- 1Division of Life Science, Graduate School of Sciences and Technology for Innovation, Yamaguchi University, Ube, Japan
- 2Department of Biological Chemistry, Faculty of Agriculture, Yamaguchi University, Yamaguchi, Japan
- 3Research Center for Thermotolerant Microbial Resources, Yamaguchi University, Yamaguchi, Japan
- 4Faculty of Agro-Industrial Technology, Rajamangala University of Technology Isan, Kalasin, Thailand
- 5Department of Biotechnology, Faculty of Technology, Khon Kaen University, Khon Kaen, Thailand
Thermotolerant genes, which are essential for survival at a high temperature, have been identified in three mesophilic microbes, including Zymomonas mobilis. Contrary to expectation, they include only a few genes for reactive oxygen species (ROS)-scavenging enzymes and heat shock proteins, which are assumed to play key roles at a critical high temperature (CHT) as an upper limit of survival. We thus examined the effects of increased expression of these genes on the cell growth of Z. mobilis strains at its CHT. When overexpressed, most of the genes increased the CHT by about one degree, and some of them enhanced tolerance against acetic acid. These findings suggest that ROS-damaged molecules or unfolded proteins that prevent cell growth are accumulated in cells at the CHT.
Introduction
Microorganisms intrinsically have an upper temperature limit for survival called a critical high temperature (CHT) (Matsushita et al., 2015; Kosaka et al., 2019). Genome-wide analysis of three mesophiles, Escherichia coli, Acetobacter tropicalis, and Zymomonas mobilis, by screening thermosensitive mutants either with a single-knockout mutant library or with a transposon-inserted mutant library has revealed that about 1.5% of genomic genes, called thermotolerant genes, are responsible for cell survival at a CHT (Charoensuk et al., 2017; Murata et al., 2018), but there is no sufficient information to conclude that a mesophile with a larger number of genomic genes has a larger number of thermotolerant genes and thus tends to be more temperature-resistant. Other factors including gene expression of key proteins may also contribute to the degree of thermotolerance. Thermotolerant genes are categorized into nine groups, including genes for metabolism, membrane stabilization, transporter, DNA repair, tRNA modification, protein quality control, translation control, cell division, and transcriptional regulation (Murata et al., 2011, 2018; Soemphol et al., 2011; Charoensuk et al., 2017), which are mostly related to fundamental activities of cells. The CHT differs to some extent from strain to strain even in the same species such that relatively thermotolerant strains can be isolated in tropical areas, assuming that thermotolerant strains have adapted to the environmental temperature. The thermotolerance of mesophiles, E. coli W3110, Z. mobilis CP4, and Z. mobilis TISTR 548, has been improved by in vivo thermal adaptation, suggesting that they have a genomic capacity for adaptation to higher temperature environments (Kosaka et al., 2019). The capacity, however, is limited to 2–3°, and the variation of thermal adaptation is also restricted. The change of only a few degrees is physiologically important, which was determined by an accurate method, called a two-step cultivation assay that eliminates the effects of start temperature and can distinguish CHT differences between two different strains of the same species (Kosaka et al., 2019).
The thermotolerance of microbes is remarkably beneficial for stable fermentation. Ethanologenic microbes, for example, are exposed to heat stress in the ethanol fermentation process (Attfield, 1997; Wang et al., 2007) due to its exothermic reaction (van Uden and da Craz Duarte, 1981; Ghose and Bandyopadhyay, 1982). Heat stress has a negative impact on their growth or viability (Basso et al., 2008; Babiker et al., 2010) and prevents their fermentation ability. These negative effects are enhanced in the presence of other factors, including a low pH, a high ethanol concentration, or a high osmolarity (Piper, 1995; Carmelo et al., 1998; Ciani et al., 2006; Coleman et al., 2007; Gibson et al., 2007; Pizarro et al., 2007). Thus, heat stress should be avoided for stable and effective fermentation by chilling fermentation reactors. Thermotolerant microbes enable high-temperature fermentation (HTF) to be performed, thus reducing the costs for chilling reactors in the bioconversion process of biomass to fuels or chemicals and for biomass hydrolysis in simultaneous saccharification fermentation and thus preventing contamination of other microbes (Murata et al., 2015).
A higher temperature results in accumulation of more oxidative stress in E. coli (Noor et al., 2009), and oxidative stress is involved in heat-induced cell death as has been shown for Saccharomyces cerevisiae (Davidson et al., 1996), being consistent with findings that overexpression of genes for catalase and superoxide dismutase is able to increase the degree of thermotolerance (Nantapong et al., 2019) and that the thermotolerance increases under anaerobic conditions (Davidson et al., 1996; Davidson and Schiestl, 2001). It is thus assumed that the impact of the CHT causes intracellular oxidative stress to elicit harmful effects on cells as a secondary stress. However, only one gene and no gene for reactive oxygen species (ROS)-scavenging enzymes (RSEs) was found as a thermotolerant gene in A. tropicalis and Z. mobilis, respectively (Soemphol et al., 2011; Charoensuk et al., 2017). The CHT would also cause damage of proteins to be unfolded or denatured. Surprisingly, no genes for general heat shock proteins (HSPs), except for degP, dnaK, and dnaJ in E. coli (Murata et al., 2011, 2018), degP in A. tropicalis (Soemphol et al., 2011), and degP in Z. mobilis (Charoensuk et al., 2017), have been identified as thermotolerant genes.
In this study, we thus examined the effects of increased expression of genes for RSEs and HSPs on cell survival at the CHT in Z. mobilis. Increased expression of most of the genes tested raised the CHT and reduced ROS compared to the controls. These findings together with previous findings suggest that the CHT is determined by functional contributions of several factors that prevent the accumulation of damaged macromolecules in cells in addition to fundamental activities by thermotolerant genes identified previously.
Materials and Methods
Materials
Oligonucleotide primers were purchased from Greiner Bio-One (Japan). A DNA purification kit, gel extraction kit, and one-step RT-PCR kit were from Qiagen (Japan). Restriction enzymes were from Biolabs (Japan) and Takara (Japan). PrimeSTAR DNA polymerase and an In-Fusion HD cloning kit were purchased from Takara (Japan). E. coli DH5α was from Toyobo (Japan). Yeast extract, peptone, tryptone, and agar were from Nacalai Tesque (Japan). Glucose and NaCl were from Sigma-Aldrich (United States). Chloramphenicol was from Boehringer Mannheim GmbH (Germany). Other chemicals used in this study were of analytical grade.
Bacterial Strains, Media, and Culture Conditions
Plasmids used in this study are listed in Supplementary Table S1. Z. mobilis TISTR 548 (Sootsuwan et al., 2007) was cultured in YPD medium [0.5% (w/v) yeast extract, 0.3% (w/v) peptone, and 3% (w/v) glucose]. Recombinant plasmids with targeted genes were introduced into E. coli DH5α (Toyobo, Japan) cells, and the cells were incubated in LB medium (1% tryptone, 0.5% yeast extract, and 0.5% NaCl) for 2 h and spread onto agar plates containing 1.5% agar. When pZA22 (Misawa et al., 1986) or its derivatives were introduced, agar plates were supplemented with chloramphenicol (50 μg ml–1). A log-phase culture, 0.5–1.0 at OD550, that had been prepared at 30°C was inoculated into a liquid medium, and cultivation was conducted under a static condition. Bacterial growth was monitored by measuring the optical density of the culture on a spectrophotometer (HITACHI, U-200) at OD550. Determination of the CHT was carried out by the two-step cultivation assay as described previously (Kosaka et al., 2019). For measuring the cell length, 12-h cultivated cells in the first culture of the two-step cultivation were collected by low-speed centrifugation, washed, and resuspended in a saline solution. The morphology of the resuspended cells was then observed under a microscope (Eclipse E600, Nikon, Japan) with 400 × magnification, and images of the same sample were taken three to five times. The lengths of approximately 100 cells were manually measured.
Construction of Expression Plasmids of Genes for RSEs and HSPs
Conventional recombinant DNA techniques were applied (Sambrook et al., 1989). For increased expression of genes for RSEs and HSPs in Z. mobilis TISTR548, operon fusion genes of the pdc promoter and each of these genes were constructed and incorporated into pZA22 (Misawa et al., 1986) as an expression vector for Z. mobilis. The pdc promoter fragment including the Shine–Dalgarno sequence of pdc (514 bp) and each gene fragment from its initiation codon to 20 bp downstream from its stop codon were amplified by PCR using the genomic DNA of TISTR548 as a template. pZA22 was linearized by PCR. The primers (Supplementary Table S2) used for PCR were designed according to the In-Fusion HD cloning method. The PCR fragments were purified using a QIAquick gel extraction kit and connected by an In-Fusion HD cloning kit. The constructed plasmids were confirmed by PCR and restriction mapping.
RT-PCR
To examine the degree of increased expression of targeted genes, RT-PCR was performed as described previously (Murata et al., 2011, 2018). Precultured cells were inoculated and cultivated in YPD medium containing chloramphenicol at 30°C for 12 h. The cells were then harvested, and total RNA was prepared by the hot phenol method (Aiba et al., 1981). RT-PCR was performed using a One-Step RNA PCR Kit with two specific primers for each gene (Supplementary Table S3) and 0.5 μg of total RNA according to the protocol from the kit supplier. After RT reaction at 50°C for 30 min, PCR consisting of denaturing at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min was carried out. The PCR products after 10, 15, 20, and 25 cycles for each gene were taken and analyzed by 1.2% agarose gel electrophoresis, followed by staining with ethidium bromide. The intensity of bands of RT-PCR products was quantitatively determined using ImageJ. The linearity of the amplification was observed up to the 25th or 35th cycle. Under our conditions, the RT-PCR was able to specifically detect mRNA because no band was observed when reverse transcriptase was omitted.
Determination of ROS
The level of intracellular ROS was determined using a fluorescence probe, 2′,7′-dichlorofluorescin diacetate (H2DCFDA) (Pérez-Gallardo et al., 2013), as described previously (Kosaka et al., 2019). Cells that were grown for 12 h at 38°C in the first culture of the two-step cultivation assay were mixed with H2DCFDA at the final concentration of 5 μM, incubated for 30 min, and collected as a pellet by centrifugation (14,000 rpm) for 1 min. The pellet was then washed with saline solution, resuspended in 10 mM potassium phosphate buffer (pH 7.0), and disrupted by sonic oscillation (Cosmo Bio Japan). The fluorescence was measured using a POWERSCAN HT microplate reader (DS Pharma Biomedical Osaka Japan) with excitation at 485 nm and emission at 582 nm. Emission values were normalized by protein concentration, which was determined by the Lowry method (Dulley and Grieve, 1975).
Observation of Stress Resistance
To examine the effects of increased expression of genes on resistance to stresses other than high temperatures, cells were precultured at 30°C until a mid-log phase, diluted (100–104), and spotted on YPD agar plates supplemented with 6–2% glucose, 3–5% ethanol, or 0.03–0.3% acetic acid. The plates were then incubated at 30°C for 48 h. To examine the effect of H2O2 on cell growth, precultured cells were inoculated and cultivated at 30°C in YPD liquid medium containing H2O2 at a final concentration of 0.1 mM. The optical density of the culture at OD550 was then measured at 12 h. All experiments were triplicated.
Results
Effects of Increased Expression of Genes for RSEs and HSPs on Growth at the CHT
To examine the contribution of RSEs and HSPs to the survival of Z. mobilis at the CHT and to the improvement of the CHT, the genes coding for these enzymes and proteins (Table 1) were individually cloned into pZA22 under the control of the pdc promoter, which is a relatively strong promoter derived from Z. mobilis, and the effects of expression of these genes were evaluated by the two-step cultivation assay, which enables determination of the CHT of mesophiles (Kosaka et al., 2019). The expression of cloned genes was confirmed by RT-PCR, indicating 1.4-to 4.0-fold increases compared to the expression of intrinsic genes in the genome (Supplementary Figure S1). Two-step cultivation assays for RSE genes were conducted at 37.5, 38, 38.5, and 39°C (Figure 1). At 38°C, all transformants with pZA-Ppdc-sod, pZA-Ppdc-cat, pZA-Ppdc-cyt, pZA-Ppdc-ahpC1, pZA-Ppdc-ahpC2, or pZA-Ppdc-ZMO1573, which bear sod, cat, cyt, ahpC1, ahpC2 [corresponding to ahpC (Charoensuk et al., 2011)], or ZMO1573, respectively, exhibited growth in the second culture, but the transformant with an empty vector as a control hardly grew. Among the genes tested, sod, cat, ZMO1573, and ZZ6-0186 had relatively stronger effects on cell growth, especially after 36 h. At 38.5°C, transformants with pZA-Ppdc-sod, pZA-Ppdc-cat, pZA-Ppdc-ZMO1573, or pZA-Ppdc-ZZ6-0186 showed clearly stronger growth than those with pZA-Ppdc-cyt, pZA-Ppdc-ahpC1, pZA-Ppdc-ahpC2, or pZA-Ppdc-ZMO1573 in the second culture. The two-step cultivation data indicated that the CHTs of transformants with pZA-Ppdc-sod, pZA-Ppdc-cat, pZA-Ppdc-ZMO1573, or pZA-Ppdc-ZZ6-0186 were 38.5°C and that the CHTs of the remaining transformants were between 38 and 38.5°C. The improvement of CHT in the former group was 1°C compared to the CHT, 37.5°C, of the transformant with an empty vector and that of the latter was more than 0.5°C and less than 1°C. These findings suggest that all of the tested genes are able to improve the CHT of Z. mobilis TISTR 548 and that sod, cat, and ZMO1573 are more effective.
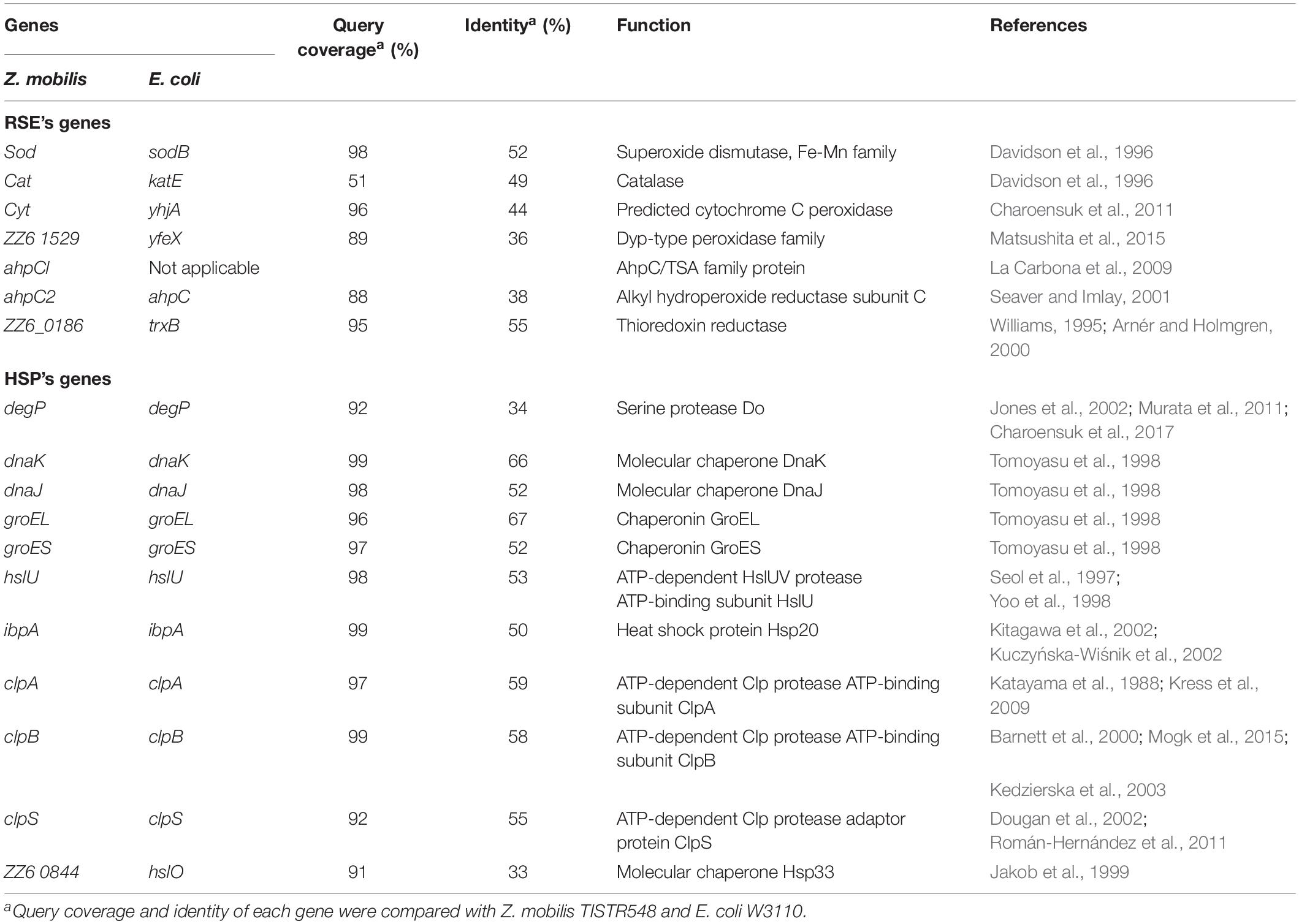
Table 1. Genes for reactive oxygen species (ROS)-scavenging enzymes (RSEs) and heat shock proteins (HSPs) that were examined in this study.
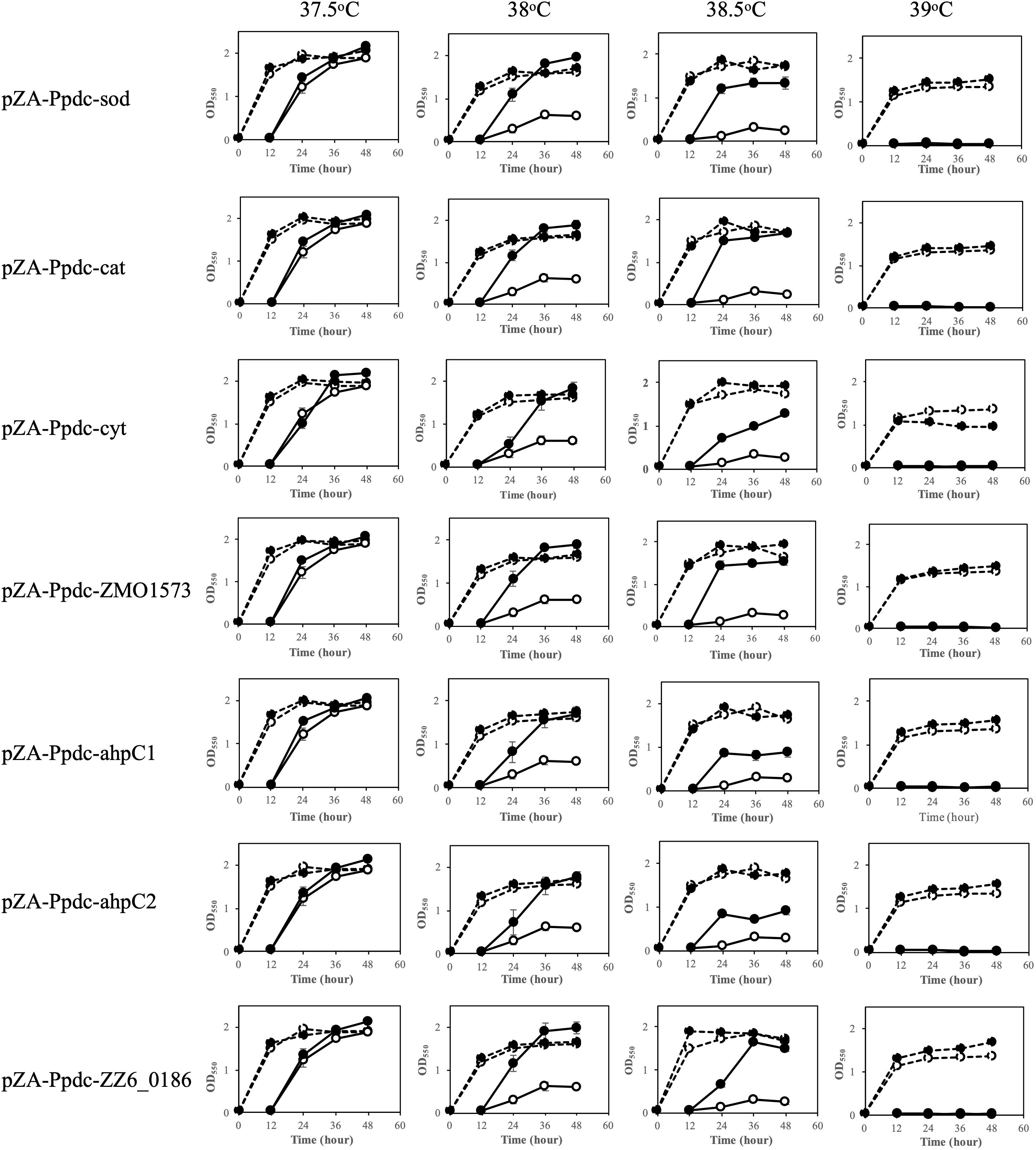
Figure 1. Effects of increased expression of genes for reactive oxygen species (ROS)-scavenging enzymes (RSEs) on growth at the critical high temperature (CHT). Growth of cells harboring a recombinant plasmid with a gene for one of the RSEs (closed circles) or an empty plasmid (open circles) was examined by two-step cultivation at 37.5–39°C in YPD medium containing chloramphenicol. After 12 h in the first cultivation (dotted lines) at each temperature, an aliquot of the culture was transferred to a fresh medium, and the second cultivation was carried out (straight lines). At the times indicated, cell density was estimated by measuring OD550. Bars represent ± SD for three independent experiments.
The effects of increased expression of genes for HSPs were also examined by two-step cultivation in the range of 37.5–39°C, in which transformants with pZA-Ppdc-degP, pZA-Ppdc-dnaKJ, pZA-Ppdc-groELS, pZA-Ppdc-hsp20, pZA-Ppdc-hisU, pZA-Ppdc-clpB, or pZA-Ppdc-clpPX, which bear degP, dnaKJ, groELS, hsp20, hisU, clpB, or clpPX, respectively, were used (Figure 2). At 38°C, all transformants except for a transformant with pZA-Ppdc-degP exhibited higher growth in the second culture than the growth of the transformant with an empty vector. Of these, transformants with pZA-Ppdc-dnaKJ, pZA-Ppdc-Hsp20, pZA-Ppdc-clpB, pZA-Ppdc-clpA, or pZA-Ppdc-clpS exhibited higher turbidity than that of the other transformants. On the other hand, cells containing pZA-Ppdc-degP showed only slightly higher turbidity than that of cells harboring an empty vector. At 38.5°C, in comparison with cells harboring an empty vector, cells harboring pZA-Ppdc-dnaKJ, pZA-Ppdc-Hsp20, or pZA-Ppdc-clpS showed relatively high turbidity in the second culture, followed by cells harboring pZA-Ppdc-groELS, pZA-Ppdc-clpB, or pZA-Ppdc-clpA. In the case of transformants with pZA-Ppdc-hslU or pZA-Ppdc-hsp33, their growth in the second culture was greatly repressed until 36 h compared to the control, suggesting that overexpression of hslU or hsp33 somehow hampers the cell growth. Notably, the transformant with pZA-Ppdc-hsp33 showed lower turbidity even in the first culture. These findings suggested that increased expression of all of the genes tested except for degP, hslU, and hsp33 can improve the thermotolerance of Z. mobilis TISTR 548 and that dnaKJ, hsp20, and clpS can up-shift the CHT by 1°C and groELS, clpB, and clpA can up-shift the CHT by 0.5–1°C.
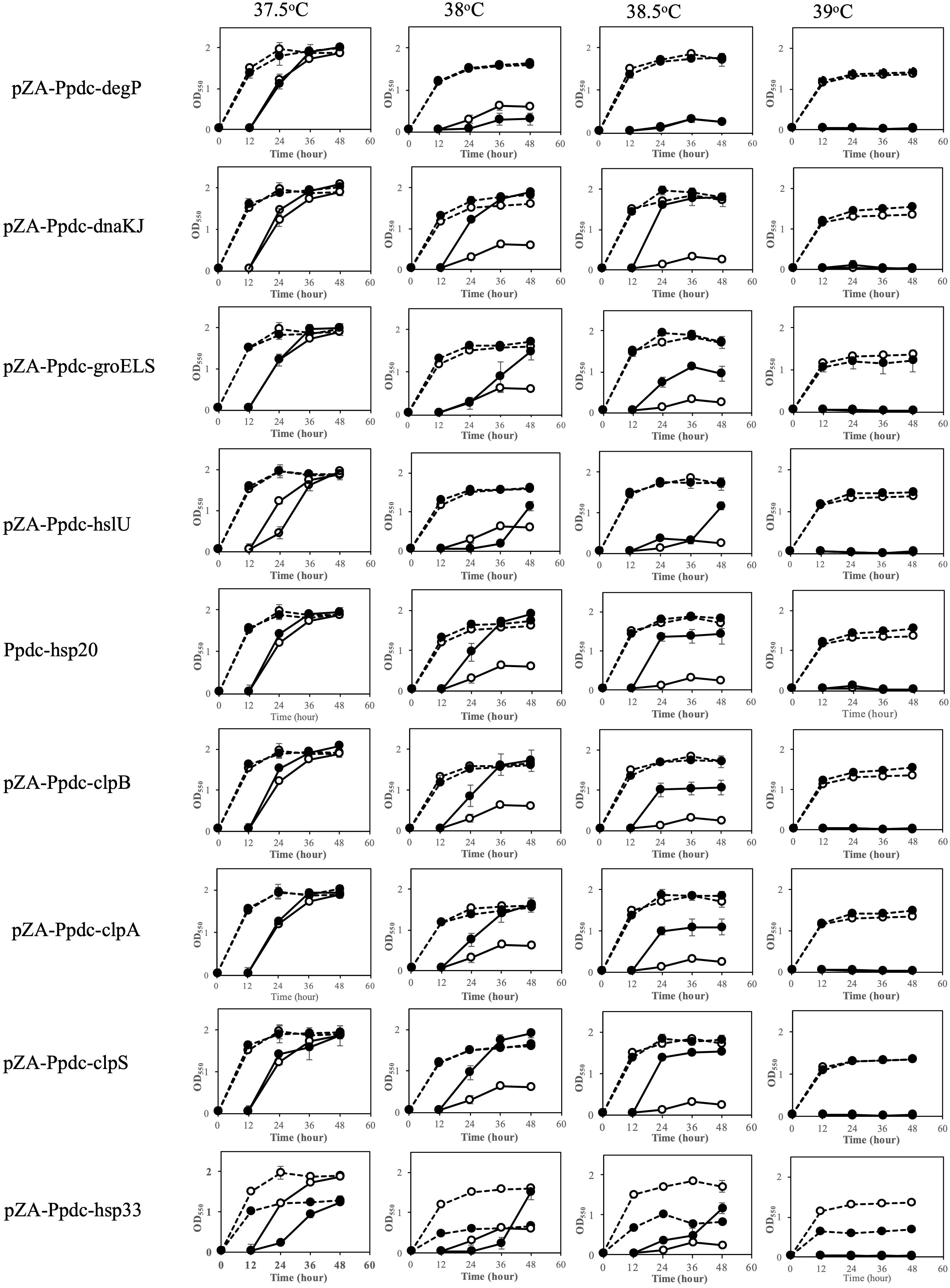
Figure 2. Effects of increased expression of genes for heat shock proteins (HSPs) on growth at the critical high temperature (CHT). Growth of cells harboring a recombinant plasmid with a gene for one of the HSPs (closed circles) or an empty plasmid (open circles) was examined by two-step cultivation at 37.5–39°C in YPD medium containing chloramphenicol as shown in Figure 1. Bars represent ± SD for three independent experiments.
Effects of Increased Expression of Genes for RSEs and HSPs on Accumulation of ROS at the CHT
When cells are exposed to a temperature close to the CHT, the level of intracellular ROS increases (Kosaka et al., 2019). Since enhancement of the expression of genes for RSEs and HSPs caused an up-shift of the CHT, it was assumed that increased expression of these genes prevented the accumulation of ROS. We thus examined the assumption at 38°C after 12 h in the first culture (Figure 3) because accumulation of ROS in the first culture has been reported to have an impact on the growth of cells in the second culture (Kosaka et al., 2019). As a result, the level of ROS of all of the transformants with a plasmid bearing one of the genes for RSEs and HSPs was lower than that of the control. However, the ROS level at the mid-log phase in the first culture was not always consistent with the level of improvement in the CHT because transformants with pZA-Ppdc-dnaKJ and pZA-Ppdc-clpA showed the lowest level of ROS but improved the CHT by 1°C and less than 1°C, respectively. This inconsistency may be due to other contribution of the genes, especially HSP genes, to the up-shift of the CHT in addition to the reduction of ROS.
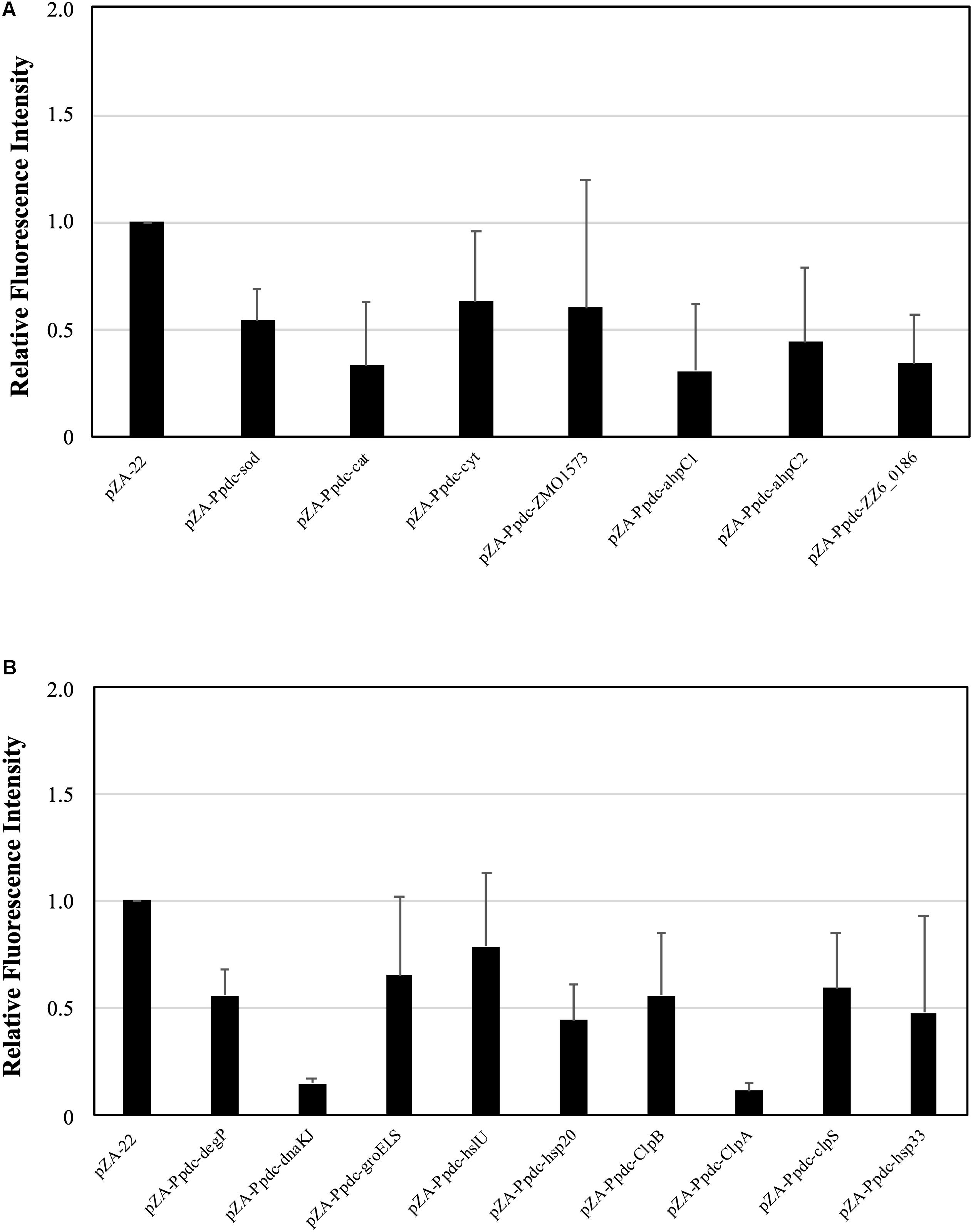
Figure 3. Effects of increased expression of genes for reactive oxygen species (ROS)-scavenging enzymes (RSEs) (A) and heat shock proteins (HSPs) (B) on accumulation of ROS at the critical high temperature (CHT). Cells were grown at 38°C in YPD medium containing chloramphenicol. Using the culture at 10 h, ROS were detected with H2DCFDA, and the fluorescent intensity reflects the level of accumulation of ROS. Bars represent ± SD for three independent experiments.
Effects of Increased Expression of Genes for RSEs and HSPs on Cell Morphology at the CHT
Cells become elongated when exposed to a temperature close to the CHT (Kosaka et al., 2019). The morphological change may be due to accumulation of stress including stress caused by ROS in cells, which presumably influences cell division. As mentioned above, the enhanced expression of genes for RSEs and HSPs reduced the intracellular level of ROS. We thus assumed that their enhanced expression prevents the morphological change. The assumption was examined by measurement of cell length at 38°C after 12 h in the first culture (Figure 4). As expected, all transformants with a plasmid bearing one of the genes for RSEs were shorter in cell length than the transformant with an empty vector, being consistent with the impact of the genes on cell growth at the CHT. While the transformants with dnaKJ, groELS, clpB, clpA, clpS, and hsp33 for HSPs were relatively short in cell length, the transformant with hsp20 showed no change, and the transformant with degP or hslU were much larger. Considering the lower levels of ROS in the transformants, increased expression of the latter three genes may somehow hamper cell division.
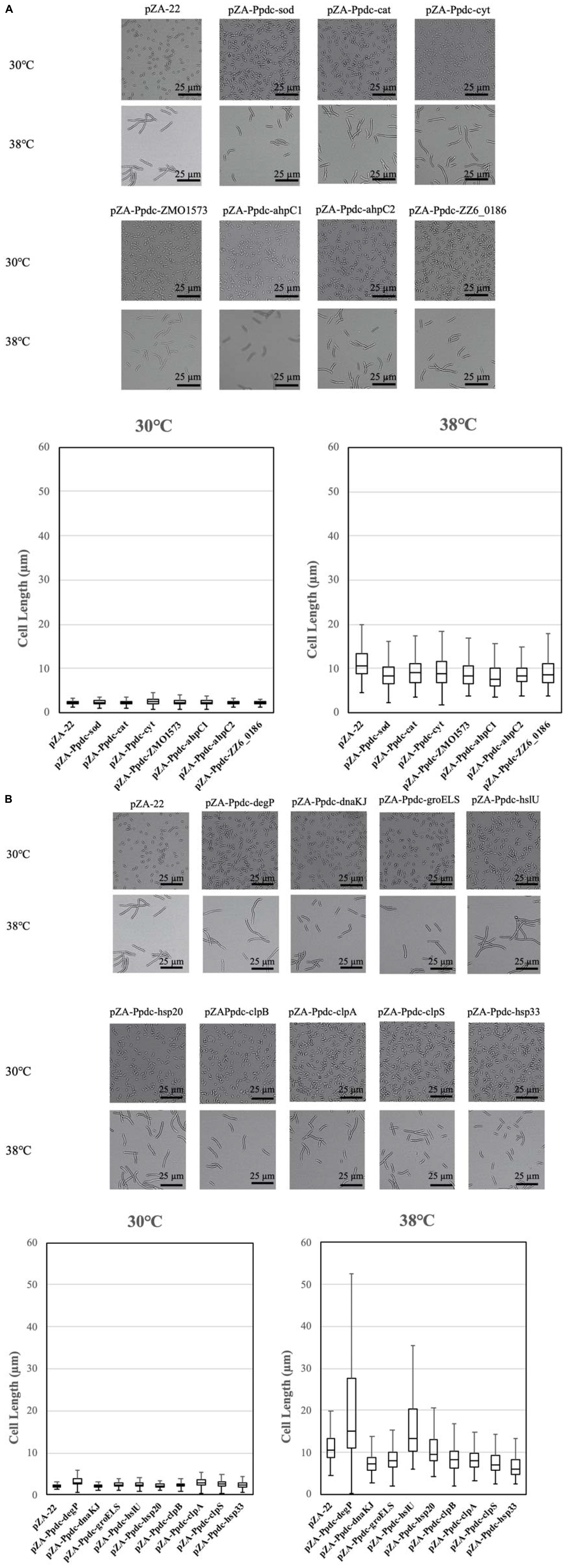
Figure 4. Effects of increased expression of genes for reactive oxygen species (ROS)-scavenging enzymes (RSEs) (A) and heat shock proteins (HSPs) (B) on cell morphology at the critical high temperature (CHT). Cells were grown at 38°C in YPD medium containing chloramphenicol. Using the culture at 12 h, cell morphology was observed, and lengths of 100 cells were measured.
Effects of Increased Expression of Genes for RSEs and HSPs on Growth Under Conditions of Stress
The experiments described above, in which the effects of increased expression of genes for RSEs and HSPs were examined, revealed that most of the genes were effective for improvement of the CHT, accumulation of ROS, and change in cell morphology. We thus examined whether these genes allow cells to be resistant to various types of stress in addition to heat (Figure 5 and Supplementary Figure S2). When tested on plates containing different concentrations of glucose or ethanol, no transformant with any gene for RSEs and HSPs showed growth that was different from that of the control transformant. After addition of 0.03% acetic acid (about pH 5.0) and 0.3% acetic acid (about pH 4.0), all transformants including those with an empty vector grew better than those without the addition of acetic acid, which may be because the host strain has the optimal pH in the acidic range. Interestingly, transformants with some genes of RSEs, sod, cat, ahpC1, ahpC2, and ZZ6_0186, and transformants with some genes of HSPs, dnaK, hsp20, clpA, clpB, and clpS, exhibited better growth on a medium containing acetic acid, being almost consistent with their up-shift of the CHT. Therefore, it is likely that the presence of acetic acid causes the macromolecule damage by a similar mechanism to that at the CHT, but it might be more than such damage when cells are challenged by acetic acid, for example, ATP deprivation and lower pH inside the cell (Lawford and Rousseau, 1993; Ullah et al., 2013). Moreover, exogenous oxidative stress was examined by cultivation at 30°C in YPD liquid medium containing 0.1 mM H2O2. Most of the transformants with genes for RSEs and HSPs except for degP showed better growth than that with an empty vector (Supplementary Figure S3), suggesting that increased expression of these genes alleviated the oxidative stress by H2O2. These findings are essentially consistent with the results of measurement of ROS in these transformants, as shown in Figure 3.
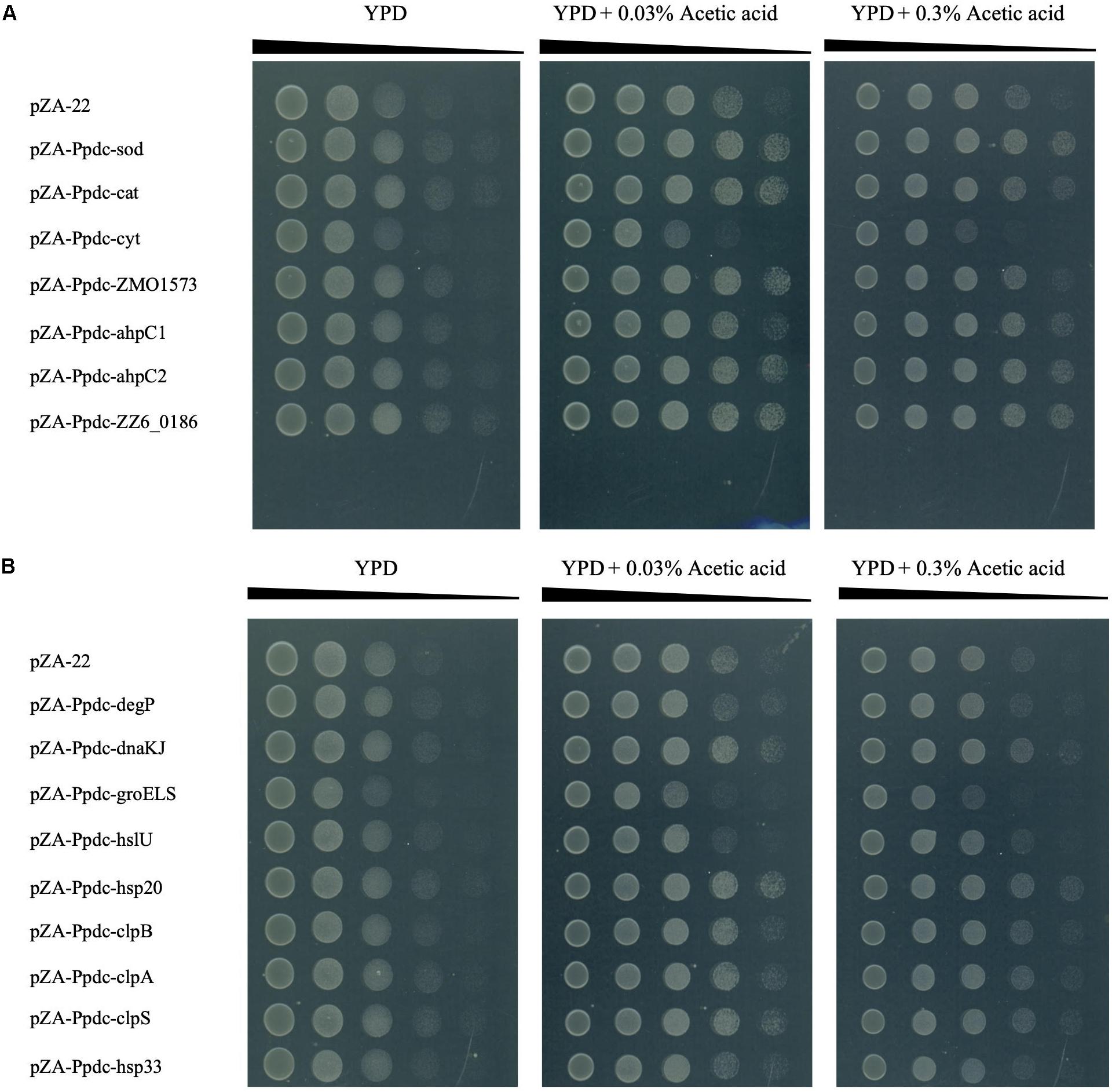
Figure 5. Effects of increased expression of genes for reactive oxygen species (ROS)-scavenging enzymes (RSEs) (A) and HSPs (B) on tolerance to acetic acid. Cells were grown at 30°C in YPD medium containing chloramphenicol overnight. The cell culture was serially diluted, spotted on YPD agar plates containing 0.03 or 0.3% acetic acid, and incubated at 30°C for 48 h.
Discussion
When exposed to a CHT, mesophiles exhibit several characteristic phenotypes, including cell elongation and accumulation of ROS (Kosaka et al., 2019). To determine the CHT of mesophiles, we had developed a two-step cultivation assay (Kosaka et al., 2019), which is much clearer and more precise than a general cultivation assay that shows an increase in turbidity even at a temperature above the CHT. On the other hand, a temperature around the CHT may cause instability of the membrane, resulting in leakage of electrons to generate ROS and may give rise to unfolding or denaturing of proteins or oxidation of proteins by accumulated ROS (Jan-Ulrik et al., 2015). In this study, we thus applied the two-step cultivation method to evaluate the contribution of genes for RSEs and HSPs to the CHT of Z. mobilis TISTR548. Their contribution was further examined by observation of the levels of ROS accumulation and cell elongation.
Among the genes tested, enhanced expression of sod, cat, ZMO1573, and ZZ6-0186 for RSEs and dnaKJ, hsp20, and clpS for HSPs up-shifted the CHT by 1°C, and most of the remaining genes up-shifted the CHT by 0.5–1.0°C. However, the contribution of these genes is less than the effect of thermal adaptation, which is able to increase the CHT by 2–3°C (Kosaka et al., 2019). Consistent with the weaker contribution to the up-shift of the CHT, the extents of reduction of ROS and of cell size seem to be relatively low. Nonetheless, it is likely that prevention of the accumulation of ROS is an effective way to improve the CHT.
The HSP genes tested in this study except for degP, hslU, and hsp33 were able to raise the CHT of Z. mobilis when their expression was increased (Figure 2). DnaKJ and GroELS function as molecular chaperones, which are involved in refolding of unfolded or denatured proteins (Narberhaus et al., 1998; Tomoyasu et al., 1998; Thanonkeo et al., 2007; Al Refaii and Alix, 2009). Hsp20 and ClpB as Hsp100 members prevent protein aggregation and solubilize aggregated proteins, respectively (Zolkiewski et al., 2012; Mogk et al., 2015). These chaperones or Hsps deal with denaturated or aggregated proteins that are expected to appear at the CHT. ClpA and ClpS are ATP-binding subunits of ClpAP protease and its adapter, respectively (Dougan et al., 2002; Maglica et al., 2008). They may contribute to the removal of denatured or aggregated proteins at the CHT. On the other hand, increased expression of hslU and hsp33 seems to inhibit cell growth even at 37.5 and 38°C. The former product is an ATP-binding subunit of HslUV protease (Yoo et al., 1998; Baytshtok et al., 2017), and the latter is a redox-regulated chaperone, which is activated by dimerization via disulfide bonds (Graf and Jakob, 2002). Considering these functions and activation process, the negative effects of HslU and Hsp33 on growth at the CHT might be due to proteolysis of and abnormal interaction with crucial proteins for cellular activities. In contrast to the HSP genes tested except for hslU and hsp33, degP for a periplasmic chaperone hardly elevated the CHT when overexpressed. A knockout mutant of degP, however, reduced the CHT by 1–2°C, and the gene has thus been categorized as a thermotolerant gene, which is shared by E. coli, Z. mobilis, and A. tropicalis (Murata et al., 2011, 2018). The inconsistency between degP and effective HSP genes may be due to the different cellular localization of their gene products in cells: DegP localizes in the periplasmic space, but other gene products localize in the cytoplasm. Alternatively, DegP might be sufficient to perform its activity in the parental strain. The increased expression of most of the cytoplasmic HSPs, but not DegP, may thus contribute to the maintenance of homeostasis inside cells at a CHT.
All seven of the RSE genes tested decreased the level of ROS and increased the CHT of Z. mobilis by their increased expression (Figures 1, 3). Four of the genes are involved in degradation of H2O2, but regulation of their expression may be distinct under different temperature conditions: the expression levels of cytC and ZMO1573 are higher at 37°C than at 30°C, whereas cat and ahpC exhibit the opposite expression pattern to that of cytC and ZMO1573 (Charoensuk et al., 2011). Such up-regulation of cytC and ZMO1573 at a high temperature may reflect insufficient H2O2-degrading activities for survival at the CHT, being in agreement with the finding that all of the four genes are able to raise the CHT when overexpressed. On the other hand, the reduced form of thioredoxin plays an important role as an antioxidant, and its reduction requires NADPH in addition to the corresponding reductase. It is likely that NADPH is not limited in Z. mobilis cells at the CHT because increased expression of ahp1 for peroxiredoxin or ZZ6_0186 for thioredoxin reductase leads to an increase in the CHT.
This study was motivated by the surprising fact that only a few genes for RSEs and HSPs have been identified as thermotolerant genes (Charoensuk et al., 2017; Murata et al., 2018), which are essential for survival at a CHT. The finding presented suggests that many genes for RSEs and HSPs have the potential to improve the CHT, although the range of improvement is within 1°C. It is thus likely that there are several genes that have overlapping functions in cells, and thus, the disruption of one gene is complemented by another gene. In relation to this, a knockout mutant of cytC for a peroxidase involved in the respiratory chain exhibited filamentous shapes and reduction in growth under a shaking condition at a high temperature, and under the same condition, sod, ahpC, and ZMO1573 are complementarily expressed to the cytC mutation (Charoensuk et al., 2011). Such robustness by the existence of complementing genes would have hindered the identification of genes for RSEs and HSPs as thermotolerant genes.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
SA, MM, KK, and KS carried out the experiments. SA, MM, TK, and PT analyzed the data. MY, SA, and MM wrote the manuscript. All authors conceived this study.
Funding
This study was supported by the Advanced Low Carbon Technology Research and Development Program, which was granted by the Japan Science and Technology Agency (JPMJAL1106) (MM, TK, and MY), and e-ASIA Joint Research Program, which was granted by Japan Science and Technology Agency (JPMJSC16E5) (MM, TK, and MY), Ministry of Research, Technology and Higher Education of the Republic of Indonesia, Agricultural Research Development Agency of Thailand, Ministry of Science and Technology of Laos, and partially supported by the Core to Core Program A. Advanced Research Networks, which was granted by the Japan Society for the Promotion of Science, the National Research Council of Thailand, Ministry of Science and Technology in Vietnam, National University of Laos, University of Brawijaya, and Beuth University of Applied Sciences Berlin (MM, TK, KS, PT, and MY), and the Japan Society for the Promotion of Science, MEXT/JSPS Kakenhi (25250028 and 16H02485 to MY). SA was supported by the government of Indonesia as a BUDI-LN LPDP Scholarship, collaboration of Ministry of Finance and Ministry of Research, Technology and Higher Education.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. Matsushita, T. Yakushi, and N. Kataoka for their helpful discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03073/full#supplementary-material
FIGURE S1 | Checking of increased expression of genes for RSEs and HSPs from recombinant plasmids. Strains are shown by introduced gene names. Total RNA was prepared from cells cultured at 30°C as described in Materials and Methods. RT-PCR was performed with primers specific for each gene to amplify approximately 500-bp DNA fragments. After RT reaction, PCR was performed for 10, 15, 20, and 25 cycles and the products were analyzed. Each PCR product and total RNA (10 μg) as a control were subjected to 1.2% agarose gel electrophoresis and staining with ethidium bromide (A,B). Intensity of stained bands was determined by using ImageJ, and the ratio of the intensity of bands from cells harboring a recombinant plasmid to that from cells harboring an empty vector, pZA22, was calculated. White columns represent the relative intensities as an expression ratio (%) (C).
FIGURE S2 | Effects of increased expression of genes for RSEs (A,B) and HSPs (C,D) on tolerance to various concentrations of glucose and ethanol. Cells were grown at 30°C in YPD medium containing chloramphenicol overnight. The cell culture at OD550 of about 1.0 was serially diluted (100–104-times dilution) and spotted on YPD agar plates containing 3–12% glucose or 3–5% ethanol, and incubated at 30°C for 48 h.
FIGURE S3 | Effects of increased expression of genes for RSEs (A) and HSPs (B) on tolerance to H2O2. Cells were grown at 30°C in YPD medium containing 0.1 mM H2O2, and optical density at OD550 of the culture was measured at 12 h.
TABLE S1 | Plasmids used in this study.
TABLE S2 | Primers used in this study.
TABLE S3 | Primers used for RT-PCR primers in this study.
References
Aiba, H., Adhya, S., and de Cromburgghe, B. (1981). Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256, 11905–11910.
Al Refaii, A., and Alix, J. H. (2009). Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK or DnaJ. Mol. Microbiol. 71, 748–762. doi: 10.1111/j.1365-2958.2008.06561.x
Arnér, E. S., and Holmgren, A. (2000). Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x
Attfield, P. V. (1997). Stress tolerance: the key to effective strains of industrial baker’s yeast. Nat. Biotechnol. 15, 1351–1357. doi: 10.1038/nbt1297-1351
Babiker, M. A., Banat, A., Hoshida, H., Ano, A., Nonklang, S., and Akada, R. (2010). High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl. Microbiol. Biotechnol. 85, 861–867. doi: 10.1007/s00253-009-2248-5
Barnett, M. E., Zolkiewska, A., and Zolkiewski, M. (2000). Structure and activity of ClpB from Escherichia coli. Role of the amino-and -carboxyl-terminal domains. J. Biol. Chem. 275, 37565–37571. doi: 10.1074/jbc.M005211200
Basso, L. C., de Amorim, H. V., de, Oliveira, A. J., and Lopes, M. L. (2008). Yeast selection for fuel ethanol production in Brazil. FEMS. Yeast Res. 8, 1155–1163. doi: 10.1111/j.1567-1364.2008.00428.x
Baytshtok, V., Chen, J., Glynn, S. E., Nager, A. R., Grant, R. A., Baker, T. A., et al. (2017). Covalently linked HslU hexamers support a probabilistic mechanism that links ATP hydrolysis to protein unfolding and translocation. J. Biol. Chem. 292, 5695–5704. doi: 10.1074/jbc.M116.768978
Carmelo, V., Santos, R., Viegas, C. A., and Sa′-Correia, I. (1998). Modification of Saccharomyces cerevisiae thermotolerance following rapid exposure to acid stress. Int. J. Food Microbiol. 42, 225–230. doi: 10.1016/s0168-1605(98)00089-0
Charoensuk, K., Irie, A., Lertwattanasakul, N., Sootsuwan, K., Thanonkeo, P., and Yamada, M. (2011). Physiological Importance of Cytochrome c Peroxidase in Ethanologenic Thermotolerant Zymomonas mobilis. J. Mol. Microbiol. Biotechnol. 20, 70–82. doi: 10.1159/000324675
Charoensuk, K., Sakurada, T., Tokiyama, A., Murata, M., Kosaka, T., Thanonkeo, P., et al. (2017). Thermotolerant genes essential for survival at a critical high temperature in thermotolerant ethanologenic Zymomonas mobilis TISTR 548. Biotechnol. Biofuels 10:204. doi: 10.1186/s13068-017-0891-0
Ciani, M., Beco, L., and Comitini, F. (2006). Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 108, 239–245. doi: 10.1016/j.ijfoodmicro.2005.11.012
Coleman, M. C., Fish, R., and Block, D. E. (2007). Temperature-dependent kinetic model for nitrogen-limited wine fermentations. Appl. Environ. Microb. 73, 5875–5884. doi: 10.1128/AEM.00670-07
Davidson, J. F., and Schiestl, R. H. (2001). Cytotoxic and Genotoxic Consequences of Heat Stress Are Dependent on the Presence of Oxygen in Saccharomyces cerevisiae. J. Bacteriol. 183, 4580–4587. doi: 10.1128/JB.183.15.4580-4587.2001
Davidson, J. F., Whyte, B., Bissinger, P. H., and Schiestl, R. H. (1996). Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 93, 5116–5121. doi: 10.1073/pnas.93.10.5116
Dougan, D. A., Reid, B. G., Horwich, A. L., and Bukau, B. (2002). ClpS, a substrate modulator of the ClpAP machine. Mol. Cell 9, 673–683. doi: 10.1016/s1097-2765(02)00485-9
Dulley, J. R., and Grieve, P. A. (1975). A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal. Biochem. 64, 136–141. doi: 10.1016/0003-2697(75)90415-7
Ghose, T. K., and Bandyopadhyay, K. K. (1982). Studies on immobilized Saccharomyces cerevisiae. II. Effect of temperature distribution on continuous rapid ethanol formation in molasses fermentation. Biotechnol. Bioeng. 24, 797–804. doi: 10.1002/bit.260240404
Gibson, B. R., Lawrence, S. J., Leclaire, J. P., Powell, C. D., and Smart, K. A. (2007). Yeast responses to stresses associated with industrial brewery handling. FEMS.Microbiol. Rev. 31, 535–569. doi: 10.1111/j.1574-6976.2007.00076.x
Graf, P. C., and Jakob, U. (2002). Redox-regulated molecular chaperones. Cell Mol. Life Sci. 59, 1624–1631. doi: 10.1021/bi9003556
Jakob, U., Muse, W., Eser, M., and Bardwell, J. C. (1999). Chaperone activity with a redox switch. Cell 96, 341–352. doi: 10.1016/s0092-8674(00)80547-4
Jan-Ulrik, D., Michael, J. G., and Ursula, J. (2015). Protein Quality Control Under Oxidative Stress Conditions. J. Mol. Biol. 427, 1549–1563. doi: 10.1016/j.jmb.2015.02.014
Jones, C. H., Dexter, P., Evans, A. K., Liu, C., Hultgren, S. J., and Hruby, D. E. (2002). Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: the PapA pilin. J. Bacteriol. 184, 5762–5771. doi: 10.1128/jb.184.20.5762-5771.2002
Katayama, Y., Gottesman, S., Pumphrey, J., Rudikoff, S., Clark, W. P., and Maurizi, M. R. (1988). The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning, and mutational analysis of the ATP-binding component. J. Biol. Chem. 263, 15226–15236.
Kedzierska, S., Akoev, V., Barnett, M. E., and Zolkiewski, M. (2003). Structure and function of the middle domain of ClpB from Escherichia coli. Biochemistry 42, 14242–14248. doi: 10.1021/bi035573d
Kitagawa, M., Miyakawa, M., Matsumura, Y., and Tsuchido, T. (2002). Escherichia coli small heat shock proteins, IbpA and IbpB, protect enzymes from inactivation by heat and oxidants. Eur. J. Biochem. 269, 2907–2917. doi: 10.1046/j.1432-1033.2002.02958.x
Kosaka, T., Nakajima, Y., Ishii, A., Yamashita, M., Yoshida, S., Murata, M., et al. (2019). Capacity for survival in global warming: adaptation of mesophiles to the temperature upper limit. PLoS. One 14:e0215614. doi: 10.1371/journal.pone.02156
Kress, W., Mutschler, H., and Weber-Ban, E. (2009). Both ATPase domains of ClpA are critical for processing of stable protein structures. J. Biol. Chem. 284, 31441–31452. doi: 10.1074/jbc.M109.022319
Kuczyńska-Wiśnik, D., Kedzierska, S., Matuszewska, E., Lund, P., Taylor, A., Lipińska, B., et al. (2002). The Escherichia coli small heat-shock proteins IbpA and IbpB prevent the aggregation of endogenous proteins denatured in vivo during extreme heat shock. Microbiology 148, 1757–1765. doi: 10.1099/00221287-148-6-1757
La Carbona, S., Sauvageot, N., Giard, J. C., Benachour, A., Posteraro, B., Auffray, Y., et al. (2009). Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. J. Microbiol. 47, 220–224. doi: 10.1007/s12275-008-0108-0
Lawford, H. G., and Rousseau, J. D. (1993). Effect of pH and acetic acid on glucose and xylose metabolism by a genetically engineered ethanologenic Escherichia coli. Appl. Biochem. Biotechnol. 3, 301–322. doi: 10.1007/bf02918999
Maglica, Z., Striebel, F., and Weber-Ban, E. (2008). An intrinsic degradation tag on the ClpA C-terminus regulates the balance of ClpAP complexes with different substrate specificity. J. Mol. Biol. 384, 503–511. doi: 10.1016/j.jmb.2008.09.046
Matsushita, K., Azuma, Y., Kosaka, T., Yakushi, T., Hoshida, H., Akada, R., et al. (2015). Genomic analyses of thermotolerant microorganisms used for high-temperature fermentations. Biosci. Biotechnol. Biochem. 80, 655–668. doi: 10.1080/09168451.2015.1104235
Misawa, N., Okamoto, T., Nakamura, K., Kitamura, K., Yanase, H., and Tonomura, K. (1986). Construction of a new shuttle vector for Zymomonas mobilis. Agric. Biol. Chem. 50, 3201–3203.
Mogk, A., Kummer, E., and Bukau, B. (2015). Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2:22. doi: 10.3389/fmolb.2015.00022
Murata, M., Fujimoto, H., Nishimura, K., Charoensuk, K., Nagamitsu, H., Raina, S., et al. (2011). Molecular strategy for survival at a critical high temperature in Eschierichia coli. PLoS. One 6:e20063. doi: 10.1371/journal.pone.0020063
Murata, M., Nitiyon, S., Lertwattanasakul, N., Sootsuwan, K., Kosaka, T., Thanonkeo, P., et al. (2015). High-temperature fermentation technology for low-cost bioethanol. J Jpn. Inst. Energy. 94, 1154–1212. doi: 10.3775/jie.94.1154
Murata, M., Ishii, A., Fujimoto, H., Nishimura, K., Kosaka, T., Mori, H., et al. (2018). Update of thermotolerant genes essential for survival at a critical high temperature in Escherichia coli. PLoS. One 13:e0189487. doi: 10.1371/journal.pone.0189487
Nantapong, N., Murata, R., Trakulnaleamsai, S., Kataoka, N., Yakushi, T., and Matsushita, K. (2019). The effect of reactive oxygen species (ROS) and ROS-scavenging enzymes, superoxide dismutase and catalase, on the thermotolerant ability of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 103, 5355–5366. doi: 10.1007/s00253-019-09848-2
Narberhaus, F., Kaser, R., Nocker, A., and Hennecke, H. (1998). A novel DNA element that controls bacterial heat-shock gene expression. Mol. Microbiol. 28, 315–323. doi: 10.1046/j.1365-2958.1998.00794.x
Noor, R., Murata, M., and Yamada, M. (2009). Oxidative stress as a trigger for growth phase-specific sigmaE-dependent cell lysis in Escherichia coli. J. Mol. Microbiol. Biotechnol. 17, 177–187. doi: 10.1159/000236029
Pérez-Gallardo, R. V., Briones, L. S., Díaz-Pérez, A. L., Gutiérrez, S., Rodríguez-Zavala, J. S., and Campos-García, J. (2013). Reactive oxygen species production induced by ethanol in Saccharomyces cerevisiae increases because of a dysfunctional mitochondrial iron-sulfur cluster assembly system. FEMS. Yeast Res. 13, 804–819. doi: 10.1111/1567-1364.12090
Piper, P. W. (1995). The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS.Microbiol. Lett. 134, 121–127. doi: 10.1111/j.1574-6968.1995.tb07925.x
Pizarro, F., Varela, C., Martabit, C., Bruno, C., Pe′rez-Correa, J. R., and Agosin, E. (2007). Coupling kinetic expressions and metabolic networks for predicting wine fermentations. Biotechnol. Bioeng. 98, 986–998. doi: 10.1002/bit.21494
Román-Hernández, G., Hou, J. Y., Grant, R. A., Sauer, R. T., and Baker, T. A. (2011). The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease. Mol. Cell 43, 217–228. doi: 10.1016/j.molcel.2011.06.009
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular cloning: a laboratory manual, 2nd Edn. New.York: Cold Spring Harbor Laboratory.
Seaver, L. C., and Imlay, J. A. (2001). Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183, 7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001
Soemphol, W., Deeraksa, A., Matsutani, M., Yakushi, T., Toyama, H., Adachi, O., et al. (2011). Global Analysis of the Genes Involved in the Thermotolerance Mechanism of Thermotolerant Acetobacter tropicalis SKU1100. Biosci. Biotechnol. Biochem. 75, 1921–1928. doi: 10.1271/bbb.110310
Seol, J. H., Yoo, S. J., Shin, D. H., Shim, Y. K., Kang, M. S., Goldberg, A. L., et al. (1997). The heat-shock protein HslVU from Escherichia coli is a protein-activated ATPase as well as an ATP-dependent proteinase. Eur. J. Biochem. 247, 1143–1150. doi: 10.1111/j.1432-1033.1997.01143.x
Sootsuwan, K., Irie, A., Murata, M., Lertwattanasakul, N., Thanonkeo, P., and Yamada, M. (2007). Thermotolerant Zymomonas mobilis: comparison of ethanol fermentation capability with that of an efficient type strain. Open Biotechnol. J. 1, 59–65. doi: 10.2174/1874070700701010059
Thanonkeo, P., Sootsuwan, K., Leelavacharamas, V., and Yamada, M. (2007). Cloning and transcriptional analysis of groES and groEL in ethanol-producing bacterium Zymomonas mobilis TISTR 548. Pak. J. Biol. Sci. 10, 13–22. doi: 10.3923/pjbs.2007.13.22
Tomoyasu, T., Ogura, T., Tatsuta, T., and Bukau, B. (1998). Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol. Microbiol. 30, 567–581. doi: 10.1046/j.1365-2958.1998.01090.x
van Uden, N., and da Craz Duarte, H. (1981). Effects of ethanol on the temperature profile of Saccharomyces cerevisiae. Z. Allg. Mikrobiol. 21, 743–750. doi: 10.1002/jobm.19810211006
Ullah, A., Chandrasekaran, G., Brul, S., and Smits, G. J. (2013). Yeast adaptation to weak acids prevents futile energy expenditure. Front. Microbiol. 4:142. doi: 10.3389/fmicb.2013.00142
Wang, Y., Gong, L., Liang, J., and Zhang, Y. (2007). Effects of alcohol on expressions of apoE in mice livers and brains. Wei. Sheng. Yan. Jiu. 36, 737–740.
Williams, C. H. (1995). Mechanism and structure of thioredoxin reductase from Escherichia coli. FASEB.J. 9, 1267–1276. doi: 10.1096/fasebj.9.13.7557016
Yoo, S. J., Kim, H. H., Shin, D. H., Lee, C. S., Seong, I. S., Seol, J. H., et al. (1998). Effects of the cys mutations on structure and function of the ATP-dependent HslVU protease in Escherichia coli. The Cys287 to Val mutation in HslU uncouples the ATP-dependent proteolysis by HslvU from ATP hydrolysis. J. Biol. Chem. 273, 22929–22935. doi: 10.1074/jbc.273.36.22929
Keywords: critical high temperature, thermotolerance, reactive oxygen species-scavenging enzyme, heat shock protein, Zymomonas mobilis
Citation: Anggarini S, Murata M, Kido K, Kosaka T, Sootsuwan K, Thanonkeo P and Yamada M (2020) Improvement of Thermotolerance of Zymomonas mobilis by Genes for Reactive Oxygen Species-Scavenging Enzymes and Heat Shock Proteins. Front. Microbiol. 10:3073. doi: 10.3389/fmicb.2019.03073
Received: 30 September 2019; Accepted: 19 December 2019;
Published: 30 January 2020.
Edited by:
Shihui Yang, Hubei University, ChinaReviewed by:
Xinqing Zhao, Shanghai Jiao Tong University, ChinaGloria Soberón-Chávez, National Autonomous University of Mexico, Mexico
Copyright © 2020 Anggarini, Murata, Kido, Kosaka, Sootsuwan, Thanonkeo and Yamada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masayuki Murata, bXVyYXRhbUB5YW1hZ3VjaGktdS5hYy5qcA==
 Sakunda Anggarini
Sakunda Anggarini Masayuki Murata
Masayuki Murata Keisuke Kido2
Keisuke Kido2 Tomoyuki Kosaka
Tomoyuki Kosaka Kaewta Sootsuwan
Kaewta Sootsuwan Pornthap Thanonkeo
Pornthap Thanonkeo Mamoru Yamada
Mamoru Yamada