- 1School of Life Sciences, Beijing University of Chinese Medicine, Beijing, China
- 2Key Laboratory of Cell Proliferation and Regulation Biology, College of Life Sciences, Beijing Normal University, Beijing, China
- 3Institute of Molecular and Cell Biology, Agency for Science, Technology and Research, Singapore, Singapore
- 4Depatment of Biochemistry, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
PP2A-like phosphatases share high homology with PP2A enzymes and are composed of a catalytic subunit and a regulatory subunit. In Candida albicans, the PP2A-like catalytic subunit SIT4 regulates cell growth, morphogenesis, and virulence. However, the functions of its regulatory subunits remain unclear. Here, by homology analysis and co-IP experiments, we identified two regulatory subunits of SIT4 in C. albicans, SAP155 (orf19.642) and SAP190 (orf19.5160). We constructed sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ mutants and found that deleting SAP155 had no apparent phenotypic consequence, while deleting SAP190 caused slow growth, hypersensitivity to cell wall stress, abnormal morphogenesis in response to serum or genotoxic stress (HU and MMS), less damage to macrophages, and attenuated virulence in mice. However, deleting both SAP155 and SAP190 caused significantly stronger defects, which was similar to deleting SIT4. Together, our results suggest that SAP190 is required for the function of SIT4 and that SAP155 can partially compensate for the loss of SAP190 in C. albicans. Given the vital role of these regulatory subunits of SIT4 in C. albicans physiology and virulence, they could serve as potential antifungal targets.
Introduction
Candida albicans (Ca) is a commensal organism of the oral cavity, gastrointestinal tract, and vagina (Arendrup, 2013; Hebecker et al., 2014). When the host immune system is compromised, such as under conditions of long-term antibiotic treatment, immunodeficiency, or chemotherapy, C. albicans can cause mucocutaneous and life-threatening disseminated infections (Romani, 2011; Goulart et al., 2018). According to statistics, C. albicans is the fourth most common cause of hospital-acquired systemic infections with a crude mortality rate of more than 50% in the United States (Lai et al., 2008; Pfaller and Diekema, 2010). C. albicans can grow as several cell types, including yeast, pseudohyphae, and true hyphae (Sudbery et al., 2004). Yeast form helps its spread, while hyphae have strong ability of tissue adhesion and invasion (Berman and Sudbery, 2002; Zhu and Filler, 2010). Furthermore, hyphae can avoid recognition and phagocytosis by host macrophages and neutrophils, thus enabling it to escape from the killing of the host immune system (Erwig and Gow, 2016). The transformation between different cell types is closely related to C. albicans pathogenicity (Lo et al., 1997; Saville et al., 2003), suggesting that the identification of proteins involved in morphogenesis may provide new targets for developing antifungal agents.
Reversible protein phosphorylation plays a crucial role in the control of nearly all cellular processes, and dephosphorylation is equally important to phosphorylation. Most phosphorylation events in eukaryotes involve the transfer of phosphate to serine (Ser) or threonine (Thr) residues. Removal of the phosphate is catalyzed by Ser/Thr protein phosphatases. According to the enzymological criteria, Ser/Thr protein phosphatases can be classified into two groups: type 1 (PP1) and type 2 (PP2); PP2 phosphatases can be further classified into several groups based on the requirement for metal ions: PP2A and PP2A-like enzymes do not require metal ions, PP2B is activated by calcium, and 2C is Mg2+ dependent (Arino et al., 2011; Albataineh and Kadosh, 2016). There are three PP2A-like phosphatases in fungi: Sit4, Pph3, and Ppg1 (Albataineh and Kadosh, 2016). In Saccharomyces cerevisiae, Sit4 plays a critical role in cell growth, proliferation, and the regulation of the Pkc1-MAPK and Tor signaling pathways (Ronne et al., 1991; Sutton et al., 1991; Angeles et al., 2002; Rohde et al., 2004). Four regulatory subunits of Sit4 has been identified, and they are named Sit4 association proteins (SAPs) and divided into two groups based on sequence similarity, the SAP4/SAP155 group and the SAP185/SAP190 group (Luke et al., 1996). Studies have shown that the SAPs have diverse functions, such as the regulation of cell growth, K+ efflux, and drug resistance (Luke et al., 1996; Manlandro et al., 2005; Miranda et al., 2010). In C. albicans, Sit4 has been identified as the catalytic subunit of PP2A-like protein phosphatase, and deletion of SIT4 causes a significant reduction in growth rate, morphogenesis, and virulence in mice (Lee et al., 2004; Noble et al., 2010). However, the functions of its regulatory subunits remain unclear. According to a search of C. albicans genome database1, we identified two regulatory subunits of Sit4, orf19.642, and orf19.5160.
In this study, we constructed sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ mutants in C. albicans SC5314 background and conducted comprehensive phenotypic characterizations and comparisons. We found that Sap190 is the main regulatory subunit of Sit4 that plays critical roles in cell growth, cell wall integrity, hyphal morphogenesis, and virulence. Sap155 is a redundant regulatory subunit, but it is functional and can partially compensate for the absence of Sap190.
Materials and Methods
Strains and Growth Conditions
The Candida albicans strains used in this study are listed in Table 1. C. albicans was routinely grown at 30°C in YPD medium (1% yeast extract, 2% peptone, and 2% glucose). For growth on plates, 2% agar was added to the medium. To select for nourseothricin-resistant transformants, 200 μg/mL of nourseothricin (Werner Bioagents, Jena, Germany) was added to the YPD agar plates (YPD-Nou plates). To obtain nourseothricin-sensitive derivatives in which the SAT1-flipper was excised by FLP-mediated recombination, transformants were grown overnight in YCB–BSA medium (2.34% w/v yeast carbon base, 0.4% w/v bovine serum albumin, pH 4.0) to induce the SAP2 promoter controlling CaFLP expression, and then streak-inoculated onto YPD plates containing 25 μg/mL nourseothricin and incubated at 30°C at least 2 days.
Hyphal growth was induced by supplementing YPD medium with 10% fetal calf serum or DMEM, and incubating at 37°C with shaking at 200 rpm, or steaking yeast cells onto Spider agar plates (1% w/v beef extract, 1% w/v mannitol, 0.2% w/v K2HPO4, and 2% w/v agar, pH 7.2) to incubate at 30°C for 7 days. Pseudohyphal growth was induced by supplementing YPD medium with 15 mM hydroxyurea or 0.02% methyl methanesulfonate (MMS), and incubating at 30°C with shaking at 200 rpm.
Strain Construction
Gene deletion was done in C. albicans SC5314 using the SAT-flipper method as described previously (Reuss et al., 2004). Briefly, the SAT1-flipper cassette flanked by 60 bp of upstream and downstream sequences of the target gene was amplified by PCR. Then, the PCR products were transformed into SC5314 cells using the lithium acetate protocol. After transformation, cells were recovered by culturing in fresh YPD medium at 30°C for 4 h with shaking at 200 rpm before spreading onto YPD-Nou plates. Two round of the transformation were required to obtained homozygous deletion mutants. Genomic DNA and total RNA were isolated from selected transformants to verify the mutations by PCR and RT-PCR analysis.
The plasmid pAG6 was constructed by Vylkova and Lorenz (2014), which is a SAT1-marked version of CIp10 and used to integrate a gene into the RP10 locus by linearizing with StuI. We cloned SAP190 into the KpnI–XhoI sites of pAG6, then linearized the plasmid with StuI, and transformed it into the sap190Δ/Δ mutant to obtain SAP190 complemented strain.
To construct SAT1-marked version of GFP or Flag-tagging vectors, GFP or Flag gene sequence followed by the URA3 terminator was inserted into the ApaI–XhoI sites of pSFS1. To tag protein with GFP or Flag at the C-terminus, the GFP or Flag-SAT1-flipper cassette flanked by 60 bp of the coding sequence 5′ to the stop codon (without the stop codon) and 60 bp of the non-coding sequence 3′ to the stop codon was amplified by PCR. The PCR products were transformed into appropriate strains. Correct tagging was verified by PCR and Western blotting analysis. The oligonucleotide primers used to construct deletion cassette and fusion protein are shown in Table 2.
Growth Curves
Late-log phase C. albicans yeast cells were diluted to OD600 = 0.01 in 10 mL of YPD medium and were cultured at 30°C with shaking at 200 rpm. 100 μL of the culture was collected every 2 h, and OD600 was measured using a microplate reader. The experiment was performed in triplicate.
Susceptibility Tests
Candida albicans cells grown to the late-log phase in YPD medium were harvested and washed twice with sterile water. The cell suspensions were 10-fold serially diluted to generate suspensions containing 106 to 103 cells/mL, and 5 μL of each dilution was spotted onto YPD plates containing the indicated concentrations of chemicals or drugs. Growth was assessed by incubating the plates at 30°C for the indicated time. All experiments were performed at least thrice.
Fluorescence Microscopy
Log-phase C. albicans yeast cells were stained with 10 μg/mL DAPI to visualize nuclei. Cells were examined by differential interference contrast (DIC) and fluorescence microscopy.
Co-immunoprecipitation (Co-IP) and Western Blotting (WB)
Co-IP and WB was performed as described previously by Han et al. (2019).
Macrophage Cytotoxicity Assay
Candida albicans toxicity on macrophages was assessed by using a Cytotoxicity LDH Assay Kit-WST (Dojindo Molecular Technologies, Inc). RAW264.7 macrophages were seeded at 2.5 × 105 cells per well of a 96-well tissue culture plate in phenol red-free DMEM and maintained for 6 h in a humidified incubator in 5% CO2 at 37°C. C. albicans cells were grown to the mid-log phase in YPD medium and washed twice with sterile PBS, and these cell suspensions were co-cultured with macrophages at a 3:1 ratio for 5 h. Supernatants were transferred into new plates and the absorbance at 490 nm was measured by a microplate reader. Cytotoxicity was calculated according to the average absorbance from each triplicate set of infected host cells relative to the maximum LDH release from lysed host cells following the manufacturer’s protocol. The experiment was performed in triplicates.
Murine Model of Disseminated Candidiasis
Mid-log phase C. albicans yeast cells were washed twice and diluted to 5 × 106 cells/mL with PBS. Ten female BALB/c mice per strain were injected via the tail vein with 200 μL of the cell suspension. The mice were monitored twice daily for survival for 20 days. To determine the organ fungal burden, five mice were infected with each strain as described above and sacrificed at 48 h after the injection to surgically remove the kidney. One kidney from each mouse was removed, weighed, and homogenized. The homogenate was serially diluted in PBS and spread onto YPD plates for counting colony forming units (CFUs) per gram of kidney. Another kidney was fixed with formaldehyde followed by 70% ethanol and then embedded in paraffin. Thin sections were cut and stained with periodic acid-Schiff staining for microscopic examination. Animal experiments were carried out in accordance to National Advisory Committee for Laboratory Animal Research Guidelines, and all procedures were approved by the IACUC of the Agency for Science, Technology and Research of Singapore.
Statistical Analyses
In this study, all data are presented as mean ± SD based on results from at least 3 independent experiments. The results of the in vitro experiments were analyzed with Student’s t-test. The results of survival curves and fungal burdens were analyzed using Kaplan-Meier test and Mann-Whitney test, respectively. A p value less than 0.05 was considered statistically significant.
Results
CaSap155 or CaSap190 Interacts With CaSit4 in Co-IP Experiments
In the C. albicans genome database, orf19.642 and orf19.5160 are designated as the regulatory subunits of Sit4, and their amino-acid sequence homologies with ScSap4/cSap155/ScSap185/ScSap190 are 23.3%/26.5%/23.8%/24.9% and 26.2%/26.5%/ 30.9%/35.0%, respectively (Supplementary Figure S1). Thus, we named them CaSap155 (orf19.642) and CaSap190 (orf19.5160) in this study.
To further investigate whether Sap155 and Sap190 are regulatory subunits of Sit4 in C. albicans, we tested whether Sap155 and Sap190 physically interact with Sit4. We tagged Sap155 and Sap190 with GFP and Sit4 with Flag all at the C-terminus. We then performed co-IP experiments using the anti-GFP-antibody conjugated beads to pull down Sap155-GFP and Sap190-GFP and then detected Sit4-Flag in western blotting analysis. The results showed that both Sap155 and Sap190 physically associate with Sit4 (Figure 1A). Furthermore, we tagged Sap155 C-terminus with Flag and Sap190 C-terminus with GFP, pulled down Sap190-GFP, and then detected Sap155-Flag in western blotting analysis. We did not detect physical interaction between Sap155 and Sap190 (Figure 1B). Thus, like S. cerevisiae, Sap155 and Sap190 independently associate with Sit4 in separate complexes in C. albicans (Luke et al., 1996).
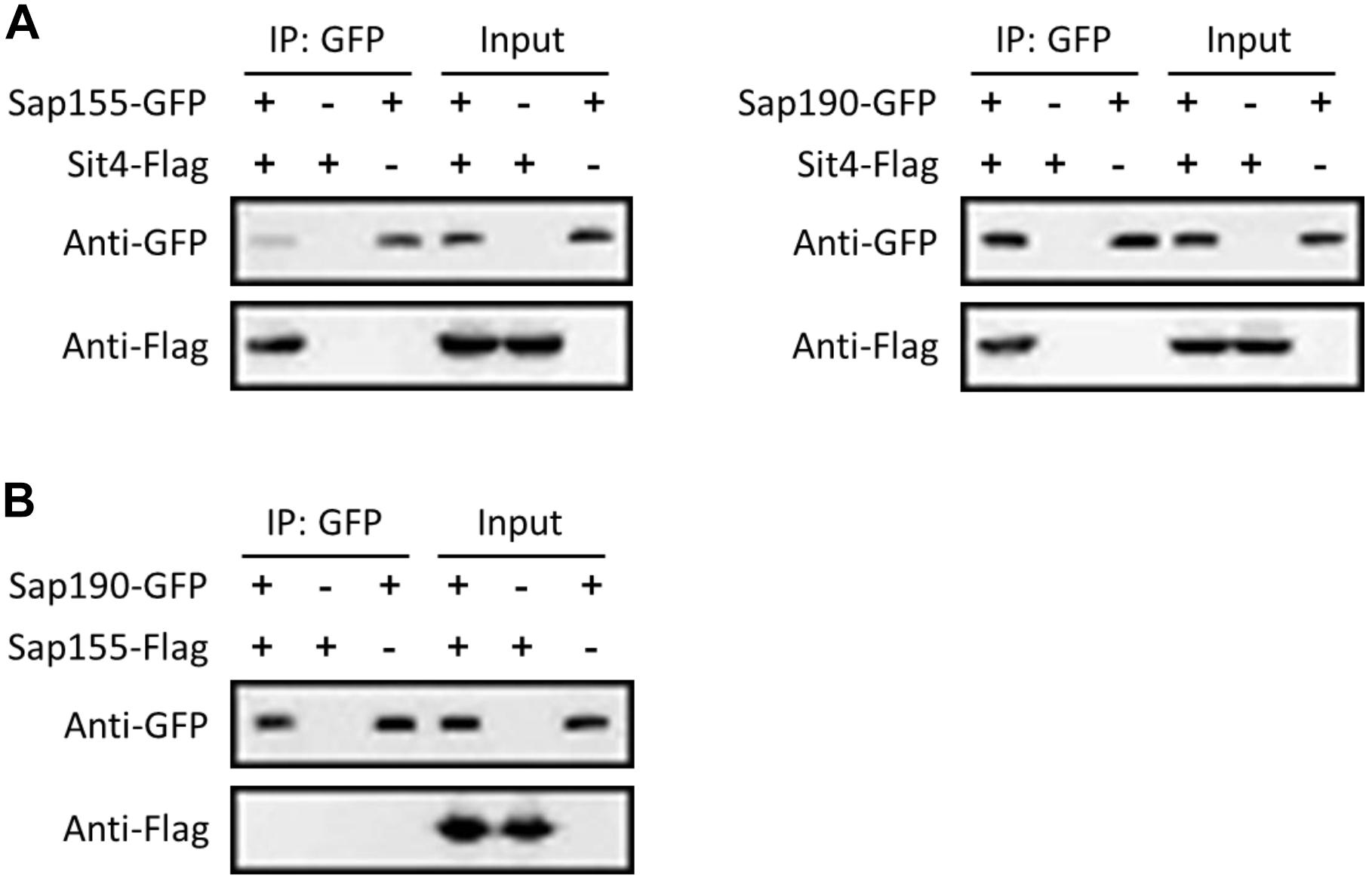
Figure 1. CaSit4 interact with either CaSap155 or CaSap190. (A) Co-IP of Sit4 with Sap155 and Sap190, and (B) co-IP of Sap155 with Sap190. Anti-GFP conjugated beads were used for pull-down from log-phase yeast cell lysates, and the pull-down products were probed with anti-GFP or anti-Flag in WB analysis. Total cell lysates were also probed similarly (input). The experiments were repeated 3 times.
Characterization and Comparison of sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ Mutants During Yeast Growth
To avoid the undesirable effects of having auxotrophic markers, we used the wild-type strain SC5314 as the parent and the SAT1-flipper method to delete the two copies of SIT4, SAP155, or SAP190, yielding the sit4Δ/Δ, sap155Δ/Δ, and sap190Δ/Δ mutants.
To investigate the functions of the regulatory subunits of Sit4 during yeast growth, wild-type (WT; SC5314), sit4Δ/Δ, sap155Δ/Δ, and sap190Δ/Δ cells were cultured in YPD liquid medium at 30°C and the growth was monitored by measuring OD600 at timed intervals (Figure 2A). Furthermore, yeast cultures of the same strains were serially diluted and spotted onto YPD plates (Figure 3). The results showed that the sap155Δ/Δ mutant exhibited normal growth, while the sap190Δ/Δ mutant grew much more slowly than WT cells. Introducing one copy of WT SAP190 at the RP10 locus of the sap190Δ/Δ mutant (sap190Δ/Δ + SAP190) fully restored the growth whereas introducing the empty vector pAG6 (sap190Δ/Δ + pAG6) had no effect, indicating that the slower growth of the sap190Δ/Δ mutant was due to the deletion of SAP190 (Figure 2B). Also, the growth of the sit4Δ/Δ mutant was slightly slower than the sap190Δ/Δ mutant (Figures 2A, 3). To further determine the roles of SAP155, we deleted SAP155 from the sap190Δ/Δ mutant to obtain the sap155Δ/Δ sap190Δ/Δ mutant. We found that the double mutant also grew little more slowly than the sap190Δ/Δ mutant while it grew at a similar rate to the sit4Δ/Δ mutant (Figures 2A, 3). These results suggest that Sap190 is the main regulatory subunit of Sit4 and is required for normal yeast growth of C. albicans, while SAP155 can partially maintain cell growth in the absence of SAP190.
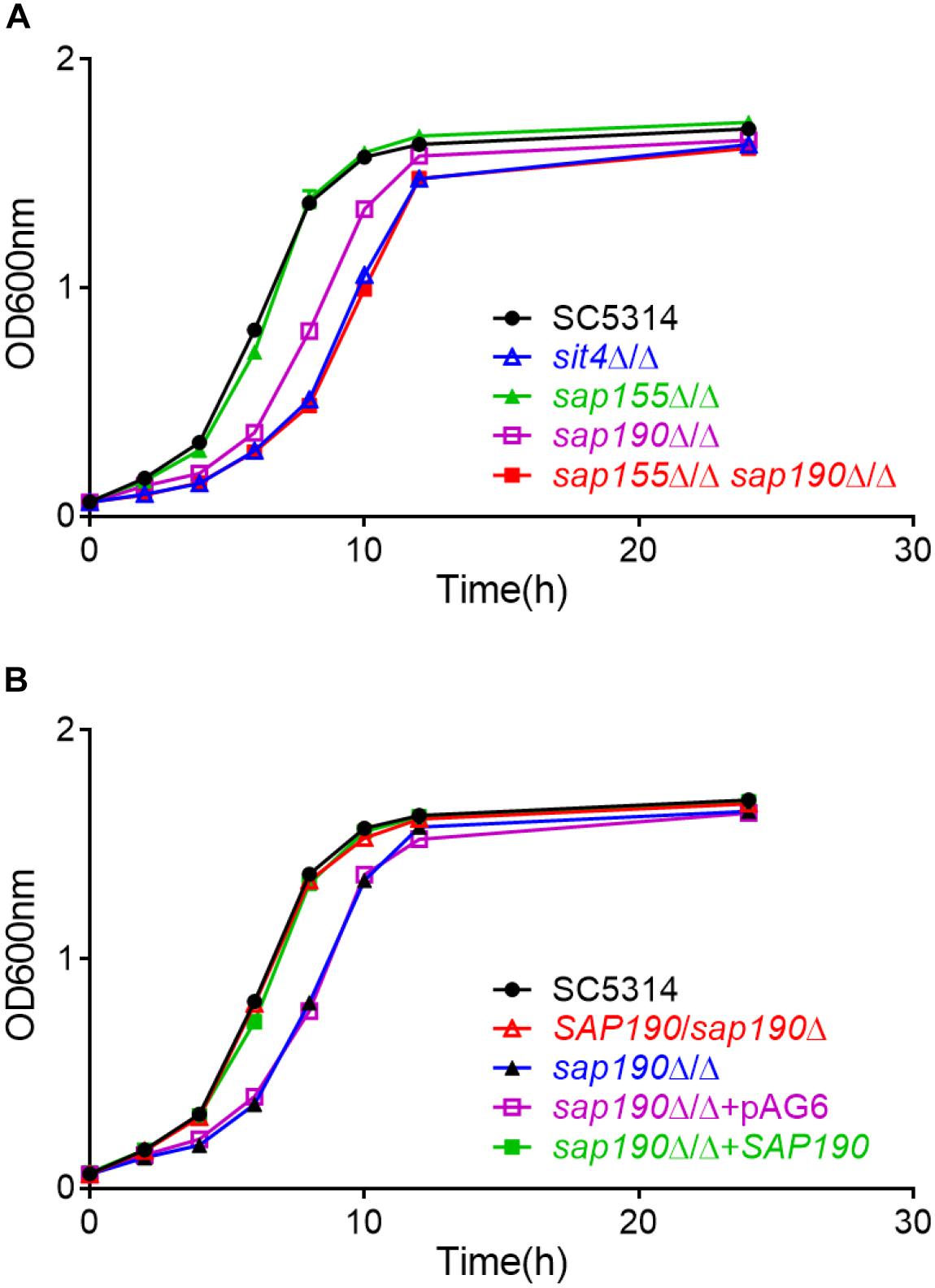
Figure 2. The growth of Candida albicans strains of the indicated genotype in YPD medium. Growth curves of (A) C. albicans SC5314, sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ strains and (B) C. albicans SC5314, SAP190/sap190Δ, sap190Δ/Δ, sap190Δ/Δ + pAG6, and sap190Δ/Δ + SAP190 strains in liquid YPD medium at 30°C with shaking at 200 rpm. All experiments were repeated using at least 3 independent clones for each mutant.
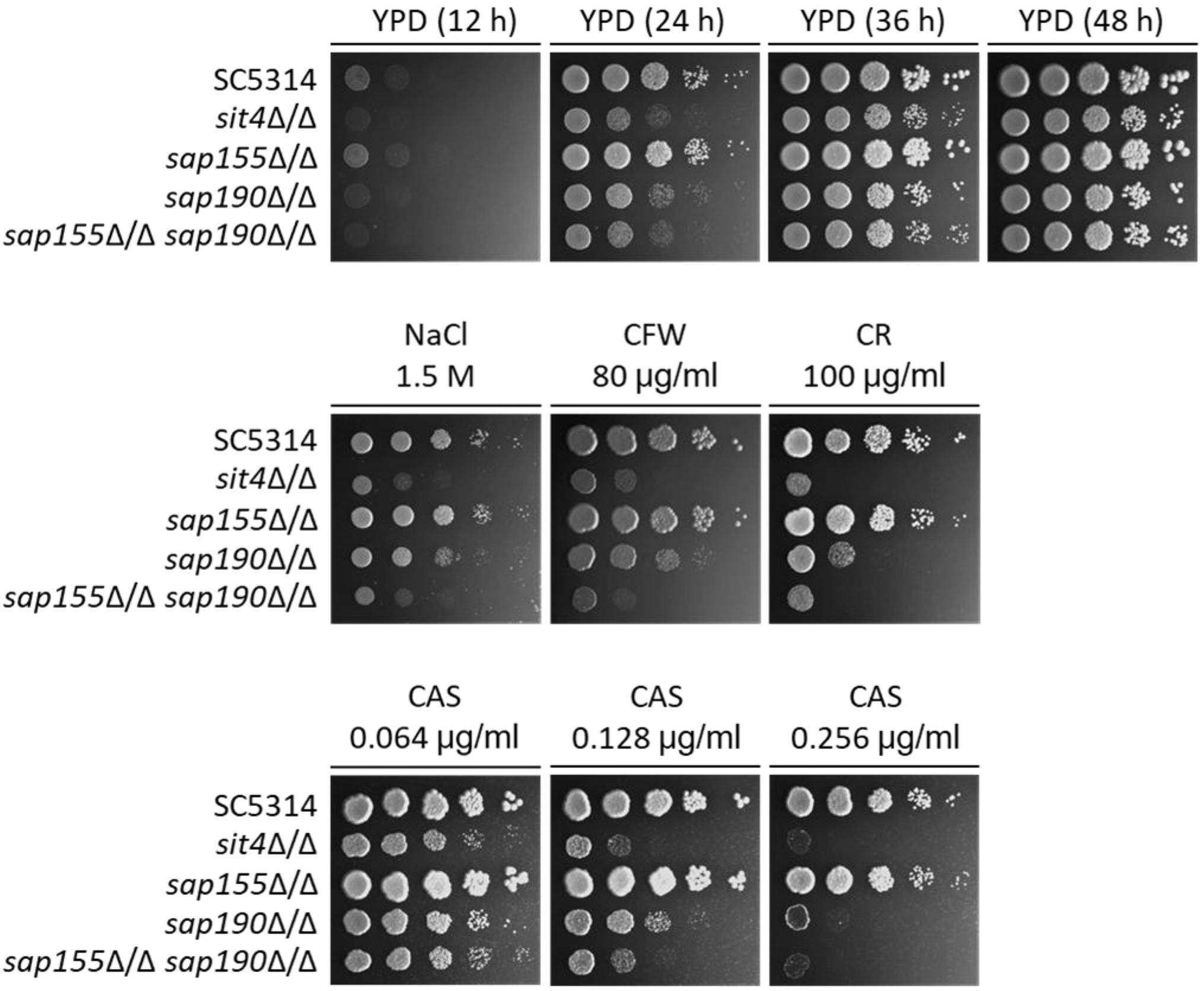
Figure 3. The sensitivity to several stress agents of sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ mutants. Log-phase yeast cells of SC5314, sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ strains were serially diluted 10-fold and spotted onto YPD plates, or YPD plates supplemented with 1.5 M NaCl, 80 μg/mL calcofluor white (CFW), 100 μg/mL congo red (CR), 0.064/0.128/0.256 μg/mL caspofungin (CAS). The YPD plates were incubated for the indicated times, and the YPD plates supplemented with stress agents were incubated at 30°C for 48 h. All experiments were repeated using at least 3 independent clones for each mutant.
Next, we stained the nucleus with DAPI and found normal nuclear localization in sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ cells (Supplementary Figure S2), and these mutants had normal cytoplasmic division, suggesting that the slow growth of cells lacking SIT4 or its regulatory subunits is not due to abnormal cell division.
Deletion of SAP155 Renders the sap190Δ/Δ Mutant More Sensitive to Cell Wall Stress
A previous study has shown that the deletion of SIT4 led to hypersensitivity to osmotic stress (Lee et al., 2004). We found that the sap155Δ/Δ sap190Δ/Δ and sit4Δ/Δ mutants grew at similar rates but both significantly more slowly than SC5314 (WT) on YPD plates containing 1.5 M NaCl (Figure 3). To investigate the functions of the regulatory subunits SAP155 and SAP190 in maintaining cell wall integrity, WT, sit4Δ/Δ, sap155Δ/Δ, and sap190Δ/Δ cells were spotted onto YPD plates containing 80 μg/ml Calcofluor White (CFW) or 100 μg/ml Congo Red (CR). We found that the growth of the sap190Δ/Δ mutant was slower than WT and the sap155Δ/Δ mutant, while the sap155Δ/Δ sap190Δ/Δ and sit4Δ/Δ mutants grew much more slowly than the sap190Δ/Δ mutant, and sit4Δ/Δ and sap155Δ/Δ sap190Δ/Δ mutants grew at similar rates (Figure 3). These results suggest that the deletion of SAP155 further sensitizes the sap190Δ/Δ mutant to osmotic and cell wall stress.
The antifungal drug caspofungin (CAS) is a non-competitive inhibitor of β-1,3-glucan synthase and commonly used clinically to treat a variety of fungal infections, including C. albicans infections (Walker et al., 2010). We next determined whether lacking Sit4 or its regulatory subunits also alters the sensitivity to CAS. We found that the sensitivity of sap190Δ/Δ mutant gradually increased with increasing concentrations of CAS in a range between 0.064 and 0.256 μg/ml and the deletion of SAP155 in the sap190Δ/Δ mutant increased the sensitivity further, particularly at the concentration of 0.128 μg/ml (Figure 3). sit4Δ/Δ and sap155Δ/Δ sap190Δ/Δ mutants showed similar sensitivities to CAS under these conditions. These results indicate that sit4Δ/Δ and sap190Δ/Δ mutants are hypersensitive to CAS, consistent with their sensitivity to osmotic and cell wall stress. The results also show that SAP155 is functional and can partially compensate for the absence of SAP190.
The Functions of SAP155 and SAP190 in Filamentous Growth Caused by Different Inducing Factors
We next examined the hyphal growth of the mutants of SIT4 and its regulatory subunits under various inducing conditions. We found that although sit4Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ cells could grow hyphae, their hyphae were much shorter compared with those of WT cells in YPD medium containing 10% serum at 37°C (Figure 4A). Interestingly, the hyphal length of sap190Δ/Δ cells was longer than that of sit4Δ/Δ and sap155Δ/Δ sap190Δ/Δ mutants. The same phenotype was also observed when hyphal growth was induced in DMEM medium (Supplementary Figure S3). sap155Δ/Δ cells did not exhibit significant defects in hyphal growth under these tested conditions. On Spider plates, WT and sap155Δ/Δ cells formed colonies with long filaments radiating from the colony periphery, while the edge of sit4Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ colonies were smooth (Figure 4A). Furthermore, it is interesting that both sit4Δ/Δ and sap155Δ/Δ sap190Δ/Δ colonies show wrinkling at the colony center, which is very different from the morphology of the other strains. These results indicate that SAP190 is required for hyphal growth and that SAP155 can partially compensate for the loss of SAP190 under some inducing conditions.
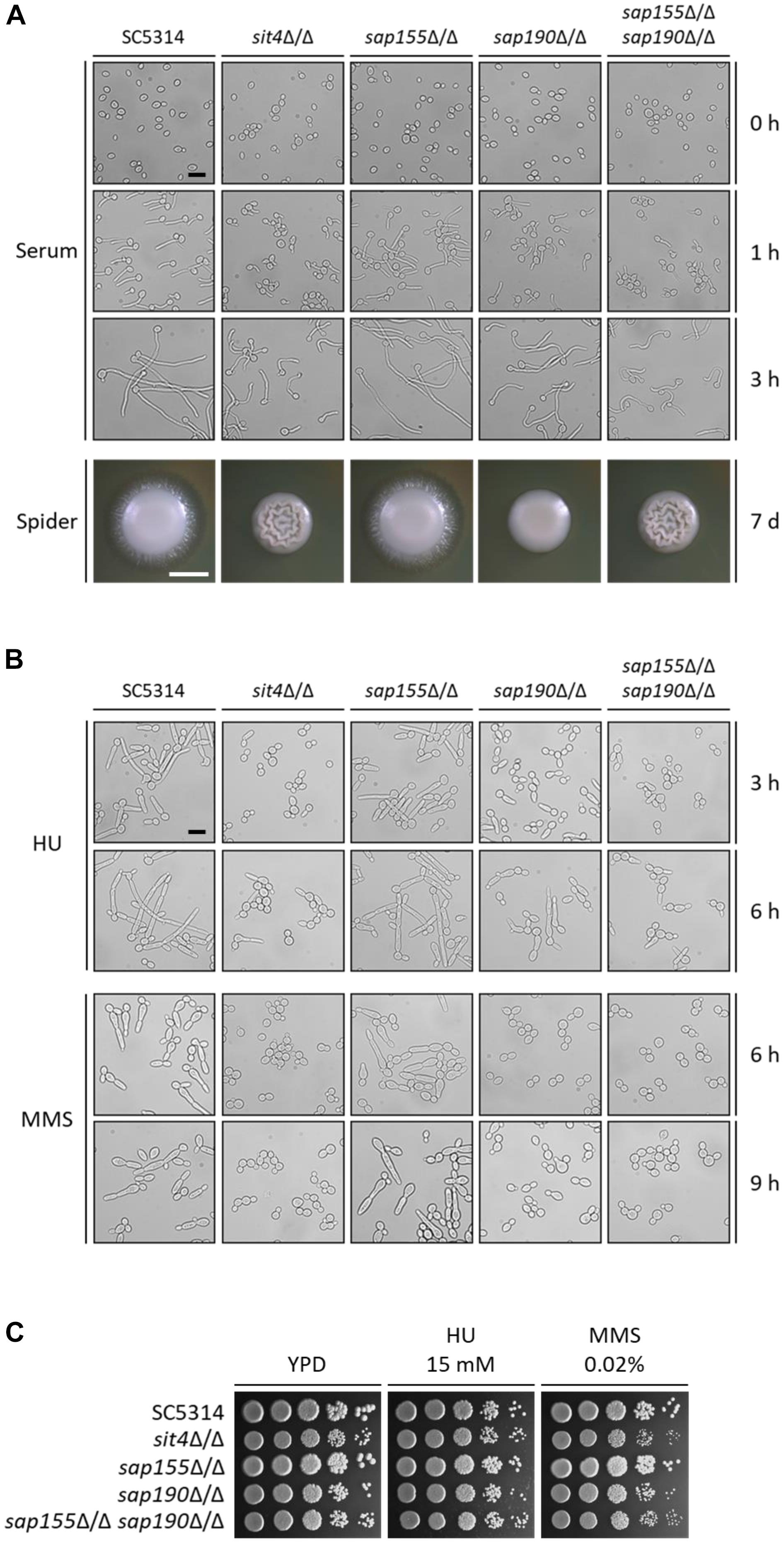
Figure 4. The morphogenesis of sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ mutants. (A) Late-log phase yeast cells of SC5314, sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ strains were re-inoculated at 1:20 dilution into fresh YPD medium containing 10% serum and incubated at 37°C with shaking at 200 rpm. Photos were taken at the indicated times. Size bars = 16 μm. Colony morphology of the same strains grown on Spider plates at 30°C for 2 or 7 days, respectively. Size bars = 1 mm. (B) Late-log phase yeast cells of the same strains as described in (A) were re-inoculated at 1:20 dilution into fresh YPD medium containing 15 mM HU or 0.02% MMS and incubated at 30°C with shaking at 200 rpm. Photos were taken at the indicated times. Size bars = 16 μm. (C) Log-phase yeast cells of the same strains as described in (A) were serially diluted 10-fold and spotted onto YPD plates supplemented with 15 mM hydroxyurea (HU) or 0.02% methyl methanesulfonate (MMS). The YPD plates were incubated at 30°C for 48 h.
Genotoxic stress, such as DNA replication inhibition and DNA damage, can cause C. albicans cell cycle arrest, leading to filamentous growth and the formation of pseudohyphae (Gow et al., 2011). Hydroxyurea (HU) is an inhibitor of DNA replication, and MMS causes DNA methylation, leading to DNA damage. To investigate the roles of Sit4 and its regulatory subunits in response to the genotoxic stress, we grew C. albicans in the presence of 15 mM HU and 0.02% MMS. We observed that, under HU treatment, while nearly all WT cells grew into long filaments, the majority of sit4Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ cells remained in the yeast form with a small number of cells showing slight elongation (Figure 4B). The phenotypes of sit4Δ/Δ and sap155Δ/Δ sap190Δ/Δ cells were similar, both exhibiting more severe defects than sap190Δ/Δ cells. Under MMS treatment, sit4Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ cells did not undergo filamentous growth (Figure 4B). After a 9-h MMS treatment, a small number of sap190Δ/Δ cells exhibited a slightly elongated yeast morphology, while all sit4Δ/Δ and sap155Δ/Δ sap190Δ/Δ cells remained in the typical yeast form. In spite of the defects in the genotoxic stress-induced filamentous growth, none of the mutants of SIT4 and its regulatory subunits showed altered sensitivity to either HU or MMS (Figure 4C). These results indicate that, firstly, Sit4 with its regulatory subunits plays an important role in regulating DNA-replication-inhibition and DNA-damage-induced filamentous growth. Secondly, Sit4 and its regulatory subunits have different roles in filamentous growth in response to different types of genotoxic stress. Thirdly, SAP190 plays a critical role in the filamentous growth induced by genotoxic stress, while SAP155 partially compensates for the loss of SAP190.
Characterization and Comparison of the Virulence of sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ Mutants in vitro and in vivo
Macrophages are the first line of host defense against C. albicans infection, but C. albicans can escape through its hyphal growth, which can penetrate and cause the lysis of macrophages (Erwig and Gow, 2016). Next, we co-cultured C. albicans with RAW264.7 macrophages and then measured the activity of lactate dehydrogenase (LDH) released by macrophages into the supernatant to determine the macrophage cytotoxicity of C. albicans. We found that cells lacking SAP155 did not alter the macrophage cytotoxicity, while cells lacking SAP190 resulted in less damage to macrophages (Figure 5). Furthermore, sap155Δ/Δ sap190Δ/Δ and sit4Δ/Δ mutants exhibited similar macrophage cytotoxicity while both caused less damage to macrophages than the sap190Δ/Δ mutant.
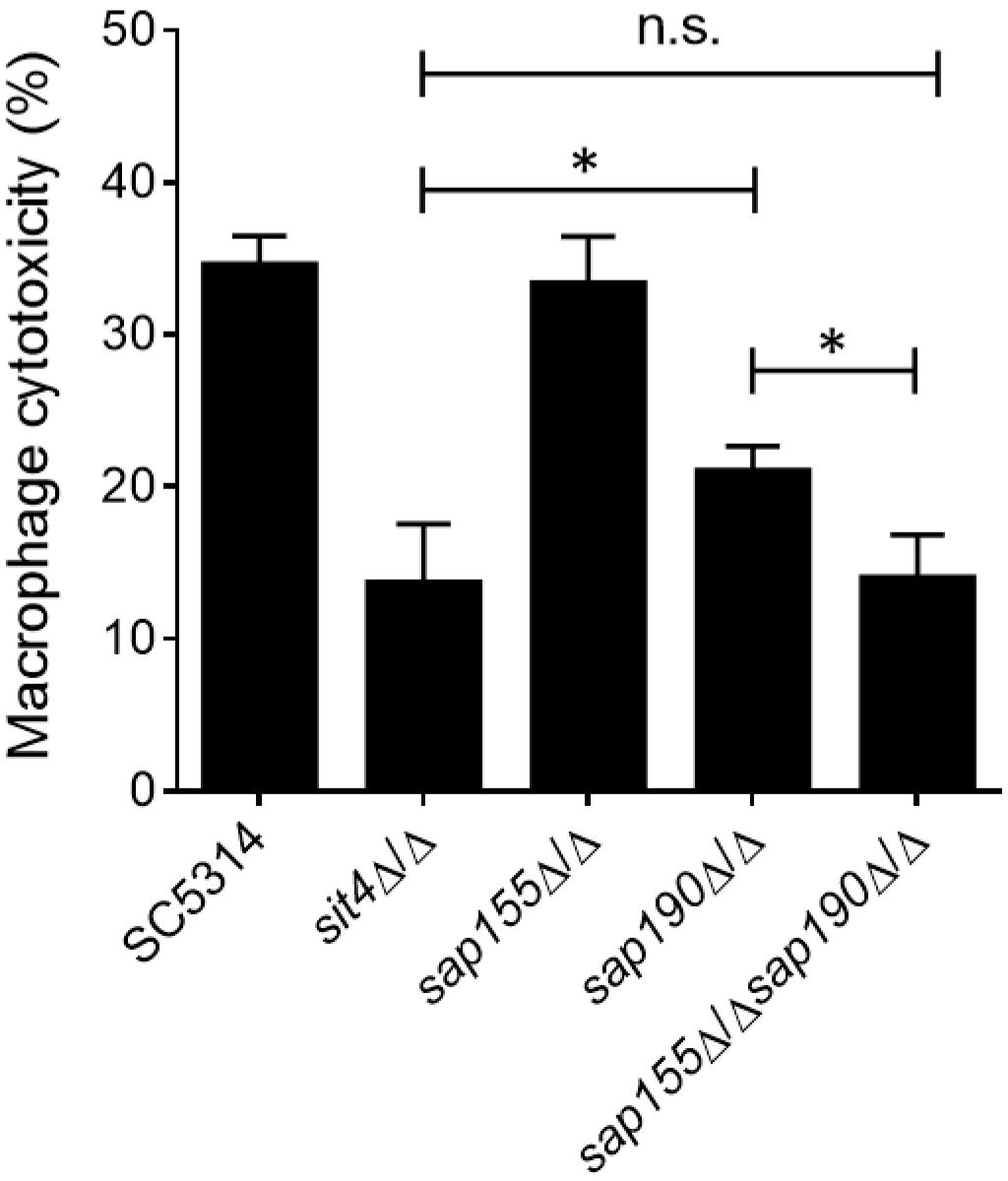
Figure 5. The ability of sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ mutants to damage macrophages in vitro. Yeast cultures of SC5314, sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ strains were co-cultured with RAW264.7 macrophages for 5 h. Cytotoxicity was determined using an LDH release assay. Results are the mean ± SD of three independent experiments, each performed in triplicates. The results were analyzed with Student’s t-test. ∗p < 0.05 compared between the indicated two groups. n.s.: no significance.
We next investigated the role of Sit4 and its regulatory subunits in virulence using a mouse model of systemic infection. Mice were injected with the WT (SC5314), sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, or sap155Δ/Δ sap190Δ/Δ strains via the tail vein, and their survival was monitored for 20 days. The results showed that all mice injected with WT or sap155Δ/Δ strains died within 9 days, and their survival median was 4–6 days. Mice infected with the sap190Δ/Δ mutant all died within 18 days, and the survival median was 10–11 days. However, ≥50% of mice injected with the sit4Δ/Δ or sap155Δ/Δ sap190Δ/Δ mutant survived for at least 20 days (Figure 6A). To assess the ability of these mutants to colonize the kidney, Five mice in each group were sacrificed 48 h post-infection to quantify CFUs and perform PAS staining of kidney sections. The results showed that CFUs in the kidneys of mice infected with the sit4Δ/Δ, sap190Δ/Δ, or sap155Δ/Δ sap190Δ/Δ mutant were much less than that in mice infected with the WT strain or the sap155Δ/Δ mutant. There was no statistically significant difference between the CFUs of sit4Δ/Δ and sap155Δ/Δ sap190Δ/Δ mutants, but the CFUs of both mutants were marked more than that of the sap190Δ/Δ mutant (Figure 6B). Also, the PAS staining of kidney sections revealed many long filaments of C. albicans cells in mice injected with SC5314 or the sap155Δ/Δ strain. In contrast, a small number of C. albicans cells were found in the kidney of mice infected with the sap190Δ/Δ mutant, and none was found in the kidney of mice infected with the sit4Δ/Δ or sap155Δ/Δ sap190Δ/Δ mutant (Figure 6C). The results demonstrate that SAP190 is required for the virulence of C. albicans and that SAP155 can partially compensate for the loss of virulence in the absence of SAP190 during co-cultivation with macrophages and systemic infection of mice.
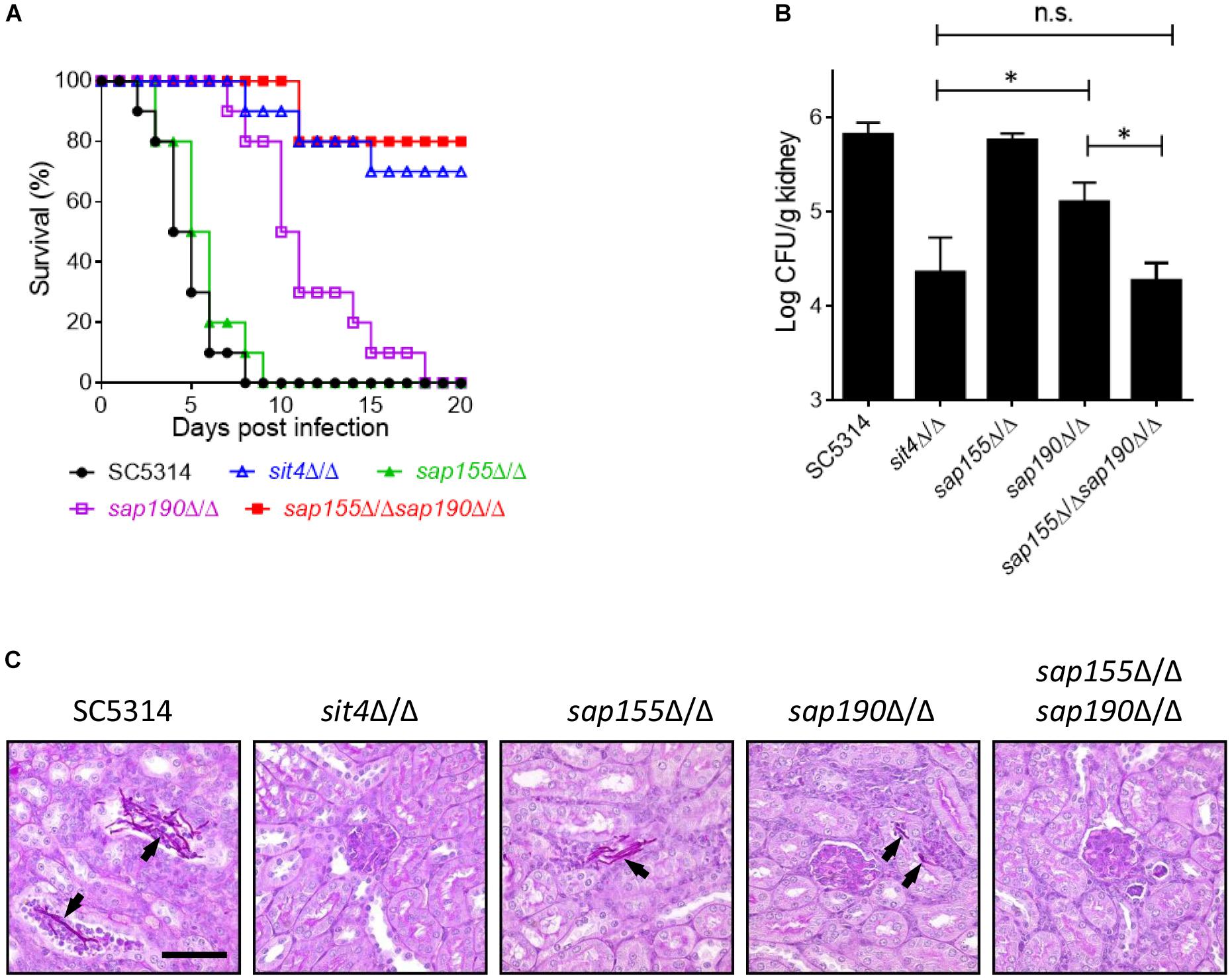
Figure 6. The virulence of sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ mutants in a murine model of disseminated candidiasis BALB/c mice were injected via the tail vein with 106 yeast cells of SC5314, sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, or sap155Δ/Δ sap190Δ/Δ strains (15 mice per C. albicans strain) and monitored for (A) survival over a period of 20 days. The results of survival curves were analyzed using Kaplan-Meier test. The statistical analysis showed that the survival curves of sap190Δ/Δ strain are significantly different compared to SC5314 or sap155Δ/Δ strain, and the survival curves of sit4Δ/Δ or sap155Δ/Δ sap190Δ/Δ strain are significantly different compared to SC5314, sap155Δ/Δ, or sap190Δ/Δ strain. Five mice were sacrificed after 48 h of the injection to (B) determine the fungal load in the kidney (the results of fungal burdens were analyzed with Mann-Whitney test. ∗p < 0.05 compared between the indicated two groups. n.s: no significance) and (C) conduct histological examinations of kidney sections. Size bars = 0.13 mm. Arrows indicate C. albicans cells in the renal tissues.
Discussion
PP2A-like phosphatases share high homology with PP2A enzymes which contain a catalytic subunit and a regulatory subunit (Albataineh and Kadosh, 2016). C. albicans Sit4 has been identified as a PP2A-like catalytic subunit (Lee et al., 2004). In this study, according to the amino-acid sequence analyses, we identified two proteins in C. albicans encoded by orf19.642 and orf19.5160 which are homologous to Sit4 regulatory subunits SAPs in S. cerevisiae. They share the relatively high homology of 26.5% and 35.0% with ScSap155 and ScSap190, respectively. Furthermore, co-IP experiments showed that like in S. cerevisiae, both proteins physically associate with Sit4, forming separate complexes in C. albicans (Luke et al., 1996). We show here that Sap190 is the main regulatory subunit of Sit4 and plays critical roles in cell growth, cell wall integrity, morphogenesis, and virulence in mice. In the SC5314 background, deleting SAP155 does not produce any apparent defects, but deleting it in the sap190Δ/Δ background leads to more severe defects, indicating that Sap155 is functionally redundant and can partially compensate for the absence of Sap190. These findings also indicate that C. albicans retains redundant regulatory subunits of Sit4, which may enhance its adaptability to some adverse environmental factors.
Cytokinesis and nuclear localization in sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ mutants do not exhibit any discernable defects. In S. cerevisiae, SIT4 is required for the G1/S transition (Sutton et al., 1991), and in Debaryomyces hansenii, deletion of SIT4 causes an increased number of G1 phase cells (Chawla et al., 2017). Therefore, like in other fungal species, the regulatory subunits of Sit4 may affect cell growth though the G1/S transition in C. albicans.
The yeast-to-hyphae morphological transition is recognized as the most important trait for C. albicans infection. Previous studies have shown that SIT4 is involved in the morphogenesis of C. albicans (Lee et al., 2004; Noble et al., 2010). Our results show that sap190Δ/Δ cells exhibited slower hyphal formation with shorter hyphal length, and sap155Δ/Δ sap190Δ/Δ cells exhibit more severe defects, which was similar to sit4Δ/Δ mutant, although the sap155Δ/Δ mutant did not show any discernible defects. The results suggest that both regulatory subunits of Sit4 are involved in regulating morphogenesis. In C. albicans, cell wall integrity is closely correlated with morphogenesis. For example, the deletion of cell wall protein-coding genes ECM33 and CSF4 led to abnormal hyphal growth (Alberti-Segui et al., 2004; Martinez-Lopez et al., 2004). Thus, SAP155 and SAP190 may regulate morphogenesis partially through their roles in cell growth and cell wall integrity. However, the targets of SIT4 and its regulatory subunits are remain unclear in C. albicans, thus their roles in the morphogenesis may be also through other unknown mechanisms.
Genotoxic stress, such as DNA replication inhibition and DNA damage, can activate cell cycle checkpoints via the phosphorylation of the checkpoint protein kinase Rad53, causing C. albicans to form pseudohyphae (Gow et al., 2011). Deletion of RAD53 not only led to a defect in filamentous growth, but also caused hypersensitivity to genotoxic stress (Shi et al., 2007). Interestingly, we show here that under HU treatment, only a small percentage of sit4Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ cells could undergo filamentous growth forming short filaments, and under MMS treatment, these mutant cells remained in the yeast form. However, deletion of SIT4 or its regulatory subunits did not alter the sensitivity to HU and MMS, suggesting that Sit4 with its regulatory subunits are involved in DNA-replication and DNA-damage checkpoint pathways to specifically regulate the filamentous growth. In future studies, we will explore the relationship between SIT4 phosphatase complexes and Rad53 under different genotoxic stresses.
Although the exact mechanism by which Sap155 and Sap190 regulate the functions of Sit4 remains unclear, deleting SAP190 causes reduced macrophage cytotoxicity in vitro and impaired virulence in mice, and deleting SAP155 in sap190Δ/Δ background resulted in more severe defects. Therefore, the regulatory subunits of Sit4 could serve as targets for developing new antifungal drugs.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The animal experiments were carried out in accordance to National Advisory Committee for Laboratory Animal Research Guidelines, and all procedures were approved by the IACUC of the Agency for Science, Technology and Research of Singapore.
Author Contributions
QH, JS, and YW conceived and created the experimental design. QH, CP, and YQW conducted the experiments. QH, YW, and LZ prepared the manuscript.
Funding
This work was supported by the research grants by JS (National Natural Science Foundation of China, Nos. 31470182 and 31270113), YW (National Medical Research Council of Singapore Grants NMRC/OFIRG16may122/2016 and NMRC/OFIRG17nov021/2018), and QH (Starting Funds of Beijing University of Chinese Medicine).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Michael C. Lorenz and Prof. Joachim Morschhauserfor for plasmid pAG6 and pSFS1.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02943/full#supplementary-material
FIGURE S1 | Sequence alignment of S. cerevisiae Sap155 and Candida albicans Sap155 (orf19.642), S. cerevisiae Sap190 and C. albicans Sap190 (orf19.5160) Completely conserved residues are colored cyan.
FIGURE S2 | The location of nucleus in sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ cells. Yeast cells of C. albicans strains of the indicated genotype were stained with DAPI to visualize the nucleus. Size bars = 12 μm.
FIGURE S3 | The hyphal growth of sit4Δ/Δ, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ mutants in DMEM. Late-log phase yeast cells of SC5314, sap155Δ/Δ, sap190Δ/Δ, and sap155Δ/Δ sap190Δ/Δ strains were re-inoculated at 1:20 dilution into fresh DMEM and incubated at 37°C with shaking at 200 rpm. Photos were taken at 1 h and 3 h. Size bars = 16 μm.
Footnotes
References
Albataineh, M. T., and Kadosh, D. (2016). Regulatory roles of phosphorylation in model and pathogenic fungi. Med. Mycol. 54, 333–352. doi: 10.1093/mmy/myv098
Alberti-Segui, C., Morales, A. J., Xing, H., Kessler, M. M., Willins, D. A., Weinstock, K. G., et al. (2004). Identification of potential cell-surface proteins in Candida albicans and investigation of the role of a putative cell-surface glycosidase in adhesion and virulence. Yeast 21, 285–302. doi: 10.1002/yea.1061
Angeles, D. L. T. M., Torres, J., Arino, J., and Herrero, E. (2002). Sit4 is required for proper modulation of the biological functions mediated by Pkc1 and the cell integrity pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277, 33468–33476. doi: 10.1074/jbc.M203515200
Arendrup, M. C. (2013). Candida and candidaemia. Susceptibility and epidemiology. Dan. Med. J. 60, B4698.
Arino, J., Casamayor, A., and Gonzalez, A. (2011). Type 2C protein phosphatases in fungi. Eukaryot Cell 10, 21–33. doi: 10.1128/EC.00249-10
Berman, J., and Sudbery, P. E. (2002). Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3, 918–930. doi: 10.1038/nrg948
Chawla, S., Kundu, D., Randhawa, A., and Mondal, A. K. (2017). The serine/threonine phosphatase DhSIT4 modulates cell cycle, salt tolerance and cell wall integrity in halo tolerant yeast Debaryomyces hansenii. Gene 606, 1–9. doi: 10.1016/j.gene.2016.12.022
Erwig, L. P., and Gow, N. A. (2016). Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 14, 163–176. doi: 10.1038/nrmicro.2015.21
Fonzi, W. A., and Irwin, M. Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728.
Goulart, L. S., Souza, W., Vieira, C. A., Lima, J. S., Olinda, R. A., and Araujo, C. (2018). Oral colonization by Candida species in HIV-positive patients: association and antifungal susceptibility study. Einstein 16:eAO4224. doi: 10.1590/S1679-45082018AO4224
Gow, N. A., van de Veerdonk, F. L., Brown, A. J., and Netea, M. G. (2011). Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat. Rev. Microbiol. 10, 112–122. doi: 10.1038/nrmicro2711
Han, Q., Pan, C., Wang, Y., Wang, N., Wang, Y., and Sang, J. (2019). The PP2A regulatory subunits, Cdc55 and Rts1, play distinct roles in Candida albicans’ growth, morphogenesis, and virulence. Fungal Genet. Biol. 131:103240. doi: 10.1016/j.fgb.2019.103240
Hebecker, B., Naglik, J. R., Hube, B., and Jacobsen, I. D. (2014). Pathogenicity mechanisms and host response during oral Candida albicans infections. Expert Rev. Anti Infect. Ther. 12, 867–879. doi: 10.1586/14787210.2014.916210
Lai, C. C., Tan, C. K., Huang, Y. T., Shao, P. L., and Hsueh, P. R. (2008). Current challenges in the management of invasive fungal infections. J. Infect. Chemother. 14, 77–85. doi: 10.1007/s10156-007-0595-7
Lee, C. M., Nantel, A., Jiang, L., Whiteway, M., and Shen, S. H. (2004). The serine/threonine protein phosphatase SIT4 modulates yeast-to-hypha morphogenesis and virulence in Candida albicans. Mol. Microbiol. 51, 691–709. doi: 10.1111/j.1365-2958.2003.03879.x
Lo, H. J., Kohler, J. R., DiDomenico, B., Loebenberg, D., Cacciapuoti, A., and Fink, G. R. (1997). Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939–949. doi: 10.1016/s0092-8674(00)80358-x
Luke, M. M., Della, S. F., Di Como, C. J., Sugimoto, H., Kobayashi, R., and Arndt, K. T. (1996). The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16, 2744–2755. doi: 10.1128/mcb.16.6.2744
Manlandro, C. M., Haydon, D. H., and Rosenwald, A. G. (2005). Ability of Sit4p to promote K+ efflux via Nha1p is modulated by Sap155p and Sap185p. Eukaryot Cell 4, 1041–1049. doi: 10.1128/EC.4.6.1041-1049.2005
Martinez-Lopez, R., Monteoliva, L., Diez-Orejas, R., Nombela, C., and Gil, C. (2004). The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence in Candida albicans. Microbiology 150, 3341–3354. doi: 10.1099/mic.0.27320-0
Miranda, M. N., Masuda, C. A., Ferreira-Pereira, A., Carvajal, E., Ghislain, M., and Montero-Lomeli, M. (2010). The serine/threonine protein phosphatase Sit4p activates multidrug resistance in Saccharomyces cerevisiae. FEMS Yeast Res. 10, 674–686. doi: 10.1111/j.1567-1364.2010.00656.x
Noble, S. M., French, S., Kohn, L. A., Chen, V., and Johnson, A. D. (2010). Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42, 590–598. doi: 10.1038/ng.605
Pfaller, M. A., and Diekema, D. J. (2010). Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36, 1–53. doi: 10.3109/10408410903241444
Reuss, O., Vik, A., Kolter, R., and Morschhauser, J. (2004). The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341, 119–127. doi: 10.1016/j.gene.2004.06.021
Rohde, J. R., Campbell, S., Zurita-Martinez, S. A., Cutler, N. S., Ashe, M., and Cardenas, M. E. (2004). TOR controls transcriptional and translational programs via Sap-Sit4 protein phosphatase signaling effectors. Mol. Cell. Biol. 24, 8332–8341. doi: 10.1128/MCB.24.19.8332-8341.2004
Romani, L. (2011). Immunity to fungal infections. Nat. Rev. Immunol. 11, 275–288. doi: 10.1038/nri2939
Ronne, H., Carlberg, M., Hu, G. Z., and Nehlin, J. O. (1991). Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol. Cell. Biol. 11, 4876–4884. doi: 10.1128/mcb.11.10.4876
Saville, S. P., Lazzell, A. L., Monteagudo, C., and Lopez-Ribot, J. L. (2003). Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2, 1053–1060. doi: 10.1128/ec.2.5.1053-1060.2003
Shi, Q. M., Wang, Y. M., Zheng, X. D., Lee, R. T., and Wang, Y. (2007). Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol. Biol. Cell 18, 815–826. doi: 10.1091/mbc.e06-05-0442
Sudbery, P., Gow, N., and Berman, J. (2004). The distinct morphogenic states of Candida albicans. Trends Microbiol. 12, 317–324. doi: 10.1016/j.tim.2004.05.008
Sutton, A., Immanuel, D., and Arndt, K. T. (1991). The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 11, 2133–2148. doi: 10.1128/mcb.11.4.2133
Vylkova, S., and Lorenz, M. C. (2014). Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog. 10:e1003995. doi: 10.1371/journal.ppat.1003995
Walker, L. A., Gow, N. A., and Munro, C. A. (2010). Fungal echinocandin resistance. Fungal Genet. Biol. 47, 117–126. doi: 10.1016/j.fgb.2009.09.003
Keywords: SAP155, SAP190, SIT4, Candida albicans, morphogenesis, virulence
Citation: Han Q, Pan C, Wang Y, Zhao L, Wang Y and Sang J (2019) PP2A-Like Protein Phosphatase (Sit4) Regulatory Subunits, Sap155 and Sap190, Regulate Candida albicans’ Cell Growth, Morphogenesis, and Virulence. Front. Microbiol. 10:2943. doi: 10.3389/fmicb.2019.02943
Received: 26 September 2019; Accepted: 06 December 2019;
Published: 20 December 2019.
Edited by:
James Bernard Konopka, Stony Brook University, United StatesReviewed by:
David Kadosh, The University of Texas Health Science Center at San Antonio, United StatesRebeca Alonso-Monge, Complutense University of Madrid, Spain
Copyright © 2019 Han, Pan, Wang, Zhao, Wang and Sang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Wang, bWNid2FuZ3lAaW1jYi5hLXN0YXIuZWR1LnNn; Jianli Sang, amxzYW5nQGJudS5lZHUuY24=
 Qi Han
Qi Han Chaoying Pan2
Chaoying Pan2 Yueqing Wang
Yueqing Wang Yue Wang
Yue Wang Jianli Sang
Jianli Sang