- 1Faculty of Science, The ithree Institute, University of Technology Sydney, Sydney, NSW, Australia
- 2Faculty of Science, Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University, Singapore, Singapore
In the aquatic environment, Vibrio spp. interact with many living organisms that can serve as a replication niche, including heterotrophic protists, or protozoa. Protozoa engulf bacteria and package them into phagosomes where the cells are exposed to low pH, antimicrobial peptides, reactive oxygen/nitrogen species, proteolytic enzymes, and low concentrations of essential metal ions such as iron. However, some bacteria can resist these digestive processes. For example, Vibrio cholerae and Vibrio harveyi can resist intracellular digestion. In order to survive intracellularly, bacteria have acquired and/or developed specific factors that help them to resist the unfavorable conditions encountered inside of the phagosomes. Many of these intra-phagosomal factors used to kill and digest bacteria are highly conserved between eukaryotic cells and thus are also expressed by the innate immune system in the gastrointestinal tract as the first line of defense against bacterial pathogens. Since pathogenic bacteria have been shown to be hypervirulent after they have passed through protozoa, the resistance to digestion by protist hosts in their natural environment plays a key role in enhancing the infectious potential of pathogenic Vibrio spp. This review will investigate the current knowledge in interactions of bacteria with protozoa and human host to better understand the mechanisms used by both protozoa and human hosts to kill bacteria and the bacterial response to them.
Introduction
Vibrio spp. are metabolically versatile bacteria that inhabit the aquatic environment. They can be found in association with living organisms as well as with abiotic sediments and surfaces. Vibrio spp. have been associated with an array of organisms, including zooplankton and phytoplankton, crustaceans such as copepods, bivalves such as oysters and mussels, plants, fishes, and even water birds (Halpern et al., 2008; Senderovich et al., 2009; Vezzulli et al., 2010; Lutz et al., 2013). Vibrio spp. in their environment interact with heterotrophic protozoa, which are specialized eukaryotic cells that can be found in a wide variety of environments. Phagotrophic protozoa are competent grazers, consuming large numbers of prey, sometimes ingesting several times their own body weight (Vickerman, 1992).
Different environmental niches accommodate different predators and prey. For example, protozoa that are mainly surface-attached specialize on feeding on attached bacteria or biofilms, while suspension feeding protozoa consume planktonic bacteria, and some can feed on planktonic or biofilm cells (Finlay, 2001; Parry, 2004; Sigee, 2005). Some bacterivorous protozoa feed selectively on prey using a variety of different mechanisms (Caron et al., 1982; Sibille et al., 1998; Hahn and Höfle, 2001). For example, amoeba use specific protein receptors for recognition of bacterial prey (Pan et al., 2018) and internalize bacteria into phagosomes using actin microfilament-dependent engulfment (Clarke and Maddera, 2006). Other protists, such as ciliates, do not discriminate prey and package them into food vacuoles using ciliary motion (Gray et al., 2012). Flagellates draw their prey toward the base of the flagellum and into an oral groove by creating a strong current. Some filter-feeding flagellates use a collar of tentacles located at the base of the flagellum that allows the smallest prey particles to pass (Chrzanowski and Simek, 1990; Matz et al., 2002; Parry, 2004).
Once bacteria have been ingested, the expression of various factors can result in resistance to digestion and allows for intracellular growth, followed by escape to the extracellular environment. Many of the bacterial factors involved in digestion resistance and intracellular survival, growth, and escape from protozoa are also factors used by pathogenic bacteria during infection of other hosts (Cirillo et al., 1999; Al-Khodor et al., 2008; Adiba et al., 2010; Sun et al., 2018). This evidence supports the coincidental evolution hypothesis that states the virulence factors used by bacteria during in vivo infection are the result of adaptation to other ecological niches (Levin, 1996). Therefore, the study of the environmental factors that select for the emergence of virulence traits becomes relevant for better understanding of the emergence of pathogenic bacteria, including Vibrio spp.
In order to avoid protozoan predation, Vibrio spp. display various anti-grazing strategies, including the formation of biofilms (Matz et al., 2005; Sun et al., 2015), production of QS-regulated proteases such as PrtV (Vaitkevicius et al., 2006), secretion of ammonium and pyomelanin (Sun et al., 2015; Noorian et al., 2017), expression of the type VI virulence-associated secretion system (VAS) of V. cholerae first identified by Pukatzki et al. (2006) and the MARTX type III of V. vulnificus involved in the lysis of a wide range of eukaryotic cells, including amoebae (Lee et al., 2013).
Reports have shown that V. cholerae and V. harveyi can resist the intracellular environment in the amoeba, Acanthamoeba castellanii (Abd et al., 2004, 2005, 2007; Saeed et al., 2007; Shanan et al., 2016; Van der Henst et al., 2016, 2018) and the ciliate, Cryptocaryon irritans (Qiao et al., 2017) respectively. In addition, the release of V. cholerae in expelled food vacuoles (EFVs) has recently been demonstrated to increase fitness in vitro and in vivo (Espinoza-Vergara et al., 2019). The fact that the passage and release of pathogenic bacteria from the intracellular protozoan environment results in increased infectivity suggests that the exposure to intra-phagosomal factors may enhance virulence phenotypes.
Low pH, antimicrobial peptides (AMPs), and proteolytic enzymes, as well as reactive oxygen and nitrogen species (ROS/RNS) are examples of the factors encountered by Vibrio spp. when in the phagosome in predatory protozoa and during the innate immune defense in the gastrointestinal (GI) tract. Thus, the intracellular environment may serve as a pre-adaptive ecosystem for Vibrio spp. before entering a human host. This review will describe the similarities in the strategies used by protozoa and human hosts to kill bacteria and the molecular factors used by Vibrio spp. to overcome such stressors. The impact of exposure to the intra-protozoal environment on the infectivity of V. cholerae and other bacterial pathogens will also be discussed.
Intracellular Survival of Vibrio spp.
Intracellular survival of Vibrio spp. has been demonstrated in various eukaryotic cells, including in the amoebae, A. castellanii, Acanthamoeba polyphaga, and Naegleria gruberi (Thom et al., 1992; Abd et al., 2007). V. cholerae O139 and O1 strains were shown to survive and grow within the cytoplasm of trophozoites and in cysts of A. castellanii (Thom et al., 1992; Abd et al., 2005, 2007). Furthermore, it has been shown that V. cholerae can access the contractile vacuole in A. castellanii and escape to the extracellular environment (Van der Henst et al., 2016). V. harveyi survives in the cytoplasm of the obligate parasitic marine ciliated protozoan, C. irritans (Qiao et al., 2017). Vibrio splendidus and Vibrio parahaemolyticus invade and survive intracellularly in other hosts such as oyster hemocytes (Duperthuy et al., 2011) and human epithelial cells (de Souza Santos and Orth, 2014), respectively.
Recent studies have investigated the intracellular mechanisms that mediate the survival and escape of V. cholerae from eukaryotic cells. Interestingly, virulence factors related to hemolytic activity and motility had a role in the intracellular survival of V. cholerae in A. castellanii (Van der Henst et al., 2018). In addition, OmpU, a major outer membrane protein that is needed for resistance to many stressors such as organic acids, bile, and AMPs as well as being a critical factor for the in vivo colonization of V. cholerae (Sperandio et al., 1995; Provenzano and Klose, 2000), plays a role in survival in protozoa. It was recently shown that OmpU is important for the expulsion of V. cholerae within food vacuoles of ciliate hosts, a fact that suggests that this protein might confer resistance to V. cholerae to the intra-phagosomal factors required for digestion (Espinoza-Vergara et al., 2019). Interestingly, it is also reported that OmpU is essential for V. splendidus host invasion and resistance to AMPs and is required for virulence in the oyster, Crassostrea gigas (Duperthuy et al., 2011). Thus, the factors that mediate the intracellular survival of V. cholerae in protozoa and their link with the pathogenic lifestyle of this bacterium are being revealed.
Intra-phagosomal Factors in Protozoa and the Innate Immune Defense of the Gastrointestinal Tract: Similar Stressors Encountered in Both Environments
Phagosomes of bacterivorous protozoa use mechanisms of killing and digestion of bacteria that are highly conserved in eukaryotic cells. Intracellular digestion begins with a reduction in the pH in order to create an acidic environment required for the proper activity of various antibacterial compounds that are vital for the digestion of bacteria. Many of these compounds have been described: (1) AMPs, amphipathic peptides that disrupt the integrity of the cell membrane, (2) ROS/RNS, also involved in the loss of membrane integrity as well as DNA damage in bacteria, and (3) proteolytic enzymes such as endopeptidases and lipases, required for the digestion of macromolecules (Flannagan et al., 2009). Here we will describe how the factors used by the innate immune system of the GI tract: the acidic environment of the stomach, ROS/RNS compounds produced after the breakdown of macromolecules in the presence of low pH and bile, proteolytic enzymes such as proteases, peptidases, lipases, amylases, and nucleases, and AMPs that are synthesized by the GI epithelium are also encountered inside phagocytic cells (Figure 1). It is likely that key factors used by Vibrio spp. to resist the intracellular environment in protozoa might also serve to protect cells against stressors in the GI tract. Here the mechanisms displayed by Vibrio spp. to resist such stressors and how this can affect the infective cycle of the model pathogen V. cholerae will be explored.
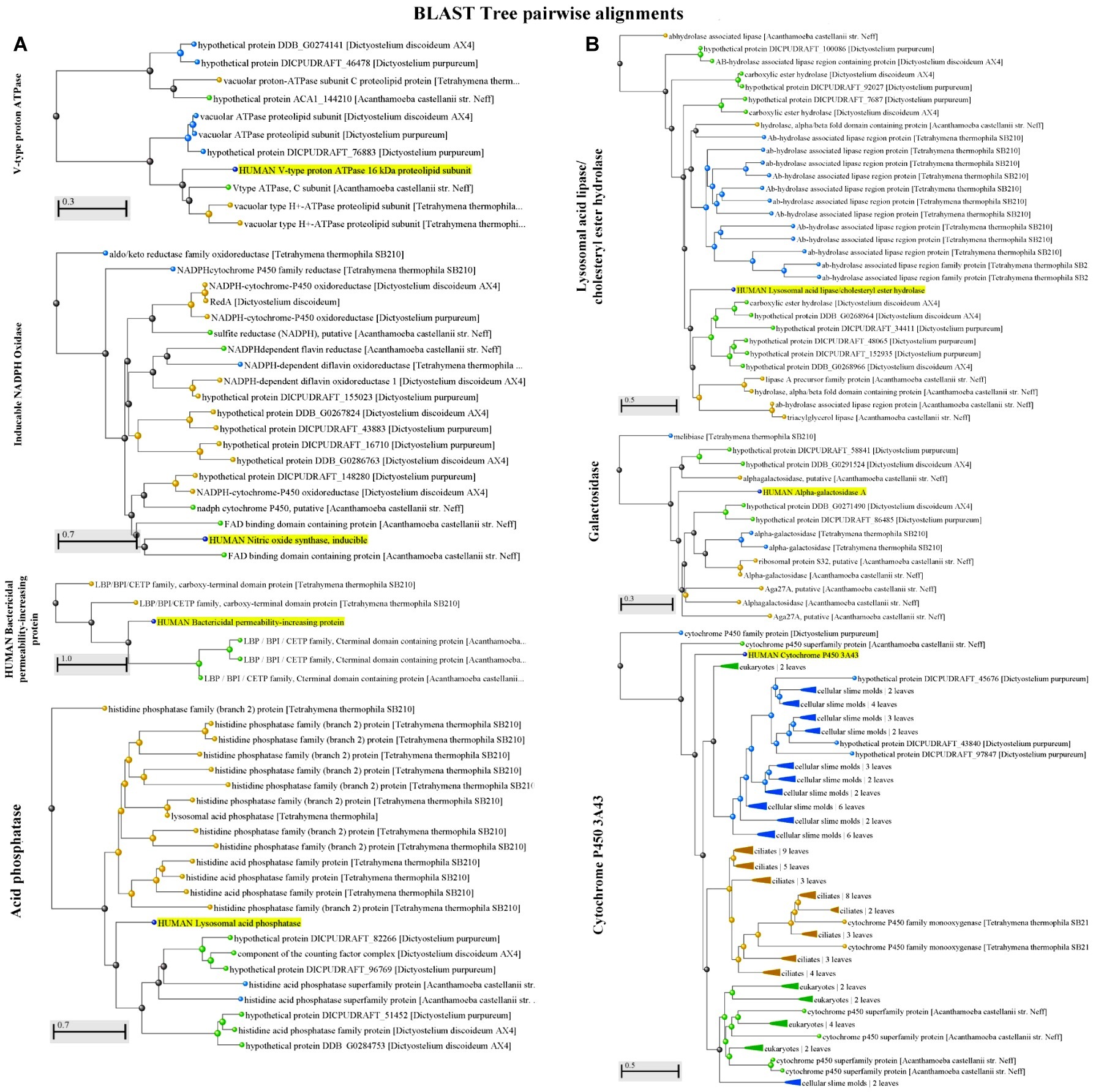
Figure 1. (A) A Blast Tree View of potential factors [V-type proton ATPase, inducible NADPH oxidase, and the human bactericidal permeability-increasing protein (BPI)] encountered in both protozoa and humans (highlighted) that contribute to the killing of Vibrio spp. shows pairwise alignment between human proteins and those found in protozoa. Produced by NCBI Tree Viewer. (B) A Blast Tree View of potential factors [acid phosphatase (lysosomal acid lipase/ cholesteryl ester hydrolase), galactosidase, cytochrome P450 3A43, and acid phosphatase] encountered in both protozoa and humans (highlighted) that contribute to the killing of Vibrio spp. shows pairwise alignment between human proteins and those found in protozoa. Produced by NCBI Tree Viewer.
Low pH
The acidification of phagosomes containing bacteria is a critical step for intracellular digestion in phagocytic cells. Under normal conditions, early phagosomes become acidified by the action of specific proteins located on the surface of phagosomes. The vacuolar V-ATPase is a highly conserved enzyme that transports H+ ions (Forgac, 1999; Toei et al., 2010) and is present in the phagosomes of protozoa and also on the surface of human GI cells (Figure 1). At the late stages of phagosome maturation, the low pH of the phagosome enables the fusion with the lysosome, an acidic organelle that contains enzymes that are crucial for the complete digestion of bacteria and macromolecules. Similarly, part of the initial steps in the digestion of macromolecules as well as the inactivation of pathogenic microorganisms in mammals takes place in the stomach, an environment that is characterized by a low pH. Here, the exposure to the acidic environment (due to hydrochloric acid) and the production of ROS/RNS in the gastric environment causes loss of membrane integrity and DNA damage in bacteria (Conner et al., 2016). Together, these facts highlight that acidification is an important conserved strategy used by different organisms to inactivate and digest bacteria.
V. cholerae expresses several survival strategies to adapt to acidic and oxidative conditions. Aggregation (or suspended biofilms) and biofilm formation have been reported to physically protect V. cholerae from acid stress (Zhu and Mekalanos, 2003) due to the strong protection given by the biofilm matrix that protects V. cholerae from various stressors, including antibiotics and ROS (Mankere et al., 2018; Wang et al., 2018). Another mechanism providing resistance to low pH is the activation of the acid tolerance response (ATR). In V. cholerae, the ATR is controlled by the modulation of the cadBA operon that is activated by the ToxR-like protein CadC (Merrell and Camilli, 2000). cadA is an infection-induced gene in V. cholerae that encodes a lysine decarboxylase (CadA) required for the active efflux of H+ ions from the bacterial cytoplasm to the extracellular space (Merrell and Camilli, 1999). The lysine cadaverine/antiporter (CadB) works together with CadA in the presence of high concentrations of H+ to provide resistance to acidic environments (Merrell and Camilli, 2000). Under acidic conditions, CadB catalyzes the uptake of lysine, which in combination with H+ ions forms cadaverine in the cell cytoplasm, a polyamine that is excreted outside of the cell by the same antiporter. In addition, the activation of ATR in V. cholerae can be mediated by ToxR in the presence of organic acids. It was shown that the ectopic expression of the ToxR-regulated outer membrane OmpU is sufficient to overcome the reduction in ATR that occurs in a ∆toxR mutation (Merrell et al., 2001).
Reports have shown that the adaptation of V. cholerae to low pH before infection causes a significant induction of the ATR system resulting in improved intestinal colonization (Merrell et al., 2002). However, it was shown that this colonization advantage is not due to increased survival of V. cholerae to the stomach environment or to the expression of colonization or virulence factors (Angelichio et al., 2004). It is believed that acid-adapted V. cholerae have a growth advantage over non-adapted cells and that this growth advantage is responsible of the hyperinfective phenotype in vivo, since the fitness advantage of acid-adapted V. cholerae could not be confirmed in vitro (Angelichio et al., 2004). Interestingly, it was recently shown that V. cholerae shows an increased resistance to low pH when contained in EFVs released by ciliated protozoa and also displays an in vitro growth advantage in high nutrient and temperature conditions and in vivo colonization advantage in the infant mouse colonization model (Espinoza-Vergara et al., 2019).
Reactive Oxygen and Nitrogen Species
Under normal physiological conditions, the human body produces small amounts of ROS/RNS in the GI tract due to chemical reactions between oxygen and nitrogen components in the presence of acids or bile (Davies et al., 2011; Aviello and Knaus, 2018). ROS/RNS are oxidative species that can directly damage the DNA of microorganisms, thereby acting as a natural antimicrobial barrier. Human professional phagocytes and amoeba are known to produce ROS/RNS such as nitric oxide (NO) and hydrogen peroxide (H2O2) inside phagosomes as an antibacterial strategy (Zhang and Soldati, 2013; Di Meo et al., 2016). Indeed, the production of ROS in the phagosome of the amoeba Dictyostelium discoideum has been visualized and quantified (Zhang and Soldati, 2013). Furthermore, it is known that the intestinal epithelial layer also produces NO by the induction of oxide synthases (iNOS) (Eckmann et al., 2000). To resist RNS/ROS, pathogenic bacteria such as V. cholerae display specific factors such as hmpA and nnrS, two genes under the control of the σ54-dependent transcriptional regulator NorR (Stern et al., 2012). Deletion of either hmpA or nnrS causes a significant reduction in long-term colonization of V. cholerae in the adult mouse model, showing that RNS is an important barrier to V. cholerae infection in vivo (Stern et al., 2012). Another strategy to resist ROS used by many microorganisms as well as eukaryotic cells is the expression of catalases, superoxide dismutase (SOD), and alkyl superoxide reductase subunit C’s (Imlay, 2008). These enzymes break down ROS into non-damaging sub-products such as H2 and O2. In V. cholerae, OxyR, and two catalases KatG and KatB are involved in the resistance to ROS (Wang et al., 2013). Thus, the factors used by V. cholerae to resist RNS/ROS may facilitate the survival of this bacterium inside phagotrophic protozoa as well as within the human intestinal tract.
Antimicrobial Peptides
Another cause of mortality for pathogenic bacteria in the host is the presence of host-derived AMPs. Production of these molecules can be mediated in the human host by several phagocytes and epithelial cells (Diamond et al., 2009), e.g., macrophages and the GI epithelium. In mammals, the two main classes of AMPs are defensins and cathelicidins (Dorin et al., 2015). Most of the AMPs act by disrupting and permeabilizing the cell membrane, causing loss of viability. Some examples of AMPs produced in the human intestinal tract are: α-defensins: human neutrophil peptides 1-4 (HNP1-4), and human defensin 5 and 6 (HD5 and HD6); β-defensins: beta defensin 1 to 4 (hBD1-4); cathelicidin: LL-37/h-CAP18 (human cathelicidin antimicrobial peptide 18 kDa); other AMPs: bactericidal/permeability-increasing protein (BPI), chemokines CCL14, CCL15, and CCL20/macrophage-immflamatory-protein-3α (Muniz et al., 2012). Importantly, AMPs are also produced by phagocytic cells in order to arrest the growth and inactivate bacteria. As shown in Figure 1, heterotrophic protozoa such as Tetrahymena spp., Dictyostelium spp., and Acanthamoeba spp. encode proteins with high similarities to BPI, an important bactericidal and LPS neutralizing AMP released by human neutrophils (Calafat et al., 2000) and GI epithelial cells (Canny and Colgan, 2005).
Polymyxin B as well as other cationic AMPs (CAMPs) has been widely used to screen for AMP resistance in V. cholerae and other Gram-negative bacteria. Genes related to the modification of the lipid A portion of LPS (Henderson et al., 2014), the outer membrane porin B, and OmpT (Mathur and Waldor, 2004) as well as the VexAB system (Bina et al., 2006) are important for V. cholerae resistance to CAMPs. Lipid A acylation has been reported to play an important role in the resistance to CAMPs in bacterial pathogens such as V. cholerae, Salmonella enterica, Escherichia coli, and Helicobacter pylori (Guo et al., 1998; Band and Weiss, 2014). In V. cholerae, mutation in the acyltransferase gene, msbB, resulted in a significant reduction in the resistance to polymyxin B and impairment in colonization of the small intestinal tract of the infant mouse (Matson et al., 2010), indicating that CAMPs are an important line of defense against V. cholerae. In addition, genes involved in the aminoacylation of lipid A encoded within the almEFG operon, are essential for resistance to CAMPs (Henderson et al., 2014). Recently, AlmG, a glycosyltransferase, positively regulated by the response regulator, CarR (Bilecen et al., 2015), has been identified to be responsible for polymyxin B resistance in pandemic V. cholerae (Henderson et al., 2017). It is known that a reduction in the aminoacylation/phosphorylation of lipid A results in a decrease in the negative charge surface of the bacterial outer membrane, causing an increased affinity for CAMPs with target molecules (Steimle et al., 2016).
Similar to the modification of lipid A, the expression of major outer membrane proteins in V. cholerae is critical for resistance to CAMPs. OmpU, the major outer membrane protein of V. cholerae, plays a key role in the resistance to polymyxin B and other CAMPs such as P2, an active peptide derived from BPI (Mathur and Waldor, 2004). It has been proposed that the interaction between OmpU and AMPs leads to the activation of the stress response mediated by the sigma factor σE, resulting in increased survival (Mathur et al., 2007). In general terms, stress responses in bacteria lead to the activation of specific pathways in response to different stressors such as starvation, biocides, and temperature in order to maintain cell viability. Another mechanism for resistance to AMPs is the activation of the VexAB system. As described previously, VexAB is an efflux system in V. cholerae involved in the resistance to antibiotics, such as polymyxin B and also tensoactive molecules such as SDS and Triton-X 100 (Bina et al., 2006). Interestingly, deletion of ∆vexAB reduced CT production, expression of virulence factors, and colonization (Bina et al., 2008), suggesting that this systems like the VexAB might be important to V. cholerae survival in the presence of AMPs inside phagosomes/food vacuoles in protozoa and also in the human intestinal tract.
Digestive and Other Enzymes
Lysosomal acid lipase (gastric lipase in the stomach), acid phosphatase and galactosidase are three digestive enzymes present in both protozoa and the GI tract (Figure 1). Although there is a lack of information about the impact of these enzymes on the pathogenicity of bacteria, the bacterial resistance to these factors in their primary aquatic habitat might promote pathogen’s growth in the intestinal tract. This idea is supported by the fact that the maintenance of normal levels of digestive enzymes such as alkaline phosphatase in the gut contributes to the growth of beneficial commensal bacteria and prevents the growth of pathogenic microorganisms (Malo et al., 2010).
Interestingly, the presence of cytochrome P450, an enzyme involved in the production of steroid hormones, cholesterol, fatty acids, and bile acids in humans is also present in protozoa (Zimniak and Waxman, 1993). It is known that the presence of bile acids induces the expression of virulence factors such as the cholera toxin in V. cholerae (Hung and Mekalanos, 2005). Despite the fact that the biosynthesis of cholesterol (bile acids precursor) has not been reported in protozoa such as Tetrahymena spp., similar organic acids potentially produced by protist hosts might induce the expression of virulence factors in pathogenic Vibrio spp. and other bacteria.
The resistance of Vibrio spp. to the factors encountered inside of the phagosomes/food vacuoles in heterotrophic protozoa might serve as a pre-adaptation niche before entering a host. In addition to the physical protection that a protozoa might confer to intracellular pathogens, as has been previously suggested, the passage of bacteria within protozoa might activate specific factors used to resist the strategies that also contribute to the inactivation of bacteria within mammalian hosts. Thus, the adaptation and resistance to the intracellular environment in protozoa may positively impact on the infective cycle of pathogenic Vibrio spp., possibly by increasing the number of viable cells that reach the site of infection or by enhanced pathogenicity.
The Impact of Protozoan Predation on Virulence
The interaction of bacteria with protozoa has been correlated with increased pathogenicity, and thus, protists hosts have been suggested to be “Trojan horses” protecting and disseminating pathogens in the environment (Barker and Brown, 1994; Denoncourt et al., 2014). For example, Gram-negative pathogens such as S. enterica, E. coli, and Listeria monocytogenes can survive and remain inside A. castellanii cysts where they are more tolerant to antibiotics and low pH (Lambrecht et al., 2015). Similar protection was shown for Legionella pneumophila, with increased resistance to chlorine when inside of A. polyphaga cysts (Kilvington and Price, 1990). It is known that eukaryotic membranes might act as a physical barrier to biocides, a fact that might explain this effect. In contrast, bacterial adaptation to the intracellular environment can also result in increased pathogenicity through improved resistance to antimicrobials and induction of virulence factors. For example, the use of divalent metals such as copper and zinc is a conserved antimicrobial mechanism against bacteria in both amoeba and macrophages (German et al., 2013). Thus, resistance to copper and/or zinc might lead to increased bacterial virulence in vivo. In E. coli and Pseudomonas aeruginosa, genes encoding copper resistance are related to grazing resistance against D. discoideum (Hao et al., 2016). In Campylobacter jejuni, the efflux system CmeABC that confers resistance to antibiotics, might also be involved in metal detoxification and increased virulence (Vieira et al., 2017).
The transit of bacteria through protozoa has been linked to increased hyperinfectivity in pathogenic bacteria. For example, S. enterica and L. pneumophila recovered after exposure to A. castellanii display hyperinvasive phenotypes during in vivo infection (Cirillo et al., 1994; Rasmussen et al., 2005). Similarly, mice infections performed with Mycobacterium ulcerans previously co-incubated with A. polyphaga led to enhanced pathogenicity (Azumah et al., 2017). In the case of Vibrio spp., it has recently been shown that their release in EFVs to the extracellular environment results in bacterial growth and colonization advantage in vitro and in vivo, respectively (Espinoza-Vergara et al., 2019). In addition, the use of a critical virulence factor in V. cholerae, OmpU, involved in colonization and resistance to low pH, AMPs, and bile, was shown to be involved in the release of EFVs from protozoa. This fact illustrates how a factor involved in the resistance to stressors encountered in the protozoan phagosome and within the human host enhances the survival and potentially increases the infectivity of V. cholerae (Figure 2). Thus, the release of Vibrio spp. in EFVs as well as the intracellular adaptation to the presence of biocides and induction of virulence in bacteria can lead to fitness advantages during infection of a host.
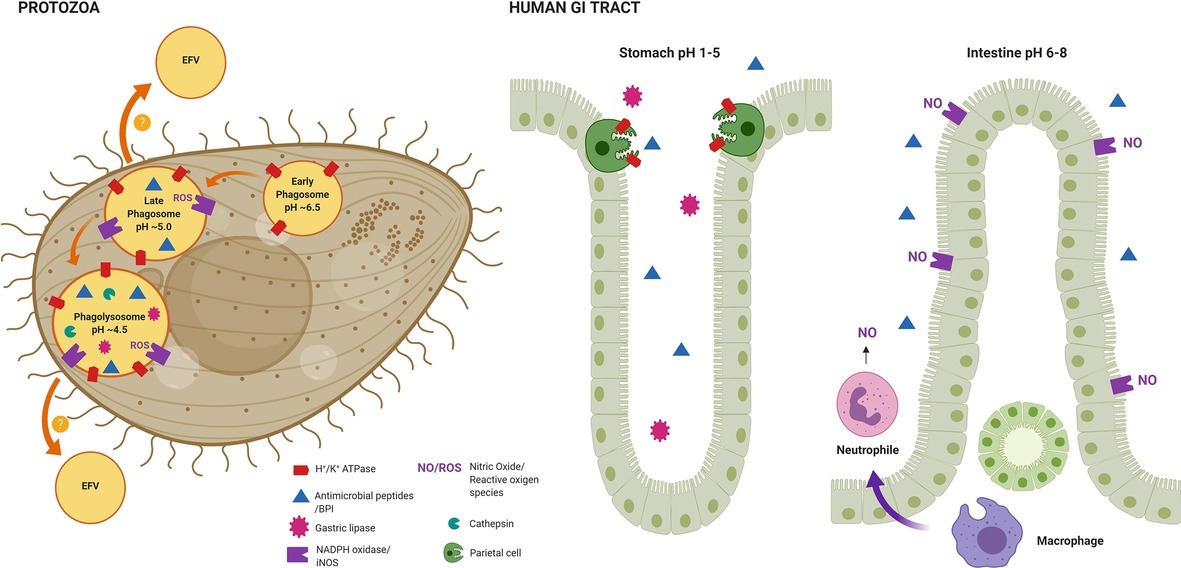
Figure 2. Representation of the conserved factors required for the inactivation and digestion of bacteria used by both protozoa and the innate defense system of the human GI tract. The maturation of bacterial-containing phagosomes in Tetrahymena (Protozoa), a process that depends on acidification. As shown, different factors are recruited at different stages of the phagosome maturation process. Some pathogenic bacteria, such as Vibrio spp., are able to resist the digestion process and are expelled in EFVs to the extracellular environment, a condition where they show a hyperinfective phenotype in vivo. From left to right in human GI tract, the antimicrobial strategies deployed by the stomach and the intestinal tract. The factors represented here are highly similar to the ones encountered in phagocytic protozoa, thus, the pre-adaptation to such stressors within protozoa might be crucial for pathogenic bacteria to survive and multiply within the human GI tract. The figure was created with BioRender.com.
Conclusions and Future Perspective
Taken together, this review highlights that the strategies used to digest and inactivate bacteria in both protozoa and the GI tract of the human host are highly conserved and further emphasize how the resistance to the intracellular digestion in protozoa might enhance the pathogenicity of Vibrio spp. More research regarding the impact on the infection cycle of intracellular exposed Vibrio spp. is fundamental to further understanding the mechanisms that result in hyperinfectivity. This will not only enable us to identify key environmental clues that enable the pathogenicity of important pathogens such as V. cholerae but also will contribute to understanding whether the activation of hyperinfectivity is a conserved response in other pathogenic bacteria that interact with protozoa.
The packaging of multiple bacteria into phagosomes by protozoa might explain how pathogenic Vibrio spp. have acquired specific factors that increase the fitness during infection. In the past, increased conjugation rates have been shown for E. coli contained in Tetrahymena phagosomes (Schlimme et al., 1997; Matsuo et al., 2010). Thus, horizontal gene transfer in protozoa might be a crucial step for pathogenic bacteria to increase fitness in both the environment and during infection of a host. As next-generation sequencing continues to become more affordable, the evaluation of horizontal gene transfer of pathogenic Vibrio spp. in protozoa becomes possible, thus adding another layer of potential selection pressure on the development of virulence.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was funded by the Australian Research Council Discovery Project DP170100453, the CONICYT Becas Chile doctoral (72140329), and by the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Program to the Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge support from the ithree Institute at the University of Technology Sydney, Sydney, Australia; the Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University, Singapore; and the Australian Research Council Discovery Project (DP170100453).
References
Abd, H., Saeed, A., Weintraub, A., Nair, G. B., and Sandström, G. (2007). Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol. Ecol. 60, 33–39. doi: 10.1111/j.1574-6941.2006.00254.x
Abd, H., Weintraub, A., and Sandström, G. (2004). Interaction between Vibrio cholerae and Acanthamoeba castellanii. Microb. Ecol. Health Dis. 16, 51–57. doi: 10.1080/08910600410029190
Abd, H., Weintraub, A., and Sandström, G. (2005). Intracellular survival and replication of Vibrio cholerae O139 in aquatic free-living amoebae. Environ. Microbiol. 7, 1003–1008. doi: 10.1111/j.1462-2920.2005.00771.x
Adiba, S., Nizak, C., van Baalen, M., Denamur, E., and Depaulis, F. (2010). From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS One 5:e11882. doi: 10.1371/journal.pone.0011882
Al-Khodor, S., Price, C. T., Habyarimana, F., Kalia, A., and Abu Kwaik, Y. (2008). A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70, 908–923. doi: 10.1111/j.1365-2958.2008.06453.x
Angelichio, M. J., Merrell, D. S., and Camilli, A. (2004). Spatiotemporal analysis of acid adaptation-mediated Vibrio cholerae hyperinfectivity. Infect. Immun. 72, 2405–2407. doi: 10.1128/IAI.72.4.2405-2407.2004
Aviello, G., and Knaus, U. G. (2018). NADPH oxidases and ROS signaling in the gastrointestinal tract. Mucosal Immunol. 11, 1011–1023. doi: 10.1038/s41385-018-0021-8
Azumah, B. K., Addo, P. G., Dodoo, A., Awandare, G., Mosi, L., Boakye, D. A., et al. (2017). Experimental demonstration of the possible role of Acanthamoeba polyphaga in the infection and disease progression in Buruli ulcer (BU) using ICR mice. PLoS One 12:e0172843. doi: 10.1371/journal.pone.0172843
Band, V. I., and Weiss, D. S. (2014). Mechanisms of antimicrobial peptide resistance in gram-negative bacteria. Antibiotics 4, 18–41. doi: 10.3390/antibiotics4010018
Barker, J., and Brown, M. R. W. (1994). Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140, 1253–1259. doi: 10.1099/00221287-140-6-1253
Bilecen, K., Fong, J. C. N., Cheng, A., Jones, C. J., Zamorano-Sánchez, D., and Yildiz, F. H. (2015). Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect. Immun. 83, 1199–1209. doi: 10.1128/IAI.02700-14
Bina, X. R., Provenzano, D., Nguyen, N., and Bina, J. E. (2008). Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 76, 3595–3605. doi: 10.1128/IAI.01620-07
Bina, J. E., Provenzano, D., Wang, C., Bina, X. R., and Mekalanos, J. J. (2006). Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch. Microbiol. 186, 171–181. doi: 10.1007/s00203-006-0133-5
Calafat, J., Janssen, H., Knol, E. F., Malm, J., and Egesten, A. (2000). The bactericidal/permeability-increasing protein (BPI) is membrane-associated in azurophil granules of human neutrophils, and relocation occurs upon cellular activation. APMIS 108, 201–208. doi: 10.1034/j.1600-0463.2000.d01-45.x
Canny, G., and Colgan, S. P. (2005). Events at the host-microbial interface of the gastrointestinal tract. I. Adaptation to a microbial world: role of epithelial bactericidal/permeability-increasing protein. Am. J. Physiol. Gastrointest. Liver Physiol. 288, G593–G597. doi: 10.1152/ajpgi.00506.2004
Caron, D. A., Davis, P. G., Madin, L. P., and Sieburth, J. M. (1982). Heterotrophic bacteria and bacterivorous protozoa in oceanic macroaggregates. Science 218, 795–797. doi: 10.1126/science.218.4574.795
Chrzanowski, T. H., and Simek, K. (1990). Prey-size selection by freshwater flagellated protozoa. Limnol. Oceanogr. 35, 1429–1436. doi: 10.4319/lo.1990.35.7.1429
Cirillo, J. D., Cirillo, S. L., Yan, L., Bermudez, L. E., Falkow, S., and Tompkins, L. S. (1999). Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67, 4427–4434. doi: 10.1128/IAI.67.9.4427-4434.1999
Cirillo, J. D., Falkow, S., and Tompkins, L. S. (1994). Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62, 3254–3261. doi: 10.1128/IAI.62.8.3254-3261.1994
Clarke, M., and Maddera, L. (2006). Phagocyte meets prey: uptake, internalization, and killing of bacteria by Dictyostelium amoebae. Eur. J. Cell Biol. 85, 1001–1010. doi: 10.1016/j.ejcb.2006.05.004
Conner, J. G., Teschler, J. K., Jones, C. J., and Yildiz, F. H. (2016). Staying alive: Vibrio cholerae’s cycle of environmental survival, transmission, and dissemination. Microbiol. Spectr. 4, 593–633. doi: 10.1128/microbiolspec.VMBF-0015-2015
Davies, B. W., Bogard, R. W., Dupes, N. M., Gerstenfeld, T. A. I., Simmons, L. A., and Mekalanos, J. J. (2011). DNA damage and reactive nitrogen species are barriers to Vibrio cholerae colonization of the infant mouse intestine. PLoS Pathog. 7:e1001295. doi: 10.1371/journal.ppat.1001295
de Souza Santos, M., and Orth, K. (2014). Intracellular Vibrio parahaemolyticus escapes the vacuole and establishes a replicative niche in the cytosol of epithelial cells. MBio 5, e01506–e01514. doi: 10.1128/mBio.01506-14
Denoncourt, A. M., Paquet, V. E., and Charette, S. J. (2014). Potential role of bacteria packaging by protozoa in the persistence and transmission of pathogenic bacteria. Front. Microbiol. 5:240. doi: 10.3389/fmicb.2014.00240
Di Meo, S., Reed, T. T., Venditti, P., and Victor, V. M. (2016). Role of ROS and RNS sources in physiological and pathological conditions. Oxidative Med. Cell. Longev. 2016:1245049. doi: 10.1155/2016/1245049
Diamond, G., Beckloff, N., Weinberg, A., and Kisich, K. O. (2009). The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 15, 2377–2392. doi: 10.2174/138161209788682325
Dorin, J. R., McHugh, B. J., Cox, S. L., and Davidson, D. J. (2015). “Mammalian antimicrobial peptides; Defensins and cathelicidins” in Molecular medical microbiology. 2nd Edn. eds. Y.-W. Tang, M. Sussman, D. Liu, I. Poxton, and J. Schwartzman (Boston: Academic Press), 539–565.
Duperthuy, M., Schmitt, P., Garzón, E., Caro, A., Rosa, R. D., Le Roux, F., et al. (2011). Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc. Natl. Acad. Sci. USA 108, 2993–2998. doi: 10.1073/pnas.1015326108
Eckmann, L., Laurent, F., Langford, T. D., Hetsko, M. L., Smith, J. R., Kagnoff, M. F., et al. (2000). Nitric oxide production by human intestinal epithelial cells and competition for arginine as potential determinants of host defense against the lumen-dwelling pathogen Giardia lamblia. J. Immunol. 164, 1478–1487. doi: 10.4049/jimmunol.164.3.1478
Espinoza-Vergara, G., Noorian, P., Silva-Valenzuela, C. A., Raymond, B. B. A., Allen, C., Hoque, M. M., et al. (2019). Vibrio cholerae residing in food vacuoles expelled by protozoa are more infectious in vivo. Nat. Microbiol. 4, 2466–2474. doi: 10.1038/s41564-019-0563-x
Finlay, B. J. (2001). “Protozoa” in Encyclopedia of biodiversity. ed. S. Levin (Academic Press: San Diego), 901–915.
Flannagan, R. S., Cosio, G., and Grinstein, S. (2009). Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7, 355–366. doi: 10.1038/nrmicro2128
Forgac, M. (1999). Structure and properties of the vacuolar (H+)-ATPases. J. Biol. Chem. 274, 12951–12954. doi: 10.1074/jbc.274.19.12951
German, N., Doyscher, D., and Rensing, C. (2013). Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiol. 8, 1257–1264. doi: 10.2217/fmb.13.100
Gray, R., Gray, A., Fite, J. L., Jordan, R., Stark, S., and Naylor, K. (2012). A simple microscopy assay to teach the processes of phagocytosis and exocytosis. CBE Life Sci. Educ. 11, 180–186. doi: 10.1187/cbe.11-07-0060
Guo, L., Lim, K. B., Poduje, C. M., Daniel, M., Gunn, J. S., Hackett, M., et al. (1998). Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95, 189–198. doi: 10.1016/S0092-8674(00)81750-X
Hahn, M. W., and Höfle, M. G. (2001). Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35, 113–121. doi: 10.1111/j.1574-6941.2001.tb00794.x
Halpern, M., Senderovich, Y., Izhaki, I., and Manchester, M. (2008). Waterfowl - the missing link in epidemic and pandemic cholera dissemination? PLoS Pathog. 4:e1000173. doi: 10.1371/journal.ppat.1000173
Hao, X., Lüthje, F., Rønn, R., German, N. A., Li, X., Huang, F., et al. (2016). A role for copper in protozoan grazing–two billion years selecting for bacterial copper resistance. Mol. Microbiol. 102, 628–641. doi: 10.1111/mmi.13483
Henderson, J. C., Fage, C. D., Cannon, J. R., Brodbelt, J. S., Keatinge-Clay, A. T., and Trent, M. S. (2014). Antimicrobial peptide resistance of Vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases. ACS Chem. Biol. 9, 2382–2392. doi: 10.1021/cb500438x
Henderson, J. C., Herrera, C. M., and Trent, M. S. (2017). AlmG, responsible for polymyxin resistance in pandemic Vibrio cholerae, is a glycyltransferase distantly related to lipid A late acyltransferases. J. Biol. Chem. 292, 21205–21215. doi: 10.1074/jbc.RA117.000131
Hung, D. T., and Mekalanos, J. J. (2005). Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc. Natl. Acad. Sci. USA 102, 3028–3033. doi: 10.1073/pnas.0409559102
Imlay, J. A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776. doi: 10.1146/annurev.biochem.77.061606.161055
Kilvington, S., and Price, J. (1990). Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 68, 519–525. doi: 10.1111/j.1365-2672.1990.tb02904.x
Lambrecht, E., Baré, J., Chavatte, N., Bert, W., Sabbe, K., and Houf, K. (2015). Protozoan cysts act as a survival niche and protective shelter for foodborne pathogenic bacteria. Appl. Environ. Microbiol. 81, 5604–5612. doi: 10.1128/AEM.01031-15
Lee, C. T., Pajuelo, D., Llorens, A., Chen, Y. H., Leiro, J. M., Padrós, F., et al. (2013). MARTX of Vibrio vulnificus biotype 2 is a virulence and survival factor. Environ. Microbiol. 15, 419–432. doi: 10.1111/j.1462-2920.2012.02854.x
Levin, B. R. (1996). The evolution and maintenance of virulence in microparasites. Emerg. Infect. Dis. 2, 93–102. doi: 10.3201/eid0202.960203
Lutz, C., Erken, M., Noorian, P., Sun, S., and McDougald, D. (2013). Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front. Microbiol. 4:375. doi: 10.3389/fmicb.2013.00375
Malo, M. S., Alam, S. N., Mostafa, G., Zeller, S. J., Johnson, P. V., Mohammad, N., et al. (2010). Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut 59, 1476–1484. doi: 10.1136/gut.2010.211706
Mankere, B., Gupta, P., Chekkoora Keloth, S., Tuteja, U., Chelvam, K. T., and Pandey, P. (2018). Increased antibiotic resistance exhibited by the biofilm of Vibrio cholerae O139. J. Antimicrob. Chemother. 73, 1841–1847. doi: 10.1093/jac/dky127
Mathur, J., Davis, B. M., and Waldor, M. K. (2007). Antimicrobial peptides activate the Vibrio cholerae σE regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 63, 848–858. doi: 10.1111/j.1365-2958.2006.05544.x
Mathur, J., and Waldor, M. K. (2004). The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect. Immun. 72, 3577–3583. doi: 10.1128/IAI.72.6.3577-3583.2004
Matson, J. S., Yoo, H. J., Hakansson, K., and DiRita, V. J. (2010). Polymyxin B resistance in el tor Vibrio cholerae requires lipid acylation catalyzed by MsbB. J. Bacteriol. 192, 2044–2052. doi: 10.1128/JB.00023-10
Matsuo, J., Oguri, S., Nakamura, S., Hanawa, T., Fukumoto, T., Hayashi, Y., et al. (2010). Ciliates rapidly enhance the frequency of conjugation between Escherichia coli strains through bacterial accumulation in vesicles. Res. Microbiol. 161, 711–719. doi: 10.1016/j.resmic.2010.07.004
Matz, C., Boenigk, J., Arndt, H., and Jürgens, K. (2002). Role of bacterial phenotypic traits in selective feeding of the heterotrophic nanoflagellate Spumella sp. Aquat. Microb. Ecol. 27, 137–148. doi: 10.3354/ame027137
Matz, C., McDougald, D., Moreno, A. M., Yung, P. Y., Yildiz, F. H., and Kjelleberg, S. (2005). Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 102, 16819–16824. doi: 10.1073/pnas.0505350102
Merrell, D. S., Bailey, C., Kaper, J. B., and Camilli, A. (2001). The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J. Bacteriol. 183, 2746–2754. doi: 10.1128/JB.183.9.2746-2754.2001
Merrell, D. S., and Camilli, A. (1999). The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34, 836–849. doi: 10.1046/j.1365-2958.1999.01650.x
Merrell, D. S., and Camilli, A. (2000). Regulation of Vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J. Bacteriol. 182, 5342–5350. doi: 10.1128/JB.182.19.5342-5350.2000
Merrell, D. S., Hava, D. L., and Camilli, A. (2002). Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43, 1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x
Muniz, L. R., Knosp, C., and Yeretssian, G. (2012). Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front. Immunol. 3:310. doi: 10.3389/fimmu.2012.00310
Noorian, P., Hu, J., Chen, Z., Kjelleberg, S., Wilkins, M. R., Sun, S., et al. (2017). Pyomelanin produced by Vibrio cholerae confers resistance to predation by Acanthamoeba castellanii. FEMS Microbiol. Ecol. 93:fix147. doi: 10.1093/femsec/fix147
Pan, M., Neilson, M. P., Grunfeld, A. M., Cruz, P., Wen, X., Insall, R. H., et al. (2018). A G-protein-coupled chemoattractant receptor recognizes lipopolysaccharide for bacterial phagocytosis. PLoS Biol. 16:e2005754. doi: 10.1371/journal.pbio.2005754
Parry, J. D. (2004). Protozoan grazing of freshwater biofilms. Adv. Appl. Microbiol. 54, 167–196. doi: 10.1016/s0065-2164(04)54007-8
Provenzano, D., and Klose, K. E. (2000). Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 97, 10220–10224. doi: 10.1073/pnas.170219997
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 103, 1528–1533. doi: 10.1073/pnas.0510322103
Qiao, Y., Wang, J., Mao, Y., Liu, M., Chen, R., Su, Y., et al. (2017). Pathogenic bacterium Vibrio harveyi: an endosymbiont in the marine parasitic ciliate protozoan Cryptocaryon irritans. Acta Oceanol. Sin. 36, 115–119. doi: 10.1007/s13131-017-1050-y
Rasmussen, M. A., Carlson, S. A., Franklin, S. K., McCuddin, Z. P., Wu, M. T., and Sharma, V. K. (2005). Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect. Immun. 73, 4668–4675. doi: 10.1128/IAI.73.8.4668-4675.2005
Saeed, A., Abd, H., Edvinsson, B., and Sandstrom, G. (2007). Vibrio cholerae-Acanthamoeba castellanii interaction showing endosymbiont-host relation. Symbiosis 44, 153–158.
Schlimme, W., Marchiani, M., Hanselmann, K., and Jenni, B. (1997). Gene transfer between bacteria within digestive vacuoles of protozoa. Microb. Ecol. 23, 239–247. doi: 10.1111/j.1574-6941.1997.tb00406.x
Senderovich, Y., Izhaki, I., and Halpern, M. (2009). Fish as reservoirs and vectors of Vibrio cholerae. PLoS One 5:e8607. doi: 10.1371/journal.pone.0008607
Shanan, S., Bayoumi, M., Saeed, A., Sandström, G., and Abd, H. (2016). Swedish isolates of Vibrio cholerae enhance their survival when interacted intracellularly with Acanthamoeba castellanii. Infect. Ecol. Epidemiol. 6:31060. doi: 10.3402/iee.v6.31060
Sibille, I., Sime-Ngando, T., Mathieu, L., and Block, J. C. (1998). Protozoan bacterivory and Escherichia coli survival in drinking water distribution systems. Appl. Environ. Microbiol. 64, 197–202. doi: 10.1128/AEM.64.1.197-202.1998
Sigee, D. C. (2005). Freshwater microbiology: Biodiversity and dynamic interactions of microorganisms in the aquatic environment. University of Manchester, UK: John Wiley & Sons, Ltd.
Sperandio, V., Girón, J. A., Silveira, W. D., and Kaper, J. B. (1995). The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect. Immun. 63, 4433–4438. doi: 10.1128/IAI.63.11.4433-4438.1995
Steimle, A., Autenrieth, I. B., and Frick, J.-S. (2016). Structure and function: lipid A modifications in commensals and pathogens. Int. J. Med. Microbiol. 306, 290–301. doi: 10.1016/j.ijmm.2016.03.001
Stern, A. M., Hay, A. J., Liu, Z., Desland, F. A., Zhang, J., Zhong, Z., et al. (2012). The NorR regulon is critical for Vibrio cholerae resistance to nitric oxide and sustained colonization of the intestines. MBio 3, e00013–e00012. doi: 10.1128/mBio.00013-12
Sun, S., Noorian, P., and McDougald, D. (2018). Dual role of mechanisms involved in resistance to predation by protozoa and virulence to humans. Front. Microbiol. 9:1017. doi: 10.3389/fmicb.2018.01017
Sun, S., Tay, Q. X. M., Kjelleberg, S., Rice, S. A., and McDougald, D. (2015). Quorum sensing-regulated chitin metabolism provides grazing resistance to Vibrio cholerae biofilms. ISME J. 9, 1812–1820. doi: 10.1038/ismej.2014.265
Thom, S., Warhurst, D., and Drasar, B. (1992). Association of Vibrio cholerae with fresh water amoebae. J. Med. Microbiol. 36, 303–306. doi: 10.1099/00222615-36-5-303
Toei, M., Saum, R., and Forgac, M. (2010). Regulation and isoform function of the V-ATPases. Biochemistry 49, 4715–4723. doi: 10.1021/bi100397s
Vaitkevicius, K., Lindmark, B., Ou, G., Song, T., Toma, C., Iwanaga, M., et al. (2006). A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc. Natl. Acad. Sci. USA 103, 9280–9285. doi: 10.1073/pnas.0601754103
Van der Henst, C., Scrignari, T., Maclachlan, C., and Blokesch, M. (2016). An intracellular replication niche for Vibrio cholerae in the amoeba Acanthamoeba castellanii. ISME J. 10, 897–910. doi: 10.1038/ismej.2015.165
Van der Henst, C., Vanhove, A. S., Drebes Dörr, N. C., Stutzmann, S., Stoudmann, C., Clerc, S., et al. (2018). Molecular insights into vibrio cholerae’s intra-amoebal host-pathogen interactions. Nat. Commun. 9:3460. doi: 10.1038/s41467-018-05976-x
Vezzulli, L., Pruzzo, C., Huq, A., and Colwell, R. R. (2010). Environmental reservoirs of Vibrio cholerae and their role in cholera. Environ. Microbiol. Rep. 2, 27–33. doi: 10.1111/j.1758-2229.2009.00128.x
Vickerman, K. (1992). The diversity and ecological significance of protozoa. Biodivers. Conserv. 1, 334–341. doi: 10.1007/BF00693769
Vieira, A., Ramesh, A., Seddon, A. M., and Karlyshev, A. V. (2017). CmeABC multidrug efflux pump contributes to antibiotic resistance and promotes Campylobacter jejuni survival and multiplication in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 83, e01600–e01617. doi: 10.1128/aem.01600-17
Wang, H., Chen, S., Zhang, J., Rothenbacher, F. P., Jiang, T., Kan, B., et al. (2013). Catalases promote resistance of oxidative stress in Vibrio cholerae. PLoS One 7:e53383. doi: 10.1371/journal.pone.0053383
Wang, H., Xing, X., Wang, J., Pang, B., Liu, M., Larios-Valencia, J., et al. (2018). Hypermutation-induced in vivo oxidative stress resistance enhances Vibrio cholerae host adaptation. PLoS Pathog. 14:e1007413. doi: 10.1371/journal.ppat.1007413
Zhang, X., and Soldati, T. (2013). Detecting, visualizing and quantitating the generation of reactive oxygen species in an amoeba model system. J. Vis. Exp. 81:e50717. doi: 10.3791/50717
Zhu, J., and Mekalanos, J. J. (2003). Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5, 647–656. doi: 10.1016/S1534-5807(03)00295-8
Keywords: protozoan predation, virulence, Vibrio, adaptation, pathogenicity, heterotrophic protist
Citation: Espinoza-Vergara G, Hoque MM, McDougald D and Noorian P (2020) The Impact of Protozoan Predation on the Pathogenicity of Vibrio cholerae. Front. Microbiol. 11:17. doi: 10.3389/fmicb.2020.00017
Edited by:
Jens Andre Hammerl, Federal Institute for Risk Assessment (BfR), GermanyReviewed by:
Shin-ichi Miyoshi, Okayama University, JapanNidia León-Sicairos, Autonomous University of Sinaloa, Mexico
Jyl S. Matson, University of Toledo, United States
Copyright © 2020 Espinoza-Vergara, Hoque, McDougald and Noorian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diane McDougald, ZGlhbmUubWNkb3VnYWxkQHV0cy5lZHUuYXU=
 Gustavo Espinoza-Vergara
Gustavo Espinoza-Vergara M. Mozammel Hoque
M. Mozammel Hoque Diane McDougald
Diane McDougald Parisa Noorian
Parisa Noorian