- 1Program of Basic Biology, Graduate School of Integrated Sciences for Life, Hiroshima University, Higashihiroshima, Japan
- 2Department of Biological Science, Graduate School of Science, Hiroshima University, Higashihiroshima, Japan
- 3Division for Health Service Promotion, University of Tokyo, Tokyo, Japan
The conjugal transfer is a major driving force in the spread of antibiotic resistance genes. Nevertheless, an effective approach has not yet been developed to target conjugal transfer to prevent the acquisition of antibiotic resistance by this mechanism. This study aimed to identify potential targets for plasmid transfer blockade by isolating mutants defective in the completion of the acquisition of antibiotic resistance via conjugal transfer. We performed genome-wide screening by combining an IncP1α-type broad host range plasmid conjugation system with a comprehensive collection of Escherichia coli gene knockout mutants (Keio collection; 3884 mutants). We followed a six-step screening procedure to identify the mutants showing conjugation deficiency precisely. No mutants defective in the conjugal transfer were isolated, strongly suggesting that E. coli cannot escape from being a recipient organism for P1α plasmid transfer. However, several mutants with low viability were identified, as well as mutants defective in establishing resistance to chloramphenicol, which was used for transconjugant selection. These results suggest that developing drugs capable of inhibiting the establishment of antibiotic resistance is a better approach than attempting to prevent the conjugal transfer to block the spread of antibiotic resistance genes. Our screening system based on the IncP1α-type plasmid transfer can be extended to isolation of target genes for other drugs. This study could be the foundation for further research to understand its underlying molecular mechanism through functional analysis of the identified genes.
Introduction
The threat of antibiotic resistance is an ongoing source of serious concern for human health (Asaduzzaman, 2018; Zumla et al., 2018). Antibiotic overuse, in both medical treatments and farming, has caused a strong selective pressure allowing the emergence of drug-resistant pathogens. Insufficient sewage treatment and the progress of globalization have also aggravated the spread of drug-resistant bacteria worldwide (Bengtsson-Palme et al., 2014; Brown and Wright, 2016; Thanner et al., 2016). Although efforts to develop new antibiotics are strenuously being pursued, there may be a possibility that a multi-drug resistant pathogen could emerge that is resistant to all existing antibiotics. Furthermore, the emergence of a resistance gene against colistin, which is used as a last resort antibiotic, has been reported in recent years (Liu et al., 2016; McGann et al., 2016; Davies and Walsh, 2018).
Antibiotic resistance that arises in a single bacterium can easily spread to other bacteria via the conjugal transfer of the resistance gene, which is a major driving force for the transfer of antibiotic resistance genes. These genes are usually transferred through vectors such as plasmids or integrative conjugative elements (ICE) (Thomas and Nielsen, 2005; Amábile-Cuevas, 2013). Various methods have been tested to block conjugal transfer to control the antibiotic resistance crisis. Three groups of conjugation factors have been proposed: those encoded on the donor chromosome, those encoded on plasmids, and those encoded on the recipient chromosome. If these conjugation-related factors are evolutionarily well-conserved and can be characterized, and if effective inhibitory drugs capable of targeting these factors could be developed, the risk of multi-antibiotic resistance could be decreased.
Several factors in the first group, for example, Sfr, have been reported to be involved in the transfer of F and F-like plasmids (Beutin and Achtman, 1979; Kato and Katayama, 2001; Starcic et al., 2003; Williams and Schildbach, 2007). Recently, a non-peer reviewed article reported the existence of more than 50 novel candidates identified by a genome-wide screening of a comprehensive collection of knockout Escherichia coli mutants and the F plasmid, although these have not been fully confirmed (Alalam et al., 2018). In the case of the second group, although the factors such as Tra and Trb and their functions are well characterized, the discovery of effective inhibitory molecules is only now underway and promising results have been reported (Shaffer et al., 2016). For the third group of factors, a genome-wide screening was performed in E. coli using conjugation of the IncW plasmid R388; however, no factors essential for conjugation were isolated, except for enzymes in the lipopolysaccharide (LPS) synthesis pathway, although these had only modest effects (6–32% of wild type). Based on this, the authors of this latter study concluded that recipient bacterial cells cannot avoid being used as recipients in bacterial conjugation (Pérez-Mendoza and de la Cruz, 2009). However, the universality of this conclusion with respect to conjugal transfer, and alternative methods, which can protect against the emergence of resistance in plasmid-incorporated cells, have not been well studied, despite reports in the 1970s showing that defects in LPS and the outer membrane protein OmpA cause a conjugation deficiency for the F plasmid (Watanabe et al., 1970; Skurray et al., 1974).
In order to identify drug targets that would be effective in blocking the emergence of antibiotic resistance by conjugal transfer, we aimed to isolate recipient mutants via conjugal transfer, by combining the IncP1α plasmid transfer system (derived from RP4) with a comprehensive collection of E. coli gene knockout mutants (the Keio collection; 3884 mutants). IncP plasmids are a group of broad host range, low copy number plasmids found in Enterobacteriaceae as well as Pseudomonas spp. They have been mainly isolated from E. coli and Klebsiella pneumoniae in human samples (Rozwandowicz et al., 2018). RP4 (also known as RK2 or RP1) was found in the clinically isolated strain S8 of Pseudomonas aeruginosa (Datta et al., 1971; Saunders and Grinsted, 1972). This plasmid has a broad host range among gram-negative bacteria and has broad transfer range even in eukaryotes and archaea (Heinemann and Sprague, 1989; Dodsworth et al., 2010; Moriguchi et al., 2013a, b; Karas et al., 2015). This characteristic of the RP4 plasmid strongly suggests that recipient organisms have little choice over whether or not conjugation occurs.
Materials and Methods
Bacterial Strains and Plasmids
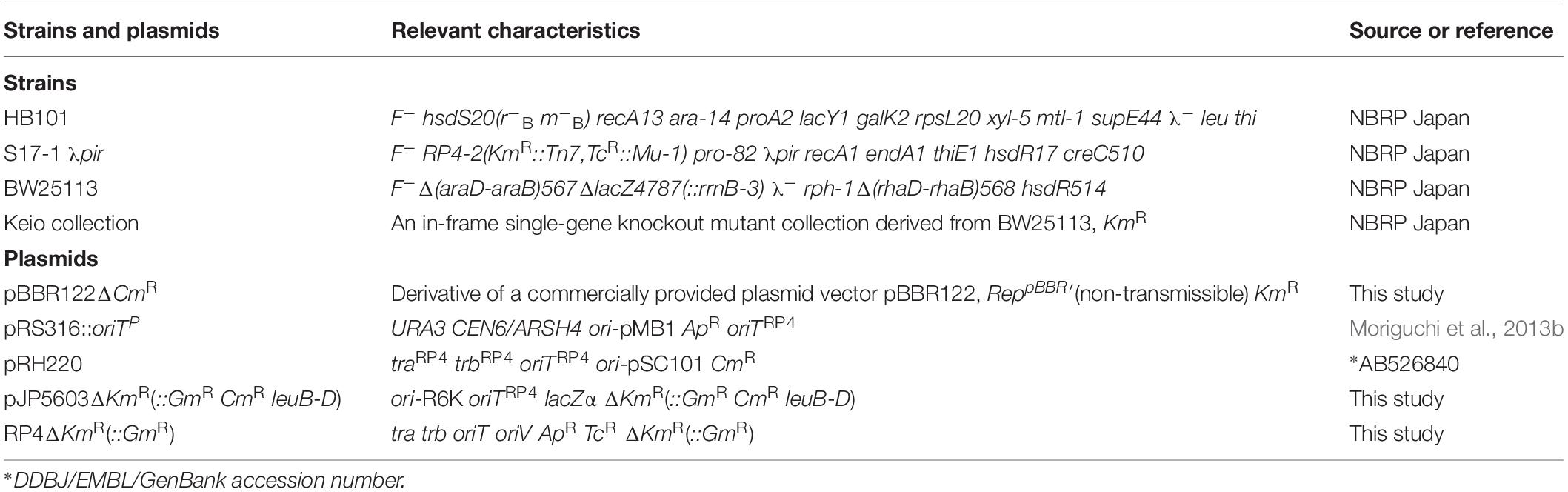
The E. coli strains and plasmids used are listed in Table 1. The complete set of E. coli deletion clones (Keio collection) was provided by the National BioResource Project (NBRP) of Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. Although the total number of strains provided was 3909, 25 strains were removed from screening because precise gene disruption had not occurred in these strains (Yamamoto et al., 2009). BW25113 (pBBR122ΔCmR) was used as the control strain.
Donor and Recipient Cell Culture
LB Lennox (LB: 1% tryptone, 0.5% yeast extract, 0.5% NaCl, 1.5% agar if necessary) medium was routinely used for E. coli culture. The antibiotics chloramphenicol (Cam; 30 μg/mL), ampicillin (Amp; 50 μg/mL), kanamycin (Kan; 50 μg/mL), tetracycline (Tet; 7.5 μg/mL), and gentamycin (Gen; 30 μg/mL) were supplemented as necessary.
For the donor culture, in the primary and secondary screenings, HB101 (pRH220 pRS316::oriTP) was cultured with medium supplemented with Cam and Amp at 5 mL scale using glass tubes for 16 to 18 h at 37°C following inoculation from a solid culture. For the third and fourth screenings, S17-1 λpir [pJP5603ΔKmR(::GmR CmR leuB-D)] and HB101 (RP4ΔKmR::GmR) were cultured in a similar manner, and the cultures were supplemented with Cam and Amp, respectively. For the fifth and sixth screenings (i.e., the final confirmation steps), HB101 (RP4ΔKmR::GmR) and HB101 (pRH220) were cultured in media supplemented with Amp and Cam, respectively.
For the recipient culture, the Keio mutants were inoculated from 96-well frozen stock plates and cultured at 100 μL scale using 96-well flat-bottom plates at 37°C for 22–24 h for primary screening, and for 20–22 h for the second to fourth screenings. In the fifth and sixth screenings, the culture scale was increased up to 600 μL, and cultured at 37°C for 20–22 h using 5 mL disposable plastic tubes.
Conjugation Reaction
For the primary screening, to provide sufficient statistical power in our calculation of the median value for the conjugation efficiency, we defined two 96-well Keio mutant strain plates (comprising 48 plates in total) as one conjugation experiment set, and 50 μL each of the donor and recipient cell cultures were mixed and incubated for 24 h at 28°C (conjugation reaction). After the conjugation reaction was complete, the reaction mixtures were well suspended and then 10 μL of each mixture was diluted with 90 μL of TNB [80 mM Tris–HCl (pH 7.5) and 0.05% NaCl]. Subsequently, 10 μL of each diluted mixture was inoculated into 90 μL of fresh medium containing Amp, Cam, and Kan in 96-well flat-bottom plates. After incubation for 24 h at 37°C using a plate shaker, the growth of transconjugants was assessed by measuring the turbidity at a wavelength of 600 nm, using a microtiter-plate reader MTP-310 (CORONA, Ibaraki, Japan).
For the second to fourth screening steps, donor cells were collected by centrifugation and re-suspended in TNB at an OD 660 nm value of 1.8. Recipient cultures in a 96-well flat-bottom plate were then moved to a 96-well v-bottom plate, and the cells were collected by centrifugation and re-suspended in 100 μL of TNB. Following this, 50 μL each of the donor and recipient cells were mixed and incubated for 1 h at 28°C. After the conjugation reaction, the mixtures were well suspended, and then diluted with TNB to the appropriate concentration for each screening step as follows: 200-fold for the second screening step, 10-fold for the third screening step, and 2000-fold for the fourth screening step. Following this, 10 μL of each of the diluted mixtures was spotted onto a solid medium plate containing the appropriate antibiotics as follows: Cam + Kan for the second and third screening steps, and Amp + Kan for the fourth screening step. Each screening step was performed in triplicate.
For the fifth and sixth screening steps, the donor and recipient cells were collected by centrifugation, and each bacterial pellet was re-suspended in TNB at an OD 660 nm value of 1.8. The protocol used for the second to fourth screenings steps was then followed with some modifications as follows: the dilution ratio was optimized for each mutant, and 10 μL of each diluted suspension was streaked onto the appropriate selection plate, Tet + Kan for the fifth screening step, and Cam + Kan for the sixth screening step.
For the second to sixth screening steps, transconjugants were detected by incubating for 18–22 h at 28°C, and colony numbers were determined by either the naked eye or using a stereomicroscope. At the sixth screening step, for slower growing mutants, an additional incubation was performed either at 37°C for up to 6 h, or at 28°C overnight. In addition, the Δrnt, ΔpriA, and ΔdnaT mutants were incubated for 24 h at 37°C at the fifth screening step, and the presence or absence of colonies was confirmed.
Data Analysis
For the initial screening step, the relative turbidity value of each transconjugant culture [defined as the transconjugant growth value (TGV)] divided by the median TGV value (MTGV) in the conjugation experiment set was calculated. The log2 value of the relative TGV (RTGV = TGV/MTGV) was defined as an arbitrary unit and calculated.
For the second to fourth screening steps, the number of transconjugant colonies for each mutant was divided by the relative turbidity value of the corresponding input recipient culture and defined as the transconjugant colony value (TCV). Then, by using the control strain BW25113 carrying pBBR122ΔCmR shown in Table 1, the average TCV (ATCV) of seven control reactions in each conjugation experiment set (defined as the ATCVctrl) was calculated. By summarizing the results of the triplicate experiments, the log2 value of the relative TCV [RTCV = Average (TCV1, TCV2, and TCV3)/Average (ATCVctrl1, ATCVctrl2, and ATCVctrl3)] was defined as an arbitrary unit and calculated.
For the fifth and sixth screening steps, the absolute value of the conjugation efficiency (transconjugants/output recipient) was calculated for each mutant.
Statistical analyses were performed using either Microsoft Excel (version 16.21) or the public domain R program (version 3.3.3).
Results
Refinement and Characterization of the Recipient Mutants for IncP1α Conjugation
An overall flowchart of the screening is shown in Supplementary Figure S1. In the initial screening step, we detected transconjugants that had accepted both the plasmids, namely pRS316:oriTP and pRH220. The growth of most transconjugants, which arose from the mutant strains, reached saturation. As shown in Figure 1, the distribution pattern of the log2(RTGV) values deviated from a Gaussian distribution with a tail skewed to lower values (average = −0.193, median = 0.000), but was closer to a Gaussian distribution when the samples that had a log2 value greater than −2, were selected (average = −0.030, median = 0.012). Since the latter distribution was expected to represent the distribution of non-conjugation-deficient mutants, 156 mutant strains, which showed log2 values equal to or less than −2 (i.e., one-fourth of the median value), were selected for analysis in the second screening step. Among them, ΔpriA mutant was included, since the log2 value of this mutant was −5.06 (i.e., a 0.030-fold change compared to the median value). A deficiency in priA causes a deficiency in replication by ColE1-like ori (Lee and Kornberg, 1991), and pRS316:oriTP was replicated using an identical origin of replication (Table 1). Therefore, we determined that this would be a useful indicator of the success of this screening system, since recipient chromosomal factors, which are critical for the acceptance of RP4, have not been identified.

Figure 1. Distribution pattern of the conjugation efficiency of Keio mutants at the primary screening step. Log2 values of each relative transconjugant growth value (RTGV) for 3884 mutants are plotted in ascending order. For one mutant (ΔholC), the log2(RTGV) could not be calculated since transconjugant growth was less than the detectable limit, and the “×” symbol shows this. Mutants with a log2(RTGV) ≤−2 were subjected to the next round of screening. HB101 (pRH220 pRS316:oriTP) was used as the donor.
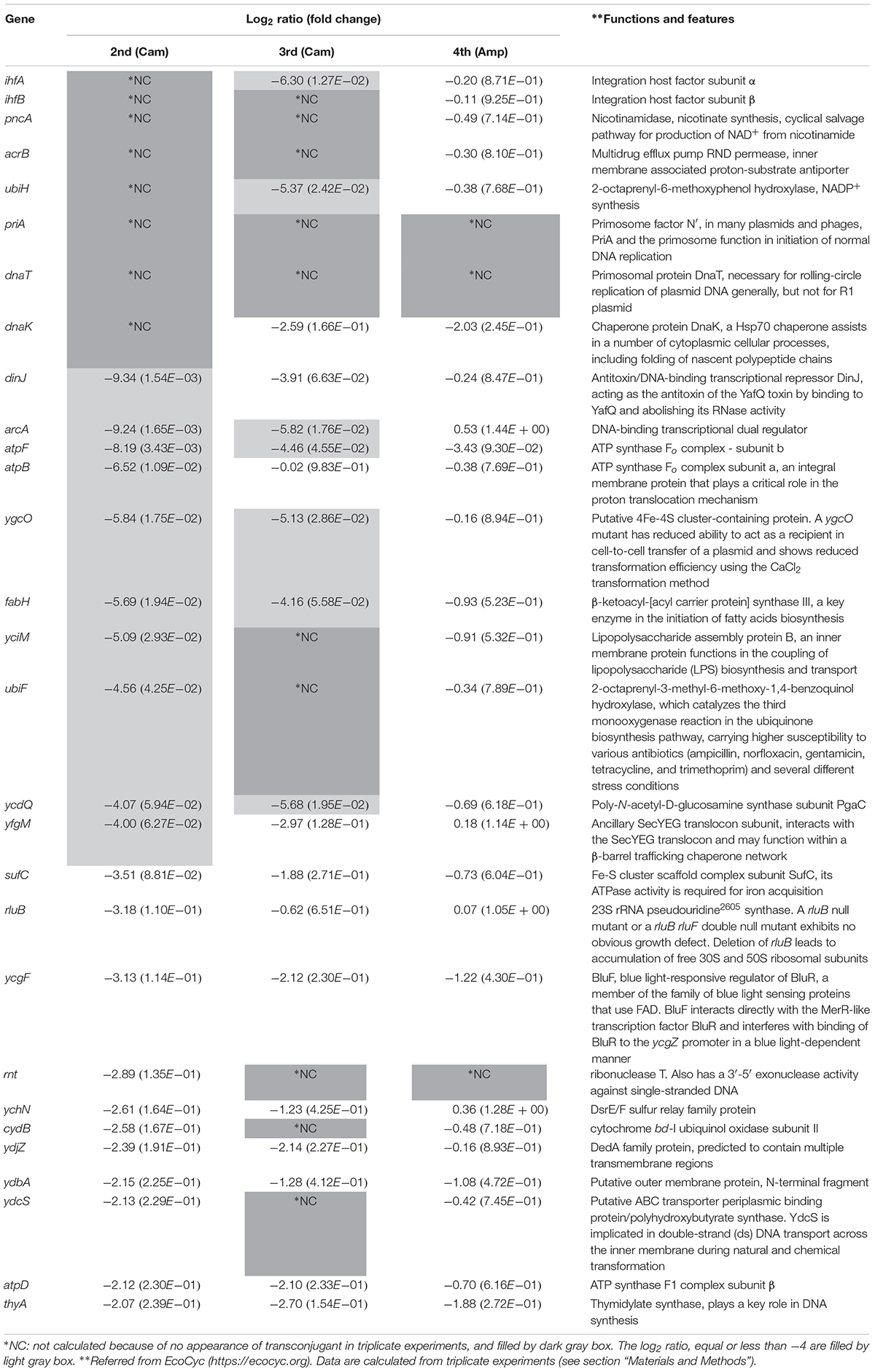
The method used in the initial screening step measured the cell growth of the transconjugants included in each conjugation reaction mixture. A different method was required to select the mutant strains that showed low conjugation efficiency as well as those that exhibited a slow growth phenotype. As a result, 156 candidates were further screened by counting the number of transconjugant colonies based on the transfer of pRH220. Twenty-nine mutant strains, which showed log2(RTCV) values of less than −2 (i.e., one-fourth of the control), were identified by this method. These 29 mutants, in which the reason for the relatively low conjugation efficiency was not clear, were termed the “down” mutant pool, and third and fourth screenings were performed to determine the reason for this dip in conjugation efficiency (Table 2).
To characterize this mutant pool further, we tried to select factors affecting the transfer from donor to recipient (the third screening step). For this, a donor strain, S17-1 λpir [pJP5603ΔKmR(::GmR CmR leuB-D)], in which RP4 was integrated into the chromosome, was used. The plasmid in this strain has an RP4 oriT and an R6K ori, and its replication is pir-dependent. As a result, the transferred plasmid is non-replicable in the recipient Keio mutants, and so transconjugants, arising as a result of homologous recombination at the leuB-D locus on the recipient chromosome, could be detected. In this assay system, mutants related to plasmid replication were expected to show normal conjugation efficiency, whereas mutants related to the transfer from donor to recipient were expected to show low conjugation efficiency. As a result, 17 mutant strains were isolated as low conjugation efficiency mutants [log2(RTCV) values < −4; Table 2].
In the third screening step, not only mutants related to transfer but also those related to the establishment of Cam resistance were included. To distinguish these two phenotypic differences, we used HB101 [RP4ΔKmR(::GmR)] as the donor strain and assessed for conjugation efficiency using Amp (fourth screening step). As a result, three mutant strains, namely Δrnt, ΔpriA, and ΔdnaT, were isolated as possible mutants affecting transfer from donors to recipients [log2(RTCV) values < −4; Table 2].
Absence of Recipient Mutants Affecting the Transfer
Interestingly, the functions of the deleted genes in the three mutants were ribonuclease (rnt) and DNA replication (priA and dnaT). These functions appear to be unrelated to the donor to recipient transfer. We observed that the appearance of transconjugant colonies in the ΔatpF mutant was delayed and that the Δrnt mutant formed extremely small colonies in one of the triplicate experiments performed during the second screening step, suggesting that the conjugation deficiency of these three mutants might be superficially derived from the extremely slow growth phenotype. In addition, the analyses described above were based on relative conjugation efficiencies calculated compared to a control. Thus, to finally identify true conjugation mutants, the absolute conjugation efficiency values were measured using a more sensitive technique (fifth screening step). It is noteworthy that in the fourth screening step, detection of RP4ΔKmR(::GmR) transfer was assessed using Amp. However, to further eliminate the possibility that the low conjugation efficiency in the three mutants was dependent on Cam or Amp, we used Tet instead, and assessed whether the low conjugation efficiency of these three mutants was superficial and caused by the extremely slow growth of the transconjugants.
The transconjugants were visualized after incubation for 24 h at 37°C using a stereomicroscope, and for 48 h at 37°C by the naked eye, all three mutants showed no reduction in conjugation efficiency (Figure 2A). Thus, these data strongly suggest that the low conjugation efficiency observed was indeed superficial and was mainly caused by the extremely slow growth of the transconjugants. Interestingly, despite the fact that the turbidity was adjusted among the recipient cell suspensions, the ΔpriA and ΔdnaT mutants showed a lower number of recipient colonies, approximately 6 and 7% of the control strains, respectively (Figure 2B). Therefore, the living cell (capable of proliferation) ratio should be low in these two mutant strains.
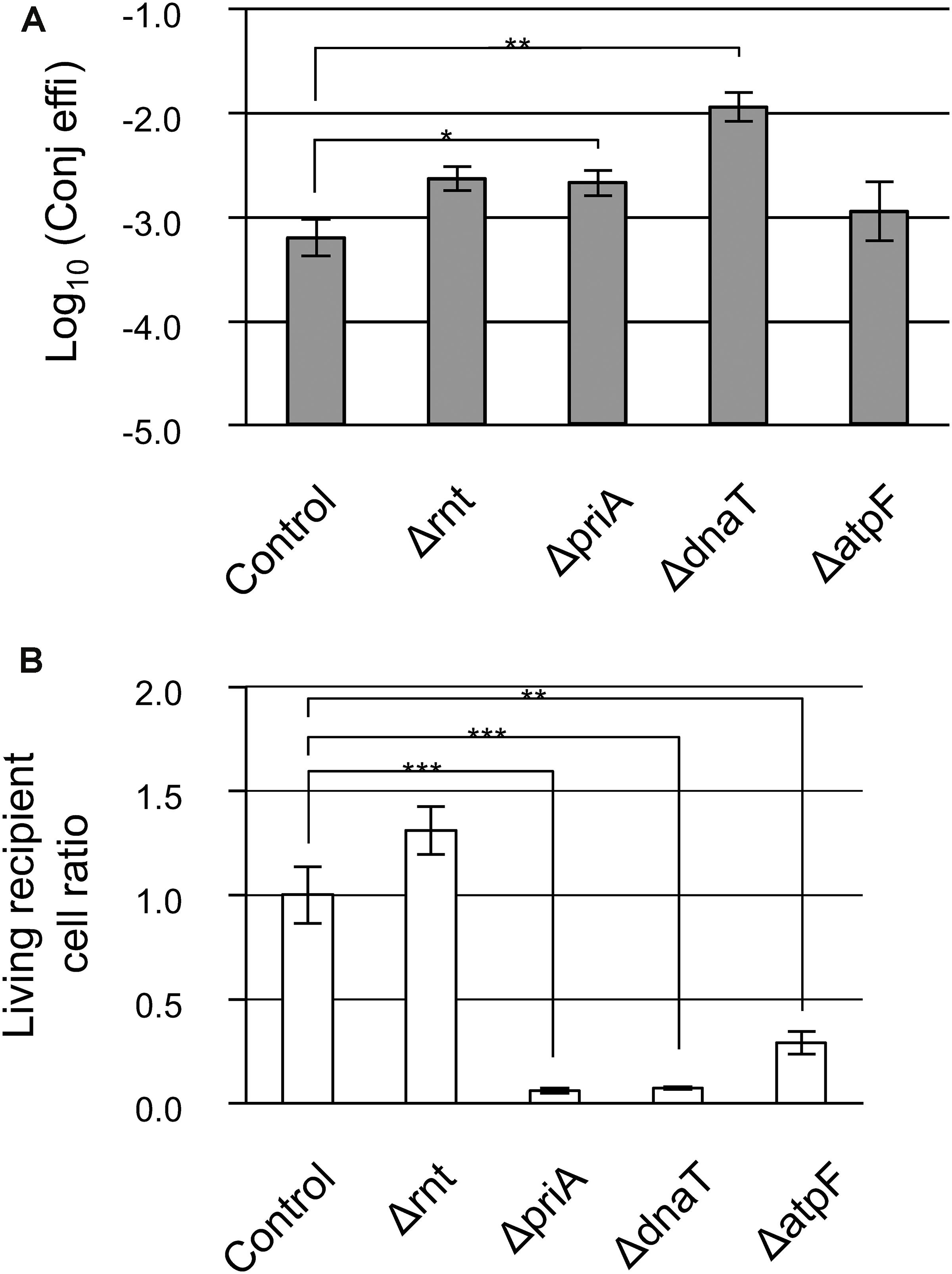
Figure 2. Confirmation analysis of the low conjugation efficiency in Δrnt, ΔpriA, and ΔdnaT mutants. (A) Conjugation efficiency of the candidate mutants. Bars represent the log10 converted values of the conjugation efficiency (transconjugants/recipient cell) and are shown as “log10(Conj effi).” (B) Relative recipient cell ratio of mutants in the conjugation reaction. Data are presented as mean ± standard error (SE). Asterisks indicate statistically significant differences: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (two-tailed t-test). Conjugation experiments for each mutant were performed at least thrice. HB101 (RP4ΔKmR::GmR) was used as the donor, and BW25113 (pBBR122ΔCmR) was used as the control.
Taken together, we conclude that there is no apparent defective recipient mutant for IncP1α conjugal transfer in these Keio mutants, and even if one existed, it appears that it would only have a mild phenotype.
Inhibition of the Establishment of Antibiotic Resistance in a Recipient Cell Could Be an Important Step in Blocking the Spread of Antibiotic Resistance Genes
Next, we looked for the existence of mutants that showed low conjugation efficiency as a result of the inhibition of Cam resistance induced by the transferred resistance gene. Except for ΔpriA and ΔdnaT, mutants that formed no transconjugant colonies in the second screening step were selected. These were ΔihfA, ΔihfB, ΔpncA, ΔacrB, ΔubiH, and ΔdnaK (Table 2).
With the exception of ΔubiH, although the six mutants grew almost normally, their transconjugants tended to show a delay in colony formation. Furthermore, the ΔubiH mutant showed delayed growth both in the recipient and in the transconjugants. Therefore, the incubation period was extended longer than that used during the second and third screenings. Except for the ΔdnaK mutant, an approximately 5–50-fold decrease in conjugation efficiency was observed in these mutants. The ΔdnaK mutant instead showed a lower living cell ratio that was approximately 40% that of the control strain (Figures 3A,B). Among these, while the conjugation efficiency of the control strain was 1.24 × 10–3 transconjugants/recipient, ΔihfA showed the lowest conjugation efficiency (2.24 × 10–5 transconjugants/recipient) and a severe delay in colony formation.
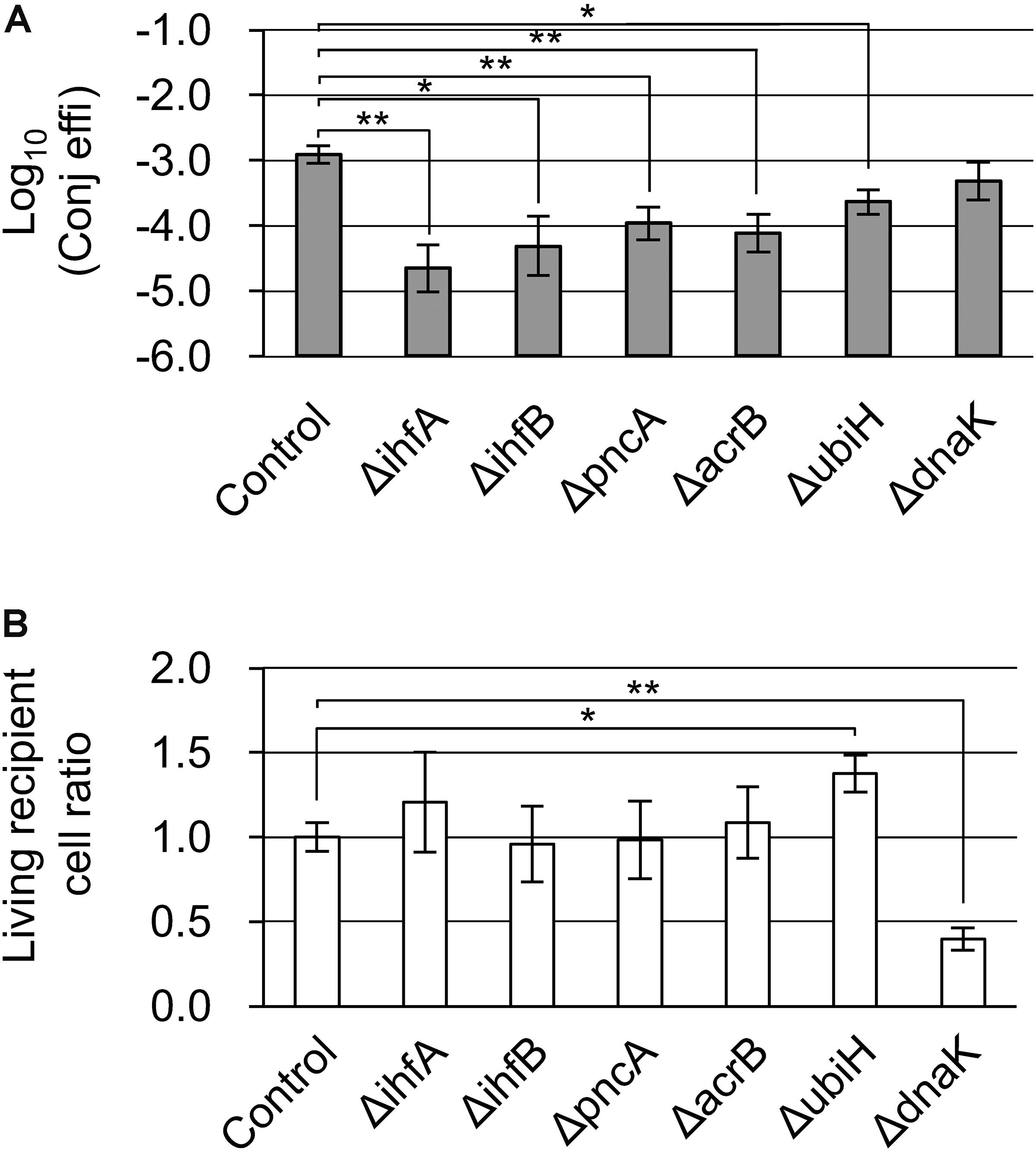
Figure 3. Confirmation analysis of Cam-dependent conjugation deficiency in the ΔihfA, ΔihfB, ΔpncA, ΔacrB, ΔubiH, and ΔdnaK mutants. (A) Conjugation efficiency of the candidate mutants. Bars represent the log10 converted value of the conjugation efficiency (transconjugants/recipient cell) and are shown as “log10(Conj effi).” (B) Relative recipient cell ratio of the mutants in conjugation reaction. Data are presented as mean ± SE. Asterisks indicate statistically significant differences: ∗p < 0.05, ∗∗p < 0.01 (two-tailed t-test). Conjugation experiments in each mutant were performed in quadruplicate. HB101 (pRH220) was used as the donor, and BW25113 (pBBR122ΔCmR) was used as the control.
Discussion
This study examined the methods to identify effective drug targets capable of blocking antibiotic resistance arising from the conjugal transfer. While no mutants defective in transfer, including mutants defective in LPS synthesis isolated in the IncW plasmid analysis were identified, mutants with low viability and capability of establishing resistance against Cam, which was used for transconjugant selection, were isolated.
One important finding is that mutations in recipient factors that enhance plasmid transfer, such as the receptor for the RP4-pilus and factors related to the synthesis of cell surface components, which aid the interaction between donor and recipient cells, were not isolated. At the beginning of this study, we expected that we would isolate the latter factors, although they only have a moderate effect, as reported in a study using the IncW conjugal transfer system (factors for LPS synthesis; Pérez-Mendoza and de la Cruz, 2009). This is because RP4 is a plasmid with a broad host range and is expected to recognize a common structure among gram-negative bacteria. However, only the ΔyciM mutant, which lacked the gene for LPS assembly protein B, remained in the “down” mutant pool. This showed a severe conjugation deficiency in the third screening step (Cam selection) but was quite limited in the fourth screening (Amp selection). Therefore, the LPS assembly protein is not likely to be involved in the plasmid transfer step or the previous steps. We further attempted to reconfirm the conjugation efficiency of the ΔrfaC and Δhns mutants. The rfaC gene is an LPS biosynthetic gene, and ΔrfaC mutant has been reported to show the strongest conjugation deficiency among all the Keio mutants in an IncW plasmid analysis (Pérez-Mendoza and de la Cruz, 2009), while hns was reported to function as a silencer of horizontally acquired genes (Lucchini et al., 2006; Navarre, 2006). As we expected, neither significant increase nor decrease in conjugation efficiency was observed in our IncP1α plasmid transfer analysis (Supplementary Figure S2). Thus, the IncP1α conjugal transfer system recognizes cell surface components differently from that of IncW, and the related gene(s) are probably essential genes whose knockout mutants are not included in Keio collection. As IncP1α type IV secretion system (T4SS) can transfer even to eukaryotic cells, highly conserved and essential surface structure(s) among organisms might be recognized by IncP1α T4SS. In addition, IncP1α and IncW (as Δhns was not isolated from the IncW study) plasmids are adapted to escape from silencing by H-NS. All these results suggest that E. coli cannot escape from being a recipient organism for IncP1α plasmid transfer, and that furthermore E. coli does not possess any positive mechanism for incorporating genetic information by IncP1α plasmid transfer. Therefore, it is impractical to develop drugs that inhibit the transfer step in conjugation by blocking a biological process in the recipient cell.
The Δrnt, ΔpriA, and ΔdnaT mutants were isolated in our screening process. Theoretically, these mutants could be isolated from both the conjugation and transformation mutant screens, as they showed a severe growth deficiency. In fact, Δrnt and ΔpriA mutants were isolated in a genome-wide screening of the cell-to-cell transfer of non-conjugative plasmids (Matsuda et al., 2012). Drugs that inhibit the products of the rnt, priA, and dnaT genes may, therefore, be potential new antibiotics, although they will not repress conjugation.
When Cam was used for the selection of transconjugants, the ΔihfA, ΔihfB, ΔpncA, ΔacrB, and ΔubiH mutants showed decreased conjugation efficiency as well as deficiencies for the transconjugants (Table 2, Figure 3, and Supplementary Figure S3). Based on a previous report by Liu et al. (2010), we focused on 46 Cam-sensitive Keio mutants in advance; however, only the ΔacrB mutant among the five conjugation-deficient mutants and only four mutants in our basic conjugation mutant pool (29 mutants), namely ΔacrB, ΔcydB, ΔyciM, and ΔygcO, were included in the 46 mutants (Liu et al., 2010; Table 2). These results indicated that the majority of Cam-dependent conjugation-deficient mutants were not related to Cam susceptibility in the recipient cells, although such susceptible mutants were significantly enriched in the mutant pool [p = 0.0006 (two-tailed), Fisher’s exact test]. Therefore, although AcrB is known to be a multidrug efflux exporter (Zwama et al., 2018), the majority of genes isolated in our screening method under Cam selection probably function in the establishment of Cam resistance by the cat gene. Developing inhibitory drugs for the function of ihfA, ihfB, pncA, and acrB could potentially not only block the spread of the Cam resistance gene but also maintain the effect of Cam even in the presence of the resistance gene as the transconjugants derived from their knockout mutants tended to show a delay in colony formation. Such drugs could also block the spread of antibiotic resistance mediated by bacterial transduction and transformation. Future studies assessing the applicability of this type of protection method toward other antibiotics, such as fluoroquinolones, carbapenems, aminoglycosides, cephalosporins, and β-lactams with or without β-lactamase inhibitors, are anticipated. Currently, gram-negative Enterobacteriaceae with resistance to colistin, which is conferred by the plasmid-encoded mcr-1 gene, is arising as a threat to human health. Moreover, recent reports have shown the presence of mcr-1 gene in IncP plasmids (Lu et al., 2017; Saavedra et al., 2017; Zhao et al., 2017), adding to the broad transfer range and broad host range of IncP plasmids among gram-negative bacteria, poses a great threat. Hence, blocking the spread of these Enterobacteriaceae is urgently necessary. The data from this study might help to address this serious issue. In addition, the IncP1α transfer-based screening system developed in the present study could be applied for isolation of target genes relating to the establishment of resistance to other antibiotics.
In summary, the six-step genome-wide screening of E. coli recipient factors involved in IncP1α plasmid transfer revealed that blocking the function of an antibiotic resistance gene is a more practical approach than blocking conjugal transfer by targeting recipient factors to prevent the spread of antibiotic resistance genes.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
KM, SY, and KS conceived and designed the experiments. KM performed the experiments. KM analyzed the data. KM, FZ, and KK contributed the reagents and materials. KM and MA wrote the manuscript. KM, FZ, MA, SY, and KK revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported in part by the Japan Society for the Promotion of Science KAKENHI (Grant Number JP16K07200).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our gratitude to the National BioResource Project (NBRP) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, for providing E. coli strains, including the Keio collection and the pJP5603 plasmid vector. We are grateful to Dr. Hiromi Tsubota (Hiroshima University) for his thoughtful discussion on statistical analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02939/full#supplementary-material
References
Alalam, H., Graf, F. E., Palm, M., Abadikha, M., Zackrisson, M., Mattsson, M., et al. (2018). Conjugation factors controlling F-plasmid antibiotic resistance transmission. bioRxiv [preprint]. doi: 10.1101/271254
Amábile-Cuevas, C. F. (2013). Antibiotic resistance: from darwin to lederberg to keynes. Microb. Drug Resist. 19, 73–87. doi: 10.1089/mdr.2012.0115
Asaduzzaman, M. (2018). Antimicrobial resistance: an urgent need for a planetary and ecosystem approach. Lancet Planet. Heal. 2, e99–e100. doi: 10.1016/S2542-5196(18)30019-6
Bengtsson-Palme, J., Boulund, F., Fick, J., Kristiansson, E., and Larsson, D. G. J. (2014). Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front. Microbiol. 5:648. doi: 10.3389/fmicb.2014.00648
Beutin, L., and Achtman, M. (1979). Two Escherichia coli chromosomal cistrons, sfrA and sfrB, which are needed for expression of F factor tra functions. J. Bacteriol. 139, 730–737.
Brown, E. D., and Wright, G. D. (2016). Antibacterial drug discovery in the resistance era. Nature 529, 336–343. doi: 10.1038/nature17042
Datta, N., Hedges, R. W., Shaw, E. J., Sykes, R. B., and Richmond, M. H. (1971). Properties of an R factor from Pseudomonas aeruginosa. J. Bacteriol. 108, 1244–1249. doi: 10.1099/00221287-84-1-199
Davies, M., and Walsh, T. R. (2018). A colistin crisis in India. Lancet Infect. Dis. 18, 256–257. doi: 10.1016/S1473-3099(18)30072-0
Dodsworth, J. A., Li, L., Wei, S., Hedlund, B. P., Leigh, J. A., and de Figueiredo, P. (2010). Interdomain conjugal transfer of DNA from bacteria to archaea. Appl. Environ. Microbiol. 76, 5644–5647. doi: 10.1128/AEM.00967-10
Heinemann, J. A., and Sprague, G. F. (1989). Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340, 205–209. doi: 10.1038/340205a0
Karas, B. J., Diner, R. E., Lefebvre, S. C., McQuaid, J., Phillips, A. P. R., Noddings, C. M., et al. (2015). Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6:6925. doi: 10.1038/ncomms7925
Kato, J., and Katayama, T. (2001). Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20, 4253–4262. doi: 10.1093/emboj/20.15.4253
Lee, E. H., and Kornberg, A. (1991). Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n’ protein. Proc. Natl. Acad. Sci. U.S.A. 88, 3029–3032. doi: 10.1073/pnas.88.8.3029
Liu, A., Tran, L., Becket, E., Lee, K., Chinn, L., Park, E., et al. (2010). Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob. Agents Chemother. 54, 1393–1403. doi: 10.1128/AAC.00906-09
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lu, X., Hu, Y., Luo, M., Zhou, H., Wang, X., Du, Y., et al. (2017). MCR-1.6, a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica serovar typhimurium isolate from a healthy individual. Antimicrob. Agents Chemother. 61:e2632-16. doi: 10.1128/AAC.02632-16
Lucchini, S., Rowley, G., Goldberg, M. D., Hurd, D., Harrison, M., and Hinton, J. C. D. (2006). H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. doi: 10.1371/journal.ppat.0020081
Matsuda, A., Kurono, N., Kawano, C., Shirota, K., Hirabayashi, A., Horino, M., et al. (2012). Genome-wide screen for Escherichia coli genes involved in repressing cell-to-cell transfer of non-conjugative plasmids. Biochem. Biophys. Res. Commun. 428, 445–450. doi: 10.1016/j.bbrc.2012.10.098
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/AAC.01103-16
Moriguchi, K., Edahiro, N., Yamamoto, S., Tanaka, K., Kurata, N., and Suzuki, K. (2013a). Transkingdom genetic transfer from Escherichia coli to Saccharomyces cerevisiae as a simple gene introduction tool. Appl. Environ. Microbiol. 79, 4393–4400. doi: 10.1128/AEM.00770-13
Moriguchi, K., Yamamoto, S., Tanaka, K., Kurata, N., and Suzuki, K. (2013b). Trans-kingdom horizontal DNA transfer from bacteria to yeast is highly plastic due to natural polymorphisms in auxiliary nonessential recipient genes. PLoS One 8:e74590. doi: 10.1371/journal.pone.0074590
Navarre, W. W. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238. doi: 10.1126/science.1128794
Pérez-Mendoza, D., and de la Cruz, F. (2009). Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any? BMC Genomics 10:71. doi: 10.1186/1471-2164-10-71
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Saavedra, S. Y., Diaz, L., Wiesner, M., Correa, A., Alejandra Arévalo, S., Reyes, J., et al. (2017). Genomic and molecular characterization of clinical isolates of Enterobacteriaceae harboring mcr-1 in Colombia, 2002 to 2016. Antimicrob. Agents Chemother. 61:e841-17. doi: 10.1128/AAC.00841-17
Saunders, J. R., and Grinsted, J. (1972). Properties of RP4, an R factor which originated in Pseudomonas aeruginosa S8. J. Bacteriol. 112, 690–696.
Shaffer, C. L., Good, J. A. D., Kumar, S., Krishnan, K. S., Gaddy, J. A., Loh, J. T., et al. (2016). Peptidomimetic small molecules disrupt type IV secretion system activity in diverse bacterial pathogens. mBio 7:e221-16. doi: 10.1128/mBio.00221-16
Skurray, R. A., Hancock, R. E., and Reeves, P. (1974). Con-mutants: class of mutants in Escherichia coli K-12 lacking a major cell wall protein and defective in conjugation and adsorption of a bacteriophage. J. Bacteriol. 119, 726–735.
Starcic, M., Zgur-Bertok, D., Jordi, B. J. A. M., Wösten, M. M. S. M., Gaastra, W., and van Putten, J. P. M. (2003). The cyclic AMP-cyclic AMP receptor protein complex regulates activity of the traJ promoter of the Escherichia coli conjugative plasmid pRK100. J. Bacteriol. 185, 1616–1623. doi: 10.1128/JB.185.5.1616-1623.2003
Thanner, S., Drissner, D., and Walsh, F. (2016). Antimicrobial resistance in agriculture. mBio 7, e2227–e2215. doi: 10.1128/mBio.02227-15
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Watanabe, T., Arai, T., and Hattori, T. (1970). Effects of cell wall polysaccharide on the mating ability of Salmonella typhimurium. Nature 225, 70–71. doi: 10.1038/225070a0
Williams, S. L., and Schildbach, J. F. (2007). TraY and integration host factor oriT binding sites and F conjugal transfer: sequence variations, but not altered spacing, are tolerated. J. Bacteriol. 189, 3813–3823. doi: 10.1128/JB.01783-06
Yamamoto, N., Nakahigashi, K., Nakamichi, T., Yoshino, M., Takai, Y., Touda, Y., et al. (2009). Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5:335. doi: 10.1038/msb.2009.92
Zhao, F., Feng, Y., Lü, X., McNally, A., and Zong, Z. (2017). IncP plasmid carrying colistin resistance gene mcr-1 in Klebsiella pneumoniae from hospital sewage. Antimicrob. Agents Chemother. 61:e2229-16. doi: 10.1128/AAC.02229-16
Zumla, A., Azhar, E. I., Hui, D. S., Shafi, S., Petersen, E., and Memish, Z. A. (2018). Global spread of antibiotic-resistant bacteria and mass-gathering religious events. Lancet Infect. Dis. 18, 488–490. doi: 10.1016/S1473-3099(18)30242-1
Keywords: IncP1α-type plasmid, spread of antibiotic resistance genes, broad host range plasmid, conjugal transfer, genome-wide screening, recipient mutants
Citation: Moriguchi K, Zoolkefli FIRM, Abe M, Kiyokawa K, Yamamoto S and Suzuki K (2020) Targeting Antibiotic Resistance Genes Is a Better Approach to Block Acquisition of Antibiotic Resistance Than Blocking Conjugal Transfer by Recipient Cells: A Genome-Wide Screening in Escherichia coli. Front. Microbiol. 10:2939. doi: 10.3389/fmicb.2019.02939
Received: 26 September 2019; Accepted: 06 December 2019;
Published: 08 January 2020.
Edited by:
Koji Yahara, National Institute of Infectious Diseases (NIID), JapanReviewed by:
Shizuo Kayama, National Institute of Infectious Diseases (NIID), JapanKoichi Tanimoto, Gunma University, Japan
Copyright © 2020 Moriguchi, Zoolkefli, Abe, Kiyokawa, Yamamoto and Suzuki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuki Moriguchi, a21vcmlndWNAaGlyb3NoaW1hLXUuYWMuanA=
 Kazuki Moriguchi
Kazuki Moriguchi Fatin Iffah Rasyiqah Mohamad Zoolkefli
Fatin Iffah Rasyiqah Mohamad Zoolkefli Masanobu Abe3
Masanobu Abe3 Katsunori Suzuki
Katsunori Suzuki