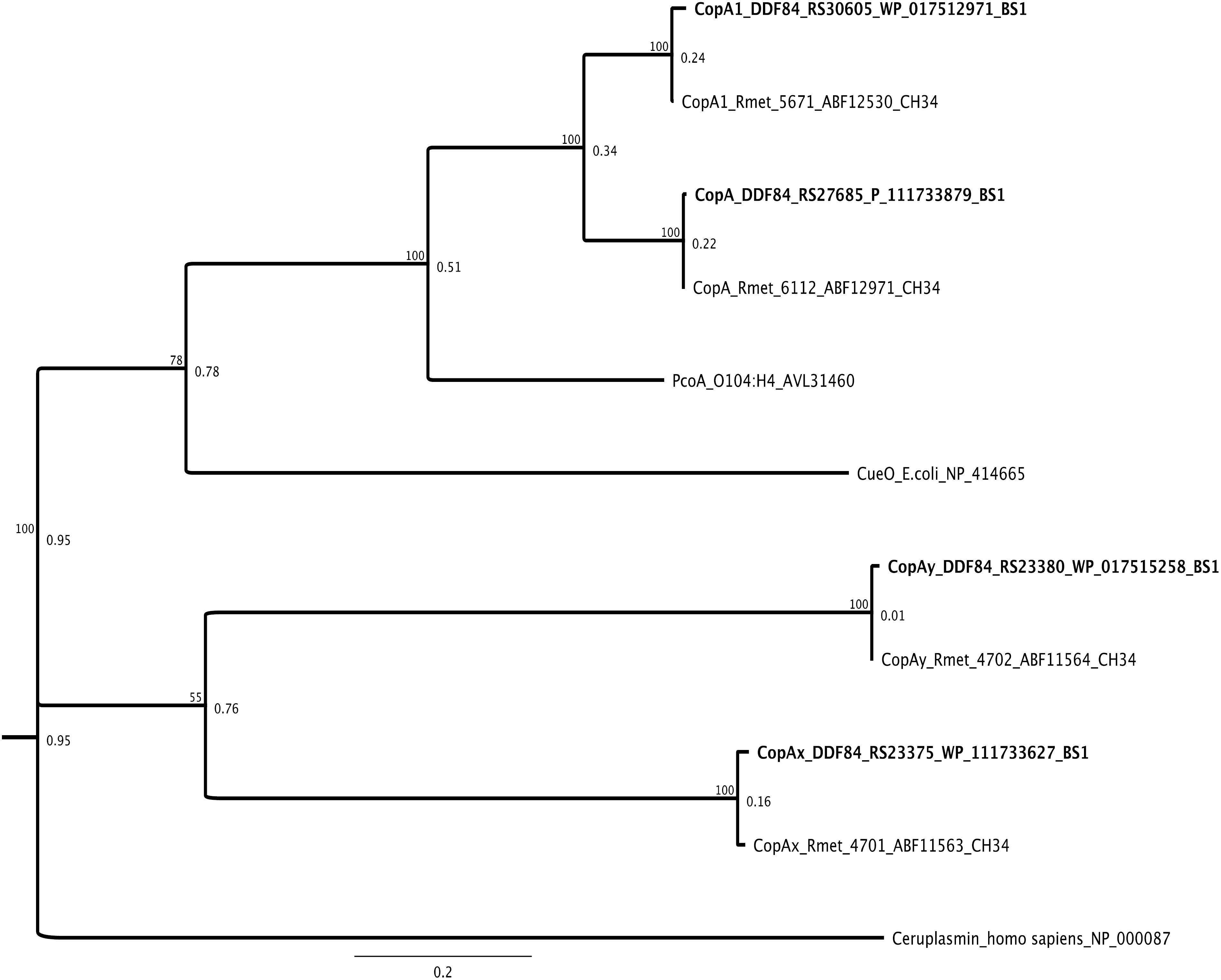
Figure 11. Molecular Phylogenetic tree based on the full amino acid sequences of the CopA (multi copper oxidases) protein encoded on the genome of C. metallidurans CH34 and the homologs as well as paralogous of both strains. In addition to the gene names, the respective locus tags and NCBI GenBank protein ID numbers of both strains are indicated, multicopper oxidases from BS1 are displayed in bold. The evolutionary history was inferred by using the Geneious prime 2020 0.4. (Geneious Tree Builder) (https://www.geneious.com), global alignment with free gaps, Identity (1.0/0.0), gap open penalty 12, gap extension penalty 3, 2 refinement iterations, Genetic Distance Model _ Jukes-Cantor, Neighbor-Joining, Resampling Method – Bootstrap; 100. The CueOE. coli, PcoAE. coli and Ceruplasmin (multi copper oxidase family) amino acid sequence from Escherichia coli strain K-12 substr. MG1655 and O104:H4 and Homo sapiens was used as reference. The tree is drawn to scale, with branch lengths calculated using the average pathway method; the scale bar corresponds to the number of substitutions per site, the branch labels are consensus Support (%) and Node Heights (Kearse et al., 2012).
The Czc determinant mediating resistance to Co, Zn, and Cd is encoded on pMOL30 in C. metallidurans CH34 (Getaneh and Alemayehu, 2006; Rea et al., 2016) starting from MgtC until CzcP from (Rmet_5985 - Rmet_5970). Duplication of this czcNICBADRSJE_czcP cluster in BS1 was shown to have led to one copy being present on chromid and pBS1, respectively, and generated by mobile elements as shown in Figure 7. Some of the czc determinant encoded proteins, such as CzcB, CzcD, CzcR, CzcS, CzcE, CzcJ, and CzcF displayed up to 100% similarity between C. metallidurans BS1 and CH34 while czc1 cluster on chromid was up to 100% similar (Supplementary Table S4). CzcA, and CzcC, CzcP were also found to have up to 99% AA sequence identity in both strains. Above mentioned components were located on different loci, as BS1 harbors them on chromid and plasmid while in CH34 they were only present on pMOL30 (Supplementary Table S6 and Figure 6). The duplicate on plasmid is less similar to czc on pMOL30 present on type strain C. metallidurans CH34, as compared to czc on chromid in C. metallidurans BS1 (Supplementary Table S4). Sequence of the gene on the megaplasmid pMOL30 of CH34 encoding CzcP was only 76% similar to CzcP encoded on the plasmid (czcP1) in BS1 which has a nonsense mutation in the middle of the reading-frame.
These observations suggested that the czc cluster present on chromid of BS1 was more preserved and similar to the czc cluster on pMOL30 in CH34 than czc present on the pBS1 in BS1. This phenomenon of a czc determinant duplication was observed in C. metallidurans BS1 while previously a similar phenomenon was also reported in C. metallidurans NA4 where a large part of the resistance systems was found not to be plasmid-encoded. C. metallidurans NA4 was shown to have similar genome architecture in that the main large metal resistance clusters on pMOL30 were syntenic with gene clusters on NA4’s chromid instead of NA4’s plasmids (pNA4_A-D) as was also the case in BS1 (Ali et al., 2019) (Supplementary Figure S2). The zntA_czcI2B2A2_S2R2 core-determinant was present (DDF84_RS21985 - DDF84_RS22020) comprising structural and regulatory genes as well as encoding the PIB2-type ATPase, ZntA (Legatzki et al., 2003; Scherer and Nies, 2009). The czcICBA cluster was referred here as a core determinant, as it is highly conserved in the genus Cupriavidus (Nies, 2016). These determinants have their corresponding components on the genome of CH34 with similarity of up to 99% and both determinants were present on chromid in respective strains. A recombination event (between to copies of Tn6050) rearranged the gene regions from Rmet_4598 – Rmet4687 and insert CMGI-B and D in strain CH34 led to a sequence break from locus czcB′′ at Rmet_4469 and starting again with locus czcB′_czcC (Rmet_4596) with same pattern onward (czcI Rmet_4595, PIB2-type ATPase Rmet_4594; ZntA) as the determinant was present in strain BS1 with 98–99% similarity index (Figure 7 and Supplementary Table S4). CMGI-B and the largest part of CMGI-D (CMGI-D′) are absent in BS1 and left the zntA – czcI2C2B2A2 intact. An interesting phenomenon of genome plasticity was pointed out in C. metallidurans BS1, as another or forth czc3 cluster could be found on the genome of BS1 with a gene encoding a Fe-CDF member (CdfY) in same direction and close to the mer2 cluster on the chromid (Figures 7, 9). CzcA3 is closely related to CzcA, CzcA1 as well as CzcA2 and shared 92.8, 92.9, and 79.2% similarity on AA level. All CzcAs clustered in one branch of the phylogenetic tree in comparison to other HME-RND proteins and the czc3 cluster showed another modifying duplication during evolution (Figure 7).
The cobalt and nickel resistance (cnr) operon in CH34 was similar to that found in BS1. Both the probable structural (cnrCBAT) and regulatory (cnrYXH) genes were present on chromid but with only 93, 89, 89, 93, 85, 83, and 61% AA sequence similarity to CnrA, CnrB, CnrC, CnrT, CnrH, CnrX, and CnrY from C. metallidurans CH34, respectively, indicating that this determinant might have undergone an accelerated mutation rate during evolutionary adaptation (Supplementary Table S5). From the complete genome sequence of BS1, we observed that nearly all those heavy metal resistance determinants harbored on pMOL30 and pMOL28 (Monchy et al., 2007) were present on the chromid in C. metallidurans BS1 displaying nearly the same pattern. Four operons, czc, cnr, chr, and mer on the chromid in C. metallidurans BS1 were present side by side with one mobile genetic element (MGEs) between czc and cnr, while in type strain CH34, czc is present on pMOL30 and cnr and chr are present on pMOL28 (Nies, 1995; Grass et al., 2000; Monchy et al., 2007) from region DDF84_RS23090 – DDF84_RS23305 (Figure 6). The similarity of these metal determinants between C. metallidurans BS1 and C. metallidurans CH34 was up to 100% except cnr but with same pattern (synteny) and location upstream of the chr cluster as it is in strain CH34 (Supplementary Table S5). The chromid also contained cop, sil and ncc determinants from 2,055,539 bp to 2,099,418 bp on the C. metallidurans BS1 genome surrounded by MGEs (Figure 6). The components of the ncc resistance determinant such as NreB, NccC, NccB, and NccA (Nickel-Cobalt-Cadmium Resistance Protein), showed a 100% identical AA sequence in both strains, CH34 and BS1. A nonsense/frameshift mutation in NccB, indicated to be present in strain CH34 and NA1, NA4, H1130 (Van Houdt et al., 2018) was also observed to be present in BS1. The smaller segment of NccB is 100% similar to the NccB segment in strain CH34 both by nucleotide and amino acid BLAST analysis but the larger locus was 99% similar, with one arginine replaced with isoleucine in BS1. The nickel resistance protein sequence, NreB, in C. metallidurans CH34 (Rmet_6144) on pMOL30 displayed 100% sequence identity to the protein found on the chromid of C. metallidurans BS1, which also encoded cnr putative structural genes, cnrCBA. The presence of the czc and cnr operons encoded on the same locus supported the idea of an ancestral cobalt or cobalt-zinc resistance operon that evolved divergently by duplication and acquired additional specificities to become cnr and czc (Janssen et al., 2010). Similar to C. metallidurans CH34, the chromate resistance (chr) determinant was located upstream of the cnr operon in C. metallidurans BS1. Using the RAST functional based comparison, it was determined that both C. metallidurans BS1 and CH34 contained genes encoding all the proteins (ChrABCEF) responsible for resistance to chromium compounds. A Blast search of the ChrI sequence of C. metallidurans CH34 obtained from NCBI against the genome of C. metallidurans BS1 in RAST showed that ChrI in C. metallidurans BS1 had only 44% AA sequence similarity to ChrI from C. metallidurans CH34.
In addition, the nimBAC locus encoding an RND-system was suggested to putatively play a role in resistance to Ni2+ and Co2+. The similarity index of nimBAC is 100%-96% as indicated in Supplementary Table S6. The nimBAC operon is suspected to be inactivated by a transposon insertion in CH34 but it is intact in NA1, NA4 and H1130 (Van Houdt et al., 2018).
Three mercury (Hg2+) resistance operon are encoded on the genome of BS1 and four in CH34, including six genes coding for MerR < – > MerTPC/ADE (Supplementary Figure S3 and Supplementary Table S6). Eleven genes encoding proteins associated with arsenic (AsO43–/AsO33–) resistance including ArsHR, 2 ACR3 (arsenical resistance protein), and 5 ArsC while 13 proteins were found associated with chromium (CrO42–) resistance including ChrABCEF (Supplementary Table S6 and Figure 6) could be detected on the genome of C. metallidurans BS1. In the genome of C. metallidurans BS1 there was a comparable number of MerR encoding genes with 97–100% similarity at the amino acid level to CH34 (Supplementary Figure S3). These included CupR (activator of CupA) and PbrR (activator of PbrA) and a not further characterized MerR regulator (locus tag: DDF84_RS1704_BS1/Rmet_3456_CH34) which clustered in the phylogenetic tree with the zinc-, cadmium- and lead-dependent homologs PbrR, CadR from C. metallidurans CH34 and CadR from Paraburkholderia xenovorans LB400 as well as ZntR from E. coli (Brocklehurst et al., 1999; Taghavi et al., 2009; Van Houdt et al., 2009). The three encoded homologous MerR regulators (locus tag: DDF84_RS08325, DDF84_RS22830 and DDF84_RS23300) of the mercury resistance clusters (MerR < - > MerTPC/ADE) are transposon duplicates with 100% similarity on the amino acid level including MerR2 (locus tag: Rmet_6171) of CH34. Two further mer clusters including the merR3 and merR4 genes from CH34 were missing in strain BS1 due to the absence of CMGI-1 and a transposon insertion on pMOL30 (Table 4). The Mer-type regulator SoxR, involved in the oxidative stress response, was present in both strains with a similarity of 98% in both strains, as well as homologs of as yet uncharacterized members of this protein family (Supplementary Figure S3).
Plasmids in C. metallidurans CH34 are of prime importance as most metal resistance determinants are encoded on pMOL30 and pMOL28 (Nies, 1999) but this is not the case in C. metallidurans BS1. The plasmid in BS1 only contains one duplicate copy of czcNICBADRSEJ_czcP cluster and abundant hypothetical proteins and mobile genetic elements. Genes encoding putative transfer functions TraW∗, TrU∗, TrbC∗, (∗orthologs of IncF plasmids) TraD, and TraN are predicted to be involved in plasmid transfer, were present on plasmid –pBS1 DNA displaying the same pattern as on pMOL30 in CH34. The pBS1 plasmid displayed more similarities with pMOL30 by having similar plasmid maintenance systems while, the pBS1 plasmid showed no similarities with pMOL28 with the sole exception of a 3.5 kbp region [mobile element; Rmet_6337 (DDF84_031600) – Rmet_6334 (DDF84_031615)]. Three copies of Polymyxin resistance protein amino-arabinose transferase ArnT were found on the chromid in BS1 and one copy on plasmid out of four ArnT locus in total while it is a component of pMOL30 in CH34. This last-resort antibiotic is affective against Gram negative bacteria by permeabilizing its outer membrane using electrostatic interactions between amino group of polymyxins and negatively charged glucosamine and 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) sugars of lipid A (Tseng et al., 1999). We believe that ArnT present in BS1 might be involved in outer membrane modifications in response to heavy metal stress. Genome plasticity might be responsible for relocating many heavy metal resistance determinants to the chromid. It could be suspected these determinants were fixed on the chromid as strain BS1 might be inhabiting an environment with high levels of heavy metals for a long time. It is predicted that transformation frequency and conjugation frequency is influenced and possibly induced by metals to select for increased survival in the environment. Mobile genetic elements (MGEs) play a vital role in evolutionary processes and are responsible for transferring genetic information among and within bacterial species. A significant role of MGEs is to harbor putative virulence factors and genes encoding molecules and functions that confer resistance to heavy metals and antibiotics while providing opportunities for Horizontal Gene Transfer.
This study presents the complete genome sequence and annotation of C. metallidurans BS1, a highly heavy metal resistant bacterium. The 7,164,523 bp complete genome sequences of C. metallidurans BS1 contains three replicons present as one chromosome, one chromid and one plasmid (pBS1). Putative heavy metal resistance determinants of C. metallidurans BS1 were largely similar to those present on the genome of C. metallidurans CH34. A noticeable difference was observed in C. metallidurans BS1 as major determinants of metal detoxification systems were located on the chromid instead of the plasmids (pMOL30 and pMOL28) in C. metallidurans CH34. Known gold–copper detoxification systems (CupR/CueR, CupA, CupC and CopA) found in C. metallidurans CH34 were also present in C. metallidurans BS1. Two prophages detected on the genome are probably intact and one very likely intact. Metal resistance determinants from different origins in BS1 displayed critical differences between C. metallidurans BS1 and CH34. Future studies would be aimed to explore the mechanisms involved in the interplay between gold and copper in gold–copper detoxification systems. In addition, one future aim will be to better understand the evolutionary adaptation to the auriferous environment of isolation on the genetic level and further reconstruct genomic rearrangements.
The data used to support the findings of this study are included within the article. Please check https://www.ncbi.nlm.nih.gov/assembly/GCA_003260185.2 for more details.
SM and SB performed the laboratory experiments, analyzed the annotated genome sequence, submitted genome to NCBI, wrote the draft manuscript, and participated in the design of the experiments. MH carried out the genomic characterization and constructed the phylogenetic trees. IF, YL, JS, and JX performed the description of the sampling environment, sampling, isolation and microbiological characterization of the bacteria. CZ, SM, and SB carried out the isolation and purification of the genomic DNA. CR, RF, SZ, and IF participated in the design of the study and revised the manuscript. All authors read and approved the final manuscript.
This research was funded by National Natural Science Foundation of China (NSFC) grant number 31770123, and State administration of Foreign Expert Project numbers GDT20173600005 and 110000214620170006.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge all the staff of Zijin gold–copper mine for their technical assistance during sample collection.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00047/full#supplementary-material
Ali, M. M., Provoost, A., Maertens, L., Leys, N., Monsieurs, P., Charlier, D., et al. (2019). Genomic and transcriptomic changes that mediate increased platinum resistance in Cupriavidus metallidurans. Genes 10:E63. doi: 10.3390/genes10010063
Altschul, S. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anton, A., Grass, G., Rensing, C., and Nies, D. H. (2004). Characteristics of zinc transport by two bacterial cation diffusion facilitators from Ralstonia metallidurans CH34 and Escherichia coli. J. Bacteriol. 186, 7499–7507. doi: 10.1128/JB.186.22.7499
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Aziz, R. K., Bartels, D., Best, A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Blondel, V. D., Guillaume, J. L., Lambiotte, R., and Lefebvre, E. (2008). Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008:10008. doi: 10.1088/1742-5468/2008/10/P10008
Brocklehurst, K. R., Hobman, J. L., Lawley, B., Blank, L., Marshall, S. J., Brown, N. L., et al. (1999). ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31, 893–902. doi: 10.1046/j.1365-2958.1999.01229.x
Brown, N. L., Stoyanov, J. V., Kidd, S. P., and Hobman, J. L. (2003). The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27, 145–163. doi: 10.1016/s0168-6445(03)00051-2
Bütof, L., Schmidt-Vogler, C., Herzberg, M., Große, C., and Nies, D. H. (2017). The components of the unique Zur regulon of Cupriavidus metallidurans mediate cytoplasmic Zinc handling. J. Bacteriol. 199:e00372-17. doi: 10.1128/JB.00372-17
Bütof, L., Wiesemann, N., Herzberg, M., Altzschner, M., Holleitner, A., Reith, F., et al. (2018). Synergistic gold-copper detoxification at the core of gold biomineralisation in: Cupriavidus metallidurans. Metallomics 10, 278–286. doi: 10.1039/c7mt00312a
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chandrangsu, P., Huang, X., Gaballa, A., and Helmann, J. D. (2019). Bacillus subtilis FolE is sustained by the ZagA zinc metallochaperone and the alarmone ZTP under conditions of zinc deficiency. Mol. Microbiol. 112, 751–765. doi: 10.1111/mmi.14314
Checa, S. K., Espariz, M., Audero, M. E. P., Botta, P. E., Spinelli, S. V., and Soncini, F. C. (2007). Bacterial sensing of and resistance to gold salts. Mol. Microbiol. 63, 1307–1318. doi: 10.1111/j.1365-2958.2007.05590.x
Collard, J. M., Provoost, A., Taghavi, S., and Mergeay, M. (1993). A new type of Alcaligenes eutrophus CH34 zinc resistance generated by mutations affecting regulation of the cnr cobalt-nickel resistance system. J. Bacteriol. 175, 779–784. doi: 10.1128/jb.175.3.779-784.1993
Cromie, M. J., and Groisman, E. A. (2010). Promoter and riboswitch control of the Mg2+ transporter MgtA from Salmonella enterica. J. Bacteriol. 192, 604–607. doi: 10.1128/JB.01239-09
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Diels, L., and Mergeay, M. A. X. (1990). DNA probe-mediated detection of resistant bacteria from soils highly polluted by heavy metals. Appl. Environ. Microbiol. 56, 1485–1491. doi: 10.1128/aem.56.5.1485-1491.1990
Edward, W., Yu, Q. Z., and Baugh, M. H. (eds) (2013). “RND-efflux pumps for metal cations,” in Microbial Efflux Pumps: Current Research, (Berlin: Springer), 1–13.
Fagan, M. J., and Saier, M. H. (1994). P-type ATPases of eukaryotes and bacteria: sequence analyses and construction of phylogenetic trees. J. Mol. Evol. 38, 57–99. doi: 10.1007/BF00175496
Fairbrother, L., Etschmann, B., Brugger, J., Shapter, J., Southam, G., and Reith, F. (2013). Biomineralization of gold in biofilms of Cupriavidus metallidurans. Environ. Sci. Technol. 47, 2628–2635. doi: 10.1021/es302381d
Fan, B., and Rosen, B. P. (2002). Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. J. Biol. Chem. 277, 46987–46992. doi: 10.1074/jbc.M208490200
Fang, L.-C., Chen, Y.-F., Zhou, Y.-L., Wang, D.-S., Sun, L.-N., Tang, X.-Y., et al. (2016). Complete genome sequence of a novel chlorpyrifos degrading bacterium, Cupriavidus nantongensis X1. J. Biotechnol. 227, 1–2. doi: 10.1016/j.jbiotec.2016.04.012
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
Gardner, P. P., Daub, J., Tate, J. G., Nawrocki, E. P., Kolbe, D. L., Lindgreen, S., et al. (2009). Rfam: updates to the RNA families database. Nucleic Acids Res. 37, D136–D140. doi: 10.1093/nar/gkn766
Gent, U., Mol, B., Goris, J., De Vos, P., Coenye, T., Hoste, B., et al. (2001). Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int. J. Syst. Evol. Microbiol. 51, 1773–1782. doi: 10.1099/00207713-51-5-1773
Getaneh, W., and Alemayehu, T. (2006). Metal contamination of the environment by placer and primary gold mining in the Adola region of southern Ethiopia. Environ. Geol. 50, 339–352. doi: 10.1007/s00254-006-0213-5
González-Guerrero, M., Raimunda, D., Cheng, X., and Argüello, J. M. (2010). Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 78, 1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x
Grass, G., Große, C., and Nies, D. H. (2000). Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J. Bacteriol. 182, 1390–1398. doi: 10.1128/JB.182.5.1390-1398.2000
Grass, G., Otto, M., Fricke, B., Haney, C. J., Rensing, C., Nies, D. H., et al. (2005). FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch. Microbiol. 183, 9–18. doi: 10.1007/s00203-004-0739-4
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, 52–57. doi: 10.1093/nar/gkm360
Grosse, C., Friedrich, S., and Nies, D. H. (2007). Contribution of extracytoplasmic function sigma factors to transition metal homeostasis in Cupriavidus metallidurans strain CH34. J. Mol. Microbiol. Biotechnol. 12, 227–240. doi: 10.1159/000099644
Große, C., Poehlein, A., Blank, K., Schwarzenberger, C., Schleuder, G., Herzberg, M., et al. (2019). The third pillar of metal homeostasis in Cupriavidus metallidurans CH34: preferences are controlled by extracytoplasmic function sigma factors. Metallomics 11, 291–316. doi: 10.1039/C8MT00299A
Heimann, J. D. (2002). The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46, 47–110. doi: 10.1016/S0065-2911(02)46002-X
Herzberg, M., Bauer, L., Kirsten, A., and Nies, D. H. (2016). Interplay between seven secondary metal uptake systems is required for full metal resistance of Cupriavidus metallidurans. Metallomics 8, 313–326. doi: 10.1039/C5MT00295H
Herzberg, M., Bauer, L., and Nies, D. H. (2014a). Deletion of the zupT gene for a zinc importer influences zinc pools in Cupriavidus metallidurans CH34. Metallomics 6, 421–436. doi: 10.1039/c3mt00267e
Herzberg, M., Dobritzsch, D., Helm, S., Baginsky, S., and Nies, D. H. (2014b). The zinc repository of Cupriavidus metallidurans. Metallomics 6, 2157–2165. doi: 10.1039/C4MT00171K
Herzberg, M., Schüttau, M., Reimers, M., Große, C., Hans-Günther-Schlegel, H.-G.-S., and Nies, D. H. (2015). Synthesis of nickel–iron hydrogenase in Cupriavidus metallidurans is controlled by metal-dependent silencing and un-silencing of genomic islands. Metallomics 7, 632–649. doi: 10.1039/C4MT00297K
Hobman, J. L., Wilkie, J., and Brown, N. L. (2005). A design for life: prokaryotic metal-binding MerR family regulators. Biometals 18, 429–436. doi: 10.1007/s10534-005-3717-7
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). EGGNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Ibáñez, M. M., Checa, S. K., and Soncini, F. C. (2015). A single serine residue determines selectivity to monovalent metal ions in metalloregulators of the MerR family. J. Bacteriol. 197, 1606–1613. doi: 10.1128/JB.02565-14
Janssen, P. J., Van Houdt, R., Moors, H., Monsieurs, P., Morin, N., Michaux, A., et al. (2010). The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One 5:e10433. doi: 10.1371/journal.pone.0010433
Jian, X., Wasinger, E. C., Lockard, J. V., Chen, L. X., and He, C. (2009). Highly sensitive and selective gold(I) recognition by a metalloregulator in Ralstonia metallidurans. J. Am. Chem. Soc. 131, 10869–10871. doi: 10.1021/ja904279n
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, E.-H., Nies, D. H., McEvoy, M. M., and Rensing, C. (2011). Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J. Bacteriol. 193, 2381–2387. doi: 10.1128/JB.01323-10
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Lagesen, K., Hallin, P., Rødland, E. A., Stærfeldt, H.-H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lee, S. (2002). Functional analysis of the Escherichia coli zinc transporter ZitB. FEMS Microbiol. Lett. 215, 273–278. doi: 10.1111/j.1574-6968.2002.tb11402.x
Lee, S., Khanal, A., Cho, A.-H., Lee, H., Kang, M.-S., Unno, T., et al. (2019). Cupriavidus sp. strain Ni-2 resistant to high concentration of nickel and its genes responsible for the tolerance by genome comparison. Arch. Microbiol. 201, 1323–1331. doi: 10.1007/s00203-019-01700-5
Legatzki, A., Grass, G., Anton, A., Rensing, C., and Nies, D. H. (2003). Interplay of the Czc system and two P-type ATPases in conferring metal resistance to Ralstonia metallidurans. J. Bacteriol. 185, 4354–4361. doi: 10.1128/JB.185.15.4354
Long, F., Su, C.-C., Lei, H.-T., Bolla, J. R., Do, S. V., and Yu, E. W. (2012). Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex. Philos. Trans. R. Soc. B Biol. Sci. 367, 1047–1058. doi: 10.1098/rstb.2011.0203
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 0955–0964. doi: 10.1093/nar/25.5.0955
Lu, M., and Fu, D. (2007). Structure of the Zinc transporter YiiP. Science 317, 1746–1748. doi: 10.1126/science.1143748
Maguire, M. E. (2006). Magnesium transporters: properties, regulation and structure. Front. Biosci. 11, 3149–3163.
Makarova, K., Wolf, Y., and Koonin, E. (2015). Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life 5, 818–840. doi: 10.3390/life5010818
Mascher, T. (2013). Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Curr. Opin. Microbiol. 16, 148–155. doi: 10.1016/j.mib.2013.02.001
Mergeay, M. (2015). “The history of Cupriavidus metallidurans strains isolated from anthropogenic environments,” in Metal Response in Cupriavidus metallidurans: Volume I: From Habitats to Genes and Proteins, eds M. Mergeay and R. Van Houdt, (Cham: Springer International Publishing), 1–19. doi: 10.1007/978-3-319-20594-6_1
Mergeay, M., Houba, C., and Gerits, J. (1978). Extrachromosomal inheritance controlling resistance to cadmium, cobalt, copper and zinc ions: evidence from curing in a Pseudomonas [proceedings]. Arch. Int. Physiol. Biochim. 86, 440–442.
Mergeay, M., Monchy, S., Vallaeys, T., Auquier, V., Benotmane, A., Bertin, P., et al. (2003). Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27, 385–410. doi: 10.1016/s0168-6445(03)00045-7
Mergeay, M., Nies, L. D., Schlegel, H. G., Gerits, J., Charles, P., and Gijsegem, F. V. A. N. (1985). Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162, 328–334. doi: 10.1128/jb.162.1.328-334.1985
Millacura, F., Janssen, P., Monsieurs, P., Janssen, A., Provoost, A., Van Houdt, R., et al. (2018). Unintentional genomic changes endow Cupriavidus metallidurans with an augmented heavy-metal resistance. Genes 9:551. doi: 10.3390/genes9110551
Missiakas, D., and Raina, S. (1998). The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28, 1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x
Molina-Henares, A. J., Godoy, P., Duque, E., and Ramos, J. L. (2009). A general profile for the MerR family of transcriptional regulators constructed using the semi-automated provalidator tool. Environ. Microbiol. Rep. 1, 518–523. doi: 10.1111/j.1758-2229.2009.00067.x
Monchy, S., Benotmane, M. A., Janssen, P., Vallaeys, T., Taghavi, S., van der Lelie, D., et al. (2007). Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 189, 7417–7425. doi: 10.1128/jb.00375-07
Monsieurs, P., Mijnendonckx, K., Provoost, A., Venkateswaran, K., Ott, C. M., Leys, N., et al. (2014). Genome sequences of Cupriavidus metallidurans strains NA1, NA4, and NE12, isolated from space equipment. Genome Announc. 2:e00719-14. doi: 10.1128/genomeA.00719-14
Monsieurs, P., Provoost, A., Mijnendonckx, K., Leys, N., Gaudreau, C., and Van Houdt, R. (2013). Genome sequence of Cupriavidus metallidurans strain H1130, isolated from an invasive human infection. Genome Announc. 1:e01051-13. doi: 10.1128/genomeA.01051-13
Montanini, B., Blaudez, D., Jeandroz, S., Sanders, D., and Chalot, M. (2007). Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8:107. doi: 10.1186/1471-2164-8-107
Munkelt, D., Grass, G., and Nies, D. H. (2004). The chromosomally encoded cation diffusion facilitator proteins DmeF and FieF from Wautersia metallidurans CH34 are transporters of broad metal specificity Doreen. J. Bacteriol. 186, 8036–8043. doi: 10.1128/JB.186.23.8036
Nawrocki, E. P., Burge, S. W., Bateman, A., Daub, J., Eberhardt, R. Y., Eddy, S. R., et al. (2015). Rfam: updates to the RNA families database. Nucleic Acids Res. 43, D130–D137. doi: 10.1093/nar/gku1063
Nies, A., Nies, D. H., and Silver, S. (1989). Cloning and expression of plasmid genes encoding resistances to chromate and cobalt in Alcaligenes eutrophus. J. Bacteriol. 171, 5065–5070. doi: 10.1128/jb.171.9.5065-5070.1989
Nies, D., Mergeay, M., Friedrich, B., and Schlegel, H. G. (1987). Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J. Bacteriol. 169, 4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987
Nies, D. H. (1992). CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J. Bacteriol. 174, 8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992
Nies, D. H. (1995). The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J. Bacteriol. 177, 2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995
Nies, D. H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51, 730–750. doi: 10.1007/s002530051457
Nies, D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi: 10.1016/s0168-6445(03)00048-2
Nies, D. H. (2016). The biological chemistry of the transition metal “transportome” of Cupriavidus metallidurans. Metallomics 8, 481–507. doi: 10.1039/C5MT00320B
Nies, D. H., Rehbein, G., Hoffmann, T., Baumann, C., and Grosse, C. (2006). Paralogs of genes encoding metal resistance proteins in Cupriavidus metallidurans strain CH34. J. Mol. Microbiol. Biotechnol. 11, 82–93. doi: 10.1159/000092820
Nikaido, H. (2018). RND transporters in the living world. Res. Microbiol. 169, 363–371. doi: 10.1016/j.resmic.2018.03.001
Okkeri, J., and Haltia, T. (2006). The metal-binding sites of the zinc-transporting P-type ATPase of Escherichia coli. Lys693 and Asp714 in the seventh and eighth transmembrane segments of ZntA contribute to the coupling of metal binding and ATPase activity. Biochim. Biophys. Acta Bioenerg. 1757, 1485–1495. doi: 10.1016/j.bbabio.2006.06.008
Ondov, B. D., Treangen, T. J., Melsted, P., Mallonee, A. B., Bergman, N. H., Koren, S., et al. (2016). Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17:132. doi: 10.1186/s13059-016-0997-x
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Paulsen, I. T., and Saier, M. H. (1997). A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 156, 99–103. doi: 10.1007/s002329900192
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Raimunda, D., González-Guerrero, M., Leeber, B. W., and Argüello, J. M. (2011). The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals 24, 467–475. doi: 10.1007/s10534-010-9404-3
Rea, M. A., Zammit, C. M., and Reith, F. (2016). Bacterial biofilms on gold grains—implications for geomicrobial transformations of gold. FEMS Microbiol. Ecol. 92:fiw082. doi: 10.1093/femsec/fiw082
Reith, F., Etschmann, B., Grosse, C., Moors, H., Benotmane, M. A., Monsieurs, P., et al. (2009). Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. Proc. Natl. Acad. Sci. U.S.A. 106, 17757–17762. doi: 10.1073/pnas.0904583106
Reith, F., Lengke, M. F., Falconer, D., Craw, D., and Southam, G. (2007). The geomicrobiology of gold. ISME J. 1, 567–584. doi: 10.1038/ismej.2007.75
Rensing, C., Fan, B., Sharma, R., Mitra, B., and Rosen, B. P. (2000). CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 97, 652–656. doi: 10.1073/pnas.97.2.652
Rensing, C., Mitra, B., and Rosen, B. P. (1997). The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 94, 14326–14331. doi: 10.1073/pnas.94.26.14326
Rojas, L. A., Yáñez, C., González, M., Lobos, S., Smalla, K., and Seeger, M. (2014). Characterization of the metabolically modified heavy metal-resistant Cupriavidus metallidurans strain MSR33 generated for mercury bioremediation. PLoS One 6:e17555. doi: 10.1371/journal.pone.0017555
Scherer, J., and Nies, D. H. (2009). CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol. Microbiol. 73, 601–621. doi: 10.1111/j.1365-2958.2009.06792.x
Silver, S., Nucifora, G., and Phung, L. T. (1993). Human Menkes X-chromosome disease and the staphylococcal cadmium-resistance ATPase: a remarkable similarity in protein sequences. Mol. Microbiol. 10, 7–12. doi: 10.1111/j.1365-2958.1993.tb00898.x
Stoyanov, J. V., and Brown, N. L. (2003). The Escherichia coli copper-responsive copA promoter is activated by gold. J. Biol. Chem. 278, 1407–1410. doi: 10.1074/jbc.C200580200
Ta, C., Reith, F., Brugger, J., Pring, A., and Lenehan, C. E. (2014). Analysis of gold(I/III)-complexes by HPLC-ICP-MS demonstrates gold(III) stability in surface waters. Environ. Sci. Technol. 48, 5737–5744. doi: 10.1021/es404919a
Taghavi, S., Lesaulnier, C., Monchy, S., Wattiez, R., Mergeay, M., and van der Lelie, D. (2009). Lead(II) resistance in Cupriavidus metallidurans CH34: interplay between plasmid and chromosomally-located functions. Antonie Van Leeuwenhoek 96, 171–182. doi: 10.1007/s10482-008-9289-0
Tatusov, R. L. (2001). The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29, 22–28. doi: 10.1093/nar/29.1.22
Tottey, S., Harvie, D. R., and Robinson, N. J. (2005). Understanding how cells allocate metals using metal sensors and metallochaperones. Acc. Chem. Res. 38, 775–783. doi: 10.1021/ar0300118
Tseng, T. T., Gratwick, K. S., Kollman, J., Park, D., Nies, D. H., Goffeau, A., et al. (1999). The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1, 107–125.
Van Houdt, R., Monchy, S., Leys, N., and Mergeay, M. (2009). New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie Van Leeuwenhoek 96, 205–226. doi: 10.1007/s10482-009-9345-4
Van Houdt, R., Monsieurs, P., Mijnendonckx, K., Provoost, A., Janssen, A., Mergeay, M., et al. (2012). Variation in genomic islands contribute to genome plasticity in Cupriavidus metallidurans. BMC Genomics 13:111. doi: 10.1186/1471-2164-13-111
Van Houdt, R., Provoost, A., Van Assche, A., Leys, N., Lievens, B., Mijnendonckx, K., et al. (2018). Cupriavidus metallidurans strains with different mobilomes and from distinct environments have comparable phenomes. Genes 9:E507. doi: 10.3390/genes9100507
Vandamme, P., and Coenye, T. (2004). Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. 54, 2285–2289. doi: 10.1099/ijs.0.63247-0
Wang, K., Sitsel, O., Meloni, G., Autzen, H. E., Andersson, M., Klymchuk, T., et al. (2014). Structure and mechanism of Zn2+-transporting P-type ATPases. Nature 514, 518–522. doi: 10.1038/nature13618
Wiesemann, N., Bütof, L., Herzberg, M., Hause, G., Berthold, L., Etschmann, B., et al. (2017). Synergistic toxicity of copper and gold compounds in Cupriavidus metallidurans. Appl. Environ. Microbiol. 83:e01679-17. doi: 10.1128/AEM.01679-17
Wiesemann, N., Mohr, J., Grosse, C., Herzberg, M., Hause, G., Reith, F., et al. (2013). Influence of copper resistance determinants on gold transformation by Cupriavidus metallidurans strain CH34. J. Bacteriol. 195, 2298–2308. doi: 10.1128/JB.01951-12
Zammit, C. M., Weiland, F., Brugger, J., Wade, B., Winderbaum, L. J., Nies, D. H., et al. (2016). Proteomic responses to gold(III)-toxicity in the bacterium Cupriavidus metallidurans CH34. Metallomics 8, 1204–1216. doi: 10.1039/C6MT00142D
Keywords: Cupriavidus metallidurans, gold–copper mine, heavy metal efflux systems, cation diffusion facilitators, complete genome
Citation: Mazhar SH, Herzberg M, Ben Fekih I, Zhang C, Bello SK, Li YP, Su J, Xu J, Feng R, Zhou S and Rensing C (2020) Comparative Insights Into the Complete Genome Sequence of Highly Metal Resistant Cupriavidus metallidurans Strain BS1 Isolated From a Gold–Copper Mine. Front. Microbiol. 11:47. doi: 10.3389/fmicb.2020.00047
Received: 01 October 2019; Accepted: 10 January 2020;
Published: 07 February 2020.
Edited by:
Robert Duran, Université de Pau et des Pays de l’Adour, FranceReviewed by:
Rob Van Houdt, Belgian Nuclear Research Centre, BelgiumCopyright © 2020 Mazhar, Herzberg, Ben Fekih, Zhang, Bello, Li, Su, Xu, Feng, Zhou and Rensing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shungui Zhou, c2d6aG91QHNvaWwuZ2QuY24=; Christopher Rensing, cmVuc2luZ0BmYWZ1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.