- 1Department of Oral Microbiology and Immunology, DRI, and BK21 Plus Program, School of Dentistry, Seoul National University, Seoul, South Korea
- 2Department of Agricultural Biotechnology, Research Institute for Agriculture and Life Sciences, Center for Food and Bioconvergence, Seoul National University, Seoul, South Korea
Enterococcus faecalis is a Gram-positive, facultative anaerobic bacterium frequently found in the gastrointestinal tract, oral cavity, and periodontal tissue. Although it is considered a commensal, it can cause bacteremia, endocarditis, endodontic infections, and urinary tract infections. Because antibiotics are cytotoxic not only to pathogens, but also to health-beneficial commensals, phage therapy has emerged as an alternative strategy to specifically control pathogenic bacteria with minimal damage to the normal flora. In this study, we isolated a novel phage, Enterococcus phage vB_EfaS_HEf13 (phage HEf13), with broad lytic activity against 12 strains of E. faecalis among the three laboratory strains and 14 clinical isolates of E. faecalis evaluated. Transmission electron microscopy showed that phage HEf13 has morphological characteristics of the family Siphoviridae. Phage HEf13 was stable at a wide range of temperature (4–60°C) and showed tolerance to acid or alkaline (pH 3–12) growth conditions. Phage HEf13 had a short latent period (25 min) with a large burst size (approximately 352 virions per infected cell). The lytic activity of phage HEf13 at various multiplicities of infection consistently inhibited the growth of diverse clinical isolates of E. faecalis without any lysogenic process. Moreover, phage HEf13 showed an effective lytic activity against E. faecalis on human dentin ex vivo infection model. Whole genome analysis demonstrated that the phage HEf13 genome contains 57,811 bp of double-stranded DNA with a GC content of 40.1% and 95 predicted open reading frames (ORFs). Annotated functional ORFs were mainly classified into four groups: DNA replication/packaging/regulation, phage structure, host cell lysis, and additional functions such as RNA transcription. Comparative genomic analysis demonstrated that phage HEf13 is a novel phage that belongs to the Sap6virus lineage. Furthermore, the results of multiple sequence alignment showed that polymorphism of phage infection protein of E. faecalis (PIPEF) contributes to determine the host specificity of phage HEf13 against various E. faecalis strains. Collectively, these results suggest that phage HEf13 has characteristics of a lytic phage, and is a potential therapeutic agent for treatment or prevention of E. faecalis-associated infectious diseases.
Introduction
Enterococcus is a genus of Gram-positive, non-spore-forming bacteria that are frequently found in the oral cavity, gastrointestinal tract, and vagina (Jett et al., 1994). Although most Enterococcus species are considered to be commensal in humans and animals (Gu et al., 2018), Enterococcus faecalis can act as an opportunistic pathogen causing bacteremia, meningitis, endocarditis, or urinary tract- and wound-related infections (Arias et al., 2010; Weiner et al., 2016). In humans, E. faecalis has been predominantly detected in the root canals of patients with post-treatment apical periodontitis or refractory apical periodontitis, suggesting an etiological role in progression of these diseases (Souto and Colombo, 2008; Vidana et al., 2011).
Antibiotics are widely used to treat and prevent diseases caused by various pathogenic bacteria. However, the emergence of antibiotic resistant E. faecalis, such as vancomycin-resistant E. faecalis (VRE), has diminished the clinical usefulness of antibiotics (O’Driscoll and Crank, 2015). On the one hand, because most strains of E. faecalis isolated from root canal and periodontal infections show a high level of resistance to antibiotics including tetracycline and erythromycin used in dental procedures, antibiotic options for endodontic treatment are limited (Pinheiro and Mayer, 2014). On the other hand, indiscriminate use of antibiotics can result in disruption of normal gut microflora, and can negatively affect human health by promoting the evolution of antibiotic resistance in the microbiome and compromising immune homeostasis (Cho and Blaser, 2012; Francino, 2015).
There is currently renewed interest in phage therapy using lytic phages, also called viruses, as a potential alternative strategy to control pathogenic bacteria as this strategy overcomes the limitations of antibiotics (Golkar et al., 2014). Unlike antibiotics, phages are highly specific for their target pathogen and thus do not damage the normal flora (Moelling et al., 2018). Phages have many other advantages, including ease of isolation and better efficacy than antibiotics at destroying bacteria in biofilms (Khalifa et al., 2016). Moreover, many clinical trials have shown that the use of phages is safe without side-effects in both humans and animals (Burrowes et al., 2011).
Based on previously published studies, a total of 22 lytic phages targeting E. faecalis (14 Siphoviridae, 6 Myoviridae, and 2 Podoviridae phages) have been isolated from various sources, including sewage, animal farmyard effluents, and human feces (Bonilla et al., 2010; Bolocan et al., 2019; Lossouarn et al., 2019). Although previous studies have demonstrated the therapeutic potency of newly isolated phages for targeting E. faecalis, practical use of these phages has been limited by their relatively narrow host range against clinically isolated E. faecalis strains (Rigvava et al., 2013; Zhang et al., 2013; Ladero et al., 2016; Cheng et al., 2017; Rahmat Ullah et al., 2017; Wang et al., 2018). In addition, despite the importance of E. faecalis in the pathogenesis of recurrent and incurable periodontitis, curative efficiency of E. faecalis phages has not been evaluated against E. faecalis strains isolated from the oral cavity. Therefore, we focused on isolation, characterization, and genomic analysis of a novel E. faecalis lytic phage against clinically isolated E. faecalis strains from the oral cavity.
Materials and Methods
Bacterial Strains and Culture Conditions
A total of 17 strains of E. faecalis, including 14 clinical isolates and three laboratory strains, were used to determine the host range of phage HEf13. As shown in Table 1, three laboratory strains of E. faecalis and two laboratory strains of E. faecium were obtained from the Korean Agricultural Culture Collection (KACC, Wanju, Republic of Korea) and 14 clinically isolated E. faecalis strains, recovered from blood, dental plaques, mucormycosis, pus, urine (VRE), and the vagina were provided by the Korean Collection for Oral Microbiology (KCOM, Gwangju, Republic of Korea), Korean Collection for Type Cultures (KCTC, Jeongeup, Republic of Korea), or the National Culture Collection of Pathogens (NCCP, Chungju, Republic of Korea). All strains of E. faecalis were grown in brain heart infusion broth (BHI broth, Difco Laboratories Inc., Franklin Lakes, NJ, United States) at 37°C with shaking under aerobic conditions.
Phage Isolation
Raw sewage water was collected from a local sewerage system (Seoul, Republic of Korea) and used as a source of E. faecalis phages. Phage isolation was performed as previously described (Lee et al., 2016). Briefly, sewage sample (10 mL) was homogenized with 40 mL sodium chloride–magnesium sulfate buffer [SM buffer; 100 mM NaCl, 10 mM MgSO4⋅7H2O, and 50 mM Tris–HCl (pH 7.5)] containing 1% chloroform. After centrifugation at 10,000 × g and 4°C for 10 min, supernatants were collected and then filtered through 0.22 μm pore-size membrane filters (Corning Inc., Corning, NY, United States). Supernatants (10 mL) were mixed with an equal volume of 2× concentrated BHI broth and incubated with 1% of overnight culture of 12 E. faecalis strains mixture with shaking at 37°C for 12 h. The suspension was harvested by centrifugation at 10,000 × g at 4°C for 10 min and filtered to remove bacterial debris. Then, supernatants diluted 10–1,000 times with BHI broth were spotted on molten 0.4% BHI soft agar containing 1 × 108 CFU/mL of E. faecalis. The next day, clear phage plaques were picked from the BHI agar plates and the phages were resuspended in SM buffer. These phage plaque isolation and phage resuspension steps were repeated three times to obtain individual phages with high purity.
Phage Propagation, Purification, and Titration
Propagation and purification of the phages were performed as described previously with minor modifications (Sambrook and Russell, 2001). For phage propagation, early log phase cultures of E. faecalis KCOM 1162 (OD600 = 0.35) were infected with phage and incubated with vigorous shaking at 37°C for 5 h. After centrifugation at 12,000 × g at 4°C for 10 min, supernatants containing phages were filtered using bottle-top vacuum filters with a 0.22-μm pore-size membrane (Merck Millipore, Burlington, MA, United States). Following the propagation step, filtrates were mixed with 1 M NaCl containing 10% polyethylene glycol 6000 (Junsei Chemical Co., Tokyo, Japan) and incubated overnight at 4°C. Precipitated phages were then harvested by centrifugation at 12,000 × g for 20 min at 4°C and resuspended in 2 mL SM buffer. Resuspended phages were subjected to density gradient ultracentrifugation (CP100NX, Hitachi, Tokyo, Japan) with different cesium chloride steps (density for each step = 1.3, 1.45, 1.5, and 1.7 g/mL) at 111,000 × g at 4°C for 2 h. Finally, purified phages were collected from the band corresponding to viral particles and dialyzed in standard dialysis buffer [10 mM NaCl, 10 mM MgCl2, 50 mM Tris–HCl (pH 8.0)] at 4°C for 2 h. To titrate the purified phages, serially diluted phages (10–1–10–9 folds) were dropped on an E. faecalis KCOM 1162 (1 × 109 CFU/mL) lawn. Plaques were examined the next day and plaque-forming units (PFUs) were calculated based on the minimal dilution rate of phages generating plaques.
Determination of Host Range
The host range of purified phages was determined by spotting assay (Kropinski et al., 2009) against the 17 strains of E. faecalis (Table 1). Briefly, 100 μL of each freshly cultured E. faecalis strain was mixed with 6 mL of warm 0.4% BHI soft agar and overlaid on previously prepared 1.5% BHI agar plates. After agar hardening, a 10 μL aliquot of purified phages at 1 × 109 PFU/mL was spotted on each agar plate and the plates were incubated at 37°C overnight. The next day, bacterial sensitivity to phages was estimated by the clarity of plaques on each plate.
Bacterial Challenge Assay
Among the 17 strains of E. faecalis tested in the host range assay, 12 strains sensitive to the purified phages based on the results of host range analysis were chosen for the bacterial challenge assay (Xie et al., 2018). One percent of an overnight culture of each E. faecalis strain was inoculated in triplicate on a 96-well plate and the plate was cultured at 37°C with shaking until the optical density at 600 nm (OD600) reached 0.2 (∼1 × 108 CFU/mL). Bacteria were infected with the purified phages at various multiplicities of infection (MOI) ranging from 0.01 to 1, and incubated at 37°C with shaking for 0–12 h. Bacterial growth was assessed by measuring OD600 using a spectrophotometer every 2 h for up to 12 h. E. faecalis strains not infected with phages were used as controls for this assay. To determine whether phage HEf13 is a temperate or lytic phage, E. faecalis KCOM 2816 strain (1 × 108 CFU/mL) at early mid log phase was infected with phage HEf13 (MOI 1) for 16 h. The infected bacteria were inoculated on 96-well plate by 1% (w/v) and incubated at 37°C for an additional 2 h. After the incubation, cultured bacteria were reinfected with phage HEf13 (MOI 0.1 and 1) in the presence or absence of mitomycin C (MMC) at 5 μg/mL (Sigma–Aldrich, St. Louis, MO, United States) (Stevens et al., 2009). Bacterial growth was measured every 2 h for up to 12 h using a spectrophotometer.
Preparation of Human Dentin Slices
Experiments using human dentin slices were approved by the Institutional Review Board of Seoul National University Dental Hospital, Seoul, Republic of Korea (CRI 17010). Extracted human single-rooted pre-molars were sonicated in 0.5% sodium azide with an ultrasonic scaler (SH-2140; Saehan-Sonic, Seoul, Republic of Korea). The roots were sliced into a size of 500 μm with an Isomet precision saw (Buehler, Lake Bluff, IL, United States). The slices were sequentially treated with 17% EDTA (Sigma–Aldrich) for 5 min, 2.5% sodium hypochlorite (Sigma–Aldrich) for 5 min, and 5% sodium thiosulfate (Sigma–Aldrich) for 5 min, followed by autoclave-sterilization for 15 min at 121°C.
Ex vivo Analysis of Phage Lytic Activity
The lytic activity of phage HEf13 against E. faecalis on human dentin slice was examined by scanning electron microscope (SEM) analysis as previously described (Kim et al., 2019). Briefly, E. faecalis (1 × 108CFU/mL) was grown on dentin slices in the presence or absence of phage HEf13 (MOI 0.1) in BHI medium at 37°C for 12 h. Attached bacteria on dentin slice were rinsed with phosphate-buffered saline (PBS) and prefixed with a 2.5% glutaraldehyde and 2% paraformaldehyde (pH 7) at 4°C overnight. After washing with PBS, the dentin slices were fixed with 1% osmium tetroxide for 90 min, then washed with distilled water, and subsequently dehydrated by gradual increase of ethanol concentrations (70–100% for 15 min). After drying with hexamethyldisilazane and coating with gold sputter, samples were visualized by SEM (S-4700, Hitachi, Tokyo, Japan).
Morphological Analysis by Electron Microscopy
Morphology of the purified phages was examined by transmission electron microscopy (TEM) analysis. The glow-discharge step to prepare Formvar/carbon-coated copper grids was performed at 15 mA and 0.26 mBar for 30 s using a glow discharge cleaning system (PELCO easiGlow; Ted Pella Inc., Redding, CA, United States). Purified phage stock (approximately 3 × 107 PFU) was dropped onto the prepared Formvar/carbon-coated copper grids and incubated at room temperature for 1 min. E. faecalis KCOM 1162 (1 × 108 CFU/mL) incubated with phages (1 × 1010 PFU/mL) at room temperature for 10 min was collected by centrifugation and resuspended in SM buffer. Bacteria were also dropped onto the prepared Formvar/carbon-coated copper grids and after a 1 min incubation, the grids were negatively stained with 2% uranyl acetate and electron micrographs of phages were obtained by TEM (Libra 120 model; Zeiss, Oberkochen, Germany operated at an accelerating voltage of 80 kV). Identification and classification of phages was conducted according to the guidelines of the International Committee on the Taxonomy of Viruses (ICTVs).
One-Step Growth Curve Analysis
One-step growth experiments were conducted as described previously with a slight modification (Parasion et al., 2012). When the culture of E. faecalis KCOM 1162 reached early log phase (OD600 = 0.35), bacteria were harvested by centrifugation at 10,000 × g at 4°C for 10 min and resuspended in fresh BHI broth. Bacteria were infected with purified phages at a MOI of 0.01 and incubated at room temperature for 5 min. After centrifugation and removal of the supernatant containing unbound phages, pellets were resuspended in 50 mL fresh BHI medium and incubated at 37°C with shaking for up to 90 min. Samples were taken every 5 min, serially diluted, and plated on E. faecalis KCOM 1162 lawn for phage titration. Burst sizes of phages were calculated as the ratio of the average phage titer value at plateau phase to that at latent phase (Svab et al., 2018).
Thermal and pH Stability
Stability of the phages to temperature and pH was assessed by phage titration after incubation in SM buffer at different temperature or pH values (Park et al., 2017). Briefly, 1.5 mL tubes containing the same amount of the purified phages (1 × 108 PFU/mL) were incubated at various temperatures ranging from 4 to 70°C, or under different pH conditions (SM buffer at pH 2–12 adjusted by NaOH or HCl) at 37°C for 12 h. After incubation, phage titers were determined using E. faecalis KCOM 1162 as described above.
Whole Genome Sequencing and Bioinformatic Analysis
Genomic DNA from purified phages was extracted using the Norgen phage DNA isolation kit (Norgen Biotek Corp., ON, Canada) according to the manufacturer’s instruction. Sequencing libraries with single index were prepared using the TruSeq Nano DNA library prep kit (Illumina, San Diego, CA, United States) and then sequenced on the Illumina MiSeq sequencing platform (Illumina, San Diego, CA, United States) with paired-end 300 nucleotide reads. Raw reads were trimmed and de novo assembled using the CLC Genomic Workbench program (Version 10.0.1; QIAGEN Inc., Hilden, Germany). Annotation of the specific function of ORFs was conducted using rapid annotations of subsystems technology (RAST) and the BLASTP database (Altschul et al., 1997; Aziz et al., 2008). Artemis (version 16; Sanger Institute, Cambridge, United Kingdom) was used to visualize and browse the genome and annotate ORFs (Carver et al., 2008). The presence of tRNA-encoding genes was determined using the tRNAscan-SE database (Lowe and Chan, 2016). A circular representation of the genome of phage HEf13 was visualized using the CGView server database (Stothard and Wishart, 2005). Phage virulence factor analysis was conducted using the Virulence Searcher database (Underwood et al., 2005). Comparative analysis of genome sequences and visualization of the genomes of phage HEf13 and other Enterococcus phages was performed using BLASTN and BLASTP databases and the EasyFig program (Version 2.2; Beatson Microbial Genomic Lab, Brisbane, Australia) (Sullivan et al., 2011).
Comparative Phylogenetic Analysis
Full length amino acid (aa) sequences of the phage portal protein (511 aa), tail fiber protein (1,330 aa), and DNA methyltransferase (150 aa) of phage HEf13 were aligned with those of nine Enterococcus phages in the Sap6virus genus and Listeria phages showing high similarity using the ClustalW Multiple sequence alignment (MSA) module in the BioEdit Sequence Alignment Editor program (Ibis Therapeutics, Carlsbad, CA, United States) (Larkin et al., 2007). After alignment, phylogenetic trees for each of the three phage proteins were generated by the Neighbor-joining method using Molecular Evolutionary Genetics Analysis 7 (MEGA7) software (Pennsylvania State University, State College, PA, United States) (Tamura et al., 2013) and groupings were estimated by bootstrap analysis (1,000 replication).
Analysis of Phage Infection Protein of E. faecalis (PIPEF) Polymorphism
Polymorphism of the variable region of PIPEF for E. faecalis strains used in the current study was analyzed to examine correlation between the host specificity of phage HEf13 and PIPEF as previously described (Duerkop et al., 2016). Briefly, genomic DNA was extracted from 17 strains of E. faecalis using a commercially available kit according to the manufacturer’s instruction (iNtRON Biotechnology, Seongnam, Republic of Korea). The genomic DNAs were amplified by PCR under the following conditions: denaturation at 95°C for 30 s and amplification via 28 cycles of 50°C for 3 min and 72°C for 3 min. PCR products were purified using a PCR/Gel purification kit (Bioneer, Daejeon, Republic of Korea) and sequenced at Cosmo Genetech (Seoul, Republic of Korea). MSAs of homologs were analyzed using Geneious 6.0.5. software (Biomatters Ltd., Auckland, New Zealand). Phylogenetic tree of PIPEF homologs was constructed by Neighbor-Joining method (1,000 bootstrap) (Tamura et al., 2013).
Results
Isolation, Host Range Determination, and Lytic Activity of Phage HEf13
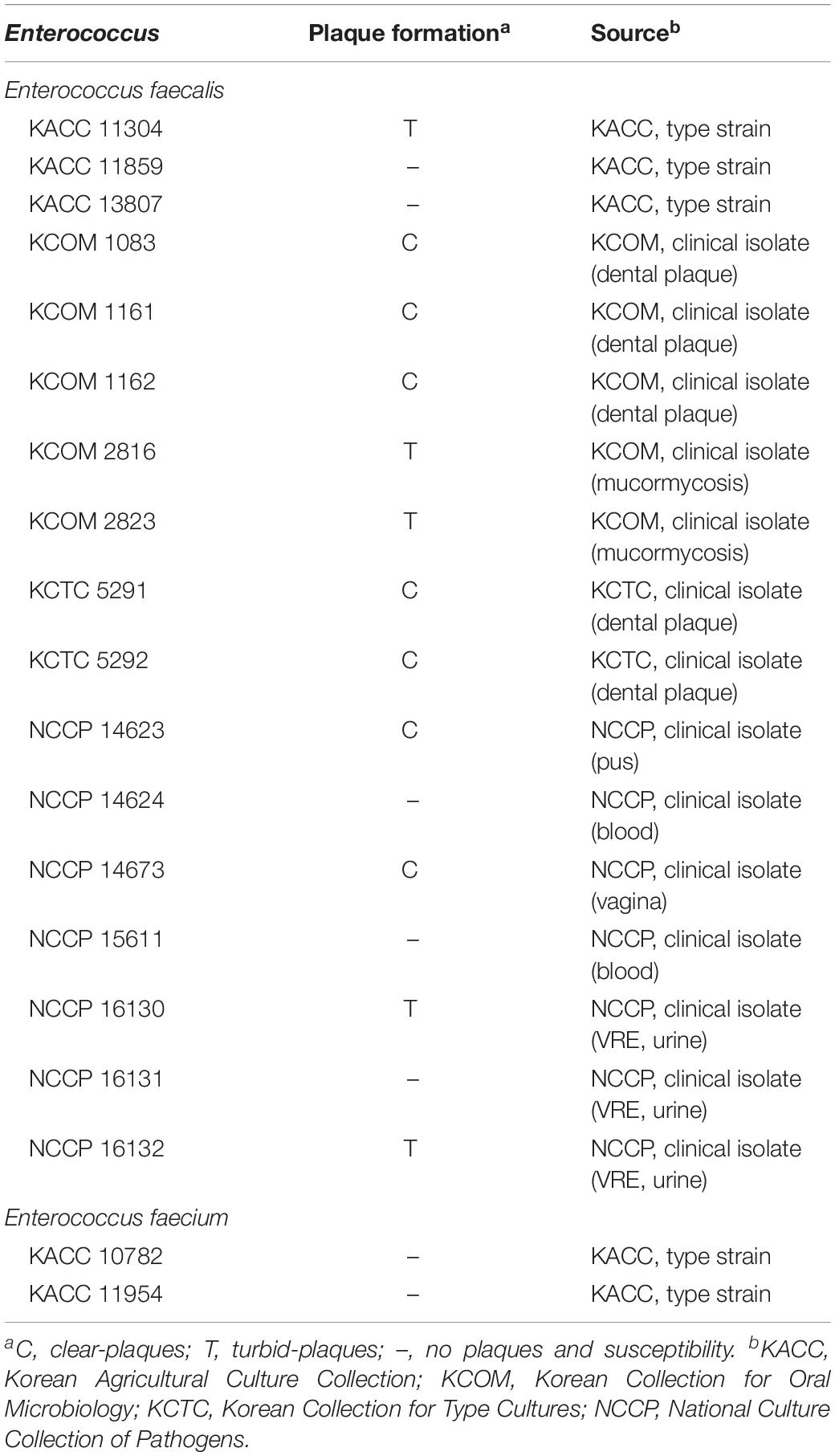
E. faecalis phage vB_EfaS_HEf13 (phage HEf13) was newly isolated from sewage water of a local sewerage system in Seoul, Republic of Korea. The host range of phage HEf13 was assessed using a spotting assay against a total 17 strains of E. faecalis (three laboratory strains and 14 clinically isolated strains including three VRE strains), and two laboratory strains of E. faecium. Phage HEf13 formed clear- or turbid-plaques in 12 E. faecalis strains, including all dental isolates of E. faecalis (Figure 1A and Table 1). However, phage HEf13 cannot recognize and lyse E. faecium, suggesting that phage HEf13 is specific only to E. faecalis, but not to other Enterococcus species. To examine the lytic activity of phage HEf13, E. faecalis strains where clear- or turbid-plaques were observed in the spotting assay were subjected to bacterial challenge assays at various MOI for up to 12 h. Similar to the pattern of host range determination, phage HEf13 showed strong lytic activity against E. faecalis where clear-plaque formation was observed in the previous spotting assay, but not the strains where turbid-plaque formation was observed (Figures 1B,C and Supplementary Figures S1A–J). However, the results showed a possibility that turbid-plaque formation by phage HEf13 may result from its lysogenic property. Therefore, in order to determine whether the phage is lytic or lysogenic, we examined the growth of E. faecalis infected with phage HEf13 in the presence of MMC, which is a lysogenic phage inducer (Stevens et al., 2009). As shown in Figure 1D. E. faecalis infected with phage HEf13 was not affected by MMC treatment, implying that the turbid-plaque formation by phage HEf13 was not due to a lysogenic process of phage HEf13. Furthermore, we evaluated the effect of phage HEf13 against E. faecalis using an ex vivo human dentin infection model. When E. faecalis on dentin slice infected with phage HEf13 was visualized using a SEM, E. faecalis was completely eradicated (Figure 1E). These results suggest that phage HEf13 is a potential therapeutic agent to treat diseases associated with E. faecalis infection such as refractory apical periodontitis.
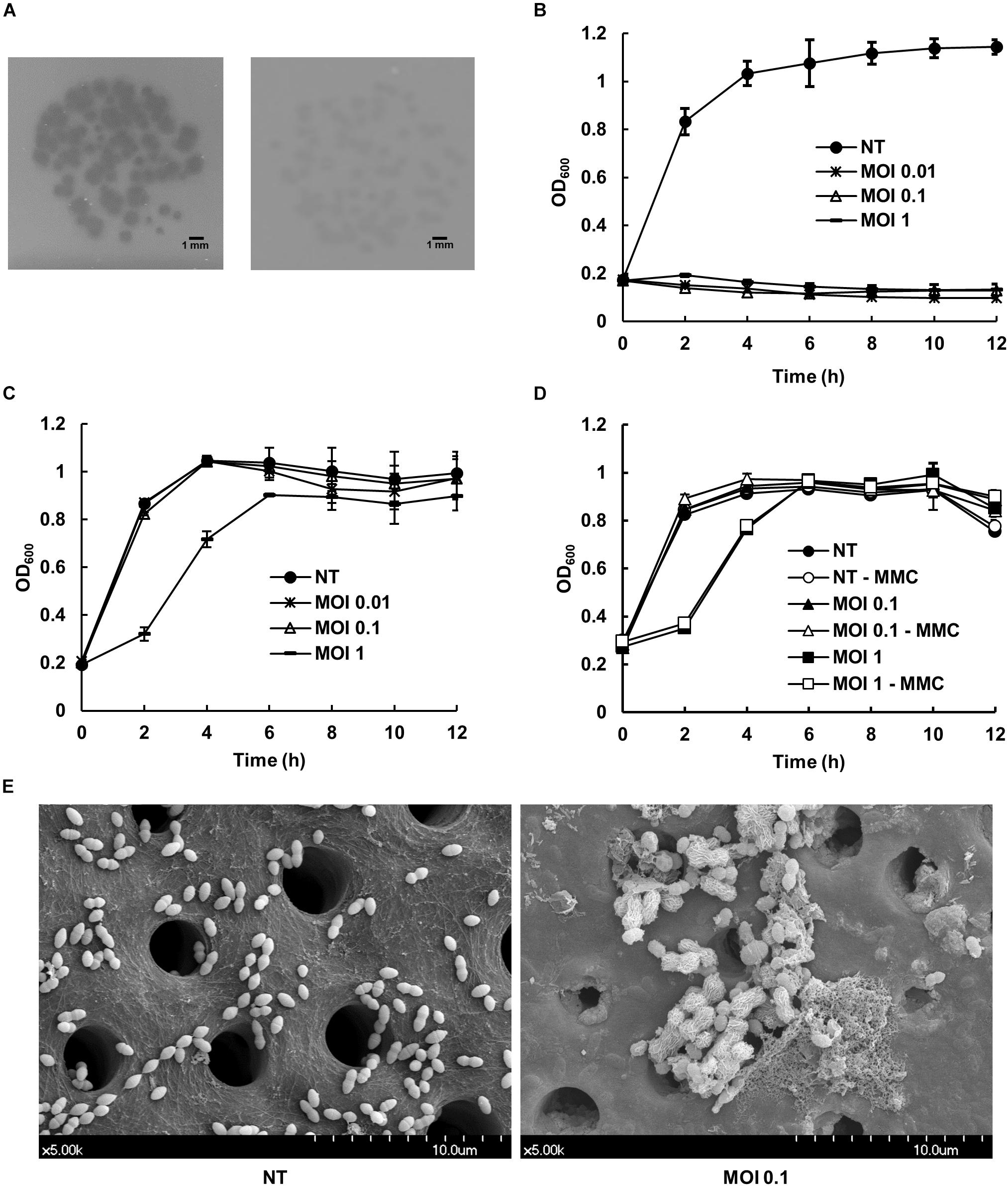
Figure 1. Effect of newly isolated Enterococcus phage vB_EfaS_HEf13 (phage HEf13) on the growth of E. faecalis. (A) Phage HEf13 suspension (1 × 104 PFU/mL) was spotted on E. faecalis KCOM 1162 (clear-plaque, left) or KACC 11304 (turbid-plaque, right) lawn and incubated at 37°C overnight. Plaque formation by phage HEf13 was photographed. Scale bar indicates 1 mm. (B,C) Representative E. faecalis strains showing clear- (B; KCOM 1162) or turbid-plaque formation (C; KCOM 2816) by phage HEf13 were chosen for the bacterial challenge assay. Each E. faecalis strain was infected with phage HEf13 at various MOI (0.01–1) and incubated at 37°C for 0–12 h. (D) Turbid-plaque-forming E. faecalis strain (KCOM 2816) was infected with phage HEf13 (MOI 1) for 16 h and the infected bacteria were then grown to early mid-log phase. After the culture, the infected bacteria were cultured in the presence or absence of phage HEf13 (MOI 0.1 or 1) and MMC at 5 μg/mL. Bacterial growth at each indicated time point was measured by spectrophotometer at 600 nm. Values are means ± standard deviations from triplicates of each treatment. (E) Dental isolate of E. faecalis strain (KCOM 1162) was inoculated on human dentin slices in the presence or absence of phage HEf13 (MOI 0.1) for 12 h. Then, the dentin slices were fixed and images were obtained by SEM (magnification: 5,000×). Scale bar indicates 10 μm with 1 μm ticks. MMC, mitomycin C; MOI, multiplicity of infection; NT, non-treatment.
Morphology of Phage HEf13
Morphological analysis of phage HEf13 and its adsorption to E. faecalis was examined by TEM. Phage HEf13 has a prolate head with a long non-contractile tail (Figures 2A–C). Head diameter is 38.4 ± 2.4 nm with a length of 95.6 ± 1.7 nm, and tail length is 135.2 ± 4.7 nm (Figures 2A–C). TEM images of E. faecalis KCOM 1162 infected with phage HEf13 demonstrated adsorption of phages to the surface of E. faecalis as marked by the arrows in Figure 2D. Taken together, these results indicate that phage HEf13 belongs to the Siphoviridae family in the order Caudovirales according to the ICTV guidelines (Fauquet et al., 2005).
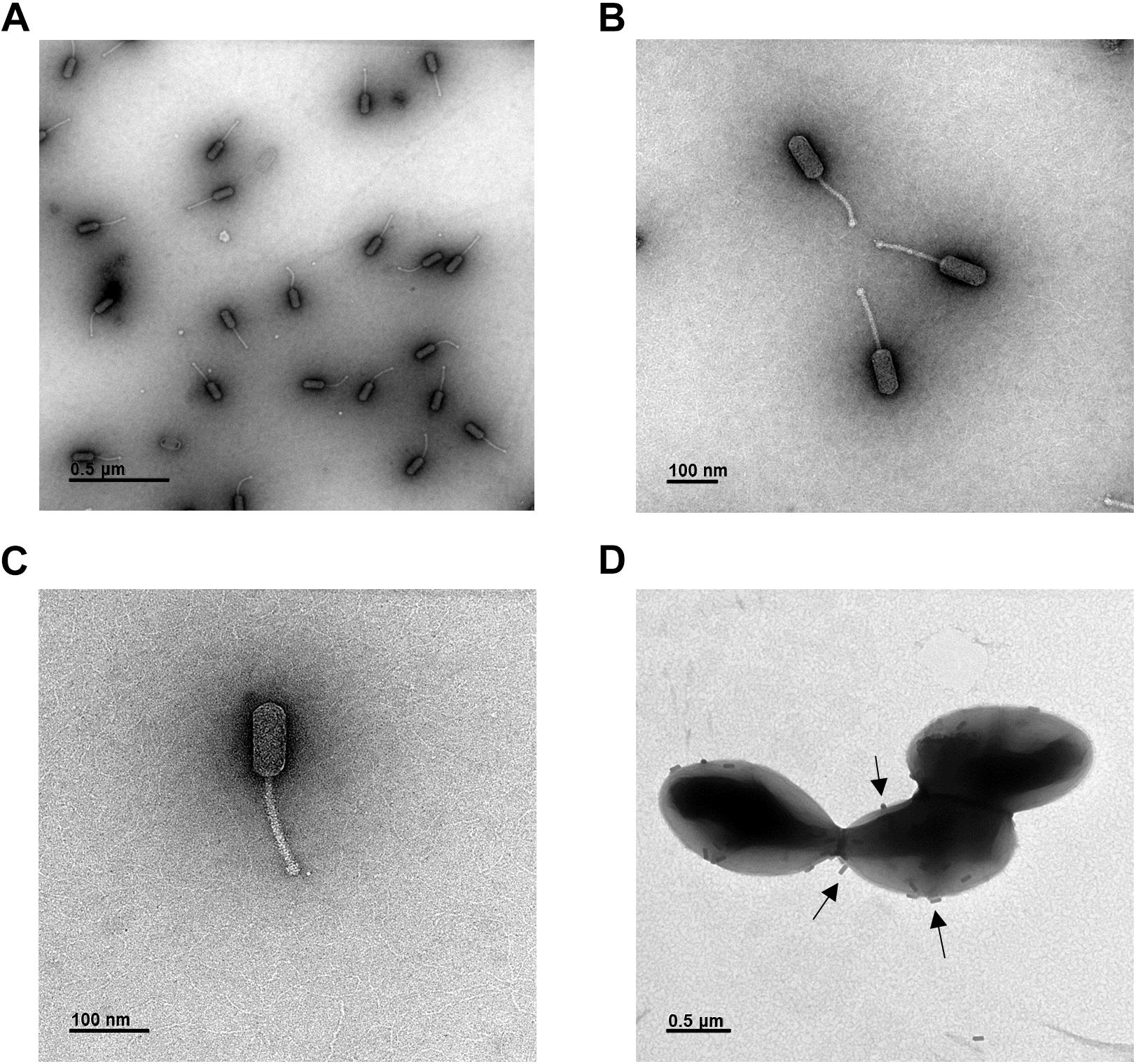
Figure 2. Phage HEf13 belongs to the Siphoviridae family. (A–C) Phage HEf13 (1 × 108 PFU) was dropped onto a Formvar/carbon-coated copper grid and then negatively stained with 2% uranyl acetate. (D) E. faecalis KCOM 1162 (1 × 108 CFU/mL) incubated with phage HEf13 (1 × 1010 PFU/mL) was dropped onto copper grids and stained with 2% uranyl acetate. Electron micrographs of phage HEf13 were obtained by TEM at (A) 35,000×, (B,C) 100,000×, and (D) 35,000× magnification. Identification and classification of phages was conducted according to ICTV guidelines. Scale bar indicates 100 nm (B,C) or 0.5 μm (A,D). Arrows in panel (D) show phage HEf13 binding to the cell wall membrane of E. faecalis KCOM 1162.
Latent Period and Burst Size of Phage HEf13
The latent period and burst size of phage HEf13 were measured by one-step growth curve analysis against E. faecalis KCOM 1162. The latent period of phage HEf13, calculated as the time interval between adsorption and the beginning of the first burst, was approximately 25 min (Figure 3). The rise period of phage HEf13 was about 30 min and mean burst size, defined as the mean phage titer value at plateau phase divided by that of the latent phase, was approximately 352 virions per infected cell. These results indicate that phage HEf13 has a short latent period and long rise period with a large burst size, accounting for its strong lytic activity against E. faecalis.
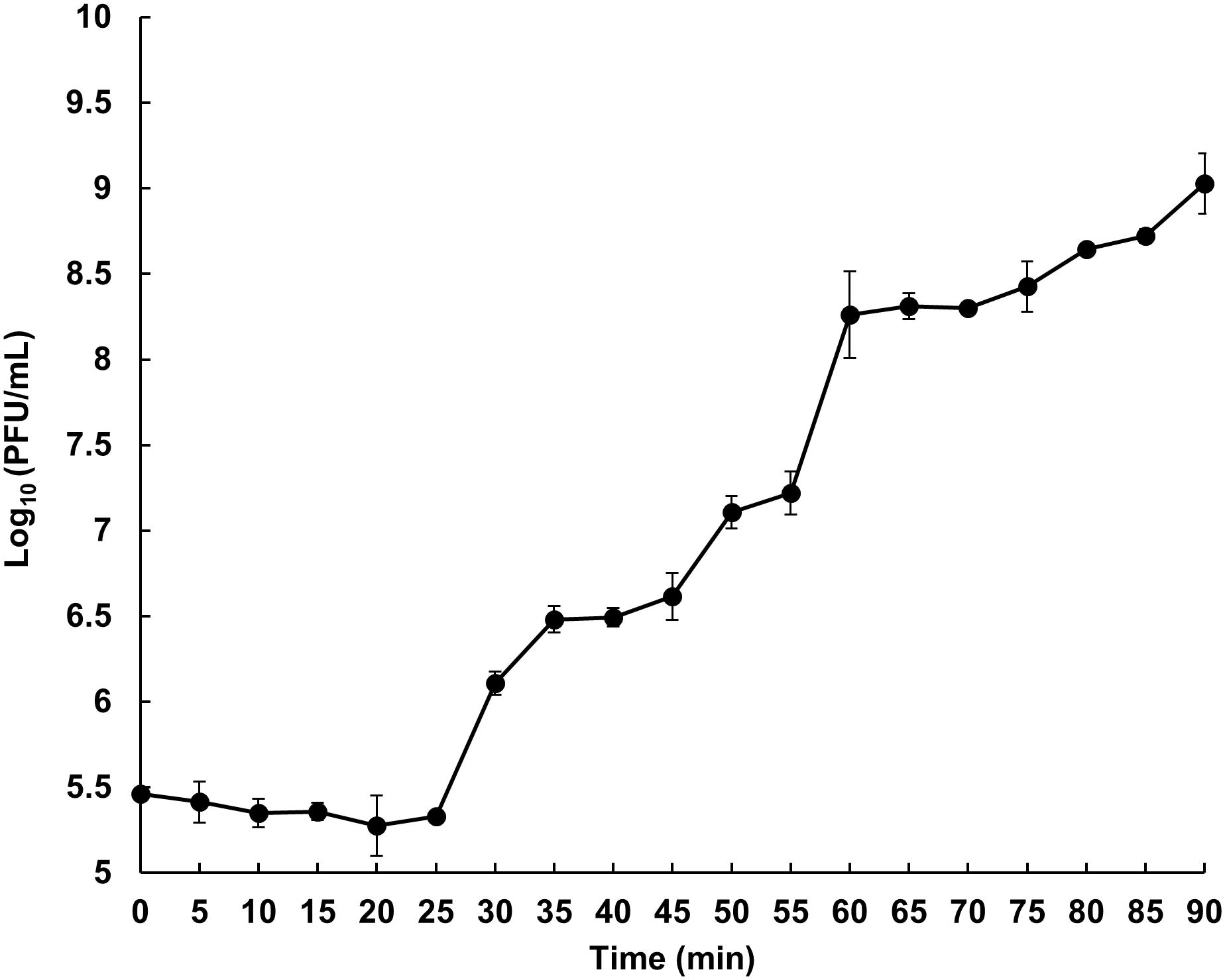
Figure 3. Phage HEf13 has a relatively short latent period and enhanced large burst size against E. faecalis. E. faecalis KCOM 1162 was infected with Phage HEf13 at an MOI of 0.01 and incubated for 5 min. After removing unbound phages by centrifugation, bacteria were incubated at 37°C up to 90 min. Samples were taken every 5 min and subjected to phage titration by spotting assay as described in the section “Materials and Methods.” Values are means ± standard deviations from triplicates of each time point.
Stability of Phage HEf13 to Different Temperature and pH Conditions
The thermal stability of phage HEf13 was evaluated by phage titration assay after phage incubation at various temperatures ranging from 4 to 70°C for 12 h. While no loss was observed in phage titers at temperatures ranging from 4 to 50°C, a phage titer of <60% was observed at 60°C. Furthermore, no phage titer was detected when phage HEf13 was incubated at 70°C (Figure 4A). Phage HEf13 tolerated a broad range of pH values from 3 to 12. However, no phage activity was noted under extremely acidic conditions (pH 2) (Figure 4B). These data show that phage HEf13 can tolerate a broad range of temperature and pH values.
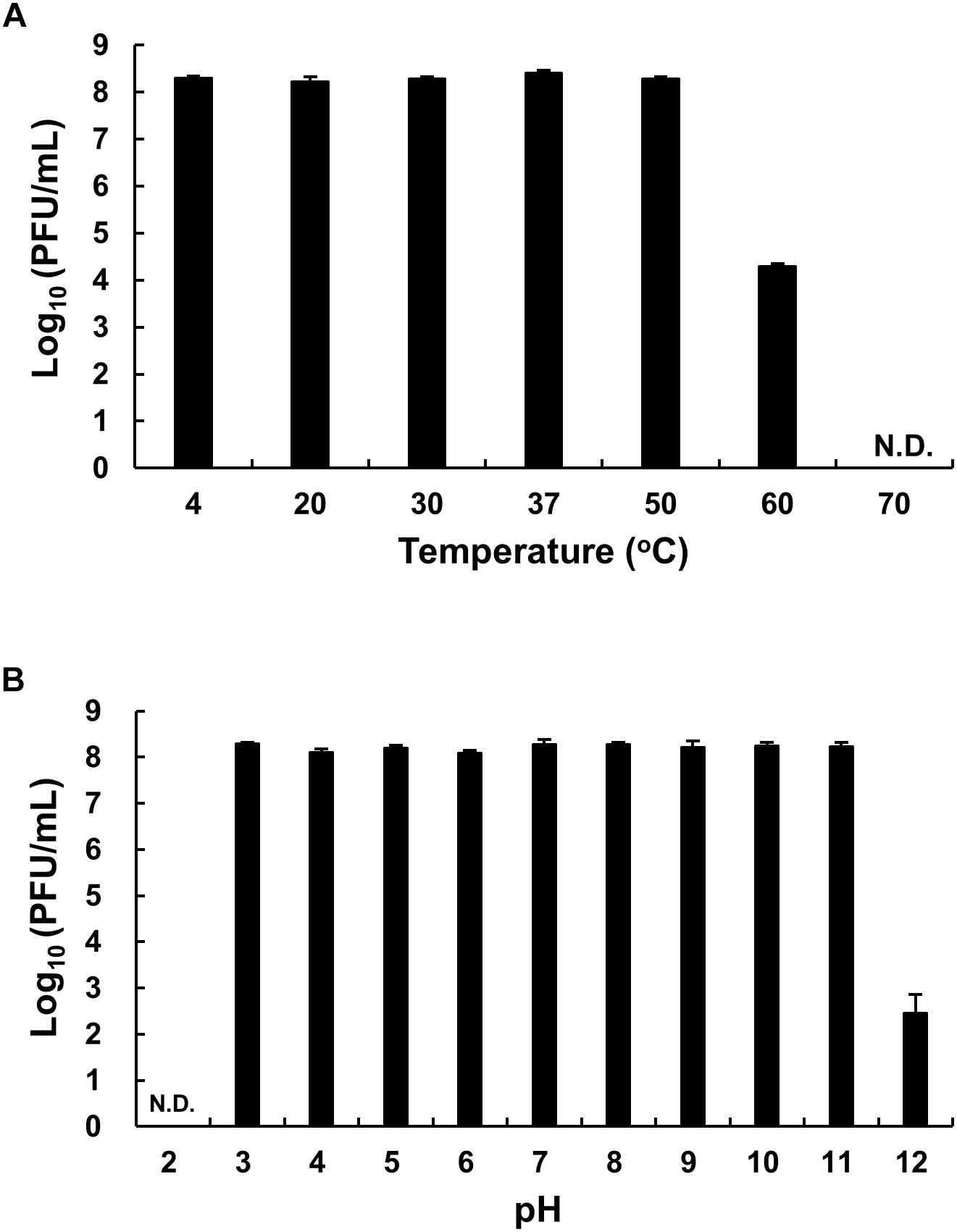
Figure 4. Phage HEf13 is stable over a broad range of temperature and pH values. (A) Phage HEf13 (1 × 108 PFU/mL) was incubated at various temperatures ranging from 4 to 70°C for 12 h. (B) Phage HEf13 (1 × 108 PFU/mL) was also incubated at different pH values ranging from 2 to 12 at 37°C for 12 h. Then, phage titers were determined by spotting assay as described in the section “Materials and Methods.” Values are presented as log values of the ratio of phage titers in each treatment to that of the initial phage titer. Values are means ± standard deviations from triplicates of each treatment. N.D., not detected.
Genomic Characterization of Phage HEf13
Whole genome sequence analysis showed that the genome of phage HEf13 (GenBank Accession Number: MH618488) is 57,811 bp in length with 95 predicted ORFs and one tRNA gene, a GC content of 40.03%, and the following nucleotide composition: G (10,737 bp, 18.57%), C (12,405 bp, 21.45%), A (15,765 bp, 27.26%), and T (18,904 bp, 32.69%) (Figure 5). Average gene length was 532 bp, with a range of 116–3,992 nucleotides, and gene coding percentage was 87.4%. Among the 95 putative ORFs, 15 ORFs were on the positive strand while the other 80 ORFs were on the negative strand. Only 30 ORFs (31.5%) were predicted to be functional proteins, whereas 65 ORFs (68.4%) were annotated as hypothetical proteins (Supplementary Table S1). Annotation of the function of the 30 ORFs predicted as functional proteins by RAST and BLASTP analyses revealed four major functional groups: DNA replication/packaging/regulation, phage structure, host cell lysis, and additional function. The DNA replication/packaging/regulation module comprised 16 genes, suggesting that phage HEf13 has a host-independent DNA replication/packaging/regulation system with the following components: DNA replication proteins (DNA polymerase I, DNA primase, DNA replication protein, and replicative DNA helicase), packaging proteins (glutaredoxin-like protein, phage portal protein, phage terminase large subunit, and phage terminase small subunit), and regulation proteins (crossover junction endodeoxyribonuclease RuvC, cytidine deaminase, deoxynucleoside monophosphate kinase, DNA binding protein, DNA methyltransferase, HNH endonuclease, and HNH homing endonuclease). The phage structural module comprised seven genes required for host recognition and phage structural assembly: tail-associated proteins (phage tail protein, phage tail tube protein, tail fiber, and tail tape measure protein) and head-associated proteins (head morphogenesis protein, head-tail connector family protein, and major capsid protein). The host cell lysis gene module encoded cell-wall binding and cell lysis proteins (depolymerase, endolysin, and holin). In addition, four functional proteins had annotations of RNA transcription (RNA ligase and transcriptional regulator) or unidentified function (ATP-dependent metalloprotease and LPS glycosyltransferase). No lysogenic genes such as lytic repressor proteins, recombinases, excisionases, or integrases were identified, suggesting that phage HEf13 might be an obligate lytic phage only with a lytic cycle. Furthermore, virulence factor analysis of the phage HEf13 genome using the Virulence Searcher database revealed that none of the ORFs encode any functional proteins that can act as human virulence factors. Thus, our results suggest that phage HEf13 does not contain any possible pathogenic factors, and is therefore safe to use to treat E. faecalis-associated diseases.
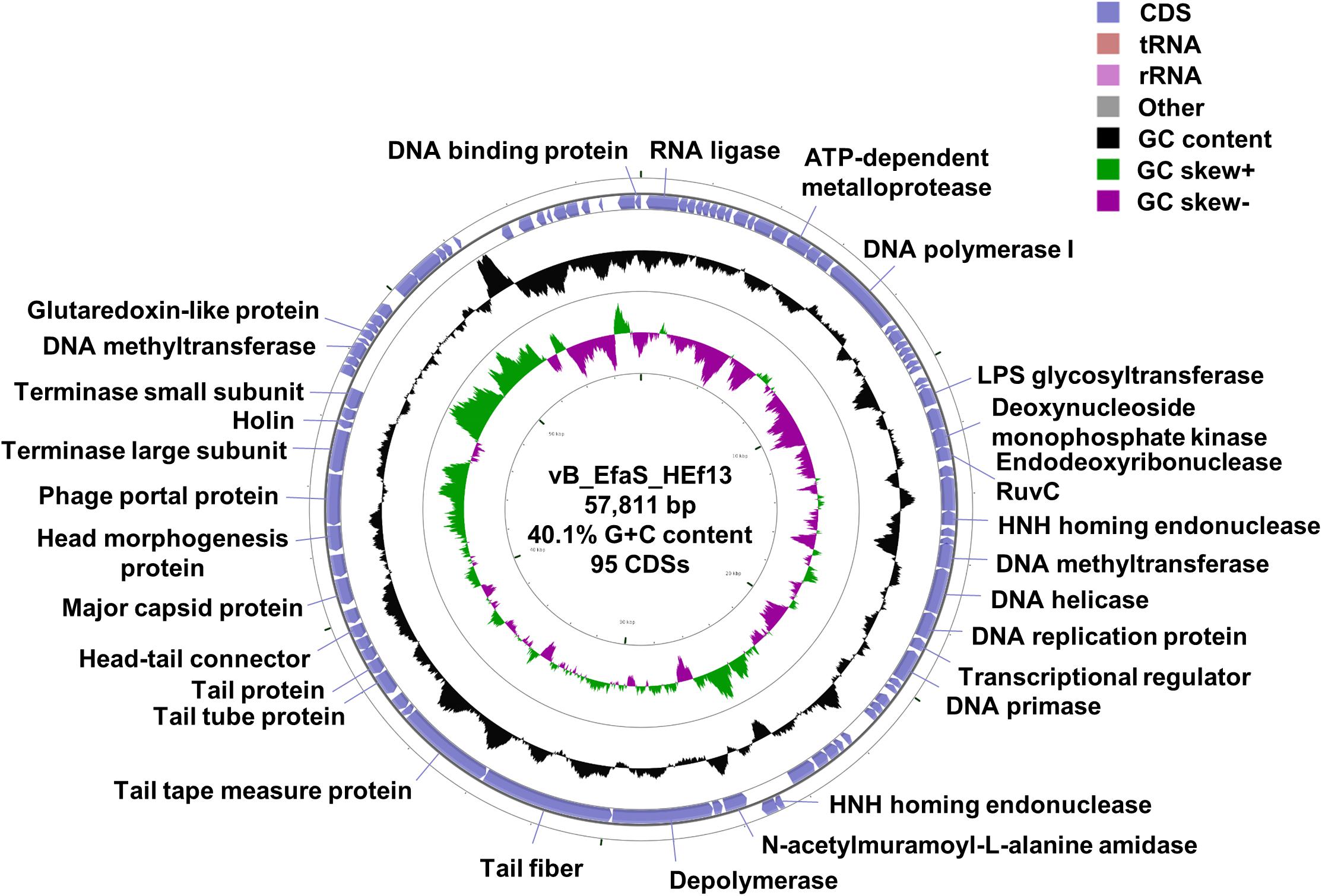
Figure 5. Genome map of phage HEf13 and its genetic characteristics. The whole genome sequence of phage HEf13 was determined using the Illumina Miseq platform and circular genome visualization of phage HEf13 was performed using the CGView server database. Annotation of the specific function of ORFs was conducted using RAST and the BLASTP database, and the complete genome sequence and annotation of ORFs were handled using the Artemis program. The second inner circle with the green and purple histogram shows the GC skew +/–, while the fourth inner circle with the black histogram indicates the GC content. The outer circle indicates the predicted ORFs of phage HEf13 together with their putative functions.
Comparative Genome Analysis of Phage HEf13 and Other Enterococcus Phages
Comparison of the nucleotide sequence of phage HEf13 with that of other E. faecalis phages using the BLASTN database revealed an average similarity of 95% between vB_EfaS_IME198 and phage EF-P29 (84–85% sequence coverage) belonging to the Sap6virus genus (Supplementary Table S1). To further analyze similarities and differences between phage HEf13 and other Sap6virus phages, we visualized comparative genome maps to assess similarities and differences in ORF organization. Most of the genes from phage HEf13 and other relative Sap6virus phages had a similar gene arrangement and were highly homologous. However, some genetic differences were observed in genes in the DNA replication/packing/regulation module (indicated in purple in Figure 6). Based on comparison of amino acid sequences using BLASTP, 77 of 95 ORFs in phage HEf13 shared 90–100% identity with ORFs of the other compared phages, but 18 ORFs, including the four unidentified ORFs, shared 0–89% identity (Supplementary Table S2). Interestingly, the four unidentified ORFs had no significant homology with any other sequences in E. faecalis phages. To determine the taxonomic affinity of phage HEf13, phylogenetic analysis using previously published genome sequences of eight other phages belonging to the Sap6virus genus was conducted. Phylogenetic trees constructed based on the amino acid sequences of phage portal protein, tail fiber protein, and DNA methyltransferase demonstrated that phage HEf13 clustered within the Sap6virus genus together with phages EF-P29, VD13, BC-611, SAP6, IME-EF1, vB_EfaS_IME198, SPQ-S1, and EF-P10 (Figures 7A–C). Collectively, the results of the phylogenetic analysis together with results of morphological – and comparative genome – analyses indicate that phage HEf13 belongs to the genus Sap6virus in the family Siphoviridae, order Caudovirales.
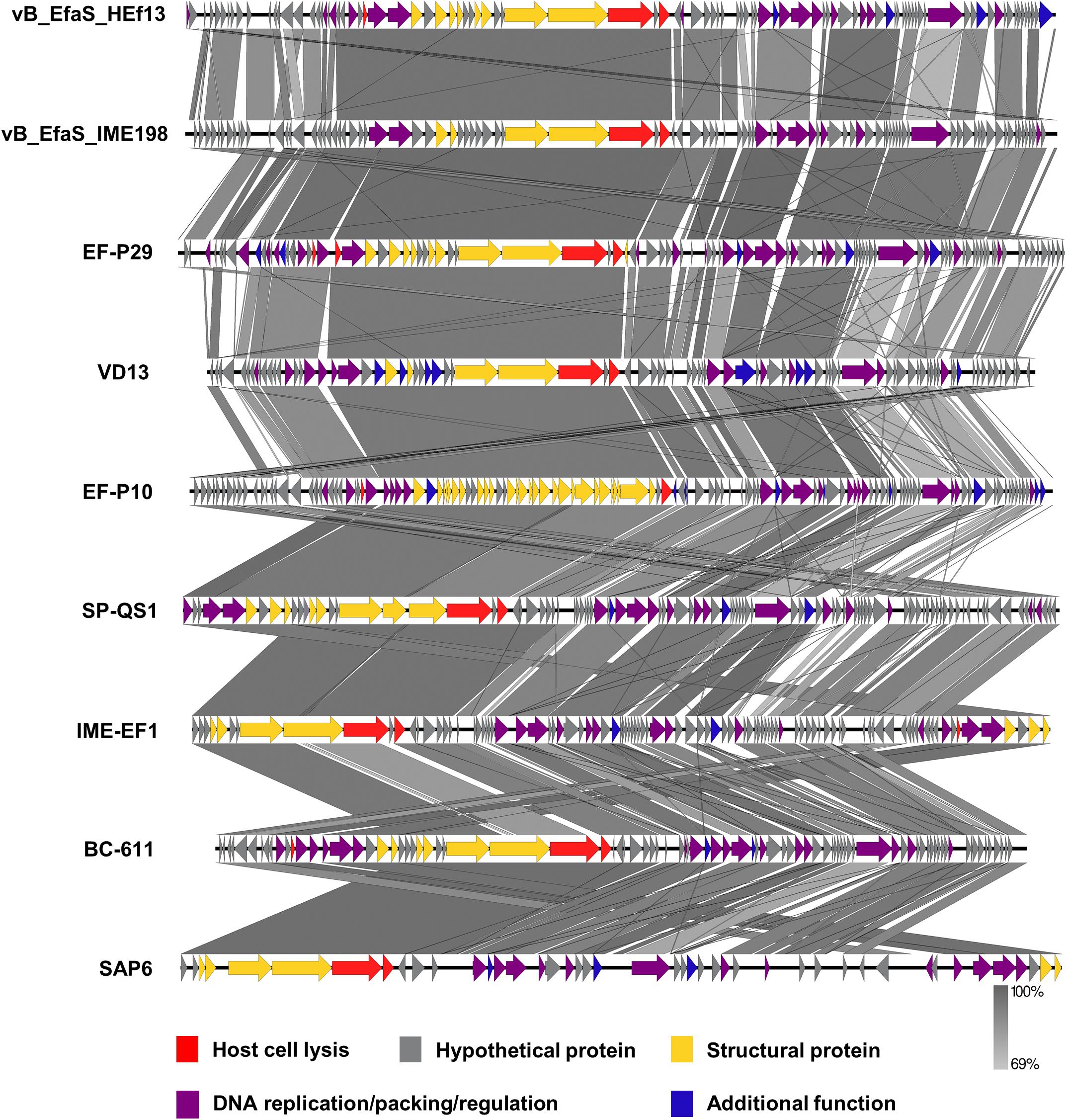
Figure 6. Phage HEf13 has high homology with phages in the Sap6virus genus of Enterococcus phages. Comparative genome analysis of phage HEf13 and eight other Enterococcus phages was performed using the EasyFig program. ORFs of phages were color-coded according to their predicted functions: host cell lysis (red), hypothetical protein (gray), structural protein (yellow), DNA replication/packing/regulation (purple), and additional function (blue). Genetic similarity profiles between phage HEf13 and other phages are presented in grayscale (percent homology).
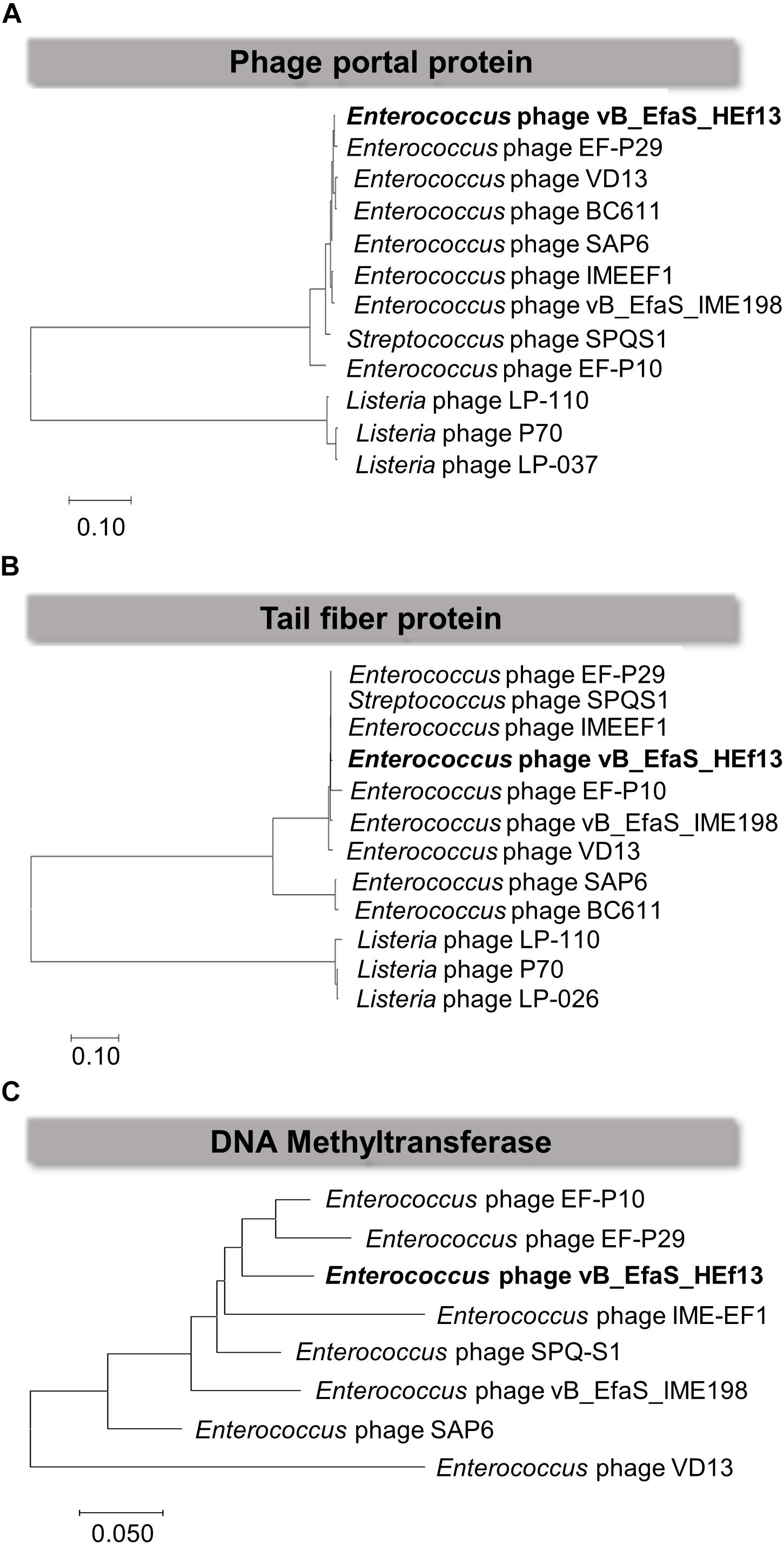
Figure 7. Phage HEf13 was classified as the Sap6virus lineage. The amino acid sequences of the phage portal protein, tail fiber protein, and DNA methyltransferase of phage HEf13 were compared with those of other phages using the BioEdit Sequence Alignment Editor program. A phylogenetic tree for each phage protein was generated using MEGA7 software. Scale bars represent a 10 (A,B) or 5% (C) difference in sequence similarity between phages.
Analysis of Polymorphism in the Potential Phage Receptor PIPEF
To understand the host specificity of phage HEf13, we investigated host cell receptors for phage HEf13. Among various bacterial cell wall membrane proteins, the PIPEF has recently been identified as a specific host cell receptor of E. faecalis Siphoviridae phages. Moreover, it has been reported that polymorphism in the variable region (covering amino acids 342–494) of PIPEF affects the binding affinity of phages to the host bacteria, subsequently determining the host range of the phage (Duerkop et al., 2016). Therefore, we examined polymorphism in the variable region of PIPEF by MSA for all E. faecalis strains used in the current study. As shown in Figure 8A, all of clear-plaque-forming strains have the same amino acid sequence in the variable region of PIPEF. However, most of turbid-plaque-forming strains and phage-resistant strains (7 of 10 strains), except for one turbid-plaque-forming strain and two phage-resistant strains, showed different sequences with those of clear-plaque-forming strains. Furthermore, phylogenetic tree of PIPEF based on the results of MSA showed that cluster of clear-plaque-forming strains were clearly distinguished from that of turbid-plaque-forming strains and phage-resistant strains, except for three strains (NCCP 15611, 16131, and 16132) (Figure 8B). These results demonstrated that polymorphism of PIPEF might be closely related with the host specificity of phage HEf13 to E. faecalis.

Figure 8. Polymorphism in the variable region of PIPEF in E. faecalis strains is related with phage HEf13 host specificity. (A) Multiple amino acid sequence alignment of the PIPEF variable region in 17 strains of E. faecalis was generated by Geneious 6.0.5 software. Difference in amino acid sequence of E. faecalis strains is presented in different colors. (B) Phylogenetic tree of PIPEF homologs was constructed by Neighbor-joining method with 1,000 replicates. Scale bar represents a 10% difference in sequence similarity between homologs. Strains are indicated by color as follows: clear-plaque-forming strain (red), turbid-plaque-forming strains (blue), and phage-resistant strains (black).
Discussion
Antibiotic-resistant E. faecalis strains have decreased the utility of antibiotics in clinical settings, making the phage therapy a highly attractive alternative therapeutic strategy. In this study, we isolated a novel lytic E. faecalis phage belonging to the genus Sap6virus in the family Siphoviridae. Our results demonstrated that phage HEf13 has a relatively broader host spectrum than previously isolated E. faecalis lytic phages. Notably, the host range of phage HEf13 was 70.5% against all tested E. faecalis strains (12 out of 17 tested strains), which is broader than other E. faecalis lytic phages with the host range from 7.6 to 42.5% (Rigvava et al., 2013; Zhang et al., 2013; Ladero et al., 2016; Cheng et al., 2017; Rahmat Ullah et al., 2017; Wang et al., 2018).
Regarding burst size, whereas the average burst size of E. faecalis phages reported previously was typically 36–122 PFU/infected bacteria, phage HEf13 has an approximately threefold higher burst size of 352 PFU/infected bacteria (Rigvava et al., 2013; Cheng et al., 2017; Wang et al., 2018). Moreover, phage HEf13 has an average latent time of 25 min after infection, which is shorter than those of other E. faecalis phages (30–50 min) (Uchiyama et al., 2008; Cheng et al., 2017; Wang et al., 2018). In addition, phage HEf13 showed strong lytic activities against all tested dental clinical isolates of E. faecalis at a broad range of MOI from 0.01 to 1 (Figure 1B and Supplementary Figures S1A,B,E,F). Earlier E. faecalis phage studies tested the lytic activity of phages against laboratory strains or clinically isolated antibiotic resistant strains recovered from various clinical specimens, but not against clinical isolates from oral specimens (Parasion et al., 2012; Rigvava et al., 2013; Zhang et al., 2013; Cheng et al., 2017; Wang et al., 2018). Thus, to the best of our knowledge, this is the first study to demonstrate the potential utility of E. faecalis phages as dental therapeutic reagents for post-treatment apical or refractory apical periodontitis, which are conditions closely related to E. faecalis infection.
Phage HEf13 appeared to be highly stable to a broad range of temperatures (4–60°C) and pH values (3–12). While phage HEf13 maintained over 50% of its titer for a long-term incubation period (after 12 h) at 60°C (Figure 4A), other previously isolated E. faecalis phages maintained about 40% of their titer for a short-term incubation period (even after 1 h) at 45–60°C (Rigvava et al., 2013; Rahmat Ullah et al., 2017). Since little is known about the pH stability of E. faecalis phages, we compared the pH stability of phage HEf13 with those of other Gram-positive bacteria phages. Phage HEf13 titers were not affected by incubation at pH 11 and remained at around 30% at pH 12 (Figure 4B), whereas reported titers of other phages were almost completely abolished at pH 11 (Li and Zhang, 2014; Chen et al., 2016; Phothichaisri et al., 2018). These comparisons demonstrated that phage HEf13 is relatively more stable to high temperatures and strongly basic conditions than other phages. Furthermore, these findings suggest that phage HEf13 may be highly potent in clinical settings considering that E. faecalis can survive and grow under harsh environments such as a pH of 11 and temperature of 45°C (McHugh et al., 2004; Foulquie Moreno et al., 2006). These characteristics of phage HEf13 make it potentially ideal to use in combinatory treatment with alkaline disinfectants such as calcium hydroxide and sodium hypochlorite, which are commonly used to treat endodontic infection.
Comparative genome analysis revealed that the genome sequence of phage HEf13 is highly conserved with those of other tested Sap6virus phages. Among 77 ORFs that showed high similarity to the ORFs of other phages in this genus, we found two ORFs of phage HEf13 that may generate different host specificity and phage protection based on differences in amino acid sequence. While the receptor binding protein (RBP) in the phage tail apparatus of phage HEf13 (ORF55) has a negatively charged amino acid (443D), the other eight Sap6virus phages evaluated have an uncharged amino acid at same position (443N). Because RBP plays an important role in phage adsorption to host bacteria, this may result in a different host specificity of phage HEf13 to other phages in same genus. In fact, a recent study using point-mutated phages demonstrated that point mutation of an amino acid in the RBP (K653N) generated significant differences in host specificity (Le et al., 2013). Additionally, the sequence of DNA methyltransferase (ORF75), which protects phages from bacterial restriction modification system, differed by 8–16% between phage HEf13 and the other Sap6virus phages evaluated, suggesting differences in phage protection.
The host specificity of HEf13 against E. faecalis appears to be associated with the PIPEF in light of the fact that all of the clear-plaque-forming strains possess the same amino acids sequence in the variable region of PIPEF even though turbid-plaque-forming strains and phage-resistant strains were not differentially clustered in the phylogenetic tree. Remarkably, three strains belonging to turbid-plaque-forming strain (NCCP 16132) or phage-resistant strains (NCCP 15611 and 16131) were also clustered in the same clade with clear-plaque-forming strains (Figure 8B). Although further study is needed, it can be explained by two potential mechanisms based on the previous studies. First, Gram-positive bacterial cell-wall components, such as capsular polysaccharides, wall-teichoic acids, and lipoteichoic acids, could hide the phage receptor by masking them, subsequently making phages inaccessible to its receptor (Munsch-Alatossava and Alatossava, 2013; Ozaki et al., 2017). Second, the aforementioned bacterial cell wall components could also compete with the receptors for phage adsorption, thereby interfering with a successful phage infection (Seed, 2015).
So far, a total of 22 E. faecalis lytic phages have been isolated and their stability and efficacy against various clinical and non-clinical strains have been evaluated. The phages identified previously, however, have a relatively narrow host spectrum and low stability to broad temperature and pH ranges, limiting their practical usefulness. Here, we characterized a newly isolated phage, HEf13, and demonstrated that this phage is highly potent against various E. faecalis strains and has high lytic ability and stability. Furthermore, the high lytic activity of phage HEf13 against dental clinical isolates suggests that phage HEf13 may be effective as a dental therapeutic agent to treat recurrent or refractory apical periodontitis related to E. faecalis infection.
Data Availability Statement
The raw datasets generated in this study can be found in GenBank, Accession Number: MH618488.
Author Contributions
DL and SH designed the research. DL and HN carried out the experiments. DL, JI, and SH analyzed and interpreted the data. DL, JI, SR, C-HY, and SH prepared and reviewed the manuscript. All authors have read and accepted the final manuscript.
Funding
This work was supported by grants from the National Research Foundation of Korea, which is funded by the Korean Government (NRF-2019R1A2C2007041 and NRF-2018R1A5A2024418).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the National Culture Collection for Pathogens for providing the pathogen resources (NCCP 14623, 14624, 14673, 15611, 16130, 16131, and 16132) of this study. We also thank the Korean Agricultural Culture Collection, Korean Collection for Oral Microbiology, and Korean Collection for Type Cultures for kindly providing E. faecalis strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02877/full#supplementary-material
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Arias, C. A., Contreras, G. A., and Murray, B. E. (2010). Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 16, 555–562. doi: 10.1111/j.1198-743X.2010.03214.x
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bolocan, A. S., Upadrasta, A., Bettio, P. H. A., Clooney, A. G., Draper, L. A., Ross, R. P., et al. (2019). Evaluation of phage therapy in the context of Enterococcus faecalis and its associated diseases. Viruses 11:E366. doi: 10.3390/v11040366
Bonilla, N., Santiago, T., Marcos, P., Urdaneta, M., Domingo, J. S., and Toranzos, G. A. (2010). Enterophages, a group of phages infecting Enterococcus faecalis, and their potential as alternate indicators of human faecal contamination. Water Sci. Technol. 61, 293–300. doi: 10.2166/wst.2010.815
Burrowes, B., Harper, D. R., Anderson, J., McConville, M., and Enright, M. C. (2011). Bacteriophage therapy: potential uses in the control of antibiotic-resistant pathogens. Expert Rev. Anti. Infect. Ther. 9, 775–785. doi: 10.1586/eri.11.90
Carver, T., Berriman, M., Tivey, A., Patel, C., Bohme, U., Barrell, B. G., et al. (2008). Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–2676. doi: 10.1093/bioinformatics/btn529
Chen, X., Xi, Y., Zhang, H., Wang, Z., Fan, M., Liu, Y., et al. (2016). Characterization and adsorption of Lactobacillus virulent phage P1. J. Dairy Sci. 99, 6995–7001. doi: 10.3168/jds.2016-11332
Cheng, M., Liang, J., Zhang, Y., Hu, L., Gong, P., Cai, R., et al. (2017). The Bacteriophage EF-P29 efficiently protects against lethal vancomycin-resistant Enterococcus faecalis and alleviates gut microbiota imbalance in a murine bacteremia model. Front. Microbiol. 8:837. doi: 10.3389/fmicb.2017.00837
Cho, I., and Blaser, M. J. (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270. doi: 10.1038/nrg3182
Duerkop, B. A., Huo, W., Bhardwaj, P., Palmer, K. L., and Hooper, L. V. (2016). Molecular basis for lytic bacteriophage resistance in enterococci. mBio 7:e01304–16. doi: 10.1128/mBio.01304-1316
Fauquet, C., Mayo, M. A., Maniloff, J., Desselberger, U., and Ball, L. A. (2005). Virus Taxonomy: Classification and Nomenclature of Viruses: Eighth Report of the International Committee on Taxonomy of Viruses. Cambridge, MA: Elsevier/Academic Press.
Foulquie Moreno, M. R., Sarantinopoulos, P., Tsakalidou, E., and De Vuyst, L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026
Francino, M. P. (2015). Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol. 6:1543. doi: 10.3389/fmicb.2015.01543
Golkar, Z., Bagasra, O., and Pace, D. G. (2014). Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries 8, 129–136. doi: 10.3855/jidc.3573
Gu, Y. H., Yamasita, T., and Kang, K. M. (2018). Subchronic oral dose toxicity study of Enterococcus faecalis 2001 (EF 2001) in mice. Toxicol. Res. 34, 55–63. doi: 10.5487/TR.2018.34.1.055
Jett, B. D., Huycke, M. M., and Gilmore, M. S. (1994). Virulence of enterococci. Clin. Microbiol. Rev. 7, 462–478. doi: 10.1128/cmr.7.4.462
Khalifa, L., Shlezinger, M., Beyth, S., Houri-Haddad, Y., Coppenhagen-Glazer, S., Beyth, N., et al. (2016). Phage therapy against Enterococcus faecalis in dental root canals. J. Oral Microbiol. 8:32157. doi: 10.3402/jom.v8.32157
Kim, A. R., Ahn, K. B., Yun, C. H., Park, O. J., Perinpanayagam, H., Yoo, Y. J., et al. (2019). Lactobacillus plantarum lipoteichoic acid inhibits oral multispecies biofilm. J. Endod. 45, 310–315. doi: 10.1016/j.joen.2018.12.007
Kropinski, A. M., Mazzocco, A., Waddell, T. E., Lingohr, E., and Johnson, R. P. (2009). Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 501, 69–76. doi: 10.1007/978-1-60327-164-6_7
Ladero, V., Gomez-Sordo, C., Sanchez-Llana, E., Del Rio, B., Redruello, B., Fernandez, M., et al. (2016). Q69 (an E. faecalis-infecting bacteriophage) as a biocontrol agent for reducing tyramine in dairy products. Front. Microbiol. 7:445. doi: 10.3389/fmicb.2016.00445
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Le, S., He, X., Tan, Y., Huang, G., Zhang, L., Lux, R., et al. (2013). Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PLoS One 8:e68562. doi: 10.1371/journal.pone.0068562
Lee, J. H., Bai, J., Shin, H., Kim, Y., Park, B., Heu, S., et al. (2016). A novel bacteriophage targeting Cronobacter sakazakii is a potential biocontrol agent in foods. Appl. Environ. Microbiol. 82, 192–201. doi: 10.1128/AEM.01827-1815
Li, L., and Zhang, Z. (2014). Isolation and characterization of a virulent bacteriophage SPW specific for Staphylococcus aureus isolated from bovine mastitis of lactating dairy cattle. Mol. Biol. Rep. 41, 5829–5838. doi: 10.1007/s11033-014-3457-3452
Lossouarn, J., Briet, A., Moncaut, E., Furlan, S., Bouteau, A., Son, O., et al. (2019). Enterococcus faecalis countermeasures defeat a virulent picovirinae bacteriophage. Viruses 11:48. doi: 10.3390/v11010048
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
McHugh, C. P., Zhang, P., Michalek, S., and Eleazer, P. D. (2004). pH required to kill Enterococcus faecalis in vitro. J. Endod. 30, 218–219. doi: 10.1097/00004770-200404000-200404008
Moelling, K., Broecker, F., and Willy, C. (2018). A wake-up call: we need phage therapy now. Viruses 10:E688. doi: 10.3390/v10120688
Munsch-Alatossava, P., and Alatossava, T. (2013). The extracellular phage-host interactions involved in the bacteriophage LL-H infection of Lactobacillus delbrueckii ssp. lactis ATCC 15808. Front. Microbiol. 4:408. doi: 10.3389/fmicb.2013.00408
O’Driscoll, T., and Crank, C. W. (2015). Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 8, 217–230. doi: 10.2147/IDR.S54125
Ozaki, T., Abe, N., Kimura, K., Suzuki, A., and Kaneko, J. (2017). Genomic analysis of Bacillus subtilis lytic bacteriophage varphiNIT1 capable of obstructing natto fermentation carrying genes for the capsule-lytic soluble enzymes poly-gamma-glutamate hydrolase and levanase. Biosci. Biotechnol. Biochem. 81, 135–146. doi: 10.1080/09168451.2016.1232153
Parasion, S., Kwiatek, M., Mizak, L., Gryko, R., Bartoszcze, M., and Kocik, J. (2012). Isolation and characterization of a novel bacteriophage phi4D lytic against Enterococcus faecalis strains. Curr. Microbiol. 65, 284–289. doi: 10.1007/s00284-012-0158-158
Park, E. A., Kim, Y. T., Cho, J. H., Ryu, S., and Lee, J. H. (2017). Characterization and genome analysis of novel bacteriophages infecting the opportunistic human pathogens Klebsiella oxytoca and K. pneumoniae. Arch. Virol. 162, 1129–1139. doi: 10.1007/s00705-016-3202-3203
Phothichaisri, W., Ounjai, P., Phetruen, T., Janvilisri, T., Khunrae, P., Singhakaew, S., et al. (2018). Characterization of bacteriophages infecting clinical isolates of Clostridium difficile. Front. Microbiol. 9:1701. doi: 10.3389/fmicb.2018.01701
Pinheiro, E. T., and Mayer, M. P. M. (2014). Enterococcus faecalis in oral infections. JBR J. Interdiscipl. Med. Dent. Sci. 3:160. doi: 10.4172/2376-032X.1000160
Rahmat Ullah, S., Andleeb, S., Raza, T., Jamal, M., and Mehmood, K. (2017). Effectiveness of a lytic phage SRG1 against vancomycin-resistant Enterococcus faecalis in compost and soil. Biomed. Res. Int. 2017:9351017. doi: 10.1155/2017/9351017
Rigvava, S., Tchgkonia, I., Jgenti, D., Dvalidze, T., Carpino, J., and Goderdzishvili, M. (2013). Comparative analysis of the biological and physical properties of Enterococcus faecalis bacteriophage vB_EfaS_GEC-EfS_3 and Streptococcus mitis bacteriophage vB_SmM_GEC-SmitisM_2. Can. J. Microbiol. 59, 18–21. doi: 10.1139/cjm-2012-2385
Sambrook, J., and Russell, D. W. (eds) (2001). “Bacteriophage lambda and its vectors,” in Molecular Cloning: A Laboratory Manual, 3rd edn (New York: Cold, Spring Harbor Laboratory Press), 2.1–2.117.
Seed, K. D. (2015). Battling phages: how bacteria defend against viral attack. PLoS Pathog. 11:e1004847. doi: 10.1371/journal.ppat.1004847
Souto, R., and Colombo, A. P. (2008). Prevalence of Enterococcus faecalis in subgingival biofilm and saliva of subjects with chronic periodontal infection. Arch. Oral. Biol. 53, 155–160. doi: 10.1016/j.archoralbio.2007.08.004
Stevens, R. H., Porras, O. D., and Delisle, A. L. (2009). Bacteriophages induced from lysogenic root canal isolates of Enterococcus faecalis. Oral Microbiol. Immunol. 24, 278–284. doi: 10.1111/j.1399-302X.2009.00506.x
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Svab, D., Falgenhauer, L., Rohde, M., Szabo, J., Chakraborty, T., and Toth, I. (2018). Identification and characterization of T5-Like bacteriophages representing two novel subgroups from food products. Front. Microbiol. 9:202. doi: 10.3389/fmicb.2018.00202
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Uchiyama, J., Rashel, M., Maeda, Y., Takemura, I., Sugihara, S., Akechi, K., et al. (2008). Isolation and characterization of a novel Enterococcus faecalis bacteriophage phiEF24C as a therapeutic candidate. FEMS Microbiol. Lett. 278, 200–206. doi: 10.1111/j.1574-6968.2007.00996.x
Underwood, A. P., Mulder, A., Gharbia, S., and Green, J. (2005). Virulence searcher: a tool for searching raw genome sequences from bacterial genomes for putative virulence factors. Clin. Microbiol. Infect. 11, 770–772. doi: 10.1111/j.1469-0691.2005.01210.x
Vidana, R., Sullivan, A., Billstrom, H., Ahlquist, M., and Lund, B. (2011). Enterococcus faecalis infection in root canals - host-derived or exogenous source? Lett. Appl. Microbiol. 52, 109–115. doi: 10.1111/j.1472-765X.2010.02972.x
Wang, R., Xing, S., Zhao, F., Li, P., Mi, Z., Shi, T., et al. (2018). Characterization and genome analysis of novel phage vB_EfaP_IME195 infecting Enterococcus faecalis. Virus Genes 54, 804–811. doi: 10.1007/s11262-018-1608-1606
Weiner, L. M., Webb, A. K., Limbago, B., Dudeck, M. A., Patel, J., Kallen, A. J., et al. (2016). Antimicrobial-Resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011-2014. Infect. Control Hosp. Epidemiol. 37, 1288–1301. doi: 10.1017/ice.2016.174
Xie, Y., Wahab, L., and Gill, J. J. (2018). Development and validation of a microtiter plate-based assay for determination of bacteriophage host range and virulence. Viruses 10:E189. doi: 10.3390/v10040189
Keywords: phage, Enterococcus phage HEf13, Enterococcus faecalis, phage infection protein of Enterococcus faecalis, whole genome analysis, human dentin
Citation: Lee D, Im J, Na H, Ryu S, Yun C-H and Han SH (2019) The Novel Enterococcus Phage vB_EfaS_HEf13 Has Broad Lytic Activity Against Clinical Isolates of Enterococcus faecalis. Front. Microbiol. 10:2877. doi: 10.3389/fmicb.2019.02877
Received: 10 September 2019; Accepted: 28 November 2019;
Published: 17 December 2019.
Edited by:
Robert Czajkowski, University of Gdańsk, PolandReviewed by:
Ahmed Askora, Zagazig University, EgyptJean-Christophe Giard, University of Caen Normandy, France
Copyright © 2019 Lee, Im, Na, Ryu, Yun and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seung Hyun Han, c2hoYW4tbWlAc251LmFjLmty
 Dongwook Lee
Dongwook Lee Jintaek Im
Jintaek Im Hongjun Na
Hongjun Na Sangryeol Ryu
Sangryeol Ryu Cheol-Heui Yun
Cheol-Heui Yun Seung Hyun Han
Seung Hyun Han