
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 10 December 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02852
This article is part of the Research TopicAntimicrobial Resistance As a Global Public Health Problem: How Can We Address It?View all 50 articles
We investigated antimicrobial resistance trends and characteristics of ESBL-producing Escherichia coli isolates from pets and whether this correlates with antibiotic usage in the clinic. Clinical samples containing E. coli from diseased cats and dogs were screened for antibiotic sensitivity and associated genotypic features. We identified 127 E. coli isolates from 1886 samples from dogs (n = 1565) and cats (n = 321) with the majority from urinary tract infections (n = 108, 85%). High rates of resistance were observed for β-lactams and fluoroquinolones and resistance to > 3 antibiotic classes (MDR) increased from 67% in 2012 to 75% in 2017 (P < 0.0001). This was especially true for strains resistant to 6–9 antibiotics that increased from 26.67 to 60.71%. Increased rates in β-lactam use for clinical treatment accompanied these increasing resistance rates. Accordingly, the most frequently encountered subtypes were blaCTX–M (n = 44, 34.65%), blaCTX–M–65 (n = 19) and blaCTX–M–15 (n = 18) and qnrB (n = 119, 93.70%). The blaCTX–M-isolates possessed 36 unique pulsed field electrophoretic types (PFGEs) and 28 different sequence types (STs) in ST405 (7, 15.9%), ST131 (3, 6.8%), ST73, ST101, ST372, and ST827 (2, 4.5% each) were the most prevalent. This data demonstrated a high level of diversity for the blaCTX–M-positive E. coli isolates. Additionally, blaNDM–5 was detected in three isolates (n = 3, 2.36%), comprised of two ST101 and one ST405 isolates, and mcr-1 was also observed in three colistin-resistant E. coli with three different STs (ST6316, ST405, and ST46). Our study demonstrates an increasing trend in MDR and ESBL-producing E. coli and this correlated with β-lactam antibiotic usage for treatment of these animals. This data indicates that there is significant risk for the spread of resistant bacteria from pets to humans and antibiotic use for pets should be more strictly regulated.
Antimicrobial resistance has become one of the most challenging problems for public health and results in 700,000 deaths annually (O’Neill, 2016). Antibiotic misuse has led to the spread of antibiotic resistance genes (ARGs) in humans, food animals, pets, songbirds, water, and soil and even agricultural plants, and this represents a significant threat to public health security (Carter et al., 2018; Hartantyo et al., 2018; Anderson et al., 2019; Chen et al., 2019; Gros et al., 2019; Sanchez et al., 2019; Vikesland et al., 2019). However, novel ARGs have emerged that encode resistance to carbapenems (blaNDM, blaIMP, blaVIM, and blaKPC) (Perez and Bonomo, 2019) colistin (mcr-1 to 9) and tigecycline (tetX3 and tetX4) (Carroll et al., 2019; He et al., 2019; Wang et al., 2019). The current limited development of novel drugs and substitutes makes the use of ARGs monitoring even more important to develop comprehensive and integrative measures for antimicrobial resistance.
Over the past two decades, there has been a significant number of infections caused by bacteria expressing extended-spectrum-β-lactamase (ESBL) and carbapenemases (Logan and Weinstein, 2017). In particular, ESBL isolates have been found in humans (Pitout and Laupland, 2008), animals, the environment (water and soil) (Runcharoen et al., 2017), meat and even vegetables (Yang et al., 2019). ESBL are becoming more common because this phenotype is being selected for by the use and exposure to β-lactams, especially the cephalosporins. This has generated a vicious cycle of drug resistance and decreased therapeutic effects. The increasing use of cephalosporin has been linked to Escherichia coli infections in pigs (Hammerum et al., 2014) and a high frequency of ESBL-producing E. coli was directly linked with a high consumption of third- or fourth- cephalosporins (Andersen et al., 2015).
One area that has not been thoroughly investigated is ARG presence in companion animals. This group comes in intimate contact with humans and pet contact can lead to bacterial spread to humans (Lloyd, 2007). In particular, E. coli is a common pathogenic agent isolated from pets (Mathers et al., 2015) and is often present in dogs and cats with urinary tract infections (UTIs), pyometra and respiratory tract infections (Karkaba et al., 2019; Moyaert et al., 2019). Multidrug resistance (MDR) E. coli isolates have emerged in companion animals in the United States and Europe (Morrissey et al., 2016), but data for China is lacking.
Here, we explored the effects of antibiotic dose on resistance phenotypes in pet bacterial isolates over a 6-year period. The aim of this study was to investigate the dissemination of ESBL-producing multidrug-resistant pathogens in diseased pets and the correlation between resistance rates and consumption of β-lactam antibiotics.
Animal samples were collected at the Veterinary Teaching Hospital of China Agricultural University (VTH-CAU), between January 2012 and June 2017. This study was approved by the China Agricultural University Animal Ethics Committee and the approval document (No. AW08104102-2) (see Supplementary Information). We collected 1886 samples from dogs (1565) and cats (321) and where some samples were gathered from different infections in the same pet. All samples consisted of urine (UTI) samples (1398, 74.1%) and samples from pyoderma (125, 6.7%), ear swabs (6.1%), effusions (115, 6.1%) and other specimens (132, 7.0%), which included pus from the uterus and soft tissue infections and several trachea lavage fluids.
Bacterial strains were recovered using blood agar and MacConkey agar plates that were incubated at 37°C for 24 h and single pink colonies were collected from each isolation plate. Subsequently, the DNA of individual clones was extracted by Fast Pure Bacteria DNA Isolation Mini Kit (Vazyme Biotech, Nanjing, China) and used as templates for PCR. PCR amplification of the 16S rDNA gene were performed for all isolates as previously described (Brianna et al., 2013), and amplicons were sequenced to confirm bacterial genus using the BLAST algorithm1.
Antimicrobial susceptibility of isolates was performed using the broth microdilution method according to Clinical and Laboratory Standards Institute guidelines (CLSI, 2015a). The breakpoints for other antimicrobials used the CLSI (M100-S25 or Vet01-A4/Vet01-S2) and EUCAST (CLSI, 2015b; EUCAST, 2019), where the breakpoints (R) for tigecycline, orbifloxacin, enrofloxacin, and marbofloxacin were recommended as ≥0.5, ≥4, ≥4, and ≥8 μg/ml, respectively. The screening panel consisted of 17 antibiotics that included ampicillin, cefazolin, cefotaxime, ceftriaxone, meropenem, amoxicillin-clavulanic acid, aztreonam, ciprofloxacin, enrofloxacin, marbofloxacin, orbifloxacin, chloramphenicol, amikacin, gentamicin, doxycycline, colistin, and tigecycline. Isolates with resistance to three or more categories of antimicrobial agents were classified as MDR (Magiorakos et al., 2012). E. coli ATCC 25922 was used as the quality control strain. Resistance was categorized according to Standardized International Terminology and our 17 test antibiotics were contained within nine categories (Magiorakos et al., 2012).
Antibiotic usage at VTH-CAU was recorded between January 2014 and September 2017 to correlate antibiotic usage and the resistance of E. coli isolates for each study animal.
Screening of E. coli isolates for ARG types was conducted using PCR for each isolate depending on the antibiotic resistance phenotype. The ARGs we examined were (1) carbapenemase genes blaNDM, blaIMP blaKPC, blaVIM, blaOXA, blaAIM, blaBIC, blaDIM, blaGIM, blaSIM, and blaSPM including blaNDM and blaCTX–M subtyping (Poirel et al., 2011), (2) β-lactamase genes blaSHV, blaTEM, and blaCTX–M (Casella et al., 2018), (3) plasmid-mediated AmpC β-lactamase genes blaMOX, blaCMY, blaLAT, blaDHA, blaACC, blaMIR, blaACT, and blaFOX (Perez-Perez and Hanson, 2002), (4) colistin resistance genes mcr-1-8 (Rebelo et al., 2018; Wang X. et al., 2018; Yang et al., 2018; Carroll et al., 2019), and (5) plasmid-mediated quinolone resistance (PMQR) genes qnrA, qnrB and qnrS including whether the gyrA and parC genes in the quinolone resistance determining region (QRDR) were mutated (Komp et al., 2003; Kraychete et al., 2016; Onseedaeng and Ratthawongjirakul, 2016). PCR primers used for screening are shown in Supplementary Table S1. All PCR amplicons were sequenced to confirm gene identity.
Conjugation assays were performed between clinical isolates and E. coli J53 to evaluate whether blaNDM and mcr-1 were mobilizable. Transconjugants were selected on MacConkey agar containing 100 mg/L sodium azide and 1 mg/L meropenem or colistin. Presumptive transconjugants were identified using PCR screening for blaNDM and mcr-1.
Transconjugant DNA was extracted and used for whole genome sequencing (WGS). A library of 250-bp paired-end was constructed by using a NEXT Ultra DNA Library Prep kit (New England Biolabs, Beverley, MA, United States) and sequenced using an Illumina HiSeq 2500 system at Bionova Biotech (Beijing, China). Raw data was de novo assembled using the SPAdes algorithm v.3.10.0. ARGs and plasmid incompatibility groups were analyzed using ResFinder v.3.22, RASTtk v.2.03 and PlasmidFinder 2.14, respectively.
The clonal relatedness of blaCTX–M positive was determined using pulsed-field gel electrophoresis (PFGE) typing conducted as previously described (Tenover et al., 1995). PFGE patterns were visually inspected and gel images were analyzed using InfoQuest FP software (Biorad, Hercules, CA, United States). Group analysis of PFGE profiles was performed using the Dice coefficient and the unweighted pair group method with arithmetic means. Simultaneously, multilocus sequence typing (MLST) analysis was conducted using the following E. coli gene set: recA, adk, fumC, icd, mdh, purA, and gyrB. The results were interpreted using the MLST database5. Sequence types (STs) of 131 clades in E. coli isolates were also screened using multiplex conventional PCR assays as previously described (Matsumura et al., 2017).
Tests of statistical significance was determined using Fisher’s exact test with Yates continuity correction in GraphPad Prism 6 (San Diego, CA, United States) and the level of significance was set at P < 0.05. All figures were designed by ggplot26 and GraphPad Prism 6.
We isolated 127 E. coli strains that included 108 (85.0%) urine and 5 (3.9%) uterus, 4 (3.1%) abdominal fluid, 3 (2.4%) pyoderma, 2 (1.6%) soft tissue infectious sites, and 1 (0.8%) from synovial fluid. The E. coli isolation rate was 6.73% (127/1886) and ranged from 4.42 to 9.41% per year. Resistance to β-lactams was extremely high for these isolates and included resistance to ampicillin (77.9%), cefotaxime (58.3%) and ceftriaxone (58.3%) and cefazolin (65.35%) with the exception of amoxicillin + clavulanic acid (22.8%) (Supplementary Tables S2, S3).
The range of resistance rates was narrow for ampicillin (68.42–92.86%) compared with cefotaxime (36.67–72.73%), ceftriaxone and cefotaxime (36.67–72.73%), cefazolin (43.33–86.36%), and amoxicillin + clavulanic acid (10.00–27.27%). These data demonstrated a significant increase in resistance rates from 2012 to 2017 and the cefotaxime, ceftriaxone, cefazolin, and amoxicillin + clavulanic acid rates more than doubled (Supplementary Figure S1 and Supplementary Table S3). The aztreonam resistance rate showed the greatest variability and was maximal in 2014 (71.43%) and minimal in 2012 (13.33%). We also found high rates of resistance to the four fluoroquinolones we tested (52.76–57.48%) except for 2013 (36.84%) (Supplementary Table S3). The other resistance groups we examined displayed irregular trends or slight increases over the 2012–2017 study period. Notably, almost all isolates showed high susceptibility to the “last-resort” antibiotics colistin, meropenem, and tigecycline and only 5 (3.94%) of our isolates were resistant to colistin and 3 (2.4%) to meropenem (Figure 1 and Supplementary Table S3).
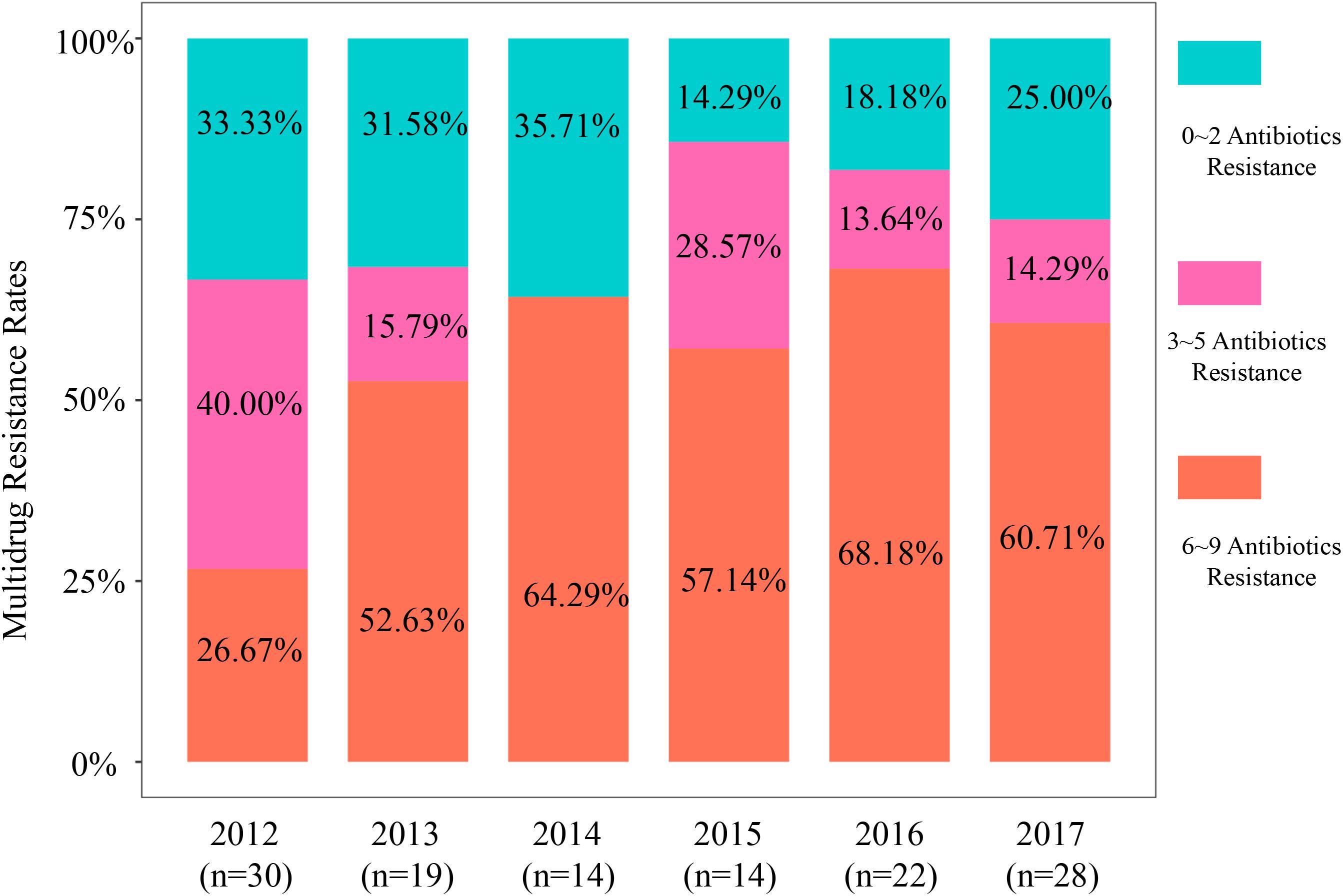
Figure 1. Multidrug resistance rates of 127 isolates based on antimicrobial category from 2012 to 2017.
The MDR values for our isolates from these pets were high with an overall MDR rate that increased 73.2% from 2012 to 2017 (n = 93). These MDR rates from 2012 to 2017 were 66.67, 68.42, 64.29, 85.71, 81.82, and 75.00% in 2017, respectively (P < 0.0001) (Supplementary Table S4). Specifically, MDR prevalence in 6–9 antimicrobial categories exhibited an obvious increase (Figure 1 and Supplementary Table S4). The MDR of our 44 CTX-M-producing E. coli was greater than for the non-CTX-M-expressing isolates (Supplementary Figures S5a,b). Moreover, all blaCTX–M-positive E. coli were MDR strains possessing resistance to the penicillins [ampicillin (AMP)], non-extended spectrum cephalosporins [cefazoline (CZO)], extended-spectrum cephalosporins [cefotaxime (CTX) and ceftriaxone (CRO)]. The most common MDR pattern was resistance to quinolones (ciproxacin, enrofloxacin, orbifloxacin, and marbofloxacin) and the tetracyclines (doxycycline) and aminoglycosides (gentamycin) + β-lactams (AMP, CZO, CTX, and CRO). When this data was viewed solely by the number of antibiotics per MDR isolate, resistance rates to 11–17 antibiotics increased significantly over the study period from 13.3, 31.6, 35.7, 35.7, 50.0, and 53.6% from 2012 to 2017, respectively (Supplementary Figure S2 and Supplementary Table S5).
We performed a correlation analysis between antibiotic usage and the rate of antibiotic resistance between 2014 and 2017. Amoxicillin-clavulanic acid (1.3 × 107 mg/yr), doxycycline (5.7 × 106 mg/yr), ampicillin (1.9 × 106 mg/yr), and enrofloxacin (8.1 × 105 mg/yr) were the most widely used antibiotics at the animal facility. With the widespread use of FQs in VTH-CAU, >50% of the clinical E. coli isolates showed resistance to FQs since 2012. Interestingly, even though fluoroquinolone usage had decreased dramatically, resistance rates to this antibiotic class remained high. In addition, the use of β-lactams, including amoxicillin-clavulanic acid, ampicillin, and ceftriaxone, had increased over the study period and was positively correlated with an increase in β-lactam resistance that was also related to dosage. Meropenem and imipenem were not used at the facility and we found no resistance from 2012∼2015 although three resistant E. coli were detected in 2016 (Supplementary Figure S3).
In our study, we identified 10 ARG types and mutations in gyrA and parC. The qnrB gene was present in 119 (93.70%) of the isolates (Supplementary Figure S4). In the gyrA gene of 77 quinolone-resistant isolates, 38 carried the mutations S83L and D87N, 9 S83L and D87Y, 2 D87N, 3 D87G, and 14 S83L (Supplementary Table S6). In the parC gene we found the mutations S80I (22 isolates), S80I and E84G (14 isolates) and S80I and E84V (8 strains). The ESBL and pAmpC-containing isolates harbored blaCTX–M (n = 44, 34.65%), blaSHV (n = 21, 16.55%), blaOXA (n = 9, 7.09%), blaCMY (n = 12, 9.45%), blaFOX (n = 4, 3.15%), and blaNDM (n = 3, 2.36%). CTX-M alleles were assigned to two main clusters; CTX-M-14 and -15 including seven CTX-M genotypes (-14, -15, -64, -65, -116, -127, and -174) where CTX-M-65 (43.18%, n = 19) and CTX-M-5 (40.91%, n = 18) predominated. All 44 blaCTX–M-positive isolates were resistant to cefotaxime and ceftriaxone and were classified as MDR. When compared with blaCTX-negative isolates, the blaCTX-positive isolates showed significantly greater resistance to second and third-generation cephalosporins, the fluoroquinolones, aztreonam, doxycycline, gentamicin, and chloramphenicol (P < 0.05) (Table 1 and Supplementary Figure S5). The only observed carbapenemase gene we identified was NDM-5 that was present in three isolates 16DU02, 16DF03, and 16XXI8 and all showed resistance to meropenem and all were collected from dogs in 2016. Of note, 16DU02 and 16DF03 were isolated from same dog but the samples were isolated from urine and an abdominal effusion, respectively, and they carried same genes blaNDM–5, blaTEM, blaCTX–M–65, and qnrB. The 16XXI8 isolate recovered from dog urine possessed blaNDM–5, blaTEM, blaCTX–M–15, blaOXA, and qnrB. Additionally, the colistin resistance gene mcr-1 was detected in three blaCTX–M positive E. coli while other mcr variants were undetected.
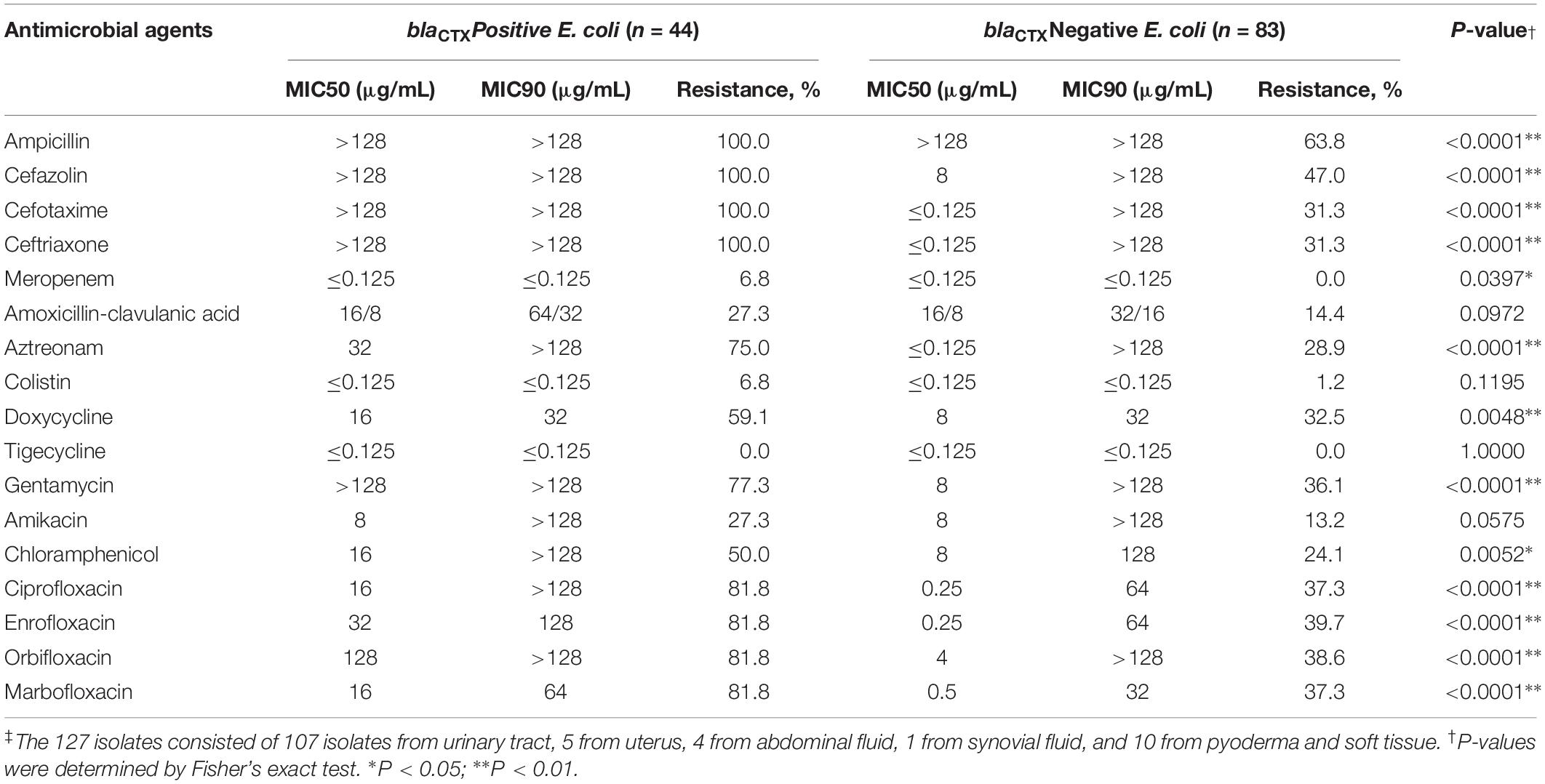
Table 1. Minimum inhibitory concentration (MIC) of antimicrobial agents for clinical E. coli isolates from cats and dogs in Beijing, China, 2012–2017 (n = 127)‡.
We further examined our mcr-1 and blaNDM isolates and tested whether these genes were present on mobile elements. In our conjugation tests, only blaNDM in three E. coli isolates were successfully transferred at frequencies of 4.86 × 10–8–8.02 × 10–7. Two complete 46,161 bp blaNDM–5-harboring plasmids pP16NDM-502 (MN701974) and pP16NDM-503 (MN701975) were obtained from strains 16DU02 and 16DF03 transconjugants, respectively. The backbone sequences were assembled and contigs and gaps were identified by additional PCR and sequence analyses. The other NDM-1-carrying plasmid was unsuccessfully assembled because of fragmentary and short contigs (Supplementary Figure S6). The two completely assembled plasmids were all in the IncX3 replication group. In addition, blaNDM–5 was contained within an insertion sequence (IS) cassette (ΔISAba125-IS5-blaNDM-ble-trpF-dsbC-IS26). blaNDM–5 and bleMBL were the only ARGs present in the two plasmids. Homology analysis revealed that pP16NDM-502 and pP16NDM-503 were ≥ 99% identical to the following IncX3 blaNDM plasmids: (i) pNDM_MGR194 (KF220657) from a Klebisella pneumoniae human isolate in India, (ii) p1079-NDM (MG825384) from a chicken E. coli isolate in China. (iii) pL65-9 (CP034744) from E. coli in goose in China, (iv) pZHDC40 (KY041843) from E. coli human isolate in China, (v) pQDE2-NDM (MH917280) from K. pneumoniae human isolate in China, (vi) pCRCB-101_1 (CP024820) from a Citrobacter freundii human isolate in Korea (vii) p128379-NDM (MF344560) from Enterobacter hormaechei from a human sample in China and (viii) pAD-19R (KX833071) from a chicken E. coli isolate in China (Supplementary Figure S6). We were unable to transfer the mcr-1 gene by conjugation in three separate tests and we could not assemble a complete plasmid sequence. However, no complete plasmid sequence of mcr-1 was successfully assembled by using three clinical isolates genomes. Although the analysis of incompatibility group in three clinical isolates genomes by analyzed PlasmidFinder-2.0 Serve revealed they possessed IncFIB, IncFIC, IncFII, and IncHI2 typical fragments, all mcr-1 genes weren’t located on those fragments (data not showed).
In our group of 127 E. coli isolates, most (n = 109, 85.8%) were successfully characterized by PFGE typing and included 38/44 blaCTX–M-positive isolates. These 38 isolates were obtained from cats (6) and dogs (32) and could be subdivided into 36 unique PFGE patterns. Interestingly, two isolates obtained from urine (15cu184) and perirenal effusion (15cu186) samples showed identical PFGE patterns and carried the same blaCTX–M–127 subtype but were recovered from different animals (Figure 2).
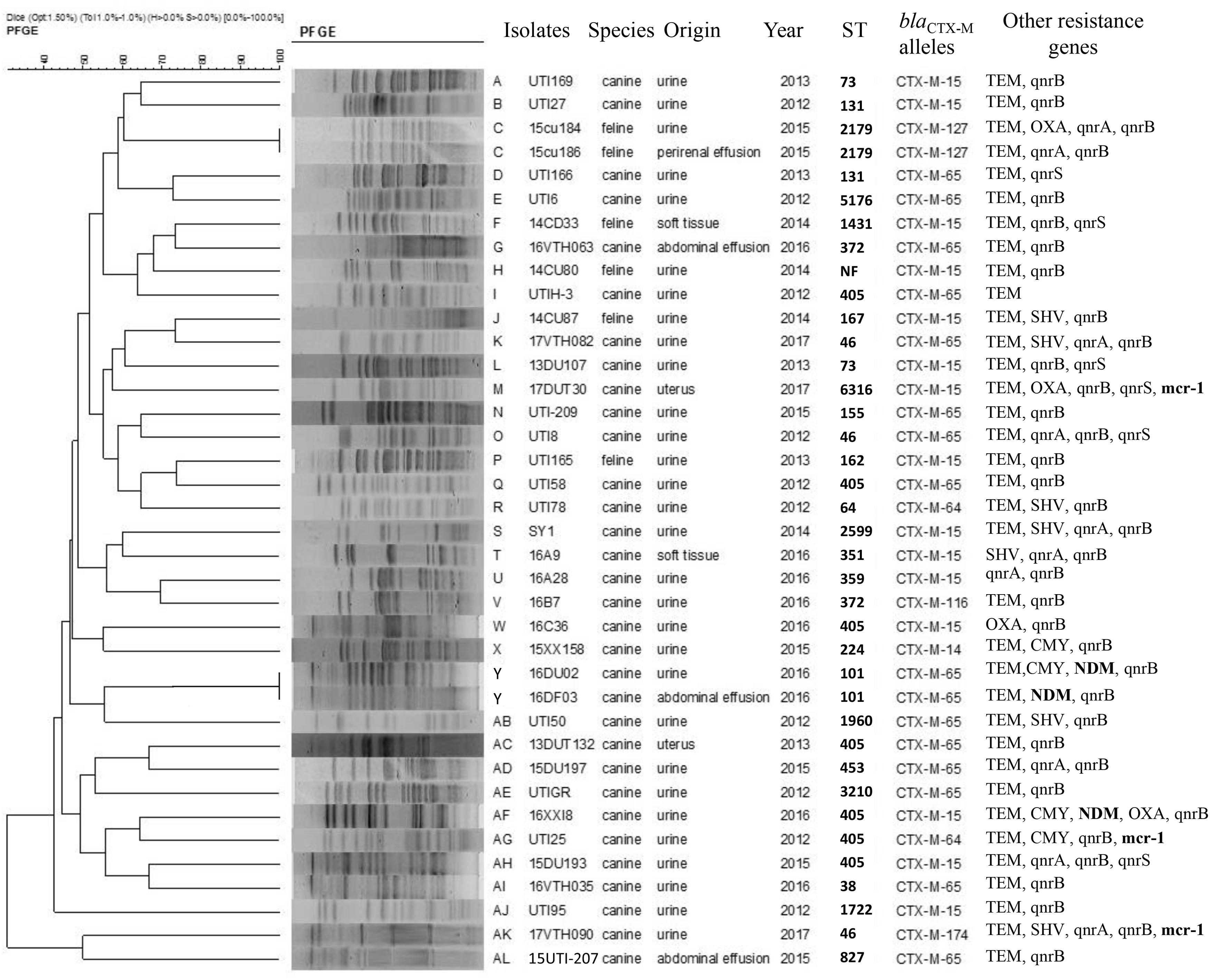
Figure 2. XbaI pulsed-field gel electrophoresis (PFGE) analysis of E. coli containing blaCTX variants.
We also found universality in MLST types and identified 28 different STs although 1 isolate failed to type. The most prevalent were ST405 (7, 15.9%), ST131 (3, 6.8%) ST73, ST101, ST372, and ST827 (2, 4.5% each). All ST405 strains were collected from dogs but in different years. Three ST131 isolates (UTI-27, UTI-166, and DU40) belonged to clade C1, in which UTI-27 and DU40 were assigned as C1-nM27, and UTI-166 as C1-M27. The blaCTX–M–65 and blaCTX–M–15 positive E. coli exhibited the greatest ST diversity and contained 13 and 14 STs, respectively. Additionally, three blaNDM–5-positive isolates belonged to ST405 (n = 1) and ST101 (n = 2) and three carrying-mcr-1 E. coli were classified as three different STs: ST6316, ST405, and ST46 (Figure 3).
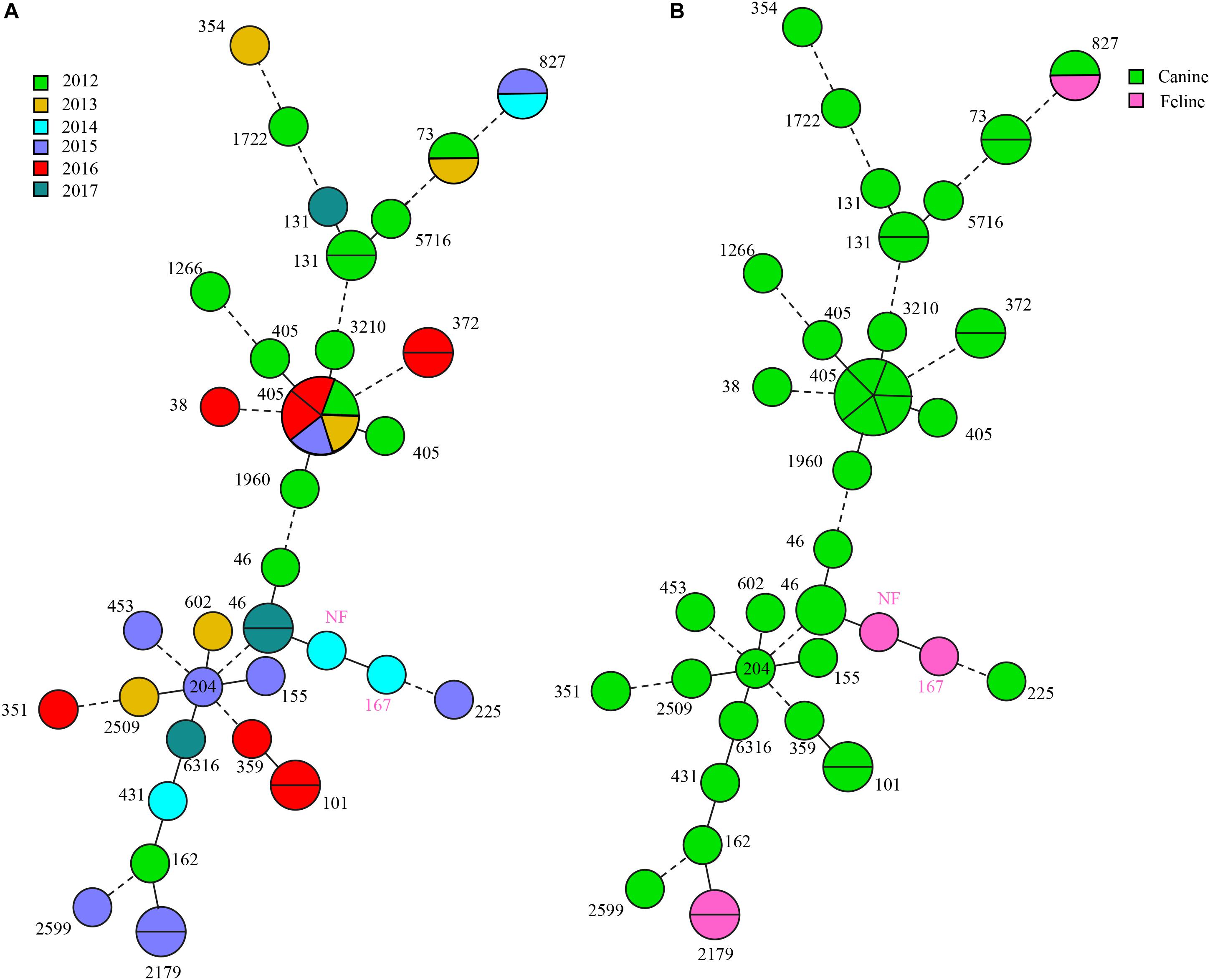
Figure 3. blaCTX–M-positive E. coli MLST typing. (A) MLST type colored based on different years. (B) MLST type colored based on different origins.
In this study, we investigated the antibiotic resistance profiles and trends in MDR E. coli isolates collected from diseased dogs and cats. We identified 127 E. coli isolates and most were associated with UTI (84.2%) accounting for 20.4% (108/529) of the confirmed bacterial UTI cases. In contrast, in the United States nearly 30% of UTI isolates from pets were E. coli (Ling et al., 2001; Hall et al., 2013). Previous studies in European countries based on 22,256 isolates from dogs and cats with UTI between 2008 and 2013 showed that E. coli was the most common pathogen in both dogs (59.50%) and cats (59.31%) (Marques et al., 2016). Isolation of E. coli from the respiratory tract is much less frequent and occurred in 10–15% of dog respiratory tract infections (Rheinwald et al., 2015; Morrissey et al., 2016).
Our 127 clinical E. coli isolates showed high prevalence rates of resistance to β-lactams (58.3–77.9%) and quinolones (52.8–57.4%). This pattern was quite different from the data of dog and cat E. coli isolates from United States that identified resistance to ampicillin at 40%, cephalexin 98% and doxycycline at 100% (Thungrat et al., 2015). An Australian study revealed that canine clinical E. coli isolates had low rates of resistance to quinolones (9.1–9.3%) and among 392 canine UTI isolates, 9.9–10.2% were resistant to third-generation cephalosporins (Saputra et al., 2017).
In the current study, the overall MDR frequency of 73.2% was higher than that observed in studies from the United States (52%) and Poland (66.8%) (Rzewuska et al., 2015; Thungrat et al., 2015). In contrast, a European multi-centre study on AMR of various bacteria isolated from companion animals with UTIs showed a much lower frequency of MDR among E. coli isolates (1.4–29.7%). The high MDR rates observed in the current study indicated that currently available antimicrobial treatment options for E. coli infections in companion animals are limited. We also identified an increase in E. coli MDR prevalence between 2012 and 2017 and increased rates of third-generation cephalosporins and amoxicillin-clavulanic acid usage was accompanied by increased AMR and MDR rates. Based on these findings, we speculate that the rising incidence of multidrug-resistant E. coli might be associated with the heavy use of these antibiotics in companion animals at the hospital.
Among the 44 CTX-M-producing E. coli isolates in this study, blaCTX–M–65 (43.2%) and blaCTX–M–15 (40.9%) were the most blaCTX–M variants. Currently, more than 220 CTX-M-lactamases have been reported and clustered into five subgroups, containing CTX-M-1, -2, -8, -9, and -25 depending on amino acid sequence homology (Peirano and Pitout, 2019). CTX-M-15 (CTX-M-1) is the most frequent CTX-M variant worldwide (Karim et al., 2001) especially in South-East Asia, China, South Korea, and Japan (Peirano and Pitout, 2019). Since 2000, blaCTX–M–15 has emerged worldwide and is the most prevalent ESBL globally (Peirano and Pitout, 2019). The spread of blaCTX–M–15 contributed to the dissemination of MDR bacterial isolates in animals but also in humans; a significant public health concern (Mugnaioli et al., 2006; Canton et al., 2012; Liu et al., 2016b). While the blaCTX–M–15 variant was also common in canine E. coli isolates from both Shanxi, China (n = 40) and the United States (n = 50), blaCTX–M–123 predominated in those studies (Liu et al., 2016a, b). The blaCTX–M–65 was the most prevalent subtype we found and is frequently detected in Salmonella isolates. In a previous study, the blaCTX–M–65 gene (n = 131) was identified among 153 ESBL-positive Salmonella isolates from poultry slaughterhouses (n = 121) and humans (n = 10) (Bai et al., 2016). Similarly, a study on chickens and pigs in China surveyed seven blaCTX–M–65 among Salmonella isolates (Zhang et al., 2016). All these studies indicated more frequent occurrences in China and is supported by data from 2005 to 2017 in another study (Bevan et al., 2017). Additionally, blaCTX–M–55 prevalence has increased in recent years in China among animal and human isolates, although not in our present study. Conversely, blaCTX–M–14 has a remarkable reduction based on data for China (Bevan et al., 2017). We found only one blaCTX–M–14 isolate while this gene is globally present (Bevan et al., 2017). Although other blaCTX–M variants, such as blaCTX-M-64, -174, -116, and -127 rarely occur, their presence in diseased pets suggests that diversity and evolution of blaCTX–M had occurred in these companion animals.
All blaCTX-carrying isolates in the current study had MDR profiles and showed high resistance rates to β-lactams, quinolones, doxycycline, gentamycin, and chloramphenicol. This suggested that other resistance genes may be co-transferred with blaCTX, making it even more difficult to eliminate the spread of MDR. Most blaCTX–M positive strains in other studies were also resistant to quinolones due to topoisomerase modifications of qnr genes (Lahlaoui et al., 2014) as we found in the present study. Additionally, mutations in gyrA and parC also can be responsible for quinolone resistance. Our results showed S83L and D87 alterations to N, Y, G generated in gyrA were commonly associated with resistance to fluoroquinolones and mostly generate high level resistance (Basu and Mukherjee, 2019).
Only three E. coli isolates containing blaNDM–5 were identified in our study animals with UTI in late 2016. Five (3.94%) were colistin resistance and three carried mcr-1. Previous reports implied that the transmissible blaNDM and mcr-1-carrying plasmids play a major role in the dissemination of these genes (Wang et al., 2017). For example, mcr-1, initially named as mobile colistin resistance gene, was generally considered as mediating the rapid spread of bacterial colistin resistance worldwide due to its mobile plasmid association. In contrast, we found that mcr-1 could not be mobilized by conjugation. In a previous study, 14/23 mcr-1-positive isolates were successfully transferred and six plasmids were non-transferable (Zhou et al., 2017). The mcr-1 gene was located in the chromosome of E. coli from a goose isolate (Lu et al., 2019). Furthermore, ISApl1 transposon can mediate mcr-1 transfer from chromosome to plasmids and this is the reason for its current global distribution (Wang R. et al., 2018). So, mcr-1-carried in a non-transferable plasmid or chromosome may be spread by ISApl1 or others transposons. In our study, we could not assemble a complete plasmid sequence for the mcr-1 genomic data; and mcr-1 genes were not present on those plasmid fragments and may be chromosomal. There are nine variants of mcr and we identified only mcr-1 (Carroll et al., 2019). This gene is the most prevalent variant globally and has been detected in almost 40 countries/regions across five continents in both involving developed and non-developed countries. Moreover, the mcr-1 gene was found in more than 11 bacterial species and in diverse locations such as rivers, public beaches, well water, wastewater, hospital sewage, foods (vegetables and meats), animals (wild birds, housefly/blowfly, cattle, pigs, poultry, and companion animals) (Feng, 2018). Our study provided new evidence for the above claim and we identified three mcr-1 positive E. coli in diseased pets. Recently, carbapenem-resistant Enterobacteriaceae (CRE) have posed a threat to humans and animals because they exhibited resistance to most β-lactams including carbapenems, further compromising treatment of MDR infections (Gupta et al., 2011; Potter et al., 2016).
Carbapenem-resistant Enterobacteriaceae is mediated largely by the production of carbapenemase especially for NDM isolates. NDM-5, an NDM-1 variant, exhibited increased enzyme activity to carbapenems (Rogers et al., 2013) and its gene blaNDM–5 has been reported worldwide (Khan et al., 2017) and is the most prevalent variant in China (Shen et al., 2018). In support of this, 84 (52%) NDM-5-producing E. coli were collected from 161 blaNDM carrying CRE in chickens (Wang et al., 2017). Our study revealed that pets have become a reservoir of NDM-5-producing E. coli. The blaNDM–5 gene is also associated with different plasmids such as IncFIA/B, IncFII, IncN, and IncX3 (Sun et al., 2015; Tyson et al., 2019; Zhang et al., 2019) and IncX3 was the dominant type (Li et al., 2018; Zhang et al., 2019). Similarly, the blaNDM–5-harboring plasmid was assigned to the IncX3 type, and their transferability was confirmed in our study indicating a risk of blaNDM–5 plasmid transfer between bacteria. However, further studies should be conducted to determine the origins of blaNDM and mcr-1 in the E. coli isolates from companion animals at the hospital. This will be helpful in designing measures to control the spread of MDR isolates.
All CTX-M-producing E. coli in our study displayed a diversity of PFGE patterns and STs demonstrating that blaCTX–M encoding ESBLs are present in diverse E. coli. Additionally, our diseased pet samples were from animals having no prior contact suggesting that clonal spread had a low frequency and it was not likely that clonal spread of blaCTX–M-positive E. coli occurred between the pets. But ESBL or other resistance genes can transfer between different E. coli by transferable genetic elements, as previously reported (Kim et al., 2019). However, two E. coli isolates (15cu184 and 15cu186) from same cat but different sample sources shared the same PFGE, ST, and blaCTX–M–127 types, indicating that clonal spread may be occurring. It was reported that dissemination of Enterobacteriaceae can occur between pets and their owners by both horizontal transfer and clonal expansion (Yao et al., 2016).
In our study, ST405 E. coli (n = 7) was the most prevalent isolates carrying blaCTX–M. They presented different blaCTX–M subtype genes including blaCTX–M–15 (n = 3), blaCTX–M–65 (n = 3), and blaCTX–M–64 (n = 1). In a clinical study from King Abdulaziz Medical City (KAMC) in Riyadh (Alghoribi et al., 2015), six ST405 E. coli harboring blaCTX–M were detected in UTI and ST405 (P ≤ 0.02) were significantly associated with ESBL production. ST405 extended-spectrum β-lactamase-producing E. coli (ESBL-EC) was also detected in animals, in which one ST405 ESBL-EC was screened in barbary macaques (Macaca sylvanus) in Algeria, notably, it held the mcr-1, blaTEM–1, and qnrB19 genes and blaCTX–M–15 (Bachiri et al., 2017). We observed a similar pattern and a UTI25 (ST405 ESBL-EC) isolate harbored mcr-1, blaTEM, qnrB, and blaCTX–M–64. Additionally, ST405 ESBL-EC (16XXI8) carried blaNDM–5, blaOXA, blaTEM, and qnrB. These data indicated ST405 ESBL-EC has become the vector of multiple resistance genes, containing carbapenem- and colistin-resistance genes. In a Sweden project, ST405 ESBL-EC was also found in UTI from a diseased cat (Bogaerts et al., 2015). We found seven ST405 ESBL-EC that were all recovered from dogs but in different years. Thus, companion animals already contain the hosts of ST405 ESBL-EC. In addition, except for ST405, we also identified two ST101 E. coli carrying blaNDM–5 (16DU02 and 16DF03). They were from the same animal source and possessed the same ST, PFGE, plasmid type and blaNDM–5, suggesting a clonal origin. ST101 was reported to be strongly associated with the NDM genotype although most were NDM-1 rather than NDM-5. Recently, five blaNDM–5-positive E. coli were identified associated with ST101 and ST1196 (Ranjan et al., 2016; Aung et al., 2018). In contrast, only two ST101 NDM-5-producing E. coli were observed in another study and most were ST167 and ST410 but were clonally spread in a hospital (Sun et al., 2019). This was similar to our study and suggested that ST101 may be becoming the most important clone for dissemination of blaNDM–5, similar to ST167 and ST410.
The clone ST131 was observed in three E. coli isolates. This is the predominant E. coli lineage in extraintestinal pathogenic E. coli (ExPEC) isolates worldwide and are associated with global community and nosocomial dissemination (Marie-Hélène et al., 2014). Currently, ST131 is classified as three clades: A, B, and C (Peirano and Pitout, 2019). In our study, all ST131 belonged to clade C and ST131 is in clade C 80% of the time (Peirano and Pitout, 2019). Clade C evolved from clade B and further evolved into C1 and C2. Recently, a new C1 subclade C1-M27 was identified in animal and human (Matsumura et al., 2016), including companion animals (Melo et al., 2019), and we found one C1-M27 subclades that are rare in China, but prevalent in Europe (Ghosh et al., 2017; Merino et al., 2018). The other ST131 belonged to subclade C1-nM27. C1 is commonly associated with quinolone resistance, in agreement with our results. The ESBL-EC isolates were commonly associated with ST131, where 40–80% of ESBL ExPEC belong to ST131 E. coli (Marie-Hélène et al., 2014), and CTX-15 was the most prevalent ESBL enzyme in ST131 ESBL-EC (Alghoribi et al., 2015). Our study demonstrated that blaCTX–M–65 was found in two ST131 ESBL-EC and one blaCTX–M–15 isolate. Almost all ST131 isolates were resistant to fluoroquinolones, especially for ESBL CTX-M-15 isolates (Marie-Hélène et al., 2014). We found similar results that almost all CTX-M ESBL isolates (40/44) were resistant to quinolones. ST131 ESBL E. coli possessed more frequent resistance to amikacin than non-ST131 ESBL isolates, but showed more frequent susceptibility to gentamicin or trimoxazole. On the other hand, the non-ESBL isolates such as E. coli ST131 were more frequently resistant to fluoroquinolones than non-ST131 isolates. Therefore, the resistance to quinolones may be associated with ST131 rather than ESBL presence (Marie-Hélène et al., 2014). These findings indicated that quinolones resistance may have been the predecessor of ESBL enzymes. We found that ST131 and ST405 were correlated to ESBL production as previously found (Alghoribi et al., 2015). Other STs in our study occurred infrequently. Recently, a water sample study revealed that ESBL-producing E. coli isolates were present 15 different STs (ST10, ST46, ST48, ST58, ST69, ST101, ST117, ST131, ST141, ST288, ST359, ST399, ST405, ST617, and ST4530) (Said et al., 2016), We detected ST46, ST101, ST131, ST359, and ST405 in our study. Multiple ST types (ST46, ST1286, ST10, ST29, ST101, and ST354) have also been found in chickens and they carried mcr-1 and produced ESBLs (Wu et al., 2018). In our work, 28 diverse STs in ESBL-EC were found.
We found a high prevalence of MDR E. coli isolates from diseased cats and dogs in Beijing and this rate has markedly increased over the last 6 years. The widespread use of third-generation cephalosporins and amoxicillin-clavulanic acid at the veterinary teaching hospital has likely contributed to the increasing frequency of β-lactam resistance in these isolates. These strains carried blaCTX, blaNDM–5, and mcr-1 and most possessed an MDR profile. Diversity analysis of the PFGE patterns and STs of these clinical E. coli isolates from different origins suggested that the dissemination of blaCTX have broad reservoirs of E. coli. Future studies should be undertaken to identify the MDR transmission mechanisms and establish national standards for the rational use of antibiotics in companion animals.
All datasets generated for this study are included in the article/Supplementary Material.
This study was approved by the Agricultural University Animal Ethics Committee, China. The animals were given the best practice veterinary care and informed consent was granted by the owners.
ZX was responsible for the study design. YC, ZL, YZ, and ZZ assisted in the data collection. ZL, YC, and LL interpreted the data. ZL, YC, and ZX completed the report writing. All authors revised, reviewed, and approved the final report.
This work was supported by the Beijing Science and Technology Plan Project (Z171100001517008) and the National Natural Science Foundation of China (31422055).
YZ was employed by the company of Shandong New Hope Liuhe Group Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02852/full#supplementary-material
Alghoribi, M. F., Gibreel, T. M., Farnham, G., Al Johani, S. M., Balkhy, H. H., and Upton, M. (2015). Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J. Antimicrob. Chemother. 70, 2757–2762. doi: 10.1093/jac/dkv188
Anderson, M., Clift, C., Schulze, K., Sagan, A., Nahrgang, S., Ait Ouakrim, D., et al. (2019). Averting the AMR Crisis: What are the Avenues for Policy Action for Countries in Europe? Copenhagen: European Observatory on Health Systems and Policies.
Andersen, V. D., Jensen, V. F., Vigre, H., Andreasen, M., and Agersø, Y. (2015). The use of third and fourth generation cephalosporins affects the occurrence of extended-spectrum cephalosporinase-producing Escherichia coli in Danish pig herds. Vet. J. 204, 345–350. doi: 10.1016/j.tvjl.2015.03.014
Aung, M. S., San, N., Maw, W. W., San, T., Urushibara, N., Kawaguchiya, M., et al. (2018). Prevalence of extended-spectrum beta-lactamase and carbapenemase genes in clinical isolates of Escherichia coli in Myanmar: dominance of blaNDM-5 and emergence of blaOXA-181. Microb. Drug. Resist. 24, 1333–1344. doi: 10.1089/mdr.2017.0387
Bachiri, T., Lalaoui, R., Bakour, S., Allouache, M., Belkebla, N., Rolain, J. M., et al. (2017). First report of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli ST405 isolated from wildlife in Bejaia, Algeria. Microb. Drug. Resist. 24, 2017–2026. doi: 10.1089/mdr.2017.0026
Bai, L., Zhao, J., Gan, X., Wang, J., Zhang, X., Cui, S., et al. (2016). Emergence and diversity of Salmonella enterica serovar indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime from patients and food-producing animals in China. Antimicrob. Agents. Chemother. 60, 3365–3371. doi: 10.1128/AAC.02849-15
Basu, S., and Mukherjee, M. (2019). Conjugal transfer of PMQR from uropathogenic E. coli under high ciprofloxacin selection pressure generates gyrA mutation. Microbial. Pathog. 132, 26–29. doi: 10.1016/j.micpath.2019.04.021
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M-beta-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Bogaerts, P., Huang, T. D., Bouchahrouf, W., Bauraing, C., Berhin, C., El, G. F., et al. (2015). Characterization of ESBL- and AmpC-producing Enterobacteriaceae from diseased companion animals in Europe. Microb. Drug. Resist. 21:643. doi: 10.1089/mdr.2014.0284
Brianna, L., Mihai, P., Martin, A., Walker, A. W., Volker, M., Ahmed, D., et al. (2013). Survey of culture, goldengate assay, universal biosensor assay, and 16s rRNA gene sequencing as alternative methods of bacterial pathogen detection. J. Clin. Microbiol. 51, 3263–3269. doi: 10.1128/JCM.01342-13
Canton, R., Gonzalez-Alba, J. M., and Galan, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., Wiedmann, M., et al. (2019). Identification of novel mobilized colistin resistance gene in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. MBio 10:e853-19. doi: 10.1128/mBio.00853-19
Carter, D. L., Docherty, K. M., Gill, S. A., Baker, K., Teachout, J., and Vonhof, M. J. (2018). Antibiotic resistant bacteria are widespread in songbirds across rural and urban environments. Sci. Total. Environ. 627, 1234–1241. doi: 10.1016/j.scitotenv
Casella, T., Haenni, M., Madela, N. K., Andrade, L. K. D., Pradela, L. K., Andrade, L. N. D., et al. (2018). Extended-spectrum cephalosporin-resistant Escherichia coli isolated from chickens and chicken meat in Brazil is associated with rare and complex resistance plasmids and pandemic ST lineages. J. Antimicrob. Chemother. 73, 3293–3297. doi: 10.1093/jac/dky335
Chen, Q. L., Cui, H. L., Su, J. Q., Penuelas, J., and Zhu, Y. G. (2019). Antibiotic resistomes in plant microbiomes. Trends. Plant. Sci. 24, 530–541. doi: 10.1016/j.tplants.2019.02.010
CLSI, (2015a). Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 25th Informational Supplement, M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI, (2015b). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacterial Isolated From Animals. Approved Standard-Fourth Edition and Supplement. CLSI documents VET01A4E and VET01S3E. Wayne, PA: Clinical and Laboratory Standards Institute.
EUCAST, (2019). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Växjö: The European Committee on Antimicrobial Susceptibility Testing.
Feng, Y. (2018). Transferability of mcr-1/2 polymyxin resistance: complex dissemination and genetic mechanism. ACS. Infect. Dis. 4, 291–300. doi: 10.1021/acsinfecdis.7b00201
Ghosh, H., Doijad, S., Falgenhauer, L., Fritzenwanker, M., Imirzalioglu, C., and Chakraborty, T. (2017). blaCTX-M-27–Encoding Escherichia coli sequence type 131 lineage C1-M27 clone in clinical isolates. Germany. Emerg. Infect. Dis. 23, 1754–1756. doi: 10.3201/eid2310.170938
Gros, M., Marti, E., Balcázar, J. L., Boy-Roura, M., Busquets, A., Colón, J., et al. (2019). Fate of pharmaceuticals and antibiotic resistance genes in a full-scale on-farm livestock waste treatment plant. J. Hazard. Mater. 378:120716. doi: 10.1016/j.jhazmat.2019.05.109
Gupta, N., Limbago, B. M., Patel, J. B., and Kallen, A. J. (2011). Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53, 60–67. doi: 10.1093/cid/cir202
Hall, J. L., Holmes, M. A., and Baines, S. J. (2013). Prevalence and antimicrobial resistance of canine urinary tract pathogens. Vet. Rec. 173:549. doi: 10.1136/vr.101482
Hammerum, A. M., Larsen, J., Andersen, V. D., Lester, C. H., Skovgaard Skytte, T. S., Hansen, F., et al. (2014). Characterization of extended-spectrum beta-lactamase (ESBL)-producing E. coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third- or fourth-generation cephalosporins. J. Antimicrob. Chemother. 69, 2650–2657. doi: 10.1093/jac/dku180
Hartantyo, S. H. P., Chau, M. L., Fillon, L., Ariff, A. Z. B. M., Kang, J. S. L., Aung, K. T., et al. (2018). Sick pets as potential reservoirs of antibiotic-resistant bacteria in Singapore. Antimicrob. Resist. Infect. Control. 7:106. doi: 10.1186/s13756-018-0399-9
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-442
Karim, A., Poirel, L., Nagarajan, S., and Nordmann, P. (2001). Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS. Microbiol. Lett. 201, 237–241. doi: 10.1111/j.1574-6968.2001.tb10762.x
Karkaba, A., Hill, K., Benschop, J., Pleydell, E., and Grinberg, A. (2019). Carriage and population genetics of extended spectrum β-lactamase-producing E. coli in cats and dogs in new zealand. Vet. Microbiol. 233, 61–67. doi: 10.1016/j.vetmic.2019.04.015
Khan, A. U., Maryam, L., and Zarrilli, R. (2017). Structure, Genetics and worldwide spread of New Delhi Metallo-beta-lactamase (NDM): a threat to public health. BMC. Microbiol. 17:101. doi: 10.1186/s12866-017-1012-8
Kim, K. G., Jeong, J., Kim, M. J., Park, D. W., Shin, J. H., Park, H. J., et al. (2019). Prevalence and molecular epidemiology of ESBLs, plasmid-determined AmpC-type β-lactamases and carbapenemases among diarrhoeagenic E. coli isolates from children in gwangju, korea: 2007-16. J. Antimicrob. Chemother. 74, 2181–2187. doi: 10.1093/jac/dkz175
Komp, L. P., Karlsson, A. D., and Hughes, D. (2003). Mutation rate and evolution of fluoroquinolone resistance in E. coli isolates from patients with urinary tract infections. Antimicrob. Agents. Chemother. 47:3222. doi: 10.1128/AAC.47.10.3222-3232.2003
Kraychete, G. B., Botelho, L. A. B., Campana, E. H., Picão, R. C., and Bonelli, R. R. (2016). Updated multiplex PCR for detection of all six plasmid-mediated qnr gene families. Antimicrob. Agents. Chemother. 60, 1416–1447. doi: 10.1128/AAC.01447-16
Lahlaoui, H., Khalifa, A. B. H., and Moussa, M. B. (2014). Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum beta-lactamase (ESBL). Med. Mal. Infect. 44, 400–404. doi: 10.1016/j.medmal.2014.03.010
Li, X., Fu, Y., Shen, M., Huang, D., Du, X., Hu, Q., et al. (2018). Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal E. coli in China. Antimicrob. Resist. Infect. Control. 7:59. doi: 10.1186/s13756-018-0349-6
Ling, G. V., Norris, C. R., Franti, C. E., Eisele, P. H., Johnson, D. L., Ruby, A. L., et al. (2001). Interrelations of organism prevalence, specimen collection method, and host age, sex, and breed among 8,354 canine urinary tract infections (1969-1995). J. Vet. Intern. Med. 15, 341–347. doi: 10.1111/j.1939-1676.2001.tb02327.x
Liu, X., Liu, H., Li, Y., and Hao, C. (2016a). High prevalence of beta-lactamase and plasmid-mediated quinolone resistance genes in extended-spectrum cephalosporin-resistant Escherichia coli from dogs in shaanxi, china. Front. Microbiol. 7:1843. doi: 10.3389/fmicb.2016.01843
Liu, X., Thungrat, K., and Boothe, D. M. (2016b). Occurrence of OXA-48 carbapenemase and other beta-lactamase genes in ESBL-producing multidrug resistant Escherichia coli from dogs and cats in the United States, 2009-2013. Front. Microbiol. 7:1057. doi: 10.3389/fmicb.2016.01057
Lloyd, H. D. (2007). Reservoirs of antimicrobial resistance in pet animals. Clin. Infect. Dis. 45, 148–152. doi: 10.1086/519254
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
Lu, X., Xiao, X., Liu, Y., Li, Y., Li, R., and Wang, Z. (2019). Chromosome-mediated mcr-1 in E. coli strain L73 from a goose. Int. J. Antimicrob. Agents. 54, 99–101. doi: 10.1016/j.ijantimicag.2019.03.003
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570
Marie-Hélène, N. C., Xavier, B., and Jean-Yves, M. (2014). E. coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Marques, C., Gama, L. T., Belas, A., Bergstrom, K., Beurlet, S., Briend-Marchal, A., et al. (2016). European multicenter study on antimicrobial resistance in bacteria isolated from companion animal urinary tract infections. BMC. Vet. Res. 12:213. doi: 10.1186/s12917-016-0840-3
Mathers, A. J., Peirano, G., and Pitout, J. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
Matsumura, Y., Pitout, J. D. D., Gomi, R., Matsuda, T., Noguchi, T., Yamamoto, M., et al. (2016). Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg. Infect. Dis. 22:1900. doi: 10.3201/eid2211.160519
Matsumura, Y., Pitout, J. D. D., Peirano, G., DeVinney, R., Noguchi, T., Yamamoto, M., et al. (2017). Rapid identification of different Escherichia coli sequence type 131 clades. Antimicrob. Agents. Chemother. 61:e179-17. doi: 10.1128/AAC.00179-17
Melo, L. C., Haenni, M., Saras, E., Duprilot, M., Nicolas-Chanoine, M. H., and Madec, J. Y. (2019). Emergence of the C1-M27 cluster in ST131 Escherichia coli from companion animals in France. J. Antimicrob. Chemother. 74, 3111–3113. doi: 10.1093/jac/dkz304
Merino, I., Hernández-García, M., Turrientes, M. C., Pérez-Viso, B., López-Fresneña, N., and Diaz-Agero, C. (2018). Emergence of ESBL-producing Escherichia coli ST131-C1-M27 clade colonizing patients in Europe. J. Antimicrob. Chemother. 73, 2973–2980. doi: 10.1093/jac/dky296
Morrissey, I., Moyaert, H., de Jong, A., El, G. F., Klein, U., Ludwig, C., et al. (2016). Antimicrobial susceptibility monitoring of bacterial pathogens isolated from respiratory tract infections in dogs and cats across europe: compath results. Vet. Microbiol. 191, 44–51. doi: 10.1016/j.vetmic.2016.05.020
Moyaert, H., Jong, D. A., Simjee, S., Rose, M., Youala, M., Garch, E. F., et al. (2019). Survey of antimicrobial susceptibility of bacterial pathogens isolated from dogs and cats with respiratory tract infections in europe: compath results. J. Appl. Microbiol. 127, 29–46. doi: 10.1111/jam.14274
Mugnaioli, C., Luzzaro, F., De Luca, F., Brigante, G., Perilli, M., Amicosante, G., et al. (2006). CTX-M-type extended-spectrum beta-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob. Agents. Chemother. 50, 2700–2706. doi: 10.1128/AAC.00068-06
O’Neill, J. (2016). Tackling drug-resistant infections globally: final report and recommendations. The review on antimicrobial resistance. Available at: http://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed August, 2019).
Onseedaeng, S., and Ratthawongjirakul, P. (2016). Rapid detection of genomic mutations in gyrA and parC genes of E. coli by multiplex allele specific polymerase chain reaction. J. Clin. Lab. Anal. 30, 947–955. doi: 10.1002/jcla.21961
Peirano, G., and Pitout, J. D. (2019). Extended spectrum beta-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79, 1529–1541. doi: 10.1007/s40265-019-01180-3
Perez, F., and Bonomo, R. A. (2019). Carbapenem-resistant Enterobacteriaceae: global action required. Lancet. Infect. Dis. 19:561. doi: 10.1016/S1473-3099(19)30210-5
Perez-Perez, F. J., and Hanson, N. D. (2002). Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153. doi: 10.1128/JCM.40.6.2153-2162.2002
Pitout, J. D., and Laupland, K. B. (2008). Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet. Infect. Dis. 8, 159–166. doi: 10.1016/S1473-3099(08)70041-0
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Potter, R. F., D’Souza, A. W., and Dantas, G. (2016). The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug. Resist. Updates. 29, 30–46. doi: 10.1016/j.drup.2016.09.002
Ranjan, A., Shaik, S., Mondal, A., Nandanwar, N., Hussain, A., Semmler, T., et al. (2016). Molecular epidemiology and genome dynamics of New Delhi Metallo-β-lactamase-producing extra intestinal pathogenic Escherichia coli strains from India. Antimicrob. Agents. Chemother. 60, 6795–6805. doi: 10.1128/AAC.01345-16
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eur. Surveill. 23:17–00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672672
Rheinwald, M., Hartmann, K., Hahner, M., Wolf, G., Straubinger, R. K., and Schulz, B. (2015). Antibiotic susceptibility of bacterial isolates from 502 dogs with respiratory signs. Vet. Rec. 176:357. doi: 10.1136/vr.102694
Rogers, B. A., Sidjabat, H. E., Anna, S., Anderson, T. L., Shalini, P., Jian, L., et al. (2013). Treatment options for New Delhi metallo-beta-lactamase-harboring Enterobacteriaceae. Microb. Drug. Resist. 19, 100–103. doi: 10.1089/mdr.2012.0063
Runcharoen, C., Raven, K. E., Reuter, S., Kallonen, T., Paksanont, S., Thammachote, J., et al. (2017). Whole genome sequencing of ESBL-producing Escherichia coli isolated from patients, farm waste and canals in Thailand. Genome. Med. 9:81. doi: 10.1186/s13073-017-0471-8
Rzewuska, M., Czopowicz, M., Kizerwetter-Swida, M., Chrobak, D., Blaszczak, B., and Binek, M. (2015). Multidrug resistance in Escherichia coli strains isolated from infections in dogs and cats in poland (2007-2013). Sci. World. J. 2015:408205. doi: 10.1155/2015/408205
Said, L. B., Jouini, A., Alonso, C. A., Klibi, N., Dziri, R., Boudabous, A., et al. (2016). Characteristics of extended-spectrum beta-lactamase (ESBL)-and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci. Total. Environ. 550, 1103–1109. doi: 10.1016/j.scitotenv.2016.01.042
Sanchez, M. L., Vallina-Victorero, M. J., Bachiller, M. R., Arbizu, R., Llaneza, E., Rozada, S., et al. (2019). Variability in the community consumption of antibiotics: a problem in Europe. Spain Asturias. Infez. Med. 27, 134–140.
Saputra, S., Jordan, D., Mitchell, T., Wong, H. S., Abraham, R. J., Kidsley, A., et al. (2017). Antimicrobial resistance in clinical Escherichia coli isolated from companion animals in Australia. Vet. Microbiol. 211, 43–50. doi: 10.1016/j.vetmic.2017.09.014
Shen, Z., Hu, Y., Sun, Q., Hu, F., Zhou, H., Shu, L., et al. (2018). Emerging carriage of blaNDM-5 and mcr-1 in Escherichia coli from healthy people in multiple regions in china: a cross sectional observational study. EClinicalMedicine 6, 11–20. doi: 10.1016/j.eclinm.2018.11.003
Sun, P., Xia, W., Liu, G., Huang, X., Tang, C., Liu, C., et al. (2019). Characterization of blaNDM-5-positive Escherichia coli prevalent in a university hospital in eastern China. Infect. Drug. Resist. 12:3029. doi: 10.2147/IDR.S225546
Sun, Y. C., Huh, H. J., Jin, Y. B., Na, Y. C., Ryu, J. G., Ki, C. S., et al. (2015). Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg. Infect. Dis. 21:1088. doi: 10.3201/eid2106.150048
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Thungrat, K., Price, S. B., Carpenter, D. M., and Boothe, D. M. (2015). Antimicrobial susceptibility patterns of clinical Escherichia coli isolates from dogs and cats in the United States: January 2008 through January 2013. Vet. Microbiol. 179, 287–295. doi: 10.1016/j.vetmic.2015.06.012
Tyson, G. H., Li, C., Ceric, O., Reimschuessel, R., Cole, S., Peak, L., et al. (2019). Complete genome sequence of a carbapenem-resistant Escherichia coli isolate with blaNDM-5 from a dog in the United States. Microbiol. Resour. Announc. 8:e00872-19. doi: 10.1128/MRA.00872-19
Vikesland, P., Garner, E., Gupta, S., Kang, S., Maile-Moskowitz, A., and Zhu, N. (2019). Differential drivers of antimicrobial resistance across the world. Acc. Chem. Res. 52, 916–924. doi: 10.1021/acs.accounts.8b00643
Wang, R., Van, D. L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. doi: 10.1038/s41467-018-03205-z
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes. Infect. 7:122. doi: 10.1038/s41426-018-0124-z
Wang, X., Wang, Y., Zhou, Y., Wang, Z., Wang, Y., Zhang, S., et al. (2019). Emergence of colistin resistance gene and its variant in Raoultella ornithinolytica. Front. Microbiol. 10:228. doi: 10.3389/fmicb.2019.00228
Wang, Y., Zhang, R., Li, J., Wu, Z., Yin, W., Schwarz, S., et al. (2017). Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microb. 2:16260. doi: 10.1038/nmicrobiol.2016.260
Wu, C., Wang, Y., Shi, X., Wang, S., Ren, H., Shen, Z., et al. (2018). Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008–2014. Emerg. Microbes. Infect. 7:30. doi: 10.1038/s41426-018-0033-1
Yang, F., Zhang, K., Zhi, S., Li, J., Tian, X., Gu, Y., et al. (2019). High prevalence and dissemination of beta-lactamase genes in swine farms in northern China. Sci. Total. Environ. 651, 2507–2513. doi: 10.1016/j.scitotenv.2018.10.144
Yang, Y., Li, Y., Lei, C., Zhang, A., and Wang, H. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1791–1795. doi: 10.1093/jac/dky111
Yao, H., Wu, D., Lei, L., Shen, Z., Wang, Y., and Liao, K. (2016). The detection of fosfomycin resistance genes in Enterobacteriaceae from pets and their owners. Vet. Microbiol. 193, 67–71. doi: 10.1016/j.vetmic.2016.07.019
Zhang, Q., Lv, L., Huang, X., Huang, Y., Zhuang, Z., and Lu, J. (2019). Rapid increase in carbapenemase-producing Enterobacteriaceae in retail meat driven by the spread of the blaNDM-5-carrying IncX3 plasmid in China from 2016 to 2018. Antimicrob. Agents. Chemother. 63:e00573-19. doi: 10.1128/AAC.00573-19
Zhang, W., Lin, X., Xu, L., Gu, X., Yang, L., and Li, W. (2016). CTX-M-27 producing Salmonella enterica serotypes typhimurium and indiana are prevalent among food-producing animals in China. Front. Microbiol. 7:436. doi: 10.3389/fmicb.2016.00436
Keywords: multidrug resistance, antimicrobial drug usage, companion animals, ESBL, Escherichia coli
Citation: Chen Y, Liu Z, Zhang Y, Zhang Z, Lei L and Xia Z (2019) Increasing Prevalence of ESBL-Producing Multidrug Resistance Escherichia coli From Diseased Pets in Beijing, China From 2012 to 2017. Front. Microbiol. 10:2852. doi: 10.3389/fmicb.2019.02852
Received: 23 August 2019; Accepted: 25 November 2019;
Published: 10 December 2019.
Edited by:
Leonardo Neves de Andrade, University of São Paulo, BrazilReviewed by:
Tiago Casella, Faculdade de Medicina de São José do Rio Preto, BrazilCopyright © 2019 Chen, Liu, Zhang, Zhang, Lei and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaofei Xia, emhhb2ZlaXhpYWNhdUAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.