
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 January 2020
Sec. Microbiotechnology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.03031
 Yilin Pang1,2†
Yilin Pang1,2† Xiaojun Ren1†
Xiaojun Ren1† Jianghui Li3†
Jianghui Li3† Feng Liang1
Feng Liang1 Xiaoyu Rao1
Xiaoyu Rao1 Yang Gao1
Yang Gao1 Wenhe Wu1
Wenhe Wu1 Dong Li3
Dong Li3 Juanjuan Wang3
Juanjuan Wang3 Jianguo Zhao3
Jianguo Zhao3 Xufen Hong3
Xufen Hong3 Fengying Jiang3
Fengying Jiang3 Wu Wang3
Wu Wang3 Huaibin Zhou1
Huaibin Zhou1 Jianxin Lyu1,4*
Jianxin Lyu1,4* Guoqiang Tan3*
Guoqiang Tan3*The whole-cell bioreporters based on the cop-operon sensing elements have been proven specifically useful in the assessment of bioavailable copper ions in water environments. In this study, a series of experiments was conducted to further improve the sensitivity and robustness of bioreporters. First, an Escherichia coli △copA△cueO△cusA mutant with three copper transport genes knocked out was constructed. Then, the copAp::gfpmut2 sensing element was inserted into the chromosome of E. coli △copA△cueO△cusA by gene knock-in method to obtain the bioreporter strain E. coli WMC-007. In optimized assay conditions, the linear detection range of Cu2+ was 0.025–5 mg/L (0.39–78.68 μM) after incubating E. coli WMC-007 in Luria–Bertani medium for 5 h. The limit of detection of Cu2+ was 0.0157 mg/L (0.25 μM). Moreover, fluorescence spectrometry and flow cytometry experiments showed more environmental robustness and lower background fluorescence signal than those of the sensor element based on plasmids. In addition, we found that the expression of GFPmut2 in E. coli WMC-007 was induced by free copper ions, rather than complex-bound copper, in a dose-dependent manner. Particularly, the addition of 40 mM 3-(N-Morpholino)propanesulfonic acid buffer to E. coli WMC-007 culture enabled accurate quantification of bioavailable copper content in aqueous solution samples within a pH range from 0.87 to 12.84. The copper recovery rate was about 95.88–113.40%. These results demonstrate potential applications of E. coli WMC-007 as a bioreporter to monitor copper contamination in acidic mine drainage, industrial wastewater, and drinking water. Since whole-cell bioreporters are relatively inexpensive and easy to operate, the combination of this method with other physicochemical techniques will in turn provide more specific information on the degree of toxicity in water environments.
Copper, one of the most common heavy metal elements, is an essential trace element for cellular metabolism. Research show that excessive copper for all living organisms is toxic (Karlsson et al., 2008). For instance, an excess of copper in the human body substantially contributes to the pathogenesis of Indian childhood cirrhosis (Freed et al., 2012), endemic Tyrolean infantile cirrhosis (Muller et al., 1996), and Wilson disease (Xu et al., 2019). In addition, accumulation of copper in the human brain can cause pathological changes in the nervous system, cerebellar motor dysfunction, and even Parkinson’s or Alzheimer’s disease (Emerit et al., 2004; White et al., 2006). It also has adverse effects on male fertility (Wirth and Mijal, 2010). In recent years, studies show that excessive copper may disrupt the labile [4Fe–4S] clusters in dehydrogenases (Macomber and Imlay, 2009) and block iron–sulfur cluster biogenesis in Escherichia coli (Fung et al., 2013; Tan et al., 2017) and Bacillus subtilis (Chillappagari et al., 2010).
Because the scale and pace of urbanization and industrialization have rapidly increased with the reform and opening-up policy of China, metal pollution has become a noticeable problem nowadays (Yang et al., 2018; Zhao Y. et al., 2018). In many cases, water environments close to human activity suffer from various degrees of copper pollution (Huang et al., 2013; Tang et al., 2013; Li F. et al., 2018), which threatens the water safety of local residents and causes huge economic losses. Therefore, prevention and control of copper pollution cannot be ignored, which is particularly important for detecting and monitoring toxic copper in the environment.
Currently, detection of heavy metal pollutants in the environment mainly relies on physicochemical methods, like atomic absorption spectroscopy (AAS) (Thongsaw et al., 2019) and inductively coupled plasma mass spectrometry (ICP-MS) (Fu et al., 2019). Although these methods are accurate, sensitive, and specific, they require expensive instrumentation and professional operation. Hence, it is difficult to apply at the grassroots level or in the field. Besides, quantification by these methods reflects the total copper content, including both bioavailable and non-bioavailable fractions (Martin-Betancor et al., 2015), making it difficult to objectively assess the toxicity of heavy metals. Therefore, developing an inexpensive, sensitive, selective, and portable method to determine the bioavailability of copper is necessary.
The academic definition of bioavailability is originally derived from the use of drugs, often associated with drug absorption in the human body or organs. It also applies to environmental pollutants (Impellitteri et al., 2003), but there is no clear definition so far (Li et al., 2014). In general, bioavailability of heavy metals is defined as those forms of metal that are absorbed and elicit an adverse effect on organisms (Deaver and Rodgers, 1996). As differences in metabolism and resistance mechanism of heavy metals exist among different species, bioavailability determined with one organism does not necessarily account for bioavailability for other organisms (Kang et al., 2018).
Whole-cell bioreporters have gained worldwide attention because they are sensitive, selective, portable, reasonably priced, and easy to handle. In addition, they have a relatively long life and a short response time (Fan et al., 2017; Jiang et al., 2017). Typically, a whole-cell bioreporter is consisted of a promoter–operator, which acts as the sensing element for heavy metal ions, with one or more reporter genes that encode easily detectable proteins (Jiang et al., 2017). However, to survive in high levels of heavy metal and reduce its toxic effect, bacteria have evolved a variety of heavy metal resistance mechanisms involved in the extrusion of metal ions from the cell (Leedjarv et al., 2008). These mechanisms lead to decrease in sensitivity of detection and unstable detection range of whole-cell bioreporters. Thereby, it is essential to elucidate the genetic mechanism of the response of host to specific metal to construct whole-cell bioreporters (Xu et al., 2013). In addition, the selectivity of typical whole-cell bioreporters is poor because of the non-specific cellular response to metal ions (Martin-Betancor et al., 2015; Din et al., 2019). For these reasons, the application of whole-cell bioreporters is restricted and in slow development.
Escherichia coli is one of the most common host strains used to construct whole-cell bioreporters for detecting copper ions (Riether et al., 2001; Hakkila et al., 2004; Ivask et al., 2009; Kang et al., 2018). E. coli has two copper efflux systems, the cue and cus system, to maintain a cellular demand level of copper: CopA, a P-type ATPase that pumps Cu(I) from the cytoplasm into periplasm (Rensing et al., 2000); CueO, an oxidase that oxidizes Cu(I) to Cu(II) in the periplasm to prevent adventitious Cu(I) entry into the cytoplasm again (Outten et al., 2001); and CusA, a subunit of a copper pump that transports copper ion from the periplasm to the extracellular environment (Munson et al., 2000). The cue system is controlled by the transcription factor CueR (Outten et al., 2000). As a member of the mercury resistance regulator MerR family, CueR contains two copper-binding cysteines (C112 and C120, as shown in Figure 1A) in its C-terminal metal-binding domain and forms homodimers. The activated CueR dimer binding two monovalent copper ions (holo-CueR) activates transcription of the copA and cueO genes by binding to their promoter regions, which induces torsional transformations in the DNA conformation. The apo-CueR (copper-free dimer) is also able to binding to the promotor region resulting in a tight DNA conformation, which represses copA and cueO expression (Figure 1B) (Outten et al., 2001; Philips et al., 2015; Bittner et al., 2017). Moreover, similar operons exist in Pseudomonas putida (Li et al., 2014) and Pseudomonas fluorescens (Kakinen et al., 2011).
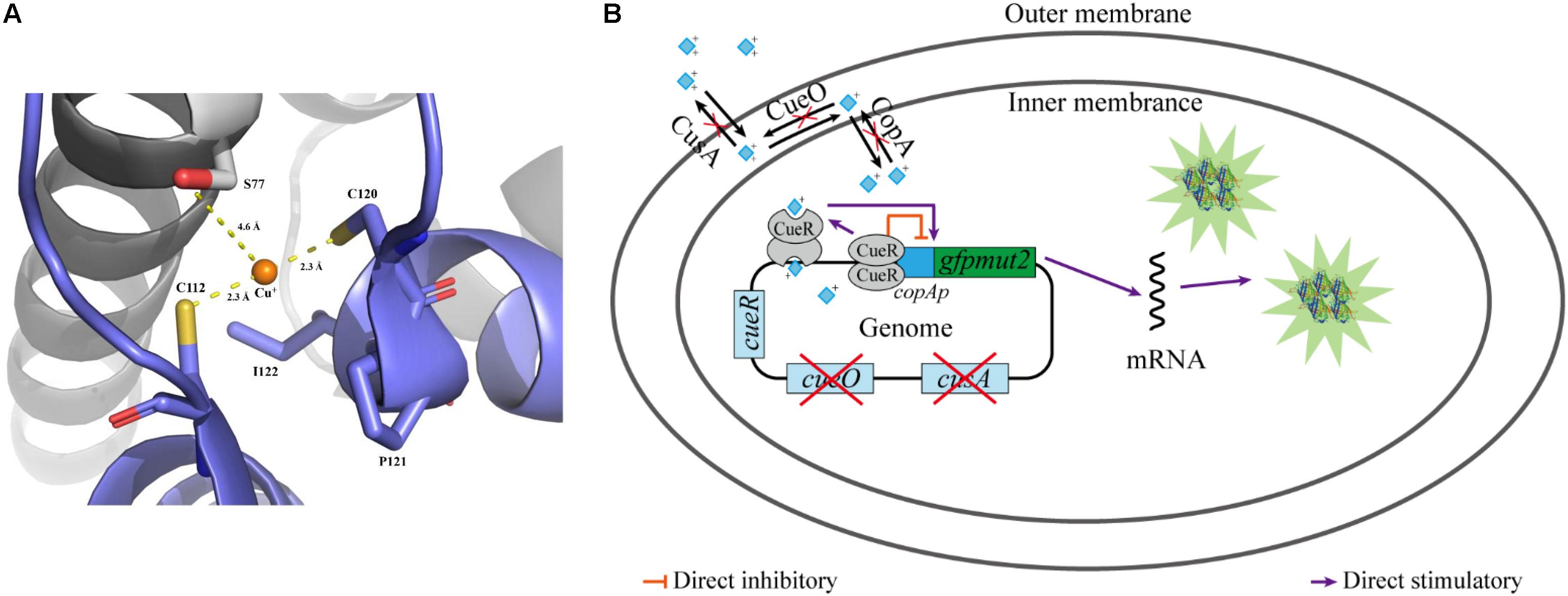
Figure 1. Diagram of working mechanism of the bioreporter E. coli WMC-007. (A) Cartoon representation of the binding site of Cu+ in CueR, and the copper-binding residues shown in stick representation. Polar interactions, stabilizing the position of Cu+ in the binding site, are indicated by yellow dashed lines. CueR is from E. coli K-12. This image was rendered using PyMOL (http://www.pymol.org); (B) schematic illustration of the bioreporter E. coli WMC-007.
Based on the above copper homeostatic systems, researchers have tried to construct various sensing elements, including copAp::luxCDABE (the linear detection range of Cu2+ is 3–30 μM) (Riether et al., 2001), pSLcueR/copAp::luxCDABE [limit of detection (LOD), 0.15 μM CuSO4] (Ivask et al., 2009), pSLcueR/copAp::PpyWT luciferase (LOD, 0.15 μM Cu2+) (Roda et al., 2011), and pPROBEcueR/cueAp::gfp (the detection range of Cu2+ is 1–70 mg/L) (Li et al., 2014), which were incorporated into the plasmids and transformed into the wild-type E. coli or P. putida to detect bioavailable copper. Recently, Kang et al. (2018) coexpressed the pCopAp-eGFP and pCDF-Duet-CueR plasmids in E. coli △copA△cueR mutant strain, significantly increasing its copper ion sensitivity. In particular, effectively reduced the background signal of whole-cell bioreporters by overexpressing CueR, thereby further enhancing sensitivity in response to copper ions (the detection range of Cu is 0–10 μM), compared with pCopAp-eGFP/E. coli △copA△cueR bioreporter. Our previous work reported a dual-functional E. coli cell that could detect and remove copper ions, which selectively responded to copper ions with a linear detection range of 0.01–25 μM (Wang et al., 2019).
The function of plasmid replication and segregation plays a key role in ensuring plasmid stability. Multiple factors can affect the stability of plasmid segregation, including the plasmid copy number, its replication mechanism, the levels of recombinant gene expression, the type of host strain, and the metabolic burden associated with all these factors (Sukhija et al., 2012; Kramer, 2016). Previous studies show that most recombinant plasmids are unstable and can be divided into two types. One is that the instability of segregation leads to the loss of plasmids; the other is the loss of heterologous genes caused by structural instability (Imanaka et al., 1980; Sukhija et al., 2012).
Even though a number of copper-induced whole-cell bioreporters with high sensitivity and/or strong specificity already exist, there are some drawbacks. In most cases, the reporter genes are harbored by the host strain in a medium or high-copy-number plasmid (Magrisso et al., 2008). Thus, in the process of bacterial cell division, it is potentially prone to copy number variability and loss of plasmids (Tom-Petersen et al., 2001). Moreover, high-copy-number plasmids could lead to unnatural and deleterious dosing of foreign genes, causing significant metabolic burden to host cells and leading to risks of horizontal gene transfer (Wrobel and Wegrzyn, 1998; Sukhija et al., 2012; Shemer et al., 2017). In particular, the background fluorescence value is very high without the addition of exogenous copper ions and overexpression of CueR (Kang et al., 2018). Nevertheless, overproduction of CueR and reporter proteins within cells potentially increases the metabolic burden. Therefore, it is necessary to construct a more effective bioreporter to minimize the influence of adverse factors mentioned above for more reliable test results.
In this report, we constructed a bioreporter strain E. coli WMC-007 with two genetic modifications. One of them was an E. coli △copA△cueO△cusA mutant strain with three Cu resistance-conferring genes knocked out, giving rise to its high sensitivity to copper ions. The other one was a copAp::gfpmut2 transcriptional fusion integrated in the chromosome of E. coli △copA△cueO△cusA (Figure 1B). The optimal growth conditions of E. coli WMC-007 and its selectivity and the optimum detection linear range parameters were determined. Preliminarily, E. coli WMC-007 was applied to detect copper ions in water environment samples. The results suggest that the constructed E. coli WMC-007 may have the potential to supplement other physicochemical techniques for detecting bioavailable copper ions in acidic mine drainage, industrial wastewater, and drinking water.
DNA polymerase, restriction endonucleases, T4 DNA ligase, and all primers were purchased from/synthesized by Takara corporation (Dalian, China). Sucrose, ZnSO4⋅7H2O, MnSO4⋅H2O, and AgNO3 were obtained from Xilong Chemical Industry Corporation (Shanghai, China). Ni3SO4⋅6H2O, MgCl2⋅6H2O, and CuSO4⋅5H2O were purchased from Xinbao Fine Chemical Factory (Shanghai, China). HgCl2 was purchased from Beisite Chemical Reagent Corporation (Chengdu, China). 3-(N-Morpholino)propanesulfonic acid (MOPS), Pb(NO3)2, FeC6H5O7, (NH4)2Fe(SO4)2, and CoCl2⋅6H2O were purchased from Sigma-Aldrich (Shanghai) Trading Corporation. The standard copper solution (1,000 μg/ml) was obtained from the National Standard Substances Center (Beijing, China). Other chemicals used were of analytical grade. All the medium and buffer solutions were prepared using deionized distilled water (Millipore, United States). All tube cultures of bacteria were soaked in potassium dichromate solution for at least 24 h to remove residual heavy metals.
Strains and plasmids used in this study are described in Supplementary Table S1. Genes encoding three major proteins (CpoA, CueO, and CusA) regulating intracellular copper homeostasis were deleted following the protocol developed by Datsenko and Wanner (2000). The gene deletion was confirmed by PCR. All primers for the deletion and confirmation can be found in Supplementary Table S2.
All experiments, unless stated otherwise, were conducted with the following protocol. E. coli WMC-007 was grown in Luria–Bertani (LB) at 37°C and rotated at 250 rpm. Overnight cultures were diluted to OD600≈0.02 using fresh LB medium and simultaneously adding the standard copper ion solution. The optical density at 600 nm (OD600) was determined with a Beckman Coulter DU800 spectrophotometer.
The fused reporter gene, copAp::gfpmut2-soxSCo, was constructed with crossover PCR using three pairs of primers (Supplementary Table S2). The 3′ end of PcopA::gfpmut2 was fused with the transcription terminator of soxS. The PCR products were ligated into pET28a vector. The ligation mixture was transformed into DH5α. The bioreporter strains were constructed by transforming copAp::gfpmut2-pET28a into wild-type MC4100, E. coli △copA△cueO△cusA, and other single- and double-mutant strains.
The gfpmut2 reporter gene knock-in was achieved following the procedure developed by Church’s group (Link et al., 1997). Briefly, the fused reporter gene, copAp::gfpmut2, was constructed with crossover PCR using three pairs of primers (Supplementary Table S2). The PCR products were cloned into a low-copy number and temperature-sensitive vector pKOV with chloramphenicol-resistant and sucrose-sensitive markers (Link et al., 1997). The cloned plasmid was introduced into E. coli △copA△cueO△cusA cells. The transformants were selected on LB plates containing 20 μg/ml chloramphenicol at 43°C. The obtained colonies were immediately plated on LB plates containing 5% (w/v) sucrose at 30°C to select the colonies that underwent the second recombination event. The colonies which were sucrose resistant and chloramphenicol sensitive were selected for further analysis. The positive colonies were confirmed with PCR using the primers flanking the target genes.
First, overnight cultures were grown in fresh liquid M9 minimal medium at 37°C and rotated at 250 rpm for 2 h, then diluted to OD600≈0.005 using fresh M9 minimal medium before preparing 5-ml aliquots. Simultaneously, different concentrations of standard copper ion solutions were added to each aliquot to have a final concentration of 0, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, and 32 μM. The mixture was incubated at 37°C and 250 rpm for 2–10 h and monitored by cell density measurements at 600 nm every 2 h for three aliquots from each concentration.
E. coli WMC-007 cells were grown in LB medium and incubated with 100 μM standard copper ion solutions at 37°C for 1–8 h. An equal volume of MilliQ H2O was added as blank control. All experiments, unless stated otherwise, were conducted with the following protocol. One milliliter of each sample was harvested, washed with pH 8.0 desalting buffer (contained 20 μg/ml chloramphenicol to block new GFPmut2 protein synthesis) twice, and diluted 20 times with the same buffer. Subsequently, the cell density of each sample was measured at 600 nm. The GFPmut2 assay was performed using a RF-5301 fluorescence spectrometer (Shimadzu Corporation, Japan) at an excitation wavelength of 481 nm and an emission wavelength of 507 nm.
E. coli WMC-007 culture was used to prepare 1-ml aliquot. Each aliquot was incubated with different concentrations of standard copper ion solutions at 37°C for 2–5 h. An equal volume of MilliQ H2O was added as blank control. Twelve aliquots were prepared for each concentration, and three of them were monitored every 1 h. The OD600 and GFPmut2 fluorescence intensity were measured as described above.
E. coli WMC-007 cells were harvested after incubation with different copper concentrations in LB medium for 5 h. The fluorescence of GFPmut2-producing cells was measured using a FluoView FV1000 laser confocal microscope (Olympus Corporation, Japan) with the following settings: excitation wavelength, 488 nm; voltage, 598 V; 1,024 × 1,024 pixel; magnification 600 times.
The cell extracts were prepared by passing the cells through NS1001L2K high-pressure cell cracker twice. Cytosolic protein extracts (30 μg total protein per lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis before blotting onto polyvinylidene fluoride membrane. GFPmut2 was detected using specific polyclonal antibodies from rabbit and goat antirabbit immunoglobulin G-conjugated with alkaline phosphatase as secondary antibody (Beyotime Institute of Biotechnology, China).
The induction of the detection component by a variety of metal ions, including Ag+, Mg2+, Zn2+, Fe2+, Fe3+, Pb2+, Mn2+, Ca2+, Co2+, Ni2+, and Cu2+, was studied by measuring the green fluorescence produced. Each metal ion (5 μM) was added to E. coli WMC-007 cultures at a cell density of OD600 = 0.02. An equal volume of MilliQ H2O was added as blank control. The cells were incubated for 5 h at 37°C, and the specific fluorescence intensity was measured as described above. At least three independent experiments were performed for each kind of metal ion and mixtures of metal ion assays.
The experimental conditions of E. coli WMC-007 were as described above, except that MilliQ H2O was added instead of copper ion solution. For bioreporter copAp::gfpmut2-pET28a/E. coli △copA△cueO△cusA, namely, E. coli WMC-006, all experiments, unless stated otherwise, were conducted with the following protocol. The overnight cultures were diluted to OD600≈0.02 with LB medium, and cells were grown to OD600 = 0.5–0.6, which was used to prepare three aliquots of 1 ml. Ten microliters MilliQ H2O was added to each aliquot as blank control. Then, it was shake incubated at 37°C and 250 rpm for 3 h. Cells were washed with desalting buffer after centrifugation and resuspended to OD600 ≈ 0.25. The OD600 and the background fluorescence intensity was measured as described above.
For flow cytometric analysis of gfpmut2 expression stability, the following procedure developed by Stiner and Halverson was carried out (Stiner and Halverson, 2002). The cultured cells were diluted with filtered (0.22-μm pore size filters) normal saline (NS) to approximately 5 × 106 CFU/ml before analysis with a FACScan flow cytometer (BD, United States). The instrument was set to detect 10,000 cells for each sample.
The standard copper ion solution was added to each aliquot to reach a concentration of 1 mg/L. An equal volume of ethylenediaminetetraacetic acid (EDTA) in different concentrations was added simultaneously, giving final concentrations of 200, 400, and 800 μM. An equal volume of MilliQ H2O was added as blank control. The copper ion content in the sample was calculated according to the linear regression equation of the standard curve.
Generally, water environments are highly acidic or alkaline when polluted by heavy metal ions. To minimize the influence of this environmental condition on bioreporter analyses, LB medium containing MOPS with a final concentration of 40 mM was used in this experiment (pH 7.2). One-tenth volume of aqueous solution with pH ranging from 0.87 to 12.57 was added to each culture aliquot. Simultaneously, standard copper solution was added to have a final concentration of 0.97 mg/L. The mixture was incubated for 5 h before analyzing the influence of pH on the copper recovery rate. The copper concentration in the sample was calculated based on the linear regression equation of the standard curve.
Unless otherwise stated, all data are presented as means ± SD.
Relative fluorescence unit (RFU) is defined as the ratio of the fluorescence intensity of bioreporter cells to the cell density at OD600; absolute fluorescence unit (AFU) is defined as the RFU of blank control subtracted from the RFU of the copper-exposed sample; induction coefficient (IC) is defined as the ratio of the relative fluorescence unit induced with copper ions to the background fluorescence unit of bioreporter cells without added heavy metal (Li et al., 2014; Kang et al., 2018).
The signal-to-noise (S/N) ratio was calculated as follows: S/N = (FM−FDB/OD600M)/(FW−FDB/OD600W), where FM is the fluorescence intensity of bioreporter cells induced with metals, FW is the background fluorescence intensity of bioreporter cells induced with MilliQ H2O, and FDB is the fluorescence intensity induced with desalting buffer.
The limit of determination (LODAFU) of copper for the bioreporter strains was determined following the procedure developed by Ivask et al. (2004). The calculation was based on this equation: LODAFU = 2(XW + 3SD)/XW, where XW is the mean background luminescence value of the bioreporter strains (10 blanks included in each assay) and SD is the standard deviation. The LOD as mol/L by the regression equation of the standard curve.
Bioavailable Cu2+ in water environment samples was determined using the Cu-inducible bioreporter E. coli WMC-007. The total Cu content was analyzed by AAS. Bioavailability was then calculated as the ratio of the copper concentration detected in bioreporters to the total copper content in water.
To improve the sensitivity of copper detection of E. coli MC4100, we constructed mutant strains of E. coli MC4100, in which one or several of copper response genes were knocked out. Minimal inhibitory concentration (Grass and Rensing, 2001) of the wild-type E. coli MC4100 and its mutants showed that E. coli △copA△cueO△cusA was the most sensitive to Cu2+. Its sensitivity was 64 times higher than that of the wild-type E. coli MC4100. The copA gene played a dominant role among the copper transport genes in E. coli (Table 1). Furthermore, when E. coli △copA△cueO△cusA cells were transformed with the copAp::gfpmut2-pET28a sensing element and induced by various concentrations of Cu2+, bright green color could be observed in the cell resuspension (Figure 2A). With increasing copper concentrations, the green color tended to be darker in a dose-dependent manner. To confirm that the accumulation of green color was due to the expression of GFPmut2, sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis was carried out using the above whole cells and there were visible bands at ∼27 kDa in copper-induced samples (10–3–10–8 M), except for the blank control (Figure 2B). The amount of protein expression increased with copper concentration, which was consistent with the naked eye observation. These results indicated that the bioreporters were successfully constructed with high sensitivity.
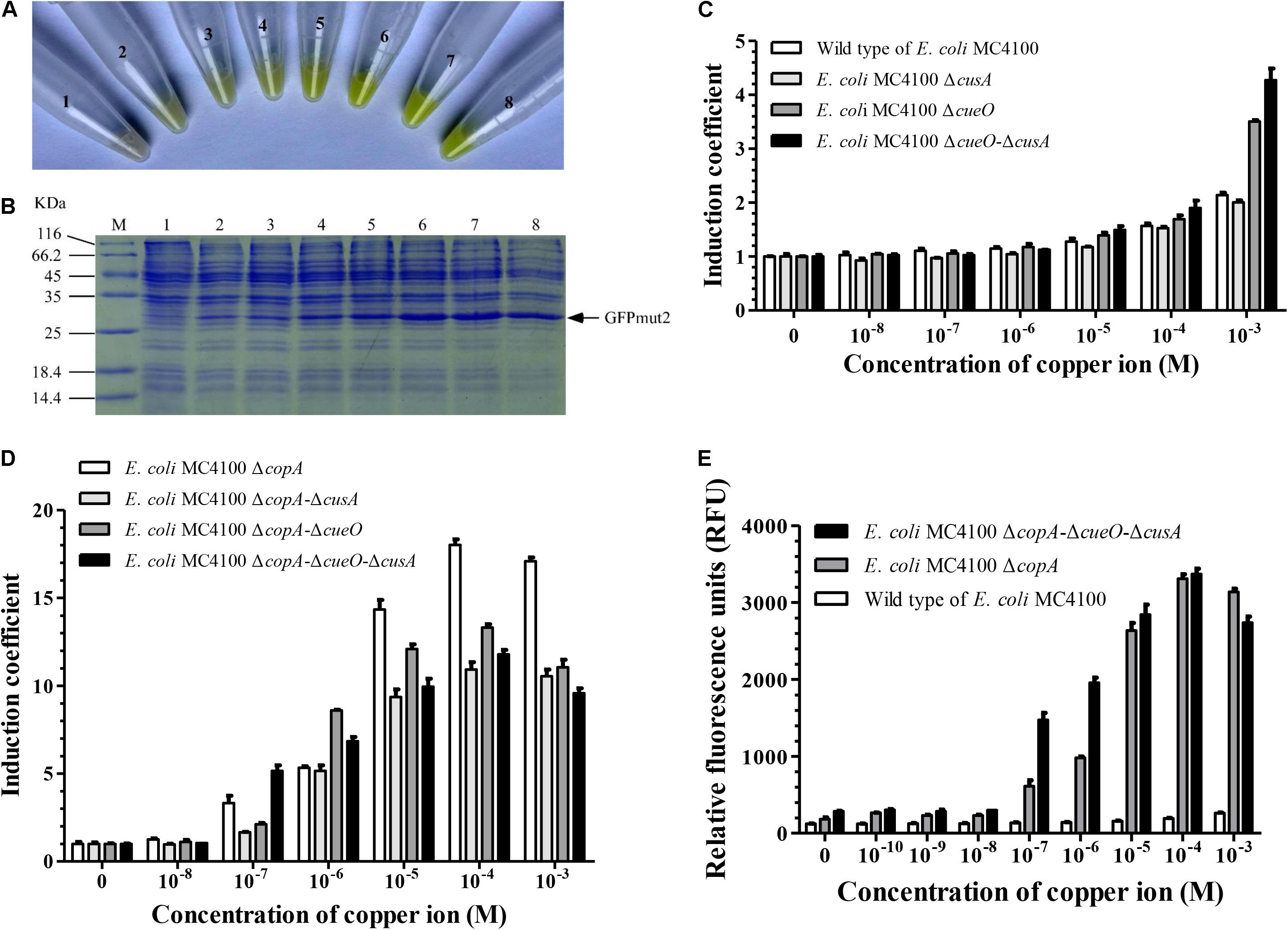
Figure 2. Effects of knockout of genes involved in copper transport in wild-type E. coli MC4100 on copper-sensing capability of bioreporters harboring copAp::gfpmut2 sensing element. (A) The copAp::gfpmut2-pET28a/ΔcusA-ΔcopA-ΔcueO (E. coli WMC-006) bioreporters induced by a series of copper concentration (numbers 2–8: 0, 1.0 × 10–8, 1.0 × 10–8, 1.0 × 10–7, 1.0 × 10–6, 1.0 × 10–5, 1.0 × 10–4, and 1.0 × 10–3 M copper), harvested and resuspended in Tris buffer. Number 1: pET28a/ΔcusA-ΔcopA-ΔcueO as negative control. (B) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (12%) analysis of whole-cell samples described in (A). (C,D) Induction coefficient (IC) of copper-induced bioreporters with copper transport genes in the cop-operon knocked out. (E) Comparison of relative fluorescence intensity from wild-type E. coli MC4100, ΔcopA, and ΔcopA-ΔcueO-ΔcusA mutated strains harboring copAp::gfpmut2 sensing element treated with different concentrations of copper ions. All the experiments producing Figure 2, were repeated at least once.
The effects of the E. coli △copA△cueO△cusA mutant host cell on the response of copAp::gfpmut2 sensing element to copper was compared with copA single-mutant strain. We constructed eight whole-cell bioreporters with different mutant strains and tested their sensitivity to copper ions. As shown in Figures 2C–E, 1 mM copper ions induced weak fluorescence in bioreporter based on wild type of E. coli MC4100, as previously reported (Kang et al., 2018; Wang et al., 2019). The IC values suggested that the effect of copper transport genes on the sensitivity of these whole-cell bioreporters was ranked: △copA > △cueO > △cusA (Figures 2C,D). In the copper concentration range of 10–7–10–6 M, the IC of bioreporters based on E. coli △copA△cueO△cusA was significantly higher than that of E. coli △copA. However, when the concentration of copper was above 1 μM, the IC of bioreporters based on E. coli △copA was significantly higher than E. coli △copA△cueO△cusA (Figures 2C,D). To explain this contradiction, the responses of bioreporters based on E. coli △copA and E. coli △copA△cueO△cusA to copper within 0–1 mM were compared. The result indicated that the relative fluorescence signal derived from E. coli △copA△cueO△cusA was significantly stronger than that of E. coli △copA, while its background fluorescence intensity was 1.56 times higher than that of bioreporter copAp::gfpmut2-pET28a/E. coli △copA (Figure 2E). Similarly, our previous work showed that the fluorescent signal generated by the copper-sensing element in E. coli △copA△cueO was significantly stronger than that of E. coli △copA (Wang et al., 2019). Therefore, the mutant strain E. coli △copA△cueO△cusA was selected as the host strain to construct a new bioreporter by inserting the copAp::gfpmut2 sensing element into its chromosome. This bioreporter was named E. coli WMC-007 and had the potential to reduce background fluorescence signal and accelerate the reaction rate.
The upper LOD is a key parameter limiting the application of whole-cell bioreporters. Before optimizing the kinetics of copper-induced GFPmut2 expression, it is necessary to determine the resistance limit of E. coli WMC-007. As shown in Figure 3A, the growth curves of the E. coli △copA△cueO△cusA and the E. coli WMC-007 were basically consistent with those of the wild-type E. coli MC4100. Cu2+ (100 μM; 6.35 mg/L) inhibited the growth of the E. coli WMC-007 slightly, while 200 μM Cu2+ significantly inhibited the growth of the E. coli WMC-007.
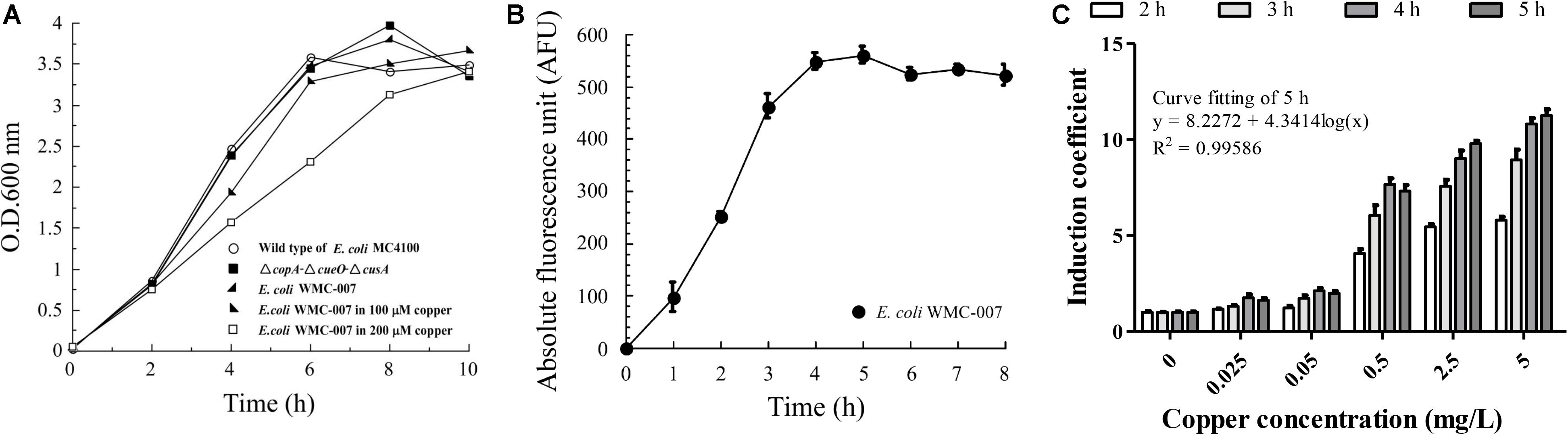
Figure 3. Optimization of incubation time for trace copper detection with bioreporter E. coli WMC-007. (A) Growth curves of the wild-type E. coli MC4100 (open circle), the E. coli ΔcopA-ΔcueO-ΔcusA (closed squares), and the bioreporter E. coli WMC-007 incubated with (closed right oblique triangles and open squares) or without (closed left oblique triangles) different concentrations of copper in LB medium at 37°C for 0–10 h. (B) Induction kinetics of GFPmut2 treated with 100 μM (6.35 mg/L) copper. (C) Effect of induction time on the sensibility and the linear range of the bioreporter E. coli WMC-007. The data were obtained from three independent experiments.
As shown in Figure 3B, under the condition of a fixed Cu2+ concentration, the trend of fluorescence accumulation in E. coli WMC-007 was almost consistent with its growth curve. After incubation for 4 h, GFPmut2 fluorescence intensity and the growth of E. coli WMC-007 entered the stationary phase. Further analysis showed that E. coli WMC-007 was able to produce detectable fluorescence signal after being exposed to copper ions for 2 h (Figure 3C). Moreover, the linear range and the IC of E. coli WMC-007 were significantly enlarged with the extension of incubation time. When induced for 5 h, the curve fitting coefficient R2 was closest to 0.9999, corresponding to the maximum detection sensitivity (Figure 3C). Therefore, 5 h incubation was selected as the best copper ion induction time, and the detection range of E. coli WMC-007 under this condition was 0.025–5 mg/L.
As shown in Figure 4A, 10 common metal salts were used to test the specificity of E. coli WMC-007. Cu2+ was the only metal resulting in a statistically significant increase in the fluorescence signal. Ag+ could induce weak expression of GFPmut2 (the AFU value of Ag+ ≤ 20% of the AFU value of Cu2+) between 0.1 and 5 μM (data not shown), while other heavy metals demonstrated no obvious induction. The specificity of E. coli WMC-007 was consistent with those reported previously (Ivask et al., 2009; Kang et al., 2018; Wang et al., 2019).
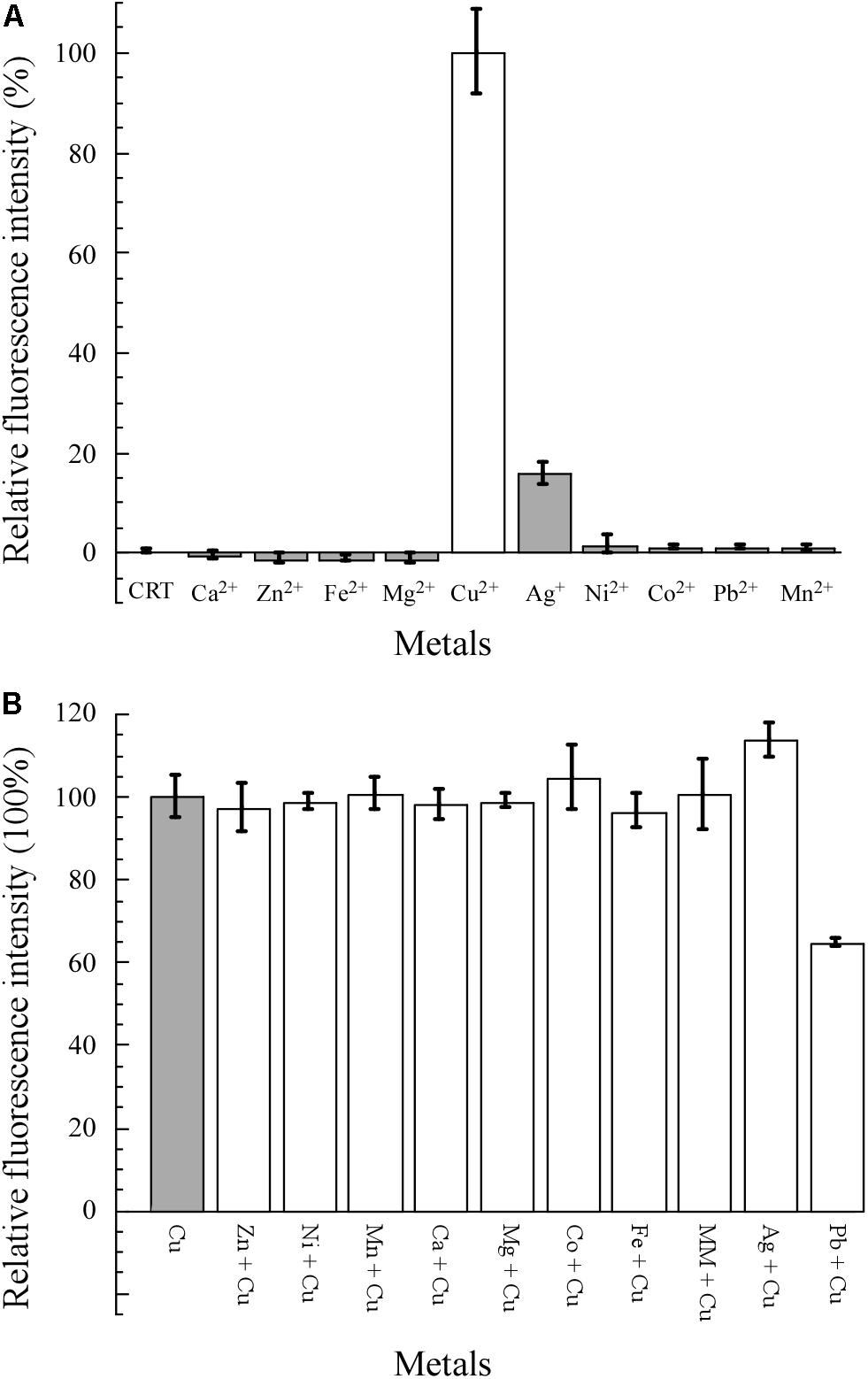
Figure 4. Specificity and selectivity of E. coli WMC-007 in response to different metals. MM, mixed metal, including all the metal ions shown in the figure except for lead and silver. The specificity (A) and selectivity (B) of E. coli WMC-007 for different metal ions (5 μM) were expressed in absolute fluorescence unit (AFU) ratio. The AFU value of Cu2+ was defined as 100% for reference. Data represent mean ± SD of three independent measurements.
The selectivity of E. coli WMC-007 for different metal ions was evaluated by performing pairwise metal assays. The final concentration of metal ions was 5 μM. As shown in Figure 4B, no statistically significant change in green fluorescence intensity was observed when mixing Mg2+, Zn2+, Fe3+, Mn2+, Ca2+, Co2+, or Ni2+ with Cu2+ separately, comparing with the control group (Cu2+). Same was observed when mixing all the metal ions. Interestingly, cross-reactions were detected when mixing Pb2+ and Cu2+, which gave rise to a significant reduction in the fluorescence intensity. By contrast, the green fluorescence was strengthened when mixing Ag+ and Cu2+. However, there was no such obvious cross-reaction in our previously reported bioreporter E. coli pBV-LOCG/ΔcopA/cueO (Wang et al., 2019). It is speculated that the fluorescence signal generated by E. coli pBV-LOCG/ΔcopA/cueO is 10 times as much as that of copper-induced E. coli WMC-007, so that the influence of Pb2+ and Ag+ on the fluorescence signal of pBV-LOCG/ΔcopA/cueO can be ignored. Overall, the constructed E. coli WMC-007 has a high specificity and selectivity for Cu2+.
As shown in Figure 5A, under the excitation wavelength of 481 nm, the E. coli WMC-007 treated with 10 μM Cu2+ could emit bright green fluorescence, which was visualized through a confocal laser scanning microscope with blue light, further indicating a successful bioreporter construction. Fluorescence intensity of the cells also had dose-dependent effect with copper concentration, and the location of the E. coli WMC-007 cells emitting green fluorescence are consistent with the microscope’s FOV under natural light. Moreover, the fluorescence intensity of the E. coli WMC-007 was very uniform under the same copper ion concentration. These results demonstrated the reporter gene harbored by the host strain in the form of chromosomal integration enhancing the genetic stability of sensor element. On the other hand, when without addition of exogenous copper ions, the RFU produced by E. coli WMC-006 cells is about 12 times of the E. coli WMC-007, and the SD of three parallel measurements (n = 3) is also significantly larger than the E. coli WMC-007 (Figure 5B).
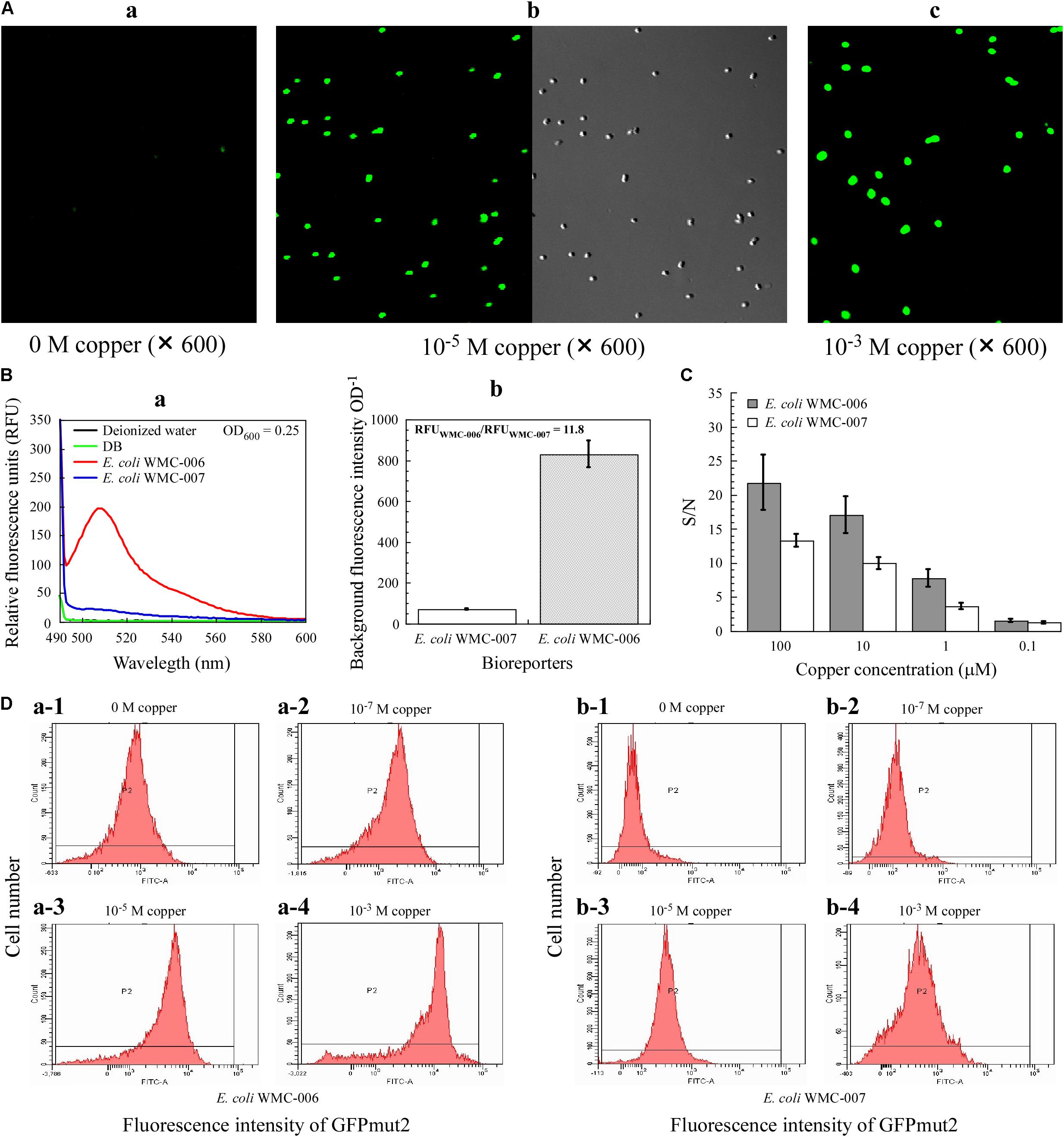
Figure 5. Phenotypic characteristics of E. coli bioreporters. (A) Laser confocal microscopy analysis of fluorescence intensity of the bioreporter E. coli WMC-007. (B) Comparison of background fluorescence produced by E. coli WMC-007 and E. coli WMC-006. (C) The signal-to-noise ratio (S/N) of bioreporters E. coli WMC-007 and E. coli WMC-006 in response to various copper concentrations. (D) Flow cytometry analysis of fluorescence signal homogeneity of the bioreporters. (a,b) Different types of peaks showing the distribution of log-transformed fluorescence intensity per cell upon exposure to different copper concentrations. The y-axis represents the number of fluorescent cells detected. The results presented in (A–D) were obtained from two and three independent experiments, respectively.
Furthermore, the S/N ratio for the detection of copper was compared between E. coli WMC-006 and E. coli WMC-007. As shown in Figure 5C, the SD value in E. coli WMC-007 experiment was significantly smaller than that in E. coli WMC-006 experiment. Nevertheless, the S/N value of E. coli WMC-007 was significantly lower than that of E. coli WMC-006 in the range of 1–100 μM copper. In addition, the difference in the S/N value became larger as the concentration of Cu2+ increased. Notably, when the concentration of Cu2+ dropped to 0.1 μM, there was no significant difference in the S/N value between the two bioreporters, while the SD in E. coli WMC-006 experiment was larger than that in the E. coli WMC-007 experiment.
As shown in Figure 5D, the fluorescence intensity emitted by copper-exposed E. coli WMC-007 cells (5 h) exhibited a narrow unimodal distribution without long tails. By contrast, the fluorescence from E. coli WMC-006 cells followed a left-skewed unimodal distribution, particularly at 10–3 M copper. In addition, flow cytometry results showed that the fluorescence intensity of bioreporters increased with copper ion concentration. Moreover, the background fluorescence value of E. coli WMC-007 was significantly lower than that of E. coli WMC-006 (Figure 5D). These results demonstrated that the sensing element integrated into the chromosome is more robust than that integrated into plasmids.
As shown in Figures 6A,B, the fluorescence intensity and expression level of GFPmut2 in E. coli WMC-007 cells were positively related to the free Cu2+ concentration. It is noted that only properly folded GFPmut2 can emit fluorescence. As a result, the linear relationship of GFPmut2 protein expression level after incubated with concentration gradient copper ions was not as good as the fluorescence intensity.
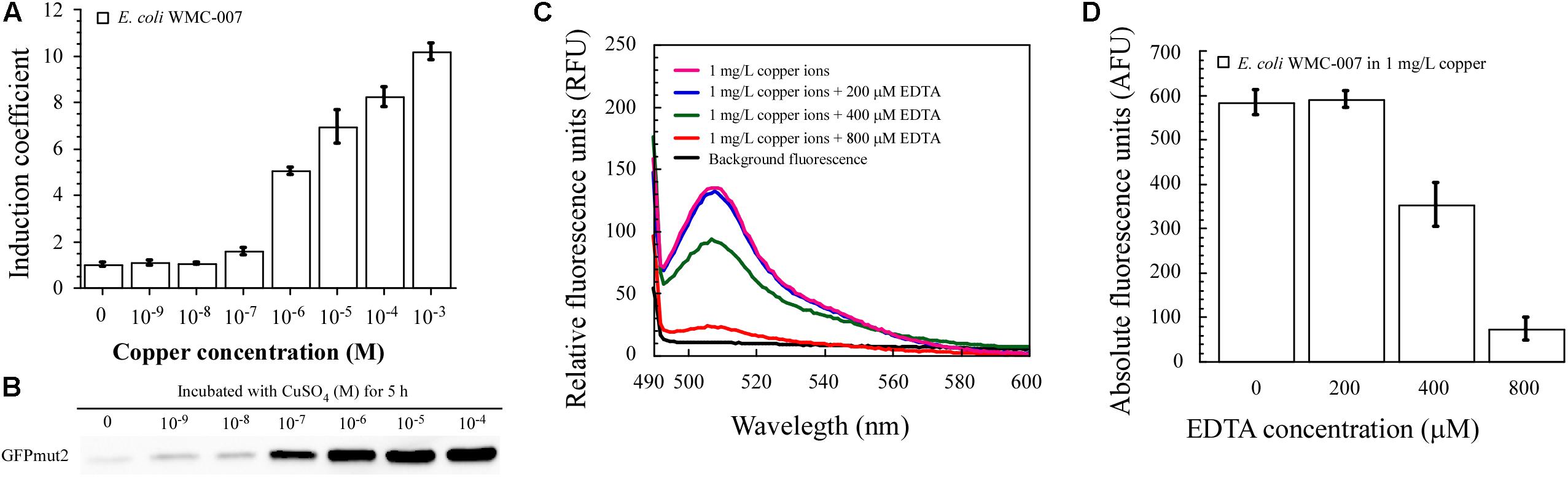
Figure 6. Dose-dependent induction of green fluorescence with free copper ion. (A) Induction coefficient (IC) of constructed bioreporter E. coli WMC-007 in response to 5-h incubation with Cu2+. (B) Western blot of GFPmut2 expressed by copper-induced E. coli WMC-007 after incubation for 5 h. (C) Fluorescence quenching effect of excess EDTA under the condition of a fixed copper concentration (1 mg/L). (D) Effect of excess EDTA on bioavailable Cu2+ content in aqueous solutions. At least two independent experiments were carried out for figure.
In conclusion, these results demonstrated that E. coli WMC-007 could accurately quantify the content of free copper ions in aqueous solution under laboratory conditions. However, copper can exist in many forms in water environment (Li A. et al., 2018). Our research findings suggest that the relationship between the total copper and the bioavailable copper in the aqueous solution can be dramatically changed by adding the chelator EDTA. As shown in Figures 6C,D, the fluorescence emitted by E. coli WMC-007 was not influenced by EDTA in low concentrations (0–200 μM). When the EDTA concentration was further increased, a large quantity of free Cu2+ were chelated in the medium. Consequently, the sensing element could not respond to EDTA-bound Cu since the chelated Cu could neither enter nor being available to E. coli cells. This experiment reveals that complex-bound copper affects the bioavailability of copper in water environment.
In optimized assay conditions, the fluorescence intensity of E. coli WMC-007 exhibited a linear relation with the log10-transformed copper concentration at 0.025–5 mg/L (0.39–78.68 μM, R2 = 0.99786), and the LOD of Cu2+ was 0.0157 mg/L (0.245 μM), as shown in Figure 7. In comparison, the linear range in E. coli WMC-006 experiment was 0.005–2 mg/L (0.078–31.53 μM), and the LOD of Cu2+ was 0.0049 mg/L (0.0766 μM) (data not shown). These results show that the upper limit for copper detection with E. coli WMC-007 is about 2.5 times higher than that with E. coli WMC-006, while its lower limit is 5 times higher than that with E. coli WMC-006.
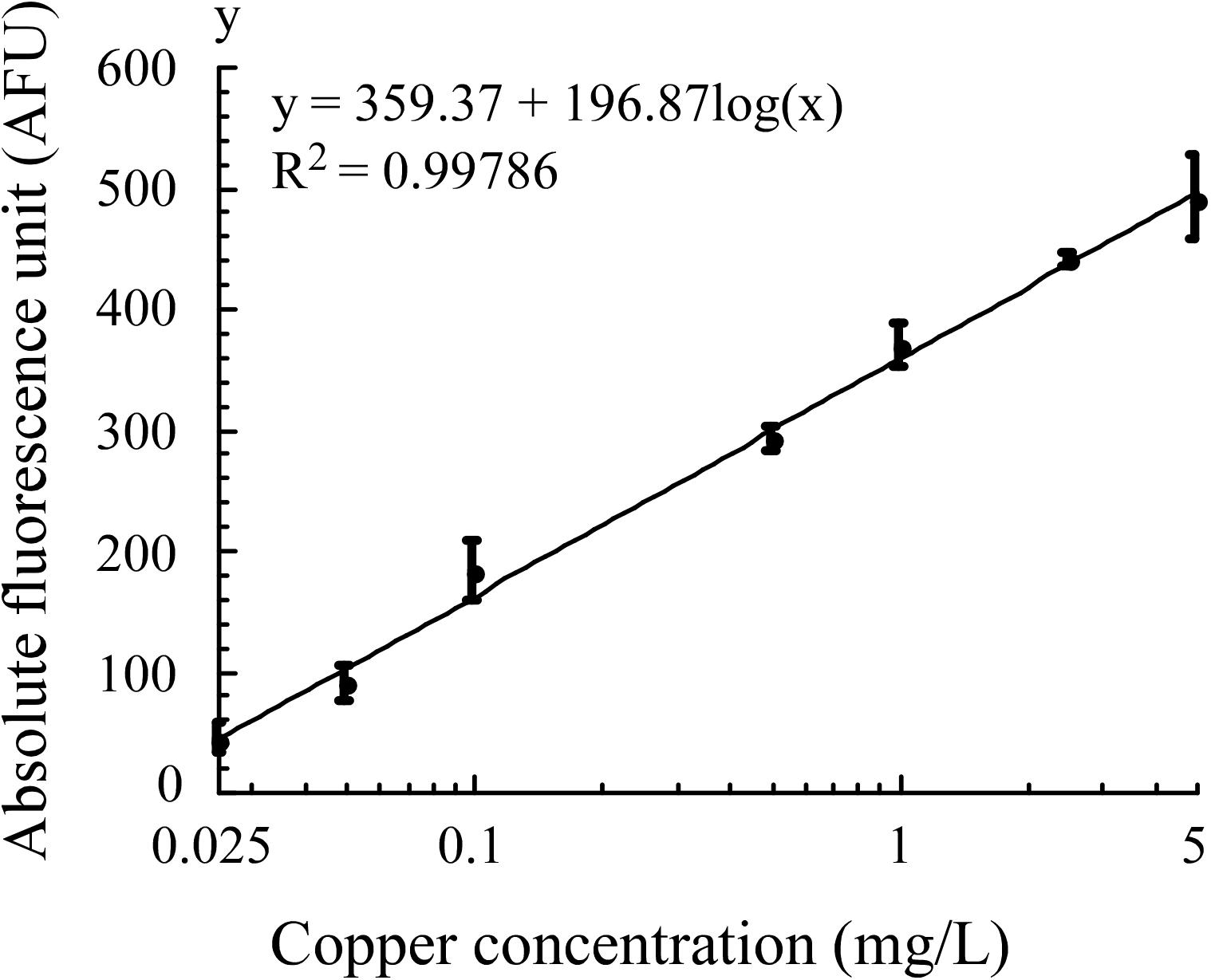
Figure 7. Fitted curve of absolute fluorescence unit produced by E. coli WMC-007 in response to copper in Luria–Bertani (LB) medium. Curve fitting was performed using KaleidaGraph software, with data obtained from at least three independent experiments.
To demonstrate the utility of E. coli WMC-007, actual water environment samples with a known concentration of contaminant were examined. Heavy metals are commonly found in extremely acidic environments, like mine drainages and acidizing tailings (pH < 3). To reduce the influence of extreme environmental conditions on the performance of E. coli WMC-007, pH buffer MOPS was added to LB medium at a final concentration of 40 mM (Riether et al., 2001; Hansen et al., 2019). Comparing with the copper concentration in aqueous solution samples within a pH range from 0.87 to 12.84 measured by AAS method, the copper recovery rate measured by E. coli WMC-007 turned out to be 95.88–113.40%, as listed in Table 2. Sample 2 (pH = −0.30) and sample 6 (pH = 14.70) had the highest and the lowest copper recovery, respectively.
In this study, to reduce the detection threshold, three major copper efflux transporters were knocked out from the wild-type E. coli MC4100, so that trace copper ions could accumulate in the cell. The copAp::gfpmut2 sensing element was inserted into the chromosome of E. coli △copA△cueO△cusA by knock-in method, producing a new bioreporter superior in a few aspects. To begin with, the gene knockout technique increased the sensitivity of wild-type E. coli MC4100 to copper ions by 64 times (Table 1), which improved the copper detection accuracy at low concentrations (nmol–μmol); second, there is no antibiotic marker gene in the single-copy sensor element, which reduces the background fluorescence signal under non-induced conditions (Figures 5A,B,D); Finally, the sensor element can be stably inherited with bacterial reproduction, which enhanced the robustness and reproducibility of the detection of copper ions (Figures 5A,C,D). Our results collectively demonstrated that the bioreporter E. coli WMC-007 has potential applications in determining Cu2+ in drinking water, irrigation water, and industrial wastewater according to the national standards of water quality in China (Figure 7 and Table 3).
Green fluorescent protein (GFP) is a generally fluorescent protein and has a variety of advantages for applications. It is non-toxic, stable, and convenient for detection. Furthermore, its fusion expression is easy to achieve and can be monitored using flow cytometry and fluorescence microscopy (Stiner and Halverson, 2002; van der Meer et al., 2004; Chalfie, 2009; Roda, 2010). Therefore, whole-cell bioreporters based on GFP have been applied to detect a number of environmental toxicants, such as genotoxic compounds (Kostrzynska et al., 2002), toluene and related compounds (Li et al., 2008), endotoxin (Goh et al., 2002), and heavy metals (Liao et al., 2006; Cerminati et al., 2015). However, the wild-type GFP has weak fluorescence. If lux or luc reporter genes are replaced by gfp reporter gene, the sensitivity of bioreporters will be reduced even in the same conditions (Hakkila et al., 2002; Stocker et al., 2003).
In this study, we used gfpmut2 (GFPmut2, S65A, V68L, S72A) reporter gene for detecting copper ions because the fluorescence signal produced by GFPmut2 was 100 times higher than that of the wild-type GFP. At high expression levels, GFPmut2 produces bright green fluorescence when stimulated by blue light (481 nm) alone. Therefore, it is possible to lower of the detection threshold of bioreporters, compared with GFP. This is confirmed by our study, demonstrating that E. coli WMC-006 and E. coli WMC-007 cells displayed yellow-green color after incubation with trace copper for 1–5 h (Figure 2A).
However, there are some drawbacks, including relatively weak fluorescence intensity, large S/N ratio, and higher LOD of copper. This is attributed to the single-copy sensor element in the chromosome. With the same incubation time, the amount of GFP produced is significantly less than that in E. coli WMC-006. A previous study showed that it took about 1–2 h for the cells to complete posttranslational fluorophore formation once GFP was formed (Stiner and Halverson, 2002). Therefore, we simultaneously added the standard copper ion solutions to the medium while inoculating. This method is not only convenient and time saving but also facilitates the accumulation of fluorescence signals. Moreover, the induction of GFPmut2 expression with copper ions was time and dose dependent, which indicated that the IC of bioreporters was positively correlated with the expression level of GFPmut2, in addition to the background fluorescence intensity (Figure 3C).
In conclusion, we speculate that increasing the copy number of the sensing element in the chromosome could effectively increase the S/N value of bioreporters and lower the detection threshold, under the condition that the background fluorescence signal of E. coli WMC-007 and the metabolic burden to host cells are not significantly increased, so that LOD of bioreporters can cover the copper detection limit in any circumstances according to Chinese national standards for water quality (as shown in Table 3).
Water environmental samples, especially wastewater effluents, are complex mixtures. Some components, such as EDTA and other “EDTA-like” substances, are known to bind metal ions, which may influence metal bioavailability in an unpredictable manner (Knepper et al., 2005; Peters et al., 2014; Constantino et al., 2017). Recent data from 162 wastewater treatment effluents in the United Kingdom reveal the median EDTA concentration of 128 mg/L (0.44 mM), which is 5.3 and 22.9 times greater than those of dissolved zinc and copper, respectively (Constantino et al., 2017). We also confirmed that EDTA-bound copper affects the bioavailability of copper in aqueous solution (Figures 6C,D). Moreover, Riether et al. (2001) reported that the luminescence of bioreporter MG1655 (copAp::lux) was significantly inhibited, when the EDTA/Cu2+ molar ratio exceeded 1. However, when the Na2S/Cu2+ molar ratio was above 1.25, the IC of MG1655 (copAp::lux) increased significantly.
In addition, Zhao C. et al. (2018) conclude that dissolved organic matter in urban stormwater runoff has a strong binding affinity with Cu2+, which may further lead to potentially significant influence on migration, transformation, and bioavailability of copper. Hu et al. (2017) also show that the changing composition of impoundment-derived dissolved organic matter can decrease the bioavailability and toxicity of Cu in Hongze Lake in China. Effluent organic matter from industrial and sewage wastewaters can be separated into hydrophobic, transphilic, and hydrophilic fractions depending on its polarity. Yoo et al. (2016) suggest that the binding and toxicity of copper are largely dependent on the polarity of effluent organic matter. Käkinen et al. (2016) also proved the solubility-driven toxicity of CuO nanoparticles to Caco2 and E. coli.
It is worth noting that the organic matter in LB medium may form complex with copper ions. We speculate that copper is easily released from the complex under acidic conditions, so that the concentration of free copper ions increases. When the medium becomes alkaline, copper ions are easily precipitated and difficult for bacteria to uptake, thereby reducing its bioavailability (as show in Table 2). These results demonstrate the potential application of the constructed E. coli WMC-007 in determining the bioavailability of copper ions with high sensitivity and accuracy in water environment samples. Most importantly, these results suggest that the results quantified by physicochemical analysis methods reflect the total copper content, making it difficult to objectively evaluate the toxicity to bacteria and human.
Generally, the composition of environmental pollutants is very complex. Nutrients, toxicants, and particles in the sample matrix may stimulate or quench the fluorescence of the bioreporters (Ivask et al., 2004). Therefore, in a follow-up experiment, we will construct a constitutively fluorescence control strain (Liao et al., 2006; Ivask et al., 2009) to cancel out the effects of the above factors on bacterial growth and the expression of GFPmut2. Like E. coli WMC-007, this constitutively fluorescence control strain is derived from E. coli △copA△cueO△cusA host cell with the expression of GFPmut2 or other fluorescent proteins. However, it lacks the metal recognizing protein and its corresponding promoter (Ivask et al., 2004). Its reporter gene is controlled by constitutive promoters, and if the promoter is a strong promoter will be perfect. Such as lacp::gfpmut2/MC4100 △lacI△lacZYA△copA△cueO△cusA whole-cell bioreporter.
Despite the fact that many whole-cell bioreporters have been constructed for measuring heavy metals, they have been rarely applied to monitor water environmental samples (Hynninen et al., 2010). Another obstacle to apply this technology is that even in highly contaminated samples, the amount of bioavailable metal in complex matrices mostly ranges between 0.1 and 50% (Magrisso et al., 2008). In our previous work, the bioreporter E. coli pBV-LOCG/ΔcopA/cueO was applied to detect copper in 117 water environmental samples. The average accuracy of the whole cell for copper quantification was 93.8 ± 30.1% compared to ICP-MS, despite the fact that the samples were acidified to pH of 2 using nitric acid (Wang et al., 2019). Therefore, these results prove that the whole-cell bioreporter is a reliable tool for high-throughput screening of copper contamination in environmental samples, although further research is needed to verify whether the bioavailable copper content in environmental samples can be truly reflected. In addition, the metal-responding element CueR is not specific for Cu, as it can be induced by Ag in relatively high concentrations (Ivask et al., 2009), which is supported by our finding in this study (Figure 4).
To solve these problems, we make several suggestions to improve E. coli WMC-007. (1) Like the Cd-specific mutants of mercury-sensing regulatory protein MerR constructed by Hakkila et al. (2011), oligonucleotideite-directed mutagenesis (Hakkila et al., 2011; Shemer et al., 2017) can be applied on the key metal-binding regions of Cu/Ag-sensing transcriptional regulatory protein CueR (mutated CueR, CueR-m). Besides, a double-promoter model will be used in the construction of the sensing element copAp::cueR-m-copAp::gfpmut2. This strategy may eliminate the response of the sensing element to Ag and further decrease the background fluorescence signal, thereby improving the specificity and sensitivity of the bioreporter. (2) Because the LOD and S/N ratio of E. coli WMC-007 are not as good as the E. coli WMC-006, one to two copies of copAp::cueR-m-copAp::gfpmut2 sensing elements can be inserted into the position of cueO and/or cusA in the chromosome of E. coli WMC-007 to further improve the detection thresholds. (3) As mentioned above, a cell viability internal control bioreporter that constitutively expresses GFPmut2 can be constructed (Roda et al., 2011). (4) An optimal medium should be selected for ecotoxicological and microbiological tests (Kakinen et al., 2011). For example, our previous work demonstrated that the limit of whole-cell copper detection in M9S medium [M9 minimal medium supplemented with 0.2% glucose, 5 μg/ml thiamin, and the 20 amino acids (10 μg/ml each)] was significantly lower than that in LB medium (Wang et al., 2019). (5) The bioassay may be performed in 96- or 384-well black polystyrene microtiter plates for high-throughput testing. (6) The whole-cell bioreporter with a physicochemical transducer (for example, a digital microfluidic diluter chip) and a detector element can be combined to construct microbial biosensors for the application of in situ and real-time detection or monitor of copper in water environments (Shemer et al., 2017). (7) To be recognized by third-party evaluation agencies, it is necessary to standardize the testing process and establish a classification system and grading standard for bioavailable copper in water environments with the combination of AAS or ICP-MS methods.
The datasets generated for this study are available on request to the corresponding author.
GT and JXL conceived and supervised the study. GT, JXL, YP, DL, and WWu designed the experiments. YP, XJR, FL, XYR, YG, JW, JZ, JHL, and XH performed the experiments. YP, XJR, and FJ analyzed and interpreted the data. YP, GT, JHL, WWa, and HZ wrote and revised the manuscript. GT reviewed the manuscript.
This study was supported by the Chinese National Natural Science Foundation Grants (81671124 and 31870775), the National High Technology Research and Development Program of China (863 Program) (2014AA06A514), and Key Discipline of Zhejiang Province in Medical Technology (first class, category A).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Dr. Huangen Ding (Louisiana State University, United States) for kindly providing the suggestive advices for this manuscript. We would also like to thank Dr. Lirui Liu (The University of Hong Kong) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03031/full#supplementary-material
Bittner, L. M., Kraus, A., Schakermann, S., and Narberhaus, F. (2017). The copper efflux regulator CueR is subject to ATP-dependent proteolysis in Escherichia coli. Front. Mol. Biosci. 4:9. doi: 10.3389/fmolb.2017.00009
Cerminati, S., Soncini, F. C., and Checa, S. K. (2015). A sensitive whole-cell biosensor for the simultaneous detection of a broad-spectrum of toxic heavy metal ions. Chem. Commun. 51, 5917–5920. doi: 10.1039/c5cc00981b
Chalfie, M. (2009). GFP: lighting up life (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 48, 5603–5611. doi: 10.1002/anie.200902040
Chillappagari, S., Seubert, A., Trip, H., Kuipers, O. P., Marahiel, M. A., and Miethke, M. (2010). Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 192, 2512–2524. doi: 10.1128/JB.00058-10
Constantino, C., Comber, S. D., and Scrimshaw, M. D. (2017). The effect of wastewater effluent derived ligands on copper and zinc complexation. Environ. Sci. Pollut. Res. Int. 24, 8363–8374. doi: 10.1007/s11356-016-8332-3
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Deaver, E., and Rodgers, J. H. (1996). Measuring bioavailable copper using anodic stripping voltammetry. Environ. Toxicol. Chem. 15, 1925–1930. doi: 10.1002/etc.5620151110
Din, G., Hasan, F., Conway, M., Denney, B., Ripp, S., and Shah, A. A. (2019). Engineering a bioluminescent bioreporter from an environmentally sourced mercury-resistant Enterobacter cloacae strain for the detection of bioavailable mercury. J. Appl. Microbiol. 23:14399. doi: 10.1111/jam.14399
Emerit, J., Edeas, M., and Bricaire, F. (2004). Neurodegenerative diseases and oxidative stress. Biomed. Pharmacother. 58, 39–46.
Fan, Y., Makar, M., Wang, M. X., and Ai, H. W. (2017). Monitoring thioredoxin redox with a genetically encoded red fluorescent biosensor. Nat. Chem. Biol. 13, 1045–1052. doi: 10.1038/nchembio.2417
Freed, E. F., Prieto, J. L., McCann, K. L., McStay, B., and Baserga, S. J. (2012). NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet. 8:e1002892. doi: 10.1371/journal.pgen.1002892
Fu, L., Lu, X., Niu, K., Tan, J., and Chen, J. (2019). Bioaccumulation and human health implications of essential and toxic metals in freshwater products of Northeast China. Sci. Total Environ. 673, 768–776. doi: 10.1016/j.scitotenv.2019.04.099
Fung, D. K., Lau, W. Y., Chan, W. T., and Yan, A. (2013). Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron-sulfur cluster enzymes and biogenesis. J. Bacteriol. 195, 4556–4568. doi: 10.1128/JB.00543-13
Goh, Y. Y., Ho, B., and Ding, J. L. (2002). A novel fluorescent protein-based biosensor for gram-negative bacteria. Appl. Environ. Microbiol. 68, 6343–6352. doi: 10.1128/aem.68.12.6343-6352.2002
Grass, G., and Rensing, C. (2001). Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183, 2145–2147. doi: 10.1128/JB.183.6.2145-2147.2001
Hakkila, K., Green, T., Leskinen, P., Ivask, A., Marks, R., and Virta, M. (2004). Detection of bioavailable heavy metals in EILATox-Oregon samples using whole-cell luminescent bacterial sensors in suspension or immobilized onto fibre-optic tips. J. Appl. Toxicol. 24, 333–342. doi: 10.1002/jat.1020
Hakkila, K., Maksimow, M., Karp, M., and Virta, M. (2002). Reporter genes lucFF, luxCDABE, gfp, and dsred have different characteristics in whole-cell bacterial sensors. Anal. Biochem. 301, 235–242. doi: 10.1006/abio.2001.5517
Hakkila, K. M., Nikander, P. A., Junttila, S. M., Lamminmaki, U. J., and Virta, M. P. (2011). Cd-specific mutants of mercury-sensing regulatory protein MerR, generated by directed evolution. Appl. Environ. Microbiol. 77, 6215–6224. doi: 10.1128/AEM.00662-11
Hansen, M. G., Marcussen, H., Holm, P. E., and Brandt, K. K. (2019). Evaluation of pH-dependent metal speciation artifacts in whole-cell bioreporter analysis. J. Environ. Qual. 48, 199–204. doi: 10.2134/jeq2018.10.0352
Hu, B., Wang, P., Wang, C., Qian, J., Hou, J., Cui, X., et al. (2017). The effect of anthropogenic impoundment on dissolved organic matter characteristics and copper binding affinity: insights from fluorescence spectroscopy. Chemosphere 188, 424–433. doi: 10.1016/j.chemosphere.2017.09.023
Huang, L., Pu, X., Pan, J. F., and Wang, B. (2013). Heavy metal pollution status in surface sediments of Swan Lake lagoon and Rongcheng Bay in the northern Yellow Sea. Chemosphere 93, 1957–1964. doi: 10.1016/j.chemosphere.2013.06.080
Hynninen, A., Tonismann, K., and Virta, M. (2010). Improving the sensitivity of bacterial bioreporters for heavy metals. Bioeng. Bugs 1, 132–138. doi: 10.4161/bbug.1.2.10902
Imanaka, T., Tsunekawa, H., and Aiba, S. (1980). Phenotypic stability of trp operon recombinant plasmids in Escherichia coli. J. Gen. Microbiol. 118, 253–261. doi: 10.1099/00221287-118-1-253
Impellitteri, C. A., Saxe, J. K., Cochran, M., Janssen, G. M., and Allen, H. E. (2003). Predicting the bioavailability of copper and zinc in soils: modeling the partitioning of potentially bioavailable copper and zinc from soil solid to soil solution. Environ. Toxicol. Chem. 22, 1380–1386. doi: 10.1002/etc.5620220626
Ivask, A., Francois, M., Kahru, A., Dubourguier, H. C., Virta, M., and Douay, F. (2004). Recombinant luminescent bacterial sensors for the measurement of bioavailability of cadmium and lead in soils polluted by metal smelters. Chemosphere 55, 147–156. doi: 10.1016/j.chemosphere.2003.10.064
Ivask, A., Rolova, T., and Kahru, A. (2009). A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotechnol. 9:41. doi: 10.1186/1472-6750-9-41
Jiang, B., Li, G., Xing, Y., Zhang, D., Jia, J., Cui, Z., et al. (2017). A whole-cell bioreporter assay for quantitative genotoxicity evaluation of environmental samples. Chemosphere 184, 384–392. doi: 10.1016/j.chemosphere.2017.05.159
Kakinen, A., Bondarenko, O., Ivask, A., and Kahru, A. (2011). The effect of composition of different ecotoxicological test media on free and bioavailable copper from CuSO4 and CuO nanoparticles: comparative evidence from a Cu-selective electrode and a Cu-biosensor. Sensors 11, 10502–10521. doi: 10.3390/s111110502
Käkinen, A., Kahru, A., Nurmsoo, H., Kubo, A. L., and Bondarenko, O. M. (2016). Solubility-driven toxicity of CuO nanoparticles to Caco2 cells and Escherichia coli: effect of sonication energy and test environment. Toxicol. In Vitro 36, 172–179. doi: 10.1016/j.tiv.2016.08.004
Kang, Y., Lee, W., Kim, S., Jang, G., Kim, B. G., and Yoon, Y. (2018). Enhancing the copper-sensing capability of Escherichia coli-based whole-cell bioreporters by genetic engineering. Appl. Microbiol. Biotechnol. 102, 1513–1521. doi: 10.1007/s00253-017-8677-7
Karlsson, H. L., Cronholm, P., Gustafsson, J., and Moller, L. (2008). Copper oxide nanoparticles are highly toxic: a comparison between metal oxide nanoparticles and carbon nanotubes. Chem. Res. Toxicol. 21, 1726–1732. doi: 10.1021/tx800064j
Knepper, T. P., Werner, A., and Bogenschutz, G. (2005). Determination of synthetic chelating agents in surface and waste water by ion chromatography-mass spectrometry. J. Chromatogr. A 2, 240–246. doi: 10.1016/j.chroma.2005.06.045
Kostrzynska, M., Leung, K. T., Lee, H., and Trevors, J. T. (2002). Green fluorescent protein-based biosensor for detecting SOS-inducing activity of genotoxic compounds. J. Microbiol. Methods 48, 43–51. doi: 10.1016/s0167-7012(01)00335-9
Kramer, M. G. (2016). Determination of plasmid segregational stability in a growing bacterial population. Methods Mol. Biol. 1409, 125–133. doi: 10.1007/978-1-4939-3515-4_11
Leedjarv, A., Ivask, A., and Virta, M. (2008). Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J. Bacteriol. 190, 2680–2689. doi: 10.1128/jb.01494-07
Li, A., Zhang, Y., Zhou, B., Xin, K., Gu, Y., Xu, W., et al. (2018). Ultrafiltration for the determination of cu complexed with dissolved organic matters of different molecular weight from a Eutrophic River, China. Bull. Environ. Contam. Toxicol. 101, 131–136. doi: 10.1007/s00128-018-2351-3
Li, F., Mao, L., Jia, Y., Gu, Z., Shi, W., Chen, L., et al. (2018). Distribution and risk assessment of trace metals in sediments from Yangtze River estuary and Hangzhou Bay, China. Environ. Sci. Pollut. Res. Int. 25, 855–866. doi: 10.1007/s11356-017-0425-0
Li, P. S., Peng, Z. W., Su, J., and Tao, H. C. (2014). Construction and optimization of a Pseudomonas putida whole-cell bioreporter for detection of bioavailable copper. Biotechnol. Lett. 36, 761–766. doi: 10.1007/s10529-013-1420-2
Li, Y. F., Li, F. Y., Ho, C. L., and Liao, V. H. (2008). Construction and comparison of fluorescence and bioluminescence bacterial biosensors for the detection of bioavailable toluene and related compounds. Environ. Pollut. 152, 123–129. doi: 10.1016/j.envpol.2007.05.002
Liao, V. H., Chien, M. T., Tseng, Y. Y., and Ou, K. L. (2006). Assessment of heavy metal bioavailability in contaminated sediments and soils using green fluorescent protein-based bacterial biosensors. Environ. Pollut. 142, 17–23. doi: 10.1016/j.envpol.2005.09.021
Link, A. J., Phillips, D., and Church, G. M. (1997). Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179, 6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997
Macomber, L., and Imlay, J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349. doi: 10.1073/pnas.0812808106
Magrisso, S., Erel, Y., and Belkin, S. (2008). Microbial reporters of metal bioavailability. Microb. Biotechnol. 1, 320–330. doi: 10.1111/j.1751-7915.2008.00022.x
Martin-Betancor, K., Rodea-Palomares, I., Munoz-Martin, M. A., Leganes, F., and Fernandez-Pinas, F. (2015). Construction of a self-luminescent cyanobacterial bioreporter that detects a broad range of bioavailable heavy metals in aquatic environments. Front. Microbiol. 6:186. doi: 10.3389/fmicb.2015.00186
Muller, T., Feichtinger, H., Berger, H., and Muller, W. (1996). Endemic tyrolean infantile cirrhosis: an ecogenetic disorder. Lancet 347, 877–880. doi: 10.1016/s0140-6736(96)91351-3
Munson, G. P., Lam, D. L., Outten, F. W., and O’Halloran, T. V. (2000). Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182, 5864–5871. doi: 10.1128/jb.182.20.5864-5871.2000
Outten, F. W., Huffman, D. L., Hale, J. A., and O’Halloran, T. V. (2001). The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276, 30670–30677. doi: 10.1074/jbc.m104122200
Outten, F. W., Outten, C. E., Hale, J., and O’Halloran, T. V. (2000). Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J. Biol. Chem. 275, 31024–31029. doi: 10.1074/jbc.M006508200
Peters, A., Merrington, G., Leverett, D., Ellor, B., Lofts, S., and Gravell, A. (2014). The effect of advanced treatment of sewage effluents on metal speciation and (bio)availability. Bull. Environ. Contam. Toxicol. 92, 248–252. doi: 10.1007/s00128-013-1143-z
Philips, S. J., Canalizo-Hernandez, M., Yildirim, I., Schatz, G. C., Mondragon, A., and O’Halloran, T. V. (2015). TRANSCRIPTION. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science 349, 877–881. doi: 10.1126/science.aaa9809
Rensing, C., Fan, B., Sharma, R., Mitra, B., and Rosen, B. P. (2000). CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 97, 652–656. doi: 10.1073/pnas.97.2.652
Riether, K. B., Dollard, M. A., and Billard, P. (2001). Assessment of heavy metal bioavailability using Escherichia coli zntAp::lux and copAp::lux-based biosensors. Appl. Microbiol. Biotechnol. 57, 712–716. doi: 10.1007/s00253-001-0852-0
Roda, A. (2010). Discovery and development of the green fluorescent protein, GFP: the 2008 Nobel Prize. Anal. Bioanal. Chem. 396, 1619–1622. doi: 10.1007/s00216-010-3452-y
Roda, A., Roda, B., Cevenini, L., Michelini, E., Mezzanotte, L., Reschiglian, P., et al. (2011). Analytical strategies for improving the robustness and reproducibility of bioluminescent microbial bioreporters. Anal. Bioanal. Chem. 401, 201–211. doi: 10.1007/s00216-011-5091-3
Shemer, B., Koshet, O., Yagur-Kroll, S., and Belkin, S. (2017). Microbial bioreporters of trace explosives. Curr. Opin. Biotechnol. 45, 113–119. doi: 10.1016/j.copbio.2017.03.003
Stiner, L., and Halverson, L. J. (2002). Development and characterization of a green fluorescent protein-based bacterial biosensor for bioavailable toluene and related compounds. Appl. Environ. Microbiol. 68, 1962–1971. doi: 10.1128/aem.68.4.1962-1971.2002
Stocker, J., Balluch, D., Gsell, M., Harms, H., Feliciano, J., Daunert, S., et al. (2003). Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 37, 4743–4750. doi: 10.1021/es034258b
Sukhija, K., Pyne, M., Ali, S., Orr, V., Abedi, D., Moo-Young, M., et al. (2012). Developing an extended genomic engineering approach based on recombineering to knock-in heterologous genes to Escherichia coli genome. Mol. Biotechnol. 51, 109–118. doi: 10.1007/s12033-011-9442-2
Tan, G., Yang, J., Li, T., Zhao, J., Sun, S., Li, X., et al. (2017). Anaerobic copper toxicity and iron-sulfur cluster biogenesis in Escherichia coli. Appl. Environ. Microbiol. 83:e00867-17.
Tang, W., Zhao, Y., Wang, C., Shan, B., and Cui, J. (2013). Heavy metal contamination of overlying waters and bed sediments of Haihe Basin in China. Ecotoxicol. Environ. Saf. 98, 317–323. doi: 10.1016/j.ecoenv.2013.09.038
Thongsaw, A., Udnan, Y., Ross, G. M., and Chaiyasith, W. C. (2019). Speciation of mercury in water and biological samples by eco-friendly ultrasound-assisted deep eutectic solvent based on liquid phase microextraction with electrothermal atomic absorption spectrometry. Talanta 197, 310–318. doi: 10.1016/j.talanta.2019.01.018
Tom-Petersen, A., Hosbond, C., and Nybroe, O. (2001). Identification of copper-induced genes in Pseudomonas fluorescens and use of a reporter strain to monitor bioavailable copper in soil. FEMS Microbiol. Ecol. 38, 59–67. doi: 10.1111/j.1574-6941.2001.tb00882.x
van der Meer, J. R., Tropel, D., and Jaspers, M. (2004). Illuminating the detection chain of bacterial bioreporters. Environ. Microbiol. 6, 1005–1020. doi: 10.1111/j.1462-2920.2004.00655.x
Wang, W., Jiang, F., Wu, F., Li, J., Ge, R., Tan, G., et al. (2019). Biodetection and bioremediation of copper ions in environmental water samples using a temperature-controlled, dual-functional Escherichia coli cell. Appl. Microbiol. Biotechnol. 103, 6797–6807. doi: 10.1007/s00253-019-09984-9
White, A. R., Du, T., Laughton, K. M., Volitakis, I., Sharples, R. A., Xilinas, M. E., et al. (2006). Degradation of the Alzheimer disease amyloid beta-peptide by metal-dependent up-regulation of metalloprotease activity. J. Biol. Chem. 281, 17670–17680. doi: 10.1074/jbc.M602487200
Wirth, J. J., and Mijal, R. S. (2010). Adverse effects of low level heavy metal exposure on male reproductive function. Syst. Biol. Reprod. Med. 56, 147–167. doi: 10.3109/19396360903582216
Wrobel, B., and Wegrzyn, G. (1998). Replication regulation of ColE1-like plasmids in amino acid-starved Escherichia coli. Plasmid 39, 48–62. doi: 10.1006/plas.1997.1319
Xu, M. B., Rong, P. Q., Jin, T. Y., Zhang, P. P., Liang, H. Y., and Zheng, G. Q. (2019). Chinese Herbal Medicine for Wilson’s disease: a systematic review and meta-analysis. Front. Pharmacol. 10:277. doi: 10.3389/fphar.2019.00277
Xu, T., Close, D. M., Sayler, G. S., and Ripp, S. (2013). Genetically modified whole-cell bioreporters for environmental assessment. Ecol. Indic. 28, 125–141. doi: 10.1016/j.ecolind.2012.01.020
Yang, Q., Li, Z., Lu, X., Duan, Q., Huang, L., and Bi, J. (2018). A review of soil heavy metal pollution from industrial and agricultural regions in China: pollution and risk assessment. Sci. Total Environ. 642, 690–700. doi: 10.1016/j.scitotenv.2018.06.068
Yoo, J., Shim, T., Hur, J., and Jung, J. (2016). Role of polarity fractions of effluent organic matter in binding and toxicity of silver and copper. J. Hazard. Mater 317, 344–351. doi: 10.1016/j.jhazmat.2016.06.009
Zhao, C., Wang, C. C., Li, J. Q., Wang, P., Ou, J. Q., and Cui, J. R. (2018). Interactions between copper(II) and DOM in the urban stormwater runoff: modeling and characterizations. Environ. Technol. 39, 120–129. doi: 10.1080/09593330.2017.1296893
Zhao, Y., Xu, M., Liu, Q., Wang, Z., Zhao, L., and Chen, Y. (2018). Study of heavy metal pollution, ecological risk and source apportionment in the surface water and sediments of the Jiangsu coastal region, China: a case study of the Sheyang Estuary. Mar. Pollut. Bull. 137, 601–609. doi: 10.1016/j.marpolbul.2018.10.044
Keywords: whole-cell bioreporter, detecting bioavailable copper, gene knock-in, GFPmut2, Escherichia coli
Citation: Pang Y, Ren X, Li J, Liang F, Rao X, Gao Y, Wu W, Li D, Wang J, Zhao J, Hong X, Jiang F, Wang W, Zhou H, Lyu J and Tan G (2020) Development of a Sensitive Escherichia coli Bioreporter Without Antibiotic Markers for Detecting Bioavailable Copper in Water Environments. Front. Microbiol. 10:3031. doi: 10.3389/fmicb.2019.03031
Received: 16 July 2019; Accepted: 17 December 2019;
Published: 24 January 2020.
Edited by:
Qiaoyun Huang, Huazhong Agricultural University, ChinaReviewed by:
Koshy Philip, University of Malaya, MalaysiaCopyright © 2020 Pang, Ren, Li, Liang, Rao, Gao, Wu, Li, Wang, Zhao, Hong, Jiang, Wang, Zhou, Lyu and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianxin Lyu, anhsdTMxM0AxNjMuY29t; Guoqiang Tan, dGdxQHdtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.