
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 14 January 2020
Sec. Microbiotechnology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02937
This article is part of the Research TopicDevelopment and Application of Molecular Technologies for Monitoring Microbial Contaminants in WaterView all 6 articles
Hepatitis E virus (HEV), particularly zoonotic genotype 3, is present in environmental waters worldwide, especially in industrialized countries. Thus, monitoring the presence of HEV in wastewater treatment plants (WWTPs) is an emerging topic due to the importance of reusing water on a global level. Given the limited data, this study aimed to monitor the occurrence of HEV in influent and effluent water in waste- and drinking-water treatment plants (WWTPs and DWTPs). To this end, different procedures to concentrate HEV in influent and effluent water from WWTPs and DWTPs were initially evaluated. The evaluated procedures resulted in average HEV recoveries of 15.2, 19.9, and 16.9% in influent, effluent, and drinking water samples, respectively, with detection limits ranging from 103 to 104 international units (IU)/L. Then, a one-year pilot study was performed to evaluate the performance of the selected concentration method coupled with three RT-qPCR assays in influent and effluent water samples from four different WWTPs. HEV prevalence in influent water varied based on both the RT-qPCR assay and WWTP, while HEV was not detected in effluent water samples. In addition, HEV prevalence using only RT-qPCR3 was evaluated in influent (n = 62) and effluent samples (n = 52) from four WWTPs as well as influent (n = 28) and effluent (n = 28) waters from two DWTPs. The present study demonstrated that HEV circulated in the Valencian region at around 30.65% with average concentrations of 6.3 × 103 IU/L. HEV was only detected in influent wastewater samples, effluent samples from WWTPs and influent and effluent samples from DWTPs were negative. However, given that the infective dose in waterborne epidemics settings is not yet known and the low sensibility of the assay, unfortunately, no direct conclusion could be achieved on the risk assessment of environmental contamination.
Hepatitis E virus (HEV) is a human enteric virus that mainly causes self-limiting acute viral hepatitis. According to the World Health Organization, 20 million cases of hepatitis E and 44,000 deaths occur worldwide every year1. HEV is an emerging foodborne pathogen (Harrison and DiCaprio, 2018), and the incidence of confirmed cases in the European Union has steadily increased over the last decade (Kupferschmidt, 2016; Ricci et al., 2017).
Hepatitis E infections are caused by a small (27–34 nm), positive-sense, single-stranded RNA virus (approx. 7.2 kb size) that belongs to the Hepeviridae family (Sooryanarain and Meng, 2019; Van der Poel and Rzezutka, 2019). HEV is excreted in feces as non-enveloped virions but circulates in the blood in a membrane-associated, quasi-enveloped form (Yin et al., 2016). HEV is classified into eight genotypes, of which genotype 1 (G1) and G2 are specific to humans. HEV G3, G4, and G7 are zoonotic genotypes that infect humans and animals and have been isolated in different animal species, especially in pigs (Van der Poel, 2014; Sooryanarain and Meng, 2019). The different HEV genotypes have different geographical distributions2. For example, HEV G1 and G2 are predominantly transmitted via the fecal-oral route in Asia, Africa, and Central America, usually through the consumption of contaminated drinking water (Khuroo et al., 2016; Van der Poel and Rzezutka, 2019). In contrast, HEV G3 and G4 are endemic in industrialized countries and transmitted primarily via the consumption of animal meats or direct contact with infected animals (Sooryanarain and Meng, 2019).
Hepatitis E virus transmission to humans through water has been largely demonstrated for HEV G1 and G2, primarily in developing countries, but transmission is also suspected for the zoonotic genotypes since HEV G3 and G4 have been detected in different types of environmental waters (Miura et al., 2016; Haramoto et al., 2018; Fenaux et al., 2019; Van der Poel and Rzezutka, 2019). Given the authorities’ concerns, several surveillance studies conducted in different geographic regions have assessed the presence of HEV in urban wastewater with highly variable occurrence (Fenaux et al., 2019). However, few studies have focused on effluent wastewater or drinking water (Fenaux et al., 2019; Purpari et al., 2019; Van der Poel and Rzezutka, 2019). In addition, available data must be interpreted with caution due to the lack of standardized HEV detection procedures and the substantial differences among studies in terms of volume of samples, concentration methods, and RT-qPCR (Fenaux et al., 2019).
To overcome these challenges, this study initially evaluated the performances of different concentration methods, RNA extraction kits, and RT-qPCR protocols in detecting and quantifying HEV in influent and effluent wastewater samples as well as in drinking water samples (Graphical Abstract). After method evaluation, the presence of HEV was monitored in influent and effluent waters from four municipal wastewater treatment plants (WWTPs) and two drinking water treatment plants (DWTPs) in the metropolitan region of Valencia (Spain).
Fecal sample containing HEV genotype 3f was used in the study. Fecal sample (10% wt/vol) was suspended in phosphate-buffered saline (PBS) containing 2 M NaNO3 (Panreac), 1% beef extract (Conda), and 0.1% Triton X-100 (Thermo Fisher Scientific) (pH 7.2). The mix was then vigorously vortexed and centrifuged at 1,000 × g for 5 min to obtain a final 10% (wt/vol) fecal suspension. The supernatant was stored at −80°C in aliquots. The first WHO international standard for HEV nucleic acid amplification technique (NAT)-based assays (code 6329/10) was purchased from Paul-Ehrlich-Institut (Germany). This standard corresponds to HEV genotype 3a positive plasma measured in international units (IU) and containing 250,000 IU/mL and it was used for RT-qPCR quantification, as detailed below (Baylis et al., 2013). Mengovirus (MgV) vMC0 (CECT 100000) was used as a process control.
Influent and effluent water samples were collected from four WWTPs and two DWTPs located in the Valencian region, Spain (Figure 1). The collected samples were transferred to the laboratory immediately, and subsequently concentrated as described below.
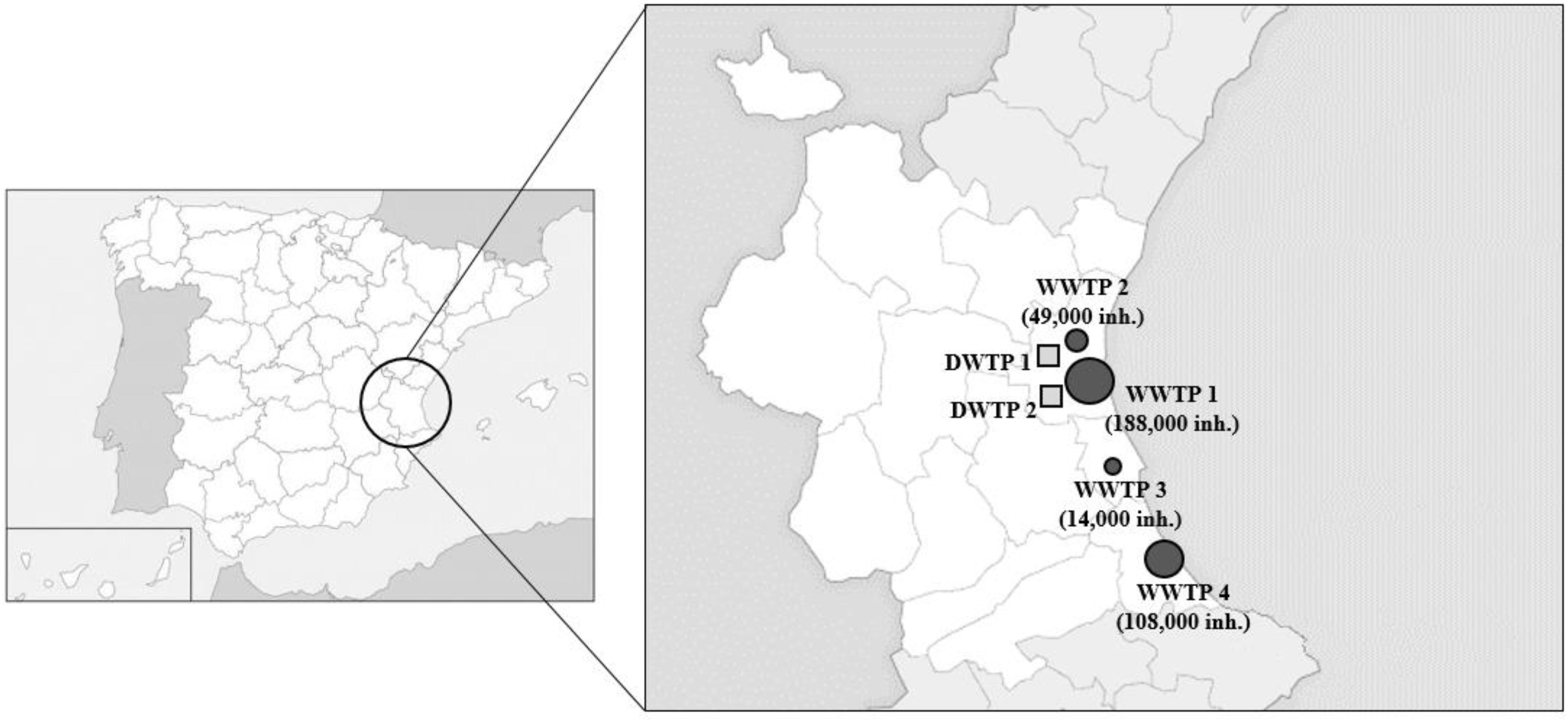
Figure 1. Map of the sampling locations. WWTP, wastewater treatment plant (squares); DWTP, drinking water treatment plants (circles). Symbols are sized according to the number of inhabitants.
Influent water samples collected from WWTP1 were artificially inoculated with 5 log IU/L of HEV and 7 log PCRU/L of MgV, spiked as process control.
Initially, the performance of two concentration methods was evaluated: an ultracentrifugation-based method (referred as UC) and an aluminum hydroxide adsorption-precipitation method (referred as Al). For UC method, 35 mL of influent water were centrifuged at 141,000 × g for 2 h 30 min at 4°C. The pellet was then incubated on ice for 30 min with 5 mL of 0.25 N glycine buffer (pH 9.5) and then the solution neutralized with 19 mL of PBS. Suspended solids were removed by centrifugation at 12,000 × g for 15 min. Viruses were finally recovered by ultracentrifugation at 505,000 × g for 1 h at 4°C and subsequently eluted in 1 mL of PBS (Rodríguez-Díaz et al., 2009).
For Al method, 35 mL of influent water were adjusted to pH 6.0 and Al(OH)3 precipitate formed by adding 1 part 0.9N AlCl3 solution to 100 parts of sample. The pH was readjusted to 6.0 and sample mixed using an orbital shaker at 150 rpm for 15 min at room temperature. Then, viruses were collected by centrifugation at 1,700 × g for 20 min. The pellet was resuspended in 1.75 mL of 3% beef extract pH 7.4, and samples were shaken for 10 min at 150 rpm. Concentrate was recovered by centrifugation at 1,900 × g for 30 min and pellet resuspended in 1 mL of PBS (AAVV, 2018; Randazzo et al., 2019) and stored at −80°C. Experiments were performed in duplicate.
The limit of detection (LoD95%) was obtained by artificially inoculating HEV at 5, 4, 3, and 2 log IU/L in 35 mL for influent water or in 200 mL for effluent water.
Samples were spiked with MgV (7 log PCRU/L) as a process control. Then, virus particles were concentrated by the previously described Al method and RNA extracted using two kits and analyzed by RT-qPCR1 and RT-qPCR2 (detailed below). For each method and contamination level, a PBS sample without influent or effluent water were included to assess potential matrix effects. Experiments were performed in duplicate by concentrating two independent samples for each condition tested.
Drinking water samples (20 L) were artificially inoculated with HEV at 7, 6, 5, and 4 log IU/L. In addition, MgV was spiked and used as process control. HEV primary concentration was performed by a Dead End Hollow Fiber Ultrafiltration (DEUF) using single-use Rexeed-25A dialysis filters (Asahi Kasei Medical Co., Ltd.) with a molecular mass cutoff of 30 kDa, a surface area of 2.5 m2, a fiber inner diameter of 185 μm and a priming volume of 137 mL (Borgmästars et al., 2017). A peristaltic pump (model FH100, Thermo Fisher Scientific) was used for all experiments.
In brief, the Rexeed-25A filters were blocked with 6.25% fetal bovine serum by circulating the blocking solution for 5 min followed by 2 h incubation at room temperature. Afterward, filter was properly assembled and flushed with 1 L of sterile water at 2,900 mL/min and then with the 20 L of inoculated drinking water samples. Subsequently, filter was assembled for a back-flush elution with 500 mL of sterile water supplemented with 0.001% Antifoam, 0.01% NaPP, and 0.01% Tween 80.
Two different approaches were evaluated for secondary concentration: a precipitation with polyethylene glycol (PEG) and a centrifuge filtration procedure by Amicon® Ultra-15 tubes (Merck Millipore Ltd.). For PEG precipitation, 300 mL of concentrate were transferred to two 250 mL centrifugation tubes, 150 mL of eluate for each tube. Then, 2 g of beef extract (Laboratorio Conda) were added into each tube and shaken until completely dissolved. Then, 50 mL of PEG/NaCl 5× were added and incubated overnight at 4°C in an orbital shaker at 150 rpm. Finally, the samples were centrifuged at 10,000 × g for 30 min and resulting pellets resuspended in 1 mL PBS.
For secondary concentration by centrifuge filtration, 15 mL volume was added to Amicon® Ultra-15 tube and concentrated via centrifugation at 4,000 × g for 15 min. This step was repeated three times using the same ultrafilter for a total of 45 mL sample processed. Then the concentrated viruses were recovered in 1 mL PBS. The viral concentrates were stored at −80°C until further processed. Experiments were performed in duplicate by concentrating two independent samples for each condition tested.
Two different commercial extraction kits were used for RNA extraction. The extraction using the NucleoSpin®RNA virus kit (Macherey-Nagel GmbH & Co.) (referred as MN) was performed according to the manufacturer’s instructions with some modifications. Briefly, 150 μL of each concentrated sample was mixed with 25 μL Plant RNA Isolation Aid (Ambion) and 600 μL of lysis buffer from the NucleoSpin® RNA virus kit and subjected to pulse-vortexing for 1 min. Afterward, the homogenate was centrifuged for 5 min at 10,000 × g to remove the debris. The supernatant was subsequently processed according to the manufacturer’s instructions. An additional extraction was carried out using the NucliSENS® miniMAG® system (BioMérieux SA) (referred as NS) and according to manufacturer instructions. In particular, the sample volume was 500 μL and the elution volume was 100 μL. Resultant RNA was analyzed using the RNA UltraSense One-Step kit (Invitrogen SA) and RT-qPCR performed as described in Schlosser et al. (2014) for HEV (referred as RT-qPCR1) and as in ISO 15216-1:2017 for MgV (Supplementary Table 5). For both RT-qPCR assays, undiluted and 1/10 diluted RNA was tested to check for RT-qPCR inhibitors.
Moreover, RNAs were also quantified using the ceeramTOOLS® Hepatitis E Virus Detection KHEV commercial kit (BioMérieux SA) (referred as RT-qPCR2) provided with an internal amplification control.
In all experiments, all samples were run in duplicate and different controls were used, including negative process, extraction and RT-qPCR controls, and controls for extraction efficiency.
Hepatitis E virus was quantified by plotting the quantification cycles (Cqs) to an external standard curve built with the International Standard WHO HEV RNA (code 6329/10). Moreover, extraction efficiencies were calculated and used as quality assurance parameters according to ISO 15216-1:2017 (2017).
A total of 62 influent and 52 effluent wastewater samples were investigated for the occurrence of HEV as hereafter detailed.
Initially, influent (n = 32) and effluent (n = 32) water samples were collected from four municipal WWTPs located in the Valencian region (eastern Spain), from May 2018 to March 2019 (Figure 1). Two-hundred milliliters of influent and effluent water samples were processed using the Al procedure. Mengovirus was used as process control. RNA extraction was performed using the NucleoSpin® RNA Virus kit (MN kit) and HEV RNA quantified by both RT-qPCR1 and RT-qPCR2. In addition, RNA samples were analyzed by a third RT-qPCR assay (referred as RT-qPCR3, Supplementary Table 5; Girón-Callejas et al., 2015). Additional influent (n = 30) and effluent (n = 20) samples were further collected in June, August, and October 2018 and from April 2019 to August 2019 and analyzed by RT-qPCR3 only.
A total of 28 influent and 28 effluent water samples were collected from two municipal DWTPs (Figure 1) in October and November 2018. The samples were maintained under refrigeration (4°C) for transportation and processed within 24 h. Water samples (20 L) were dechlorinated with sodium thiosulphate (10% wt/vol) after collection, added with mengovirus and concentrated using the Rexeed-25A filters and PEG precipitation, as detailed above. Resultant RNA was extracted by the NucleoSpin® RNA Virus kit (MN kit) and detected by RT-qPCR3.
Results were statistically analyzed and significance of differences was determined on the ranks with a one-way analysis of variance (ANOVA) and Tukey’s multiple comparison tests. In all cases, a value of p < 0.05 was deemed significant. Spearman’s rank-order correlation coefficient (ρS) was determined between inhabitants and HEV positive samples by using Statistica software (StatSoft Inc., Tulsa, OK, United States). The estimated probability of detection with 95% confidence (LoD95%) was calculated by using the PODLOD calculation program (version9) (Wilrich and Wilrich, 2009) for all water samples.
Fecal samples were collected at Hospital Clínico Universitario de Valencia (Valencia, Spain). The study was approved by the Comisión de Ética en Investigación Experimental of the University of Valencia (Spain), in accordance with the World Medical Association’s Declaration of Helsinki and the relevant European and Spanish guidelines and regulations.
One major limitation in understanding HEV transmission in contaminated waters is the lack of standardized and validated methods (Ricci et al., 2017). Thus, to provide data on the performance of the HEV detection methods in environmental waters, an ultracentrifugation-based protocol (UC) was compared to an aluminum precipitation procedure (Al) using artificially inoculated influent water samples. The mean HEV recoveries obtained with the UC concentration procedure ranged from 7.98 to 16.83% using MN kit and from 10.24 to 55.08% using NS kit. The Al procedure resulted in mean HEV recovery values ranging from 7.00 to 20.54% and from 10.18 to 90.19% using MN and NS kits, respectively (Table 1).

Table 1. Performance of concentration methods (ultracentrifugation and aluminum precipitation), RNA extraction kits and RT-qPCR assays for HEV detection in artificially inoculated influent water samples.
The Al procedure was selected for the determination of LoD95% since an ultracentrifuge is not required. To determine LoD95%, influent water was artificially inoculated with MgV together with four levels of HEV and samples concentrated according to the Al procedure. RNA extraction from concentrates was performed using MN and NS kits and subsequently analyzed by RT-qPCR1 and RT-qPCR2.
The mean HEV recovery values obtained using the MN and NS kits ranged from 8.81 to 36.8% and from 8.90 to 41.45%, respectively (Figure 2 and Supplementary Table 1), and no statistically significant differences were observed (P > 0.05). On average, LoD95% was 2.9 × 105 IU/L for MN kit and 2.2 × 106 IU/L for NS kit, calculated according to Wilrich and Wilrich (2009). Accordingly, LoD95% increased approximately 10-fold when NS was compared to MN extraction procedure. Overall, the MN kit combined with RT-qPCR1 provided the best LoD95%, which was similar to or slightly higher than those previously reported for other enteric viruses in influent waters (approx. 104–105 genome copies/L) (Nordgren et al., 2009; Randazzo et al., 2019).
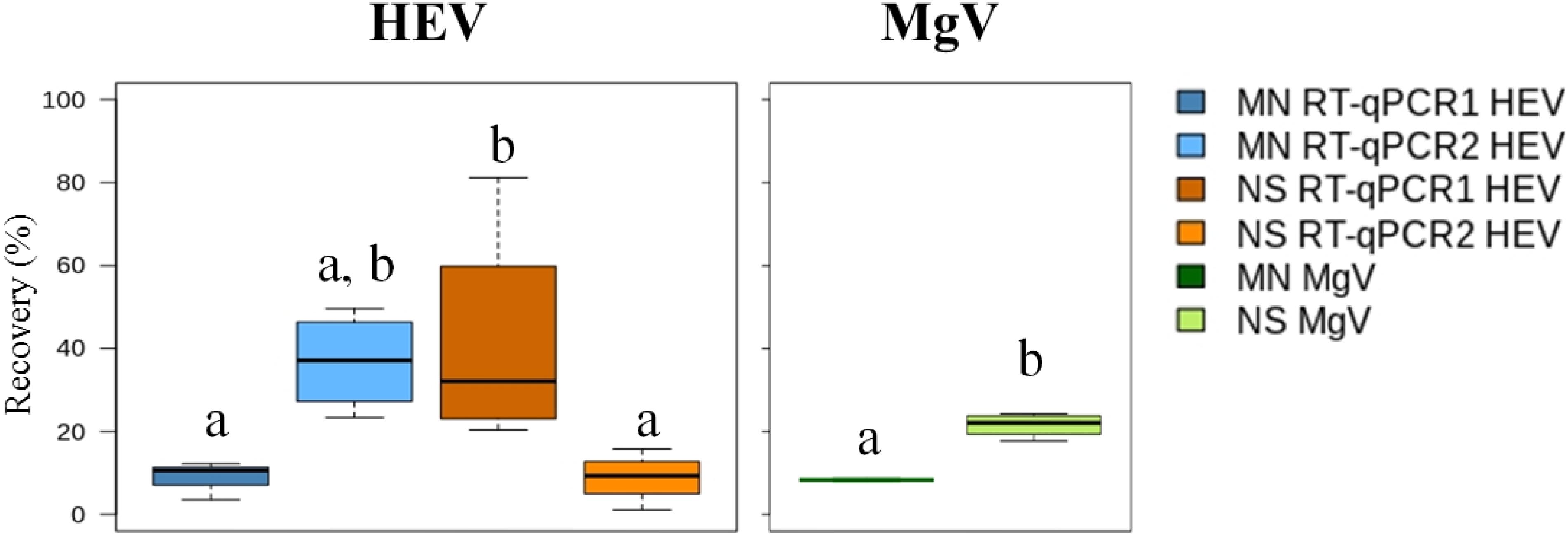
Figure 2. Median HEV recovery (%) in influent water samples using the aluminum protocol and comparing two extraction kits and two RT-qPCRs assays. MN: NucleoSpin®RNA virus kit (Macherey-Nagel GmbH & Co.); NS: NucliSENS® miniMag® system (BioMérieux SA); RT-qPCR1: Schlosser et al., 2014; RT-qPCR2: ceeramTOOLS® Hepatitis E Virus Detection KHEV kit (BioMérieux SA). Within each virus, different letters denote significant differences among methods (P < 0.05).
The MgV recovered using the MN and NS kits ranged from 7.92 to 8.72% (8.34% mean) and from 17.76 to 24.29% (21.56% mean), respectively (Figure 2 and Supplementary Table 1). These results support previously reported MgV recoveries in influent waters (Miura et al., 2016). Because only 35 mL of sample are needed for the analysis and ultracentrifugation is not required, the procedure is a potential alternative method for routine influent water screening.
Few studies over the last decade have assessed the presence of HEV in effluent water samples due in part to the lack of validated procedures (Fenaux et al., 2019). Therefore, the performance of the Al concentration method was analyzed using effluent water samples that were collected downstream from WWTP1 and artificially spiked with four levels of HEV and with MgV, as process control. The MN and NS extraction kits and RT-qPCR1 and RT-qPCR2 were used for sample processing. Viral recovery and HEV LoD95% were determined and the results are shown in Figure 3 and Supplementary Table 2.
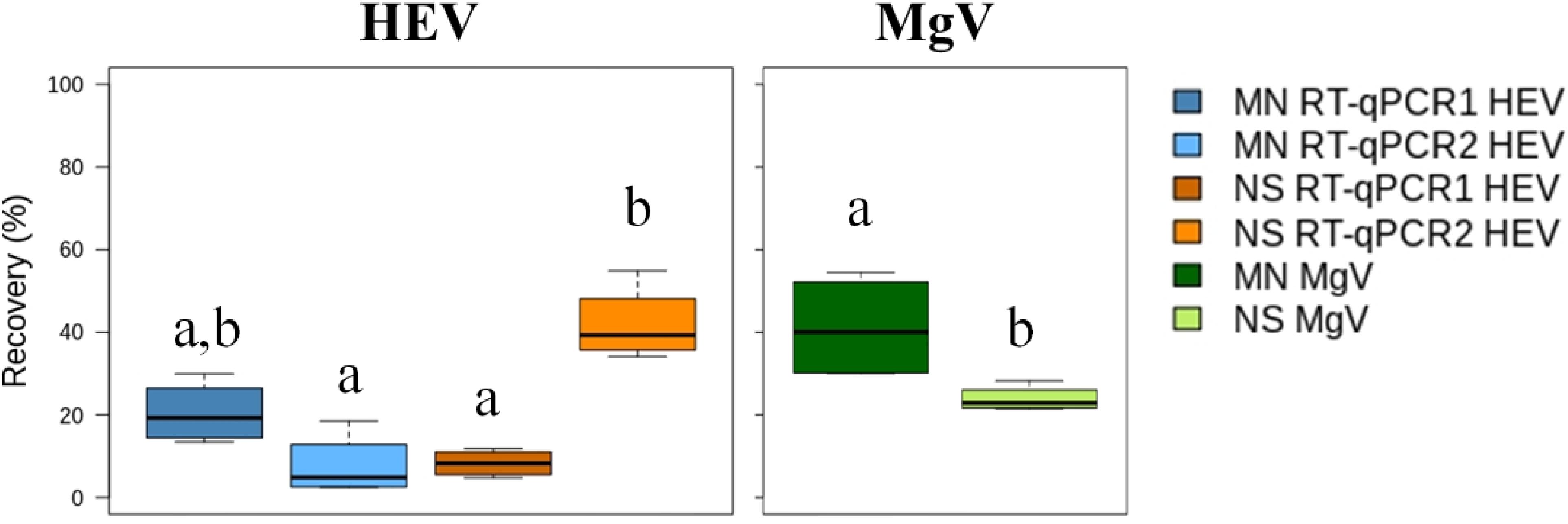
Figure 3. Median HEV recovery (%) in effluent water samples using the aluminum protocol and comparing two extraction kits and two RT-qPCRs assays. MN: NucleoSpin®RNA virus kit (Macherey-Nagel GmbH & Co.); NS: NucliSENS® miniMag® system (BioMérieux SA); RT-qPCR1: Schlosser et al., 2014; RT-qPCR2: ceeramTOOLS® Hepatitis E Virus Detection KHEV kit (BioMérieux SA). Within each virus, different letters denote significant differences among methods (P < 0.05).
MgV recoveries using the MN and NS kit ranged from 30.08 to 54.50% (41.17% mean) and from 21.52 to 28.32% (23.90% mean), respectively, which are slightly higher than the 8–13% recovery rates of cross-flow ultrafiltration reported previously (Miura et al., 2016). The mean recovery of HEV ranged from 8.33 to 30.01% using the MN kit and from 7.72 to 41.90% with the NS kit. LoD95% was 1.25 × 104 IU/L regardless of the RNA extraction and RT-qPCR used (Supplementary Table 2).
Prior to concentration, 20 L tap water samples were added with four different concentrations of HEV, and MgV, as a whole process control. Primary virus concentration was performed using DEUF with Rexeed-25A filters, resulting in an average eluate volume of 605 ± 38.22 mL. Then, the secondary concentration was evaluated comparing in parallel a PEG precipitation and a centrifuge filtration. DEUF ultrafiltration combined with PEG precipitation and MN kit resulted in HEV mean recovery of 16.6 to 36.6%, while recoveries ranged from 7.2 to 8.3% for NS kit (Figure 4A and Supplementary Table 3). The centrifuge filtration procedure using MN and NS resulted in mean HEV recovery values ranging from 1.8 to 4.9% and 23.7 to 35.7%, respectively (Figure 4B and Supplementary Table 4). A minimum recovery rate of 1% MgV was achieved from all procedures, validating the results. For all the tested procedures, the HEV LoD95% in drinking water was of 6.2 × 103 IU/L (Supplementary Tables 3, 4).
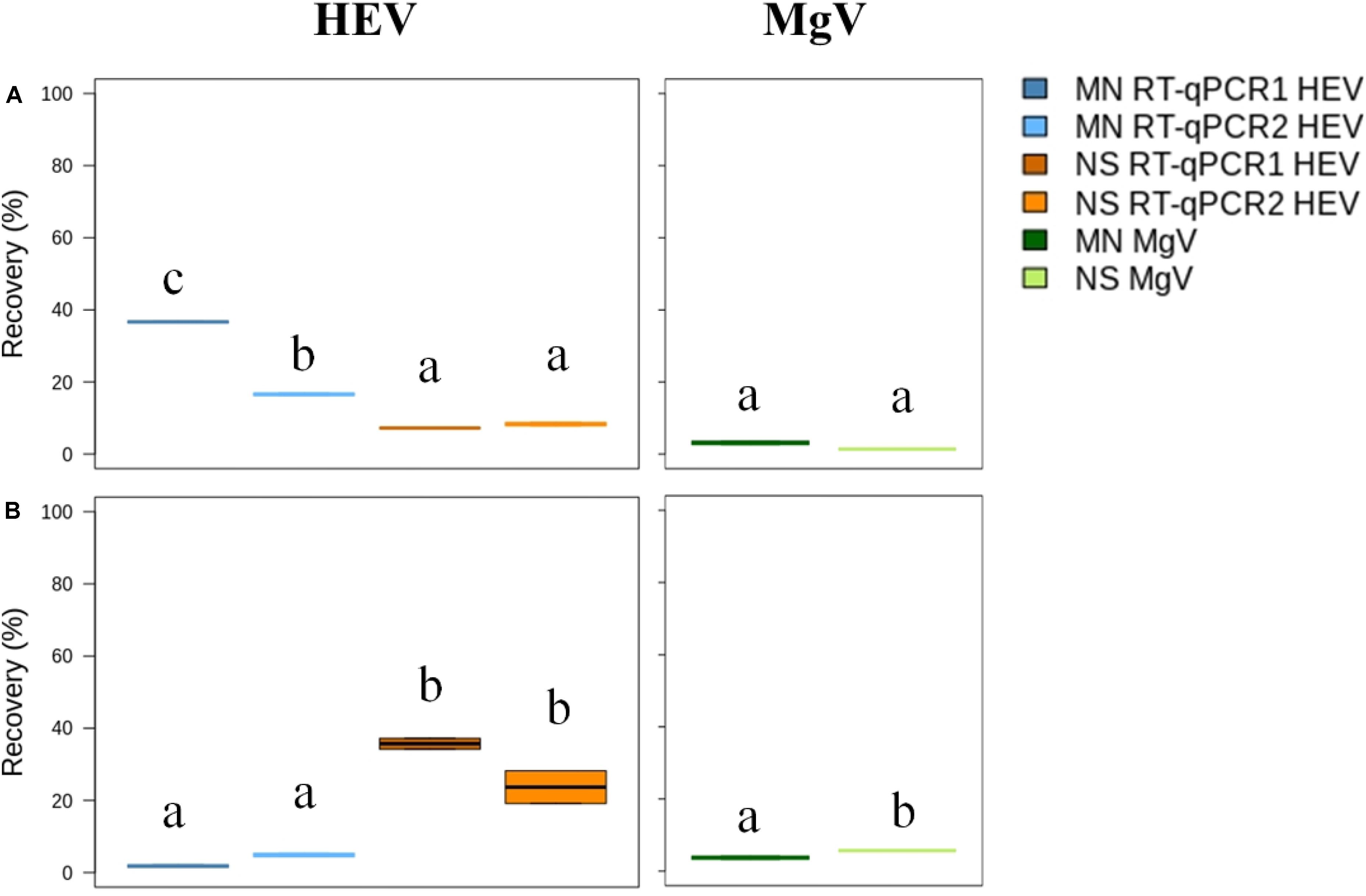
Figure 4. Median HEV recovery (%) in drinking water by Rexeed 25AX ultrafiltration followed by precipitation with polyethylene glycol (A) or centrifuge filtration with Amicon filters (B). MN: NucleoSpin®RNA virus kit (Macherey-Nagel GmbH & Co.); NS: NucliSENS® miniMag® system (BioMérieux SA); RT-qPCR1: Schlosser et al., 2014; RT-qPCR2: ceeramTOOLS® Hepatitis E Virus Detection KHEV kit (BioMérieux SA). Within each virus, different letters denote significant differences among methods (P < 0.05).
A lack of information on HEV viral loads before and after treatments applied in WWTPs has been identified (Fenaux et al., 2019). In the present study, a total of 64 samples were collected upstream (n = 32) and downstream (n = 32) of four WWTPs, and these samples were concentrated according to the Al procedure combined with the MN kit and analyzed by different RT-qPCRs assays. To improve the sensibility of the RT-qPCR assays, the initial 35 mL influent water sample volume was increased to 200 mL. Initially, two different RT-qPCRs were applied to assess HEV occurrence. Surprisingly, RT-qPCR1 showed a limited number of positives compared to RT-qPCR2 (Figure 5) despite a previous study reported similar performance of these assays in influent water samples (Randazzo et al., 2018). Suspecting that a different HEV genotype was circulating, a third RT-qPCR assay was included in the study. In particular, a method widely used in clinical and environmental virology firstly described by Jothikumar et al. (2006) and modified by Girón-Callejas et al. (2015) (RT-qPCR3) was applied to retest samples. All the samples had a minimum recovery rate of 1% MgV, validating the results. Overall, out of 32 influent water samples, 12 were positive for at least one of the three HEV RT-qPCR assays, and an overall HEV prevalence of 37.5% was found. Different numbers of positive samples and different prevalence rates were recorded during the comparison of the three RT-qPCR assays (Figure 5). In particular, prevalence rates of 12.5, 28.5, and 37.5% in influent waters were recorded for the RT-qPCR1, RT-qPCR2, and RT-qPCR3 assays, respectively. Although RT-qPCR1 fail to detect HEV in several samples, lower Cq values were observed in samples collected from January 2019 on (Figure 5). The observed differences may be due to HEV genotype variability. Unfortunately, conducted genotyping analyses did not solve the question because of the negative results, likely due to the low genome titers in the samples. Therefore, due to the high variability of the HEV genotypes (Smith et al., 2013, 2016), the RT-qPCR assays used for environmental analyses must be carefully checked to avoid false negative results.
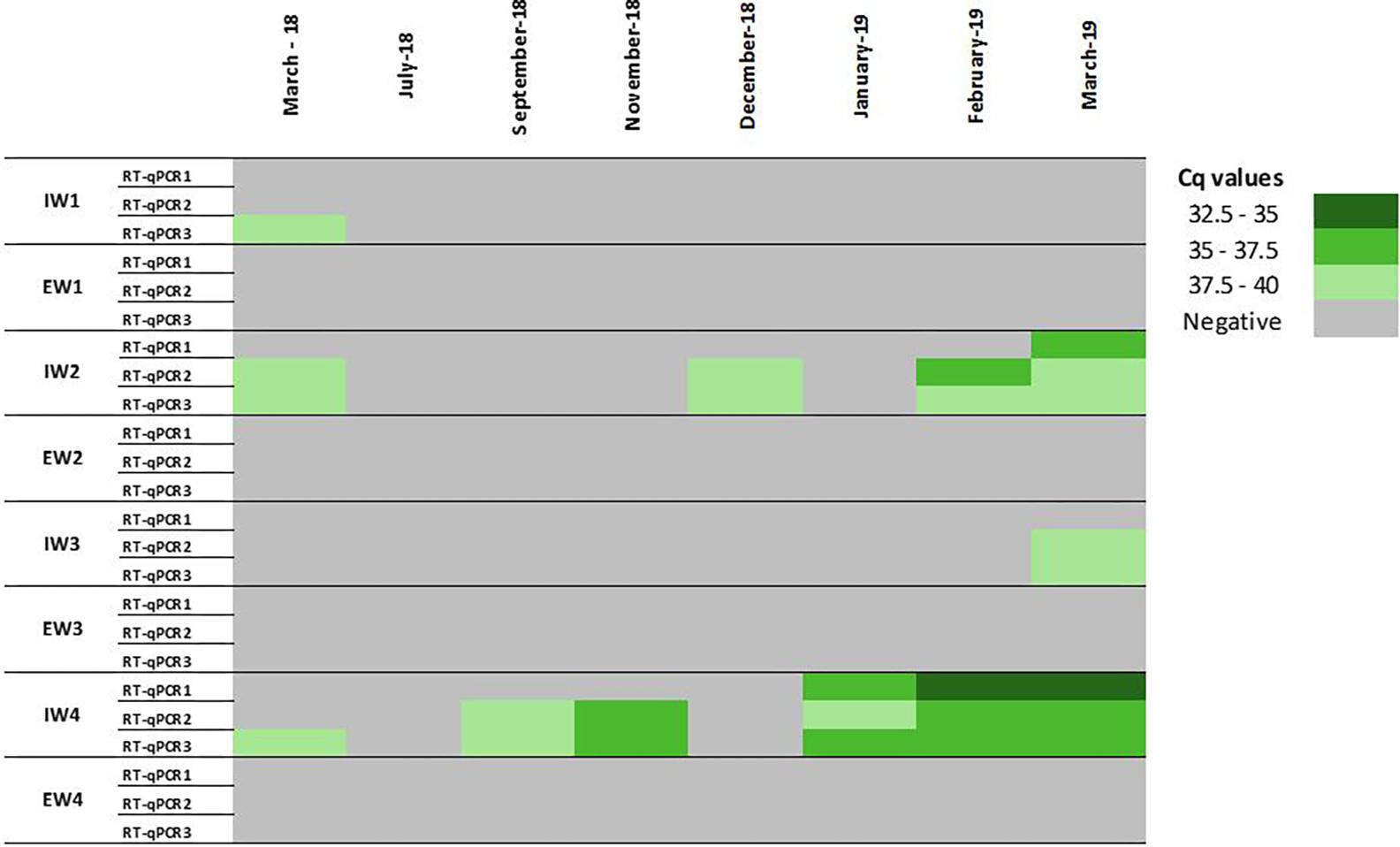
Figure 5. Occurrence of HEV in influent (IW) and effluent (EW) waters by comparing three RT-qPCR assays. RT-qPCR1: Schlosser et al. (2014); RT-qPCR2: ceeramTOOLS Hepatitis E Virus Detection KHEV kit (BioMérieux SA); RT-qPCR3: Girón-Callejas et al. (2015) (cf., Supplementary Table 5 for details of assays).
Data about the occurrence of HEV in Spanish wastewaters are scarce, therefore the number of influent and effluent samples were expanded and 62 influent and 52 effluent water samples were analyzed by RT-qPCR3 (Table 2). In the current study HEV is widely disseminated (30.65%) in Valencian influent waters even though the prevalence rates among WWTPs varies widely (Supplementary Figure 1). For example, WWTP4 had a prevalence rate of 73.33% (11/15) using the RT-qPCR3 assay. As a public health concern, this WWTP receives domestic sewage from several municipalities, accounting for 108,000 inhabitants, even though we cannot exclude streams from pig farms or other agricultural run-offs. In contrast, WWTP1 (14,000 inhabitants) and WWTP2 (188,000 inhabitants) showed only 12.5 and 13.33% prevalence, respectively. These data show no correlation between HEV prevalence and the number of inhabitants served by WWTPs (ρS = 0.26).
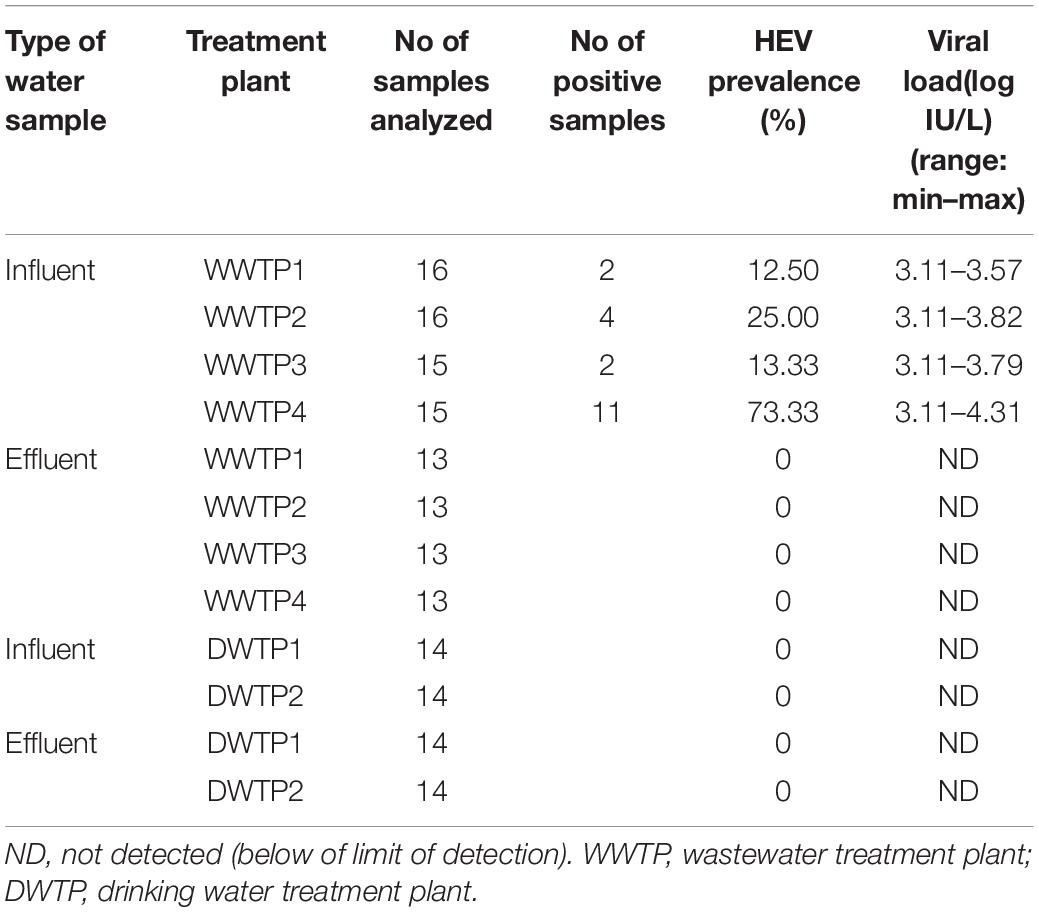
Table 2. Prevalence and HEV loads (IU/L) from four WWTPs (n = 114) and two drinking water treatment plants (n = 56) using RT-qPCR3.
Studies conducted in Barcelona (Spain) have shown similar prevalence (from 13.5 to 43.5% in influent waters, with absence or low detection of HEV in effluent waters (Clemente-Casares et al., 2003; Rodriguez-Manzano et al., 2010; Rusiñol et al., 2015).
The present study showed HEV contamination in influent waters ranging from approximately 1.3 × 103–3.5 × 104 IU/L using the RT-qPCR3 assay (Table 2), which is consistent with previously reported levels (Fenaux et al., 2019). HEV genomes were not detected in effluent waters (Table 2). These results are consistent with most of the studies published in Europe (Fenaux et al., 2019), even those done after a confirmed outbreak (Miura et al., 2016). This suggests that treatments applied at WWTPs (Supplementary Figure 1) are efficient in removing HEV despite the fact that a reduction of 1–2 log would result in concentrations below the LoD95%. Thus, further improvements are needed to increase the sensitivity of the methods applied for virus concentration in effluent waters.
Additionally, a total of 56 samples were collected upstream (n = 28) and downstream (n = 28) of two DWTPs, and 20 L water samples were concentrated by DEUF using Rexeed-25A filters combined with PEG precipitation, the MN kit and analyzed by RT-qPCR3. None of the influent and effluent samples were positive for HEV despite all the samples had a minimum recovery rate of 1% MgV (Table 2).
Hepatitis E virus is considered an emerging pathogen in industrialized countries, especially in Europe, and analytical procedures for estimating HEV concentrations in water samples are required. Among the different methodologies evaluated in this study, HEV concentration with aluminum hydroxide was able to detect HEV in influent and effluent water samples. However, the limited sensitivity of the method could be improved, for example by increasing the sample volume. The procedure for drinking water includes a DEUF step using a 30 kDa membrane to reduce the sample volume from 20 to 200 L to approximately 500 mL. Overall, the results showed that HEV is efficiently recovered from spiked drinking water samples processed using a PEG secondary concentration and the MN extraction kit.
This study also confirms that the selection of the RT-qPCR assays is critical since the overall performance of the methods varied considerably, most likely based on the circulating strains. In particular, this aspect remarkably affects genotyping results and thus epidemiology and traceability investigations.
Wastewater is an important environmental source for studying the epidemiology of viral pathogens transmitted via the fecal-oral route, and the current study demonstrated that HEV circulated in the Valencian region at around 30.6% during 2018–2019. No HEV was detected in effluent samples from WWTP and influent and effluent samples from DWTP. However, given that the infective dose in waterborne epidemics settings is not yet known and the low sensibility of the assay, unfortunately, no direct conclusion could be achieved on the risk assessment of environmental contamination.
All datasets generated for this study are included in the article/Supplementary Material.
EC-F, WR, and AP-C performed the assays, compiled the data, and wrote the draft of the manuscript. WR and GS conceived the original idea, interpreted the results, and drafted the manuscript. All authors contributed to the final manuscript.
This work was financially supported by the projects GV/2019/156 funded by Generalitat Valenciana and the “VIRIDIANA” project AGL2017-82909 (AEI/FEDER, UE) supported by Spanish Ministry of Science, Innovation and Universities (MICINN). WR was supported by a postdoctoral fellowship (APOSTD/2018/150) and EC-F by an APOTI grant (APOTIP/2018/007) both funded by the Generalitat Valenciana. EC-F is recipient of a predoctoral contract from the MICINN, Call 2018. The authors acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We gratefully acknowledge Agustín Garrido (IATA-CSIC) for providing the technical assistance. We also acknowledge the “Entidad de Saneamiento de Aguas Residuales” (EPSAR) and the Global Omnium S.L. for authorizing the sampling and Dr. M. J. Alcaraz for providing the HEV clinical samples. Names of specific vendors, manufacturers, or products are included for informational purposes only and does not imply endorsement by authors or their affiliations.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02937/full#supplementary-material
AAVV (2018). “Section 9510 D. Virus concentration by aluminum hydroxide adsorption-precipitation, chapter detection of enteric viruses,” in Standard Methods for the Examination of Water and Wastewater, 23rd Edn, eds E. W. Rice, R. B. Baird, and A. D. Eaton (Denver, CO: American Water Works Association).
Baylis, S. A., Sakata, H., Okada, Y., Mizusawa, S., Hanschmann, K.-M. O., Nübling, C. M., et al. (2013). World Health Organization international standard to harmonize assays for detection of Hepatitis E Virus RNA - Volume 19, Number 5—May 2013 - emerging infectious disease journal - CDC. Emerg. Infect. Dis. 19, 729–735. doi: 10.3201/eid1905.121845
Borgmästars, E., Jazi, M. M., Persson, S., Jansson, L., Rådström, P., Simonsson, M., et al. (2017). Improved detection of norovirus and hepatitis a virus in surface water by applying pre-pcr processing. Food Environ. Virol. 9, 395–405. doi: 10.1007/s12560-017-9295-3
Clemente-Casares, P., Pina, S., Buti, M., Jardi, R., MartIn, M., Bofill-Mas, S., et al. (2003). Hepatitis E virus epidemiology in industrialized countries. Emerg. Infect. Dis. 9, 448–454. doi: 10.3201/eid0904.020351
Fenaux, H., Chassaing, M., Berger, S., Gantzer, C., Bertrand, I., and Schvoerer, E. (2019). Transmission of hepatitis E virus by water: an issue still pending in industrialized countries. Water Res. 151, 144–157. doi: 10.1016/j.watres.2018.12.014
Girón-Callejas, A., Clark, G., Irving, W. L., and McClure, C. P. (2015). In silico and in vitro interrogation of a widely used HEV RT-qPCR assay for detection of the species Orthohepevirus A. J. Virol. Methods 214, 25–28. doi: 10.1016/j.jviromet.2014.11.025
Haramoto, E., Kitajima, M., Hata, A., Torrey, J. R., Masago, Y., Sano, D., et al. (2018). A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 135, 168–186. doi: 10.1016/j.watres.2018.02.004
Harrison, L., and DiCaprio, E. (2018). Hepatitis E Virus: an emerging foodborne pathogen. Front. Sustain. Food Syst. 2:14. doi: 10.3389/fsufs.2018.00014
ISO 15216-1:2017, (2017). Microbiology of the Food Chain – Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR – Part 1: Method for Quantification. Geneva:ISO.
Jothikumar, N., Cromeans, T. L., Robertson, B. H., Meng, X. J., and Hill, V. R. (2006). A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 131, 65–71. doi: 10.1016/j.jviromet.2005.07.004
Khuroo, M. S., Khuroo, M. S., and Khuroo, N. S. (2016). Transmission of Hepatitis E Virus in developing countries. Viruses 8:253. doi: 10.3390/v8090253
Kupferschmidt, K. (2016). Europe’s new hepatitis problem. Science 353, 862–863. doi: 10.1126/science.353.6302.862
Miura, T., Lhomme, S., Le Saux, J.-C., Le Mehaute, P., Guillois, Y., Couturier, E., et al. (2016). Detection of Hepatitis E Virus in sewage after an outbreak on a French Island. Food Environ. Virol. 8, 194–199. doi: 10.1007/s12560-016-9241-9
Nordgren, J., Matussek, A., Mattsson, A., Svensson, L., and Lindgren, P.-E. (2009). Prevalence of norovirus and factors influencing virus concentrations during one year in a full-scale wastewater treatment plant. Water Res. 43, 1117–1125. doi: 10.1016/j.watres.2008.11.053
Purpari, G., Macaluso, G., Di Bella, S., Gucciardi, F., Mira, F., Di Marco, P., et al. (2019). Molecular characterization of human enteric viruses in food, water samples, and surface swabs in Sicily. Int. J. Infect. Dis. 80, 66–72. doi: 10.1016/j.ijid.2018.12.011
Randazzo, W., Piqueras, J., Evtoski, Z., Sastre, G., Sancho, R., Gonzalez, C., et al. (2019). Interlaboratory comparative study to detect potentially infectious human enteric viruses in influent and effluent waters. Food Environ. Virol. 11, 350–363. doi: 10.1007/s12560-019-09392-2
Randazzo, W., Vásquez-García, A., Bracho, M. A., Alcaraz, M. J., Aznar, R., and Sánchez, G. (2018). Hepatitis E virus in lettuce and water samples: a method-comparison study. Int. J. Food Microbiol. 277, 34–40. doi: 10.1016/j.ijfoodmicro.2018.04.008
Ricci, A., Allende, A., Bolton, D., Chemaly, M., Davies, R., Fernandez Escamez, P. S., et al. (2017). Public health risks associated with hepatitis E virus (HEV) as a food-borne pathogen. EFSA J. 15:e4886. doi: 10.2903/j.efsa.2017.4886
Rodríguez-Díaz, J., Querales, L., Caraballo, L., Vizzi, E., Liprandi, F., Takiff, H., et al. (2009). Detection and characterization of waterborne gastroenteritis viruses in urban sewage and sewage-polluted river waters in Caracas, Venezuela. Appl. Environ. Microbiol. 75, 387–394. doi: 10.1128/AEM.02045-08
Rodriguez-Manzano, J., Miagostovich, M., Hundesa, A., Clemente-Casares, P., Carratala, A., Buti, M., et al. (2010). Analysis of the evolution in the circulation of HAV and HEV in eastern Spain by testing urban sewage samples. J. Water Health 8, 346–354. doi: 10.2166/wh.2009.042
Rusiñol, M., Fernandez-Cassi, X., Timoneda, N., Carratalà, A., Abril, J. F., Silvera, C., et al. (2015). Evidence of viral dissemination and seasonality in a Mediterranean river catchment: implications for water pollution management. J. Environ. Manage. 159, 58–67. doi: 10.1016/j.jenvman.2015.05.019
Schlosser, J., Eiden, M., Vina-Rodriguez, A., Fast, C., Dremsek, P., Lange, E., et al. (2014). Natural and experimental hepatitis E virus genotype 3-infection in European wild boar is transmissible to domestic pigs. Vet. Res. 45:121. doi: 10.1186/s13567-014-0121-8
Smith, D. B., Purdy, M. A., and Simmonds, P. (2013). Genetic variability and the classification of hepatitis E virus. J. Virol. 87, 4161–4169. doi: 10.1128/JVI.02762-12
Smith, D. B., Simmonds, P., Izopet, J., Oliveira-Filho, E. F., Ulrich, R. G., Johne, R., et al. (2016). Proposed reference sequences for hepatitis E virus subtypes. J. Gen. Virol. 97, 537–542. doi: 10.1099/jgv.0.000393
Sooryanarain, H., and Meng, X.-J. (2019). Hepatitis E virus: reasons for emergence in humans. Curr. Opin. Virol. 34, 10–17. doi: 10.1016/j.coviro.2018.11.006
Van der Poel, W. (2014). Food and environmental routes of Hepatitis E virus transmission. Curr. Opin. Virol. 4, 91–96. doi: 10.1016/j.coviro.2014.01.006
Van der Poel, W., and Rzezutka, A. (2019). “Hepatitis E,” in Global Water Pathogen Project, eds J. S. Meschke, and R. Girones. Lansing, MI: UNESCO. doi: 10.14321/waterpathogens.13
Wilrich, C., and Wilrich, P.-T. (2009). Estimation of the POD function and the LOD of a qualitative microbiological measurement method. J. AOAC Int. 92, 1763–1772.
Keywords: Hepatitis E virus, wastewater, drinking water, water quality, RT-qPCR, occurrence
Citation: Cuevas-Ferrando E, Randazzo W, Pérez-Cataluña A and Sánchez G (2020) HEV Occurrence in Waste and Drinking Water Treatment Plants. Front. Microbiol. 10:2937. doi: 10.3389/fmicb.2019.02937
Received: 30 September 2019; Accepted: 06 December 2019;
Published: 14 January 2020.
Edited by:
Eric N. Villegas, United States Environmental Protection Agency, United StatesReviewed by:
Feng Ju, Westlake Institute for Advanced Study (WIAS), ChinaCopyright © 2020 Cuevas-Ferrando, Randazzo, Pérez-Cataluña and Sánchez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Walter Randazzo, d2FsdGVyLnJhbmRhenpvQHV2LmVz; d2FsdGVycmFuZGF6em9AaG90bWFpbC5pdA==; Gloria Sánchez, Z2xvcmlhc2FuY2hlekBpYXRhLmNzaWMuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.