
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 28 January 2020
Sec. Fungi and Their Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00038
 Svyatoslav S. Sokolov1*
Svyatoslav S. Sokolov1* Margarita A. Vorobeva2
Margarita A. Vorobeva2 Alexandra I. Smirnova1,2
Alexandra I. Smirnova1,2 Ekaterina A. Smirnova1
Ekaterina A. Smirnova1 Nataliya I. Trushina1,3
Nataliya I. Trushina1,3 Kseniia V. Galkina1,2
Kseniia V. Galkina1,2 Fedor F. Severin1
Fedor F. Severin1 Dmitry A. Knorre1,4
Dmitry A. Knorre1,4Lam proteins transport sterols between the membranes of different cellular compartments. In Saccharomyces cerevisiae, the LAM gene family consists of three pairs of paralogs. Because the function of paralogous genes can be redundant, the phenotypes of only a small number of LAM gene deletions have been reported; thus, the role of these genes in yeast physiology is still unclear. Here, we surveyed the phenotypes of double and quadruple deletants of paralogous LAM2(YSP2)/LAM4 and LAM1(YSP1)/LAM3(SIP3) genes that encode proteins localized in the junctions of the plasma membrane and endoplasmic reticulum. The quadruple deletant showed increased sterol content and a strong decrease in ethanol, heat shock and high osmolarity resistance. Surprisingly, the quadruple deletant and LAM2/LAM4 double deletion strain showed increased tolerance to the azole antifungals clotrimazole and miconazole. This effect was not associated with an increased rate of ABC-transporter substrate efflux. Possibly, increased sterol pool in the LAM deletion strains postpones the effect of azoles on cell growth. Alternatively, LAM deletions might alleviate the toxic effect of sterols as Lam proteins can transport toxic sterol biosynthesis intermediates into membrane compartments that are sensitive to these compounds. Our findings reveal novel biological roles of LAM genes in stress tolerance and suggest that mutations in these genes may confer upregulation of a mechanism that provides resistance to azole antifungals in pathogenic fungi.
Ergosterol is a primary sterol found in the plasma membrane of Ascomycota fungi (Weete et al., 2010). Inhibition of the upstream reactions of ergosterol biosynthesis abrogates cell growth and division (Giaever et al., 2002). While cells can proliferate without the genes ERG2, ERG3, ERG4, ERG5, and ERG6, which are required for the later steps of the ergosterol biosynthesis pathway (Giaever et al., 2002), deletions of these genes have been shown to decrease yeast fitness under non-optimal conditions (Abe and Hiraki, 2009; Jakubkova et al., 2016; Liu et al., 2017). However, the deletion of ERG genes increases the resistance of yeast cells to some stresses, including high osmolarity (Bard et al., 1978) and high tetramethylammonium concentrations (Kodedová and Sychrová, 2015). These effects are linked to hyperpolarization of the plasma membrane in ergosterol-deficient strains (Bard et al., 1978; Welihinda et al., 1994). Moreover, ergosterol plays a major role in the ethanol tolerance of yeast cells (Aguilera et al., 2006), and inhibiting ergosterol biosynthesis at earlier stages of the pathway can increase yeast resistance to some stresses. For instance, partial inhibition of C-14 demethylation of lanosterol (Erg11p) by fluconazole increases the growth rate of yeast cells in the presence of 400 mM NaCl (Montañés et al., 2011); deletion of ERG genes increases the growth rate under elevated temperatures of 39.5°C (Liu et al., 2017). Therefore, while being essential for survival in some stressful conditions, high ergosterol content in the plasma membrane can be detrimental in other conditions.
Ergosterol biosynthesis takes place in the endoplasmic reticulum (ER). Ergosterol is subsequently transported to the plasma membrane (PM), by (mostly) non-vesicular mechanisms (Baumann et al., 2005; Alli-Balogun and Levine, 2019; Sokolov et al., 2019). Lam proteins with sterol-binding StART-like domains contribute to ER/PM ergosterol turnover (Gatta et al., 2015). StART domains bind ergosterol and facilitate its transport between membranes (Horenkamp et al., 2018; Tong et al., 2018); therefore, the deletion of LAM genes can alter sterol distribution in cells and influence the sterol concentration in PMs.
The phenotypes resulting from mutations in the LAM genes remain uncertain. The redundancy of some LAM genes further complicates the study. The Saccharomyces cerevisiae genome contains three pairs of paralogous LAM genes: LAM1 (YSP1)/LAM3 (SIP3), LAM2 (YSP2)/LAM4, and LAM5/LAM6 (Wong and Levine, 2016). While LAM1/LAM3 and LAM2/LAM4 paralogs are localized in the contact sites of ER and PM, the localization of LAM5/LAM6 is not adjacent to the PM (Gatta et al., 2015). It has been shown that Lam6 resides in the mitochondrial/ER and mitochondrial/vacuole contact sites as well as in the nuclear vacuolar junction (Elbaz-Alon et al., 2015). In our study, we focused on Lam1–Lam4 proteins, which appear to have similar intracellular localization.
A single-gene deletion in any of the pairs can be compensated for by the function of the paralog. Nonetheless, single-mutant knockouts of LAM genes produce specific phenotypes: (1) the deletion of either LAM1 (YSP1) (Pozniakovsky et al., 2005) or LAM2 (YSP2) (Sokolov et al., 2006) increases the survival of yeast cells treated with high concentrations of amiodarone. Amiodarone is an antiarrhythmic drug that induces PM hyperpolarization, calcium influx, and acidification of the cytoplasm in yeast cells (Maresova et al., 2009). It also inhibits ABC-transporter-mediated drug efflux in yeasts (Knorre et al., 2009). Importantly, ergosterol biosynthesis mutants are hypersensitive to amiodarone (Gupta et al., 2003). Together, this indicates a strong interaction between LAM and ERG genes. (2) We also found that the Δlam2 (ysp2) strain is resistant to acidification of the cytoplasm induced by acetic acid (Sokolov et al., 2006). (3) The LAM2 gene confers resistance to antifungal amphotericin B (Gatta et al., 2015) and LAM3 (SIP3) to miconazole (François et al., 2009). Given that amphotericin B specifically disrupts ergosterol-enriched membranes, LAM mutant strains may carry increased sterol concentrations in the PM. On the other hand, it has been reported that amphotericin B aggregates extracted ergosterol from the yeast PM and that presaturation of amphotericin with ergosterol prevents its antifungal activity (Anderson et al., 2014). Therefore, it is impossible to draw definitive conclusions solely based on amphotericin B sensitivity. To the best of our knowledge, the changes in sterol content observed in yeast LAM mutants have not been measured by any direct methods so far. (4) While the deletion of either LAM2 (YSP2) or LAM3 (SIP3) gene increases resistance to quinine, an agent that disturbs membrane integrity, ERG3 and ERG6 deletion strains were found to be hypersensitive to quinine (Dos Santos et al., 2014). Nonetheless, the biological role of LAM genes is still unclear and the phenotypes of LAM mutant strains remain poorly characterized.
In this study, we surveyed the phenotypes of S. cerevisiae strains with double deletions of LAM1/LAM3 and LAM2/LAM4 paralog pairs. We found that the cells of the quadruple deletant, Δlam1Δlam2Δlam3Δlam4, have increased sterol content and decreased resistance to some environmental stresses, such as heat shock, increased salinity, and high ethanol concentrations. During salt stress, we detected a redundancy of gene function, and the LAM2 gene contributed highly to the major part of the effect on ethanol resistance. Surprisingly, in the Δlam2Δlam4 strain we observed an increased resistance to azole antifungals. We showed that this effect was not associated with the increased drug efflux by the pleiotropic drug resistance transporters (PDR), although the deletions induced Pdr5 accumulation in the cell PM and the vacuole. We discuss several possible mechanisms which could mediate this increase in azole resistance upon deletion of LAM genes.
Deletion and GFP-fusion strains were obtained by homologous recombination of the PCR product with heterologous selection markers, as listed in Table 1. All strains in this study are derivatives of the W303 strain. As templates for PCR, the DNA of yeast strains from Euroscarf collection, pUG27 (Gueldener et al., 2002) and pFA6a-His3MX6 (Longtine et al., 1998) plasmids were used. All newly generated strains were verified by PCR with independently designed primers (Table 2) and RT-qPCR (Supplementary Text S1, Table S1, Figure S1). We used standard yeast-rich and synthetic mediums described by Sherman (2002). We obtained yeast extract from BD and D-glucose from Helicon. Clotrimazole, miconazole, nigericin, amphotericin B, propanol, butanol, and FM4–64 were obtained from Thermo Fisher Scientific. Bacto Agar, peptone, NaCl, KCl, and NaN3 were obtained from Amresco, and 2-Deoxy-D-glucose was obtained from Chem-Impex Int’l Inc.
Cells were grown overnight in 20 ml of YPD in 100-ml flasks upto the stationary phase and then centrifuged and resuspended in 20 ml of water. Fifteen milliliters of the cell suspension were transferred onto a drying filter and dried to a constant weight at 95°C. Thus, the dry weight of 15 ml of cell suspension was determined. Ergosterol content in 1 ml of cell suspension was determined as described in by Arthington-Skaggs et al. (1999) with modifications. One milliliter of the cells was concentrated by brief centrifugation and resuspended in 10 μl of water. The cells were then resuspended in 500 μl of a 25% solution of KOH in ethanol (1 g of KOH was dissolved in 1.4 ml of water and adjusted to 4 ml with ethanol) for 1 h at 85°C. The solution was cooled to room temperature, and sterol was extracted into a mixture of 500 μl of heptane with 100 μl of water by vortexing for 3 min. The absorption spectrum of sterol was recorded using spectrophotometer (SpectrostarNANO) in the Microplate UV–VIS, 96/F (Eppendorf) in 200 μl of sample.
Filipin staining was performed according to the method described by Grossmann et al. (2007). Living cells were washed in 50 mM potassium phosphate buffer, pH 5.5, diluted to A600 = 0.3, stained with 5 μg/ml filipin (Sigma) for 5 min, washed again in the same buffer, concentrated by brief centrifugation, and analyzed using a fluorescent microscope with a U-MNU2 filter set (λexitation = 360–370 nm; dichroic mirror λ = 400 nm; λemission > 420 nm). Because filipin is very prone to photobleaching, we started fluorescence exposure of each field of view simultaneously with accumulation of the camera signal.
Cells were grown overnight to exponential phase, then diluted to an optical density of OD550 = 0.2 (Thermo Genesys2). Two microliters of cell suspension were transferred onto 0.3% agar YNB complete plates. Colony morphology was analyzed on the fifth day of growth.
Exponentially growing cells were diluted to an optical density of OD550 = 0.2 and inoculated into a 48-well plate (Greiner). Plates were incubated in a spectrophotometer (SpectrostarNANO) with the following settings: orbital shaking at 500 rpm for 2 min at 30°C before measurements; measurements were performed at 5-min intervals. To quantify the results in non-stressful conditions, we compared the growth rates (μ) between the control and mutant strains. In stressful conditions, we compared the increase in OD between the first and tenth hour of growth. We excluded first-hour measurements from the analysis due to artifacts in the first measurements, which could be attributed to fogging of the plate lid.
Cells were grown overnight to the logarithmic growth phase, then diluted to OD550 = 0.2. We measured the survival of cells as the number of colony forming units (CFU) in the mutant strain normalized to unstressed cells. We evaluated the number of colonies after 24 h of growth at 30°C on solid YPD medium. Hundred percent represented the number of CFU at the moment of stress exposure (t = 0). Cells were subjected to either 12% (v/v) ethanol for 1 h, 2.3 M NaCl for 1 h, heat shock (47°C for 30 min), or freezing (−20°C for 18 h).
Fluorescence of GFP was assessed with a Beckman Coulter FC 500 flow cytometer using an excitation wavelength of 488 nm on the emission filter (525/40 nm). The accumulation of Nile red and rhodamine 6G was measured with an emission filter (575/20 nm). At least 30,000 events were analyzed in each experiment. We calculated the average fluorescence intensity in populations for each separate-day biological replicate. Cells with GFP expression were grown overnight in solid YPD medium and then resuspended to a density of 2 × 105 cells/ml in the same medium. Fluorescence was assessed after 1 h of preincubation with clotrimazole or solvent at 30°C.
Cells were grown overnight on solid YPD medium, resuspended to a density of 2 × 105 cells/ml in phosphate-buffered saline (PBS, Gibco), and supplemented with 5 mM 2-Deoxy-D-glucose and 10 mM NaN3. After 2 h of incubation with Nile red (3.5 μM) or rhodamine 6G (400 nM) at 30°C, cells were washed twice with PBS and resuspended in PBS with 0.1% glucose. The amount of Nile red and rhodamine 6G in the yeast cells was measured by flow cytometry.
To study the accumulation of GFP, yeast cells were visualized using an Olympus BX41 microscope with the U-MNIBA3 filter (excitation wavelength 470–495 nm; beam splitter filter 505 nm; emission 510–550 nm) for GFP, and the U-MNG2 filter (excitation wavelength 530–550 nm, beam splitter filter 570 nm; emission >590 nm) for FM4–64. Photographs were taken with a DP30BW CCD camera. All results were reproduced in at least three biological replicates. To visualize vacuolar membranes, yeast cells were stained with 0.8 μM FM4–64 (Thermo Fisher Scientific) at 30°C for 1 h in YPD with clotrimazole or solvent, then washed twice with PBS.
We compared different strains and conditions using the Wilcoxon rank-sum test or Wilcoxon signed-rank test with the Bonferroni adjustment for multiple comparisons. To visualize the data, we used R software (R Core Team, 2014) and provided individual data points when possible.
We generated a set of S. cerevisiae strains with deleted pairs of LAM paralogs (see Table 1). LAM1/LAM3 and LAM2/LAM4 genes represent the paralog pairs that arose as a result of whole-genome duplication and were preserved in the S. cerevisiae genome during evolution. The quadruple deletion strain showed increased total sterol concentration (Figure 1A). To determine the intracellular distribution of sterol, we stained yeast cells with filipin, a sterol-sensitive fluorescent dye. Filipin interacts with sterol but not with esterified sterol [see Wilhelm et al., 2019]. To validate the usage of filipin, we took a strain with conditionally regulated ERG9 gene and that with a dominant gain of function mutation in sterol-sensitive transcription factor UPC2–1 and demonstrated that 24 h of preincubation of PGAL-ERG9 strain in YPD plates (the condition of PGAL-ERG9 repression) significantly reduced filipin staining of yeast cells (Figure 1B). Furthermore, the UPC2-1 allele increased filipin staining in both the cytoplasm and PM (Figures 1B–D). Therefore, we concluded that in our experimental system, filipin can be used to visualize sterol. We quantified filipin fluorescence in the PMs (Figure 1C) and cytosol (Figure 1D) of the wild-type and LAM mutant cells. Deletion of either LAM gene pairs increased filipin staining, suggesting an increase in sterol accumulation. Surprisingly, in the UPC2-1 and LAM double and quadruple deletion strains, we detected cytoplasmic staining (Figure 1E). It appeared that despite an increase in the plasma membrane sterol levels, the UPC2-1 and LAM deletants showed redistribution of non-esterified sterol from PM into the inner cellular compartments.
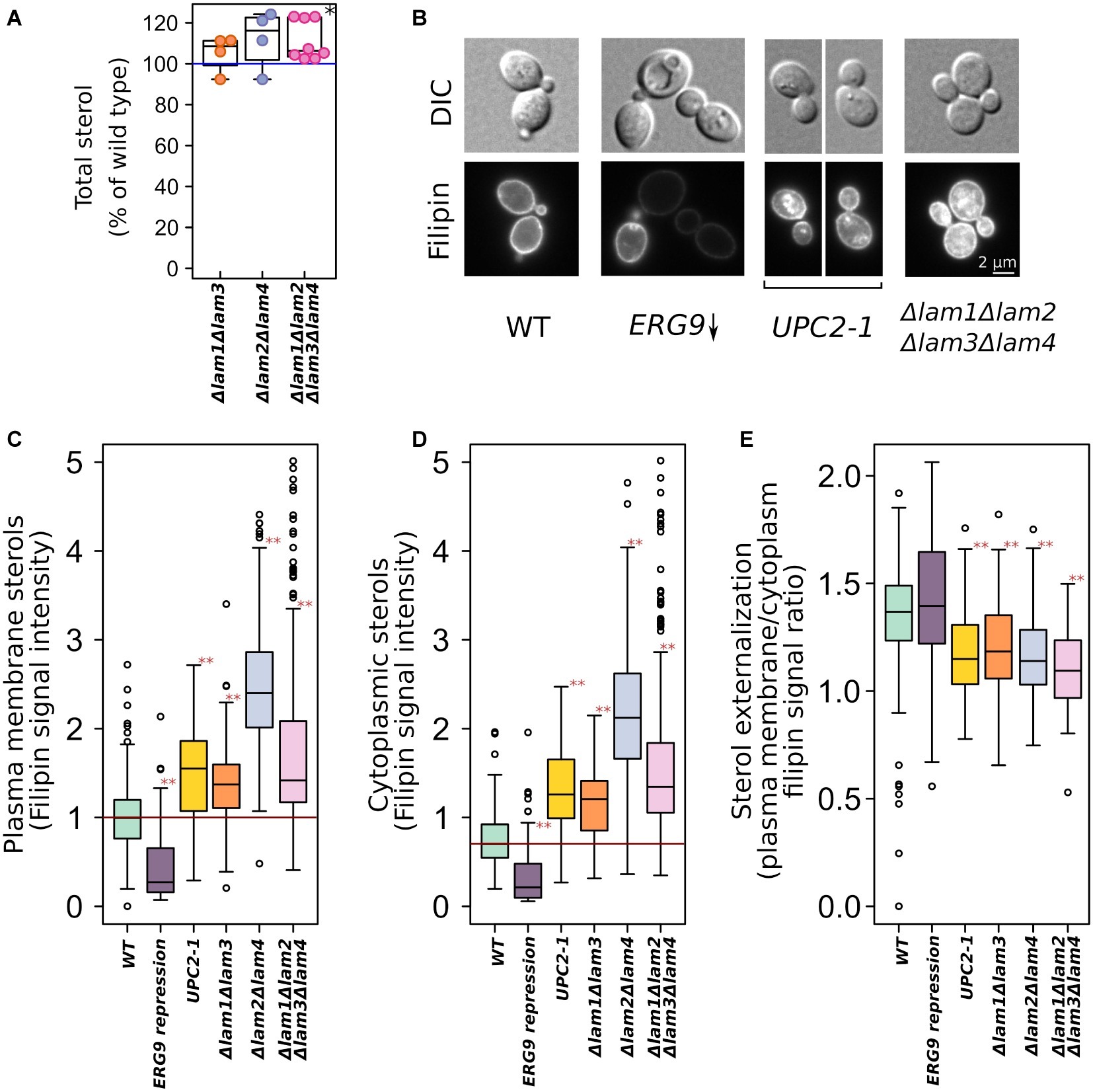
Figure 1. Increased sterol concentrations in the strains with deleted LAM genes. (A) Total sterol content was measured via light absorbance at λ = 282 nm of the wild-type and deletion strain cells. Plasma membrane ergosterol levels were assessed via fluorescent microscopy using filipin, a sterol-binding fluorescent dye. (B) Representative photographs of the wild-type and mutant yeast cells stained with filipin. Yeast cells were grown in YPD plates and then analyzed using a fluorescent microscope. This condition ensured repression of PGAL-ERG9 gene. Quantification of filipin plasma membrane (C) and cytoplasmic (D) staining for strains from B as well for mutant strains with double deletions. (E) Ratio of PM/inner compartments filipin staining. *p < 0.01 for comparisons with the WT, paired Wilcoxon signed-rank test; **p < 10−7 for comparisons with the WT, paired Wilcoxon signed-rank test with Bonferroni adjustment.
When analyzing growth kinetics in rich medium with glucose as carbon source (YPD), we detected a small decrease in growth rate for Δlam1Δlam2Δlam3Δlam4 and Δlam2Δlam4 mutant strains (Figure 2A). Moreover, although LAM deletions conferred no effect of LAM deletions on colony size in solid YPD medium, the quadruple mutant strain showed decreased ability to invade low-agar media (Figure 2B). Next, for the preliminary screening of phenotypes, we assessed the effects of alcohols (ethanol and propanol) and hyperosmolarity conditions. The resistance to these stresses depends on the PM sterol concentrations, according to previously published reports (see section “Introduction”). We found that all tested conditions, with the exception of high KCl concentration, inhibited the growth of the mutants that lacked LAM2 and LAM4, while the effect of the double deletion of LAM1 and LAM3 was less pronounced in all tested cases (Figures 2C,D).
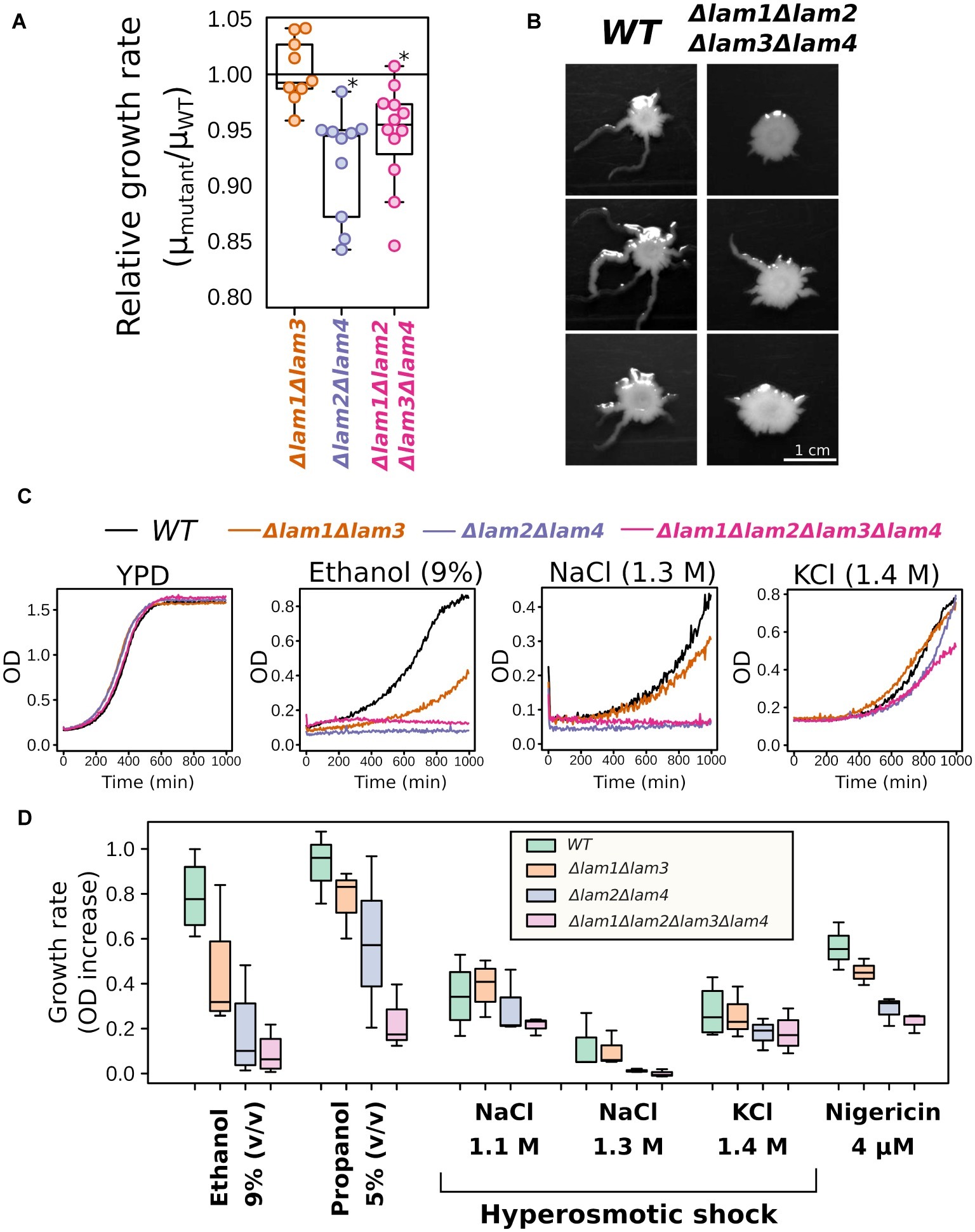
Figure 2. Growth rate phenotypes of LAM1-4 deletion mutants. (A) Deletion of LAM2 and LAM4 paralogs decreases relative growth rate in YPD; *p < 0.05, Wilcoxon signed-rank test. (B) Yeast colonies after 5 days of growth in low-agar solid medium, representative photographs (n = 3). (C,D) LAM2 and LAM4 significantly contribute to growth rate in stressful conditions. (C) Representative growth curves. (D) Quantification of the results. (n = 3–4).
We tested the survival of the double and quadruple LAM mutants under selected extreme environmental stresses: high osmolarity, high ethanol concentration, heat shock, and freezing. In line with growth experiments, we observed decreased survival of Δlam1Δlam2Δlam3Δlam4 and Δlam2Δlam4 cells in the presence of high concentrations of ethanol and under lethal heat shock (Figures 3A–D). However, both strains showed no significant change in survival after high hyperosmolarity stress or freezing (Figures 3B,D).
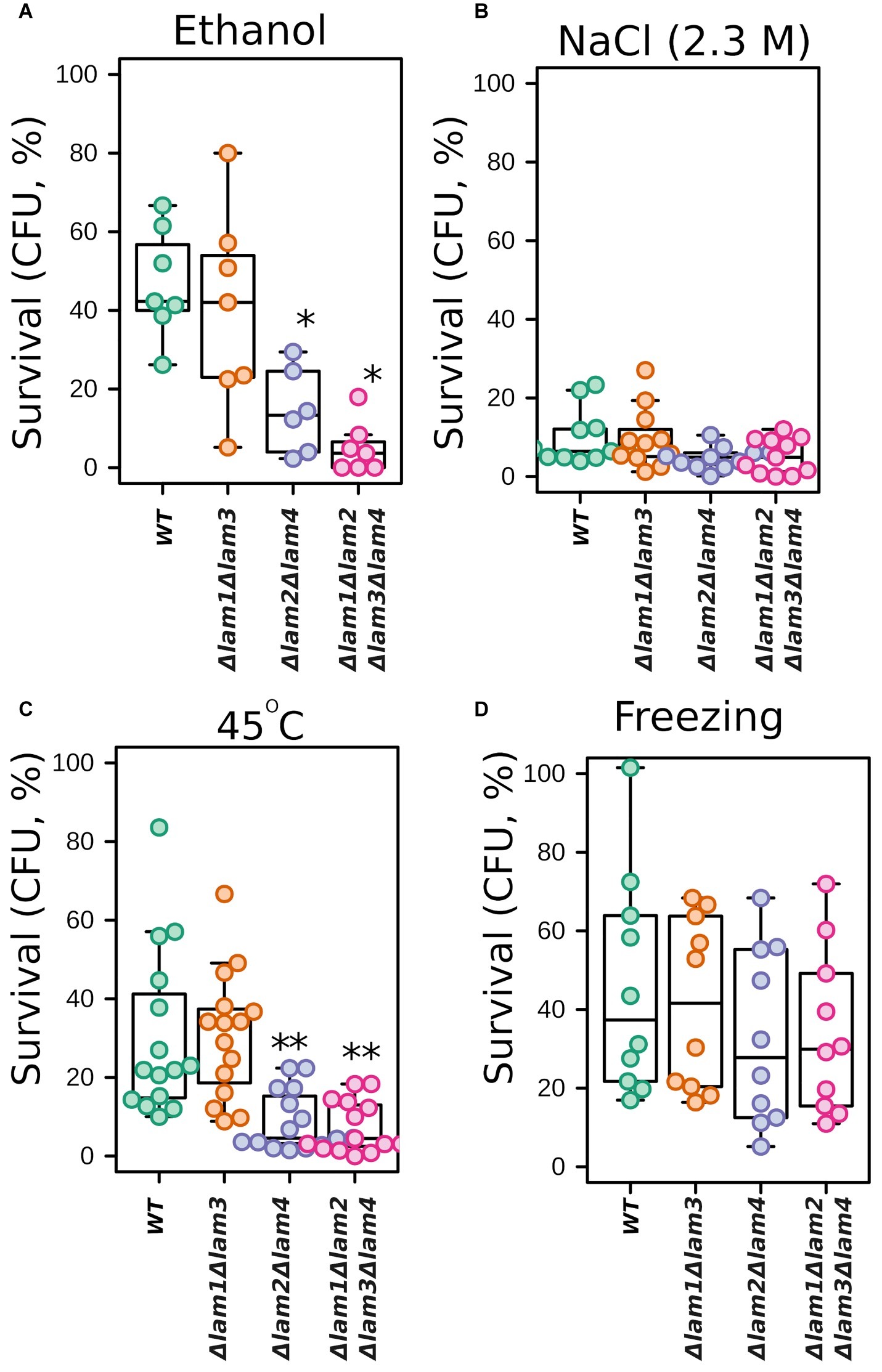
Figure 3. Survival of LAM deletion mutants exposed to stresses. The survival was measured as the number of colony forming units (CFU) in the mutant strain normalized to unstressed cells. 100% represents the number of CFU at the beginning of stress exposure. Cells were subjected to 12% v/v ethanol for 1 h (A); 2.3 M NaCl for 1 h (B); heat shock (47°C for 30 min) (C); or freezing (−20°C for 18 h) (D). Experiments conducted in YPD with cells grown to exponential phase. *p < 0.05, **p < 0.005 for comparisons with WT, unpaired Wilcoxon-Mann-Whitney test.
The Δlam2Δlam4 knockout strain showed more pronounced phenotypes than the Δlam1Δlam3 strain. Thus, we decided to test the relative contributions of LAM2 and LAM4 to ethanol/high salinity tolerance. We compared the growth kinetics of Δlam2Δlam4 with two single-gene knockout strains, Δlam2 and Δlam4 (Figure 4). We found that the effect on the growth rate in the presence of 9% ethanol is determined mainly by the LAM2 gene (Figures 4A,B). Meanwhile, the functions of LAM2 and LAM4 appeared to be redundant for the high osmolarity growth phenotype (Figures 4C,D).
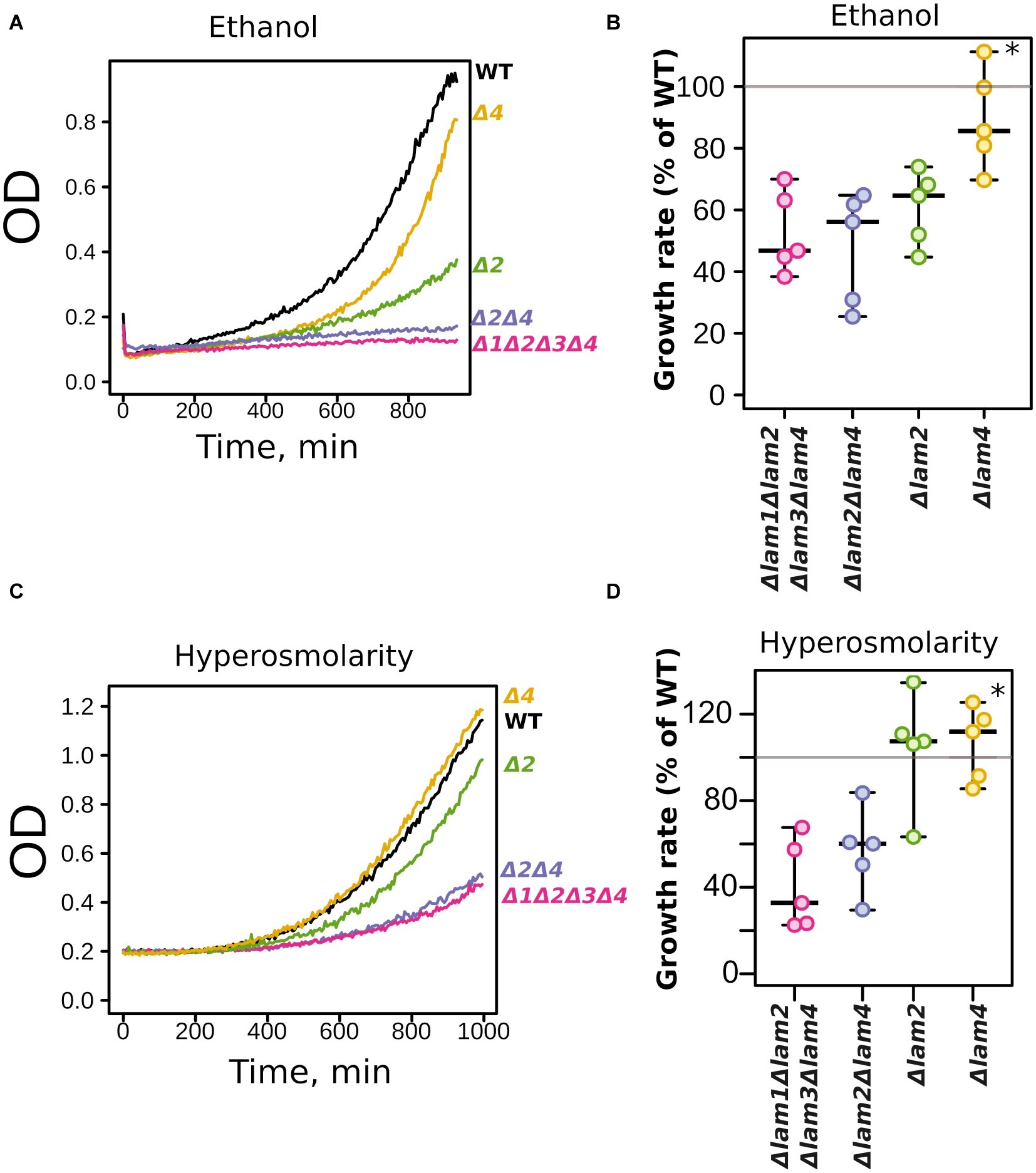
Figure 4. Relative contribution of LAM2 and LAM4 genes to ethanol (9% v/v) (A,B) and hyperosmolarity tolerance (1.1 M NaCl) (B,C). Representative growth curves (A,C) and quantification of OD during 9 h of growth (B,D). 100% represents the growth of the WT (parental) strain in the same conditions. *p < 0.02 for comparisons with Δlam2Δlam4, unpaired Mann-Whitney test with Bonferroni adjustment.
Yeast response to high osmolarity stress relies on multiple proteins and systems (Hohmann, 2015). These systems include PM osmosensor proteins, which sense the changes in membrane properties and transduce the signal to the high osmolarity glycerol pathway (Tatebayashi et al., 2015). Thus, it is possible that the deletion of LAM genes interferes with the ability of yeast cells to adapt to increased osmolarity. In this case, the deletion of LAM genes would have prevented the preadaptation response to increased NaCl concentration in the incubation medium. To test this theory, we incubated yeast cells in the presence of 0.4 M NaCl and then increased the concentration to 1.1 М. Such preadaptation increased the growth rate of both the wild-type strain and the quadruple deletion strain, Δlam1Δlam2Δlam3Δlam4, in the presence of 1.1 M NaCl (Figure 5). Therefore, the absence of Lam proteins in PM-ER junctions did not prevent the ability of yeast cells to induce a hyperosmolarity response.
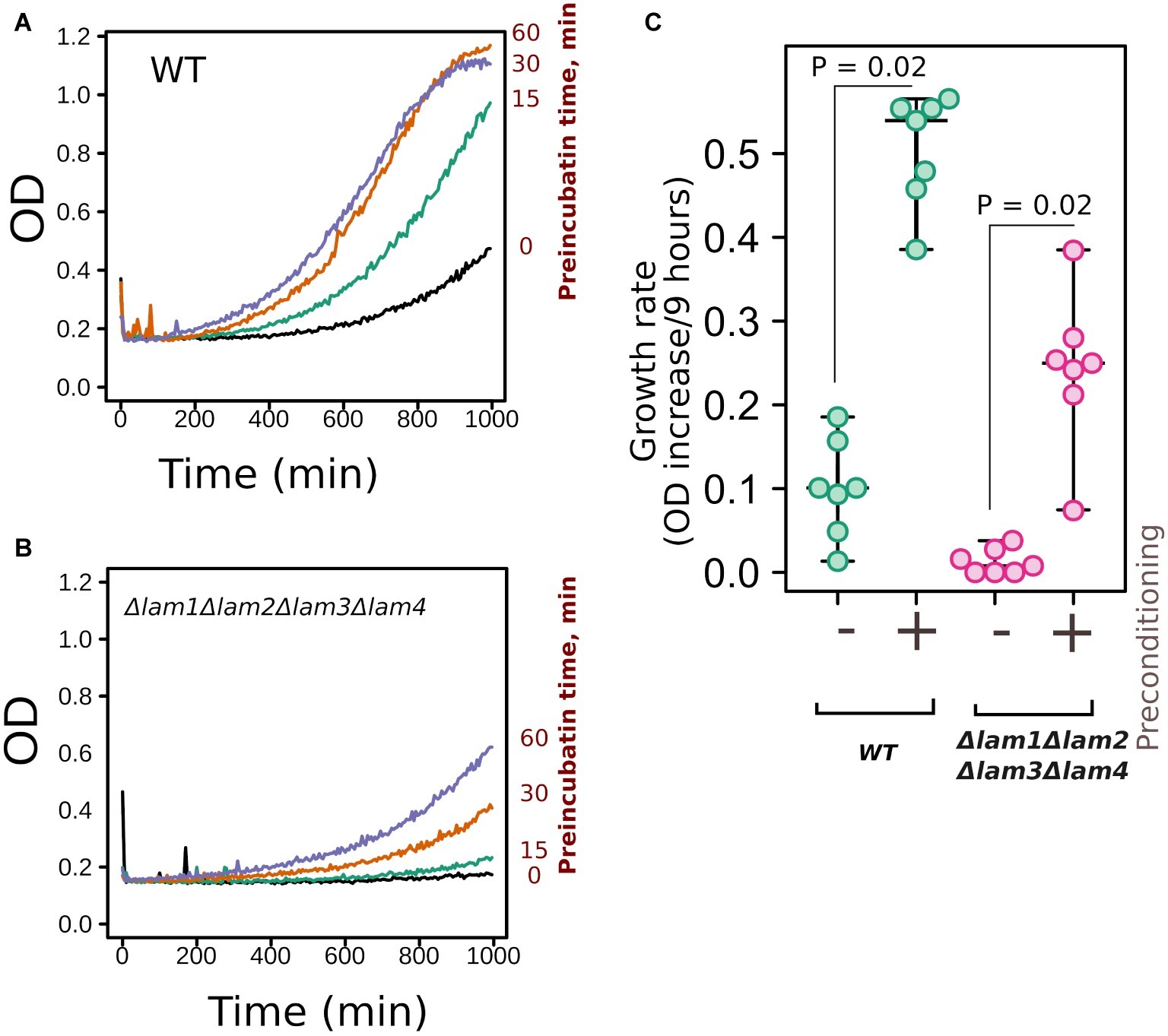
Figure 5. Deletion of LAM genes did not prevent yeast preadaptation to high osmolarity. (A,B) Yeast cells of control (WT) or quadruple mutant Δlam1Δlam2Δlam3Δlam4 strains were incubated with 0.4 M of NaCl for different time intervals (indicated in the figure) and then the concentration of NaCl was increased to 1.1 M. We measured yeast growth kinetics starting from this point. (C) Quantification of the results presented in Figures 5A,B. Increases in OD during first 9 h of growth are indicated. Experiments were conducted in YPD with yeast cultures in exponential growth phase. Values of p were calculated according to Wilcoxon signed-rank test.
While the deletion of LAM genes decreased the resistance to environmental stresses, it increased the resistance to azole antifungals (Figure 6). In line with the results published by Gatta et al. (2015), LAM genes provided resistance to polyene antimycotic amphotericin B (Figure 6A). Amphotericin targets ergosterol-rich membranes: it absorbs ergosterol from the PM and consequently disrupts its barrier function (Anderson et al., 2014). Surprisingly, the addition of an azole antifungal (clotrimazole or miconazole) significantly decreased the survival of the wild-type cells but not LAM mutant cells (Figures 6B–D).
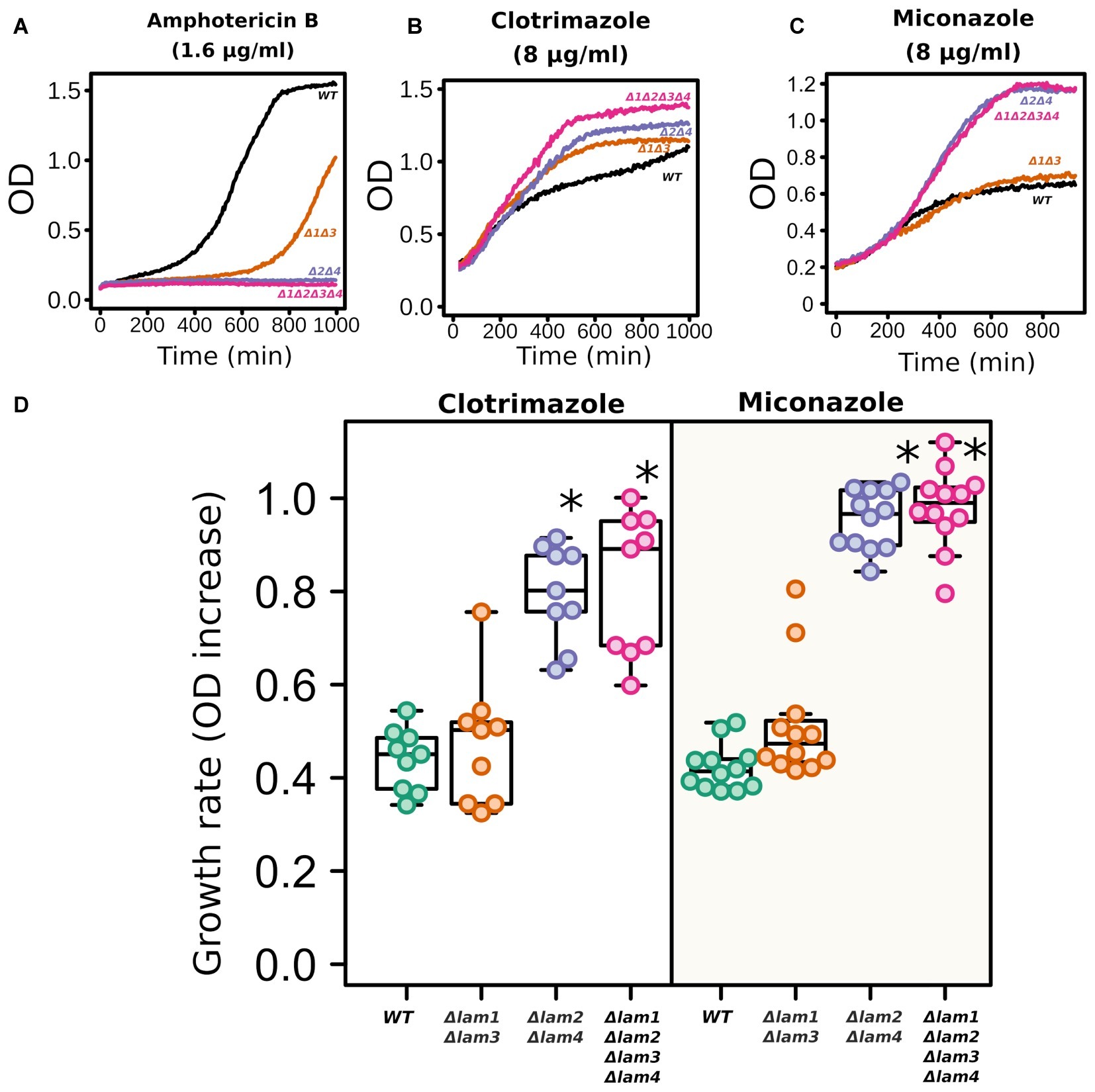
Figure 6. LAM deletion strains are resistant to azole antifungals. Amphotericin B (1.6 μg/ml), clotrimazole (8 μg/ml), and miconazole (8 μg/ml). Representative growth curves (A–C) and quantification of the results (D). For quantification, we used the increase in ODλ550 during 9 h of growth. *p < 0.001 for comparisons with WT strain, unpaired Mann–Whitney test with Bonferroni adjustment.
One of the key mechanisms that provide azole tolerance is the upregulation of PDR transporters that extrude xenobiotic compounds from cells (Prasad et al., 2015). An increase in azole resistance can be attributed to the upregulation or activation of these transporter proteins, which is triggered by sterol deficiency (Vu et al., 2019). To test whether azole tolerance in the LAM quadruple mutant is mediated by PDR transporters, we measured the accumulation of PDR substrates and the level of main ABC-transporter in the wild-type strain and in the Δlam1Δlam2Δlam3Δlam4 strain. First, we measured the level of Pdr5-GFP by flow cytometry. We produced the mutant strain by fusing GFP to the genomic copy of the PDR5 gene. Pdr5 is one of the major pleiotropic drug resistance ABC-transporters in S. cerevisiae. We found that the level of Pdr5-GFP was increased in the Δlam1Δlam2Δlam3Δlam4 strain (Figures 7A,B). We used clotrimazole as a positive control with increased Pdr5 levels (see Galkina et al. (2018)). However, we did not detect an increase in PDR activity in the Δlam1Δlam2Δlam3Δlam4 mutant strain. First, there was no pronounced difference in the resistance to cycloheximide between the LAM mutant strains. Cycloheximide is a protein synthesis inhibitor and a substrate of the Pdr5 transporter (Leppert et al., 1990). Surprisingly, despite an increase in Pdr5 concentration in the LAM quadruple mutant strain, we did not detect a pronounced difference between LAM knockout strains (Figure 7C). Moreover, we did not detect an increase in efflux of the PDR fluorescent substrates Nile red (Figure 7D) and rhodamine 6G (Figure 7E) in the LAM quadruple mutant.
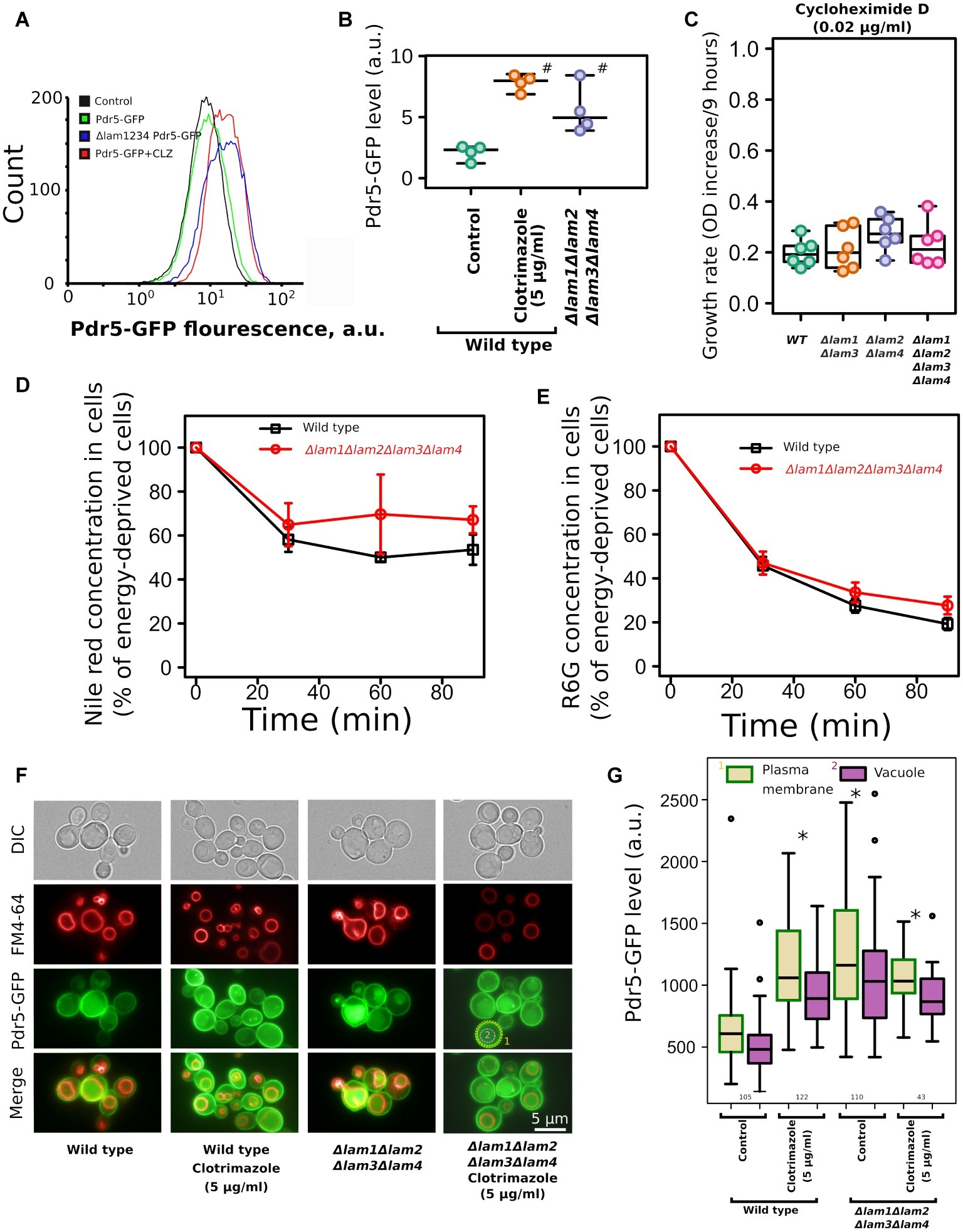
Figure 7. Deletion of LAM genes increases the level of ABC-transporter Pdr5-GFP but does not increase the efflux rate of ABC-transporter substrates. (A) Flow cytometry analysis of Pdr5-GFP wild-type and Δlam1Δlam2Δlam3Δlam4 strains. As a positive control, to induce Pdr5-GFP accumulation, we treated the wild-type strain with clotrimazole (CLZ, 5 μg/ml); a control strain lacking the PDR5-GFP gene was used as a negative control. (B) Quantification of (A), #corresponds to p = 0.057, for comparisons with WT strain, unpaired Mann–Whitney test with Bonferroni adjustment. (C) The increase of OD during first 9 h of growth of the mutant strain with cycloheximide D, a protein synthesis inhibitor and a substrate of PDR transporters (0.02 μg/ml). Experiments were conducted in YPD with yeast cultures in exponential growth phase. (D,E) The dynamics of pumping the ABC-transporter substrates Nile red (D) and rhodamine 6G (E) out of wild-type and Δlam1Δlam2Δlam3Δlam4 cells (n = 3). (F) Photograph of yeast cells expressing Pdr5-GFP stained with FM4–64. The average signal intensity of plasma membrane (outlined by yellow dashed lines) was compared to the average signal in vacuole (red line). (G) Quantification of the plasma membrane/vacuolar signal intensity ratio. Numbers of analyzed cells pooled from two biological replicates are indicated below the boxplots. *p < 10−8 for comparisons to PM Pdr5-GFP level in control cells, unpaired Mann–Whitney test with Bonferroni adjustment.
Alteration of membrane sterols could affect the targeting of membrane proteins to the PM. For example, in Candida albicans deletion of the ergosterol biosynthesis pathway genes Δerg24, Δerg6, and Δerg4 induces mistargeting of СaCdr1, a Pdr5 ortholog, to vacuoles instead of the PM (Pasrija et al., 2008). To test if the deletion of LAM genes changes the localization of Pdr5, we analyzed the intracellular localization of Pdr5-GFP in wild-type and mutant strains. We found that quadruple deletion of LAM genes did not induce intracellular redistribution of Pdr5-GFP and that in Δlam1Δlam2Δlam3Δlam4 strain, Pdr5-GFP was predominantly localized in the PM (Supplementary Figure S2). In agreement with the flow cytometry data, we detected increased concentration of Pdr5-GFP in clotrimazole-treated and Δlam1Δlam2Δlam3Δlam4 cells. Moreover, the signal from GFP increased in the PM as well as in the vacuole (Figures 7F,G). However, we did not detect an additional increase in PM Pdr5-GFP levels upon the treatment of yeast cells with clotrimazole (Figures 7F,G). These results suggest that increase in azole tolerance in the Δlam1Δlam2Δlam3Δlam4 mutant strain is associated with increased Pdr5 levels, but the quadruple deletion does not provide a pronounced increase in drug efflux activity.
In mammalian cells, membrane-bound sterol-transporting proteins internalize high-density lipoprotein cholesterol to intracellular compartments (Sandhu et al., 2018). However, the biological role of these proteins is still unclear due to the absence of phenotypes for single-gene knockdowns. The genome of S. cerevisiae contains three pairs of paralogous membrane-anchored sterol-transporter genes (LAM genes). These paralogous genes arose because of whole-genome duplication. In this study, we produced a quadruple knockout strain, Δlam1Δlam2Δlam3Δlam4 (Table 1), that lacks all four genes that encode the PM-ER-tethered Lam proteins. Lam5p and Lam6p are localized in ER-mitochondria and ER-vacuolar junctions and were not analyzed in our study.
In the quadruple deletion strain, we assessed total sterol content via spectrophotometry and analyzed the intracellular distribution of non-esterified sterols using the sterol-binding fluorescent dye filipin. We found that quadruple deletion increased total sterol content by a factor of approximately 1.11 (Figure 1A). Because Lam2 protein mediates retrograde sterol transport from the PM to the inner compartments (see Gatta et al., 2015), the deletion of LAM1-4 genes may increase the PM sterol concentration relative to that in the inner compartments. Meanwhile, we detected sterol redistribution from the PM to the cytoplasm. While the wild-type parental strain showed strong PM staining and a clear cytoplasm, in LAM mutants, we detected cytoplasmic staining (Figure 1B), reflecting either increased ER or lipid droplets sterol levels or decreased sterol esterification levels in the inner compartments. Strikingly, the filipin-staining phenotype of LAM deletions was similar to that of the gain of function dominant mutation, UPC2-1, of the sterol-sensing transcription factor (Crowley et al., 1998). We speculate that the deletion of LAM genes decreases the sterol available for Upc2-mediated sensing, resulting in its activation. We are currently investigating this possibility. Nonetheless, filipin staining revealed increased sterol concentrations in the PM (Figures 1B,C).
We found that the double and quadruple mutants show slightly decreased growth rates under standard fermentable conditions (Figure 2A). High growth rate under optimal conditions is usually antagonistic to stress resistance due to reallocation of resources between proliferation and stress tolerance. Indeed, many strains with low growth rates show increased stress tolerance (Zakrzewska et al., 2011). Meanwhile, in our study, the quadruple LAM deletion mutant was sensitive to certain environmental stresses: high salinity, alcohols (Figures 2, 3), and heat shock (Figure 3). It is known that ergosterol levels determine a cell’s resistance to these stresses. For example, sterol deficiency in yeast cells decreases ethanol tolerance and heat shock tolerance, but this effect is not due to changes in trehalose or heat shock-protein levels (Swan and Watson, 1998). Moreover, the stability of the PM depends on its sterol composition: a high sterol concentration makes membranes more resistant to different surfactants (Apel-Paz et al., 2005). Therefore, we suggest that the phenotypes of the Δlam1Δlam2Δlam3Δlam4 strain are explained directly by altered PM sterol content rather than by indirect effect on stress-response protein machinery. It is also possible that the deletion of LAM genes prevents activation of cell stress-response signaling and activation of the generalized stress-response pathway. However, in our experiments, the deletion strain retained the ability to increase NaCl tolerance in response to preconditioning to moderate salinity (Figure 4).
The conservation of four paralogous genes (LAM1–LAM4) in fungi, which underwent whole-genome duplication, suggests that, despite the redundancy in function, each Lam protein has its own unique function. Our study revealed that LAM2 (YSP2) plays a major role in ethanol tolerance: the effect of LAM2 deletion was the same order of magnitude as the effect of quadruple deletion (Figures 4A,B). At the same time, a copy of LAM4 gene was sufficient to confer growth in increased salinity (Figures 4C,D). This differential response is unlikely to be a result of different activities or concentrations of Lam2p and Lam4p. Indeed, according to available global proteomic analysis, Lam2p (Ysp2p) is approximately twice as abundant in yeast cells compared to Lam4p (Ho et al., 2018). However, it is also possible that the low-abundance protein, Lam4p, transports minor sterol biosynthesis intermediates, while Lam2p is responsible for ergosterol transport or has wider substrate specificity. The substrate specificity of Lam proteins is still unknown; however sterol-binding pockets in their StART-like domains are strongly conserved within the paralogous pair Lam2/Lam4 (Horenkamp et al., 2018).
Deletion of LAM genes did not always decrease cell fitness under the conditions that we tested. The absence of LAM2 and LAM4 increased the cell’s tolerance to azoles (Figure 6). We considered four possible mechanisms of this effect:
1. The deletion of LAM genes can upregulate PDR PM transporters. These transporters extrude azoles from the cytoplasm at the cost of ATP hydrolysis (Prasad and Goffeau, 2012). Indeed, we detected an increase in Pdr5-GFP in yeast PMs in the quadruple LAM mutant (Figure 7). However, despite the increased concentration of the protein, we did not observe an increase in ABC-transporter substrate efflux rate in this strain (Figure 7). This discrepancy suggests that the absence of LAM genes increases Pdr5 concentration and simultaneously decreases the activity of this protein. Indeed, the activity of PDR ABC-transporters is sensitive to the sterol composition of the PM (Kodedová and Sychrová, 2015). We suggest that a decrease in ABC-transporter activity could induce Pdr5 accumulation as a compensatory mechanism.
2. Azole-induced ergosterol depletion (Watson et al., 1989) can also contribute to the cytostatic effect on yeasts. Therefore, a mutation that increases sterol concentration in cells may make additional divisions by cells exposed to azoles possible. If ER-PM Lam proteins facilitate retrograde ergosterol traffic from the PM to the ER (Gatta et al., 2015), then their deletion can increase PM ergosterol concentration. Indeed, deletion of LAM genes increased total and PM sterol content (Figure 1), supporting this explanation of azole-resistant phenotype.
3. Azole antifungals inhibit lanosterol demethylase Erg11p, which catalyzes the reaction of lanosterol demethylation in the ergosterol biosynthesis pathway (Sant et al., 2016). The inhibition of Erg11p decreases the PM sterol level and induces the accumulation of toxic sterol intermediates (Kelly et al., 1995; Martel et al., 2010). Here, we speculate that Lam proteins can contribute to the transport of these intermediates to a compartment sensitive to these intermediates, i.e., the PM. Indeed, ergosterol is synthesized in the ER and only then redistributed to other cellular membranes (Zweytick et al., 2000).
4. Deletion of LAM genes could have affected the lipid composition of the PM. These changes can theoretically selectively increase PM permeability to one compound, while decrease it to others. However, although we cannot rule out this possibility, it is unlikely that the deletion of LAM genes decreases permeability specifically to azoles but not the other compounds tested in our study (cycloheximide, nigericin, Nile red, and rhodamine 6G). Our data did not allow us to discriminate between the above-discussed mechanisms. Moreover, being non-exclusive, these mechanisms may collectively contribute to the observed effects.
To summarize, we have characterized the phenotype of a strain of S. cerevisiae that had four LAM genes deleted that encode membrane-anchored sterol transporters localized in PM-ER junctions. We found that the deletions increased yeast sensitivity to environmental stresses but alleviated azole sensitivity. In all tested cases, the effect of LAM2 and LAM4 deletion was more pronounced than the effect of LAM1 and LAM3 deletion. Altered sensitivity of LAM deletants to azole antifungals suggests that the mutations in these genes can be an additional route for evolution of azole tolerance in pathogenic fungi.
All datasets generated for this study are included in the article/Supplementary Material.
SS, FS, and DK designed the study. SS and FS acquired funding. SS, NT, and AS generated mutant strains. SS, NT, ES, MV, and KG measured growth rate kinetics and cell survival in stressful conditions. AS, SS, and DK performed microscopy experiments. KG performed flow cytometry and fluorescent substrate efflux experiments. SS, KG, AS, and DK analyzed the data. DK prepared the illustrations and drafted the manuscript. All authors contributed to manuscript editing and approved the final version.
Experiments with yeast growth on low agar (Figure 2B) were supported by Russian Foundation for Basic Research grant N 18–04-01183. The rest of the work (Figures 1, 2A,C,D, 3–7) was supported by Russian Science Foundation grant N 18-14-00151.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00038/full#supplementary-material
Abe, F., and Hiraki, T. (2009). Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1788, 743–752. doi: 10.1016/j.bbamem.2008.12.002
Aguilera, F., Peinado, R. A., Millán, C., Ortega, J. M., and Mauricio, J. C. (2006). Relationship between ethanol tolerance, H+ -ATPase activity and the lipid composition of the plasma membrane in different wine yeast strains. Int. J. Food Microbiol. 110, 34–42. doi: 10.1016/j.ijfoodmicro.2006.02.002
Alli-Balogun, G. O., and Levine, T. P. (2019). Regulation of targeting determinants in interorganelle communication. Curr. Opin. Cell Biol. 57, 106–114. doi: 10.1016/j.ceb.2018.12.010
Anderson, T. M., Clay, M. C., Cioffi, A. G., Diaz, K. A., Hisao, G. S., Tuttle, M. D., et al. (2014). Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat. Chem. Biol. 10, 400–406. doi: 10.1038/nchembio.1496
Apel-Paz, M., Doncel, G. F., and Vanderlick, T. K. (2005). Impact of membrane cholesterol content on the resistance of vesicles to surfactant attack. Langmuir 21, 9843–9849. doi: 10.1021/la050568r
Arthington-Skaggs, B. A., Jradi, H., Desai, T., and Morrison, C. J. (1999). Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J. Clin. Microbiol. 37, 3332–3337. doi: 10.1128/JCM.37.10.3332-3337.1999
Bard, M., Lees, N. D., Burrows, L. S., and Kleinhans, F. W. (1978). Differences in crystal violet uptake and cation-induced death among yeast sterol mutants. J. Bacteriol. 135, 1146–1148. doi: 10.1128/JB.135.3.1146-1148.1978
Baumann, N. A., Sullivan, D. P., Ohvo-Rekilä, H., Simonot, C., Pottekat, A., Klaassen, Z., et al. (2005). Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry 44, 5816–5826. doi: 10.1021/bi048296z
Crowley, J. H., Leak, F. W. Jr., Shianna, K. V., Tove, S., and Parks, L. W. (1998). A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180, 4177–4183. doi: 10.1128/JB.180.16.4177-4183.1998
Dos Santos, S. C., Teixeira, M. C., Dias, P. J., and Sá-Correia, I. (2014). MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: understanding their physiological function through post-genomic approaches. Front. Physiol. 5:180. doi: 10.3389/fphys.2014.00180
Elbaz-Alon, Y., Eisenberg-Bord, M., Shinder, V., Stiller, S. B., Shimoni, E., Wiedemann, N., et al. (2015). Lam6 regulates the extent of contacts between organelles. Cell Rep. 12, 7–14. doi: 10.1016/j.celrep.2015.06.022
François, I. E. J. A., Bink, A., Vandercappellen, J., Ayscough, K. R., Toulmay, A., Schneiter, R., et al. (2009). Membrane rafts are involved in intracellular miconazole accumulation in yeast cells. J. Biol. Chem. 284, 32680–32685. doi: 10.1074/jbc.M109.014571
Galkina, K. V., Besedina, E. G., Zinovkin, R. A., Severin, F. F., and Knorre, D. A. (2018). Penetrating cations induce pleiotropic drug resistance in yeast. Sci. Rep. 8:8131. doi: 10.1038/s41598-018-26435-z
Gatta, A. T., Wong, L. H., Sere, Y. Y., Calderón-Noreña, D. M., Cockcroft, S., Menon, A. K., et al. (2015). A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife 4:e07253. doi: 10.7554/eLife.07253
Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Véronneau, S., et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391. doi: 10.1038/nature00935
Grossmann, G., Opekarová, M., Malinsky, J., Weig-Meckl, I., and Tanner, W. (2007). Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 26, 1–8. doi: 10.1038/sj.emboj.7601466
Gueldener, U., Heinisch, J., Koehler, G. J., Voss, D., and Hegemann, J. H. (2002). A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23. doi: 10.1093/nar/30.6.e23
Gupta, S. S., Ton, V.-K., Beaudry, V., Rulli, S., Cunningham, K., and Rao, R. (2003). Antifungal activity of amiodarone is mediated by disruption of calcium homeostasis. J. Biol. Chem. 278, 28831–28839. doi: 10.1074/jbc.M303300200
Ho, B., Baryshnikova, A., and Brown, G. W. (2018). Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 6, 192–205.e3. doi: 10.1016/j.cels.2017.12.004
Hohmann, S. (2015). An integrated view on a eukaryotic osmoregulation system. Curr. Genet. 61, 373–382. doi: 10.1007/s00294-015-0475-0
Horenkamp, F. A., Valverde, D. P., Nunnari, J., and Reinisch, K. M. (2018). Molecular basis for sterol transport by StART-like lipid transfer domains. EMBO J. 37:e98002. doi: 10.15252/embj.201798002
Jakubkova, M., Dzugasova, V., Truban, D., Abelovska, L., Bhatia-Kissova, I., Valachovic, M., et al. (2016). Identification of yeast mutants exhibiting altered sensitivity to valinomycin and nigericin demonstrate pleiotropic effects of ionophores on cellular processes. PLoS One 11:e0164175. doi: 10.1371/journal.pone.0164175
Kelly, S. L., Lamb, D. C., Corran, A. J., Baldwin, B. C., and Kelly, D. E. (1995). Mode of action and resistance to azole antifungals associated with the formation of 14 alpha-methylergosta-8,24(28)-dien-3 beta,6 alpha-diol. Biochem. Biophys. Res. Commun. 207, 910–915. doi: 10.1006/bbrc.1995.1272
Knorre, D. A., Krivonosova, T. N., Markova, O. V., and Severin, F. F. (2009). Amiodarone inhibits multiple drug resistance in yeast Saccharomyces cerevisiae. Arch. Microbiol. 191, 675–679. doi: 10.1007/s00203-009-0493-8
Kodedová, M., and Sychrová, H. (2015). Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One 10:e0139306. doi: 10.1371/journal.pone.0139306
Leppert, G., McDevitt, R., Falco, S. C., Van Dyk, T. K., Ficke, M. B., and Golin, J. (1990). Cloning by gene amplification of two loci conferring multiple drug resistance in Saccharomyces. Genetics 125, 13–20.
Liu, G., Chen, Y., Færgeman, N. J., and Nielsen, J. (2017). Elimination of the last reactions in ergosterol biosynthesis alters the resistance of Saccharomyces cerevisiae to multiple stresses. FEMS Yeast Res. 17:fox063. doi: 10.1093/femsyr/fox063
Longtine, M. S., McKenzie, A. 3rd., Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., et al. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U
Maresova, L., Muend, S., Zhang, Y.-Q., Sychrova, H., and Rao, R. (2009). Membrane hyperpolarization drives cation influx and fungicidal activity of amiodarone. J. Biol. Chem. 284, 2795–2802. doi: 10.1074/jbc.M806693200
Martel, C. M., Parker, J. E., Bader, O., Weig, M., Gross, U., Warrilow, A. G. S., et al. (2010). Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 54, 4527–4533. doi: 10.1128/AAC.00348-10
Montañés, F. M., Pascual-Ahuir, A., and Proft, M. (2011). Repression of ergosterol biosynthesis is essential for stress resistance and is mediated by the Hog1 MAP kinase and the Mot3 and Rox1 transcription factors. Mol. Microbiol. 79, 1008–1023. doi: 10.1111/j.1365-2958.2010.07502.x
Pasrija, R., Panwar, S. L., and Prasad, R. (2008). Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob. Agents Chemother. 52, 694–704. doi: 10.1128/AAC.00861-07
Pozniakovsky, A. I., Knorre, D. A., Markova, O. V., Hyman, A. A., Skulachev, V. P., and Severin, F. F. (2005). Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J. Cell Biol. 168, 257–269. doi: 10.1083/jcb.200408145
Prasad, R., Banerjee, A., Khandelwal, N. K., and Dhamgaye, S. (2015). The ABCs of Candida albicans multidrug transporter Cdr1. Eukaryot. Cell 14, 1154–1164. doi: 10.1128/EC.00137-15
Prasad, R., and Goffeau, A. (2012). Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu. Rev. Microbiol. 66, 39–63. doi: 10.1146/annurev-micro-092611-150111
R Core Team (2014). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Sandhu, J., Li, S., Fairall, L., Pfisterer, S. G., Gurnett, J. E., Xiao, X., et al. (2018). Aster proteins facilitate nonvesicular plasma membrane to ER cholesterol transport in mammalian cells. Cell 175, 514–529.e20. doi: 10.1016/j.cell.2018.08.033
Sant, D. G., Tupe, S. G., Ramana, C. V., and Deshpande, M. V. (2016). Fungal cell membrane-promising drug target for antifungal therapy. J. Appl. Microbiol. 121, 1498–1510. doi: 10.1111/jam.13301
Sherman, F. (2002). Getting started with yeast. Methods Enzymol. 350, 3–41. doi: 10.1016/S0076-6879(02)50954-X
Sokolov, S., Knorre, D., Smirnova, E., Markova, O., Pozniakovsky, A., Skulachev, V., et al. (2006). Ysp2 mediates death of yeast induced by amiodarone or intracellular acidification. Biochim. Biophys. Acta 1757, 1366–1370. doi: 10.1016/j.bbabio.2006.07.005
Sokolov, S. S., Trushina, N. I., Severin, F. F., and Knorre, D. A. (2019). Ergosterol turnover in yeast: an interplay between biosynthesis and transport. Biochemistry 84, 346–357. doi: 10.1134/S0006297919040023
Swan, T. M., and Watson, K. (1998). Stress tolerance in a yeast sterol auxotroph: role of ergosterol, heat shock proteins and trehalose. FEMS Microbiol. Lett. 169, 191–197.
Tatebayashi, K., Yamamoto, K., Nagoya, M., Takayama, T., Nishimura, A., Sakurai, M., et al. (2015). Osmosensing and scaffolding functions of the oligomeric four-transmembrane domain osmosensor Sho1. Nat. Commun. 6:6975. doi: 10.1038/ncomms7975
Tong, J., Manik, M. K., and Im, Y. J. (2018). Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites. Proc. Natl. Acad. Sci. USA 115, E856–E865. doi: 10.1073/pnas.1719709115
Vu, B. G., Thomas, G. H., and Moye-Rowley, W. S. (2019). Evidence that ergosterol biosynthesis modulates activity of the Pdr1 transcription factor in Candida glabrata. MBio 10:e00934-19. doi: 10.1128/mBio.00934-19
Watson, P. F., Rose, M. E., Ellis, S. W., England, H., and Kelly, S. L. (1989). Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem. Biophys. Res. Commun. 164, 1170–1175. doi: 10.1016/0006-291X(89)91792-0
Weete, J. D., Abril, M., and Blackwell, M. (2010). Phylogenetic distribution of fungal sterols. PLoS One 5:e10899. doi: 10.1371/journal.pone.0010899
Welihinda, A. A., Beavis, A. D., and Trumbly, R. J. (1994). Mutations in LIS1 (ERG6) gene confer increased sodium and lithium uptake in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1193, 107–117.
Wilhelm, L. P., Voilquin, L., Kobayashi, T., Tomasetto, C., and Alpy, F. (2019). Intracellular and plasma membrane cholesterol labeling and quantification using filipin and GFP-D4. Methods Mol. Biol. 1949, 137–152. doi: 10.1007/978-1-4939-9136-5_11
Wong, L. H., and Levine, T. P. (2016). Lipid transfer proteins do their thing anchored at membrane contact sites… but what is their thing? Biochem. Soc. Trans. 44, 517–527. doi: 10.1042/BST20150275
Zakrzewska, A., van Eikenhorst, G., Burggraaff, J. E. C., Vis, D. J., Hoefsloot, H., Delneri, D., et al. (2011). Genome-wide analysis of yeast stress survival and tolerance acquisition to analyze the central trade-off between growth rate and cellular robustness. Mol. Biol. Cell 22, 4435–4446. doi: 10.1091/mbc.E10-08-0721
Keywords: azoles, drug resistance, LAM genes, sterol, stress tolerance, yeast
Citation: Sokolov SS, Vorobeva MA, Smirnova AI, Smirnova EA, Trushina NI, Galkina KV, Severin FF and Knorre DA (2020) LAM Genes Contribute to Environmental Stress Tolerance but Sensibilize Yeast Cells to Azoles. Front. Microbiol. 11:38. doi: 10.3389/fmicb.2020.00038
Received: 03 September 2019; Accepted: 09 January 2020;
Published: 28 January 2020.
Edited by:
Dominique Sanglard, Université de Lausanne, SwitzerlandReviewed by:
Miguel Cacho Teixeira, University of Lisbon, PortugalCopyright © 2020 Sokolov, Vorobeva, Smirnova, Smirnova, Trushina, Galkina, Severin and Knorre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Svyatoslav S. Sokolov, c3ZpYXRvc2xhdi5zb2tvbG92QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.